
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 17 September 2024
Sec. Microbiotechnology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1442797
This article is part of the Research Topic Assessment and Application of Microbial Additives in Unconventional Feeds Fermentation View all 7 articles
 Xuerui Wan1
Xuerui Wan1 Yongjie SunKang1
Yongjie SunKang1 Yijun Chen2
Yijun Chen2 Zhao Zhang1
Zhao Zhang1 Huitian Gou1
Huitian Gou1 Yu Xue3
Yu Xue3 Chuan Wang1*
Chuan Wang1* Yaqin Wei4*
Yaqin Wei4* Yuze Yang2*
Yuze Yang2*Introduction: Endoglucanase (EG) and cellobiohydrolase (CBH) which produced by microorganisms, have been widely used in industrial applications.
Methods: In order to construct recombinant bacteria that produce high activity EG and CBH, in this study, eg (endoglucanase) and cbh (cellobiohydrolase) were cloned from the rumen microbial genome of yak and subsequently expressed independently and co-expressed within Lactococcus lactis NZ9000 (L. lactis NZ9000).
Results: The recombinant strains L. lactis NZ9000/pMG36e-usp45-cbh (L. lactis-cbh), L. lactis NZ9000/pMG36e-usp45-eg (L. lactis-eg), and L. lactis NZ9000/pMG36e-usp45-eg-usp45-cbh (L. lactis-eg-cbh) were successfully constructed and demonstrated the ability to secrete EG, CBH, and EG-CBH. The sodium carboxymethyl cellulose activity of the recombinant enzyme EG was the highest, and the regenerated amorphous cellulose (RAC) was the specific substrate of the recombinant enzyme CBH, and EG-CBH. The optimum reaction temperature of the recombinant enzyme CBH was 60°C, while the recombinant enzymes EG and EG-CBH were tolerant to higher temperatures (80°C). The optimum reaction pH of EG, CBH, and EG-CBH was 6.0. Mn2+, Fe2+, Cu2+, and Co2+ could promote the activity of CBH. Similarly, Fe2+, Ba2+, and higher concentrations of Ca2+, Cu2+, and Co2+ could promote the activity of EG-CBH. The addition of engineered strains to whole-plant corn silage improved the nutritional quality of the feed, with the lowest pH, acid detergent fiber (ADF), and neutral detergent fiber (NDF) contents observed in silage from the L. lactis-eg group (p < 0.05), and the lowest ammonia nitrogen (NH3-N), and highest lactic acid (LA) and crude protein (CP) contents in silage from the L. lactis-eg + L. lactis-cbh group (p < 0.05), while the silage quality in the L. lactis-cbh group was not satisfactory.
Discussion: Consequently, the recombinant strains L. lactis-cbh, L. lactis-eg, and L. lactis-eg-cbh were successfully constructed, which could successfully expressed EG, CBH, and EG-CBH. L. lactis-eg promoted silage fermentation by degrading cellulose to produce sugar, enabling the secretory expression of EG, CBH, and EG-CBH for potential industrial applications in cellulose degradation.
Cellulase is a complex enzyme system involving a variety of hydrolases. Cellobiohydrolase (exoglucanase), endo-glucanase, and β-glucosidase (cellobiase) work in a coordinated manner to degrade cellulose into small molecules such as glucose that can be absorbed and utilized by the animal intestine (Segato et al., 2014). Cellulases are naturally found in lower animals, insects, bacteria, fungi, plants, and the large intestine and rumen of ruminants (Himmel et al., 2010). The structure and enzymatic activity of cellulases from different sources vary greatly. To construct engineered bacteria with high enzymatic activity, it is necessary first to screen to obtain select genes that express enzyme with excellent activities. The rumen is inhabited by a complex and diverse microbial community and is a natural system that is highly efficient in the conversion and utilization of cellulosic substances. The microbial community metabolizes and efficiently degrades grazing to provide the host with energy and nutrients for survival (Huws et al., 2018), making the rumen a habitat for highly active lignocellulose hydrolases (An et al., 2005; Wei et al., 2016). Sequenced and analyzed the DNA from bovine rumen microorganisms and identified 27,755 putative carbohydrate-active genes, with 57% being enzymatically active against cellulose substrates (Hess et al., 2011). Tiwari et al. (2021) investigations had revealed that over 99% of prokaryotic microorganisms remained elusive through existing pure culture isolation techniques. Directly cloning genes sequences from metagenome of yak rumen microorganisms would be a fast and potent avenue to access cellulase genes.
The cellulase-expressing systems mainly include bacteria, yeast, and plants; each with its own advantages and disadvantages (Garvey et al., 2013). The expression of multiple protein enzymes with complementary activities, preferably in a single strain, is essential for the development of engineered strains that degrade complex substrates (Tarraran and Mazzoli, 2018). The genes expressing endoglucanase and cellobiohydrolase were co-expressed with higher cellulose degradation and biodegradation activities in Trichoderma reesei (T. reevi) than those obtained from the expression of a single enzyme gene (Wang et al., 2014). The egl II (endoglucanase II) and cbh II (cellobiohydrolase II) genes from T. reevi were co-expressed in Pichia pastoris and had the same catalytic properties as the enzymes produced separately (de Amorim Araújo et al., 2015). Lactococcus lactis (L. lactis), a microorganism with probiotic properties, is usually considered safe for expressing heterologous genes (Zegers et al., 1999; Gaya et al., 2020). Some cellulase genes have been successfully expressed in L. lactis to obtain highly active enzymes, which had a widely application (Liu et al., 2005; Ozkose et al., 2009; Zhou et al., 2015). However, the expression level and activity of enzyme proteins in engineered bacteria are still the bottleneck limiting the industrial application of cellulase. Therefore, more attempts are needed, such as selecting more potential enzyme genes when constructing engineered bacteria and co-expressing enzyme genes from different sources in the same expression system.
Silage used lactic acid bacteria to ferment carbohydrates in raw materials to produce organic acids, reduce the pH value of the feed system, inhibit the propagation of harmful microorganisms in the feed, and thus prevent feed deterioration (Fabiszewska et al., 2019). Adding lactic acid bacteria to whole-plant corn silage could enhance the fermentation efficiency and improve the silage quality (Guo et al., 2019). When fermentation substrate was insufficient, fermentation efficiency would be reduced. Adding cellulase could degrade lignocellulose in silage raw materials into monosaccharides, providing more substrates to promote lactic acid bacteria fermentation (Liu et al., 2024; Zhao et al., 2024). Therefore, lactic acid bacteria and cellulase are the two most commonly used silage additives. A large number of studies have focused on the addition of lactic acid bacteria and cellulase in silage alone or in combination to improve the quality of silage (Du et al., 2023, Yu et al., 2024). However, the cost of cellulase hampers its extensive use as feed supplement. Recombinant lactic acid bacteria expressing cellulase can be applied to silage to achieve the effect of adding lactic acid bacteria and cellulase together. Moreover, it can secrete cellulase continuously with the reproduction of recombinant lactic acid bacteria, and exert the synergistic effect of lactic acid bacteria and cellulase. The effects of transgenically engineered L. lactis strains on enhancing the conversion of lignocellulose to sugars were better than the combination of cellulase and wild-type L. lactis subsp (Liu et al., 2019). At present, there are few reports on the application of recombinant lactic acid bacteria expressing cellulase in silage fermentation (Rossi et al., 2001; Guo et al., 2019). It is likely to be a hotspot of research on silage additives in the future.
The microbial community in yak rumen produced a large number of highly active lignocellulose hydrolases (An et al., 2005; Wei et al., 2016). In previous studies, we expressed β-glucosidase (bg) in Lactococcus lactis NZ9000 (L. lactis NZ9000) from the yak rumen (Wang et al., 2023). Cellulase has been widely used in many industrial applications. To develop new strategies to minimize enzyme production costs, in this study, cbh and eg genes from metagenome of rumen microorganisms from yak were cloned in the lactobacillus food-grade expression vector. Ribosome binding sites (RBS) were introduced before each gene to prepare a secretion-type expression vector and transformed into L. lactis NZ9000 through electroporation to obtain the recombinant bacteria co-expressing cellobiohydrolase and endoglucanase. The recombinant enzymatic activity and optimal growth conditions of the recombinant strains were analyzed. Subsequently, the recombinant strains were added to whole-plant corn for laboratory silage, and those roles in the fermentation of silage were assessed. This study laid a foundation for the application of lignocellulose hydrolases and recombinant LAB in feed preparation.
Table 1 is the bacterial strains and plasmids for this study. Luria-Bertani (LB) medium (both agar and broth) was used to culture Escherichia coli DH5α at 37°C, and GM17 medium was used to culture L. lactis NZ9000 grown in at 30°C. Erythromycin (0.1 μg/mL; Solarbio Science & Technology Co., Ltd., Beijing, China) was utilized used to screen the positive recombinants of L. lactis NZ9000 with the pMG36e backbone.
