- 1Bamboo Diseases and Pests Control and Resources Development Key Laboratory of Sichuan Province, College of Life Science, Leshan Normal University, Leshan, Sichuan, China
- 2College of Resources, Sichuan Agricultural University, Chengdu, Sichuan, China
- 3College of Bioscience and Biotechnology, Hunan Agricultural University, Changsha, Hunan, China
- 4Aba Prefecture Ecological Protection and Development Research Institute, Wenchuan, Sichuan, China
- 5Institute of Nature Conservation Area Planning, Sichuan Forestry and Grassland Survey and Planning Institute, Chengdu, Sichuan, China
Levulinic acid, a hydrolysis product of lignocellulose, can be metabolized into important compounds in the field of medicine and pesticides by engineered strains of Saccharomyces cerevisiae. Levulinic acid, as an intermediate product widely found in the conversion process of lignocellulosic biomass, has multiple applications. However, its toxicity to Saccharomyces cerevisiae reduces its conversion efficiency, so screening Saccharomyces cerevisiae genes that can tolerate levulinic acid becomes the key. By creating a whole-genome knockout library and bioinformatics analysis, this study used the phenotypic characteristics of cells as the basis for screening and found the HMX1 gene that is highly sensitive to levulinic acid in the oxidative stress pathway. After knocking out HMX1 and treating with levulinic acid, the omics data of the strain revealed that multiple affected pathways, especially the expression of 14 genes related to the cell wall and membrane system, were significantly downregulated. The levels of acetyl-CoA and riboflavin decreased by 1.02-fold and 1.44-fold, respectively, while the content of pantothenic acid increased. These findings indicate that the cell wall-membrane system, as well as the metabolism of acetyl-CoA and riboflavin, are important in improving the resistance of Saccharomyces cerevisiae to levulinic acid. They provide theoretical support for enhancing the tolerance of microorganisms to levulinic acid, which is significant for optimizing the conversion process of lignocellulosic biomass to levulinic acid.
1 Introduction
Levulinic acid (LA) is a kind of green platform compound with high additional value derived from the hydrolysis of lignocellulose (Ennaert et al., 2016; Chen and Lee, 2020). LA is a γ-ketonic acid containing five carbon atoms (Pileidis and Titirici, 2016) and has a wide range of applications in fuel additives (Lange et al., 2010; Alonso et al., 2013), liquid fuels, medicine, agriculture, and other fields (Han et al., 2019). Since lignocellulosic biomass can produce LA without the use of expensive hydrolytic enzymes, this greatly reduces the cost of utilizing lignocellulosic biomass. LA can be converted into useful chemicals, such as LA esters, γ-valerolactone (GVL), and alkanes of various molecular weights (Pileidis and Titirici, 2016; Adeleye et al., 2019). Studies have shown that (Habe et al., 2020) certain microorganisms, such as Pseudomonas putida KT2440, Brevibacterium epidermidis LA39-2, Rhodopseudomonas sp. No. 7, and Pseudomonas sp. LA18T, can effectively convert LA into polyhydroxyalkanoates (PHA). PHA is a natural macromolecular biomaterial with good biocompatibility, biodegradability, and thermoplastic processing performance similar to plastics. Therefore, it can be used as both a biomedical material and a biodegradable packaging material. In addition, scientists also use engineered brewer's yeast to produce 5-aminolevulinic acid (5-ALA) (Hara et al., 2019) through efficient fermentation in the biosynthetic pathway of heme, which is essential for organisms to synthesize chlorophyll, heme, vitamin B12, and other substances. It is often used in agriculture to increase photosynthesis efficiency and promote color change. In medicine, 5-ALA is a new type of photodynamic drug that is not only used for the treatment of local or generalized skin cancer, but also for the diagnosis of bladder cancer, digestive tract cancer, lung cancer, and other types of cancer.
Despite its ability to produce PHA at a certain level, Pseudomonas putida poses a potential threat to human health due to its pathogenic properties. Therefore, it is of utmost importance to select a healthy and safe fermentation strain. Currently, Saccharomyces cerevisiae stands out as a preferred strain for genetic engineering, owing to its advantages. Furthermore, it is widely acknowledged by the U.S. Food and Drug Administration as a Generally Recognized As Safe (GRAS) microbial strain. Although Saccharomyces cerevisiae lacks the innate ability to directly convert LA into PHA, its straightforward and modifiable genome renders it a prime candidate for genetic engineering. Through this process, it can be transformed into a strain proficient in utilizing LA for PHA production. As an example, researchers have successfully identified and characterized a seven-gene operon in Pseudomonas putida KT2440 that facilitates the catabolism of LA. By introducing this operon into Saccharomyces cerevisiae, a synthetic pathway for PHA production can be established. However, we must acknowledge that technical hurdles persist, primarily stemming from the toxic effects of LA, an acidic compound, on Saccharomyces cerevisiae, which can hinder fermentation efficiency (Li et al., 2017). The toxicity of LA is predominantly exhibited through its ability to induce acidification within the intracellular environment. Cells need to consume ATP to exclude excess protons and maintain intracellular pH stability, which will affect the energy supply of cells (Hyland et al., 2013), and then affect the growth and fermentation process of Saccharomyces cerevisiae. Consequently, a thorough investigation into the tolerance mechanism of Saccharomyces cerevisiae toward LA is imperative for developing yeast strains that are better suited for industrial-scale applications.
Previous studies have shown that the general catabolic repressor Mig1pΔ can enhance the tolerance and fermentation efficiency of Saccharomyces cerevisiae to toxic concentrations of acetic acid, formic acid, and LA (Balderas-Hernández et al., 2018). After adaptive laboratory evolution (ALE), the S. cerevisiae F3 strain has gained enhanced tolerance to formic acid and unexpectedly demonstrated higher tolerance to acetic acid. In addition, in yeast cells treated with formic acid, the levels of aromatic amino acids have increased, nucleotide synthesis has slowed down, and energy consumption has decreased. These changes may have enhanced the yeast's tolerance to formic acid. However, the intricate mechanisms that contribute to the LA tolerance in Saccharomyces cerevisiae remain unexplored.
This study aims to leverage the Saccharomyces cerevisiae whole-genome knockout library and SGAtool software to systematically screen for genes that are crucial for LA tolerance. Comprehensive analyses using KEGG and GO pathways, coupled with spot plate experiments, have revealed that the deletion of the HMX1 gene markedly heightens the sensitivity of Saccharomyces cerevisiae to LA, thereby suggesting that the HMX1 gene plays a pivotal role in enhancing the yeast's resilience under LA stress. To further elucidate this mechanism, we have employed cutting-edge transcriptomics and metabolomics techniques to unravel the stress response mechanism exhibited by the HMX1 knockout strain in the presence of LA, ultimately confirming the regulatory function of HMX1 in modulating the yeast's tolerance to LA.
2 Experimental materials and methods
2.1 Fungal strains, culture medium, and culture conditions
The knockout library of non-essential genes of Saccharomyces cerevisiae used in this experiment was donated by Professor Beidong Liu from Gothenburg University. LA, agar, peptone, glucose, yeast extract, geneticin (G418), sodium chloride, etc. were purchased from Chengdu Wanke Co., Ltd. The fluorescent dyes, Mito Tracker Green FM, Yeast Vacuole Membrane Marker MDY-64, and ER-Tracker Red dye, were all purchased from Thermo Scientific, while 2'7'-dichlorofluorescein diacetate was purchased from Sigma. YPD solid medium was prepared by adding 2 g of glucose, 2 g of agar, 1 g of yeast extract, and 2 g of peptone per 100 mL. The YPD liquid medium is prepared by removing 2 g of agar from the above-mentioned basis. YPD+G418 medium was prepared by adding G418 to a final concentration of 100 mg/L to the YPD medium. Before conducting spot plate experiments, the knockout strains were first inoculated on YPD+G418 plates by streaking to obtain single colonies. A single colony was then inoculated into a 100 mL Erlenmeyer flask containing 30 mL of YPD+G418 liquid medium and incubated at 30°C with shaking at 200 r/min for 18–24 h.
2.2 Spot plating
The cultured fungal suspensions were uniformly calibrated to a cell concentration (OD600) of 1.0 and then diluted in a 10-fold concentration gradient to obtain suspensions of different concentrations. A multi-channel pipette was used to aspirate 5 μL of the diluted fungal suspension and spot it onto YPD+G418 solid medium containing different concentrations of inhibitors. After incubation for 3–4 days, the plates were observed and photographed.
