- Faculty of Agricultural, Environmental and Food Sciences, Free University of Bolzano, Bolzano, Italy
Biofertilizers based on plant growth promoting rhizobacteria (PGPR) are nowadays gaining increasingly attention as a modern tool for a more sustainable agriculture due to their ability in ameliorating root nutrient acquisition. For many years, most research was focused on the screening and characterization of PGPR functioning as nitrogen (N) or phosphorus (P) biofertilizers. However, with the increasing demand for food using far fewer chemical inputs, new investigations have been carried out to explore the potential use of such bacteria also as potassium (K), sulfur (S), zinc (Zn), or iron (Fe) biofertilizers. In this review, we update the use of PGPR as biofertilizers for a smarter and more sustainable crop production and deliberate the prospects of using microbiome engineering-based methods as potential tools to shed new light on the improvement of plant mineral nutrition. The current era of omics revolution has enabled the design of synthetic microbial communities (named SynComs), which are emerging as a promising tool that can allow the formulation of biofertilizers based on PGPR strains displaying multifarious and synergistic traits, thus leading to an increasingly efficient root acquisition of more than a single essential nutrient at the same time. Additionally, host-mediated microbiome engineering (HMME) leverages advanced omics techniques to reintroduce alleles coding for beneficial compounds, reinforcing positive plant-microbiome interactions and creating plants capable of producing their own biofertilizers. We also discusses the current use of PGPR-based biofertilizers and point out possible avenues of research for the future development of more efficient biofertilizers for a smarter and more precise crop fertilization. Furthermore, concerns have been raised about the effectiveness of PGPR-based biofertilizers in real field conditions, as their success in controlled experiments often contrasts with inconsistent field results. This discrepancy highlights the need for standardized protocols to ensure consistent application and reliable outcomes.
1 Introduction
It is now a fact that the global request of agricultural food products is progressively rising because of the increase in the World population. In fact, as far as FAO projections are considered, human population inhabiting the planet is expected to reach more than 10 billion people by 2070 (Daniel et al., 2022). In this scenario, the agricultural sector is urged to increase crops’ yield to meet the standard of food security. Indeed, since the green revolution (middle of 20th century), the increased inputs of chemical fertilizer proved itself as an efficient strategy to enhance agricultural productivity (Fasusi et al., 2021). Unfortunately, due to the limited nutrient use efficiency, crops are able to exploit only a portion of the applied fertilizers, while the remaining part (generally much more than half) remains in the soil being then, very often, transformed in not available forms through different mechanisms (e.g., precipitation, adsorption, leaching, volatilization) (Zhang et al., 2018). It is evident that this phenomenon, in addition to be less sustainable, particularly in a long period vision, poses serious risks for both the environment and human beings. In this regard as well as in the framework of the 17 sustainable development goals launched by the EU,1 the implementation of innovative solutions for a more sustainable agriculture appears to be mandatory in the coming years. In this context, the exploitation of the potentialities lying in the so termed rhizosphere management seems extremely promising. The concept rhizosphere management refers to a series of actions aimed at increasing in this specific volume of soil the available fractions of the different nutrients for an equilibrate crop development and, then, production (Zia et al., 2021; Kumawat et al., 2022). Interestingly, beneficial soil microorganisms, also accounted as plant growth-promoting rhizobacteria (PGPR), are known to be a promising tool in this sense thanks to their capability to significantly ameliorate the edaphic conditions of the rhizosphere soil (Pii et al., 2015a). In this regard, it should be highlighted that for crops a more balanced availability of nutrients, specifically in the rhizosphere, is not only strategic to guarantee the production (food security) in a more sustainable way, but also crucial for both the quality of primary production (food safety) (Tomasi et al., 2015; Sambo et al., 2019) and the composition of its epiphytic microbial community (Valentinuzzi et al., 2021), being this latter fundamental for the post-harvest characteristics of the edible plants tissues and their shelf life.
In a previous review, we have already postulated the need of deepening our understanding about the molecular and biochemical mechanisms underpinning plant-microbe interactions (Pii et al., 2015a) as a condition for a possible wider and more effective use of PGPRs in agriculture, particularly in a context of precision agriculture and its greater environmental sustainability. In these last years, many PGPR strains have been isolated and characterized for their potential usefulness in the management of the rhizosphere (Fasusi et al., 2021). It is interesting to highlight that some of them are nowadays commonly used at the field scale as biofertilizers to achieve the objectives of higher food security and greater agricultural sustainability. Moreover, the results achievable with the application of these biofertilizers fit perfectly into the concept of a smart agriculture characterized by higher crop resilience and low external inputs that, even when essential, can be also tailored to the actual needs of the crops and to the variability of the agricultural land area (Cesco et al., 2023).
Thus, considering recent scientific advancements in microbiology and biotechnology, this review incorporates insights from 158 relevant research papers published between 1996 and 2024. Notably, the majority of these papers (84%) were published in the last decade (2014–2024), reflecting the latest developments in the field. Accordingly, the review is organized into sections that highlight the crucial roles of plant-PGPR interactions in crop fertilization. It begins with a comprehensive exploration of PGPR as biofertilizers, focusing on their ability to enhance soil fertility and crop productivity by improving nutrient availability. The review then examines how essential nutrients (e.g., N, P, K, S, Zn, Fe) are mobilized and used by plants, discussing innovative biological strategies to enhance their availability, nutrient roles in plant metabolism, their soil forms, and uptake mechanisms. Additionally, the review highlights the potential of PGPR-based biofertilizers to boost nutrient availability and plant growth. It emphasizes the successes and challenges of these biofertilizers, underscoring the need for technologies like SynComs and genetic engineering to develop high-performing biofertilizers and engineered plants for enhanced nutrient acquisition and improved soil conditions. Finally, the review addresses skepticism about PGPR-based biofertilizers due to inconsistent field results compared to controlled experiments, and it calls for standardized protocols to ensure consistent application and reliability, facilitating their transition to practical agriculture.
In a view of an ever more efficient and eco-friendly agriculture, this research is especially important for farmer communities committed to sustainable agriculture systems. Adopting PGPR-based biofertilizers can enhance nutrient use efficiency, minimize environmental impacts, and boost both crop yield and quality (Kumawat et al., 2023). This research supports the development of innovative and sustainable agricultural methods that address increasing food demand while protecting the environment and ensuring the long-term viability of farming operations.
2 Plant growth-promoting rhizobacteria as biofertilizers
About 20 years ago, the term biofertilizer was meant to define “a substance containing living microorganisms which, when applied to seeds, plant surfaces, or soil, colonizes the rhizosphere or the interior of the plant and promotes growth by increasing the supply or availability of primary nutrients to the host plant” (Vessey, 2003). Through the years, several pieces of research have been carried out and, thanks to the knowledge acquired, one of the most recent definition, that is more in line with the content of this review, describes biofertilizer as “a product containing beneficial microorganisms with the potential to improve soil fertility and crop productivity by enhancing nutrient availability” (Atieno et al., 2020). Since PGPR have been broadly reported as beneficial bacteria, they have been used worldwide as biofertilizers and characterized for their beneficial properties. The most reported effects are mainly ascribed to the involvement of members of the genera Azospirillum, Rhizobium, Bacillus, Pseudomonas, Burkholderia, Enterobacter, Herbaspirillum, Bradyrhizobium, Pantoea, Paenibacillus, Azotobacter, and Serratia (Tabassum et al., 2017; Bhadrecha et al., 2023; Kumawat et al., 2024). However, to be considered an ideal biofertilizer with the potential to enhance plant nutrition and development upon inoculation, a PGPR strain should present the following characteristics:
1. It should possess a high rhizosphere colonization rate upon inoculation (Backer et al., 2018).
2. It should be able to establish a stable and long-term colonization with the host plant (Liu et al., 2019).
3. It should show a maximal, consistent and reproducible efficacy under a range of field conditions (Herrmann and Lesueur, 2013).
4. It should be able to face intense competition with indigenous soil microbes (Backer et al., 2018).
5. It should promote plant growth by improving nutrient availability at rhizosphere level (Aloo et al., 2022), similarly to a localized fertilization.
6. It should present broad-spectrum versatility that encompasses a wide range of environmental adaptations (Mitter et al., 2021).
7. It must be environmentally friendly and consistent with the requirements of sustainable practices (Keswani et al., 2019).
8. It should be classified as safe, posing low threat to human health, soil and live communities (Keswani et al., 2019) as desired in a context of One Health.
It is interesting to note that PGPR can also directly promote plant growth by providing phytohormones or signaling molecules, or they can indirectly influence plant health by synthesizing bioactive compounds with potential as antimicrobials or stress tolerance, hence earning the designation of biostimulants (Kaushal et al., 2023). These compounds can modulate the root system architecture development, improve the photosynthetic capacity of a plant, or activate the antioxidant defense system (Rosier et al., 2018; Grover et al., 2021). However, since the focus of this review is primarily on PGPR that can turn sparingly available fractions of essential nutrients into more accessible forms for plants, PGPR with biostimulating actions will not be considered and discussed.
For this reason, PGPR-based biofertilizers can be classified based on the specific mineral nutrient they contribute to promote the availability for plants (Table 1). For instance, some PGPR strains are known to facilitate different processes including nitrogen (N) fixation (i.e., transforming atmospheric N2 into NH3 or NH4+), solubilization of nutrients (i.e., turning insoluble forms of P into H2PO4−, insoluble K pools into the free ionic form [K+], sparingly available Zn sources into the free ionic species [Zn2+]), oxidation of substrates (i.e., producing SO42− via oxidation of S0-containing sources), and/or metal chelation/complexation (e.g., exuding siderophores, phenolic compounds and/or organic acids to scavenge Fe3+ and other nutrients) (Colombo et al., 2014; Malusá and Vassilev, 2014; Mitter et al., 2021; Aloo et al., 2022). Moreover, it is important to mention that some strains of PGPR acting as biofertilizers, may have the ability to increase at the same time the supply to plants of more than one essential nutrient. Therefore, it is interesting to highlight that the action of these bacteria, whether single- or multi-element, is specifically localized in the rhizosphere, making them an extremely valuable tool for a precision agriculture context. In fact, the increasingly chance to use the endogenous nutritional resources of soil locally significantly reduces the need to broadcast synthetic fertilizers (Vacheron et al., 2013; Pii et al., 2015a, 2016; Rosier et al., 2018). Furthermore, neglecting or not giving appropriate attention to the PGPR strains that exert direct effects on crop growth and health, particularly in the context of more efficient and more sustainable agriculture, would certainly represent a missed opportunity.
3 Plant nutrient acquisition in a context of plant-biofertilizer interaction
3.1 The case study of nitrogen
Nitrogen, accounting for 1–5% of the whole plant dry biomass, is a key component of amino acids, proteins, and chlorophyll being, therefore, essential for plant growth and development (Jiang et al., 2020). The largest reservoir of N in the world is found in the atmosphere, mostly as nitrogen gas (N2), which can only be used by plants after transformation through chemical [via lightning action, CNF, (Barth et al., 2023)] or biological N2 fixation (BNF). This latter process can be performed by diazotrophs prokaryotes (N2 fixers), including symbiotic bacteria, as well as free-living or associative bacteria (Figure 1A). These bacteria encompass nif genes encoding the nitrogenase enzyme, which catalyzes the reduction of N2 to NH4+ (Soumare et al., 2020). Indeed, the bacteria-derived NH4+, if available in the rhizosphere, can then be also taken up by roots through plasma membrane transporters belonging to the AMT1 subfamily of the ammonium transporter/methylammonium permease family by exploiting either NH4+-uniport or NH3/H+ co-transport (Loque and Wirén, 2004; Neuhauser and Ludewig, 2014). In this regard, it should be mentioned that pieces of evidence showing alternative routes for NH4+ movement across the plasma membrane have been gathered through the years. In particular, it was observed that NH4+ can be also taken up by plants through non-selective cation channels (NSCC) and K+ specific channels, as for instance AKT1 (Coskun et al., 2013; Esteban et al., 2016). Nevertheless, it is worth mentioning that these not specific mechanisms have been suggested to play a predominant role in NH4+ acquisition only when the external concentration of the nutrient is higher than 1–2 mM (Muratore et al., 2021), being thus responsible for the onset of NH4+ toxicity in plants.
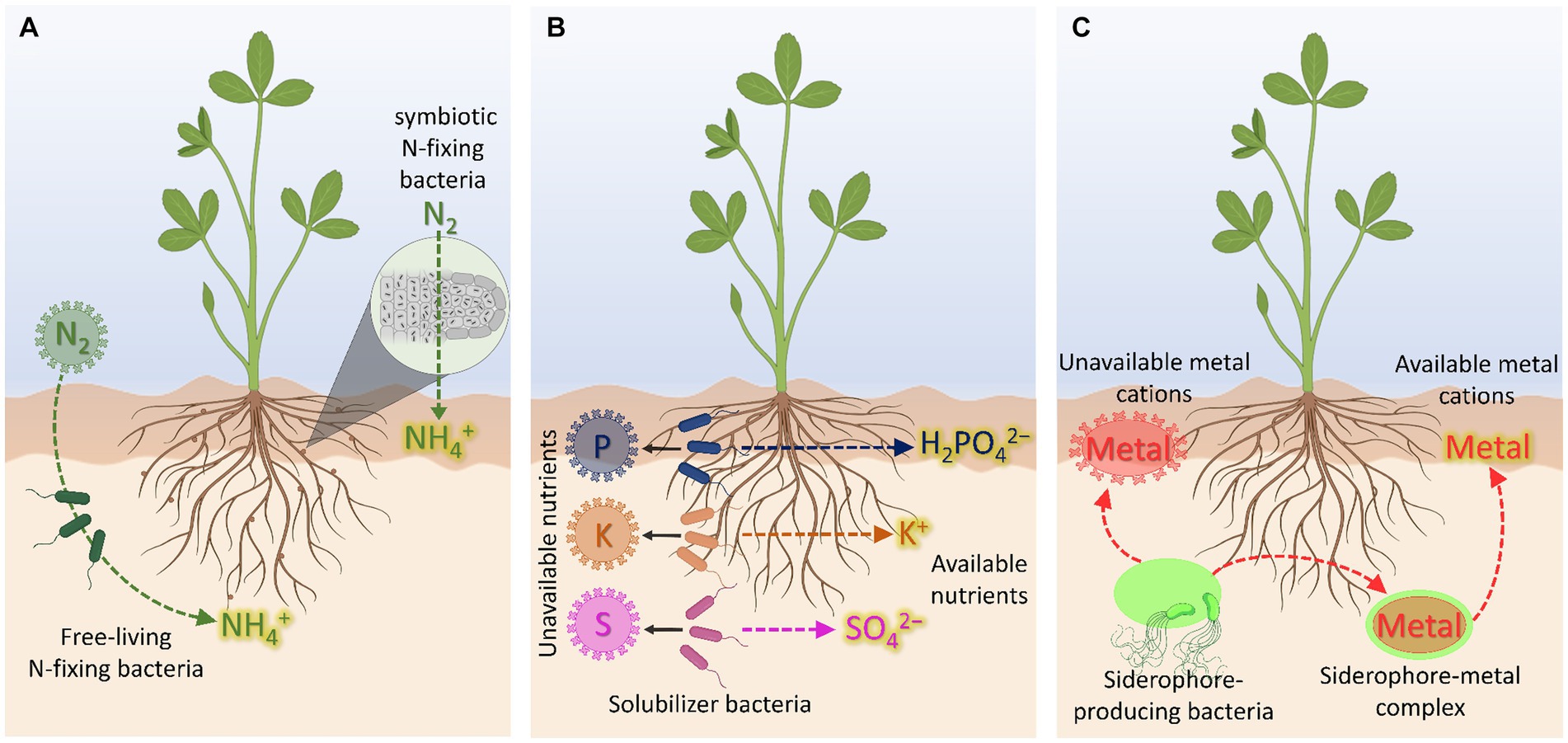
Figure 1. Biofertilizer-mediated nutrient availability. (A) N-biofertilizers. (B) P-, K-, and S-biofertilizers. (C) Cation-chelating biofertilizers based on siderophore-producing bacteria. Created with BioRender (https://biorender.com/).
