- 1Institute of Molecular Medicine, National Tsing Hua University, Hsinchu City, Taiwan
- 2School of Chinese Medicine, China Medical University, Taichung City, Taiwan
The transcription factor PsrA regulates fatty acid metabolism, the type III secretion system, and quinolone signaling quorum sensing system in Pseudomonas aeruginosa. To explore additional roles of PsrA in P. aeruginosa, this study engineered a P. aeruginosa PAO1 strain to carry a recombinant plasmid with the psrA gene (pMMBpsrA) and examined the impact of elevated psrA expression to the bacterium. Transcriptomic analysis revealed that PsrA significantly downregulated genes encoding the master quorum-sensing regulators, RhlR and LasR, and influenced many quorum-sensing-associated genes. The role of PsrA in quorum sensing was further corroborated by testing autoinducer synthesis in PAO1 [pMMBpsrA] using two reporter bacteria strains Chromobacterium violaceum CV026 and Escherichia coli [pSB1075], which respond to short- and long-chain acyl homoserine lactones, respectively. Phenotypic comparisons of isogenic ΔpsrA, ΔlasR, and ΔpsrAΔlasR mutants revealed that the reduced elastase, caseinase, and swarming activity in PAO1 [pMMBpsrA] were likely mediated through LasR. Additionally, electrophoretic mobility shift assays demonstrated that recombinant PsrA could bind to the lasR promoter at a 5’-AAACGTTTGCTT-3′ sequence, which displays moderate similarity to the previously reported consensus PsrA binding motif. Furthermore, the PsrA effector molecule oleic acid inhibited PsrA binding to the lasR promoter and restored several quorum sensing-related phenotypes to wild-type levels. These findings suggest that PsrA regulates certain quorum-sensing phenotypes by negatively regulating lasR expression, with oleic acid acting as a crucial signaling molecule.
1 Introduction
Pseudomonas aeruginosa is a Gram-negative, rod-shaped pathogen that primarily infects immunocompromised individuals and cystic fibrosis patients (Rowe et al., 2005). The bacterium is notorious for its multidrug resistance and biofilm formation, which often complicates treatment (Livermore, 2002). In addition to delivering proteins into host cells via the type III secretion system, P. aeruginosa produces a plethora of pathogenicity factors, including various adhesins and secreted toxins such as exotoxin A, elastases, and hemolysins (Wick et al., 1990; Hong and Ghebrehiwet, 1992; Van Delden and Iglewski, 1998; Kipnis et al., 2006). In addition, most P. aeruginosa strains produce pyocyanin, a redox-active blue-green phenazine pigment, which also acts as a terminal signal in the quorum-sensing systems in P. aeruginosa (Dietrich et al., 2006). This pigment affects the respiratory epithelium by disrupting electron transport and metabolism (Denning et al., 1998; Rada and Leto, 2013), and is crucial in P. aeruginosa-mediated pathogenesis in a mouse lung infection model of cystic fibrosis (Lau et al., 2004b).
The expression of many virulence genes in P. aeruginosa is governed by a complex cell-density responding mechanism named quorum sensing (Smith and Iglewski, 2003; Jimenez et al., 2012). Three major quorum-sensing systems have been well characterized in P. aeruginosa: the Lux family Las and Rhl systems, which use acyl-homoserine lactone signals, and the PQS (Pseudomonas Quinolone Signal) system, which use 2-alkyl-4-quinolones as its cognate signals. The transcription factor LasR is considered the master regulator of quorum sensing. In concert with the autoinducer N-3-oxo-dodecanoyl-L-homoserine lactone (3-oxo-C12-HSL) synthesized by synthase LasI, LasR coordinates the expression of other quorum sensing regulators and many virulence genes, including rhlR (Schuster and Greenberg, 2006), pqsR (Schuster and Greenberg, 2006), vqsR (Li et al., 2007), lasB (elastase) (Gambello and Iglewski, 1991), lasA (elastase) (Toder et al., 1991; Passador et al., 1993), aprA (alkaline protease) (Gambello et al., 1993), toxA (exotoxin A) (Storey et al., 1998), and phz1 and phz2 operons, which encode enzymes involved in pyocyanin synthesis (Gallagher et al., 2002; Lau et al., 2004a). LasR is also regulated by several transcription factors including AlgR2 (Ledgham et al., 2003), GacA/GacS (Parkins et al., 2001), MvaT (Diggle et al., 2002), RpoN (Heurlier et al., 2003; Thompson et al., 2003), and Vfr (Albus et al., 1997), thus forming a complex virulence regulatory network in P. aeruginosa.
PsrA, a TetR family transcription factor, has been shown to bind a palindromic sequence (C/GAAACN2–4GTTTG/C) present in numerous promoters including its own and that of the stationary-phase sigma factor gene rpoS (Kojic and Venturi, 2001; Kojic et al., 2002). Multiple studies have identified PsrA as a regulator of the type III secretion system (T3SS) operon exsCEBA (Shen et al., 2006; Kang et al., 2009), the integron integrase gene LexA (Novovic et al., 2019), and the fatty acid catabolism operon fadBA5 (Kojic et al., 2005; Son et al., 2007; Kang et al., 2008). Notably, specific long-chain fatty acids can relieve PsrA repression on the acyl-CoA dehydrogenase gene fadE (PA0506), leading to increased production of 2-heptyl-3-hydroxy-4-quinolone, the autoinducer of the PQS quorum-sensing system, an extracellular compound whose production peaks in the late stationary phase (Kojic et al., 2005; Wells et al., 2017). Similar roles for PsrA in regulating quorum sensing and phenazine synthesis have been observed in Pseudomonas syringae and Pseudomonas chlororaphis (Chin et al., 2005; Chatterjee et al., 2007), respectively. Collectively, these findings strongly imply PsrA’s intricate involvement in regulating various bacterial virulence factors via the quorum sensing system.
It has been demonstrated that the expression of psrA is induced in vivo in CF patients (Son et al., 2007) and in vitro by long-chain fatty acids (Kang et al., 2008, 2009; Wells et al., 2017), the natural quorum sensing inhibitor protoanemonin (Bobadilla Fazzini et al., 2013), and lung surfactant (LaBauve and Wargo, 2014). However, the impact of high psrA expression levels on P. aeruginosa physiology remains unclear. To provide complementary insights to studies using gene knockout strains and to replicate the condition of psrA upregulation, this study constructed a psrA overexpression clone to investigate its effect on various virulence-related properties of P. aeruginosa PAO1. Extensive transcriptomic and phenotypic analyses demonstrated that PsrA overexpression triggers several quorum sensing-related phenotypes in P. aeruginosa PAO1, most likely through direct downregulation of lasR gene expression.
2 Materials and methods
2.1 Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Supplementary Table S1. All bacterial strains were routinely propagated in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, and 1% NaCl). The bacterial strains carrying pMMB66EH-derivative plasmids were grown in LB supplemented with 500 μg/mL carbenicillin. M8 medium for the motility assay was prepared with the following components: 24 mM Na2HPO4, 22 mM KH2PO4, 8 mM NaCl, 0.5% casamino acid, 0.4% glucose, 0.2 mM MgSO4, and 0.1 mM CaCl2. The overnight bacterial culture was prepared as follows: a single colony of each strain was inoculated from the LB agar plate to LB broth and incubated at 37°C for 18 h at 200 rpm. Subsequently, it was diluted 1:100 in fresh LB broth and incubated for 18 h at 37°C. All chemicals used in this study were reagent grade and purchased from Sigma-Aldrich unless indicated otherwise. A stock solution of sodium oleate is prepared at 200 mM.
2.2 Generation of deletion mutants, complementary, and overexpression strains
Single and double deletion mutants of psrA and lasR were constructed in P. aeruginosa PAO1 using the allelic exchange approach (Hoang et al., 1998). The genomic sequence of P. aeruginosa PAO1 was obtained from the Pseudomonas genome database1. DNA of fragments approximately 1 kb in size flanking the coding regions were amplified using PCR and cloned in tandem into the suicide vector pEX18Tc (Supplementary Table S1). The generated plasmids were transformed separately into Escherichia coli S17 λpir and subsequently introduced into P. aeruginosa by conjugation. The transconjugants were selected on LB plates containing tetracycline (100 μg/mL), and the second crossover strains were selected positively on LB plates supplemented with 10% sucrose. The generation of the psrA overexpression clone was performed by cloning the psrA coding region into EcoRI-HindIII sites of the wide-host-range, low-copy-number plasmid pMMB66EH (Fürste et al., 1986) (Supplementary Table S1). The resulting plasmid was then mobilized into P. aeruginosa PAO1 via conjugation. The deletion of psrA or lasR in the bacterial clones and the presence of the pMMB66EH-derived psrA containing complementation plasmid were confirmed by PCR (Supplementary Table S2) and DNA sequencing.
2.3 Determination of P. aeruginosa biological properties
2.3.1 Pyocyanin production
The quantity of pyocyanin was determined as described (Palumbo, 1972). Briefly, two volumes of chloroform were added into the overnight bacterial culture supernatant and mixed by shaking for 1 hour. The mixture was then centrifuged, and the bottom solvent layer was acidified with 0.2 N HCl. The pyocyanin quantity was spectrophotometrically determined at 520 nm.
2.3.2 Elastase activity
The elastase activity was determined by mixing ten microliters of the filtrate prepared from overnight P. aeruginosa culture with 990 μL of a solution consisting of 100 mM Tris–HCl (pH 7.2), 5 mg/mL elastin-Congo red, and 1 mM CaCl2. The reaction mixture was incubated at 37°C with gentle shaking for four hours before the terminated addition of ten microliters of 0.12 M ethylenediaminetetraacetic acid (EDTA). The dissolved Congo red was measured at 495 nm after centrifugation (Bjorn et al., 1979).
2.3.3 Hemolytic activity
The hemolytic activity of different P. aeruginosa strains were assayed using tryptic soy agar with 5% sheep blood. Three microliters of P. aeruginosa overnight culture were spotted on the plates and incubated at 30°C for 24 h. The size of the clear zone that represented bacterial hemolytic activity was scored.
2.3.4 Caseinase activity
Caseinase activity was determined according to the method described by Sokol et al. (1979). Three microliters of P. aeruginosa overnight culture were inoculated on the casein agar plates, which contained 3% (w/v) skimmed milk powder, 1% (w/v) tryptone, 0.5% (w/v) yeast extract, 1% (w/v) NaCl, and 2% (w/v) agar, and incubated at 30°C for 24 h. The size of the clear zone represented the bacterial caseinolytic ability.
2.3.5 Exopolysaccharide production
Culture plates containing 1% (w/v) tryptone, 0.004% (w/v) Congo red, and 0.5% (w/v) agar were used to determine the exopolysaccharide production. Three microliters of overnight cultured bacteria were spotted on the agar plates and incubated at 30°C for 24 h before examination. The higher the production of exopolysaccharides by bacteria, the more intense the red color of the bacterial colony is observed.
