
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 12 December 2024
Sec. Phage Biology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1455710
This article is part of the Research TopicBacteriophages, Prophages, and Their Products: Regulating Bacterial PopulationsView all 12 articles
The emergence of the multidrug-resistant (MDR) Escherichia coli ST131 clone has significantly impacted public health. With traditional antibiotics becoming less effective against MDR bacteria, there is an urgent need for alternative treatment options. This study aimed to isolate and characterize four lytic phages (EC.W2-1, EC.W2-6, EC.W13-3, and EC.W14-3) from hospital sewage water and determine their effectiveness against the ST131 clone. These phages demonstrated a broad host range, effectively lysing 94.4% of highly pathogenic E. coli ST131 isolates. Morphological observations and phylogenetic analysis indicate that EC.W2-1, and EC.W13-3 belong to the Tequatrovirus genus in the Straboviridae family, while EC.W2-6 and EC.W14-3 are part of the Kuravirus genus in the Podoviridae family. Phages remained stable at pH 2–10 for 4 h and below 80°C for 1 h. These four phages showed in vitro bacterial lytic activity at various MOIs (0.1–0.001). The one-step growth curve of phages exhibited a short latent period of approximately 10–20 min and a moderate burst size of 50–80 (pfu/cell). Phages’ genome size ranged from 46,325–113,909 bp, with G + C content of 35.1 –38.3%. No virulence or drug resistance genes were found, which enhanced their safety profile. In vivo, EC.W2-6 and EC.W13.3, along with their cocktail, fully protected against the ESBL-producing E. coli ST131 infection model in vivo. Combining these phages and a 3-day repeated single phage, EC.W13-3 significantly enhanced the survival rate of E. coli ST131 infected mice at low MOI (0.01–0.001). The in vivo effectiveness of the isolated phages and the EC.W2-6 and EC.W14-3 cocktail in highly reducing bacterial load CFU/g in multiple organs strongly supports their potential efficacy. Based on in vivo, in vitro, and genomic analyses, phages have been proposed as novel and suitable candidates for killing the pandemic ST131 clone.
Escherichia coli sequence type 131 (ST131) is a clonal group of concern, known for causing antimicrobial-resistant infections predominantly in community settings (Rogers et al., 2011). The ST131 pandemic was initially detected in 2008 using multilocus sequence typing (MLST) to identify CTX-M-15 extended-spectrum β-lactamase-producing E. coli strains across various continents. E. coli ST131 is mostly known as a major cause of urinary tract infections (UTIs). Besides causing UTIs, this extraintestinal pathogenic bacterium can also lead to serious infections, including bloodstream infections, sepsis, and intra-abdominal infections (Whelan et al., 2023). These infections affect individuals of all ages and range from cystitis to life-threatening sepsis. Treatment requires a tailored approach considering antimicrobial resistance and infection site. Although phenotypic detection of ST131 is not possible, DNA-based methods like MLST and PCR are used for identification (Rogers et al., 2011; Nicolas-Chanoine et al., 2014).
The clone ST131, which is a major cause of fluoroquinolone-resistant E. coli infections, has a high prevalence ranging from 60 to 90% (Peirano et al., 2020). Fluoroquinolone resistance in ST131 is caused by amino acid substitutions in the QRDR genes (gyrA, parC, and parE), which reduce the effectiveness of fluoroquinolone in inhibiting bacterial DNA replication and transcription (Shariati et al., 2022). Recently, the plasmid-mediated colistin resistance gene mcr-1 was detected in ST131. The presence of mcr-1 in ST131 raises concerns about the spread of colistin resistance (Majewski et al., 2021). The ST131 clone of E. coli harbors different β-lactamase genes (Miyoshi-Akiyama et al., 2016). β-lactamases are enzymes produced by bacteria that can deactivate β-lactam antibiotics such as penicillins and cephalosporins. The most common types of β-lactamase genes in the ST131 clone are CTX-M family β-lactamases, with TEM, SHV, and CMY β-lactamases being less frequent. Approximately 40–80% of extraintestinal pathogenic E. coli strains with extended-spectrum β-lactamases (ESBLs) belong to the ST131 clone (Hibstu et al., 2022).
The geographical distribution of ST131 is not fully known; however, it is commonly found in antimicrobial-resistant E. coli infections in Europe, North America, Canada, Japan, and South Korea (Bevan et al., 2017). In a national survey in Japan, the ST131 clones were found in 38.1% of ESBL-producing E. coli isolates between 2013 and 2014. Notably, in this region, there is higher genetic diversity within the ST131 clone and a greater variety of accompanying CTX-M ESBL genes than in other areas (Miyoshi-Akiyama et al., 2016). Similarly, in South Korea, from 2016 to 2017, 27% of ESBL-producing E. coli isolates were ST131, with only 57% carrying an ESBL gene (Baek et al., 2021). E. coli ST131, an antimicrobial-resistant clone, has been found in humans, companion animals, non-companion animals, and food sources. The extensive presence of ST131 in various populations raises concerns regarding its potential transmission and its impact on public health (Banerjee and Johnson, 2014).
As mentioned previously, the ST131 clone of E. coli possesses various antimicrobial resistance mechanisms. However, limited information is available regarding the specific antimicrobial therapies used to treat the infections caused by this clone. This situation underscores the pressing need for innovative therapeutic approaches and development of new antimicrobial agents to effectively combat these infections (Kim et al., 2018; Rehman et al., 2019). The limited availability of new antibiotics has renewed interest in phage therapy as a potential solution, increasing the search for novel phages (Reardon, 2014). Phages are viruses that infect and kill bacteria, offering a potential alternative to antibiotics in the face of growing antimicrobial resistance. Clinical approaches to phage therapy have been reported in several countries, including the United States, Georgia, Poland, and Russia (Hibstu et al., 2022). Phages exhibit selective tropism, targeting specific bacteria via surface receptors while minimizing harm to the host microbiome. Generally safe for mammals, potential risks include immune responses and endotoxin release. Lytic phages directly kill bacteria and can be engineered for targeted drug delivery (Emencheta et al., 2024). Scientific evidence supporting the benefits of phage therapy in humans and animals has steadily increased in the recent decades (Hon et al., 2022). Additionally, they have been used as indicators of water safety (de Almeida Kumlien et al., 2021). As the availability of phages is limited, there is an urgent need to isolate safe, highly lytic, and well-characterized phages for phage therapy.
Current research focuses on isolating, characterizing, and evaluating the efficacy of novel E. coli phages in combating ST131 E. coli infections in vitro and in vivo. The goal of this study was to explore phages as an alternative strategy to combat the growing issue of MDR E. coli clone ST131. However, phage therapy is still in its early stages and faces challenges, such as regulatory barriers and manufacturing limitations. Nonetheless, advancements in phage therapy have provided hope for addressing antibiotic resistance and improving the treatment options for serious bacterial infections.
In vivo experiments were conducted on BALB/c mice strain from Yeangnam Bio (166 Palgong-ro, Dong-gu Daegu) with six-week-old female mice receiving sterile food and water. The procedures followed guidelines set by the National Ethics Committee and approved by the Kyungpook National University Animal Care and Use Committee (KNU: 2023–0478).
E. coli isolates were obtained from Kyungpook National University Hospital Culture Collection for Pathogens (KNUH-NCCP). The isolates were initially cultured on 5% sheep blood agar at 37°C for 24 h. Then, the samples were transferred to Brain Heart Infusion broth (BHI) and incubated at 37°C for 24 h. Finally, the isolates were preserved at −70°C with a 50% glycerol stock for future experiments.
E. coli hosts were cultured on blood agar plates. Sewage water samples were collected from Kyungpook National University Hospital in Daegu, South Korea. Sewage water was treated by centrifugation at 12,000 × g for 10 min to remove the debris. The resulting supernatant was filtered using 0.22 μm pore-sized membrane filters. The filtrate was used to infect E. coli cultures in the early exponential phase. After overnight incubation at 30°C with shaking, infected cultures were stored at 4°C for 48 h. The supernatants were centrifuged again and filtered through a 0.22 μm pore-size membrane. Phage titers were determined using the double layer method. The purified phages were stored at −70°C in glycerol-supplemented medium (Kim et al., 2022).
Antibiotic susceptibility of the 18 E. coli isolates was tested against 21 antibiotics from 10 families. These families include cephalosporins, monobactams, aminopenicillin, fluoroquinolones, aminoglycosides, penicillin, sulfonamide-trimethoprim, tetracycline, carbapenem, and extended-spectrum beta-lactamases. To determine the minimum inhibitory concentration (MIC), antibiotic disks and culture materials were purchased from Becton Dickinson and Company. E. coli ATCC25922 was used as the control. Each isolate was tested in triplicate, according to the Clinical and Laboratory Standards Institute guidelines (CLSI, 2020; Shamsuzzaman et al., 2023). According to these guidelines, isolates were classified as resistant, intermediate, or susceptible. The MLST analysis described in our study was previously conducted by the Department of Microbiology at Kyungpook National University (Shamsuzzaman et al., 2024).
Eighteen E. coli clinical isolates (ST131) were tested for susceptibility to phage lytic activity by spot tests following the method outlined by Kim et al. (2018). To prepare the bacterial lawn, 10 mL of 0.75% soft agar with 100 μL of bacterial culture in the stationary phase was poured onto a BHA agar plate. Once the overlay solidified, a 15 μL aliquot of the phage, with a concentration of approximately 1011–13 PFU/ml, was directly spotted on the bacterial lawn. The plates were dried and incubated overnight at 37°C. Bacterial susceptibility was determined using the clear lytic zones on the plates.
