- Jilin Provincial Key Laboratory of Plant Resource Science and Green Production, Jilin Normal University, Siping, China
Introduction: Scleroderma bovista can form symbiotic ectomycorrhizal fungi with hazel roots. The mechanism through which S. bovista promotes hazelnut growth remains unclear.
Methods: This study aimed to evaluate the effect of ectomycorrhizal fungus S. bovista on the growth and development of hazel roots and gene expression changes through comparative transcriptome analysis.
Results: After inoculation with S. bovista, the fungus symbiotically formed ectomycorrhiza with hazel roots. The fresh weights of the aboveground and underground parts of My treatment (inoculated with S. bovista and formed mycorrhiza) were much higher than those of the control, respectively. The length, project area, surface area, volume, forks, and diameter of the inoculated seedlings root were 1.13 to 2.48 times higher than those of the control. In the paired comparison, 3,265 upregulated and 1,916 downregulated genes were identified. The most significantly enriched Gene Ontology term for the upregulated Differentially Expressed Genes was GO:0005215 (transporter activity). Immunohistochemical analysis suggested that the expression levels of auxin and Auxin Response Factor9 were significantly increased by S. bovista after the formation of mycorrhizal fungi in hazelnut root tips.
Discussion: These results indicate that genes related to auxin biosynthesis, transport and signaling, and transport of nutrients may contribute to root development regulation in hazel ectomycorrhiza.
1 Introduction
Mycorrhizal fungi are highly evolved mutualistic symbionts formed between soil fungi and plant roots. In this mutually beneficial symbiosis, the host plant absorbs mineral nutrients from outside the root system through symbiotic fungal hyphae, and the fungi assimilate photosynthetic carbon through the host plant (Harley, 1988; Cheng et al., 2021; Zou et al., 2021). Approximately 5 billion tons of photosynthetic products are fixed in the soil yearly through mycorrhizal fungi worldwide, making it significant in the carbon and nitrogen balance of the entire ecosystem (Jiang et al., 2017). The role of mycorrhizal symbionts in improving plant uptake and accumulation of soil mineral nutrients, promoting plant resistance to drought, waterlogging, salt, disease, and heavy metal stress in ecosystems has been recognized. The families and genera of important woody plants with ectomycorrhizal fungi include Betulaceae, Rhamnaceae, Rosaceae, Casuarinaceae, and Dipterocarpaceae (Corrales et al., 2022). Ectomycorrhizal fungi have been found in plants such as Alnus, Betula, Carpinus, Ostrya, Ostryopsis, and Corylus in the Betulaceae. Hazelnut is a species of the Corylus genus in the family Betulaceae and is one of the world’s four major nuts. The hazelnut industry is significant in supporting the economic development of Northeast China, Shandong and Hebei mountainous areas (Liu et al., 2014). Although the existence of ectomycorrhizal fungi in hazelnuts has been confirmed, studies on hazelnut mycorrhizal fungi in China are lacking, and there is no corresponding microbial agent for production practices, which is not conductive to hazel industrial development.
Hazelnut mycorrhizal fungi in Northeast China were recently discovered (Cheng et al., 2023). Though inoculation of hazel mycorrhiza samples and repeatedly colonies culture, a pure colony of Scleroderma bovista was obtained. Microbial diversity analysis of the rhizosphere soil, paraffin sectioning, sequencing, and sequence comparison showed that S. bovista can form symbiotic ectomycorrhizal fungi with hazel roots. Outdoor potted cultivation of hazel seedlings and inoculation with S. bovista and were performed, and it was found that S. bovista can strongly promote the growth of the aboveground and underground parts of hazel seedlings (Cheng et al., 2023). Based on these results, it is assumed that the natural distribution of S. bovista and hazel production areas in China overlap. It is speculated that S. bovista can form a stable symbiotic relationship with the hazel root system in production areas and may be vital in promoting root development, plant growth, and improving stress tolerance. S. bovista is an edible medicinal fungus with hemostatic and detoxifying effects and a high medicinal value. Its tender fruiting body is a delicious mushroom. S. bovista needs to form symbiotic relationships with trees to form fruiting bodies (Zhang et al., 2021). Therefore, establishing a symbiotic relationship between S. bovista and hazel trees benefits the growth and development of hazel trees, the development of a hazel understory economy, and improves cultivation. Thus, S. bovista is a potentially essential ectomycorrhizal fungus urgently required for developing the hazel industry.
The formation of ectomycorrhiza is a common ecological phenomenon in nature. Ectomycorrhizal fungi hyphae can extend into the root cortex cells to form a fungal mesh, while spreading on the root surface to form a hyphal sheath, replacing the role of root hairs and absorbing nutrients and water. This structure not only improves the water and nutrient metabolism of plants, but also increases the absorption of trace elements such as copper and zinc by the host plant roots, decomposes organic matter in the soil, accelerates soil nutrient cycling, improves soil physicochemical properties, and enhances the effectiveness of nutrients in the soil (Kuyper and Suz, 2023). Ectomycorrhizal fungi can also promote plant growth by enhancing the absorption of nutrients by trees, which is one of the important mechanisms for promoting plant growth. Especially in low soil phosphorus concentrations, ectomycorrhizal fungi enable plants to more effectively absorb phosphorus from the soil, thereby promoting plant growth (Almeida et al., 2023). However, the mechanism through which S. bovista promotes hazelnut growth remains unclear. Therefore, this study aimed to evaluate the effects of the ectomycorrhizal fungus S. bovista on the growth and development of hazelnuts and gene expression changes through comparative transcriptome analysis.
2 Materials and methods
2.1 Scleroderma bovista inoculation and measurement of root growth indicators
In 2023, seeds of the hazel (Corylus heterophylla × Corulys avellana) cultivar “Yuzhui” and our self-isolated and purified strain of S. bovista were chosen for this study (Cheng et al., 2023). Hazelnut seeds were first stratified in a 4°C refrigerator for 2 months and soaked overnight with 500 mg/L GA3 (Gibberellin A3) to further break dormancy. Seed accelerating germination was performed at 25°C in an illuminating incubator for 2 weeks, and 50 seeds with a root length of 3–5 cm were used for sowing in early May. Hazel seeds were sown in plastic pots containing 6 kg of sterilized humus soil, with 25 pots of the control (inoculated with autoclaved S. bovista) and 25 of My treatments (inoculated with S. bovista and formed mycorrhiza) in May. The sowing depth was 5 cm. In My treatment, S. bovista was cultured in dishes containing MMN medium (Modified Melin-Norkrans Medium) according to the description of a previous reference, and 10 S. bovista cubes with a side length of 1.0 cm were inoculated into each pot (Cheng et al., 2023). Potted seedlings were cultivated outdoors at Jilin Normal University (43°9′18″N, 124°20′11″E). Among the 25 treated seedlings, eight were used to measure growth indicators, and six to extract root RNA. Subsequently, 4 months after sowing, the seedlings were carefully removed from plastic pots in September. After removing the soil adhering to the roots, a WinRhino root analysis system (Regent Instruments Inc., Canada) was used to measure root indicator differences between the control and My treatments. Paraffin sections and mycorrhiza staining were performed according to a previously described method (Cheng et al., 2023).
2.2 RNA sequencing
Six seedlings each in the control and My treatments were used to extract RNA in late July. At this moment, the growth-promoting effect of ectomycorrhizal fungi was already evident. The roots of control and My treatments were sampled, and six digital gene expression libraries were constructed to investigate gene expression differences, including Ck-1, Ck-2, Ck-3, My-1, My-2, and My-3. There were two treatments (Ck and S. bovista inoculation) in our experiments, three biological replicates in each treatment, and two seedlings in each biological replicate. The RNA EasySpin Isolation System (AiDLab Biotech, Beijing, China) was used to extract total RNA from fresh root samples following the product manual. Post-processing and sequencing of the RNA samples were performed according to our previous reports (Cheng et al., 2023). Sequencing was performed on an Illumina NovaSeq 6000 platform, and raw transcriptome data were deposited in the Sequence Read Archive1 under the access number of PRJNA1061797.
2.3 Bioinformatic analysis of transcriptome data
Clean data were generated by filtering the raw data, removing adapters, sequences containing non-AGCT bases at the 5′ end, sequences with a quality value less than those of Q20, reads with N content of more than 10%, and small fragments with a length less than 25 bp. HISAT2,2 an alignment program for mapping next-generation sequencing reads, was used to align clean data with the hazel reference genome3 (Liu et al., 2021; Zhang et al., 2021). The obtained RNA-Seq alignments were assembled into transcripts using StringTie software4 (Shumate et al., 2022). The obtained transcripts were aligned to six public databases to acquire annotation information, including the NR, Swiss Prot, Pfam, STRING, gene oncology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. StringTie was used to calculate the fragments per kilobase of transcript per million fragments mapped (FPKM) value of each gene in the sample based on the alignment results of the HISAT2 software, representing the expression level of the gene in the samples, and DESeq2 was used to identify differentially expressed genes (DEGs) (Love et al., 2014; Zhang et al., 2021; Shumate et al., 2022). The criteria for determining DEGs were set as |Log2Fold Change| >1.00 and false discovery rate (FDR) <0.05. Fold Change refers to the value obtained by dividing the expression level of sample My by the expression level of sample Ck, and FDR refers to the false discovery rate. Hierarchical clustering analysis of the DEGs was performed using Multi Experiment Viewer.5 ClusterProfiler software (3.4.4) was used to perform GO and KEGG pathway enrichment analysis of DEGs, and the criteria for determining significantly enriched GO terms and DEGs KEGG pathway were set as p-value <0.05 and adjusted p-value <0.05.
2.4 qRT-PCR
Twenty DEGs (10 each of upregulated and downregulated) were randomly chosen to perform further qRT-PCR analysis, and the detailed primer sequences used in the analysis are listed in Supplementary Table S1. Beta actin was used as a reference gene in the analysis, and the fold changes (FCs) of the chosen genes were calculated using 2−ΔΔCt method (Schmittgen and Livak, 2008).
2.5 Immunohistochemical analysis
S. bovista cultured in vitro in auxin free medium and medium containing 2.0 mg/L auxin, the ectomycorrhiza and non-mycorrhiza root tips of hazelnuts were fixed in a 4% paraformaldehyde solution. Subsequently, the root tip samples were subjected to ethanol gradient dehydration, paraffin embedding, and sectioning, with a slice thickness of 8 μM. The paraffin slices were spread flat at 36°C and dried at 25°C. After confirming the slice integrity through microscopic examination, immunohistochemical analysis was performed. The primary process of immunohistochemical analysis was performed as previously described (Wei et al., 2021). A monoclonal anti-auxin antibody (SIGMA, United States) was used for immunohistochemical analysis of auxin. A specific antibody against a 390 amino acid (aa) long peptide sequence (10–400 aa) containing unique regions in ARF9 (Cor0156840.1) was synthesized by ABclonal Biotechnology Co., Ltd. (Wuhan, China). The secondary antibody in the negative control was distilled water, and that in the experimental group was labeled with Horseradish Peroxidase. The chromogenic agent used for immunohistochemical analysis of the auxin and ARF9 was 3, 3-N-Diaminobenzidine Tertrahydrochloride DAB. Horseradish peroxidase can catalyze DAB to produce brown precipitates, indicating the expression position and levels of auxin and ARF9 in the tissues. Finally, a light microscope (Axio Imager M2; Carl Zeiss, Jena, Germany) was used to observe and record the expression and distribution of auxins and ARF9.
2.6 Statistical analysis
Analysis of variance using SAS software version 8.01 (SAS Institute, Inc., Cary, NC, United States) was used to determine significant differences between the control and My treatments. The means of the data acquired from the control and My treatments were compared using the least significant difference test at a 5% significance level.
