- 1Medical Department, Sias University, Zhengzhou, Henan, China
- 2Medical Department, Zhengzhou University of Industry Technology, Zhengzhou, Henan, China
- 3Ultrasound Department, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
Background: An escalating body of clinical trials and observational studies hints at a plausible link between gut flora and postpartum depression (PPD). The definitive causal dynamics between these two entities remain shrouded in ambiguity. Therefore, in this study, we employed the two-sample Mendelian randomization approach to ascertain the causal link between gut microbiota and PPD.
Methods: Summary-level GWAS data related to the human gut microbiota were obtained from the international consortium MiBioGen and the Dutch Microbiome Project (species). For PPD, GWAS data were derived from the FinnGen biobank, consisting 57,604 cases and 596,601 controls. The inverse variance weighted method (IVW) as the cornerstone of our analytical approach. Subsequent to this, a comprehensive suite of tests for pleiotropy and heterogeneity were conducted to ensure the reliability and robustness of our findings.
Results: We identified 12 bacterial taxa associated with the risk of PPD. Veillonellaceae, Ruminococcaceae UCG 011, Bifidobacterium adolescentis, Paraprevotella clara, Clostridium leptum, Eubacterium siraeum, Coprococcus catus exhibited an inversely associated with the risk of PPD. Alphaproteobacteria, Roseburia, FamilyXIIIAD3011group, Alistipes onderdonkii, Bilophila wadsworthia showed a positive correlation with the risk of PPD.
Limitations: The GWAS data derived from the MiBioGen consortium, DMP, and FinnGen consortium, may introduce selection bias. Moreover, the data primarily originates from European populations, hence extrapolating these results to diverse populations should be approached with caution. The etiological factors behind PPD remain enigmatic, alluding to the existence of potential undisclosed confounders.
Conclusion: Based on this MR analysis, we found a causal relationship between certain gut microbial communities and PPD. Future clinical studies can further explore the treatment of PPD through the combined use of microorganisms. This not only offers insights into the pathogenesis of PPD but also lays the foundation for utilizing gut microbiota as biotherapeutics in treating neurological disorders.
1 Introduction
Postpartum depression (PPD) represents a grave mental health concern, typically manifesting within a week to several weeks following childbirth. Beyond the evident physical and psychological toll on the mother, PPD has far-reaching implications for the child, potentially hampering their emotional, behavioral, cognitive, and intellectual trajectory in ways that can span their lifetime (Josefsson et al., 2002; Sohr-Preston and Scaramella, 2006; van der Waerden et al., 2017). Globally, it’s estimated that 10–15% of new mothers grapple with PPD, although the prevalence fluctuates across regions (Palumbo et al., 2017). Alarmingly, the prevalence is heightened in developing nations. Countries with the lowest incidence rates of PPD include: Singapore (3%), the Netherlands (8%), the United States (8.4%), and Switzerland (11%). Countries with higher incidence rates include: Chile (38%), South Africa (37%), Turkey (28%), and China (21.4%) (Hahn-Holbrook et al., 2017; Liu et al., 2022).
The human gut is a bustling microcosm, In the case of an average adult weighing 70 kg, the gut microbiota comprises approximately 3.8 × 1013 microorganisms, collectively possessing genomic content that dwarfs our own by over 100-fold (Gill et al., 2006; Sender et al., 2016). The human gut microbiota is composed of four major phyla: Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria (Eckburg et al., 2005). In some cohort studies, compared to healthy controls (HC), patients with Major Depressive Disorder (MDD) exhibit higher abundances of Bacteroidetes, Proteobacteria, and Actinobacteria (Jiang et al., 2015). In patients with PPD, a relatively lower abundance of the Firmicutes phylum is observed (Zhou et al., 2020). A burgeoning body of evidence points to intricate bidirectional communication between our brains and gut, with the microbial denizens and their metabolic by-products playing pivotal roles. This intricate dialogue is called the brain-gut-microbiota axis (Mayer et al., 2014; Cryan et al., 2019). In patients with depression, the homeostasis of the gut microbiota is disrupted, leading to impaired gut function. This, in turn, results in intestinal barrier dysfunction and various inflammatory responses (Liu et al., 2023). These inflammatory responses are correlated with the pathogenesis of depression. The brain-gut axis is bidirectional, with the vagus nerve being the primary regulatory pathway between the brain and the gut microbiota. Some studies suggest that subdiaphragmatic vagotomy may reduce inflammatory responses, thereby alleviating depressive symptoms (Zhang et al., 2020; Pu et al., 2021). Clinically, vagal nerve stimulation (VNS) is an FDA-approved neuromodulation therapy for the treatment of severe treatment-resistant depression (TRD). VNS increases the levels of serotonin (5-hydroxytryptamine, 5-HT) and norepinephrine, and through the anti-inflammatory pathway of the vagus nerve, it reduces systemic inflammatory responses, thereby improving depressive symptoms (Kamel et al., 2022). Short-chain fatty acids (SCFAs) act as intermediary products between the gut microbiota and the brain, primarily including acetate, propionate, and butyrate. SCFA produced by gut microbiota inhibit histone deacetylases and activate G-protein-coupled receptors, influencing systemic physiological responses (Tan et al., 2014). Among these, propionate can reduce levels of γ-aminobutyric acid (GABA), and the indoleamine serotonin (El-Ansary et al., 2012). During pregnancy, alterations in the GABA(A) receptor may increase neuronal excitability in the brain. However, the sharp decrease in neuroactive steroids in the brain after childbirth has a causal relationship with the expression of GABA(A) receptors in the hippocampus. This dynamic could contribute to the development of postpartum neurological and psychiatric disorders (Maguire et al., 2009). Regulating the gut microbiota can improve intestinal barrier function, reduce systemic inflammatory responses, and increase the abundance of beneficial bacteria in the gut. This enhances their ability to produce beneficial metabolites such as SCFAs and GABA, which have positive effects on alleviating and treating postpartum depression (Ramsteijn et al., 2020; Tian et al., 2021).
Mendelian Randomization (MR), rooted in epidemiology, leverages single nucleotide polymorphisms (SNPs) identified in genome-wide association studies (GWAS) as instrumental variables (IVs) to estimate the causal relationship between exposure factors and disease outcomes. These SNPs are genetically associated with the exposure factors but are not influenced by confounding factors and reverse causation. The advantage of MR is that these instrumental variables are predetermined conceptually, greatly minimizing the impact of potential confounders and reverse causality (Ellervik et al., 2019). Recent studies have applied MR to analyze the causal relationship between psychiatric disorders and the gut microbiome. Actinobacteria, Bifidobacterium, Ruminococcus1, and Streptococcaceae have been associated with MDD (Chen et al., 2022), Prevotellaceae with autism spectrum disorder, Betaproteobacteria with bipolar disorder (Ni et al., 2021). Despite the presence of symptomatic and genetic parallels between PPD and MDD, substantial disparities in their etiology and treatment methodologies are evident (Jairaj and Rucker, 2022).
Therefore, this study aims to use the MR method to analyze the potential causal relationships between various levels of gut microbiota and PPD, to explore the differences in the gut microbiota between PPD and MDD, and other psychiatric disorders, thus providing new insights into treatment approaches for PPD.
2 Materials and methods
2.1 Mendelian randomization analysis
MR provides one method for assessing the causal nature of some exposures (Smith and Ebrahim, 2003; Lawlor et al., 2008). In epidemiological research, MR harnesses genetic variants as instrumental variables (IVs) to decipher the causal interplay between exposure and its consequent outcome (Bowden and Holmes, 2019).
