
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 23 August 2024
Sec. Microbiotechnology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1457628
Phenyllactic acid (PhLA), an important natural organic acid, can be used as a biopreservative, monomer of the novel polymeric material poly (phenyllactic acid), and raw material for various medicines. Herein, we achieved a high-level production of PhLA in Escherichia coli through the application of metabolic engineering and fermentation optimization strategies. First, the PhLA biosynthetic pathway was established in E. coli CGSC4510, and the phenylalanine biosynthetic pathway was disrupted to improve the carbon flux toward PhLA biosynthesis. Then, we increased the copy number of the key genes involved in the synthesis of the PhLA precursor phenylpyruvic acid. Concurrently, we disrupted the tryptophan biosynthetic pathway and enhanced the availability of phosphoenolpyruvate and erythrose 4-phosphate, thereby constructing the genetically engineered strain MG-P10. This strain was capable of producing 1.42 ± 0.02 g/L PhLA through shake flask fermentation. Furthermore, after optimizing the dissolved oxygen feedback feeding process and other conditions, the PhLA yield reached 52.89 ± 0.25 g/L in a 6 L fermenter. This study successfully utilized metabolic engineering and fermentation optimization strategies to lay a foundation for efficient PhLA production in E. coli as an industrial application.
Phenyllactic acid (PhLA) is a small-molecule natural organic acid found in pickles and natural honey (Armaforte et al., 2006). It can be polymerized into poly (phenyllactic acid), a novel material with good thermal stability, strong ductility, excellent ultraviolet absorption, and chemical resistance, making it a research focus in fields such as biology, chemistry, and energy (Nguyen et al., 2011; Erickson et al., 2012). Moreover, PhLA is a novel and effective antibacterial and antifungal preservative, providing a viable solution to microbial contamination and reducing food safety-related hazards (Schnürer and Magnusson, 2005; Valerio et al., 2016). It is a crucial raw material for producing drugs such as englitazone, nonprotein amino acids, hypoglycemic agents, and anti-human immunodeficiency virus agents (Urban and Moore, 1992; Weckwerth et al., 2000). Additionally, PhLA inhibits platelet aggregation in blood vessels, dilating them to increase coronary flow and combat myocardial hypoxia (Wu et al., 2007). These characteristics of PhLA provide promising application prospects in the pharmaceutical industry.
Currently, there are two main approaches of PhLA synthesis: chemical and microbial syntheses. Chemical synthesis of PhLA generates many by-products, such as phenylpyruvic acid (PhPA), and also involves complex technical routes, harsh reaction conditions, serious environmental pollution, and difficulties in subsequent separation (Bustos et al., 2018). Microbial synthesis of PhLA offers several advantages, such as mild reaction conditions, easy availability of fermentation substrates, lesser environmental pollution, relatively low cost, and good development prospects (Kawaguchi et al., 2015). Therefore, microbial production has become an attractive alternative method. In lactic acid bacteria, PhLA is a by-product of phenylalanine metabolism. Phenylalanine undergoes transamination to produce PhPA under the catalysis of deaminase, with α-ketoglutaric acid as an amino acceptor. Then, PhPA is converted to PhLA by a dehydrogenase (Mu et al., 2009; Prasuna et al., 2012).
With the development of metabolic engineering and synthetic biology, biocatalysis technology has been widely applied, and constructing microbial cell factories for the heterologous synthesis of PhLA has attracted increasing attention (Liu et al., 2015). Phenylalanine, PhPA, and α-ketoglutaric acid are precursors for synthesizing PhLA. By exogenously adding these precursors, microbial conversion can be used to produce PhLA and thus increase its yield (Weber and Zannoni, 1966; Valerio et al., 2016). Phenylalanine is converted into PhLA through transamination and reduction, with the former reaction being the probable rate-limiting step in PhLA biosynthesis. In a previous study, α-ketoglutaric acid was added during the culture of Lactobacillus plantarum TMW1.468, improving the transamination of phenylalanine and markedly enhancing the PhLA yield (Vermeulen et al., 2006). According to another study, lactate dehydrogenase and formate dehydrogenase were co-expressed to engineer Escherichia coli for PhLA production; this strain could convert 82.8 mmol/L PhPA to 79.6 mmol/L PhLA (Zheng et al., 2015). Some researchers utilized E. coli to co-express L-lactate dehydrogenase (L-LDH) derived from L. plantarum and glucose dehydrogenase (GDH) derived from Bacillus megaterium, using PhPA as the substrate for whole-cell catalytic synthesis of L-PhLA, ultimately achieving a final product concentration of 103.8 mmol/L, approximately 17.2 g/L (Zhu et al., 2017). Similarly, PhPA was used as a substrate for bioconversion, achieving a PhLA yield of 20.5 g/L (Luo et al., 2020). However, there are limited recent reports of the de novo synthesis of PhLA, with most reports focusing on whole-cell catalytic synthesis using PhPA as a substrate (Zheng et al., 2015; Zhu et al., 2017). The high cost of PhPA makes biocatalysis more expensive. In a previous study, phenylpyruvate reductase (encoded by pprA) from Wickerhamia fluorescens TK1 was expressed in a high-phenylalanine—producing E. coli strain, and the engineered strain produced 29 g/L PhLA under optimized fermentation conditions using glucose as the substrate (Fujita et al., 2013). The heterologous expression of phenylpyruvate reductase in E. coli to construct a PhLA biosynthetic pathway was considered feasible (Figure 1). However, in addition to the enhanced expression of phenylpyruvate reductase, the supply levels of initial substrates—phosphoenolpyruvate (PEP) and erythrose-4-phosphate (E4P)—are critical for the efficient synthesis of PhLA in E. coli (Figure 1). Exploring efficient de novo synthesis of PhLA from low-cost raw materials is an urgent issue for future research.