2.3 Subcellular structure staining
To investigate the growth rate and survival rate of the gene knockout strain relative to its parent strain BY4741, both strains were overnight cultured in a biological shaker at 30°C and 200 rpm. During this process, the initially cultured strain samples were defined as 0 h. Subsequently, the overnight cultured strains were transferred to liquid YPD+G418 medium containing 100 mM LA (at this point, the OD600 value of the samples was 0.8) and further cultured for 3 h under the same conditions. Samples defined as 3 h were also collected during this period. After that, the 0-h and 3-h strain samples were transferred to 1.5 mL EP tubes (at this point, the OD600 value of the samples was 1.0). The structural integrity of the cells was observed using a fluorescence confocal microscope equipped with DIC, GFP, Rhod, and DAPI filters. The accumulation of reactive oxygen species, nuclear chromatin disorders, mitochondrial structure, and the morphology of the endoplasmic reticulum and vacuoles in the BY4741 standard strain (non-knockout strain) and the HMX1 gene knockout strain were evaluated, and the proportions of different morphologies were counted (Madeo et al., 1999).
2.4 Screening of LA-tolerant or sensitive knockout strains
After overnight culturing, the strains BY4741 and the HMX1 gene knockout strain were adjusted to a cell concentration of OD600 = 0.1, and 20 mL samples were taken for transcriptome and metabolome analysis, generating the datasets YOK202W_Q_LA and YLR205CC_Q_LA. Subsequently, 100 mM LA was added to the medium, and the cells were further cultured for 3 h. Another 20 mL samples were taken to obtain the datasets YOK202W_H_LA and YLR205C_H_LA. Transcriptome analysis was performed in triplicate, and metabolome analysis was also repeated three times. In addition, the remaining culture was further incubated, and cell density was measured every 6 h to plot a growth curve, assess the impact of gene knockout on strain tolerance, and ensure sampling accuracy. Among them, the datasets YOK202W_Q_LA and YOK202W_H_LA generated from the strain without HMX1 gene knockout were set as the control group.
2.5 Complementation experiment for LA-tolerant or sensitive knockout strains
From the molecular barcoded yeast (MoBY) open reading frame (ORF) library, Escherichia coli strains harboring complementary plasmids containing shared tolerant or sensitive knockout genes were carefully selected. Following extensive cultivation, the plasmids were extracted and subsequently transformed into knockout strains on SD-Ura agar plates for rigorous screening. A spot assay was conducted to systematically compare the growth patterns of the non-knockout strain, the complemented strain, and the knockout strain on YPD+G418 solid medium, including medium supplemented with 100 mM LA. Photographic documentation was utilized to capture the growth performance, and a thorough analysis of these images was performed to validate the tolerance of the knockout gene in Saccharomyces cerevisiae to levulinic acid. The comprehensive results of this complementation experiment are presented in Supplementary Figure S8.
2.6 Transcriptome analysis
Total RNA was extracted using the TRIzol reagent (Thermo Fisher, batch number 15,596,018) and then assessed for quantity and integrity using the RNA Nano 6000 Assay Kit on the Bioanalyzer 2100 system (Agilent Technologies, California, USA). Gene expression levels were estimated using FPKM (Fragments Per Kilobase of transcript per Million mapped reads). Differential expression analysis was performed using the DESeq2 R package. The criterion for significant differential expression was set as |log2 (foldchange)| ≥ 1 (padj ≤ 0.05). KEGG and GO enrichment analysis of differentially expressed genes (DEGs) was performed using the ClueGo program in Cytoscape. All parameters used in the RNA-seq experiments are presented in Supplementary material 1.
2.7 Metabolome analysis
In non-targeted metabolome analysis, UHPLC-MS/MS analysis was performed on the samples using a Vanquish UHPLC system (ThermoFisher, Germany) coupled with an Orbitrap Q Exactive™ HF mass spectrometer (Thermo Fisher, Germany). Raw data files were processed using Compound Discoverer 3.1 (CD3.1, ThermoFisher) to perform peak alignment, peak detection, and quantitative analysis of each metabolite. Statistical analysis was performed using statistical software R and Python. These metabolites were annotated using the KEGG database.
2.8 Integrated analysis of transcriptome and metabolome
A network diagram can visually represent the relationship between metabolites and genes. Selected top 10 differentially expressed genes and top 5 differentially expressed metabolites were used for plotting. All differentially expressed genes and metabolites obtained were mapped to the KEGG pathway database to obtain their common pathway information, determining the major biochemical and signal transduction pathways that both differentially expressed metabolites and genes participate in. iPath (interactive Pathways Explorer [https://pathways.embl.de/]) is an online analysis tool for visualizing metabolic pathways. It summarizes various metabolic pathways in biological systems, with nodes representing various biochemical molecules and lines representing biochemical reactions. The enriched pathways shared by both differentially expressed metabolites and genes were mapped to the iPath website. Based on the above enrichment results, a metabolic pathway map was presented for the metabolic and transcriptomic enrichment.
3 Results and analysis
3.1 Screening of key genes and spot plating experiment verification
More than 4,400 gene knockout strains of Saccharomyces cerevisiae were inoculated on solid plates containing LA using a large-scale cell manipulation platform. Solid plate photography, SGAtool software scanning analysis, and weight value ≥0.2 screening were conducted. KEGG and GO enrichment analysis of differential phenotypic genes using Cytoscape software revealed that the genes HMX1, YLR225C, XBP1, YBL055C, YJR096W, PRX1, NCL1, and YHB1 on the membrane fluidity pathway exhibited sensitivity to LA (as shown in Supplementary Table S1). The selected weight values were 0.33, 0.63, 0.30, 0.41, 0.30, 0.40, 0.36, and 0.32, respectively. Figure 1A illustrates the eight key genes enriched on the membrane fluidity pathway. The genes XBP1, YBL055C, YJR096W, PRX1, NCL1, and YHB1 are all associated with cellular oxidative stress responses, while HMX1 and YLR225C are only related to oxidative stress.

Figure 1. Screening of knockout strains sensitive to levulinic acid (LA) in S. cerevisiae. (A) KEGG and GO enrichment analysis of genes sensitive to levulinic acid (LA) in Saccharomyces cerevisiae. (B) Spot test of strain BY4741 on YPD medium with varying concentrations of LA. Spot test validation of the key knockout strain of Saccharomyces cerevisiae in YPD medium containing 100 mM LA.
In the screening experiment of knockout strains for the eight key genes in the oxidative stress pathway—HMX1, YLR225C, XBP1, YBL055C, YJR096W, PRX1, NCL1, and YHB1—these strains formed colony spot at dilution of 10−4 and colony spot at dilution of 10−5 colonies on solid media containing YPD+G418, displaying consistent growth patterns with the BY4741 standard strain. On YPD+G418 solid media containing 100 mM LA, both the BY4741 standard strain and the NCL1 and PRX1 knockout strains formed colony spot at dilution of 10−2, showing no significant differences in growth. The knockout strains of YHB1, YJR096, YLR225C, and YBL055C also formed colony spot at dilution of 10−3 and colony spot at dilution of 10−4 under the same conditions, exhibiting consistent growth patterns with the BY4741 standard strain. The XBP1 knockout strain showed no significant difference in growth compared to the non-knockout BY4741 strain, both forming colony spot at dilution of 10−5. Notably, on YPD+G418 solid media with 100 mM LA, the BY4741 standard strain formed colony spot at dilution of 10−3 and colony spot at dilution of 10−4, while the HMX1 knockout strain failed to form any colonies, indicating a significant difference in growth compared to the BY4741 standard strain (Figure 1B). In this experiment, dot blot tests revealed that seven of the knockout strains (YLR225C, XBP1, YBL055C, YJR096W, PRX1, NCL1, and YHB1) showed no significant difference in LA tolerance compared to the non-knockout BY4741 strain. However, the tolerance to LA significantly weakened after knocking out the HMX1 gene, suggesting that this gene plays a role in promoting the tolerance of Saccharomyces cerevisiae under LA stress.
