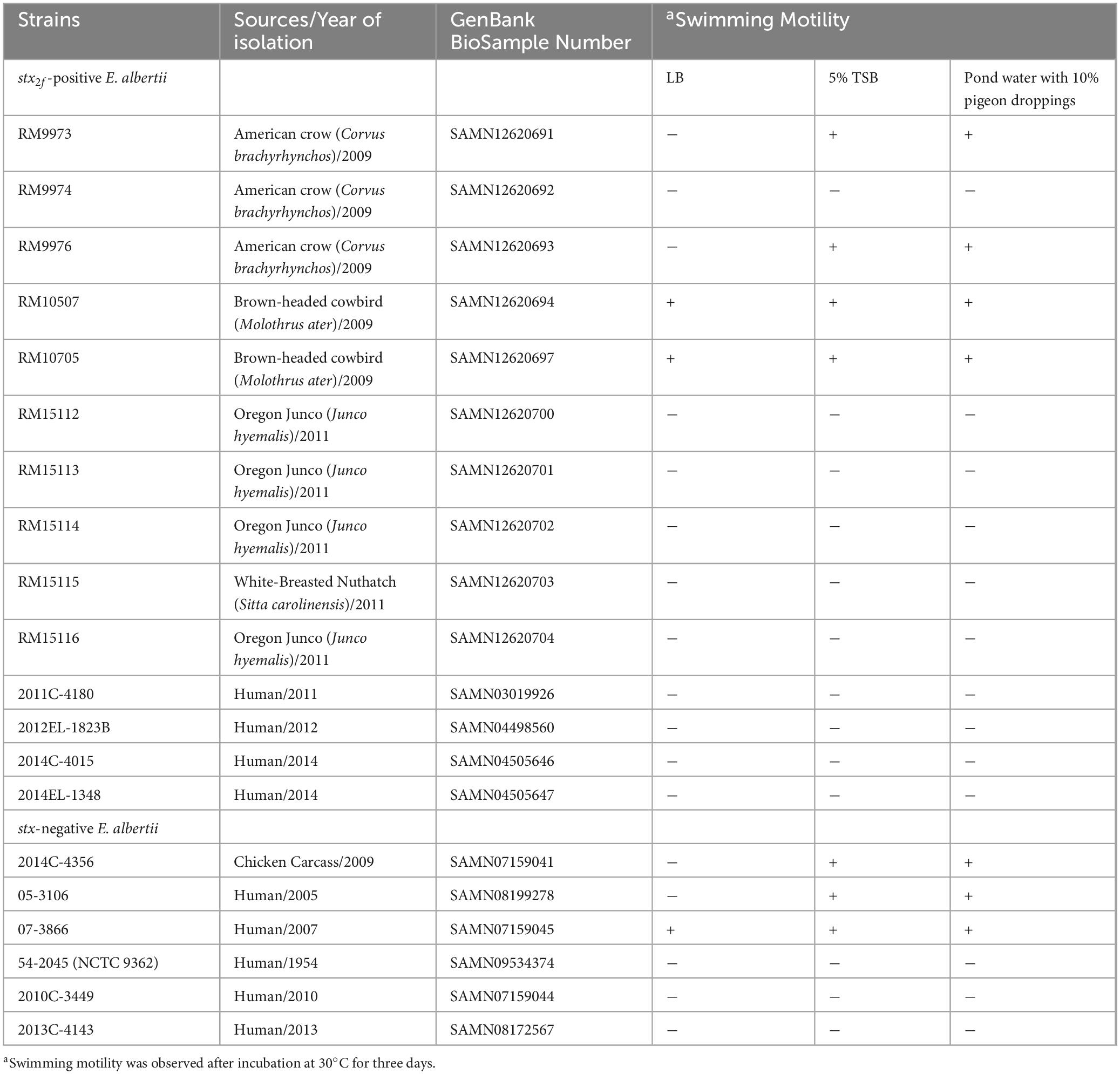
- 1Produce Safety and Microbiology Research Unit, U.S. Department of Agriculture, Agricultural Research Service, Western Regional Research Center, Albany, CA, United States
- 2Enteric Diseases Laboratory Branch, Centers for Disease Control and Prevention, Atlanta, GA, United States
Escherichia albertii is an emerging foodborne pathogen. We previously reported that some avian Shiga toxin-producing E. albertii strains exhibited higher or comparable cytotoxicity in Vero-d2EGFP cells with several enterohemorrhagic E. coli (EHEC) outbreak strains. To better understand the environmental persistence of this pathogen, comparative genomics and phenotypic assays were applied to assess adhesion capability, motility, and biofilm formation in E. albertii. Among the 108 adherence-related genes, those involved in biogenesis of curli fimbriae, hemorrhagic E. coli pilus, type 1 fimbriae, and Sfm fimbriae were conserved in E. albertii. All 20 E. albertii strains carried a complete set of primary flagellar genes that were organized into four gene clusters, while five strains possessed genes related to the secondary flagella, also known as lateral flagella. Compared to EHEC strain EDL933, the eight chemotaxis genes located within the primary flagellar gene clusters were deleted in E. albertii. Additional deletion of motility genes flhABCD and motBC was identified in several E. albertii strains. Swimming motility was detected in three strains when grown in LB medium, however, when grown in 5% TSB or in the pond water-supplemented with 10% pigeon droppings, an additional four strains became motile. Although all E. albertii strains carried curli genes, curli fimbriae were detected only in four, eight, and nine strains following 24, 48, and 120 h incubation, respectively. Type 1 fimbriae were undetectable in any of the strains grown at 37°C or 28°C. Strong biofilms were detected in strains that produced curli fimbriae and in a chicken isolate that was curli fimbriae negative but carried genes encoding adhesive fimbriae K88, a signature of enterotoxigenic E. coli strains causing neonatal diarrhea in piglets. In all phenotypic traits examined, no correlation was revealed between the strains isolated from different sources, or between the strains with and without Shiga toxin genes. The phenotypic variations could not be explained solely by the genetic diversity or the difference in adherence genes repertoire, implying complex regulation in expression of various adhesins. Strains that exhibited a high level of cytotoxicity and were also proficient in biofilm production, may have potential to emerge into high-risk pathogens.
1 Introduction
Escherichia albertii, an emerging foodborne pathogen, is the most divergent lineage among the other Escherichia species and clades (Walk et al., 2009; Ooka et al., 2015). Due to similar biochemical properties and possession of the intimin gene located on the locus of enterocyte effacement (LEE) pathogenicity island, many E. albertii isolates have been misidentified as enteropathogenic E. coli (EPEC), or enterohemorrhagic E. coli (EHEC) (Ooka et al., 2012). E. albertii causes diarrhea, abdominal pain, and high fever in humans, although bacteremia and extraintestinal infections were also reported (Gomes et al., 2020). The well-known virulence factors in E. albertii include LEE encoded intimin and its Tir receptor, responsible for the initial adherence of pathogen cells to the host epithelial cell surfaces, as well as the LEE-encoded type three secretion system (T3SS) and the effector proteins. Other common virulence factors include Shiga toxin (Stx), cytolethal distending toxin (CDT), type six secretion systems (T6SS), and the vacuolating autotransporter toxin Vat (Carter et al., 2023a). Sporadic infections and outbreaks of foodborne gastroenteritis caused by E. albertii have been reported worldwide (Konno et al., 2012; Ooka et al., 2013; Ori et al., 2018; Masuda et al., 2020; Bengtsson et al., 2023; Iguchi et al., 2023). Transmission of E. albertii is thought to occur via contaminated food or water although in most outbreaks the transmission vehicles were not identified (Masuda et al., 2020; Muchaamba et al., 2022).
Growing evidence supports that E. albertii has a wide habitat range. E. albertii strains have been isolated from domestic and wild animals, various foods, and aquatic environments (Muchaamba et al., 2022). Among the reported animal hosts, birds appear to be one of the main reservoirs/carriers (Oaks et al., 2010; Hinenoya et al., 2021; Hinenoya et al., 2022; Wang et al., 2022; Barmettler et al., 2023; Xu et al., 2024). Presence of E. albertii in various water bodies and food products including chicken, pork, duck meat, mutton, and oysters has been reported (Felfoldi et al., 2010; Maheux et al., 2014; Lindsey et al., 2015; Maeda et al., 2015; Wang et al., 2016; Arai et al., 2022), however, little is known about the contamination routes and the environmental prevalence and persistence of this emerging foodborne pathogen. A recent study investigating the survival of E. albertii in foods and water revealed that E. albertii grew faster in chicken than in pork or in oysters but had low viability in warm environmental water (Hirose et al., 2024). Induction of flagellar biosynthesis and swimming motility was observed in some strains when cells were exposed to hypoosmotic pressure or at ambient temperature, suggesting a role of flagellar motility in the survival of E. albertii in aquatic environments (Ikeda et al., 2020).
Biofilm is a common microbial lifestyle in natural environments (Watnick and Kolter, 2000). Compared with planktonic cells, biofilm-associated cells are better at coping with environmental stresses and have increased resistance to toxic substances including antibiotics and chemical sanitizers. Therefore, biofilm formation by enteropathogenic bacteria would increase their survival and persistence in natural environments and may serve as a source of contamination. Biofilm formation involves multiple steps, including initial surface contact, transient association, attachment, maturation, and dispersion (O’Toole et al., 2000). Numerous bacterial adherence factors including surface adhesive appendages and autotransporter proteins play a role in biofilm formation. In E. coli K-12 strains, flagellar motility, curli fimbriae, as well as FimH adhesin were found to be important for initial surface contact and attachment. Additionally, flagellar motility was found playing a role in biofilm dispersion (Pratt and Kolter, 1998; Reisner et al., 2003; Karatan and Watnick, 2009).
