
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 16 July 2024
Sec. Microbial Physiology and Metabolism
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1409359
This article is part of the Research TopicBacteriocins and other Ribosomally Synthesised and Post-translationally modified Peptides (RiPPs) as modulators of the microbiomeView all 9 articles
 John A. Renye*
John A. Renye* George A. Somkuti†
George A. Somkuti† Phoebe X. Qi
Phoebe X. Qi Dennis H. Steinberg
Dennis H. Steinberg Michael J. McAnulty
Michael J. McAnulty Amanda L. Miller
Amanda L. Miller Giselle K. P. Guron
Giselle K. P. Guron Adam M. Oest
Adam M. OestStreptococcus thermophilus strain B59671 naturally produces thermophilin 110, a broad-spectrum bacteriocin encoded within the bacteriocin-like peptide (blp) gene cluster, and thermophilin 13 from a separate chromosomal locus. Analysis of the blp gene cluster revealed two genes, blpU and blpK, as potentially encoding bacteriocins. Deletion of blpK from the B59671 chromosome did not result in a loss of antimicrobial activity against either S. thermophilus ST113 or Pediococcus acidilactici F. A deletion mutant of blpU could not be generated in B59671, but the mature BlpU peptide obtained through overexpression in E. coli BL21 or chemical synthesis inhibited the growth of S. thermophilus strains, Streptococcus mutans UA159, P. acidilactici F, and Listeria innocua GV9 L-S, evidencing as a broad-spectrum bacteriocin that does not require modification for activity. This study also showed that the transcription of blpU was approximately 16-fold higher in B59671 than in an induced culture of S. thermophilus LMD-9, which produces a blp-encoded bacteriocin. The increased expression of BlpU in B59671 may explain the unique antimicrobial spectrum associated with this strain. Additionally, it was shown that a blpC deletion mutant of B59671, which prevents expression of BlpU and BlpK, inhibited the growth of other S. thermophilus strains and Bacillus cereus, suggesting that thermophilin 13 produced by B59671 possessed both intra- and interspecies antimicrobial activity. While this study confirmed that BlpU can function as an independent antimicrobial peptide, further studies are required to determine if BlpK can function independently as a broad-spectrum antimicrobial.
Streptococcus thermophilus (S. thermophilus) is a thermophilic lactic acid bacterium, commonly used as a starter culture in the production of yogurt and cheeses. Several strains of S. thermophilus have been reported to naturally produce antimicrobial peptides called bacteriocins. These ribosomally encoded peptides have been shown to display a narrow and/or broad spectrum of activity with the potential to inhibit the growth of spoilage and food-borne pathogenic bacteria (Villani et al., 1995; Ward and Somkuti, 1995; Marciset et al., 1997; Ivanova et al., 1998; Mathot et al., 2003; Gilbreth and Somkuti, 2005; Kabuki et al., 2007; Khalil, 2009; Rossi et al., 2013).
The genes required for the production of some thermophilins have been characterized, including thermophilin 13, a two-component bacteriocin (Marciset et al., 1997), and thermophilin 1277, a lantibiotic (Kabuki et al., 2007). Additionally, a bacteriocin-like peptide (blp) gene cluster that resembles the blp gene cluster from Streptococcus pneumoniae (de Saizieu et al., 2000), has been identified in several strains of S. thermophilus and shown to encode functional bacteriocins (Hols et al., 2005; Fontaine and Hols, 2008; Somkuti et al., 2015; Renye et al., 2019). The blp gene cluster in S. thermophilus was found to contain a three-gene operon (blpABC) that encoded the components of an ABC-transporter (blpAB) required for processing and secretion of a mature 30-mer quorum-sensing induction peptide (BlpC30), encoded by blpC (Fontaine et al., 2007). Immediately downstream of the blpABC operon, was a two-gene operon (blpRH) that encoded a histidine kinase (BlpH) and a response regulator (BlpR) required for sensing the BlpC30 induction peptide and inducing the expression of other genes within the cluster (Blomqvist et al., 2006). The remaining components of each gene cluster differ among strains and contain genes believed to encode bacteriocins based on the presence of a double glycine leader sequence within the small proteins. In strain LMD-9, the potential bacteriocin genes included blpD, U, E, and F (Hols et al., 2005); and in strains ST106 and ST109 these genes included blpD, U, and K (Somkuti et al., 2015; Renye et al., 2019).
Strains LMD-9 and ST106 required the addition of a synthetic BlpC30 to the growth medium to induce bacteriocin production (Fontaine et al., 2007). However, strain ST109 was capable of naturally producing its broad-spectrum bacteriocin (Renye et al., 2019). The molecular mechanism that prevents strains, such as LMD-9 and ST106, from naturally producing their respective bacteriocins remains uncharacterized. Other strains, including LMG81311 and CNRZ1066, have been shown to possess a truncation within BlpB which prevents them from producing bacteriocins, even in the presence of exogenous BlpC (Hols et al., 2005). Further analysis of LMD-9 revealed that BlpD alone was sufficient to inhibit the growth of most target bacteria, with BlpU, E, and F identified as accessory peptides that broaden the activity spectrum of BlpD (Fontaine and Hols, 2008). The presence of BlpD in strains ST106 and ST109 suggests it is also the primary bacteriocin within these strains (Somkuti et al., 2015; Renye et al., 2019). Additional variations within this gene cluster have been reported (Rossi et al., 2013), and more may arise as the number of fully sequenced strains of S. thermophilus increases.
Previous work from our laboratory described the natural production of an anti-pediococcal bacteriocin, thermophilin 110, by S. thermophilus B59671 (Gilbreth and Somkuti, 2005), and showed that bacteriocin production was regulated by BlpC (Renye and Somkuti, 2013). In silico analysis of the sequenced B59671 genome later revealed the presence of the thermophilin 13 gene cluster in addition to the blp gene cluster (Renye et al., 2017; Salini et al., 2023). McAnulty et al. (2024) showed that thermophilin 13 was expressed in B59671, and transcription increased when BlpC was deleted (STB59671C−), suggesting cross-talk between the bacteriocin regulatory systems. STB59671C− still inhibited the growth of S. thermophilus 113, confirming the intraspecies antimicrobial activity of thermophilin 13. However, when the genes encoding thermophilin 13 (thmA/B) were knocked-out, the strain still inhibited the growth of P. acidilactici F and Cutibacterium acnes ATCC6919, suggesting that interspecies bacteriocin activity was due to the production of thermophilin 110.
The purpose of this study was to determine which genes within the blp gene cluster encoded the active bacteriocin through the use of gene knockout mutants, and the overexpression or synthesis of targeted peptides for use in in vitro antimicrobial assays.
S. thermophilus strains B59671 (NRRL, Agricultural Research Service Culture Collection, NCAUR, USDA, Peoria, IL), ST109, ST113, ST128 (in-house collection), LMG18311, LMD-9, and Streptococcus salivarius K12 (ATCC BAA1024) were grown in tryptone-yeast extract-lactose (TYL) broth at 37°C. Pediococcus acidilactici strain F (gift from B. Ray, University of Wyoming) and Lactobacillus helveticus 1942 (in-house collection) were grown in deMan, Rogosa, and Sharpe (MRS) medium. Escherichia coli strains DH5α and BL21, Enterococcus faecalis VA797, Enterococcus faecium H3, Bacillus cereus ATCC1457, and Listeria innocua GV9-LS were grown in Brain Heart Infusion (BHI) medium, and Streptococcus mutans strains GS-5 and UA159 were grown in Todd Hewitt Broth (THB) at 37°C. All strains were grown aerobically with E. coli, B. cereus and L. innocua broth cultures agitated at 200 rpm. Gene expression within the LMD-9 blp locus was induced by adding a final concentration of 250 ng/mL of synthesized BlpC30 (stock solution 1 mg/mL in H2O) to the growth medium. BlpC30 (30 mer induction peptide) was prepared by microwave-assisted solid phase peptide synthesis with Fmoc-protected amino acids, using a CEM Liberty synthesizer (CEM Corp., Matthews, NC). The purified peptide was analyzed by HPLC and the molecular mass was confirmed by MALDI-TOF MS.
