- 1Institute of Medical Microbiology and Hygiene/National Reference Laboratory for Listeria, Division for Public Health, Austrian Agency for Health and Food Safety, Graz, Austria
- 2FoodHub – Centre of Excellence for Digitalization of Microbial Food Safety Risk Assessment and Quality Parameters for Accurate Food Authenticity Certification, University of Donja Gorica, Podgorica, Montenegro
- 3Diagnostic Veterinary Laboratory, Podgorica, Montenegro
Introduction: Listeria monocytogenes is an ubiquitous foodborne pathogen that represents a serious threat to public health and the food industry.
Methods: In this study Whole Genome Sequencing (WGS) was used to characterize 160 L. monocytogenes isolates obtained from 22,593 different food sources in Montenegro during the years 2014–2022.
Results: Isolates belonged to 21 different clonal complexes (CCs), 22 sequence types (STs) and 73 core genome multilocus sequence types (cgMLST) revealing a high diversity. The most prevalent STs were ST8 (n = 29), ST9 (n = 31), ST121 (n = 19) and ST155 (n = 20). All isolates carried virulence genes (VGs), 111 isolates carried mobile genetic elements (MGEs) (ranging from 1 to 7 MGEs) and 101 isolates carried plasmids (ranging from 1 to 3 plasmids). All isolates carried the intrinsic resistance genes fosX and lin. None of the isolates carried acquired antimicrobial resistance genes (ARGs).
Discussion/conclusion: Continuous monitoring and surveillance of L. monocytogenes is needed for improving and ameliorating the public health.
1 Introduction
Listeria monocytogenes (L. monocytogenes) is the etiological agent of human and animal listeriosis (Chlebicz and Slizewska, 2018), with three major invasive clinical presentations in humans: bloodstream infection, infection of the central nervous system, and maternal foetal listeriosis (Hernandez-Milian and Payeras-Cifre, 2014). Strains of L. monocytogenes are often isolated from ready-to-eat (RTE) food, including meat, milk, dairy, fish and other seafood, but also ice cream, fresh frozen end processed vegetables, fruits, composite vegetables food and spices (Gambarin et al., 2012; Mateus et al., 2013). This pathogenic bacterium is ubiquitous in different habitats, such as: soil, water, vegetation, animal feed, farms and industrial environment (Kimura, 2006). As a contaminant of food production facilities, L. monocytogenes can be found in raw materials, in the environment of the facilities itself, on equipment and the final product. From all these spots, L. monocytogenes may spread throughout the facility via aerosols, personnel, food workflows and contaminated contact materials leading to its persistent presence and frequent resistance to hygiene measures for cleaning and disinfection (Zuber et al., 2019). Along with the ability to adapt to different niches and tolerate adverse environmental conditions better than all other non-sporulating bacteria (Liu et al., 2002), the ability of L. monocytogenes to form biofilms (Lakicevic and Nastasijevic, 2016) is another property which leads to long-term persistence and repeated cross-contamination of food products (Ferreira et al., 2014).
Listeria monocytogenes monitoring programs using molecular sub-typing techniques have been helpful in identifying persistent isolates within whole food chains (Leong et al., 2017). Microbial whole-genome sequencing (WGS) provides identification, characterization and subtyping of a microorganism at the ultimate level of resolution (Ronholm, 2018) making it a superior molecular typing method for monitoring foodborne outbreaks. L. monocytogenes is differentiated into 13 serotypes and four phylogenetic lineages of which most human listeriosis cases are associated with L. monocytogenes from lineage I, while L. monocytogenes from lineage II is commonly found in natural and farm environments (Orsi et al., 2011; Pyz-Lukasik et al., 2022; Benjamin et al., 2023).
The abuse or misuse of antibiotics, in both human and veterinary medicine, has led to an increase in the number of multidrug-resistant strains in recent decades (Maung et al., 2023). Although L. monocytogenes is normally susceptible to many antibiotics, some multidrug-resistant strains of L. monocytogenes have been isolated from food and the environment (Maung et al., 2023), following its initial discovery in France in 1988 (Poyart-Salmeron et al., 1990). In the food industry, the resistance of L. monocytogenes strains to disinfectants complicates the control and eradication of the bacteria in environments where hygiene is crucial (Oloketuyi and Khan, 2017). This poses a significant challenge that many groups address on a daily basis. Even though major advances in this field have been made, it remains crucial to develop safe and effective disinfectants against L. monocytogenes.
Virulence factors, encoded by virulence genes (VGs), are the key factor for L. monocytogenes to adapt and spread in the environment (Disson et al., 2021; Quereda et al., 2021). Listeria Pathogenicity Islands (LIPIs) are regions in the L. monocytogenes genomes playing a crucial role in virulence and pathogenicity (Quereda et al., 2021; Wiktorczyk-Kapischke et al., 2023). The four known LIPIs (LIPI-1 – LIPI-4) contain genes involved in various stages of infection like adhesion, invasion, and evasion of the immune response of the host. The key virulence factors encoded by LIPIs are internalins, listeriolysins and phospholipases (Wiktorczyk-Kapischke et al., 2023). LIPI-1 is essential for the intracellular lifecycle of L. monocytogenes and encodes the virulence factors InlA, InlB and listeriolysin O (LLO). LIPI-2 enables the movement within host cells and encodes ActA. LIPI-3 is found in certain strains and responsible for the increased virulence (Pyz-Lukasik et al., 2022), carrying lls genes (llsA, llsB, llsD, llsG, llsH, llsP, llsX, and llsY) which encodes listeriolysin S (LLS), a hemolytic/cytolytic factor impacting potential virulence. LIPI-4 encodes proteins involved in carbohydrate metabolism contributing to the ability of L. monocytogenes to survive and persist in the environment (Wiktorczyk-Kapischke et al., 2023). ST1 and ST4 L. monocytogenes have been previously reported of carrying both, LIPI-3 and LIPI-4 and are therefore regarded as hypervirulent clones (Shen et al., 2022). On the other hand, several types of Mobile Genetic Elements (MGEs) like transposons, plasmids and bacteriophages have been identified, that contribute to resistance and enhanced virulence (Parra-Flores et al., 2022).
In this study, we utilized WGS to characterize L. monocytogenes isolates collected over an eight-year period (2014–2022) as part of pathogen monitoring in Montenegro’s food industry with the aim to examine similarity and possible common origin sources between isolates. The absence of documented cases of human listeriosis in Montenegro underscores the importance for greater attention regarding this pathogen. Identification and association of identical/similar isolates circulating through the food chain will enhance monitoring efforts significantly. This approach can facilitate the establishment of more effective surveillance programs, helping food operators to successfully meet food safety regulations. Finally, these measures are crucial to improve consumer safety and public health.
2 Materials and methods
2.1 Origin and sample collection
A total of 22,593 different samples (environmental/food/other origin) were sampled from production facilities, retail shops and control at the borders. The samples were collected by analysing different food products of animal origin, i.e., raw milk and fresh meat during a period of 8 years (2014–2022), through programs of regular monitoring of the presence of L. monocytogenes in products on the market (8,698 samples), through programs of official controls (7,373 samples), but also within the framework of regular self-control by producers and food business operators (6,522 samples). The sampling that was done as part of regular monitoring programs as well as official control was carried out in accordance with the sampling plan in compliance with the principles of Commission Regulation (EC) no. 2073/2005 including the sampling of five units of the same production lot. The plan of sampling and testing of products carried out by manufacturers is most often also based on Commission Regulation (EC) no. 2073/2005. Tissue swabs were collected from different surfaces, including those in contact with food in production facilities (n = 798), the surface around the facility (n = 55) and workers’ hand (n = 115). Additionally, samples from raw milk (n = 14) and fresh meat (n = 34) were collected. For downstream analysis using Whole Genome Sequencing (WGS), one L. monocytogenes colony per positive sample was selected.
2.2 Cultural detection of Listeria monocytogenes
Samples were tested according to the EN ISO 11290-1 standard (ISO, 2022). Thus, samples were enriched (first in half Fraser broth at 30°C for 24 h, then in Fraser broth at 35°C for 48 h) and were then plated on Agar Listeria according to Ottaviani and Agosti (ALOA, Oxoid Ltd., Basingstoke, United Kingdom) and on Palcam agar (Oxoid Ltd., Basingstoke, United Kingdom) and incubated at 37°C for up to 48 h. Colonies which retained typical L. monocytogenes appearance were transferred to Tryptone Soya Agar plates supplemented with 0.6% yeast extract (TSYEA, Oxoid, Basingstoke, United Kingdom) and incubated at 37°C for 24 h. All isolates were identified with API Listeria system (BioMérieux SA, France) and all were stored in Brain Heart Infusion Broth (BHI, Oxoid Ltd., Basingstoke, United Kingdom) with 15% glycerol at −80°C. The isolates were refreshed once, every year, and then returned to storage at −80°C.
2.3 DNA extraction and whole genome sequencing
For whole genome sequencing (WGS) bacterial isolates were grown on blood agar plates supplemented with 5% sheep blood (COS) (Biomerieux, Vienna, Austria) at 37°C/24 h/aerobiosis. High molecular weight DNA was isolated from overnight cultures using the KingFisher Apex System (Thermofisher, Vienna, Austria) with the MagMAX Viral/Pathogen Ultra Nucleic Acid Isolation Kit (Thermofisher, Vienna, Austria) following the manufacturer’s instruction for Gram-positive bacteria with the following modifications: 2 hours of lysis with 60 μL enzyme mix. DNA was quantified using Dropsense 16 (Trinean NV/SA, Gentbrugge, Belgium) and Qubit Flex Fluorometer (Thermofisher, Vienna, Austria) with 1x dsDNA High Sensitivity (HS) kit (Thermofisher, Vienna, Austria). Genomic libraries were prepared using the DNA Illumina Prep kit (Illumina, San Diego, United States) according to the manufacturer’s instructions. Paired-end sequencing was performed on a NextSeq2000 instrument (Illumina, San Diego, United States) with a read length of 2 × 150 bp (Illumina, San Diego, CA, United States) aiming for a minimum coverage of 30 -fold.
2.4 Sequence data analysis
Raw reads were de novo assembled using SPAdes (version 3.11.1) (Bankevich et al., 2012). Contigs were filtered for a minimum coverage of 5 and a minimum length of 200 base pairs using SeqSphere+ software v8.5.1 (Ridom, Münster, Germany).
Subtyping of isolates was conducted in SeqSphere+ software v8.5.1 with classical multilocus sequence typing (MLST) based on the seven housekeeping genes as described by Salcedo et al. (2003), Clonal Complex (CC) (Ragon et al., 2008), core genome (cg)MLST (Werner et al., 2015) using a cluster threshold of 10 allelic differences.
