- 1Laboratory of Agro-Alimentary and Health, Faculty of Sciences and Techniques, Hassan First University of Settat, Settat, Morocco
- 2Department of Biology, Faculty of Biology, Alexandru Ioan Cuza University of Iasi, Iași, Romania
- 3Center of Biomedical Research, Romanian Academy, Iasi Branch, Iași, Romania
- 4Academy of Romanian Scientists, Bucharest, Romania
- 5CENEMED Platform for Interdisciplinary Research, “Grigore T. Popa” University of Medicine and Pharmacy of Iasi, Iasi, Romania
- 6Regional Center of Agronomic Research of Settat, Settat, Morocco
- 7Laboratory of Biochemistry, Neurosciences, Natural Resources and Environment, Faculty of Sciences and Techniques, Hassan First University of Settat, Settat, Morocco
- 8Higher Institute of Nursing Professions and Health Technical (ISPITS), Marrakech, Morocco
- 9Laboratory of Physical Chemistry of Processes and Materials, Faculty of Sciences and Techniques, Hassan First University, Settat, Morocco
- 10Department of Chemistry, “Ioan Haulica” Institute, Apollonia University, Iași, Romania
- 11Urology Department, Grigore T. Popa University of Medicine and Pharmacy, Iași, Romania
- 12Family Medicine Department, Grigore T. Popa University of Medicine and Pharmacy, Iași, Romania
The alarming rise in antimicrobial resistance (AMR) has created a significant public health challenge, necessitating the discovery of new therapeutic agents to combat infectious diseases and oxidative stress-related disorders. The Lentzea flaviverrucosa strain E25-2, isolated from Moroccan forest soil, represents a potential avenue for such research. This study aimed to identify the isolate E25-2, obtained from soil in a cold Moroccan ecosystem, and further investigate its antimicrobial and antioxidant activities. Phylogenetic analysis based on 16S rRNA gene sequences revealed the strain’s classification within the Lentzea genus, with a sequence closely resembling that of Lentzea flaviverrucosa AS4.0578 (96.10% similarity). Antimicrobial activity in solid media showed moderate to strong activity against Staphylococcus aureus ATCC 25923, Bacillus cereus strain ATCC 14579, Escherichia coli strain ATCC 25922, Candida albicans strain ATCC 60193 and 4 phytopathogenic fungi. In addition, ethyl acetate extract of this isolate demonstrated potent antimicrobial activity against 7 clinically multi-drug resistant bacteria. Furthermore, it demonstrated antioxidant activity against 2,2-diphenyl-1-picrylhydrazyl (DPPH) and 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) free radicals, as well as a significant increase in ferric reducing antioxidant power. A significant positive correlation was observed between antioxidant activities and total content of phenolic compounds (p < 0.0001), along with flavonoids (p < 0.0001). Furthermore, gas chromatography-mass spectrometry (GC-MS) analysis revealed the presence of amines, hydroxyl groups, pyridopyrazinone rings, esters and pyrrolopyrazines. The Lentzea genus could offer promising prospects in the fight against antibiotic resistance and in the prevention against oxidative stress related diseases.
1 Introduction
The advent of antibiotics in the 20th century revolutionized the treatment of infectious diseases, marking a new era in medical science (Browne et al., 2020). These “magic bullets” have saved countless lives by effectively targeting and eliminating bacterial pathogens (da Cunha et al., 2019). However, the widespread and often indiscriminate use of antibiotics has created a significant public health challenge with antimicrobial resistance (AMR). Nowadays, antimicrobial-resistant infections have become the third leading cause of death, following cardiovascular diseases and cancer (Meštrović et al., 2023). AMR occurs when microorganisms such as bacteria, viruses, fungi, and parasites evolve mechanisms to withstand the drugs designed to kill them (World Health Organization, 2023). This phenomenon not only renders standard treatments ineffective but also increases the risk of severe illness and death (Tang et al., 2023). In 2019, approximately 1.27 million deaths were attributed to infections caused by antimicrobial resistance (AMR), and nearly 5 million deaths were linked to multidrug-resistant (MDR) infections (Salam et al., 2023). By 2050, this number is expected to rise to 10 million annually, greatly exceeding the mortality rates of cancer (Will 10 Million People Die a Year due to Antimicrobial Resistance by 2050?). One of the earliest known “superbugs,” methicillin-resistant Staphylococcus aureus (MRSA), is linked to a high death toll from AMR infections globally (World Health Organization, 2023). Presently, 3.5% of active tuberculosis cases and 18% of previously treated cases are categorized as multidrug-resistant tuberculosis (MDR-TB) worldwide, with increasing concern over extensively drug-resistant tuberculosis (XDR-TB) in many MDR-TB cases (Uddin et al., 2021).
Given the alarming rise in infectious diseases and bacterial resistance, it is essential to discover new natural bioactive compounds as alternatives to antibiotics to combat the progression toward multidrug resistance (MDR). In this context, it was reported that the most effective source for discovering powerful new bioactive compounds is natural products derived from Microorganisms including bacteria (Hug et al., 2020). Bacteria produce secondary metabolites, which are not necessary for survival but provide advantages under specific conditions (Hug et al., 2020). These secondary metabolites exhibit diverse biological functions and are used as antifungals, anticancer, immunosuppressive, and antibiotics compounds (Barka et al., 2016; Jones and Elliot, 2017). Actinobacteria are massive antibiotic producers, accounting for almost two-thirds of all known antibiotics used in medicine (Lu et al., 2019). They are an important and inexhaustible source for the natural production of antibiotics, and they remain a source of interest for the discovery of new antibiotics (Genilloud, 2017). Furthermore, the Actinobacteria are recognized for their biocontrol potential due to their ability to produce bioactive compounds (Barka et al., 2016).
The Lentzea genus is a Gram-positive group, aerobic, non-motile, filamentous Actinobacteria characterized by branching aerial mycelia that fragment into rod-like elements (Cao et al., 2015). The Lentzea genome contains between 68.6 and 79.6% guanine and cytosine (G + C) (Cao et al., 2015). As part of our program focused on isolating Actinobacteria from the soils of the Azrou forest in the Fez-Meknes region of Morocco, aiming to explore their chemical diversity and potential in antibiotic discovery (Rammali et al., 2022), we came across a rare Actinobacteria strain identified as Lentzea sp. strain E25-2. Remarkably, the bioactive compounds produced by this genus, except for Lentzea violacea, have not been widely documented (Hussain et al., 2017). In this study, we first identified the genus of isolate E25-2, obtained from the soil of a cold Moroccan ecosystem. Secondly, our aim is to explore its biological properties, particularly focusing on its antimicrobial and antioxidant activities.
2 Materials and methods
2.1 Soil sampling, isolation and conservation
In February and early March 2019, a pure strain, designated E25-2, was isolated from a soil sample from an unexploited cold ecosystem in the Azrou forest, in the Fez-Meknes region of Morocco (GPS: 33° 26′ 28″ N 5° 13′ 22″ W). The soil sample was collected at five separate points over an area of 400 m2 following the described sampling method by Sengupta et al. (2015) and underwent pretreatment by the enrichment method (Kitouni et al., 2005). For Actinobacteria isolation, M2 medium supplemented with 50 mg/L actidione (cycloheximide) (Bouaziz et al., 2016) was used. Strain E25-2 was grown on ISP2 agar medium, incubated at 28°C, and subsequently stored temporarily in inclined tubes at 4°C and long-term in a 20% glycerol solution at −20°C (Marimuthu et al., 2020).
2.2 Genotypic identification
The genomic DNA extraction from isolate E25-2, the amplification and sequencing reactions, was carried out following the protocol described in our previous work (Rammali et al., 2022, 2024). The obtained 16S rRNA gene sequence for isolate E25-2 was aligned with related Lentzea genus sequences using MEGA-X software (Kumar et al., 2018). Phylogenetic tree construction employed the neighbor-joining tree method (Saitou and Nei, 1987). Sequence similarity was assessed using GenBank and EzBioCloud genomic databases.
2.3 Phenotypic characteristics of E25-2 isolate
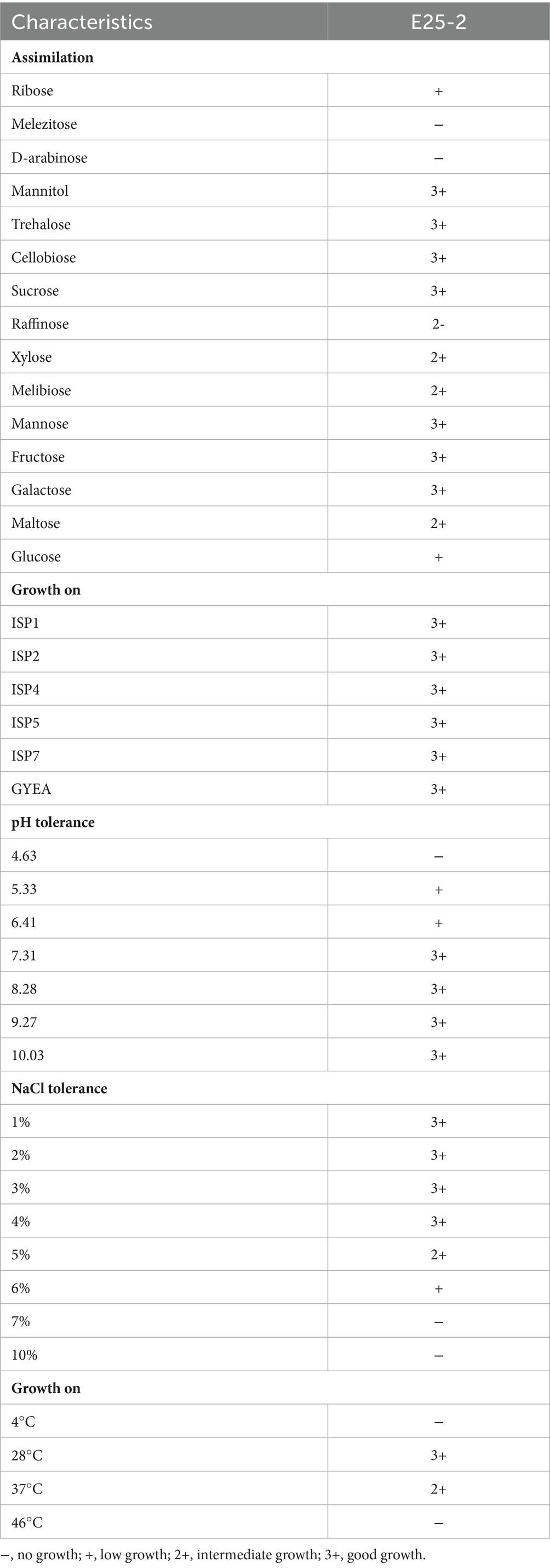
According to Shirling and Gottlieb (1966), the cultural characteristics of isolate E25-2 were observed on ISP (International Streptomyces Project) media (ISP-1, ISP-2, ISP-4, ISP-5 and GYEA). The cell morphology of the isolate was determined by light microscopy with a digital camera (Olympus CX43RF) in its native condition and subsequent to Gram staining, employing the slide culture method (Williams and Cross, 1971). Melanoid pigment production, tolerance to various concentrations of NaCl (1, 2, 3, 4, 5, 7 and 10%), tolerance to different pH values (4.63, 5.33, 6.41, 7.31, 8.28, 9.27 and 10.03), growth at different temperature levels (4, 28, 37 and 46°C) and assimilation of carbohydrates and their derivatives as the sole carbon source were evaluated in this study (Singh et al., 2016).
