- 1School of Biotechnology and Biomolecular Sciences, University of New South Wales, Sydney, NSW, Australia
- 2Birling Laboratories, Bringelly, NSW, Australia
- 3Centre for Infectious Diseases and Microbiology-Laboratory Services, Institute of Clinical Pathology and Medical Research, NSW Health Pathology, Westmead Hospital, Westmead, NSW, Australia
- 4Marie Bashir Institute for Infectious Diseases and Biosecurity, Sydney Medical School, University of Sydney, Sydney, NSW, Australia
Contamination of poultry products by Salmonella enterica serovar Typhimurium (STm) is a major cause of foodborne infections and outbreaks. This study aimed to assess the diversity and antimicrobial resistance (AMR) carriage of STm in three chicken processing plants using genomic sequencing. It also aimed to investigate whether any particular strain types were associated with cases of human illness. Multilevel genome typing (MGT) was used to analyze 379 STm isolates from processed chicken carcasses. The diversity of chicken STm sequence types (STs) increased from MGT1 (2 STs) to MGT9 (257 STs). STs at MGT5 to MGT9 levels that were unique to one processing plant and shared among the processing plants were identified, likely reflecting the diversity of STm at their farm source. Fifteen medium resolution MGT5 STs matched those from human infections in Australia and globally. However, no STs matched between the chicken and human isolates at high resolution levels (MGT8 or MGT9), indicating the two STm populations were phylogenetically related but were unlikely to be directly epidemiologically linked. AMR genes were rare, with only a blaTEM-1 gene carried by a 95 kb IncI1 Alpha plasmid being identified in 20 isolates. In conclusion, subpopulations that were widespread in processing plants and had caused human infections were described using MGT5 STs. In this STM population, AMR was rare with only sporadic resistance to a single drug class observed. The genomic analysis of STm from chicken processing plants in this study provided insights into STm that contaminate meat chickens early in the food production chain.
1 Introduction
Non-Typhoidal Salmonella (NTS) is a common foodborne pathogen across the globe. Salmonella enterica serovar Typhimurium (STm) is the predominant NTS serovar causing foodborne infections in Australia. In recent decades, the escalation in reported instances of human salmonellosis cases and outbreaks has created a pressing public health concern (Ford et al., 2016; McLure et al., 2022). Salmonella primarily disseminates through contaminated food and contact with animal excrement or contaminated surfaces (Arnold et al., 2015; Moffatt et al., 2016). Infiltration of this pathogen into the food supply chain, especially during chicken processing, underscores the significance of investigating sources of contamination events (Fearnley et al., 2011; Ferrari et al., 2019; Stewart and Pavic, 2023). Strategies to prevent these events can then be developed to improve food safety and protect public health since the demand for chicken meat in Australia has increased significantly during the recent decades (Wong et al., 2015).
Integrated surveillance methods are important to understand and investigate outbreaks of Salmonella (Galanis et al., 2012; Sintchenko et al., 2012; Hu et al., 2021). Therefore, one of the most effective methodologies would be to detect the pathogen through the food production chain before causing infections and illness (Delgado-Suarez et al., 2018). Rapid, effective, and accurate identification of Salmonella populations is important to control outbreaks and disease transmission (Payne et al., 2020; Hu et al., 2021), and is vital for public health surveillance.
Multilevel genome typing (MGT) of STm is a publicly available whole genome sequencing (WGS) typing system (Payne et al., 2020; Kaur et al., 2022). It can be used to track STm spread and to understand evolutionary history of the pathogen (Payne et al., 2020). This typing method is based on multilocus sequence typing (MLST) (Maiden et al., 1998) and consisted of a set of nine MLST schemes progressively increasing in size and resolution (Figure 1) (Payne et al., 2020). Sequence types (STs) assigned at each MGT level can then be used to study the epidemiological distribution of STm populations (Payne et al., 2020). MGT1 is the traditional S. enterica MLST and has the lowest resolution whereas MGT8 is the core genome MLST (cgMLST) of the species, and MGT9 is the STm core genome MLST (serovar cgMLST) scheme (Payne et al., 2020). The highest resolution level, MGT9, is useful for outbreak investigation. By providing multiple levels of resolutions within a single typing system, MGT can accurately differentiate and type STm strains for both short and long-term epidemiology (Payne et al., 2020).
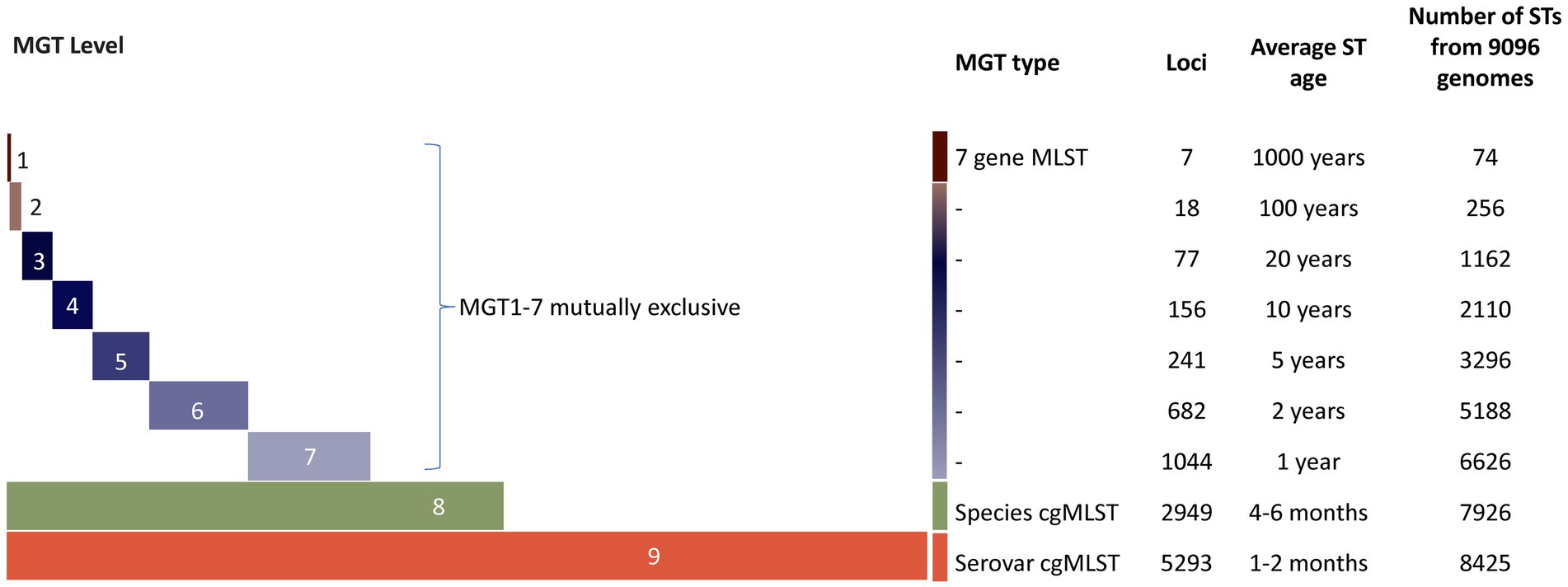
Figure 1. Illustrations of the multilevel genome typing (MGT) system from Payne et al. (2020). STm MGT scheme of 9 levels (represented by colored horizontal bars); the number of loci is increasing from MGT1 to MGT9 (represented by increasing length of the bars). The lowest resolution level, MGT1 is the classic seven gene MLST scheme and the highest resolution level, MGT9 made up of the cgMLST scheme of STm. MGT1–GT7 are mutually exclusive and composed of species cgMLST (MGT8). MGT8 (species cgMLST) is a subset of MGT9. The average ST age was defined as the average time for a new allele change to give rise to a new ST at a given MGT level. The increasing resolution was demonstrated using 9,096 genomes.
The emergence and dissemination of antimicrobial resistance (AMR) in NTS against several crucial antimicrobials like fluoroquinolones and extended spectrum cephalosporins are a significant threat to public health worldwide (Crump et al., 2011; WHO, 2014). The prevalence of ciprofloxacin resistance among Salmonella isolated from broilers was found to be high across 19 European Union member states (European Food Safety et al., 2021). In a recent international review, colistin resistance in Salmonella from poultry was found to be prevalent in both developing and developed countries, frequently surpassing 5% of isolated cases (Apostolakos and Piccirillo, 2018). Importantly, the rapid acquisition of AMR genes within plasmids potentially gives rise to multidrug-resistant bacteria that challenge treatment strategies (Ingle et al., 2021; Castaneda-Barba et al., 2023).
