- National Key Laboratory of Intelligent Tracking and Forecasting for Infectious Diseases, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
The concept of “enterotypes” in microbiome research has attracted substantial interest, particularly focusing on the abundance of Prevotella spp. in the human gut. In this study, the intricate dynamics of Prevotella spp. in the human gut microbiota was investigated, based on the metagenomic method. First, 239 fecal samples from individuals across four regions of China revealed a bimodal distribution, highlighting the abundance and variability in Prevotella spp. within the Chinese population. Second, the longitudinal cohort study included 184 fecal samples from 52 time points collected from seven individuals who demonstrated either the outbreaks or disappearances of Prevotella spp., emphasizing the transient nature of Prevotella abundance levels and suggesting shifts in Prevotella “enterotypes.” Furthermore, a turnover of the dominant Prevotella spp. was observed, indicating the potential presence of diverse subtypes of Prevotella enterotype. Notably, the genomic analysis demonstrated the persistence of specific Prevotella strains within individuals over extended periods, highlighting the enduring presence of Prevotella in the human gut. In conclusion, by integrating the temporal and geographical scales in our research, we gained deeper insights into the dynamics of Prevotella, emphasizing the importance of considering the dynamics at the time and species level in gut microbiota studies and their implications on human health.
Introduction
The characterization of the gut microbiota has sparked debate, particularly regarding the concept of “enterotypes” (Arumugam et al., 2011), akin to blood types, suggesting distinct gut microbiota types that could be correlated with overall health (Siezen and Kleerebezem, 2011; Wu et al., 2011; Costea et al., 2018). Prevotella, one of the most prevalent genera in the gut, is a critical type in these distinct enterotypes (Arumugam et al., 2011; Siezen and Kleerebezem, 2011; Wu et al., 2011; Costea et al., 2018). Prevotella has been the subject of extensive research because of its complex relationship with human health (De Filippis et al., 2019; Metwaly and Haller, 2019; Abdelsalam et al., 2023). Prevotella has been associated with positive health outcomes, promoting the fermentation of dietary fiber and the production of short-chain fatty acids (Bedarf et al., 2017). These fatty acids have been linked to improved metabolic health and reduced risk of certain diseases, such as inflammatory bowel disease (Bajer et al., 2017). Prevotella has also been linked to a balanced immune response and maintenance of gut barrier function, both of which are essential for overall health. On the other hand, an overabundance of Prevotella spp. in the gut has been associated with negative health outcomes, including Parkinson’s disease (Stefanis, 2012), and certain types of cancer (Gunathilake et al., 2019). Some studies have also suggested that Prevotella spp. may contribute to the development of obesity (Moran-Ramos et al., 2023) and metabolic disorders by promoting carbohydrate fermentation and the production of potentially harmful metabolites (Metwaly and Haller, 2019).
The conflicting research findings on the impacts of Prevotella spp. on human health underscore the critical need for further investigation to enhance our comprehension of its role within the gut microbiota and its implications on human health. The latest taxonomic findings offer a potential explanation for the intricacies of Prevotella. These recent taxonomic studies from the International Journal of Systematic and Evolutionary Microbiology (Oren and Göker, 2023) have reclassified the genus Prevotella into seven distinct genera, introducing four novel genera: Segatella, Hoylesella, Leyella, and Palleniella (Hitch et al., 2022). This new classification hints at the diverse relationships that different microbes under the original Prevotella classification may have with human health, necessitating a more nuanced approach to research.
However, previous analyses of gut microbiota structures have predominantly focused on the level of the original Prevotella genus. There has been a lack of resolution at the species level and neglect of research on the distribution patterns of the various taxa within the updated Prevotella. Consequently, there has been a dearth of in-depth investigations linking the latest taxonomic insights with outcomes in gut microbiota. Numerous inquiries persist regarding the existence of Prevotella enterotypes, the potential for additional subtypes, whether they consist of multiple species or a select few key species, the interrelationships among species across the seven Prevotella genera, their stability within the human body, and the underlying reasons for any fluctuations.
Drawing on this taxonomic framework, we have the opportunity to reassess the concept of Prevotella enterotypes through the use of temporal and geographical scales, thereby corroborating its presence and impact on human health in the Chinese population.
Materials and methods
Sample collection and cohort description
As a part of the Chinese Microbiome Project (CMP), 239 fecal samples were collected from individuals residing in four provinces across China: Beijing, Jiangsu, Henan, and Sichuan, representing diverse geographical regions within the country. Prior to the sample collection, the participants underwent a pretest and completed a comprehensive pre-questionnaire, which included demographic details, personal and familial medical histories, and lifestyle practices such as smoking, physical activity, and dietary patterns (Supplementary Table S1). These data were used to evaluate the general health status of the participants. Detailed information on the sampling procedure in the CMP can be found in our previous publication (Zhang et al., 2022).
In conjunction with the CMP initiative, fecal samples were procured monthly from seven healthy individuals residing in Beijing, China, spanning the timeline from March 2017 to October 2022. To ensure a longitudinal study, samples from individuals with fewer than 10 data points were excluded. During each sampling session, the participants completed a detailed questionnaire regarding their medical history over the preceding month, and physical metrics, including height, weight, blood pressure, and blood glucose levels, were documented on-site. Ultimately, 184 fecal samples were obtained from these 7 individuals, resulting in a dataset comprising 52 data points. The study was approved by the bioethical committee (ICDC-2022-001) of the Chinese Centre for Disease Control and Prevention (CDC), and all procedures were conducted per the relevant guidelines and regulations, with written informed consent obtained from all participants.
DNA extraction, sequencing, and quality control
The fecal samples were collected within 24 h of completion of the pretest and pre-questionnaire using disposable bedpans, to minimize the risk of contamination from toilet water.
