- 1Department of Food Biotechnology, Branch for Northwest and West Region, Agricultural Biotechnology Research Institute of Iran, Agricultural Research, Education and Extension Organization (AREEO), Tabriz, Iran
- 2Regenerative Medicine Research Center (RMRC), Health Technology Institute, Kermanshah University of Medical Sciences, Kermanshah, Iran
Bifidobacteria are a prominent type of bacteria that have garnered significant research attention for their exceptional probiotic properties and capacity to produce exopolysaccharides (EPSs). These compounds exhibit diverse physical, chemical, and biological characteristics, prompting numerous investigations into their potential applications. Researchers have noted their beneficial effects as immune modulators within the host’s body across various industries. Extensive research has been conducted on the immunomodulatory effects of bifidobacteria-derived EPSs, with emerging engineering strategies aimed at enhancing their immune-modulating capabilities. Understanding the structure, physicochemical properties, and biological activities of these compounds is crucial for their effective utilization across different industries. Our review encompassed numerous studies exploring Bifidobacterium and its metabolites, including EPSs, across various sectors, drawing from diverse databases. The distinctive properties of EPSs have spurred investigations into their applications, revealing their potential to bolster the immune system, combat inflammation, and treat various ailments. Additionally, these compounds possess antioxidant and antimicrobial properties, making them suitable for incorporation into a range of products spanning food, health, and medicine.
1 Introduction
Bifidobacteria, a genus within the Actinobacteria branch, are Gram-positive, anaerobic bacteria shaped like bacilli, acknowledged as a pivotal bacterial group in the intestinal microbial community, particularly during natural childbirth and infancy. First isolated in 1899 by Tissier from the feces of breastfed babies, this genus encompasses over 50 different species, including notable examples such as Bifidobacterium bifidum, Bifidobacterium longum, Bifidobacterium breve, Bifidobacterium animal, Bifidobacterium adolescentis, Bifidobacterium pseudostreptococcus, and Bifidobacterium pseudolongum (Schell et al., 2002; Fanning et al., 2012b; Eshaghi et al., 2017; Ruiz et al., 2017; Hidalgo-Cantabrana et al., 2018; Turroni et al., 2019; Yao et al., 2021; Li et al., 2023; Nicolescu et al., 2023; Pacyga-Prus et al., 2023).
In infants, bifidobacteria typically account for about 90% of intestinal bacteria, while in adults, this proportion decreases to 3%–5%. The mode of infant feeding significantly impacts the establishment of bifidobacteria in the gut, with breastfed infants showing higher levels compared to formula-fed counterparts (Liu et al., 2014; Ruiz et al., 2017; Choi et al., 2022; Li et al., 2023). These bacteria have the capability to produce metabolites known as exopolysaccharides (EPSs) during fermentation. Traditionally, EPSs play a vital role in fermented dairy products owing to its gelling and thickening properties, which also offer potential health benefits (Salazar et al., 2009; Sørensen et al., 2022). Due to their unique physiological and biological properties, these compounds can help strengthen the body’s immune system and effectively combat inflammation and various diseases, as well as serving as biological additives in various products such as food, pharmaceuticals, and health products (Xu et al., 2019; Angelin and Kavitha, 2020; Korcz and Varga, 2021; Netrusov et al., 2023).
In particular, EPSs, with their diverse chemical and structural composition, perform various functions across industries including agriculture, dairy, biofilm formation, cosmetics, etc., demonstrating their biotechnological significance (Netrusov et al., 2023). EPSs are a type of polysaccharide (Lim et al., 2020) and carbohydrate polymer widely distributed in various organisms, including plants, animals, microorganisms, and others (Yue et al., 2023a). EPS produced by lactic acid bacteria typically consists of glucose, galactose, and rhamnose sugar units in varying ratios, commonly found in genera such as bifidobacteria and lactobacilli (Kaur and Dey, 2023). These polysaccharides can either form a capsule when covalently attached to the cell surface, known as capsular polysaccharides (CPSs; Chen et al., 2017; Angelin and Kavitha, 2020; Alessandri et al., 2021), or be secreted into the surrounding environment of the cell, easily released in the growth environment, creating a slimy coating, and are then referred to as EPSs, which are crucial for the formation of bacterial biofilms (Alessandri et al., 2021; Xie et al., 2023).