The most studied symbiotic N2-fixing bacteria are rhizobia, which can establish symbiotic relationships with leguminous plants by forming specialized root nodule organs. Rhizobia, once enclosed in these nodules, experience a microaerobic environment, where nanomolar levels of oxygen (20–50 nM) are necessary for the nif gene expression and consequently, ensure high amounts of fixed N2 (Ledermann et al., 2021). It is estimated that the overall contribution of BNF in rhizobia-legumes systems is between 50 and 80% accounting for at least 70 million metric tons of N fixed per year (Sindhu et al., 2019). For this reason, highly effective strains of rhizobia such as Rhizobium, Bradyrhizobium, and Sinorhizobium are often used as biofertilizers in most major legume crops thanks to their ability to fix N2 in amounts as high as the amounts provided by synthetic N-fertilizers. In 2022, the global biofertilizer market was valued at USD 2.67 billion, whereby USD 1.92 billion was represented rhizobia-based biofertilizers, while the remaining USD 750 million constituted biofertilizers based on free-living N-fixing bacteria, P-solubilizing bacteria, K-solubilizing bacteria, and other PGPR. Moreover, their demand is estimated to grow by a further 12.3% compounding annual growth rate (CAGR) to reach about USD 6.97 billion by 2030.2
Differently of symbiotic rhizobia, free-living associative bacteria inhabit the surfaces or the interstitial spaces of the host plant and use the root exudates as carbon sources to fix N2 for their own use, providing the excess to the host (Guo et al., 2023). Several free-living N-fixing bacteria, including members from the genera Azospirillum, Azotobacter, Herbaspirillum, Bacillus, Clostridium, Burkholderia, and Gluconobacter, have been reported to fix N2 in the rhizosphere and bulk soil. These bacteria are particularly important for N nutrition in non-leguminous plants. Nonetheless, their contribution on BNF is more limited, providing up to 25% (13–22 kg N ha−1 yr.−1) of the total N in crops like rice, wheat and maize (Ladha et al., 2016). In this context, an attractive avenue to improve BNF in non-leguminous crops has been the use of genetic approaches to engineer root nodulation with symbiotic bacteria also in cereals. However, none of the attempts performed to date has succeeded (Pankievicz et al., 2019). On the other hand, notably progress has been made in genetic engineering strategies toward better N-fixing bacteria. Thus, by understanding the heterologous expression of the nif gene cluster in Klebsiella oxytoca, Azotobacter vinelandii, and Paenibacillus polymyxa, many scientists have been able to transfer their N-fixation trait to non-diazotrophic hosts (Li and Chen, 2020). For instance, the metabolic engineering of A. vinelandii resulted in a constitutive expression of nif genes with an increased production of NH4+. Thanks to this improvement, an enhanced growth of not N-fertilized cucumber plants has been recorded in plants when root colonized by the engineered A. vinelandii strain (Ambrosio et al., 2017). It is interesting to highlight that increased concentrations of NH4+ were also reported by (Pii et al., 2019) in the association between Azospirillum brasilense and maize plants. The authors also described that Azospirillum counteracted the plant response to NO3− uptake as a result of the activation of processes linked with NH4+ fluxes and assimilation.
Recently, Rago et al. (2019) proposed to use N-fixing bacteria in association with electrochemical systems for the reduction of N2 to NH4+ at low energy cost, in a process called bioelectrochemical N fixation (e-BNF). This process aims at the production of microbial biomass through simultaneous N2 and CO2 fixation using electricity as only energy input, thus opening a new path toward local on-demand N-biofertilizer production, based on renewable power. In this regard, it is worth to mention that by using genetic manipulation in combination with e-BNF, (Dong et al., 2021) integrated a modified nif gene cluster into the genome of the non-diazotrophic cyanobacterium Synechococcus elongatus and, thereafter an e-BNF system was used for NH4+ production. The authors reported that the e-BNF led to an increased production of NH4+ which was 21 times higher than that generated from solely photosynthesis-driven N fixation. Accordingly, these strategies provide novel technological feasibility to raise BNF inputs while alleviating the negative impacts of synthetic N-fertilizers (e.g., low crop N-efficiency, leaching, volatilization, use of not renewable energy for the synthesis).
3.2 The case study of phosphorus
Phosphorus is an essential nutrient representing the 0.1–0.8% of the dry matter and playing crucial roles in photosynthesis, energy transfer, respiration, and biosynthesis of molecules (Vance et al., 2003). In soil, P is present in both inorganic (i.e., free ionic species and minerals) and organic forms (i.e., phosphates monoesters and diesters, polyphosphates, and phosphonates), being the mineral and organic fractions not usable by plants as they are (Ducousso-Détrez et al., 2022). Moreover, it should be noted that the free ionic species have a strong affinity for soil particles being, therefore, readily retained in soil. As a consequence, only a minimal part of P in soil (about the 0.1%) is available for plant acquisition (Lambers, 2022). At root level, P is taken up as inorganic phosphate (H2PO4−/HPO42−, Pi) against the electrochemical gradient established at the plasma membrane (Hinsinger, 2001). The acquisition of Pi from the soil solution is mediated by high affinity transporters, encoded by the gene family PHT1 (Naureen et al., 2018), whose energy demand is sustained by the activity of plasma membrane H+-ATPase (Młodzińska and Zboińska, 2016). On the other hand, it is well known that, with respect to the amount of P commonly supplied to a crop with chemical fertilizers, the majority undergoes precipitation and leaching processes, limiting then the fraction actually used by the crop to only 10–20% (Ibrahim et al., 2022). In this context, the use of P-solubilizing bacteria (PSB) can nowadays represent a valid opportunity not only for an efficient mining and solubilization of P sources endogenously present in soil but also to limit the application of fertilizers characterized by such low use efficiency (Figure 1B). Moreover, if the localized effect of the strain (depending essentially on the root colonization by the bacteria) is considered, the sustainability of the entire process appears even more evident.
From the point of view of the mechanisms, the P solubilization mediated by PSB include the production and release of organic acids, phenolic compounds, siderophores, hydroxyl ions, protons, and CO2 leading to soil acidification and/or ion exchange of P by carboxylates (Alori et al., 2017). In fact, significant advancements including the isolation and characterization of bacterial genes have demonstrated that the glucose dehydrogenase (GDH, encoded by the gcd gene) and its cofactor pyrroloquinoline quinone (PQQ, encoded by the pqq operon) mediate the secretion of gluconic acid associated with P solubilization in Gram-negative bacteria (Rasul et al., 2019). Other mechanisms governing the inorganic P-solubilization by PSB include the production of hydrolytic enzymes encoded by the pyrophosphatase (ppa) and exopolyphosphatase (ppx) genes (Wu et al., 2022). However, with respect to the component of the mechanism connected with the organic acids exudation, it has been recently demonstrated, by using Leonard’s jars and Enterobacter sp., that the organic acids mainly involved in a soil–plant system (i.e., oxalic and citric acids) did not correspond to those released by the same bacterium in the highest concentrations under in vitro conditions (i.e., ketoglutaric and malic acids) (Zuluaga et al., 2023b). This experience clearly shows that the combination of the information acquired in in vitro experiments with that in conditions closer to the cultivated soil (i.e., in vivo) is crucial for the identification of the bacteria with the best P-solubilizing attributes for the field level application (Elhaissoufi et al., 2023).
Once solubilized, the crops benefit from the increased P availability enhancing their total biomass accumulation and productivity (Zuluaga et al., 2021; Wang and Song, 2022; Elhaissoufi et al., 2023). A large number of PSB has been described in this sense, including species of Burkholderia, Enterobacter, Bacillus, Pseudomonas, Rhizobium, Ralstonia, Serratia, Azotobacter, Paenibacillus, and Erwinia (Zuluaga et al., 2020; Timofeeva et al., 2022). However, to better exploit the abilities of these bacteria, it becomes necessary a deeper understanding of their modes of action (including the regulation and elicitation of the interesting traits) and the development of strategies capable to boost their biofertilization properties. In this scenario, considering that the repeated (year after year) application of P fertilizers has led in the long period to a P accumulation in many agricultural soils (also known as legacy P, Doydora et al., 2020), the current strategies are focusing on the ability of PSB to mobilize legacy P supplying the nutrient to plants, either alone or in combination with external P inputs. The results have shown that PSB can enhance P uptake of plant by increasing the immediately plant-available P pool in rhizosphere soil (Long and Wasaki, 2023). However, the supply of limited doses of exogenous P can potentiate the effect of PSB by solubilizing higher amounts of endogenous native Ca-, Fe- and Al-bound P (Alam et al., 2023). The need for supplemental P with PSB is expected, since these bacteria also require P and other essential elements for their own metabolic processes (Doydora et al., 2020).
Finally, it is important to mention what highlighted in a meta-analysis conducted by (Zutter et al., 2022) considering 104 articles published between April 1976 and May 2021 related to “phosphate solubilizing bacteria” and “plant growth.” The authors raised the following issues:
i. biomass is generally chosen as the best indicative parameter to evaluate a plant P-status. However, the meta-analysis showed that plant growth promotion is not always related to improved P-uptake. Therefore, the use of multiple imaging techniques could also be implemented as a useful tool to assess P supply by PSB;
ii. the use of PSB consortia does not significantly provide an added benefit to plant growth and P-uptake compared to single strains. Thus, the combination of several PSB should be designed with care;
iii. strains of Burkholderia sp. and Enterobacter sp. have shown better effects on plant P-uptake and biomass than Bacillus sp. and Pseudomonas sp., but the use of these two strains is restricted due to their danger to human health. However, it has been assured that some species among these genera are unlikely to harm humans and, therefore, they should be used as safe P-biofertilizers in agriculture (Sawana et al., 2014).
With respect to biotechnology application, few experiences of bioengineering PSB are described in literature. For instance, the incorporation of a pqq gene cluster in Herbaspirillum seropedicae resulted in the activation of gluconic acid secretion, which conferred the ability of P solubilization and the improvement of N and P status in inoculated rice plants (Wagh et al., 2014, 2016). In addition to its intrinsic ability to solubilize inorganic P, the phosphobacterium 9320-SD was transformed with a bacterial gene encoding phytase and this allowed the mineralization of organic phosphate into plant-available P (Liu et al., 2015). In this regard, the manipulation of key genes through genome editing approaches could be a good strategy for enhancing the potential of PGPB as biofertilizers. Nonetheless, oncoming investigations should also include field trials under variable conditions to assess the real potential of genetically modified bacterial strains.
3.3 The case study of potassium
Potassium, ranging from 0.5 to 6% of the plant dry matter, is the third more represented essential nutrient in plants and it is implicated in crucial metabolic functions including photosynthesis, transport of nutrients and photosynthates, ATP production, and protein synthesis (Szczerba et al., 2009; Hasanuzzaman et al., 2018). In soil, despite being one of the most abundant elements, only 2–3% is directly available for plant uptake in the free ionic form K+ (Olaniyan et al., 2022). At root level, K+ uptake is achieved by the activity of High Affinity Transport Systems (HATS) and Low Affinity Transport Systems (LATS) (Britto and Kronzucker, 2008). The high-affinity K+ uptake is achieved through the activity of either the KT/HAK/KUP transporter family or the HKT transporter family when the external concentration of K+ is in the μM range (Santa-Maria et al., 2000). In this case, K+ is taken up exploiting a symport mechanism along with H+, in an energy-dependent process (Maathuis et al., 1997). As in the case of other mineral nutrients, the proton motive force across the plasma membrane is generated by the activity of ATPase enzymes, which hydrolyze ATP to extrude H+ toward the rhizosphere (Palmgren, 2001; Pardo et al., 2006). Conversely, the LATS is active when the external K+ concentration is higher than 0.3–0.5 mM (Wang and Wu, 2013), whereby it is taken up by non-selective and selective ion channels, as in the case of AKT1 transporter (Hirsch et al., 1998). With respect to the soil-microorganism-plant interface, it is well known that one of the very effective strategies to increase the K available fraction is that relying on the use of K-solubilizing bacteria (KSB, Etesami et al., 2017, Figure 1B). The action of these bacteria is rather complex and involves the release of organic acids (OAs), the secretion of exopolysaccharides (EPS), and the formation of biofilms on mineral surfaces (Sattar et al., 2019). The release of organic acids at the mineral surfaces, as well as the acidolysis and the carbonic acid production microbial respiration-dependent (Basak et al., 2017), can favor the chemical weathering of K-bearing minerals, like mica, biotite, muscovite, feldspar, illite, and orthoclase (Sattar et al., 2019). Differently, the secreted EPS can strongly adsorb organic acids, and the adhesion on the surface of K-bearing minerals will result in a locally higher concentration of OAs, with clear impact on weathering effects (Liu et al., 2012). Similarly, when biofilm formation is considered, microbial cells are concentrated within a self-produced extracellular polymers matrix (composed by proteins, DNA and polysaccharides). These conditions offer the chance to boost the biochemical reactions underpinning K mobilization (Nagaraju et al., 2017).
