
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 October 2024
Sec. Microbe and Virus Interactions with Plants
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1415700
This article is part of the Research Topic Tobacco Disease and Biological Control View all 13 articles
 Qipeng Jiang1†
Qipeng Jiang1† Yong Wang2†
Yong Wang2† Jiamin Yu3†
Jiamin Yu3† Jinfeng Wang1
Jinfeng Wang1 Shiping Guo3
Shiping Guo3 Dongyang Liu2
Dongyang Liu2 Xiangwen Yu3
Xiangwen Yu3 Lianqiang Jiang2
Lianqiang Jiang2 Gang Long2
Gang Long2 Daojiang Xi1
Daojiang Xi1 Shuhong Chen2
Shuhong Chen2 Yue Wang1
Yue Wang1 Wei Ding1*
Wei Ding1*Rhizosphere microbial communities strongly affect outbreaks of root-knot nematode (RKN) disease. However, little is known about the interactions among fungi, bacteria and RKN. The bacterial and fungal community compositions in the rhizospheres of four representative tobacco varieties, both resistant and susceptible to RKN, were characterized using 16S rRNA gene sequencing for bacteria and internal transcribed spacer gene sequencing for fungi. Our findings revealed that the fungi played crucial roles in facilitating the cross-kingdom and symbiotic fungal–bacterial interactions to suppress RKN. Moreover, our investigation suggested Microbacterium as a potential microbial antagonist against RKN based on its enhanced presence in RKN-resistant tobacco genotypes, and the relative abundance of Microbacterium was 34.49% greater in the rhizosphere of resistant tobacco than that of susceptible tobacco significantly. Notably, the richness of fungal community enhanced tobacco’s microbe-associated resistance to RKN through the positive regulation of the richness and diversity of bacterial community and the relative abundance of Microbacterium. This study underscores the critical role of the fungus–dominated fungal–bacterial community in bolstering tobacco resistance against RKN. The potential antagonistic role of Microbacterium presents promising avenues for innovative RKN management strategies.
Root-knot nematodes (RKNs, Meloidogyne spp.) are the most destructive and widespread type of plant-parasitic nematode (PPN) and have been shown to significantly hinder nutrient and water uptake in plants, leading to developmental delays (Jones et al., 2013). RKN infection not only causes wounds in plant roots but also predisposes plants to further damage by other soil-borne pathogens, thereby leading to complex plant diseases (Kyndt et al., 2017; Zhang Y. et al., 2020). This has led to substantial economic losses globally, particularly affecting the production of Solanaceae crops, such as tomato (Du et al., 2022; El-Sappah et al., 2019), eggplant (Khan and Siddiqui, 2017; Zhou et al., 2019) and tobacco (Cao et al., 2023; Xu et al., 2023). In the context of the urgent demand for environmentally friendly strategies to manage RKNs, biocontrol is gaining increasing attention as a potential and sustainable strategy (Sun et al., 2021). This approach is recognized for maintaining the One Health of humans, soil, plants and animals (Berg et al., 2020).
The pivotal role of microbial communities associated with plants, often referred to as the ‘second genome’ of plants (Afzal et al., 2019; Berendsen et al., 2012), significantly contributes to the fitness and performance of the holobiont (Fadiji et al., 2023; Singh et al., 2020). Soils, considered as the foundation of the ‘One Health’ concept, harbor the most diverse and complex microbiomes within terrestrial ecosystems (Banerjee and van der Heijden, 2023). The rhizosphere, a narrow zone of soil influenced by root secretions, plays a crucial role in plant health. The rhizosphere microbiome acts as a primary line of defense for plants against pathogens (Banerjee and van der Heijden, 2023), and numerous microbial strains have been identified as antagonists of RKN through a range of direct and indirect interactions, including antagonism and disease suppression (Antil et al., 2023; Forghani and Hajihassani, 2020). When faced with pathogen, healthy resistant plant varieties recruit and receive a protective soil microbiome subset in the rhizosphere. This subset suppresses pathogens by competing nutrients (Du et al., 2022), producing antimicrobial substances (Luo et al., 2022), regulating plant growth (Xiang et al., 2018), and inducing plant defensive reactions (Antil et al., 2023). Previous studies have proven the potential of the soil microbiome to suppress plant-parasitic nematodes (Adam et al., 2014; Liu et al., 2020; Zhou et al., 2018), highlighting that variations in rhizospheric microbial communities can explain, to some extent, the variable infestation success of RKNs (Elhadyl et al., 2017; Lu et al., 2014). Numerous rhizospheric bacteria and fungi, such as Bacillus spp. (Bui et al., 2020; Liu et al., 2020), Pseudomonas spp. (Liu et al., 2020), Microbacterium spp. (Zhao J. et al., 2019; Zhao Z. B. et al., 2019), Paecilomyces spp. (Kiriga et al., 2018), and Trichoderma spp. (Khan et al., 2020; Martínez-Medina et al., 2017), have been reported to effectively control RKNs. Therefore, the rhizosphere represents one of the most promising and valuable sources of biocontrol microbiomes that remains to be explored and utilized for RKNs management.
Plant genotypes are involved in the assembly process of the rhizosphere microbiome, primarily in the form of diverse plant-derived compounds (Oyserman et al., 2022; Sasse et al., 2018), and significantly contribute to the development of specific microbiome-associated disease resistance (Lee et al., 2021; William et al., 2018). Variations in the severity of RKN diseases among different tobacco varieties in the same field suggest the presence of a specific composition and interaction of the microbiome in the rhizosphere, which plays a regulatory role in RKN. Although studies have revealed the role of rhizospheric microbes in antagonistic interactions with RKNs in great detail (Antil et al., 2023; Zhou et al., 2019), efforts to identify the core microbiome interacting with RKNs among resistant and susceptible plants in the field are still limited.
In recent years, culture-independent high-throughput sequencing has greatly expanded the repertoire of the plant-associated microbiome and its role in disease-suppression (Liu et al., 2016; Zhang S. et al., 2020). This advancement has enabled us to unravel complex microbial communities, discover novel microbial indicators, and investigate the interactions between the core microbiota and RKN in the plant rhizosphere. We hypothesized that the bacterial–fungal community in the rhizosphere contributed to tobacco’s resistance to RKN and specific microbiomes played critical antagonistic roles in manage root-knot nematodes. The aims of this study were to (1) characterize the bacterial and fungal communities in the rhizosphere of tobacco varieties that resistant and susceptible to RKN, and (2) identify bacteria and fungi potentially interact with RKN and explore potential antagonists for RKN management. The results of this study provide promising avenues for innovative RKN management strategies.
The field experiment (26°17′38″N, 102°1′10″E, elevation: 1,892 m) was conducted from May 1st to August 30th, 2021, in Liangshan Yi Autonomous Prefecture, Sichuan Province (Supplementary Figure S1). The experimental field had been subjected to continuous tobacco cropping for years and was induced with root-knot nematode (RKN) disease. The field experiment was repeated at the same location in 2022 (Supplementary Figure S2). The RKN infecting tobacco was identified as Meloidogyne incongnita in our previous study (Jiang et al., 2021).
Four representative tobacco varieties, Zhongchuan208 (ZC208), Yunyan87 (YY87), Yunyan85 (YY85), and Honghuadajinyuan (HD), were used in the field experiment. Tobacco plants were bred and cultured under identical conditions by the Liangshan Prefecture Branch of Sichuan Tobacco Corporation. The experimental field was divided evenly into 12 plots, each variety of tobacco was planted in 3 randomly arranged plots (each plot size was 4.8 m × 14 m), and each plot had 100 plants (4 lines × 25 plants/line, the line spacing is 1.2 m, the spacing between two close plants in one line is 0.55 m).
To evaluate the progression of RKN disease in different tobacco varieties, the disease grades of the aboveground and root parts of each plant were investigated (Fan et al., 2023). The criteria for disease grade refer to the national standard of the tobacco pest classification survey method of China (GBT-23222-2008, Table 1). The aboveground disease grade for each plant was recorded every 20 days within 80 days after transplanting. The criteria ranked the degree of aboveground disease as grades 0 to 9 depending on plant growth and symptoms on leaves. Plants were uprooted with the whole root ball, the soil around the roots was carefully removed under running tap water 100 days after transplanting, and the root disease grade for each root was assessed. The criteria ranked the degree of root disease as grades 0 to 9 depending on the proportion of root knots that infected the root.
To evaluate the difference in the degree of RKN disease among different tobacco varieties, disease incidence rate, disease index and area under the disease progression curve (AUDPC) were calculated using the following formulas:
where Nd is the number of disease-infected plants, and N is the total number of investigated plants (Ali et al., 2022).
where Ni is the number of plants with the respective disease grade, vi is the disease grade (0, 1, 3, 5, 7, and 9), and N is the total number of investigated plants (Ali et al., 2022).
where Vi and Vi−1 are the disease indices on ti (20, 40, 60, 80, 100) and ti−1 (0, 20, 40, 60, 80, 100), respectively, and ti-ti−1 is the number of days between ti and ti−1 (Dyer et al., 2022).
To investigate the density of RKNs and identify the characteristics of the microbial community in the rhizosphere of different tobacco varieties, samples of the tobacco rhizosphere and bulk soil were collected 100 days after transplanting in the experimental field. Three rhizosphere soil samples for each tobacco variety were collected following the methods described in our previous study (Liu et al., 2016). Five individual rhizosphere soils from five random plants in each plot were collected and mixed into one sample. Five samples of representative bulk soil from five randomly arranged sites in the experimental field were collected, and the sampling depth was 10–20 cm. Each soil sample was separated into two even samples (Sample A and Sample B). Samples A and B were transported to Southwest University, Chongqing, for further testing within 3 days. Sample A was stored at room temperature (24°C) to determine the density of RKN. Sample B was stored at −20°C for DNA extraction.
RKN-infected tobacco roots were observed and photographed under a stereomicroscope. The density of second-stage juveniles of Meloidogyne incongnita (M. incongnita J2) in each soil sample was determined, considering that M. incongnita J2 is the infectious stage during which nematodes puncture the root tissues, resulting in knots or swellings (Abad et al., 2003). Hatched M. incongnita J2s in the soil samples were isolated according to the Baermann funnel method, and the populations were counted using a stage micrometer under a Nikon microscope (Tokyo, Japan). The density of M. incongnita J2 is expressed per 100 g of dry soil.
