- 1Division of Microbiology, National Center for Toxicological Research, United States Food and Drug Administration, Jefferson, AR, United States
- 2Office of Applied Science, Center for Veterinary Medicine, U.S. Food and Drug Administration, Laurel, MD, United States
The pervasive environmental metal contamination has led to selection of heavy-metal resistance genes in bacteria. The pco and sil clusters are located on a mobile genetic element and linked to heavy-metal resistance. These clusters have been found in Salmonella enterica serovars isolated from human clinical cases and foods of animal origin. This may be due to the use of heavy metals, such as copper, in animal feed for their antimicrobial and growth promotion properties. The sil cluster can be found alone or in combination with pco cluster, either in the chromosome or on a plasmid. Previous reports have indicated that sil, but not pco, cluster contributes to copper resistance in S. enterica Typhimurium. However, the role of the pco cluster on the physiology of non-typhoidal S. enterica remains poorly understood. To understand the function of the pco gene cluster, a deletion mutant of pcoABCD genes was constructed using allelic exchange mutagenesis. Deletion of pcoABCD genes inhibited growth of S. enterica in high-copper medium, but only under anaerobic environment. Complementation of the mutant reversed the growth phenotype. The survival of S. enterica in RAW264.7 macrophages was not affected by the loss of pcoABCD genes. This study indicates that the acquired pco cluster is crucial for copper detoxification in S. enterica, but it is not essential for intracellular replication within macrophages.
Introduction
Salmonella enterica serovars are among the most common causes of bacterial gastrointestinal disease worldwide and pose a major public health concern with an estimated 1.35 million Salmonella infections including 26,000 leading to hospitalization, and 420 deaths each year in the United States (European Food Safety Authority and European Centre for Disease Prevention and Control, 2019; Centers for Disease Control and Prevention, 2023). Globally, it is estimated that over 93 million cases of Salmonella gastroenteritis and 155,000 deaths occur annually, leading to the exacerbation of the global burden of Salmonellosis (Majowicz et al., 2010). Antimicrobial resistance in S. enterica is a public health concern due to the rapid spread of the multidrug-resistant (MDR) strains, which is linked to the overuse of antibiotics (Punchihewage-Don et al., 2022). These MDR strains cause approximately 410,000 infection every year in the U.S. (Punchihewage-Don et al., 2022).
Horizontal gene transfer mediates antimicrobial resistance among different bacterial species through the exchange of mobile genetic cassettes, which are often on transferrable plasmids or transposons (Sun et al., 2019). Notably, natural transfer of conjugative plasmids is initiated when bacteria are exposed to environmental stresses, which can induce the DNA uptake system of the recipient cell (Sun et al., 2019). Non-typhoidal Salmonella have acquired heavy-metal resistance genes for copper (Cu) and silver (Ag) to cope with the high level of heavy metals pollutants in the environment and this metal tolerance was found to be co-occur with antibiotic resistance (Mourao et al., 2016). MDR cassettes that carry antimicrobial resistance genes are commonly associated with heavy-metal resistance genes, suggesting that co-selection of antimicrobial resistance can occur in the presence of heavy metals, even at sub-inhibitory concentrations of these metals. The pco and sil clusters have also been detected in association with the arsenic (ars) resistance genes in the epidemic clone of S. Typhimurium, suggesting that these clones can survive in environments with various types of heavy-metal contaminants (Joana Mourao et al., 2020; Branchu et al., 2019). More recently, a whole genome sequence (WGS) analysis using long-read sequencing of 134 MDR S. enterica isolated from retail meat revealed that 60% of these MDR strains carry heavy-metal resistance genes along with AMR genes, which raise the concern of co-selection of heavy metal and antibiotic resistance (Li et al., 2021).
Copper is an essential trace element for all bacteria. The redox-active property of copper is crucial for cuproenzymes, such as the cytochrome c oxidase respiratory super complexes and superoxide dismutase, which protect bacteria from superoxide radicals generated by macrophages and neutrophils as a defense mechanism against invading pathogen (Samanovic et al., 2012). However, excess copper is detrimental to cells as it replaces native metal cofactors in proteins leading to their damage. Therefore, in response to copper, bacteria have evolved sophisticated mechanisms to evade copper toxicity either by extruding or sequestering copper, or by oxidizing the toxic Cu (I) to less toxic Cu (II) (Giachino and Waldron, 2020). For example, S. enterica serovar Typhimurium encodes CopA and GolT, metal-transporting P-type ATPase proteins, which export copper outside the cell to maintain copper homeostasis (Osman et al., 2010). In addition to copA and golT, S. Typhimurium carries the gene that encodes CueO, a periplasmic multicopper oxidase, which oxidizes Cu (I) to Cu (II); deletion of cueO gene resulted in severe attenuation in mouse model (Achard MET et al., 2010). Essential trace metals, such as copper, zinc, molybdenum, selenium, manganese, iron, and cobalt are extensively used in animal and poultry feed as nutritional additives to promote growth (Hejna et al., 2018). The recommended copper concentration in poultry feed by the National Research Council (NRC) is 8 mg/kg (Subcommittee on Poultry Nutrition CoAN, 1994), whereas the maximum concentration of copper allowed in poultry and animal feed by the European Union (EU) is ranging from 15–35 mg/kg, except in piglets (up to 12 weeks) feed, where 170 mg/kg can be permitted (EFSA Panel on Additives and Products or Substances used in Animal Feed, 2016). But in fact, copper is over-supplemented in animals diet (up to 800 mg/kg) (Korish and Attia, 2020). Despite its insidious toxicity, copper was found to be over supplemented in animals feed compared to other trace metals (Lopez-Alonso and Miranda, 2020). Consequently, this overuse has potentially led to increase the minimal inhibitory concentration (MIC) by opportunistic bacteria such as Enterococcus spp. to high levels of copper, which is associated with the acquisition of copper-resistance genes (Yazdankhah et al., 2014).
In Escherichia coli, the pco cluster contains seven genes, pcoABCDERS (Brown et al., 1995). pcoA encodes the multicopper oxidase, PcoA, which oxidizes Cu (I) to the less toxic Cu (II) form in the periplasm; whereas, PcoC is a periplasmic protein that binds one ion of Cu (II) (Huffman et al., 2002). PcoE is also a periplasmic protein, which is thought to act as a “metal sponge” that scavenges free Cu ions from the periplasmic space (Zimmermann et al., 2012). On the other hand, PcoB is an outer membrane protein and predicted to act as a Cu (II) importer that may facilitate Cu uptake in Cu-scarce environment or sequester excess Cu (Li et al., 2022). PcoD is a predicted inner membrane protein of unknown function. The pco and sil gene clusters (frequently found in Enterobacteriaceae species) are co-transferred from one bacterium to another by a Tn7-like transposon element (32.54 kb), which is located between the integrons (Fang et al., 2016; Billman-Jacobe et al., 2018; Lee et al., 2002). Additionally, two studies have shown that the sil gene cassette alone, without the pco cluster, conferred resistance to high levels of Cu under anaerobic conditions (Mourao et al., 2016; Mourao et al., 2015). These results suggest that sil genes can handle copper toxicity without the need of the pco cassette. The pco and sil clusters were recently detected in the Salmonella genomic island 4 (SGI-4), an integrative conjugative element (ICE), in monophasic S. Typhimurium; only deletion of silABC, but not pcoABC, reduced the minimum inhibitory concentration (MIC) to CuSO4 under anaerobic conditions (Branchu et al., 2019). A similar study also found the cus (renamed to sil) and pco clusters in SGI-3 in S. Typhimurium; the loss of pco cluster did not result in growth defects in copper-supplemented medium under anaerobic environment (Arai et al., 2019). However, the role of pco cluster in tolerance to toxic levels of Cu (I) is still unclear.
The present study examines the role of the pco gene cluster in non-typhoidal Salmonella for copper resistance and survival in macrophages. Our work shows that the acquired pco cluster contributes to Cu (I) resistance. However, our study reveals that the acquisition of the pco cluster has no impact on replication efficiency within macrophages.