Shiga toxin-producing E. coli (STEC) produces diverse fimbrial and nonfimbrial adhesins that facilitate the attachment to and/or colonization by STEC cells in diverse ecological niches (McWilliams and Torres, 2014; Vogeleer et al., 2014). In STEC O157:H7 strains, curli fimbriae were found to mediate binding to, and invasion of epithelial cells, and promote the attachment of pathogens to plant and abiotic surfaces (Gophna et al., 2001; Fink et al., 2012; Carter et al., 2016). The hemorrhagic E. coli pilus (HCP), originally identified in STEC O157:H7 as a colonization factor (Xicohtencatl-Cortes et al., 2007), contributed to the biofilm formation of STEC O157:H7 strains on abiotic surfaces (Xicohtencatl-Cortes et al., 2009). In E. coli and other enteric pathogens, expression of type 1 fimbriae is controlled by a phase variation mechanism, which reversibly switches between the “ON” and “OFF” state of fim genes transcription (Abraham et al., 1985). This switch is mediated by an invertible DNA element, fimS, and two site-specific recombinases. Inversion of fimS abolishes the transcription of fimA, which encodes the major subunit of type 1 fimbriae. Expression of type 1 fimbriae were detected in STEC non-O157 strains, but not in O157:H7 strains (Roe et al., 2001). In STEC O157:H7 strains, transcription of fimA is locked at the “OFF” state due to a 16-bp deletion within the fimS (Iida et al., 2001). Type 1 fimbriae contributed to the attachment of STEC cells to abiotic surfaces in a O128:H2 strain and contributed to the biofilm formation when the fim genes of the STEC O157:H7 strain Sakai were expressed in a nonpathogenic E. coli strain (Cookson et al., 2002; Elpers and Hensel, 2020).
Knowledge about environmental persistence of E. alberti is scarce. Biofilm formation by E. albertii was reported in only a few clinical strains at 37°C although the efficiency of biofilm formation was much lower than that of E. coli strain 042 (Lima et al., 2019). Understanding prevalence and persistence of E. albertii in nonhost environments will provide valuable information for risk assessment and to bridge gaps in understanding the epidemiology of this emerging human pathogen. We previously reported genomic features and virulence genes repertoire of Shiga toxin-producing E. albertii strains isolated from wild birds in an agricultural region in California and revealed that some bird strains exhibited higher or comparable cytotoxicity with several EHEC outbreak strains (Carter et al., 2023a). To gain insight into the persistence of E. albertii in nonhost environments, we systematically evaluated the adhesion capability and several phenotypic traits known to contribute to bacterial biofilm formation in a set of E. albertii avian and clinical strains. Our study revealed great genetic diversity in genes encoding fimbrial and nonfimbrial adhesins in E. albertii as well as vast strain variations in expression of curli fimbriae, swimming motility, and in biofilm formation. Our study provides a foundation into further understanding how E. albertii senses and responds to environmental stimuli for improved survival in the changing environments.
2 Materials and methods
2.1 Bacterial strains and growth media
Bacterial strains and their sources are listed in Table 1. The complete genome sequences of E. albertii strains were reported previously (Carter et al., 2023a). The strains were grown routinely in Luria-Bertani (LB) broth (10 g tryptone, 5 g yeast extract, and 5 g NaCl per liter) unless noted.
2.2 Sequence analysis
The flagellar genes in E. albertii strains were identified by using BLASTn searches with the flagellar genes of the E. coli K-12 sub-strain MG1655 and the EHEC strain EDL933 (Supplementary Table 1). Additional flagellar genes were identified from the E. albertii genome annotations as described previously (Carter et al., 2023a) (Supplementary Table 2). E. coli genes related to fimbriae and pili biogenesis and genes encoding protein adhesins (Supplementary Table 3) were used as queries of BLASTn to identify homologs of adherence-related genomic loci in E. albertii strains. The BLASTn was performed in Geneious Prime® with a threshold of 65% for gene coverage and 70% or 25% for sequence identity at nucleotides or amino acids level, respectively. Homologs of each gene or the entire operons were extracted from the corresponding bacterial genomes. DNA sequences were aligned using Clustal Omega in Geneious Prime® (2024.0.3) and neighbor-joining consensus trees were constructed with the following parameters: Genetic Distance Model, Jukes-Cantor; Resampling Method, bootstrap; and number of replicates, 10,000.
2.3 Motility tests
Swimming motility was examined for each strain grown on soft agar (0.25%) in rich medium (LB), diluted TSB (5%), and sterile pond water containing 10% pigeon droppings as described previously (Murakami et al., 2020) with modification. The pond water was collected from a public accessible creek in Albany, California (37°53′43.86′′N, 122°18′16.68′′W). The pigeon droppings were collected near a train station in El Cerrito, California (37°54′9.63′′N, 122°17′56.17′′W). To prepare the 10% pigeon-droppings suspension, pigeon droppings were first suspended in nine volumes of pond water and then filtered through a 0.22-μm filter followed by adding agar to 0.25% prior to autoclaving. Single colonies of each E. albertii strain were point-inoculated on soft agar plates using sterile toothpicks. The plates were incubated at 30°C for three days prior to observing the motility.
2.4 Detection of curli fimbriae
Curli fimbriae were examined by growing each strain at 26°C for 1, 2, and 5 days on Congo Red indicator (CRI) plates, consisting of LB agar plates without sodium chloride (LBNS) and supplemented with 40 μg/ml of Congo Red dye and 10 μg/ml of Coomassie Brilliant Blue, as described previously (Carter et al., 2011). Curli-producing strains were indicated by red colonies whereas curli-deficient strains were indicated by white colonies on CRI plates.
2.5 Detection of type 1 fimbriae
Production of type 1 fimbriae was examined by hemagglutination for each strain grown in LBHS broth statically at 37°C or in LBNS broth statically at 28°C for two days. Cells were collected by centrifugation at 8,000 g for 3 min and resuspended in 1x PBS buffer at a final concentration about 3 x 108 cells/ml. Fifty μl of bacterial suspension was then mixed with 50 μl of guinea pig red blood cells (Innovative Research Inc) at room temperature in the presence or absence of 1% D mannose as previously described (Biscola et al., 2011). E. coli strain DH5a was used as a positive control and EHEC strain EDL933 was used as a negative control.
2.6 Biofilm formation and quantification
Biofilm assays were carried out as described previously (Carter et al., 2023b). Briefly, overnight cultures of E. albertii grown in LB at 37°C were inoculated in LBNS broth at a final concentration of 1x106 cells/ml. One ml of inoculated LBNS broth was aliquoted into a borosilicate glass tube and then incubated statically at 28°C for 1, 2, and 5 days. At the end of each incubation, the planktonic cells were removed carefully, and the tubes were rinsed twice with one ml sterile distilled water and then stained with one ml 0.1% crystal violet at room temperature for 30 min. The dye was then removed gently, and the tubes were washed twice with sterile distilled water. The crystal violet that bound to the glass tube was solubilized in 0.5 ml of 33% acetic acid and the absorbance was determined at 570 nm using a microplate reader (SpectraMax 340; Molecular Devices, Sunnyvale, CA). Tubes with uninoculated media served as negative controls. Each data set was the average of results from at least three biological replicates. All data were first evaluated for normal distribution by the Shapiro-Wilk test using Graph Pad Prism 10 Version 10.2.3 (Dotmatics). The differences in biofilm formation, represented by the absorbance at 570 nm, among the strains were assessed by the adjusted P-value of the Tukey’s multiple comparisons test after a One-way ANOVA test (P ≤ 0.05). Similarly, the differences in biofilm formation of each strain at various incubation times were assessed by the adjusted P-value of the Tukey’s multiple comparisons test after a One-way ANOVA test.
3 Results
3.1 E. albertii flagellar genes
Of the 48 genes related to flagella biosynthesis and motility in strain EDL933, homologs of 40 and 35 genes were identified in 13 and seven E. albertii strains, respectively (Supplementary Table 1). In strain EDL933, these 48 flagellar genes are distributed at four genomic locations, with a size of 11.5 Kb, 15.6 Kb, 6.7 Kb, and 11.0 Kb for Regions 1–4, respectively. Examining the genomic locations of the flagellar genes in E. albertii revealed a similar genes organization as in strain EDL933 (Figure 1A). Among the four genomic locations, the greatest sequence variation was detected in Region 2. In strain EDL933, Region 2 contained seven flagellar genes and eight chemotaxis genes (cheZYBR, tap, tar, and cheWA). Unlike strain EDL933, the eight chemotaxis genes were deleted in all E. albertii strains examined. Furthermore, an additional deletion of genes flhBCD and motBA was detected in a subset of E. albertii strains including five avian and two clinical strains. This deletion appeared to be mediated by a recombination between the sites within genes flhA and otsA since a 183-bp otsA gene fragment was located immediately upstream of a truncated flhA gene. In contrast, the flagellar genes located in the other three regions in strain EDL933 were all conserved in E. albertii strains.
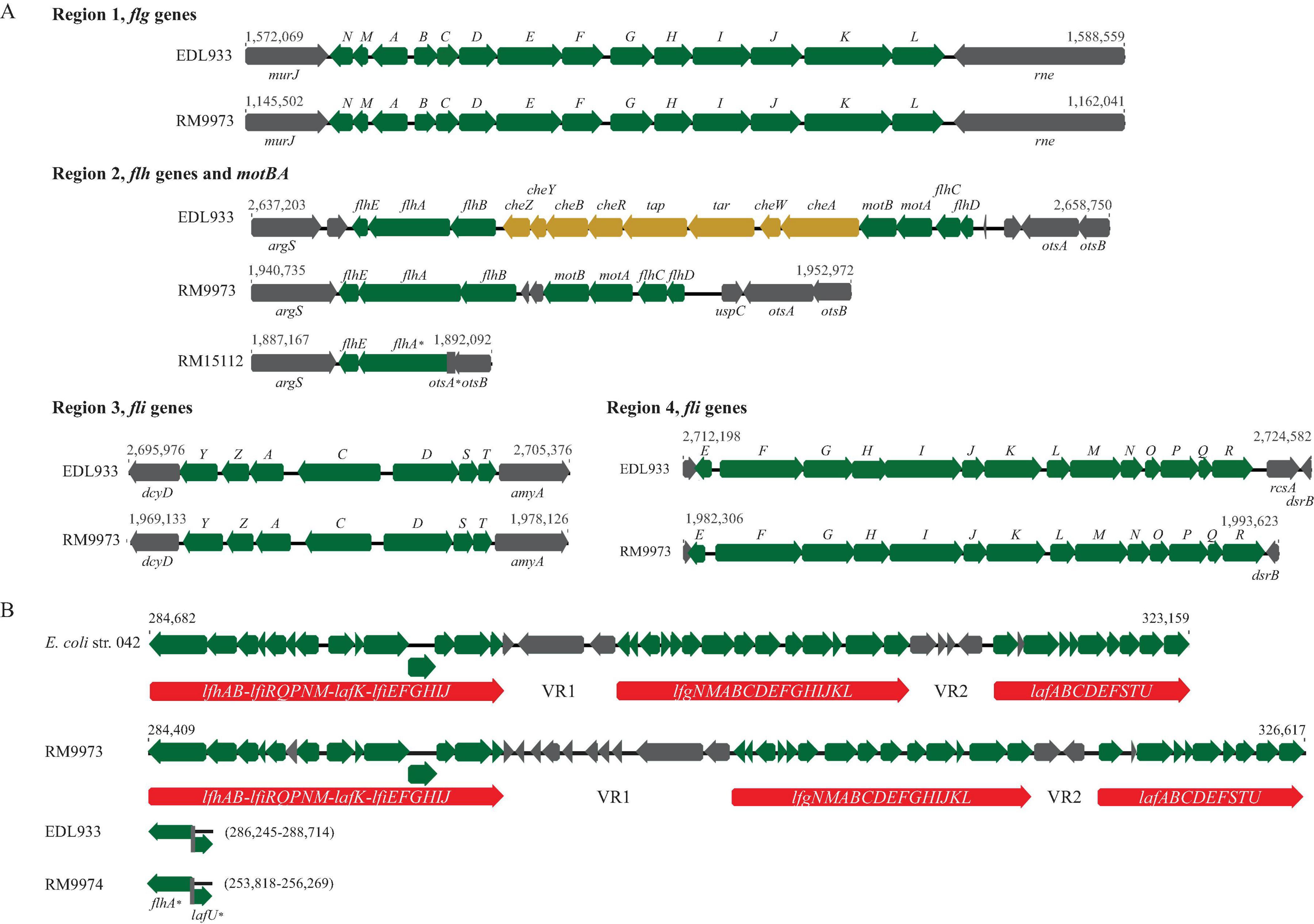
Figure 1. Sequence analyses of E. albertii flagellar genes. (A) Gene organization and genomic locations of the primary flagellar genes. Numbers indicate the corresponding chromosomal positions of the four flagellar gene clusters in E. coli O157:H7 strain EDL933 and E. albertii avian strains RM9973 and RM15112. Green arrows represent the flagellar genes detected in E. albertii and grey arrows represent the bordering genes or the hypothetical genes. Yellow arrows represent E. coli genes that are missing in E. albertii strains. (B) Gene organization and genomic locations of the secondary flagellar genes (Flag-2). Numbers indicate the corresponding chromosomal positions in EAEC strain 042 and avian strain RM9973. The three flagellar gene clusters are indicated by the red arrows. In the strains lacking a Flag-2, such as EDL933 and RM9974, the corresponding chromosomal sites were uniformly occupied by the two truncated genes, flhA and lafU. Genes labeled with an “*” indicate those carrying mutations within the coding sequences.