2.3.6 Swarming and swimming activities
The plates for the swarming activity assay contain M8 medium and 0.5% (w/v) Bacto agar. For the swimming assay, plates that consist of 1% (w/v) tryptone, 1% (w/v) NaCl, and 0.3% (w/v) agar were prepared (Kohler et al., 2000). Three microliters of overnight cultured bacteria were spotted on the swarming and swimming agar plates and incubated for 24 h at 30°C before examination.
2.4 Total RNA extraction and transcriptome analysis
Overnight grown P. aeruginosa strains were diluted 100-fold into fresh LB broth supplemented with 500 μg/mL carbenicillin and incubated at 37°C with agitation for 9 h till the late log phase at which point the cell densities of all bacteria were comparable (Supplementary Figure S1). The bacterial RNA was extracted using the TRIzol® reagent (Invitrogen, United States) and further purified using the PureLink™ RNA Mini kit (Thermo-Fisher Scientific, United States) as previously described (Lo et al., 2018) according to the manufacturer’s instructions. The contaminating DNA was removed with DNase I (TakaraBio, Japan) along with an RNase inhibitor (GeneDireX, Taiwan). Then the RNA concentration was determined using a NanoDrop ND-1000 spectrophotometer. Complementary DNA was synthesized with GeneScript First-Strand Synthesis Kit with 2 μg RNA and random hexamers following the manufacturer’s instructions. RNA-Seq was performed duplicates using Hiseq 4,000 (Illumina, Inc., San Diego, CA). About 86.5 million and 82.9 million reads were generated for RNA samples from PAO1 [pMMB66EH] and PAO1 [pMMBpsrA], respectively. The relative gene expression level was determined and normalized based on reads per kilobase per million mapped reads (RPKM).
2.5 Quantitative real-time PCR analysis
One microliter of synthesized cDNA was used for qRT-PCR analysis according to the instruction of the Power SYBR Green PCR Master Mix (Applied Biosystems, Warrington, United Kingdom) was used for qRT-PCR analysis. The primers used in the analysis are listed in Supplementary Table S2. The mRNA levels relative to the internal controls (16S rRNA and proC) were calculated from triplicate independent experiments using the 2−ΔΔCt approach (Yuan et al., 2006).
2.6 Purification of the recombinant PsrA protein
Recombinant PsrA was generated by cloning the psrA open reading frame into the expression vector pET30a (Novagen) to obtain plasmid pET30psrA and overexpressed in E. coli BL21(DE3) under the induction of 1.0 mM isopropyl-ß-D-thiogalactopyranoside (IPTG) for 3 h at 37°C. The cells were harvested by centrifugation at 5,000 g for 10 min at 4°C, resuspended in buffer A (20 mM Tris–HCl, pH 7.8, 500 mM NaCl, and 10% glycerol), and disrupted by sonication. Subsequently, the supernatant containing soluble protein lysate obtained from centrifugation at 14,000 g for 30 min at 4°C was filtered through a 0.45 μm filter to avoid any remaining cell debris and loaded onto a Ni-charged column. Purification of the C-terminal hexahistidine-tagged PsrA was conducted under conditions recommended by the manufacturer (Thermo-Fisher Scientific, United States). The protein was eluted from the column with buffer A containing 300 mM imidazole and dialyzed against buffer containing 20 mM Tris–HCl, pH 7.8, 200 mM NaCl, and 20% glycerol at 4°C overnight. The concentration of PsrA was determined by the Bradford assay.
2.7 Electrophoretic mobility shift assay
A modified electrophoretic mobility assay (EMSA) was performed as described previously (Kang et al., 2009), approximately one microgram of PsrA was incubated with 100 ng of testing DNA fragment in a binding buffer containing 25 mM 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid, pH 7.6, 15 mM NaCl, 1 mM dithiothreitol, 20 mM bovine serum albumin, 0.1 mM EDTA at room temperature for 30 min. The PsrA-DNA mixture was resolved on a 6% polyacrylamide gel in Tris-borate-EDTA buffer under a constant voltage of 120 V for 90 min at 4°C. The gel was stained with SYBR™Safe DNA gel stain (Invitrogen, United States) and examined under UV illumination.
2.8 Western blot analysis
A 723-bp fragment of DNA sequence containing the psrA coding sequence and a FLAG (DYKDDDK) tag was generated by PCR using P. aeruginosa PAO1 as the template and cloned into the plasmid pMMB66EH. The resulting construct was conjugated to P. aeruginosa PAO1 and grown overnight with or without IPTG induction. The cells were harvested by centrifugation, and aliquots of protein lysate were resolved by electrophoresis on a 4–20% SDS-polyacrylamide gel and electrophoretically transferred to a nitrocellulose membrane (GE HealthCare, Chicago, United States). After incubation with 5% skim milk to block non-specific binding of antibodies, the membrane underwent a two-step immunoreaction. First, it was incubated with rabbit polyclonal anti-FLAG antibody (Elabscience, Texas, United States) diluted 1:5,000 in TBST (20 mM Tris–HCl, pH 7.6, 150 mM NaCl, and 0.2%(v/v) Tween-20) followed by horseradish peroxidase-conjugated goat anti-rabbit antibody (GeneTex, Taiwan) diluted 1:5,000 in TBST. The presence of PsrA-FLAG was detected using the chemiluminescence method with ECL substrate (GeneTex, Taiwan) and visualized with a biomolecular imager ImageQuant LAS4000 Mini (GE HealthCare, United States).
2.9 N-acyl homoserine lactone assays
The short-chain autoinducer C4-HSL bioassay was conducted using Chromobacterium violaceum CV026 as a reporter (McClean et al., 1997). The assay plates consisted of a 10 mL bottom layer of 1.5% LB agar and a 10 mL top layer of 0.3% LB agar containing 200 μL of overnight grown C. violaceum CV026. A well 7.5 mm in diameter was made at the center of the upper agar layer, and 3 μL of overnight-grown P. aeruginosa were added to the well. The plate was then incubated at 30°C and the production of the purple pigment violacein around the wells in response to C4-HSL was examined at 24 h.
The detection of long-chain autoinducer 3-oxo-C12-HSL was performed using E. coli JM109 [pSB1075] as a reporter (Winson et al., 1998). Briefly, the testing P. aeruginosa culture filtrate was collected by centrifugation at 5,000 g for 10 min, and the supernatant was filtered through a 0.45 μm filter. In a 96-well microtiter plate, 100 μL of 10× LB-diluted JM109 [pSB1075] was mixed with 100 μL 10× LB-diluted culture filtrate, and the plate was incubated at 30°C for 4 h with rocking at 150 rpm. The bioluminescence signal was measured using a fluorescence multilabel reader (Wallac VICTOR3) and calculated as a relative light unit per optical density at 595 nm of the culture (RLU/OD595).
2.10 Statistical methods
All data were presented as mean ± standard error of the mean for each group. Statistical analyses were performed using student’s t-test, one-way analysis of variance (ANOVA), or two-way ANOVA with Tukey’s multiple comparison test. p values of <0.05 were considered statistically significant. The statistical significance was set to *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. All error bars represent the standard error of the mean. The analyses were performed using Prism 6 (GraphPad software).
3 Results
3.1 PsrA influenced quorum sensing in P. aeruginosa
To bypass the self-repressing nature of PsrA, we cloned the psrA gene into pMMB66EH, replacing its native promoter with an IPTG-inducible promoter. Quantitative RT-PCR analysis of the P. aeruginosa harboring the plasmid pMMBpsrA confirmed that this resulted in significantly higher psrA expression in the P. aeruginosa strain harboring pMMBpsrA compared to wild-type PAO1 (Figure 1A). A Western blot analysis was also performed to confirm that PsrA was produced in a higher quantity in PAO1 [pMMBpsrA] than its parental counterpart (Supplementary Figure S2). To evaluate the effect of the increased expression of psrA on P. aeruginosa PAO1, we conducted RNA-Seq analyses comparing the transcriptomes of PAO1 [pMMB66EH] and PAO1 [pMMBpsrA]. Approximately 550 genes were upregulated, and 1,200 genes were downregulated more than twofold in PAO1 [pMMBpsrA] compared to PAO1 [pMMB66EH] (Supplementary Table S3). Notably, the expression of several quorum sensing-related genes was altered in PAO1 [pMMBpsrA]. Examples of psrA-upregulated genes included the PQS quorum sensing operon (pqsABCDE/phnAB) and most genes of the pyocyanin synthesis operons (phz1 and phz2). Conversely, major virulence genes with reduced expression in PAO1 [pMMBpsrA] included those encoding siderophore synthesis (fepCBDG), pyoverdine synthesis (PA2384-PA2415), the type III secretion system (T3SS; PA1694-PA1721), alginate synthesis (PA3544-PA3551), lipid A modification enzymes (PA3552-PA3559), the major elastase LasB, and the caseinase Piv. The detailed sequencing data of RNA-Seq has been deposited to Gene Expression Omnibus (GEO) under the accession number GSE249518.