Bacteriocin activity was measured using an agar diffusion method (Gilbreth and Somkuti, 2005). Cultures of S. thermophilus B59671 and LMD-9 were grown to an OD660 of ~0.8 in TYL and pelleted at 10,000×g for 10 min at 4°C for collection of cell-free supernatants (CFS). CFS (~50 μL; ~320 AU/mL) was loaded into precast wells in an agar medium (1.5%) seeded with the target bacteria (0.5% v/ v). Bacterial targets included: S. thermophilus ST109, ST113 (positive control strain), 128, and LMG18311, S. salivarius K12 in TYL; P. acidilactici (positive control strain) and L. helveticus in MRS; B. cereus, E. faecalis and E. faecium in BHI; and S. mutans GS-5 and UA159 in THB. Plates were allowed to equilibrate at 4°C overnight, and then incubated at 37°C and examined for the appearance of zones of inhibition. ST113 was routinely run as a negative control to show that inhibition zones were not due to acid production.
P. acidilactici F cells were grown overnight at 37°C, and then diluted in fresh MRS (25 fold) and grown until mid-log phase (O.D.600 nm = 0.5). Cells were harvested by centrifugation (10,000×g for 5 min at 4°C) and resuspended in 0.1% peptone water at the same optical density. Cells were then incubated at 37°C in the absence or presence of 510 AU/mL thermophilin 110. Culture O.D. was monitored for up to 3 h, and cells were collected at various times with 50 μL of bacteria placed on acetone-cleaned 12 mm Micro-cover glass slides (Thermo Scientific, Portsmouth, NH, United States) and allowed to adhere for 30 min. Samples were covered with 2 mL of 2.5% glutaraldehyde, (Electron Microscopy Sciences, Hatfield, PA, United States) and allowed to fix for 30 min. Samples were rinsed twice (30 min each) with 2–3 mL 0.1 M imidazole, (Electron Microscopy Sciences, Hatfield, PA, United States), followed by washing with 50, 80, and 90% ethanol (The Warner-Graham Company, Cockeysville, MD, USA) at 30-min intervals (2–3 mL each). The samples were then washed three times with 2 mL of 100% ethanol (30-min per wash) before critical point drying. The samples were stacked in a wire basket, separated by cloth, and placed in a Critical Point Drying Apparatus (Denton Vacuum, Inc., Cherry Hill, NJ, United States), using liquid carbon dioxide (Welco Co, Allentown, PA, United States) for approximately 20 min. The samples were mounted on stubs and sputter gold coated for 1 min (EMS 150R ES, EM Sciences, Hatfield, PA). Samples were then viewed with an FEI Quanta 200 F Scanning Electron Microscope (Hillsboro, OR, United States) with an accelerating voltage of 10 KV in high vacuum mode.
Sequencing of the blp gene cluster in S. thermophilus B59671 was performed by chromosome walking using purified genomic DNA as a template. The initial PCR primers were designed based on the nucleic acid sequence of LMD-9, corresponding to the 5′ end of blpA (5′-ATGTTTCGTTTTCGTAGG-3′) and the 3′ end of blpX (5′-AATTTTTACGATTCATTT-3′). After each round of sequencing, a new primer was designed based on the obtained sequence and walking was continued until the entire gene cluster was sequenced in both directions. Nucleic acid sequencing was performed using a 3730 PRISM® DNA analyzer (Applied Biosystems, Foster City, CA) with the ABI PRISM Big Dye terminator cycle sequencing reagent. Sequences were analyzed and contigs were created using Sequencher 4.9 (Gene Codes Corp, Ann Arbor, MI). Open Reading frame (ORF) identification and promoter analysis were performed using Clone Manager software (Sci-ed Software, Cary, NC).
S. thermophilus strains were grown in TYL broth until the culture reached an OD600 between 0.8 and 1.0. RNA was extracted using the RiboPure™-Bacteria kit (Ambion-Life Technologies, Grand Island, NY). To eliminate any residual DNA, RNA was treated with DNase I (Ambion) for 30 min at 37°C. RT-PCR was performed using a Lightcycler® 96 (Roche Diagnostics Corp., Indianapolis, IN). Primers for blpC, blpD, blpU and 16S rRNA (control) are listed in Table 1. Cycling conditions were selected as the following: 40 cycles of 95°C for 30 s, 45°C for 30 s, and 60°C for 30 s. The melt curve analysis was performed from 60 to 95°C with fluorescence readings taken continuously after a 1% temperature increase. cDNA synthesis was performed with Transcriptor Universal cDNA Master (Roche) with 1 μg of total RNA. RT-PCR analysis was conducted with 0.015 ng cDNA using the Fast Start Essential DNA Green Master (Roche). Lightcycler® 96 software was used to determine Cq values and relative quantification. Results were reported from a minimum of three independent RT-PCR reactions with the 16S rRNA gene used as a reference and shown to be stably expressed.
An integrative vector for knocking out blpK in S. thermophilus B59671 was constructed as previously described (Renye and Somkuti, 2013). The primers used to amplify 600 bp fragments of blpK flanking genes were designed as follows: orf6 Fwd (SpeI): CCCACTAGTCGCTAAGGGCTTACTTGA and orf6 Rev. (PstI): CCCCTGCAGGG-AAATCTTCCTCTAATT; and ISB Fwd (XhoI): CCCCTCGAGGCTTTGTTAGGTAATATC and ISB Rev. (KpnI): CCCGGTACCGAGGACAACATTCTCAATCG. The amplified fragments were cloned into pKS1 (Shatalin and Neyfakh, 2005) at the corresponding restriction sites, resulting in the plasmid, pSTKOK, and subsequently transformed and propagated in E. coli DH5α. The integrative vector pSTKOK was then electroporated into strain B59671 using a previously described protocol (Somkuti and Steinberg, 1988), with transformants selected on TYL containing 15 μg/mL erythromycin (erm) at 30°C. Transformants were sub-cultured in TYL+ erm at 37°C to allow for chromosomal integration of pSTKOK, and then sub-cultured in TYL at 30°C to allow for the second homologous recombination event to occur. A B59671 mutant with blpK replaced by a kanamycin resistance marker was confirmed by PCR using the primers: orf5Fwd: GAAAACAAAGATATGTATATTCGCCTA and ISB2Rev: AACGTTTTCAAGAGCGCAAT. S. thermophilus transformants were maintained in TYL containing 150 μg/mL kanamycin.
The nucleic acid sequence (162 base pairs) corresponding to the mature BlpU peptide (excluding the 23 amino acid leader peptide) was amplified from S. thermophilus B59671 with the primers: BlpU Fwd (BamHI): ATAGGATCCATCGAAGGTCGTGGATGTAGCTGGGGAGGT (nucleotides encoding the Factor Xa recognition sequence (isoleucine-glutamic acid-glycine-arginine) are italicized) and BlpU Rev. (XhoI): ATCTCGAGTCACCACCAGCATGTTGCTC. PCR was performed using the high-fidelity Pfx Taq polymerase (Life Technologies, Grand Island, NY); and the resulting DNA fragment was cloned into pGEX 6P-1 (GE Healthcare Life Sciences, Piscataway, NJ) at the corresponding restriction sites, downstream of the isopropyl β-D-1-thiogalactopyranoside (IPTG) inducible promoter. Cloning was confirmed by PCR using primers specific to blpU.
