- 1Department of Biomedical Sciences, School of BioSciences and Technology, Vellore Institute of Technology, Vellore, India
- 2Department of Biological Sciences, SRM University-AP, Amaravathi, India
- 3Department of Biochemistry, School of Biological Sciences, Madurai Kamaraj University, Madurai, India
- 4School of Chemical Engineering, Vellore Institute of Technology, Vellore, India
Endophytic Streptomyces sp. are recognized as a potential resource for valuable natural products but are less explored. This study focused on exploring endophytic Streptomyces species residing within tomato plants (Solanum lycopersicum) harboring genes for the production of a novel class of antibiotics. Our research involved the isolation and characterization of Streptomyces sp. VITGV156, a newly identified endophytic Streptomyces species that produces antimicrobial products. VITGV156 harbors a genome of 8.18 mb and codes 6,512 proteins, of which 4,993 are of known function (76.67%) and 1,519 are of unknown function (23.32%). By employing genomic analysis, we elucidate the genome landscape of this microbial strain and shed light on various BGCs responsible for producing polyketide antimicrobial compounds, with particular emphasis on the antibiotic kendomycin. We extended our study by evaluating the antibacterial properties of kendomycin. Overall, this study provides valuable insights into the genome of endophytic Streptomyces species, particularly Streptomyces sp. VITGV156, which are prolific producers of antimicrobial agents. These findings hold promise for further research and exploitation of pharmaceutical compounds, offering opportunities for the development of novel antimicrobial drugs.
Introduction
Antibiotics are a cornerstone of modern medicine and have become inevitable in healthcare. However, the global emergence of antimicrobial resistance (AMR) has restricted the use of many antibiotics and initiated the quest for the discovery of novel antibiotic compounds (Antimicrobial Resistance Collaborators et al., 2022; Laxminarayan, 2022). Several plant and microbial sources have been screened for the presence of bioactive metabolites, and microbial sources have gained increasing attention owing to their resourcefulness and ease of production (Tanaka and Omura, 1990; Kekuda et al., 2015). Among the microbial sources, Actinobacteria stands atop in the discovery of antimicrobial compounds, and Streptomyces is the most prominent. Streptomyces is a large class of Gram-positive bacteria well recognized for its ability to produce antimicrobial compounds.
As of July 12, 2023, the Streptomyces genus encompasses 1,179 species and 73 subspecies distributed across three genera. Among these, 590 Streptomyces genomes have been sequenced, supported by 4,903 SRA data entries in the NCBI database. More than 45% of the identified antimicrobial compounds are attributed to several species of Streptomyces (Kinkel et al., 2014; Law et al., 2017). In addition, they are prolific producers of bioactive compounds with antifungal, antiviral, antioxidant, and anticancer properties (Loganathan et al., 2013; Karthik et al., 2014; Ayswaria et al., 2020; Alam et al., 2022; Lacey and Rutledge, 2022). Therefore, the isolation and characterization of Streptomyces sp. from diverse ecosystems and niches have been adopted worldwide for bioprospecting (Chhetri et al., 2024). Screening of Streptomyces sp. from previously unexplored areas not only paves the way for the isolation of novel species but also provides interesting bioactive compounds (Law et al., 2019).
Streptomyces spp. are classified as saprophytes that help in the recycling of dead and decaying organic matter in soil. Thus, soil has become the major habitat for all Streptomyces species. However, several species have been reported to exhibit endophytic relationships with plant hosts (Seipke et al., 2012; Olanrewaju and Babalola, 2019). They enter the plant host through natural openings, root colonization, association with other endophytic organisms, and seed transmission and colonize the roots, stem, and leaves of the host plant (Coombs and Franco, 2003; Chen et al., 2016; Kandel et al., 2017; Khare et al., 2018). The endophytic association of Streptomyces with plant hosts benefits them in many ways, including conferring resistance to infection and plant growth promotion (Quecine et al., 2008; Verma et al., 2011; Vurukonda et al., 2018; Worsley et al., 2020). In turn, endophytic relationships also have a significant impact on Streptomyces. Several studies have shown that endophytic relationships promote the evolution of secondary metabolic pathways (Ayswaria et al., 2020; Al-Quwaie, 2024) in Streptomyces. This results in the production of novel secondary metabolites that have ecological and biotechnological implications. Emerging advances in genome mining techniques have further revealed the presence of latent BGCs in Streptomyces genomes, underscoring their potential for novel drug discovery.
Polyketide antibiotics, derived from Streptomyces bacteria, represent a vital class of antimicrobial compounds. Many known antibiotics, including renowned examples such as erythromycin, tetracycline, and streptomycin, are polyketide antibiotics produced by various Streptomyces sp. strains. These antibiotics are synthesized through polyketide synthase enzymes, which assemble diverse chemical structures. These antibiotics exhibit broad-spectrum activity against various bacterial pathogens, making them indispensable in clinical medicine and agriculture. Kendomycin, a macrocyclic polyketide, is produced by various Streptomyces species, including S. violaceoruber (Yuan et al., 2004). Recent studies have detailed multiple synthesis methods for kendomycin, underscoring its importance in biological investigations. This compound has cytotoxic effects on various human tumor cell lines, such as MCF-7, HMO2, and HepG2. Moreover, previous findings indicate that kendomycin disrupts mammalian proteasome functions and triggers apoptosis in U937 cells. Its potent antibacterial activity extends to both Gram-negative and Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MRSA) (Bode and Zeeck, 2000; Elnakady et al., 2007; Duggal et al., 2011; Hesje et al., 2011; Tattevin, 2011). The identification of novel Streptomyces spp. capable of producing a new class of antibiotics is valuable for both exploring the biosynthetic pathways and commercial production of antimicrobial compounds.
Tomatoes stand out for their exceptional nutritional value, boasting an array of bioactive compounds, including carotenoids, polyphenols, ascorbate, and folate. This nutritional richness positions them as paramount agricultural products, providing vital nutrients. Over its lifecycle, the tomato plant interacts with diverse microorganisms. Notably, endophytic actinobacteria find sanctuaries within plants, actively contributing to plant defense mechanisms by synthesizing pharmaceutical compounds (Kiran et al., 2022). Endophytic Streptomyces colonizing tomato plants act as biocontrol agents and enhance growth in the host plant (Goudjal et al., 2016; Passari et al., 2019; Ling et al., 2020; Devi et al., 2022). Among many biocontrol agents, kendomycin is a small natural molecule that has gained significant attention due to its strong antimicrobial activity.
Despite the beneficial effects of endophytic Streptomyces and the metabolites they produce, their BGCs remain poorly understood. Therefore, this study aimed to isolate and characterize endophytic Streptomyces strains capable of producing novel antimicrobial compounds from tomato plants from the perspective of the genome landscape. In this study, a Streptomyces species (VITGV156) was isolated and shown to have strong antimicrobial activity against Escherichia coli. The whole-genome sequence was determined, and the taxonomic identity was determined. Mining of the VITGV156 genome revealed the presence of BGCs for several polyketide antibiotics. The antimicrobial activity of a polyketide antibiotic was also analyzed.
Materials and methods
Chemicals and media
The chemicals, media components, and standard antibiotics used in the study were purchased from HiMedia Pvt. Ltd., Mumbai, India and Sigma-Aldrich Corporation, Bangalore, India. All solvents used were of analytical/HPLC grade and were purchased from SD Fine Chem Limited, India. The antibiotics and fine chemicals used were purchased from SD Fine Chem Limited, India. The antibiotic kendomycin (BIA-K1143) was purchased from BioAustralis Fine Chemicals, Australia.
Sample collection
Healthy tomato plants (cv. Arka Rakshak) were collected from tomato field fields in and around Madurai, India. The aerial regions of 10 plants were carefully removed and washed with tap water. The samples were placed in airtight bags, taken to the laboratory, and processed within 4–6 h.
Isolation of endophytic Streptomyces sp. from tomato plants
The collected plant samples were processed aseptically in Biological Safety Cabinet Class II. The samples were sterilized by sequential immersion in 70% (v/v) ethanol for 5 min and sodium hypochlorite solution (0.9%, w/v, available chlorine) for 20 min. The surface-sterilized root samples were washed in sterile water three times to remove surface sterilization agents. Next, the root samples were soaked in 10% (w/v) NaHCO3 solution for 10 min to retard the growth of endophytic fungi. The sample was divided into small fragments (0.5 cm × 0.5 cm) and placed on IS2 media (0.4% yeast extract powder, 1% malt extract powder, 0.4% dextrose, and 2% agar). Fifteen ppm (w/v) nalidixic acid was added to suppress the growth of contaminating bacteria, and the mixture was then incubated at room temperature for 15 days at 30°C. The colonies that emerged were subsequently purified by further streaking on ISP2 plates two times. A pure culture was maintained in glycerol (20%, v/v) at −80°C.
Identification and characterization of Streptomyces sp. VITGV156
The strain Streptomyces sp. VITGV156 was identified via phenotypic and genotypic characterization. The morphology of the strain was examined using both phase contrast microscopy (MT5210/MEIJI) and scanning electron microscopy (Carl Zeiss-Evo 18). The preparation of the samples for scanning electron microscopy was performed according to Chanama et al. (2023) and Veilumuthu and Christopher (2022a). These imaging techniques seamlessly unveiled the intricate morphological attributes of the strain, rendering a vivid depiction of its distinctive characteristics.
Genome annotation and comparative genomic analysis
Gene prediction and functional annotation were performed by Rapid Annotation using Subsystem Technology (RAST) (Brettin et al., 2015). This annotation was further complemented with the COG (Cluster of Orthologous Groups of Proteins) (Tatusov et al., 2000) database1 with an E-value cutoff of 1e−02 to gain insights into biological, molecular, and cellular function-encoding genes. For the analysis of 16S rRNA sequences within the Streptomyces genus, 16S rRNA sequences were retrieved from NCBI. Alignment and model selection were performed using MUSCLE v3.8.1551 (Edgar, 2004) and MEGAX v11.0.13 (Tamura et al., 2021), respectively. Maximum likelihood phylogenies were generated using RAxML v8.2.12 (Stamatakis, 2014) with the GTRGAMMA (mtREV24 + G + I + F) nucleotide substitution model and a bootstrap value of 100. The resulting phylogenetic tree was visualized using the iTOL platform (Letunic and Bork, 2021). The average nucleotide identity scores for the selected genomes were calculated using pyani-0.2.10. GGDC 3.0 was used for genome-to-genome comparisons. A circular map of Streptomyces sp. VITGV156 was generated using the Proksee tool (Grant et al., 2023). Orthology was determined within protein datasets from 10 genomes using the Reciprocal Best Hits (RBH) BLAST method implemented in ProteinOrtho (Klemm et al., 2023), with an E-value threshold set at 1e−5, along with default parameters. The biosynthetic clusters involved in the synthesis of secondary metabolites were analyzed using antiSMASH 7.0 (Blin et al., 2023).
Biomass production and metabolite extraction
The Streptomyces strain VITGV156 is an endophyte in tomato plants that is cultured in ISP2 broth. Once the culture reached the lag phase, it was divided equally into 10 500-ml culture flasks, each containing 300 mL of ISP2 medium (Veilumuthu et al., 2023). These culture flasks were kept in a laboratory shaking incubator at 150 rpm for 21 days at 30°C. Following incubation, the cells were removed by centrifugation (2,000 rpm, 5 min, 4°C), and the supernatant was further filtered through Whatman paper to obtain a cell-free filtrate. The secondary metabolite was recovered from the broth using a two-phase solvent extraction system with ethyl acetate (1:1). Then this mixture was vigorously shaken for 10 min and kept in a shaker for 24 h at 200 rpm. This was allowed to settle for 30 min. Organic phases were collected and concentrated in a rotary evaporator (model RE100-Pro) at 54°C and 80 rpm (Veilumuthu et al., 2022).
Chemical analysis of LC-HRMS
The ethyl acetate extract derived from the culture filtrate of Streptomyces sp. VITGV156 was subjected to LC-HRMS analysis. Data analysis was conducted using the LC-Solution tools provided by Shimadzu Corporation, Kyoto, Japan. A Waters® Micromass® Q-Tof micro™ mass spectrometer was used for the LC-HRMS-ESI.
