- 1Institute of Marine Biology and Pharmacology, Ocean College, Zhejiang University, Zhoushan, China
- 2Xianghu Laboratory, Hangzhou, China
- 3Trend Biotech Co., Ltd., Hangzhou, China
- 4Zhoushan Sewerage Treatment Co., Ltd., Zhoushan, China
- 5Environmental Sanitation Management Office, Zhoushan, China
- 6Ocean Research Center of Zhoushan, Zhejiang University, Zhoushan, China
N,N-dimethylformamide (DMF) is an organic solvent with stable chemical properties and high boiling point. Based on its good solubility, DMF is widely used in synthetic textile, leather, electronics, pharmaceutical and pesticide industries. However, the DMF pollutes the environment and does harm to human liver function, kidney function, and nerve function. Herein, an efficient DMF-degrading strain, DM175A1-1, was isolated and identified as Paracoccus sulfuroxidans. This strain can use DMF as the sole source of carbon and nitrogen. Whole-genome sequencing of strain DM175A1-1 revealed that it has a 3.99-Mbp chromosome a 120-kbp plasmid1 and a 40-kbp plasmid2. The chromosome specifically harbors the dmfA1 and dmfA2 essential for the initial steps of DMF degradation. And it also carries the some part of genes facilitating subsequent methylotrophic metabolism and glutathione-dependent pathway. Through further DMF tolerance degradation experiments, DM175A1-1 can tolerate DMF concentrations up to 10,000 mg/L, whereas the majority of Paracoccus strains could only show degradation activity below 1,000 mg/L. And the efficiency of organic nitrogen conversion to NH3-N in DMF can reach 99.0% when the hydraulic retention time (HRT) is controlled at 5 days. Meanwhile, it showed a significant degradation effect at a pharmaceutical enterprise in Zhejiang Province with high concentration of DMF wastewater. This study provides a new strain Paracoccus sulfuroxidans DM175A1-1 which shows a significant influence on DMF degradation, and reveals the characterization on its DMF degradation. It lays the foundation for the application of biological method in the efficient degradation of DMF in industrial wastewater.
Introduction
N,N-dimethylformamide (DMF) is a common multifunctional solvent that is widely used in pesticide, pharmaceutical, petrochemical, and other production industries (Ding and Jiao, 2012; Bromley-Challenor et al., 2000; Nisha et al., 2015). In recent years, with the expansion of the scale of chemical production, DMF has aggravated the pollution of the environment with the increase of industrial wastewater (Chen et al., 2016). DMF can enter the human body through breathing and skin contact, which has greater harm to human liver function, kidney function, and nerve function (Wang et al., 2014; Li and Zeng, 2019). At present, there are physical, chemical, and biological methods for treating DMF wastewater, and biological processes have been applied in industrial wastewater treatment because of their advantages of low cost and low pollution (Das et al., 2006; Sun et al., 2008; Dziewit et al., 2010). When the concentration of pollutants in wastewater is high, it will influence the growth and degradation effect of microorganisms (Astals et al., 2015). At present, the treatment of the DMF wastewater is mainly relying on activated sludge. However, the activated sludge could only tolerant about 200 mg/L DMF concentration compared to the 2,000 mg/L DMF concentration of factory wastewater (Zhou et al., 2018; Astals et al., 2015). The low degradation rate of DMF will cause the exceedance of total nitrogen released, which increase the difficulty in subsequent treatment of nitrogen content. Hence, it needs to be domesticated and isolated from the environment to get some strains that can degrade DMF efficiently, which is essential for practical industrial production applications.
Several bacteria that degrade DMF have been isolated and identified, such as Alcaligenes (Hasegawa et al., 1997), Paracoccus (Nisha et al., 2015; Doronina et al., 1998; Czarnecki et al., 2017), Pseudomonas (Ghisalba et al., 1985), Methylobacterium (Doronina et al., 2000), Mycobacterium (Urakami et al., 1990), Ochrobactrum (Veeranagouda et al., 2006), etc. Paracoccus is the main genus for degrading DMF, and there are several Paracoccus strains were reported that have the capability of degrading DMF effectively (Zhou et al., 2018; Astals et al., 2015). Swaroop et al. (2009) found that the Paracoccus sp. strain DMF bacteria would briefly accumulate dimethylamine and methylamine during the growth process using DMF, which would eventually be converted into ammonia and carbon dioxide and degrade DMF.
In previous study, they found that some of the strains Paracoccus aminophilus JCM 7686 and Methylobacterium sp. Strain DM1 contained enzymes that could degrade DMF, named DMFase (Dziewit et al., 2010; Lu et al., 2019). In contrast, others did not include this enzyme, so they speculated that there might be two pathways of DMF degradation. The critical enzyme in one of the pathways was DMFase, which degraded DMF to dimethylamine (DMA) and formic acid (Dziewit et al., 2010). Subsequently, DMA forms methylamine and formaldehyde by the action of dimethyl dehydrogenase. Then methylamine will form ammonia and formaldehyde by the activity of methylamine dehydrogenase (Ghisalba et al., 1985; Veeranagouda et al., 2006). Another pathway of DMF degradation is achieved through repeated oxidative demethylation of DMF, which is speculated to be possessed by some Pseudomonas species (Hans-Peter et al., 2010). The two methyl groups on N are removed by methyl formamide oxidase and formamide oxidase, respectively, to form formamide, which is further hydrolyzed by formamidase to produce ammonia and formic acid (Ghisalba et al., 1985). The DMF degradation mechanisms of different genera may differ significantly, and there are relatively few studies on DMF degradation mechanisms in terms of genes. Therefore, it is essential to identify the DMF degradation mechanisms of Paracoccus sulfuroxidans.
In this study, we isolated four efficient DMF degrading strains by acclimatization, which were able to utilize DMF as the sole carbon and nitrogen source. Among them, DM175A1-1 showed the best degradation effect, and the whole gene sequencing data of this strain were analyzed to elucidate its relevant genes and degradation pathways responsible for DMF degradation. The DM175A1-1 can achieve efficient ammonification of DMF, which could solve the difficulty of converting organic nitrogen to ammonia nitrogen during the degradation then, the nitrogen removal can be achieved in combination with nitrification or denitrification processes subsequently. Finally, in order to enhance the tolerance of the high DMF concentration about DM175A1-1, we further investigated its degradation ability by tolerance test.
Materials and methods
Culture medium
The inorganic salt medium with DMF as the sole source of carbon and nitrogen (1.2 g/L KH2PO4, 6.8 g/L K2HPO4, 0.5 g/L NaCl, 0.1 g/L MgSO4·7H2O, 0.1 g/L MnSO4·H2O, 0.1 g/L CaCl2, 0.1 g/L FeSO4·7H2O, 0.006 g/L Na2MoO4·2H2O, 0.006 g/L CuSO4·5H2O, 0.007 g/L ZnSO4·7H2O, 0.0001 g/L CoCl2·6H2O, 0.0124 g/L H3BO3, 0.00001 g/L Vitamin B1, pH 6.0) was used to domesticate the isolates. LB medium as the seed solution medium for strains. DMF1 medium as the purification medium for strains (additional 0.2 g/L DMF and 0.2 g/L dimethylamine were added on top of the ingredients in LB medium).
