
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 June 2024
Sec. Virology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1424489
The expansion of betel palm cultivation is driven by rising demand for betel nut, yet this growth is accompanied by challenges such as decreased agricultural biodiversity and the spread of infectious pathogens. Among these, Yellow Leaf Disease (YLD) emerges as a prominent threat to betel palm plantation. Areca Palm Velarivirus 1 (APV1) has been identified as a primary causative agent of YLD, precipitating leaf yellowing, stunted growth, and diminished yield. However, the precise mechanisms underlying APV1-induced damage remain elusive. Our study elucidates that APV1 infiltrates chloroplasts, instigating severe damage and consequential reductions in chlorophyll a/b and carotene levels, alongside notable declines in photosynthetic efficiency. Moreover, APV1 infection exerts broad regulatory effects on gene expression, particularly suppressing key genes implicated in chloroplast function and photosynthesis. These disruptions correlate with growth retardation, yield diminishment, and compromised nut quality. Intriguingly, the paradoxical destruction of the host's photosynthetic machinery by APV1 prompts inquiry into its evolutionary rationale, given the virus's dependence on host resources for replication and proliferation. Our findings reveal that APV1-induced leaf yellowing acts as a beacon for transmission vectors, hinting at a nuanced “host-pathogen-vector co-evolutionary” dynamic.
The betel palm (Areca catechu L.) is one of the most economically significant crops cultivated in Southeast Asia, with its commercial value being highest in China and India due to its widespread popularity as a chewable product (Khan et al., 2023). In 2020, the total output value of betel nut fruit in Hainan province, China, reached 14.68 billion yuan (2 billion USD), becoming a vital source of income for 2.2 million farmers, accounting for ca. 40% of the agricultural population in Hainan (Khan et al., 2022). Presently, there are 41,800 enterprises related to betel nut processing, employing nearly 300,000 people. The annual output value of China's betel nut industry has reached 60 billion yuan (8.3 billion USD). The increasing betel nut consumption has drew global attention to the health issue of this additive substance (Chen et al., 2023). Additionally, the increasing demand for betel nuts has driven the continuous expansion of betel palm cultivation. Widespread monoculture has led to a decrease in agricultural biodiversity, resulting in an increasing threat from insects and diverse phytopathogens. Among these threats, Yellowing Leaf Disease (YLD) stands out as the most devastating (Johnson et al., 2015; Wang et al., 2020; Khan et al., 2022, 2023). As the name suggests, leaf chlorosis is the most prominent symptom, and in the later stages, the palm's crown size decreases, and “bunchy top” symptoms appear, causing a sharp decline in the yield and quality of betel nut (Yu et al., 2015; Cao et al., 2021; Zhang et al., 2022).
Although YLD has long been associated with phytoplasma (Nayar and Seliskar, 1978; Kanatiwela-de Silva et al., 2015), the causal agent of phytoplasma remains controversial due to the lack of convincing evidence (Purushothama et al., 2007). In recent years, Areca palm velarivirus 1 (APV1) was identified in YLD samples through RNA sequencing (Yu et al., 2015; Cao et al., 2021; Zhang et al., 2022). Field investigations have shown a strong association between APV1 and YLD (Wang et al., 2020). Furthermore, APV1 can be transmitted by both Pseudococcus cryptus and Ferrisia virgata, causing typical YLD symptoms in betel palm seedlings, suggesting that APV1 might be the causal agent of YLD (Zhang et al., 2022; Zhao et al., 2024). APV1 has been identified as a member of the genus Velarivirus in the family Closteroviridae according to phylogenetic analysis and genome structure (Yu et al., 2015; Wang et al., 2020).
Viruses in the Closteroviridae family have non-enveloped filamentous particles ranging from 650 to 2200 nm in length and 12 nm in diameter, with mono-, bi-, or tripartite positive-sense RNA genomes ranging from 13 to 19 kb (Karasev, 2000; Martelli, 2019). According to the 2020 ICTV virus taxonomy profile, the Closteroviridae family consists of four genera: Closterovirus, Ampelovirus, Crinivirus, and Velarivirus (Martelli, 2019; Fuchs et al., 2020). Recently, three new genera (Bluvavirus, Menthavirus, and Olivavirus) were added to the Closteroviridae family (https://ictv.global/taxonomy). As the names of these viruses suggest, many species of closterovirus, such as Carrot yellow leaf virus, Wheat yellow leaf virus, Cucurbit yellow stunting disorder virus (Nagendran et al., 2023), and Sweet potato chlorotic stunt virus (Zhao et al., 2022), as well as Tomato chlorosis virus (Liu et al., 2022), cause yellowing symptoms. However, there is limited articles published about the interactions between closterovirus and chloroplasts, and further investigation is needed to understand the mechanism underlying the leaf yellowing symptom.
Damage to the chloroplast and modifications to photosynthesis are common and conserved strategies employed by plant viruses to establish an optimal niche for infection (Zhao et al., 2016). Understanding the interactions between chloroplasts and viruses is crucial for unraveling the mode of infection and viral pathogenicity (Bhattacharyya and Chakraborty, 2018). APV1 infection causes leaf yellowing, growth retardation, yield loss, decreased nut quality and even tree death. However, the precise mechanisms underlying remain elusive, the aim of the work is to investigate the APV1-induced damage of ultrastructure and related cytopathology. The data presented in this work indicate that APV1 infection induces chloroplast disassembly, degradation of chlorophyll and carotene, and decreased photosynthesis efficiency, which may contribute to the loss of yield and decreased quality of betel nut products.
Mealybugs (F. virgata) were originally isolated from betel palm and were reared on pumpkin fruit in nylon net cages (75 × 75 × 75 cm). One-year old betel palm seedlings and mealybugs were cultivated in chamber as previous report (Zhang et al., 2022). First instars of F. virgata were transferred from pumpkin fruit to APV1 infected plants for 48-h acquisition access period (AAP) then further transferred onto test seedlings for 7 days inoculation access period (IAP). The seedlings were sprayed with insecticide to kill the mealybugs after inoculation. A mock inoculation was performed using non-viruliferous mealybugs. The inoculated seedlings were detected by RT-PCR using APV1 specific primers 2 months after inoculation.