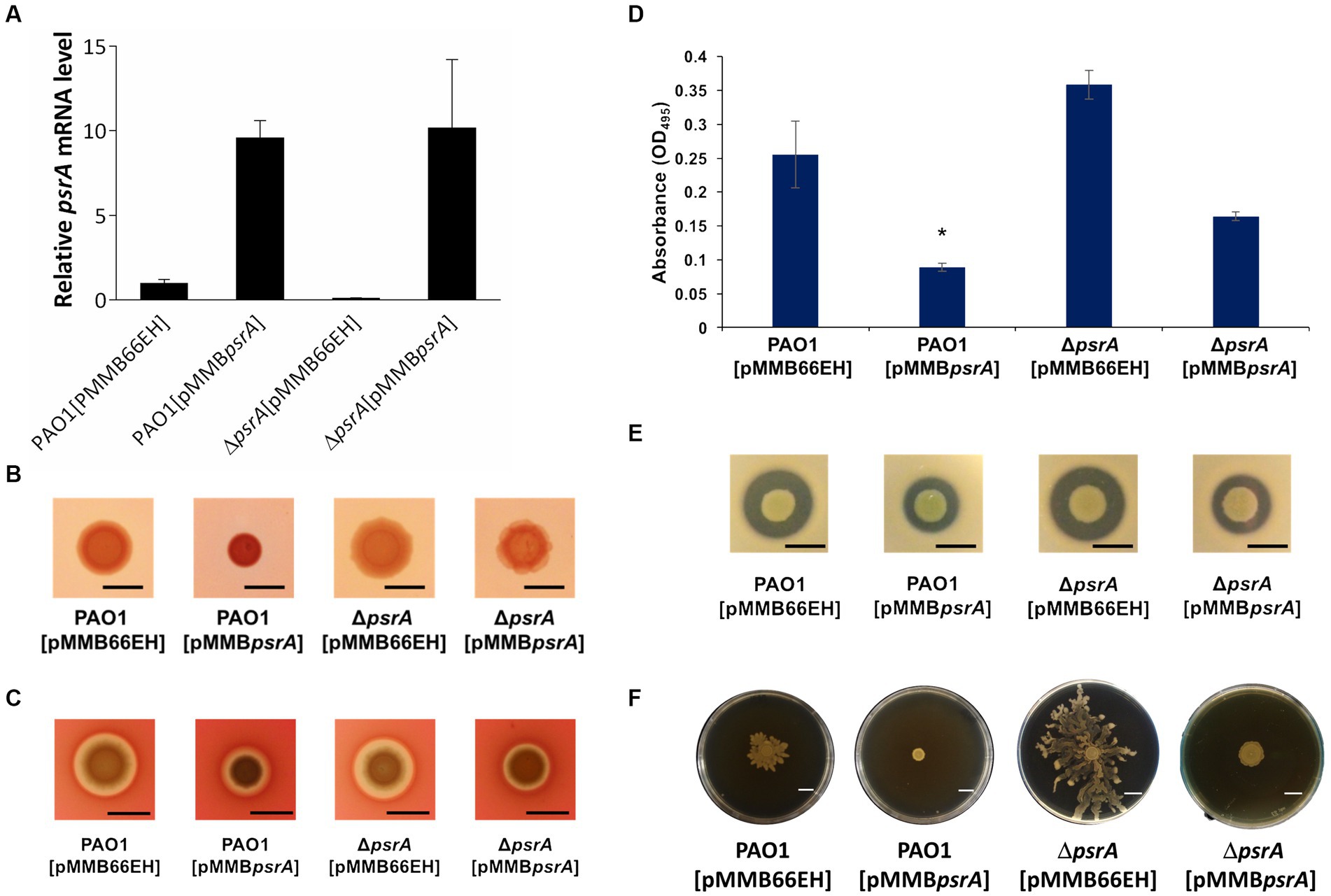
Figure 1. Effects of PsrA on gene expression of psrA, exopolysaccharide production, hemolysis, elastase, caseinase activity and swarming activity in P. aeruginosa PAO1. (A) Real-time quantitative PCR analysis of the effect of PsrA on psrA gene expression. The relative quantity of psrA transcript levels was compared with that of PAO1. The data represents means of triplicates ± standard errors. (B) Exopolysaccharide production. Three microliters overnight bacterial culture of each strain was inoculated on Congo red agar and incubated at 37°C for 24 h. Scale bar = 1 cm. (C) Hemolysis activity. Three microliters overnight bacterial culture of each strain was inoculated on blood agar plates, and the hemolytic zone was examined after incubation of the plates at 37°C for 24 h. Scale bar = 1 cm. (D) Elastase activity. The supernatant of the overnight bacterial culture was incubated with a solution containing elastin-Congo red with shaking at 37°C for 4 h. The release of Congo red by elastase was measured at OD495. The data represent means of quadruplicates ± standard errors. *p < 0.05. (E) Caseinase activity. Three microliters overnight bacterial culture of each strain was inoculated on casein agar plates, and the casein hydrolytic activity was examined after incubation of the plates at 37°C for 24 h. Scale bar = 1 cm. (F) Swarming activity. M8 medium containing 0.5% agar was used in the swarming motility assay. Three microliters overnight bacterial culture of each strain was inoculated on swarming plates and observed after incubation at 30°C for 24 h. Scar bar = 1 cm.
We validated the transcriptomic findings by examining several quorum sensing-regulated phenotypes. In line with the gene expression data, pyocyanin synthesis significantly increased in PAO1 [pMMBpsrA] compared to the wild-type strain up to ~6 folds (Figure 2). Meanwhile, the ∆psrA mutant displayed decreased pyocyanin production. These findings are aligned with an earlier study reporting that deletion of psrA decreased the production of PQS and pyocyanin (Wells et al., 2017). Introducing pMMBpsrA into the ∆psrA strain also significantly enhanced pyocyanin production (Figure 2). Contrasting patterns emerged for other phenotypes: exopolysaccharide, elastase, hemolysis, caseinase, and swarming activities all decreased in PAO1 [pMMBpsrA] but showed subtle increases in the psrA knockout strain (Figures 1B–F).
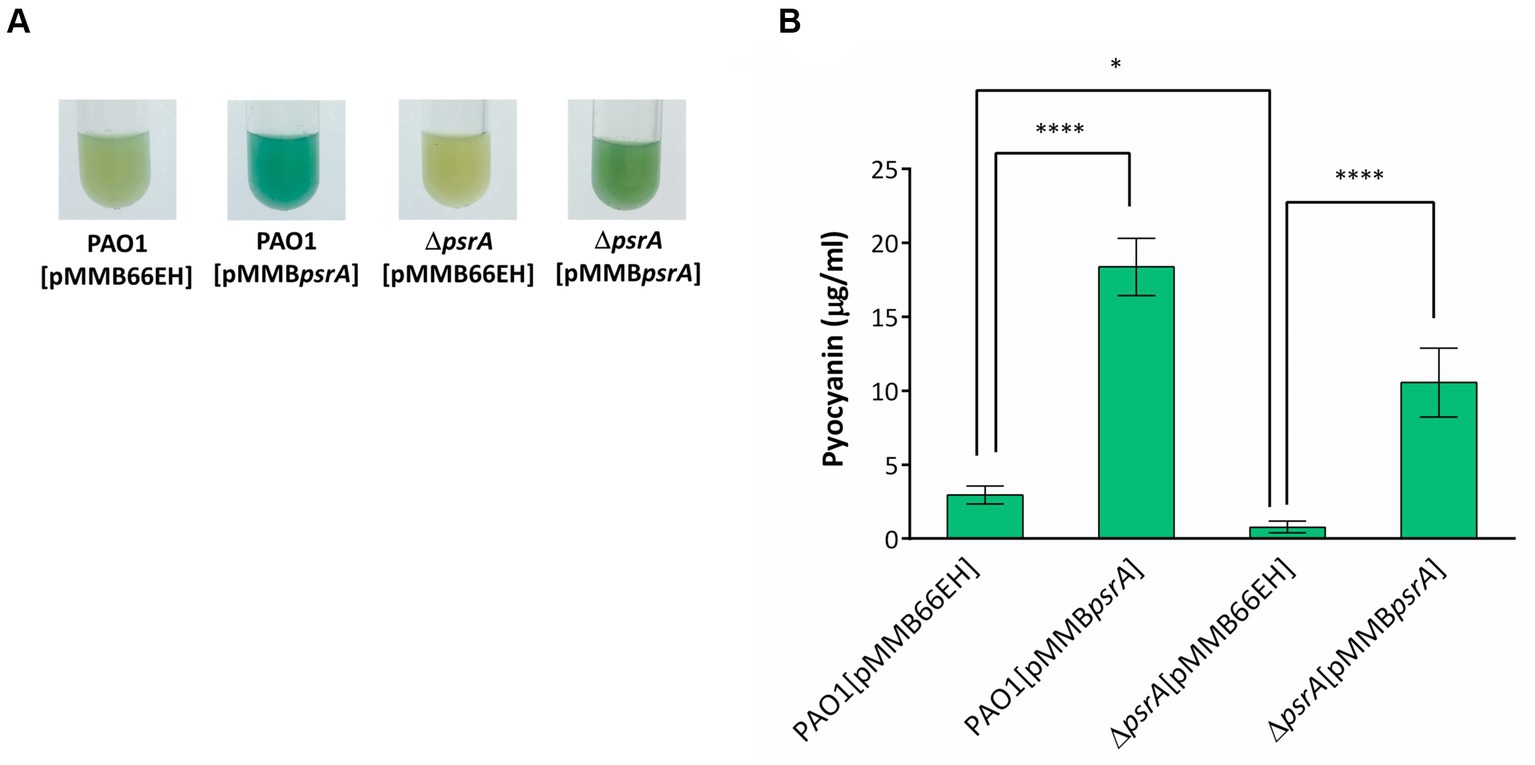
Figure 2. Effects of PsrA on pyocyanin production of P. aeruginosa PAO1. (A) Pyocyanin production, reflected by the intensity of the blue-greenish color, of different P. aeruginosa recombinant strains after growth in LB broth for 18 h. (B) Spectrophotometric analysis of pyocyanin produced by different P. aeruginosa PAO1 recombinant strains. The values represent mean ± S.D. from three biological repeats. Multiple comparisons between groups were assessed by one-way ANOVA followed by Tukey’s comparison test (*p < 0.05 and ****p < 0.001).
3.2 PsrA affected lasR and rhlR expression in P. aeruginosa
Interestingly, downregulated expression of lasR and rhlR, two master regulators of quorum sensing in P. aeruginosa PAO1 [pMMBpsrA], was noted (Supplementary Table S3), though the decrease of lasR is more significant than rhlR (9.3- and 2.5-fold respectively). The decreased expression of lasR was again validated using qRT-PCR confirming that the lasR RNA in PAO1 [pMMBpsrA] was reduced to approximately one-fourth of that in PAO1 [pMMB66EH] (Figure 3). The repression of these key signaling systems indicates that PsrA acts as a negative regulator of lasR, regulating cell-to-cell communication in P. aeruginosa PAO1.
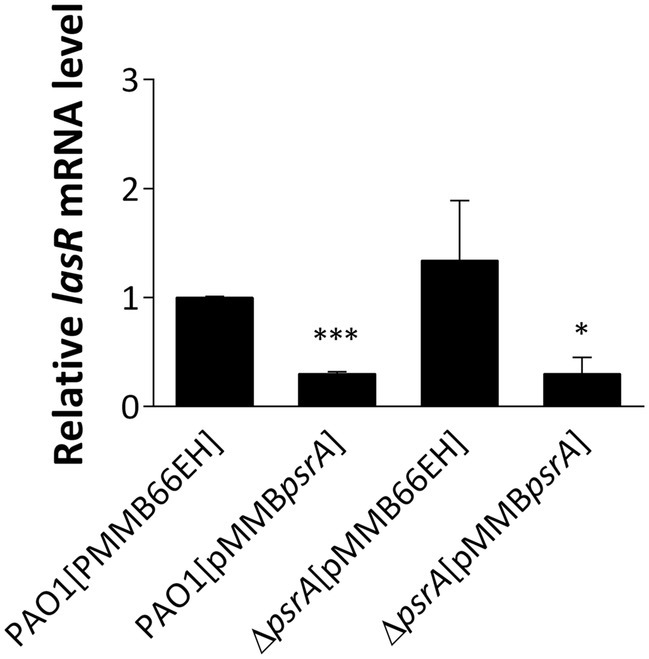
Figure 3. Real-time quantitative PCR analysis of the effect of PsrA on lasR gene expression. The relative quantity of lasR transcript levels was compared with that of PAO1. The data represent means ± standard errors from three independent experiments, p-value was supported by Student’s t-test (*p < 0.05 and ***p < 0.001).