Microbial DNA from soil samples was extracted using the FastDNA Spin Kit (MP Biomedicals, United States kits) following the standard protocol. Microbial DNA was stored at −80°C. The amplification and purification of soil microbial DNA were conducted according to methods described in our previous study (Zhang et al., 2017). The 515 forward (5′-GTGCCAGCMGCCGCGG-3′) and 806 reverse (5′-GGACTACHVGGGTWTCTAAT-3′) primers were used to amplify the V4 region of the bacterial 16S rDNA gene. ITS1 forward (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 reverse (5′-GCTGCGTTCTTCATCGATGC-3′) were used to amplify the ITS1 region of the fungal ITS gene.
The 16S rRNA and ITS gene fragments were sequenced by Shanghai Majorbio Co., Ltd., China, using the Illumina MiSeqPE250 platform.1 The quality control and annotation of the raw sequencing data were conducted to generate high-quality reads, and amplicon sequence variants (ASVs) with a consistency of 100% were assigned to taxa at the phylum, class, order, family and genus levels using QIIME2.2 The taxonomic assignments of each bacterial and archaeal ASV were performed using the Silva138 database, and the taxonomic assignment of each fungal high-quality read was performed using the Unite8.0 database.
Microbial α diversity and β diversity analyses were performed using the free online platform of the Majorbio Cloud Platform.3 Specifically, the Chao1, Simpson, and Simpson evenness indices were calculated as microbial α diversity based on Faith’s phylogenetic metric at the ASV level. Specifically, Higher Chao1 index and Simpson evenness index suggested higher richness and evenness of the microbial communities respectively, additionally, higher Simpson index suggested lower diversity of the microbial communities. To determine the dissimilarity of the β diversity of the microbial communities, principal coordinate analysis (PCoA) was performed based on the Bray–Curtis distance according to the phylogenetic tree. To identify discriminative taxa between groups, linear discriminate analysis (LDA) effect size (LEfSe) was performed (Segata et al., 2011). A factorial Kruskal–Wallis sum-rank test (α = 0.05) was used in LEfSe to identify taxa from the phylum to genus level with significant differential abundances between categories (using all-against-all comparisons, LDA score > 2.0).
To determine the effect of the soil microbiome on RKNs in the tobacco rhizosphere, the underlying co-occurrence of bacteria, fungi and RKNs was determined through network analysis via the Molecular Ecological Network analysis pipeline (MEAN4) and Grephi software (Sun et al., 2023). Network analysis was performed at the ASV level (relative abundance greater than 0.01%) for the bacterial–fungal communities to ensure the accuracy of interactions, and network analysis was performed at the genus level to reduce the complexity of calculations and ensure the accuracy of taxonomic information. Data filtering was performed prior to avoiding zero values that could result in spurious correlations, and the taxa represented in 50% of the samples were retained (Zhao J. et al., 2019; Zhao Z. B. et al., 2019). To determine the direct effects of the α diversity of bacterial and fungal communities on RKNs, a structural equation model (SEM) was constructed in IBM SPSS AMOS software.
All statistical significance was assessed by ANOVA in SPSS Statistics software. To evaluate the correlation between variables, linear regression analysis was performed with SPSS statistics software. Other figures were generated in Origin software.
The resistance levels of four different tobacco varieties to RKNs were evaluated in a field experiment (Figures 1A–D; Supplementary Figure S1). The results indicated significant differences in resistance to RKNs among the four tobacco varieties. Specifically, the resistance level of HD plants was significantly lower than that of the ZC208, YY85, and YY87 plants, whose AUDPC was 96% lower than that of the HD plants (Figure 1E). The density of M. incongnita J2 in both bulk soil (25 ~ 101 per 100 g dry soil) and rhizosphere soil (89 ~ 301 per 100 g dry soil) was assessed. The density of M. incongnita J2 in the rhizosphere was 195% greater than that in the bulk soil (p < 0.05), suggesting an enrichment of M. incongnita J2 in the tobacco rhizosphere (Figure 1F). Notably, the proportion of M. incongnita J2 in the rhizosphere of the HD treatment group was 11 ~ 22% greater than that in the rhizosphere of the other treatment groups. Furthermore, the density of M. incongnita J2 was positively correlated with the incidence rate (Figure 1G, r = 0.90, p < 0.001) and disease index (Figure 1F, r = 0.85, p < 0.001), confirming that M. incongnita J2 in the tobacco rhizosphere contributes positively to RKN disease outbreaks. These results suggested that HD exhibited stronger attraction to M. incongnita J2 and lower resistance against RKNs than ZC208, YY85, and YY87. Thus, HD was identified as a susceptible variety, while ZC208, YY85, and YY87 were categorized as resistant varieties in the following analysis.
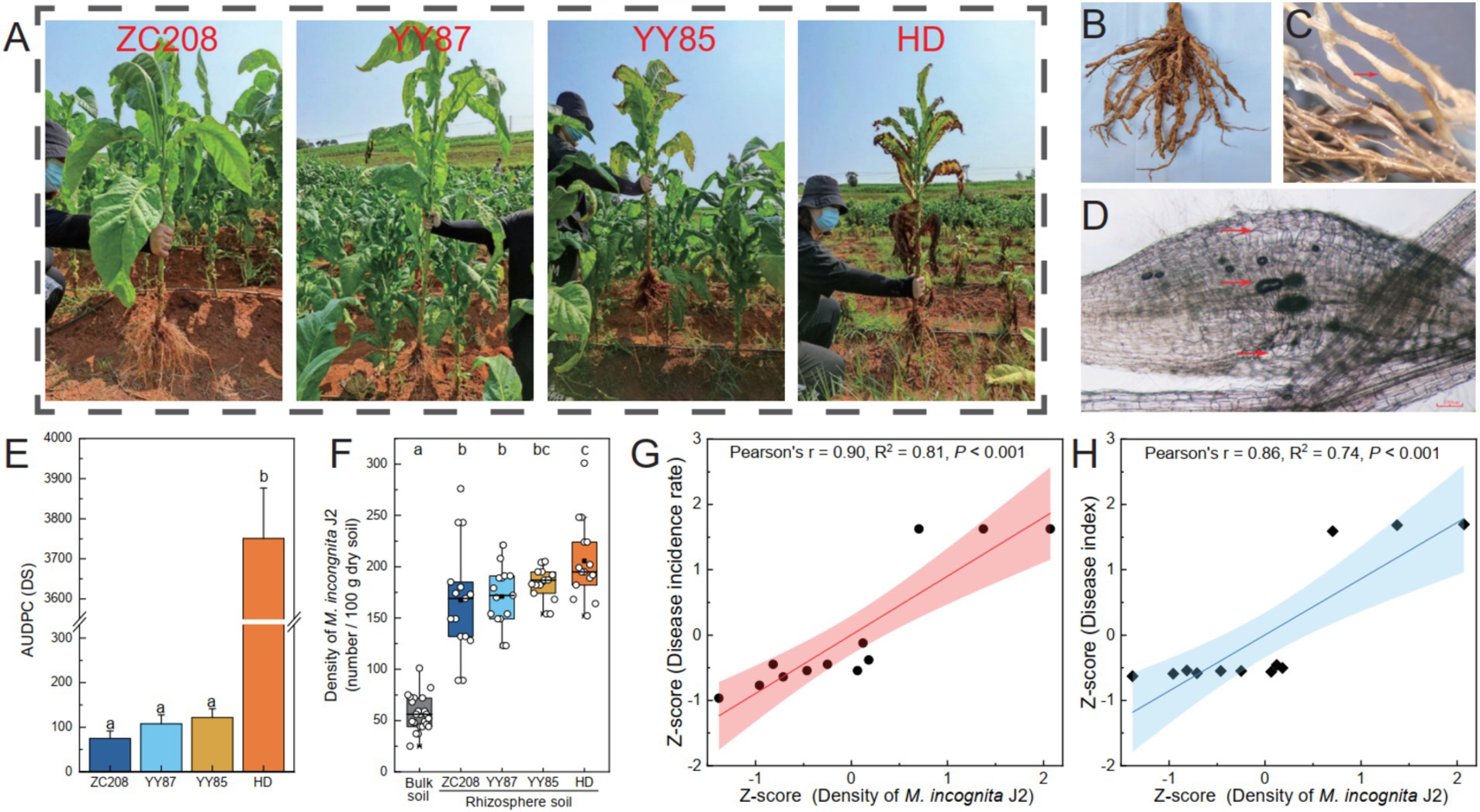
Figure 1. Variation in resistance to root-knot nematode disease among different tobacco varieties. (A) Disease-induced tobacco. The disease-induced tobacco roots (B) and root-knots (C). (D) The nematodes in disease-induced tobacco roots and the giant cells on the roots. (E) Area under disease progression curve (AUDPC). (F) Density of M. incongnita J2 populations. Relationship between disease incidence rate (G) and disease index (H) and the density of M. incongnita J2 populations.
To reveal the effects of the soil microbiome on root-knot nematodes, the bacterial and fungal communities of the bulk soil and tobacco rhizosphere were characterized by high-throughput sequencing. For the bacterial communities, a total of 4,472,837 reads with an average read length of 256 bp were obtained through 16S rRNA high-throughput sequencing analysis. These reads were clustered into 37,313 ASVs, which were assigned to 45 bacterial phyla and six archaeal phyla. For fungal communities, 4,739,497 reads with an average read length of 241 bp were detected through ITS high-throughput sequencing analysis. These reads were clustered into 5,611 ASVs, which were assigned to five fungal phyla.
To evaluate the alpha diversity of the bacterial and fungal communities among the different sample groups, the Chao1 index, Simpson diversity index and Simpson evenness index were calculated to assess the richness, diversity and evenness of the microbial communities (Table 2). The results showed that the bacterial richness and diversity in the tobacco rhizosphere were greater than those in the bulk soil. Conversely, the richness and diversity of fungi in the rhizosphere soil were lower than those in the bulk soil, although these differences were statistical insignificance (p > 0.05). Our findings suggested that tobacco roots may possess an active effect on soil bacteria in the rhizosphere. Specifically, soil bacteria were generally recruited and enriched in the rhizosphere based on its higher alpha diversity of bacterial community. However, the rhizosphere effect on soil fungi was negative, and only specific fungi were selected and prosper in the tobacco rhizosphere based on its lower alpha diversity of fugal community.