Three groups of betel palm seedlings were chosen for host choosing experiment, including APV1 positive seedlings with yellowing symptom (APV1+ yellow), APV1 positive seedling without yellowing symptom (APV1+ green), and the healthy seedlings (healthy control). After 4 h starvation, 100 adult F. virgate mealybugs were released from the cage laying between the two types of A. catechu seedlings, the number of mealybugs freely distributed onto each type of seedlings was calculated at 6 h after releasing, respectively.
Leaves from APV1 infected plants and control plants were collected and ground in liquid nitrogen for Western-blot analysis. Total proteins were isolated and separated by SDS-PAGE. The protein was transferred from gel onto PVDF membrane using a Mini Trans-Blot Electrophoretic Transfer system (Bio-Rad, #1703930). APV1 was detected using a mouse anti-APV1 CP monoclonal antibody (1:5,000 dilution) and horseradish peroxidase-conjugated goat anti-mouse IgG (1:2,000 dilution) as the secondary antibody (Solarbio, catalog no. SE131). The blot was visualized using a chemiluminescence film (Thermo Fisher, catalog no. 34577).
Leaf samples from the APV1-inoculated and mock-inoculated betel palm seedlings were collected for RNA-Seq and de novo assembly were conducted as previously described (Wang et al., 2020). All differential expressed genes (DEGs) were subjected to gene ontology (GO) and KEGG enrichment analysis using GOseq and KOBAS software, respectively (Li et al., 2020). qRT-PCR was carried out as previously described (Livak and Schmittgen, 2001; Zhang et al., 2022). Briefly, total RNA was purified from leave samples using a plant RNA extraction kit (Tiangen Biotech, Beijing, China), and cDNA was synthesized with a one-step reverse transcription kit (Thermo Fisher Scientific, Waltham, MA, USA). Actin gene was chosen for the normalization. The primer sequences used in this experiment are listed in Supplementary Table S1.
The penultimate fully unfolded leaves of YLD betel palm and healthy betel palm were collected, and 0.2 g of each leaf sample was cut into small pieces. These pieces were then homogenized in 25 ml of 95% ethanol and kept in darkness for 24 h. The supernatants were analyzed at 470, 649, and 665 nm, respectively. Chlorophyll was calculated according to Arnon's equation. The grand means of the values were subjected to analysis of variance (ANOVA) by SPSS version 19.0 (Li et al., 2020).
The penultimate fully expanded leaves of the YLD betel palm and healthy betel palm were measured using a portable open gas exchange system with a 3 × 3 cm leaf chamber (LI6800-12A, Li-COR Inc., Lincoln, NE, USA). Measurements were conducted between 9:00 and 11:00 in the morning on clear days with natural fluctuations in air temperature and vapor pressure deficit. The temperature in the climate chamber was set at 28°C and 55% relative humidity. The CO2 level was ~400 μmol mol−1. Each sample was measured five times (Tseliou et al., 2021).
Chlorophyll fluorescence of the YLD betel palm and healthy betel palm were analyzed using a MINI-PAM-II fluorometer (Walz, Germany). The analyses were repeated three times. Induction curves were performed using a MINI-PAM-II fluorometer following the manufacturer's protocol. The fluorescence parameters of the light response curve and induction curve were converted using WinControl-3 software (Walz, Germany), and the graphs were drawn using Excel software (Ivanov and Bernards, 2016).
Transmission electron microscopy (TEM) was conducted as previously described (Jin et al., 2018). Ultrathin sections were cut to 70–90 nm thickness using a Leica EM UC7 ultramicrotome. The sections were sequentially stained with uranyl acetate for 20 min and Reynolds' lead citrate for 5 min. The sections were then viewed with a Hitachi H-7650 or a JEM-1230 transmission electron microscope operated at 80 kV.
Immune Electron Microscopy (IEM) was conducted as previously report (Folimonova et al., 2008). The leaf samples were fixed using phosphate buffer containing 1.8% paraformaldehyde and 0.25% glutaraldehyde, and imbedded in LR White resin (London Resin Company, Hampshire, United Kingdom). Immunogold labeling of ultrathin sections (70- to 80-nm) was performed using a purified mouse monoclonal antibody against APV1-CP (concentration: 2 mg/ml, diluted 1:500) as the primary antibody, followed by incubation with goat anti-mouse 12-nm gold conjugate (diluted 1:25; Sigma). The specimens were meticulously observed using a HT7800/HT7700 transmission electron microscope (HITACHI, Japan).
To investigate the mechanism underlying the yellowing symptoms induced by APV1 in betel palm, Betel palm seedlings were inoculated with through APV1 through F. virgate mealybugs. At 60 days post inoculation (IS-1), leaf yellowing symptoms initially appeared at the leaflet tips. Over time, the yellowing gradually extended toward the petiole, resulting in most of the leaves turning yellow, while the midribs remained green, forming a distinct green-yellow border at 180 days post-inoculation (IS-2) (Figure 1A). This observation aligns with previous reports (Wang et al., 2020; Zhang et al., 2022). In addition to leaf yellowing, APV1-infected seedlings exhibited significant growth retardation compared to the control (Figure 1B; Supplementary Figure S1). Western blot analysis using a monoclonal antibody against APV1-CP revealed the accumulation of APV1 in the inoculated betel palm seedlings (Figure 1C). Measurement results indicated a significant decrease in photosynthesis-related pigments. The contents of chlorophyll a, chlorophyll b, and carotenoids, decreased from 0.693, 0.374, and 0.128 mg/g in control sample to 0.318, 0.209, and 0.067 mg/g in APV1-infected samples, respectively (Figure 1D).
