- 1Inflammation Biology, QIMR Berghofer Medical Research Institute, Brisbane, QLD, Australia
- 2Molecular Nutrition, QIMR Berghofer Medical Research Institute, Brisbane, QLD, Australia
- 3School of Biomedical Sciences and Pharmacy, Faculty of Health and Medicine, Hunter Medical Research Institute, University of Newcastle, Callaghan, NSW, Australia
- 4UMR0703 APEX, INRAE, Oniris, Nantes, France
- 5GVN Centre of Excellence, Australian Infectious Disease Research Centre, Brisbane, QLD, Australia
Introduction: The severity of Coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is often dictated by a range of comorbidities. A considerable literature suggests iron deficiency and iron overload may contribute to increased infection, inflammation and disease severity, although direct causal relationships have been difficult to establish.
Methods: Here we generate iron deficient and iron loaded C57BL/6 J mice by feeding standard low and high iron diets, with mice on a normal iron diet representing controls. All mice were infected with a primary SARS-CoV-2 omicron XBB isolate and lung inflammatory responses were analyzed by histology, immunohistochemistry and RNA-Seq.
Results: Compared with controls, iron deficient mice showed no significant changes in lung viral loads or histopathology, whereas, iron loaded mice showed slightly, but significantly, reduced lung viral loads and histopathology. Transcriptional changes were modest, but illustrated widespread dysregulation of inflammation signatures for both iron deficient vs. controls, and iron loaded vs. controls. Some of these changes could be associated with detrimental outcomes, whereas others would be viewed as beneficial.
Discussion: Diet-associated iron deficiency or overload thus induced modest modulations of inflammatory signatures, but no significant histopathologically detectable disease exacerbations.
Introduction
In mammals hundreds of proteins use iron in a multitude of cellular activities (Galy et al., 2024) including inflammation and immunity (Ni et al., 2022). Iron levels and distributions in different tissues and cells, under different conditions, are regulated by a complex network of processes (Vogt et al., 2021). Iron studies have been conducted for a range of diseases, with a diverse spectrum of findings (Ali et al., 2017; Charlebois and Pantopoulos, 2023). For instance, for tuberculosis and salmonella, anaemia is linked to poor outcomes, but iron supplementation can exacerbate infections (Hoffmann et al., 2021; Nienaber et al., 2023). For hepatitis B and hepatitis C virus infections, iron overload has been associated with poor prognosis (Schmidt, 2020), whereas for pediatric HIV cases, iron supplementation has been associated with disease progression (Andersen et al., 2022).
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiological agent of Coronavirus disease 2019 (COVID-19) (Worobey et al., 2022; Crits-Christoph et al., 2023) and has caused a global pandemic involving ~775 million cases and ~ 7 million deaths worldwide (WHO, 2024a). COVID-19 is often associated with a ‘cytokine storm’ and the life threatening, acute respiratory distress syndrome (ARDS). The severity of COVID-19 is influenced by a range of comorbidities (Chenchula et al., 2023; Finnerty et al., 2023; Silaghi-Dumitrescu et al., 2023), with a large body of literature suggesting that iron deficiency and iron overload may also contribute to disease severity (Cavezzi et al., 2020; Campione et al., 2021; Girelli et al., 2021; Habib et al., 2021; Lanser et al., 2021a; Biasiotto and Ferrari, 2022; Gupta et al., 2022; Sonnweber et al., 2022; Zhou et al., 2022; Chaubey et al., 2023; Naidu et al., 2023; Hanson et al., 2024; Liao et al., 2024).
Multiple studies have shown that SARS-CoV-2 infection disrupts iron homeostasis and/or modulates iron-associated biomarkers (Dahan et al., 2020; Taneri et al., 2020; Girelli et al., 2021; Onur et al., 2021; Tojo et al., 2021; Wojciechowska et al., 2021; Bastin et al., 2022; Hegelund et al., 2022; Mohus et al., 2022; Sana and Avneesh, 2022; Aslan et al., 2023; Naidu et al., 2023). This is not unique to SARS-CoV-2, as many infections modulate iron biomarkers (Ward et al., 2022), with clinical determinations of iron status thus generally unreliable in patients presenting with infectious and/or inflammatory diseases (Suchdev et al., 2017; Camaschella, 2019). Establishing the iron status of a patient presenting with COVID-19 is thus similarly complicated. How patients’ iron status prior to SARS-CoV-2 infection affects the severity of COVID-19 after infection is also often difficult to explore in clinical settings, as patients tend to present only after they have developed disease.
Iron deficiency is a widespread problem and is associated with a range of clinical issues, primarily anemia (Gattermann et al., 2021; Pasricha et al., 2021). Pre-existing anemia has been associated with increased mortality risk for hospitalized COVID-19 patients (Lanser et al., 2021a), perhaps due to SARS-CoV-2 infection further exacerbating the anemia (Bergamaschi et al., 2021). Dietary iron deficiency is responsible for about half the ≈2 billion cases of anemia globally (Pasricha et al., 2021; Hussien et al., 2023); however, anemia can have a variety of causes; globally this primarily involves thalassemias, sickle cell trait, and malaria (WHO, 2024b). Other conditions are also associated with anemia, including alcoholism (Manrai et al., 2022), diabetes (Praveen et al., 2020), cardiovascular disease (Lanser et al., 2021b) and chronic obstructive pulmonary disease (Alisamir et al., 2022). Whether the anemia arising from an iron deficient diet, or the comorbidity giving rise to the anemia, is responsible for the increase in COVID-19 severity remains unclear (Ko et al., 2020; Gerayeli et al., 2021; Gregory et al., 2021; Papadopoulos et al., 2021; Andreen et al., 2022). Iron dysregulation and inflammatory stress erythropoiesis have also been associated with long-COVID (Hanson et al., 2024). However, cause and affect are again difficult to verify, as increased iron dysregulation and stress erythropoiesis may be the result of more severe SARS-CoV-2 infections (Bergamaschi et al., 2021; Huerga Encabo et al., 2021), which then predispose to more pronounced long-COVID (Rosa et al., 2023; Zhao et al., 2023).
A number of publications have speculated on a connection between iron overload and increased severity of COVID-19, largely based on iron biomarker studies (Cavezzi et al., 2020; Campione et al., 2021; Habib et al., 2021; Biasiotto and Ferrari, 2022; Gupta et al., 2022; Naidu et al., 2023; Liao et al., 2024). In addition, iron loaded mice, injected with a pseudovirus dis-laying the spike protein, showed an increase in serum CCL4, IL1β, IL-6 and TNFα levels (Chaubey et al., 2023). Iron overload can arise from a number of conditions, most famously hereditary hemochromatosis or thalassemias (Hsu et al., 2022), which lead to a unique pattern of body and cellular iron distributions (Queiroz-Andrade et al., 2009; Cavey et al., 2019). However, iron overload can also arise from excessive dietary iron intake, which is primarily associated with excessive iron supplement consumption (Bell et al., 2000; Deugnier et al., 2002; Barton et al., 2006; Lands and Isang, 2017), but can also be associated with a high iron diet (Kasvosve et al., 2000). Whether pre-existing iron loading due to a high iron diet (in the absence of co-morbidities) exacerbates acute COVID-19 severity remains largely unexplored.
Here we use adult wild-type C57BL/6 J mice and established models of diet-induced iron deficiency and iron overload (Mirciov et al., 2017; Rishi et al., 2018; Ali et al., 2020; Helman et al., 2022), and SARS-CoV-2 infection (Bishop et al., 2024; Carolin et al., 2024). Iron deficient and iron loaded mice were compared with control mice (fed a normal iron diet) after infection with a primary human omicron XBB isolate of SARS-CoV-2 (Stewart et al., 2023). Unlike the original strain isolates, omicron variants effectively utilize the mouse Angiotensin-Converting Enzyme 2 as an entry receptor (Zhang et al., 2022), allowing use of wild-type mice. The impacts of the iron modified diets on viral replication and inflammatory disease in the infected mouse lungs were characterized using histology (H&E staining), immunohistochemistry and RNA-Seq at 2 days post infection (dpi) (peak viral load) and 6 dpi (peak of acute immune pathology) (Bishop et al., 2024; Carolin et al., 2024). Although widespread modulations in transcriptional signatures associated with inflammatory responses were observed, neither iron deficiency nor iron overload resulted in significant increases in viral replication or histopathology.
Results
Iron deficient vs. control diet; mouse weights and viral loads
Male C57BL/6 J mice were fed either a control diet (normal iron) or a diet deficient in iron for 7 weeks (Figure 1A). The iron deficient diet resulted in a significant reduction in growth rates so that mice were ≈5 grams (≈17%) lighter than controls just prior to infection (0 dpi) (Figure 1B). XBB infection had no significant effects on the weight of mice in either the control or the iron deficient groups (Supplementary Figure S1). Lungs were harvested on 2 dpi (peak viral titers) and 6 dpi (day of peak lung pathology) (Bishop et al., 2022; Bishop et al., 2024). There were no significant differences in lung tissue titers (Figure 1C), or in lung viral read counts as determined by RNA-Seq (Figure 1D). Viral titers in the nasal turbinates were slightly, but significantly, lower (0.62 log10CCID50/g, p = 0.009) at 2 dpi in mice fed the iron deficient diet (Supplementary Figure 2A).
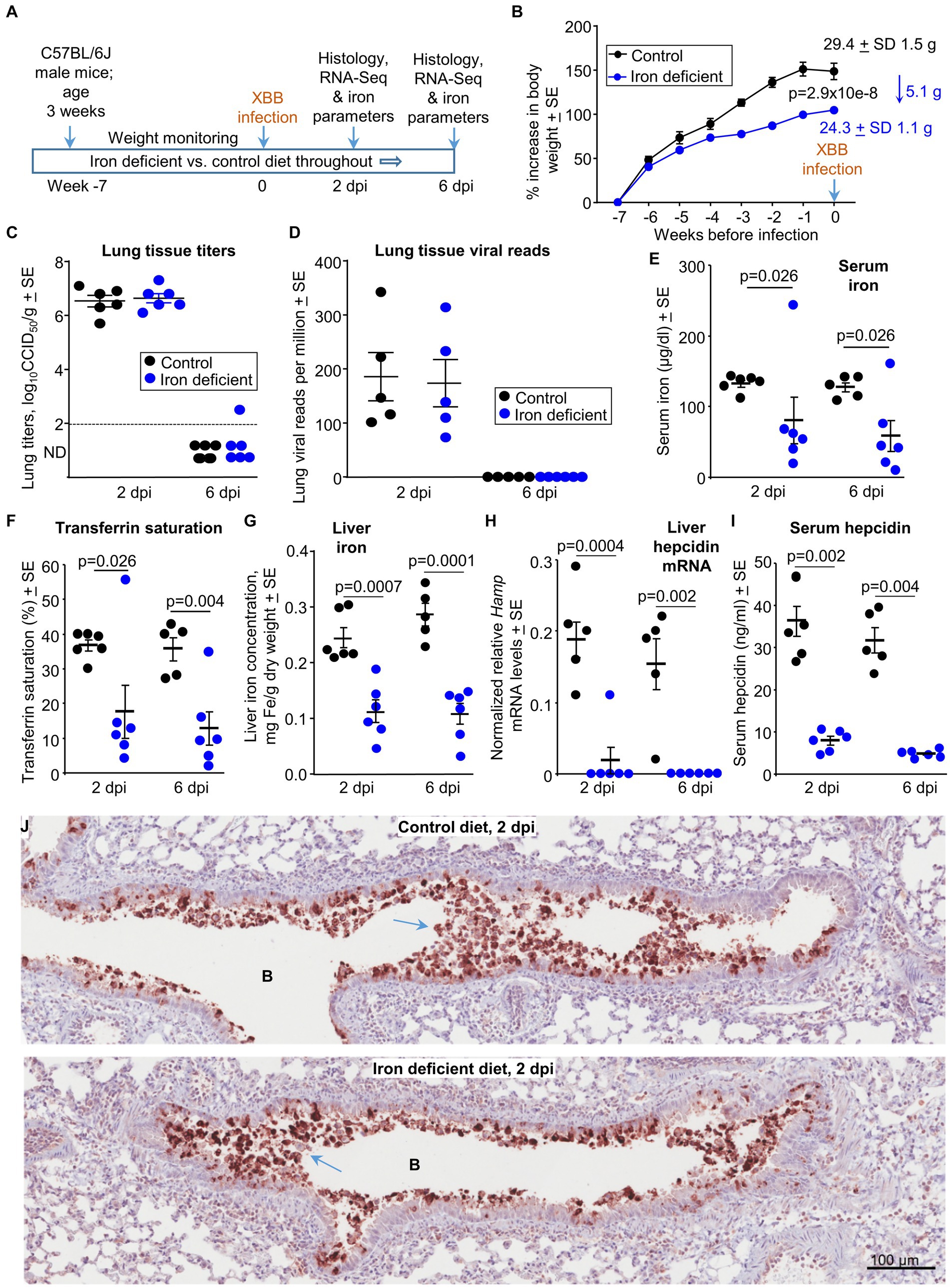
Figure 1. Iron deficient vs. control diets; weights, viral loads and iron parameters. (A) Time line of experiment. (B) Mean percentage increases in mouse body weights prior to XBB infection. The mean body weights ± SD in grams are also provided for week 0 (also 0 dpi) in text at the top right of the graph. Statistics by t test for weight differences at week zero, n = 11/12 mice per group. There were no significant weight changes after infection (Supplementary Figure S1). (C) Lung tissue XBB virus titers. Limit of detection ≈2 log10CCID50/g (dashed line); ND, not detected. (D) XBB viral read counts obtained from RNA-Seq of lung tissues. (E–I) Iron parameters from the same mice described in b-d; Control—black symbols, Iron deficient—blue symbols. (E) Serum iron levels. Statistics by Kolmogorov–Smirnov exact tests. (F) Transferrin saturation. Statistics by Kolmogorov–Smirnov exact test (2 dpi) and t test (6 dpi). (G) Liver iron levels. Statistics by t tests. (H) Liver Hamp mRNA levels; the Hamp gene encodes hepcidin. Statistics by Kolmogorov–Smirnov exact tests. (I) Serum hepcidin levels. Statistics by Kolmogorov–Smirnov exact tests. (J) IHC of mouse lung at 2 dpi for mice on the control diet compared with mice on an iron deficient diet. Staining was undertaken using an anti-SARS-CoV-2 spike protein monoclonal antibody. B—bronchial air space. Blue arrows—sloughing of virus infected bronchial epithelial cells (and associated cell debris) into the bronchial lumen.
Thus, although an iron deficient diet moderately reduced the growth of the mice, SARS-CoV-2 tissue titers were not significantly different in lungs, and were slightly lower in nasal turbinates.
Iron parameters for mice on iron deficient vs. control diets at 2 and 6 dpi
Standard iron parameters were evaluated to confirm the effects of the iron deficient diet. As expected, serum (non-heme) iron levels (Figure 1E) and transferrin saturation levels (Figure 1F) were significantly lower in mice on the iron deficient diet. We could not identify any reasons for high serum iron and transferrin saturation levels in two iron deficient mice (Figures 1E,F, one at 2 dpi and one at 6 dpi), with results confirmed by repeat assays. They perhaps reiterate the unreliable nature of such iron studies during active inflammatory disease (Suchdev et al., 2017; Camaschella, 2019). Mice on the iron deficient diet showed significantly lower liver iron levels (Figure 1G), with assessments of liver iron arguably the most reliable method (Kohgo et al., 2008) to confirm that the diet had successfully reduced iron levels. Liver hepcidin mRNA levels and serum hepcidin levels were also significantly lower in mice on the iron deficient diet (Figures 1H,I), consisted with the role of hepcidin in iron sequestration (Ginzburg, 2019). Although inflammation can up-regulate hepcidin, the robust iron deficiency likely plays the dominant role in suppressing hepcidin levels in this setting (Figure 1I) (Darshan et al., 2010).
In summary, taken together, the measured iron parameters confirmed that the iron deficient diet had successfully reduced iron levels in the mice.
Iron deficient vs. control diet; immunohistochemistry at 2 dpi
Lung sections were stained with a SARS-CoV-2 spike specific monoclonal antibody (Morgan et al., 2023). Staining was primarily associated with the bronchial epithelium and cellular debris in the bronchial lumen (Figure 1J). The latter likely represents bronchial epithelial cells sloughed-off into the airways after infection-induced cytopathic effects (CPE). No overt differences in staining was observed for mice on the iron deficient vs. control diets, consistent with data in Figures 1C,D. IHC staining of an uninfected mouse lung, illustrating the low level of background staining, is shown in Supplementary Figure S3A.
Iron deficient vs. control diet; histochemistry and histopathology
Lung sections taken at 2 and 6 dpi were stained with H&E and scanned slides were examined by a European board-certified veterinary pathologist for histopathological lesions. Lung lesions were scored using 6 criteria with examples shown; (i) emphysema (Figure 2A), (ii) bronchial epithelium damage (Figure 2B), (iii) bronchial content (Figure 2C), which occasionally included red blood cells (RBC) (Supplementary Figure S3B), (iv) vascular changes comprising leukostasis (Figures 2D,E), perivascular hemorrhage (Figure 2D) and/or leukocytoclasis (Figure 2F), (v) perivascular edema (Figure 2G) and (vi) perivascular and/or peribronchial cuffing (Figure 2H). Scores were summed to provide a cumulative score for each mouse (Supplementary Figure S4A), with no significant differences emerging between groups (Figure 2I).
