- 1Institute of Vegetable, Hunan Academy of Agricultural Sciences, Changsha, Hunan, China
- 2Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China
- 3Key Laboratory for Vegetable Biology of Hunan Province, College of Horticulture, Hunan Agricultural University, Changsha, Hunan, China
Capsaicinoids are produced uniquely in pepper fruits, and its level determines the commercial quality and health-promoting properties of pepper. So, it is particularly important to increase capsaicinoids content in pepper. Rhizosphere microbiota is critical to plant growth and performance, and affected by plant varieties. However, the impact of pepper varieties with different capsaicinoids yields on the rhizosphere microbiota is poorly understood. Using high-throughput sequencing of the 16S rRNA and internal transcribed spacer (ITS) region, we investigated the rhizosphere microbial community among five pepper varieties containing different capsaicinoids. Our results demonstrated that pepper variety significantly influenced the diversity and structure of rhizosphere microbial community. Bacterial diversity in varieties with high capsaicinoids content was significantly higher than in varieties with low capsaicinoids content, while fungal diversity was opposite to bacterial diversity. The correlation analysis revealed that 19 dominant bacterial genera (e.g., Chujaibacter, Rhodanobacter, and Gemmatimonas) were significantly correlated with capsaicinoids content, and nine of them were also significantly associated with soil nutrients, whereas only one fungal genus (Podospora) was significantly correlated with capsaicinoids content. Additionally, almost all genera which significantly correlated to capsaicinoids content were biomarkers of the five pepper varieties and the correlation was well corresponding to the capsaicinoids content. Overall, our results confirmed that the variety of pepper significantly affected the rhizosphere microbial community in the fields, and bacteria and fungi responded differently to capsaicinoids, which may affect the biosynthesis of capsaicinoids and contribute to further improvement of capsaicinoids production in pepper fruits.
Introduction
Pepper (Capsicum) is native to the tropical and temperate Americas (Carrizo García et al., 2016). The genus Capsicum contains approximately 35 species, of which the major five cultivated species are Capsicum annuum, Capsicum baccatum, Capsicum chinense, Capsicum frutescens, and Capsicum pubescens. Among them, Capsicum annuum is the most widely cultivated species in the world-wide (Pickersgill, 1997; Carrizo García et al., 2016). Pepper displays a special pungency, which is formed by the accumulation of a kind of alkaloids synthesized in the placenta, called capsaicinoids, whose major representatives are capsaicin and dihydrocapsaicin, accounting for almost 90% of all capsaicinoids (Kozukue et al., 2005; Aza-González et al., 2011). Capsaicinoids show the potential applications range from food flavorings to therapeutics. For instance, capsaicinoids have antimicrobial properties and might be useful as biopesticides (Xing et al., 2006). It has been described that capsaicinoids protect Capsicum chacoense seeds against Fusarium, which is a major cause of predispersal seed mortality (Tewksbury et al., 2008). Additionally, capsaicinoids possess the biological properties of antitumor, antioxidant and anti-obesity, and are used for food, pharmaceutical, medical, cosmetic and dietary (Aza-González et al., 2011; Luo et al., 2011; Wang F. Z. et al., 2022).
Capsaicinoids accumulation is determined by genotype or cultivar, node position, fruiting and maturity stages, and environmental growth conditions such as light, temperature, water, and mineral nutrition (Naves et al., 2019; Uarrota et al., 2021). The degree of pungency is variable between different Capsicum cultivars. The 12 different varieties of Capsicum cultivars belonging to three species (Capsicum annuum, Capsicum chinense, and Capsicum frutescens) showed considerable variation in capsaicinoids content (Giuffrida et al., 2013). The biosynthesis of capsaicinoids is related to the convergence of the phenylpropanoid and branched-chain fatty acid pathways, which involve three nitrogenous molecules: phenylalanine, valine, and leucine (Bennett and Kirby, 1968; Leete and Louden, 1968). Currently, the raw materials for industrial production of capsaicinoids are mainly derived from the fruit, which is high cost and low yield (Zhu et al., 2019). It is necessary to improve capsaicinoids yield. Many studies have been reported that the capsaicinoids content in Capsicum fruits has been associated positively with available N (Medina-Lara et al., 2008; Aldana-Iuit et al., 2015; Zhang et al., 2020). Some reports have shown that microorganisms can promote plant growth and the accumulation of metabolites (Lv et al., 2024). The inoculation of pepper fruit with Streptomyces pactum Act12 has been demonstrated to increase the content of vitamins, phenolic acids, alkaloids, and flavonoids (Zhao et al., 2023). To date, there have been no reports of the use of microorganisms to increase capsaicinoids content in pepper.
The rhizosphere, the soil adjacent to roots, provides a niche for the interaction between plant roots and microorganisms (Singh et al., 2004; Philippot et al., 2013). The rhizosphere microbiomes provide a number of beneficial functions for plant host, including growth promotion, nutrient uptake, stress tolerance and resistance to pathogens. So, it is also referred to as the plant’s second genome, and is crucial for plant health (Berendsen et al., 2012). Plant secondary metabolites play an important role in the interaction between plants and microorganisms, which can protect plants against microbial pathogens (Cipollini, 2000). Recent studies have shown that plant secondary metabolites affect microbiome composition and function. Prominent among those metabolites are the glucosinolates (Siebers et al., 2018), flavonoids (Kudjordjie et al., 2021), coumarins (Stringlis et al., 2018; Voges et al., 2019), benzoxazinoids (Hu et al., 2018; Cotton et al., 2019; Kudjordjie et al., 2019), and triterpenes (Huang et al., 2019). Benzoxazinoids are indole-derived compounds exuded by the roots (de Bruijn et al., 2018). Three reports reveal that maize mutants unable to synthesize BXs showed significant alterations in composition of both bacterial and fungal communities (Hu et al., 2018; Cotton et al., 2019; Kudjordjie et al., 2019). The phenolic compounds coumarins are relatively abundant in the rhizosphere, and have function in iron acquisition. Two recent studies have shown that coumarins also play a key role in shaping the root microbiome composition, as shown by analyses of microbiota assemblies of coumarin-deficient Arabidopsis mutants (Stringlis et al., 2018; Voges et al., 2019). In terms of the triterpenes thalianin and arabidin, these two metabolites contribute substantially to the assembly of Arabidopsis-specific root microbiota (Huang et al., 2019). It is worth noting that many metabolites shown to affect plant microbiome also trigger changes in the composition of gut microbiome. These include the glucosinolates (Kellingray et al., 2017), terpenoids (Chen et al., 2021), flavonoids (Baky et al., 2022), and phenolic compounds (Domínguez-Avila et al., 2021).
Capsaicinoids is a kind of secondary metabolite from pepper, and many evidences suggest that capsaicinoids can influence the composition, abundance, and function of the gut microbiota (Baboota et al., 2014; Wang et al., 2020). Moreover, capsaicinoids concentration influence microbial communities and kimchi metabolites during kimchi fermentation (Park et al., 2019). Yet, whether capsaicinoids affect plant microbiome has not yet been addressed. In addition, the effects of rhizosphere microbiota on root metabolite composition and exudation have also been reported. Korenblum et al. (2020) showed that different microbial communities induce systemic changes in metabolite exudation of tomato root. For instance, bacteria affiliated with the genus Bacillus induced synthesis of acylsugars secondary metabolites. However, the relationship between rhizosphere microorganisms and the level of capsaicinoids is unknown. It has been reported that plant genotype and soil type have a significant impact on their rhizosphere microbiomes (Berg and Smalla, 2009; Lundberg et al., 2013; Yeoh et al., 2017; Veach et al., 2021). Soil available nutrient concentrations dominate the assembly of rhizosphere bacterial community (Ren et al., 2020). Furthermore, differences between plant genotypes influence the composition and function of the rhizosphere microbiomes in many plants, such as rice (Hardoim et al., 2013), corn (Peiffer et al., 2013), wheat (Meyer et al., 2010), potato (Weinert et al., 2011), and soybean (Zhong et al., 2019). However, it is largely lacking how pepper varieties affect the structure and functions in rhizosphere bacterial and fungal communities.
Here, we collected rhizosphere soil from five pepper varieties with different pungency, to analyze the rhizosphere fungal and bacterial communities by 16S rRNA gene and ITS amplicon sequencing technologies. The objective of this study was to investigate the effect of pepper varieties and its capsaicinoids content on the rhizosphere microbial community. We demonstrated that pepper variety and its capsaicinoids content significantly influenced the structure and composition of rhizosphere communities under field conditions, which may affect the biosynthesis of capsaicinoids. This will help us understand the influence of the pepper genotype and trait on the interaction between microorganisms and plants.
Materials and methods
Plant materials and field trials
Seeds of five pepper varieties, XY6, XY21, XY39, XY40, and XY42, were obtained from Hunan Xiangyan Seed Industry Co., Ltd. All of them belong to Capsicum annuum, except XY40, which is a hybrid line of Capsicum annuum and Capsicum chinense. The surface-sterilized seeds were sown in nutrient substrate. At eight leaf stage, the seedlings were transplanted to the experimental field. Field trials were performed from April to August in 2021 at the Gaoqiao experimental field of Hunan Xiangyan Seed Industry CO., LTD. Samples of pepper rhizosphere soil and pepper fruit were collected at the mature green stage. Four biological duplicates were obtained for each pepper variety, either soil or fruit samples. Soil samples were collected using the root shaking method (Inceoǧlu et al., 2010). Each soil sample was divided into two parts, one part of was stored at 4°C for physicochemical properties, the other part was stored at −80°C for sequencing analysis. The placenta tissues were isolated from fruits for capsaicinoids content determination (Tanaka et al., 2017).
Determination of capsaicinoids contents
The dried placental tissues were powdered and 0.1 g were treated with 20 mL acetonitrile using an ultrasonic machine at the temperature of 65°C for 20 min. After low-speed centrifugation, the extract solution was filtered through 0.2 μm filters (Millipore, USA). The analysis of capsaicinoids were performed by high-performance liquid chromatography (HPLC, LC-20AT, SHIMADZU, Japan). The capsaicin (C) and dihydrocapsaicin (DHC) standards or extracted samples were injected into a Shim-pack GIST C18 column (dimension 250 × 4.6 mm, particle size 5 μm). An isocratic mixture of water: acetonitrile was used as the mobile phases. The injection volume, flow rate, run time, and temperature were 10 μL, 1.0 mL/min, 30 min, and 30°C, respectively. The C and DHC were detected with the UV detector at 222 nm. The capsaicinoids content (CAPs) was calculated as (C + DHC)/0.91 (Bennett and Kirby, 1968).
