
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 30 July 2024
Sec. Microbiotechnology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1435674
This article is part of the Research Topic Microorganisms in Dehalogenation: Regulation and Enhancement View all 7 articles
Halogenated organic compounds are persistent pollutants that pose a serious threat to human health and the safety of ecosystems. Cobamides are essential cofactors for reductive dehalogenases (RDase) in organohalide-respiring bacteria (OHRB), which catalyze the dehalogenation process. This review systematically summarizes the impact of cobamides on organohalide respiration. The catalytic processes of cobamide in dehalogenation processes are also discussed. Additionally, we examine OHRB, which cannot synthesize cobamide and must obtain it from the environment through a salvage pathway; the co-culture with cobamide producer is more beneficial and possible. This review aims to help readers better understand the importance and function of cobamides in reductive dehalogenation. The presented information can aid in the development of bioremediation strategies.
Organohalides exist on earth with volcanic activity, forest fire, organic oxidation, and animal or plant activity (Chandra and Kumar, 2015). With the development of the chemical industry, many artificially complex organohalides have been synthesized and produced for commercial use. The long-term use and improper disposal of organohalides (e.g., pesticides, chemical reagents, and industrial activities) cause widespread pollution (Gribble, 1998; Jin and Chen, 2017; Kallenborn et al., 2021; Xu et al., 2021). Typical organohalides include perchloroethylene (PCE), hexachlorobenzene (HCB), trichloroethane (TCA), polychlorinated biphenyls (PCBs), pentachlorophenol (PCP), hexachlorocyclohexanes (HCHs), dichlorodiphenyltrichloroethanes (DDTs), polybrominated diphenyl ethers (PBDEs), poly-fluoroalkyl substances (PFASs), and so on (Huang et al., 2014; Lu et al., 2019; He et al., 2021; Abbasian Chaleshtari and Foudazi, 2022; Cheng et al., 2023). Besides the persistence and stability in the environment, its carcinogenicity, teratogenicity, and mutagenicity may irreversibly damage the balance of the ecosystem and pose a carcinogenic risk to humans (Yankovych et al., 2023). Moreover, most organohalides are lipophilic, which allows them to stay in fat-rich organs, and are difficult to metabolically eliminate. Their long-term accumulation may damage the nervous and immune systems (Bennett et al., 2021).
Among the various remediation strategies for organohalide contamination, traditional physical and chemical methods, such as incineration, adsorption, advanced oxidation, and electrochemical techniques, can effectively remove or degrade organohalide contamination (Xu et al., 2021; Femina Carolin et al., 2023). However, these methods pose challenges due to the by-products, high costs, and energy consumption. In contrast, biotransformation has emerged as an economically and environmentally friendly method for eliminating organohalides from the environment rather than transferring the organohalide contamination to another location (Zhu et al., 2022). Microorganisms play a crucial role in conducting bioremediation processes, with many having the ability to transform organohalide pollutants, e.g., Dehaloococcoides, Dehalobacter, Desulfitobacterium, Geobacter, and Sulfurospirillum (Villemur et al., 2006; Maillard and Holliger, 2016; Zinder, 2016; Reguera and Kashefi, 2019; Jin et al., 2023), which possess a unique reductive dehalogenase (RDase); they are known as organohalide-respiring bacteria (OHRB). During the transformation process, OHRB use organohalides as an electron acceptor and drives free electron transport to RDase, which leads to the bond cleavage of C-Cl. RDase functions as the terminal enzyme in the organohalide dehalogenation process and is crucial for halogen removal (Fincker and Spormann, 2017; Wang et al., 2018).
Previous studies confirmed that cobamide is integral to RDase. It enables RDase to react with organohalides and is important in OHRB metabolism (Yan et al., 2018). Cobamide, whose basic structure is a corrin ring, combines with an upper and lower ligand, forming a larger family of cobamide, such as cobalamin (Deery et al., 2022). Cobalamin is the most specific subunit of cobamide that is directly involved in removing halogen atoms (Kunze et al., 2017; Schubert et al., 2019). However, it is interesting to note that not all OHRB can synthesize cobamide to assemble functional RDase; therefore, the ability to synthesize cobamide can be considered a characteristic to distinguish corrinoid-auxotrophic OHRB (Maphosa et al., 2010). Similar to the distinction between facultative OHRB and obligate OHRB, most obligate OHRB are also corrinoid-auxotrophic OHRB; this characteristic is related to energy conservation. The more flexible metabolism of facultative OHRB allows them to grow on a variety of electron acceptors and easily establish co-metabolic relationships with other strains. In contrast, obligate OHRB are more restricted in obtaining energy, and organohalide respiration is their only energy-converse pathway (Zhang et al., 2021). Both obligate and facultative OHRB share the feature of using organohalide for their growth via RDase. Still, they have different suitability to cobamide and completely distinct ways of obtaining cobamide.
Facultative OHRB, such as Geobacter lovleyi, Desulfovibrio, Sulfurospirillum multivorans, and Desulfitobacterium, are described as containing a set of cobamide synthesis pathways, as well as genes for the transport/uptake (e.g., btuBFCD) of cobamide synthetic (e.g., cbiZ, cbiB, cobU, cobS, cobT) (Men et al., 2012; Reinhold et al., 2012; Schubert, 2017; Nakamura et al., 2018). However, most obligate OHRB are classified as corrinoid auxotrophs, including Dehalobacter restrictus spp., Dehalococcoides mccartyi spp., and Dehalogenimonas spp. In contrast, Dehalobacter restrictus Y51 has been identified to have a complete de novo corrinoid synthesis gene. However, the absence of the cbiH gene in Dehalobacter restrictus Y51 significantly affects corrinoid synthesis and is classified as a corrinoid-auxotrophic obligate OHRB. Similarly, Dehalogenimonas lykanthroporepellens BL-DC-9 has been found to lack the cbi gene, which prevents it from synthesizing corrinoid de novo. In addition, according to the Genbank data, species Dehalococcoides mccartyi, including strain 195, CBDB1, BAV1, VS, FL2, and GT, are characterized as typical corrinoid-auxotrophic OHRB without de novo corrinoid synthesis (Löffler et al., 2013). They cannot synthesize corrinoid, the basic structure of cobalamin for organohalide respiration (Yan et al., 2013). However, some studies have also suggested that corrinoid-auxotrophic OHRB have dehalogenation activity without producing cobalamin by themselves (Yan et al., 2016). It is plausible that the cobamides were synthesized by other symbiotic partners who provided these cobamides to OHRB for maintaining the dehalogenation process in a low-energy mode (Men et al., 2013; Yan et al., 2016).
The cobamide in biocatalysis has been studied for many years, and the effect of cobamides on OHRB has been reported for decades (Guo and Chen, 2018). However, the related information of cobamide on catalytic dehalogenation is still fragmented. In this review, the role of cobamide in OHRB is systematically summarized, including the impact of cobamides on organohalide respiration. In addition, cobamide as a catalyst in the dehalogenation process is discussed. We have also summarized the cobamide salvage pathway of OHRB and the synergy of cobamides in the microbial community. Additionally, we provide several suggestions for further investigations on cobamides for reductive dehalogenation and the applications of OHRB. This review also summarizes the role of cobamide in reductive dehalogenation and provides a reference for the study of reductive dehalogenation.
The structure of cobamide consists of a corrin ring containing a central cobalt ion, a lower ligand, and an upper ligand; the ligand can be characteristic of various reported cobamide types in the natural environment. The typical lower ligand includes benzimidazole, purines, and phenol, and there are four common upper ligand types (Figure 1). Although the function of cobamide is mainly controlled by the lower ligand, previous research has shown that even cobamide forms with the same lower ligand type may not be functionally equivalent. Additionally, the upper ligand can also affect cobamide function (Zhai et al., 2012). It is widely accepted that cobalamin, with 5,6-dimethylbenzimidazole (DMB) as the lower ligand, is the most common cobamide type and is widely used (Yi et al., 2012).
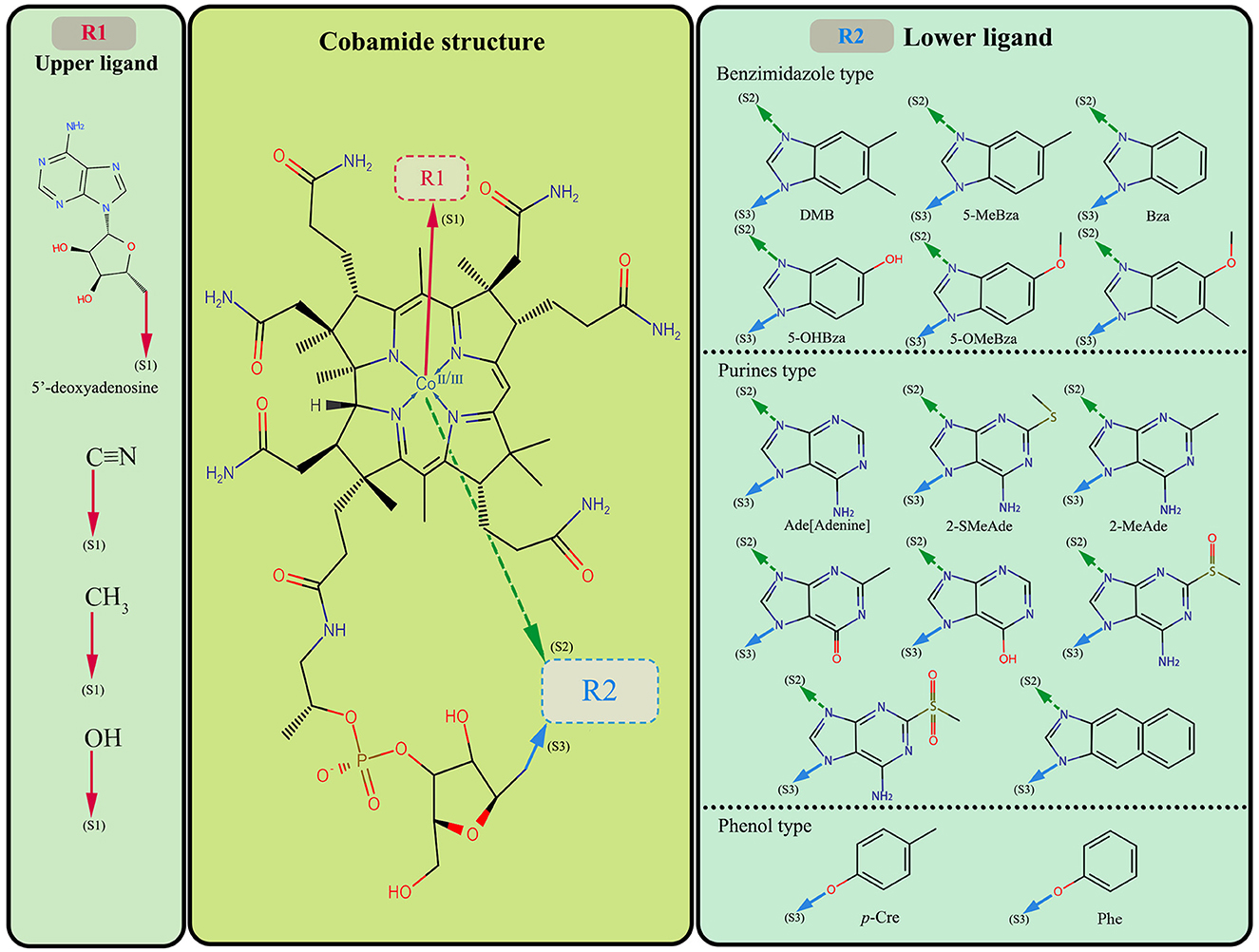
Figure 1. The middle part is the cobamide paradigm; R1 and R2 are the upper and lower ligands of the variable substructure, respectively. The left part lists the variable types of upper ligands and the right part accounts for the lower ligands of available cobamide. The red arrow is the connection position of the upper ligand and the cobamide structure, and the green and blue arrow represents the connection position of the lower ligand and the cobamide structure. DMB, 5,6-dimethylbenzimidazole; 5-MeBza, 5-methylbenzimidazole; Bza, benzimidazole; 5-OHBza, 5-hydroxybenzimidazole; 5-OMeBza, 5-methoxybenzimidazole; Ade, adenine; 2-SMeAde, 2-methylmercaptoadenine; 2-MeAde, 2-methyladenine; p-Cre, p-Cresol; Phe, Phenol.