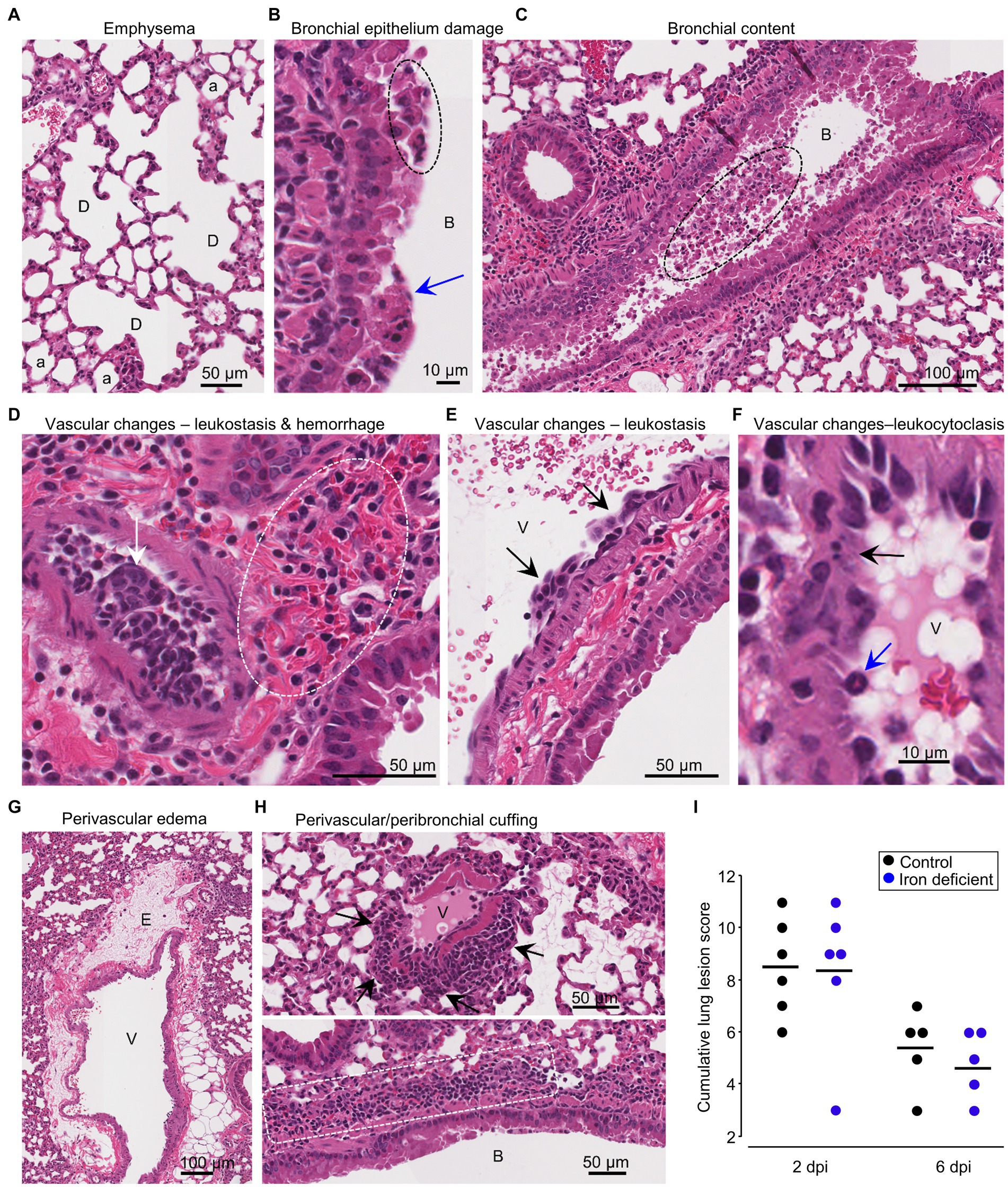
Figure 2. Iron deficient vs. control diets; lung histopathology. H&E staining of infected lungs. (A) Iron deficient diet 2 dpi showing dilated coalescing alveoli (D) illustrating emphysema. a, alveoli. Whole slide score for emphysema = 1 on a 0–2 scale. (B) Control diet 2 dpi showing necrotic bronchial epithelia cells (blue arrow) and partial loss of bronchial epithelial architecture (dashed oval). B, bronchial lumen. Whole slide score for bronchial epithelial damage = 2 on a 0–3 scale. (C) Control diet 2 dpi showing cellular debris in bronchial air space (dashed oval); this material stains positive for viral antigen. B, bronchial air space. Whole slide score for bronchial content = 2 on a 0–3 scale. (D) Control diet 2 dpi showing occlusion of the vascular lumen by clumps of leukocytes (leukostasis) (white arrow), with perivascular hemorrhage (dashed oval). Whole slide score for vascular wall changes = 3 on a 0–3 scale. (E) Iron deficient diet 6 dpi showing clumps of leukocytes adherent to the blood vessel intima (arrows). V, vascular lumen. Whole slide score for vascular wall changes = 2. (F) Control diet 2 dpi showing leukocytoclasis (vascular damage caused by nuclear debris from infiltrating neutrophils) (black arrow). Blue arrow—neutrophil. Whole slide score for vascular wall changes = 3. (G) Control diet 2 dpi showing perivascular edema (E). V, vascular lumen. Whole slide score for edema = 2 on a 0–2 scale. (H) (Top) Iron deficient diet 2 dpi showing perivascular cuffing (arrows). V, vascular lumen. Whole slide score for perivascular/peribronchial cuffing =2. (Bottom) Control diet showing peribronchial cuffing (white dashed box showing leukocytes). B, bronchial lumen. Whole slide score for perivascular/peribronchial cuffing =3 on a 0–3 scale. (I) Cumulative lung lesion scores for each mouse; raw data is shown in Supplementary Figure S4A.
White space analysis, an approximate measure of lung consolidation, showed the expected (Rawle et al., 2021; Yan et al., 2022; Dumenil et al., 2023) significant reduction in virus-infected mice compared with uninfected mice (Supplementary Figure S5). However, no significant differences emerged between mice on the different diets, although white space reductions appeared to have occurred slightly earlier (2 dpi) in some iron deficient mice (Supplementary Figure S5).
Using the same H&E stained sections, a pixel count analysis was undertaken to generate a ratio of nuclear (purple) to cytoplasmic (red) staining, which provides an approximate measure of leukocyte infiltration (Prow et al., 2019; Dumenil et al., 2023). As expected, infected mice showed significantly more infiltrates than naïve mice; however, no significant differences emerged between infected mice on the different diets (Supplementary Figures S6A,B).
Iron deficient vs. control diet; lung RNA-Seq at 2 dpi
RNA-Seq analysis of lungs at 2 dpi from mice fed an iron deficient vs. a control diet identified only 109 DEGs, with all but 5 of these showing low (<1 log2) fold change (Supplementary Table S1). (PC2/PC1 plots for all the RNA-Seq data are shown in Supplementary Figure S7). A heat map of the top 100 genes that provided the greatest contribution to the segregation between iron deficient vs. control groups, further illustrated that gene expression differences between these groups was low and not particularly focused on any specific set of genes (Supplementary Figures S8A,B). The results from the bioinformatic analyses are shown in Supplementary Table S1 and are summarized in Figure 3A.
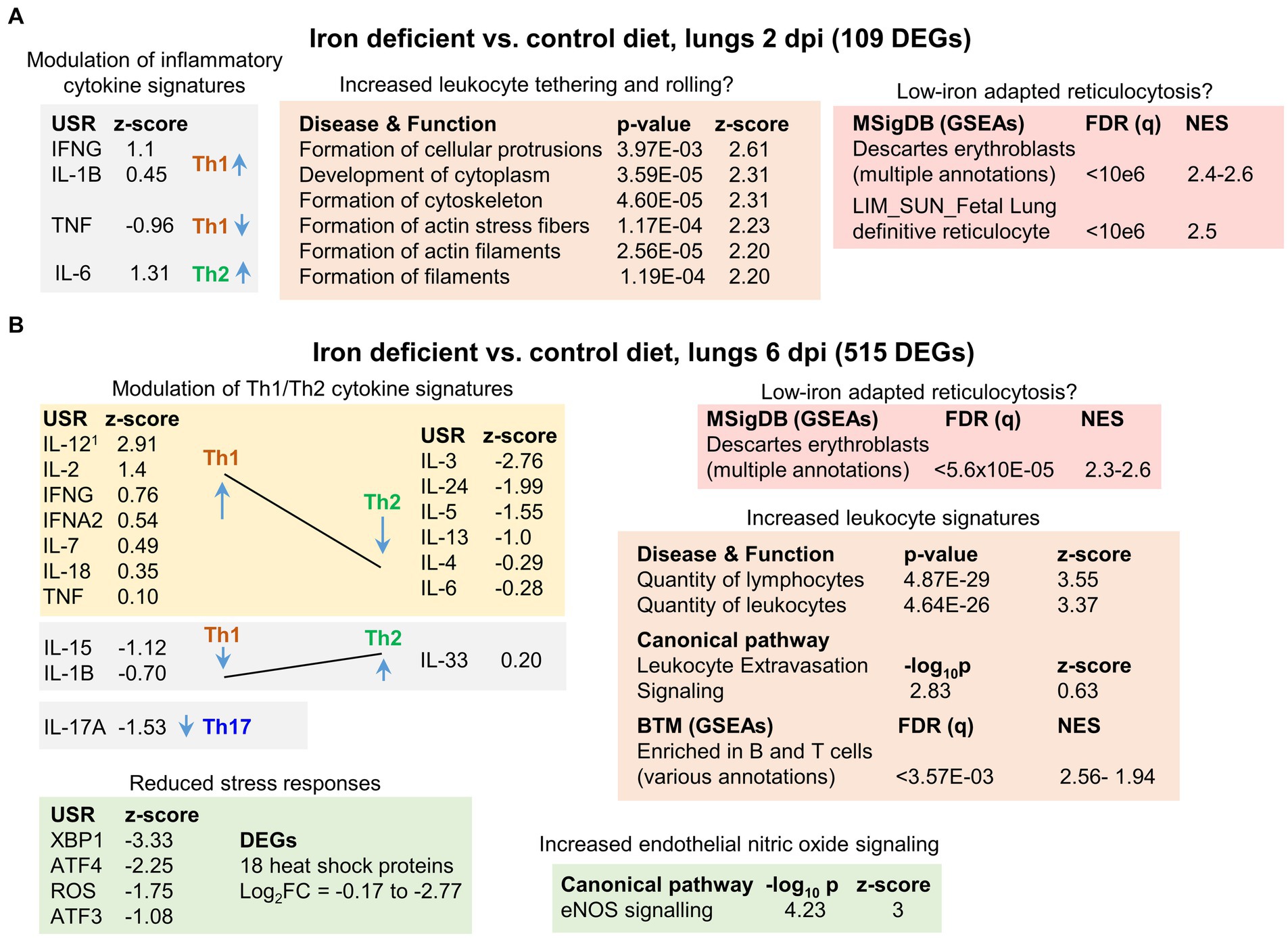
Figure 3. Iron deficient vs. control diets; bioinformatic summary. (A) RNA-Seq analysis of lungs from mice fed an iron deficient vs. control diet at 2 dpi yielded 109 DEGs. DEGs were analyzed by IPA using the Up-Steam Regulator (USR) (cytokine annotations shown), and Disease and Function features. The All genes list was interrogated using GSEAs and gene sets provided by MSigDB. Full data sets are provided in Supplementary Table S1. 1USR annotation is “IL-12 (family)” and Molecular Type is “group” rather than “cytokine.” (B) RNA-Seq analysis of lungs from mice fed an iron deficient vs. control diet at 6 dpi yielded 515 DEGs. Bioinformatic summary as for a, with the addition of USRs associated with stress responses, and IPA Canonical pathway annotations. Full data sets are provided in Supplementary Table S2.
The 109 DEGs were analyzed by Ingenuity Pathway Analysis (IPA). Using the Up Stream Regulator (USR) feature, mild modulation of inflammatory cytokine signatures was identified (Figure 3A). As iron deficiency has been associated with changes in the Th1/Th2 cytokine balance (Roth-Walter et al., 2017; Ni et al., 2022; Roth-Walter, 2022), the Th1 or Th2 association for each cytokine is shown (Figure 3A), although 2 dpi is generally too early for significant adaptive T cell responses. The IPA Diseases or Functions feature identified a series of top annotations associated with cellular protrusions, cytoskeleton and actin (Figure 3A). These annotations may indicate increased leukocyte tethering and rolling (Kameritsch and Renkawitz, 2020) in the iron deficient mice. However, significant histological differences were not apparent (Supplementary Figure S9), as might be expected with only 109 DEGs.
Gene set enrichment analyses (GSEAs) were undertaken using the ‘All genes’ list (ranked by fold change) (Supplementary Table S1) and gene sets from the Molecular Signatures Database (MSigDB) (Liberzon et al., 2015; Castanza et al., 2023). This analysis identified a series of annotations, with high Normalized Enrichment Scores (NES), that were associated with erythroblasts (Figure 3A, NES ≈ 2.4–2.6). Erythroblasts are ordinarily restricted to bone marrow, with the loss of the nucleus from these cells preceding release of the resulting reticulocytes, immature red blood cells (RBC), into the circulation. Reticulocytes retain the mRNA profile of erythroblasts in the final stages of maturation (Goh et al., 2007). These erythroblast annotations thus likely reflect an increase in gene signatures associated with reticulocytosis in iron deficient mice. Viral infections, including COVID-19, can result in significant damage to RBC (Al-Kuraishy et al., 2022; Russo et al., 2022), with COVID-19 also able to infect RBC progenitors (Kronstein-Wiedemann et al., 2022). Erythropoiesis is thus stimulated (Hanson et al., 2024), with erythroblasts adapting to iron deficient conditions by modulating gene expression (Liu et al., 2008; Wollmann et al., 2014; Kobayashi et al., 2017), with such modulation likely giving rise to these erythroblast annotations.
In summary, at 2 dpi iron deficiency imparted only mild transcriptional changes, which were associated with minor modulation of inflammatory cytokine, and perhaps tethering and rolling, signatures. Erythroblast/reticulocyte signatures also indicated erythroblast adaptation to low iron conditions.
Iron deficient vs. control diet; lung RNA-Seq at 6 dpi
RNA-Seq analysis of lungs at 6 dpi from mice fed an iron deficient diet vs. a control diet identified 515 DEGs, with all but 23 of these showing low (<1 log2) fold change (Supplementary Table S2). The results from the bioinformatic analyses are shown in Supplementary Table S2 and are summarized in Figure 3B (see below).
IPA USR analysis again identified modulation of cytokine response signatures that were generally associated with up-regulation of Th1 signatures and down-regulation of Th2 signatures (Figure 3B). This contrasts with previous reports that suggest immune activation under iron-deficient conditions results in the expansion of Th2, but not Th1 cells (Roth-Walter et al., 2017; Roth-Walter, 2022). However, such Th1/Th2 modulation is likely to be setting dependent (Ni et al., 2022). For instance, iron deficiency is reported to blunt IL-6 responses (Ekiz et al., 2005; Darshan et al., 2010) in some settings, but not others (Nakagawa et al., 2014). Consistent with the observations herein (Figure 3B), iron deficiency has been reported to reduce IL-4 (Kuvibidila et al., 2012) and IL-17A responses (Li et al., 2021; Teh et al., 2021). In addition, iron deficiency has been reported to cause non-proliferating, altruistic T cells to produce IL-2 (Berg et al., 2020).
Top IPA Diseases or Functions annotations indicated an increase in the quantity of lymphocytes/leukocytes in infected iron-deficient mouse lungs (Figure 3B; Increased leukocyte signatures). BTM GSEAs suggest these increases are primarily associated with B cells (Supplementary Table S2), consistent with identification of CXCR5 as an upregulated DEG (log2FC = 0.88) (Supplementary Table S2). CXCR5 is the receptor for CXCL13, which is the key chemokine for B cell recruitment to sites of inflammation (Kowarik et al., 2012; Armas-González et al., 2018). However, histological analyses indicated only marginal, non-significant, increases in leukocytes in iron deficient mice (Supplementary Figure S6A, 6 dpi; Supplementary Figure S4A, cuffing 6 dpi), arguing the transcriptional modulations (Figure 3B) do not reflect an overall overt increase in inflammatory infiltrates. Instead, they may reflect transcription changes associated with (i) modest increases in infiltrates, with, for instance, the extravasation annotation indicating a relatively low z-score (Figure 3B, Z-score = 0.63), and/or (ii) Th1/Th2 modulation changing leukocyte transcriptional profiles, and/or (iii) changes in the type of cells infiltrating the infected lungs in iron deficient mice (see below).
The top upregulated Canonical pathway was endothelial nitric oxide synthase (Figure 3B, eNOS), with increased endothelial nitric oxide (NO) signaling previously associated with iron deficiency (Dunaway et al., 2023). eNOS has a central role in endothelial homeostasis and is generally viewed as serving a beneficial role in lung inflammation (Qi et al., 2016; Guimarães et al., 2021; Ren et al., 2021) and ARDS (Albertine et al., 1999; Vassiliou et al., 2021). eNOS uncoupling can lead to generation of reactive oxygen species (ROS) and lung injury (Gross et al., 2015). However, this was not indicated in this setting as the ROS signature was reduced in iron deficient mice (Figure 3B, ROS). A number of other stress responses signatures were also lower in iron deficient mice; specifically, XBP1 (endoplasmic reticulum stress) (Cohen et al., 2017), ATF3 and ATF4 (stress-induced transcription factors) (Bradley et al., 2021; Shahriari-Felordi et al., 2022; Li et al., 2023; Niethamer et al., 2023) (Figure 3B). In addition, expression of 18 heat shock protein mRNAs was lower (Figure 3B, DEGs), with Hspa1a and Hspa1b (Hsp70 family members) (Bishop et al., 2024) the most down-regulated DEGs (Supplementary Table S2).