The DNA was extracted within 24 h of sample collection using the QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Subsequently, shotgun metagenomic libraries were prepared using the TruePrep DNA Library Prep Kit from Illumina, and sequencing was performed on an Illumina HiSeq platform to generate paired-end reads of 150 bp. Quality control analysis involved the use of FastQC1 and Fastp (Chen et al., 2018) to filter out low-quality reads. This included trimming of low-quality bases (< Q20) and retention of reads with a length ≥ 100 bp. After the quality control of raw reads, the Bowtie2 tool was used to remove the human sequences (Langmead and Salzberg, 2012).
The public fecal microbiome data was downloaded from the Human Microbiome Project (HMP) database (Human Microbiome Project, 2012; Lloyd-Price et al., 2017) and it included shotgun metagenomics data from 153 healthy individuals.
Metagenomic data analysis pipeline
The analysis pipeline utilized in this study is available on GitHub.2
The MetaPhIAn2 tool was used to calculate the relative abundance of Prevotella spp. for the qualified samples (Truong et al., 2015, 2017), and an evolutionary tree of Prevotella strains was constructed using StrainPhiAn tool (Truong et al., 2017). Another strain-level analysis software, SameStr (Podlesny et al., 2022), was used to determine whether any two positive samples contained the same strain. The metagenome bins were assembled using SPAdes v3.13.0 (Bankevich et al., 2012) and MetaBAT v2.12.1 (Kang et al., 2019), followed by taxonomic classification using the GTDB-Tk toolkit (Chaumeil et al., 2019, 2022). A phylogenetic tree was constructed using the ANIclustermap3 for the metagenome bins identified as Prevotella, based on the average nucleotide value, with the additional inclusion of 88 strain genomes from National Center for Biotechnology Information (NCBI). The final phylogenetic tree was annotated using the iTOL tool (Letunic and Bork, 2021). The identification of single nucleotide polymorphism (SNP) between the genomes was achieved using the MUMmer software (Marçais et al., 2018). The line, bar, and circular plots in this study were generated by R scripts using the ggplot2 package (Wickham, 2009). The co-occurrence network of the gut microbiota was constructed using the R package igraph (Csardi and Nepusz, 2006). The correlation between bacterial species was calculated using the Spearman algorithm. The correlations with a coefficient greater than 0.6 or less than −0.6, as well as a p-value of <0.001, were retained in the analysis. The enterotype of each sample was assigned using an online tool4 and R packages biotypes.5
Results
Abundance of Prevotella on genus and species level