The cyanogen group is the upper ligand of cobalamin (also called cyanocobalamin or vitamin B12), while the other upper ligand type includes hydroxyl, methyl, and 5′-deoxyadenosine, which can make up vitamin B12 analogs such as hydroxocobalamin, methylcobalamin, and 5′-deoxyadenosylcobalamin (Fang et al., 2017). The upper ligand also influence the cobamide function. Specifically, Zhai et al. (2012) suggested that cobalamin and hydroxocobalamin have no direct biological activity before biotransformation, and 5′-deoxyadenosylcobalamin is often applied as a coenzyme in microbial metabolism as it is the active coenzyme B12 form of cobalamin. The vitamin B12 analogues would be utilized after upper ligand biotransformation. It is more likely that metabolism converts the upper ligand to adenosine, enabling the cyanocobalamin into coenzyme B12, thus engaging in metabolic processes (Guo and Chen, 2018; Dulay et al., 2020). Here, we point out that the upper ligand may affect biological selectivity, but these effects are minor and not substantial enough to mask the influence of the lower ligand on biological selectivity. Therefore, we did not delve into the combined effects of cobamide upper ligand and lower ligand. Instead, we shifted our focus to discussing the lower ligand, and the following cobalamin generally refers to the cobalamin analogs.
The combination of the upper and lower ligands determines the variety of cobamide forms. It has been reported that 29 forms of cobamide exist in the environment (Brown, 2005), and at least 16 forms of lower ligand have been identified (Allen and Stabler, 2008; Yan et al., 2018). Each form of cobamide has its unique function in microbial metabolism. The lower ligand affects the transformation of cobamide into an active state, which causes different selections of cobamides in organisms (Yan et al., 2016). Based on the demand and synthesis of cobamide by community members, it can be divided into three types: those who do not use cobamide or produce it, those who use and produce cobamide alone, and those who need cobamide but are unable to produce it (Sokolovskaya et al., 2020). Shelton et al. (2019) estimate the proportion of these microorganism types in the environment: approximately 86% of bacteria require cobamides metabolism, but merely 37% produce cobamides de novo. The cobamide is assembled in many catalytic processes, including intramolecular rearrangements, methyl transfer, ribonucleotide reduction, and reductive dehalogenation (Dickman, 1977; Kunze et al., 2017; Farnberger et al., 2019). For example, the Dehalococcoides mccartyi is combined with the cobalamin catalysis. Cobalamin can be directly involved in the RDase assembly of Dehalococcoides mccartyi, whereas other cobamides require further remodeling through the cobamide salvage pathway (Yi et al., 2012). In addition, studies have reported that some OHRB have a preference for specific cobamide structures. For instance, Sulfurospirillum multivorans requires [Ade]cobamide (adenine as lower ligand) while the Dehalococcoides mccartyi strain prefers the cobalamin determined by the spatial structure of cobamide and RDase (Yi et al., 2012; Johannissen et al., 2017; Kunze et al., 2017).
The dehalogenation capability and the reaction rate are mostly affected by the lower ligand of cobamide, and specific lower ligand leads to high dehalogenation activity (Schubert et al., 2018). The activity of PceA from Sulfurospirillum multivorans was decreased, and the ability of organohalide respiration was also reduced after the DMB was replaced by adenine (Keller et al., 2014). Similarly, Dehalococcoides mccartyi did not show activity in dehalogenation during the cultivation without cobamide, while growth occurs when cobalamin or DMB structures is available (Yi et al., 2012; Men et al., 2015).
The cobamides in the natural environment are mainly produced by microbes (Men et al., 2015; Fang et al., 2017; Shelton et al., 2019), indicating that the forms of cobamides can be diverse in environments with complex microbial communities. As mentioned above, most cobamide cannot be assembled into RDase by OHRB directly, and suitable cobamide is limited. The endogenous production and exogenous utilization of cobamide can be the sources for OHRB. Endogenous cobamide biosynthesis is a complex process; except for some facultative OHRB, such as Geobacter lovleyi, that can synthesize cobamide to support dehalogenation, most OHRB cannot (Wagner et al., 2012; Deery et al., 2022).
For obligate OHRB, the lack of suitable cobamides restricts the application of exogenous cobamides. Even though they are the key factor for the RDase assembly process, not all available ones are used directly. The OHRB possess a set remodeling function that aims to transform these unsuitable cobamides into a directly usable form (Moore and Escalante-Semerena, 2016; Balabanova et al., 2021). This function involves the uptake of cobamides from the external environment and modification of the lower ligand to adapt to organohalide respiration, which is a complex process that may reduce dehalogenation efficiency (Men et al., 2014; Balabanova et al., 2021). The lower ligand mainly determines the effectiveness of cobamide function and influences the bioavailability of OHRB to different forms of cobamide. Dehalococcoides mccartyi strains have been discovered to modify at least seven cobamide lower ligands, including adenine, 2-SMeAde, 5-OHBza, Bza, 5-MeBza, p-Cresol, and corrinoid (Men et al., 2015). However, it should be noted that the remodeling function needs a lower ligand structure, such as DMB. The above remodeling function can only be achieved while DMB is present, even when the non-cobalamin forms are available. Generally, the addition of DMB enables guided cobalamin synthesis that supports RDase activity and OHRB growth (Yan et al., 2013, 2016).
The concentration and suitable structure of cobamides are both important for dehalogenation (Sokolovskaya Olga et al., 2019). According to previous reports, the concentration required by Dehalococcoides sp. for maintaining dehalogenation is 1 μg·L−1 (He et al., 2007). When the cobalamin concentration increased to 25 μg·L−1, the dehalogenation rate nearly doubled, the cell growth yield increased by 2.8–9.1 fold, and 50 μmol TCE was completely degraded to ethene in 1 month. The TCE degradation rates did not increase when the concentration of cobalamin was higher than 25 μg·L−1, which is also difficult to achieve in the environment. In the microcosm that contains cobalamin producers, approximately 10 μg·L−1 cobalamin was detected after cultivation, suggesting that the laboratory-calculated amount of minimum concentration (>1 μg·L−1) for OHRB growth is easy to achieve (Men et al., 2012). However, it should be noted that the concentrations of cobamide produced in the environment may differ from laboratory measurement; it may contain other available cobamides (such as 5-MeBza, 5-OHBza, etc.) that can also be used in dehalogenation. Thus, it is plausible that the growth of OHRB in the environment is not limited by cobamide but by the cobamide producers. Furthermore, to apply all the available cobamides for OHRB growth, a strategy is to use the remodeling function to transform these available cobamides into suitable ones. Yan et al. (2012) added 10 mM DMB into the culture to guide biosynthesis and generate cobalamin; compared to the control, the Dehalococcoides mccartyi strain BAV1, strain GT, and strain FL2 cell densities increased 31, 41, and 37 fold, respectively. In contrast, negligible Dehalococcoides growth was observed in the non-cobalamin cultures without DMB, which indicates the remodeling function of OHRB does not have sufficient DMB to catalyze the conversion of cobamide to cobalamin, leading to deficient RDase synthesis. Consequently, OHRB could not successfully obtain growth energy from organohalide respiration.
To conclude, in pure culture, the impact of cobamide on the dehalogenation process depends on two key factors: the lower ligand and the concentration of available cobamides. Alternatively, in the mixed culture, corrinoid-auxotrophic OHRB, especially, can perform more complete dehalogenation processes benefiting from the presence of other microorganisms that produce available cobamides and usable ligands.
Since DeWeerd isolated and cultured the first OHRB in 1984, more than 70 OHRB strains have been identified in varied environments (Shelton and Tiedje, 1984; Atashgahi et al., 2016). The dechlorinating capability of different OHRB depends on the type of RDase that relies on the catalysis of cobamide (Ji et al., 2017). Different RDases obtain reducing power through the combination of cobamides, allowing RDase to undergo reductive dehalogenation. The high reducing power of cobamide also makes the OHRB vulnerable to oxygen, which requires strictly anaerobic conditions for their cultivation.
The potential application of OHRB for biotransformation is limited by their strict cultivation requirements and low energy yield (Wang et al., 2022). These factors also hinder the isolation and characterization of pure OHRB strains as well as the identification of the RDase. Many RDases were identified from Dehalococcoides, Sulfurospirillum, Dehalobacter, and so on, such as PceA, VcrA, and BvcA, were identified as degrading chlorinated ethenes (Neumann et al., 1996; Magnuson et al., 1998; Parthasarathy et al., 2015). DcaA, DcpA, and DcrA which catalyze the reductive dehalogenated of chlorinated hydrocarbons chlorinated hydrocarbons (Marzorati et al., 2007; Padilla-Crespo et al., 2013; Tang and Edwards, 2013); PcbA, PteA, and PbrA of reductive dehalogenated PBDEs or PCB (Ding et al., 2017; Zhao et al., 2021, 2022), and CbrA of reductive dehalogenated chlorobenzene (Monteagudo-Cascales et al., 2019).
Dehalogenation by different RDases involves a conservative electron transfer chain that is essential for organohalide respiration (Richardson, 2013). The hydrogenase catalyzes the oxidation of hydrogen or other electron donors to produce free electrons, which are then transferred through a series of electron-transport enzymes to the RDase (Kunze et al., 2017; Wang et al., 2018). For example, the free electrons through the quinone or CISM (Complex iron-sulfur molybdo) enzyme system to the RdhB (The membrane anchor protein), then the free electrons transmit to the RdhA (The membrane peripheral protein). In the internal structural electron transport of RDase (Figure 2), two pairs of 4Fe–4S clusters are identified in the RdhA. These were identified by electron paramagnetic resonance and UV-Vis spectroscopy (Parthasarathy et al., 2015; Nakamura et al., 2018). These two 4Fe-4S clusters are the final transit of the electron chain that transfers electrons to activate the cobamide. The corrin ring structure of cobamide provides six coordination bonds connecting the cobalt atoms, providing high reducibility for RDase. The substrate channel of RDase allows the organohalide to pass through and bind to the high-reducible cobamide (Jugder et al., 2015; Parthasarathy et al., 2015; Payne et al., 2015; Fincker and Spormann, 2017).
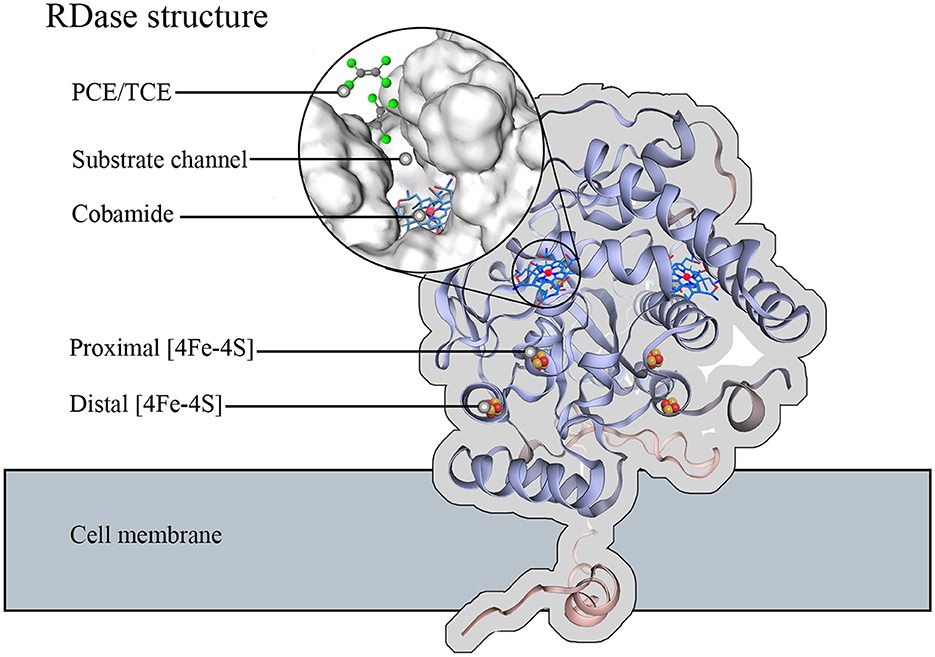
Figure 2. Function structure of most RDase. The RDase structure was used and simulated on SWISS-MODEL using the Dehalococcoides mccartyi dehalogenase gene provided by NCBI (GenBank: KSV18274.1) (Bienert et al., 2017; Waterhouse et al., 2018; Studer et al., 2020). The cobamide is embedded in the core of the RDase and accessible from substrate channels with two pair 4Fe-4S clusters that mediate electron transfer from the exterior to the cobamide.
