
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 February 2025
Sec. Microbiotechnology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1417919
This article is part of the Research Topic Microbial Co-cultures: A New Era of Synthetic Biology and Metabolic Engineering, Volume II View all 7 articles
 Antonio Azzollini1,2,3*†
Antonio Azzollini1,2,3*† Barbara Sgorbini4†
Barbara Sgorbini4† Nicole Lecoultre5
Nicole Lecoultre5 Carlo Bicchi4
Carlo Bicchi4 Jean-Luc Wolfender1,2*
Jean-Luc Wolfender1,2* Patrizia Rubiolo4*
Patrizia Rubiolo4* Katia Gindro5
Katia Gindro5Co-cultivation of microorganisms has emerged as a promising methodology for deciphering the intricate molecular interactions between species. This approach facilitates the replication of natural niches of ecological or clinical relevance where microbes consistently interact. In this context, increasing attention has been addressed toward elucidating the molecular crosstalk within fungal co-cultures. However, a major challenge in this area of research is determining the fungal origin of metabolites induced in co-cultivation systems. Molecules elicited in co-cultures may not be detectable in the individual cultures, making it challenging to establish which microorganism is responsible for their induction. For agar-diffused metabolites, imaging mass spectrometry can help overcome this obstacle by localizing the induced molecules during fungal confrontations. For volatile metabolites, however, this remains an open problem. To address this issue, in this study, a three-head-to-head co-culture strategy was developed, specifically focusing on the exploration of volatile interactions between fungi via headspace solid-phase microextraction combined with gas chromatography mass spectrometry. This methodology was applied to study the volatile molecular interactions of three fungal species: Fusarium culmorum, Aspergillus amstelodami, and Cladosporium cladosporioides. The adopted strategy revealed a Fusarium-specific induction of three volatile molecules: γ-terpinene and two unidentified sesquiterpene compounds. Interestingly, γ-terpinene showed antifungal activity in a bioassay against the other two fungal species: Aspergillus amstelodami and Cladosporium cladosporioides. The proposed methodology could help to investigate volatile molecular interactions and highlight metabolite induction specific to a particular fungus involved in in vitro fungal confrontations. This is relevant for a better understanding of the complex biosynthetic responses of fungi in consortia and for identifying volatile molecules with antifungal activity.
Microorganisms play an essential role in sustaining life in both plant and animal organisms (Zilber-Rosenberg and Rosenberg, 2008; Singh et al., 2020; Suman et al., 2022). Microbes interact with each other and their environment to form complex ecosystems, forming intricate networks of ecological relationships that are essential to ecosystem functioning (Moënne-Loccoz et al., 2015; Cosetta and Wolfe, 2019; Gralka, 2023). Their interactions can be mediated by sophisticated molecular communication that regulates key processes such as competition, cooperation, and symbiosis (Braga et al., 2016; Netzker et al., 2020; Weiland-Bräuer, 2021). A thorough understanding of these molecular dynamics is crucial for deciphering the functioning of microbial consortia and their impact not only on the health of host organisms but also on the ecological balances in which they are involved (Rapp et al., 2020; Kapoore et al., 2022; Liu and Xu, 2022).
In this context, microbial co-cultivation has emerged as a promising strategy for the activation of silent biosynthetic clusters (genes that are typically dormant under standard laboratory or environmental conditions) and the stimulation of the production of new bioactive molecules. Furthermore, this approach has the potential to facilitate the study of the intricate molecular dialog among different microbial species, including fungi (Tan et al., 2019; Liu and Kakeya, 2020; Xu et al., 2023). In practice, this strategy involves in vitro co-cultivation of microorganisms in a confined environment (e.g., a Petri dish) (Arora et al., 2020; Selegato and Castro-Gamboa, 2023). In particular, this technique has been employed to study fungal-fungal interactions, including studies between different endophytic fungi (Chagas et al., 2013; Li et al., 2020), as well as interactions involving fungi from the marine environments (Wang et al., 2015; Ding et al., 2017) and from the extreme environments (Stierle et al., 2017). The co-cultivation of fungi has enabled the synthesis of numerous structural classes, including alkaloids (Zhu and Lin, 2006; Zhu et al., 2011; Zhu et al., 2013; Afiyatullov et al., 2018; Sadahiro et al., 2020), terpenes (Zhou et al., 2018), polyketides (Chagas et al., 2013; Li et al., 2020) and peptides (Huang et al., 2014; Li et al., 2014). Moreover, these metabolites showed a broad spectrum of biological properties, including antifungal (Chagas et al., 2013), antibacterial (Zhu et al., 2011), and antiviral activities (Yang et al., 2018; Knowles et al., 2022).
Fungal co-cultures can be cultivated on solid media in Petri dishes ranging from 9 to 15 cm in diameter (Bertrand et al., 2014b). This setup requires protracted extraction and sample preparation phases, leading to significantly prolonged experimentation durations. In this regard, for a more efficient experimental setting, the miniaturization of fungal co-cultivation has proven to be an appropriate way to rapidly collect metabolite profiles from an extensive set of replicates required for metabolomic studies. For example, multi-well 2-cm Petri dishes have been used to co-cultivate Fusarium and Aspergillus strains and highlight their dynamic agar-diffused molecular crosstalk (Bertrand et al., 2014a). To investigate fungal interactions also at the volatile level, this experimental methodology was adapted to a miniaturized setup based on the co-cultivation of fungi directly in 20-mL headspace vials, which were then sampled via headspace solid-phase microextraction (HS-SPME). This approach was used to study an ecologically relevant fungal co-culture of Eutypa lata (Pers.) Tul. & C. Tul. (Diatrypaceae) and Botryosphaeria obtusa (Schwein.) Shoemaker (Botryosphaeriaceae) and revealed dynamically induced molecules at both volatile and non-volatile levels (Azzollini et al., 2018).
Overall, in the field of fungal co-cultures, numerous investigations have been conducted to identify induced metabolites using liquid chromatography mass spectrometry (LC–MS)-based methodologies (Bertrand et al., 2013; Bohni et al., 2016; Xu et al., 2018; Murakami et al., 2020). Such strategies enable MS-guided purification of selected metabolites, possibly streamlining the complete identification of targeted molecules (Azzollini et al., 2016).
Unlike LC–MS-based approaches, which have been broadly used for the profiling of non-volatile compounds, limited studies have explored the use of gas chromatography–mass spectrometry (GC–MS) for the profiling of fungal volatile molecules (Knowles et al., 2022).
Fungi are abundant producers of various volatile organic compounds (VOCs) encompassing a diverse range of chemicals, such as aliphatic and aromatic hydrocarbons, aldehydes, ketones, esters, alcohols, and mono-, sesqui-, and diterpenes (Effmert et al., 2012; Kramer and Abraham, 2012). Interestingly, certain VOCs emitted by fungi display antifungal and antibacterial properties, thereby underlining their potential biological relevance (Hung et al., 2015; Inamdar et al., 2020; Karsli and Şahin, 2021). Fungal VOCs may play a role in influencing interactions within microbial consortia, influencing not only fungal-fungal interactions but also cross-kingdom relationships, such as those between fungi and bacteria (Khalid and Keller, 2021; Zhou et al., 2022), plants (Russo et al., 2022; Razo-Belmán et al., 2023), and even human hosts (Inamdar et al., 2020; El Jaddaoui et al., 2023). For example, in plant-fungal interactions, VOCs from fungi such as Trichoderma can promote plant growth, illustrating their possible role in plant-fungal ecosystems (Lee et al., 2016; You et al., 2022). From a human health standpoint, the study of fungal VOCs is of particular importance in the light of the growing recognition of the lung and oral human mycobiome (Ghannoum et al., 2010; Nilsson et al., 2019; Tiew et al., 2020). In regions where gas-phase interactions are possible, such as the oral cavity and lungs, fungal VOCs may play a role in shaping both local microbial ecology and influencing human health (Nguyen et al., 2015; Kong and Morris, 2017; Vitte et al., 2022). Overall, the variation of the emission of VOCs during fungal interactions has been documented (Guo et al., 2019; Speckbacher et al., 2021; Escudero-Leyva et al., 2023), however, investigation of VOC production within fungal co-cultures remains an emerging field, opening a vast landscape for further scientific investigations.
Furthermore, a critical challenge associated with investigation of volatile interactions in fungal co-cultures is that volatile metabolites detected in the co-culture, but not in the corresponding single cultures (de novo induced), cannot be linked to any of the two interacting fungi (Bertrand et al., 2014b; Arora et al., 2020). Thus, the species producers of these metabolites remain unknown. For agar-diffused molecules, imaging mass spectrometry can provide a valuable solution by accurately localizing the induced molecules involved in interspecies interactions (Watrous and Dorrestein, 2011; Sica et al., 2014; Müller et al., 2022). However, this remains an unresolved issue for volatile molecules induced during microbial confrontations. Accurate identification of the fungus responsible for the induction of volatile molecules is essential to deepen our understanding of the ecological dynamics and behavior of fungi within a particular co-culture or community. Therefore, in this study, a strategy was developed to address this issue.
In particular, this study investigated three fungal species: Cladosporium cladosporioides (Fresen.) G.A. de Vries, Aspergillus amstelodami (L. Mangin) Thom & Church and Fusarium culmorum (Wm. G. Sm.) Sacc. These fungi have been selected as they have been possibly associated with the human oral mycobiome (Ghannoum et al., 2010), where gas-phase interactions relevant to both human microbiota and host physiology can take place (Hertel et al., 2018; Monedeiro-Milanowski et al., 2022; Kiss et al., 2023; Weber et al., 2023).
As part of this work and as a proof-of-concept, these three fungal species were studied in a co-cultivation system that enabled sampling of the metabolome headspace and allowed for the identification of species-specific inductions in a three-head-to-head confrontation model.
A specific bioassay was used to evaluate the biological activity of an identified induced molecule and to understand its potential role within the fungal consortia (Azzollini et al., 2018). This bioactivity test was designed to allow only the migration of volatile compounds within the test environment.
