
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 19 June 2024
Sec. Extreme Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1410024
This article is part of the Research TopicInsights in Extreme Microbiology: 2023View all 9 articles
The Deinococcus genus is renowned for its remarkable resilience against environmental stresses, including ionizing radiation, desiccation, and oxidative damage. This resilience is attributed to its sophisticated DNA repair mechanisms and robust defense systems, enabling it to recover from extensive damage and thrive under extreme conditions. Central to Deinococcus research, the D. radiodurans strains ATCC BAA-816 and ATCC 13939 facilitate extensive studies into this remarkably resilient genus. This study focused on delineating genetic discrepancies between these strains by sequencing our laboratory’s ATCC 13939 specimen (ATCC 13939K) and juxtaposing it with ATCC BAA-816. We uncovered 436 DNA sequence differences within ATCC 13939K, including 100 single nucleotide variations, 278 insertions, and 58 deletions, which could induce frameshifts altering protein-coding genes. Gene annotation revisions accounting for gene fusions and the reconciliation of gene lengths uncovered novel protein-coding genes and refined the functional categorizations of established ones. Additionally, the analysis pointed out genome structural variations due to insertion sequence (IS) elements, underscoring the D. radiodurans genome’s plasticity. Notably, ATCC 13939K exhibited a loss of six ISDra2 elements relative to BAA-816, restoring genes fragmented by ISDra2, such as those encoding for α/β hydrolase and serine protease, and revealing new open reading frames, including genes imperative for acetoin decomposition. This comparative genomic study offers vital insights into the metabolic capabilities and resilience strategies of D. radiodurans.
The order Deinococcales, part of the Deinococcus-Thermus phylum, consists of two families: Deinococcaceae and Trueperaceae (Ho et al., 2016). The Deinococcaceae family encompasses two genera: Deinococcus, which currently has 89 species with validly published names, and Deinobacterium, which only comprises one species, Deinobacterium chartae (Parte et al., 2020). Deinococcus radiodurans (D. radiodurans), the representative species of the Deinococcus genus, exhibits remarkable resilience to various stresses, including ionizing radiation (IR), UV exposure, DNA-damaging reagents, desiccation, and oxidative stress (Cox and Battista, 2005).
Since its discovery in 1956 as a contaminant in food sterilized by gamma radiation (γ-radiation) (Brooks and Murray, 1981), D. radiodurans (previously known as Micrococcus radiodurans) has been widely studied as a model organism to explore mechanisms underlying the extreme resistance to radiation and oxidative stress (Slade and Radman, 2011; Lim et al., 2019). Double-strand DNA breaks (DSBs), the most lethal form of DNA damage caused by IR, are repaired through the homologous recombination (HR) pathway. D. radiodurans utilizes synthesis-dependent strand annealing (SDSA), a primary HR subpathway for DSB repair (Zahradka et al., 2006; Elbakry and Löbrich, 2021). However, the presence of HR proteins with unconventional features, such as RecA and UvrD, combined with Deinococcus-specific proteins like DdrA and DdrB, amplifies the effectiveness of DSB repair (Bentchikou et al., 2010; Timmins and Moe, 2016). These combined actions potentially render the D. radiodurans version of SDSA, known as extended-SDSA (ESDSA), distinct from those of standard organisms. Besides traditional enzymatic antioxidants like catalase and superoxide dismutase, D. radiodurans’ radiation resistance is thought to be fortified by antioxidant complexes that combine manganese ions with metabolites, shielding proteins from oxidative damage (Daly et al., 2010). These findings suggest that the primary determinant of radiation resistance is proteome protection rather than genome preservation (Sharma et al., 2017). The distinct characteristics of this bacterium position it as an invaluable model for exploring various biological processes, including DNA repair, oxidative defense, and radiation biology.
Deinococcus radiodurans has also demonstrated potential applications in fields like bioremediation and biotechnology. This bacterium has been genetically tailored to remove heavy metals from radioactive locations or to possess enhanced biosorption capacity for uranium (Brim et al., 2000; Manobala et al., 2019). Biomaterials from D. radiodurans, including carotenoids (deinoxanthin), exopolysaccharides (DeinoPol), and membrane vesicles, exhibit significant industrial promise. Due to their antioxidant and radioprotective properties, these derivatives can suit food, cosmetics, and pharmaceutical industries (Farci et al., 2017; Jeong et al., 2020; Park et al., 2022; Han et al., 2023). The genetic materials of D. radiodurans have gained interest among researchers. By introducing the D. radiodurans regulators PprI (also called IrrE) and DR_1558 into industrial microbes, these microbes exhibit increased resistance to environmental stresses and produce more valuable natural compounds (Appukuttan et al., 2015; Park et al., 2020; Wang et al., 2024). A new RNA-guided nuclease suitable for genome editing was recently discovered in the D. radiodurans genome (Karvelis et al., 2021).
The D. radiodurans strain isolated in 1956 was named R1 and was designated ATCC 13939 at the ATCC. Its genome was sequenced in 1999 (White et al., 1999). However, it was discovered that the sequenced genome was not actually from the ATCC 13939 strain. Therefore, the sequenced R1 was re-designated as D. radiodurans R1 ATCC BAA-816 (White et al., 2004). Over the past two decades, several studies have noted genetic discrepancies between ATCC BAA-816 and ATCC 13939, resulting in a frameshift (Southworth and Perler, 2002; Mennecier et al., 2004; Cao and Julin, 2009; Niiranen et al., 2015). These genetic variations might have emerged due to separate laboratory cultivation (White et al., 2004). Since both strains have been instrumental in Deinococcus research, identifying the nucleotide sequence variations between ATCC 13939 and ATCC BAA-816 would be of significant interest to both the Deinococcus research community and those researchers studying DNA repair and oxidative stress. In this study, we sequenced our lab’s stock strain of D. radiodurans R1 ATCC 13939, referred to as ATCC 13939K, and compared it with the published ATCC BAA-816 genome to pinpoint genetic variations.
The genomic characteristics of ATCC 13939K were analyzed in comparison with the ATCC BAA-816 strain and three additional D. radiodurans R1 genome sequences available in the National Center for Biotechnology Information’s (NCBI) Genome database. These sequences, referred to as R1-2016 (Hua and Hua, 2016), ATCC 13939E (Repar et al., 2021), and ATCC 13939O (Eugénie et al., 2021), were included to provide a comprehensive comparison of genomic attributes across strains (Table 1). Each strain encompasses two circular chromosomes and two plasmids, pMP and pCP. The aggregate genome length of ATCC 13939K stands at 3,285,071 bp, accompanied by a G + C content of 66.65%. Bioinformatic predictions have identified a sum of 3,150 protein-coding sequences (CDSs), in addition to 50 tRNA genes and nine rRNA genes (with three copies each of 5S, 16S, and 23S) within its genome (Table 1). The genome magnitude of ATCC 13939K aligns closely with that of BAA-816 (3,284,156 bp) and is roughly six kbp in excess compared to ATCC 13939E and ATCC 13939O. This disparity is ascribed mainly to variations in the length of chromosome 1 among the strains (Table 1). Prominent insertions discerned in chromosome 2, as well as the two plasmids of R1-2016 (Hua and Hua, 2016), were absent in other R1 strains (Table 1). In pairwise alignments, ATCC 13939K exhibited a 99.98% nucleotide-level identity with BAA-816 and manifested considerable synteny conservation, underscoring its collinearity and the absence of pronounced genomic structural variations.
In the genomic comparison between BAA-816 and ATCC 13939K, a total of 436 DNA sequence differences were identified, including 100 single nucleotide variations (SNVs), 278 insertions with sizes ranging from 1 to 6 base pairs (bp), and 58 short deletions (Table 2 and Supplementary Table S1). Previous studies have shown that R1-2016 and ATCC 13939E exhibit 577 and 559 genetic variations compared to BAA-816 (Hua and Hua, 2016; Repar et al., 2021). The genomic alignment of these differences with the BAA-816 genome revealed that 107 are located in intergenic regions, while the majority, 329 (75.7%), occur within gene regions, including pseudogenes and the 23S rRNA genes (Table 2 and Supplementary Table S1). These variations are observed across 259 distinct genes, with several genes harboring multiple differences. For instance, gene DR_1922, encoding exonuclease SbcC, presented with 4 SNVs and 5 insertional events (Supplementary Table S1). Among the 76 SNVs detected in CDSs, 21 were synonymous substitutions, while 53 led to amino acid (aa) changes. Notably, 2 SNVs lead to the elimination of stop codons in genes DR_1333 and DR_2250 (Supplementary Table S1). Furthermore, 241 insertion or deletion (InDel) events were identified, resulting in frameshifts in 164 CDSs and alterations in the reading frames of 46 pseudogenes (Supplementary Table S1).
Predominantly, InDel events lead to modifications in the length of CDSs or cause the fusion of genes. In the latter case, two or three adjacent genes may merge to form a singular CDS. For a comparative analysis of alterations in the CDSs, we reconfigured the initiation positions of each chromosome and plasmid in ATCC 13939K to coincide with those of BAA-816. Additionally, the designation of the predicted CDSs prefixed with ‘KDR’ was synchronized with the inaugural annotation of the BAA-816 genes. Within the BAA-816 genome, the first gene of chromosome 1, designated DR_0001, is characterized to encode the DNA polymerase III β clamp (DnaN) consisting of 393 aa, as per the initial annotation provided by the authors (GenBank genome AE000513.1). However, this CDS is omitted in the annotation crafted by NCBI (NC_001263.1) (Supplementary Table S2). This absence suggests that the protein homology in particular segments might be insufficient to validate protein-coding designations robustly (Tatusova et al., 2016). In the ATCC 13939K genome, the dnaN gene is newly annotated and assigned the locus tag KDR_0001. This gene spans 1,086 nucleotides and encodes a β-clamp of 361 aa (Supplementary Table S2), a consequence of a 1-bp deletion corresponding to guanine (G) at the 1,037th position in DR_0001 (Supplementary Table S1). This observation is consistent with a previous study (Niiranen et al., 2015).
A comparative genomic analysis among Deinococcus species revealed several annotation inaccuracies in the foundational genomes of BAA-816 (AE000513.1, AE001825.1, AE001826.1, and AE00001827.1). Many of these misannotations were rectified in the reannotated genomes (NC_001263.1, NC_001264.1, NC_000958.1, and NC_000959.1) using the annotation protocols from NCBI. More information on this can be found at https://ncbi.nlm.nih.gov/refseq/about/prokaryotes/reannotation/. An illustrative example is DR_0003. When its orientation is reversed, this gene encodes the DNA damage response protein DdrC (de Groot et al., 2009). Within the reference sequence NC_001263.1, DdrC is appropriately annotated as DR_RS00015 and corresponds to KDR_0003r from ATCC 13939K (Supplementary Table S2). Consequently, the features of the predicted CDSs from ATCC 13939K were also juxtaposed with those in the NCBI-curated annotation (Supplementary Table S2).
Upon examining the CDS across the ATCC 13939 strains, we observed that 2,557 CDSs possess the same length. In addition, 2,629 ATCC 13939K CDSs align with those in ATCC 13939E, while 2,584 match ATCC 13939O (Supplementary Table S3). Within the ATCC 13939K genome, the second gene on chromosome 1, labeled KDR_0002, is annotated as encoding a DnaA protein with a length of 454 aa. In contrast, the corresponding DnaA proteins in ATCC 13939E and ATCC 13939O are annotated as having 466 aa (Supplementary Table S3). This discrepancy arises from different annotation procedures, as the gene exhibits no sequence differences (Supplementary Table S1). Considering the three CDSs, DR_RS00010, E5E91_RS00010, and DRO_RS00010, annotated by NCBI share consistency (Supplementary Tables S2, S3), there is a possibility that the length of KDR_0002 might undergo revision in future NCBI annotations. In addition, among the newly identified CDSs in ATCC 13939K, several with a length of less than 50 aa, such as KDR_0131n, 0333n, and 0347n, are absent in other 13,939 strains (Supplementary Table S3), indicating that NCBI may exclude these short CDSs in their annotation processes. To better understand the genetic variations across the sequenced R1 strains, we juxtaposed the ATCC 13939K genes that exhibit sequence differences (Supplementary Table S1) against their counterparts annotated in R1-2016, ATCC 13939E, and ATCC 13939O (Supplementary Table S4).