As summarized in previous studies, the cobamide in RDase plays a crucial role in the reductive dehalogenation reaction (Table 1). The cobalamin receives free electrons transferred from the 4Fe-4S clusters to the cobalamin site, activating cobalamin and maintaining it in a highly reductive state, which activates the bond cleavage of the halogenated substrate in three ways: The first mechanism is the free radical-catalyzed dehalogenation mechanism (Figure 3b). The abstraction of an electron from the Co(I)balamin to substrate R-X forms the R-X radical state, and Co(II)balamin, the R-X radical, eliminates the halogen by further protonation/dehalogenation. Another free electron transfers from 4Fe-4S clusters to Co(II)balamin, leaving an unpaired electron on the cobalt center and reflash to Co(I)balamin. This exposes the cobalt's reducing ability, which allows it to react with various halogen radicals. The second mechanism is the cobamide-organic adduct mechanism (Figure 3a). The Co(I)balamin forms a bond with the carbon atom of the R-X, resulting in Co(III)balamin with the simultaneous elimination of halide ions and the subsequent cleavage of the C-Co bond under the electron transfer. At the same time, Co(III)balamin is reduced to Co(I)balamin by the electron transfer from the 4Fe-4S clusters. The third mechanism is the cobamide-halide adduct mechanism (Figure 3c). The Co(I)balamin directly interacts with a halogen atom of the substrate R-X, forming a Co(I)balamin-halide-carbon state. The halide-carbon bond then cleaves by protonation/radical action, forming a Co(III)/Co(II)balamin-halide state and reverting to the Co(I)balamin state through electron transfer.
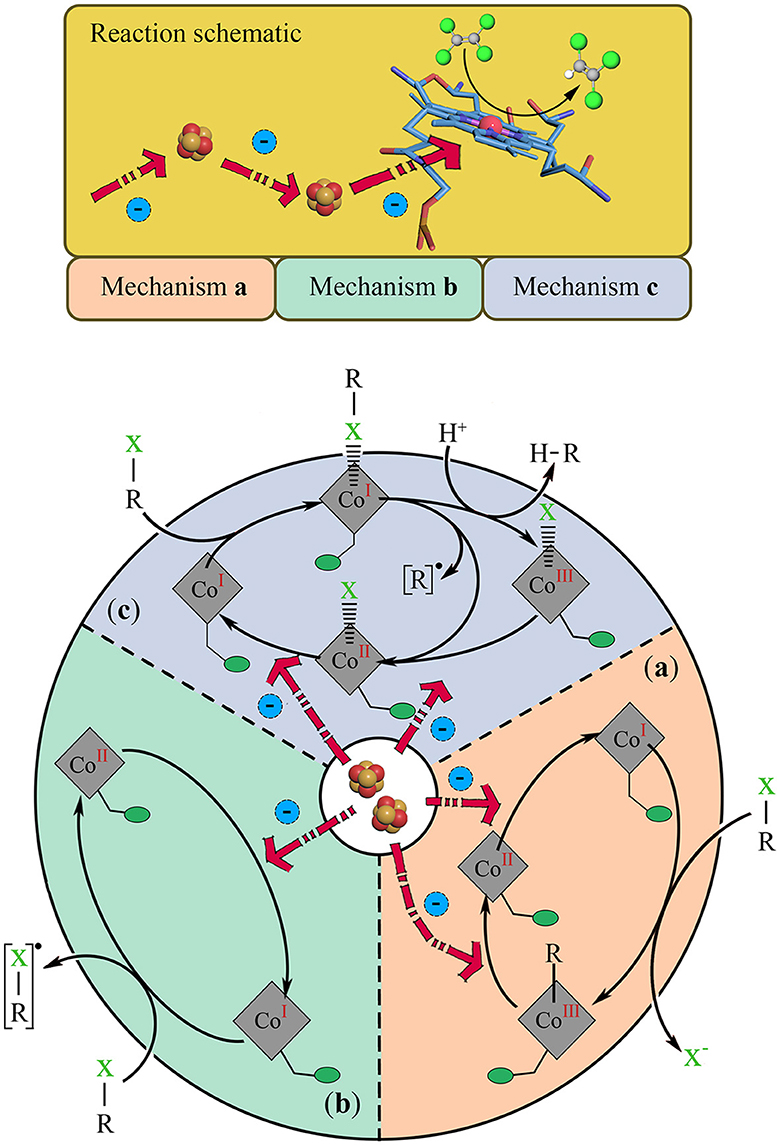
Figure 3. Cobamide structure in the dehalogenation reaction mechanism. The top part is the cobamide structure activated by the free electron transfer chain; and the bottom side are the possible mechanisms of cobalamin reacting with Organohalide (R-X), (a) the cobamide-organic adduct mechanism, (b) the free radical-catalytic mechanism, and (c) the cobamide-halide adduct mechanism. The cobamide state follows the reaction change and backtrack with the free electron activate, and finally form a closed cycle. The structure of 4Fe-4S cluster is shown by red and yellow balls, while PCE is shown as the green and gray balls.
When the cobalt atom is transformed into the activated state of Co(I) through free electrons, it can provide a high reduction potential, which is crucial for reducing organohalides and is known as Co(I)balamin. Only this state is reactive as a low potential electron donor or nucleophilic reaction center, while the oxidation state of Co(II)balamin or Co(III)balamin is the oxidative state and possesses no reducibility. Furthermore, during the dehalogenation process, the standard potential of the cobalamin center depends on the RDase type. Wang et al. (2018) reviewed the redox potential of Co(II/I) in most of OHRB, and found that Co(II/I) transition Em is often lower than −350 mV; the PceA of Sulfurospirillum multivorans has an estimated E° = −570 mV, similar to the CprA of Dehalobacter restrictus with Em ≈ −480 mV; and the state of cobalamin is transformed from (I) to (II) and even to (III). This variation in the redox potential of cobalamin in different OHRB is significant as it influences the reactivity of cobalamin and, consequently, the dehalogenation process, and depends on the standard redox potentials of the organohalide substrates (Em: 260 to 570 mV) and the electron donors (e.g., of H2/H+ = −414 mV) (Wang et al., 2018; Yu et al., 2023). This requires the Fe-S clusters (Em of 4Fe-4S = −440 mV) to transfer free electrons to revert the state back to Co(I)balamin (Fincker and Spormann, 2017). It should be noted that the actual electron transport process is more complex and requires various enzymes (Hase, dehydrogenase; quinone, intermediate electron shuttle; CISM; RdhB, etc.) to establish the electron transfer chain in the organohalide respiration. The bond cleavage mechanism and electron transfer chain in the organohalide respiration process still need further study.
As discussed, specific cobamide form is an essential cofactor of RDase (e.g., cobalamin for Dehalococcoides mccartyi RDases, [Ade]cobamide for Sulfurospirillum multivorans PceA). It is crucial for the growth and energy conversion of these corrinoid-auxotrophic OHRB (Keller et al., 2018). The Dehalococcoides dehalogenation process seems to favor the cobalamin as the optimal cofactor, and these corrinoid-auxotrophic OHRB are more likely to take up the cobalamin than other cobamide when both are present. However, the corrinoid-auxotrophic OHRB are often deficient in the available cobalamin environment, or the main cobamide types are unsuitable for use. Compared to obligate OHRB, facultative OHRB are not restricted by organohalide respiration, which allows getting energy from other energy conversion metabolisms in the environment (Maphosa et al., 2010; Liu and Häggblom Max, 2018; Yang et al., 2020b; Liang et al., 2021; Zhang et al., 2022). Various metabolism pathways enable facultative OHRB to retain the function of corrinoid de novo synthesis and the organohalide respiration ability, such as Geobacter lovleyi (Nonaka et al., 2006; Wagner et al., 2012) and Desulfitobacterium hafniense Y51 (Reinhold et al., 2012; Schubert, 2017). In contrast, the low concentration of organohalides in the environment makes it hard to maintain obligate OHRB growth, and there is no extra energy to support the synthesis of corrinoids de novo. Consequently, some microorganisms, such as the species Dehalococcoides mccartyi, have been found to lack the gene for the synthesis of corrinoid de novo during evolution (Seshadri et al., 2005; Türkowsky et al., 2018). Shelton et al. (2019) suggests that the corrinoid de novo synthesis pathway consists of about 30 synthesis steps, which is more complex and redundant than the salvage pathway, consisting of several steps. Since these corrinoid-auxotrophic OHRB lack the genes for cobamide synthesis, they need alternative pathways to acquire cobamide for organohalide respiration (Rupakula et al., 2015; Moore and Escalante-Semerena, 2016).
The genome of species Dehalococcoides mccartyi has lost the genes for cobamide biosynthesis and replaced them with genes for cobamide modification and transport (Löffler et al., 2013; Yan et al., 2016; Men et al., 2017). In other words, the cobamide needed for its dehalogenation metabolism must be obtained from outside sources, and obtaining the necessary cobamide from other members of the dehalogenation community is often the most energy-efficient way. A previous study reported that the cobamide transport gene was detected in over 90% of OHRB (Zhang et al., 2009), and functional genes for cobamide uptake and salvage have been detected in typical corrinoid-auxotrophic Dehalococcoides species (Men et al., 2013). Further studies have shown that corrinoid-auxotrophic OHRB regulate genes involved in cobamide uptake and salvage when performing organohalide respiration (Men et al., 2014). Such as the BtuFCD protein responsible for the transport of cobalamin, the DMB phosphoribosyl transferase (CobT) and the adenosylcobinamidephosphate guanylyltransferase (CobU) to remodel the other cobamide to cobalamin (Escalante-Semerena, 2007; Balabanova et al., 2021; Ewald et al., 2022; Mathur et al., 2022). Additionally, all species of Dehalococcoides mccartyi possess genes such as cbiP, cbiB, cobU, cobC, cobT, and cobS, which are involved in cobalamin remodeling (Scott and Roessner, 2002; Wang et al., 2022). Furthermore, the polymerase chain reaction (PCR) amplification research showed that the defect of the cobamide synthesis gene will trigger the activation of the gene that regulates the transport and remedial pathway of cobamide (Moore and Escalante-Semerena, 2016). The transporter proteins are assembled in the cell and are used to identify and uptake the extracellular available cobamides for the microorganisms to maintain their dehalogenation ability. This pattern may be a common strategy that helps them sustain their normal metabolic activity and avoid the negative effects of cobamide deficiency for corrinoid-auxotrophic OHRB.
The biosynthetic cobamide has been found to show many structures. The function of cobamide-dependent enzymes depends on the core of cobamide upper ligands, lower ligands, corrin ring, and the nucleotide loop (Shelton et al., 2019). The corrinoid-auxotrophic OHRB can only use the special cobalamin for dehalogenation, while other cobamides cannot be used directly and need further remodeling. Compared with the direct use of cobalamin, the structure modification process is longer and limits the dechlorination rate, which leads to RDase activity at a minimal level (Keller et al., 2014; Men et al., 2014). As a result, these OHRB express the salvage genes that strengthen the use of available cobamides (Figure 4). This mode indicates that species Dehalococcoides mccartyi can assemble cobalamin if the precursors (e.g., corrinoid, DMB) are present. Men et al. (2015) detected the salvage of Dehalococcoides mccartyi from [p-Cre]Cobamide (p-Cresol as a lower ligand) and further confirmed that the Dehalococcoides mccartyi up-regulates the salvage genes in cobalamin deficiency environment, and use DMB to modify other cobamides forms (Men et al., 2017). However, cobamide remodeling is a complex metabolic pathway that leads obligate OHRB to use optimum cobamide to conserve energy for growth (Men et al., 2017; Balabanova et al., 2021). This suggests that direct uptake of cobalamin offers more advantages, such as energy saving, higher efficiency, and shorter durations, than modifying other cobamide structures through the remodeling function. We believe these cobamide salvage genes and related functions assemble a complete dehalogenation function of corrinoid-auxotrophic OHRB.
