
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 November 2024
Sec. Aquatic Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1432042
 Yuhui Xie1,2†
Yuhui Xie1,2† Fenfa Li1†
Fenfa Li1† Qingyi Xie3
Qingyi Xie3 Fandong Kong3
Fandong Kong3 Yun Xu1
Yun Xu1 Qingyun Ma3
Qingyun Ma3 Wenqiang Wu1
Wenqiang Wu1 Dongyi Huang1
Dongyi Huang1 Xinqiang Xie4
Xinqiang Xie4 Shuangqing Zhou1,5*
Shuangqing Zhou1,5* Youxing Zhao3*
Youxing Zhao3* Xiaolong Huang1*
Xiaolong Huang1*Marine actinomycetes exhibit a high level of biodiversity and possess significant potential for the production of high-value secondary metabolites. During the course of investigation of marine actinobacteria from corals, two Saccharopolyspora strains, namely, HNM0983T and HNM0986T, were isolated from stony corals collected from the coastal area of Hainan Island. The 16S ribosomal RNA (rRNA) gene sequence analysis revealed that these two strains are putative novel taxa of the genus Saccharopolyspora. Whole-genome sequencing comparisons further confirmed the two strains as belonging to two novel Saccharopolyspora species, which can be distinguished phenotypically and chemically from their current closest phylogenetic relatives. Some genomic information of the genus Saccharopolyspora was compared for evaluating the production capacity of secondary metabolites. A total of 519 biosynthetic gene clusters (BGCs) from the genus Saccharopolyspora were used for analysis, and terpene BGCs were found to be widespread and most abundant in this genus. In addition, abundant novel BGCs in the genus Saccharopolyspora are not clustered with the known BGCs in the database, indicating that the metabolites of the genus Saccharopolyspora deserve further exploration. On the basis of these presented results, Saccharopolyspora montiporae sp. nov. (type strain = HNM0983T = CCTCC AA 2020014T = KCTC 49526T) and Saccharopolyspora galaxeae sp. nov. (type strain = HNM0986T = CCTCC AA 2020011T = KCTC 49524T) are proposed as the names for the new strains, respectively.
The genus Saccharopolyspora, a member of the family Pseudonocardiaceae, was first proposed in 1975 (Lacey and Goodfellow, 1975) and revised in 1989 (Korn-Wendisch et al., 1989). The genus Saccharopolyspora currently comprises 39 species with validly published and correct names1 (Sayed et al., 2020). The genus Saccharopolyspora has a wide distribution and is often found in some extreme environments, such as mangrove sediments (Suksaard et al., 2018), marine invertebrates (Pimentel-Elardo et al., 2008; Souza et al., 2017), deep-sea sediments (Jiang et al., 2016), hypersaline lakes (Lv et al., 2014; Xia et al., 2017), and deserts (Yang et al., 2018; Saygin et al., 2021).
The genus Saccharopolyspora has a high application potential and needs further exploration, especially in the field of novel or active natural product discovery. Some crucial drugs, agricultural antibiotics, and active compounds have been found in the genus Saccharopolyspora, such as erythromycin A, spinosyn A and D (Kim and Goodfellow, 2015), cebulantin (Moon et al., 2019), and compound KR21-0001A (Janthanom et al., 2024). More importantly, the majority of members of the genus are regarded as valuable sources of many secondary metabolites with diverse chemical classes, such as glycolipids, alkaloids, macrolides, peptides, oligosaccharides, and quinones, which possess different biological activities, including cytotoxic, insecticidal, and antimicrobial (Sayed et al., 2020). Recent genome-wide comparative analysis results further support the metabolic potential of Saccharopolyspora taxa (Saygin et al., 2021).
As a common marine organism, coral is widely distributed in the ocean. Corals provide a relatively stable living environment for marine microorganisms, and a large number of novel microorganisms have been isolated from corals in recent years, such as Saccharopolyspora coralli (Zhou et al., 2020), Nocardiopsis coralli (Li et al., 2021), and Alkalimarinus coralli (Li et al., 2023). In this study, strains HNM0983T and HNM0986T were isolated from the stony corals Montipora foliosa and Galaxea astreata, respectively, which were collected from the sea of Wenchang, Hainan, China. These two strains were identified as two novel species of the genus Saccharopolyspora by polyphasic analysis. In addition, we compared their genomic differences with other Saccharopolyspora strains. Comprehensive genome analyses showed that strains HNM0983T and HNM0986T have the potential for novel bioactive compounds.
The coral samples were collected from the coast of the eastern suburbs of Wenchang City, Hainan Province, China (110.88 E, 19.53 N). The collected coral samples were immediately placed in sterile sample bags and stored in the freezer (−80°C). The Coral Reef Protection Laboratory, School of Oceanography, Hainan University completed coral identification and classification. Coral samples were ground into powder and diluted with sterile seawater. The diluted coral homogenate was spread on Gause’s synthetic medium, which was prepared using 50% (v/v) seawater and supplemented with 75 mg of potassium dichromate per liter (Gao et al., 2012). All plates were incubated at 28°C for 4 weeks, and single colonies of actinomycetes were purified continuously on ATCC 172 medium (Hong et al., 2009). Purified cultures were stored in 20% (v/v) glycerol and frozen at −80°C.
PCR amplification of the 16S rRNA sequence was described in a previous study (Huang et al., 2012). Almost complete 16S rRNA sequences from strains HNM0983T (1,464 bp) and HNM0986T (1,502 bp) were compared with the database on the EzBioCloud website (Yoon et al., 2017a). The 16S rRNA phylogenetic tree was constructed using the neighbor-joining method (Saitou and Nei, 1987), the maximum-likelihood method (Felsenstein, 1981), and the maximum-parsimony method (Fitch, 1981) in MEGA 7.0 software, respectively (Kumar et al., 2016). The topologies of the 16S rRNA phylogenetic trees were evaluated with 1,000 iterations for bootstrap support (Felsenstein, 1985). The genomic DNA was extracted by using the Wizard® Genomic DNA Purification Kit (Promega Beijing Biotech Co., Ltd). All sequencing services were performed by Shanghai Majorbio Biopharm Technology Co., Ltd. (Shanghai, China). The Illumina HiSeq X Ten was used as a sequencing platform and assembled by SOAPdenovo (version 2.04; Luo et al., 2012). Genome quality assessment, genome circle map construction, and genome annotation were performed by the Bacterial and Viral Bioinformatics Resource Center (BV-BRC) server (Olson et al., 2023). The whole-genome phylogenomic tree was constructed on the Type Strain Genome Server (TYGS; Meier-Kolthoff and Göker, 2019).
ISP 1-ISP 7 agar (Shirling and Gottlieb, 1966) and Czapek agar (Kinderlerer, 1995) were used to test the state of growth, with all culture mediums supplemented with 16.5 g/L sea salt and cultured at 28°C for 3 weeks. The designation of colony color was determined by comparison with the American Inter-Society Color Council–National Bureau of Standards (ISCC–NBS) color charts (Kelly, 1964). NaCl tolerance (concentrations ranged from 0 to 25% with 1% intervals), pH tolerance (3.0–11.0, in intervals of 1.0 pH units), and growth temperature range (0°C, 4°C, 10°C, 28°C, 37°C, 40°C, and 45°C) were tested on International Streptomyces Project-2 (ISP 2) medium. Carbon and nitrogen source utilization tests were consistent with previous reports (Xie et al., 2020; Williams et al., 1983). Hydrolysis of starch, production of H2S and melanin, milk coagulation, and nitrate reduction were carried out as described previously (Goodfellow, 1971; Athalye et al., 1985a). To ensure the accuracy of the test results, the above tests were repeated three times.
For chemotaxonomic analyses, strains HNM0983T and HNM0986T were cultured in ATCC172 broth at 28°C for 7 days. Whole-cell hydrolysate determination was performed according to previous reports (Lechevalier and Lechevalier, 1970). Polar lipids were extracted and examined according to published procedures (Minnikin et al., 1984). Menaquinones were analyzed using high-performance liquid chromatography (HPLC; Collins et al., 1977; Tamaoka et al., 1983). Extraction and analysis of cellular fatty acids were performed by using the method of Athalye et al. (1985b).