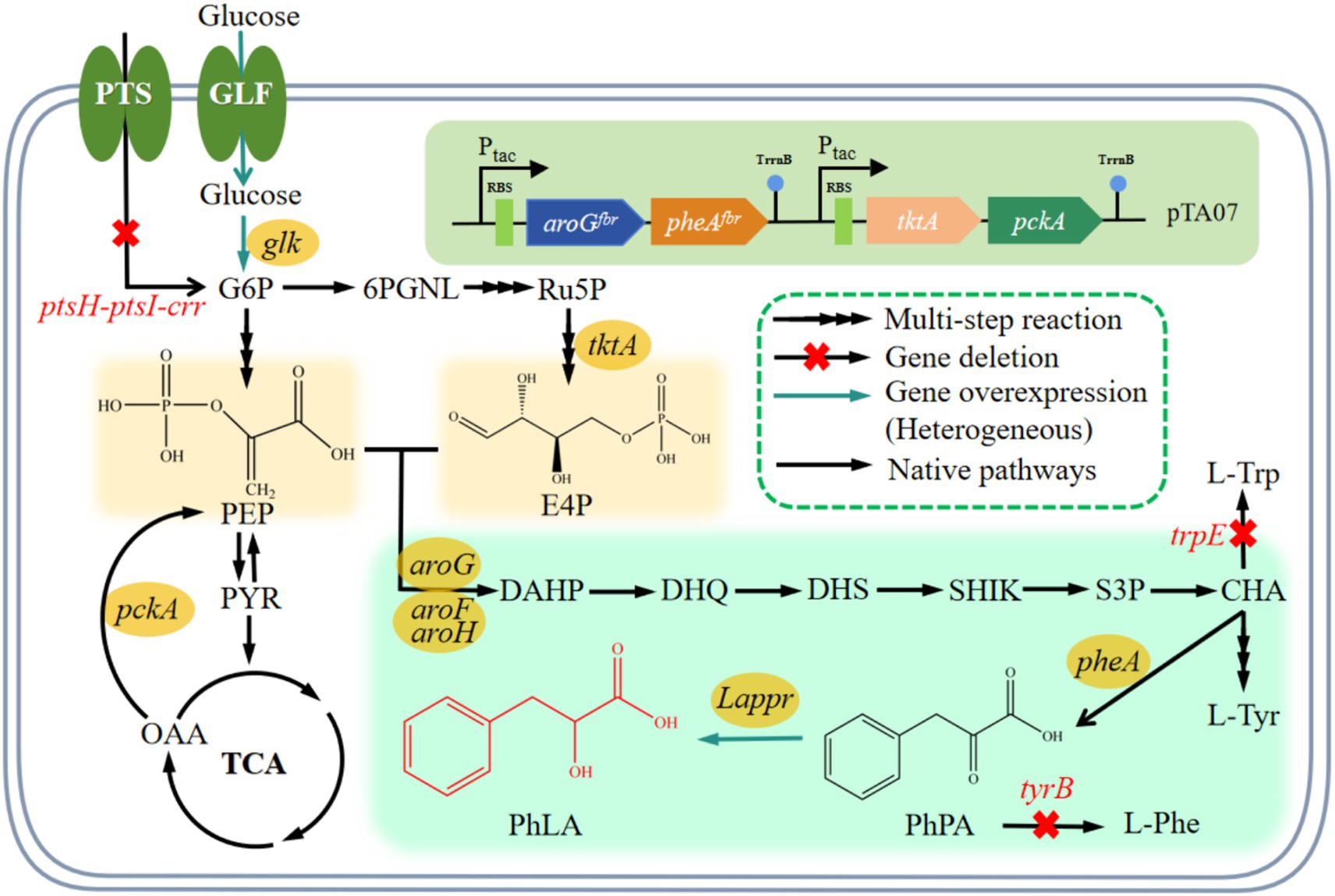
Figure 1. PhLA biosynthetic pathway and relevant metabolic reactions in E. coli. The conversion reaction of PhPA into PhLA was constructed by introducing the Lactobacillus ppr gene encoding phenylpyruvate reductase in E. coli. Abbreviations for metabolites: G6P, glucose-6-phosphate; 6PGNL, glucose 6-phosphate endolipid; Ru5P, ribulose-5-phosphate; E4P, erythrose-4-phosphate; PEP, phosphoenolpyruvate; PYR, pyruvate; TCA, tricarboxylic acid; OAA, oxaloacetic acid; DAHP, 3-deoxy-D-arabinoheptulosonic acid 7-phosphate; DHQ, 3-dehydroquinate; DHS, 3-dehydroshikimate; SHIK, shikimate; S3P, SHK-3-phosphate; CHA, chorismate; L-Trp, L-tyrptophan; L-Tyr, L-tyrosine; L-Phe, L-phenylalanine; PhPA, phenylpyruvate; PhLA; phenyllactic acid. Genes and enzymes: glf, glucose facilitator; glk, glucokinase; tktA, transketolase; pckA, PEP carboxykinase; aroF, aroG, and aroH, DAHP synthase AroF, AroG, and AroH, respectively; pheA, chorismate mutase/prephenate dehydratase; trpE, anthranilate synthase; tyrB, tyrosine aminotransferase; Lappr, phenylpyruvate reductase. PTS, glucose phosphotransferase system encoded by the ptsG, ptsH, ptsI, and crr genes.
A PhLA biosynthetic pathway was established in the phenylalanine-producing E. coli CGSC4510 for the efficient de novo synthesis of PhLA. The genes aroGfbr and pheAfbr, crucial for the PhLA synthesis pathway, were overexpressed in engineered E. coli, and the tryptophan synthesis pathway was weakened to increase metabolic flux and enhance PhLA yield. Additionally, the central metabolic pathway and glucose transport system were modified to improve the supply of the precursors PEP and E4P. Finally, the glucose-feeding process was optimized based on the dissolved oxygen (DO)-feedback, and the potential of the engineered E. coli to produce PhLA in a 6 L fermenter was evaluated. This study provides a successful example of applying E. coli for the efficient synthesis of PhLA.
The media used in this study include Luria-Bertani (LB) medium (10 g/L peptone, 5 g/L yeast extract, 10 g/L NaCl), seed medium (10 g/L NaCl, 15 g/L peptone, 10 g/L yeast extract, 2 g/L glucose), and fermentation medium (20 g/L glucose, 4 g/L yeast extract, 3 g/L MgSO4·7H2O, 5.5 g/L (NH4)2SO4, 3 g/L K2HPO4, 0.7 g/L tyrosine, 0.7 g/L phenylalanine, 0.35 g/L tryptophan, 0.5 g/L sodium citrate, 0.6 g/L sodium glutamate, 0.05 g/L FeSO4, 0.004 g/L MnSO4, 0.5 g/L fumaric acid, 0.001 g/L VB1, 0.001 g/L VB3, 0.0003 g/L VB8).
For shake flask fermentation, a single colony was inoculated into 5 mL of LB medium containing appropriate antibiotics and cultivated overnight at 37°C and 220 rpm. The culture was then inoculated into 20 mL of seed medium at a 5% (v/v) ratio and cultivated overnight at 37°C and 220 rpm. The seed culture was then inoculated into fermentation medium at a 10% (v/v) ratio and cultivated at 37°C and 200 rpm for 48 h. Ampicillin (100 mg/L) was added during cultivation to promote plasmid retention. When the culture reached an OD600 of 0.6, IPTG was added to a final concentration of 0.1 mM to induce recombinant protein expression. Each strain was cultured in triplicate.
All chemicals and reagents were of analytical grade and HPLC grade, purchased from Sigma-Aldrich Inc. (Shanghai, China) or Beijing Chemical Works (Beijing, China). Q5 High-Fidelity DNA Polymerase, Gibson Assembly Cloning Kit, and restriction enzymes were purchased from New England BioLabs (Beijing, China). Plasmid extraction and purification kits were purchased from Shandong Sparkjade Biotechnology Co., Ltd. (Shandong, China). Primers for PCR amplification were synthesized by Biosune (Fuzhou, China).
All genes used in this study were either amplified from genomic DNA or synthesized by General Biosystems (Anhui, China) with codon optimization. Genes were amplified using primer pairs tac-aroGfbr-F/R, tac-pheAfbr-F/R, and tac-GA-F/R, and then ligated with linearized plasmid pTrc99a to construct plasmids pTA03, pTA04, and pTA05 to enhance the carbon flux of PhLA. To further optimize expression, the tktA gene was amplified with the primer pair tac-tktA-F/R and ligated with the linearized plasmid pTA05 using the Gibson assembly method, resulting in plasmid pTA06. Similarly, plasmid pTA07 was constructed to overexpress pckA. The primers used for plasmid construction in this study are listed in Supplementary Table S1, and all constructed plasmids are shown in Table 1.
The CRISPR/Cas9 technique was performed mainly according to previously reported methods (Shen et al., 2022). Phenylpyruvate reductase (encoded by ppr) from Lactobacillus sp. CGMCC 9967, glycolaldehyde reductase (encoded by yiaE) from E. coli, and phenylpyruvate reductase (encoded by pprA) from W. fluorescens TK1 were integrated into the amino transferase (encoded by tyrB) site of the E. coli CGSC4510 genome and expressed under the inducible promoter tac, resulting in recombinant strains MG-P1, MG-P2, and MG-P3. Additionally, anthranilate synthase (encoded by trpE) was knocked out to weaken the competing pathway. Furthermore, to increase the availability of PEP, a precursor for phenylalanine lactate biosynthesis, glucose permease (encoded by glf) and glucose kinase (encoded by glk) from Zymomonas mobilis were integrated in tandem into the critical gene loci (ptsH-ptsI-crr) of the carbohydrate phosphotransferase system (PTS) in the E. coli genome and expressed under the inducible promoter tac. Accordingly, all constructed strains in this study are named and listed in Table 1.
Feedback feeding fermentation strategy is often used to achieve high-concentration production of the target product. During the feedback fed-batch fermentation process, when glucose is used as the sole carbon source, dissolved oxygen (DO) directly reflects glucose utilization (Park et al., 2011; Lv et al., 2020). When glucose is consumed, the oxygen uptake rate accompanied by a decrease in cell growth rate will lead to an increase in DO value. When glucose is supplemented, the increase in oxygen uptake rate that accompanies the increased cell growth rate will lead to a decrease in the DO value. It is commonly believed that the advantage of controlling the glucose supplementation rate through DO-feedback fed-batch control lies in its ability to prevent both glucose excess and DO limitation. By employing this method, an increase in cell growth and product yield can be achieved (Jing et al., 2018).
