
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 July 2024
Sec. Infectious Agents and Disease
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1409593
This article is part of the Research Topic Diagnosis and Treatment Strategies of Tick-borne Diseases View all 8 articles
Neoehrlichia mikurensis (N. mikurensis) is an emerging tick-borne pathogen that can cause neoehrlichiosis. Rodents are considered the major host for N. mikurensis. Currently, N. mikurensis has been detected in rodents in several studies from China and other countries. However, no research on N. mikurensis infection in rodents has been reported in the Liupan mountain region. The region of Liupan Mountain, located in northwestern China, is the center of the triangle formed by the cities of Xi’an, Yinchuan, and Lanzhou, with multiple tourist sites in the region. To survey whether there is N. mikurensis in hosts, rodents were captured in this region in September 2020. A nested polymerase chain reaction was used to detect the DNA of N. mikurensis, followed by nucleotide sequencing and phylogenetic analysis. In the region, among 88 rodents, 3 rodents were detected positive for N. mikurensis, a detection rate of 3.4%. Based on phylogenetic analysis of the partial groEL gene sequences, N. mikurensis from rodents in Liupan Mountain clustered in the same evolutionary branch with those found in rodents from Japan, Russia, and northeastern China, and also in ticks and clinical cases from Heilongjiang Province in northeastern China.
Neoehrlichia mikurensis (N. mikurensis), an emerging pathogen belonging to the family Anaplasmataceae (Kawahara et al., 2004), was first detected in Ixodes ricinus in the Netherlands in 1999 (Schouls et al., 1999). Currently, N. mikurensis has been reported among a variety of animals, such as rodents (Obiegala et al., 2014), hedgehogs (Földvári et al., 2014), and a dog (Diniz et al., 2011). Of these, rodents can be considered to be the major host for N. mikurensis.
N. mikurensis in rodents has been reported in several countries, such as South Korea (Jha et al., 2018), Germany (Galfsky et al., 2019), Sweden (Andersson and Råberg, 2011), Japan (Kenji et al., 2007), Hungary (Szekeres et al., 2015), and Russia (Rar et al., 2010). In 2003, Pan et al. (2003) found the pathogen in Rattus norvegicus from Guangzhou, China. Subsequently, it was also found in rodents from other provinces in China, such as Yunnan (Du et al., 2018), Jilin (Li et al., 2013) and Liaoning provinces (Wang et al., 2019). Since the first human case of neoehrlichiosis was reported in 2010 (Welinder-Olsson et al., 2010), more and more human infections with N. mikurensis have been reported globally (Pekova et al., 2011; Li et al., 2012; Welc-Faleciak et al., 2014; Quarsten et al., 2017; Boyer et al., 2021; Hoper et al., 2021; Lenart et al., 2021; Markowicz et al., 2021; Gonzalez-Carmona et al., 2023; Gynthersen et al., 2023).
However, no research on neoehrlichiosis or N. mikurensis infection in hosts has been reported in the region of Liupan Mountain, China. So, detecting whether there is N. mikurensis in rodents is of great significance for assessing the risk of N. mikurensis infection in residents. Silaghi et al. (2012) detected N. mikurensis in the liver, spleen, kidney, blood, transudate, and skin tissues of rodents, with the highest detection rate in the kidney. Therefore, in this study, we used nested PCR to detect the presence of N. mikurensis in rodent kidney organs in the region.
The Liupan Mountain Nature Reserve (106°09′-106°30′E, 35°15′-35°41′N), located in northwestern China, is the center of the triangle formed by the cities of Xi’an, Yinchuan, and Lanzhou, with multiple tourist sites in the region.
In September 2020, rodents were captured by night trapping method at Laolongtan Scenic Spot, Huanghua Town, and Xingsheng Town in the region. After the captured rodents were brought back to the laboratory, they were species-identified based on their morphological characteristics (Lu, 1982), and then dissection was performed to obtain the kidney tissues, which were stored in 2 mL cryotubes at −80°C.
Approximately 25 mg of kidney tissue was taken under aseptic operation. DNA was extracted according to the instructions of a Tissue DNA Isolation Kit (Magnetic Beads) (Jiangsu Bioperfectus Technologies Co., Ltd., Jiangsu, China) and stored at −40°C.