The digital DNA–DNA hybridization (dDDH) analysis was performed using the DSMZ (German Collection of Microorganisms and Cell Cultures) Genome-to-Genome Distance Calculator platform (Meier-Kolthoff et al., 2013). Average nucleotide identity (ANI) values of the genomes of strains with closely related type strains were calculated using the ANI Calculator (Yoon et al., 2017b). Pan-genome analysis was performed using the Pan-Genome Analysis Web Server (PGAP; Chen et al., 2018). Protein clustering analysis was performed using OrthoVenn2 (Xu et al., 2019). Visual comparisons of the whole genomes of strains HNM0983T and HNM0986T and other most closely related type strains were performed using the BV-BRC server (Olson et al., 2023). The biosynthetic gene clusters (BGCs) were annotated using antiSMASH 7.0 (Blin et al., 2023). The similarity network of BGCs was constructed using BiG-SCAPE version 1.1.2, employing the cutoff threshold of 0.6 (Navarro-Muñoz et al., 2020), and visualized using Cytoscape version 3.10.2 (Otasek et al., 2019).
Strain HNM0983T had the highest 16S rRNA similarity with S. rosea IMMIB L-1070T (96.80%), Saccharopolyspora spinosa NRRL 18395T (96.75%), Saccharopolyspora phatthalungensis SR8.15T (96.75%), and Saccharopolyspora hattusasensis CR3506T (96.41%). Strain HNM0986T had the highest 16S rRNA similarity with Saccharopolyspora gloriosae DSM 45582T (97.85%), Saccharopolyspora gregorii NCIMB 12823T (97.70%), S. cebuensis SPE 10-1T (96.05%), and S. rosea IMMIB L-1070T (96.02%). Obviously, all of the aforementioned similarities are lower than the threshold suggested for species demarcation (98.7%; Chun et al., 2018). In the 16S rRNA phylogenetic tree (constructed by the neighbor-joining method), strain HNM0983T and S. rosea IMMIB L-1070T formed a distinct monophyletic lineage, while strain HNM0986T formed another distinct monophyletic lineage with S. gloriosae DSM 45582T and S. gregorii NCIMB 12823T (Figure 1). Similar 16S rRNA phylogenetic analysis results also appeared in the maximum-likelihood (Supplementary Figure S1) and maximum-parsimony (Supplementary Figure S2 phylogenetic trees). Obviously, based on the result of 16S rRNA phylogenies, strains HNM0983T and HNM0986T were found to belong to the genus Saccharopolyspora, and they were all different from the known type strains. Furthermore, the phylogenetic tree constructed by whole-genome sequences also obtained similar results. Based on the available genomic information for the genus Saccharopolyspora (a total of 22 type strains), strains HNM0983T, HNM0986T, S. gloriosae DSM 45582T, and S. erythraea DSM 40517T were most closely related at the genomic level (Figure 2). Overall genomic relatedness indices (OGRI) were used to further determine the relationship among all these Saccharopolyspora strains.
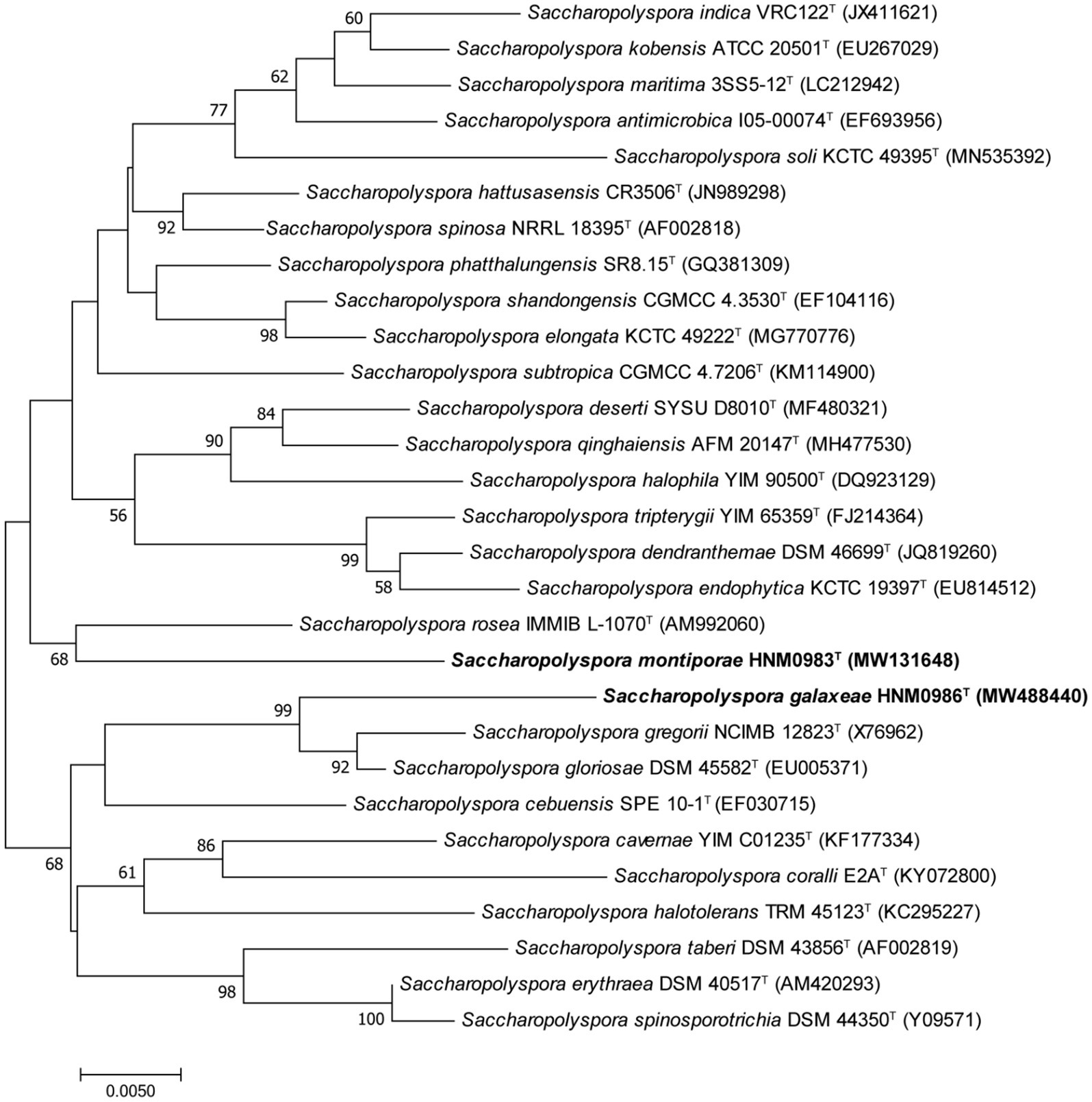
Figure 1. Neighbor-joining phylogenetic tree derived from 16S rRNA sequences, showing the relationships between HNM0983T, HNM0986T, and other type strains of the genus Saccharopolyspora. Only values above 50% are shown. Bar indicates 5 nucleotide substitutions per 1000 nucleotides.