Soil physicochemical analysis
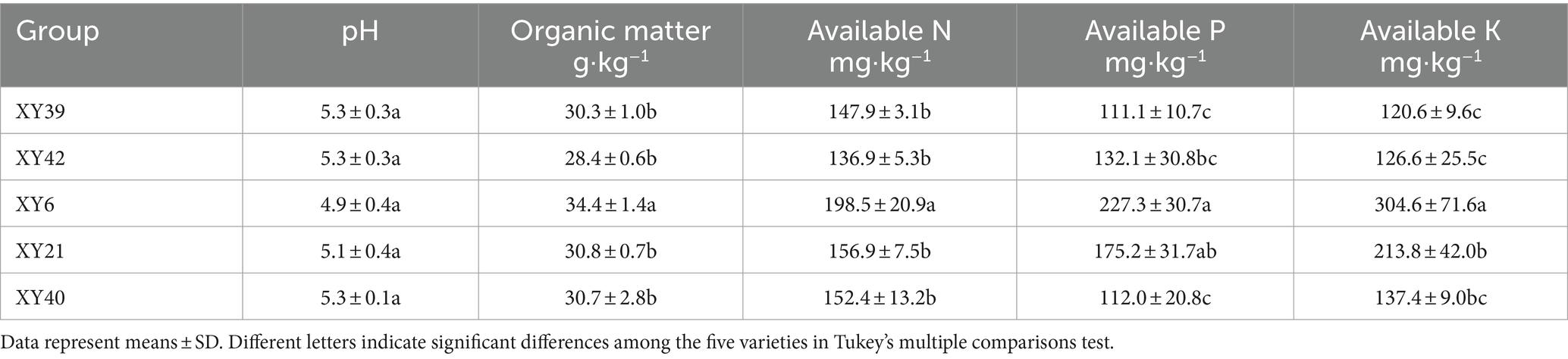
Soil pH was measured in water with a soil/water ratio of 1:2.5 (w/v) by a FE20 pH meter (Mettler-Toledo International Inc., China) (Liu et al., 2020). Soil available nitrogen (AN) was determined using the alkaline hydrolysis diffusion method (Hu et al., 2008). Soil available phosphorus (AP) and available potassium (AK) were tested according to methods described by Fan et al. (2019). Soil organic matter (OM) was determined using the K2CrO7-H2SO4 method (Bremner and Jenkinson, 1960).
16S rDNA and ITS sequencing
Total DNA from rhizosphere soil samples was extracted using the E.Z.N.A.® Soil DNA Kit (Omega, Inc., USA) according to the manufacturer’s instructions. DNA concentration and purity were determined with NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, CA, USA). The primer pairs 341F/805R (5′-CCTACGGGNGGCWGCAG-3′/5′-GACTACHVGGGTATCTAATCC-3′) and ITS1FI2/ITS2 (5′-GTGARTCATCGAATCTTTG-3′/5′-TCCTCCGCTTATTGATATGC-3′) were used to amplify the bacterial 16S V3-V4 region and fungal ITS2 region, respectively. The PCR products were confirmed with 2% agarose gel electrophoresis and purified by AMPure XT beads (Beckman Coulter Genomics, Danvers, MA, USA) and quantified by Qubit (Invitrogen, USA). Sequencing was performed at S LC-Bio Technologies (Hangzhou, China) Co., Ltd., using an Illumina NovaSeq platform.
Bioinformatic analysis
Paired-end reads was assigned to samples based on their unique barcode and truncated by cutting off the barcode and primer sequence. Paired-end reads of 16S rRNA gene and ITS gene were merged using FLASH version 1.2.8 (Magoč and Salzberg, 2011) and PEAR version 0.9.6 (Zhang et al., 2014), respectively. Quality filtering on the raw reads were performed under specific filtering conditions to obtain the high-quality clean tags according to the FQTRIM version 0.94 (Pertea, 2018). Chimeric sequences were filtered using VSEARCH software version 2.3.4 (Rognes et al., 2016). After dereplication using DADA2 (Callahan et al., 2016), we obtained feature table and feature sequence. Taxonomic assignment of 16S rRNA gene and ITS gene was performed using QIIME2 plugin feature-classifier against the SILVA v132 database (Quast et al., 2013) and UNITE 8.2 database (Nilsson et al., 2019), respectively. The relative abundance (X bacteria or fungi count/total count) is used in bacteria or fungi taxonomy. Alpha diversity and Beta diversity were analyzed by QIIME2 process (Bolyen et al., 2019), and pictures were drawn by R package (v3.5.2) (Dixon, 2003). Alpha diversity included Chao1 and Shannon indices. Beta diversity was calculated using unweighted UniFrac distance (Lozupone et al., 2007) and visualized through a principal coordinate analysis (PCoA) (Ramette, 2007). Analysis of similarities (ANOSIM) was carried out using R (version: 3.4.3) and the Vegan package in R (version: 2.3.0). The R value and p value of the ANOSIM were calculated using a permutation test with 999 permutations. Linear discriminant analysis effect size (LEfSe) analysis was performed using the OmicStudio tools1 (Kruskal–Wallis test p < 0.05, and LDA score > 3.5) (Segata et al., 2011). Spearman’s rank correlation coefficients analysis was also performed using the OmicStudio tools (|r| > 0.5 and p < 0.05).2
Statistical analysis
Means and standard deviation values were calculated using SPSS software version 19.0. Tukey’s multiple comparison testing was used to evaluate the significant differences among varieties. Figures were constructed in GraphPad Prism version 8.0 (GraphPad Software Inc., San Diego, CA, USA).
Results
Capsaicinoids content of different pepper varieties
Five varieties of pepper, XY6, XY21, XY39, XY40 and XY42, were used in this study (Figure 1A). The capsaicinoids contents (CAPs) were detected in the placenta of the five varieties (Figure 1B). XY40 and XY21 contained the highest capsaicinoids content (86.54 mg/g and 70.2 mg/g) in the placenta, and showed significantly higher capsaicinoids than XY6 (43.41 mg/g). Meanwhile, XY40, XY21 and XY6 showed significantly higher capsaicinoids than XY42 (12.99 mg/g) and XY39 (2.53 mg/g). The contents of capsaicin (C) and dihydrocapsaicin (DHC) in the placenta were in line with that of capsaicinoids.
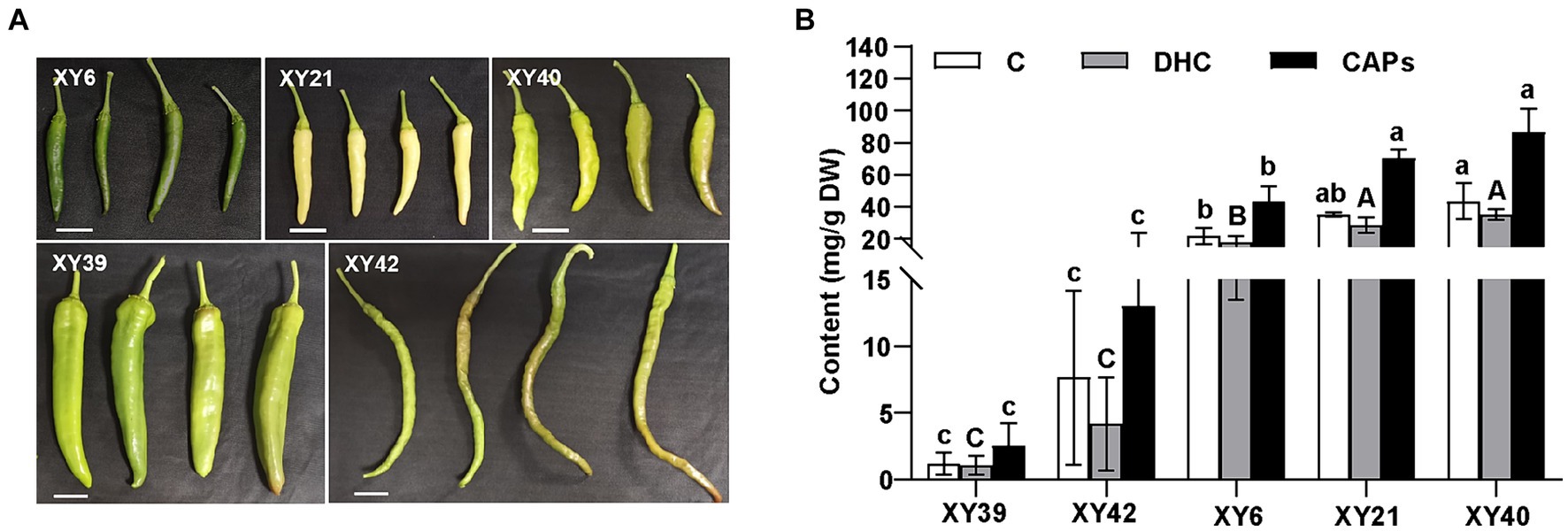
Figure 1. The capsaicinoids accumulation of pepper at the mature green stage. (A) The phenotypes of fruits of five pepper varieties. (B) The capsaicin (C), dihydrocapsaicin (DHC) and capsaicinoids (CAPs) contents in pepper placenta. Tukey’s multiple comparisons tests were performed to identify the significant differences; the same letter indicates a nonsignificant difference between the means (p > 0.05).
Soil physicochemical properties
The physicochemical characteristics of the soil from different pepper varieties were summarized in Table 1. The pH of the five different soils ranged from 4.9 to 5.3, showing no significant differences. The AK and AP contents of soils from XY6 and XY21 markedly increased compared to that of XY39 and XY42. The soil from XY6 exhibited significantly OM and AN contents compared to soils from XY21, XY40, XY39 and XY42.
Diversity of microbial community in rhizosphere soil of different pepper varieties
A total of 1,474,543 raw bacterial sequences and 1,689,023 fungal sequences were identified using Illumina MiSeq analysis. After a series of preprocessing steps, 1,158,561 bacterial and 1,633,413 fungal qualified reads were classified into 19,459 bacterial and 2,799 fungal operational taxonomic units (OTUs), respectively. Among them, 6,715 (34.51%) bacterial OTUs and 1,712 (61.16%) fungal OTUs failed to be identified to any known phylum based on the UNITE database. The amplicon sequence data for the 16S and ITS in the paper have been uploaded to the NCBI Sequence Read Archive (SRA) database with accession number PRJNA1145089.