Information on serogroups was extracted from genome data based on the 5-plex PCR (lmo118, lmo0737, ORF2110, ORF2829 and prs) as described (Michel et al., 2004; Lee et al., 2012).
NCBI AMRFinder+ v3.11.2. was used with the EXACT method at 100% setting together with the BLAST alignment at >90% of protein sequences against the AMRFinder+ database to detect antimicrobial resistance genes (ARG) (Michael et al., 2019). The Virulence Factor Database (VFDB) (http://www.mgc.ac.cn/cgi-bin/VFs/genus.cgi?Genus=Listeria; accessed on 6th June 2023) was used to detect VGs (Liu et al., 2022). Thresholds were set for the target scanning procedure to ≥85% identity with the reference sequence and ≥ 99% with the aligned reference sequence. Pathogenicity islands LIPI-3 and LIPI-4 were extracted from genome data. The CGE Mobile Element Finder v1.0.5. was used with the <90% identity and > 95% alignment method to detect Mobile Genetic Elements (MGE) (Johansson et al., 2021). Chromosome & Plasmid Finder v1.0. through MOB-suite v3.1.4. available in SeqSphere+ v8.5.1. was used with the Mash Neighbor Distance >0.06 to detect plasmids (Robertson and Nash, 2018). Isolates of the most prevalent STs of L. monocytogenes of our study (ST8, ST9, ST121, ST155, ST204) were compared with those from pubMLST database1 and the Austrian Reference Laboratory for Listeria, which currently includes more than 20,000 genomes.
3 Results
3.1 Detection of Listeria monocytogenes
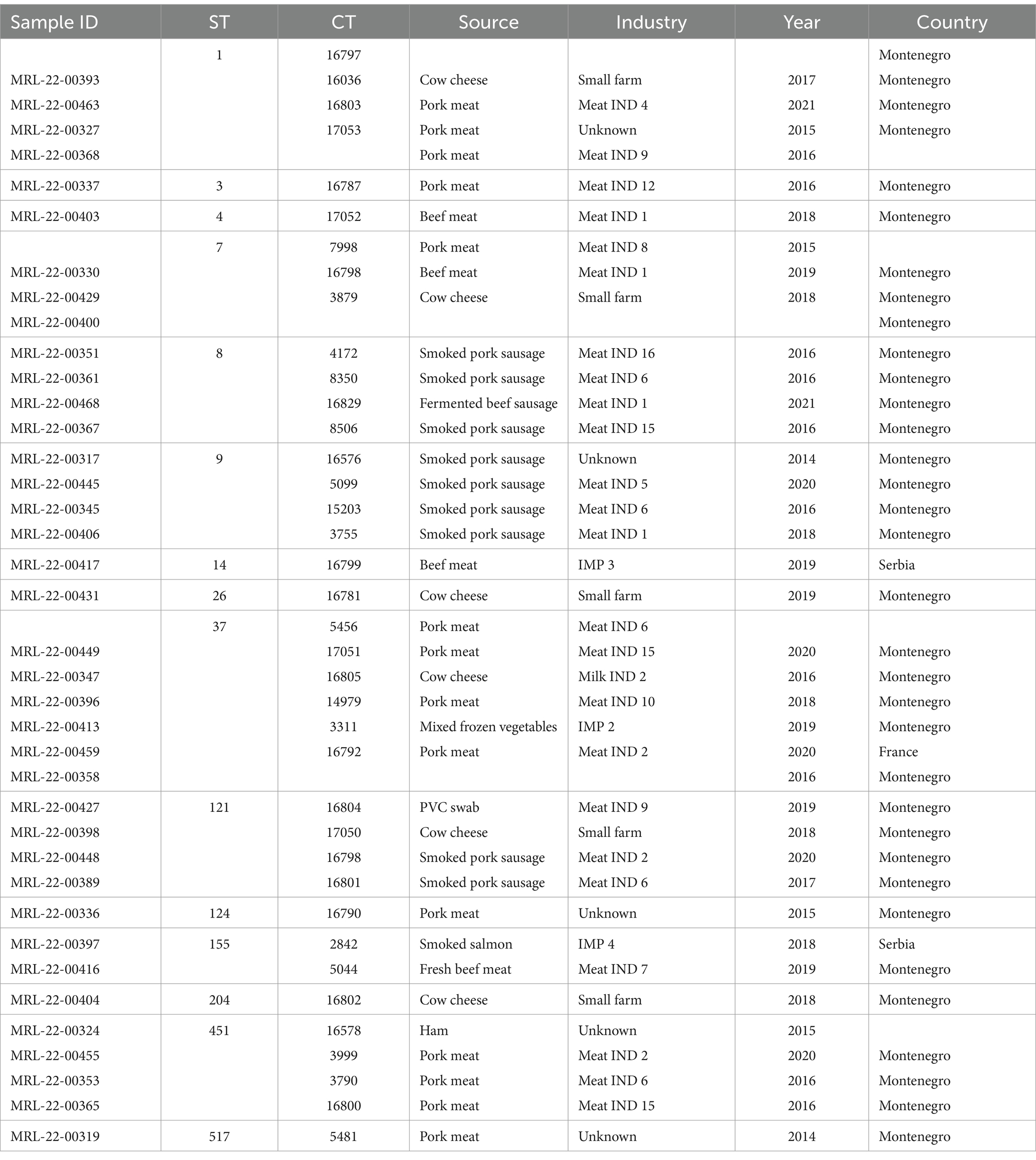
Out of 22,593 samples tested, L. monocytogenes was detected in 160 (0.7%) of them. From each positive sample, a single colony of L. monocytogenes was isolated, resulting in a collection of isolates consisting of 147 isolates from Montenegro and 13 isolates from food products imported from other European countries. During 8 years of monitoring the presence of L. monocytogenes in the food chain, a total of 986 environmental swabs, 48 raw material samples and 21,559 RTE food samples were examined. From all 147 isolates with Montenegrin origin, 131 isolates were from food products from 19 different industries, five isolates were isolated from small farms (households) and 11 L. monocytogenes isolates were isolated from unknown companies from Montenegro between 2014 and 2015. In addition, 13 isolates were obtained from food imported from different European countries: France = 1, Serbia = 2, Italy = 9 and Bosnia and Hercegovina = 1 (Tables 1, 2). From the total of isolates included in the study two L. monocytogenes isolates were obtained from environmental swabs, eight L. monocytogenes isolates originated from raw material and 150 L. monocytogenes isolates originated from food. 80% of the isolates originated from meat (pork, beef, chicken, and mixed meat), 16% from dairy products (mozzarella, cow cheese, butter, fresh milk), 0.7%, from ready-to-eat fish, 0.7% from frozen vegetables, 0.7% from food contact surfaces in production plants, and 1.8% were of unknown origin.
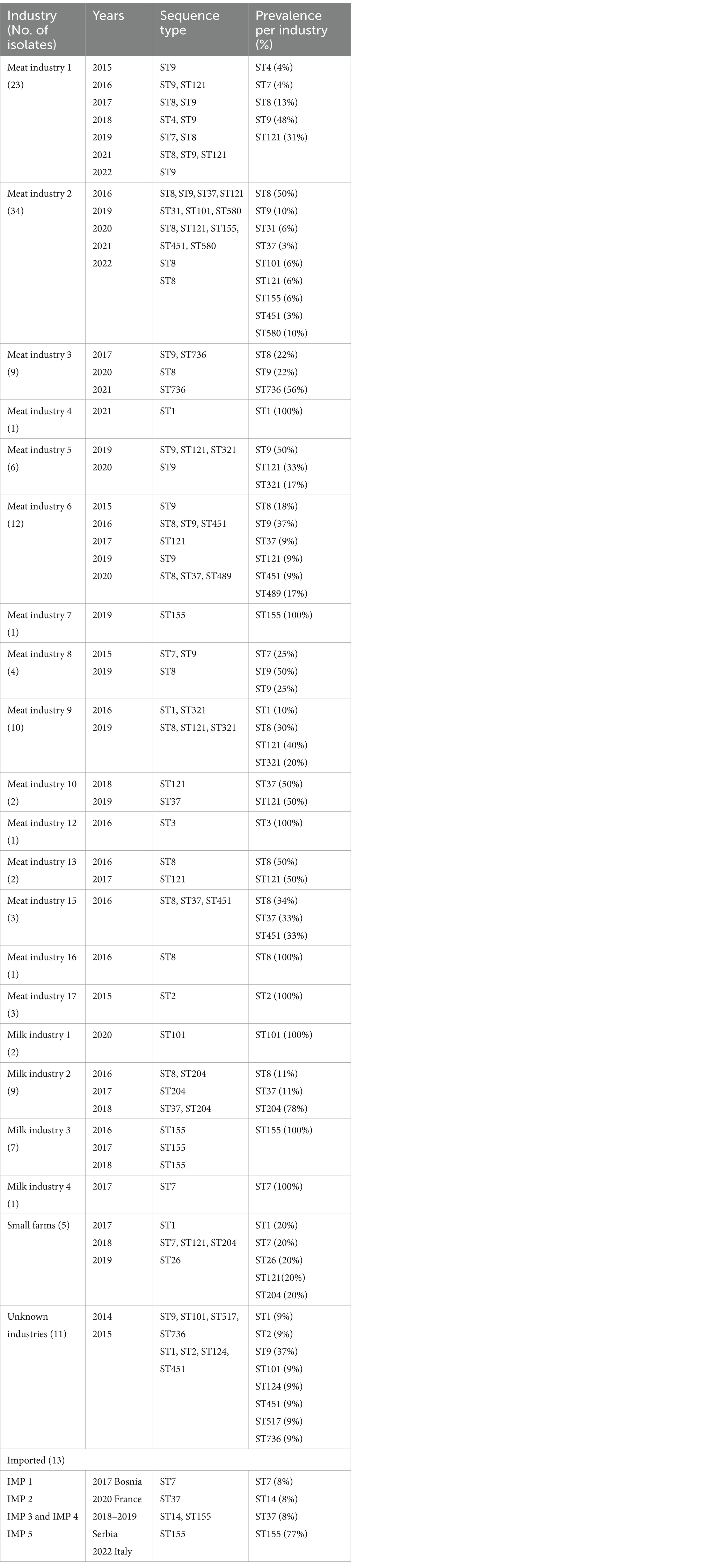
Table 1. Information on diversity of industries/producers, years, and sequence types (STs) in the L. monocytogenes isolates included in this study.