In D. radiodurans, one of the most critical stress response mechanisms is the DNA repair process. Numerous research articles published in the last few decades have focused on the composition of DNA repair systems and found discrepancies in the BAA-816 genome sequence. For example, DR_0099 is predicted to code for a single-stranded DNA-binding (SSB) protein consisting of 143 aa in strain BAA-816 (Supplementary Table S2). However, in ATCC 13939, a continuous 906-bp ORF that includes DR_0099 and DR_0100 has been identified, encoding a larger deinococcal SSB protein (Eggington et al., 2004), which matches the SSB proteins observed in ATCC 13939K and other R1 strains examined in this study (Supplementary Table S4).
In the ATCC 13939K strain, the genes DR_1258 and DR_1259, initially believed to code for the SNF2/Rad54 helicase-related protein and SNF2/Rad54 helicase, respectively, were identified as a single ORF (KDR_1258m) encoding Snf2 intein (Supplementary Table S4). The existence and splicing activity of this Snf2 intein were empirically validated (Southworth and Perler, 2002). Frameshifts were identified in several DNA replication and repair genes, such as dnaN (DR_0001), mutS1 (DR_1039), and recJ (DR_1126) in strain ATCC 13939K (Supplementary Table S1), aligning with previous research (Mennecier et al., 2004; Bentchikou et al., 2010; Niiranen et al., 2015). These CDSs, absent in the BAA-816 genome NC_001263.1, have been restored in ATCC 13939K and other R1 strains (Supplementary Table S4).
The protein DR_2566, which includes the domain of unknown function (DUF) 2,726 (Supplementary Table S2), is presumed to be a very short patch repair (VSR)-like nuclease (Slade and Radman, 2011; Steczkiewicz et al., 2012). While DR_2566 and KDR_2566 have comparable lengths of 168 aa and 172 aa, respectively (Figure 1A and Supplementary Table S4), they show variations in the 1 to 60 aa residue region at their N-termini (Supplementary Figure S1). An examination of the KDR_2566 nucleotide sequence indicates the presence of an additional cytosine (C) at position 182, leading to a frameshift in the N-terminal region (Supplementary Figure S1 and Supplementary Table S1).
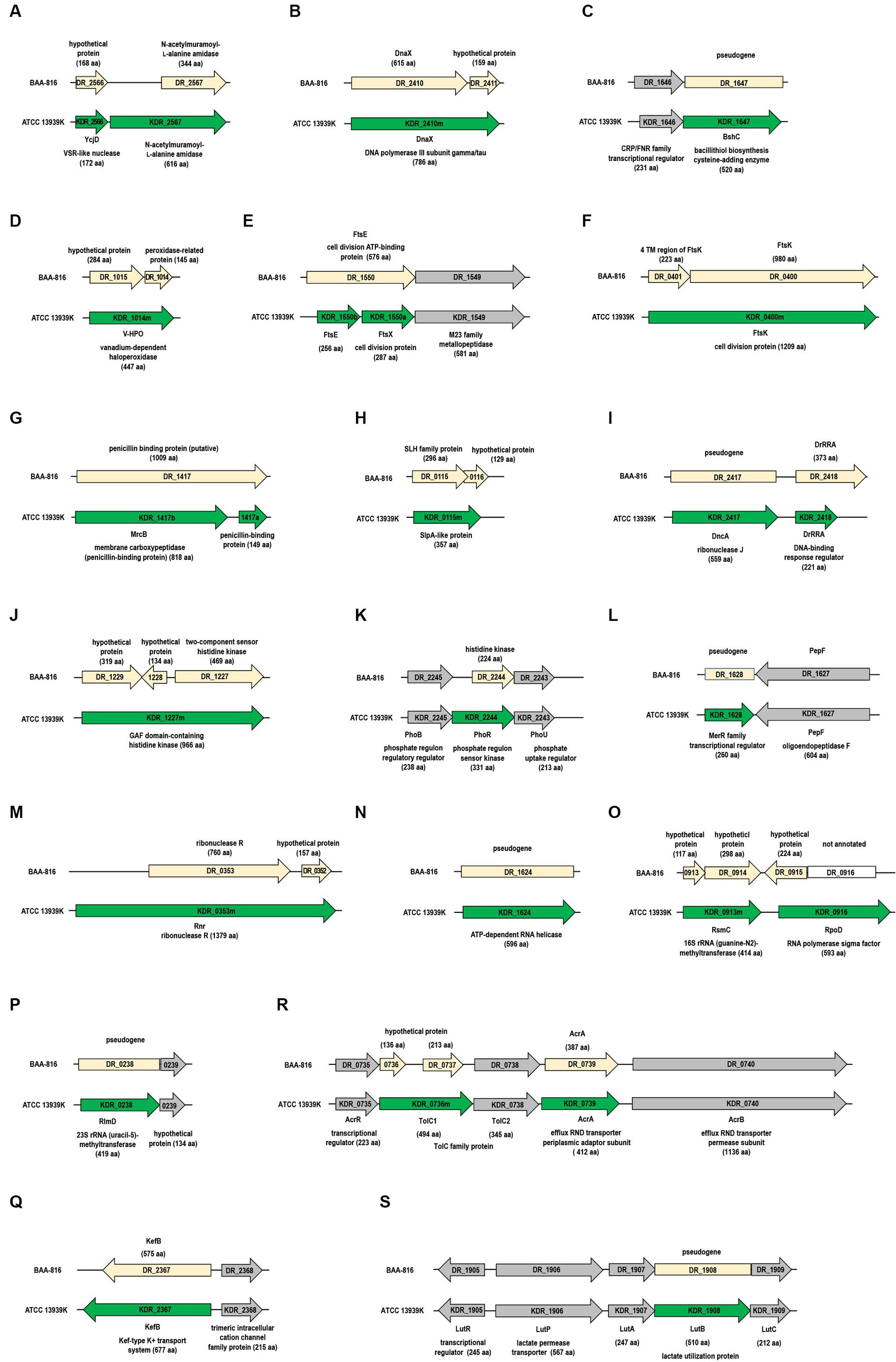
Figure 1. Differences in gene configurations between the BAA-816 and ATCC 13939K strains. Light yellow arrows represent genes from the BAA-816 strain, while genes from the ATCC 13939K strain are depicted with green arrows. Genes are categorized by their protein functions: (A,B) DNA repair proteins, (C,D) antioxidant proteins, (E,F) cell division proteins, (G,H) cell wall proteins, (I–K) two-component regulatory systems, (L,M) transcriptional regulators, (N–P) RNA metabolism proteins, and (Q to S) transporters. Conserved genes that are present in both strains are indicated with gray arrows. The direction of transcription for each gene is marked with arrowheads. Each gene is labeled with its locus tag, corresponding to the description of its associated protein.
The DNA polymerase III holoenzyme is composed of two dimerized β subunits, a core Pol III dimer, and a unique γ complex (consisting of γ, τ, δ, δ’, χ, ψ) that assists in loading the β processivity clamp to the DNA template. Both γ and τ proteins are encoded from the dnaX gene (Hejna and Moses, 2009). While the dnaX ORF aligns with the τ protein’s size, the shorter γ protein, which aligns with τ’s N-terminal fragment, is generated through programmed translational frameshifting (Farabaugh, 1996). Although DR_2410 was predicted to produce both proteins, a significant size discrepancy was observed in τ (Makarova et al., 2001). Our observations indicate that strain ATCC 13939K lacks the thymine (T) at position 1,825 in DR_2410 (Supplementary Figure S2). As a result, DR_2410 and DR_2411 merge into a unified ORF in ATCC 13939K, termed KDR_2410m (Figure 1B). The length of DnaX in other R1 strains is also 786 aa (Supplementary Table S4), suggesting a need for further experimental validation to determine the synthesis and accurate size of γ and τ.
In D. radiodurans, the thiol known as bacillithiol (BSH) plays a role in neutralizing hydrogen peroxide (H2O2) in collaboration with the unique bacilliredoxin AbxC (Jeong et al., 2021). The enzyme BshC, which is integral to the final stage of BSH synthesis by adding cysteine, is commonly observed to have a length exceeding 500 aa in BSH-producing bacteria (Gaballa et al., 2010). In the KDR_1647 DNA sequence, an extra C at the 954th position leads to coding for a 520-aa BshC (Figure 1C and Supplementary Figure S3). In contrast, DR_1647 is annotated as a pseudogene (Supplementary Table S2).
Vanadium-dependent haloperoxidases (V-HPOs) are part of a group of non-heme enzymes that utilize peroxide alongside manganese catalases and thiol peroxidases, such as peroxiredoxins and glutathione peroxidases (Bernroitner et al., 2009). V-HPOs facilitate the oxidation of halides (including I−, Br−, and Cl−) when H2O2 is present (Leblanc et al., 2015). In the BAA-816 strain, the genes DR_1015 and DR_1014, coding for a hypothetical protein and a peroxidase-related protein, respectively, are separate (Figure 1D). But, in the ATCC 13939K strain, these genes combine into a single 1,344 bp gene, KDR_1014m, encoding V-HPO. This combination is due to G found at the 778th position (Figure 1D and Supplementary Figure S4).
The Ddr (DNA damage response) and Ppr (pleiotropic protein promoting DNA repair) proteins in D. radiodurans are crucial for the bacterium’s remarkable ability to withstand and repair extreme DNA damage (Lim et al., 2019). Similarly to ddrC, ddrH is transcribed from the reverse strand at locus DR_0438 (de Groot et al., 2009). The ddrH gene is correctly annotated as KDR_0438r in ATCC 13939K (Supplementary Table S5). In the BAA-816 strain, DR_0997 (ddrI) is predicted to encode a 260-aa cyclic AMP receptor protein (CRP), which acts as a global transcriptional regulator. Deletion of G at position 543 in DR_0997 leads to a truncated version of the DdrI protein in ATCC 13939 (Yang et al., 2016). This deletion is noted in KDR_0997, resulting in a DdrI protein of 203 aa in length (Supplementary Tables S1, S5). The insertion of G into DR_1440 restored the complete ORF for KDR_1440, along with its functional protein form (Supplementary Tables S1, S5). KDR_1440, known as DdrM, potentially functions as an iron efflux protein, contributing to iron homeostasis and enhancing resistance to oxidative stress in D. radiodurans (Dai et al., 2018).
PprI is a protease that activates DNA repair genes by cleaving DdrO. This trans-acting repressor binds to RDRM (radiation/desiccation response motif) sequences in gene promoters (Lim et al., 2019). These RDRM sequences, acting as cis-regulatory elements, are integral to the radiation desiccation response (RDR) genes in D. radiodurans (Eugénie et al., 2021). Frameshifts were detected in several RDR proteins, including SSB, MutS1, HelD, DR_C0017, and DR_C0023 (Supplementary Tables S1, S5).
The cell division proteins of D. radiodurans, FtsA (DR_0630) and FtsZ (DR_0631), have been examined through both in vitro and in vivo studies (Modi and Misra, 2014; Maurya et al., 2018). For other Fts proteins like FtsE (DR_1550), FtsW (DR_2497), and FtsQ (DR_0629), their interactions with chromosome partitioning Par proteins have been investigated (Maurya et al., 2016). FtsE acts as the cytoplasmic ATP-binding component, while FtsX is the membrane-bound counterpart. These two proteins form a membrane-associated complex resembling an ATP-binding cassette (ABC)-type transporter. The ftsE and ftsX genes are located side-by-side in an operon (Pichoff et al., 2019). In the BAA-816 strain, DR_1550 is identified as a 576-aa FtsE. But, in the ATCC 13939K strain, this is divided into FtsE (KDR_1550b) and FtsX (KDR_1550a) (Figure 1E). This FtsEX complex plays a role in controlling the activities of the periplasmic peptidoglycan (PG) hydrolases (Pichoff et al., 2019). Notably, in the ATCC 13939K strain, the ftsE and ftsX genes are co-located with the KDR_1549 gene (Figure 1E), which encodes for enzymes from the M23 metallopeptidase family, commonly recognized for their PG hydrolase activity (Razew et al., 2022).