The primary function of Bifidobacterium EPS is to shield these bacteria from acidity and bile salts during transit through the digestive system, thereby enhancing their adhesion to the intestinal mucosa (Llamas-Arriba et al., 2019). Consequently, Bifidobacteria are commonly utilized for the direct production of EPS in fermented products (Xu et al., 2019). Additionally, EPSs play various roles in protecting bacteria against osmotic stress (Bhagat et al., 2021), desiccation (Carezzano et al., 2023), extreme temperatures, salinity, UV rays, chemical agents such as antibiotics and heavy metals (Shukla et al., 2017), phagocytosis, and bacteriophage attacks (Laiño et al., 2016; Schmid, 2018; Angelin and Kavitha, 2020; Korcz and Varga, 2021; Netrusov et al., 2023). Hence, they serve as vital biological components in the interaction between microorganisms and the host (Laiño et al., 2016).
Certain species of Bifidobacterium, such as B. longum, B. breve, B. bifidum, B. adolescentis, B. catenulatum, and B. infantis, possess the capability to produce EPSs (Salazar et al., 2009; Hidalgo-Cantabrana et al., 2014b). These EPSs varieties exhibit a wide array of properties, serving as preservatives in food products, enhancing the immune system, acting as antimicrobial agents, and functioning as antioxidants (Fanning et al., 2012b; Laiño et al., 2016; Yue et al., 2023a). Moreover, they demonstrate antitumor potential (Wang et al., 2019), along with properties such as anti-diabetic, anti-inflammatory bowel, anti-aging, immune modulation, wound healing, and blood cholesterol reduction (Laiño et al., 2016; Llamas-Arriba et al., 2019; Yue et al., 2023a).
For instance, EPS derived from B. longum w11 has exhibited antioxidant activity in vitro and has been shown to regulate cellular oxidative stress (Inturri et al., 2017a). Furthermore, research indicates that bifidobacteria can mitigate the progression or symptoms of various diseases, including colorectal cancer, diarrhea, necrotizing enterocolitis, and inflammatory bowel disease (Liu et al., 2019). In a particular study, EPS isolated from lactobacilli and bifidobacteria demonstrated efficacy in attenuating the inflammatory response of enterotoxigenic E. coli on pig intestinal enterocytes (Wachi et al., 2014). Additionally, bifidobacteria have been observed to interact with human immune cells and modulate specific pathways involved in both innate and adaptive immune responses (Ruiz et al., 2017). By employing strategies such as optimizing cultivation conditions, as well as genetic and metabolic engineering, it becomes feasible to tailor the performance, structural, and functional characteristics of bacterial EPSs (Hidalgo-Cantabrana et al., 2014b).
2 Composition of Bifidobacterium EPS
EPS produced by bifidobacteria are recognized as potentially biologically active compounds. They exhibit a diverse range of structures and are primarily synthesized in response to various environmental stimuli (Nicolescu et al., 2023). Research indicates that the production rate of EPS by these bacteria is influenced by factors such as strain variation, environmental composition, and culture conditions, including temperature, pH, and carbon-nitrogen ratio (Angelin and Kavitha, 2020; Mohd Nadzir et al., 2021).
EPS can be categorized into two groups based on their properties, namely homopolysaccharides (HoPSs), composed of a single monosaccharide, and heteropolysaccharides (HePSs), consisting of one or more types of monosaccharides (Salazar et al., 2016; Angelin and Kavitha, 2020; Netrusov et al., 2023; Salimi and Farrokh, 2023). While homopolysaccharides are produced by certain lactic acid bacteria, their production in bifidobacteria remains unidentified (Hidalgo-Cantabrana et al., 2012; Castro-Bravo et al., 2018b; Lynch et al., 2018). In bifidobacteria, HePs consist of various repeating monosaccharide units, predominantly D-glucose, D-galactose, L-rhamnose, and occasionally, N-acetylated monosaccharides such as N-acetyl-glucosamine (GluNAc) and N-acetyl-galactosamine (GalNAc), as well as fucose, glucuronic acid, glycerol, or mannose, which may be branched or unbranched (Castro-Bravo et al., 2018b; Angelin and Kavitha, 2020; Jurášková et al., 2022; Netrusov et al., 2023).
Examples of heteropolysaccharides produced by bacteria include xanthan, alginate, valan, kefir, golan, and hyaluronic acid (Mohd Nadzir et al., 2021). The structure of the repeating units is elucidated using nuclear magnetic resonance and other chromatographic methods (Hidalgo-Cantabrana et al., 2012; Castro-Bravo et al., 2018b).