2.4 Preliminary evaluation of antimicrobial activity
Primary screening to evaluate the antimicrobial activity of the E25-2 isolate was performed using the double-layer method described by Badji et al. (2005), utilizing four agar media (Bennett, ISP1, ISP2, and GYEA) against various microorganisms: Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 25923, Bacillus cereus ATCC 14579, and Candida albicans ATCC 60193 (obtained from the Institut Pasteur collection in Casablanca, Morocco), as well as Fusarium sp. MN944575, Fusarium sp. MN944576, and Trichoderma longibrachiatum M37 (these phytopathogenic fungi were obtained from the Laboratory of Agro-Alimentary and Health, Faculty of Sciences and Techniques, Hassan First University of Settat, Morocco).
2.5 Secondary evaluation of antimicrobial activity
2.5.1 Preparation of the extract of E25-2 isolate
The fermentation and secondary metabolite extraction of the E25-2 strain were conducted according to the protocol outlined in our previous work (Rammali et al., 2022, 2023, 2024). Antimicrobial activity in organic extracts from isolate E25-2.
The assessment of the antimicrobial efficacy of extracts derived from the organic phase of the E25-2 isolate was conducted using the disk diffusion method (Badji et al., 2005). Evaluation involved testing against Escherichia coli strain ATCC 25922, Staphylococcus aureus strain ATCC 25923, Bacillus cereus ATCC 14579, Candida albicans ATCC 60193, and 5 clinical MDR strains (Enterococcus strain 18k1386, Staphylococcus aureus strain 18k1052, Proteus vulgaris strain 16C1737, Neisseria gonorrhoeae strain 16D1170, Escherichia coli strain 16D1150). The antibiotic resistance profile of the clinical MDR strains tested was verified against 16 antibiotics (Supplementary Table S3). These MDR strains were obtained from the collection of the Pasteur Settat Morocco Medical Analysis Laboratory. Prior to antimicrobial testing, bacterial cells were harvested and adjusted to an optical density (OD) of 0.08–0.13 at 625 nm, approximately corresponding to 106 CFU/mL, using a spectrophotometer (Selectra VR2000, Barcelona, Spain) (CASFM, 2020). Similarly, for antifungal activity assessments, inoculum optical densities were maintained within the 0.18–0.20 range at 623 nm, corresponding to a concentration of approximately 106 spores/mL (Bastide et al., 1986). DMSO-impregnated discs of equivalent volume served as negative controls. Streptomycin was used for antibacterial activity assessment, while cycloheximide served as the positive control for antifungal activities.
2.6 Total phenolic and flavonoid contents
The total phenolic contents quantification in the ethyl acetate extract of the E25-2 isolate was performed using the Folin–Ciocalteu method, as outlined by Bensadón et al. (2010). The absorbance was recorded at 760 nm, with gallic acid used as standard. Additionally, the total flavonoid contents were determined according to Bahorun et al. (2006). The absorbance was recorded at 415 nm, quercetin as standard.
2.7 In vitro antioxidant activity tests
The DPPH assay of ethyl acetate extract of E25-2 isolate was carried out following the protocol of Blois (1965). The absorbance was measured at 517 nm. Ascorbic acid was used as a positive antioxidant. The ABTS assay of ethyl acetate extract of E25-2 isolate was assessed following the method previously adopted by Re et al. (1999). The result of the reaction was recorded by measuring absorbance at 734 nm. Trolox was used as a positive antioxidant. Concerning FRAP, it was determined according to Oyaizu (1986). The absorbance was measured at 700 nm. With ascorbic acid used as the positive antioxidant. In all assays, a spectrophotometer (Selectra VR2000, Barcelona, Spain) was employed to measure the absorbance.
2.8 Evaluating toxicity of ethyl acetate extract of E25-2 isolate
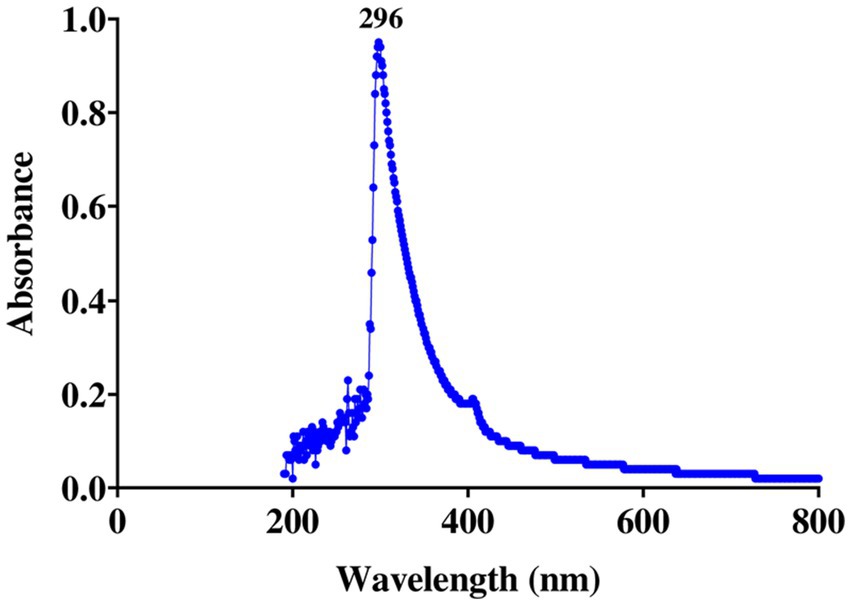
The UV–visible spectrophotometer was used to measure the absorption spectra of the ethyl acetate extract from the E25-2 isolate, solubilized in DMSO, across the wavelength range of 190 to 850 nm (HACH lange DR6000) (Bastide et al., 1986).
2.9 Evaluating toxicity of E25-2 isolate using in vitro method hemolysis
The hemolytic activity of ethyl acetate extract of E25-2 isolate was performed following the methods described by our group (Rammali et al., 2024). The degree of hemolysis was expressed as the hemolysis rate, calculated according to the following formula:
2.10 Gas chromatography-mass spectrometry analysis
The gas chromatography-mass spectrometry (GC-MS) analysis was performed according to the protocol detailed in our previous work (Rammali et al., 2022, 2024).
2.11 Statistical analysis
The experiments were conducted in triplicate, and the results were presented as the mean value along with the standard deviation (SD). To assess differences between groups in phenolic compound, flavonoid compound, and antioxidant activity assays, the GraphPad Prism 8.4.3 software was used. Specifically, we performed a standard two-way ANOVA followed by Tukey’s multiple comparisons test. Statistical significance was set at p < 0.05. Pearson correlation analysis was conducted using GraphPad Prism 8.4.3 software to assess the relationship between total phenolic and flavonoid compounds and antioxidant activity.
3 Results
3.1 16S rRNA PCR and phylogenetic approaches
The sequencing analysis revealed that the almost complete sequence of the 16S rRNA gene of isolate E25-2 was 1,103 bp (GenBank accession number: OR865862) (Supplementary Table S1). By comparing the aligned sequence of the 16S rRNA gene of isolate E25-2 with the corresponding partial sequences of the 16S rRNA gene from type strains accessible in the GenBank genomic database, we were able to determine the taxonomic classification of isolate E25-2. According to the results, strain E25-2 demonstrates approximately 97% similarity with the genus Lentzea (Supplementary Table S1). The phylogenetic tree indicated that the E25-2 strain exhibited a 16S rRNA sequence similarity of 96.10% with Lentzea flaviverrucosa AS4.0578 (Figure 1).

Figure 1. The phylogenetic tree, based on the 16S rRNA gene, was constructed using the Neighbour-Joining Tree statistical method using MEGA X software. The 16S rRNA gene sequence analysis illustrates the evolutionary relationship between Lentzea sp. strain E25-2, indicated by a red circle, and its closest known taxa. The bar (0.02) represents the number of substitutions per nucleotide position. GenBank accession numbers are provided in brackets. Cryptobacterium curtum was used as the outgroup in the analysis. T, type strain.
3.2 Phenotypic analysis of Lentzea sp. E25-2 isolate
The culture characteristics analysis revealed that isolate E25-2 exhibits growth on all tested agar culture media (ISP1, ISP2, ISP4, ISP5, ISP7, and GYEA) (Table 1). Colonies of isolate E25-2 are star-shaped, ranging from 1 to 6 mm in diameter (Supplementary Figure S1). The aerial mycelium, observed on the surface of the medium, has a whitish, non-powdery appearance, while the substrate mycelium on the underside of the medium, has an orange-yellow color (Supplementary Table S2). Under light microscopy in the fresh state and after Gram staining, isolate E25-2 was identified as a filamentous, immobile Gram-positive bacterium. It displayed segmented, branched aerial mycelium under observation (Supplementary Figure S2). The isolate exhibits robust growth on all tested agar types (ISP1, ISP2, ISP4, ISP5, ISP7 and GYEA) after 1 week of aerobic incubation at 28°C. The isolate E25-2 is negative for melanoid pigment production. In fact, no diffusible pigments were observed on the culture media. Moreover, it grows in a pH range from 5.33 to 10.03, with optimal growth at pH 8.28, and is unable to grow at pH below 5 (Table 1). The isolate E25-2 tolerates NaCl concentrations from 1 to 6%, but no growth above 7%. The carbon source identification test showed that this isolate can assimilate all carbohydrate compounds: D-xylose, mannitol, D-raffinose, cellobiose, sucrose, D-galactose, melibiose, ribose, D-mannose, D-fructose, trehalose, maltose, and glucose except Melezitose and D-arabinose (Table 1).
3.3 Antimicrobial potential of isolate E25-2
Isolate E25-2 exhibited antimicrobial activity on all the agar media tested, with significantly higher activity observed on ISP2 and Bennett media. Conversely, ISP1 and GYEA provided less favorable conditions for secondary metabolite production (Figures 2, 3). The antimicrobial assays revealed significant activity of isolate E25-2 against both Gram-positive bacteria (Staphylococcus aureus ATCC 25923, Bacillus cereus ATCC 14579) and Gram-negative bacteria (Escherichia coli ATCC 25922), as well as Candida albicans ATCC 60193. Furthermore, isolate E25-2 demonstrated notable antimicrobial activity against two phytopathogenic fungi Fusarium sp. MN944576 and Fusarium sp. MN944575, as well as Trichoderma longibrachiatum (M37), which is known to cause invasive lung infections.