Isolation and identification of domesticated strains
Fresh sludge of 10 mL from each pharmaceutical plant was inoculated into a triangular flask containing 100 mL of inorganic salt medium (DMF concentration of 1,000 mg/L) and incubated in an incubator at 30°C with 130 r/min oscillation, during which the total nitrogen (TN) and NH3-N values in the system were sampled every one day. The culture was incubated until the ammonification rate of TN reached more than 90% (ammonification refers to the conversion of organic nitrogen in TN into NH3-N), then transferred to fresh inorganic salt medium (DMF concentration of 2,000 mg/L) at 10% inoculum, monitored the TN and NH3-N values in the system and continued to incubate. The DMF concentration was gradually increased to 5,000 mg/L by transferring five times consecutively.
One millilitre of bacterial broth from the culture system with good growth condition of the strain was taken for high-throughput sequencing. At the same time, the above bacteria were at a dilution of 10−2 to 10−8 spread onto a solid medium containing 1,000 mg/L DMF and incubated at 30°C for 5 days single colonies were picked into DMF1 medium, incubated at 30°C for 3 days, and then purified by scribing, and the single purified strains were identified by DNA sequencing. The morphology of the bacterium was observed by light microscopy and cultured with Petri dishes. The isolated and purified strains were subjected to Gram staining and physiological and biochemical characterization experiments, and the identification results of 16S rDNA were used as the species name.
Genome sequencing and genetic information analysis
The version of MEGA was MEGA X. The evolutionary using the Neighbor-Joining method, Kimura 2-parameter method, Numbers at branch points indicate bootstrap percentages (based on 1,000 replications). Only values above 50% are shown. Bar, 0.01 substitutions per nucleotide position. The 16S rRNA gene sequence of Tropicibacter naphthalenivorans CECT 7648 was used as the outgroup.
Genomic DNA was extracted using the Wizard genomic DNA purification kit (product no. A1125; Promega). Whole-genome sequencing was performed on the Illumina MiSeq and PacBio sequencing platforms. Functional gene predictions and annotations were performed using GeneMarkS,1 the Rapid Annotation Subsystem Technology (RAST) database, BLAST,2 and UniProt.3 Insertion elements were predicted using Isfinder.
Degradation of the DMF
The strains were amplified and cultured in LB medium until the OD600 reached about 1.5, centrifuged at 6,000 r/min for 3 min, the supernatant was discarded, and the pellet was used further. The pellet was then resuspend with sterile distilled water to prepare the seed solution. The feed water was an inorganic salt medium containing 5,000 mg/L DMF, adjusted pH = 6.0–7.0, which was artificially configured wastewater. The inorganic salt medium was added to the strain treatment tank, adjusted pH = 6.0–7.0, and various sub-liquids were inoculated in the strain treatment tank according to 20% inoculum, and granular activated carbon was added as the strain carrier to enrich and retain the strain. The hydraulic retention time (HRT) of the influent water in the strain treatment tank was controlled to be 3 days, dissolved oxygen (DO) >2.0 mg/L, and water temperature 30–35°C. The primary water quality data in the system were monitored daily, such as COD, NH3-N, and TN.
Statistical analysis
Every group of the experiments repeated for 4 times, n = 4. Data were expressed as the mean ± standard error of the mean (SEM), visualized using GraphPad Prism (8.3.0), one-way analysis of variance (ANOVA) of SPSS (version 25.0), and Dunnett’s multiple comparison test for statistical analysis, with significant differences between groups (p < 0.05).
Results and discussion
The acclimatization and isolation of the strains
Fresh sludge was used for acclimatization, in which the strains were cultured in an inorganic medium with a DMF concentration of 1,000 mg/L for 1 week, the NH3-N in the system increased significantly to about 160 mg/L, and the TN ammonification rate reached more than 90%. Four strains of bacteria with DMF degradation effect were obtained by acclimatization culture.
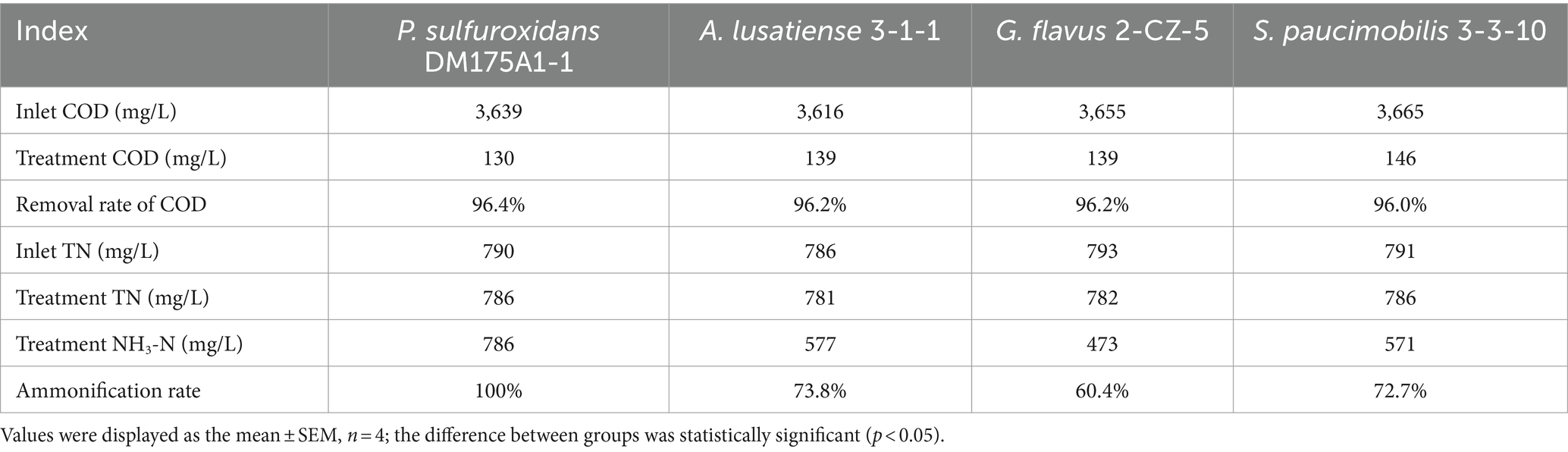
As shown in the results of Figure 1 and Table 1, in the group system of P. sulfuroxidans DM175A1-1, both NH3-N and TN showed a slowly increasing trend in the first 6 days, and the proportion of NH3-N to TN was basically above 95%; as shown in Table 1, after the 7th day, both NH3-N and TN data in the treated system were stable at about 780 mg/L, and the proportion of NH3 to TN was at above 98%. In the system of the A. lusatiense 3-1-1, after the 7th day, the NH3-N data in the treatment system were stable at about 575 mg/L, the TN data were about 780 mg/L, and the proportion of NH3-N to TN was maintained at about 74%.