The dissolved oxygen feedback fed-batch control strategy in this study: 3 L fermentation medium was fermented in a 6 L bioreactor (Bioflo310, New Brunswick Scientific) for 48 h. The medium used was the same as the aforementioned fermentation medium, and 300 mL seed culture was added to the 3 L fermentation medium. The initial stirring speed was 200 rpm, with an aeration rate of 0.5 vvm, and the pH of the culture was maintained at 7.0 by automatically adding 25% ammonia water. The cultivation temperature was 37°C, and the dissolved oxygen was maintained at 5–10%, 10–20%, 20–30%, and 30–40% during fermentation. Sampling from the bioreactor is done by placing a centrifuge tube at the fermenter sampling port and using the fermenter tank pressure to discharge the fermentation broth out of the sampling port. Glucose was used as the carbon source in the fermentation medium, and the glucose concentration was controlled at 0.1–1.0% by feeding 60% glucose solution, with an initial glucose concentration of 2%. When the DO concentration exceeded the set value, the fermentation control system would feed 60% glucose solution into the fermentation medium. When the DO concentration dropped to the set saturation value, the automatic feeding system would stop, indicating the restoration of cell viability due to nutrient supplementation.
The yield of PhLA was detected and analyzed using high-performance liquid chromatography (HPLC, Waters Alliance e2695, United States). An Inert Sustain AQ-C18 column (5 μm, 4.6 × 250 mm) was used, with a column temperature of 30°C, an injection volume of 20 μL, and a flow rate of 1.0 mL/min. The detection wavelength was 210 nm. The mobile phase A is 0.05% trifluoroacetic acid aqueous solution, and mobile phase B is 0.05% trifluoroacetic acid methanol solution. The gradient was 40% B to 80% B from 0 to 15 min, maintained at 80% B from 15 to 16 min, and decreased from 80% B to 40% B from 16 to 18 min. Glucose concentration in the culture was measured using an enzyme electrode glucose sensor (BF-4, Oji Scientific Instruments, Hyogo, Japan). OD600 was measured using a UV–visible spectrophotometer (UV-1800, Shimadzu, Japan) to monitor cell growth, and dry cell weight (DCW) was calculated based on a pre-calibrated relationship (OD = 0.33 g/L DCW) (Wu et al., 2018).
PhPA can be converted to PhLA catalyzed by phenylpyruvate reductase (Xu et al., 2016). In this study, we constructed a PhLA biosynthetic pathway in E. coli by overexpressing phenylpyruvate reductase (encoded by ppr) from Lactobacillus sp. CGMCC 9967, glycolaldehyde reductase (encoded by yiaE) from E. coli, and phenylpyruvate reductase (encoded by pprA) from W. fluorescens TK1 (Figure 2A). From our laboratory collection, we utilized E. coli CGSC4510, which is capable of producing phenylalanine as the starting strain. In the phenylalanine biosynthetic pathway of E. coli, PhPA is converted to phenylalanine by aminotransferase (TyrB), whose expression is regulated by TyrR (Liu et al., 2018). Therefore, to enable E. coli to biosynthesize PhLA, we employed the CRISPR/Cas9 technology to integrate the genes Lappr, EcyiaE, and WfpprA under the control of the tac promoter into the TyrB locus of the E. coli CGSC4510 chromosome, resulting in the strains MG-P1, MG-P2, and MG-P3, respectively (Figure 2B). This transformation disrupted the phenylalanine biosynthetic pathway while constructing the PhLA biosynthetic pathway. In shake flask fermentation, MG-P1 expressing Lappr had the highest PhLA yield, reaching 0.251 ± 0.02 g/L, which was 24.7 and 15.5% more than that of MG-P2 (0.189 ± 0.01 g/L) and MG-P3 (0.212 ± 0.02 g/L), respectively (Figure 2C). Additionally, the residual quantity of phenylalanine is presented in Supplementary Figure S1. These results indicate that LaPPR had enhanced activity than the other two enzymes, showing greater catalytic activity toward PhPA, thus converting more PhPA to PhLA. Consequently, we successfully constructed a PhLA biosynthetic pathway in E. coli, resulting in the MG-P1 strain, which has a robust capability to produce PhLA and was used for further experiments.
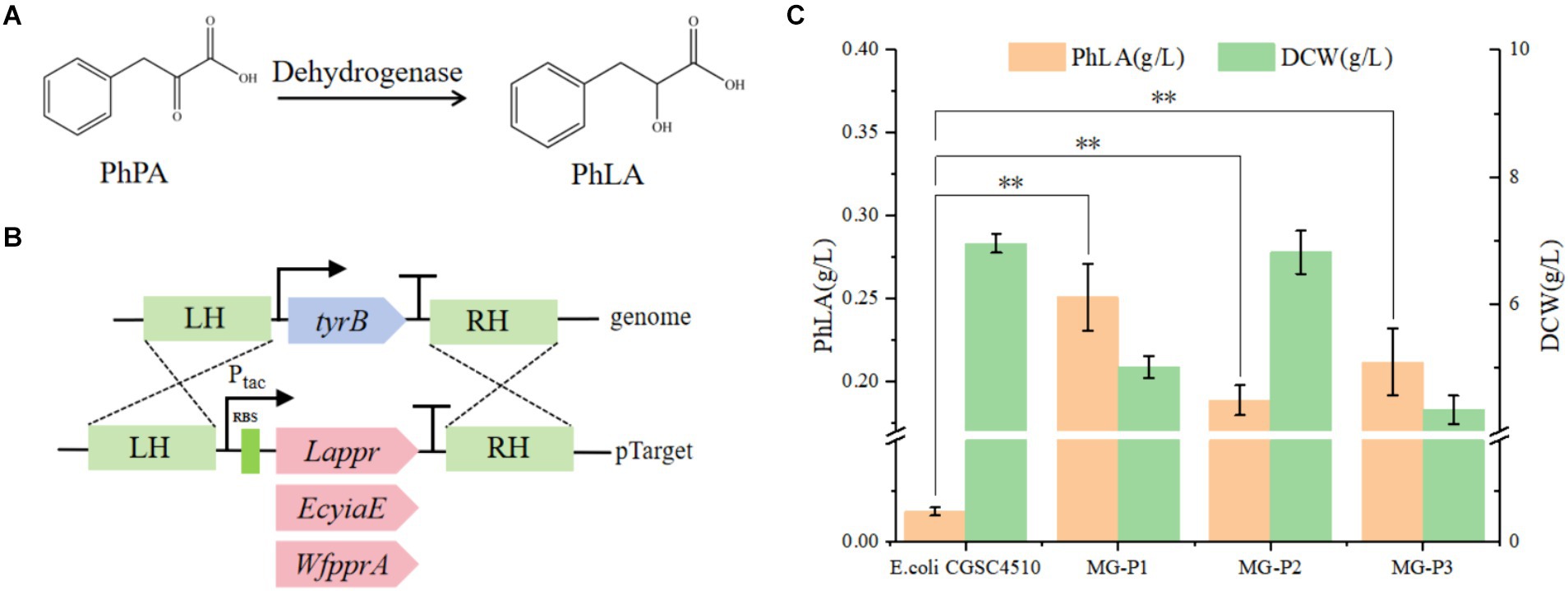
Figure 2. Construction of the PhLA biosynthesis pathway in E. coli. (A) The reduction of phenylpyruvate to PhLA catalyzed by dehydrogenase. (B) Schematic representation of homologous recombination of dehydrogenases from different sources into the TyrB locus. (C) PhLA production titers in recombinant E. coli strains MG-P1, MG-P2, and MG-P3, expressing Lappr, EcyiaE, and WfpprA, respectively. **p < 0.001. The data shown are the averages of three independent experiments with the standard deviations.
