
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 June 2024
Sec. Aquatic Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1403964
Beibu Gulf is an important semi-enclosed bay located in the northwestern South China Sea, and is famous for its high bio-productivity and rich bio-diversity. The fast development along the Beibu Gulf Economical Rim has brought pressure to the environment, and algal blooms occurred frequently in the gulf. In this study, surface water samples and micro-plankton samples (20–200 μm) were collected in the northern Beibu Gulf coast. Diversity and distribution of eukaryotic planktonic microalgae were analyzed by both metabarcoding and microscopic analyses. Metabarcoding revealed much higher diversity and species richness of microalgae than morphological observation, especially for dinoflagellates. Metabarcoding detected 144 microalgal genera in 8 phyla, while microscopy only detected 40 genera in 2 phyla. The two methods revealed different microalgal community structures. Dinoflagellates dominated in microalgal community based on metabarcoding due to their high copies of 18 s rRNA gene, and diatoms dominated under microscopy. Altogether 48 algal bloom and/or toxic species were detected in this study, 34 species by metabarcoding and 19 species by microscopy. Our result suggested a high potential risk of HABs in the Beibu Gulf. Microalgal community in the surface water samples demonstrated significantly higher OTU/species richness, alpha diversity, and abundance than those in the micro-plankton samples, although more HAB taxa were detected by microscopic observations in the micro-plankton samples. Furthermore, nano-sized taxa, such as those in chlorophytes, haptophytes, and chrysophyceans, occurred more abundantly in the surface water samples. This study provided a comprehensive morphological and molecular description of microalgal community in the northern Beibu Gulf.
Microalgae are the most important primary productors of the marine ecosystems, and play a key role in the global biogeochemical processes (Vogt, 2015; Worden et al., 2015). Microalgal community structure is regulated by a combination of physical, chemical, and biological factors, and fluctuates greatly to environmental variations (Raymont, 1983). Therefore, microalgal community structure can act as an indicator for environmental assessment of the marine ecosystems. However, some microalgal taxa form harmful algal blooms (HABs) and even produce toxins, which have negative impacts on marine organisms, cause serious economic losses to fisheries, aquaculture and tourism activities, as well as produce significant impacts on the environment and human health (Hallegraeff et al., 2004). About 177 marine microalgal species are listed as harmful microalgae, 117 of which belong to dinoflagellates (Lundholm et al., 2009). HABs occurred frequently in the worldwide coastal areas in the recent decades due to the global warming and eutrophication caused by the increase nutrient loading from human activities (Gilbert, 2017), especially in the Chinese coastal sea areas (Xiao et al., 2019; Sakamoto et al., 2021). Blooms caused by toxic flagellates especially those of dinoflagellates increased greatly in the China seas (Li et al., 2021; Tian et al., 2021; Wang et al., 2023).
Beibu Gulf (16°00′–21°30′N, 105°40′–111°00′E) is an important semi-enclosed bay located in the northwestern South China Sea (SCS). It connects to the open SCS by the Qiongzhou Strait in the northeast and to the open sea in the south (Figure 1). The gulf is famous for its high bio-productivity and rich bio-diversity (Gan et al., 2013; Chen et al., 2018). Although water quality in the Beibu Gulf is generally good, the fast development along the Beibu Gulf Economical Rim has brought pressure to the environment (Kaiser et al., 2013; Lai et al., 2014; Lao et al., 2019). Some coastal sea areas, such as Qinzhou Bay, Fangchenggang Bay and Lianzhou Bay, have been undertaking severe environmental problems, such as eutrophication (Lai et al., 2014; Lao et al., 2021; Pan et al., 2021) and HABs (Xu et al., 2019; Wang et al., 2021; Niu et al., 2022; Xie et al., 2023). Before 2000, the cyanobacterium Microcystis aeruginosa was the dominant bloom species in the Beibu Gulf, and blooms of cyanobacteria, dinoflagellates, and diatoms co-occurred during the period 2001–2010, and the haptophyte Phaeocystis globosa has become a major causative species of HABs after 2011 (Xu et al., 2019; Wang et al., 2021; Gibson et al., 2022; Zhu et al., 2023). A massive bloom caused by the dinoflagellate Noctiluca scintillans with total coverage area over 20,000 km2 occurred in February 2021 in Beibu Gulf (Xie et al., 2023). Frequent HABs have pressed adverse impacts on local ecosystems, and Beibu Gulf has become a hot area for HAB study in China (Xu et al., 2019; Wang et al., 2021; Gibson et al., 2022; Xie et al., 2023; Zhu et al., 2023).
Many previous studies have reported microalgal community structure in Beibu Gulf based on microscopic analysis (Xu et al., 2014; Wang et al., 2015; Qin et al., 2023), pigment analysis combined with high performance liquid chromatography (HPLC)-CHEMTAX analysis (Wang et al., 2021; Pan et al., 2023), flow cytometry (Wang J. X. et al., 2022; Xu et al., 2022), and metabarcoding during the Phaeocystis blooms (Gibson et al., 2022; Zhu et al., 2023). However, most of these studies are based on one method alone, which cannot fully reflect microalgal diversity and distribution of HAB taxa.
A large proportion of HAB taxa belong to the micro-sized microalgae (20–200 μm), for example, most of the harmful dinoflagellates and all of the harmful raphidophyceans (Hallegraeff et al., 2004). Although diatom cells are usually <20 μm, most of the harmful and/or bloom diatoms can form chains with the length > 20 μm such as Chaetoceros spp., Pseudo-nitzschia spp., Skeletonema spp., Thalassiosira spp., and Thalassionema spp. (Hallegraeff et al., 2004). Therefore, a plankton net with mesh size of 20 μm is suggested to qualitatively concentrate microalgae especially those of HAB taxa (Anderson and Throndsen, 2004). In this study, surface water samples and micro-plankton samples (20–200 μm) were collected in the northern Beibu Gulf coast. Metabarcoding of the V4 region of 18S rRNA gene was performed to investigate diversity and distribution of eukaryotic microalgae with the focus on HAB species, and the results were compared with those based on microscopic observation. The purpose of this study is to assess microalgal diversity in the northern Beibu Bay by the combination of microscopic and metabarcoding analyses, and to understand the distribution of HAB species and the potential risks of HABs.
The northern Beibu Gulf covers an area of 1.3 × 105 km2, with an average water depth of 38 m and a 1,629 km coastline (Chen et al., 2011; Li et al., 2014). The average annual water temperature is ca. 24.5°C. The warm weather together with abundant nutrient inputs from coastal rivers, such as the Nanliu and Qinjiang rivers, makes this region one of the most abundant fishing grounds in China (Chen et al., 2011).
Samples were collected during the autumn survey supported by Guangxi Zhuang Autonomous Region Marine Environment Monitoring Center in November 2020. Surface water samples were collected at 10 stations (Figure 1) from a depth of 2.0 m by a submerged pump. Surface water was filtered through a 200 μm mesh to remove large zooplankton. One liter filtered water sample were fixed in 1.5% Lugol’s iodine solution, concentrated to a final volume of 20 mL before microscopic observation. Triplicate 1.0 L filtered water samples were filtered by 0.45 μm polycarbonate membrane (Millipore, United States) for metabarcoding analysis, and stored at −80°C until DNA extraction. And the filtrate was used for nutrient analysis.
Microplankton samples were collected at a depth of 2 m using a double-layered net with mesh sizes between 20 and 200 μm. A known volume of seawater (20 L) was pumped through the double-layered net and concentrated to 500 mL, 100 mL of which was fixed with 1.5% Lugol’s iodine solution for microscopic observation after concentration to a final volume of 20 mL, and the other three 100 mL samples were filtered through 0.45 μm polycarbonate membranes for metabarcoding analysis.
Water temperature, salinity, pH, dissolved oxygen (DO), and chlorophyll a (Chl a) were measured by in situ a Multiparameter Sonde YSI 6600 (Yellow Spring Inc., United States). Dissolved inorganic nutrients were measured in triplicate via the spectrophotometric method (Strickland and Parsons, 1972).
Microalgae were analyzed and counted in triplicate under an inverted microscope (Nikon, TE2000-U, Japan) with 0.1 mL of the fixed samples at 400–600 times magnification. Morphological identification is performed at the lowest possible taxonomic level based on the taxonomic literatures (Tomas et al., 1997; Hallegraeff et al., 2004; Omura et al., 2012; Lassus et al., 2016).
Environmental DNA (eDNA) is extracted and purified from filtered samples using the HiPure Plant DNA Mini Kit (Majorbio Bio-Pharm Technology Co. Ltd., China) according to the manufacturer’s protocol. The DNA concentration and quality were assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, United States) with OD260/280 ≈ 1.8 and concentration > 10 ng/μL. The extracted eDNA samples were stored at −80°C for PCR amplification and sequencing.
Primer 3NDf 5′-GGCAAGTCTGGTGCCAG-3′ (Cavalier-Smith et al., 2009) and V4_euk_R2 5′-ACGGTATCTATCTCTTCG-3′ (Bråte et al., 2010) were used to amplify the V4 region of 18S rRNA gene by an ABI GeneAmp® 9700 PCR thermocycler (ABI, CA, United States). PCR system included 4 uL 5 × FastPfu Buffer, 2 uL 2.5 mM dNTPs, 0.8 uL forward primer (5 μM), 0.8 uL reverse primer (5 μM), 0.4 uL FastPfu polymerase, 0.2 uL bovine Serum albumin (BSA), 10 ng DNA template, and ddH2O to a final volume of 20 μL. The PCR cycle conditions were as following: (1) pre-denatured at 95°C for 3 min; (2) denatured at 95°C for 30 s, annealed at 55°C for 30 s, extended at 72°C for 45 s, 30 cycles; (3) extended at 72°C for 10 min and end at 4°C. The PCR products were run on 2% agarose gel, purified using the AxyPrep DNA gel extraction kit (Axygen Biosciences, Union City, CA, United States) according to the manufacturer’s instructions, and quantified using the Quantus™ fluorometer (Promega, United States). The purified amplifiers were collected in equimolar quantities. Sequencing was performed on the Illumina MiSeq PE300 platform (San Diego, United States) according to the standard protocol of Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). Raw data was deposited to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA)1 with the accession number PRJNA1069115.