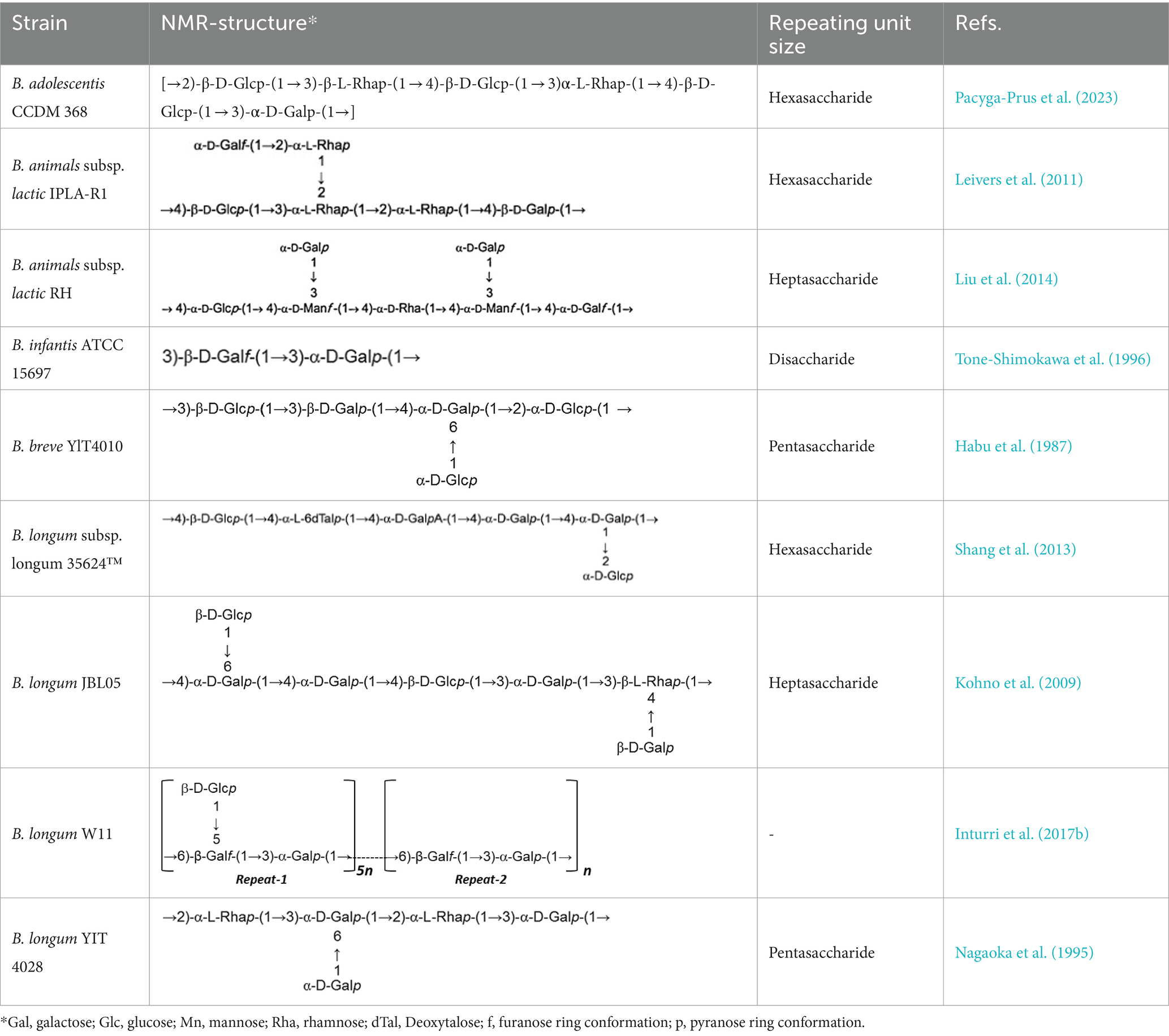
In a study investigating approximately 30 EPS from Bifidobacterium strains using various chromatographic methods, the main monosaccharides identified were D-galactose, found in all Bifidobacterium EPS, followed by D-glucose, present in over half of them, and finally L-rhamnose, found in half of the Bifidobacterium EPS. However, exceptions exist where the proportion of rhamnose is higher in certain B. animalis subsp. EPS (Hidalgo-Cantabrana et al., 2012). Additionally, studies indicate that Bifidobacterium strains containing D-mannose exhibit a higher rate of EPS production compared to L-mannose strains (Chen et al., 2017). Table 1 illustrating the structure of EPS units produced by several bifidobacteria strains.