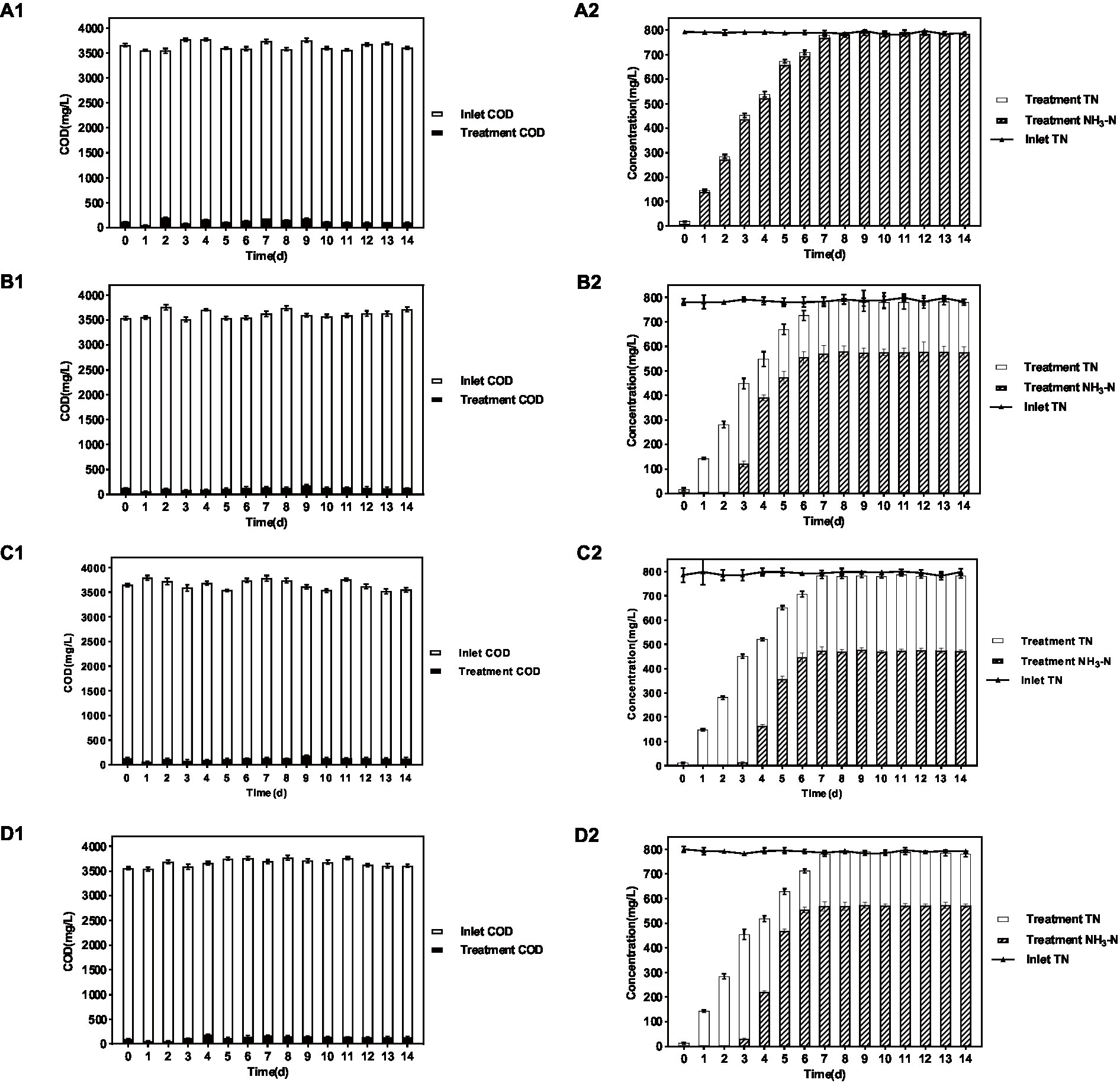
Figure 1. Treatment effect of 4 strains on various indicators of wastewater. (A–D) Represent P. sulfuroxidans DM175A1-1, A. lusatiense 3-1-1, G. flavus 2-CZ-5, S. paucimobilis 3-3-10, respectively. The number 1 after alphabet represents the COD treatment effect of the strain, number 2 represents the TN treatment effect of the strain and the ammonification effect. Values were displayed as the mean ± SEM, n = 4; the difference between groups was statistically significant (p < 0.05).
In the G. flavus 2-CZ-5 experimental system, after the 7th day, the NH3-N data in the treated system were stable at about 473 mg/L, the TN data at about 780 mg/L, and the proportion of NH3-N to TN was maintained at about 60%. in the S. paucimobilis 3-3-10 experimental system, after the 7th day, the NH3-N data in the treated system were stable at about 575 mg/L, the TN data at about 780 mg/L, and the proportion of NH3-N to TN was maintained at about 60%. The NH3-N data were stable at about 572 mg/L, and the TN data were about 780 mg/L, and the proportion of NH3-N to TN was maintained at about 73%.
The experimental results of DMF degradation by single bacteria showed that in the four single bacteria experimental systems, all strains could take advantage of DMF effectively and their OD600 is over 1.50, pH in all four systems increased significantly to more than 8.0, and a certain amount of acid should be added daily to adjust pH to about 6. The DMF degradation effect of P. sulfuroxidans DM175A1-1 was better than the other three monocultures, and the DMF degradation rate reached more than 90%.
Identification of the strains
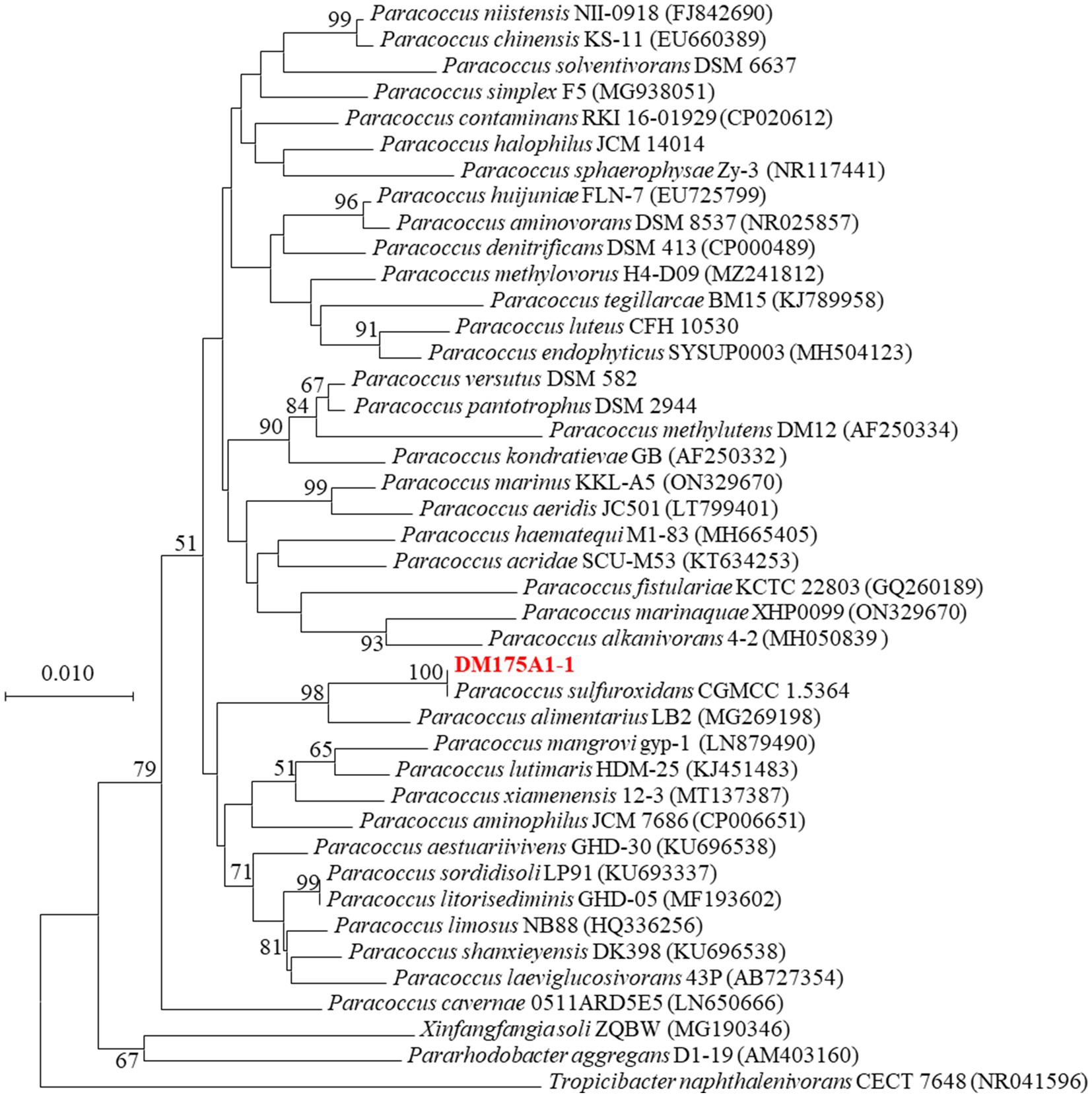
After 3 days incubation on LB Petri dishes, the colony morphology was round and raised. The colony surface was smooth and moist. The edge was neat and beige in color, Gram negative, and the physiological and biochemical characteristics were initially identified as Paracoccus. 16S rDNA sequence comparison, the similarity between strain DM175A1-1 and Paracoccus sulfuroxidans CGMCC 1.5364T was 99%, so the phylogenetic tree was constructed by MEGA software, and strain DM175A1-1 was also closely clustered with Paracoccus sulfuroxidans CGMCC 1.5364T. As shown in Figure 2, the phylogenetic tree was constructed by MEGA software, and strain DM175A1-1 was also closely clustered with Paracoccus sulfuroxidans CGMCC 1.5364T, so it was identified as Paracoccus sulfuroxidans strain.