Alpha diversity of rhizosphere microbial community of pepper was estimated by both the Chao1 index (richness) and Shannon index (diversity). The results showed that the richness and diversity of bacteria and fungi in rhizosphere soil exhibited significantly differences among different pepper varieties (Figure 2). Specifically, for bacteria, the Shannon index of XY6, XY21 and XY40 was significantly higher than that of XY39 and XY42, whereas the Chao1 index in XY6, XY21 and XY40 was significantly higher than in XY39 alone. Moreover, the Chao1 index of XY40 was significantly higher than that of XY42 (Figures 2A,B). In terms of the fungal community, the Chao1 index of XY6 and XY21 was significantly lower than that of XY39 and XY42, whereas the Shannon index in XY6 alone was significantly lower than in XY39 and XY42 (Figures 2C,D), which suggested that fungal richness and diversity were contrary to bacterial richness and diversity among different pepper varieties.
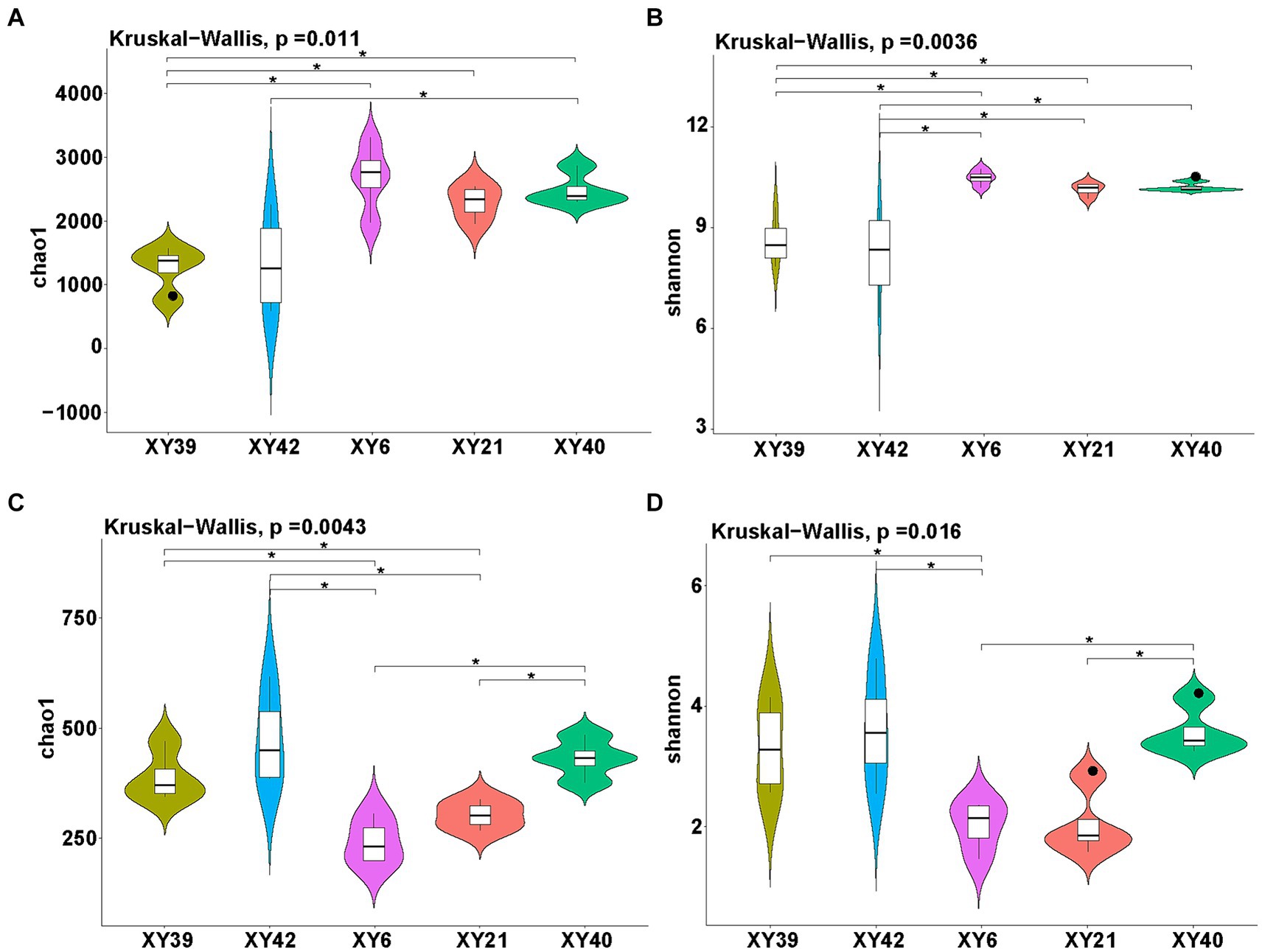
Figure 2. The diversity evaluation of rhizosphere microbial communities of five pepper varieties. (A,B) Comparison of bacterial communities among five varieties using the Chao1 index (A) and Shannon index (B). (C,D) Comparison of fungal communities among five varieties using the Chao1 index (C) and Shannon index (D). Asterisk indicates significant differences (p < 0.05).
Beta diversity of rhizosphere microbial community of pepper was also calculated by unweighted UniFrac distance. PCoA revealed significant variations in the bacterial and fungal communities among the different varieties. PCoA1 and PCoA2 of the bacterial community explained 18.39 and 6.95% of the variations, respectively (Figure 3A). Meanwhile, PCoA1 and PCoA2 of the fungal community explained 16.78 and 7.76% of the variations, respectively (Figure 3B). Furthermore, results from ANOSIM comparisons demonstrated that varieties had significant effects on the bacterial community (R = 0.4379; p = 0.001) and fungal community (R = 0.7254; p = 0.001) (Figure 3).
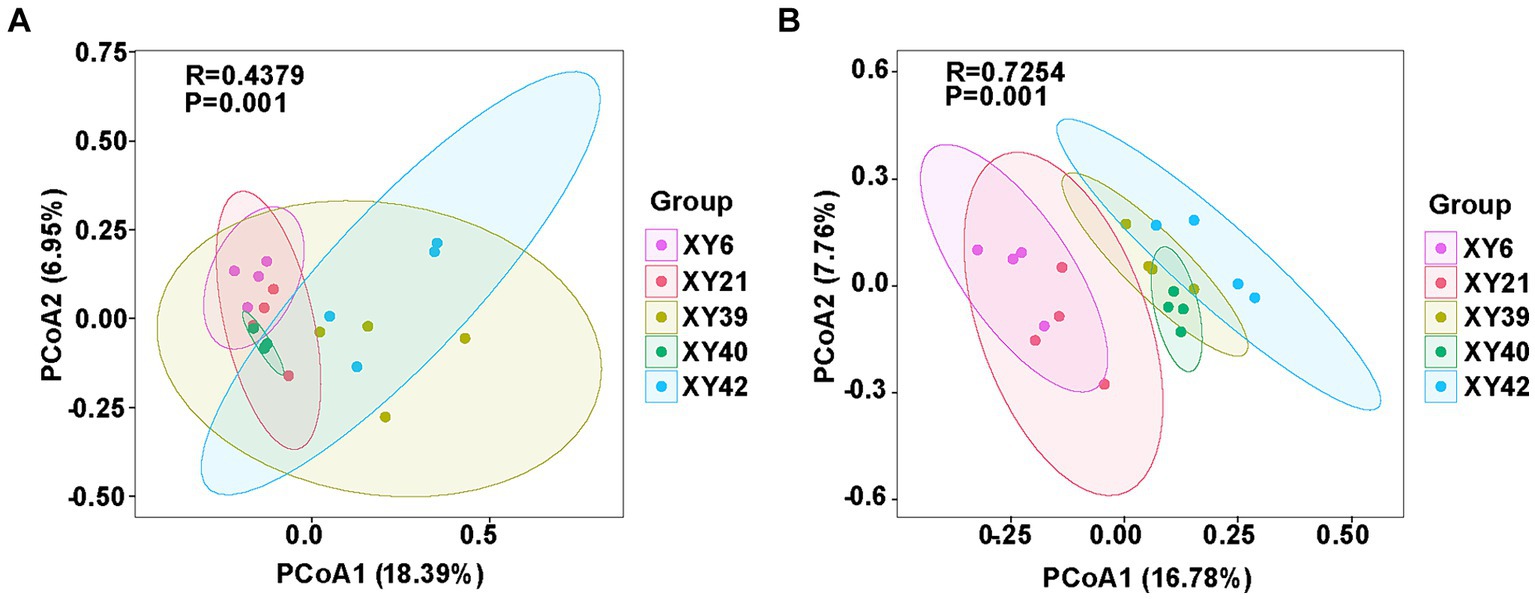
Figure 3. The comparison of rhizosphere microbial communities [(A) bacteria; (B) fungi] among five pepper varieties using PCoA and ANOSIM. PCoA, principal coordinate analysis; ANOSIM, analysis of similarities.
Microbial community composition in rhizosphere soil of different pepper varieties
At the phylum level, the dominant bacterial phyla of rhizosphere soils across all varieties were Proteobacteria, Acidobacteria, Actinobacteria, Firmicutes, and Gemmatimonadetes (Figure 4A). Proteobacteria accounted for the most significant proportion reaching to 32.93–55.46% of all OTUs. The dominant fungal phyla of rhizosphere soils across all varieties were Ascomycota and Zygomycota (Figure 4C). In term of the genus level, the most abundant bacterial genera were Chujaibacter, Bacillus, Gemmatimonadaceae_unclassified, Rhodanobacter, Acidobacteriales_unclassified, and Gemmatimonas (Figure 4B), and the predominant fungal genera were Chaetomiaceae_unclassified, Mortierella, Ascomycota_unclassified, and Thielavia in all varieties (Figure 4D).