The primers designed based on NCBI eg and cbh sequences are provided in Table 2. Twenty 5–6 year-old gibbed and male Tianzhu yaks were used to extract the whole genome from their yak rumen fluid using the Cetyltrimethylammonium Bromide (CTAB) methods. Each yak which weighed about 250 kg was raised on wild grass at Wushaoling Pasture in the Tibetan Autonomous County of Tianzhu (Gansu Province, China) (Popova et al., 2010; Wang et al., 2023). cbh-F/R and eg-F/R were used as primers to amplify the cbh and eg by PCR. PCR was conducted in a total volume of 50 μL, which contained the following items: EasyPfu Polymerase buffer, 2 μL (TransGen Biotech Co., Ltd. Beijing, China); 10 × EasyPfu buffer 5 μL; 2.5 mmol/L dNTPs, 5 μL; the whole genome of yak rumen, 2 μL; nuclease-free water, 26 μL; each PCR primer 2.5 μL. The operation was carried out as follows: pre-denaturation at 95°C for 5 min and 30 cycles of 95°C for 30 s, 56.5°C for 30 s, 72°C for 90 s, and 72°C extension for 10 min (Wang et al., 2023). Easy Pure PCR Purification Kit (TransGen Biotech Co., Ltd., Beijing, China) was used to purify the PCR products of cbh and eg.
The purified eg and cbh PCR productswere digested with SacI/Xba Iand Xba I/Pst I (TakaRa Biotechnology Co., Ltd. Dalian, China), respectively, which was then linked with the digested amplicon using T4-DNA ligase (TakaRa Biotechnology Co., Ltd. Dalian, China) to produce the recombinant vectors pMG36e-usp45-eg and pMG36e-usp45-cbh. Subsequently, the purified eg PCR products and pMG36e-usp45-cbh were, respectively, digested with SacIand Xba I, and the digestion product of eg and pMG36e-usp45-cbh were linked using T4-DNA ligase to produce the recombinant vectors pMG36e-usp45-eg-usp45-cbh which were the introduced into E. coli DH5α. The positive recombinants of E. coli DH5α/pMG36e-usp45-eg, E.coli DH5α/pMG36e-usp45-cbh, and E. coli DH5α/pMG36e-usp45-eg-usp45-cbh were verified by PCR and double enzymes digestion (Wang et al., 2023). After the verification, the correct recombinant plasmids were selected for sequencing, the results of which were compared with NCBI Basic Local Alignment Search Tool (BLAST).
A Gene Pulser Xcell (Times-Legend Bioscientific Co., Ltd.) was used to transform the verified correct recombinant plasmids into the L. lactis NZ9000 via electroporation under the following conditions: at a pulse voltage of 2,200 V, pulse resistance of 100 Ω, pulse capacitance of 25 μF, and a pulse time of 3 s. The verified correct recombinant plasmids included pMG36e-usp45-cbh, pMG36e-usp45-eg, and pMG36e-usp45-eg-usp45-cbh. 0.1 μg/mL erythromycin was used to select the positive transformants of L. lactis/pMG36e-usp45-eg (L. lactis-eg), L. lactis/pMG36e-usp45-cbh (L. lactis-cbh), L. lactis/pMG36e-usp45-eg-usp45-cbh (L. lactis-eg-cbh), which were then verified by PCR (Wang et al., 2023).
The inoculum of L. lactis-eg, L. lactis-cbh, and L. lactis-eg-cbh were added to the sample in GM17 plates whichcontained 1 g/L of CMC-Na and 0.1 μg/mL of erythromycin (pH 7.0) to culture for 48 h at 30°C. After that, they were exposed to 1 g/L Congo red solution. Then the plates were incubated for 30 min and washed with 1 mol/L NaCl to discover the clear zones against a red background which developed via hydrolysis of CMC-Na (Wang et al., 2023).
The engineered strains were incubated at 30°C for 36 h to collect the supernatant. The precipitation method (An et al., 2005; Niu et al., 2018) was used to purify recombinant proteins from the supernatant. By sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), the molecular masses of the enzymes of recombinant were estimated. By measuring the amount of reduced sugars released by recombinant enzymes using the 3,5-dinitrosalicylic acid method, the activity of all enzymes was determined with Glucose as a standard (Geem et al., 2021). Regenerated amorphous cellulose (RAC), CMC-Na, dried cotton, microcrystalline cellulose (MCC), filter paper, and acid were used as substrates to test the cellulase activity of individual and recombinant fusion enzymes, and recombinant proteins inactivated by boiling inactivation were used as blank controls (Zhao et al., 2021).
To measure the optimum reaction temperature for the recombinant enzyme, the supernatant of the recombinant enzyme was used to react with the optimum substrate at different temperatures (30–90°C) for 30 min. To measure the optimum pH for recombinase activity, the recombinase supernatant in several buffers of different pH (pH 3.0–9.0) was sued to react with the optimum substrate at the optimum temperature for 30 min to determine (Wang et al., 2023).
Regenerated amorphous cellulose at various concentrations (0.1–10 nM) was used as the substrate to determine the activity of recombinant enzymes at 80°C and pH 6.0. The data were fitted in GraphPad Prism 8.4.2 with Michaelis–Menten equation fitting and Lineweaver-Burk plots (double-reciprocal plots) to estimate the enzyme kinetic parameters, such as maximum enzymatic activity for substrate conversion (Vmax), Michaelis–Menten constant (Km), turnover number (Kcat), and Kcat/Km.
To ensure the final concentrations of the solutions reached 1 mmol/mL and 5 mmol/mL, 1% RAC was used as substrate at 80°C and pH 6.0, and K+, Mg2+, such elements as Mn2+, Zn2+, Hg2+, Fe2+, Co2+, Ca2+, and Cu2+ were added to the reconstituted enzyme supernatant and optimal substrate for the reaction. To ensure the final concentrations of the solutions were 1 and 10% at optimal temperature and pH for 30 min, Tween20 and Tween80 were added to the reconstituted enzyme supernatant and optimal substrate for the reaction. Different concentrations of ions and solutions affected the activity of the recombinant enzyme. The effects were measured and compared to the solutions without metal ions and chemical reagents used as blank controls (Wang et al., 2023).
Recombinant L. lactis NZ9000 were added to whole-plant corn silage in polyethylene bags in the laboratory. The experiment included six treatments, namely, L. lactis-eg, L. lactis-cbh, L. lactis-eg-cbh, L. lactis-eg + L. lactis-cbh, L. lactis NZ9000, and GM17 liquid medium as the control. Recombinant L. lactis NZ9000 was cultured in GM17 liquid medium to the logarithmic growth stage, after which the concentration of the culture was adjusted to 1 ×109 CFU/mL and the bacteria were added to whole-plant corn with a 70% water content and 2–4 cm at a ratio of 5 mL/kg, and mixed evenly. The evenly mixed silage raw materials were then packed into polyethylene bags (500 g/bag) and compacted, after which the silage bags were sealed using a vacuum sealer. Each treatment was repeated three times. The sealed silage bags were placed in a constant temperature environment (20°C) for fermentation. On day 30, samples were taken from the silage and assessed by measuring their pH values, the levels of water soluble carbohydrate (WSC), neutral detergent fiber (NDF), acid detergent fiber (ADF), crude protein (CP), lactic acid (LA), acetic acid (AA), and ammonia nitrogen (NH3-N) (Wan et al., 2016).
To assess how varied temperatures, pH, ions, and solution concentrations affected the enzymatic activity of the recombinant enzymes, GraphPad Prism 9.5 two-way ANOVA was used to analyze the data. The means were compared using Tukey’s test for significance at p < 0.05. To evaluate the influence of recombinant L. lactis NZ9000 on the quality of whole-plant corn silage, the data were statistically analyzed using SPSS 18.0, and the mean values were compared by Duncan method for significance at p < 0.05.
Three recombinant plasmid maps designed for this study are shown in Figure 1. The recombinant plasmid contained the ribosomal binding site (RBS), the signal peptide usp45, and the promoter P32. pMG36e-usp45-eg, pMG36e-usp45-cbh, and pMG36e-usp45-eg-usp45-cbh were successfully constructed and verified via double digestion. Sequencing showed the size of eg and cbh were 1,500 and 1,599 bp (Figure 1D). We performed BLAST alignment of the obtained eg and cbh gene sequences in NCBI and then selected sequences with high similarity to perform multiple sequence alignment and genetic evolution by the MegAlign software in DNAstar. The results showed a 100% homology of the eg gene with the eg of Bacillus subtilis (GenBank Accession No. AB695293.1) and a 100% homology of the cbh gene with the cbh of Aspergillus fumigatus (GenBank Accession No. XM_745951.1) (Figure 2).