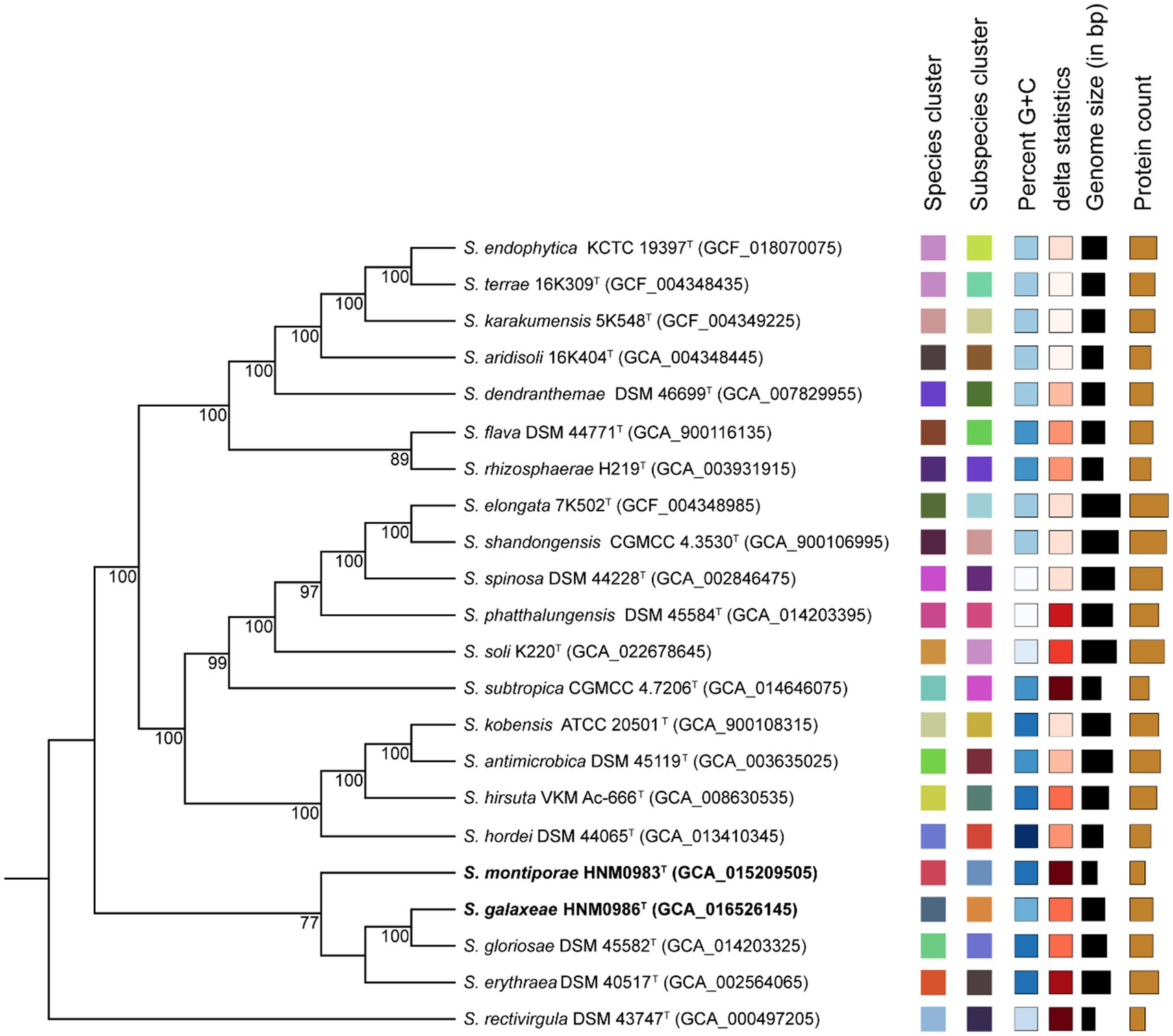
Figure 2. Genomic phylogenetic tree constructed by TYGS. Tree inferred with FastME 2.1.6.1 from Genome-Basic Local Alignment Search Tool (BLAST) Distance Phylogeny (GBDP) distances calculated from genome sequences. The branch lengths are scaled according to the GBDP distance formula d5. The numbers above branches are GBDP pseudo-bootstrap support values >60% from 100 replications, with an average branch support of 84.6%. Leaf labels with different colors indicate species and subspecies clusters.
For strain HNM0983T, dDDH values compared with other strains ranged from 20.4% (Saccharopolyspora rectivirgula DSM 43747T) to 22.8% (Saccharopolyspora antimicrobica DSM 45119T; Supplementary Table S1), which were significantly lower than the recommended thresholds of 70% for species demarcation (Goris et al., 2007). Similarly, ANI values varied from 77.2% (Saccharopolyspora aridisoli 16K404T, Saccharopolyspora dendranthemae DSM 46699T, and Saccharopolyspora rhizosphaerae H219T) to 79.0% (Saccharopolyspora hirsuta VKM Ac-666T; Supplementary Table S1), remaining well below the recommended cutoff value of 95.0–96.0% for species recognition (Richter and Rosselló-Móra, 2009). For strain HNM0986T, dDDH values ranged from 21.2% (S. dendranthemae DSM 46699T) to 24.6% (S. gloriosae DSM 45582T; Supplementary Table S1), and ANI values ranged from 76.6% (S. rectivirgula DSM 43747T) to 81.3% (S. gloriosae DSM 45582T). Independent dDDH and ANI tests confirmed that strains HNM0983T and HNM0986T are distinct from existing type strains. Consequently, strains HNM0983T and HNM0986T represent novel species of the genus Saccharopolyspora.
The novel strains HNM0983T and HNM0986T showed different growth states on a variety of media, but neither was able to grow on ISP 4 agar. The growth status of HNM0983T was superior to that of HNM0986T on the majority of the tested media, and the growth differences between the two strains are detailed in Supplementary Table S2. The optimal NaCl concentration of strain HNM0983T was 0–1% (w/v), but the maximum NaCl tolerance was 20%, the pH tolerance range was 5.0–10.0, and the optimal growth pH was 8.0–9.0. The growth temperature of HNM0983T ranged from 10°C to 37°C, and the optimal temperature was 28°C. Strain HNM0986T had the same growth temperature range as HNM0983T but was different in NaCl concentration tolerance and pH tolerance. The optimal NaCl concentration of strain HNM0986T was 0–7% (w/v), but the maximum NaCl tolerance was 15%, the pH tolerance range was 5.0–10.0, and the optimal growth pH was 7.0. In addition, there were some differences in the utilization of single carbon and nitrogen sources between these two strains, such as sucrose, raffinose, maltose, cellobiose, D-galactose, D-sorbitol, D-mannose, D-melibiose, L-asparagine, and L-arginine. Strains HNM0983T and HNM0986T can also be distinguished from their phylogenetic neighbors by biochemical reactions and degradability tests, the results of which are shown in Table 1.
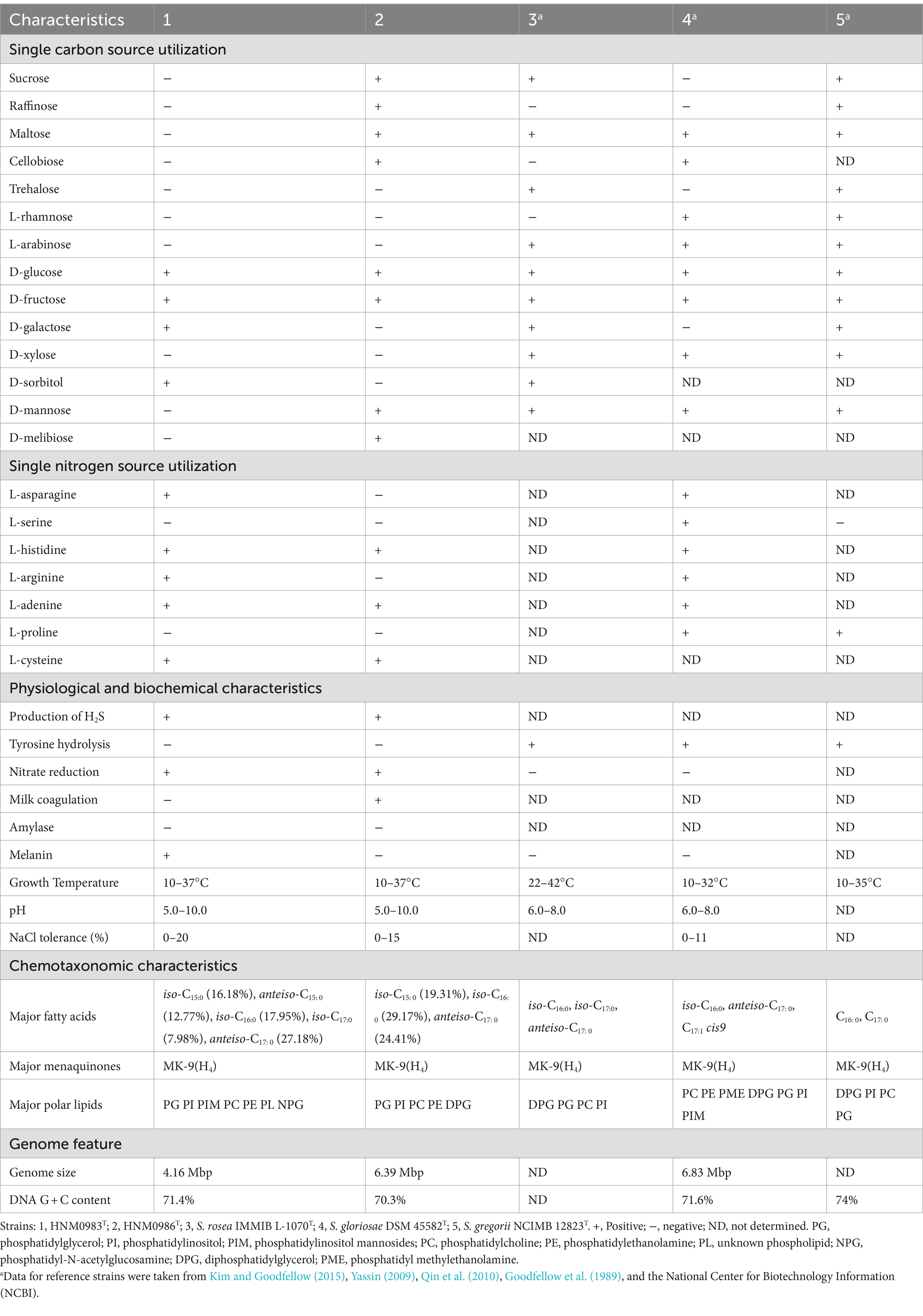
Table 1. Comparison of physiological properties and chemotaxonomic characteristics between two new strains and their closely related strains.