Potato Dextrose Agar (PDA, Difco) was used as the culture medium. The pure reference standard of γ-terpinene and the solvent dibutylphthalate (DBP) were purchased from Sigma–Aldrich.
Three fungal species were selected for this study. The strains were purchased from the collection of CBS (Central Utrecht, The Netherlands): specimen numbers 119376, 134912, 120098, respectively, Aspergillus amstelodami, Cladosporium cladosporioides and Fusarium culmorum. These three fungal strains were stored at 4°C in the Agroscope dynamic mycotheca in vials containing a diluted potato dextrose broth solution (1:4) (Sigma-Aldrich).1
Fusarium culmorum, Aspergillus amstelodami, and Cladosporium cladosporioides pure cultures were prepared by placing 2-mm PDA plugs of fungal pre-cultures in the center of the headspace vial. The co-cultures were prepared by placing two 2-mm agar plugs of a pre-culture of the two different fungal species on opposite sides of the headspace vial (Supplementary Figure S1). Immediately after inoculation, each vial was closed with an appropriate cap (HDSP cap 18 mm magnetic PTFE/Sil, Agilent Technologies, United States). Blank samples (PDA only) were prepared. Cultures were incubated at 21°C in the dark. To study the induction of volatile molecules in these three fungal co-cultures, nine replicates (n = 9) of each single culture and co-culture were sampled via HS-SPME and analyzed via GC–MS after 9 days of incubation.
The volatile fraction (headspace) of each individual single culture and co-culture was sampled using a 2-cm DVB/CAR/PDMS SPME fiber (Supelco, Bellefonte, PA, United States) introduced directly into the headspace vial used for cultivation. Sampling was performed using an MPS2 autosampler (Gerstel, Mülheim a/d Ruhr, Germany) at 40°C (no pre-equilibrium, 30 min of sampling). The fiber was automatically injected into an Agilent 7890 GC coupled to an Agilent 5975C MS (Agilent, Little Falls, DE, United States). Internal standard [C13 diluted in dibutylphthalate (DBP)] was preloaded onto the fiber using the in-fiber standardization technique (Wang et al., 2005). Sampling was performed only once for each culture to avoid any perturbation of the volatile fraction over time. The GC–MS conditions were as follows: inlet temperature 250°C, desorption time: 5 min, split injection (1/10 split ratio), carrier gas (helium at a constant flow rate of 1 mL/min); column: HP5MS (5% phenyl, 95% polysiloxane - 30 m × 0.25 mm i.d. × 0.25 μm; Agilent Technologies). Oven temperature program: 50°C (1 min) - 3°C/min - 200°C - 15°C/min - 250°C (4 min). MS was operated in the EI mode at 70 eV with a mass range of 35–350 amu in full scan mode.
Data were processed using an Agilent MSD ChemStation version F.01.03.2357. GC peaks were identified by comparing their linear retention indices (calculated versus a C9-C25 hydrocarbon mixture) and their mass spectra in comparison with those present in commercially available libraries (Wiley, Adams, NIST). Standard deviation was employed in the evaluation of normalized area calculations for induced volatile molecules. γ-Terpinene identity was confirmed by analyzing the pure reference standard (Sigma-Aldrich).
To evaluate the antifungal activity of the volatile induced compound γ-terpinene, a 9-cm Petri dish, divided in two by a plastic septum, was used. This partitioning allowed only the diffusion of volatile molecules between the two sectors of the dish. Distinct pure cultures of Aspergillus amstelodami and Cladosporium cladosporioides were inoculated on PDA within the first sector of the Petri dish using a 2-mm agar plug from their respective pre-cultures. The compound γ-terpinene was injected into a filter paper posed in the second sector of the Petri dish. The activity of γ-terpinene was assessed at the following concentrations: 917 μL/L, 524 μL/L, and 131 μL/L. The concentrations reported for this bioassay were expressed as liquid volume of each volatile compound per dish volume (for simplification, the dish volume was calculated without considering the plastic septum). For the control samples, no compound was injected into the second sector of the Petri dish. Mycelial growth was monitored after 7 days. Five replicate experiments were performed for each concentration.
In the context of this study, a strategy has been devised for the purpose of investigating the volatile interactions that are mediated by VOCs (volatile organic compounds) in fungal co-cultures, and for identifying the fungus responsible for inducing volatile molecules within such co-cultures. This approach involves culturing both individual cultures and co-cultures in 20 mL SMPE vials within a three-head-to-head co-cultivation system (Figure 1). The fungal co-cultures and single cultures were further analyzed via gas chromatography–mass spectrometry (GC–MS) to investigate the volatile fractions of these samples. This experimental design was conceived to highlight species-specific induction of secondary metabolites, as in all confrontations, one partner is always opposed to the others in separate head-to-head co-cultures. For example, if a particular metabolite is induced in the co-culture of species A with B (AxB) and in the co-culture of species A with C (AxC), but absent in the co-culture of species B with C (BxC), this suggests that species A is responsible for inducing the molecule in question (Figure 1). This setup allows for the precise identification of the fungal species responsible for the induction of volatile secondary metabolites in a co-culture environment, facilitating the understanding of species-specific molecular interactions.
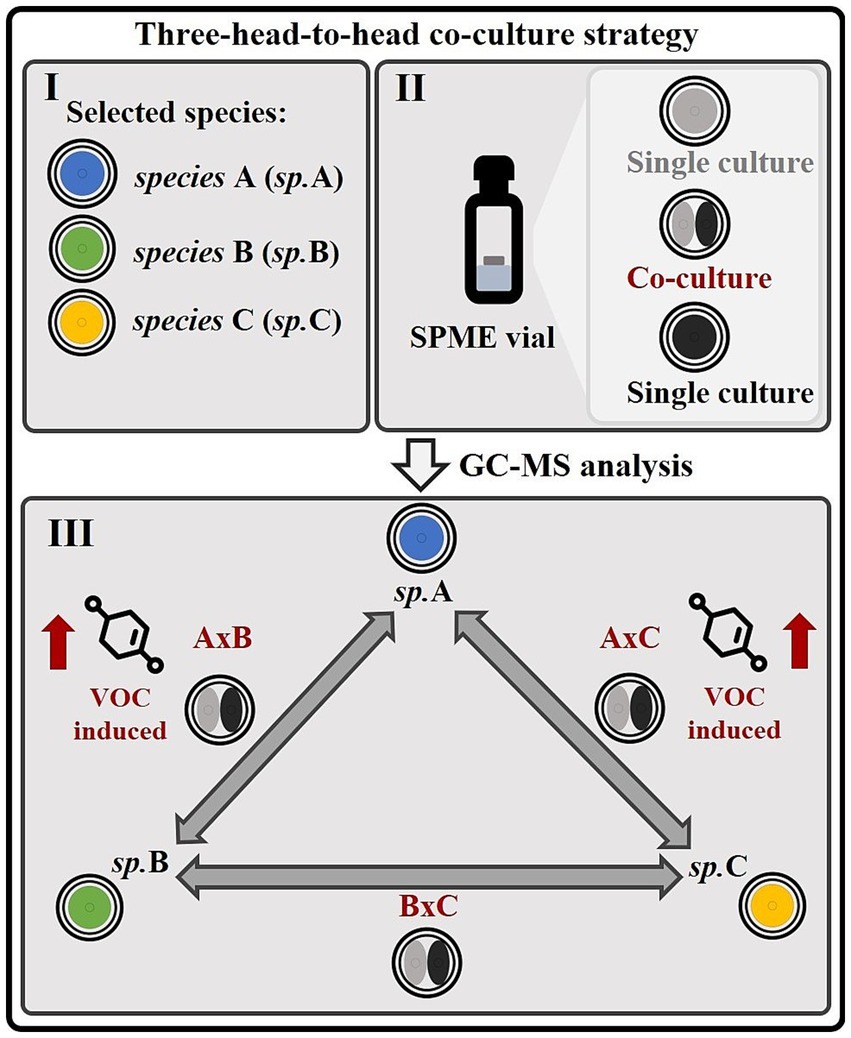
Figure 1. Strategy for investigating volatile interactions and species-specific metabolite induction in microbial or fungal co-cultures. (I) This experimental strategy is designed to investigate volatile interactions among three microbial or fungal species (A, B, C) and identify the species responsible for inducing specific metabolites within co-cultures. (II) The organisms are cultivated directly in SPME vials, allowing sampling of the volatile metabolome. (III) Three distinct co-cultures (A × B, A × C, and B × C), constituting a “head-to-head” system, as well as their corresponding monocultures, are analyzed using GC–MS. This setup enables the identification of species-specific inductions of volatile molecules. For instance, if a particular metabolite is detected in co-cultures A × B and A × C, but not in B × C, as illustrated in this figure, it can be inferred that species A is responsible for inducing the production of that volatile organic compound (VOC).
In this work, the aforementioned methodological approach was employed to investigate the volatile interspecies molecular crosstalk among Fusarium culmorum (Fus), Aspergillus amstelodami (Asp), and Cladosporium cladosporioides (Cla). These microbial fungi were selected because, interestingly, they may be related to the human oral mycobiome, where gas-phase interactions that are relevant to host physiology may occur (Ghannoum et al., 2010; Monedeiro-Milanowski et al., 2022; Kiss et al., 2023). In addition, Fusarium spp., Aspergillus spp., and Cladosporium spp. have also been notably associated to lung and respiratory health (Carneiro et al., 2011; Richardson et al., 2019; Ma et al., 2021). The aforementioned strains were grown in 2-mL of solid media in SPME (headspace) vials. To improve the statistical robustness of the experimental design, nine replicates of the three different co-cultures (Fusarium culmorum vs. Aspergillus amstelodami, Fusarium culmorum vs. Cladosporium cladosporioides, Aspergillus amstelodami vs. Cladosporium cladosporioides, respectively referred to as FusxAsp, FusxCla and AspxCla) and of the corresponding single cultures (Fus, Asp, Cla) were incubated for 9 days and sampled at this time point via HS-SPME (Figure 2). All experiments were performed concomitantly and under identical culture conditions. Cultivation in a 20-mL headspace vial was found to be appropriate for the analysis of the volatile metabolome in multiple confrontation studies (Azzollini et al., 2018). The two fungal species were simultaneously co-cultured in headspace vials to analyze variations in the volatile metabolome resulting from their interactions. This format allows for the rapid generation of a large number of replicates. Moreover, this cultivation setup is specifically designed to identify species-specific secondary metabolite induction, as each confrontation pairs one fungal species with another in the aforementioned three-head-to-head co-cultivation system. For instance, metabolite induction in FusxAsp and FusxCla co-cultures, with absence in AspxCla, can suggest a Fusarium-specific expression of the elicited molecule (Figure 2).
