- 1Department of Civil and Mineral Engineering, University of Toronto, Toronto, ON, Canada
- 2Department of Earth and Planetary Science, University of California, Berkeley, Berkeley, CA, United States
- 3School of Mathematical and Physical Sciences, University of Technology Sydney, Ultimo, NSW, Australia
- 4Commonwealth Scientific Industrial and Research Organization, Black Mountain, ACT, Australia
- 5EcoReg Solutions, Guelph, ON, Canada
- 6Commonwealth Scientific Industrial and Research Organization, Clayton, VIC, Australia
Sulfur oxidizing bacteria (SOB) play a key role in sulfur cycling in mine tailings impoundment (TI) waters, where sulfur concentrations are typically high. However, our understanding of SOB sulfur cycling via potential S oxidation pathways (sox, rdsr, and S4I) in these globally ubiquitous contexts, remains limited. Here, we identified TI water column SOB community composition, metagenomics derived metabolic repertoires, physicochemistry, and aqueous sulfur concentration and speciation in four Canadian base metal mine, circumneutral-alkaline TIs over four years (2016 – 2019). Identification and examination of genomes from nine SOB genera occurring in these TI waters revealed two pH partitioned, metabolically distinct groups, which differentially influenced acid generation and sulfur speciation. Complete sox (csox) dominant SOB (e.g., Halothiobacillus spp., Thiomonas spp.) drove acidity generation and S2O32- consumption via the csox pathway at lower pH (pH ~5 to ~6.5). At circumneutral pH conditions (pH ~6.5 to ~8.5), the presence of non-csox dominant SOB (hosting the incomplete sox, rdsr, and/or other S oxidation reactions; e.g. Thiobacillus spp., Sulfuriferula spp.) were associated with higher [S2O32-] and limited acidity generation. The S4I pathway part 1 (tsdA; S2O32- to S4O62-), was not constrained by pH, while S4I pathway part 2 (S4O62- disproportionation via tetH) was limited to Thiobacillus spp. and thus circumneutral pH values. Comparative analysis of low, natural (e.g., hydrothermal vents and sulfur hot springs) and high (e.g., Zn, Cu, Pb/Zn, and Ni tailings) sulfur systems literature data with these TI results, reveals a distinct TI SOB mining microbiome, characterized by elevated abundances of csox dominant SOB, likely sustained by continuous replenishment of sulfur species through tailings or mining impacted water additions. Our results indicate that under the primarily oxic conditions in these systems, S2O32- availability plays a key role in determining the dominant sulfur oxidation pathways and associated geochemical and physicochemical outcomes, highlighting the potential for biological management of mining impacted waters via pH and [S2O32-] manipulation.
1 Introduction
Biological sulfur oxidation can present significant risks to the environment via acid generation, contaminant mobilization, and oxygen consumption (i.e., acid mine drainage) (Edwards et al., 1999; Elberling and Damgaard, 2001; Baker and Banfield, 2003; Druschel et al., 2003; Johnson and Hallberg, 2003; Korehi et al., 2014). This biological acidity production occurs in both natural (termed acid rock drainage, found in alpine catchments; Lacelle et al., 2007; Zarroca et al., 2021) and anthropogenic (e.g., mine tailings impoundments and waste rock piles; Akcil and Koldas, 2006) environments, though the scale to which it occurs in anthropogenic environments is typically much larger.
Base metal mine tailings impoundment (TI) wastewaters often have elevated sulfur concentrations due to the dominance of sulfide minerals in base metal ores [e.g., chalcopyrite (CuFeS2), sphalerite ((Zn, Fe)S), etc.]. Sulfides can be partially oxidized during the grinding, flotation, and leaching steps of sulfide mineral ore extraction (Liljeqvist et al., 2011), resulting in the production and subsequent release of sulfur oxidation intermediate compounds (SOI) from tailings streams to TIs. In addition to sulfide (ΣH2S), SOI commonly found in mining impacted waters include thiosulfate (S2O32-), tetrathionate (S4O62-), elemental sulfur (S0), sulfite (SO32-), and a range of other polythionates (SxOy2-) (Makhija and Hitchen, 1979; Miranda-Trevino et al., 2013; Whaley-Martin et al., 2020). The concentrations of individual SOI in TI wastewater can vary significantly spatially, seasonally, and amongst mining operations. SOI may be present in relatively high concentrations in tailings slurries, but are typically much lower in the TI waters due to dilution by other water inputs collected in these actively managed systems (Thamdrup et al., 1994), though they are typically higher than concentrations observed in natural environments (Foucher et al., 2001; Canfield and Farquhar, 2009; Silva et al., 2012; Camacho et al., 2020b; Vincent et al., 2021). SOI can be reduced, oxidized, and disproportionated, resulting in differential SOI speciation and pH outcomes, via both abiotic and biotic reactions (Philippot et al., 2007; Klatt and Polerecky, 2015; Houghton et al., 2016) further contributing to the complexity of the sulfur cycle in these environments. As mine TI systems continue to grow in number and size around the world, an understanding of the biogeochemical cycling of sulfur compounds occurring within these contexts, the SOB involved, and the influencing factors determining water quality outcomes is increasingly important.
Few studies have addressed the coupling of sulfur oxidation metabolic pathways in TIs to SOB taxonomy, physicochemistry, and sulfur geochemistry (Whaley-Martin et al., 2019, 2023; Miettinen et al., 2021), contrasting more well-studied extremophilic acid mine drainage environments (Bond et al., 2000; Johnson and Hallberg, 2003; Dold, 2014). Recent research has highlighted a divergence of microbial communities found in circumneutral mining impacted TI waters from those of acid mine drainage environments (Whaley-Martin et al., 2019, 2023; Camacho et al., 2020b; Lopes et al., 2020; Miettinen et al., 2021). A number of sulfur oxidation pathways have been identified as being used by SOB potentially found in mining environments, though the physicochemical, geochemical, and/or ecological parameters governing which S oxidation pathway(s) occur are not well defined (Han and Perner, 2015; Wang et al., 2016; Watanabe et al., 2019). These pathways include the sulfur oxidation (sox), reverse dissimilatory sulfite reductase (rdsr), and tetrathionate intermediate (S4I) pathways (Friedrich et al., 2001; Han and Perner, 2015; Klatt and Polerecky, 2015; Wang et al., 2016; Watanabe et al., 2019; Whaley-Martin et al., 2023). The sox pathway has seven structural genes which encode four proteins (soxXA, soxYZ, soxB, and soxCD) allowing this pathway to mediate S2O32-, SO32-, S0, and ΣH2S dependent cytochrome c reduction (Friedrich et al., 2001). SoxAX catalyzes the attachment of S2O32- to cysteine residue on the soxY of the soxYZ complex (forming soxZY-Cys-S-) (Friedrich et al., 2000, 2001; Bamford et al., 2002; Watanabe et al., 2019). The soxCD complex then oxidizes the sulfate sulfur from SoxZY-Cys-S- to produce the sulfonate group as soxZY-Cys-SO3- (Quentmeier et al., 2000; Zander et al., 2011; Watanabe et al., 2019). SoxB further hydrolyzes the sulfane sulfur from SoxZY-Cys-SO3- to a free sulfate ion (Quentmeier et al., 2000; Zander et al., 2011; Watanabe et al., 2019). Free SOI are not produced by the sox pathway when found in its complete form (complete sox; csox) as they are covalently attached to soxYZ until complete oxidation to sulfate (SO42-; Grabarczyk and Berks, 2017). An incomplete form of the sox pathway (lacking soxCD, isox) has also been identified which can form S0 (Frigaard and Dahl, 2008; Watanabe et al., 2019). This S0 can be subsequently oxidized by rdsr, sulfur-oxidizing heterodisulfide reductase-like (shdr), or sulfur dioxygenase (sdo) to SO32- (Klatt and Polerecky, 2015). The rdsr pathway is composed of the same proteins as the dsr pathway (sat, aprAB, dsrAB) though the reductive and oxidative varieties are phylogenetically discernible (van Vliet et al., 2021). SOI including S2O32-, S0, and ΣH2S, can be oxidized through the rdsr pathway and generate free SO32- (Klatt and Polerecky, 2015). Sulfane sulfur generated from the isox pathway can then be transported to the cytoplasm in the form of persulfides (R-S-S-) to be further oxidized to HSO3- by dsrAB or shdr (Pott and Dahl, 1998; Frigaard and Dahl, 2008; Cao et al., 2018; Koch and Dahl, 2018). The HSO3- can be further oxidized to APS (adenosine 5’-phosphosulfate) by aprBA with electron transfer by aprM or hdrAACB and then to SO42- by sat (Meyer and Kuever, 2007; Loy et al., 2009). Rdsr pathway presence is typically associated with high energy efficiency compared to the sox pathway (Klatt and Polerecky, 2015). Several beta- and gammaproteobacteria have been found to utilize the S4I pathway (or Kelly-Trudinger pathway) which generates free S4O62- (Dam et al., 2007). The S4I pathway oxidizes S2O32- to S4O62- by thiosulfate dehydrogenase (tsdA) or thiosulfate:quinol oxioreductase (TQO or doxD; Brito et al., 2015; Wang et al., 2016; Hutt et al., 2017). Subsequent processing of free S4O62- can be catalyzed by tetrathionate reductase (ttrABC) to produce S2O32- or tetrathionate hydrolase (tetH) to produce S2O32-, S0, and SO42-, which are both common in SOB (Wang et al., 2016; Camacho et al., 2020a; Miettinen et al., 2021). A recent study (Whaley-Martin et al., 2023) identified oxygen as a control on whether the csox (high O2) or rdsr (low O2) pathway dominated in one TI with differing water quality outcomes, indicating that physicochemical and/or SOI substrate partitioning of SOB within TI may happen more broadly.
The objectives of this cross-mine study were to identify mining TI associated SOB, examine functional differences in SOB communities, and align their associated S oxidizing repertoires to geochemical and physicochemical characteristics and outcomes in circumneutral TI waters of four base metal mines located across Canada (Manitoba, Newfoundland, Ontario). A better understanding of these genetic, geochemical, and/or physicochemical connections will inform biological management strategies as well as further understanding of S biogeochemical cycling more broadly. Four years (2016 – 2019) of S geochemistry, physicochemistry, and genus level community structure and function data from the four base metal mine TIs were examined. To determine if the findings of this study were site specific or reflective of broader environmental trends, comparisons were made to published studies on other mines and industrial environments, as well as natural environments.
2 Materials and methods
2.1 Site descriptions
Four base metal mine TI waters were sampled from 2016 to 2019 resulting in a total of 42 water samples (see Supplementary Figure S1). These four mines are located across central and eastern Canada and consist of: Mine 1 in Flin Flon, Manitoba (Cu, Zn, Au, Ag), Mine 2 in Sudbury, Ontario (Ni, Cu, Co, Pt, Pd), Mine 3 in Snow Lake, Manitoba (Cu, Zn, Se, Te, Ag), and Mine 4 in Baie Verte, Newfoundland (Cu, Au). The mines range in size, age, and stage of development with Mine 1 and Mine 2 being the oldest (operating on and off since 1927 and 1928, respectively), Mine 3 opened in 1979 and Mine 4 as the youngest with mining originating on the property around 1997, with the commissioning of the TI in 2009 (Table 1). The TI facilities across sites also vary in size and depth with Mine 2 being both the largest and deepest at ~38 m, followed by Mine 1 at ~7 to 10 m depth and finally Mine 3 and Mine 4 at ~1.5 to 2.5 m depth. TI water cover sampling depths for Mine 1, Mine 2, Mine 3, and Mine 4 ranged 5 to 10 m, 0.5 to 10 m, 0.5 to 3 m, and 0.5 to 1.5 m, respectively, and this investigation targeted seasonal open water collection (early spring to late fall).
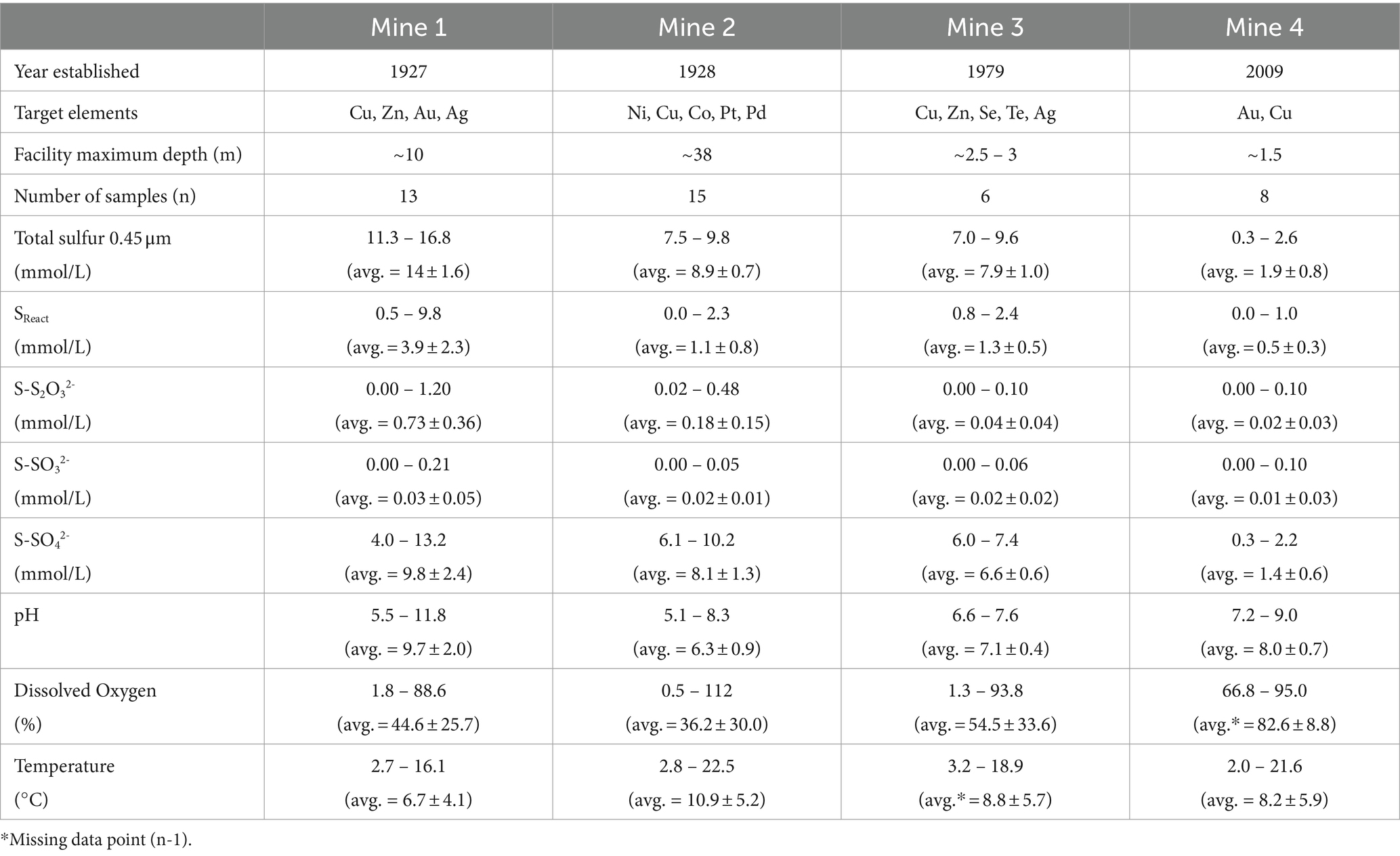
Table 1. Mine information, sulfur geochemistry, and physicochemistry data for Mine 1, Mine 2, Mine 3, and Mine 4 tailings impoundment waters (2016 – 2019) presented as average (± standard deviation).