Raw FASTQ files were de-multiplexed using an in-house perl script, and then quality-filtered by fastp (version 0.23.4, https://github.com/OpenGene/fastp) and merged by FLASH (version 1.2.11, http://www.cbcb.umd.edu/software/flash). Only overlapping sequences > 10 bp were assembled with the maximum mismatch ratio of 0.2. The optimized sequences were clustered into operational taxonomic units (OTUs) using USEARCH (version 11.0.667)-UPARSE (Edgar, 2013) with 97% sequence similarity level. The most abundant sequence for each OTU was selected as a representative sequence. Probable chimeric sequences were identified and removed using the UCHIME2 software package (Edgar, 2016). Chloroplast and mitochondria DNA reads were removed via QIIME2 (version 2023.5, https://qiime2.org/). The classification of each OTU was annotated by RDP Classifier (version 2.14) using a confidence threshold of 0.7 against the SILVA 18S rRNA gene database (version 138.1, https://www.arb-silva.de/). Algal OTUs were confirmed and selected by the taxonomic information in AlgaeBase (Guiry and Guiry, 2024, https://www.algaebase.org/), and the algal sequences were used for further analysis.
Those algal OTUs, unannotated against SILVA, were further searched against the NCBI database (https://www.ncbi.nlm.nih.gov/, searched in September 2023) using BLAST with a percent identity cutoff 97% and an e-value cutoff close to zero (≤10−30). The classification of algae was according to the up-to-date system suggested by Guiry (2024) and the AlgaeBase (Guiry and Guiry, 2024). HAB and/or bloom species were confirmed in the online version of the IOC-UNESCO Taxonomic Reference List of Harmful Micro Algae (Lundholm et al., 2009) or had been reported as a HAB or bloom species in previous studies.
In order to understand the bias between our primer and the classic V4 primer from Stoeck et al. (2010), and also the influence of different similarity and database on the analysis results, we have compared the two pairs of primers by metabarcoding four samples (S2, S4, S6, and S9) collected from this study using OTU assignment with 97% similarity against SILVA database and ASV assignment with 100% similarity against PR2 database. The results are listed in Supplementary Table S1. The results showed that the primer 3NDf and V4_euk_R2 increased the diversity and relative abundance of algal sequences, especially for those of dinoflagellates. While the classic V4 primers from Stoeck et al. (2010) obtained more relative abundance of diatoms. The results were comparable using SILVA database and PR2 database for the same primer, and also OTU assignment with 97% similarity and ASV assignment with 100% similarity. As the purpose of our study is to understand the diversity and distribution of HAB species, e.g., dinoflagellates, so we chose the primer 3NDf and V4_euk_R2 (Cavalier-Smith et al., 2009; Bråte et al., 2010).
DNA reads in each sample were normalized to the fewest reads in this study (88,072 reads). Alpha diversity indices of microalgal community, including richness, Shannon, Simpson, and Pielou indices were calculated. Venn diagrams were calculated and plotted based on different samples. Bray-Curtis (BC) index was measured using vegdist package, and non-metric multidimensional scaling (NMDS) was constructed based on BC index using vegan and ggplot2 packages. Permutation multivariate analysis of variance (PERMANOVA) was performed using the adonis function to test group differences. The bubble charts of dominant taxa were drawn with the R packages ggplot2, reshape2 and forcats. All these analyses were performed using the R4.1.0.
Altogether 1,167 eukaryotic algal OTUs were recorded, including 151 OTUs of diatoms (those in classes of Bacillariophyceae, Mediophyceae, and Coscinodiscophyceae), 831 OTUs in Dinophyta (dinoflagellates), 48 OTUs in Chlorophyta, and 35 OTUs in Haptophyta. The microalgal OTU richness ranged from 65 to 539 OTUs, and was generally lower in the micro-plankton samples than in the surface water samples (Figure 2A).

Figure 2. Microalgal community structure revealed by metabarcoding and microscopy. (A) OTU richness in each sample by metabarcoding, (B) Species richness in each sample by microscopy, (C) Cell number in each sample by microscopy, (D) Proportion of microalgal DNA reads to the eukaryotic community and cell number in the surface water samples (S) and micro-plankton samples (M), (E) Relative abundance of different groups of microalgae to the microalgal community, (F) Richness of different groups of microalgae in the surface water samples (S) and micro-plankton samples (M). S1–S10, Surface water samples; M1–M10, Micro-plankton samples; Diat, Diatoms including those in Bacillariophyceae, Mediophyceae, and Coscinodiscophyceae; Dino, Dinophyta; Chlo, Chlorophyta; Hapt, Haptophyta; Dict, Dictyochophyceae; Others, All other microalgal groups.
The eukaryotic algae detected in this study included 144 genera in 23 classes of 8 phyla (Supplementary Table S2). There were 308 OTUs (26.39%) identified at the species level, and 632 OTUs (54.16%) at the genus level. Altogether 202 species were detected in this study, including 74 species of diatoms, 65 species of dinoflagellates, 27 species of chlorophytes, 7 species of haptophytes, 15 species of cryptophyceans, 5 species of chrysophyceans, 4 species of raphidophyceans, and 5 species in other five classes (Supplementary Table S3). There were 198 and 166 species recorded in surface water (S) and micro-plankton (M) samples, respectively.
Dinoflagellates dominated in both surface water and micro-plankton samples, which accounted for about 60% of the algal DNA reads (Figure 2E). The microalgal community structure differed between the surface water and micro-plankton samples, reflected by lower relative DNA reads to the eukaryotic community, lower cell number (Figure 2D), and lower OTU richness (Figure 2F) in micro-plankton samples compared to those in surface water samples. Although contribution of diatom reads to the microeukaryotic community was much higher in surface water samples (7.48% on average) than that in micro-plankton samples (3.89% on average; Figure 2D), the percentage of diatom reads to microalgal reads in surface water samples (16.09%) was lower than that in micro-plankton samples (36.30%; Figure 2E). In addition, the relative reads of the nano-sized microalgae, such as chlorophytes and haptophytes, were significantly higher in surface water samples (8.01%–10.06%) than those in micro-plankton samples (mostly <1%) (Figure 2E). There were 1,068 and 747 OTUs of the eukaryotic microalgae detected in the surface water and micro-plankton samples, respectively (Figure 2F). The species compositions were similar in surface water and micro-plankton samples, and dinoflagellates were most diverse group, followed by diatoms, chlorophytes, and haptophytes (Figure 2F).
A total of 62 taxa (40 taxa at the species level) were identified in 40 genera of six classes in two microalgal phyla in this study, including 43 taxa of diatoms, 18 taxa of dinoflagellates, one taxon in Dictyochophyceae (Table 1). The number of microalgal species observed in each sample ranged from 13 to 32 taxa (Figure 2B), with an average of 21 taxa. Species richness was slightly higher in the micro-plankton samples (53 taxa) than in the surface water samples (44 taxa) (Figure 2F). Species richness of diatoms was the highest in all the samples, ranging from 9 to 27 taxa per sample (Figure 2B). Microalgal cell number ranged from 1.55 × 103 cells/L to 5.47 × 104 cells/L per sample (Figure 2C). On the whole, microalgal cell numbers were significantly higher in surface water samples than in microplankton samples (p < 0.01) (Figure 2C), with averages of 2.59 × 104 and 1.43 × 104 cells/L in surface water and micro-plankton samples, respectively (Figure 2D). Diatoms dominated in microalgal community with the relative abundances of 85.37%–100% and an average of 96.65% (Figure 2E).
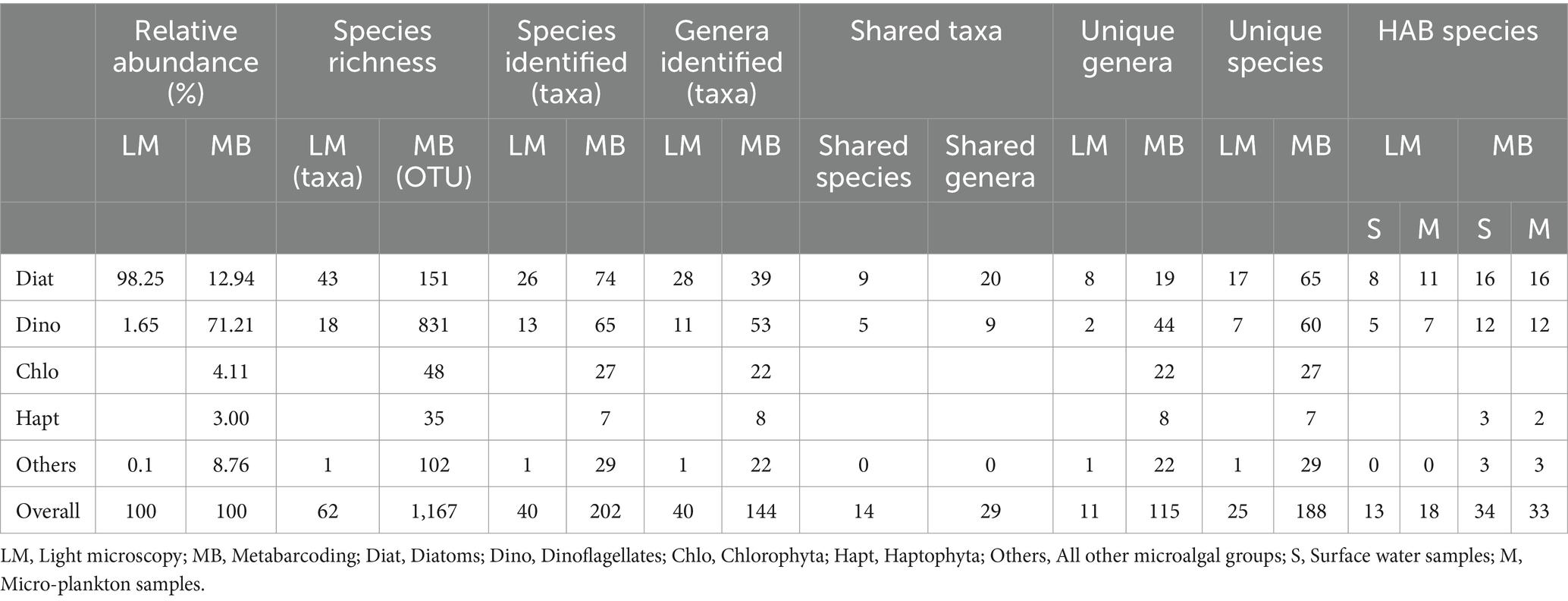
Table 1. Comparison of microalgal community structure between light microscopy (LM) and metabarcoding (MB) analyses.