3.2 Transcriptomic and metabolomic analysis of HMX1 knockout strain under LA stress conditions
To investigate the tolerance mechanism of HMX1 gene in Saccharomyces cerevisiae under LA stress, this experiment conducted a differential analysis of the transcriptomic and metabolomic levels between the HMX1 knockout strain and BY4741 under LA stress. As shown in Figure 2A, there was no significant difference in the growth rate between the HMX1 knockout strain and the BY4741 strain in YPD+G418 media, indicating that the gene knockout did not have a significant impact on cell proliferation. However, as shown in Figure 2B, in media containing 100 mM LA, the growth rate of the HMX1 knockout strain was significantly lower than that of the BY4741 strain, suggesting that the knockout of the HMX1 gene significantly affected the strain's tolerance to LA. To further explore the molecular mechanism of this phenomenon, cells from the HMX1 knockout strain and the BY4741 strain treated with LA stress for 3 h were collected and subjected to transcriptomic and metabolomic analyses.
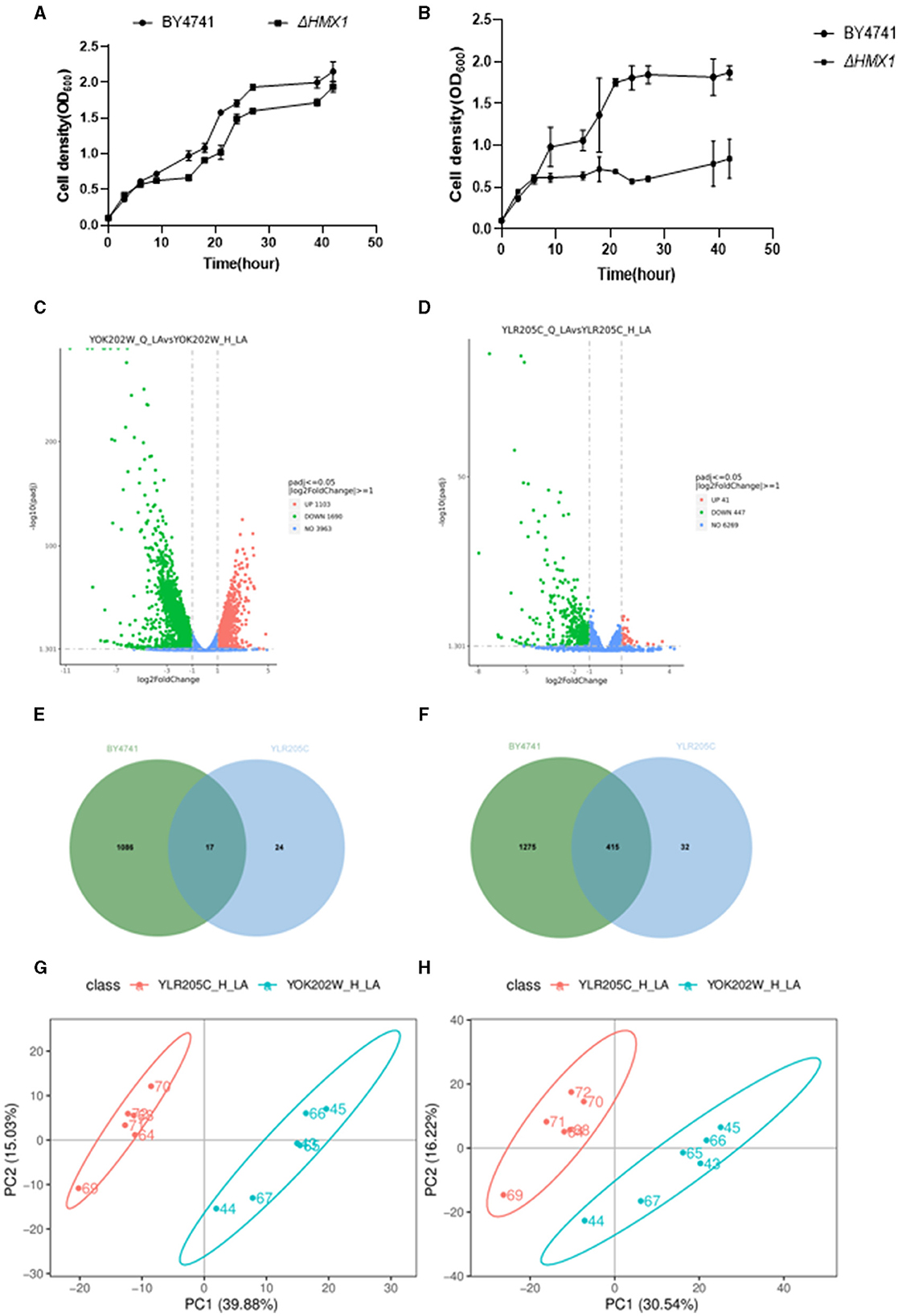
Figure 2. Analysis of growth, transcriptome, and metabolome of BY4741 strain and HMX1Δ knockout strain under 100mM levulinic acid (LA) conditions. The growth trajectory of BY4741 strain and HMX1Δ knockout strain in YPD medium (A) and YPD medium containing 100Mm LA (B). Volcano plot illustrating the genes with differential expression in the BY4741 strain (C) and HMX1Δ (D) after LA treatment for 3 h. (E) Venn diagram analysis of up-regulated genes in the HMX1Δ strain compared to the BY4741 strain under LA stress. (F) Venn diagram analysis of down-regulated genes in the HMX1Δ strain compared to the BY4741 strain under LA stress. Principal component analysis of positively charged (G) and negatively charged (H) metabolites between strain HMX1Δ and BY4741.
Based on the data analysis in Supplementary Table S2, the total amount of sequencing clean bases for all samples significantly exceeded 6.0 G. Given that the Saccharomyces cerevisiae genome contains approximately 6,000 genes, with an average gene length of about 1,000 base pairs (bp), and a transcript containing approximately 6 × 106 bp, it can be concluded that the sequencing depth of this experiment exceeded 1,000 × . Typically, a sequencing depth of 100 × is sufficient for analysis, but the sequencing depth of this experiment far exceeds this standard, indicating an ample amount of data. In addition, the Q20 and Q30 quality scores of the sequencing data were both above 90%, indicating a very low sequencing error rate and high data quality. In summary, this sequencing produced sufficient and high-quality data, fully meeting the needs for in-depth analysis. As shown in Supplementary Figure S1, this figure demonstrates the results of the correlation analysis between samples. The analysis revealed a high positive correlation among biologically replicated samples, validating the consistency of data obtained from each replicate experiment and providing a reliable data foundation for subsequent research.
After performing background removal analysis on the sequencing data of the BY4741 (YOK202W) standard strain and the HMX1 (YLR205C) knockout strain, we compared the gene expression profiles before and after LA treatment. The study found that in the BY4741 reference strain, LA treatment resulted in altered expression of 2,793 genes, with 1,086 genes upregulated and 1,275 genes downregulated (Figure 2E). In contrast, in the HMX1 knockout strain, a total of 488 genes showed altered expression, with 24 genes upregulated and 32 genes downregulated (Figure 2F). The volcano plots (Figures 2C, D) displayed the differentially expressed genes in the three strains, laying a foundation for further analysis.
A Venn diagram (Figure 2G) was used to compare the downregulated genes between the HMX1 knockout strain and the BY4741 reference strain. It was found that the BY4741 strain had 1,275 uniquely downregulated genes, while the HMX1 knockout strain had 32 uniquely downregulated genes. There were 1,307 downregulated genes shared between the two strains. Cytoscape software was employed to perform gene enrichment analysis on the 32 uniquely downregulated genes in the HMX1 knockout strain. The results indicated that these genes were mainly enriched in three pathways: organic anion transport, carboxylic acid transport, and monocarboxylic acid transport.
Pearson correlation analysis (Supplementary Figures S2, S3) demonstrated a high correlation between replicate samples in the metabolomics sequencing, confirming the reliability of the data and providing a solid foundation for subsequent analysis. Principal component analysis of the metabolomic profiles after LA treatment (Figures 2G, H) revealed significant differences between the HMX1 knockout strain and the BY4741 reference strain, indicating differences in their metabolite composition.