PhPA serves as the primary precursor of PhLA, with its intracellular concentration directly affecting the production of PhLA. In E. coli, aromatic amino acids (such as phenylalanine) are naturally produced via the shikimate pathway (Figure 1). The first reaction in this pathway is the condensation of PEP and E4P, catalyzed by 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthases. This reaction is the first rate-limiting step during the synthesis of aromatic amino acids. The three isoenzymes of DAHP synthases—AroG, AroH, and AroF—are subject to feedback inhibition by their respective end products—phenylalanine, tryptophan, and tyrosine. The activity of DAHP synthases determines the carbon flux into the shikimate pathway (Bongaerts et al., 2001). AroF and AroG contribute to 20 and 80% of the total enzyme activity, respectively (Tribe et al., 1976). The second rate-limiting step occurs during the synthesis of PhPA, catalyzed by the bifunctional enzyme chorismate mutase—prephenate dehydratase (PheA), which plays a crucial role in the production of PhPA and is subject to feedback inhibition by phenylalanine (Liu et al., 2018). Therefore, optimizing these rate-limiting steps is essential for improving the production of PhLA. For efficient PhLA production, it is necessary to overexpress aroGfbr (Kikuchi et al., 1997) and pheAfbr (Zhou et al., 2010) in E. coli.
We constructed the plasmids pTA03, pTA04, and pTA05 (Figure 3A) to overexpress aroGfbr and pheAfbr individually or simultaneously. These plasmids were transformed into the MG-P1 strain, resulting in the recombinant strains MG-P4, MG-P5, and MG-P6. In shake flask fermentation, compared with MG-P1 strain, PhLA production by MG-P4, MG-P5, and MG-P6 increased by 67, 131, and 186%, respectively. Overexpressing pheAfbr alone was more effective than overexpressing aroGfbr alone, and their combined overexpression had the most pronounced effect on the PhLA yield, with its titer reaching 0.72 ± 0.03 g/L (Figure 3C). The overexpression of aroGfbr and pheAfbr increased the metabolic flux into the phenylalanine biosynthesis pathway, thereby enhancing the production of PhLA. Additionally, in the aromatic amino acid metabolic pathway, chorismic acid can diverge into two metabolic branches: one leading to phenylalanine and tyrosine and the other to tryptophan. Inhibition of the tryptophan biosynthetic pathway can direct more carbon flux toward the phenylalanine metabolic branch (Bang et al., 2016). Therefore, to further enhance PhLA production, we knocked out trpE from the MG-P6 strain, resulting in the MG-P7 strain (Figure 3B). In shake flask fermentation, PhLA production was enhanced by 22%, achieving a titer of 0.88 ± 0.03 g/L in MG-P7 strain (Figure 3C). This result demonstrates that inhibiting the tryptophan biosynthetic pathway prevents carbon flux loss, thereby further enhancing PhLA production.
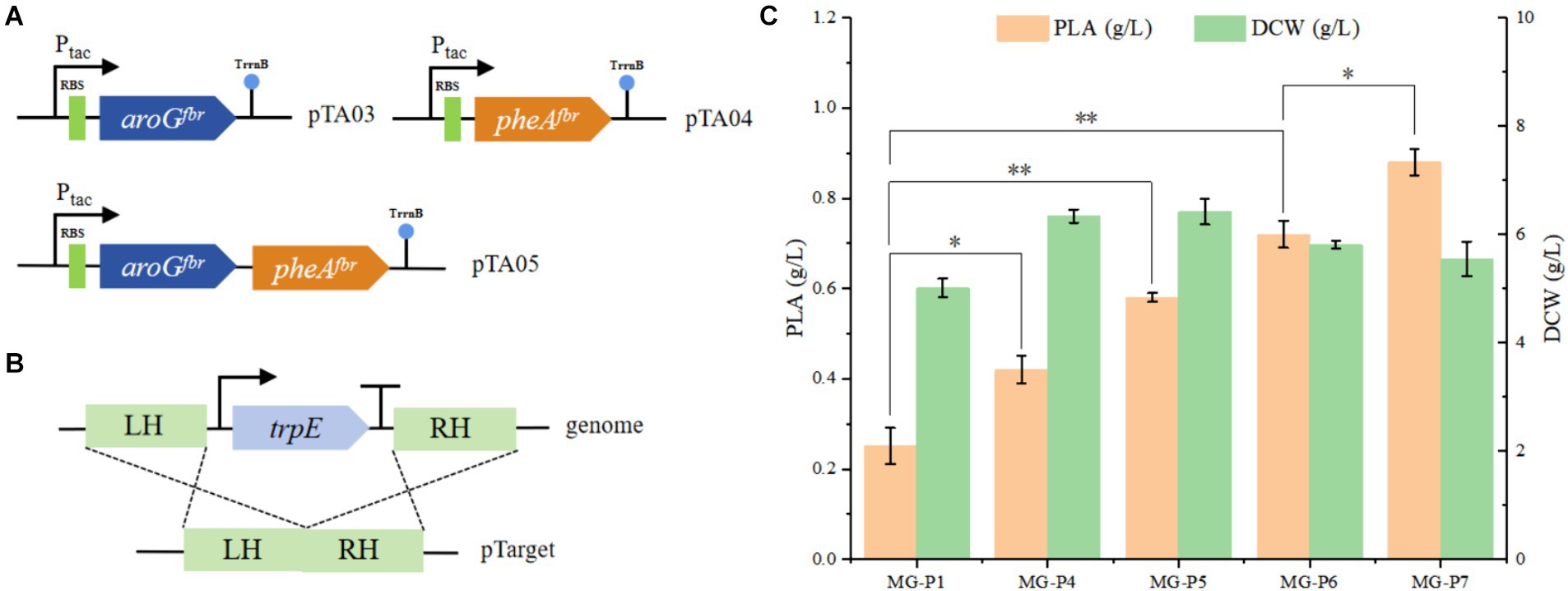
Figure 3. To enhance the production of PhLA, the rate-limiting enzymes of the PhLA synthesis pathway were overexpressed, and the tryptophan synthesis pathway was depleted. (A) Plasmids pTA03, pTA04, and pTA05 were constructed to overexpress the aroGfbr and pheAfbr individually and simultaneously, respectively. (B) Diagram of the trpE knockout. (C) Titers of PhLA produced by the recombinant E. coli strains MG-P1, MG-P4, MG-P5, MG-P6, and MG-P7. *p < 0.05 and **p < 0.001. The data shown are the averages of three independent experiments with the standard deviations.
The availability of PEP and E4P as initial substrates is a crucial factor for achieving efficient production of PhLA. In E. coli, PEP is a key intermediate in glucose transport and a precursor for the biosynthesis of pyruvate and oxaloacetate (Figure 1). The systems and enzymes involved in the production and consumption of PEP mainly include the PTS system, PEP synthase, PEP carboxykinase, PEP carboxylase, and pyruvate carboxylase (Bongaerts et al., 2001). The PTS system consumes approximately 50% of the PEP, leaving only a small amount for the synthesis of aromatic amino acids (Gosset, 2005). Replacing the PTS system with the glucose transport system that does not rely on PEP can improve the efficiency of aromatic amino acid synthesis by reducing PEP consumption and thus increasing the carbon flux into the aromatic amino acid biosynthetic pathway (Liu et al., 2017; Chen et al., 2018). Glucose permease (Glf) from Pseudomonas putida has a excellent glucose uptake efficiency (Parker et al., 1995). The synthesis of one molecule of phenylalanine requires the consumption of two molecules of PEP and one molecule of E4P. E4P can be produced under the catalysis of transketolase (encoded by tktA) or transaldolase (encoded by talB), with TktA being more effective than TalB in directing carbon flux toward the synthesis of aromatic amino acids (Lu and Liao, 1997).