Multireplicon genome of strain DM175A1-1
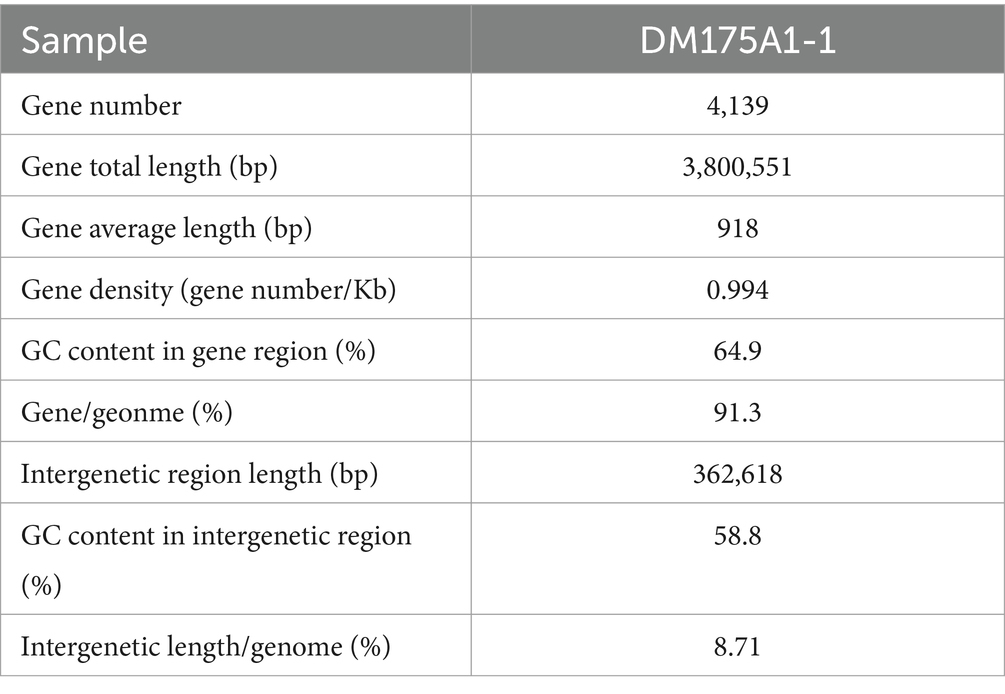
Whole-genome sequencing was performed, and analysis of the data revealed that the genome of strain DM175A1-1 has a multireplicon structure, with a single circular chromosome and two plasmids (Figure 3). The circular chromosome is 3,993,573 bp in size; the plasmid1 is 123,272 bp in size; and the plasmid2 is 46,324 bp in size. The whole length of the genome sequence is 4,163,169, with a G + C content of 64.40%. After the analysis of the function, the CDS results showed that the Gene number is 4,139, and the gene total length is 3,800,551 bp, with a coding percentage of 91.3% (Table 2).
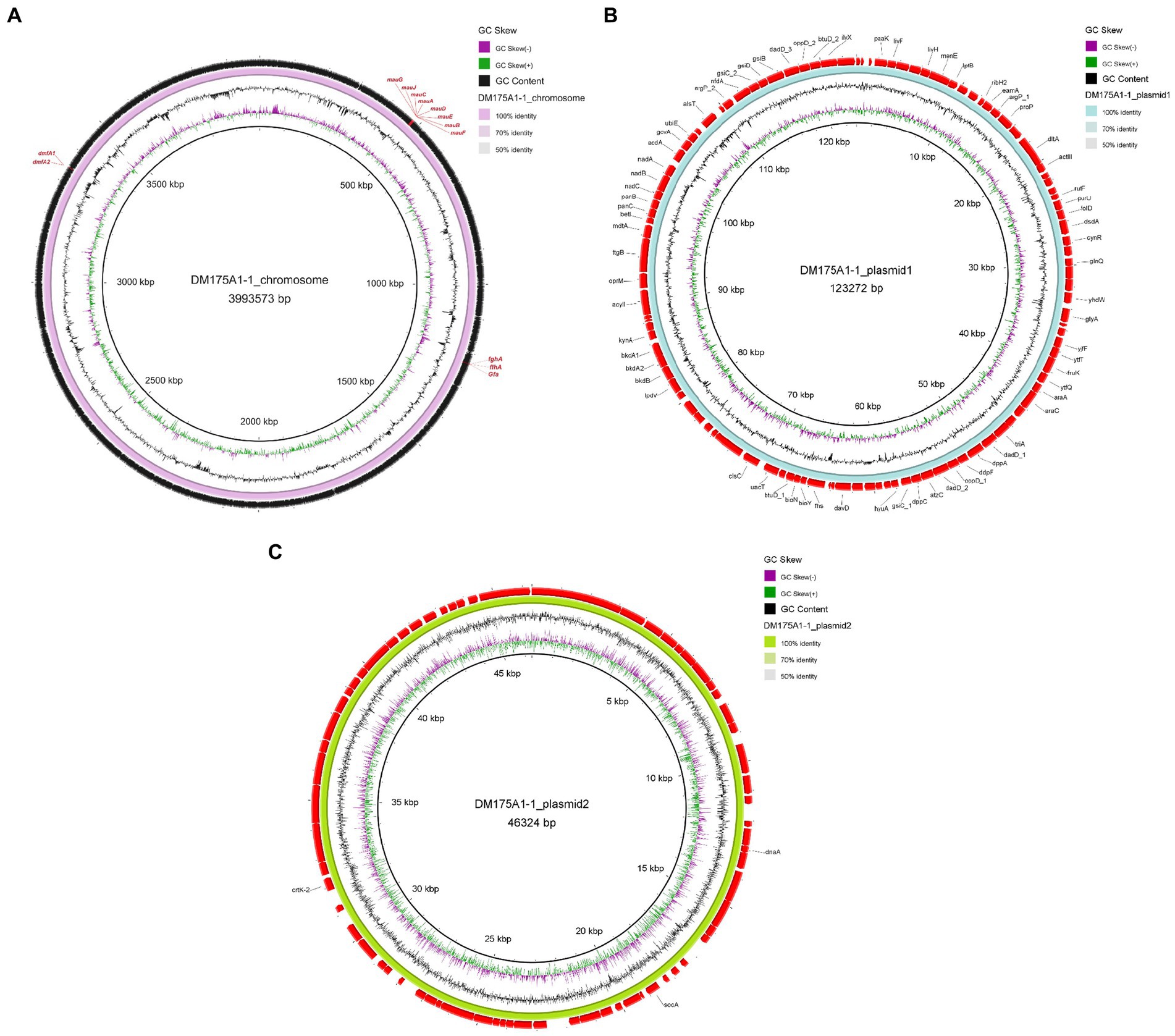
Figure 3. Genomic analysis revealing the circular replicons in the genome of strain DM175A1-1. (A) The chromosome. (B) The plasmid1. (C) The plasmid2.