3 Results
3.1 Effect of Scleroderma bovista inoculation on seedling growth
S. bovista inoculation significantly promotes hazelnut seedling growth (Figure 1). After growing outdoors for 4 months, My treatment (inoculated with S. bovista and formed mycorrhiza) seedlings had significantly higher plant height, larger leaf area, and more developed root systems (Figure 1A). In September, many roots showed mycorrhiza formation at the bottom of the potted soil during the My treatment. Ectomycorrhiza is white-colored, and S. bovista only coexists with the hazelnut root system at the root tip, forming a white mantle with a length of only 3–5 mm (Figures 1B,C). Paraffin sectioning and staining of mycorrhiza revealed that the mantle was wrapped around the root tip of hazel, with a thickness of approximately 100 μm (Figure 1D). In addition, we observed Hartig nets in the intercellular areas of the cortex (Figure 1D). Mantles and Hartig nets are typical ectomycorrhizal structures.

Figure 1. The effect of inoculation with Scleroderma bovista on the growth and root development of hazelnut seedlings. (A) Inoculated seedlings with S. bovista and non-inoculated control. My, seedling with mycorrhiza. Ck, control. (B) Mycorrhiza of seedling inoculated with S. bovista. This picture was taken from the pot bottom. The red ellipse indicates white mycorrhiza. (C) Mycorrhiza of seedling inoculated with S. bovista. This picture was taken with a light microscope of Axio Imager M2. (D) Longitudinal section of mycorrhizal fungi. The green arrow indicates the root cap, while the red arrow indicates the ectomycorrhizal fungi mantle wrapped outside the root tip. Ha, Hartig net; Ma, mantle; Eh, external hyphae.
3.2 Effect of Scleroderma bovista inoculation on root development
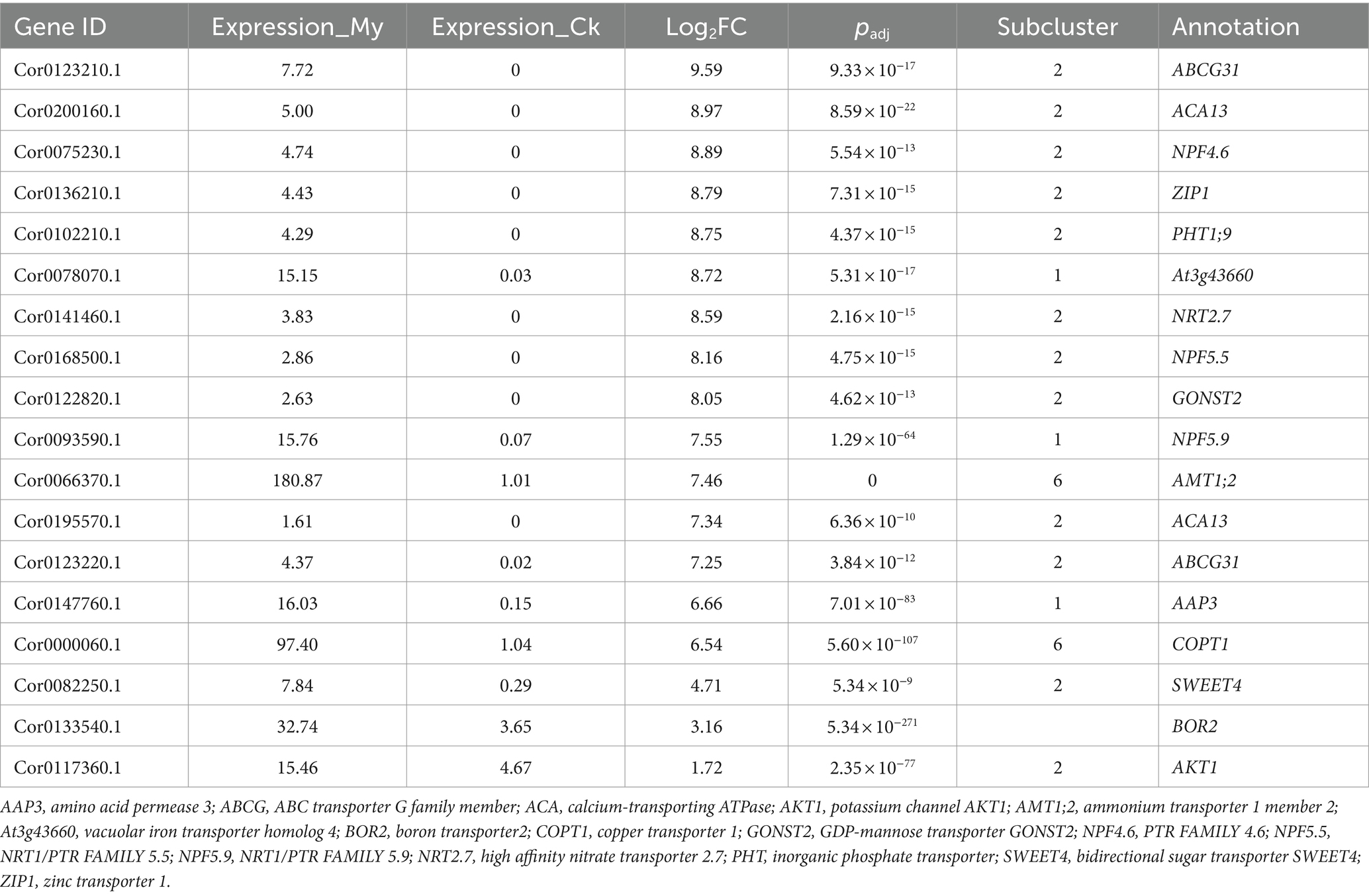
The fresh weights of the aboveground and underground parts of the inoculated seedlings were 2.73 and 2.15 times higher than those of the control (Figures 2A,B), respectively, and there was a significant difference between the two treatments, indicating that the inoculated seedlings grew faster. The length, project area, surface area, volume, forks, and diameter of the inoculated seedling roots were 2.48, 1.88, 1.88, 1.90, 2.06, and 1.13 times higher than those of the control, respectively (Figures 2C–H), and the root indicators between these two treatments reached a significant level (p < 0.05). These results indicate that S. bovista inoculation significantly affects root development. After S. bovista inoculation, the absorption area of hazel seedlings increased, and the root system branched more. These changes significantly enhanced the water and nutrient absorption capacities of the hazelnut seedlings.
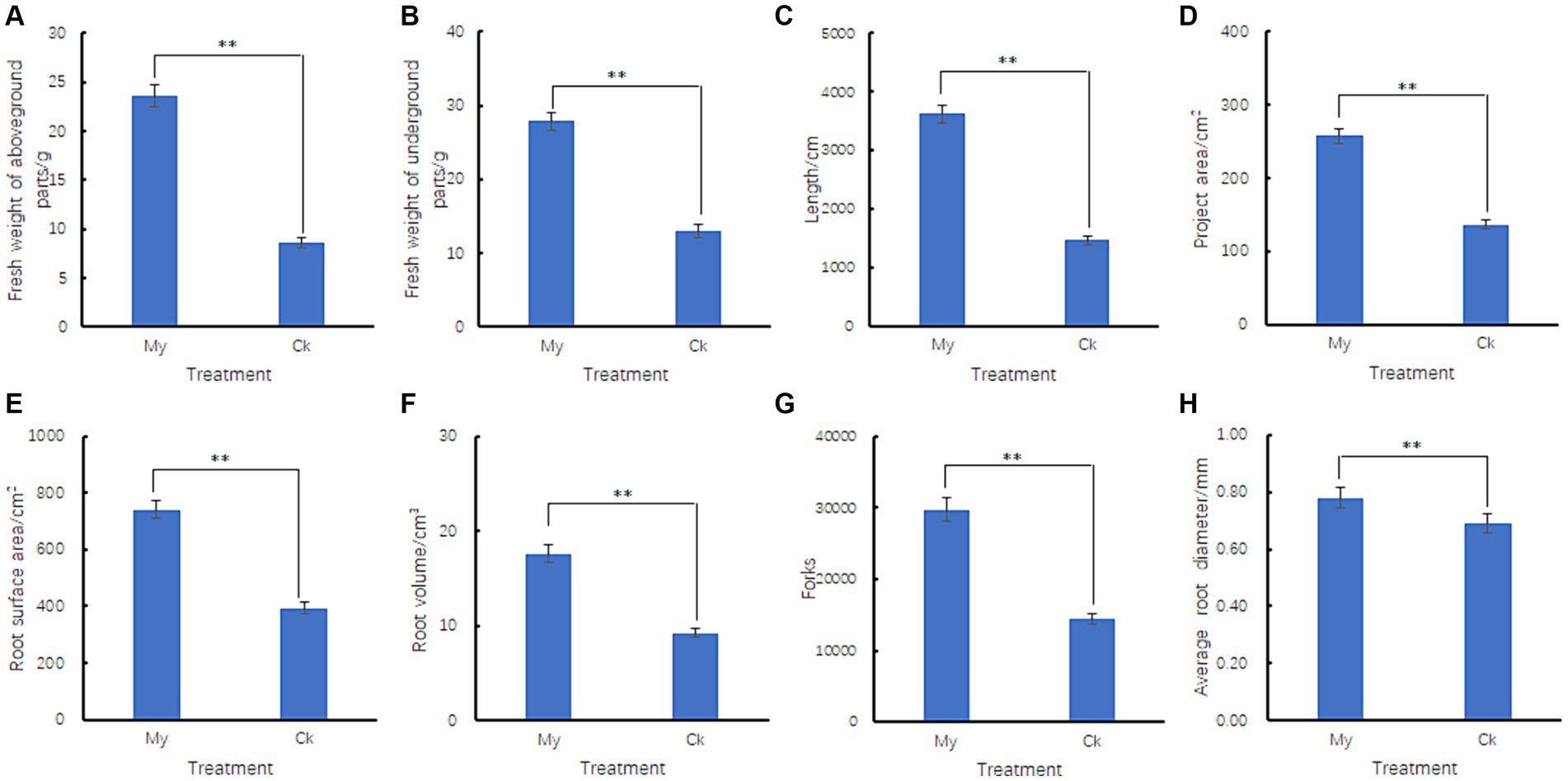
Figure 2. Effect of Scleroderma bovista inoculation on the root indexes of hazel. Comparison of fresh mass of aboveground parts (A), underground parts (B), root length (C), root project area (D), root surface area (E), root volume (F), root forks (G) and root average diameter (H) between control and S. bovista inoculation treatment. CK, non-inoculated control. My, treatments inoculated with S. bovista and formed mycorrhiza. **Indicates significant difference at p ≤ 0.05 as indicated by LSD test.
3.3 RNA sequencing, sequence mapping, and transcript expression
Six biological samples were sequenced using the Illumina NovaSeq 6000 platform. A total of 712,889,086 raw reads were generated, covering a length of 106,933,362,900 bp. All raw sequencing data were deposited in the sequence read archive under accession number PRJNA1061797. After filtering for low-quality sequences, we obtained 712,369,050 clean reads covering 102,081,099,845 bp. Clean reads were mapped to the hazel reference genomes (Liu et al., 2021), and 564,217,598 reads could be mapped to the reference genome. Among the 166,493,090 clean reads from the three control samples, 149,071,936 were mapped to the reference genome, accounting for 89.53% of all clean reads. Among the 397,724,508 clean reads from the three My treatment samples, 229,725,428 reads were mapped to the reference genome, accounting for 57.75% of all the clean reads. There were 22,319 protein-coding genes in the hazel genome. In the Ck treatments, 18,527 genes were expressed in the roots, with an average FPKM value of 42.26 (Supplementary Table S2). In My treatments, 19,292 genes were expressed in the roots, with an average FPKM value of 44.18 (Supplementary Table S2). Therefore, My treatments resulted in the expression of more genes and higher expression levels than Ck treatments. We drew a violin chart of the expression levels of various samples to explore the overall effect of S. bovista inoculation on the FPKM distribution of root genes in hazels. The median FPKM of My treatments was very close to that of Ck, and the lower quartile of My treatments was significantly higher than that of Ck (Figure 3). These results indicate that S. bovista inoculation treatment increased the number of expressed genes in hazel roots and significantly increased the expression levels of low-abundance genes.