The groEL gene of N. mikurensis was amplified by nested PCR. The primers were used as reported by Li et al. (2013) and synthesized by Shanghai BioGerm Medical Technology Co., Ltd. (Shanghai, China). The final amplification product target fragment size was 891 bp. For the first round of the PCR amplification, the reaction mix was prepared as follows: 12.5 μL of Premix Taq™ (TaKaRa Taq™ Version 2.0), 0.5 μL each of CNM-out 1 and CNM-out 2 (10 μmol/L), DNA template 3 μL, ultrapure water 8.5 μL. The reaction conditions were: 94°C pre-denaturation 5 min; 94°C denaturation 30 s, 53°C annealing 30 s, 72°C extension 1 min 30 s, for a total of 40 cycles; 72°C final extension 5 min. For the second round of PCR amplification, the reaction mix was prepared as follows: Premix Taq™ (TaKaRa Taq™ Version 2.0) 12.5 μL, 1 μL each of CNM-in 1 and CNM-in 2 (10 μmol/L), 0.5 μL of first-round product, and 10 μL of ultrapure water. The reaction conditions were the same as those of the first-round reaction, except for the final extension at 72°C for 1 min. The PCR products were separated using electrophoresis on 1.5% agarose gels, with GelRed™ Nucleic Acid Gel Stain (Biotium, America) and visualized under Gel Doc™ XR+ Imaging System (Bio-Rad, United States).
The positive products were sequenced bidirectionally by Shanghai BioGerm Medical Technology Co., Ltd. (Shanghai, China). Sequence assembly and manual correction were performed using SeqMan software (Version 7.1, Lasergene, DNASTAR, Madison, WI, United States). One of the assembled sequences was uploaded to GenBank database, and used for homology and phylogenetic analyses. After performing blast alignment of the selected sequence at the National Center for Biotechnology Information, sequences with high homology were downloaded. In addition, sequences from selected geographical origins and hosts were also downloaded in this study to better analyze the evolutionary information of the groEL of N. mikurensis. The groEL sequences of Anaplasma phagocytophilum (AF033101) and Ehrlichia chaffeensis (L10917) were used as outgroups. Based on MEGA X (Kumar et al., 2018), model with the lowest BIC scores (Bayesian Information Criterion) was considered as the best-fit substitution model. The phylogenetic tree was constructed by the maximum likelihood method based on the best-fit substitution model (Tamura 3-parameter+G + I) (Tamura, 1992) using MEGA X (Kumar et al., 2018) and tested with 1,000 bootstrap replications. Sequences were aligned with MegAlign software (Version 7.1, Lasergene, DNASTAR, Madison, WI, United States), and the percent identities were calculated with the same software.
Among 88 rodents captured in the region, 42 were form Laolongtan Scenic Spot, 27 from Huanghua Town, and 19 from Xingsheng Town. Apodemus peninsulae (36.4%, 32/88), Cricetus migratorius (26.1%, 23/88), Tscherskia triton (21.6%, 19/88), Apodemus agrarius (11.4%, 10/88), Niviventer confucianus (2.3%, 2/88), Eozapus setchuanus (1.1%, 1/88), and Mus musculus (1.1%, 1/88) were identified.
Three samples were positive for N. mikurensis positive, including two A. peninsulae (LPM-R14 and LPM-R25) and one T. triton (LPM-R37).
The similarity of the three N. mikurensis sequences obtained in this study was greater than 99%. The sequence of sample LPM-R14 was uploaded to GenBank database (accession number, PP818814). Based on phylogenetic analysis of the partial groEL gene sequences, four evolutionary branches of N. mikurensis were reconstructed (Figure 1). The overall tree topology was consistent with Li et al. (2013), in particular the subdivision of N. mikurensis into four main clades (Clusters I-IV). The sequence (PP818814) from A. peninsulae clustered in Cluster I with those found in rodents from Japan (LC717483, LC717487), Russia (MN701627, FJ966365), and northwestern China, such as Inner Mongolia Autonomous Region (JQ359076), Heilongjiang (JQ359069, JQ359070, JQ359071, KC108717) and Jilin provinces (JQ359072, JQ359073, JQ359074). Moreover, Cluster I also included N. mikurensis infection in ticks from Russia (MG182157, KX980039, FJ966359) and Heilongjiang Province (JQ359077, JQ359078), and clinical cases of neoehrlichiosis from Heilongjiang Province (JQ359062). The homologies of PP818814 and sequences of Clusters I-IV in the phylogenetic tree were shown in Supplementary Tables S1–S3.