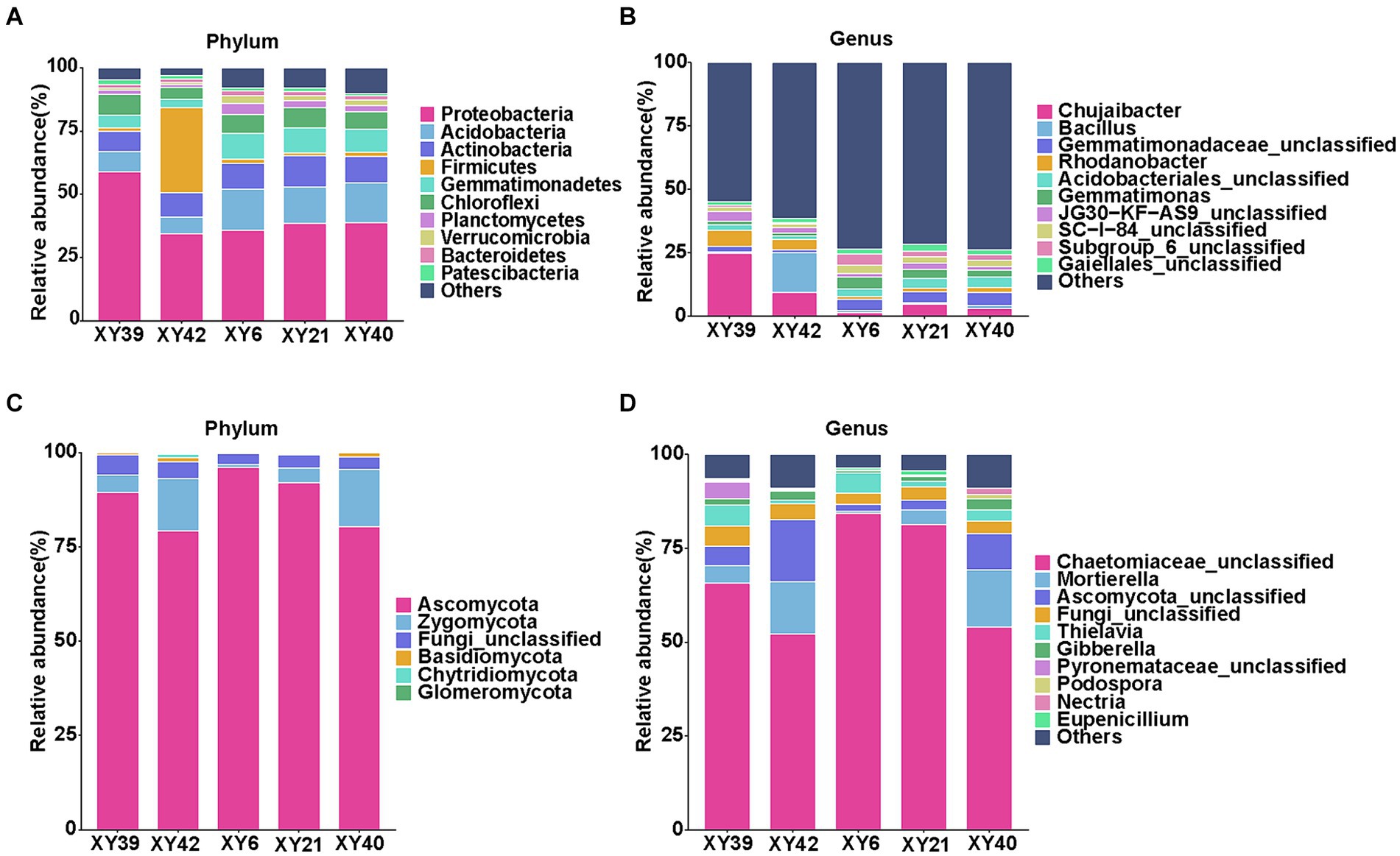
Figure 4. Community composition in the rhizosphere soil of five varieties. (A,B) Relative abundances of bacterial phylum (A) and genus (B). (C,D) Relative abundances of fungal phylum (C) and genus (D).
Then we compared the differences in bacterial phyla and genera between rhizosphere soils of five varieties. At the phylum level, Acidobacteria, Gemmatimonadetes, and Verrucomicrobia were significantly more abundant in XY6, XY21 and XY40 than XY39 and XY42 (p < 0.05, Figure 4A). The relative abundance of Firmicutes in XY42 was significantly higher than that of the other varieties (p < 0.01, Figure 4A). At the genus level, the relative abundance of Chujaibacter and Rhodanobacter was significantly decreased in XY6, XY21 and XY40 compared to XY39 and XY42. The relative abundance of Gemmatimonas in XY6 and XY21 was significantly higher than that in XY39 and XY42 (p < 0.05, Figure 4B). The relative abundance of Bacillus in XY42 was significantly higher than that in other varieties (p < 0.01, Figure 4B). Next, we compared the differences in fungal phyla and genera between rhizosphere soils of five varieties. It was found that Zygomycota was the phylum with higher relative abundance in XY42 than that in XY6 and XY21, and Glomeromycota is unique to XY42 (p < 0.01, Figure 4C). Mortierella and Gibberella were the genera with lower relative abundance in XY6 than that in other varieties (p < 0.05, Figure 4D). Compared with bacteria, the fungal differences were not particularly significant.
To further screen the rhizosphere bacteria and fungi affected by variety, the LEfSe was employed to identify biomarkers among varieties at the genus level. The results showed that there were 30 bacterial biomarkers and 7 fungal biomarkers in the rhizosphere soil of the five varieties (Figure 5). In XY39, the main bacterial biomarkers were Chujaibacter and Rhodanobacter. In XY42, Pullulanibacillus and Ammoniphilus were the main bacterial biomarkers. In XY6, Gemmatimonas, Haliangium and Nitrospira were the prominent bacterial biomarkers. In XY21, the genera that differed significantly from other varieties were Ellin6067 and Candidatus_Solibacter (Figure 5A). For XY40, the main bacterial biomarker was Anaeromyxobacter, and fungal biomarkers were Mortierella, Gibberella, and Podospora (Figures 5A,B). These results suggested that varieties markedly impacted bacterial and fungal communities in pepper.
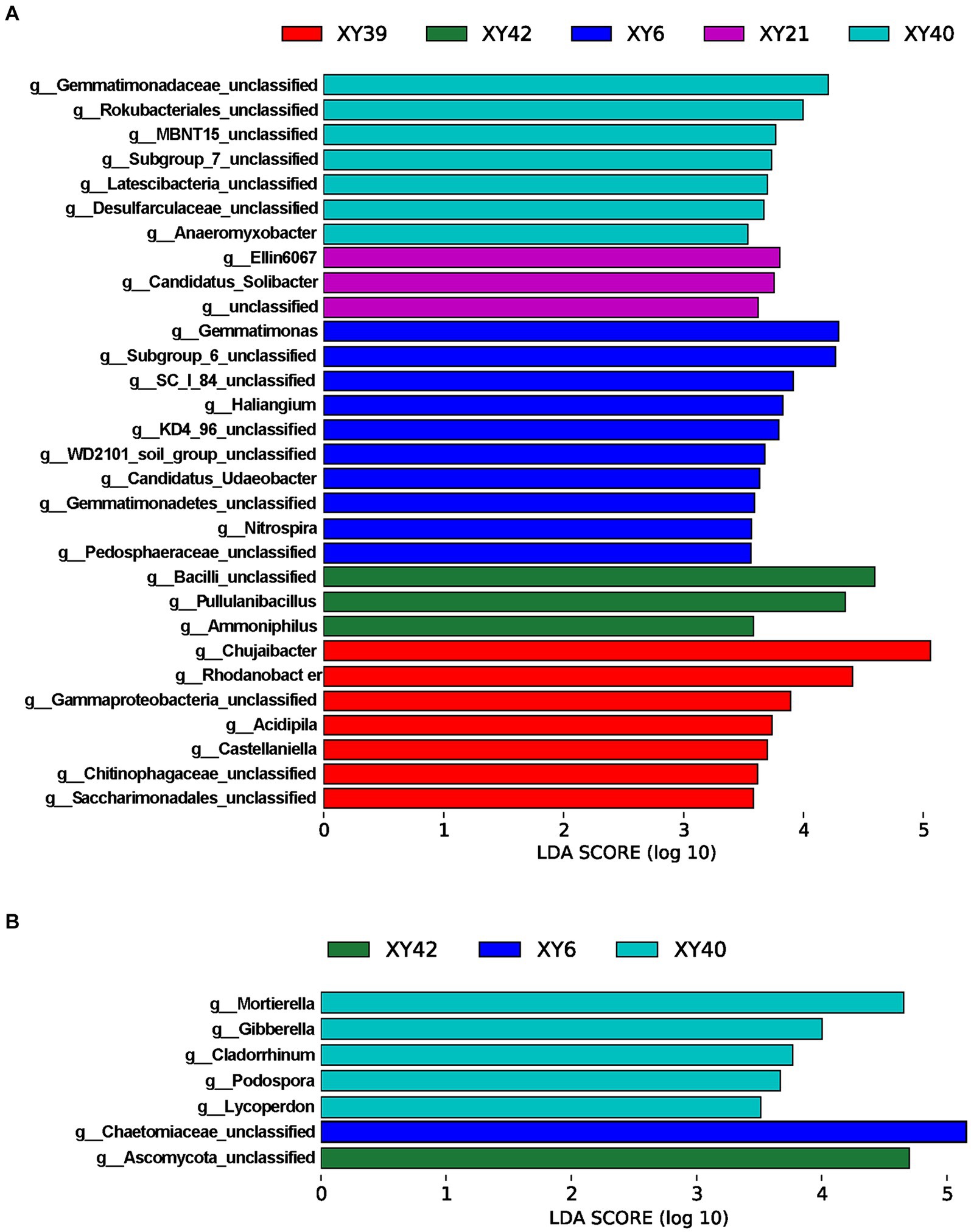
Figure 5. LDA scores of the genera biomarkers in the rhizosphere soil of five pepper varieties. (A) Bacteria. (B) Fungi. LDA scores are shown as horizontal bars for the genus biomarkers with an LDA score > 3.5 as listed, Kruskal–Wallis rank sum test, p < 0.05. LDA, linear discriminant analysis.
Relationships between soil factors, capsaicinoids content and microbial communities
To explore relationships between soil factors (shown in Table 1), capsaicinoids content (CAPs) and bacterial and fungal composition at the genus level (top 30 microbial genera), Spearman correlation analysis was carried out (Figure 6). The results showed that CAPs, AP, AK, AN and OM had significant effects on bacteria and fungi. The top 30 bacterial genus clusters could be divided into five subclusters. Subcluster 1 and subcluster 2 were negatively correlated with CAPs, AP, AK, AN and OM. Subcluster 3 and subcluster 4 were positively correlated with CAPs, AP, AK, AN and OM. Specially, Ellin6067, Gemmatimonas, Haliangium, and Candidatus_Solibacter were significantly positively correlated with CAPs, AP, AK, AN and OM, whereas Chujaibacter and Rhodanobacter were significantly negatively correlated with CAPs, AP, AK, AN and OM. Candidatus_Udaeobacter was significantly positively correlated with CAPs, AK and AN. Pullulanibacillus was significantly negatively correlated with AN and OM, and Bryobacter was significantly negatively correlated with AK (Figure 6A, p < 0.05).