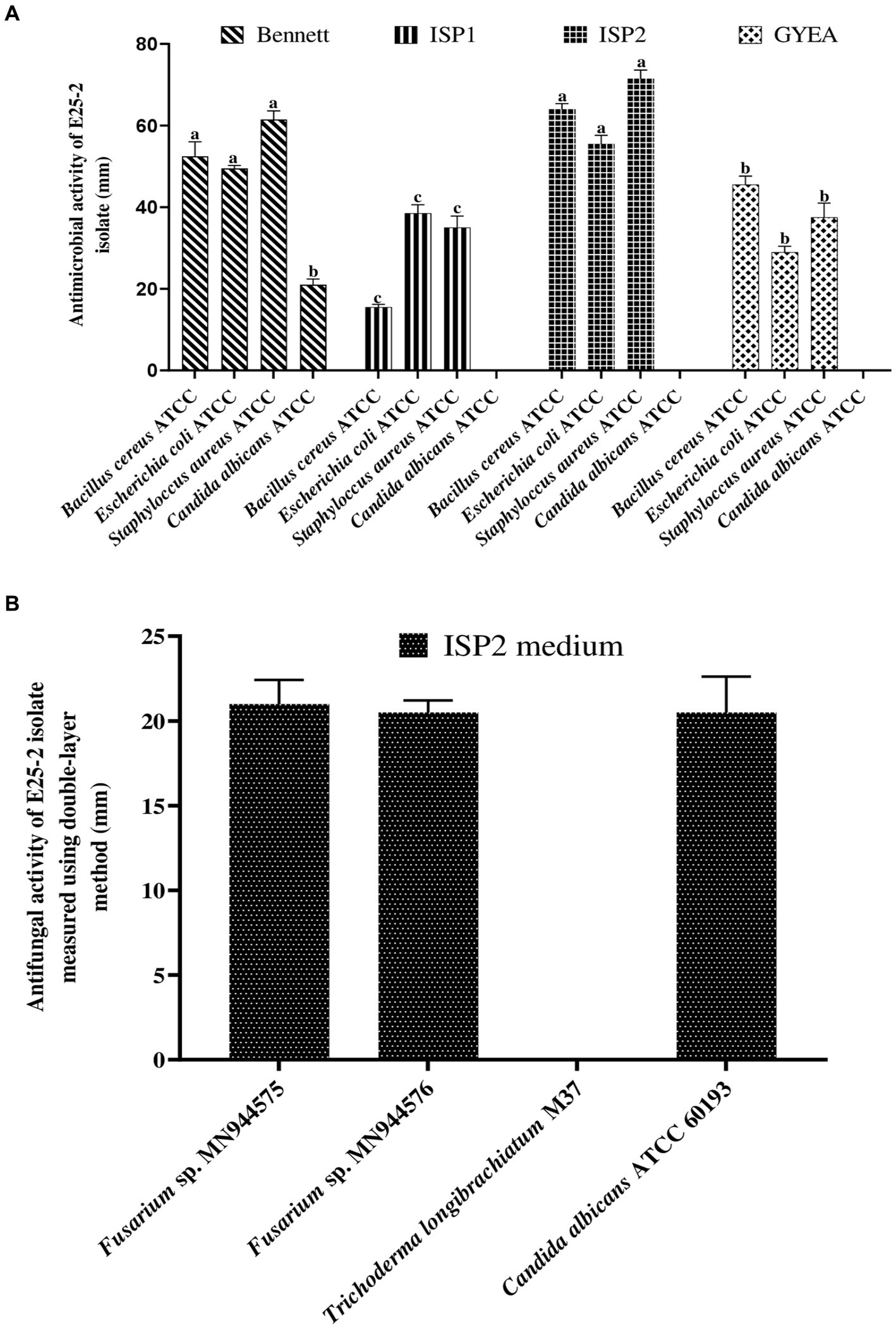
Figure 2. Antibacterial (A) and antifungal (B) activities of isolate E25-2 assessed via double layer method on solid media.
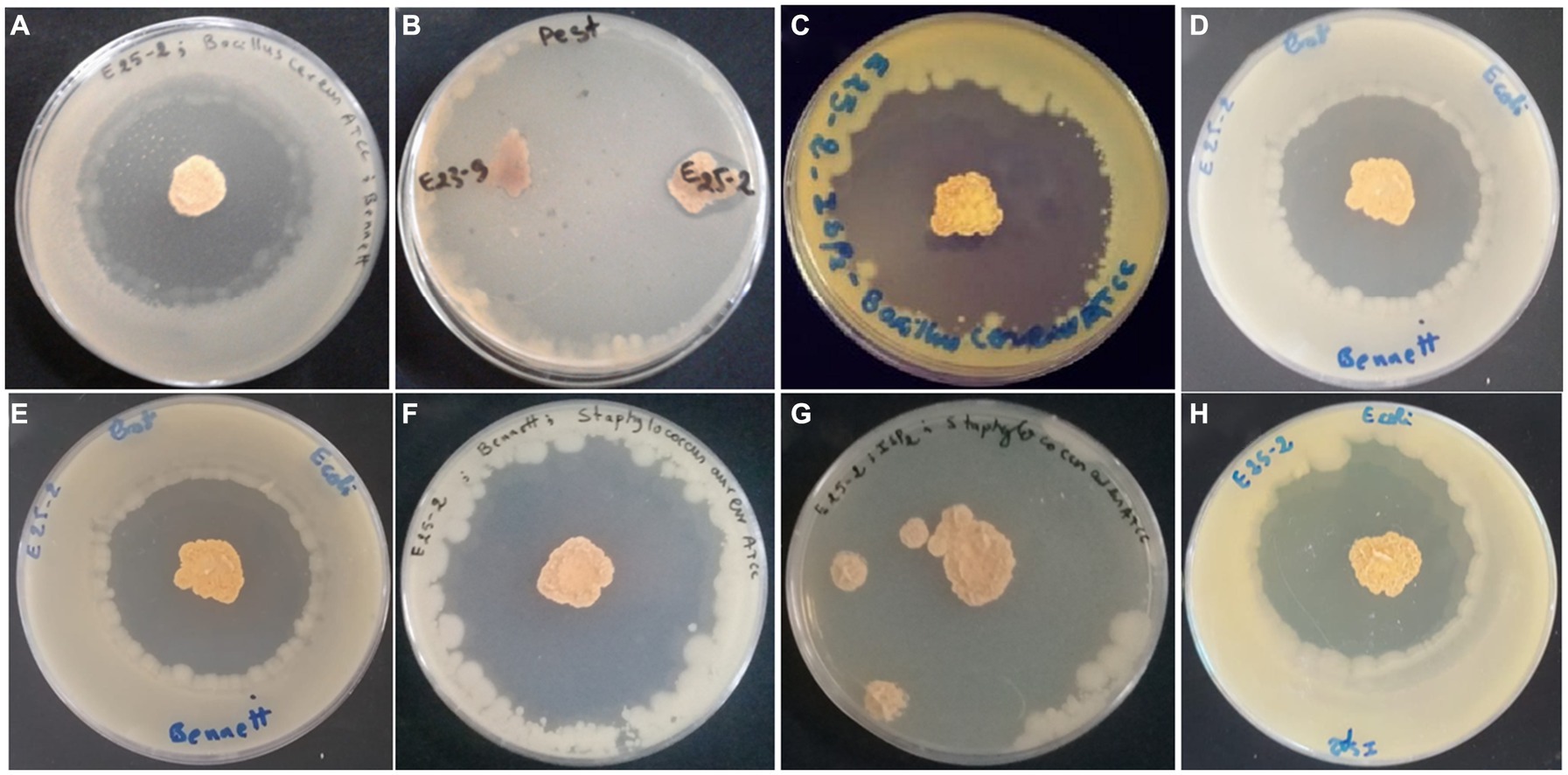
Figure 3. Antimicrobial activity of Lentzea sp. isolate E25-2 using the double-layer method on ISP1, ISP2, Bennet and GYEA media. (A) Antibacterial activity of E25-2 against Bacillus cereus ATCC 14579 using Bennett medium. (B) Antifungal activity of E25-2 against Fusarium sp. strain MN944575. (C) Antibacterial activity of E25-2 against Bacillus cereus ATCC 14579 using ISP2 medium. (D,E) Antibacterial activity of E25-2 against Escherichia coli ATCC 25922 using Bennett medium. (F) Antibacterial activity of E25-2 against Staphylococcus aureus ATCC 25923 using Bennett medium. (G) Antibacterial activity of E25-2 against Staphylococcus aureus ATCC 25923 using ISP2 medium. (H) Antibacterial activity of E25-2 against Escherichia coli ATCC 25922 using ISP2 medium.
Antimicrobial activity, assessed using n-hexane, dichloromethane, ethyl acetate and n-butanol, revealed that ethyl acetate was the most effective solvent for extracting the main antimicrobial agents (with zones of inhibition ranging between 6.5 ± 0.71 and 25 ± 1.41 mm). In addition, isolate E25-2 underwent specific secondary screening, involving MDR pathogenic bacteria and phytopathogenic fungi (Figure 4; Table 2; Supplementary Table S3). Both primary and secondary screening results indicated a more pronounced activity of this isolate against Gram-positive bacteria, compared to Gram-negative bacteria.
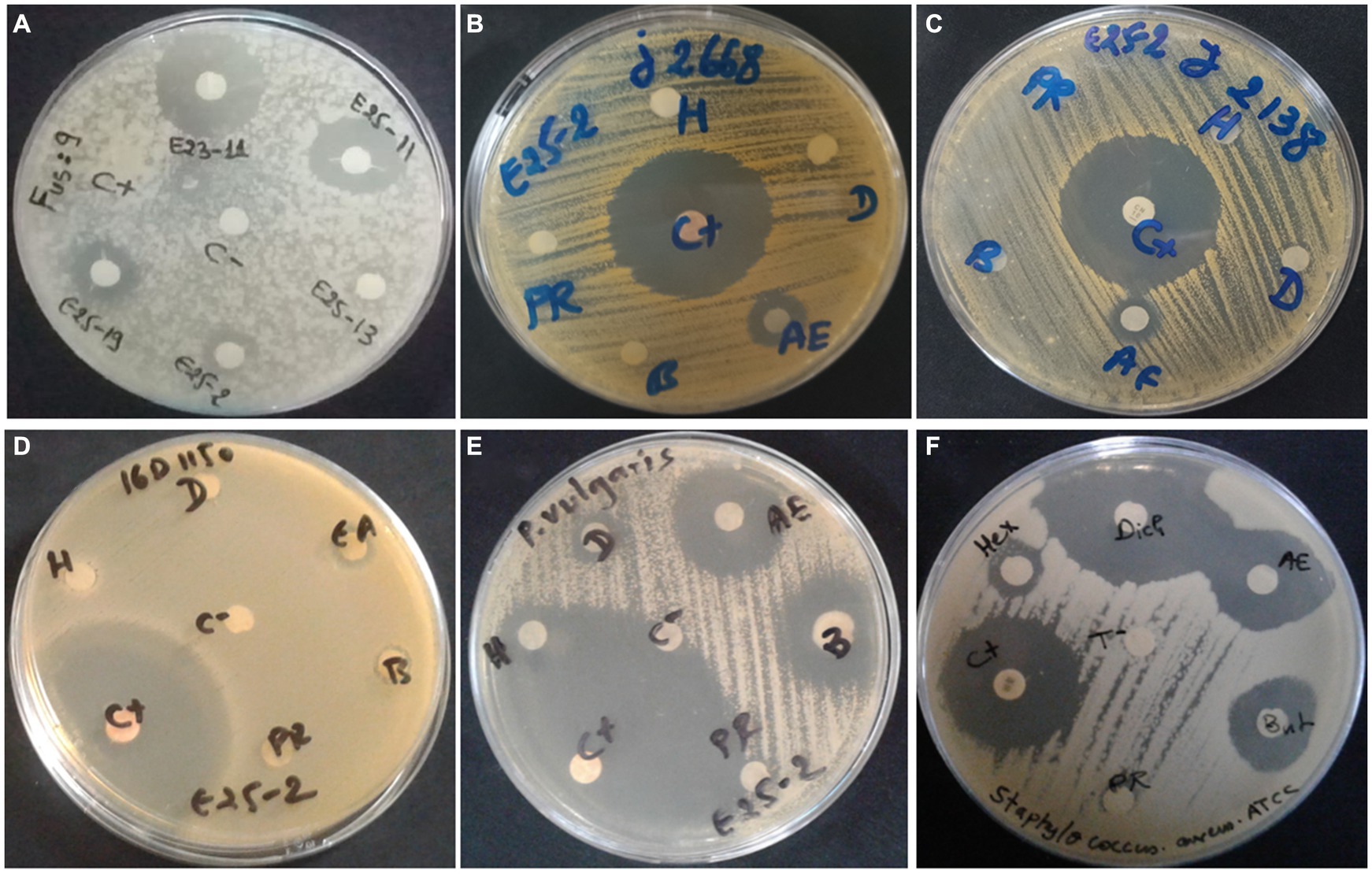
Figure 4. Antimicrobial activity of Lentzea sp. isolate E25-2 against MDR clinical bacteria and plant pathogenic fungi using disk diffusion method. C (+), Streptomycin (positive control); C (−), negative control; H, hexane; D, dichloromethane; EA, ethyl acetate; B, butanol; PR, residual phase. (A) Antifungal activity of E25-2 and other isolates against Fusarium sp. strain MN944575. (B) Antibacterial activity of E25-2 against clinical Staphylococcus aureus 23J2668. (C) Antibacterial activity of E25-2 against clinical Staphylococcus saprophyticus 23J2138. (D) Antibacterial activity of E25-2 against clinical Escherichia coli 16D1150. (E) Antibacterial activity of E25-2 against clinical Proteus vulgaris 16C1737. (F) Antibacterial activity of E25-2 against Staphylococcus aureus ATCC 25923.