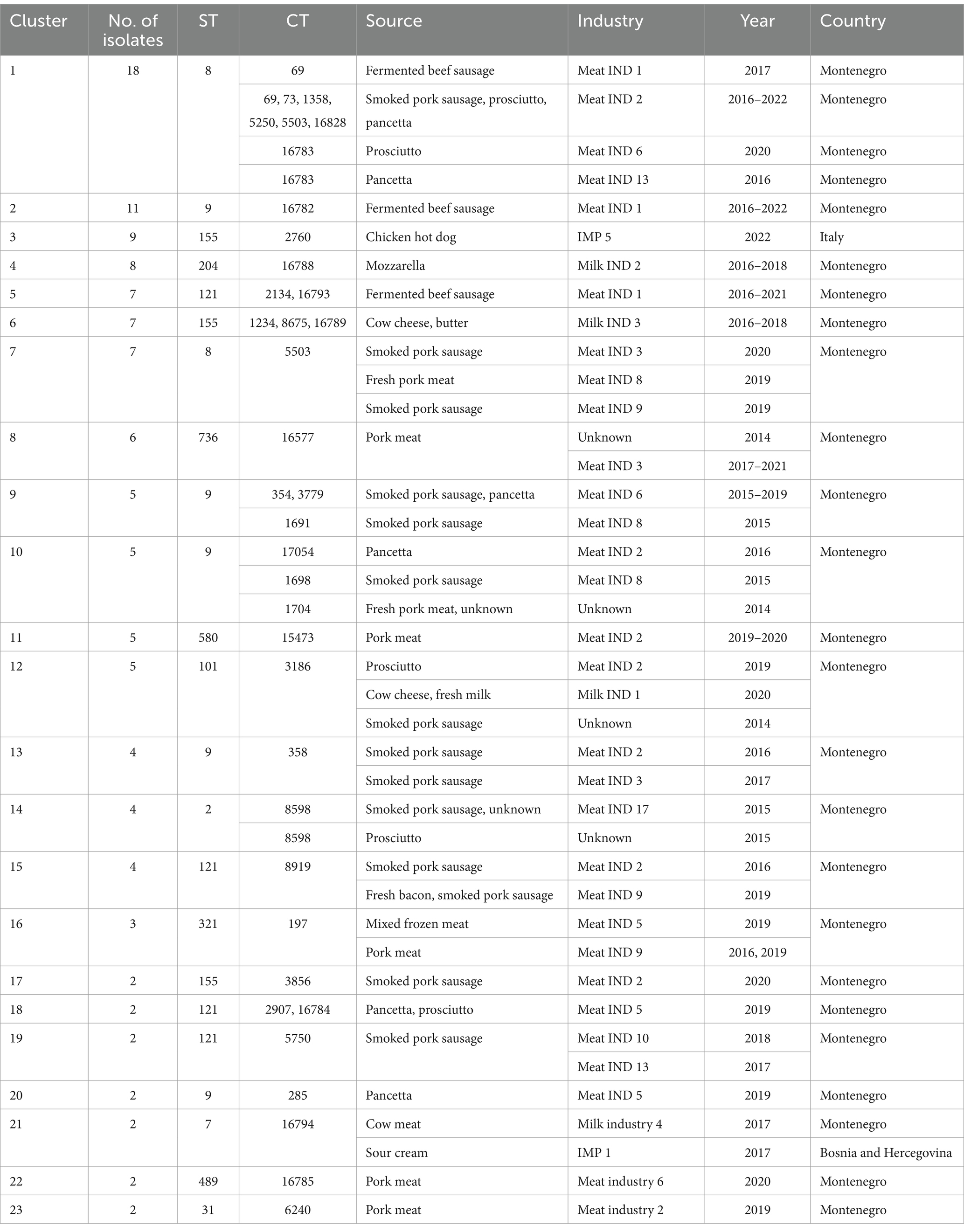
Table 2. Information on L. monocytogenes isolates belonging to different clusters included in this study.
3.2 Whole genome sequence based subtyping, description of clusters, and description of serogroups
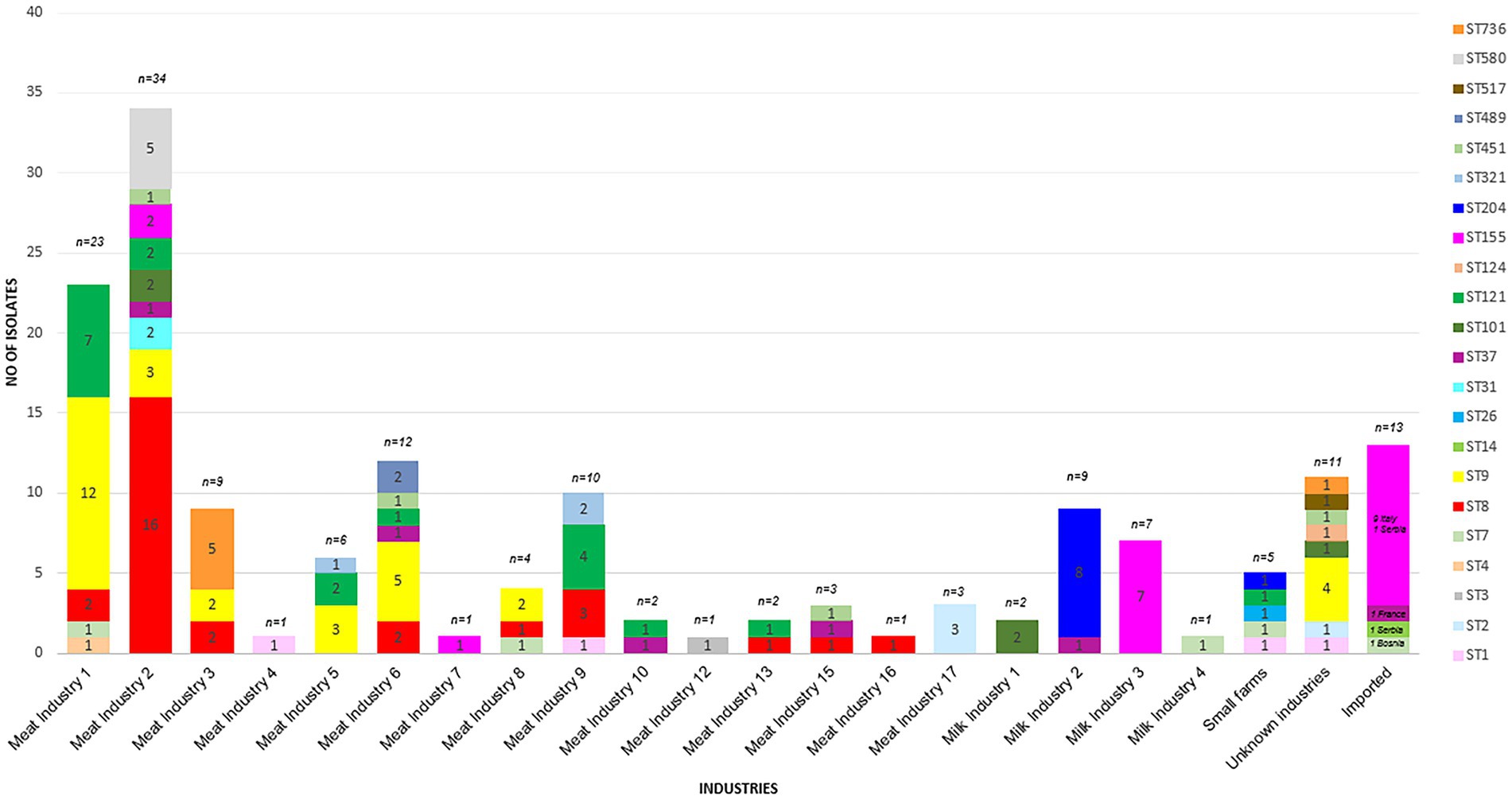
WGS based characterization of the 160 L. monocytogenes isolates revealed a high diversity. Isolates belonged to four different serogroups [IIa (n = 105), IIb (n = 10), IIc (n = 36) and IVb (n = 9)], 21 different clonal complexes (CCs), 22 different sequence types (STs) and 73 cgMLST complex types (CTs). One hundred twenty-two isolates belonged to a total of 23 cluster and 38 isolates were singletons (Figure 1; Table 2). Number of isolates per industry ranged from one isolate to 34 isolates (Table 1).

Figure 1. Minimum spanning tree (MST) of 160 L. monocytogenes isolates included in the study based on the cgMLST defined by Ruppitsch et al. (2015). Cluster definition was set in 10 allelic differences. Isolates are coloured by company/industry. Information in every isolate: ST, alias, year of isolation.
Listeria monocytogenes isolates belonging to serogroup IIa encompassed 13 different STs (ST7, ST8, ST14, ST26, ST31, ST37, ST101, ST121, ST124, ST155, ST204, ST321, and ST451), isolates belonging to serogroup IIb encompassed 4 different STs (ST3, ST489, ST517, and ST736), isolates belonging to serogroup IIc encompassed 2 different STs (ST9 and ST580), and L. monocytogenes isolates belonging to serogroup IVb encompassed 3 different STs (ST1, ST2, and ST4).
The most prevalent STs were ST8 (n = 29), ST9 (n = 31), ST121 (n = 19), ST155 (n = 20, including 9 isolates assigned to the Italian hotdog outbreak), and ST204 (n = 9) from the period 2015–2020 (Figure 2). The industries with the highest prevalence of isolates were Meat IND 1 (n = 23), Meat IND 2 (n = 34) and Meat IND 6 (n = 12).
L. monocytogenes isolates from Meat IND 1 (n = 23) encompassed five different STs (Table 1; Figures 2, 3). ST9 was the most prevalent with twelve isolates (52%) being detected every year from 2015–2022 (except 2019). All isolates belonged to cluster 2 (Figure 1). ST121 was the second most prevalent with seven isolates (31%). ST121 was the most prevalent type in the year 2016.
Listeria monocytogenes isolates from Meat IND 2 (n = 34) encompassed nine different STs (Table 1; Figures 2, 3). ST8 was the most prevalent with sixteen isolates (50%). ST8 was detected every year from 2016 to 2022 (except in 2019). All isolates belonged to cluster 1 (Figure 1).
L. monocytogenes isolates from Meat IND 6 (n = 12) encompassed six different STs (Table 1; Figures 2, 3). ST9 was the most prevalent with five isolates (37%) detected in 2015, 2016 and 2019. Four isolates belonged to cluster 9. One ST9 isolate differed by 1,129 alleles from cluster 9 (Figure 1).
Listeria monocytogenes isolates from Milk IND 1, 2, 3 and 4 encompassed five different STs (Table 1; Figures 2, 3) from years 2016–2020. ST155 was the most prevalent with seven isolates (35% of the ST155) in Milk IND 3. All isolates belonged to cluster 6 (Figure 1). ST204 was the most prevalent with nine isolates (100% of the total ST204) in Milk IND 2 and one small farm. All isolates belonged to cluster 4 (Figure 1).