To elucidate the structure of the gut microbiota and correlate the results with enterotypes, we conducted analyses using the original and the latest taxonomic information. Under the original classification framework, Prevotella was considered as a genus. Our findings revealed a wide variation in the abundance of Prevotella among individuals across China. While the average abundance of Prevotella was 27.91% according to the MetaPhIAn method, certain samples exhibited unexpectedly high levels of approximately 97.31% (Figure 1A). This distribution indicates a bimodal pattern of the abundance of Prevotella within the Chinese population, with some individuals showing high abundance while others showed low levels (Figure 1B). We utilized the same methodology to analyze the publicly available HMP database (Human Microbiome Project, 2012; Lloyd-Price et al., 2017). Similar patterns were observed in the analysis of the HMP public dataset, providing further evidence for the existence of a subset of individuals with elevated Prevotella levels in the gut microbiota.
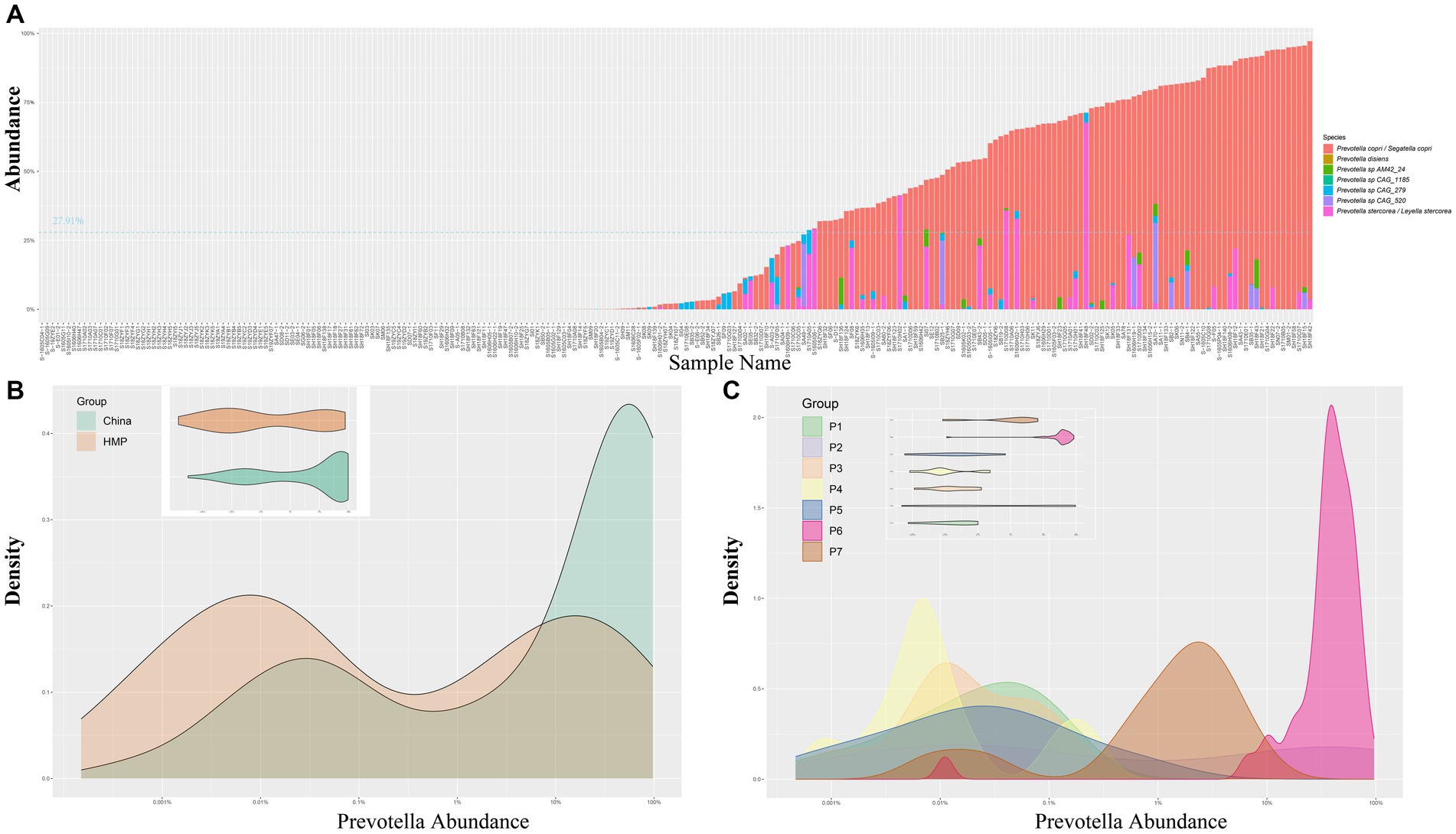
Figure 1. (A) Bar chart representing the abundance of Prevotella in 239 samples from the CMP dataset. (B) Density plot and violin plot for the abundance of Prevotella in the CMP and HMP datasets. (C) Density plot and violin plot displaying the distribution of Prevotella abundance in 7 individuals from the time cohort in this study. CMP, Chinese Microbiome Project; HMP, Human Microbiome Project.
Furthermore, we utilized the latest taxonomic classification to analyze the gut microbiota. Recent taxonomic research (2023) has subdivided the genus Prevotella into seven genera, introducing four new genera named Segatella, Hoylesella, Leyella, and Palleniella (Hitch et al., 2022). Prevotella copri was reclassified as Segatella copri, detected in 66.94% of the 239 samples, showing the highest detection rate (Figure 1A). Prevotella stercorea was reclassified as Leyella stercorea, detected in 25.10% of the 239 samples, ranking second. While other species were also detected (Figure 1A), their abundance and positivity rates were comparatively lower.
Short-term outbreaks or disappearances of Prevotella over time
In our study, we further investigated the temporal dynamics of Prevotella species under both the original and the latest taxonomic frameworks. In the original classification framework, the longitudinal study of up to 5 years demonstrated short-term blooms or decreases in the abundance of Prevotella over time (Figures 1C, 2A). Short-term blooms, defined as instances where the abundance of Prevotella exceeded 5 times the individual’s average abundance, and short-term decreases, defined as abundance dropping to more than 5 times lower than the previous time point (Zhang et al., 2022; Han et al., 2024), were observed. These fluctuations were recorded at 17 time points over the 5 years and were distributed