Within Australia, the restricted usage of antimicrobials in livestock and close monitoring of all fresh meat products (Barlow et al., 2015; Abraham et al., 2019) means that AMR incidence is minimal in Salmonella isolated from Australian farm animals (Barlow et al., 2015; Abraham et al., 2019, 2022). However, although there is a public health Salmonella surveillance program in Australia (OzFoodNetWorkingGroup, 2015), there is always a need to monitor AMR in farm animals for one health (Williamson et al., 2018; Abraham et al., 2019, 2022).
In this study, we were focused on early detection of STm along the chicken production chain. Therefore, the objective of this study was to assess the diversity and AMR carriage of S. Typhimurium in three chicken processing plants using whole genome sequencing and to investigate whether any particular strain types were associated with cases of human illness.
2 Materials and methods
2.1 Source of Salmonella Typhimurium isolates
In Australia there are two major meat chicken breeds, Cobb and Ross, and the Australian poultry industry harvest birds from 1,600 to 3,200 grams in dress weight. Microbiological screening of processed chicken carcasses is routinely performed at Birling Laboratory, this includes isolation of Salmonella using the ISO 6579 culture method with PCR confirmation for STm (Mooijman, 2018). The STm isolates were from three chicken processing plants in New South Wales (NSW) and were isolated from post chill carcasses as part of routine testing as required by the jurisdiction. A total of 379 STm isolates were collected as routine surveillance between January 2021 to December 2022 (Supplementary Table S2).
All samples tested were whole bird rinsates in accordance to AS 5013.20-2017. The standard Australian processing sampling plan is based upon number of samples processed with small plants (<100,000 samples) sampling 2 carcasses pre shift and large plants (>100,000 samples) sampling 1 in 25,000 carcasses. Whole carcasses were sent to service laboratories and the carcass rinse method was used to test for Salmonella presence/absence.
Carcasses were rinsed in 500 mL of buffered peptone water (BPW), the BPW was incubated overnight at 37°C. The non-selective BPW culture was then transferred into selective enrichment in Rappaport Vassiliades Broth (RVS) (Edwards Group),100 μL in 9.9 mL and Tetrathionate Hajna broth (Edwards Group), 1 mL in 9 mL broth. These broths were then incubated overnight at 42°C and 37°C, respectively. The selective broths were plated out on Hektoen and Xylose Lysine Deoxycholate (XLD) plates (Edwards Group), and incubated overnight at 37°C. Typical colonies were transferred onto ChromID Salmonella agar plates (Edwards Group), and incubated overnight at 37°C. Colonies with typical Salmonella features were then transferred to Nutrient Agar and incubated overnight at 37°C. The isolates were then confirmed as STm by serological grouping with O:5 and H:i antisera (Cell Biosciences). Further confirmation was carried out by a proprietary in-house amplicon based Next-generation sequencing (NGS) typing scheme. All 4,713 publicly available raw reads sets from Australian STm isolates were downloaded (5/4/2024) from Enterobase and NCBI (National Center for Biotechnology Information) and compared with S. Typhimurium isolated from chicken in this study.
2.2 DNA extraction, library preparation and sequencing
The genomic DNA was extracted using DNeasy Blood & Tissue Kits (QIAGEN) as per the manufacturer’s instructions. The sequencing libraries were prepared using Nextera XT DNA Library Prep Kit (Illumina) and sequenced on NextSeq 500 instrument using NextSeq 500/550 v2 mid output Kits (Illumina).
2.3 Multilevel genome typing
Raw reads of STm isolates were processed using MGT-Reads to Alleles as described in Payne et al. (2020). This script performs genome assembly, quality filtering, serotype confirmation (SISTR) and initial allele calling.1 Allele call files were then uploaded onto the MGT website to assign final allele numbers and MGT STs. Kraken2 (Wood et al., 2019) was used to identify any contamination with other species. The public STm genome data and MGT types were accessed through MGTdb (Kaur et al., 2022).2
2.4 Phylogenetic analysis
Phylogenetic tree of 379 STm isolates was built from allelic profiles of MGT9 STs using GrapeTree 1.5.0 (Zhou et al., 2018) using the RapidNJ algorithm (Simonsen et al., 2008). The visualization was executed utilizing the interactive mode provided by Grapetree.
Raw reads were used to build phylogenetic trees using core genome single nucleotide polymorphisms (SNPs). The reference genome sequence used was STm strain LT2 with complete genome (NC_003197.2) obtained from NCBI public databases in FASTA format. Snippy version 4.6.03 was used to align the sequence reads to the reference genome and variants were identified. IQ-TREE4 multicore version 2.2.0.3 (COVID-edition for Linux 64-bit built Aug 2.2022) was used to construct the phylogenetic tree using the resulting core alignment generated from Snippy with 10,000 bootstrap pseudo replicates (Hoang et al., 2018). The results were visualized and annotated with metadata with iTol (version 6) interactive online tool (Letunic and Bork, 2021).
2.5 Antimicrobial resistance and plasmid specific genes
Antimicrobial resistance genes and mutations were detected from 379 assembled genomes using AbritAMR version 1.0.145 using the AMRFinder Plus database (version 3.10.42) (Feldgarden et al., 2021; Sherry et al., 2023) and ResFinder (version 4.5.0) (Florensa et al., 2022).6 To identify plasmid specific AMR genes, assembly contigs were further annotated using Prokka (rapid prokaryotic genome annotation) version 1.14.6 (Seemann, 2014).7 The 379 STm isolates were screened for plasmids genes using plasmidfinder (Carattoli et al., 2014).
2.6 Nanopore sequencing
For isolates with AMR genes (20 isolates), nanopore sequencing was performed using a MinION [Oxford Nanopore Technologies, Oxford, UK (United Kingdom)]. Input genomic DNA was quantified using a Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, United States) and a NanoDrop spectrophotometer (Thermo Fisher Scientific). Then the long reads were assembled using microPIPE8 in combination with Illumina sequencing reads. Nucleotide blast (Camacho et al., 2009) analysis was performed using the NCBI nucleotide blast tool9 to identify similarities between the contigs with AMR gene and sequences available in the NCBI nucleotide database. Following the assembly of plasmid contigs, a phylogenetic analysis was conducted to elucidate the evolutionary relationship among the identified plasmids. The analysis was performed using Parsnp (Treangen et al., 2014)10 and the results were visualized using iTol (version 6) interactive online tool and Gingr (Treangen et al., 2014) Multiple plasmids were aligned using Clustal Omega (Sievers et al., 2011), and plasmid comparisons were visualized using BRIG (Alikhan et al., 2011). SNPs were identified by alignments within the plasmids. Repetitive regions were manually excluded. The isolates with AMR were screened for plasmids genes using plasmidfinder (Carattoli et al., 2014). Finally, BLAST was performed with identified plasmids gene sequences with the rest of the 373 genomic DNA sequences to detect the presence of plasmids.
3 Results
3.1 Multilevel genome typing and phylogenetic clustering of Salmonella Typhimurium chicken isolates
The 379 STm isolates from processed chicken carcasses were typed using the STm MGT scheme (Table 1). At MGT1 (seven-gene MLST) they were typed into two STs, with MGT1 ST19 and ST2066 representing 87.1% (330 isolates) and 12.9% (49 isolates) of the isolates, respectively. These isolates were further typed at higher MGT levels (Table 1). At MGT8 (species cgMLST), the isolates were divided into 204 ST with 1 to 23 isolates per ST. At MGT9 (serovar cgMLST), the isolates were divided into 257 STs with 1 to 16 isolates per ST.