The plasmid was transformed into E. coli BL21 for recombinant expression of the fusion protein. E. coli containing pGEX 6P-1/blpU was grown overnight in BHI broth containing 100 μg/mL ampicillin (amp) at 37°C under agitation at 200 rpm. The culture was then diluted 1:100 into 100 mL BHI amp and grown at 37°C until the optical density (OD600) reached 0.8–1.0, at which point IPTG was added at a final concentration of 0.1 mM. The culture was incubated for an additional 6 h, after which cells were collected by centrifugation at 10,000×g for 10 min, washed twice with phosphate-buffered saline (PBS) prior to resuspension in 20 mL PBS, and lysed by sonication (10 pulses of 30 s) using a microtip cell disruptor (Heat Systems Ultrasonics, Farmingdale, NY, United States). The lysate was centrifuged to remove cellular debris, then mixed with 1x bed volume of a 50% slurry glutathione Sepharose 4B resin (GE Healthcare) and incubated at 25°C for 2 h and 4°C overnight. The GST-BlpU fusion protein (~31 kDa) was visualized on a 12% Bis-Tris SDS-PAGE with Simply Blue™ SafeStain (ThermoFisher Scientific, Waltham, MA), was eluted from the Sepharose 4B resin as described by the manufacturer and dialyzed (12–14 kDa molecular weight cut off) against Factor Xa digestion buffer (20 mM Tris/HCl-100 mM NaCl-2 mM CaCl, pH 8.0) at 4°C overnight. The solution within the dialysis bag was recovered and analyzed by SDS-PAGE and then digested with 50 μL (1 mg/mL) of Factor Xa protease. The digested mixture was analyzed by 12% Bis-Tris SDS-PAGE with silver nitrate staining and analyzed for antimicrobial activity against P. acidilactici using a gel overlay method (Gilbreth and Somkuti, 2005).
The mature BlpU peptide (53 aa: GCSWGGFAKQGVATGVGNGLRLGIKTRTWQGA-VAGAAGGAIVGGVGYGATCWW) was synthesized and purified by high-performance liquid chromatography (Lifetein LLC, South Plainfield, NJ) to a purity of 85.9%. The peptide was dissolved in dimethyl sulfoxide (DMSO) at a stock concentration of 4.6 mg/mL. Further dilution to a working stock of 100 μg/mL was carried out in ddH2O. Antimicrobial activity was determined in a well-diffusion assay using 50 μL of the working stock solution.
Chromosome walking was used to sequence the blp gene cluster within strain B59671, leading to a single 10.7 kb contig shown to contain 14 open reading frames (ORFs) (Figure 1). The resulting contig was aligned with the circularized whole genome sequence (WGS) of strain B59671 (Renye et al., 2017) to identify the location of the blp locus and confirm the accuracy of the WGS sequencing results. As reported for other S. thermophilus strains, the three-gene operon containing homologs of blpA, blpB, and blpC was directly adjacent to the two-gene operon encoding for the histidine kinase (BlpH) and response regulator (BlpR) required for inducing the expression of genes within the blp locus (Bolotin et al., 2004; Fontaine et al., 2007; Sun et al., 2011; Kang et al., 2012). When compared with the blp locus in LMD-9 (Accession number: CP000419), it was determined that the predicted proteins, BlpR and BlpH, differed by 7 and 11 amino acids, respectively, in B59671.
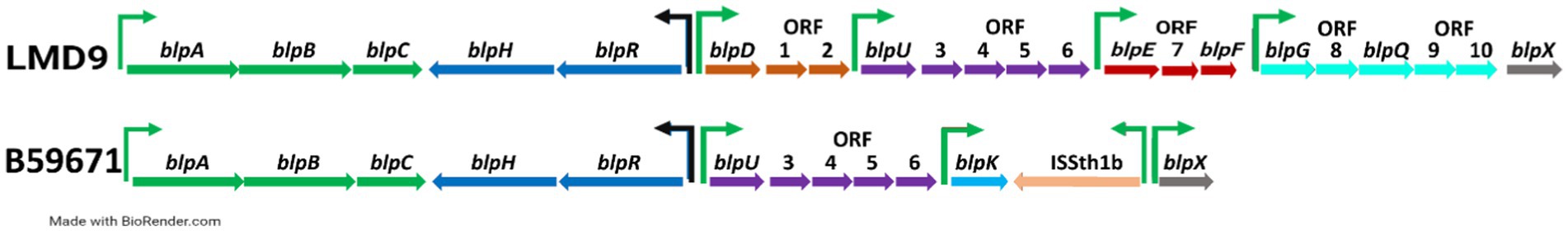
Figure 1. Map of the Streptococcus thermophilus bacteriocin-like peptide (blp) gene cluster in strains LMD-9 (top) and B59671 (bottom). Homologous gene or operons are grouped by color, and constitutive promoters (black arrows) and BlpC30 inducible promoters (green arrows) are shown. Gene nomenclature was taken from Fontaine et al. (2007), and gene maps were created with BioRender.com.
In strain B59671, the region between the blpRH operon and blpX contained two ORFs which encode putative antimicrobial peptides based on the presence of a double glycine (GG) leader sequence. The predicted mature peptide encoded by blpU showed 100% identity with the corresponding peptides in LMD-9, LMG18311 (Accession number: CP000023) and ST106, but differed with the peptide expressed in ST109 by a single amino acid substitution, valine to glycine at the N-terminus (Renye et al., 2019). In all strains blpU was part of a two-gene operon with orf3, which was predicted to encode the immunity protein required for protection against BlpU (Fontaine and Hols, 2008). The second ORF within the B59671 blp gene cluster was predicted to encode an 81 amino acid peptide with 98% identity to BlpK in strain CNRZ1066 (Hols et al., 2005), but differed more with the BlpK homologs in ST106 and ST109 due to the presence of a four-amino acid repeat (IGGA) insertion at positions 20–23 of the mature peptide (Renye et al., 2019).
The nucleic acid sequences upstream of the blpABC, blpU-orf3, and blpK operons were compared to the previously characterized blp-inducible promoters within the LMD-9 gene cluster (Fontaine et al., 2007). The blpABC promoter in strain B59671 was identical to the characterized region in LMD-9, including the presence of two imperfect direct repeats (DR), the left repeat (LR): ACCGTTTGGGACG and right repeat (RR): ACTTTTTGGGACG, shown to be essential for BlpC30-induced gene expression (Blomqvist et al., 2006). The promoters upstream regions of blpU-orf3 and blpK matched the promoter region upstream of the blpE/orf7/blpF operon in LMD-9. This promoter sequence differed from the blpU promoter in LMD-9 by two nucleotides in the LR sequence: ACCATTCGGGACA (B59671) and ACTACTCGGGACA (LMD-9) (Fontaine et al., 2007).