Interestingly, a Flag-2 locus, which encodes a secondary flagellar system that resembles the lateral flagella in Aeromonas hydrophila and Vibrio parahaemolyticus (Ren et al., 2005), was identified in one avian (RM9973) and four clinical E. albertii strains. The Flag-2 loci in E. albertii varied in size from 33 Kb in the clinical strain 2010C-3449 to 44 Kb in the clinical strain 05-3106. Like the Flag-2 locus in the enteroaggregative E. coli (EAEC) strain 042, Flag-2 genes were organized into three gene clusters, separated by the two variable regions, VR1 and VR2 (Figure 1B). In strain 042, the first gene cluster contains 14 genes that are involved in regulation and expression of flagellar basal body components. Homologs of these 14 genes were detected in the avian strain RM9973 and in the clinical strain 05-3106. Genes lfhB and lfiR were deleted in the strain 07-3866, while genes lafK and lfiEFGHIJ were deleted in both strains 54-2045 and 2010C-3449 (Supplementary Table 2). The second gene cluster in strain 042 also contains 14 genes encoding flagellar structural proteins and the third gene cluster carries nine genes that are mainly involved in flagellar filament synthesis. Homologs of all genes within the second and the third gene clusters were detected in the Flag-2 positive E. albertii strains. In the Flag-2 negative E. albertii strains, this region was about 2.5 Kb, containing the truncated two border genes, flhA and lafU (Figure 1B). A highly similar truncated Flag-2 locus was detected in E. coli strains EDL933 and K-12 strain MG1655 (% Identity > 90).
3.2 Motility in E. albertii
When grown in LB at 30°C for three days, motility was observed in two avian strains, RM10507 and RM10705, and one clinical strain 07-3866 (Table 1). Both strains RM10507 and RM10705 were isolated from brown-headed cowbird and were Flag-2 negative. These three strains remained motile when grown in 5% TSB or in pond water supplemented with 10% pigeon droppings (Table 1). Interestingly, four nonmotile strains when grown in LB, became motile when grown in 5% TSB or in the pond water supplemented with 10% pigeon droppings (Table 1). These four strains included two avian strains, RM9973 and RM9976 that were both isolated from American crow, the chicken isolate 2014C-4356, and the clinical strain 05-3106. The majority of nonmotile phenotypes could be explained by the mutations identified in the flagellar genes, including the deletion of motAB and flhBCD in avian strains RM15112-RM15116 and in clinical strains 2014C-4015 and 2014EL-1348 (Figure 1B), point deletions in fliF of the strains RM9974 and 2011C-4180 and in motA of the strain 2013C-4143, and an amber mutation in flgG and flhA of the strains 54-2045 and 2010C-3449, respectively (Supplementary Table 1).
3.3 E. albertii fimbrial genes
Homologs of genes encoding 12 fimbriae and pili implicated in adherence, biofilm formation, and pathogenesis in diverse E. coli pathotypes were examined in E. albertii. All genes are listed in Supplementary Table 3. Homologs of genes encoding curli fimbriae, type 1 fimbriae, and hemorrhagic E. coli pilus (HCP) were detected in all strains while homologs of genes encoding adhesive fimbriae, Sfm fimbriae, and P fimbriae were detected in a subset of E. albertii strains examined.
3.3.1 Curli genes and expression of curli fimbriae
Like E. coli, genes related to biogenesis of curli fimbriae in E. albertii are organized in two divergent operons, csgDEFG and csgBAC, and located upstream of tRNA gene serX (Figure 2A). Sequence analysis revealed that all E. albertii curli genes were placed in the same clade that was separated from the E. coli curli genes (Figure 2B). The curli genes of the E. albertii strains shared a high sequence similarity ( > 95%) with each other, except for the clinical strain 2010C-3449, in which, both csgE and csgD were truncated due to an IS insertion, while csgA carried an amber mutation.
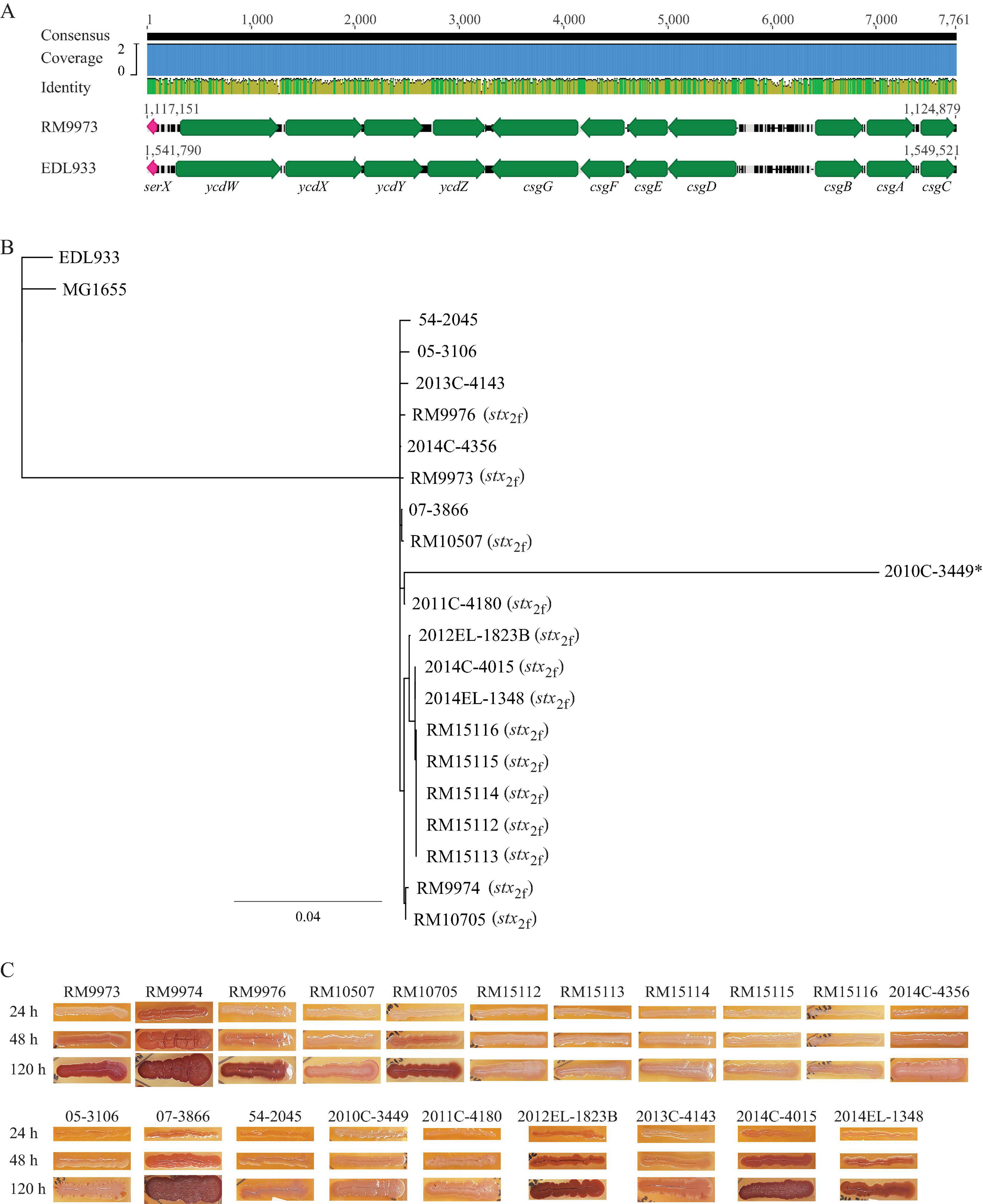
Figure 2. Sequence analyses of curli genes and detection of curli fimbriae in E. albertii. (A) Chromosomal locations of curli operons and pairwise comparison of the curli genes between the E. albertii avian strain RM9973 and E. coli O157:H7 strain EDL933. Numbers indicate the corresponding chromosomal positions in each strain. Green arrows represent the annotated genes, and the pink arrows represents tRNA gene serX. (B) Sequence analysis of curli genes. The curli operons were identified by BLASTn search of a database containing all genomes examined using a 4.4-Kb DNA fragment containing the seven curli genes of the E. coli strain MG1655 as a query in Geneious Prime®. The sequences of the curli genes were extracted from corresponding genomes and aligned using Clustal Omega alignment in Geneious Prime®. A consensus tree was constructed with the following parameters: Genetic Distance Model, Jukes-Cantor; Resampling tree method: Bootstrap; Number of Replicates: 10,000; Support Threshold: 50%. The stx2f positive strains are indicated in parentheses. The strain marked with an “*” indicates presence of mutations within the coding sequences of curli genes. (C) Detection of curli fimbriae on CRI plates. Curli fimbriae were examined by growing each strain on the CRI plates at 26°C for 24 h, 48 h, and 120 h. Production of curli fimbriae is indicated by red colonies which resulted from the binding of CR dye supplemented in growth medium.