Both quantitative and qualitative methods are utilized to detect and identify EPS produced by bifidobacteria. Qualitative techniques encompass electron microscopy (EM) and confocal laser scanning microscopy (CLSM), providing visual insights into EPS structure and morphology. On the quantitative front, various methods are employed to analyze EPS composition including liquid chromatography (HPLC), gas chromatography (Han et al., 2016), colorimetric methods, size exclusion chromatography (SEC), ion exclusion chromatography (Konieczna et al., 2012), nuclear magnetic resonance spectroscopy (NMR), and Fourier transforms infrared spectroscopy (FTIR). These methods, either individually or in combination, enable comprehensive analyses of EPS composition, thereby contributing to a deeper understanding of their properties and potential applications (Korcz and Varga, 2021).
3 Pathway of EPS biosynthesis in bifidobacteria
The mechanism underlying EPS synthesis in the Bifidobacterium genus remains incompletely understood due to the elusive structure and composition of Bifidobacterium EPS. Previous studies have revealed that most bifidobacteria lack genes associated with HoPSs synthesis, which encode enzymes like glycan sucrase and fructan sucrase. However, a pathway for HePSs synthesis in bifidobacteria has been postulated based on the predicted functions of eps genes (Hidalgo-Cantabrana et al., 2014b; Castro-Bravo et al., 2018b).
HePSs polymers possess a complex composition, and their synthesis involves multiple enzymes and proteins, rendering the process intricate (Castro-Bravo et al., 2018b; Whitfield et al., 2020). Enzymes involved in HePSs biosynthesis can be categorized into four groups:
1. Hexokinase: These enzymes activate glucose to glucose-6-phosphate.
2. Uridine-5′diphosphate (UDP)-glucose pyrophosphorylase: They catalyze the conversion of glucose-1-phosphate to UDP-glucose, a critical molecule in EPS synthesis.
3. Glycosyltransferases: These enzymes transfer sugar nucleotides to a glycosyl carrier lipid.
4. Wzx protein (flipase) and ABC transporters: These groups of enzymes are involved in the polymerization and transport of EPS units across the cytoplasmic membrane. Wzx protein acts as a flipase, ejecting EPS repeat units bound to a lipid carrier across the membrane, while ABC transporters transport single repeating units attached to the lipid carrier UDP-C55 (Willis and Whitfield, 2013).
The synthesis of HePSs in bifidobacteria involves several steps:
1. Synthesis of repeating sugar units within the cytoplasm.
2. Cytoplasmic assembly of the EPS unit.
3. Export of the repeating EPS units to the extracellular side.
4. Polymerization and determination of the length of the final skeleton chain, with all steps except polymerization occurring in the cytoplasm (Cuthbertson et al., 2009; Zannini et al., 2016; Yang et al., 2019; Angelin and Kavitha, 2020).
Initially, glucose is converted into glucose-6-phosphate by intracellular hexokinase enzymes. Subsequently, glucose-6-phosphate is converted into glucose-1-phosphate by the enzyme phosphoglucomutase. UDP-glucose, essential for EPS synthesis, is then formed from glucose-1-phosphate by uridine diphosphate glucose pyrophosphorylase.
In the subsequent step, glycosyltransferase priming enzymes link the first monosaccharide from Pischas or the activated sugar nucleotide to a membrane-bound isoprenoid lipid carrier [Undecaprenyl phosphate (C55)]. Successive glycosyltransferases catalyze the glycosidic bond between new nucleotide sugars and the initial monosaccharide, leading to the addition of more sugar fragments. The structure of each oligopolysaccharide varies depending on the number and characteristics of Glycosyltransferases (Castro-Bravo et al., 2018a,b; Wang et al., 2019; Angelin and Kavitha, 2020).
Carrier lipids, identified as isoprenoid alcohols, have their terminal alcohol groups connected to remaining monosaccharides via a pyrophosphate bridge. These carrier lipids may undergo modifications such as acetylation, acylation, sulfosylation, and methylation, if necessary.