Previously it was reported that thermophilin 110 was naturally produced by S. thermophilus strain B5967; however, bacteriocin production by strain LMD-9 required induction with the BlpC quorum sensing peptide (BlpC30) at 250 ng/mL (Renye et al., 2016). Semiquantitative RT-PCR was used to compare the expression of blp components known to be upregulated by BlpC30 when strains B59671 and LMD-9 reached late exponential phase (OD660 of ~0.8). The level of blpC mRNA was substantially higher in B59671 when compared to both non-induced and induced cultures of LMD-9 (Figure 2), similar to what was observed when transcription levels were compared between B59671 and ST106 (McAnulty et al., 2024). Following induction with the BlpC30 peptide, BlpC expression increased by 6-fold in LMD-9, ~16-fold lower than the expression level in B59671. Transcription of the blpU-orf3 operon was more than 45-fold higher in B59671 when compared to both induced and non-induced cultures of LMD-9 (Figure 2). Consistent with a previous report (Fontaine et al., 2007), transcription of the blpuU-orf3 operon responded poorly to induction with BlpC30 in LMD-9 (~3-fold). However, the expression of BlpD increased by ~45 fold following induction with BlpC30 in LMD-9 (Figure 2), a level of expression similar to what was observed for BlpU in B5967. In B59671, the expression level of BlpK was comparable to that of BlpU, which was expected since their promoter regions were identical (data not shown).
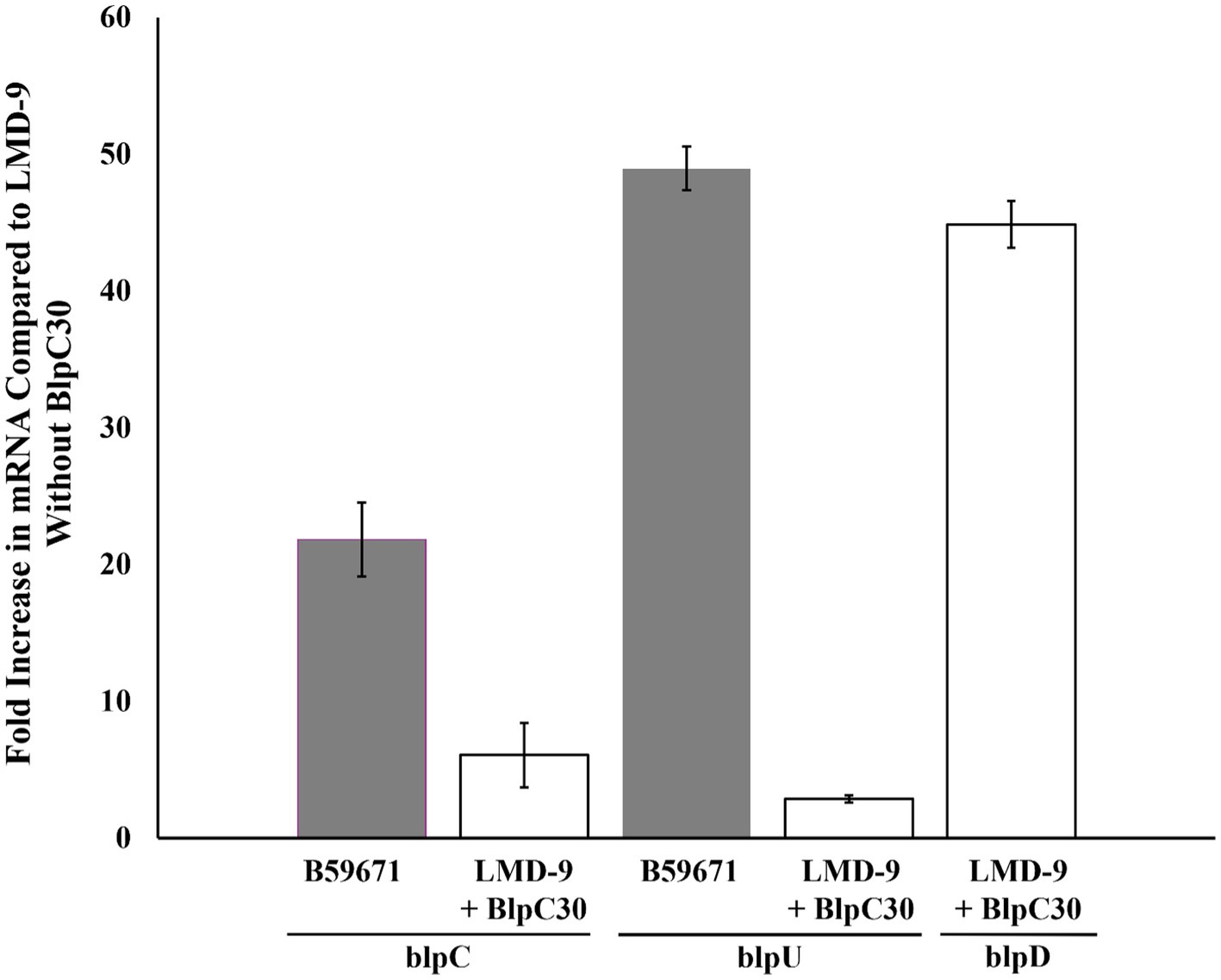
Figure 2. Real-time PCR analysis of blpC and blpU expression in B59671 (gray bars) and LMD-9 + (white bars); and blpD in LMD-9 + BlpC30 (white bar) in tryptone yeast extract (TYL) medium. Expression is presented as the fold-increase in mRNA levels as compared to the LMD-9 control (non-induced). Data are the average of at least three independent experiments (± Standard Deviation).
Thermophilin 110 was previously reported to induce cell lysis of P. acidilactici (Gilbreth and Somkuti, 2005) and Cutibacterium acnes (Renye et al., 2023) based on a drop in culture optical density (OD600nm). In this study, when P. acidilactici cells were exposed to 512 AU/mL thermophilin 110, the optical density of the culture dropped from 1.06 (OD600nm) to 0.299 (OD600nm) by 2 h; and viable counts dropped from 8.49 log CFU/mL (T0) to 5.96 log CFU/mL (T2). Additionally, lysed cells were observed by SEM within 0.5 h (Figure 3B, arrows), with several fully lysed cells observed after 1.5 h (Figure 3C, arrows). These results confirmed the bactericidal activity of thermophilin 110.
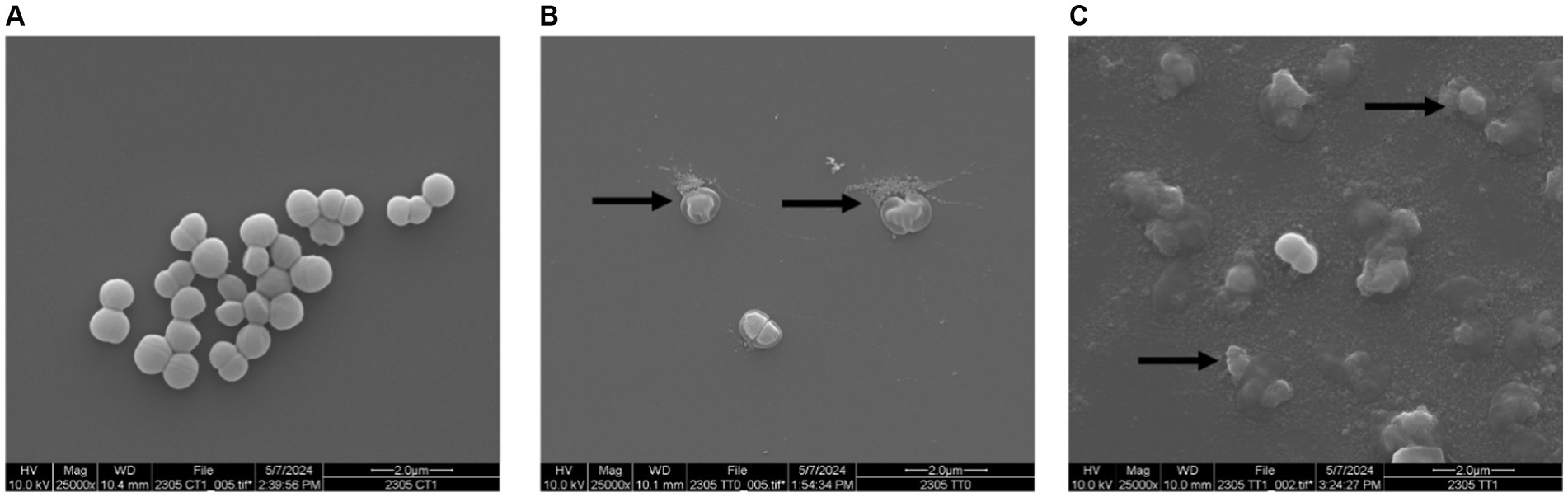
Figure 3. Lysis of P. acidilactici by thermophilin 110. P. acidilactici cells grown in MRS until mid-exponential phase, and then resuspended in 0.1% peptone water in the absence (A) or presence of 512 AU/mL thermophilin 110 (B, C). SEM images were collected after P. acidilactici cells were in peptone water without thermophilin 110 (A) or exposed to thermophilin 110 for 0.5 h (B) or 1.5 h (C), with images shown at 25,000X magnification.