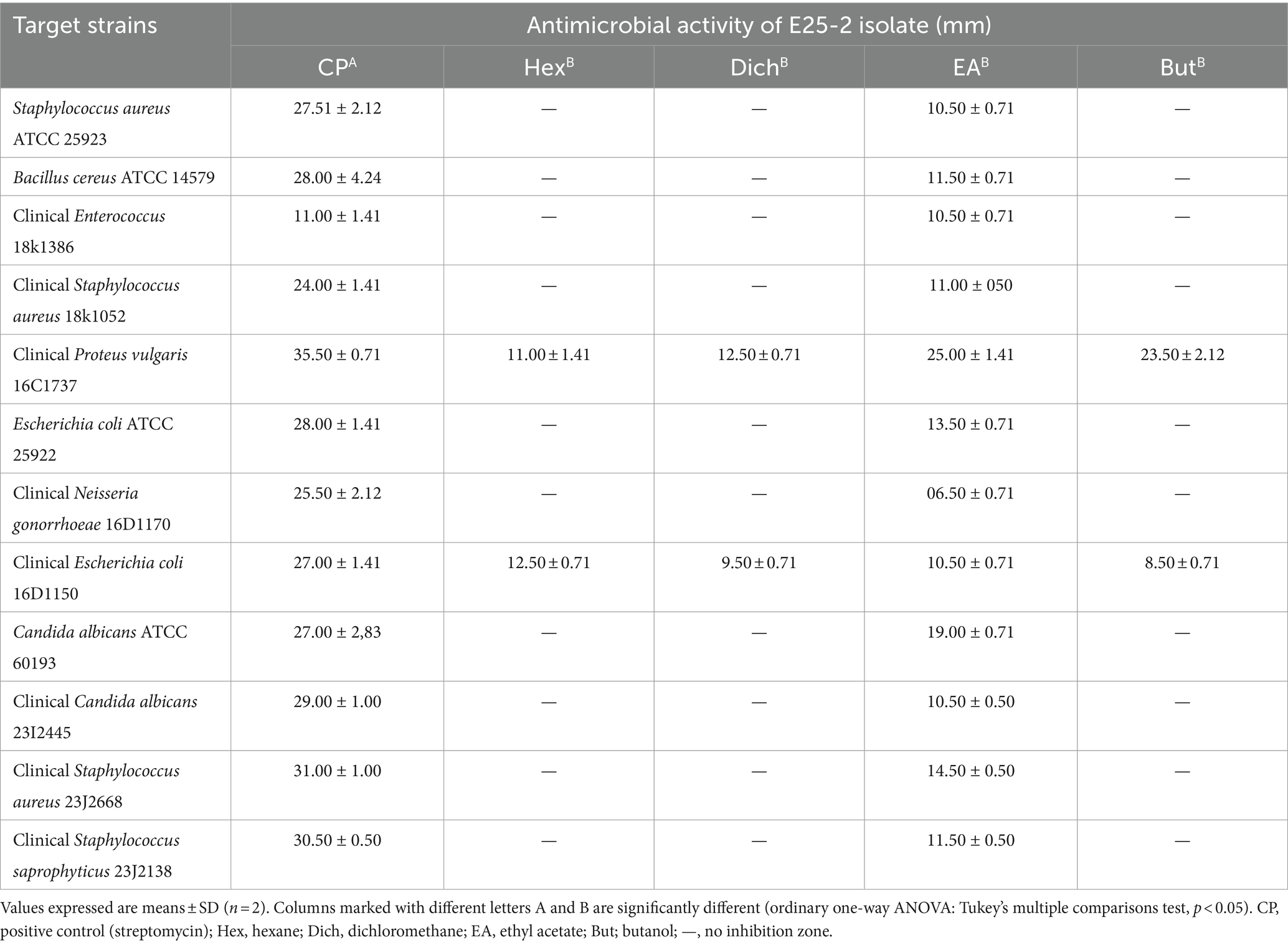
Table 2. Antimicrobial activity of E25-2 isolate using disk diffusion method according to extraction solvents.
3.4 The phenolic and flavonoid content of ethyl acetate extract from E25-2 isolate
The findings of the flavonoid and total phenolic contents of the ethyl acetate extract of isolate E25-2 are presented in Table 3. Pearson’s statistical analysis revealed a positive correlation between the concentrations of ethyl acetate extract and total phenolic content (r = 0.997; p < 0.001), as well as a positive correlation with flavonoid content (r = 0.834; p < 0.05). These correlations were calculated as the mean of three repetitive measurements.
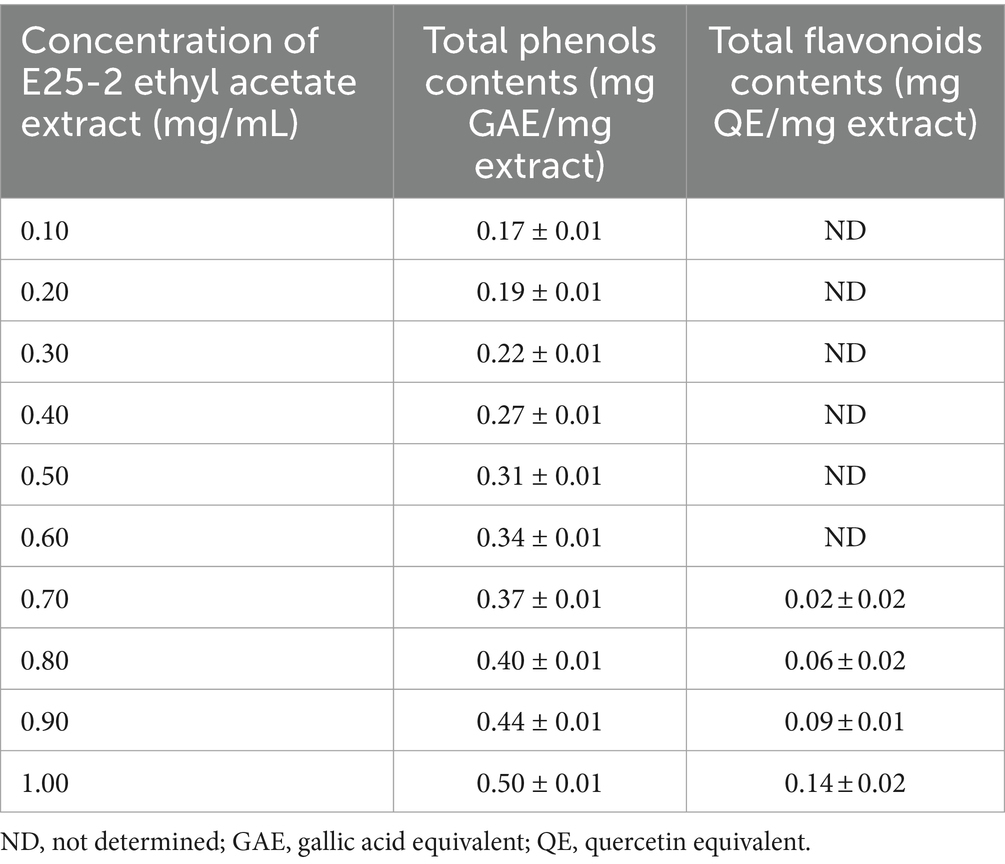
Table 3. Flavonoid and total phenolic contents of ethyl acetate extract of Lentzea sp. E25-2 strain.
3.5 In-vitro antioxidant potential of E25-2 isolate
The ethyl acetate extract of isolate E25-2 exhibited lower DPPH and ABTS activities com-pared to the standards (ascorbic acid and Trolox) (p < 0.0001) (Figures 5A,B). The results presented in the Figure 5A indicate that the ethyl acetate extract from the E25-2 isolate exhibited significant DPPH free radical scavenging activity between the different tested concentrations (p < 0.0001), with inhibition percentages ranging from 2.91 ± 0.92 to 37.78 ± 1.93% for concentrations between 0.2 and 1 mg/mL. In addition, the extract demonstrated high ABTS radical scavenging activity between the different tested concentrations (p < 0.0001), with inhibition percentages ranging from 1.32 ± 1.96 to 30.58 ± 1.78% over the same concentration range (Figure 5B).
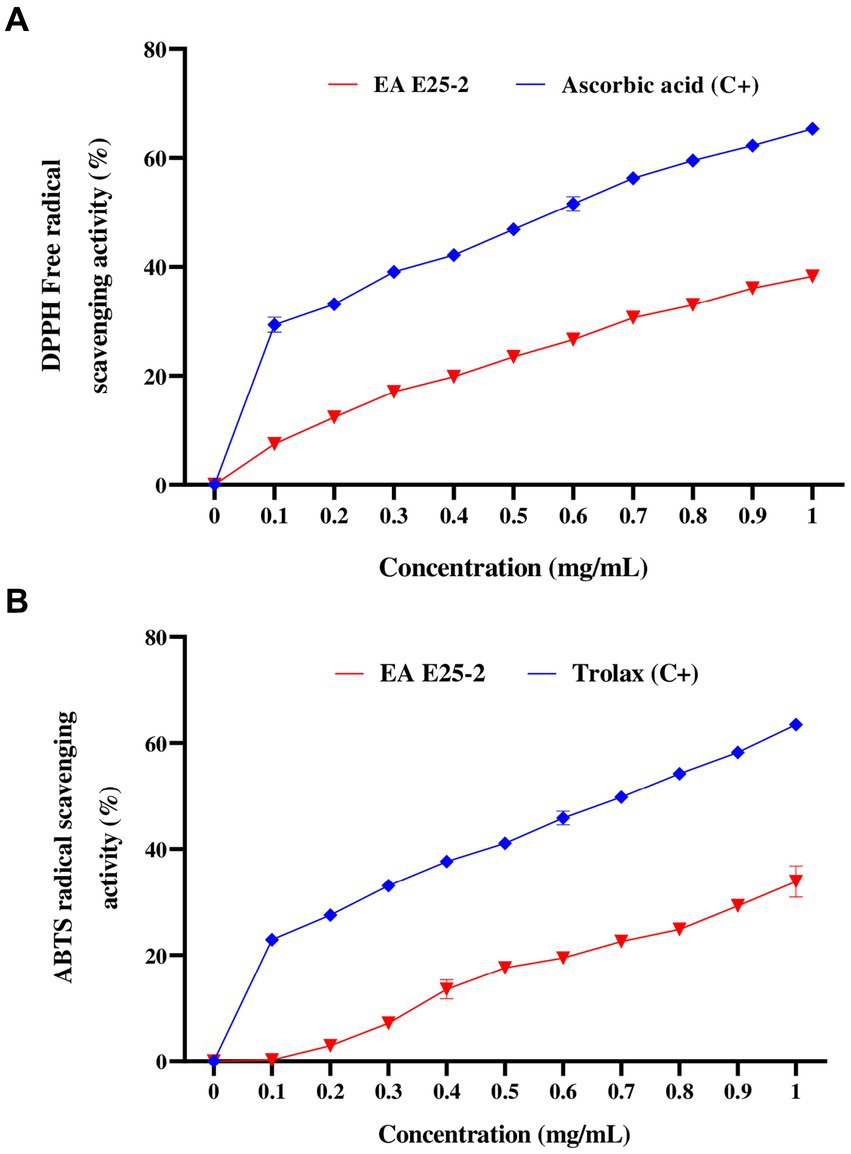
Figure 5. Antioxidant activity of ethyl acetate extract of Lentzea sp. E25-2 strain in various antioxidant assays. (A) antioxidant activity by DPPH assay and (B) antioxidant activity by ABTS assay. Values expressed are means ± SD (n = 3). Symbol (****) indicates p < 0.0001 significant difference between ethyl acetate extract of Lentzea sp. E25-2 isolate and controls. (C+), positive control; (EA), ethyl acetate.