3.2.1 Listeria monocytogenes ST8 (n = 29)
Twenty-nine L. monocytogenes ST8 isolates (18% of the total) were obtained from ten industries in Montenegro during the period 2016–2022. ST8 isolates were the most prevalent in Meat IND 2 (n = 16, 55,2% of ST8), Meat IND 13 (n = 1, 50%) and Meat IND 16 (n = 1, 100%). L. monocytogenes ST8 isolates were distributed to cluster 1 (n = 18), cluster 7 (n = 7) and four singletons. Cluster 1 comprised one isolate from Meat IND 1 (3.4% of the ST8), fifteen isolates from Meat IND 2 (48%), one isolate from Meat IND 6 (3.4%) and one isolate from Meat IND 13 (3.4%) with a maximum of eight allelic differences within the cluster (Figure 1). Cluster 7 comprised seven isolates differing by a maximum of three alleles. One isolate was obtained from Meat IND 2 (3.4%), two isolates from Meat IND 3 (7%), one isolate from Meat IND 8 (3.4%) and three isolates from Meat IND 9 (10%) (Figure 1). Cluster 1 and cluster 7 isolates differed by a minimum of 11 allelic differences (Figure 1). The four ST8 singletons were isolated from Meat IND 1 from 2021, Meat IND 6 from 2016, Meat IND 15 from 2016 and Meat IND 16 from 2016 (Figure 1; Supplementary Figure S1). They differed from each other by a minimum of 20 and a maximum of 35 alleles and from cluster 1 and cluster 7 isolates by a minimum of 12 and 19 alleles, respectively.
ST8 isolates from Meat IND 2 (cluster 1) were differing by 12 alleles from a salmon isolate from Poland (CC8/ST8/CT1269) (accession no. SRR975366) (Moura et al., 2016) and by 18 alleles from a human isolate from France (CC8/ST8/CT2873) (accession no. ERR1100940) (Maury et al., 2016; Moura et al., 2016).
3.2.2 Listeria monocytogenes ST9 (n = 31)
Thirty-one L. monocytogenes ST9 isolates (19% of the total) were obtained from ten different industries in Montenegro from the period 2014 to 2022. ST9 isolates were the most prevalent type in Meat IND 1 (n = 12, representing the 38% of ST9), Meat IND 5 (n = 3, 10%), Meat IND 6 (n = 5, 16%), and Meat IND 8 (n = 2, 6%) (Table 1; Figure 2). L. monocytogenes ST9 isolates were distributed to five clusters (cluster 2, 9, 10, 13 and 20) and four isolates were singletons. Cluster 2 isolates (n = 11, 35.48% of ST9) were obtained from beef meat from Meat IND 1 from 2015 to 2022, whereas isolates from the other clusters were derived from pork meat. Cluster 9 isolates (n = 5, 16.12% of ST9) were obtained from Meat IND 6 and Meat IND 8, with a maximum of eight allelic difference. Isolates within Cluster 10 (n = 5, 16.12% of ST9) were obtained from Meat IND2 (n = 1, 3.22%), Meat IND 8 (n = 1, 3.22%) and from and unknown industries (n = 3, 9.67%), with a maximum of ten allelic differences within the cluster. Cluster 13 isolates (n = 4; 12.90% of ST9) were obtained from Meat IND 2 and Meat IND 3 with a maximum difference of nine alleles. Isolates in Cluster 20 (n = 2, 6.45% of ST9) derived from Meat IND 5 showed no allelic difference in their core genome.
A ST9 singleton (MRL-22-00345) from Meat IND 6 differed by 18 and 23 alleles from two human isolates from France, (CC9/ST9/CT7213) (accession no. ERR1100975) and (CC9/ST9/CT7272) (accession no. ERR1100950) (Maury et al., 2016; Moura et al., 2016), respectively. ST9 isolates from Meat IND 6 (cluster 9) differed by 50 alleles from a human isolate from France (CC9/ST9/CT7208) (accession no. ERR1100972) (Maury et al., 2016; Moura et al., 2016). ST9 isolates from Meat IND 2 (cluster 13) differed by 25 alleles from a human isolate from France (CC9/ST9/CT7211) (accession no. ERR1100973) (Maury et al., 2016; Moura et al., 2016). Strains with the same cgMLST profile (CC9/ST9/CT354) and (CC9/ST9/CT3779), mainly obtained from meat samples, were found in the database of the Austrian National Reference Laboratory (NRL) for Listeria (personal communication).
3.2.3 Listeria monocytogenes ST121 (n = 19)
Nineteen L. monocytogenes ST121 isolates (12% of the total) were obtained from seven industries and one small farm from 2016 to 2021. L. monocytogenes ST121 were distributed to four clusters (cluster 5, 15, 18 and 19) and four singletons. Cluster 5 (n = 7, representing 37% of ST121) comprised isolates obtained from Meat IND 1 from the years 2016 (n = 6) and 2021 (n = 1) differing by a maximum of six alleles (Table 1; Figure 2). Cluster 15 (n = 4; representing 21% of ST121) isolates were obtained from Meat IND 2 in 2016 (n = 1) and from Meat IND 9 in 2019 (n = 3, representing 16% of ST121) with a maximum of three allelic differences. Cluster 18 (n = 2) isolates were obtained from Meat IND 5 in 2019. Cluster 19 (n = 2; representing 10% of ST121) isolates were obtained from Meat IND 10 in 2018 and Meat IND 13 in 2019. Four ST121 singletons were obtained from three industries (Meat IND 2, 6, 9) and a small farm differing by 23 to 61 alleles from each other and by 17 to 51 alleles from the clusters (Figure 1; Supplementary Figure S2).
ST121 isolates from Meat IND 5 (cluster 17) differed by 9, 21 and 68 alleles from French isolates originating from human and food-associated sources (CC121/ST121/CT2907) (accession no. 1100947), (CC121/ST121/CT7267) (accession no. ERR1100919) and (CC121/ST121/CT7212) (accession no. 1100974), respectively, (Maury et al., 2016; Moura et al., 2016).
3.2.4 Listeria monocytogenes ST155 (n = 20)
Twenty L. monocytogenes isolates ST155 were obtained from three industries in Montenegro, one industry from Serbia and one from Italy from 2016 to 2022. ST155 isolates were distributed to three clusters (cluster 3, 6 and 17) and two isolates were singletons. Cluster 3 (n = 9, representing 45% of ST155) comprised identical isolates obtained from chicken sausages from industry IMP 5 from Italy in 2022. Cluster 6 (n = 7, representing 35% of ST155) isolates were obtained from Milk IND 3 from 2016 and 2018 differing by a maximum of three alleles. Cluster 17 (n = 2; 10% of ST155) isolates were obtained from pork meat from Meat IND 2 in 2020 showing one allelic difference. Two singletons were obtained from industry IMP 4 (Serbia, 2018) and beef meat from meat IND 7 in 2019 differing by a minimum of 62 alleles from each other (Figure 1; Supplementary Figure S3).
ST155 isolates from Milk IND 3 (cluster 6) differed by 32, 42 and 51 alleles from one isolate from Finland (CC155/ST155/CT15) (accession no. NC_017547) (den Bakker et al., 2013; Maury et al., 2016), two isolates from France [one from human (CC155/ST155/CT2885) (accession no. ERR1100937) (Maury et al., 2016; Moura et al., 2016) and one from salmon (CC155/ST155/CT7342) (accession no. ERR10849967) (Moura et al., 2024)].
A clinical isolate from Serbia (unpublished, personal communication) from the year 2015 (CC155/ST155/CT2760) available through the NRL for Listeria database differed by 5 alleles from cluster 3 food isolates (Italian outbreak; EpiPulse ID: 2022-FWD-00053). In addition, nine clinical isolates from Serbia and eight isolates from Austria isolated from fish (CC155/ST155/CT2842) from the NRL for Listeria database clustered with a singleton from our study (MRL-22-00397, CC155/ST155/CT2842), which was obtained from smoked salmon (personal communication).
3.2.5 Listeria monocytogenes ST204 (n = 9)
A total of nine L. monocytogenes ST204 isolates were obtained from Milk IND 2 (n = 8, representing 88% of ST204) and one small farm in Montenegro (n = 1, 12%). All Milk IND 2 isolates belonged to cluster 4 differing by a maximum of one allele within the cluster, whereas the small farm isolate differed by a minimum of 37 alleles from cluster 4 (Figure 1; Supplementary Figure S4).
3.2.6 Other Listeria monocytogenes STs
Six ST736 isolates (3.75% of the total) obtained from Meat IND 3 from 2017 to 2021 and from an unknown industry in 2014 belonged to cluster 8. Five ST580 isolates (3.125% of the total) from Meat IND 2 belonged to cluster 11. Five ST101 isolates (3.125% of the total of isolates) obtained from Milk IND 1 in 2020, Meat IND 2 in 2019, and an unknown industry in 2014 a belonged to cluster 12 (Table 2; Figure 1). Twenty-three isolates were singletons obtained from different sources, industries, and years (Table 3). Singletons belonging to ST1 (n = 4, 2.5% of the total), ST37 (n = 6, 3.75%) and ST451 (n = 4, 2.5%) were the most prevalent and were obtained from different sources and from different industries (Table 3).
Some singletons obtained from pork meat with less prevalent STs from our study showed a close genetic relationship with strains from the Austrian NRL for Listeria database (unpublished, personal communication). Singleton CC1/ST1/CT16036 (MRL-22-00463) was identical to two Austrian strains obtained from ice-cream. Singleton CC7/ST7/CT7998 (MRL-22-00330) differed by 4 and 9 alleles, from two Austrian isolates obtained from sausage and sheep meat. Singleton CC37/ST37/CT5456 (MRL-22-00449) differed by one allele to four Austrian strains obtained from meat and meat products. Singleton CC11/ST451/CT3790 (MRL-22-00353) clustered with 40 Austrian strains which were obtained from meat and meat products. Singleton CC517/ST517/CT5481 (MRL-22-00319) clustered with 44 Austrian isolates, which were obtained from meat and meat products.
Cluster 16 isolates (CC321/ST321/CT197) from our study differed by nine alleles from a clinical isolate (accession no SRR13744450) from Queens, United States of America.
Detailed information about isolates included in this study are available in the supplements (Supplementary Table S1).