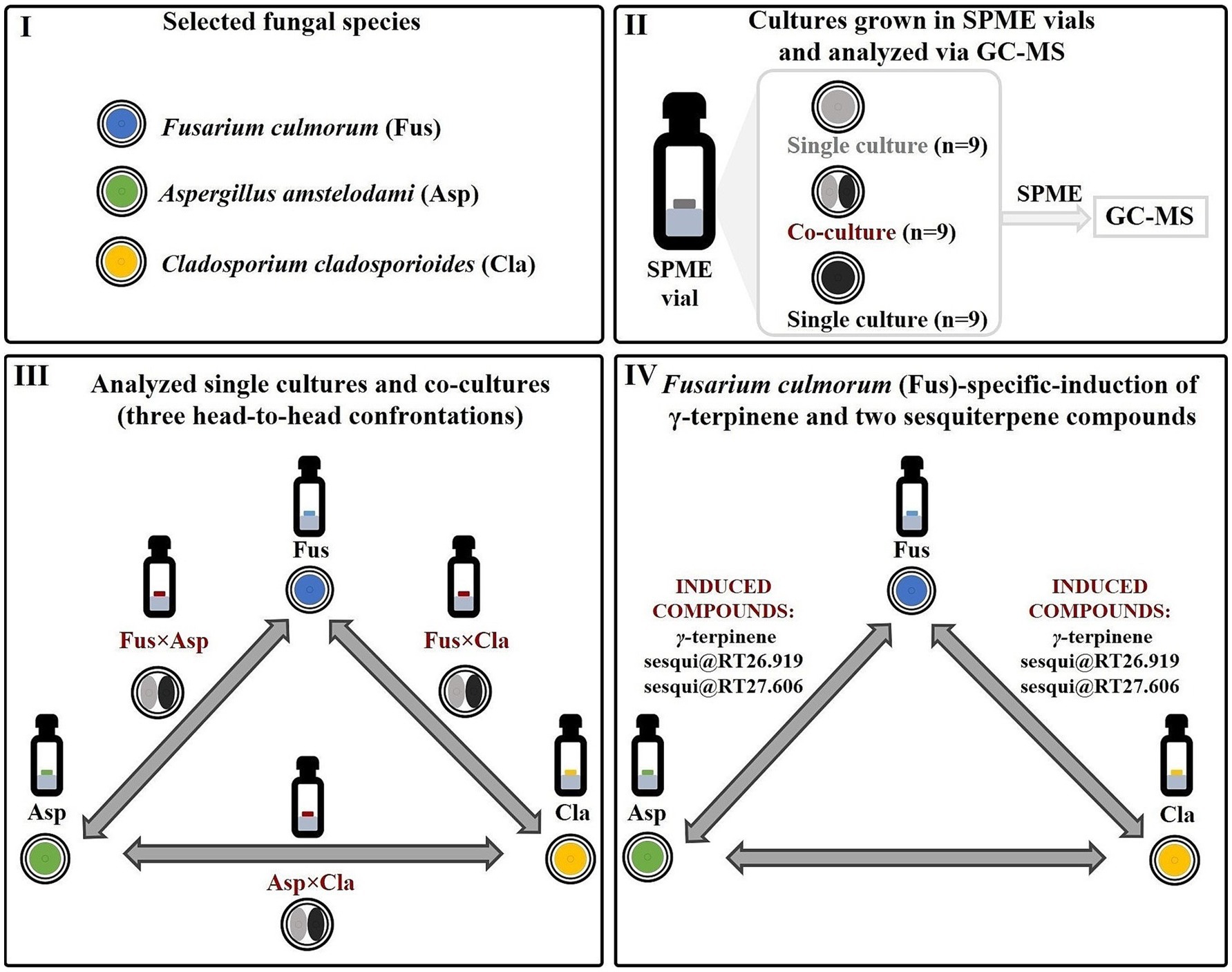
Figure 2. Implementation of the proposed strategy and detection of Fusarium-specific induction of γ-terpinene and two unidentified sesquiterpene compounds. (I) Volatile molecular interactions between three fungal species (Fusarium culmorum, Aspergillus amstelodami and Cladosporium cladosporioides) were investigated. (II) Fungal co-cultures, as well as the respective single cultures, were grown directly in SPME (headspace) vials, sampled using HS-SPME and analyzed using GC–MS. For each head-to-head fungal confrontation experiment, nine co-cultures, as well nine single cultures of each of the confronting fungi were studied. (III) Three different fungal co-cultures (constituting a three-head-to-head system) were analyzed: Fusarium culmorum vs. Aspergillus amstelodami, Fusarium culmorum vs. Cladosporium cladosporioides, Aspergillus amstelodami vs. Cladosporium cladosporioides (respectively referred to as FusxAsp, FusxCla and AspxCla) and of the corresponding single cultures (Fus, Asp, and Cla). (IV) A Fusarium-specific induction of the three volatile induced molecules (γ-terpinene, sesqui@RT26.919, and sesqui@RT27.606) was highlighted. It was observed that these molecules were elicited only in co-cultures where Fusarium culmorum (Fus) is one of the confronting fungi (FusxAsp and FusxCla). No metabolite induction was observed in the Aspergillus amstelodami vs. Cladosporium cladosporioides (AspxCla) co-cultures.
After 9 days of growth, each single culture and co-culture was sampled and analyzed using HS-SPME-GC–MS. Volatile compounds released into the headspace by the single cultures and co-cultures were sampled by introducing a SPME fiber, which allowed their accumulation on the fiber polymer at the end of the incubation period. Direct thermal desorption of the SPME fiber into the GC injector provided GC–MS profiling of the metabolites from both the single culture and co-culture volatile fractions.
After the GC–MS analysis of all single cultures and co-cultures, the compounds detected in the HS-SPME-GC–MS profiles were identified by comparing the acquired spectra with those of commercial libraries (Wiley, Adams, Nist) or by injecting a reference standard and using linear retention indices calculated versus a mixture of C9-C25 alkanes. The GC peaks of the total ion chromatogram were integrated, and the obtained areas were used to compare the profiles, thereby highlighting features induced in co-cultures that were not detected in single cultures. Three features were detected as induced in all the nine replicates of the two different co-cultures of FusxAsp and FusxCla: γ-terpinene and two unidentified sesquiterpene compounds.
The identity of the γ-terpinene was confirmed by co-injection of a commercially available standard (Supplementary Figure S2). The two unidentified sesquiterpene compounds @RT 26.919 and @RT 27.606 had both a molecular weight of 204 Da and a retention index of 1,413 and 1,431, respectively. In the absence of available standards and reference spectra in the available EI-MS libraries, it was not possible to achieve full de novo identification of these compounds. Furthermore, the scale at which the HS-SPME-GC–MS experiments were conducted did not permit the isolation and subsequent structural elucidation of these molecules via NMR. These two sesquiterpene compounds were referred to as sesqui@RT26.919 and sesqui@RT27.606, respectively, and only the compound type could be attributed based on the fragmentation pattern of their associated EI-MS spectra (see Supplementary Figure S3).
The presence of these specific features in the previously mentioned Fusarium co-cultures (FusxAsp and FusxCla), and their absence in the single cultures and in the AspxCla co-culture are depicted in Figure 3. In this representation, the variation in the absolute peak area (vertical axis) of these features across single cultures and co-cultures is displayed. Each point on the horizontal axis represents a single culture or co-culture (nine points for each single culture or co-culture represent replicates). For easier interpretation, co-cultures are colored in red, whereas single cultures of Fusarium, Aspergillus and Cladosporium are shown in blue, green, and yellow, respectively. As shown in Figure 3, the induced molecules consistently appeared in all the nine replicates of the Fusarium-related co-cultures (FusxAsp and FusxCla), while these molecules were not detected in any of the co-cultures of AspxCla and in any of the single cultures. This persistent induction is remarkable (Supplementary Figure S4), as the literature often reports cases in which molecules are predominantly expressed in co-cultures, but are still present, albeit at basal or lower levels, in single cultures (Bertrand et al., 2014b; Arora et al., 2020). The induction of these compounds suggests that certain silent biosynthetic gene clusters may be activated by the interaction of Fusarium with the other fungi.