among 7 individuals (Supplementary Figure S1). In the two instances of short-term blooms, the participants reported antibiotic drug usage, while there were no reports of antibiotic use related to the other fluctuations or time points. This suggests that high or low Prevotella abundance may only be temporary, indicating potential variability in Prevotella “enterotypes” based on single time point samples. Under the latest taxonomic framework, our findings similarly support the occurrence of short-term blooms or decreases in abundance for various Prevotella/Segatella/Leyella species (Supplementary Figure S1).
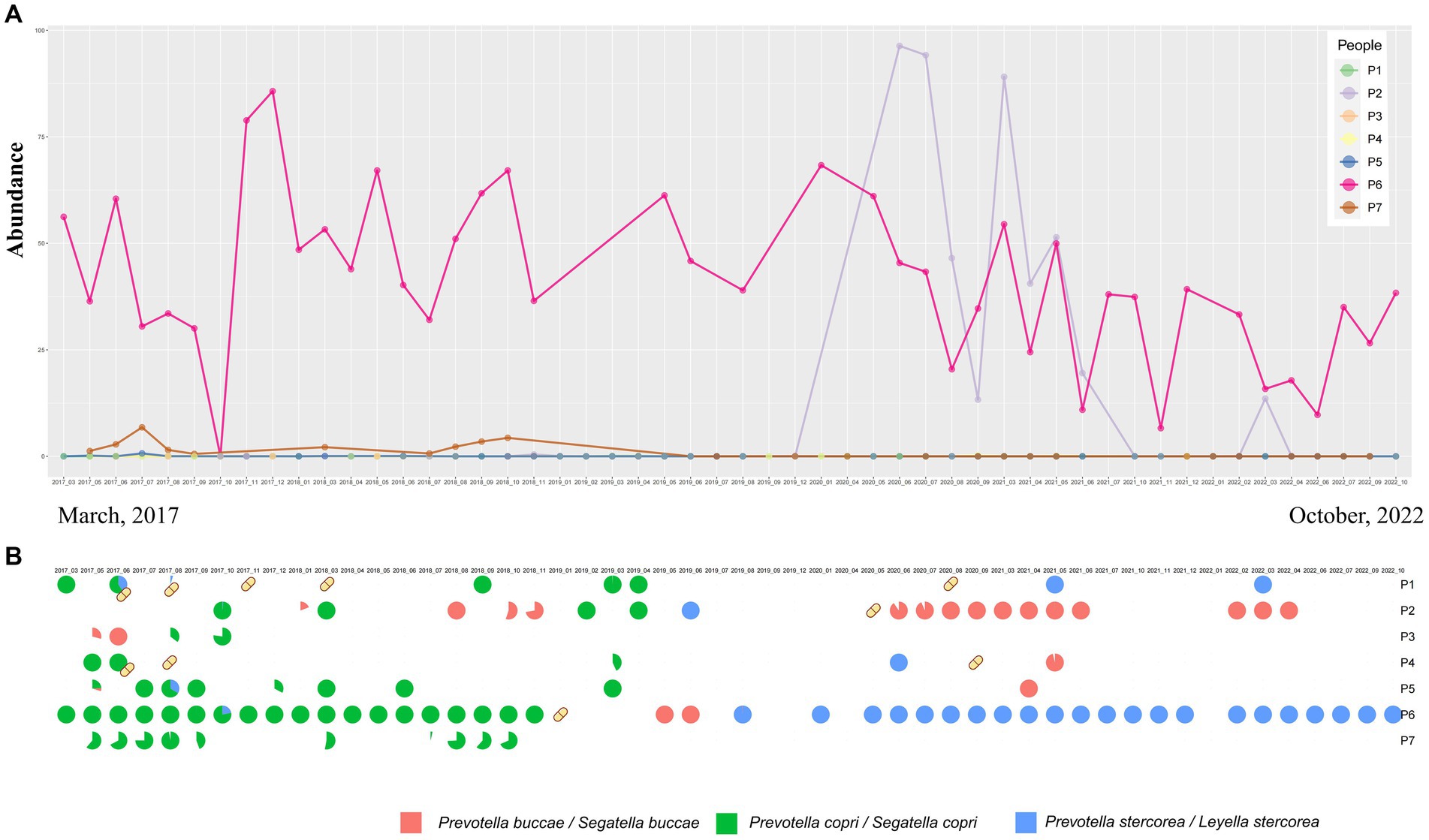
Figure 2. (A) Genus-level abundance of Prevotella from samples at different time points sourced from seven different participants. (B) Temporal changes in the proportion of abundance of Prevotella at the species level over time. Yellow capsule icons represent participants who were on antibiotics during that month.
Dominant species turnover within the Prevotella genus
The definition of “dominant species” in this study is as follows: within the samples if only one species of Prevotella is detected, or if the abundance of this species exceeds 80% of the total abundance of all species within the same genus, then this is referred to as the dominant species.
Prevotella copri (Segatella copri) was the most abundant species in the CMP cohort study and was identified as the dominant species in 51.46% of samples (Figure 1A). Other species such as Prevotella stercorea (Leyella stercorea) were also identified as dominant species in 5 samples (2.09%). The results obtained from the time dataset also show that Prevotella copri (Segatella copri) is the most prevalent among all Prevotella species. Prevotella copri (Segatella copri) was detected in 53 of 240 samples, and it was the dominant species in 34 samples. However, changes in prevalence were observed over time (Supplementary Figures S1A–F). Furthermore, there was a turnover of the dominant Prevotella species over time, indicating that Prevotella not only changes in abundance but also exhibits turnover at the species level (Figure 2B). Upon reviewing the responses to the questionnaire regarding antibiotic use during each sampling, we found a correlation between antibiotic usage and the turnover of dominant species (Figure 2B). For instance, in the case of individual P6, who had a record of antibiotic use in January 2019, the dominant Prevotella species shifted from Prevotella copri (Segatella copri) before antibiotic treatment to Prevotella buccae (Segatella buccae) after treatment. However, antibiotic use alone could not explain the turnover of all the dominant species. For the same individual, after July 2019, the dominant species reverted to Prevotella stercorea (Leyella stercorea) without any antibiotic usage. These results suggest that besides antibiotic use, other unknown factors also play a role in driving changes in Prevotella within the human gut.