The impact of enhanced precursor supply on PhLA production was investigated by further overexpressing tktA in the pTA05 plasmid, resulting in the pTA06 plasmid (Figure 4A). This plasmid was employed to transform the MG-P7 strain, producing the MG-P8 strain. Shake flask fermentation showed that elevating E4P supply during MG-P8 culture effectively enhanced PhLA production, resulting in a titer of 1.14 ± 0.01 g/L, a 30% enhancement, compared to the control strain (Figure 4C). Similarly, to increase PEP supply, we overexpressed phosphoenolpyruvate carboxykinase (encoded by the pckA) in the MG-P8 strain (Figure 4A), generating the MG-P9 strain, which produced 1.21 ± 0.03 g/L PhLA, indicating an increase of 6.1%. Additionally, we knocked out ptsH-ptsI-crr in MG-P9 to disrupt the native PTS system in E. coli. We then complemented these loci with the genes encoding Glf and glucose kinase (Glk) from Z. mobilis to restore the growth capability, creating the MG-P10 strain (Figure 4B). After culturing this strain in a fermentation medium with 40 g/L glucose for 48 h, the PhLA yield reached 1.42 ± 0.02 g/L, with a glucose consumption of 39.8 g/L. Compared to the control strain, the PhLA titer and the conversion rate of glucose to PhLA were remarkably improved (Figure 4C; Supplementary Figure S2). This finding indicates that the co-expression of glf and glk enhanced the glucose uptake efficiency, thereby improving the growth and PhLA production abilities of the strain. These results suggest that the combined action of TktA, PckA, Glk, and Glf elevated the intracellular availability of PEP and E4P, significantly boosting PhLA production.
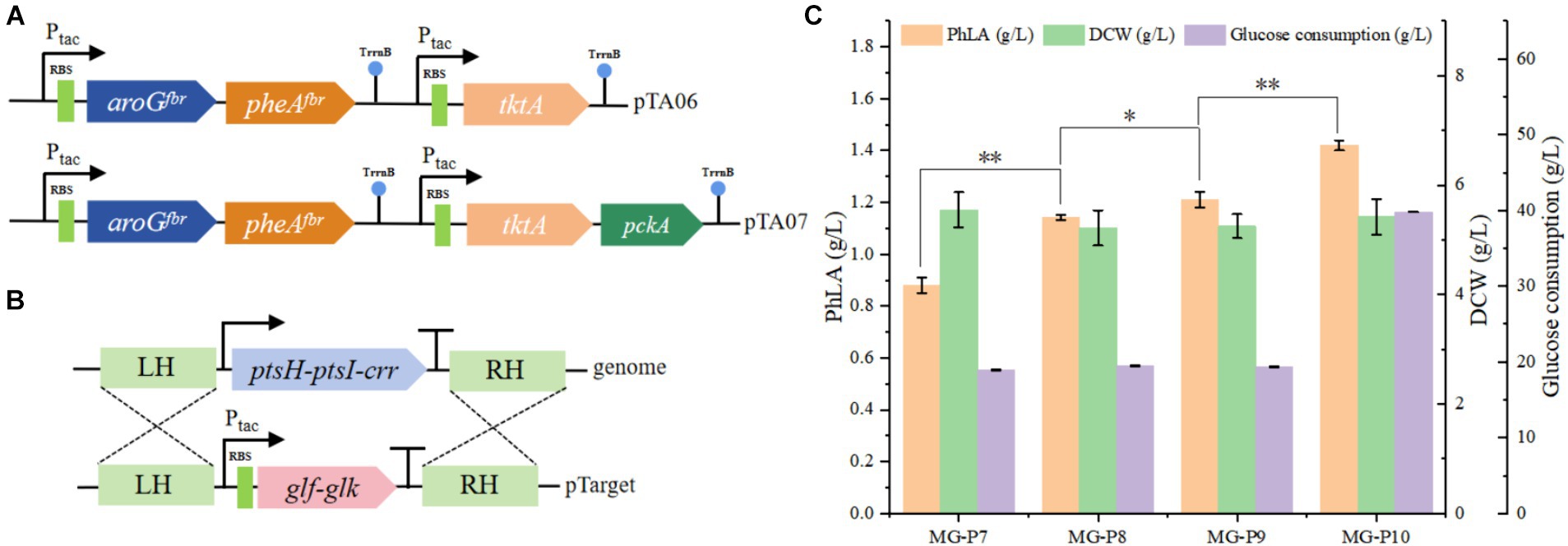
Figure 4. Enhancing the utilization efficiency of the precursor substances PEP and E4P in E. coli to improve PhLA production. (A) Construction of plasmids pTA06 and pTA07 for overexpressing the tktA and pckA genes. (B) Diagram of the homologous recombination of Glk and Glf from Z. mobilis into the ptsH-ptsI-crr locus. (C) PhLA production by recombinant E. coli strains MG-P8, MG-P9, and MG-P10, with MG-P10 cultured in fermentation medium containing 40 g/L glucose. *p < 0.05 and **p < 0.001. The data shown are the averages of three independent experiments with the standard deviations.
Based on the results of the shake flask fermentation of the engineered strain, MG-P10 was selected for fed-batch fermentation in a 6 L fermenter to evaluate its productivity and optimize the fermentation process. Previous reports have described the advantages of DO-feedback feeding in process control, which can suppress substrate feedback inhibition and DO supply limitations, thereby increasing the yield of the target product (Jing et al., 2018). Although providing excess glucose under oxygen-limited conditions during fermentation could improve PhLA yield (Kawaguchi et al., 2019), detailed reports on the effects of different levels of DO-feedback feeding on PhLA production in E. coli are still lacking. Therefore, we further explored the DO-feedback feeding strategy in a 6 L bioreactor to enhance the PhLA yield. Figure 5 and Table 2 show the changes in glucose concentration, DCW, PhLA titer, and acetic acid accumulation under various DO control levels. At a DO range of 20–30%, the PhLA titer, DCW, and glucose-to-PhLA yield reached 52.89 ± 0.25 g/L, 48.32 g/L, and 0.225 g/g (Figure 5C). Its PhLA titer is 6% higher than when the DO value is set in the range of 10–20% (Figure 5B). As the DO setpoint increased, there was a marked decline by up to 82% in the formation of the by-product acetic acid after 48 h of cultivation (Figures 5A,D). In E. coli, metabolic overflow occurs with excess glucose in the medium due to increased glucose consumption, leading to more by-products like acetic acid. This observation might be due to two reasons: first, the 20–30% DO-feedback feeding strategy avoided excessive glucose intake and insufficient DO supply; second, some acetic acid might have been metabolized by the strain to activate the TCA cycle, meeting its growth needs (Park et al., 2011; Kawaguchi et al., 2019). Compared to the other by-products, acetic acid severely inhibits E. coli growth and target product synthesis, thus reducing the fermentation performance (Kawaguchi et al., 2019). Under the enhanced DO-feedback feeding strategy of 30–40%, the DCW, PhLA titer, and glucose-to-PhLA yield of the engineered strain were 56.89 g/L, 45.68 ± 0.21 g/L, and 0.177 g/g, respectively (Figure 5D). Under these conditions, although the acetic acid content was only 0.81 g/L, the glucose conversion rate was 0.177, 21.3% lower than at the 20–30% DO setting, despite a 17.7% increase in DCW. These observations suggest that a part of the glucose was used for cell growth, and the relatively high DO level might have partially oxidized the raw materials and reduced productivity (Zhu et al., 2015). Additionally, this phenomenon could be due to the inhibition of glycolysis by the 20–30% DO level, restricting the energy required for PhLA biosynthesis. Under oxygen-limited conditions, the expression of tktA, aroF, aroG, and aroH in the shikimate pathway is upregulated, providing more E4P as a starting material for this pathway (Kawaguchi et al., 2019). Therefore, in this study, an appropriate DO-feedback feeding strategy could also address the problem of maintaining DO levels as cell density increases in the later stages of fermentation. These results indicate that maintaining DO-feedback feeding at 20–30% is more suitable for fermentation-based PhLA production, and optimizing the DO level is crucial for high PhLA yield.