We submitted the genome of the DM175A1-1 to the blast server. P. aminovorans JCM 7685, which has a complete DMF degradation pathway, carries two genes, dmfA1 and dmfA2, responsible for encoding the DMFase that initially degrades DMF to DMA and MA (Czarnecki and Bartosik, 2019). The DM175A1-1 also has the dmfA1 and dmfA2 genes encoding the DMFase, as shown in Figure 3, so the initial pathway of DM175A1-1 to degrade DMF to DMA and MA may be the same as that of the P. aminovorans JCM 7685. Then P. aminovorans JCM 7685 degrades DMA to formate by the NMG pathway, in which key genes in the NMG pathway are not found in DM175A1-1, so it was hypothesised that other DMA degradation pathways may exist in DM175A1-1 (Czarnecki and Bartosik, 2019). The MA degradation pathway has been reported in Paracoccus spp. P. denitrificans Pd 1222, MA is degraded to formaldehyde by the action of MA dehydrogenase encoded by mauA and mauB, and P. denitrificans Pd 1222 oxidises formaldehyde to formate by a glutathione-dependent pathway requiring three enzymes, namely S-(hydroxymethyl) glutathione synthetase (gfa), S-(hydroxymethyl) glutathione synthase, S (hydroxymethyl) glutathione dehydrogenase (flhA) and S-formylglutathione hydrolase (fghA) (Czarnecki and Bartosik, 2019; Lu et al., 2019). As shown in Figure 3, the chromosome of DM175A1-1 carries the mau gene cluster encoding formaldehyde dehydrogenase, where the cluster contains mauA and mauB, and three genes in the pathway for further breakdown of formaldehyde to formate, as shown in the Figure 3, flhA, fghA and gfa, respectively. Therefore, the product of the initial degradation of DM175A1-1, MA, can be converted to formaldehyde through the MA dehydrogenase encoded by mauA and mauB, which in turn is oxidised to formate through the glutathione-dependent pathway. As DM175A1-1 contains some of the genes in the DMF degradation pathway that have been studied, but also lacks some of the genes in the degradation pathway, new degradation pathways may exist in DM175A1-1 and are the next direction for further research.
Tolerance test of the Paracoccus sulfuroxidans DM175A1-1
To further verify the practical effect of strain DM175A1-1, the DMF concentration in the influent water was increased to 10,000 mg/L for the experiment. The experimental procedure was the same as 1.4.2. The system without the strain was set as the blank group, and the system with the activated sludge (MLVSS of 6,000 mg/L) was set as the control group, and the treatment effects of HRT 3 days and 5 days were investigated. The purpose was to check the tolerance of the strain P. sulfuroxidans DM175A1-1 to DMF and compare it with the treatment effect of activated sludge.
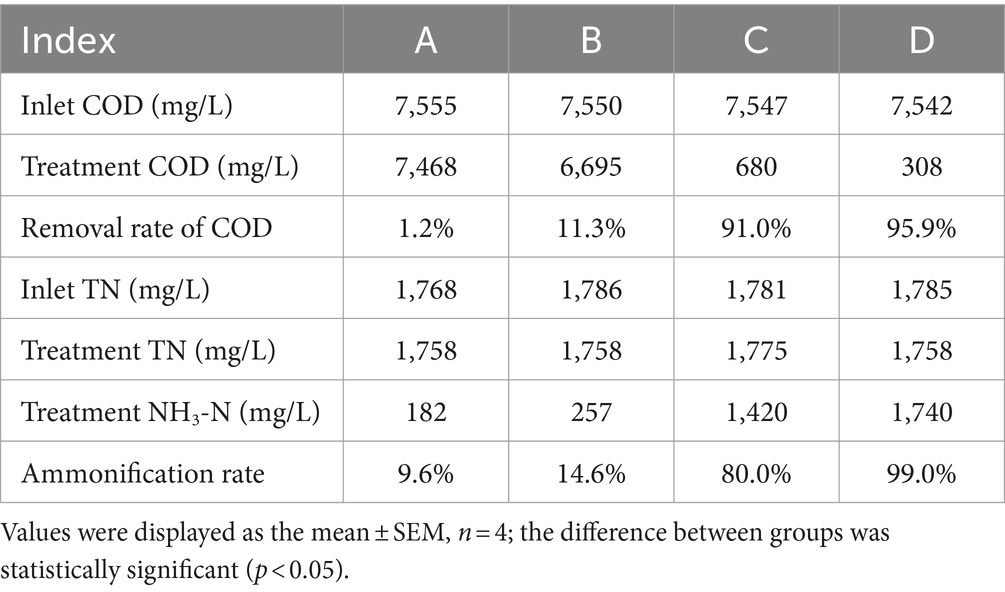
The results are shown in Figure 4 and Table 3. When the influent DMF concentration reached 10,000 mg/L, the activated sludge group decreased in the late ammonification period, probably due to poisoning, and the final ammonification rate was as low as 14.6%. In contrast, the P. sulfuroxidans DM175A1-1 strain treatment group (groups C and D) had apparent growth and better activity in the treatment system, with specific higher COD removal capacity and ammonification capacity, and when the HRT was 3 days, the COD removal rate was 91.0%, and ammonification rate was 80.0%; when the HRT was extended to 5 days, the COD removal rate continued to rise to 91.0% and When the HRT was extended to 5 days, the COD removal rate continued to increase to 91.0%, and the ammonification rate reached 99.0%. In conclusion, P. sulfuroxidans DM175A1-1 can tolerate DMF concentrations up to 10,000 mg/L, and the conversion efficiency of organic nitrogen to NH3-N in DMF can reach 99.0% when the HRT is controlled at 5 days.
