
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 11 December 2024
Sec. Terrestrial Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1428517
This article is part of the Research TopicInsights in Terrestrial Microbiology: 2023/2024View all 10 articles
Peatlands are invaluable but threatened ecosystems that store huge amounts of organic carbon globally and emit the greenhouse gasses carbon dioxide (CO2) and methane (CH4). Trophic interactions of microbial groups essential for methanogenesis are poorly understood in such systems, despite their importance. Thus, the present study aimed at unraveling trophic interactions between fermenters and methanogens in a nitrogen-limited, subarctic, pH-neutral fen. In situ CH4 emission measurements indicated that the fen is a source of CH4, and that CH4 emissions were higher in plots supplemented with ammonium compared to unsupplemented plots. The amino sugar N-acetylglucosamine was chosen as model substrate for peat fermenters since it can serve as organic carbon and nitrogen source and is a monomer of chitin and peptidoglycan, two abundant biopolymers in the fen. Supplemental N-acetylglucosamine was fermented to acetate, ethanol, formate, and CO2 during the initial incubation of anoxic peat soil microcosms without preincubation. Subsequently, ethanol and formate were converted to acetate and CH4. When methanogenesis was inhibited by bromoethanesulfonate, acetate and propionate accumulated. Long-term preincubation considerably increased CH4 production in unsupplemented microcosms and microcosms supplemented with methanogenic substrates. Supplemental H2-CO2 and formate stimulated methanogenesis the most, whereas acetate had an intermediary and methanol a minor stimulatory effect on methane production in preincubated microcosms. Activity of acetogens was suggested by net acetate production in microcosms supplemented with H2-CO2, formate, and methanol. Microbial community analysis of field fresh soil indicated the presence of many physiologically unresolved bacterial taxa, but also known primary and secondary fermenters, acetogens, iron reducers, sulfate reducers, and hydrogenotrophic methanogens (predominately Methanocellaceae and Methanoregulaceae). Aceticlastic methanogens were either not abundant (Methanosarcinaceae) or could not be detected due to limited coverage of the used primers (Methanotrichaceae). The collective results indicate a complex interplay of synergy and competition between fermenters, methanogens, acetogens, and potentially iron as well as sulfate reducers. While acetate derived from fermentation or acetogenesis in this pH-neutral fen likely plays a crucial role as carbon source for the predominant hydrogenotrophic methanogens, it remains to be resolved whether acetate is also converted to CH4 via aceticlastic methanogenesis and/or syntrophic acetate oxidation coupled to hydrogenotrophic methanogenesis.
Arctic and boreal peatlands are long-term carbon stores which have sequestered carbon dioxide (CO2) over millennia, and as a consequence have accumulated thick organic layers that can span several meters (Tarnocai et al., 2009). Northern peatlands are considered especially affected by climate warming and may switch from net C sinks to net C sources if greenhouse gas (methane (CH4) and CO2) emissions exceed sequestration with climate warming and changes in precipitation (Gallego-Sala et al., 2018). Moreover, peatland carbon may be lost by leaching of organic matter into streams, lakes, and nearshore ocean systems that may substantially affect water quality and nutrient regimes of typically nutrient-limited aquatic systems in the Arctic (Limpens et al., 2008).
Microorganisms are drivers of greenhouse gas production and consumption in peatland ecosystems (Conrad, 1996; Kolb and Horn, 2012), and microbial activity thus determines whether peatlands are net sources or sinks for greenhouse gasses. In water-saturated anoxic peat with low availability of alternative electron acceptors such as nitrate or sulfate, mineralization of organic matter will be driven primarily by fermentations and methanogenesis (Limpens et al., 2008; Drake et al., 2009). Carbohydrate monomers like glucose, xylose, and N-acetylglucosamine (NAG), which are derived from complex polymers such as cellulose, hemicellulose, and chitin, respectively, are degraded via primary and secondary fermentations to alcohols, short chain fatty acids like acetate, formate or propionate as well as CO2 and molecular hydrogen (H2) (Drake et al., 2009). These fermentation products are eventually consumed by methanogens, which form CH4 and CO2 (Drake et al., 2009).
Chitin (a component of fungal cell walls and the exoskeleton of many invertebrates) is the second most abundant biopolymer on earth, and together with ligno-cellulose, the two refractory compounds constitute the main source of organic matter in wetlands (Mann, 1988; Mitsch, 1994). NAG is the single monomer of chitin, and it has been shown for anoxic river sediments that chitin-derived NAG is typically consumed by chitinolytic fermenters although many non-chitinolytic fermenters have the ability to degrade NAG as well under the premise that it becomes bioavailable (Gooday, 1994; Wörner and Pester, 2019). NAG is also a major monomer in peptidoglycan, a component of bacterial cell walls, and since dead microbial cells constitute an abundant and readily degradable source of organic carbon in peatlands, peptidoglycan-derived NAG can be assumed to be an important substrate for non-chitinolytic fermenters in these ecosystems (Pazos and Peters, 2019; Tveit et al., 2015).
Since NAG is an amino sugar, the decomposition of NAG-containing polymers from organic matter could be important for both carbon and nitrogen cycling in peatlands (Kang et al., 2005; Beier and Bertilsson, 2013). However, information on how NAG is fermented and what trophic links between NAG-consuming fermenters and methanogens exist in peatlands is scarce (Wüst et al., 2009).
Communities of methanogens in peatlands are typically dominated by hydrogenotrophic methanogens that use H2 (and often also formate) as electron donor to reduce CO2 to CH4 and/or aceticlastic methanogens that split acetate into CH4 and CO2 (Kotsyurbenko et al., 2019; Bräuer et al., 2020). Methanol disproportionation to CH4 and CO2 or complete reduction to CH4 with H2 as electron donor are well known. Some peat methanogens utilize methanol, but it is considered to be less important for the overall CH4 production in peatlands (Conrad, 2020). The methanogenic potentials and the contribution of hydrogenotrophic versus aceticlastic methanogenesis in peatlands are variable and depend on various factors such as temperature, pH, hydrology, and vegetation (Kotsyurbenko et al., 2007; Rooney-Varga et al., 2007; Hines et al., 2008; Conrad, 2020). As an example, the deposition of easily degradable organic carbon compounds from Carex roots especially during the growing season has been conceptualized to stimulate methanogenesis and favor aceticlastic methanogens in fens whereas hydrogenotrophic methanogenesis is often more important in acidic bogs covered predominately by Sphagnum mosses that generally produce less methane (Kelly et al., 1992; Kotsyurbenko et al., 2019). Furthermore, CH4 production and the methanogenic communities in one peatland often change seasonally, with depth (i.e., the recalcitrance of the peat organic matter), and based on the microtopography of the sampling site (Galand et al., 2003; Reiche et al., 2008; Hodgkins et al., 2014; Zalman et al., 2018).
In the present study, NAG was chosen as an easily degradable model amino sugar substrate providing both organic carbon and nitrogen to stimulate primary fermentation processes and subsequent processes that are trophically linked to methanogenesis in Puukkosuo fen (Northern Finland), a nitrogen-limited, subarctic, meso-eutrophic and pH-neutral peatland dominated by Carex sedges and Sphagnum mosses (Palmer and Horn, 2015). The stimulatory effect of methanogenic substrates was evaluated to investigate potential bottle necks for the methanogens inhabiting the fen soil. In addition to soil incubation experiments, in situ measurements of CO2 and CH4 emissions as well as microbial community analysis of field fresh soil were performed to unravel bacterial taxa, methanogens and H2-metabolizers present at Puukkosuo fen (Supplementary Figure S1).
The study site, Puukkosuo fen, is located in northeastern Finland (66°22′38″N, 29°18′28″E; elevation 200 m NN). The mean annual air temperature and mean annual precipitation at the site are −0.43 ± 0.09°C and 772 ± 12 mm, respectively (average of years 1966–2011, measured at Oulanka research station). Puukkosuo fen is pH-neutral (pH ~ 6.9), meso-eutrophic and water saturated with a vegetation consisting mainly of mosses (Sphagnum spp.) and graminoids (e.g., Carex spp.). Four replicate soil cores from layers 0 to 20 cm were taken on July 28th, 2010. These four cores were pooled on site and mixed thoroughly. The mixed sample was divided into subsamples for incubation studies and molecular analyses, and the subsamples were transported on ice to the laboratory where they were stored at 4°C for up to 11 months for microcosm analyses or at −80°C for approximately 3 weeks for nucleic acid extractions. The sampled surface soil had a soil moisture content of 90%, soil carbon, soil nitrogen and the carbon-to-nitrogen ratio were 434 g kgDW−1, 29 g/kgDW−1 and 15, respectively (Palmer and Horn, 2015).
In situ gas emissions of fen soil were determined by the closed-chamber technique as described previously (Palmer and Horn, 2015). Briefly, closed poly(methyl methacrylate) (PMMA) chambers were placed onto the soil surface and gas samples (5 mL per sampling timepoint) were taken from the chambers via gas outlets at 4 timepoints (0–3 h). Prior to installation of the chambers, plots were watered with 2 L fen porewater without supplements (control) and with 20 mM supplemented ammonium (4 replicate plots per treatment). Gas samples were injected into gas tight evacuated containers (Exetainers, Labco Limited, High Wycombe, UK), and CO2 and CH4 concentrations were determined using Hewlett-Packard 5980 series II gas chromatographs equipped with thermal conductivity and flame ionization detectors, respectively, as described in Küsel and Drake (1995).
Two separate sets of anoxic microcosms experiments were conducted to assess fermentative and methanogenic potentials of the sampled surface soil (0–20 cm depth), respectively. All incubations were set up in triplicates of 1:10 dilutions of fen soil (20 g soil +180 mL anoxic water or 10 g of soil +90 mL of anoxic water) in gas-tight 0.5 L infusion flasks (Müller & Krempel, Bülach, Switzerland) that were sealed with screw caps and rubber stoppers (Glasgerätebau Ochs, Bovenden, Germany). Headspaces were flushed with sterile argon. For the fermentation potential experiments, the following treatments were set up in May 2011 and incubated at 20°C in the dark for 10 days: (i) unsupplemented control microcosms, (ii) microcosms supplemented with 500 μM N-acetylglucosamine (NAG), (iii) microcosms with 500 μM NAG and bromoethanesulfonate (BES; 20 mM), and (iv) unsupplemented control microcosms with BES. In the latter two microcosms, BES was used as an inhibitor for methanogenesis (Oremland and Capone, 1988), thus allowing for the detection of fermentation products that might be used by methanogens in microcosms without BES. Fermentation potentials were tested without preincubation of the microcosms. Net turnover of carbon and reductant for the time frames day 0–4, 4–10, and 0–10 was calculated as follows: The initial concentrations of compounds measured at the beginning of a time frame were subtracted from final concentrations measured at the end of a time frame in both, NAG treatments and unsupplemented controls. Then, the resulting concentrations in unsupplemented controls with and without BES were subtracted from those in NAG supplemented microcosms with and without BES, respectively, to get net turnover concentrations for each compound. Finally, net turnover concentrations of compounds were multiplied with 8/32 for NAG, 1/2 for formate, 2/12 for ethanol, 2/8 for acetate, 3/14 for propionate, 1/8 for methane, and 1/0 for CO2 to get net turnovers of carbon/reductant for each compound. Positive and negative values indicate net production and consumption of a compound in a given time frame, respectively.
For the methanogenic potential experiments, one subset of microcosms was set up in May 2011 and preincubated for 120 days at 20°C in the dark, and a second subset of microcosms was set up in July 2011 without preincubation. The preincubation served to reduce potentially existing alternative electron acceptors in the soil and thus stimulate peat methanogens. 1 mM formate, 1 mM acetate, 1 mM methanol or 8 vol % H2 (i.e., 8.4 mmol L−1 microcosm slurry) and 2 vol % CO2 (i.e., 2.1 mmol L−1 microcosm slurry) were supplemented at the beginning of the main incubation, which was 11 days at 20°C in the dark. Methanogenic substrates were resupplemented after 8 days of incubation (note that formate was not resupplemented in preincubated microcosms initially supplemented with formate). In addition to microcosms supplemented with potential methanogenic substrates, unsupplemented control microcosms with or without 20 mM BES were set up (note that BES was added at the beginning of the preincubation for the respective control of the preincubated microcosms).