In Escherichia coli (E. coli), three N-acetylmuramoyl-L-alanine amidases, AmiA, AmiB, and AmiC, primarily facilitate PG hydrolysis (Pichoff et al., 2019). Putative amidase proteins, including DR_1387, DR_1632, DR_2394, DR_2567, and DR_C0013, have been identified in the BAA-816 genome through KEGG mapping. The G to C change, positioned 25 nucleotides upstream of the GTG start codon for DR_2567, results in a long ORF termed KDR_2567 in strain ATCC 13939K (Supplementary Figure S5). This modified ORF encodes a protein of 616 aa, in contrast to the 344 aa of the DR_2567 protein predicted from the BAA-8166 genome sequence, featuring an extension of 272 aa at its N-terminal end (Figure 1A). FtsK, involved in cell division and chromosome segregation, possesses transmembrane (TM) domains at its N-terminus (Bisicchia et al., 2013). In the strain ATCC 13939K, the proteins DR_0400 (annotated as FtsK) and DR_0401 (representing the 4TM domains of FtsK) are consolidated into a single protein, KDR_0400m (Figure 1F), corroborating a recent finding (Mishra et al., 2022).
The mrcB gene, corresponding to locus tag DR_1417 in BAA-816, encodes the penicillin-binding protein 1b (PBP1b), which is crucial for preserving PG integrity and structure (Vigouroux et al., 2020). In BAA-816, this protein is 1,009 aa long, whereas it spans 807 to 818 aa in other R1 strains (Figure 1G and Supplementary Table S4). SlpA (DR_2577) is a primary S-layer protein responsible for sustaining the integrity of the D. radiodurans cell envelope (Farci et al., 2015). Other proteins like DR_0383, DR_1115, DR_1124, and DR_1185 are also identified as potential S-layer proteins (Makarova et al., 2001). Notably, in the ATCC 13939K strain, DR_0115 and DR_0116, which are separate in BAA-816, merge to form a singular gene, KDR_0115m (Figure 1H), which encodes for a SlpA-like protein (von Kügelgen et al., 2022). KDR_2572m, a fusion of DR_2572 and DR_2573, encodes a bactofilin with potential cytoskeletal functions (Supplementary Table S4). However, no reported studies have explored the functional role of bactofilins in D. radiodurans.
The two-component systems (TCSs), which comprise a sensor histidine kinase (HK) and a cytoplasmic response regulator (RR), are pivotal in the radioresistance of Deinococcus species (Lim et al., 2019). DrRRA is a novel RR essential for the radioresistance of D. radiodurans. Mutations in this gene reduce γ-radiation resistance and induce widespread transcriptional changes in numerous genes, predominantly those associated with DNA repair (Wang et al., 2008). Insertion of a G at the 647th position results in a frameshift in the DrRRA gene of ATCC 13939K (Supplementary Table S1). Consequently, the DrRRA protein (KDR_2418) is shortened to 221 aa, compared to the 373 aa previously described for BAA_816 (Figure 1I). Expression analysis confirms that the actual size of DrRRA is 221 aa (Liu et al., 2012). KDR_1227m features two consecutive GAF domains in its N-terminal region and an HK domain at the C-terminus. This configuration appears as a merged product of DR_1227, DR_1228, and DR_1229 (Figure 1J).
In bacterial cells, the PhoBR TCS, comprising PhoB (RR) and PhoR (HK), triggers the transcription of genes that encode the PstSCAB system. This ABC transporter system is essential for the high-affinity uptake of phosphate ions, particularly under phosphate-deprived conditions (Santos-Beneit, 2015). In D. radiodurans, the PstSCAB complex is encoded by the genes DR_A0157 through DR_A0160 (Supplementary Table S2). In ATCC 13939K, a 996-bp ORF spans DR_2244 and its upstream region, labeled as KDR_2244, encoding a 331 aa PhoR protein (Figure 1K). This ORF contains an additional C at position 2,238,476, situated 150 nucleotides upstream of the GTG translation initiation codon of DR_2244 (Supplementary Table S1). In other R1 strains, the predicted size of this protein is 305 aa (Supplementary Table S4). Mutations in phoR have been observed to increase γ-radiation sensitivity (Im et al., 2013). Furthermore, when exposed to H2O2, D. radiodurans accumulates polyphosphate granules (Dai et al., 2021), suggesting a role for the PhoBR TCS in its oxidative stress response.
PerR, a homolog of the Fur (ferric uptake repressor), acts as a transcriptional regulator in response to peroxide (Lim et al., 2019). A Fur homolog, designated DrPerR, has been newly annotated in ATCC 13939 (Liu et al., 2014). The DrPerR gene sequence (GeneBank accession number KJ817356) features a G insertion at position 1:2,340,150 compared to the BAA-816 genome. This insertion creates an ORF that encodes an 80 aa DrPer protein (Liu et al., 2014). The same G insertion is found in ATCC 13939K (Supplementary Table S1), leading to the assignment of the locus tag KDR_2341n to DrPerR (Supplementary Table S2).
The MerR and SmtB/ArsR families represent two general classes of transcriptional regulatory proteins that play crucial roles in heavy metal stress response (Busenlehner et al., 2003). In the genome AE000513.1, DR_1628 is a pseudogene. However, in NC_001263.1, it is divided into two genes: DR_RS08320, which codes for the MerR-type helix-turn-helix (HTH) domain, and DR_RS16830, encoding the TipAS antibiotic-recognition domain (Supplementary Table S2). In the ATCC 13939K strain, a nucleotide insertion at position 1,651,397 disrupted the stop codon of DR_RS08320 (Supplementary Tables S1, S2), leading to the fusion of these separate genes under the tag KDR_1628, which represents a MerR family transcriptional regulator (Figure 1L).
The merged gene from DR_0233 and DR_0234 produces the ArsR transcriptional regulator, denoted as KDR_0233m (Supplementary Table S4). The N-terminal region of KDR_0233m includes a DNA-binding HTH domain typical of the ArsR family, while the C-terminus encompasses DUF2087 (Supplementary Table S2). A further experimental investigation is required to elucidate the functions of these newly identified full-length proteins.
Ribonucleases (RNases) are a set of enzymes involved in all aspects of RNA metabolism. Specifically, RNase J is distinguished by its capability to serve as both an endonuclease and a processive 5′ exonuclease, crucial for RNA processing and breakdown (Richards and Belasco, 2011). While the DR_2417 ORF is labeled as a pseudogene, KDR_2417 is recognized to encode the functional RNase J (Figure 1I and Supplementary Table S2) due to a missing adenine (A) at the 995th position (Supplementary Table S1). Sequencing of the PCR product amplified from ATCC 13939 confirmed the absence of frameshift (Das and Misra, 2012).
RNase II and RNase R belong to the RNR (ribonucleotide reductase) superfamily, characterized by nonspecific, 3′ to 5′ processive exoribonuclease activity. DR_0020 is classified as an RNase II-type enzyme (Schmier et al., 2012). In the BAA-816 genome, the DR_0353 gene encodes an RNase R protein of 760 aa (Supplementary Table S2). In contrast, ATCC 13939K features a single ORF, KDR_0353m, extending across DR_0352, DR_0353, and the upstream region, which produces a 1,379 aa RNase R protein (Figure 1M). Typically, deinococcal RNase R proteins range in size from 1,000 to 1,500 aa (data not shown).
RNA helicases, which modify RNA secondary structures and RNA-protein interactions, play essential roles in RNA metabolism (Owttrim, 2013). Identified within the DEAD-box protein family is a presumptive RNA helicase, KDR_1624, which manifests as a full-length protein comprising 596 aa (Figure 1N). A two-nucleotide deletion in DR_1624, previously annotated as a pseudogene in BAA-816 (Supplementary Table S1), restored its ORF.
A two-nucleotide deletion in DR_1624, previously annotated as a pseudogene in BAA-816, corrected the frameshift error (Supplementary Table S1).
S-adenosylmethionine-dependent methyltransferases (MTases) play a role in RNA post-transcriptional modifications by adding a methyl group to ribosomal RNA (rRNA) nucleotides (Mosquera-Rendón et al., 2014). One such MTase, RsmC, modifies G to 2-methylguanosine (m2G) at the 1,207 position of 16S rRNA and can function as an RNA chaperone protein, facilitating ribosome assembly (Keshav et al., 2020). In ATCC 13939K, KDR_0913m, a unified ORF of DR_0913 and DR_0914, is identified as RsmC (Figure 1O). RlmD catalyzes the formation of 5-methyl-uridine at position 1939 (m5U 1939) in 23S rRNA, a function widely observed in bacteria and eukaryotes (McCown et al., 2020). While KDR_0238 encodes this protein in ATCC 13939K, DR_0238 is classified as a pseudogene in BAA-816 (Figure 1P).
The translocation and assembly module (TAM) plays a role in the transporting and secretion of outer membrane proteins. It has been reported that the three ORFs, DR_1460, DR1461, and DR_1462, merge to form a single ORF that encodes a 4,002 aa long TamB homolog (Yu et al., 2017). Comparison between BAA-816 and ATCC 13939K reveals five individual G insertions and one SNV in the KDR_1460m locus (Supplementary Tables S1, S2), aligning with the previous report (Yu et al., 2017). DR_2367 encodes the potassium-efflux system protein KefB, which is 575 aa long. In ATCC 13939K, a G insertion at DR_2367’s stop codon creates a frameshift, extending the KefB protein (KDR_2367) by 100 aa, with an additional G insertion at position 2,365,232 (Figure 1Q and Supplementary Table S1). The resulting 677-aa KefB protein includes a Zn-finger-like domain at its C-terminus.
Resistance-nodulation-cell division (RND) transporters are drug efflux pumps known for removing various toxic substances, including antibiotics. Initially believed to be unique to gram-negative (GN) bacteria, genes that encode proteins with the structural hallmarks of RND systems have been identified in gram-positive (GP) organisms like Corynebacterium glutamicum and Bacillus subtilis (Schindler and Kaatz, 2016). A well-known example is the AcrAB-TolC system in E. coli. The tripartite RND systems are typically composed of the transmembrane AcrB protein (1,049 aa), the periplasmic AcrA protein (397 aa), and the outer membrane TolC protein (493 aa) (Jang, 2023). The local repressor AcrR is also upstream of the acrAB operon (Alvarez-Ortega et al., 2013). In ATCC 13939K, a unified ORF combining DR_0736 and DR_0737, designated as KDR_0736m, encodes TolC1 (494 aa) (Supplementary Table S4). The KDR_0735-KDR_0740 operon, which codes for AcrR, AcrA, AcrB, and TolC (Figure 1R), suggests that the RND complex in D. radiodurans is operational.
In D. radiodurans, fructose is the preferred carbohydrate source (Venkateswaran et al., 2000). The bacterial phosphoenolpyruvate phosphotransferase system (PTS), responsible for carbohydrate transport and phosphorylation, includes cytoplasmic energy-coupling proteins such as enzyme I (EI) and HPr, along with sugar-specific enzyme II (EII) complexes (Comas et al., 2008). In Pseudomonas putida, the fruR-fruBKA operon is involved in fructose uptake, where fruB codes for a unique multi-phosphoryl transfer protein that integrates EIIAFru-HPr-EI domains (Chavarría et al., 2016). Upon resequencing, a complete coding sequence for FruB (KDR_B0075) was identified in ATCC-13939K (Supplementary Tables S1, S4). Additionally, the entire fruR-fruBKA operon is preserved in the ATCC 13939 strains (Supplementary Table S3).
In B. subtilis, lactate utilization, specifically L-lactate conversion to pyruvate, is facilitated by the lutABC operon, which encodes three iron–sulfur-containing proteins. Typically, this conserved operon is situated alongside the lutR and lutP genes responsible for coding a transcriptional regulator and lactate permease, respectively (Chai et al., 2009). However, in the case of DR_1908, which aligns with lutB, it’s noted as a pseudogene, with only a partial LutB protein (DR_RS09770) present in the NCBI-annotated genome NC_001263.1 (Supplementary Table S2). A previously identified frameshift in DR_1908 (Hwang et al., 2013) has been corrected by a C insertion at position 1,928,139 in ATCC-13939K, restoring the ORF (Figure 1S and Supplementary Table S1). It is worth noting that D. radiodurans can use lactate as its sole carbon source (Venkateswaran et al., 2000).
Gene gain and loss often happen through the addition and removal of various-sized genome segments, frequently involving mobile genetic elements (MGEs) (Iranzo et al., 2019). Intercellular MGEs like plasmids and phages facilitate DNA transfer between bacterial cells. On the other hand, DNA movement within cells is primarily mediated by specific MGEs such as transposons (Tns) and insertion sequences (ISs). IS elements represent the most basic type of Tn, containing only essential genes for their transposition process (Bennett, 2004). Notably, IS elements are more abundant in D. radiodurans than E. coli and B. subtilis, which serve as model organisms for GN and GP bacteria (Makarova et al., 2001).