The chemical taxonomic analyses of strains HNM0983T and HNM0986T showed that they had characteristics of the genus Saccharopolyspora (Kim and Goodfellow, 2015). They all contained MK-9(H4) as the major menaquinone and meso-diaminopimelic acid as the diagnostic diamino acid. The main differences in the chemical classification of the genus Saccharopolyspora were focused on the composition of fatty acids and polar lipids. The fatty acid composition of HNM0983T, HNM0986T, and closely related strain species is shown in Table 1. Iso-C15:0 is a unique fatty acid component, only detected in HNM0983T and HNM0986T. The major polar lipids of strain HNM0983T contained phosphatidylglycerol, phosphatidylinositol, phosphatidylinositol mannosides, phosphatidylcholine, phosphatidylethanolamine, phosphatidyl-N-acetylglucosamine, and some unknown phospholipids (Supplementary Figure S3). The major polar lipids of strain HNM0986T were composed of phosphatidylinositol, phosphatidylcholine, phosphatidylglycerol, phosphatidylethanolamine, and diphosphatidylglycerol (Supplementary Figure S4). Phosphatidylcholine and phosphatidylglycerol were detected in all of the closely related strains, while diphosphatidylglycerol was detected in all closely related strains except strain HNM0983T (Table 1). In summary, HNM0983T and HNM0986T can be differentiated from closely related strains within the genus Saccharopolyspora through chemical classification.
Almost complete genomes of strains HNM0983T and HNM0986T were deposited in GenBank (the accession numbers JADEYC000000000 and JADDUE000000000, respectively). The sizes of the genomes of strains HNM0983T and HNM0986T were 4.16 and 6.39 Mbp, respectively. The G + C contents of strains HNM0983T and HNM0986T were 71.4 and 70.3%, respectively. The genome quality assessment and genome circular maps are shown in Supplementary Figure S5. The pan-genome analysis showed that with the addition of new genomes, the size of the pan-genome was further increased, indicating that the structure of this pan-genome is open type (Figure 3). In the open type of pan-genome, strains may contain some unique genes, so we compared the genomic composition of strains closely related to HNM0983T and HNM0986T. The clustering of proteins of five related strains is shown in Figure 4. A total of 1729 homologous proteins were annotated in these strains, and each strain also contained some unique protein-coding genes. It is worth noting that based on the visual genome map, a large number of gene rearrangements occur in the genus Saccharopolyspora (Figure 5), even in closely related strains, which is consistent with the previous report (Saygin et al., 2021).
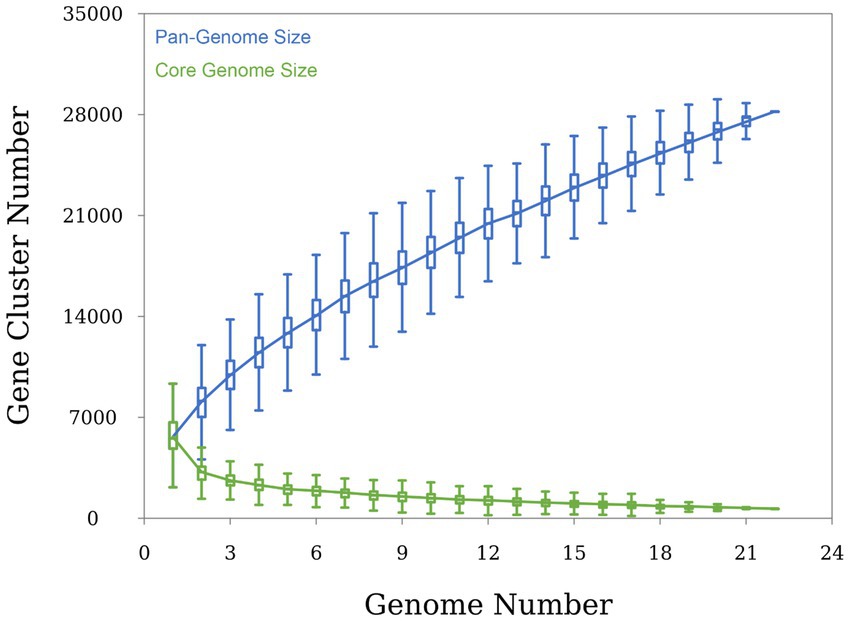
Figure 3. Pan-genome analysis of the genus Saccharopolyspora (conducted on 22 type strains of the genus Saccharopolyspora).
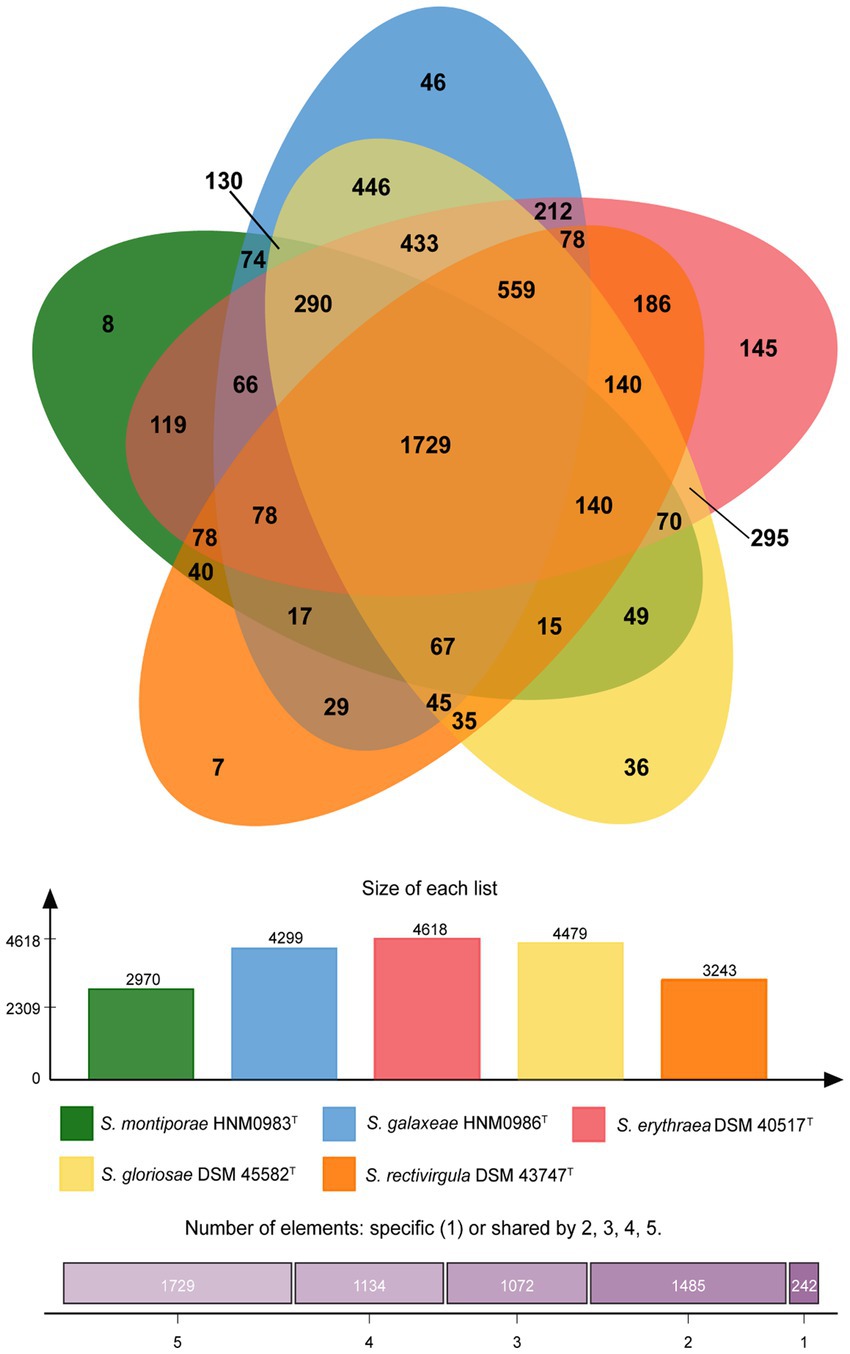
Figure 4. Protein cluster analysis of strains HNM0983T, HNM0986T, and three closely related strains of Saccharopolyspora.