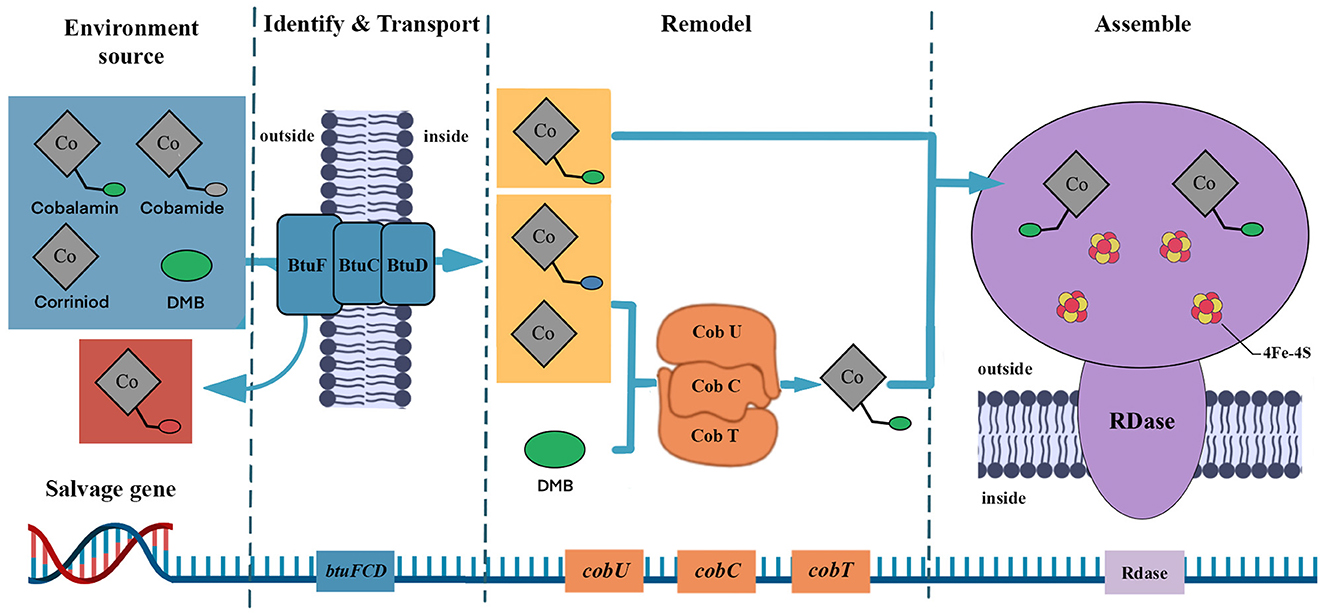
Figure 4. The salvage gene schematic. The blue section represents various cobamide types, the gray section represents the non-remodel cobamides type, and the red sphere is the immutable lower ligand, which cannot be remodeled. The yellow section represents remodeled cobamides, and the green sphere is the variable lower ligand, which can be directly used for RDase assembly after remodeling.
In conclusion, cobamide participates in the organohalide respiration process and is regarded as a key coenzyme for halogen removal. The obligate OHRB possess the functional genes that include cobamide uptake, transfer, and salvage, as well as remodeling functions that assist in the acquisition of cobamide from the environment and modify cobamide into a suitable structure; we summarize these examples in Table 2. This avoids synthesizing corrinoid de novo, which allows these obligate OHRBs to maintain dehalogenation and save energy efficiently. While facultative OHRB integrates many functions, efficient growth but low cell yield leads to lower dehalogenation efficiency and longer dehalogenation duration.
Cobamides, H2, and carbon sources are the microbial interact substances in dehalogenating microbial communities. Cobamide is also an essential cofactor for several important enzymes catalyzing transmethylation and rearrangement reactions in bacteria and archaea. Cobamide can be key in facilitating cross-feeding and symbiosis (Feng et al., 2018; Sokolovskaya et al., 2020). Many bacteria and archaea can synthesize cobamide de novo and contain the cobamide salvage pathway (Fang et al., 2017; Agarwal et al., 2019; Kipkorir et al., 2021).
In the dehalogenation community, OHRB play the main role in reductive dehalogenation. The electron acceptor, electron donor, and electron transfer chain have been extensively studied, and cobamides are considered to be directly in charge of the removal of halogens (Wang et al., 2018, 2019; Cui et al., 2021). The corrinoid-auxotrophic OHRB relies on cobamide exchange with other species to sustain organohalide respiration, as a cobamide deficiency can disrupt the electron transport chain. This underscores the importance of a credible dehalogenation co-culture model, where microorganisms collaborate to provide essential substances, such as carbon sources, hydrogen, and cobamides. This forms a synergistic effect to enhance the dehalogenation performance and extent. Numerous studies have demonstrated that the OHRB and specific microbes, such as Desulfovibrio and Methanosarcina (Men et al., 2012; Wang et al., 2019), can establish an interspecies electron transfer mechanism that promotes the process of reductive dehalogenation, which is summarized in Table 3.
Facultative OHRB interacting with obligate OHRB is a common form. The dehalogenation community consists of OHRB with different metabolic patterns and niches. Some facultative OHRB can synthesize cobamide and dehalogenate organohalides, forming a more self-sufficient co-culture system than only obligate OHRB (Lai and Becker, 2013; Fincker and Spormann, 2017; Ning et al., 2022). While obligate OHRB has a stricter metabolism pathway, it depends on organohalides as electron acceptors, hydrogen or simple organic compounds (e.g., acetate) as electron donors, and other factors for their organohalide respiration. Although they have specific and efficient dehalogenation capabilities, the growth rate of the cell culture is slow, and the cultivation conditions are strict. As the genus Dehalococcoides lacks the gene to synthesize cobalamin, the growth of isolated Dehalococcoides is relatively slow, with a long doubling time (Men et al., 2013). In contrast, previous studies have demonstrated that Dehalococcoides in microbial enrichments or with sufficient cobalamin have a faster and more robust growth rate, reaching two times the cell density of cultures with limited cobalamin (Yan et al., 2013). When Dehalococcoides are co-cultured with other OHRB, their cell yield increases by 1.5 times compared to the control (Amos et al., 2009; Hug et al., 2012). These results suggest that Dehalococcoides can benefit from the interactions with other microorganisms and the supplementation of cobalamin from the environment.
As previously discussed, cobalamin is the most popular cofactor compared to other types of cobamides for most OHRB; therefore, microorganisms that have the ability to synthesize cobalamin are more likely to be effective partners in the dehalogenation process. It has been observed that both G.lovleyi and G.sulfurreducens can produce distinct cobamides. However, when these two were co-cultured independently with Dehalococcoides, only G.lovleyi supported reductive dehalogenation activity (Yan et al., 2012). The G.lovleyi directly released cobalamin as a suitable cofactor for reductive dehalogenation, whereas G.sulfurreducen released an unavailable cobamide, which could not support reductive dehalogenation activity. Providing cobalamin to Dehalococcoides is a highly effective way to promote reductive dehalogenation, and the co-culture strategy has been extended to combine the facultative OHRB to provide cobalamin. Some facultative OHRB, such as Geobacter sp. and Dehalobacter sp. can synthesize cobalamin and perform organohalide respiration. Wagner et al. (2012) suggested that an active Geobacter lovleyi community could provide Dehalococoides specific cobamide to establish a co-culture system. Consequently, obligate and facultative OHRB can potentially form a stronger dehalogenation community and eliminate the cobalamin restriction on obligate OHRB with simultaneous coupling of the dehalogenation process. From the perspective of cofactors, cobalamin is crucial for RDase due to its high reducibility, which facilitates the energy cycle of OHRB.
However, similar dehalogenation activity was observed in the OHRB community without exogenous cobalamin (Men et al., 2013). This confirms that other microorganisms provide cobalamin directly or other cobamide to remodel for Dehalococcoides during organohalide respiration. Despite the Dehalococcoides strain's incapacity to biosynthesis cobalamin, the cobalamin gap in OHRB supply and demand can be filled by other microorganisms.
Another microbial interaction involves non-OHRB cobamide producers, such as fermentors, acetogens, and methanogens. Known cobamide producers include Clostridium spp., Desulfovibrio spp., Acetobacterium woodii, and Methanosarcina barkeri, but their interspecific cobalamin transfer ability needs to be further confirmed (i.e., the ability to release cobalamin to the environment) (Hazra et al., 2015; Shelton et al., 2019). These anaerobic microorganisms can synthesize corrinoids de novo and export the cobamide to narrow the demand gap. Therefore, the corrinoid or cobamide supplier for corrinoid-auxotrophic OHRB within co-culture microbial communities must be explored further.
Furthermore, the co-culture system can enhance the dehalogenation performance of OHRB by modulating the interactions among dehalogenating communities (Min et al., 2019). Maphosa et al. (2012) reported that the Sedimentibacter strain provides corrinoid to Dehalobacter strain E1, addressing the deficiency in corrinoid synthesis. This suggests that these microorganisms play a key role in sustaining high rates of dehalogenation functions of OHRB. For example, methanogens can produce cobalamin during energy metabolism, which may be released from the cell and applied by corrinoid-auxotrophic OHRB for dechlorination (Wen et al., 2015). However, it should be noted that both methanogens and obligate OHRB utilize hydrogen in their metabolic processes and potentially can be competitors of each other. Meanwhile, interspecific competition is inevitable. Previous studies have reported that only about 5% of hydrogen was used as electron donors for organohalide respiration (Ma et al., 2003; Yang et al., 2020a), and large amounts of hydrogen were used for methanogenesis. In addition, it has been quantified that methanogenesis consumes about 80% of the hydrogen (Kuroda and Watanabe, 1995; Jiang et al., 2023). Although the hydrogen competitions actually happen between methanogens and OHRB, it has been accepted that the hydrogen demand between them is not at the same level (Feldewert et al., 2020). Previous studies have suggested that the dehalogenation of OHRB will be affected only when the hydrogen concentration in water is lower than 2 nM, but methanogens cannot consume hydrogen to this extent (Yang et al., 2020a). It is more likely that, compared to the disadvantage of hydrogen competition, methanogens have a greater positive impact by supporting reductive dehalogenation by providing cobalamin and other cofactors (Maymó-Gatell et al., 1995; Jin et al., 2020). Furthermore, other research shows that the methanogen F430 enzyme is similar to cobamide; the core Ni+ ion also contains high reducibility. Yuan et al. (2021) suggest that the MCR enzyme (Methyl-coenzyme M reductase) can reduce the activation barriers for dichlorination, which is a cobamide-similar structure. Therefore, methanogens are more beneficial for reductive dehalogenation than disadvantages.
Similarly, Li et al. (2021) reported a tri-culture system with Shewanella oneidensis MR-1, methanogens, and Dehalococcoides mccartyi strain 195 (Dhc 195) that established a high-efficiency electron transport network to assist TCE degradation (Li et al., 2019). MR-1 facilitates direct interspecies electron transfer (DIET) between community members, promoting methanogens and other members to synthesize cobalamin and accelerating the process of electron transfer to RDase. It is similar to the electron shuttles, which assist the interspecies electron transfer process. This indicates a feasible scheme for supporting dehalogenation, the free electron thought DIET combined with high-valence cobalamin (Co [II/III] state) to revert the high reducibility and finally supporting the Dhc 195 synthetic/activation RDase to enhance the organohalide respiration. In addition, the methanogens (e.g., Methanosarcina barkeri, Methanobacterium formicicum, Methanobrevibacter ruminantium, etc.) can synthesize 5′-hydroxybenzimidazolyl-cobamide (5-OHBza) and [Ade]cobamide, respectively, which could support the reductive dehalogenation of Dehalococcoides as well, they are potential cobalamin providers (Stupperich and Kräutler, 1988; Wagner et al., 2016). Studies have evaluated the association between methanogens and OHRB, and methanogens are not the only source of cobamide for OHRB (Yoshikawa et al., 2021; Yuan et al., 2021). However, there are reports that Dehalococcoides compete with methanogens for free cobamide (Wen et al., 2020), they may have other beneficial effects on the dehalogenation process, such as electron transfer, sustaining the low redox potential, or reducing energy barriers. Therefore, methanogens play a positive role in the dehalogenation community.