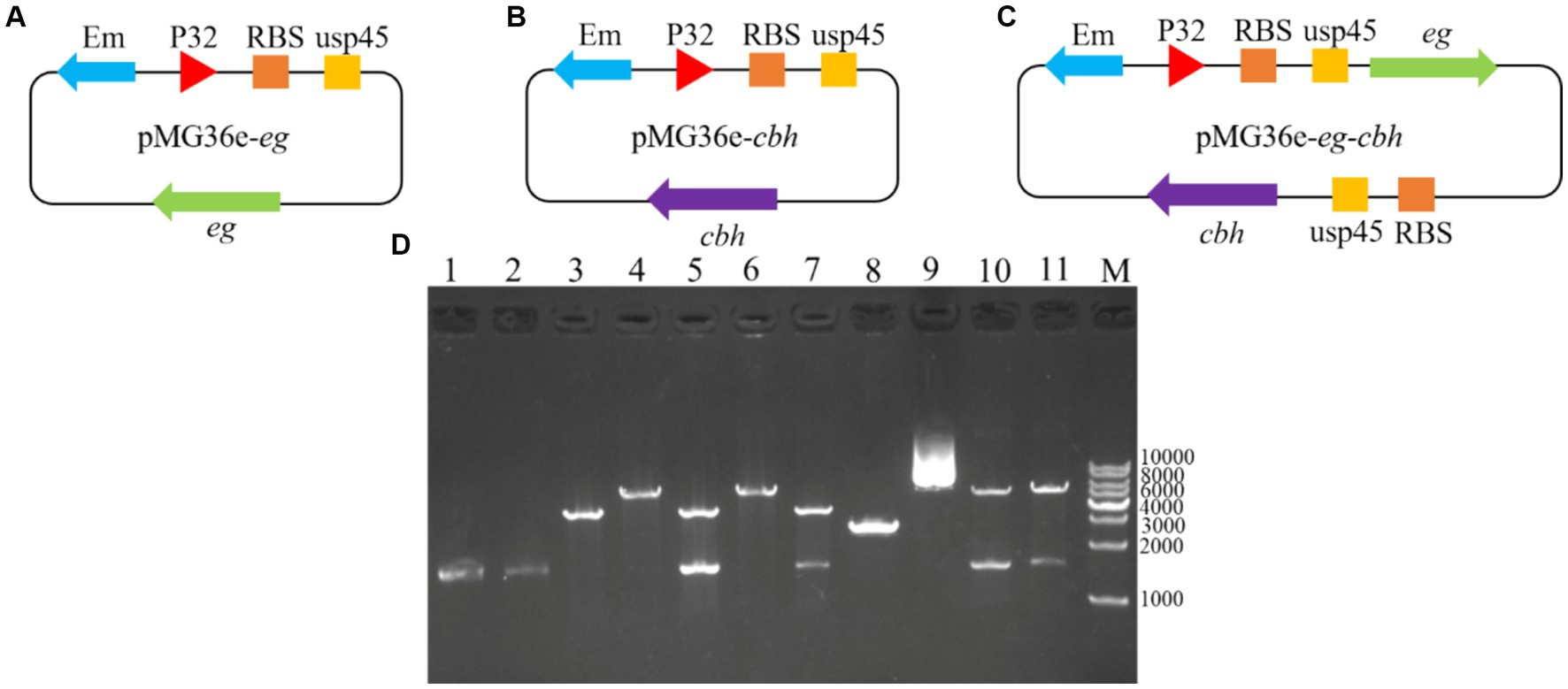
Figure 1. Recombinant plasmids construction strategy and double enzyme digestion verification. (A) Recombinant plasmid construction strategy of pMG36e-usp45-eg; (B) Recombinant plasmid construction strategy of pMG36e-usp45-cbh; (C) Recombinant plasmid construction strategy of pMG36e-usp45-eg-usp45-cbh; (D) lane 1: Colony PCR of eg; lane 2: Colony PCR of cbh; lane 3: Single enzyme digestion of empty vector pMG36e; lane 4: Single enzyme digestion of pMG36e-usp45-eg; lane 5: Double enzyme digestion of pMG36e-usp45-eg use SacIand XbaI; lane 6: Single enzyme digestion of pMG36e-usp45-cbh; lane 7: Double enzyme digestion of pMG36e-usp45-eg use Xba I and Pst I; lane 8: Colony PCR of eg-cbh; lane 9: Single enzyme digestion of pMG36e-usp45-eg-usp45-cbh; lane 10: Double enzyme digestion of pMG36e-usp45-eg-usp45-cbh use SacIand XbaI; lane 11: Double enzyme digestion of pMG36e-usp45-eg-usp45-cbh use Xba I and Pst I; M: DL10000 DNA Marker.
Lactococcus lactis-eg and L. lactis-cbh could secretory express EG and CBH proteins, respectively. The molecular weights of EG and CBH were 54.9 and 56.4 kDa by analysising use Quantity One 1-D software (version 4.6.2). In addition, via SDS-PAGE showed that L. lactis-eg-cbh had two bands of similar size between 55 and 70 kDa, indicating that L. lactis-eg-cbh could co-express and secrete recombinant protein EG and CBH (Figure 3).
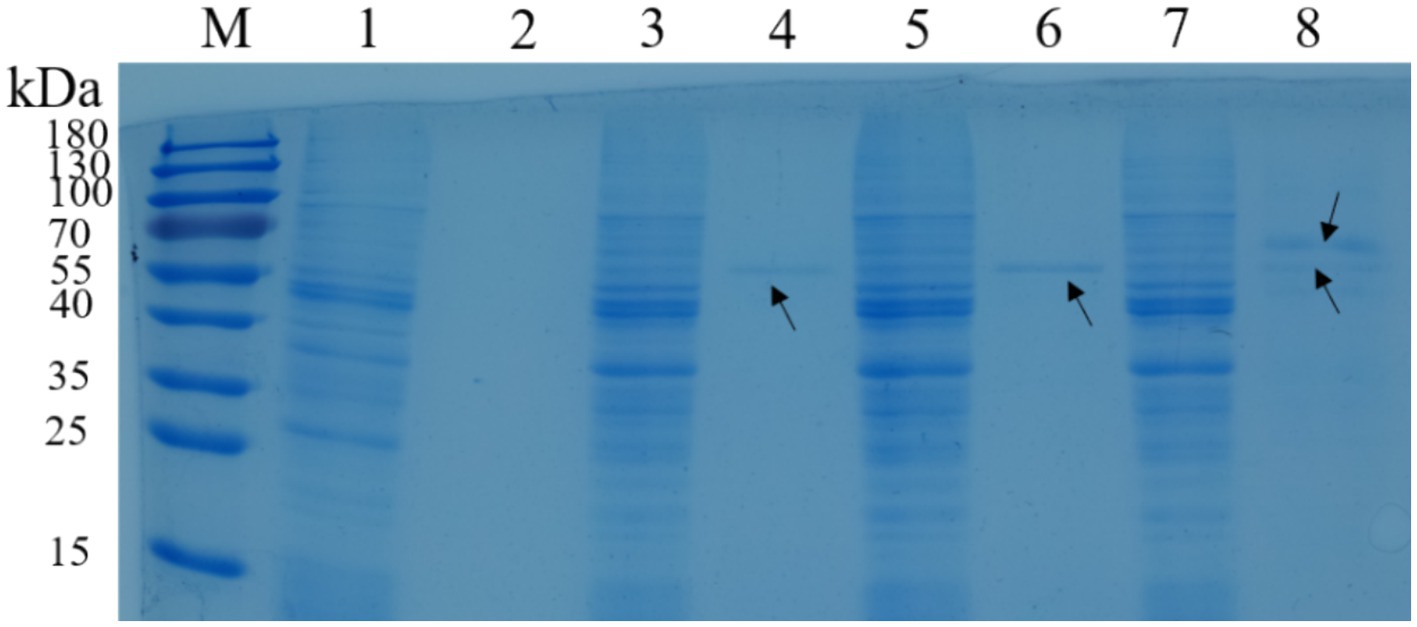
Figure 3. SDS-PAGE of recombinant strains. M: protein molecular weight marker; lane 1: intracellular protein of Lactococcus lactis NZ9000; lane 2: supernatant of L. lactis NZ9000 via TCA/acetone precipitation; lane 3: intracellular protein of L. lactis-eg; lane 4: supernatant of L. lactis-eg via TCA/acetone precipitation; lane 5: intracellular protein of L. lactis-cbh; lane 6: supernatant of L. lactis-cbh via TCA/acetone precipitation; lane 7: intracellular protein of L. lactis-eg-cbh; and lane 8: supernatant of L. lactis-eg-cbh via TCA/acetone precipitation.
As shown in Figure 4, L. lactis-eg had significant hydrolysis circles around them. Conversely, L. lactis-cbh did not have hydrolysis circles around them. The supernatant and bacterial liquid of L. lactis-eg-cbh showed a slight hydrolysis circle, but the supernatant showed an obvious hydrolysis circle after concentration.