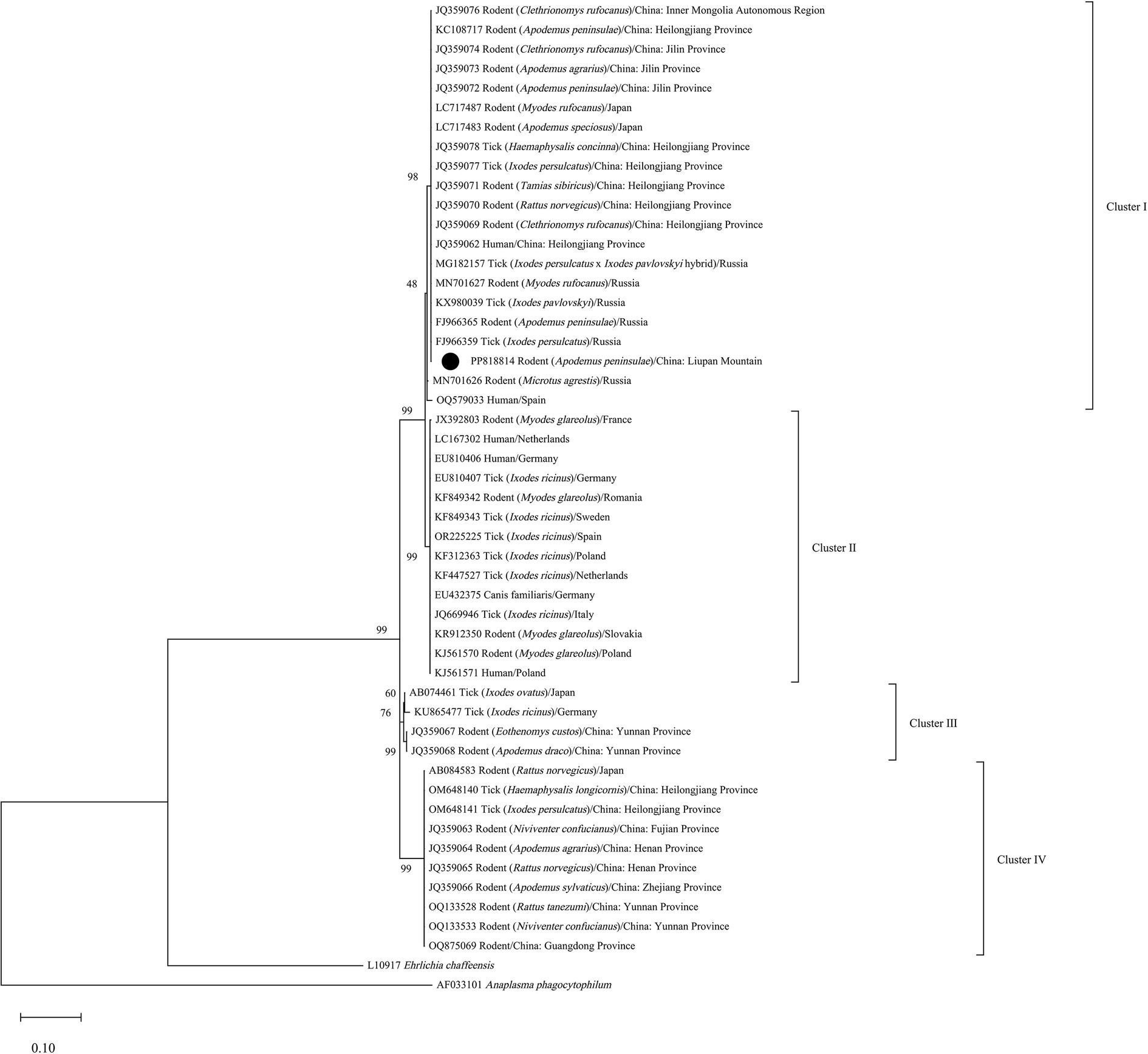
Figure 1. Phylogenetic tree of N. mikurensis inferred from partial fragments of the groEL gene sequences. •, Sequence obtained in the study.