2.2 Physicochemical characterization and sampling scheme
Samples collected from Mine 2 TI were taken from two different points in the reservoir (see Supplementary Figure S2) which include a floating platform at the deepest point of the reservoir and the outflow dam. The outflow dam represents an approximate average of the overall water column and is ~2 m deep at the sampling location (samples taken at 0.5 m or 1 m). Samples from Mine 3 and Mine 1 TIs were collected off the end of docks ~4 m from shore. Mine 4 TI samples were collected from a boat ~20 m from shore.
Physicochemical and geochemical water samples from Mine 2 were collected and processed on-site within 1 h of collection, while water samples from Mine 1, Mine 3, and Mine 4 were shipped to the University of Toronto for processing, taking two to ten days potential shipping impacts addressed in Whaley-Martin et al. (2019). Depth samples from Mine 1 and Mine 2 were collected using a sterilized Van Dorn sampler as described in Whaley-Martin et al. (2019) and Mine 3 and Mine 4 samples were collected using a sterilized surface grab sampler. Water samples from Mine 1 and Mine 3 were collected in polyethylene liners (Uline S-1379) that were 70% ethanol sterilized and rinsed with target sample water three times prior to filling, using water directly from the Van Dorn sampler or surface grab sampler. Once 10 – 20 L were collected, liners were sealed without headspace, and placed in clean 20 L containers for shipping. Containers and liners remained unopened until arrival at the laboratory at the University of Toronto. Mine 2 water sample collection for microbial analysis followed the same protocol. Mine 4 water samples were collected into 70% ethanol sterilized and thrice rinsed (with target sample water) polyethylene containers before being submerged and filled directly leaving no headspace.
Each TI water sample was characterized for sulfur geochemistry and microbial community composition. Geochemical characterization included analyses of total sulfur (Total S), sulfate (SO42-), thiosulfate (S2O32-), and sulfite (SO32-) concentrations and determination of reactive sulfur (SReact; all S atoms capable of oxidation; determined by [Total S] – [SO42-]; Whaley-Martin et al., 2020). Microbial community structure and function assessment included 16S rRNA and metagenomic analyses. Field measured TI water physicochemical parameters including temperature, pH, dissolved oxygen (DO; concentration and % saturation) and conductivity or salinity data were collected using a YSI 600 XLM (Mine 2), ProDSS water quality meter (Mine 1 and Mine 3), or a ThermoScientific Orion Star A329 Multiprobe (Mine 4).
Samples collected for 16S rRNA and metagenomic analyses were vacuum filtered (~2 to ~5 L) in triplicate through 0.1 μm and 0.2 μm filter units (Thermo Scientific™ Nalgene™ Rapid-Flow™ Sterile Disposable Filter Units with aPES Membrane) until clogged. Filters were immediately excised in a sterile Biological Safety Cabinet and kept frozen at -80°C until future extraction.
2.3 Geochemical analyses
Total sulfur samples were collected in triplicate by filtering 40 mL (per replicate) through a 0.45 μm filter (Pall Acrodisc® 25 mm 0.45 μm Supor® membrane filters) with polypropylene syringes into 50 mL Falcon™ tubes pre-spiked with 80 μL of HNO3 (Optima grade, Fisher Chemical). Unfiltered (UF) and 0.2 μm filtered total S samples were also collected, however, one-way ANOVA analysis revealed no statistically significant difference between the three filter fractions (p = 0.998) from these TI waters, therefore the 0.45 μm filter fraction data were used as these values represented the most complete dataset. These samples were stored at 4°C until they were shipped to the Commonwealth Scientific and Industrial Research Organization (CSIRO) for analyses using an inductively coupled plasma atomic emission spectroscopy (ICP-AES) on a Varian730 ES (Mulgrave, Australia). The limit of detection (LOD) for sulfur was 0.03 mM and concentrations were calculated by measuring intensity at the 181.972 nm sulfur emission line. Fast Automated Curve-fitting Technique (FACT) was used to correct for background and inter-element interferences.
100 μL aliquots of sample waters were preserved in triplicate for S2O32- and SO32- analysis using scaled derivatization methods described in Rethmeier et al. (1997). Samples were frozen until analysis. [S2O32-] and [SO32-] were quantified using a Shimadzu LC-20 AD prominence liquid chromatography (LC) system coupled with a fluorescence UV/VIS detector. An Alltima™ HP C18 reversed phase column (150 mm × 4.6 mm × 5 μm, Grace™) was used at 35°C using an isocratic mobile phase comprising 35% HPLC grade methanol and 65% of 0.25% acetic acid v/v (filtered, pH adjusted to 3.5 using NaOH) at 1 mL/min. The total run time was 12 min and the SO32- peak eluted at ~3.1 min and S2O32- eluted at ~3.4 min. The excitation wavelength used was 380 nm and the emission wavelength was 478 nm. Calibration curves were prepared using Na2SO3 (Sigma Aldrich, ≥ 98% purity) and Na2S2O3 (Sigma Aldrich, 99% purity). Limits of quantification for both S2O32- and SO32- were 0.01 mM.
Prior to 2019, SO42- samples were collected into clean sample bottles that were thrice rinsed (with target sample water) and filled leaving no headspace. Samples were stored at 4°C until analyzed. Aqueous dissolved SO42- was quantified by spectrophotometry using a HACH DR2800 (HACH Company, Loveland, CO, United States) using USEPA SulfaVer 4 Method 8051. SO42- samples collected in 2019 were 0.2 μm filtered (Pall Acrodisc® 25 mm 0.2 μm Supor® membrane filters) in triplicate using polypropylene syringes into 1.7 mL polypropylene vials and stored at 4°C until analyzed. Waters were analyzed for anions following USEPA Method 300.0/300.1 (only SO42- is reported here) on a Dionex™ ICS-6000 Capillary HPIC™ (high pressure ion chromatography; Thermo Scientific™, Part Number: ICS6000-003) System with a Dionex™ ICS-6000 CD Conductivity Detector (Thermo Scientific™, Part Number: 079829) and a Dionex IonPac™ AS18-Fast, 4 × 150 mm AS18 anion-exchange column (Thermo Scientific™, Part Number: 072062) and a Dionex IonPac™ AG18-Fast, 4 × 30 mm guard column (Thermo Scientific™, Part Number: 075762). A Dionex™ ICS-6000 EG Eluent Generator was used to produce the isocratic mobile phase of 23 mM KOH. Sample injection volume was 10 μL with sample aliquots diluted using 18.2 MΩ·cm deionized water when required. Matrix spikes and check standards were run every 20 samples and were also expected to fall withing ±15% of the expected value. The limit of detection for these samples was determined to be 0.008 mM dissolved SO42- with an instrument error of ±0.1 mM (Whaley-Martin et al., 2020; Yan et al., 2022).
Previous comparisons of the HACH spectrophotometry and IC methodology were completed on these waters by Whaley-Martin et al. (2020) identifying that dissolved [SO42-] obtained by HACH spectroscopy and IC methods were consistent between methods for measured dissolved [SO42-] between 0 and 250 mg/L.
2.4 16S rRNA sampling and statistical analyses
Genomic DNA was extracted from filters using Qiagen DNEasy PowerWater DNA Isolation Kits and the sample extracts were submitted to the McMaster University Genome Facility (Hamilton, Ontario, Canada) for subsequent analysis. Genomic DNA was quantified using quantitative PCR (polymerase chain reaction). Aliquots of purified DNA were used to amplify the V4 region of the 16S rRNA gene following the methods from Bartram et al. (2011) using Illumina primers and standards protocols from the Earth Microbiome Project (Caporaso et al., 2011, 2012). Briefly, the primers amplified the 515f (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806r (5′-GGACTACNVGGGTWTCTAAT-3′) V4-variable regions of bacterial and archaeal 16S rRNA gene. 50 ng of DNA template was used for PCR and the PCR mix contained 1 U of recombinant Taq DNA Polymerase (Invitrogen™), 1x buffer, 1.5 mM MgCl2, 0.4 mg/mL bovine serum albumin (BSA), 0.2 mM deoxynucleotide triphosphate (dNTPs) and 5 pM of each primer. In accordance with Bates et al. (2010), the PCR reaction was as follows: initial denaturing at 98°C for 5 min, 35 cycles of denaturing at 98°C for 30 s, annealing at 50°C for 30 s and an extension at 72°C for 30 s with a final extension at 72°C for 10 min. The PCR products were confirmed using electrophoresis and sent for sequencing. A SequalPrep normalization kit (ThemoFisher #A1051001) was used to normalize all amplicons to 1.25 ng/L and then sequenced using the Illumina Mi-Seq.
DADA2 (v. 1.6.0) was used to check data for bimeras; 3 – 5% of the reads were determined to be bimeras and excluded from the dataset. Sequences which have undergone DADA2 denoising are referred to as amplicon sequence variants (ASVs). Cutadapt was used to filter and trim raw sequences using a minimum read length of 100 bp and used a minimum quality score of 30 (Martin, 2011). ASV tables were then merged and combined for each Illumina run and the SILVA database (v. 138.1) was used for taxonomic assignment using RStudio v. 1.4.1. While 16S rRNA calculated relative abundance is a widely employed method for assessing microbial community composition (e.g., Seth et al., 2024; Yu et al., 2024) due to its decreased cost, speed, and scalability (Durazzi et al., 2021), it is important to acknowledge potential sources of error inherent in this approach. These potential sources of errors include variability in 16S gene copy number and amplification/sequencing bias and error (Suzuki and Giovannoni, 1996; Hong et al., 2009; Kembel et al., 2012). However, their combination with other independent lines of evidence, i.e., metagenome and geochemical data, can assist in robust understanding of microbial community composition, function, and environmental preferences (Louca et al., 2018; Campa et al., 2022; Whaley-Martin et al., 2023).
Shannon Diversity index and sample richness values were calculated in RStudio v. 1.4.1 based on the number of unique amplicon sequence variant (ASVs) and sequence abundances. Data visualization and linear regression (Pearson’s correlations and associated ANOVA tests) were performed in OriginPro v. 2022. Additional ANOVA and post-doc Tukey pairwise statistical analyses were performed in RStudio v. 1.4.1.
2.5 Metagenomic sequencing, reads processing, and assembly
The genomic DNA was extracted from the sample filters individually and used for subsequent metagenomic sequencing. The DNA extracts were dried and resuspended in 25 μL aliquots of water. The Illumina library preparation kits were used in the construction of sequencing libraries with an insert length of ~500 bp. The libraries were sequenced using the Illumina HiSeq-1500 platform with paired-end 150 bp sequencing kits by the Farncombe Metagenomics Facility at McMaster University (Hamilton, Ontario, Canada), as previously described in Whaley-Martin et al. (2023).
The raw sequencing reads were filtered to remove Illumina adapters, PhiX and other Illumina trace contaminants using BBTools (Bushnell et al., 2017) and low quality bases and reads were removed using Sickle (v. 1.33). All reads with both ends remained were used for subsequent de novo assembly via IDBA_UD (parameters: --mink 20, --maxk 140, --step 20, --pre_correction; Peng et al., 2012) or metaSPAdes (parameters: -k 21,33,55,77,99,127; Nurk et al., 2017). Sequencing coverage for each scaffold from a given sample was individually mapped using quality paired-end reads to the full assembly using Bowtie2 with default parameters (Langmead and Salzberg, 2012). Generated sam files were converted to bam format and sorted using samtools (Li et al., 2009). Then the coverage of each scaffold was calculated using the script of jgi_summarize_bam_contig_depths from MetaBAT (Kang et al., 2015). Using MetaBAT, all scaffolds with a minimum length of 2,500 bp were assigned to genome bins with both tetranucleotide frequency and sequencing coverage profiles from all samples considered. Both binned and unbinned scaffolds with a minimum length of 1,000 bp were uploaded to ggKbase1 for manual genome bin refinement, which was based on GC content, sequencing coverage, and taxonomic information of each scaffold as previously described in Chen et al. (2020) and was primarily based on patterns and distribution of GC content and sequencing coverage of contigs in each bin. Contamination contigs/scaffolds were removed from metagenome assembled genomes using ggKbase if the contig/scaffold had divergent sequencing coverage (those with <0.5x or > 2x coverage relative to most other contigs/scaffolds in the metagenome assembled genome), GC content, and/or divergent taxonomic assignment. Taxonomic assignment of metagenome assembled genomes was determined based on taxonomic classification of protein-coding genes within each contig or scaffold. This process involved identifying the best taxonomic matches of the protein-coding genes to references proteins and determining the last common ancestor where ≥50% of the matches align with a particular taxonomic group. Overall taxonomic assignment of a metagenome assembled genome was then derived from the cumulative taxonomic assignments of all its contigs/scaffolds.
2.5.1 Gene prediction and annotation
Gene prediction and subsequent analyses were completed using assembled scaffolds with a minimum length of 1,000 bp (herein “1k_scaffolds) to assist in obtaining metagenome assembled genomes with minimal contamination and high completeness. The protein-coding genes were predicted using Prodigal v. 2.6.3 from 1k_scaffolds (parameters: -m -p meta; Hyatt et al., 2010). 16S rRNA genes were predicted from 1k_scaffolds based on an HMM database in accordance with methods presented by Brown et al. (2015). The tRNAs on all 1k_scaffolds were predicted using tRNAscanSE (v. 2.0.3; Chan and Lowe, 2019). For functional annotation, the predicted protein-coding genes were searched against the Kyoto Encyclopedia of Genes and Genomes databases (KEGG; Kanehisa et al., 2017), UniRef100 (Suzek et al., 2007), and UniProt (Apweiler et al., 2004) via Usearch (v.10.0.240_i86linux64; Edgar and Bateman, 2010). HMM databases (Anantharaman et al., 2016) and KOfam HMM database (Kanehisa and Sato, 2020) were used to search and identify the predicted protein-coding genes for specific metabolic potentials of interest. DiSCo (Neukirchen and Sousa, 2021) was used to distinguish dsrC and its homologs. BLASTp was used to search predicted proteins against the shdr protein sequences encoded by Acidithiobacillus caldus SM-1, Thioalkalivibrio sp. K90mix, and Hyphomicrobium denitrificans (Koch and Dahl, 2018) followed by manual confirmation. Screening for hdrAACB genes was completed by checking the annotations from KEGG, UniRef100, and UniProt using the key word “hdr” and the acquired list of genes was manually confirmed. The aprM and aprBA genes were identified by comparing all protein-coding genes (from corresponding metagenomes) against aprM/aprBA identified in Thiobacillus denitrificans (WP_011312796.1) using BLASTp with an e-value threshold of 1e-10 and hits were manually verified. The tetH gene was identified using a BLASTp search as previously described in Watanabe et al. (2019) followed by manual verification. Failure to detect genes which require annotated reference genomes (shdr, hdrAACB, aprM, aprBA, and tetH) for detection, rather than KO values (or other database values), may result in under-reporting of those genes until more annotated reference genomes are reported in the literature.
3 Results and discussion
3.1 SOB community 16S rRNA composition and metagenomic inferred function
Microbial community composition was determined using 16S rRNA relative abundances for each TI sample (n = 13 from Mine 1, n = 15 from Mine 2, n = 6 from Mine 3, and n = 8 from Mine 4). Nine major genera of SOB were identified in these 42 TI samples including: Halothiobacillus spp. (12.7 ± 20.5%), Sediminibacterium spp. (4.8 ± 9.1%), Thiobacillus spp. (3.9 ± 7.8%), Sulfuricurvum spp. (3.7 ± 11.5%), Thiovirga spp. (1.5 ± 3.2%), Sulfuritalea spp. (0.7 ± 2.9%), Sulfurimonas spp. (0.4 ± 1.3%), Sulfuriferula spp. (0.4 ± 0.7%), and Thiomonas spp. (0.1 ± 0.2%). Each genus occurred at >1% abundance in at least one sample (Table 2). The presence and relative abundance of these nine SOB genera differed among mines and over time.