Additionally, a tri-culture of cobamide-producing bacteria with OHRB has also been reported; Desulfovibrio vulgaris Hildenborough (DVH) can produce cobalamin and establish a tri-culture system containing Dhc 195, DVH, and methanogens (Men et al., 2012). The Dhc 195 cell density in the tri-culture was approximately twice as high as in the isolated culture, and the expression of corrinoid transport and salvage function genes was decreased. This could be attributed to DVH, which provides cobalamin directly to Dhc 195, decreases structure remodeling energy consumption, and improves the effective corrinoid transfer, resulting in more energy conversion to support OHRB and a high cell yield. Moreover, Yan et al. (2013) used acetogens to establish a co-culture with Dehalococcoides: acetogens Sporomusa ovata and Sporomusa sp. KB-1, which can synthesize [Phe]cobamide and [p-Cre]cobamide, respectively. The [Phe]cobamide successfully activates organohalide respiration of Dehalococcoides, while [p-Cre]cobamide cannot. This indicates that the dehalogenation metabolism requires a specific cobamide type that the OHRB can selectively utilize before resorting to the remodeling function. However, this selection is strain-dependent and may not always occur. In that case, OHRB will start to transform cobamides and activate the remodeling function.
There are also microbial interaction modes that provide the lower ligand for remodeling. The remodeling function mechanism is designed to convert the lower ligand and produce sufficient cobalamin. It is plausible that the DMB is another key factor in the reductive dehalogenation of obligate OHRB. The remodeling function operates only when DMB is added to the co-culture microcosms as the lower ligand to guide cobalamin biosynthesis. However, DMB is mainly synthesized artificially, and there are few reported anaerobic biosynthetic pathways. Hazra et al. (2015) reported that Eubacterium limosum has a complete pathway of DMB biosynthesis. The anaerobic biosynthesis of DMB requires additional modification through the bzaABCDE genes. Shelton et al. (2019) suggest that this complete gene set is found only in a few species. It is still unclear which bacteria can biosynthesize or release DMB in an anaerobic dehalogenation community, but it was observed that DMB is the key lower ligand of cobalamin and can be applied to remodel cobamide into cobalamin. In addition, the DMB related utilization gene has been confirmed in the Dehalococcoides genome. Dehalococcoides can assemble cobalamin in vitro or in vivo by remodeling function, and the DMB synthesizer can be an efficient co-culture partner (Men et al., 2017; Esken et al., 2020; Mathur et al., 2020; Sokolovskaya et al., 2020).
In conclusion, we have summarized the interactions within dehalogenation communities, emphasizing the irreplaceable role of cobamide in symbiosis. We have noted the capacity of numerous bacteria and archaea to synthesize cobamides and the demand within the co-culture. Furthermore, we have discussed the positive impact of symbiotic partners in promoting reductive dehalogenation, such as methanogens, facultative OHRB, DVH, and acetogens. The significance of DMB in the remodeling function as the key ligand of cobalamin is emphasized. Despite the limited knowledge regarding DMB biosynthesis, it is confirmed that the incorporation of DMB enhances dehalogenation capacity, and further study is required.
Obligate and facultative OHRB play a vital role in microbial ecosystems, occupying an irreplaceable role in dehalogenation. The reductive dehalogenation process has been accepted as the optimal pathway for reducing halogenated compounds, with cobamide identified as the core component. Except the Sulfurospirillum multivorans directly use the [Ade]cobamide, cobalamin has been proposed as the optimal structure for reductive dehalogenation. Cobalamin-mediated mechanisms have a similar electron flow in reductive dehalogenation. Furthermore, obligate and facultative OHRB have different adaptations in corrinoid-auxotrophic environments. These corrinoid-auxotrophic OHRBs, through salvage and remodel pathway to narrow the gap of cobalamin deficiency, and establish a co-culture system with other cobamide producers for continuous reductive dehalogenation.
Current research is focused on determining the biosynthetic pathways of cobalamin in most OHRB, gene regulatory networks, and the effects of cobalamin on dehalogenation capacity under specific environmental conditions. However, the distinct degrees and rates of dehalogenation in the obligate and facultative OHRB within microbial ecosystems have been observed. Not only cobalamin, such as organohalide type, oxygen, temperature, and pH, competition, and interaction with other microorganisms also significant influence the rate and degree of dehalogenation. Therefore, it is important to further explore the factors that influence its species and abundance in specific environments. These factors can also guide the design and optimization of microbial communities to avoid the disadvantageous conditions encountered in the field. However, to forecast such complex microbial communities, which require significant amounts of data and suitable deep learning models, metatranscriptomics, and metaproteomics are necessary; more specifically, experimental information on microbial interaction should be provided. Furthermore, previous studies of the cobamide selectivity in OHRB are still in the laboratory stage. Field tests of community interactions for cobamides require further verification, and understanding the specific cobamide preferences of these microorganisms is crucial. However, the challenges of the lengthy culture time and scarcity of purified OHRB are significant. UsingAI-enabled environmental computing to narrow the scope of experiments is a viable option, which requires several of environmrntal data sets (e.g., substrate property, environmental and species information, interaction network, etc) for structured learning and deep learing to predict interspecific substrate exchange. The effect of different cobamides and their lower ligands on reductive dehalogenation may be elucidated through machine learning. The recently reported AlphaFold 3 accurately predicts macromolecules, and this may be used to predict the complex cobamides family and the ligand role in RDase which would promote the study of the binding and catalytic mechanisms of cobalamin.
In conclusion, future research in the field of OHRB should delve into their characteristics and mechanisms, focusing on cobalamin-related processes, interactions between microorganisms, symbiotic relationships, and their interactions with environmental factors. These can be facilitating by artificial intelligence and deep learning. The results of these studies will contribute to improving the application efficiency of OHRB in environmental remediation, thus facilitating the development of environmental remediation technologies.
YLu: Conceptualization, Methodology, Visualization, Writing – original draft, Writing – review & editing. FL: Visualization, Writing – review & editing. JZ: Supervision, Writing – review & editing. QT: Writing – review & editing. DY: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. YLi: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China under Grant Number 42377012; the Guangxi Natural Science Foundation under Grant Number 2021GXNSFBA196092 and AD22080067; and the Guangxi Education Agency (Gui Teachers [2021] No. 22).
We would like to thank all the reviewers for their helpful comments and thank the editor and the reviewers for their useful feedback that improved this paper. The authors thank Shuai Yang, Yashi Lin, and other colleagues for their suggestions for the figures and contributions to the content.
DY was employed by Guangxi Yuhuacheng Environmental Protection Technology Co.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbasian Chaleshtari, Z., and Foudazi, R. (2022). A review on per- and polyfluoroalkyl substances (PFAS) remediation: separation mechanisms and molecular interactions. ACS ESandT Water 2, 2258–2272. doi: 10.1021/acsestwater.2c00271
Agarwal, S., Dey, S., Ghosh, B., Biswas, M., and Dasgupta, J. (2019). Mechanistic basis of vitamin B12 and cobinamide salvaging by the Vibrio species. Biochim. Biophys. Acta 1867, 140–151. doi: 10.1016/j.bbapap.2018.11.004
Allen, R. H., and Stabler, S. P. (2008). Identification and quantitation of cobalamin and cobalamin analogues in human feces1. Am. J. Clin. Nutr. 87, 1324–1335. doi: 10.1093/ajcn/87.5.1324
Amos, B., Suchomel, J., Pennell, K., and Löffler, F. (2009). Spatial and temporal distributions of Geobacter lovleyi and Dehalococcoides spp. during bioenhanced PCE-NAPL dissolution. Environ. Sci. Technol. 43:1977–1985. doi: 10.1021/es8027692
Atashgahi, S., Lu, Y., and Smidt, H. (2016). “Overview of known organohalide-respiring bacteria—phylogenetic diversity and environmental distribution,” in Organohalide-Respiring Bacteria, eds. L. Adrian, and F.E. Löffler (Berlin, Heidelberg: Springer Berlin Heidelberg), 63–105. doi: 10.1007/978-3-662-49875-0_5
Balabanova, L. A., Averianova, L., Marchenok, M., Son, O. M., and Tekutyeva, L. A. (2021). Microbial and genetic resources for cobalamin (Vitamin B12) biosynthesis: from ecosystems to industrial biotechnology. Int. J. Mol. Sci. 22:4522. doi: 10.3390/ijms22094522
Bennett, K. A., Robinson, K. J., Armstrong, H. C., Moss, S. E. W., Scholl, G., Tranganida, A., et al. (2021). Predicting consequences of POP-induced disruption of blubber glucose uptake, mass gain rate and thyroid hormone levels for weaning mass in grey seal pups. Environ. Int. 152:106506. doi: 10.1016/j.envint.2021.106506
Bienert, S., Waterhouse, A., de Beer, T. A. P., Tauriello, G., Studer, G., Bordoli, L., et al. (2017). The SWISS-MODEL Repository - new features and functionality. Nucleic Acids Res. 45, W313–W319. doi: 10.1093/nar/gkw1132
Bommer, M., Kunze, C., Fesseler, J., Schubert, T., Diekert, G., and Dobbek, H. (2014). Structural basis for organohalide respiration. Science 346, 455–458. doi: 10.1126/science.1258118
Brown, K. L. (2005). Chemistry and enzymology of vitamin B12. Chem. Rev. 105, 2075–2150. doi: 10.1021/cr030720z
Bulka, O., Webb, J., Dworatzek, S., Mahadevan, R., and Edwards, E. A. (2023). A multifunctional dehalobacter? Tandem chloroform and dichloromethane degradation in a mixed microbial culture. Environ. Sci. Technol. 57, 19912–19920. doi: 10.1021/acs.est.3c06686
Chen, C., Xu, G., Rogers, M. J., and He, J. (2024). Metabolic synergy of Dehalococcoides populations leading to greater reductive dechlorination of polychlorinated biphenyls. Environ. Sci. Technol. 58, 2384–2392. doi: 10.1021/acs.est.3c08473
Cheng, J., Liu, M., Su, X., Rittmann, B. E., Lu, Z., Xu, J., et al. (2023). Conductive materials on biocathodes altered the electron-transfer paths and modulated γ-HCH dechlorination and CH4 production in microbial electrochemical systems. Environ. Sci. Technol. 57, 2739–2748. doi: 10.1021/acs.est.2c06097
Cooper, M., Wagner, A., Wondrousch, D., Sonntag, F., Sonnabend, A., Brehm, M., et al. (2015). Anaerobic microbial transformation of halogenated aromatics and fate prediction using electron density modeling. Environ. Sci. Technol. 49, 6018–6028. doi: 10.1021/acs.est.5b00303
Cui, Y., Yang, Y., Yan, J., and Xiuying, L. (2021). Advances of using Dehalogenimonas in anaerobic degradation of chlorinated compounds and bioremediation of contaminated sites. Chin. J. Biotechnol. 37, 3565–3577.