To perform an MR study, three main assumptions must be fulfilled (Konig and Greco, 2018). (1) There must be a strong correlation between the genetic variant and the exposure; (2) the genetic instrument is independent of potential confounders of the exposure-outcome association; (3) the genetic variant should be solely associated with the outcome through the exposure, without being influenced by other confounders (Haycock et al., 2016; Konig and Greco, 2018; Hirtz et al., 2022).
2.2 Study design
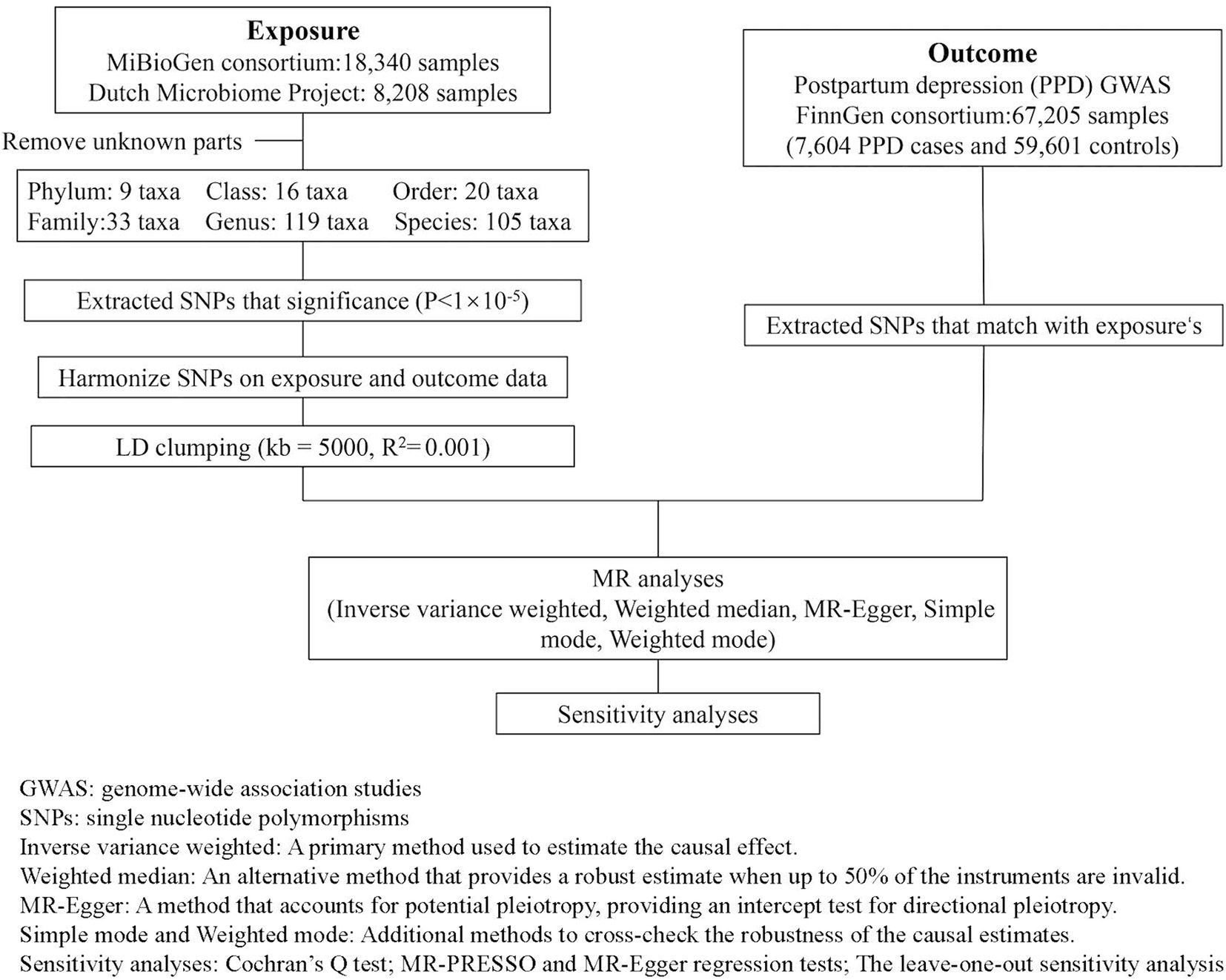
In this research design, gut microbiota SNPs are taken as the exposure variable, and PPD is considered as the outcome variable. Figure 1 illustrates the detailed flowchart.
Figure 1. Overview of the analysis process of the causal relationship between the gut microbiota and PPD through MR analysis.
2.3 Data source
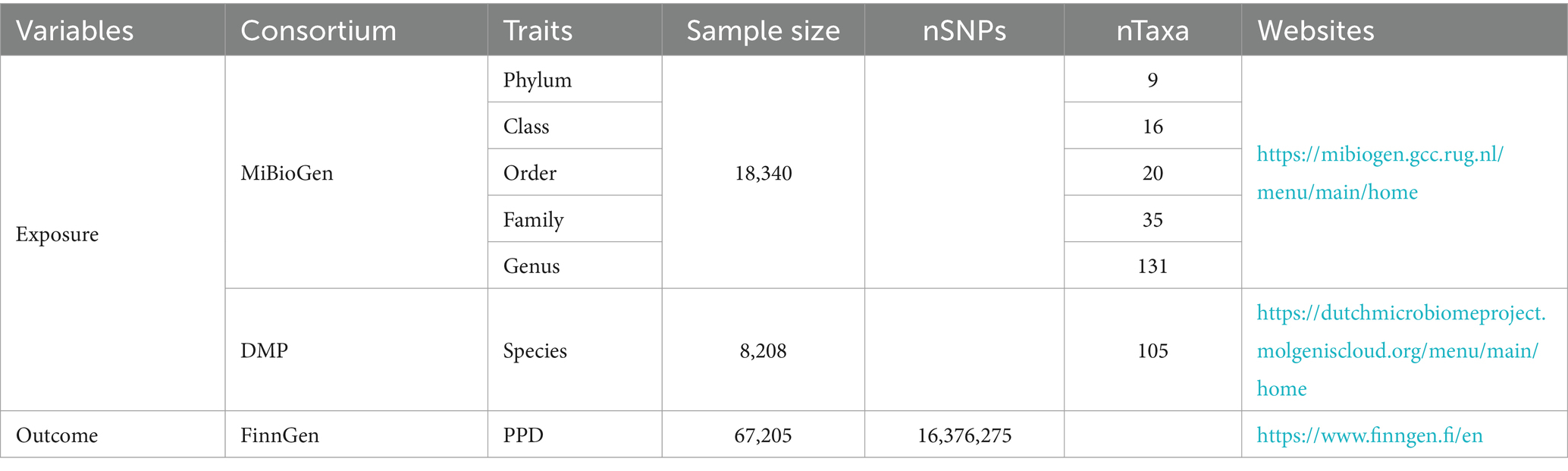
Summary-level GWAS data related to the human gut microbiota from the international consortium MiBioGen (Kurilshikov et al., 2021) and the Dutch Microbiome Project (DMP) (Lopera-Maya et al., 2022). GWAS summary-level data on PPD were obtained from the FinnGen biobank, comprising a total of 67,205 individuals of European ancestry, including 7,604 cases, 59,601 controls. The detailed descriptions of exposure and outcome, including the data source are presented in Table 1.
2.4 Selection of instrumental variable
To ensure the accuracy and reliability of the causal effect between gut microbiota and PPD, we adopt the following quality control methods to select microbiota-related IVs:
1. Based on previous experience and to obtain more comprehensive results, we select IVs with a significance threshold of p < 10−5.
2. To rule out the influence of linkage disequilibrium on the results, we used PLINK software with threshold values set at clump_kb = 5,000 and clump_r2 = 0.001.