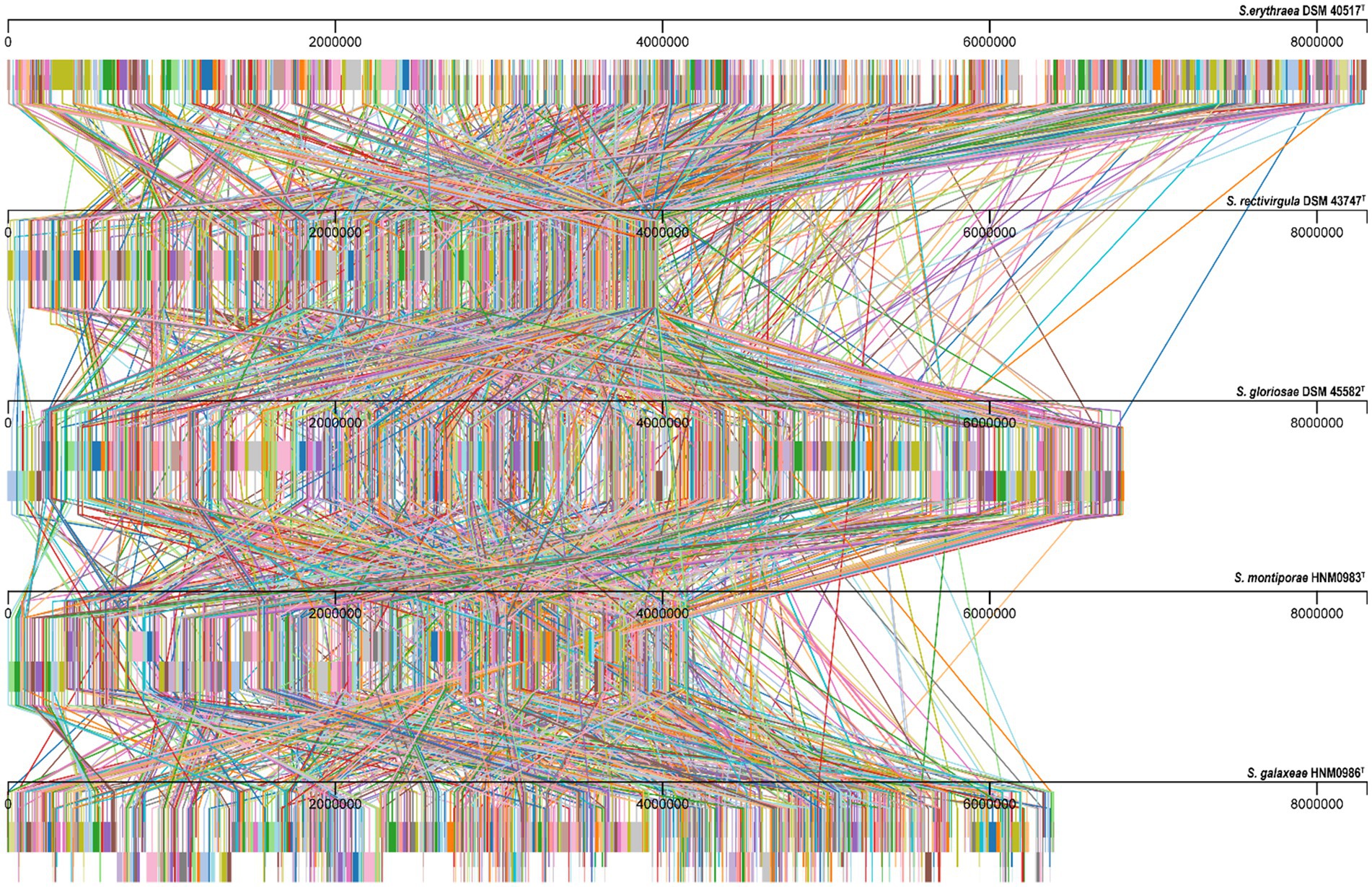
Figure 5. Alignment results of genome homologous regions of strains closely related to strains HNM0983T and HNM0986T. Homologous regions are connected by lines.
The marine environment has unfavorable factors such as high salinity, low temperature, and oligotrophic nutrition. Marine microorganisms may have some special metabolic pathways to enhance environmental resistance and interspecific competitiveness (Chen et al., 2022). The genome annotation of strains HNM0983T and HNM0986T showed a large number of genes related to habitat adaptation, such as heat shock proteins, cold shock proteins, betaine production, Na+ transport, heavy metal resistance, and transport (Supplementary Tables S3, S4). These genes provide important support for the homeostasis of the strain itself. In addition, secondary metabolites may also be important defensive weapons against potential threats.
The antiSMASH 7.0 analysis results showed that strain HNM0983T harbored a total of 10 BGCs for the biosynthesis of secondary metabolites, while HNM0986T had 26 BGCs (Supplementary Table S5). The annotation also showed that some gene clusters shared homology with BGCs of known active compounds, including cinnapeptin (Zhang and Seyedsayamdost, 2020), borregomycin (Chang and Brady, 2013), pyralomicin 1a (Kawamura et al., 1996), and ectoine (Chen et al., 2023). However, the lower gene cluster similarity indicates that these gene clusters may encode some new compounds. Furthermore, the composition of 519 BGCs from 22 strains of the genus Saccharopolyspora was analyzed by the BiG-SCAPE server. The results showed that terpene BGC was the most abundant in the genus Saccharopolyspora, and these strains widely had good terpene synthesis capacity (Figure 6). In general, marine bacteria are not the main contributors of natural terpene products, but multiple active terpenes have still been found in marine bacteria, especially in marine actinomycetes (Gozari et al., 2021). Due to the special environment of the ocean, the terpenoids from the marine bacteria may involve some special detoxification reactions, such as chlorination and bromination (Gozari et al., 2021). In addition, various types of polyketide synthase (PKS), non-ribosomal peptide synthetase (NRPS), and hybrid gene clusters were also widely distributed in Saccharopolyspora (Figure 6), and dozens of related products have been found in Saccharopolyspora (Sayed et al., 2020). A total of 395 BGCs from the Saccharopolyspora were found in the BGCs network. This finding showed that there remains a significant number of novel BGCs within this genus (Figure 6). Meanwhile, among the 2,735 reference BGCs in the MIBiG database (Terlouw et al., 2023), 534 of them were detected. Interestingly, the majority of the reference BGCs were only associated with a small number of BGCs from the Saccharopolyspora (Figure 6). The BGCs from the genus Saccharopolyspora tended to cluster closely together in the BGCs network, possibly indicating that these BGCs were of a conservative type in the evolution of this genus.