Furthermore, to explore the effect of the α diversity of microbial communities on root-knot nematodes, linear regression analysis between the α diversity of microbial communities and the density of M. incongnita J2 populations was conducted; the independent variables were the Chao1 index, Simpson diversity index and Simpson evenness index, and the dependent variable was the density of M. incongnita J2 populations (Figure 2). The results indicated that the Chao1 index (Spearman’s r = 0.57, p < 0.05), 1/Simpson diversity index (Spearman’s r = 0.64, p < 0.05) and Simpson evenness index (Spearman’s r = 0.59, p < 0.05) of the bacterial community were positively associated with the density of M. incongnita J2. In contrast, the Chao1 index (Spearman’s r = −0.09, p > 0.05), 1/Simpson diversity index (Spearman’s r = −0.21, p > 0.05) and Simpson evenness index (Spearman’s r = −0.32, p > 0.05) of the fungal community showed a negative relationship with the density of M. incongnita J2, although these correlations did not reach statistical significance. The results suggested that the effects of soil bacterial and fungal communities on RKN were different and that the soil bacterial community had a stronger and more direct effect on RKN.
To clarify the effect of the rhizosphere on the soil microbial community, the bacterial–fungal community compositions of the bulk soil and rhizosphere of different tobacco varieties were characterized. The results suggested that the bacterial community compositions at the phylum level among the different groups were similar. Specifically, over 43% of the tested soil microbiomes were bacteria, and over 52% were fungi (Supplementary Figure S1). Actinobacteriota, Proteobacteria and Chloroflexi were the most abundant bacterial phylum, and their total relative abundances (RAs) were greater than 65% (Figure 3A). The fungal community composition at the phylum level differed significantly among the different groups. Ascomycota was the most abundant fungal phylum, whose RAs were greater than 50%. Additionally, the RAs of Ascomycota in the rhizosphere of the HD were lower than those in the bulk soil and rhizosphere of the resistant plants (Figure 3B). Sphingomonas, Gemmatimonas, Nocardioides, Streptomyces, and Conexibacter were the most abundant bacterial genus, remarkably, the RA of Nocardioides in the rhizosphere of the ZC208 were 60.85, 31.97, and 80.83% higher than those in rhizosphere of HD, YY85, and YY87, respectively (Figure 3C). Additionally, Gibberella and Xenoacremonium were the most abundant fungal genus and the RA of Gibberella in the bulk soil were much higher than that in the rhizosphere (Figure 3D).
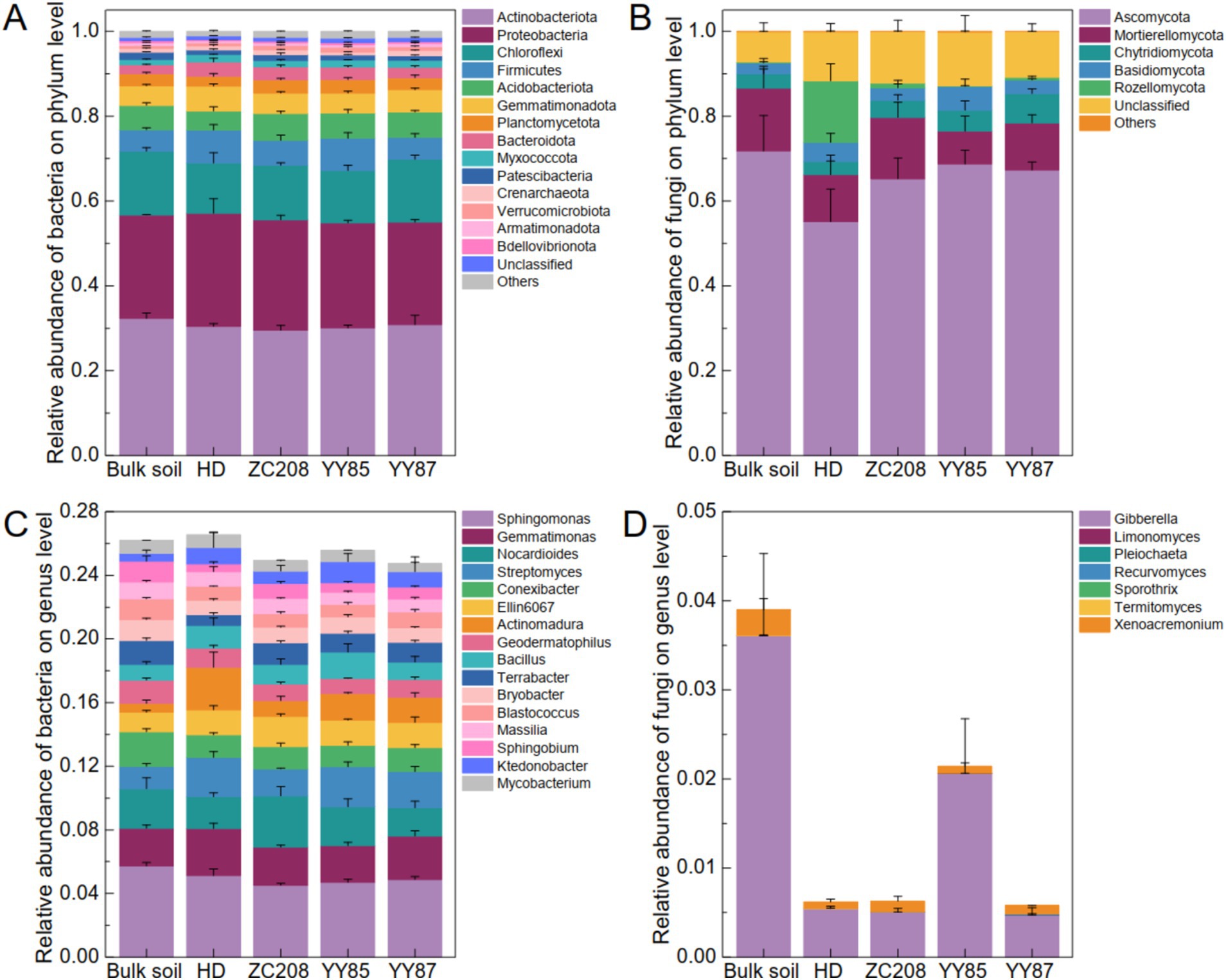
Figure 3. Soil microbial community composition. (A) Bacterial community composition at the phylum. (B) Fungal community composition at the phylum. The dominated genuses of bacterial (C) and fungal (D) community.
The variations in the microbial communities were analyzed using principal coordinate analysis (PCoA). The results showed a remarkable divergence in the soil bacterial communities between the bulk soil and rhizosphere (Figures 4A,B, Adonis, R2 = 0.32, p < 0.05). However, the soil fungal communities of the soil samples were not significantly different between the bulk soil and rhizosphere (Figures 4D,E, Adonis, R2 = 0.27, p > 0.05). Additionally, the difference in the bacterial community between the rhizosphere of HD and bulk soil was more pronounced than that observed for the resistant varieties (Figure 4B, ANOVA, p > 0.05). However, the variation in the fungal community between the bulk soil and rhizosphere of HD was lower than that between the bulk soil and rhizosphere of the resistant variety (Figure 4E, ANOVA, p > 0.05).
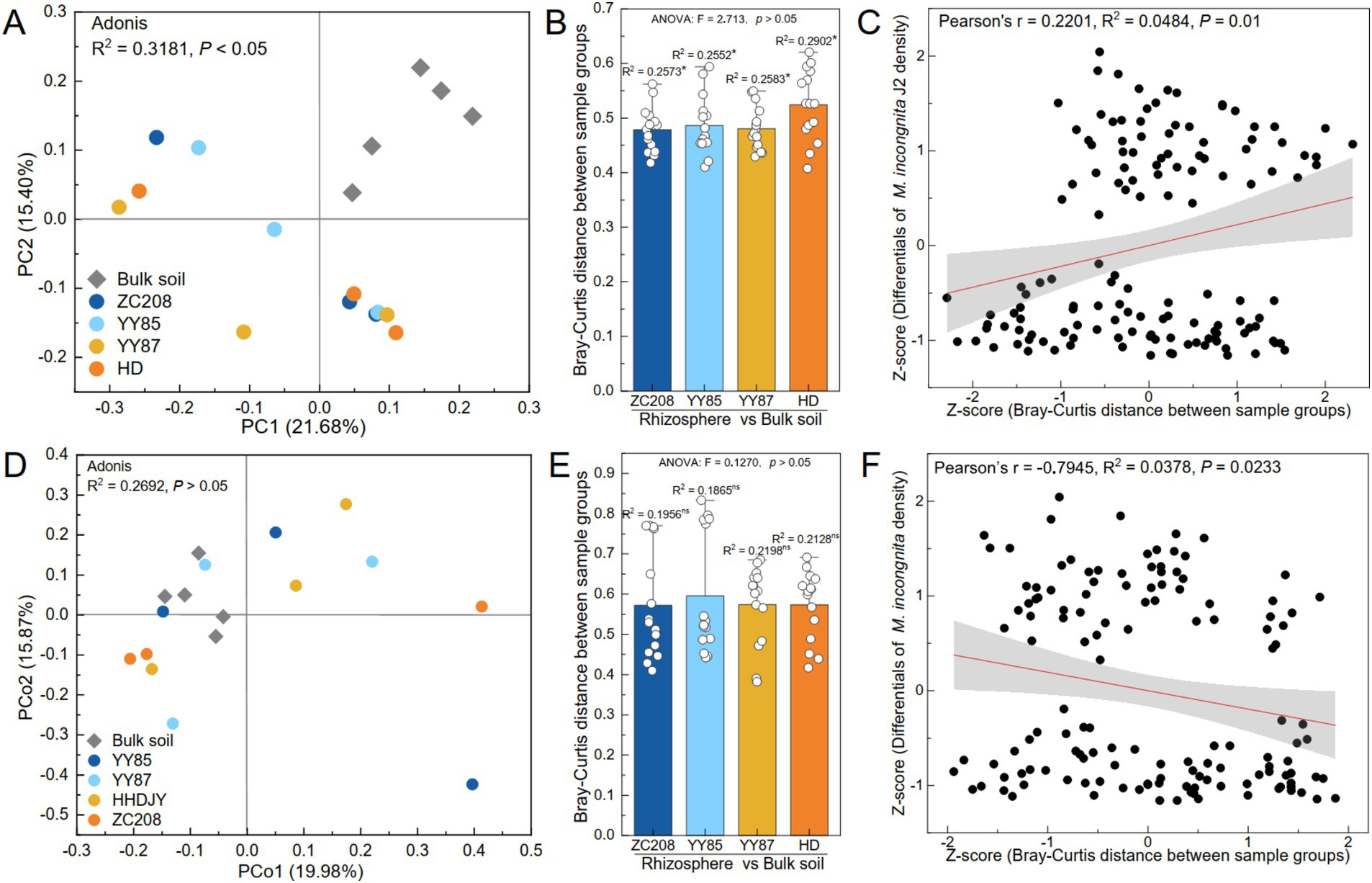
Figure 4. Effect of bacterial and fungal communities on the density of M. incongnita J2. Principal coordinate analysis (PCoA) and permutational multivariate analysis of variance (PERMANOVA) of the bacterial (A) and fungal (D) communities of the samples at the ASV level. Bray–Curtis distance of bacterial (B) and fungal (E) communities between the bulk soil and rhizosphere of the susceptible variety (HD) and resistant variety (ZC208, YY85, and YY87). The independent variables of the linear regression analyses were the Bray–Curtis distances of the bacterial (C) and fungal (F) communities, and the dependent variable was the difference in M. incongnita J2 density.