In summary, iron deficient mice at 6 dpi showed modest transcriptional changes when compared to controls. Bioinformatic analyses indicated signatures associated with increased Th1/Th2 ratios, modulated leukocyte expression patterns, and elevated eNOS and reduced stress responses.
Iron loading vs. control diet; weights and lung viral loads
Male C57BL/6 J mice were fed either a control diet or an iron loading diet for 7 weeks starting at 4 weeks of age (Figure 4A). The iron loading diet resulted in a small but significant reduction in body weight (mean 1.4 g reduction) at 0 dpi (Figure 4B). No significant weight changes were observed post XBB infection in either the control or the iron loading groups (Supplementary Figure S1). Lung tissue titers at 2 dpi showed a modest, but significant, 0.65 log10CCID50/g reduction in lung viral titers from mice fed the iron loading diet (Figure 4C). Nasal turbinate viral titers showed no significant differences at 2 dpi (Supplementary Figure S2B). Viral lung read counts (from RNA-Seq analysis) also showed a reduction (of 0.39 log10), but this did not reach significance (Figure 4D).
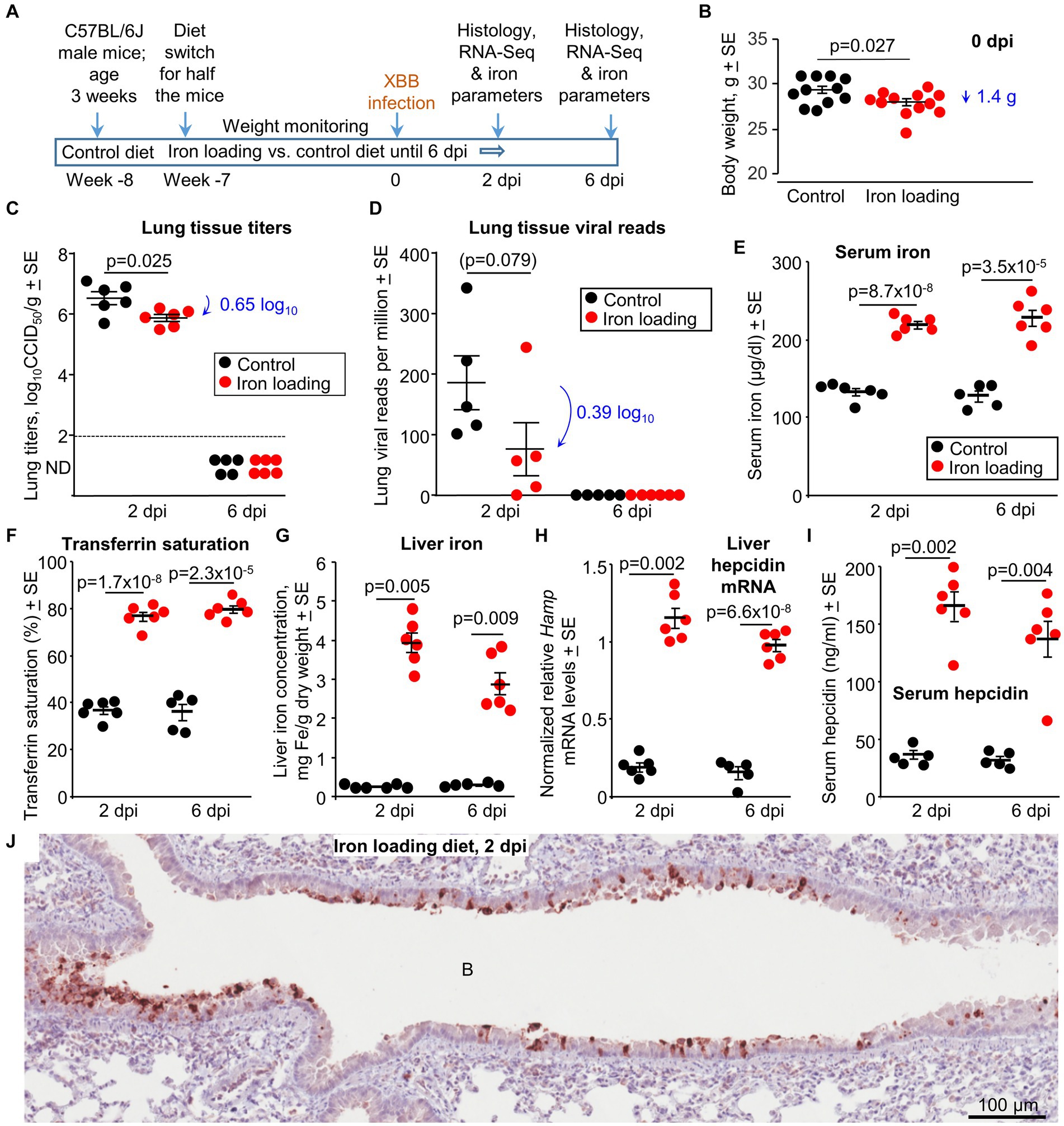
Figure 4. Iron loading vs. control diets; weights, viral loads and iron prarameters. (A) Time line of experiment. (B) Mouse body weights at 0 dpi, prior to XBB infection. Statistics by t test, n = 11/12 mice per group. There were no significant weight changes after infection (Supplementary Figure S1). (C) Lung tissue XBB virus titers. Limit of detection ≈2 log10CCID50/g (dashed line); ND – not detected. (D) XBB viral read counts obtained from RNA-Seq of lung tissues. (E–I) Iron parameters from the same mice described in b-d; Controls—black symbols, Iron loaded—red symbols. (E) Serum iron levels. Statistics by t tests. (F) Transferrin saturation. Statistics by t tests. (G) Liver iron levels. Statistics by Kolmogorov–Smirnov exact tests. (H) Liver Hamp mRNA levels. Statistics by Kolmogorov–Smirnov exact test (2 dpi) and t test (6 dpi). (I) Serum hepcidin levels. Statistics by Kolmogorov–Smirnov exact tests. (Data for control mice in b-i is the same as in Figure 1). (J) IHC of lung at 2 dpi for a mouse on the iron loading diet, using an anti-SARS-CoV-2 spike protein monoclonal antibody. B, bronchial air space. IHC of lung from a control mouse is shown in Figure 1J (top image). Staining of an uninfected lung is shown in Supplementary Figure S3A.
Thus, the iron loading diet marginally reduced the mean mouse body weight, with lungs showing modest, but significant, viral titer reductions at 2 dpi. The latter is consistent with some (Singh et al., 2023), but not other (Chaubey et al., 2023), in vitro studies.
Iron parameters for mice on iron loading vs. control diets at 2 and 6 dpi
Standard iron parameters were again evaluated to confirm the effects of the iron loading diet. As expected, serum iron levels (Figure 4E) and transferrin saturation levels (Figure 4F) were significantly higher in mice on the iron loading diet. Mice on the iron loading diet showed significantly higher liver iron levels (Figure 4G). mRNA levels for liver Hamp (the gene that codes for hepcidin) (Figure 4H) and serum hepcidin protein levels (Figure 4I) were also significantly higher in mice on the iron loading diet.
In summary, the iron parameters all confirmed that the iron loading diet had successfully increased iron levels in the mice.
Iron loading vs. control diet; immunohistochemistry at 2 dpi
Lung sections were stained with a SARS-CoV-2 specific monoclonal antibody. Staining was primarily associated with the bronchial epithelium, with minimal stained material in the bronchial airways (Figure 4J). Staining was less abundant (compared with controls, Figure 1J), consistent with the lower viral load (Figure 4C). IHC staining of an uninfected mouse lung is shown in Supplementary Figure S3A.
Iron loading vs. control diet; histochemistry and lung lesions
Lung sections taken at 2 and 6 dpi were stained by H&E and scanned slides were examined by a veterinary pathologist. Lung lesions were scored as above, with lesions at 6 dpi emerging to be slightly less severe across the scoring criteria (Supplementary Figure S4B, Figure 5A), with the cumulative lesion score significantly lower for iron loaded mice when compared with mice on the control diet (Figure 5B). This observation likely reflects the lower viral loads in lungs from iron loaded mice (Figure 4C).
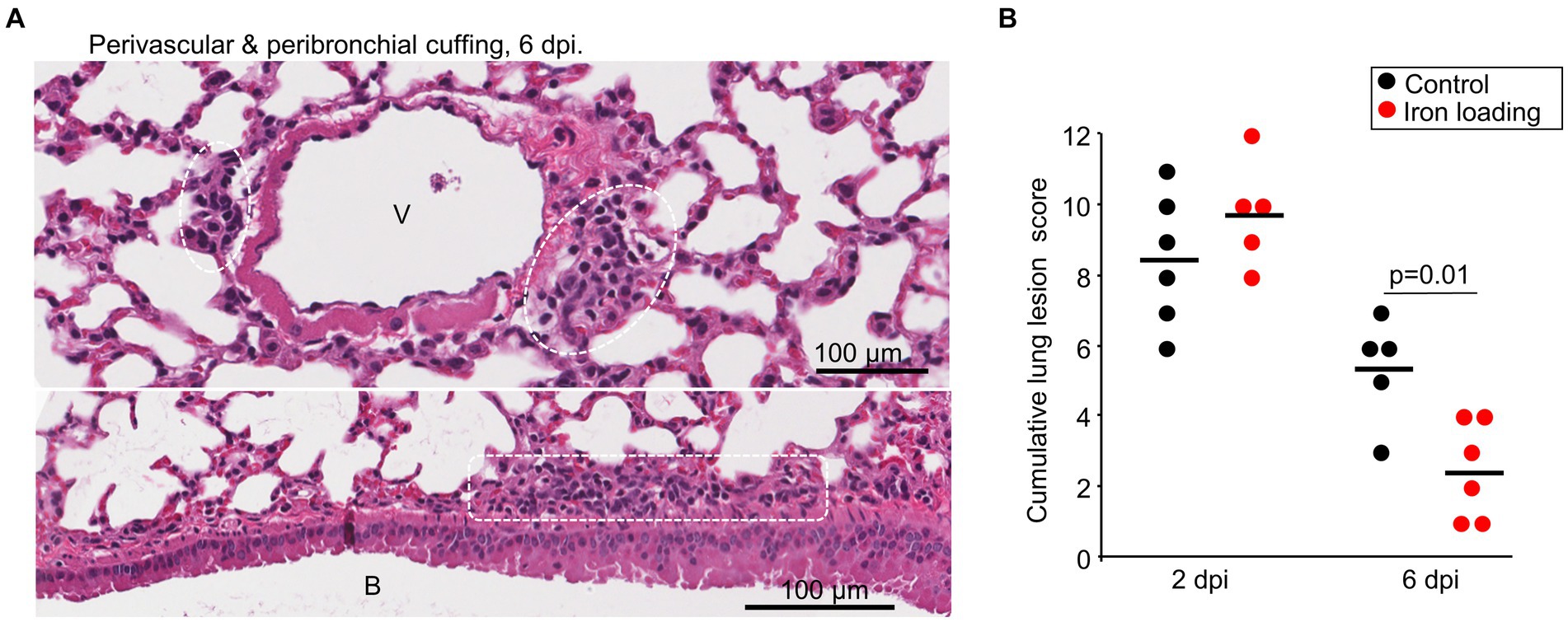
Figure 5. Iron loading vs. control diets; histology. (A) Example of perivascular cuffing (top) and peribronchial cuffing (bottom) at 6 dpi for iron loaded mice. Leukocytes indicated by white dashed box/ovals. V, vascular lumen. B, bronchial lumen. Whole slide score for perivascular/peribronchial cuffing =1 on a 0–3 scale. (B) Cumulative lung lesion scores for each mouse, raw data is shown in Supplementary Figure S4B. Statistics by t test.
White space analysis (Supplementary Figure S10) and ratios of nuclear (purple) to cytoplasmic (red) staining (Supplementary Figure S6C) showed no significant differences between infected mice on iron loading vs. control diets.
Iron loading vs. control diet; lung RNA-Seq at 2 dpi
RNA-Seq analysis of lungs at 2 dpi from mice fed an iron loading vs. control diet identified only 1 DEG, insufficient for meaningful pathway analysis. Full gene lists and bioinformatic analyses are provided in Supplementary Table S3 and are summarized in Figure 6A (see below).

Figure 6. Iron loading vs. control diets; bioinformatic summary. (A) RNA-Seq analysis of lungs from mice fed an iron loading vs. control diet at 2 dpi yielded only 1 DEG, insufficient for pathway analysis. The ‘All genes’ list was interrogated using GSEAs and the gene sets provided by MSigDB. Full data sets are available in Supplementary Table S3. (B) RNA-Seq analysis of lungs from mice fed an iron loading vs. control diet at 6 dpi yielded 497 DEGs. DEGs were analyzed by IPA as in Figure 3. Full data sets are available in Supplementary Table S4.
GSEAs using the MSigDB gene sets provided a number of IFN annotations with high negative NES values (Figure 6A). These likely reflect the lower lung viral loads (Figures 4C,D), with less virus replication in iron loaded mice resulting in less stimulation of IFN responses. MSigDB GSEAs also identified complement activation with high positive NES values (Figure 6A). SARS-CoV2 is known to activate complement via the lectin (Ali et al., 2024) and alternative pathways (Yu et al., 2020). Complement activation can mediate antiviral effects against SARS-CoV-2 (Santiesteban-Lores et al., 2021), providing a potential explanation for the reduction in viral loads. Iron infusions are reported to trigger complement via the lectin and alternative pathways (Hempel et al., 2016; Faria et al., 2019). One might thus speculate that iron loading reduces the threshold for complement activation during SARS-CoV-2 infection. The one DEGs (Per1) may also be linked to complement activation (Shivshankar et al., 2020). However, it should be noted that complement activation primarily involves proteolytic processes that are not directly detectable by RNA-Seq. Complement activation by adaptive immune responses (mediated by IgM/IgG and the classical pathway) comes later during the course of infection, and can also (Ali et al., 2024) contribute to COVID-19 severity (Georg et al., 2022). However, complement activation was not identified in iron loaded mice at 6 dpi (Supplementary Table S4), with virus largely cleared in the current model at this time (Figures 4C,D).
GSEAs using the BTM gene sets suggested an increased number of B cells infiltrating the infected lungs in iron loaded mice (Figure 6A).
Iron loading vs. control diet; lung RNA-Seq at 6 dpi
RNA-Seq analysis of lungs at 6 dpi from mice fed an iron loading vs. a control diet identified 497 DEGs. Fold changes were again modest, with only 2 genes showing a log2 fold change >2. Full gene lists and bioinformatics are provided in Supplementary Table S4 and are summarized in Figure 6B.
Modulation of Th1/Th2 cytokines was again apparent with mostly increased Th1 and reduced Th2 USR cytokine z scores in mice on an iron loading diet (Figure 6B). This result contrasts with Salmonella typhimurium infection of mice on a high iron diet where Th1 responses were inhibited (Pfeifhofer-Obermair et al., 2021). However, the effects of high iron on the Th1/Th2 balance may be setting specific, with, for instance, iron promoting M1 differentiation of macrophages (Ni et al., 2022), and lung macrophages playing a central role in COVID-19 inflammation (Olivier et al., 2023). Several anti-inflammatory USR cytokine signatures provided negative z-scores (Figure 6B), indicating less anti-inflammatory activity in iron loaded mice. However, the IFNA2 USR signature had a negative z-score (Figure 6B, Anti-viral), suggesting reduced inflammation, with type I IFNs major drivers of inflammation (Ji et al., 2023). A reduced type I IFN signature is consistent with the lower viral loads at 2 dpi (Figure 4C) and the negative NES for IFN-associated GSEAs at 2 dpi (Figure 6A).
The annotations associated with increased leukocyte signatures (Figure 6B) do not reflect significant, histologically observable, increases in leukocytes infiltrates (Figure 5A, Supplementary Figures S4B, S6C). This again suggests they are associated with modest infiltrate changes, Th1/Th2 response changes and/or changes in the cell types infiltrating the lungs (see below). A series of BTMs suggested an increase in B cells (Figure 6B), although no chemokines or chemokine receptors that would readily explain migration of B cells into the lungs were present in the DEG list (Supplementary Table S4).
Iron has often been associated with ROS production (Chen et al., 2020; Nousis et al., 2023), and ROS-associated annotations were identified amongst the Canonical pathway annotations. However, z-scores were modest (≤1.3) (Figure 6B, ROS annotations), perhaps ameliorated by the lower viral loads. Increased ROS in iron overload setting is often ascribe to the Fenton reaction (e.g., (Mizumura and Gon, 2021)); however, the physiological relevance of this reaction in vivo is not without controversy (Muranov, 2024).
The negative z-score for the hemorrhagic disease annotation (Figure 6B) likely reflects the reductions in viral load, with lung hemorrhage well documented in COVID-19 mouse models (Sun et al., 2020; Ali et al., 2024). A number of transcription factor USRs also showed negative z-scores in iron loaded mice (Supplementary Table S4). These include, FOXC1 (Ahmed et al., 2022), XBP1 (Fernández et al., 2024a), EIF4E (Korneeva et al., 2023) and SREBF1 (aka SREBP1) (Soares et al., 2023), which are induced by SARS-CoV-2 infection. FOXC1 (Ma et al., 2020), XBP1 (Wang et al., 2023), and EIF4E (Schwarz et al., 2002), as well as TCF3 (Miao et al., 2014), are also involved in wound repair. Reduced infection (Figures 4C,D) and/or an ensuing reduced requirement for tissue repair, may explain these negative z-scores.