Additionally, the activity spectrum of thermopilin 110 was confirmed using a selection of previously reported susceptible bacteria (Gilbreth and Somkuti, 2005; Renye and Steinberg, 2021), since a recent study revealed that S. thermophilus B59671 produces a second active bacteriocin, thermophilin 13 (McAnulty et al., 2024). Cell-free supernatants from B59671C− maintained intraspecies activity against four S. thermophilus strains and showed broad-spectrum activity against B. cereus (Table 2), most likely due to the production of thermophilin 13. Antimicrobial activity was not observed against enterococci, Lb. helveticus, S. mutans, and S. salivarius when CFS from B59671C− was tested, suggesting that these unique activities were dependent on the production of thermophilin 110.
To determine which genes encoded active bacteriocins within the B59671 blp gene cluster, the integrative vector pKS1 was used to construct deletion mutants for both blpU and blpK. A plasmid was successfully constructed to replace blpU with a kanamycin resistance maker (kanr), but the knock out mutant was not obtained after several attempts (data not shown). However, the vector pSTKOK was generated and successfully used to delete the chromosomal copy of blpK from B59671. The mutant, S. thermophilus B59671K−, was sensitive to erythromycin and resistant to kanamycin, confirming a double crossover recombination event where kanr was inserted within the chromosome. The mutant was further confirmed by PCR analysis using primers corresponding to the genes orf5 and ISSB, which flank blpK. Insertion of kanr increased the PCR product size from 1.3 kB to 2.8 kB. Cell-free supernatant from an overnight culture of S. thermophilus B59671K− (blpK knockout) was shown to inhibit the growth of P. acidilactici PAF (Figure 4, well 3), with a zone similar in size to the parent culture (well 1). However, ST B59671C− did not inhibit pediococcal growth (Figure 4, well 2), validating that the active bacteriocin was encoded within the blp locus. These results suggested that BlpU was a bacteriocin with anti-pediococcal activity.
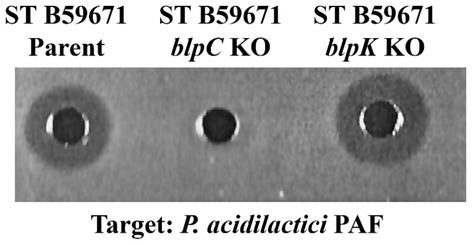
Figure 4. Antimicrobial activity of S. thermophilus B59671 and knock out mutants against Pediococcus acidilactici PAF. Cell free supernatants were collected after 24 h of growth and loaded into precast wells. Anti-pediococcal activity was evident by zones of inhibition around wells. (Left to Right) Well 1: CFS from ST B59671 Parent strain; Well 2: ST B59671 blpC knock out mutant; Well 3: B59671 blpK knock out mutant.
Since the creation of a blpU knockout strain was unsuccessful, overexpression of the predicted mature peptide (without the 23 amino acid leader sequence) was demonstrated using E. coli BL21 as the host bacterium. The plasmid pGEX 6P-1/blpU was constructed to express a GST-BlpU fusion protein, which also contained a Factor Xa cleavage sequence at the N-terminus of BlpU. The recombinant gene was located downstream of an IPTG inducible promoter, and transcription was induced when the culture reached an OD600 ~ 0.8, with the culture incubated for an additional 6 h. After lysis of the induced E. coli cells, the fusion protein was recovered using 50% slurry of glutathione Sepharose 4B resin. The eluted protein was run on an SDS-PAGE gel which showed the presence of an expected fusion protein, ~31 kDa (Figure 5A, lane 2). The band corresponding to the fusion protein was not observed in uninduced cells (Figure 5A, lane 3), or in a lysate from E. coli BL21 without the expression plasmid, attesting that the protein band was not a host cell protein. The GST-BlpU fusion protein did not show antimicrobial activity against P. acidilactici, most likely due to the presence of the GST tag on its N-terminus (data not shown). After Factor Xa cleavage, a small peptide (~5 kDa) was observed by SDS-PAGE analysis after staining with silver nitrate (Figure 5B, lane 2), and a faint inhibition zone corresponding to the released peptide was observed when a washed SDS-PAGE was overlayed onto an agar medium inoculated with P. acidilactici (Figure 5C, lane 2). This confirmed that the GST tag must be removed to render BlpU active; and the location of the active BlpU on gel overlay corresponded to the antimicrobial peptide present in CFS from S. thermophilus B59671 (Figure 5C, lane 3). The inhibition zone results strongly suggested that BlpU is a broad-spectrum bacteriocin produced by this strain.