To identify the relationship between the microbial community composition and the density of M. incongnita J2, linear regression analyses were conducted, and the independent variable was the Bray–Curtis distance between samples, while the dependent variable was the difference in the density of M. incongnita J2 between the respective samples. The differences in the bacterial communities were positively associated with differences in the density of M. incongnita J2 (Figure 4C, Pearson’s r = 0.22, p < 0.05). However, the differences in fungal communities were negatively associated with the difference in the density of M. incongnita J2 (Figure 4F, Pearson’s r = −0.79, p < 0.05). Our findings further indicated that the soil bacterial community had a more direct effect on RKN; however, the soil fungal community demonstrated an indirect interaction with RKN.
To identify distinctive microbial taxa in both the tobacco rhizosphere and bulk soil, linear discriminant analysis (LDA) effect size (LEfSe) from phylum to genus was performed. Additionally, linear regression analyses between the RAs of the distinctive taxa and M. incongnita J2 density were conducted. Our analysis revealed 26 distinctive bacterial taxa, six of which were prevalent in bulk soil and exhibited a negative relationship with M. incongnita J2 density. In contrast, 20 distinctive bacterial taxa were predominant in the tobacco rhizosphere and displayed a positive correlation with M. incongnita J2 density (Figure 5A). Moreover, 17 distinctive fungal taxa were identified, 11 of which were more abundant in bulk soil and were negatively correlated with M. incongnita J2 density, while six of the identified taxa were more prevalent in the tobacco rhizosphere and were positively correlated with M. incongnita J2 density (Figure 5C).
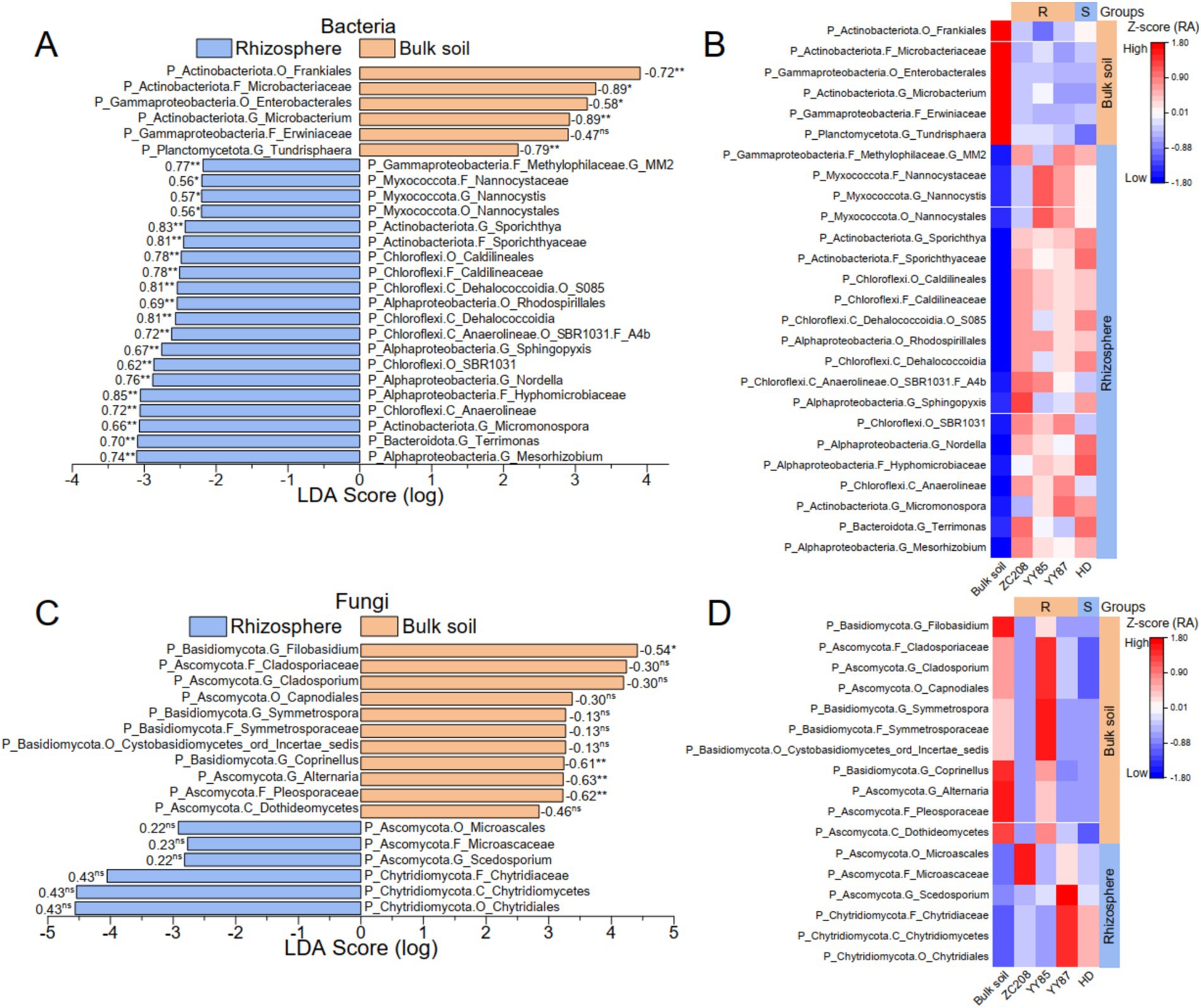
Figure 5. Discriminant microbial taxa associated with the density of M. incongnita J2. The discriminative bacterial (A) and fungal (C) taxa based on LEfSe (LDA score ≥ 2.0, p < 0.05) from phylum to genus between the bulk soil and the tobacco rhizosphere. Heatmaps of discriminative bacterial (B) and fungal (D) taxa based on the RA (Z score) in the rhizosphere of resistant (ZC208, YY87, and YY85) and susceptible (HD) tobacco plants. The numbers on the LDA score bars in (A,C) are Pearson’s r values associated with the M. incongnita J2 density. The p values from Pearson’s analysis are indicated as “ns” for p > 0.05, “*” for p < 0.05, and “**” for p < 0.01. The groups on the right side of the heatmaps in (B,C) highlighted the taxa enriched in the bulk soil or the rhizosphere.
To reveal the relationship between the distinctive taxa and tobacco resistance to RKNs, the RAs of distinctive bacterial (Figure 5B) and fungal (Figure 5D) taxa in resistant (ZC208, YY87, and YY85) and susceptible (HD) tobacco varieties were compared. The results showed that taxa enriched in resistant tobacco varieties, such as Microbacterium (Pearson’s r with J2 = −0.89, p < 0.01) and Tundrisphaera (Pearson’s r with J2 = −0.79, p < 0.01), were positively correlated with M. incongnita J2 density, and their RAs were more than 55% greater than that of HD (susceptible). Conversely, taxa prevalent in HD, such as Terrimonas (Pearson’s r with J2 = 0.70, p < 0.01), Nordella (Pearson’s r with J2 = 0.76, p < 0.01) and Hyphomicrobiaceae (Pearson’s r with J2 = 0.85, p < 0.01), were positively correlated with M. incongnita J2 density, and their RAs in the rhizosphere of HD were 21 to 53% greater than those of the resistant varieties.
To evaluate the assembly processes of the bacterial and fungal communities, the β-nearest taxon index (β-NTI) based on the null model and Sloan neutral model was calculated, and the differences in assembly processes between the rhizosphere and bulk soil, as well as between the resistant and susceptible tobacco varieties, were investigated (Figure 6A). The results revealed a predominant influence of stochastic processes (|β-NTI| < 2) in shaping the bacterial community assembly, both in the rhizosphere and bulk soil (85%) and in resistant and susceptible varieties (85%). However, for the variation in the fungal community, stochastic processes only contributed 45% of the variation between the rhizosphere and bulk soil and 63% of the variation between the resistant and susceptible varieties. These findings suggested a notable distinction between bacterial and fungal community assemblies in soil. Specifically, the fungal communities appeared to be more significantly influenced by tobacco roots than bacteria, implying a more targeted recruitment of fungal taxa by tobacco in the rhizosphere rather than a stochastic assembly.