Under stress conditions, the differential expression of metabolites in each strain was analyzed. It was found that there were 71 and 111 metabolites upregulated and downregulated in the HMX1 knockout strain, respectively, while 83 and 95 metabolites were upregulated and downregulated in the BY4741 strain, respectively. Venn diagram analysis showed that there were 144 and 197 metabolites that were commonly upregulated and downregulated in both the HMX1 knockout strain and the BY4741 strain (Supplementary Figures S4, S5). To gain a deeper understanding of the specific expression of metabolites in the HMX1 knockout strain, a metabolite enrichment analysis was conducted. The results showed that the specifically upregulated metabolites in the HMX1 knockout strain were enriched in pathways such as Fructose and mannose metabolism, Purine metabolism, and Pyrimidine metabolism (Supplementary Figure S6). The specifically downregulated metabolites in the HMX1 knockout strain were enriched in pathways such as Fatty acid biosynthesis, Riboflavin metabolism, and Pantothenate and COA biosynthesis (Supplementary Figure S7).
3.3 Knockout of HMX1 affects the formation of cell wall-membrane system
To clarify the effect of HMX1 knockout on the membrane system, this experiment observed the endoplasmic reticulum, mitochondria, and vacuoles, which have membrane structures (Figure 3). As shown in Figures 3A, C, under LA stress, both mitochondria and the endoplasmic reticulum exhibited different degrees of damage. From Figure 3B, it can be seen that after 3 h of treatment, the proportion of cells with mitochondrial damage in the BY4741 reference strain increased from 6.73% to 16.77%, while the proportion of cells with mitochondrial damage in the HMX1 knockout strain increased from 7.26% to 37.95%. Additionally, after 3 h of treatment, the proportion of cells with endoplasmic reticulum damage in the BY4741 reference strain increased from 11.30% to 19.21%, while the proportion of cells with endoplasmic reticulum damage in the HMX1 knockout strain increased from 10.75% to 51.86%. Regarding vacuole damage, the proportion of cells with vacuole damage in the BY4741 reference strain increased from 20.59% to 46.34%, while the proportion of cells with vacuole damage in the HMX1 knockout strain increased from 11.76% to 69.43%. From the above results, it can be seen that under LA stress, the mitochondria, endoplasmic reticulum, and vacuoles with membrane structures in the HMX1 knockout strain exhibited significant damage.
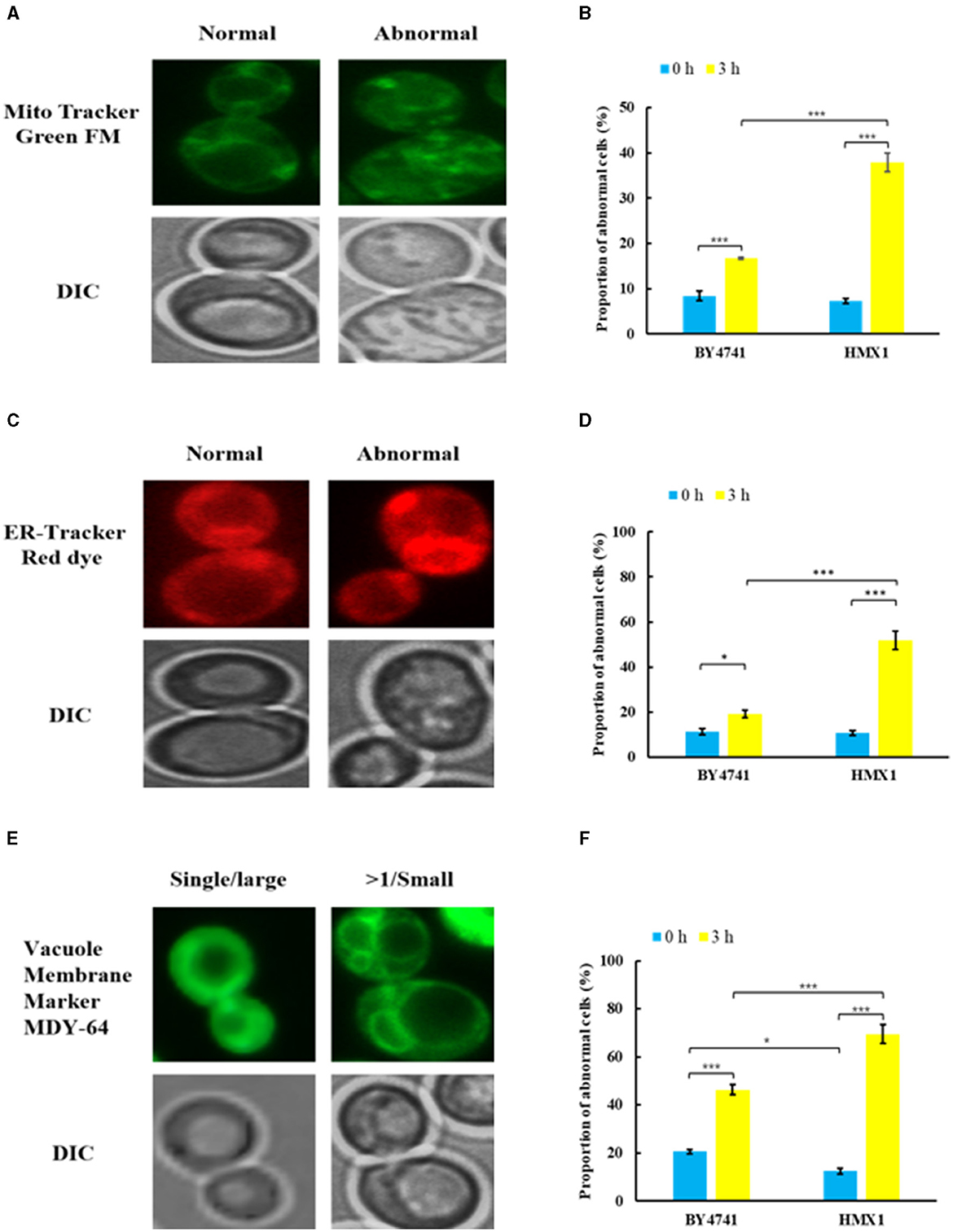
Figure 3. Morphological changes of mitochondria (MT), endoplasmic reticulum (ER), and vacuoles (VC) in the strain BY4741 and HMX1Δ under LA stress. Different morphologies of MT (A), ER (C), VC (E) in cells. The proportion of cells that displayed abnormal MT (B), abnormal ER (D), and more than single small VC (F) after LA treatment for 0 and 3 h. Mito Tracker Green FM: the mitochondria-specific dye. ER-Tracker Red dye: Endoplasmic reticulum stain. Vacuole Membrane Marker MDY-64: vacuole dyeing agent. Single/large: single large vacuole. >1/ Small: more than single small vacuole. DIC, differential interference microscope. *p < 0.05, ***p < 0.001 indicates significant differences. The data represents averages of three experiments. At least 100 cells were examined on each bright-field image.
Analysis of transcriptome sequencing data revealed that genes specifically downregulated in the HMX1 knockout strain were enriched in the monocarboxylic acid transport pathway (Supplementary Table S3). These results confirm, at the gene expression level, that the knockout of HMX1 caused damage to the membrane system. Through the enrichment and collation of downregulated genes, it was found that the genes ADY2, ATO3, and FAT3 responsible for organic anion transport and carboxylic acid transport were specifically downregulated by 1.28, 1.35, and 1.59 times, respectively, in the HMX1 knockout strain (Figure 4). All these genes are related to the energy metabolism and cell membrane of Saccharomyces cerevisiae cells. In addition, since HMX1 is closely related to sterol synthesis, this experiment also investigated other genes related to cell wall system synthesis, such as the HLR1 gene, which showed significant differences in the HMX1 knockout strain (Figure 4). As can be seen from Figure 4, nine genes related to the plasma membrane were downregulated to some extent in the HMX1 knockout strain, while the downregulation was much less pronounced in the BY4741 strain. Overall, the knockout of HMX1 is likely to result in insufficient iron accumulation, which inhibits sterol acquisition, thereby reducing the stability of the cell wall and weakening the resistance of the membrane system to LA.