In D. radiodurans, the ISDra2 element, originally termed IS8301 and belonging to the IS200/IS605 family, comprises two genes: tnpA and tnpB. These genes code for transposase and RNA-guided DNA nuclease (Islam et al., 2003; Karvelis et al., 2021). ISDra2’s transposition activity significantly increases under γ-radiation or UV radiation exposure (Mennecier et al., 2006). In the ATCC 13939 strain, ISDra2 exists as a single functional copy (DR_1652-DR_1651) and one inactive, degenerate copy (DR_0177-DR_0178), while BAA-816 contains seven complete and one incomplete ISDra2 copies (Pasternak et al., 2010). Comparative genomic analysis revealed the loss of six ISDra2 elements in ATCC 13939K (Figures 2A–F) and other examined 13,939 strains (Supplementary Table S3). This loss led to the reconstitution of genes disrupted by ISDra2 insertion. For instance, KDR_1930m and KDR_2322m, coding for α/β-hydrolase fold enzyme and serine protease were restored (Figures 2D,E). In the ATCC 13939K strain, the ISDra2 segment DR_0979-DR_0978 was replaced by five ORFs (Figure 2F). Among these ORFs, KDR_0977n2 is not defined in other 13939 strains (Supplementary Table S3). The enzyme acetoin dehydrogenase, produced by the genes KDR_0977n3 (acoA) and KDR_0977n4 (acoB), plays a critical role in the breakdown of acetoin (Xiao and Xu, 2007). Additionally, the genes KDR_0980n and KDR_0980 produce glutamate dehydrogenase (Gdh), which facilitates the reversible conversion of glutamate into α-ketoglutarate and ammonia (Miñambres et al., 2000). In a unique case of gene disruption by ISDra2, the segment KDR_1963n1-KDR_1963n2 is integrated into the pilT gene, which codes for the pilus retraction ATPase (Figure 2G). This disruption is exclusive to the ATCC 13939K strain and does not occur in other 13939 strains (Supplementary Table S3). Given that the DNA translocation system in D. radiodurans is associated with type IV pili, mutations in the pilT gene lead to a decrease in the efficiency of natural transformation (Ithurbide et al., 2020). The reduced transformation efficiency was observed in the ATCC 13939K strain (Supplementary Figure S6). This disruption in the 13939K strain is a notable genomic difference that affects the strain’s phenotype.
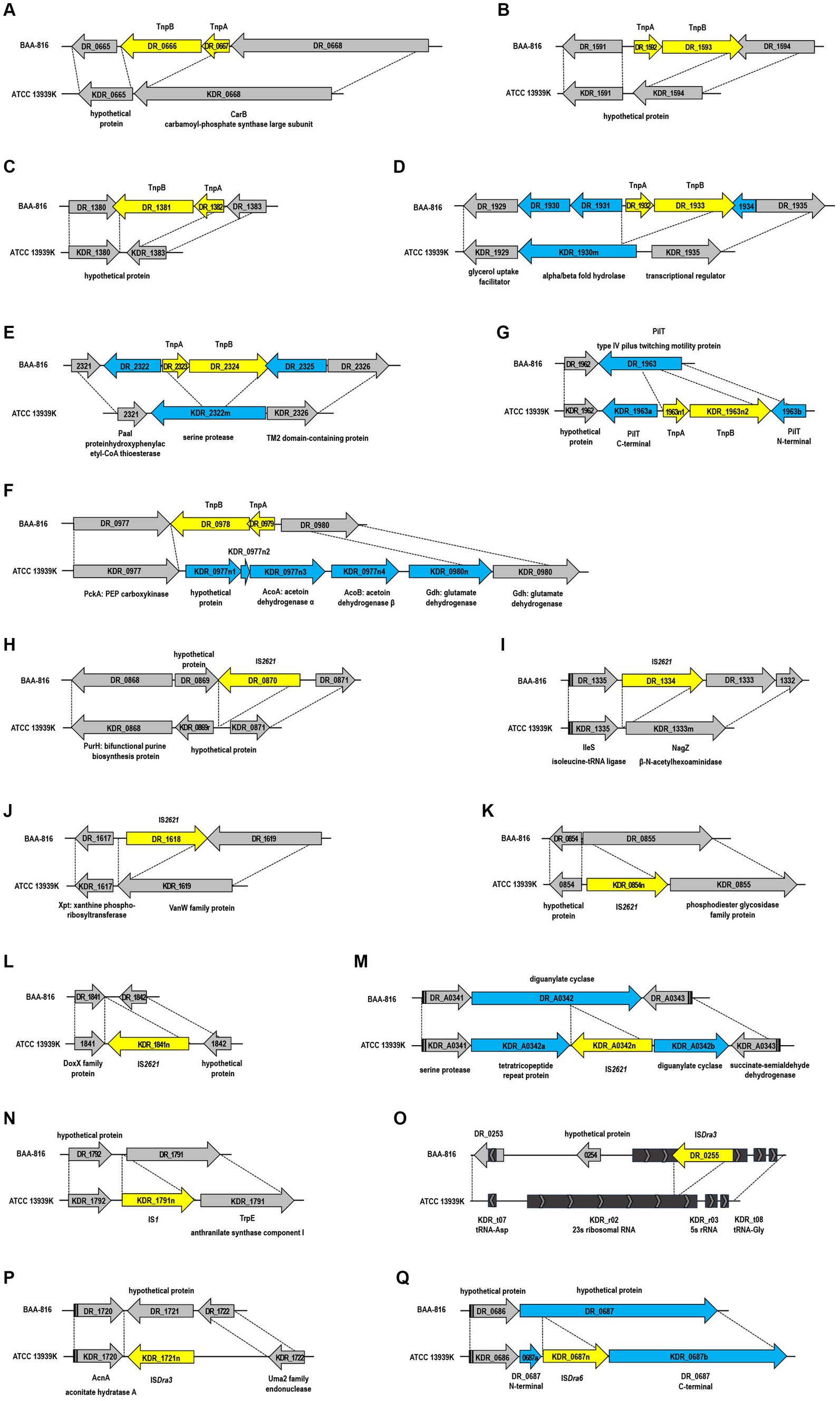
Figure 2. Structural genomic variations mediated by insertion sequences (IS) between the BAA-816 and ATCC 13939K strains. The IS elements are represented by yellow arrows and are categorized as follows: (A–F) ISDra2, (H–M) IS2621, (N) IS1, (O,P) ISDra3, and (Q) ISDra6. Genes that are either restored, disrupted, or newly identified by the transposition of IS elements are denoted by blue arrows. Genes that are conserved across both strains are indicated by gray arrows. Dashed lines delineate homologous regions shared between the genomes of the two strains. Each gene is labeled with its locus tag.
IS2621, spanning 1,322 bp and enclosed by 19-bp perfect inverted terminal repeats, represents another transpositionally active IS in D. radiodurans (Narumi et al., 1997; Mennecier et al., 2006). The BAA-816 strain originally contained five copies of IS2621, identified as DR_0870, DR_1,334, DR_1618, DR_2222, and DR_B0059 (Long et al., 2015). However, in the ATCC 13939K strain, three of these copies were lost (Figures 2H–J), aligning with findings from previous research (Long et al., 2015). Additionally, ATCC 13939K acquired three new IS2621 copies (Figures 2K–M). Among these, two copies, KDR_0854n and KDR_1841n, were exclusively found in ATCC 13939K, while the third copy, KDR_A0342n, was present in other 13939 strains (Supplementary Table S3). The occurrence of IS elements, specifically in ATCC 13939K, including KDR_0854n, KDR_1841n, and KDR_1963n1-1963n2, was confirmed through PCR assay (Supplementary Figure S7). Cyclic-di-GMP (c-di-GMP) acts as a crucial second messenger in the signal transduction pathways of many bacteria, regulating various cellular processes. The cellular levels of c-di-GMP are controlled by a balance between its production, catalyzed by diguanylate cyclases that contain GGDEF domains, and its breakdown, mediated by phosphodiesterases that possess either EAL or HD-GYP domains (Petchiappan et al., 2020). DR_A0342 is notable for harboring both HD-GYP and GGDEF domains within its C-terminal region, illustrating its role in c-di-GMP homeostasis (Galperin et al., 2001; Pei and Grishin, 2001). The insertion of IS2621 into DR_A0342 splits it into two parts, KDR_A0342a and KDR_A0342b (Figure 2M). Importantly, KDR_A0342b retains the intact domains (data not shown), suggesting its potential functionality in regulating c-di-GMP levels.
In the genome NC_001263.1, DR_RS16465 is identified as IS1, equivalent to KDR_0835r in the ATCC 13939K strain (Supplementary Table S2). Additionally, an extra copy of IS1 has been inserted in both ATCC 13939K (Figure 2N) and other 13939 strains (Supplementary Table S3). ISDra3, ISDra4, and ISDra6 have been classified as members of the IS630 family in the ISFinder database.1 These elements are characterized by sequences containing stretches of A residues. For instance, ISDra3, which includes DR_0255, DR_B0139, and DR_C0004, features a run of nine As, and ISDra6, incorporating DR_1523, DR_B0056, and DR_B0113, is marked by a sequence of eight As (Baranov et al., 2005). In the BAA-816 strain, the 23S rRNA genes were not fully characterized: rrnaA23S (DR_r05) and rrnaB23S (DR_r09) had a length of only 876 nucleotides each, while rrnaC23S (DR_r02) extended to 1,943 nucleotides, a result of the insertion by ISDra3 element DR_0255 (Pei et al., 2009; Supplementary Table S2). In ATCC 13939K, re-annotation and the absence of DR_0255 allowed for the identification of KDR_r02, KDR_r05, and KDR_r09, each at 2,876 nucleotides, harmonizing the lengths of all three 23S rRNA genes (Figure 2O and Supplementary Table S2). Furthermore, KDR_1721n and KDR_0687n were found as additional insertions of ISDra3 and ISDra6, respectively, in the ATCC 13939K strain (Figures 2P,Q), consistent with the previous report (Long et al., 2015).
Gene gain or loss in bacteria can occur without MGEs due to improper recombination processes, such as homologous recombination or inaccurate non-homologous repair activities. Errors in DNA replication or the repair process, mainly when fixing aberrant replication forks, can result in the deletion or duplication of genes. Consequently, this leads to either the loss or gain of genetic material (Periwal and Scaria, 2015). The genomic comparison between 13939K and BAA-816 reveals that the genes DR_A0268 and DR_A0269, present in BAA-816, are absent in the 13939K strain (Figure 3A). Examination of a 1,697 nucleotide sequence that includes DR_A0268 and DR_A0269 shows that a portion of DR_A0268, between coordinates 287,484 and 288,083, is nearly identical to the C-terminal sequence of DR_A0270, found between coordinates 289,001 and 289,600, with only three bases differing (Figure 3B). Additionally, the sequence spanning 288,083–288,973 on chromosome 2 aligns perfectly with the segment of chromosome 1 between coordinates 2,463,286–2,464,176 (Figure 3B). This alignment suggests that recombination between homologous sequences of DR_A0270 and DR_A0268 could have resulted in the loss of the DR_A0269 region in strain ATCC 13939K. Furthermore, DR_A0268 has been updated to DR_RS17005 in annotations (Supplementary Table S2), and the C-terminal aa sequences of KDR_A0270 match those of DR_RS17005 (Supplementary Figure S8). In the ATCC 13939K strain, there was a notable presence of two newly inserted genes, KDR_1221n1 and KDR_1221n2 (Figure 3C). Proteins in the drug/metabolite transporter (DMT) superfamily are characterized by a unique structure that includes varying numbers of TM α-helices, commonly at counts of 4, 5, 9, or 10 per protein. The DMT proteins with 10 TM segments are formed by duplicating a fundamental five-segment precursor within the gene (Jack et al., 2001). Specifically, the DMT protein encoded by gene KDR_1221n2 possesses 10 helical segments (data not shown), implying that it may have evolved through a gene duplication event.