Microcosms’ headspaces and liquid phases were sampled in 1–2 day intervals. Concentrations of CH4 and CO2 in gas samples were analyzed with a gas chromatograph (Hewlett Packard 5890 series II equipped with FID or TCD detector); reliable concentrations of H2 could not be quantified due to technical problems; concentrations of sugars, organic acids, and alcohols in liquid samples were analyzed via high pressure liquid chromatography equipped with an Aminex HPX-87H ion exclusion column and refractive index and UV detectors (Küsel and Drake, 1995). Amounts of gaseous and dissolved compounds in microcosms are given as μM concentrations and can be converted to μmol per g dry weight of soil by dividing with a conversion factor of 10.5.
Nucleic acids were extracted from a single replicate of fresh homogenized pooled surface layer fen soil as previously described using a bead-beating protocol (Peršoh et al., 2008; Palmer and Horn, 2015). Bacterial 16S rRNA genes as well as the functional gene markers mcrA, [FeFe]-hydrogenase and group 4 [NiFe]-hydrogenase genes were amplified using the primer pairs 341F (5′-CCT ACG GGA GGC AGC AG-3′)/907RM (5′-CCG TCA ATT CMT TTG AGT TT-3′) (Muyzer et al., 1993), ME1 (5′-GCM ATG CAR ATH GGW ATG TC-3′)/ME2 (5′-TCA TKG CRT AGT TDG GRT AGT-3′) (Hales et al., 1996), NiFe-gF (5′-GAY CGI RTI TGY GGI ATY TGY GG-3′)/NiFe-gR (5′-GTR CAI GAR TAR CAI GGR TC-3′), and HydH1f (5′-TIA CIT SIT GYW SYC CIG SHT GG-3′)/HydH3r (5′-CAI CCI YMI GGR CAI SNC AT-3′) (Schmidt et al., 2010, 2011), respectively. Each primer was preceded by a 6-bp-long barcode (5′-ACTATC - gene specific primer-3′), to allow for separation of target sequences after batch sequencing as the sequencing run also included samples that were not part of the present study. Barcoded amplicon pyrosequencing of PCR products was conducted in 2011 as previously described at the Göttingen Genomics Laboratory using the Roche GS-FLX 454/Titanium technology (Palmer et al., 2012; Palmer and Horn, 2015). Sequence analysis of forward reads was done at a later date in QIIME 2 version 2022.2.0 (Bolyen et al., 2019). Imported sequences were demultiplexed using the q2-cutadapt plugin version 2022.2.0 by searching for the barcode + primer with a maximum error rate of 0.1 and removing them as a well as any preceding bases (Martin, 2011).
Quality controlled amplicon sequence variants (ASVs) were generated using the q-dada2 plugin for 454 sequencing (“denoise-pyro”; Callahan et al., 2016) with the following parameters: Sequences were trimmed to a length of 300 bp (“—p-trunc-len” 300), excluding any shorter sequences, and the “consensus” method was used for chimera detection and removal. Taxonomic assignment of ASVs was conducted by subjecting ASVs representative sequences of 16S rRNA gene and functional gene amplicons to a BLAST search of nucleotide (BlastN) and in silico translated protein sequences (BlastX), respectively, to identify the most closely related cultured relatives for each sequence (Altschul et al., 1990). Alpha diversity metrics [observed ASVs, Faith phylogenetic diversity (Faith, 1992), Shannon diversity (Shannon, 1948)] were calculated for reads subsampled at different sequencing depths using the “alpha-rarefaction” command of the q2-diversity plugin version 2022.2.0 in QIIME 2. Amplicon sequence reads have been deposited in the European Nucleotide Archive (ENA) under accession number PRJEB58427.
In Puukkosuo fen, CO2 and CH4 emissions of plots irrigated with unamended porewater were approx. 400 μmol m−2 h−1 and 35 μmol m−2 h−1, respectively (Figures 1A,B). Amendment with ammonium affected CO2 and CH4 emissions: Both CO2 and CH4 emissions were higher from plots with ammonium amendment, indicating that ammonium might stimulate methanogenesis as well as general microbial activity via alleviation of nitrogen limitation. The resulting CO2:CH4 ratios were approx. 12.5 and 18 for unamended and ammonium amended plots, respectively (Figure 1C).
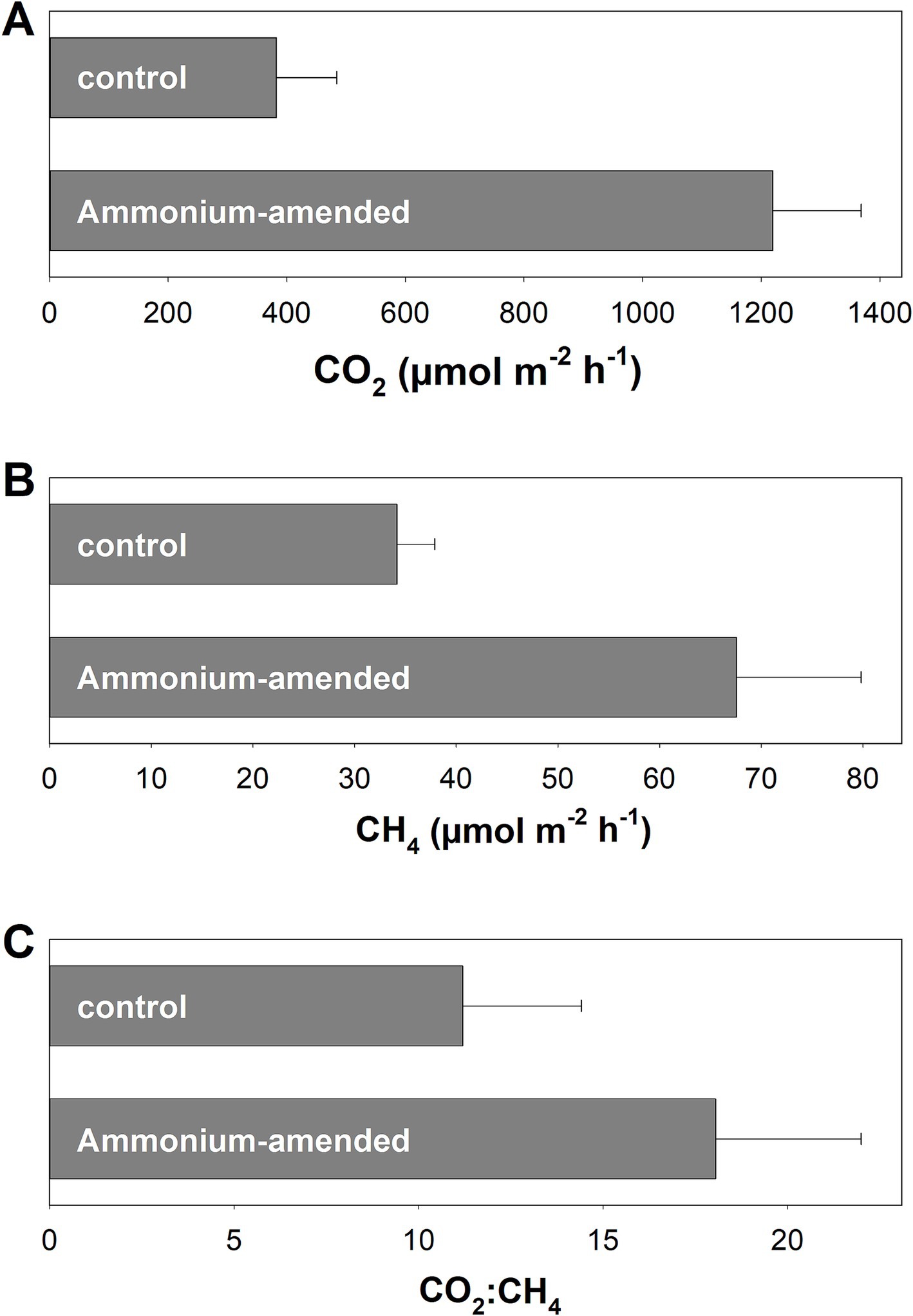
Figure 1. In situ emission of CO2 (A) and CH4 (B) as well as CO2:CH4 ratios (C) in control plots and plots amended with 2 L of fen porewater containing 20 mM ammonium. Measurements were conducted in August 2010. Mean values and standard errors of 4 replicates per treatment are displayed.
Amplicon pyrosequencing of bacterial 16S rRNA genes yielded a total of 3,459 quality filtered reads forming 239 ASVs. Alpha diversity parameters indicated that the sequencing depth was sufficient to cover most of the bacterial diversity present in the fen (Supplementary Figure S2). Blast search revealed low (<90%) identity of some ASV representatives to sequences of cultured representatives (Supplementary Table S1), indicating that there is a high degree of uncultured bacterial diversity in Puukkosuo fen. The bacterial community was dominated by Proteobacteria, Actinobacteriota, Acidobacteria and Firmicutes (Figure 2A), microbial taxa that are commonly found to be abundant in peatlands (Pankratov et al., 2005; Dedysh, 2011; Tveit et al., 2013). Some of the detected ASVs were affiliated to genera known to comprise fermenters (e.g., Clostridium, Pelosinus, Opitutus, and Spirochaeta), acetogens (Clostridium), iron reducers (e.g., Aciditherimonas, Anaeromyxobacter, and Geobacter), and sulfate reducers (e.g., Desulfosporosinus and Desulfomonile) (Supplementary Table S1).
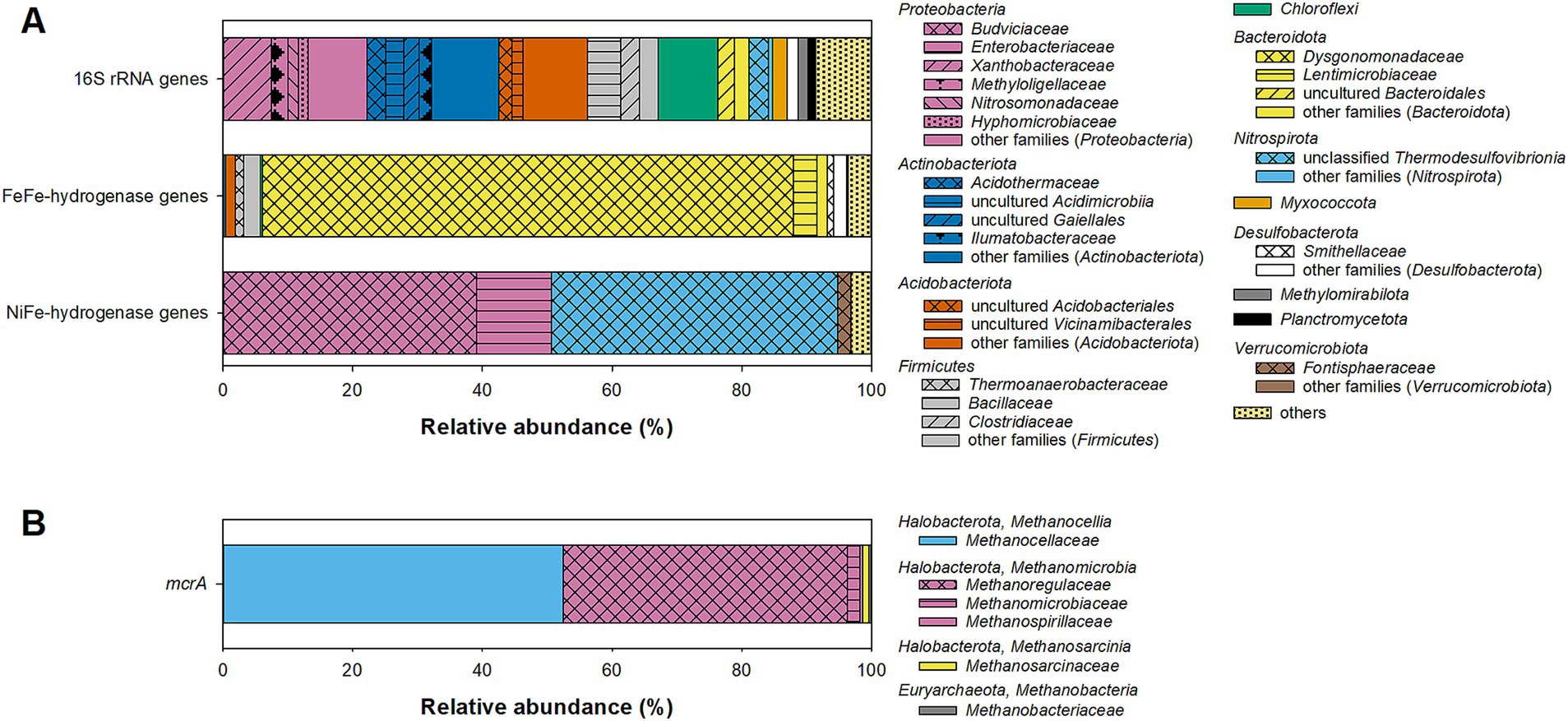
Figure 2. Relative abundance of bacterial (A) and archaeal (B) taxa in forward reads of (A) 16S rRNA genes, [FeFe]-hydrogenase genes, and group 4 [NiFe]-hydrogenase genes and (B) mcrA genes in field-fresh pH-neutral fen soil. Taxonomic classification was done on family-level based on the classifications in Supplementary Table S1. Families with relative abundances <1.5% were grouped as “other families (phylum).” Phyla with a relative abundance <1% and unclassified sequences were grouped as “others” in (A).