Regarding iron antioxidant reducing power (FRAP), the results revealed a significant iron-reducing activity for ethyl acetate extract across the tested concentrations (p < 0.0001). This activity showed values from 0.43 ± 0.01 to 0.89 ± 0.02 mg AAE (ascorbic acid equivalent) per milligram of extract (Figure 6).
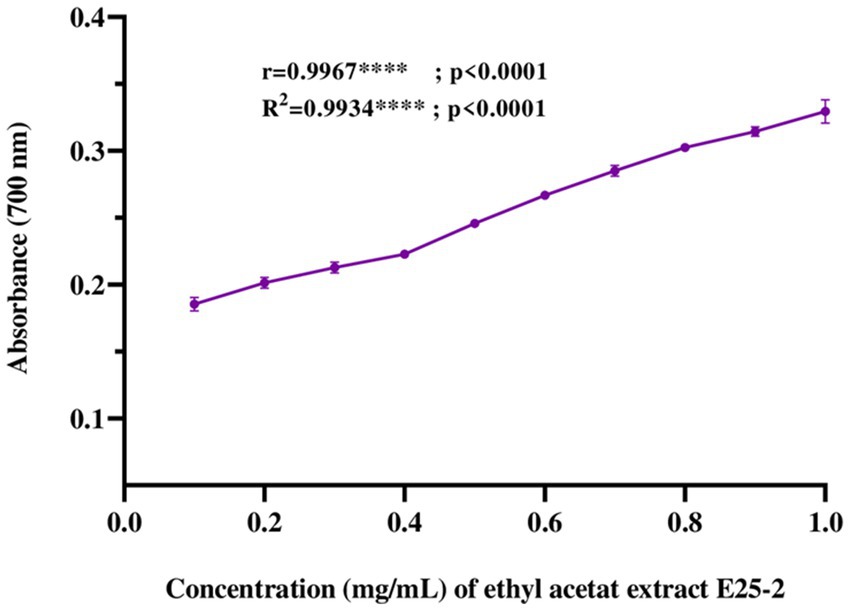
Figure 6. Ferric reducing antioxidant power (FRAP) of ethyl acetate extract of Lentzea sp. E25-2: standard deviation analysis for 10 doses (0.1 mg–1 mg) in triplicate (n = 3).
3.6 Correlation between antioxidant activity and total phenol and flavonoid contents
To evaluate the relationship between the antioxidant capacity of E25-2 ethyl acetate extract and its total phenol and flavonoid contents, a correlation analysis was also carried out. Figures 7A–F presents the Pearson correlation coefficients between the variables. The findings demonstrated a remarkably strong positive association between antioxidant activity, as measured by DPPH, ABTS and FRAP assays, and the polyphenol and flavonoid contents of ethyl acetate extract of Lentzea sp. isolate E25-2. A significantly positive correlation was observed between the total polyphenol content (TPC) and the ferric reducing antioxidant power (FRAP) of the E25-2 extract (r = 0.987, p < 0.0001), as well as between the total flavonoid content (TFC) and FRAP of this extract (r = 0.821, p < 0.05).
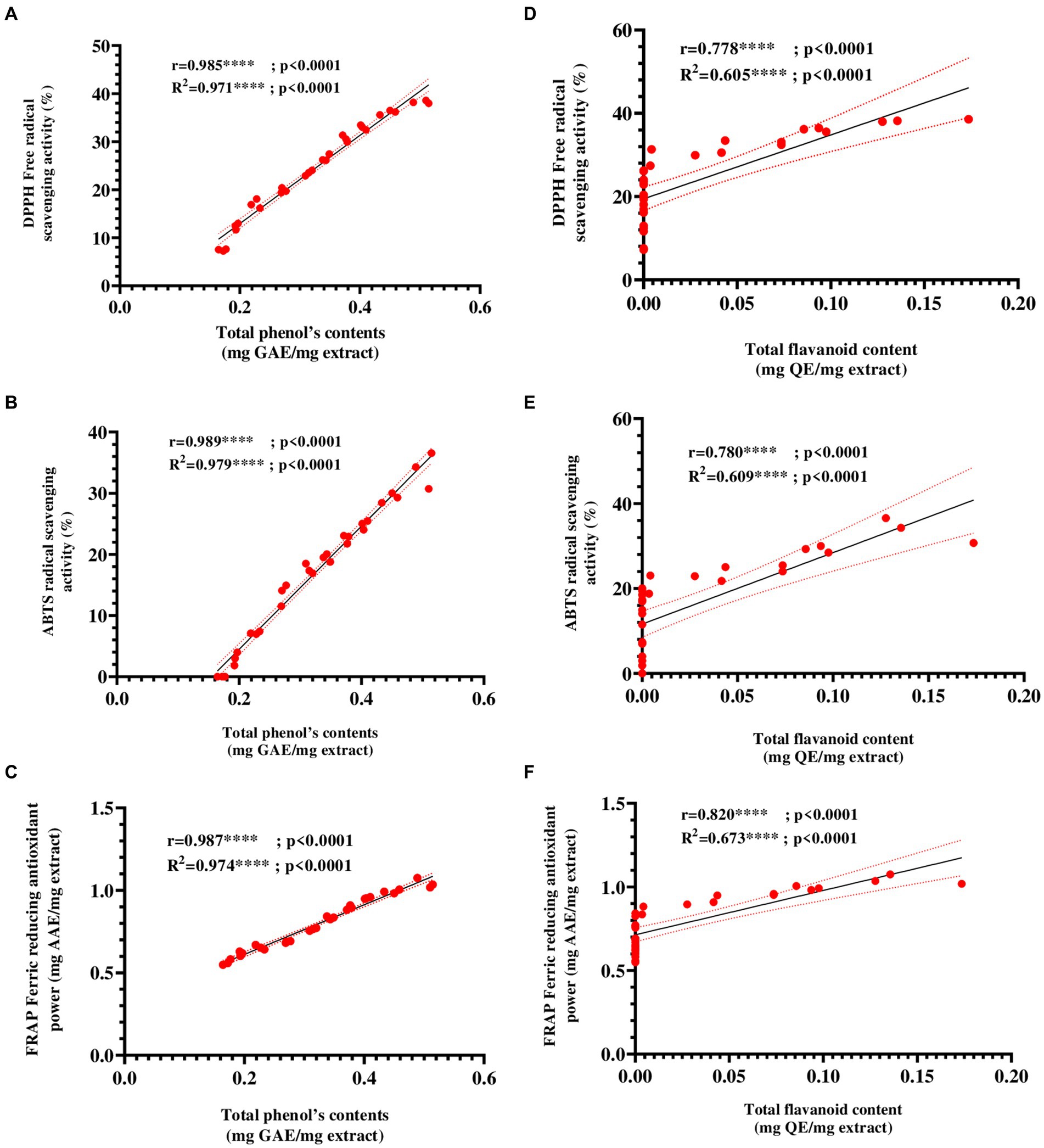
Figure 7. Pearson’s correlation coefficients between total phenolic and flavonoid contents and antioxidant activities ethyl acetate extract of Lentzea sp. E25-2 isolate. (A) Pearson correlation between DPPH and total phenolic content. (B) Pearson correlation between ABTS and total phenolic content. (C) Pearson correlation between FRAP and total phenolic content. (D) Pearson correlation between DPPH and flavonoids content. (E) Pearson correlation between ABTS and flavonoids content. (F) Pearson correlation between FRAP and flavonoids content. Symbol (****) indicates p < 0.0001 highly significant between tests.
3.7 Evaluating toxicity of ethyl acetate extract from isolate E25-2 using UV-visible spectral assay
Spectral examination of the ethyl acetate extract from isolate E25-2 indicated the absence of polyene molecules (Figure 8). Polyene molecules are generally defined by their tendency to exhibit three distinct absorption maxima in the UV-visible range, located between 291 and 405 nm.
3.8 Evaluating toxicity of E25-2 isolate using in vitro method hemolysis
As shown in Figure 9, no visible hemolysis was observed at different concentrations (0.125 to 1.25 mg/mL) of the ethyl acetate extract from isolate E25-2. Additionally, quantitative determination of hemoglobin by UV-vis spectroscopy also demonstrated negligible hemolytic activity of the ethyl acetate extract from isolate E25-2.
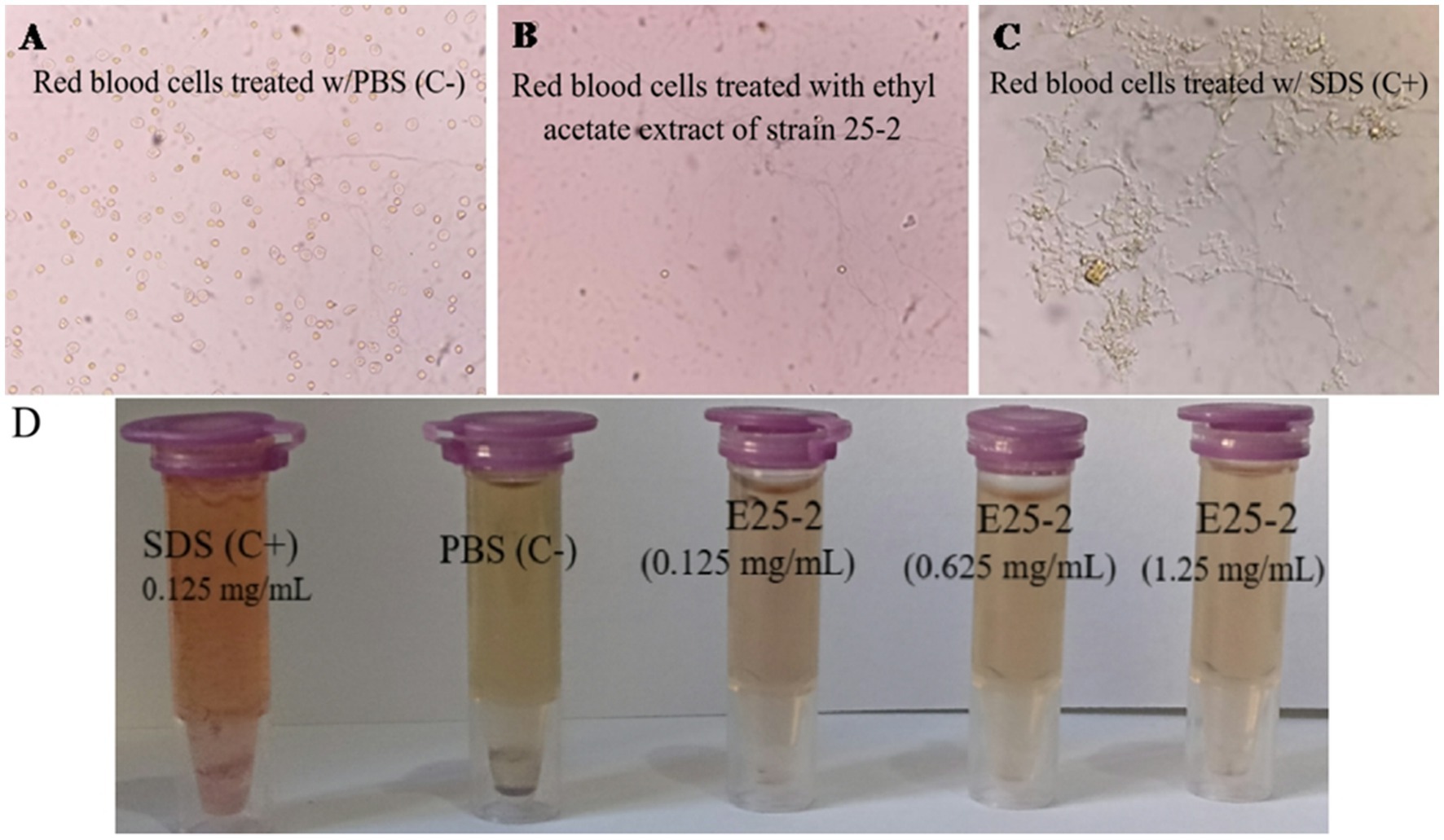
Figure 9. Evaluation of the PBS, ethyl acetate extract E25-2, and SDS effect on red blood cells: a comparative microscopic study. (A) Microscopic image showing the effect of PBS (negative control), (B) ethyl acetate extract, and (C) SDS (positive control) on red blood cells treated in different concentrations. (D) The ethyl acetate extract toxicity evaluation of strain E25-2 using the hemolysis test on human red blood cells.