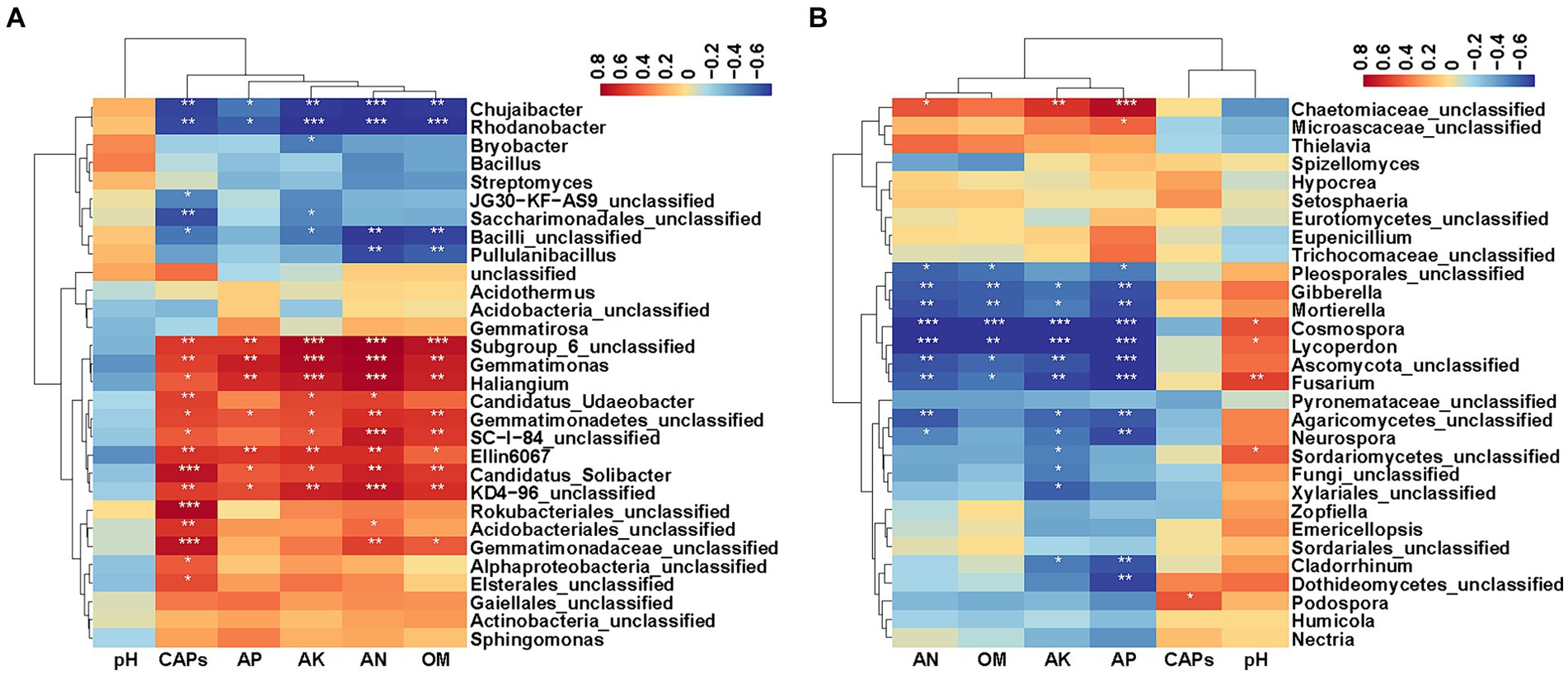
Figure 6. Heatmaps of the Spearman correlation coefficient. (A) Heatmap of the correlation between soil factors, capsaicinoids content and the top 30 abundant bacterial genera. (B) Heatmap of the correlation between soil factors, capsaicinoids content and the top 30 abundant fungal genera. Asterisks indicate significant differences, *p < 0.05; **p < 0.01; ***p < 0.001.
The top 30 fungal genus clusters could also be divided into five subclusters (Figure 6B). Subcluster 1 was positively correlated with AP, AK, AN and OM, but it was negatively correlated with pH. Subcluster 3 and subcluster 4 were negatively correlated with AP, AK, AN and OM, and positively correlated with pH. Specially, Gibberella, Mortierella, Cosmospora, Lycoperdon, and Fusarium were significantly negatively correlated with AP, AK, AN and OM. Cosmospora, Lycoperdon, and Fusarium were significantly positively correlated with pH. Neurospora was significantly negatively correlated with AP, AK, AN and Cladorrhinum was significantly negatively correlated with AP and AK. In addition, Podospora showed a significant positive correlation with CAPs (Figure 6B, p < 0.05).
Discussion
The interactions between roots and their rhizosphere microbiota are critical to plant growth and performance (Lu et al., 2018; Kwak et al., 2018). Root microbiota drive plant growth (Qu et al., 2020) and play a significant role in nutrient acquisition (Chibucos and Tyler, 2009) and soil nutrient dynamics (Hou et al., 2018). Currently, multiple reports demonstrated that the rhizosphere microbial communities are genetically regulated by host genotype and related traits (Bulgarelli et al., 2012; Bouffaud et al., 2014; Lebeis et al., 2015). At present, studies of the effect of capsaicin on microbial communities have particular focused on gut microbiota (Baboota et al., 2014; Wang et al., 2020) and pepper fermentation (Shi et al., 2023). There is currently very little evidence of the effect of capsaicin on plant inter-root microbial communities. In this study, five pepper varieties with different pungency were investigated for influences of genotypes on rhizosphere microbial communities. The Chao1 and Shannon indices revealed that there were differences in the richness and diversity of rhizosphere microorganisms among different pepper varieties (Figure 2). Furthermore, both PCoA and ANOSIM results showed that pepper varieties had significant effects on the structure of rhizosphere microorganisms (Figure 3). Those findings were in agreement with previous studies, which revealed that there were significant differences in rhizosphere microbial communities among different potato, tea and alfalfa varieties (Marques et al., 2014; Zhang et al., 2022; Qi et al., 2023). Moreover, the richness and diversity of the bacteria in XY40, XY21 and XY6 were significantly higher than that in XY39 and XY42 (Figure 2A), and XY40, XY21 and XY6 also exhibited significantly higher CAPs than XY42 and XY39 (Figure 1B), suggesting that CAPs may increase bacterial richness and diversity. However, similar results were not obtained for fungal. Only for XY6, the richness and diversity of fungal were different and were significantly lower compared to XY42 and XY39 (Figure 2B), indicating that CAPs may decrease fungal richness and diversity. These results demonstrated that bacteria and fungi showed different response to CAPs. Capsaicin has been reported to have antifungal activity against pathogens (Vuerich et al., 2023). We hypothesize that this is related to the fact that capsaicin has an antifungal effect, and that the higher the capsaicin content, the lower the abundance of the fungus, leading to this result. Side by side, this illustrates the potential of capsaicin as a bio-antifungal agent.
Proteobacteria, Acidobacteria, and Actinobacteria were the dominant bacterial phyla in rhizosphere of pepper (Figure 4A), which was consistent with previous studies on maize (Kong et al., 2019; Li et al., 2022) and Vaccinium cultivated soils (Yurgel et al., 2017; Li et al., 2020; Ramirez-Villacis et al., 2023). This result was expected because Proteobacteria members are well adapted to plant rhizosphere across diverse plant species (Philippot et al., 2013; Yang et al., 2023), and Acidobacteria is one of the most abundant bacterial phyla in the rhizosphere soil (Lee et al., 2008; Cheng et al., 2020). For fungal community, the detected fungal phyla were dominantly composed of Ascomycota in rhizosphere (Figure 4C), as reported in previous studies (Swift et al., 2021; Zahid et al., 2021; Darriaut et al., 2022; Marasco et al., 2022; Qi et al., 2023). The composition of rhizosphere community was also different among different pepper varieties (Figure 4). Our study found that Acidobacteria, Gemmatimonadetes and Verrucomicrobia were significantly higher in the rhizosphere soil from XY40, XY21 and XY6 than XY39 and XY42 (Figure 4A). Interestingly, XY40, XY21 and XY6 also exhibited significantly higher CAPs than XY42 and XY39 (Figure 1B). Therefore, we hypothesized that the difference in CAPs content may be the reason for the differences in rhizosphere bacterial community composition among different varieties. Acidobacteria, Gemmatimonadetes and Verrucomicrobia belong to oligotrophic bacteria. According to oligotrophic-copiotrophic theory, oligotrophic bacteria can maintain growth and be prevalent under nutrient-poor conditions. In contrast, copiotrophic bacteria (e.g., Proteobacteria and Firmicutes) usually are more prevalent in nutrient-rich conditions (Fierer et al., 2007). In our results, the nutrient contents (AK and AP) of soils from XY6 and XY21 were higher compared to soils from XY39 and XY42 (Table 1), which was contrary to the oligotrophic-copiotrophic theory. Additionally, the copiotrophic phylum Firmicutes also did not follow the predicted patterns (Figure 4A), suggesting that the differences in bacterial community composition of pepper were not due to soil nutrient contents. These conclusions validated our hypothesis that the changes in rhizosphere bacterial community composition of different pepper varieties were at least partially due to CAPs content. For fungal community, only phylum Zygomycota in XY42 with lower CAPs content was higher than in XY21 and XY6 with higher CAPs content (Figure 4C). The changes were not significant between different varieties, which suggested that CAPs had a greater effect on the bacterial community than fungal community.
Many bacterial taxa responded to CAPs, whereas less fungal taxa responded to CAPs. For example, seven bacterial genera, including three predominant bacterial genera (Gemmatimonas, Chujaibacter and Rhodanobacter), were significantly correlated with CAPs (Figure 6A). For the fungi, only Podospora showed a significant correlation with CAPs (Figure 6B). Moreover, the relative abundance of the three predominant bacterial genera were significantly different between high CAPs varieties (XY21 and XY6) and low CAPs varieties (XY39 and XY42) (Figure 4B). However, the relative abundance of Podospora was only significantly higher in XY40 (high CAPs) than in XY42 (low CAPs) (Figure 4D). These results further supported our hypothesis that CAPs affected microbial communities among different pepper varieties, and demonstrated the relative differences in response to CAPs of bacteria versus fungi. In addition, the seven bacterial genera were also significantly correlated with soil factors such as AP, AK, AN and OM (Figure 6A), demonstrating both CAPs and soil factors might be the reasons for the change of bacterial community, however, which factor has a greater effect on bacterial community needs to be further verified. In terms of fungal genera, Mortierella and Gibberella were significantly negatively correlated with AP, AK, AN and OM but not CAPs (Figure 6B). Furthermore, compared with other varieties, the relative abundance of Mortierella and Gibberella were lower in XY6 with the highest soil nutrient contents (AP, AK, AN and OM) (Figure 4D). However, similar results were not obtained in bacteria. These results indicated that fungal communities were more susceptible to soil environment compared to bacterial communities in the rhizosphere of pepper, which was consistent with the results in young grapevines (Darriaut et al., 2022), but contrary to the results of the study on blueberries was (Ramirez-Villacis et al., 2023).