3.3 Mobile genetic elements and plasmids
One hundred eleven out of 160 isolates carried at least one MGE. Nine isolates (5.6% of the total) carried over five MGEs. The most prevalent MGE was the unit transposon Tn5422, which was found in eighty-four isolates (52.5% of the total) followed by the insertion sequences ISLmo1 and ISLmo9 found in forty-three (26.8% of the total) and forty-seven (29.3% of the total) isolates, respectively (Supplementary Table S1). All L. monocytogenes ST8, ST9 and ST121 isolates carried MGE Tn5422. ISLmo1, ISLmo7, ISLmo8, and ISLmo9 were present in 32, 39, 39, and 55% of L. monocytogenes ST9, respectively. All ST204 and two ST155 isolates carried ISLmo1. The remaining eighteen ST155 isolates and ST1, ST2, ST4, ST7, ST451, ST489, ST517, ST736 isolates carried no MGEs. ST3, ST9, ST31, and ST580 carried 5–7 MGEs (Supplementary Table S1).
One hundred one out of 160 L. monocytogenes isolates carried at least one plasmid. The plasmid sizes ranged from 25.6–432.1 kb. All L. monocytogenes isolates belonging to ST3, ST8, ST9, ST31, ST121, ST321, ST517 and ST580 carried plasmids (Supplementary Table S1; Supplementary Figure S5). L. monocytogenes isolates belonging to those ST profiles were obtained from pork or beef sausages from different meat industries. Seven out of twenty ST155 isolates (35% of ST155), one out of four ST451 isolates (0.6%) and one out of six ST736 isolates (3.75%) carried plasmids (Supplementary Table S1). All ST155 plasmid carrying isolates were obtained from butter or cow cheese from Milk IND 3. ST451 and ST736 plasmid carrying isolates were obtained from pork meat from unknown and Meat IND 3, respectively.
3.4 Antimicrobial genes, pathogenicity islands, and virulence genes
The fosX and lin genes were detected in all 160 L. monocytogenes isolates included in this study.
All known Listeria VGs were detected in the Montenegrin isolates (Supplementary Table S1). Eight out of 160 L. monocytogenes isolates (5% of the total) carried pathogenicity island LIPI-3. Specifically, four ST1 (2.5% of the total) isolates, one ST3 (0.6%) isolate, one ST4 (0.6%) isolate and two ST489 (1.25%) isolates obtained from seven different industries carried LIPI-3. Two out of 160 isolates carried LIPI-4 (1.25% of the total). Specifically, one ST4 (0.6%) isolate and one ST517 (0.6%) isolate obtained from different type of meat and different industries carried LIPI-4. One ST4 (0.6%) isolate obtained from beef meat from Meat IND 1 carried both, LIPI-3 and LIPI-4. L. monocytogenes ST1 isolates lacked the actA, ami, aut, gtcA, inlF, inlJ, inlK, and vip. L. monocytogenes ST2 lacked the ami, aut, gtcA, inlF, inlJ, inlK, lapB, and vip. L. monocytogenes ST7, ST37, ST204 and ST321 lacked vip. L. monocytogenes ST8, ST9, ST155, ST451 and ST580 had the complete spectra of VGs. One ST101 isolate lacked VGs aut, fbpA, iap/cwhA, inlF, inlA, inlJ, inlP, lplA1, plcB, and prfA, all other ST101 isolates carried all known VGs. L. monocytogenes ST121 lacked actA, iap/cwhA, inlF, and inlJ.
Briefly, one hundred eighteen out of 160 L. monocytogenes isolates carried InlF, one hundred twenty L. monocytogenes isolates carried InlJ, one hundred twenty-five L. monocytogenes isolates carried ActA, one hundred forty-five L. monocytogenes isolates carried aut, one hundred forty-six isolates carried ami and InlK, one hundred fifty-one L. monocytogenes isolates carried GtcA, one hundred fifty-two L. monocytogenes isolates carried lapB, one hundred fifty-seven L. monocytogenes isolates carried InlA, InlB and InlP, one hundred fifty-eight L. monocytogenes isolates carried PdgA and LplA1, one hundred fifty-nine L. monocytogenes isolates carried FbpA, Hyl, plcA, plcB, PrfA and Bsh, one hundred sixty L. monocytogenes isolates carried OatA and LntA. ClpC, ClpE, ClpP, InlC, lap, Mpl, LspA, LpeA, PrsA2, and Iap genes were present in all L. monocytogenes isolates.
4 Discussion
4.1 Genetic diversity
Controlling L. monocytogenes in the food chain is an ongoing and important challenge due to its resilience in different environments. Over a period of 8 years, samples from food products and food production environments of different origin and composition were examined, showing a prevalence rate of 0.7% among all samples analysed. In a similar study conducted in the USA from the period 2005–2017, the prevalence of L. monocytogenes in 150,000 RTE meat samples was 0.4% (Mamber et al., 2020), which is similar to the prevalence in RTE meat samples in our study (0.6%). The incidence of L. monocytogenes in RTE pork in our study (69%) is significantly higher compared to similar studies (Gamboa-Marin et al., 2012; Wang et al., 2018), where percentages ranged from 13.5–33.9%. The presence of L. monocytogenes in RTE milk products was 12%, which is consistent with findings from other studies (Chen et al., 2020). The occurrence in RTE fish was 0.5%, which is lower than reported in recent studies (Korsak et al., 2012; European Food Safety Authority, and European Centre for Disease Prevention and Control, 2017).
In our study, all isolates belonged either to lineage I or II, both of which are known sources of listeriosis outbreaks, as previously described (Fretz et al., 2010; Orsi et al., 2011; Ruppitsch et al., 2015; Gelbíčová et al., 2018; Pietzka et al., 2019; Matle et al., 2020; Benjamin et al., 2023). Genetic characterization of L. monocytogenes isolates from Montenegro is crucial for microbiological safety because both, lineage I and lineage II L. monocytogenes strains pose a serious threat to public health.
Based on the WGS characterization, the 160 isolates of our study were grouped into four serogroups (IIa, IIb, IIc, and IVb), being serogroup IIa the most prevalent (66.4%), followed by serogroup IIc (21.7%), serogroup IIb (6.2%) and lastly, serogroup IVb (5.6%). Similar findings were reported by Korsak et al. (2012), where 471 L. monocytogenes strains isolated from different foods had the following serogroup distribution: IIa (54.4%), IIc (25.5%), IIb (12.5%) and IVb (7.6%). Serogroup IIa was isolated from a wide variety of milk and meat products from several industries mainly from Montenegro, but also from other countries such as France, Spain, Serbia, and Bosnia. These results are in concordance with previous studies characterizing L. monocytogenes from different food sources, which showed the higher prevalence of serogroup IIa in comparison to others (Clémentine et al., 2016; Pyz-Lukasik et al., 2022). Some of the characteristics that may contribute to a higher prevalence of serogroup IIa in the food and food processing environment are a more efficient biofilm formation and predomination in biofilms of mixed cultures (Youwen et al., 2009), a greater resistance to bacteriocins (Korsak et al., 2012) and the possession of a great number of plasmids (60% of L. monocytogenes serogroup IIa isolates of our study carried at least one plasmid) that confer resistance to toxic metals and possibly other compounds found in the environment (Orsi et al., 2011). Nevertheless, not only group IIa is a contaminant in food since previous studies also reported that these four groups (groups IIa, IIb, IIc, and IVb) are involved in listeriosis worldwide (Gorski, 2021). For instance, the study carried out by Espinosa-Mata et al. (2022) reported that serogroup IVb was the most prevalent among L. monocytogenes isolates coming from cheese, which is in the opposite direction from our study since our isolates coming from cheese belongs mainly to serogroup IIa.
The most prevalent STs among our isolates were ST8, ST9, ST121 and ST155, which is in concordance with other studies (Cabal et al., 2019; Shen et al., 2022). In our study, we observed the spreading of the same ST in different meat and dairy industries during different years. ST9 is a common food associated (Althaus et al., 2014; Martín et al., 2014; Maury et al., 2016, 2019) one in most European countries and worldwide (Bespalova et al., 2021). ST9 has been shown to persist in the same facility for more than 9 years successfully overcoming hygienic barriers within the factory (Annette et al., 2020). A possible hypothesis for the occurrence of the exact same ST9 clone in the two industries in Montenegro is that the strain entered the facilities with the raw meat imported from Spain (personal communication).
Although there are no reports on cases of human listeriosis in Montenegro, the fact that hypervirulent ST8 strains are among the most prevalent STs in food samples in this study is of concern. ST8 isolates were detected in almost all meat and dairy industries. A previous study showed that in Germany, thirteen out of thirty-nine listeriosis cases caused by L. monocytogenes ST8 were detected at health-care facilities where patients had consumed ready-to-eat meat products from the same manufacturer (Lachmann et al., 2021). L. monocytogenes ST8 and ST121 are described to contain plasmids that contribute to a higher tolerance to high temperature, salinity, acidic environments, oxidative stress and disinfectants (Naditz et al., 2019), contributing to their ubiquity and persistence capabilities. These results agree with ours, since all ST8 and ST121 carried plasmids. In our study ST121 is predominantly associated with the meat food industry, which does not contradict other studies where ST121 is predominantly associated with food plant environments (Schmitz-Esser et al., 2015). The L. monocytogenes ST155 isolates examined in this study were isolated from milk products and chicken meat from Montenegro and Italy, respectively, as well as from an unknown source from Serbia. Previous studies have documented the involvement of ST155 in clinical cases (Zhao et al., 2021) and outbreaks (Stessl et al., 2022) in recent years. Other STs reported as hypervirulent, such as ST1, ST2, and ST4 were detected in nine isolates coming mainly from meat products but one isolate was derived from cowmilk cheese. Detection of the hypervirulent ST1, ST2 and ST4 strains (Guidi et al., 2021) in our study represents a public health concern. Our ST1 strains differed by a minimum of 40 alleles to ST1 strains from recent studies (Maury et al., 2016; Guidi et al., 2021). National surveillance data from France and the Netherlands indicate this clonal complex as predominant in systemic infections in humans with neurological forms of listeriosis (Maury et al., 2016; Koopmans et al., 2017).