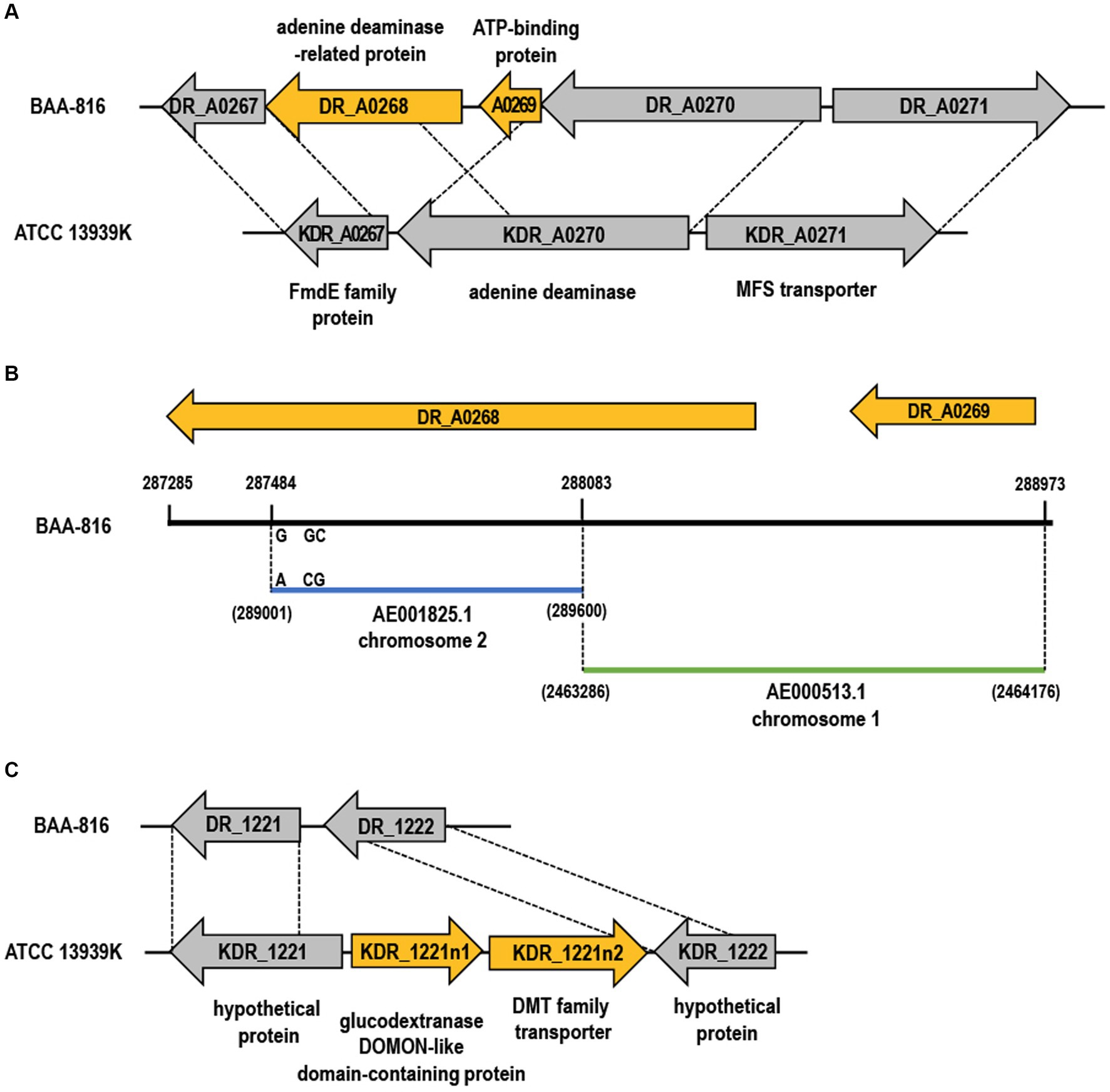
Figure 3. Gene gain and loss in the ATCC 13939K strain. (A,C) The genes lost and gained in ATCC 13939K are indicated by yellow arrows. Genes that are conserved across both strains are indicated by gray arrows. Homologous regions present in the BAA-816 strain are connected with dashed lines, and each gene is assigned a locus tag. (B) Chromosomal comparison of genes DR_A0268 and DR_A0269. A segment of DR_A0268 (yellow arrow) aligns nearly identically with the C-terminal end of DR_A0270 (blue bar), with a minor divergence of only three base pairs. The segment encompassing DR_A0269 (288,083–288,973) matches the region from 2,463,286 to 2,464,176 on chromosome 1 (green bar).
Identified initially as strain R1 in 1956 and cataloged as ATCC 13939, the D. radiodurans strain was subject to a later correction when it was determined that the sequenced genome belonged to a different strain, thereby reclassifying it as ATCC BAA-816. In this study, we sequenced the D. radiodurans strain ATCC 13939K and unveiled genetic differences relative to ATCC BAA-816. The comparative analysis uncovered many genetic variations, including SNVs and InDels, that substantially influence gene functions and structures. These variations induced alterations in CDS lengths, the restoration of pseudogenes, and occurrences of gene fusion and splits. Despite these genetic differences, phenotypic assays for resistance to γ-radiation, UV, H2O2, and mitomycin C showed similar survival rates between strains 13939K and BAA-816 (Supplementary Figure S9). The polymorphisms largely concur with those in other analyzed D. radiodurans strains (R1-2016, 13939E, and 13939O) (Supplementary Table S4). These findings suggest that many genetic discrepancies between the strains are likely attributable to sequencing errors in the BAA-816 reference genome rather than variations from different laboratory cultivation practices. This assumption is substantiated by proteomic analyses that have corrected previously identified frameshifts in the D. radiodurans genome (Willems et al., 2020). While resequencing of the R1 ATCC BAA-816 genome is needed for confirmation, updating the D. radiodurans genome sequence promises substantial insights for Deinococcus-focused research and broader studies into microbial resilience and DNA repair mechanisms.
In D. radiodurans, gene gain and loss are facilitated by IS elements. These elements can change their insertion location and copy numbers, introduce novel genetic functionalities, or alter gene activity, substantially impacting bacterial fitness (Vale et al., 2022). This dynamic reshaping of the genome is exemplified in the ATCC 13939K strain, where the ISDra2 element’s transposition resulted in the introduction of the genes necessary for acetoin metabolism (Figure 2F). Acetoin (3-hydroxy-2-butanone) is crucial in bacterial metabolism, acting as an essential intermediary for energy production from nutrients (Xiao and Xu, 2007). The pathway begins with α-acetolactate synthase (ALS) combining two pyruvate molecules into α-acetolactate (AL), which is converted to acetoin via α-acetolactate decarboxylase (ALD) or to diacetyl through non-enzymatic oxidative decarboxylation. Acetoin reductase (AR) then converts diacetyl to acetoin, which the acetoin dehydrogenase complex (ADC) transforms into acetyl-CoA for energy production via the tricarboxylic acid cycle (Xiao and Xu, 2007; Supplementary Figure S10).
The ilvBN genes encode α-acetohydroxy acid synthase (AHAS), another enzyme involved in AL formation, differing from ALS by its structure (Benson et al., 1996). ALS consists of a single approximately 60 kDa subunit. In contrast, AHAS is a two-subunit enzyme comprising a larger catalytic subunit (59–66 kDa) and a smaller regulatory subunit (9–35 kDa) (Eram et al., 2015). In the BAA-816 strain, the genes DR_1516 and DR_1517 are designated as ilvB and a pseudogene, respectively (Supplementary Table S2). However, a complete set of ilvBN genes is identified in the ATCC 13939K strain due to a C insertion within the sequence of KDR_1517 (Supplementary Tables S1, S2). The loss of the ISDra2 segment DR_0979-DR_0978 revealed the presence of ADC, encoded by the genes acoA (KDR_0977n3) and acoB (KDR_0977n4) (Figure 2F). These genes coding for AHAS and ADC are also present in other strains of ATCC 13939 (Supplementary Table S3). Moreover, ARs from Thermococcus (Ying and Ma, 2011) and Mycobacterium (Takeda et al., 2014) show about 30% similarity to the protein DR_A0005 (data not shown). Although a detailed functional analysis of DR_A0005 is necessary, D. radiodurans ATCC 13939 strains likely encompass the complete acetoin metabolism pathway (Supplementary Figure S10). This metabolic flexibility, alternating between acetoin production and consumption, allows the bacteria to adjust to variable environmental conditions, enhancing their growth and survivability. Acetoin is produced and excreted during the exponential growth phase to prevent cytoplasmic and extracellular over-acidification. In the absence of preferred carbon sources, as the culture transitions to the stationary phase, acetoin serves as a substrate to sustain culture density (Xiao and Xu, 2007). Hence, our findings enrich the comprehension of D. radiodurans’ adaptive mechanisms and pave the way for novel biotechnological applications.
The main pathways for integrating ammonia into biomolecules involve converting ammonia into glutamine (Gln) and glutamate (Glu) through three essential enzymes: glutamine synthetase (GS), glutamate synthase (also known as Gln:2-oxoglutarate aminotransferase, GOGAT), and glutamate dehydrogenase (GDH) (Reitzer, 2014). GS converts Glu into Gln, and GOGAT catalyzes the transfer of the amido group from Gln to 2-oxoglutarate (2OG), yielding two molecules of Glu. In the absence of GOGAT, GDH facilitates the direct synthesis of Glu from 2OG and ammonia. The GS-GOGAT pathway, therefore, represents a primary and widely conserved way of ammonia assimilation in bacteria (Pengpeng and Tan, 2013). In D. radiodurans, there are two versions of GS, identified as DR_2033 (GlnA3), which has been reannotated to DR_RS10425, and DR_0451 (GlnA1), along with GOGAT, encoded by DR_0183 (gltB) and DR_0182 (gltD) (Venkateswaran et al., 2000; Ghosal et al., 2005; Supplementary Table S2). An insertion and deletion of G nucleotide occurred in DR_RS10425, but this variation did not result in a frameshift (Supplementary Tables S1, S2). Instead, it caused a substitution of serine for isoleucine at position 496 in KDR_2033 (data not shown), maintaining the length of both GlnA3 proteins at 719 aa (Supplementary Table S2).
Given that the glutamate dehydrogenase (GDH)-dependent pathway for ammonia assimilation is energy-efficient and exhibits low ammonium affinity, it assumes a significant role in nitrogen metabolism, particularly in environments with abundant nitrogen (Pengpeng and Tan, 2013; Reitzer, 2014). GDH is categorized into three types based on its coenzyme specificity and role: NAD-dependent GDH2 (EC 1.4.1.2) primarily facilitates the production of 2-oxoglutarate (2OG) through glutamate (Glu) catabolism, while NADP-dependent GDH4 (EC 1.4.1.4) is crucial for Glu synthesis during ammonia assimilation. GDH3 (EC 1.4.1.3) is versatile, supporting both 2OG formation and the reverse process, and shows specificity for both NAD(P)H coenzymes (Jaspard, 2006). In D. radiodurans (BAA-816), GDH3 is identified as DR_0980 and GDH4 as DR_1718 in the KEGG Enzyme database. Across 21 Deinococcus species cataloged in KEGG, their genomes typically exhibit a pair of GDH3 enzymes arranged in tandem (data not shown). Based on their genomic positioning within Deinococcus species, it’s inferred that KDR_0980n and KDR_0980 represent forms of GDH3. Notably, KDR_0980 is 45 aa longer than DR_0980 due to a frameshift caused by C insertions (Supplementary Tables S1, S2). While it is known that D. radiodurans cannot directly assimilate ammonia (Venkateswaran et al., 2000), ATCC 13939 strains retain classic ammonia assimilation pathways, GDH and GS-GOGAT, within their genomes. The observed genetic variance between strains BAA-816 and ATCC 13939 suggests that research involving ATCC 13939 could offer alternative perspectives on the process of ammonia assimilation.
The ATCC 13939 strains uniformly exhibit deletions and insertions of IS elements, with ISDra2, IS2621, and ISDra3 losing six, three, and one copies each, compared to the BAA-816 genome. Moreover, one insertion each of IS2621, IS1, ISDra3, and ISDra6 is common among these strains. Unique to the ATCC 13939K strain, additional transpositions were observed: an increase of one and two copies for ISDra2 (KDR_1963n1-1963n2) and IS2621 (KDR_0854n and KDR_1841n), respectively (Figure 2 and Supplementary Table S3). These IS element transpositions among the ATCC 13939 strains indicate evolutionary developments that set them apart from BAA-816, along with genetic differences stemming from laboratory cultivation. The estimated transposition rate of IS elements is 2.50 × 10−3 per genome per generation in wild-type D. radiodurans (Long et al., 2015), influencing strain evolution. Such transpositions can significantly alter phenotypes through genetic variations in protein-coding genes. In ATCC 13939 strains, the loss of ISDra2 reinstated genes coding for α/β fold hydrolase and serine protease (Figures 2D,E). The disruption of pilT by ISDra2 notably reduced transformation efficiency in strain 13939K (Supplementary Figure S6). These highlight the significant role of IS elements in microbial research, demonstrating their ability to alter both the genome and specific characteristics, including metabolic processe of certain strains.