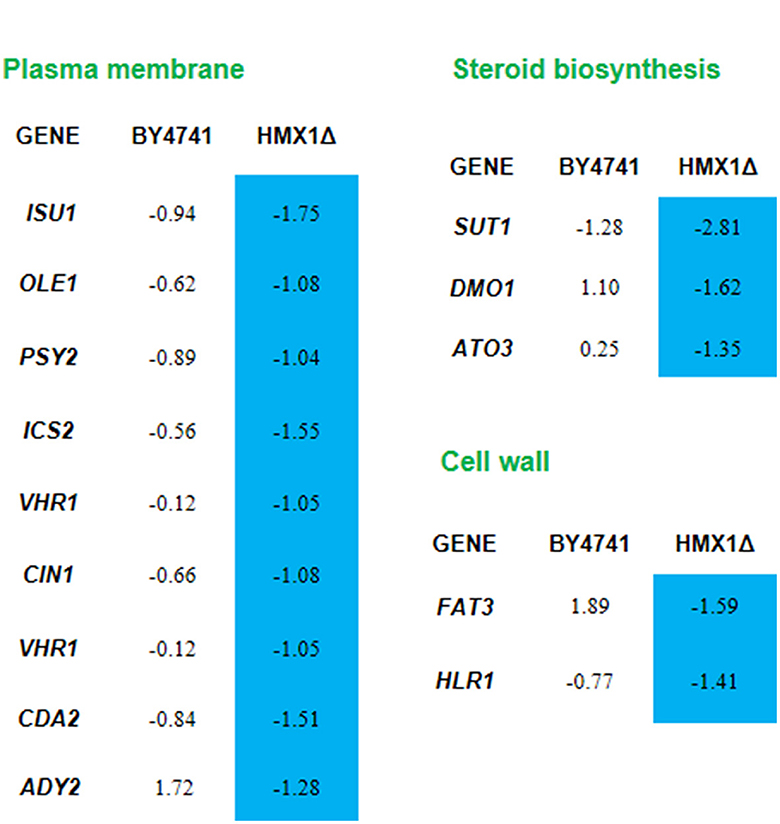
Figure 4. Response mechanism of the cell wall-membrane system to levulinic acid (LA) in the HMX1Δ strain. Expression levels of genes in the BY4741 and HMX1Δ strains in response to 100 mM LA, represented by the values of log2(fold change). Blue indicates significant downregulation of the gene.
3.4 Knockout of HMX1 inhibits coenzyme-A synthesis
Metabolomic analysis of the HMX1 knockout strain and BY4741 strain exposed to LA revealed that specifically downregulated metabolites in the HMX1 knockout strain were enriched in the Pantothenate and COA biosynthesis pathway. In the HMX1 knockout strain, metabolites related to pantothenate and COA synthesis, such as Dephospho-COA and Coenzyme-A, exhibited specific downregulation by 1.01 and 1.02 times, respectively, whereas in the BY4741 strain, they were downregulated by only 0.35 times. Additionally, the precursor of Coenzyme-A, Pantothenate, underwent a specific downregulation of 1.18 times in the BY4741 strain, which shifted to a specific downregulation of only 0.60 times in the HMX1 knockout strain. These findings suggest that the knockout of the HMX1 gene has a certain degree of influence on the biosynthesis of pantothenate and COA (Figure 5A). Simultaneously, it was observed that there were no significant differences in the expression levels of genes CAB1, CAB2, and CAB3 related to the synthesis of these metabolites in both the HMX1 knockout strain and BY4741 strain under LA stress. However, CAB4 and CAB5 exhibited specific downregulation by 1.48 and 1.12 times, respectively. This indicates that the changes in Dephospho-COA and related metabolites are associated with changes in gene transcription levels. Intracellularly, Dephospho-COA reduces oxidative stress by transferring acetyl groups, protecting cells from damage caused by free radicals. As shown in Figures 5B, C, the proportion of cells with reactive oxygen species (ROS) in the BY4741 strain was 22.48% and 23.15% at 0 h and 3 h after LA treatment, respectively, indicating that LA does not stimulate the accumulation of intracellular ROS in BY4741 cells. However, in the HMX1 knockout strain, the proportion of cells with ROS increased significantly from 21.37% to 35.73% after 3 h of LA treatment, suggesting that under LA stress and with the absence of the HMX1 gene, the accumulation of ROS is significantly elevated. This demonstrates that the efficiency of ROS clearance is significantly reduced in the HMX1 knockout strain, which is likely related to the decrease in Coenzyme-A (Figure 5A). Previous studies have shown that other precursors of acetyl-CoA, such as pyruvate or acetate, can enter other intracellular compartments, resulting in reduced availability of acetyl-CoA in the cytoplasm (Schadeweg and Boles, 2016). Our experimental findings revealed that while the content of Pantothenate, a precursor for Coenzyme-A synthesis, showed a certain degree of accumulation, the content of Coenzyme-A in the HMX1 knockout strain under LA stress increased by 1.02 times compared to that in the BY4741 strain under LA stress. We speculate that the knockout of the HMX1 gene under LA stress leads to reduced Coenzyme-A synthesis.
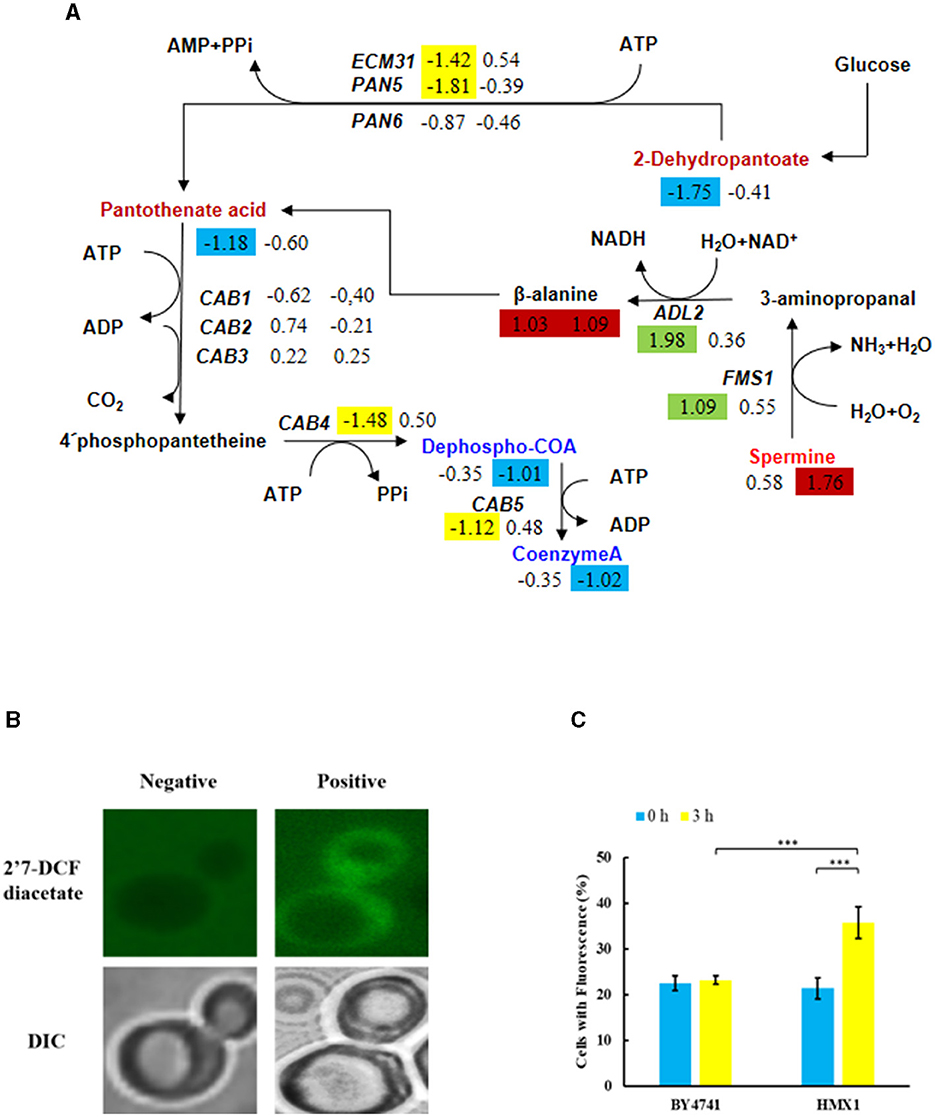
Figure 5. Under LA stress, the decrease in pantothenate-mediated Acetyl-CoA (CoA) in BY4741 and HMX1Δ strains leads to an increase in reactive oxygen species (ROS). (A) Changes in the expression levels of key genes and the contents of key metabolites in the pantothenate synthesis and metabolism pathway. The metabolite contents and gene expression levels in BY4741 strain (left column) and HMX1Δ strain (right column) after 3 h of treatment with 100 mM LA compared to 0 h treatment are represented by log2 (fold change) values. Green indicates significant upregulation of genes. Yellow indicates significant downregulation of genes. Red indicates a significant increase in metabolite content. Blue indicates a significant decrease in metabolite content. (B) Accumulation of ROS in cells. Changes in ROS content (C) and necrotic cell ratio in BY4741 and HMX1Δ strains after 0 and 3 h of LA treatment. 2′7′-Dichlorodihydrofluorescein diacetate: ROS indicator dye. DAPI, DNA-specific dye diamidino-2-phenylindole. ***p < 0.001 indicates a significant difference. Data represent the mean of three experiments. At least 100 cells were examined per bright-field image.