Unexpectedly, production of curli fimbriae varied greatly among the E. albertii strains (Figure 2C). Among the 10 avian strains, production of curli fimbriae was observed in strains RM9973, RM9974, RM9976 and RM10705, although all avian strains carried intact coding sequences for all curli genes. As expected, no curli fimbriae were observed for clinical strain 2010C-3449. Among the other clinical strains, production of curli fimbriae was detected in three out of four stx2f positive strains (2012EL-1823B, 2014C-4015, 2014EL-1348) and in the stx2f negative strain 07-3866. Colonies of strain 2013C-4143 exhibited pink and light red color following 48 h and 120 h incubation, respectively, suggesting that this strain could produce curli fimbriae under the condition examined but with less amount compared with the other curli-positive strains (Figure 2C).
3.3.2 Type 1 fimbriae genes and expression of type 1 fimbriae
In E. coli, the type 1 fimbriae genes (fimB, fimE, and fimAICDFGH) are located on an 8.8-Kb DNA fragment. Expression of the type 1 fimbriae is controlled by a phase variation mechanism, in which, transcription of fimA is switched to “ON” or “OFF” by an invertible DNA element, fimS, and two site-specific recombinases encoded by genes fimB and fimE, respectively (Figure 3A). Homologs of the nine fim genes were identified in all 20 E. albertii strains examined. Additionally, the invertible element, fimS, flanked by two 9-bp inverted repeats (IRs), was also detected in all E. albertii strains. The IRs (5′-TTGGGGCCA-3′) in E. albertii strains were identical to the IRs in E. coli strains EDL933 and K-12 strain MG1655, except for the avian strain RM9973, in which a single base substitution of G to A occurred at position 6.
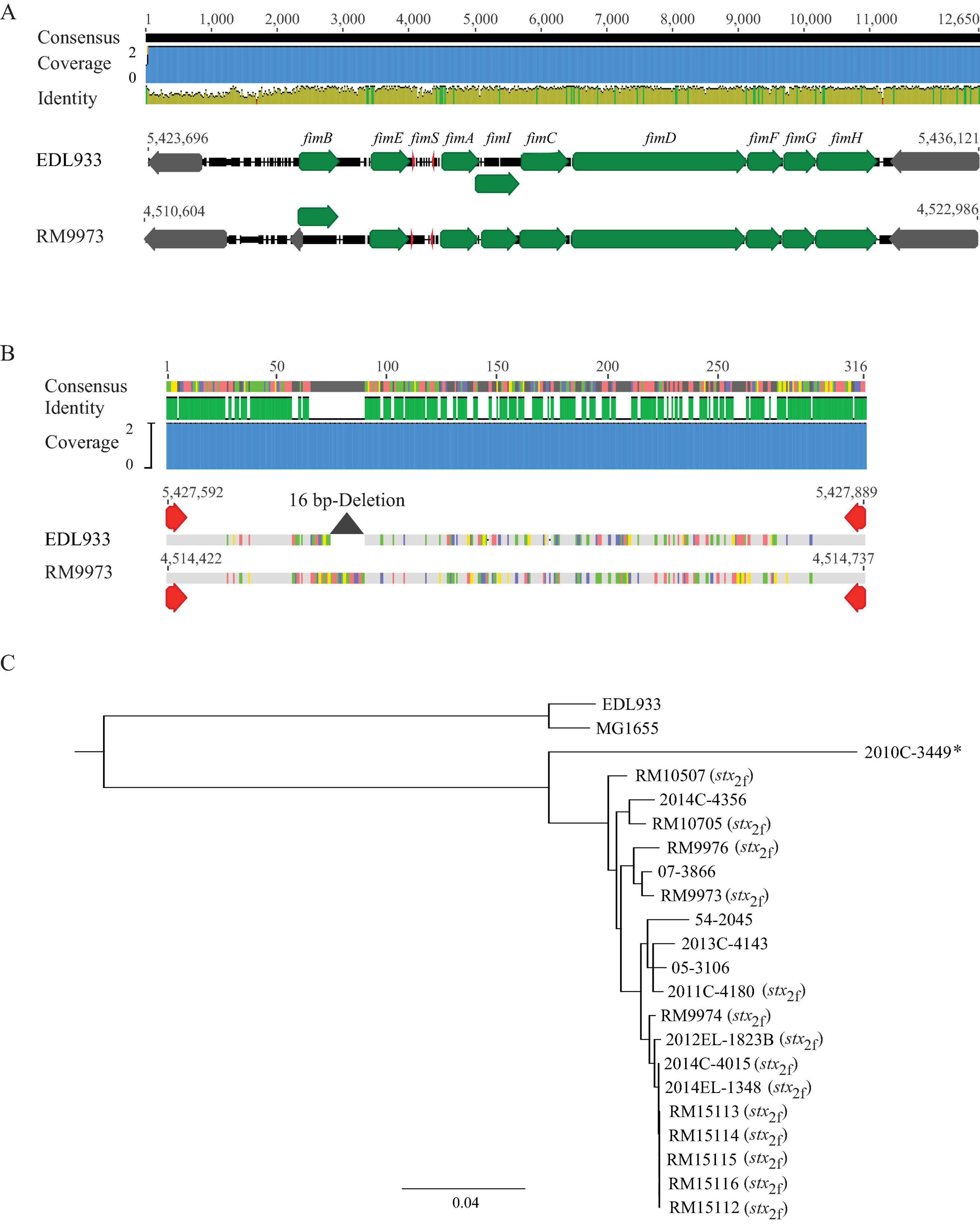
Figure 3. Sequence analyses of E. albertii type 1 fimbriae genes. (A) Chromosomal locations and pairwise comparison of the type 1 fimbriae genes between the E. albertii avian strain RM9973 and E. coli O157:H7 strain EDL933. Numbers indicate the chromosomal positions in each strain. Green arrows represent the fim genes; Gray arrows represent the neighbor genes; and the red arrows represents right and left inverted repeats (IRs) within fimS. The 9-bp IR in strain EDL933 is 5′-ttggggcca-3′ while in strain RM9973, the 9-bp IR is 5′-ttgggacca-3′. (B) Pairwise alignment of the cis element fimS of the E. albertii avian strain RM9973 and E. coli O157:H7 strain EDL933. Red arrows represent the IRs. The grey triangle represents the 16 bp-deletion in EDL933 fimS. (C) Sequence analysis of E. albertii type 1 fimbriae genes with the fim genes in E. coli strains EDL933 and MG1655. The fim genes were identified by BLASTn search of a database containing all genomes examined in this study using an 8.7-Kb DNA fragment containing the nine fim genes of the E. coli strain EDL933 as a query in Geneious Prime®. The sequences of the fim genes were extracted from corresponding genomes and aligned using Clustal Omega alignment in Geneious Prime®. A consensus tree was constructed using Geneious Tree Builder with the following parameters: Genetic Distance Model, Jukes-Cantor; Resampling tree method: Bootstrap; Number of Replicates: 10,000; Support Threshold: 50%. The stx2f positive strains are indicated in parentheses. The strain marked with an “*” carries mutations within the coding sequences of type 1 fimbriae genes.
Expression of type 1 fimbriae in E. coli O157:H7 strains including EDL933 is silenced due to a 16 bp deletion in fimS (Roe et al., 2001). This deletion locks the transcription of fimA at the “OFF” orientation. Comparative analyses of E. albertii fimS genes with the EDL933 fimS revealed an intact fimS in E. albertii strains, like the fimS in E. coli K-12 strain MG1655 (Figure 3B). Sequence analyses of other fim genes placed all E. albertii strains in the same clade, separated from the fim genes in the E. coli strains (Figure 3C). Various mutations including point deletions and IS insertions were revealed in the fimA, fimC, and fimD genes of clinical strain 2010C-3449, the fimD of the clinical strain 54-2045, and the fimI of the clinical strain 2013C-4143 (Supplementary Table 3).
However, a mannose-sensitive hemagglutination assay failed to detect type 1 fimbriae in any of the 20 E. albertii strains examined when they were grown in LBHS at 37°C or in LBNS at 28°C, like strain EDL933. Production of type 1 fimbriae was detected in E. coli DH5a cells under both testing conditions.
3.3.3 Other fimbriae genes
Homologs of genes encoding hemorrhagic E. coli pilus (hcpABC) were identified in all 20 E. albertii strains, and mutations in hcpB (annotated as gspE in E. albertii), encoding the type II secretion system protein GspE, were present in seven out of the 20 strains examined (Supplementary Table 3). Similarly, homologs of genes encoding the Sfm fimbriae (sfmACDHF and sfmZ) were identified in the 18 out of the 20 strains examined. Mutations were most common in sfmD, encoding a fimbrial biogenesis usher protein (Supplementary Table 3). In the clinical strains 54-2045 and 2010C-3449, only a homolog of sfmA was present. Among the adhesive fimbriae genes examined, homologs of cfaABCD genes, which are often present on the chromosomes of EHEC strains, were identified in 10 out of the 14 stx2f-positive E. albertii strains, while homologs of faeCDEFGHIJ genes, which are often present on the plasmids of enterotoxigenic E. coli (ETEC) strains, were identified in the clinical strain 07-3866 and in the chicken isolate 2014C-4356 (Supplementary Table 3). In E. coli, there are 12 genes (papXGFEKJDCHABI) related to biogenesis of P fimbriae. Homologs of seven genes, papEKJDCHA, were identified in seven out of the 20 E. albertii strains, homologs of five genes, papJDCHA, were identified in the chicken isolate 2014C-4356, and homologs of four genes, papDCHA, were identified in the clinical strain 2012EL-1823B.