The Venn diagram highlighted the differences of algal communities among the 20 samples (Figures 3A,B). There were only 8 OTUs and 3 taxa shared among all samples by metabarcoding and morphological observation, respectively. The results suggested different microalgal species composition among samples.
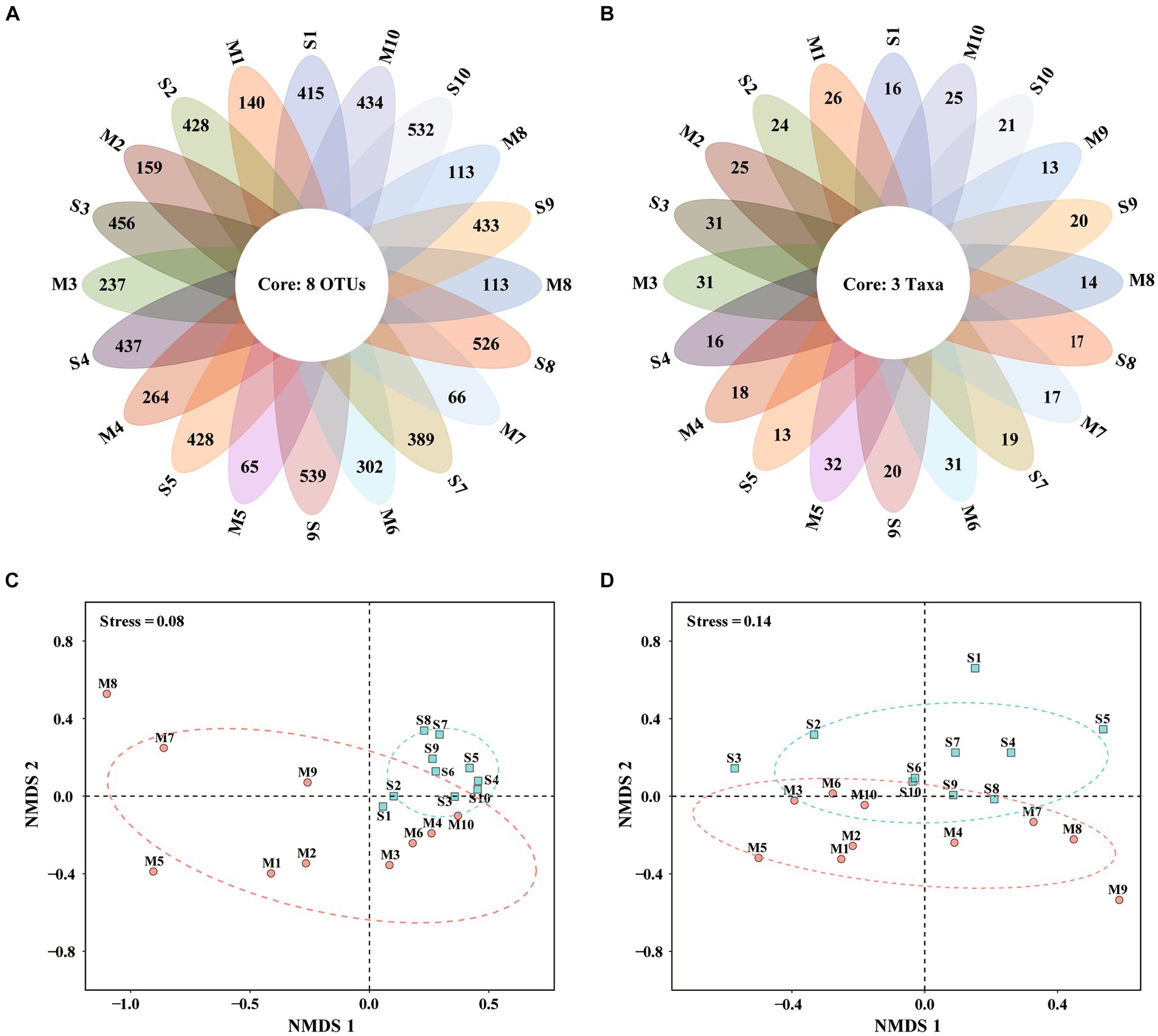
Figure 3. Venn diagrams and non-metric multidimensional scaling (NMDS) based on eukaryotic microalgal community revealed by metabarcoding (A,C) and microscopy (B,D). S1–S10, Surface water samples; M1–M10, Micro-plankton samples.
Non-metric multidimensional scaling (NMDS) well differentiated surface water and micro-plankton samples analyzed by metabarcoding (Figure 3C) and microscopic observation (Figure 3D). Based on metabarcoding data (Figure 3C), 10 surface water samples clustered together in the positive axis of NMDS1, indicating similar community structure among them. While micro-plankton samples scatteredly distributed at the second, third and fourth quadrants (Figure 3C), suggesting more diverse communities. Based on microscopic data (Figure 3D), surface water samples were mostly located in the first and second quadrants, while micro-plankton samples distributed in the third and fourth quadrants. There were significant differences between surface water samples and micro-plankton samples based on data revealed by both analyses (R2 = 0.321, p < 0.001 for metabarcoding, R2 = 0.123, p < 0.05 for microscopy, PERMANOVA; Table 2). The results suggested that the differences of microalgal community structure between different cell size fractions (all sizes vs. 20–200 μm) were much greater than those among different stations.

Table 2. Permutational multivariate analysis of variance (PERMANOVA) to evaluate the distances between surface water samples (S) and micro-plankton samples (M) in the Non-metric multidimensional scaling (NMDS).
Figure 4 illustrates the distribution of the top 30 dominant genera based on metabarcoding (Figure 4A) and microscopic analyses (Figure 4B). Pyrophacus ranked the first abundant genus in metabarcoding dataset, and occurred abundantly in the micro-plankton samples (Figure 4A). The parasitic dinoflagellate Amoebophrya occurred abundantly in all samples, and ranked the top 4th genus. Gyrodinium, Neoceratium, Prorocentrum, Gymnodinium, Karlodinium, Heterocapsa were the common dominant dinoflagellate genera, and they were present in all samples. Other dominant dinoflagellate genera, such as Gonyaulax, Alexandrium, Protoperidinium, and Dinophysis were found in most samples. Chaetoceros, Rhizosolenia, and Thalassiosira were the most dominant diatoms, which ranked the top 2nd, 3rd, and 7th abundant genera, respectively. Most of the dominant taxa occurred more widely in surface water samples, however some micro-sized dinoflagellates and colony diatoms were more abundant in the micro-plankton samples, for example the dinoflagellates Pyrophacus, Gyrodinium, and Neoceratium, and the diatoms Chaetoceros, and Rhizosolenia. On the other hand, the nano-sized microalgae such as the haptophytes Chrysochromulina, Scyphosphaera, and Phaeocystis, the chlorophytes Ostreococcus, Micromonas, and Nephroselmis, the cryptophyceans Teleaulax and Geminigera, occurred abundantly in surface water samples (Figure 4A).