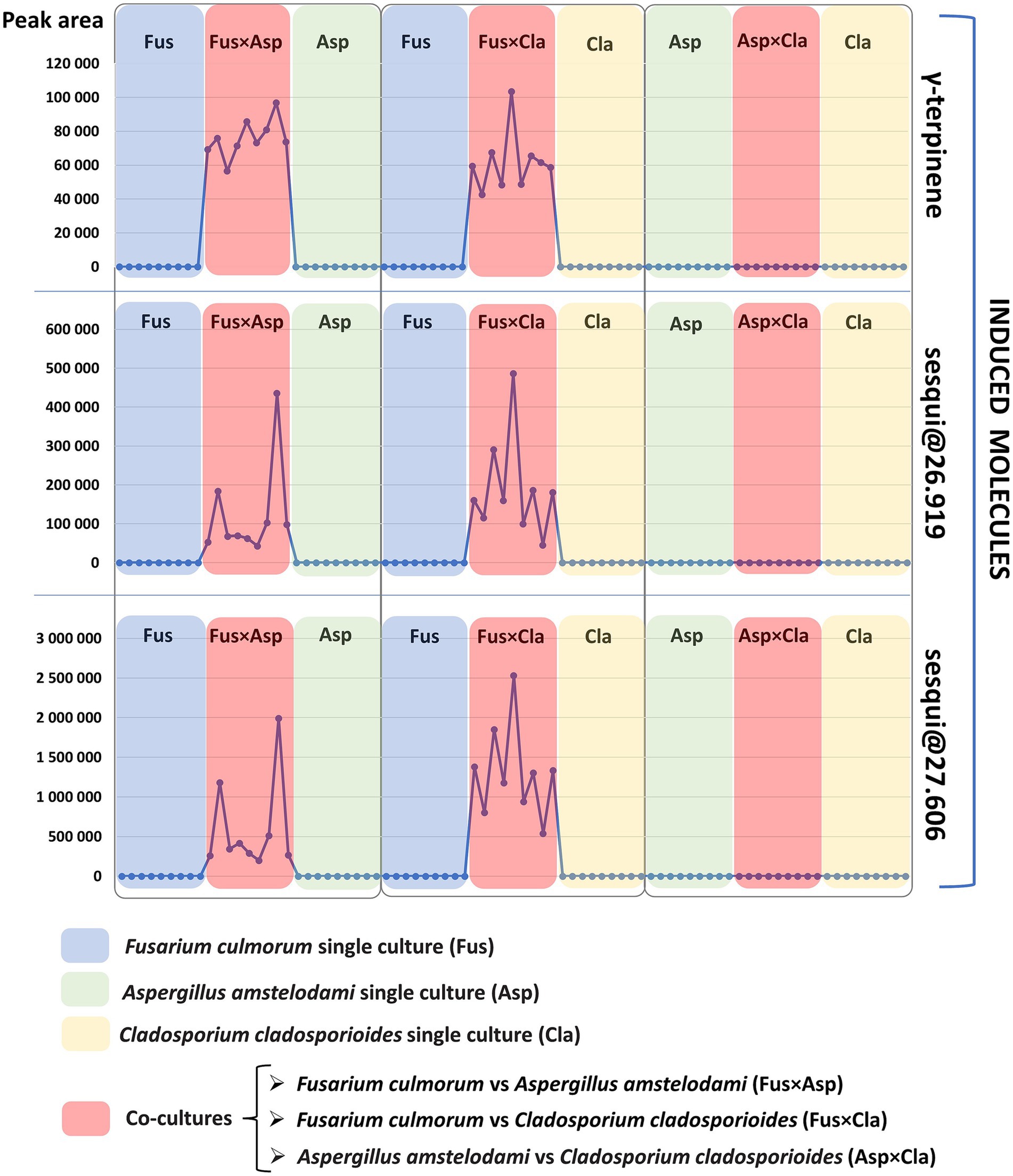
Figure 3. Induction pattern of the three volatile induced molecules (starting from the top of the figure: γ-terpinene, sesqui@RT26.919, and sesqui@RT27.606). Each single culture and co-culture sample is represented by a dot on the horizontal axis. The peak area detected for each volatile molecule is marked on the vertical axis. Co-cultures of Fusarium culmorum vs. Aspergillus amstelodami, Fusarium culmorum vs. Cladosporium cladosporioides, Aspergillus amstelodami vs. Cladosporium cladosporioides are indicated by the red color in this figure and are, respectively, referred to as FusxAsp, FusxCla and AspxCla. Fusarium (Fus), Aspergillus (Asp) and Cladosporium (Cla) single cultures are indicated by the following colors, respectively: blue for Fusarium, green for Aspergillus and yellow for Cladosporium. As evidenced here, induced molecules are detected only in co-cultures were Fusarium culmorum is one of the co-cultivated fungi: Fusarium culmorum vs. Aspergillus amstelodami (FusxAsp) and Fusarium culmorum vs. Cladosporium cladosporioides (FusxCla).
Contrary to this persistent induction in all co-culture replicates of FusxAsp and FusxCla, very high variability was observed in terms of the peak area of the induced metabolites, especially for the two sesquiterpene compounds (Tables 1, 2 reports the average normalized peak area and standard deviation for the induced compounds). Given that biosynthetic gene clusters are activated in the context of fungal co-cultures to produce these induced molecules, it might be possible that epigenetic mechanisms contribute to the variability in metabolite expression as fungi dynamically prioritize resource allocation between growth and defense (Pfannenstiel and Keller, 2019; Pillay et al., 2022). Additionally, it can be hypothesized that fluctuations in metabolite production may play a role in preventing resistance in competitors. By varying their metabolite profiles, fungi may prevent other microorganisms from adapting to their chemical defenses. This variability in compound expression is well-described in the literature and explains the importance of conducting these studies with several biological replicates (Bertrand et al., 2014a), as was done in the context of this work.
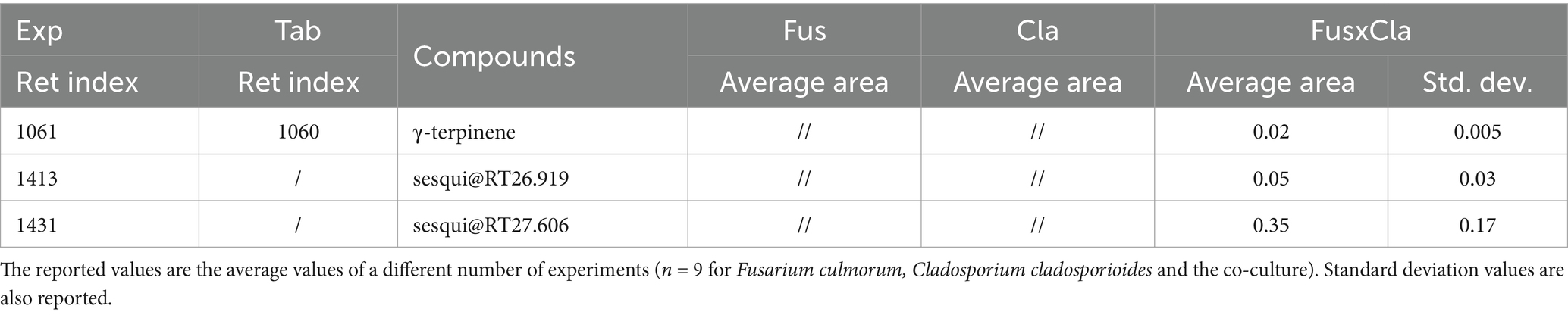
Table 1. Normalized areas HS-SPME-GC-MS (versus internal standard, i.e., C13 in the dibutyl phthalate [DBP] solution) of the compounds emitted by single cultures and the co-cultures of Fusarium culmorum vs. Cladosporium cladosporioides (referred to as FusxCla in this table).
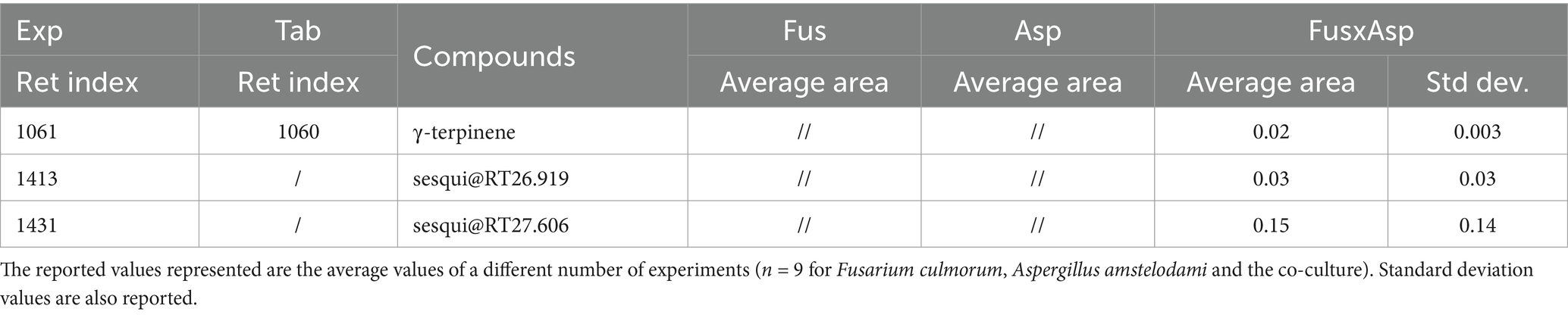
Table 2. Normalized areas HS-SPME-GC-MS (versus internal standard, i.e., C13 in the DBP solution) of the compounds emitted by single cultures and the co-cultures of Fusarium culmorum vs. Aspergillus amstelodami (referred to as FusxAsp in this table).
The concurrent induction of γ-terpinene, sesqui@RT26.919, and sesqui@RT27.606 in two of the three co-cultures (FusxAsp and FusxCla) underscores a Fusarium-specific induction of these three molecules. This suggests that Fusarium could be responsible for eliciting these compounds in the two aforementioned co-cultures. Supplementary Figure S5 shows the detection of γ-terpinene, sesqui@RT26.919 and sesqui@RT27.606 only in the HS-SPME GC–MS metabolite profiles of FusxAsp and FusxCla.
Moreover, these results indicate the effectiveness of this experimental model in highlighting species-specific induction of volatile molecules. Thus, this methodology could be helpful in pinpointing the fungus responsible for the de novo production of metabolites in co-cultures.
Prior studies on the investigation of volatile interactions in microbial co-cultures have utilized solid-phase micro-extraction gas chromatography mass spectrometry (SPME-GCMS) (Spraker et al., 2014), or have employed gas chromatography ion mobility spectrometry (GC-IMS) (Speckbacher et al., 2021), or have been conducted in combination with ultrahigh-performance liquid chromatography coupled to high-resolution mass spectrometry (UHPLC-HRMS/MS) for the concomitant analysis of the non-volatile compounds involved in the interactions (Azzollini et al., 2018; Escudero-Leyva et al., 2023). However, despite these advances, the fundamental question of which fungus is responsible for the induction of specific volatile molecules has not been fully addressed in previous studies. The implementation of the proposed methodology could help to fill this gap and provide clearer insights into the identification of the species responsible for the volatile compound induction in co-cultures.