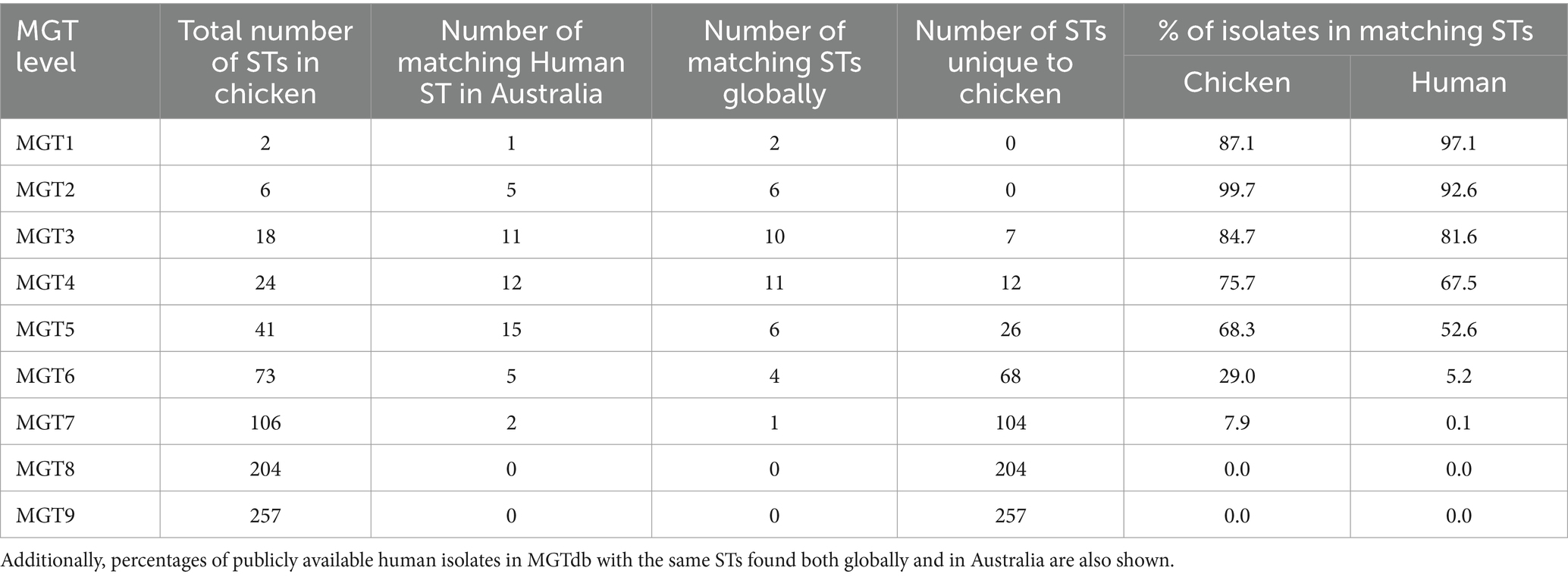
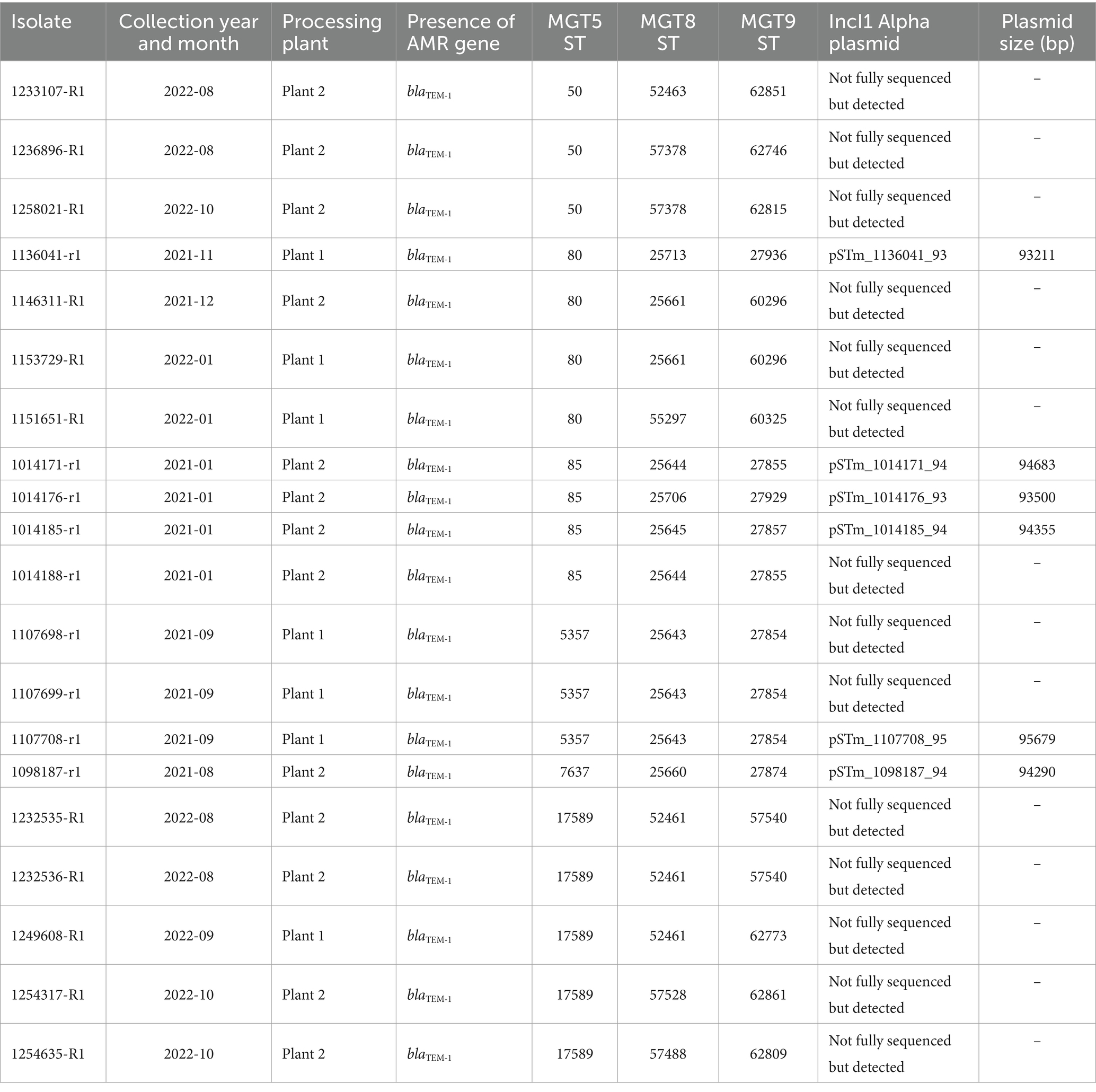
Table 1. Multilevel genome typing of chicken S. Typhimurium isolates and comparison with human isolates.
Phylogenetic clustering of the STm isolates was performed using MGT9 allele profiles. STs at different MGT levels were mapped onto the tree (Figure 2; Supplementary Figures S3, S4) and MGT5 STs were found to best reflect the major phylogenetic clusters (Figure 2), compared to other MGT levels. Of the 41 MGT5 STs, 22 STs with more than one isolate contained 94.9% of the isolates, with MGT5 ST342 being the largest at 18.5% along with MGT5 ST50 at 12.9%, and MGT5 ST9241 at 11.3%. MGT8 and MGT9 STs were consistent with whole genome SNP analysis in phylogenetic grouping. At MGT8 and MGT9, 75.88 and 80.33% of STs were found to be in the same SNP type with identical resolution (Supplementary Table S3).
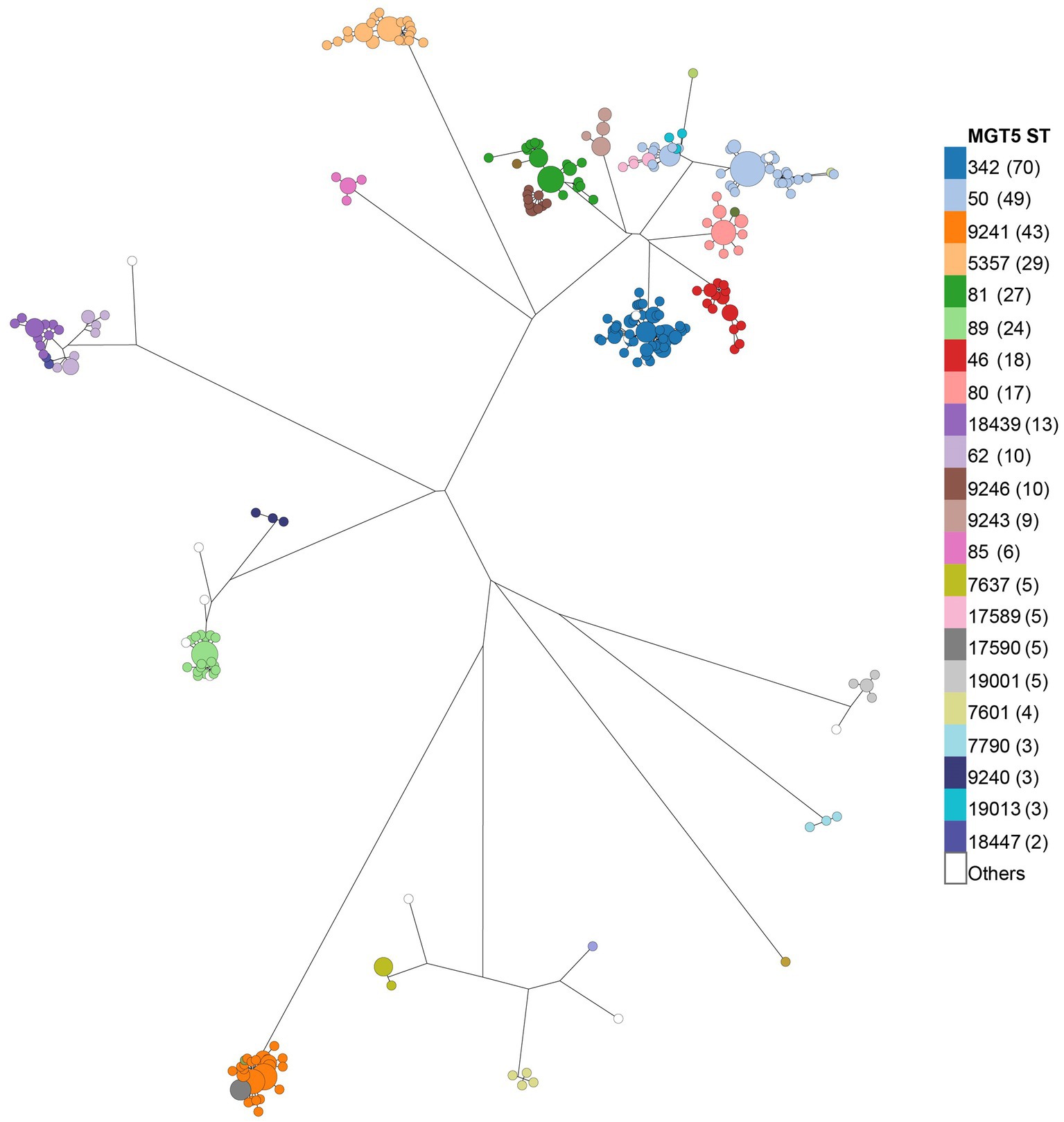
Figure 2. Population structure of chicken STm isolates at the MGT5 level. The phylogenetic tree was constructed using MGT9 allelic profiles. Each round dot at the tip of the tree represents an MGT9 ST. MGT5 STs were overlaid onto the tree per color legend to visualize MGT9 STs grouped by MGT5 STs and phylogenetic clustering of MGT5 STs. The Number in brackets after each ST in the color legend is the number of isolates of a given MGT5 ST.
