- 1Department of Population Health and Reproduction, School of Veterinary Medicine, University of California, Davis, Davis, CA, United States
- 2100K Pathogen Genome Project, School of Veterinary Medicine, University of California, Davis, Davis, CA, United States
- 3Veterinary Medicine Teaching and Research Center, School of Veterinary Medicine, University of California, Davis, Tulare, CA, United States
Food-producing animals such as dairy cattle are potential reservoirs of antimicrobial resistance (AMR), with multidrug-resistant (MDR) organisms such as Escherichia coli observed in higher frequency in young calves compared to older cattle. In this study, we characterized the genomes of enteric MDR E. coli from pre-weaned dairy calves with and without diarrhea and evaluated the influence of host-level factors on genomic composition. Whole genome sequence comparative analysis of E. coli (n = 43) revealed substantial genomic diversity that primarily clustered by sequence type and was minimally driven by calf diarrheal disease status (healthy, diarrheic, or recovered), antimicrobial exposure, and dietary zinc supplementation. Diverse AMR genes (ARGs)—including extended-spectrum beta-lactamase genes and quinolone resistance determinants—were identified (n = 40), with unique sets of ARGs co-occurring in gene clusters with large AMR plasmids IncA/C2 and IncFIB(AP001918). Zinc supplementation was not significantly associated with the selection of individual ARGs in E. coli, however analysis of ARG and metal resistance gene pairs identified positive associations between certain aminoglycoside, beta-lactam, sulfonamide, and trimethoprim ARGs with acid, tellurium and mercury resistance genes. Although E. coli in this study lacked the typical virulence factors of diarrheagenic strains, virulence genes overlapping with those in major pathotypes were identified. Among the 103 virulence genes detected, the highest abundance and diversity of genes corresponded to iron acquisition (siderophores and heme uptake). Our findings indicate that the host-level factors evaluated in this study were not key drivers of genomic variability, but that certain accessory genes in enteric MDR E. coli may be enriched. Collectively, this work provides insight into the genomic diversity and host-microbe interface of MDR E. coli from pre-weaned dairy calves.
1 Introduction
Escherichia coli is a diverse and ubiquitous organism present in the healthy enteric microbiome of humans and animals and as a pathogen responsible for various diarrheagenic and extraintestinal diseases (Jackson et al., 2011; Braz et al., 2020). The occurrence of antimicrobial resistant (AMR) E. coli in food-producing animals, such as dairy cattle, has been identified across various cattle groups in farm environmental matrices, feces, food products (e.g., milk and cheese), and clinical samples (e.g., diarrhea and clinical mastitis) (Ombarak et al., 2018; Formenti et al., 2021; Jeamsripong et al., 2021; Majumder et al., 2021; Imre et al., 2022). The prevalence and persistence of drug-resistant E. coli is both a veterinary and human medicine concern, with pathogenic strains compromising animal health and safety of food products, and commensals serving as important reservoirs for the dissemination of AMR.
Multidrug-resistant (MDR) E. coli have been observed in higher frequency in younger cattle, particularly in calves around 2 weeks in age (Berge et al., 2005, 2010). This age-dependent and transient increase in AMR of dairy calves is thought to be driven by the early developing gut microbiome, in which initial exposure to the environment, antibiotic therapy, dietary changes, and other factors collectively contribute to the rapid establishment of the bovine resistome (Khachatryan et al., 2004; Noyes et al., 2016; Liu et al., 2019; Springer et al., 2019; Oh et al., 2020). Previous studies have demonstrated the dynamic nature of AMR selection and enrichment in calves, with the acquisition of AMR occurring beyond influences of antibiotic exposure (Liu et al., 2019; Haley and Van Kessel, 2022) and calves harboring greater diversity in AMR than the potential sources (e.g., dam) seeding their resistome (Haley et al., 2020; Massé et al., 2021). Additionally, studies have suggested that biocides used as disinfectants and heavy metal additives in feed may contribute to the co-selection of AMR with biocide and metal resistance (Wales and Davies, 2015; Cheng et al., 2019).
In pre-weaned dairy calves, diarrhea is the leading cause of morbidity and mortality, which frequently results in antimicrobial treatment (Berchtold and Constable, 2009; Habing et al., 2017). To reduce AMR without compromising animal health, antimicrobial alternatives such as dietary zinc supplementation have been explored and shown to be effective in preventing diarrhea and expediting diarrheal recovery (Glover et al., 2013; Feldmann et al., 2019; Chang et al., 2020; Ma et al., 2020; Wo et al., 2022). In this work, we evaluated fecal MDR E. coli isolates from pre-weaned dairy calves in a zinc supplementation clinical trial using whole genome sequencing (WGS) comparative analysis. The objective of this study was to characterize AMR and virulence genes and to evaluate calf diarrheal disease status, dietary zinc supplementation, and antimicrobial treatment as potential drivers of genomic variability in MDR E. coli. We hypothesize that these host-level factors will contribute to differences in genomic AMR, virulence, and metal resistance profiles, and that the presence of certain genes will provide insight into the persistence of enteric MDR E. coli in calves.
2 Materials and methods
2.1 Isolate source
Fecal E. coli isolates in this study were obtained from pre-weaned dairy calves enrolled in a double-blind, block-randomized, placebo-controlled zinc supplementation clinical trial assessing dietary zinc supplementation on diarrhea prevention and calf health. Details on the original trial procedures were previously described (Feldmann et al., 2019). Briefly, all calves were under the same management practices (e.g., housing and diet) and standard on-farm treatment protocols. The repository of 43 E. coli isolates correspond to pre-weaned dairy calves 2 weeks in age (range: 14–16 days). One representative fecal E. coli isolate per calf was used for analysis, with each isolate corresponding to a calf after 14 consecutive days of dietary zinc sulfate or placebo treatment. Treatments were administered during morning milk feeding with calves in the zinc group receiving 0.22 g zinc sulfate monohydrate (80 mg of elemental zinc) (Sigma-Aldrich Company, St. Louis, MO, United States) with 0.44 g milk replacer powder, and calves in the placebo group receiving only 0.44 g milk replacer powder (Feldmann et al., 2019). At the time of isolate collection, calves were in various stages of diarrheal disease (pre-diarrheic/healthy, diarrheic, or recovered) and exposure to antimicrobial treatment for diarrhea (0, 1, or 2 doses of 31.5 mL (1,575 mg) spectinomycin administered once daily, SpectoGard, Bimeda, Inc., Le Sueur, MN, United States). Other antimicrobial exposures included tetracycline and neomycin administered through daily milk, which were consistent in dosage and duration over time for all calves throughout the study. Calf-level data corresponding to isolates were collected from daily assessment records for individual calves. All isolates were confirmed as E. coli using conventional PCR and underwent antimicrobial susceptibility testing (AST) using broth microdilution and the NARMS Gram Negative panel (YCMV3AGNF) as previously described (Lee et al., 2024).
2.2 DNA extraction and whole genome sequencing (WGS)
Genomic DNA was extracted from pure overnight E. coli cultures per manufacturer’s protocol using the Qiagen’s DNeasy Blood and Tissue kit (Qiagen, Valencia, CA, United States). WGS was conducted using methods from the 100 K Pathogen Genome Project as previously described (Weis et al., 2017; Bandoy and Weimer, 2020; Aguilar-Zamora et al., 2022; Hurtado et al., 2022; Woerde et al., 2023; Hernández-Juárez et al., 2021). Briefly, genomic DNA purity and integrity were assessed using the Nanodrop and the Agilent 2200 TapeStation with the Genomic DNA ScreenTape Assay (Agilent Technologies, Inc., Santa Clara, CA, United States), respectively. Sequencing libraries were constructed using the KAPA HyperPlus library preparation kit (Roche Sequencing Solutions, Pleasanton, CA, United States). Double-stranded genomic DNA was fragmented and indexed using Weimer 384 TS-LT DNA barcodes (Integrated DNA Technologies, Coralville, IA, United States), followed by dual-SPRI size selection and PCR amplification. Final library sizes were confirmed on the LabChip GX using the HT DNA 1 K kit (PerkinElmer, Waltham, MA, United States). Library quantification was conducted using the KAPA Library Quantification Kit (Roche Sequencing Solutions, Pleasanton, CA, United States) to ensure normalized concentrations for sequencing pooling. Final libraries were sequenced using the Illumina HiSeq X Ten with PE150.
2.3 Whole genome assembly and comparison
Genomic sequence data was processed as previously described (Bandoy and Weimer, 2020; Higdon et al., 2020; Flores-Valdez et al., 2021; Miller et al., 2021; Depenbrock et al., 2024). Briefly, Trimmomatic was used to remove low-quality sequence and adapters, and FastQC was used to review sequence quality. Paired-end reads from WGS were assembled using Shovill with the SPAdes assembler and a Kmer size of 31. Quality of assemblies was then evaluated using CheckM. Genome similarity was measured using Sourmash with Minhash signatures with a Kmer length of 31 and scaled sketch size of 100,000 per megabase (Brown and Irber, 2016). The matrix output from Sourmash was visualized in R using the pheatmap package (RDocumentation, 2024).
2.4 Multilocus sequence typing (MLST) and pangenome analysis
The sequence type (ST) for each genome was determined based on the Achtman seven-locus scheme (adk, fumC, gyrB, icd, mdh, purA, and recA) using the PubMLST database (Kaas et al., 2012; PubMLST, 2024). Pangenome analysis was conducted using Roary as described previously (Page et al., 2015; Bandoy and Weimer, 2020; Miller et al., 2021). Pangenome composition and gene diversity estimation were then visualized using open source python script ‘roary_plots.py’ and native Rscript (create_pan_genome_plots.R), respectively (Higdon et al., 2020).
2.5 Identification of antimicrobial resistance genes (ARGs), virulence genes, metal resistance genes, and plasmid replicons
Genetic determinants for antimicrobial resistance (ARGs), virulence, metal resistance, and plasmid replicons were determined using Abricate and the ResFinder, VFDB, BacMet, and PlasmidFinder databases, respectively (Zankari et al., 2012; Carattoli et al., 2014; Pal et al., 2014; Chen et al., 2016; Seemann, 2024). Additionally, SNP based resistance for quinolones was identified using RGI with the CARD database (Alcock et al., 2020a,b). Hits were determined if meeting the criteria of ≥90% coverage and ≥ 95% identity. For metal resistance genes, only experimentally confirmed genes were included in the analysis.
2.6 Data analyses
Descriptive statistics on the distribution of ARGs, virulence factors, metal resistance genes, and plasmid replicons were conducted in SAS OnDemand for Academics. Differences in the mean number of ARGs and virulence genes by factors of treatment group, diarrhea status, and number of therapeutic antibiotic doses were evaluated using a t-test/ANOVA or Mann–Whitney U test.
Proportions of E. coli genomes with presence of ARGs and virulence factors were plotted as heatmaps in R using the pheatmap package. Rows of the heatmaps were clustered using the Euclidean distance metric and complete linkage method. Bar plots and violin plots of the distribution of ARGs and virulence factors, respectively, were visualized in R using ggplot2 (Wickham et al., 2023).
To investigate the differences in antimicrobial resistance, virulence, metal resistance, and pangenome composition amongst isolates, clustering based on the presence and absence matrices for each were assessed by grouping factors of treatment group, diarrhea status, sequence type, and antibiotic exposure as previously described (Lee et al., 2023). A PERMDISP2 procedure was conducted to evaluate if dispersions of groups for each grouping factor were homogenous (Anderson, 2006; Anderson et al., 2006). Permutational analysis of variance (PERMANOVA) and ANOSIM (analysis of similarity) were then performed to evaluate equivalence of centroids of groups and average of ranks of within-group to between-group distances, respectively (Anderson and Walsh, 2013). Additionally, non-metric multidimensional scaling was performed by grouping factor of sequence type for AMR and virulence genes. All tests were performed using 10,000 permutations and a Jaccard distance metric in R using the vegan package (Oksanen et al., 2022).
Logistic regression models were constructed to assess the association between the presence of ARGs with calf-level factors. Models were constructed with outcomes specified as the presence or absence of individual ARGs, quinolone resistance determinants (presence of any point mutations or plasmid-mediated quinolone resistance determinants), and extended spectrum beta-lactamase (ESBL) resistance genes. ARGs which were present in all E. coli genomes were omitted from analysis. Calf-level factors included in model building included treatment group (isolate from zinc – or placebo – treated calf), therapeutic spectinomycin exposure at the time of isolate collection, and diarrhea status of the calf at the time of isolate collection. Antibiotic exposure and calf diarrhea status were evaluated based on individual calf-level data collected through daily assessments. Specifically, spectinomycin treatment was coded as a binary variable (received treatment or not), number of doses received (0, 1, or 2 doses), or days from the last spectinomycin dose received, and diarrhea status was coded as days on or from diarrhea or a categorical variable (healthy/pre-diarrheic, diarrheic, or recovered). Final models were selected based on the lowest AIC after inclusion of confounders (antimicrobial exposure for all ARG models) and any other significant predictors. Given their public health significance, the association between the presence of extended spectrum beta-lactamase (ESBL) genes and other ARGs were also evaluated using Fisher’s exact test.