3.3 PsrA affected the production of acyl-homoserine lactone autoinducers
To further verify whether PsrA regulates quorum sensing in P. aeruginosa and given that the Rhl system produces C4-HSL while the Las system produces 3-oxo-C12-AHL, we utilized two acyl homoserine lactone (AHL) reporters to specifically determine each AHL type. Chromobacterium violaceum CV026 produces a purple pigment called violacein in a dose-dependent manner when exposed to C4- to C8-HSL, making it a suitable reporter for detecting C4-HSL produced by P. aeruginosa PAO1 (McClean et al., 1997). As shown in Figure 4A, the strain PAO1 [pMMBpsrA] showed significantly diminished production of C4- to C8-HSL compared to control strains, as evidenced by less purple pigmentation from C. violaceum CV026. Conversely, the deletion of psrA triggered augmented violacein production from C. violaceum CV026, further indicating the negative effect of PsrA on C4-HSL production in P. aeruginosa PAO1. This finding aligns with RNA-Seq data revealing that PsrA overexpression represses rhlR, a key regulator that governs short-chain AHL synthesis. Additionally, culture supernatants were tested using the E. coli JM109 [pSB1075] bioluminescent sensor specific for the 3-oxo-C12-HSL signal (Winson et al., 1998). Consistent with the transcriptomic results indicating lasR repression in PAO1 [pMMBpsrA], the higher expression of PsrA also reduced 3-oxo-C12-HSL levels, which is normally regulated by LasR. Together, these findings substantiate that PsrA perturbs multiple quorum sensing circuit nodes, congruent with its transcriptionally regulating cognate signaling genes (Figure 4B).
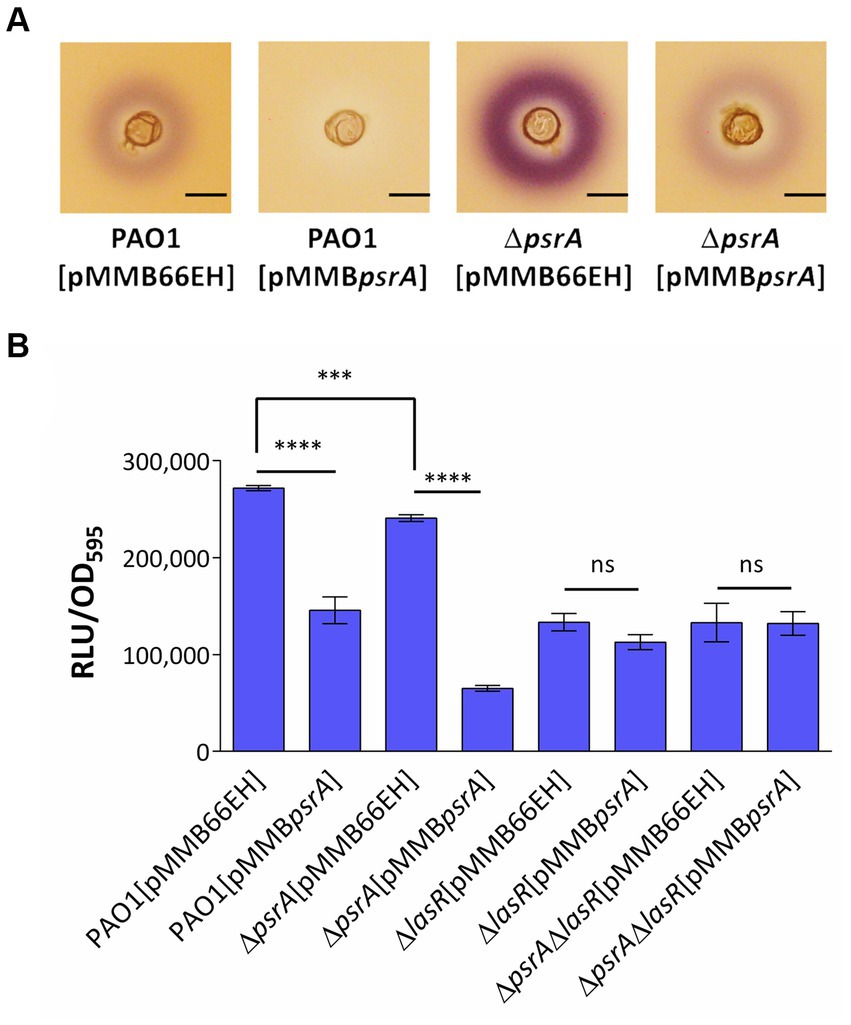
Figure 4. Assessment of PsrA effects on the production of AHL signals. (A) Chromabacterium violaceum CV026 plate assay. The plate was prepared by overlaying a top LB layer containing C. violaceum CV026 on a pre-solidifying LB agar layer. A well was made in the middle of the top layer. Then three microliters of overnight bacterial culture were added to wells. The purple violacein zone surrounding the well generated by C. violaceum CV026 in response to the presence of C4- to C8-HSL was examined. Scale bar = 1 cm. (B) Escherichia coli JM109 [pSB1075] biosensor assay. Overnight bacterial culture supernatant was co-cultured with E. coli harboring N-acyl homoserine lactone sensor plasmids, pSB1075 for 4 h. Bioluminescent signals in relative light units (RLU) conferred by pSB1075 in the presence of 3-oxo-C12-HSL and the culture optical density at 590 nm (OD595) were recorded. All error bars represent the standard error of the mean. Multiple comparisons were evaluated by one-way ANOVA with Tukey’s test. ***p < 0.001 and ****p < 0.001; ns, not significant, n = 5.
3.4 The effects of PsrA on AHL production were likely mediated through LasR
Given LasR’s integral role governing quorum sensing, we examined if LasR mediates the autoinducer reductions by PsrA. Using the 3-oxo-C12-HSL bioreporter again, lasR deletion in PAO1 expectedly decreased 3-oxo-C12-HSL levels. The strain PAO1 [pMMBpsrA] also showed a similar lowering. However, the increased psrA expression levels in a ∆lasR background did not further diminish 3-oxo-C12-HSL (Figure 4B), indicating the absence of additive effects. This lack of synergy suggests the impact of PsrA on 3-oxo-C12-HSL occurs through LasR. Overall, the data fit a model where PsrA may act through LasR repression to achieve downstream quorum sensing phenotypes.
3.5 LasR also mediated several PsrA phenotypes
We next investigated if PsrA influences additional LasR-regulated phenotypes like virulence factor production. Both PAO1 [pMMBpsrA] and the ΔlasR [pMMB66EH] showed reduced elastase activity (Figure 5A). However, introducing pMMBpsrA into the ΔlasR mutant did not enhance the decreased levels of elastase, indicating that PsrA requires LasR to manifest repression. Moreover, the ΔlasRΔpsrA double knockout displayed similar elastase levels as the ΔlasR strain. This LasR-dependent pattern held for other quorum sensing related phenotypes including swarming motility and caseinase activity (Figures 5B,C). A subtly different trend emerged for pyocyanin synthesis; the psrA overexpression ΔlasR strain retained some pyocyanin not seen in the single ΔlasR mutant (Figure 5D). This suggests that while PsrA predominantly utilizes LasR to modulate virulence outputs, additional regulatory pathways are likely to contribute to pyocyanin production.
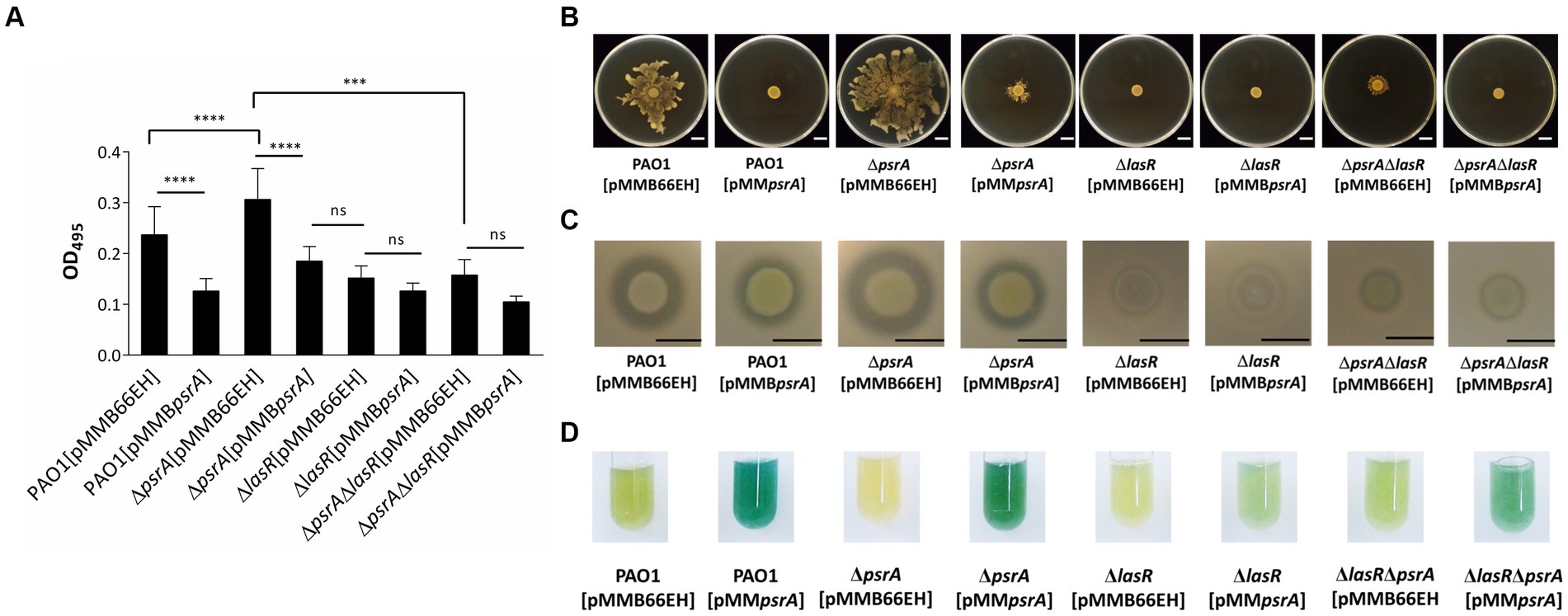
Figure 5. Effects of PsrA in P. aeruginosa PAO1 lasR deletion on quorum-sensing related phenotypes. (A) Elastase activity. The supernatant from the overnight bacterial culture was incubated with an elastin-Congo red solution, shaking at 37°C for 4 h. The release of Congo red by elastase was measured at OD495. The data represent ± standard errors from independent quadruplicates. Multiple comparisons were evaluated by one-way ANOVA with Tukey’s test. ***p < 0.001 and ****p < 0.0001; ns, not significant. (B) Swarming activity. Three microliters overnight bacterial culture of each strain was inoculated on M8 medium plates containing 0.5% agar and observed after incubation at 30°C for 24 h. Scar bar = 1 cm. (C) Caseinase activity. Three microliters overnight bacterial culture of each strain was inoculated on casein agar plates and examined after incubation at 37°C for 24 h. Scale bar = 1 cm. (D) Pyocyanin production. Pyocyanin production of each bacterial strain was examined after 18 h incubation at 37°C.