3.4 Nonfimbrial adhesin genes and their genetic diversity
The most common autotransporter adhesin genes detected in E. albertii were paa, ehaC, eaeH, ehaB, and sinB (Supplementary Table 3). paa encodes an AcfC family adhesin. A homolog of paa was present in nearly all E. albertii strains examined and exhibited > 80% sequence identity with the paa gene in EHEC strain EDL933. The gene ehaC encodes an AIDA-I family autotransporter adhesin. A homolog of ehaC was detected in all E. albertii strains examined, although a point deletion and a point insertion were present in the avian strain RM10705 and the clinical strain 2010C-3449, respectively (Supplementary Table 3). The E. albertii ehaC genes exhibited ∼ 75% sequence identity with the ehaC gene in strain EDL933. eaeH encodes an intimin-like adhesin FdeC. A distant homolog (∼ 80% length in CDS and 27.6% identity in amino acids) was identified in all E. albertii strains examined, although mutations were detected in the clinical strain 2011C-4180 (IS insertion), and avian strains RM10507 (Insertion of 5’-GTCTG-3’) and RM10705 (a point deletion). A homolog of ehaB, ranging in size from 2430 bp to 2979 bp was detected in E. albertii strains. Interestingly, the ehaB genes in avian strains RM9973 and RM9976, and in clinical strains 05-3106, 54-2045, and 2014C-4356, displayed higher sequence similarity with the EDL933 ehaB gene compared with the ehaB genes in other E. albertii strains. The gene sinH, encoding an intimin-like inverse autotransporter, was present in all E. albertii strains examined. In fact, the gene sinH appeared to be widespread in E. albertii but only present in a subset of E. coli strains. Homologs of sinH were not identified in EHEC strain EDL933.
3.5 Biofilm formation
Following the initial 24 h incubation, a visible ring was observed for avian strain RM9974 and four clinical strains, 2012EL-1823B, 2014C-4015, 2014EL-1348, and 07-3866 (Figure 4A). Consistently, quantitative analysis revealed that the attached biomass for the above five strains were all significantly greater than the rest of the strains except the comparison between strains 2014EL-1348 and RM15112 (One-way ANOVA, adjust P < 0.05) (Supplementary Table 4). Among the five biofilm producing strains, strains RM9974, 2014C-4015, and 07-3866 produced significantly greater amounts of biofilm than the other two strains (Figure 4B). Following 48 h incubation, the attached biomass for the five biofilm producing strains were all significantly greater than the corresponding biofilms at 24 h (Figure 4C) (One way ANOVA test, adjust P < 0.05). Among the strains that did not produce any detectable biofilm at 24 h, a visible ring was observed for strains RM9973 and RM10705 (Figure 4A). Quantitative analysis revealed that strains 07-3866, 2014C-4015, 2014EL-1348, and RM9974 produced significantly greater amounts of biofilm than that of the strain 2012EL-1823B (One way ANOVA test, adjust P < 0.05) (Figure 4C). Among the strains that did not produce any visible biofilms at 48 h, strain 2014C-4356 produced a considerable amount of biomass on the glass surface following 120 h incubation (Figure 4A). Among the strains that produced biofilms following 48 h incubation, a significant increase in attached biomass was observed for all strains following 120 h incubation (One-way ANOVA, adjust P < 0.05) (Figure 4D). At 120 h post inoculation, quantitative analysis revealed that avian strain RM9974, chicken isolate 2014C-4356 along with the clinical strain 2014C-4015 were the strongest biofilm producers, followed by the clinical strains 2014EL-1348 and 2012EL-1823B, and the avian strain RM9973, which all produced significantly greater amounts of biofilm than the clinical strain 07-3866 (One-Way ANOVA, adjust P < 0.05). For avian strains RM9976 and RM10705, although a visible ring was detected on glass surfaces, they were not significantly different from those of non-biofilm producing strains (Figure 4D).
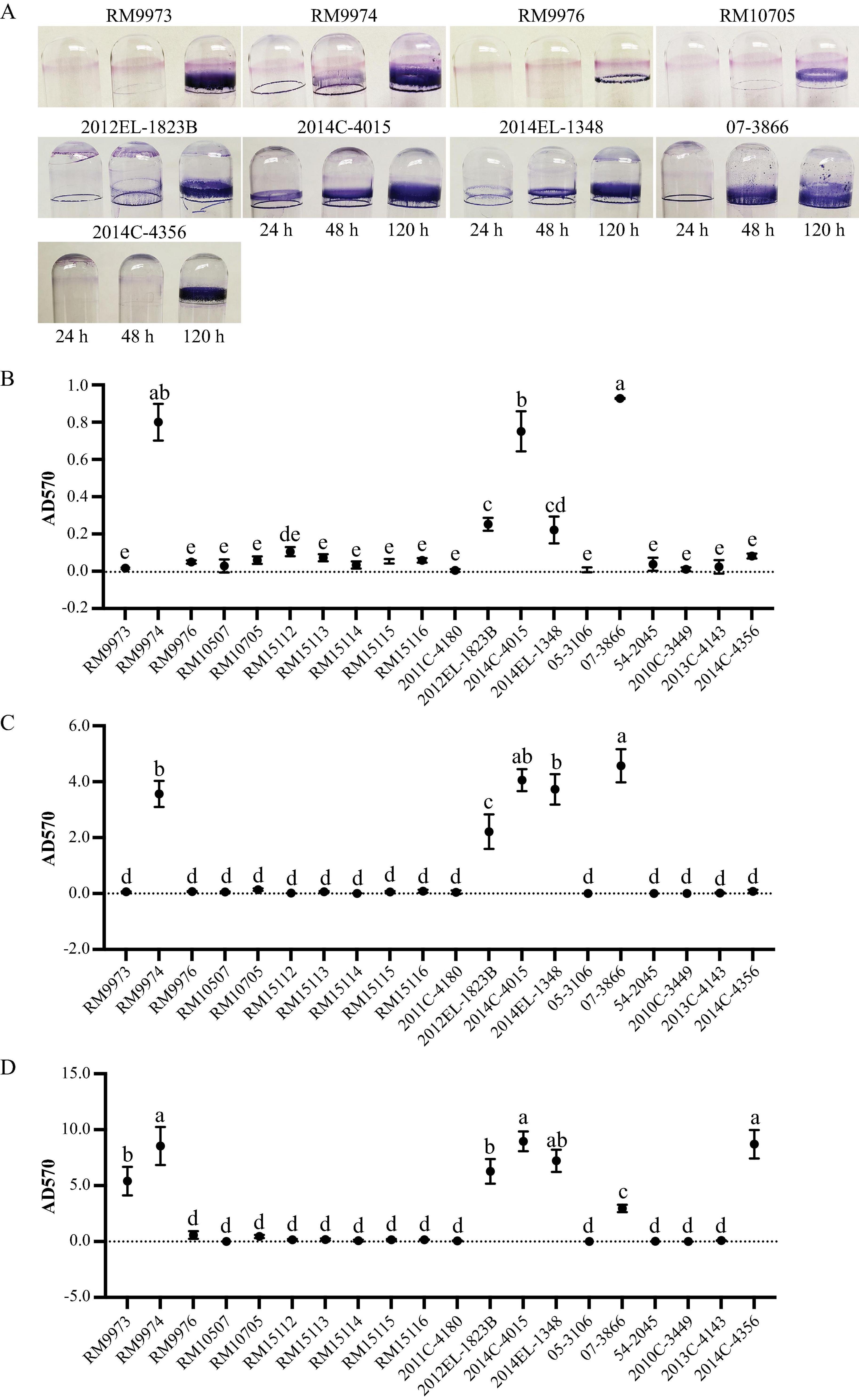
Figure 4. Biofilm formation by E. albertii strains on glass surfaces. (A) Crystal violet staining the attached biomass on the glass surfaces under a static growth condition for 24 h, 48 h, and 120 h. Only strains that can produce visible rings are shown here. (B–D) quantitative analyses of biofilms under a static growth condition for 24 h (B), 48 h (C), and 120 h (D). Each data set represents the mean and SD of three biological replicates. Differences that are statistically significant (One-way ANOVA followed by a Tukey’s multiple comparisons test, adjust P < 0.05) are indicated by different letters. The detailed results of the statistical analyses are presented in Supplementary Table 4.
4 Discussion
Flagellar motility allows bacteria to move rapidly towards nutrients and away from toxic substances, thus it plays an essential role in bacteria to explore new niches and to establish colonization. Moreover, flagella also serve as a virulence factor in many enteric pathogens, contributing to adhesion, invasion, and host colonization (Moens and Vanderleyden, 1996; Colin et al., 2021). In E. coli, nearly 50 genes are involved in flagella assembly and function. Expression of the flagellar genes is tightly regulated by a three-tiered transcriptional hierarchy to ensure production of flagellum at the right time and under the applicable conditions (Khan et al., 2020). The group I genes, flhDC, encode the master transcriptional regulator FlhDC that activates the expression of the group II genes. In E. coli, there are nearly 30 genes belonging to the group II and many of these genes encode components of the flagellum basal body and hook and the sigma factor 28 FliA. FliA regulates the expression of group III genes, which are involved in synthesis of complete flagellum and chemotaxis systems. The regulation of flagellar gene expression in E. albertii is unknown, although a similar hierarchical regulation fashion is expected considering the close phylogenetic relationship between the two species. Originally, E. albertii was thought nonmotile and lacked flagella although 74% E. albertii strains were reported to carry a complete set of flagellar biosynthesis genes (Abbott et al., 2003; Ooka et al., 2015). Induction of flagellar motility by low osmotic pressure was observed in 27 out of the 59 E. albertii strains tested (Ikeda et al., 2020); similarly, induction of swimming motility by nutrients derived from pigeon droppings was observed in six out of the 12 strains examined (Murakami et al., 2020), implying strain variation in expression of flagellar motility in E. albertii. Consistently, our study revealed great diversity in flagellar genes repertoire and conditionally expressing swimming motility in E. albertii. Among the 20 E. albertii strains examined, three were motile regardless of the growth conditions, while four were motile only when grown in pond water supplemented with pigeon droppings or in the diluted TSB medium. Deletion of a large DNA fragment containing genes flhAB, motBA, and flhCD is likely the molecular basis of nonmotile phenotypes observed in five avian and two clinical strains. Furthermore, a loss-of-function mutation in genes fliC, fliF, fliJ, flgD, flgG, flhA, and motA may explain some but not all non-motile phenotypes observed in our study. Considering the highly complex regulation in flagellar gene expression, comparative transcriptomic studies may provide insight into the molecular basis of the strain variation in expression of flagellar motility.