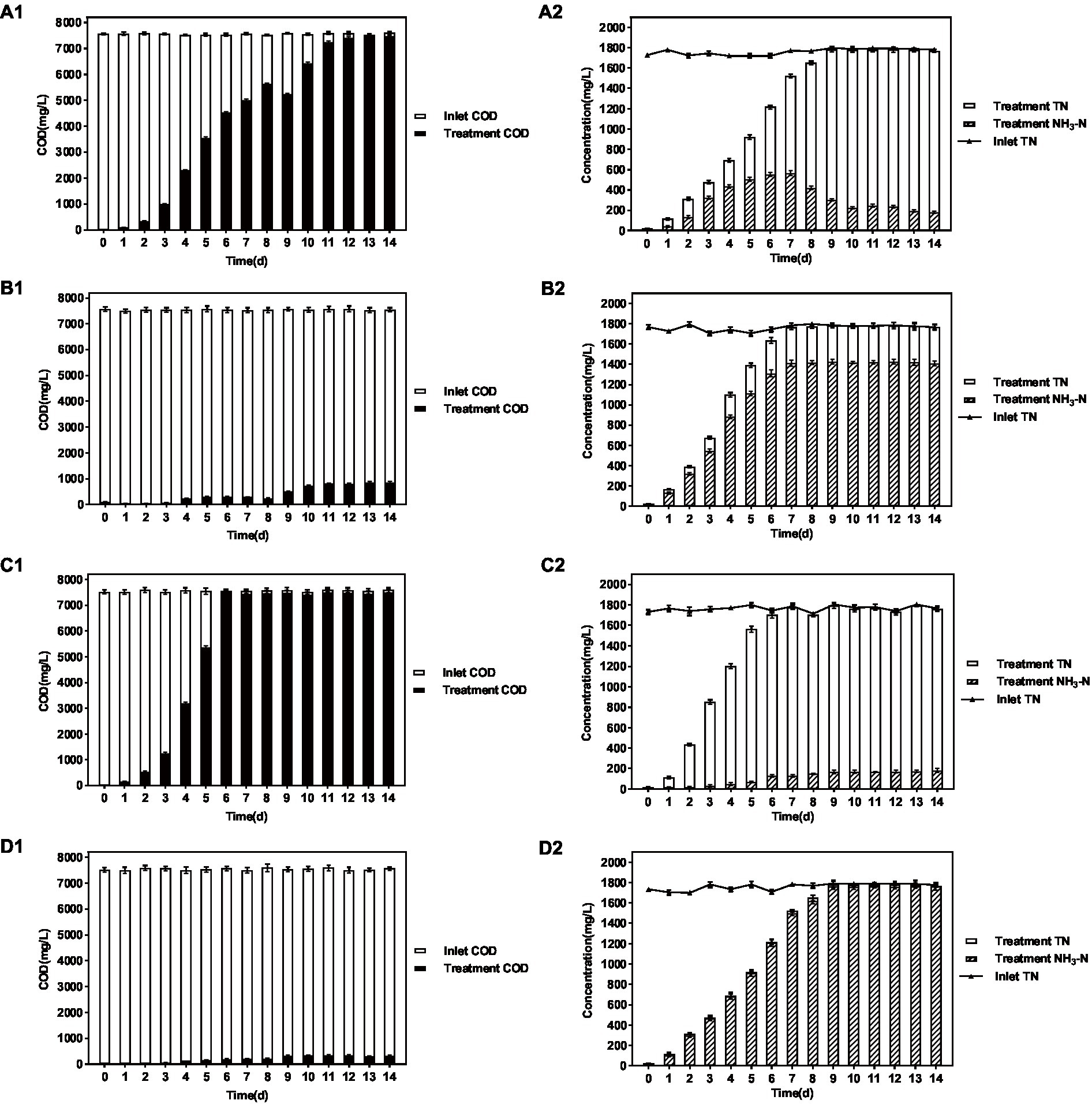
Figure 4. Effect of P. sulfuroxidans DM175A1-1 on the treatment of high concentration DMF. (A–D) Space Team (HRT 3 days), Action Pollution Team (HRT 3 days), P. sulfuroxidans DM175A1-1 Team (HRT 3 days), P. sulfuroxidans DM175A1-1 Teams (HRT 5 days), 1.2 represents the COD treatment effect of the strain, the TN treatment effect of the strain, and the ammonification effect. Values were displayed as the mean ± SEM, n = 4; the difference between groups was statistically significant (p < 0.05).
The treatment of the wastewater based on Paracoccus sulfuroxidans DM175A1-1
The wastewater came from a pharmaceutical enterprise in Zhejiang, with a DMF content of about 1.2% (w/v), a water volume of 60 t/d, and wastewater quality, as shown in Table 4. In order to reduce the introduction of nitrogenous substances from the strain medium into the wastewater, the centrifuges are not provided in the factory, so the strain was fermented with 1/5 concentration of LB medium and was pitched when the fermentation reached about 1.5 OD600 value. The wastewater was supplemented with KH2PO4 0.5 g/L, and H2SO4 was used to adjust the pH of the wastewater = 6.0.
The device is used to handle the wastewater, a filler layer is set at 1/3 of the distance from the bottom of the device, and the filler is preferably granular activated carbon, with a dosage of 10–13% (the volume ratio of filler to the device). The aerobic process was adopted, controlling HRT about 5 days, DO> 2 mg/L, water temperature 30–33°C, and 20% of strain injection. During the strain treatment, the pH of the strain treatment pool will rise to 9.0, and H2SO4 needs to be added to consistently control the pH of the strain treatment pool = 6.0–7.0 (optimal 6.0). When the treatment effect is not good, additional carbon sources such as glucose 2–3 g/L can be supplemented or supplemented strains. The primary water quality data in the system were monitored daily.
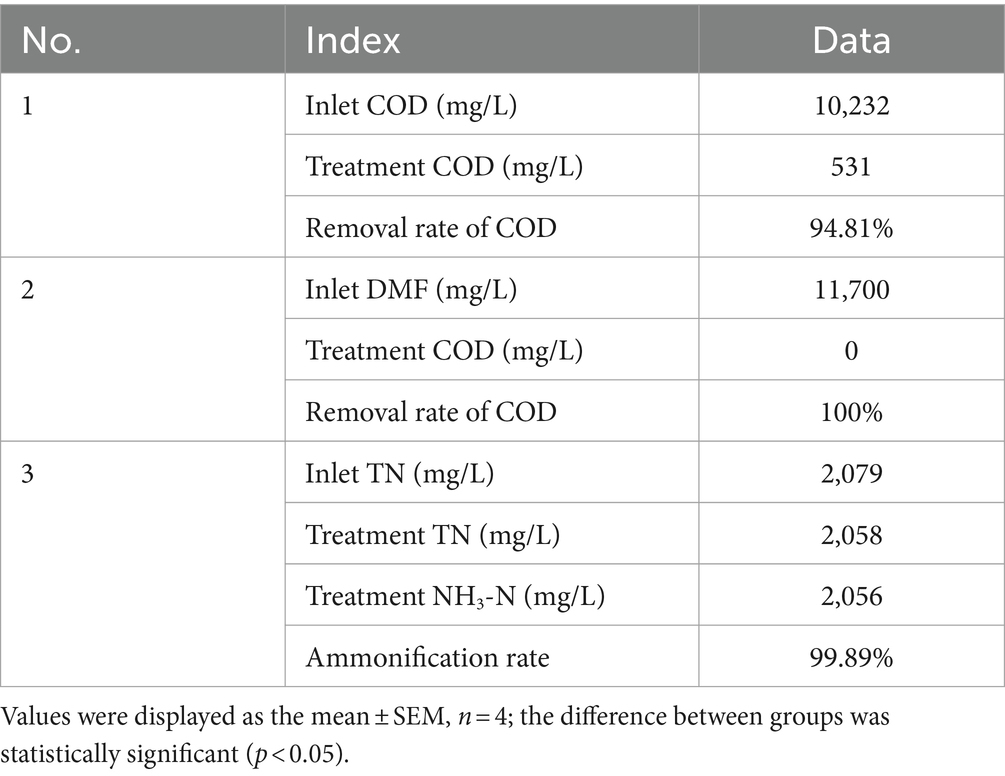
The results are shown in Figure 5 and Table 5. P. sulfuroxidans DM175A1-1 strain treated wastewater with DMF content up to 1.2% and TN about 2,000 mg/L well, and the COD removal rate was nearly 95%, which could basically achieve mineralization; DMF removal rate reached 100%, and combined with COD data, it was presumed that the remaining material was the intermediate product in the process of DMF degradation; TN Basically, all of the TN was converted into NH3-N, and the ammonification rate reached 99.89%.