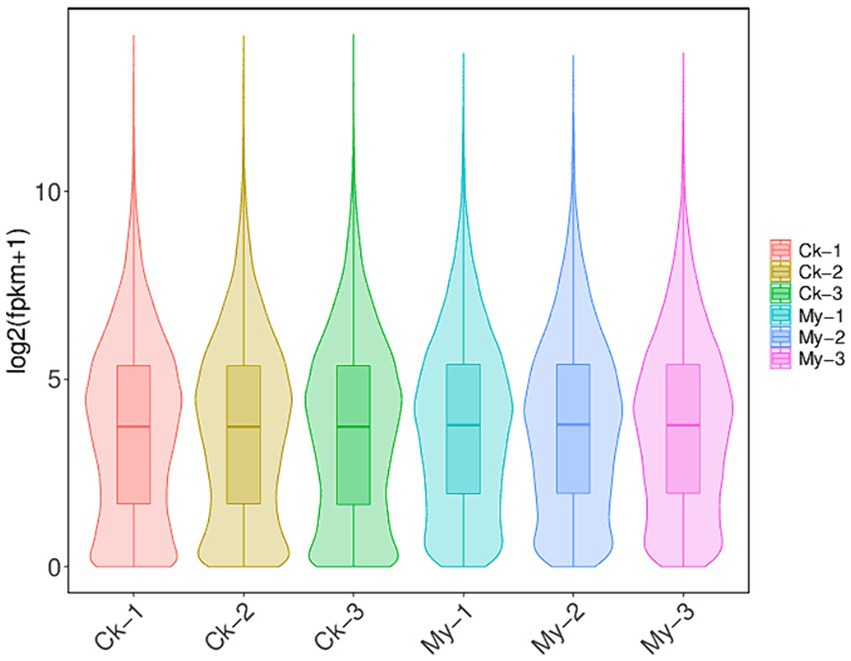
Figure 3. Violin chart of gene expression levels. CK, non-inoculated control. My, treatments inoculated with S. bovista and formed mycorrhiza. The numbers after CK and My represent biological replicates.
3.4 DEGs
The criteria |Log2Fold Change| >1.00 and FDR <0.05 were set to determine DEGs in paired comparison of My vs. Ck. Compared with the Ck treatment, 3,265 upregulated and 1,916 downregulated genes were identified in the My treatment (Figure 4). The number of upregulated genes was 1.70 times that of the downregulated, suggesting that S. bovista inoculation improves gene expression levels. All DEGs and their corresponding annotations are presented in Supplementary Table S3.
Figure 4. A volcanic map of differentially expressed genes. CK, non-inoculated control. My, treatments inoculated with S. bovista and formed mycorrhiza. The horizontal axis represents the gene expression difference between two samples, which is the value obtained by dividing the expression level of sample My by the expression level of sample Ck. The vertical axis represents the p-value, and the higher the p-value, the more significant the expression difference. The values in the horizontal and vertical axes have been logarithmized. Each dot in the graph represents a specific gene, with red dots indicating significantly upregulated genes, blue dots indicating significantly downregulated genes, and black dots indicating non significantly differentially expressed genes.
3.5 Hierarchical clustering analysis of DEGs
Hierarchical clustering analysis of DEGs was used to group genes with the same or similar expression patterns, revealing the function of unknown genes or the unknown functions of known genes. A gene expression heatmap was used to visualize the differences in the expression levels of the DEGs between the control and My samples (Figure 5A). All 5,181 DEGs were divided into eight subclusters (Supplementary Table S4), each containing 229 (Figure 5B, subcluster 1), 3,017 (Figure 5C, subcluster 2), 1,840 (Figure 5D, subcluster 3), 2 (Figure 5E, subcluster 4), 71 (Figure 5F, subcluster 5), 16 (Figure 5G, subcluster 6), 3 (Figure 5H, subcluster 7), and 3 (Figure 5I, subcluster 8) DEGs. Therefore, Sub-cluster 2 contained the most genes, followed by subclusters 3 and 1. The three subclusters accounted for 98.17% of the total DEGs and exhibited similar expression patterns. In subclusters 1 and 2, the gene expression level of the My treatments was higher than that of the control, whereas in subcluster 3, the gene expression level of the My treatments was lower than that of the control. Similarly, we deduced that these differences in gene expression levels might cause the promotion of hazel root growth through S. bovista inoculation.
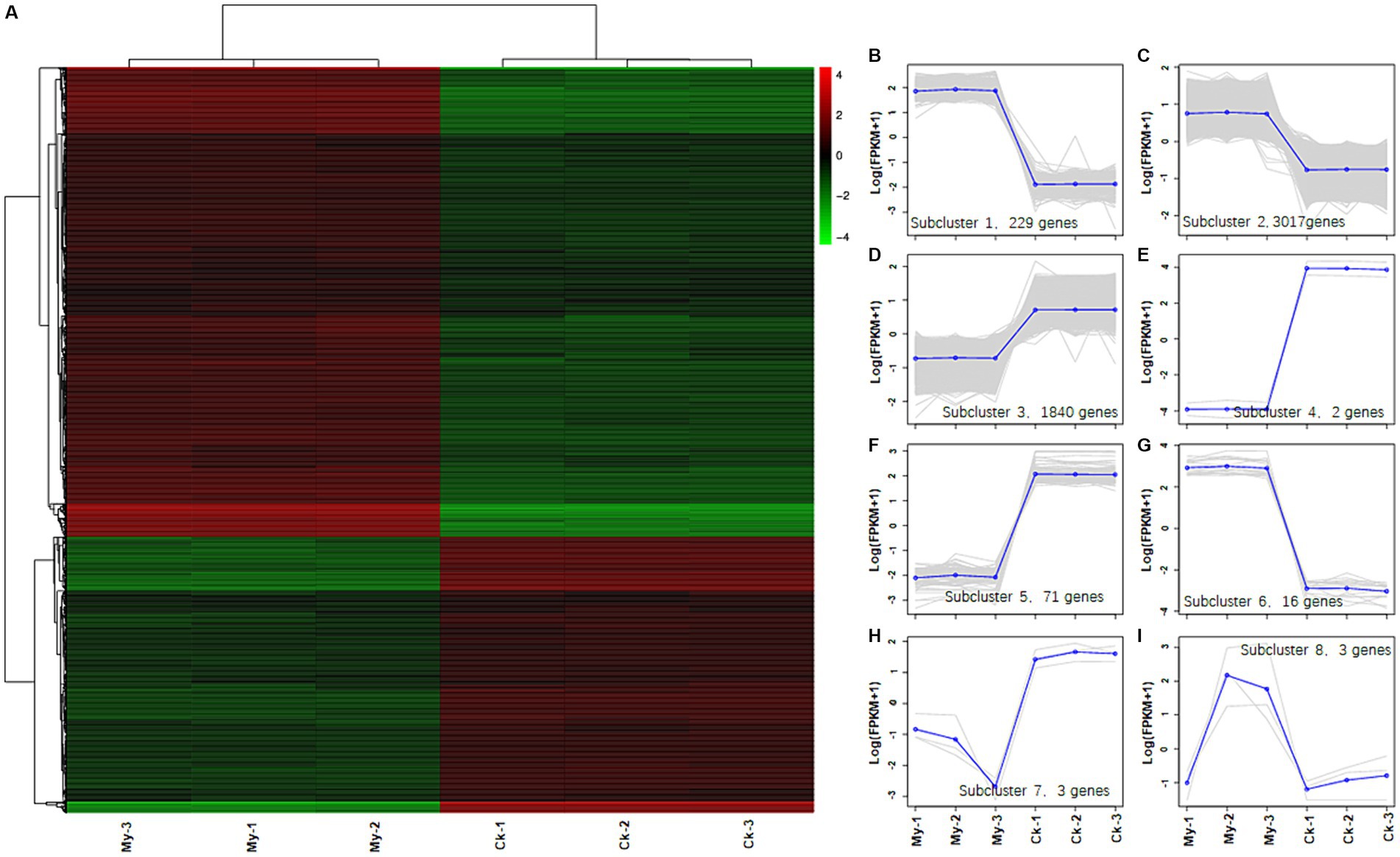
Figure 5. Heatmap analysis of differentially expressed genes between control, Scleroderma bovista inoculation treatment and trend analysis of eight subclusters. (A) Heatmap analysis of differentially expressed genes between control and S. bovista inoculation. Each column in the left graph represents a sample, and each row represents a gene. The color in the graph represents the expression level of the gene in the group of samples (Log10FPKM), while red represents a higher expression level of the gene in the sample and green represents a lower expression level. Each line in the right figure represents a gene, and the blue line represents the average expression level of all genes in the subcluster. Each right graph displays a type of expression pattern, which reflects the trend of changes in the expression level of this group of genes. (B) Subcluster 1. (C) Subcluster 2. (D) Subcluster 3. (E) Subcluster 4. (F) Subcluster 5. (G) Subcluster 6. (H) Subcluster 7. (I) Subcluster 8. CK, non-inoculated control. My, treatments inoculated with S. bovista and formed mycorrhiza. The numbers after CK and My represent biological replicates.
3.6 GO enrichment analysis of upregulated and downregulated DEGs
GO enrichment analysis of 3,265 upregulated and 1,916 downregulated genes was performed to study the general functions of the DEGs. The GO enrichment histogram of DEGs intuitively reflects DEG distribution in GO terms enriched in biological processes, molecular functions, and cellular components (Figure 6). Regarding biological process, the most significantly enriched GO terms for the upregulated DEGs were GO:0007018 (microtubule-based movement), followed by GO:0006811 (ion transport) and GO:0009698 (phenylpropanoid metabolic process), etc. (Figure 6A and Supplementary Table S5). The most significantly enriched GO terms for the downregulated DEGs were GO:0009800 (cinnamic acid biosynthetic process), followed by GO:0009803 (cinnamic acid metabolic process) and GO:0009072 (aromatic amino acid family metabolic process), etc. (Figure 6B and Supplementary Table S6).