Bacterial strains and culture conditions
The bacterial strains used in this research were Escherichia coli K12 strain MG1655, Pseudomonas aeruginosa (PA14), and methicillin-resistant Staphylococcus aureus (MRSA: ATCC 43300). For the growth of all bacterial strains, Luria–Bertani Broth (LB) broth and agar (composition: 1% tryptone (w/v), 0.5% yeast extract (w/v), and 1% NaCl (w/v), 2% agar/pH 7.2) were used. Cultures for all experiments were maintained at 37°C. The bacteriological chemicals utilized in this study were procured from Sisco Research Laboratories (SRL), Mumbai, India.
Determination of the minimum inhibitory concentrations
The minimum inhibitory concentrations (MIC) of the antibiotic kendomycin were determined using the broth microdilution method as described previously by Wiegand et al. (2008). The microbroth dilution method was performed with transparent 96-well plate. Kendomycin was prepared at different working concentrations using a stock solution (0.5 mg/mL). A saline suspension of the test strain equivalent to 2.0 McFarland standard (containing 1 × 107 to 1 × 108 cfu mL−1) was prepared. A volume of 1–10 μL of the suspension was added to each well (to attain a final density of ~1 × 105 cfu mL−1), and the cultures were incubated at 37°C for 12–16 h. The MIC was defined as the lowest concentration of a test antibiotic that completely inhibited visible bacterial growth.
Microscopic analysis
Live cell imaging was performed as described previously (Govindarajan and Amster-Choder, 2017; Nagarajan et al., 2023). Actively growing cells were used for treatment and imaging. Overnight cultures were subcultured (1:100 dilution), and the cells were grown for 2 h at 37°C and then exposed to kendomycin (50 μg/mL) for 3–4 h before imaging. For the control, the cells were grown at 37°C for 5–6 h and imaged. The cells were harvested and stained with Synapto Red C2 (4 mg/mL) and DAPI (5 mg/mL) for 10 min. The cells were washed with 1× phosphate-buffered saline (PBS) and resuspended in 100–200 μL of PBS before imaging. In total, 10–20 μL of stained cells were spotted on 1% agarose pads (in 0.85% saline) with uncoated coverslips before visualization under a microscope. The cells were visualized and photographed using a Nikon Eclipse Ti2-E instrument equipped with a 100× CFI Plan Apochromat oil objective and a DSQi-2 Monochrome Camera (Nikon). Images were processed using NIS Elements AR software (Nikon).
ADMAT analysis
The SMILES of each molecule were extracted from the PubChem database and converted into a 3D structure in PDB format using the online SMILES converter NCI/CADD. After that, the metabolites were produced by adding Gasteiger charges, non-polar hydrogen atoms, allocating aromatic carbons, and identifying the root using AutoDock Vina. After that, the ligands were stored in pdbqt format (Aragón-Muriel et al., 2021; Melinda et al., 2021). The SwissADME database was subsequently used to assess the drug-like characteristics of the active compounds. This is predicated on Christopher Lipinski’s discovery of Lipinski’s rule of 5. There are five parameters to it. The molecular weight should be within 500 Dalton, the value of logP should be <5, the number of bond donors should be H < 5, and the number of bond acceptors should be H > 10 (Naqvi et al., 2018).
Results
Isolation and characterization
Our previous study (Veilumuthu and Christopher, 2022b) on analyzing the diversity of Streptomyces sp. endophytes in tomato plants revealed the hidden microbial treasures dwelling within these plants (Chanama et al., 2023). Endophytic Streptomyces sp. The production of diverse secondary metabolites was the focal point of this investigation. The practiced sample processing methodologies ensured the viability of endophytic species within the samples. A total of 240 endophytic actinomycetes were observed from the tomato plants taken for analysis, which is very evident from their distinct colony morphology. A visual representation of the endophytic Streptomyces sp. isolated from tomato plants on ISP2 agar plates is given in Figure 1A. Overall, Figure 1 shows the distinctive cultural attributes and colony morphology. Individual colonies were selected and purified by further streaking. A broad range of compounds produced by Streptomyces sp. were reported to exhibit antimicrobial activity. Therefore, a preliminary screening was performed by assessing the antimicrobial activity of the isolated strains by visualizing their activity against Escherichia coli. One of the isolates exhibited significant activity, exhibiting a large zone of clearance (Figures 1B,C). The strain was named Streptomyces sp. VITGV156 and characterized further.
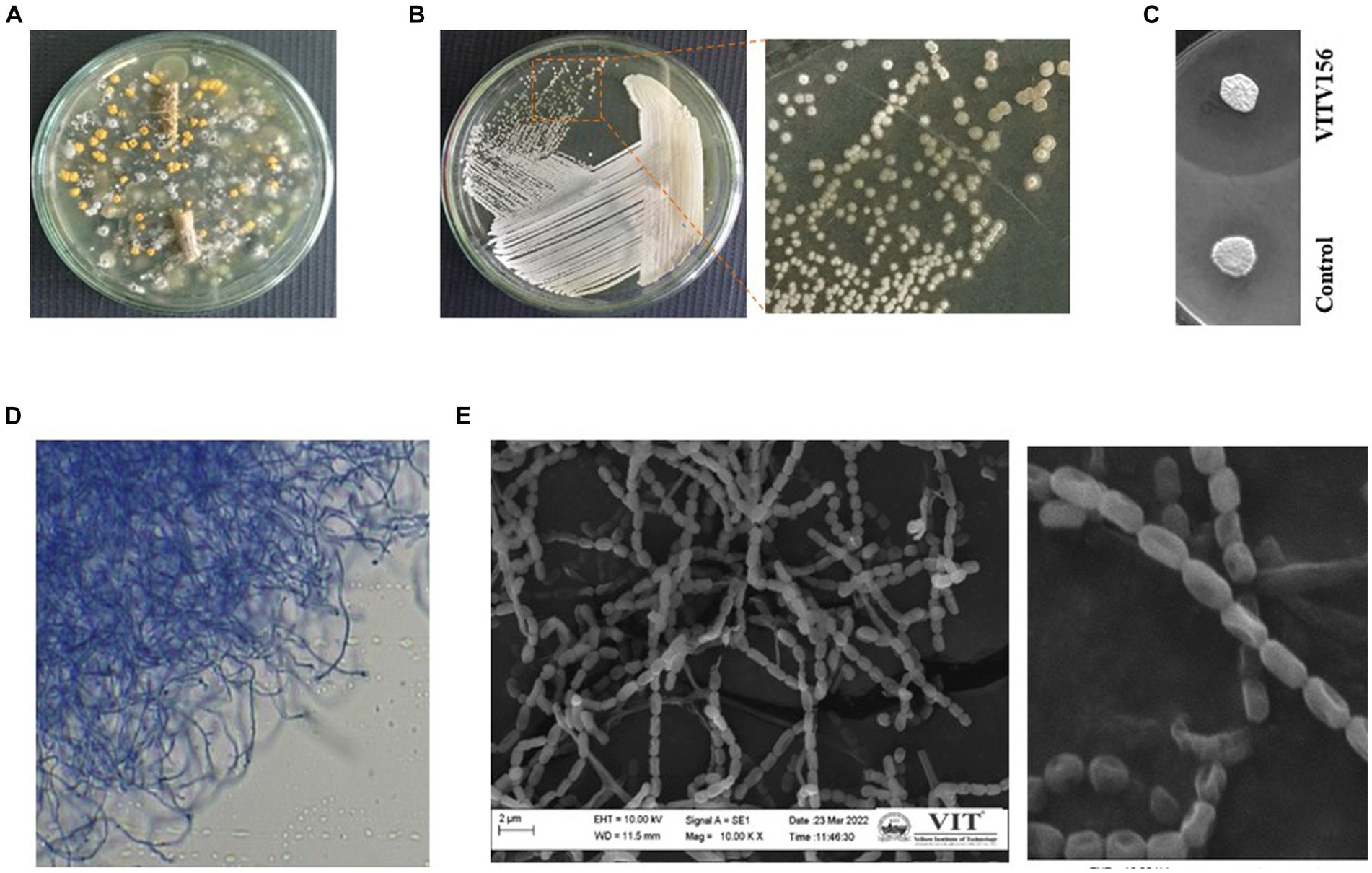
Figure 1. Isolation of Streptomyces sp. VITGV156. (A) Petri plate with a 7-day-old culture on ISP2 agar plate exhibiting diverse colony morphologies. (B) Colony morphology of Streptomyces sp. VITGV156. (C) VITGV156 and the control (another isolate with no antibacterial activity) were screened for antibacterial activity. Compared with the control, VITGV16 inhibited the growth of Escherichia coli. (D) Long, discontinuous hyphal growth was observed via phase contrast microscopy. (E) SEM image of the spore chain.
Phenotypic characterization was performed to support the novelist VITGV156. Upon growth, they form aerial mycelia on the surface. The color of the substrate mycelium is pale yellow on ISP2. This complex mycelial composition was further characterized by clusters comprising both elongated and abbreviated fragments, as visually demonstrated in Figure 1D. A closer examination revealed an interconnected network of aerial hyphae. SEM images of the hyphae revealed delicate and weak chains of spores, with each spore featuring a prominent cavity on one side (Figure 1E). The spores measured approximately 1 μm in size.
Genome features of Streptomyces sp. VITGV156
Whole-genome sequencing was performed with genomic DNA extracted from Streptomyces sp. VITGV156 grown on ISP2 media for 15 days. High-quality genome sequencing was performed with the Illumina NextSeq 500 platform. The genome of Streptomyces sp. VITGV156 was successfully assembled into a complete chromosome measuring 7,207,566 bp, with a G + C content of 72.6% (Figure 2A). The genomic features of VITGV156 are detailed in Table 1. Within this genome, 6,583 predicted genes were identified, comprising 6,512 protein-coding genes and 20 rRNA-associated genes, including 67 tRNA genes (Supplementary material S1). Among the 6,512 encoded proteins, functions were assigned to 4,993 proteins (76.67%), while the functions of the remaining 1,519 proteins (23.32%) remained unknown and were annotated as hypothetical proteins. Notably, only 64.81% (4221) of the proteins from the VITGV156 genome were mapped to functional categories within the Clusters of Orthologous Groups (COGs) (Figure 2B), as detailed in Supplementary material S1. The genes were categorized into three functional categories: information storage and processing, cellular processes, and signaling metabolism.
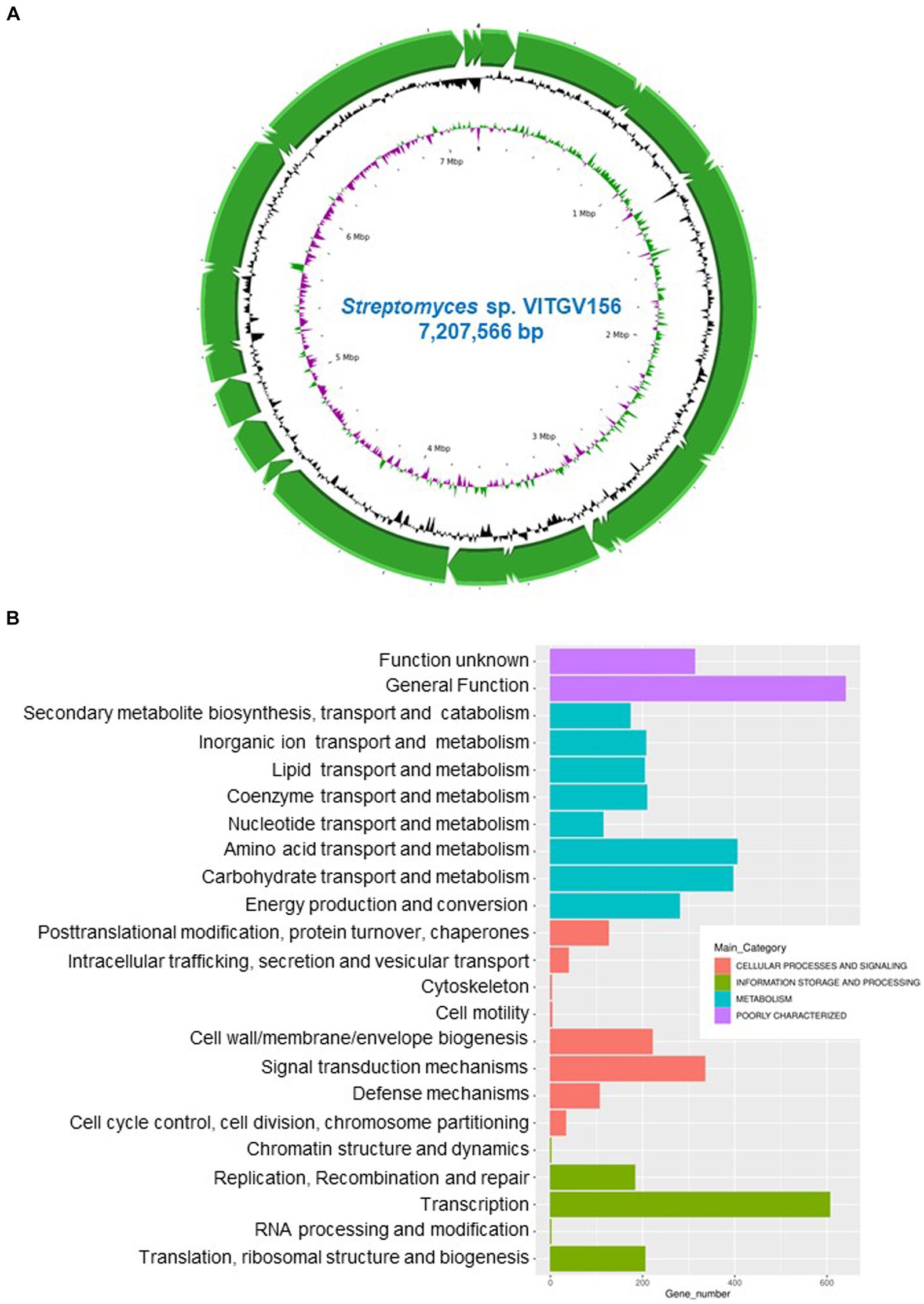
Figure 2. Genomic landscape of VITGV156. (A) Circular genomic map of Streptomyces sp. VITGV156. Assembled contigs are depicted as green arrows, accompanied by positive and negative GC skews indicated in the inner circle. (B) Cluster of Orthologous Gene (COG) Database annotation of Streptomyces sp. VITGV156. The graph shows the functional categorization of the Streptomyces sp. VITGV156-encoded ORFs into four main functional categories: information storage and processing, cellular processes and signaling, metabolism, and poorly uncharacterized. These categories are further subdivided into subcategories, as depicted by similarly colored bars along with their corresponding numbers.