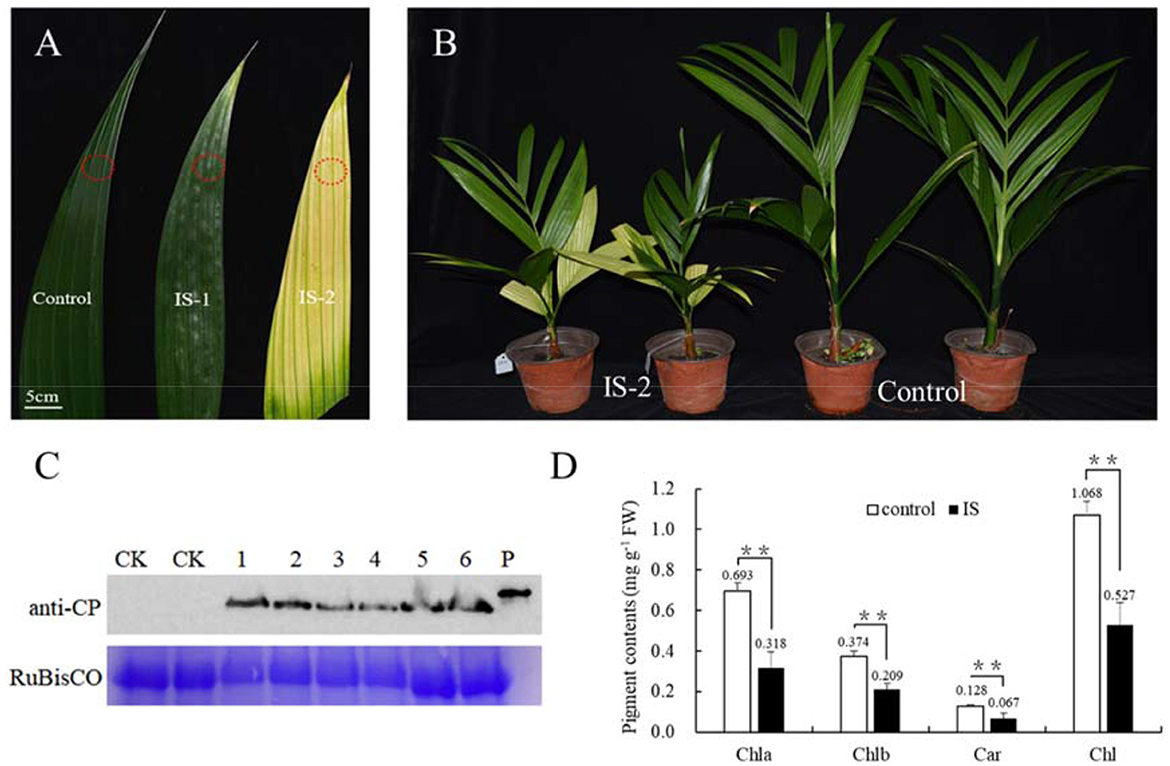
Figure 1. APV1 infection caused yellowing symptom and decreased levels of pigments. (A) Leaf yellowing symptoms in infected sample at 60 days (IS-1) and 180 days post inoculation (IS-2) after APV1 inoculation by F. virgate. Red rings indicated the positions of collected samples for tissue sections. (B) Seedlings symptoms of areca palm at 180 dpi (left) and mock control (right); (C) Western-blot indicated the APV1 accumulation after APV1 inoculation using monoclonal antibody against APV1-CP. 1-3: three samples of IS-1, 4-6: three samples of IS-2, CK: mock inoculation, P: His-CP fusion protein isolated from prokaryotic expression in E. coli; (D) Pigment content in APV1-infected sample (IS-2) or healthy (control) A. catechu seedlings. Chla, chlorophyll a; Chlb, chlorophyll b; Car, carotenoid; Chl, chlorophyll a + chlorophyll b. Data are the averages from three independent experiments. The small bars represent standard deviation. Asterisks indicate significant differences between IS and control (**P < 0.01).
Chlorophyll fluorescence can serve as an indicator of plant photosynthesis efficiency and can be used to detect early-stage biotic and abiotic stresses (Moustaka and Moustakas, 2023). Measurement results showed that four crucial indicators of chlorophyll fluorescence, e.g., photosynthetic electron transport rate (ETR), actual photochemical efficiency in photosystem II (YII), photochemical quenching (qP), and non-photochemical quenching (NPQ), were significantly lower in APV1-infected samples (IS-2) than that in the mock control (Figure 2). Furthermore, under conditions of saturated light intensity (1,000 μmol m−2 s−1), the net photosynthetic rate (Pn) and stomatal conductance (Gs) were significantly lower in APV1-infected samples (IS-2), while the intercellular CO2 concentration (Ci) was higher than that of the control (Figure 3). A higher Ci value indicates lower photosynthesis efficiency (Tominaga et al., 2018). In summary, APV1 infection reduced chlorophyll pigmentation, resulted in fluctuation of chlorophyll fluorescence, and led to decreased photosystem efficiency.
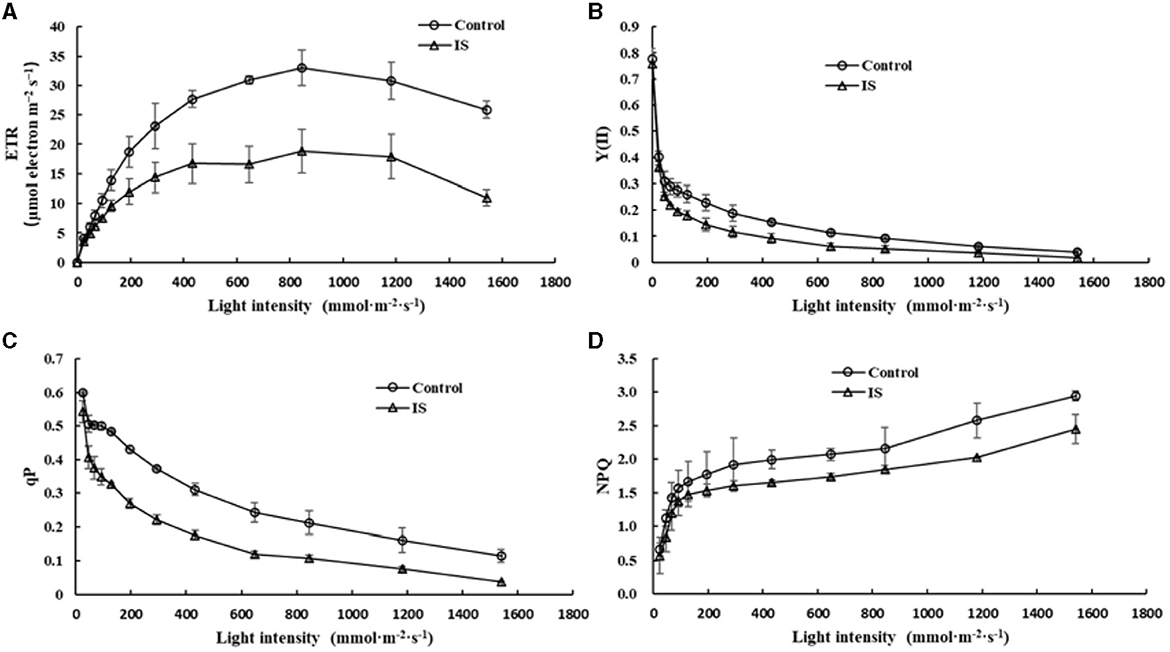
Figure 2. Photosynthesis efficiency affected by APV1 infection. Change of photosynthetic electron transport rate (ETR, A), actual photochemical efficiency in photosystem II (YII, B), photochemical quenching (qP, C), and non-photochemical quenching (NPQ, D) were recorded in APV1-infected (IS-2) sample or healthy (control) of areca palm. Data are the averages from three independent experiments.