3.2 Epidemiological trends of the chicken STm isolates
The spatial and temporal trends of chicken STm isolates were examined using MGT5 (Supplementary Figure S2), MGT8 and MGT9 STs (Figure 3). The STm isolates were obtained from three different chicken processing plants which are from three different geographical locations with most of the isolates coming from plant 1 (52.2% of the isolates) (Supplementary Table S2). Of the 41 MGT5 STs, four with 42.2% of the total isolates (ST342, ST5357, ST46, and ST9241) were found in all three plants (Supplementary Figure S2A). The highest number of isolates and STs were between October 2021 to January 2022, which coincided with the hottest months of the year in Australia. There were persistent STs such as MGT5 ST342, MGT5 ST46, and MGT5 ST9241 which were found for more than 12 months while some MGT5 STs were only found in a single month (Supplementary Figure S2B).
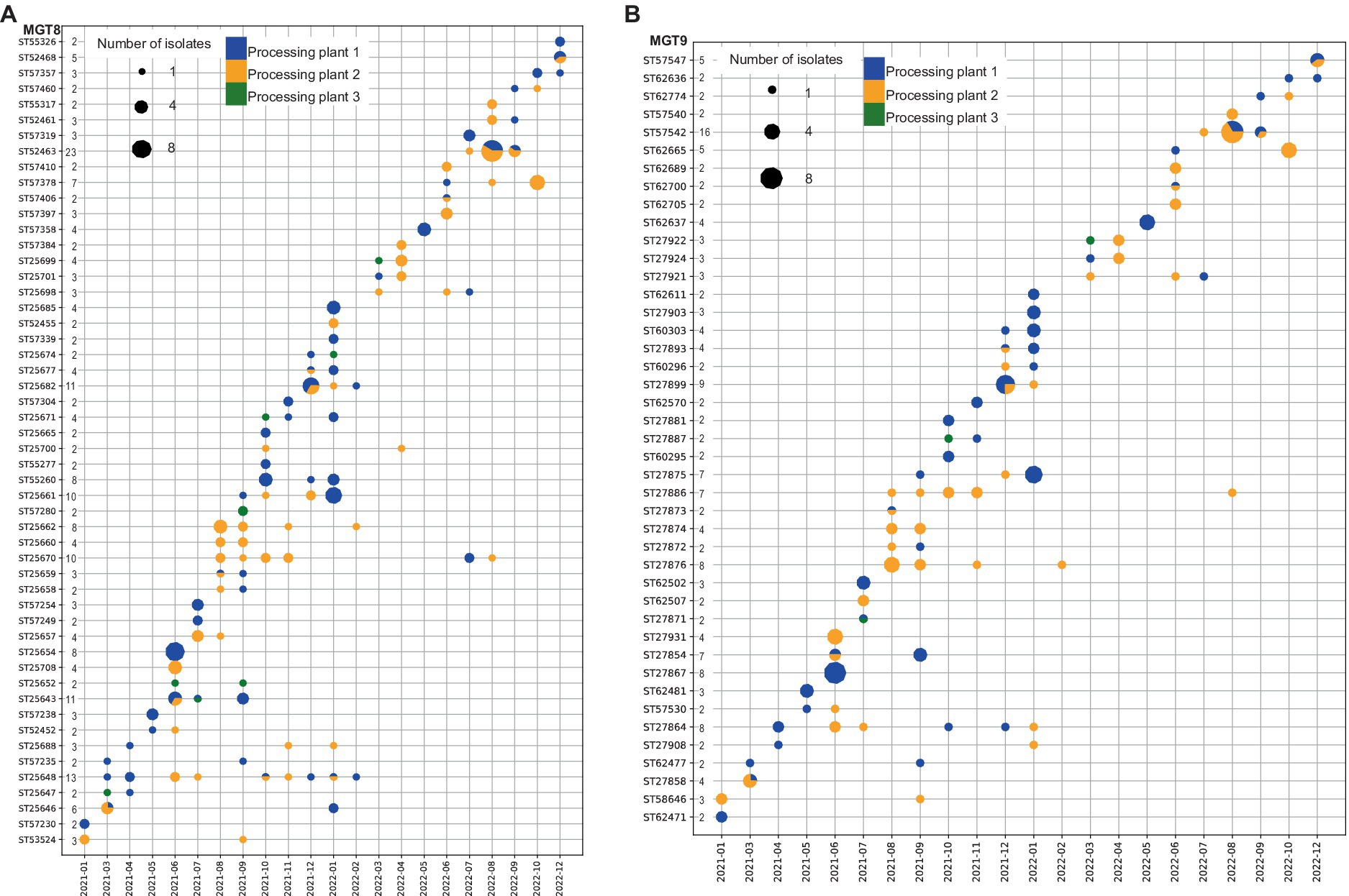
Figure 3. Temporal and spatial dynamics of isolates at MGT8 and MGT9 levels. Distribution of STs with multiple isolates per ST across the three processing plants and temporal patterns throughout the collection period. (A) At MGT8 level; (B) at MGT9 level. Y axis lists STs and the number of isolates while X axis marks the year and month of isolate collection. Note the discontinuity of months as months without isolates are not shown. The colors of the dots represent processing plants, and the size of the dots represents the number of isolates as shown in the legends.
At MGT8 (Figure 3A), 52 (59.9% of the isolates) of the 204 STs had more than 1 isolate and 1 ST (MGT9 ST25643) was shared across all the processing plants. Twenty-one MGT8 STs with 32.7% of the isolates were shared among two plants and 18 MGT8 STs with 29.5% of the isolates were shared between processing plant 1 and 2. Of these shared STs, MGT8 ST25670 and MGT8 ST25648 were sampled across 12 to 13 months from March 2021 to August 2022 while the rest were sampled across one to 11 months from March 2021 to December 2022. Plant specific STs were sampled across 1 to 4 months during the collection period.
At MGT9 (Figure 3B) only 43 (43.5% of the isolates) of the 257 STs had more than 1 isolate and similar to MGT8 STs, 21 MGT9 STs with 24.3% of the isolates were shared between two plants and none of the STs was shared among all the processing plants. At MGT9 20 STs represent a subset of their respective MGT8 STs demonstrating further resolution of MGT9 while the remaining STs contained the same isolates as their respective MGT8 STs. Of the 21 MGT9 STs found in multiple processing plants 18 STs, containing 22.4% of all isolates, were shared between processing plant 1 and 2. Of the shared STs, MGT9 ST27908, MGT9 ST27864, and MGT9 ST27866 were sampled across 10 to 13 months from April 2021 to August 2022 while the remaining 18 STs were sampled across 1 to 8 months throughout the collection period. All plant specific STs were sampled across 1 to 4 months.