Streptomyces sp. VITGV156—comparative genome analyses
The 16S rRNA sequence of Streptomyces sp. VITGV156 exhibited the highest similarity with the 16S rRNA sequence of Streptomyces rochei strain JK, followed by Streptomyces huasconensis strain D23 and Streptomyces alfalfae strain XN-04 (Figure 3A). Phylogenetic analysis based on 16S rRNA sequences also demonstrated that it forms a distinct clade alongside Streptomyces fodineus strain TW1S1, Streptomyces spinosirectus strain CRSS-Y-16, Streptomyces caniscabiei strain ID03-3A, Streptomyces akebiae strain MG28, Streptomyces avermitilis MA-4680 NBRC 14893, Streptomyces alboniger strain ATCC 12461, and Streptomyces qaidamensis, whereas Escherichia coli str K12 substrain MG1655 was taken as an outgroup to denote one of the most distant relatives of the isolated species of Streptomyces. To further validate the phylogeny, we performed multilocus sequence analysis (MLSA) (Macheras et al., 2011) using housekeeping genes. Therefore, we conducted a housekeeping gene analysis to validate the taxonomic relationships among Streptomyces species. Specifically, we analyzed the gene sequences of atpD, gyrB, recA, rpoB, and trpB, which are commonly used in multilocus sequence analysis (MLSA) (Komaki, 2022) for Streptomyces (Figure 3B). We further included sequences from nine more conserved genes for MLST analysis of this strain (tigrfam_recA, rpsJ_bact, uS11_bact, ftsZ, rpsS_bact, rplN_bact, rpoC_TIGR, rpsE_bact, and rpsL_bact) (Supplementary Table S1). Our phylogenetic analysis, which included 14 housekeeping genes from Streptomyces and five outgroups (Figure 3B), yielded a tree topology similar to that obtained using 16S rRNA. This analysis supports the assertion that strain VITGV156 is closely related to S. rochei JK.
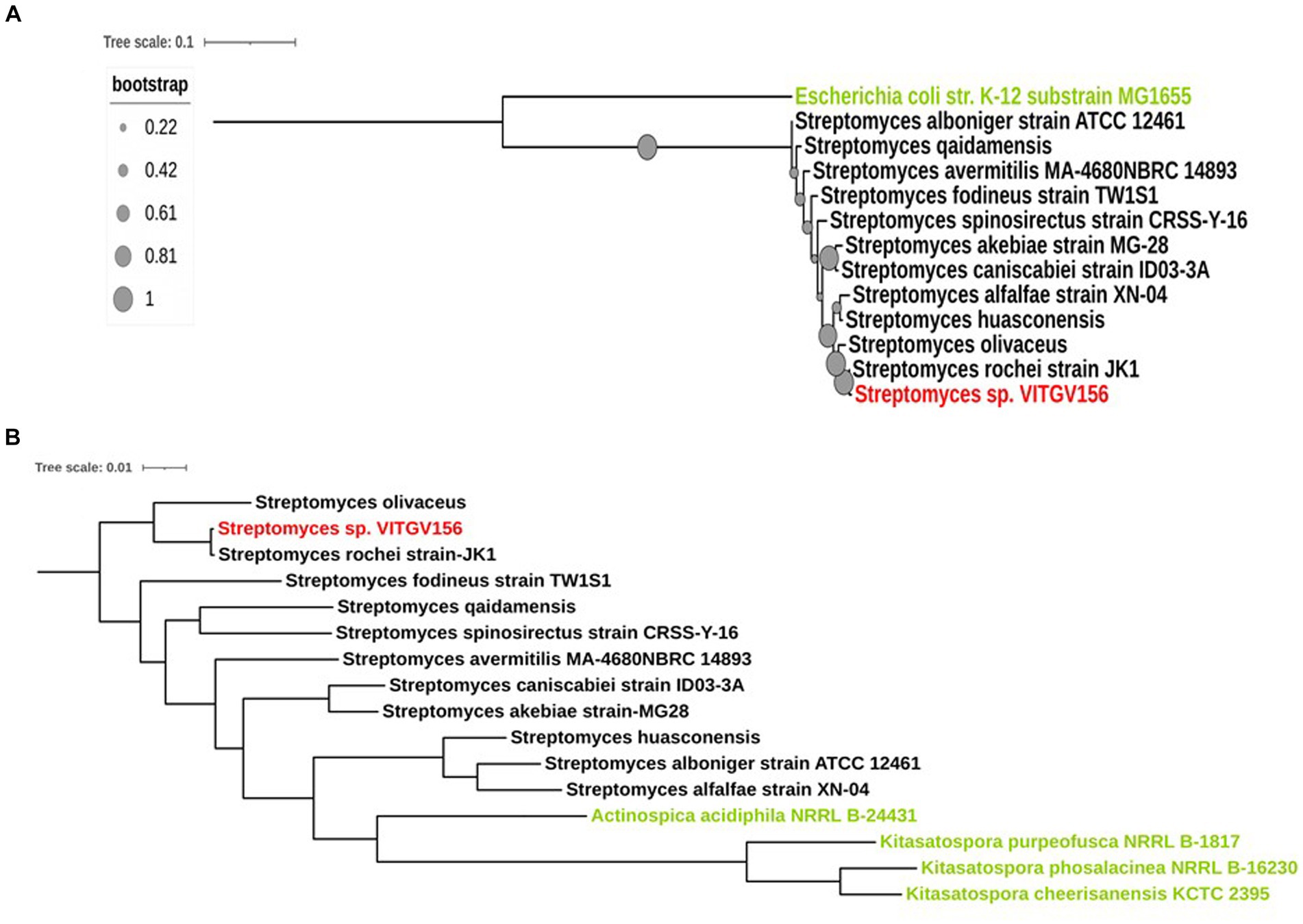
Figure 3. Phylogenetic analysis of Streptomyces sp. VITGV156. (A) 16S rRNA phylogenetic analysis of Streptomyces sp. VITGV156. The isolated strain of Streptomyces, denoted in red, shares a clade with the Streptomyces rochei strain Jk1. The outgroup is denoted in green. Bootstrap values are depicted as solid circles and annotated on the branches. (B) Phylogeny based on housekeeping proteins. Fourteen concatenated housekeeping proteins were used to generate a phylogenetic tree using AutoMLST and visualized with iTOL. Actinospica acidiphila NRRL B-24431, Kitasatospora purpeofusca NRRL B-1817, Kitasatospora phosalacinea NRRL B-16230, and Kitasatospora cheerisanensis KCTC 2395 were used as outgroup species in this study (denoted in green).
To support this analysis, we computed the in silico DDH values of Streptomyces sp. VITGV156 compared to those of these Streptomyces strains using the Genome-to-Genome Distance Calculator server. We found that the highest DDH value of 95.63 was exhibited by the Streptomyces rochei strain JK1 (Supplementary material S1). Furthermore, we calculated the average nucleotide identity (ANI) for these genomes, revealing that Streptomyces sp. VITGV156 shares 98.94% similarity with the Streptomyces rochei strain JK1 (Figure 4A). These findings are consistent with the 16S rRNA phylogenetic analysis, indicating that the isolated strain of Streptomyces is the closest relative to the species Streptomyces rochei.
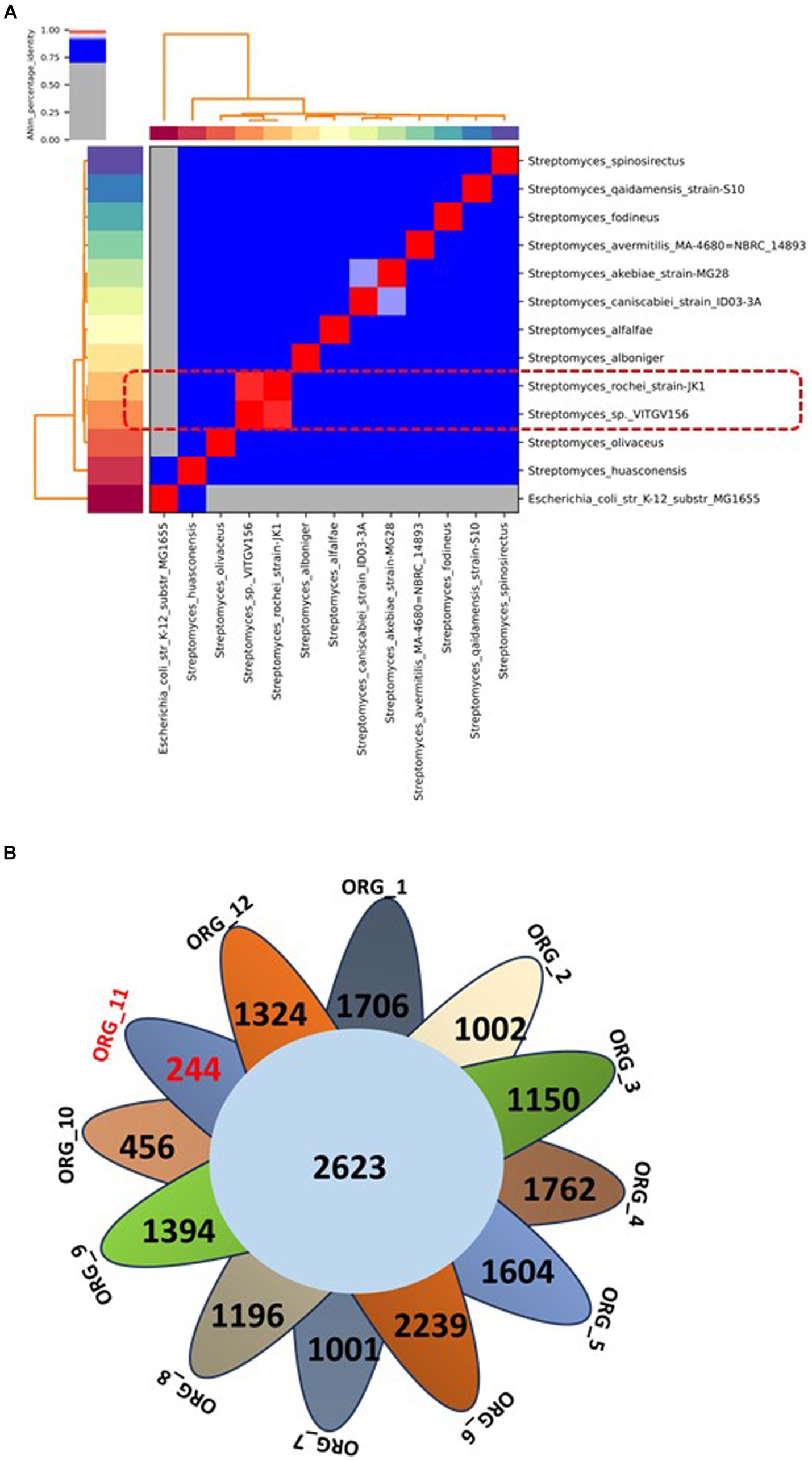
Figure 4. (A) Average nucleotide identity (ANI) of Streptomyces compared with 11 other complete genomes of Streptomyces species. The highest score is highlighted in red within the matrix. The ANI (98.94%) of the isolated strain and one of its closest relatives are indicated in the box in Red dotted line. (B) A Venn diagram showing the distribution of core and unique genes among the 12 genomes of the Streptomyces genus. The number of unique genes in Streptomyces sp. VITGV156 is highlighted in red (Org_11).