A large number of studies have found that differences in the rhizosphere microbiome led to differences in host performance (Badri et al., 2013; Wagner et al., 2014; Fitzpatrick et al., 2018). So, we speculated that rhizosphere microbial communities can influence the biosynthesis of CAPs. Our study found that Ellin6067, Gemmatimonas, Candidatus_Udaeobacter, and Candidatus_Solibacter were significantly positively correlated with both AN and CAPs (Figure 6A). Ellin6067 and Candidatus_Solibacter belong to the phylum Acidobacteria, Gemmatimonas belongs to the phylum Gemmatimonadetes, and Candidatus_Udaeobacter belongs to the phylum Verrucomicrobia (Schoch et al., 2020). At phylum level, Acidobacteria, Gemmatimonadetes and Verrucomicrobia were significantly more abundant in high CAPs varieties (XY40, XY21 and XY6) than low CAPs varieties (XY39 and XY42) (Figure 4A). Acidobacteria and Gemmatimonadetes had the function of nitrogen fixation (Li et al., 2023). Candidatus_Udaeobacter was considered to be the potential bacteria taking part in nitrogen transformation (Gao et al., 2022). On the contrary, Chujaibacter and Rhodanobacter were significantly negatively correlated with both AN and CAPs (Figure 6A). It was reported that Rhodanobacter had the ability of denitrification (Prakash et al., 2012). Chujaibacter was also considered to be the predominant population involved in nitrogen removal (Xin et al., 2021). N availability in soils directly affected CAPs production (Medina-Lara et al., 2008; Aldana-Iuit et al., 2015; Zhang et al., 2020). These conclusions supported our hypothesis that these bacteria might regulate biosynthesis of CAPs by increasing the N availability of soils. In addition, almost all bacterial genera that significantly correlated with CAPs content could be found in the biomarkers list analyzed by LEfSe (Figure 5A). Chujaibacter and Rhodanobacter, with a negative correlation with CAPs, corresponded to the biomarkers of XY39 with a lower CAPs content, while Gemmatimonas, Haliangium, Ellin6067 and Candidatus_Solibacter, with a positive correlation with CAPs, corresponded to the biomarkers of XY6 and XY21 with a higher CAPs content (Figures 1B, 5A, 6A). Previous studies have found that there were differences in the rhizosphere bacterial community structure between local varieties and artificially selected strains of pepper (Wang C. et al., 2022; Wang et al., 2023), which was consistent with our findings, but more importantly, this study is the first to demonstrate that CAPs are also the main factor affecting the rhizosphere microbial community among different pepper varieties.
In summary, we analyzed the effects of different pepper varieties on rhizosphere microbial community, and explored the role of rhizosphere microorganisms in improving CAPs content of pepper. The results reported here indicate that pepper variety significantly impacts the structure and composition of pepper rhizosphere microbial communities. Changes in microbial communities were significantly associated with CAPs among different pepper varieties, and the bacterial communities were more impacted by the CAPs than the fungi. These findings help to better understand the complex interactions between host plants and microbiome. It also offers the possibility of manipulating the plant rhizosphere microbiome as an effective strategy to improve CAPs content of pepper.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
XiL: Formal analysis, Investigation, Writing – original draft. YZ: Formal analysis, Investigation, Writing – original draft. CZ: Writing – review & editing. XuL: Writing – review & editing. XZ: Writing – review & editing. LO: Conceptualization, Data curation, Supervision, Writing – review & editing. YT: Conceptualization, Data curation, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported the National Key Research and Development Program of China (2022YFD1700100), Agricultural Science and Technology Innovation Project of Hunan Province (2022cx39), Zou Xuexiao Academician Innovation Workstation Platform Construction Support Project (TL2023YF007), and National Bulk Vegetable Industry Technology System-Improvement of Pepper Variety (CARS-23-A07).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Aldana-Iuit, J. G., Sauri-Duch, E., Miranda-Ham, M. D. L., Castro-Concha, L. A., Cuevas-Glory, L. F., and Vázquez-Flota, F. A. (2015). Nitrate promotes capsaicin accumulation in Capsicum chinense immobilized placentas. Biomed. Res. Int. 2015, 1–6. doi: 10.1155/2015/794084
Aza-González, C., Núñez-Palenius, H. G., and Ochoa-Alejo, N. (2011). Molecular biology of capsaicinoid biosynthesis in chili pepper (Capsicum spp.). Plant Cell Rep. 30, 695–706. doi: 10.1007/s00299-010-0968-8
Baboota, R. K., Murtaza, N., Jagtap, S., Singh, D. P., Karmase, A., Kaur, J., et al. (2014). Capsaicin-induced transcriptional changes in hypothalamus and alterations in gut microbial count in high fat diet fed mice. J. Nutr. Biochem. 25, 893–902. doi: 10.1016/j.jnutbio.2014.04.004
Badri, D. V., Zolla, G., Bakker, M. G., Manter, D. K., and Vivanco, J. M. (2013). Potential impact of soil microbiomes on the leaf metabolome and on herbivore feeding behavior. New Phytol. 198, 264–273. doi: 10.1111/nph.12124
Baky, M. H., Elshahed, M., Wessjohann, L., and Farag, M. A. (2022). Interactions between dietary flavonoids and the gut microbiome: a comprehensive review. Br. J. Nutr. 128, 577–591. doi: 10.1017/S0007114521003627
Bennett, D. J., and Kirby, G. W. (1968). Constitution and biosynthesis of capsaicin. J. Chem. Soc. C Org. 4, 442–446. doi: 10.1039/J39680000442
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berg, G., and Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654.x
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bouffaud, M. L., Poirier, M. A., Muller, D., and Moënne-Loccoz, Y. (2014). Root microbiome relates to plant host evolution in maize and other Poaceae. Environ. Microbiol. 16, 2804–2814. doi: 10.1111/1462-2920.12442
Bremner, J. M., and Jenkinson, D. S. (1960). Determination of organic carbon in soil. I. Oxidation by dichromate of organic matter in soil and plant materials. Eur. J. Soil Sci. 11, 394–402. doi: 10.1111/J.1365-2389.1960.TB01093.X
Bulgarelli, D., Rott, M., Schlaeppi, K., Loren, V., van Themaat, E., Ahmadinejad, N., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Callahan, B., McMurdie, P., Rosen, M., Han, A., Johnson, A., and Holmes, S. (2016). DADA2: high resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Carrizo García, C., Barfuss, M. H. J., Sehr, E. M., Barboza, G. E., Samuel, R., Moscone, E. A., et al. (2016). Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Ann. Bot. 118, 35–51. doi: 10.1093/aob/mcw079
Chen, J., Ding, X., Wu, R., Tong, B., Zhao, L., Lv, H., et al. (2021). Novel sesquiterpene glycoside from loquat leaf alleviates type 2 diabetes mellitus combined with nonalcoholic fatty liver disease by improving insulin resistance, oxidative stress, inflammation, and gut microbiota composition. J. Agric. Food Chem. 69, 14176–14191. doi: 10.1021/acs.jafc.1c05596
Cheng, Z., Lei, S., Li, Y., Huang, W., Ma, R., Xiong, J., et al. (2020). Revealing the variation and stability of bacterial communities in tomato rhizosphere microbiota. Microorganisms 8, 1–15. doi: 10.3390/microorganisms8020170
Chibucos, M. C., and Tyler, B. M. (2009). Common themes in nutrient acquisition by plant symbiotic microbes, described by the gene ontology. BMC Microbiol. 9, S6–S8. doi: 10.1186/1471-2180-9-S1-S6
Cipollini, M. L. (2000). Secondary metabolites of vertebrate-dispersed fruits: evidence for adaptive functions. Rev. Chil. Hist. Nat. 73, 421–440. doi: 10.4067/s0716-078x2000000300006
Cotton, T. E. A., Pétriacq, P., Cameron, D. D., Meselmani, M. A., Schwarzenbacher, R., Rolfe, S. A., et al. (2019). Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME J. 13, 1647–1658. doi: 10.1038/s41396-019-0375-2
Darriaut, R., Antonielli, L., Martins, G., Ballestra, P., Vivin, P., Marguerit, E., et al. (2022). Soil composition and rootstock genotype drive the root associated microbial communities in young grapevines. Front. Microbiol. 13, 1–21. doi: 10.3389/fmicb.2022.1031064
de Bruijn, W. J. C., Gruppen, H., and Vincken, J. P. (2018). Structure and biosynthesis of benzoxazinoids: plant defence metabolites with potential as antimicrobial scaffolds. Phytochemistry 155, 233–243. doi: 10.1016/j.phytochem.2018.07.005
Dixon, P. (2003). Computer program review VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Domínguez-Avila, J. A., Villa-Rodriguez, J. A., Montiel-Herrera, M., Pacheco-Ordaz, R., Roopchand, D. E., Venema, K., et al. (2021). Phenolic compounds promote diversity of gut microbiota and maintain colonic health. Dig. Dis. Sci. 66, 3270–3289. doi: 10.1007/s10620-020-06676-7
Fan, K., Delgado-Baquerizo, M., Guo, X., Wang, D., Wu, Y., Zhu, M., et al. (2019). Suppressed N fixation and diazotrophs after four decades of fertilization. Microbiome 7, 143–110. doi: 10.1186/s40168-019-0757-8
Fierer, N., Bradford, M. A., and Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Fitzpatrick, C. R., Copeland, J., Wang, P. W., Guttman, D. S., Kotanen, P. M., and Johnson, M. T. J. (2018). Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl. Acad. Sci. USA 115, E1157–E1165. doi: 10.1073/pnas.1717617115