3.9 Gas chromatography-mass spectrometry analysis
GC-MS analysis of the ethyl acetate extract from isolate E25-2 indicated the presence of amines, hydroxyl groups, pyrido-pyrazinone rings, esters, and pyrrolopyrazines. These chemical compounds were identified by comparing their mass spectra with the NIST library database. Detailed information regarding the chemical compounds, including their retention time, molecular weight, and molecular formula, is provided in Table 4. Moreover, their respective chemical structures are illustrated in Figure 10.
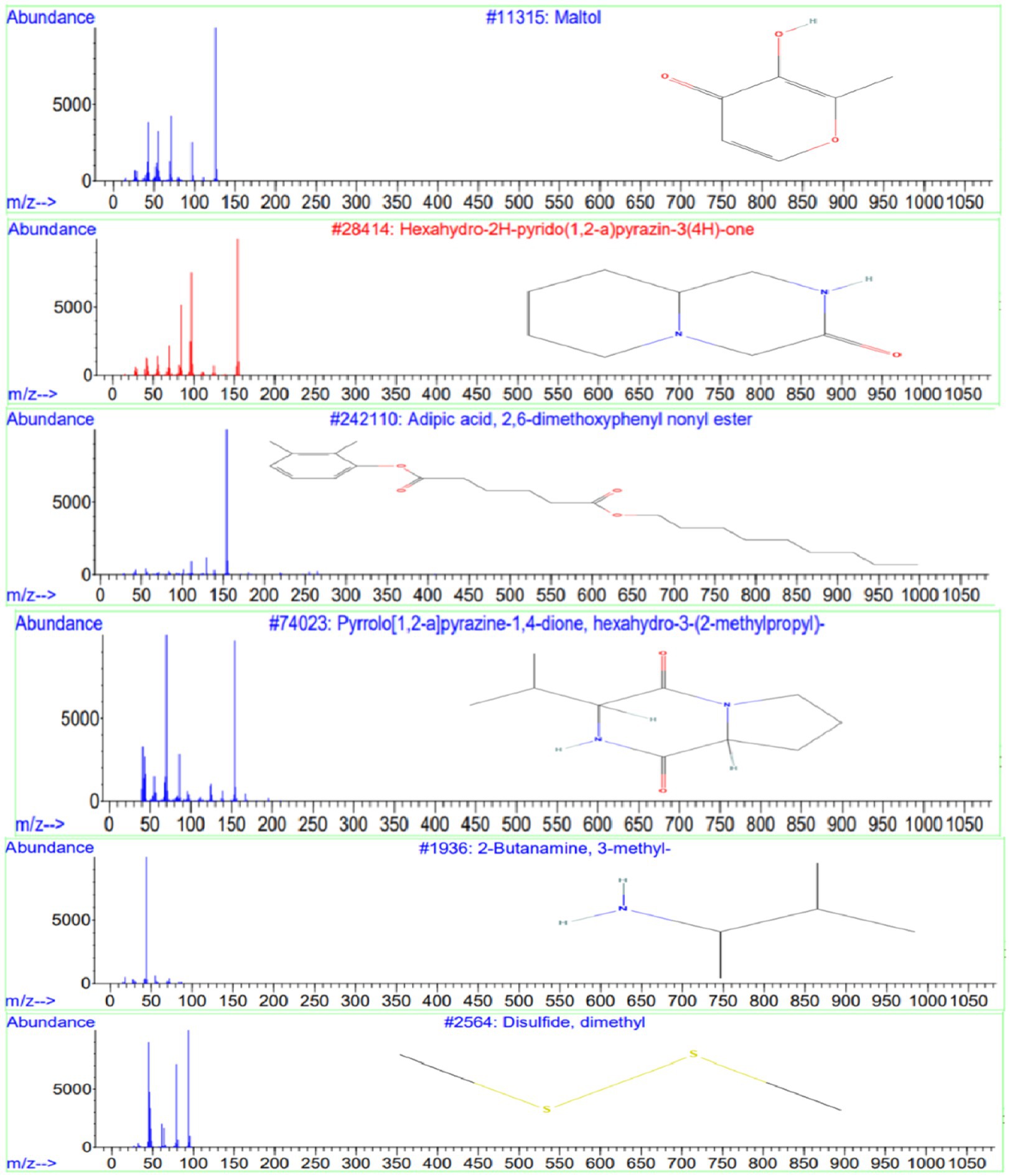
Figure 10. Chemical structures and mass spectra of 4 important secondary metabolites present in ethyl acetate extract of strain E25-2 analyzed by GC-MS.
4 Discussion
The isolate named E25-2 was classified as belonging to the genus Lentzea. Based on the results of the phylogenetic analysis, strain E25-2 has a 16S rRNA sequence similarity of 96.10% with Lentzea flaviverrucosa AS4.0578 NR_028763. Compared with the closest known strains, such as Lentzea flaviverrucosa, the sequence similarity is less than 98.7% (96.10%), suggesting the possibility that this is a new taxon according to Rossi-Tamisier et al. (2015). This result was confirmed by EzBioCloud databases, which also indicated a sequence similarity of 96.10% with the same closest taxon (Supplementary Table S4). These findings reinforce the hypothesis that strain E25-2 may represent a distinct new taxon within the genus Lentzea.
In terms of phenotype, isolate E25-2 has a segmented, branched, whitish-colored, non-powdery aerial mycelium and a yellow-orange vegetative mycelium. This study also allowed us to characterize isolate E25-2 in terms of physiological, biochemical and micromorphological characteristics, to enhance our understanding of the isolate. Moreover, E25-2 can assimilate all carbohydrate compounds but it is unable to utilize D-arabinose and melezitose. These phenotypic characteristics of isolate E25-2 are similar to the Lentzea genus, it is Gram-positive, aerobic, non-motile bacteria that form highly branched substrates and segregated aerial mycelia, and it unable to assimilate D-arabinose and melezitose (Maiti and Mandal, 2022). Consistent with the description provided by Maiti and Mandal (2022), members of the Lentzea genus, including isolate E25-2, demonstrate an inability to assimilate D-arabinose and melezitose. The majority of Actinomycetes species thrive in mesophilic conditions, displaying optimal growth temperatures ranging from 25 to 35°C, and a preferred pH level between 6.5 and 8.0 (Goodfellow et al., 2012; Barka et al., 2016). Optimum growth for isolate E25-2 is between 28 and 37°C. No growth was observed at 4°C or 46°C. These findings align closely with the outcomes reported by Singh et al. (2012).
Lentzea species, in particular those belonging to group 692 (Maiti and Mandal, 2022), appear to be a diverse source of genes involved in the production of secondary metabolites with pharmaceutical importance including, antimicrobial, antioxidant, anticancer, antibacterial, antimycobacterial, antifungal, antiviral, insecticidal, antiparasitic and other properties associated with natural products (Maiti and Mandal, 2022). Isolate E25-2 has demonstrated considerable antimicrobial activity against Gram-positive and Gram-negative bacteria, as well as phytopathogenic fungi and yeasts. According to the results of the antimicrobial test, Lentzea sp. E25-2 demonstrated antimicrobial activity against MDR pathogenic bacteria, as well as against phytopathogenic fungi and yeasts. According to the literature, the Streptomyces genus is recognized for its ability to naturally produce secondary metabolites with antimicrobial properties, and has strong biocontrol potential, acting effectively against MDR bacteria and phytopathogenic fungi such as Fusarium sp. (de Lima Procópio et al., 2012; Rammali et al., 2022). However, the antimicrobial activity of the Lentzea genus has rarely been reported in the literature. In our study, the Lentzea sp. E25-2 isolate could be considered an important source for the exploration of antimicrobial compounds, opening up promising prospects for potential applications in this field.
Reactive oxygen species (ROS), are reactive molecules that have the potential to inflict damage on nucleic acids, proteins, carbohydrates, and lipids, contributing to various disorders such as oxidative stress, aging, cancer, and immune dysfunction (Santos-Sánchez et al., 2019). Antioxidants have the ability to slow down or inhibit free radicals reducing oxidative stress and preventing cellular damage (Rammali et al., 2022). Microorganisms such as Actinomycetes efficiently produce a variety of natural bioactive molecules, including antioxidants (Chandra et al., 2020). A more recent study has shown that Lentzea genomes contain at least 53 genes for mitigating oxidative stress (Maiti and Mandal, 2022). Furthermore, existing literature suggests that relying on a solitary test is insufficient for a comprehensive evaluation of the antioxidant activity present in extracts (Tirzitis and Bartosz, 2010; Gupta, 2015; Law et al., 2019). Therefore, in order to obtain a more in-depth assessment, three in vitro tests including DPPH, ABTS and FRAP, were carried out. These three complementary methods provide a more complete picture of the antioxidant capacity of ethyl acetate extract from isolate E25-2. The ethyl acetate extract derived from Lentzea sp. E25-2 demonstrated higher antioxidant properties, suggesting the presence of bioactive compounds with diverse mechanisms of antioxidant activity. These findings imply that the Lentzea strain might possess the capability to produce one or more antioxidants, potentially valuable in preventing oxidative stress. To our knowledge, no strain of the Lentzea genus has been tested against DPPH, ABTS and FRAP free radicals to explore its antioxidant capacity. Phenolic compounds, characterized by an aromatic ring with hydroxyl groups, are recognized for their antioxidant properties (Balasundram et al., 2006). Furthermore, these substances exhibit additional advantageous bioactivities (Balasundram et al., 2006; Rammali et al., 2022). The statistical analysis conducted in this investigation substantiates this proposition by revealing a significant correlation (p < 0.0001) between the antioxidant activity of the ethyl acetate extract, assessed through DPPH, ABTS, and FRAP tests, and the concentrations of total phenolics and flavonoids.
Due to the structural similarity between human cholesterol and ergosterol, predominant in fungal cells, polyene antifungal molecules interact with cholesterol, disrupting fungal cell membranes and leading to cell death. This interaction with human cholesterol is associated with potential toxicity. As a result, researchers often avoid incorporating these molecules into research programs for new bioactive substances (Yilma et al., 2007). In this context, the strain studied, Lentzea sp. E25-2, is particularly interesting, as it is devoid of toxic polyene molecules, which are detected by three specific absorption maxima in the UV-visible range between 291 and 405 nm (Aouiche et al., 2012). This characteristic suggests that this strain could be promising for the discovery of new bioactive molecules, particularly in the antifungal field.
A hemolytic assay was conducted to assess the impact of bioactive compounds present in the ethyl acetate extract of isolate E25-2 on red blood cell membrane disruption. Several studies have demonstrated that the in vitro hemolysis test correlates well with in vivo toxicity via the hemolytic effect (Lu et al., 2009). Hemoglobin is a protein found in red blood cells, playing a crucial role in transporting oxygen from the lungs to the tissues of the body. However, the hemoglobin released by hemolysis can break down into by-products, some of which can contribute to vasoactive reactions, resulting in damage to various vital organs such as the liver, kidneys and heart (Buehler and D’Agnillo, 2010). Consequently, the extract showed no membranolytic activity on the erythrocyte membrane at all the tested concentrations. According to Saurav and Kannabiran (2012), evaluating membrane stability during exposure to new drugs is crucial, and erythrocytes serve as a suitable model for studying this stability. The impact of various bioactive compounds on the mechanical stability of the erythrocyte membrane serves as a reliable indicator of overall membrane stability.