In our study ST101 strains were detected in products from two different industries (Milk IND 1 and Meat IND 2), located within the same industrial yard and being part of the same production brand. While the production procedures of these two industries are separated, our findings suggest that cross-contamination of products with this ST101 strain consistently present in the environment is likely occurring. Recognizing the environment as a key source of L. monocytogenes, Linke et al. (2014), underscored the significance of the environment as a primary source of L. monocytogenes, as shown in their two-year study of soil and water samples, which revealed ST101 as one of the dominant strains (Linke et al., 2014). In the Listeria SEQuencing (LiSEQ) project ST101 isolates were linked with milk/milk products (Painset et al., 2019). The detection of two identical ST7/CT16794 isolates in 2017, one from fresh cow’s milk from Montenegro and the other from sour cream from Bosnia might be explained by the import of milk contaminated with listeria from Bosnia and Serbia (personal communication). Along with the identification of ST7 strains in clinical and food samples, it has been linked to listeriosis cases in cattle and sheep (Annette et al., 2020; Zhao et al., 2021). Among the 20 different STs isolated from meat products ST7 has been reported, along with others like ST1, ST2, ST3 and ST155 to persist in industrial plants for extended periods. The consistent isolation of isolates with no allelic differences from spatially distant industries suggests a possible contamination originating from the raw material. Notably, the only isolate (ST37) detected in frozen vegetables during the entire testing period was detected in a product imported from France in 2020. This same ST was also found in isolates coming from pork from various meat industries and in cow milk cheese. In addition to a frequent isolation from clinical samples, ST37 has been isolated from meat, fish as well as dairy products (Alvarez-Molina et al., 2021; Kubicová et al., 2021; Maćkiw et al., 2021). ST37 has also been isolated from mallard, pheasant and teal feces, moose carcass and wild boar (Fredriksson-Ahoma et al., 2022). In Austria in 2017, ST37 was the fifth most frequent ST in the category food including meat, dairy products, and vegetables (Cabal et al., 2019).
Several L. monocytogenes types of our study were found to be closely related to L. monocytogenes strains several other European countries (den Bakker et al., 2013; Maury et al., 2016; Moura et al., 2016, 2024). This highlights the broader European scenario of L. monocytogenes dissemination and the conservation of isolation sources across different countries, which is influenced by the international trade of food. These findings underscore the importance of stringent control measures for L. monocytogenes strains in the global food chain to prevent cross-border contamination and ensure food safety.
4.2 Virulence genes and pathogenicity islands of Listeria monocytogenes isolates
LIPIs are specific regions in the genome of L. monocytogenes that encode virulence factors playing a crucial role for its pathogenicity (Quereda et al., 2021; Wiktorczyk-Kapischke et al., 2023). Those virulence factors, as previously said, are key factors in the adaptation and spread of L. monocytogenes in the environment (Disson et al., 2021; Quereda et al., 2021; Fredriksson-Ahoma et al., 2022). de Melo Tavares et al. (2020) showed that L. monocytogenes lineage II did not carry LIPI-3, which is in contrast with our study since three L. monocytogenes lineage II in our study carried LIPI-3, although the LIPI-3 carrying L. monocytogenes were in the majority lineage IVb. Shen et al. (2022) and Lake et al. (2021) showed that L. monocytogenes CC4 was a carrier of LIPI-4, which is in accordance with our findings. However, in our study the other strain with LIPI-4 was L. monocytogenes ST517, for which there is no information whether it normally has LIPI-4. Furthermore, numerous studies agree that L. monocytogenes ST9 and ST121 do not carry pathogenicity islands of any kind (Shen et al., 2022), which also agrees with our results, since none of them carried neither LIPI-3 nor LIPI-4. Recently, Zhang et al. (2024) described L. monocytogenes isolates from RTE carrying LIPI-3 and LIPI-4, as well as ST3 isolates carrying LIPI-3, which is in concordance with the results of our study.
A special feature of L. monocytogenes lineage II is the presence of vip, inlF and inlK which has been described only to be present in this lineage and not in others (Pyz-Lukasik et al., 2022), which is in concordance with our results since, for instance all our L. monocytogenes lineage IV isolates were missing these genes (vip, inlF, inlK) in addition to the lack of others such as inlJ, gtcA, ami, and aut.
Several studies showed that aut is not present in L. monocytogenes lineage IVb (Cabanes et al., 2004) which agrees with our findings since in all of them aut gene was absent. On the other hand, the study carried out by Pyz-Lukasik et al. (2022) suggests that vip gene is only present in pathogenic Listeria species, while it is absent in non-pathogenic Listeria. Some studies revealed that CC9 L. monocytogenes is highly associated with the presence of vip gen (Wang et al., 2019) which agree with our results (present in eighty-seven isolates). In contrast, some studies reported L. monocytogenes ST9 strains (which is one of the STs included in CC9) as hypo virulent. This would contrast with the above and with our results since, if L. monocytogenes ST9 is hypo virulent it should have no association with vip and instead in our study they do.
4.3 Antimicrobial resistance genes, plasmids, and mobile genetic elements
All our isolates carried lin and fosX ARGs, which is in concordance with other studies (Olaimat, 2018; Hanes and Huang, 2022), lin and fosX are considered as natural/ intrinsic resistance factors of L. monocytogenes conferring resistance to lincosamides and fosfomycin (Olaimat, 2018; Fredriksson-Ahoma et al., 2022; Hanes and Huang, 2022). No acquired ARGs were detected.
Previous studies reported that the most prevalent MGEs found in L. monocytogenes were ISLmo3, ISLmo5, ISLmo7, ISLmo9, ISLmo8, ISS1N, cn_8625_ISS1N and cn_12410_ISS1N (Parra-Flores et al., 2022), which agrees with our findings since all were found in our strains. Tn5422 is a natural transposon of L. monocytogenes which can generate deletions being probably the reason for the size diversity of the L. monocytogenes plasmids (Lebrun et al., 1994). On the other hand, Tn6188 is structurally related to Tn554 from Staphylococcus aureus, which suggests a common origin or horizontal gene transfer within these both species (Müller et al., 2013). In the same study they found that this Tn6188 was present in some Listeria monocytogenes strains coming from food and food processing environments predominantly from serovar 1/2a. These results are in concordance with our findings.
ST1, ST2, ST3, ST155 and ST204 have been described as plasmid-carriers clones coming from food (Wagner et al., 2020; Mafuna et al., 2021; Schmitz-Esser et al., 2021). In contrast, in our study L. monocytogenes ST1, ST2, ST3, ST155 and ST204 strains did not carry any plasmids. On the other hand, ST101, ST124, ST489 were not yet reported in the literature as plasmid carriers, which is in concordance with our results.
5 Conclusion
Monitoring the presence of L. monocytogenes over a longer period is a reliable indicator of this pathogen’s opportunities in the food chain. Our results reveal a high genetic diversity and variability of L. monocytogenes in the Montenegrin food chain, underscoring the importance of ongoing surveillance. This data is crucial for improving and ameliorating the public health and food safety sector. Whole-genome sequencing (WGS) allows researchers the tracking of the trajectory and persistence of L. monocytogenes among other pathogens, and the identification of genetic markers of resistance and virulence. The combination of WGS and seeking the epidemiological link provides a comprehensive view of the pathogen’s spread and impact. Furthermore, the Austrian reference database for L. monocytogenes, alongside other extensive and continuously updated pathogen databases, has allowed us to verify that many of our isolates share a close genetic relationship with isolates of the same sequence type (ST) widely distributed across different European countries. These findings reinforce the relevance of our study in a broader European context, not only at the level of Montenegro, establishing relevant connections in the field of food safety and public health.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
BD: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. AP: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. AM: Conceptualization, Formal analysis, Funding acquisition, Investigation, Resources, Supervision, Visualization, Writing – review & editing. WR: Conceptualization, Formal analysis, Funding acquisition, Investigation, Resources, Software, Supervision, Writing – original draft, Writing – review & editing. IZ: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project was partially funded by the Alliance of International Science Organizations (ANSO) through the project: Integrated Monitoring, Bridging the Knowledge Gap and Control of Foodborne Viral and Bacterial Contamination in the Food Supply Chain. Project No.: ANSO-CR-PP-2022-07.
Acknowledgments
This research was carried out with the activity of the Diagnostic Veterinary Laboratory staff, to whom we are especially grateful to Snežana Vučinić, DVM. We extend our gratitude to Tanja Tošić from the University Clinical Centre of Serbia for her valuable assistance in the strain comparison.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1418333/full#supplementary-material
Footnotes
References
Althaus, D., Lehner, A., Brisse, S., Maury, M., Tasara, T., and Stephan, R. (2014). Characterization of Listeria monocytogenes strains isolated during 2011–2013 from human infections in Switzerland. Foodborne Pathog. Dis. 11, 753–758. doi: 10.1089/fpd.2014.1747