In this context there are several PGPR strains including Arthrobacter, Bacillus, Burkholderia, Enterobacter, Pseudomonas, Acidothiobacillus, Flavobacterium, and Thiobacillus that, exhibiting one or more of these actions at a time, can be defined as KSB bacteria (Olaniyan et al., 2022). However, although the contribution of these bacteria in improving K acquisition in crops is out of any doubt, particularly in soils with impaired fertility, it should be highlighted that in general the extent of this contribution seems to not cover the entire plant requirement for an equilibrate development (Ali et al., 2021; Aloo et al., 2022). Moreover, since the bacteria-based nutrient mobilization is generally relatively slow, the single inoculation of KSB may not be able to fulfill the nutrient plant needs. Accordingly, recent studies have shown that the use of biofertilizer bacterial consortia, composed of KSB in combination with other types of PGPR-based biofertilizers (i.e., integrating bacterial actions that are multiple, concurrent, and involving more than a single nutrient at a time), constitutes an innovative eco-friendly strategy to improve in a wider way the plant nutrient availability and agricultural production (Jain and Hari, 2021; Sherpa et al., 2021).
Although any bioengineering applications to the KSB traits has been described in the literature yet, the whole genome sequencing of potential KSB strains [e.g., Bacillus aryabhattai (Chen et al., 2022) and Priestia megaterium (Wu et al., 2023)] is increasingly being undertaken to possibly locate K solubilization-related genes that might be helpful in the development of biotechnological tools.
3.4 The case study of sulfur
Sulfur, representing up to 0.5% of the total plant dry biomass, is an essential macronutrient and crucial for the biosynthesis of the two S-containing amino acids (i.e., methionine and cysteine). These amino acids are then precursors for the synthesis of several other secondary metabolites (e.g., glutathione, phytochelatins, phytosiderphores) that are relevant for a balanced and healthy plant development (Astolfi et al., 2021). Even though the total amount of S in soils ranges from 19 to 4,000 mg kg−1, only a limited fraction represented by the inorganic sulfate form (SO42−) is readily available for plant uptake (Ranadev et al., 2023). In fact, the 90–95% of the total S pool in soil is generally present in the organic and/or mineral forms, not directly acquirable by roots unless after mobilization/oxidation of S0 to SO42− (Mitter et al., 2021). The remaining (5–10%) is mainly represented by the relatively mobile SO42− fraction, whose 95% is adsorbed to the mineral soil fraction. Consequently, generally not more than 5% of the total SO42− is present in the soil solution ranging from 0.15 to 1.2 g kg soil−1 (Scherer, 2009).
At the root cells level, the SO42− uptake is mediated by sulfate transporters (SULTR) family, which, depending on the plant species, can be formed by 12 to 16 members, subdivided in four distinct functional groups (Takahashi et al., 2011). Functional studies in Arabidopsis thaliana have shown that SULTR1;1 and SULTR1;2 mediate the high affinity transport under SO42−-limiting growth conditions (Takahashi, 2019). The uptake of SO42− is achieved through an active transport, whose motive force is suggested to be constituted by the proton gradient formed across the plasma membrane (Buchner et al., 2004). Based on this background it appears clear that the majority of S in soil is present in forms and/or redox states that cannot be directly used by the plant. If on the one hand these aspects reveal how strategic for successful crop production is the practice of S fertilization, on the other they highlight how much the biological S-oxidation by sulfur-oxidizing bacteria (SOB) represent an interesting opportunity for developing new biofertilizers able to exploit more efficiently the endogenous soil S-sources (Figure 1B). In this context, it should be noted that, typically, the chemolithotrophic bacteria are the most dominant and efficient S-oxidizers in soils, known for converting the reduced forms of S (i.e., S2−, S0, SO32−, and S2O32−) into SO42− (Tourna et al., 2014). This group comprises several species of Thiobacillus, Acidithiobacillus, Sulfolobus, Thermothrix, Paracoccus, Thiomicrospira, Thiosphaera, and Acidianus (Ranadev et al., 2023). However, the process of S oxidation can be also performed by heterotrophic bacteria belonging to the genera Burkholderia, Enterobacter, Klebsiella, Pseudomonas, and Xanthobacter, as described by (Chaudhary et al., 2022). Regarding the mechanisms of S oxidation by the SOB, three oxidation processes have been proposed: (1) the Sox pathway, in which photo- and chemolithotrophic alpha-proteobacteria can directly oxidize all reduced forms of S into SO42− (encoded by the sox operon); (2) the S4I pathway, where obligate chemolithotrophic beta- and gamma-proteobacteria produce S4O62− as an intermediate; and (3) the branched thiosulfate oxidation pathway, that involves intracellular sulfur depositing under anaerobic conditions (Ranadev et al., 2023). With reference to the field experiences, it has been well demonstrated with several agricultural crops that the inoculation of SOB in combination with elemental S sources promotes plant growth improving the yield (Pourbabaee et al., 2020; Ranadev et al., 2023). Furthermore, the application of elemental S enriched with SOB seems not only to speed up the elemental S conversion into SO42−, but to induce a rhizosphere soil acidification with the consequent effects on the availability of various other macro- and micro-nutrients (Chaudhary et al., 2022; Nadeem et al., 2022).
With respect to the bioengineering applications to the interesting traits of these SOB, up to now no experiences are reported in literature. Nonetheless, several authors are carrying out efforts aimed at investigating the whole genome of SOB strains (Koch and Dahl, 2018; Watanabe et al., 2019) in order to identify key genes and functions that might be used as valuable traits for genetically engineered strains as S biofertilizers in agriculture.
3.5 The case study of zinc
Zinc is an essential micronutrient whose concentration levels in plant tissues varies from 20 to 100 mg kg−1 (shoot dry matter). It is required for plant growth (e.g., integral component of lipids, proteins and carbohydrate synthesis via carbonic anhydrase), plant development (e.g., DNA replication, RNA biosynthesis, involvement in IAA synthesis), and plant resistance against biotic and abiotic stresses (e.g., detoxification of superoxide radicals) (Hassan et al., 2020). In soil, depending on the geochemical composition and weathering of parent rocks, it is naturally present at concentrations ranging from 10 to 300 mg kg−1 (Kabata-Pendias and Mukherjee, 2007). Sorption, complexation and, to a lesser extent, dissolution/precipitation mechanisms determine the solid–liquid partitioning of Zn in soil. When the soluble fraction is considered, Zn can occur in different forms, either as free divalent cation (Zn2+) or complexed with organic ligands (Cesco et al., 2022). Although Zn2+ is the form mainly taken up at root level, the uptake of chelated forms (e.g., Zn-phytosiderophore or zinc-organic acids) across the plasma membrane is also reported in some plants (von Wirén et al., 1996). The Zn2+ transport across the root plasma membrane is predominantly mediated by members of the ZIP (Zn-regulated transporter proteins) family (Greco et al., 2012). Nevertheless, several pieces of research have also highlighted that ZIP family protein iron-regulated transporters 1 and 3 (IRT1 and IRT3) can significantly contribute to Zn influx in root cells (Dubeaux et al., 2018; Lee et al., 2021). Furthermore, members of the Yellow-Stripe1–like (YSL) family, the heavy metal ATPases (HMAs), and the cation diffusion facilitator (CDF) have also been shown to play a role in Zn homeostasis in plants (Sinclair and Krämer, 2012; Cardini et al., 2021).
The highest values of Zn are generally found in soils rich in organic matter, clay minerals, and CaCO3. On the contrary, soils characterized by high pH values and low organic matter contents can often induce Zn deficiency phenomena, particularly in wet and cool conditions. To prevent in crops the consequences of this shortage (which include smaller leaves, chlorosis and stunted development), the exogenous application of ZnSO4 as chemical fertilizer has been extensively used due to its high solubility and low cost. However, ZnSO4 is rapidly converted into insoluble forms in soils, and only 2–5% of the total Zn applied can be actually used by plants (Beig et al., 2023). In the frame of PGPR, Zn-solubilizing bacteria (ZnSB) are nowadays emerging as a potential eco-friendly alternative to enhance Zn availability in rhizosphere soil. Similarly to what described for PSB and KSB, the proposed mechanism underlying the ZnSB’s contribution to the Zn acquisition in crops is essentially based on a Zn-solubilizing action through a series of processes including acidification of the nearby soil (e.g., thanks to the exudation of organic acids and protons), the metal–ligand exchange (e.g., via the release of carboxylates, siderophores and other organic ligands) and/or the activity of redox systems on cell membranes (Kamran et al., 2017). In this context, several pieces of literature have described that PGPR belonging to the genera Pseudomonas, Bacillus, Rhizobium, Enterobacter, Priestia and Pantoea, when applied to different crops (cereals and legumes), have demonstrated effectiveness in improving the Zn acquisition with beneficial effects both on crop development and its productivity (Kamran et al., 2017; Yadav et al., 2022; Srithaworn et al., 2023). Moreover, there is recent evidence supporting the idea of an effectively use of ZnSB also to pursue the objective of biofortification of agricultural production, such as the case of wheat grain. In this specific case, a significant seed biofortification was obtained by using ZnSB in combination with Zn chemical fertilizers (Yadav et al., 2022; Saleem et al., 2023).
With respect to innovations through genetic modifications, there is no evidence in literature describing programs to improve the Zn-solubilizing traits of these bacteria through biotechnological approaches. Furthermore, deeper insights into their Zn-solubilizing mechanisms are still necessary to maximize Zn accessibility in the rhizosphere environment.
3.6 The case study of iron and other micronutrients: bacterial siderophores
Iron (Fe) belongs to the group of essential micronutrients, being, among these, the one required in highest amounts. Due to its ability to easily switch between to oxidation states (Fe2+ and Fe3+), it is a cofactor of proteins involved in electron transfer and of many enzymes catalyzing redox reactions. For these reasons, Fe is involved in important metabolic pathways like photosynthesis, respiration, chlorophyll biosynthesis, and antioxidant defense system (Murgia et al., 2022). In soil, although present at relatively high concentrations (20–40 mg kg−1) mainly as ferric (hydro)-oxides, its availability for plants is limited due to the poor solubility of the metal in the soil solution (Zuluaga et al., 2023a). Crops display at their root level the functionality of two different strategies of Fe acquisition: in dicots the FeIII-chelate reduction-based one while in monocots a strategy dependent on chelation by phytosiderophores with high affinity for binding Fe3+ (for more details see also Morrisey and Guerinot, 2009).
With respect to the PGPR and plant Fe nutrition, several pieces of literature have demonstrated that these bacteria can significantly enhance in plants the Fe acquisition process in scenarios of restricted Fe availability. This effect is ascribed both to the synthesis and the release in the rhizosphere of huge amounts of organic acids, phenolic compounds, siderophores (favoring essentially the component of Fe solubilization), and to an enhanced activity of the plasma membrane ferric chelate reductase (FCR) of roots (enhancing the nutrient uptake step). In this regard, it is interesting to mention the experiences gained with the root inoculation with Azospirillum brasilense in cucumber plants. In these specific conditions, the PGPR’s effect appears to be ascribable to a rather complex of actions including not only an enhanced exudation of chelating compounds and an induced FCR activity as previously described, but also the modulation of the expression levels of key genes related to Fe acquisition and allocation (Pii et al., 2015b, 2016; Marastoni et al., 2019). Moreover, it is worth to highlight that this beneficial effect was also evident in cucumber plants fed adequately with Fe resulting in contents of the nutrient in the tissues much higher than those observed in plants not inoculated with the PGPRs (Pii et al., 2016).
In recent years, siderophore-producing bacteria (SPB) have been proposed as a sustainable alternative to synthetic fertilizers. Bacterial siderophores are low molecular weight organic compounds exhibiting cation-chelating properties and synthesized either under nutrient shortage in order to cope with the nutritional disorder or to alleviate the toxicity level of heavy metal (Vijay et al., 2023). Although the term siderophore is more often used for the context of Fe nutrition and to indicate specifically a ligand able to form chelates with Fe, these compounds display affinity also for other elements like Cu, Mn, Mo, and Zn, all relevant for the plant and its metabolism (Figure 1C). Therefore, the root inoculation with SPB could be also envisaged as a strategy to improve the bioavailability not only of Fe but also of these other micronutrients. In this context, regarding specifically to the SPB contribution to Fe nutrition, Ferreira et al. (2019) demonstrated that a siderophore-based product prepared from Azotobacter vinelandii culture ameliorated Fe deficiency of soybean plants, thus representing a friendly Fe-fertilizer alternative for application in calcareous soils. Furthermore, (McRose and Baars, 2017) provided evidence that, under Fe starvation, A. vinelandii produced higher amounts of vibrioferrin, an α-hydroxycarboxylate siderophore. However, under Mo limitation, this bacterial strain completely repressed the production of vibrioferrin, producing instead higher concentrations of protochelin, a siderophore belonging to the catechol class. These results suggest that the SPB A. vinelandii can selectively direct the production of siderophores depending on the element for which the limitation is sensed. However, it is important to highlight that the stability of the complex metal-siderophore (Me-S) can play a strategic role, both in a negative and positive sense. In fact, if on the one hand, when sufficiently strong, the Me-S complex stability ensures its persistence in the soil even in particularly disadvantaged conditions, on the other, when excessively strong, could represent a limit to its use by the roots, in particular for the strategy based on the ligand exchange like in monocots. In this regard it should be highlighted that the scientific debate is still wide (Colombo et al., 2014). Therefore, for a more efficient use of root inoculation with SPB (and/or alternatively directly a product containing the siderophores) at the field scale, further investigations and knowledge, particularly at the mechanism scale, appear to be fundamental for the plant Fe nutrition context.