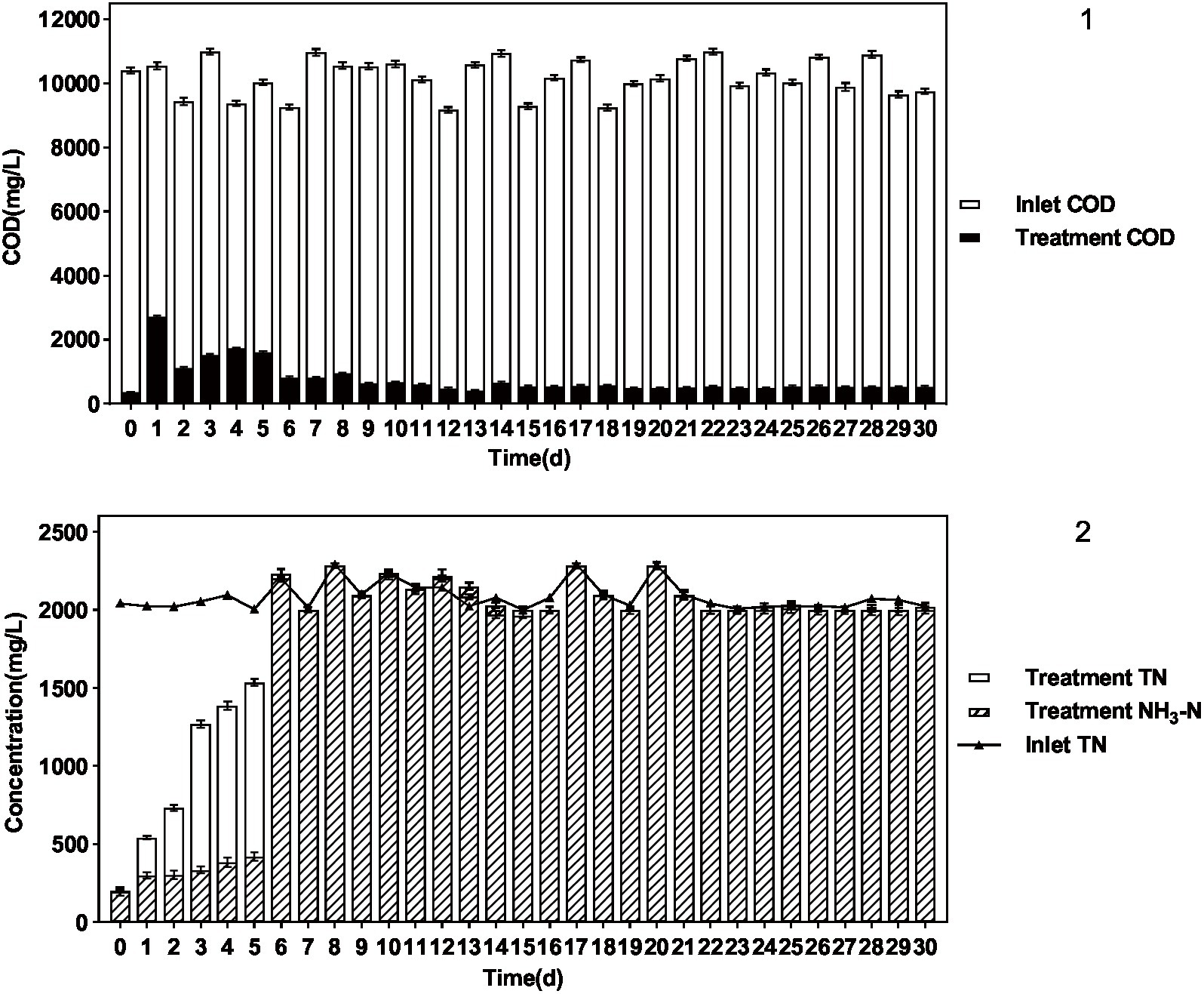
Figure 5. Treatment effect of P. sulfuroxidans DM175A1-1 on high concentration DMF wastewater. (1) The COD treatment effect of the strain. (2) The TN treatment effect of the strain and the ammonification effect. Values were displayed as the mean ± SEM, n = 4; the difference between groups was statistically significant (p < 0.05).
Conclusion
In this study, the DM175A1-1 was identified as Paracoccus sulfuroxidans, which were growing with DMF as the only carbon and nitrogen source. Whole-genome sequencing of strain DM175A1-1 revealed that it has a 3.99-Mbp chromosome a 120-kbp plasmid1 and a 40-kbp plasmid2. Through the comparison of the genome to others, we found that the chromosome has the key genes in the initial phase of methylotrophy pathways in the genus Paracoccus. Then after the further DMF tolerance degradation experiments, DM175A1-1 can tolerate DMF concentration up to 10,000 mg/L, and the efficiency of organic nitrogen conversion to NH3-N in DMF can reach 99.0% when the HRT is controlled at 5 days. Meanwhile, the application experiments were conducted for a pharmaceutical enterprise in Zhejiang Province with a high concentration of DMF wastewater, and the wastewater treatment effect was good. The prevailing treatment methods for DMF-containing wastewater are typically biochemical, which are frequently costly and environmentally benign. At present, there are few applications of microbial degradation in industry. This study has successfully demonstrated the degradation of DMF in enterprise applications, providing insights and support for future research. As the specific degradation pathway of this strain has not yet been fully elucidated, future research will focus on elucidating the mechanism and mechanism. This will further deepen the connection between the genome and degradation products, and determine the subsequent degradation mechanism and pathway.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
GZ: Writing – review & editing, Writing – original draft, Visualization, Methodology, Investigation, Formal analysis, Data curation. YS: Writing – review & editing, Resources, Investigation. WZ: Writing – review & editing, Resources, Investigation. XL: Writing – review & editing, Resources, Investigation. LS: Writing – review & editing, Resources, Investigation. ZS: Writing – review & editing, Resources, Investigation. DZ: Writing – review & editing, Resources. KW: Writing – review & editing, Validation, Investigation. ZM: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by Major Science and Technology Innovation Projects in Hangzhou City (No. 2023SZD0130) and Zhejiang Provincial Basic Public Welfare Research Program (No. LGN22C010006).