3.6 PsrA binds directly to the lasR promoter
To determine whether PsrA directly controls lasR gene expression, an affinity-purified PsrA was tested for its binding to several DNA fragments comprising the putative lasR promoter (Supplementary Figure S3) in an EMSA. Figure 6A demonstrates that PsrA is capable of binding to the lasR promoter at nucleotide position 153–285 bp. Using serial truncated DNA fragments of the lasR promoter has allowed us to further locate the PsrA binding site to a 50 bp region lasR promoter at nucleotide positions 201–250. Three DNA fragments (mutant-1, mutant-2, and mutant-3) with different nucleotide substitutions were generated based on two predicted PsrA binding sites within the 50 bp region (Figure 6B). Further analysis of these synthetic DNA fragments revealed that the sequence TTAAACGTTTGCTTAC is essential for the binding of PsrA (Figure 6C). The sequence exhibited moderate (Figure 6D) homology with the PsrA binding motif G/CAAACN(2–4)GTTTG/C reported previously (Kojic et al., 2002). A summary of the DNA fragments selected for EMSA and the binding result is depicted in Figure 6E. Attention is drawn to the question of whether PsrA might also regulate another pivotal quorum-sensing regulator, such as RhlR or the PQS regulator PqsR (MvfR), both of which play a role in pyocyanin production. However, the negative binding results between PsrA and the promoter regions of these two genes from the EMSAs (Supplementary Figure S4) suggest that PsrA may not directly regulate these regulators. Indeed, the downregulation of the quorum-sensing-related genes appears to be a result of reduced lasR expression.
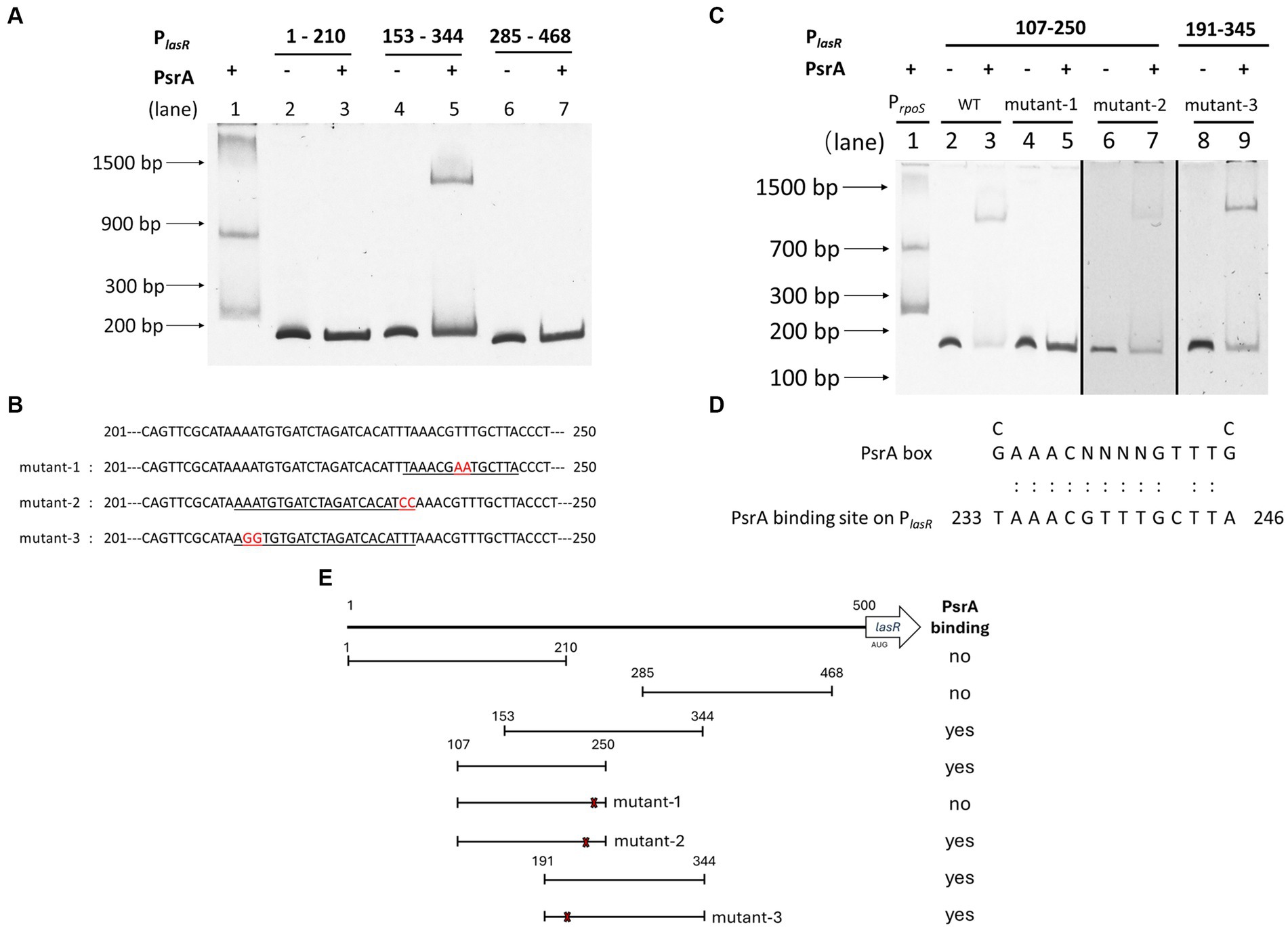
Figure 6. Identification of PsrA binding site on the lasR promoter using electrophoretic mobility shift assay (EMSA). (A) Localization of PsrA binding site to a 200 bp region. The DNA fragments used in the study areas are as follows. Lanes 1, PrpoS as a positive control; 2, PlasR 1–210 bp; 3, PlasR 1–210 bp with PsrA protein; 4, PlasR 153–344 bp; 5, PlasR 153–344 bp with PsrA protein; 6, PlasR 285–468 bp; 7, PlasR 285–468 bp with PsrA protein. (B) The nucleotide sequences of the two-nucleotide substitutions in mutant-1, mutant-2, and mutant-3 for identification of the PsrA binding sequence are marked in red. Underlined is the predicted PsrA binding sequence. (C) Identification of the specific binding site of PsrA on PlasR. DNA fragments used in each lane are 1, positive control PrpoS; 2, PlasR 107–250 bp; 3, PlasR 107–250 bp with PsrA; 4, PlasR 107–250 bp (mutant-1); 5, PlasR 107–250 bp (mutant-1) with PsrA; 6, PlasR 107–250 bp (mutant-2); 7, PlasR 107–250 bp (mutant-2) with PsrA; 8, PlasR 191–344 bp (mutant-3); 9, PlasR 191–344 bp (mutant-3) with PsrA. Negative images of the SYBR Safe-stained gel are shown. (D) Comparison of PsrA binding sequence on lasR promoter and the known PsrA box. The numbers indicate the nucleotide position of the PsrA binding site on the lasR promoter. (E) Schematic diagram showing the relative positions of different DNA fragments comprising different regions of the lasR promoter. The numbers represent the nucleotide positions of each testing truncated lasR promoter region. The locations of three site-specific nucleotide substitutions (mutant-1, mutant-2, and mutant-3) are marked with an × sign. A summary of recombinant PsrA protein binding is shown on the right.
3.7 Oleate affects PsrA binding to the lasR promoter
Building upon prior works showing PsrA responding to long-chain fatty acids in regulating the fadBA5 beta-oxidation operon, this study explored the potential influence of oleate on PsrA binding to the lasR promoter. As demonstrated in Figure 7, oleate effectively blocked PsrA binding at a concentration of 30 μM, paralleling the concentration known to inhibit PsrA binding to the fad promoter (Kang et al., 2008, 2009).
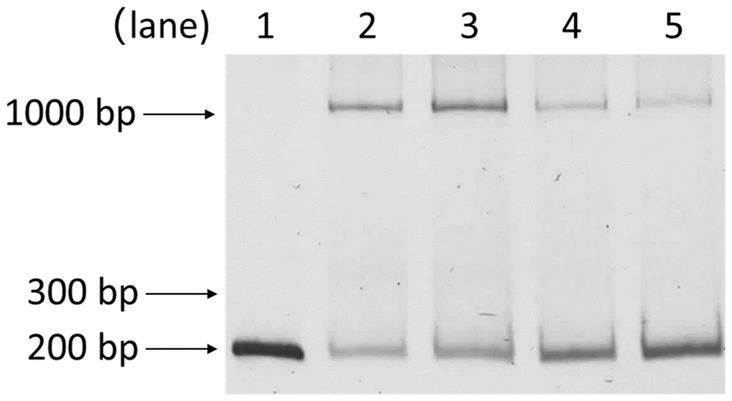
Figure 7. Effect of oleate on the binding of PsrA with PlasR 153–344 bp. Lanes 1, PlasR 153–344 bp only; 2, PlasR 153–344 bp with PsrA; 3, PlasR 153–344 bp with PsrA and 30 μM oleate; 4, PlasR 153–344 bp with PsrA and 60 μM oleate; 5, PlasR 153–344 bp with PsrA and 100 μM oleate. Negative images of the SYBR Safe-stained gel are shown.
3.8 Oleate affects PsrA related quorum sensing phenotypes
The psrA overexpressing strain exhibited a hyper pyocyanin production phenotype (Figure 2). The addition of 1 mM oleate in the culture maintained the pyocyanin synthesis of PAO1 [pMMBpsrA] and ΔpsrA [pMMBpsrA] at the wild-type level suggesting that the effect of PsrA was disrupted by oleate (Figures 8A,B). Likewise, the addition of oleate alleviated the inhibition effect of PsrA on 3-oxo-C12-HSL production in psrA overexpressing strain of PAO1 and ΔpsrA, emphasizing the tuning role of oleate in the PsrA-mediated regulation in P. aeruginosa PAO1 (Figure 8C). Taken together, these findings suggest a common mode of PsrA regulation by fatty acids across distinct operons.