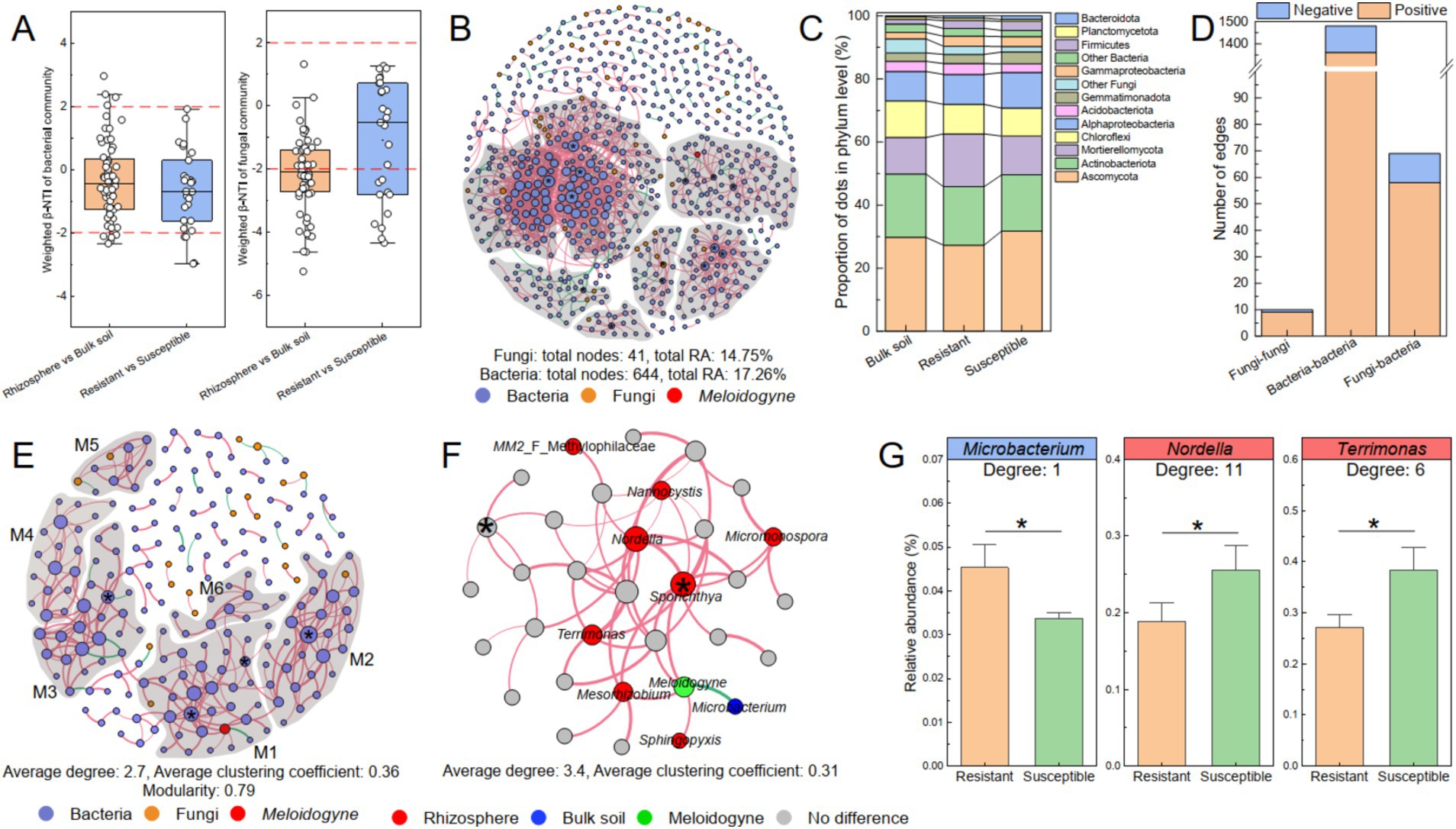
Figure 6. The co-occurrence network of the bacterial–fungal communities related to Meloidogyne. (A) β-NTI between sample pairs of rhizosphere and bulk soil and between resistant and susceptible varieties. The co-occurrence networks of the bacterial–fungal communities at the ASV level (B) and genus level (E). The nodes in the co-occurrence network are color coded: bacteria in blue, fungi in orange, and M. incongnita J2 in red. Modules were delineated with gray areas and labeled M1–M6, representing nodes with 10 or more connections. A connection between two nodes indicates a significant correlation (RA ≥ 0.01%, r ≥ 0.6, p < 0.05). Positive correlations are shown in red, whereas negative correlations are shown in green. Nodes marked with “*” represent the identified module hubs (Zi ≥ 2.50, Pi < 0.62). (C) The composition of nodes in the co-occurrence network at the phylum level. (D) The correlation of dots (ASVs) in the co-occurrence network, with distinctions between intrakingdom and interkingdom. (F) M1 of the co-occurrence network, including M. incongnita J2 (green), is depicted. Nodes enriched in the rhizosphere soil (red) and bulk soil (dark blue) were identified according to LEfSe. (G) Key microbial genera related to M. incongnita J2. Genera that were negatively correlated with J2 density are highlighted in blue boxes, while those that were positively correlated with J2 density are highlighted in red boxes. The labels indicate the degree of nodes (genus level) in module 1. * between the bars indicates significant differences at the p < 0.05 level.
To reveal the interaction patterns between bacterial and fungal communities, a co-occurrence network comprising 644 bacterial ASVs and 41 fungal ASVs was constructed. Within this network, the total RAs of the bacterial and fungal ASVs were 17.26 and 14.75%, respectively (Figure 6B). Notably, more than 80% of these ASVs were classified as key taxa, such as Ascomycota (29.54%), Actinobacteriota (18.86%), Mortierellomycota (13.53%), Chloroflexi (10.02%), and Alphaproteobacteria (9.96%). Among these, Mortierellomycota was more abundant in the rhizosphere of resistant tobacco than in that of susceptible tobacco (Figure 6C). Over 85% of the interactions of fungal ASVs were interkingdom (fungi–bacteria), however, bacterial ASVs engaged in interkingdom interactions were less than 5%a. Remarkably, 84.06% of interkingdom interactions were positive, suggesting that the dominant relationships between bacterial and fungal communities was symbiotic (Figure 6D). Moreover, the RAs of ASVs that participated in both interkingdom (fungi–bacteria) and intrakingdom (fungi–fungi) interactions were greater in the rhizosphere of resistant tobacco (10.60%) than in that of susceptible tobacco (8.15%), and most of the ASVs were classified as Mortierella, Chaetomium, or Aspergillus. This finding underscores the potential critical role of fungi in symbioticcross-kingdom interactions within the tobacco rhizosphere, but the underlying mechanism of fungal communities, such as Mortierella, Chaetomium, and Aspergillus, as pivotal for promoting tobacco resistance to RKNs needs further investigation.
To further identify potential keystone taxa associated with M. incongnita J2 density and elucidate their interaction patterns, a co-occurrence network incorporating 192 representative bacterial genera, 19 fungal genera, and M. incongnita J2 was established. The analysis revealed that the bacterial–fungal-J2 co-occurrence network was highly modular (modularity = 0.79), and the average degree of the network was 2.7, additionally, M. incongnita J2 was positioned in Modular 1 (M1, Figure 6E). To explore key genera responsive to the disease indices of J2 and RKNs, a comparative analysis was conducted between discriminative genera identified by LEfSe (distinguishing between the rhizosphere and bulk soil) and potential keystone genera within M1. Moreover, M1 was presented separately, and discriminative genera were highlighted to explore those closely related to M. incongnita J2 (degree = 5, Figure 6F). The results indicated that Microbacterium, which was significantly enriched in the bulk soil, negatively regulated M. incongnita J2 in the network (Spearman’s r = −0.89, p < 0.01). Conversely, Mesorhizobium, Sphingopyxis, and Sporichthya, which were significantly enriched in the tobacco rhizosphere, had positive interactions with M. incongnita J2 density. In addition, Sporichthya was identified as a module hub in M1 (Zi ≥ 2.50, Pi < 0.62, degree = 11), playing a crucial keystone role in maintaining the structure and function of the module. Moreover, Nordella (degree = 11), Terrimonas (degree = 6), Nannocystis (degree = 4), MM2 (Methylophilaceae, degree = 1) and Micromonospora (degree = 4) contributed greatly to the stability and activity of M1 by widely and positively interacting with other nodes, including M. incongnita J2.
To clarify the differences in the distributions of key microbial taxa between resistant (ZC208, YY87, and YY85) and susceptible (HD) tobacco rhizospheres, both the discriminative taxa identified by LEfSe and key taxa associated with M. incongnita J2 were examined. The findings showed that the RA of Microbacterium was 34.49% greater in the rhizosphere of resistant tobacco than in that of susceptible tobacco (p < 0.05, Figure 6G). Additionally, Nordella and Terrimonas were substantially enriched in the rhizosphere of the susceptible tobacco plants by 35–41% compared with those in the rhizosphere of the resistant plants (p < 0.05, Figure 6G). Conversely, the RA of Sporichthya was only slightly greater in the rhizosphere of susceptible tobacco than in that of resistant tobacco (p > 0.05).
The rhizosphere microbiome plays a critical role in determining plant health (Dastogeer et al., 2022; Yang et al., 2023), and the composition and function of microbial communities in the same soil differ between the bulk soil and rhizosphere soil and even among plant genotypes within a species (Lee et al., 2021; Oyserman et al., 2022). This study characterized the significant variations in microbial communities between the bulk soil and the rhizosphere soil of susceptible and resistant tobacco plants. Specifically, the richness and diversity of bacteria were greater in the rhizosphere soil than in the bulk soil, and the opposite trend was observed for fungi (Table 1). The results partly support that plants may actively recruit and shape their rhizosphere microbial communities, and this process is closely linked to their genotype (Berendsen et al., 2012). Similarly, Zhou et al. (2019) confirmed that noninfested rhizosphere soils from four different plants had greater microbial diversity than soils infested with RKNs, and each kind of plant hosted unique microbial communities. Furthermore, our β-NTI results revealed that deterministic processes predominantly govern the assembly of fungal communities in the rhizosphere, and tobacco genotype was one of the deterministic factors (Figure 6A). Additionally, our examination of the co-occurrence network of bacterial–fungal communities indicated the pivotal role of fungi, such as Mortierella (Borrell et al., 2017), Chaetomium (Spinelli et al., 2022) and Aspergillus (Li et al., 2023), in cross-kingdom interactions in the tobacco rhizosphere, and these fungi have been reported to play key roles in soil microecology. In conclusion, the fungal community in the tobacco rhizosphere appears to be more genotype-determined and has a substantial impact on the cross-kingdom interactions of bacterial–fungal communities. Additionally, the area of the four tobacco varieties studied was more than 80% of all tobacco-planting area in Liangshan Yi Autonomous Prefecture, Sichuan Province, and the result of variation in resistance to RKN among the four tobacco varieties were confirmed by two consecutive years of field experiment, but the generalizability of the findings in this study may be limited because of the representativeness and balance of the experimental design, since there were only one susceptible and three resistant tobacco varieties were tested. Therefore, the cross-kingdom interactions of microbial communities in the tobacco rhizosphere require further elucidation and a more balanced selection of susceptible and resistant varieties in future studies should be considered.