3.3 Carriage of antibiotic resistance genes and plasmids
The 379 STm isolates were screened for AMR genes using AbritAMR and ResFinder. A β-lactamase gene, blaTEM-1, was identified in 20 isolates that were collected in 2021 to 2022 (Table 2). They belonged to 15 MGT9 STs (Table 2). Of those MGT9 STs, ST27855, ST60296, and ST57540 had two isolates while MGT9 ST27854 had 3 isolates. To determine whether blaTEM-1 was carried by a plasmid or on the chromosome, we fully sequenced the genomes of six of the nine isolates from different MGT9 STs (Supplementary Table S2). The blaTEM-1 gene was found to be on an IncI1 Alpha plasmid in all six isolates (Table 2). All six isolates also carried an IncIFIB(S)/IncIFII(S) plasmid [similar to S. Typhimurium plasmid pSLT (AE006471)] and 3 isolates carried a Col156 plasmid [similar to Salmonella enterica Col156 plasmid (CP058808.1)]. BLAST analysis using complete sequences of all three plasmids extracted from the complete assembly of isolate 1014176-rl and confirmed that all the remaining 373 isolates with draft genomes in the study carried an IncIFIB(S)/IncIFII(S) plasmid while 40 isolates carried a Col156 plasmid and all 20 isolates with the blaTEM-1 gene carried the same IncI1 Alpha plasmid (Supplementary Table S2).
The IncI1 alpha plasmids from the six isolates were between 93,211 bp – 95,679 bp in size (Table 2) and were most similar (with more than 99% identity) to a plasmid from Escherichia coli ECP19-2498 (CP066749.1) which was 95,679 bp in size (Figure 4). Comparison of STm plasmids showed minor structural changes among them (Figure 4). pSTm_1014171_94 had an indel (89598–90332 bp) encoding an IS3 family insertion sequence (IS) IS629. pSTm_1014185_94 had an indel (89210–89845 bp) encoding an IS3 family IS IS1203. pSTm_1107708_95 had an indel (77797–78015 bp) encoding antitoxin ccdA. An indel encoding Shufflon protein D was found in all the plasmids except for pSTm_1136041_93. Colicin IA synthesis gene (cai) and colicin IA immunity protein synthesis gene (cia) were identified in six STm plasmids closely located with blaTEM-1 (less than 800 bp apart) (Figure 4). Within the E coli ECP19-2498 plasmid, there were two genes, istA (67923 bp–69458 bp) and istB (69475 bp–70230 bp) which were not found in the six STm plasmids. Apart from these indels, pairwise comparison of STm plasmids and ECP19-2498 plasmid differ by 1 to 6 SNPs.
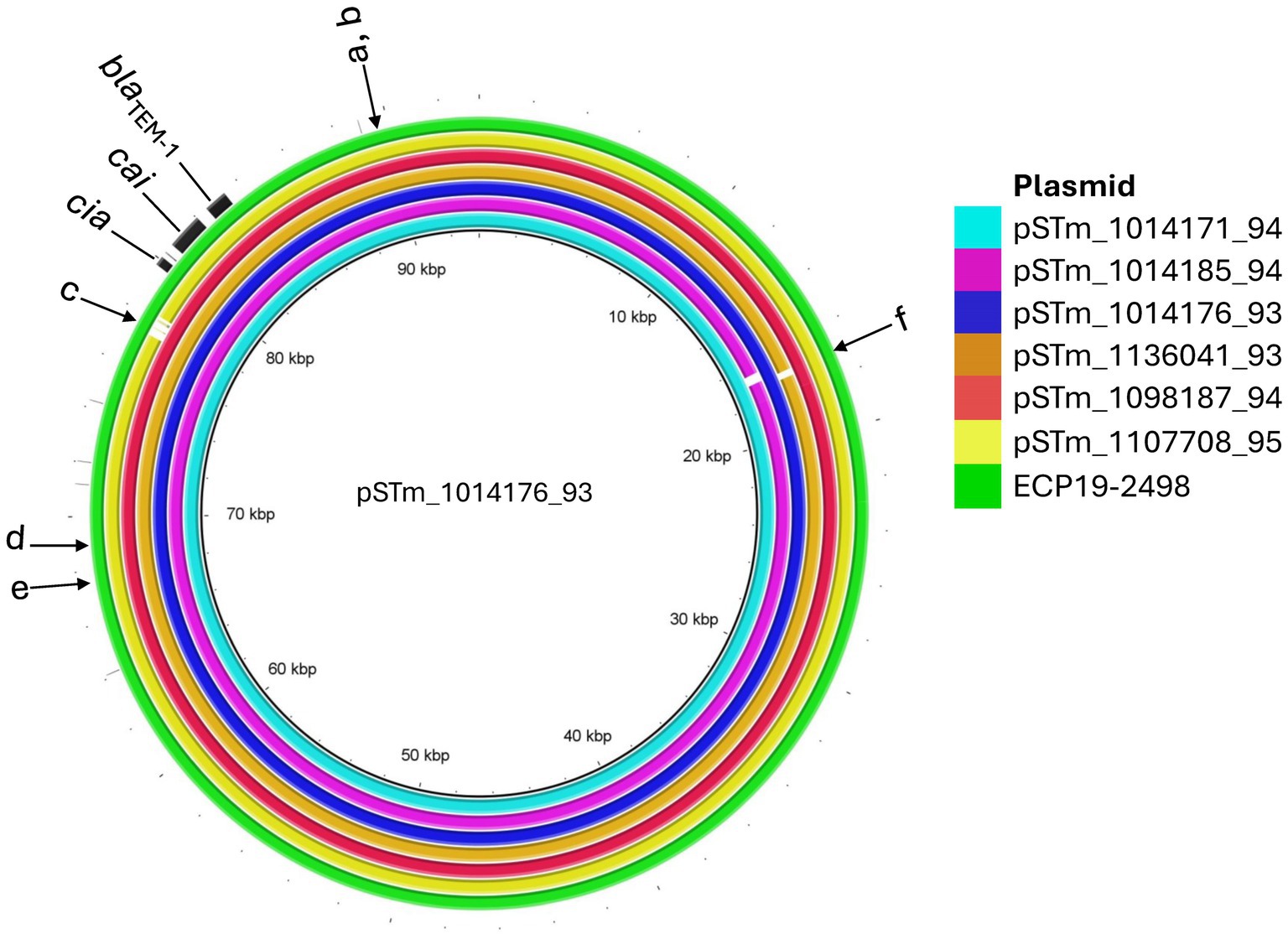
Figure 4. Multiple genome comparisons using BRIG with one of the IncI1 Alpha plasmids from this study, pSTm_1014176_93 as the reference. Colored rings indicate the sequence similarity between IncI1 Alpha plasmid genomes from the other isolates and Escherichia coli O157:H7 strain ECP19-2498 (CP066749.1) plasmid. In the figure, blaTEM-1, cai (colicin IA synthesis gene) and cia (colicin IA immunity protein synthesis gene) genes were annotated. Letters a-f indicates indels as follows: (a) IS629 (in pSTm_1014171_94), (b) IS1203 (in pSTm_1014185_94), (c) Antitoxin ccdA (in pSTm_1107708_95), (d) IS21 family transposase istA, (e) IS21-like element ISSso4 family helper ATPase istB (in ECP19-2498 plasmid) and (f) An indel region encoding Shufflon protein D was found in all the plasmid except for pSTm_1136041_93.
3.4 Comparison of chicken STm isolates with historical Australian isolates in the MGT database
The chicken isolates were compared with historical Australian isolates in the MGT database at all MGT levels (Table 1). Among the 4,714 Australian isolates in the database, 3,728 were from humans, while the remaining isolates were from fecal matter (human or animal), animals, processed foods, raw food, and surfaces. These isolates had a collection year ranging from 1992 to 2020. No STs were identical at MGT8 and MGT9 levels between chicken isolates from this study and isolates in the database. At MGT5 (Supplementary Table S1), MGT6 and MGT7, there were 15, 5 and 2 STs that matched between our STm isolates and the database isolates (Table 1). Note that at MGT5, isolates of the same ST could have diverged by many years (Payne et al., 2020). As an example, we constructed an SNP based phylogeny of both human and chicken MGT5 ST342 isolates and showed that human and chicken derived isolates were in well separated branches of the tree (Figure 5). Thus, the human isolates shared a common ancestor with chicken isolates but with no direct epidemiological link.