Materials and methods
Bacterial strains
The S. enterica isolates used in this study were isolated from retail meat in the United States (Table 1) (Li et al., 2021; McDermott et al., 2016). Growth phenotype was compared to the wild-type S. enterica ATCC13076 (lacks pco and sil clusters). Analyzing the whole genome sequence of SL-3 and SL-4 and identifying the location of the pco and sil cluster was performed using the MOB-RECON software tool in the cloud-based Galaxy bioinformatics platform.1
Construction of a pco deletion mutant and complementation
To create mutation in the pco cassette, the upstream region of pcoA was amplified from SL-4 with primers: P27 and P28 (Table 2), and the downstream region of pcoD was amplified with primers: P29 and P30 (Table 2) to create overlapping PCR products. The upstream and downstream PCR products of pcoA and pcoD were cut with restriction enzymes: SalI and SacI, respectively and inserted into the SalI and SacI sites of the temperature-sensitive plasmid pDG3 (encodes β-galactosidase gene, bgaB, for blue/white screening) (Gudeta and Foley, 2024) to generate plasmid pAH3 using NEBuilder HiFi DNA assembly kit (New England BioLabs, Ipswich, MA, USA). SL-4 was then electroporated with plasmid pAH3. To allow integration of pAH3 in SL-4 chromosome, transformants were plated onto LB agar supplemented with 0.5% glucose, X-Gal 80 μg/mL, and 25 μg/mL chloramphenicol and incubated at 42°C overnight. A single blue colony from the transformants plate was inoculated in LB broth supplemented with 1% rhamnose and incubated at 30°C for 4 h. To facilitate allelic replacement of pcoABCD with the upstream and downstream PCR products on pAH3, cells were then harvested by centrifugation at 10,000 × g, resuspended in LB broth, plated onto LB agar plates without antibiotics, and incubated at 37°C for 24 h for blue/white screening. White colonies were screened to confirm the loss of pcoD using primers P19 and P24. To complement SL-4ΔpcoABCD mutant, we amplified pcoEABCDRS genes from the parent strain using primers: P55 and P56, which contain SalI and SacI restriction enzyme sites, respectively. The pcoEABCDRS PCR product was cut with SalI and SacI restriction enzymes and inserted into the SalI and SacI sites of plasmid pBBR1-MCS2 to create pAH8 (SI 1A). Then, pAH8 was electroporated into SL-4ΔpcoABCD. To facilitate the delivery of the 12 kb pAH8 in SL-4ΔpcoABCD, cells were grown in LB supplemented with 8 μg/mL polymyxin B nonapeptide (permeabilizes bacterial outer membrane) (Qin et al., 2022) and 50 μg/mL kanamycin before electroporation. Transformants were plated onto LB agar plates supplemented with 50 μg/mL kanamycin and 3 mM CuSO4 and incubated at 37°C anaerobically for 3 days. The complemented SL-4ΔpcoABCD was verified by PCR using primer pairs: P23/P56 and P61/P24.
Growth conditions and in vitro growth curves
Strains were grown in LB only or LB supplemented with 4 mM CuSO4 or LB agar supplemented with 2 mM CuSO4. For the copper-tolerance growth curves, overnight cultures of S. enterica isolates grown in LB broth at 37°C. Kanamycin (50 μg/mL) was added to SL-4ΔpcoABCD/empty vector (pBBR1-MCS2) and the complemented strain to maintain the pBBR1-MCS2 and pAH8, respectively. Bacterial cultures were diluted to an OD600 of 0.05 in the same medium and inoculated in 96-well plates containing LB broth only or LB broth supplemented with 3 mM CuSO4. The cultures were incubated stationary at 37°C under anaerobic condition for 7 days. Anaerobic atmosphere was generated using Mitsubishi AnaeroPack-Anaero (Thermo Fisher Scientific, Waltham, MA, USA). Cultures were mixed immediately before measuring the optical density (OD600) at the designated time points.
Minimum inhibitory concentrations (MICs) to copper sulfate
Minimum inhibitory concentrations of S. enterica strains to CuSO4 were determined by the agar dilution assay. Strains were inoculated from a single colony into LB broth and incubated at 37°C overnight then diluted to approximately 10−7 CFU/mL and 5 μL spotted onto Muller-Hinton agar supplemented with CuSO4 (0.5, 0.75, 1, 1.5, 2, 3, 4, 5, 6, 7, 8, and 12 mM). Plates were incubated aerobically or anaerobically at 37°C for 24 or 72 h, respectively.
Alamar blue assay
Strains of S. enterica were inoculated to OD600 of 0.05 into 96-well plates containing LB broth alone or LB supplemented with 1–3 mM CuSO4 and incubated at 37°C aerobically or anaerobically overnight or 72 h, respectively. Next, 20 μL alamarBlue reagent (Thermo Fisher Scientific, Waltham, MA, USA) was added to each well, and plates were incubated for 4 h at 37°C. The assessment of viable bacteria was according to the reduction of the blue indicator, resazurin to resorufin (pink).
Macrophage infection
RAW264.7 macrophage cell lines were cultured in DMEM basal medium supplemented with 10% fetal bovine serum (DMEM-FBS) at 37°C and 5% CO2 and replaced daily with fresh pre-warmed medium. Cells were seeded in 24-well plates at a density of 1.5 ×106 cells per well and incubated at 37°C and 5% CO2 until reached confluency. RAW264.7 macrophage were then infected with S. enterica: WT, SL-3, SL-4, and SL-4ΔpcoABCD strains at an MOI of 1 (1 bacteria/macrophage cell) for 1 h at 37°C. At indicated time-points, cells were washed twice with phosphate buffered saline (PBS) and lysed with 1% triton X-100 for 10 min. Cell lysates were serially diluted in PBS, plated onto LB agar plates, and incubated for 24 h at 37°C for colony enumeration.
Statistical analysis
Statistical analyses of bacterial growth data were performed using Student’s unpaired t test (two-tailed). Statistically significant difference in growth curves and replication in macrophages between the groups was calculated using ANOVA with pair-wise comparisons.
Results
pco cluster structure and location
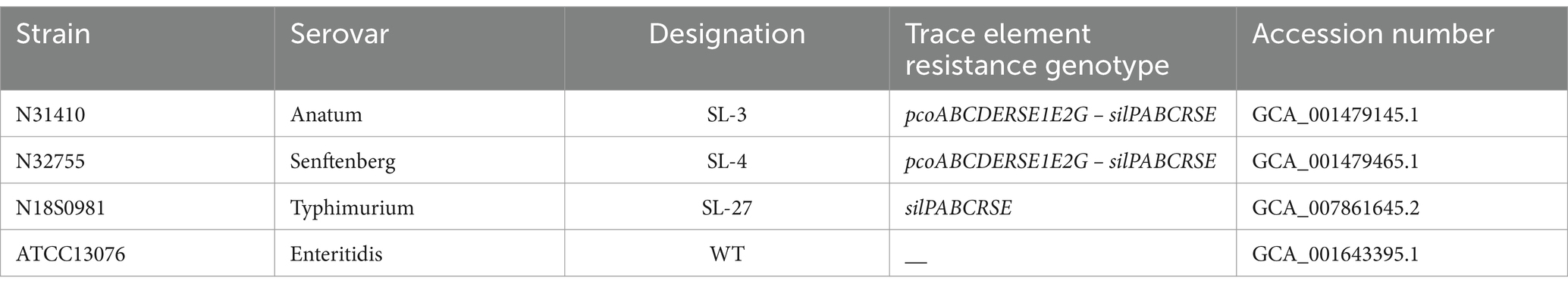
In silico analysis of the whole genome sequence showed that S. enterica serovar Anatum isolate N31410 (SL-3) and S. enterica serovar Senftenberg isolate N32755 (SL-4) contain the pcoABCDERSE1E2G genes; these genes are located adjacent to silver-resistance cassette, silPABCRSE (Figure 1A). The pco and sil clusters are located in the chromosome of S. enterica serovar Anatum isolate N31410 (SL-3) (Figure 2A). Intriguingly, S. enterica serovar Senftenberg isolate N32755 (SL-4) carries pco and sil genes cluster on a 75 kb incompatibility group (Inc) FIB plasmid (AA423) (Figure 1B), which is unusual for those genes to occur on plasmids of the IncF family. Remarkably, these gene clusters are not located in conjunction with or in the vicinity of antimicrobial-resistant (AMR) genetic elements, such as blaTEM-1, aadA, aac(6″)-Ib4, sul, and tetA as they are frequently co-located with these antibiotic resistance genes (Mourao et al., 2016).