With respect to innovations through genetic modifications, there is no evidence in literature describing programs to improve the Fe-solubilizing traits of bacteria through biotechnological approaches.
4 Microbiome engineering-based approaches in biofertilizer development
Despite the well-documented beneficial effects of using biofertilizers based on single-strains or bacterial consortia, their adaptability to agricultural practices cannot be guaranteed, mainly because of their limited persistence in soil over time (Mitter et al., 2021). Nowadays, different microbiome engineering-based approaches involving the manipulation of microbial communities have emerged as a promising technology to promote synergistic interactions that can provide collective benefits to the host plant. These approaches include the creation of synthetic communities (SynComs) and the host-mediated microbiome engineering (HMME) (Yang et al., 2023). The key advantage of these approaches is not only the expected improvement of nutrient availability in the rhizosphere (with the consequent decrease in chemical fertilizer supplementation with the related environmental benefits), but also the positive effects of the rhizosphere microbiome to the plant health, amplifying the plant’s capability to cope with biotic and abiotic stresses.
4.1 The case study of biofertilizer based on synthetic communities
Synthetic microbial communities (SynComs) refer to artificial microbial consortia designed to recreate the core microbiome of a specific plant (Mitter et al., 2021). The synthetic consortium is normally engineered with three or more strains used to get a more realistic understanding of the interactions between microorganisms, plants, and the environment (Marín et al., 2021). According to their size, SynComs can be classified as a low- or high-complex consortia. Low-complex SynComs (<10 strains) are easier to design and can reduce costs and steps when scaling up bacterial growth in industrial processes. However, the small size of these consortia may not be taxonomically representative, thus neglecting important associations which are critical at the functional level. On the other hand, high-complex SynComs (>10 strains) can more efficiently mimic the autochthonous community in the rhizosphere with better chance of keeping associations intact, despite the limitations on their design (Mitter et al., 2021).
Regarding the use of SynComs as a potential strategy to develop ecology-based biofertilizers that enhance nutrient acquisition of plants, some studies have been carried out on this area. For instance, (Kaur et al., 2022) used a low-complex SynCom (4 strains) to investigate its impact on soil nutrient availability and growth of cotton plants. They found an increased nitrate availability in soils linked not only to the presence of the N-fixing Brevibacterium in the SynCom, but also to the enrichment of Cyanobacteria members induced by the consortium itself. Furthermore, after observing that indica rice recruited a higher proportion of bacteria related to N cycle as compared to japonica rice, Zhang et al. (2019) designed both a high-complex SynCom using16 indica-derived strains and a low-complex one using three japonicum-derived strains. The effects of both SynComs on the growth and N nutrition of IR24 rice plants were investigated and it was observed that the indica-derived high-complex SynCom was able to improve the transformation of organic N into NO3− and NH4+ favoring, thus, in this crop higher values of N use efficiency (NUE). This result is perfectly coherent with historical observations of the higher NUE in indica varieties of rice than in the japonica ones (Hu et al., 2015).
Concerning the possible application of SynComs concept to plants growing in soils characterized by variable P availability, Finkel et al. (2019) designed a high-complex consortium encompassing 185 strains. Interestingly, in this experience a clear link between the presence of the genus Burkholderia and the efficient use of the P sources has been highlighted. In particular, plants colonized by a SynCom lacking genus Burkholderia displayed a higher P allocation at the shoot level than those inoculated with the complete consortium containing Burkholderia strains. These results highlight that the rhizosphere of plants can be also colonized by latent opportunistic microbial competitors, which, under a nutrient shortage, may be capable to further exacerbate the nutritional disorder of the host plant. Despite these observations, the SynComs, once inoculated in plants exposed to a nutrient shortage, have always proven themselves efficient in bringing benefits to the host plants. Similarly, (Harbort et al., 2020) developed a 115-strains SynCom to remediate Fe deficiency conditions in Arabidopsis plants. The research highlighted that the plant biosynthesis of coumarins and their exudation were required for SynCom-mediated rescue of plant development under Fe limitation. Interestingly, the same SynCom was not able to induce an over-accumulation of Fe, suggesting that the aiding activity of the consortium could be a plant-driven phenomenon, whose control was guaranteed by the root release of specific exudates. Overall, these pioneering studies, despite being at their beginning, clearly demonstrate the potential high functionality of the SynComs in enhancing the rhizosphere availability and the root acquisition of essential nutrients, yet overcoming some of the restraints of the single-strain biofertilizers currently available in the market.
4.2 The case study of host-mediated microbiome engineering
Plants generally influence the composition of the microbial community inhabiting their rhizosphere, where some microbial members are specifically recruited by the host, while other assemble opportunistically (Pantigoso et al., 2022). Hence, host-mediated microbiome engineering (HMME) is a biotechnological approach that uses the plant host to indirectly select specialized microbiome assembly colonizing the rhizosphere and, there, able to enhance plant growth, nutrient acquisition, and resilience to stresses (French et al., 2021). In some cases, such type of microbial selection has happened unintentionally after hundreds of years of domestication or during plant breeding programs, which led to the evolution of wild plant genotypes and, consequently, provoked an optimization in the rhizosphere microbiome of subsequent plants (Escudero-Martinez and Bulgarelli, 2023). For instance, corn is one of the most farmed and fertilized crops in the world, and its domestication has led to the recruitment in the rhizosphere of N-cycling functional groups as compared with ancestral corn genotypes (Favela et al., 2022). Moreover, it has been observed that, during nutrient starvation, corn (Zea mays) plants have evolved the ability to synthesize and release a variety of exudates into the rhizosphere. These organic compounds may have not only a role in influencing the biogeochemical cycles of nutrients in the rhizosphere, but also in promoting the recruitment of specific plant-beneficial taxa of microorganisms. In this context, it is interesting to mention the development of aerial roots secreting a huge amount of carbohydrate-rich mucilage observed in plants of a maize landrace grown under conditions of limited N availability (i.e., very limited or no fertilization) (Deynze et al., 2018). The authors described that the high levels of N fixation observed were supported by the abundant secretion of the mucilage, which favored the assembling of a complex N-fixing microbiome by roots. Similarly, under N deprivation, (Yu et al., 2021) observed that the flavonoids-releasing roots of maize plants specifically recruit in the rhizosphere bacteria of the taxon Oxalobacteraceae, that, in turn, facilitates N uptake through modulation of lateral root development. Based on these experiences, it appears evident that the reintroduction of alleles coding for beneficial compounds into crops by using HMME approaches could be a promising step toward the improvement of positive interactions between the plant rhizosphere and its microbiome.
Moreover, the increasing advances in plants-applied omics techniques has led to the identification of biosynthetic gene clusters (BGCs) responsible for the production of important secondary metabolites (e.g., terpenes, alkaloids, benzoxazinoids, cyanogenic glucosides, and polyketides). It is well known that these compounds are considerably involved in the assembly of rhizosphere microbiome (Polturak et al., 2022). Therefore, plant genetic transformations targeted to these BCGs clusters by using HMME based on genetic techniques, can represent a promising and futuristic step to reinforce the ability of plants to recruit specific beneficial microbiota. However, further research in this host-mediated microbiota selection by roots is still necessary. In particular, different aspects, like the choice of host trait, transferability of the recruited microbiome, and the environment component that can interfere with the selection of the most promising microbiome, might be considered and deeper addressed (French et al., 2021). Therefore, understanding the molecular and biochemical mechanisms governing the interactions between plant host and its associated microbiome, especially under nutritional disorders, would allow the design of crops with ability to produce/select “their own biofertilizers,” by reshaping specialized microbiomes that efficiently modulate (in their favor) the biogeochemical cycles of the nutrients in soil.
5 Current scenarios and future perspectives for PGPR-based biofertilizers
The global biofertilizer market has to date an estimated value of USD 3.1 billion and it is projected to increase to USD 7.0 billion in 2030 (markets&markets). Currently, most of the biofertilizers available in the market are based on either a single PGPR strain with single or multiple PGP activities, or microbial consortia with multiple PGP activities (Figures 2A,B). Both kinds of products have shown success in promoting plant growth and improving soil physiochemical properties at a relatively low production cost. In this context it is interesting to note that Bradyrhizobium-based biofertilizers applied to soybean plants are probably the most successful example of these products across the world, fully supplying the crop’s demand of N in countries like Brazil (Santos et al., 2019). However, the efficacy of these biofertilizers often shows disparities due to (i) environmental variability, (ii) the intense competition with native microbial communities in the soil, and (iii) the application methods. Amazingly, due to the outstanding effects observed after the use of PGPR-based biofertilizers, some farmers have tried to produce their own biofertilizers using rudimentary bio-factories in a process called on-farm production (Figure 2C). This process consists of using makeshift equipment and infrastructure, including fermenters, open tanks, or even water tanks, without appropriate control of contaminations, which can lead to inconsistencies in production quality and efficiency (Bocatti et al., 2022). Therefore, beyond the admirable intention of these farmers, this on-farm practice represents a real threat for the entire agriculture in a general sense since plant, animal, and human pathogens have been predominantly found in such products, with non-effective results for plant growth and health (Bocatti et al., 2022). This aspect becomes even more relevant if the concept of One Health and its application in every context that concerns our planet, are considered.
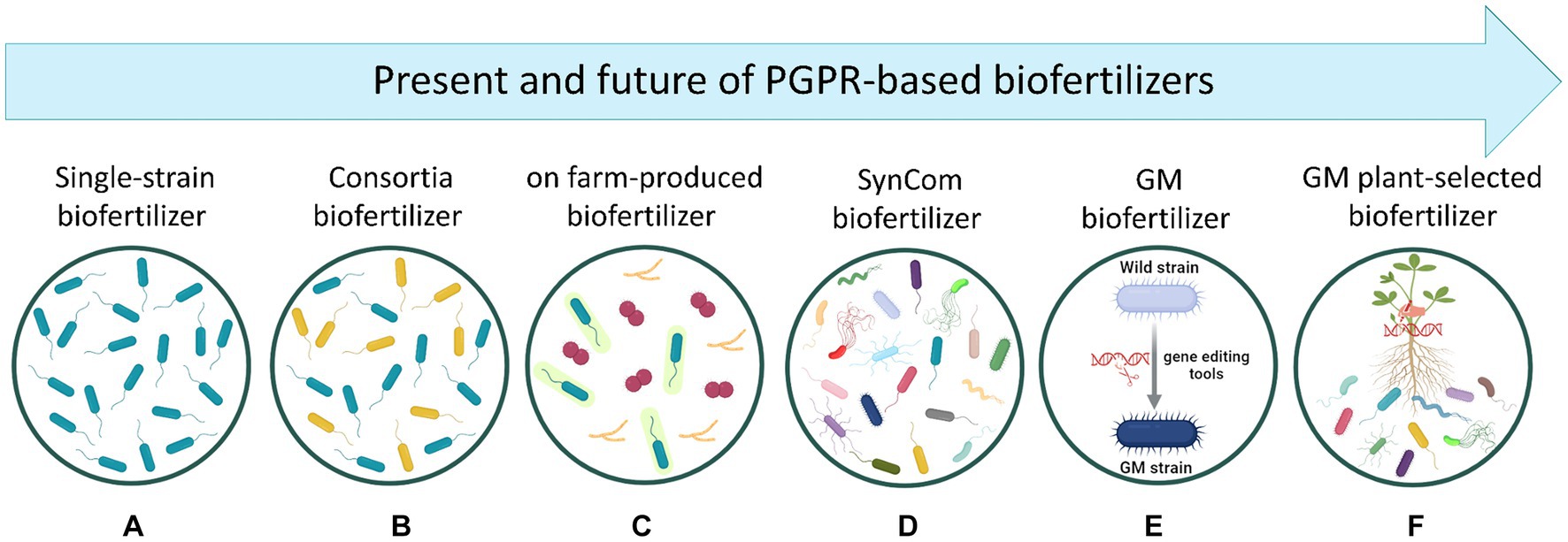
Figure 2. Representation of the present and future of PGPR-based biofertilizer development. (A) Biofertilizers based on a single bacterial strain. (B) Biofertilizers containing multiple strains (two or more) which are selected by considering their ability in enhancing plant uptake of soil nutrients. (C) Biofertilizers produced by farmers in their own farms by using rudimentary bio-factories without appropriate control of contaminations, which may result in highly contaminated, non-effective products. (D) Biofertilizers containing multiple strains which are selected after analyzing the diversity profile of plant microbiome. Thus, SynComs are designed to recreate the core microbiome containing key microbial taxa carrying essential functional genes for the host plant. (E) Genetically modified biofertilizers where bacterial genes are modified with gene editing tools (e.g., CRISPR/Cas9, RNAi) to improve plant growth and nutrition. However, the incorporation of these products into farm systems remains controversial since their efficacy, survivability, and environmental hazards are not well understood (Qiu et al., 2019). (F) Genetically modified plants driving the selection/recruitment of PGPR biofertilizers. Created with BioRender (https://biorender.com/).
Following the line of future possibilities in the development of high-performing biofertilizers, the design of SynComs represents certainly an innovative and promising technology (Figure 2D), as already detailed in the previous section. However, when producing biofertilizers based on these SynComs, some challenges shall be considered. One of them pertains to the ability of designing effective SynComs with a minimal number of bacteria. In this regard, it should be noted that for commercial uses, the industrial production of multi-strain biofertilizers is commonly performed by separately fermenting different batches and mixed them afterwards. Thus, it appears clear that, to scale up bacterial growth at industrial level, theoretically one fermenter per bacterial strain is required. However, if this approach were also applied to the production of high-complex SynComs, its economic and structural non-sustainability appears evident. In this scenario, the selection of bacterial strains displaying multifarious and synergistic traits is urgently needed to ensure a feasible and simplified way to produce SynCom-based biofertilizers.