In this study, Microbacterium was suggested as a potential microbial antagonist against RKN and showed a significant negative correlation with M. incongnita J2, moreover, it enhanced presence in RKN-resistant tobacco genotypes compared to RKN-susceptible tobacco genotypes (Figure 6G). Research has demonstrated that Microbacterium spp. can notably improve plant resistance and suppress the penetration and development of plant-parasitic nematodes by inducing plant systemic resistance (Sahu et al., 2020; Zhao J. et al., 2019) and boosting secondary metabolites in roots (Gupta et al., 2017). Our findings further revealed that Microbacterium is a promising microbial antagonist for future RKN management strategies. To illustrate the interaction patterns between the fungal–bacterial community and Meloidogyne incongnita J2 in the tobacco rhizosphere, a model based on a structural equation model (SEM) was proposed to match the hypothetical models (Figure 7). According to this model, bacterial richness had a direct and significant positive effect on Meloidogyne incongnita J2 (p < 0.001), while Microbacterium had direct and significant negative effects on Meloidogyne incongnita J2 (p < 0.001). Additionally, fungal richness had direct and positive effects on bacterial richness (p > 0.05), bacterial diversity (p > 0.05) and the abundance of Microbacterium (p > 0.05). This model roughly revealed the fungi–bacteria–nematode interaction patterns in the tobacco rhizosphere for the first time, in conclusion, the bacterial community and fungal community had different effects on the interaction of RKN, specifically, the effect of the bacterial community on RKN was direct; in contrast, the effect of the fungal community on RKN was indirect which was worked through the regulation of the bacterial community. This finding confirmed both the results of linear regression analysis between the alpha diversity of microbial communities and the density of M. incongnita J2 (Figure 2) and the results of the occurrence network of representative ASVs in this study (Figure 6). According to these interaction patterns, the enhanced presence of Microbacterium in the rhizosphere of resistant tobacco possibly contributed to root resistance against RKN. Since this study identified potential microbial antagonists Microbacterium relying on sequencing data, the direct effect of Microbacterium on RKN suppression has not been confirmed, so the supplementary experiments demonstrating the direct effect of Microbacterium on RKN in the future would make this conclusion more robust and applicable to real-world scenarios. Meanwhile, whether the microbial interactions observed in this study were specific to tobacco plants or if they could be applied to other crops affected by RKN could not be evaluated and need to be further explored because of lack of similar researches.
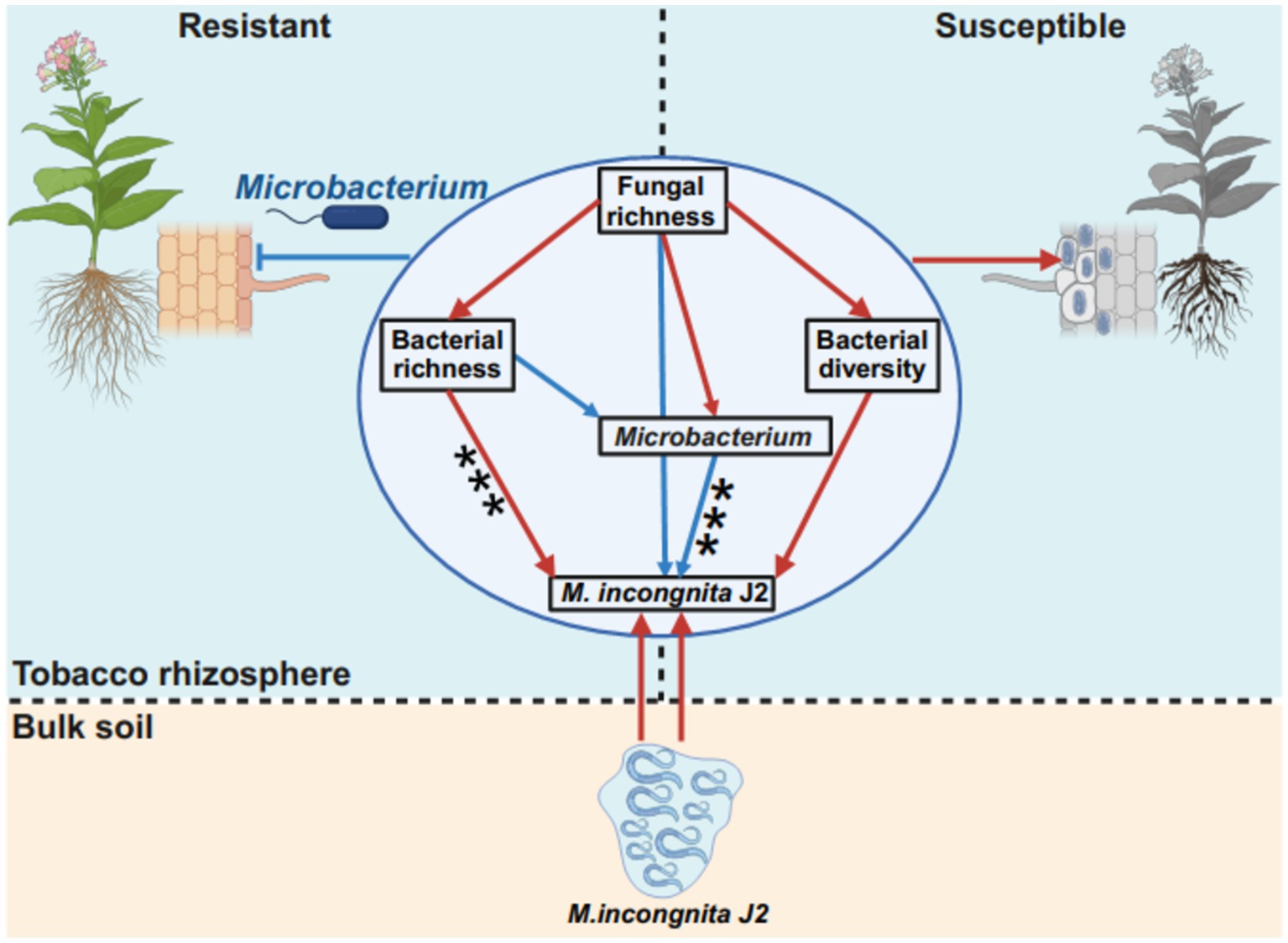
Figure 7. Interaction patterns of the fungal–bacterial community and Meloidogyne. Differences in interaction patterns between M. incongnita J2 and the fungal–bacterial community in the rhizosphere of resistant and susceptible tobacco plants are shown. The structural equation model (SEM) in the circle illustrates the effect of fungal richness on bacterial richness, bacterial diversity, the RA of Microbacterium and the density of M. incongnita J2. The low chi-square (χ2 = 0.494), nonsignificant probability level (p = 0.781), high goodness-of-fit index (GFI = 0.988), low Akaike information criterion (AIC = 26.494), and low root-mean-square error of approximation (RMSEA = 0.000) indicated that our data matched the hypothetical models. The red solid arrows indicate the promoting effects, and the blue solid arrows indicate the suppressing effects.
Plant roots actively select specific microbes to thrive in the rhizosphere, which can enhance nutrient uptake and promote plant growth (Berendsen et al., 2012). In our study, various microbes that prospered in the tobacco rhizosphere (Figure 6F), such as Nordella (Liu et al., 2023), Terrimonas (Ou et al., 2019; Wei et al., 2019; Zhang et al., 2023), Mesorhizobium (Hassen et al., 2020; Singh et al., 2021), Sphingopyxis (Verma et al., 2020), Sporichthya (Zhang et al., 2019), Nannocystis (Hüpeden et al., 2020; Sun et al., 2022), and Micromonospora (Martínez-Hidalgo et al., 2014; Martínez-Hidalgo et al., 2015) were identified as core members of the plant rhizosphere and endogenous microbiome. These microbes are typically involved in improving nutrient availability and promoting plant growth (Berendsen et al., 2012). As a result of the growth promotion by these special microbes, the quantity of root exudates released into the rhizosphere increases with plant growth (Bais et al., 2006), potentially explaining why microbes flourishing in the tobacco rhizosphere. Remarkably, the RAs of Nordella and Terrimonas were significantly greater in the rhizosphere of susceptible tobacco than in that of resistant tobacco (Figure 6G), suggesting a possible positive influence on RKNs. However, the specific roles of these microbes in the rhizosphere and their interactions with RKNs require further exploration to elucidate the underlying microbial drivers of RKN disease.
Second-stage juveniles (J2s) of RKNs aggregate around host root surfaces (Castagnone-Sereno et al., 2015) and subsequently invade plant roots and cause root knots (Forghani and Hajihassani, 2020). During the progression of RKN disease, the rhizosphere serves as a crucial zone for RKNs to locate and invade plant roots, hence, the density of RKNs in the rhizosphere is one of the most important factors related to disease (Topalovic and Geisen, 2023). Our findings verified that J2s significantly aggregated in the rhizosphere of tobacco plants grown in bulk soil. The density of J2s in the rhizosphere soil was 195% greater than that in the bulk soil (Figure 1). During the progression of RKNs aggregating around host root, root exudates may act as regulatory signals influencing the interactions between plant and soil microorganisms in the rhizosphere (Jing et al., 2023), and researchers had confirmed that some kinds of exudates, such as flavonoids, glucosinolates, terpenoids, and alkaloids, can stimulate and attract RKNs to the plant rhizosphere (Rasmann and Hiltpold, 2022). However, the underlying mechanism by which tobacco root exudates attract RKNs is still unknown. Remarkably, the densities of J2s in resistant (89 ~ 276 per 100 g dry soil) and susceptible (152 ~ 301 per 100 g dry soil) tobacco were similar (Figure 1F); however, the disease index was much greater in susceptible tobacco than in resistant tobacco (Figure 1E). This suggests that in addition to the density of RKNs in the rhizosphere, factors such as RKNs behavior, including RKNs activity and virulence, as well as the intrinsic characteristics of root tissues, are involved (Abad et al., 2003). The inherent characteristics of tobacco roots also contribute to tobacco RKN disease, which needs to be further clarified in the future.
Taken together, our study revealed that the fungal-bacterial community in the rhizosphere of tobacco are genotype-specific and may affect the microbe-associated resistance to root-knot nematodes (RKNs), and we explored the crucial roles of the fungal community in facilitating the cross-kingdom and symbiotic fungal–bacterial interactions to suppress RKNs. Moreover, Microbacterium is suggested as a potential and novel microbial antagonist against RKN based on its enhanced presence in RKN-resistant tobacco genotypes and the significant negative correlation with RKNs. Notably, the richness of fungal community enhanced tobacco’s microbe-associated resistance to RKN through the positive regulation of the richness and diversity of bacterial community and the relative abundance of Microbacterium. Our finding enhances the understanding of the interactions between RKN and soil microbiomes, provides novel insight for identifying the underlying microbial antagonists for PPN management, and highlights pathway to develop RKN biocontrol products by integrating benefit microbial communities.
Raw sequencing data have been deposited into the NCBI Sequence Read Archive (SRA) database under accession number PRJNA1069245 for 16S rRNA, and PRJNA1078088 for ITS.