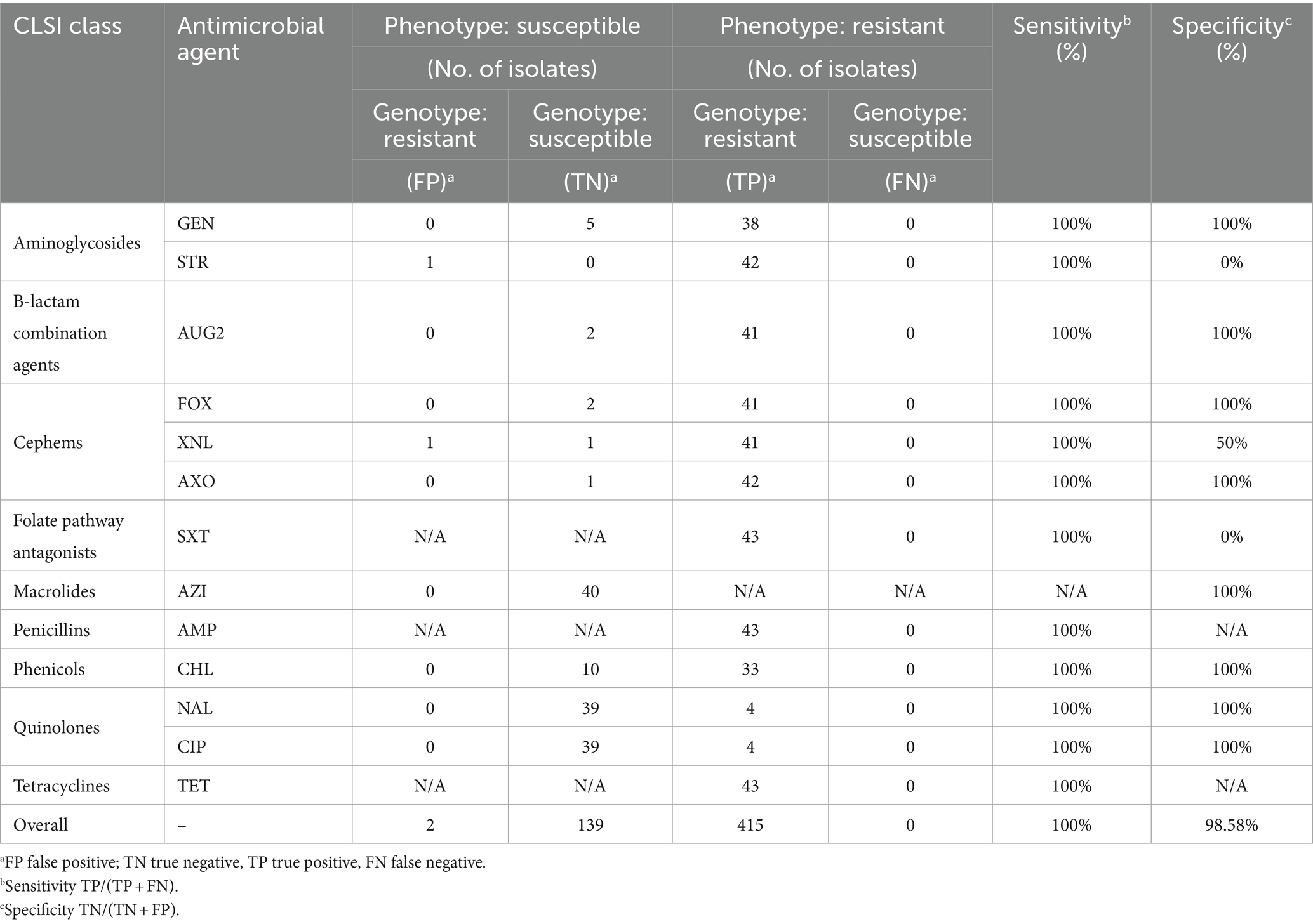
Antimicrobial susceptibility testing data previously collected on study isolates (broth microdilution using the NARMS Gram Negative panel, YCMV3AGNF) were used to assess the concordance between genotypic and phenotypic resistance (Lee et al., 2022) for the following drugs: gentamicin, streptomycin, amoxicillin-clavulanic acid, cefoxitin, ceftiofur, ceftriaxone, trimethoprim-sulfamethoxazole, azithromycin, ampicillin, chloramphenicol, nalidixic acid, ciprofloxacin, and tetracycline. Classification of isolates into susceptible, intermediate, and resistant categories were conducted using CLSI breakpoints, with the exception of streptomycin and azithromycin where NARMS breakpoints were used due to lack of CLSI breakpoints (Supplementary file). Additionally, sulfisoxazole was omitted from analysis as resistance could not be determined due to the limited range of drug dilutions in the panel. Multidrug-resistance (MDR) was defined as resistance to ≥1 drug in ≥3 antimicrobial classes (Magiorakos et al., 2012). Concordance included phenotypically resistant isolates with the corresponding ARG(s) (TP, true positive) and phenotypically susceptible isolates with absence of corresponding ARG(s) (TN, true negative). Discordance included phenotypically resistant isolates not having the corresponding ARG(s) (FN, false negative), and phenotypically susceptible isolates having the corresponding ARG(s) (FP, false positive). Sensitivity and specificity were evaluated as TP/(TP + FN) and TN/(TN + FP), respectively. For analysis, intermediate isolates were grouped with susceptible isolates.
To evaluate the co-occurrence of plasmid replicons and ARGs, a pairwise co-occurrence matrix was constructed and visualized as networks using Gephi (Bastian et al., 2009) as previously described (Lee et al., 2023). To assess the linkage patterns of ARGs and metal resistance genes, pairwise probabilistic co-occurrence analysis was conducted using default settings in the R package co-occur (Griffith et al., 2016).
3 Results
3.1 WGS of MDR Escherichia coli isolates
Escherichia coli genomes in this study had an average of 193 contigs, coverage of 112X, and quality score of 38. Additional quality metrics, AST data, and metadata of genomes in this study are available in the Supplementary file.
3.2 Concordance of AMR phenotypes with genotypes
Escherichia coli isolates in this study were previously determined to be MDR through AST. To assess AMR concordance, predictions of AMR phenotype from genotype was evaluated for 13 drugs using previously collected AST data. Across all tested drugs, genotypic AMR predicted phenotypic AMR with an overall sensitivity of 100% and specificity of 98.58% (Table 1). Discordances in specificity included a streptomycin susceptible isolate with a streptomycin resistance gene (aadA2), and a ceftiofur intermediate isolate with carriage of an AmpC beta-lactamase gene (blaCMY2).
3.3 Escherichia coli genome population structure
Whole genome analysis of the isolates revealed a large genomic diversity of E. coli genomes. All-by-all comparison identified three main clusters that exhibited minimal to no relationship to calf disease status (healthy, diarrheic, or recovered calves), treatment group (placebo or zinc), or therapeutic antimicrobial treatment (0, 1, or 2 doses of spectinomycin). A total of 20 unique sequence types (STs) based on the 7-gene allelic profile were identified among 42 isolates, with one isolate unable to be assigned to a ST. The most frequently occurring STs included ST362 (7/43, 16.28%), followed by ST10 (4/43, 9.30%), ST101 (4/43, 9.30%), and ST641 (4/43, 9.30%). STs correlated with group and individual clusters from whole genome comparisons, with distinctive variability in genome content observed within each ST (Figure 1), indicating that the genes used to define ST were stable, but the remainder of the genome contained large variations. Specifically, the most prevalent STs exhibited substantial heterogeneity in genome composition, particularly ST10, ST101, and ST641 which had variable accessory genes including those for AMR and virulence (Figures 1, 2B, 3B). This observation indicated that WGS provided higher resolution characterization of strain variation than MLST, and prompted examining the pangenome for better understanding of the gene variation among isolates in this study.
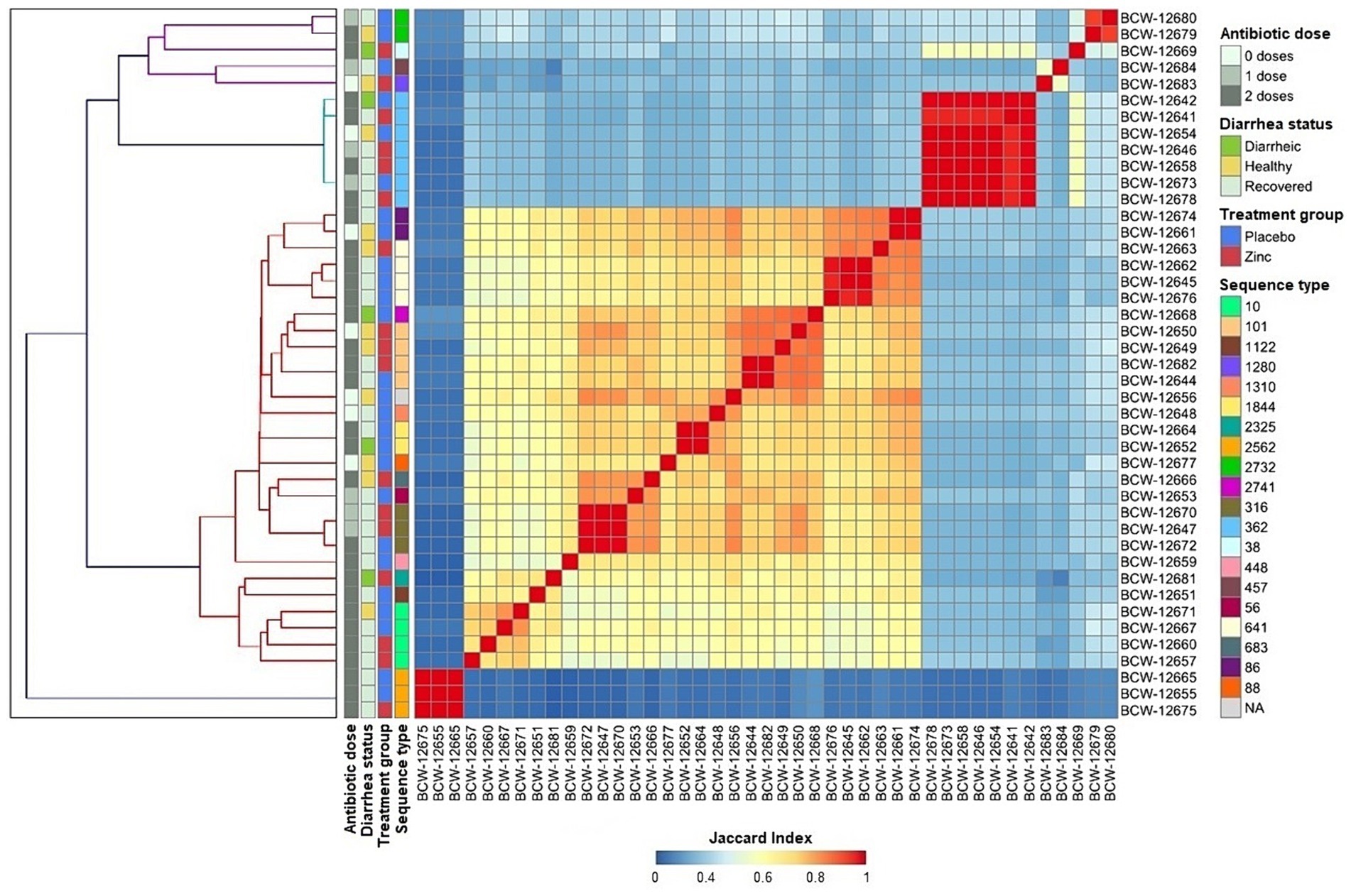
Figure 1. All-by-all comparison of genome similarity of E. coli isolates (n = 43) from pre-weaned dairy calves, generated using MinHash sketches from draft whole-genome assemblies of k-mers with a length of 31 and sketch size of 100,000. The heatmap color gradient corresponds to the Jaccard Similarity Index (JSI) for each pairwise comparison, with values close to 0 and 1 corresponding to high genome dissimilarity and similarity, respectively.