In recent times, it became possible the use of genetic engineering approaches to optimize bacterial genes, thus providing the chance to set up eco-friendly biofertilizers improved in their capacity to ameliorate the edaphic conditions of the crops’ rhizosphere (Figure 2E). The genetic editing on targeted PGPR genes holds the potential of being fast and reasonably effective, due to the direct introduction of specific traits into well-characterized bacteria. Therefore, in the field of PGPR-based biofertilizers, genetic engineering stands as a promising strategy by which a wild-type PGPR strain can be optimized to overexpress the molecular machinery underlying N, P, K, S, Fe, and/or Zn acquisition for itself and the host plant. In fact, the manipulation of nif, fix, and nod genes in N-fixing bacteria have been shown to improve their symbiotic or associative performance in chickpea plants, leading to an increased N content of the crop tissues and yield (Silva et al., 2019). One other factor that could be exploited by using genetic manipulation is the insertion of genes related to PGP traits (one or more) into a bacterium that shows only one PGP mechanism. This approach may avoid the current need of mixing two bacterial strains when used in biofertilizer formulations. A hypothetical example of such types of transformation could be represented by a natural N-fixer that has been genetically strengthened with the ability to solubilize P and/or produce Fe-chelating compounds. Considering that for many years, engineering PGP traits has mostly focussed on N fixation and P solubilization, it is critical to enhance our knowledge about the molecular and functional mechanisms behind agronomically potential bacteria including KSB, SOB, and ZnSB to perform the most appropriated genome editing strategies toward more effective biofertilizers. Nevertheless, it cannot be ignored that the introduction into the environment of genetically manipulated bacterial strains might represent a potential risk of genetic pollution (i.e., horizontal genes transfer toward autochthonous microbiomes).
Besides the genetic modification of PGPR, another promising approach could be represented by engineering plants to either (i) increase their efficiency in nutrient acquisition and use, or (ii) recruit at the root level a more efficient microbial community. Considering the first point, and despite the genetic challenge, the development of plants able to directly fix atmospheric N without the help of bacterial partners would represent a feat without precedents, not only for alleviating adverse effects of synthetic N-fertilizers but also for the economic benefits derived from the higher harvest in system with low external inputs. In this regard it is interesting to note that current attempts are using the yeast Saccharomyces cerevisiae as model organism for initial testing of the functionality of eukaryotic nif proteins. In this attempt, chloroplasts and mitochondria are selected as candidate compartments for nitrogenase assembly and functioning (Burén and Rubio, 2018). With reference instead to the second point, plants could be also manipulated to select a specific rhizosphere microbiome capable of improving the nutrient acquisition process (Figure 2F). In this respect, it is well known that different plant species with contrasting physiologies and traits can shape different microbial community structures via root exudation. Therefore, by identifying exuded molecules related to the signaling and recruitment of specialized biofertilizing microbial communities, plant genes encoding the production of these exudates could be potentially manipulated. To achieve this objective, a deeper understanding of the mechanisms underlying root exudation patterns under different nutritional status and environmental conditions is needed. In this context, the integration of multiple omics approaches including metabolomics, ionomics, genomics, and transcriptomics could help identifying and deciphering the key components necessary for successful plant manipulation to efficiently “self-select” the best biofertilizing microbiome.
6 Common scepticisms about biofertilizers: from the lab to the field
Despite their potential, there have been ongoing discussions and doubts regarding the overall efficacy of PGPR-based biofertilizers in enhancing plant growth and increasing crop yields in real-world field conditions. This skepticism arises from the significant gap between the effects demonstrated by PGPR-based biofertilizers in laboratory or greenhouse experiments and the results found in field trials. While controlled experiments offer precision and reproducibility (Zuluaga et al., 2020; Scagliola et al., 2021), they often fail to capture the complex interactions between PGPR, plants, and soil microbiota present at field scale (Thilakarathna and Raizada, 2017; Meyer et al., 2019). Moreover, several reports suggest that the viability and proliferation of PGPR-based biofertilizers introduced into fields vary and are greatly influenced by environmental factors such as temperature, rainfall, and soil composition, as well as interactions with the host plant and indigenous microorganisms (Strigul and Kravchenko, 2006; Owen et al., 2015; Antoszewski et al., 2022). In addition, agronomical practices can significantly impact the composition of crops’ rhizosphere microbial community by modifying plant metabolism (Cesco et al., 2021). Similarly, these practices influence the epiphytic microbial community on fruits, which is affected by the plant’s nutritional state (Valentinuzzi et al., 2021). These findings underscore the complexity of the interactions among soil, microbes, and crops in the field, that may partly explain the varying levels of permanence and efficiency observed in the application of biostimulants.
In a meta-analysis conducted by Schütz et al. (2018), 171 peer-reviewed field studies were evaluated to assess the impact of biofertilizers on crop yield and nutrient use efficiency of N and P. The authors found that biofertilizers performed best in dry climates, showing a yield increase of 20.0%, compared to a 14.9% increase in tropical climates. Additionally, the yield response to biofertilizers was generally small at low soil P levels, but their efficacy improved with higher soil P levels, demonstrating the high variability of microbial biofertilizers under different field and environmental conditions. Similarly, a meta-analysis of 28 research papers examined the effectiveness of different rhizobial inoculants on soybean traits under field conditions (Thilakarathna and Raizada, 2017). It was shown that the effectiveness of the inoculants differed in terms of nodule count (−28 to +178 nodules) and grain-N yield (−6 to +176%) compared to uninoculated controls. This variability was attributed to various factors, including soybean genotype, interactions between soybean and rhizobia strains, inoculant formulation, concentration, and method of application. Adding to this complexity, research by Pandey et al. (1998) demonstrated that bacterial biofertilizers effective in subtropical climates struggled in temperate alpine regions due to their inability to establish and survive in lower temperatures. Furthermore, Sanz-sáez et al. (2015) found that inoculating soybean with Bradyrhizobium japonicum did not enhance photosynthesis, growth, or seed yield under ambient or elevated CO2 levels in the field, likely due to competition from native rhizobia.
In scientific literature, the prevalence of failed field experiments is likely considerable, yet such instances are seldom disclosed. This discrepancy primarily stems from the scientific community’s focus on innovation over the repetition and validation of previously published studies (Bashan et al., 2020; O’Callaghan et al., 2022). Moreover, the lack of standardized protocols for isolating, characterizing, and formulating PGPR strains exacerbates this issue, hindering the progression of biofertilizers from development to their ultimate field application. Standardized protocols would ensure consistency in application methods, such as the concentration of inoculants, timing, and techniques (e.g., seed coating versus soil drenching) allowing researchers and farming communities to draw more definitive conclusions about the efficacy of biofertilizers (Khan et al., 2023). Recognizing this need, Neuhoff et al. (2024) recently proposed a comprehensive set of standards for agronomic field trials, summarized in Figure 3, aimed at evaluating the efficacy of PGPR-based biofertilizers in arable crops within temperate climates. Thus, by adopting standardized protocols, the scientific community can address the gap in reliable field data, ultimately improving the progression of biofertilizers from experimental stages to widespread agricultural use. This approach will not only enhance the credibility of biofertilizer research but also support farmers in making informed decisions about their use, fostering broader acceptance and implementation.
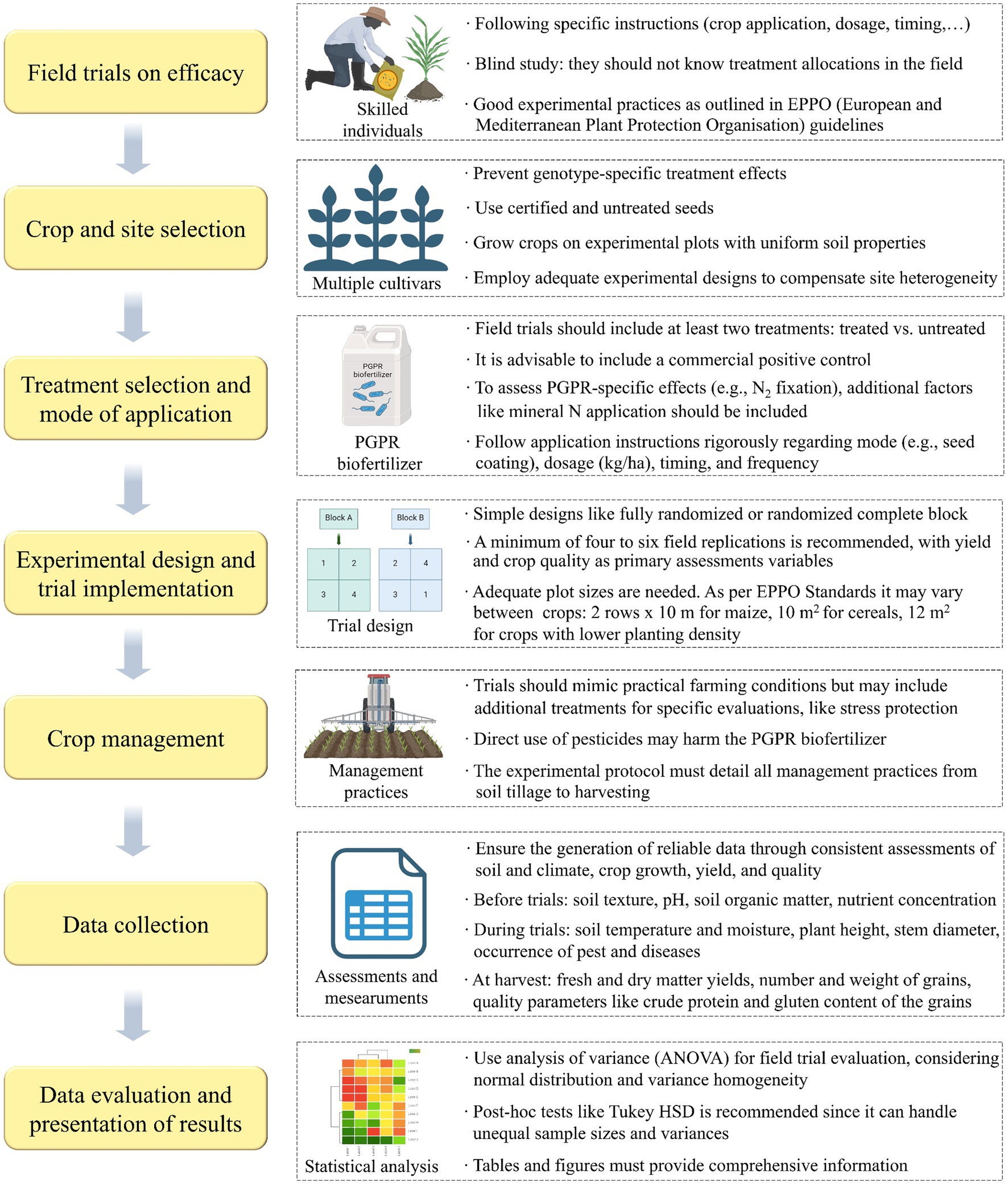
Figure 3. Guidelines for PGPR-based biofertilizers field trial design and implementation as proposed by Neuhoff et al. (2024).
7 Conclusion
It is evident that in the next future the current agriculture must face and meet important challenges whose success should consider the long-term environmental sustainability in a context of One Health paradigm and, properly of these last few years, the rapidly changing climate conditions. To achieve these objectives, all the tools and approaches that can contribute appear to be of crucial importance. The possibility of acting at a localized level at the rhizosphere (precision farming), allowing a more efficient exploitation of the endogenous nutrient resources already present in the soil as the PGPR-based biofertilizers seem to do, is certainly among the promising tools to contribute to the achievement of the general aim of a smarter and more sustainable agriculture. This review highlights how single strain and SynComs PGPR can impact soil fertility and nutrient availability for crop plants, how they influence the essential nutrient mobilization and uptake, and evaluates their effectiveness in controlled and field conditions. It also addresses skepticism about field results versus controlled experiments, advocating for standardized protocols and innovative approaches to improve biofertilizer performance. From this perspective, the potential of PGPR-based biofertilizers is evident, yet significant developments are needed regarding both the bacteria and the crops to optimize their use, especially in soils with altered fertility. Integrating advanced omics techniques, closely aligned with agricultural rhizosphere conditions, with cutting-edge bioengineering approaches could provide valuable insights. This integration is essential for advancing agriculture toward greater sustainability and resilience.
Author contributions
MA: Conceptualization, Data curation, Investigation, Visualization, Writing – original draft, Writing – review & editing. RF: Conceptualization, Data curation, Investigation, Visualization, Writing – original draft, Writing – review & editing. SC: Conceptualization, Data curation, Funding acquisition, Visualization, Writing – original draft, Writing – review & editing. YP: Conceptualization, Data curation, Funding acquisition, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the Free University of Bozen/Bolzano through the projects COMPETITIVE (coded TN202I); the Agritech National Research Center and received funding from the European Union Next-Generation EU (PIANO NAZIONALE DI RIPRESA E RESILIENZA (PNRR) – MISSIONE 4 COMPONENTE 2, INVESTIMENTO 1.4 – D.D. 1032 17/06/2022, CN00000022). In particular, our study represents an original paper related to the Spoke 4 Multifunctional and resilient agriculture and forestry systems for the mitigation of climate change risks and in particular to the Tasks 4.1.2 titled Smart phenotyping platforms for the on-farm selection of resilient varieties and rootstocks (YP and SC). This manuscript reflects only the authors’ views and opinions, neither the European Union nor the European Commission can be considered responsible for them.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Ahmed, E., and Holmström, S. J. M. (2014). Siderophores in environmental research: roles and applications. Microb. Biotechnol. 7, 196–208. doi: 10.1111/1751-7915.12117
Alam, K., Barman, M., Prasad, S., Kannepalli, D., and Livleen, A. (2023). Modification of inorganic fractions of phosphorus by phosphate-solubilising microorganisms in conjunction with phosphorus fertilisation in a tropical inceptisol. J. Soil Sci. Plant Nutr. 23, 2488–2497. doi: 10.1007/s42729-023-01206-6
Ali, A., Awad, M., Hegab, S., Gawad, A., and Eissa, M. (2021). Effect of potassium solubilizing bacteria (Bacillus cereus) on growth and yield of potato. J. Plant Nutr. 44, 411–420. doi: 10.1080/01904167.2020.1822399
Aloo, B. N., Tripathi, V., Makumba, B. A., and Mbega, E. R. (2022). Plant growth-promoting rhizobacterial biofertilizers for crop production: the past, present, and future. Front. Plant Sci. 13:1002448. doi: 10.3389/fpls.2022.1002448
Alori, E. T., Glick, B. R., and Babalola, O. O. (2017). Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 8:971. doi: 10.3389/fmicb.2017.00971
Ambrosio, R., Cesar, J., Ortiz-marquez, F., and Curatti, L. (2017). Metabolic engineering of a diazotrophic bacterium improves ammonium release and biofertilization of plants and microalgae. Metab. Eng. 40, 59–68. doi: 10.1016/j.ymben.2017.01.002
Antoszewski, M., Mierek-Adamska, A., and Dąbrowska, G. B. (2022). The importance of microorganisms for sustainable agriculture: a review. Meta 12, 1–47. doi: 10.3390/metabo12111100
Astolfi, S., Celletti, S., Vigani, G., Mimmo, T., and Romera, F. J. (2021). Interaction between sulfur and iron in plants. Front. Plant Sci. 12:670308. doi: 10.3389/fpls.2021.670308
Atieno, M., Herrmann, L., Nguyen, H., Phan, H., Nguyen, N., Srean, P., et al. (2020). Assessment of biofertilizer use for sustainable agriculture in the Great Mekong region. J. Environ. Manag. 275, 1–9. doi: 10.1016/j.jenvman.2020.111300
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9:1473. doi: 10.3389/fpls.2018.01473
Barth, P., Stüeken, E. E., Helling, C., Rossmanith, L., Peng, Y., Walters, W., et al. (2023). Isotopic constraints on lightning as a source of fixed nitrogen in Earth’s early biosphere. Nat. Geosci. 16, 478–484. doi: 10.1038/s41561-023-01187-2
Basak, B. B., Sarkar, B., Biswas, D. R., Sarkar, S., and Sanderson, P. (2017). “Bio-intervention of naturally occurring silicate minerals for alternative source of potassium: challenges and opportunities” in Advances in Agronomy. ed. D. Sparks (Elsevier Inc), 115–145.