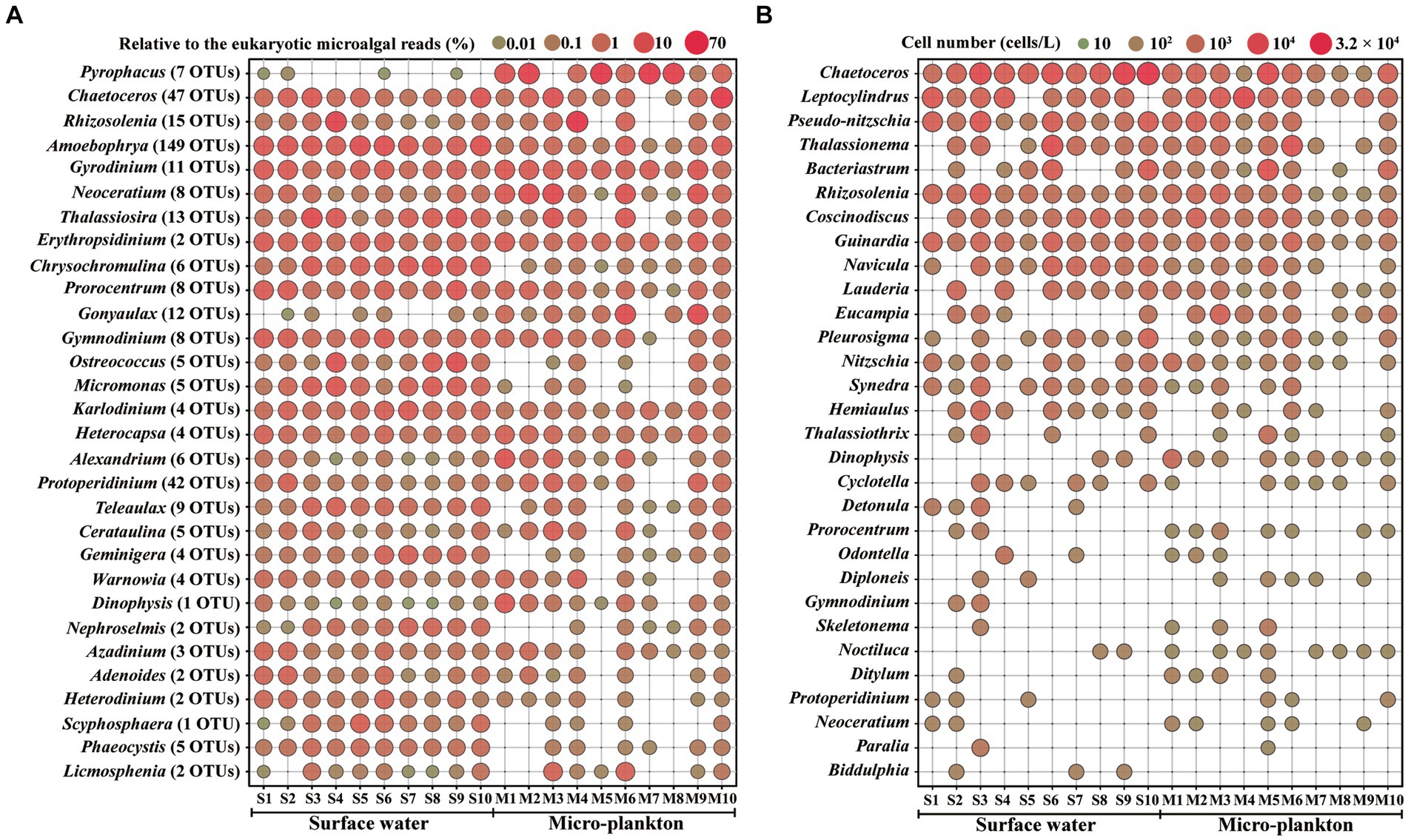
Figure 4. Distribution of the top 30 abundant genera by metabarcoding (A) and microscopy (B). S1–S10, Surface water samples; M1–M10, Micro-plankton samples.
More than one OTUs were assigned to each species, for example 9 OTUs in Rhizosolenia setigera, 7 OTUs in Pyrophacus steinii, 6 OTUs in Teleaulax amphioxeia, 5 OTUs in Gonyaulax spinifera, and 5 OTUs in Protoperidinium conicum (Supplementary Table S3). Among the dominant genera, 149 OTUs were attributed to Amoebophrya, 47 OTUs to Chaetoceros, and 42 OTUs to Protoperidinium (Figure 4A).
The top 30 dominant genera based on microscopic analysis included 25 genera of diatoms and 5 genera of dinoflagellates (Figure 4B). Diatoms occupied all of the top 16 dominant genera. Most of these diatom taxa either belong to the micro-sized diatoms with cell size > 20 μm or to the colony diatoms, which are easily to be observed by microscopy. Only three genera, Chaetoceros, Rhizosolenia, and Guinardia, were present in all samples. Dinoflagellate genera such as Dinophysis, Prorocentrum, Noctiluca, and Neoceratium, occurred more widely and abundantly in the micro-plankton samples. The dominant genera identified by both microscopy and metabarcoding were Chaetoceros, Rhizosolenia, Dinophysis, Prorocentrum, Gymnodinium, Protoperidinium, and Neoceratium.
Metabarcoding revealed significantly higher microalgal OTU/species richness and Shannon index (p < 0.05), and richness and Shannon index were significantly higher in surface water samples than the micro-plankton samples (p < 0.05; Figure 5). Simpson index was comparable among samples and between the two analysis methods, however Pielou index was significantly higher revealed by microscopic observation than by metabarcoding.
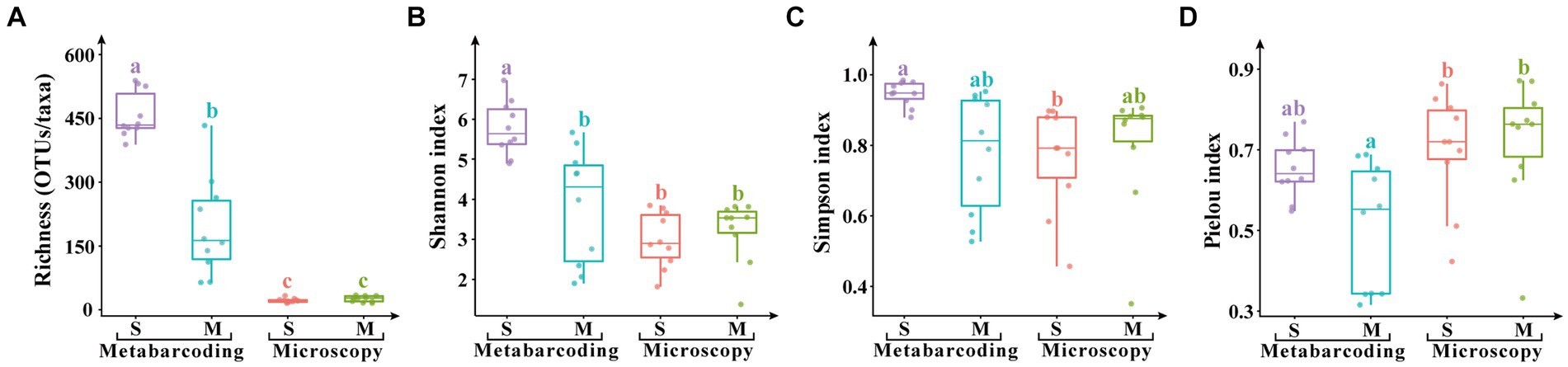
Figure 5. Alpha diversity indices of eukaryotic microalgae in the surface water samples (S) and the micro-plankton samples (M). (A) Species/ASV richness, (B) Shannon index, (C) Simpson index, (D) Pielou index. The significant difference of alpha indices among samples and analytical methods was performed by TukeyHSD test in R4.0.1. Data bars carrying different letter designations (A–C) indicated significant difference (p < 0.05), and the same and similar letter designations (e.g., a and ab, b and ab) denoted no significant difference (p > 0.05).
The microalgal community showed significant differences between the results from microscopic observation and metabarcoding analysis. Diatoms predominated in the microalgal community based on microscopic observation, and dinoflagellates dominated based on metabarcoding analysis (Table 1). Metabarcoding annotated more species (202 species) than morphospecies (40 species) observed by microscopy. On the other hand, Chlorophyta, Haptophyta, Chrysophyceae, Cryptophyceae, Chlorarachniophyceae, Prasinodermatophyceae, and Rhodophyceae were detected in metabarcoding but overlooked in microscopy (Supplementary Table S2) due to their cryptic characteristics, such as small size and fragile cell walls. There were 14 species and 29 genera shared between the two methods (Table 1). Twenty-five and 188 unique species, and 11 and 115 unique genera were recorded by microscopy and metabarcoding, respectively (Table 1).
Altogether 48 algal bloom and/or HAB species were detected in this study revealed by both methods, including 24 diatoms, 18 dinoflagellates, and each of three species in Haptophyta and Raphidophyceae (Table 3). Metabarcoding detected much more bloom and/or HAB species (34 species) than those by microscopy (19 species). Actually, some HAB taxa were only detected to the genus level rather than the species level by microscopy, which cannot be sure whether to be HAB species. More HAB species were detected in the micro-plankton samples by microscopy, however they were comparable in the surface water and micro-plankton samples detected by metabarcoding (Table 1).
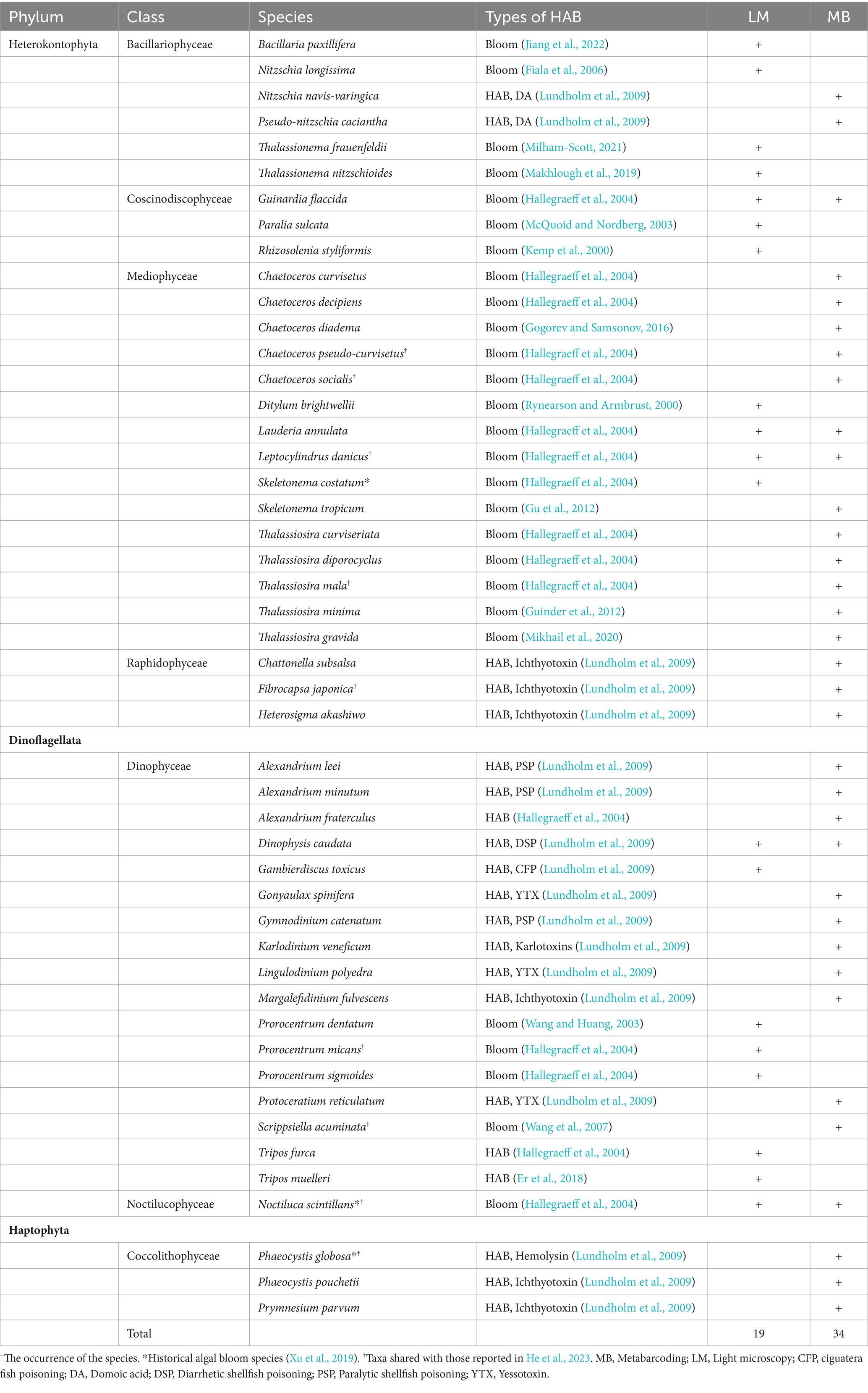
Table 3. List of algal bloom and/or harmful algal bloom (HAB) species detected in this study by light microscopy (LM) and metabarcoding (MB).