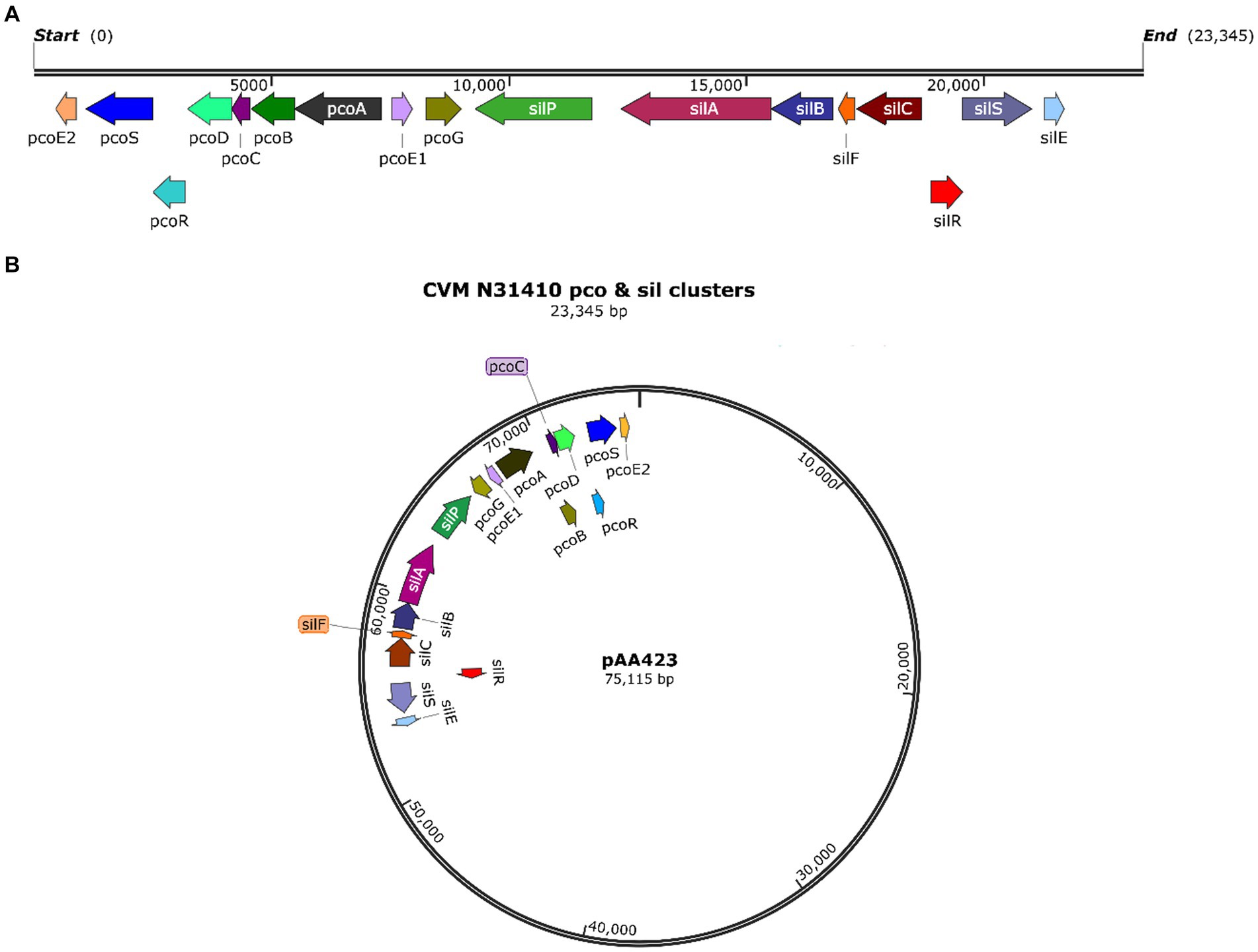
Figure 1. (A) Map of the pco and sil gene cassette in the chromosome of S. enterica CVM N31410 (SL-3) isolate. (B) Plasmid map of the pAA423 plasmid located in S. enterica CVM N32755 (SL-4) isolate. The maps were created by SnapGene 7.0. 2.
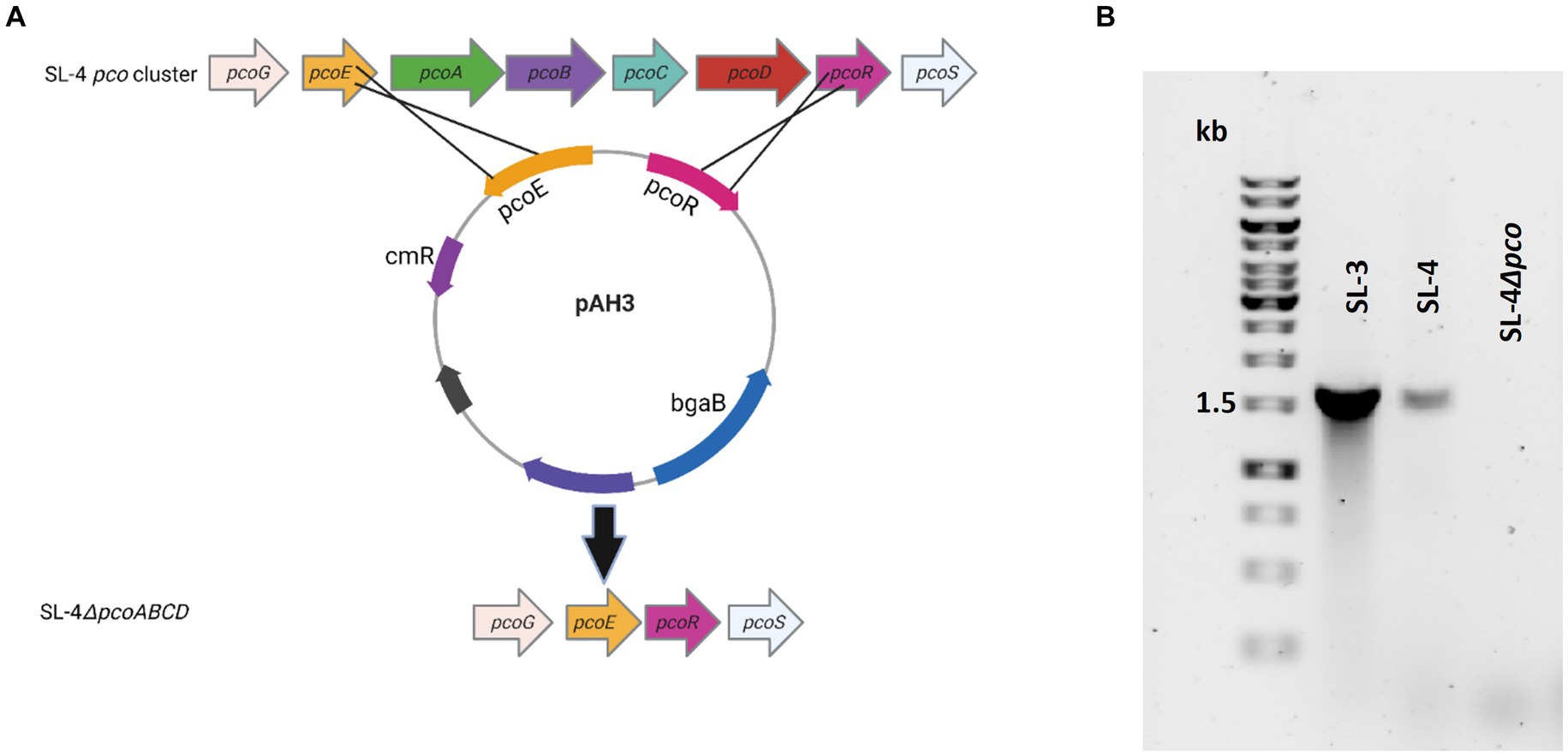
Figure 2. Confirmation of pcoABCD mutation. (A) Overview of SL-4 pcoA-D deletion. Plasmid pAH3 encompasses the upstream (pcoE) and downstream region (pcoR) regions of pcoA-D was used to replace pcoA-D. (B) PCR screening for the loss of pcoD using primers: P-19 and P-24. The expected amplicon size for the presence of the gene is 1.6 kb.