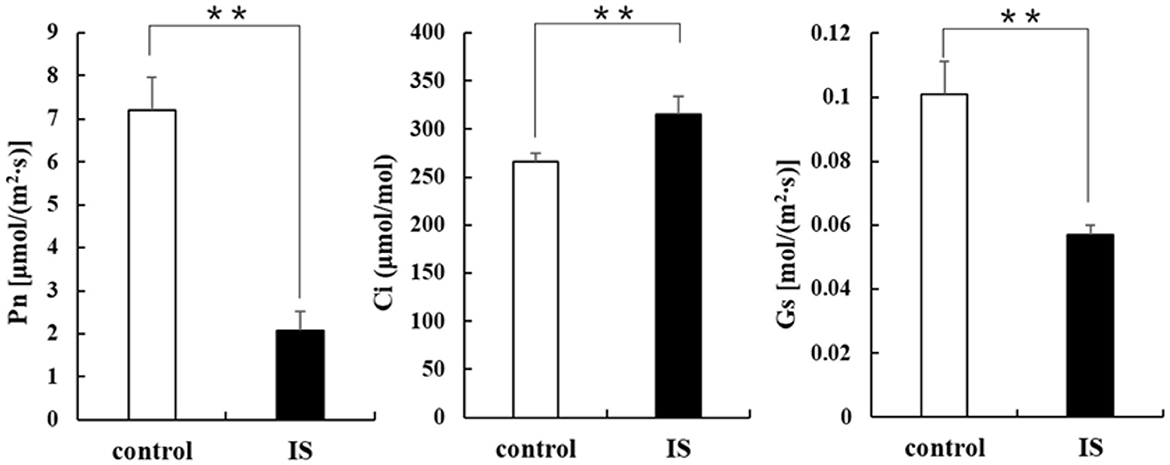
Figure 3. Photosynthesis related parameters affected by APV1 infection. Net photosynthetic rate (Pn), stomatal conductance (Gs), and intercellular carbon dioxide concentration (Ci) were determined in APV1 infected sample (IS-2) of A. catechu seedling and mock inoculation (control). Data are the averages from three independent experiments. The small bars represent standard deviation. Asterisks indicate significant differences between IS and control with (**P < 0.01).
To investigate the causes of yellowing symptoms, reduced chlorophyll, and decreased photosynthesis efficiency induced by APV1, leaves from APV1-infected and healthy control samples were collected for tissue sectioning and transmission electron microscopy (TEM) observation. In healthy control cells, chloroplasts were oval in shape and evenly distributed, with starch grains (SG) surrounded by grana lamellae (GL). In the early stages of virus infection (IS-1), the chloroplast membrane separated from the cell wall (CW), and the size and number of osmiophilic granules (OG) reduced. The GL became less organized, and SG were separated from each other, with many unknown particles and white holes appearing in the cytoplasm. In the late stage (IS-2), chloroplasts were nearly completely degraded, with a sharp reduction in size, and large starch grains (SG) accumulating in the cytoplasm (Figure 4). These findings confirm that APV1 infection severely damages the chloroplast structure and other cytoplasmic organelles in leaf cells. In this work, immune electron microscopy (IEM) was used to examine the distribution of APV1 in leaf. IEM revealed that APV1 mainly localized in chloroplast and occasionally in cytoplasm, confirming that APV1 has invaded chloroplast and caused degradation of chloroplast (Figure 5).

Figure 4. Leaf tissue sections of areca palm were observed under transmission electron microscopy (TEM). Mock control, infected leaf sample at 60 days (IS-1) and 180 days (IS-2) after APV1 inoculation were collected for tissue sections and observed under transmission electron microscopy (TEM). Sampling position for the tissue sections were indicated with red ring in Figure 1A. CW, cell wall; Chl, chloroplast; OG, osmiophilic granule; SG, starch grains; GL, grana lamella are indicated.