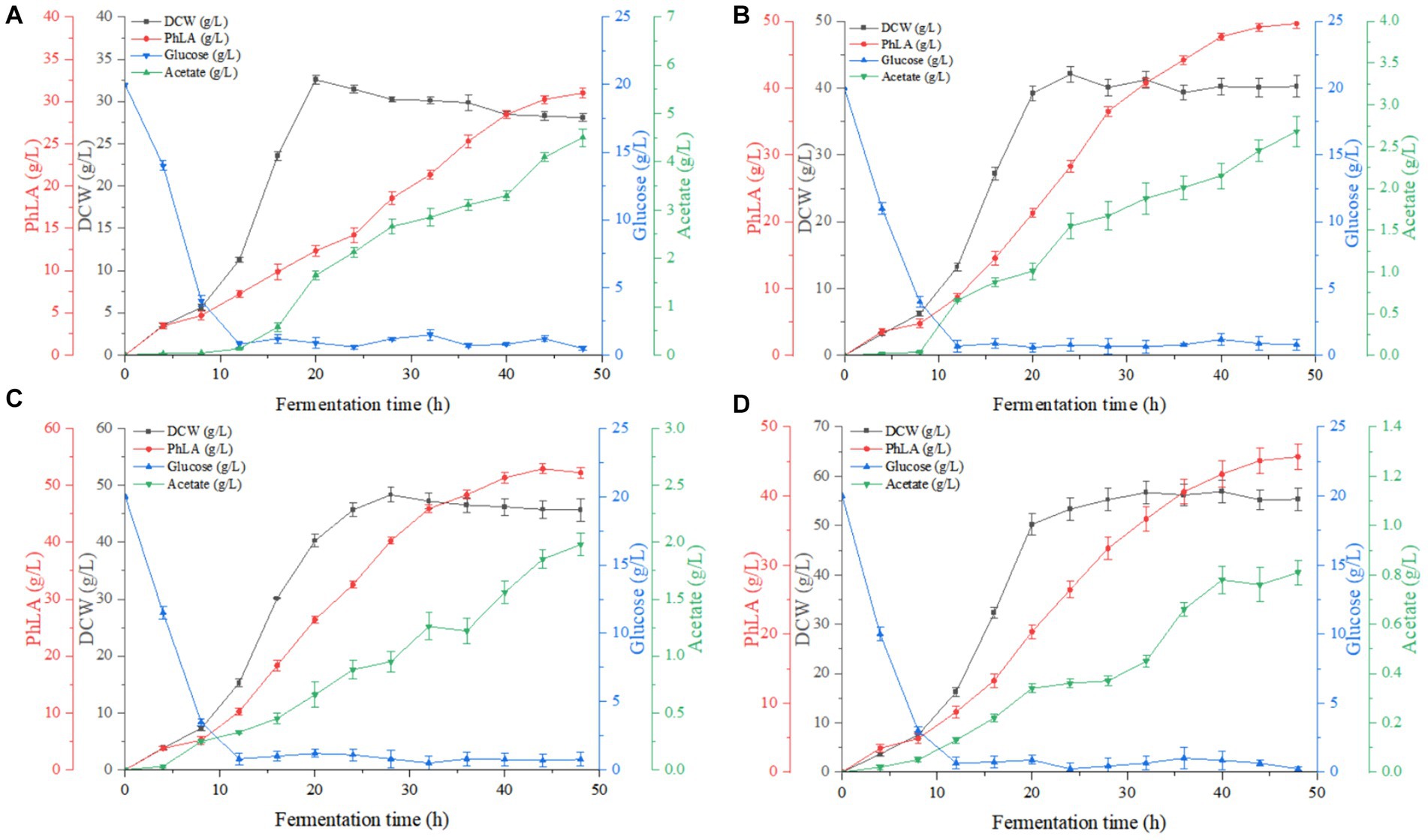
Figure 5. Fed-batch fermentation of recombinant E. coli for PhLA production based on dissolved oxygen (DO)-feedback control strategy in the 6 L bioreactor. (A) DO-feedback setting value = 5–10%. (B) DO-feedback setting value = 10–20%. (C) DO-feedback setting value = 20–30%. (D) DO-feedback setting value 30–40%. The data shown are the averages of three independent experiments with the standard deviations.
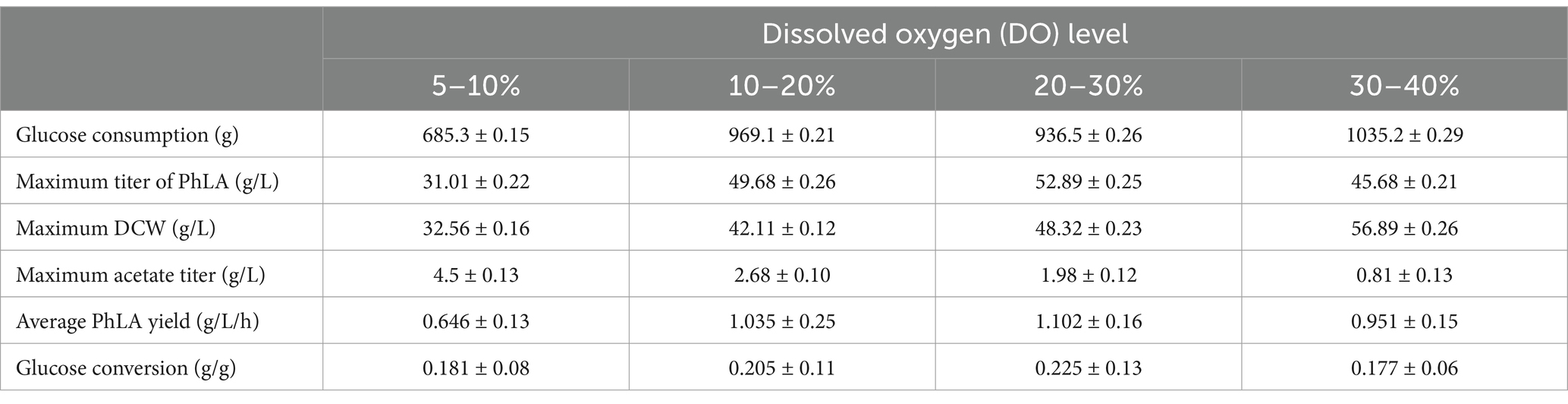
Table 2. Comparison of dissolved oxygen DO-feedback feeding strategy on fed-batch fermentation of PhLA by E. coli.
Herein, we constructed an efficient microbial cell factory using E. coli for the synthesis of PhLA. We also performed a series of rational and systematic optimizations of the PhLA biosynthetic pathway to achieve sustainable and green production. We disrupted the phenylalanine synthesis pathway in E. coli CGSC4510; heterologously expressed Lappr, EcyiaE, and WfpprA; and Reduction of PhPA to PhLA, thereby establishing the PhLA biosynthetic pathway in E. coli. Among these genes, Lappr encoded proteins exhibited the higher catalytic activity, enabling the engineered strain MG-P1 to produce 0.251 ± 0.02 g/L PhLA after 48 h of shake flask culture. Studies evaluating the effects of pprA, gxrA, ycdW, and yiaE on PhLA production indicated that pprA expression can enhance PhLA production more efficiently (Fujita et al., 2013). Here, we discovered a phenylpyruvate reductase, LaPPR, with higher enzymatic activity, which has higher catalytic efficiency than WfPPRA, possibly because the catalytic efficiency of LaPPR is greater than that of WfPPRA when using PhPA as substrate, which enables it to convert more PhPA into PhLA. The PhLA synthesis pathway is complex and tightly regulated, with DAHP synthases and PheA being the key rate-limiting enzymes. By overexpressing aroGfbr and pheAfbr and knocking out trpE, which is a crucial enzyme of the tryptophan synthesis pathway, we increased the metabolic flux. Consequently, the engineered E. coli strain MG-P7 achieved a PhLA titer of 0.88 ± 0.03 g/L via shake flask fermentation, indicating a 2.5-fold increase compared with that achieved using the MG-P1 strain. These results confirm that aroGfbr and pheAfbr are critical factors in determining the biosynthesis of PhLA, and their co-expression can significantly improve the PhLA yield, consistent with the finding of a previous study (Zhang et al., 2013). The increased PhLA yield is likely due to the overexpression of aroGfbr, which enhances the carbon flux from the central metabolism into the shikimate pathway, as well as the overexpression of pheAfbr, which channels more chorismic acid into the PhLA pathway. The disruption of trpE further prevents the loss of metabolic flux, ultimately leading to the efficient accumulation of PhLA (Kikuchi et al., 1997; Zhou et al., 2010; Bang et al., 2016). The supply of the precursors PEP and E4P as well as the intensity of the condensation reaction directly determine the metabolic flux of the shikimate pathway and yield of PhLA. We explored the impact of enhancing the supply of E4P and PEP on PhLA production by simultaneously overexpressing tktA and pckA and constructing a glucose transport system independent of PEP to replace the PTS system. Co-overexpression of glf, glk, pckA, and tktA in E. coli with a disrupted PTS system markedly enhanced the intracellular supply of PEP and E4P, resulting in a PhLA yield of up to 1.42 ± 0.02 g/L. This strategy not only improved the strain’s ability to produce PhLA but also enhanced glucose utilization. According to a previous study, the overexpression of tktA in E. coli slightly improved DAHP accumulation but not the DAHP conversion rate (Tyagi et al., 2017). Moreover, overexpression of tktA could disrupt central metabolism and glucose uptake (Ikeda and Katsumata, 1999). Overexpression of PEP synthase alone had no remarkable impact on tryptophan synthesis (Patnaik et al., 1995). These findings suggest that focusing metabolic engineering solely on increasing the accumulation of either of the substances in PEP and E4P may exacerbate the imbalance between the two, leading to the waste of carbon sources and ultimately failing to significantly improve synthesis efficiency. Therefore, synergistically elevating the supply levels of essential precursors is crucial for promoting PhLA accumulation. Finally, through the optimization of the DO-feedback feeding process, the engineered strain MG-P10 achieved a maximum PhLA titer of 52.89 ± 0.25 g/L in a 6 L fermenter when DO was maintained at a level of 20–30%, glucose-to-PhLA yield reached 0.225 g/g. This result indicates that optimal DO levels are essential for enhancing PhLA synthesis. In summary, this study successfully utilized an engineered E. coli strain for the de novo synthesis of PhLA, providing a foundation for its industrial production.