Finally, the synthesized polymers are secreted to the extracellular side using two secretory systems: ABC transporters and the flippase-polymerase complex (WZX-WZY). Most eps clusters in Bifidobacterium strains indicate the existence of both systems in this genus. In the Wzx-Wzy-dependent pathway, the protein flippase (Wzx) ejects the EPS repeat units bound to the lipid carrier across the membrane, followed by a polymerase (Wzy) that transfers the repeating units outside the cell. The final chain length is determined by protein tyrosine kinase (Wzz; Hidalgo-Cantabrana et al., 2014b; Castro-Bravo et al., 2018a). A schematic representation of the hypothetical EPS biosynthesis pathway in Bifidobacterium, dependent on the Wzx-Wzy pathway, is depicted in Figure 1.
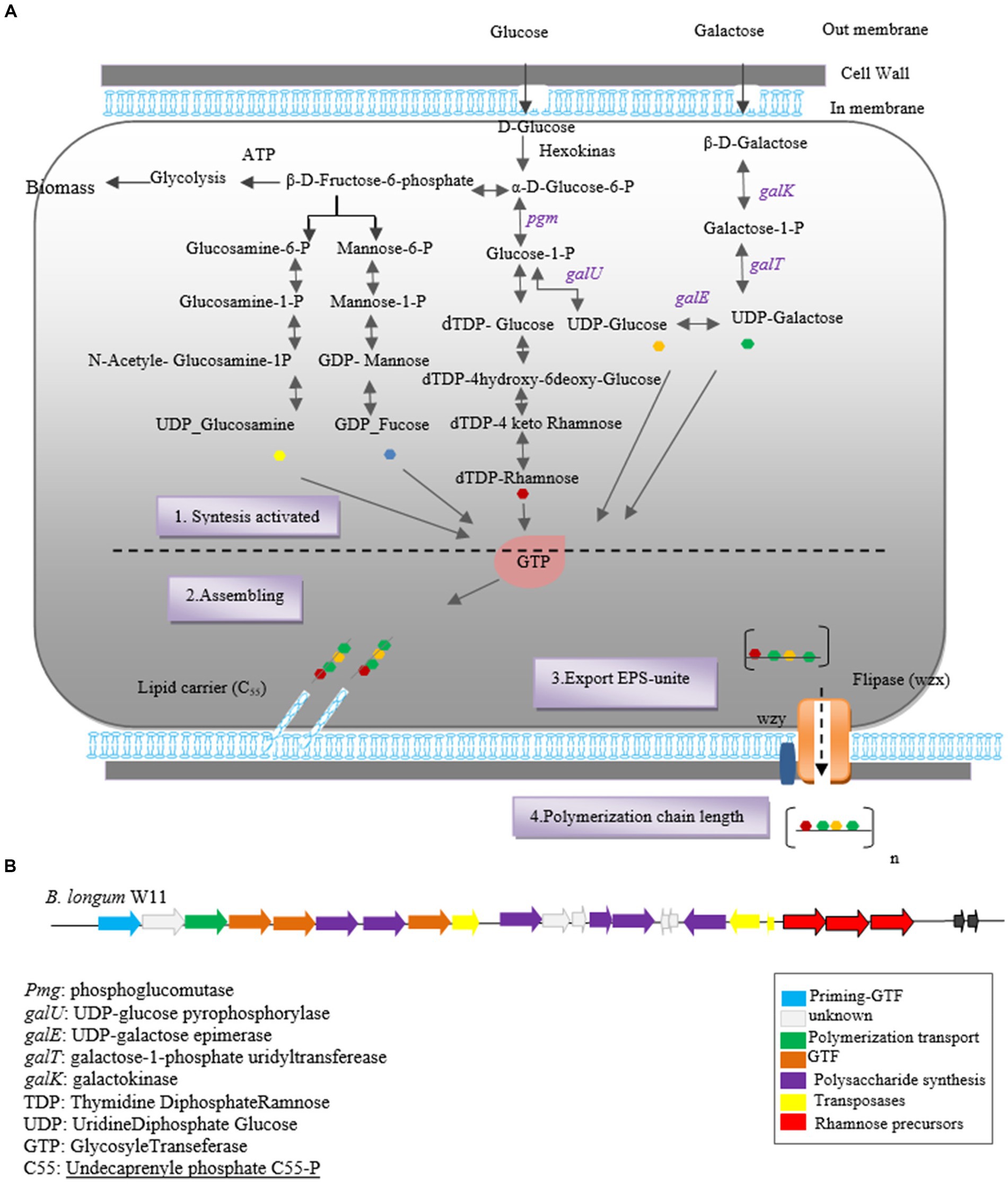
Figure 1. Heteropolysaccharide biosynthesis pathway in Bifidobacterium genus (A) and gene cluster of Bifidobacterium longum W11 (B).