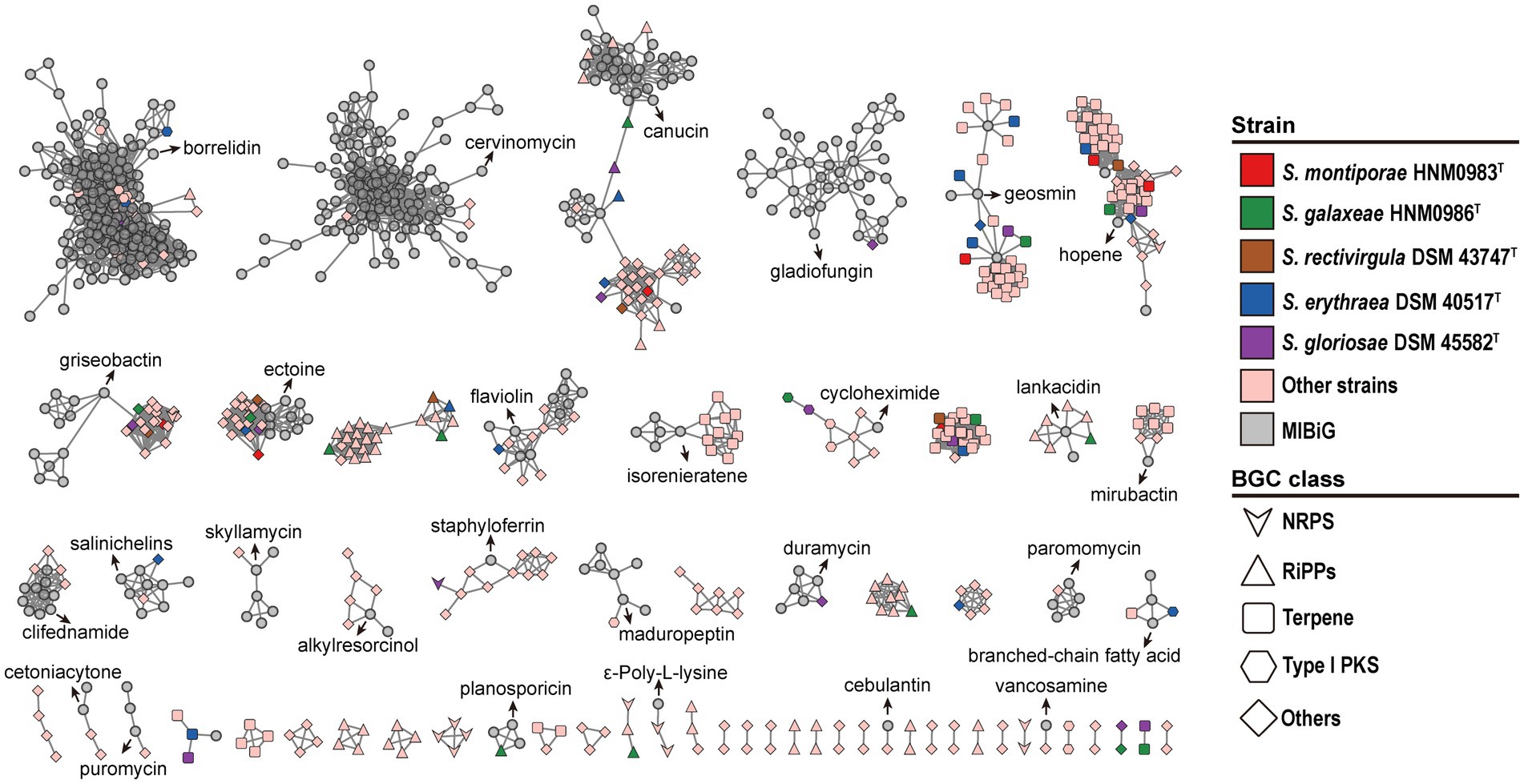
Figure 6. Sequence similarity network of all BGCs in 22 Saccharopolyspora strains. Each node represents one BGC, and the gray symbols represent reference BGCs in the MIBiG database (Terlouw et al., 2023).
With the misuse of antibiotics, the problem of drug-resistant pathogens is becoming more and more serious. The pressing need for new bioactive molecules has led researchers to focus on novel microbes in extreme environments (Carroll et al., 2023). Although the genus Saccharopolyspora has received less attention, some active molecules from strains of this genus have been discovered and reported. The strains HNM0983T and HNM0986T isolated in this study further expanded the members of the genus Saccharopolyspora and provided new research objects for exploring metabolites from the genus Saccharopolyspora.
Marine actinomycetes associated with stony corals have been highlighted as a potential hotspot for new bioactive metabolites (Siro et al., 2022). Considering the fact that a new species contains novel BGCs, encoding the novel bioactive metabolites (Shi et al., 2022), these new Saccharopolyspora species isolated from stony corals in the present study show certain potential to produce novel bioactive substances for medical or agricultural use. From the combination of phylogenetic, genomic, phenotypic, and chemotaxonomic characteristics presented, we believe that two strains should be considered as novel species of the genus Saccharopolyspora, for which Saccharopolyspora montiporae sp. nov. HNM0983T and Saccharopolyspora galaxeae sp. nov. HNM0986T are proposed.
Saccharopolyspora montiporae (mo.nti.poʼrae. N.L. gen.n. montiporae of the coral Montipora foliosa from which the type strain was isolated) is a novel Gram-stain-positive, non-motile, aerobic actinobacterium that forms well-developed and extensively branched substrate mycelium. Good growth occurs on ISP 2, moderate growth on ISP 3 and ISP 7, and poor growth on ISP 1, ISP 5, ISP 6, and Czapek agar. Growth occurs at pH 5.0–10.0 (optimum 8.0–9.0), 10–37°C (optimum 28°C), and 0–20% (w/v) NaCl (optimum 0–1%). Catalase, nitrate reductase, melanin, and H2S production tests are all positive, while tyrosine hydrolysis, milk coagulation, and amylase tests are all negative. D-glucose, D-fructose, D-galactose, and D-sorbitol are utilized as sole carbon sources but sucrose, raffinose, maltose, cellobiose, trehalose, L-rhamnose, L-arabinose, D-xylose, D-mannose, and D-melibiose are not. L-asparagine, L-histidine, L-arginine, L-adenine, and L-cysteine are used as sole nitrogen sources but L-serine and L-proline are not. The major menaquinone is MK-9(H4). Major polar lipids include phosphatidylglycerol, phosphatidylinositol, phosphatidylinositol mannosides, phosphatidylcholine, phosphatidylethanolamine, phosphatidyl-N-acetylglucosamine, and three unknown phospholipids. The cell-wall diamino acid is meso-diaminopimelic acid. Major fatty acids are iso-C15:0, iso-C16:0, iso-C17:0 and anteiso-C17:0. The type strain is HNM0983T (=CCTCC AA 2020014T = KCTC 49526T). The genome size of the strain is 4.16 Mbp, and the G + C content is 71.4%. The GenBank accession numbers for the 16S rRNA gene and the whole-genome sequence are MW131648 and JADEYC000000000, respectively.
Saccharopolyspora galaxeae (ga. laʼ xe. ae. N.L. gen.n. galaxeae of the coral Galaxea astreata from which the type strain was isolated) is a novel Gram-stain-positive, non-motile, aerobic actinobacterium that forms well-developed and extensively branched substrate mycelium. Good growth occurs on ISP 2, ATCC172, and Czapek agar, moderate growth on ISP 3, and poor growth on ISP 7. Growth occurs at pH 5.0–10.0 (optimum 7.0), 10–37°C (optimum 28°C), and 0–15% (w/v) NaCl (optimum 0–7%). Catalase, nitrate reductase, milk coagulation, and H2S production tests are all positive, while amylase, tyrosine hydrolysis, and melanin tests are all negative. Sucrose, raffinose, maltose, cellobiose, D-glucose, D-fructose, D-mannose, and D-melibiose are utilized as sole carbon sources but trehalose, L-rhamnose, L-arabinose, D-galactose, D-xylose, and D-sorbitol are not. L-histidine, L-adenine, and L-cysteine are used as sole nitrogen sources but L-asparagine, L-serine, L-arginine, and L-proline are not. The major menaquinone is MK-9(H4). The major polar lipids include phosphatidylglycerol, phosphatidylinositol, phosphatidylcholine phosphatidylethanolamine, and diphosphatidylglycerol. Cell-wall diamino acid is meso-diaminopimelic acid. The major fatty acids are iso-C15:0, iso-C16:0, and anteiso-C17:0. The type strain is HNM0986T (=CCTCC AA 2020011T = KCTC 49524T). The genome size of the strain is 6.39 Mbp, and the G + C content is 70.3%. The GenBank accession numbers for the 16S rRNA gene and the whole-genome sequence are MW488440 and JADDUE000000000, respectively.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: JADEYC000000000 JADDUE000000000.