Deery, E., Lawrence, A. D., and Warren, M. J. (2022). “Biosynthesis of cobamides: Methods for the detection, analysis and production of cobamides and biosynthetic intermediates,” in Methods in Enzymology, ed. E. N. G. Marsh (London: Academic Press), 3–23. doi: 10.1016/bs.mie.2022.01.013
Dickman, S. R. (1977). Ribonucleotide reduction and the possible role of cobalamin in evolution. J. Mol. Evol. 10, 251–260. doi: 10.1007/BF01764600
Ding, C., Rogers, M. J., Yang, K.-L., and He, J. (2017). Loss of the ssrA genome island led to partial debromination in the PBDE respiring Dehalococcoides mccartyi strain GY50. Environ. Microbiol. 19, 2906–2915. doi: 10.1111/1462-2920.13817
Drzyzga, O., and Gottschal Jan, C. (2002). Tetrachloroethene dehalorespiration and growth of Desulfitobacterium frappieri TCE1 in strict dependence on the activity of Desulfovibrio fructosivorans. Appl. Environ. Microbiol. 68, 642–649. doi: 10.1128/AEM.68.2.642-649.2002
Dulay, H., Tabares, M., Kashefi, K., and Reguera, G. (2020). Cobalt resistance via detoxification and mineralization in the iron-reducing bacterium Geobacter sulfurreducens. Front. Microbiol. 11:600463. doi: 10.3389/fmicb.2020.600463
Escalante-Semerena, J. (2007). Conversion of cobinamide into adenosylcobamide in bacteria and archaea. J. Bacteriol. 189, 4555–4560. doi: 10.1128/JB.00503-07
Esken, J., Goris, T., Gadkari, J., Bischler, T., Förstner, K., Sharma, C., et al. (2020). Tetrachloroethene respiration in Sulfurospirillum species is regulated by a two-component system as unraveled by comparative genomics, transcriptomics, and regulator binding studies. Microbiologyopen 9:e1138. doi: 10.1002/mbo3.1138
Ewald, J., Schnoor, J., and Mattes, T. (2022). Combined read- and assembly-based metagenomics to reconstruct a Dehalococcoides mccartyi genome from PCB-contaminated sediments and evaluate functional differences among organohalide-respiring consortia in the presence of different halogenated contaminants. FEMS Microbiol. Ecol. 98:fiac067. doi: 10.1093/femsec/fiac067
Fang, H., Kang, J., and Zhang, D. (2017). Microbial production of vitamin B12: a review and future perspectives. Microb. Cell Fact. 16:15. doi: 10.1186/s12934-017-0631-y
Farnberger, J. E., Hiebler, K., Bierbaumer, S., Skibar, W., Zepeck, F., and Kroutil, W. (2019). Cobalamin-dependent apparent intramolecular methyl transfer for biocatalytic constitutional isomerization of catechol monomethyl ethers. ACS Catal. 9, 3900–3905. doi: 10.1021/acscatal.8b05072
Feldewert, C., Lang, K., and Brune, A. (2020). The hydrogen threshold of obligately methyl-reducing methanogens. FEMS Microbiol. Lett. 367:fnaa137. doi: 10.1093/femsle/fnaa137
Femina Carolin, C., Kamalesh, T., Senthil Kumar, P., and Rangasamy, G. (2023). An insights of organochlorine pesticides categories, properties, eco-toxicity and new developments in bioremediation process. Environ. Pollut. 333:122114. doi: 10.1016/j.envpol.2023.122114
Feng, S., Merino, N., Okamoto, A., and Gedalanga, P. (2018). Interkingdom microbial consortia mechanisms to guide biotechnological applications. Microb. Biotechnol. 11, 833–847. doi: 10.1111/1751-7915.13300
Fincker, M., and Spormann, A. M. (2017). Biochemistry of catabolic reductive dehalogenation. Annu. Rev. Biochem. 86, 357–386. doi: 10.1146/annurev-biochem-061516-044829
Franke, S., Seidel, K., Adrian, L., and Nijenhuis, I. (2020). Dual Element (C/Cl) Isotope analysis indicates distinct mechanisms of reductive dehalogenation of chlorinated ethenes and dichloroethane in Dehalococcoides mccartyi strain BTF08 with defined reductive dehalogenase inventories. Front. Microbiol. 11:1507. doi: 10.3389/fmicb.2020.01507
Gribble, G. W. (1998). Naturally occurring organohalogen compounds. Acc. Chem. Res. 31, 141–152. doi: 10.1021/ar9701777
Guo, M., and Chen, Y. (2018). Coenzyme cobalamin: biosynthesis, overproduction and its application in dehalogenation—a review. Rev. Environ. Sci. Bio/Technol. 17, 259–284. doi: 10.1007/s11157-018-9461-6
Halliwell, T., Fisher, K., Payne, K. P., Rigby, S. E. J., and Leys, D. (2020). Catabolic reductive dehalogenase substrate complex structures underpin rational repurposing of substrate scope. Microorganisms 8:1344. doi: 10.3390/microorganisms8091344
Hazra, A. B., Han, A. W., Mehta, A. P., Mok, K. C., Osadchiy, V., Begley, T. P., et al. (2015). Anaerobic biosynthesis of the lower ligand of vitamin B12. Proc. Nat. Acad. Sci. 112, 10792–10797. doi: 10.1073/pnas.1509132112
He, H., Li, Y., Shen, R., Shim, H., Zeng, Y., Zhao, S., et al. (2021). Environmental occurrence and remediation of emerging organohalides: a review. Environ. Pollut. 290:118060. doi: 10.1016/j.envpol.2021.118060
He, J., Holmes, V., Lee, P., and Alvarez-Cohen, L. (2007). Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl. Environ. Microbiol. 73, 2847–2853. doi: 10.1128/AEM.02574-06
Heckel, B., Mcneill, K., and Elsner, M. (2018). Chlorinated ethene reactivity with vitamin B12 is governed by cobalamin chloroethylcarbanions as crossroads of competing pathways. ACS Catal. 8, 3054–3066. doi: 10.1021/acscatal.7b02945
Huang, B., Lei, C., Wei, C., and Zeng, G. (2014). Chlorinated volatile organic compounds (Cl-VOCs) in environment—sources, potential human health impacts, and current remediation technologies. Environ. Int. 71, 118–138. doi: 10.1016/j.envint.2014.06.013
Hug, L. A., Beiko, R. G., Rowe, A. R., Richardson, R. E., and Edwards, E. A. (2012). Comparative metagenomics of three Dehalococcoides-containing enrichment cultures: the role of the non-dechlorinating community. BMC Genomics 13:327. doi: 10.1186/1471-2164-13-327
Ji, L., Wang, C., Ji, S., Kepp, K. P., and Paneth, P. (2017). Mechanism of cobalamin-mediated reductive dehalogenation of chloroethylenes. ACS Catal. 7, 5294–5307. doi: 10.1021/acscatal.7b00540
Jiang, G., Zhang, X., and Luan, J. (2023). Research progress in bio-conversion of carbon dioxide to methane. Acta Microbiol. Sin. 63, 2245–2260. doi: 10.13343/j.cnki.wsxb.20230046
Jin, H., Huo, L., Yang, Y., Lv, Y., Wang, J., Maillard, J., et al. (2023). Sulfurospirillum diekertiae sp. nov., a tetrachloroethene-respiring bacterium isolated from contaminated soil. Int. J. System. Evolut. Microbiol. 73:005693. doi: 10.1099/ijsem.0.005693
Jin, H., Yang, Y., Li, X., Song, Y., and Yan, J. (2020). Progress in microbial degradation of hexachlorobutadiene. Microbiol. China 47, 3407–3418. doi: 10.13344/j.microbiol.china.200607
Jin, L., and Chen, B. (2017). Natural origins, concentration levels, and formation mechanisms of organohalogens in the environment. Progr. Chem. 29, 1093–1114. doi: 10.7536/PC170563
Johannissen, L. O., Leys, D., and Hay, S. (2017). A common mechanism for coenzyme cobalamin-dependent reductive dehalogenases. Phys. Chem. Chem. Phys. 19, 6090–6094. doi: 10.1039/C6CP08659D
Jugder, B.-E., Ertan, H., Lee, M., Manefield, M., and Marquis, C. P. (2015). Reductive dehalogenases come of age in biological destruction of organohalides. Trends Biotechnol. 33, 595–610. doi: 10.1016/j.tibtech.2015.07.004
Kallenborn, R., Hühnerfuss, H., Aboul-Enein, H. Y., and Ali, I. (2021). Chiral Environmental Pollutants: Analytical Methods, Environmental Implications and Toxicology. Cham: Springer International Publishing, 107–254. doi: 10.1007/978-3-030-62456-9
Keller, S., Kunze, C., Bommer, M., Paetz, C., Menezes, R., Svatos, A., et al. (2018). Selective utilization of benzimidazolyl-norcobamides as cofactors by the tetrachloroethene reductive dehalogenase of Sulfurospirillum multivorans. J. Bacteriol. 200:00584–00517. doi: 10.1128/JB.00584-17
Keller, S., Ruetz, M., Kunze, C., Kräutler, B., Diekert, G., and Schubert, T. (2014). Exogenous 5,6-dimethylbenzimidazole caused production of a non-functional tetrachloroethene reductive dehalogenase in Sulfurospirillum multivorans. Environ. Microbiol. 16, 3361–3369. doi: 10.1111/1462-2920.12268
Kim, M., and Han, J. (2024). Treatment techniques for removal of polybrominated diphenyl ethers (PBDEs) from real wastewater: limitations, challenges, and future research directions. J. Water Proc. Eng. 63:105463. doi: 10.1016/j.jwpe.2024.105463
Kipkorir, T., Mashabela Gabriel, T., De Wet Timothy, J., Koch, A., Wiesner, L., Mizrahi, V., et al. (2021). De Novo cobalamin biosynthesis, transport, and assimilation and cobalamin-mediated regulation of methionine biosynthesis in Mycobacterium smegmatis. J. Bacteriol. 203:e00620. doi: 10.1128/JB.00620-20
Kruse, S., Türkowsky, D., Birkigt, J., Matturro, B., Franke, S., Jehmlich, N., et al. (2021). Interspecies metabolite transfer and aggregate formation in a co-culture of Dehalococcoides and Sulfurospirillum dehalogenating tetrachloroethene to ethene. ISME J. 15, 1794–1809. doi: 10.1038/s41396-020-00887-6
Kunze, C., Bommer, M., Hagen, W. R., Uksa, M., Dobbek, H., Schubert, T., et al. (2017). Cobamide-mediated enzymatic reductive dehalogenation via long-range electron transfer. Nat. Commun. 8:15858. doi: 10.1038/ncomms15858
Kuroda, M., and Watanabe, T. (1995). CO2 reduction to methane and acetate using a bio-electro reactor with immobilized methanogens and homoacetogens on electrodes. Energy Conver. Manag. 36, 787–790. doi: 10.1016/0196-8904(95)00122-T
Lai, Y., and Becker, J. G. (2013). Compounded effects of chlorinated ethene inhibition on ecological interactions and population abundance in a Dehalococcoides- Dehalobacter coculture. Environ. Sci. Technol. 47, 1518–1525. doi: 10.1021/es3034582
Li, X., Yang, Y., Wang, J., Jin, H., Zhang, Y., Cui, Y., et al. (2022). Organohalide respiration with diclofenac by dehalogenimonas. Environ. Sci. Technol. 56, 11266–11276. doi: 10.1021/acs.est.1c08824
Li, Y., Wen, L., Zhao, H., and Zhu, L. (2019). Addition of Shewanella oneidensis MR-1 to the Dehalococcoides-containing culture enhances the trichloroethene dechlorination. Environ. Int. 133:105245. doi: 10.1016/j.envint.2019.105245
Li, Y., Zhao, H., and Zhu, L. (2021). Iron sulfide enhanced the dechlorination of trichloroethene by Dehalococcoides mccartyi Strain 195. Front. Microbiol. 12:665281. doi: 10.3389/fmicb.2021.665281
Liang, Y., Lu, Q., Liang, Z., Liu, X., Fang, W., Liang, D., et al. (2021). Substrate-dependent competition and cooperation relationships between Geobacter and Dehalococcoides for their organohalide respiration. ISME Commun. 1:23. doi: 10.1038/s43705-021-00025-z
Liao, R.-Z., Chen, S.-L., and Siegbahn, P. E. M. (2015). Which oxidation state initiates dehalogenation in the B12-dependent enzyme NpRdhA: CoII, CoI, or Co0? ACS Catal. 5, 7350–7358. doi: 10.1021/acscatal.5b01502
Liu, J., and Häggblom Max, M. (2018). Genome-guided identification of organohalide-respiring deltaproteobacteria from the marine environment. MBio 9, e02471–e02418. doi: 10.1128/mBio.02471-18
Löffler, F., Yan, J., Ritalahti, K., Adrian, L., Edwards, E., Konstantinidis, K., et al. (2013). Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. System. Evolut. Microbiol. 63, 625–635. doi: 10.1099/ijs.0.034926-0
Lu, Q., Qiu, L., Yu, L., Zhang, S., De Toledo, R. A., Shim, H., et al. (2019). Microbial transformation of chiral organohalides: distribution, microorganisms and mechanisms. J. Hazard. Mater. 368, 849–861. doi: 10.1016/j.jhazmat.2019.01.103