A number of RBC-associated annotations were identified, including hemolysis (Figure 6B). Although viral infections (Russo et al., 2022), including SARS-CoV-2 (Al-Kuraishy et al., 2022), can cause hemolysis, why this should be higher in iron loaded mice is unclear. This may be due to the toxic effects of iron on RBC (Kozlova et al., 2022), or is associated with complement activation (Figure 6A), with complement-mediated hemolysis a well-documented phenomenon (Bortolotti et al., 2023; Xiao et al., 2024). GATA1 is the master regulator of erythropoiesis (Gutiérrez et al., 2020), which might be upregulated to compensate for RBC loss.
POR (NADPH-cytochrome P450 oxidoreductase) was identified as the top scoring USR by z-score (Figure 6B, Supplementary Table S4). Amongst other functions, POR is involved in the induction of ferroptosis (Ai et al., 2021). Ferroptosis is a form of cell death promoted by iron that involves peroxidation of lipids (Zhang et al., 2023), which has been implicated in tissue damage during COVID-19 (Jacobs et al., 2020; Qiu et al., 2024). Heme oxygenase 1(HMOX1) is a crucial ferroptosis factor (Chen et al., 2023), and the transferrin receptor (TFRC) is a ferroptosis marker (Feng et al., 2020), with increased ROS/H202 and reduced glutathione (Figure 6B) crucial to the process of lipid peroxidation (Zhang et al., 2023). However, it should be noted that there is no single universal ferroptosis pathway, with many initiators, sensitizers and modulators (Dixon and Pratt, 2023); hence reliable ferroptosis annotations are often lacking in bioinformatic pathway tools such as IPA.
In summary, iron loaded mice showed modestly lower lung viral loads at 2 dpi, with RNA-Seq indicating modest transcriptional changes. Signatures at 6 dpi in iron loaded mice were associated with a general bias toward increased Th1/Th2 cytokine ratios, hemolysis, ROS and perhaps ferroptosis, but also reduced type I IFN and hemorrhage.
Dysregulated cell compositions in infected lungs of iron deficient and iron loaded mice
To gain insights into how iron deficiency and iron overload might influence the cellular compositions in the lungs after SARS-CoV-2 infection, cellular deconvolution (SpatialDecon) analysis was undertaken. This used the normalized count matrices (that provide the number of aligned reads for each gene for each mouse) and the gene expression matrices from the Lung mouse cell atlas.
In iron deficient mice at 2 dpi, a significant increase in von Willebrand factor (VwF) positive endothelia cells was identified (Figure 7A). This observation is perhaps consistent with the increased extravasation annotations (Figure 3A), as endothelial VwF promotes extravasation (Petri et al., 2010; Mojzisch and Brehm, 2021).
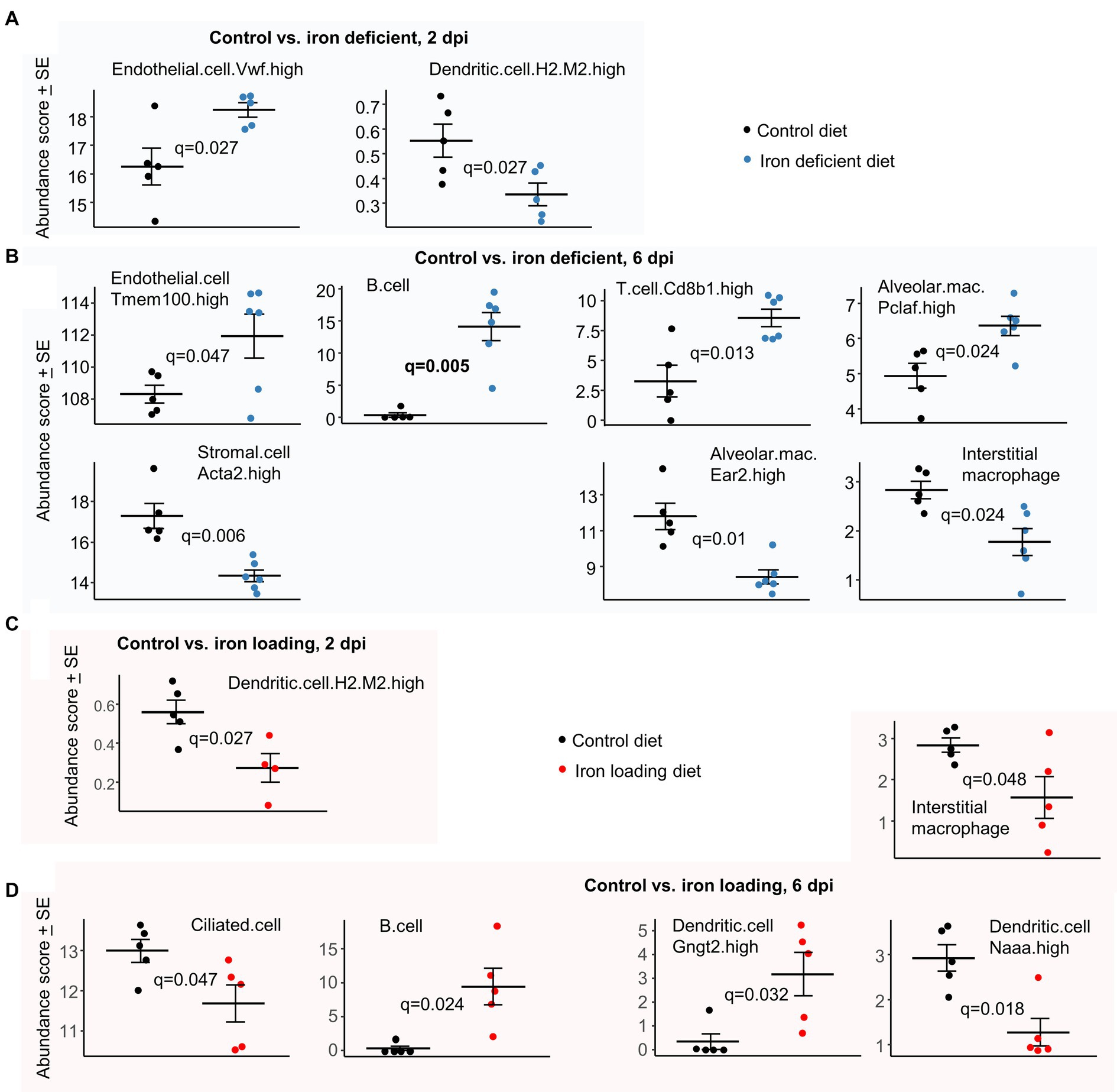
Figure 7. Cellular deconvolution analyses for iron deficient vs. control and iron loading vs. control diets. Relative abundances of cell types were estimated using SpatialDecon and cell-type expression matrices obtained from the NanoString Lung mouse cell atlas. (A) Control vs. iron deficient, 2 dpi. (B) Control vs. iron deficient, 6 dpi. (C) Control vs. iron loading, 2 dpi. (D) Control vs. iron loading, 6 dpi. Statistics by t test with FDR correction.
In iron deficient mice at 6 dpi, the increase in endothelial transcriptional signatures (Figure 7B) is consistent with the increase in the eNOS signaling pathway (Figure 3B), with eNOS a key survival factor for endothelial cells during inflammation (Dimmeler and Zeiher, 1999). Stromal cells were less abundant in iron deficient mice; these cells are involved in wound repair (Jerkic et al., 2023) and their proliferation may be impaired under iron-deficient conditions (Recalcati et al., 2019). The higher B and T cell abundance scores in iron deficient mice (Figure 7B) are consistent with the BTM GSEA results (Figure 3B). Difference in mean abundance scores for B cell represents the largest and most significant (Figure 7B, q = 0.006) observed herein. Lung B and T cells are well described for COVID-19 (Cavalli et al., 2020; Yang et al., 2024); however, iron deficiency is generally associated with impaired B and T cell responses (Jiang et al., 2019; Wideman et al., 2023). The Interstitial macrophages are generally anti-inflammatory during disease processes (Zhou et al., 2024) and Ear2 is upregulated on alveolar macrophages under Th2 conditions (Cormier et al., 2002), so reduced abundance scores for these two cell types in iron deficient mice (Figure 7B) may be associated with the elevated Th1/Th2 ratios (Figure 3B). In contrast, proliferating alveolar macrophages (Pclaf is a proliferation marker) have a higher abundance score in iron deficient mice (Figure 7B), with these cells known to self-renew and adopt a M1 pro-inflammatory phenotype when exposed to IFNγ or TNF (Olivier et al., 2023).
In iron loaded mice at 2 dpi, only Dendritic.cell.H2.M2.high (Han et al., 2018) were identified as significantly less abundant (Figure 7C), with less infection perhaps resulting in less dendritic cell activation and/or recruitment. In iron loaded mice at 6 dpi, the abundance score for ciliated (epithelial) cells was significantly lower (Figure 7D), with these cells efficiently infected by omicron variants (Wu et al., 2023). The lower levels of infection (Figure 4C) and epithelia cell sloughing into bronchi in iron loaded mice (Figure 4J) compared with controls (Figure 1J, top), is consistent with a lower requirement for renewal and thus the reduced (transcription-based) abundance scores. The increased B cell abundance scores (Figure 7D) is consistent with BTM GSEAs (Figure 6B), with B cell abundance scores also increased in iron deficient mice (Figure 7B). A reduction in abundance scores for Interstitial macrophages at 6 dpi (Figure 7D) was similarly evident for iron deficient mice (Figure 7B), and may again be associated with the elevated Th1/Th2 ratios (Figure 6B). DCs expressing N-acylethanolamine acid amidase (Naaa) have a lower abundance score in iron loaded mice (Figure 7D). These cells are reported to play a proinflammatory role (Li et al., 2022), with reduced type I IFN responses (Figure 6B, IFNA2) perhaps contributing to their lower abundance (Tough, 2004). Gngt2 is a M1 marker (Jablonski et al., 2015) and describes a subgroup of DCs (Han et al., 2018), whose specific function has yet to be described.
Cellular deconvolution using expression matrices from the ImmGen cell family (which is based on various tissues, not just lung) further illustrates dysregulation of leukocyte subsets at 6 dpi in both iron deficient and iron loaded mice (Supplementary Figure S11).
Discussion
We provide herein detailed comparisons of lung SARS-CoV-2 omicron XBB infection and inflammatory disease in wild-type mice that were fed an iron deficient vs. a control diet, and mice fed an iron loading vs. control diet. In iron deficient and iron loaded mice, viral loads were either not significantly affected or were mildly (≈ 0.6 log10), but significantly, reduced in lungs or nasal turbinates (Figure 4C, Supplementary Figure S2A). This strongly argues that the effectiveness of the host’s anti-viral SARS-CoV-2 responses was not significantly compromised by altered iron status. Modulation/dysregulation of immune responses by iron deficiency and overload is well described in various setting (Haschka et al., 2021; Mu et al., 2021), and was also clearly evident herein (Figures 3, 6, 7; Supplementary Figure S11). However, this led neither to impaired ability to control the virus nor to overtly more severe lung histopathology.
Pleiotropic outcomes of SARS-CoV-2 infections in iron deficient and iron overloaded mice might be envisaged, given the complexity of iron regulation (Galy et al., 2024), the modulation of iron biomarkers and homeostasis during SARS-CoV-2 infections (Dahan et al., 2020; Taneri et al., 2020; Onur et al., 2021; Tojo et al., 2021; Wojciechowska et al., 2021; Bastin et al., 2022; Hegelund et al., 2022; Sana and Avneesh, 2022; Aslan et al., 2023; Naidu et al., 2023), and the robust pro-inflammatory responses associated with SARS-CoV-2 infections (Bishop et al., 2022; Dharra et al., 2023; Olivier et al., 2023; Bishop et al., 2024). However, perhaps unexpected was the increase in Th1/Th2 ratios and B cell signatures in both iron deficient and iron load mice. This observation is reminiscent of the commonly illustrated U-shaped relationships for iron status. For instance, both iron deficit and iron excess leads to impaired or dysregulated maternal immunity during pregnancy (Dewey and Oaks, 2017). A similar U shaped relationship is also reported for maternal hemoglobin and preterm birth and ARDS (Ohuma et al., 2023). Serum iron biomarkers and COVID-19 severity also show a similar relationship (Tojo et al., 2021). B cell responses are also often impaired during iron deficiency (Ni et al., 2022), with IgM responses also reported to be blunted in iron loaded mice (Omara and Blakley, 1994). However, the effect of iron deficiency and overload on adaptive T cell responses would appear to be quite diverse and setting dependent (Walker and Walker, 2000; Kuvibidila et al., 2012; Reuben et al., 2017; Roth-Walter et al., 2017; Ni et al., 2022; Roth-Walter, 2022). In addition, in humans early Th1 responses have been associated with protection against severe COVID-19 (Garcia-Gasalla et al., 2022; Gil-Etayo et al., 2022); however, in mouse models, early pro-inflammatory signatures (2–7 dpi) tend to correlate with pathogenic “cytokine storm” profiles (Bishop et al., 2022; Bishop et al., 2024). Infection in humans usually spreads from the upper respiratory track into the lower respiratory track (lungs). This progression is not recapitulated in mice where lung infection requires direct intra-pulmonary inoculation of virus (Fumagalli et al., 2022; Rawle et al., 2021; Dumenil et al., 2023). Whether the increased Th1/Th2 ratios seen herein might be associated with beneficial or detrimental activities is thus debatable.
Perhaps surprising was the wide scale reduction in stress responses in iron deficient mice at 6 dpi (Figure 3, XBP1, ATF4, ROS). The most dominant of these (by z score) was XBP1, which is associated with the endoplasmic reticulum (ER) unfolded protein response (UPR). This pathway is activated by SARS-CoV-2 infection, promotes viral replication in epithelial cells, and is associated with induction of proinflammatory responses (Fernández et al., 2024a; Fernández et al., 2024b). Why this response is blunted in iron deficient mice may be related to the requirement for iron and heme effectors and binding proteins for Ire1 clustering, a process that lies immediately upstream of XBP1 activation (Cohen et al., 2017). ATF4 is triggered by PERK, another sensor of ER stress and mediator of the UPR (Davies et al., 2024). Similarly, as iron is an important component of ROS-generating enzymes (Ying et al., 2021), iron deficiency may reduce ROS production capacity in this setting.
A limitation of this study is that we have not investigated the responses over the long-term, with a role for iron status in increasing the severity of long-COVID suggested by several studies (Sonnweber et al., 2022; Wu et al., 2023; Hanson et al., 2024). However, whether mouse models (Choi et al., 2024) can faithfully recapitulate pathological or immunopathological features of human long-COVID remains to be established (Hofer, 2022), with underlying co-morbidities (Russell et al., 2023) clearly absent in genetically identical, specific pathogen free, laboratory mice. We have also not provided insights into the sizable range of co-morbidities that can give rise to anemia or iron overload (Hsu et al., 2022; Hess et al., 2023; Sohal and Kowdley, 2024), and how these would affect COVID-19; however, this would constitute a considerable undertaking. For instance, we have not studied the ‘homeostatic iron regulator’ deficient (Hfe−/−) mouse for mouse model of hereditary hemochromatosis (Albalat et al., 2021). A counter rationale for Hfe−/− mouse studies is that compelling evidence for Hfe mutations affecting COVID-19 patient outcomes has yet to emerge (Ristić et al., 2023). We have also used herein a mouse model of relatively mild disease, as distinct from the more severe model involving infections of K18-hACE2 mice with original strain isolates. However, the latter is complicated by early mortality associated with fulminant brain infections (Dumenil et al., 2023), which are generally not a feature of human disease (Stewart et al., 2023). Thus, whether iron status would influence severe lung infection and disease is not addressed in the current study. Lastly, we have not investigated changes in iron distributions and their immunological consequences over time in different tissues (e.g., lungs, liver, spleen, lymph nodes) (Giorgi et al., 2015; Ali et al., 2020), primarily as there were no overt detrimental outcomes that would focus such studies.
In conclusion, the current study of iron deficient vs. control and iron loaded vs. control SARS-CoV-2 infected mice, finds modest transcriptional changes indicating a range of inflammatory response modulations, but no significant histopathologically detectable disease exacerbations. Some human studies have also failed to find a significant association between iron status and severity of acute COVID-19 (Achan et al., 2022; Marhaeni et al., 2023; Ristić et al., 2023). This is not to say that all diseases or conditions that lead to iron deficiency or overload are similarly benign, as they may indicate co-morbidities that can promote COVID-19 severity (Bhalla et al., 2021; Abu-Ismail et al., 2023).
Materials and methods
Ethics statements and regulatory compliance
Collection of nasal swabs from consented COVID-19 patients was approved by the University of Queensland HREC (2022/HE001492). All mouse work was conducted in accordance with the Australian code for the care and use of animals for scientific purposes (National Health and Medical Research Council, Australia). Mouse work was approved by the QIMR Berghofer MRI Animal Ethics Committee (P3600 and P3535). All infectious SARS-CoV-2 work was conducted in the BioSafety Level 3 (PC3) facility at the QIMR Berghofer MRI (Department of Agriculture, Fisheries and Forestry, certification Q2326 and Office of the Gene Technology Regulator certification 3,445). Mice were euthanized using carbon dioxide.