The pco genes cluster is required for copper tolerance
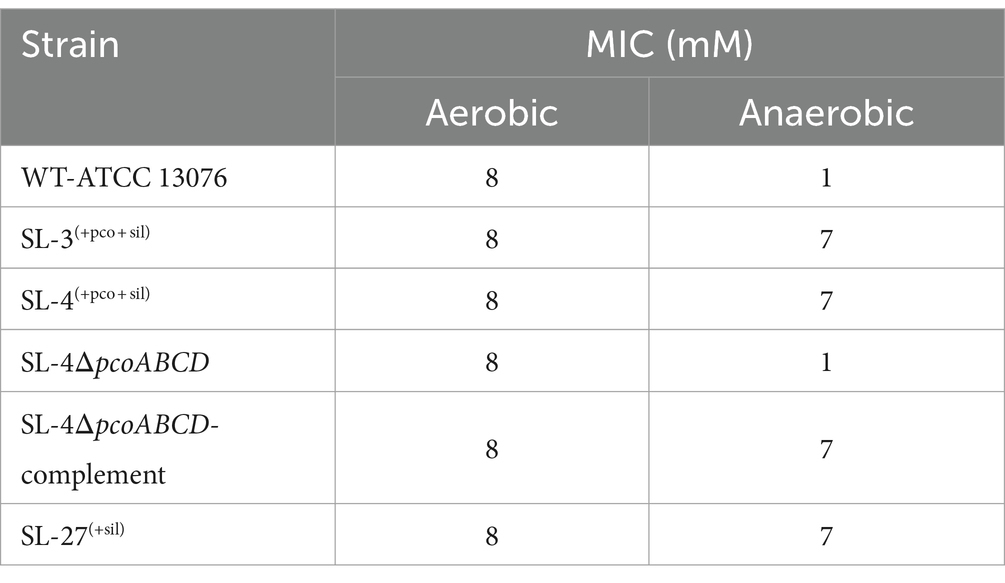
To assess roles of the pco genes cluster in copper tolerance, we created a deletion of pcoA-D cassette in SL-4 through allelic exchange mutagenesis (Figure 2A). The deletion mutant was confirmed by PCR (Figure 2B). Growth of strains was tested in LB medium only or LB medium containing high levels of copper. In this experiment, we tested copper resistance of the parent strain SL-4 and SL-3. Additionally, we compared growth rate of SL-3, SL-4, and SL-4ΔpcoABCD, and SL-4ΔpcoABCD-complement to S. enterica Typhimurium (SL-27), which carries sil cluster only, and S. enterica ATCC 13076 (negative control). All strains were grown in LB supplemented with 3 mM CuSO4. Bacterial cultures were incubated aerobically or anaerobically with or without CuSO4. The results show that there is no significant difference in growth between all the strains growing in copper-supplemented medium under aerobic condition (Figure 3A). Also, all the strains grow at the same level in LB broth only under aerobic (data not shown) or anaerobic condition (Figure 3B). Because bacterial growth rate slows down in anaerobic environment, and we also wanted to examine if Cu (I)-resistant S. enterica can tolerate long-term exposure of high levels of copper, we monitored growth of anaerobic culture for 7 days. Our data reveal that deletion of the pcoABCD cassette resulted in a significant growth reduction in Cu-supplemented medium when incubated anaerobically (Figure 4A). Growth was restored by the complemented SL-4ΔpcoABCD strain (Figure 4A). SL-3(+pco + sil), SL-4(+pco + sil), and SL-27(+sil) exhibited high resistance to copper and grew at a significantly higher growth rate than the mutant and wild-type S. enterica (Figure 4A). We also examined the MICs of S. enterica strains to copper on solid medium (Table 3). The MIC of all strains to CuSO4 was the same (8 mM) under aerobic condition. Under anaerobic environment, the MIC of SL-3(+pco + sil), SL-4(+pco + sil), SL-4ΔpcoABCD-complement, and SL-27(+sil) was 7 mM, whereas the MIC of SL-4ΔpcoABCD and WT was 1 mM, where no visible colonies were formed (Figure 4B). We did not see any difference in the MIC between the strains carrying both pco and sil clusters (SL-3 and SL-4) and SL-27, which harbors sil cassette only.
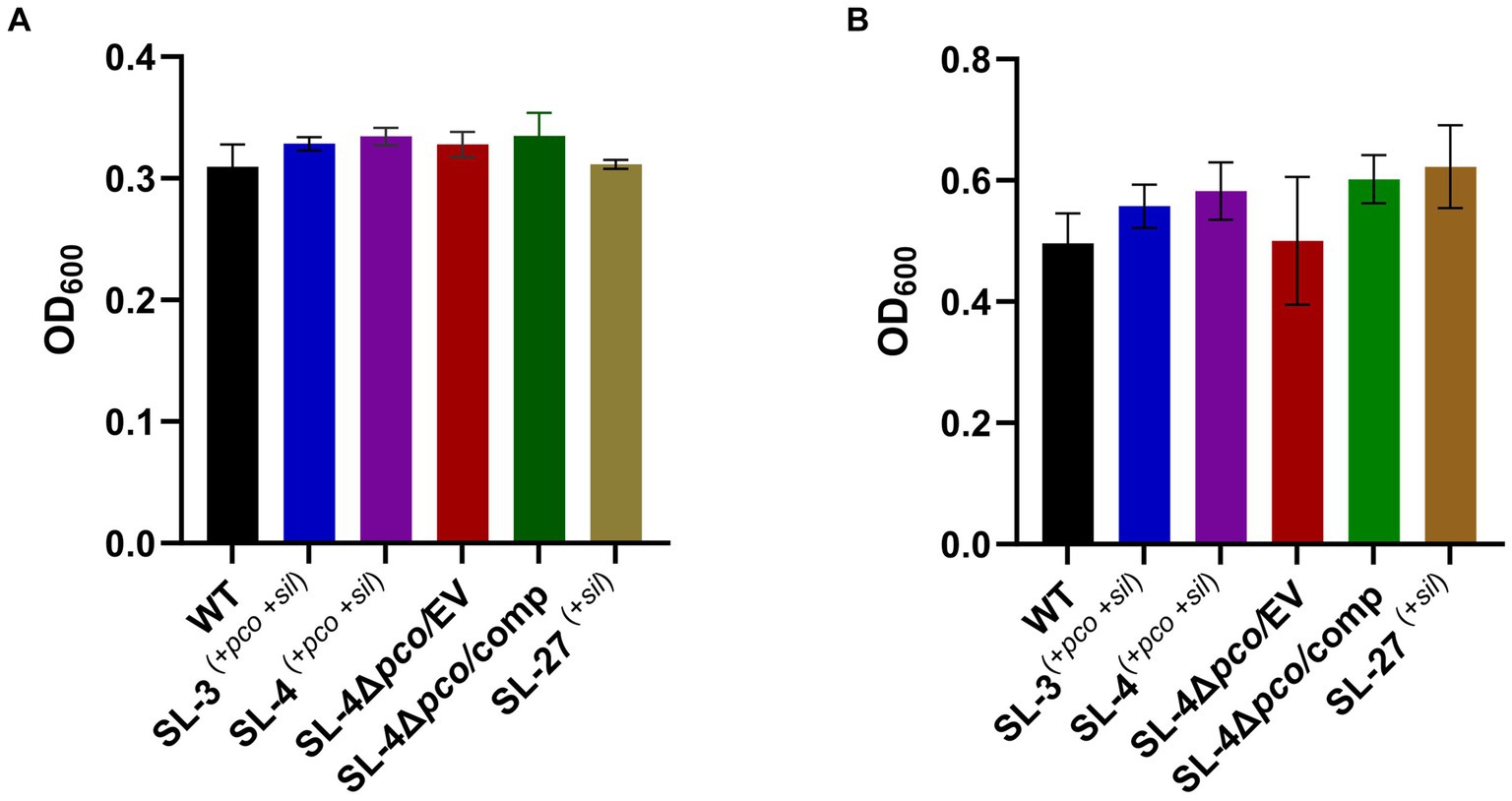
Figure 3. Assays for the effect of pcoABCD deletion on growth in LB supplemented with CuSO4 or LB only under aerobic or anaerobic conditions, respectively. WT S. enterica, SL-3, SL-4, SL-4Δpco/EV (empty vector), SL-4Δpco/comp (complement), and SL-27 strains were cultured in LB with 4 mM CuSO4 and incubated aerobically for 24 h (A) or grown in LB only and incubated anaerobically (B). Error bars represent the standard error from two independent experiments in triplicates. The p-values were calculated by unpaired two-tailed Student’s t test. All p-values are greater than 0.05.