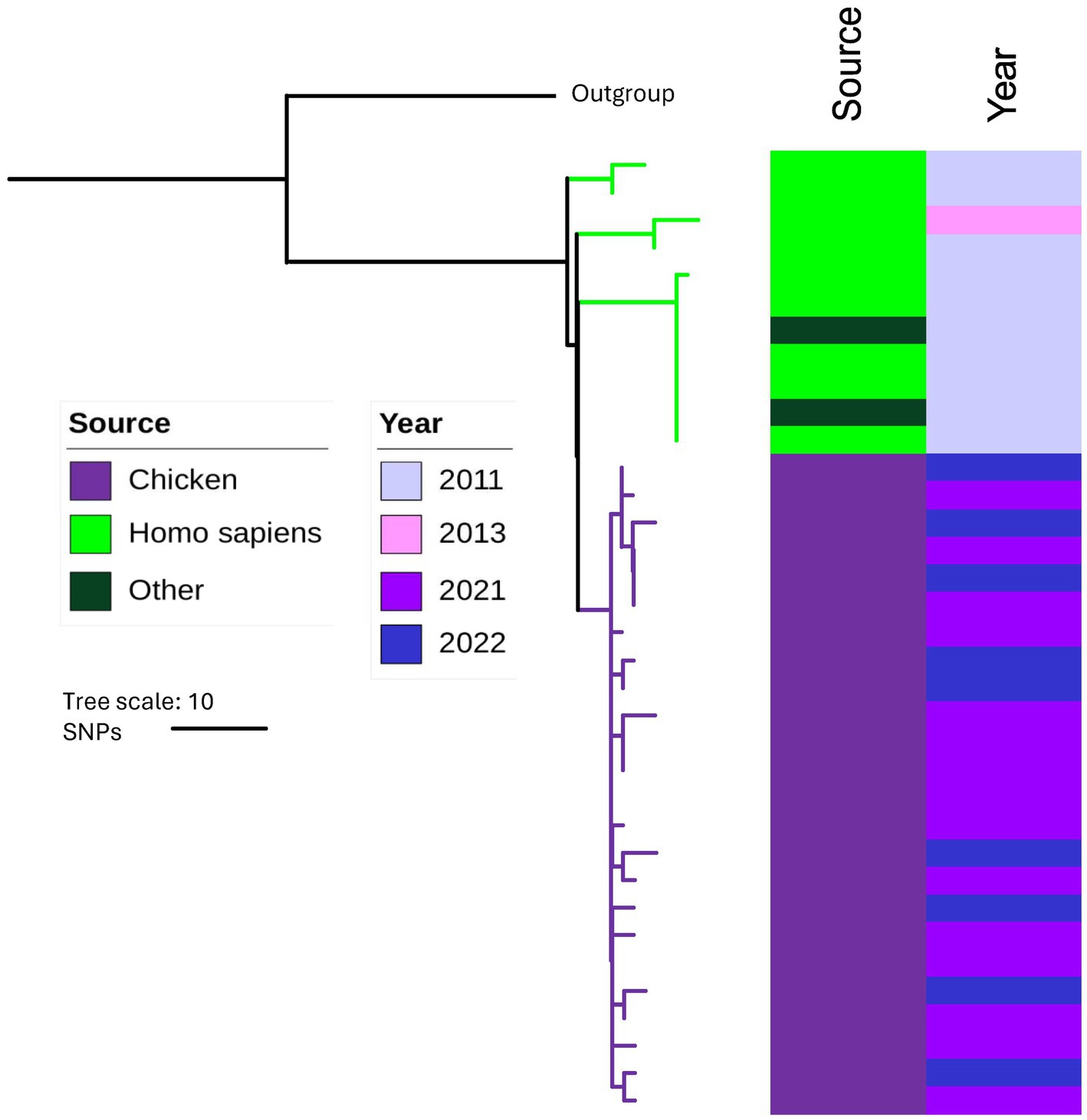
Figure 5. SNP based Phylogeny of isolates in MGT5 ST342. The colored strips on the right hand side represent source and the year of collection of the isolates. The outgroup used was an MGT5 ST5357 isolate. Chicken STm isolates (year 2021–2022) are represented by purple color and historical human isolates (year 2011–2013) in the MGT data base are represented by green color.
We also compared STs of our chicken isolates at each MGT level with historical human STm isolates from other countries in the MGT database which had 16,374 human isolates (Table 1). No STs were identical at MGT6 to MGT9 between chicken STm from this study and isolates from other countries in the database (Supplementary Figure S1). At MGT5, two chicken STs, ST46 and ST3537, shared with human STs from other countries. MGT5 ST46 was found in New Zealand and United Kingdom while ST3537 was found in the United Kingdom (Supplementary Figure S1). All STs shared with other countries were also found in human isolates from Australia.
4 Discussion
Through contamination of meat chickens, Salmonella can pass down the food production chain to cause infections in humans (Ferrari et al., 2019; Van Immerseel et al., 2019). Meat chickens have a higher prevalence of Salmonella and one of the major sources of human exposure (McLure et al., 2022). However very little is known of STm from meat chickens in Australia as many outbreaks in Australia were caused by STm that can be traced back to contaminations of eggs or egg products (McWhorter et al., 2015; Moffatt et al., 2016; Ford et al., 2018; McLure et al., 2022). In this study, we sequenced 379 isolates obtained from 3 chicken processing plants to assess the genetic diversity of STm and their relationship to retrospective human isolates in Australia. Contemporary human isolate data from Australia were not publicly available, preventing direct comparison with meat chicken isolates of this study. Previous studies in NSW carcass processing facilities have shown low level of Salmonella cross-contamination between carcasses within the processing environment (Pavic et al., 2015). All processing facilities are required by the food regulators to frequently shut down operations to conduct cleaning which is verified by a complementary hygiene swabbing program using ATPase to assure no protein matter is present on food contact surfaces prior to start up (Pavic et al., 2015). Total viable counts are used to assure that the sanitizers and contact times are fit for purpose (Pavic et al., 2015). The cross contamination between carcasses during processing is possible (Pavic et al., 2015) but would be limited to birds slaughtered on the same day. The diversity of isolates seen in this study supports this finding as contamination from within the processing plant would likely be highly genetically homogenous. A more likely scenario is that STm sampled in this study is representative of a diverse set of endemic strains from local donor farms (<100 km from plants).
MGT for STm has 9 levels with scalable resolution (Payne et al., 2020). Lower MGT levels (MGT1 to MGT7) are useful for elucidating the longer-term epidemiological patterns, while the higher MGT levels (MGT8 and MGT9) are aimed for investigating short term epidemiological trends (Payne et al., 2020). Specifically, MGT8 and MGT9 corresponds to species-level cgMLST and serovar-level cgMLST, respectively (Payne et al., 2020). To showcase the application and difference in resolution of MGT8 and MGT9, we applied both to the STm isolates in this study. MGT9 offered the highest resolution and isolates of the same MGT9 ST were genetically nearly identical.
Using the increasing resolution of MGT levels the population of STm from processing plants can be compared with human isolates to identify the degree of relatedness between the two populations. By traditional MLST (MGT1), two STs, ST19 and ST2066 were observed. MGT1 ST19 is the predominant global ST while MGT1 ST2066 is relatively rare having been isolated in Australian human infections (Lan et al., 2009; Pang et al., 2012). By MGT5 there was considerable diversity with 41 STs, 15 of which overlapped with STs from human infections in Australia. Comparisons were also made with human STm from other countries, in particular, United Kingdom and United States as there were large numbers of STm sequenced. At MGT5, 2 STs were shared, these STs caused human infections in multiple countries including Australia. At MGT8 to MGT9, no STs were shared internationally and all the chicken STs were unique to Australia. Human isolates shared a common ancestor with chicken isolates in recent past, however there was no direct epidemiological link.
MGT typing revealed temporal and spatial trends of STm within and between processing plants. At MGT5, 4 STs were more prevalent and found across the collection period and were present in all three processing plants, suggesting that these STs were widely present across different farms while the STs that were specific to one plant were likely to be restricted to certain farms which supplied chickens to only one processing plant. At MGT9, there were few isolates with identical STs in a processing plant. Again, this diversity suggests that the STms were passed down from farms with the source live chicken rather than from persistent contamination of the processing plant. However, 43 of the 257 STs MGT9 STs (43.5% of the population) were sampled 2 to 16 times. About half of these STs were sampled from one processing plant. Five MGT9 STs were found in more than 1 plant from April 2021 to August 2022. This observation suggests a common vehicle of transmission that could include common breeder flocks, feed, equipment, and services across multiple farms. At MGT8 (species cgMLST), the pattern of distribution observed was similar to MGT9, but a higher percentage of the STs (59.9% of the population) contained more than 1 isolate. These findings highlight the usefulness of MGT8 and MGT9 STs as a high-resolution typing tool to potentially trace back the contamination of chicken products or infections in humans to its source at farm level.