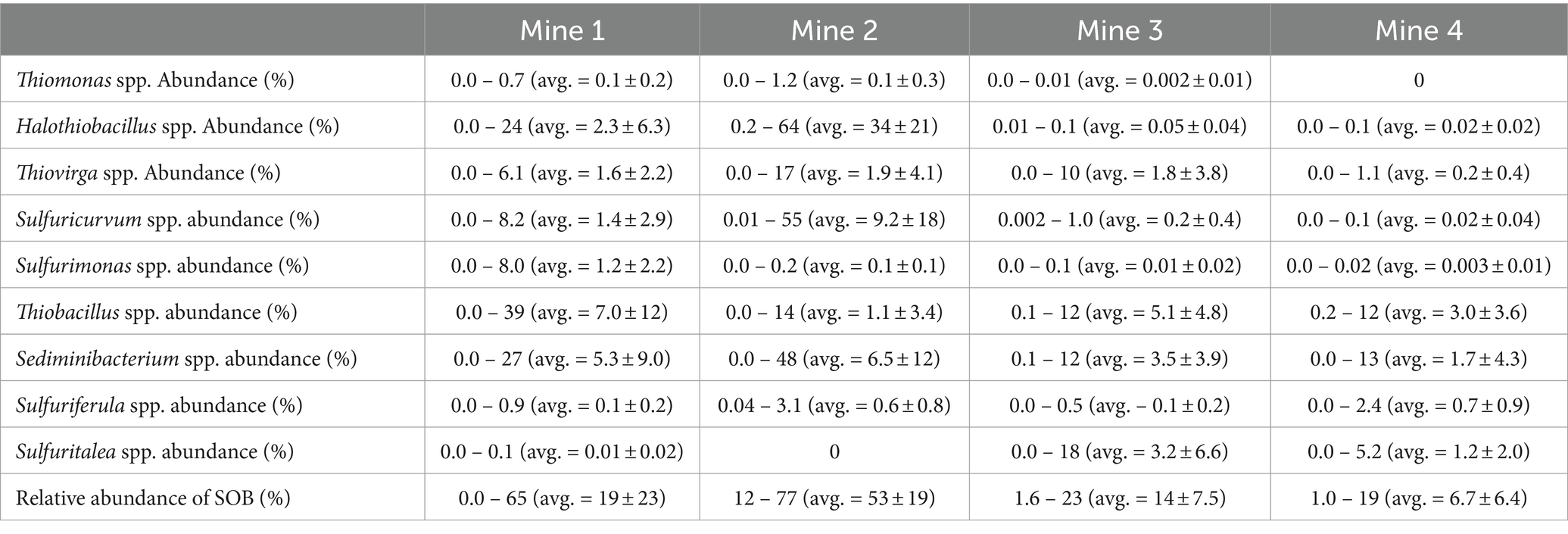
Table 2. Average (± standard deviation), minimum and maximum 16S rRNA percent relative abundances across sulfur oxidizing bacteria genera data for Mine 1, Mine 2, Mine 3, and Mine 4 tailings impoundment waters (2016 – 2019).
To identify the potential sulfur metabolizing pathways encoded by these SOB genera, 116 metagenomes were constructed from 38 samples collected at these four mines between 2016 and 2018 (Supplementary Table 2). Genomes for Sulfurimonas and Sulfuritalea could not be reconstructed and thus interpretations for these two genera relied on literature reports. Three major pathways including sox, rdsr, and S4I were examined, as well as additional sulfur oxidation genes not specific to those three pathways (Figure 1), based on the foundational understanding outlined by Watanabe et al. (2019) and expanded by Whaley-Martin et al. (2023). Genes encoding the sox pathway, either complete (csox; soxXYZABCD) or incomplete (isox; soxXYZAB and lacking soxCD) were most common – occurring in eight of the nine genera. Thiomonas, Halothiobacillus, and Thiovirga genomes encoded the csox pathway (resulting in generation of SO42-; Figure 1) which is consistent with published reports for Thiomonas and Halothiobacillus (Veith et al., 2012; Lin et al., 2015). Outside of other published works from the mines included in this study, there are very limited data available in the literature for Thiovirga spp., Miettinen et al. (2021) identified possible Thiovirga spp. in zinc and copper ore processing facilities in Portugal that lacked only the soxC subunit, indicating there may be variability within the Thiovirga genus regarding the ability for complete oxidation to SO42- via the sox pathway, unlike the findings presented here (Figure 1). Sulfurimonas spp. have been reported to often possess the soxCD genes (i.e., csox) (Lahme et al., 2020; Wang et al., 2021, 2023) however, this genus may lack soxAB genes instead, which would not allow for complete oxidation via the sox pathway (Lahme et al., 2020; Wang et al., 2021). At least one strain of Sulfurimonas (Strain NW10T) has been reported to possess genes for the csox pathway but used the isox pathway instead (Wang et al., 2021).
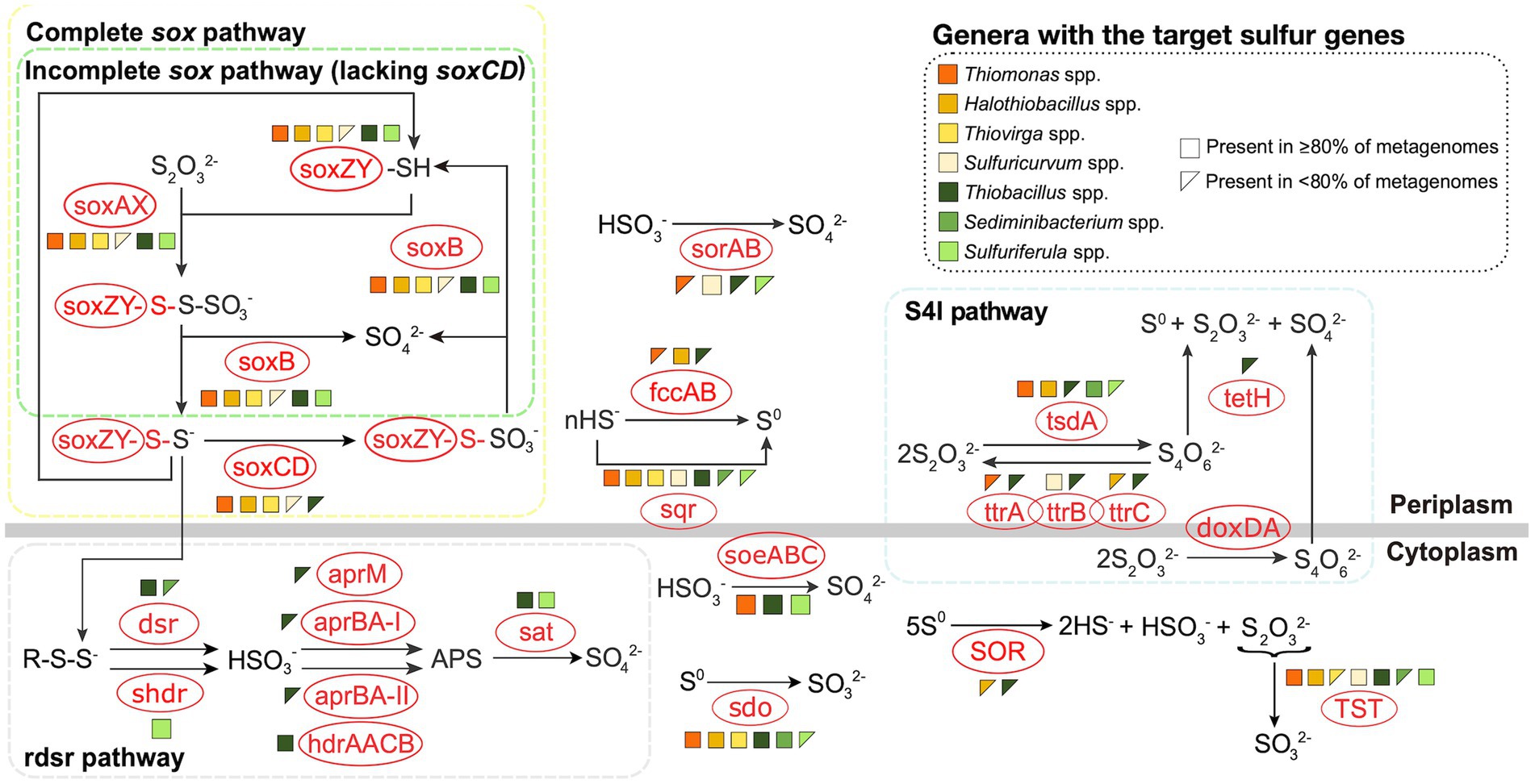
Figure 1. Metabolic potentials and pathways for sulfur oxidizing bacteria genera from the four TIs investigated here. Figure was adapted from Whaley-Martin et al. (2023) and based on Watanabe et al. (2019). Genomes were reconstructed from TI samples collected from the four target mines between 2016 and 2018. Due to low abundance of some organisms, variable quantities of genomes were used including 3 Thiomonas, 31 Halothiobacillus, 3 Thiovirga, 31 Thiobacillus, 30 Sediminibacterium, 4 Sulfuricurvum, and 14 Sulfuriferula.
Sediminibacterium was the only detected SOB in this study that did not encode any sox genes (Figure 1). Two genera were identified as possessing the isox pathway; Sulfuriferula and Thiobacillus (Figure 1). Watanabe et al. (2019) reported Sulfuritalea hydrogenivorans sk43H, which at present is the only published pure culture strain, as possessing soxAX, soxYZ, and soxB but lacking soxCD, thus indicating the presence of the isox pathway. Three of the reconstructed Sulfuricurvum spp. genomes encoded csox, while one encoded no sox genes (Figure 1), with the latter potentially due to the low quality of the genome. This finding of csox encoding Sulfuricurvum from mining TI samples described here, and also reported in Whaley-Martin et al. (2023), diverges from the current literature which typically reports Sulfuricurvum spp. found in the natural environment (terrestrial aquifer, geothermal springs) as possessing the isox pathway (lacking soxCD; Handley et al., 2014; Meziti et al., 2021).
A S4I pathway gene for the first reaction generating S4O62-, was the second most abundant identified across the seven genera with reconstructed genomes (Figure 1). Two distinct catalysts, tsdA and doxDA, are responsible for the conversion of S2O32- to S4O62- (Nguyen et al., 2022). However, doxDA was absent in the reconstructed genomes, while tsdA was present in the genomes of five of the seven genera, including >80% of Thiomonas, Halothiobacillus, and Sediminibacterium genomes and < 80% of Thiobacillus and Sulfuriferula genomes (Figure 1). Generation of tetrathionate from tsdA activity can be subsequently disproportionated via tetH to form S0, S2O32-, and SO42-, which can then potentially feed both the incomplete/complete sox pathway and/or S0 storage pathways (including sdo, SOR, and rdsr; Friedrich et al., 2005; Frigaard and Dahl, 2008; Watanabe et al., 2019). Though tsdA was found in five genomes, tetH, which would be required for the completion of the S4I pathway, was only found in five of the thirty-one Thiobacillus genomes (<80%; Figure 1). This may indicate the potential for genera with only tsdA (such as Thiomonas, Halothiobacillus, Sediminibacterium and Sulfuriferula) to couple with tetH encoding Thiobacillus to complete the S4I pathway. Limited tetH detection may also reflect current limitations in tetH reference material. TtrABC can catalyze the reduction of S4O62- to produce S2O32- though its components (ttrA, ttrB, and ttrC) were only variably present across Thiobacillus genomes (Figure 1). Thiomonas spp., Sulfuricurvum spp., and Halothiobacillus spp. were found to possess ttrA, ttrB, and ttrC respectively, but there is no available research indicating these subunits are active in sulfur reduction individually. Watanabe et al. (2019) reported that doxDA, tsdA, and tetH genes were absent in Sulfuritalea hydrogenivorans sk43H indicating this Sulfuritalea species did not encode the S4I pathway. Currently, there is a lack of specific data regarding the presence or activity of the S4I pathway (and associated genes tsdA, doxDA, and tetH) within Sulfurimonas. Consequently, it remains uncertain in results here and in available literature, whether Sulfurimonas possess the genomic capacity to support the S4I pathway.
The genes for the complete rdsr pathway were detected in 29 of the 31 Thiobacillus genomes (Figure 1). Sulfuriferula possessed the potential to express shdr (SO32- production) and sat (SO42- production through APS) but lacked the intermediate aprM/aprBA-I or aprBA-II/hdrAACB genes that mediate the reaction produce APS (Figure 1), aligning with results presented in Watanabe et al. (2019) for Sulfuriferula sp. AH1 and Sulfuriferula thiophila mst6. These missing steps could be compensated for by the cytoplasmically oriented soeABC that was present in all Sulfuriferula genomes, which generates SO42- directly from SO32- (Koch and Dahl, 2018; Figure 1). Alternatively, SO32- can be transported to the periplasm via TauE-like transporter where it can spontaneously react with hydrogen sulfide if present (Koch and Dahl, 2018). The lack of rdsr and shdr pathway genes in the SOB identified in this study, may reflect the relatively high dissolved oxygen concentrations across these water samples (averaging between 36.2 – 82.6 % saturation across the four mines; Table 1) which do not favour establishment of anaerobic and/or microaerophilic SOB which typically harbour the rdsr pathway (Klatt and Polerecky, 2015). Isolated genomes of Thiobacillus contained aprM, aprBA-I, aprBA-II, and/or hdrAACB, which would allow them to further oxidize SO32- to APS (Figure 1). Watanabe et al. (2019) reported the presence of partial (lacking aprM and aprBA-I) rdsr pathway genes (including dsrAB, dsrEFH, dsrC, dsrMKJOP, and sat) in the model species Sulfuritalea hydrogenivorans sk43H. Furthermore, evidence reported by Purcell et al. (2014) indicates Sulfuritalea hydrogenivorans sk43H can carry out the rdsr pathway within Antarctic lake sediments. Sulfuritalea hydrogenivorans sk43H also hosts a partial shdr pathway including aprBA-II and hdrAACB (Watanabe et al., 2019). Second only to the Thiobacillus genomes presented here, Sulfuritalea hydrogenivorans sk43H hosts the most diverse set of rdsr pathway genes of the nine SOB genera identified. While there are several published Sulfurimonas genomes, detailed insights into rdsr pathway occurrence remains limited. This may be due to the absence of the rdsr pathway within the Sulfurimonas genus as Wang et al. (2023) reports Sulfurimonas sp. ST-27 lacks dsrAB, dsrE, dsrCD, dsrMKJOP, and aprBA, but does contain sat. The rdsr and shdr pathways may play an important role in bacteria harbouring the isox pathway by facilitating the conversion of sulfane sulfur, produced as an intermediate, into SO42- (Xin et al., 2023). SOB that encode the isox pathway without the rdsr or shdr pathways, may produce and accumulate sulfane sulfur as a by-product of S2O32- oxidation, though the mechanism for reducing oxidative stress from the accumulation of sulfane sulfur is not well delineated and may result in the accumulation of volatile H2S (Xin et al., 2023). Additionally, sulfane sulfur has been found to exhibit toxicity in both bacteria and fungi, including when produced through the isox pathway in Cupriavidus pinatubonensis (Sato et al., 2011; Xin et al., 2023). Of the four genera in this study which contain the isox pathway, only Sulfuritalea and Thiobacillus possess the ability to detoxify sulfane sulfur using either the rdsr or shdr pathway.