Alvarez-Molina, A., Cobo-Díaz, J. F., López, M., Prieto, M., de Toro, M., and Alvarez-Ordóñez, A. (2021). Unraveling the emergence and population diversity of Listeria monocytogenes in a newly built meat facility through whole genome sequencing. Int. J. Food Microbiol. 340:109043. doi: 10.1016/j.ijfoodmicro.2021.109043
Annette, F., Solveig, L., and Trond, M. (2020). In-depth longitudinal study of Listeria monocytogenes ST9 isolates from the meat processing industry: resolving diversity and transmission patterns using whole-genome sequencing. Appl. Environ. Microbiol. 86, e00579–e00520. doi: 10.1128/AEM.00579-20
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Benjamin, F., Karine, C., Sandrine, T., Arnaud, F., Guillaume, G., Carole, F., et al. (2023). Identification by high-throughput real-time PCR of 30 major circulating Listeria monocytogenes clonal complexes in Europe. Microbiol. Spectr. 11:e0395422. doi: 10.1128/spectrum.03954-22
Bespalova, T. Y., Mikhaleva, T. V., Meshcheryakova, N. Y., Kustikova, O. V., Matovic, K., Dmitric, M., et al. (2021). Novel sequence types of Listeria monocytogenes of different origin obtained in the republic of Serbia. Microorganisms 9:1289. doi: 10.3390/microorganisms9061289
Cabal, A., Pietzka, A., Huhulescu, S., Allerberger, F., Ruppitsch, W., and Schmid, D. (2019). Isolate-based surveillance of Listeria monocytogenes by whole genome sequencing in Austria. Front. Microbiol. 10:2282. doi: 10.3389/fmicb.2019.02282
Cabanes, D., Dussurget, O., Dehoux, P., and Cossart, P. (2004). Auto, a surface associated autolysin of Listeria monocytogenes required for entry into eukaryotic cells and virulence. Mol. Microbiol. 51, 1601–1614. doi: 10.1111/j.1365-2958.2003.03945.x
Chen, Y., Chen, M., Wang, J., Wu, Q., Cheng, J., Zhang, J., et al. (2020). Heterogeneity, characteristics, and public health implications of Listeria monocytogenes in ready-to-eat foods and pasteurized Milk in China. Front. Microbiol. 11:642. doi: 10.3389/fmicb.2020.00642
Chlebicz, A., and Slizewska, K. (2018). Campylobacteriosis, salmonellosis, Yersiniosis, and Listeriosis as zoonotic foodborne diseases: a review. Int. J. Environ. Res. 15:863. doi: 10.3390/ijerph15050863
Clémentine, H., Benjamin, F., Laurent, G., Pimlapas, L., Damien, M., Jean-François, M., et al. (2016). Population genetic structure of Listeria monocytogenes strains as determined by pulsed-field gel electrophoresis and multilocus sequence typing. Appl. Environ. Microbiol. 82, 5720–5728. doi: 10.1128/AEM.00583-16
de Melo Tavares, R., da Silva, D. A., Camargo, A. C., Yamatogi, R. S., and Nero, L. A. (2020). Interference of the acid stress on the expression of llsX by Listeria monocytogenes pathogenic island 3 (LIPI-3) variants. Food Res. Int. 132:109063. doi: 10.1016/j.foodres.2020.109063
den Bakker, H. C., Desjardins, C. A., Griggs, A. D., Peters, J. E., Zeng, Q., Young, S. K., et al. (2013). Evolutionary dynamics of the accessory genome of Listeria monocytogenes. PLoS One 8:e67511. doi: 10.1371/journal.pone.0067511
Disson, O., Moura, A., and Lecuit, M. (2021). Making sense of the biodiversity and virulence of Listeria monocytogenes. Trends Microbiol. 29, 811–822. doi: 10.1016/j.tim.2021.01.008
Espinosa-Mata, E., Mejía, L., Villacís, J. E., Alban, V., and Zapata, S. (2022). Detection and genotyping of Listeria monocytogenes in artisanal soft cheeses from Ecuador. Rev. Argent Microbiol. 54, 53–56. doi: 10.1016/j.ram.2021.02.013
European Food Safety Authority, and European Centre for Disease Prevention and Control (2017). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 15:e05077. doi: 10.2903/j.efsa.2017.5077
Ferreira, V., Wiedmann, M., Teixeira, P., and Stasiewicz, M. J. (2014). Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J. Food Prot. 77, 150–170. doi: 10.4315/0362-028X.JFP-13-150
Fredriksson-Ahoma, M., Sauvala, M., Kurittu, P., Heljanko, V., Heikinheimo, A., and Paulsen, P. (2022). Characterisation of Listeria monocytogenes isolates from hunted game and game meat from Finland. Food Secur. 11:3679. doi: 10.3390/foods11223679
Fretz, R., Pichler, J., Sagel, U., Much, P., Ruppitsch, W., Pietzka, A. T., et al. (2010). Update: multinational listeriosis outbreak due to “Quargel”, a sour milk curd cheese, caused by two different L. monocytogenes serotype 1/2a strains, 2009–2010. Eur. Secur. 15:19543. doi: 10.2807/ese.15.16.19543-en
Gambarin, P., Magnabosco, C., Losio, M. N., Pavoni, E., Gattuso, A., Arcangeli, G., et al. (2012). Listeria monocytogenes in ready-to-eat seafood and potential hazards for the consumers. Int. J. Microbiol. 2012:497635. doi: 10.1155/2012/497635
Gamboa-Marin, A., Buitrago, M. S., Perey-Perey, K., Mercado, R. M., Poutou Pinales, R., and Carrascal-Camacho, A. (2012). Prevalence of Listeria monocytogenes in pork-meat and other processed products from the Colombian swine industry. Rev. MVZ Cordoba 17, 2827–2833. doi: 10.21897/rmvz.250
Gelbíčová, T., Zobaníková, M., Tomáštíková, Z., Van Walle, I., Ruppitsch, W., and Karpíšková, R. (2018). An outbreak of listeriosis linked to Turkey meat products in the Czech Republic, 2012–2016. Epidemiol. Infect. 146, 1407–1412. doi: 10.1017/S0950268818001565
Gorski, L. (2021). “Serotype assignment by Sero-agglutination, ELISA, and PCR” in Listeria Monocytogenes: methods and protocols. eds. E. M. Fox, H. Bierne, and B. Stessl (New York, NY: Springer US), 57–78.
Guidi, F., Orsini, M., Chiaverini, A., Torresi, M., Centorame, P., Acciari, V. A., et al. (2021). Hypo- and hyper-virulent Listeria monocytogenes clones persisting in two different food processing plants of Central Italy. Microorganisms 9:376. doi: 10.3390/microorganisms9020376
Hanes, R. M., and Huang, Z. (2022). Investigation of antimicrobial resistance genes in Listeria monocytogenes from 2010 through to 2021. Int. J. Environ. Res. Public Health 19:5506. doi: 10.3390/ijerph19095506
Hernandez-Milian, A., and Payeras-Cifre, A. (2014). What is new in Listeriosis? Biomed. Res. Int. 2014:358051. doi: 10.1155/2014/358051
ISO 11290-1:2017. (2022). Horizontal method for the detection and enumeration of Listeria monocytogenes and of Listeria spp. International Organization for Standardization. Available at https://www.iso.org/standard/60313.html
Johansson, M. H. K., Bortolaia, V., Tansirichaiya, S., Aarestrup, F. M., Roberts, A. P., and Petersen, T. N. (2021). Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 76, 101–109. doi: 10.1093/jac/dkaa390
Kimura, B. (2006). Recent advances in the study of the genotypic diversity and ecology of Listeria monocytogenes. Microbes Environ. 21, 69–77. doi: 10.1264/jsme2.21.69
Koopmans, M. M., Bijlsma, M. W., Brouwer, M. C., van de Beek, D., and van der Ende, A. (2017). Listeria monocytogenes meningitis in the Netherlands, 1985–2014: a nationwide surveillance study. J. Infect. 75, 12–19. doi: 10.1016/j.jinf.2017.04.004
Korsak, D., Borek, A., Daniluk, S., Grabowska, A., and Pappelbaum, K. (2012). Antimicrobial susceptibilities of Listeria monocytogenes strains isolated from food and food processing environment in Poland. Int. J. Food Microbiol. 158, 203–208. doi: 10.1016/j.ijfoodmicro.2012.07.016
Kubicová, Z., Roussel, S., Félix, B., and Cabanová, L. (2021). Genomic diversity of Listeria monocytogenes isolates from Slovakia (2010 to 2020). Front. Microbiol. 12:12. doi: 10.3389/fmicb.2021.729050
Lachmann, R., Halbedel, S., Adler, M., Becker, N., Allerberger, F., Holzer, A., et al. (2021). Nationwide outbreak of invasive listeriosis associated with consumption of meat products in health care facilities, Germany, 2014–2019. Clin. Microbiol. Infect. 27, 1035.e1–1035.e5. doi: 10.1016/j.cmi.2020.09.020
Lake, F. B., van Overbeek, L. S., Baars, J. J. P., Koomen, J., Abee, T., and den Besten, H. M. W. (2021). Genomic characteristics of Listeria monocytogenes isolated during mushroom (Agaricus bisporus) production and processing. Int. J. Food Microbiol. 360:109438. doi: 10.1016/j.ijfoodmicro.2021.109438
Lakicevic, B., and Nastasijevic, I. (2016). Food reviews international Listeria monocytogenes in retail establishments: contamination routes and control strategies Listeria monocytogenes in retail establishments: contamination routes and control strategies. Food Rev. Intl. 33, 247–269. doi: 10.1080/87559129.2016.1175017
Lebrun, M., Audurier, A., and Cossart, P. (1994). Plasmid-borne cadmium resistance genes in Listeria monocytogenes are present on Tn5422, a novel transposon closely related to Tn917. J. Bacteriol. 176, 3049–3061. doi: 10.1128/jb.176.10.3049-3061.1994
Lee, S., Ward, T. J., Graves, L. M., Wolf, L. A., Sperry, K., Siletzky, R. M., et al. (2012). Atypical Listeria monocytogenes serotype 4b strains harboring a lineage II-specific gene cassette. Appl. Environ. Microbiol. 78, 660–667. doi: 10.1128/AEM.06378-11
Leong, D., NicAogáin, K., Luque-Sastre, L., McManamon, O., Hunt, K., Alvarez-Ordóñez, A., et al. (2017). A 3-year multi-food study of the presence and persistence of Listeria monocytogenes in 54 small food businesses in Ireland. Int. J. Food. Microbiol. 249, 18–26. doi: 10.1016/j.ijfoodmicro.2017.02.015
Linke, K., Rückerl, I., Brugger, K., Karpiskova, R., Walland, J., Muri-Klinger, S., et al. (2014). Reservoirs of Listeria species in three environmental ecosystems. Appl. Environ. Microbiol. 80, 5583–5592. doi: 10.1128/AEM.01018-14
Liu, S., Graham, J. E., Bigelow, L., Morse, P. D., and Wilkinson, B. J. (2002). Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microbiol. 68, 1697–1705. doi: 10.1128/AEM.68.4.1697-1705.2002
Liu, B., Zheng, D., Zhou, S., Chen, L., and Yang, J. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917. doi: 10.1093/nar/gkab1107
Maćkiw, E., Korsak, D., Kowalska, J., Felix, B., Stasiak, M., Kucharek, K., et al. (2021). Genetic diversity of Listeria monocytogenes isolated from ready-to-eat food products in retail in Poland. Int. J. Food Microbiol. 358:109397. doi: 10.1016/j.ijfoodmicro.2021.109397
Mafuna, T., Matle, I., Magwedere, K., Pierneef, R. E., and Reva, O. N. (2021). Whole genome-based characterization of Listeria monocytogenes isolates recovered from the food chain in South Africa. Front. Microbiol. 12:12. doi: 10.3389/fmicb.2021.669287