The D. radiodurans strains, ATCC BAA-816 and ATCC 13939, originated from the ATCC repository. Cultures were grown at 30°C in TGY broth (0.5% tryptone, 0.3% yeast extract, and 0.1% glucose) or on TGY plates supplemented with 1.5% Bacto-agar. The chemicals used throughout the study were purchased from Sigma-Aldrich (St. Louis, MO, USA).
From single colonies on TGY agar, the D. radiodurans ATCC 13939K strain was cultured in TGY broth, shaking at 200 rpm at 30°C for 18 h. Post-cultivation, the cells were centrifuged, and genomic DNA was extracted using the AccuPrep® Genomic DNA Extraction Kit (Bioneer, Korea) according to the manufacturer’s instructions. Specifically, 1 × 109 cells were lysed using 180 μL of lysis buffer (20 mM Tris–HCl, 2 mM sodium EDTA, 1.2% Triton® X-100, pH 8.0), supplemented with 20 μL of lysozyme (100 mg/mL) and 10 μL of RNase A. The lysate was incubated at 37°C, followed by the addition of 20 μL of Proteinase K and 200 μL of GB Buffer, with subsequent incubation at 60°C. DNA purification was then performed using spin columns. For genome sequencing, a combination of Illumina NovaSeq 6000 and PacBio Sequel II platforms was used. DNA libraries were prepared using the TruSeq DNA Nano kit (Illumina, San Diego, CA, USA) and the SMRTbell Prep Kit 3.0 (PacBio, Menlo Park, CA, USA). The initial assembly was achieved with PacBio reads achieving 298.4× coverage, using the CANU V1.7 software (Koren et al., 2017), and was further refined using Illumina reads at 480.8× coverage through Pilon V1.21 software (Walker et al., 2014).
The genome sequence comprises four circular DNA structures totaling 3,285,071 bp. Analysis of GC content was conducted using the ANI Calculator (Richter et al., 2016). Prokka (v1.13) was employed to define the genomic profile, including CDS, tRNA, and rRNA genes (Seemann, 2014). Functional annotation of identified proteins was aligned with COG and KEGG databases, and sequence verification was performed against NCBI databases using BLASTP and BLASTX. The nucleotide sequence of ATCC 13939K was deposited in the NCBI Nucleotide database (GenBank number, CP150840 ~ CP150843).
Raw read data from Illumina sequencing was used for SNP detection. Following filtration, the clean read sequences were aligned to the reference sequence using the Burrows-Wheeler Aligner (BWA, version 0.7.17) (Li and Durbin, 2010). After mapping, duplicate reads were removed using Sambamba (version 0.6.8) (Tarasov et al., 2015), and SAMTools were used to identify variants (Danecek et al., 2021). Amino acid changes from these variants were obtained using SnpEff (version 4.3 t) (Cingolani, 2022).
For the preparation of competent cells, exponentially growing cells were collected via centrifugation and then resuspended in TGY medium at a concentration of 5 × 108 cells/mL. The medium was supplemented with 30 mM CaCl2 and 10% (v/v) glycerol, after which it was quickly frozen and stored at −80°C for preservation. Two plasmids were used for the transformation assays: pRadgro with a chloramphenicol resistance marker, which replicates in D. radiodurans, and pKatAPH3, carrying a kanamycin resistance gene, used for targeted gene disruption (Ohba et al., 2005; Misra et al., 2006). For the pKatAPH3-based assay, the upstream and downstream regions (approximately 1 kb) of bphP (DR_A0050) were PCR amplified using the primer pairs, Up-F (5′-tatctcgagtcgcgcggcctgtt-3′)/Up-R (5′-tatgatatcgatgctgggcctcttccggga-3′) and Down-F (5′-tatggatccggcaacgggtcccggctcat-3′)/Down-R (5′-tatctcgagcgacgttgcggttgtcttcgcc-3′). The PCR products were digested with XhoI/EcoRV and BamHI/PstI to produce the upstream and downstream fragments and ligated into the pKatAPH3 vector. The recombinant plasmid was named pKatAPH3-BphP. For transformation, aliquots of competent cells (100 μL each) were thawed on ice and mixed with an equal TGY medium containing 30 mM CaCl2. Subsequently, 250 ng of either plasmid DNA was added. Following a 30 min incubation at 0°C, the mixture underwent heat treatment at 32°C for 90 min. After the heat treatment, 800 μL of TGY media were added to the mixture to aid in the recovery and expression of antibiotic resistance, followed by further incubation for 5 h. Diluted samples were then plated onto TGY plates containing the appropriate antibiotics for selection.
PCR assays were utilized to validate the integration of specific IS elements within the genome of the ATCC 13939K strain. The dedicated primer pairs were used for the detection of each IS element: 0854F (5′-gtgggctcgcctagcatgtt-3′)/0855R (5′-gaacttctacccccaggcgg-3′) for KDR_0854n, 1841F (5′-atgttcggcgccatctggta-3′)/1842R (5′-cctcaacctctacgccgagg-3′) for KDR_1841n, and 1963F (5′-taggcgtccatcgtgaccatgc-3′)/1963R (5′-gatgtactcgatgatgaacgagcc-3′) for KDR_1963n1 and KDR_1963n2. Amplification was carried out using PrimeSTAR DNA polymerase (TaKaRa Bio Inc., Japan) under the following conditions: an initial denaturation step at 94°C for 40 s, followed by 20 cycles of amplification consisting of 40 s at 94°C, 40 s at 57°C, and 3 min at 72°C. A final elongation step at 72°C for 5 min was performed. The PCR products were then subjected to agarose gel electrophoresis.
Overnight cultures in the stationary phase were inoculated into fresh TGY broth at a 1:100 dilution and grown to log phase (OD600 ≈ 1.0). These cultures were then diluted to an OD600 of approximately 0.1 in TGY broth and treated with varying concentrations of mitomycin C (MMC) and hydrogen peroxide (H₂O₂) for 1 h at 30°C. Post H₂O₂ treatment, cells were treated with catalase (Sigma-Aldrich, Saint Louis, CA, USA) to degrade any residual H₂O₂, followed by 10-fold serial dilutions in distilled water, and subsequent spotting onto TGY agar plates. At room temperature, γ-radiation exposure was carried out using a 60Co gamma irradiator (AECL, IR-79; MDS Nordion International Co. Ltd., Ottawa, Canada) at the Advanced Radiation Technology Institute, Republic of Korea. Ultraviolet C (UV-C) exposure was performed using a UV crosslinker (CX-2000, UVP LLC, Upland, CA, USA), where plates with spotted cells were subjected to UV-C radiation. Post-exposure, the TGY plates were incubated at 30°C for 2–3 days, allowing colony-forming unit (CFU) enumeration.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
SJ: Methodology, Investigation, Writing – review & editing, Writing – original draft. HS: Data curation, Conceptualization, Writing – original draft. J-HJ: Writing – review & editing, Writing – original draft, Validation, Supervision, Methodology, Formal analysis. K-WJ: Visualization, Validation, Writing – review & editing. SR: Supervision, Writing – review & editing. SL: Writing – review & editing, Writing – original draft, Supervision, Resources, Funding acquisition, Conceptualization.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the KAERI institutional R&D Program (Project No. 523610-24) and the National Research Foundation of Korea (NRF) grant (NRF-2021M2E8A1047781) funded by Ministry of Science and ICT (MIST), Republic of Korea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1410024/full#supplementary-material
Alvarez-Ortega, C., Olivares, J., and Martínez, J. L. (2013). RND multidrug efflux pumps: what are they good for? Front. Microbiol. 4:7. doi: 10.3389/fmicb.2013.00007
Appukuttan, D., Singh, H., Park, S. H., Jung, J. H., Jeong, S., Seo, H. S., et al. (2015). Engineering synthetic multistress tolerance in Escherichia coli by using a deinococcal response regulator, DR1558. Appl. Environ. Microbiol. 82, 1154–1166. doi: 10.1128/AEM.03371-15
Baranov, P. V., Hammer, A. W., Zhou, J., Gesteland, R. F., and Atkins, J. F. (2005). Transcriptional slippage in bacteria: distribution in sequenced genomes and utilization in IS element gene expression. Genome Biol. 6:R25. doi: 10.1186/gb-2005-6-3-r25
Bennett, P. M. (2004). Genome plasticity: insertion sequence elements, transposons and integrons, and DNA rearrangement. Methods Mol. Biol. 266, 71–113. doi: 10.1385/1-59259-763-7:071
Benson, K. H., Godon, J. J., Renault, P., Griffin, H. G., and Gasson, M. G. (1996). Effect of ilvBN-encoded α-acetolactate synthase expression on diacetyl production in Lactococcus lactis. Appl. Microbiol. Biotechnol. 45, 107–111. doi: 10.1007/s002530050656
Bentchikou, E., Servant, P., Coste, G., and Sommer, S. (2010). A major role of the RecFOR pathway in DNA double-strand-break repair through ESDSA in Deinococcus radiodurans. PLoS Genet. 6:e1000774. doi: 10.1371/journal.pgen.1000774
Bernroitner, M., Zamocky, M., Furtmüller, P. G., Peschek, G. A., and Obinger, C. (2009). Occurrence, phylogeny, structure, and function of catalases and peroxidases in cyanobacteria. J. Exp. Bot. 60, 423–440. doi: 10.1093/jxb/ern309
Bisicchia, P., Steel, B., Mariam Debela, M. H., Löwe, J., and Sherratt, D. (2013). The N-terminal membrane-spanning domain of the Escherichia coli DNA translocase FtsK hexamerizes at midcell. MBio 4:e00800-13. doi: 10.1128/mBio.00800-13
Brim, H., McFarlan, S. C., Fredrickson, J. K., Minton, K. W., Zhai, M., Wackett, L. P., et al. (2000). Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nat. Biotechnol. 18, 85–90. doi: 10.1038/71986
Brooks, B. W., and Murray, R. G. E. (1981). Nomenclature for “Micrococcus radiodurans” and other radiation-resistant cocci: Deinococcaceae fam. nov. and Deinococcus gen. nov., including five species. Int. J. Syst. Evol. Microbiol. 31, 353–360. doi: 10.1099/00207713-31-3-353
Busenlehner, L. S., Pennella, M. A., and Giedroc, D. P. (2003). The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27, 131–143. doi: 10.1016/S0168-6445(03)00054-8
Cao, Z., and Julin, D. A. (2009). Characterization in vitro and in vivo of the DNA helicase encoded by Deinococcus radiodurans locus DR1572. DNA Repair 8, 612–619. doi: 10.1016/j.dnarep.2008.12.011
Chai, Y., Kolter, R., and Losick, R. (2009). A widely conserved gene cluster required for lactate utilization in Bacillus subtilis and its involvement in biofilm formation. J. Bacteriol. 191, 2423–2430. doi: 10.1128/JB.01464-08
Chavarría, M., Goñi-Moreno, Á., de Lorenzo, V., and Nikel, P. I. (2016). A metabolic widget adjusts the phosphoenolpyruvate-dependent fructose influx in Pseudomonas putida. mSystems 1:e00154-16. doi: 10.1128/mSystems.00154-16
Cingolani, P. (2022). Variant annotation and functional prediction: SnpEff. Methods Mol. Biol. 2493, 289–314. doi: 10.1007/978-1-0716-2293-3_19
Comas, I., González-Candelas, F., and Zúñiga, M. (2008). Unraveling the evolutionary history of the phosphoryl-transfer chain of the phosphoenolpyruvate:phosphotransferase system through phylogenetic analyses and genome context. BMC Evol. Biol. 8:147. doi: 10.1186/1471-2148-8-147