Phage adsorption rate and burst size were determined following the method outlined by Rahman et al. (2011) E. coli cells (ATCC25922) were cultured in BHI media until they reached exponential phase. The cells were then infected with the phage at a low multiplicity of infection (MOI) of 0.0001 and incubated at room temperature. Samples were collected and centrifuged at intervals of 0, 1, 3, 5, 10, 15, and 20 min. The supernatants were used for plaque assays to determine the titers of the unabsorbed phages. The burst size of the phage was determined using a single growth-curve experiment. E. coli cells were harvested by centrifugation and resuspended in 1 mL of fresh BHI medium at a concentration of 5 × 109 CFU/mL. The phage was added at an MOI of 0.0001 and allowed to adsorb for 30 min at 4°C. After centrifugation at 12,000 × g for 5 min, the pellet was resuspended in 10 mL of fresh BHI medium. The mixture was then incubated at 37°C, and samples were collected at 5-min intervals for up to 45 min. The samples were promptly diluted and analyzed for phage plaque counts. Each experiment was independently repeated thrice.
The thermal and pH stability of the phage were evaluated following the method described by Tang et al. (2023). To assess thermal stability, the phage suspension (∼108 PFU/mL) was incubation at temperatures ranging from 4 to 80°C for 1 h. The phage suspension (∼108 PFU/mL) was incubated in BHI broth with pH levels ranging from 2 to 10 at 37°C for 4 h to determine pH stability. Phage titers were determined utilizing the double-layer agar method.
This method determines the bactericidal activity of phages and phage cocktails against E. coli ATCC 25922 and E. coli ST131. Bacterial cultures (108 CFU/mL) were treated with individual phages and combination phages (1:1) at various MOIs (10, 1, 0.1, 0.01, 0.001) in BHI broth at 37°C with gentle shaking. Optical density at 600 nm was measured using a UV–Vis spectrophotometer (Molecular Devices, LLC, San Jose, CA, United States) in a 96-well plate at 1 h intervals for 12 h and again at 24 h. Bacterial culture without phages served as the positive control, while BHI broth served as the negative control. All assays were performed in triplicate (Tang et al., 2023).
Phage DNAs were extracted using the phenol-chloroform method described by Jakočiūnė and Moodley (2018). To conduct next-generation sequencing (NGS), phage genomic DNA was sequenced using an Illumina Miseq platform in San Diego, CA, USA. The sequencing reads were assembled through the Celemics pipeline based in South Korea.1 To analyze genome sequence similarity, BLASTn2 was used for alignment. The RAST online website3 was utilized to predict open reading frames (ORFs), and the predictions were cross-checked and corrected through the NCBI database. Gene function maps were created using a custom program developed in the lab and refined using Geneious Prime 2023.2.4 The tRNAscan-SE v.2.0 tool5 was employed to predict tRNA. ResFinder6 and VirulenceFinder7 were utilized to identify drug resistance genes and virulence genes, respectively. A phylogenetic tree based on the phage proteome rectangular tree circular tree were generated using VIPTree.8 For the construction of a complete genome phylogenetic tree based on the whole genome sequence of the isolated phages, the genome-BLAST distance phylogenetic approach provided by VICTOR9 was used. Phylogenetic trees of large terminase and minor capsid proteins were constructed using the neighbor-joining (NJ) method, and the Bootstrap method was employed to assess the reliability of the phylogenetic trees. Additionally, the complete genome sequences of the isolated phages were aligned with other phages using the BLASTn tool available in the NCBI database. The Mauve algorithm (v2.3.1) was used to visualize the complete genome sequence similarity between the isolated phages and their closest neighbor, E. coli phages.10
Overnight bacterial cultures were grown in BHI broth at 37°C with shaking reached an OD of approximately 2. To ensure they were in the logarithmic phase, 100 μL of the culture was reinoculated into 10 mL fresh BHI and incubated at 37°C. Once in the logarithmic phase, cultures were washed twice in 1X PBS. Viable bacteria were quantified by plating serial dilutions on bacteriological agar to calculate CFU. Following this, mice were administered an intraperitoneal injection (IP) of a 50% lethal dose (LD50) of 109 CFU/mL. One-hour post-infection, mice were administered intraperitoneal injections of purified phages at varying plaque-forming units (PFU) in 100 μL of PBS. The infected mice were divided into treatment groups for analysis, with each group containing 5 mice. The control group consisted of infected mice that did not receive any treatment. The phage-treated group included mice infected with E. coli-specific phages. The positive control group was treated with PBS, whereas the negative control group was treated with both phages and PBS. Daily survival rates were recorded for each group to evaluate the effectiveness of phage therapy in preventing mortality (Pu et al., 2022).
To determine the colony-forming units (CFU) and plaque-forming units (PFU), mice were euthanized 16–18 h post-infection. Following euthanisation, necropsies were performed under sterile conditions. Each treatment group contained 4 mice. The lungs, kidneys, liver, and spleen were weighed and homogenized in 1X PBS using sterile blades. Subsequently, organ homogenates were serially diluted and plated on LB ampicillin (100 μg/mL) plates, followed by overnight incubation at 37°C. Phage counts were quantified using a soft agar overlay assay (Green et al., 2017).
Statistical analyses were performed using GraphPad Prism (version X). Survival analysis was conducted using the Kaplan–Meier method, with differences between groups assessed through the log-rank (Mantel-Cox) test. To compare bacterial loads across different organs, one-way ANOVA was used followed by Tukey’s post-hoc test for multiple comparisons. For phage analysis, data from adsorption and burst size assays were analyzed using unpaired t-tests. Results were considered statistically significant at p-values <0.05 and are presented as mean ± standard error of the mean (SEM).
Based on the spot test results, phages EC.W2-1, EC.W2-6, EC.W13-3, and EC.W14-3 exhibited high lytic activity against pathogenic ST131 isolates, lysing 94.44% (17/18) of the isolates (Table 1). These phages also showed lytic activity against 14 different ST types, including 60 multi-drug-resistant E. coli isolates, with lytic activity percentages ranging from 50.0 to 56.6% (Shamsuzzaman et al., 2024). However, the efficiency of plating (EOP) values of phages against E. coli KBN 10PO7288 (ST131) compared to the reference E. coli ATCC25922 were approximately 2.56 × 109, 4.56 × 106, 261.53, and 372.09, respectively (Supplementary Table S1).
As shown in Figure 1, AST results revealed that all 18 E. coli ST131 isolates exhibited a multidrug-resistant profile, indicating an alarming 100% prevalence of MDR (18/18) (Supplementary Table S2). The antibiotic resistance and susceptibility patterns of 18 ST131 isolates were investigated, and results revealed significant resistance to cephalosporins, specifically cefazolin (100%, 14/14), ceftazidime (94.1%, 16/17), cefoxitin (57.1%, 8/14), ceftriaxone (100%, 2/2), cefotaxime (93.3%, 14/15), and cefepime (88.8%, 16/18). The monobactam family also exhibited notable resistance, with aztreonam showing 94.1% resistance (16/17). Aminopenicillins also showed resistant to amoxicillin (72.2%, 13/18) and ampicillin (100%, 12/12), demonstrating high resistance rates. Carbapenem-resistant strains were observed with imipenem (44.4%, 8/18) and meropenem (38.8%, 7/18). The isolates also showed high fluoroquinolone resistance, with 81.2% (13/16) to ciprofloxacin and 100% (6/6) to levofloxacin. The aminoglycoside family showed resistance to amikacin (11.1%, 2/18) and gentamicin (33.3%, 6/18). In addition, the penicillin family resisted piperacillin (40%, 2/5) and the sulfonamide-trimethoprim family resisted trimethoprim (50%, 4/8). ESBL were detected in 78.5% of the isolates (11/14).
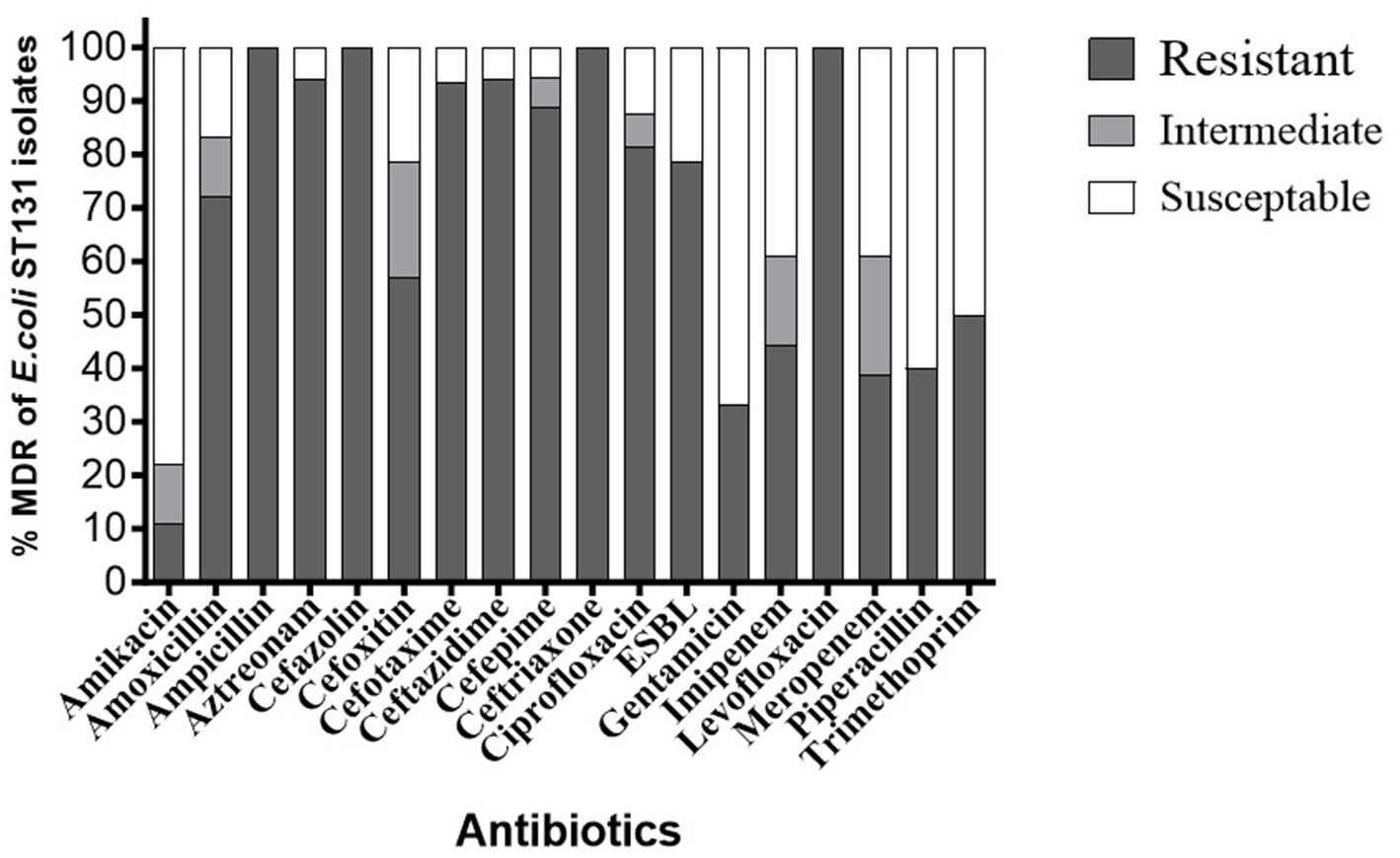
Figure 1. Antibiotic resistance ratios of different E. coli ST131 isolates. % Resistance indicates the proportion of isolates of E. coli ST131 that resist each antibiotic.