Microalgal communities in the Beibu Gulf have been studied based on microscopic observation (Xu et al., 2014; Wang et al., 2015; Qin et al., 2023). However, the nano-sized microalgae are often overlooked, and similar and cryptic taxa cannot be accurately distinguished under light microscopic observations (Esenkulova et al., 2020). To our knowledge, this is the first attempt to study the microalgal community and HAB species in the Beibu Gulf combining microscopic observation and metabarcoding data. Microscopic observations showed that diatoms dominated the microalgal community, which agreed with those in previous studies based on microscopic observations (Sun et al., 2011; Xu et al., 2014; Wang et al., 2015). The low degree of overlap of microalgal taxa in Venn diagram (Figures 3A,B) suggested differences in microalgal community structure between samples, even if they were collected from close stations due to fluctuant environmental parameters among stations (Figure 6).
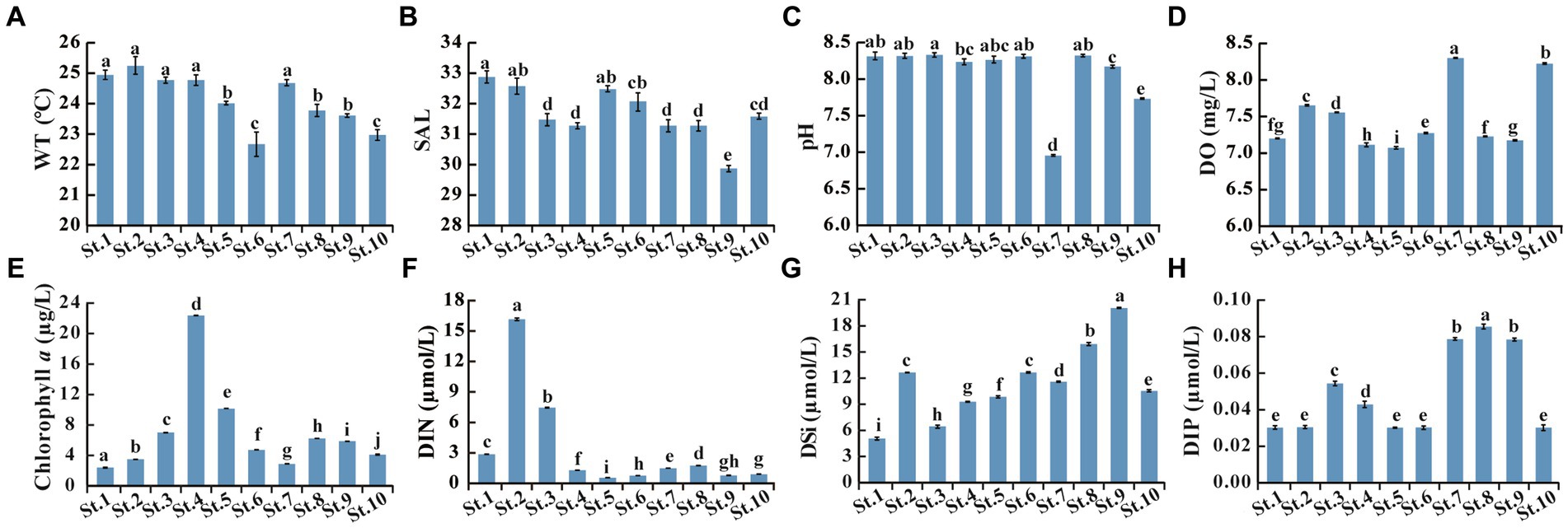
Figure 6. Environmental parameters measured in the 10 stations. (A) WT, Water temperature; (B) SAL, Salinity; (C) pH; (D) DO, Dissolved oxygen; (E) Chlorophyll a; (F) DIN, Dissolved inorganic nitrogen as the sum of NO2−-N, NO3−-N, and NH4+-N; (G) DSi, Dissolved inorganic silicate (SiO32−-Si); (H) DIP, Dissolved inorganic phosphorus (PO43−-P). Data bars carrying different letter designations (a–h) indicated significant difference (p < 0.05), and the same and similar letter designations (e.g., a and ab, b and ab) denoted no significant difference (p > 0.05).
Metabarcoding revealed an increased abundance of dinoflagellates and a reduction in diatoms. The different results between metabarcoding and microscopy may be primarily due to substantial variation in 18S rRNA gene copy number among microalgal species (Zhu et al., 2005). In general, dinoflagellates have a higher 18S rRNA gene copy number than other microalgae (Gong and Marchetti, 2019), and a high logarithmic correlation between DNA copy number and cell size was observed (Zhu et al., 2005). Since nano-sized diatoms (<20 μm), such as Chaetoceros spp., Leptocylindrus danicus, and Pseudo-nitzschia spp., dominated in this study (Figure 4B), their gene copies are lower than those of dinoflagellate cells. High variation in the copy number of the 18S rRNA gene may lead to a rapid decline in consistency between the DNA reads and the actual organism abundances (Gong and Marchetti, 2019). The predominance and bias of dinoflagellates in microalgal metabarcoding have been widely reported (Xu et al., 2017; Song et al., 2019; Liu et al., 2020; Cui et al., 2021; Wang Z. H. et al., 2022).
Microalgal community structure differed significantly between the surface water and micro-plankton samples (Figures 3C,D), reflected by higher abundance (cell number and proportion within the eukaryotic community), richness (taxa and OTUs), and alpha diversity in the surface water samples (Figures 2, 5). We collected micro-plankton samples in order to better understand distribution of the micro-sized dinoflagellates especially those of HAB taxa. Dinoflagellates occurred more abundantly and widely in micro-plankton samples based on microscopic observations (Figures 2D,E, 4B). Meanwhile, more HAB taxa were recorded in micro-plankton samples (18 taxa) than those in the surface water samples (13 taxa) based on microscopic analysis (Table 1). However, HAB taxa detected by metabarcoding were comparable in the surface water and micro-plankton samples. Furthermore, a large number of nano- and pico-sized microalgae, such as those in chlorophytes, cryptophytes, and haptophytes, are lost in the micro-plankton samples even by metabarcoding. Our results suggest that HAB species may be more abundant in the micro-plankton samples based on microscopic observations, whereas it is not necessary to collect concentrated micro-plankton samples when studying HAB species by metabarcoding.
Twenty-three classes in eight microalgal phyla were detected by metabarcoding, however only six classes in two phyla were identified by microscopy (Supplementary Table S2). Numerous taxa (species and genera) in Chlorophyta, Haptophyta, Rhodophyta, Chlorarachniophyceae, Chrysophyceae, Cryptophyceae, and Prasinodermatophyceae detected in metabarcoding were not identified by microscopy (Supplementary Table S2). The results suggest metabarcoding revealing much higher bio-diversity than that by microscopy. Metabarcoding has a strong ability to detect the tiny, morphologically similar, fragile, and rare species (Salonen et al., 2019). Plenty of nano-sized chlorophytes, chlorarachniophyceans, chrysophyceans, haptophytes, and prasinodermatophyceans (<10 μm) were detected in this study (Supplementary Table S2). Lin et al. (2021) found abundant chlorophytes (mostly Micromonas and Ostreococcus) and chrysophyceans (Paraphysomonas) in 0.45–5-μm plankton community of the Beibu Gulf based on metabarcoding. However, majority of these tiny-sized species are missed in previous microscopic studies (Xu et al., 2014; Wang et al., 2015; Qin et al., 2023). In addition, metabarcoding distinguished morphologically similar or fragile genera to many species that cannot be identified to the species level under light microscopy, such as species in diatoms Chaetoceros, Leptocylindrus, and Thalassiosira, and dinoflagellates Alexandrium, Azadinium, and taxa in naked dinoflagellate order Gymnodiniales such as Gymnodinium, Gyrodinium, and Karlodinium etc. (Supplementary Table S2).