Nine hundred and eighty-one quality-filtered [FeFe]-hydrogenase reads were obtained which grouped into 58 ASVs (see Supplementary Figure S3 for alpha diversity plots). Identities of ASV representative sequences to sequences of the next cultured representatives ranged between 56 and 89% (Supplementary Table S1). Based on a previously established family-level hydrogenase amino acid sequence similarity cut-off of 80% (Schmidt et al., 2011), 82% of the [FeFe]-hydrogenase reads could be affiliated to the Dysgonomonadaceae, a family harboring propionate fermenters that can degrade carbohydrates and/or proteins (Hahnke et al., 2016) (Figure 2A). Other [FeFe]-hydrogenase ASVs with low relative abundance had identities above or next to 80% to known syntrophic fermenters (Syntrophus and Smithella), sulfate reducers (Desulfovirgula), and iron reducers (Mesoterricola) (de Bok et al., 2001; Kaksonen et al., 2007; James et al., 2016; Itoh et al., 2023) (Supplementary Table S1). Several ASVs were only distantly related (<70% identity) to hydrogenases of cultured bacteria (Supplementary Table S1), and an adequate phylogenetic or functional affiliation of such essentially novel [FeFe]-hydrogenases is not possible.
Two hundred and sixty quality-filtered reads were obtained for group 4 [NiFe]-hydrogenase genes, and it is likely that such low sequencing depth was insufficient to capture the full diversity of group 4 [NiFe]-hydrogenase genes. However, flattening out rarefaction curves indicated that the 20 ASVs that were detected largely represent the hydrogenase diversity that could be amplified with the used primer pair, which was originally designed to detect group 4 [NiFe]-hydrogenases of Gammaproteobacteria (Schmidt et al., 2011) (Supplementary Figure S4). Identities of ASV representative sequences to sequences of the next cultured representatives ranged from 68 to 86% (Supplementary Table S1). About one half of the group 4 [NiFe]-hydrogenase reads were affiliated to the Enterobacterales-families Budviciaceae and Enterobacteriaceae that harbor facultative aerobes performing mixed acid fermentation under anoxic conditions (Brenner and Farmer, 2005; Adeolu et al., 2016) (Figure 2A). Most of the remaining group 4 [NiFe]-hydrogenase reads formed three ASVs with 71% identity to the H2-oxidizing sulfate reducer Thermodesulfovibrio thiophilus (Supplementary Table S1); however, due to the relatively low identity, a closer phylogenetic and functional affiliation of the detected hydrogenases remains elusive.
For mcrA, 1,661 quality-filtered reads were obtained, which grouped into 37 ASVs (see Supplementary Figure S5 for alpha diversity plots). Most of the mcrA reads were affiliated either to the Methanoregulaceae (44% relative abundance) or the Methanocellaceae (52% relative abundance; Figure 2B). Both families comprise hydrogenotrophic methanogens that can be commonly found in peatlands or other wetlands (Oren, 2014a; Sakai et al., 2014). Notably, only one ASV (mcrA-fw-33; 1% relative abundance) was affiliated with the Methanosarcinaceae, a family that comprise many aceticlastic methanogens (note that the closest cultured relative, Methanosarcina lacustris (81% identity), does not utilize acetate as a growth substrate) (Oren, 2014b). None of the ASVs was affiliated with Methanothrix (formerly Methanosaeta), a genus comprising exclusively aceticlastic methanogens that have been repeatedly detected in peatlands (Kotsyurbenko et al., 2019; Bräuer et al., 2020). However, the used primer pair ME1/2 has been reported to be unsuitable for the amplification of Methanothrix (e.g., Lueders et al., 2001; Shigematsu et al., 2004), which needs to be taken into consideration.
In unsupplemented microcosms without BES, CO2 steadily accumulated, and it was by far the most prominent of the detected products (Figure 3). CH4 concentrations increased steadily as well, but comparatively low amounts of the gas accumulated during the 10 days of anoxic incubation (Insert in Figure 3G). The relatively low accumulation of CH4 compared to that of CO2 is reflected by generally high CO2:CH4 ratios that were highest at day 2 (1,600) and subsequently decreased to ~300 toward the end of the incubation (Supplementary Figure S6). Thus, while methanogenesis became relatively more important over time, alternative (and unresolved) respiratory processes dominated the mineralization of endogenous peat organic carbon. Other fermentation products frequently detected in anoxic peat microcosms like acetate, formate, ethanol, and propionate did not accumulate (Figures 3B–E). In unsupplemented microcosms with added BES, CH4 accumulation was suppressed while the residual product profile was overall similar to that in unsupplemented microcosms without BES.
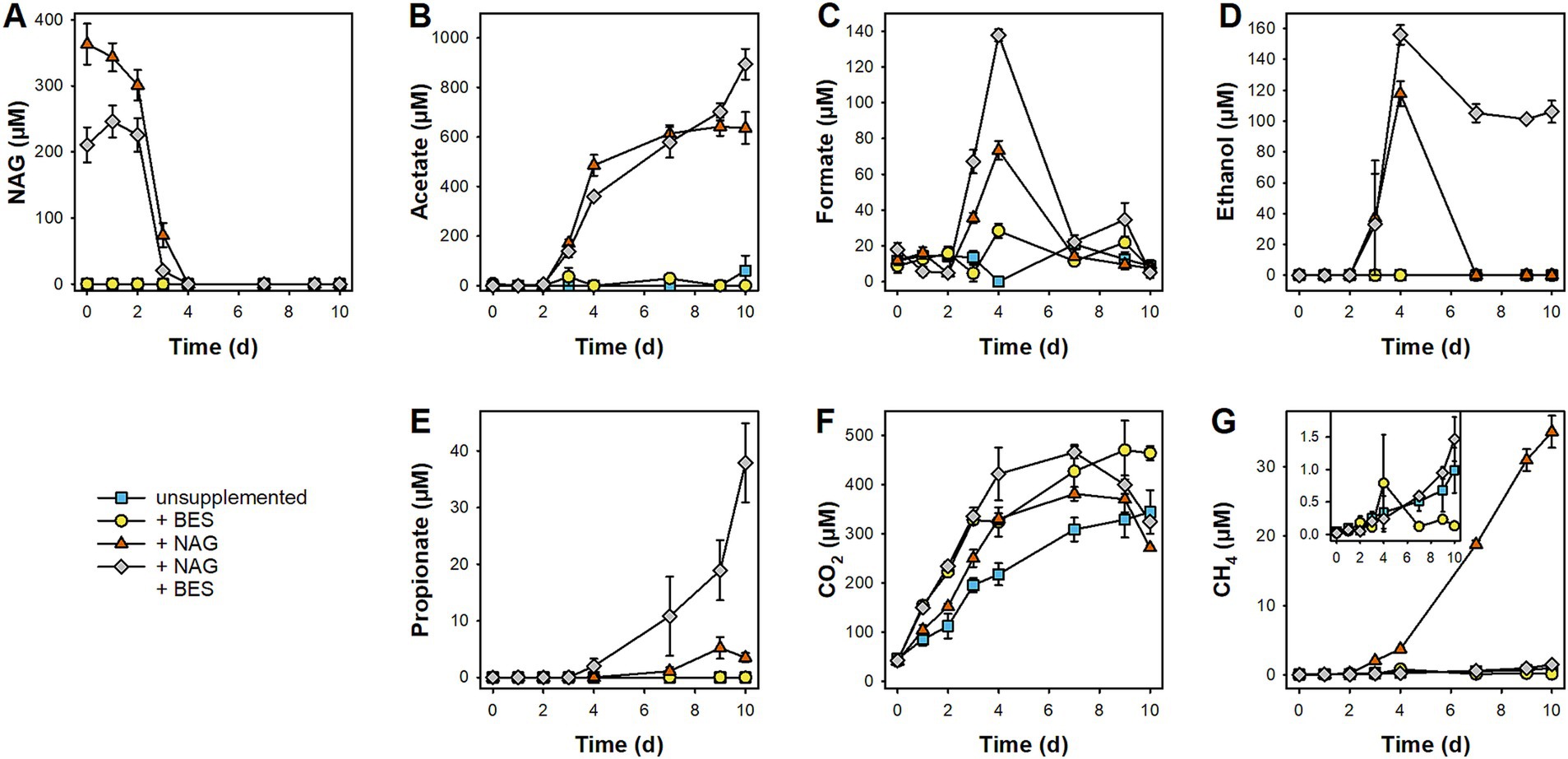
Figure 3. Effect of N-Acetylglucosamine (NAG) on production and consumption of fermentation products and methane in pH-neutral fen soil during 10 days of anoxic incubation. Microcosms were incubated in absence and presence of 20 mM BES (+ BES) as well as without and with supplemented NAG (+ NAG). Mean values of three replicates and standard errors are displayed for concentrations of NAG (A), acetate (B), formate (C), ethanol (D), propionate (E), CO2 (F), and CH4 (G). The insert in panel (G) is an enlargement of the low concentration range to better show low levels of CH4.
In NAG-supplemented microcosms without BES, NAG was consumed completely within 4 days without appreciable delay, with most rapid consumption from day 2 to 4 (Figure 3A). NAG consumption was accompanied by the accumulation of acetate, formate, ethanol, and CO2, suggesting ongoing primary fermentation (Figure 3). In the second stage of the incubation (Day 4–10), formate, ethanol, and partially CO2 were consumed while mainly CH4, some acetate, and traces of propionate accumulated. In NAG-supplemented microcosms with BES, the same fermentation products were observed as in NAG-supplemented microcosms without BES during the initial stage (Day 0–4), however, transient accumulation of formate and ethanol were more prominent with BES (Figure 3). In the subsequent stage (Day 4–10), concentrations of formate, ethanol, and CO2 decreased, whereas acetate and some propionate accumulated in NAG-supplemented microcosms with BES, in which methanogenesis was largely blocked.