Gao, Y., Yuan, L., Du, J., Wang, H., Yang, X., Duan, L., et al. (2022). Bacterial community profile of the crude oil-contaminated saline soil in the Yellow River Delta natural reserve, China. Chemosphere 289:133207. doi: 10.1016/j.chemosphere.2021.133207
Giuffrida, D., Dugo, P., Torre, G., Bignardi, C., Cavazza, A., Corradini, C., et al. (2013). Characterization of 12 Capsicum varieties by evaluation of their carotenoid profile and pungency determination. Food Chem. 140, 794–802. doi: 10.1016/j.foodchem.2012.09.060
Hardoim, P. R., Nazir, R., Sessitsch, A., Elhottová, D., Korenblum, E., Van Overbeek, L. S., et al. (2013). The new species Enterobacter oryziphilus sp. nov. and Enterobacter oryzendophyticus sp. nov. are key inhabitants of the endosphere of rice. BMC Microbiol. 13, 164–113. doi: 10.1186/1471-2180-13-164
Hou, D., Wang, R., Gao, X., Wang, K., Lin, Z., Ge, J., et al. (2018). Cultivar-specific response of bacterial community to cadmium contamination in the rhizosphere of rice (Oryza sativa L.). Environ. Pollut. 241, 63–73. doi: 10.1016/j.envpol.2018.04.121
Hu, L., Robert, C. A. M., Cadot, S., Zhang, X., Ye, M., Li, B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9, 2738–2713. doi: 10.1038/s41467-018-05122-7
Hu, X., Su, S., Wang, Z., and Zhang, M. (2008). Study on characteristics of nitrogen from slow-release compound fertilizers using alkal-i hydrolyzed reduction diffusing method. Plant Nutr. Fertil. Sci. 14, 373–378. doi: 10.11674/zwyf.2008.0227
Huang, A. C., Jiang, T., Liu, Y. X., Bai, Y. C., Reed, J., Qu, B., et al. (2019). A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 364:eaau6389. doi: 10.1126/science.aau6389
Inceoǧlu, Ö., Salles, J. F., Van Overbeek, L., and Van Elsas, J. D. (2010). Effects of plant genotype and growth stage on the betaproteobacterial communities associated with different potato cultivars in two fields. Appl. Environ. Microbiol. 76, 3675–3684. doi: 10.1128/AEM.00040-10
Kellingray, L., Tapp, H. S., Saha, S., Doleman, J. F., Narbad, A., and Mithen, R. F. (2017). Consumption of a diet rich in Brassica vegetables is associated with a reduced abundance of sulphate-reducing bacteria: a randomised crossover study. Mol. Nutr. Food Res. 61, 1–11. doi: 10.1002/mnfr.201600992
Kong, X., Han, Z., Tai, X., Jin, D., Ai, S., Zheng, X., et al. (2019). Maize (Zea mays L. Sp.) varieties significantly influence bacterial and fungal community in bulk soil, rhizosphere soil and phyllosphere. FEMS Microbiol. Ecol. 96, 1–11. doi: 10.1093/femsec/fiaa020
Korenblum, E., Dong, Y., Szymanski, J., Panda, S., Jozwiak, A., Massalha, H., et al. (2020). Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. U. S. A. 117, 3874–3883. doi: 10.1073/pnas.1912130117
Kozukue, N., Han, J. S., Kozukue, E., Lee, S. J., Kim, J. A., Lee, K. R., et al. (2005). Analysis of eight capsaicinoids in peppers and pepper-containing foods by high-performance liquid chromatography and liquid chromatography - mass spectrometry. J. Agric. Food Chem. 53, 9172–9181. doi: 10.1021/jf050469j
Kudjordjie, E. N., Sapkota, R., and Nicolaisen, M. (2021). Arabidopsis assemble distinct root-associated microbiomes through the synthesis of an array of defense metabolites. PLoS One 16, e0259171–e0259119. doi: 10.1371/journal.pone.0259171
Kudjordjie, E. N., Sapkota, R., Steffensen, S. K., Fomsgaard, I. S., and Nicolaisen, M. (2019). Maize synthesized benzoxazinoids affect the host associated microbiome. Microbiome 7, 59–17. doi: 10.1186/s40168-019-0677-7
Kwak, M. J., Kong, H. G., Choi, K., Kwon, S. K., Song, J. Y., Lee, J., et al. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1100–1109. doi: 10.1038/nbt.4232
Lebeis, S. L., Paredes, S. H., Lundberg, D. S., Breakfield, N., Gehring, J., McDonald, M., et al. (2015). Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864. doi: 10.1126/science.aaa8764
Lee, S. H., Ka, J. O., and Cho, J. C. (2008). Members of the phylum Acidobacteria are dominant and metabolically active in rhizosphere soil. FEMS Microbiol. Lett. 285, 263–269. doi: 10.1111/j.1574-6968.2008.01232.x
Leete, E., and Louden, M. C. L. (1968). Biosynthesis of capsaicin and dihydrocapsaicin in Capsicum frutescens. J. Am. Chem. Soc. 90, 6837–6841. doi: 10.1021/ja01026a049
Li, J., Mavrodi, O. V., Hou, J., Blackmon, C., Babiker, E. M., and Mavrodi, D. V. (2020). Comparative analysis of rhizosphere microbiomes of southern highbush blueberry (Vaccinium corymbosum L.), darrow’s blueberry (V. darrowii camp), and rabbiteye blueberry (V. virgatum Aiton). Front. Microbiol. 11:370. doi: 10.3389/fmicb.2020.00370
Li, Y., Qu, Z., Xu, W., Chen, W., Hu, Y., and Wang, Z. (2022). Maize (Zea mays L.) genotypes induce the changes of rhizosphere microbial communities. Arch. Microbiol. 204, 321–313. doi: 10.1007/s00203-022-02934-6
Li, Y., Xiong, L., Yu, H., Zeng, K., Wei, Y., Li, H., et al. (2023). Function and distribution of nitrogen-cycling microbial communities in the Napahai plateau wetland. Arch. Microbiol. 205, 357–316. doi: 10.1007/s00203-023-03695-6
Liu, W., Jiang, L., Yang, S., Wang, Z., Tian, R., Peng, Z., et al. (2020). Critical transition of soil bacterial diversity and composition triggered by nitrogen enrichment. Ecology 101:e03053. doi: 10.1002/ecy.3053
Lozupone, C. A., Hamady, M., Kelley, S. T., and Knight, R. (2007). Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73, 1576–1585. doi: 10.1128/AEM.01996-06
Lu, T., Ke, M., Lavoie, M., Jin, Y., Fan, X., Zhang, Z., et al. (2018). Rhizosphere microorganisms can influence the timing of plant flowering. Microbiome 6, 231–212. doi: 10.1186/s40168-018-0615-0
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2013). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90. doi: 10.1038/nature11237
Luo, X. J., Peng, J., and Li, Y. J. (2011). Recent advances in the study on capsaicinoids and capsinoids. Eur. J. Pharmacol. 650, 1–7. doi: 10.1016/j.ejphar.2010.09.074
Lv, J. Y., Yang, S. Y., Zhou, W., Liu, Z. W., Tan, J. F., and Wei, M. (2024). Microbial regulation of plant secondary metabolites: impact, mechanisms and prospects. Microbiol. Res. 283:127688. doi: 10.1016/j.micres.2024.127688
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Marasco, R., Alturkey, H., Fusi, M., Brandi, M., Ghiglieno, I., Valenti, L., et al. (2022). Rootstock–scion combination contributes to shape diversity and composition of microbial communities associated with grapevine root system. Environ. Microbiol. 24, 3791–3808. doi: 10.1111/1462-2920.16042
Marques, J. M., da Silva, T. F., Vollu, R. E., Blank, A. F., Ding, G. C., Seldin, L., et al. (2014). Plant age and genotype affect the bacterial community composition in the tuber rhizosphere of field-grown sweet potato plants. FEMS Microbiol. Ecol. 88, 424–435. doi: 10.1111/1574-6941.12313
Medina-Lara, F., Echevarría-Machado, I., Pacheco-Arjona, R., Ruiz-Lau, N., Guzmán-Antonio, A., and Martinez-Estevez, M. (2008). Influence of nitrogen and potassium fertilization on fruiting and capsaicin content in habanero pepper (Capsicum chinense Jacq.). HortScience 43, 1549–1554. doi: 10.21273/hortsci.43.5.1549
Meyer, J. B., Lutz, M. P., Frapolli, M., Péchy-Tarr, M., Rochat, L., Keel, C., et al. (2010). Interplay between wheat cultivars, biocontrol pseudomonads, and soil. Appl. Environ. Microbiol. 76, 6196–6204. doi: 10.1128/AEM.00752-10
Naves, E. R., de Ávila Silva, L., Sulpice, R., Araújo, W. L., Nunes-Nesi, A., Peres, L. E. P., et al. (2019). Capsaicinoids: pungency beyond Capsicum. Trends Plant Sci. 24, 109–120. doi: 10.1016/j.tplants.2018.11.001
Nilsson, R. H., Larsson, K. H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2019). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264. doi: 10.1093/nar/gky1022
Park, B., Yang, J. S., Moon, E. W., Seo, H. Y., and Ha, J. H. (2019). Influence of capsaicinoids content on the microbial community during kimchi fermentation. J. Microbiol. Biotechnol. 29, 1580–1590. doi: 10.4014/jmb.1907.07023
Peiffer, J. A., Spor, A., Koren, O., Jin, Z., Tringe, S. G., Dangl, J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U. S. A. 110, 6548–6553. doi: 10.1073/pnas.1302837110
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and Van Der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Pickersgill, B. (1997). Genetic resources and breeding of Capsicum spp. Euphytica 96, 129–133. doi: 10.1023/A:1002913228101
Prakash, O., Green, S. J., Jasrotia, P., Overholt, W. A., Canion, A., Watson, D. B., et al. (2012). Rhodanobacter denitrificans sp. nov., isolated from nitrate-rich zones of a contaminated aquifer. Int. J. Syst. Evol. Microbiol. 62, 2457–2462. doi: 10.1099/ijs.0.035840-0
Qi, J., Fu, D., Wang, X., Zhang, F., and Ma, C. (2023). The effect of alfalfa cultivation on improving physicochemical properties soil microorganisms community structure of grey desert soil. Sci. Rep. 13, 13747–13713. doi: 10.1038/s41598-023-41005-8
Qu, Q., Zhang, Z., Peijnenburg, W. J. G. M., Liu, W., Lu, T., Hu, B., et al. (2020). Rhizosphere microbiome assembly and its impact on plant growth. J. Agric. Food Chem. 68, 5024–5038. doi: 10.1021/acs.jafc.0c00073
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ramette, A. (2007). Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 62, 142–160. doi: 10.1111/j.1574-6941.2007.00375.x