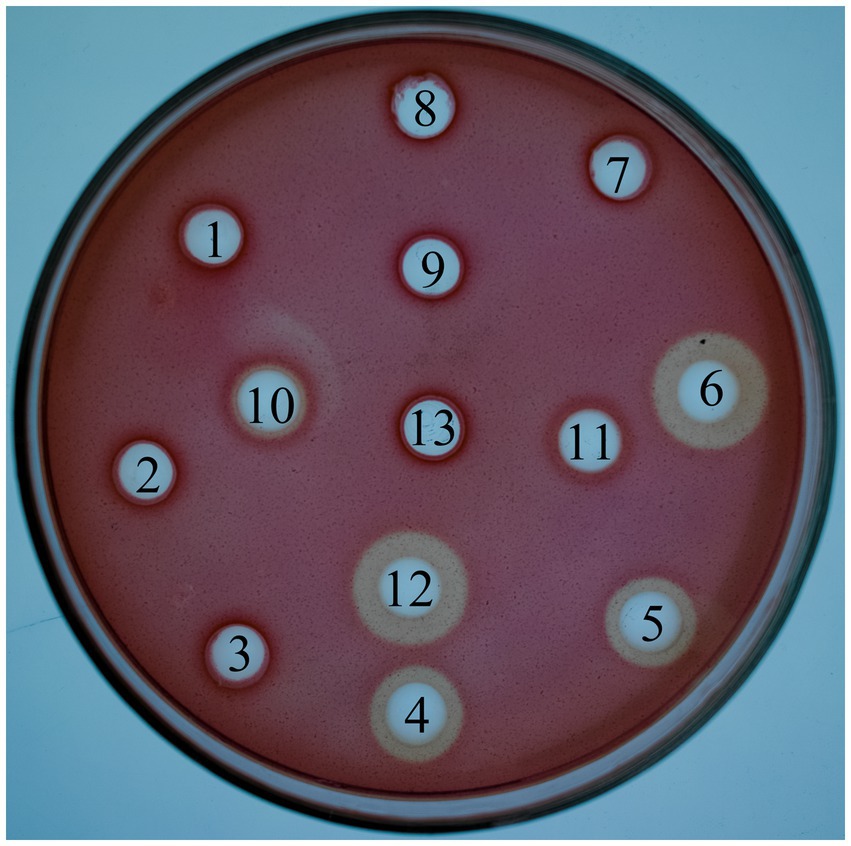
Figure 4. Congo red staining with CMC-Na. 1: Supernatant of Lactococcus lactis NZ9000; 2: The bacterial liquid of L. lactis NZ9000; 3: Supernatant of L. lactis NZ9000 via TCA/acetone precipitation; 4: Supernatant of L. lactis-eg; 5: The bacterial liquid of L. lactis-eg; 6: Supernatant of L. lactis-eg via TCA/acetone precipitation; 7: Supernatant of L. lactis-cbh; 8: The bacterial liquid of L. lactis-cbh; 9: Supernatant of L. lactis-cbh via TCA/acetone precipitation; 10: Supernatant of L. lactis-eg-cbh; 11: The bacterial liquid of L. lactis-eg-cbh; 12: Supernatant of L. lactis-eg-cbh via TCA/acetone precipitation; 13: Blank.
The crude enzyme solution of L. lactis-eg, L. lactis-cbh, and L. lactis-eg-cbh exhibited different substrate specificities. Among them, the activity of recombinant enzyme EG was the highest for CMC-Na, at 13.42 U/mL and the specific substrate of recombinant enzyme CBH was RAC, with the activity being 8.07 U/mL. The co-expression product of EG and CBH had the highest specificity for RAC, with the enzyme activity being 10.73 U/mL. The hydrolytic activity of the co-expression products of EG and CBH on RAC, cotton wool, MCC, filter paper, and CMC-Na was significantly higher than that of CBH expressed singly. Compared with the singly expressed EG, the co-expressed products of EG and CBH had significantly improved the hydrolysis activity on cotton wool but not on RAC and MCC, while the hydrolysis activity on filter paper and CMC-Na was significantly decreased (Figure 5A).
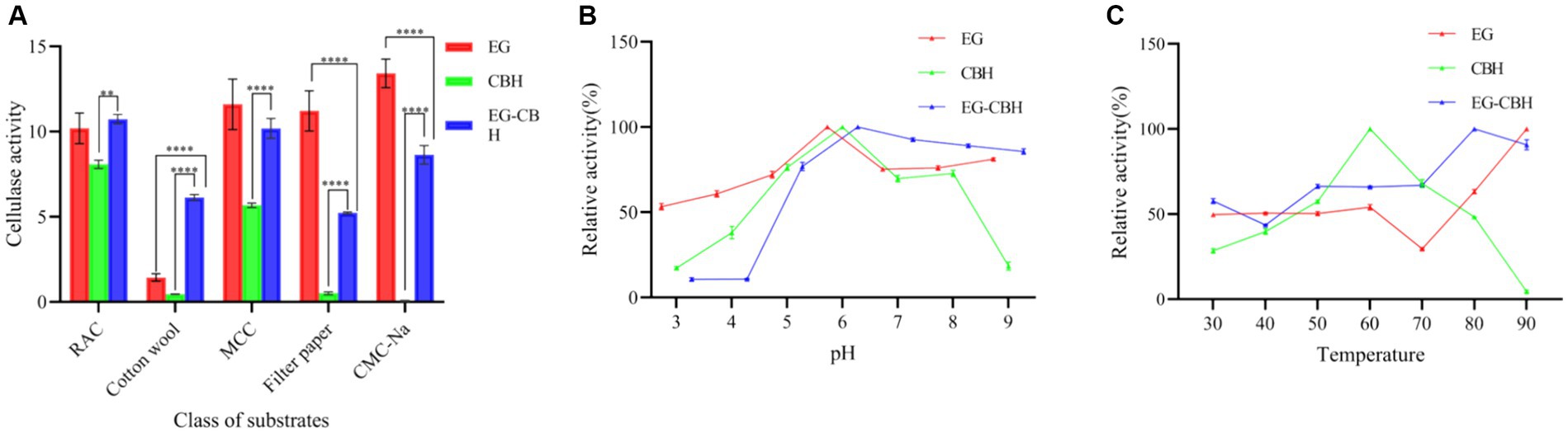
Figure 5. Effect of substrate, pH, and temperature on recombinant enzymes activity. (A) Substrate specificity of recombinant enzymes; (B) Effect of pH on recombinant enzymes activity; (C) Effect of temperature on recombinant enzymes activity; * represents the difference within experimental groups, **p < 0.01, ****p < 0.0001; Error bars in graphs represent standard deviation of the data.
The activities of recombinant enzymes EG, CBH, and EG-CBH increased with the increase of pH in the range of 3.0–6.0. The optimal pH range of enzyme activity of EG, CBH, and EG-CBH was 5.0–8.0, and the relative enzyme activity was more than 70%, especially at pH 6.0 (Figure 5B).
Within the range of 30–60°C of reaction temperature, the activity of EG, CBH, and EG-CBH enzymes increased with the temperature, and the activity of CBH enzymes peaked when the temperature was 60°C. When the temperature was higher than 60°C, the enzyme activity of EG decreased briefly at 70°C and then continued to rise. Interestingly, the co-expression product of EG and CBH showed tolerance for higher temperatures, and the enzyme activity of EG-CBH reached its peak at 80°C (Figure 5C).
At the optimal temperature and pH, RAC was used as the substrate to assay the kinetic parameters of the recombinant enzymes. Compared with Vmax, Km, Kcat, and Kcat/Km values of the recombinant enzymes in bacterial supernatant of L. lactis-eg, L. lactis-cbh, and L. lactis-eg-cbh, the results of the assessment were shown in Table 3. The bacterial supernatant of L. lactis-eg was characterized as having the highest Vmax (1,601 μmol∙min−1 mg−1), Kcat (73.09 s−1), Kcat/Km (0.44 L∙s−1∙μmol−1), and the lowest Km (167.6 μmol∙L−1), but bacterial supernatant of L. lactis-cbh and L. lactis-eg-cbh had very similar Vmax (1,491 and 1,491 μmol∙min−1 mg−1), Km (174.6 and 174.6 μmol∙L−1), Kcat (66.04 and 67.63 s−1), and Kcat/Km (0.38 and 0.39 L∙s−1∙μmol−1).
Mn2+, at 1 mmol/L was found to promote the enzyme activity of CBH. When the concentration of Mn2+ increased, it significantly promoted the enzyme activity of EG-CBH, while Mn2+ had no effect on the activity of EG. The low concentration of Zn2+ significantly inhibited the enzyme activity of EG-CBH, which became zero with the increase in ion concentration, while Zn2+ had no effect on the activity of a single recombinant enzyme. Fe2+ promoted the activity of the three recombinants, and the high concentration of Fe2+ increased the activity of CBH and EG-CBH. Ba2+ had no effect on the activity of any single recombinant enzyme and significantly promoted the activity of EG-CBH. Ca2+ at 1 mmol/L promoted the activity of EG and inhibited the activity of EG-CBH. When the Ca2+ ion concentration increased, the enzyme activity of EG-CBH was promoted. At low concentration, Cu2+ only promoted CBH. When the concentration reached 5 mmol/L, it inhibited the EG activity and significantly promoted the CBH and EG-CBH activities.