Net carbon and reductant turnover analysis was performed to determine which of the observed products could be directly linked to NAG consumption and which products were formed during the degradation of primary NAG-fermentation products. In NAG-supplemented microcosms without BES, only about 50% of the carbon and reductant theoretically derived from the amount of NAG consumed could be recovered in the detected products during the initial stage (Day 0–4, Figures 4A,C). Acetate accounted for ~75% of recovered carbon and reductant and was therefore the dominant detected fermentation product followed by ethanol, CO2, and formate. During the second stage (Day 4–10, Figures 4A,C), net consumption of ethanol, formate, and CO2 could account for the net production of acetate and CH4. Carbon and reductant recovered in acetate, ethanol, formate, and CO2 accounted for most of the NAG that was degraded during the initial incubation in NAG-supplemented microcosms with BES (Day 0–4, Figures 4B,D). In the subsequent stage (Day 4–10, Figures 4B,D), only a part of the acetate and propionate that were formed could be explained by the observed consumption of ethanol, formate, and CO2, indicating that other undetected sources of carbon and reductant were converted as well. Furthermore, when comparing the net carbon and reductant turnover in NAG-supplemented microcosms without BES to that in NAG-supplemented microcosms with BES over the whole 10 day incubation period, it was remarkable that less than half the consumed carbon and reductant could be recovered in the absence of BES (Day 0–10) whereas more carbon and reductant were recovered in the products than what was theoretically available from the substrates in the presence of BES (Figure 4). The reasons for this discrepancy were unknown.
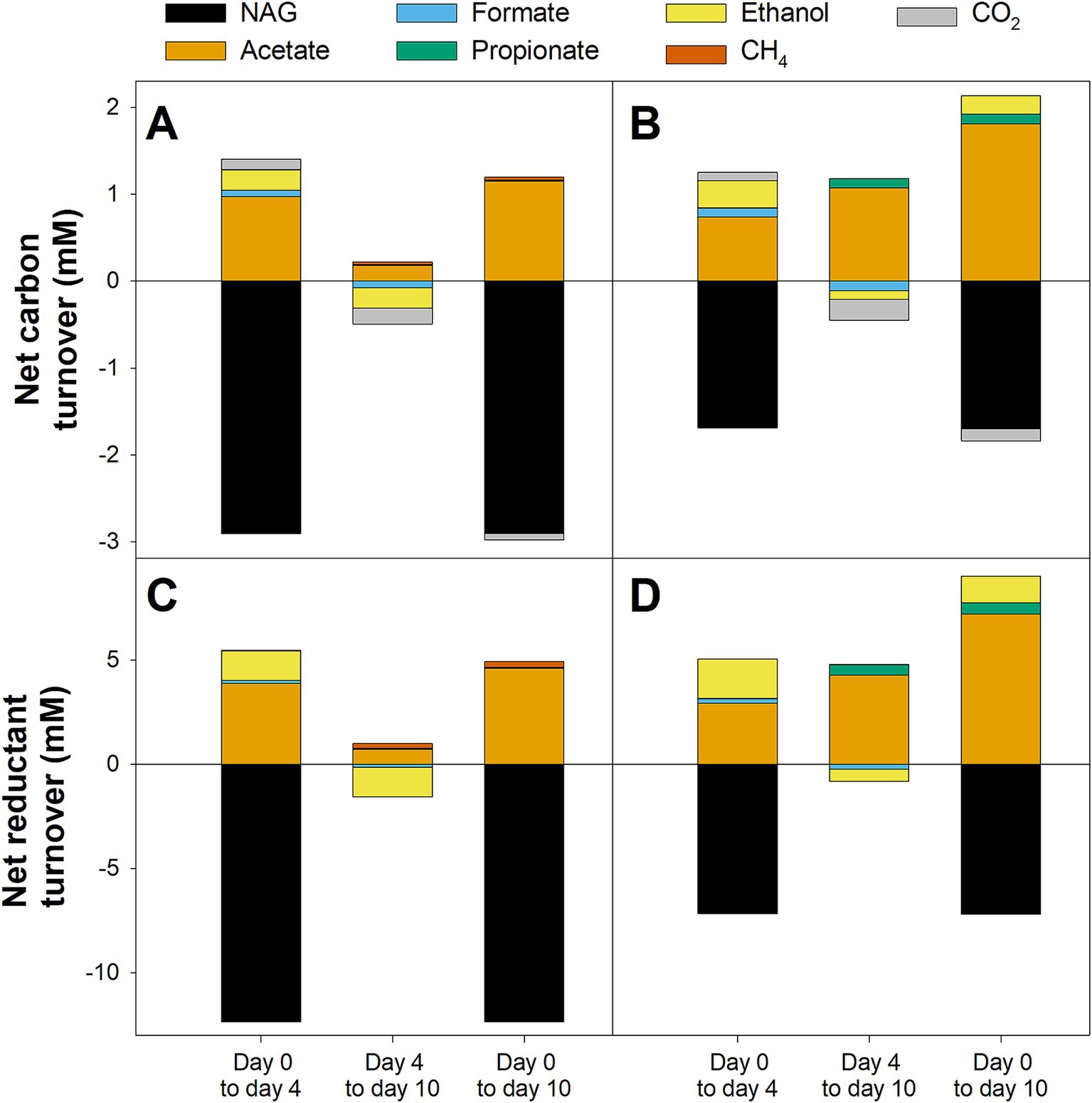
Figure 4. Net turnover of carbon (A,B) and reductant (C,D) for different stages in N-Acetylglucosamine (NAG) supplemented anoxic fen soil microcosms. Microcosms were supplemented with NAG and incubated in absence (A,C) or presence of 20 mM BES (B,D). See the materials and methods section for a detailed description of the calculation. Mean values of three replicates are displayed.
CH4 accumulation in unsupplemented microcosms without preincubation was slow (Figure 5). An equally slow CH4 accumulation was observed in unsupplemented microcosms of the fermentation experiment that were also not preincubated (Figure 3). In contrast, CH4 accumulated readily and without delay in unsupplemented microcosms preincubated for 120 d in which the final CH4 concentrations were more than 30 times higher than in unsupplemented microcosms without preincubation (Figure 5). All tested supplemented substrates (H2/CO2, formate, acetate, and methanol) stimulated CH4 production in microcosms with and without preincubation; however, the stimulatory effect was more pronounced in preincubated microcosms (Figure 5). In fact, the fast initial consumption of supplemental formate and supplemental acetate in microcosms without preincubation was largely uncoupled from CH4 accumulation, indicating that these substrates were predominately consumed by microbial oxidation processes others than methanogenesis at least at the start of the incubation (Figure 5; Supplementary Figure S7). In preincubated microcosms, the stimulatory effect on CH4 accumulation was highest for H2/CO2 and formate and lowest for methanol (Figure 5). Note that, in contrast to the other substrates, formate was not resupplemented in preincubated microcosms (Supplementary Figure S7), and therefore CH4 accumulation slowed down once the initially supplemented formate was consumed (Figure 5). Supplemental H2/CO2 did not only stimulate methane accumulation but also the accumulation of acetate, especially in microcosms without preincubation (Supplementary Figure S7), indicating that hydrogenotrophic methanogenesis and hydrogenotrophic acetogenesis were going on in parallel. Acetate also accumulated in BES-supplemented microcosms without preincubation and during the preincubation in BES-supplemented microcosms with preincubation in which methanogenesis was largely blocked (Figure 5; Supplementary Figure S7). Whether acetate accumulation in BES-supplemented microcosms was due to ongoing acetogenesis or fermentation could not be resolved. However, the accumulation of propionate, either during the incubation or during the preincubation, indicates that fermentation processes were going on in BES-supplemented microcosms (Supplementary Figure S7).
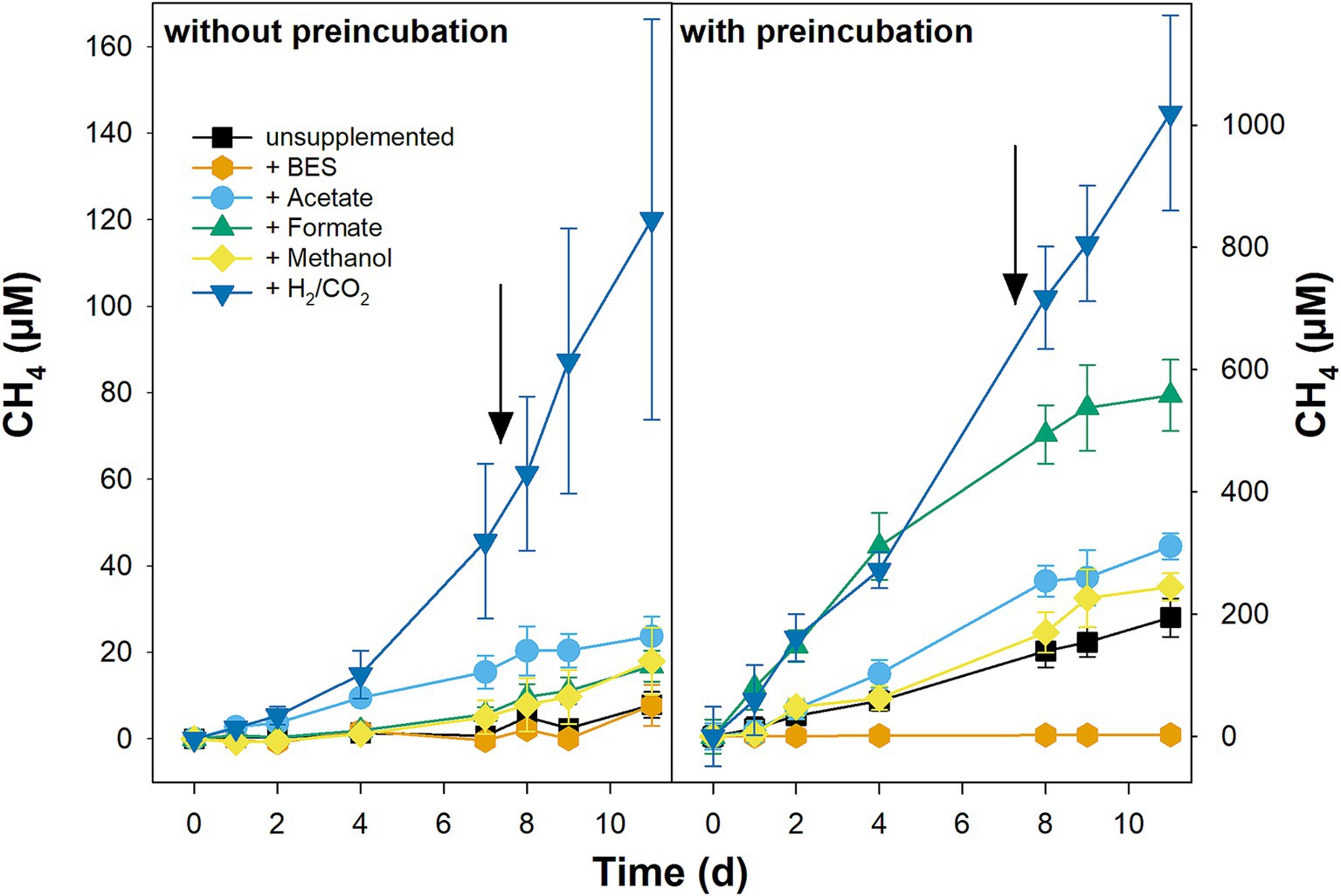
Figure 5. Effect of methanogenic substrates on the production of CH4 in pH-neutral fen soil. Microcosms were set-up without (left) and with (right) preincubation of 120 days to reduce alternative electron acceptors. Microcosm were left unsupplemented or supplemented with 20 mM BES, 1,000 μM acetate, formate, or methanol, or with 8% vol. H2 and 2% vol. CO2. Substrates were resupplemented after sampling on day 7 (indicated by arrows; formate was not resupplemented in the preincubated microcosms). Mean values of three replicates and standard errors are displayed. Methane present at t0 was subtracted to allow for better comparison of net CH4 production in microcosms without and with preincubations.
In summary, the stimulatory effect of methanogenic substrates could be better evaluated in microcosms with 120 d preincubation in which electron acceptors other than CO2 were presumably largely depleted. The potential for hydrogenotrophic methanogenesis was higher than for aceticlastic methanogenesis, while the potential for methylotrophic methanogenesis was low during the 11 d incubation.