Persistence of Prevotella at the strain level in the human gut
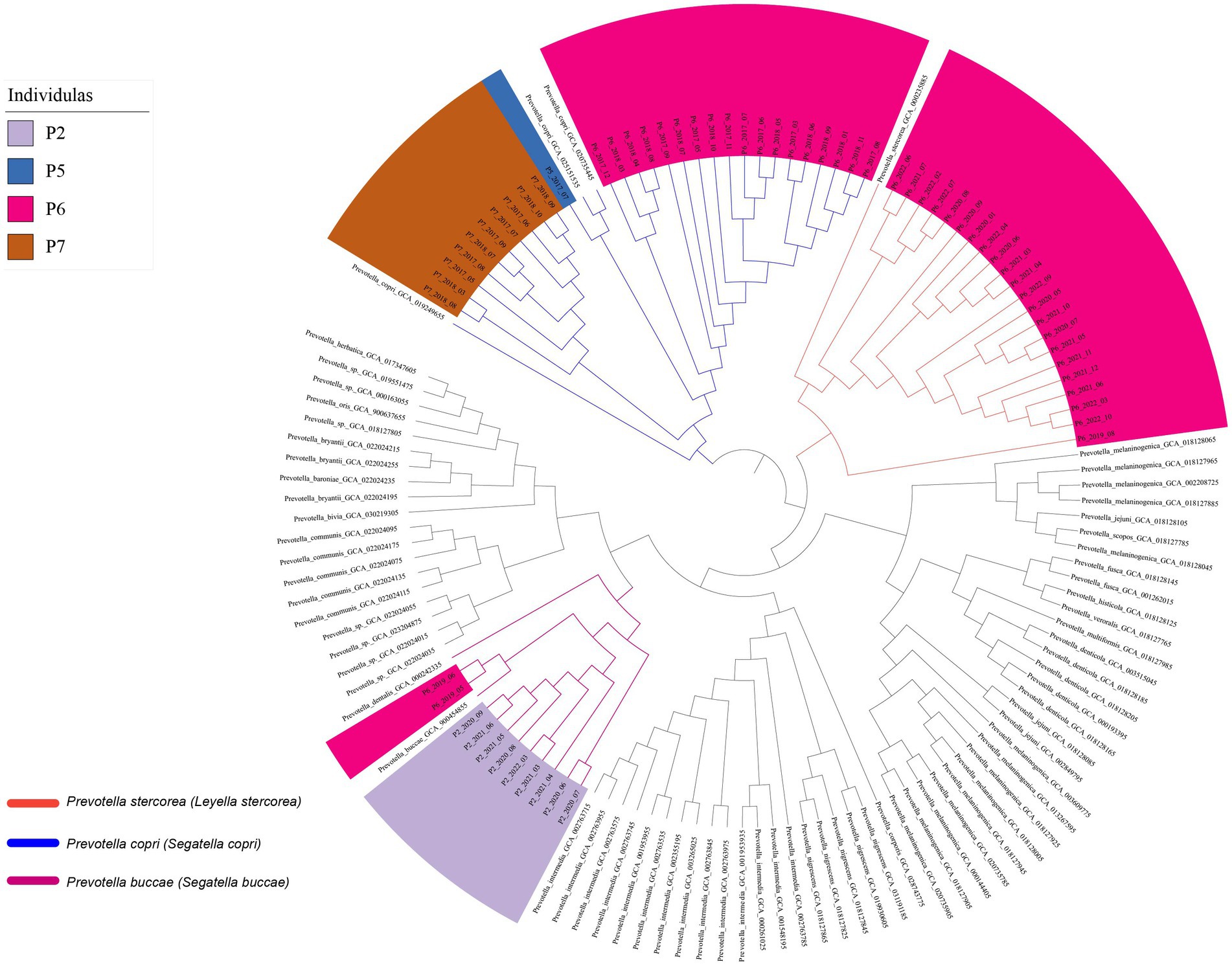
At the strain level, genome analysis revealed relatively stable Prevotella strains within samples from the same individual over time. This was corroborated by the consistent clustering of Prevotella copri (Segatella copri) strains from individual P6 across multiple time points (March 2017 to November 2018), and the genomic similarity (average nucleotide identity value) between these strains was higher than 99.95%, implying a degree of strain persistence within the gut microbiota (Figure 3). Prevotella stercorea (Leyella stercorea) was also identified in the samples collected at multiple time points from August 2019 to October 2022 from individual P6; it exhibited high similarity at the genome level, indicating its persistence within the individual at the annual level. Similar characteristics were observed for Prevotella buccae (Segatella buccae) from individual P2 and Prevotella copri (Segatella copri) from individual P7, confirming the persistence of specific Prevotella strains within individuals over time. These findings highlight the enduring presence of Prevotella strains in the human body over an extended period.
We further evaluated the single nucleotide polymorphisms (SNPs) between strains at the genome level at different time points. Despite a high degree of similarity among the strains (ANI > 99.95%, Figure 3), we observed tens to hundreds of single nucleotide polymorphisms (SNPs) at the genome level between pairs of the genome (Avg: 0.1 SNP/kb). By examining the temporal distance between the sampling points of the samples and the distribution of SNPs (Supplementary Figure S3), we did not find a significant correlation between the number of SNPs and time and distance. These results indicate that the majority of SNPs among Prevotella strains in the human gut are neutral mutations, rather than being fixed over time within the human gut Prevotella population.
In addition to genomic analysis, marker genes from each species were utilized to construct a phylogenetic tree (Supplementary Figure S2) and SameStr software was used to analyze whether they originated from the same strain. It is worth noting that the marker genes of these species were sourced from the Metaphlan2 database (Truong et al., 2015, 2017). The results, as shown in Supplementary Figure S2, are consistent with the genomic analysis (Figure 3), supporting the notion that the same strain can exist within an individual for a certain period, up to years. Replacement of Prevotella strains within individuals in adjacent months was not observed in this study.