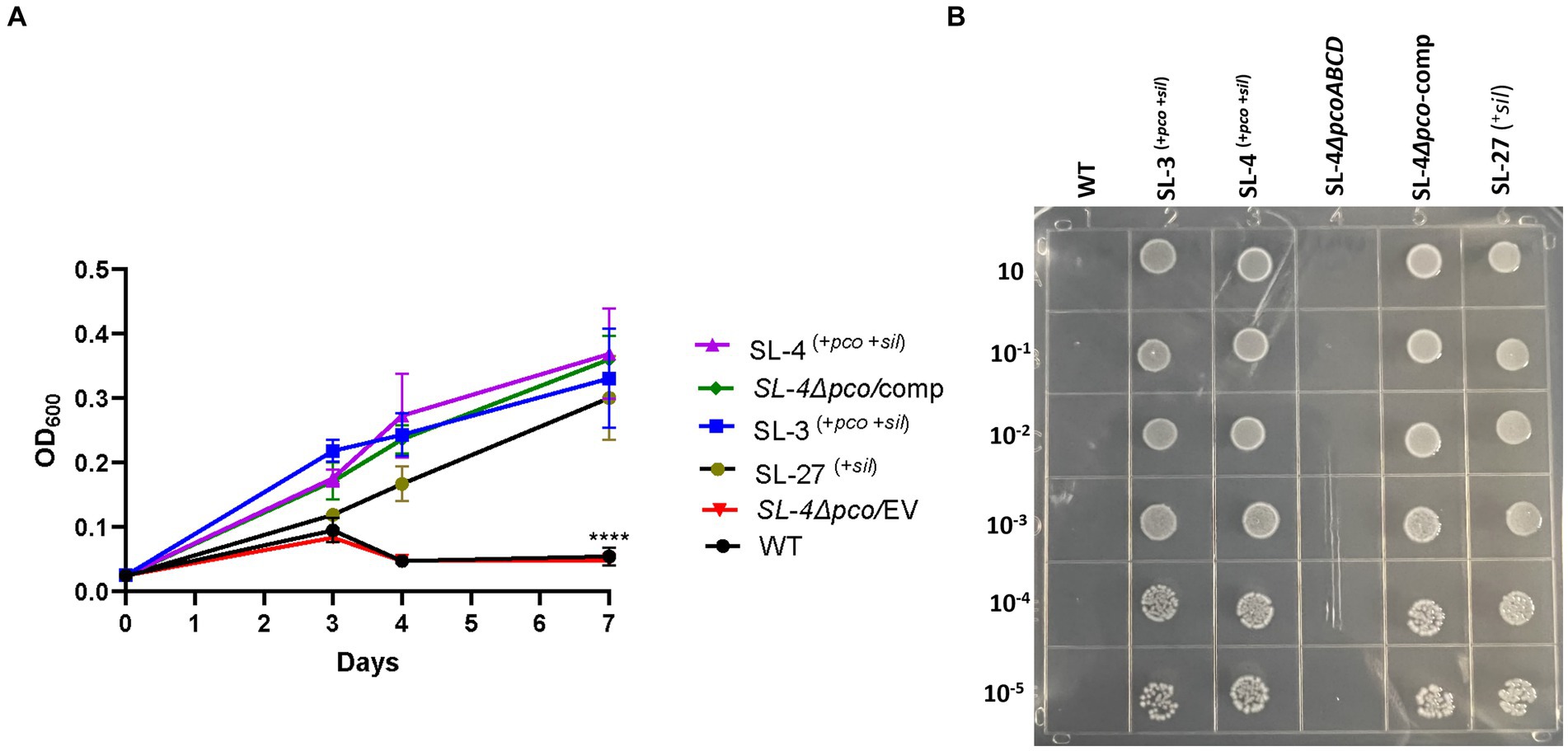
Figure 4. The pco cluster enhances tolerance to high levels of copper under anaerobic conditions. (A) Growth curve of WT S. enterica, SL-3, SL-4, SL-4Δpco/EV (empty vector), SL-4Δpco/comp (complement), and SL-27 in LB broth supplemented with 3 mM CuSO4. Cultures were incubated at 37°C anaerobically for 7 days. Cell densities shown (OD600) are the mean +/− standard error from three independent cultures assayed in triplicates. The p-values were calculated by two-way ANOVA (***p ≤ 0.0001). (B) Plating results of the same strains that were grown in LB broth then serially diluted spotted onto LB agar supplemented with 2 mM CuSO4. Plates were then incubated at 37°C anaerobically for 72 h.
Growth inhibition of the pco mutant by copper in anaerobic environment was also confirmed by Alamar Blue assay. Growth of the SL-4ΔpcoABCD mutant in Cu-supplemented medium was not impacted when incubated aerobically (Supplementary Figure S2A). However, the mutated exhibited growth inhibition (blue color) in LB supplemented with CuSO4 (1–3 mM) when compared with the parent strain and SL-3 cultures (pink color) (Supplementary Figure S2B).
Together, these data suggest that pcoABCD genes contribute to copper (I) tolerance in S. enterica.
Loss of the pcoABCD genes does not have an impact on replication in macrophages
As an intracellular pathogen, S. enterica encounters high levels of copper in macrophages, specifically in phagolysosome, which is employed by host immune response to kill intracellular pathogens (Wagner et al., 2006), we assessed whether the deletion the of the pco genes cluster has an impact on replication during macrophage infection. We hypothesize that pco genes aid in the ability of SL-4 to overcome copper toxicity in phagolysosome, which in turn enhances survival in macrophages. The survival of SL-4Δpco, SL-4, and wild-type S. enterica was examined in RAW264.7 macrophages, in which macrophages were infected with these strains at an MOI of 1. The infection experiment revealed that deletion of pcoABCD genes had no significant reduction in replication within RAW264.7 macrophages up to 72 h post-infection (Figure 5). These results indicate that S. enterica uses another mechanism to evade the macrophage intracellular killing.
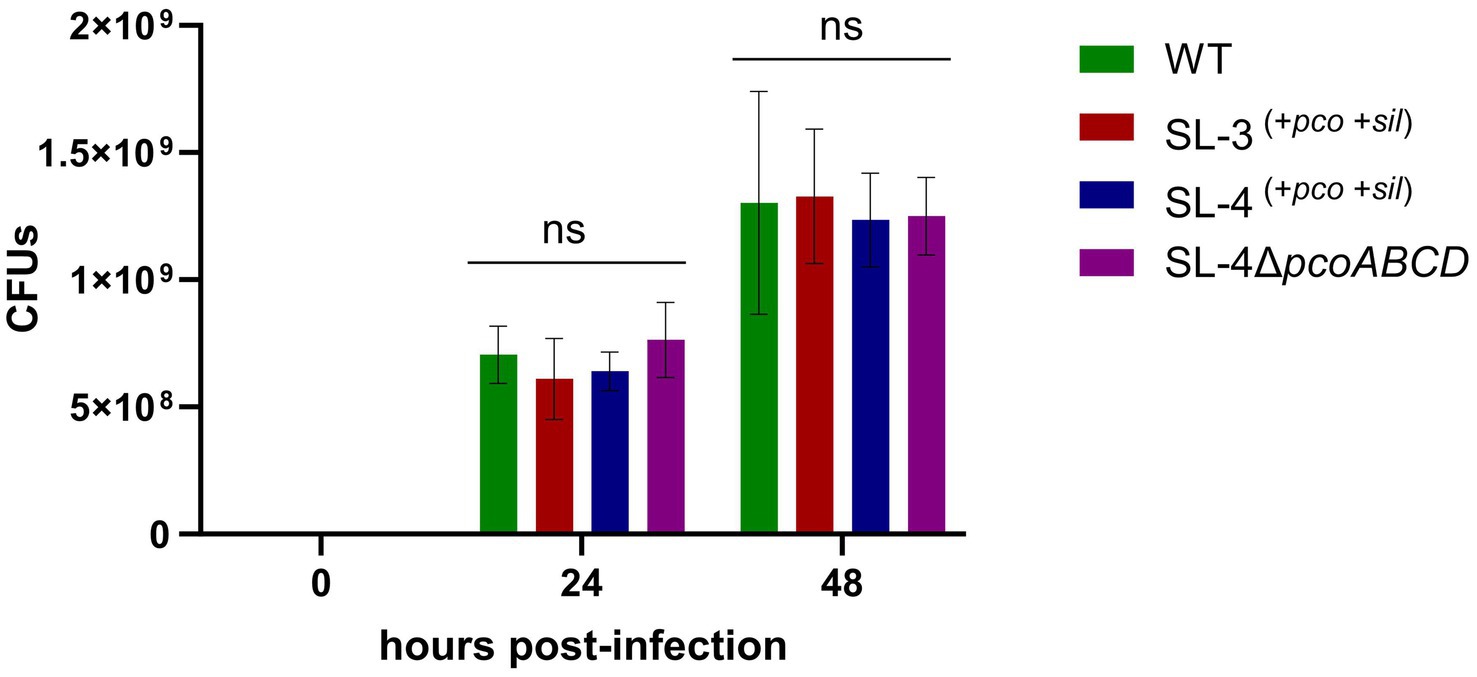
Figure 5. Loss of pcoABCD genes does not impact replication in RAW 264.7 macrophages. RAW 264.7 macrophages were infected with S. enterica (WT), SL-3, SL-4, and SL-4Δpco at MOI = 1 for 1 h in DMEM supplemented with 10% FBS. At the indicated time points, cells were lysed and plated onto LB agar medium and CFUs were enumerated. Data shown indicate the mean CFU with standard error of two independent experiments with three biological replicates. The p-values were calculated by two-way ANOVA (ns, p > 0.05).