The panproteome encompasses all proteins shared among more than two species and comprises the core proteome, dispensable proteome, and unique proteome. The collective proteome of 12 Streptomyces genomes comprises 91,301 proteins distributed into 12,293 orthologous protein clusters (see Supplementary material S2). Among these clusters, 2,623 were conserved across all genomes; constituting the core proteome for these Streptomyces strains (Figure 4B, Table 2). Additionally, 9,671 and 14,351 clusters were categorized as accessory and unique genes, respectively (see Supplementary material S2). This analysis identified 270 unique genes in the Streptomyces sp. VITGV156 strain, 207 of which are hypothetical and are documented in Supplementary material S2.
Our comparative analysis of the Streptomyces sp. VITGV156 (Org_11) strain revealed its novelty, identifying it as the closest relative to S. rochei (Org_10). Within its genome, we identified 270 unique genes, 76.66% of which were entirely novel. Annotation data indicate that these genes are hypothetical, lacking known domains, and potentially encoding novel functions that are not observed in any other organism. This isolation distinguished this newly discovered species from other Streptomyces species. We report its genomic characterization through comparative genomic analyses. The VITGV156 strain was submitted to the National Center for Microbial Resources (NCMR) under accession number MCC4965.
The genome of VITGV156 encodes a wide range of polyketide antibiotics
To identify the gene clusters responsible for the production of putative secondary metabolites, the genome sequence of VITGV156 was analyzed using the antiSMASH software. The genome of VITGV156 encodes 29 putative biosynthetic gene clusters (BGCs). Eight compounds showed relatively low similarity (<20%) to known natural products, suggesting that they are unidentified compounds. Of the 29 BGCs found to be responsible for the production of secondary metabolites, 65% (16 BGCs) corresponded to the biosynthesis of peptide compounds, which included non-ribosomal peptide synthetases (NRPSs) and ribosomally synthesized and posttranslationally modified peptides (RiPPs). NRPSs are large, multidomain enzymes found in bacteria and fungi that are responsible for the biosynthesis of complex peptides. Unlike ribosomal protein synthesis, NRPSs catalyze peptide biosynthesis in a template-independent manner. The peptides produced by NRPs exhibit a wide range of biological activities and thus have antimicrobial and anticancer properties (Sieber and Marahiel, 2005; Strieker et al., 2010; Martínez-Núñez and López, 2016). The genome of VITGV156 encodes a calcium-dependent antibiotic gene (CDA1/CDA2a). CDA1 and CDA2a are enzymes involved in the biosynthesis of CDAs. These enzymes are responsible for catalyzing specific steps in the synthesis of the CDA molecule. The exact functions of CDA1 and CDA2a may vary depending on the specific reactions they catalyze within the biosynthetic pathway. The antimicrobial activity of CDAs is well known in Gram-positive bacteria (Micklefield, 2009; Wood and Martin, 2019). RiPPs are peptides synthesized by ribosomes in a template-dependent manner and are modified posttranscriptionally by modifying enzymes. RiPPs are known for their remarkable biological activities (antimicrobial, anticancer, and immunomodulatory activities) (Papini, 2009; Ongpipattanakul et al., 2022). In addition, 17% of BGCs (4) corresponded to the synthesis of terpenes, and 10% of BGCs (3) corresponded to the synthesis of siderophores. The indole compound 5-dimethylallylindole-3-acetonitrile was also produced by the strain (Table 3).
Moreover, the genome of VITGV156 harbors genes responsible for the biosynthesis of at least 11 polyketide antibiotics (italicized in Supplementary Table S2). Polyketide compounds are a diverse class of naturally derived compounds that are prominent for their bioactivity, including antibiotics, immunosuppressants, and anticancer chemotherapeutics (Gomes et al., 2013; Tacar et al., 2013; Risdian et al., 2019). Interestingly, the BGCs encoding polyketide antibiotics were less similar to the existing BGCs, indicating that they were hitherto unidentified.
To determine the efficiency of the predicted polyketide compounds, we analyzed their drug-likeness properties, as per the AntiSMASH report. The detailed list is given in Supplementary Table S3. The molecular weights of the compounds were found to be in the range of 182–1,375 g/mol. Of the 10 compounds tested, three did not obey Lipinski’s rule, as there was more than 1 violation, and seven of these compounds obeyed Lipinski’s rule, demonstrating their drug-like properties. In this study, we further investigated the potential antimicrobial activity of the polyketide antibiotic kendomycin (Elnakady et al., 2016).
Kendomycin biosynthetic clusters of VITGV156
Kendomycin is a polyketide antibiotic known for its anticancer and antibacterial properties (Elnakady et al., 2007). The BGCs identified in the genome of VITGV156 were analyzed for kendomycin biosynthesis clusters. The VITGV156 genome encodes approximately five type I PKS gene clusters, which form the polyketide backbone of kendomycin and are similar to those of S. verrucosispora and S. violaceoruber. The genes encoding the T1PKS-kendomycin biosynthesis-related gene cluster span approximately 30.02 kb and encode 20 open reading frames (ORFs), akin to the BGCs found in S. verrucosispora and S. violaceoruber. However, we could not find any other enzymes (such as modifying, tailoring, and cycling enzymes in the kendomycin biosynthesis pathway). Notably, the presence of PKS genes and associated proteins in Streptomyces sp. VITGV156 indicates their involvement in various biosynthetic pathways, such as the 3,5-DHPG pathway, b-ketoadipate pathway, and tricarboxylic acid cycle. The kendomycin type I PKS assembly line comprises four enzymes encoded by a putative operon, with eight elongation modules incorporating extender units. Initiation involves activation of a likely benzoic acid derivative starter unit by a CoA ligase domain, while elongation is facilitated by specific enzymes assigned to different modules based on domain analysis. Further investigations are warranted to fully elucidate the complexities of kendomycin biosynthesis.
Kendomycin treatment affects membrane and nucleoid organization
We then aimed to elucidate the toxicity induced by kendomycin, a polyketide antibiotic. The antimicrobial property of kendomycin has drawn attention due to its promising activity against drug-resistant bacteria. To ascertain the toxicity of kendomycin, we assessed the minimum inhibitory concentration for Gram-positive [methicillin-resistant S. aureus (MRSA)] and Gram-negative (P. aeruginosa and E. coli) bacteria through the broth microdilution method (Wiegand et al., 2008). The inhibitory concentrations of kendomycin for MRSA, P. aeruginosa, and E. coli ranged from 10 to 15 μg/mL (Figure 5A). We further extended the study by evaluating the bacterial morphology after brief exposure (4–6 h) to high concentrations of kendomycin (50 μg/mL). Microscopic visualization of kendomycin-treated bacteria that exhibited severe damage to the membrane. The bacterial membrane exhibited abnormalities, which were visualized both by phase contrast and with a fluorescent red filter (stained with SynaptoRed C2). Phase contrast images of Gram-negative bacteria revealed severe irregularities on the cell surface, whereas SynaptoRed C2 staining revealed distinct puncta patches in the cell membrane in MRSA (Figure 5B). These results indicated possible damage to the bacterial membrane upon kendomycin exposure. Previous studies on kidney toxicity reported global changes in the proteome and transcriptome (Duangmal et al., 2016). Since bacterial nucleoids are associated with the expression patterns of different genes (Kar et al., 2005; Jin and Cabrera, 2006; Dillon and Dorman, 2010), we assessed the nucleoid morphology of the bacterial cells before and after treatment with 4′,6-diamidino-2-phenylindole (DAPI), a DNA-staining dye. The results indicated a drastic change in the nucleoid morphology in all the bacterial strains tested. We observed a reduction in the size of the nucleoid along with morphological changes in the nucleoid after treatment (Figure 5C). Taken together, our results indicated that kendomycin treatment potentially causes damage to cells by affecting various targets.
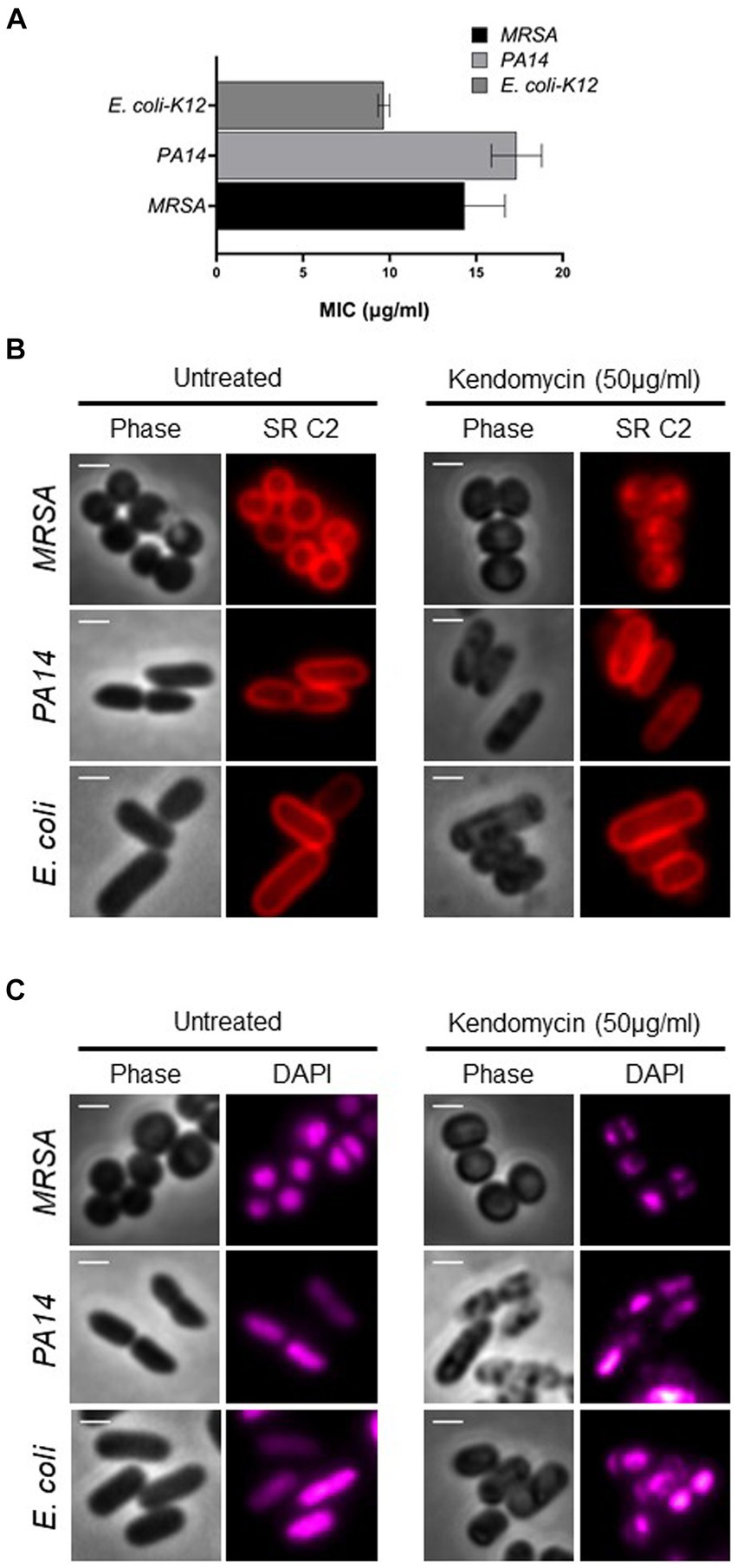
Figure 5. Antimicrobial activity of Kendomycin. (A) Minimum inhibitory concentration of kendomycin for the indicated bacteria. (B) Images of the indicated bacterial cells exposed to different concentrations of kendomycin (50 μg/mL) and membrane stained. The bacterial membrane was observed by fluorescence microscopy, and the cells were observed via phase contrast microscopy. Phase contrast (gray) and Synapto Red C2 (red) are shown. The scale bar corresponds to 2 μm. (C) Images of the indicated bacterial cells after exposure to different concentrations of kendomycin (50 μg/mL) and nucleoid staining. Bacterial nucleoids were observed by fluorescence microscopy, and the cells were observed via phase contrast microscopy. For better visualization, DAPI is shown in magenta. Phase contrast (gray) and DAPI (magenta) are shown. The scale bar corresponds to 2 μm.