However, only 14 species and 29 genera were shared in both methods (Table 1), and even among the top 30 dominant genera, only 7 genera were detected in both microscopic and metabarcoding datasets (Figure 4). The results suggested a huge gap between the two methods. The reason for this phenomenon is firstly due to the low coverage at the species level for many taxa in the reference databases, which may hinder correct interpretation of metabarcoding data (Forster et al., 2016). Only 308 OTUs (26.39%) were identified at the species level, and 632 OTUs (54.16%) at the genus level after annotated against both the SILVA and NCBI databases in our study. Anyway, as new sequencing data are continuously contributing to the existing databases, the situation is likely to improve in the future. In addition, species identification based on traditional microscopic methods is restricted to those organisms with well-documented morphological characteristics and easy to be identified (Eland et al., 2012). Diatoms identified by microscopy in this study were mostly those with cell size > 20 μm or the colony taxa (Figure 4B; Supplementary Table S2). While morphological observation provides cell number of each taxon, and better reflects the real situation in the water column. The two databases can complement each other, and the combination of the two methods better reflect planktonic microalgal community in the water column.
A total of 48 algal bloom and/or HAB taxa were identified in this study, however, only three species of them (Skeletonema costatum, Noctiluca scintillans, and Phaeocystis globosa) formed blooms previously in the Beibu Gulf (Xu et al., 2019). He et al. (2023) detected 37 HAB species by metabarcoding in one station during the eight surveys between 2016 and 2017, but only nine HABs were the same as in our study (Table 3). These results suggest rich and diverse HAB species in the Beibu Gulf, which may fluctuate greatly with sampling time.
Diatoms are the foundation of the global marine ecosystem, accounting for about 40% of the ocean’s total primary production and 20% of the Earth’s total primary production (Tréguer et al., 2018; Karlusich et al., 2020). Most diatoms do not produce toxins harmful to marine organisms. Sometimes they form algal blooms that result in basically harmless water discolorations; however, under special conditions, dense blooms may cause indiscriminate kills of fish and invertebrates through oxygen depletion (Hallegraeff et al., 2004). Two potential domoic acid (DA) producers, Nitzschia navis-varingica and Pseudo-nitzschia caciantha, were detected in our study (Table 3). Pseudonitzschia spp. are the major DA producers, with 73 species recorded so far (Guiry and Guiry, 2024), of which 33 species/variants produce DA (Lundholm et al., 2009). DA has been detected in shellfish samples collected from the Beibu Gulf, with the maximum level of 401 μg/kg and the detection rate of 17.7% (Ji et al., 2022). Pseudo-nitzschia was the third top genus observed by microscopy (Figure 4B) with the average cell numbers of 1.7 × 103 cells/L in our study. Based on metabarcoding, its relative DNA reads was outside the top 30 genera. Both abundances of Pseudo-nitzschia detected in this study and DA concentrations and detection rate reported by Ji et al. (2022) in the Beibu Gulf were much lower than those reported in the other sea areas of the southern Chinese coast (Wang Z. H. et al., 2022), suggesting relatively a low risk of DA poisoning events in this region.
Dinoflagellates are particularly important for HABs, because about 75–80% of known toxic microalgae belong to dinoflagellates (Cembella, 2003). A total of 18 harmful dinoflagellate taxa were identified in this study, and some were only found in microscopic analysis, such as Gambierdiscus toxicus, Tripos furca, and T. muelleri (Table 3). The results suggest that there are certain biases in the metabarcoding process and/or gaps in the sequence database for above genera. However, more diverse species in Alexandrium were found in metabarcoding, and more potentially harmful dinoflagellate species were detected (Gonyaulax spinifera, Gymnodinium catenatum, Karlodinium veneficum, Lingulodinium polyedra, Margalefidinium fulvescens, and Protoceratium reticulatum). Some of these HAB taxa have the potential to produce toxins that can cause toxic effects on human health through the food chain, for example, the paralytic shellfish poisoning (PSP) producers Alexandrium spp., diarrhetic shellfish poisoning (DSP) producer Dinophysis caudata, ciguatera fish poisoning (CFP) producer Gambierdiscus toxicus, yessotoxin (YTX) producers Gonyaulax spinifera, Lingulodinium polyedra, and Protoceratium reticulatum, and karlotoxins producers Karlodinium veneficum (Table 3). In addition, a large number of HAB species, which are potentially toxic to fish and shellfish and cause mass mortalities of aquaculture animals, were detected in the Beibu Gulf in our study, such as the dinoflagellate Margalefidinium fulvescens, the haptophytes Phaeocystis globosa, Phaeocystis pouchetii, and Prymnesium parvum, and the raphidophyceans Fibrocapsa japonica, Heterosigma akashiwo, and Chattonella subsalsa (Table 3). Our result suggested a high potential risk of HABs in the Beibu Gulf.
Phaeocystis globosa and Noctiluca scintillans are the main bloom-causative species in the Beibu Gulf, and their blooms have occurred frequently in the recent decades (Xu et al., 2019). A massive bloom caused by Noctiluca scintillans with a total coverage area of >20,000 km2 occurred in Qinzhou Bay of the Beibu Gulf in February 2021 (Xie et al., 2023). However, abundances of these two species were low revealed by both microscopic and metabarcoding analyses in this study. Phaeocystis globosa is a nano-sized haptophyte, which can form giant colonies during the bloom, and exists as solitary cells in the sea water during the rest of the time (Liang et al., 2023). The tiny solitary cells (3.5–4.5 μm) are easily overlooked under routine light microscopic observation, which makes them hard to be detected except during its blooms. Noctiluca scintillans is a heterotrophic dinoflagellate, which needs to feed on other small microalgae, and its outbreak usually follows the diatom blooms (Kopuz et al., 2014; Kitatsuji et al., 2019). Therefore, the low abundances of these two bloom species in our study were possibly because our sampling missed their growth seasons.
In conclusion, metabarcoding revealed much higher microalgal diversity and species richness than those by morphological observation. Microalgal community in surface water samples demonstrated significantly higher OTU/species richness, alpha diversity, and abundance than those in micro-plankton samples. Metabarcoding and microscopy demonstrated different microalgal community, which was dominated by dinoflagellates and diatoms, respectively. Metabarcoding has a strong ability to detect the small-sized, morphologically similar, fragile and rare species. While morphological observation provides cell number of each taxon, and better reflects the real situation in the water column. The combination of morphological and metabarcoding approaches comprehensively reveals the microalgal community structure and diversity of HAB species.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
CW: Formal analysis, Writing – original draft. JG: Data curation, Methodology, Writing – review & editing. WL: Data curation, Investigation, Writing – original draft. JW: Methodology, Writing – review & editing. ZW: Conceptualization, Funding acquisition, Writing – original draft. QL: Conceptualization, Data curation, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (no. 2022YFE0122100), the Science and Technology Basic Resources Investigation Program of China (no. 2018FY100200), and the Chinese National Natural Foundation of Science (no. 42076141).
Samples were collected onboard during the autumn survey supported by Guangxi Zhuang Autonomous Region Marine Environment Monitoring Center in November 2020. We are greatly appreciated to all the crew on board for their hard work and assistance in collecting samples for this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1403964/full#supplementary-material
Anderson, P., and Throndsen, J. (2004). “Estimating cell numbers” in Manual on harmful marine Miceroalgae. eds. G. M. Hallegraeff, D. M. Anderson, and A. D. Cembella (Paris: UNECSO), 99–129.
Bråte, J., Logares, R., Berney, C., Ree, D. K., Klaveness, D., Jakobsen, K. S., et al. (2010). Freshwater Perkinsea and marine-freshwater colonizations revealed by pyrosequencing and phylogeny of environmental rDNA. ISME J. 4, 1144–1153. doi: 10.1038/ismej.2010.39
Cavalier-Smith, T., Lewis, R., Chao, E. E., Oates, B., and Bass, D. (2009). Helkesimastix marina n. sp. (Cercozoa: Sainouroidea superfam. N.) a gliding zooflagellate of novel ultrastructure and unusual ciliary behaviour. Protist 160, 452–479. doi: 10.1016/j.protis.2009.03.003
Cembella, A. D. (2003). Chemical ecology of eukaryotic microalgae in marine ecosystems. Phycologia 42, 420–447. doi: 10.2216/i0031-8884-42-4-420.1
Chen, F. J., Lin, J., Qian, B. H., Wu, Z., Huang, P., Chen, K., et al. (2018). Geochemical assessment and spatial analysis of heavy metals in the surface sediments in the eastern Beibu gulf: a reflection on the industrial development of the South China coast. Int. J. Environ. Res. Public Health 15:496. doi: 10.3390/ijerph15030496
Chen, Z. Z., Qiu, Y. S., and Xu, S. N. (2011). Changes in trophic flows and ecosystem properties of the Beibu gulf ecosystem before and after the collapse of fish stocks. Ocean Coast. Manag. 54, 601–611. doi: 10.1016/j.ocecoaman.2011.06.003
Cui, Z. M., Xu, Q., Gibson, K., Liu, S. Y., and Chen, N. S. (2021). Metabarcoding analysis of harmful algal bloom species in the Changjiang estuary, China. Sci. Total Environ. 782:146823. doi: 10.1016/j.scitotenv.2021.146823
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C. (2016). UCHIME2: improved chimera prediction for amplicon sequencing. BioRxiv [Preprint], BioRxiv: 074252.