3. To ensure that SNPs for exposure and outcome are influenced by the same alleles, we are excluding palindromic SNPs from IVs.
4. The first assumption of MR can be directly validated by calculating the F-statistic. An F-statistic greater than 10 indicates the absence of weak instrument bias (F for a single SNP equals β2/SE2) (Hirtz et al., 2022).
2.5 Mendelian randomization methods
In the current study, all analyses were performed using R software version 4.3.2, utilizing the “Mendelian-Randomization” package. Five popular MR methods were used: Inverse-variance weighted (IVW), MR-Egger, weighted median estimator, simple modal-based estimation, and the weighted mode method. Each statistical method operates under its own set of methodological assumptions. The IVW method estimates the overall causal effect of an exposure on an outcome by performing a weighted average of the effects of multiple genetic variants. This approach assumes the absence of horizontal pleiotropy (Burgess et al., 2013). The MR-Egger assumes the presence of pleiotropy in >50% of SNPs (Bowden et al., 2015). When less than 50% of the information comes from invalid IVs, the weighted median estimator offers a consistent estimate of causal effects (Bowden et al., 2016). While the modal-based estimate (MBE) has a weaker ability to detect causal effects, it relaxes instrumental variable assumptions and requires a smaller sample size (Hartwig et al., 2017). Under different methodological assumptions, IVW results are more reliable in the absence of heterogeneity and pleiotropy. In the presence of heterogeneity but no pleiotropy, the WM method is more reliable. When pleiotropy exists, the MR-Egger method performs better (Burgess and Thompson, 2017).
To satisfy the third assumption of MR and ensure the accuracy and stability of the results, further sensitivity analysis was conducted:
1. Cochran’s Q test is used to quantify the heterogeneity between SNPs. A p-value less than 0.05 indicates significant heterogeneity (Bowden et al., 2019).
2. MR-PRESSO and MR-Egger regression tests are employed to detect potential horizontal pleiotropic effects. If the intercept term is significant, it means the existence of horizontal multiplicity. Compared to MR-Egger, MR-PRESSO offers higher accuracy (Verbanck et al., 2018).