Additionally, the approach outlined in this study can be valuable for investigating fungal consortia and microbial interactions, particularly those of ecological and clinical relevance, such as those observed in the oral or lung mycobiomes (Belvoncikova et al., 2022; Fan et al., 2023). For example, it offers an effective approach for in vitro studies of fungi that comprise the lung mycobiome, where their presence and abundance may vary in correlation with the progression and severity of respiratory diseases such as asthma, chronic obstructive pulmonary disease (COPD) and allergic bronchopulmonary aspergillosis (ABPA) (Yii et al., 2017; Richardson et al., 2019; de Dios Caballero et al., 2022). These fungi are not merely passive colonizers; rather, they possibly engage in active biochemical competition, notably through the production and release of VOCs (Hung et al., 2015; El Ariebi et al., 2016). It is of great importance to gain an understanding of the VOC-mediated interactions and to identify the fungal species responsible for VOC production in competitive contexts, particularly during respiratory disease exacerbations when fungal communities often shift (Tiew et al., 2021; de Dios Caballero et al., 2022). Such insights could prove pivotal in the development of non-invasive diagnostic tools based on VOC monitoring via breath analysis (Sharma et al., 2023; Diefenderfer et al., 2024).
Furthermore, the findings from this study underscore the pivotal role Fusarium culmorum plays in biosynthetic processes when in co-culture with other fungal species. The chemical signals released during co-cultures may have specifically triggered the activation of silent biosynthetic gene clusters (BGCs), in Fusarium culmorum, leading to the production of the three induced volatile molecules mentioned above, including γ-terpinene. This induced compound may act as a signaling molecule, allowing interspecies interaction thorough non-contact mechanisms. Moreover, γ-terpinene may likely be produced by Fusarium culmorum as part of a defense mechanism against the other competing fungal species present in the co-cultures. Notably, the specific induction of γ-terpinene in its co-cultures with the other two fungal species highlights the need for further investigation into the biological activity of this compound. Understanding the role of γ-terpinene is crucial for gaining deeper insights into the ecological significance of F. culmorum in these interactions. To investigate this further, the effects of γ-terpinene on the growth of Cladosporium cladosporioides and Aspergillus amstelodami were evaluated.
For a better comprehension of the possible biological role of γ-terpinene in these interactions, the antifungal activity of this induced molecule was investigated against Aspergillus amstelodami and Cladosporium cladosporioides. Briefly, this bioassay consisted of a 9-cm Petri dish divided in two by a septum that allowed for the migration of only the volatile compound from one sector to another (Azzollini et al., 2018). In the first sector, the fungus was inoculated, whereas in the other sector, the volatile compound was injected into a filter paper. In this context, volatile compounds can migrate from one sector to another and inhibit fungal growth without direct contact with the fungal thallus.
To assess the antifungal activity of γ-terpinene, a preliminary experiment was conducted in five replicates using a concentration of 917 μL/L (expressed as liquid volume of volatile compound per dish volume). Under these conditions, the growth of Aspergillus amstelodami and Cladosporium cladosporioides was inhibited after 7 days (Figure 4). To further evaluate the potency of γ-terpinene’s antifungal properties, the bioassay was executed in five replicates, also using two lower concentrations: 524 μL/L and 131 μL/L. Following a 7-day growth period, the mycelium growth inhibition occurred for both fungi also at the 524 μL/L concentration. Moreover, a slight inhibition of growth was observed for Aspergillus amstelodami at 131 μL/L (Supplementary Table S1). These findings, therefore, show the antifungal properties of γ-terpinene against both fungi Aspergillus amstelodami and Cladosporium cladosporioides. Our results, while consistent with previous studies highlighting the antifungal activity of γ-terpinene against Candida albicans (Rivera-Yañez et al., 2017) and endophytic fungi, such as Botrytis cinerea (Espinosa-García and Langenheim, 1991), contrast with the findings of a previous study where γ-terpinene did not exhibit any antifungal activity against Cladosporium cladosporioides (Agnieszka Wróblewska et al., 2019) when assessed using a diffusion disk method for fungal growth inhibition. This discrepancy in results may be attributed to differences in bioassay techniques and experimental conditions, with our setup being better suited to detect the inhibitory effect of γ-terpinene on fungal growth. In particular, our experimental design probably mimicked an environment in which γ-terpinene was more effective in exerting its antifungal activity. Factors such as the composition of the nutrient medium used for fungal growth and the exposure time to the tested molecule may have played a role in increasing the susceptibility of the fungus to γ- terpinene. Consequently, the observations in our study provide additional evidence of γ-terpinene’s broad-spectrum antifungal properties, extending its inhibitory activity to Cladosporium cladosporioides and Aspergillus amstelodami.
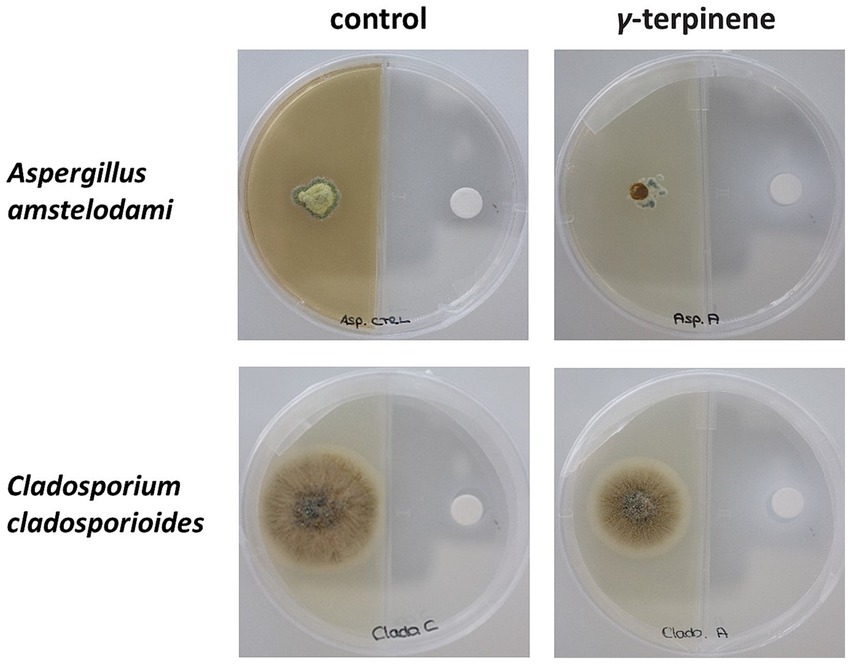
Figure 4. Antifungal bioassay performed using γ-terpinene at a concentration of 917 μL/L. The inhibition of the growth of the fungus Aspergillus amstelodami and Cladosporium cladosporioides could be noticed after 7 days (second column from left).
The production of volatile antifungal molecules during fungal co-cultures suggests a multifaceted ecological role of these induced molecules in microbial interactions. Their volatile nature allows them to function as chemical signals, facilitating interspecies communication without requiring direct physical contact (Morath et al., 2012; Schulz-Bohm et al., 2017). In such environments, volatile antifungal compounds like γ-terpinene may be synthesized as part of the biochemical arsenal to inhibit the growth of competing fungi. This inhibition may allow the producing organism to secure vital resources and establish dominance within its ecological niche, providing a competitive advantage (Hung et al., 2015; Quintana-Rodriguez et al., 2018). Additionally, induced antifungal molecules may influence growth patterns, and modulate ecological interactions within microbial communities of ecological as well of clinical significance, such as the human mycobiome (Belvoncikova et al., 2022; Hagihara et al., 2023). The role of antifungal VOCs, such as γ-terpinene, as a defense and signaling molecules underscores the importance of this type of compounds in fungal ecosystems. Moreover, it illustrates the evolutionary sophistication of fungi in developing chemical strategies to compete and communicate in dynamic microbial communities.
This study uniquely highlights the species-specific induction of two unidentified sesquiterpenes and of an antifungal volatile molecule, γ-terpinene, detected only in the Fusarium-related co-cultures FusxAsp and FusxCla. Additionally, it presents an effective methodology to investigate volatile fungal interactions and identify the species responsible for the induction of volatile molecules in co-cultures. Moreover, the proposed three-head-to-head co-culture strategy is broadly applicable and can be adapted to study co-cultures between bacteria and fungi (Moussa et al., 2020) allowing for a more comprehensive investigation of volatile interactions between different types of microorganisms (possibly part of the same microbiome or ecosystem). Detection of the specific induction of antimicrobial molecules within a microbial co-culture is of critical importance from ecological and microbiological perspectives. For example, from an ecological perspective, a better understanding of the interactions within a co-culture system can provide insights into how microbial communities function and coexist, thereby contributing to ecosystem stability and resilience (Braga et al., 2016; Gupta et al., 2021).
The methodology described in this study can be applied to investigate volatile interactions within microbial fungal consortia of ecological or clinical significance, such as those of the human mycobiome (Belvoncikova et al., 2022). It would be of interest to investigate whether VOCs produced during interactions of fungi associated with the oral or lung mycobiome can be detected not only through in vitro co-cultivation and GC–MS analysis, but also via electronic nose or via ambient mass spectrometry techniques in the breath of patients experiencing disease progression and exacerbations (Bregy et al., 2018; Finamore et al., 2019; VA et al., 2021). This could provide valuable insights into the progression of chronic lung diseases, where shifts in the microbial and mycobiome composition critically influence disease trajectory and outcomes (Tiew et al., 2021; Natalini et al., 2023; Scialò et al., 2023).
Furthermore, the presented methodology may be employed for the examination of the molecular dialog between disparate fungal taxa, thereby revealing various types of fungal interactions. For instance, it can highlight volatile molecules produced by fungi to recognize strains of the same species, or it can underscore which VOCs are produced between compatible or competing species. This exploration can also enhance our understanding of fungal mating processes through volatile compounds, shedding light on the intricate ways in which fungi communicate and interact at the molecular level (Bölker and Kahmann, 1993; Jones and Bennett, 2011; Wilson et al., 2023).
The proposed strategy may facilitate tackling these challenges, thereby enhancing our understanding of volatile molecular communication within in vitro fungal co-cultures and microbial consortia, an area that remains underexplored.
Original GC-MS datasets are available in a publicly accessible repository: https://osf.io/f3gsv/?view_only=b60f211e21d24b2ca8113cc2298ad5af.