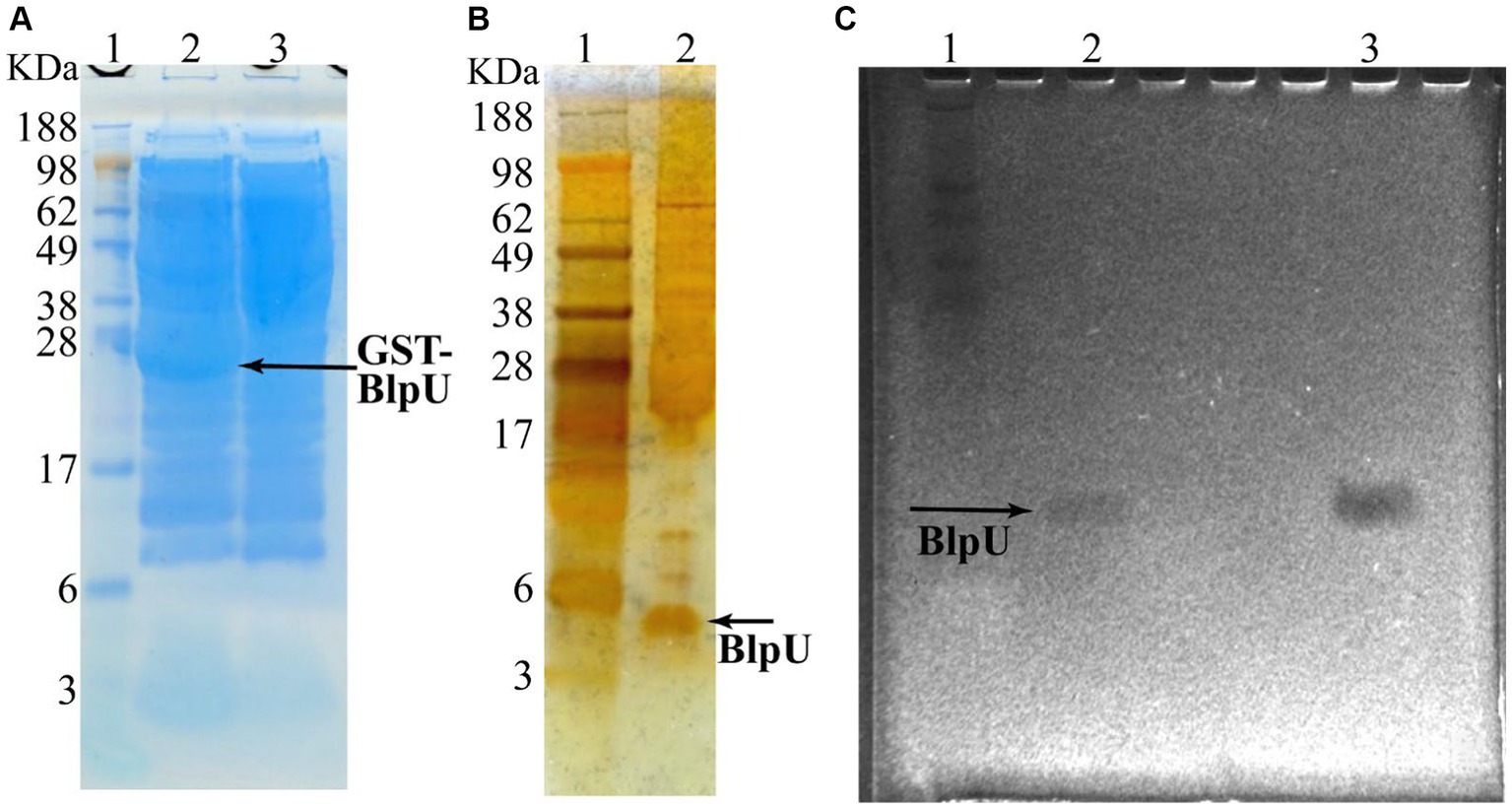
Figure 5. Expression of recombinant mature BlpU peptide in E. coli BL21. (A) Simply Blue stained SDS-PAGE of cell lysates from E. coli BL21 containing plasmid which encodes GST-BlpU fusion protein. Lane 1: Protein marker; Lane 2: Lysate from IPTG-induced cells, GST-BlpU labeled (arrow); Lane 3: Lysate from cells that were not induced (no fusion protein). (B) Silver nitrate stained SDS-PAGE of purified BlpU following Factor Xa cleavage. Lane 1: Protein marker; Lane 2: Lysate following Factor Xa cleavage, BlpU labeled (arrow). (C) Gel overlay assay showing inhibition zones with P. acidilactici PAF used as the target bacterium. Lane 1: Protein marker; Lane 2: Recombinant BlpU from E. coli; Lane 3: CFS from overnight ST B5971 culture.
To further establish that BlpU is an active bacteriocin in the absence of other components encoded within the blp gene cluster of S. thermophilus B59671, the mature peptide was chemically synthesized. The resulting peptide (purity ~86%) was resuspended in DMSO at a concentration of 4.6 mg/mL. The peptide at a working concentration of 100 μg/mL (in sterile water) was applied to inhibit the growth of S. thermophilus ST113, P. acidilactici, L. inoccua, and S. mutans (Figure 6, column 2). Inhibition zones were comparable to the zones observed using a partially purified thermophilin 110 preparation (~512 AU/mL) as a control (Figure 6, column 1). Inhibition zones were not observed around wells loaded with a diluted DMSO solution as a negative control, evidencing the activities arose from the presence of BlpU (Figure 6, column 3). S. thermophilus B59671 was shown to be resistant to all bacteriocin preparations.
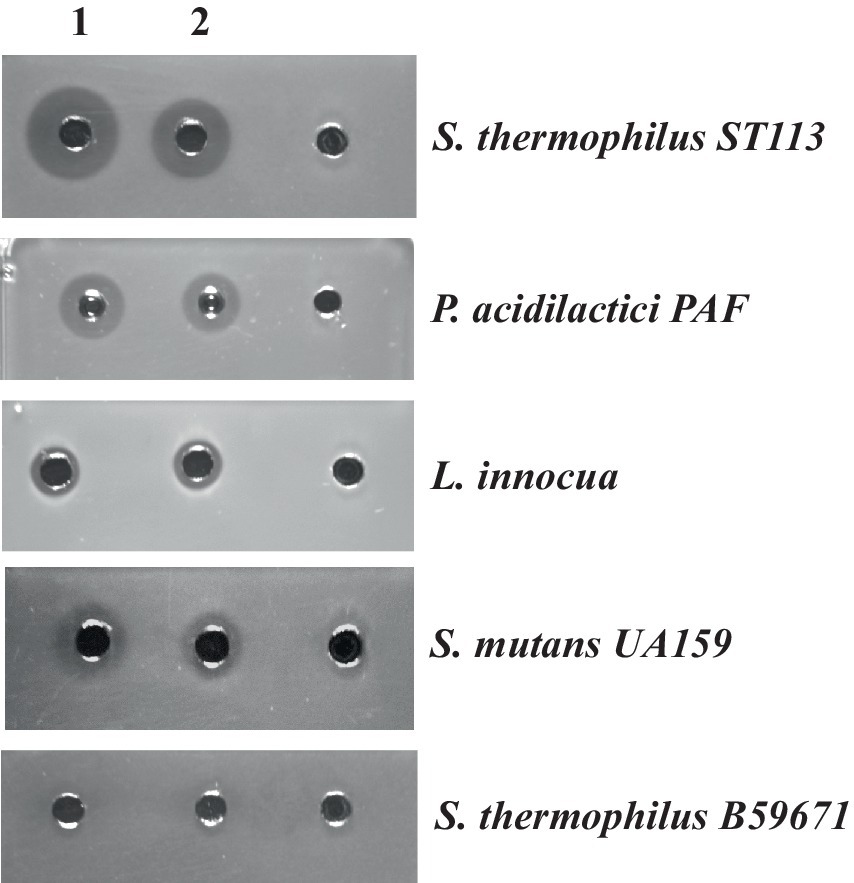
Figure 6. Antimicrobial activity of the chemically synthesized mature BlpU peptide against selected bacterial targets: S. thermophilus ST113, P. acidilactici PAF, L. innocua, S. mutans UA159 and S. thermophilus B59671. Wells in column 1: 50 μL of partially purified thermophilin 110 from B59671 CFS; column 2: 50 μL of the synthesized BlpU (100 μg/mL); column 3: 50 μL of the DMSO/water solution used to resuspend synthetic peptide.
Natural production of blp bacteriocins has only been reported in a limited number of strains, including B59671 and ST109 (Gilbreth and Somkuti, 2005; Renye and Steinberg, 2021), whereas the production of Blp bacteriocins must be induced in other strains, including LMD-9, ST106, and ST118 (Fontaine et al., 2007; Somkuti and Renye, 2015) by either overexpressing blpC or supplementing the growth medium with synthetic BlpC30. While BlpD has been shown to be essential for the antimicrobial activity in LMD-9 (Fontaine and Hols, 2008), the blp gene cluster in B59671 lacks a homolog of blpD. Since blpU and blpK are the only genes in the B59671 blp cluster that encode proteins with a conserved class II bacteriocin double-glycine motif, it is implied that BlpU and/or BlpK must be the active bacteriocin in B59671.