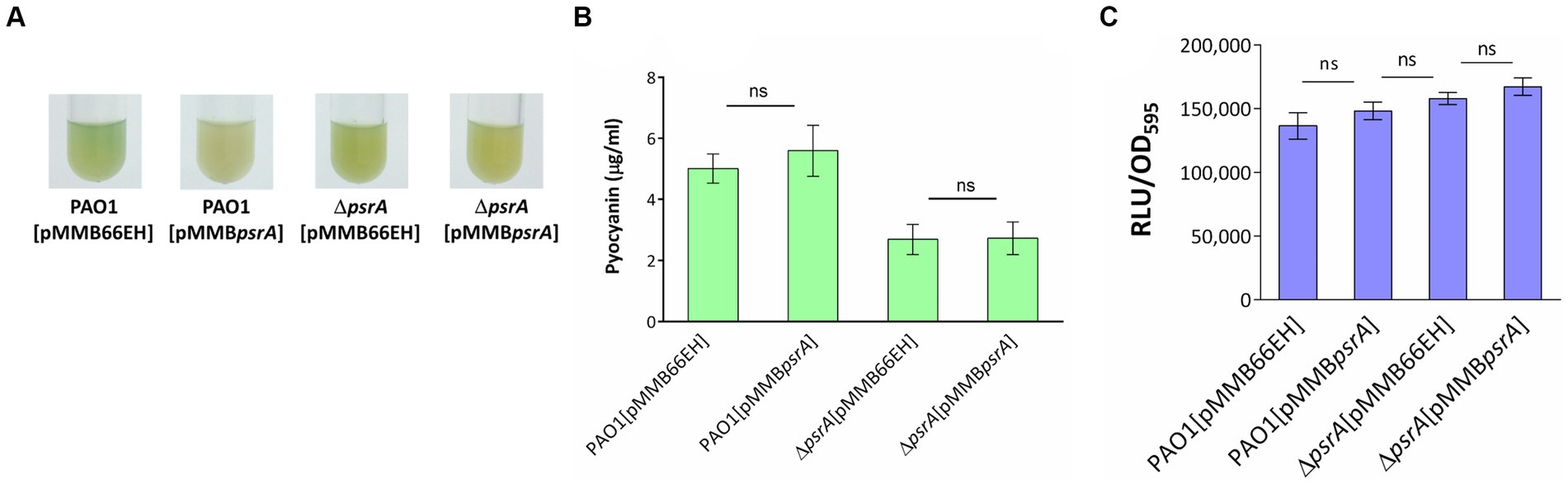
Figure 8. Effect of oleate on the PsrA-mediated pyocyanin and 3-oxo-C12-AHL production. (A) P. aeruginosa PAO1 strains were tested for their pyocyanin production in the presence of 1 mM oleate for 18 h at 37°C. (B) Spectrophotometric analysis of pyocyanin produced by different P. aeruginosa PAO1 recombinant strains under the effect of 1 mM oleate. The values represent mean ± S.D. from three biological repeats. Multiple comparisons between groups were assessed by one-way ANOVA followed by Tukey’s comparison test (ns, not significant). (C) The 3-oxo-C12-AHL released by the bacteria in response to oleate was evaluated with a bioluminescence-based JM109 [pSB1075] reporter system. The data represent the values of means independent triplicates ± standard errors. Multiple comparisons were evaluated by one-way ANOVA with Tukey’s test. ns, not significant.
4 Discussion
This study demonstrated that PsrA overexpression induced pyocyanin production (Figure 2) but suppresses multiple quorum-sensing-related phenotypes, including exopolysaccharide production, hemolysis activity, elastase activity, caseinase activity, and motility (Figure 1). In addition, this study demonstrated that PsrA repressed quorum sensing-associated phenotypes through direct regulation of lasR at the transcription level (Figures 3, 6). Consistent with the notion, the increased expression of psrA in a ΔlasR background did not further reduce 3-oxo-C12-HSL levels, suggesting that PsrA’s impact on 3-oxo-C12-HSL production operates through LasR (Figure 4). Notably, the ΔlasRΔpsrA double mutant displayed similar levels of elastase, caseinase, and swarming activities observed in the ΔlasR strain, reinforcing this dependence (Figure 5). Although PsrA disturbed the production of C4-HSL (Figure 4), there is no evidence from this study to show that the repression effects on RhlR and PqsR are direct (Supplementary Figure S4). Thus, together with AlgR2, GacA/GacS, MvaT, RpoN, and Vfr (Albus et al., 1997; Parkins et al., 2001; Diggle et al., 2003; Heurlier et al., 2003; Ledgham et al., 2003; Thompson et al., 2003), PsrA can be regarded as another regulator of quorum sensing in P. aeruginosa (Figure 9).
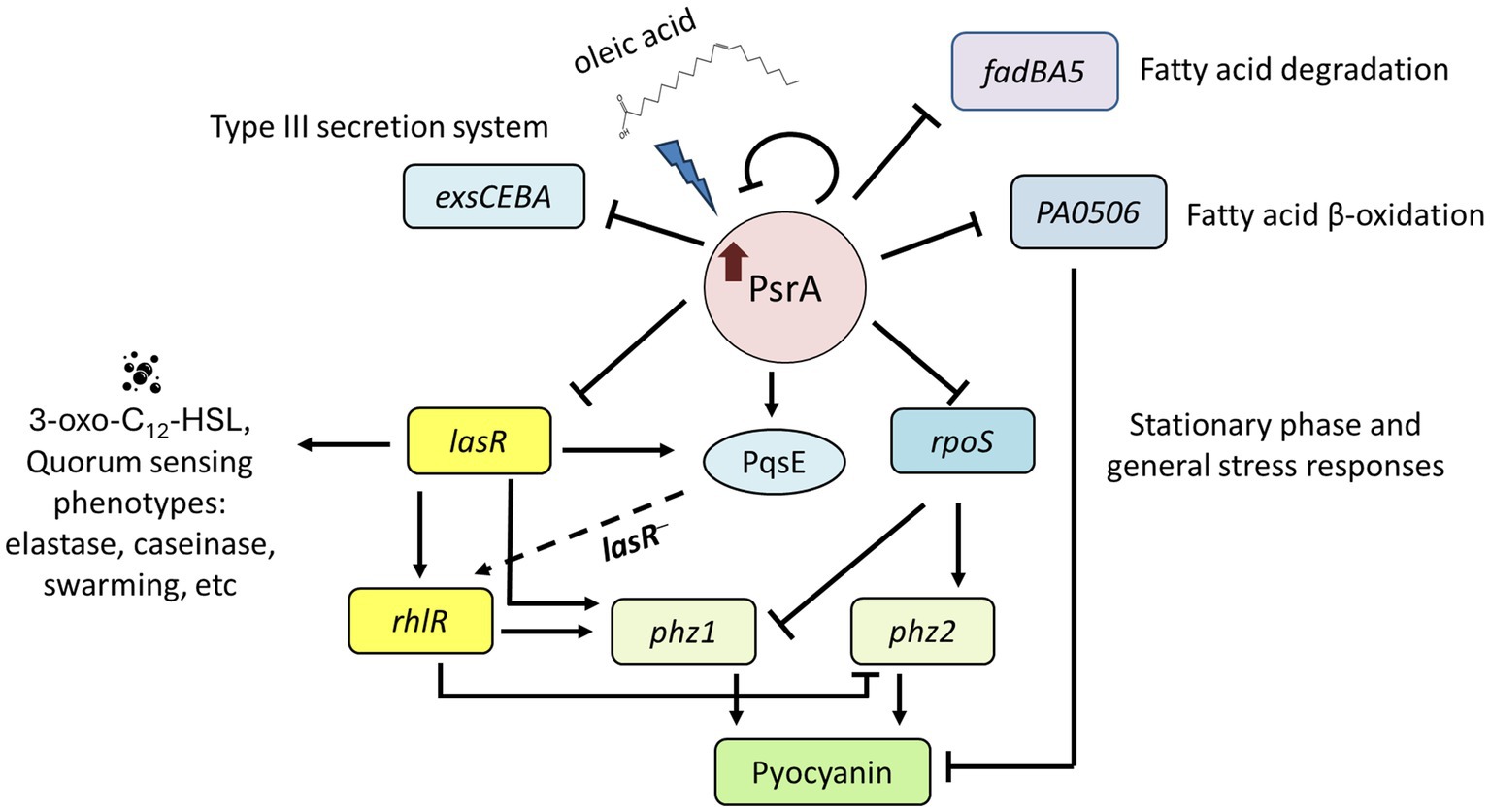
Figure 9. Proposed function model of PsrA regulon. PsrA serves as a regulator of quorum sensing through direct regulation of lasR at the transcription level. It negatively regulates 3-oxo-C12-HSL-based quorum-sensing regulator lasR and promotes LasR downstream virulence-related activities, such as pyocyanin caseinase, elastase, hemolysis, swarming activities, and RhlR-regulated genes. Meanwhile, oleic acid reverses the repression effect of PsrA. Moreover, PsrA has been reported to regulate the expression of genes encoding the general stress sigma factor RpoS, fatty acid degradation regulator FadBA5, acyl-coA dehydrogenase PA0506, and type III secretion systems. The dashed line indicates a possible regulatory role of PqsE to rhlR in the absence of LasR.
In addition to acting as a regulator of quorum sensing, PsrA has been reported to directly regulate the expression of the general stress regulator RpoS (Kojic and Venturi, 2001; Chatterjee et al., 2007; Kang et al., 2009; Wells et al., 2017). RpoS positively regulates the expression of approximately 500 genes and suppresses 270 genes in the stationary phase of P. aeruginosa (Schuster et al., 2004). How PsrA coordinately regulates the expression of RpoS and LasR regulons is intricate. For example, it has been reported that the expression of rpoS is abolished in a P. aeruginosa lasR mutant (Latifi et al., 1996). Another study revealed that the deletion of rpoS increased the expression of lasR (He et al., 2019). The coordination of RpoS and LasR regulons is critical for the bacterium to survive in hostile environments, and how this is achieved is not fully understood. This study highlights the significance of PsrA in the regulation of these two important regulons, although the detailed mechanism and the spectrum of the signal molecules sensed by PsrA regulation remain to be identified.
LasR has been reported as a positive regulator of pyocyanin production (Gallagher et al., 2002; Schuster and Greenberg, 2006). However, in our study, substantial amounts of pyocyanin were still detected in the PsrA-overexpressing strain despite the absence of LasR (Figure 5D), indicating that LasR is not the sole mediator of PsrA-mediated pyocyanin overproduction. Identifying additional factors involved in PsrA-enhanced pyocyanin production is challenging, partly due to the presence of two independently regulated pyocyanin biosynthesis systems, Phz1 and Phz2, in P. aeruginosa. One possible player is FadE, a key enzyme in the fatty acid β-oxidation pathway. Since PsrA is a known repressor of fadE (PA0506) expression, its overexpression could lead to the accumulation of acyl-CoA, which in turn promotes the synthesis of PQS and pyocyanin (Wells et al., 2017). Another potential factor is the general stress sigma factor RpoS, which downregulates phz1 expression while upregulating phz2 expression (Sun et al., 2019). An rpoS deletion mutant exhibited enhanced pyocyanin production (Suh et al., 1999), suggesting that PsrA-mediated rpoS downregulation may contribute to increased pyocyanin production. Additionally, in the absence of LasR, PqsE’s role in Rhl-dependent quorum-sensing activation becomes prominent (He et al., 2019; Groleau et al., 2020). PqsE, a thioesterase dispensable for PQS biosynthesis, was induced by PsrA (Supplementary Table S3), potentially enabling the bacteria to produce high levels of pyocyanin without LasR. Whether PsrA-induced repression of lasR will lead to the development of a LasR-independent Rhl quorum-sensing system remains to be elucidated.
Although PsrA plays a crucial role in P. aeruginosa physiology, information regarding its effectors remains limited. This transcription factor is known to respond to long-chain fatty acid signals to regulate the fadBA5 β-oxidation operon (Kang et al., 2008). However, the specificity and relative potency of different fatty acids on the promoter-binding activity of PsrA have not been fully characterized. This is particularly intriguing because certain fatty acids serve as versatile communication signals within and between bacterial species (Cortes-López et al., 2020). For example, branched-chain fatty acids and myristic acid (C14:0) influence motility and pyocyanin production in P. aeruginosa (Inoue et al., 2008; Abd-Alla and Bashandy, 2012). A recent study suggested that lauric acid (C12:0) and myristic acid might interact with PsrA to enhance virulence in a murine abscess model (Juárez-Rodríguez et al., 2020). Understanding how different fatty acids modulate PsrA activity will be an important research direction for the future.