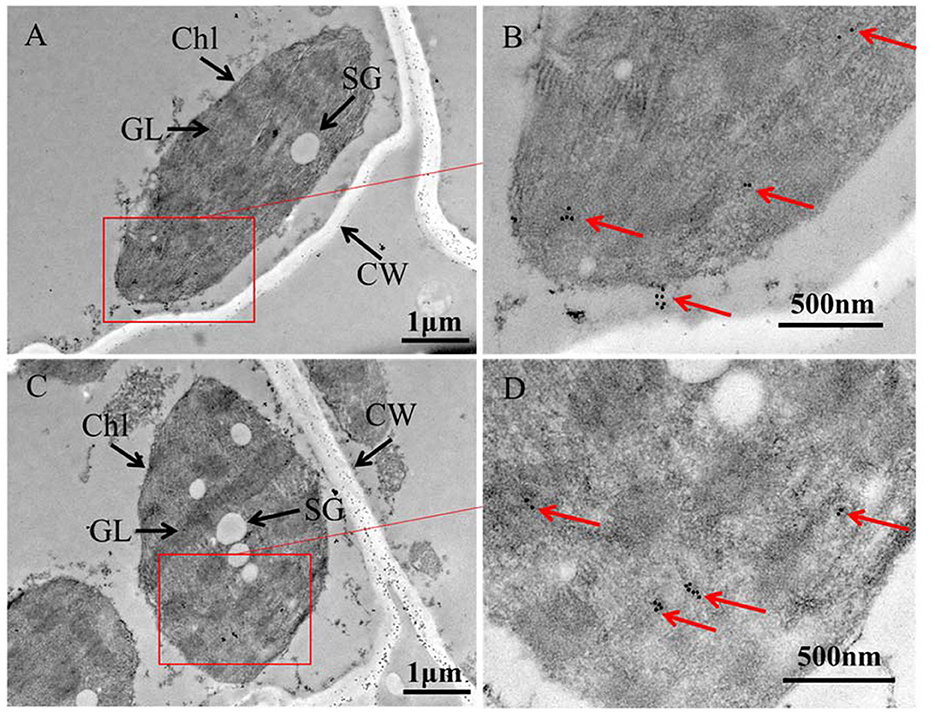
Figure 5. Immune Electron Microscopy (IEM) showing APV1 in chloroplasts of A. catechu leave. (A, C) Groups of chloroplasts at lower magnification. (B, D) Areas from the cells shown in panel at higher magnification. APV1-specific monoclonal antibody was used as primary antibodies and secondary antibodies conjugated with gold particles. CW, cell wall; Chl, chloroplast; SG, starch grains; GL, grana lamella are indicated.
To identify genes regulated by APV1 infection, we conducted RNA-seq and analyzed differentially expressed genes (DEGs) between APV1-infected samples and healthy controls. A total of 166,590 unigenes were obtained through de novo assembly from the clean RNA-seq reads. DEG analysis revealed 12,553 up-regulated DEGs and 5,698 down-regulated DEGs in response to APV1 infection (Supplementary Figure S2). Gene ontology (GO) annotation enrichment showed that DEGs were distributed across 45 GO terms, with the most DEGs associated with terms such as Binding, Protein binding, and Intracellular organelle (Supplementary Figure S3). Notably, key genes involved in chlorophyll biosynthesis, such as chlorophyll synthase (CS) and chlorophyll b reductase (CbR), as well as genes related to carotene metabolism, such as zeta-carotene desaturase (zCD) and phytoene synthase (PS), were down-regulated in APV1-infected samples (Supplementary Figure S4). This was further confirmed by qRT-PCR, with the expression of zCD, PS, CS and CbR significantly down-regulated by APV1 infection (Figure 6). These findings suggest that APV1 infection has a global impact on gene expression patterns, suppressing the expression of key genes involved in chloroplast and photosynthesis, which were nuclear-encoded.
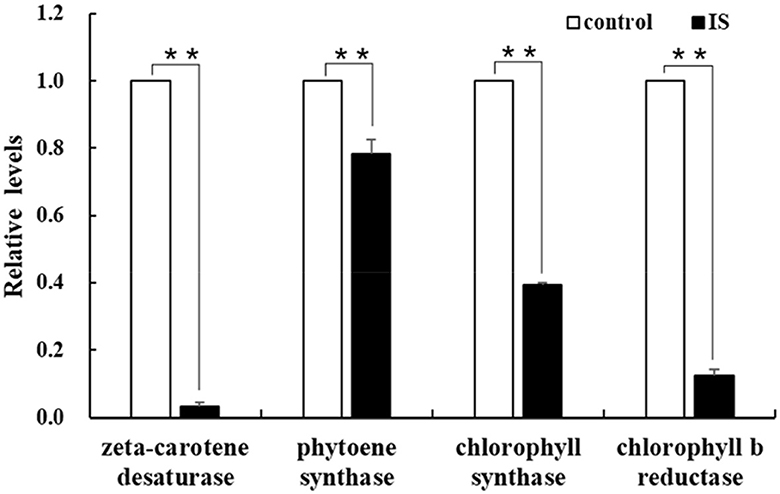
Figure 6. Verification of the differentially expressed genes (DEGs). The expression of chlorophyll and carotene biosynthesis-related genes were analyzed by qRT-PCR. Three independent repetitions have been performed. Statistical data were analyzed by paired sample t-test using SPSS software. Asterisk indicates significant difference (**P < 0.01).
To investigate whether yellowing symptoms might attract transmission vectors for the plant virus, the APV1 vector F. virgate was used in host-choosing experiments among three types of betel palm seedlings. As expected, there was no significant difference in mealybug distribution between APV1-positive seedlings without yellowing symptoms and healthy seedlings (t = 0.458, P > 0.05), or between two groups of healthy seedlings (t = 0.266, P > 0.05). However, a significant difference was observed between APV1-positive seedlings with yellowing symptoms and healthy seedlings (t = 3.800, P < 0.05), with more mealybugs choosing the seedlings displaying yellowing symptoms as hosts (Figure 7). These results support the hypothesis that yellowing symptoms act as attractants for APV1 transmission vectors.
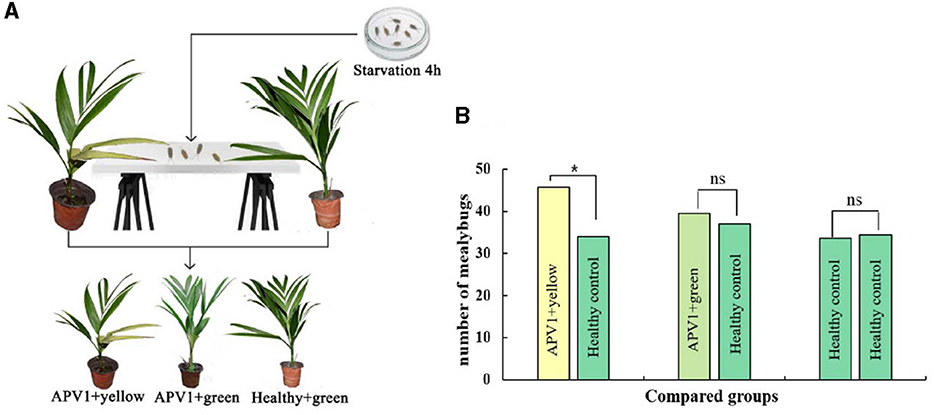
Figure 7. Leaf yellowing symptom is an attractant for Ferrisia virgate. (A) Schematic diagram for host choosing experiment; (B) Three combinations, e.g., APV1+ yellow vs. APV1+ green, APV1+green vs. healthy control, and healthy control vs. healthy control, were compared, respectively. Four independent repetitions have been performed. Statistical data were analyzed by paired sample t-test using SPSS software. Asterisk indicates significant difference (*P < 0.05). ns, no significant difference.