3.5 Knockout of HMX1 impacts riboflavin metabolism
Acetyl-CoA is an essential cofactor involved in various metabolic pathways. As mentioned earlier, the knockout of the HMX1 gene hinders the synthesis of acetyl-CoA. Therefore, this experiment found that the specifically downregulated metabolites in the HMX1 knockout strain were also enriched in the Riboflavin metabolism pathway. Riboflavin is synthesized through enzymatic reactions involving GTP and ribulose-5-phosphate catalyzed by enzymes encoded by the riboflavin biosynthetic operon. We speculate that the specific downregulation of key metabolites in this pathway may affect riboflavin synthesis. In the riboflavin metabolism, the levels of Riboflavin were significantly reduced by 1.44 times in the HMX1 knockout strain under LA stress. However, there was no specific increase or decrease in the BY4741 strain under LA stress. Notably, the precursor of Riboflavin, IGMP, exhibited a significant increase of 1.16 times in the BY4741 strain under LA stress, while it decreased by 1.71 times in the HMX1 knockout strain under the same conditions (Figure 6). Additionally, the expression levels of the key enzyme genes involved in this pathway, namely RIB1, RIB3, RIB4, RIB5, and RIB7, exhibited similar trends in both strains under LA stress (Figure 6). This suggests that the transcriptional levels of these key enzyme genes do not affect the increase or decrease in the levels of key metabolites in the Riboflavin metabolism pathway. Based on the above results, it can be concluded that the decrease in IGMP is a crucial factor leading to the reduction in Riboflavin content. Taken together, we infer that under LA stress and with the knockout of the HMX1 gene, the synthesis of riboflavin is reduced.
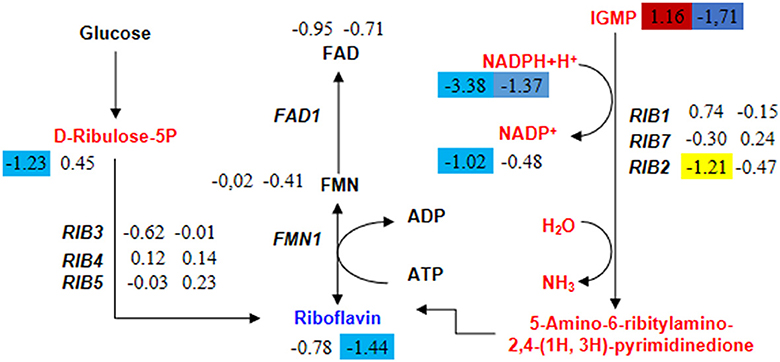
Figure 6. Changes in the expression levels of key genes and the content of key metabolites in the riboflavin metabolism pathway in BY4741 and HMX1Δ strains after 100 mM LA treatment. The metabolite content and gene expression levels [represented by log2 (fold change) values] in BY4741 strain (left column) and HMX1Δ strain (right column) after 3 h of treatment with 100 mM LA, compared to the treatment for 0 h. Yellow indicates significant downregulation of genes. Red indicates a significant increase in metabolite content. Blue indicates a significant decrease in metabolite content.
4 Conclusion and discussion
In this experiment, we confirmed through spot plate assays that the oxidative stress pathway plays an important role in LA tolerance. Additionally, the gene HMX1 identified in this study has not been previously studied in the context of LA tolerance, but we found that it can enhance the tolerance of Saccharomyces cerevisiae to levulinic acid, suggesting that it is a new gene in the oxidative stress pathway involved in LA tolerance. Through subcellular structural observations, we found that this gene has protective effects on mitochondria, endoplasmic reticulum, vacuoles, and reactive oxygen species.
According to the analysis of omics, the knockout of HMX1 has caused downregulation of genes related to the cell wall and cell membrane system under LA stress. However, there was no significant downregulation of membrane system and cell wall-related genes in the HMX1 knockout strain without LA stress. The above results indicate that the knockout of HMX1 affects the expression level of related genes, rather than being caused by LA. Therefore, it can be inferred that HMX1 has a protective effect on the cell membrane. The knockout of the HMX1 gene may weaken the ability of Saccharomyces cerevisiae to cope with oxidative stress, resulting in an increase in the level of intracellular reactive oxygen species (ROS), and thus increasing the possibility of cell damage (Collinson et al., 2011). HMX1 is a homolog of heme oxygenase, which plays a role in oxidative stress response, heme catabolism, and intracellular iron homeostasis. It has been found to be localized on the endoplasmic reticulum and outer nuclear membrane (Protchenko and Philpott, 2003). Yeast cells regulate sterol uptake through heme, where sterols are critical components of cell membranes (Jordá and Puig, 2020). Previous studies have found that the deletion of HMX1 leads to iron accumulation defects and decreased heme degradation activity in yeast cells, resulting in the accumulation of heme in yeast cells (Protchenko and Philpott, 2003). Moreover, some studies have reported that the growth of Saccharomyces cerevisiae under iron-deficient conditions requires the synthesis of ergosterol (Jordá et al., 2021). Therefore, it can be speculated that the iron accumulation defects caused by the knockout of the HMX1 gene inhibit the uptake of sterols by BY4741. The effects of weak acids on Saccharomyces cerevisiae are usually considered to be due to the accumulation of protons or acid ions in the cytoplasm, leading to acidification, which may have toxic effects on the normal metabolic functions of cells (Russell, 1992; Ullah et al., 2012). Therefore, cells need more energy and resources to deal with the reconstruction and maintenance of the cell wall, and the deletion of HMX1 may lead to the blockage of these processes, resulting in unstable exchange of substances inside and outside the cell. Knockout of the HMX1 gene may further exacerbate this situation, as HMX1 may be involved in regulating gene expression or protein function related to cell membrane permeability. Ergosterol molecules are transported in the form of sterol esters to multiple subcellular structures, such as mitochondria, vacuoles, and the plasma membrane and inner membrane systems of the endoplasmic reticulum (Girardi Piva et al., 2022). In this study, under LA stress, HMX1 knockout strains exhibited damage to their mitochondria, vacuoles, and endoplasmic reticulum (Figure 2). Consequently, it is plausible to deduce that the ergosterol content, which is indirectly regulated by HMX1 in these knockout strains, undergoes a reduction. This reduction, in turn, weakens the cell membrane-wall system, thereby compromising the resistance of Saccharomyces cerevisiae to LA.
Research results indicate that overexpression of the OLE1 gene leads to increased oleic acid content and increased unsaturation of fatty acids on the plasma membrane, which in turn enhances the tolerance of cells to acetoin (Guo et al., 2018).
However, this study found that nine genes related to the cell membrane, including ISU1 and OLE1, showed specific downregulation in the HMX1 knockout strain under LA stress, leading to cell membrane damage and reduced tolerance to LA. The cholesterol pathway, which is also included, plays a crucial role in membrane stability. Since the key gene SUT1 responsible for the cholesterol pathway shows specific downregulation in the HMX1 knockout strain under LA stress. Other synthesis-related genes such as DMO1 and ATO3 also show specific downregulation in both strains under LA stress. Therefore, it can be speculated that the cholesterol content may decrease in the HMX1 knockout under LA stress. As cholesterol is a key component for maintaining the integrity and adaptability of the cell membrane, increasing the level of cholesterol helps enhance the stability and resistance of the cell membrane (Vázquez et al., 2019). Therefore, it can be inferred that the knockout of the HMX1 gene may interfere with the production of ergosterol and cholesterol, indicating that HMX1 plays an important role in maintaining the protective function of the cell membrane and enhancing the tolerance of yeast to LA.