Bashan, Y., Prabhu, S. R., de-Bashan, L. E., and Kloepper, J. W. (2020). Disclosure of exact protocols of fermentation, identity of microorganisms within consortia, formation of advanced consortia with microbe-based products. Biol. Fertil. Soils 56, 443–445. doi: 10.1007/s00374-020-01464-x
Beig, B., Bilal, M., Niazi, K., Jahan, Z., Haider, G., Zia, M., et al. (2023). Development and testing of zinc sulfate and zinc oxide nanoparticle-coated urea fertilizer to improve N and Zn use efficiency. Front. Plant Sci. 13:1058219. doi: 10.3389/fpls.2022.1058219
Bhadrecha, P., Singh, S., and Dwibedi, V. (2023). ‘A plant’s major strength in rhizosphere’: the plant growth promoting rhizobacteria. Arch. Microbiol. 205, 165–125. doi: 10.1007/s00203-023-03502-2
Bocatti, R. C., Ferreira, E., Ribeiro, R. A., Chueire, L., Delamuta, J., Kobayashi, R., et al. (2022). Microbiological quality analysis of inoculants based on Bradyrhizobium spp. and Azospirillum brasilense produced “on farm” reveals high contamination with non - target microorganisms. Braz. J. Microbiol. 53, 267–280. doi: 10.1007/s42770-021-00649-2
Britto, D. T., and Kronzucker, H. J. (2008). Cellular mechanisms of potassium transport in plants. Physiol. Plant. 133, 637–650. doi: 10.1111/j.1399-3054.2008.01067.x
Buchner, P., Takahashi, H., and Hawkesford, M. J. (2004). Plant sulphate transporters: co-ordination of uptake, intracellular and long-distance transport. J. Exp. Bot. 55, 1765–1773. doi: 10.1093/jxb/erh206
Burén, S., and Rubio, L. (2018). State of the art in eukaryotic nitrogenase engineering. FEMS Microbiol. Lett. 365, 1–9. doi: 10.1093/femsle/fnx274
Cardini, A., Pellegrino, E., White, P. J., Mazzolai, B., Mascherpa, M. C., and Ercoli, L. (2021). Transcriptional regulation of genes involved in zinc uptake, sequestration and redistribution following foliar zinc application to Medicago sativa. Plan. Theory 10, 1–19. doi: 10.3390/plants10030476
Cesco, S., Lucini, L., Miras-Moreno, B., Borruso, L., Mimmo, T., Pii, Y., et al. (2021). The hidden effects of agrochemicals on plant metabolism and root-associated microorganisms. Plant Sci. 311:1/9. doi: 10.1016/j.plantsci.2021.111012
Cesco, S., Sambo, P., Borin, M., Basso, B., Orzes, G., and Mazzetto, F. (2023). Smart agriculture and digital twins: applications and challenges in a vision of sustainability. Eur. J. Agron. 146, 1–9. doi: 10.1016/j.eja.2023.126809
Cesco, S., Terzano, R., Astolfi, S., Brunetto, G., Vigani, G., Pii, Y., et al. (2022). “Nutrient and elemental toxicieties” in Soil constraints on crop production. eds. Y. Dang, N. Menzies, and R. Dalal (Newcastle, UK: Cambridge Scholar Publishing), 218–243.
Chaudhary, S., Dhanker, R., Singh, K., Brar, B., and Goyal, S. (2022). Characterization of sulfur-oxidizing bacteria isolated from mustard (Brassica juncea L.) rhizosphere having the capability of improving sulfur and nitrogen uptake. J. Appl. Microbiol. 133, 2814–2825. doi: 10.1111/jam.15742
Chen, Y., Yang, H., Shen, Z., and Ye, J. (2022). Whole-genome sequencing and potassium-solubilizing mechanism of Bacillus aryabhattai SK1-7. Front. Microbiol. 12:722379. doi: 10.3389/fmicb.2021.722379
Colombo, C., Palumbo, G., He, J. Z., Pinton, R., and Cesco, S. (2014). Review on iron availability in soil: interaction of Fe minerals, plants, and microbes. J. Soils Sediments 14, 538–548. doi: 10.1007/s11368-013-0814-z
Coskun, D., Britto, D. T., Li, M., Becker, A., and Kronzucker, H. J. (2013). Rapid ammonia gas transport accounts for futile transmembrane cycling under NH3/NH4+ toxicity in plant roots. Plant Physiol. 163, 1859–1867. doi: 10.1104/pp.113.225961
Daniel, A. I., Fadaka, A. O., Gokul, A., Bakare, O. O., Aina, O., Fisher, S., et al. (2022). Biofertilizer: the future of food security and food safety. Microorganisms 10, 1–16. doi: 10.3390/microorganisms10061220
Deynze, A. V., Zamora, P., Delaux, P., Heitmann, C., Jayaraman, D., Rajasekar, S., et al. (2018). Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLoS Biol. 16:e2006352. doi: 10.6084/m9.figshare.6534545
Dong, F., Lee, Y. S., Gaffney, E. M., Haddadin, H., Minteer, S. D., Chen, H., et al. (2021). An engineered, non-diazotrophic cyanobacterium and its application in bioelectrochemical nitrogen fixation. Cell Rep. Phys. Sci. 2, 1–16. doi: 10.1016/j.xcrp.2021.100444
Doydora, S., Gatiboni, L., Grieger, K., Hesterberg, D., Jones, J. L., McLamore, E. S., et al. (2020). Accessing legacy phosphorus in soils. Soil Syst. 4, 1–22. doi: 10.3390/soilsystems4040074
Dubeaux, G., Neveu, J., Zelazny, E., and Vert, G. (2018). Metal sensing by the IRT1 transporter-receptor orchestrates its own degradation and plant metal nutrition. Mol. Cell 69, 953–964. doi: 10.1016/j.molcel.2018.02.009
Ducousso-Détrez, A., Fontaine, J., Lounès-Hadj Sahraoui, A., and Hijri, M. (2022). Diversity of phosphate chemical forms in soils and their contributions on soil microbial community structure changes. Microorganisms 10:609. doi: 10.3390/microorganisms10030609
Elhaissoufi, W., Ibnyasser, A., Haddine, M., Zeroual, Y., Ghani, R., Barakat, A., et al. (2023). Screening of potential phosphate solubilizing bacteria inoculants should consider the contrast in P bio-solubilization rate along with plant growth promotion and P use efficiency. J. Appl. Microbiol. 134, 1–14. doi: 10.1093/jambio/lxac077
Escudero-Martinez, C., and Bulgarelli, D. (2023). Engineering the crop microbiota through host genetics. Annu. Rev. Phytopathol. 61, 257–277. doi: 10.1146/annurev-phyto-021621-121447
Esteban, R., Ariz, I., Cruz, C., and Fernando, J. (2016). Review: mechanisms of ammonium toxicity and the quest for tolerance. Plant Sci. 248, 92–101. doi: 10.1016/j.plantsci.2016.04.008
Etesami, H., Emami, S., and Alikhani, H. A. (2017). Potassium solubilizing bacteria (KSB): mechanisms, promotion of plant growth, and future prospects. A review. J. Soil Sci. Plant Nutr. 17, 897–911. doi: 10.4067/S0718-95162017000400005
Fasusi, O. A., Cruz, C., and Babalola, O. O. (2021). Agricultural sustainability: microbial biofertilizers in rhizosphere management. Agriculture 11, 1–19. doi: 10.3390/agriculture11020163
Favela, A., Bohn, M., and Kent, A. (2022). N-cycling microbiome recruitment differences between modern and wild Zea mays. Phytobiomes J. 6, 151–160. doi: 10.1094/PBIOMES-08-21-0049-R
Ferreira, C. M. H., López-rayo, S., Lucena, J. J., Soares, E. V., and Soares, H. M. V. M. (2019). Evaluation of the efficacy of two new biotechnological-based freeze-dried fertilizers for sustainable Fe deficiency correction of soybean plants grown in calcareous soils. Front. Plant Sci. 10:1335. doi: 10.3389/fpls.2019.01335
Finkel, O., Salas-Gonzalez, I., Castrillo, G., Spaepen, S., Law, T., Teixeira, P., et al. (2019). The effects of soil phosphorus content on plant microbiota are driven by the plant phosphate starvation response. PLoS Biol. 17:e3000534. doi: 10.1371/journal.pbio.3000534
French, E., Kaplan, I., Iyer-pascuzzi, A., Nakatsu, C. H., and Enders, L. (2021). Emerging strategies for precision microbiome management. Nat. Plants 7, 256–267. doi: 10.1038/s41477-020-00830-9
Greco, M., Chiappetta, A., Bruno, L., and Bitonti, M. B. (2012). In Posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J. Exp. Bot. 63, 695–709. doi: 10.1093/jxb/err313
Grover, M., Bodhankar, S., Sharma, A., Sharma, P., and Singh, J. (2021). PGPR mediated alterations in root traits: way toward sustainable crop production. Front. Sustain. Food Syst. 4:618230. doi: 10.3389/fsufs.2020.618230
Guo, K., Yang, J., Yu, N., Luo, L., and Wang, E. (2023). Biological nitrogen fixation in cereal crops: progress, strategies, and perspectives. Plant Commun. 4:100499. doi: 10.1016/j.xplc.2022.100499
Harbort, C. J., Hashimoto, M., Inoue, H., Sattely, E. S., Garrido-oter, R., and Schulze-lefert, P. (2020). Root-secreted coumarins and the microbiota interact to improve iron nutrition in Arabidopsis. Cell Host Microbe 28, 825–837. doi: 10.1016/j.chom.2020.09.006
Hasanuzzaman, M., Bhuyan, M., Nahar, K., Hossain, M., Mahmud, J., Hossen, M., et al. (2018). Potassium: a vital regulator of plant responses and tolerance to abiotic stresses. Agronomy 8:31. doi: 10.3390/agronomy8030031
Hassan, M. U., Aamer, M., Chattha, M. U., Haiying, T., Shahzad, B., Barbanti, L., et al. (2020). The critical role of zinc in plants facing the drought stress. Agriculture 10, 1–20. doi: 10.3390/agriculture10090396
Herrmann, L., and Lesueur, D. (2013). Challenges of formulation and quality of biofertilizers for successful inoculation. Appl. Microbiol. Biotechnol. 97, 8859–8873. doi: 10.1007/s00253-013-5228-8
Hinsinger, P. (2001). Bioavailability of soil inorganic P in the rhizosphere as affected by root-induced chemical changes: a review. Plant Soil 173, 173–195. doi: 10.1023/A:1013351617532
Hirsch, R. E., Lewis, B. D., Spalding, E. P., and Sussman, M. R. (1998). A role for the AKT1 potassium channel in plant nutrition. Science 918, 1–5. doi: 10.1126/science.280.5365.918
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838. doi: 10.1038/ng.3337
Ibrahim, M., Iqbal, M., Tang, Y. T., Khan, S., Guan, D. X., and Li, G. (2022). Phosphorus mobilization in plant–soil environments and inspired strategies for managing phosphorus: a review. Agronomy 12, 1–17. doi: 10.3390/agronomy12102539
Jain, D., and Hari, R. (2021). Effect of microbial consortia on growth and yield of wheat under typic haplustepts. Plant Physiol. Rep. 26, 570–580. doi: 10.1007/s40502-021-00607-y
Jiang, Q., Zhang, J., Xu, X., Liu, G., and Zhu, J. (2020). Effects of free-air CO2 enrichment (FACE) and nitrogen (N) supply on N uptake and utilization of indica and japonica cultivars (Oryza sativa L.). Ecol. Process. 9, 1–12. doi: 10.1186/s13717-020-00238-5
Kabata-Pendias, A., and Mukherjee, A. (2007). “Trace elements of group 12 (previously group IIb)” in Trace elements from soil to human. ed. A. Kabata-Pendias (Berlin, Heidelberg: Springer), 283–319.