With the increase in ion concentration, Hg2+ significantly inhibited the activity of the three recombinant enzymes. When the concentration of Co2+ was 1 mmol/L, it could only promote CBH activity. When the concentration reached 5 mmol/L, it significantly promoted CBH and EG-CBH activities but had no effect on EG activity. At 1 mmol/L, K+ inhibited the enzyme activity of EG-CBH, while 5 mmol/L, K+ inhibited the enzyme activity of CBH and EG-CBH. At 1 mmol/L, Mg2+ inhibited the enzyme activity of CBH, and EG-CBH, and when the concentration increased, it promoted the enzyme activity of EG while inhibiting the activity of CBH, EG-CBH; in addition, the inhibition of EG-CBH was weakened. The inhibitory effect of EDTA on the activities of three enzymes increased with the increase in concentration. Tween had different effects on EG-CBH at different concentrations. At 1% concentration, Tween promoted the activity of the EG-CBH enzyme and produced opposite results with the increase in concentration (Figures 6A,B).
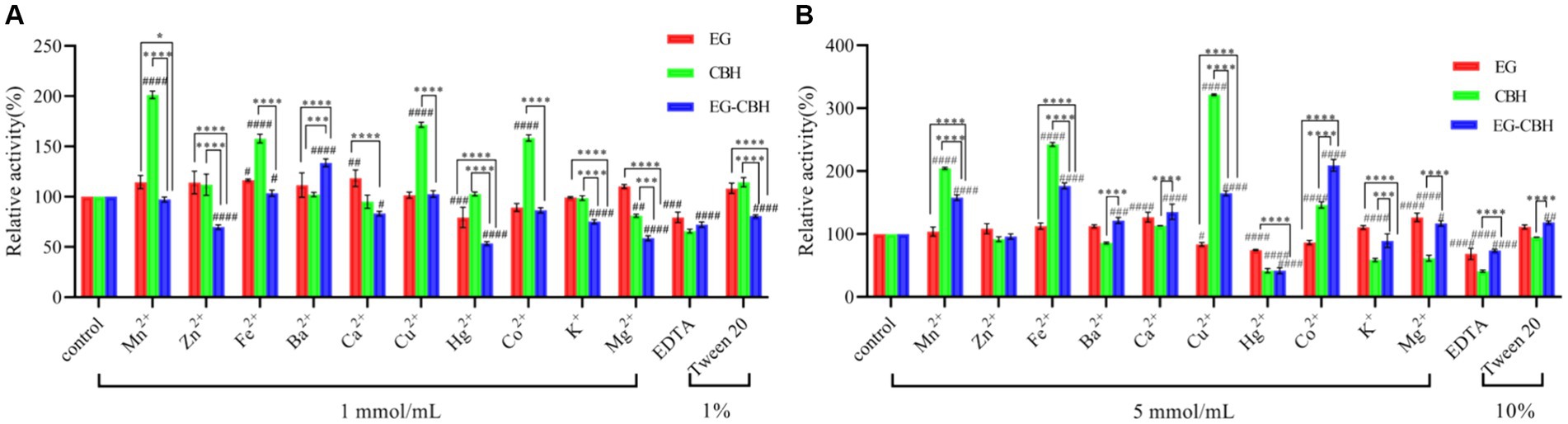
Figure 6. Effect of ions and chemical reagents on recombinant enzymes. (A) Effect of ions at and 1% solution on enzyme activity; (B) Effect of ions at 5 mmol/mL and 10% solution on enzyme activity (#represents the difference between experimental groups and control group, #0.01 < p < 0.05, ##p < 0.01, ###p < 0.001, ####p < 0.0001; * represents the difference within experimental groups, *0.01 < p < 0.05, ***p < 0.001, ****p < 0.0001; Error bars in graphs represent standard deviation of the data).
Recombinant strains were added to the whole corn, and the quality of the silage were assessed on day 30 after the addition; the results of the assessment are shown in Table 4. The pH of silage in the L. lactis-eg group was found to be the lowest, followed by the L. lactis-eg + L. lactis-eg-cbh group. The pH values were significantly lower in these two groups compared with the other groups (p < 0.05), although there was no significant difference between the two groups. The LA content in the L. lactis-eg + L. lactis-cbh group was highest, followed by the L. lactis-eg group, which were both significantly higher than the contents in other groups (p < 0.05). The AA contents of the silage in all recombinant L. lactis NZ9000 groups were significantly higher than those in the L. lactis NZ9000 and Control groups (p < 0.05), with the highest AA content observed in the L. lactis-eg-cb group, significantly higher than that in the other three recombinant L. lactis NZ9000 groups. The LA/AA ratio in the silage was highest in the L. lactis-eg + L. lactis-cbh group, followed by the L. lactis-eg group, with the L. lactis-eg-cbh group being the lowest. The NH3-N content of silage in the L. lactis-eg + L. lactis-cbh group was significantly lower than that in other groups (p < 0.05), while the WSC content was highest in the L. lactis-eg group, followed by the L. lactis-eg + L. lactis-cbh and L. lactis-eg-cbh groups, significantly higher than that in the other groups (p < 0.05). The contents of ADF and NDF in the L. lactis-eg group were the lowest, followed by the L. lactis-eg + L. lactis-cbh group, differing significantly from those in other groups (p < 0.05). The CP content of silage in the L. lactis-eg + L. lactis-cbh group was found to be highest, followed by the L. lactis-eg group, both of which were significantly higher than that observed in the other groups (p < 0.05).
Cellulose was an important carbon source for microorganisms in yak rumen, and Glycoside hydrolases (GHs) were the most abundant class of carbohydrate-active enzymes (Jiang et al., 2022). Therefore, rumen microorganisms is one of the main sources of cellulase. High throughput 16S rRNA sequencing and metagenomic analysis have indicated that the rumen microbiome of plateau yaks has more enzymes genes involved in cellulase and hemicellulase than that of cattle (Dashtban et al., 2010; Zhao et al., 2022). In addition, cellulase produced by rumen fungi is one of the most active fibrolytic enzymes (Selinger et al., 1996). Therefore, cbh and eg genes from uncultured rumen microorganisms of yak were cloned in pMG36e to construct expression vectors pMG36e-usp45-eg, pMG36e-usp45-cbh, and pMG36e-usp45-eg-usp45-cbh. Engineered strains L. lactis-eg, L. lactis-cbh, and L. lactis-eg-cbh were constructed successfully, and cellobiohydrolase and endoglucanase were successfully independently and co-expressed. pMG36e was constructed based on the transcriptional and translational signals of the protease gene of the L. lactis milk lipid subspecies; it was a constitutive expression vector for the inserted gene in L. lactis NZ9000 was a manually constructed recombinant host with the nisin-controlled gene expression system, which has advantages over other expression systems for expressing recombinant proteins and has been used widely for expressing a variety of proteins from other bacteria (Liu et al., 2022). Compared with E. coli, L. lactis has a single outer membrane and is able to secrete heterologous proteins directly into the extracellular environment; moreover, the total protein enzyme expressed in L. lactis has been shown to be higher than in E. coli (Liu et al., 2017). The ability of L. lactis to efficiently express and secrete proteins, which is closed related to the selection of effective transcriptional promoters and secretory signal peptides (Stephenson et al., 2011). Usp45 as the main signal peptide of L. lactis, which replaced the natural Nuc signal peptide, the efficiency of heterologous protein secretion has been greatly increased (Le Loi et al., 2001).
In addition to increasing the number of cellulolytic enzymes, in the future, the main focus in the construction of recombinant cellulolytic bacteria should be on improving the synergistic effect of designer cellulase system (Tarraran and Mazzoli, 2018). The complete degradation of rice straw was achieved by Bacillus subtilis SU40 β-glucosidase and B. subtilis MG7 endoglucosidase (Asha et al., 2015). Three different cellulase genes (bg, eg, and cbh) were co-expressed in Saccharomyces cerevisiae, and the enzyme activity on the filter paper was higher than each individual enzyme (Liu et al., 2018). The overexpression of NfBgl3A (thermophilic β-glucosidase from Neosartorya fischeri) by Trichoderma reesei (T. reicheni) significantly increased glucose yield and also improved the cellulose degradation performance of major cellulase components, including fibrinous disaccharide hydrolase and endoglucanase, compared with the addition of NfBgl3A to T. reicheni in vitro (Xue et al., 2016). β-glucosidase (bglZ) and endoglucanase (eglII) were expressed individually, fused (fus-eglII + eglZ), and co-expressed (coex-bglZ + bglII) in B. megaterium MS941, in the hydrolysis of bagasse. Compared with mixed-individual protein (mix-eglII + bglZ) and fus-eglII + bglZ, coex-bglZ + eglII resulted in faster and enhanced release of reducing sugar from bagasse (Kurniasih et al., 2014). In congo red staining with CMC-Na experiment, L. lactis-cbh did not have hydrolysis circles around them, and the supernatant and bacterial solution of L. lactis-eg-cbh showed only slight hydrolytic circles, but the hydrolytic circle after the concentration of its supernatant was not much different from that of the L. lactis-eg (Figure 4). It is speculated that the hydrolytic activity of supernatant after concentration may be due to the increase of EG content. In other words, the expression level of recombinant enzyme EG in L. lactis-eg-cbh is lower than that of L. lactis-eg. The same results can also be seen in Figure 5A, where recombinant enzyme CBH had no hydrolytic activity on CMC-Na, while EG-CBH had lower hydrolytic activity on CMC-Na than EG. Recombinant enzymes EG and CBH showed different specificity and hydrolytic activity for different substrates. The activity of crude enzyme solution obtained by L. lactis-eg-cbh against CMC-Na, filter paper and MCC were lower than that obtained by L. lactis-eg, higher than that by L. lactis-cbh. However, we found that EG-CBH produced by L. lactis-eg-cbh showed a synergistic effect on the degradation of RAC and cotton wool, and their enzyme activity were higher than EG produced by L. lactis-eg-cbh, even though the EG produced in L. lactis-eg-cbh might not be as much as L. lactis-eg (Figure 5A). This was consistent with the findings of Srisodsuk et al., CBHI of Trichoderma reesei alone was practically inactive toward cotton, and EGI of Trichoderma reesei slowly solubilized part of cotton. Working synergistically, EGI and the CBHI solubilized cotton more rapidly than EGI alone (Srisodsuk et al., 1998). Considering the activity of EG, CBH and EG-CBH enzymes and the synergistic effect of EG-CBH enzymes, RAC was determined as the substrate for the subsequent evaluation of enzyme activity.