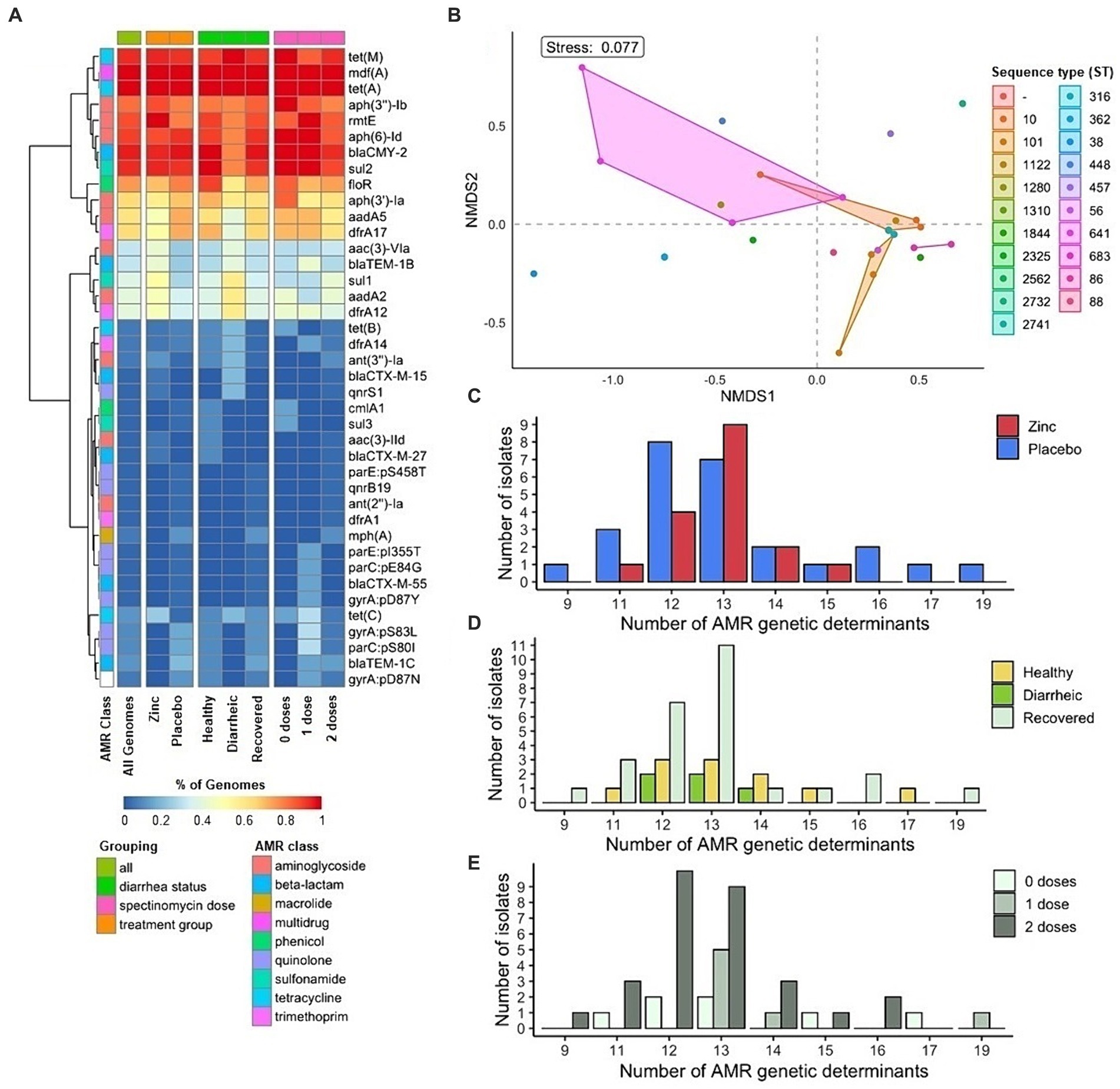
Figure 2. Antimicrobial resistance genetic determinants in fecal E. coli isolates from pre-weaned dairy calves (n = 43). (A) Heat map of ARG prevalence among isolates. (B) Non-metric multidimensional scaling of ARG composition of isolates by grouping factor of sequence type. Distribution of number of ARGs in E. coli isolates by (C) treatment group (D) diarrhea status and (E) therapeutic antibiotic exposure.
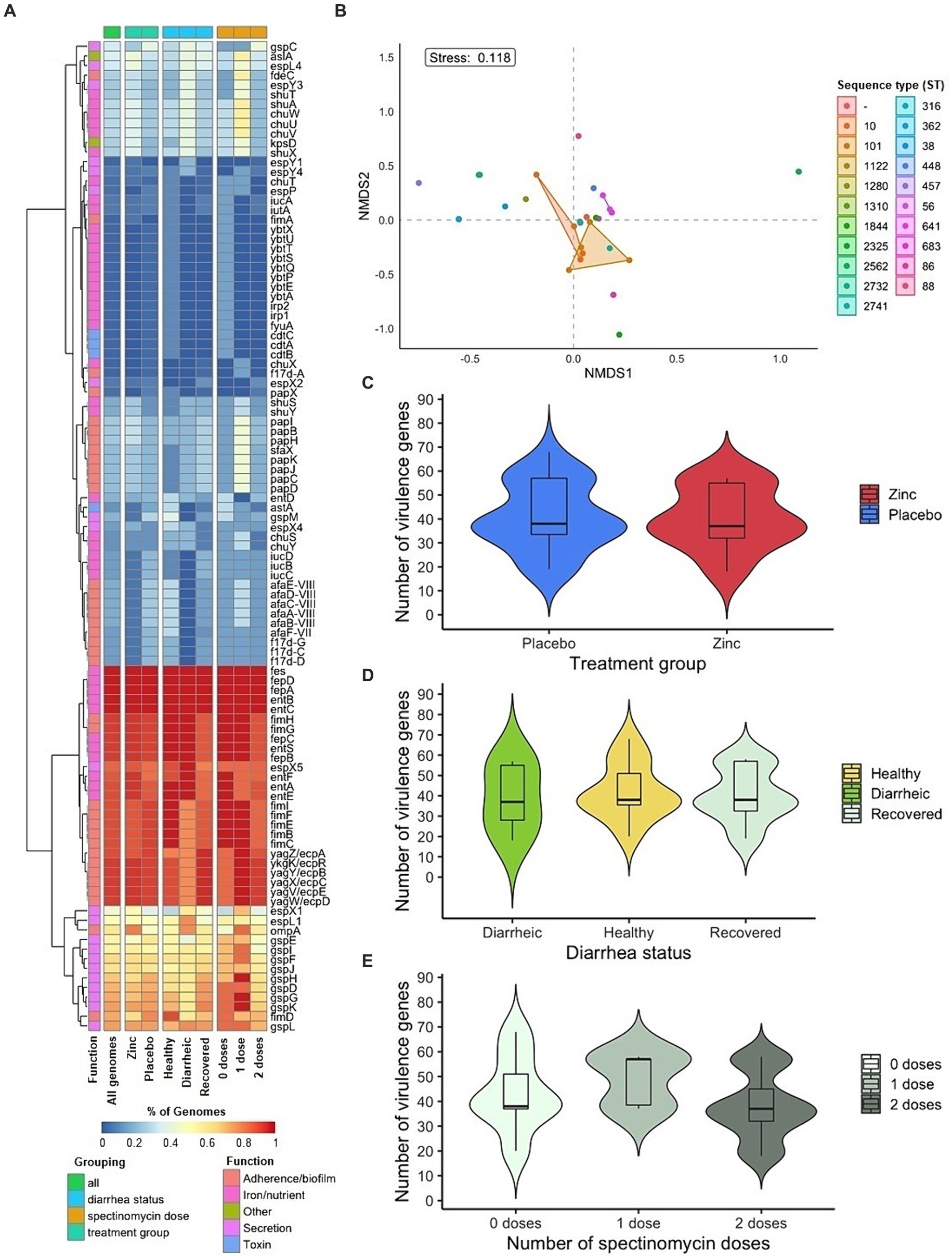
Figure 3. Virulence genes in fecal E. coli isolates from pre-weaned dairy calves (n = 43). (A) Heat map of virulence gene prevalence among isolates. (B) Non-metric multidimensional scaling of virulence gene composition in isolates by grouping factor of sequence type. Distribution of number of virulence genes in E. coli isolates by (C) treatment group (D) diarrhea status and (E) therapeutic antibiotic exposure.
3.4 Pangenome analysis of Escherichia coli isolates
The pangenome of E. coli isolates in this study was open and comprised of 14,011 genes that included a core genome with 3,117 genes and a soft-core, shell, and cloud genomes of 219, 3,076, and 7,599 genes, respectively. Analysis of the cumulative gene curve representing the number of total homologous genes and conserved homologs indicated an open pangenome that was covered with approximately 10 genomes within this population (Supplementary Figure S1). While the core was represented within a smaller portion of the isolates, genes from the variable portion of the pangenome represented 77.75% variation in the isolate population.
3.5 AMR, virulence, metal resistance, and pangenome profiles and diversity
The collective ARGs, virulence genes, metal resistance genes, and pangenome elements of E. coli were evaluated using multivariate analysis to assess if variability in these genomic profiles were driven by host-level factors. Tests for differences in E. coli genomic content for AMR, virulence, metal resistance, and pangenome elements indicated that dispersion differences were not significantly different among isolates by treatment group, diarrhea status, and therapeutic antibiotic exposure (PERMDISP2 p > 0.05, Table 2). Additionally, grouping factors evaluated in this study accounted for a low proportion of variance in AMR, virulence, metal resistance, and pangenome composition in E. coli genomes (PERMANOVA R2 = 9.39E-3-0.04), with equal or greater dissimilarities in average of ranks within group than those of between-groups across all factors (ANOSIM R = ~0 or R < 0) (Table 2). These analyses indicated that the host-level factors evaluated in this study—diarrheal disease status, dietary zinc supplementation, and antibiotic treatment—had minimal influence on the genomic composition of E. coli. These findings provided impetus to evaluate the distribution of genes individually with respect to host-level factors.
3.6 Antimicrobial resistance genetic determinants (ARGs)
Across the 43 E. coli genomes, a total of 40 ARGs among diverse antimicrobial classes were detected. The average and median number of ARGs per genome—including SNPs for quinolone resistance—was 13 ARGs with a range of 9 to 19. ARGs conferring resistance to antimicrobials of public health significance included seven SNPs in chromosomal genes—pS83L, pD87N, and pD87Y in gyrA, pS80I and pE84G in parC, and pI355T and pS458T in parE—and 2 plasmid-mediated quinolone resistance genes—qnrB19 and qnrS1—associated with quinolone resistance, and those for AmpC (blaCMY-2) and extended-spectrum (blaCTX-M-15, blaCTX-M-27, and blaCTX-M-55) beta-lactamases (ESBL). The presence of ESBL gene(s) in E. coli was significantly associated with the presence of one or more quinolone resistance determinants (p < 0.05, Fisher’s exact test).
ARGs present in more than half of the isolates included mdf(A) (43/43, 100%), aph(6)-Id (39/43, 90.7%), rmtE (38/43, 88.4%), aph(3″)-Ib (36/43, 83.7%), and aadA5 (27/43, 62.8%) for aminoglycoside resistance, blaCMY-2 (41/43, 95.3%) for beta-lactam resistance, dfrA17 (28/43, 65.1%) for trimethoprim resistance, floR (33/43, 76.7%) for phenicol resistance, sul2 (40/43, 93.0%) for sulfonamide resistance, and tet(A) (43/43, 100%) and tet(M) (40/43, 93.0%) for tetracycline resistance (Figure 2A). The average number of ARGs across all genomes and collective AMR profiles did not differ significantly by dietary zinc supplementation treatment group (zinc or placebo), diarrhea status (healthy, diarrheic, and recovered), and therapeutic antibiotic exposure (0, 1, or 2 doses) (Table 2, Figures 2C–E).
3.7 Mobile genetic elements associated with ARGs
As E. coli isolates in this study were MDR, it was of interest to investigate the mobile genetic elements associated with ARGs that may contribute to AMR co-transfer. Eighteen putative plasmids based on the presence of plasmid replicons were identified across all genomes, with a pairwise co-occurrence matrix indicating high frequency co-occurrence of AMR gene clusters with certain putative plasmids (Figure 4). The most frequently co-occurring gene network of aph(6)-Id, blaCMY-2, floR, mdf(A), sul2, and tet(A) was associated with the IncA/C2 plasmid replicon in 30 (69.8%) genomes. A second smaller network including mdf(A), rmtE, and tet(A) co-occurred with the IncFIB (AP001918) plasmid replicon at a frequency of 20 (46.5%) genomes. At a minimum threshold co-occurrence of ≥10 genomes (about 25% of the genomes), a larger network of genes including aac(3)-VIa, aadA2, dfrA12, mdf(A), rmtE, sul1, tet(A), and tet(M) were detected with IncHI2/2A plasmid replicons. Screening for plasmid replicons among genomes in this study identified unique sets of ARGs in co-occurrence with primarily large AMR plasmids.