Additional cytoplasmic sulfur oxidative genes investigated include TST (S2O32- to SO32-) and soeABC (SO32- to SO42-; Figure 1). TST was broadly present across the SOB occurring in >80% of the Thiomonas spp., Halothiobacillus spp., Thiobacillus spp., Sulfuricurvum spp., and Sulfuriferula spp. metagenomes and in <80% of the Thiovirga spp. and Sediminibacterium spp. metagenomes (Figure 1). To date, no information is available regarding the presence or absence of TST in Sulfurimonas or Sulfuritalea genomes. Wang et al. (2019) suggests TST may use S2O32- produced by SOR to further oxidize it to sulfur and SO32-. SoeABC transforms SO32- to SO42- in the cytoplasm and may even contribute to SO32- oxidation as part of the rdsr pathway (Dahl et al., 2013; Whaley-Martin et al., 2023). Greater than 80% of the Thiomonas, Thiobacillus and Sulfuriferula genomes contained soeABC (Figure 1). SoeABC has also been identified in Sulfuritalea hydrogenivorans sk43H (Watanabe et al., 2014) but no information on soeABC’s occurrence in Sulfurimonas is currently available. Additional periplasmic sulfur oxidation genes identified in this study include sqr, fccAB, and sorAB (Figure 1). Both sqr and fccAB are implicated in catalysis of hydrogen sulfide to S0 which can form S0 globules and be further oxidized to SO32- via the rdsr pathway (Nosalova et al., 2023). Sqr was widely present appearing in >80% of the genomes identified in all genera apart from Sediminibacterium and Sulfuriferula metagenomes where they were present in <80% of the metagenomes (Figure 1). Sulfuritalea hydrogenivorans sk43H and various Sulfurimonas species were found to host the sqr gene (Watanabe et al., 2019; Wang et al., 2021, 2023). Among the reconstructed genomes, >80% of the Halothiobacillus genomes harbored the fccAB gene cluster while the fccAB cluster was identified in <80% of the Thiomonas and Thiobacillus genomes (Figure 1). Sulfuritalea hydrogenivorans sk43H has been reported to contain fccAB (Watanabe et al., 2019) and currently no Sulfurimonas species have been reported to contain fccAB. SorAB, which catalyzes the conversion of SO32- to SO42- in the periplasm, was found in >80% of the Sulfuricurvum genomes and < 80% of the Thiomonas, Thiobacillus, and Sulfuriferula genomes (Figure 1). SorAB has also been reported in Sulfuritalea hydrogenivorans sk43H (Watanabe et al., 2019) as well as eight of the eleven Sulfurimonas species examined by Wang et al. (2021). The four most abundant genera in these four TI collectively possess the capacity for sulfur oxidation via all three universal pathways, csox (Halothiobacillus, Sulfuricurvum), isox + rdsr (Thiobacillus), and S4I (Halothiobacillus, Sediminibacterium, Thiobacillus) suggesting adaptation of these TI SOB communities to occupy all potential sulfur oxidizing niches that occur in these highly physicochemically and geochemically dynamic TI systems.
3.1.1 SOB functional classification
Under acid mine drainage conditions (acidic, metal rich), Kuang et al. (2016) identified metabolic function as a better predictor of microbial community structure and function than taxonomy. Similarly, here, patterns in metagenomic data were used to classify these SOB genera into csox dominant and non-csox dominant SOB genera groupings. Csox dominant SOB genera identified here included Halothiobacillus spp., Thiovirga spp., Thiomonas spp., and Sulfuricurvum spp. based on the presence and high abundance of the csox pathway (Figure 1). Non-csox dominant SOB genera were characterized by isox gene pathway presence and increased abundance of genes associated with alternative pathways such as rdsr (dsrABCEFH, aprAB, sat; Whaley-Martin et al., 2023) and/or S4I (tsdA, tetH; Ghosh and Dam, 2009; Wang et al., 2019; Whaley-Martin et al., 2023) (Figure 1). Thiobacillus spp., Sediminibacterium spp., Sulfuriferula spp., Sulfuritalea spp., and Sulfurimonas spp. were categorized as non-csox dominant SOB genera (Figure 1). Importantly, reactions catalyzed by the csox pathway favour the complete oxidation of SOI to SO42-, resulting in increased acidity and SO42- production, while more energy efficient pathways (e.g., rdsr; Klatt and Polerecky, 2015), favoured by non-csox dominant SOB commonly generate free SOI and in some cases may consume H+ (Dam et al., 2007; Klatt and Polerecky, 2015; Hutt et al., 2017). Therefore, presence or absence of csox vs. non-csox dominant pathways may reflect, and in turn, result in physicochemically and geochemically distinct waters. Mine waters with non-csox dominant SOB (i.e., Thiobacillus, which can generate free SOI) may be at greater risk of offsite acidification due to the ability of many SOI to pass through current treatment to receiving environments where their subsequent oxidation could release acidity.
3.1.2 Spatial and temporal trends in SOB community composition and function
The abundance of the nine identified SOB genera differed spatially and temporally for individual mines as well as across the four mines (Figure 2). The two oldest TIs, Mine 2 and Mine 1, had the highest SOB abundances (Figure 2). Mine 2 TI had the largest total abundance of SOB genera across all samples (54 ± 19%) and was the only mine TI where csox dominant SOB genera (predominantly Halothiobacillus spp.) had a higher average abundance than non-csox dominant SOB genera (predominantly Sediminibacterium spp.; Figure 2, Table 2). With more than 50% of the community on average consisting of SOB, Mine 2 TI had the lowest non-SOB genera abundance (38 ± 17%) and lowest unknown genera abundance (12 ± 9.4%; Figure 2). Based on evidence from this study and a previous study (Whaley-Martin et al., 2023), the Mine 2 community had the potential to express the csox pathway (via Halothiobacillus spp., Thiovirga spp., Thiomonas spp. or Sulfuricurvum spp.), isox pathway (via Thiobacillus spp. or Sulfuriferula spp.), rdsr pathway (via Thiobacillus spp.), and/or the S4I pathway (via Thiobacillus spp.). The average csox dominant SOB abundance was significantly higher in Mine 2 (p < 0.001) compared to the other three mines, while the abundance of non-csox dominant SOB were not significantly different across the four mines. Averaging a total SOB abundance 2.7 times lower than Mine 2, Mine 1 TI had the second highest total SOB abundance (20 ± 24%) (Figure 2, Table 2). SOB genera present at Mine 1 could express the csox pathway (via Halothiobacillus spp., Thiovirga spp., or Sulfuricurvum spp.), isox pathway (via Thiobacillus spp. or Sulfurimonas spp.), rdsr pathway (via Thiobacillus spp.), and/or the S4I pathway (via Thiobacillus spp.).
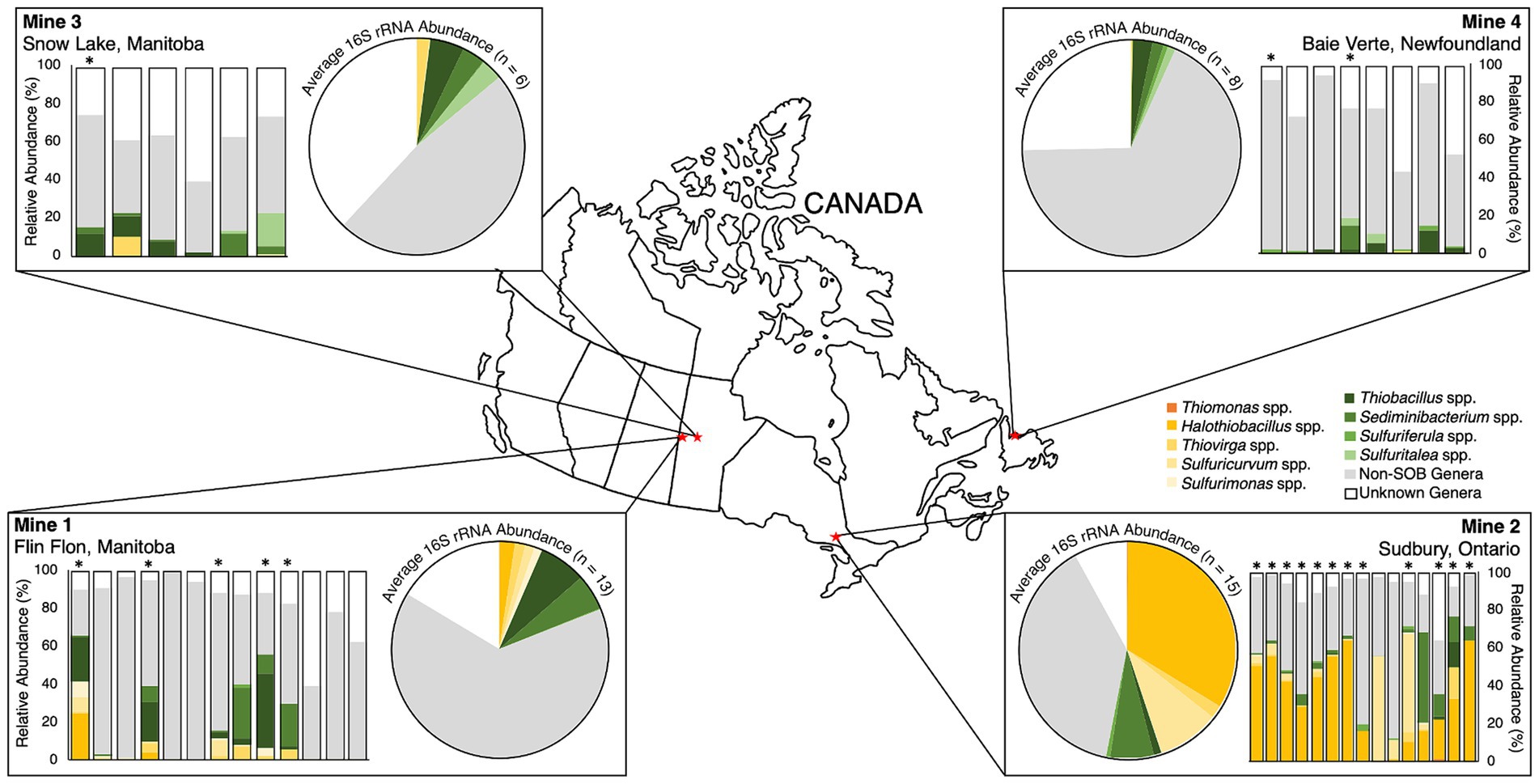
Figure 2. 16S rRNA relative abundance (%) of top nine sulfur oxidizing bacteria genera from 2016 – 2019, non-SOB genera and unknown genera for each of the four mines mapped to their sample locations across Canada. Individual samples are shown as bar graphs and mine average abundances are shown as pie graphs. “Non-SOB genera” are all other identified sequences and “unknown genera” are identified sequences not matched to genera in the Silva Database v138.1. Asterisks (*) denote samples with metagenome data included in this study.
Mine 3’s TI waters averaged 14 ± 7.5% total SOB abundance (Table 2), which was approximately 4 times lower than Mine 2 and 1.4 times lower than Mine 1. Mine 3 TI did, however, have the highest abundance of unknown genera (38 ± 12%) and the second highest abundance of identified non-SOB genera (48 ± 8.2%; Figure 2). The majority of TI SOB present in Mine 3 TI waters were identified as non-csox dominant SOB and averaged 12 ± 6.2% primarily consisting of Thiobacillus spp., Sediminibacterium spp. and Sulfuritalea spp. (Figure 2, Table 2). The average abundance of csox dominant SOB genera at Mine 3 was 2.0 ± 3.7% and was primarily Thiovirga spp. (Figure 2, Table 2). 16S rRNA abundance, metagenome data and present literature indicate that SOB genera from Mine 3 could express the csox pathway (via Thiovirga spp.) and/or the isox (via Thiobacillus spp. or Sulfuritalea spp.), rdsr (via Thiobacillus spp.), and S4I pathway (via Thiobacillus spp.).
The lowest total SOB abundance was found in the Mine 4 TI samples, with an average abundance of 7.1 ± 6.3 and > 97% of the SOB present represented by non-csox dominant SOB genera (Table 2). Non-csox dominant SOB genera averaged 6.5 ± 6.6% of the overall community abundance, while csox dominant SOB abundance was the lowest of the four mines averaging 0.3 ± 0.4%, with only Thiovirga spp. ever reaching above 1% of the overall community abundance at Mine 4 (Table 2). Mine 4 had the second highest contribution of unknown genera representing 27 ± 17% of the microbial community (Figure 2). Mine 4 had the most limited S oxidation potential with no major SOB csox abundance (Table 2). Thiobacillus spp. present at Mine 4, contained tsdA enabling part 1 of the S4I pathway (S4O62- formation), but did not contain tetH associated with the 2nd part of the S4I pathway (S4O62- disproportionation) and did contain genes for the isox and rdsr pathway.
3.2 TI wastewater sulfur geochemistry
Mean [total S0.45μm] and [SO42-] across the four mines display a pattern of increasing concentration with age, though this relationship is not proportional. Notably, Mine 1 and Mine 2 are most similar in age and Mine 2 and Mine 3 are most similar in concentrations (Table 1, Figure 3). However, [total S0.45μm] exhibited substantial variability across the four mines – ranging from 0.3 mM (Mine 4) to 16.8 mM (Mine 1) (Table 1). ANOVA and post-hoc Tukey pairwise comparison tests revealed a significantly higher average total S0.45μm concentration at Mine 1 (14 ± 1.6 mM) compared to Mine 2 (8.9 ± 0.7), Mine 3 (7.9 ± 1.0), and Mine 4 (1.9 ± 0.8 mM) (Table 1; p < 0.001). [SO42-] displayed a similar trend where Mine 1 exhibited significantly higher concentrations than Mine 2 (p < 0.05), Mine 3 (p < 0.001), and Mine 4 (p < 0.001) (Table 1, Figure 3). Elevated sulfur concentrations in Mine 1 and Mine 2 TIs can be attributed to their extensive use since the 1920s. Mine 3, while younger than Mine 1 and Mine 2, contains a comparatively higher [total S0.45μm], reflecting tailings additions from multiple mining operations and a shallow water cover depth. Mine 4, the smallest and youngest TI, has accumulated the lowest volume of tailings, resulting in the lowest [total S0.45μm].
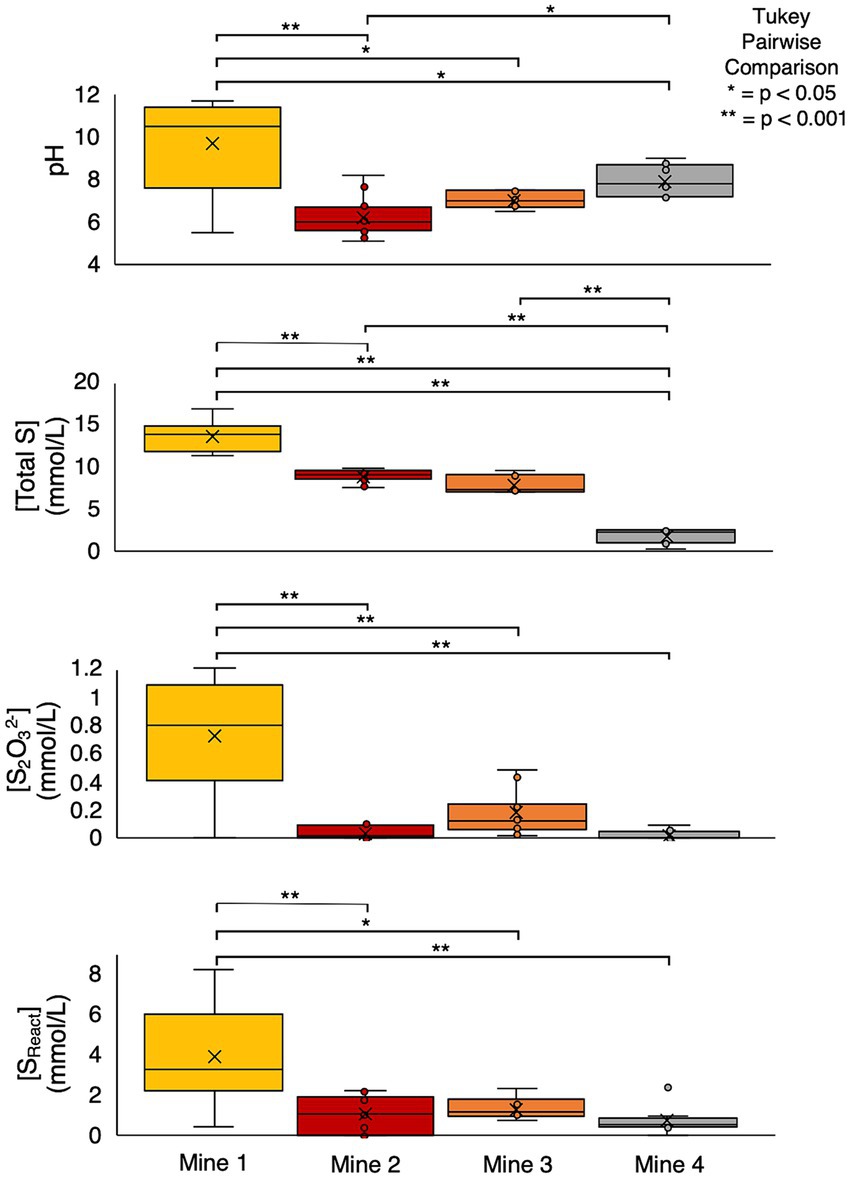
Figure 3. Box and whisker plots and statistical analysis (ANOVA and post-hoc tukey pairwise statistical comparison) of cross mine pH, total S (mmol/L), S2O32- (mmol/L), and SReact (mmol/L). Box limits represent the first and third quartile of each dataset, with a black line indicating the median value and an “x” denoting the mean.