Relationship between Prevotella and other microbes
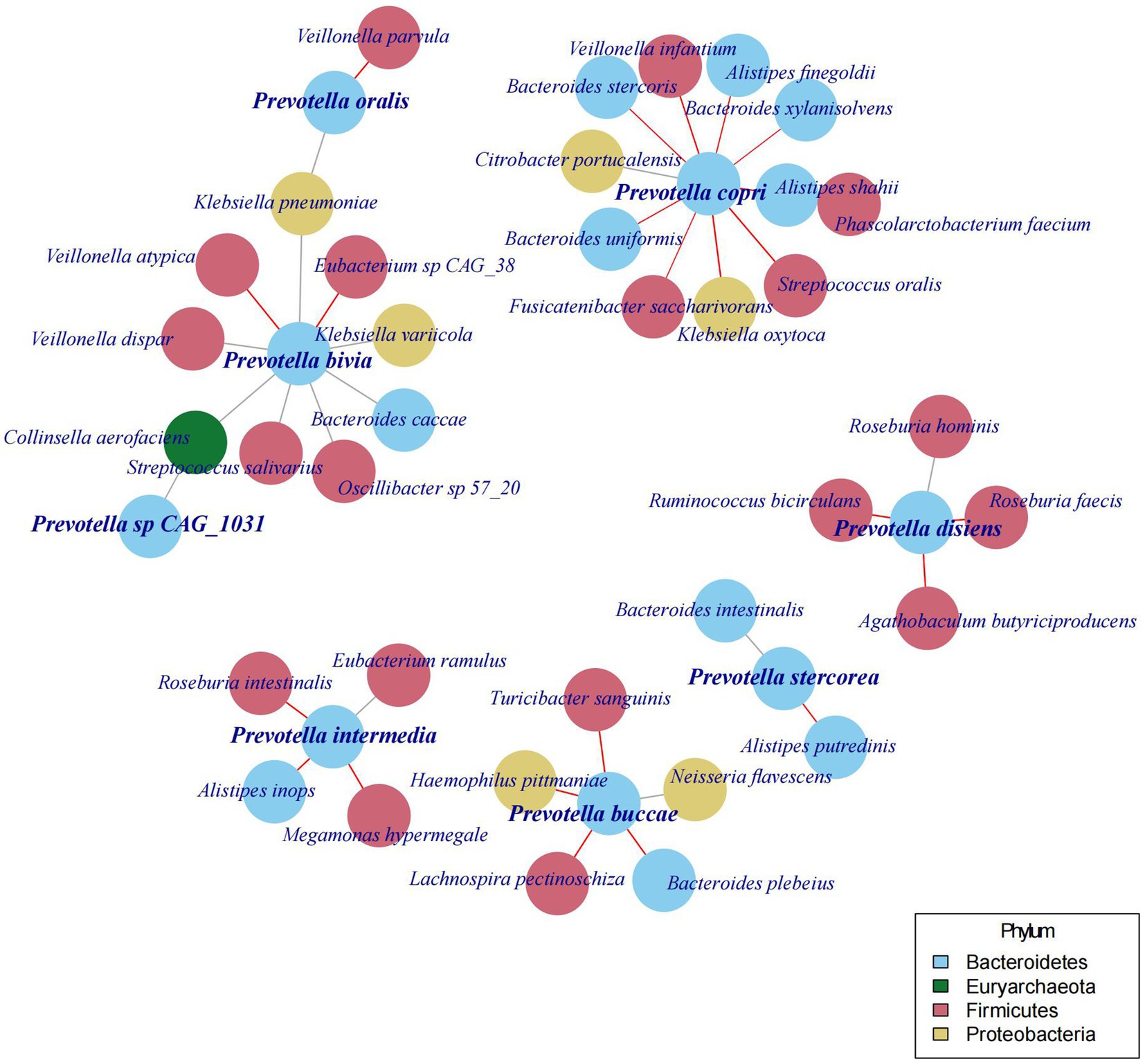
In the preceding sections of this study, we discovered various species of Prevotella existing in the human gut, and their abundance is not stable over time and may even show a turnover of the dominant species. In the following analysis, we aimed to determine whether changes in the abundance and the species of Prevotella would also lead to variations in other gut microbial species. Therefore, we constructed a co-occurrence network between Prevotella and other bacteria based on the community structure of gut microbiota and found that the abundance of Prevotella species is correlated with 37 other bacterial species (Figure 4).
Different Prevotella species are associated with distinct sets of other bacterial species. For instance, Prevotella copri (Segatella copri) is linked to 11 other species, with a positive correlation with one species (Citrobacter portucalensis) and a negative correlation with 10 species. Conversely, Prevotella stercorea (Leyella stercorea) is only associated with 2 other species, and these associated species are different from those linked to other Prevotella species. Moreover, the bacterial species associated with Prevotella copri (Segatella copri) are distinct from those associated with Prevotella bivia, Prevotella buccae, Prevotella disiens, Prevotella intermedia, and Prevotella oralis. Although further investigations are required to confirm the causal relationships between these bacterial species, the above findings underscore the diversity among the various species of Prevotella.
Discussion
In recent years, the concept of “enterotypes” has garnered attention in microbiome research, proposing distinct gut microbiota compositions based on the abundance of certain bacterial genera, including Prevotella (Arumugam et al., 2011; Siezen and Kleerebezem, 2011; Wu et al., 2011; Costea et al., 2018; Metwaly and Haller, 2019; Couch et al., 2021; Alili et al., 2022; Fu et al., 2022; Liang et al., 2022; Moran-Ramos et al., 2023). However, “enterotypes” have faced challenges and criticisms. This study, based on a comprehensive survey, confirmed that Prevotella abundance in the human gut occurs in two distinct groups, with some individuals in the population experiencing a high abundance while others experience a low abundance. Furthermore, the longitudinal analysis revealed that the abundance of Prevotella is not constant, with sudden increases or decreases observed in samples from the same individual at specific time points, which suggests that single time point assessments may capture transient fluctuations in the abundance of Prevotella, potentially leading to false classifications for its “enterotype.” This variability underscores the complexity of microbiome studies. Although the results of this study do not definitively confirm or refute the existence of a Prevotella enterotype, they emphasize the importance of considering temporal dynamics when evaluating the characteristics of gut microbiota.
Moreover, the relationship between Prevotella spp. and health needs to be evaluated at a finer level as previous studies (Zhang et al., 2019; Fu et al., 2022; Liang et al., 2022) are often based on genus-level surveys, particularly through 16S sequencing analysis, and have not fully indicated the presence of multiple Prevotella species in the gut microbiota, each potentially serving as a dominant species. This study revealed that even samples with similar abundances of Prevotella could harbor different species, with the turnover and replacement of dominant species observed in samples from the same individual at different time points (Figures 2A,B). In this research, we also confirmed that with the variation in different Prevotella species, the associated gut microbiota differed, highlighting the diversity among Prevotella species. These findings imply that the samples with a similar abundance of Prevotella may elicit different interactions with the host owing to the presence of distinct species. Therefore, if an enterotype does exist, it likely encompasses multiple subtypes at the species level, necessitating a more nuanced approach when studying the association between gut microbiota and human health, focusing on the species level. The latest taxonomic research (2023) further subdivided the genus Prevotella into seven genera, including four novel genera for which the names Segatella, Hoylesella, Leyella, and Palleniella are proposed (Hitch et al., 2022). According to this updated classification framework, our study confirms that Prevotella copri (Segatella copri) is the most abundant and widely distributed species among the mentioned taxa, serving as the dominant species in 51.46% of samples in the CMP dataset. Furthermore, other Prevotella species coexist in fecal samples, with some of these acting as dominant species in certain samples, such as Prevotella stercorea (Leyella stercorea) and Prevotella buccae (Segatella buccae). Furthermore, our study validates the temporal turnover among these species at the time scale (Figure 2).