Iron diet modifications
Male C57BL/6 J mice were bred in-house at the QIMR Berghofer MRI animal facility and were held under standard animal house conditions [for details (see Yan et al., 2022)]. Breeding pairs were maintained on standard rodent pellet diet (120 mg/kg iron; Norco Stockfeed, Lismore, Australia). Mice were allowed unlimited access to food and water at all times. Iron deficient diet. Three-week old mice were weaned onto an iron deficient diet based on AIN93G (~1 mg/kg iron, Specialty Feeds, Glen Forrest, Australia). This iron deficient diet produces a mild to moderate anemia (Mirciov et al., 2017; Zakrzewski et al., 2022). Iron loading diet. Three-week old mice were fed the control diet for 1 week, after which they were switched to an iron loading diet, consisting of the iron deficient diet supplemented with 0.5% iron as carbonyl iron (Sigma, Product no. C3518). This 1 week delay in switching mice to the iron loading diet is necessary, as moving weanling mice directly onto an iron loading diet dramatically reduces growth rates. Control diet. The control diet comprised the aforementioned iron deficient chow supplemented with 50 mg/kg iron as ferric citrate. All mice were maintained on these diets throughout until euthanasia. The aforementioned iron modified diets represent standard and ethically acceptable procedures for generating mouse models of iron deficiency and iron loading (Santos et al., 1998; Dupic et al., 2002; Aslam et al., 2014; Pereira et al., 2015).
Assays for iron parameters
Serum (non heme) iron and transferrin saturation levels were measured using the Iron/TIBC Reagent Kit (Pointe Scientific, Canton, MI). The volumes in the kit were adjusted to allow the assay to be performed in 96-well plates as described (Frazer et al., 2017). Liver (non heme) iron levels were assayed by colorimetric assay as described previously (Fuqua et al., 2014). Hamp mRNA levels were determined by RT qPCR as described (Frazer et al., 2017). Serum hepcidin levels were determined using the Hepcidin Murine-Compete™ ELISA Kit (Intrinsic Life Sciences) as per manufacturer’s instructions.
The SARS-CoV-2 omicron XBB isolate
The XBB isolate (SARS-CoV-2UQ01) was voluntarily donated to the University of Queensland (Brisbane, Australia) by a deidentified adult COVID-19 patient with degree-level education via a self-collected nasopharyngeal swab. The patient provided written consent (Stewart et al., 2023; Carolin et al., 2024). The isolate was initially grown on Vero E6-TMPRSS2 cells (Amarilla et al., 2021). The isolate is XBB.1.9.2.1.4 (Pango EG.1.4), a recombinant of BA.2.10.1 and BA.2.75; sequence deposited as hCoV-19/Australia/UQ01/2023; GISAID EPI_ISL_17784860. XBB viral stocks were propagated in Vero E6 cells (Rawle et al., 2021), and were titered using CCID50 assays (Yan et al., 2021). Medium was checked for endotoxin (Johnson et al., 2005) and cultures for mycoplasma (MycoAlert, Lonza).
Mouse model of SARS-CoV-2 infection and monitoring of disease
Mice received intrapulmonary infections delivered via the intranasal route with 5 × 104 CCID50 of XBB in 50 μL RPMI 1640 whilst under light anesthesia as described (Dumenil et al., 2023). Mice were weighed and monitored as described (Dumenil et al., 2023; Stewart et al., 2023).
Mice were euthanized using CO2, lungs were removed, with the left lung fixed in formalin for histology, the right lung inferior lobe placed in RNAlater for RNA-Seq and the remaining lobes used for tissue titers determination by CCID50 assays using Vero E6 cells as described (Rawle et al., 2021; Dumenil et al., 2023).
CCID50 assays
Tissue titers were determined as described (Rawle et al., 2021). Briefly, 5-fold serial dilutions of clarified tissue homogenates were applied in duplicates to Vero E6 cells in 96 well plates. After 6 days cytopathic effects were observed by inverted light microscope. The virus titer was determined by the method of Spearman and Karber; an Excel sheet is available at https://www.klinikum.uni-heidelberg.de/zentrum-fuer-infektiologie/molecular-virology/welcome/downloads.
Immunohistochemistry
Immunohistochemistry was undertaken using the anti-SARS-CoV-2 spike protein monoclonal antibody, SCV2-1E8, as described (Morgan et al., 2023), except that the monoclonal (IgG2a) was purified using Protein A affinity chromatography and applied to sections at 4 μg/mL for 1 h.
Histology
Lungs were fixed in 10% formalin, embedded in paraffin, and sections stained with H&E (Sigma Aldrich). Slides were scanned using Aperio AT Turbo (Aperio, Vista, CA, United States). Quantitation of white space in scanned images of H&E stained lung parenchyma (with areas greater than ≈100 μm set as a threshold) was undertaken using PixelClassifierTools in QuPath v0.3.2 (Bankhead et al., 2017), and provides an approximate measure of lung consolidation (Dumenil et al., 2023). Scanned H&E stained whole lung sections were analyzed by Aperio Positive Pixel Count Algorithm (Leica Biosystems) to generate nuclear (strong purple staining) over cytoplasmic (total red staining) pixel count ratios, providing an approximate measure of leukocyte infiltration (Prow et al., 2019; Dumenil et al., 2023).
All H&E stained whole lung sections were scanned and.svs files examined by a trained European board-certified veterinary pathologist using Qu-Path (v 0.5.1). Lung lesions were scored using 6 criteria. Emphysema was scored; 0 = no lesion, 1 = dilated and coalescent alveoli, 2 = “bullae” in the parenchyma. Bronchial epithelium damage was score; 0 = no lesion, 1 = small clusters of necrotic epithelial cells, 2 = scattered foci of epithelial degeneration with layer architecture partial loss, 3 = focal complete epithelial loss. Bronchial content was scored; 0 = empty lumen; 1 = presence of a small amount of material; 2 = partial obliteration; 3 = complete occlusion. Vascular wall changes were scored; 0 = no lesion, 1 = leukostasis, 2 = focal wall damages (including leukocytoclasis), 3 = transmural vessel wall alteration and/or vascular lumen obliteration. Perivascular edema was scored; 0 = no lesion, 1 = focal mild edema, 2 = extended marked edema with lymphoid vessel dilatation. Peribronchial/perivascular cuffing was scored; 0 = no lesion; 1 = focal inflammatory cell infiltration; 2 = circumferential inflammatory cell infiltration, 3 = coalescing inflammatory cell infiltration between bronchi and vessels. A total cumulative score was then calculated by summing all 6 parameter scores for each mouse (range 0 to 16).
RNA-Seq and bioinformatics
RNA-Seq (Illumina Nextseq 2000 platform generating 75 bp paired end reads) and bioinformatics was undertaken as described (Bishop et al., 2022; Bishop et al., 2024). Raw sequencing data (fastq files) have been deposited in the NCBI SRA, BioProject: PRJNA1102925 and are publicly available. Mean quality scores were above Q20 for all samples. Mouse RNA-Seq reads were aligned to a combined mouse (GRCm39, version M27) and SARS-CoV-2 BA.5 reference genome (Stewart et al., 2023) using STAR aligner. Viral read counts were generated using Samtools v1.16. RSEM v1.3.1 was used to generate expected counts for host genes. EdgeR was then used to generate TMM normalized count matrices, with a separate count matrix generated for iron deficient vs. control and iron loaded vs. control. Differentially expressed genes were identified using EdgeR using a FDR cut-off of 0.05.
Pathway analyses were performed using host DEGs and Ingenuity Pathway Analysis (IPA, v84978992) (QIAGEN), which provides Canonical pathways, Up-Stream Regulators (USR) and Diseases or Functions features as described (Dumenil et al., 2023; Bishop et al., 2024). Annotations without z scores or with significance (q or p) below 0.05 were removed.
Gene Set Enrichment Analyses (GSEAs) were undertaken using GSEA v4.1.0 with gene sets provided in MSigDB (≈ 45,000 gene sets) and in the Blood Transcription Modules (Li et al., 2014), and gene lists ranked by log2 fold-change. Relative abundances of cell types were estimated in R v4.1.0 from TMM normalized RSEM count matrices using SpatialDecon v1.4.3 (Danaher et al., 2022) and cell-type expression matrices obtained from the Lung mouse cell atlas1 and the ImmGen cell family.2 Statistics were undertaken by t tests with False Discovery Rate corrections using the Benjamini-Hochberg method (q).
Statistics
The t-test was used if the difference in variances was <4 fold, skewness was > − 2 and kurtosis was <2. The t test significance and variance were determined using Microsoft Excel. Skewness and kurtosis were determined using IBM SPSS Statistics for Windows v19.0 (IBM Corp., Armonk, NY, United States). Otherwise, the non-parametric Kolmogorov–Smirnov exact test was performed using GraphPad Prism 10.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was approved by QIMR Berghofer MRI Animal Ethics Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
AC: Investigation, Writing – original draft, Writing – review & editing. DF: Conceptualization, Writing – review & editing. KY: Investigation, Methodology, Writing – review & editing. CB: Data curation, Formal analysis, Methodology, Software, Supervision, Validation, Writing – review & editing. BT: Investigation, Writing – review & editing. WN: Investigation, Supervision, Writing – review & editing. SH: Investigation, Methodology, Writing – review & editing. JH: Conceptualization, Writing – review & editing. TL: Visualization, Writing – review & editing. DR: Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing. AS: Conceptualization, Data curation, Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The authors thank the Brazil Family Foundation (and others) for their generous philanthropic donations that helped set up the PC3 (BSL3) SARS-CoV-2 research facility at QIMR Berghofer MRI, as well as ongoing research into SARS-CoV-2, COVID-19 and long-COVID. A.S. was supported by the National Health and Medical Research Council (NHMRC) of Australia (Investigator grant APP1173880). D.F. was supported by the NHMRC (Ideas grant APP2030126).
Acknowledgments
The authors thank the following QIMRB staff; Dr. I. Anraku for management of the PC3 facility at QIMR Berghofer MRI, Dr. Viviana Lutzky for proof reading, Dr. Crystal Chang for histology services, the animal house staff for mouse breeding and agistment, and Dr. Gunter Hartel for assistance with statistics.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1441495/full#supplementary-material
Footnotes
1. ^https://github.com/Nanostring-Biostats/CellProfileLibrary/blob/master/Mouse/Adult/Lung_MCA.RData
2. ^https://github.com/Nanostring-Biostats/CellProfileLibrary/blob/master/Mouse/Adult/ImmuneAtlas_ImmGen_cellFamily.RData
References
Abu-Ismail, L., Taha, M. J. J., Abuawwad, M. T., Al-Bustanji, Y., Al-Shami, K., Nashwan, A., et al. (2023). COVID-19 and Anemia: what do we know so far? Hemoglobin 47, 122–129. doi: 10.1080/03630269.2023.2236546
Achan, J., Serwanga, A., Wanzira, H., Kyagulanyi, T., Nuwa, A., Magumba, G., et al. (2022). Current malaria infection, previous malaria exposure, and clinical profiles and outcomes of COVID-19 in a setting of high malaria transmission: an exploratory cohort study in Uganda. Lancet Microbe 3, e62–e71. doi: 10.1016/S2666-5247(21)00240-8
Ahmed, F. F., Reza, M. S., Sarker, M. S., Islam, M. S., Mosharaf, M. P., Hasan, S., et al. (2022). Identification of host transcriptome-guided repurposable drugs for SARS-CoV-1 infections and their validation with SARS-CoV-2 infections by using the integrated bioinformatics approaches. PLoS One 17:e0266124. doi: 10.1371/journal.pone.0266124
Ai, Y., Yan, B., and Wang, X. (2021). The oxidoreductases POR and CYB5R1 catalyze lipid peroxidation to execute ferroptosis. Mol. Cell. Oncol. 8:1881393. doi: 10.1080/23723556.2021.1881393
Albalat, E., Cavey, T., Leroyer, P., Ropert, M., Balter, V., and Loréal, O. (2021). Hfe Gene Knock-out in a mouse model of hereditary hemochromatosis affects bodily Iron isotope compositions. Front. Med. 8:711822. doi: 10.3389/fmed.2021.711822
Albertine, K. H., Wang, Z. M., and Michael, J. R. (1999). Expression of endothelial nitric oxide synthase, inducible nitric oxide synthase, and endothelin-1 in lungs of subjects who died with ARDS. Chest 116, 101S–102S. doi: 10.1378/chest.116.suppl_1.101S
Ali, Y. M., Carnell, G. W., Fumagalli, S., Mercurio, D., Seminara, S., Lynch, N. J., et al. (2024). Inhibition of the lectin pathway of complement activation reduces acute respiratory distress syndrome severity in a mouse model of SARS-CoV-2 infection. J. Infect. Dis. 229, 680–690. doi: 10.1093/infdis/jiad462
Ali, M. K., Kim, R. Y., Brown, A. C., Mayall, J. R., Karim, R., Pinkerton, J. W., et al. (2020). Crucial role for lung iron level and regulation in the pathogenesis and severity of asthma. Eur. Respir. J. 55:1901340. doi: 10.1183/13993003.01340-2019
Ali, M. K., Kim, R. Y., Karim, R., Mayall, J. R., Martin, K. L., Shahandeh, A., et al. (2017). Role of iron in the pathogenesis of respiratory disease. Int. J. Biochem. Cell Biol. 88, 181–195. doi: 10.1016/j.biocel.2017.05.003
Alisamir, M., Ebrahimi, M., and Rahim, F. (2022). Anemia in chronic obstructive pulmonary disease: A systematic review. Respir. Investig. 60, 510–521. doi: 10.1016/j.resinv.2022.03.006
Al-Kuraishy, H. M., Al-Gareeb, A. I., Kaushik, A., Kujawska, M., and Batiha, G. E. (2022). Hemolytic anemia in COVID-19. Ann. Hematol. 101, 1887–1895. doi: 10.1007/s00277-022-04907-7
Amarilla, A. A., Sng, J. D. J., Parry, R., Deerain, J. M., Potter, J. R., Setoh, Y. X., et al. (2021). A versatile reverse genetics platform for SARS-CoV-2 and other positive-strand RNA viruses. Nat. Commun. 12:3431. doi: 10.1038/s41467-021-23779-5
Andersen, C. T., Duggan, C. P., Manji, K., Seage, G. R., Iii, S. D., Perumal, N., et al. (2022). Iron supplementation and paediatric HIV disease progression: a cohort study among children receiving routine HIV care in Dar Es Salaam, Tanzania. Int. J. Epidemiol. 51, 1533–1543. doi: 10.1093/ije/dyac017
Andreen, N., Andersson, L.-M., Sundell, N., Gustavsson, L., and Westin, J. (2022). Mortality of COVID-19 is associated with comorbidity in patients with chronic obstructive pulmonary disease. Infect. Dis. 54, 508–513. doi: 10.1080/23744235.2022.2050422
Armas-González, E., Domínguez-Luis, M. J., Díaz-Martín, A., Arce-Franco, M., Castro-Hernández, J., Danelon, G., et al. (2018). Role of CXCL13 and CCL20 in the recruitment of B cells to inflammatory foci in chronic arthritis. Arthritis Res. Ther. 20:114. doi: 10.1186/s13075-018-1611-2
Aslam, M. F., Frazer, D. M., Faria, N., Bruggraber, S. F., Wilkins, S. J., Mirciov, C., et al. (2014). Ferroportin mediates the intestinal absorption of iron from a nanoparticulate ferritin core mimetic in mice. FASEB J. 28, 3671–3678. doi: 10.1096/fj.14-251520
Aslan, E. S., Aydın, H., Tekin, Y. K., Keleş, S., White, K. N., and Hekim, N. (2023). Association between iron metabolism and SARS-COV-2 infection, determined by ferritin, hephaestin and hypoxia-induced factor-1 alpha levels in COVID-19 patients. Mol. Biol. Rep. 50, 2471–2478. doi: 10.1007/s11033-022-08221-3
Bankhead, P., Loughrey, M. B., Fernández, J. A., Dombrowski, Y., Mcart, D. G., Dunne, P. D., et al. (2017). QuPath: open source software for digital pathology image analysis. Sci. Rep. 7:16878. doi: 10.1038/s41598-017-17204-5
Barton, J. C., Lee, P. L., West, C., and Bottomley, S. S. (2006). Iron overload and prolonged ingestion of iron supplements: clinical features and mutation analysis of hemochromatosis-associated genes in four cases. Am. J. Hematol. 81, 760–767. doi: 10.1002/ajh.20714
Bastin, A., Shiri, H., Zanganeh, S., Fooladi, S., Momeni Moghaddam, M. A., Mehrabani, M., et al. (2022). Iron Chelator or Iron supplement consumption in COVID-19? The role of Iron with severity infection. Biol. Trace Elem. Res. 200, 4571–4581. doi: 10.1007/s12011-021-03048-8
Bell, H., Berg, J. P., Undlien, D. E., Distante, S., Raknerud, N., Heier, H. E., et al. (2000). The clinical expression of hemochromatosis in Oslo, Norway. Excessive oral iron intake may lead to secondary hemochromatosis even in HFE C282Y mutation negative subjects. Scand. J. Gastroenterol. 35, 1301–1307. doi: 10.1080/003655200453665
Berg, V., Modak, M., Brell, J., Puck, A., Künig, S., Jutz, S., et al. (2020). Iron deprivation in human T cells induces nonproliferating accessory helper cells. Immuno Horizons 4, 165–177. doi: 10.4049/immunohorizons.2000003