Though not often reported, [total S] from various environments varies widely from ~10 μM to 800 μM observed in freshwater lakes (Vincent et al., 2021) to ~28 mM in sea water (Canfield and Farquhar, 2009; Vincent et al., 2021) with SO42- typically being the largest contributor to the overall sulfur balance (Canfield and Farquhar, 2009). Reported [SO42-] from other mining environments studies range from 3.6 mM to >250 mM (Foucher et al., 2001; Silva et al., 2012; Kinnunen et al., 2018; Camacho et al., 2020b). [Total S] (0.3 – 16.8 mM; Table 1) in this study are typically higher than the reported freshwater <1 μM value, while on the lower end of reported values for mining contexts (Foucher et al., 2001; Kinnunen et al., 2018).
Reactive sulfur concentrations ([SReact] calculated as [Total S] – [SO42-]; Whaley-Martin et al., 2020), representing all sulfur atoms capable of oxidation, were determined for each TI sample. ANOVA and post-hoc Tukey pairwise comparison tests revealed that Mine 1 had a significantly higher average SReact compared to Mine 2 (p < 0.001), Mine 3 (p < 0.05) and Mine 4 (p < 0.001) (Figure 3). Considering the potential 10-fold variation in both [total S0.45μm] and [SReact] between samples, %SReact ([SReact]/[Total S0.45μm] x 100) provides a useful metric to reflect the proportion of SReact within each system’s individual total S pool. %SReact ranged from 0 to 92.3% across the four mines, with the highest average %SReact occurring at Mine 1 (28 ± 16.2%) followed by Mine 4 (27 ± 13%), Mine 3 (17 ± 4.4%), and Mine 2 (12 ± 9.5%). SReact may include various sulfur species such as S2O32- and SO32- (quantified in this study), as well as additional unresolved sulfur species including but not limited to S4O62-, S2O42-, and S0 which were not quantified here but could potentially contribute to or support microbial sulfur cycling (Bak and Pfennig, 1987; Kelly et al., 1997). [SO32-] were generally low (often at or near the detection limit) with the highest concentrations found at Mine 1 (0.03 ± 0.05 mM; Table 1). S2O32- however, was detectable across all sites with a significantly higher (p < 0.001) average concentration at Mine 1 compared to the other three mines (Figure 3). [S2O32-] was the largest measured SReact contributor, constituting 0 – 100% (calculated %S2O32- = [S2O32-]/[SReact] x 100) of the SReact pool at each mine. While Mine 4 had the largest average %SReact, it had the smallest average %S2O32- (5.3 ± 8.2%) and therefore the largest proportion of unresolved sulfur species. On average, [S2O32-] comprised approximately one-fifth of both Mine 1 (22 ± 16%) and Mine 2’s (19 ± 27%) [SReact] pool.
3.3 TI wastewater physicochemistry
42 TI water cap samples were collected from the four mines between 2016 and 2019 during open water periods (early spring to late fall; Supplementary Figure S1) exhibited variable pH and DO values which are known to be important influencers of microbial ecological niches and associated sulfur oxidation pathways (Chen et al., 2013; Whaley-Martin et al., 2023). High temporal variability in DO (% saturation) and pH occurred for each mine as well as across mines, reflecting morphometric differences between TI facilities (particularly depth; Table 1) and the dynamic nature of actively managed TIs (Figure 3). Oxygen profiles from Mine 1, Mine 2, and Mine 3 demonstrated a steep oxygen gradient within TI water caps (Mine 4 profile data unavailable), ranging from <1 to >100% saturation (Table 1) across the four mines. Cross mine comparison revealed Mine 4 had significantly higher %DO compared to Mine 1 and Mine 2 (ANOVA and a post-hoc Tukey pairwise comparison test, p < 0.05; Figure 3) and no statistically significant differences among the remaining mines %DO.
Across the four mines, pH ranged from 5.1 to 11.8 (Table 1; Figure 3). Lime (Ca(OH)2) and alkaline tailings additions resulted in the highest average pH at Mine 1 (9.7 ± 2.0) and significantly higher pH values than Mine 2 (p < 0.001), Mine 3 (p < 0.05), and Mine 4 (p < 0.05) (Figure 3). Mine 2 TI waters had the lowest average pH (6.3 ± 0.9) with pH regularly falling below pH 6.5 (~64% of datapoints) and an observed decrease in pH from late spring to early fall. Mine 3’s TI water cap had the narrowest range of pH values (6.6 – 7.6, avg. = 7.1) with no observed seasonal fluctuations. Mine 4’s average pH was 8.0 ± 0.7 (7.2 – 9.0), with 75% of samples below pH 8.5, which was significantly higher than Mine 2 (p < 0.05) and lower than Mine 1 (p < 0.05; Figure 3). All four TIs exhibit dimictic lake mixing patterns, resulting in more homogenous dissolved oxygen and pH profiles during turnover events (spring and fall).
The acidity to SO42- ratio ([H+]/[SO42-]) can be used as a proxy for discerning direct sulfur oxidation from disproportionation (Bernier and Warren, 2007; Whaley-Martin et al., 2023). Higher [H+]/[SO42-] values observed at lower pH values indicate greater dominance of the csox pathway, indicating more direct and complete oxidation to SO42- (higher acidity generation, minimal SOI generation; Supplementary Figure S3). Lower [H+]/[SO42-] values observed at pH values >7, are consistent with more regeneration of SOI via disproportionation or partial oxidation attributed to isox, rdsr, and S4I pathways activity, resulting in lower net acidity generation (Supplementary Figure S3). While these ratio values may be underestimates at pH values >7, where buffering capacity is likely present, the observed pH dependent [H+]/[SO42-] trend is consistent with both S2O32- and SOB results. [S2O32-] was positively correlated to pH (p < 0.0001, Pearson’s r = 0.80; Supplementary Figure S4A), while total TI SOB abundance was negatively correlated with pH (p < 0.01, Pearson’s r = −0.64; Supplementary Figure S4B). Interestingly, average mining and anthropogenic (M&A) literature and environmental literature values were consistent with the pattern observed here (Supplementary Figure S4B).
3.4 pH effect on SOB community structure
Further examination of the relationship between SOB and pH for these four mine TIs, highlighted a pH dependent pattern in the occurrence of csox and non-csox dominant SOB. While clear relationships between pH and total SOB abundance, [S2O32-], and [H+]/[SO42-] ratios were observed, no discernible correlation was identified between dissolved oxygen and any of these parameters. Peak abundances of non-csox dominant SOB were observed at circumneutral pH (pH ~6 to ~8.5; Figure 4Ai) while the highest abundances of csox dominant SOB were observed below pH ~6.5 (Figure 4Aii). Samples above pH ~8.5 had low abundances of both csox and non-csox dominant SOB (Figures 4Ai,ii). Current literature indicates at least two genera of SOB (Thioalkalimicrobium and Thioalkalivibrio) are capable of growth under alkaline conditions (Sorokin et al., 2001) though neither were observed in this study’s TI samples. Lower pH value (pH < ~6.5) TI waters had the highest abundances of csox dominant SOB, lower [S2O32-] and higher [H+]/[SO42-] ratios; collectively consistent with direct oxidation via the csox pathway (Figure 4Aii). At more circumneutral pH values (pH ~6 to ~8.5), where non-csox dominant SOB dominated, higher [S2O32-] and low [H+]/[SO42-] values were also observed, indicating distinct pH dependent SOB community structure and S pathway(s) (Figures 4Ai,ii).
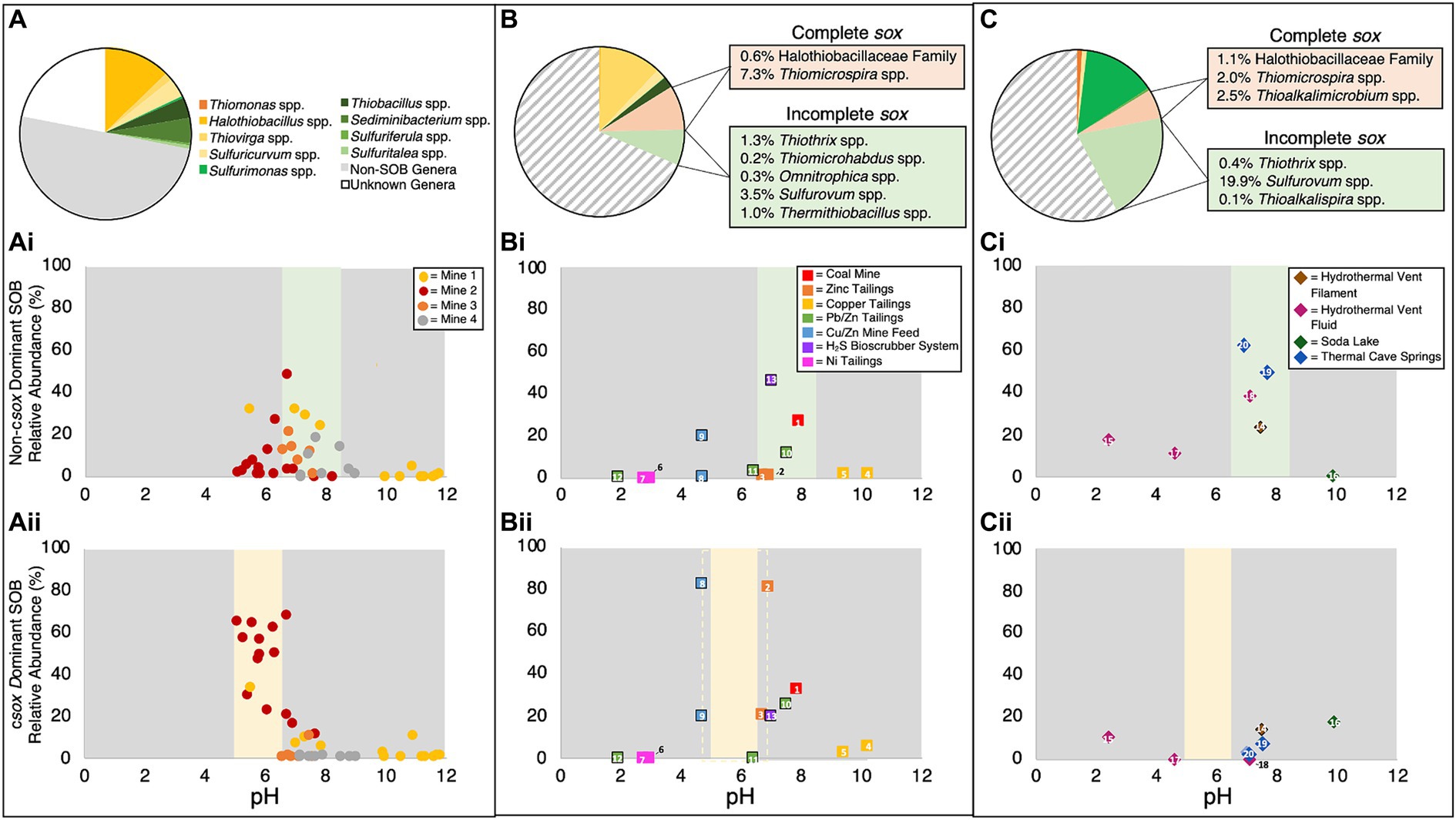
Figure 4. Genus level identification of (A–C) csox dominant SOB relative abundances, (Ai, Bi, Ci) non-csox dominant SOB relative abundances and (Aii, Bii, Cii) average 16S rRNA SOB communities identified in (A, Ai, Aii) tailings impoundment water samples from this study, (B, Bi, Bii) other mining and anthropogenic sample data from the literature and (C, Ci, Cii) environmental sample data obtained from the literature. “Non-SOB genera” are all other identified sequences in the samples and “unknown genera” are identified sequences not matched to genera in the Silva Database v138.1 (Data included in Bi, Bii, Ci, and Cii was collected from the following papers and can be identified using the superscript number: 1Kadnikov et al., 2019, 2-5Miettinen et al., 2021, 6-7Auld et al., 2017, 8-9Lopes et al., 2020, 10-12Chen et al., 2013, 13Haosagul et al., 2020, 14Patwardhan et al., 2018, 15Arce-Rodríguez et al., 2019, 16Vavourakis et al., 2019, 17-18Meier et al., 2017, 19-20Reigstad et al., 2011). Black outlines indicate non-water samples (e.g., soil, rock, tailings, biofilm, etc.).
3.4.1 Comparison to broader environments
The exploration of the pH – csox dominant/non-csox dominant SOB relationship extended across diverse contexts by integrating literature data (pH and SOB abundance) from broader mining environments, an industrial H2S bioscrubber, and a variety of natural environmental sites (e.g., hydrothermal vents, soda lakes, and thermal cave springs) (Reigstad et al., 2011; Meier et al., 2017; Patwardhan et al., 2018; Arce-Rodríguez et al., 2019; Vavourakis et al., 2019) (see Supplementary Table S1). Genus level SOB abundances were categorized into csox dominant SOB and non-csox dominant SOB. Literature identified SOB (not found in this study’s TI samples) were classified as csox dominant and non-csox dominant SOB using available genetic data and consensus of the current literature based on the reported presence of a complete or incomplete sox pathway. Additional SOB genera included three csox dominant clades Thiomicrospira spp., Thioalkalimicrobium spp., and Halothiobacilleaceae Family and seven non-csox dominant clades (Thiothrix spp., Thiomicrohabdus spp., Omnitrophica spp., Sulfurovum spp., Thermithiobacillus spp., Sulfobacillus spp., and Thioalkalispira spp.). Though class and family level abundances have been published for TI water cap samples, genus level data were not widely available in the literature. Data from various alternative sample locations (e.g., water from mill feed, solid tailings pore water), including both solid and water samples, were used. This approach aimed to broaden the range of geochemical and physicochemical characteristic investigated.
A brief summary and comparison of the relevant data for the overall groups of samples are provided in Supplementary Table S1 as more complete descriptions for individual sites can be found in their respective publications. The collective M&A literature data (n = 13) covered a pH range from 1.9 to 10.2 (Supplementary Table S1), the environmental samples (n = 7) ranged from pH 2.4 to 9.9 (Supplementary Table S1), while this study’s TI samples ranged from pH 5.1 to 11.8 (Table 1). Average total SOB abundance of this study’s TI samples (29 ± 26%) and M&A literature samples (31 ± 31%) were similar, while the average total SOB abundance for the environmental literature samples (42 ± 26%) was >10% higher (Figures 4A–C). Additional SOB genera (outside of the nine identified for TI in this study) represented ~15% (or ~ 50% of the total SOB) of the M&A literature and ~ 26% (or ~ 62% of the total SOB) of the environmental literature samples (Figures 4B,C). Both the M&A and environmental literature SOB communities diverged by at least 50% from those characterized here for base metal TI wastewaters, indicating notable disparities in microbial community composition suggesting SOB genera specific ecological niches.