Elevated temperature conditions offer manifold advantages, including heightened matrix solubility, augmented enzyme catalytic efficiency, and reduced contamination risks, so numerous industrial processes are conducted under high-temperature conditions (Schröder et al., 2014). Thermostable cellulases have emerged as pivotal candidates for industrial applications (huang et al., 2023). Cbh gene (from the Caldicellulosiruptor bescii) was expressed in Escherichia coli (BL21), its expression product CbCBH displayed peak activity against nitrobenzene cellobioside (pNPC) at 65°C and pH 6.0, and it demonstrated remarkable stability across a broad temperature range (60–80°C) for 8 h. CbCBH exhibited great thermostability making it a promising candidate for biofuel (Aqeel et al., 2024). In this study, the enzyme activity of the crude enzyme solution of recombinant strains increased with the increase in temperature. The optimum reaction temperature of the recombinant enzyme CBH was 60°C, while the recombinant enzymes EG and EG-CBH were tolerant to higher temperatures (80°C). This discovery indicated that the recombinant enzyme CBH, EG and EG-CBH holds promising potential for industrial applications, such as biofuel sector.
The lower the Km, the higher the binding affinity of the protease with a particular substrate was. Kcat/Km characterized the catalytic efficiency of different enzymes on specific substrates (Kaur et al., 2023). The determination of these parameters is important for understanding enzyme function, optimizing reaction conditions and developing new applications (McCarthy et al., 2003). In this study, Kinetic constants determined for recombinant enzymes in bacterial supernatant with RAC as substrate yielded similar Km, Vmax, Kcat, and Kcat/Km values for L. lactis-cbh and L. lactis-eg-cbh. Bacterial supernatant of L. lactis-eg exhibited highest affinity for RAC (Km value of 167.6 μmol∙L−1) and the highest catalytic efficiency (Vmax value of 1,601 μmol∙min−1 mg−1, Kcat value of 73.09 s−1, Kcat/Km of 0.44 L∙s−1∙μmol−1) (Table 3). It is speculated that the activity of the recombinant enzyme EG-CBH produced by L. lactis-eg-cbh may be limited by the low activity of CBH in addition to the influence of the expression level of EG. Therefore, in order to improve the synergistic effect of EG-CBH enzyme expressed by recombinant bacteria, it is necessary to continue screening more potential CBH genes and optimize the co-expression system in the future.
Different metal ions have different affinity with amino acid residues of cellulase, which causes the conformational change of enzyme, thus stimulating or inhibiting the activity of enzyme. Fe2+, Co2+, and Mn2+ stimulate the activities of glucanase and xylanase in the rumen of goats (Cheng et al., 2016). Hg2+ could significantly reduce the activity of Mycobacterium thermophilus β-glucosidase (Pyeon et al., 2019). In this study, metal ions and chemical reagents affected the crude enzyme liquid of L. lactis-eg-cbh in a manner nearly similar to that of L. lactis-eg. Mn2+, Fe2+, Ca2+, Cu2+, and Co2+ promoted the expression of recombinant enzyme. While Zn2+, K+, and EDTA had little effect on the activity of recombinant enzyme, Hg2+ inhibited the activity of recombinant enzymes. The results of recombinant enzyme activity and assessment of enzymatic properties showed that the characteristics of L. lactis-eg-cbh were similar to those of L. lactis-eg, indicating the potential dominant role of endoglucanase in the whole co-expression process.
Liu et al. transferred bgl1, cbh2, and egl3 genes from T. reesei into L. lactis MG1363, respectively, and obtained three engineered bacteria, which were mixed in equal proportions and added into high-moisture alfalfa silage. Compared to the control group (original lactic acid bacteria and lactic acid bacteria plus cellulase), silage showed a lower pH, ammonia nitrogen content, lack of butyric acid, and higher quality score (Liu et al., 2019). In order to evaluate the preliminary application effect of recombinants in silage fermentation, they were added to whole-plant corn silage in the laboratory. No significant differences were found between any of the assays in the groups treated with the addition of L. lactis NZ9000 to the whole-corn silage compared to the control group. The results indicated that the addition of L. lactis NZ9000 could not promote fermentation of the whole-corn silage directly. However, the use of recombinant L. lactis-eg, L. lactis-cbh, L. lactis-eg + L. lactis-cbh, and L. lactis-eg-cbh resulted in better silage potential, and especially contained recombinant bacteria that expressed the eg gene. For example, the pH and ADF and NDF contents of the silage in the L. lactis-eg group were the lowest among all the treatment groups, while the NH3-N contents of the silage in the L. lactis-eg + L. lactis-cbh group containing L. lactis-eg were lowest, together with the highest LA and CP contents. This could be attributed to the higher EG enzyme activity of the L. lactis-eg, leading to greater cellulose degradation and sugar production, and thus promoting lactic acid bacteria-mediated fermentation in the silage. Compared with L. lactis-eg group, L. lactis-cbh recombinantly expressing the cbh gene was less efficient, resulting in poorer silage quality, which might be related to the lower activity of cellobiose hydrolase produced by L. lactis-cbh. The L. lactis-eg-cbh group, in which the recombinant eg and cbh genes were co-expressed, showed higher pH, lower LA and WSC contents, and higher NDF and ADF contents of the silage compared to the L. lactis-eg + L. lactis-cbh group in which the two genes were expressed separately. This indicates that separate expression of the genes followed by mixing of the bacteria before addition to the silage resulted in better feed quality than when the genes were co-expressed. The above findings were consistent with the results of the congo red staining in the CMC-Na experiment and substrate specificity of recombinant enzymes experiment (Figures 4, 5A). It is speculated that the lower expression level of EG in L. lactis-eg-cbh affects its cellulose hydrolytic activity, providing further confirmation that the promotion of whole-corn silage fermentation by recombinant L. lactis-eg promoted was essentially dependent on the production of cellulose-degrading enzymes by the bacteria. Therefore, in order to enhance silage quality through the addition of recombinant lactic acid bacteria, it is imperative to meticulously select lactic acid bacteria with proven silage potential and proficient cellulase expression capabilities, thereby achieving a synergistic effect encompassing both lactic acid bacteria and cellulase.
Overall, the engineered strains L. lactis-cbh, L. lactis-eg, and L. lactis-eg-cbh were successfully constructed and EG, CBH, and EG-CBH were expressed. EG-CBH showed a synergistic effect on the degradation of RAC and cotton wool, and their enzyme activity were higher than EG and CBH. The enzyme activity of CBH reached its peak at 60°C and pH 6.0, while the optimum reaction conditions of EG and EG-CBH were 80°C and pH 6.0. EG exhibited higher affinity (Km value of 167.6 μmol∙L−1) for RAC and the higher catalytic efficiency (Kcat/Km of 0.44 L∙s−1∙μmol−1) than CBH and EG-CBH, with no difference between the latter two. These attributes underscore its compelling potential across industrial cellulose degradation and production realms. In addition, Fe2+ and Ca2+ could be used to promote the enzyme activity of recombinase EG, Mn2+, Fe2+, Cu2+, and Co2+ can promote the enzyme activity of recombinase CBH, Fe2+, Ba2+ and higher concentrations of Ca2+, Cu2+, and Co2+ can promote activities of co-expression products of EG and CBH. The addition of engineered strains improved whole-corn silage fermentation and feed nutritional quality, with the best effect seen after addition of L. lactis-eg. One can thus conclude that the gene of lignocellulose-degrading enzyme from the yak rumenu microorganism and its engineering bacteria have important development prospects and application value in feed silage and are worthy of further development and research in the future.
The original contributions presented in the study are publicly available. This data can be found here: https://figshare.com/articles/online_resource/The_sequence_of_endoglucanase_and_cellobiohydrolase/26918479.