Kamran, S., Shahid, I., Baig, D. N., Rizwan, M., and Malik, K. A. (2017). Contribution of zinc solubilizing bacteria in growth promotion and zinc content of wheat. Front. Microbiol. 8:2593. doi: 10.3389/fmicb.2017.02593
Kaur, S., Egidi, E., Trivedi, P., Macdonald, C. A., Prakash, J., Wang, J., et al. (2022). Synthetic community improves crop performance and alters rhizosphere microbial communities. J. Sustain. Agric. Environ. 1, 118–131. doi: 10.1002/sae2.12017
Kaushal, P., Ali, N., Saini, S., Pati, P. K., and Pati, A. M. (2023). Physiological and molecular insight of microbial biostimulants for sustainable agriculture. Front. Plant Sci. 14:1041413. doi: 10.3389/fpls.2023.1041413
Keswani, C., Prakash, O., Bharti, N., Vílchez, J. I., Sansinenea, E., Lally, R. D., et al. (2019). Re-addressing the biosafety issues of plant growth promoting rhizobacteria. Sci. Total Environ. 690, 841–852. doi: 10.1016/j.scitotenv.2019.07.046
Khan, A., Singh, A. V., Gautam, S. S., Agarwal, A., Punetha, A., Upadhayay, V. K., et al. (2023). Microbial bioformulation: a microbial assisted biostimulating fertilization technique for sustainable agriculture. Front. Plant Sci. 14:1270039. doi: 10.3389/fpls.2023.1270039
Koch, T., and Dahl, C. (2018). A novel bacterial sulfur oxidation pathway provides a new link between the cycles of organic and inorganic sulfur compounds. ISME J. 12, 2479–2491. doi: 10.1038/s41396-018-0209-7
Kumawat, K. C., Razdan, N., and Saharan, K. (2022). Rhizospheric microbiome: bio-based emerging strategies for sustainable agriculture development and future perspectives. Microbiol. Res. 254:126901. doi: 10.1016/j.micres.2021.126901
Kumawat, K. C., Sharma, B., Nagpal, S., Kumar, A., Tiwari, S., and Nair, R. M. (2023). Plant growth-promoting rhizobacteria: salt stress alleviators to improve crop productivity for sustainable agriculture development. Front. Plant Sci. 13:1101862. doi: 10.3389/fpls.2022.1101862
Kumawat, K. C., Sharma, P., Sirari, A., Sharma, B., Kumawat, G., Nair, R. M., et al. (2024). Co-existence of halo-tolerant Pseudomonas fluorescens and Enterococcus hirae with multifunctional growth promoting traits to ameliorate salinity stress in Vigna radiata. Chemosphere 349, 1–14. doi: 10.1016/j.chemosphere.2023.140953
Ladha, J. K., Tirol-pad, A., Reddy, C. K., Cassman, K. G., Verma, S., and Powlson, D. S. (2016). Global nitrogen budgets in cereals: a 50-year assessment for maize, rice, and wheat production systems. Sci. Rep. 6, 1–9. doi: 10.1038/srep19355
Lambers, H. (2022). Phosphorus acquisition and utilization in plants. Annu. Rev. Plant Biol. 73, 17–42. doi: 10.1146/annurev-arplant-102720-125738
Ledermann, R., Schulte, C., and Poole, P. (2021). How rhizobia adapt to the nodule environment. J. Bacteriol. 203, 1–18. doi: 10.1128/JB.00539-20
Lee, S., Lee, J., Ricachenevsky, F. K., Punshon, T., Tappero, R., Salt, D. E., et al. (2021). Redundant roles of four ZIP family members in zinc homeostasis and seed development in Arabidopsis thaliana. Plant J. 108, 1162–1173. doi: 10.1111/tpj.15506
Li, Q., and Chen, S. (2020). Transfer of nitrogen fixation (nif) genes to non-diazotrophic hosts. Chembiochem 21, 1717–1722. doi: 10.1002/cbic.201900784
Liu, L., Du, W., Luo, W., Su, Y., Hui, J., and Ma, S. (2015). Development of an engineered soil bacterium enabling to convert both insoluble inorganic and organic phosphate into plant available phosphate and its use as a biofertilizer. Mol. Biotechnol. 57, 419–429. doi: 10.1007/s12033-014-9834-1
Liu, X., Jiang, X., He, X., Zhao, W., Cao, Y., Guo, T., et al. (2019). Phosphate-solubilizing Pseudomonas sp. strain P34-L promotes wheat growth by colonizing the wheat rhizosphere and improving the wheat root system and soil phosphorus nutritional status. J. Plant Growth Regul. 38, 1314–1324. doi: 10.1007/s00344-019-09935-8
Liu, D., Lian, B., and Dong, H. (2012). Isolation of Paenibacillus sp. and assessment of its potential for enhancing mineral weathering. Geomicrobiol J. 29, 413–421. doi: 10.1080/01490451.2011.576602
Long, H., and Wasaki, J. (2023). Effects of phosphate-solubilizing bacteria on soil phosphorus fractions and supply to maize seedlings grown in lateritic red earths and cinnamon soils. Microbes Environ. 38, 1–8. doi: 10.1264/jsme2.ME22075
Loque, D., and Wirén, N. (2004). Regulatory levels for the transport of ammonium in plant roots. J. Exp. Bot. 55, 1293–1305. doi: 10.1093/jxb/erh147
Maathuis, F. J. M., Sanders, D., and Gradmann, D. (1997). Kinetics of high-affinity K+ uptake in plants, derived from K+ −induced changes in current-voltage relationships. A modelling approach to the analysis of carrier-mediated transport. Planta 203, 229–236. doi: 10.1007/s004250050186
Malusá, E., and Vassilev, N. (2014). A contribution to set a legal framework for biofertilisers. Appl. Microbiol. Biotechnol. 98, 6599–6607. doi: 10.1007/s00253-014-5828-y
Marastoni, L., Pii, Y., Maver, M., Valentinuzzi, F., Cesco, S., and Mimmo, T. (2019). Role of Azospirillum brasilense in triggering different Fe chelate reductase enzymes in cucumber plants subjected to both nutrient deficiency and toxicity. Plant Physiol. Biochem. 136, 118–126. doi: 10.1016/j.plaphy.2019.01.013
Marín, O., González, B., and Poupin, M. J. (2021). From microbial dynamics to functionality in the rhizosphere: a systematic review of the opportunities with synthetic microbial communities. Front. Plant Sci. 12:650609. doi: 10.3389/fpls.2021.650609
McRose, D. L., and Baars, O. (2017). Siderophore production in Azotobacter vinelandii in response to Fe-, Mo- and V-limitation. Environ. Microbiol. 19, 3595–3605. doi: 10.1111/1462-2920.13857
Meyer, G., Maurhofer, M., Frossard, E., Gamper, H. A., Mäder, P., Mészáros, É., et al. (2019). Pseudomonas protegens CHA0 does not increase phosphorus uptake from 33P labeled synthetic hydroxyapatite by wheat grown on calcareous soil. Soil Biol. Biochem. 131, 217–228. doi: 10.1016/j.soilbio.2019.01.015
Mitter, E. K., Tosi, M., Obregón, D., Dunfield, K. E., and Germida, J. J. (2021). Rethinking crop nutrition in times of modern microbiology: innovative biofertilizer technologies. Front. Sustain. Food Syst. 5:606815. doi: 10.3389/fsufs.2021.606815
Młodzińska, E., and Zboińska, M. (2016). Phosphate uptake and allocation - A closer look at arabidopsis thaliana L. and Oryza sativa L. Front. Plant Sci. 7:1198. doi: 10.3389/fpls.2016.01198
Morrisey, J., and Guerinot, M. (2009). Iron uptake and transport in plants: the good, the bad, and the ionome. Chem. Rev. 109, 4553–4567. doi: 10.1021/cr900112r
Muratore, C., Espen, L., and Prinsi, B. (2021). Nitrogen uptake in plants: the plasma membrane root transport systems from a physiological and proteomic perspective. Plan. Theory 10, 1–26. doi: 10.3390/plants10040681
Murgia, I., Marzorati, F., Vigani, G., and Morandini, P. (2022). Plant iron nutrition: the long road from soil to seeds. J. Exp. Bot. 73, 1809–1824. doi: 10.1093/jxb/erab531
Nadeem, S. M., Hanif, A., Khan, M. Y., Waqas, M. R., Ahmad, Z., Ashraf, M. R., et al. (2022). Elemental sulphur with sulphur oxidizing bacteria enhances phosphorus availability and improves growth and yield of wheat in calcareous soil. Arch. Agron. Soil Sci. 69, 1–9. doi: 10.1080/03650340.2022.2099541
Nagaraju, Y., Triveni, S., Subhashreddy, R., and Jhansi, P. (2017). Biofilm formation of zinc solubilizing, potassium releasing bacteria on the surface of fungi. Int. J. Curr. Microbiol. App. Sci. 6, 2037–2047. doi: 10.20546/ijcmas.2017.604.241
Naureen, Z., Sham, A., Al Ashram, H., Gilani, S. A., Al Gheilani, S., Mabood, F., et al. (2018). Effect of phosphate nutrition on growth, physiology and phosphate transporter expression of cucumber seedlings. Plant Physiol. Biochem. 127, 211–222. doi: 10.1016/j.plaphy.2018.03.028
Neuhauser, B., and Ludewig, U. (2014). Uncoupling of ionic currents from substrate transport in the plant ammonium transporter AtAMT1;2. J. Biol. Chem. 289, 11650–11655. doi: 10.1074/jbc.C114.552802
Neuhoff, D., Neumann, G., and Weinmann, M. (2024). Testing plant growth promoting microorganisms in the field - a proposal for standards. Front. Plant Sci. 14:1324665. doi: 10.3389/fpls.2023.1324665
O’Callaghan, M., Ballard, R. A., and Wright, D. (2022). Soil microbial inoculants for sustainable agriculture: limitations and opportunities. Soil Use Manag. 38, 1340–1369. doi: 10.1111/sum.12811
Olaniyan, F. T., Alori, E. T., Adekiya, A. O., Ayorinde, B. B., Daramola, F. Y., and Osemwegie, O. O. (2022). The use of soil microbial potassium solubilizers in potassium nutrient availability in soil and its dynamics. Ann. Microbiol. 72, 1–12. doi: 10.1186/s13213-022-01701-8
Owen, D., Williams, A. P., Griffith, G. W., and Withers, P. J. A. (2015). Use of commercial bio-inoculants to increase agricultural production through improved phosphrous acquisition. Appl. Soil Ecol. 86, 41–54. doi: 10.1016/j.apsoil.2014.09.012
Palmgren, M. G. (2001). Plant plasma membrane H+ -ATPases: powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 817–845. doi: 10.1146/annurev.arplant.52.1.817
Pandey, A., Sharma, E., and Palni, L. M. S. (1998). Influence of bacterial inoculation on maize in upland farming systems of the Sikkim Himalaya. Soil Biol. Biochem. 30, 379–384. doi: 10.1016/S0038-0717(97)00121-1
Pankievicz, V. C. S., Irving, T. B., Maia, L. G. S., and Ané, J. (2019). Are we there yet? The long walk towards the development of efficient symbiotic associations between nitrogen-fixing bacteria and non-leguminous crops. BMC Biol. 17:99. doi: 10.1186/s12915-019-0710-0
Pantigoso, H. A., Newberger, D., and Vivanco, J. M. (2022). The rhizosphere microbiome: plantmicrobial interactions for resource acquisition. J. Appl. Microbiol. 133, 2864–2876. doi: 10.1111/jam.15686
Pardo, M., Cubero, B., Leidi, E. O., and Quintero, F. J. (2006). Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. J. Exp. Bot. 57, 1181–1199. doi: 10.1093/jxb/erj114
Pii, Y., Aldrighetti, A., Valentinuzzi, F., Mimmo, T., and Cesco, S. (2019). Azospirillum brasilense inoculation counteracts the induction of nitrate uptake in maize plants. J. Exp. Bot. 70, 1313–1324. doi: 10.1093/jxb/ery433
Pii, Y., Marastoni, L., Springeth, C., Chiara, M., Maria, G., Cesco, S., et al. (2016). Modulation of Fe acquisition process by Azospirillum brasilense in cucumber plants. Environ. Exp. Bot. 130, 216–225. doi: 10.1016/j.envexpbot.2016.06.011
Pii, Y., Mimmo, T., Tomasi, N., Terzano, R., Cesco, S., and Crecchio, C. (2015a). Microbial interactions in the rhizosphere: beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol. Fertil. Soils 51, 403–415. doi: 10.1007/s00374-015-0996-1
Pii, Y., Penn, A., Terzano, R., Crecchio, C., Mimmo, T., and Cesco, S. (2015b). Plant-microorganism-soil interactions influence the Fe availability in the rhizosphere of cucumber plants. Plant Physiol. Biochem. 87, 45–52. doi: 10.1016/j.plaphy.2014.12.014
Polturak, G., Liu, Z., and Osbourn, A. (2022). New and emerging concepts in the evolution and function of plant biosynthetic gene clusters. Curr. Opin. Green Sustain. Chem. 33:100568. doi: 10.1016/j.cogsc.2021.100568
Pourbabaee, A. A., Dinekaboodi, S. K., Seyed, H. M., Alikhani, H. A., and Emami, S. (2020). Potential application of selected sulfur-oxidizing bacteria and different sources of sulfur in plant growth promotion under different moisture conditions. Commun. Soil Sci. Plant Anal. 51, 735–745. doi: 10.1080/00103624.2020.1729377
Qiu, Z., Egidi, E., Liu, H., Kaur, S., and Singh, B. K. (2019). New frontiers in agriculture productivity: optimised microbial inoculants and in situ microbiome engineering. Biotechnol. Adv. 37:107371. doi: 10.1016/j.biotechadv.2019.03.010
Rago, L., Zecchin, S., Villa, F., Goglio, A., Corsini, A., Cavalca, L., et al. (2019). Bioelectrochemical nitrogen fixation (e-BNF): electro-stimulation of enriched biofilm communities drives autotrophic nitrogen and carbon fixation. Bioelectrochemistry 125, 105–115. doi: 10.1016/j.bioelechem.2018.10.002
Ranadev, P., Ashwin, R., Bagyaraj, D. J., and Shinde, A. H. (2023). Sulfur oxidizing bacteria in agro ecosystem and its role in plant productivity - a review. J. Appl. Microbiol. 134, 1–15. doi: 10.1093/jambio/lxad161
Rasul, M., Yasmin, S., Suleman, M., Zaheer, A., Reitz, T., Tarkka, M. T., et al. (2019). Glucose dehydrogenase gene containing phosphobacteria for biofortification of phosphorus with growth promotion of rice. Microbiol. Res. 223–225, 1–12. doi: 10.1016/j.micres.2019.03.004
Rosier, A., Medeiros, F. H. V., and Bais, H. P. (2018). Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant Soil 428, 35–55. doi: 10.1007/s11104-018-3679-5
Saleem, S., Malik, A., and Khan, S. T. (2023). ZnO nanoparticles in combination with Zn biofertilizer improve wheat plant growth and grain Zn content without significantly changing the rhizospheric microbiome. Environ. Exp. Bot. 213:105446. doi: 10.1016/j.envexpbot.2023.105446
Sambo, P., Nicoletto, C., Giro, A., Pii, Y., Valentinuzzi, F., Mimmo, T., et al. (2019). Hydroponic solutions for soilless production systems: issues and opportunities in a smart agriculture perspective. Front. Plant Sci. 10:923. doi: 10.3389/fpls.2019.00923
Santa-Maria, G. E., Danna, C. H., and Czibener, C. (2000). High-affinity potassium transport in barley roots. Ammonium-sensitive and -insensitive pathways. Plant Physiol. 123, 297–306. doi: 10.1104/pp.123.1.297
Santos, M. S., Nogueira, M. A., and Hungria, M. (2019). Microbial inoculants: reviewing the past, discussing the present and previewing an outstanding future for the use of beneficial bacteria in agriculture. AMB Express 9, 1–22. doi: 10.1186/s13568-019-0932-0
Sanz-sáez, Á., Heath, K. D., Burke, P. V., and Ainsworth, E. A. (2015). Inoculation with an enhanced N2-fixing Bradyrhizobium japonicum strain (USDA110) does not alter soybean (Glycine max Merr.) response to elevated CO2. Plant Cell Environ. 38, 2589–2602. doi: 10.1111/pce.12577
Sattar, A., Naveed, M., Ali, M., Zahir, Z. A., Nadeem, S. M., Yaseen, M., et al. (2019). Perspectives of potassium solubilizing microbes in sustainable food production system: a review. Appl. Soil Ecol. 133, 146–159. doi: 10.1016/j.apsoil.2018.09.012
Sawana, A., Adeolu, M., Gupta, R. S., Nierman, W. C., and Craig, J. (2014). Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 5:429. doi: 10.3389/fgene.2014.00429
Scagliola, M., Valentinuzzi, F., Mimmo, T., Cesco, S., Crecchio, C., and Pii, Y. (2021). Bioinoculants as promising complement of chemical fertilizers for a more sustainable agricultural practice. Front. Sustain. Food Syst. 4:622169. doi: 10.3389/fsufs.2020.622169
Scherer, H. W. (2009). Sulfur in soils. J. Plant Nutr. Soil Sci. 172, 326–335. doi: 10.1002/jpln.200900037
Schütz, L., Gattinger, A., Meier, M., Müller, A., Boller, T., Mäder, P., et al. (2018). Improving crop yield and nutrient use efficiency via biofertilization. A global meta-analysis. Front. Plant Sci. 8:2204. doi: 10.3389/fpls.2017.02204
Sherpa, M., Sharma, L., Bag, N., and Das, S. (2021). Isolation, characterization, and evaluation of native rhizobacterial consortia developed from the rhizosphere of rice grown in organic state Sikkim, India, and their effect on plant growth. Front. Microbiol. 12:713660. doi: 10.3389/fmicb.2021.713660
Silva, J., Menéndez, E., Eliziário, F., Mateos, P. F., Alexandre, A., and Oliveira, S. (2019). Heterologous expression of nifA or nodD genes improves chickpea- Mesorhizobium symbiotic performance. Plant Soil 436, 607–621. doi: 10.1007/s11104-019-03950-0
Sinclair, S. A., and Krämer, U. (2012). The zinc homeostasis network of land plants. Biochim. Biophys. Acta 1823, 1553–1567. doi: 10.1016/j.bbamcr.2012.05.016
Sindhu, S., Sharma, R., Sindhu, S., and Sehrawat, A. (2019). “Soil fertility improvement by symbiotic rhizobia for sustainable agriculture” in Soil fertility management for Sustainable development. eds. D. G. Panpatte and Y. K. Jhala (Gateway East, Singapore: Springer Nature Singapore), 101–166.