This experiment found that under LA stress, the active oxygen content did not accumulate significantly in the non-knockout strain, indicating that LA does not lead to the accumulation of active oxygen in the BY4741 standard strain cells. However, the active oxygen content increased in the HMX1 knockout strain under LA stress. Previous studies have found that the genes involved in oxygen radical detoxification in Saccharomyces cerevisiae (GRX2, HMX1, SOD1, TRX3, and TSA1) include the knockout gene HMX1 (Olzhausen et al., 2009). Therefore, it can be speculated that the increase in active oxygen content is caused by the knockout of HMX1. However, it may also be related to acetyl-CoA, which exists in the cytoplasm and mitochondria of Saccharomyces cerevisiae cells. The synthesis of acetyl-CoA starts with pantothenic acid in Saccharomyces cerevisiae. Previous studies have found that the pantothenate kinase reaction limits the synthesis of CoA in yeast and is inhibited by acetyl-CoA (Vadali et al., 2004; Philpott and Protchenko, 2008). Within the cell, Dephospho-CoA reduces its own oxidative stress by transferring acetyl groups and protecting itself from free radical damage. We speculate that under LA stress, the knockout strain HMX1 allows Saccharomyces cerevisiae to quickly adjust its internal metabolism based on growth conditions. The relatively small increase in pantothenic acid in the HMX1 knockout strain under LA stress compared to the non-knockout strain HMX1 under LA stress may be a compensatory mechanism for the cell to cope with the absence of the HMX1 gene. However, the increased content is not sufficient to support the synthesis of Coenzyme-A, leading to an increase in intracellular active oxygen. This suggests that the efficiency of active oxygen scavenging is significantly reduced in the HMX1 gene knockout strain, which is likely related to the decrease in Coenzyme-A.
Additionally, this study found that the riboflavin content also decreased in the HMX1 knockout strain. Riboflavin is an essential compound for yeast growth. The production of riboflavin is significantly influenced by the supply of its precursor substances, and increasing the supply of these precursors will help improve the yield of riboflavin (You et al., 2021). Therefore, it can be inferred that the decrease in riboflavin content may be directly related to the reduction in the content of precursor substances. The trigger for overproduction of riboflavin in yeast is associated with the activation of intracellular stress signaling cascades in response to environmental changes. Related studies have shown that the results of riboflavin overproduction caused by oxidative stress in A. gossypii are related to mycelial growth and stress defense against ROS (Kato et al., 2020). In addition, iron-limiting conditions induce the overproduction of riboflavin in various yeasts, such as Candida guilliermondii, Candida flava, and Candida famata (Prokopiv et al., 2013; Andreieva et al., 2020). As mentioned earlier, the deletion of the HMX1 gene leads to defects in iron accumulation, and iron deficiency can lead to excessive synthesis of riboflavin in several yeast species. Therefore, it can be speculated that the stress mechanism that prevents riboflavin from defending against ROS accumulation is impeded in the HMX1 knockout strain under LA stress. Furthermore, the contents of NADPH and NADP+ in the HMX1 knockout strain under LA stress showed a certain increase compared to the non-knockout strain BY4741 under LA stress. Riboflavin has an antioxidant effect on oxidative stress, especially lipid peroxidation. The mechanism of riboflavin protecting the body from oxidative stress can be attributed to the glutathione redox cycle. Moreover, research results indicate that riboflavin functions as a precursor of the coenzymes flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) to alter cell metabolism and increase the NADH/NADPH ratio, thereby enhancing resistance to oxidative stress (Olfat et al., 2022). Overall, the increase in riboflavin content may improve the tolerance of the HMX1 knockout strain to LA stress.
The results of this study have revealed the response mechanism of Saccharomyces cerevisiae HMX1 knockout strain in response to LA stress. The study found that increasing the content of cell membrane-wall system, pantothenic acid, acetyl-CoA, and riboflavin is crucial for enhancing the resistance of Saccharomyces cerevisiae to LA. These findings provide a scientific basis for the modification of yeast strains that are tolerant to LA, and also open up new possibilities for the use of LA as a raw material for fermentation to produce high-value-added products. This lays a theoretical foundation for modifying strains to improve their tolerance to levulinic acid, which is of great significance for the efficient conversion of lignocellulosic biomass resources into levulinic acid.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA1088400.
Author contributions
JT: Validation, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization, Writing – original draft. YC: Validation, Software, Project administration, Methodology, Investigation, Data curation, Conceptualization, Writing – original draft. QL: Supervision, Software, Methodology, Investigation, Conceptualization, Writing – original draft, Validation, Data curation. WX: Supervision, Project administration, Investigation, Data curation, Conceptualization, Writing – original draft. XX: Writing – original draft, Validation, Data curation, Conceptualization. XC: Writing – original draft, Validation, Formal analysis, Data curation. LixY: Writing – original draft, Software, Formal analysis, Investigation. BM: Writing – original draft, Supervision, Software, Investigation, Validation. JL: Writing – original draft, Validation, Supervision, Project administration, Methodology, Formal analysis, Software. FL: Writing – original draft, Methodology, Software, Formal analysis. CF: Writing – original draft, Validation, Methodology, Supervision. WLo: Writing – original draft, Methodology, Data curation, Project administration, Formal analysis. HL: Writing – original draft, Software, Methodology, Data curation, Formal analysis. XH: Writing – original draft, Methodology, Data curation, Supervision, Software. PF: Writing – original draft, Validation, Methodology, Supervision, Project administration. WLi: Writing – original draft, Software, Methodology, Data curation, Project administration, Formal analysis. KZ: Writing – original draft, Software, Resources, Project administration. LiuY: Writing – original draft, Visualization, Resources, Formal analysis. YY: Visualization, Resources, Project administration, Writing – review & editing. MM: Visualization, Resources, Project administration, Writing – review & editing. HW: Visualization, Resources, Project administration, Formal analysis, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the National Natural Science Foundation of China (No. 32200044), the Talent Research Startup Projects of Leshan Normal University (No. RC2022003), the 2023 College Students' Innovation and Entrepreneurship Training Program (Nos. S202310649091 and S202310649093), and the Scientific Research and Cultivation Plan of Leshan Normal University (Nos. 2022SSDJS009, KYPY2024-0005, and KYPY2024-0012).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1416903/full#supplementary-material
References
Adeleye, A. T., Louis, H., Akakuru, O. U., Joseph, I., Enudi, O. C., and Michael, D. P. (2019). A review on the conversion of levulinic acid and its esters to various useful chemicals. Aims Energy 7, 165–185. doi: 10.3934/energy.2019.2.165