YXi: Writing – original draft, Writing – review & editing. FL: Writing – original draft. QX: Writing – original draft. FK: Writing – original draft. YXu: Writing – original draft. QM: Writing – original draft. WW: Writing – original draft. DH: Writing – original draft. XX: Writing – original draft. SZ: Writing – original draft, Writing – review & editing. YZ: Writing – review & editing. XH: Conceptualization, Writing – original draft, Writing – review & editing, Funding acquisition, Methodology.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Guangxi Natural Science Foundation of China (2021GXNSFAA196044), the key research and development funding program of Hainan province (ZDYF2022XDNY171), the Financial Fund of the Ministry of Agriculture and Rural Affairs, People’s Republic of China (NFZX2024), and the Project by the Department of Science and Technology of Guangdong Province (2019QN01N107).
We are grateful to Xiubao Li (College of Oceanography, Hainan University, Hainan, China) for his devotion to coral identification.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1432042/full#supplementary-material
Athalye, M., Goodfellow, M., Lacey, J., and White, R. P. (1985a). Numerical classification of Actinomadura and Nocardiopsis. Int. J. Syst. Evol. Microbiol. 35, 86–98. doi: 10.1099/00207713-35-1-86
Athalye, M., Noble, W. C., and Minnikin, D. E. (1985b). Analysis of cellular fatty acids by gas chromatography as a tool in the identification of medically important coryneform bacteria. J. Appl. Bacteriol. 58, 507–512. doi: 10.1111/j.1365-2672.1985.tb01491.x
Blin, K., Shaw, S., Augustijn, H. E., Reitz, Z. L., Biermann, F., Alanjary, M., et al. (2023). antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 51, W46–W50. doi: 10.1093/nar/gkad344
Carroll, A. R., Copp, B. R., Davis, R. A., Keyzers, R. A., and Prinsep, M. R. (2023). Marine natural products. Nat. Prod. Rep. 40, 275–325. doi: 10.1039/D2NP00083K
Chang, F. Y., and Brady, S. F. (2013). Discovery of indolotryptoline antiproliferative agents by homology-guided metagenomic screening. Natl. Acad. Sci. U S A. 110, 2478–2483. doi: 10.1073/pnas.1218073110
Chen, Y., Pan, T., Chai, G., and Li, Z. (2022). Complete genome of Mycetocola spongiae MSC19T isolated from deep-sea sponge Cacospongia mycofijiensis indicates the adaptation to deep-sea environment and sponge-microbe symbioses. Mar. Genomics 63:100955. doi: 10.1016/j.margen.2022.100955
Chen, J., Qiao, D., Yuan, T., Feng, Y., Zhang, P., Wang, X., et al. (2023). Biotechnological production of ectoine: current status and prospects. Folia Microbiol. (Praha). 69, 247–258. doi: 10.1007/s12223-023-01105-4
Chen, X., Zhang, Y., Zhang, Z., Zhao, Y., Sun, C., Yang, M., et al. (2018). PGAweb: a web server for bacterial Pan-genome analysis. Front. Microbiol. 9:1910. doi: 10.3389/fmicb.2018.01910
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., da Costa, M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
Collins, M. D., Pirouz, T., Goodfellow, M., and Minnikin, D. E. (1977). Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100, 221–230. doi: 10.1099/00221287-100-2-221
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Felsenstein, J. (1985). Phylogenies and the comparative method. Am. Nat. 125, 1–15. doi: 10.1086/284325
Fitch, W. M. (1981). The fourteenth annual numerical taxonomy conference. Syst. Zool. 30, 81–83. doi: 10.2307/2992306
Gao, X., Lu, Y., Xing, Y., Ma, Y., Lu, J., Bao, W., et al. (2012). A novel anticancer and antifungus phenazine derivative from a marine actinomycete BM-17. Microbiol. Res. 167, 616–622. doi: 10.1016/j.micres.2012.02.008
Goodfellow, M. (1971). Numerical taxonomy of some nocardioform bacteria. J. Gen. Microbiol. 69, 33–80. doi: 10.1099/00221287-69-1-33
Goodfellow, M., Lacey, J., Athalye, M., Embley, T. M., and Bowen, T. (1989). Saccharopolyspora gregorii and Saccharopolyspora hordei: two new actinomycete species from fodder. Microbiology 135, 2125–2139. doi: 10.1099/00221287-135-8-2125
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Gozari, M., Alborz, M., El-Seedi, H. R., and Jassbi, A. R. (2021). Chemistry, biosynthesis and biological activity of terpenoids and meroterpenoids in bacteria and fungi isolated from different marine habitats. Eur. J. Med. Chem. 210:112957. doi: 10.1016/j.ejmech.2020.112957
Hong, K., Gao, A., Xie, Q., Gao, H., Zhuang, L., and Lin, H. (2009). Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 7, 24–44. doi: 10.3390/md7010024
Huang, X. L., Zhuang, L., Lin, H. P., Li, J., Goodfellow, M., Hong, K., et al. (2012). Isolation and bioactivity of endophytic filamentous actinobacteria from tropical medicinal plants. Afr. J. Biotechnol. 11, 9855–9864. doi: 10.5897/AJB11.3839
Janthanom, R., Kikuchi, Y., Kanto, H., Hirose, T., Tahara, A., Ishii, T., et al. (2024). A new analog of dihydroxybenzoic acid from Saccharopolyspora sp. KR21-0001. Beilstein J. Org. Chem. 20, 497–503. doi: 10.3762/bjoc.20.44
Jiang, Y., Wei, X., Chen, X., Jiang, Y., Xue, Q., Lai, H., et al. (2016). Saccharopolyspora griseoalba sp. nov., a novel actinomycete isolated from the Dead Sea. Antonie Van Leeuwenhoek 109, 1635–1641. doi: 10.1007/s10482-016-0763-9
Kawamura, N., Sawa, R., Takahashi, Y., Sawa, T., Naganawa, H., and Takeuchi, T. (1996). Pyralomicins, novel antibiotics from Microtetraspora spiralis. III. Biosynthesis of pyralomicin 1a. J. Antibiot. (Tokyo) 49, 657–660. doi: 10.7164/antibiotics.49.657
Kelly, K. L. (1964). Color-name charts illustrated with centroid colors. Chicago: Inter-Society Color Council-National Bureau of Standards.
Kim, S. B., and Goodfellow, M. (2015). “Saccharopolyspora,” in The Bergey’s Manual of Systematics of Archaea and Bacteria. ed. W. B. Whitman (Hoboken: John Wiley and Sons), 1–30.