White, O., Eisen, J. A., Heidelberg, J. F., Hickey, E. K., Peterson, J. D., Dodson, R. J., et al. (2004). Corrections and Clarifications. Erratum: Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 303:766. doi: 10.1126/science.303.5659.766b
Cox, M. M., and Battista, J. R. (2005). Deinococcus radiodurans − the consummate survivor. Nat. Rev. Microbiol. 3, 882–892. doi: 10.1038/nrmicro1264
Dai, S., Jin, Y., Li, T., Weng, Y., Xu, X., Zhang, G., et al. (2018). DR1440 is a potential iron efflux protein involved in maintenance of iron homeostasis and resistance of Deinococcus radiodurans to oxidative stress. PLoS One 13:e0202287. doi: 10.1371/journal.pone.0202287
Dai, S., Xie, Z., Wang, B., Yu, N., Zhao, J., Zhou, Y., et al. (2021). Dynamic polyphosphate metabolism coordinating with manganese ions defends against oxidative stress in the extreme bacterium Deinococcus radiodurans. Appl. Environ. Microbiol. 87:e02785-20. doi: 10.1128/AEM.02785-20
Daly, M. J., Gaidamakova, E. K., Matrosova, V. Y., Kiang, J. G., Fukumoto, R., Lee, D. Y., et al. (2010). Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS One 5:e12570. doi: 10.1371/journal.pone.0012570
Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. doi: 10.1093/gigascience/giab008
Das, A. D., and Misra, H. S. (2012). DR2417, a hypothetical protein characterized as a novel β-CASP family nuclease in radiation resistant bacterium, Deinococcus radiodurans. Biochim. Biophys. Acta 1820, 1052–1061. doi: 10.1016/j.bbagen.2012.03.014
de Groot, A., Dulermo, R., Ortet, P., Blanchard, L., Guérin, P., Fernandez, B., et al. (2009). Alliance of proteomics and genomics to unravel the specificities of Sahara bacterium Deinococcus deserti. PLoS Genet. 5:e1000434. doi: 10.1371/journal.pgen.1000434
Eggington, J. M., Haruta, N., Wood, E. A., and Cox, M. M. (2004). The single-stranded DNA-binding protein of Deinococcus radiodurans. BMC Microbiol. 4:2. doi: 10.1186/1471-2180-4-2
Elbakry, A., and Löbrich, M. (2021). Homologous recombination subpathways: a tangle to resolve. Front. Genet. 12:723847. doi: 10.3389/fgene.2021.723847
Eram, M. S., Sarafuddin, B., Gong, F., and Ma, K. (2015). Characterization of acetohydroxyacid synthase from the hyperthermophilic bacterium Thermotoga maritima. Biochem. Biophys. Rep. 4, 89–97. doi: 10.1016/j.bbrep.2015.08.014
Eugénie, N., Zivanovic, Y., Lelandais, G., Coste, G., Bouthier de la Tour, C., Bentchikou, E., et al. (2021). Characterization of the radiation desiccation response regulon of the radioresistant bacterium Deinococcus radiodurans by integrative genomic analyses. Cells 10:2536. doi: 10.3390/cells10102536
Farabaugh, P. J. (1996). Programmed translational frameshifting. Microbiol. Rev. 60, 103–134. doi: 10.1128/mr.60.1.103-134.1996
Farci, D., Bowler, M. W., Esposito, F., McSweeney, S., Tramontano, E., and Piano, D. (2015). Purification and characterization of DR_2577 (SlpA) a major S-layer protein from Deinococcus radiodurans. Front. Microbiol. 6:414. doi: 10.3389/fmicb.2015.00414
Farci, D., Esposito, F., El Alaoui, S., and Piano, D. (2017). S-layer proteins as a source of carotenoids: isolation of the carotenoid cofactor deinoxanthin from its S-layer protein DR_2577. Food Res. Int. 99, 868–876. doi: 10.1016/j.foodres.2016.10.003
Gaballa, A., Newton, G. L., Antelmann, H., Parsonage, D., Upton, H., Rawat, M., et al. (2010). Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc. Natl. Acad. Sci. U. S. A. 107, 6482–6486. doi: 10.1073/pnas.1000928107
Galperin, M. Y., Nikolskaya, A. N., and Koonin, E. V. (2001). Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203, 11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x
Ghosal, D., Omelchenko, M. V., Gaidamakova, E. K., Matrosova, V. Y., Vasilenko, A., Venkateswaran, A., et al. (2005). How radiation kills cells: survival of Deinococcus radiodurans and Shewanella oneidensis under oxidative stress. FEMS Microbiol. Rev. 29, 361–375. doi: 10.1016/j.femsre.2004.12.007
Han, J. M., Song, H. Y., Jung, J. H., Lim, S., Seo, H. S., Kim, W. S., et al. (2023). Deinococcus radiodurans-derived membrane vesicles protect HaCaT cells against H2O2-induced oxidative stress via modulation of MAPK and Nrf2/ARE pathways. Biol. Proced. 25:17. doi: 10.1186/s12575-023-00211-4
Hejna, J. A., and Moses, R. E. (2009). “DNA replication” in Encyclopedia of microbiology, vol. 5. 3rd Edn. Ed. M. Schaechter (New York, USA: Academic Press), 113–122.
Ho, J., Adeolu, M., Khadka, B., and Gupta, R. S. (2016). Identification of distinctive molecular traits that are characteristic of the phylum “Deinococcus-Thermus” and distinguish its main constituent groups. Syst. Appl. Microbiol. 39, 453–463. doi: 10.1016/j.syapm.2016.07.003
Hua, X., and Hua, Y. (2016). Improved complete genome sequence of the extremely radioresistant bacterium Deinococcus radiodurans R1 obtained using PacBio single-molecule sequencing. Genome Announc. 4:e00886-16. doi: 10.1128/genomeA.00886-16
Hwang, W. C., Bakolitsa, C., Punta, M., Coggill, P. C., Bateman, A., Axelrod, H. L., et al. (2013). LUD, a new protein domain associated with lactate utilization. BMC Bioinformatics 14:341. doi: 10.1186/1471-2105-14-341
Im, S., Song, D., Joe, M., Kim, D., Park, D. H., and Lim, S. (2013). Comparative survival analysis of 12 histidine kinase mutants of Deinococcus radiodurans after exposure to DNA-damaging agents. Bioprocess Biosyst. Eng. 36, 781–789. doi: 10.1007/s00449-013-0904-8
Iranzo, J., Wolf, Y. I., Koonin, E. V., and Sela, I. (2019). Gene gain and loss push prokaryotes beyond the homologous recombination barrier and accelerate genome sequence divergence. Nat. Commun. 10:5376. doi: 10.1038/s41467-019-13429-2
Islam, S. M., Hua, Y., Ohba, H., Satoh, K., Kikuchi, M., Yanagisawa, T., et al. (2003). Characterization and distribution of IS8301 in the radioresistant bacterium Deinococcus radiodurans. Genes Genet. Syst. 78, 319–327. doi: 10.1266/ggs.78.319
Ithurbide, S., Coste, G., Lisboa, J., Eugénie, N., Bentchikou, E., Bouthier de la Tour, C., et al. (2020). Natural transformation in Deinococcus radiodurans: a genetic analysis reveals the major roles of DprA, DdrB, RecA, RecF, and RecO proteins. Front. Microbiol. 11:1253. doi: 10.3389/fmicb.2020.01253
Jack, D. L., Yang, N. M., and Saier, M. H. Jr. (2001). The drug/metabolite transporter superfamily. Eur. J. Biochem. 268, 3620–3639. doi: 10.1046/j.1432-1327.2001.02265.x
Jang, S. (2023). AcrAB−TolC, a major efflux pump in gram-negative bacteria: toward understanding its operation mechanism. BMB Rep. 56, 326–334. doi: 10.5483/BMBRep.2023-0070
Jaspard, E. (2006). A computational analysis of the three isoforms of glutamate dehydrogenase reveals structural features of the isoform EC 1.4.1.4 supporting a key role in ammonium assimilation by plants. Biol. Direct 1:38. doi: 10.1186/1745-6150-1-38
Jeong, S., Jung, J. H., Kim, M. K., de Groot, A., Blanchard, L., Ryu, S., et al. (2021). Atypical bacilliredoxin AbxC plays a role in responding to oxidative stress in radiation-resistant bacterium Deinococcus radiodurans. Antioxidants 10:1148. doi: 10.3390/antiox10071148
Jeong, S. W., Kim, J. H., Kim, J. W., Kim, C. Y., Kim, S. Y., and Choi, Y. J. (2020). Metabolic engineering of extremophilic bacterium Deinococcus radiodurans for the production of the novel carotenoid deinoxanthin. Microorganisms 9:44. doi: 10.3390/microorganisms9010044
Karvelis, T., Druteika, G., Bigelyte, G., Budre, K., Zedaveinyte, R., Silanskas, A., et al. (2021). TnpB is a programmable RNA-guided DNA endonuclease. Nature 599, 692–696. doi: 10.1038/s41586-021-04058-1
Keshav, G. C., Gyawali, P., Balci, H., and Abeysirigunawardena, S. (2020). Ribosomal RNA methyltransferase RsmC moonlights as an RNA chaperone. Chembiochem 21, 1885–1892. doi: 10.1002/cbic.201900708
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Leblanc, C., Vilter, H., Fournier, J. B., Delage, L., Potin, P., Rebuffet, E., et al. (2015). Vanadium haloperoxidases: from the discovery 30 years ago to X-ray crystallographic and V K-edge absorption spectroscopic studies. Coord. Chem. Rev. 301-302, 134–146. doi: 10.1016/j.ccr.2015.02.013
Li, H., and Durbin, R. (2010). Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Lim, S., Jung, J. H., Blanchard, L., and de Groot, A. (2019). Conservation and diversity of radiation and oxidative stress resistance mechanisms in Deinococcus species. FEMS Microbiol. Rev. 43, 19–52. doi: 10.1093/femsre/fuy037
Liu, Y., Gao, Z. Q., She, Z., Qu, K., Wang, W. J., Shtykova, E. V., et al. (2012). The structural basis of the response regulator DrRRA from Deinococcus radiodurans. Biochem. Biophys. Res. Commun. 417, 1206–1212. doi: 10.1016/j.bbrc.2011.12.110
Liu, C., Wang, L., Li, T., Lin, L., Dai, S., Tian, B., et al. (2014). A PerR-like protein involved in response to oxidative stress in the extreme bacterium Deinococcus radiodurans. Biochem. Biophys. Res. Commun. 450, 575–580. doi: 10.1016/j.bbrc.2014.06.015
Long, H., Kucukyildirim, S., Sung, W., Williams, E., Lee, H., Ackerman, M., et al. (2015). Background mutational features of the radiation-resistant bacterium Deinococcus radiodurans. Mol. Biol. Evol. 32, 2383–2392. doi: 10.1093/molbev/msv119
Makarova, K. S., Aravind, L., Wolf, Y. I., Tatusov, R. L., Minton, K. W., Koonin, E. V., et al. (2001). Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65, 44–79. doi: 10.1128/MMBR.65.1.44-79.2001
Manobala, T., Shukla, S. K., Rao, T. S., and Kumar, M. D. (2019). A new uranium bioremediation approach using radio-tolerant Deinococcus radiodurans biofilm. J. Biosci. 44:122. doi: 10.1007/s12038-019-9942-y
Maurya, G. K., Modi, K., Banerjee, M., Chaudhary, R., Rajpurohit, Y. S., and Misra, H. S. (2018). Phosphorylation of FtsZ and FtsA by a DNA damage-responsive Ser/Thr protein kinase affects their functional interactions in Deinococcus radiodurans. mSphere 3:e00325-18. doi: 10.1128/mSphere.00325-18
Maurya, G. K., Modi, K., and Misra, H. S. (2016). Divisome and segrosome components of Deinococcus radiodurans interact through cell division regulatory proteins. Microbiology 162, 1321–1334. doi: 10.1099/mic.0.000330
McCown, P. J., Ruszkowska, A., Kunkler, C. N., Breger, K., Hulewicz, J. P., Wang, M. C., et al. (2020). Naturally occurring modified ribonucleosides. Wiley Interdiscip. Rev. RNA. 11:e1595. doi: 10.1002/wrna.1595
Mennecier, S., Coste, G., Servant, P., Bailone, A., and Sommer, S. (2004). Mismatch repair ensures fidelity of replication and recombination in the radioresistant organism Deinococcus radiodurans. Mol. Gen. Genomics. 272, 460–469. doi: 10.1007/s00438-004-1077-6
Mennecier, S., Servant, P., Coste, G., Bailone, A., and Sommer, S. (2006). Mutagenesis via IS transposition in Deinococcus radiodurans. Mol. Microbiol. 59, 317–325. doi: 10.1111/j.1365-2958.2005.04936.x