In this study, N. mikurensis was detected for the first time in rodents from the region of Liupan Mountain. The detection rate of N. mikurensis was 3.4% in the region, which was similar to Zhejiang (3.7%) and Henan provinces (3.9%), but lower than Jilin Province (13.8%), China (Li et al., 2013). Our findings indicate that sequences of N. mikurensis from rodents in Liupan Mountain clustered in the same evolutionary branch with those found in rodents, ticks, and human cases from Heilongjiang Province. Liupan Mountain is the center of the triangle formed by the cities of Xi’an, Yinchuan, and Lanzhou, and there are multiple tourist attractions in the region. Therefore, they indicate that there is a potential risk of N. mikurensis infection to the residents in Liupan Mountain. Currently, N. mikurensis is mainly detected by nested PCR or qPCR, and there is a lack of methods for its serologic detection (Wass et al., 2019). Due to the few reported human cases in China, limited detection methods, and insufficient understanding of the clinical manifestations of neoehrlichiosis by physicians, misdiagnosis or missed diagnosis may occur.
Based on phylogenetic analysis of the groEL and 16S rRNA genes, Li et al. (2013) concluded that N. mikurensis has four evolutionary branches (Clusters I-IV) that are related to geographic origins, with sequences from northeastern China and the Asian part of Russia clustering in Cluster I. Subsequently, Sun et al. (2023) detected sequences in ticks from northeastern China clustered in Cluster IV. The phylogenetic tree inferred in this study showed that the sequence detected in the case from Spain clustered in Cluster I. This further indicate that geographical distribution and genetic variation are not necessarily concordant, and there are at least two genetic lineages of N. mikurensis currently in China.
In conclusion, N. mikurensis was found in A. peninsulae and T. triton in Liupan Mountain, and PP818814 sourced from the region clustered into the same evolutionary branch with N. mikurensis infection in rodents from Japan, Russia, and northeastern China, also with the pathogen in ticks and human cases from Heilongjiang Province. They indicate that there is a potential risk of N. mikurensis infection to residents in the region. Further studies should be conducted to detect the presence and abundance of N. mikurensis in ticks from the same area. And the ability of physicians to diagnose and differential diagnose neoehrlichiosis should be improved to identify whether there is the infection of N. mikurensis in local residents.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
According to Regulation on the Management of Vector Prevention and Control issued by National Health Commission of the People’s Republic of China and Integrated Strategy for Vector Prevention and Control included in Healthy China 2030 Planning Framework, rats trapped is an activity for vector control. Therefore, ethical approval was not required in the study.
XW: Conceptualization, Data curation, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. BP: Resources, Writing – original draft. ZK: Writing – review & editing, Supervision. JZ: Writing – original draft, Investigation. YY: Writing – original draft, Resources. TC: Resources, Writing – original draft. LY: Writing – review & editing, Supervision.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1409593/full#supplementary-material
Andersson, M., and Råberg, L. (2011). Wild rodents and novel human pathogen Candidatus Neoehrlichia mikurensis, southern Sweden. Emerg. Infect. Dis. 17, 1716–1718. doi: 10.3201/eid1709.101058