Eland, L. E., Davenport, R., and Mota, C. R. (2012). Evaluation of DNA extraction methods for freshwater eukaryotic microalgae. Water Res. 46, 5355–5364. doi: 10.1016/j.watres.2012.07.023
Er, H. H., Lee, L. K., Lim, Z. F., Teng, S. T., Leaw, C. P., and Lim, P. T. (2018). Responses of phytoplankton community to eutrophication in Semerak lagoon (Malaysia). Environ. Sci. Pollut. Res. 25, 22944–22962. doi: 10.1007/s11356-018-2389-0
Esenkulova, S., Sutherland, B. G., Tabata, A., Haigh, N., Pearce, C. M., and Miller, K. M. (2020). Comparing metabarcoding and morphological approaches to identify phytoplankton taxa associated with harmful algal blooms. FACETS 5, 784–811. doi: 10.1139/facets-2020-0025
Fiala, M., Kuosa, H., Kopczyńska, E. E., Oriol, L., and Delille, D. (2006). Spatial and seasonal heterogeneity of sea ice microbial communities in the first-year ice of Terre Adélie area (Antarctica). Aquat. Microb. Ecol. 43, 95–106. doi: 10.3354/ame043095
Forster, D., Dunthorn, M., Mahe, F., Dolan, J. R., Audic, S., Bass, D., et al. (2016). Benthic protists: the under-charted majority. FEMS Microbiol. Ecol. 92:fiw120. doi: 10.1093/femsec/fiw120
Gan, H. Y., Lin, J. Q., Liang, K., and Xia, Z. (2013). Selected trace metals (as, cd and hg) distribution and contamination in the coastal wetland sediment of the northern Beibu gulf, South China Sea. Mar. Pollut. Bull. 66, 252–258. doi: 10.1016/j.marpolbul.2012.09.020
Gibson, K., Song, H. Y., and Chen, N. S. (2022). Metabarcoding analysis of microbiome dynamics during a Phaeocystis globosa bloom in the Beibu gulf, China. Harmful Algae 114:102217. doi: 10.1016/j.hal.2022.102217
Gilbert, P. M. (2017). Eutrophication, harmful algae and biodiversity—challenging paradigms in a world of complex nutrient changes. Mar. Pollut. Bull. 124, 591–606. doi: 10.1016/j.marpolbul.2017.04.027
Gogorev, R. M., and Samsonov, N. I. (2016). The genus Chaetoceros (Bacillariophyta) in Arctic and Antarctic. Novosti Sist. Nizsh. Rast. 50, 56–111. doi: 10.31111/nsnr/2016.50.56
Gong, W., and Marchetti, A. (2019). Estimation of 18S gene copy number in marine eukaryotic plankton using a next-generation sequencing approach. Front. Mar. Sci. 6:219. doi: 10.3389/fmars.2019.00219
Gu, H. F., Zhang, X. D., Sun, J., and Luo, Z. H. (2012). Diversity and seasonal occurrence of Skeletonema (Bacillariophyta) species in Xiamen harbour and surrounding seas, China. Cryptogam. Algol. 33, 245–263. doi: 10.7872/crya.v33.iss3.2012.245
Guinder, V. A., Molinero, J. C., Popovich, C. A., Marcovecchio, J. E., and Sommer, U. (2012). Dominance of the planktonic diatom Thalassiosira minima in recent summers in the Bahía Blanca estuary, Argentina. J. Plankton Res. 34, 995–1000. doi: 10.1093/plankt/fbs060
Guiry, M. D. (2024). How many species of algae are there? A reprise. Four kingdoms, 14 phyla, 63 classes and still growing. J. Phycol. 60:13431. doi: 10.1111/jpy.13431
Guiry, M. D., and Guiry, G. M. (2024). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. Available at: https://www.algaebase.org (Accessed 15 May 2024).
Hallegraeff, G. M., Anderson, D. M., and Cembella, A. D. (2004). Manual on Harmful Marine Microalgae. Paris: UNESCO Publishing.
He, L. Y., Yu, Z. M., Xu, X., Zhu, J. N., Yuan, Y. Q., Cao, X. H., et al. (2023). Metabarcoding analysis identifies high diversity of harmful algal bloom species in the coastal waters of the Beibu gulf. Ecol. Evol. 13:e10127. doi: 10.1002/ece3.10127
Ji, Y., Yan, G. W., Wang, G. X., Liu, J. W., Tang, Z. X., Yan, Y. J., et al. (2022). Prevalence and distribution of domoic acid and cyclic imines in bivalve mollusks from Beibu gulf, China. J. Hazard. Mater. 423:127078. doi: 10.1016/j.jhazmat.2021.127138
Jiang, T., Wu, G. N., Niu, P. L., Cui, Z. G., Bian, X. D., Xie, Y. X., et al. (2022). Short-term changes in algal blooms and phytoplankton community after the passage of super typhoon Lekima in a temperate and inner sea (Bohai Sea) in China. Ecotox. Environ. Safe. 232:113223. doi: 10.1016/j.ecoenv.2022.113223
Kaiser, D., Unger, D., Qiu, G. L., Zhou, H. L., and Gan, H. Y. (2013). Natural and human influences on nutrient transport through a small subtropical Chinese estuary. Sci. Total Environ. 450, 92–107. doi: 10.1016/j.scitotenv.2013.01.096
Karlusich, J. J. P., Ibarbalz, F. M., and Bowler, C. (2020). Phytoplankton in the Tara Ocean. Ann. Rev. Mar. Sci. 12, 233–265. doi: 10.1146/annurev-marine-010419-010706
Kemp, A. E., Pike, J., Pearce, R. B., and Lange, C. B. (2000). The “fall dump”—a new perspective on the role of a “shade flora” in the annual cycle of diatom production and export flux. Deep Sea Res. Part II-Top. Stud. Oceanogr. 47, 2129–2154. doi: 10.1016/S0967-0645(00)00019-9
Kitatsuji, S., Yamaguchi, H., Asahi, T., Ichimi, K., Onitsuka, G., and Tada, K. (2019). Does Noctiluca scintillans end the diatom bloom in coastal water? J. Exp. Mar. Biol. Ecol. 510, 10–14. doi: 10.1016/j.jembe.2018.09.006
Kopuz, U., Feyzioglu, A. M., and Valente, A. (2014). An unusual red-tide event of Noctiluca scintillans (Macartney) in the southeastern Black Sea. Turk. J. Fish. Quat. Sci. 14, 261–268. doi: 10.4194/1303-2712-v14_1_28
Lai, J. X., Jiang, F. J., Ke, K., Xu, M. B., Lei, F., and Chen, B. (2014). Nutrients distribution and trophic status assessment in the northern Beibu gulf, China. Chinese J. Oceanol. Limnol. 32, 1128–1144. doi: 10.1007/s00343-014-3199-y
Lao, Q. B., Liu, G. Q., Shen, Y. L., Su, Q. Z., and Lei, X. T. (2021). Biogeochemical processes and eutrophication status of nutrients in the northern Beibu gulf, South China. J. Earth Syst. Sci. 130:199. doi: 10.1007/s12040-021-01706-y
Lao, Q. B., Su, Q. Z., Liu, G. Q., Shen, Y. L., Chen, F. J., Lei, X. T., et al. (2019). Spatial distribution of and historical changes in heavy metals in the surface seawater and sediments of the Beibu gulf, China. Mar. Pollut. Bull. 146, 427–434. doi: 10.1016/j.marpolbul.2019.06.080
Lassus, P., Chomérat, N., Hess, P., and Nézan, E. (2016). Toxic and harmful Micro-algae of the World Ocean, IOC manuals Guides: Intergovernmental oceanographic commission of UNESCO, 68, Paris.
Li, S. S., Xu, S. Y., Liang, M. Z., and He, G. Y. (2014). Research on marine environmental protection and the management of the Beibu gulf in Guangxi (in Chinese, with English abstract). J. Qinzhou Univ. 29, 1–5.
Li, X. Y., Yu, R. C., Geng, H. X., and Li, Y. F. (2021). Increasing dominance of dinoflagellate red tides in the coastal waters of Yellow Sea, China. Mar. Pollut. Bull. 168:112439. doi: 10.1016/j.marpolbul.2021.112439
Liang, D. Y., Wang, X. D., Huo, Y. P., Xiang, H., Li, S. S., and Wang, Y. (2023). Morphological, molecular, and life cycle characteristics of Phaeocystis globosa Scherffel (Prymnesiophyceae) in the Southeast China Sea. Harmful Algae 127:102477. doi: 10.1016/j.hal.2023.102477
Lin, L. Z., Gu, H. F., Luo, Z. H., and Wang, N. (2021). Size-dependent spatio-temporal dynamics of eukaryotic plankton community near nuclear power plant in Beibu gulf, China. J. Oceanol. Limnol. 39, 1910–1925. doi: 10.1007/s00343-020-0248-6
Liu, S. Y., Gibson, K., Cui, Z. M., Chen, Y., Sun, X. X., and Chen, N. S. (2020). Metabarcoding analysis of harmful algal species in Jiaozhou Bay. Harmful Algae 92, 1–12. doi: 10.1016/j.hal.2020.101772
Lundholm, N., Churro, C., Fraga, S., Hoppenrath, M., Iwataki, M., Larsen, J., et al. (2009). IOC-UNESCO Taxonomic Reference List of Harmful Micro Algae. Available at: https://www.marinespecies.org/hab (Accessed 7 May 2022).
Makhlough, A., Saravi, H. N., Eslami, F., and Keyhansani, A. R. (2019). Scale characteristics of the bloom event: a case study in the Iranian coastal waters of the southern Caspian Sea. Iran. J. Fish. Sci. 18, 124–139. doi: 10.22092/ijfs.2018.117600
McQuoid, M. R., and Nordberg, K. (2003). The diatom Paralia sulcata as an environmental indicator species in coastal sediments. Estuar. Coast. Shelf Sci. 56, 339–354. doi: 10.1016/S0272-7714(02)00187-7
Mikhail, S., Khalil, N., and El-Hadary, M. (2020). A new recorded of red tide forming species; Heterocapsa triquetra, Gymnodinium impudicum, Heterosigma akashiwo and Thalassiosira rotula in Alexandria waters, Egypt. Egypt. J. Aquat. Biol. Fish. 24, 207–223. doi: 10.21608/ejabf.2020.117258
Milham-Scott, D. P. (2021). Phytoplankton response to changes in water quality and a flood event in Hervey Bay, Queensland. UNET J. Sci. Soc. 2, 35–54. doi: 10.52042/UNETJOSS010201
Niu, Z., Guan, W. B., Wang, J. X., Yuan, Y. Q., Kong, F. Z., Liu, C., et al. (2022). Dynamics of Phaeocystis globosa bloom and implications for its seed sources in the Beibu gulf, China. J. Oceanol. Limnol. 40, 2385–2400. doi: 10.1007/s00343-022-1447-0
Omura, T., Iwataki, M., Borja, V. M., Takayama, H., and Fukuyo, Y. (2012). Marine phytoplankton of the Western Pacific. Tokyo: Kouseisha Kouseikaku Co., Ltd.