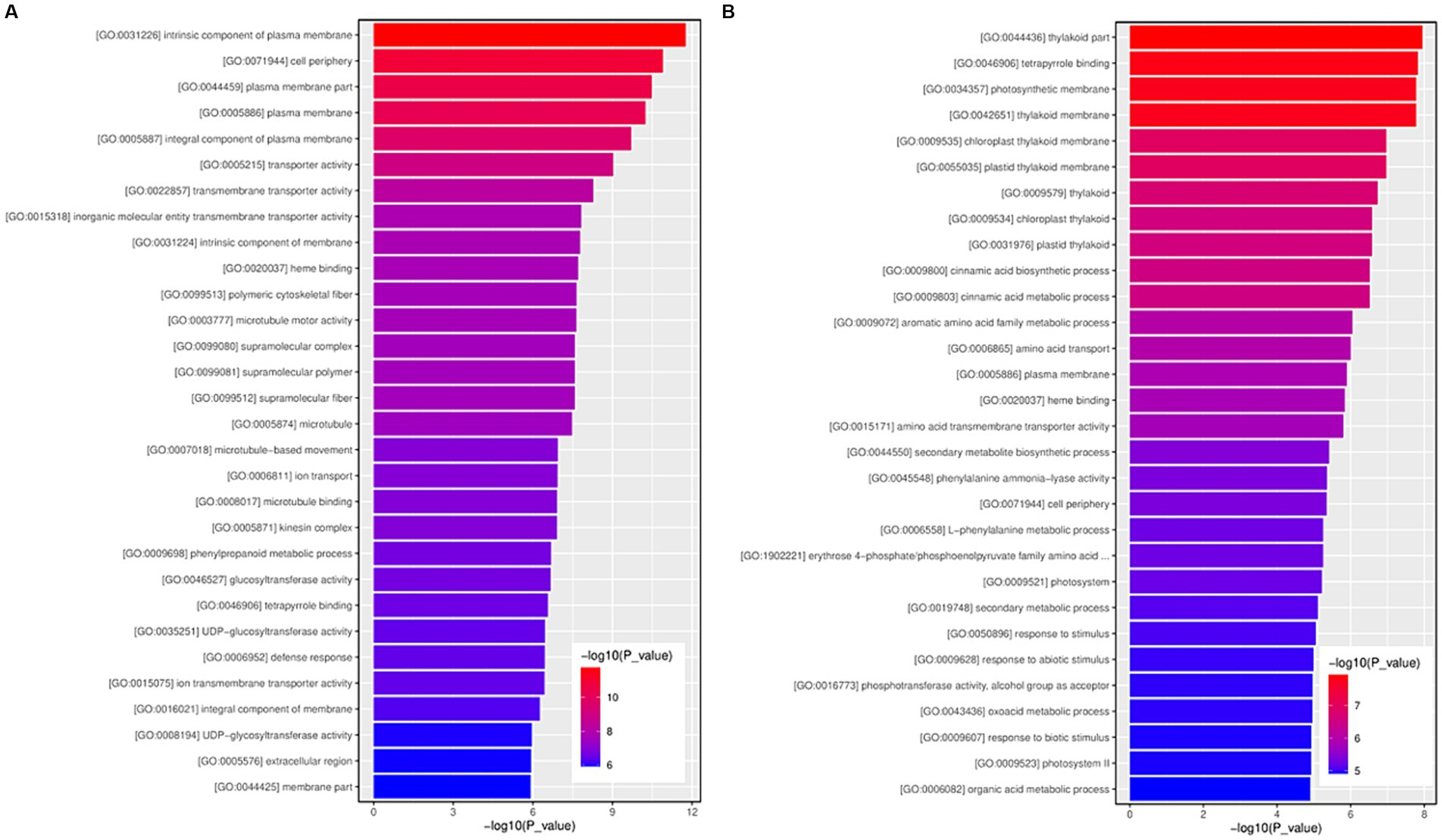
Figure 6. The GO function enrichment histogram of significantly differentially expressed genes. (A) The GO function enrichment histogram of up-regulated genes. (B) The GO function enrichment histogram of down-regulated genes. Each column in the figure is a GO term, and the horizontal coordinate text represents the name of the GO. The height of the column, also known as the abscissa, represents the significance of enrichment, known as p-value. The darker the color, the more significant the enrichment of the function. The color gradient on the right represents the size of p-value.
Concerning molecular function, the most significantly enriched GO terms for the upregulated DEGs were GO:0005215 (transporter activity), followed by GO:0022857 (transmembrane transporter activity) and GO:0015318 (inorganic molecular entity transmembrane transporter activity), etc. (Figure 6A and Supplementary Table S5). The most significantly enriched GO terms for the downregulated DEGs were GO:0046906 (tetrapyrrole binding), followed by GO:0020037 (heme binding) and GO:0015171 (amino acid transmembrane transporter activity), etc. (Figure 6B and Supplementary Table S6).
For cellular components, the most significantly enriched GO terms for the upregulated DEGs were GO:0031226 (intrinsic component of plasma membrane), followed by GO:0071944 (cell periphery) and GO:0044459 (plasma membrane part), etc. (Supplementary Table S4) (Figure 6A and Supplementary Table S5). The most significantly enriched GO terms for the downregulated DEGs were GO:0044436 (thylakoid part), followed by GO:0034357 (photosynthetic membrane) and GO:0042651 (thylakoid membrane), etc. (Figure 6B and Supplementary Table S6).
3.7 KEGG pathway enrichment analysis of DEGs
Adjusted p-values <0.05 were set as the threshold to determine the significant KEGG pathways of DEGs. Nine and 22 KEGG pathways of upregulated and downregulated DEGs, respectively, were significantly enriched (Supplementary Tables S7, S8). For the upregulated DEGs, the most significantly enriched DEGs KEGG pathways were associated with ko00941 (flavonoid biosynthesis), ko01110 (biosynthesis of secondary metabolites), and ko04075 (plant hormone signal transduction). (Figure 7A and Supplementary Table S7). Among the downregulated DEGs, the most significantly enriched DEGs KEGG pathways were ko01130 (biosynthesis of antibiotics), ko01110 (biosynthesis of secondary metabolites), and ko01200 (carbon metabolism) (Figure 7B and Supplementary Table S8). Among the 22 significantly enriched KEGG pathways, 20 were associated with metabolism and the remaining pathways were related to environmental information processing.
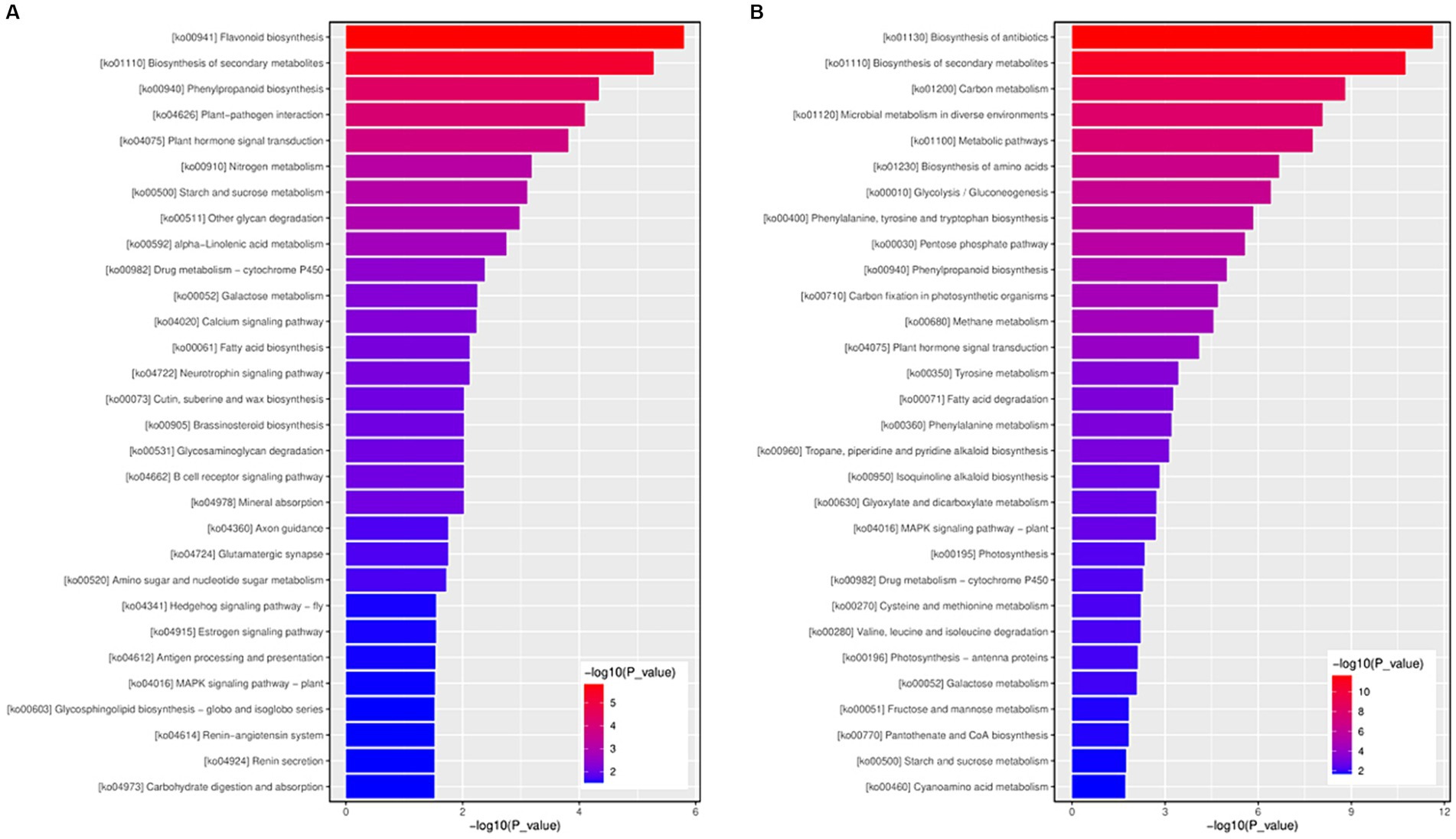
Figure 7. The KEGG function enrichment histogram of significantly differentially expressed genes. (A) The KEGG function enrichment histogram of up-regulated genes. (B) The KEGG function enrichment histogram of down-regulated genes. Each column in the figure represents a KEGG path, and the horizontal axis text represents the name of the KEGG. The height of the column, the abscissa, represents the significance of enrichment (p-value). The darker the color, the more significant the enrichment of the KEGG path. The right color gradient represents the size of p-value.
3.8 qRT-PCR of DEGs
Twenty DEGs (10 each of upregulated and downregulated) were randomly chosen for qRT-PCR analysis. Among the 20 DEGs, the fold changes (FCs) of those obtained from RNA-seq and qRT-PCR were well correlated (Figures 8A,B), suggesting that the expression patterns from the qRT-PCR analysis were consistent with those obtained from RNA sequencing. Thus, the reliability of the RNA-seq data was validated.
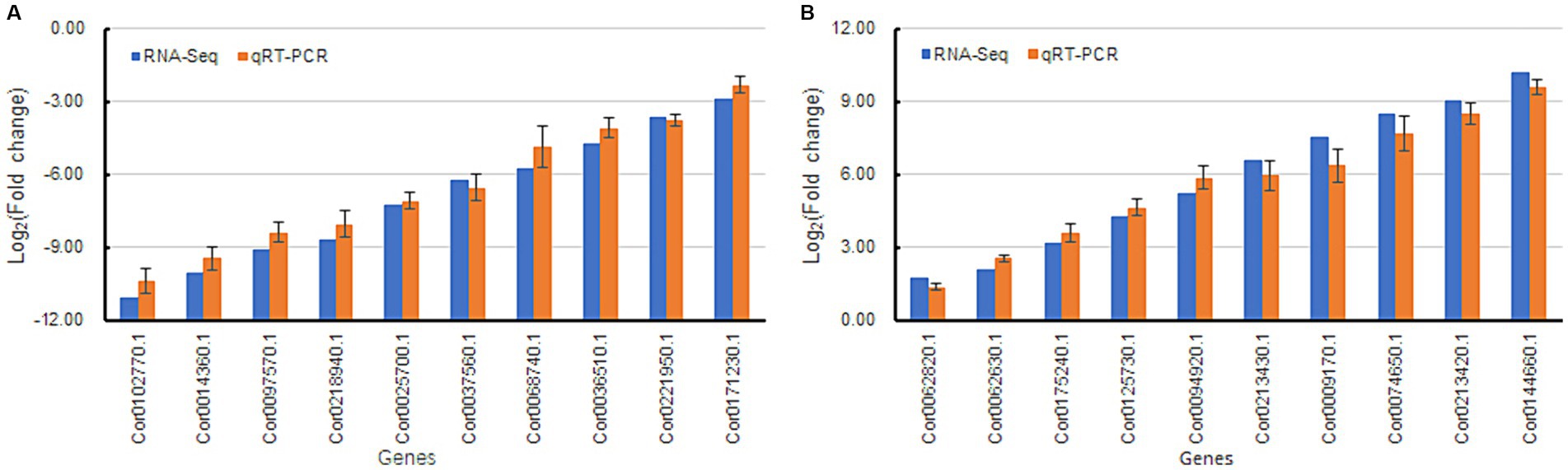
Figure 8. Validation of differentially expressed genes by qRT-PCR analysis. Beta actin was used as a reference gene in the analysis. Fold change refers to the value obtained by dividing the expression level of sample My by the expression level of sample Ck. Bars represent mean ± standard deviation (n = 3). RNA-seq and qRT-PCR indicated that the gene fold change obtained was acquired by transcriptome sequencing and real-time quantitative PCR, respectively. (A) indicates downregulated genes, and (B) represents upregulated genes.