Miñambres, B., Olivera, E. R., Jensen, R. A., and Luengo, J. M. (2000). A new class of glutamate dehydrogenases (GDH). Biochemical and genetic characterization of the first member, the AMP-requiring NAD-specific GDH of Streptomyces clavuligerus. J. Biol. Chem. 275, 39529–39542. doi: 10.1074/jbc.M005136200
Mishra, S., Misra, H. S., and Kota, S. (2022). FtsK, a DNA motor protein, coordinates the genome segregation and early cell division processes in Deinococcus radiodurans. mBio 13:e0174222. doi: 10.1128/mbio.01742-22
Misra, H. S., Khairnar, N. P., Kota, S., Shrivastava, S., Joshi, V. P., and Apte, S. K. (2006). An exonuclease I-sensitive DNA repair pathway in Deinococcus radiodurans: a major determinant of radiation resistance. Mol. Microbiol. 59, 1308–1316. doi: 10.1111/j.1365-2958.2005.05005.x
Modi, K., and Misra, H. S. (2014). Dr-FtsA, an actin homologue in Deinococcus radiodurans differentially affects Dr-FtsZ and Ec-FtsZ functions in vitro. PLoS One 9:e115918. doi: 10.1371/journal.pone.0115918
Mosquera-Rendón, J., Cárdenas-Brito, S., Pineda, J. D., Corredor, M., and Benítez-Páez, A. (2014). Evolutionary and sequence-based relationships in bacterial AdoMet-dependent non-coding RNA methyltransferases. BMC. Res. Notes 7:440. doi: 10.1186/1756-0500-7-440
Narumi, I., Cherdchu, K., Kitayama, S., and Watanabe, H. (1997). The Deinococcus radiodurans uvrA gene: identification of mutation sites in two mitomycin-sensitive strains and the first discovery of insertion sequence element from deinobacteria. Gene 198, 115–126. doi: 10.1016/s0378-1119(97)00301-6
Niiranen, L., Lian, K., Johnson, K. A., and Moe, E. (2015). Crystal structure of the DNA polymerase III β subunit (β-clamp) from the extremophile Deinococcus radiodurans. BMC Struct. Biol. 15:5. doi: 10.1186/s12900-015-0032-6
Ohba, H., Satoh, K., Yanagisawa, T., and Narumi, I. (2005). The radiation responsive promoter of the Deinococcus radiodurans pprA gene. Gene 363, 133–141. doi: 10.1016/j.gene.2005.07.035
Owttrim, G. W. (2013). RNA helicases: diverse roles in prokaryotic response to abiotic stress. RNA Biol. 10, 96–110. doi: 10.4161/rna.22638
Park, H. R., Lee, J. H., Ji, H. J., Lim, S., Ahn, K. B., and Seo, H. S. (2022). Radioprotection of deinococcal exopolysaccharide BRD125 by regenerating hematopoietic stem cells. Front. Oncol. 12:898185. doi: 10.3389/fonc.2022.898185
Park, S. H., Sohn, Y. J., Park, S. J., and Choi, J. I. (2020). Effect of DR1558, a Deinococcus radiodurans response regulator, on the production of GABA in the recombinant Escherichia coli under low pH conditions. Microb. Cell Factories 19:64. doi: 10.1186/s12934-020-01322-3
Parte, A. C., Carbasse, J. S., Meier-Kolthoff, J. P., Reimer, L. C., and Göker, M. (2020). List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612. doi: 10.1099/ijsem.0.004332
Pasternak, C., Ton-Hoang, B., Coste, G., Bailone, A., Chandler, M., and Sommer, S. (2010). Irradiation-induced Deinococcus radiodurans genome fragmentation triggers transposition of a single resident insertion sequence. PLoS Genet. 6:e1000799. doi: 10.1371/journal.pgen.1000799
Pei, J., and Grishin, N. V. (2001). GGDEF domain is homologous to adenylyl cyclase. Proteins 42, 210–216. doi: 10.1002/1097-0134(20010201)42:2<210::aid-prot80>3.0.co;2-8
Pei, A., Nossa, C. W., Chokshi, P., Blaser, M. J., Yang, L., Rosmarin, D. M., et al. (2009). Diversity of 23S rRNA genes within individual prokaryotic genomes. PLoS One 4:e5437. doi: 10.1371/journal.pone.0005437
Pengpeng, W., and Tan, Z. (2013). Ammonia assimilation in rumen bacteria: a review. Anim. Biotechnol. 24, 107–128. doi: 10.1080/10495398.2012.756402
Periwal, V., and Scaria, V. (2015). Insights into structural variations and genome rearrangements in prokaryotic genomes. Bioinformatics 31, 1–9. doi: 10.1093/bioinformatics/btu600
Petchiappan, A., Naik, S. Y., and Chatterji, D. (2020). Tracking the homeostasis of second messenger cyclic-di-GMP in bacteria. Biophys. Rev. 12, 719–730. doi: 10.1007/s12551-020-00636-1
Pichoff, S., Du, S., and Lutkenhaus, J. (2019). Roles of FtsEX in cell division. Res. Microbiol. 170, 374–380. doi: 10.1016/j.resmic.2019.07.003
Razew, A., Schwarz, J. N., Mitkowski, P., Sabala, I., and Kaus-Drobek, M. (2022). One fold, many functions-M23 family of peptidoglycan hydrolases. Front. Microbiol. 13:1036964. doi: 10.3389/fmicb.2022.1036964
Reitzer, L. (2014). “Amino acid synthesis” in Reference module in biomedical sciences. Ed. M. J. Caplan (Amsterdam: Elsevier).
Repar, J., Zahradka, D., Sović, I., and Zahradka, K. (2021). Characterization of gross genome rearrangements in Deinococcus radiodurans recA mutants. Sci. Rep. 11:10939. doi: 10.1038/s41598-021-89173-9
Richards, J., and Belasco, J. G. (2011). Ribonuclease J: how to lead a double life. Structure 19, 1201–1203. doi: 10.1016/j.str.2011.08.004
Richter, M., Rosselló-Móra, R., Oliver Glöckner, F., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Santos-Beneit, F. (2015). The pho regulon: a huge regulatory network in bacteria. Front. Microbiol. 6:402. doi: 10.3389/fmicb.2015.00402
Schindler, B. D., and Kaatz, G. W. (2016). Multidrug efflux pumps of gram-positive bacteria. Drug Resist. Updat. 27, 1–13. doi: 10.1016/j.drup.2016.04.003
Schmier, B. J., Seetharaman, J., Deutscher, M. P., Hunt, J. F., and Malhotra, A. (2012). The structure and enzymatic properties of a novel RNase II family enzyme from Deinococcus radiodurans. J. Mol. Biol. 415, 547–559. doi: 10.1016/j.jmb.2011.11.031
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sharma, A., Gaidamakova, E. K., Grichenko, O., Matrosova, V. Y., Hoeke, V., Klimenkova, P., et al. (2017). Across the tree of life, radiation resistance is governed by antioxidant Mn2+, gauged by paramagnetic resonance. Proc. Natl. Acad. Sci. U. S. A. 114, E9253–E9260. doi: 10.1073/pnas.1713608114
Slade, D., and Radman, M. (2011). Oxidative stress resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 75, 133–191. doi: 10.1128/MMBR.00015-10
Southworth, M. W., and Perler, F. B. (2002). Protein splicing of the Deinococcus radiodurans strain R1 Snf2 intein. J. Bacteriol. 184, 6387–6388. doi: 10.1128/JB.184.22.6387-6388.2002
Steczkiewicz, K., Muszewska, A., Knizewski, L., Rychlewski, L., and Ginalski, K. (2012). Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 40, 7016–7045. doi: 10.1093/nar/gks382
Takeda, M., Anamizu, S., Motomatsu, S., Chen, X., and Thapa, C. R. (2014). Identification and characterization of a mycobacterial NAD+-dependent alcohol dehydrogenase with superior reduction of diacetyl to (S)-acetoin. Biosci. Biotechnol. Biochem. 78, 1879–1886. doi: 10.1080/09168451.2014.943649
Tarasov, A., Vilella, A. J., Cuppen, E., Nijman, I. J., and Prins, P. (2015). Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034. doi: 10.1093/bioinformatics/btv098
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Timmins, J., and Moe, E. (2016). A decade of biochemical and structural studies of the DNA repair machinery of Deinococcus radiodurans: major findings, functional and mechanistic insight and challenges. Comput. Struct. Biotechnol. J. 14, 168–176. doi: 10.1016/j.csbj.2016.04.001
Vale, F. F., Lehours, P., and Yamaoka, Y. (2022). Editorial: the role of mobile genetic elements in bacterial evolution and their adaptability. Front. Microbiol. 13:849667. doi: 10.3389/fmicb.2022.849667
Venkateswaran, A., McFarlan, S. C., Ghosal, D., Minton, K. W., Vasilenko, A., Makarova, K., et al. (2000). Physiologic determinants of radiation resistance in Deinococcus radiodurans. Appl. Environ. Microbiol. 66, 2620–2626. doi: 10.1128/AEM.66.6.2620-2626.2000
Vigouroux, A., Cordier, B., Aristov, A., Alvarez, L., Özbaykal, G., Chaze, T., et al. (2020). Class-a penicillin binding proteins do not contribute to cell shape but repair cell-wall defects. Elife 9:e51998. doi: 10.7554/eLife.51998
von Kügelgen, A., van Dorst, S., Alva, V., and Bharat, T. A. M. (2022). A multidomain connector links the outer membrane and cell wall in phylogenetically deep-branching bacteria. Proc. Natl. Acad. Sci. U. S. A. 119:e2203156119. doi: 10.1073/pnas.2203156119
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wang, L., Tan, Y. S., Chen, K., Ntakirutimana, S., Liu, Z. H., Li, B. Z., et al. (2024). Global regulator IrrE on stress tolerance: a review. Crit. Rev. Biotechnol., 1–21. doi: 10.1080/07388551.2023.2299766
Wang, L., Xu, G., Chen, H., Zhao, Y., Xu, N., Tian, B., et al. (2008). DrRRA: a novel response regulator essential for the extreme radioresistance of Deinococcus radiodurans. Mol. Microbiol. 67, 1211–1222. doi: 10.1111/j.1365-2958.2008.06113.x
White, O., Eisen, J. A., Heidelberg, J. F., Hickey, E. K., Peterson, J. D., Dodson, R. J., et al. (1999). Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286, 1571–1577. doi: 10.1126/science.286.5444.1571
Willems, P., Fijalkowski, I., and Van Damme, P. (2020). Lost and found: re-searching and re-scoring proteomics data aids genome annotation and improves proteome coverage. mSystems 5:e00833-20. doi: 10.1128/mSystems.00833-20
Xiao, Z., and Xu, P. (2007). Acetoin metabolism in bacteria. Crit. Rev. Microbiol. 33, 127–140. doi: 10.1080/10408410701364604
Yang, S., Xu, H., Wang, J., Liu, C., Lu, H., Liu, M., et al. (2016). Cyclic AMP receptor protein acts as a transcription regulator in response to stresses in Deinococcus radiodurans. PLoS One 11:e0155010. doi: 10.1371/journal.pone.0155010
Ying, X., and Ma, K. (2011). Characterization of a zinc-containing alcohol dehydrogenase with stereoselectivity from the hyperthermophilic archaeon Thermococcus guaymasensis. J. Bacteriol. 193, 3009–3019. doi: 10.1128/JB.01433-10
Yu, J., Li, T., Dai, S., Weng, Y., Li, J., Li, Q., et al. (2017). A tamB homolog is involved in maintenance of cell envelope integrity and stress resistance of Deinococcus radiodurans. Sci. Rep. 7:45929. doi: 10.1038/srep45929
Keywords: Deinococcus radiodurans, comparative genomics, ATCC 13939, ATCC BAA-816, gene annotation, insertion sequence elements
Citation: Jeong S, Singh H, Jung J-H, Jung K-W, Ryu S and Lim S (2024) Comparative genomics of Deinococcus radiodurans: unveiling genetic discrepancies between ATCC 13939K and BAA-816 strains. Front. Microbiol. 15:1410024. doi: 10.3389/fmicb.2024.1410024
Received: 31 March 2024; Accepted: 04 June 2024;
Published: 19 June 2024.
Edited by:
Andreas Teske, University of North Carolina at Chapel Hill, United StatesReviewed by:
Haitham Sghaier, National Center for Nuclear Science and Technology, TunisiaCopyright © 2024 Jeong, Singh, Jung, Jung, Ryu and Lim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sangyong Lim, c2F5bGltQGthZXJpLnJlLmty
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.