Bergamaschi, G., Borrelli De Andreis, F., Aronico, N., Lenti, M. V., Barteselli, C., Merli, S., et al. (2021). Anemia in patients with Covid-19: pathogenesis and clinical significance. Clin. Exp. Med. 21, 239–246. doi: 10.1007/s10238-020-00679-4
Bhalla, S., Sharma, B., Smith, D., Boley, R., Mccluskey, C., Ilyas, Y., et al. (2021). Investigating unhealthy alcohol use as an independent risk factor for increased COVID-19 disease severity: observational cross-sectional study. JMIR Public Health Surveill. 7:e33022. doi: 10.2196/33022
Biasiotto, G., and Ferrari, F. (2022). Covidin, a possible new player between hepcidin and ferroportin in hypoxia and inflammation caused by COVID-19. J. Cell. Biochem. 123, 1701–1703. doi: 10.1002/jcb.30246
Bishop, C. R., Dumenil, T., Rawle, D. J., Le, T. T., Yan, K., Tang, B., et al. (2022). Mouse models of COVID-19 recapitulate inflammatory pathways rather than gene expression. PLoS Pathog. 18:e1010867. doi: 10.1371/journal.ppat.1010867
Bishop, C. R., Yan, K., Nguyen, W., Rawle, D. J., Tang, B., Larcher, T., et al. (2024). Microplastics dysregulate innate immunity in the SARS-CoV-2 infected lung. Front. Immunol. 15:1382655. doi: 10.3389/fimmu.2024.1382655
Bortolotti, M., Barcellini, W., and Fattizzo, B. (2023). Molecular pharmacology in complement-mediated hemolytic disorders. Eur. J. Haematol. 111, 326–336. doi: 10.1111/ejh.14026
Bradley, K. L., Stokes, C. A., Marciniak, S. J., Parker, L. C., and Condliffe, A. M. (2021). Role of unfolded proteins in lung disease. Thorax 76, 92–99. doi: 10.1136/thoraxjnl-2019-213738
Campione, E., Lanna, C., Cosio, T., Rosa, L., Conte, M. P., Iacovelli, F., et al. (2021). Lactoferrin against SARS-CoV-2: in vitro and in silico evidences. Front. Pharmacol. 12:666600. doi: 10.3389/fphar.2021.666600
Carolin, A., Yan, K., Bishop, C. R., Tang, B., Nguyen, W., Rawle, D. J., et al. (2024). Tracking inflammation resolution signatures in lungs after SARS-CoV-2 omicron BA.1 infection of K18-hACE2 mice. Biorxiv. doi: 10.1101/2024.03.12.584739
Castanza, A. S., Recla, J. M., Eby, D., Thorvaldsdóttir, H., Bult, C. J., and Mesirov, J. P. (2023). Extending support for mouse data in the molecular signatures database (MSigDB). Nat. Methods 20, 1619–1620. doi: 10.1038/s41592-023-02014-7
Cavalli, E., Petralia, M. C., Basile, M. S., Bramanti, A., Bramanti, P., Nicoletti, F., et al. (2020). Transcriptomic analysis of COVID-19 lungs and bronchoalveolar lavage fluid samples reveals predominant B cell activation responses to infection. Int. J. Mol. Med. 46, 1266–1273. doi: 10.3892/ijmm.2020.4702
Cavey, T., Latour, C., Island, M. L., Leroyer, P., Guggenbuhl, P., Coppin, H., et al. (2019). Spleen iron, molybdenum, and manganese concentrations are coregulated in hepcidin-deficient and secondary iron overload models in mice. FASEB J. 33, 11072–11081. doi: 10.1096/fj.201801381RR
Cavezzi, A., Troiani, E., and Corrao, S. (2020). COVID-19: hemoglobin, iron, and hypoxia beyond inflammation a narrative review. Clin. Pract. 10:1271. doi: 10.4081/cp.2020.1271
Charlebois, E., and Pantopoulos, K. (2023). Nutritional aspects of Iron in health and disease. Nutrients 15:2441. doi: 10.3390/nu15112441
Chaubey, G. K., Dilawari, R., Modanwal, R., Talukdar, S., Dhiman, A., Raje, C. I., et al. (2023). Excess iron aggravates the severity of COVID-19 infection. Free Radic. Biol. Med. 208, 186–193. doi: 10.1016/j.freeradbiomed.2023.08.011
Chen, Y., Fang, Z.-M., Yi, X., Wei, X., and Jiang, D.-S. (2023). The interaction between ferroptosis and inflammatory signaling pathways. Cell Death Dis. 14:205. doi: 10.1038/s41419-023-05716-0
Chen, X., Yu, C., Kang, R., and Tang, D. (2020). Iron metabolism in Ferroptosis. Front. Cell Dev. Biol. 8:590226. doi: 10.3389/fcell.2020.590226
Chenchula, S., Vidyasagar, K., Pathan, S., Sharma, S., Chavan, M. R., Bhagavathula, A. S., et al. (2023). Global prevalence and effect of comorbidities and smoking status on severity and mortality of COVID-19 in association with age and gender: a systematic review, meta-analysis and meta-regression. Sci. Rep. 13:6415. doi: 10.1038/s41598-023-33314-9
Choi, C. Y., Gadhave, K., Villano, J., Pekosz, A., Mao, X., and Jia, H. (2024). Generation and characterization of a humanized ACE2 mouse model to study long-term impacts of SARS-CoV-2 infection. J. Med. Virol. 96:e29349. doi: 10.1002/jmv.29349
Cohen, N., Breker, M., Bakunts, A., Pesek, K., Chas, A., Argemi, J., et al. (2017). Iron affects Ire1 clustering propensity and the amplitude of endoplasmic reticulum stress signaling. J. Cell Sci. 130, 3222–3233. doi: 10.1242/jcs.201715
Cormier, S. A., Yuan, S., Crosby, J. R., Protheroe, C. A., Dimina, D. M., Hines, E. M., et al. (2002). T(H)2-mediated pulmonary inflammation leads to the differential expression of ribonuclease genes by alveolar macrophages. Am. J. Respir. Cell Mol. Biol. 27, 678–687. doi: 10.1165/rcmb.4882
Crits-Christoph, A., Levy, J. I., Pekar, J. E., Goldstein, S. A., Singh, R., Hensel, Z., et al. (2023). Genetic tracing of market wildlife and viruses at the epicenter of the COVID-19 pandemic. BioRxiv. doi: 10.1101/2023.09.13.557637
Dahan, S., Segal, G., Katz, I., Hellou, T., Tietel, M., Bryk, G., et al. (2020). Ferritin as a marker of severity in COVID-19 patients: A fatal correlation. Isr. Med. Assoc. J. 22, 494–500.
Danaher, P., Kim, Y., Nelson, B., Griswold, M., Yang, Z., Piazza, E., et al. (2022). Advances in mixed cell deconvolution enable quantification of cell types in spatial transcriptomic data. Nat. Commun. 13:385. doi: 10.1038/s41467-022-28020-5
Darshan, D., Frazer, D. M., Wilkins, S. J., and Anderson, G. J. (2010). Severe iron deficiency blunts the response of the iron regulatory gene Hamp and pro-inflammatory cytokines to lipopolysaccharide. Haematologica 95, 1660–1667. doi: 10.3324/haematol.2010.022426
Davies, J. P., Sivadas, A., Keller, K. R., Roman, B. K., Wojcikiewicz, R. J. H., and Plate, L. (2024). Expression of SARS-CoV-2 nonstructural proteins 3 and 4 can tune the unfolded protein response in cell culture. J. Proteome Res. 23, 356–367. doi: 10.1021/acs.jproteome.3c00600
Deugnier, Y., Loreal, O., Carre, F., Duvallet, A., Zoulim, F., Vinel, J. P., et al. (2002). Increased body iron stores in elite road cyclists. Med. Sci. Sports Exerc. 34, 876–880. doi: 10.1097/00005768-200205000-00023
Dewey, K. G., and Oaks, B. M. (2017). U-shaped curve for risk associated with maternal hemoglobin, iron status, or iron supplementation. Am. J. Clin. Nutr. 106, 1694S–1702S. doi: 10.3945/ajcn.117.156075
Dharra, R., Kumar Sharma, A., and Datta, S. (2023). Emerging aspects of cytokine storm in COVID-19: the role of proinflammatory cytokines and therapeutic prospects. Cytokine 169:156287. doi: 10.1016/j.cyto.2023.156287
Dimmeler, S., and Zeiher, A. M. (1999). Nitric oxide–an endothelial cell survival factor. Cell Death Differ. 6, 964–968. doi: 10.1038/sj.cdd.4400581
Dixon, S. J., and Pratt, D. A. (2023). Ferroptosis: A flexible constellation of related biochemical mechanisms. Mol. Cell 83, 1030–1042. doi: 10.1016/j.molcel.2023.03.005
Dumenil, T., Le, T. T., Rawle, D. J., Yan, K., Tang, B., Nguyen, W., et al. (2023). Warmer ambient air temperatures reduce nasal turbinate and brain infection, but increase lung inflammation in the K18-hACE2 mouse model of COVID-19. Sci. Total Environ. 859:160163. doi: 10.1016/j.scitotenv.2022.160163
Dunaway, L. S., Nyshadham, S., Luse, M. A., Loeb, S. A., Goldfarb, A., and Isakson, B. (2023). Endothelial hemoglobin alpha mediates Iron regulation of endothelial nitric oxide. Blood 142:502. doi: 10.1182/blood-2023-181150
Dupic, F., Fruchon, S., Bensaid, M., Loreal, O., Brissot, P., Borot, N., et al. (2002). Duodenal mRNA expression of iron related genes in response to iron loading and iron deficiency in four strains of mice. Gut 51, 648–653. doi: 10.1136/gut.51.5.648
Ekiz, C., Agaoglu, L., Karakas, Z., Gurel, N., and Yalcin, I. (2005). The effect of iron deficiency anemia on the function of the immune system. Hematol. J. 5, 579–583. doi: 10.1038/sj.thj.6200574
Faria, B., Gaya Da Costa, M., Poppelaars, F., Franssen, C. F. M., Pestana, M., Berger, S. P., et al. (2019). Administration of Intravenous Iron Formulations Induces Complement Activation in-vivo. Front. Immunol. 10:1885. doi: 10.3389/fimmu.2019.01885
Feng, H., Schorpp, K., Jin, J., Yozwiak, C. E., Hoffstrom, B. G., Decker, A. M., et al. (2020). Transferrin receptor is a specific Ferroptosis marker. Cell Rep. 30, 3411–3423.e7. doi: 10.1016/j.celrep.2020.02.049
Fernández, J. J., Mancebo, C., Garcinuño, S., March, G., Alvarez, Y., Alonso, S., et al. (2024a). Innate IRE1α-XBP1 activation by viral single-stranded RNA and its influence on lung cytokine production during SARS-CoV-2 pneumonia. Genes & Immunity 25, 43–54. doi: 10.1038/s41435-023-00243-6
Fernández, J. J., Marín, A., Rosales, R., Penrice-Randal, R., Mlcochova, P., Alvarez, Y., et al. (2024b). The IRE1α-XBP1 arm of the unfolded protein response is a host factor activated in SARS-CoV-2 infection. Biochim. Biophys. Acta 1870:167193. doi: 10.1016/j.bbadis.2024.167193
Finnerty, J. P., Hussain, A. B. M. A., Ponnuswamy, A., Kamil, H. G., and Abdelaziz, A. (2023). Asthma and COPD as co-morbidities in patients hospitalised with Covid-19 disease: a global systematic review and meta-analysis. BMC Pulm. Med. 23:462. doi: 10.1186/s12890-023-02761-5
Frazer, D. M., Wilkins, S. J., Darshan, D., Mirciov, C. S. G., Dunn, L. A., and Anderson, G. J. (2017). Ferroportin is essential for Iron absorption during suckling, but is Hyporesponsive to the regulatory hormone Hepcidin. Cell. Mol. Gastroenterol. Hepatol. 3, 410–421. doi: 10.1016/j.jcmgh.2016.12.002
Fumagalli, V., Ravà, M., Marotta, D., Di Lucia, P., Laura, C., Sala, E., et al. (2022). Administration of aerosolized SARS-CoV-2 to K18-hACE2 mice uncouples respiratory infection from fatal neuroinvasion. Sci. Immunol. 7:eabl9929. doi: 10.1126/sciimmunol.abl9929
Fuqua, B. K., Lu, Y., Darshan, D., Frazer, D. M., Wilkins, S. J., Wolkow, N., et al. (2014). The multicopper ferroxidase hephaestin enhances intestinal iron absorption in mice. PLoS One 9:e98792. doi: 10.1371/journal.pone.0098792
Galy, B., Conrad, M., and Muckenthaler, M. (2024). Mechanisms controlling cellular and systemic iron homeostasis. Nat. Rev. Mol. Cell Biol. 25, 133–155. doi: 10.1038/s41580-023-00648-1
Garcia-Gasalla, M., Berman-Riu, M., Pons, J., Rodríguez, A., Iglesias, A., Martínez-Pomar, N., et al. (2022). Hyperinflammatory state and low T1 adaptive immune response in severe and critical acute COVID-19 patients. Front. Med. 9:828678. doi: 10.3389/fmed.2022.828678
Gattermann, N., Muckenthaler, M. U., Kulozik, A. E., Metzgeroth, G., and Hastka, J. (2021). The evaluation of Iron deficiency and Iron overload. Dtsch. Arztebl. Int. 118, 847–856. doi: 10.3238/arztebl.m2021.0290
Georg, P., Astaburuaga-García, R., Bonaguro, L., Brumhard, S., Michalick, L., Lippert, L. J., et al. (2022). Complement activation induces excessive T cell cytotoxicity in severe COVID-19. Cell 185, 493–512.e25. doi: 10.1016/j.cell.2021.12.040
Gerayeli, F. V., Milne, S., Cheung, C., Li, X., Yang, C. W. T., Tam, A., et al. (2021). COPD and the risk of poor outcomes in COVID-19: A systematic review and meta-analysis. eClinicalMedicine 33:100789. doi: 10.1016/j.eclinm.2021.100789
Gil-Etayo, F. J., Garcinuño, S., Utrero-Rico, A., Cabrera-Marante, O., Arroyo-Sanchez, D., Mancebo, E., et al. (2022). An early Th1 response is a key factor for a favorable COVID-19 evolution. Biomedicines 10:296. doi: 10.3390/biomedicines10020296
Ginzburg, Y. Z. (2019). “Chapter two - Hepcidin-ferroportin axis in health and disease” in Vitamins and hormones. ed. G. Litwack (London: Academic Press), 17–45.
Giorgi, G., D'anna, M. C., and Roque, M. E. (2015). Iron homeostasis and its disruption in mouse lung in iron deficiency and overload. Exp. Physiol. 100, 1199–1216. doi: 10.1113/ep085166
Girelli, D., Marchi, G., Busti, F., and Vianello, A. (2021). Iron metabolism in infections: focus on COVID-19. Semin. Hematol. 58, 182–187. doi: 10.1053/j.seminhematol.2021.07.001
Goh, S. H., Josleyn, M., Lee, Y. T., Danner, R. L., Gherman, R. B., Cam, M. C., et al. (2007). The human reticulocyte transcriptome. Physiol. Genomics 30, 172–178. doi: 10.1152/physiolgenomics.00247.2006
Gregory, J. M., Slaughter, J. C., Duffus, S. H., Smith, T. J., Lestourgeon, L. M., Jaser, S. S., et al. (2021). COVID-19 severity is tripled in the diabetes community: A prospective analysis of the Pandemic's impact in type 1 and type 2 diabetes. Diabetes Care 44, 526–532. doi: 10.2337/dc20-2260
Gross, C. M., Rafikov, R., Kumar, S., Aggarwal, S., Ham Iii, P. B., Meadows, M. L., et al. (2015). Endothelial nitric oxide synthase deficient mice are protected from lipopolysaccharide induced acute lung injury. PLoS One 10:e0119918. doi: 10.1371/journal.pone.0119918
Guimarães, L. M. F., Rossini, C. V. T., and Lameu, C. (2021). Implications of SARS-Cov-2 infection on eNOS and iNOS activity: consequences for the respiratory and vascular systems. Nitric Oxide 111-112, 64–71. doi: 10.1016/j.niox.2021.04.003
Gupta, Y., Maciorowski, D., Medernach, B., Becker, D. P., Durvasula, R., Libertin, C. R., et al. (2022). Iron dysregulation in COVID-19 and reciprocal evolution of SARS-CoV-2: Natura nihil frustra facit. J. Cell. Biochem. 123, 601–619. doi: 10.1002/jcb.30207
Gutiérrez, L., Caballero, N., Fernández-Calleja, L., Karkoulia, E., and Strouboulis, J. (2020). Regulation of GATA1 levels in erythropoiesis. IUBMB Life 72, 89–105. doi: 10.1002/iub.2192
Habib, H. M., Ibrahim, S., Zaim, A., and Ibrahim, W. H. (2021). The role of iron in the pathogenesis of COVID-19 and possible treatment with lactoferrin and other iron chelators. Biomed. Pharmacother. 136:111228. doi: 10.1016/j.biopha.2021.111228
Han, X., Wang, R., Zhou, Y., Fei, L., Sun, H., Lai, S., et al. (2018). Mapping the mouse cell atlas by microwell-Seq. Cell 172, 1091–1107.e17. doi: 10.1016/j.cell.2018.02.001
Hanson, A. L., Mulè, M. P., Ruffieux, H., Mescia, F., Bergamaschi, L., Pelly, V. S., et al. (2024). Iron dysregulation and inflammatory stress erythropoiesis associates with long-term outcome of COVID-19. Nat. Immunol. 25, 471–482. doi: 10.1038/s41590-024-01754-8
Haschka, D., Hoffmann, A., and Weiss, G. (2021). Iron in immune cell function and host defense. Semin. Cell Dev. Biol. 115, 27–36. doi: 10.1016/j.semcdb.2020.12.005
Hegelund, M. H., Glenthøj, A., Ryrsø, C. K., Ritz, C., Dungu, A. M., Sejdic, A., et al. (2022). Biomarkers for iron metabolism among patients hospitalized with community-acquired pneumonia caused by infection with SARS-CoV-2, bacteria, and influenza. APMIS 130, 590–596. doi: 10.1111/apm.13259
Helman, S. L., Wilkins, S. J., Mckeating, D. R., Perkins, A. V., Cuffe, J. S. M., Hartel, G., et al. (2022). A novel ferritin-Core analog is a safe and effective alternative to Oral ferrous Iron for treating Iron deficiency during pregnancy in mice. J. Nutr. 152, 714–722. doi: 10.1093/jn/nxab363
Hempel, J. C., Poppelaars, F., Gaya Da Costa, M., Franssen, C. F. M., De Vlaam, T. P. G., Daha, M. R., et al. (2016). Distinct in vitro complement activation by various intravenous Iron preparations. Am. J. Nephrol. 45, 49–59. doi: 10.1159/000451060
Hess, S. Y., Owais, A., Jefferds, M. E. D., Young, M. F., Cahill, A., and Rogers, L. M. (2023). Accelerating action to reduce anemia: review of causes and risk factors and related data needs. Ann. N. Y. Acad. Sci. 1523, 11–23. doi: 10.1111/nyas.14985
Hofer, U. (2022). Animal model of long COVID? Nat. Rev. Microbiol. 20:446. doi: 10.1038/s41579-022-00761-6
Hoffmann, A., Haschka, D., Valente De Souza, L., Tymoszuk, P., Seifert, M., Von Raffay, L., et al. (2021). Baseline iron status and presence of anaemia determine the course of systemic Salmonella infection following oral iron supplementation in mice. EBioMedicine 71:103568. doi: 10.1016/j.ebiom.2021.103568
Hsu, C. C., Senussi, N. H., Fertrin, K. Y., and Kowdley, K. V. (2022). Iron overload disorders. Hepatology. Communications 6, 1842–1854. doi: 10.1002/hep4.2012
Huerga Encabo, H., Grey, W., Garcia-Albornoz, M., Wood, H., Ulferts, R., Aramburu, I. V., et al. (2021). Human erythroid progenitors are directly infected by SARS-CoV-2: implications for emerging erythropoiesis in severe COVID-19 patients. Stem Cell Rep. 16, 428–436. doi: 10.1016/j.stemcr.2021.02.001
Hussien, R. S., Jabuk, S. I., Altaee, Z. M., and Al-Maamori, A. M. (2023). Review of anemia: types and causes. Eur. J. Res. Dev. Sustain. 4, 1–3.