To date, peatlands are well recognized for their complex importance for the global climate since they can function as sinks or sources for the greenhouse gasses CO2, CH4, and N2O (Frolking and Roulet, 2007; Strack et al., 2008; Harris et al., 2022). Previously, the potential to function as N2O sink has been reported for the pH-neutral Puukkosuo fen (Palmer and Horn, 2015). In the present study, in situ gas measurements at Puukkosuo fen showed that CH4 was emitted alongside CO2, indicating that at the time point of the measurements methanogenesis was ongoing and exceeded or bypassed methane oxidation (Figure 1B). The 35 μmol CH4 m−2 h−1 that have been emitted on average were in the lower range of what has been observed in other peatlands (Horn et al., 2003; Hamberger et al., 2008; Salmon et al., 2022). However, the values given in the aforementioned references are maximum emissions for the respective sites measured over a much longer period than in the present study, and prolonged measurement campaigns would be needed to determine the temporal changes and the range in CH4 emissions at Puukkosuo fen over different seasons and years. The finding that in situ CH4 emissions were higher when ammonium was amended indicated that methanogenesis was limited by N-availability (Figure 1B), which is in line with the low concentrations of inorganic nitrogen reported for the fen (Palmer and Horn, 2015). Puukkosuo fen is a pristine peatland and anthropogenic N-influx or -deposition is likely limited. Thus, methanogens and other microbes rely on natural reactive nitrogen input like precipitation and groundwater flow, energy intensive nitrogen fixation, and/or the release of nitrogen from the decomposition of peat organic matter.
As discussed above, the overall microbial activity and methanogenesis in particular are limited by the availability of nitrogen in Puukkosuo fen. Therefore, NAG as an easily degradable carbon and nitrogen source was chosen as model substrate to study primary fermentation and subsequent degradation processes in peat soil microcosms. The immediate fermentation of NAG indicates that fen microbes are poised to consume NAG (Figure 3). Acetate, ethanol and formate were the primary fermentation products that were observed during NAG consumption in the presence and absence of BES (Figures 3, 4). The fermentation profile is indicative of mixed acid fermentation as performed by facultative aerobic enterobacteria (Müller, 2008) which were abundant in group 4 [NiFe]-hydrogenase gene libraries of fresh peat soil (families Budviciaceae and Enterobacteriaceae in Figure 2). However, the detected fermentation products can be also produced by other fermentative taxa like many members of the Clostridiaceae (Wiegel, 2015), a family that accounted for 3% of the 16S rRNA gene sequences retrieved from fresh peat soil (Figure 2). A time-resolved community analysis on transcript or protein level would be necessary to better identify active fermenters in NAG-supplemented peat soil microcosms.
Acetate and ethanol were also observed in NAG-supplemented microcosms with peat soil from a temperate, moderately acidic fen (Wüst et al., 2009). While acetate is also commonly observed as major fermentation product in peat soil microcosms supplemented with glucose or xylose, ethanol was not detected so frequently when these sugar monomers were fermented (Kotsyurbenko et al., 1996; Hamberger et al., 2008; Wüst et al., 2009; Hunger et al., 2015). Nevertheless, ethanol was detected in fresh and incubated peat soil and was a potential product of the fermentation of root-derived organic carbon (Metje and Frenzel, 2005, 2007; Tveit et al., 2015; Meier et al., 2021). Butyrate and propionate are other fermentation products that can transiently or permanently accumulate in sugar-supplemented peat soil microcosms (Kotsyurbenko et al., 1996; Hamberger et al., 2008; Wüst et al., 2009; Hunger et al., 2015). However, propionate accumulation was largely uncoupled from NAG consumption and butyrate was not detected in the present study (Figure 3). Interestingly, butyrate was observed during NAG fermentation when the initial concentrations of the substrate were increased to 0.9 mM or higher (i.e., at least twice as high as in the present study), suggesting a tight coupling of butyrate production and consumption in the fen (Wüst et al., 2009).
While propionate accumulation was not observed in unsupplemented microcosms with BES (without preincubation) in the NAG experiment (Figure 3E), propionate accumulated after day 7 in the respective treatment of the methanogenic substrates experiment (Supplementary Figure S7). In addition, propionate accumulated during the preincubation of microcosms with BES (Supplementary Figure S7). These findings indicated that propionate was produced during the fermentation of endogenous organic matter derived from Puukkosuo fen soil. In this regard, (transient) propionate accumulation was observed in microcosms with unsupplemented peat soil before (Schmidt et al., 2015; Tveit et al., 2015), suggesting that propionate is a common product of the fermentation of peat-derived organic carbon. Taxa known to comprise propionate fermenters dominated in [FeFe]-hydrogenase gene libraries (Dysgonomonadaceae and Lentimicrobiaceae (Chen and Dong, 2005; Hahnke et al., 2016; Sun et al., 2016), Figure 2) and were also present in 16S rRNA gene libraries (ASV r85, 97.3% identity to Psychrosinus fermentans (Sattley et al., 2008), and ASV r238, 98.3% identity to Opitutus sp. VeSm13 (Janssen et al., 1997); Supplementary Table S1) of fresh Puukkosuo fen soil.
The term secondary fermenter is mostly used when describing a syntrophic association in which an alcohol- or organic acid-oxidizing bacterium (i.e., the secondary fermenter) depends on H2 (or formate) removal by a methanogenic partner (Schink, 1997). However, ethanol can be converted to propionate and acetate independent of interspecies hydrogen transfer between a fermenter and a methanogen (Schink et al., 1987), and such a conversion of ethanol was proposed to occur in anoxic incubations with arctic, pH-neutral peat soil (Tveit et al., 2015). In the current study, ethanol consumption in parallel to propionate accumulation was observed especially in the second stage (between day 4 and day 10) of NAG treatments with BES in which methanogenesis was inhibited (Figure 3). Propionate accumulation was less pronounced in NAG treatments without BES in which the transiently accumulated ethanol was completely consumed between day 4 and day 7 (Figure 3). This suggested that ethanol oxidation was largely performed in syntrophic association without the formation of propionate [as known for Pelobacter carbinolicus (Schmidt et al., 2014)] when methanogenesis was not inhibited. Alternatively, syntrophic oxidation of propionate might have prevented the accumulation of propionate derived from ethanol oxidation in NAG treatments without BES. In this respect, [FeFe]-hydrogenase genes affiliated with the family Smithellaceae were detected in fresh peat soil (Figure 2) and the type species of this family, Smithella propionica, is capable of syntrophic propionate oxidation (de Bok et al., 2001). Syntrophic propionate oxidation likely also prevented the accumulation of propionate derived from the fermentation of endogenous organic carbon in unsupplemented microcosms with or without preincubation (Supplementary Figure S7). Syntrophic propionate oxidation was studied in a moderately acidic fen in Germany in which Smithella and Syntrophobacter, two genera with contrasting metabolic pathways of propionate oxidation, were identified as active propionate oxidizers (Schmidt et al., 2016). However, the responsible propionate oxidizers in the neutral Puukkosuo fen still need to be identified.
Hydrogenotrophic methanogens are well known partners of syntrophic fermenters. mcrA-gene libraries of fresh peat soil were largely dominated by ASVs that were affiliated to either Methanoregulaceae or Methanocellaceae (Figure 2), both of which comprise hydrogenotrophic methanogens (Oren, 2014a; Sakai et al., 2014). Both families together represent the dominant groups of hydrogenotrophic methanogens in various peatlands (Galand et al., 2002, 2005; Hunger et al., 2011, 2015; Yavitt et al., 2012; Blake et al., 2015; Schmidt et al., 2016; Bräuer et al., 2020). In line with the high abundance of hydrogenotrophic methanogens in fresh Puukkosuo fen soil, CH4 production was highest in H2/CO2-supplemented microcosms with or without preincubation (Figure 5). Except for Methanoregula boonei, all other cultured members of the Methanoregulaceae and Methanocellaceae can utilize formate in addition to H2 as electron donor during methanogenesis (Oren, 2014a; Sakai et al., 2014). Thus, it was not surprising that supplemental formate stimulated CH4 production equally well as supplemental H2/CO2 until formate got depleted in microcosms with preincubation (Figure 5). Interestingly, the stimulatory effect of formate was much weaker compared to that of H2/CO2 in microcosms without preincubation although formate was available throughout the incubation (Figure 5; Supplementary Figure S7). The lower availability of acetate might have caused weaker methane production in formate-supplemented microcosms without preincubation compared to those supplemented with H2/CO2 (Supplementary Figure S7), since all cultured members of the Methanoregulacea and Methanocellaceae require acetate for growth (Oren, 2014a; Sakai et al., 2014).
While supplemental H2/CO2 and formate commonly stimulate methanogenesis in peat microcosms, supplemental acetate can have a positive, neutral or negative effect (Williams and Crawford, 1984; Kotsyurbenko et al., 1996; Horn et al., 2003; Bräuer et al., 2004; Wüst et al., 2009; Hunger et al., 2011, 2015; Blake et al., 2015). The inhibitory effect of acetate on methanogens is observed exclusively in acidic peatlands in which undissociated acetic acid permeating into the cell can be toxic to methanogens at elevated concentrations (Russell, 1991; Horn et al., 2003; Bräuer et al., 2004). A stimulatory effect of supplemental acetate on CH4 production, as observed in the present study (Figure 5) and in other peatlands (Bräuer et al., 2004; Hunger et al., 2015), indicates that the potential of acetate consumption by the complex microbial community involved in methanogenic degradation exceeds the acetate production from endogenous organic carbon. As discussed above, the stimulatory effect of acetate in the present study could be explained by the acetate requirements for growth of Methanoregulaceae and Methanocellaceae, the two dominant methanogenic taxa observed in fresh peat soil (Figure 2) (Oren, 2014a; Sakai et al., 2014). In addition, acetate might have been converted to CH4 either in syntrophic cooperation between acetate oxidizers and hydrogenotrophic methanogens (Nusslein et al., 2001; Hattori, 2008) or by aceticlastic methanogens. It must be pointed out that aceticlastic methanogens of the family Methanosarcinaceae were largely absent in the mcrA-gene libraries of field fresh fen soil (Figure 2). However, in a slightly alkaline fen in Germany Methanosarcinaceae were not detected in field fresh soil, but represented 4–27% of the mcrA-gene sequences after 3 weeks of incubation in microcosms with contrasting substrate supply (see Mire 1 in Hunger et al., 2015), underlining that rare aceticlastic methanogens can considerably increase in relative abundance within comparatively short incubation times. Furthermore, the relative abundance of aceticlastic Methanotrichaceae in field fresh soil could not be evaluated due to the insufficient coverage of their mcrA genes by the primer set ME1/2 (Lueders et al., 2001; Shigematsu et al., 2004).
Methanol, which is released by living plants and the degradation of plant material (Galbally and Kirstine, 2002), is conceptualized to be only of minor importance as a substrate for methanogens in peatlands (Drake et al., 2009; Kotsyurbenko et al., 2019). Nevertheless, methanol can have a stimulatory effect on methanogenesis in peat soil microcosms (Bräuer et al., 2004; Wüst et al., 2009; Jiang et al., 2010). In the present study, a relatively weak stimulation of methanogenesis was observed in Methanol-supplemented microcosms with and without preincubation (Figure 5), suggesting that methanol is probably not a major substrate for methanogenesis in Puukkosuo fen.