In this study, we investigated the lytic activity, growth curve, thermal stability, and pH stability of the phages EC.W2-1, EC.W2-6, EC.W13-3, and EC.W14-3. According to the phage adsorption assay, 90% of phages could adsorb onto E. coli ATCC25922 within 10 min (Figures 2a–d). The results of the one-step growth curve analysis showed that the phage displayed a latent period of approximately 10–20 min, followed by rapid release of virus particles. The final titer reached a range –107.5 to 108 PFU/mL after a burst period of 30–40 min, with a burst size of approximately 50–80 PFUs/cell (Figures 2e–h). Regarding thermal stability, there were no significant changes in the titers of the four phages after incubation at temperatures ranging from 4 to 60°C for 60 min. However, phage titers showed a substantial decrease at 70°C, and were completely inactivated at 80°C. Furthermore, these phages were stable over a pH range of 2–10 within 4 h, with the optimal pH for stability being between 6 and 8 (Figures 3a–h). Four phages were isolated from the hospital sewage water. After incubation at 37°C for 18 h on a double-layer agar plate, clear plaques with diameters of approximately 1–2 mm. Transmission electron microscopy (TEM) images revealed that EC.W2-1, EC.W13-3, and EC.W14-3 had an icosahedral head measuring 100 ± 2 to 115 ± 5 nm in diameter, along with a contracted tail measuring approximately 100 ± 4 to 113 ± 2 nm in length. In contrast, the EC.W2-6 phage had a head size of 60 ± 3 nm and tail size of 8 ± 2 nm (Shamsuzzaman et al., 2024).
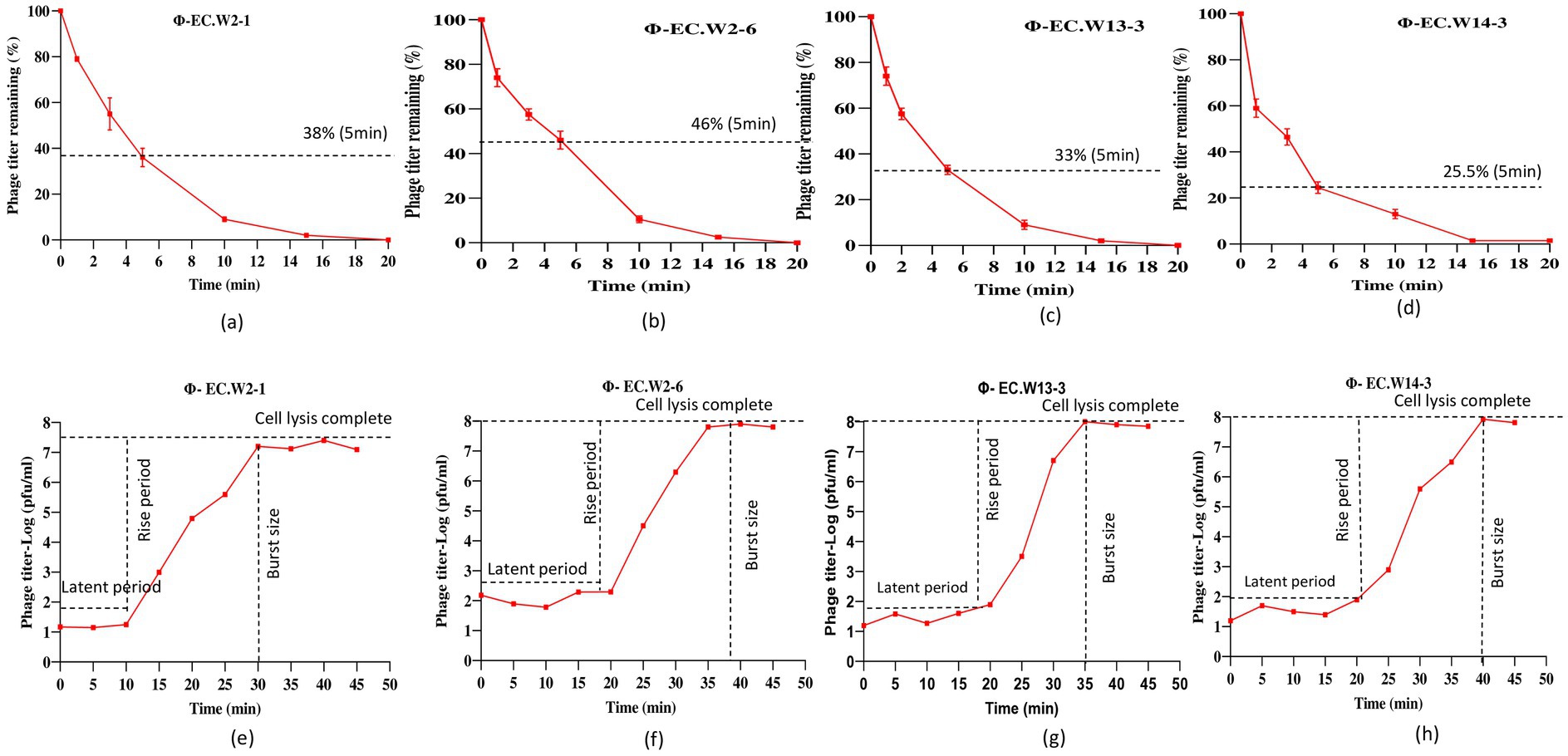
Figure 2. Adsorption rate and burst size of four phages to the E. coli ATCC25922. (a–d) Adsorption assay measuring the percentage of remaining free phages over 20 min at MOI of 0.0001 and (e–h) one-step growth curve showing the latent period and burst size of four novel E. coli bacteriophages in BHI medium at MOI of 0.0001. Values represent means ± standard deviations from the duplication of each treatment.
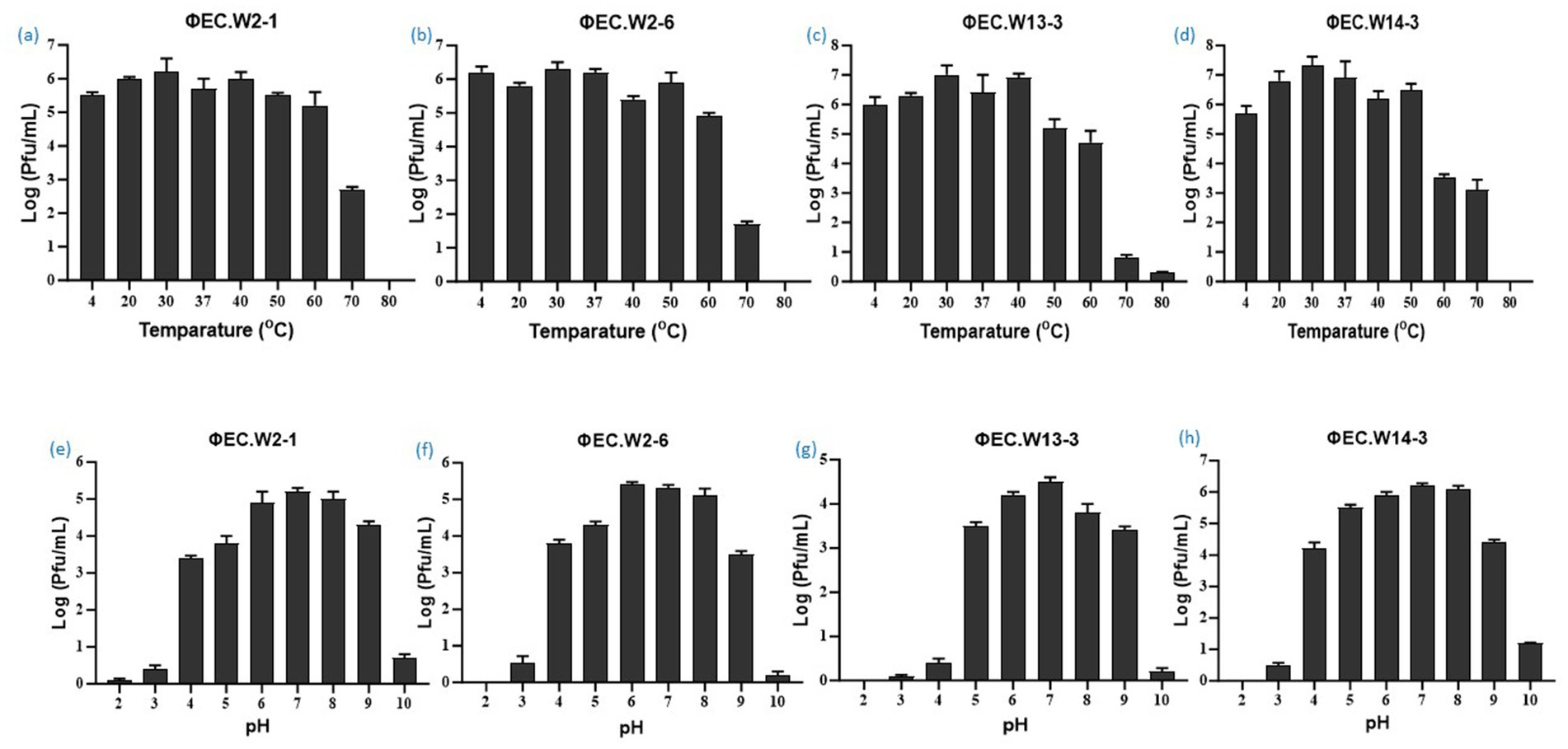
Figure 3. Stability of four novel E. coli bacteriophages. (a–d) Effect of temperature on phage stability, with phages treated at 4, 20, 30, 37, 40, 50, 60, 70, and 80°C for 1 h, and surviving phage titers calculated. (e–f) pH stability of phages, with concentrations (~108 PFU/mL) incubated at pH 2 to 10 at 37°C for 4 h. Values represent means ± standard deviations from triplicate treatments.