3. The leave-one-out sensitivity analysis is adopted to assess the stability of the results.
3 Results
3.1 Instrumental variables selection
Detailed information for 3,509 IVs is presented in Supplementary Table S1 (p-value < 1e−5). These IVs are categorized into 9 phyla (121 SNPs, range 10–18), 20 orders (275 SNPs, range 5–19), 16 classes (219 SNPs, range 8–22), 33 families (438 SNPs, range 5–21), 119 genera (1,511 SNPs, range 3–26), 105 species (945 SNPs, range 1–17). The F-statistic of each IV was greater than 10 (Supplementary Table S1).
3.2 Two-sample MR
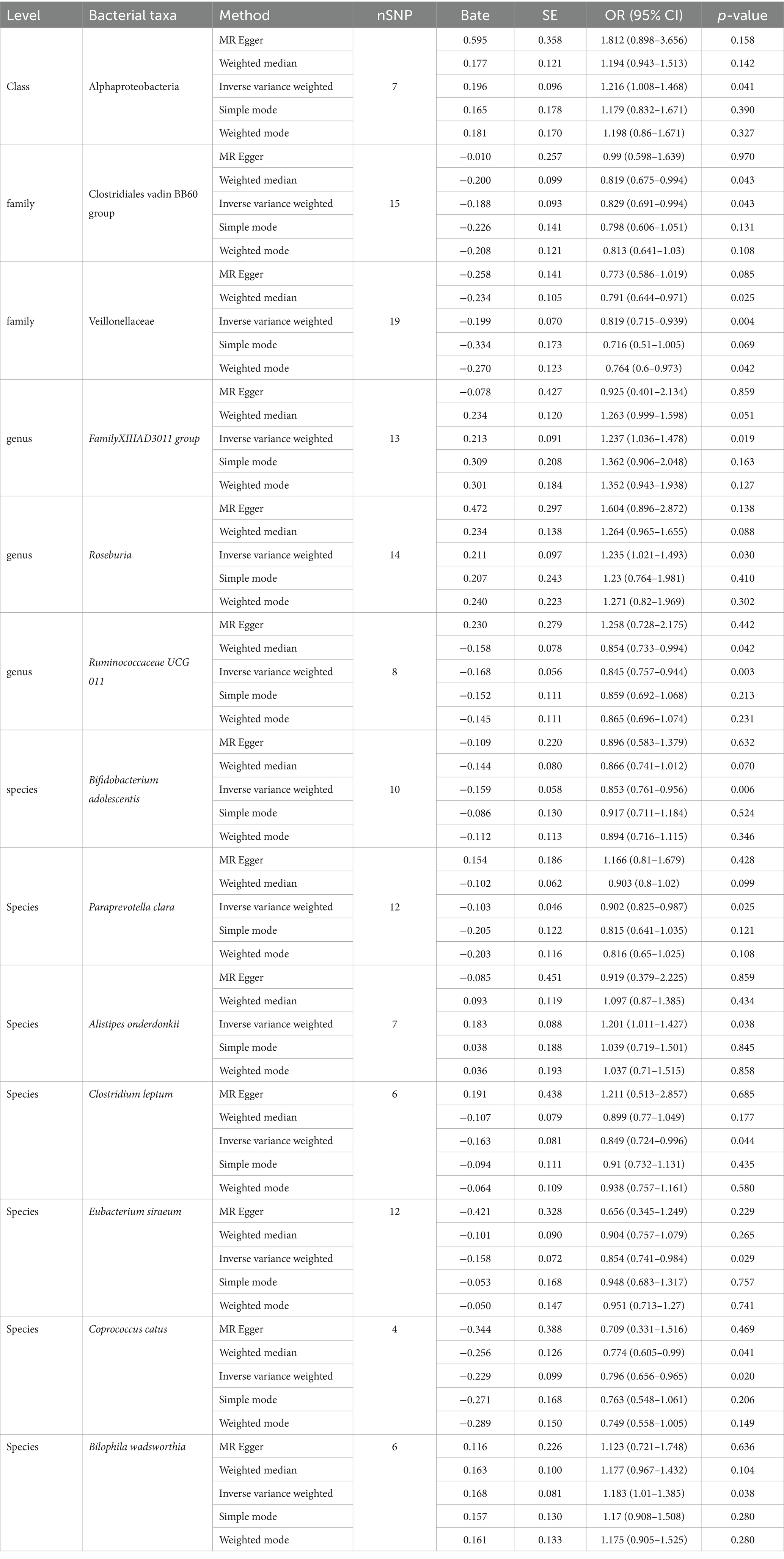
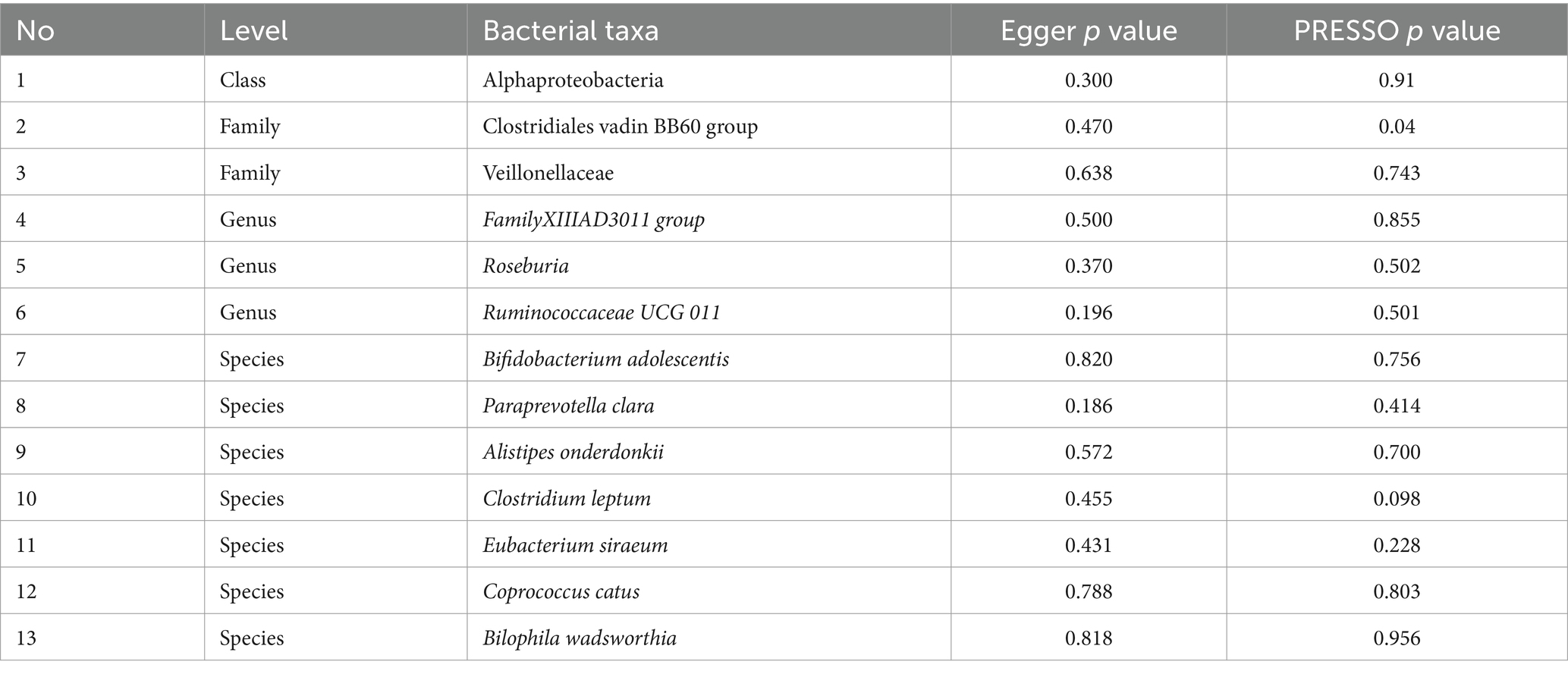
We employed IVW as the primary analysis method for MR. The results indicate an association between the risk of PPD and 13 genetically predicted bacterial taxa (Figure 2). The results are presented in Table 2.
Using the IVW method as the primary analysis, the results indicated that 13 bacterial taxa were associated with the risk of PPD when the significance level was less than 0.05. Specifically, the following taxa were inversely associated with the risk of PPD: Clostridiales vadin BB60 group, Veillonellaceae (at family level); Ruminococcaceae UCG 011 (at genus level); Bifidobacterium adolescentis, Paraprevotella clara, Clostridium leptum, Eubacterium siraeum, Coprococcus catus (at species level). Whereas the following taxa may be associated with a higher risk of PPD: Alphaproteobacteria (at class level); FamilyXIIIAD3011 group, Roseburia (at genus level); Alistipes onderdonkii, Bilophila wadsworthia (at species level) (Figure 2; Table 2). The scatter plots (Supplementary Figure S1) and forest plots (Supplementary Figure S2) of the above results demonstrate the stability of the findings.
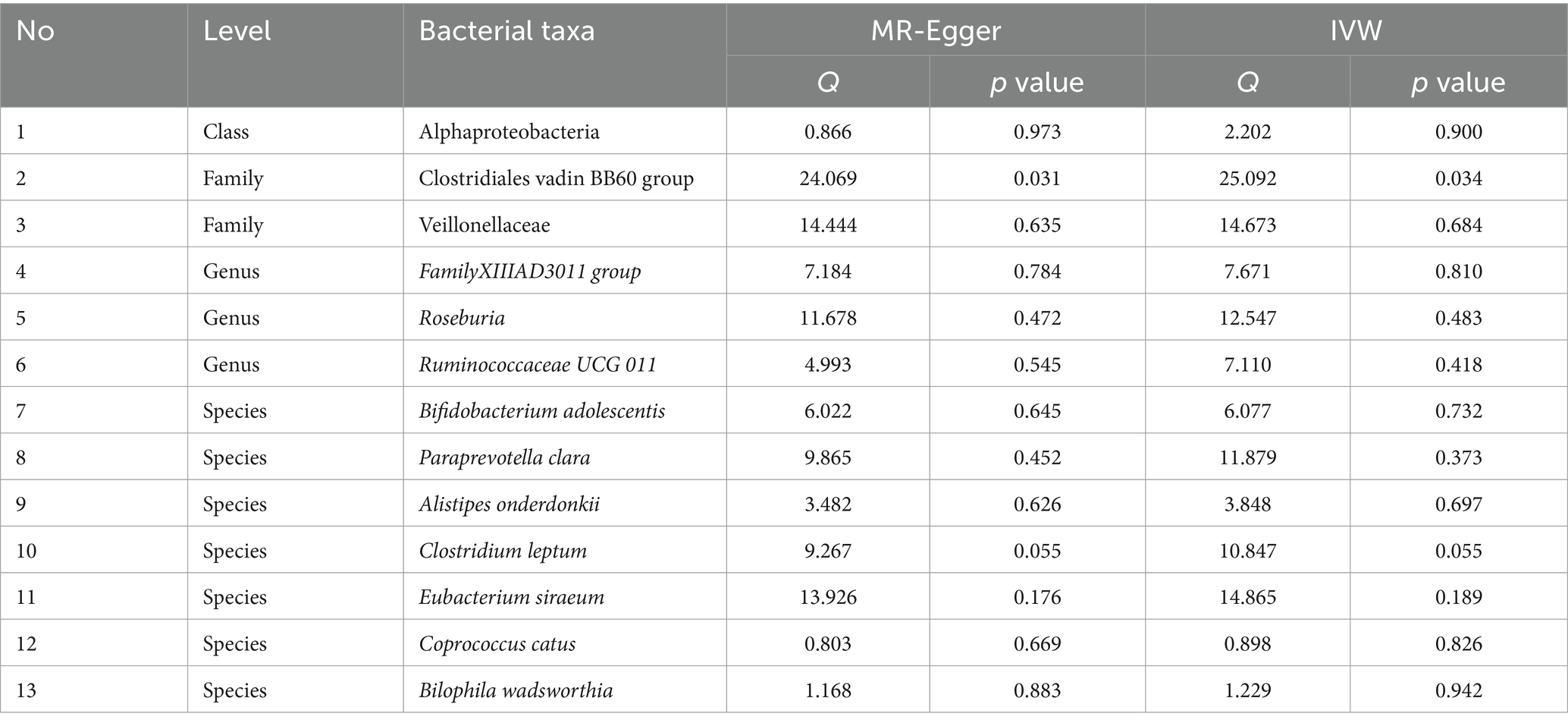
Table 3 shows that the instrumental variable of Clostridiales vadin BB60 group has a significant heterogeneity with the outcome by Cochran’s Q test (p < 0.05), and the others were no significant heterogeneity identified among the SNPs (p > 0.05).
Table 4 shows that there was no demonstrated pleiotropy detected in the MR Egger test (p < 0.05). MR-PRESSO suggests that Clostridiales vadin BB60 group has horizontal pleiotropy with outcome (p = 0.04, Table 3). According to the findings of the leave-one-out analysis, no SNPs were found to have affected causal association (Supplementary Figure S3).