On average, this study’s samples had ~64% of the total SOB abundance consisting of csox dominant SOB, while ~36% were classified as non-csox dominant (Figure 4A). The M&A literature samples averaged slightly higher csox dominant SOB contribution with ~72% of the total SOB abundance being attributed to csox dominant SOB (Figure 4B). Conversely, the environmental literature samples averaged a much lower contribution of csox dominant SOB, contributing only ~18% of the total SOB abundance (Figure 4C). Thiovirga spp. and Thiomicrospira spp., both classified as csox dominant SOB, were the two most abundant SOB in the M&A literature samples, while the two most abundant SOB in the environmental samples were both classified as non-csox dominant SOB (Sulfurimonas spp. and Sulfurovum spp.) (Figures 4B,C; Supplementary Table S1). In contrast, the two most abundant SOB from this study’s TI waters were Halothiobacillus spp. (csox dominant) and Sediminibacterium spp. (non-csox dominant). Thiomicrohabdus spp., Omnitrophica spp., and Thermithiobacillus spp. were unique to M&A samples with Thiomicrohabdus spp. and Omnitrophica spp. only being found in a single sample (Kadnikov et al., 2019) and Thermithiobacillus spp. identified in two samples from the same mine (Lopes et al., 2020). Thioalkalispira spp. and Thioalkalimicrobium spp. were unique to one environmental sample (Vavourakis et al., 2019).
Four M&A literature samples were found to have elevated abundances (>10%) of non-csox dominant SOB including three solid samples and one water sample (Figure 4Bii). Non-csox dominant SOB abundances of the M&A literature samples were found to be highest in the only non-mining sample (13 SPM Swine Farm H2S Bioscrubber System; Figure 4Bii). Three of the four M&A literature samples with elevated SOB abundances fell between pH ~6.5 and ~ 8, covering a similar range to the samples included in this study (Figure 4Bi). Among the seven M&A literature samples displaying elevated (>10%) csox dominant SOB abundances, five were found to occur within a pH range of ~5 and ~ 7 (Figure 4Bii); a slightly higher upper range compared to this study’s TI results (< pH 6.5; Figure 4Aii). The highest csox dominant SOB abundances in M&A literature samples were identified in samples at pH ~5 and ~ 7 (Figure 4Bii) though no data between those two pH values are presently available in the literature. At pH >8, there were two M&A literature samples which both showed low abundances of both csox dominant and non-csox dominant SOB (Figures 4Bi,ii), aligned with the pattern of low SOB abundance at more alkaline pH observed in this study’s TI samples. Samples below pH 4 (closer to acid mine drainage conditions) from the M&A literature samples had very low SOB abundances (Figures 4Bi,ii) in both solid and water samples.
Differences observed between the M&A literature data and this study’s TI data may reflect differences in S substrate availability due to the types or locations of samples included. The M&A mining-related solid sample’s primary source of sulfur would be sulfide-containing ores whereas SOI, aqueous dissolved species, prevalent in the TI waters investigated here, would be limited in solid samples. Solid tailings and feed samples may similarly exhibit different patterns of SOB community structure and abundance and associated sulfur oxidation genes due to these differences in available sulfur species, as only one of the three main pathways highlighted here (sox) utilizes sulfide (Klatt and Polerecky, 2015). This may account for the elevated csox dominant SOB abundances observed in some of the circumneutral solid samples included in Figure 4Bii. Extrapolating the pattern identified in this study’s samples to other anthropogenically impacted environments would require more data (both genus level abundance and pH data) for TI water caps which are not currently available. However, these comparative results, across a range of systems where data are available, show some consistencies in patterns with those observed for base metal TI wastewaters, namely low SOB abundances at elevated pH values (pH >8) and similar pH ranges for the peak functional pathway abundances of both non-csox dominant (pH ~6.5 to ~8.5) and csox dominant (~4.5 to ~7) SOB (Figures 4Bi,ii).
The environmental literature samples that fell between pH ~6.5 and ~ 8.5 had elevated non-csox dominant SOB abundances (Figure 4Ci) consistent with results for this study’s TI waters. No environmental literature samples were found between pH ~5 and ~ 6.5 precluding a direct comparison to the peak csox dominant SOB abundances observed for TI here. Similar to the observed TI results here, however, three environmental literature samples (two samples <pH ~5 and one sample > pH ~8.5) had lower abundances of non-csox dominant SOB than those within the more circum-neutral range (pH ~6.5 – ~8.5) (Figure 4Ci). The environmental literature group of samples was the only group where all samples had >1% non-csox dominant SOB (Figure 4Ci). Unlike the previous two groups of samples, none of the environmental samples had high abundances of csox dominant SOB (all <20% csox dominant SOB abundance; Figure 4Cii). This may reflect typically much lower environmental SOI concentrations (O’Brien and Birkner, 1977) and therefore metabolisms that exhaust SOI by completely oxidizing them to SO42- (i.e., csox) may not be favoured over those that disproportionate or recycle SOI such as the S4I or rdsr pathways (ie. non-csox dominant SOB). The divergence of mining impacted environments (both in this study (oxic TI wastewaters) and literature data) from natural environments suggests a specialized mining specific microbiome with higher abundance(s) of csox dominant SOB favoured by higher concentrations of S species in these contexts.
3.5 Factors influencing microbial community structure and function
The isox pathway or the isox + rdsr pathway generates more ATP and is more efficient due to energy conservation via S0 production and storage (Klatt and Polerecky, 2015). While energy efficiency is important, it is not the sole determinant of success in any given environment (Klatt and Polerecky, 2015). This is exemplified across the diverse set of environments presented here, where both the isox or isox + rdsr pathway, as well as the csox pathway appear at different times. Microbial success can be determined by growth rate, which considers both the substrate uptake rate and the growth yield (Klatt and Polerecky, 2015). Both growth yield and efficiency are directly related, but lower efficiency (such as that associated with csox) can be made up for by speed, resulting in equal or greater success (Sorokin and Kuenen, 2005; Klatt and Polerecky, 2015). Whaley-Martin et al. (2023) provided laboratory enrichment data from the same four mines included in this study to showcase that csox SOB (Halothiobacillus spp. dominant) enrichments exhibit a considerably faster S2O32- oxidation rate compared to the enrichments with rdsr containing SOB. Growth strategies which rely on speed are only likely to dominate in environments with unlimited substrates (Klatt and Polerecky, 2015). Though mining and other industrial environments technically do not have unlimited SOI, there are consistent replenishments of SOI through tailings additions or continuous wastewater treatment/flow. The observed proliferation of SOB with the csox pathway in mining and industrial contexts (Figures 4Aii,Bii), aligns with their demonstrated capability for rapid S2O32- oxidation under primarily oxic conditions. This highlights their competitive advantage in these substrate-rich environments where consistent SOI inputs help sustain their growth. The transition from non-csox dominant SOB (isox or isox + rdsr) to csox dominant SOB occurs within the range of 0.1 to 0.3 mmol/L [S2O32-] (Supplementary Figure S4A), suggesting a potential SOI threshold for the initiation of this metabolic shift. The associated pH decrease with the increased presence of csox dominant SOB may occur as a by-product of the activity of the csox pathway which produces SO42- and acidity and may slowly decrease pH. Further, under natural environmental conditions, where SOI are not as readily replenished, we do not see the proliferation of the csox pathway (Figure 4Cii).
Across the four TIs investigated here, evidence of pH and [S2O32-] dependent occurrence of all four key metabolic pathways, csox pathway, S4I pathway, isox pathway, and rdsr pathway emerged (Figure 5). pH partitioned three of these pathways: namely csox pathway dominated at lower pH values (pH ~5 to ~6.5) while the isox and rdsr pathways were more prevalent at circumneutral pH values (pH ~6.5 to ~8.5; Figure 5). The shift from circumneutral to more acidic pH may represent the onset of a positive feedback loop led by csox dominant SOB such as Halothiobacillus spp. which has previously been implicated in catalyzing the shift to net acid generation in laboratory scale experiments (Whaley-Martin et al., 2019). The csox pathway, responsible for the direct oxidation of S2O32- to SO42-, which generates more acidity than the S4I, isox, or rdsr pathway results in decreasing [S2O32-] and increasing [H+]/[SO42-] values (Figure 5). Uniquely, portions of the S4I pathway, namely the tsdA gene, were observed in SOB genera from across the entire pH range (Figure 5).
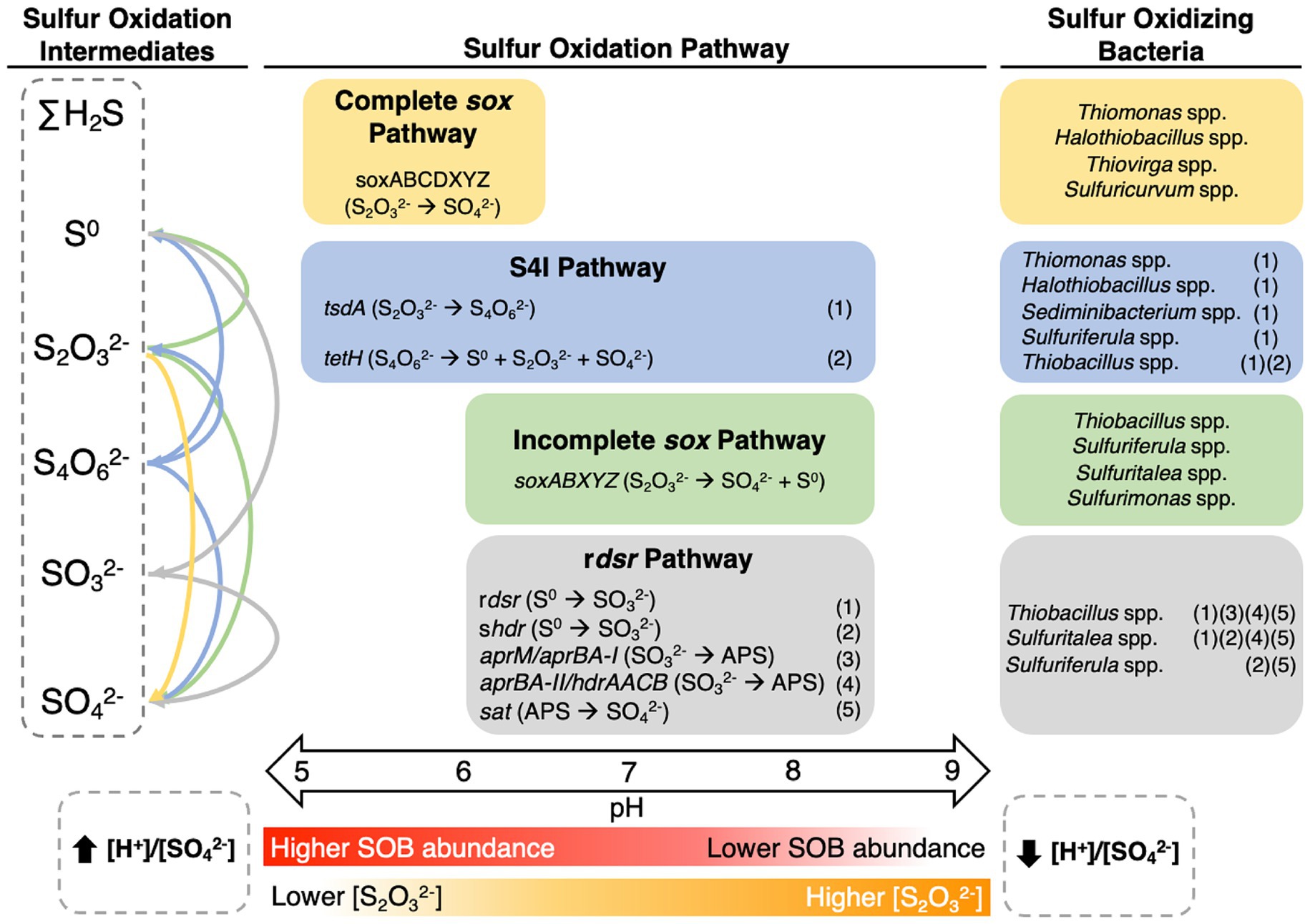
Figure 5. Conceptual model of pH bound metabolic activity with the associated responsible genera and geochemical pathway and outcomes in metal mining TI waters.
4 Conclusion
Results here reveal new insights into the interplay between pH, SOB community composition and function (csox dominant vs. non-csox dominant), S cycling and acidity generation within primarily oxic mine TI waters over seasonal, annual, and mine operation scales. Our results highlight two key pH dependent niches characterized by the presence of either (i) csox dominant SOB (e.g., Thiomonas spp., Halothiobacillus spp.) associated with lower pH values and lower [S2O32-] or (ii) non-csox dominant SOB (isox and/or rdsr pathways; e.g. Thiobacillus spp., Sulfuriferula spp.) associated with more circumneutral pH conditions and higher [S2O32-] (Figure 5). The presence of the first part of the S4I pathway (tsdA; S2O32- to S4O62-) was ubiquitous across pH and [S2O32-] niches; while possible subsequent processing of S4O62- via tetH which can lead to S0, and S2O32- regeneration, as well as SO42-, was limited to Thiobacillus spp., observed more prevalently in circumneutral pH values (Figure 5). Extrapolation of these results to broader environments via comparative analysis using M&A and environmental literature data revealed a specialized mining SOB microbiome characterized by elevated abundances of csox dominant SOB. Elevated SOI concentrations, typical of mining environments, support the establishment and sustainment of csox dominant communities unique to these environments. Our comprehensive study into SOB community dynamics, sulfur oxidation pathways, and the influence of geochemical and physicochemical factors in mining impacted waters, highlight the importance of thiosulfate availability, and pH constraints on sulfur oxidizing metabolism under oxic conditions, prevalent in TI contexts. This study highlights opportunities to manipulate TI SOB communities through pH adjustment and/or [S2O32-] management, offering potential avenues to reduce the risk of SOI discharge into receiving waters.
Data availability statement
The 16S rRNA sequencing data generated from this study have been deposited in the NCBI database under accession codes PRJNA510600, PRJNA882497, and PRJNA930015. The metagenome-assembled genomes generated in this study are available at Z2dLYmFzZS5iZXJrZWxleS5lZHU= via https://ggkbase.berkeley.edu/SOB_across_mines/organisms.
Author contributions
LT: Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing, Resources, Software. KW-M: Investigation, Methodology, Project administration, Writing – review & editing. L-XC: Data curation, Formal analysis, Software, Writing – review & editing. TC: Investigation, Methodology, Project administration, Supervision, Writing – review & editing. JA: Formal analysis, Investigation, Writing – original draft, Writing – review & editing. CJ: Methodology, Resources, Writing – review & editing. JK: Methodology, Resources, Writing – review & editing. LR: Methodology, Resources, Writing – review & editing. HS: Methodology, Resources, Writing – review & editing. JB: Conceptualization, Funding acquisition, Resources, Writing – review & editing. SA: Resources, Writing – review & editing, Methodology. LW: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Research activities and personnel were supported by Genome Canada Large Scale Applied Research Program 2015 (Grant Number OGI-124) and Ontario Research Fund–Research Excellence (Round 8).
Acknowledgments
The authors thank Laura Rossi for her laboratory assistance on this work. Additional thanks go out to on-site mine personnel who supported and aided in sample collection and processing including Stephanie Marshall, Sam McGarry, Nicole Gisby, Shirley Neault, Peter Mercer, Sean Breen, Landice Yestrau, Lana Leclerc, Ryan Trudeau, and Joel Nilsen. Research was made possible through the support of the Genome Canada Large Scale Applied Research Program and Ontario Research Fund – Research Excellence grants awarded to LW. The National Sciences and Engineering Research Council (NSERC) CREATE Mine of Knowledge Graduate Travel and Research Grants, Northern Scientific Training Program Travel Grant and QEII-GSST are also gratefully acknowledged.