Our study revealed the presence of the Flag-2 locus in E. albertii. The Flag-2 locus appears to be widespread among the Enterobacterales (De Maayer et al., 2020) and serve as a hot spot for gene insertions and deletions. Consistently, great sequence variation was observed among the five Flag-2 loci identified, including large deletions, point mutations and transposon insertions. However, several VR1 genes that are predicated on having a role in posttranslational regulation of flagellar biosynthesis are conserved in E. albertii, including the glycosyltransferase gene and the lysine-N-methylase gene. Unlike the primary flagellar system, the function of the Flag-2 locus is not fully understood. Expression of Flag-2 genes was observed in Yersinia enterocolitica with a maximal level at 20°C and, in Plesiomonas shigelloides, the Flag-2 locus encoded lateral flagella appeared to be essential for swarming motility (Bresolin et al., 2008; Merino et al., 2015). Systematic analyses of cargo genes located in the VR2 in Flag-2 loci suggested a role in secretion of virulence factor and in inter-bacterial competition (De Maayer et al., 2020). Searching other E. albertii genomes deposited in public databases as of April 2024 revealed that about 25% of genomes carry a Flag-2 locus. Additional studies are needed to elucidate any physiological roles or ecological benefits conferred by this secondary flagellar system in E. albertii.
Among the 12 fimbriae/pili that are commonly present in E. coli, genes encoding curli fimbriae, hemorrhagic E. coli pilus, type 1 fimbriae, and Sfm fimbriae were identified in most of the E. albertii strains examined, while genes related to biogenesis of adhesive fimbriae, or P fimbriae were only present in a subset of strains. Curli, also known as bacterial amyloid, is an important colonization factor involved in initial surface attachment, biofilm formation, and induction of the host inflammatory response (Barnhart and Chapman, 2006). Although 19 out of the 20 E. albertii strains examined in our study carried intact curli genes, production of curli fimbriae was detected in only nine strains. This strain variation could not be explained solely by the differences in the coding sequences of the curli genes or the differences in the intergenic regions between the two curli operons, including the promoters of csgD and csgB, since some curli-deficient strains shared the identical intergenic sequences with the curli expressing strains (Data not shown). Strain variation in curli production were reported in E. coli and Salmonella enterica (Romling et al., 1998; Dyer et al., 2007), which both have served as the model organisms for studying curli biogenesis and regulation. In both E. coli and S. enterica, expression of curli is regulated by a complex regulation network involving multiple transcriptional regulators, two-component regulatory systems, and in some isolates cyclic dinucleotide 3’,5’-cyclic di-GMP (Barnhart and Chapman, 2006; Blomfield and van der Woude, 2007). Therefore, mutations in any of these regulators or the target sequences that interact with the regulators directly or indirectly could have an impact on the expression of curli fimbriae. For example, mutations in genes encoding the transcriptional regulators rpoS or rcsB were reported to be the molecular bases of strain variation in curli production in EHEC O157:H7 (Carter et al., 2012; Carter et al., 2014) and mutations in the csgD promoter could lead to overproduction of curli fimbriae (Uhlich et al., 2002). Knowledge about the regulation of curli fimbriae in E. albertii is limited. Examining genes encoding putative transcriptional regulators of the curli genes in E. albertii revealed a loss-of-function mutation in the rpoS gene in strains 05-3106 and 54-2045, while no mutations were identified in genes encoding Crl, MlrA, CpxRA, OmpR-EnvZ, or RcsBC. Additional studies are required to gain a comprehensive understanding of curli regulation network in E. albertii as well as the environmental and physiological signals that may induce or repress the expression of curli fimbriae.
Although the majority of E. albertii strains examined in our study carried functional fim genes and an intact fimS, none of them displayed a mannose-sensitive hemagglutination (MSHA) phenotype under the growth conditions examined. Expression of type 1 fimbriae was reported to be dependent on the growth conditions. For example, optimal production of a predominantly type 1 fimbriae positive population in Shigella required serial passage every 48 to 72 h in unshaken brain heart infusion broth at 37°C (Snellings et al., 1997). Since the goal of our study was to reveal if the type 1 fimbriae contributed to the biofilm formation in E. albertii, the conditions tested for production of type 1 fimbriae were the conditions used for examining biofilm formation, which may not be optimal for expression of type 1 fimbriae. Additionally, the phase variation of type 1 fimbriation is regulated at multiple levels. In E. coli, switch of fimS is required but not sufficient for biosynthesis of type 1 fimbriae. Besides FimB and FimE, other transcriptional regulators including IHF, Lrp, and H-NS were reported to be involved in fimS switch (Blomfield and van der Woude, 2007). Variations in the activities of FimB and FimE, cross talks between fimbrial operons, as well as the presence of other recombinases can all contribute to variation in expression of type 1 fimbriae. Additional studies are required to understand growth conditions, physiological cues, and environmental signals for induction of type 1 fimbriae in E. albertii.
Among the nonfimbrial adhesins examined, genes encoding the autotransporter (AT) adhesins were predominant in E. albertii. For example, homologs of ehaA, ehaB, ehaC, and upaH that all encode an AIDA-I type autotransporter (AT) adhesin were identified in all or most of the strains examined and a homolog of ehaG, encoding a trimeric AT adhesin was identified in all strains. Other AIDA-1 type adhesins genes in E. albertii included aatA, aidA, agn43, and cah. In E. coli, Ag43 is the most prevalent AIDA-1 type AT adhesin, however, in E. albertii, the agn43 was identified only in a clinical strain. Other commonly detected non-fimbrial adhesin genes were eaeH, paa, and sinH. The gene eaeH encodes an intimin-like adhesin that facilitates adhesion of bacterial cells, delivery of heat-labile toxin, and colonization of the small intestine in ETEC (Sheikh et al., 2014). The gene paa, encoding an AcfC family adhesin, is widespread in both EHEC and ETEC strains. Paa contributes to the formation of A/E lesions in animal hosts and thus is an important virulence factor in various E. coli pathotypes (An et al., 1999; Batisson et al., 2003). The paa gene appears to be widespread in E. albertii and in some strains, there are two paa loci, including the bird strains isolated in Poland (BioSample numbers: SAMN33094111, SAMN33094114, SAMN33094099, SAMN33094102, and SAMN33094112), and a poultry strain isolated in China (SAMN17525956). The gene sinH encodes an intimin-like inverse autotransporter. The inverse autotransporters were reported to play a role in biofilm formation in E. coli and contributed to biofilm formation and virulence in Yersinia ruckeri (Martinez-Gil et al., 2017; Goh et al., 2019; Wrobel et al., 2020). However, deletion of sinH in the UPEC strain CFT073 did not impact the biofilm formation significantly, rather, the mutant displayed a significant fitness reduction during UTI in a murine model (Shea et al., 2022). Like paa, sinH appears to be conserved in E. albertii. BLASTn search of additional 57 complete E. albertii genomes deposited in GenBank as of April 2024 identified a sinH in all of them.
Like curli production, E. albertii strains differed greatly in biofilm formation on glass surfaces. Consistent with our previous report that, in STEC, curli fimbriae are important for biofilm formation on abiotic surfaces (Carter et al., 2016; Carter et al., 2019), all curli-producing E. albertii strains produced moderate or strong biofilms under the condition examined. Interestingly, some curli-positive strains produced visible biofilm following a 24-h incubation, while others did not produce biofilms until a 120-h incubation, implying a difference in biofilm development among the E. albertii strains. Furthermore, although no curli fimbriae were detected in chicken isolate 2014C-4356, strong biofilms were observed on day 5 of incubation, suggesting a role of other adhesins in biofilm formation. Strains 2014C-4356 and 07-3866 were the only strains carrying genes encoding the adhesive fimbriae that are located on a large plasmid commonly found in ETEC strains, such as pUMNK88_Hly (GenBank accession # NC_017643.1). It requires further investigation to determine if the plasmid-borne adhesive fimbriae are expressed in E. albertii and whether it contributes to biofilm formation in strains that do not produce curli fimbriae.
Biofilms of foodborne pathogens can enhance their survival and persistence in diverse ecological niches and serve as sources of contamination in food production environments and of infection in health-care environments. Adhesion is the first step in biofilm development and in establishing colonization in animal hosts. Strong adherence often implies enhanced surface attachment and biofilm formation, leading to increased fitness and pathogenic potential. Therefore, understanding the adhesion capability and the underlining factors in E. albertii would provide valuable information for development of effective control strategies. Our study revealed that curli fimbriae, Type 1 fimbriae, Sfm fimbriae, and HCP appear to be the common fimbrial adhesins in E. albertii, while adhesive fimbriae was a strain-specific trait. Among the numerous nonfimbrial adhesins identified in E. albertii, autotransporter adhesins EhaA, EhaB, EhaC, EhaG, and SinH, and the adherence factors EaeH and Paa are common, while Agn43, Cah, and Iha that are widespread in E. coli, are only associated with a few strains. E. albertii strains carry different combinations of fimbrial and nonfimbrial adhesins that may facilitate colonization of E. albertii in diverse niches. Our study further revealed great variations in expression of curli fimbriae and in biofilm production, suggesting complex regulation in expression of adhesins in E. albertii. Studies are needed to identify environmental cues that induce the adhesions expression and the receptors specifically interacted with each adhesin to gain insight into molecular basis of niche selection for E. albertii, an emerging human and avian pathogen.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Table 1.
Author contributions
MC: Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review and editing. DC: Data curation, Methodology, Validation, Writing – review and editing. RL: Conceptualization, Methodology, Resources, Writing – review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of the article. This work was supported by the USDA-ARS CRIS project 2030-42000-052-000D and the Advanced Molecular Detection (AMD) Initiative grant number AMD-21 at the Centers for Disease Control and Prevention.
Acknowledgments
We wish to thank Nicole Laniohan for providing technical assistance in curli detection and biofilm assay.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention, the Department of Health and Human Services, or the United States government. Furthermore, the use of any product names, trade names, images, or commercial sources is for identification purposes only, and does not imply endorsement or government sanction by the U.S. Department of Health and Human Services.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1456637/full#supplementary-material
Supplementary Table 1 | E. albertii primary flagellar genes.
Supplementary Table 2 | E. albertii secondary flagellar genes.
Supplementary Table 3 | E. albertii fimbrial and non-fimbrial adhesin genes.
Supplementary Table 4 | Statistical analyses of E. albertii biofilms.