GC-MS analysis was conducted to identify the bioactive compounds present in the ethyl acetate extract of isolate E25-2. The compounds identified in this study include phenolic compounds, characterized by an aromatic ring carrying one or more hydroxyl (OH) groups. These compounds are renowned for their well-documented antioxidant properties. The phenolic compound detected in the ethyl acetate extract is maltol, whose structure includes a phenol group, represented by the aromatic ring. This compound has been shown to have antioxidant, anti-inflammatory and anti-tumor activity. The compounds identified encompass phenolic compounds characterized by an aromatic ring containing one or more hydroxyl (OH) groups, renowned for their antioxidant properties (Balasundram et al., 2006). Within the ethyl acetate extract, maltol, a phenolic compound with a structure featuring a phenol group within the aromatic ring, was detected. Previous study has demonstrated that maltol exhibits antioxidant, anti-inflammatory, and anti-tumor activities (Han H. et al., 2015).
Phenolic compounds could be a natural source of physiological properties, such as antioxidant effects and other bioactivities (Balasundram et al., 2006). Heterocyclic compounds were also identified in the ethyl acetate extract, including pyrazines and pyrrolopyrazines. Pyrazines, heterocyclic compounds commonly found in nature, are generally produced by microorganisms (Schulz and Dickschat, 2007). Some pyrazines have been associated with beneficial activities such as antioxidants, anticancer and antimicrobial properties (Ser et al., 2016). The compound Hexahydro-2H-pyrido (1,2-a) pyrazin-3(4H)-one has been identified in the ethyl acetate extract of isolate E25-2 and has been reported in the literature to exhibit various properties, such as antifungal, diuretic, antidiabetic, anticancer, antibacterial, antiviral, hypnotic, analgesic activities, and antimycobacterial (Dolezal and Zitko, 2015).
Pyrrolopyrazines are known for their diverse range of bioactivities, which include antioxidant, antitumor, antibacterial, antifungal, and anti-angiogenic properties (Ser et al., 2015). Additionally, the compound “3-isobutyl-hexahydro-pyrrolo[1,2-a]pyrazine-1,4-dione-” was detected in the ethyl acetate extract and has been reported in the literature to exhibit various properties, such as antibacterial (Ser et al., 2015; Sanjenbam and Kannabiran, 2016; Raharja et al., 2019), fungicidal (Basavanna et al., 2021), antioxidant and insecticidal (Ho, 2012). Furthermore, the compound disulfide, dimethyl detected in the ethyl acetate extract has previously been reported to exhibit various biological activities, including antioxidant, antifungal, and analgesic effects (Morales-López et al., 2017; Pozsgai et al., 2017; Tyagi et al., 2020). Similarly, the compound 2-Butanamine, 3-methyl- detected in the extract was previously reported to possess antimicrobial activities (Hameed et al., 2018).
Overall, the chemical compounds identified in the current study are well known for their antimicrobial and antioxidant activities, indicating that these components could be responsible for the antioxidant and antimicrobial activity of the ethyl acetate extract of Lentzea sp. E25-2. Consequently, this study provides further evidence of the potential of Lentzea sp. E25-2 derived from Moroccan forest soil as a promising source of antimicrobial and antioxidant agents. However, further research is needed to precisely identify the individual compound or combination of compounds responsible for the observed activities.
5 Conclusion
In conclusion, the isolate E25-2, derived from Moroccan forest soil, was classified as belonging to the genus Lentzea based on 16S rRNA sequencing and phylogenetic analysis. Notably, E25-2 exhibited substantial antimicrobial activity against a range of Gram-positive and Gram-negative bacteria, phytopathogenic fungi, and yeast, with ethyl acetate identified as the most effective solvent for extracting antimicrobial agents. The ethyl acetate extract of E25-2 showed significant antioxidant properties, as evidenced by DPPH, ABTS, and FRAP assays, which correlated strongly with its phenolic and flavonoid contents. The ethyl acetate extract analysis by GCMS identified various bioactive compounds, including pyrazines, phenolics, and pyrrolopyrazines, known for their antimicrobial and antioxidant activities. Importantly, the extract was found to be non-toxic, as indicated by the absence of polyene molecules and negligible hemolytic activity. Our results suggest that Lentzea sp. E25-2 is a promising source of novel bioactive compounds with prospective pharmaceutical uses, particularly as antimicrobial and antioxidant agents. This highlights the soil-derived microorganisms’ value in the new natural products discovery. Further research is needed to isolate and characterize the specific compounds responsible for the bioactivities observed in Lentzea sp. E25-2, which could lead to new therapeutic agents’ development.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
SR: Writing – original draft, Writing – review & editing. AC: Writing – original draft, Writing – review & editing. MA: Writing – original draft, Writing – review & editing. AR: Writing – original draft, Writing – review & editing. FK: Writing – original draft, Writing – review & editing. KD: Writing – original draft, Writing – review & editing. AK: Writing – original draft, Writing – review & editing. LR: Writing – original draft, Writing – review & editing. BN: Writing – original draft, Writing – review & editing. AP: Writing – original draft, Writing – review & editing. BB: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1429035/full#supplementary-material
References
Aouiche, A., Sabaou, N., Meklat, A., Zitouni, A., Mathieu, F., and Lebrihi, A. (2012). Activité antimicrobienne de Streptomyces sp. PAL111 d’origine saharienne contre divers microorganismes cliniques et toxinogènes résistants aux antibiotiques. J. Mycol. Med. 22, 42–51. doi: 10.1016/j.mycmed.2011.12.077
Badji, B., Riba, A., Mathieu, F., Lebrihi, A., and Sabaou, N. (2005). Activité antifongique d’une souche d’Actinomadura d’origine saharienne sur divers champignons pathogènes et toxinogènes. J. Mycol. Med. 15, 211–219. doi: 10.1016/j.mycmed.2005.07.001
Bahorun, T., Soobrattee, M. A., Luximon-ramma, M. V., and Aruoma, P. O. I. (2006). Free radicals and antioxidants in cardiovascular health and disease. Internet J. Med. Update 1, 24–40. doi: 10.4314/ijmu.v1i2.39839
Balasundram, N., Sundram, K., and Samman, S. (2006). Phenolic compounds in plants and agri-industrial by-products: antioxidant activity, occurrence, and potential uses. Food Chem. 99, 191–203. doi: 10.1016/j.foodchem.2005.07.042
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Klenk, H.-P., et al. (2016). Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/MMBR.00019-15
Basavanna, V., Ningaiah, S., Chandramouli, M., Sobha, A., and Doddamani, S. (2021). Quinolinyl-pyrazoles: synthesis and pharmacological evolution in the recent decennial. J. Iran. Chem. Soc. 18, 1479–1522. doi: 10.1007/s13738-020-02152-1
Bastide, A., De Méo, M., Andriantsoa, M., Laget, M., and Duménil, G. (1986). Isolement et sélection de souches d’actinomycètes productrices de substances antifongiques de structure non-polyénique. MIRCEN J. Appl. Microbiol. Biotechnol. 2, 453–466. doi: 10.1007/BF00933368
Bensadón, S., Hervert-Hernández, D., Sáyago-Ayerdi, S. G., and Goñi, I. (2010). By-products of Opuntia ficus-indica as a source of antioxidant dietary fiber. Plant Foods Hum. Nutr. 65, 210–216. doi: 10.1007/s11130-010-0176-2
Blois, M. (1965). Antioxidant determinations by the use of a stable free radical. Nature 181, 1199–1200. doi: 10.1038/1811199a0
Bouaziz, S., Messis, A., Bettache, A., El Hadj, M. D. O., and Benallaoua, E. S. (2016). Antifungal activity of Streptomyces sp. 14 strain isolated from Ouargla (Southeast of Algeria): identification, production and characterization of the active substance. Int. J. Biosci. 9, 45–56. doi: 10.12692/ijb/9.5.45-56
Browne, K., Chakraborty, S., Chen, R., Willcox, M. D. P., Black, D. S., Walsh, W. R., et al. (2020). A new era of antibiotics: the clinical potential of antimicrobial peptides. Int. J. Mol. Sci. 21:7047. doi: 10.3390/ijms21197047
Buehler, P. W., and D’Agnillo, F. (2010). Toxicological consequences of extracellular hemoglobin: biochemical and physiological perspectives. Antioxid. Redox Signal. 12, 275–291. doi: 10.1089/ars.2009.2799
Cao, C.-L., Zhou, X.-Q., Qin, S., Tao, F.-X., Jiang, J.-H., and Lian, B. (2015). Lentzea guizhouensis sp. nov., a novel lithophilous actinobacterium isolated from limestone from the karst area, Guizhou, China. Antonie Van Leeuwenhoek 108, 1365–1372. doi: 10.1007/s10482-015-0589-x
CASFM (2020). Comité de l’antibiogramme de la Société Française de Microbiologie Recommandations 2020 : Comité de l’antibiogramme de la Société Française de Microbiologie. Paris, France, 6–82. Available at: https://www.sfm-microbiologie.org/wp-content/uploads/2020/04/CASFM2020_Avril2020_V1.1.pdf
Chandra, P., Sharma, R. K., and Arora, D. S. (2020). Antioxidant compounds from microbial sources: a review. Food Res. Int. 129:10889. doi: 10.1016/j.foodres.2019.108849
da Cunha, B. R., Fonseca, L. P., and Calado, C. R. C. (2019). Antibiotic discovery: where have we come from, where do we go? Antibiotics 8:45. doi: 10.3390/antibiotics8020045
de Lima Procópio, R. E., da Silva, I. R., Martins, M. K., de Azevedo, J. L., and de Araújo, J. M. (2012). Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 16, 466–471. doi: 10.1016/j.bjid.2012.08.014
Dolezal, M., and Zitko, J. (2015). Pyrazine derivatives: a patent review (June 2012—present). Expert Opin. Ther. Pat. 25, 33–47. doi: 10.1517/13543776.2014.982533
Genilloud, O. (2017). Actinomycetes: still a source of novel antibiotics. Nat. Prod. Rep. 34, 1203–1232. doi: 10.1039/c7np00026j
Goodfellow, M., Peter, K. Ã., Busse, H.-J., Trujillo, M. E., Ludwig, W., Suzuki, K., et al. (2012). Bergey’s manual of systematic bacteriology. Volume 5: the Actinobacteria. New York, NY: Springer.
Gupta, D. (2015). Methods for determination of antioxidant capacity: a review. Int. J. Pharm. Sci. Res. 6:546. doi: 10.13040/IJPSR.0975-8232.6(2).546-66
Hameed, R. H., Al-Shareefi, E., and Hameed, I. H. (2018). Analysis of methanolic fruit extract of Citrus aurantifolia using gas chromatography—mass spectrum and FT-IR techniques and evaluation of its anti-bacterial activity. Indian J. Public Health Res. Dev. 9, 480–486. doi: 10.5958/0976-5506.2018.00490.4
Han, H., Iakovenko, L., and Wilson, A. C. (2015). Loss of homogentisate 1,2-dioxygenase activity in Bacillus anthracis results in accumulation of protective pigment. PLoS One 10:e0128967. doi: 10.1371/journal.pone.0128967
Han, Y., Xu, Q., Hu, J. N., Han, X. Y., Li, W., and Zhao, L. C. (2015). Maltol, a food flavoring agent, attenuates acute alcohol-induced oxidative damage in mice. Nutrients 7, 682–696. doi: 10.3390/nu7010682
Ho, Y. W. (2012). 5-(1-Pyrrolyl)-thieno[2,3-d]pyrimidine as building block in heterocyclic synthesis: synthesis of some new 6-substituted 5- (1-pyrrolyl)- thieno[2,3-d]pyrimidines and 5(4)-Substituted pyrimido- [2,3:4,5]thieno[2,3-e]pyrrolo[1,2-a]pyrazine derivatives. Chem. Sci. Rev. Lett. 5, 245–255.