Conflict of interest
LR and HS were employed by EcoReg Solutions.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1426584/full#supplementary-material
Footnotes
References
Akcil, A., and Koldas, S. (2006). Acid mine drainage (AMD): causes, treatment and case studies. J. Clean. Prod. 14, 1139–1145. doi: 10.1016/J.JCLEPRO.2004.09.006
Anantharaman, K., Brown, C. T., Hug, L. A., Sharon, I., Castelle, C. J., Probst, A. J., et al. (2016). Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 7:13219. doi: 10.1038/ncomms13219
Apweiler, R., Bairoch, A., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., et al. (2004). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 32, D115–D119. doi: 10.1093/NAR/GKH131
Arce-Rodríguez, A., Puente-Sánchez, F., Avendaño, R., Martínez-Cruz, M., de Moor, J. M., Pieper, D. H., et al. (2019). Thermoplasmatales and sulfur-oxidizing bacteria dominate the microbial community at the surface water of a CO 2-rich hydrothermal spring located in Tenorio volcano national park. Costa Rica. Extremophiles 23, 177–187. doi: 10.1007/s00792-018-01072-6
Auld, R. R., Mykytczuk, N. C. S., Leduc, L. G., and Merritt, T. J. S. (2017). Seasonal variation in an acid mine drainage microbial community. Can. J. Microbiol. 63, 137–152. doi: 10.1139/cjm-2016-0215
Bak, F., and Pfennig, N. (1987). Chemolithotrophic growth of Desulfovibrio sulfodismutans sp. nov. by disproportionation of inorganic sulfur compounds. Arch. Microbiol. 147, 184–189. doi: 10.1007/BF00415282
Baker, B. J., and Banfield, J. F. (2003). Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44, 139–152. doi: 10.1016/S0168-6496(03)00028-X
Bamford, V. A., Bruno, S., Rasmussen, T., Appia-Ayme, C., Cheesman, M. R., Berks, B. C., et al. (2002). Structural basis for the oxidation of thiosulfate by a sulfur cycle enzyme. EMBO J. 21, 5599–5610. doi: 10.1093/EMBOJ/CDF566
Bartram, A. K., Lynch, M. D. J., Stearns, J. C., Moreno-Hagelsieb, G., and Neufeld, J. D. (2011). Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl. Environ. Microbiol. 77, 3846–3852. doi: 10.1128/AEM.02772-10
Bates, S. T., Berg-Lyons, D., Caporaso, J. G., Walters, W. A., Knight, R., and Fierer, N. (2010). Examining the global distribution of dominant archaeal populations in soil. ISME J. 5, 908–917. doi: 10.1038/ismej.2010.171
Bernier, L., and Warren, L. A. (2007). Geochemical diversity in S processes mediated by culture-adapted and environmental-enrichments of Acidithiobacillus spp. Geochim. Cosmochim. Acta 71, 5684–5697. doi: 10.1016/J.GCA.2007.08.010
Bond, P. L., Druschel, G. K., and Banfield, J. F. (2000). Comparison of acid mine drainage microbial communities in physically and geochemically distinct ecosystems. Appl. Environ. Microbiol. 66, 4962–4971. doi: 10.1128/AEM.66.11.4962-4971.2000
Brito, J. A., Denkmann, K., Pereira, I. A. C., Archer, M., and Dahl, C. (2015). Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum. J. Biol. Chem. 290, 9222–9238. doi: 10.1074/jbc.m114.623397
Brown, C. T., Hug, L. A., Thomas, B. C., Sharon, I., Castelle, C. J., Singh, A., et al. (2015). Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523, 208–211. doi: 10.1038/nature14486
Bushnell, B., Rood, J., and Singer, E. (2017). BBMerge–accurate paired shotgun read merging via overlap. PLoS One 12:e0185056. doi: 10.1371/JOURNAL.PONE.0185056
Camacho, D., Frazao, R., Fouillen, A., Nanci, A., Lang, B. F., Apte, S. C., et al. (2020a). New insights into Acidithiobacillus thiooxidans sulfur metabolism through coupled gene expression, solution chemistry, microscopy, and spectroscopy analyses. Front. Microbiol. 11:411. doi: 10.3389/fmicb.2020.00411
Camacho, D., Jessen, G. L., Mori, J. F., Apte, S. C., Jarolimek, C. V., and Warren, L. A. (2020b). Microbial succession signals the initiation of acidification in mining wastewaters. Mine Water Environ. 39, 669–683. doi: 10.1007/S10230-020-00711-9/FIGURES/5
Campa, M. F., Chen See, J. R., Unverdorben, L. V., Wright, O. G., Roth, K. A., Niles, J. M., et al. (2022). Geochemistry and multiomics data differentiate streams in Pennsylvania based on unconventional oil and gas activity. Microbiol. Spect. 10:22. doi: 10.1128/spectrum.00770-22
Canfield, D. E., and Farquhar, J. (2009). Animal evolution, bioturbation, and the sulfate concentration of the oceans. Proc. Natl. Acad. Sci. 106, 8123–8127. doi: 10.1073/PNAS.0902037106
Cao, X., Koch, T., Steffens, L., Finkensieper, J., Zigann, R., Cronan, J. E., et al. (2018). Lipoate-binding proteins and specific lipoate-protein ligases in microbial sulfur oxidation reveal an atpyical role for an old cofactor. eLife 7:e37439. doi: 10.7554/ELIFE.37439
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Int. Soc. Microb. Ecol. J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chan, P. P., and Lowe, T. M. (2019). tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 1962, 1–14. doi: 10.1007/978-1-4939-9173-0_1
Chen, L. X., Anantharaman, K., Shaiber, A., Murat Eren, A., and Banfield, J. F. (2020). Accurate and complete genomes from metagenomes. Genome Res. 30, 315–333. doi: 10.1101/GR.258640.119
Chen, L., Li, J., Chen, Y., Huang, L., Hua, Z., Hu, M., et al. (2013). Shifts in microbial community composition and function in the acidification of a lead/zinc mine tailings. Environ. Microbiol. 15, 2431–2444. doi: 10.1111/1462-2920.12114
Dahl, C., Franz, B., Hensen, D., Kesselheim, A., and Zigann, R. (2013). Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology 159, 2626–2638. doi: 10.1099/mic.0.071019-0
Dam, B., Mandal, S., Ghosh, W., Das Gupta, S. K., and Roy, P. (2007). The S4-intermediate pathway for the oxidation of thiosulfate by the chemolithoautotroph Tetrathiobacter kashmirensis and inhibition of tetrathionate oxidation by sulfite. Res. Microbiol. 158, 330–338. doi: 10.1016/J.RESMIC.2006.12.013
Dold, B. (2014). Evolution of acid mine drainage formation in Sulphidic mine tailings. Fortschr. Mineral. 4, 621–641. doi: 10.3390/MIN4030621
Druschel, G. K., Hamers, R. J., and Banfield, J. F. (2003). Kinetics and mechanism of polythionate oxidation to sulfate at low pH by O2 and Fe3+. Geochim. Cosmochim. Acta 67, 4457–4469. doi: 10.1016/S0016-7037(03)00388-0
Durazzi, F., Sala, C., Castellani, G., Manfreda, G., Remondini, D., and De Cesare, A. (2021). Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 11, 1–10. doi: 10.1038/s41598-021-82726-y
Edgar, R. C., and Bateman, A. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/BIOINFORMATICS/BTQ461
Edwards, K. J., Gihring, T. M., and Banfield, J. F. (1999). Seasonal variations in microbial populations and environmental conditions in an extreme acid mine drainage environment. Appl. Environ. Microbiol. 65, 3627–3632. doi: 10.1128/AEM.65.8.3627-3632.1999
Elberling, B., and Damgaard, L. R. (2001). Microscale measurements of oxygen diffusion and consumption in subaqueous sulfide tailings. Geochim. Cosmochim. Acta 65, 1897–1905. doi: 10.1016/S0016-7037(01)00574-9
Foucher, S., Battaglia-Brunet, F., Ignatiadis, I., and Morin, D. (2001). Treatment by sulfate-reducing bacteria of Chessy acid-mine drainage and metals recovery. Chem. Eng. Sci. 56, 1639–1645. doi: 10.1016/S0009-2509(00)00392-4
Friedrich, C. G., Bardischewsky, F., Rother, D., Quentmeier, A., and Fischer, J. (2005). Prokaryotic sulfur oxidation. Curr. Opin. Microbiol. 8, 253–259. doi: 10.1016/J.MIB.2005.04.005
Friedrich, C. G., Quentmeier, A., Bardischewsky, F., Rother, D., Kraft, R., Kostka, S., et al. (2000). Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J. Bacteriol. 182, 4677–4687. doi: 10.1128/jb.182.17.4677-4687.2000
Friedrich, C. G., Rother, D., Bardischewsky, F., Ouentmeier, A., and Fischer, J. (2001). Oxidation of reduced inorganic sulfur compounds by Bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 67, 2873–2882. doi: 10.1128/AEM.67.7.2873-2882.2001
Frigaard, N. U., and Dahl, C. (2008). Sulfur metabolism in phototrophic sulfur Bacteria. Adv. Microb. Physiol. 54, 103–200. doi: 10.1016/S0065-2911(08)00002-7
Ghosh, W., and Dam, B. (2009). Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol. Rev. 33, 999–1043. doi: 10.1111/J.1574-6976.2009.00187.X
Grabarczyk, D. B., and Berks, B. C. (2017). Intermediates in the sox sulfur oxidation pathway are bound to a sulfane conjugate of the carrier protein SoxYZ. PLoS One 12:173395. doi: 10.1371/JOURNAL.PONE.0173395
Han, Y., and Perner, M. (2015). The globally widespread genus Sulfurimonas: versatile energy metabolisms and adaptations to redox clines. Front. Microbiol. 6:989. doi: 10.3389/fmicb.2015.00989
Handley, K. M., Bartels, D., O’Loughlin, E. J., Williams, K. H., Trimble, W. L., Skinner, K., et al. (2014). The complete genome sequence for putative H2- and S-oxidizer Candidatus Sulfuricurvum sp., assembled de novo from an aquifer-derived metagenome. Environ. Microbiol. 16, 3443–3462. doi: 10.1111/1462-2920.12453
Haosagul, S., Prommeenate, P., Hobbs, G., and Pisutpaisal, N. (2020). Sulfide-oxidizing bacteria community in full-scale bioscrubber treating H2S in biogas from swine anaerobic digester. Renew. Energy 150, 973–980. doi: 10.1016/J.RENENE.2019.11.139
Hong, S., Bunge, J., Leslin, C., Jeon, S., and Epstein, S. S. (2009). Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J. 3, 1365–1373. doi: 10.1038/ISMEJ.2009.89
Houghton, J. L., Foustoukos, D. I., Flynn, T. M., Vetriani, C., Bradley, A. S., and Fike, D. A. (2016). Thiosulfate oxidation by Thiomicrospira thermophila: metabolic flexibility in response to ambient geochemistry. Environ. Microbiol. 18, 3057–3072. doi: 10.1111/1462-2920.13232
Hutt, L. P., Huntemann, M., Clum, A., Pillay, M., Palaniappan, K., Varghese, N., et al. (2017). Permanent draft genome of Thiobacillus thioparus DSM 505T, an obligately chemolithoautotrophic member of the Betaproteobacteria. Stand. Genomic Sci. 12, 1–8. doi: 10.1186/s40793-017-0229-3
Hyatt, D., Chen, G. L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformat. 11:119. doi: 10.1186/1471-2105-11-119
Johnson, D. B., and Hallberg, K. B. (2003). The microbiology of acidic mine waters. Res. Microbiol. 154, 466–473. doi: 10.1016/S0923-2508(03)00114-1
Kadnikov, V. V., Mardanov, A. V., Beletsky, A. V., Antsiferov, D. V., Kovalyova, A. A., Karnachuk, O. V., et al. (2019). Sulfur-oxidizing Bacteria dominate in the water from a flooded coal mine shaft in Kuzbass. Microbiology 88, 120–123. doi: 10.1134/S0026261719010107
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y., and Morishima, K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361. doi: 10.1093/NAR/GKW1092
Kanehisa, M., and Sato, Y. (2020). KEGG mapper for inferring cellular functions from protein sequences. Prot. Sci. 29, 28–35. doi: 10.1002/PRO.3711
Kang, D. D., Froula, J., Egan, R., and Wang, Z. (2015). MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015:e1165. doi: 10.7717/peerj.1165
Kelly, D. P., Shergill, J. K., Lu, W. P., and Wood, A. P. (1997). Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Van Leeuwenhoek 71, 95–107. doi: 10.1023/A:1000135707181
Kembel, S. W., Wu, M., Eisen, J. A., and Green, J. L. (2012). Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLoS Comput. Biol. 8:e1002743. doi: 10.1371/JOURNAL.PCBI.1002743
Kinnunen, P., Kyllönen, H., Kaartinen, T., Mäkinen, J., Heikkinen, J., and Miettinen, V. (2018). Sulphate removal from mine water with chemical, biological and membrane technologies. Water Sci. Technol. 2017, 194–205. doi: 10.2166/WST.2018.102
Klatt, J. M., and Polerecky, L. (2015). Assessment of the stoichiometry and efficiency of CO2 fixation coupled to reduced sulfur oxidation. Front. Microbiol. 6:484. doi: 10.3389/fmicb.2015.00484
Koch, T., and Dahl, C. (2018). A novel bacterial sulfur oxidation pathway provides a new link between the cycles of organic and inorganic sulfur compounds. ISME J. 12, 2479–2491. doi: 10.1038/s41396-018-0209-7
Korehi, H., Blöthe, M., and Schippers, A. (2014). Microbial diversity at the moderate acidic stage in three different sulfidic mine tailings dumps generating acid mine drainage. Res. Microbiol. 165, 713–718. doi: 10.1016/J.RESMIC.2014.08.007
Kuang, J., Huang, L., He, Z., Chen, L., Hua, Z., Jia, P., et al. (2016). Predicting taxonomic and functional structure of microbial communities in acid mine drainage. Int. Soc. Microb. Ecol. J. 10, 1527–1539. doi: 10.1038/ismej.2015.201
Lacelle, D., Doucet, A., Clark, I. D., and Lauriol, B. (2007). Acid drainage generation and seasonal recycling in disturbed permafrost near Eagle Plains, northern Yukon territory Canada. Chem. Geol. 243, 157–177. doi: 10.1016/J.CHEMGEO.2007.05.021
Lahme, S., Callbeck, C. M., Eland, L. E., Wipat, A., Enning, D., Head, I. M., et al. (2020). Comparison of sulfide-oxidizing Sulfurimonas strains reveals a new mode of thiosulfate formation in subsurface environments. Environ. Microbiol. 22, 1784–1800. doi: 10.1111/1462-2920.14894
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/BIOINFORMATICS/BTP352
Liljeqvist, M., Sundkvist, J. E., Saleh, A., and Dopson, M. (2011). Low temperature removal of inorganic sulfur compounds from mining process waters. Biotechnol. Bioeng. 108, 1251–1259. doi: 10.1002/BIT.23057