Although a turnover was observed in the dominant Prevotella species and sudden fluctuations in their abundance, our investigation demonstrated that Prevotella strains can persist in the human body for an extended period. The 5-year longitudinal study revealed that Prevotella copri (Segatella copri) was consistently present in samples from P6 (March 2017 to November 2018), with genomic analysis indicating the same strain, without a turnover at the strain level. This finding is in contradiction with our previous finding based on Escherichia coli (Han et al., 2024), as this species experiences a rapid monthly turnover rate (87.5% within a month), whereas Prevotella persists in the gut for a longer duration. We refer to Escherichia coli and other rapidly turning over bacteria, similar to it, as transient gut microbes. In contrast, for Prevotella, which can persist in the gut for a certain period, we are inclined to believe they can establish colonization within the gut. In this study, we also observed the evolutionary trajectory of Prevotella colonization in the gut. Over the years of persistence of Prevotella in the human gut, we did not detect a rise in the number of SNPs with prolonged colonization. This observation implies the potential predominance of neutral evolution within Prevotella’s evolutionary trajectory in the human gut.
In summary, our investigation of Prevotella spp., an important component of the gut microbiota, confirmed sudden fluctuations in abundance at the genus level, turnover of dominant species at the species level, and the persistence of the same strain for years. We gained deeper insights into Prevotella by combining the temporal and geographical scales. This underscores the need to consider the dynamics at the time and species level when studying the gut microbiota and its implications on human health. Further research is warranted to elucidate the complex interactions of Prevotella with the host, as well as the factors (lifestyles, age, and drug usage) influencing its abundance and activity in the gut, ultimately advancing our understanding of the role of Prevotella on human health.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/sra/PRJNA1116923, PRJNA1116923.
Ethics statement
The studies involving humans were approved by our analysis involved fecal samples obtained from healthy Chinese individuals who provided written informed consent prior to enrolment in this study. Furthermore, the study was approved by the China CDC, and all experiments were performed in accordance with relevant guidelines and regulations. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
NH: Data curation, Investigation, Methodology, Software, Writing – original draft. XP: Formal analysis, Software, Writing – review & editing. TZ: Data curation, Software, Visualization, Writing – review & editing. YQ: Investigation, Validation, Writing – review & editing. XL: Investigation, Validation, Writing – review & editing. WZ: Methodology, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program of China (2018YFC1200100) and the Major Infectious Diseases such as AIDS and Viral Hepatitis Prevention and Control Technology Major Projects (2018ZX10305410 and 2018ZX10733402) and 2021SKLID509.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1414000/full#supplementary-material
Footnotes
1. ^ https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
2. ^ https://github.com/zhangwencdc/Prevotella
3. ^ https://github.com/moshi4/ANIclustermap
References
Abdelsalam, N. A., Hegazy, S. M., and Aziz, R. K. (2023). The curious case of Prevotella copri. Gut Microbes 15:2249152. doi: 10.1080/19490976.2023.2249152
Alili, R., Belda, E., Fabre, O., Pelloux, V., Giordano, N., Legrand, R., et al. (2022). Characterization of the gut microbiota in individuals with overweight or obesity during a real-world weight loss dietary program: a focus on the Bacteroides 2 Enterotype. Biomedicines 10:10016. doi: 10.3390/biomedicines10010016
Arumugam, M., Raes, J., Pelletier, E., Le Paslier, D., Yamada, T., Mende, D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180. doi: 10.1038/nature09944
Bajer, L., Kverka, M., Kostovcik, M., Macinga, P., Dvorak, J., Stehlikova, Z., et al. (2017). Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J. Gastroenterol. 23, 4548–4558. doi: 10.3748/wjg.v23.i25.4548
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bedarf, J. R., Hildebrand, F., Coelho, L. P., Sunagawa, S., Bahram, M., Goeser, F., et al. (2017). Functional implications of microbial and viral gut metagenome changes in early stage L-DOPA-naive Parkinson's disease patients. Genome Med. 9:39. doi: 10.1186/s13073-017-0428-y
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2019). GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927. doi: 10.1093/bioinformatics/btz848
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2022). GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics 38, 5315–5316. doi: 10.1093/bioinformatics/btac672
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Costea, P. I., Hildebrand, F., Arumugam, M., Bäckhed, F., Blaser, M. J., Bushman, F. D., et al. (2018). Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 3, 8–16. doi: 10.1038/s41564-017-0072-8
Couch, C. E., Stagaman, K., Spaan, R. S., Combrink, H. J., Sharpton, T. J., Beechler, B. R., et al. (2021). Diet and gut microbiome enterotype are associated at the population level in African buffalo. Nat. Commun. 12:2267. doi: 10.1038/s41467-021-22510-8
Csardi, G., and Nepusz, T. (2006). The igraph software package for complex network research. Interjournal Complex Syst. 1695, 1–9.