Lu, Y., Liang, F., Qin, F., Zhong, L., Jiang, J., Liu, Q., et al. (2024). Tourmaline guiding the electric field and dechlorination pathway of 2,3-dichlorophenol by Desulfitobacterium hafniense. J. Environ. Sci. 135, 262–273. doi: 10.1016/j.jes.2022.12.033
Ma, X., Novak, P. J., Clapp, L. W., Semmens, M. J., and Hozalski, R. M. (2003). Evaluation of polyethylene hollow-fiber membranes for hydrogen delivery to support reductive dechlorination in a soil column. Water Res. 37, 2905–2918. doi: 10.1016/S0043-1354(03)00111-8
Magnuson, J. K., Stern, R. V., Gossett, J. M., Zinder, S. H., and Burris, D. R. (1998). Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl. Environ. Microbiol. 64, 1270–1275. doi: 10.1128/AEM.64.4.1270-1275.1998
Maillard, J., and Holliger, C. (2016). “The genus Dehalobacter,” in Organohalide-Respiring Bacteria, eds. L. Adrian, and F. E. Löffler (Berlin, Heidelberg: Springer Berlin Heidelberg), 153–171. doi: 10.1007/978-3-662-49875-0_8
Maphosa, F., De Vos, W. M., and Smidt, H. (2010). Exploiting the ecogenomics toolbox for environmental diagnostics of organohalide-respiring bacteria. Trends Biotechnol. 28, 308–316. doi: 10.1016/j.tibtech.2010.03.005
Maphosa, F., Van Passel, M. W. J., De Vos, W. M., and Smidt, H. (2012). Metagenome analysis reveals yet unexplored reductive dechlorinating potential of Dehalobacter sp. E1 growing in co-culture with Sedimentibacter sp. Environ. Microbiol. Rep. 4, 604–616. doi: 10.1111/j.1758-2229.2012.00376.x
Marzorati, M., De Ferra, F., Van Raemdonck, H., Borin, S., Allifranchini, E., Carpani, G., et al. (2007). A novel reductive dehalogenase, identified in a contaminated groundwater enrichment culture and in Desulfitobacterium dichloroeliminans strain DCA1, is linked to dehalogenation of 1,2-dichloroethane. Appl. Environ. Microbiol. 73, 2990–2999. doi: 10.1128/AEM.02748-06
Mathur, Y., Sreyas, S., Datar, P., Sathian, M., and Hazra, A. (2020). CobT and BzaC catalyze the regiospecific activation and methylation of the 5-hydroxybenzimidazole lower ligand in anaerobic cobamide biosynthesis. J. Biol. Chem. 295, 10522–10534. doi: 10.1074/jbc.RA120.014197
Mathur, Y., Vartak, A. R., and Hazra, A. B. (2022). “Guardian of cobamide diversity: probing the role of CobT in lower ligand activation in the biosynthesis of vitamin B12 and other cobamide cofactors,” in Methods in Enzymology, ed. E.N.G. Marsh (London: Academic Press), 25–59. doi: 10.1016/bs.mie.2022.01.001
Maymó-Gatell, X., Tandoi, V., Gossett, J., and Zinder, S. (1995). Characterization of an H2-utilizing enrichment culture that reductively dechlorinates tetrachloroethene to vinyl chloride and ethene in the absence of methanogensis and acetogenesis. Appl. Environ. Microbiol. 61, 3928–3933. doi: 10.1128/aem.61.11.3928-3933.1995
Men, Y., Feil, H., Verberkmoes, N. C., Shah, M. B., Johnson, D. R., Lee, P. K. H., et al. (2012). Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with Desulfovibrio vulgaris Hildenborough and Methanobacterium congolense: global transcriptomic and proteomic analyses. ISME J. 6, 410–421. doi: 10.1038/ismej.2011.111
Men, Y., Lee, P. K. H., Harding, K. C., and Alvarez-Cohen, L. (2013). Characterization of four TCE-dechlorinating microbial enrichments grown with different cobalamin stress and methanogenic conditions. Appl. Microbiol. Biotechnol. 97, 6439–6450. doi: 10.1007/s00253-013-4896-8
Men, Y., Seth Erica, C., Yi, S., Allen Robert, H., Taga Michiko, E., and Alvarez-Cohen, L. (2014). Sustainable growth of Dehalococcoides mccartyi 195 by corrinoid salvaging and remodeling in defined lactate-fermenting consortia. Appl. Environ. Microbiol. 80, 2133–2141. doi: 10.1128/AEM.03477-13
Men, Y., Seth, E. C., Yi, S., Crofts, T. S., Allen, R. H., Taga, M. E., et al. (2015). Identification of specific corrinoids reveals corrinoid modification in dechlorinating microbial communities. Environ. Microbiol. 17, 4873–4884. doi: 10.1111/1462-2920.12500
Men, Y., Yu, K., Bælum, J., Gao, Y., Tremblay, J., Prestat, E., et al. (2017). Metagenomic and metatranscriptomic analyses reveal the structure and dynamics of a dechlorinating community containing Dehalococcoides mccartyi and corrinoid-providing microorganisms under cobalamin-limited conditions. Appl. Environ. Microbiol. 83, e03508–03516. doi: 10.1128/AEM.03508-16
Min, Z., Zhang, L., Franks, A. E., Feng, X., Brookes, P. C., Xu, J., et al. (2019). Improved synergistic dechlorination of PCP in flooded soil microcosms with supplementary electron donors, as revealed by strengthened connections of functional microbial interactome. Soil Biol. Biochem. 136:107515. doi: 10.1016/j.soilbio.2019.06.011
Monteagudo-Cascales, E., García-Mauriño, S. M., Santero, E., and Canosa, I. (2019). Unraveling the role of the CbrA histidine kinase in the signal transduction of the CbrAB two-component system in Pseudomonas putida. Sci. Rep. 9:9110. doi: 10.1038/s41598-019-45554-9
Moore, T. C., and Escalante-Semerena, J. C. (2016). “Corrinoid metabolism in dehalogenating pure cultures and microbial communities,” in Organohalide-Respiring Bacteria, eds. L. Adrian, and F.E. Löffler (Berlin, Heidelberg: Springer Berlin Heidelberg), 455–484. doi: 10.1007/978-3-662-49875-0_19
Nakamura, R., Obata, T., Nojima, R., Hashimoto, Y., Noguchi, K., Ogawa, T., et al. (2018). Functional expression and characterization of tetrachloroethene dehalogenase from Geobacter sp. Front. Microbiol. 9:1774. doi: 10.3389/fmicb.2018.01774
Neumann, A., Wohlfarth, G., and Diekert, G. (1996). Purification and characterization of tetrachloroethene reductive dehalogenase from Dehalospirillum multivorans*. J. Biol. Chem. 271, 16515–16519. doi: 10.1074/jbc.271.28.16515
Ning, Z., Zhang, M., Zhang, N., Guo, C., Hao, C., Zhang, S., et al. (2022). Metagenomic characterization of a novel enrichment culture responsible for dehalogenation of 1,2,3-trichloropropane to allyl chloride. J. Environ. Chem. Eng. 10:108907. doi: 10.1016/j.jece.2022.108907
Nonaka, H., Keresztes, G., Shinoda, Y., Ikenaga, Y., Abe, M., Naito, K., et al. (2006). Complete genome sequence of the dehalorespiring bacterium Desulfitobacterium hafniense Y51 and comparison with Dehalococcoides ethenogenes 195. J. Bacteriol. 188, 2262–2274. doi: 10.1128/JB.188.6.2262-2274.2006
Padilla-Crespo, E., Yan, J., Swift, C., Wagner, D., Chourey, K., Hettich, R., et al. (2013). Identification and environmental distribution of dcpA, which encodes the reductive dehalogenase catalyzing the dichloroelimination of 1,2-dichloropropane to propene in organohalide-respiring Chloroflexi. Appl. Environ. Microbiol. 80, 808–818. doi: 10.1128/AEM.02927-13
Palau, J., Trueba-Santiso, A., Yu, R., Mortan, S. H., Shouakar-Stash, O., Freedman, D. L., et al. (2023). Dual C–Br isotope fractionation indicates distinct reductive dehalogenation mechanisms of 1,2-dibromoethane in dehalococcoides- and dehalogenimonas-containing cultures. Environ. Sci. Technol. 57, 1949–1958. doi: 10.1021/acs.est.2c07137
Parthasarathy, A., Stich, T. A., Lohner, S. T., Lesnefsky, A., Britt, R. D., and Spormann, A. M. (2015). Biochemical and EPR-spectroscopic investigation into heterologously expressed vinyl chloride reductive dehalogenase (VcrA) from Dehalococcoides mccartyi Strain VS. J. Am. Chem. Soc. 137, 3525–3532. doi: 10.1021/ja511653d
Payne, K. P., Quezada, C. P., Fisher, K., Dunstan, M. S., Collins, F. A., Sjuts, H., et al. (2015). Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation. Nature 517, 513–516. doi: 10.1038/nature13901
Reguera, G., and Kashefi, K. (2019). The electrifying physiology of Geobacter bacteria, 30 years on. Adv. Microbial Physiol. 74, 1–96. doi: 10.1016/bs.ampbs.2019.02.007
Reinhold, A., Westermann, M., Seifert, J., Von Bergen, M., Schubert, T., and Diekert, G. (2012). Impact of vitamin B12 on formation of the tetrachloroethene reductive dehalogenase in Desulfitobacterium hafniense strain Y51. Appl. Environ. Microbiol. 78, 8025–8032. doi: 10.1128/AEM.02173-12
Richardson, R. E. (2013). Genomic insights into organohalide respiration. Curr. Opin. Biotechnol. 24, 498–505. doi: 10.1016/j.copbio.2013.02.014
Rupakula, A., Kruse, T., Boeren, S., Holliger, C., Smidt, H., and Maillard, J. (2013). The restricted metabolism of the obligate organohalide respiring bacterium Dehalobacter restrictus: lessons from tiered functional genomics. Philos. Trans. R. Soc. 368:20120325. doi: 10.1098/rstb.2012.0325
Rupakula, A., Lu, Y., Kruse, T., Boeren, S., Holliger, C., Smidt, H., et al. (2015). Functional genomics of corrinoid starvation in the organohalide-respiring bacterium Dehalobacter restrictus strain PER-K23. Front. Microbiol. 5:751. doi: 10.3389/fmicb.2014.00751
Salom, D., Fernández-Verdejo, D., Moral-Vico, J., Font, X., and Marco-Urrea, E. (2023). Combining nanoscale zero-valent iron and anaerobic dechlorinating bacteria to degrade chlorinated methanes and 1,2-dichloroethane. Environ. Sci. Pollut. Res. 30, 45231–45243. doi: 10.1007/s11356-023-25376-z
Sanford Robert, A., Cole James, R., and Tiedje James, M. (2002). Characterization and description of anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl-halorespiring facultative anaerobic myxobacterium. Appl. Environ. Microbiol. 68, 893–900. doi: 10.1128/AEM.68.2.893-900.2002
Schubert, T. (2017). The organohalide-respiring bacterium Sulfurospirillum multivorans: a natural source for unusual cobamides. World J. Microbiol. Biotechnol. 33:93. doi: 10.1007/s11274-017-2258-x
Schubert, T., Adrian, L., Sawers, R. G., and Diekert, G. (2018). Organohalide respiratory chains: composition, topology and key enzymes. FEMS Microbiol. Ecol. 94:fiy035. doi: 10.1093/femsec/fiy035
Schubert, T., Von Reu,ß, S. H, Kunze, C., Paetz, C., Kruse, S., Brand-Schön, P., et al. (2019). Guided cobamide biosynthesis for heterologous production of reductive dehalogenases. Microb. Biotechnol. 12, 346–359. doi: 10.1111/1751-7915.13339
Scott, A. I., and Roessner, C. A. (2002). Biosynthesis of cobalamin (vitamin B12). Biochem. Soc. Trans. 30, 613–620. doi: 10.1042/bst0300613
Seshadri, R., Adrian, L., Fouts, D. E., Eisen, J. A., Phillippy, A. M., Methe, B. A., et al. (2005). Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307, 105–108. doi: 10.1126/science.1102226
Shelton, A. N., Seth, E. C., Mok, K. C., Han, A. W., Jackson, S. N., Haft, D. R., et al. (2019). Uneven distribution of cobamide biosynthesis and dependence in bacteria predicted by comparative genomics. ISME J. 13, 789–804. doi: 10.1038/s41396-018-0304-9
Shelton, D. R., and Tiedje, J. M. (1984). Isolation and partial characterization of bacteria in an anaerobic consortium that mineralizes 3-chlorobenzoic acid. Appl. Environ. Microbiol. 48, 840–848. doi: 10.1128/aem.48.4.840-848.1984
Sokolovskaya Olga, M., Mok Kenny, C., Park Jong, D., Tran Jennifer, L. A., Quanstrom Kathryn, A., and Taga Michiko, E. (2019). Cofactor selectivity in methylmalonyl coenzyme a mutase, a model cobamide-dependent enzyme. mBio. 10:01319. doi: 10.1128/mBio.01303-19
Sokolovskaya, O., Shelton, A., and Taga, M. (2020). Sharing vitamins: cobamides unveil microbial interactions. Science 369:eaba0165. doi: 10.1126/science.aba0165
Song, J.-X., Li, L., Sheng, F.-F., Guo, C.-X., Zhang, Y.-M., Li, Z.-Y., et al. (2015). 2, 4, 6-trichlorophenol mineralization promoted by anaerobic reductive dechlorination of acclimated sludge and extracellular respiration dechlorination pathway. Huan Jing Ke Xue 36, 3764–3770.