Lin, K. H., Liao, B. Y., Chang, H. W., Huang, S. W., Chang, T. Y., Yang, C. Y., et al. (2015). Metabolic characteristics of dominant microbes and key rare species from an acidic hot spring in Taiwan revealed by metagenomics. BMC Genom. 16, 1–16. doi: 10.1186/s12864-015-2230-9
Lopes, A. R., Madureira, D., Diaz, A., Santos, S., Vila, M. C., and Nunes, O. C. (2020). Characterisation of bacterial communities from an active mining site and assessment of its potential metal solubilising activity. J. Environ. Chem. Eng. 8:104495. doi: 10.1016/J.JECE.2020.104495
Louca, S., Polz, M. F., Mazel, F., Albright, M. B. N., Huber, J. A., O’Connor, M. I., et al. (2018). Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2, 936–943. doi: 10.1038/s41559-018-0519-1
Loy, A., Duller, S., Baranyi, C., Mußmann, M., Ott, J., Sharon, I., et al. (2009). Reverse dissimilatory sulfite reductase as phylogenetic marker for a subgroup of sulfur-oxidizing prokaryotes. Environ. Microbiol. 11, 289–299. doi: 10.1111/J.1462-2920.2008.01760.X
Makhija, R., and Hitchen, A. (1979). The titrimetric determination of sulphate, thiosulphate and polythionates in mining effluents. Anal. Chim. Acta 105, 375–382. doi: 10.1016/S0003-2670(01)83769-7
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/EJ.17.1.200
Meier, D. V., Pjevac, P., Bach, W., Hourdez, S., Girguis, P. R., Vidoudez, C., et al. (2017). Niche partitioning of diverse sulfur-oxidizing bacteria at hydrothermal vents. ISME J. 11, 1545–1558. doi: 10.1038/ismej.2017.37
Meyer, B., and Kuever, J. (2007). Molecular analysis of the distribution and phylogeny of dissimilatory adenosine-5′-phosphosulfate reductase-encoding genes (aprBA) among sulfur-oxidizing prokaryotes. Microbiology 153, 3478–3498. doi: 10.1099/MIC.0.2007/008250-0
Meziti, A., Nikouli, E., Hatt, J. K., Konstantinidis, K. T., and Kormas, K. A. (2021). Time series metagenomic sampling of the Thermopyles, Greece, geothermal springs reveals stable microbial communities dominated by novel sulfur-oxidizing chemoautotrophs. Environ. Microbiol. 23, 3710–3726. doi: 10.1111/1462-2920.15373
Miettinen, H., Bomberg, M., Le, T. M. K., and Kinnunen, P. (2021). Identification and metabolism of naturally prevailing microorganisms in zinc and copper mineral processing. Fortschr. Mineral. 11:30. doi: 10.3390/MIN11020156
Miranda-Trevino, J. C., Pappoe, M., Hawboldt, K., and Bottaro, C. (2013). The importance of Thiosalts speciation: review of analytical methods, kinetics, and treatment. Crit. Rev. Environ. Sci. Technol. 43, 2013–2070. doi: 10.1080/10643389.2012.672047
Neukirchen, S., and Sousa, F. L. (2021). DiSCo: a sequence-based type-specific predictor of Dsr-dependent dissimilatory Sulphur metabolism in microbial data. Microbial Genom. 7:603. doi: 10.1099/MGEN.0.000603
Nguyen, P. M., Do, P. T., Pham, Y. B., Doan, T. O., Nguyen, X. C., Lee, W. K., et al. (2022). Roles, mechanism of action, and potential applications of sulfur-oxidizing bacteria for environmental bioremediation. Sci. Total Environ. 852:158203. doi: 10.1016/J.SCITOTENV.2022.158203
Nosalova, L., Piknova, M., Kolesarova, M., and Pristas, P. (2023). Cold Sulfur Springs—neglected niche for autotrophic sulfur-oxidizing Bacteria. Microorganisms 11:1436. doi: 10.3390/MICROORGANISMS11061436
Nurk, S., Meleshko, D., Korobeynikov, A., and Pevzner, P. A. (2017). MetaSPAdes: a new versatile metagenomic assembler. Genome Res. 27, 824–834. doi: 10.1101/gr.213959.116
O’Brien, D. J., and Birkner, F. B. (1977). Kinetics of oxygenation of reduced sulfur species in aqueous solution. Environ. Sci. Technol. 11, 1114–1120. doi: 10.1021/es60135a009
Patwardhan, S., Foustoukos, D. I., Giovannelli, D., Yücel, M., and Vetriani, C. (2018). Ecological succession of sulfur-oxidizing epsilon- and gammaproteobacteria during colonization of a shallow-water gas vent. Front. Microbiol. 9:2970. doi: 10.3389/fmicb.2018.02970
Peng, Y., Leung, H. C. M., Yiu, S. M., and Chin, F. Y. L. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428. doi: 10.1093/BIOINFORMATICS/BTS174
Philippot, P., Van Zuilen, M., Lepot, K., Thomazo, C., Farquhar, J., and Van Kranendonk, M. J. (2007). Early archaean microorganisms preferred elemental sulfur, not sulfate. Science 317, 1534–1537. doi: 10.1126/science.1145861
Pott, A. S., and Dahl, C. (1998). Sirohaem sulfite reductase and other proteins encoded by genes at the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology 144, 1881–1894. doi: 10.1099/00221287-144-7-1881
Purcell, A. M., Mikucki, J. A., Achberger, A. M., Alekhina, I. A., Barbante, C., Christner, B. C., et al. (2014). Microbial sulfur transformations in sediments from subglacial Lake Whillans. Front. Microbiol. 5:594. doi: 10.3389/FMICB.2014.00594
Quentmeier, A., Kraft, R., Kostka, S., Klockenkämper, R., and Friedrich, C. G. (2000). Characterization of a new type of sulfite dehydrogenase from Paracoccus pantotrophus GB17. Arch. Microbiol. 173, 117–125. doi: 10.1007/s002039900118
Reigstad, L. J., Jorgensen, S. L., Lauritzen, S. E., Schleper, C., and Urich, T. (2011). Sulfur-oxidizing chemolithotrophic proteobacteria dominate the microbiota in high arctic thermal springs on Svalbard. Astrobiology 11, 665–678. doi: 10.1089/AST.2010.0551
Rethmeier, J., Rabenstein, A., Langer, M., and Fischer, U. (1997). Detection of traces of oxidized and reduced sulfur compounds in small samples by combination of different high-performance liquid chromatography methods. J. Chromatogr. A 760, 295–302. doi: 10.1016/S0021-9673(96)00809-6
Sato, I., Shimatani, K., Fujita, K., Abe, T., Shimizu, M., Fujii, T., et al. (2011). Glutathione reductase/glutathione is responsible for cytotoxic elemental sulfur tolerance via polysulfide shuttle in fungi. J. Biol. Chem. 286, 20283–20291. doi: 10.1074/jbc.M111.225979
Seth, N., Vats, S., Lakhanpaul, S., Arafat, Y., Mazumdar-Leighton, S., Bansal, M., et al. (2024). Microbial community diversity of an integrated constructed wetland used for treatment of sewage. Front. Microbiol. 15:1355718. doi: 10.3389/FMICB.2024.1355718
Silva, A. M., Lima, R. M. F., and Leão, V. A. (2012). Mine water treatment with limestone for sulfate removal. J. Hazard. Mater. 221–222, 45–55. doi: 10.1016/J.JHAZMAT.2012.03.066
Sorokin, D. Y., and Kuenen, J. G. (2005). Haloalkaliphilic sulfur-oxidizing bacteria in soda lakes. FEMS Microbiol. Rev. 29, 685–702. doi: 10.1016/J.FEMSRE.2004.10.005
Sorokin, D. Y., Lysenko, A. M., Mityushina, L. L., Tourova, T. P., Jones, B. E., Rainey, F. A., et al. (2001). Thioalkalimicrobium aerophilum gen. Nov., sp. nov. and Thioalkalimicrobium sibericum sp. nov., and Thioalkalivibrio versutus gen. Nov., sp. nov., Thioalkalivibrio nitratis sp. nov. and Thioalkalivibrio denitrificans sp. nov., novel obligately alkaliphilic and obligately chemolithoautotrophic sulfur-oxidizing bacteria from soda lakes. Int. J. Syst. Evol. Microbiol. 51, 565–580. doi: 10.1099/00207713-51-2-565
Suzek, B. E., Huang, H., McGarvey, P., Mazumder, R., and Wu, C. H. (2007). UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23, 1282–1288. doi: 10.1093/BIOINFORMATICS/BTM098
Suzuki, M. T., and Giovannoni, S. J. (1996). Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62, 625–630. doi: 10.1128/AEM.62.2.625-630.1996
Thamdrup, B., Fossing, H., and Jørgensen, B. B. (1994). Manganese, iron and sulfur cycling in a coastal marine sediment, Aarhus bay Denmark. Geochim. Cosmochim. Acta 58, 5115–5129. doi: 10.1016/0016-7037(94)90298-4
van Vliet, D. M., von Meijenfeldt, F. A. B., Dutilh, B. E., Villanueva, L., Sinninghe Damsté, J. S., Stams, A. J. M., et al. (2021). The bacterial sulfur cycle in expanding dysoxic and euxinic marine waters. Environ. Microbiol. 23, 2834–2857. doi: 10.1111/1462-2920.15265
Vavourakis, C. D., Mehrshad, M., Balkema, C., Van Hall, R., Andrei, A. Ş., Ghai, R., et al. (2019). Metagenomes and metatranscriptomes shed new light on the microbial-mediated sulfur cycle in a Siberian soda lake. BMC Biol. 17, 1–20. doi: 10.1186/s12915-019-0688-7
Veith, A., Botelho, H. M., Kindinger, F., Gomes, C. M., and Kletzin, A. (2012). The sulfur oxygenase reductase from the mesophilic bacterium Halothiobacillus neapolitanus is a highly active Thermozyme. J. Bacteriol. 194:677. doi: 10.1128/JB.06531-11
Vincent, S. G. T., Jennerjahn, T., and Ramasamy, K. (2021). “Biogeocycling of nutrients (C, N, P, S, and Fe) and implications on greenhouse gas emissions” in Microbial communities in coastal sediments. eds. S. G. T. Vincent, T. C. Jennerjahn, and K. Ramasamy (Amsterdam: Elsevier), 119–145.
Wang, S., Jiang, L., Hu, Q., Cui, L., Zhu, B., Fu, X., et al. (2021). Characterization of Sulfurimonas hydrogeniphila sp. nov., a novel bacterium predominant in Deep-Sea hydrothermal vents and comparative genomic analyses of the genus Sulfurimonas. Front. Microbiol. 12:626705. doi: 10.3389/fmicb.2021.626705
Wang, S., Jiang, L., Xie, S., Alain, K., Wang, Z., Wang, J., et al. (2023). Disproportionation of inorganic sulfur compounds by mesophilic Chemolithoautotrophic Campylobacterota. MSystems 8:22. doi: 10.1128/MSYSTEMS.00954-22
Wang, Z. B., Li, Y. Q., Lin, J. Q., Pang, X., Liu, X. M., Liu, B. Q., et al. (2016). The two-component system RsrS-RsrR regulates the tetrathionate intermediate pathway for thiosulfate oxidation in Acidithiobacillus caldus. Front. Microbiol. 7:1755. doi: 10.3389/fmicb.2016.01755
Wang, R., Lin, J. Q., Liu, X. M., Pang, X., Zhang, C. J., Yang, C. L., et al. (2019). Sulfur oxidation in the acidophilic autotrophic Acidithiobacillus spp. Front. Microbiol. 9:3290. doi: 10.3389/fmicb.2018.03290
Watanabe, T., Kojima, H., and Fukui, M. (2014). Complete genomes of freshwater sulfur oxidizers Sulfuricella denitrificans skB26 and Sulfuritalea hydrogenivorans sk43H: genetic insights into the sulfur oxidation pathway of betaproteobacteria. Syst. Appl. Microbiol. 37, 387–395. doi: 10.1016/J.SYAPM.2014.05.010
Watanabe, T., Kojima, H., Umezawa, K., Hori, C., Takasuka, T. E., Kato, Y., et al. (2019). Genomes of neutrophilic sulfur-oxidizing chemolithoautotrophs representing 9 proteobacterial species from 8 genera. Front. Microbiol. 10:316. doi: 10.3389/fmicb.2019.00316
Whaley-Martin, K. J., Chen, L.-X., Nelson, T. C., Gordon, J., Kantor, R., Twible, L. E., et al. (2023). O2 partitioning of sulfur oxidizing bacteria drives acidity and thiosulfate distributions in mining waters. Nat. Commun. 14:2006. doi: 10.1038/s41467-023-37426-8
Whaley-Martin, K., Jessen, G. L., Nelson, T. C., Mori, J. F., Apte, S., Jarolimek, C., et al. (2019). The potential role of Halothiobacillus spp. in sulfur oxidation and acid generation in circum-neutral mine tailings reservoirs. Front. Microbiol. 10:297. doi: 10.3389/FMICB.2019.00297/BIBTEX
Whaley-Martin, K., Marshall, S., Nelson, T. E. C., Twible, L., Jarolimek, C. V., King, J. J., et al. (2020). A mass-balance tool for monitoring potential dissolved sulfur oxidation risks in mining impacted waters. Mine Water Environ. 39, 291–307. doi: 10.1007/s10230-020-00671-0
Xin, Y., Wang, Y., Zhang, H., Wu, Y., Xia, Y., Li, H., et al. (2023). Cupriavidus pinatubonensis JMP134 alleviates Sulfane sulfur toxicity after the loss of Sulfane dehydrogenase through oxidation by Persulfide dioxygenase and hydrogen sulfide release. Meta 13:218. doi: 10.3390/METABO13020218/S1
Yan, Y., Colenbrander Nelson, T. E., Twible, L., Whaley-Martin, K., Jarolimek, C. V., King, J. J., et al. (2022). Sulfur mass balance and speciation in the water cap during early-stage development in the first pilot pit lake in the Alberta Oil Sands. Environ. Chem. 19, 236–253. doi: 10.1071/EN22057
Yu, X., Li, Y., Wu, Y., Gao, H., Liu, W., Liu, H., et al. (2024). Seasonal changes of prokaryotic microbial community structure in Zhangjiayan reservoir and its response to environmental factors. Sci. Rep. 14:5513. doi: 10.1038/s41598-024-55702-5
Zander, U., Faust, A., Klink, B. U., De Sanctis, D., Panjikar, S., Quentmeier, A., et al. (2011). Structural basis for the oxidation of protein-bound sulfur by the sulfur cycle molybdohemo-enzyme sulfane dehydrogenase SoxCD. J. Biol. Chem. 286, 8349–8360. doi: 10.1074/jbc.M110.193631
Keywords: sulfur oxidizing bacteria (SOB), pH, tailings impoundments, sox genes, thiosulfate
Citation: Twible LE, Whaley-Martin K, Chen L-X, Colenbrander Nelson T, Arrey JLS, Jarolimek CV, King JJ, Ramilo L, Sonnenberg H, Banfield JF, Apte SC and Warren LA (2024) pH and thiosulfate dependent microbial sulfur oxidation strategies across diverse environments. Front. Microbiol. 15:1426584. doi: 10.3389/fmicb.2024.1426584
Edited by:
Gordon T. Taylor, Stony Brook University, United StatesReviewed by:
Costantino Vetriani, Rutgers, The State University of New Jersey, United StatesShun Li, Shandong University, China
Copyright © 2024 Twible, Whaley-Martin, Chen, Colenbrander Nelson, Arrey, Jarolimek, King, Ramilo, Sonnenberg, Banfield, Apte and Warren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lesley A. Warren, bGVzbGV5LndhcnJlbkB1dG9yb250by5jYQ==
 Lauren E. Twible
Lauren E. Twible Kelly Whaley-Martin
Kelly Whaley-Martin Lin-Xing Chen
Lin-Xing Chen Tara Colenbrander Nelson
Tara Colenbrander Nelson James L.S. Arrey
James L.S. Arrey Chad V. Jarolimek3
Chad V. Jarolimek3 Lisa Ramilo
Lisa Ramilo Helga Sonnenberg
Helga Sonnenberg Jillian F. Banfield
Jillian F. Banfield Simon C. Apte
Simon C. Apte Lesley A. Warren
Lesley A. Warren