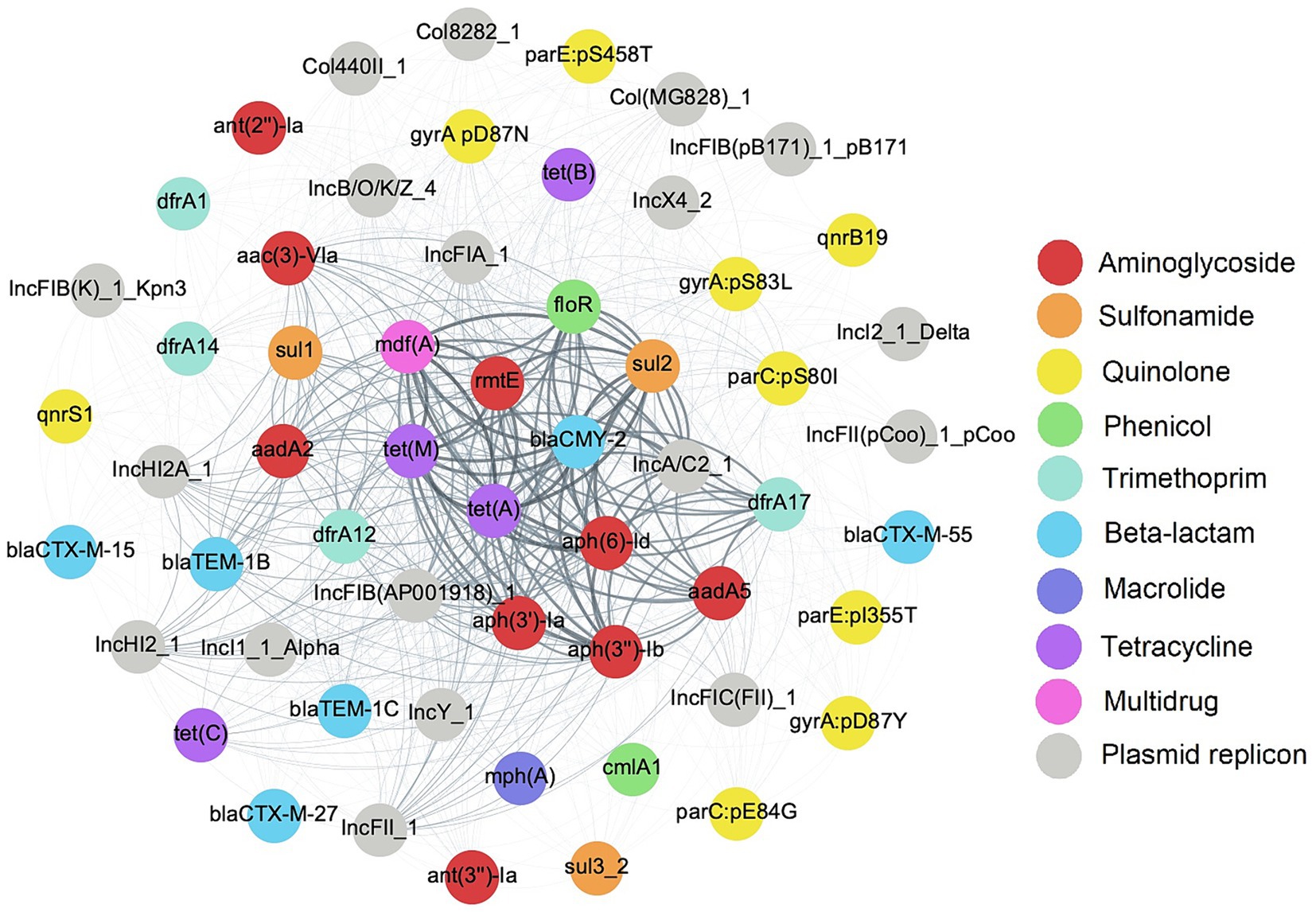
Figure 4. Co-occurrence network of plasmid replicons and antimicrobial resistance genetic determinants (ARGs) in E. coli isolates. Nodes representing ARGs are color coded by antimicrobial class and edges representing low to high frequency of co-occurrence are depicted in a light to dark color gradient.
3.8 Association between dietary zinc supplementation and genotypic AMR
The relationship between genotypic AMR and calf zinc treatment group of isolates was examined to determine the association between dietary zinc supplementation in pre-weaned dairy calves and the selection of specific ARGs. From descriptive analysis, SNPs in genes for quinolone resistance were exclusively detected in isolates from placebo calves. Antibiotic exposure-adjusted logistic regression models identified higher odds of certain ARGs in E. coli isolates from zinc-treated compared to placebo calves (dfrA12, aadA2, sul2, aac(3)-VIa, aph(3″)-Ib, blaTEM-1B, sul1, and alleles of blaCTX-M), though none of these associations were significant (OR = 1.60–2.92, p > 0.05). Conversely, there were non-significant lower odds for other ARGs and point mutations associated with quinolone resistance for isolates from zinc-treated to placebo calves [aadA5, dfrA17, floR, aph(3′)-Ia, blaCMY-2, aph(6)-Id (OR = 0.23–0.82, p > 0.05)] (Figure 5, Table 3, Supplementary Tables S1–S15).
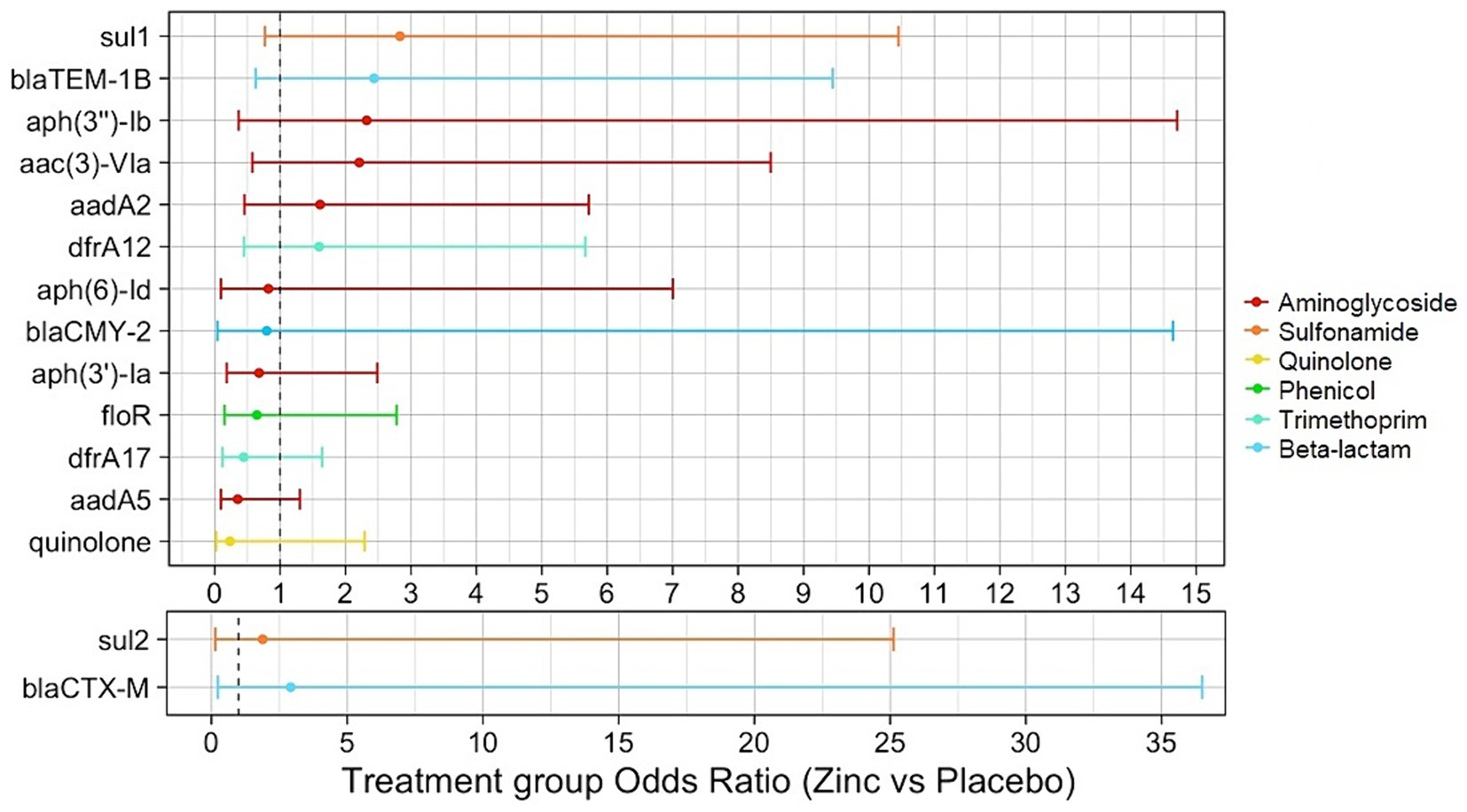
Figure 5. Antibiotic exposure-adjusted logistic regression models evaluating the association between presence of antimicrobial resistance genetic determinants (ARGs) and calf treatment group of E. coli isolates. Point estimates for each model are color-coded by antimicrobial class. Binary outcomes for quinolone and blaCTX-M models were specified as the presence/absence of any quinolone resistance mechanism (plasmid-mediated genes or point mutations) and the presence/absence of any blaCTX-M alleles, respectively.
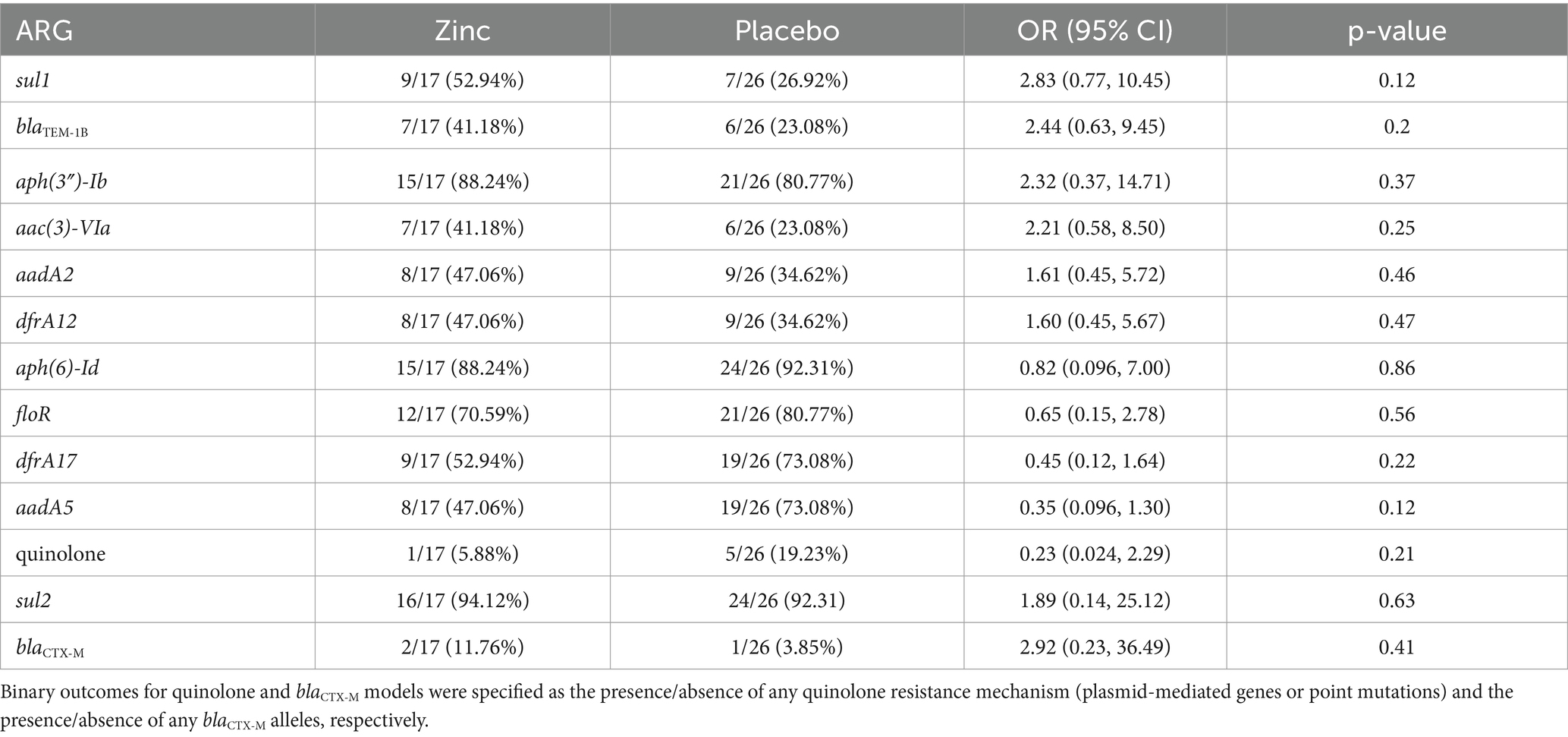
Table 3. Distribution of antimicrobial resistance genetic determinants (ARGs) in E. coli by calf treatment group and treatment group point estimates (zinc vs. placebo) from antibiotic exposure-adjusted logistic regression models.
3.9 Virulence genes
A total of 103 virulence genes corresponding to adherence/biofilm formation (n = 36), iron/nutrient acquisition (n = 40), secretion (n = 21), toxin (n = 4), and other functions (n = 2) were detected across E. coli genomes. The average and median number of virulence genes were 40.58 and 38, respectively (range of 18 to 68). Five virulence genes related to enterobactin (entB, entC, fepA, fepD and fes) were detected across all isolates (Figure 3A). The number of virulence genes and collective virulence profiles across genomes did not differ significantly by dietary zinc supplementation treatment group, diarrhea status, and therapeutic antibiotic exposure (Table 2, Figures 3C–E).