Ramirez-Villacis, D. X., Pinos-Leon, A., Vega-Polo, P., Salas-González, I., Jones, C. D., and Torres, M. D. L. (2023). Untangling the effects of plant genotype and soil conditions on the assembly of bacterial and fungal communities in the rhizosphere of the wild andean blueberry (Vaccinium floribundum Kunth). Microorganisms 11:399. doi: 10.3390/microorganisms11020399
Ren, Y., Xun, W., Yan, H., Ma, A., Xiong, W., Shen, Q., et al. (2020). Functional compensation dominates the assembly of plant rhizospheric bacterial community. Soil Biol. Biochem. 150:107968. doi: 10.1016/j.soilbio.2020.107968
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Schoch, C. L., Ciufo, S., Domrachev, M., Hotton, C. L., Kannan, S., Khovanskaya, R., et al. (2020). NCBI taxonomy: a comprehensive update on curation, resources and tools. Database 2020, 1–21. doi: 10.1093/database/baaa062
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Shi, Q., Tang, H. H., Mei, Y., Chen, J. F., Wang, X. R., Liu, B. Q., et al. (2023). Effects of endogenous capsaicin stress and fermentation time on the microbial succession and flavor compounds of chili paste (a Chinese fermented chili pepper). Food Res. Int. 168:112763. doi: 10.1016/j.foodres.2023.112763
Siebers, M., Rohr, T., Ventura, M., Schütz, V., Thies, S., Kovacic, F., et al. (2018). Disruption of microbial community composition and identification of plant growth promoting microorganisms after exposure of soil to rapeseed-derived glucosinolates. PLoS One 13:e0200160. doi: 10.1371/journal.pone.0200160
Singh, B. K., Millard, P., Whiteley, A. S., and Murrell, J. C. (2004). Unravelling rhizosphere-microbial interactions: opportunities and limitations. Trends Microbiol. 12, 386–393. doi: 10.1016/j.tim.2004.06.008
Stringlis, I. A., Yu, K., Feussner, K., De Jonge, R., Van Bentum, S., Van Verk, M. C., et al. (2018). MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. U. S. A. 115, E5213–E5222. doi: 10.1073/pnas.1722335115
Swift, J. F., Hall, M. E., Harris, Z. N., Kwasniewski, M. T., and Miller, A. J. (2021). Grapevine microbiota reflect diversity among compartments and complex interactions within and among root and shoot systems. Microorganisms 9, 1–20. doi: 10.3390/microorganisms9010092
Tanaka, Y., Nakashima, F., Kirii, E., Goto, T., Yoshida, Y., and Yasuba, K. (2017). Difference in capsaicinoid biosynthesis gene expression in the pericarp reveals elevation of capsaicinoid contents in chili peppers (Capsicum chinense). Plant Cell Rep. 36, 267–279. doi: 10.1007/s00299-016-2078-8
Tewksbury, J. J., Carlo, A., Haak, D. C., Reagan, K. M., Machnicki, N. J., Levey, D. J., et al. (2008). Evolutionary ecology of pungency in wild chilies. Proc. Natl. Acad. Sci. USA 105, 11808–11811. doi: 10.1073/pnas.0802691105
Uarrota, V. G., Maraschin, M., de Bairros, Â. F. M., and Pedreschi, R. (2021). Factors affecting the capsaicinoid profile of hot peppers and biological activity of their non-pungent analogs (Capsinoids) present in sweet peppers. Crit. Rev. Food Sci. Nutr. 61, 649–665. doi: 10.1080/10408398.2020.1743642
Veach, A. M., Morris, R., Yip, D. Z., Yang, Z. K., Engle, N. L., Cregger, M. A., et al. (2021). Rhizosphere microbiomes diverge among Populus trichocarpa plant-host genotypes and chemotypes, but it depends on soil origin. Microbiome 9, 1–15. doi: 10.1186/s40168-021-01003-2
Voges, M. J. E. E. E., Bai, Y., Schulze-Lefert, P., and Sattely, E. S. (2019). Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc. Natl. Acad. Sci. U. S. A. 116, 12558–12565. doi: 10.1073/pnas.1820691116
Vuerich, M., Petrussa, E., Filippi, A., Cluzet, S., Fonayet, J. V., Sepulcri, A., et al. (2023). Antifungal activity of chili pepper extract with potential for the control of some major pathogens in grapevine. Pest Manag. Sci. 79, 2503–2516. doi: 10.1002/ps.7435
Wagner, M. R., Lundberg, D. S., Coleman-Derr, D., Tringe, S. G., Dangl, J. L., and Mitchell-Olds, T. (2014). Natural soil microbes alter flowering phenology and the intensity of selection on flowering time in a wild Arabidopsis relative. Ecol. Lett. 17, 717–726. doi: 10.1111/ele.12276
Wang, F., Huang, X., Chen, Y., Zhang, D., Chen, D., Chen, L., et al. (2020). Study on the effect of capsaicin on the intestinal flora through high-throughput sequencing. ACS Omega 5, 1246–1253. doi: 10.1021/acsomega.9b03798
Wang, C., Qu, Y., Wang, S., Zhao, Q., Xie, Z., and Zhang, X. (2022). Analysis of rhizosphere bacterial community characteristics in different genotypes Capsicum frutescens seedlings. Trop. Agric. Sci. Technol. 45, 23–28. doi: 10.16005/j.cnki.tast.2022.04.006
Wang, C., Su, T., Zhang, X., Li, Z., Xu, J., Wang, S., et al. (2023). Rhizosphere bacterial community structure in different pepper varieties (strains) at seedling stage. Southwest China J. Agric. Sci. 36, 1636–1643. doi: 10.16213/j.cnki.scjas.2023.8.007
Wang, F. Z., Xue, Y., Fu, L., Wang, Y. T., He, M. X., Zhao, L., et al. (2022). Extraction, purification, bioactivity and pharmacological effects of capsaicin: a review. Crit. Rev. Food Sci. Nutr. 62, 5322–5348. doi: 10.1080/10408398.2021.1884840
Weinert, N., Piceno, Y., Ding, G. C., Meincke, R., Heuer, H., Berg, G., et al. (2011). PhyloChip hybridization uncovered an enormous bacterial diversity in the rhizosphere of different potato cultivars: many common and few cultivar-dependent taxa. FEMS Microbiol. Ecol. 75, 497–506. doi: 10.1111/j.1574-6941.2010.01025.x
Xin, X., Liu, S., Qin, J., Ye, Z., Liu, W., Fang, S., et al. (2021). Performances of simultaneous enhanced removal of nitrogen and phosphorus via biological aerated filter with biochar as fillers under low dissolved oxygen for digested swine wastewater treatment. Bioprocess Biosyst. Eng. 44, 1741–1753. doi: 10.1007/s00449-021-02557-z
Xing, F., Cheng, G., and Yi, K. (2006). Study on the antimicrobial activities of the capsaicin microcapsules. J. Appl. Polym. Sci. 102, 1318–1321. doi: 10.1002/app.23766
Yang, Y., Chai, Y., Xie, H., Zhang, L., Zhang, Z., Yang, X., et al. (2023). Responses of soil microbial diversity, network complexity and multifunctionality to three land-use changes. Sci. Total Environ. 859:160255. doi: 10.1016/j.scitotenv.2022.160255
Yeoh, Y. K., Dennis, P. G., Paungfoo-Lonhienne, C., Weber, L., Brackin, R., Ragan, M. A., et al. (2017). Evolutionary conservation of a core root microbiome across plant phyla along a tropical soil chronosequence. Nat. Commun. 8, 215–219. doi: 10.1038/s41467-017-00262-8
Yurgel, S. N., Douglas, G. M., Comeau, A. M., Mammoliti, M., Dusault, A., Percival, D., et al. (2017). Variation in bacterial and eukaryotic communities associated with natural and managed wild blueberry habitats. Phytobiomes J. 1, 102–113. doi: 10.1094/PBIOMES-03-17-0012-R
Zahid, M. S., Li, D., Javed, H. U., Sabir, I. A., Wang, L., Jiu, S., et al. (2021). Comparative fungal diversity and dynamics in plant compartments at different developmental stages under root-zone restricted grapevines. BMC Microbiol. 21, 1–16. doi: 10.1186/s12866-021-02376-y
Zhang, Z., Ge, S. B., Fan, L. C., Guo, S., Hu, Q., Ahammed, G. J., et al. (2022). Diversity in rhizospheric microbial communities in tea varieties at different locations and tapping potential beneficial microorganisms. Front. Microbiol. 13:1027444. doi: 10.3389/fmicb.2022.1027444
Zhang, J., Kobert, K., Flouri, T., and Stamatakis, A. (2014). PEAR: a fast and accurate Illumina paired-end reAd mergeR. Bioinformatics 30, 614–620. doi: 10.1093/bioinformatics/btt593
Zhang, J., Lv, J., Xie, J., Gan, Y., Coulter, J. A., Yu, J., et al. (2020). Nitrogen source affects the composition of metabolites in pepper (Capsicum annuum L.) and regulates the synthesis of capsaicinoids through the GOGAT–GS pathway. Food Secur. 9, 1–21. doi: 10.3390/foods9020150
Zhao, Y. S., Sun, C. Y., Wang, S. Z., Zhang, M. L., Li, Y. L., Xue, Q. H., et al. (2023). Widely targeted metabolomic, transcriptomic, and metagenomic profiling reveal microbe-plant-metabolic reprogramming patterns mediated by Streptomyces pactum Act12 enhance the fruit quality of Capsicum annuum L. Food Res. Int. 166:112587. doi: 10.1016/j.foodres.2023.112587
Zhong, Y., Yang, Y., Liu, P., Xu, R., Rensing, C., Fu, X., et al. (2019). Genotype and rhizobium inoculation modulate the assembly of soybean rhizobacterial communities. Plant Cell Environ. 42, 2028–2044. doi: 10.1111/pce.13519
Keywords: pepper, varieties, capsaicinoids, microbial community, rhizosphere
Citation: Li X, Zhang Y, Zhou C, Li X, Zou X, Ou L and Tao Y (2024) The changes of rhizosphere microbial communities in pepper varieties with different capsaicinoids. Front. Microbiol. 15:1430682. doi: 10.3389/fmicb.2024.1430682
Edited by:
Xiancan Zhu, Anhui Normal University, ChinaReviewed by:
Jun Zhao, Nanjing Normal University, ChinaDevendra Jain, Maharana Pratap University of Agriculture and Technology, India
Zhiyong Pan, Huazhong Agricultural University, China
Copyright © 2024 Li, Zhang, Zhou, Li, Zou, Ou and Tao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Tao, dGFveXUxMjM0NTZAMTI2LmNvbQ==; Lijun Ou, b3U5NTcyQGh1bmF1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Xin Li
Xin Li Yan Zhang
Yan Zhang Chi Zhou
Chi Zhou Xuefeng Li1
Xuefeng Li1 Xuexiao Zou
Xuexiao Zou Lijun Ou
Lijun Ou Yu Tao
Yu Tao