Studer, G., Rempfer, C., Waterhouse, A. M., Gumienny, R., Haas, J., and Schwede, T. (2020). QMEANDisCo - distance constraints applied on model quality estimation. Bioinformatics 36, 1765–1771. doi: 10.1093/bioinformatics/btz828
Stupperich, E., and Kräutler, B. (1988). Pseudo vitamin B12 or 5-hydroxybenzimidazolyl-cobamide are the corrinoids found in methanogenic bacteria. Arch. Microbiol. 149, 268–271. doi: 10.1007/BF00422016
Tang, S., and Edwards, E. A. (2013). Identification of Dehalobacter reductive dehalogenases that catalyse dechlorination of chloroform, 1,1,1-trichloroethane and 1,1-dichloroethane. Philos. Trans. R. Soc. 368:20120318. doi: 10.1098/rstb.2012.0318
Türkowsky, D., Jehmlich, N., Diekert, G., Adrian, L., von Bergen, M., and Goris, T. (2018). An integrative overview of genomic, transcriptomic and proteomic analyses in organohalide respiration research. FEMS Microbiol. Ecol. 94:fly013. doi: 10.1093/femsec/fiy013
Villemur, R., Lanthier, M., Beaudet, R., and Lépine, F. (2006). The Desulfitobacterium genus. FEMS Microbiol. Rev. 30, 706–733. doi: 10.1111/j.1574-6976.2006.00029.x
Wagner, D., Hug, L., Hatt, J., Spitzmiller, M., Padilla-Crespo, E., Ritalahti, K., et al. (2012). Genomic determinants of organohalide-respiration in Geobacter lovleyi, an unusual member of the Geobacteraceae. BMC Genomics 13:200. doi: 10.1186/1471-2164-13-200
Wagner, T., Ermler, U., and Shima, S. (2016). MtrA of the sodium ion pumping methyltransferase binds cobalamin in a unique mode. Sci. Rep. 6:28226. doi: 10.1038/srep28226
Wang, J., Li, X., Song, Y., Yan, J., and Yang, Y. (2022). Effects of environmental factors on anaerobic microbial dehalogenation: a review. Microbiol. China 49, 4357–4381.
Wang, S., Chen, C., Zhao, S., and He, J. (2019). Microbial synergistic interactions for reductive dechlorination of polychlorinated biphenyls. Sci. Total Environ. 666, 368–376. doi: 10.1016/j.scitotenv.2019.02.283
Wang, S., Qiu, L., Liu, X., Xu, G., Siegert, M., Lu, Q., et al. (2018). Electron transport chains in organohalide-respiring bacteria and bioremediation implications. Biotechnol. Adv. 36, 1194–1206. doi: 10.1016/j.biotechadv.2018.03.018
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Wen, L., Li, Y., Zhu, L., and Zhao, H. (2020). Influence of non-dechlorinating microbes on trichloroethene reduction based on vitamin B12 synthesis in anaerobic cultures. Environ. Pollut. 259:113947. doi: 10.1016/j.envpol.2020.113947
Wen, L., Zhang, Y., Pan, Y., Wu, W., Meng, S., Zhou, C., et al. (2015). The roles of methanogens and acetogens in dechlorination of trichloroethene using different electron donors. Environ. Sci. Pollut. Res. 22, 19039–19047. doi: 10.1007/s11356-015-5117-z
Xu, G., Zhao, X., Zhao, S., Rogers, M. J., and He, J. (2023). Salinity determines performance, functional populations, and microbial ecology in consortia attenuating organohalide pollutants. ISME J. 17, 660–670. doi: 10.1038/s41396-023-01377-1
Xu, R., Xie, Y., Tian, J., and Chen, L. (2021). Adsorbable organic halogens in contaminated water environment: a review of sources and removal technologies. J. Clean. Prod. 283:124645. doi: 10.1016/j.jclepro.2020.124645
Yan, J., Bi, M., Bourdon, A. K., Farmer, A. T., Wang, P.-H., Molenda, O., et al. (2018). Purinyl-cobamide is a native prosthetic group of reductive dehalogenases. Nat. Chem. Biol. 14, 8–14. doi: 10.1038/nchembio.2512
Yan, J., Im, J., Yang, Y., and Löffler, F. E. (2013). Guided cobalamin biosynthesis supports Dehalococcoides mccartyi reductive dechlorination activity. Philos. Trans. R. Soc. 368:20120320. doi: 10.1098/rstb.2012.0320
Yan, J., Ritalahti, K., Wagner, D., and Löffler, F. (2012). Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to organohalide-respiring Dehalococcoides mccartyi strains. Appl. Environ. Microbiol. 78, 6630–6636. doi: 10.1128/AEM.01535-12
Yan, J., Simşir, B., Farmer A T, Bi, M., Yang, Y., Campagna, S. R., and Löffler, F. E. (2016). The corrinoid cofactor of reductive dehalogenases affects dechlorination rates and extents in organohalide-respiring Dehalococcoides mccartyi. ISME J. 10, 1092–1101. doi: 10.1038/ismej.2015.197
Yang, Y., Higgins, S. A., Yan, J., Simşir, B., Chourey, K, Iyer, R., Hettich, R. L., et al. (2017). Grape pomace compost harbors organohalide-respiring Dehalogenimonas species with novel reductive dehalogenase genes. ISME J. 11, 2767–2780. doi: 10.1038/ismej.2017.127
Yang, Y., Sanford, R., Yan, J., Chen, G., Cápiro, N., Xiuying, L., et al. (2020a). Roles of organohalide-respiring Dehalococcoidia in carbon cycling. mSystems 5, e00757–e00719. doi: 10.1128/mSystems.00757-19
Yang, Y., Zhang, Y., Cápiro, N., and Yan, J. (2020b). Genomic characteristics distinguish geographically distributed Dehalococcoidia. Front. Microbiol. 11:546063. doi: 10.3389/fmicb.2020.546063
Yankovych, H., Vaclavikova, M., and Melnyk, I. (2023). A review on adsorbable organic halogens treatment technologies: approaches and application. Sustainability 15:9601. doi: 10.3390/su15129601
Yi, S., Seth, E., Men, Y., Stabler, S., Allen, R., Alvarez-Cohen, L., et al. (2012). Versatility in corrinoid salvaging and remodeling pathways supports corrinoid-dependent metabolism in Dehalococcoides mccartyi. Appl. Environ. Microbiol. 78, 7745–7752. doi: 10.1128/AEM.02150-12
Yoshikawa, M., Zhang, M., Kawabe, Y., and Katayama, T. (2021). Effects of ferrous iron supplementation on reductive dechlorination of tetrachloroethene and on methanogenic microbial community. FEMS Microbiol. Ecol. 97:fiab069. doi: 10.1093/femsec/fiab069
Yu, Y., Zhang, Y., Liu, Y., Lv, M., Wang, Z., Wen, L.-L., et al. (2023). In situ reductive dehalogenation of groundwater driven by innovative organic carbon source materials: Insights into the organohalide-respiratory electron transport chain. J. Hazardous Mater. 452:131243. doi: 10.1016/j.jhazmat.2023.131243
Yuan, J., Li, S., Cheng, J., Guo, C., Shen, C., He, J., et al. (2021). Potential role of methanogens in microbial reductive dechlorination of organic chlorinated pollutants in situ. Environ. Sci. Technol. 55, 5917–5928. doi: 10.1021/acs.est.0c08631
Zhai, W., Fang, H., Zhuge, B., Zhang, C., Xue, Y., and Zhuge, J. (2012). Facilitated expression and function identification of key genes converting cyanocobalamin to adenosylcobalamin. Chinese J. Apppl. Environ. Biol. 18:267. doi: 10.3724/SP.J.1145.2012.00267
Zhang, Y., Jin, H., Xiuying, L., Song, Y., Yan, J., and Yang, Y. (2021). Advances in degradation mechanisms of 1,2,3-trichloropropane and remediation technology of contaminated sites. Chin. J. Biotechnol. 37, 3578–3590. doi: 10.13345/j.cjb.210417
Zhang, Y., Rodionov, D., Gelfand, M., and Gladyshev, V. (2009). Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genomics 10:78. doi: 10.1186/1471-2164-10-78
Zhang, Z., Ali, M., Tang, Z., Sun, Q., Wang, Q., Liu, X., et al. (2024). Unveiling complete natural reductive dechlorination mechanisms of chlorinated ethenes in groundwater: Insights from functional gene analysis. J. Hazard. Mater. 469:134034. doi: 10.1016/j.jhazmat.2024.134034
Zhang, Z., Zhang, C., Yang, Y., Zhang, Z., Tang, Y., Su, P., et al. (2022). A review of sulfate-reducing bacteria: metabolism, influencing factors and application in wastewater treatment. J. Clean. Prod. 376:134109. doi: 10.1016/j.jclepro.2022.134109
Zhao, S., Ding, C., Xu, G., Rogers, M. J., Ramaswamy, R., and He, J. (2022). Diversity of organohalide respiring bacteria and reductive dehalogenases that detoxify polybrominated diphenyl ethers in E-waste recycling sites. ISME J. 16, 2123–2131. doi: 10.1038/s41396-022-01257-0
Zhao, S., Rogers, M. J., Cao, L., Ding, C., and He, J. (2021). Identification of reductive dehalogenases that mediate complete debromination of penta- and tetrabrominated diphenyl ethers in Dehalococcoides spp. Appl. Environ. Microbiol. 87:e0060221. doi: 10.1128/AEM.00602-21
Zhong, H., Lyu, H., Wang, Z., Tian, J., and Wu, Z. (2024). Application of dissimilatory iron-reducing bacteria for the remediation of soil and water polluted with chlorinated organic compounds: Progress, mechanisms, and directions. Chemosphere 352:141505. doi: 10.1016/j.chemosphere.2024.141505
Zhu, X., Wang, X., Li, N., Wang, Q., and Liao, C. (2022). Bioelectrochemical system for dehalogenation: a review. Environ. Pollut. 293:118519. doi: 10.1016/j.envpol.2021.118519
Zinder, S. H. (2016). “The genus dehalococcoides,” in Organohalide-Respiring Bacteria, eds. L. Adrian, and F. E. Löffler (Berlin, Heidelberg: Springer Berlin Heidelberg), 107–136. doi: 10.1007/978-3-662-49875-0_6
Keywords: OHRB, reductive dehalogenation, cobamides, salvage pathway, co-culture
Citation: Lu Y, Lu F, Zhang J, Tang Q, Yang D and Liu Y (2024) Understanding the sources, function, and irreplaceable role of cobamides in organohalide-respiring bacteria. Front. Microbiol. 15:1435674. doi: 10.3389/fmicb.2024.1435674
Received: 20 May 2024; Accepted: 25 June 2024;
Published: 30 July 2024.
Edited by:
Guofang Xu, National University of Singapore, SingaporeReviewed by:
Wenjing Qiao, Nanjing Agricultural University, ChinaCopyright © 2024 Lu, Lu, Zhang, Tang, Yang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dan Yang, eWFuZ19kYW4xOTY5QDE2My5jb20=; Yaqing Liu, eWFxaW5nLmxpdUBneHUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.