4 Discussion
Our MR analysis further illuminated a nominal causal bond linking PPD with 13 distinct microbial taxa. To ensure the transparency and completeness of our Mendelian Randomization study report, we adopted the STROBE-MR guidelines (Supplementary Table S2). STROBE-MR is a set of reporting guidelines specifically designed for Mendelian Randomization studies, consisting of 20 items. We have provided the complete STROBE-MR checklist in the appendix for readers’ reference.
Observational research findings indicate a decrease in the abundance of the Faecalibacterium genus in patients with PPD (Zhou et al., 2020), a phenomenon that aligns with the negative correlation we observed with Ruminococcaceae UCG 011 through our MR analysis. Ruminococcaceae portray a noticeable reduction in individuals with pronounced depression (Radjabzadeh et al., 2022). In the realm of animal studies, they exhibit a positive association with vital sugar metabolic cascades, such as gluconeogenesis, glycolysis, and the pentose phosphate trajectory (Zhang et al., 2019). Given that obesity is one of the comorbidities of depression, we hypothesize that it might indirectly affect PPD. Within Ruminococcaceae, Faecalibacterium prausnitzii is one of the main bacteria in the human gut and converts acetate to butyrate through the butyryl-CoA: acetate CoA-transferase (Duncan et al., 2002). This mechanism could potentially account for the diminished abundance of the Faecalibacterium genus observed in patients with PPD.
Previous MR studies on the gut microbiota and MDD indicated that the Actinobacteria class, Bifidobacterium genus, and Ruminococcus1 genus had a protective effect against MDD (Chen et al., 2022). This aligns with our findings for Ruminococcaceae UCG 011 (OR 0.845, 95%CI 0.757–0.944) and Bifidobacterium adolescentis (OR 0.853, 95%CI 0.761–0.956). Bifidobacterium, the most common microbe within Actinobacteria in the human gut, is also a widely-used probiotic. A study demonstrated that Bifidobacterium can alleviate depression and gut-related diseases by altering the gut microbiota and its tryptophan metabolism (Tian et al., 2022). Postpartum, women experience an increase in the abundance of Bifidobacterium, a change that may also enhance the production of short-chain fatty acids, thereby aiding in the establishment and refinement of the neonatal gut microbiota environment (Qin et al., 2022). Given the symptomatic similarities between PPD and MDD, Bifidobacterium may also have the potential to treat or prevent PPD.
Research has shown that good mental health in mothers can reduce anxiety and mitigate stress responses, which is crucial for the successful delivery and upbringing of offspring (Hillerer et al., 2011). The hippocampus, a limbic structure involved in emotion and cognition, plays a critical role in these processes. Within the hippocampus, brain-derived neurotrophic factor (BDNF) and its receptor, TrkB (tyrosine kinase receptor B), are directly involved in various physiological functions of the central nervous system, such as neuronal survival, synaptic plasticity, and learning and memory (Leibrock et al., 1989; von Bohlen und Halbach, 2010). During pregnancy, if a mother is subjected to chronic stress or chronic restraint stress (CRS), the expression of BDNF and TrkB in the hippocampus can be significantly reduced. This reduction is believed to be one of the contributing factors to the high incidence of depression during pregnancy and postpartum (Ye et al., 2011). Notably, repeated restraint stress in the final week of pregnancy can induce depressive-like behaviors in mothers (O'Mahony et al., 2006).
The administration of Bifidobacterium adolescentis in mice subjected to CRS not only reduces anxiety and depressive-like behaviors and increases BDNF expression levels, but also reverses the dysbiosis of the gut microbiota induced by CRS (Guo et al., 2019). Additionally, Bifidobacterium longum has been shown to significantly reduce depressive-like behaviors in the forced swim test (FST) and decrease anxiety-like behaviors in the open field test (OFT) (Savignac et al., 2014). The combined use of Bifidobacterium longum and Lactobacillus helveticus has been found to alleviate symptoms of anxiety and depression in healthy individuals (Radford-Smith and Anthony, 2023).
Within the MR assessment, particular bacterial groups, Veillonellaceae (OR 0.819, 95%CI 0.715–0.939), Paraprevotella clara (OR 0.902, 95%CI 0.825–0.987), Clostridium leptum (OR 0.849, 95%CI 0.724–0.996), Eubacterium siraeum (OR 0.854, 95%CI 0.741–0.984), Coprococcus catus (OR 0.796, 95%CI 0.656–0.965), emerged as potentially protective entities.
Veillonellaceae, as non-fermentative microbes, distinguish themselves with their propensity to modulate the enzymatic activity of methylmalonyl-CoA decarboxylase, thereby playing a pivotal role in fatty acid metabolism. Noteworthy is their marked depletion in autism-afflicted individuals, potentially influencing lactate fermentation and specific short-chain fatty acid synthesis (Gronow et al., 2010; Wang et al., 2023). They also show a negative correlation with psychological stress (Ma et al., 2023).
Paraprevotella clara can reduce the levels of trypsin in the colon, thereby decreasing the likelihood of developing inflammatory bowel diseases, while also offering protection against certain viral infections in the colon (Li et al., 2022). Intestinal inflammation may reduce gut permeability, leading to the translocation of microbes originally present in the intestinal tract into the peripheral circulation, thereby disrupting normal brain function and affecting specific behaviors (D'Mello et al., 2015). This is consistent with our research findings that Paraprevotella clara exhibits a negative correlation with PPD.
Due to a lack of experimental studies, the mechanisms by which Clostridium leptum, Coprococcus catus, and Eubacterium siraeum affect PPD remain unclear. In a survey study of 1,070 patients with depression, Coprococcus was found to be nearly depleted in individuals with depression. Coprococcus possesses biosynthetic pathways related to dopamine synthesis and participates in the synthesis of GABA, improving the quality of psychological life (Valles-Colomer et al., 2019). Clostridium leptum is associated with glucose metabolism (Palmnas-Bedard et al., 2022), leading us to speculate that Clostridium leptum may be related to GABA produced during the body’s glucose metabolic processes. However, this hypothesis has not yet been validated.
Five positive results were identified: Alphaproteobacteria (OR 1.216, 95%CI 1.008–1.468), Roseburia (OR 1.235, 95%CI 1.021–1.493), FamilyXIIIAD3011group (OR 1.237, 95%CI 1.036–1.478), Alistipes onderdonkii (OR 1.201, 95%CI 1.011–1.427), Bilophila wadsworthia (OR 1.183, 95%CI 1.01–1.385). A surge in Alphaproteobacteria’s presence typically heralds a perturbation in the gut’s ecological equilibrium (Litvak et al., 2017). Previous research insights spotlight Rhodospirillaceae, a member of the Alphaproteobacteria phylum, underscoring its positive correlation with the genesis of Alzheimer’s disease (Zhou et al., 2021). Therefore, we infer that an increase in the abundance of Alphaproteobacteria may signal changes in the gut ecological environment, thereby affecting the normal growth of other bacterial communities and the composition of their metabolic products.