Several studies have demonstrated that PsrA plays dual regulatory roles: it activates biofilm formation, swarming motility, and T3SS genes (Kojic and Venturi, 2001; Shen et al., 2006; Gooderham et al., 2008; Kang et al., 2009), while repressing the fadBA5 β-oxidation operon (Kang et al., 2008). Activation of rpoS expression was observed in the PsrA mutant in some studies (Kojic and Venturi, 2001; Shen et al., 2006) but not in others (Gooderham et al., 2008; Kang et al., 2008). Intriguingly, augmented expression of PsrA due to the addition of oleate led to decreased expression of T3SS genes and rpoS (Kang et al., 2009). This aligns with our transcriptomic results, indicating that PsrA can downregulate rpoS at higher dosages (Figure 9; Supplementary Table S3). These findings strongly suggest that PsrA’s regulatory role is highly nuanced, involving a balance between free PsrA and oleic acid-bound PsrA, additional protein factors, and bacterial growth stages. Considering the culture conditions of our study, the availability of long-chain fatty acids cannot be ruled out, complicating the assessment of free PsrA levels when only a single copy is present. Therefore, the use of a promoterless PsrA in the low-copy plasmid pMMB66EH in our study might help elucidate the role of PsrA, simulating an environment favorable for its expression.
Because of the abundance of fatty acids in living organisms, analysis of PsrA-fatty acids interaction will greatly facilitate our understanding of bacterial pathogenesis. The specificity and affinity of different fatty acids in regulating PsrA binding to the promoter of key regulatory genes can be further determined. This can be followed by studying the physiological impacts, including virulence gene expression, influenced by PsrA-fatty acid interaction. Finally, the roles of PsrA-fatty acid interaction in tissue tropism, species specificity, and competition with other microbes during P. aeruginosa infections will need to be elucidated.
Disabling quorum sensing has garnered interest as a strategy for treating bacterial infections. Some natural and synthetic quorum-sensing inhibitors, such as the fungi-derived component chrysophanol, can suppress quorum-sensing signals and prevent the biofilm formation of P. aeruginosa and E. coli. Another example of a natural quorum-sensing inhibitor is curcumin. When coupled with or coated onto different nanoparticles, these inhibitors exhibit a synergistic antifouling effect, eradicating biofilm formation from P. aeruginosa on urinary catheter surfaces (Prateeksha et al., 2019, 2021, 2023; Kumar et al., 2022). These promising approaches represent potential alternatives for the development of new antimicrobial strategies. Given PsrA’s regulation of quorum sensing and many related virulence factors, it is a viable candidate for an anti-virulence therapy target (Rémy et al., 2018; Scoffone et al., 2019).
In conclusion, we demonstrated that PsrA acted as a regulator of quorum sensing through direct regulation of lasR, with oleic acid acting as a crucial signaling molecule in this regulatory process. The interplay between PsrA and LasR provides a novel insight into the regulatory mechanism of quorum sensing in P. aeruginosa revealing the potential of PsrA as a target for therapeutic interventions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
L-CK: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. C-CT: Investigation, Methodology, Validation, Writing – original draft. Y-HL: Data curation, Investigation, Methodology, Writing – original draft. Y-LL: Conceptualization, Data curation, Validation, Writing – original draft. N-WC: Investigation, Writing – original draft. C-TL: Resources, Supervision, Writing – original draft, Writing – review & editing. H-YC: Conceptualization, Data curation, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Science and Technology Council, Taiwan under grant no. MOST-109-2320-B-007-007-MY3 and MOST-112-2320-B-007-012-MY3.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1424330/full#supplementary-material
Footnotes
References
Abd-Alla, M. H., and Bashandy, S. R. (2012). EfProduction of quorum sensing inhibitors in growing onion bulbs Iinfected with Pseudomonas aeruginosa E (HQ324110). ISRN Microbiol. 2012:161890. doi: 10.5402/2012/161890
Albus, A. M., Pesci, E. C., Runyen-Janecky, L. J., West, S. E., and Iglewski, B. H. (1997). Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179, 3928–3935. doi: 10.1128/jb.179.12.3928-3935.1997
Bjorn, M. J., Sokol, P. A., and Iglewski, B. H. (1979). Influence of iron on yields of extracellular products in Pseudomonas aeruginosa cultures. J. Bacteriol. 138, 193–200. doi: 10.1128/jb.138.1.193-200.1979
Bobadilla Fazzini, R. A., Skindersoe, M. E., Bielecki, P., Puchałka, J., Givskov, M., and Martins sos Santos, V. A. (2013). Protoanemonin: a natural quorum sensing inhibitor that selectively activates iron starvation response. Environ. Microbiol. 15, 111–120. doi: 10.1111/j.1462-2920.2012.02792.x
Chatterjee, A., Cui, Y., Hasegawa, H., and Chatterjee, A. K. (2007). PsrA, the Pseudomonas sigma regulator, controls regulators of epiphytic fitness, quorum-sensing signals, and plant interactions in Pseudomonas syringae pv. Tomato strain DC3000. Appl. Environ. Microbiol. 73, 3684–3694. doi: 10.1128/aem.02445-06
Chin, A. W. T. F., van den Broek, D., Lugtenberg, B. J., and Bloemberg, G. V. (2005). The Pseudomonas chlororaphis PCL1391 sigma regulator psrA represses the production of the antifungal metabolite phenazine-1-carboxamide. Mol. Plant-Microbe Interact. 18, 244–253. doi: 10.1094/mpmi-18-0244
Cortes-López, H., Juárez-Rodríguez, M., García-Contreras, R., Soto-Hernández, M., and Castillo-Juárez, I. (2020). “Old acquaintances in a new role: regulation of bacterial communication systems by fatty acids,” in Regulation of Bacterial Communication Systems by Fatty Acids. eds. V. R. Rai and J. A. Bai 1st ed. (Boca Raton: Taylor & Francis Group), 47–58.
Denning, G. M., Railsback, M. A., Rasmussen, G. T., Cox, C. D., and Britigan, B. E. (1998). Pseudomonas pyocyanine alters calcium signaling in human airway epithelial cells. Am. J. Phys. 274, L893–L900. doi: 10.1152/ajplung.1998.274.6.L893
Dietrich, L. E., Price-Whelan, A., Petersen, A., Whiteley, M., and Newman, D. K. (2006). The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61, 1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x
Diggle, S. P., Winzer, K., Chhabra, S. R., Worrall, K. E., Cámara, M., and Williams, P. (2003). The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 50, 29–43. doi: 10.1046/j.1365-2958.2003.03672.x
Diggle, S. P., Winzer, K., Lazdunski, A., Williams, P., and Cámara, M. (2002). Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J. Bacteriol. 184, 2576–2586. doi: 10.1128/jb.184.10.2576-2586.2002
Fürste, J. P., Pansegrau, W., Frank, R., Blöcker, H., Scholz, P., Bagdasarian, M., et al. (1986). Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48, 119–131. doi: 10.1016/0378-1119(86)90358-6
Gallagher, L. A., McKnight, S. L., Kuznetsova, M. S., Pesci, E. C., and Manoil, C. (2002). Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 184, 6472–6480. doi: 10.1128/jb.184.23.6472-6480.2002
Gambello, M. J., and Iglewski, B. H. (1991). Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000. doi: 10.1128/jb.173.9.3000-3009.1991
Gambello, M. J., Kaye, S., and Iglewski, B. H. (1993). LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect. Immun. 61, 1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993
Gooderham, W. J., Bains, M., McPhee, J. B., Wiegand, I., and Hancock, R. E. (2008). Induction by cationic antimicrobial peptides and involvement in intrinsic polymyxin and antimicrobial peptide resistance, biofilm formation, and swarming motility of PsrA in Pseudomonas aeruginosa. J. Bacteriol. 190, 5624–5634. doi: 10.1128/jb.00594-08
Groleau, M. C., de Oliveira Pereira, T., Dekimpe, V., and Déziel, E. (2020). PqsE is essential for RhlR-dependent quorum sensing regulation in Pseudomonas aeruginosa. mSystems 5:e20. doi: 10.1128/mSystems.00194-20
He, Q., Feng, Z., Wang, Y., Wang, K., Zhang, K., Kai, L., et al. (2019). LasR might act as an intermediate in overproduction of phenazines in the absence of RpoS in Pseudomonas aeruginosa. J. Microbiol. Biotechnol. 29, 1299–1309. doi: 10.4014/jmb.1904.04029
Heurlier, K., Dénervaud, V., Pessi, G., Reimmann, C., and Haas, D. (2003). Negative control of quorum sensing by RpoN (sigma54) in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185, 2227–2235. doi: 10.1128/jb.185.7.2227-2235.2003
Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J., and Schweizer, H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. doi: 10.1016/s0378-1119(98)00130-9
Hong, Y. Q., and Ghebrehiwet, B. (1992). Effect of Pseudomonas aeruginosa elastase and alkaline protease on serum complement and isolated components C1q and C3. Clin. Immunol. Immunopathol. 62, 133–138. doi: 10.1016/0090-1229(92)90065-v
Inoue, T., Shingaki, R., and Fukui, K. (2008). Inhibition of swarming motility of Pseudomonas aeruginosa by branched-chain fatty acids. FEMS Microbiol. Lett. 281, 81–86. doi: 10.1111/j.1574-6968.2008.01089.x
Jimenez, P. N., Koch, G., Thompson, J. A., Xavier, K. B., Cool, R. H., and Quax, W. J. (2012). The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 76, 46–65. doi: 10.1128/mmbr.05007-11
Juárez-Rodríguez, M. M., Cortes-López, H., García-Contreras, R., González-Pedrajo, B., Díaz-Guerrero, M., Martínez-Vázquez, M., et al. (2020). Tetradecanoic acids with anti-virulence properties increase the pathogenicity of Pseudomonas aeruginosa in a murine cutaneous infection model. Front. Cell Infect. Microbiol. 10:597517. doi: 10.3389/fcimb.2020.597517