References
Abbott, S. L., O’Connor, J., Robin, T., Zimmer, B. L., and Janda, J. M. (2003). Biochemical properties of a newly described Escherichia species, Escherichia albertii. J. Clin. Microbiol. 41, 4852–4854. doi: 10.1128/JCM.41.10.4852-4854.2003
Abraham, J. M., Freitag, C. S., Clements, J. R., and Eisenstein, B. I. (1985). An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 82, 5724–5727. doi: 10.1073/pnas.82.17.5724
An, H., Fairbrother, J. M., Desautels, C., and Harel, J. (1999). Distribution of a novel locus called Paa (porcine attaching and effacing associated) among enteric Escherichia coli. Adv. Exp. Med. Biol. 473, 179–184. doi: 10.1007/978-1-4615-4143-1_17
Arai, S., Yamaya, S., Ohtsuka, K., Konishi, N., Obata, H., Ooka, T., et al. (2022). Detection of Escherichia albertii in Retail Oysters. J. Food Prot. 85, 173–179. doi: 10.4315/JFP-21-222
Barmettler, K., Biggel, M., Treier, A., Muchaamba, F., and Stephan, R. (2023). Livestock as possible reservoir of Escherichia albertii in Switzerland. Schweiz. Arch. Tierheilkd. 165, 299–306. doi: 10.17236/sat00393
Barnhart, M. M., and Chapman, M. R. (2006). Curli biogenesis and function. Annu. Rev. Microbiol 60, 131–147. doi: 10.1146/annurev.micro.60.080805.142106
Batisson, I., Guimond, M. P., Girard, F., An, H., Zhu, C., Oswald, E., et al. (2003). Characterization of the novel factor paa involved in the early steps of the adhesion mechanism of attaching and effacing Escherichia coli. Infect. Immun 71, 4516–4525. doi: 10.1128/IAI.71.8.4516-4525.2003
Bengtsson, R. J., Baker, K. S., Cunningham, A. A., Greig, D. R., John, S. K., Macgregor, S. K., et al. (2023). The genomic epidemiology of Escherichia albertii infecting humans and birds in Great Britain. Nat. Commun. 14:1707. doi: 10.1038/s41467-023-37312-3
Biscola, F. T., Abe, C. M., and Guth, B. E. (2011). Determination of adhesin gene sequences in, and biofilm formation by, O157 and non-O157 Shiga toxin-producing Escherichia coli strains isolated from different sources. Appl. Environ. Microbiol. 77, 2201–2208. doi: 10.1128/AEM.01920-10
Blomfield, I., and van der Woude, M. (2007). Regulation of fimbrial expression. EcoSal Plus 2:2. doi: 10.1128/ecosal.2.4.2.2
Bresolin, G., Trcek, J., Scherer, S., and Fuchs, T. M. (2008). Presence of a functional flagellar cluster Flag-2 and low-temperature expression of flagellar genes in Yersinia enterocolitica W22703. Microbiology (Reading) 154, 196–206. doi: 10.1099/mic.0.2007/008458-0
Carter, M. Q., Brandl, M. T., Louie, J. W., Kyle, J. L., Carychao, D. K., Cooley, M. B., et al. (2011). Distinct acid resistance and survival fitness displayed by Curli variants of enterohemorrhagic Escherichia coli O157:H7. Appl. Environ. Microbiol. 77, 3685–3695. doi: 10.1128/AEM.02315-10
Carter, M. Q., Feng, D., and Li, H. H. (2019). Curli fimbriae confer Shiga toxin-producing Escherichia coli a competitive trait in mixed biofilms. Food Microbiol. 82, 482–488. doi: 10.1016/j.fm.2019.03.024
Carter, M. Q., Louie, J. W., Feng, D., Zhong, W., and Brandl, M. T. (2016). Curli fimbriae are conditionally required in Escherichia coli O157:H7 for initial attachment and biofilm formation. Food Microbiol. 57, 81–89. doi: 10.1016/j.fm.2016.01.006
Carter, M. Q., Louie, J. W., Huynh, S., and Parker, C. T. (2014). Natural rpoS mutations contribute to population heterogeneity in Escherichia coli O157:H7 strains linked to the 2006 US spinach-associated outbreak. Food Microbiol. 44, 108–118. doi: 10.1016/j.fm.2014.05.021
Carter, M. Q., Parker, C. T., Louie, J. W., Huynh, S., Fagerquist, C. K., and Mandrell, R. E. (2012). RcsB contributes to the distinct stress fitness among Escherichia coli O157:H7 curli variants of the 1993 hamburger-associated outbreak strains. Appl. Environ. Microbiol. 78, 7706–7719. doi: 10.1128/AEM.02157-12
Carter, M. Q., Quinones, B., He, X., Pham, A., Carychao, D., Cooley, M. B., et al. (2023a). Genomic and phenotypic characterization of shiga toxin-producing Escherichia albertii strains isolated from wild birds in a major agricultural region in California. Microorganisms 11:2803. doi: 10.3390/microorganisms11112803
Carter, M. Q., Quinones, B., Laniohan, N., Carychao, D., Pham, A., He, X., et al. (2023b). Pathogenicity assessment of Shiga toxin-producing Escherichia coli strains isolated from wild birds in a major agricultural region in California. Front. Microbiol. 14:1214081. doi: 10.3389/fmicb.2023.1214081
Colin, R., Ni, B., Laganenka, L., and Sourjik, V. (2021). Multiple functions of flagellar motility and chemotaxis in bacterial physiology. FEMS Microbiol. Rev. 45:fuab038. doi: 10.1093/femsre/fuab038
Cookson, A. L., Cooley, W. A., and Woodward, M. J. (2002). The role of type 1 and curli fimbriae of Shiga toxin-producing Escherichia coli in adherence to abiotic surfaces. Int. J. Med. Microbiol. 292, 195–205. doi: 10.1078/1438-4221-00203
De Maayer, P., Pillay, T., and Coutinho, T. A. (2020). Flagella by numbers: Comparative genomic analysis of the supernumerary flagellar systems among the Enterobacterales. BMC Genom. 21:670. doi: 10.1186/s12864-020-07085-w
Dyer, J. G., Sriranganathan, N., Nickerson, S. C., and Elvinger, F. (2007). Curli production and genetic relationships among Escherichia coli from cases of bovine mastitis. J. Dairy Sci. 90, 193–201. doi: 10.3168/jds.S0022-0302(07)72620-6
Elpers, L., and Hensel, M. (2020). Expression and functional characterization of various chaperon-usher fimbriae, Curli Fimbriae, and Type 4 Pili of enterohemorrhagic Escherichia coli O157:H7 Sakai. Front. Microbiol. 11:378. doi: 10.3389/fmicb.2020.00378
Felfoldi, T., Heeger, Z., Vargha, M., and Marialigeti, K. (2010). Detection of potentially pathogenic bacteria in the drinking water distribution system of a hospital in Hungary. Clin. Microbiol. Infect. 16, 89–92. doi: 10.1111/j.1469-0691.2009.02795.x
Fink, R. C., Black, E. P., Hou, Z., Sugawara, M., Sadowsky, M. J., and Diez-Gonzalez, F. (2012). Transcriptional responses of Escherichia coli K-12 and O157:H7 associated with lettuce leaves. Appl. Environ. Microbiol. 78, 1752–1764. doi: 10.1128/AEM.07454-11
Goh, K. G. K., Moriel, D. G., Hancock, S. J., Phan, M. D., and Schembri, M. A. (2019). Bioinformatic and molecular analysis of inverse autotransporters from Escherichia coli. mSphere 4, e00572–19. doi: 10.1128/mSphere.00572-19
Gomes, T. A. T., Ooka, T., Hernandes, R. T., Yamamoto, D., and Hayashi, T. (2020). Escherichia albertii pathogenesis. EcoSal Plus 9:15. doi: 10.1128/ecosalplus.ESP-0015-2019
Gophna, U., Barlev, M., Seijffers, R., Oelschlager, T. A., Hacker, J., and Ron, E. Z. (2001). Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect. Immun. 69, 2659–2665. doi: 10.1128/IAI.69.4.2659-2665.2001
Hinenoya, A., Li, X. P., Zeng, X., Sahin, O., Moxley, R. A., Logue, C. M., et al. (2021). Isolation and characterization of Escherichia albertii in poultry at the pre-harvest level. Zoon. Public Health 68, 213–225. doi: 10.1111/zph.12812
Hinenoya, A., Wang, H., Patrick, E. M., Zeng, X., Cao, L., Li, X. P., et al. (2022). Longitudinal surveillance and comparative characterization of Escherichia albertii in wild raccoons in the United States. Microbiol. Res. 262:127109. doi: 10.1016/j.micres.2022.127109
Hirose, S., Konishi, N., Sato, M., Suzumura, K., Obata, H., Ohtsuka, K., et al. (2024). Growth and survival of Escherichia albertii in food and environmental water at various temperatures. J. Food Prot. 87:100249. doi: 10.1016/j.jfp.2024.100249
Iguchi, A., Takemura, T., Ogura, Y., Nguyen, T. T. H., Kikuchi, T., Okuno, M., et al. (2023). Genomic characterization of endemic diarrheagenic Escherichia coli and Escherichia albertii from infants with diarrhea in Vietnam. PLoS Negl. Trop. Dis. 17:e0011259. doi: 10.1371/journal.pntd.0011259
Iida, K., Mizunoe, Y., Wai, S. N., and Yoshida, S. (2001). Type 1 fimbriation and its phase switching in diarrheagenic Escherichia coli strains. Clin. Diagn. Lab. Immunol. 8, 489–495. doi: 10.1128/CDLI.8.3.489-495.2001
Ikeda, T., Shinagawa, T., Ito, T., Ohno, Y., Kubo, A., Nishi, J., et al. (2020). Hypoosmotic stress induces flagellar biosynthesis and swimming motility in Escherichia albertii. Commun. Biol. 3:87. doi: 10.1038/s42003-020-0816-5
Karatan, E., and Watnick, P. (2009). Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73, 310–347. doi: 10.1128/MMBR.00041-08
Khan, F., Tabassum, N., Pham, D. T. N., Oloketuyi, S. F., and Kim, Y. M. (2020). Molecules involved in motility regulation in Escherichia coli cells: A review. Biofouling 36, 889–908. doi: 10.1080/08927014.2020.1826939
Konno, T., Yatsuyanagi, J., Takahashi, S., Kumagai, Y., Wada, E., Chiba, M., et al. (2012). Isolation and identification of Escherichia albertii from a patient in an outbreak of gastroenteritis. Jpn. J. Infect. Dis. 65, 203–207. doi: 10.7883/yoken.65.203