Jablonski, K. A., Amici, S. A., Webb, L. M., Ruiz-Rosado Jde, D., Popovich, P. G., Partida-Sanchez, S., et al. (2015). Novel markers to delineate murine M1 and M2 macrophages. PLoS One 10:e0145342. doi: 10.1371/journal.pone.0145342
Jacobs, W., Lammens, M., Kerckhofs, A., Voets, E., Van San, E., Van Coillie, S., et al. (2020). Fatal lymphocytic cardiac damage in coronavirus disease 2019 (COVID-19): autopsy reveals a ferroptosis signature. ESC Heart Fail 7, 3772–3781. doi: 10.1002/ehf2.12958
Jerkic, M., Szaszi, K., Laffey, J. G., Rotstein, O., and Zhang, H. (2023). Key role of mesenchymal stromal cell interaction with macrophages in promoting repair of lung injury. Int. J. Mol. Sci. 24:3376. doi: 10.3390/ijms24043376
Ji, L., Li, T., Chen, H., Yang, Y., Lu, E., Liu, J., et al. (2023). The crucial regulatory role of type I interferon in inflammatory diseases. Cell Biosci. 13:230. doi: 10.1186/s13578-023-01188-z
Jiang, Y., Li, C., Wu, Q., An, P., Huang, L., Wang, J., et al. (2019). Iron-dependent histone 3 lysine 9 demethylation controls B cell proliferation and humoral immune responses. Nat. Commun. 10:2935. doi: 10.1038/s41467-019-11002-5
Johnson, B. J., Le, T. T., Dobbin, C. A., Banovic, T., Howard, C. B., Flores Fde, M., et al. (2005). Heat shock protein 10 inhibits lipopolysaccharide-induced inflammatory mediator production. J. Biol. Chem. 280, 4037–4047. doi: 10.1074/jbc.M411569200
Kameritsch, P., and Renkawitz, J. (2020). Principles of leukocyte migration strategies. Trends Cell Biol. 30, 818–832. doi: 10.1016/j.tcb.2020.06.007
Kasvosve, I., Gangaidzo, I., Gomo, Z., and Gordeuk, V. (2000). African Iron overload. Acta Clin. Belg. 55, 88–93. doi: 10.1080/17843286.2000.11754276
Ko, J. Y., Danielson, M. L., Town, M., Derado, G., Greenlund, K. J., Kirley, P. D., et al. (2020). Risk factors for coronavirus disease 2019 (COVID-19)–associated hospitalization: COVID-19–associated hospitalization surveillance network and behavioral risk factor surveillance system. Clin. Infect. Dis. 72, e695–e703. doi: 10.1093/cid/ciaa1419
Kobayashi, M., Kato, H., Hada, H., Itoh-Nakadai, A., Fujiwara, T., Muto, A., et al. (2017). Iron-heme-Bach1 axis is involved in erythroblast adaptation to iron deficiency. Haematologica 102, 454–465. doi: 10.3324/haematol.2016.151043
Kohgo, Y., Ikuta, K., Ohtake, T., Torimoto, Y., and Kato, J. (2008). Body iron metabolism and pathophysiology of iron overload. Int. J. Hematol. 88, 7–15. doi: 10.1007/s12185-008-0120-5
Korneeva, N., Khalil, M. I., Ghosh, I., Fan, R., Arnold, T., and De Benedetti, A. (2023). SARS-CoV-2 viral protein Nsp2 stimulates translation under normal and hypoxic conditions. Virol. J. 20:55. doi: 10.1186/s12985-023-02021-2
Kowarik, M. C., Cepok, S., Sellner, J., Grummel, V., Weber, M. S., Korn, T., et al. (2012). CXCL13 is the major determinant for B cell recruitment to the CSF during neuroinflammation. J. Neuroinflammation 9:93. doi: 10.1186/1742-2094-9-93
Kozlova, E., Sherstyukova, E., Sergunova, V., Kozlov, A., Gudkova, O., Inozemtsev, V., et al. (2022). The toxic influence of excess free Iron on red blood cells in the biophysical experiment: An in vitro study. J. Toxicol. 2022, 7113958–7113916. doi: 10.1155/2022/7113958
Kronstein-Wiedemann, R., Stadtmuller, M., Traikov, S., Georgi, M., Teichert, M., Yosef, H., et al. (2022). SARS-CoV-2 infects red blood cell progenitors and dysregulates hemoglobin and Iron metabolism. Stem Cell Rev. Rep. 18, 1809–1821. doi: 10.1007/s12015-021-10322-8
Kuvibidila, S. R., Velez, M., Gardner, R., Penugonda, K., Chandra, L. C., and Yu, L. (2012). Iron deficiency reduces serum and in vitro secretion of interleukin-4 in mice independent of altered spleen cell proliferation. Nutr. Res. 32, 107–115. doi: 10.1016/j.nutres.2011.12.005
Lands, R., and Isang, E. (2017). Secondary hemochromatosis due to chronic Oral Iron supplementation. Case Rep. Hematol. 2017, 2494167–2494163. doi: 10.1155/2017/2494167
Lanser, L., Burkert, F. R., Bellmann-Weiler, R., Schroll, A., Wildner, S., Fritsche, G., et al. (2021a). Dynamics in Anemia development and dysregulation of Iron homeostasis in hospitalized patients with COVID-19. Meta 11:653. doi: 10.3390/metabo11100653
Lanser, L., Fuchs, D., Scharnagl, H., Grammer, T., Kleber, M. E., Marz, W., et al. (2021b). Anemia of chronic disease in patients with cardiovascular disease. Front. Cardiovasc. Med. 8:666638. doi: 10.3389/fcvm.2021.666638
Li, D., Jing, J., Dong, X., Zhang, C., Wang, J., and Wan, X. (2023). Activating transcription factor 3: A potential therapeutic target for inflammatory pulmonary diseases. Immunity Inflamm. Dis. 11:e1028. doi: 10.1002/iid3.1028
Li, Y., Li, Y., Xu, S., Chen, Y., Zhou, P., Hu, T., et al. (2022). N-Acylethanolamine acid amidase (NAAA) exacerbates psoriasis inflammation by enhancing dendritic cell (DCs) maturation. Pharmacol. Res. 185:106491. doi: 10.1016/j.phrs.2022.106491
Li, S., Rouphael, N., Duraisingham, S., Romero-Steiner, S., Presnell, S., Davis, C., et al. (2014). Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat. Immunol. 15, 195–204. doi: 10.1038/ni.2789
Li, L., Xia, Y., Yuan, S., Li, F., Xie, X., Luo, Y., et al. (2021). Iron deprivation restrains the differentiation and pathogenicity of T helper 17 cell. J. Leukoc. Biol. 110, 1057–1067. doi: 10.1002/JLB.3MA0821-015R
Liao, Z., Wang, C., Tang, X., Yang, M., Duan, Z., Liu, L., et al. (2024). Human transferrin receptor can mediate SARS-CoV-2 infection. Proc. Natl. Acad. Sci. 121:e2317026121. doi: 10.1073/pnas.2317026121
Liberzon, A., Birger, C., Thorvaldsdóttir, H., Ghandi, M., Mesirov, J. P., and Tamayo, P. (2015). The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425. doi: 10.1016/j.cels.2015.12.004
Liu, S., Bhattacharya, S., Han, A., Suragani, R. N. V. S., Zhao, W., Fry, R. C., et al. (2008). Haem-regulated eIF2α kinase is necessary for adaptive gene expression in erythroid precursors under the stress of iron deficiency. Br. J. Haematol. 143, 129–137. doi: 10.1111/j.1365-2141.2008.07293.x
Ma, H., Lu, L., Xia, H., Xiang, Q., Sun, J., Xue, J., et al. (2020). Circ0061052 regulation of FoxC1/snail pathway via miR-515-5p is involved in the epithelial-mesenchymal transition of epithelial cells during cigarette smoke-induced airway remodeling. Sci. Total Environ. 746:141181. doi: 10.1016/j.scitotenv.2020.141181
Manrai, M., Dawra, S., Kapoor, R., Srivastava, S., and Singh, A. (2022). Anemia in cirrhosis: An underestimated entity. World J. Clin. Cases 10, 777–789. doi: 10.12998/wjcc.v10.i3.777
Marhaeni, W., Felicia, F. V., Sumadi Jap, A. L., Hartoyo, E., and Andayani, P. (2023). Comparing serum ferritin levels during COVID-19 infection and recovery period in pediatric patients with transfusion-dependent thalassemia, a single-center study. Front. Med. 10:1056599. doi: 10.3389/fmed.2023.1056599
Miao, Q., Ku, A. T., Nishino, Y., Howard, J. M., Rao, A. S., Shaver, T. M., et al. (2014). Tcf3 promotes cell migration and wound repair through regulation of lipocalin 2. Nat. Commun. 5:4088. doi: 10.1038/ncomms5088
Mirciov, C. S., Wilkins, S. J., Dunn, L. A., Anderson, G. J., and Frazer, D. M. (2017). Characterization of putative erythroid regulators of Hepcidin in mouse models of Anemia. PLoS One 12:e0171054. doi: 10.1371/journal.pone.0171054
Mizumura, K., and Gon, Y. (2021). Iron-regulated reactive oxygen species production and programmed cell death in chronic obstructive pulmonary disease. Antioxidants 10:1569. doi: 10.3390/antiox10101569
Mohus, R. M., Flatby, H., Liyanarachi, K. V., Dewan, A. T., Solligård, E., Damås, J. K., et al. (2022). Iron status and the risk of sepsis and severe COVID-19: a two-sample Mendelian randomization study. Sci. Rep. 12:16157. doi: 10.1038/s41598-022-20679-6
Mojzisch, A., and Brehm, M. A. (2021). The manifold cellular functions of von Willebrand factor. Cells 10:2351. doi: 10.3390/cells10092351
Morgan, M. S., Yan, K., Le, T. T., Johnston, R. A., Amarilla, A. A., Muller, D. A., et al. (2023). Monoclonal antibodies specific for SARS-CoV-2 spike protein suitable for multiple applications for current variants of concern. Viruses 15:139. doi: 10.3390/v15010139
Mu, Q., Chen, L., Gao, X., Shen, S., Sheng, W., Min, J., et al. (2021). The role of iron homeostasis in remodeling immune function and regulating inflammatory disease. Sci. Bull. 66, 1806–1816. doi: 10.1016/j.scib.2021.02.010
Muranov, K. O. (2024). Fenton reaction in vivo and in vitro possibilities and limitations. Biochemistry 89, S112–S126. doi: 10.1134/S0006297924140074
Naidu, S. A. G., Clemens, R. A., and Naidu, A. S. (2023). SARS-CoV-2 infection dysregulates host Iron (Fe)-redox homeostasis (Fe-R-H): role of Fe-redox regulators, Ferroptosis inhibitors, anticoagulants, and Iron-chelators in COVID-19 control. J. Dietary Suppl. 20, 312–371. doi: 10.1080/19390211.2022.2075072
Nakagawa, H., Tamura, T., Mitsuda, Y., Goto, Y., Kamiya, Y., Kondo, T., et al. (2014). Inverse correlation between serum interleukin-6 and iron levels among Japanese adults: a cross-sectional study. BMC Hematol. 14:6. doi: 10.1186/2052-1839-14-6
Ni, S., Yuan, Y., Kuang, Y., and Li, X. (2022). Iron metabolism and immune regulation. Front. Immunol. 13:816282. doi: 10.3389/fimmu.2022.816282
Nienaber, A., Uyoga, M. A., Dolman-Macleod, R. C., and Malan, L. (2023). Iron status and supplementation during tuberculosis. Microorganisms 11:785. doi: 10.3390/microorganisms11030785
Niethamer, T. K., Levin, L. I., Morley, M. P., Babu, A., Zhou, S., and Morrisey, E. E. (2023). Atf3 defines a population of pulmonary endothelial cells essential for lung regeneration. eLife 12:e83835. doi: 10.7554/eLife.83835
Nousis, L., Kanavaros, P., and Barbouti, A. (2023). Oxidative stress-induced cellular senescence: is labile Iron the connecting link? Antioxidants 12:1250. doi: 10.3390/antiox12061250
Ohuma, E. O., Jabin, N., Young, M. F., Epie, T., Martorell, R., Peña-Rosas, J. P., et al. (2023). Association between maternal haemoglobin concentrations and maternal and neonatal outcomes: the prospective, observational, multinational, INTERBIO-21st fetal study. Lancet Haematol. 10, e756–e766. doi: 10.1016/S2352-3026(23)00170-9
Olivier, T., Blomet, J., and Desmecht, D. (2023). Central role of lung macrophages in SARS-CoV-2 physiopathology: a cross-model single-cell RNA-seq perspective. Front. Immunol. 14:1197588. doi: 10.3389/fimmu.2023.1197588
Omara, F. O., and Blakley, B. R. (1994). The IgM and IgG antibody responses in iron-deficient and iron-loaded mice. Biol. Trace Elem. Res. 46, 155–161. doi: 10.1007/BF02790075
Onur, S. T., Altın, S., Sokucu, S. N., Fikri, B., Barça, T., Bolat, E., et al. (2021). Could ferritin level be an indicator of COVID-19 disease mortality? J. Med. Virol. 93, 1672–1677. doi: 10.1002/jmv.26543
Papadopoulos, V. P., Koutroulos, M.-V., Zikoudi, D.-G., Bakola, S.-A., Avramidou, P., Touzlatzi, N., et al. (2021). Diabetes-related acute metabolic emergencies in COVID-19 patients: a systematic review and meta-analysis. Diabetol. Int. 12, 445–459. doi: 10.1007/s13340-021-00502-9
Pasricha, S.-R., Tye-Din, J., Muckenthaler, M. U., and Swinkels, D. W. (2021). Iron deficiency. Lancet 397, 233–248. doi: 10.1016/S0140-6736(20)32594-0
Pereira, D. I., Aslam, M. F., Frazer, D. M., Schmidt, A., Walton, G. E., Mccartney, A. L., et al. (2015). Dietary iron depletion at weaning imprints low microbiome diversity and this is not recovered with oral Nano Fe(III). Microbiology 4, 12–27. doi: 10.1002/mbo3.213
Petri, B., Broermann, A., Li, H., Khandoga, A. G., Zarbock, A., Krombach, F., et al. (2010). von Willebrand factor promotes leukocyte extravasation. Blood 116, 4712–4719. doi: 10.1182/blood-2010-03-276311
Pfeifhofer-Obermair, C., Tymoszuk, P., Nairz, M., Schroll, A., Klais, G., Demetz, E., et al. (2021). Regulation of Th1 T cell differentiation by Iron via upregulation of T cell immunoglobulin and mucin containing Protein-3 (TIM-3). Front. Immunol. 12:637809. doi: 10.3389/fimmu.2021.637809
Praveen, M., Jain, N., Raizada, N., Sharma, S., Narang, S., and Madhu, S. V. (2020). Anaemia in patients with type 2 diabetes mellitus without nephropathy is related to iron deficiency. Diabetes Metab. Syndr. Clin. Res. Rev. 14, 1837–1840. doi: 10.1016/j.dsx.2020.09.020
Prow, N. A., Hirata, T. D. C., Tang, B., Larcher, T., Mukhopadhyay, P., Alves, T. L., et al. (2019). Exacerbation of chikungunya virus rheumatic immunopathology by a high Fiber diet and butyrate. Front. Immunol. 10:2736. doi: 10.3389/fimmu.2019.02736
Qi, D., Tang, X., He, J., Wang, D., Zhao, Y., Deng, W., et al. (2016). Omentin protects against LPS-induced ARDS through suppressing pulmonary inflammation and promoting endothelial barrier via an Akt/eNOS-dependent mechanism. Cell Death Dis. 7, –e2360. doi: 10.1038/cddis.2016.265
Qiu, B., Zandkarimi, F., Saqi, A., Castagna, C., Tan, H., Sekulic, M., et al. (2024). Fatal COVID-19 pulmonary disease involves ferroptosis. Nat. Commun. 15:3816. doi: 10.1038/s41467-024-48055-0
Queiroz-Andrade, M., Blasbalg, R., Ortega, C. D., Rodstein, M. A., Baroni, R. H., Rocha, M. S., et al. (2009). MR imaging findings of iron overload. Radiographics 29, 1575–1589. doi: 10.1148/rg.296095511
Rawle, D. J., Le, T. T., Dumenil, T., Yan, K., Tang, B., Nguyen, W., et al. (2021). ACE2-lentiviral transduction enables mouse SARS-CoV-2 infection and mapping of receptor interactions. PLoS Pathog. 17:e1009723. doi: 10.1371/journal.ppat.1009723