Roseburia stands as a significant regulator in the realms of gut microbiota ecology, immune modulation, and neurological afflictions. Within this genus, five species notably dominate: Roseburia intestinalis, R. hominis, R. inulinivorans, R. faecis, and R. cecicola (Tamanai-Shacoori et al., 2017). Of these, R. intestinalis holds a particular distinction, constituting between 0.9 and 5.0% of the entire microbial community and acting as the chief butyrate producer (Hold et al., 2003). In an insightful study, colitis-afflicted rats treated with R. intestinalis demonstrated a diminished display of anxiety and depressive-like symptoms. The underlying mechanism could be R. intestinalis’ influence on 5-hydroxytryptamine (5-HT) expression in colonic regions. As a neurotransmitter, 5-HT plays a pivotal role in mediating the gut-brain axis, influencing brain functionality (Xu et al., 2021). While the abundance of R. intestinalis is lower in patients with depression compared to the healthy population (Zheng et al., 2016), other studies have shown a higher abundance of R. intestinalis in patients with MDD compared to healthy individuals (Jiang et al., 2015). These findings highlight that while Roseburia does play a role in neurological diseases, its interaction mechanisms are intricate. Furthermore, Reserpine can inhibit the central nervous system’s regular functions by altering catecholamines and 5-HT levels in brain tissues, leading to depressive behaviors (Yan et al., 2023). Taking into account Roseburia’s position as a positive determinant in our MR analysis, it’s plausible to hypothesize that an overabundance of Roseburia might interfere with levels of serotonergic depression markers like 5-HT, thereby affecting the functionality of the nervous system.
An increase in the abundance of Bilophila wadsworthia can disrupt hippocampal synaptic plasticity, neurogenesis, and gene expression. Ketogenic diets and hypoxic environments can alter the abundance of Bilophila wadsworthia, thereby impairing cognitive behavior (Olson et al., 2021). However, some studies have found that ketogenic diets can increase GABA levels. Currently, there is no definitive clinical data to indicate whether ketogenic diets can improve or exacerbate PPD (Wlodarczyk et al., 2021).
Alistipes onderdonkii is a relatively new bacterium identified to increase in abundance under conditions of high stress or prolonged fatigue. PPD often involves extended periods of battling fatigue or stress. The increase in Alistipes decreases serotonin availability and disrupts the gut-brain axis (Parker et al., 2020).
Previous research has conducted numerous analyses on the association between the gut microbiome and psychiatric disorders. Although PPD shares certain symptomatic and genetic similarities with other mental health conditions, the pathogenesis of PPD is influenced by multiple factors, particularly fluctuations in hormone levels. Therefore, our MR analysis has the potential to reveal more causal relationships, paving the way for future research into bacterial biosensor detection and therapeutic interventions for related diseases.
This study possesses several strengths and limitations. Employing the MR method to analyze the causal relationship between PPD and the gut microbiota minimizes interference from confounding factors. Techniques such as MR-PRESSO and MR-Egger were utilized to eliminate the effects of horizontal pleiotropy and heterogeneity, ensuring the accuracy of the results. This study refined the classification of the gut microbiome down to the species level, making the results more comprehensive and detailed compared to previous research. However, the study also has its limitations; the GWAS data derived from the MiBioGen consortium, DMP, and FinnGen consortium, may introduce selection bias. Moreover, the data primarily originates from European populations, hence extrapolating these results to diverse populations should be approached with caution. The etiological factors behind PPD remain enigmatic, alluding to the existence of potential undisclosed confounders. Despite employing various methods for sensitivity analysis, the impact of horizontal pleiotropy could not be fully assessed. Lastly, in vitro experimental validation was not conducted. In future studies, we will strengthen the functional validation through related experiments to further substantiate our research findings.
In conclusion, with the continuous development of medical statistical techniques and the refinement of GWAS data, future studies should focus on the integrated development of multiple disciplines and omics, exploring complex diseases, genetic variations, and environmental changes at various levels of interaction.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
WJ: Writing – review & editing, Data curation, Formal analysis. BL: Conceptualization, Writing – original draft, Formal analysis, Writing – review & editing. LW: Supervision, Writing – original draft. LZ: Writing – review & editing, Visualization. SC: Methodology, Writing – review & editing. RH: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the 2019 Henan Province Private Higher Education Brand Professional Support Project.
Acknowledgments
We thank the FinnGen consortium, MiBioGen consortium and the Dutch Microbiome Project for sharing the genetic data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1415237/full#supplementary-material
References
Bowden, J., Davey Smith, G., and Burgess, S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525. doi: 10.1093/ije/dyv080
Bowden, J., Davey Smith, G., Haycock, P. C., and Burgess, S. (2016). Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314. doi: 10.1002/gepi.21965
Bowden, J., Del Greco, M. F., Minelli, C., Zhao, Q., Lawlor, D. A., Sheehan, N. A., et al. (2019). Improving the accuracy of two-sample summary-data Mendelian randomization: moving beyond the NOME assumption. Int. J. Epidemiol. 48, 728–742. doi: 10.1093/ije/dyy258
Bowden, J., and Holmes, M. V. (2019). Meta-analysis and Mendelian randomization: a review. Res. Synth. Methods 10, 486–496. doi: 10.1002/jrsm.1346
Burgess, S., Butterworth, A., and Thompson, S. G. (2013). Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665. doi: 10.1002/gepi.21758
Burgess, S., and Thompson, S. G. (2017). Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32, 377–389. doi: 10.1007/s10654-017-0255-x
Chen, M., Xie, C. R., Shi, Y. Z., Tang, T. C., and Zheng, H. (2022). Gut microbiota and major depressive disorder: a bidirectional Mendelian randomization. J. Affect. Disord. 316, 187–193. doi: 10.1016/j.jad.2022.08.012
Cryan, J. F., O'Riordan, K. J., Cowan, C. S. M., Sandhu, K. V., Bastiaanssen, T. F. S., Boehme, M., et al. (2019). The microbiota-gut-brain axis. Physiol. Rev. 99, 1877–2013. doi: 10.1152/physrev.00018.2018
D'Mello, C., Ronaghan, N., Zaheer, R., Dicay, M., Le, T., MacNaughton, W. K., et al. (2015). Probiotics improve inflammation-associated sickness behavior by altering communication between the peripheral immune system and the brain. J. Neurosci. 35, 10821–10830. doi: 10.1523/JNEUROSCI.0575-15.2015