3.9 Immunohistochemical analysis of auxin and ARF9
Differential transcriptome analysis of Ck and My revealed many DEGs related to auxin synthesis and signal transduction, suggesting that the regulation of auxin ectomycorrhizal fungi S. bovista on root development may be related to auxin. To support this, immunohistochemical analyses of Ck and My treatments were conducted using antibodies against auxin and ARF9. We conducted immunohistochemical analysis of auxin in vitro cultured S. bovista. When there was no auxin in the culture medium, the growth of S. bovista was very slow, and the presence of auxin could not be observed in the hypha (Supplementary Figures S1A, B1,B2); when auxin was present in the culture medium, the growth rate of S. bovista was significantly promoted, and high concentrations of auxin could be observed in the hypha (Supplementary Figures S1A, C1,C2). These results indicated that the S. bovista cannot synthesize auxin itself. Auxins in the root tips of non-inoculated plants were primarily distributed on the cell walls (Figures 9A1,A2, B1,B2). Compared with the non-mycorrhizal root tips, the distribution and expression levels of auxin in the mycorrhizal roots were significantly different. S. bovista formed a mantle at the root tip of the hazel, and auxin was primarily distributed in the mantle, followed by the cell wall, with the lowest concentration within the cell (Figures 9C1,C2, D1,D2). Immunohistochemical analysis of ARF9 showed that in the non-mycorrhizal control, ARF9 was expressed in the cell walls of the epidermis and cortex (Figures 9E1,E2, F1,F2). The expression level of ARF9 in the mycorrhizae was significantly higher than that in the non-mycorrhizal control. Compared with the epidermal and cortical cells of mycorrhiza, ARF9 was primarily distributed in the mantle, followed by the cell wall (Figures 9G1,G2, H1,H2). After the formation of mycorrhiza at hazelnut root tips, the expression levels of auxin and ARF9 significantly increased in S. bovista. In the My treatment, auxin and ARF9 secreted by the root tips actively entered the outside mantle.
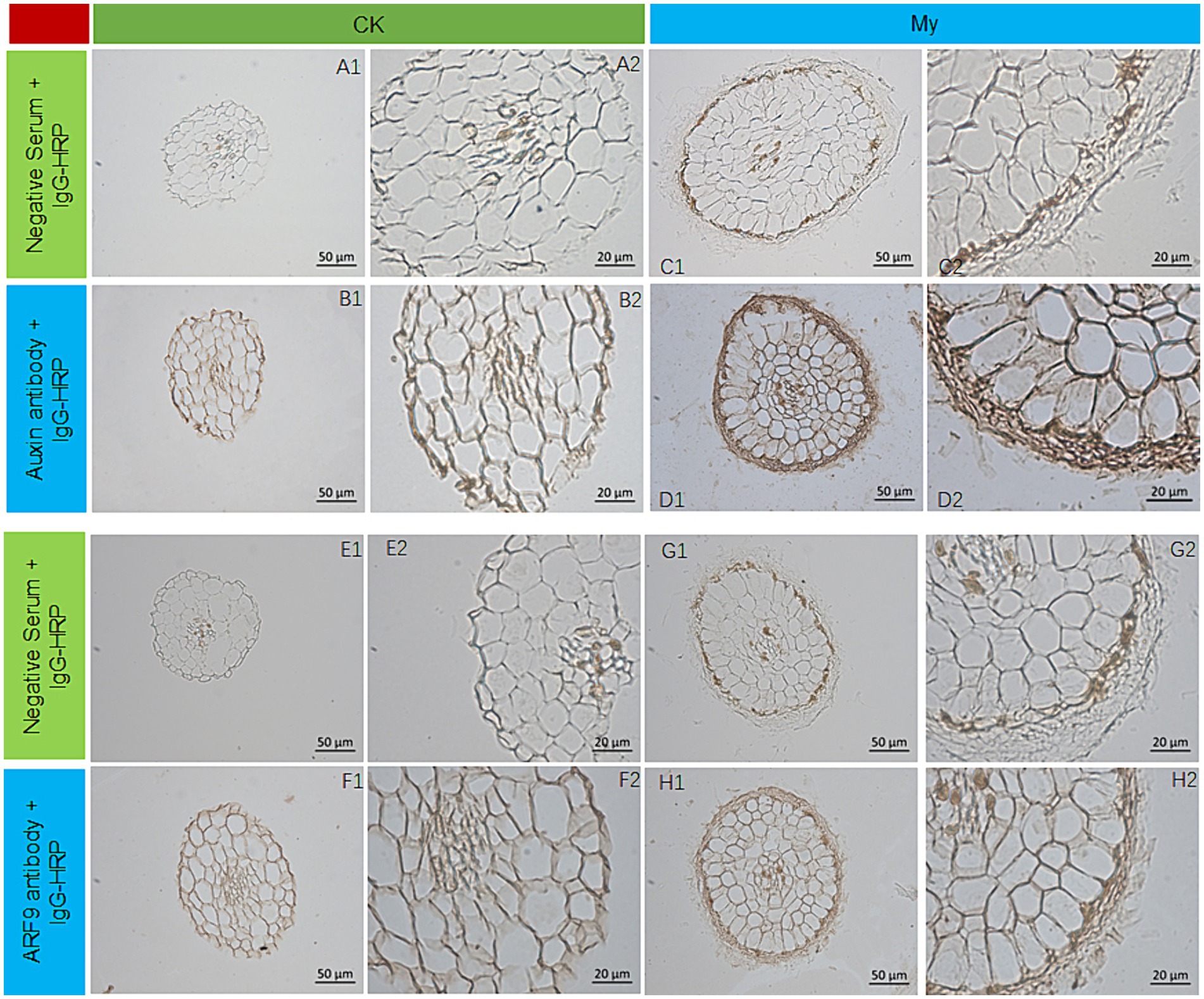
Figure 9. Auxin and ARF9 expression by immunohistochemical analysis. (A1) CK, negative control. (A2) Enlarged view of A1. (B1) CK, the first antibody is auxin antibody. (B2) Enlarged view of B1. (C1) My, negative control. (C2) Enlarged view of C1. (D1) My, the first antibody is auxin antibody. (D2) Enlarged view of D1. (E1) CK, negative control. (E2) Enlarged view of E1. (F1) CK, the first antibody is ARF9 antibody. (F2) Enlarged view of F1. (G1) My, negative control. (G2) Enlarged view of G1. (H1) My, the first antibody is ARF9 antibody. (H2) Enlarged view of H1. CK, non-inoculated control. My, treatments inoculated with S. bovista and formed mycorrhiza.
4 Discussion
4.1 Scleroderma bovista colonizes hazelnut plants’ roots and thus boosts their growth behavior
Most ectomycorrhizal fungi are macro fungi. S. bovista is an edible medicinal fungus with hemostatic and detoxifying effects and a high medicinal value. Its tender fruiting body is a delicious mushroom. To form edible fruiting bodies, S. bovista needs to form a symbiotic relationship with host trees (Zhang et al., 2021). Therefore, the symbiotic relationship established between the S. bovista and the hazelnut tree benefits the growth and development of the hazelnut tree, benefits the development of the under-forest economy in the hazelnut orchard, and increases cultivation benefits. Previously, we discovered naturally formed ectomycorrhizal fungi in the hazelnut production area of Northeast China. Outdoor-potted hazelnut seedlings were inoculated with S. bovista, which promoted the growth of their aboveground and underground parts (Cheng et al., 2023). It is believed that S. bovista and hazelnut trees in the hazelnut production area of Northeast China have similar habitats (Cheng et al., 2023). S. bovista can form a stable symbiotic relationship with the hazelnut root system in the production area. It has been speculated that it may be crucial in promoting root development and plant growth and improving plant stress resistance. We inoculated potted hazelnut seedlings with S. bovista, and confirmed that the inoculation resulted in a significant increase in root biomass compared to non-inoculated controls. The length, projection area, surface area, volume, forks, and diameter of the inoculated seedlings root were 1.13–2.48 times higher than those of the control, suggesting a more developed root system in S. bovista inoculated treatments, consistent with our previous study (Cheng et al., 2023). Transcriptomic analysis of the non-mycorrhiza control and mycorrhiza treatment was compared in this study to elucidate the possible underlying mechanism by which S. bovista promotes the growth and development of hazelnut seedlings and to provide a scientific basis for developing novel cultivation techniques and ectomycorrhizal agents for hazelnuts.
4.2 Candidate DEGs that may be involved in transportation in hazel ectomycorrhiza
Transcriptome sequencing and DEGs identification revealed that the FPKM values of genes in the My treatments were higher than those in the control. Among the 5,181 DEGs, the number of upregulated DEGs was 1.7 times higher than that of the downregulated DEGs. These results indicate that symbiosis between S. bovista and hazelnut roots was more conducive to the promotion of gene expression. The GO enrichment analysis for the upregulated genes indicated that the most significantly enriched GO terms for the regulated DEGs included GO: 007018 (microtubule-based movement) and GO: 006811 (ion transport), suggesting that the symbiosis between S. bovista and hazelnut roots benefits cell division and ion transport. The most significantly enriched GO terms for the downregulated DEGs were GO: 0044436 (thylakoid part) and GO:0034357 (photosynthetic membrane), suggesting that the symbiosis between S. bovista and hazelnut roots caused the downregulation of genes related to photosynthesis. The root system grows in the dark soil and cannot undergo photosynthesis. The downregulation of photosynthesis-related genes in the root system benefits plants to save energy and promote root growth.
ATP-binding cassette (ABC) proteins contain ATP-binding domains (NBD/NBF). The ABC protein family is large, and the ABC proteins carrying transmembrane domain are called ABC transporter proteins (Theodoulou, 2000; Uroz and Oger, 2017). There are eight plant ABCA, ABCG, and ABCI subtypes, which are primarily involved in the active transport of heavy metals, antibiotics, and small-molecule hormones (Mentewab and Stewart, 2005; Kretzschmar et al., 2011; Borghi et al., 2015; Hwang et al., 2016; Ofori et al., 2018). In Arabidopsis, ABCG31 is involved in regulating sterol accumulation on pollen surfaces (Geisler et al., 2017). Brassinosteroids are a class of plant-specific steroid hormones, and they are reportedly crucial in plant root development (Choi et al., 2014). Significantly, two DEGs encoding ABCG31 were upregulated 9.59 and 7.25 folds in My treatment, and their orthologs in other plants were critical in sterol and small molecule hormone transport (Table 1). This suggests that ABCG31 may affect the root development in My treatment by regulating the steroid (Brassino) transport.
In Arabidopsis, calcium-transporting ATPase 13 (ACA13) is involved in vegetative growth and immune processes, and its mutation causes seedling death upon bolting, suggesting an essential role in plant growth (Nolan et al., 2023). Two DEGs encoding ACA13 were upregulated 8.97 and 7.34 folds in the My treatment, and their orthologs in other plants were crucial regulators in calcium transport and plant growth (Table 1). This suggests that ACA13 may affect root development during My treatment by regulating calcium transport, vegetative growth, and immune processes.
Nitrate is the primary form of nitrogen absorbed by most terrestrial plants. It is a nutrient that makes up biomolecules and a signaling molecule that regulates plant growth and developmental processes. NPF5.5, NPF4.6, and NRT2.7 are nitrate transporters (Mentewab and Stewart, 2005; Borghi et al., 2015). NPF4.6 transports abscisic acid and nitrate (David et al., 2014; Léran et al., 2015; Yu et al., 2018). Numerous studies have shown that abscisic acid (ABA) promotes root growth, increases stress resistance under moderate stress, and is essential for maintaining root growth under normal conditions (Kanno et al., 2013). Three DEGs encoding NPF5.5, NPF4.6, and NRT2.7 were highly upregulated in the My treatment, and their orthologs in other plants had critical regulatory roles in nitrate and ABA transport (Table 1), suggesting that they may affect root development in the My treatment through this transport regulation mechanism.