The presence of AMR genes in the STm isolates of this study was very low, with 20 isolates carrying blaTEM-1, a β lactamase gene conferring resistance to ampicillin. Rigorous regulation of antimicrobial usage in food animals within Australia has contributed to the low levels of AMR observed in bacteria originating from Australian livestock (Barlow et al., 2015; Abraham et al., 2019). The blaTEM-1 gene was found to be carried by an IncI1 Alpha plasmid in all blaTEM-1 positive isolates and the plasmid was highly similar to an E. coli plasmid. Gaining plasmids with resistance genes provides a competitive advantage in conditions with high antimicrobial usage (Alikhan et al., 2022). However, given the low antibiotic usage in Australia it is possible that the selection for this plasmid may be due to other plasmid encoded factors. The plasmid also carried colicin IA synthesis gene (cai) and colicin IA immunity protein synthesis gene (cia). It is possible that colicin production conferred a competitive advantage when competing with other microorganisms (Rendueles et al., 2014). IncI1 plasmids are frequently found in Salmonella from food animal sources as well as human infections (Smith et al., 2015; Kaldhone et al., 2019; Foley et al., 2021) and their capacity to disseminate AMR genes within enteric pathogens is well recognized (Smith et al., 2015; Kaldhone et al., 2019; Foley et al., 2021).
5 Conclusion
Genomic analysis of STm isolates from chicken processing plants in NSW provided insights into STm populations that contaminate chickens in the food production pipeline. This study also showcased the application of MGT in food production STm surveillance. There was a high diversity of STm with most isolates belonging to unique MGT9 STs. The diversity of STm from chicken processing plants was most likely a reflection of STm diversity at farm level. However, there were isolates from different sampling times or processing plants belonging to the same MGT9 STs, suggesting contamination of chickens by the same ST at its initial source or at the processing plants. Comparison of retrospective human isolates from Australia and other countries revealed that the chicken and human STm STs, in particular those from Australia, overlapped at MGT5. While the two STm populations were not identical, they were related and shared the most recent common ancestor. These findings will be useful for developing intervention strategies to reduce the transmission of STm down the food production chain to cause infections in humans.
Data availability statement
The data presented in the study are deposited in the NCBI repository, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1119266.
Author contributions
SB: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Visualization, Writing – original draft, Writing – review & editing. SW: Resources, Supervision, Validation, Writing – review & editing. JS: Resources, Writing – review & editing. MP: Conceptualization, Methodology, Project administration, Software, Supervision, Validation, Writing – review & editing. SK: Software, Validation, Writing – review & editing. QW: Resources, Writing – review & editing. VS: Resources, Writing – review & editing. AP: Resources, Supervision, Validation, Writing – review & editing. RL: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Health and Medical Research Council Ideas grant (2011806). SB was supported by an Australian Government Research Training Program Scholarship.
Acknowledgments
The authors thank the staff at Birling Laboratories for supplying the samples and NSW Enteric Reference Laboratory at ICPMR for chicken STm isolate processing and routine serotyping.
Conflict of interest
SW, JS, and AP were employed by Birling Laboratories and provided poultry isolates and metadata.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1440777/full#supplementary-material
Footnotes
1. ^https://github.com/LanLab/MGT_reads2alleles
2. ^https://mgtdb.unsw.edu.au/typhimurium
3. ^https://github.Com/tseemann/snippy
4. ^https://github.com/iqtree/iqtree2
5. ^https://github.com/MDU-PHL/abritamr
6. ^https://bitbucket.org/genomicepidemiology/resfinder.git
7. ^https://github.com/tseemann/prokka
8. ^https://github.com/BeatsonLab-MicrobialGenomics/micropipe
References
Abraham, S., O'Dea, M., Sahibzada, S., Hewson, K., Pavic, A., Veltman, T., et al. (2019). Escherichia coli and Salmonella spp. isolated from Australian meat chickens remain susceptible to critically important antimicrobial agents. PLoS One 14:e0224281. doi: 10.1371/journal.pone.0224281
Abraham, R., Sahibzada, S., Jordan, D., O'Dea, M., Hampson, D. J., McMillan, K., et al. (2022). Antimicrobial resistance and genomic relationships of Salmonella enterica from Australian cattle. Int. J. Food Microbiol. 371:109672. doi: 10.1016/j.ijfoodmicro.2022.109672
Alikhan, N. F., Moreno, L. Z., Castellanos, L. R., Chattaway, M. A., McLauchlin, J., Lodge, M., et al. (2022). Dynamics of Salmonella enterica and antimicrobial resistance in the Brazilian poultry industry and global impacts on public health. PLoS Genet. 18:e1010174. doi: 10.1371/journal.pgen.1010174
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 1–10. doi: 10.1186/1471-2164-12-402
Apostolakos, I., and Piccirillo, A. (2018). A review on the current situation and challenges of colistin resistance in poultry production. Avian Pathol. 47, 546–558. doi: 10.1080/03079457.2018.1524573
Arnold, M. E., Gosling, R. J., Martelli, F., Mueller-Doblies, D., and Davies, R. H. (2015). Evaluation of the sensitivity of faecal sampling for detection of monophasic Salmonella Typhimurium and other Salmonella in cattle and pigs. Epidemiol. Infect. 143, 1681–1691. doi: 10.1017/S0950268814002453
Barlow, R. S., McMillan, K. E., Duffy, L. L., Fegan, N., Jordan, D., and Mellor, G. E. (2015). Prevalence and antimicrobial resistance of Salmonella and Escherichia coli from Australian cattle populations at slaughter. J. Food Prot. 78, 912–920. doi: 10.4315/0362-028X.JFP-14-476
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform 10:421. doi: 10.1186/1471-2105-10-421
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Castaneda-Barba, S., Top, E. M., and Stalder, T. (2023). Plasmids, a molecular cornerstone of antimicrobial resistance in the one health era. Nat. Rev. Microbiol. 22, 18–32. doi: 10.1038/s41579-023-00926-x
Crump, J. A., Medalla, F. M., Joyce, K. W., Krueger, A. L., Hoekstra, R. M., Whichard, J. M., et al. (2011). Antimicrobial resistance among invasive nontyphoidal Salmonella enterica isolates in the United States: National Antimicrobial Resistance Monitoring System, 1996 to 2007. Antimicrob. Agents Chemother. 55, 1148–1154. doi: 10.1128/AAC.01333-10
Delgado-Suarez, E. J., Selem-Mojica, N., Ortiz-Lopez, R., Gebreyes, W. A., Allard, M. W., Barona-Gomez, F., et al. (2018). Whole genome sequencing reveals widespread distribution of typhoidal toxin genes and VirB/D4 plasmids in bovine-associated nontyphoidal Salmonella. Sci. Rep. 8:9864. doi: 10.1038/s41598-018-28169-4
European Food Safety, A., and European Centre for Disease, P.Control (2021). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 19:e06490. doi: 10.2903/j.efsa.2021.6490
Fearnley, E., Raupach, J., Lagala, F., and Cameron, S. (2011). Salmonella in chicken meat, eggs and humans; Adelaide, South Australia, 2008. Int. J. Food Microbiol. 146, 219–227. doi: 10.1016/j.ijfoodmicro.2011.02.004
Feldgarden, M., Brover, V., Gonzalez-Escalona, N., Frye, J. G., Haendiges, J., Haft, D. H., et al. (2021). AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 11:12728. doi: 10.1038/s41598-021-91456-0
Ferrari, R. G., Rosario, D. K. A., Cunha-Neto, A., Mano, S. B., Figueiredo, E. E. S., and Conte-Junior, C. A. (2019). Worldwide epidemiology of Salmonella serovars in animal-based foods: a Meta-analysis. Appl. Environ. Microbiol. 85:e00591-19. doi: 10.1128/AEM.00591-19
Florensa, A. F., Kaas, R. S., Clausen, P., Aytan-Aktug, D., and Aarestrup, F. M. (2022). ResFinder - an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb Genom 8:000748. doi: 10.1099/mgen.0.000748