The phylogenetic relationship of the novel phages was determined through a comparative analysis of their whole genome sequences with the closest known species of Straboviridae and Podoviridae E. coli phages. The analysis revealed that EC.W2-1 and EC.W13-3 exhibited the highest DNA sequence similarities to the Tequatrovirus genus in the Straboviridae family, and phages Escherichia phage vB_EcoM_G4498 (complete genome) and Escherichia phage vB_EcoM_NBG2 (complete genome). For these isolates, the query coverage and percentage identity were recorded as 99/96.45% and 92/95.40%, respectively. On the other hand, EC.W2-6 and EC.W14-3 displayed the highest DNA sequence similarities to the Kuravirus genus in the Podoviridae family, and Escherichia phage vB_EcoP_SU10 (complete genome) and Escherichia phage 172-1(complete genome), with a query coverage and percentage identity of 94/96.21% and 89/89.62%, respectively (Table 2). Based on the analysis of their sequences and phylogenetic relationships, the phages were identified as new members of the Straboviridae and Podoviridae families.
Whole-genome sequence analysis indicated that EC.W2-1, EC.W2-6, EC.W13-3, and EC.W14-3 had circular double-stranded DNA structures, with genome sizes of 46,325, 67,000, 113,909, and 69,164 bp, respectively. Additionally, the G + C contents were found to be 35.5, 38.3, 35.3, and 35.1%, respectively (Table 2). Genome analysis revealed the absence of virulence or drug resistance genes, which enhanced their safety profiles. The CGView server and Geneious Prime generated visual representations of circular genomes, showcasing various aspects such as sequence characteristics, base composition plots, analysis outcomes, and sequences. These visual images provide a comprehensive and intuitive visualization of the genomes, allowing for a better understanding of their structural and compositional features (Supplementary Figures S1a–d). Phylogenetic analysis carried out using the VIPtree showed that EC.W2-1 and EC.W13-3 phages were most closely related to Escherichia phage vB_EcoM_G4498 (NC_054918) Escherichia phage ECO4 (NC_054911), Escherichia phage vB_EcoM_G9062 (NC_054920), Escherichia phage MN03 (NC_070990), and Escherichia phage vB_EcoP-(NC_070989). On the other hand, EC.W2-6, and EC.W14-3 close related to Escherichia phage ECO4 (NC_054911) Escherichia phage 172–1 (NC_028903). All phages belong to the novel genera Tequatrovirus and Kuravirus within the families Straboviridae and Podoviridae (Supplementary Figure S2). In addition, alignment with their closest neighbor using the MAUVE alignment tool suggested that these isolates are closely related and share recent common ancestors. However, small localized regions were identified, indicating functional differences between these phages. These findings suggest unique characteristics and capabilities of genetic makeup (Figure 4). Phylogenetic analysis conducted through genome comparisons using the TYGS server also supported EC.W2-1, EC.W2-6, EC.W13-3, and EC.W14-3, which are novel members of the genera Tequatrovirus and Kuravirus (Supplementary Figure S3). Analysis of the genome sequences of EC.W2-1, EC.W2-6, EC.W13-3, and EC.W14-3 revealed that these phages contained 71, 125, 189, and 113 predicted open reading frames (ORFs), respectively. All these ORFs were located on the positive strand of the genome. There are no potential tRNA-coding genes in the genomes of these phages. Among all the ORFs identified in these phages, the predicted proteins have known potential functions related to various processes such as endolysin, holin, tail fiber protein, DNA replication/transcription/packaging, and cell lysis. On the other hand, the remaining ORFs have been classified as hypothetical proteins.
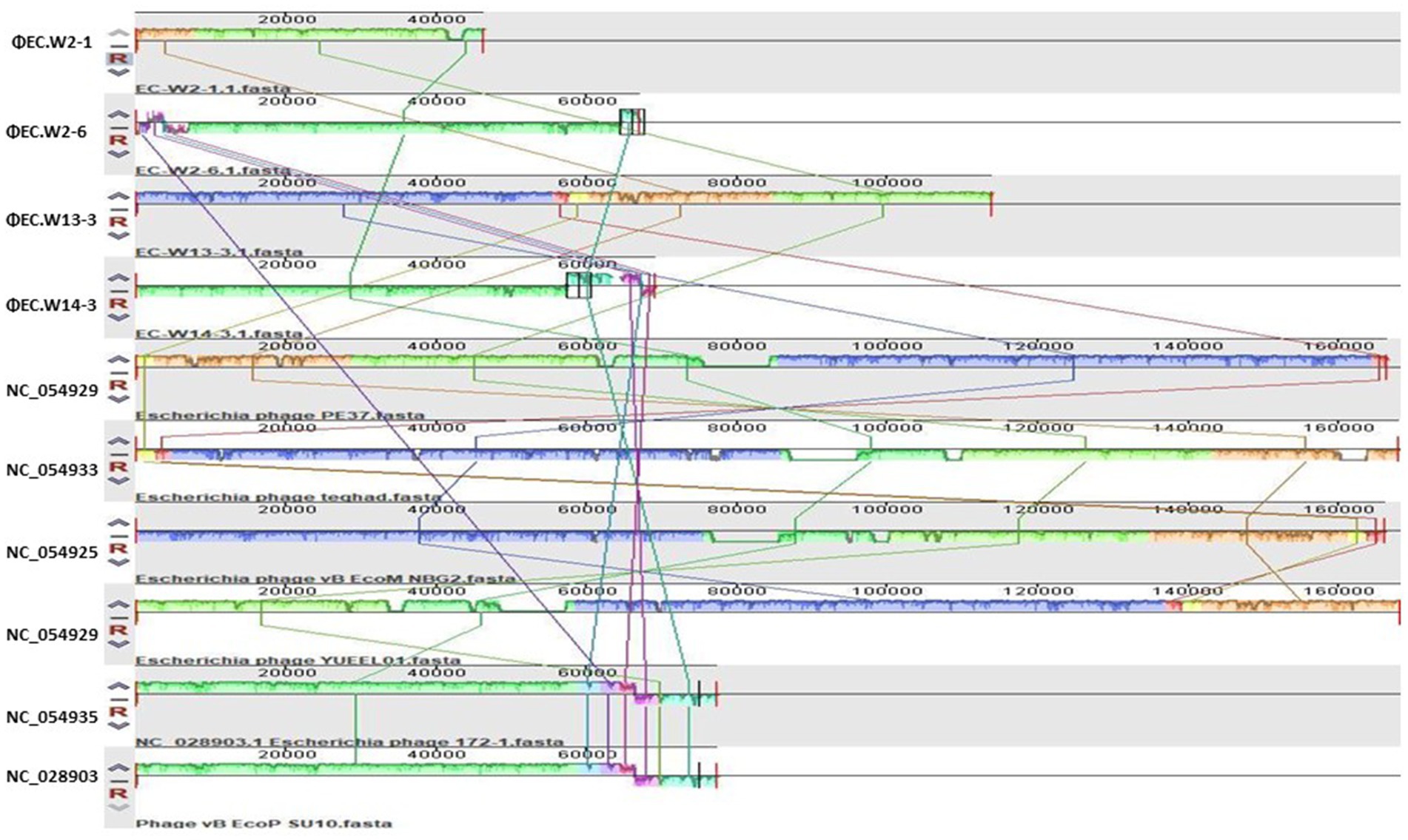
Figure 4. This study employed the progressive Mauve algorithm (v2.3.1) to perform multiple alignments of the genomes of 4 novel E. coli bacteriophages and the six closest genomes of E. coli bacteriophages. The objective was to investigate the rearrangement patterns and synteny in these genomes. The genomes were laid out horizontally, with homologous segments represented by colored blocks. The regions outside the blocks lacked homology and were represented by white areas, which were unique to each genome and not aligned.
In vitro bactericidal activity of phages against E. coli ATCC25922 at various MOIs (Supplementary Figure S4). Within 24 h, the OD600 value of the positive control increased continuously from 0.05 to 1.25, whereas that of the negative control remained unchanged. Phage treatment significantly inhibited E. coli ATCC25922 growth at all MOIs within the first 8 h. However, the OD600 values gradually increased after 8 h of phage treatment. After 24 h, there was a significant difference in the OD600 values compared to the positive control and a considerable difference in the MOI values. The OD600 values continued to rise gradually until 24 h. Although the bacterial counts for all MOIs exceeded 108 CFU/mL at 24 h, there was a significant difference between MOI 10 and 0.001 compared to the positive control. Interestingly, MOI 10 exhibited better lytic activity in the first 12 h than other MOI values. However, after 24 h, an MOI of 0.001 resulted in higher lytic activity. These findings demonstrated that these phages could significantly inhibit bacterial growth at all MOIs, with the highest bactericidal activity observed at an MOI of 0.001 after 24 h.