AA: Writing – original draft, Writing – review & editing, Conceptualization, Formal analysis, Investigation, Project administration, Supervision, Visualization. BS: Writing – original draft, Writing – review & editing, Formal analysis, Investigation, Supervision, Visualization. NL: Writing – review & editing, Writing – original draft, Investigation. CB: Writing – review & editing, Writing – original draft. J-LW: Writing – original draft, Writing – review & editing, Supervision. PR: Writing – review & editing, Writing – original draft, Supervision. KG: Writing – review & editing, Writing – original draft, Supervision.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
AA thanks Pierre-Marie Allard for his advice. Figure 1 contains the following icons: “molecule” icon by Ilham Fitrotul Hayat from thenounproject.com CC BY 3.0, “PETRI DISH” icon by Barbara Marsillac from thenounproject.com CC BY 3.0 and “Injection vial” icon by Arosh Khan from thenounproject.com CC BY 3.0. Figure 2 contains the following icons: “PETRI DISH” icon by Barbara Marsillac from thenounproject.com CC BY 3.0 and “Injection vial” icon by Arosh Khan from thenounproject.com CC BY 3.0. Supplementary Figure S1 contains the following icons: “molecule” icon by Ilham Fitrotul Hayat from thenounproject.com CC BY 3.0, “PETRI DISH” icon by Barbara Marsillac from thenounproject.com CC BY 3.0, “Injection vial” icon by Arosh Khan from thenounproject.com CC BY 3.0, “liquid chromatography-mass spectrometry” icon by Julie Ko from thenounproject.com CC BY 3.0, “Analysis” icon by Ifanicon from thenounproject.com CC BY 3.0 and “petri dish” icon by Azam Ishaq from thenounproject.com CC BY 3.0.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1417919/full#supplementary-material
Afiyatullov, S. S., Zhuravleva, O. I., Antonov, A. S., Berdyshev, D. V., Pivkin, M. V., Denisenko, V. A., et al. (2018). Prenylated indole alkaloids from co-culture of marine-derived fungi aspergillus sulphureus and Isaria felina. J. Antibiot. 71, 846–853. doi: 10.1038/s41429-018-0072-9
Agnieszka Wróblewska, A., Retajczyk, M., Kądziołka, D., and Markowska-Szczupak, A. (2019). Microbiological tests of natural limonene and the compounds obtained after isomerization of limonene in the presence of Ti-SBA-15 catalyst--α-Terpinene, γ-Terpinene, Terpinolene, and p-cymene. J. Cosmet. Sci. 70, 137–147.
Arora, D., Gupta, P., Jaglan, S., Roullier, C., Grovel, O., and Bertrand, S. (2020). Expanding the chemical diversity through microorganisms co-culture: current status and outlook. Biotechnol. Adv. 40:107521. doi: 10.1016/j.biotechadv.2020.107521
Azzollini, A., Boggia, L., Boccard, J., Sgorbini, B., Lecoultre, N., Allard, P.-M., et al. (2018). Dynamics of metabolite induction in fungal co-cultures by metabolomics at both volatile and non-volatile levels. Front. Microbiol. 9:72. doi: 10.3389/fmicb.2018.00072
Azzollini, A., Favre-Godal, Q., Zhang, J., Marcourt, L., Ebrahimi, S. N., Wang, S., et al. (2016). Preparative scale MS-guided isolation of bioactive compounds using high-resolution flash chromatography: antifungals from Chiloscyphus polyanthos as a case study. Planta Med. 82, 1051–1057. doi: 10.1055/s-0042-108207
Belvoncikova, P., Splichalova, P., Videnska, P., and Gardlik, R. (2022). The human mycobiome: colonization, composition and the role in health and disease. J. Fungi 8:1046. doi: 10.3390/jof8101046
Bertrand, S., Azzollini, A., Schumpp, O., Bohni, N., Schrenzel, J., Monod, M., et al. (2014a). Multi-well fungal co-culture for de novo metabolite-induction in time-series studies based on untargeted metabolomics. Mol. BioSyst. 10, 2289–2298. doi: 10.1039/C4MB00223G
Bertrand, S., Bohni, N., Schnee, S., Schumpp, O., Gindro, K., and Wolfender, J.-L. (2014b). Metabolite induction via microorganism co-culture: a potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 32, 1180–1204. doi: 10.1016/j.biotechadv.2014.03.001
Bertrand, S., Schumpp, O., Bohni, N., Bujard, A., Azzollini, A., Monod, M., et al. (2013). Detection of metabolite induction in fungal co-cultures on solid media by high-throughput differential ultra-high pressure liquid chromatography–time-of-flight mass spectrometry fingerprinting. J. Chromatogr. A 1292, 219–228. doi: 10.1016/j.chroma.2013.01.098
Bohni, N., Hofstetter, V., Gindro, K., Buyck, B., Schumpp, O., Bertrand, S., et al. (2016). Production of fusaric acid by Fusarium spp. in pure culture and in solid medium co-cultures. Molecules 21:370. doi: 10.3390/molecules21030370
Bölker, M., and Kahmann, R. (1993). Sexual pheromones and mating responses in fungi. Plant Cell 5, 1461–1469
Braga, R. M., Dourado, M. N., and Araújo, W. L. (2016). Microbial interactions: ecology in a molecular perspective. Braz. J. Microbiol. 47, 86–98. doi: 10.1016/j.bjm.2016.10.005
Bregy, L., Nussbaumer-Ochsner, Y., Sinues, P. M.-L., García-Gómez, D., Suter, Y., Gaisl, T., et al. (2018). Real-time mass spectrometric identification of metabolites characteristic of chronic obstructive pulmonary disease in exhaled breath. Clin. Mass Spectrom. 7, 29–35. doi: 10.1016/j.clinms.2018.02.003
Carneiro, H. A., Coleman, J. J., Restrepo, A., and Mylonakis, E. (2011). Fusarium infection in lung transplant patients: report of 6 cases and review of the literature. Medicine 90, 69–80. doi: 10.1097/MD.0b013e318207612d
Chagas, F. O., Dias, L. G., and Pupo, M. T. (2013). A mixed culture of endophytic fungi increases production of antifungal polyketides. J. Chem. Ecol. 39, 1335–1342. doi: 10.1007/s10886-013-0351-7
Cosetta, C. M., and Wolfe, B. E. (2019). Causes and consequences of biotic interactions within microbiomes. Curr. Opin. Microbiol. 50, 35–41. doi: 10.1016/j.mib.2019.09.004
de Dios Caballero, J., Cantón, R., Ponce-Alonso, M., García-Clemente, M. M., De La, G. G., Pedrosa, E., et al. (2022). The human mycobiome in chronic respiratory diseases: current situation and future perspectives. Microorganisms 10:810. doi: 10.3390/microorganisms10040810
Diefenderfer, J., Bean, H. D., and Higgins Keppler, E. A. (2024). New breath diagnostics for fungal disease. Curr. Clin. Microbiol. Rep. 11, 51–61. doi: 10.1007/s40588-024-00216-x
Ding, W., Lu, Y., Feng, Z., Luo, S., and Li, C. (2017). A new nonadride derivative from the co-culture broth of two mangrove fungi. Chem. Nat. Compd. 53, 691–693. doi: 10.1007/s10600-017-2092-2
Effmert, U., Kalderás, J., Warnke, R., and Piechulla, B. (2012). Volatile mediated interactions between bacteria and fungi in the soil. J. Chem. Ecol. 38, 665–703. doi: 10.1007/s10886-012-0135-5
El Ariebi, N., Hiscox, J., Scriven, S. A., Müller, C. T., and Boddy, L. (2016). Production and effects of volatile organic compounds during interspecific interactions. Fungal Ecol. 20, 144–154. doi: 10.1016/j.funeco.2015.12.013
El Jaddaoui, I., Rangel, D. E., and Bennett, J. W. (2023). Fungal volatiles have physiological properties. Fungal Biol. 127, 1231–1240. doi: 10.1016/j.funbio.2023.03.005
Escudero-Leyva, E., Quirós-Guerrero, L., Vásquez-Chaves, V., Pereira-Reyes, R., Chaverri, P., and Tamayo-Castillo, G. (2023). Differential volatile organic compound expression in the interaction of Daldinia eschscholtzii and Mycena citricolor. ACS Omega 8, 31373–31388. doi: 10.1021/acsomega.3c03865
Espinosa-García, F. J., and Langenheim, J. H. (1991). Effects of sabinene and γ-terpinene from coastal redwood leaves acting singly or in mixtures on the growth of some of their fungus endophytes. Biochem. Syst. Ecol. 19, 643–650. doi: 10.1016/0305-1978(91)90080-J
Fan, Y., Wu, L., and Zhai, B. (2023). The mycobiome: interactions with host and implications in diseases. Curr. Opin. Microbiol. 75:102361. doi: 10.1016/j.mib.2023.102361
Finamore, P., Scarlata, S., and Incalzi, R. A. (2019). Breath analysis in respiratory diseases: state-of-the-art and future perspectives. Expert. Rev. Mol. Diagn. 19, 47–61. doi: 10.1080/14737159.2019.1559052
Ghannoum, M. A., Jurevic, R. J., Mukherjee, P. K., Cui, F., Sikaroodi, M., Naqvi, A., et al. (2010). Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 6:e1000713. doi: 10.1371/journal.ppat.1000713
Gralka, M. (2023). Searching for principles of microbial ecology across levels of biological organization. Integr. Comp. Biol. 63, 1520–1531. doi: 10.1093/icb/icad060
Guo, Y., Ghirardo, A., Weber, B., Schnitzler, J.-P., Benz, J. P., and Rosenkranz, M. (2019). Trichoderma species differ in their volatile profiles and in antagonism toward ectomycorrhiza Laccaria bicolor. Front. Microbiol. 10:448972. doi: 10.3389/fmicb.2019.00891