Similarly, ZIP1 participates in zinc transport, accumulation, and partitioning (Teng et al., 2023). PHT1;9 is involved in inorganic phosphate acquisition and transport in the root system (Gaitán-Solís et al., 2015). At3g43660 and NPF5.9 are involved in iron transport (Gollhofer et al., 2011; Remy et al., 2012). COPT1 is involved in copper transport, and its overexpression slightly increases the endogenous iron concentration in rice grains (Chen et al., 2021). GONST2 is a nucleotide sugar transporter that provides GDP-mannose for glycosyl inositol phosphoryl ceramide glycosylation (Andrés-Bordería et al., 2017). AMT1;2 is involved in ammonium uptake by Arabidopsis roots (Jing et al., 2021). AAP3 is essential in amino acid transport (Giehl et al., 2017). In soybean, GsBOR2 is crucial in the efflux of boron from cells, and its overexpression in Arabidopsis generates longer roots than those in wild-type plants (Marella et al., 2013). SWEET4 is a bidirectional sugar transporter (Duan et al., 2018; Andargie and Li, 2019). AKT1 is a potassium channel protein, and the overexpression of GmAKT1 in Arabidopsis promotes root length and K+ concentration (Liu et al., 2016). DEGs encoding ZIP1, PHT1;9, At3g43660, COPT1, GONST2, AMT1;2, and SWEET4 were highly upregulated in the My treatment, and their orthologs in other plants had critical regulatory roles in zinc, phosphate, nucleotide sugar, ammonium, amino acid, and sugar transport (Table 1), suggesting that they may significantly affect plant growth and root development in the My treatment by regulating elements, ABA, and sugar transport processes.
4.3 Candidate DEGs that may be involved in root development and branching in ectomycorrhiza of hazel
Auxins are vital phytohormones with essential effects on plant growth, development, and rooting. The Arabidopsis WALLS ARE THIN1 (WAT1) gene family comprises 46 members, and WAT1 is believed to be involved in auxin signaling and secondary cell wall formation in stem fibers. wat1-1 mutation causes a significant downregulation of auxin-related gene expression, decreasing auxin content in the stem (Feng et al., 2012). WAT1 is an auxin transporter protein on the vacuole membrane, which is crucial in regulating plant vacuolar auxin homeostasis (Ranocha et al., 2010). Significantly, 14 DEGs encoding WAT1 were >1 fold upregulated in My treatment, and their orthologs in other plants are critical in auxin homeostasis, auxin signaling, and secondary cell wall formation. The three most highly regulated DEGs are listed in Table 2. This suggests that WAT1 may affect root development during My treatment by regulating auxin homeostasis and signaling.
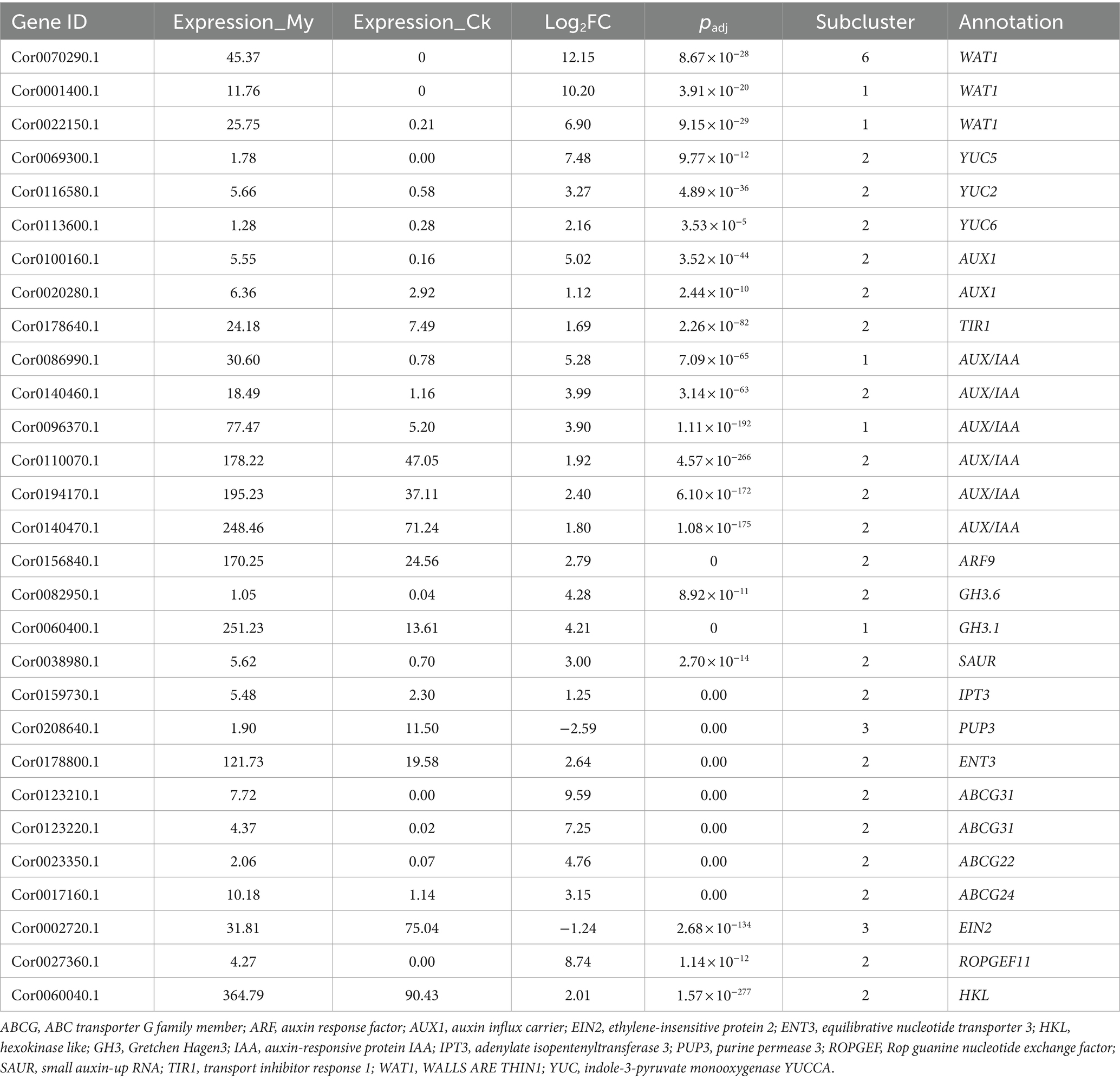
Table 2. Genes of interest that may be involved in root development and branching in ectomycorrhiza of hazel.
Auxin synthesis involves tryptophan-and non-tryptophan-dependent pathways. Plants primarily complete the synthesis of tryptophan to auxin through the tryptamine, indole-3-pyruvate (IPA), and indole-3-acetonitrile pathways, depending on the intermediate products. In the IPA pathway, the rate-limiting step for converting IPA to indoleacetic acid (IAA) is categorized by YUCCA (Kim et al., 2013; Ranocha et al., 2013; Chen et al., 2014). Simultaneous loss of mutations in YUC5, YUC3, YUC7, YUC8, and YUC9 caused short and agravitropic primary roots, implying vital regulatory roles for YUC5 in root development (Kim et al., 2013). Auxin regulates stem growth rate, inhibits lateral buds, and promotes rooting. Three DEGs encoding YUC5, YUC2, and YUC6 were highly upregulated in the My treatment. We have completed genome sequencing of S. bovista, and its genome sequence was uploaded to the public database NCBI with the accession number of PRJNA1048296. We did not find any genes related to auxin synthesis in its genome. Consistent with these, after S. bovista were cultured on auxin free medium, no auxin could not be detected in the hypha of S. bovista using immunohistochemical analysis. After symbiosis with hazelnut roots, immunohistochemical analysis showed that auxin expression levels in the mycorrhiza complex increased significantly, further supporting that S. bovista promotes auxin biosynthesis in hazelnut roots. Thus, the orthologs of YUC5, YUC2, and YUC6 in other plants significantly regulate auxin biosynthesis and root development (Table 2), suggesting that they may promote root development during My treatment by regulating auxin biosynthesis.
In Arabidopsis, AUX1 (auxin influx carrier) functions as a permease-like regulator of root gravitropism (Bennett et al., 1996; Li et al., 2015). The Aux/IAA-TIR1-ARF complex is crucial in activating auxin signal transduction. TIR1 is in the nucleus and acts as an auxin receptor, binding to auxin molecules to initiate auxin signaling (Vandenbussche et al., 2010). When auxin is absent or at a low concentration, AUX/IAA forms heterodimers with ARF, preventing ARF from binding to the promoters of downstream auxin-induced genes and inhibiting gene transcription (Tian et al., 2002; Hayashi et al., 2008). However, when plant cells are stimulated by external or internal auxins, IAA binds to TIR1 to form a TIR1-IAA complex. IAA binding stabilizes the interaction between the second domain of TIR1 and Aux/IAA proteins, causing their degradation (Tian et al., 2002; Hayashi et al., 2008). This releases the ARF protein and promotes the transcription of downstream genes, activating the auxin response. The GH3 (Gretchen Hagen3) bind auxin and amino acids, controlling plant auxin levels (Hagen and Guilfoyle, 2002). The 20 GH3 genes in Arabidopsis can be divided into three subfamilies, with GH3.1 and GH3.6 belonging to Subfamily II. Changes in free auxin content caused by the expression of AtGH3s affect plant architecture and root development in Arabidopsis. AtGH3.6 overexpression caused Arabidopsis dfl1-D dwarf lines with shorter stems and fewer lateral roots than the wild type (Wang et al., 2023). Auxin regulates the expression of many genes, including three types of early auxin-responsive gene families: Aux/IAA, GH3, and small auxin-up RNA (SAUR) (Nakazawa et al., 2008). Among these gene families, SAURs are the most rapidly responsive to auxin. SAURs are primarily involved in regulating auxin synthesis and transportation (Liscum and Reed, 2002). DEGs encoding AUX1, Aux/IAA, TIR1, ARF, GH3, and SAURs were differentially expressed in My treatment, with most genes being upregulated. Consistent with this, immunohistochemical analysis of ARF9 showed that its expression in the mycorrhiza complex was promoted by symbiosis between S. bovista and hazel roots. The orthologs of AUX1, Aux/IAA, TIR1, ARF, GH3, and SAURs in other plants regulate auxin influx and its signal transduction, level, synthesis, and transportation (Table 2), suggesting that they may significantly affected root development in the My treatment by regulating auxin synthesis, transportation, and signal transduction processes. These data suggest that auxin biosynthesis and signal transduction pathways are activated in symbiosis with S. bovista, benefiting hazel root development.
We found that ARF9 had the highest expression level in the mantle. ARF was not in the genome of S. bovista. All hazelnut ARF sequences were obtained from the hazelnut genome database using SignalP software (v5.0)6 (Wang et al., 2020). Signal peptide prediction was conducted, and potential signal peptide cleavage sites were found in the coding sequence of ARF (Cor0105760.1). The sequence of ARF9 is homologous to it. It is speculated that after the expression of Cor0105760.1, the protein in the hazelnut root system is guided by the signal peptide into the extracellular space and enters the symbiotic mantle to regulate the symbiotic process between S. bovista and hazelnut. This discovery may mean that plant ARF proteins enter the microbial hyphae through a mechanism that regulates symbiosis between microorganisms and plant roots, providing a novel idea for analyzing the symbiosis mechanism between ectomycorrhizal fungi and plant hosts.
Cytokinins are primarily synthesized at the root tip and transported to other parts of the root and aboveground parts. The key rate-limiting step in cytokinin synthesis is catalyzed by IPT (Petersen et al., 2011). Purine permeaminase (PUP) and equilibrative nucleotide transporter 3 (ENT3) are transporters that can actively transport cytokinin (Galichet et al., 2008). The three primary types of cytokinin transporter proteins belong to the PUP, ENT, and ABCG families and are membrane-localized (Wang et al., 2023). DEGs encoding IPT3, PUP3, ENT3, ABCG31, ABCG22, and ABCG24 were differentially expressed in the My treatment, and most were upregulated. The orthologs of IPT3, PUP3, ENT3, ABCG31, ABCG22, and ABCG24 in other plants are essential regulators of cytokinin biosynthesis and transport, implying that they markedly affected the root branch in the My treatment by regulating cytokinin biosynthesis and transport processes.