This study compared individual phages and cocktails to determine their ability to inhibit bacterial growth in two E. coli strains (ATCC25922-ST73 and KBN7288-ST131). Analysis of bacterial growth curves over 24 h revealed that both individual phages and phage cocktails significantly inhibited E. coli ATCC25922 growth throughout the study. Specifically, from 0 to 12 h, the individual phages and cocktail significantly suppressed the bacterial growth. However, during the 6–10-h timeframe, the cocktail composed of EC.W2-1 and EC.W2-6 showed slightly lower inhibition than individual phages. Notably, after 24 h, the phage cocktail consisting of EC.W2-6 and EC.W13-3 showed a high level of bacterial growth inhibition (Figure 5a). Similarly, when evaluating the lytic activity of individual phages and the phage cocktail against E. coli KBN 7288 (ST131), no significant changes in growth inhibition were observed within the first 4 h. However, after 4 h, the phages displayed varying levels of lytic activity. Consistent with these findings, the phage cocktail consisting of EC.W2-6 and ECW13-3 exhibited high inhibition of E. coli KBN 7288 growth after 24 h (Figure 5b). This study also analyzed the lytic activity against E. coli KBN 7288 by using various MOI of phage cocktails and compared with their single phage, where the cocktail (EC.W2-6 and ECW13-3) at MOI 10 and MOI 1 provided better lytic activity against E. coli KBN 7288. However, there were no significant differences among the other MOIs of the phage cocktail (Supplementary Figure S5).
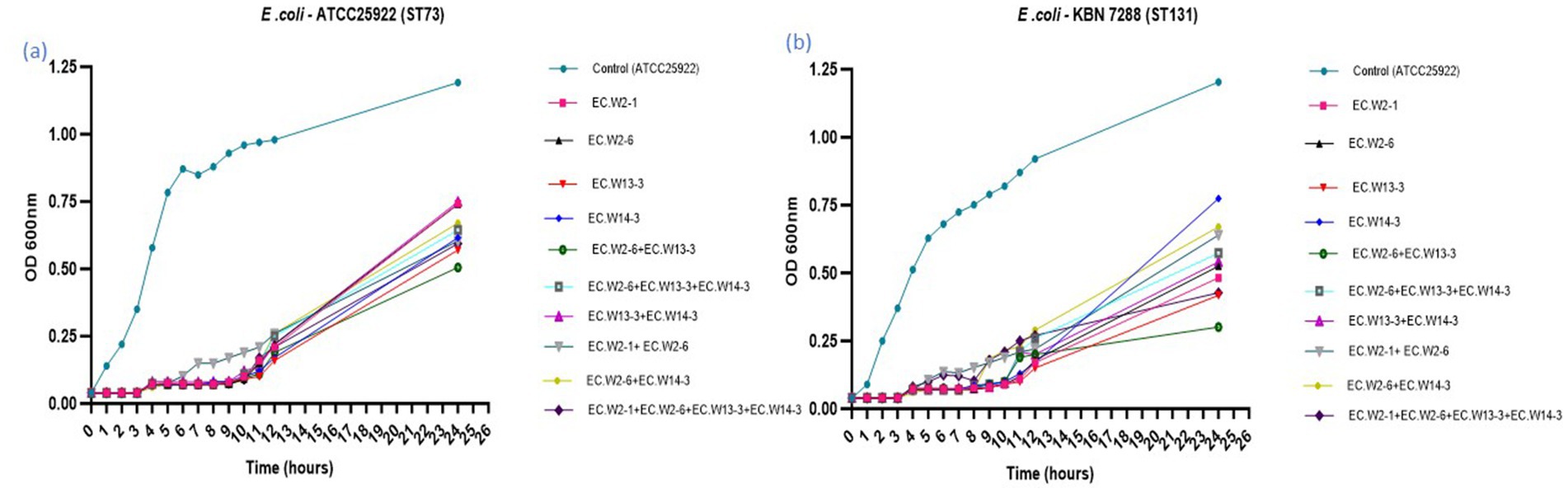
Figure 5. Lytic activity of four E. coli phages and their cocktail at MOI 0.001 against E. coli KBN 7288 (ST131) and E. coli ATCC 25922 (ST73).
The aim of this study was to assess the effectiveness of a phage-and-phage combination against E. coli ST131. Mice treated with phage EC.W13-3 at MOIs of 10 and 1 exhibited approximately 80% survival rates within 7 days. In contrast, the untreated positive control group experienced 100% mortality within 3 days, whereas the negative control group showed complete survival (Figure 6a). The study also monitored changes in the body weight of mice, revealing weight gains in mice treated with EC.W13-3 at MOIs of 10 and 0.01, indicating positive therapeutic effects. Conversely, mice exposed to a lower MOI of 0.001 or the control group displayed gradual weight loss, suggesting disease progression or ineffective treatment (Figure 6b). Additionally, a 3-day repeated treatment of EC.W13-3 at MOI 0.1 and MOI 0.001 significantly increased the survival rate by approximately 40% (Figure 6c). When investigating the effects of single phages EC.W2-6 and EC.W13-3 and their combination at MOI 01, the combination demonstrated a 100% survival rate in mice, whereas the survival rates for single phages were 60 and 80% (Figure 6d). This study evaluated the impact of individual phage treatments, including EC.W2-1, EC.W2-6, EC.W13-3, and EC.W14-3, on bacterial burden in various organs of a mouse model. Treatment with each phage substantially reduced the bacterial load across multiple organs, and phage counts (PFU/g) were calculated from the plaques on E. coli 10P0 KBN7288. Specifically, treatment with EC.W2-1 resulted in a significant reduction in bacterial load in the kidneys (2.5 log10 cfu/g), lungs (2.5 log10 cfu/g), liver (1.9 log10 cfu/g), and spleen (1.7 log10 cfu/g) (Figure 7a). Similarly, EC.W2-6 exhibited efficacy, demonstrating significant reductions in bacterial burden in the kidneys (3.2 log10 cfu/g), lungs (1.8 log10 cfu/g), liver (2.20 log10 cfu/g), and spleen (1.75 log10 cfu/g) (Figure 7b). Furthermore, treatment with EC.W13-3 resulted in notable reductions in log10 cfu/g, approximately 3 in the kidneys, 2.5 in the lungs, 2.55 in the liver, and 1.5 in the spleen (Figure 7c). In addition, EC.W14-3 significantly reduced log10 cfu/g by 2.7 in the lungs, 2.5 in the kidneys, 1.9 in the liver, and approximately 2.4 in the spleen (Figure 7d). Moreover, combining EC.W2-6 and EC.W13-3 in a cocktail showed enhanced effectiveness compared with individual phage treatments. This combination led to a remarkable reduction in bacterial burden in the kidneys (3.3 log10 cfu/g), lungs (3 log10 cfu/g), liver (2.5 log10 cfu/g), and spleen (2.5 log10 cfu/g) (Figure 7e).
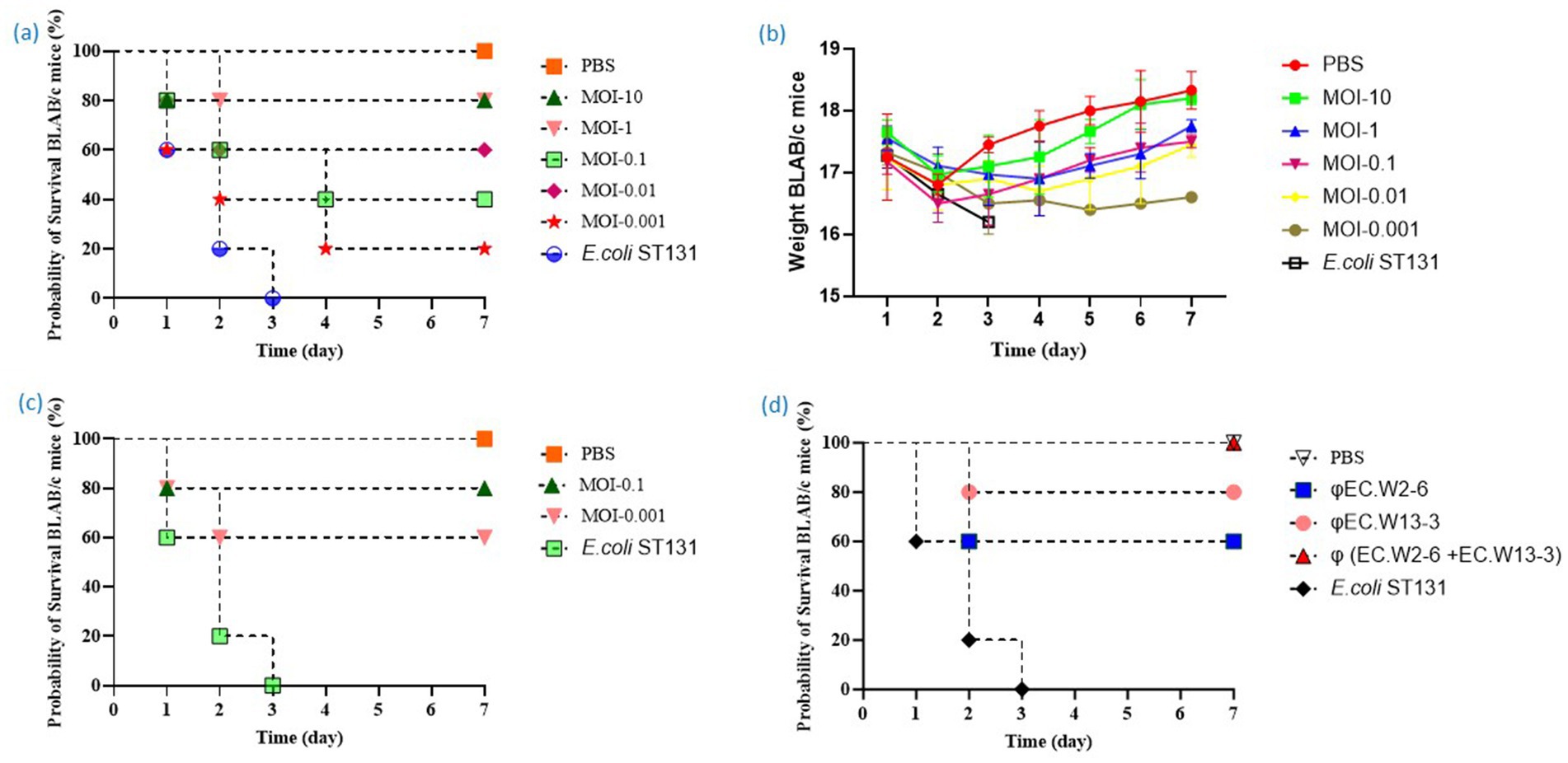
Figure 6. This study analyzes the toxicity of E. coli ST131 (KBN 7288) in BALB/c mice, identifies the optimal MOI for effective survival in phage therapy, and evaluates the efficacy of single phage and phage cocktails. (a) Survival curve of mice treated with phage EC.W13-3 at various MOIs or PBS. (b) Weight changes in mice one-week post E. coli administration. The weights of mice in the free phage group, mice treated with phage EC.W13-3 at various MOIs, and negative control groups were monitored for 2 week. (c) Evaluation of the efficacy of repeated phage therapy against E. coli ST131. The survival curve of mice given phage EC.W13-3 for 3 days at MOI-0.1 and MOI-0.001. (d) The study evaluates the efficacy of single phage and phage cocktail therapy against E. coli ST131 at MOI-0.1 in a mice model. The bacterial dose of 109 CFU was injected at 0 h, and after 1 h, the mice were infected with 109 PFU of phage.