Discussion
In this study, we have clearly demonstrated that the acquired pco cluster functions in copper tolerance in non-typhoidal S. enterica, specifically under anaerobic conditions. Our in silico analysis reveals that pco and sil clusters are found in the chromosome (in SL-3) or on an IncFIB conjugative plasmid (in SL-4) and are not associated with AMR genes. Pco and sil clusters are mainly located on IncH-type plasmids and are co-localized with AMR genes (Li et al., 2021; Billman-Jacobe et al., 2018). To our knowledge, the integration of these clusters on IncFIB plasmid and the lack of association with AMR genes have not been reported previously. Our results show that a mutant lacking pcoABCD genes is sensitive to high levels of copper only under anaerobic condition, and that the sil cluster was not able to rescue the pco mutant. These results are consistent with the conversion of the non-toxic form of copper, Cu (II), to the toxic form, Cu (I) in anoxic environments (Beswick et al., 1976). Complementation of the SL-4ΔpcoABCD reversed growth phenotype to the same level of the parent strain. On the other hand, loss of these genes, had no impact on the capability of non-typhoidal S. enterica to replicate in macrophages. These data indicate that S. enterica may use the copper exporting P-type ATPase, GolT, as a surrogate for the pco cluster to overcome copper toxicity in macrophages (Pontel et al., 2014). The macrophage infection data is consistent with a previous study which showed that although deletion of golT resulted in accumulation of copper in the mutant, it did not impact replication in macrophages; however, survival in macrophages was compromised when cells were infected with the golT and copA double mutant (Osman et al., 2010).
The previous studies on the role of the pco cluster in copper tolerance in S. enterica are somewhat equivocal. Mourao et al., has reported that regardless of the presence or absence of pcoA-D genes along with silA-E genes in S. enterica, they are resistant to high levels of CuSO4 in anaerobic cultures (Mourao et al., 2016; Mourao et al., 2015). Both studies suggested that the sil cassette alone could manipulate copper homeostasis inside the cell. Our results show that S. Typhimurium (SL-27), which carries sil cluster only, was able to tolerate high levels of Cu (I) as shown in the growth curves and MIC data. These data indicate that this cluster can handle copper toxicity under anaerobic environment and are in accord with earlier studies (Mourao et al., 2016; Branchu et al., 2019; Mourao et al., 2015; Arai et al., 2019). Contrary to Aria’s and Branchu’s studies, where they did not detect MIC differences between the pco mutants and parent strains (Branchu et al., 2019; Arai et al., 2019), our data clearly show growth inhibition of the pco deletion mutant when grown in copper-supplemented media, which was restored by the complemented strain. The fact that S. Typhimurium encodes a multicopper oxidase (CueO), which protects from copper toxicity through the conversion of Cu (I) to Cu (II) (Achard et al., 2010), makes it possible that the copper resistance activity of the Pco proteins might be masked by CuO. Although the deletion of silCBA genes in Branchu’s study rendered the mutant sensitive to Cu (I), it was still able to tolerate up to 6–7 mM CuSO4 under anaerobic condition, and the MIC dropped dramatically below 2 mM upon deletion of both sil and pco cluster (Branchu et al., 2019). Indeed, their data support our hypothesis that pco cluster is required for full Cu (I) resistance when both clusters present together. Selection of copper resistance genes is attributed to the extensive use of copper as growth-promoter beyond the recommended concentrations in animal and poultry diet. Consistent with the level of copper (approximately 1–5 mM) detected in chickens manure fed on copper-supplemented diet (Korish and Attia, 2020), the copper-resistant strains were able to tolerate up to 7 mM, whereas the pco deletion mutant failed to grow in the presence of 1 mM CuSO4 under anaerobic conditions. Expression of pcoABCDRSE genes from the plasmid pPA173 in E. coli lacking the Cu (I)-translocating P-type ATPase (CopA) did not protect the ΔcopA from copper toxicity but rescued the parent strain grown under the same condition (Lee et al., 2002). This study suggests that Pco proteins maintain copper homeostasis at the periplasmic level, and that CopA is required to further export excess copper across the outer membrane (Lee et al., 2002). Additionally, transformation of E. coli with plasmid pRJ1004, which contains the pco cluster, resulted in reduced accumulation of Cu (II) during initial exposure to 64Cu (II); however, ironically, the same strain accumulated more Cu (II) during the stationary phase, which may suggest that some of the Pco proteins transport Cu (II) across the inner membrane to the periplasmic space (Brown et al., 1995). In accordance with its role in E. coli, our seven-day growth curve results indicated that the pco cluster protects S. enterica from copper toxicity, but it remains unclear if copper is accumulated in the periplasmic space or is pumped across the outer membrane through PcoB. Interestingly, genome mining revealed that SL-3 and SL-4 harbor a gene that encodes a copper-transporting ATPase protein, which has homology to CopA that may act as an outer membrane efflux pump. Because we did not create a double mutation of pcoABCD and copA, we still do not know if copA is required for full copper tolerance in S. enterica. Thus, based on our data, we suggest that the Pco proteins function as a Cu (I) efflux system. In accordance with previous studies (Mourao et al., 2016; Mourao et al., 2015; Medardus et al., 2014), our data demonstrate that the loss of pcoABCD genes did not result in growth retardation in an aerobic environment, which is ascribed to the absence of the toxic cuprous form, Cu (I), in this condition. The copper resistance mechanism of the pco cluster stems from its ability to detoxify Cu (I) and extrude Cu (II) outside the bacterial cell (Zimmermann et al., 2012; Lawaree et al., 2016) as shown in our model (Figure 6).