Gupta, G., Ndiaye, A., and Filteau, M. (2021). Leveraging experimental strategies to capture different dimensions of microbial interactions. Front. Microbiol. 12:700752. doi: 10.3389/fmicb.2021.700752
Hagihara, M., Kato, H., Shibata, Y., Umemura, T., Ariyoshi, T., Hirai, J., et al. (2023). Mycobiome and Mycobiome-associated diseases. Med. Mycol. J. 64, 55–62. doi: 10.3314/mmj.23-002
Hertel, M., Schuette, E., Kastner, I., Hartwig, S., Schmidt-Westhausen, A. M., Preissner, R., et al. (2018). Volatile organic compounds in the breath of oral candidiasis patients: a pilot study. Clin. Oral Investig. 22, 721–731. doi: 10.1007/s00784-017-2147-6
Huang, S., Ding, W., Li, C., and Cox, D. G. (2014). Two new cyclopeptides from the co-culture broth of two marine mangrove fungi and their antifungal activity. Pharmacogn. Mag. 10, 410–414. doi: 10.4103/0973-1296.141781
Hung, R., Lee, S., and Bennett, J. W. (2015). Fungal volatile organic compounds and their role in ecosystems. Appl. Microbiol. Biotechnol. 99, 3395–3405. doi: 10.1007/s00253-015-6494-4
Inamdar, A. A., Morath, S., and Bennett, J. W. (2020). Fungal volatile organic compounds: more than just a funky smell? Ann. Rev. Microbiol. 74, 101–116. doi: 10.1146/annurev-micro-012420-080428
Jones, S. K. Jr., and Bennett, R. J. (2011). Fungal mating pheromones: choreographing the dating game. Fungal Genet. Biol. 48, 668–676. doi: 10.1016/j.fgb.2011.04.001
Kapoore, R. V., Padmaperuma, G., Maneein, S., and Vaidyanathan, S. (2022). Co-culturing microbial consortia: approaches for applications in biomanufacturing and bioprocessing. Crit. Rev. Biotechnol. 42, 46–72. doi: 10.1080/07388551.2021.1921691
Karsli, A., and Şahin, Y. S. (2021). The role of fungal volatile organic compounds (FVOCs) in biological control. Türkiye Biyolojik Mücadele Dergisi 12, 79–92. doi: 10.31019/tbmd.818701
Khalid, S., and Keller, N. P. (2021). Chemical signals driving bacterial–fungal interactions. Environ. Microbiol. 23, 1334–1347. doi: 10.1111/1462-2920.15410
Kiss, H., Örlős, Z., Gellért, Á., Megyesfalvi, Z., Mikáczó, A., Sárközi, A., et al. (2023). Exhaled biomarkers for point-of-care diagnosis: recent advances and new challenges in Breathomics. Micromachines 14:391. doi: 10.3390/mi14020391
Knowles, S. L., Raja, H. A., Roberts, C. D., and Oberlies, N. H. (2022). Fungal–fungal co-culture: a primer for generating chemical diversity. Nat. Prod. Rep. 39, 1557–1573. doi: 10.1039/D1NP00070E
Kong, H. H., and Morris, A. (2017). The emerging importance and challenges of the human mycobiome. Virulence 8, 310–312. doi: 10.1080/21505594.2017.1279780
Kramer, R., and Abraham, W.-R. (2012). Volatile sesquiterpenes from fungi: what are they good for? Phytochem. Rev. 11, 15–37. doi: 10.1007/s11101-011-9216-2
Lee, S., Yap, M., Behringer, G., Hung, R., and Bennett, J. W. (2016). Volatile organic compounds emitted by Trichoderma species mediate plant growth. Fungal Biol. Biotechnol. 3, 1–14. doi: 10.1186/s40694-016-0025-7
Li, H.-T., Liu, T., Yang, R., Xie, F., Yang, Z., Yang, Y., et al. (2020). Phomretones A–F, C12polyketides from the co-cultivation ofPhomasp. YUD17001 andArmillariasp. RSC Adv. 10, 18384–18389. doi: 10.1039/D0RA02524K
Li, C., Wang, J., Luo, C., Ding, W., and Cox, D. G. (2014). A new cyclopeptide with antifungal activity from the co-culture broth of two marine mangrove fungi. Nat. Prod. Res. 28, 616–621. doi: 10.1080/14786419.2014.887074
Liu, C., and Kakeya, H. (2020). Cryptic chemical communication: secondary metabolic responses revealed by microbial co-culture. Chem. Asian J. 15, 327–337. doi: 10.1002/asia.201901505
Liu, Y., and Xu, P. (2022). Quantitative and analytical tools to analyze the spatiotemporal population dynamics of microbial consortia. Curr. Opin. Biotechnol. 76:102754. doi: 10.1016/j.copbio.2022.102754
Ma, X., Hu, J., Yu, Y., Wang, C., Gu, Y., Cao, S., et al. (2021). Assessment of the pulmonary adaptive immune response to Cladosporium cladosporioides infection using an experimental mouse model. Sci. Rep. 11:909. doi: 10.1038/s41598-020-79642-y
Moënne-Loccoz, Y., Mavingui, P., Combes, C., Normand, P., and Steinberg, C. (2015). “Microorganisms and biotic interactions” in Environmental Microbiology: Fundamentals and Applications: Microbial Ecology, 395–444.
Monedeiro-Milanowski, M., Monedeiro, F., Ligor, T., and Buszewski, B. (2022). “Saliva and related specimens as a source of volatile biomarkers” in Volatile Biomarkers for Human Health: From Nature to Artificial Senses, 100.
Morath, S. U., Hung, R., and Bennett, J. W. (2012). Fungal volatile organic compounds: a review with emphasis on their biotechnological potential. Fungal Biol. Rev. 26, 73–83. doi: 10.1016/j.fbr.2012.07.001
Moussa, M., Ebrahim, W., Kalscheuer, R., Liu, Z., and Proksch, P. (2020). Co-culture of the bacterium Pseudomonas aeruginosa with the fungus Fusarium tricinctum induces bacterial antifungal and quorum sensing signaling molecules. Phytochem. Lett. 36, 37–41. doi: 10.1016/j.phytol.2020.01.013
Müller, W. H., McCann, A., Arias, A. A., Malherbe, C., Quinton, L., De Pauw, E., et al. (2022). Imaging metabolites in agar-based bacterial co-cultures with minimal sample preparation using a DIUTHAME membrane in surface-assisted laser desorption/ionization mass spectrometry. ChemistrySelect 7:e202200734. doi: 10.1002/slct.202200734
Murakami, S., Hayashi, N., Inomata, T., Kato, H., Hitora, Y., and Tsukamoto, S. (2020). Induction of secondary metabolite production by fungal co-culture of Talaromyces pinophilus and Paraphaeosphaeria sp. J. Nat. Med. 74, 545–549. doi: 10.1007/s11418-020-01400-1
Natalini, J. G., Singh, S., and Segal, L. N. (2023). The dynamic lung microbiome in health and disease. Nat. Rev. Microbiol. 21, 222–235. doi: 10.1038/s41579-022-00821-x
Netzker, T., Shepherdson, E. M., Zambri, M. P., and Elliot, M. A. (2020). Bacterial volatile compounds: functions in communication, cooperation, and competition. Ann. Rev. Microbiol. 74, 409–430. doi: 10.1146/annurev-micro-011320-015542
Nguyen, L. D., Viscogliosi, E., and Delhaes, L. (2015). The lung mycobiome: an emerging field of the human respiratory microbiome. Front. Microbiol. 6:89. doi: 10.3389/fmicb.2015.00089
Nilsson, R. H., Anslan, S., Bahram, M., Wurzbacher, C., Baldrian, P., and Tedersoo, L. (2019). Mycobiome diversity: high-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 17, 95–109. doi: 10.1038/s41579-018-0116-y
Pfannenstiel, B. T., and Keller, N. P. (2019). On top of biosynthetic gene clusters: how epigenetic machinery influences secondary metabolism in fungi. Biotechnol. Adv. 37:107345. doi: 10.1016/j.biotechadv.2019.02.001
Pillay, L. C., Nekati, L., Makhwitine, P. J., and Ndlovu, S. I. (2022). Epigenetic activation of silent biosynthetic gene clusters in endophytic fungi using small molecular modifiers. Front. Microbiol. 13:815008. doi: 10.3389/fmicb.2022.815008
Quintana-Rodriguez, E., Rivera-Macias, L. E., Adame-Alvarez, R. M., Torres, J. M., and Heil, M. (2018). Shared weapons in fungus-fungus and fungus-plant interactions? Volatile organic compounds of plant or fungal origin exert direct antifungal activity in vitro. Fungal Ecol. 33, 115–121. doi: 10.1016/j.funeco.2018.02.005
Rapp, K. M., Jenkins, J. P., and Betenbaugh, M. J. (2020). Partners for life: building microbial consortia for the future. Curr. Opin. Biotechnol. 66, 292–300. doi: 10.1016/j.copbio.2020.10.001
Razo-Belmán, R., Ángeles-López, Y. I., García-Ortega, L. F., León-Ramírez, C. G., Ortiz-Castellanos, L., Yu, H., et al. (2023). Fungal volatile organic compounds: mechanisms involved in their sensing and dynamic communication with plants. Front. Plant Sci. 14:1257098. doi: 10.3389/fpls.2023.1257098
Richardson, M., Bowyer, P., and Sabino, R. (2019). The human lung and aspergillus: You are what you breathe in? Med. Mycol. 57, S145–S154. doi: 10.1093/mmy/myy149
Rivera-Yañez, C. R., Terrazas, L. I., Jimenez-Estrada, M., Campos, J. E., Flores-Ortiz, C. M., Hernandez, L. B., et al. (2017). Anti-Candida activity of Bursera morelensis Ramirez essential oil and two compounds, α-pinene and γ-terpinene—an in vitro study. Molecules 22:2095. doi: 10.3390/molecules22122095
Russo, A., Pollastri, S., Ruocco, M., Monti, M. M., and Loreto, F. (2022). Volatile organic compounds in the interaction between plants and beneficial microorganisms. J. Plant Interact. 17, 840–852. doi: 10.1080/17429145.2022.2107243