The manuscript presents research on animals that do not require ethical approval for their study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
QJ: Conceptualization, Data curation, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. YoW: Conceptualization, Project administration, Resources, Supervision, Writing – review & editing. JY: Methodology, Project administration, Supervision, Writing – review & editing. JW: Investigation, Validation, Visualization, Writing – original draft. SG: Conceptualization, Resources, Supervision, Writing – review & editing. DL: Resources, Supervision, Writing – review & editing. XY: Methodology, Project administration, Resources, Supervision, Writing – review & editing. LJ: Investigation, Methodology, Project administration, Resources, Supervision, Writing – review & editing. GL: Project administration, Resources, Supervision, Writing – review & editing. DX: Investigation, Visualization, Writing – review & editing. SC: Resources, Supervision, Writing – review & editing. YuW: Investigation, Writing – review & editing. WD: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Key Project from Sichuan Company of China National Tobacco Corporation (202051340024416 and SCYC202010).
JY, SG, and XY were employed by Sichuan Branch of China Tobacco Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1415700/full#supplementary-material
Abad, P., Favery, B., Rosso, M. N., and Castagnone-Sereno, P. (2003). Root-knot nematode parasitism and host response: molecular basis of a sophisticated interaction. Molecular Plant Pathology 4, 217–224. doi: 10.1046/j.1364-3703.2003.00170.x
Adam, M., Westphal, A., Hallmann, J., and Heuer, H. (2014). Specific microbial attachment to root knot nematodes in suppressive soil. Appl. Environ. Microbiol. 80, 2679–2686. doi: 10.1128/AEM.03905-13
Afzal, I., Shinwari, Z. K., Sikandar, S., and Shahzad, S. (2019). Plant beneficial endophytic bacteria: mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 221, 36–49. doi: 10.1016/j.micres.2019.02.001
Ali, A., Elrys, A. S., Liu, L. L., Iqbal, M., Zhao, J., Huang, X. Q., et al. (2022). Cover plants-mediated suppression of fusarium wilt and root-knot incidence of cucumber is associated with the changes of rhizosphere fungal microbiome structure-under plastic shed system of North China. Front. Microbiol. 13:697815. doi: 10.3389/fmicb.2022.697815
Antil, S., Kumar, R., Pathak, D. V., and Kumari, A. (2023). Recent advances in utilizing bacteria as biocontrol agents against plant parasitic nematodes emphasizing Meloidogyne spp. Biol. Control 183:105244. doi: 10.1016/j.biocontrol.2023.105244
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The role of root exudates in rhizosphere interations with plants and other organisms. Annual Review of Plant Biology 57, 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Banerjee, S., and van der Heijden, M. G. A. (2023). Soil microbiomes and one health. Nat. Rev. Microbiol. 21, 6–20. doi: 10.1038/s41579-022-00779-w
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berg, G., Rybakova, D., Fischer, D., Cernava, T., Vergès, M. C. C., Charles, T., et al. (2020). Microbiome definition re-visited: old concepts and new challenges. Microbiome 8:103. doi: 10.1186/s40168-020-00875-0
Borrell, A. N., Shi, Y., Gan, Y., Bainard, L. D., Germida, J. J., and Hamel, C. (2017). Fungal diversity associated with pulses and its influence on the subsequent wheat crop in the Canadian prairies. Plant Soil 414, 13–31. doi: 10.1007/s11104-016-3075-y
Bui, H. X., Hadi, B. A. R., Oliva, R., and Schroeder, N. E. (2020). Beneficial bacterial volatile compounds for the control of root-knot nematode and bacterial leaf blight on rice. Crop Prot. 135:104792. doi: 10.1016/j.cropro.2019.04.016
Cao, Y., Lu, N., Yang, D., Mo, M., Zhang, K.-Q., Li, C., et al. (2023). Root-knot nematode infections and soil characteristics significantly affected microbial community composition and assembly of tobacco soil microbiota: a large-scale comparison in tobacco-growing areas. Front. Microbiol. 14:1282609. doi: 10.3389/fmicb.2023.1282609
Castagnone-Sereno, P., Mulet, K., and Iachia, C. (2015). Tracking changes in life-history traits related to unnecessary virulence in a plant-parasitic nematode. Ecol. Evol. 5, 3677–3686. doi: 10.1002/ece3.1643
Dastogeer, K. M. G., Yasuda, M., and Okazaki, S. (2022). Microbiome and pathobiome analyses reveal changes in community structure by foliar pathogen infection in rice. Front. Microbiol. 13:949152. doi: 10.3389/fmicb.2022.949152
Du, J. F., Gao, Q. X., Ji, C., Song, X., Liu, Y., Li, H. Y., et al. (2022). Bacillus licheniformis JF-22 to control Meloidogyne incognita and its effect on tomato rhizosphere microbial community. Front. Microbiol. 13:863341. doi: 10.3389/fmicb.2022.863341
Dyer, D. R., Newman, M., and Lawrence, K. S. (2022). Diversity and temporal distribution of fusarium oxysporum f. sp. vasinfectum races and genotypes as influenced by Gossypium cultivar. Front. Fungal Biol. 3:1022761. doi: 10.3389/ffunb.2022.1022761
Elhadyl, A., Giné, A., Topalovic, O., Jacquiod, S., Sorensen, S. J., Sorribas, F. J., et al. (2017). Microbiomes associated with infective stages of root-knot and lesion nematodes in soil. PLoS One 12:e0177145. doi: 10.1371/journal.pone.0177145
El-Sappah, A. H., Islam, M. M., El-awady, H. H., Yan, S., Qi, S. M., Liu, J. Y., et al. (2019). Tomato natural resistance genes in controlling the root-knot nematode. Genes 10:925. doi: 10.3390/genes10110925
Fadiji, A. E., Yadav, A. N., Santoyo, G., and Babalola, O. O. (2023). Understanding the plant-microbe interactions in environments exposed to abiotic stresses: an overview. Microbiol. Res. 271:127368. doi: 10.1016/j.micres.2023.127368
Fan, M. M., Li, J. Z., Dai, K., Liu, M. J., Zhou, W. B., Zhang, L. M., et al. (2023). Root-knot density as a new index can quantitatively diagnose the damage of root nematodes to plant growth. Agronomy-Basel 13:136. doi: 10.3390/agronomy13010136
Forghani, F., and Hajihassani, A. (2020). Recent advances in the development of environmentally benign treatments to control root-knot nematodes. Front. Plant Sci. 11:1125. doi: 10.3389/fpls.2020.01125
Gupta, R., Singh, A., Ajayakumar, P. V., and Pandey, R. (2017). Chitinolytic microbes confer Meloidogyne incognita resistance and augment secondary metabolites in Bacopa monnieri (L.) Pennell. Arch. Phytopathol. Plant Protect. 50, 178–196. doi: 10.1080/03235408.2017.1285522
Hassen, A. I., Swanevelder, Z. H., Bopape, F. L., and Lamprecht, S. C. (2020). Draft genome sequence of Mesorhizobium sp. strain SARCC-RB16n, an effective nodulating and nitrogen-fixing symbiont of rooibos Aspalathus linearis (Burm. F.) in South Africa. Microbiol. Resour. Announc. 9, e01187–e01119. doi: 10.1128/MRA.01187-19
Hüpeden, J., Wemheuer, B., Indenbirken, D., Schulzd, C., and Spieck, E. (2020). Taxonomic and functional profiling of nitrifying biofilms in freshwater, brackish and marine RAS biofilters. Aquac. Eng. 90:102094. doi: 10.1016/j.aquaeng.2020.102094
Jiang, Q. P., Jiang, L. Q., Gong, J., Yu, J. M., Yang, C., Liu, D. Y., et al. (2021). Exploring the key factors causing the outbreak of tobacco root-knot nematodes in Liangshan prefecture of Sichuan Province. Acta Tabacaria Sin. 27, 89–98. doi: 10.16472/j.chinatobacco.2021.067
Jing, H., Wang, H. L., Wang, G. L., Liu, G. B., and Cheng, Y. (2023). The mechanism effects of root exudate on microbial community of rhizosphere soil of tree, shrub, and grass in forest ecosystem under N deposition. ISME Commun. 3:120. doi: 10.1038/s43705-023-00322-9
Jones, J. T., Haegeman, A., Danchin, E. G. J., Gaur, H. S., Helder, J., Jones, M. G. K., et al. (2013). Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 14, 946–961. doi: 10.1111/mpp.12057
Khan, R. A. A., Najeeb, S., Hussain, S., Xie, B. Y., and Li, Y. (2020). Bioactive secondary metabolites from Trichoderma spp. against phytopathogenic fungi. Microorganisms 8:817. doi: 10.3390/microorganisms8060817
Khan, M., and Siddiqui, Z. A. (2017). Effects of fly ash amendments, Ralstonia solanacearum, Meloidogyne incognita and Phomopsis vexans on the growth of Solanum melongena. Acta Phytopathol. Entomol. Hungar. 52, 145–156. doi: 10.1556/038.52.2017.017
Kiriga, A. W., Haukeland, S., Kariuki, G. M., Coyne, D. L., and Beek, N. V. (2018). Effect of Trichoderma spp. and Purpureocillium lilacinum on Meloidogyne javanica in commercial pineapple production in Kenya. Biol. Control 119, 27–32. doi: 10.1016/j.biocontrol.2018.01.005
Kyndt, T., Yimer, H., Haeck, A., Singh, R., De Vleesschauwer, D., Denil, S., et al. (2017). Below-ground attack by the root knot nematode Meloidogyne graminicola predisposes rice to blast disease. Phytopathology 107:119. doi: 10.1094/MPMI-11-16-0225-R
Lee, S. M., Kong, H. G., Song, G. C., and Ryu, C. M. (2021). Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease. ISME J. 15, 330–347. doi: 10.1038/s41396-020-00785-x
Li, X. N., Zhang, T., Xue, Y., Xu, X., Cui, X. Y., and Fu, J. M. (2023). Aspergillus aculeatus enhances nutrient uptake and forage quality in bermudagrass by increasing phosphorus and potassium availability. Front. Plant Sci. 14:1165567. doi: 10.3389/fpls.2023.1165567