Kang, Y., Lunin, V. V., Skarina, T., Savchenko, A., Schurr, M. J., and Hoang, T. T. (2009). The long-chain fatty acid sensor, PsrA, modulates the expression of rpoS and the type III secretion exsCEBA operon in Pseudomonas aeruginosa. Mol. Microbiol. 73, 120–136. doi: 10.1111/j.1365-2958.2009.06757.x
Kang, Y., Nguyen, D. T., Son, M. S., and Hoang, T. T. (2008). The Pseudomonas aeruginosa PsrA responds to long-chain fatty acid signals to regulate the fadBA5 beta-oxidation operon. Microbiology 154, 1584–1598. doi: 10.1099/mic.0.2008/018135-0
Kipnis, E., Sawa, T., and Wiener-Kronish, J. (2006). Targeting mechanisms of Pseudomonas aeruginosa pathogenesis. Med. Mal. Infect. 36, 78–91. doi: 10.1016/j.medmal.2005.10.007
Kohler, T., Curty, L. K., Barja, F., van Delden, C., and Pechere, J. C. (2000). Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182, 5990–5996. doi: 10.1128/JB.182.21.5990-5996.2000
Kojic, M., Aguilar, C., and Venturi, V. (2002). TetR family member psrA directly binds the Pseudomonas rpoS and psrA promoters. J. Bacteriol. 184, 2324–2330. doi: 10.1128/jb.184.8.2324-2330.2002
Kojic, M., Jovcic, B., Vindigni, A., Odreman, F., and Venturi, V. (2005). Novel target genes of PsrA transcriptional regulator of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 246, 175–181. doi: 10.1016/j.femsle.2005.04.003
Kojic, M., and Venturi, V. (2001). Regulation of rpoS gene expression in Pseudomonas: involvement of a TetR family regulator. J. Bacteriol. 183, 3712–3720. doi: 10.1128/jb.183.12.3712-3720.2001
Kumar, S., Paliya, B. S., and Singh, B. N. (2022). Superior inhibition of virulence and biofilm formation of Pseudomonas aeruginosa PAO1 by phyto-synthesized silver nanoparticles through anti-quorum sensing activity. Microb. Pathog. 170:105678. doi: 10.1016/j.micpath.2022.105678
LaBauve, A. E., and Wargo, M. J. (2014). Detection of host-derived sphingosine by Pseudomonas aeruginosa is important for survival in the murine lung. PLoS Pathog. 10:e1003889. doi: 10.1371/journal.ppat.1003889
Latifi, A., Foglino, M., Tanaka, K., Williams, P., and Lazdunski, A. (1996). A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21, 1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x
Lau, G. W., Hassett, D. J., Ran, H., and Kong, F. (2004a). The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 10, 599–606. doi: 10.1016/j.molmed.2004.10.002
Lau, G. W., Ran, H., Kong, F., Hassett, D. J., and Mavrodi, D. (2004b). Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect. Immun. 72, 4275–4278. doi: 10.1128/iai.72.7.4275-4278.2004
Ledgham, F., Soscia, C., Chakrabarty, A., Lazdunski, A., and Foglino, M. (2003). Global regulation in Pseudomonas aeruginosa: the regulatory protein AlgR2 (AlgQ) acts as a modulator of quorum sensing. Res. Microbiol. 154, 207–213. doi: 10.1016/s0923-2508(03)00024-x
Li, L. L., Malone, J. E., and Iglewski, B. H. (2007). Regulation of the Pseudomonas aeruginosa quorum-sensing regulator VqsR. J. Bacteriol. 189, 4367–4374. doi: 10.1128/jb.00007-07
Livermore, D. M. (2002). Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34, 634–640. doi: 10.1086/338782
Lo, Y. L., Chen, C. L., Shen, L., Chen, Y. C., Wang, Y. H., Lee, C. C., et al. (2018). Characterization of the role of global regulator FliA in the pathophysiology of Pseudomonas aeruginosa infection. Res. Microbiol. 169, 135–144. doi: 10.1016/j.resmic.2018.02.001
McClean, K. H., Winson, M. K., Fish, L., Taylor, A., Chhabra, S. R., Camara, M., et al. (1997). Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143, 3703–3711. doi: 10.1099/00221287-143-12-3703
Novovic, K. D., Malesevic, M. J., Filipic, B. V., Mirkovic, N. L., Miljkovic, M. S., Kojic, M. O., et al. (2019). PsrA regulator connects cell physiology and class 1 integron integrase gene expression through the regulation of lexA gene expression in Pseudomonas spp. Curr. Microbiol. 76, 320–328. doi: 10.1007/s00284-019-01626-7
Palumbo, S. A. (1972). Role of iron and sulfur in pigment and slime formation by Pseudomonas aeruginosa. J. Bacteriol. 111, 430–436. doi: 10.1128/jb.111.2.430-436.1972
Parkins, M. D., Ceri, H., and Storey, D. G. (2001). Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 40, 1215–1226. doi: 10.1046/j.1365-2958.2001.02469.x
Passador, L., Cook, J. M., Gambello, M. J., Rust, L., and Iglewski, B. H. (1993). Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260, 1127–1130. doi: 10.1126/science.8493556
Prateeksha, P., Bajpai, R., Rao, C. V., Upreti, D. K., Barik, S. K., and Singh, B. N. (2021). Chrysophanol-functionalized silver nanoparticles for anti-adhesive and anti-biofouling coatings to prevent urinary catheter-associated infections. ACS Appl Nano Mater 4, 1512–1528. doi: 10.1021/acsanm.0c03029
Prateeksha, R. C. V., Das, A. K., Barik, S. K., and Singh, B. N. (2019). ZnO/curcumin nanocomposites for enhanced inhibition of Pseudomonas aeruginosa virulence via LasR-RhlR quorum sensing systems. Mol. Pharm. 16, 3399–3413. doi: 10.1021/acs.molpharmaceut.9b00179
Prateeksha, P., Sharma, V., Nagpoore, N., Jadaun, V., Rao, C., and Singh, B. (2023). Bacteria-responsive multidrug delivery nanosystem for combating long-term biofilm-associated infections. Adv. Funct. Mater. 33:2214852. doi: 10.1002/adfm.202214852
Rada, B., and Leto, T. L. (2013). Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol. 21, 73–81. doi: 10.1016/j.tim.2012.10.004
Rémy, B., Mion, S., Plener, L., Elias, M., Chabrière, E., and Daudé, D. (2018). Interference in bacterial quorum sensing: a biopharmaceutical perspective. Front. Pharmacol. 9:203. doi: 10.3389/fphar.2018.00203
Rowe, S. M., Miller, S., and Sorscher, E. J. (2005). Cystic fibrosis. N. Engl. J. Med. 352, 1992–2001. doi: 10.1056/NEJMra043184
Schuster, M., and Greenberg, E. P. (2006). A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296, 73–81. doi: 10.1016/j.ijmm.2006.01.036
Schuster, M., Hawkins, A. C., Harwood, C. S., and Greenberg, E. P. (2004). The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51, 973–985. doi: 10.1046/j.1365-2958.2003.03886.x
Scoffone, V. C., Trespidi, G., Chiarelli, L. R., Barbieri, G., and Buroni, S. (2019). Quorum sensing as antivirulence target in cystic fibrosis pathogens. Int. J. Mol. Sci. 20:81838. doi: 10.3390/ijms20081838
Shen, D. K., Filopon, D., Kuhn, L., Polack, B., and Toussaint, B. (2006). PsrA is a positive transcriptional regulator of the type III secretion system in Pseudomonas aeruginosa. Infect. Immun. 74, 1121–1129. doi: 10.1128/iai.74.2.1121-1129.2006
Smith, R. S., and Iglewski, B. H. (2003). P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 6, 56–60. doi: 10.1016/s1369-5274(03)00008-0
Sokol, P. A., Ohman, D. E., and Iglewski, B. H. (1979). A more sensitive plate assay for detection of protease production by Pseudomonas aeruginosa. J. Clin. Microbiol. 9, 538–540. doi: 10.1128/jcm.9.4.538-540.1979
Son, M. S., Matthews, W. J. Jr., Kang, Y., Nguyen, D. T., and Hoang, T. T. (2007). In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 75, 5313–5324. doi: 10.1128/iai.01807-06
Storey, D. G., Ujack, E. E., Rabin, H. R., and Mitchell, I. (1998). Pseudomonas aeruginosa lasR transcription correlates with the transcription of lasA, lasB, and toxA in chronic lung infections associated with cystic fibrosis. Infect. Immun. 66, 2521–2528. doi: 10.1128/iai.66.6.2521-2528.1998
Suh, S. J., Silo-Suh, L., Woods, D. E., Hassett, D. J., West, S. E., and Ohman, D. E. (1999). Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181, 3890–3897. doi: 10.1128/JB.181.13.3890-3897.1999
Sun, L., Chi, X., Feng, Z., Wang, K., Kai, L., Zhang, K., et al. (2019). phz1 contributes much more to phenazine-1-carboxylic acid biosynthesis than phz2 in Pseudomonas aeruginosa rpoS mutant. J. Basic Microbiol. 59, 914–923. doi: 10.1002/jobm.201900165
Thompson, L. S., Webb, J. S., Rice, S. A., and Kjelleberg, S. (2003). The alternative sigma factor RpoN regulates the quorum sensing gene rhlI in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 220, 187–195. doi: 10.1016/s0378-1097(03)00097-1
Toder, D. S., Gambello, M. J., and Iglewski, B. H. (1991). Pseudomonas aeruginosa LasA: a second elastase under the transcriptional control of lasR. Mol. Microbiol. 5, 2003–2010. doi: 10.1111/j.1365-2958.1991.tb00822.x
Van Delden, C., and Iglewski, B. H. (1998). Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg. Infect. Dis. 4, 551–560. doi: 10.3201/eid0404.980405
Wells, G., Palethorpe, S., and Pesci, E. C. (2017). PsrA controls the synthesis of the Pseudomonas aeruginosa quinolone signal via repression of the FadE homolog, PA0506. PLoS One 12:e0189331. doi: 10.1371/journal.pone.0189331
Wick, M. J., Hamood, A. N., and Iglewski, B. H. (1990). Analysis of the structure-function relationship of Pseudomonas aeruginosa exotoxin A. Mol. Microbiol. 4, 527–535. doi: 10.1111/j.1365-2958.1990.tb00620.x
Winson, M. K., Swift, S., Fish, L., Throup, J. P., Jørgensen, F., Chhabra, S. R., et al. (1998). Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163, 185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x
Keywords: fatty acid, gene expression, LASR, oleic acid, Pseudomonas aeruginosa , PSRA, quorum sensing, transcription factor
Citation: Kok L-C, Tsai C-C, Liao Y-H, Lo Y-L, Cheng N-W, Lin C-T and Chang H-Y (2024) Roles of transcriptional factor PsrA in the regulation of quorum sensing in Pseudomonas aeruginosa PAO1. Front. Microbiol. 15:1424330. doi: 10.3389/fmicb.2024.1424330
Edited by:
Tomohiro Shimada, Meiji University, JapanReviewed by:
B. N. Singh, National Botanical Research Institute (CSIR), IndiaWael Abdel Halim Hegazy, Zagazig University, Egypt
Nadim Majdalani, National Institutes of Health (NIH), United States
Wulf Blankenfeldt, Helmholtz Center for Infection Research (HZ), Germany
Copyright © 2024 Kok, Tsai, Liao, Lo, Cheng, Lin and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hwan-You Chang, aHljaGFuZ0BsaWZlLm50aHUuZWR1LnR3
 Li-Ching Kok
Li-Ching Kok Chia-Chun Tsai1
Chia-Chun Tsai1 Ching-Ting Lin
Ching-Ting Lin Hwan-You Chang
Hwan-You Chang