LC-HRMS analysis of isolated compounds
The task of isolating metabolites presents significant challenges. To identify these metabolites, LC-HRMS analysis was done for VITGV156 crude extracts. The LC-HRMS data were scrutinized utilizing m/z, retention time, and molecular formula, with additional databases utilized to search for and assign formulas and compound structures. Following evaluation and interpretation, the compound identified at a retention time (RT) value of 3.63 was determined to be kendomycin (Figure 6). The molecular formula and molecular mass of these identified compounds are C29H42O5 and 470.3007, respectively (Figure 7).
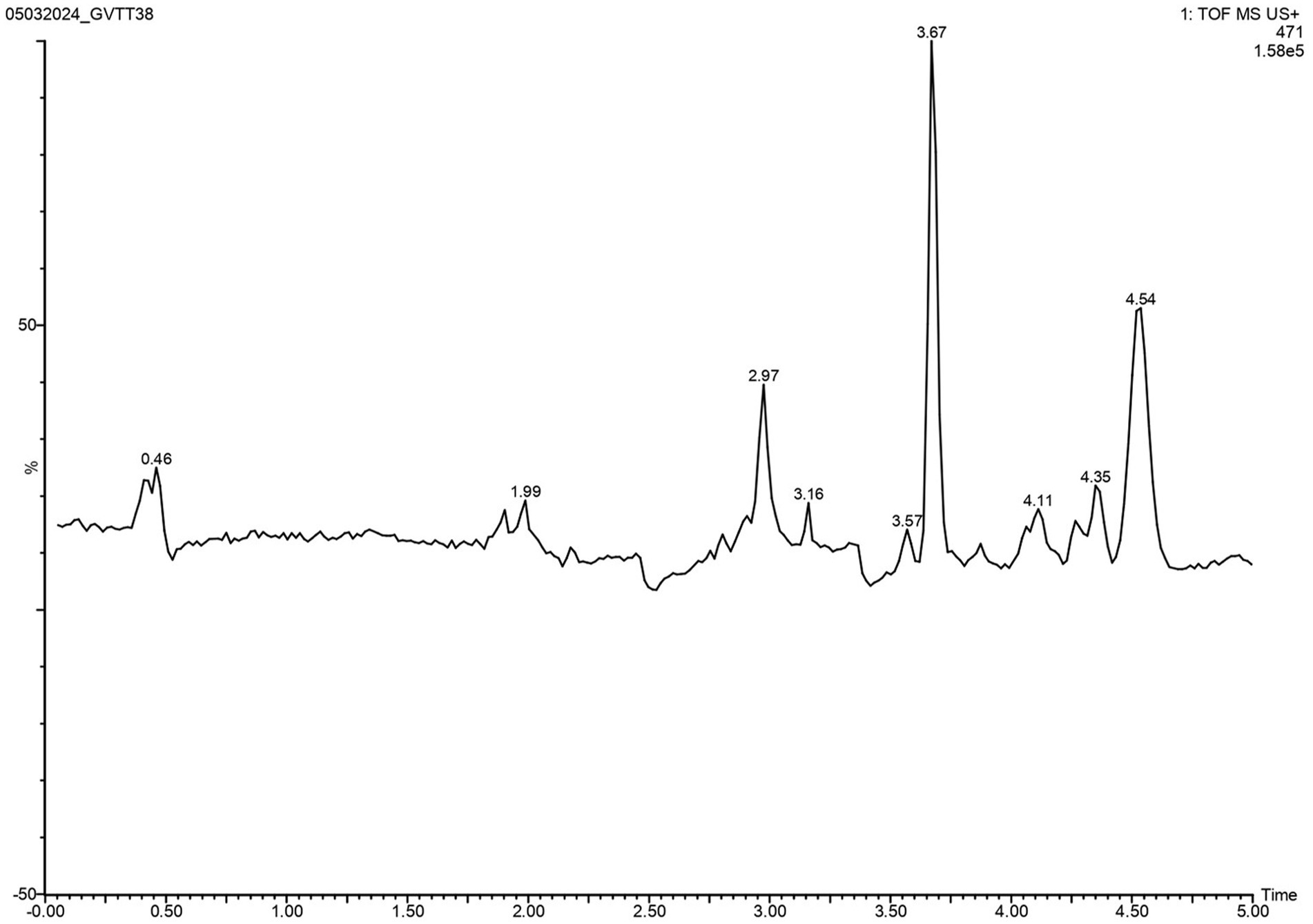
Figure 6. LC–MS/MS chromatogram of the secondary metabolite of Streptomyces sp. VITGV156. The peak corresponds to the bioactive molecule kendomycin (peak at 3.637, m/z = 470.3007).
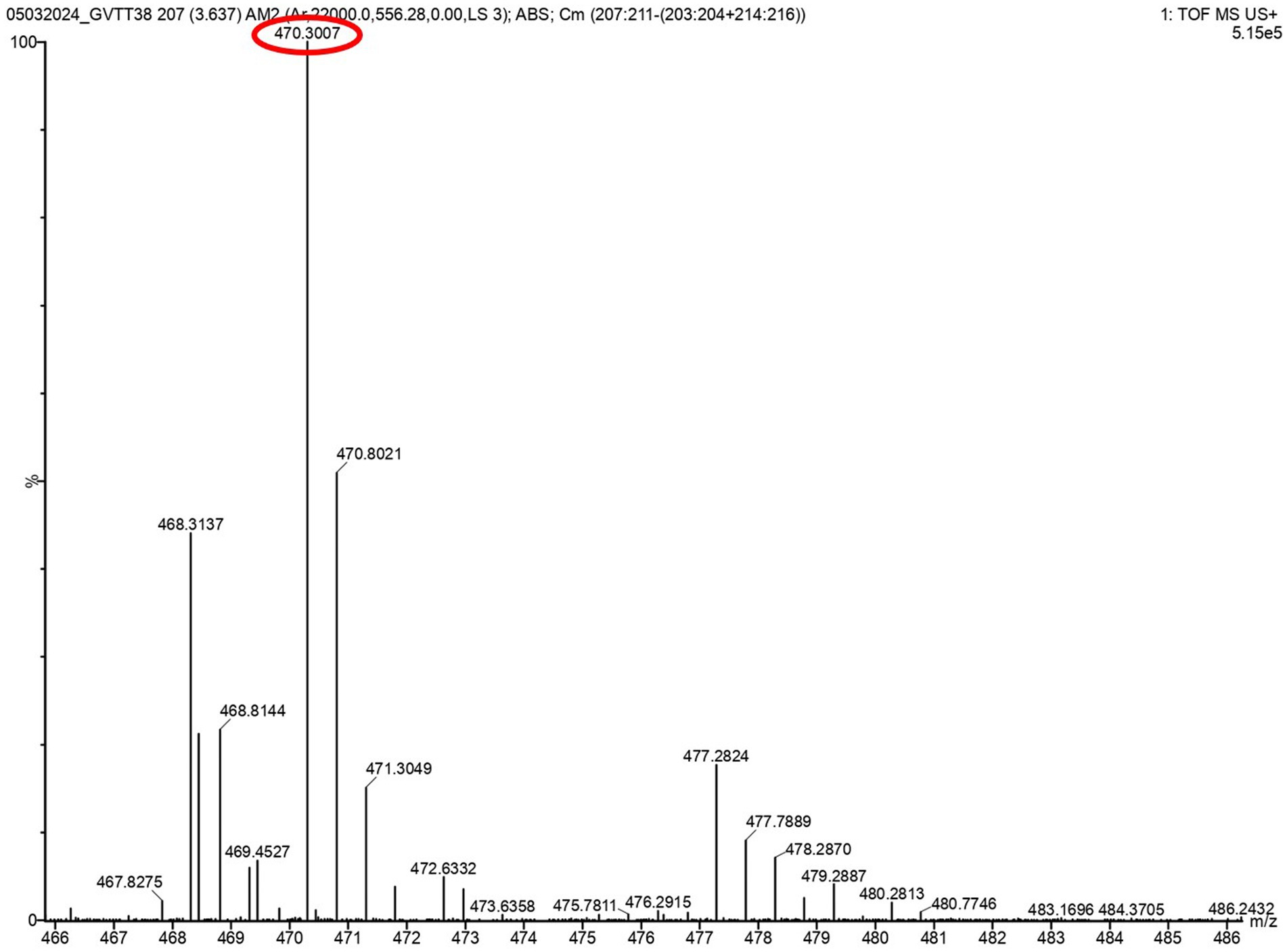
Figure 7. LC-HRMS-ESI spectrum of the compound kendomycin. The retention time was 3.637 min, and the mass of the spectrum is 470.3007.
Discussion
Streptomyces sp. are ubiquitously present in the soil and are associated with different flora. Endophytic Streptomyces, which are symbiotic with the host, offer many benefits to the host, including the production of plant growth-promoting factors and the inhibition of the growth of pathogenic bacteria (as biocontrol agents) (Vurukonda et al., 2018). Additionally, endophytic associations confer endophytic bacteria with the ability to produce host metabolites by establishing deeper connections (Li et al., 2022). Therefore, the isolation and characterization of endophytic Streptomyces are more advantageous than the use of free-living Streptomyces.
In terms of global dietary preferences, tomatoes are a prominent and widely consumed plant-based food. Renowned for their rich repository of clinically significant bioactive compounds, these fruits hold immense nutritional value. An intriguing facet of tomato plants lies in their symbiotic association with an array of endophytic microbes (Chopra and Roberts, 2001; Martínez-Núñez and López, 2016; Santoyo et al., 2016; Li et al., 2022). This study aimed to isolate Streptomyces sp. endophytic from tomato plants, which have high potential to produce secondary metabolites of nutritional and commercial value. Our attempt to isolate Streptomyces endophytes from tomato plants resulted in the identification of a novel strain, Streptomyces sp. VITGV156. The strain exhibited significant antibacterial activity and exhibited potential for secondary metabolites. The observed colony and spore of VITGV156 resemble the characteristics of Streptomyces sp. and are on par with the findings of various other studies (Aouar et al., 2012; Bhandari et al., 2022).