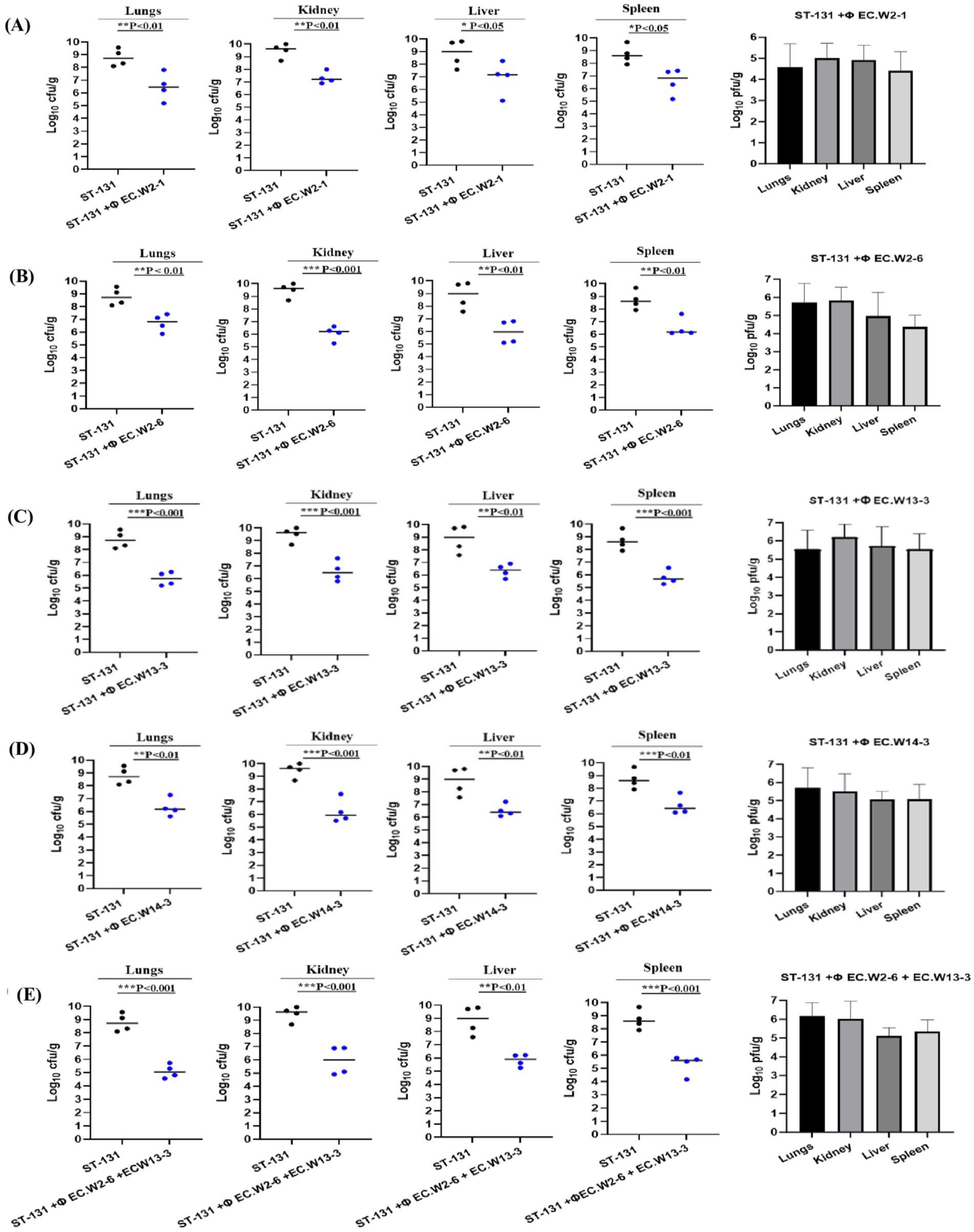
Figure 7. Phage therapy in a mouse model of ST131 MDR bacteraemia. BALB/c, 6-week-old female mice, were infected with an intraperitoneal (IP) injection of 109 CFU of E. coli KBN7288. One hour post-infection, mice received an IP with 109 PFU of four purified E. coli phages individually (A) EC.W2-1, (B) EC.W2-6, (C) EC.W13-3, (D) EC.W14-3, and as a cocktail (E) EC.W2-6 + EC.W13-3. Disease severity was evaluated, and organ samples were collected and plated to determine bacterial CFU and phage PFU. The efficacy of each treatment was assessed based on the CFU of E. coli KBN7288 in various organs. Black dots represent individual mice given bacteria alone, and blue dots represent the phage-treated, infected mice. Black bars represent the mean values for each group. The p-values were generated using a T-test, with one, two, or three stars denoting the significance level. One star (*) indicates p < 0.05, two stars (**) indicate p < 0.01, and three stars (***) indicate p < 0.001. Error bars denote standard deviation (n = 4).
In this study, four E. coli phages (EC.W2-1, EC.W2-6, EC.W13-3, and EC.W14-3) were isolated in the hospital sewage water. Morphological features and sequence analysis revealed that phages have strong similarities to the Straboviridae and Podoviridae families, positioning them as promising candidates for phage-based therapies against E. coli infections because of their ability to effectively target and eliminate E. coli bacteria and combat antibiotic resistance (Koonjan et al., 2021; Wolfram-Schauerte et al., 2022). Our isolated phages displayed a broad host range, effectively targeting 94.44% (17/18) of E. coli ST131 isolates. Additionally, they exhibited lytic activity against 14 different ST types, including 60 MDR E. coli isolates, with lytic percentages ranging from 50.0 to 56.6% (Shamsuzzaman et al., 2024). The phages remained stable over a wide pH range (2–10) and at temperatures ranging from 4 to 60°C, indicating their ability to survive under various environmental conditions. The extremely low optimal incubation period and moderate burst size highlighted the high proliferation efficiency and lytic activity of these phages. In an in vivo experiment, phage treatment significantly reduced the bacterial load, making them potential candidates for phage therapy against E. coli ST131 and other ST types of E. coli (Dufour et al., 2016; Green et al., 2017).
Whole-genome analysis revealed that none of the four phages contained virulence, antibiotic resistance, or bacterial toxin-related genes, suggesting a minimal risk of promoting antibiotic resistance and making them suitable for treating bacterial infections (Chaudhary et al., 2022). Depolymerases are enzymes that are encoded by phages infecting encapsulated bacteria, typically within open reading frames linked to structural proteins, often found in tail fibers, base plates, and neck regions (Guo et al., 2023). The analysis of isolated phage genomes revealed the presence of several tail fiber proteins with a high degree of similarity, ranging from 94 to 100%, to those found in other E. coli phages (Supplementary Table S3). By recognizing specific ligands on bacterial surfaces, these depolymerases specifically target capsular polysaccharides (CPSs) and lipopolysaccharides (LPSs), resulting in the breakdown of their repeating units of polysaccharides (Cai et al., 2023). Instead of directly killing bacteria, phage depolymerase strips away the protective polysaccharide layers, rendering the bacteria vulnerable to the immune system and antibacterial agents (Topka-Bielecka et al., 2021). Based on this information, we can assume that these phages could produce these enzymes, resulting in the high killing ability of E. coli ST131 isolates. Phage EC.W2-6 is an interesting bacteriophage that contains an endolysin with a remarkable 98.77 and 96.93% similarity to the endolysins found in Escherichia phages (YP_001671762, YP_00920818). Endolysins are enzymes that target bacterial cell wall peptidoglycans and cause cell lysis (Murray et al., 2021). Their specificity, unique mode of action, and lack of resistance mechanisms make them promising alternatives to antibiotics, particularly for combating multidrug-resistant infections (Nelson et al., 2012). Analysis of phage genomes revealed several proteins potentially crucial for combating MDR E. coli, especially ST131. These include holins, membrane proteins forming pores to release phage progeny (Fernandes and São-José, 2016); putative peptidases, involved in protein breakdown (Matsumi et al., 2005); replication, recombination, and repair proteins, maintaining phage genome integrity (Poteete, 2004); and packaging machinery proteins for DNA packaging and phage assembly (Isidro et al., 2004). Baseplate proteins facilitate host recognition and infection (Leiman et al., 2003). While the roles of several hypothetical proteins remain unclear, a full understanding of these proteins is essential for optimizing phage therapy (Shang et al., 2023).