De Filippis, F., Pasolli, E., Tett, A., Tarallo, S., Naccarati, A., De Angelis, M., et al. (2019). Distinct genetic and functional traits of human intestinal Prevotella copri strains are associated with different habitual diets. Cell Host Microbe 25:e443, 444–453.e3. doi: 10.1016/j.chom.2019.01.004
Fu, T., Zhou, L., Fu, Z., Zhang, B., Li, Q., Pan, L., et al. (2022). Enterotype-specific effect of human gut microbiota on the fermentation of marine algae oligosaccharides: a preliminary proof-of-concept in vitro study. Polymers 14:770. doi: 10.3390/polym14040770
Gunathilake, M. N., Lee, J., Choi, I. J., Kim, Y. I., Ahn, Y., Park, C., et al. (2019). Association between the relative abundance of gastric microbiota and the risk of gastric cancer: a case-control study. Sci. Rep. 9:13589. doi: 10.1038/s41598-019-50054-x
Han, N., Peng, X., Zhang, T., Qiang, Y., Li, X., and Zhang, W. (2024). Rapid turnover and short-term blooms of Escherichia coli in the human gut. J. Bacteriol. 206:e0023923. doi: 10.1128/jb.00239-23
Hitch, T. C. A., Bisdorf, K., Afrizal, A., Riedel, T., Overmann, J., Strowig, T., et al. (2022). A taxonomic note on the genus Prevotella: description of four novel genera and emended description of the genera Hallella and Xylanibacter. Syst. Appl. Microbiol. 45:126354. doi: 10.1016/j.syapm.2022.126354
Human Microbiome Project (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214.
Kang, D. D., Li, F., Kirton, E., Thomas, A., Egan, R., An, H., et al. (2019). MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. doi: 10.7717/peerj.7359
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Liang, Y., Shen, Y., Li, G., Yuan, Y., Zhang, M., and Gao, J. (2022). Schizophrenia patients with Prevotella-Enterotype have a higher risk of obesity. Front. Psych. 13:864951. doi: 10.3389/fpsyt.2022.864951
Lloyd-Price, J., Mahurkar, A., Rahnavard, G., Crabtree, J., Orvis, J., Hall, A. B., et al. (2017). Strains, functions and dynamics in the expanded human microbiome project. Nature 550, 61–66. doi: 10.1038/nature23889
Marçais, G., Delcher, A. L., Phillippy, A. M., Coston, R., Salzberg, S. L., and Zimin, A. (2018). MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 14:e1005944. doi: 10.1371/journal.pcbi.1005944
Metwaly, A., and Haller, D. (2019). Strain-level diversity in the gut: the P. copri case. Cell Host Microbe 25, 349–350. doi: 10.1016/j.chom.2019.02.006
Moran-Ramos, S., Cerqueda-Garcia, D., Lopez-Contreras, B., Larrieta-Carrasco, E., Villamil-Ramirez, H., Molina-Cruz, S., et al. (2023). A metagenomic study identifies a Prevotella copri enriched microbial profile associated with non-alcoholic steatohepatitis in subjects with obesity. J. Gastroenterol. Hepatol. 38, 791–799. doi: 10.1111/jgh.16147
Oren, A., and Göker, M. (2023). Validation list no. 209. Valid publication of new names and new combinations effectively published outside the IJSEM. Int. J. Syst. Evol. Microbiol. 73:5709. doi: 10.1099/ijsem.0.005709
Podlesny, D., Arze, C., Dorner, E., Verma, S., Dutta, S., Walter, J., et al. (2022). Metagenomic strain detection with SameStr: identification of a persisting core gut microbiota transferable by fecal transplantation. Microbiome 10:53. doi: 10.1186/s40168-022-01251-w
Siezen, R. J., and Kleerebezem, M. (2011). The human gut microbiome: are we our enterotypes? Microb. Biotechnol. 4, 550–553. doi: 10.1111/j.1751-7915.2011.00290.x
Stefanis, L. (2012). Alpha-Synuclein in Parkinson's disease. Cold Spring Harb. Perspect. Med. 2:a009399. doi: 10.1101/cshperspect.a009399
Truong, D. T., Franzosa, E. A., Tickle, T. L., Scholz, M., Weingart, G., Pasolli, E., et al. (2015). MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 12, 902–903. doi: 10.1038/nmeth.3589
Truong, D. T., Tett, A., Pasolli, E., Huttenhower, C., and Segata, N. (2017). Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 27, 626–638. doi: 10.1101/gr.216242.116
Wu, G. D., Chen, J., Hoffmann, C., Bittinger, K., Chen, Y. Y., Keilbaugh, S. A., et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108. doi: 10.1126/science.1208344
Zhang, W., Han, N., Zhang, T., Qiang, Y., Peng, X., Li, X., et al. (2022). The spatial features and temporal changes in the gut microbiota of a healthy Chinese population. Microbiol. Spectr. 10:e0131022. doi: 10.1128/spectrum.01310-22
Keywords: microbiome, Prevotella spp., human gut, enterotypes, genome
Citation: Han N, Peng X, Zhang T, Qiang Y, Li X and Zhang W (2024) Temporal dynamics and species-level complexity of Prevotella spp. in the human gut microbiota: implications for enterotypes and health. Front. Microbiol. 15:1414000. doi: 10.3389/fmicb.2024.1414000
Edited by:
Edoardo Pasolli, University of Naples Federico II, ItalyReviewed by:
Diego Armando Esquivel Hernandez, Metropolitan Autonomous University, MexicoYulin Wang, Shandong University, China
Copyright © 2024 Han, Peng, Zhang, Qiang, Li and Zhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen Zhang, emhhbmd3ZW5AaWNkYy5jbg==
 Na Han
Na Han Xianhui Peng
Xianhui Peng Wen Zhang
Wen Zhang