Whole-genome sequencing helps in the comprehensive analysis of the VITGV156 genome, the identification and classification of the isolate, and bioprospecting for potential secondary metabolite gene clusters. The isolated Streptomyces sp. VITGV156 is phylogenetically most closely related to S. rochei and has a remarkable resemblance in terms of cellular morphology (Pazhanimurugan et al., 2016). Consistent with these results, another endophytic species, Streptomyces sp. PT2, which is isolated from tomato plants, exhibits high similarity with S. rochei (Singh and Dubey, 2018). Several species of S. rochei have been isolated from various niches, including the rhizosphere and terrestrial ecosystem, and have wide applications (Kanini et al., 2013; Hamdan et al., 2021; Zhou et al., 2024). Bioprospecting for antimicrobial secondary metabolites revealed a staggering count of 29 biosynthesis-related gene clusters, of which 17 gene clusters exhibited noteworthy resemblances to other existing BGCs (similarity above 50%). The genome encompasses several BGCs for the production of polyketide antimicrobial compounds, such as vicenistatin, the fluostatin M-Q, stambomycin, streptovaricin, kanamycin, kendomycin B, methylenomycin A, prejadomycin, rabelomycin, gaudimycin C, granaticin, versipelostatin, and istamycin (Table 2). This finding indicates that VITGV156 is fertile ground for the emergence of novel derivatives and bioactive compounds. These compounds have been previously identified in various vicenistatin strains of Streptomyces halstedii (Chopra and Roberts, 2001); the fluostatin M-Q in Streptomyces sp. PKU-MA00045 (Jin et al., 2018); the stambomycin in Streptomyces ambofaciens (Laureti et al., 2011); the kendomycin B in Streptomyces sp. Cl 58–27 (Paulus et al., 2021); the methylenomycin A in Streptomyces violaceusruber SANK 95570 (Aguilar and Hopwood, 1982); the buyrolactone compounds in Streptomyces chattanoogensis (Du et al., 2011); the versipelostatin in Streptomyces versipellis 4,083-SVS6 (Ueda et al., 2008); and the istamycin in Streptomyces tenjimariensis ATCC31603 (Hoang et al., 2016). Notably, many BGCs exhibit minimal similarity, below 50% (4), and a significant number of BGCs exhibit similarities, below 20% (8), further indicating that Streptomyces sp. VITGV156 is a potent producer of diverse bioactive secondary metabolites. Remarkably, most of the BGCs encoding polyketide antibiotics are <20%, indicating that they could be derivates of this class of antibiotics, which is worth investigating. The genome of VITGV156 contains genes responsible for the production of a different class of antimicrobial compounds, which includes non-ribosomal peptide synthetases (NRPSs) and ribosomally synthesized and posttranslationally modified peptides (RiPPs). NRPSs are complex multimodular enzymes that produce non-ribosomal peptides (NRPs), which have diverse biological applications (Strieker et al., 2010; Martínez-Núñez and López, 2016). Many non-ribosomal peptides have antimicrobial activity, and many serve as last-resort antimicrobial agents (Liu et al., 2019). Notably, at least five different NRPSs (Table 2) encode VITGV156, one of which is a well-known antibacterial agent, Streptothricin (Morgan et al., 2023). The products of other NRPSs remain elusive. RiPPs are ribosomal-synthesized posttranslationally modified antimicrobial peptides of commercial value. Owing to their broad range of antibacterial activity against Gram-positive and Gram-negative bacteria, they are commonly used in the food and pharmaceutical industries (Van Kraaij et al., 1999; Arnison et al., 2013). The genome of VITGV156 encodes two RiPPs, two of which exhibit low sequence similarity with known compounds (Table 2). These results are consistent with the results of previous studies on Streptomyces sp. Babs14, which exhibited 29 biosynthetic secondary metabolite gene clusters, of which eight gene clusters displayed 100% congruence with known clusters (Zerouki et al., 2021). An earlier investigation (Cruz-Morales et al., 2017) identified secondary metabolites such as siderophores in Streptomyces sp. CC77. This finding adds to the broader understanding of secondary metabolite diversity within the Streptomyces genus. Analogous findings have been reported across several Streptomyces species; for instance, Streptomyces sp. ICC1 boasts a compilation of 37 BGCs (Gosse et al., 2019). Additionally, the genome of VITGV156 also revealed the presence of genes responsible for the biosynthesis of butyrolactone, siderophores, indoles, ectoine, and terpenes.
Notably, Streptomyces sp. VITGV156 has BGCs for many polyketide antibiotics. They are characterized by their complex structures, which contain multiple cyclic rings, conjugated double bonds, and various functional groups (John, 1991). Interestingly, these compounds exhibit broad-spectrum antimicrobial activity and have been shown to be effective even against drug-resistant pathogens (Kizhakkekalam et al., 2020; Chakraborty et al., 2021). They are synthesized by multimodal enzyme complexes called polyketide synthases (PKSs). Simple building blocks such as acetyl-CoA and malonyl-CoA are iteratively condensed to form a polyketide chain, which is subsequently processed by a group of tailoring enzymes. These enzymes include ketoreductases (KRs), dehydratases (DHs), enoyl reductases (ERs), and methyltransferases (MTs), which modify the polyketide backbone. This process results in the production of a vast array of structurally diverse molecules. Finally, the modified polyketide chain undergoes a cyclization reaction to form the characteristic pentacyclic core structure (Chen and Du, 2016; Ray and Moore, 2016; Xu and Arimoto, 2016; Wang et al., 2020). Among these antibiotics, the antibiotic class kendomycin has attracted attention for the following reasons. Kendomycin has broad-spectrum antimicrobial activity and has been shown to affect various cellular processes upon exposure (Elnakady et al., 2007, 2016; Tranter et al., 2020), making it a suitable candidate for the treatment of multidrug-resistant pathogens. However, kendomycin is produced by a relatively small number of Streptomyces sp. strains, making it rare (Xu and Arimoto, 2016; Chen et al., 2021; Paulus et al., 2021). Additionally, the biosynthetic pathway of kendomycin biosynthesis has not been fully elucidated. The genome of VITGV156 harbors five Type I PKSs corresponding to the biosynthesis of kendomycin. However, the details of their structure and enzymatic capability remain less explored. The identification of kendomycin B BGC through whole-genome sequencing and subsequent bioinformatics analyses has provided a foundational understanding of the genetic architecture driving kendomycin synthesis in Streptomyces sp. VITGV156. Notably, the presence of a type I polyketide synthase (PKS) gene cluster, sharing significant similarity with known kendomycin BGCs in Verrucosispora sp. and Streptomyces violaceoruber, underscores the conservation of kendomycin biosynthetic pathways across different strains. In addition, other enzymes of the kendomcyin biosynthesis pathway, such as modifying and cyclization enzymes need to be identified, which forms the future scope of this study.
To correlate the relevance of this study to plant pathogen control, many citations are there: Streptomyces griseus H7602 and Streptomyces griseorubens E44G are associated with the control of Phytophthora capsici and Fusarium oxysporum f. sp. lycopersici, both devastating pathogens of tomato plants (Khan et al., 2023). The ecological significance of these findings lies in the potential of these Streptomyces strains to offer sustainable solutions for disease management in tomato cultivation. By harnessing the beneficial effects of these endophytic bacteria, farmers can reduce their reliance on chemical pesticides and promote environmentally friendly agricultural practices.
Our attempt to understand the molecular basis of the antimicrobial activity of kendomcyin B revealed that kendomycin caused multifaceted damage in both Gram-positive and Gram-negative bacteria. Our studies revealed a striking pattern of damage induced by kendomycin B in bacteria, with membrane damage accompanied by nucleoid structure remodeling. Bacterial cellular processes are intricately associated with the cell membrane owing to the lack of cellular organelles (Govindarajan and Amster-Choder, 2016). Therefore, the bacterial membrane is highly dynamic, heterogeneous in composition, and allows lateral diffusion of membrane proteins to facilitate cellular processes (Vereb et al., 2003). Therefore, it is plausible that kendomycin-induced toxicity is due to the inhibition of various cellular processes associated with the bacterial membrane and nucleoid. Kendomycin exposure disrupts the cell division process by interfering with cell septum formation in a manner similar to penicillin (Elnakady et al., 2016). Overall, our exploration of endophytic Streptomyces resulted in the isolation of Streptomyces sp. VITGV156, which has remarkable bioprospecting potential for antimicrobial compounds.
Conclusion
In this study, we endeavored to isolate endophytes. Streptomyces spp. are capable of producing novel antibiotics from tomato plants. Our studies resulted in the isolation of Streptomyces sp. VITGV156 from healthy tomato plants. The whole-genome sequencing of VITGV156 revealed its genomic landscape and its striking similarity to that of S. rochei. Additional analysis revealed the presence of several BGCs capable of producing polyketide antibiotics. Our analysis of kendomycin biosynthetic clusters revealed five Type I polyketide synthases: S. verrucosispora and S. violaceoruber. Furthermore, we extended our analysis and showed that kendomycin B causes damage to the membrane, changes the nucleoid morphology, and possibly can bind to membrane proteins during bacterial killing.
Data availability statement
The whole genome of Streptomyces sp. VITGV156 was submitted to the NCBI SRA repository under BioProject accession number PRJNA750872 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA750872), BioSample accession number SAMN20499087 (https://www.ncbi.nlm.nih.gov/biosample/20499087), SRA accession number SRS9645416 and Accession ID: 20499087 & NCMR (Culture Deposit Accession no)—MCC4965.
Author contributions
PV: Writing – original draft, Investigation, Methodology, Writing – review & editing. TN: Writing – review & editing, Data curation, Formal analysis, Resources. SM: Data curation, Formal analysis, Writing – review & editing, Validation. SS: Validation, Writing – review & editing, Conceptualization, Investigation, Methodology. LM: Investigation, Methodology, Writing – review & editing, Resources. TT: Writing – review & editing, Conceptualization, Supervision, Validation. JC: Conceptualization, Supervision, Writing – review & editing, Resources, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by VIT SEED GRANT (Nos. SG20220080 and SPL/SG20230175) of the Vellore Institute of Technology, Vellore, India.
Acknowledgments
VP acknowledges the Indian Council of Medical Research (ICMR) for the SRF (File No: 45/36/2022/-DDI/BMS). TN acknowledges SRM University AP for financial assistance through UPDF. VP thanks Sutharsan Govindarajan, SRM University AP, for his help with the microscopy analysis. All the authors acknowledge the Central Instrumentation Facilities of SRM University AP for their support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1407289/full#supplementary-material
Footnotes
References
Aguilar, A., and Hopwood, D. A. (1982). Determination of methylenomycin a synthesis by the pSV1 plasmid from Streptomyces violaceus-ruber SANK 95570. J. Gen. Microbiol. 128, 1893–1901. doi: 10.1099/00221287-128-8-1893