The blp gene cluster in B59671, as well as in LMD-9 and ST106, encodes a fully functional quorum sensing system, including the peptide processing and transport proteins (BlpAB), quorum sensing induction peptide (BlpC), and a histidine kinase and response regulator (BlpHR). However, strains LMD-9 and ST106 do not produce their respective bacteriocins naturally (Fontaine et al., 2007; Somkuti and Renye, 2015), most likely a result of the quorum sensing peptide, BlpC, never reaching a threshold concentration required to induce gene expression from within the blp loci. This study showed that transcription of blpC was more than 20-fold higher in B59671 than in a BlpC30-induced culture of LMD-9. A similar result was reported in a transcriptomic study of B59671 and ST106 (McAnulty et al., 2024), again suggesting the constitutive expression of BlpC was not sufficient to induce transcription of other components within the blp gene cluster.
In strain LMG18311, a truncation in BlpB explains why the blp quorum sensing system does not function, but comparative genomic studies have not established why the blpABC operon is expressed significantly higher in strains B59671 and ST109. The promoter sequence upstream of the blpABC operon is identical in strains B59671 and LMD-9, and so is the nucleic acid sequence of the blpABC and blpRH operons, which reside on opposite DNA strands. It was previously hypothesized that a leaky terminator between the blpABC and blpRH operons may allow for the formation of a double-stranded RNA molecule that could be degraded by RNase III (Lamontagne and Elela, 2004). However, it remains unclear why this would only occur in selected strains. Another possibility is that transcriptional regulators located outside the blp gene cluster may regulate the expression of the blp quorum sensing system. Cross talk via ComR (Fontaine et al., 2013), or other bacteriocin signaling peptides (thmX) or global transcriptional regulators within S. thermophilus (McAnulty et al., 2024) are potential candidates and need to be explored further.
Homologs of blpU are present in LMD-9, ST106, and ST109, yet these strains have not been reported to inhibit the growth of some bacterial targets sensitive to thermophilin 110, including S. mutans, C. acnes 6,919, and P. acidilactici F. This may be attributed to a lowered level of BlpU production in these strains. In LMD-9, BlpU was characterized as an accessory peptide that extends the activity spectrum of BlpD (Fontaine and Hols, 2008). The expression of blpU was induced by the presence of BlpC30, however, induction was lower than what was observed for the blpD-orf2 or blpE-blpF operons in LMD-9 (Fontaine et al., 2007). Analysis of the promoter upstream regions of blpU and blpK in B59671 showed that they are identical to the promoter upstream of the blpE-blpF operon in LMD-9, which was the second most responsive promoter to BlpC30 induction in LMD-9 (Fontaine et al., 2007). The promoter upstream of blpU in other strains may limit peptide expression so that the unique antimicrobial activities associated with the production of BlpU are not observed. Additionally, strain B59671 does not possess a homolog of blpG, which encodes a thiol-disulfide oxidase that is required for peptide modification and subsequent antimicrobial activities described for strain LMD-9 (Fontaine and Hols, 2008). The absence of BlpG in B59671 may prevent modifications to BlpU which are detrimental to the unique antimicrobial activities described in this study.
Characterization of the antimicrobial activity associated with S. thermophilus B59671 is complicated by the presence of two functional bacteriocin gene clusters (Salini et al., 2023; McAnulty et al., 2024). Acquiring the ability to produce two separate bacteriocins may have provided a competitive advantage for B59671 to outcompete closely related species within its natural niche, and now may make it an ideal candidate for use in food and non-food applications. When compared to traditional antibiotics, bacteriocins have a limited spectrum of activity and their use is not expected to result in the selection of microorganisms resistant to clinically relevant antimicrobials. This has allowed bacteriocin-producing lactic acid bacteria to be investigated for improving the safety and shelf-life of foods, and as probiotics. However, potential applications for S. thermophilus strains as bioprotective cultures remains scarce. This is predominantly due to most strains not being able to produce their bacteriocins naturally; however, this study showed that B59671 exhibits a broad spectrum of antimicrobial activity due to the natural production of both thermophilin 110 and thermophilin 13, but additional studies are needed to assess the efficacy of these bacteriocins within food matrices or in vivo.
Results from this study have shown for the first time that blpU from strain B59671 encodes a functional bacteriocin, BlpU, with unique broad-spectrum antimicrobial activities against P. acidilactici PAF, L. innocua, and S. mutans UA159. The unsuccessful attempts to knock out blpU in B59671 suggested it could have an essential function in this strain, or that the cloning strategy may have impaired the immunity protein encoded by orf3. With the immunity protein for BlpK currently unknown, the possibility exists that orf3 is required for host immunity against both BlpU and BlpK. Mutants resulting in the deletion of blpU were obtained in strain LMD-9 (Fontaine and Hols, 2008), but this may have been possible due to the substantial differences between the blp gene clusters in LMD-9 and B59671, specifically the absence of a BlpK homolog in LMD-9. Further studies are required to determine if BlpK can function as an independent broad-spectrum bacteriocin, similar to what was shown for BlpU in this study.
The genome sequence for S. thermophilus B59671, which includes the blp gene cluster shown, is deposited in GenBank with the accession number CP022547.
JR: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. GS: Conceptualization, Formal analysis, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. PQ: Methodology, Formal analysis, Writing –review & editing. DS: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. MM: Formal analysis, Methodology, Writing – review & editing. AM: Formal analysis, Methodology, Writing – review & editing. GG: Writing – review & editing. AO: Data curation, Formal analysis, Methodology, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the in-house project: 8072-41000-114-00D, “Reclaiming Value from Coproducts of Dairy Food Manufacture.”
We thank J. Uknalis (USDA-ARS-ERRC) for SEM imaging, M. Toht (USDA-ARS-ERRC) for technical assistance and D. Needleman (USDA-ARS-ERRC) for assistance with nucleic acid sequencing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1409359/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | RP-HPLC chromatograph for synthesized BlpU peptide (Peak 4) from S. thermphilus B59671 (LifeTein, Hillsborough, NJ). Column: 4.6 × 250 mm, PLRP-S 100A; Mobile Phase: A=0.1% TFA/Acetonitrile, B = 0.1% TFA/water; Flow rate: 1.0 mL/min, UV wavelength: 220 nm; injection volume was 5 μL.