To use phages to treat human bacterial infections, translation from in vitro activity to in vivo efficacy is not guaranteed, despite a high success rate (Dufour et al., 2016). Our investigation into the potential of the four isolated phages revealed that they could infect targeted ST131 isolates and reduce the bacterial load in the survival of mice models. Using animal pneumonia, septicemia, and urinary tract infection models, isolated phages LM33_P1 and HP3 showed in vivo efficacy in reducing bacterial load in several organs (Dufour et al., 2016; Green et al., 2017). This study found that a combination of phages (phage cocktail) and repeated phage therapy was much more effective than a single-dose phage therapy. Repeated phage therapy using the EC.W13-3 phage at an MOI of 0.1 and MOI 0.001 significantly increased the survival rate. The survival rate increased from 40 to 80% at MOI 0.1 and from 20 to 60% at MOI 0.001. Additionally, using the phage cocktails EC.W2-6 and EC.W13-3 resulted in 100% survival, whereas single-phage therapy achieved 60 and 80% survival rates at MOI-01, respectively. A previous study showed that the application of phage cocktails and repeated phage treatments can remarkably reduce the bacterial load of nosocomial pathogens in hospital wastewater (Weissfuss et al., 2023). According to this study, phage treatment effectively reduced bacterial counts in different organs. Specifically, phages EC.W2-1 and EC.W2-6 were found to have lower bacterial counts in the kidneys, lungs, liver, and spleen. Additionally, phages EC.W13-3 and EC.W14-3 also showed significant reductions in the bacterial load. This study also found that combining EC.W2-6 and EC.W13-3 in a cocktail enhanced the effectiveness of treatment, resulting in reduced bacterial levels across all organs. Although these results are promising, it is essential to note that animal experiments have limitations and caution should be exercised when translating these findings to the clinical setting (Henry et al., 2013). Bacterial resistance to phage infection is a significant challenge (Örmälä and Jalasvuori, 2013), compounded by the impact of the human immune system (Łusiak-Szelachowska et al., 2014), including the production of anti-phage antibodies (Bearden et al., 2005). Previous research has shown that the vital antibody response to φX-174 serves as a crucial indicator of immune responsiveness in humans (Bearden et al., 2005), with significant implications for individuals with conditions such as HIV (Bernstein et al., 1985), genetic immunodeficiencies (Shearer et al., 2001) and those undergoing immunosuppressive treatments (Hayashi et al., 2018). In a clinical study involving immunocompromised 60% patients receiving phage therapy for bacterial infections, the local administration of phage resulted in low antibody neutralization in serum, suggesting potential efficacy in high-risk patient populations (Green et al., 2017).
This study demonstrates a significant advancement in phage therapy against MDR E. coli ST131. Four novel phages, isolated from hospital sewage, exhibited high individual lytic efficacy (>90%), surpassing the effectiveness of previously reported phages such as ΦLM33_P1 (70% efficacy against ST131 and ST69), ΦHP3, ΦEC1, and ΦCF2 (Dufour et al., 2016; Green et al., 2017). The increased efficacy of our phages is likely due to their greater potency and taxonomic diversity. Unlike earlier phages from the Autographiviridae and Myoviridae families, our phages belong to the Straboviridae and Podoviridae families, indicating distinct mechanisms of action and host interactions (Koonjan et al., 2021; Wolfram-Schauerte et al., 2022). Most importantly, our phage cocktail showed superior in vitro and in vivo performance, highlighting the promise of combination phage therapy as a more effective treatment for E. coli ST131 in clinical settings (Weissfuss et al., 2023).
Finally, this research highlights the serious global health threat of multidrug-resistant bacterial strains, specifically the E. coli ST131 clone. This study isolated four lytic phages (EC.W2-1, EC.W2-6, EC.W13-3, EC.W14-3) from hospital sewage water. These phages showed a broad host range and effectively lysed 94.4% of highly pathogenic E. coli ST131 isolates. They exhibited stability under different pH and temperature conditions and demonstrated strong lytic activity in vitro. Genomic analysis indicated the lack of virulence or drug-resistance genes. In vivo experiments confirmed that the phages effectively protected against MDR E. coli ST131 colon. In the injectional mice model, single phage and phage cocktail administrations improved survival rates and significantly reduced bacterial load in various organs. Based on this comprehensive evaluation, we propose phage EC.W2-1. EC.W2-6, EC.W13-3 and EC.W14-3 are potential solutions for targeting the ST131 pandemic. They provide hope amidst the growing threat of multidrug-resistant bacterial infections.
The complete genome sequences of phages EC.W2-1, EC.W2-6, EC.W13-3, and EC.W14-3 have been deposited in GenBank with the NCBI accession numbers PP445228, PP445229, PP496997 and PP496998, respectively.
The animal studies were approved by the Kyungpook National University Animal Care and Use Committee. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
MS: Conceptualization, Formal analysis, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. SK: Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing – review & editing. JK: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by a grant from the Korea Disease Control and Prevention Agency (Grant No. 2022-ER2202-00).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1455710/full#supplementary-material
1. ^https://btseq.celemics.com/
2. ^https://blast.ncbi.nlm.nih.gov/Blast.cgi
3. ^https://rast.nmpdr.org/rast.cgi
5. ^http://lowelab.ucsc.edu/cgi-bin/tRNAscan-SE2.cgi
6. ^https://cge.cbs.dtu.dk/services/ResFinder/
7. ^https://cge.cbs.dtu.dk/services/VirulenceFinder/
8. ^https://www.genome.jp/viptree/
Baek, Y. J., Kim, Y. A., Kim, D., Shin, J. H., Uh, Y., Shin, K. S., et al. (2021). Risk factors for extended-Spectrum-β-lactamase-producing Escherichia coli in community-onset bloodstream infection: impact on long-term care hospitals in Korea. Ann. Lab. Med. 41, 455–462. doi: 10.3343/ALM.2021.41.5.455
Banerjee, R., and Johnson, J. R. (2014). A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob. Agents Chemother. 58, 4997–5004. doi: 10.1128/AAC.02824-14
Bearden, C. M., Agarwal, A., Book, B. K., Vieira, C. A., Sidner, R. A., Ochs, H. D., et al. (2005). Rituximab inhibits the in vivo primary and secondary antibody response to a Neoantigen, bacteriophage phiX174. Am. J. Transplant. 5, 50–57. doi: 10.1111/j.1600-6143.2003.00646.x
Bernstein, L. J., Ochs, H. D., Wedgwood, R. J., and Rubinstein, A. (1985). Defective humoral immunity in pediatric acquired immune deficiency syndrome. J. Pediatr. 107, 352–357. doi: 10.1016/S0022-3476(85)80505-9
Bevan, E. R., Jones, A. M., and Hawkey, P. M. (2017). Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/JAC/DKX146
Cai, R., Ren, Z., Zhao, R., Lu, Y., Wang, X., Guo, Z., et al. (2023). Structural biology and functional features of phage-derived depolymerase Depo32 on Klebsiella pneumoniae with K2 serotype capsular polysaccharides. Microbiol. Spectr. 11:e0530422. doi: 10.1128/SPECTRUM.05304-22
Chaudhary, N., Singh, D., Maurya, R. K., Mohan, B., Mavuduru, R. S., and Taneja, N. (2022). Whole genome sequencing and in vitro activity data of Escherichia phage NTEC3 against multidrug-resistant Uropathogenic and extensively drug-resistant Uropathogenic E. coli isolates. Data Brief 43:108479. doi: 10.1016/J.DIB.2022.108479
CLSI. (2020). Performance standards for antimicrobial susceptibility testing: 20th informational supplement. Available at: cir.nii.ac.jp (Accessed November 5, 2024).