Duncan, S. H., Hold, G. L., Harmsen, H. J. M., Stewart, C. S., and Flint, H. J. (2002). Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. Nov., comb. nov. Int. J. Syst. Evol. Microbiol. 52, 2141–2146. doi: 10.1099/00207713-52-6-2141
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
El-Ansary, A. K., Ben Bacha, A., and Kotb, M. (2012). Etiology of autistic features: the persisting neurotoxic effects of propionic acid. J. Neuroinflammation 9:74. doi: 10.1186/1742-2094-9-74
Ellervik, C., Roselli, C., Christophersen, I. E., Alonso, A., Pietzner, M., Sitlani, C. M., et al. (2019). Assessment of the relationship between genetic determinants of thyroid function and atrial fibrillation: a Mendelian randomization study. JAMA Cardiol. 4, 144–152. doi: 10.1001/jamacardio.2018.4635
Gill, S. R., Pop, M., Deboy, R. T., Eckburg, P. B., Turnbaugh, P. J., Samuel, B. S., et al. (2006). Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359. doi: 10.1126/science.1124234
Gronow, S., Welnitz, S., Lapidus, A., Nolan, M., Ivanova, N., Glavina Del Rio, T., et al. (2010). Complete genome sequence of Veillonella parvula type strain (Te3). Stand. Genomic Sci. 2, 57–65. doi: 10.4056/sigs.521107
Guo, Y., Xie, J. P., Deng, K., Li, X., Yuan, Y., Xuan, Q., et al. (2019). Prophylactic effects of Bifidobacterium adolescentis on anxiety and depression-like phenotypes after chronic stress: a role of the gut microbiota-inflammation axis. Front. Behav. Neurosci. 13:126. doi: 10.3389/fnbeh.2019.00126
Hahn-Holbrook, J., Cornwell-Hinrichs, T., and Anaya, I. (2017). Economic and health predictors of national postpartum depression prevalence: a systematic review, meta-analysis, and meta-regression of 291 studies from 56 countries. Front. Psych. 8:248. doi: 10.3389/fpsyt.2017.00248
Hartwig, F. P., Davey Smith, G., and Bowden, J. (2017). Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 46, 1985–1998. doi: 10.1093/ije/dyx102
Haycock, P. C., Burgess, S., Wade, K. H., Bowden, J., Relton, C., and Davey Smith, G. (2016). Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. Am. J. Clin. Nutr. 103, 965–978. doi: 10.3945/ajcn.115.118216
Hillerer, K. M., Reber, S. O., Neumann, I. D., and Slattery, D. A. (2011). Exposure to chronic pregnancy stress reverses peripartum-associated adaptations: implications for postpartum anxiety and mood disorders. Endocrinology 152, 3930–3940. doi: 10.1210/en.2011-1091
Hirtz, R., Hars, C., Naaresh, R., Laabs, B. H., Antel, J., Grasemann, C., et al. (2022). Causal effect of age at menarche on the risk for depression: results from a two-sample multivariable Mendelian randomization study. Front. Genet. 13:918584. doi: 10.3389/fgene.2022.918584
Hold, G. L., Schwiertz, A., Aminov, R. I., Blaut, M., and Flint, H. J. (2003). Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl. Environ. Microbiol. 69, 4320–4324. doi: 10.1128/AEM.69.7.4320-4324.2003
Jairaj, C., and Rucker, J. J. (2022). Postpartum depression: a role for psychedelics? J. Psychopharmacol. 36, 920–931. doi: 10.1177/02698811221093793
Jiang, H., Ling, Z., Zhang, Y., Mao, H., Ma, Z., Yin, Y., et al. (2015). Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 48, 186–194. doi: 10.1016/j.bbi.2015.03.016
Josefsson, A., Angelsioo, L., Berg, G., Ekstrom, C. M., Gunnervik, C., Nordin, C., et al. (2002). Obstetric, somatic, and demographic risk factors for postpartum depressive symptoms. Obstet. Gynecol. 99, 223–228. doi: 10.1016/s0029-7844(01)01722-7
Kamel, L. Y., Xiong, W., Gott, B. M., Kumar, A., and Conway, C. R. (2022). Vagus nerve stimulation: an update on a novel treatment for treatment-resistant depression. J. Neurol. Sci. 434:120171. doi: 10.1016/j.jns.2022.120171
Konig, I. R., and Greco, F. M. D. (2018). Mendelian randomization: progressing towards understanding causality. Ann. Neurol. 84, 176–177. doi: 10.1002/ana.25293
Kurilshikov, A., Medina-Gomez, C., Bacigalupe, R., Radjabzadeh, D., Wang, J., Demirkan, A., et al. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165. doi: 10.1038/s41588-020-00763-1
Lawlor, D. A., Harbord, R. M., Sterne, J. A., Timpson, N., and Davey Smith, G. (2008). Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med. 27, 1133–1163. doi: 10.1002/sim.3034
Leibrock, J., Lottspeich, F., Hohn, A., Hofer, M., Hengerer, B., Masiakowski, P., et al. (1989). Molecular cloning and expression of brain-derived neurotrophic factor. Nature 341, 149–152. doi: 10.1038/341149a0
Li, Y., Watanabe, E., Kawashima, Y., Plichta, D. R., Wang, Z., Ujike, M., et al. (2022). Identification of trypsin-degrading commensals in the large intestine. Nature 609, 582–589. doi: 10.1038/s41586-022-05181-3
Litvak, Y., Byndloss, M. X., Tsolis, R. M., and Baumler, A. J. (2017). Dysbiotic proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr. Opin. Microbiol. 39, 1–6. doi: 10.1016/j.mib.2017.07.003
Liu, L., Wang, H., Chen, X., Zhang, Y., Zhang, H., and Xie, P. (2023). Gut microbiota and its metabolites in depression: from pathogenesis to treatment. EBioMedicine 90:104527. doi: 10.1016/j.ebiom.2023.104527
Liu, X., Wang, S., and Wang, G. (2022). Prevalence and risk factors of postpartum depression in women: a systematic review and meta-analysis. J. Clin. Nurs. 31, 2665–2677. doi: 10.1111/jocn.16121
Lopera-Maya, E. A., Kurilshikov, A., van der Graaf, A., Hu, S., Andreu-Sanchez, S., Chen, L., et al. (2022). Effect of host genetics on the gut microbiome in 7,738 participants of the Dutch microbiome project. Nat. Genet. 54, 143–151. doi: 10.1038/s41588-021-00992-y
Ma, L., Yan, Y., Webb, R. J., Li, Y., Mehrabani, S., Xin, B., et al. (2023). Psychological stress and gut microbiota composition: a systematic review of human studies. Neuropsychobiology 82, 247–262. doi: 10.1159/000533131
Maguire, J., Ferando, I., Simonsen, C., and Mody, I. (2009). Excitability changes related to GABAA receptor plasticity during pregnancy. J. Neurosci. 29, 9592–9601. doi: 10.1523/JNEUROSCI.2162-09.2009
Mayer, E. A., Knight, R., Mazmanian, S. K., Cryan, J. F., and Tillisch, K. (2014). Gut microbes and the brain: paradigm shift in neuroscience. J. Neurosci. 34, 15490–15496. doi: 10.1523/JNEUROSCI.3299-14.2014
Ni, J. J., Xu, Q., Yan, S. S., Han, B. X., Zhang, H., Wei, X. T., et al. (2021). Gut microbiota and psychiatric disorders: a two-sample Mendelian randomization study. Front. Microbiol. 12:737197. doi: 10.3389/fmicb.2021.737197
Olson, C. A., Iniguez, A. J., Yang, G. E., Fang, P., Pronovost, G. N., Jameson, K. G., et al. (2021). Alterations in the gut microbiota contribute to cognitive impairment induced by the ketogenic diet and hypoxia. Cell Host Microbe 29, 1378–1392.e6. doi: 10.1016/j.chom.2021.07.004
O'Mahony, S. M., Myint, A. M., van den Hove, D., Desbonnet, L., Steinbusch, H., and Leonard, B. E. (2006). Gestational stress leads to depressive-like behavioural and immunological changes in the rat. Neuroimmunomodulation 13, 82–88. doi: 10.1159/000096090
Palmnas-Bedard, M. S. A., Costabile, G., Vetrani, C., Aberg, S., Hjalmarsson, Y., Dicksved, J., et al. (2022). The human gut microbiota and glucose metabolism: a scoping review of key bacteria and the potential role of SCFAs. Am. J. Clin. Nutr. 116, 862–874. doi: 10.1093/ajcn/nqac217