The chloroplast is a vital organelle in plants responsible for photosynthesis and the biosynthesis of essential biochemical components. Chloroplasts also play a crucial role in defense responses, triggering immune responses against viral pathogens. Paradoxically, chloroplasts are often prime targets for viruses during infections (Zhao et al., 2016; Bhattacharyya and Chakraborty, 2018). Understanding the interactions between chloroplasts and viruses is crucial for unraveling the viral pathogenicity (Bhattacharyya and Chakraborty, 2018). Viruses from the Closteroviridae family, such as Citrus tristeza virus (CTV) and Grapevine leafroll-associated viruses (GLRaVs), have been extensively studied (Karasev, 2000; Albiach-Marti et al., 2010; Dawson et al., 2013, 2015; Naidu et al., 2015). However, the interaction between closteroviruses and chloroplasts has been relatively unexplored. Previous studies showed that citrus tristeza virus (CTV) accumulated preferentially in the phloem parenchyma and to a lesser degree in companion cells and occasionally in sieve elements (Zhou et al., 2002; Folimonova et al., 2008). Ultrastructural analysis also revealed that Beet yellows closterovirus (BYV) preferentially accumulated phloem and occasionally in leaf mesophyll cells (Medina et al., 1999). Data from this study demonstrated that APV1 invades chloroplasts and causes the disassembly of chloroplasts and extensive cell structure damage (Figures 4, 5). Additionally, APV1 globally regulated chloroplast photosynthesis-related genes (CPRGs) (Supplementary Figure S4, Figure 6) and reduced the concentrations of chlorophyll a/b, carotene, and photosynthetic efficiency (Figures 2–4). These findings confirm for the first time a central role for chloroplasts in the APV1-plant interaction. Virus infections can cause various degrees of damage to chloroplast structure and lead to malformations, including a reduction in chloroplast numbers (Zechmann et al., 2003; Zhao et al., 2016; Lei et al., 2017; DeBlasio et al., 2018), atypical chloroplast appearances, swollen chloroplasts (Lehto et al., 2003), changes in chloroplast content (Li et al., 2006), decreased grana stack formation, increased starch granule size and number, irregular outmembrane structures (Cowan et al., 2012), and chloroplast disruption (Zhao et al., 2016). APV1 infection completely disrupted the chloroplast structure, resulting in the reduction of chlorophyll a/b, which was exhibited by leaf yellowing symptom. APV1-induced chloroplast disassembly and reduced photosynthesis efficiency should account for such as the growth retardation, yield loss, and decreased nut product quality. This finding points out a direction for future research of YLD.
To fully understand the interaction between APV1 and betel palm, the interaction between the APV1 and chloroplasts should be further investigated. The interaction between chloroplasts and viruses is highly complex and involves the identification of virus-host-interacting proteins (Li et al., 2016; Zhao et al., 2016; Bhattacharyya and Chakraborty, 2018). A preliminary yeast-two-hybrid result showed that APV1-CP interacted with betel palm small heat shock protein (sHSP), a chaperone protein that involved in protein folding (unpublished data), indicating that APV1 might affect chloroplast through the unfolded protein response (Fu et al., 2018; Li et al., 2021). However, this hypothesis requires further investigation. Closing the knowledge gaps in the APV1-chloroplast interaction will aid in utilizing chloroplast-based antiviral resistance and developing management technologies to prevent betel palm chloroplast from APV1 damage.
Finally, plant viruses are obligate biotrophic pathogens reliant on the plant's apparatus and photosynthetic products for replication and proliferation. However, virus infections often result in chloroplast disruption and leaf yellowing symptoms. This raises the question of how infected viruses benefit from these symptoms. Host-preference experiments carried out in this work demonstrated that APV1-induced leaf yellowing symptoms act as an attractant for the APV1 vector, F. virgate (Figure 7), suggesting a complex “host-pathogen-vector co-evolution”.
YLD has a devastating effect on betel palm, causing leaf yellowing, growth retardation, yield loss, decreased nut quality and even death. However, the mechanism underlying is so far rarely reported. Study of cytopathology and ultrastructure of APV1 infected betel palm unraveled that APV1 invaded chloroplasts and caused serious damage of the chloroplasts, resulting reduction of pigments and photosynthesis efficiency. APV1 globally impacted on gene expression patterns and suppressed the expression of nuclear-encoded genes involved in chloroplast and photosynthesis. APV1 induced reduction of photosynthesis efficiency contributed to yield loss and reduced quality of betel nut. Leaf yellowing symptom act as an attractant for F. virgata, the APV1 vector, demonstrating a co-evolution.
Sequence Read Archive (SRA) raw data are available through the central BioProject database at NCBI under project accession PRJNA1120749 and BioSamples accession SAMN41706791.