Virulence genes from the afa-7 and afa-8 clusters (afaA-E) encoding afimbrial adhesins were detected primarily in isolates from placebo calves (85.71%, 6/7), with the full gene set present in six isolates. Other virulence genes detected related to colonization included those encoding F17 fimbriae (f17d-A, f17d-C, f17d-D, and f17d-G) in six isolates, and P fimbriae in 12 isolates (pap genes) (Bertin et al., 2000; Bihannic et al., 2014; Ryu et al., 2020). Additionally, genes in the fim cluster (fimA-I) encoding type 1 fimbriae were present in the majority of isolates, though only one isolate harbored the fimA structural gene and three isolates the fimH adhesin gene. Major virulence genes related to secretion included those corresponding to Type II (gsp) and Type III (esp) secretion systems. Virulence genes for toxins, astA (enteroaggregative heat-stable enterotoxin) and/or cdtABC (cytolethal distending toxin), were identified in isolate(s) from pre–and post-diarrheic calves. Overall, virulence genes were interspersed in the population across calf zinc treatment group and diarrhea status. The largest number and diversity of virulence genes identified corresponded to iron/nutrient acquisition, including genes chuSTUVWXY (heme uptake), entA-F (enterobactin), fepA-D (enterobactin), fyuA (yersiniabactin receptor), iucABCD-iutA (aerobactin), and those in the ybt operon (yersiniabactin) (Figure 3A).
3.10 Association between AMR and metal resistance genes
A total of 153 metal resistance genes (MRGs) were identified across all E. coli genomes examined, with the average and median number of MRGs per genome being 128.42 and 127, respectively, with a range of 123 to 135. Co-occurrence analysis of ARGs and metal resistance genes included 16,585 gene pairs and identified 96 positive and 77 negative co-occurrences. Positive associations including both ARGs and metal resistance were observed between aminoglycoside (aac(3)-VIa, aadA2), beta-lactam (blaTEM-1B), sulfonamide (sul1), and trimethoprim (dfrA12) resistance and acid (gadA and gadB), tellurium (terZ and terW) and mercury (merT) resistance genes (Figure 6).
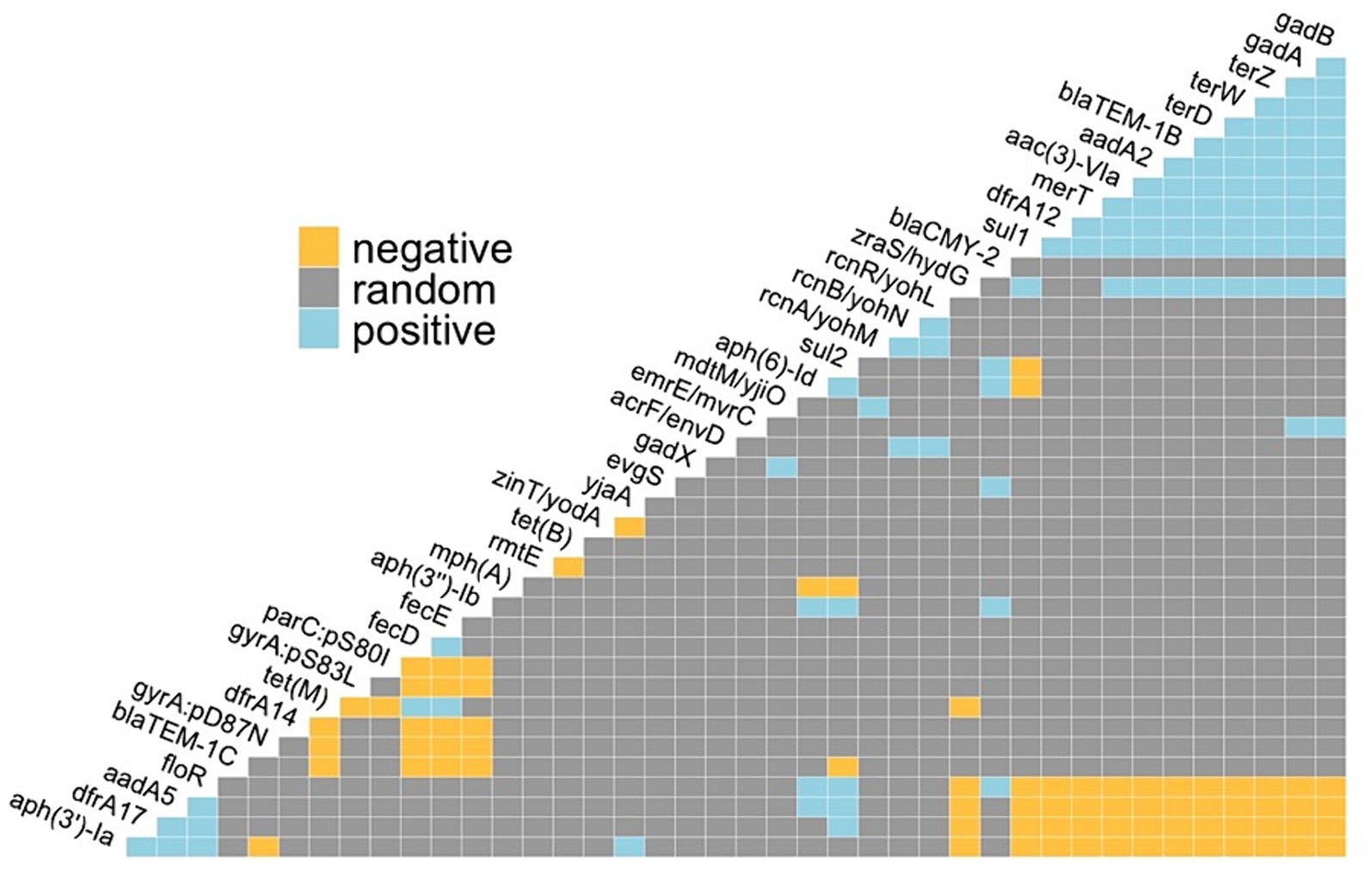
Figure 6. Pairwise probabilistic co-occurrence analysis for positive, negative, or random associations between antimicrobial resistance genetic determinants and metal resistance genes in E. coli isolates.
4 Discussion
The enteric microbiota serves as a symbiotic partner of the host, with crucial roles in intestinal health, metabolism, and host immune response (Casadevall and Pirofski, 2000; Kaiko and Stappenbeck, 2014; Jandhyala et al., 2015). The acquisition and loss of genes—such as those for AMR—in enteric microbes like E. coli can occur as adaptive responses to environmental (e.g., dysbiosis) and host changes (e.g., diet and disease). In this study, we investigated the host-microbe interface of enteric MDR E. coli from pre-weaned dairy calves to evaluate potential contributing factors to MDR persistence and better understand the relationship between genomic composition and host-level factors of antimicrobial exposure, dietary zinc supplementation, and calf diarrheal disease.
Whole genome sequence analysis revealed high genome variability and an open pangenome of multidrug-resistant (MDR) E. coli from dairy calves in this study. The diverse population structure of E. coli has been well-documented, with the frequent acquisition, loss, and modification of genes contributing to its large gene pool, fitness, and competitive ability to thrive in widespread geographical and host environments (Horesh et al., 2021). From all-by-all comparisons of the WGS, the isolates in this study clustered by sequence type (ST) but not host-level factors of disease status, dietary influences, or antimicrobial exposure. Common STs identified included ST362, a frequently occurring ST in calves that has been associated with extra intestinal infections (Falgenhauer et al., 2017; Vieille et al., 2019; Homeier-Bachmann et al., 2022). Other prevalent STs were those with zoonotic potential, such as ST641 which has been isolated from poultry and goat sources (Cortés et al., 2010; Zhuge et al., 2021; Treilles et al., 2023), ST10, a widespread lineage of pathogenic and commensal E. coli which are prominently MDR in animal populations (Haley et al., 2023; Silva et al., 2023; Wang et al., 2023), and ST101, another frequently occurring MDR clonal group frequently detected in food, water, food animal, and human matrices (Umpiérrez et al., 2017; Zhong et al., 2019; Sauget et al., 2023; Silva et al., 2023).
The accessory genome of E. coli encodes various characteristics for survival and reproduction, including those related to AMR (Hall et al., 2021). The early developing microbiota of calves has been observed to harbor high prevalence and diversity of ARGs (Liu et al., 2019; Haley et al., 2023), which is corroborated by the large assortment of ARGs detected in our E. coli genomes. In this study, the number of ARGs in E. coli did not correlate with antimicrobial use. However, the presence of ARGs corresponding to certain antimicrobial classes were consistent with the AMR selection pressures in our study; the high prevalence and diversity of tetracycline and aminoglycoside ARGs detected in E. coli genomes was reflective of the tetracycline and neomycin administered in dietary milk and spectinomycin for the therapeutic treatment of diarrhea. The same tetracycline and aminoglycoside ARGs in our study are frequently present in E. coli from calves (Jeamsripong et al., 2021; Salaheen et al., 2023), even from those with no previous antibiotic exposure (Liu et al., 2019). Similarly, other ARGs identified in our study including those encoding beta-lactam, phenicol, trimethoprim, and/or sulfonamide resistance have also been previously reported in calves (Haley et al., 2023; Salaheen et al., 2023), suggesting that the collection of ARGs in our study is representative of the enteric calf resistome during early life.
A major mechanism of third-generation cephalosporin resistance in Salmonella and E. coli from food and food-producing animals is AmpC-type beta-lactamase blaCMY-2, which was detected in almost every E. coli genome in this study, despite the lack of beta-lactam use in calves. The occurrence of blaCMY-2 in dairy cattle has been presumed to be from frequent use of ceftiofur for the intramammary treatment of mastitis and parenteral treatment of acute metritis and bacterial pneumonia (Durel et al., 2019). However, studies have found limited evidence for the direct dissemination of blaCMY-2 through ceftiofur use (Daniels et al., 2009; Schmidt et al., 2013) or associations between recent ceftiofur treatment and reduced-susceptible E. coli at the individual cow level (Tragesser et al., 2006). We found a high frequency of a co-occurrence networks with blaCMY-2, ARGs corresponding to aminoglycoside, phenicol, sulfonamide, and tetracycline resistance, and the IncA/C2 plasmid replicon in our study isolates. These data support observations from other studies, in which the occurrence of blaCMY-2 in absence of cephalosporin use has been postulated to be from its acquisition on large MDR plasmids, followed by clonal expansion and/or the presence of indirect and co-selective AMR pressures that maintain these plasmids at the herd-level (Alcaine et al., 2005; Subbiah et al., 2011; Martin et al., 2012; Schmidt et al., 2013; Deng et al., 2015). Other beta-lactam ARGs conferring resistance to cephalosporins found in this study included ESBL genes from the blaCTX-M family (blaCTX-M-15, blaCTX-M-27, and blaCTM-M-55) from three E. coli genomes. In addition to being resistant to third-generation cephalosporins, ESBL – producing E. coli have important clinical consequences as they are frequently MDR to other critically important antimicrobials such as quinolones (Zurfluh et al., 2014; Azargun et al., 2018; Furmanek-Blaszk et al., 2023), a finding that is corroborated through the significant association observed between the presence of ESBL and quinolone resistance determinants among E. coli in this study.
While antimicrobial use is perceived as a main driver of AMR, non-antimicrobial factors such as heavy metal exposure have also been recognized to influence AMR selection. Heavy metals such as zinc are frequently used as growth promoters or therapeutic agents in livestock species (Yazdankhah et al., 2014); for example, dietary zinc supplementation in pre-weaned calves may be used to reduce the burden of diarrheal disease and promote calf growth (Glover et al., 2013; Feldmann et al., 2019; Chang et al., 2020; Liu et al., 2023; Yu et al., 2023). Little is known on the influence of dietary zinc on genomic AMR in cattle, however a previous study in swine found that high zinc in feed (2.5 g/kg) significantly increased intestinal abundance of tetracycline and sulfonamide ARGs (Vahjen et al., 2015). As all E. coli genomes in our study had tet genes, we were unable to evaluate the selection of tetracycline ARGs. Adjusted logistic regression models found higher odds ratios for the presence of sulfonamide genes— sul1 (OR = 2.83, 95% CI 0.77–10.45) and sul2 (OR = 1.89, 95% CI 0.14–25.12)—in E. coli from zinc compared to those from placebo calves. Although these findings were not statistically significant, the direction of associations support the aforementioned findings of potential sulfonamide ARG selection from dietary zinc (Vahjen et al., 2015). We also found unique SNPs in the genes conferring quinolone resistance in isolates from placebo treated calves, suggesting an antagonistic effect of zinc on certain classes of ARGs. However, the lower odds ratio for the presence of quinolone ARGs from logistic regression in isolates from zinc compared to placebo treated calves (OR = 0.23 95% CI 0.02–2.29) was also not statistically significant. These non-significant findings may be attributed to the small sample size of isolates in our study that may have resulted in inadequate power to detect differences in addition to other uncharacterized variables. Hence, future studies employing larger sample sizes are needed to ascertain the relationship between zinc exposure and ARG selection, particularly for those in our study (sul2, blaCMY-2, aph(3″)-Ib, and blaCTX-M alleles) with large confidence intervals for point estimates.