Alam, K., Mazumder, A., Sikdar, S., Zhao, Y.-M. M., Hao, J., Song, C., et al. (2022). Streptomyces: the biofactory of secondary metabolites. Front. Microbiol. 13:968053. doi: 10.3389/fmicb.2022.968053
Al-Quwaie, D. A. (2024). The role of Streptomyces species in controlling plant diseases: a comprehensive review. Australas. Plant Pathol. 53, 1–14. doi: 10.1007/s13313-023-00959-z
Antimicrobial Resistance CollaboratorsMurray, C. J., Ikuta, K. S., Sharara, F., Swetschinski, L., Robles Aguilar, G., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Aouar, L., Lerat, S., Ouffroukh, A., Boulahrouf, A., and Beaulieu, C. (2012). Taxonomic identification of rhizospheric actinobacteria isolated from Algerian semiarid soil exhibiting antagonistic activities against plant fungal pathogens. Can. J. Plant Pathol. 34, 165–176. doi: 10.1080/07060661.2012.681396
Aragón-Muriel, A., Liscano, Y., Upegui, Y., Robledo, S. M., Ramírez-Apan, M. T., Morales-Morales, D., et al. (2021). In vitro evaluation of the potential pharmacological activity and molecular targets of new Benzimidazole-based schiff base metal complexes. Antibiotics 10, 1–27. doi: 10.3390/antibiotics10060728
Arnison, P. G., Bibb, M. J., Bierbaum, G., Bowers, A. A., Bugni, T. S., Bulaj, G., et al. (2013). Ribosomally synthesized and posttranslationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. doi: 10.1039/C2NP20085F
Ayswaria, R., Vasu, V., and Krishna, R. (2020). Diverse endophytic Streptomyces species with dynamic metabolites and their meritorious applications: a critical review. Crit. Rev. Microbiol. 46, 750–758. doi: 10.1080/1040841X.2020.1828816
Blin, K., Shaw, S., Augustijn, H. E., Reitz, Z. L., Biermann, F., Alanjary, M., et al. (2023). antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualization. Nucleic Acids Res. 51, W46–W50. doi: 10.1093/nar/gkad344
Bhandari, S., Bhattarai, B. R., Adhikari, A. A., Aryal, B., Shrestha, A., Aryal, N., et al. (2022). Characterization of Streptomyces Species and Validation of Antimicrobial Activity of Their Metabolites through Molecular Docking. Processes 10, 2149. doi: 10.3390/pr10102149
Bode, H. B., and Zeeck, A. (2000). Structure and biosynthesis of kendomycin, a carbocyclic ansa-compound from Streptomyces. J. Chem. Soc. Perkin. 1, 323–328. doi: 10.1039/A908387A
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Chakraborty, K., Kizhakkekalam, V. K., and Joy, M. (2021). Macrocyclic polyketides with siderophore mode of action from marine heterotrophic Shewanella algae: prospective anti-infective leads attenuate drug-resistant pathogens. J. Appl. Microbiol. 130, 1552–1570. doi: 10.1111/jam.14875
Chanama, M., Suriyachadkun, C., and Chanama, S. (2023). Streptomyces antimicrobicus sp. nov., a novel clay soil-derived actinobacterium producing antimicrobials against drug-resistant bacteria. PLoS One 18:e0286365. doi: 10.1371/JOURNAL.PONE.0286365
Chen, H., and Du, L. (2016). Iterative polyketide biosynthesis by modular polyketide synthases in bacteria. Appl. Microbiol. Biotechnol. 100, 541–557. doi: 10.1007/s00253-015-7093-0
Chen, X., Pizzatti, C., Bonaldi, M., Saracchi, M., Erlacher, A., Kunova, A., et al. (2016). Biological control of lettuce drop and host plant colonization by Rhizospheric and endophytic Streptomycetes. Front. Microbiol. 7:714. doi: 10.3389/fmicb.2016.00714
Chen, J., Zhang, S., Chen, Y., Tian, X., Gu, Y., and Ju, J. (2021). Identification and heterologous expression of the Kendomycin B biosynthesis-related gene cluster from Verrucosispora sp. SCSIO 07399. Mar. Drugs 19, 1–14. doi: 10.3390/md19120673
Chhetri, G., Kim, M. J., Kim, I., Tran, D. V. H., Kim, Y.-W., Kim, H. W., et al. (2024). Streptomyces tagetis sp. nov., a chromomycin producing bacteria isolated from the roots of Tagetes patula. Front. Microbiol. 15:1361583. doi: 10.3389/fmicb.2024.1361583
Chopra, I., and Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260. doi: 10.1128/MMBR.65.2.232-260.2001
Coombs, J. T., and Franco, C. M. M. (2003). Visualization of an endophytic Streptomyces species in wheat seed. Appl. Environ. Microbiol. 69, 4260–4262. doi: 10.1128/AEM.69.7.4260-4262.2003
Cruz-Morales, P., Ramos-Aboites, H. E., Licona-Cassani, C., Selem-Mójica, N., Mejía-Ponce, P. M., Souza-Saldívar, V., et al. (2017). Actinobacteria phylogenomics, selective isolation from an iron oligotrophic environment and siderophore functional characterization, unveil new desferrioxamine traits. FEMS Microbiol. Ecol. 93, 1–12. doi: 10.1093/FEMSEC/FIX086
Devi, S., Sharma, M., and Manhas, R. K. (2022). Investigating the plant growth promoting and biocontrol potentiality of endophytic Streptomyces SP. SP5 against early blight in Solanum lycopersicum seedlings. BMC Microbiol. 22:285. doi: 10.1186/s12866-022-02695-8
Dillon, S. C., and Dorman, C. J. (2010). Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195. doi: 10.1038/nrmicro2261
Du, Y.-L., Shen, X.-L., Yu, P., Bai, L.-Q., and Li, Y.-Q. (2011). Gamma-butyrolactone regulatory system of Streptomyces chattanoogensis links nutrient utilization, metabolism, and development. Appl. Environ. Microbiol. 77, 8415–8426. doi: 10.1128/AEM.05898-11
Duangmal, K., Muangham, S., Mingma, R., Yimyai, T., Srisuk, N., Kitpreechavanich, V., et al. (2016). Kineococcus mangrovi sp. nov., isolated from mangrove sediment. Int. J. Syst. Evol. Microbiol. 66, 1230–1235. doi: 10.1099/ijsem.0.000860
Duggal, P., Naseri, I., and Sobol, S. E. (2011). The increased risk of community-acquired methicillin-resistant Staphylococcus aureus neck abscesses in young children. Laryngoscope 121, 51–55. doi: 10.1002/LARY.21214
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Elnakady, Y. A., Chatterjee, I., Bischoff, M., Rohde, M., Josten, M., Sahl, H.-G., et al. (2016). Investigations to the antibacterial mechanism of action of Kendomycin. PLoS One 11:e0146165. doi: 10.1371/journal.pone.0146165
Elnakady, Y. A., Rohde, M., Sasse, F., Backes, C., Keller, A., Lenhof, H.-P., et al. (2007). Evidence for the mode of action of the highly CytotoxicStreptomycesPolyketide Kendomycin. Chembiochem 8, 1261–1272. doi: 10.1002/cbic.200700050
Gomes, E. S., Schuch, V., and de Macedo Lemos, E. G. (2013). Biotechnology of polyketides: new breath of life for the novel antibiotic genetic pathways discovery through metagenomics. Braz. J. Microbiol. 44, 1007–1034. doi: 10.1590/s1517-83822013000400002
Gosse, J. T., Ghosh, S., Sproule, A., Overy, D., Cheeptham, N., and Boddy, C. N. (2019). Whole genome sequencing and metabolomic study of cave Streptomyces isolates ICC1 and ICC4. Front. Microbiol. 10:1020. doi: 10.3389/fmicb.2019.01020
Goudjal, Y., Zamoum, M., Sabaou, N., Mathieu, F., and Zitouni, A. (2016). Potential of endophytic Streptomyces spp. for biocontrol of fusarium root rot disease and growth promotion of tomato seedlings. Biocontrol Sci. Tech. 26, 1691–1705. doi: 10.1080/09583157.2016.1234584
Govindarajan, S., and Amster-Choder, O. (2016). Where are things inside a bacterial cell? Curr. Opin. Microbiol. 33, 83–90. doi: 10.1016/j.mib.2016.07.003
Govindarajan, S., and Amster-Choder, O. (2017). The bacterial sec system is required for the organization and function of the MreB cytoskeleton. PLoS Genet. 13:e1007017. doi: 10.1371/journal.pgen.1007017
Grant, J. R., Enns, E., Marinier, E., Mandal, A., Herman, E. K., Chen, C., et al. (2023). Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51, W484–W492. doi: 10.1093/nar/gkad326
Hamdan, A. M., Abd-El-Mageed, H., and Ghanem, N. (2021). Biological treatment of hazardous heavy metals by Streptomyces rochei ANH for sustainable water management in agriculture. Sci. Rep. 11:9314. doi: 10.1038/s41598-021-88843-y
Hesje, C. K., Sanfilippo, C. M., Haas, W., and Morris, T. W. (2011). Molecular epidemiology of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolated from the eye. Curr. Eye Res. 36, 94–102. doi: 10.3109/02713683.2010.534229
Hoang, N. H., Huong, N. L., Kim, B., Sohng, J. K., Yoon, Y. J., and Park, J. W. (2016). Istamycin aminoglycosides profiling and their characterization in Streptomyces tenjimariensis ATCC 31603 culture using high-performance liquid chromatography with tandem mass spectrometry. J. Sep. Sci. 39, 4712–4722. doi: 10.1002/jssc.201600925
Jin, D. J., and Cabrera, J. E. (2006). Coupling the distribution of RNA polymerase to global gene regulation and the dynamic structure of the bacterial nucleoid in Escherichia coli. J. Struct. Biol. 156, 284–291. doi: 10.1016/j.jsb.2006.07.005
Jin, J., Yang, X., Liu, T., Xiao, H., Wang, G., Zhou, M., et al. (2018). Fluostatins M-Q featuring a 6-5-6-6 ring skeleton and high oxidized A-rings from marine Streptomyces sp. PKU-MA00045. Mar. Drugs 16, 1–14. doi: 10.3390/MD16030087
John, A. R. (1991). Polyketide synthase com plexes: their structure and function in antibiotic biosynthesis. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 332, 107–114. doi: 10.1098/rstb.1991.0038
Kandel, S. L., Joubert, P. M., and Doty, S. L. (2017). Bacterial endophyte colonization and distribution within plants. Microorganisms 5, 1–26. doi: 10.3390/microorganisms5040077
Kanini, G. S., Katsifas, E. A., Savvides, A. L., and Karagouni, A. D. (2013). Streptomyces rochei ACTA1551, an indigenous Greek isolate studied as a potential biocontrol agent against fusarium oxysporum f.sp. lycopersici. Biomed. Res. Int. 2013:387230. doi: 10.1155/2013/387230
Kar, S., Edgar, R., and Adhya, S. (2005). Nucleoid remodeling by an altered HU protein: reorganization of the transcription program. Proc. Natl. Acad. Sci. USA 102, 16397–16402. doi: 10.1073/pnas.0508032102
Karthik, L., Kumar, G., Kirthi, A. V., Rahuman, A. A., and Bhaskara Rao, K. V. (2014). Streptomyces sp. LK3 mediated synthesis of silver nanoparticles and its biomedical application. Bioprocess Biosyst. Eng. 37, 261–267. doi: 10.1007/s00449-013-0994-3
Kekuda, P., Onkarappa, R., and Jayanna, N. (2015). Characterization and antibacterial activity of a glycoside antibiotic from Streptomyces variabilis PO-178. Sci. Technol. Arts Res. J. 3:116. doi: 10.4314/star.v3i4.17
Khan, S., Srivastava, S., Karnwal, A., and Malik, T. (2023). Streptomyces as a promising biological control agents for plant pathogens. Front. Microbiol. 14:1285543. doi: 10.3389/FMICB.2023.1285543
Khare, E., Mishra, J., and Arora, N. K. (2018). Multifaceted interactions between endophytes and plant: developments and prospects. Front. Microbiol. 9:2732. doi: 10.3389/fmicb.2018.02732
Kinkel, L. L., Schlatter, D. C., Xiao, K., and Baines, A. D. (2014). Sympatric inhibition and niche differentiation suggest alternative coevolutionary trajectories among Streptomycetes. ISME J. 8, 249–256. doi: 10.1038/ismej.2013.175
Kiran, G., Karthik, L., Thumma, G., and Vishnukirthi, A. (2022). Genome data mining, chemistry and bioactivity of Sesquiterpenes from Actinobacteria. Actinobacteria 1, 101–120. doi: 10.1007/978-981-16-5835-8_6
Kizhakkekalam, V. K., Chakraborty, K., and Joy, M. (2020). Oxygenated elansolid-type of polyketide spanned macrolides from a marine heterotrophic Bacillus as prospective antimicrobial agents against multidrug-resistant pathogens. Int. J. Antimicrob. Agents 55:105892. doi: 10.1016/j.ijantimicag.2020.105892
Klemm, P., Stadler, P. F., and Lechner, M. (2023). Proteinortho6: pseudoreciprocal best alignment heuristic for graph-based detection of (co-)orthologs. Front. Bioinform. 3:1322477. doi: 10.3389/fbinf.2023.1322477
Komaki, H. (2022). Resolution of housekeeping gene sequences used in MLSA for the genus Streptomyces and reclassification of Streptomyces anthocyanicus and Streptomyces tricolor as heterotypic synonyms of Streptomyces violaceoruber. Int. J. Syst. Evol. Microbiol. 72:005370. doi: 10.1099/ijsem.0.005370
Lacey, H. J., and Rutledge, P. J. (2022). Recently, discovered secondary metabolites from Streptomyces species. Molecules 27, 1–16. doi: 10.3390/molecules27030887