de Almeida Kumlien, A. C. M., Borrego, C. M., and Balcázar, J. L. (2021). Antimicrobial resistance and bacteriophages: an overlooked intersection in water disinfection. Trends Microbiol. 29, 517–527. doi: 10.1016/j.tim.2020.12.011
Dufour, N., Clermont, O., La Combe, B., Messika, J., Dion, S., Khanna, V., et al. (2016). Bacteriophage LM33_P1, a fast-acting weapon against the pandemic ST131-O25b: H4 Escherichia coli clonal complex. J. Antimicrob. Chemother. 71, 3072–3080. doi: 10.1093/JAC/DKW253
Emencheta, S. C., Onugwu, A. L., Kalu, C. F., Ezinkwo, P. N., Eze, O. C., Vila, M. M. D. C., et al. (2024). Bacteriophages as nanocarriers for targeted drug delivery and enhanced therapeutic effects. Mater. Adv. 5, 986–1016. doi: 10.1039/D3MA00817G
Fernandes, S., and São-José, C. (2016). More than a hole: the holin lethal function may be required to fully sensitize bacteria to the lytic action of canonical endolysins. Mol. Microbiol. 102, 92–106. doi: 10.1111/MMI.13448
Green, S. I., Kaelber, J. T., Li, M., Trautner, B. W., Ramig, R. F., and Maresso, A. W. (2017). Bacteriophages from ExPEC reservoirs kill pandemic multidrug-resistant strains of clonal group ST131 in animal models of bacteremia. Sci. Rep. 7:46151. doi: 10.1038/srep46151
Guo, Z., Liu, M., and Zhang, D. (2023). Potential of phage depolymerase for the treatment of bacterial biofilms. Virulence 14:2273567. doi: 10.1080/21505594.2023.2273567
Hayashi, N., Akamatsu, H., Iwatsuki, K., Shimada-Omori, R., Kaminaka, C., Kurokawa, I., et al. (2018). Japanese dermatological association guidelines: guidelines for the treatment of acne vulgaris 2017. J. Dermatol. 45, 898–935. doi: 10.1111/1346-8138.14355
Henry, M., Lavigne, R., and Debarbieux, L. (2013). Predicting in vivo efficacy of therapeutic bacteriophages used to treat pulmonary infections. Antimicrob. Agents Chemother. 57, 5961–5968. doi: 10.1128/AAC.01596-13
Hibstu, Z., Belew, H., Akelew, Y., and Mengist, H. M. (2022). Phage therapy: a different approach to fight bacterial infections. Biologics 16, 173–186. doi: 10.2147/BTT.S381237
Hon, K., Liu, S., Camens, S., Bouras, G. S., Psaltis, A. J., Wormald, P. J., et al. (2022). APTC-EC-2A: a lytic phage targeting multidrug Resistant E. coli planktonic cells and biofilms. Microorganisms 10:102. doi: 10.3390/microorganisms10010102
Isidro, A., Henriques, A. O., and Tavares, P. (2004). The portal protein plays essential roles at different steps of the SPP1 DNA packaging process. Virology 322, 253–263. doi: 10.1016/J.VIROL.2004.02.012
Jakočiūnė, D., and Moodley, A. (2018). A rapid bacteriophage DNA extraction method.Methods and protocols 1:27. doi: 10.3390/mps1030027
Kim, S., Kim, S. H., Rahman, M., and Kim, J. (2018). Characterization of a Salmonella Enteritidis bacteriophage showing broad lytic activity against gram-negative enteric bacteria. J. Microbiol. 56, 917–925. doi: 10.1007/s12275-018-8310-1
Kim, S., Song, H., Jin, J. S., Lee, W. J., and Kim, J. (2022). Genomic and phenotypic characterization of Cutibacterium acnes bacteriophages isolated from acne patients. Antibiotics 11:1041. doi: 10.3390/antibiotics11081041
Koonjan, S., Cooper, C. J., and Nilsson, A. S. (2021). Complete genome sequence of vb_ecop_su7, a podoviridae coliphage with the rare c3 morphotype. Microorganisms 9:1576. doi: 10.3390/microorganisms9081576
Leiman, P. G., Shneider, M. M., Kostyuchenko, V. A., Chipman, P. R., Mesyanzhinov, V. V., and Rossmann, M. G. (2003). Structure and location of gene product 8 in the bacteriophage T4 baseplate. J. Mol. Biol. 328, 821–833. doi: 10.1016/S0022-2836(03)00366-8
Łusiak-Szelachowska, M., Zaczek, M., Weber-Dabrowska, B., Miȩdzybrodzki, R., Kłak, M., Fortuna, W., et al. (2014). Phage neutralization by sera of patients receiving phage therapy. Viral Immunol. 27, 295–304. doi: 10.1089/VIM.2013.0128
Majewski, P., Gutowska, A., Smith, D. G. E., Hauschild, T., Majewska, P., Hryszko, T., et al. (2021). Plasmid mediated mcr-1.1 Colistin-resistance in clinical Extraintestinal Escherichia coli strains isolated in Poland. Front. Microbiol. 12:547020. doi: 10.3389/fmicb.2021.547020
Matsumi, R., Atomi, H., and Imanaka, T. (2005). Biochemical properties of a putative signal peptide peptidase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 187, 7072–7080. doi: 10.1128/JB.187.20.7072-7080.2005
Miyoshi-Akiyama, T., Sherchan, J. B., Doi, Y., Nagamatsu, M., Sherchand, J. B., Tandukar, S., et al. (2016). Comparative genome analysis of extended-Spectrum-β-lactamase-producing Escherichia coli sequence type 131 strains from Nepal and Japan. mSphere 1:e00289-16. doi: 10.1128/mSphere.00289-16
Murray, E., Draper, L. A., Ross, R. P., and Hill, C. (2021). The advantages and challenges of using Endolysins in a clinical setting. Viruses 13:680. doi: 10.3390/V13040680
Nelson, D. C., Schmelcher, M., Rodriguez-Rubio, L., Klumpp, J., Pritchard, D. G., Dong, S., et al. (2012). Endolysins as antimicrobials. Adv. Virus Res. 83, 299–365. doi: 10.1016/B978-0-12-394438-2.00007-4
Nicolas-Chanoine, M. H., Bertrand, X., and Madec, J. Y. (2014). Escherichia coli st131, an intriguing clonal group. Clin. Microbiol. Rev. 27, 543–574. doi: 10.1128/CMR.00125-13
Örmälä, A.-M., and Jalasvuori, M. (2013). Phage therapy. Bacteriophage 43, 122–125. doi: 10.4161/BACT.24219
Peirano, G., Lynch, T., Matsumara, Y., Nobrega, D., Finn, T. J., DeVinney, R., et al. (2020). Trends in population dynamics of Escherichia coli sequence type 131, Calgary, Alberta, Canada, 2006–2016- volume 26, number 12—December 2020- emerging infectious diseases journal - CDC. Emerg. Infect. Dis. 26, 2907–2915. doi: 10.3201/eid2612.201221
Poteete, A. R. (2004). Modulation of DNA repair and recombination by the bacteriophage λ Orf function in Escherichia coli K-12. J. Bacteriol. 186, 2699–2707. doi: 10.1128/JB.186.9.2699-2707.2004
Pu, M., Li, Y., Han, P., Lin, W., Geng, R., Qu, F., et al. (2022). Genomic characterization of a new phage BUCT541 against Klebsiella pneumoniae K1-ST23 and efficacy assessment in mouse and galleria mellonella larvae. Front. Microbiol. 13:950737. doi: 10.3389/fmicb.2022.950737
Rahman, M., Kim, S., Kim, S. M., Seol, S. Y., and Kim, J. (2011). Characterization of induced Staphylococcus aureus bacteriophage SAP-26 and its anti-biofilm activity with rifampicin. Biofouling 27, 1087–1093. doi: 10.1080/08927014.2011.631169
Rehman, S., Ali, Z., Khan, M., Bostan, N., and Naseem, S. (2019). The dawn of phage therapy. Rev. Med. Virol. 29:e2041. doi: 10.1002/RMV.2041
Rogers, B. A., Sidjabat, H. E., and Paterson, D. L. (2011). Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66, 1–14. doi: 10.1093/JAC/DKQ415
Shamsuzzaman, M., Dahal, R. H., Kim, S., and Kim, J. (2023). Genome insight and probiotic potential of three novel species of the genus Corynebacterium. Front. Microbiol. 14:1225282. doi: 10.3389/fmicb.2023.1225282
Shamsuzzaman, M., Kim, S., Choi, Y.-J., Kim, B., Dahal, R. H., Shin, M., et al. (2024). Therapeutic phage candidates for targeting prevalent sequence types of Carbapenem-resistant Escherichia coli. Foodborne Pathog. Dis. doi: 10.1089/FPD.2024.0023
Shang, J., Peng, C., Tang, X., and Sun, Y. (2023). PhaVIP: phage VIrion protein classification based on chaos game representation and vision transformer. Bioinformatics 39, i30–i39. doi: 10.1093/BIOINFORMATICS/BTAD229
Shariati, A., Arshadi, M., Khosrojerdi, M. A., Abedinzadeh, M., Ganjalishahi, M., Maleki, A., et al. (2022). The resistance mechanisms of bacteria against ciprofloxacin and new approaches for enhancing the efficacy of this antibiotic. Front. Public Health 10:1025633. doi: 10.3389/fpubh.2022.1025633
Shearer, W. T., Lugg, D. J., Rosenblatt, H. M., Nickolls, P. M., Sharp, R. M., Reuben, J. M., et al. (2001). Antibody responses to bacteriophage φX-174 in human subjects exposed to the Antarctic winter-over model of spaceflight. J. Allergy Clin. Immunol. 107, 160–164. doi: 10.1067/MAI.2001.112269
Tang, Z., Tang, N., Wang, X., Ren, H., Zhang, C., Zou, L., et al. (2023). Characterization of a lytic Escherichia coli phage CE1 and its potential use in therapy against avian pathogenic Escherichia coli infections. Front. Microbiol. 14:1091442. doi: 10.3389/fmicb.2023.1091442
Topka-Bielecka, G., Dydecka, A., Necel, A., Bloch, S., Nejman-Faleńczyk, B., Węgrzyn, G., et al. (2021). Bacteriophage-derived Depolymerases against bacterial biofilm. Antibiotics 10:175. doi: 10.3390/ANTIBIOTICS10020175
Weissfuss, C., Wienhold, S. M., Bürkle, M., Gaborieau, B., Bushe, J., Behrendt, U., et al. (2023). Repetitive exposure to bacteriophage cocktails against Pseudomonas aeruginosa or Escherichia coli provokes marginal humoral immunity in Naïve mice. Viruses 15:387. doi: 10.3390/v15020387
Whelan, S., Lucey, B., and Finn, K. (2023). Uropathogenic Escherichia coli (UPEC)-associated urinary tract infections: the molecular basis for challenges to effective treatment. Microorganisms 11:2169. doi: 10.3390/MICROORGANISMS11092169
Keywords: Escherichia coli, clone ST131, antibiotic resistance, bacteriophages, therapeutic efficiency, phage cocktail
Citation: Shamsuzzaman M, Kim S and Kim J (2024) Bacteriophage as a novel therapeutic approach for killing multidrug-resistant Escherichia coli ST131 clone. Front. Microbiol. 15:1455710. doi: 10.3389/fmicb.2024.1455710
Received: 27 June 2024; Accepted: 30 October 2024;
Published: 12 December 2024.
Edited by:
Gamaliel López-Leal, Center for Research in Cellular Dynamics-UAEM, MexicoReviewed by:
Md Mozammel Hoque, University of Technology Sydney, AustraliaCopyright © 2024 Shamsuzzaman, Kim and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jungmin Kim, bWlua2ltQGtudS5hYy5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.