Soumare, A., Boubekri, K., Lyamlouli, K., Hafidi, M., Ouhdouch, Y., and Kouisni, L. (2020). From isolation of phosphate solubilizing microbes to their formulation and use as biofertilizers: status and needs. Front. Bioeng. Biotechnol. 7:425. doi: 10.3389/fbioe.2019.00425
Srithaworn, M., Jaroenthanyakorn, J., Tangjitjaroenkun, J., Suriyachadkun, C., and Chunhachart, O. (2023). Zinc solubilizing bacteria and their potential as bioinoculant for growth promotion of green soybean (Glycine max L. Merr.). PeerJ 11, 1–17. doi: 10.7717/peerj.15128
Strigul, N. S., and Kravchenko, L. V. (2006). Mathematical modeling of PGPR inoculation into the rhizosphere. Environ. Model Softw. 21, 1158–1171. doi: 10.1016/j.envsoft.2005.06.003
Szczerba, M. W., Britto, D. T., and Kronzucker, H. J. (2009). K+ transport in plants: physiology and molecular biology. J. Plant Physiol. 166, 447–466. doi: 10.1016/j.jplph.2008.12.009
Tabassum, B., Khan, A., Tariq, M., Ramzan, M., Saleem, M., Khan, I., et al. (2017). Bottlenecks in commercialisation and future prospects of PGPR. Appl. Soil Ecol. 121, 102–117. doi: 10.1016/j.apsoil.2017.09.030
Takahashi, H. (2019). Sulfate transport systems in plants: functional diversity and molecular mechanisms underlying regulatory coordination. J. Exp. Bot. 70, 4075–4087. doi: 10.1093/jxb/erz132
Takahashi, H., Kopriva, S., Giordano, M., Saito, K., and Hell, R. (2011). Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62, 157–184. doi: 10.1146/annurev-arplant-042110-103921
Thilakarathna, M. S., and Raizada, M. N. (2017). A meta-analysis of the effectiveness of diverse rhizobia inoculants on soybean traits under field conditions. Soil Biol. Biochem. 105, 177–196. doi: 10.1016/j.soilbio.2016.11.022
Timofeeva, A., Galyamova, M., and Sedykh, S. (2022). Prospects for using phosphate-solubilizing microorganisms as natural fertilizers in agriculture. Plan. Theory 11, 1–23. doi: 10.3390/plants11162119
Tomasi, N., Pinton, R., Dalla, L., Cortella, G., Terzano, R., Mimmo, T., et al. (2015). New ‘solutions’ for floating cultivation system of ready-to-eat salad: a review. Trends Food Sci. Technol. 46, 267–276. doi: 10.1016/j.tifs.2015.08.004
Tourna, M., Maclean, P., Condron, L., Callaghan, M. O., and Wakelin, S. A. (2014). Links between sulphur oxidation and sulphur-oxidising bacteria abundance and diversity in soil microcosms based on soxB functional gene analysis. FEMS Microbiol. Ecol. 88, 538–549. doi: 10.1111/1574-6941.12323
Vacheron, J., Desbrosses, G., Bouffaud, M., Touraine, B., Moënne-Loccoz, Y., Muller, D., et al. (2013). Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 4:356. doi: 10.3389/fpls.2013.00356
Valentinuzzi, F., Pii, Y., Borruso, L., Mimmo, T., Puglisi, E., Trevisan, M., et al. (2021). Epiphytic microbial community and post-harvest characteristics of strawberry fruits as affected by plant nutritional regime with silicon. Agronomy 11, 1–13. doi: 10.3390/agronomy11122407
Vance, C. P., Uhde-Stone, C., and Allan, D. L. (2003). Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 157, 423–447. doi: 10.1046/j.1469-8137.2003.00695.x
Vessey, J. K. (2003). Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255, 571–586. doi: 10.1023/A:1026037216893
Vijay, K., Shibasini, M., Sivasakthivelan, P., and Kavitha, T. (2023). Microbial siderophores as molecular shuttles for metal cations: sources, sinks and application perspectives. Arch. Microbiol. 205, 1–17. doi: 10.1007/s00203-023-03644-3
von Wirén, N., Marschner, H., and Romheld, V. (1996). Roots of iron-efficient maize also absorb phytosiderophore-chelated zinc. Plant Physiol. 111, 1119–1125. doi: 10.1104/pp.111.4.1119
Wagh, J., Chanchal, K., Sonal, S., Praveena, B., Archana, G., Kumar, G. N., et al. (2016). Inoculation of genetically modified endophytic Herbaspirillum seropedicae Z67 endowed with gluconic and 2-ketogluconic acid secretion, confers beneficial effects on rice (Oriza sativa). Plant Soil 409, 51–64. doi: 10.1007/s11104-016-2937-7
Wagh, J., Shah, S., and Bhandari, P. (2014). Heterologous expression of pyrroloquinoline quinone (pqq) gene cluster confers mineral phosphate solubilization ability to Herbaspirillum seropedicae Z67. Appl. Microbial. Cell Physiol. 98, 5117–5129. doi: 10.1007/s00253-014-5610-1
Wang, Z., and Song, Y. (2022). Toward understanding the genetic bases underlying plant - mediated “cry for help” to the microbiota. iMeta 1, 1–12. doi: 10.1002/imt2.8
Wang, Y., and Wu, W. (2013). Potassium transport and signaling in higher plants. Annu. Rev. Plant Biol. 64, 451–476. doi: 10.1146/annurev-arplant-050312-120153
Watanabe, T., Kojima, H., Umezawa, K., Hori, C., and Scott, K. (2019). Genomes of neutrophilic sulfur-oxidizing chemolithoautotrophs representing 9 proteobacterial species from 8 genera. Front. Microbiol. 10:316. doi: 10.3389/fmicb.2019.00316
Wu, X., Rensing, C., Han, D., Xiao, K., Dai, Y., Tang, Z., et al. (2022). Genome-resolved metagenomics reveals distinct phosphorus acquisition strategies between soil microbiomes. mSystems 7, 1–13. doi: 10.1128/msystems.01107-21
Wu, X., Zhao, Z., Zhao, Z., Zhang, Y., Li, M., and Yu, Q. (2023). Analysis of the potassium-solubilizing Priestia megaterium strain NK851 and its potassium feldspar-binding proteins. Int. J. Mol. Sci. 24, 1–12. doi: 10.3390/ijms241814226
Yadav, R. C., Sharma, S. K., Varma, A., Rajawat, M., Khan, M., Sharma, P., et al. (2022). Modulation in biofertilization and biofortification of wheat crop by inoculation of zinc-solubilizing rhizobacteria. Front. Plant Sci. 13:777771. doi: 10.3389/fpls.2022.777771
Yang, S., Liu, H., Xie, P., Wen, T., Shen, Q., and Yuan, J. (2023). Emerging pathways for engineering the rhizosphere microbiome for optimal plant health. J. Agric. Food Chem. 71, 4441–4449. doi: 10.1021/acs.jafc.2c08758
Yu, P., He, X., Baer, M., Beirinckx, S., Tian, T., Moya, Y. A. T., et al. (2021). Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation. Nat. Plants 7, 481–499. doi: 10.1038/s41477-021-00897-y
Zhang, J., Liu, Y., Zhang, N., Hu, B., Jin, T., Xu, H., et al. (2019). NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 37, 676–684. doi: 10.1038/s41587-019-0104-4
Zhang, L., Yan, C., Guo, Q., Zhang, J., and Ruiz-Menjivar, J. (2018). The impact of agricultural chemical inputs on environment: global evidence from informetrics analysis and visualization. Int. J. Low-Carbon Technol. 13, 338–352. doi: 10.1093/ijlct/cty039
Zia, R., Shoib, M., Jawad, M., Hakim, S., and Imran, A. (2021). Plant survival under drought stress: implications, adaptive responses, and integrated rhizosphere management strategy for stress mitigation. Microbiol. Res. 242, 1–16. doi: 10.1016/j.micres.2020.126626
Zuluaga, M. Y. A., Cardarelli, M., Rouphael, Y., Cesco, S., Pii, Y., and Colla, G. (2023a). Iron nutrition in agriculture: from synthetic chelates to biochelates. Sci. Hortic. 312:111833. doi: 10.1016/j.scienta.2023.111833
Zuluaga, M. Y. A., De Oliveira, A. L. M., Valentinuzzi, F., Jayme, N. S., Monterisi, S., Fattorini, R., et al. (2023b). An insight into the role of the organic acids produced by Enterobacter sp. strain 15S in solubilizing tricalcium phosphate: in situ study on cucumber. BMC Microbiol. 23:184. doi: 10.1186/s12866-023-02918-6
Zuluaga, M. Y. A., Milani, K. M. L., Gonçalces, L. S. A., and Oliveira, A. L. M. (2020). Diversity and plant growth-promoting functions of diazotrophic/N-scavenging bacteria isolated from the soils and rhizospheres of two species of Solanum. PLoS One 15:e0227422. doi: 10.1371/journal.pone.0227422
Zuluaga, M. Y. A., Oliveira, A. L. M., Valentinuzzi, F., Tiziani, R., Pii, Y., Mimmo, T., et al. (2021). Can inoculation with the bacterial biostimulant Enterobacter sp. strain 15S be an approach for the smarter P fertilization of maize and cucumber plants? Front. Plant Sci. 12:719873. doi: 10.3389/fpls.2021.719873
Keywords: beneficial bacteria, crop nutrition, nitrogen biofertilizer, phosphate biofertilizer, bacterial siderophores
Citation: Alzate Zuluaga MY, Fattorini R, Cesco S and Pii Y (2024) Plant-microbe interactions in the rhizosphere for smarter and more sustainable crop fertilization: the case of PGPR-based biofertilizers. Front. Microbiol. 15:1440978. doi: 10.3389/fmicb.2024.1440978
Edited by:
Vincenzo Lionetti, Sapienza University of Rome, ItalyReviewed by:
Kailash Chand Kumawat, Sam Higginbottom University of Agriculture, Technology and Sciences, IndiaAbhishek Joshi, Mohanlal Sukhadia University, India
Copyright © 2024 Alzate Zuluaga, Fattorini, Cesco and Pii. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Monica Yorlady Alzate Zuluaga, bW9uaWNheW9ybGFkeS5hbHphdGV6dWx1YWdhQHVuaWJ6Lml0; Youry Pii, eW91cnkucGlpQHVuaWJ6Lml0
†These authors have contributed equally to this work
 Monica Yorlady Alzate Zuluaga
Monica Yorlady Alzate Zuluaga Roberto Fattorini†
Roberto Fattorini† Stefano Cesco
Stefano Cesco Youry Pii
Youry Pii