Palumbo, G., Mirabella, F., and Gigantesco, A. (2017). Positive screening and risk factors for postpartum depression. Eur. Psychiatry 42, 77–85. doi: 10.1016/j.eurpsy.2016.11.009
Parker, B. J., Wearsch, P. A., Veloo, A. C. M., and Rodriguez-Palacios, A. (2020). The genus Alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front. Immunol. 11:906. doi: 10.3389/fimmu.2020.00906
Pu, Y., Tan, Y., Qu, Y., Chang, L., Wang, S., Wei, Y., et al. (2021). A role of the subdiaphragmatic vagus nerve in depression-like phenotypes in mice after fecal microbiota transplantation from Chrna7 knock-out mice with depression-like phenotypes. Brain Behav. Immun. 94, 318–326. doi: 10.1016/j.bbi.2020.12.032
Qin, S., Liu, Y., Wang, S., Ma, J., and Yang, H. (2022). Distribution characteristics of intestinal microbiota during pregnancy and postpartum in healthy women. J. Matern. Fetal Neonatal Med. 35, 2915–2922. doi: 10.1080/14767058.2020.1812571
Radford-Smith, D. E., and Anthony, D. C. (2023). Prebiotic and probiotic modulation of the microbiota-gut-brain axis in depression. Nutrients 15:1880. doi: 10.3390/nu15081880
Radjabzadeh, D., Bosch, J. A., Uitterlinden, A. G., Zwinderman, A. H., Ikram, M. A., van Meurs, J. B. J., et al. (2022). Gut microbiome-wide association study of depressive symptoms. Nat. Commun. 13:7128. doi: 10.1038/s41467-022-34502-3
Ramsteijn, A. S., Jasarevic, E., Houwing, D. J., Bale, T. L., and Olivier, J. D. (2020). Antidepressant treatment with fluoxetine during pregnancy and lactation modulates the gut microbiome and metabolome in a rat model relevant to depression. Gut Microbes 11, 735–753. doi: 10.1080/19490976.2019.1705728
Savignac, H. M., Kiely, B., Dinan, T. G., and Cryan, J. F. (2014). Bifidobacteria exert strain-specific effects on stress-related behavior and physiology in BALB/c mice. Neurogastroenterol. Motil. 26, 1615–1627. doi: 10.1111/nmo.12427
Sender, R., Fuchs, S., and Milo, R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. doi: 10.1371/journal.pbio.1002533
Smith, G. D., and Ebrahim, S. (2003). ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 32, 1–22. doi: 10.1093/ije/dyg070
Sohr-Preston, S. L., and Scaramella, L. V. (2006). Implications of timing of maternal depressive symptoms for early cognitive and language development. Clin. Child. Fam. Psychol. Rev. 9, 65–83. doi: 10.1007/s10567-006-0004-2
Tamanai-Shacoori, Z., Smida, I., Bousarghin, L., Loreal, O., Meuric, V., Fong, S. B., et al. (2017). Roseburia spp.: a marker of health? Future Microbiol. 12, 157–170. doi: 10.2217/fmb-2016-0130
Tan, J., McKenzie, C., Potamitis, M., Thorburn, A. N., Mackay, C. R., and Macia, L. (2014). The role of short-chain fatty acids in health and disease. Adv. Immunol. 121, 91–119. doi: 10.1016/B978-0-12-800100-4.00003-9
Tian, P., Chen, Y., Zhu, H., Wang, L., Qian, X., Zou, R., et al. (2022). Bifidobacterium breve CCFM1025 attenuates major depression disorder via regulating gut microbiome and tryptophan metabolism: a randomized clinical trial. Brain Behav. Immun. 100, 233–241. doi: 10.1016/j.bbi.2021.11.023
Tian, X. Y., Xing, J. W., Zheng, Q. Q., and Gao, P. F. (2021). 919 syrup alleviates postpartum depression by modulating the structure and metabolism of gut microbes and affecting the function of the hippocampal GABA/glutamate system. Front. Cell. Infect. Microbiol. 11:694443. doi: 10.3389/fcimb.2021.694443
Valles-Colomer, M., Falony, G., Darzi, Y., Tigchelaar, E. F., Wang, J., Tito, R. Y., et al. (2019). The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 4, 623–632. doi: 10.1038/s41564-018-0337-x
van der Waerden, J., Bernard, J. Y., De Agostini, M., Saurel-Cubizolles, M. J., Peyre, H., Heude, B., et al. (2017). Persistent maternal depressive symptoms trajectories influence children’s IQ: the EDEN mother-child cohort. Depress. Anxiety 34, 105–117. doi: 10.1002/da.22552
Verbanck, M., Chen, C. Y., Neale, B., and Do, R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698. doi: 10.1038/s41588-018-0099-7
von Bohlen und Halbach, O. (2010). Involvement of BDNF in age-dependent alterations in the hippocampus. Front. Aging Neurosci. 2:36. doi: 10.3389/fnagi.2010.00036
Wang, F., Yang, H., Wu, Y., Peng, L., and Li, X. (2023). SAELGMDA: identifying human microbe-disease associations based on sparse autoencoder and LightGBM. Front. Microbiol. 14:1207209. doi: 10.3389/fmicb.2023.1207209
Wlodarczyk, A., Cubala, W. J., and Stawicki, M. (2021). Ketogenic diet for depression: a potential dietary regimen to maintain euthymia? Prog. Neuro-Psychopharmacol. Biol. Psychiatry 109:110257. doi: 10.1016/j.pnpbp.2021.110257
Xu, F., Cheng, Y., Ruan, G., Fan, L., Tian, Y., Xiao, Z., et al. (2021). New pathway ameliorating ulcerative colitis: focus on Roseburia intestinalis and the gut-brain axis. Ther. Adv. Gastroenterol. 14:17562848211004469. doi: 10.1177/17562848211004469
Yan, X., Song, X., Chen, W., Jia, Y., Gao, J., Wang, X., et al. (2023). Frizzled 6 mutation regulates reserpine-induced depression-like behavior and Wnt signaling pathway in mice. Eur. J. Pharmacol. 957:175996. doi: 10.1016/j.ejphar.2023.175996
Ye, Y., Wang, G., Wang, H., and Wang, X. (2011). Brain-derived neurotrophic factor (BDNF) infusion restored astrocytic plasticity in the hippocampus of a rat model of depression. Neurosci. Lett. 503, 15–19. doi: 10.1016/j.neulet.2011.07.055
Zhang, J., Ma, L., Chang, L., Pu, Y., Qu, Y., and Hashimoto, K. (2020). A key role of the subdiaphragmatic vagus nerve in the depression-like phenotype and abnormal composition of gut microbiota in mice after lipopolysaccharide administration. Transl. Psychiatry 10:186. doi: 10.1038/s41398-020-00878-3
Zhang, F., Zheng, W., Xue, Y., and Yao, W. (2019). Suhuai suckling piglet hindgut microbiome-metabolome responses to different dietary copper levels. Appl. Microbiol. Biotechnol. 103, 853–868. doi: 10.1007/s00253-018-9533-0
Zheng, P., Zeng, B., Zhou, C., Liu, M., Fang, Z., Xu, X., et al. (2016). Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host's metabolism. Mol. Psychiatry 21, 786–796. doi: 10.1038/mp.2016.44
Zhou, Y., Chen, C., Yu, H., and Yang, Z. (2020). Fecal microbiota changes in patients with postpartum depressive disorder. Front. Cell. Infect. Microbiol. 10:567268. doi: 10.3389/fcimb.2020.567268
Keywords: gut microbiota, postpartum depression, Mendelian randomization, instrumental variables (IVs), single nucleotide polymorphisms (SNPs)
Citation: Jin W, Li B, Wang L, Zhu L, Chai S and Hou R (2024) The causal association between gut microbiota and postpartum depression: a two-sample Mendelian randomization study. Front. Microbiol. 15:1415237. doi: 10.3389/fmicb.2024.1415237
Edited by:
Wei Peng, Chengdu University of Traditional Chinese Medicine, ChinaReviewed by:
Nadia Andrea Andreani, Max Planck Institute for Evolutionary Biology, GermanyRaju Baskar, University of Texas Southwestern Medical Center, United States
Manigandan Venkatesan, The University of Texas Health Science Center at San Antonio, United States
Copyright © 2024 Jin, Li, Wang, Zhu, Chai and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Li, bWlsZHNpcjkzQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Wenjun Jin
Wenjun Jin Bo Li
Bo Li Lijun Wang
Lijun Wang Lin Zhu1
Lin Zhu1