XC: Conceptualization, Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. BG: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. JL: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. HW: Project administration, Supervision, Writing – original draft, Writing – review & editing. RZ: Conceptualization, Project administration, Supervision, Writing – original draft, Writing – review & editing. XH: Funding acquisition, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research was supported by the Project of Sanya Yazhou Bay Science and Technology City (Grant No. SCKJ-JYRC-2022-71) and the earmarked fund for Agriculture Research System in Hainan Province (Grant No. HNARS-1-G4-1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1424489/full#supplementary-material
Albiach-Marti, M. R., Robertson, C., Gowda, S., Tatineni, S., Belliure, B., Garnsey, S. M., et al. (2010). The pathogenicity determinant of Citrus tristeza virus causing the seedling yellows syndrome maps at the 3'-terminal region of the viral genome. Mol. Plant Pathol. 11, 55–67. doi: 10.1111/j.1364-3703.2009.00572.x
Bhattacharyya, D., and Chakraborty, S. (2018). Chloroplast: the Trojan horse in plant-virus interaction. Mol. Plant Pathol. 19, 504–518. doi: 10.1111/mpp.12533
Cao, X., Zhao, R., Wang, H., Zhang, H., Zhao, X., Khan, L. U., et al. (2021). Genomic diversity of Areca Palm Velarivirus 1 (APV1) in Areca palm (Areca catechu) plantations in Hainan, China. BMC Genom. 22:725. doi: 10.1186/s12864-021-07976-6
Chen, S., Dai, S., and Hou, Y. (2023). Learn from tobacco to reduce betel nut use. Science 382, 777–778. doi: 10.1126/science.adk7903
Cowan, G. H., Roberts, A. G., Chapman, S. N., Ziegler, A., Savenkov, E. I., and Torrance, L. (2012). The potato mop-top virus TGB2 protein and viral RNA associate with chloroplasts and viral infection induces inclusions in the plastids. Front. Plant Sci. 3:290. doi: 10.3389/fpls.2012.00290
Dawson, W. O., Bar-Joseph, M., Garnsey, S. M., and Moreno, P. (2015). Citrus tristeza virus: making an ally from an enemy. Annu. Rev. Phytopathol. 53, 137–155. doi: 10.1146/annurev-phyto-080614-120012
Dawson, W. O., Garnsey, S. M., Tatineni, S., Folimonova, S. Y., Harper, S. J., and Gowda, S. (2013). Citrus tristeza virus-host interactions. Front. Microbiol. 4:88. doi: 10.3389/fmicb.2013.00088
DeBlasio, S. L., Rebelo, A. R., Parks, K., Gray, S. M., and Heck, M. C. (2018). Disruption of chloroplast function through downregulation of phytoene desaturase enhances the systemic accumulation of an aphid-borne, phloem-restricted virus. Mol. Plant Microbe Interact. 31, 1095–1110. doi: 10.1094/MPMI-03-18-0057-R
Folimonova, S. Y., Folimonov, A. S., Tatineni, S., and Dawson, W. O. (2008). Citrus tristeza virus: survival at the edge of the movement continuum. J. Virol. 82, 6546–6556. doi: 10.1128/JVI.00515-08
Fu, S., Xu, Y., Li, C., Li, Y., Wu, J., and Zhou, X. (2018). Rice stripe virus interferes with S-acylation of remorin and induces its autophagic degradation to facilitate virus infection. Mol. Plant 11, 269–287. doi: 10.1016/j.molp.2017.11.011
Fuchs, M., Bar-Joseph, M., Candresse, T., Maree, H. J., Martelli, G. P., Melzer, M. J., et al. (2020). ICTV virus taxonomy profile: closteroviridae. J. Gen. Virol. 101, 364–365. doi: 10.1099/jgv.0.001397
Ivanov, D. A., and Bernards, M. A. (2016). Chlorophyll fluorescence imaging as a tool to monitor the progress of a root pathogen in a perennial plant. Planta 243, 263–279. doi: 10.1007/s00425-015-2427-9
Jin, X., Jiang, Z., Zhang, K., Wang, P., Cao, X., Yue, N., et al. (2018). Three-dimensional analysis of chloroplast structures associated with virus infection. Plant Physiol. 176, 282–294. doi: 10.1104/pp.17.00871
Johnson, P. T., Ostfeld, R. S., and Keesing, F. (2015). Frontiers in research on biodiversity and disease. Ecol. Lett. 18, 1119–1133. doi: 10.1111/ele.12479
Kanatiwela-de Silva, C., Damayanthi, M., de Silva, R., Dickinson, M., de Silva, N., and Udagama, P. (2015). Molecular and scanning electron microscopic proof of phytoplasma associated with areca palm yellow leaf disease in Sri Lanka. Plant Dis. 99, 1641–1641. doi: 10.1094/PDIS-01-15-0072-PDN
Karasev, A. V. (2000). Genetic diversity and evolution of closteroviruses. Annu. Rev. Phytopathol. 38, 293–324. doi: 10.1146/annurev.phyto.38.1.293
Khan, L. U., Cao, X., Zhao, R., Tan, H., Xing, Z., and Huang, X. (2022). Effect of temperature on yellow leaf disease symptoms and its associated areca palm velarivirus 1 titer in areca palm (Areca catechu L.). Front. Plant Sci. 13:1023386. doi: 10.3389/fpls.2022.1023386
Khan, L. U., Zhao, R., Wang, H., and Huang, X. (2023). Recent advances of the causal agent of yellow leaf disease (YLD) on areca palm (Areca catechu L.). Trop. Plants 2, 1–7. doi: 10.48130/TP-2023-0007
Lehto, K., Tikkanen, M., Hiriart, J. B., Paakkarinen, V., and Aro, E. M. (2003). Depletion of the photosystem II core complex in mature tobacco leaves infected by the flavum strain of tobacco mosaic virus. Mol. Plant Microbe Interact. 16, 1135–1144. doi: 10.1094/MPMI.2003.16.12.1135
Lei, R., Jiang, H., Hu, F., Yan, J., and Zhu, S. (2017). Chlorophyll fluorescence lifetime imaging provides new insight into the chlorosis induced by plant virus infection. Plant Cell Rep. 36, 327–341. doi: 10.1007/s00299-016-2083-y
Li, C., Xu, Y., Fu, S., Liu, Y., Li, Z., Zhang, T., et al. (2021). The unfolded protein response plays dual roles in rice stripe virus infection through fine-tuning the movement protein accumulation. PLoS Pathog. 17:e1009370. doi: 10.1371/journal.ppat.1009370
Li, J., Jia, X., Liu, L., Cao, X., Xiong, Y., Yang, Y., et al. (2020). Comparative biochemical and transcriptome analysis provides insights into the regulatory mechanism of striped leaf albinism in arecanut (Areca catechu L.). Ind. Crops Prod. 154:112734. doi: 10.1016/j.indcrop.2020.112734
Li, Y., Cui, H., Cui, X., and Wang, A. (2016). The altered photosynthetic machinery during compatible virus infection. Curr. Opin. Virol. 17, 19–24. doi: 10.1016/j.coviro.2015.11.002
Li, Y. H., Hong, J., Xue, L., Yang, Y., Zhou, X. P., and Jiang, D. A. (2006). Effects of Broad bean wilt virus 2 isolate infection on photosynthetic activities and chloroplast ultrastructure in broad bean leaves. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 32, 490–496.