After symbiosis formation between exogenous fungi and their hosts, the root structure of plants undergoes significant changes, including the formation of multiple thick and short lateral roots to increase the surface area for ectomycorrhizal fungi colonization. The root hairs of plant roots decrease because their function is replaced by the fungal sheath formed by ectomycorrhizal fungi (Romanov and Schmülling, 2021). The complex PIRF1, formed by a combination of Phytochrome and ROP guanine nucleotide exchange factor (RopGEF1), regulates root development by activating the Rho-like GTPases of plants in the cytoplasm. Mutations in PIRF1 cause delayed root elongation and irregular root hair formation, suggesting that RopGEF1 negatively regulates primary root development (Tikhonovich and Provorov, 2007). Transgenic lines that overexpress hexokinase-like1 (HKL1) suggest that hexokinase is a negative regulator of root hair development (Shin et al., 2010). ein2 mutants cause shorter root hairs phenotype (Karve and Moore, 2009). After symbiosis formation between S. bovista and hazel root, the function of root hairs is replaced by a fungal sheath, and mycorrhizal roots are characterized by more short roots, fewer long roots, and fewer root hairs. The orthologs of ROPGEF1 and HKL1 in other plants are vital regulators in the development of prime root and root hair (Table 2); as negative regulators, its upregulation may inhibit the development of long root formation and root hair of My treatment; ein2 is a positive regulator in root formation and root hair, it was upregulated in the My treatment. Expression changes in these sets of DEGs caused fewer root-hair phenotypes.
In the symbiotic establishment stage, hyphae physically contact the surface of the plant roots, wrap around the root tip, form hyphal sheaths, and invade the plant root cortex cells to form a Hartig network. During this process, the symbiotic fungi avoid host immune rejection. Recent studies have shown that the genetic engineering of individual plant host genes can selectively inhibit or allow the colonization of specific fungi (Yu et al., 2015; Qiao et al., 2021; Wang et al., 2022). It has been determined that the G-type lectin receptor-like kinase (PtLecRLK1) in Populus tomentosa determines the colonization of Laccaria bicolor in the roots. When PtLecRLK1 was transferred to the non-host plants Arabidopsis and switchgrass, the plant allowed Laccaria bicolor to invade the root system and form a Hartig network, establishing a symbiotic relationship (Labbé et al., 2019; Qiao et al., 2021). This result suggests that PtLecRLK1 is a positive regulatory factor of the symbiosis between Laccaria bicolor and plants. DEGs encoding G-type lectin S-receptor-like serine/threonine-protein kinase were differentially expressed in My treatment, including 11 upregulated and 28 downregulated DEGs (Supplementary Table S3). Among these, Cor0108530.1, Cor0099180.1, and Cor0133230.1 were highly upregulated. The orthologs of G-type lectin S-receptor-like serine/threonine-protein kinases in other plants are crucial in regulating the symbiosis between ectomycorrhizal fungi and plants, suggesting their potential significance in the ectomycorrhiza development of My treatment.
5 Conclusion
After inoculation with S. bovista, the hazelnut root system formed an ectomycorrhizal symbiosis with the fungus. Ectomycorrhizal fungus S. bovista can increase the lateral root branching of hazelnuts, increase the root absorption area, and strongly promote the growth of hazelnut seedlings. Genes related to auxin synthesis, transportation, and signal transduction have been identified, including YUC5, AUX1, Aux/IAA, TIR1, ARF, GH3, and SAURs. They are upregulated in mycorrhiza and may be beneficial for root growth and development. Genes related to nutrient transport has been identified, including ZIP1, PHT1;9, At3g43660, COPT1, GONST2, AMT1;2, and SWEET4, and they may be are involved in zinc, phosphate, nucleotide sugar, ammonium, amino acid, and sugar transport. They are upregulated in mycorrhizal fungi, and may improve plant nutrient absorption. Finally, a set of G-type lectin S-receptor-like serine/threonine-protein kinase was identified, which was upregulated in My treatment and may be involved in the symbiotic regulation of S. bovista and host root systems. The conclusion of this study is mainly based on transcriptome data and has not been further validated for gene mutations and overexpression, therefore the conclusion has limitations. Nevertheless, this study provides a preliminary and reasonable explanation for the promotion of hazelnut seedling growth by S. bovista, which provides a scientific basis for further research and development of hazelnut mycorrhizal preparations in the future.
Scleroderma bovista can form symbiotic ectomycorrhizal fungi with hazel roots. The mechanism through which S. bovista promotes hazelnut growth remains unclear. This study aimed to evaluate the effects of the ectomycorrhizal fungus S. bovista on the growth and development of hazelnuts and gene expression changes through comparative transcriptome analysis. After inoculation with S. bovista, the fungus symbiotically formed ectomycorrhiza with hazel roots. The fresh weights of the aboveground and underground parts of My treatment (inoculated with S. bovista and formed mycorrhiza) were much higher than those of the control, respectively. The length, project area, surface area, volume, forks, and diameter of the inoculated seedlings root were 1.13 to 2.48 times higher than those of the control. In the paired comparison, 3,265 upregulated and 1,916 downregulated genes were identified. The most significantly enriched Gene Ontology term for the upregulated differentially expressed genes was GO:0005215 (transporter activity). Immunohistochemical analysis suggested that the expression levels of auxin and auxin response factor 9 were significantly increased by S. bovista after the formation of mycorrhizal fungi in hazelnut root tips. These results indicate that genes related to auxin biosynthesis, transport and signaling, and transport of nutrients may contribute to root development regulation in hazel ectomycorrhiza.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
YC: Conceptualization, Writing – review & editing. SS: Writing – review & editing, Methodology. HL: Methodology, Writing – review & editing. YD: Methodology, Writing – review & editing. HH: Methodology, Writing – review & editing. QM: Methodology, Writing – review & editing. JL: Writing – review & editing, Conceptualization, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by National Natural Science Foundation of China (Grant No. 32171840) and Jilin Provincial Department of Science and Technology (Grant No. 20210509033RQ).
Acknowledgments
The authors would like to thank all donors who supported this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1431120/full#supplementary-material
Footnotes
1. ^https://www.ncbi.nlm.nih.gov/SRA
2. ^https://daehwankimlab.github.io/hisat2/
4. ^https://ccb.jhu.edu/software/stringtie/index.shtml
5. ^http://mev.tm4.org/#/welcome
6. ^https://services.healthtech.dtu.dk/service.php?SignalP-5.0
References
Almeida, J. P., Menichetti, L., Ekblad, A., Rosenstock, N. P., and Wallander, H. (2023). Phosphorus regulates ectomycorrhizal fungi biomass production in a Norway spruce forest. Biogeosciences 20, 1443–1458. doi: 10.5194/bg-20-1443-2023
Andargie, M., and Li, J. X. (2019). Expression of the Arabidopsis SWEET genes during rice false smut infection in the transgenic Arabidopsis thaliana containing increased levels of ATP and sucrose. J. Plant Biochem. Biotechnol. 28, 509–520. doi: 10.1007/s13562-019-00512-4
Andrés-Bordería, A., Andrés, F., Garcia-Molina, A., Perea-García, A., Domingo, C., Puig, S., et al. (2017). Copper and ectopic expression of the Arabidopsis transport protein COPT1 alter iron homeostasis in rice (Oryza sativa L.). Plant Mol. Biol. 95, 17–32. doi: 10.1007/s11103-017-0622-8
Bennett, M. J., Marchant, A., Green, H. G., May, S. T., Ward, S. P., Millner, P. A., et al. (1996). Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273, 948–950. doi: 10.1126/science.273.5277.948
Borghi, L., Kang, J., Ko, D., Lee, Y., and Martinoia, E. (2015). The role of ABCG-type ABC transporters in phytohormone transport. Biochem. Soc. Trans. 43, 924–930. doi: 10.1042/BST20150106
Chen, Q., Dai, X., De-Paoli, H., Cheng, Y., Takebayashi, Y., Kasahara, H., et al. (2014). Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 55, 1072–1079. doi: 10.1093/pcp/pcu039
Chen, S., Gu, T., Qi, Z., Yan, J., Fang, Z. J., Lu, Y. T., et al. (2021). Two NPF transporters mediate iron long-distance transport and homeostasis in Arabidopsis. Plant Commun. 2:100244. doi: 10.1016/j.xplc.2021.100244
Cheng, X. F., Wu, H. H., Zou, Y. N., Wu, Q. S., and Kuča, K. (2021). Mycorrhizal response strategies of trifoliate orange under well-watered, salt stress, and waterlogging stress by regulating leaf aquaporin expression. Plant Physiol. Biochem. 162, 27–35. doi: 10.1016/j.plaphy.2021.02.026
Cheng, Y., Yang, B., Miao, R., Zhang, X., He, H., Zhao, Y., et al. (2023). Isolation, identification, and evaluation of an ectomycorrhizal fungus from a hazel orchard in China. Sci. Hortic. 309:111679. doi: 10.1016/j.scienta.2022.111679
Choi, H., Ohyama, K., Kim, Y. Y., Jin, J. Y., Lee, S. B., Yamaoka, Y., et al. (2014). The role of Arabidopsis ABCG9 and ABCG31 ATP binding cassette transporters in pollen fitness and the deposition of steryl glycosides on the pollen coat. Plant Cell 26, 310–324. doi: 10.1105/tpc.113.118935
Corrales, A., Koch, R. A., Vasco-Palacios, A. M., Smith, M. E., Ge, Z. W., and Henkel, T. W. (2022). Diversity and distribution of tropical ectomycorrhizal fungi. Mycologia 114, 919–933. doi: 10.1080/00275514.2022.2115284
David, L. C., Dechorgnat, J., Berquin, P., Routaboul, J. M., Debeaujon, I., Daniel-Vedele, F., et al. (2014). Proanthocyanidin oxidation of Arabidopsis seeds is altered in mutant of the high-affinity nitrate transporter NRT2.7. J. Exp. Bot. 65, 885–893. doi: 10.1093/jxb/ert481
Duan, X., Yu, Y., Zhang, Y., Chen, C., Duanmu, H., Cao, L., et al. (2018). A potential efflux boron transporter gene GsBOR2, positively regulates Arabidopsis bicarbonate tolerance. Plant Sci. 274, 284–292. doi: 10.1016/j.plantsci.2018.05.032
Feng, C., He, C., Wang, Y., Xu, H., Xu, K., Zhao, Y., et al. (2012). Genome-wide identification of soybean Shaker K+ channel gene family and functional characterization of GmAKT1 in transgenic Arabidopsis thaliana under salt and drought stress. J. Plant Physiol. 266:153529. doi: 10.1016/j.jplph.2021.153529
Gaitán-Solís, E., Taylor, N. J., Siritunga, D., Stevens, W., and Schachtman, D. P. (2015). Overexpression of the transporters AtZIP1 and AtMTP1 in cassava changes zinc accumulation and partitioning. Front. Plant Sci. 6:492. doi: 10.3389/fpls.2015.00492