The ecological significance of acetogens in peatlands is still uncertain despite recent findings that indicated the contribution of acetogens to acetate production in some peatlands (Drake et al., 2009; Hunger et al., 2011, 2015; Hädrich et al., 2012; Ye et al., 2014; Kotsyurbenko et al., 2019; Meier et al., 2022). In the 16S rRNA gene library of field fresh peat soil, ASV r72 (0.8% relative abundance) and ASV r73 (0.3% relative abundance) were affiliated with the two acetogens Clostridium magnum (98.3% identity) and Clostridium muellerianum (94.3% identity), respectively (Supplementary Table S1) (Bomar et al., 1991; Doyle et al., 2022). In this respect, distinct acetate accumulation in H2/CO2-supplemented microcosms without preincubation and to a lesser extent in H2/CO2-supplemented microcosms with preincubation as well as in formate-supplemented microcosms with and without preincubation suggested that acetogens successfully competed with methanogens for both substrates under the experimental conditions (Supplementary Figure S7). Similarly, acetate accumulation was observed in H2/CO2 or formate supplemented microcosms in various peatlands, suggesting that acetogens successfully compete with methanogens at elevated concentrations of these substrates (Kotsyurbenko et al., 1996; Horn et al., 2003; Bräuer et al., 2004; Wüst et al., 2009; Hunger et al., 2011, 2015, 2016; Hädrich et al., 2012; Meier et al., 2022). Some acetate accumulated in methanol-supplemented microcosms with or without preincubation (Supplementary Figure S7), and it is possible that the weak stimulatory effect of methanol on methanogenesis was due to trophic links between acetogens and methanogens rather than direct consumption of methanol by methanogens (Jiang et al., 2010). Furthermore, ethanol can be a substrate for acetogens (Bertsch et al., 2016), and the finding that ethanol was consumed parallel to acetate accumulation during the second stage of the NAG-supplemented microcosms with or without BES might indicate that acetogens were involved in the degradation of NAG-derived ethanol (Figures 3, 4). In this regard, it has been hypothesized that acetogens could be involved in the degradation of ethanol released during the fermentation of root organic carbon in peatlands (Meier et al., 2021, 2022).
As discussed above, syntrophic acetate oxidation to CH4 might play a role in Puukkosuo fen. In such a trophic cooperation acetate conversion to H2/CO2 or formate by the acetate oxidizer can be conducted via the oxidative citric acid cycle or the oxidative acetyl-CoA cycle (Müller et al., 2015; Manzoor et al., 2016). The latter resembles the reversal of H2/CO2- or formate-dependent acetogenesis (Zinder, 1994), and some acetogens can indeed reverse their metabolism according to the thermodynamic conditions and substrate availability (Lee and Zinder, 1988; Oehler et al., 2012). However, if acetogens function as acetate oxidizers when H2 concentrations are low and acetogenesis becomes thermodynamically unfavorable in Puukkosuo fen remains to be elucidated.
In theory, CO2 and CH4 are produced in a 1:1 ratio during the complete anaerobic degradation of organic carbon at the redox level of glucose if methanogenesis is the sole terminal electron accepting process (C6H12O6 → 3CO2 + 3CH4) (Zinder, 1993), whereas ratios >1 are usually attributed to the use of other inorganic electron acceptors like O2, nitrate, ferric iron, or sulfate in addition to CO2 (McCarty, 1972). Humic substances that are abundant in peat can be used as organic electron acceptors, and thus also contribute to high CO2:CH4 ratios in peat soil in situ or in vitro (Heitmann et al., 2007; Roden et al., 2010; Gao et al., 2019). In this study, average CO2:CH4 ratios measured in situ were 12.4 ± 3.1 (Figure 1C), suggesting that methanogenesis was not the sole terminal electron accepting process in Puukkosuo fen at the time point of sampling. In this respect, a high denitrification potential has been observed in Puukkosuo fen although the availability of nitrate is low (Palmer and Horn, 2015). Notable sulfate concentrations were detected in the soil (53 and 23 μg/gDW in 0–20 cm and 20–40 cm soil, respectively), and such a relatively small pool of sulfate can sustain comparatively high rates of sulfate reduction when reduced sulfur compounds are continuously reoxidized (Pester et al., 2012). 16S rRNA gene and [FeFe]-hydrogenase gene ASVs related to sulfate reducers of the genera Desulfosporosinus, Desulfomonile, and Desulfovirgula of fresh peat soil underlined the possibility of sulfate reducers contributing to the anaerobic degradation of organic matter in Puukkosuo fen (Supplementary Table S1). Furthermore, some 16S rRNA gene ASVs were related to iron reducers from the genera Aciditherimonas, Anaeromyxobacter, and Geobacter (Supplementary Table S1), indicating that iron reduction, which can be important in peatlands (Reiche et al., 2008), might play a role in anaerobic degradation of organic carbon in Puukkosuo fen. Moreover, aerobic or anaerobic methane oxidation might have contributed to CO2:CH4 ratios greater than 1 as well (Smemo and Yavitt, 2011; Zalman et al., 2018). In this respect, ASV r154 and ASV r229 of the 16S rRNA gene library were affiliated to the methanotrophic genera Methylocystis and Methylobacter (Supplementary Table S1).
CO2:CH4 ratios in unsupplemented fresh peat soil microcosms were much higher than those observed in situ (Figure 1C; Supplementary Figure S6), supporting the assumption that sample handling, storage, and incubation procedures can obscure methanogenic conditions when peat soil is incubated in vitro, supporting the need for a pre-incubation to better mimic in situ near conditions (Yavitt and Seidman-Zager, 2006).
The accumulation of fermentation products in NAG supplemented microcosms indicated a high fermentation potential of the microbial community in the fen, and formate as well as ethanol as important intermediates (Figure 3). Such fermentations were performed by a fen community harboring phylogenetically novel hydrogenases and organisms, thus the fen represents a reservoir of hitherto unknown microbial diversity.
In the present study, NAG, a monomer of the second most abundant biopolymer chitin, was used as a model compound. Since polymer hydrolysis often is the rate-limiting step resulting in low concentrations of NAG, chitin supplementation, and treatments with NAG in the lower μM range with refeeding coupled to time-resolved mRNA analysis of peat soil microcosms is recommended to identify key organisms of chitin and NAG degradation. Future research should likewise address the fate of acetate at Puukkosuo fen as a model system for pH neutral subarctic fens.
In conclusion, the microbial community of the CH4-emitting Puukkosuo fen showed a pronounced response to supplemental substrates for fermentation and hydrogenotrophic methanogenesis, indicating a high potential activity of both processes. Elucidating active primary and secondary fermenters, the role of acetogens, as well as the pathways of anaerobic acetate conversion to methane and involved taxa are key challenges for future research. Our current study provides a good starting point for conducting such subsequent studies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, PRJEB58427.
KK: Conceptualization, Formal analysis, Funding acquisition, Investigation, Project administration, Visualization, Writing – original draft, Writing – review & editing. OS: Formal analysis, Visualization, Writing – original draft, Writing – review & editing. MH: Conceptualization, Supervision, Writing – review & editing, Funding acquisition, Resources.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Financial support for this study was provided by LAPBIAT (part of the Sixth EU Framework Programme Infrastructures), the German Academic Exchange Service (DAAD), the German Research Foundation (DFG Research Unit 562 “Dynamics of soil processes under extreme meteorological boundary conditions,” FOR562, HO4020/2-2), the University of Bayreuth, and the Research Council of Finland (project 322753 awarded to KK).
The authors would like to thank Harold L. Drake for continuous support, Jyrki Manninen for organizational help, the team at Oulanka research station for excellent on-site support, Sonja Perras Mertens for help with laboratory experiments, the Central Analytics Department of BayCEER for analyses of nitrate, nitrite, ammonium, TC, TN, and TOC, as well as Rolf Daniel and Andrea Thürmer for pyrosequencing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1428517/full#supplementary-material
Adeolu, M., Alnajar, S., Naushad, S., and Gupta, R. S. (2016). Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales Ord. Nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. Nov., Pectobacteriaceae fam. Nov., Yersiniaceae fam. Nov., Hafniaceae fam. Nov., Morganellaceae fam. Nov., and Budviciaceae fam. Nov. Int. J. Syst. Evol. Microbiol. 66, 5575–5599. doi: 10.1099/ijsem.0.001485
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Beier, S., and Bertilsson, S. (2013). Bacterial chitin degradation—mechanisms and ecophysiological strategies. Front. Microbiol. 4:149. doi: 10.3389/fmicb.2013.00149
Bertsch, J., Siemund, A. L., Kremp, F., and Müller, V. (2016). A novel route for ethanol oxidation in the acetogenic bacterium Acetobacterium woodii: the acetaldehyde/ethanol dehydrogenase pathway. Environ. Microbiol. 18, 2913–2922. doi: 10.1111/1462-2920.13082
Blake, L. I., Tveit, A., Øvreås, L., Head, I. M., and Gray, N. D. (2015). Response of methanogens in Arctic sediments to temperature and methanogenic substrate availability. PLoS One 10:e0129733. doi: 10.1371/journal.pone.0129733
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bomar, M., Hippe, H., and Schink, B. (1991). Lithotrophic growth and hydrogen metabolism by Clostridium magnum. FEMS Microbiol. Lett. 83, 347–350. doi: 10.1111/j.1574-6968.1991.tb04488.x
Bräuer, S. L., Basiliko, N., Siljanen, H. M. P., and Zinder, S. H. (2020). Methanogenic archaea in peatlands. FEMS Microbiol. Lett. 367:fnaa172. doi: 10.1093/femsle/fnaa172
Bräuer, S. L., Yavitt, J. B., and Zinder, S. H. (2004). Methanogenesis in McLean bog, an acidic peat bog in upstate New York: stimulation by H2/CO2 in the presence of rifampicin, or by low concentrations of acetate. Geomicrobiol J. 21, 433–443. doi: 10.1080/01490450490505400
Brenner, D. J., and Farmer, J. J. (2005). “Family I. Enterobacteriaceae” in Bergey’s manual of systematics of Archaea and Bacteria. eds. G. M. Garrity, D. J. Brenner, N. R. Krieg, and J. T. Staley (New York: Springer), 587–850.
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chen, S., and Dong, X. (2005). Proteiniphilum acetatigenes gen. Nov., sp. nov., from a UASB reactor treating brewery wastewater. Int. J. Syst. Evol. Microbiol. 55, 2257–2261. doi: 10.1099/ijs.0.63807-0
Conrad, R. (1996). Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60, 609–640. doi: 10.1128/mr.60.4.609-640.1996
Conrad, R. (2020). Importance of hydrogenotrophic, aceticlastic and methylotrophic methanogenesis for methane production in terrestrial, aquatic and other anoxic environments: a mini review. Pedosphere 30, 25–39. doi: 10.1016/S1002-0160(18)60052-9
de Bok, F. A. M., Stams, A. J. M., Dijkema, C., and Boone, D. R. (2001). Pathway of propionate oxidation by a syntrophic culture of Smithella propionica and Methanospirillum hungatei. Appl. Environ. Microbiol. 67, 1800–1804. doi: 10.1128/AEM.67.4.1800-1804.2001
Dedysh, S. N. (2011). Cultivating uncultured bacteria from northern wetlands: knowledge gained and remaining gaps. Front. Microbiol. 2:184. doi: 10.3389/fmicb.2011.00184
Doyle, D. A., Smith, P. R., Lawson, P. A., and Tanner, R. S. (2022). Clostridium muellerianum sp. nov., a carbon monoxide-oxidizing acetogen isolated from old hay. Int. J. Syst. Evol. Microbiol. 72:005297. doi: 10.1099/ijsem.0.005297
Drake, H. L., Horn, M. A., and Wüst, P. K. (2009). Intermediary ecosystem metabolism as a main driver of methanogenesis in acidic wetland soil. Environ. Microbiol. 1, 307–318. doi: 10.1111/j.1758-2229.2009.00050.x
Faith, D. P. (1992). Conservative evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. doi: 10.1016/0006-3207(92)91201-3
Frolking, S., and Roulet, N. T. (2007). Holocene radiative forcing impact of northern peatland carbon accumulation and methane emissions. Glob. Change Biol. 13, 1079–1088. doi: 10.1111/j.1365-2486.2007.01339.x
Galand, P. E., Fritze, H., and Yrjälä, K. (2003). Microsite-dependent changes in methanogenic populations in a boreal oligotrophic fen. Environ. Microbiol. 5, 1133–1143. doi: 10.1046/j.1462-2920.2003.00520.x
Galand, P. E., Juottonen, H., Fritze, H., and Yrjälä, K. (2005). Methanogen communities in a drained bog: effect of ash fertilization. Microb. Ecol. 49, 209–217. doi: 10.1007/s00248-003-0229-2
Galand, P. E., Saarnio, S., Fritze, H., and Yrjälä, K. (2002). Depth related diversity of methanogen Archaea in Finnish oligotrophic fen. FEMS Microbiol. Ecol. 42, 441–449. doi: 10.1111/j.1574-6941.2002.tb01033.x
Galbally, I. E., and Kirstine, W. V. (2002). The production of methanol by flowering plants and the global cycle of methanol. J. Atmos. Chem. 43, 195–229. doi: 10.1023/A:1020684815474
Gallego-Sala, A. V., Charman, D. J., Brewer, S., Page, S. E., Prentice, I. C., Friedlingstein, P., et al. (2018). Latitudinal limits to the predicted increase of the peatland carbon sink with warming. Nat. Clim. Chang. 8, 907–913. doi: 10.1038/s41558-018-0271-1
Gao, C., Sander, M., Agethen, S., and Knorr, K.-H. (2019). Electron accepting capacity of dissolved and particulate organic matter control CO2 and CH4 formation in peat soils. Geochim. Cosmochim. Acta 245, 266–277. doi: 10.1016/j.gca.2018.11.004
Gooday, G. W. (1994). “Physiology of microbial degradation of chitin and chitosan” in Biochemistry of microbial degradation. ed. C. Ratledge (Dordrecht: Springer Netherlands), 279–312.