Boyer, P. H., Baldinger, L., Degeilh, B., Wirth, X., Kamdem, C. M., Hansmann, Y., et al. (2021). The emerging tick-borne pathogen Neoehrlichia mikurensis: first French case series and vector epidemiology. Emerg. Microb. Infect. 10, 1731–1738. doi: 10.1080/22221751.2021.1973347
Diniz, P. P. V. P., Schulz, B. S., Hartmann, K., and Breitschwerdt, E. B. (2011). "Candidatus Neoehrlichia mikurensis" infection in a dog from Germany. J. Clin. Microbiol. 49, 2059–2062. doi: 10.1128/JCM.02327-10
Du, C.-H., Liu, H.-B., Wei, R., Jongejan, F., Gao, Z.-H., Shao, Z.-T., et al. (2018). Investigation on Ehrlichia infection in small mammals and ticks from Tengchong, Yunnan Province, southern China. Vector-Borne Zoonotic Dis. 18, 563–566. doi: 10.1089/vbz.2017.2205
Földvári, G., Jahfari, S., Rigó, K., Jablonszky, M., Szekeres, S., Majoros, G., et al. (2014). Candidatus Neoehrlichia mikurensis and Anaplasma phagocytophilum in urban hedgehogs. Emerg. Infect. Dis. 20, 496–498. doi: 10.3201/eid2003.130935
Galfsky, D., Król, N., Pfeffer, M., and Obiegala, A. (2019). Long-term trends of tick-borne pathogens in regard to small mammal and tick populations from Saxony, Germany. Parasit. Vectors 12:131. doi: 10.1186/s13071-019-3382-2
Gonzalez-Carmona, P., Portillo, A., Cervera-Acedo, C., Gonzalez-Fernandez, D., and Oteo, J. A. (2023). Candidatus Neoehrlichia mikurensis infection in patient with antecedent. Emerg. Infect. Dis. 29, 1659–1662. doi: 10.3201/eid2908.230428
Gynthersen, R. M. M., Stensvold, C. R., Nielsen, S. L., Moller, H. J., Nielsen, H. V., Lebech, A.-M., et al. (2023). Neoehrlichia mikurensis-an emerging opportunistic tick-borne infection in immunosuppressed patients. J. Intern. Med. 293, 782–790. doi: 10.1111/joim.13638
Hoper, L., Skoog, E., Stenson, M., Grankvist, A., Wass, L., Olsen, B., et al. (2021). Vasculitis due to Candidatus Neoehrlichia mikurensis: a cohort study of 40 Swedish patients. Clin. Infect. Dis. 73, E2372–E2378. doi: 10.1093/cid/ciaa1217
Jha, P., Kim, C. M., Kim, D. M., Yoon, N. R., Jha, B., Park, J. W., et al. (2018). First detection and identification of Candidatus Neoehrlichia mikurensis in South Korea. PLoS One 13:e0209685. doi: 10.1371/journal.pone.0209685
Kawahara, M., Rikihisa, Y., Isogai, E., Takahashi, M., Misumi, H., Suto, C., et al. (2004). Ultrastructure and phylogenetic analysis of 'Candidatus Neoehrlichia mikurensis' in the family Anaplasmataceae, isolated from wild rats and found in Ixodes ovatus ticks. Int. J. Syst. Evol. Microbiol. 54, 1837–1843. doi: 10.1099/ijs.0.63260-0
Kenji, T., Satoru, A., Takako, K., Asao, I., Chiaki, I., Hiroshi, S., et al. (2007). Molecular survey of Babesia microti, Ehrlichia species and Candidatus neoehrlichia mikurensis in wild rodents from Shimane prefecture, Japan. Microbiol. Immunol., 51, 359–367. doi: 10.1111/j.1348-0421.2007.tb03923.x
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lenart, M., Simoniti, M., Strašek-Smrdel, K., Špik, V. C., Selič-Kurinčič, T., and Avšič-Županc, T. (2021). Case report: first symptomatic Candidatus Neoehrlichia mikurensis infection in Slovenia. BMC Infect. Dis. 21:579. doi: 10.1186/s12879-021-06297-z
Li, H., Jiang, J.-F., Liu, W., Zheng, Y.-C., Huo, Q.-B., Tang, K., et al. (2012). Human infection with Candidatus Neoehrlichia mikurensis, China. Emerg. Infect. Dis. 18, 1636–1639. doi: 10.3201/eid1810.120594
Li, H., Jiang, J., Tang, F., Sun, Y., Li, Z., Zhang, W., et al. (2013). Wide distribution and genetic diversity of "Candidatus Neoehrlichia mikurensis" in rodents from China. Appl. Environ. Microbiol. 79, 1024–1027. doi: 10.1128/AEM.02917-12
Lu, B. (1982). Handbook for the identification of important medical animals in China. Beijing: People's Medical Publishing House.