XW: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. YS: Data curation, Software, Writing – original draft. YC: Formal analysis, Resources, Writing – review & editing. ZZ: Data curation, Investigation, Writing – original draft. HG: Data curation, Formal analysis, Writing – review & editing. YX: Data curation, Formal analysis, Writing – original draft. CW: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. YW: Conceptualization, Methodology, Project administration, Resources, Validation, Writing – review & editing. YY: Resources, Software, Writing – original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the Natural Science Foundation of Gansu Province (22JR5RA856), the Youth Mentor Fund of Gansu Agricultural University (GAU-QDFC-2023-04), the Key Research and Development Project Plan of Gansu Academy of Sciences (No. 2024ZDYF-02), the Key Research and Development Project Plan of Gansu Provincial Science and Technology Department (18YF1NA077 and 20YF3NA020), and the National Natural Science Foundation of China (No. 31500067).
The authors would like to thank all the reviewers who participated in the review, as well as MJEditor (www.mjeditor.com) for providing English editing services during the preparation of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
An, D., Dong, X., and Dong, Z. (2005). Prokaryote diversity in the rumen of yak (Bos grunniens) and Jinnan cattle (Bos taurus) estimated by 16S rDNA homology analyses. Anaerobe 11, 207–215. doi: 10.1016/j.anaerobe.2005.02.001
Aqeel, A., Ahmed, Z., Akram, F., Abbas, Q., and Ikram-Ul-Haq,. (2024). Cloning, expression and purification of cellobiohydrolase gene from Caldicellulosiruptor bescii for efficient saccharification of plant biomass. Int. J. Biol. Macromol. 271:132525. doi: 10.1016/j.ijbiomac.2024.132525
Asha, B. M., Pathma, J., and Sakthivel, N. (2015). Isolation and characterization of a novel thermostable β-glucosidase from Bacillus subtilis SU40. Prikl. Biokhim. Mikrobiol. 51, 24–29. doi: 10.7868/s0555109915010031
Cheng, J., Huang, S., Jiang, H., Zhang, Y., Li, L., Wang, J., et al. (2016). Isolation and characterization of a non-specific endoglucanase from a metagenomic library of goat rumen. World J. Microbiol. Biotechnol. 32:12. doi: 10.1007/s11274-015-1957-4
Dashtban, M., Maki, M., Leung, K. T., Mao, C., and Qin, W. (2010). Cellulase activities in biomass conversion: measurement methods and comparison. Crit. Rev. Biotechnol. 30, 302–309. doi: 10.3109/07388551.2010.490938
De Amorim Araújo, J., Ferreira, T. C., Rubini, M. R., Duran, A. G., De Marco, J. L., de Morae, L. M., et al. (2015). Coexpression of cellulases in pichia pastoris as a self-processing protein fusion. AMB Express 5:84. doi: 10.1186/s13568-015-0170-z
Du, Z. M., Yamasaki, S., Oya, T., and Cai, Y. (2023). Cellulase-lactic acid bacteria synergy action regulates silage fermentation of woody plant. Biotechnol. Biofuels Bioprod. 16:125. doi: 10.1186/s13068-023-02368-2
Fabiszewska, A. U., Zielińska, K. J., and Wróbel, B. (2019). Trends in designing microbial silage quality by biotechnological methods using lactic acid bacteria inoculants: a minireview. World J. Microbiol. Biotechnol. 35:76. doi: 10.1007/s11274-019-2649-2
Garvey, M., Klose, H., Fischer, R., Lambertz, C., and Commandeur, U. (2013). Cellulases for biomass degradation: comparing recombinant cellulase expression platforms. Trends Biotechnol. 31, 581–593. doi: 10.1016/j.tibtech.2013.06.006
Gaya, P., Peirotén, Á., and Landete, J. M. (2020). Expression of a β-glucosidase in bacteria with biotechnological interest confers them the ability to deglycosylate lignans and flavonoids in vegetal foods. Appl. Microbiol. Biotechnol. 104, 4903–4913. doi: 10.1007/s00253-020-10588-x
Geem, K. R., Song, Y., Hwang, I., Bae, H. J., and Lee, D. W. (2021). Production of gloeophyllum trabeum endoglucanase Cel12A in Nicotiana benthamiana for cellulose degradation. Front. Plant Sci. 12:696199. doi: 10.3389/fpls.2021.696199
Guo, J. G., Xie, Y. X., Yu, Z., Meng, G., and Wu, Z. (2019). Effect of Lactobacillus plantarum expressing multifunctional glycoside hydrolases on the characteristics of alfalfa silage. Appl. Microbiol. Biotechnol. 103, 7983–7995. doi: 10.1007/s00253-019-10097-6
Hess, M., Sczyrba, A., Egan, R., Kim, T. W., Chokhawala, H., Schroth, G., et al. (2011). Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331, 463–467. doi: 10.1126/science.1200387
Himmel, M. E., Xu, Q., Luo, Y., Ding, S. Y., Lamed, R., and Bayer, E. A. (2010). Microbial enzyme systems for biomass conversion: emerging paradigms. Biofuels. 1, 323–341. doi: 10.4155/bfs.09.25
Huang, Y. Y., Lv, Z. H., Zheng, H. Z., Zhu, Q., Liu, M. T., Sang, P., et al. (2023). Characterization of a thermophilic and glucose-tolerant GH1 β-glucosidase from hot springs and its prospective application in corn Stover degradation. Front. Microbiol. 14:1286682. doi: 10.3389/fmicb.2023.1286682
Huws, S. A., Creevey, C. J., Oyama, L. B., Mizrahi, I., Denman, S. E., Popova, M., et al. (2018). Addressing global ruminant agricultural challenges through understanding the rumen microbiome: past, present, and future. Front. Microbiol. 9:2161. doi: 10.3389/fmicb.2018.02161
Jiang, H., Cao, H. W., Chai, Z. X., Chen, X. Y., Zhang, C. F., Zhu, Y., et al. (2022). Dynamic alterations in yak (Bos grunniens) rumen micro-biome in response to seasonal variations in diet. Physiol. Genomics 54, 514–525. doi: 10.1152/physiolgenomics.00112.2022
Kaur, S., Vasiljevic, T., and Huppertz, T. (2023). Milk protein hydrolysis by actinidin-kinetic and thermodynamic characterisation and comparison to bromelain and papain. Food Secur. 12:4248. doi: 10.3390/foods12234248
Kuipers, O. P., de Ruyter, P. G., Kleerebezem, M., and de Vos, W. M. (1998). Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64, 15–21. doi: 10.1016/S0168-1656(98)00100-X
Kurniasih, S. D., Alfi, A., Natalia, D., Radjasa, O. K., and Nurachman, Z. (2014). Construction of individual, fused, and co-expressed proteins of endoglucanase and β-glucosidase for hydrolyzing sugarcane bagasse. Microbiol. Res. 169, 725–732. doi: 10.1016/j.micres.2014.02.002
Le Loi, Y., Nouaille, S., Commissaire, J., Brétigny, L., Gruss, A., and Langella, P. (2001). Signal peptide and propeptide optimization for heterologous protein secretion in Lactococcus lactis. Appl. Environ. Microbiol. 67, 4119–4127. doi: 10.1128/AEM.67.9.4119-4127.2001
Liu, Z. L., Li, H. N., Song, H. T., Xiao, W. J., Xia, W. C., Liu, X. P., et al. (2018). Construction of a trifunctional cellulase and expression in Saccharomyces cerevisiae using a fusion protein. BMC Biotechnol. 18:43. doi: 10.1186/s12896-018-0454-x
Liu, Q. H., Li, J. F., Zhao, J., Wu, J. X., and Shao, T. (2019). Enhancement of lignocellulosic degradation in high-moisture alfalfa via anaerobic bioprocess of engineered Lactococcus lactis with the function of secreting cellulose. Biotechnol. Biofuels 12:88. doi: 10.1186/s13068-019-1429-4
Liu, Q. H., Shao, T., Dong, Z. H., and Bai, Y. F. (2017). Solution for promoting egl3 gene of Trichoderma reesei high-efficiency secretory expression in Escherichia coli and Lactococcus lactis. Process Biochem. 62, 135–143. doi: 10.1016/j.procbio.2017.07.031
Liu, F., van Heel, A. J., Chen, J., and Kuipers, O. P. (2022). Functional production of clostridial circularin a in Lactococcus lactis NZ9000 and mutational analysis of its aromatic and cationic residues. Front. Microbiol. 13:1026290. doi: 10.3389/fmicb.2022.1026290
Liu, X. D., Wang, A. F., Zhu, L. Q., Guo, W., Guo, X. J., Zhu, B. C., et al. (2024). Effect of additive cellulase on fermentation quality of whole-plant corn silage ensiling by a Bacillus inoculant and dynamic microbial community analysis. Front. Microbiol. 14:1330538. doi: 10.3389/fmicb.2023.1330538