Lima, M. P., Yamamoto, D., Santos, A. C. M., Ooka, T., Hernandes, R. T., Vieira, M. A. M., et al. (2019). Phenotypic characterization and virulence-related properties of Escherichia albertii strains isolated from children with diarrhea in Brazil. Pathog. Dis. 77:ftz014. doi: 10.1093/femspd/ftz014
Lindsey, R. L., Fedorka-Cray, P. J., Abley, M., Turpin, J. B., and Meinersmann, R. J. (2015). Evaluating the occurrence of Escherichia albertii in chicken carcass rinses by PCR, Vitek analysis, and sequencing of the rpoB gene. Appl. Environ. Microbiol. 81, 1727–1734. doi: 10.1128/AEM.03681-14
Maeda, E., Murakami, K., Sera, N., Ito, K., and Fujimoto, S. (2015). Detection of Escherichia albertii from chicken meat and giblets. J. Vet. Med. Sci. 77, 871–873. doi: 10.1292/jvms.14-0640
Maheux, A. F., Boudreau, D. K., Bergeron, M. G., and Rodriguez, M. J. (2014). Characterization of Escherichia fergusonii and Escherichia albertii isolated from water. J. Appl. Microbiol. 117, 597–609. doi: 10.1111/jam.12551
Martinez-Gil, M., Goh, K. G. K., Rackaityte, E., Sakamoto, C., Audrain, B., Moriel, D. G., et al. (2017). YeeJ is an inverse autotransporter from Escherichia coli that binds to peptidoglycan and promotes biofilm formation. Sci. Rep. 7:11326. doi: 10.1038/s41598-017-10902-0
Masuda, K., Ooka, T., Akita, H., Hiratsuka, T., Takao, S., Fukada, M., et al. (2020). Epidemiological aspects of Escherichia albertii Outbreaks in Japan and genetic characteristics of the causative pathogen. Foodborne Pathog. Dis 17, 144–150. doi: 10.1089/fpd.2019.2654
McWilliams, B. D., and Torres, A. G. (2014). Enterohemorrhagic Escherichia coli Adhesins. Microbiol. Spectr. 2:3. doi: 10.1128/microbiolspec.EHEC-0003-2013
Merino, S., Aquilini, E., Fulton, K. M., Twine, S. M., and Tomas, J. M. (2015). The polar and lateral flagella from Plesiomonas shigelloides are glycosylated with legionaminic acid. Front. Microbiol. 6:649. doi: 10.3389/fmicb.2015.00649
Moens, S., and Vanderleyden, J. (1996). Functions of bacterial flagella. Crit. Rev. Microbiol. 22, 67–100. doi: 10.3109/10408419609106456
Muchaamba, F., Barmettler, K., Treier, A., Houf, K., and Stephan, R. (2022). Microbiology and epidemiology of Escherichia albertii-an emerging elusive foodborne pathogen. Microorganisms 10:875. doi: 10.3390/microorganisms10050875
Murakami, K., Kimura, S., Nagafuchi, O., Sekizuka, T., Onozuka, D., Mizukoshi, F., et al. (2020). Flagellum expression and swimming activity by the zoonotic pathogen Escherichia albertii. Environ. Microbiol. Rep. 12, 92–96. doi: 10.1111/1758-2229.12818
Oaks, J. L., Besser, T. E., Walk, S. T., Gordon, D. M., Beckmen, K. B., Burek, K. A., et al. (2010). Escherichia albertii in wild and domestic birds. Emerg. Infect. Dis. 16, 638–646. doi: 10.3201/eid1604.090695
Ooka, T., Ogura, Y., Katsura, K., Seto, K., Kobayashi, H., Kawano, K., et al. (2015). Defining the genome features of Escherichia albertii, an emerging enteropathogen closely related to Escherichia coli. Genome Biol. Evol. 7, 3170–3179. doi: 10.1093/gbe/evv211
Ooka, T., Seto, K., Kawano, K., Kobayashi, H., Etoh, Y., Ichihara, S., et al. (2012). Clinical significance of Escherichia albertii. Emerg. Infect. Dis. 18, 488–492. doi: 10.3201/eid1803.111401
Ooka, T., Tokuoka, E., Furukawa, M., Nagamura, T., Ogura, Y., Arisawa, K., et al. (2013). Human gastroenteritis outbreak associated with Escherichia albertii, Japan. Emerg. Infect. Dis. 19, 144–146. doi: 10.3201/eid1901.120646
Ori, E. L., Takagi, E. H., Andrade, T. S., Miguel, B. T., Cergole-Novella, M. C., Guth, B. E. C., et al. (2018). Diarrhoeagenic Escherichia coli and Escherichia albertii in Brazil: Pathotypes and serotypes over a 6-year period of surveillance. Epidemiol. Infect. 147:e10. doi: 10.1017/S0950268818002595
O’Toole, G., Kaplan, H. B., and Kolter, R. (2000). Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79. doi: 10.1146/annurev.micro.54.1.49
Pratt, L. A., and Kolter, R. (1998). Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30, 285–293. doi: 10.1046/j.1365-2958.1998.01061.x
Reisner, A., Haagensen, J. A., Schembri, M. A., Zechner, E. L., and Molin, S. (2003). Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48, 933–946. doi: 10.1046/j.1365-2958.2003.03490.x
Ren, C. P., Beatson, S. A., Parkhill, J., and Pallen, M. J. (2005). The Flag-2 locus, an ancestral gene cluster, is potentially associated with a novel flagellar system from Escherichia coli. J. Bacteriol. 187, 1430–1440. doi: 10.1128/JB.187.4.1430-1440.2005
Roe, A. J., Currie, C., Smith, D. G., and Gally, D. L. (2001). Analysis of type 1 fimbriae expression in verotoxigenic Escherichia coli: A comparison between serotypes O157 and O26. Microbiology (Reading) 147, 145–152. doi: 10.1099/00221287-147-1-145
Romling, U., Sierralta, W. D., Eriksson, K., and Normark, S. (1998). Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28, 249–264. doi: 10.1046/j.1365-2958.1998.00791.x
Shea, A. E., Stocki, J. A., Himpsl, S. D., Smith, S. N., and Mobley, H. L. T. (2022). Loss of an intimin-like protein encoded on a uropathogenic E. coli pathogenicity island reduces inflammation and affects interactions with the urothelium. Infect. Immun. 90:e0027521. doi: 10.1128/IAI.00275-21
Sheikh, A., Luo, Q., Roy, K., Shabaan, S., Kumar, P., Qadri, F., et al. (2014). Contribution of the highly conserved EaeH surface protein to enterotoxigenic Escherichia coli pathogenesis. Infect. Immun. 82, 3657–3666. doi: 10.1128/IAI.01890-14
Snellings, N. J., Tall, B. D., and Venkatesan, M. M. (1997). Characterization of Shigella type 1 fimbriae: Expression, FimA sequence, and phase variation. Infect. Immun. 65, 2462–2467. doi: 10.1128/iai.65.6.2462-2467.1997
Uhlich, G. A., Keen, J. E., and Elder, R. O. (2002). Variations in the csgD promoter of Escherichia coli O157:H7 associated with increased virulence in mice and increased invasion of HEp-2 cells. Infect. Immun. 70, 395–399. doi: 10.1128/IAI.70.1.395-399.2002
Vogeleer, P., Tremblay, Y. D., Mafu, A. A., Jacques, M., and Harel, J. (2014). Life on the outside: Role of biofilms in environmental persistence of Shiga-toxin producing Escherichia coli. Front. Microbiol. 5:317. doi: 10.3389/fmicb.2014.00317
Walk, S. T., Alm, E. W., Gordon, D. M., Ram, J. L., Toranzos, G. A., Tiedje, J. M., et al. (2009). Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 75, 6534–6544. doi: 10.1128/AEM.01262-09
Wang, H., Li, Q., Bai, X., Xu, Y., Zhao, A., Sun, H., et al. (2016). Prevalence of eae-positive, lactose non-fermenting Escherichia albertii from retail raw meat in China. Epidemiol. Infect. 144, 45–52. doi: 10.1017/S0950268815001120
Wang, H., Zhang, L., Cao, L., Zeng, X., Gillespie, B., and Lin, J. (2022). Isolation and characterization of Escherichia albertii originated from the broiler farms in Mississippi and Alabama. Vet. Microbiol. 267:109379. doi: 10.1016/j.vetmic.2022.109379
Watnick, P., and Kolter, R. (2000). Biofilm, city of microbes. J. Bacteriol. 182, 2675–2679. doi: 10.1128/JB.182.10.2675-2679.2000
Wrobel, A., Saragliadis, A., Perez-Ortega, J., Sittman, C., Gottig, S., Liskiewicz, K., et al. (2020). The inverse autotransporters of Yersinia ruckeri, YrInv and YrIlm, contribute to biofilm formation and virulence. Environ. Microbiol. 22, 2939–2955. doi: 10.1111/1462-2920.15051
Xicohtencatl-Cortes, J., Monteiro-Neto, V., Ledesma, M. A., Jordan, D. M., Francetic, O., Kaper, J. B., et al. (2007). Intestinal adherence associated with type IV pili of enterohemorrhagic Escherichia coli O157:H7. J. Clin. Invest. 117, 3519–3529. doi: 10.1172/JCI30727
Xicohtencatl-Cortes, J., Monteiro-Neto, V., Saldana, Z., Ledesma, M. A., Puente, J. L., and Giron, J. A. (2009). The type 4 pili of enterohemorrhagic Escherichia coli O157:H7 are multipurpose structures with pathogenic attributes. J. Bacteriol. 191, 411–421. doi: 10.1128/JB.01306-08
Keywords: Escherichia albertii, foodborne pathogen, biofilm, adhesins, fimbriae, flagella, motility
Citation: Carter MQ, Carychao D and Lindsey RL (2024) Conditional expression of flagellar motility, curli fimbriae, and biofilms in Shiga toxin- producing Escherichia albertii. Front. Microbiol. 15:1456637. doi: 10.3389/fmicb.2024.1456637
Received: 28 June 2024; Accepted: 14 August 2024;
Published: 09 September 2024.
Edited by:
Renmao “Tim” Tian, Illinois Institute of Technology, United StatesReviewed by:
Steven L. Foley, National Center for Toxicological Research (FDA), United StatesSalvatore Walter Papasergi, National Research Council (CNR), Italy
Copyright © 2024 Carter, Carychao and Lindsey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michelle Qiu Carter, bWljaGVsbGUuY2FydGVyQHVzZGEuZ292
 Michelle Qiu Carter
Michelle Qiu Carter Diana Carychao
Diana Carychao Rebecca L. Lindsey
Rebecca L. Lindsey