Alonso, D. M., Wettstein, S. G., and Dumesic, J. A. (2013). Gamma-valerolactone, a sustainable platform molecule derived from lignocellulosic biomass. Green Chem. 15, 584–595. doi: 10.1039/c3gc37065h
Andreieva, Y., Petrovska, Y., Lyzak, O., Liu, W., Kang, Y., Dmytruk, K., et al. (2020). Role of the regulatory genes SEF1, VMA1 and SFU1 in riboflavin synthesis in the flavinogenic yeast Candida famata (Candida flareri). Yeast. 37, 497–504. doi: 10.1002/yea.3503
Balderas-Hernández, V. E., Correia, K., and Mahadevan, R. (2018). Inactivation of the transcription factor mig1 (YGL035C) in Saccharomyces cerevisiae improves tolerance towards monocarboxylic weak acids: acetic, formic and levulinic acid. J. Ind. Microbiol. Biotechnol. 45, 735–751. doi: 10.1007/s10295-018-2053-1
Chen, Y. W., and Lee, H. V. (2020). Recent progress in homogeneous Lewis acid catalysts for the transformation of hemicellulose and cellulose into valuable chemicals, fuels, and nanocellulose. Rev. Chem. Eng. 36, 215–235. doi: 10.1515/revce-2017-0071
Collinson, E. J., Wimmer-Kleikamp, S., Gerega, S. K., Yang, Y. H., Parish, C. R., Dawes, I. W., et al. (2011). The yeast homolog of heme oxygenase-1 affords cellular antioxidant protection via the transcriptional regulation of known antioxidant genes. J. Biol. Chem. 286, 2205–2214. doi: 10.1074/jbc.M110.187062
Ennaert, T., Van Aelst, J., Dijkmans, J., De Clercq, R., Schutyser, W., Dusselier, M., et al. (2016). Potential and challenges of zeolite chemistry in the catalytic conversion of biomass. Chem. Soc. Rev. 45, 584–611. doi: 10.1039/C5CS00859J
Girardi Piva, G., Casalta, E., Legras, J.-L., Tesnière, C., Sablayrolles, J.-M., Ferreira, D., et al. (2022). Characterization and role of sterols in Saccharomyces cerevisiae during white wine alcoholic fermentation. Fermentation 8:90. doi: 10.3390/fermentation8020090
Guo, Z. P., Khoomrung, S., Nielsen, J., and Olsson, L. (2018). Changes in lipid metabolism convey acid tolerance in Saccharomyces cerevisiae. Biotechnol. Biofuels. 11:297. doi: 10.1186/s13068-018-1295-5
Habe, H., Sato, Y., and Kirimura, K. (2020). Microbial and enzymatic conversion of levulinic acid, an alternative building block to fermentable sugars from cellulosic biomass. Appl. Microbiol. Biotechnol. 104, 7767–7775. doi: 10.1007/s00253-020-10813-7
Han, Y., Ye, L., Gu, X., Zhu, P., and Lu, X. (2019). Lignin-based solid acid catalyst for the conversion of cellulose to levulinic acid using γ-valerolactone as solvent. Industr. Crops Prod. 127, 88–93. doi: 10.1016/j.indcrop.2018.10.058
Hara, K. Y., Saito, M., Kato, H., Morikawa, K., Kikukawa, H., Nomura, H., et al. (2019). 5-Aminolevulinic acid fermentation using engineered Saccharomyces cerevisiae. Microb. Cell Fact. 18:194. doi: 10.1186/s12934-019-1242-6
Hyland, P. B., Mun, S. L.-S., and Mahadevan, R. (2013). Prediction of weak acid toxicity in Saccharomyces cerevisiae using genome-scale metabolic models. Industr. Biotechnol. 9, 229–235. doi: 10.1089/ind.2013.0004
Jordá, T., and Puig, S. (2020). Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae. Genes (Basel). 11:795. doi: 10.3390/genes11070795
Jordá, T., Rozès, N., and Puig, S. (2021). Sterol composition modulates the response of saccharomyces cerevisiae to iron deficiency. J. Fungi. 7:901. doi: 10.3390/jof7110901
Kato, T., Azegami, J., Yokomori, A., Dohra, H., El Enshasy, H. A., and Park, E. Y. (2020). Genomic analysis of a riboflavin-overproducing Ashbya gossypii mutant isolated by disparity mutagenesis. BMC Gen. 21:319. doi: 10.1186/s12864-020-6709-7
Lange, J. P., Price, R., Ayoub, P. M., Louis, J., Petrus, L., Clarke, L., et al. (2010). Valeric biofuels: a platform of cellulosic transportation fuels. Angew Chem. Int. Ed. Engl. 49, 4479–4483. doi: 10.1002/anie.201000655
Li, Y.-C., Gou, Z.-X., Zhang, Y., Xia, Z.-Y., Tang, Y.-Q., and Kida, K. (2017). Inhibitor tolerance of a recombinant flocculating industrial Saccharomyces cerevisiae strain during glucose and xylose co-fermentation. Brazil. J. Microbiol. 48, 791–800. doi: 10.1016/j.bjm.2016.11.011
Madeo, F., Fröhlich, E., Ligr, M., Grey, M., Sigrist, S. J., Wolf, D. H., et al. (1999). Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 145, 757–767. doi: 10.1083/jcb.145.4.757
Olfat, N., Ashoori, M., and Saedisomeolia, A. (2022). Riboflavin is an antioxidant: a review update. Br. J. Nutr. 128, 1887–1895. doi: 10.1017/S0007114521005031
Olzhausen, J., Schübbe, S., and Schüller, H. J. (2009). Genetic analysis of coenzyme A biosynthesis in the yeast Saccharomyces cerevisiae: identification of a conditional mutation in the pantothenate kinase gene CAB1. Curr. Genet. 55, 163–173. doi: 10.1007/s00294-009-0234-1
Philpott, C. C., and Protchenko, O. (2008). Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell. 7, 20–27. doi: 10.1128/EC.00354-07
Pileidis, F. D., and Titirici, M. M. (2016). Levulinic acid biorefineries: new challenges for efficient utilization of biomass. ChemSusChem. 9, 562–582. doi: 10.1002/cssc.201501405
Prokopiv, T. M., Fedorovych, D. V., Boretsky, Y. R., and Sibirny, A. A. (2013). Oversynthesis of riboflavin in the yeast Pichia guilliermondii is accompanied by reduced catalase and superoxide dismutases activities. Curr. Microbiol. 66, 79–87. doi: 10.1007/s00284-012-0242-0
Protchenko, O., and Philpott, C. C. (2003). Regulation of intracellular heme levels by HMX1, a homologue of heme oxygenase, in Saccharomyces cerevisiae. J. Biol. Chem. 278, 36582–36587. doi: 10.1074/jbc.M306584200
Russell, J. (1992). Another explanation for the toxicity of fermentation acids at low pH: anion accumulation versus uncoupling. J. Appl. Bacteriol. 73, 363–370. doi: 10.1111/j.1365-2672.1992.tb04990.x
Schadeweg, V., and Boles, E. (2016). Increasing n-butanol production with Saccharomyces cerevisiae by optimizing acetyl-CoA synthesis, NADH levels and trans-2-enoyl-CoA reductase expression. Biotechnol. Biofuels. 9:257. doi: 10.1186/s13068-016-0673-0
Ullah, A., Orij, R., Brul, S., and Smits, G. J. (2012). Quantitative analysis of the modes of growth inhibition by weak organic acids in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 78, 8377–8387. doi: 10.1128/AEM.02126-12
Vadali, R. V., Bennett, G. N., and San, K. Y. (2004). Cofactor engineering of intracellular CoA/acetyl-CoA and its effect on metabolic flux redistribution in Escherichia coli. Metab. Eng. 6, 133–139. doi: 10.1016/j.ymben.2004.02.001
Vázquez, J., Grillitsch, K., Daum, G., Mas, A., Beltran, G., and Torija, M. J. (2019). The role of the membrane lipid composition in the oxidative stress tolerance of different wine yeasts. Food Microbiol. 78, 143–154. doi: 10.1016/j.fm.2018.10.001
Keywords: saccharomyces cerevisiae, levulinic acid, response mechanism, transcriptome, metabolome
Citation: Tang J, Chen Y, Li Q, Xin W, Xiao X, Chen X, Yang L, Mou B, Li J, Lu F, Fu C, Long W, Liao H, Han X, Feng P, Li W, Zhou K, Yang L, Yang Y, Ma M and Wang H (2024) The response mechanism analysis of HMX1 knockout strain to levulinic acid in Saccharomyces cerevisiae. Front. Microbiol. 15:1416903. doi: 10.3389/fmicb.2024.1416903
Received: 13 April 2024; Accepted: 10 June 2024;
Published: 26 June 2024.
Edited by:
Jiong Hong, University of Science and Technology of China, ChinaReviewed by:
Rishabh Sharma, Temple University, United StatesHuadong Peng, University of Queensland, Australia
Copyright © 2024 Tang, Chen, Li, Xin, Xiao, Chen, Yang, Mou, Li, Lu, Fu, Long, Liao, Han, Feng, Li, Zhou, Yang, Yang, Ma and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Menggen Ma, bWdlbkBzaWNhdS5lZHUuY24=; Yaojun Yang, cnN5eWpAMTI2LmNvbQ==; Hanyu Wang, SGFueXUud2FuZzYxNDNAb3V0bG9vay5jb20=
†These authors have contributed equally to this work
 Jiaye Tang
Jiaye Tang Yulei Chen
Yulei Chen Qian Li1,2†
Qian Li1,2† Wenli Xin
Wenli Xin Chun Fu
Chun Fu Hong Liao
Hong Liao Peng Feng
Peng Feng Menggen Ma
Menggen Ma