Beyond the selection of individual ARGs, co-selection of both ARGs and metal resistance genes may occur through co-resistance, a phenomenon where dissimilar mechanisms for both resistances are selected due to their genetic linkage (Wales and Davies, 2015). The linkage of ARGs and metal resistance genes has been well documented (Baker-Austin et al., 2006; Wales and Davies, 2015; Nguyen et al., 2019). Patterns in ARG and metal resistance gene co-occurrence have been observed to vary depending on the organism, host, and location (Poole, 2017). In our study, we identified several positive co-occurrences between ARGs encoding resistance to beta-lactam, sulfonamide, aminoglycoside and trimethoprim and metal resistance genes encoding mercury and tellurium resistance. Positive co-occurrences between these ARGs from the same classes of antimicrobials and mercury and tellurium resistance genes were previously reported in fecal E. coli from dairy herds from Pennsylvania (Haley et al., 2023). These data suggest that specific AMR and metal resistance genes are pervasive and selected for in dairy cattle and their farm environments irrespective of geographical location.
In addition to evaluating potential host-level drivers of AMR in calf E. coli, this study compared genotypic AMR—the presence of ARGs and point mutations conferring quinolone resistance—with phenotypic AMR data from antimicrobial susceptibility testing. Genotypic AMR exhibited a high degree of concordance with phenotypic AMR for genomically heterogeneous MDR isolates in this study. Despite the small sample size of isolates (n = 43) from one host (dairy calves) and source (single dairy farm), our findings are consistent with previous work evaluating genotypic and phenotypic concordance in E. coli and Salmonella from cattle and/or food animal sources (Tyson et al., 2015; McDermott et al., 2016; Carroll et al., 2021; Lee et al., 2022, 2023). Discrepancies for streptomycin and ceftiofur as observed in two isolates in this study have been frequently reported (Tyson et al., 2015; McDermott et al., 2016; Lee et al., 2023), and may be a result of lack of CLSI breakpoints for these veterinary drugs, technical variability in AST/WGS processes (e.g., 2-fold variations in MIC from AST at intermediate and resistant cut-off thresholds), and choice of classifying intermediate isolates. As an example of the latter, grouping of intermediate and susceptible isolates for analysis resulted in discrepancy of a ceftiofur immediate isolate in this study; the genotypic and phenotypic AMR for this isolate would have been congruent if intermediate isolates were instead treated as resistant.
Diarrheal disease status of calves was not significantly associated with genomic variability in this study, including virulence profiles. E. coli can be categorized into various pathotypes depending on the presence of certain virulence attributes (Kaper et al., 2004), with common pathotypes associated with neonatal calf diarrhea including enteropathogenic (EPEC), Shiga toxin-producing (STEC), enterotoxigenic (ETEC), and enteroaggregative (EAEC) E. coli (Awad et al., 2020). While MDR E. coli isolates in this study lacked the comprehensive virulence markers of these diarrheagenic pathotypes, they encoded a wide diversity of virulence genes that overlap with those in pathogenic strains. For instance, adhesin virulence genes observed in our study, fim and pap genes encoding Type I fimbriae and P fimbriae respectively, are associated with various pathotypes in both humans and animals (Bertin et al., 2000; Sarowska et al., 2019), and f17 genes encoding F17 fimbriae and afa-7 and afa-8 gene clusters encoding afimbrial adhesion appear to be more host-specific and predominant in bovine E. coli associated with diarrhea and septicemia (Lalioui and Le Bouguénec, 2001; Bihannic et al., 2014; Shahrani et al., 2014). Additionally, detected in a few isolates were cdtABC and astA encoding cytolethal distending toxin (CDT) and enteroaggregative heat-stable enterotoxin (EAST1), which are typically present in EPEC and ETEC, respectively (Yamamoto and Echeverria, 1996; Osek, 2003; Gomes et al., 2016; Meza-Segura et al., 2017). The presence of some but not all pathotype-determining virulence genes in E. coli from our study suggests that the acquisition and loss of genes (e.g., from host and environmental influences) may dynamically contribute to the virulence potential of E. coli and their ability to cause diarrheal disease.
The most abundant virulence genes in MDR E. coli in this study were those involved in iron acquisition (e.g., sideophores and heme uptake). Iron plays a critical role in microbial metabolic processes and cell division, and its acquisition is an important host–microbe interaction that contributes to bacterial survival and pathogen infection (Caza and Kronstad, 2013; Nairz and Weiss, 2020). Previous studies identified several iron acquisition systems— some of which were also identified in our study (e.g., iucABCD-iutA)— to be significantly enriched in MDR bovine E. coli (Haley et al., 2023, 2024). Virulence factors and ARGs are essential for bacteria to overcome host immune responses and antimicrobial exposure, respectively. The simultaneous carriage of both in MDR E. coli may confer a fitness advantage in adverse conditions, promoting the co-selection and maintenance of these genes in MDR isolates as opposed to their susceptible counterparts (Beceiro et al., 2013). Moreover, the pre-weaned calf diet is primarily composed of milk, which is nutritionally negligible in iron and may contribute to a low-iron environment in the calf gut that has been hypothesized to favor the selection of MDR E. coli with more extensive repertoires of iron acquisition systems (Haley et al., 2023, 2024). During infection and disease, host-driven iron sequestration occurs as an immune defense strategy to inhibit the growth of pathogens (Beceiro et al., 2013; Nairz and Weiss, 2020). As E. coli genomes in our study were from pre-weaned calves in various stages of diarrheal disease (pre-diarrheic, diarrheic, and recovered), we hypothesize that host-mediated iron withdrawal is another factor which may further favor the survival of MDR E. coli with high iron-scavenging capacity.
In conclusion, our analysis indicated that the genomes of MDR E. coli from pre-weaned dairy calves were highly diverse and minimally driven by the host-level factors evaluated in this study (dietary zinc supplementation, therapeutic antimicrobial treatment, and diarrhea disease status). Key limitations include the relatively small sample size of isolates and the absence of a susceptible and/or non-MDR group of E. coli genomes for comparison. Future work that evaluates longitudinal effects would provide greater insight on the relationship between genomic diversity and factors such as disease—which may occur in progressive stages—and antimicrobial exposure, which can rapidly and transiently impact the gut microbiome. Our findings corroborate previous reports of MDR E. coli from calves harboring diverse ARGs conferring resistance to clinically important drugs, enriched abundance of virulence factors for iron acquisition systems, and linkage of certain metal resistance genes and ARGs. These data suggest that the selection and persistence of MDR E. coli in calves are adaptive and attributed to the presence of these and/or other unidentified genes that confer a fitness advantage in the calf gut.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material. WGS data for isolates in this study are available at the 100K Pathogen Genome Project BioProject (PRJNA1142533).
Author contributions
KL: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft, Writing – review & editing. CS: Data curation, Formal analysis, Methodology, Writing – review & editing, Software. SA: Conceptualization, Data curation, Investigation, Methodology, Resources, Writing – review & editing. BH: Writing – review & editing, Data curation, Methodology. XL: Writing – review & editing, Conceptualization, Resources. EA: Writing – review & editing, Conceptualization, Resources. BW: Writing – review & editing, Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. KL was supported by the Austin Eugene Lyons fellowship from the University of California, Davis. This study was additionally made possible in part by the Western Center for Agricultural Health and Safety (WCAHS) Small Grant Program (Project PI: KL) through a larger CDC/NIOSH grant #U54OH007550 and the 100K Pathogen Genome Project (PI: BW).
Acknowledgments
We would like to thank the Weimer lab for their helpful feedback and intellectual discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1420300/full#supplementary-material
References
Aguilar-Zamora, E., Weimer, B. C., Torres, R. C., Gómez-Delgado, A., Ortiz-Olvera, N., Aparicio-Ozores, G., et al. (2022). Molecular epidemiology and antimicrobial resistance of Clostridioides difficile in hospitalized patients from Mexico. Front. Microbiol. 12:787451. doi: 10.3389/fmicb.2021.787451
Alcaine, S. D., Sukhnanand, S. S., Warnick, L. D., Su, W. L., McGann, P., McDonough, P., et al. (2005). Ceftiofur-resistant Salmonella strains isolated from dairy farms represent multiple widely distributed subtypes that evolved by independent horizontal gene transfer. Antimicrob. Agents Chemother. 49, 4061–4067. doi: 10.1128/AAC.49.10.4061-4067.2005
Alcock, B. P., Huynh, W., Chalil, R., Smith, K., Raphenya, A., Wlodarski, M., et al. (2020a). CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res 51, D690–D699. doi: 10.1093/nar/gkac920
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020b). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Anderson, M. J. (2006). Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62, 245–253. doi: 10.1111/j.1541-0420.2005.00440.x
Anderson, M. J., Ellingsen, K. E., and McArdle, B. H. (2006). Multivariate dispersion as a measure of beta diversity. Ecol. Lett. 9, 683–693. doi: 10.1111/j.1461-0248.2006.00926.x
Anderson, M. J., and Walsh, D. C. I. (2013). PERMANOVA, ANOSIM, and the mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol. Monogr. 83, 557–574. doi: 10.1890/12-2010.1
Awad, W. S., El-Sayed, A. A., Mohammed, F. F., Bakry, N. M., Abdou, N.-E. M. I., and Kamel, M. S. (2020). Molecular characterization of pathogenic Escherichia coli isolated from diarrheic and in-contact cattle and buffalo calves. Trop. Anim. Health Prod. 52, 3173–3185. doi: 10.1007/s11250-020-02343-1
Azargun, R., Sadeghi, M. R., Soroush Barhaghi, M. H., Samadi Kafil, H., Yeganeh, F., Ahangar Oskouee, M., et al. (2018). The prevalence of plasmid-mediated quinolone resistance and ESBL-production in Enterobacteriaceae isolated from urinary tract infections. Infect Drug Resist. 11, 1007–1014. doi: 10.2147/IDR.S160720
Baker-Austin, C., Wright, M. S., Stepanauskas, R., and McArthur, J. V. (2006). Co-selection of antibiotic and metal resistance. Trends Microbiol. 14, 176–182. doi: 10.1016/j.tim.2006.02.006
Bandoy, D. D. R., and Weimer, B. C. (2020). Biological machine learning combined with Campylobacter population genomics reveals virulence gene allelic variants cause disease. Microorganisms 8:549. doi: 10.3390/microorganisms8040549
Bastian, M., Heymann, S., and Jacomy, M. (2009). Gephi: An open source software for exploring and manipulating networks. Proc. Int. AAAI Conf Web Soc. Media 3, 361–362. doi: 10.1609/icwsm.v3i1.13937
Beceiro, A., Tomás, M., and Bou, G. (2013). Antimicrobial resistance and virulence: a successful or deleterious Association in the Bacterial World? Clin. Microbiol. Rev. 26, 185–230. doi: 10.1128/CMR.00059-12
Berchtold, J. F., and Constable, P. D. (2009). Antibiotic treatment of diarrhea in Preweaned calves. Food Animal Practice 2009, 520–525. doi: 10.1016/B978-141603591-6.10103-4
Berge, A. C. B., Atwill, E. R., and Sischo, W. M. (2005). Animal and farm influences on the dynamics of antibiotic resistance in faecal Escherichia coli in young dairy calves. Prev. Vet. Med. 69, 25–38. doi: 10.1016/j.prevetmed.2005.01.013
Berge, A. C., Hancock, D. D., Sischo, W. M., and Besser, T. E. (2010). Geographic, farm, and animal factors associated with multiple antimicrobial resistance in fecal Escherichia coli isolates from cattle in the western United States. J. Am. Vet. Med. Assoc. 236, 1338–1344. doi: 10.2460/javma.236.12.1338
Bertin, Y., Girardeau, J.-P., Darfeuille-Michaud, A., and Martin, C. (2000). Epidemiological study of pap genes among Diarrheagenic or septicemic Escherichia coli strains producing CS31A and F17 Adhesins and characterization of Pap31A fimbriae. J. Clin. Microbiol. 38, 1502–1509. doi: 10.1128/JCM.38.4.1502-1509.2000
Bihannic, M., Ghanbarpour, R., Auvray, F., Cavalié, L., Châtre, P., Boury, M., et al. (2014). Identification and detection of three new F17 fimbrial variants in Escherichia coli strains isolated from cattle. Vet. Res. 45:76. doi: 10.1186/s13567-014-0076-9
Braz, V. S., Melchior, K., and Moreira, C. G. (2020). Escherichia coli as a multifaceted pathogenic and versatile bacterium. Front. Cell. Infect. Microbiol. 10:548492. doi: 10.3389/fcimb.2020.548492
Brown, C. T., and Irber, L. (2016). Sourmash: a library for MinHash sketching of DNA. J. Open Source Softw. 1:27. doi: 10.21105/joss.00027
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/aac.02412-14
Carroll, L. M., Buehler, A. J., Gaballa, A., Siler, J. D., Cummings, K. J., Cheng, R. A., et al. (2021). Monitoring the microevolution of Salmonella enterica in healthy dairy cattle populations at the individual farm level using whole-genome sequencing. Front. Microbiol. 12:763669. doi: 10.3389/fmicb.2021.763669