Galichet, A., Hoyerová, K., Kamínek, M., and Gruissem, W. (2008). Farnesylation directs AtIPT3 subcellular localization and modulates cytokinin biosynthesis in Arabidopsis. Plant Physiol. 146, 1155–1164. doi: 10.1104/pp.107.107425
Geisler, M., Aryal, B., di Donato, M., and Hao, P. (2017). A critical view on ABC transporters and their interacting partners in auxin transport. Plant Cell Physiol. 58, 1601–1614. doi: 10.1093/pcp/pcx104
Giehl, R. F. H., Laginha, A. M., Duan, F., Rentsch, D., Yuan, L., and von Wirén, N. (2017). A critical role of AMT2;1 in root-to-shoot translocation of ammonium in Arabidopsis. Mol. Plant 10, 1449–1460. doi: 10.1016/j.molp.2017.10.001
Gollhofer, J., Schläwicke, C., Jungnick, N., Schmidt, W., and Buckhout, T. J. (2011). Members of a small family of nodulin-like genes are regulated under iron deficiency in roots of Arabidopsis thaliana. Plant Physiol. Biochem. 49, 557–564. doi: 10.1016/j.plaphy.2011.02.011
Hagen, G., and Guilfoyle, T. (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. doi: 10.1023/A:1015207114117
Harley, J. L. (1988). The significance of mycorrhiza. Mycol. Res. 92, 129–139. doi: 10.1016/S0953-7562(89)80001-2
Hayashi, K., Tan, X., Zheng, N., Hatate, T., Kimura, Y., Kepinski, S., et al. (2008). Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc. Natl. Acad. Sci. U.S.A. 105, 5632–5637. doi: 10.1073/pnas.0711146105
Hwang, J. U., Song, W. Y., Hong, D., Ko, D., Yamaoka, Y., Jang, S., et al. (2016). Plant ABC transporters enable many unique aspects of a terrestrial plant's lifestyle. Mol. Plant 9, 338–355. doi: 10.1016/j.molp.2016.02.003
Jiang, Y., Wang, W., Xie, Q., Liu, N., Liu, L., Wang, D., et al. (2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356, 1172–1175. doi: 10.1126/science.aam9970
Jing, B., Ishikawa, T., Soltis, N., Inada, N., Liang, Y., Murawska, G., et al. (2021). The Arabidopsis thaliana nucleotide sugar transporter GONST2 is a functional homolog of GONST1. Plant Direct 5:e00309. doi: 10.1002/pld3.309
Kanno, Y., Kamiya, Y., and Seo, M. (2013). Nitrate does not compete with abscisic acid as a substrate of AtNPF4.6/NRT1.2/AIT1 in Arabidopsis. Plant Signal. Behav. 8:e26624. doi: 10.4161/psb.26624
Karve, A., and Moore, B. D. (2009). Function of Arabidopsis hexokinase-like1 as a negative regulator of plant growth. J. Exp. Bot. 60, 4137–4149. doi: 10.1093/jxb/erp252
Kim, J. I., Baek, D., Park, H. C., Chun, H. J., Oh, D. H., Lee, M. K., et al. (2013). Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. Mol. Plant 6:337. doi: 10.1093/mp/sss100
Kretzschmar, T., Burla, B., Lee, Y., Martinoia, E., and Nagy, R. (2011). Functions of ABC transporters in plants. Essays Biochem. 50, 145–160. doi: 10.1042/bse0500145
Kuyper, T. W., and Suz, L. M. (2023). Do ectomycorrhizal trees select ectomycorrhizal fungi that enhance phosphorus uptake under nitrogen enrichment? Forests 14:467. doi: 10.3390/f14030467
Labbé, J., Muchero, W., Czarnecki, O., Wang, J., Wang, X., Bryan, A. C., et al. (2019). Mediation of plant-mycorrhizal interaction by a lectin receptor-like kinase. Nat. Plants 5, 676–680. doi: 10.1038/s41477-019-0469-x
Léran, S., Garg, B., Boursiac, Y., Corratgé-Faillie, C., Brachet, C., Tillard, P., et al. (2015). AtNPF5.5, a nitrate transporter affecting nitrogen accumulation in Arabidopsis embryo. Sci. Rep. 5:7962. doi: 10.1038/srep07962
Li, Q., Yin, M., Li, Y., Fan, C., Yang, Q., Wu, J., et al. (2015). Expression of Brassica napus TTG2, a regulator of trichome development, increases plant sensitivity to salt stress by suppressing the expression of auxin biosynthesis genes. J. Exp. Bot. 66, 5821–5836. doi: 10.1093/jxb/erv287
Liscum, E., and Reed, J. W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400. doi: 10.1007/978-94-010-0377-3_10
Liu, J., Wei, H., Zhang, X., He, H., Cheng, Y., and Wang, D. (2021). Chromosome-level genome assembly and HazelOmics database construction provides insights into unsaturated fatty acid synthesis and cold resistance in hazelnut (Corylus heterophylla). Front. Plant Sci. 12:766548. doi: 10.3389/fpls.2021.766548
Liu, J., Zhang, H., Cheng, Y., Wang, J., Zhao, Y., and Geng, W. (2014). Comparison of ultrastructure, pollen tube growth pattern and starch content in developing and abortive ovaries during the progamic phase in hazel. Front. Plant Sci. 5:528. doi: 10.3389/fpls.2014.00528
Liu, X., Zhang, Y., Yang, C., Tian, Z., and Li, J. (2016). AtSWEET4, a hexose facilitator, mediates sugar transport to axial sinks and affects plant development. Sci. Rep. 6:24563. doi: 10.1038/srep24563
Love, M., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Marella, H., Nielsen, E., Schachtman, D., and Taylor, C. G. (2013). The amino acid permeases AAP3 and AAP6 are involved in root-knot nematode parasitism of Arabidopsis. Mol. Plant-Microbe Interact. 26, 44–54. doi: 10.1094/MPMI-05-12-0123-FI
Mentewab, A., and Stewart, C. N. (2005). Overexpression of an Arabidopsis thaliana ABC transporter confers kanamycin resistance to transgenic plants. Nat. Biotechnol. 23, 1177–1180. doi: 10.1038/nbt1134
Nakazawa, M., Yabe, N., Ichikawa, T., Yamamoto, Y. Y., Yoshizumi, T., Hasunuma, K., et al. (2008). DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 25, 213–221. doi: 10.1111/j.1365-313X.2001.00957.x
Nolan, T. M., Vukašinović, N., Hsu, C. W., Zhang, J., Vanhoutte, I., Shahan, R., et al. (2023). Brassinosteroid gene regulatory networks at cellular resolution in the Arabidopsis root. Science 379:eadf4721. doi: 10.1126/science.adf4721
Ofori, P., Mizuno, A., Suzuki, M., Martinoia, E., Reuscher, S., Aoki, K., et al. (2018). Genome-wide analysis of ATP binding cassette (ABC) transporters in tomato. PLoS One 13:e0200854. doi: 10.1371/journal.pone.0200854
Petersen, T., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Qiao, Z., Yates, T., Shrestha, H., Engle, N. L., Flanagan, A., Morrell-Falvey, J. L., et al. (2021). Towards engineering ectomycorrhization into switchgrass bioenergy crops via a lectin receptor-like kinase. Plant Biotechnol. J. 19, 2454–2468. doi: 10.1111/pbi.13671
Ranocha, P., Denancé, N., Vanholme, R., Freydier, A., Martinez, Y., Hoffmann, L., et al. (2010). Walls are thin 1 (WAT1), an Arabidopsis homolog of Medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers. Plant J. 63, 469–483. doi: 10.1111/j.1365-313X.2010.04256.x
Ranocha, P., Dima, O., Nagy, R., Felten, J., Corratgé-Faillie, C., Novák, O., et al. (2013). Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 4:2625. doi: 10.1038/ncomms3625
Remy, E., Cabrito, T., Batista, R., Teixeira, M. C., Sá-Correia, I., and Duque, P. (2012). The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 195, 356–371. doi: 10.1111/j.1469-8137.2012.04167.x
Romanov, G., and Schmülling, T. (2021). Opening doors for cytokinin trafficking at the ER membrane. Trends Plant Sci. 26, 305–308. doi: 10.1016/j.tplants.2021.02.006
Schmittgen, T., and Livak, K. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shin, D., Cho, M., Kim, T., Yoo, J., Kim, J. I., Han, Y. J., et al. (2010). A small GTPase activator protein interacts with cytoplasmic phytochromes in regulating root development. J. Biol. Chem. 285, 32151–32159. doi: 10.1074/jbc.M110.133710
Shumate, A., Wong, B., Pertea, G., and Pertea, M. (2022). Improved transcriptome assembly using a hybrid of long and short reads with StringTie. PLoS Comput. Biol. 18:e1009730. doi: 10.1101/2021.12.08.471868
Teng, Z., Lyu, J., Chen, Y., Zhang, J., and Ye, N. (2023). Effects of stress-induced ABA on root architecture development: positive and negative actions. Crop J. 11, 1072–1079. doi: 10.1016/j.cj.2023.06.007
Theodoulou, F. L. (2000). Plant ABC transporters. Biochim. Biophys. Acta 1465, 79–103. doi: 10.1016/S0005-2736(00)00132-2
Tian, Q., Uhlir, N. J., and Reed, J. W. (2002). Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14, 301–319. doi: 10.1105/tpc.010283
Tikhonovich, I. A., and Provorov, N. A. (2007). Beneficial plant-microbe interactions. Compr. Mol. Phytopathology 50, 365–420. doi: 10.1002/0471263397.env212
Uroz, S., and Oger, P. (2017). Caballeronia mineralivorans sp. nov., isolated from oak-Scleroderma citrinum mycorrhizosphere. Syst. Appl. Microbiol. 40, 345–351. doi: 10.1016/j.syapm.2017.05.005
Vandenbussche, F., Petrasek, J., Zadnikova, P., Hoyerová, K., Pesek, B., Raz, V., et al. (2010). The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137, 597–606. doi: 10.1242/dev.040790
Wang, P., Lu, S., Xie, M., Wu, M., Ding, S., Khaliq, A., et al. (2020). Identification and expression analysis of the small auxin-up RNA (SAUR) gene family in apple by inducing of auxin. Gene 750:144725. doi: 10.1016/j.gene.2020.144725
Wang, N., Tang, C., Fan, X., He, M., Gan, P., Zhang, S., et al. (2022). Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rust fungi. Cell 185, 2961–2974.e19. doi: 10.1016/j.cell.2022.06.027
Wang, Y., Yuan, Z., Wang, J., Xiao, H., Wan, L., Li, L., et al. (2023). The nitrate transporter NRT2.1 directly antagonizes PIN7-mediated auxin transport for root growth adaptation. Proc. Natl. Acad. Sci. U.S.A. 120:e2221313120:e2221313120. doi: 10.1073/pnas.2221313120
Wei, H., Cheng, Y., Sun, Y., Zhang, X., He, H., and Liu, J. (2021). Genome-wide identification of the ARF gene family and ARF3 target genes regulating ovary initiation in hazel via ChIP sequencing. Front. Plant Sci. 12:766548. doi: 10.3389/fpls.2021.715820
Yu, C., Sun, C., Shen, C., Wang, S., Liu, F., Liu, Y., et al. (2015). The auxin transporter, OsAUX1, is involved in primary root and root hair elongation and in cd stress responses in rice (Oryza sativa L.). Plant J. 83, 818–830. doi: 10.1111/tpj.12929
Yu, H., Yan, J., Du, X., and Hua, J. (2018). Overlapping and differential roles of plasma membrane calcium ATPases in Arabidopsis growth and environmental responses. J. Exp. Bot. 69, 2693–2703. doi: 10.1093/jxb/ery073
Zhang, Y., Park, C., Bennett, C., Thornton, M., and Kim, D. (2021). Rapid and accurate alignment of nucleotide conversion sequencing reads with HISAT-3N. Genome Res. 31, 1290–1295. doi: 10.1101/gr.275193.120
Keywords: hazel, ectomycorrhizal fungus, comparative transcriptome, Scleroderma bovista , growth
Citation: Cheng Y, Sun S, Lou H, Dong Y, He H, Mei Q and Liu J (2024) The ectomycorrhizal fungus Scleroderma bovista improves growth of hazelnut seedlings and plays a role in auxin signaling and transport. Front. Microbiol. 15:1431120. doi: 10.3389/fmicb.2024.1431120
Edited by:
Tao Zhang, Northeast Normal University, ChinaCopyright © 2024 Cheng, Sun, Lou, Dong, He, Mei and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianfeng Liu, amlhbmZlbmdsaXUxOTc2QDE2My5jb20=
†ORCID: Jianfeng Liu, orcid.org/0000-0003-3220-8941
 Yunqing Cheng
Yunqing Cheng Siyu Sun
Siyu Sun Hongli He
Hongli He Qi Mei
Qi Mei Jianfeng Liu
Jianfeng Liu