This study produced PhLA in E. coli via metabolic engineering and a DO-feedback feeding control strategy. By introducing phenylpyruvate reductase into E. coli, we constructed the biosynthetic pathway for PhLA. This pathway was enhanced by overexpressing the vital rate-limiting enzymes, weakening the tryptophan biosynthetic pathway, and increasing the supply of key precursors. Finally, PhLA accumulation in the engineered E. coli was further improved through optimization of the DO-feedback feeding control strategy. These results indicate that E. coli is an efficient chassis microorganism for the production of PhLA. This study demonstrates the potential of metabolic engineering and fermentation optimization strategies for efficient PhLA synthesis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
WW: Funding acquisition, Writing – review & editing, Investigation, Methodology, Writing – original draft. MC: Writing – original draft, Writing – review & editing. CL: Writing – original draft, Writing – review & editing, Formal analysis, Investigation, Methodology. JZ: Formal analysis, Investigation, Methodology, Writing – original draft. RX: Formal analysis, Data curation, Writing – review & editing. ZP: Formal analysis, Methodology, Writing – original draft. JL: Writing – original draft, Data curation, Methodology. FQ: Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by National Natural Science Foundation of China (32272287), Natural Science Foundation of Fujian Province of China (2021J01348), Fujian Vocational College of Bio-engineering (2022MS03), China Agriculture Research System of MOF and MARA (CARS-170501), and Science and Technology Planning Project of Fuzhou (2022-P-015).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1457628/full#supplementary-material
Armaforte, E., Carri, S., Ferri, G., and Caboni, M. F. (2006). High-performance liquid chromatography determination of phenyllactic acid in MRS broth. J. Chromatogr. A 1131, 281–284. doi: 10.1016/j.chroma.2006.07.095
Bang, H. B., Lee, Y. H., Kim, S. C., Sung, C. K., and Jeong, K. J. (2016). Metabolic engineering of Escherichia coli for the production of cinnamaldehyde. Microb. Cell Factories 15:16. doi: 10.1186/s12934-016-0415-9
Bongaerts, J., Krämer, M., Müller, U., Raeven, L., and Wubbolts, M. (2001). Metabolic engineering for microbial production of aromatic amino acids and derived compounds. Metab. Eng. 3, 289–300. doi: 10.1006/mben.2001.0196
Bustos, A. Y., de Valdez, G. F., and Gerez, C. L. (2018). Optimization of phenyllactic acid production by Pediococcus acidilactici CRL 1753. Application of the formulated bio-preserver culture in bread. Biol. Control 123, 137–143. doi: 10.1016/j.biocontrol.2018.05.017
Chen, Y., Liu, Y., Ding, D., Cong, L., and Zhang, D. (2018). Rational design and analysis of an Escherichia coli strain for high-efficiency tryptophan production. J. Ind. Microbiol. Biotechnol. 45, 357–367. doi: 10.1007/s10295-018-2020-x
Erickson, B., Nelson,, and Winters, P. (2012). Perspective on opportunities in industrial biotechnology in renewable chemicals. Biotechnol. J. 7, 176–185. doi: 10.1002/biot.201100069
Fujita, T., Nguyen, H. D., Ito, T., Zhou, S., Osada, L., Tateyama, S., et al. (2013). Microbial monomers custom-synthesized to build true bio-derived aromatic polymers. Appl. Microbiol. Biotechnol. 97, 8887–8894. doi: 10.1007/s00253-013-5078-4
Gosset, G. (2005). Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate:sugar phosphotransferase system. Microb. Cell Factories 4:14. doi: 10.1186/1475-2859-4-14
Ikeda, M., and Katsumata, R. (1999). Hyperproduction of tryptophan by Corynebacterium glutamicum with the modified pentose phosphate pathway. Appl. Environ. Microbiol. 65, 2497–2502. doi: 10.1128/AEM.65.6.2497-2502.1999
Jing, K., Tang, Y., Yao, C., del Rio-Chanona, E. A., Ling, X., and Zhang, D. (2018). Overproduction of L-tryptophan via simultaneous feed of glucose and anthranilic acid from recombinant Escherichia coli W3110: kinetic modeling and process scale-up. Biotechnol. Bioeng. 115, 371–381. doi: 10.1002/bit.26398
Kawaguchi, H., Miyagawa, H., Nakamura-Tsuruta, S., Takaya, N., Ogino, C., and Kondo, A. (2019). Enhanced phenyllactic acid production in Escherichia coli via oxygen limitation and shikimate pathway gene expression. Biotechnol. J. 14:e1800478. doi: 10.1002/biot.201800478
Kawaguchi, H., Teramura, H., Uematsu, K., Hara, K. Y., Hasunuma, T., Hirano, K., et al. (2015). Phenyllactic acid production by simultaneous saccharification and fermentation of pretreated sorghum bagasse. Bioresour. Technol. 182, 169–178. doi: 10.1016/j.biortech.2015.01.097
Kikuchi, Y., Tsujimoto, K., and Kurahashi, O. (1997). Mutational analysis of the feedback sites of phenylalanine-sensitive 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase of Escherichia coli. Appl. Environ. Microbiol. 63, 761–762. doi: 10.1128/AEM.63.2.761-762.1997
Liu, L., Chen, S., and Wu, J. (2017). Phosphoenolpyruvate:glucose phosphotransferase system modification increases the conversion rate during L-tryptophan production in Escherichia coli. J. Ind. Microbiol. Biotechnol. 44, 1385–1395. doi: 10.1007/s10295-017-1959-3
Liu, Y., Xu, Y., Ding, D., Wen, J., Zhu, B., and Zhang, D. (2018). Genetic engineering of Escherichia coli to improve L-phenylalanine production. BMC Biotechnol. 18, 5–12. doi: 10.1186/s12896-018-0418-1
Liu, S. P., Zhang, L., Mao, J., Ding, Z. Y., and Shi, G. Y. (2015). Metabolic engineering of Escherichia coli for the production of phenylpyruvate derivatives. Metab. Eng. 32, 55–65. doi: 10.1016/j.ymben.2015.09.007
Lu, J. L., and Liao, J. C. (1997). Metabolic engineering and control analysis for production of aromatics: role of transaldolase. Biotechnol. Bioeng. 53, 132–138. doi: 10.1002/(SICI)1097-0290(19970120)53:2<132::AID-BIT2>3.0.CO;2-P
Luo, X., Zhang, Y., Yin, F., Hu, G., Jia, Q., Yao, C., et al. (2020). Enzymological characterization of a novel d-lactate dehydrogenase from Lactobacillus rossiae and its application in d-phenyllactic acid synthesis. 3 Biotech 10, 101–112. doi: 10.1007/s13205-020-2098-5
Lv, P. J., Qiang, S., Liu, L., Hu, C. Y., and Meng, Y. H. (2020). Dissolved-oxygen feedback control fermentation for enhancing β-carotene in engineered Yarrowia lipolytica. Sci. Rep. 10:17114. doi: 10.1038/s41598-020-74074-0