Mamber, S. W., Mohr, T. B., Leathers, C., Mbandi, E., Bronstein, P. A., Barlow, K., et al. (2020). Occurrence of Listeria monocytogenes in ready-to-eat meat and poultry product verification testing Samples from U.S. Department of Agriculture–regulated producing establishments, 2005 through 2017. J. Food Prot. 83, 1598–1606. doi: 10.4315/JFP-20-010
Martín, B., Perich, A., Gómez, D., Yangüela, J., Rodríguez, A., Garriga, M., et al. (2014). Diversity and distribution of Listeria monocytogenes in meat processing plants. Food Microbiol. 44, 119–127. doi: 10.1016/j.fm.2014.05.014
Mateus, T., Silva, J., Maia, R. L., and Teixeira, P. (2013). Listeriosis during pregnancy: a public health concern. ISRN Obstet. Gynecol. 2013:851712. doi: 10.1155/2013/851712
Matle, I., Mbatha, K. R., and Madoroba, E. (2020). A review of Listeria monocytogenes from meat and meat products: epidemiology, virulence factors, antimicrobial resistance and diagnosis. Onderstepoort J. Vet. Res. 87:1869. doi: 10.4102/ojvr.v87i1.1869
Maung, A. T., Abdelaziz, M. N. S., Mohammadi, T. N., Zhao, J., EI-Telbany, M., Nakayama, M., et al. (2023). Comparison of prevalence, characterization, antimicrobial resistance and pathogenicity of foodborne Listeria monocytogenes in recent 5 years in Japan. Microb. Pathog. 183:106333. doi: 10.1016/j.micpath.2023.106333
Maury, M. M., Bracq-Dieye, H., Huang, L., Vales, G., Lavina, M., Thouvenot, P., et al. (2019). Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 10:2488. doi: 10.1038/s41467-019-10380-0
Maury, M. M., Tsai, Y. H., Charlier, C., Touchon, M., Chenal-Francisque, V., Leclercq, A., et al. (2016). Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313. doi: 10.1038/ng.3501
Michael, F., Vyacheslav, B., HD, H., PA, B., SD, J., Igor, T., et al. (2019). Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 63:e00483-19. doi: 10.1128/aac.00483-19
Michel, D., Carmen, B., Philippe, G., Christine, J., and Paul, M. (2004). Differentiation of the major Listeria monocytogenes Serovars by multiplex PCR. J. Clin. Microbiol. 42, 3819–3822. doi: 10.1128/jcm.42.8.3819-3822.2004
Moura, A., Criscuolo, A., Pouseele, H., Maury, M. M., Leclercq, A., Tarr, C., et al. (2016). Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2:16185. doi: 10.1038/nmicrobiol.2016.185
Moura, A., Leclercq, A., Vales, G., Tessaud-Rita, N., Bracq-Dieye, H., Thouvenot, P., et al. (2024). Phenotypic and genotypic antimicrobial resistance of Listeria monocytogenes: an observational study in France. Lancet Reg. Health Eur. 37:100800. doi: 10.1016/j.lanepe.2023.100800
Müller, A., Rychli, K., Muhterem-Uyar, M., Zaiser, A., Stessl, B., Guinane, C. M., et al. (2013). Tn6188 – a novel transposon in Listeria monocytogenes responsible for tolerance to Benzalkonium chloride. PLoS One 8:e76835. doi: 10.1371/journal.pone.0076835
Naditz, A. L., Dzieciol, M., Wagner, M., and Schmitz-Esser, S. (2019). Plasmids contribute to food processing environment–associated stress survival in three Listeria monocytogenes ST121, ST8, and ST5 strains. Int. J. Food Microbiol. 299, 39–46. doi: 10.1016/j.ijfoodmicro.2019.03.016
Olaimat, A. (2018). Emergence of antibiotic resistance in Listeria monocytogenes isolated from food products: a comprehensive review. Compr. Rev. Food Sci. Food Saf. 17, 1277–1292. doi: 10.1111/1541-4337.12387
Oloketuyi, S. F., and Khan, F. (2017). Inhibition strategies of Listeria monocytogenes biofilms—current knowledge and future outlooks. J. Basic Microbiol. 57, 728–743. doi: 10.1002/jobm.201700071
Orsi, R. H., Den Bakker, H. C., and Wiedmann, M. (2011). Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301, 79–96. doi: 10.1016/j.ijmm.2010.05.002
Painset, A., Björkman, J. T., Kiil, K., Guillier, L., Mariet, J. F., Félix, B., et al. (2019). LiSEQ – whole-genome sequencing of a cross-sectional survey of Listeria monocytogenes in ready-to-eat foods and human clinical cases in Europe. Microb. Genom. 5:e000257. doi: 10.1099/mgen.0.000257
Parra-Flores, J., Holý, O., Bustamante, F., Lepuschitz, S., Pietzka, A., Contreras-Fernández, A., et al. (2022). Virulence and antibiotic resistance genes in Listeria monocytogenes strains isolated from ready-to-eat foods in Chile. Front. Microbiol. 12:796040. doi: 10.3389/fmicb.2021.796040
Pietzka, A., Allerberger, F., Murer, A., Lennkh, A., Stöger, A., Cabal Rosel, A., et al. (2019). Whole genome sequencing based surveillance of L. monocytogenes for early detection and investigations of Listeriosis outbreaks. Front. Public Health 7:139. doi: 10.3389/fpubh.2019.00139
Poyart-Salmeron, C., Carlier, C., Trieu-Cuot, P., Courvalin, P., and Courtieu, A. L. (1990). Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet 335, 1422–1426. doi: 10.1016/0140-6736(90)91447-I
Pyz-Lukasik, R., Paszkiewicz, W., Kielbus, M., Ziomek, M., Gondek, M., Domaradzki, P., et al. (2022). Genetic diversity and potential virulence of Listeria monocytogenes isolates originating from polish artisanal cheeses. Food Secur. 11:2805. doi: 10.3390/foods11182805
Quereda, J. J., Morón-García, A., Palacios-Gorba, C., Dessaux, C., García-del Portillo, F., Pucciarelli, M. G., et al. (2021). Pathogenicity and virulence of Listeria monocytogenes: a trip from environmental to medical microbiology. Virulence 12, 2509–2545. doi: 10.1080/21505594.2021.1975526
Ragon, M., Wirth, T., Hollandt, F., Lavenir, R., Lecuit, M., Le Monnier, A., et al. (2008). A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 4:e1000146. doi: 10.1371/journal.ppat.1000146
Robertson, J., and Nash, J. H. E. (2018). MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 4:e000206. doi: 10.1099/mgen.0.000206
Ronholm, J. (2018). Editorial: game changer – next generation sequencing and its impact on food microbiology. Front. Microbiol. 9:363. doi: 10.3389/fmicb.2018.00363
Ruppitsch, W., Prager, R., Halbedel, S., Hyden, P., Pietzka, A., Huhulescu, S., et al. (2015). Ongoing outbreak of invasive listeriosis, Germany, 2012 to 2015. Euro Surveill. 20:30094. doi: 10.2807/1560-7917.ES.2015.20.50.30094
Salcedo, C., Arreaza, L., Alcalá, B., de la Fuente, L., and Vázquez, J. A. (2003). Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41, 757–762. doi: 10.1128/jcm.41.2.757-762.2003
Schmitz-Esser, S., Anast, J. M., and Cortes, B. W. (2021). A large-scale sequencing-based survey of plasmids in Listeria monocytogenes reveals global dissemination of plasmids. Front. Microbiol. 12:653155. doi: 10.3389/fmicb.2021.653155
Schmitz-Esser, S., Müller, A., Stessl, B., and Wagner, M. (2015). Genomes of sequence type 121 Listeria monocytogenes strains harbor highly conserved plasmids and prophages. Front. Microbiol. 6:380. doi: 10.3389/fmicb.2015.00380
Shen, J., Zhang, G., Yang, J., Zhao, L., Jiang, Y., Guo, D., et al. (2022). Prevalence, antibiotic resistance, and molecular epidemiology of Listeria monocytogenes isolated from imported foods in China during 2018 to 2020. Int. J. Food Microbiol. 382:109916. doi: 10.1016/j.ijfoodmicro.2022.109916
Stessl, B., Ruppitsch, W., and Wagner, M. (2022). Listeria monocytogenes post-outbreak management – when could a food production be considered under control again? Int. J. Food Microbiol. 379:109844. doi: 10.1016/j.ijfoodmicro.2022.109844
Wagner, E., Zaiser, A., Leitner, R., Quijada, N. M., Pracser, N., Pietzka, A., et al. (2020). Virulence characterization and comparative genomics of Listeria monocytogenes sequence type 155 strains. BMC Genomics 21:847. doi: 10.1186/s12864-020-07263-w
Wang, Y., Li, X., Osmundson, T., Shi, L., and Yan, H. (2019). Comparative genomic analysis of a multidrug-resistant Listeria monocytogenes ST477 isolate. Foodborne Pathog. Dis. 16, 604–615. doi: 10.1089/fpd.2018.2611
Wang, H., Luo, L., Zhang, Z., Deng, J., Wang, Y., Miao, Y., et al. (2018). Prevalence and molecular characteristics of Listeria monocytogenes in cooked products and its comparison with isolates from listeriosis cases. Front. Med. 12, 104–112. doi: 10.1007/s11684-017-0593-9
Werner, R., Ariane, P., Karola, P., Stefan, B., Lasa, F. H., Franz, A., et al. (2015). Defining and evaluating a Core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J. Clin. Microbiol. 53, 2869–2876. doi: 10.1128/jcm.01193-15
Wiktorczyk-Kapischke, N., Skowron, K., and Wałecka-Zacharska, E. (2023). Genomic and pathogenicity islands of Listeria monocytogenes—overview of selected aspects. Front. Mol. Biosci. 10:1161486. doi: 10.3389/fmolb.2023.1161486
Youwen, P., Frederick, B., and Sophia, K. (2009). Competition of Listeria monocytogenes serotype 1/2a and 4b strains in mixed-culture biofilms. Appl. Environ. Microbiol. 75, 5846–5852. doi: 10.1128/AEM.00816-09
Zhang, P., Ji, L., Wu, X., Chen, L., Yan, W., and Dong, F. (2024). Prevalence, genotypic characteristics, and antibiotic resistance of Listeria monocytogenes from retail foods in Huzhou, China. J. Food Prot. 87:100307. doi: 10.1016/j.jfp.2024.100307
Zhao, Q., Hu, P., Li, Q., Zhang, S., Li, H., Chang, J., et al. (2021). Prevalence and transmission characteristics of Listeria species from ruminants in farm and slaughtering environments in China. Emerg. Microbes Infect. 10, 356–364. doi: 10.1080/22221751.2021.1888658
Keywords: Listeria monocytogenes , whole-genome sequencing, food products, antimicrobial resistance genes, Montenegro
Citation: Daza Prieto B, Pietzka A, Martinovic A, Ruppitsch W and Zuber Bogdanovic I (2024) Surveillance and genetic characterization of Listeria monocytogenes in the food chain in Montenegro during the period 2014–2022. Front. Microbiol. 15:1418333. doi: 10.3389/fmicb.2024.1418333
Edited by:
Dario De Medici, National Institute of Health (ISS), ItalyReviewed by:
Francesco Pomilio, Istituto Zooprofilattico Sperimentale dell’Abruzzo e del Molise, ItalyGuerrino Macori, University College Dublin, Ireland
Copyright © 2024 Daza Prieto, Pietzka, Martinovic, Ruppitsch and Zuber Bogdanovic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ivana Zuber Bogdanovic, SXZhbmEuWnViZXJAdWRnLmVkdS5tZQ==; Ariane Pietzka, YXJpYW5lLnBpZXR6a2FAYWdlcy5hdA==
 Beatriz Daza Prieto
Beatriz Daza Prieto Ariane Pietzka
Ariane Pietzka Aleksandra Martinovic
Aleksandra Martinovic Werner Ruppitsch
Werner Ruppitsch Ivana Zuber Bogdanovic
Ivana Zuber Bogdanovic