Casadevall, A., and Pirofski, L. (2000). Host-pathogen interactions: basic concepts of microbial commensalism, colonization, infection, and disease. Infect. Immun. 68, 6511–6518. doi: 10.1128/IAI.68.12.6511-6518.2000
Caza, M., and Kronstad, J. (2013). Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front. Cell. Infect. Microbiol. 3:80. doi: 10.3389/fcimb.2013.00080
Chang, M. N., Wei, J. Y., Hao, L. Y., Ma, F. T., Li, H. Y., Zhao, S. G., et al. (2020). Effects of different types of zinc supplement on the growth, incidence of diarrhea, immune function, and rectal microbiota of newborn dairy calves. J. Dairy Sci. 103, 6100–6113. doi: 10.3168/jds.2019-17610
Chen, L., Zheng, D., Liu, B., Yang, J., and Jin, Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 44, D694–D697. doi: 10.1093/nar/gkv1239
Cheng, G., Ning, J., Ahmed, S., Huang, J., Ullah, R., An, B., et al. (2019). Selection and dissemination of antimicrobial resistance in Agri-food production. Antimicrob. Resist. Infect. Control 8:158. doi: 10.1186/s13756-019-0623-2
Cortés, P., Blanc, V., Mora, A., Dahbi, G., Blanco, J. E., Blanco, M., et al. (2010). Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl. Environ. Microbiol. 76, 2799–2805. doi: 10.1128/AEM.02421-09
Daniels, J. B., Call, D. R., Hancock, D., Sischo, W. M., Baker, K., and Besser, T. E. (2009). Role of Ceftiofur in selection and dissemination of blaCMY-2-mediated cephalosporin resistance in Salmonella enterica and commensal Escherichia coli isolates from cattle. Appl. Environ. Microbiol. 75, 3648–3655. doi: 10.1128/AEM.02435-08
Deng, H., Si, H.-B., Zeng, S.-Y., Sun, J., Fang, L.-X., Yang, R.-S., et al. (2015). Prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in a farrowing farm: ST1121 clone harboring IncHI2 plasmid contributes to the dissemination of blaCMY-2. Front. Microbiol. 6:1210. doi: 10.3389/fmicb.2015.01210
Depenbrock, S., Schlesener, C., Aly, S., Williams, D., ElAshmawy, W., McArthur, G., et al. (2024). Antimicrobial resistance genes in respiratory Bacteria from weaned dairy heifers. Pathogens 13:300. doi: 10.3390/pathogens13040300
Durel, L., Gallina, G., and Pellet, T. (2019). Assessment of ceftiofur residues in cow milk using commercial screening test kits. Vet. Rec. Open 6:e000329. doi: 10.1136/vetreco-2018-000329
Falgenhauer, L., Ghosh, H., Doijad, S., Yao, Y., Bunk, B., Spröer, C., et al. (2017). Genome analysis of the Carbapenem- and Colistin-resistant Escherichia coli isolate NRZ14408 reveals horizontal gene transfer pathways towards Panresistance and enhanced virulence. Antimicrob. Agents Chemother. 61, e02359–e02316. doi: 10.1128/AAC.02359-16
Feldmann, H. R., Williams, D. R., Champagne, J. D., Lehenbauer, T. W., and Aly, S. S. (2019). Effectiveness of zinc supplementation on diarrhea and average daily gain in pre-weaned dairy calves: a double-blind, block-randomized, placebo-controlled clinical trial. PLoS One 14:e0219321. doi: 10.1371/journal.pone.0219321
Flores-Valdez, M., Ares, M. A., Rosales-Reyes, R., Torres, J., Girón, J. A., Weimer, B. C., et al. (2021). Whole genome sequencing of pediatric Klebsiella pneumoniae strains reveals important insights into their virulence-associated traits. Front. Microbiol. 12:711577. doi: 10.3389/fmicb.2021.711577
Formenti, N., Martinelli, C., Vitale, N., Giovannini, S., Salogni, C., Tonni, M., et al. (2021). Antimicrobial resistance of Escherichia coli in dairy calves: a 15-year retrospective analysis and comparison of treated and untreated animals. Animals 11:2328. doi: 10.3390/ani11082328
Furmanek-Blaszk, B., Sektas, M., and Rybak, B. (2023). High prevalence of plasmid-mediated quinolone resistance among ESBL/AmpC-producing Enterobacterales from free-living birds in Poland. Int. J. Mol. Sci. 24:12804. doi: 10.3390/ijms241612804
Glover, A. D., Puschner, B., Rossow, H. A., Lehenbauer, T. W., Champagne, J. D., Blanchard, P. C., et al. (2013). A double-blind block randomized clinical trial on the effect of zinc as a treatment for diarrhea in neonatal Holstein calves under natural challenge conditions. Prev. Vet. Med. 112, 338–347. doi: 10.1016/j.prevetmed.2013.09.001
Gomes, T. A. T., Elias, W. P., Scaletsky, I. C. A., Guth, B. E. C., Rodrigues, J. F., Piazza, R. M. F., et al. (2016). Diarrheagenic Escherichia coli. Braz. J. Microbiol. 47, 3–30. doi: 10.1016/j.bjm.2016.10.015
Griffith, D. M., Veech, J. A., and Marsh, C. J. (2016). Cooccur: probabilistic species co-occurrence analysis in R. J. Stat. Soft. 69, 1–17. doi: 10.18637/jss.v069.c02
Habing, G., Harris, K., Schuenemann, G. M., Piñeiro, J. M., Lakritz, J., and Clavijo, X. A. (2017). Lactoferrin reduces mortality in preweaned calves with diarrhea. J. Dairy Sci. 100, 3940–3948. doi: 10.3168/jds.2016-11969
Haley, B. J., Kim, S.-W., Salaheen, S., Hovingh, E., and Van Kessel, J. A. S. (2020). Differences in the microbial community and Resistome structures of feces from Preweaned calves and lactating dairy cows in commercial dairy herds. Foodborne Pathog. Dis. 17, 494–503. doi: 10.1089/fpd.2019.2768
Haley, B. J., Kim, S. W., Salaheen, S., Hovingh, E., and Van Kessel, J. A. S. (2023). Genome-wide analysis of Escherichia coli isolated from dairy animals identifies virulence factors and genes enriched in multidrug-resistant strains. Antibiotics 12:1559. doi: 10.3390/antibiotics12101559
Haley, B. J., Salaheen, S., Kim, S. W., and Kessel, J. A. V. (2024). Virulome analysis of Escherichia coli ST117 from bovine sources identifies similarities and differences with strains isolated from other food animals. PLoS One 19:e0296514. doi: 10.1371/journal.pone.0296514
Haley, B. J., and Van Kessel, J. A. S. (2022). The resistome of the bovine gastrointestinal tract. Curr. Opin. Biotechnol. 73, 213–219. doi: 10.1016/j.copbio.2021.07.025
Hall, R. J., Whelan, F. J., Cummins, E. A., Connor, C., McNally, A., and McInerney, J. O. (2021). Gene-gene relationships in an Escherichia coli accessory genome are linked to function and mobility. Microb. Genom. 7:000650. doi: 10.1099/mgen.0.000650
Hernández-Juárez, L. E., Camorlinga, M., Méndez-Tenorio, A., Calderón, J. F., Huang, B. C., Bandoy, D. D. R., et al. (2021). Analyses of publicly available Hungatella hathewayi genomes revealed genetic distances indicating they belong to more than one species. Virulence 12, 1950–1964. doi: 10.1080/21505594.2021.1950955
Higdon, S. M., Huang, B. C., Bennett, A. B., and Weimer, B. C. (2020). Identification of nitrogen fixation genes in Lactococcus isolated from maize using population genomics and machine learning. Microorganisms 8:2043. doi: 10.3390/microorganisms8122043
Homeier-Bachmann, T., Kleist, J. F., Schütz, A. K., and Bachmann, L. (2022). Distribution of ESBL/AmpC-Escherichia coli on a dairy farm. Antibiotics 11:940. doi: 10.3390/antibiotics11070940
Horesh, G., Blackwell, G. A., Tonkin-Hill, G., Corander, J., Heinz, E., and Thomson, N. R. (2021). A comprehensive and high-quality collection of Escherichia coli genomes and their genes. Microb. Genom. 7:000499. doi: 10.1099/mgen.0.000499
Hurtado, R., Barh, D., Weimer, B. C., Viana, M. V. C., Profeta, R., Sousa, T. J., et al. (2022). WGS-based lineage and antimicrobial resistance pattern of Salmonella Typhimurium isolated during 2000–2017 in Peru. Antibiotics 11:1170. doi: 10.3390/antibiotics11091170
Imre, K., Ban-Cucerzan, A., Herman, V., Sallam, K. I., Cristina, R. T., Abd-Elghany, S. M., et al. (2022). Occurrence, pathogenic potential and antimicrobial resistance of Escherichia coli isolated from raw Milk cheese commercialized in Banat region, Romania. Antibiotics 11:721. doi: 10.3390/antibiotics11060721
Jackson, S. A., Patel, I. R., Barnaba, T., LeClerc, J. E., and Cebula, T. A. (2011). Investigating the global genomic diversity of Escherichia coli using a multi-genome DNA microarray platform with novel gene prediction strategies. BMC Genomics 12:349. doi: 10.1186/1471-2164-12-349
Jandhyala, S. M., Talukdar, R., Subramanyam, C., Vuyyuru, H., Sasikala, M., and Reddy, D. N. (2015). Role of the normal gut microbiota. World J. Gastroenterol. 21, 8787–8803. doi: 10.3748/wjg.v21.i29.8787
Jeamsripong, S., Li, X., Aly, S. S., Su, Z., Pereira, R. V., and Atwill, E. R. (2021). Antibiotic resistance genes and associated phenotypes in Escherichia coli and Enterococcus from cattle at different production stages on a dairy farm in Central California. Antibiotics 10:1042. doi: 10.3390/antibiotics10091042
Kaas, R. S., Friis, C., Ussery, D. W., and Aarestrup, F. M. (2012). Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes. BMC Genomics 13:577. doi: 10.1186/1471-2164-13-577
Kaiko, G. E., and Stappenbeck, T. S. (2014). Host–microbe interactions shaping the gastrointestinal environment. Trends Immunol. 35, 538–548. doi: 10.1016/j.it.2014.08.002
Kaper, J. B., Nataro, J. P., and Mobley, H. L. T. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Khachatryan, A. R., Hancock, D. D., Besser, T. E., and Call, D. R. (2004). Role of calf-adapted Escherichia coli in maintenance of antimicrobial drug resistance in dairy calves. Appl. Environ. Microbiol. 70, 752–757. doi: 10.1128/AEM.70.2.752-757.2004
Lalioui, L., and Le Bouguénec, C. (2001). Afa-8 gene cluster is carried by a Pathogenicity Island inserted into the tRNAPhe of human and bovine pathogenic Escherichia coli isolates. Infect. Immun. 69, 937–948. doi: 10.1128/IAI.69.2.937-948.2001
Lee, K. Y., Atwill, E. R., Li, X., Feldmann, H. R., Williams, D. R., Weimer, B. C., et al. (2024). Impact of zinc supplementation on phenotypic antimicrobial resistance of fecal commensal bacteria from pre-weaned dairy calves. Sci. Rep. 14:4448. doi: 10.1038/s41598-024-54738-x
Lee, K. Y., Atwill, E. R., Pitesky, M., Huang, A., Lavelle, K., Rickard, M., et al. (2022). Antimicrobial resistance profiles of non-typhoidal Salmonella from retail meat products in California, 2018. Front. Microbiol. 13:835699. doi: 10.3389/fmicb.2022.835699
Lee, K. Y., Lavelle, K., Huang, A., Atwill, E. R., Pitesky, M., and Li, X. (2023). Assessment of prevalence and diversity of antimicrobial resistant Escherichia coli from retail meats in Southern California. Antibiotics 12:782. doi: 10.3390/antibiotics12040782
Liu, J., Ma, F., Degen, A., and Sun, P. (2023). The effects of zinc supplementation on growth, diarrhea, antioxidant capacity, and immune function in Holstein dairy calves. Animals 13:2493. doi: 10.3390/ani13152493
Liu, J., Taft, D. H., Maldonado-Gomez, M. X., Johnson, D., Treiber, M. L., Lemay, D. G., et al. (2019). The fecal resistome of dairy cattle is associated with diet during nursing. Nat. Commun. 10:4406. doi: 10.1038/s41467-019-12111-x
Ma, F. T., Wo, Y. Q. L., Shan, Q., Wei, J. Y., Zhao, S. G., and Sun, P. (2020). Zinc-methionine acts as an anti-diarrheal agent by protecting the intestinal epithelial barrier in postnatal Holstein dairy calves. Anim. Feed Sci. Technol. 270:114686. doi: 10.1016/j.anifeedsci.2020.114686
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Majumder, S., Jung, D., Ronholm, J., and George, S. (2021). Prevalence and mechanisms of antibiotic resistance in Escherichia coli isolated from mastitic dairy cattle in Canada. BMC Microbiol. 21:222. doi: 10.1186/s12866-021-02280-5
Martin, L. C., Weir, E. K., Poppe, C., Reid-Smith, R. J., and Boerlin, P. (2012). Characterization of blaCMY-2 plasmids in Salmonella and Escherichia coli isolates from food animals in Canada. Appl. Environ. Microbiol. 78, 1285–1287. doi: 10.1128/AEM.06498-11
Massé, J., Lardé, H., Fairbrother, J. M., Roy, J.-P., Francoz, D., Dufour, S., et al. (2021). Prevalence of antimicrobial resistance and characteristics of Escherichia coli isolates from fecal and manure pit samples on dairy farms in the province of Québec, Canada. Front. Vet. Sci. 8:654125. doi: 10.3389/fvets.2021.654125
McDermott, P. F., Tyson, G. H., Kabera, C., Chen, Y., Li, C., Folster, J. P., et al. (2016). Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob. Agents Chemother. 60, 5515–5520. doi: 10.1128/AAC.01030-16
Meza-Segura, M., Zaidi, M. B., Maldonado-Puga, S., Huerta-Cantillo, J., Chavez-Dueñas, L., Navarro-Garcia, F., et al. (2017). Cytolethal distending toxin-producing Escherichia coli strains causing severe diarrhoea in young Mexican children. JMM Case Rep. 4:e005079. doi: 10.1099/jmmcr.0.005079
Miller, J. J., Weimer, B. C., Timme, R., Lüdeke, C. H. M., Pettengill, J. B., Bandoy, D. J. D., et al. (2021). Phylogenetic and biogeographic patterns of Vibrio parahaemolyticus strains from North America inferred from whole-genome sequence data. Appl. Environ. Microbiol. 87, e01403–e01420. doi: 10.1128/AEM.01403-20
Nairz, M., and Weiss, G. (2020). Iron in infection and immunity. Mol. Asp. Med. 75:100864. doi: 10.1016/j.mam.2020.100864
Nguyen, C. C., Hugie, C. N., Kile, M. L., and Navab-Daneshmand, T. (2019). Association between heavy metals and antibiotic-resistant human pathogens in environmental reservoirs: a review. Front. Environ. Sci. Eng. 13:46. doi: 10.1007/s11783-019-1129-0
Noyes, N. R., Yang, X., Linke, L. M., Magnuson, R. J., Cook, S. R., Zaheer, R., et al. (2016). Characterization of the resistome in manure, soil and wastewater from dairy and beef production systems. Sci. Rep. 6:24645. doi: 10.1038/srep24645
Oh, S.-I., Ha, S., Roh, J.-H., Hur, T.-Y., and Yoo, J. G. (2020). Dynamic changes in antimicrobial resistance in fecal Escherichia coli from neonatal dairy calves: An individual follow-up study. Animals 10:1776. doi: 10.3390/ani10101776
Oksanen, J., Simpson, G. L., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., et al. (2022). Vegan: Community Ecology Package. Available at: https://CRAN.R-project.org/package=vegan (Accessed February 6, 2023).