Mu, W., Liu, F., Jia, J., Chen, C., Zhang, T., and Jiang, B. (2009). 3-Phenyllactic acid production by substrate feeding and pH-control in fed-batch fermentation of Lactobacillus sp. SK007. Bioresour. Technol. 100, 5226–5229. doi: 10.1016/j.biortech.2009.05.024
Nguyen, H. D., Jin, X., Kaneko, D., and Kaneko, T. (2011). Syntheses of high molecular weight poly (L-phenyllactic acid) s by a direct polycondensation in the presence of stable Lewis acids. Chem. Lett. 40, 584–585. doi: 10.1246/cl.2011.584
Park, J. H., Kim, T. Y., Lee, K. H., and Lee, S. Y. (2011). Fed-batch culture of Escherichia coli for L-valine production based on in silico flux response analysis. Biotechnol. Bioeng. 108, 934–946. doi: 10.1002/bit.22995
Parker, C., Barnell, W. O., Snoep, J. L., Ingram, L. O., and Conway, T. (1995). Characterization of the Zymomonas mobilis glucose facilitator gene product (glf) in recombinant Escherichia coli: examination of transport mechanism, kinetics and the role of glucokinase in glucose transport. Mol. Microbiol. 15, 795–802. doi: 10.1111/j.1365-2958.1995.tb02350.x
Patnaik, R., Spitzer, R. G., and Liao, J. C. (1995). Pathway engineering for production of aromatics in Escherichia coli: confirmation of stoichiometric analysis by independent modulation of AroG, TktA, and Pps activities. Biotechnol. Bioeng. 46, 361–370. doi: 10.1002/bit.260460409
Prasuna, M. L., Mujahid, M., Sasikala, C., and Ramana, C. V. (2012). L-phenylalanine catabolism and L-phenyllactic acid production by a phototrophic bacterium, Rubrivivax benzoatilyticus JA2. Microbiol. Res. 167, 526–531. doi: 10.1016/j.micres.2012.03.001
Schnürer, J., and Magnusson, J. (2005). Antifungal lactic acid bacteria as biopreservatives. Trends Food Sci. Technol. 16, 70–78. doi: 10.1016/j.tifs.2004.02.014
Shen, P., Gu, S., Jin, D., Su, Y., Wu, H., Li, Q., et al. (2022). Engineering metabolic pathways for cofactor self-sufficiency and serotonin production in Escherichia coli. ACS Synth. Biol. 11, 2889–2900. doi: 10.1021/acssynbio.2c00298
Tribe, D. E., Camakaris, H., and Pittard, J. (1976). Constitutive and repressivle enzymes of the common pathway of aromatic biosynthesis in Escherichia coli K-12: regulation of enzyme synthesis at different growth rates. J. Bacteriol. 127, 1085–1097. doi: 10.1128/JB.127.3.1085-1097.1976
Tyagi, N., Saini, D., Guleria, R., and Mukherjee, K. J. (2017). Designing an Escherichia coli strain for phenylalanine overproduction by metabolic engineering. Mol. Biotechnol. 59, 168–178. doi: 10.1007/s12033-017-9999-5
Urban, F. J., and Moore, B. S. (1992). Synthesis of optically active 2-benzyldihydrobenzopyrans for the hypoglycemic agent englitazone. J. Heterocyclic Chem. 29, 431–438. doi: 10.1002/jhet.5570290223
Valerio, F., Di Biase, M., Lattanzio, V. M., and Lavermicocca, P. (2016). Improvement of the antifungal activity of lactic acid bacteria by addition to the growth medium of phenylpyruvic acid, a precursor of phenyllactic acid. Int. J. Food Microbiol. 222, 1–7. doi: 10.1016/j.ijfoodmicro.2016.01.011
Vermeulen, N., Gánzle, M. G., and Vogel, R. F. (2006). Influence of peptide supply and cosubstrates on phenylalanine metabolism of Lactobacillus sanfranciscensis DSM20451T and Lactobacillus plantarum TMW1. 468. J. Agric. Food Chem. 54, 3832–3839. doi: 10.1021/jf052733e
Weber, W. W., and Zannoni, V. G. (1966). Reduction of phenylpyruvic acids to phenyllactic acids in mammalian tissues. J. Biol. Chem. 241, 1345–1349. doi: 10.1016/S0021-9258(18)96780-3
Weckwerth, W., Miyamoto, K., Iinuma, K., Krause, M., Glinski, M., Storm, T., et al. (2000). Biosynthesis of PF1022A and related cyclooctadepsipeptides. J. Biol. Chem. 275, 17909–17915. doi: 10.1074/jbc.M001084200
Wu, W. B., Guo, X. L., Zhang, M. L., Huang, Q. G., Qi, F., and Huang, J. Z. (2018). Enhancement of l-phenylalanine production in Escherichia coli by heterologous expression of Vitreoscilla hemoglobin. Biotechnol. Appl. Biochem. 65, 476–483. doi: 10.1002/bab.1605
Wu, L., Qiao, H., Li, Y., and Li, L. (2007). Protective roles of puerarin and Danshensu on acute ischemic myocardial injury in rats. Phytomedicine 14, 652–658. doi: 10.1016/j.phymed.2007.07.060
Xu, G.-C., Zhang, L.-L., and Ni, Y. (2016). Enzymatic preparation of D-phenyllactic acid at high space-time yield with a novel phenylpyruvate reductase identified from Lactobacillus sp. CGMCC 9967. J. Biotechnol. 222, 29–37. doi: 10.1016/j.jbiotec.2015.12.011
Zhang, C., Zhang, J., Kang, Z., Du, G., Yu, X., Wang, T., et al. (2013). Enhanced production of L-phenylalanine in Corynebacterium glutamicum due to the introduction of Escherichia coli wild-type gene aroH. J. Ind. Microbiol. Biotechnol. 40, 643–651. doi: 10.1007/s10295-013-1262-x
Zheng, Z., Zhao, M., Zang, Y., Zhou, Y., and Ouyang, J. (2015). Production of optically pure L-phenyllactic acid by using engineered Escherichia coli coexpressing L-lactate dehydrogenase and formate dehydrogenase. J. Biotechnol. 207, 47–51. doi: 10.1016/j.jbiotec.2015.05.015
Zhou, H., Liao, X., Wang, T., Du, G., and Chen, J. (2010). Enhanced L-phenylalanine biosynthesis by co-expression of pheAfbr and aroFwt. Bioresour. Technol. 101, 4151–4156. doi: 10.1016/j.biortech.2010.01.043
Zhu, Y., Liu, Y., Li, J., Shin, H. D., Du, G., Liu, L., et al. (2015). An optimal glucose feeding strategy integrated with step-wise regulation of the dissolved oxygen level improves N-acetylglucosamine production in recombinant Bacillus subtilis. Bioresour. Technol. 177, 387–392. doi: 10.1016/j.biortech.2014.11.055
Keywords: metabolic engineering, Escherichia coli, phenyllactic acid, synthetic biology, dissolved oxygen feedback fed-batch
Citation: Wu W, Chen M, Li C, Zhong J, Xie R, Pan Z, Lin J and Qi F (2024) Efficient production of phenyllactic acid in Escherichia coli via metabolic engineering and fermentation optimization strategies. Front. Microbiol. 15:1457628. doi: 10.3389/fmicb.2024.1457628
Received: 01 July 2024; Accepted: 14 August 2024;
Published: 23 August 2024.
Edited by:
Qian Wang, Shandong University, ChinaCopyright © 2024 Wu, Chen, Li, Zhong, Xie, Pan, Lin and Qi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weibin Wu, d2VpYmlud3VreEBob3RtYWlsLmNvbQ==; Feng Qi, Zi5xaUBmam51LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.