Hug, J. J., Krug, D., and Müller, R. (2020). Bacteria as genetically programmable producers of bioactive natural products. Nat. Rev. Chem. 4, 172–193. doi: 10.1038/s41570-020-0176-1
Hussain, A., Rather, M. A., Dar, M. S., Aga, M. A., Ahmad, N., Manzoor, A., et al. (2017). Novel bioactive molecules from Lentzea violacea strain AS 08 using one strain-many compounds (OSMAC) approach. Bioorg. Med. Chem. Lett. 27, 2579–2582. doi: 10.1016/j.bmcl.2017.03.075
Jones, S. E., and Elliot, M. A. (2017). Streptomyces exploration: competition, volatile communication and new bacterial behaviours. Trends Microbiol. 25, 522–531. doi: 10.1016/j.tim.2017.02.001
Kitouni, M., Boudemagh, A., Oulmi, L., Reghioua, S., Boughachiche, F., Zerizer, H., et al. (2005). Isolation of actinomycetes producing bioactive substances from water, soil and tree bark samples of the north-east of Algeria. J. Mycol. Med. 15, 45–51. doi: 10.1016/j.mycmed.2004.12.005
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Law, J. W. F., Chan, K. G., He, Y. W., Khan, T. M., Ab Mutalib, N. S., Goh, B. H., et al. (2019). Diversity of Streptomyces spp. from mangrove forest of Sarawak (Malaysia) and screening of their antioxidant and cytotoxic activities. Sci. Rep. 9, 15262–15215. doi: 10.1038/s41598-019-51622-x
Lu, S., Duffin, R., Poland, C., Daly, P., Murphy, F., Drost, E., et al. (2009). Efficacy of simple short-term in vitro assays for predicting the potential of metal oxide nanoparticles to cause pulmonary inflammation. Environ. Health Perspect. 117, 241–247. doi: 10.1289/ehp.11811
Lu, Q. P., Ye, J. J., Huang, Y. M., Liu, D., Liu, L. F., Dong, K., et al. (2019). Exploitation of potentially new antibiotics from mangrove Actinobacteria in Maowei sea by combination of multiple discovery strategies. Antibiotics 8:236. doi: 10.3390/antibiotics8040236
Maiti, P. K., and Mandal, S. (2022). Comprehensive genome analysis of Lentzea reveals repertoire of polymer-degrading enzymes and bioactive compounds with clinical relevance. Sci. Rep. 12, 8409–8424. doi: 10.1038/s41598-022-12427-7
Marimuthu, S., Karthic, C., Mostafa, A. A., Al-Enazi, N. M., Abdel-Raouf, N., and Sholkamy, E. N. (2020). Antifungal activity of Streptomyces sp. SLR03 against tea fungal plant pathogen Pestalotiopsis theae. J. King Saud Univ. 32, 3258–3264. doi: 10.1016/j.jksus.2020.08.027
Meštrović, T., Ikuta, K. S., Swetschinski, L., Gray, A., Aguilar, G. R., Han, C., et al. (2023). The burden of bacterial antimicrobial resistance in Croatia in 2019: a country-level systematic analysis. Croat. Med. J. 64, 272–283. doi: 10.3325/cmj.2023.64.272
Morales-López, J., Centeno-Álvarez, M., Nieto-Camacho, A., López, M. G., Pérez-Hernández, E., Pérez-Hernández, N., et al. (2017). Evaluation of antioxidant and hepatoprotective effects of white cabbage essential oil. Pharm. Biol. 55, 233–241. doi: 10.1080/13880209.2016.1258424
Oyaizu, M. (1986). Studies on products of browning reaction. Antioxidative activities of products of browning reaction prepared from glucosamine. Japan. J. Nutr. Diet. 44, 307–315. doi: 10.5264/eiyogakuzashi.44.307
Pozsgai, G., Payrits, M., Sághy, É., Sebestyén-Bátai, R., Steen, E., Szőke, É., et al. (2017). Analgesic effect of dimethyl trisulfide in mice is mediated by TRPA1 and sst4 receptors. Nitric Oxide 65, 10–21. doi: 10.1016/j.niox.2017.01.012
Raharja, N. I., Widanarni,, and Wahyudi, A. T. (2019). Marine sponge-associated bacteria as biocontrol agents of vibriosis on whiteleg shrimp caused by Vibrio parahaemolyticus. Biodiversitas 20, 3164–3169. doi: 10.13057/biodiv/d201108
Rammali, S., El Aalaoui, M., Sbaghi, M., Dari, K., Bencharki, B., Azeroual, A., et al. (2023). Insecticidal potential of Streptomyces sp. dichloromethane extracts against the cactus cochineal Dactylopius opuntiae (Cockerell). Not. Sci. Biol. 13:11574. doi: 10.15835/nsb15411574
Rammali, S., Hilali, L., Dari, K., Bencharki, B., Rahim, A., Timinouni, M., et al. (2022). Antimicrobial and antioxidant activities of Streptomyces species from soils of three different cold sites in the Fez-Meknès Morocco. Sci. Rep. 12, 17233–17222. doi: 10.1038/s41598-022-21644-z
Rammali, S., Rahim, A., El Aalaoui, M., Bencharki, B., Dari, K., Habach, A., et al. (2024). Antimicrobial potential of Streptomyces coeruleofuscus SCJ isolated from microbiologically unexplored garden soil in Northwest Morocco. Sci. Rep. 14:3359. doi: 10.1038/s41598-024-53801-x
Re, R., Nicoletta, P., Anna, P., Ananth, P., Min, Y., and Catherine, R.-E. (1999). Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 26, 1231–1237. doi: 10.1016/S0891-5849(98)00315-3
Rossi-Tamisier, M., Benamar, S., Raoult, D., and Fournier, P. E. (2015). Cautionary tale of using 16s rRNA gene sequence similarity values in identification of human-associated bacterial species. Int. J. Syst. Evol. Microbiol. 65, 1929–1934. doi: 10.1099/ijs.0.000161
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Salam, M. A., Al-Amin, M. Y., Salam, M. T., Pawar, J. S., Akhter, N., Rabaan, A. A., et al. (2023). Antimicrobial resistance: a growing serious threat for global public health. Healthcare 11:1946. doi: 10.3390/healthcare11131946
Sanjenbam, P., and Kannabiran, K. (2016). Bioactivity of pyrrolo[1,2-a]pyrazine-1,4-dione,hexahydro-3-(phenylmethyl)- extracted from Streptomyces sp. VITPK9 Isolated from the Salt Spring Habitat of Manipur, India. Asian J. Pharm. 10, 265–270. doi: 10.22377/ajp.v10i04.865
Santos-Sánchez, N. F., Salas-Coronado, R., Villanueva-Cañongo, C., and Hernández-Carlos, B. (2019). “Antioxidant compounds and their antioxidant mechanism” in Antioxidants (IntechOpen), 1–29. doi: 10.5772/intechopen.85270
Saurav, K., and Kannabiran, K. (2012). Cytotoxicity and antioxidant activity of 5-(2, 4-dimethylbenzyl) pyrrolidin-2-one extracted from marine Streptomyces VITSVK5 spp. Saudi J. Biol. Sci. 19, 81–86. doi: 10.1016/j.sjbs.2011.07.003
Schulz, S., and Dickschat, J. S. (2007). Bacterial volatiles: the smell of small organisms. Nat. Prod. Rep. 24, 814–842. doi: 10.1039/b507392h
Sengupta, S., Pramanik, A., Ghosh, A., and Bhattacharyya, M. (2015). Antimicrobial activities of actinomycetes isolated from unexplored regions of Sundarbans mangrove ecosystem. BMC Microbiol. 15:170. doi: 10.1186/s12866-015-0495-4
Ser, H.-L., Palanisamy, U. D., Yin, W.-F., Abd Malek, S. N., Chan, K.-G., Goh, B.-H., et al. (2015). Presence of antioxidative agent, Pyrrolo [1, 2-a] pyrazine-1, 4-dione, hexahydro-in newly isolated Streptomyces mangrovisoli sp. nov. Front. Microbiol. 6:854. doi: 10.3389/fmicb.2015.00854
Ser, H. L., Tan, L. T. H., Palanisamy, U. D., Abd Malek, S. N., Yin, W. F., Chan, K. G., et al. (2016). Streptomyces antioxidans sp. nov., a novel mangrove soil actinobacterium with antioxidative and neuroprotective potentials. Front. Microbiol. 7:899. doi: 10.3389/fmicb.2016.00899
Shirling, E. B., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Singh, V., Haque, S., Singh, H., Verma, J., Vibha, K., Singh, R., et al. (2016). Isolation, screening, and identification of novel isolates of actinomycetes from India for antimicrobial applications. Front. Microbiol. 7:1921. doi: 10.3389/fmicb.2016.01921
Singh, A. P., Singh, R. B., and Mishra, S. (2012). Microbial and biochemical aspects of antibiotic producing microorganisms from soil samples of certain industrial area of India-an overview. Open Nutraceuticals J. 5, 107–112. doi: 10.2174/1876396001205010107
Tang, K. W. K., Millar, B. C., and Moore, J. E. (2023). Antimicrobial resistance (AMR). Br. J. Biomed. Sci. 80, 1–11. doi: 10.3389/bjbs.2023.11387
Tirzitis, G., and Bartosz, G. (2010). Determination of antiradical and antioxidant activity: basic principles and new insights. Acta Biochim. Pol. 57:139. doi: 10.18388/abp.2010_2386
Tyagi, S., Lee, K. J., Shukla, P., and Chae, J. C. (2020). Dimethyl disulfide exerts antifungal activity against Sclerotinia minor by damaging its membrane and induces systemic resistance in host plants. Sci. Rep. 10, 6547–6512. doi: 10.1038/s41598-020-63382-0
Uddin, T. M., Chakraborty, A. J., Khusro, A., Zidan, B. M. R. M., Mitra, S., Emran, T. B., et al. (2021). Antibiotic resistance in microbes: history, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 14, 1750–1766. doi: 10.1016/j.jiph.2021.10.020
World Health Organization. (2023). Antimicrobial resistance. Available at: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (Accessed November 21, 2023).
Williams, S. T., and Cross, T. (1971). Chapter XI Actinomycetes. Methods Microbiol. 4, 295–334. doi: 10.1016/S0580-9517(09)70016-9
Keywords: Lentzea flaviverrucosa, antimicrobial activity, antioxidant activity, 16S rRNA gene sequences, hemolytic activity, GC-MS
Citation: Rammali S, Ciobică A, El Aalaoui M, Rahim A, Kamal FZ, Dari K, Khattabi A, Romila L, Novac B, Petroaie A and Bencharki B (2024) Exploring the antimicrobial and antioxidant properties of Lentzea flaviverrucosa strain E25-2 isolated from Moroccan forest soil. Front. Microbiol. 15:1429035. doi: 10.3389/fmicb.2024.1429035
Edited by:
Palaniyandi Velusamy, Sree Balaji Medical College and Hospital, IndiaReviewed by:
Gamal Mohamed El-Said El-Sherbiny, Al-Azhar University, EgyptDexi Li, Henan Agricultural University, China
Copyright © 2024 Rammali, Ciobică, El Aalaoui, Rahim, Kamal, Dari, Khattabi, Romila, Novac, Petroaie and Bencharki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura Romila, bGF1cmEuZGFydHVAZ21haWwuY29t
 Said Rammali
Said Rammali Alin Ciobică
Alin Ciobică Mohamed El Aalaoui
Mohamed El Aalaoui Abdellatif Rahim
Abdellatif Rahim Fatima Zahra Kamal
Fatima Zahra Kamal Khadija Dari1
Khadija Dari1