Hädrich, A., Heuer, V. B., Herrmann, M., Hinrichs, K.-U., and Küsel, K. (2012). Origin and fate of acetate in an acidic fen. FEMS Microbiol. Ecol. 81, 339–354. doi: 10.1111/j.1574-6941.2012.01352.x
Hahnke, S., Langer, T., Koeck, D. E., and Klocke, M. (2016). Description of Proteiniphilum saccharofermentans sp. nov., Petrimonas mucosa sp. nov. and Fermentimonas caenicola gen. Nov., sp. nov., isolated from mesophilic laboratory-scale biogas reactors, and amended description of the genus Proteiniphilum. Int. J. Syst. Evol. Microbiol. 66, 1466–1475. doi: 10.1099/ijsem.0.000902
Hales, B. A., Edwards, C., Ritchie, D. A., Hall, G., Pickup, R. W., and Saunders, J. R. (1996). Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62, 668–675. doi: 10.1128/aem.62.2.668-675.1996
Hamberger, A., Horn, M. A., Dumont, M. G., Murrell, J. C., and Drake, H. L. (2008). Anaerobic consumers of monosaccharides in a moderately acidic fen. Appl. Environ. Microbiol. 74, 3112–3120. doi: 10.1128/AEM.00193-08
Harris, L. I., Richardson, K., Bona, K. A., Davidson, S. J., Finkelstein, S. A., Garneau, M., et al. (2022). The essential carbon service provided by northern peatlands. Front. Ecol. Environ. 20, 222–230. doi: 10.1002/fee.2437
Hattori, S. (2008). Syntrophic acetate-oxidizing microbes in methanogenic environments. Microb. Environ. 23, 118–127. doi: 10.1264/jsme2.23.118
Heitmann, T., Goldhammer, T., Beer, J., and Blodau, C. (2007). Electron transfer of dissolved organic matter and its potential significance for anaerobic respiration in a northern bog. Glob. Change Biol. 13, 1771–1785. doi: 10.1111/j.1365-2486.2007.01382.x
Hines, M. E., Duddleston, K. N., Rooney-Varga, J. N., Fields, D., and Chanton, J. P. (2008). Uncoupling of acetate degradation from methane formation in Alaskan wetlands: connections to vegetation distribution. Glob. Biogeochem. Cycles 22:GB2017. doi: 10.1029/2006GB002903
Hodgkins, S. B., Tfaily, M. M., McCalley, C. K., Logan, T. A., Crill, P. M., Saleska, S. R., et al. (2014). Changes in peat chemistry associated with permafrost thaw increase greenhouse gas production. Proc. Natl. Acad. Sci. 111, 5819–5824. doi: 10.1073/pnas.1314641111
Horn, M. A., Matthies, C., Küsel, K., Schramm, A., and Drake, H. L. (2003). Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 69, 74–83. doi: 10.1128/AEM.69.1.74-83.2003
Hunger, S., Gößner, A. S., and Drake, H. L. (2015). Anaerobic trophic interactions of contrasting methane-emitting mire soils: processes versus taxa. FEMS Microbiol. Ecol. 91:fiv045. doi: 10.1093/femsec/fiv045
Hunger, S., Schmidt, O., Gößner, A. S., and Drake, H. L. (2016). Formate-derived H2, a driver of hydrogenotrophic processes in the root-zone of a methane-emitting fen. Environ. Microbiol. 18, 3106–3119. doi: 10.1111/1462-2920.13301
Hunger, S., Schmidt, O., Hilgarth, M., Horn, M. A., Kolb, S., Conrad, R., et al. (2011). Competing formate- and carbon dioxide-utilizing prokaryotes in an anoxic methane-emitting fen soil. Appl. Environ. Microbiol. 77, 3773–3785. doi: 10.1128/AEM.00282-11
Itoh, H., Sugisawa, Y., Mise, K., Xu, Z., Kuniyasu, M., Ushijima, N., et al. (2023). Mesoterricola silvestris gen. Nov., sp. nov., Mesoterricola sediminis sp. nov., Geothrix oryzae sp. nov., Geothrix edaphica sp. nov., Geothrix rubra sp. nov., and Geothrix limicola sp. nov., six novel members of Acidobacteriota isolated from soils. Int. J. Syst. Evol. Microbiol. 73:006073. doi: 10.1099/ijsem.0.006073
James, K. L., Ríos-Hernández, L. A., Wofford, N. Q., Mouttaki, H., Sieber, J. R., Sheik, C. S., et al. (2016). Pyrophosphate-dependent ATP formation from acetyl coenzyme a in Syntrophus aciditrophicus, a new twist on ATP formation. MBio 7, e01208–e01216. doi: 10.1128/mBio.01208-16
Janssen, P. H., Schuhmann, A., Mörschel, E., and Rainey, F. A. (1997). Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl. Environ. Microbiol. 63, 1382–1388. doi: 10.1128/aem.63.4.1382-1388.1997
Jiang, N., Wang, Y., and Dong, X. (2010). Methanol as the primary methanogenic and acetogenic precursor in the cold Zoige wetland at Tibetan plateau. Microb. Ecol. 60, 206–213. doi: 10.1007/s00248-009-9602-0
Kaksonen, A. H., Spring, S., Schumann, P., Kroppenstedt, R. M., and Puhakka, J. A. (2007). Desulfovirgula thermocuniculi gen. Nov., sp. nov., a thermophilic sulfate-reducer isolated from a geothermal underground mine in Japan. Int. J. Syst. Evol. Microbiol. 57, 98–102. doi: 10.1099/ijs.0.64655-0
Kang, H., Freeman, C., Soon Park, S., and Chun, J. (2005). N-Acetylglucosaminidase activities in wetlands: a global survey. Hydrobiologia 532, 103–110. doi: 10.1007/s10750-004-9450-3
Kelly, C. A., Dise, N. B., and Martens, C. S. (1992). Temporal variations in the stable carbon isotopic composition of methane emitted from Minnesota peatlands. Glob. Biogeochem. Cycles 6, 263–269. doi: 10.1029/92GB01478
Kolb, S., and Horn, M. A. (2012). Microbial CH4 and N2O consumption in acidic wetlands. Front. Microbiol. 3:78. doi: 10.3389/fmicb.2012.00078
Kotsyurbenko, O. R., Friedrich, M. W., Simankova, M. V., Nozhevnikova, A. N., Golyshin, P. N., Timmis, K. N., et al. (2007). Shift from acetoclastic to H2-dependent methanogenesis in a west Siberian peat bog at low pH values and isolation of an acidophilic Methanobacterium strain. Appl. Environ. Microbiol. 73, 2344–2348. doi: 10.1128/AEM.02413-06
Kotsyurbenko, O. R., Glagolev, M. V., Merkel, A. Y., Sabrekov, A. F., and Terentieva, I. E. (2019). “Methanogenesis in soils, wetlands, and peat” in Biogenesis of hydrocarbons. eds. A. J. M. Stams and D. Z. Sousa (Cham: Springer), 211–228.
Kotsyurbenko, O. R., Nozhevnikova, A. N., Soloviova, T. I., and Zavarzin, G. A. (1996). Methanogenesis at low temperatures by microflora of tundra wetland soil. A. Van Leeuw. J. Microb. 69, 75–86. doi: 10.1007/BF00641614
Küsel, K., and Drake, H. L. (1995). Effects of environmental parameters on the formation and turnover of acetate by forest soils. Appl. Environ. Microbiol. 61, 3667–3675. doi: 10.1128/aem.61.10.3667-3675.1995
Lee, M. J., and Zinder, S. H. (1988). Isolation and characterization of a thermophilic bacterium which oxidizes acetate in syntrophic association with a methanogen and which grows acetogenically on H2-CO2. Appl. Environ. Microbiol. 54, 124–129. doi: 10.1128/aem.54.1.124-129.1988
Limpens, J., Berendse, F., Blodau, C., Canadell, J. G., Freeman, C., Holden, J., et al. (2008). Peatlands and the carbon cycle: from local processes to global implications – a synthesis. Biogeosciences 5, 1475–1491. doi: 10.5194/bg-5-1475-2008
Lueders, T., Chin, K. J., Conrad, R., and Friedrich, M. (2001). Molecular analyses of methyl-coenzyme M reductase α-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3, 194–204. doi: 10.1046/j.1462-2920.2001.00179.x
Mann, K. H. (1988). Production and use of detritus in various freshwater, estuarine, and coastal ecosystems. Limnol. Oceanogr. 33, 910–930. doi: 10.4319/lo.1988.33.4_part_2.0910
Manzoor, S., Bongcam-Rudloff, E., Schnürer, A., and Müller, B. (2016). Genome-guided analysis and whole transcriptome profiling of the mesophilic syntrophic acetate oxidising bacterium Syntrophaceticus schinkii. PLoS One 11:e0166520. doi: 10.1371/journal.pone.0166520
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. doi: 10.14806/ej.17.1.200
McCarty, P. L. (1972). “Energetics of organic matter degradation” in Water pollution microbiology. ed. R. Mitchell (New York: John Wiley), 91–118.
Meier, A. B., Oppermann, S., Drake, H. L., and Schmidt, O. (2021). Organic carbon from graminoid roots as a driver of fermentation in a fen. FEMS Microbiol. Ecol. 97:fiab143. doi: 10.1093/femsec/fiab143
Meier, A. B., Oppermann, S., Drake, H. L., and Schmidt, O. (2022). The root zone of graminoids: a niche for H2-consuming acetogens in a minerotrophic peatland. Front. Microbiol. 13:978296. doi: 10.3389/fmicb.2022.978296
Metje, M., and Frenzel, P. (2005). Effect of temperature on anaerobic ethanol oxidation and methanogenesis in acidic peat from a northern wetland. Appl. Environ. Microbiol. 71, 8191–8200. doi: 10.1128/AEM.71.12.8191-8200.2005
Metje, M., and Frenzel, P. (2007). Methanogenesis and methanogenic pathways in a peat from subarctic permafrost. Environ. Microbiol. 9, 954–964. doi: 10.1111/j.1462-2920.2006.01217.x
Müller, V. (2008). “Bacterial fermentation” in Encyclopedia of life sciences (Chichester: John Wiley).