Sadahiro, Y., Kato, H., Williams, R. M., and Tsukamoto, S. (2020). Irpexine, an isoindolinone alkaloid produced by coculture of endophytic fungi, Irpex lacteus and Phaeosphaeria oryzae. J. Nat. Prod. 83, 1368–1373. doi: 10.1021/acs.jnatprod.0c00047
Schulz-Bohm, K., Martín-Sánchez, L., and Garbeva, P. (2017). Microbial volatiles: small molecules with an important role in intra-and inter-kingdom interactions. Front. Microbiol. 8:2484. doi: 10.3389/fmicb.2017.02484
Scialò, F., Vitale, M., D'Agnano, V., Mariniello, D. F., Perrotta, F., Castaldo, A., et al. (2023). Lung microbiome as a treatable trait in chronic respiratory disorders. Lung 201, 455–466. doi: 10.1007/s00408-023-00645-3
Selegato, D. M., and Castro-Gamboa, I. (2023). Enhancing chemical and biological diversity by co-cultivation. Front. Microbiol. 14:1117559. doi: 10.3389/fmicb.2023.1117559
Sharma, A., Kumar, R., and Varadwaj, P. (2023). Smelling the disease: diagnostic potential of breath analysis. Mol. Diagn. Ther. 27, 321–347. doi: 10.1007/s40291-023-00640-7
Sica, V. P., Raja, H. A., El-Elimat, T., and Oberlies, N. H. (2014). Mass spectrometry imaging of secondary metabolites directly on fungal cultures. RSC Adv. 4, 63221–63227. doi: 10.1039/C4RA11564C
Singh, B. K., Liu, H., and Trivedi, P. (2020). Eco-holobiont: a new concept to identify drivers of host-associated microorganisms. Environ. Microbiol. 22, 564–567. doi: 10.1111/1462-2920.14900
Speckbacher, V., Zeilinger, S., Zimmermann, S., Mayhew, C. A., Wiesenhofer, H., and Ruzsanyi, V. (2021). Monitoring the volatile language of fungi using gas chromatography-ion mobility spectrometry. Anal. Bioanal. Chem. 413, 3055–3067. doi: 10.1007/s00216-021-03242-6
Spraker, J. E., Jewell, K., Roze, L. V., Scherf, J., Ndagano, D., Beaudry, R., et al. (2014). A volatile relationship: profiling an inter-kingdom dialogue between two plant pathogens, Ralstonia Solanacearum and aspergillus Flavus. J. Chem. Ecol. 40, 502–513. doi: 10.1007/s10886-014-0432-2
Stierle, A. A., Stierle, D. B., Decato, D., Priestley, N. D., Alverson, J. B., Hoody, J., et al. (2017). The berkeleylactones, antibiotic macrolides from fungal coculture. J. Nat. Prod. 80, 1150–1160. doi: 10.1021/acs.jnatprod.7b00133
Suman, J., Rakshit, A., Ogireddy, S. D., Singh, S., Gupta, C., and Chandrakala, J. (2022). Microbiome as a key player in sustainable agriculture and human health. Front. Soil Sci. 2:821589. doi: 10.3389/fsoil.2022.821589
Tan, Z. Q., Leow, H. Y., Lee, D. C. W., Karisnan, K., Song, A. A. L., Mai, C. W., et al. (2019). Co-culture systems for the production of secondary metabolites: current and future prospects. Open Biotechnol. J. 13, 18–26. doi: 10.2174/1874070701913010018
Tiew, P. Y., Dicker, A. J., Keir, H. R., Poh, M. E., Pang, S. L., Mac Aogáin, M., et al. (2021). A high-risk airway mycobiome is associated with frequent exacerbation and mortality in COPD. Eur. Respir. J. 57:2002050. doi: 10.1183/13993003.02050-2020
Tiew, P. Y., Mac Aogain, M., Ali, N. A. B. M., Thng, K. X., Goh, K., Lau, K. J., et al. (2020). The mycobiome in health and disease: emerging concepts, methodologies and challenges. Mycopathologia 185, 207–231. doi: 10.1007/s11046-019-00413-z
VA, B., Subramoniam, M., and Mathew, L. (2021). Noninvasive detection of COPD and lung Cancer through breath analysis using MOS sensor array based e-nose. Expert. Rev. Mol. Diagn. 21, 1223–1233. doi: 10.1080/14737159.2021.1971079
Vitte, J., Michel, M., Malinovschi, A., Caminati, M., Odebode, A., Annesi-Maesano, I., et al. (2022). Fungal exposome, human health, and unmet needs: a 2022 update with special focus on allergy. Allergy 77, 3199–3216. doi: 10.1111/all.15483
Wang, J., Huang, S., Li, C., Ding, W., She, Z., and Li, C. (2015). A new coumarin produced by mixed fermentation of two marine fungi. Chem. Nat. Compd. 51, 239–241. doi: 10.1007/s10600-015-1252-5
Wang, Y., O’Reilly, J., Chen, Y., and Pawliszyn, J. (2005). Equilibrium in-fibre standardisation technique for solid-phase microextraction. J. Chromatogr. A 1072, 13–17. doi: 10.1016/j.chroma.2004.12.084
Watrous, J. D., and Dorrestein, P. C. (2011). Imaging mass spectrometry in microbiology. Nat. Rev. Microbiol. 9, 683–694. doi: 10.1038/nrmicro2634
Weber, R., Streckenbach, B., Welti, L., Inci, D., Kohler, M., Perkins, N., et al. (2023). Online breath analysis with SESI/HRMS for metabolic signatures in children with allergic asthma. Front. Mol. Biosci. 10:1154536. doi: 10.3389/fmolb.2023.1154536
Weiland-Bräuer, N. (2021). Friends or foes—microbial interactions in nature. Biology 10:496. doi: 10.3390/biology10060496
Wilson, A. M., Wingfield, M. J., and Wingfield, B. D. (2023). Structure and number of mating pheromone genes is closely linked to sexual reproductive strategy in Huntiella. BMC Genomics 24:261. doi: 10.1186/s12864-023-09355-9
Xu, S., Li, M., Hu, Z., Shao, Y., Ying, J., and Zhang, H. (2023). The potential use of fungal co-culture strategy for discovery of new secondary metabolites. Microorganisms 11:464. doi: 10.3390/microorganisms11020464
Xu, X.-Y., Shen, X.-T., Yuan, X.-J., Zhou, Y.-M., Fan, H., Zhu, L.-P., et al. (2018). Metabolomics investigation of an association of induced features and corresponding fungus during the co-culture of Trametes versicolor and Ganoderma applanatum. Front. Microbiol. 8:2647. doi: 10.3389/fmicb.2017.02647
Yang, S.-Q., Li, X.-M., Li, X., Li, H.-L., Meng, L.-H., and Wang, B.-G. (2018). New citrinin analogues produced by coculture of the marine algal-derived endophytic fungal strains aspergillus sydowii EN-534 and Penicillium citrinum EN-535. Phytochem. Lett. 25, 191–195. doi: 10.1016/j.phytol.2018.04.023
Yii, A. C., Koh, M. S., Lapperre, T. S., Tan, G. L., and Chotirmall, S. H. (2017). The emergence of aspergillus species in chronic respiratory disease.
You, J., Li, G., Li, C., Zhu, L., Yang, H., Song, R., et al. (2022). Biological control and plant growth promotion by volatile organic compounds of Trichoderma koningiopsis T-51. J. Fungi 8:131. doi: 10.3390/jof8020131
Zhou, Y., Wang, H., Xu, S., Liu, K., Qi, H., Wang, M., et al. (2022). Bacterial-fungal interactions under agricultural settings: from physical to chemical interactions. Stress Biol. 2:22. doi: 10.1007/s44154-022-00046-1
Zhou, Q.-Y., Yang, X.-Q., Zhang, Z.-X., Wang, B.-Y., Hu, M., Yang, Y.-B., et al. (2018). New azaphilones and tremulane sesquiterpene from endophytic Nigrospora oryzae cocultured with Irpex lacteus. Fitoterapia 130, 26–30. doi: 10.1016/j.fitote.2018.07.018
Zhu, F., Chen, G., Chen, X., Huang, M., and Wan, X. (2011). Aspergicin, a new antibacterial alkaloid produced by mixed fermentation of two marine-derived mangrove epiphytic fungi. Chem. Nat. Compd. 47, 767–769. doi: 10.1007/s10600-011-0053-8
Zhu, F., Chen, G., Wu, J., and Pan, J. (2013). Structure revision and cytotoxic activity of marinamide and its methyl ester, novel alkaloids produced by co-cultures of two marine-derived mangrove endophytic fungi. Nat. Prod. Res. 27, 1960–1964. doi: 10.1080/14786419.2013.800980
Zhu, F., and Lin, Y. (2006). Marinamide, a novel alkaloid and its methyl ester produced by the application of mixed fermentation technique to two mangrove endophytic fungi from the South China Sea. Chin. Sci. Bull. 51, 1426–1430. doi: 10.1007/s11434-006-1426-4
Keywords: volatile molecules, mass spectrometry, co-culture, VOC-mediated interactions, Fusarium, antifungal, Aspergillus, Cladosporium
Citation: Azzollini A, Sgorbini B, Lecoultre N, Bicchi C, Wolfender J-L, Rubiolo P and Gindro K (2025) A mass spectrometry-based strategy for investigating volatile molecular interactions in microbial consortia: unveiling a Fusarium-specific induction of an antifungal compound. Front. Microbiol. 15:1417919. doi: 10.3389/fmicb.2024.1417919
Received: 22 April 2024; Accepted: 26 December 2024;
Published: 25 February 2025.
Edited by:
Jay Prakash Verma, Banaras Hindu University, IndiaReviewed by:
Manoj Kumar Solanki, University of Silesia in Katowice, PolandCopyright © 2025 Azzollini, Sgorbini, Lecoultre, Bicchi, Wolfender, Rubiolo and Gindro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonio Azzollini, YW50b25pby5henpvbGxpbmlAY2h1di5jaA==; Jean-Luc Wolfender, SmVhbi1MdWMuV29sZmVuZGVyQHVuaWdlLmNo; Patrizia Rubiolo, cGF0cml6aWEucnViaW9sb0B1bml0by5pdA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.