Laureti, L., Song, L., Huang, S., Corre, C., Leblond, P., Challis, G. L., et al. (2011). Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc. Natl. Acad. Sci. USA 108, 6258–6263. doi: 10.1073/pnas.1019077108
Law, J. W. F., Ser, H. L., Ab Mutalib, N. S., Saokaew, S., Duangjai, A., Khan, T. M., et al. (2019). Streptomyces monashensis sp. nov., a novel mangrove soil actinobacterium from East Malaysia with antioxidative potential. Sci. Rep. 9, 3056–3018. doi: 10.1038/s41598-019-39592-6
Law, J. W.-F. F., Ser, H.-L. L., Duangjai, A., Saokaew, S., Bukhari, S. I., Khan, T. M., et al. (2017). Streptomyces colonosanans sp. nov., a novel Actinobacterium isolated from Malaysia mangrove soil exhibiting Antioxidative activity and cytotoxic potential against human colon cancer cell lines. Front. Microbiol. 8:877. doi: 10.3389/fmicb.2017.00877
Laxminarayan, R. (2022). The overlooked pandemic of antimicrobial resistance. Lancet 399, 606–607. doi: 10.1016/S0140-6736(22)00087-3
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, Z., Wen, W., Qin, M., He, Y., Xu, D., and Li, L. (2022). Biosynthetic mechanisms of secondary metabolites promoted by the interaction between endophytes and plant hosts. Front. Microbiol. 13:928967. doi: 10.3389/FMICB.2022.928967
Ling, L., Han, X., Li, X., Zhang, X., Wang, H., Zhang, L., et al. (2020). A Streptomyces sp. NEAU-HV9: isolation, identification, and potential as a biocontrol agent against Ralstonia Solanacearum of tomato plants. Microorganisms 8, 1–15. doi: 10.3390/microorganisms8030351
Liu, Y., Ding, S., Shen, J., and Zhu, K. (2019). Nonribosomal antibacterial peptides that target multidrug-resistant bacteria. Nat Prod Rep 36, 573–592. doi: 10.1039/C8NP00031J
Loganathan, K., Kumar, G., Kirthi, A. V., Rao, K. V. B., and Rahuman, A. A. (2013). Entomopathogenic marine actinomycetes as potential and low-cost biocontrol agents against bloodsucking arthropods. Parasitol. Res. 112, 3951–3959. doi: 10.1007/s00436-013-3585-y
Macheras, E., Roux, A.-L., Bastian, S., Leão, S. C., Palaci, M., Sivadon-Tardy, V., et al. (2011). Multilocus sequence analysis and rpoB sequencing of Mycobacterium abscessus (sensu lato) strains. J. Clin. Microbiol. 49, 491–499. doi: 10.1128/JCM.01274-10
Martínez-Núñez, M. A., and López, V. E. L. (2016). Nonribosomal peptides synthetases and their applications in industry. Sustain. Chem. Process 4, 1–8. doi: 10.1186/S40508-016-0057-6
Melinda, Y. N., Widada, J., Wahyuningsih, T. D., Febriansah, R., Damayanti, E., and Mustofa, M. (2021). Metabologenomics approach to the discovery of novel compounds from Streptomyces sp. GMR22 as anti-SARS-CoV-2 drugs. Heliyon 7:e08308. doi: 10.1016/j.heliyon.2021.e08308
Micklefield, J. (2009). Biosynthesis and biosynthetic engineering of calcium-dependent lipopeptide antibiotics. Pure Appl. Chem. 81, 1065–1074. doi: 10.1351/PAC-CON-08-08-29
Morgan, C. E., Kang, Y. S., Green, A. B., Smith, K. P., Dowgiallo, M. G., Miller, B. C., et al. (2023). Streptothricin F is a bactericidal antibiotic effective against highly drug-resistant gram-negative bacteria that interacts with the 30S subunit of the 70S ribosome. PLoS Biol. 21:e3002091. doi: 10.1371/JOURNAL.PBIO.3002091
Naqvi, A. A. T., Mohammad, T., Hasan, G. M., and Hassan, M. I. (2018). Advancements in docking and molecular dynamics simulations toward ligand–receptor interactions and structure–function relationships. Curr. Top. Med. Chem. 18, 1755–1768. doi: 10.2174/1568026618666181025114157
Olanrewaju, O. S., and Babalola, O. O. (2019). Streptomyces: implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol. 103, 1179–1188. doi: 10.1007/s00253-018-09577-y
Ongpipattanakul, C., Desormeaux, E. K., Dicaprio, A., Van Der Donk, W. A., Mitchell, D. A., and Nair, S. K. (2022). Mechanism of action of Ribosomally synthesized and post-translationally modified peptides. Chem. Rev. 122, 14722–14814. doi: 10.1021/ACS.CHEMREV.2C00210
Veilumuthu, P., Nagarajan, T., Sasikumar, S., Siva, R., Jose, S., and Christopher, J. G. (2022). Streptomyces sp. VITGV100: an endophyte from Lycopersicon esculentum as new source of indole type compounds. Biochem. Syst. Ecol. 105:104523. doi: 10.1016/J.BSE.2022.104523
Papini, A. M. (2009). The use of posttranslationally modified peptides for detection of biomarkers of immune-mediated diseases. J. Pept. Sci. 15, 621–628. doi: 10.1002/PSC.1166
Passari, A. K., Upadhyaya, K., Singh, G., Abdel-Azeem, A. M., Thankappan, S., Uthandi, S., et al. (2019). Enhancement of disease resistance, growth potential, and photosynthesis in tomato (Solanum lycopersicum) by inoculation with an endophytic actinobacterium, Streptomyces thermocarboxydus strain BPSAC147. PLoS One 14:e0219014. doi: 10.1371/journal.pone.0219014
Paulus, C., Gromyko, O., and Luzhetskyy, A. (2021). New Kendomycin derivative isolated from Streptomyces sp. cl 58-27. Molecules 26:6834. doi: 10.3390/molecules26226834
Pazhanimurugan, R., Radhakrishnan, M., Shanmugasundaram, T., Gopikrishnan, V., and Balagurunathan, R. (2016). Terpenoid bioactive compound from Streptomyces rochei (M32): taxonomy, fermentation and biological activities. World J. Microbiol. Biotechnol. 32:161. doi: 10.1007/s11274-016-2121-5
Quecine, M. C., Araujo, W. L., Marcon, J., Gai, C. S., Azevedo, J. L., and Pizzirani-Kleiner, A. A. (2008). Chitinolytic activity of endophytic Streptomyces and potential for biocontrol. Lett. Appl. Microbiol. 47, 486–491. doi: 10.1111/j.1472-765X.2008.02428.x
Ray, L., and Moore, B. S. (2016). Recent advances in the biosynthesis of unusual polyketide synthase substrates. Nat. Prod. Rep. 33, 150–161. doi: 10.1039/c5np00112a
Risdian, C., Mozef, T., and Wink, J. (2019). Biosynthesis of polyketides in Streptomyces. Microorganisms 7, 1–18. doi: 10.3390/microorganisms7050124
Santoyo, G., Moreno-Hagelsieb, G., del Carmen Orozco-Mosqueda, M., and Glick, B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. doi: 10.1016/J.MICRES.2015.11.008
Seipke, R. F., Kaltenpoth, M., and Hutchings, M. I. (2012). Streptomyces as symbionts: an emerging and widespread theme? FEMS Microbiol. Rev. 36, 862–876. doi: 10.1111/j.1574-6976.2011.00313.x
Sieber, S. A., and Marahiel, M. A. (2005). Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem. Rev. 105, 715–738. doi: 10.1021/CR0301191
Singh, R., and Dubey, A. K. (2018). Diversity and applications of endophytic Actinobacteria of plants in special and other ecological niches. Front. Microbiol. 9:1767. doi: 10.3389/fmicb.2018.01767
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and postanalysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Strieker, M., Tanović, A., and Marahiel, M. A. (2010). Nonribosomal peptide synthetases: structures and dynamics. Curr. Opin. Struct. Biol. 20, 234–240. doi: 10.1016/J.SBI.2010.01.009
Nagarajan, T., Govindarajan, S., and Munavar, M. H. (2023). Trans-translation system is important for maintaining genome integrity during DNA damage in bacteria. Res. Microbiol. 174:104136. doi: 10.1016/j.resmic.2023.104136
Tacar, O., Sriamornsak, P., and Dass, C. R. (2013). Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 65, 157–170. doi: 10.1111/j.2042-7158.2012.01567.x
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tanaka, Y., and Omura, S. (1990). Metabolism and products of actinomycetes. An introduction. Actinomycetologica 4, 13–14. doi: 10.3209/saj.4_13
Tattevin, P. (2011). Community-acquired methicillin-resistant Staphylococcus aureus (MRSA) infections. Med. Mal. Infect. 41, 167–175. doi: 10.1016/J.MEDMAL.2010.11.017
Tatusov, R. L., Galperin, M. Y., Natale, D. A., and Koonin, E. V. (2000). The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. doi: 10.1093/nar/28.1.33
Tranter, D., Filipuzzi, I., Lochmann, T., Knapp, B., Kellosalo, J., Estoppey, D., et al. (2020). Kendomycin cytotoxicity against bacterial, fungal, and mammalian cells is due to cation chelation. J. Nat. Prod. 83, 965–971. doi: 10.1021/acs.jnatprod.9b00826
Ueda, J.-Y., Chijiwa, S., Takagi, M., and Shin-Ya, K. (2008). A novel versipelostatin analog, versipelostatin F isolated from Streptomyces versipellis 4083-SVS6. J. Antibiot. 61, 752–755. doi: 10.1038/ja.2008.89
Van Kraaij, C., De Vos, W. M., Siezen, R. J., and Kuipers, O. P. (1999). Lantibiotics: biosynthesis, mode of action and applications. Nat. Prod. Rep. 16, 575–587. doi: 10.1039/A804531C
Veilumuthu, P., and Christopher, G. J. (2022a). Antimicrobial compounds produced by Streptomyces sp. VITGV01 against selected human pathogens. Res. J. Biotechnol. 17, 16–28. doi: 10.25303/1712RJBT16028
Veilumuthu, P., and Christopher, J. G. (2022b). Diversity of Actinomycetes in tomato plants. Ind. J. Agric. Res. 57, 95–102. doi: 10.18805/IJARe.A-5913
Veilumuthu, P., Nandana, K., and Christopher, J. G. (2023). Genomic report on Lycopersene producing Streptomyces sp. VITGV38. Curr. Trends Biotechnol. Pharm. 17, 1245–1251. doi: 10.5530/CTBP.2023.3S.60
Vereb, G., Szöllosi, J., Matkó, J., Nagy, P., Farkas, T., Vigh, L., et al. (2003). Dynamic, yet structured: the cell membrane three decades after the singer-Nicolson model. Proc. Natl. Acad. Sci. USA 100, 8053–8058. doi: 10.1073/pnas.1332550100
Verma, V. C., Singh, S. K., and Prakash, S. (2011). Bio‐control and plant growth promotion potential of siderophore producing endophyticStreptomycesfromAzadirachta indicaA. Juss. Z. Allg. Mikrobiol. 51, 550–556. doi: 10.1002/jobm.201000155
Vurukonda, S. S. K. P., Giovanardi, D., and Stefani, E. (2018). Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 19, 1–26. doi: 10.3390/ijms19040952
Wang, J., Zhang, R., Chen, X., Sun, X., Yan, Y., Shen, X., et al. (2020). Biosynthesis of aromatic polyketides in microorganisms using type II polyketide synthases. Microb. Cell Factories 19:110. doi: 10.1186/s12934-020-01367-4
Wiegand, I., Hilpert, K., and Hancock, R. E. W. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175. doi: 10.1038/nprot.2007.521
Wood, T. M., and Martin, N. I. (2019). The calcium-dependent lipopeptide antibiotics: structure, mechanism, & medicinal chemistry. Fortschr. Med. 10, 634–646. doi: 10.1039/C9MD00126C
Worsley, S. F., Newitt, J., Rassbach, J., Batey, S. F. D., Holmes, N. A., Murrell, J. C., et al. (2020). Streptomyces endophytes promote host health and enhance growth across plant species. Appl. Environ. Microbiol. 86, 1–17. doi: 10.1128/AEM.01053-20
Xu, S., and Arimoto, H. (2016). Strategies for construction of the all-carbon macrocyclic skeleton of the ansamycin antibiotic-kendomycin. J. Antibiot. 69, 203–212. doi: 10.1038/ja.2016.5
Yuan, Y., Men, H., and Lee, C. (2004). Total synthesis of Kendomycin: a macro–C–Glycosidation approach. J. Am. Chem. Soc. 126, 14720–14721. doi: 10.1021/JA0447154
Zerouki, C., Bensalah, F., Kuittinen, S., Pappinen, A., and Turunen, O. (2021). Whole-genome sequencing of two Streptomyces strains isolated from the sand dunes of Sahara. BMC Genomics 22, 578–521. doi: 10.1186/S12864-021-07866-X
Keywords: Streptomyces sp. VITGV156, endophytes, tomato plant, kendomycin, MRSA, antiSMASH, biosynthetic gene cluster
Citation: Veilumuthu P, Nagarajan T, Magar S, Sundaresan S, Moses LJ, Theodore T and Christopher JG (2024) Genomic insights into an endophytic Streptomyces sp. VITGV156 for antimicrobial compounds. Front. Microbiol. 15:1407289. doi: 10.3389/fmicb.2024.1407289
Edited by:
Karthik Loganathan, Salem Microbes Pvt. Ltd., IndiaReviewed by:
Joseph Selvin, Pondicherry University, IndiaGokul Shankar Sabesan, Melaka Manipal Medical College, India
Copyright © 2024 Veilumuthu, Nagarajan, Magar, Sundaresan, Moses, Theodore and Christopher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John Godwin Christopher, Z29kd2luakB2aXQuYWMuaW4=
†These authors share first authorship
‡These authors have contributed equally to this work
 Pattapulavar Veilumuthu1†
Pattapulavar Veilumuthu1† Lenus Joy Moses
Lenus Joy Moses John Godwin Christopher
John Godwin Christopher