Foley, S. L., Kaldhone, P. R., Ricke, S. C., and Han, J. (2021). Incompatibility group I1 (IncI1) plasmids: their genetics, biology, and public health relevance. Microbiol. Mol. Biol. Rev. 85:e00031-20. doi: 10.1128/MMBR.00031-20
Ford, L., Glass, K., Veitch, M., Wardell, R., Polkinghorne, B., Dobbins, T., et al. (2016). Increasing incidence of Salmonella in Australia, 2000-2013. PLoS One 11:e0163989. doi: 10.1371/journal.pone.0163989
Ford, L., Wang, Q., Stafford, R., Ressler, K. A., Norton, S., Shadbolt, C., et al. (2018). Seven Salmonella Typhimurium outbreaks in Australia linked by trace-Back and whole genome sequencing. Foodborne Pathog. Dis. 15, 285–292. doi: 10.1089/fpd.2017.2353
Galanis, E., Parmley, J., and De With, N. (2012). Integrated surveillance of Salmonella along the food chain using existing data and resources in British Columbia, Canada. Food Res Int 45, 795–801. doi: 10.1016/j.foodres.2011.04.015
Hoang, D. T., Chernomor, O., Von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hu, L., Cao, G., Brown, E. W., Allard, M. W., Ma, L. M., and Zhang, G. (2021). Whole genome sequencing and protein structure analyses of target genes for the detection of Salmonella. Sci. Rep. 11:20887. doi: 10.1038/s41598-021-00224-7
Ingle, D. J., Ambrose, R. L., Baines, S. L., Duchene, S., Goncalves da Silva, A., Lee, D. Y. J., et al. (2021). Evolutionary dynamics of multidrug resistant Salmonella enterica serovar 4,[5],12:i:- in Australia. Nat. Commun. 12:4786. doi: 10.1038/s41467-021-25073-w
Kaldhone, P. R., Carlton, A., Aljahdali, N., Khajanchi, B. K., Sanad, Y. M., Han, J., et al. (2019). Evaluation of incompatibility group I1 (IncI1) plasmid-containing Salmonella enterica and assessment of the plasmids in Bacteriocin production and biofilm development. Front Vet Sci 6:298. doi: 10.3389/fvets.2019.00298
Kaur, S., Payne, M., Luo, L., Octavia, S., Tanaka, M. M., Sintchenko, V., et al. (2022). MGTdb: a web service and database for studying the global and local genomic epidemiology of bacterial pathogens. Database (Oxford) 2022:baac094. doi: 10.1093/database/baac094
Lan, R., Reeves, P. R., and Octavia, S. (2009). Population structure, origins and evolution of major Salmonella enterica clones. Infect. Genet. Evol. 9, 996–1005. doi: 10.1016/j.meegid.2009.04.011
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Maiden, M. C., Bygraves, J. A., Feil, E., Morelli, G., Russell, J. E., Urwin, R., et al. (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. 95, 3140–3145. doi: 10.1073/pnas.95.6.3140
McLure, A., Shadbolt, C., Desmarchelier, P. M., Kirk, M. D., and Glass, K. (2022). Source attribution of salmonellosis by time and geography in New South Wales, Australia. BMC Infect Dis 22:14. doi: 10.1186/s12879-021-06950-7
McWhorter, A. R., Davos, D., and Chousalkar, K. K. (2015). Pathogenicity of Salmonella strains isolated from egg shells and the layer farm environment in Australia. Appl. Environ. Microbiol. 81, 405–414. doi: 10.1128/AEM.02931-14
Moffatt, C. R., Musto, J., Pingault, N., Miller, M., Stafford, R., Gregory, J., et al. (2016). Salmonella Typhimurium and outbreaks of egg-associated disease in Australia, 2001 to 2011. Foodborne Pathog. Dis. 13, 379–385. doi: 10.1089/fpd.2015.2110
Mooijman, K. A. (2018). The new ISO 6579-1: a real horizontal standard for detection of Salmonella, at last! Food Microbiol. 71, 2–7. doi: 10.1016/j.fm.2017.03.001
OzFoodNetWorkingGroup (2015). Monitoring the incidence and causes of diseases potentially transmitted by food in Australia: annual report of the OzFoodNet network, 2011. Commun. Dis. Intell. Q. Rep. 39, E236–E264.
Pang, S., Octavia, S., Reeves, P. R., Wang, Q., Gilbert, G. L., Sintchenko, V., et al. (2012). Genetic relationships of phage types and single nucleotide polymorphism typing of Salmonella enterica Serovar Typhimurium. J. Clin. Microbiol. 50, 727–734. doi: 10.1128/jcm.01284-11
Pavic, A., Cox, J. M., and Chenu, J. W. (2015). Effect of extending processing plant operating time on the microbiological quality and safety of broiler carcasses. Food Control 56, 103–109. doi: 10.1016/j.foodcont.2014.11.048
Payne, M., Kaur, S., Wang, Q., Hennessy, D., Luo, L., Octavia, S., et al. (2020). Multilevel genome typing: genomics-guided scalable resolution typing of microbial pathogens. Euro Surveill. 25:1900519. doi: 10.2807/1560-7917.ES.2020.25.20.1900519
Rendueles, O., Beloin, C., Latour-Lambert, P., and Ghigo, J. M. (2014). A new biofilm-associated colicin with increased efficiency against biofilm bacteria. ISME J. 8, 1275–1288. doi: 10.1038/ismej.2013.238
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sherry, N. L., Horan, K. A., Ballard, S. A., Gonҫalves da Silva, A., Gorrie, C. L., Schultz, M. B., et al. (2023). An ISO-certified genomics workflow for identification and surveillance of antimicrobial resistance. Nat. Commun. 14:60. doi: 10.1038/s41467-022-35713-4
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Simonsen, M., Mailund, T., and Pedersen, C. N. S. (2008). Rapid neighbour-joining. Alg Bioinform 5251, 113–122. doi: 10.1007/978-3-540-87361-7_10
Sintchenko, V., Wang, Q., Howard, P., Ha, C. W., Kardamanidis, K., Musto, J., et al. (2012). Improving resolution of public health surveillance for human Salmonella enterica serovar Typhimurium infection: 3 years of prospective multiple-locus variable-number tandem-repeat analysis (MLVA). BMC Infect. Dis. 12, 1–10. doi: 10.1186/1471-2334-12-78
Smith, H., Bossers, A., Harders, F., Wu, G., Woodford, N., Schwarz, S., et al. (2015). Characterization of epidemic IncI1-Igamma plasmids harboring ambler class a and C genes in Escherichia coli and Salmonella enterica from animals and humans. Antimicrob. Agents Chemother. 59, 5357–5365. doi: 10.1128/AAC.05006-14
Stewart, J., and Pavic, A. (2023). Advances in enteropathogen control throughout the meat chicken production chain. Compr. Rev. Food Sci. Food Saf. 22, 2346–2407. doi: 10.1111/1541-4337.13149
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15, 1–15. doi: 10.1186/s13059-014-0524-x
Van Immerseel, F., De Zutter, L., Houf, K., Pasmans, F., Haesebrouck, F., and Ducatelle, R. (2019). Strategies to control Salmonella in the broiler production chain. Worlds Poult. Sci. J. 65, 367–392. doi: 10.1017/s0043933909000270
Williamson, D. A., Lane, C. R., Easton, M., Valcanis, M., Strachan, J., Veitch, M. G., et al. (2018). Increasing antimicrobial resistance in Nontyphoidal Salmonella isolates in Australia from 1979 to 2015. Antimicrob. Agents Chemother. 62:e02012-17. doi: 10.1128/AAC.02012-17
Wong, L., Selvanathan, E. A., and Selvanathan, S. (2015). Modelling the meat consumption patterns in Australia. Econ. Model. 49, 1–10. doi: 10.1016/j.econmod.2015.03.002
Wood, D. E., Lu, J., and Langmead, B. (2019). Improved metagenomic analysis with kraken 2. Genome Biol. 20:257. doi: 10.1186/s13059-019-1891-0
Keywords: Salmonella Typhimurium, sequence type, multilevel genome typing, meat chicken, carcass
Citation: Bandaranayake S, Williamson S, Stewart J, Payne M, Kaur S, Wang Q, Sintchenko V, Pavic A and Lan R (2024) Genomic diversity of Salmonella enterica serovar Typhimurium isolated from chicken processing facilities in New South Wales, Australia. Front. Microbiol. 15:1440777. doi: 10.3389/fmicb.2024.1440777
Edited by:
Narjol González-Escalona, United States Food and Drug Administration, United StatesReviewed by:
Andrea Isabel Moreno Switt, Pontificia Universidad Católica de Chile, ChilePrasad Thomas, Indian Veterinary Research Institute (IVRI), India
Copyright © 2024 Bandaranayake, Williamson, Stewart, Payne, Kaur, Wang, Sintchenko, Pavic and Lan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruiting Lan, ci5sYW5AdW5zdy5lZHUuYXU=
 Samitha Bandaranayake
Samitha Bandaranayake Sarah Williamson2
Sarah Williamson2 Michael Payne
Michael Payne Sandeep Kaur
Sandeep Kaur Qinning Wang
Qinning Wang Vitali Sintchenko
Vitali Sintchenko Ruiting Lan
Ruiting Lan