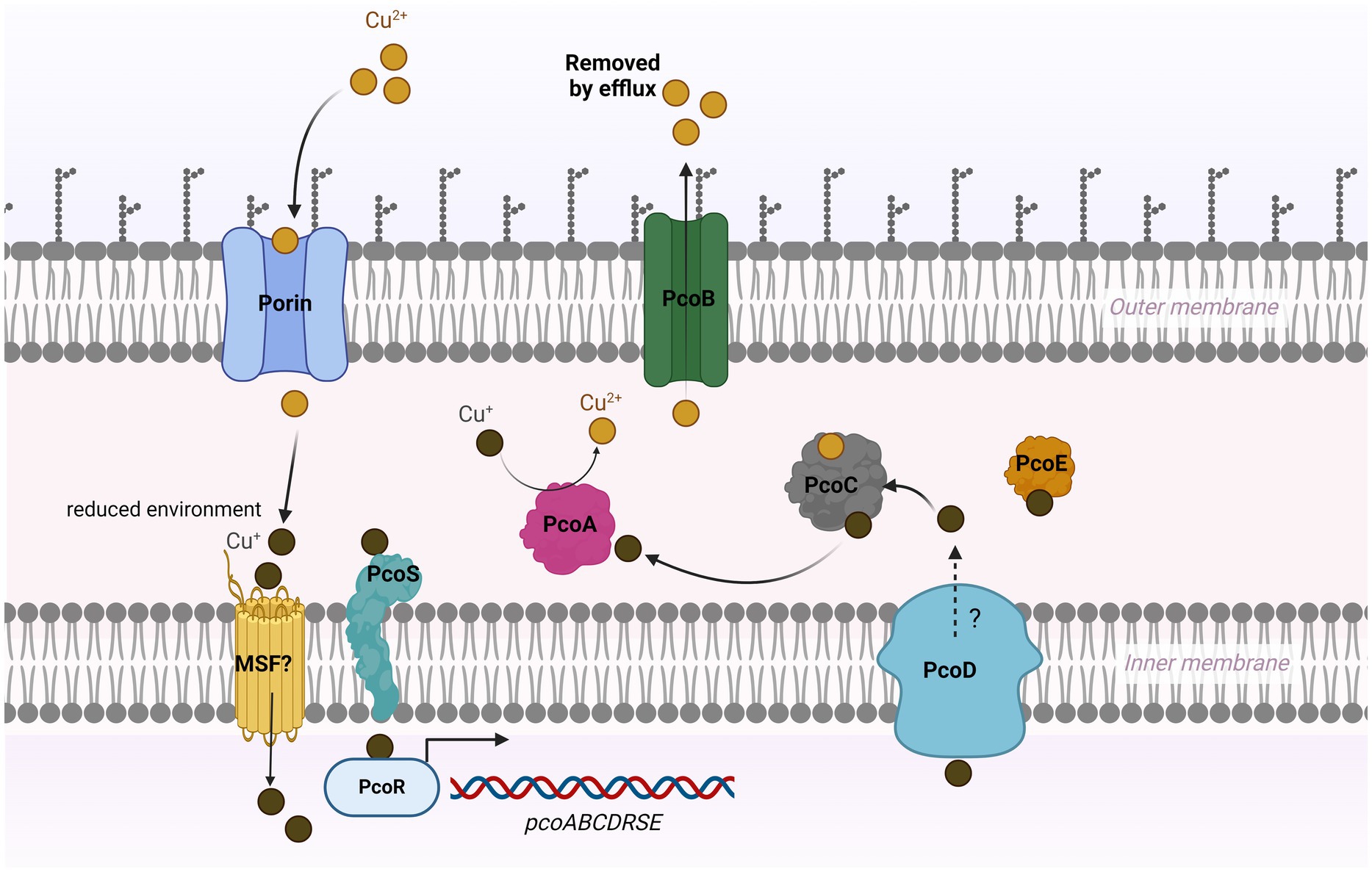
Figure 6. Model for copper resistance mediated by pco cluster in Salmonella. In a high-copper environment, copper enters bacterial cell via unknown mechanism that may involve outer membrane porin-like proteins and major facilitator super family (MSF). Cu (II) is converted to Cu (I) under reduced condition in the periplasm. The probable sensor protein PcoS may activate PcoR by phosphorylation, which then triggers the expression of the pco genes. PcoC binds both forms of Cu and transfers Cu (I) to the multicopper oxidase PcoA, where it is oxidized to the less reactive Cu (II). The Cu (I)-binding protein PcoE stores the excess (I) in the periplasm. PcoD may function as an efflux pump to extrude the intracellular Cu (I) to the periplasm. The outer membrane protein PcoB exports Cu (II) outside the cell.
The widespread usage of heavy metals potentially induces a selection pressure to allow pathogens thrive in hostile environment (Vats et al., 2022). Notably, copper has been widely used in food processing plants as a bactericide (Borkow and Gabbay, 2005), which may help explain the increased prevalence of the acquired copper-resistance genes in Enterobacteriaceae (Li et al., 2021; Mustafa et al., 2021).
Overall, this study highlights the role of the pco cluster in copper tolerance in non-typhoidal S. enterica.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics statement
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
AH: Writing – review & editing, Writing – original draft, Methodology, Data curation, Conceptualization. SH: Writing – review & editing, Methodology. DG: Methodology, Writing – review & editing. SZ: Writing – review & editing. SF: Writing – review & editing, Project administration, Funding acquisition. AK: Writing – review & editing, Supervision, Resources, Investigation, Funding acquisition, Conceptualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The research is funded by the U.S Food and Drug Administration (FDA) and The Oak Ridge Institute for Science and Education (ORISE).
Acknowledgments
We thank Kidon Sung and Jing Han for critical review of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
This manuscript reflects the views of the authors and does not necessarily reflect those of the Food and Drug Administration.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1454763/full#supplementary-material
Footnotes
References
Achard, M. E. S., Tree, J. J., Holden, J. A., Simpfendorfer, K. R., Wijburg, O. L. C., Strugnell, R. A., et al. (2010). The multi-copper-ion oxidase CueO of Salmonella enterica serovar typhimurium is required for systemic virulence. Infect. Immun. 78, 2312–2319. doi: 10.1128/IAI.01208-09
Arai, N., Sekizuka, T., Tamamura, Y., Kusumoto, M., Hinenoya, A., Yamasaki, S., et al. (2019). Salmonella Genomic Island 3 is an integrative and conjugative element and contributes to copper and arsenic tolerance of Salmonella enterica. Antimicrob. Agents Ch. 63:e00429-19. doi: 10.1128/AAC.00429-19
Beswick, P. H., Hall, G. H., Hook, A. J., Little, K., Mcbrien, D. C. H., and Lott, K. A. K. (1976). Copper toxicity-evidence for conversion of cupric to cuprous copper Invivo under anaerobic conditions. Chem. Biol. Interact. 14, 347–356. doi: 10.1016/0009-2797(76)90113-7
Billman-Jacobe, H., Liu, Y. H., Haites, R., Weaver, T., Robinson, L., Marenda, M., et al. (2018). pSTM6-275, a conjugative IncHI2 plasmid of Salmonella enterica that confers antibiotic and heavy-metal resistance under changing physiological conditions. Antimicrob Agents Ch. 62:e02357-17. doi: 10.1128/AAC.02357-17
Borkow, G., and Gabbay, J. (2005). Copper as a biocidal tool. Curr. Med. Chem. 12, 2163–2175. doi: 10.2174/0929867054637617
Branchu, P., Charity, O. J., Bawn, M., Thilliez, G., Dallman, T. J., Petrovska, L., et al. (2019). SGI-4 in monophasic typhimurium ST34 is a novel ICE that enhances resistance to copper. Front. Microbiol. 10:01118. doi: 10.3389/fmicb.2019.01118
Brown, N. L., Barrett, S. R., Camakaris, J., Lee, B. T. O., and Rouch, D. A. (1995). Molecular-genetics and transport analysis of the copper-resistance determinant (Pco) from Escherichia-Coli plasmid Prj 1004. Mol. Microbiol. 17, 1153–1166. doi: 10.1111/j.1365-2958.1995.mmi_17061153.x
EFSA Panel on Additives and Products or Substances used in Animal Feed (2016). Scientific opinion on the revision of the currently authorised maximum copper content in complete feed. EFSA J. 14:4563. doi: 10.2903/j.efsa.2016.4563
European Food Safety Authority and European Centre for Disease Prevention and Control (2019). The European Union one health 2018 Zoonoses report. EFSA J. 17:5926. doi: 10.2903/j.efsa.2019.5926
Fang, L. X., Li, X. P., Li, L., Li, S. M., Liao, X. P., Sun, J., et al. (2016). Co-spread of metal and antibiotic resistance within ST3-IncHI2 plasmids from E. coli isolates of food-producing animals. Sci Rep-Uk. 6:6. doi: 10.1038/srep25312
Giachino, A., and Waldron, K. J. (2020). Copper tolerance in bacteria requires the activation of multiple accessory pathways. Mol. Microbiol. 114, 377–390. doi: 10.1111/mmi.14522
Gudeta, D. D., and Foley, S. L. (2024). Versatile allelic replacement and self-excising integrative vectors for plasmid genome mutation and complementation. Microbiology. Spectrum 12:e03387-23. doi: 10.1128/spectrum.03387-23
Hejna, M., Gottardo, D., Baldi, A., Dell'Orto, V., Cheli, F., Zaninelli, M., et al. (2018). Review: nutritional ecology of heavy metals. Animal 12, 2156–2170. doi: 10.1017/S175173111700355X
Centers for Disease Control and Prevention. Salmonella (2023). Available at: https://www.cdc.gov/salmonella/index.html#print (Accessed October 20, 2023).