Conflict of interest
YS and WZ were employed by Trend Biotech Co., Ltd. XL and LS were employed by Zhoushan Sewerage Treatment Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1419461/full#supplementary-material
Footnotes
References
Astals, S., Musenze, R. S., Bai, X., Tannock, S., Tait, S., Pratt, S., et al. (2015). Anaerobic co-digestion of pig manure and algae: impact of intracellular algal products recovery on co-digestion performance. Bioresour. Technol. 181, 97–104. doi: 10.1016/j.biortech.2015.01.039
Bromley-Challenor, K., Caggiano, N., and Knapp, J. (2000). Bacterial growth on N,N-dimethylformamide: implications for the biotreatment of industrial wastewater. J. Ind. Microbiol. Biotechnol. 25, 8–16. doi: 10.1038/sj.jim.7000015
Chen, Y., Li, B., Qiu, Y., Xu, X., and Shen, S. (2016). A novel chemical/biological combined technique for N,N-dimethylformamide wastewater treatment. Environ. Technol. 37, 1088–1093. doi: 10.1080/09593330.2015.1102328
Czarnecki, J., and Bartosik, D. (2019). Diversity of methylotrophy pathways in the genus Paracoccus (Alphaproteobacteria). Curr. Issues Mol. Biol. 33, 117–132. doi: 10.21775/cimb.033.117
Czarnecki, J., Dziewit, L., Puzyna, M., Prochwicz, E., Tudek, A., Wibberg, D., et al. (2017). Lifestyle-determining extrachromosomal replicon pAMV1 and its contribution to the carbon metabolism of the methylotrophic bacterium Paracoccus aminovorans JCM 7685. Environ. Microbiol. 19, 4536–4550. doi: 10.1111/1462-2920.13901
Das, S., Banthia, A. K., and Adhikari, B. (2006). Pervaporation separation of DMF from water using a crosslinked polyurethane urea-PMMA IPN membrane. Desalination 197, 106–116. doi: 10.1016/j.desal.2005.12.019
Ding, S., and Jiao, N. (2012). N,N-dimethylformamide: a multipurpose building block. Angew. Chem. Int. Ed. Engl. 51, 9226–9237. doi: 10.1002/anie.201200859
Doronina, N. V., Trotsenko, Y. A., Krausova, V. I., and Suzina, N. E. (1998). Paracoccus methylutens sp. nov.—a new aerobic facultatively methylotrophic bacterium utilizing dichloromethane. Syst. Appl. Microbiol. 21, 230–236. doi: 10.1016/S0723-2020(98)80027-1
Doronina, N. V., Trotsenko, Y. A., Tourova, T. P., Kuznetsov, B. B., and Leisinger, T. (2000). Methylopila helvetica sp. nov. and Methylobacterium dichloromethanicum sp. nov.—novel aerobic facultatively methylotrophic bacteria utilizing dichloromethane. Syst. Appl. Microbiol. 23, 210–218. doi: 10.1016/S0723-2020(00)80007-7
Dziewit, L., Dmowski, M., Baj, J., and Bartosik, D. (2010). Plasmid pAMI2 of Paracoccus aminophilus JCM 7686 carries N,N-dimethylformamide degradation-related genes whose expression is activated by a LuxR family regulator. Appl. Environ. Microbiol. 76, 1861–1869. doi: 10.1128/AEM.01926-09
Ghisalba, O., Cevey, P., Kiienzi, M., and Schgir, H. P. (1985). Biodegradation of chemical waste by specialized methylotrophs, an alternative to physical methods of waste disposal. Conserv. Recycl. 8, 47–71. doi: 10.1016/0361-3658(85)90025-6
Hans-Peter, S., Holzmann, W., Tombo, G. M. R., and Ghisalba, O. (2010). Purification and characterization of N,N-dimethylformamidase from Pseudomonas DMF 3/3. Eur. J. Biochem. 158, 469–475. doi: 10.1111/j.1432-1033.1986.tb09778.x
Hasegawa, Y., Matsuo, M., Sigemoto, Y., Sakai, T., and Tokuyama, T. (1997). Purification and characterization of N,N-dimethylformamidase from Alcaligenes sp. KUFA-1. J. Ferment. Bioeng. 84, 543–547. doi: 10.1016/S0922-338X(97)81909-5
Li, M. J., and Zeng, T. (2019). The deleterious effects of N,N-dimethylformamide on liver: a mini-review. Chem. Biol. Interact. 298, 129–136. doi: 10.1016/j.cbi.2018.12.011
Lu, X. Y., Wang, W. W., Zhang, L. G., Hu, H. Y., Xu, P., Wei, T., et al. (2019). Molecular mechanism of N,N-dimethylformamide degradation in Methylobacterium sp. strain DM1. Appl. Environ. Microbiol. 85:e00275. doi: 10.1128/AEM.00275-19
Nisha, K. N., Devi, V., Varalakshmi, P., and Ashokkumar, B. (2015). Biodegradation and utilization of dimethylformamide by biofilm forming Paracoccus sp. strains MKU1 and MKU2. Bioresour. Technol. 188, 9–13. doi: 10.1016/j.biortech.2015.02.042
Sun, G., Xu, A., He, Y., Yang, M., Du, H., and Sun, C. (2008). Ruthenium catalysts supported on high-surface-area zirconia for the catalytic wet oxidation of N,N-dimethyl formamide. J. Hazard. Mater. 156, 335–341. doi: 10.1016/j.jhazmat.2007.12.023
Swaroop, S., Sughosh, P., and Ramanathan, G. (2009). Biomineralization of N,N-dimethylformamide by Paracoccus sp. strain DMF. J. Hazard. Mater. 171, 268–272. doi: 10.1016/j.jhazmat.2009.05.138
Urakami, T., Kobayashi, H., and Araki, H. (1990). Isolation and identification of N,N-dimethylformamide-biodegrading bacteria. J. Ferment. Bioeng. 70, 45–47. doi: 10.1016/0922-338X(90)90029-V
Veeranagouda, Y., Emmanuel Paul, P. V., Gorla, P., Siddavattam, D., and Karegoudar, T. B. (2006). Complete mineralisation of dimethylformamide by Ochrobactrum sp. DGVK1 isolated from the soil samples collected from the coalmine leftovers. Appl. Microbiol. Biotechnol. 71, 369–375. doi: 10.1007/s00253-005-0157-9
Wang, C., Huang, C., Wei, Y., Zhu, Q., Tian, W., and Zhang, Q. (2014). Short-term exposure to dimethylformamide and the impact on digestive system disease: an outdoor study for volatile organic compound. Environ. Pollut. 190, 133–138. doi: 10.1016/j.envpol.2014.03.026
Keywords: N,N-dimethylformamide, Paracoccus sulfuroxidans, wastewater treatment, efficient degradation, biological method
Citation: Zheng G, Su Y, Zhang W, Liu X, Shao L, Shen Z, Zhang D, Wang K and Miao Z (2024) Characterization and genome sequence of N,N-dimethylformamide degradation in Paracoccus sulfuroxidans DM175A1-1 isolated from activated sludge of wastewater. Front. Microbiol. 15:1419461. doi: 10.3389/fmicb.2024.1419461
Edited by:
Arpit Shukla, Indian Institute of Advanced Research, IndiaReviewed by:
Dignesh Khunt, Gujarat Technological University, IndiaSaba Miri, York University, Canada
Copyright © 2024 Zheng, Su, Zhang, Liu, Shao, Shen, Zhang, Wang and Miao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhoudi Miao, NDg5NjY0MzA2QHFxLmNvbQ==
 Gang Zheng1,2
Gang Zheng1,2 Dongdong Zhang
Dongdong Zhang Zhoudi Miao
Zhoudi Miao