Recalcati, S., Gammella, E., Buratti, P., Doni, A., Anselmo, A., Locati, M., et al. (2019). Macrophage ferroportin is essential for stromal cell proliferation in wound healing. Haematologica 104, 47–58. doi: 10.3324/haematol.2018.197517
Ren, Y., Li, L., Wang, M.-M., Cao, L.-P., Sun, Z.-R., Yang, Z.-Z., et al. (2021). Pravastatin attenuates sepsis-induced acute lung injury through decreasing pulmonary microvascular permeability via inhibition of Cav-1/eNOS pathway. Int. Immunopharmacol. 100:108077. doi: 10.1016/j.intimp.2021.108077
Reuben, A., Chung, J. W., Lapointe, R., and Santos, M. M. (2017). The hemochromatosis protein HFE 20 years later: An emerging role in antigen presentation and in the immune system. Immunity Inflamm. Dis. 5, 218–232. doi: 10.1002/iid3.158
Rishi, G., Secondes, E. S., and Nathan Subramaniam, V. (2018). Hemochromatosis: evaluation of the dietary iron model and regulation of hepcidin. Biochim. Biophys. Acta Mol. basis Dis. 1864, 2550–2556. doi: 10.1016/j.bbadis.2018.05.005
Ristić, S., Milić, S., Tatalović, T., Bilobrk, M., Rončević, D., Ćurko-Cofek, B., et al. (2023). The influence of hemochromatosis gene (HFE) mutations on SARS-CoV-2 susceptibility and COVID-19 severity. Balkan Med. J. 40, 229–231. doi: 10.4274/balkanmedj.galenos.2023.2023-1-39
Rosa, R. G., Cavalcanti, A. B., Azevedo, L. C. P., Veiga, V. C., De Souza, D., Dos Santos, R. D. R. M., et al. (2023). Association between acute disease severity and one-year quality of life among post-hospitalisation COVID-19 patients: coalition VII prospective cohort study. Intensive Care Med. 49, 166–177. doi: 10.1007/s00134-022-06953-1
Roth-Walter, F. (2022). Iron-deficiency in atopic diseases: innate immune priming by allergens and Siderophores. Front. Allergy 3:859922. doi: 10.3389/falgy.2022.859922
Roth-Walter, F., Pacios, L. F., Bianchini, R., and Jensen-Jarolim, E. (2017). Linking iron-deficiency with allergy: role of molecular allergens and the microbiome. Metallomics 9, 1676–1692. doi: 10.1039/c7mt00241f
Russell, C. D., Lone, N. I., and Baillie, J. K. (2023). Comorbidities, multimorbidity and COVID-19. Nat. Med. 29, 334–343. doi: 10.1038/s41591-022-02156-9
Russo, A., Tellone, E., Barreca, D., Ficarra, S., and Lagana, G. (2022). Implication of COVID-19 on erythrocytes functionality: red blood cell biochemical implications and Morpho-functional aspects. Int. J. Mol. Sci. 23:2171. doi: 10.3390/ijms23042171
Sana, A., and Avneesh, M. (2022). Identification of hematological and inflammatory parameters associated with disease severity in hospitalized patients of COVID-19. J. Family Med. Prim. Care 11, 260–264. doi: 10.4103/jfmpc.jfmpc_941_21
Santiesteban-Lores, L. E., Amamura, T. A., Da Silva, T. F., Midon, L. M., Carneiro, M. C., Isaac, L., et al. (2021). A double edged-sword - the complement system during SARS-CoV-2 infection. Life Sci. 272:119245. doi: 10.1016/j.lfs.2021.119245
Santos, M., Clevers, H., De Sousa, M., and Marx, J. J. (1998). Adaptive response of iron absorption to anemia, increased erythropoiesis, iron deficiency, and iron loading in beta2-microglobulin knockout mice. Blood 91, 3059–3065. doi: 10.1182/blood.V91.8.3059.3059_3059_3065
Schmidt, S. M. (2020). The role of iron in viral infections. Front. Biosci. 25, 893–911. doi: 10.2741/4839
Schwarz, K. W., Murray, M. T., Sylora, R., Sohn, R. L., and Dulchavsky, S. A. (2002). Augmentation of wound healing with translation initiation factor eIF4E mRNA. J. Surg. Res. 103, 175–182. doi: 10.1006/jsre.2002.6360
Shahriari-Felordi, M., Alikhani, H. K., Hashemian, S.-M. R., Hassan, M., and Vosough, M. (2022). Mini review ATF4 and GRP78 as novel molecular targets in ER-stress modulation for critical COVID-19 patients. Mol. Biol. Rep. 49, 1545–1549. doi: 10.1007/s11033-021-07071-9
Shivshankar, P., Fekry, B., Eckel-Mahan, K., and Wetsel, R. A. (2020). Circadian clock and complement immune system—complementary control of physiology and pathology? Front. Cell. Infect. Microbiol. 10:418. doi: 10.3389/fcimb.2020.00418
Silaghi-Dumitrescu, R., Patrascu, I., Lehene, M., and Bercea, I. (2023). Comorbidities of COVID-19 patients. Medicina 59:1393. doi: 10.3390/medicina59081393
Singh, J., Anantharaj, A., Panwar, A., Rani, C., Bhardwaj, M., Kumar, P., et al. (2023). BA.1, BA.2 and BA.2.75 variants show comparable replication kinetics, reduced impact on epithelial barrier and elicit cross-neutralizing antibodies. PLoS Pathog. 19:e1011196. doi: 10.1371/journal.ppat.1011196
Soares, V. C., Dias, S. S. G., Santos, J. C., Azevedo-Quintanilha, I. G., Moreira, I. B. G., Sacramento, C. Q., et al. (2023). Inhibition of the SREBP pathway prevents SARS-CoV-2 replication and inflammasome activation. Life Sci. Alliance 6:e202302049. doi: 10.26508/lsa.202302049
Sohal, A., and Kowdley, K. V. (2024). A review of new concepts in Iron overload. Gastroenterol. Hepatol. 20, 98–107.
Sonnweber, T., Grubwieser, P., Sahanic, S., Böhm, A. K., Pizzini, A., Luger, A., et al. (2022). The impact of Iron Dyshomeostasis and Anaemia on long-term pulmonary recovery and persisting symptom burden after COVID-19: A prospective observational cohort study. Meta 12:546. doi: 10.3390/metabo12060546
Stewart, R., Yan, K., Ellis, S. A., Bishop, C. R., Dumenil, T., Tang, B., et al. (2023). SARS-CoV-2 omicron BA.5 and XBB variants have increased neurotropic potential over BA.1 in K18-hACE2 mice and human brain organoids. Front. Microbiol. 14:1320856. doi: 10.3389/fmicb.2023.1320856
Suchdev, P. S., Williams, A. M., Mei, Z., Flores-Ayala, R., Pasricha, S.-R., Rogers, L. M., et al. (2017). Assessment of iron status in settings of inflammation: challenges and potential approaches†‡§. Am. J. Clin. Nutr. 106, 1626S–1633S. doi: 10.3945/ajcn.117.155937
Sun, S.-H., Chen, Q., Gu, H.-J., Yang, G., Wang, Y.-X., Huang, X.-Y., et al. (2020). A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe 28, 124–133.e4. doi: 10.1016/j.chom.2020.05.020
Taneri, P. E., Gómez-Ochoa, S. A., Llanaj, E., Raguindin, P. F., Rojas, L. Z., Roa-Díaz, Z. M., et al. (2020). Anemia and iron metabolism in COVID-19: a systematic review and meta-analysis. Eur. J. Epidemiol. 35, 763–773. doi: 10.1007/s10654-020-00678-5
Teh, M. R., Frost, J. N., Armitage, A. E., and Drakesmith, H. (2021). Analysis of Iron and Iron-interacting protein dynamics during T-cell activation. Front. Immunol. 12:714613. doi: 10.3389/fimmu.2021.714613
Tojo, K., Sugawara, Y., Oi, Y., Ogawa, F., Higurashi, T., Yoshimura, Y., et al. (2021). The U-shaped association of serum iron level with disease severity in adult hospitalized patients with COVID-19. Sci. Rep. 11:13431. doi: 10.1038/s41598-021-92921-6
Tough, D. F. (2004). Type I interferon as a link between innate and adaptive immunity through dendritic cell stimulation. Leuk. Lymphoma 45, 257–264. doi: 10.1080/1042819031000149368
Vassiliou, A. G., Zacharis, A., Keskinidou, C., Jahaj, E., Pratikaki, M., Gallos, P., et al. (2021). Soluble angiotensin converting enzyme 2 (ACE2) is upregulated and soluble endothelial nitric oxide synthase (eNOS) is downregulated in COVID-19-induced acute respiratory distress syndrome (ARDS). Pharmaceuticals 14:695. doi: 10.3390/ph14070695
Vogt, A. C. S., Arsiwala, T., Mohsen, M., Vogel, M., Manolova, V., and Bachmann, M. F. (2021). On Iron metabolism and its regulation. Int. J. Mol. Sci. 22:4591. doi: 10.3390/ijms22094591
Walker, E. M., and Walker, S. M. (2000). Effects of iron overload on the immune system. Ann. Clin. Lab. Sci. 30, 354–365.
Wang, J. M., Li, H., Xu, L., Kim, H., Qiu, Y., and Zhang, K. (2023). Boosting UPR transcriptional activator XBP1 accelerates acute wound healing. PNAS Nexus 2:pgad050. doi: 10.1093/pnasnexus/pgad050
Ward, J. L., Torres-Gonzalez, M., and Ammons, M. C. B. (2022). The influence of viral infections on Iron homeostasis and the potential for Lactoferrin as a therapeutic in the age of the SARS-CoV-2 pandemic. Nutrients 14:3090. doi: 10.3390/nu14153090
WHO (2024a). Anaemia. Available at: https://www.who.int/news-room/fact-sheets/detail/anaemia (Accessed August 29, 2024).
WHO (2024b). COVID-19 dashboard. Available at: https://data.who.int/dashboards/covid19/cases?n=c (Accessed August 29, 2024).
Wideman, S. K., Frost, J. N., Richter, F. C., Naylor, C., Lopes, J. M., Viveiros, N., et al. (2023). Cellular iron governs the host response to malaria. PLoS Pathog. 19:e1011679. doi: 10.1371/journal.ppat.1011679
Wojciechowska, M., Wisniewski, O. W., Kolodziejski, P., and Krauss, H. (2021). Role of hepcidin in physiology and pathophysiology. Emerging experimental and clinical evidence. J. Physiol. Pharmacol. 72, 23–33. doi: 10.26402/jpp.2021.1.03
Wollmann, M., Gerzson, B. M. C., Schwert, V., Figuera, R. W., and De Oliveira Ritzel, G. (2014). Reticulocyte maturity indices in iron deficiency anemia. Rev. Bras. Hematol. Hemoter. 36, 25–28. doi: 10.5581/1516-8484.20140009
Worobey, M., Levy, J. I., Malpica Serrano, L., Crits-Christoph, A., Pekar, J. E., Goldstein, S. A., et al. (2022). The Huanan seafood wholesale market in Wuhan was the early epicenter of the COVID-19 pandemic. Science 377, 951–959. doi: 10.1126/science.abp8715
Wu, C. T., Lidsky, P. V., Xiao, Y., Cheng, R., Lee, I. T., Nakayama, T., et al. (2023). SARS-CoV-2 replication in airway epithelia requires motile cilia and microvillar reprogramming. Cell 186:e120, 112–130.e20. doi: 10.1016/j.cell.2022.11.030
Wu, J. Y., Liu, M.-Y., Hung, K.-C., Hsu, W.-H., Tsai, Y.-W., Liu, T.-H., et al. (2023). Nutritional deficiency anemia and post-acute sequelae in patients with severe acute respiratory syndrome coronavirus 2 infection: A six-month retrospective cohort analysis of 30 892 patients. J. Med. Virol. 95:e29246. doi: 10.1002/jmv.29246
Xiao, M. T., Ellsworth, C. R., and Qin, X. (2024). Emerging role of complement in COVID-19 and other respiratory virus diseases. Cell. Mol. Life Sci. 81:94. doi: 10.1007/s00018-024-05157-8
Yan, K., Dumenil, T., Tang, B., Le, T. T., Bishop, C. R., Suhrbier, A., et al. (2022). Evolution of ACE2-independent SARS-CoV-2 infection and mouse adaption after passage in cells expressing human and mouse ACE2. Virus Evol. 8:63. doi: 10.1093/ve/veac063
Yan, K., Rawle, D. J., Le, T. T., and Suhrbier, A. (2021). Simple rapid in vitro screening method for SARS-CoV-2 anti-virals that identifies potential cytomorbidity-associated false positives. Virol. J. 18:123. doi: 10.1186/s12985-021-01587-z
Yang, Y., Miller, H., Byazrova, M. G., Cndotti, F., Benlagha, K., Camara, N. O. S., et al. (2024). The characterization of CD8+ T-cell responses in COVID-19. Emerg. Microb. Infect. 13:2287118. doi: 10.1080/22221751.2023.2287118
Ying, J. F., Lu, Z. B., Fu, L. Q., Tong, Y., Wang, Z., Li, W. F., et al. (2021). The role of iron homeostasis and iron-mediated ROS in cancer. Am. J. Cancer Res. 11, 1895–1912
Yu, J., Yuan, X., Chen, H., Chaturvedi, S., Braunstein, E. M., and Brodsky, R. A. (2020). Direct activation of the alternative complement pathway by SARS-CoV-2 spike proteins is blocked by factor D inhibition. Blood 136, 2080–2089. doi: 10.1182/blood.2020008248
Zakrzewski, M., Wilkins, S. J., Helman, S. L., Brilli, E., Tarantino, G., Anderson, G. J., et al. (2022). Supplementation with Sucrosomial® iron leads to favourable changes in the intestinal microbiome when compared to ferrous sulfate in mice. Biometals 35, 27–38. doi: 10.1007/s10534-021-00348-3
Zhang, X.-D., Liu, Z.-Y., Wang, M.-S., Guo, Y.-X., Wang, X.-K., Luo, K., et al. (2023). Mechanisms and regulations of ferroptosis. Front. Immunol. 14:1269451. doi: 10.3389/fimmu.2023.1269451
Zhang, W., Shi, K., Geng, Q., Ye, G., Aihara, H., and Li, F. (2022). Structural basis for mouse receptor recognition by SARS-CoV-2 omicron variant. Proc. Natl. Acad. Sci. 119:e2206509119. doi: 10.1073/pnas.2206509119
Zhao, Y., Shi, L., Jiang, Z., Zeng, N., Mei, H., Lu, Y., et al. (2023). The phenotype and prediction of long-term physical, mental and cognitive COVID-19 sequelae 20 months after recovery, a community-based cohort study in China. Mol. Psychiatry 28, 1793–1801. doi: 10.1038/s41380-023-01951-1
Zhou, S., Li, H., and Li, S. (2022). The associations of Iron related biomarkers with risk, clinical severity and mortality in SARS-CoV-2 patients: A Meta-analysis. Nutrients 14:3406. doi: 10.3390/nu14163406
Keywords: iron deficiency, iron loading, SARS-CoV-2, omicron XBB, C57BL/6J mice, inflammation, lung, RNA-Seq
Citation: Carolin A, Frazer D, Yan K, Bishop CR, Tang B, Nguyen W, Helman SL, Horvat J, Larcher T, Rawle DJ and Suhrbier A (2024) The effects of iron deficient and high iron diets on SARS-CoV-2 lung infection and disease. Front. Microbiol. 15:1441495. doi: 10.3389/fmicb.2024.1441495
Edited by:
Claudia Maria Trombetta, University of Siena, ItalyReviewed by:
Gaurav Kumar Chaubey, Institute of Microbial Technology (CSIR), IndiaMilan Surjit, Translational Health Science and Technology Institute (THSTI), India
Copyright © 2024 Carolin, Frazer, Yan, Bishop, Tang, Nguyen, Helman, Horvat, Larcher, Rawle and Suhrbier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel J. Rawle, RGFuaWVsLlJhd2xlQHFpbXJiZXJnaG9mZXIuZWR1LmF1; Andreas Suhrbier, QW5kcmVhcy5TdWhyYmllckBxaW1yYmVyZ2hvZmVyLmVkdS5hdQ==
†These authors share first authorship
‡These authors share last authorship
 Agnes Carolin
Agnes Carolin David Frazer
David Frazer Kexin Yan1
Kexin Yan1 Cameron R. Bishop
Cameron R. Bishop Wilson Nguyen
Wilson Nguyen Jay Horvat
Jay Horvat Thibaut Larcher
Thibaut Larcher Daniel J. Rawle
Daniel J. Rawle Andreas Suhrbier
Andreas Suhrbier