Blomqvist, T., Steinmoen, H., and Havarstein, L. S. (2006). Pheromone-induced expression of recombinant proteins in Streptococcus thermophilus. Arch. Microbiol. 186, 465–473. doi: 10.1007/s00203-006-0162-0
Bolotin, A., Quinquis, B., Renault, P., Sorokin, A., Ehrlich, S. D., Kulakauskas, S., et al. (2004). Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22, 1554–1558. doi: 10.1038/nbt1034
De Saizieu, A., Gardes, C., Flint, N., Wagner, C., Kamber, M., Mitchell, T. J., et al. (2000). Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182, 4696–4703. doi: 10.1128/JB.182.17.4696-4703.2000
Fontaine, L., Boutry, C., Guedon, E., Guillot, A., Ibrahim, M., Grossiord, B., et al. (2007). Quorum-sensing regulation of the production of Blp bacteriocins in Streptococcus thermophilus. J. Bacteriol. 189, 7195–7205. doi: 10.1128/JB.00966-07
Fontaine, L., Goffin, P., Dubout, H., Delplace, B., Baulard, A., Lecat-Guillet, N., et al. (2013). Mechanism of competence activation by the ComRS signalling system in streptococci. Mol. Microbiol. 87, 1113–1132. doi: 10.1111/mmi.12157
Fontaine, L., and Hols, P. (2008). The inhibitory spectrum of thermophilin 9 from Streptococcus thermophilus LMD-9 depends on the production of multiple peptides and the activity of BlpG(St), a thiol-disulfide oxidase. Appl. Environ. Microbiol. 74, 1102–1110. doi: 10.1128/AEM.02030-07
Gilbreth, S. E., and Somkuti, G. A. (2005). Thermophilin 110: a bacteriocin of Streptococcus thermophilus ST110. Curr. Microbiol. 51, 175–182. doi: 10.1007/s00284-005-4540-7
Hols, P., Hancy, F., Fontaine, L., Grossiord, B., Prozzi, D., Leblond-Bourget, N., et al. (2005). New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 29, 435–463. doi: 10.1016/j.femsre.2005.04.008
Ivanova, I., Miteva, V., Stefanova, T., Pantev, A., Budakov, I., Danova, S., et al. (1998). Characterization of a bacteriocin produced by Streptococcus thermophilus 81. Int. J. Food Microbiol. 42, 147–158. doi: 10.1016/S0168-1605(98)00067-1
Kabuki, T., Uenishi, H., Watanabe, M., Seto, Y., and Nakajima, H. (2007). Characterization of a bacteriocin, Thermophilin 1277, produced by Streptococcus thermophilus SBT1277. J. Appl. Microbiol. 102, 971–980. doi: 10.1111/j.1365-2672.2006.03159.x
Kang, X., Ling, N., Sun, G., Zhou, Q., Zhang, L., and Sheng, Q. (2012). Complete genome sequence of Streptococcus thermophilus strain MN-ZLW-002. J. Bacteriol. 194, 4428–4429. doi: 10.1128/JB.00740-12
Khalil, R. (2009). Evidence for probiotic potential of a capsular-producing Streptococcus thermophilus CHCC 3534 strain. Pol. J. Microbiol. 58, 49–55
Lamontagne, B., and Elela, S. A. (2004). Evaluation of the RNA determinants for bacterial and yeast RNase III binding and cleavage. J. Biol. Chem. 279, 2231–2241. doi: 10.1074/jbc.M309324200
Marciset, O., Jeronimus-Stratingh, M. C., Mollet, B., and Poolman, B. (1997). Thermophilin 13, a nontypical antilisterial poration complex bacteriocin, that functions without a receptor. J. Biol. Chem. 272, 14277–14284. doi: 10.1074/jbc.272.22.14277
Mathot, A. G., Beliard, E., and Thuault, D. (2003). Streptococcus thermophilus 580 produces a bacteriocin potentially suitable for inhibition of Clostridium tyrobutyricum in hard cheese. J. Dairy Sci. 86, 3068–3074. doi: 10.3168/jds.S0022-0302(03)73906-X
McAnulty, M. J., Guron, G. K., Oest, A. O., Miller, A. L., and Renye, J. A. Jr. (2024). The quorum sensing peptide BlpC regulates the transcription of genes outside its associated gene cluster and impacts the growth of Streptococcus thermophilus. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1304136
Renye, J. A. Jr., Mendez-Encinas, M., White, A., Miller, A. L., McAnulty, M. J., Yadav, M. P., et al. (2023). Antimicrobial activity of thermophilin 110 against the opportunistic pathogen Cutibacterium acnes. Biotechnol. Lett. 45, 1365–1379. doi: 10.1007/s10529-023-03419-2
Renye, J. A. Jr., Needleman, D. S., Somkuti, G. A., and Steinberg, D. H. (2017). Complete genome sequence of Streptococcus thermophilus B59671, which naturally produces the broad spectrum bacteriocin thermophilin 110. Genome Announc. 5:pii: e01213-17. doi: 10.1128/genomeA.01213-17
Renye, J. A. Jr., Needleman, D. S., and Steinberg, D. H. (2019). Complete genome sequences of bacteriocin-producing Streptococcus thermophilus strains ST106 and ST109. Microbiol. Resour. Announc. 8:pii: e01336-18. doi: 10.1128/MRA.01336-18
Renye, J. A. Jr., and Somkuti, G. A. (2013). BlpC-regulated bacteriocin production in Streptococcus thermophilus. Biotechnol. Lett. 35, 407–412. doi: 10.1007/s10529-012-1095-0
Renye, J. A. Jr., Somkuti, G. A., Garabal, J. I., and Steinberg, D. H. (2016). Bacteriocin production by Streptococcus thermophilus in complex growth media. Biotechnol. Lett. 38, 1947–1954. doi: 10.1007/s10529-016-2184-2
Renye, J. A. Jr., and Steinberg, D. H. (2021). Thermopilin 110 inhibits growth and biofilm formation of Streptococcus mutans. Biotechnol. Rep. (Amst.) 31:e00647. doi: 10.1016/j.btre.2021.e00647
Rossi, F., Marzotto, M., Cremonese, S., Rizzotti, L., and Torriani, S. (2013). Diversity of Streptococcus thermophilus in bacteriocin production; inhibitory spectrum and occurrence of thermophilin genes. Food Microbiol. 35, 27–33. doi: 10.1016/j.fm.2013.02.006
Salini, F., Iacumin, L., Comi, G., and Dicks, L. M. T. (2023). Thermophilin 13: in silico analysis provides new insights in genes involved in bacteriocin production. Microorganisms 11, 1–11. doi: 10.3390/microorganisms11030611
Shatalin, K. Y., and Neyfakh, A. A. (2005). Efficient gene inactivation in Bacillus anthracis. FEMS Microbiol. Lett. 245, 315–319. doi: 10.1016/j.femsle.2005.03.029
Somkuti, G. A., and Renye, J. A. Jr. (2015). Effect of a BlpC-based quorum-sensing induction peptide on bacteriocin production in Streptococcus thermophilus. J. Food Res. 4, 88–96. doi: 10.5539/jfr.v4n1p88
Somkuti, G. A., and Steinberg, D. H. (1988). Genetic transformation of Streptococcus thermophilus by electroporation. Biochemie 70, 579–585. doi: 10.1016/0300-9084(88)90095-8
Sun, Z., Chen, X., Wang, J., Zhao, W., Shao, Y., Wu, L., et al. (2011). Complete genome sequence of Streptococcus thermophilus strain ND03. J. Bacteriol. 193, 793–794. doi: 10.1128/JB.01374-10
Villani, F., Pepe, O., Mauriello, G., Salzano, G., Moschetti, G., and Coppola, S. (1995). Antilisterial activity of thermophilin 347, a bacteriocin produced by Streptococcus thermophilus. Int. J. Food Microbiol. 25, 179–190. doi: 10.1016/0168-1605(94)00153-W
Keywords: Streptococcus thermophilus, bacteriocin, quorum sensing, blp operon, thermophilin
Citation: Renye JA, Somkuti GA, Qi PX, Steinberg DH, McAnulty MJ, Miller AL, Guron GKP and Oest AM (2024) BlpU is a broad-spectrum bacteriocin in Streptococcus thermophilus. Front. Microbiol. 15:1409359. doi: 10.3389/fmicb.2024.1409359
Received: 29 March 2024; Accepted: 11 June 2024;
Published: 16 July 2024.
Edited by:
Harsh Mathur, Moorepark Food Research Centre, Teagasc Food Research Centre, IrelandReviewed by:
Vineet Kumar, The University of Texas at Austin, United StatesCopyright © 2024 Renye, Somkuti, Qi, Steinberg, McAnulty, Miller, Guron and Oest. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John A. Renye, am9obi5yZW55ZUB1c2RhLmdvdg==
†Deceased
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.