Ombarak, R. A., Hinenoya, A., Elbagory, A.-R. M., and Yamasaki, S. (2018). Prevalence and molecular characterization of antimicrobial resistance in Escherichia coli isolated from raw Milk and raw Milk cheese in Egypt. J. Food Prot. 81, 226–232. doi: 10.4315/0362-028X.JFP-17-277
Osek, J. (2003). Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1) gene and its relationship with fimbrial and enterotoxin markers in E. coli isolates from pigs with diarrhoea. Vet. Microbiol. 91, 65–72. doi: 10.1016/S0378-1135(02)00262-6
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Pal, C., Bengtsson-Palme, J., Rensing, C., Kristiansson, E., and Larsson, D. G. J. (2014). BacMet: antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 42, D737–D743. doi: 10.1093/nar/gkt1252
Poole, K. (2017). At the Nexus of antibiotics and metals: the impact of cu and Zn on antibiotic activity and resistance. Trends Microbiol. 25, 820–832. doi: 10.1016/j.tim.2017.04.010
PubMLST (2024). Public databases for molecular typing and microbial genome diversity. Available at: https://pubmlst.org/ (Accessed February 2, 2024).
RDocumentation (2024). Pheatmap function – RDocumentation. Available at: https://www.rdocumentation.org/packages/pheatmap/versions/1.0.12/topics/pheatmap (Accessed February 2, 2024).
Ryu, J. H., Kim, S., Park, J., and Choi, K. S. (2020). Characterization of virulence genes in Escherichia coli strains isolated from pre-weaned calves in the Republic of Korea. Acta Vet. Scand. 62:45. doi: 10.1186/s13028-020-00543-1
Salaheen, S., Kim, S. W., Springer, H. R., Hovingh, E. P., Van Kessel, J. A. S., and Haley, B. J. (2023). Characterization of antimicrobial resistance genes and virulence factors in the genomes of Escherichia coli ST69 isolates from Preweaned dairy calves and their phylogenetic relationship with poultry and human clinical strains. Microb. Drug Resist. 29, 249–255. doi: 10.1089/mdr.2022.0266
Sarowska, J., Futoma-Koloch, B., Jama-Kmiecik, A., Frej-Madrzak, M., Ksiazczyk, M., Bugla-Ploskonska, G., et al. (2019). Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathog. 11:10. doi: 10.1186/s13099-019-0290-0
Sauget, M., Atchon, A. K., Valot, B., Garch, F. E., Jong, A., Moyaert, H., et al. (2023). Genome analysis of third-generation cephalosporin-resistant Escherichia coli and Salmonella species recovered from healthy and diseased food-producing animals in Europe. PLoS One 18:e0289829. doi: 10.1371/journal.pone.0289829
Schmidt, J. W., Griffin, D., Kuehn, L. A., and Brichta-Harhay, D. M. (2013). Influence of therapeutic Ceftiofur treatments of feedlot cattle on fecal and Hide Prevalences of commensal Escherichia coli resistant to expanded-Spectrum Cephalosporins, and molecular characterization of resistant isolates. Appl. Environ. Microbiol. 79, 2273–2283. doi: 10.1128/AEM.03592-12
Seemann, T. (2024). tseemann/abricate. Available at: https://github.com/tseemann/abricate (Accessed April 16, 2024).
Shahrani, M., Dehkordi, F. S., and Momtaz, H. (2014). Characterization of Escherichia coli virulence genes, pathotypes and antibiotic resistance properties in diarrheic calves in Iran. Biol. Res. 47:28. doi: 10.1186/0717-6287-47-28
Silva, A., Silva, V., Pereira, J. E., Maltez, L., Igrejas, G., Valentão, P., et al. (2023). Antimicrobial resistance and clonal lineages of Escherichia coli from food-producing animals. Antibiotics 12:1061. doi: 10.3390/antibiotics12061061
Springer, H. R., Denagamage, T. N., Fenton, G. D., Haley, B. J., Van Kessel, J. A. S., and Hovingh, E. P. (2019). Antimicrobial resistance in fecal Escherichia coli and Salmonella enterica from dairy calves: a systematic review. Foodborne Pathog. Dis. 16, 23–34. doi: 10.1089/fpd.2018.2529
Subbiah, M., Top, E. M., Shah, D. H., and Call, D. R. (2011). Selection pressure required for long-term persistence of blaCMY-2-positive IncA/C plasmids ▿. Appl. Environ. Microbiol. 77, 4486–4493. doi: 10.1128/AEM.02788-10
Tragesser, L. A., Wittum, T. E., Funk, J. A., Winokur, P. L., and Rajala-Schultz, P. J. (2006). Association between ceftiofur use and isolation of Escherichia coli with reduced susceptibility to ceftriaxone from fecal samples of dairy cows. Am. J. Vet. Res. 67, 1696–1700. doi: 10.2460/ajvr.67.10.1696
Treilles, M., Châtre, P., Drapeau, A., Madec, J.-Y., and Haenni, M. (2023). Spread of the mcr-1 colistin-resistance gene in Escherichia coli through plasmid transmission and chromosomal transposition in French goats. Front. Microbiol. 13:1023403. doi: 10.3389/fmicb.2022.1023403
Tyson, G. H., McDermott, P. F., Li, C., Chen, Y., Tadesse, D. A., Mukherjee, S., et al. (2015). WGS accurately predicts antimicrobial resistance in Escherichia coli. J. Antimicrob. Chemother. 70, 2763–2769. doi: 10.1093/jac/dkv186
Umpiérrez, A., Bado, I., Oliver, M., Acquistapace, S., Etcheverría, A., Padola, N. L., et al. (2017). Zoonotic potential and antibiotic resistance of Escherichia coli in neonatal calves in Uruguay. Microbes Environ. 32, 275–282. doi: 10.1264/jsme2.ME17046
Vahjen, W., Pietruszyńska, D., Starke, I. C., and Zentek, J. (2015). High dietary zinc supplementation increases the occurrence of tetracycline and sulfonamide resistance genes in the intestine of weaned pigs. Gut Pathog. 7:23. doi: 10.1186/s13099-015-0071-3
Vieille, T., Winiszewski, H., Chirouze, C., Bertrand, X., and Fournier, D. (2019). Escherichia coli endocarditis in an hemodialysis patient. Med. Mal. Infect. 49, 478–479. doi: 10.1016/j.medmal.2019.03.004
Wales, A. D., and Davies, R. H. (2015). Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics 4, 567–604. doi: 10.3390/antibiotics4040567
Wang, J., Huang, Y., Guan, C., Li, J., Yang, H., Zhao, G., et al. (2023). Characterization of an Escherichia coli isolate Coharboring the virulence gene astA and Tigecycline resistance gene tet(X4) from a dead piglet. Pathogens 12:903. doi: 10.3390/pathogens12070903
Weis, A. M., Clothier, K. A., Huang, B. C., Kong, N., and Weimer, B. C. (2017). Draft genome sequence of multidrug-resistant abortive Campylobacter jejuni from northern California. Genome Announc. 5, e00171–e00117. doi: 10.1128/genomeA.00171-17
Wickham, H., Chang, W., Henry, L., Pedersen, T. L., Takahashi, K., Wilke, C., et al. (2023). ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. Available at: https://cran.r-project.org/web/packages/ggplot2/index.html (Accessed February 2, 2024).
Wo, Y., Jin, Y., Gao, D., Ma, F., Ma, Z., Liu, Z., et al. (2022). Supplementation with zinc Proteinate increases the growth performance by reducing the incidence of diarrhea and improving the immune function of dairy calves during the first month of life. Front. Vet. Sci. 9:911330. doi: 10.3389/fvets.2022.911330
Woerde, D. J., Reagan, K. L., Byrne, B. A., Weimer, B. C., Epstein, S. E., Schlesener, C., et al. (2023). Characteristics of extended-Spectrum β-lactamase producing Enterobacterales isolated from dogs and cats, 2011–2021. Vet. Sci. 10:178. doi: 10.3390/vetsci10030178
Yamamoto, T., and Echeverria, P. (1996). Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect. Immun. 64, 1441–1445. doi: 10.1128/iai.64.4.1441-1445.1996
Yazdankhah, S., Rudi, K., and Bernhoft, A. (2014). Zinc and copper in animal feed – development of resistance and co-resistance to antimicrobial agents in bacteria of animal origin. Microb. Ecol. Health Dis. 25:25862. doi: 10.3402/mehd.v25.25862
Yu, X., Wo, Y., Ma, F., Shan, Q., Wei, J., and Sun, P. (2023). Zinc methionine improves the lipid metabolism that is associated with the alteration of intestine mucosal proteomes and microbiota of newborn Holstein dairy calves. Anim. Res. One Health 2, 71–85. doi: 10.1002/aro2.10
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zhong, Y.-M., Liu, W.-E., and Zheng, Z.-F. (2019). Epidemiology and molecular characterization of mcr-1 in Escherichia coli recovered from patients with bloodstream infections in Changsha, Central China. Infect. Drug Resist. 12, 2069–2076. doi: 10.2147/IDR.S209877
Zhuge, X., Zhou, Z., Jiang, M., Wang, Z., Sun, Y., Tang, F., et al. (2021). Chicken-source Escherichia coli within phylogroup F shares virulence genotypes and is closely related to extraintestinal pathogenic E. coli causing human infections. Transbound. Emerg. Dis. 68, 880–895. doi: 10.1111/tbed.13755
Keywords: antimicrobial resistance (AMR), multidrug resistance (MDR), Escherichia coli, dairy calves, calf diarrhea, whole genome sequencing
Citation: Lee KY, Schlesener CL, Aly SS, Huang BC, Li X, Atwill ER and Weimer BC (2024) Whole genome sequence analysis reveals high genomic diversity and potential host-driven adaptations among multidrug-resistant Escherichia coli from pre-weaned dairy calves. Front. Microbiol. 15:1420300. doi: 10.3389/fmicb.2024.1420300
Edited by:
Kristina Kadlec, Independent Researcher, Wunstorf, GermanyReviewed by:
Hazem Ramadan, Mansoura University, EgyptKeri N. Norman, Texas A&M University, United States
Copyright © 2024 Lee, Schlesener, Aly, Huang, Li, Atwill and Weimer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bart C. Weimer, YmN3ZWltZXJAdWNkYXZpcy5lZHU=
†ORCID: Bart C. Weimer, https://orcid.org/0000-0002-7471-1978
 Katie Y. Lee
Katie Y. Lee Cory L. Schlesener1,2
Cory L. Schlesener1,2 Sharif S. Aly
Sharif S. Aly Xunde Li
Xunde Li Edward R. Atwill
Edward R. Atwill Bart C. Weimer
Bart C. Weimer