Müller, B., Manzoor, S., Niazi, A., Bongcam-Rudloff, E., and Schnürer, A. (2015). Genome-guided analysis of physiological capacities of Tepidanaerobacter acetatoxydans provides insights into environmental adaptations and syntrophic acetate oxidation. PLoS One 10:e0121237. doi: 10.1371/journal.pone.0121237
Muyzer, G., de Waal, E. C., and Uitterlinden, A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700. doi: 10.1128/aem.59.3.695-700.1993
Nusslein, B., Chin, K.-J., Eckert, W., and Conrad, R. (2001). Evidence for anaerobic syntrophic acetate oxidation during methane production in the profundal sediment of subtropical Lake Kinneret (Israel). Environ. Microbiol. 3, 460–470. doi: 10.1046/j.1462-2920.2001.00215.x
Oehler, D., Poehlein, A., Leimbach, A., Müller, N., Daniel, R., Gottschalk, G., et al. (2012). Genome-guided analysis of physiological and morphological traits of the fermentative acetate oxidizer Thermacetogenium phaeum. BMC Genomics 13:723. doi: 10.1186/1471-2164-13-723
Oremland, R. S., and Capone, D. G. (1988). “Use of ‘specific’ inhibitors in biogeochemistry and microbial ecology” in Advances in microbial ecology advances in microbial ecology. ed. K. C. Marshall (Boston, MA: Springer US), 285–383.
Oren, A. (2014a). “The family Methanoregulaceae” in The prokaryotes. eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer Berlin Heidelberg), 253–258.
Oren, A. (2014b). “The family Methanosarcinaceae” in The prokaryotes. eds. E. Rosenberg, E. F. Delong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer Berlin Heidelberg), 259–281.
Palmer, K., Biasi, C., and Horn, M. A. (2012). Contrasting denitrifier communities relate to contrasting N2O emission patterns from acidic peat soils in arctic tundra. ISME J. 6, 1058–1077. doi: 10.1038/ismej.2011.172
Palmer, K., and Horn, M. A. (2015). Denitrification activity of a remarkably diverse fen denitrifier community in Finnish Lapland is N-oxide limited. PLoS One 10:e0123123. doi: 10.1371/journal.pone.0123123
Pankratov, T. A., Belova, S. E., and Dedysh, S. N. (2005). Evaluation of the phylogenetic diversity of prokaryotic microorganisms in Sphagnum peat bogs by means of fluorescence in situ hybridization (FISH). Microbiology 74, 722–728. doi: 10.1007/s11021-005-0130-8
Pazos, M., and Peters, K. (2019). “Peptidoglycan” in Bacterial cell walls and membranes subcellular biochemistry. ed. A. Kuhn (Cham: Springer International Publishing), 127–168.
Peršoh, D., Theuerl, S., Buscot, F., and Rambold, G. (2008). Towards a universally adaptable method for quantitative extraction of high-purity nucleic acids from soil. J. Microbiol. Methods 75, 19–24. doi: 10.1016/j.mimet.2008.04.009
Pester, M., Knorr, K.-H., Friedrich, M. W., Wagner, M., and Loy, A. (2012). Sulfate-reducing microorganisms in wetlands – fameless actors in carbon cycling and climate change. Front. Microbiol. 3:72. doi: 10.3389/fmicb.2012.00072
Reiche, M., Torburg, G., and Küsel, K. (2008). Competition of Fe(III) reduction and methanogenesis in an acidic fen. FEMS Microbiol. Ecol. 65, 88–101. doi: 10.1111/j.1574-6941.2008.00523.x
Roden, E. E., Kappler, A., Bauer, I., Jiang, J., Paul, A., Stoesser, R., et al. (2010). Extracellular electron transfer through microbial reduction of solid-phase humic substances. Nat. Geosci. 3, 417–421. doi: 10.1038/ngeo870
Rooney-Varga, J. N., Giewat, M. W., Duddleston, K. N., Chanton, J. P., and Hines, M. E. (2007). Links between archaeal community structure, vegetation type and methanogenic pathway in Alaskan peatlands. FEMS Microbiol. Ecol. 60, 240–251. doi: 10.1111/j.1574-6941.2007.00278.x
Russell, J. B. (1991). Intracellular pH of acid-tolerant ruminal bacteria. Appl. Environ. Microbiol. 57, 3383–3384. doi: 10.1128/aem.57.11.3383-3384.1991
Sakai, S., Conrad, R., and Imachi, H. (2014). “The family Methanocellaceae” in The prokaryotes. eds. E. Rosenberg, E. F. Delong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer Berlin Heidelberg), 209–214.
Salmon, E., Jégou, F., Guenet, B., Jourdain, L., Qiu, C., Bastrikov, V., et al. (2022). Assessing methane emissions for northern peatlands in ORCHIDEE-PEAT revision 7020. Geosci. Model Dev. 15, 2813–2838. doi: 10.5194/gmd-15-2813-2022
Sattley, W. M., Jung, D. O., and Madigan, M. T. (2008). Psychrosinus fermentans gen. Nov., sp. nov., a lactate-fermenting bacterium from near-freezing oxycline waters of a meromictic Antarctic lake. FEMS Microbiol. Lett. 287, 121–127. doi: 10.1111/j.1574-6968.2008.01300.x
Schink, B. (1997). Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61, 262–280. doi: 10.1128/mmbr.61.2.262-280.1997
Schink, B., Kremer, D. R., and Hansen, T. A. (1987). Pathway of propionate formation from ethanol in Pelobacter propionicus. Arch. Microbiol. 147, 321–327. doi: 10.1007/BF00406127
Schmidt, O., Drake, H. L., and Horn, M. A. (2010). Hitherto unknown [Fe-Fe]-hydrogenase gene diversity in anaerobes and anoxic enrichments from a moderately acidic fen. Appl. Environ. Microbiol. 76, 2027–2031. doi: 10.1128/AEM.02895-09
Schmidt, A., Frensch, M., Schleheck, D., Schink, B., and Müller, N. (2014). Degradation of acetaldehyde and its precursors by Pelobacter carbinolicus and P. acetylenicus. PLoS One 9:e115902. doi: 10.1371/journal.pone.0115902
Schmidt, O., Hink, L., Horn, M. A., and Drake, H. L. (2016). Peat: home to novel syntrophic species that feed acetate- and hydrogen-scavenging methanogens. ISME J. 10, 1954–1966. doi: 10.1038/ismej.2015.256
Schmidt, O., Horn, M. A., Kolb, S., and Drake, H. L. (2015). Temperature impacts differentially on the methanogenic food web of cellulose-supplemented peatland soil. Environ. Microbiol. 17, 720–734. doi: 10.1111/1462-2920.12507
Schmidt, O., Wüst, P. K., Hellmuth, S., Borst, K., Horn, M. A., and Drake, H. L. (2011). Novel [NiFe]- and [FeFe]-hydrogenase gene transcripts indicative of active facultative aerobes and obligate anaerobes in earthworm gut contents. Appl. Environ. Microbiol. 77, 5842–5850. doi: 10.1128/AEM.05432-11
Shannon, C. E. (1948). A mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
Shigematsu, T., Tang, Y., Kobayashi, T., Kawaguchi, H., Morimura, S., and Kida, K. (2004). Effect of dilution rate on metabolic pathway shift between aceticlastic and nonaceticlastic methanogens in chemostat cultivation. Appl. Environ. Microbiol. 70, 4048–4052. doi: 10.1128/AEM.70.7.4048-4052.2004
Smemo, K. A., and Yavitt, J. B. (2011). Anaerobic oxidation of methane: an underappreciated aspect of methane cycling in peatland ecosystems? Biogeosciences 8, 779–793. doi: 10.5194/bg-8-779-2011
Strack, M., Waddington, J., Turetsky, M., Roulet, N., and Byrne, K. (2008). “Northern peatlands, greenhouse gas exchange and climate change” in Peatlands and climate change. ed. M. Strack (Jyväskylä, Finland: International Peat Society), 44–69.
Sun, L., Toyonaga, M., Ohashi, A., Tourlousse, D. M., Matsuura, N., Meng, X.-Y., et al. (2016). Lentimicrobium saccharophilum gen. Nov., sp. nov., a strictly anaerobic bacterium representing a new family in the phylum Bacteroidetes, and proposal of Lentimicrobiaceae fam. Nov. Int. J. Syst. Evol. Microbiol. 66, 2635–2642. doi: 10.1099/ijsem.0.001103
Tarnocai, C., Canadell, J. G., Schuur, E., Kuhry, P., Mazhitova, G., and Zimov, S. (2009). Soil organic carbon pools in the northern circumpolar permafrost region. Global Biogeochem. Cycles 23:GB2023. doi: 10.1029/2008GB003327
Tveit, A., Schwacke, R., Svenning, M. M., and Urich, T. (2013). Organic carbon transformations in high-Arctic peat soils: key functions and microorganisms. ISME J. 7, 299–311. doi: 10.1038/ismej.2012.99
Tveit, A. T., Urich, T., Frenzel, P., and Svenning, M. M. (2015). Metabolic and trophic interactions modulate methane production by Arctic peat microbiota in response to warming. Proc. Natl. Acad. Sci. 112, E2507–E2516. doi: 10.1073/pnas.1420797112
Wiegel, J. (2015). “Clostridiaceae,” in Bergey’s manual of systematics of Archaea and Bacteria. Bergey’s Manual of Systematics of Archaea and Bacteria. eds. M. E. Trujillo, S. Dedysh, P. DeVos, B. Hedlund, P. Kämpfer, and F. A. Rainey, et al. (Hoboken, New Jersey: John Wiley & Sons, Inc.).
Williams, R. T., and Crawford, R. L. (1984). Methane production in Minnesota peatlands. Appl. Environ. Microbiol. 47, 1266–1271. doi: 10.1128/aem.47.6.1266-1271.1984
Wörner, S., and Pester, M. (2019). Microbial succession of anaerobic chitin degradation in freshwater sediments. Appl. Environ. Microbiol. 85, e00963–e00919. doi: 10.1128/AEM.00963-19
Wüst, P. K., Horn, M. A., and Drake, H. L. (2009). Trophic links between fermenters and methanogens in a moderately acidic fen soil. Environ. Microbiol. 11, 1395–1409. doi: 10.1111/j.1462-2920.2009.01867.x
Yavitt, J. B., and Seidman-Zager, M. (2006). Methanogenic conditions in northern peat soils. Geomicrobiol J. 23, 119–127. doi: 10.1080/01490450500533957
Yavitt, J. B., Yashiro, E., Cadillo-Quiroz, H., and Zinder, S. H. (2012). Methanogen diversity and community composition in peatlands of the central to northern Appalachian Mountain region, North America. Biogeochemistry 109, 117–131. doi: 10.1007/s10533-011-9644-5
Ye, R., Jin, Q., Bohannan, B., Keller, J. K., and Bridgham, S. D. (2014). Homoacetogenesis: a potentially underappreciated carbon pathway in peatlands. Soil Biol. Biochem. 68, 385–391. doi: 10.1016/j.soilbio.2013.10.020
Zalman, C., Keller, J. K., Tfaily, M., Kolton, M., Pfeifer-Meister, L., Wilson, R. M., et al. (2018). Small differences in ombrotrophy control regional-scale variation in methane cycling among Sphagnum-dominated peatlands. Biogeochemistry 139, 155–177. doi: 10.1007/s10533-018-0460-z
Zinder, S. H. (1993). “Physiological ecology of methanogens” in Methanogenesis: Ecology, physiology, Biochemistry & Genetics. ed. J. G. Ferry, Chapman & Hall microbiology series (Boston, MA: Springer US), 128–206.
Keywords: N-acetylglucosamine, intermediary ecosystem metabolisms, anaerobic feed chain, intermediates, methyl-CoM-reductase genes, hydrogenase genes, microbial community
Citation: Kujala K, Schmidt O and Horn MA (2024) Synergy and competition during the anaerobic degradation of N-acetylglucosamine in a methane-emitting, subarctic, pH-neutral fen. Front. Microbiol. 15:1428517. doi: 10.3389/fmicb.2024.1428517
Received: 06 May 2024; Accepted: 29 November 2024;
Published: 11 December 2024.
Edited by:
Jeanette M. Norton, Utah State University, United StatesReviewed by:
Carl-Eric Wegner, Heinrich Heine University of Düsseldorf, GermanyCopyright © 2024 Kujala, Schmidt and Horn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marcus A. Horn, aG9ybkBpZm1iLnVuaS1oYW5ub3Zlci5kZQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.