Huffman, D. L., Huyett, J., Outten, F. W., Doan, P. E., Finney, L. A., Hoffman, B. M., et al. (2002). Spectroscopy of cu(II)-PcoC and the multicopper oxidase function of PcoA, two essential components of Escherichia coli pco copper resistance operon. Biochemistry 41, 10046–10055. doi: 10.1021/bi0259960
Joana Mourao, A. R., Ribeiro, S., Peixe, L., Novais, C., and Antunes, P. (2020). Tolerance to arsenic contaminant among multidrugresistant and copper-tolerant Salmonella successful clones is associated with diverse ars operons and genetic contexts. Environ. Microbiol. 22, 2829–2842. doi: 10.1111/1462-2920.15016
Korish, M. A., and Attia, Y. A. (2020). Evaluation of heavy metal content in feed, litter, meat, meat products, liver, and Table eggs of chickens. Animals (Basel). 10:727. doi: 10.3390/ani10040727
Lawaree, E., Gillet, S., Louis, G., Tilquin, F., Le Blastier, S., Cambier, P., et al. (2016). Caulobacter crescentus intrinsic dimorphism provides a prompt bimodal response to copper stress. Nat. Microbiol. 1:16098. doi: 10.1038/nmicrobiol.2016.98
Lee, S. M., Grass, G., Rensing, C., Barrett, S. R., Yates, C. J. D., Stoyanov, J. V., et al. (2002). The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem Bioph Res Co. 295, 616–620. doi: 10.1016/S0006-291X(02)00726-X
Li, P., Nayeri, N., Gorecki, K., Becares, E. R., Wang, K., Mahato, D. R., et al. (2022). Pco B is a defense outer membrane protein that facilitates cellular uptake of copper. Protein Sci. 31:4364. doi: 10.1002/pro.4364
Li, C., Tyson, G. H., Hsu, C. H., Harrison, L., Strain, E., Tran, T. T., et al. (2021). Long-read sequencing reveals evolution and Acquisition of Antimicrobial Resistance and Virulence Genes in Salmonella enterica. Front. Microbiol. 12:777817. doi: 10.3389/fmicb.2021.777817
Lopez-Alonso, M., and Miranda, M. (2020). Copper supplementation, a challenge in cattle. Animals (Basel). 10:1890. doi: 10.3390/ani10101890
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O’Brien, S. J., et al. (2010). The global burden of Nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
McDermott, P. F., Tyson, G. H., Kabera, C., Chen, Y. S., Li, C., Folster, J. P., et al. (2016). Whole-genome sequencing for detecting antimicrobial resistance in Nontyphoidal Salmonella. Antimicrob. Agents Ch. 60, 5515–5520. doi: 10.1128/AAC.01030-16
Medardus, J. J. M., Nicol, M., Morrow, W. M., Rajala-Schultz, P. J., Kazwala, R., and Gebreyes, W. A. (2014). In-feed use of heavy metal micronutrients in US swine production systems and its role in persistence of multidrug-resistant salmonellae. Appl. Environ. Microb. 80, 2317–2325. doi: 10.1128/AEM.04283-13
Mourao, J., Marcal, S., Ramos, P., Campos, J., Machado, J., Peixe, L., et al. (2016). Tolerance to multiple metal stressors in emerging non-typhoidal MDR Salmonella serotypes: a relevant role for copper in anaerobic conditions. J. Antimicrob. Chemoth. 71, 2147–2157. doi: 10.1093/jac/dkw120
Mourao, J., Novais, C., Machado, J., Peixe, L., and Antunes, P. (2015). Metal tolerance in emerging clinically relevant multidrug-resistant Salmonella enterica serotype 4,[5],12: i: - clones circulating in EuropeJoana. Int. J. Antimicrob. Ag. 45, 610–616. doi: 10.1016/j.ijantimicag.2015.01.013
Mustafa, G. R., Zhao, K., He, X., Chen, S., Liu, S., Mustafa, A., et al. (2021). Heavy metal resistance in Salmonella typhimurium and its association with disinfectant and antibiotic resistance. Front. Microbiol. 12:702725. doi: 10.3389/fmicb.2021.702725
Osman, D., Waldron, K. J., Denton, H., Taylor, C. M., Grant, A. J., Mastroeni, P., et al. (2010). Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. Front. Microbiol. 285, 25259–25268. doi: 10.1074/jbc.M110.145953
Pontel, L. B., Scampoli, N. L., Porwollik, S., Checa, S. K., McClelland, M., and Soncini, F. C. (2014). Identification of a Salmonella ancillary copper detoxification mechanism by a comparative analysis of the genome-wide transcriptional response to copper and zinc excess. Microbiology 160, 1659–1669. doi: 10.1099/mic.0.080473-0
Punchihewage-Don, A. J., Hawkins, J., Adnan, A. M., Hashem, F., and Parveen, S. (2022). The outbreaks and prevalence of antimicrobial resistant Salmonella in poultry in the United States: an overview. Heliyon 8:e11571. doi: 10.1016/j.heliyon.2022.e11571
Qin, J., Hong, Y., Pullela, K., Morona, R., Henderson, I. R., and Totsika, M. (2022). A method for increasing electroporation competence of gram-negative clinical isolates by polymyxin B nonapeptide. Sci. Rep. 12:11629. doi: 10.1038/s41598-022-15997-8
Samanovic, M. C. D., Dennis, T., and Heran, D. (2012). Copper in microbial pathogenesis: meddling with the metal. Cell Host Microbe 11, 106–115. doi: 10.1016/j.chom.2012.01.009
Subcommittee on Poultry Nutrition CoAN (1994). Board on Agriculture, National Research Council. Nutrient Requirements of Poultry, vol. 1994. Washington, DC: National Academy Press.
Sun, D. C., Jeannot, K., Xiao, Y. H., and Knapp, C. W. (2019). Editorial: horizontal gene transfer mediated bacterial antibiotic resistance. Front. Microbiol. 10:01933. doi: 10.3389/fmicb.2019.01933
Vats, P., Kaur, U. J., and Rishi, P. (2022). Heavy metal-induced selection and proliferation of antibioti c resistance: a review. J. Appl. Microbiol. 132, 4058–4076. doi: 10.1111/jam.15492
Wagner, D., Maser, J., Moric, I., Vogt, S., Kern, W. V., and Bermudez, L. E. (2006). Elemental analysis of the Mycobacterium avium phagosome in Balb/c mouse macrophages. Biochem. Bioph. Res. Co. 344, 1346–1351. doi: 10.1016/j.bbrc.2006.04.048
Yazdankhah, S., Rudi, K., and Bernhoft, A. (2014). Zinc and copper in animal feed - development of resistance and co-resistance to antimicrobial agents in bacteria of animal origin. Microb Ecol Health D. 25:25. doi: 10.3402/mehd.v25.25862
Zimmermann, M., Udagedara, S. R., Sze, C. M., Ryan, T. M., Howlett, G. J., Xiao, Z. G., et al. (2012). PcoE-A metal sponge expressed to the periplasm of copper resistance Escherichia coli. Implication of its function role in copper resistance. J. Inorg. Biochem. 115, 186–197. doi: 10.1016/j.jinorgbio.2012.04.009
Keywords: copper resistance, pco cluster, Salmonella enterica, heavy metal resistance, pcoABCD genes
Citation: Hikal AF, Hasan S, Gudeta D, Zhao S, Foley S and Khan AA (2024) The acquired pco gene cluster in Salmonella enterica mediates resistance to copper. Front. Microbiol. 15:1454763. doi: 10.3389/fmicb.2024.1454763
Edited by:
Muhammad Kamruzzaman, Westmead Institute for Medical Research, AustraliaReviewed by:
Guoqiang Zhu, Yangzhou University, ChinaVineet Kumar, The University of Texas at Austin, United States
Copyright © 2024 Hikal, Hasan, Gudeta, Zhao, Foley and Khan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashraf A. Khan, YXNocmFmLmtoYW5AZmRhLmhocy5nb3Y=
 Ahmed F. Hikal
Ahmed F. Hikal Sameer Hasan
Sameer Hasan Dereje Gudeta
Dereje Gudeta Shaohua Zhao
Shaohua Zhao Steven Foley
Steven Foley Ashraf A. Khan
Ashraf A. Khan