Pan, H. L., Liu, D. S., Shi, D. L., Yang, S. Y., and Pan, W. (2021). Analysis of the spatial and temporal distributions of ecological variables and the nutrient budget in the Beibu gulf. Acta Oceanol. Sin. 40, 14–31. doi: 10.1007/s13131-021-1794-2
Pan, H. Z., Xu, M. B., Lan, C. B., Ma, J. X., Li, J., Lu, J. C., et al. (2023). Determination of phytoplankton community structure and biomass with HPLC-CHEMTAX and microscopic methods during winter and summer in the Qinzhou Bay of the Beibu gulf. Front. Mar. Sci. 10:1186106. doi: 10.3389/fmars.2023.1186106
Qin, X. L., Chen, X. Y., Chen, C. P., Sun, L., Su, X. Y., Liang, J. R., et al. (2023). Succession and controlling factors of phytoplankton assemblages during a period with recurrent outbreaks of Phaeocystis globosa blooms in Qinzhou Bay, China. Front. Mar. Sci. 10:1120161. doi: 10.3389/fmars.2023.1120161
Rynearson, T. A., and Armbrust, E. V. (2000). DNA fingerprinting reveals extensive genetic diversity in a field population of the centric diatom Ditylum brightwellii. Limnol. Oceanogr. 45, 1329–1340. doi: 10.4319/lo.2000.45.6.1329
Sakamoto, S., Lim, W. A., Lu, D. D., Dai, X. F., Orlova, T., and Iwataki, M. (2021). Harmful algal blooms and associated fisheries damage in East Asia: current status and trends in China, Japan, Korea and Russia. Harmful Algae 102:101787. doi: 10.1016/j.hal.2020.101787
Salonen, I. S., Chronopoulou, P. M., Leskinen, E., and Koho, K. A. (2019). Metabarcoding successfully tracks temporal changes in eukaryotic communities in coastal sediments. FEMS Microbiol. Ecol. 95:fiy226. doi: 10.1093/femsec/fiy226
Song, L., Wu, J., Du, J., Li, N., Song, G. J., Wang, K., et al. (2019). The characteristics and distribution of eukaryotic phytoplankton Community in Liaodong bay, China. Ocean Sci. J. 54, 183–203. doi: 10.1007/s12601-019-0007-9
Stoeck, T., Bass, D., Nebel, M., Christen, R., Jones, M., Breiner, H.-W., et al. (2010). Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 19, 21–31. doi: 10.1111/j.1365-294X.2009.04480.x
Strickland, J. D., and Parsons, T. R. (1972). “Internationale Revue Der Gesamten Hydrobiologie Und Hydrographie” in A practical handbook of seawater analysis, vol. 167 eds. J. C. Stevenson, J. Watson, and J. M. Reinhart (Ottawa: Fisheries Research Board of Canada, Bulletin), 293.
Sun, J., Li, X., and Tian, W. (2011). Summer phytoplankton community in the Beibu Gulf. Trans. Oceanol. Limnol. 3, 1–7. doi: 10.13984/j.cnki.cn37-1141.2011.03.020
Tian, Y., Hu, S. M., Lin, X. Z., Huang, H., Song, X. Y., Yan, Y., et al. (2021). Mechanisms of high-frequency dinoflagellate blooms of Scrippsiella trochoidea in Daya bay, South China Sea. J. Oceanol. Limnol. 39, 1293–1304. doi: 10.1007/s00343-020-9082-0
Tomas, C. R., Syvertsen, E. E., Staidinger, K. A., Tangen, K., Throndsen, J., and Heimdal, B. R. (1997). Identifying marine phytoplankton. San Diego: Academic Press.
Tréguer, P., Bowler, C., Moriceau, B., Dutkiewicz, S., Gehlen, M., Aumont, O., et al. (2018). Influence of diatom diversity on the ocean biological carbon pump. Nat. Geosci. 11, 27–37. doi: 10.1038/s41561-017-0028-x
Wang, J. H., and Huang, X. (2003). Ecological characteristics of Prorocentrum dentatum and the cause of harmful algal bloom formation in China Sea (in Chinese, with English abstract). Chin. J. Appl. Ecol. 14, 1065–1069.
Wang, J. X., Kong, F. Z., Geng, H. X., Zhang, Q. C., Yuan, Y. Q., and Yu, R. C. (2021). CHEMTAX analysis of phytoplankton assemblages revealed potential indicators for blooms of haptophyte Phaeocystis globosa. Ecol. Indic. 131:108177. doi: 10.1016/j.ecolind.2021.108177
Wang, J. X., Kong, F. Z., Geng, H. X., Zhao, Y., Guan, W. B., He, C., et al. (2022). Pigment characterization of the Giant-Colony-forming haptophyte Phaeocystis globosa in the Beibu gulf reveals blooms of different origins. Appl. Environ. Microbiol. 88, e01654–e01621. doi: 10.1128/aem.01654-21
Wang, F. J., Lin, Y. S., Cao, W., Zhang, W. J., Zheng, L. M., Yang, W. D., et al. (2015). The relationship between nutrients and phytoplankton community structure in northern Beibu gulf. J. Trop. Oceanogr. 34, 73–85. doi: 10.11978/2014134
Wang, Z. H., Liu, L., Tang, Y. L., Li, A. F., Liu, C., Xie, C. L., et al. (2022). Phytoplankton community and HAB species in the South China Sea detected by morphological and metabarcoding approaches. Harmful Algae 118:102297. doi: 10.1016/j.hal.2022.102297
Wang, X., Liu, C., Zhang, Q. C., Chen, J. F., Wang, J. X., Zhao, Q. Y., et al. (2023). A dinoflagellate bloom caused by multiple species of Kareniaceae in the coastal waters of Fujian in June 2022 and its adverse impacts on Brachionus plicatilis and Artemia salina. Mar. Pollut. Bull. 196:115685. doi: 10.1016/j.marpolbul.2023.115685
Wang, Z. H., Qi, Y. Z., and Yang, Y. F. (2007). Cyst formation: an important mechanism for the termination of Scrippsiella trochoidea (Dinophyceae) bloom. J. Plankton Res. 29, 209–218. doi: 10.1093/plankt/fbm008
Worden, A. Z., Follows, M. J., Giovannoni, S. J., Wilken, S., Zimmerman, A. E., and Keeling, P. J. (2015). Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science 347:1257594. doi: 10.1126/science.1257594
Xiao, X., Agustacuı, S., Pan, Y. R., Yu, Y., Wu, J. P., and Duarte, C. M. (2019). Warming amplifies the frequency of harmful algal blooms with eutrophication in Chinese coastal waters. Environ. Sci. Technol. 53, 13031–13041. doi: 10.1021/acs.est.9b03726
Xie, Q. L., Yan, N. Y., Yang, X., Gao, R. C., Chen, X. L., Wu, H. J., et al. (2023). Synoptic view of an unprecedented red Noctiluca scintillans bloom in the Beibu gulf, China. Sci. Total Environ. 863:160980. doi: 10.1016/j.scitotenv.2022.160980
Xu, Q. S., Huang, M. Q., Yang, S., Li, X. L., Zhao, H. X., Tang, J. L., et al. (2022). Ecological stoichiometry influences phytoplankton alpha and beta diversity rather than the community stability in subtropical bay. Ecol. Evol. 12:e9301. doi: 10.1002/ece3.9301
Xu, S. N., Lin, H. J., Dai, M., Xiao, Y. Y., and Chen, Z. Z. (2014). Ecological characteristics of phytoplankton community in Guangxi coastal area. Chin. J. Ecol. 33, 2733–2739. doi: 10.13292/j.1000-4890.2014.0238
Xu, X., Yu, Z. M., Cheng, F. J., He, L. Y., Cao, X. H., and Song, X. X. (2017). Molecular diversity and ecological characteristics of the eukaryotic phytoplankton community in the coastal waters of the Bohai Sea, China. Harmful Algae 61, 13–22. doi: 10.1016/j.hal.2016.11.005
Xu, Y. X., Zhang, T., and Zhou, J. (2019). Historical occurrence of algal blooms in the northern Beibu gulf of China and implications for future trends. Front. Microbiol. 10:451. doi: 10.3389/fmicb.2019.00451
Zhu, F., Massana, R., Not, F., Marie, D., and Vaulot, D. (2005). Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol. Ecol. 52, 79–92. doi: 10.1016/j.femsec.2004.10.006
Keywords: microalgae, phytoplankton, Beibu Gulf, metabarcoding, microscopy, harmful algal blooms (HABs)
Citation: Wang C, Gu J, Li W, Wang J, Wang Z and Lin Q (2024) Metabarcoding reveals a high diversity and complex eukaryotic microalgal community in coastal waters of the northern Beibu Gulf, China. Front. Microbiol. 15:1403964. doi: 10.3389/fmicb.2024.1403964
Received: 20 March 2024; Accepted: 24 May 2024;
Published: 05 June 2024.
Edited by:
Jean-David Grattepanche, Temple University, United StatesCopyright © 2024 Wang, Gu, Li, Wang, Wang and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaohui Wang, dHd6aEBqbnUuZWR1LmNu; Qiuqi Lin, dGxpbnFxQGpudS5lZHUuY24=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.