Liu, G. Y., Lin, X., Xu, S. Y., Liu, G., Liu, F., and Mu, W. (2020). Screening, identification and application of soil bacteria with nematicidal activity against root-knot nematode (Meloidogyne incognita) on tomato. Pest Manag. Sci. 76, 2217–2224. doi: 10.1002/ps.5759
Liu, J. T., Xu, H. Y., Wang, Z. C., Liu, J., and Gong, X. (2023). Core endophytic bacteria and their roles in the coralloid roots of cultivated Cycas revoluta (Cycadaceae). Microorganisms 11:2364. doi: 10.3390/microorganisms11092364
Liu, X., Zhang, S., Jiang, Q., Bai, Y., Shen, G., Li, S., et al. (2016). Using community analysis to explore bacterial indicators for disease suppression of tobacco bacterial wilt. Sci. Rep. 6:36773. doi: 10.1038/srep36773
Lu, H., Wang, X., Zhang, K. Q., Xu, Y. Y., Zhou, L., and Li, G. H. (2014). Identification and nematicidal activity of bacteria isolated from cow dung. Ann. Microbiol. 64, 407–411. doi: 10.1007/s13213-013-0660-7
Luo, L., Zhao, C. Z., Wang, E. T., Raza, A., and Yin, C. Y. (2022). Bacillus amyloliquefaciens as an excellent agent for biofertilizer and biocontrol in agriculture: an overview for its mechanisms. Microbiol. Res. 259:127016. doi: 10.1016/j.micres.2022.127016
Martínez-Hidalgo, P., Galindo-Villardón, P., Igual, J. M., and Martínez-Molina, E. (2014). Micromonospora from nitrogen fixing nodules of alfalfa (Medicago sativa L.). A new promising plant probiotic Bacteria. Sci. Rep. 4:6389. doi: 10.1038/srep06389
Martínez-Hidalgo, P., García, J. M., and Pozo, M. J. (2015). Induced systemic resistance against Botrytis cinerea by Micromonospora strains isolated from root nodules. Front. Microbiol. 6:922. doi: 10.3389/fmicb.2015.00922
Martínez-Medina, A., Fernandez, I., Lok, G. B., Pozo, M. J., Pieterse, C. M. J., and Van Wees, S. C. M. (2017). Shifting from priming of salicylic acid- to jasmonic acid-regulated defences by Trichoderma protects tomato against the root knot nematode Meloidogyne incognita. New Phytol. 213, 1363–1377. doi: 10.1111/nph.14251
Ou, Y. N., Penton, C. R., Geisen, S., Shen, Z. Z., Sun, Y. F., Lv, N. N., et al. (2019). Deciphering underlying drivers of disease suppressiveness against pathogenic fusarium oxysporum. Front. Microbiol. 10:2535. doi: 10.3389/fmicb.2019.02535
Oyserman, B. O., Flores, S. S., Griffioen, T., Pan, X. Y., van der Wijk, E., Pronk, L., et al. (2022). Disentangling the genetic basis of rhizosphere microbiome assembly in tomato. Nat. Commun. 13:3228. doi: 10.1038/s41467-022-30849-9
Rasmann, S., and Hiltpold, I. (2022). Root exudation of specialized molecules for plant-environment interaction. Chimia 76, 922–927. doi: 10.2533/chimia.2022.922
Sahu, K. P., Kumar, A., Patel, A., Kumar, M., Gopalakrishnan, S., Prakash, G., et al. (2020). Rice blast lesions: an unexplored Phyllosphere microhabitat for novel antagonistic bacterial species against Magnaporthe oryzae. Microb. Ecol. 81, 731–745. doi: 10.1007/s00248-020-01617-3
Sasse, J., Martinoia, E., and Northen, T. (2018). Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41. doi: 10.1016/j.tplants.2017.09.003
Segata, N., Abubucker, S., Goll, J., Schubert, A. M., Izard, J., Cantarel, B. L., et al. (2011). Microbial community function and biomarker discovery in the human microbiome. Genome Biol. 12, 24–25. doi: 10.1186/1465-6906-12-S1-P47
Singh, B. K., Liu, H. W., and Trivedi, P. (2020). Eco-holobiont: a new concept to identify drivers of host-associated microorganisms. Environ. Microbiol. 22, 564–567. doi: 10.1111/1462-2920.14900
Singh, Z., Singh, G., Aggarwal, N., Virk, H. K., and Sharma, P. (2021). Symbiotic efficiency Vis-a-Vis chickpea performance as affected by seed inoculation with Mesorhizobium, phosphorus-solubilizing bacteria, and phosphorus application. J. Plant Nutr. 45, 246–258. doi: 10.1080/01904167.2021.1943435
Spinelli, V., Brasili, E., Sciubba, F., Ceci, A., Giampaoli, O., Miccheli, A., et al. (2022). Biostimulant effects of Chaetomium globosum and Minimedusa polyspora culture filtrates on Cichorium intybus plant: growth performance and metabolomic traits. Front. Plant Sci. 13:879076. doi: 10.3389/fpls.2022.879076
Sun, Y., Teng, Y., Li, R., Wang, X., and Zhao, L. (2023). Microbiome resistance mediates stimulation of reduced graphene oxide to simultaneous abatement of 2,2′,4,4′,5-pentabromodiphenyl ether and 3,4-dichloroaniline in paddy soils. J. Hazard. Mater. 465:133121. doi: 10.1016/j.jhazmat.2023.133121
Sun, R. X., You, X. W., Cheng, Y. D., Gan, D. P., Suo, F. Y., Wang, B., et al. (2022). Response of microbial compositions and interactions to biochar amendment in the peanut-planted soil of the Yellow River Delta, China. Front. Environ. Sci. 10:924358. doi: 10.3389/fenvs.2022.924358
Sun, X. W., Zhang, R., Ding, M. J., Liu, Y. X., and Li, L. (2021). Biocontrol of the root-knot nematode Meloidogyne incognita by a nematicidal bacterium Pseudomonas simiae MB751 with cyclic dipeptide. Pest Manag. Sci. 77, 4365–4374. doi: 10.1002/ps.6470
Topalovic, O., and Geisen, S. (2023). Nematodes as suppressors and facilitators of plant performance. New Phytol. 238, 2305–2312. doi: 10.1111/nph.18925
Verma, H., Dhingra, G. G., Sharma, M., Gupta, V., Negi, R. K., Singh, Y., et al. (2020). Comparative genomics of Sphingopyxis spp. unravelled functional attributes. Genomics 112, 1956–1969. doi: 10.1016/j.ygeno.2019.11.008
Wei, Z., Gu, Y., Friman, V. P., Kowalchuk, G. A., Xu, Y. C., Shen, Q. R., et al. (2019). Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 5:eaaw0759. doi: 10.1126/sciadv.aaw0759
William, M. L., Rodrigo, M., Raaijmakers, J. M., and Mui, T. S. (2018). Breeding for soil-borne pathogen resistance impacts active rhizosphere microbiome of common bean. ISME J. 12:5. doi: 10.1038/s41396-018-0234-6
Xiang, N., Lawrence, K. S., and Donald, P. A. (2018). Biological control potential of plant growth-promoting rhizobacteria suppression of Meloidogyne incognita on cotton and Heterodera glycines on soybean: a review. J. Phytopathol. 166, 449–458. doi: 10.1111/jph.12712
Xu, S. X., Tian, P., Jiang, Z. M., Chen, X. X., Li, B., Sun, J., et al. (2023). Transcriptome analysis of two tobacco varieties with contrast resistance to Meloidogyne incognita in response to PVY MSNR infection. Front. Plant Sci. 14:1213494. doi: 10.3389/fpls.2023.1213494
Yang, K., Fu, R., Feng, H., Jiang, G., Finkel, O., Sun, T., et al. (2023). RIN enhances plant disease resistance via root exudate-mediated assembly of disease-suppressive rhizosphere microbiota. Mol. Plant 16, 1379–1395. doi: 10.1016/j.molp.2023.08.004
Zhang, Y., Du, H. M., Chen, Y. L., Wei, H. H., Dai, Q. G., Liu, J. M., et al. (2023). Influence of biochar-based urea substituting urea on rice yield, bacterial commnity and nitrogen cycling in paddy fields. J. Sci. Food Agric. 103, 2794–2805. doi: 10.1002/jsfa.12333
Zhang, S., Jiang, Q., Liu, X., Liu, L., and Ding, W. (2020). Plant growth promoting rhizobacteria alleviate aluminum toxicity and ginger bacterial wilt in acidic continuous cropping soil. Front. Microbiol. 11:569512. doi: 10.3389/fmicb.2020.569512
Zhang, Y., Li, S. S., Li, H. X., Wang, R. R., Zhang, K. Q., and Xu, J. P. (2020). Fungi-nematode interactions: diversity, ecology, and biocontrol prospects in agriculture. J. Fungi 6:206. doi: 10.3390/jof6040206
Zhang, S., Liu, X., Jiang, Q., Shen, G., and Ding, W. (2017). Legacy effects of continuous chloropicrin-fumigation for 3-years on soil microbial community composition and metabolic activity. AMB Express 7:178. doi: 10.1186/s13568-017-0475-1
Zhang, L. N., Wang, D. C., Hu, Q., Dai, X. Q., Xie, Y. S., Li, Q., et al. (2019). Consortium of plant growth-promoting rhizobacteria strains suppresses sweet pepper disease by altering the rhizosphere microbiota. Front. Microbiol. 10:1668. doi: 10.3389/fmicb.2019.01668
Zhao, Z. B., He, J. Z., Geisen, S., Han, L. L., Wang, J. T., Shen, J. P., et al. (2019). Protist communities are more sensitive to nitrogen fertilization than other microorganisms in diverse agricultural soils. Microbiome 7:33. doi: 10.1186/s40168-019-0647-0
Zhao, J., Liu, D., Wang, Y. Y., Zhu, X. F., Xuan, Y. H., Liu, X. Y., et al. (2019). Biocontrol potential of Microbacterium maritypicum Sneb159 against Heterodera glycines. Pest Manag. Sci. 75, 3381–3391. doi: 10.1002/ps.5546
Zhou, D. M., Feng, H., Schuelke, T., De Santiago, A., Zhang, Q. M., Zhang, J. F., et al. (2019). Rhizosphere microbiomes from root knot nematode non-infested plants suppress nematode infection. Microb. Ecol. 78, 470–481. doi: 10.1007/s00248-019-01319-5
Keywords: root-knot nematodes, Microbacterium , fungal-bacterial interaction, tobacco resistance, co-occurrence network
Citation: Jiang Q, Wang Y, Yu J, Wang J, Guo S, Liu D, Yu X, Jiang L, Long G, Xi D, Chen S, Wang Y and Ding W (2024) Using fungal–bacterial community analysis to explore potential microbiomes to manage Meloidogyne incongnita. Front. Microbiol. 15:1415700. doi: 10.3389/fmicb.2024.1415700
Received: 11 April 2024; Accepted: 10 October 2024;
Published: 22 October 2024.
Edited by:
Jinliang Liu, Jilin University, ChinaReviewed by:
Mohamed Mannaa, Pusan National University, Republic of KoreaCopyright © 2024 Jiang, Wang, Yu, Wang, Guo, Liu, Yu, Jiang, Long, Xi, Chen, Wang and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Ding, ZHdpbmc4MThAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.