Liu, X., Sun, X., Wang, S., Liu, Y., Dai, H., and Zhu, X. (2022). Tomato chlorosis virus infecting Luffa cylindrica and Citrullus lanatus in China. Plant Dis. 106:3004. doi: 10.1094/PDIS-01-22-0086-PDN
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Martelli, G. P. (2019). A brief historical account of the family closteroviridae. Methods Mol. Biol. 2015, 7–13. doi: 10.1007/978-1-4939-9558-5_2
Medina, V., Peremyslov, V. V., Hagiwara, Y., and Dolja, V. V. (1999). Subcellular localization of the HSP70-homolog encoded by beet yellows closterovirus. Virology 260, 173–181. doi: 10.1006/viro.1999.9807
Moustaka, J., and Moustakas, M. (2023). Early-stage detection of biotic and abiotic stress on plants by chlorophyll fluorescence imaging analysis. Biosensors 13:796. doi: 10.3390/bios13080796
Nagendran, K., Kumari, S., Pandey, S., Karmakar, P., Chaubey, T., Kumar, R., et al. (2023). Emergence of yellowing disease in cucurbitaceous vegetables caused by Crinivirus and Polerovirus in India. Virology 587:109876. doi: 10.1016/j.virol.2023.109876
Naidu, R. A., Maree, H. J., and Burger, J. T. (2015). Grapevine leafroll disease and associated viruses: a unique pathosystem. Annu. Rev. Phytopathol. 53, 613–634. doi: 10.1146/annurev-phyto-102313-045946
Nayar, R., and Seliskar, C. E. (1978). Mycoplasma like organisms associated with yellow leaf disease of Areca catechu L. Euro J. For. Path 8, 125–128. doi: 10.1111/j.1439-0329.1978.tb00625.x
Purushothama, C. R. A., Ramanayaka, J. G., Sano, T., Casati, P., and Bianco, P. A. (2007). Are phytoplasmas the etiological agent of yellow leaf disease of Areca catechu in India? Bull. Insectol. 60, 413–414.
Tominaga, J., Shimada, H., and Kawamitsu, Y. (2018). Direct measurement of intercellular CO2 concentration in a gas-exchange system resolves overestimation using the standard method. J. Exp. Bot. 69, 1981–1991. doi: 10.1093/jxb/ery044
Tseliou, E., Chondrogiannis, C., Kalachanis, D., Goudoudaki, S., Manoussopoulos, Y., and Grammatikopoulos, G. (2021). Integration of biophysical photosynthetic parameters into one photochemical index for early detection of Tobacco Mosaic Virus infection in pepper plants. J. Plant Physiol. 267:153542. doi: 10.1016/j.jplph.2021.153542
Wang, H., Zhao, R., Zhang, H., Cao, X., Li, Z., Zhang, Z., et al. (2020). Prevalence of yellow leaf disease (YLD) and its associated areca palm velarivirus 1 (APV1) in betel palm (Areca catechu) plantations in Hainan, China. Plant Dis. 104, 2556–2562. doi: 10.1094/PDIS-01-20-0140-RE
Yu, H., Qi, S., Chang, Z., Rong, Q., Akinyemi, I. A., and Wu, Q. (2015). Complete genome sequence of a novel velarivirus infecting areca palm in China. Arch. Virol. 160, 2367–2370. doi: 10.1007/s00705-015-2489-9
Zechmann, B., Muller, M., and Zellnig, G. (2003). Cytological modifications in zucchini yellow mosaic virus (ZYMV)-infected Styrian pumpkin plants. Arch. Virol. 148, 1119–1133. doi: 10.1007/s00705-003-0005-0
Zhang, H., Zhao, X., Cao, X., Khan, L. U., Zhao, R., Wang, H., et al. (2022). Transmission of areca palm velarivirus 1 (APV1) by mealybugs causes yellow leaf disease (YLD) in betel palm (Areca catechu). Phytopathology 112, 700–707. doi: 10.1094/PHYTO-06-21-0261-R
Zhao, F., Liu, H., Qiao, Q., Wang, Y., Wang, S., Tian, Y. T., et al. (2022). Calystegia hederacea: a new natural host of Sweet potato latent virus in China. Plant Dis. 106:2765. doi: 10.1094/PDIS-11-21-2613-PDN
Zhao, J., Zhang, X., Hong, Y., and Liu, Y. (2016). Chloroplast in plant-virus interaction. Front. Microbiol. 7:1565. doi: 10.3389/fmicb.2016.01565
Zhao, X., Zhao, R., Cao, X., Wang, Y., Wang, H., and Huang, X. (2024). Transmission of areca palm velarivirus 1 (APV1) by Pseudococcus cryptus. Phytopathhol. Res. 6, 10. doi: 10.1186/s42483-024-00228-x
Keywords: Areca catechu, co-evolution, cytopathology, ultrastructure, Yellow Leaf Disease
Citation: Cao X, Gao B, Lu J, Wang H, Zhao R and Huang X (2024) Areca palm velarivirus 1 infection caused disassembly of chloroplast and reduction of photosynthesis in areca palm. Front. Microbiol. 15:1424489. doi: 10.3389/fmicb.2024.1424489
Received: 28 April 2024; Accepted: 27 May 2024;
Published: 13 June 2024.
Edited by:
Leiliang Zhang, Shandong First Medical University and Shandong Academy of Medical Sciences, ChinaReviewed by:
Ho-jong Ju, Jeonbuk National University, Republic of KoreaCopyright © 2024 Cao, Gao, Lu, Wang, Zhao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruibai Zhao, emhhb3JiaG5AMTYzLmNvbQ==; Xi Huang, eGlodWFuZ0BoYWluYW51LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.