Liu, J. R., Yu, B., Liu, F. H., Cheng, K. J., and Zhao, X. (2005). Expression of rumen microbial Fibrolytic enzyme genes in probiotic Lactobacillus reuteri. Appl. Environ. Microbiol. 71, 6769–6775. doi: 10.1128/AEM.71.11.6769-6775.2005
McCarthy, T., Hanniffy, O., Savage, A. V., and Tuohy, M. G. (2003). Catalytic properties and mode of action of three endo-beta-glucanases from Talaromyces emersonii on soluble beta-1,4-and beta-1,3; 1, 4-linked glucans. Int. J. Biol. Macromol. 33, 141–148. doi: 10.1016/s0141-8130(03)00080-1
Niu, L. J., Zhang, H., Wu, Z. K., Wang, Y. B., Liu, H., Wu, X. L., et al. (2018). Modified TCA/acetone precipitation of plant proteins for proteomic analysis. PLoS One 13:e0202238. doi: 10.1371/journal.pone.0202238
Ozkose, E., Akyol, I., Kar, B., Comlekcioglu, U., and Ekinci, M. S. (2009). Expression of fungal cellulase gene in Lactococcus lactis to construct novel recombinant silage inoculants. Folia Microbiol. 54, 335–342. doi: 10.1007/s12223-009-0043-4
Popova, M., Martin, C., and Morgavi, D. P. (2010). Improved protocol for high-quality co-extraction of DNA and RNA from rumen digesta. Folia Microbiol. 55, 368–372. doi: 10.1007/s12223-010-0060-3
Pyeon, H. M., Lee, Y. S., and Choi, Y. L. (2019). Cloning, purification, and characterization of GH3 β-glucosidase, MtBgl85, from Microbulbifer thermotolerans DAU221. PeerJ. 7:e7106. doi: 10.7717/peerj.7106
Rossi, F., Rudella, A., Marzotto, M., and Dellaglio, F. (2001). Vector-free cloning of a bacterial endo-1,4-beta-glucanase in lactobacillus plantarum and its effect on the acidifying activity in silage: use of recombinant cellulolytic Lactobacillus plantarum as silage inoculant. Antonie Van Leeuwenhoek 80, 139–147. doi: 10.1023/a:1012223220427
Schröder, C., Elleuche, S., Blank, S., and Antranikian, G. (2014). Characterization of a heat-active archaeal β-glucosidase from a hydrothermal spring metagenome. Enzym. Microb. Technol. 57, 48–54. doi: 10.1016/j.enzmictec.2014.01.010
Segato, F., Damásio, A. R., de Lucas, R. C., Squina, F. M., and Prade, R. A. (2014). Genomics review of holocellulose deconstruction by aspergilli. Microbiol. Mol. Biol. Rev. 78, 588–613. doi: 10.1128/MMBR.00019-14
Selinger, L. B., Forsberg, C. W., and Cheng, K. J. (1996). The rumen: a unique source of enzymes for enhancing livestock production. Anaerobe 2, 263–284. doi: 10.1006/anae.1996.0036
Srisodsuk, M., Kleman-Leyer, K., Keränen, S., Kirk, T. K., and Teeri, T. T. (1998). Modes of action on cotton and bacterial cellulose of a homologous endoglucanase-exoglucanase pair from Trichoderma reesei. Eur. J. Biochem. 251, 885–892. doi: 10.1046/j.1432-1327.1998.2510885.x
Stephenson, D. P., Moore, R. J., and Allison, G. E. (2011). Transformation of, and heterologous protein expression in, lactobacillus agilis and Lactobacillus vaginalis isolates from the chicken gastrointestinal tract. Appl. Environ. Microbiol. 77, 220–228. doi: 10.1128/AEM.02006-10
Tarraran, L., and Mazzoli, R. (2018). Alternative strategies for lignocellulose fermentation through lactic acid bacteria: the state of the art and perspectives. FEMS Microbiol. Lett. 365, 1–14. doi: 10.1093/femsle/fny126
Tiwari, N., Bansal, M., and Sharma, J. G. (2021). “14-Metagenomics: A powerful lens viewing the microbial world, in Wastewater Treatment Reactors. 309–339.
Van de Guchte, M., van der Vossen, J. M., Kok, J., and Venema, G. (1989). Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. Appl. Environ. Microbiol. 55, 224–228. doi: 10.1128/aem.55.1.224-228.1989
Wan, X. R., Wu, J. P., Lei, Z. M., He, Y. Q., and Wu, R. (2016). Effect of lactic acid bacteria on corn silage quality and stability after aerobic exposure. Acta Pratacul. Sin. 25, 204–211.
Wang, W., Meng, F. J., Liu, P., Yang, S. L., and Wei, D. Z. (2014). Construction of a promoter collection for genes co-expression in filamentous fungus Trichoderma reesei. J. Ind. Microbiol. Biotechnol. 41, 1709–1718. doi: 10.1007/s10295-014-1508-2
Wang, C., Yang, Y. Z., Ma, C. J., Sunkang, Y. J., Tang, S. Q., and Zhang, Z. (2023). Expression of β-glucosidases from the yak rumen in lactic acid bacteria: a genetic engineering approach. Microorganisms 11:1387. doi: 10.3390/microorganisms11061387
Wei, Y. Q., Yang, H. J., Luan, Y., Long, R. J., Wu, Y. J., and Wang, Z. Y. (2016). Isolation, identification and fibrolytic characteristics of rumen fungi grown with indigenous methanogen from yaks (Bos grunniens) grazing on the Qinghai-Tibetan plateau. J. Appl. Microbiol. 120, 571–587. doi: 10.1111/jam.13035
Xue, X. L., Wu, Y. L., Qin, X., Ma, R., Luo, H. Y., and Su, X. Y. (2016). Revisiting overexpression of a heterologous β-glucosidase in Trichoderma reesei: fusion expression of the Neosartorya fischeri Bgl3A to cbh1 enhances the overall as well as individual cellulase activities. Microb. Cell Factories 15:122. doi: 10.1186/s12934-016-0520-9
Yu, Q., Xu, J. J., Li, M. X., Xi, Y. L., Sun, H., Xie, Y. X., et al. (2024). Synergistic effects of ferulic acid esterase-producing lactic acid bacteria, cellulase and xylanase on the fermentation characteristics, fibre and nitrogen components and microbial community structure of Broussonetia papyrifera during ensiling. J. Sci. Food Agric. 104, 3543–3558. doi: 10.1002/jsfa.13239
Zegers, N. D., Kluter, E., van Der Stap, H., van Dura, E., van Dalen, P., and Shaw, M. (1999). Expression of the protective antigen of Bacillus anthracis by Lactobacillus casei: towards the development of an oral vaccine against anthrax. J. Appl. Microbiol. 87, 309–314. doi: 10.1046/j.1365-2672.1999.00900.x
Zhang, F., Wang, X., Lu, W., Li, F., and Ma, C. (2019). Improved quality of corn silage when combining cellulose-decomposing bacteria and Lactobacillus buchneri during silage fermentation. Biomed. Res. Int. 2019, 1–11. doi: 10.1155/2019/4361358
Zhao, J., Shao, T., Chen, S., Tao, X., and Li, J. (2021). Characterization and identification of cellulase-producing Enterococcus species isolated from Tibetan yak (Bos grunniens) rumen and their application in various forage silages. J. Appl. Microbiol. 131, 1102–1112. doi: 10.1111/jam.15014
Zhao, C. C., Wang, L. M., Ke, S. L., Chen, X. H., Kenéz, Á., and Xu, W. (2022). Yak rumen microbiome elevates fiber degradation ability and alters rumen fermentation pattern to increase feed effi-ciency. Anim. Nutr. 11, 201–214. doi: 10.1016/j.aninu.2022.07.014
Zhao, G. Q., Wu, H., Li, Y. Y., Li, L., He, J. J., and Yang, X. J. (2024). Fermentation characteristics and microbial community composition of wet brewer’s grains and corn Stover mixed silage prepared with cellulase and lactic acid bacteria supplementation. Anim Biosci. 37, 84–94. doi: 10.5713/ab.23.0177
Keywords: cellobiohydrolase, endoglucanase, Lactococcus lactis, secretory expression, co-expression, silage fermentation
Citation: Wan X, SunKang Y, Chen Y, Zhang Z, Gou H, Xue Y, Wang C, Wei Y and Yang Y (2024) Co-expression of endoglucanase and cellobiohydrolase from yak rumen in lactic acid bacteria and its preliminary application in whole-plant corn silage fermentation. Front. Microbiol. 15:1442797. doi: 10.3389/fmicb.2024.1442797
Received: 02 June 2024; Accepted: 22 July 2024;
Published: 17 September 2024.
Edited by:
Yushan Jia, Inner Mongolia Agricultural University, ChinaReviewed by:
Yixiao Xie, Guizhou University, ChinaCopyright © 2024 Wan, SunKang, Chen, Zhang, Gou, Xue, Wang, Wei and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuan Wang, d2FuZ2NodWFuQGdzYXUuZWR1LmNu; Yaqin Wei, d2VpeXExMUBsenUuZWR1LmNu; Yuze Yang, eXl6ODQ5MjkwNTZAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.