Kinderlerer, J. L. (1995). Czapek casein 50% glucose (CZC50G): a new medium for the identification of foodborne Chrysosporium spp. Lett. Appl. Microbiol. 21, 131–136. doi: 10.1111/j.1472-765X.1995.tb01024.x
Korn-Wendisch, F., Kempf, A., Grund, E., Kroppenstedt, R. M., and Kutzner, H. J. (1989). Transfer of Faenia rectivirgula Kurup and agre 1983 to the genus Saccharopolyspora Lacey and Goodfellow 1975, elevation of Saccharopolyspora hirsuta subsp. taberi Labeda 1987 to species level, and emended description of the genus Saccharopolyspora. Int. J. Syst. Evol. Microbiol. 39, 430–441. doi: 10.1099/00207713-39-4-430
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lacey, J., and Goodfellow, M. (1975). A novel actinomycete from sugar-cane bagasse: Saccharopolyspora hirsuta gen. Et. Sp. nov. J. Gen. Microbiol. 88, 75–85. doi: 10.1099/00221287-88-1-75
Lechevalier, M. P., and Lechevalier, H. (1970). Chemical composition as a criterion in the classification of aerobic actinomycetes. Int. J. Syst. Evol. Microbiol. 20, 435–443. doi: 10.1128/jb.94.4.875-883.1967
Li, Q., Liu, C., Zhang, S., and Li, J. (2023). Description of Alkalimarinus coralli sp. nov., isolated from tissue of the coral Pocillopora damicornis. Int. J. Syst. Evol. Microbiol. 73:722. doi: 10.1099/ijsem.0.005722
Li, F., Xie, Q., Zhou, S., Kong, F., Xu, Y., Ma, Q., et al. (2021). Nocardiopsis coralli sp. nov. a novel actinobacterium isolated from the coral Galaxea astreata. Int. J. Syst. Evol. Microbiol. 71:004817. doi: 10.1099/ijsem.0.004817
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 1:18. doi: 10.1186/2047-217X-1-18
Lv, L. L., Zhang, Y. F., Xia, Z. F., Zhang, J. J., and Zhang, L. L. (2014). Saccharopolyspora halotolerans sp. nov., a halophilic actinomycete isolated from a hypersaline lake. Int. J. Syst. Evol. Microbiol. 64, 3532–3537. doi: 10.1099/ijs.0.063792-0
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf. 14:60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. doi: 10.1038/s41467-019-10210-3
Minnikin, D. E., O’donnell, A. G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., et al. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241. doi: 10.1016/0167-7012(84)90018-6
Moon, K., Xu, F., and Seyedsayamdost, M. R. (2019). Cebulantin, a cryptic Lanthipeptide antibiotic uncovered using bioactivity-coupled HiTES. Angew. Chem. Int. Ed. Eng. 58, 5973–5977. doi: 10.1002/anie.201901342
Navarro-Muñoz, J. C., Selem-Mojica, N., Mullowney, M. W., Kautsar, S. A., Tryon, J. H., Parkinson, E. I., et al. (2020). A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 16, 60–68. doi: 10.1038/s41589-019-0400-9
Olson, R. D., Assaf, R., Brettin, T., Conrad, N., Cucinell, C., Davis, J. J., et al. (2023). Introducing the bacterial and viral bioinformatics resource center (BV-BRC): a resource combining PATRIC. IRD and ViPR. Nucleic. Acids Res. 51, D678–D689. doi: 10.1093/nar/gkac1003
Otasek, D., Morris, J. H., Bouças, J., Pico, A. R., and Demchak, B. (2019). Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 20:185. doi: 10.1186/s13059-019-1758-4
Pimentel-Elardo, S. M., Tiro, L. P., Grozdanov, L., and Hentschel, U. (2008). Saccharopolyspora cebuensis sp. nov., a novel actinomycete isolated from a Philippine sponge (Porifera). Int. J. Syst. Evol. Microbiol. 58, 628–632. doi: 10.1099/ijs.0.64971-0
Qin, S., Chen, H. H., Klenk, H. P., Kim, C. J., Xu, L. H., and Li, W. J. (2010). Saccharopolyspora gloriosae sp. nov., an endophytic actinomycete isolated from the stem of Gloriosa superba L. Int. J. Syst. Evol. Microbiol. 60, 1147–1151. doi: 10.1099/ijs.0.015792-0
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 106, 19126–19131. doi: 10.1073/pnas.0906412106
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sayed, A. M., Abdel-Wahab, N. M., Hassan, H. M., and Abdelmohsen, U. R. (2020). Saccharopolyspora: an underexplored source for bioactive natural products. J. Appl. Microbiol. 128, 314–329. doi: 10.1111/jam.14360
Saygin, H., Ay, H., Guven, K., Inan-bektas, K., Cetin, D., and Sahin, N. (2021). Saccharopolyspora karakumensis sp. nov., Saccharopolyspora elongata sp. nov., Saccharopolyspora aridisoli sp. nov., Saccharopolyspora terrae sp. nov. and their biotechnological potential revealed by genome analysis. Syst. Appl. Microbiol. 44:126270. doi: 10.1016/j.syapm.2021.126270
Shi, S., Cui, L., Zhang, K., Zeng, Q., Li, Q., Ma, L., et al. (2022). Streptomyces marincola sp. nov., a novel marine actinomycete, and its biosynthetic potential of bioactive natural products. Front. Microbiol. 13:860308. doi: 10.3389/fmicb.2022.860308
Shirling, E. T., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Evol. Microbiol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Siro, G., Pipite, A., Christi, K., Srinivasan, S., and Subramani, R. (2022). Marine actinomycetes associated with stony corals: a potential hotspot for specialized metabolites. Microorganisms 10:1349. doi: 10.3390/microorganisms10071349
Souza, D. T., Silva, F. S. P. D., Silva, L. J. D., Crevelin, E. J., Moraes, L. A. B., Zucchi, T. D., et al. (2017). Saccharopolyspora spongiae sp. nov., a novel actinomycete isolated from the marine sponge Scopalina ruetzleri (Wiedenmayer, 1977). Int. J. Syst. Evol. Microbiol. 67, 2019–2025. doi: 10.1099/ijsem.0.001912
Suksaard, P., Srisuk, N., and Duangmal, K. (2018). Saccharopolyspora maritima sp. nov., an actinomycete isolated from mangrove sediment. Int. J. Syst. Evol. Microbiol. 68, 3022–3027. doi: 10.1099/ijsem.0.002941
Tamaoka, J., Katayama-Fujimura, Y., and Kuraishi, H. (1983). Analysis of bacterial menaquinone mixtures by high performance liquid chromatography. J. Appl. Bacteriol. 54, 31–36. doi: 10.1111/j.1365-2672.1983.tb01297.x
Terlouw, B. R., Blin, K., Navarro-Muñoz, J. C., Avalon, N. E., Chevrette, M. G., Egbert, S., et al. (2023). MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Res. 51, D603–D610. doi: 10.1093/nar/gkac1049
Williams, S. T., Goodfellow, M., Alderson, G., Wellington, E. M., Sneath, P. H., and Sackin, M. J. (1983). Numerical classification of Streptomyces and related genera. J. Gen. Microbiol. 129, 1743–1813. doi: 10.1099/00221287-129-6-1743
Xia, Z. F., Luo, X. X., Wan, C. X., and Zhang, L. L. (2017). Saccharopolyspora aidingensis sp. nov., an actinomycete isolated from a salt lake. Int. J. Syst. Evol. Microbiol. 67, 687–691. doi: 10.1099/ijsem.0.001696
Xie, Y., Zhou, S., Xu, Y., Wu, W., Xia, W., Zhang, R., et al. (2020). Gordonia mangrovi sp. nov., a novel actinobacterium isolated from mangrove soil in Hainan. Int. J. Syst. Evol. Microbiol. 70, 4537–4543. doi: 10.1099/ijsem.0.004310
Xu, L., Dong, Z., Fang, L., Luo, Y., Wei, Z., Guo, H., et al. (2019). OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 47, W52–W58. doi: 10.1093/nar/gkz333
Yang, Z. W., Salam, N., Asem, M. D., Fang, B. Z., Lan, L., Xiao, M., et al. (2018). Saccharopolyspora deserti sp. nov., a novel halotolerant actinobacterium isolated from a desert. Int. J. Syst. Evol. 68, 860–864. doi: 10.1099/ijsem.0.002598
Yassin, A. F. (2009). Saccharopolyspora rosea sp. nov., isolated from a patient with bronchial carcinoma. Int. J. Syst. Evol. Microbiol. 59, 1148–1152. doi: 10.1099/ijs.0.005249-0
Yoon, S., Ha, S., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017a). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Yoon, S. H., Ha, S. M., Lim, J. M., Kwon, S. J., and Chun, J. (2017b). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Zhang, C., and Seyedsayamdost, M. R. (2020). Discovery of a cryptic Depsipeptide from Streptomyces ghanaensis via MALDI-MS-guided high-throughput elicitor screening. Angew. Chem. Int. Ed. Eng. 59, 23005–23009. doi: 10.1002/anie.202009611
Keywords: marine actinobacterium, Saccharopolyspora, polyphasic taxonomy, comparative genomics, coral commensal microbes
Citation: Xie Y, Li F, Xie Q, Kong F, Xu Y, Ma Q, Wu W, Huang D, Xie X, Zhou S, Zhao Y and Huang X (2024) Comprehensive genome analysis of two novel Saccharopolyspora strains—Saccharopolyspora montiporae sp. nov. and Saccharopolyspora galaxeae sp. nov. isolated from stony corals in Hainan. Front. Microbiol. 15:1432042. doi: 10.3389/fmicb.2024.1432042
Received: 14 May 2024; Accepted: 21 October 2024;
Published: 13 November 2024.
Edited by:
Michael Rappe, University of Hawaii at Manoa, United StatesReviewed by:
R. Anburaj, National College, Tiruchirappalli, IndiaCopyright © 2024 Xie, Li, Xie, Kong, Xu, Ma, Wu, Huang, Xie, Zhou, Zhao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolong Huang, aHVhbmd4aWFsb25nQGhhaW5hbnUuZWR1LmNu; Shuangqing Zhou, enNxMjAyMEBnbG1jLmVkdS5jbg==; Youxing Zhao, emhhb3lvdXhpbmdAaXRiYi5vcmcuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.