Markowicz, M., Schötta, A.-M., Höss, D., Kundi, M., Schray, C., Stockinger, H., et al. (2021). Infections with Tickborne pathogens after tick bite, Austria, 2015-2018. Emerg. Infect. Dis. 27, 1048–1056. doi: 10.3201/eid2704.203366
Obiegala, A., Pfeffer, M., Pfister, K., Tiedemann, T., Thiel, C., Balling, A., et al. (2014). Candidatus Neoehrlichia mikurensis and Anaplasma phagocytophilum: prevalences and investigations on a new transmission path in small mammals and ixodid ticks. Parasit. Vectors 7:563. doi: 10.1186/s13071-014-0563-x
Pan, H., Liu, S., Ma, Y., Tong, S., and Sun, Y. (2003). Ehrlichia-like organism gene found in small mammals in the suburban district of Guangzhou of China. Ann. N. Y. Acad. Sci. 990, 107–111. doi: 10.1111/j.1749-6632.2003.tb07346.x
Pekova, S., Vydra, J., Kabickova, H., Frankova, S., Haugvicova, R., Mazal, O., et al. (2011). Candidatus Neoehrlichia mikurensis infection identified in 2 hematooncologic patients: benefit of molecular techniques for rare pathogen detection. Diagn. Microbiol. Infect. Dis. 69, 266–270. doi: 10.1016/j.diagmicrobio.2010.10.004
Quarsten, H., Grankvist, A., Høyvoll, L., Myre, I. B., Skarpaas, T., Kjelland, V., et al. (2017). Candidatus Neoehrlichia mikurensis and Borrelia burgdorferi sensu lato detected in the blood of Norwegian patients with erythema migrans. Ticks Tick Borne Dis. 8, 715–720. doi: 10.1016/j.ttbdis.2017.05.004
Rar, V. A., Livanova, N. N., Panov, V. V., Doroschenko, E. K., Pukhovskaya, N. M., Vysochina, N. P., et al. (2010). Genetic diversity of Anaplasma and Ehrlichia in the Asian part of Russia. Ticks Tick Borne Dis. 1, 57–65. doi: 10.1016/j.ttbdis.2010.01.002
Schouls, L. M., Van De Pol, I., Rijpkema, S. G., and Schot, C. S. (1999). Detection and identification of Ehrlichia, Borrelia burgdorferi sensu lato, and Bartonella species in Dutch Ixodes ricinus ticks. J. Clin. Microbiol. 37, 2215–2222. doi: 10.1128/JCM.37.7.2215-2222.1999
Silaghi, C., Woll, D., Mahling, M., Pfister, K., and Pfeffer, M. (2012). Candidatus Neoehrlichia mikurensis in rodents in an area with sympatric existence of the hard ticks Ixodes ricinus and Dermacentor reticulatus, Germany. Parasit. Vectors 5:285. doi: 10.1186/1756-3305-5-285
Sun, J., Liu, H., Yao, X. Y., Zhang, Y. Q., Lv, Z. H., and Shao, J. W. (2023). Circulation of four species of Anaplasmataceae bacteria in ticks in Harbin, northeastern China. Ticks Tick Borne Dis. 14:102136. doi: 10.1016/j.ttbdis.2023.102136
Szekeres, S., Claudia Coipan, E., Rigó, K., Majoros, G., Jahfari, S., Sprong, H., et al. (2015). Candidatus Neoehrlichia mikurensis and Anaplasma phagocytophilum in natural rodent and tick communities in southern Hungary. Ticks Tick Borne Dis. 6, 111–116. doi: 10.1016/j.ttbdis.2014.10.004
Tamura, K. (1992). Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 9, 678–687. doi: 10.1093/oxfordjournals.molbev.a04075
Wang, Z., Wang, J., Yu, M., Feng, L., Wang, X., Xu, Z., et al. (2019). Investigation on Candidatus Neoehrlichia mikurensis in rodents collected in forest area in the northeastern region of China. Chin. J. Zoon. 35, 330–333. doi: 10.3969/j.issn.1002-2694.2019.00.032
Wass, L., Grankvist, A., Bell-Sakyi, L., Bergström, M., Ulfhammer, E., Lingblom, C., et al. (2019). Cultivation of the causative agent of human neoehrlichiosis from clinical isolates identifies vascular endothelium as a target of infection. Emerg. Microb. Infect. 8, 413–425. doi: 10.1080/22221751.2019.1584017
Welc-Faleciak, R., Sinski, E., Kowalec, M., Zajkowska, J., and Pancewicz, S. A. (2014). Asymptomatic "Candidatus Neoehrlichia mikurensis" infections in immunocompetent humans. J. Clin. Microbiol. 52, 3072–3074. doi: 10.1128/JCM.00741-14
Keywords: Neoehrlichia mikurensis, rodents, detection, phylogenetic analysis, Liupan Mountain
Citation: Wang X, Pang B, Kou Z, Zhao J, Yan Y, Chen T and Yang L (2024) Detection and phylogenetic classification of Neoehrlichia mikurensis in rodents from the region of Liupan Mountain, China. Front. Microbiol. 15:1409593. doi: 10.3389/fmicb.2024.1409593
Received: 30 March 2024; Accepted: 10 June 2024;
Published: 04 July 2024.
Edited by:
Jinyu Shan, University of Leicester, United KingdomReviewed by:
Qiaoyun Ren, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2024 Wang, Pang, Kou, Zhao, Yan, Chen and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liping Yang, eWxpcGluZ0BzZHUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.