The biosynthesis of EPSs in Bifidobacterium species involves two key stages: the synthesis of precursor sugar nucleotides and the EPS cluster, which includes genes responsible for sugar nucleotide production and EPS biosynthesis enzymes. Notable genes involved in precursor sugar nucleotide synthesis include galK, galE, galT, galU, rmlA, rmlB1, and rmlCD, along with early glycosyltransferases. These genes have been identified in B. longum subspecies and strains NCC2705, DJO10A, and B. longum subsp. longum CRC 002 (Audy et al., 2010).
The eps gene cluster comprises genes encoding EPS biosynthesis enzymes and proteins. In silico analysis of the eps cluster has revealed a conserved gene set, including the predicted glycosyltransferase Primary (p_gtf), which catalyzes the synthesis of the EPS unit in the initial step. This gene is typically present in all eps clusters and is crucial for EPS production in the Bifidobacterium genus (Ferrario et al., 2016). For instance, the eps cluster of B. longum W11 has been described (Inturri et al., 2017b) as predicted in Figure 1B.
The first identification of EPS genes within the Bifidobacterium genome was reported by Lee and O’Mallivan (2010), uncovering predicted genes involved in EPS synthesis within these clusters (Castro-Bravo et al., 2018a). Subsequently, the sequence of the eps cluster responsible for EPS synthesis in B. animalis subsp. lactis was elucidated, along with the structure of the high molecular weight (HMW) rhamnose-rich repeating unit EPS produced by strain IPLA-R1 (Leivers et al., 2011).
Subsequent research and genetic analysis by Hidalgo et al. in 2014, focusing on 28 completed Bifidobacterium genomes, shed light on the diversity of the eps cluster among Bifidobacterium strains. For instance, B. animalis subsp. IPLA R1 was found to harbor a 54.3 kb eps cluster containing 42 genes, exhibiting variation across Bifidobacterium subspecies (Hidalgo-Cantabrana et al., 2014b; Ferrario et al., 2016). On the other hand, B. bifidum E3 strain’s eps cluster comprised 20 genes, indicating variation even within species (Yue et al., 2023a).
Moreover, higher diversity was observed in B. adolescentis and B. breve strains, highlighting strain-dependent variation in eps clusters. Notably, B. breve UCC2003 strain was found to possess three eps clusters, namely eps1, eps2a, and eps2b (Fanning et al., 2012a; Ferrario et al., 2016). However, despite the presence of a conserved eps2 cluster in the genome of B. pullorum LMG21816, this strain exhibited a negative EPS phenotype under tested conditions. It was suggested that the absence of p-gtf indicates the incompleteness of the eps cluster in this strain, resulting in the lack of EPS production (Ferrario et al., 2016).
In a study conducted by Ferrario et al. (2016), the genomes of 48 bifidobacteria strains available in the gene bank were analyzed to identify potential eps clusters. The researchers utilized the p-gtf sequence as a molecular marker to retrieve eps genome sequences, except for B. bifidum LMG11041. This study corroborated findings from a previous study by Hidalgo et al. in 2014, which indicated a lack of common structural organization in the eps biosynthesis clusters of bifidobacteria. However, the study revealed consistent interspecies diversity among strains possessing eps clusters, particularly in terms of cluster length, number, and predicted gene functions. The size of eps gene clusters varied significantly among Bifidobacterium strains, ranging from 9 genes identified in the eps region of B. mongoliense to 55 genes in B. dentium (Ferrario et al., 2016).
Notably, the glycosyltransferase enzyme, catalyzing the initial step of EPS synthesis, was found encoded within all eps clusters of the studied Bifidobacterium strains (Hidalgo-Cantabrana et al., 2014b; Castro-Bravo et al., 2018b; Zhou et al., 2019). However, interspecies variation in the genetic content responsible for EPS synthesis in Bifidobacterium indicates the flexibility of the genome within this genus (Zhou et al., 2019).
It’s noteworthy that there’s no common structural organization observed among species and strains within the Bifidobacterium genus. Furthermore, the G + C content of most EPS clusters in this genus are lower than that of the entire genome, suggesting horizontal acquisition of these genes. The likely donors of these genes are inhabitants with which bifidobacteria share a common environment, such as members of Lactobacillaceae and Lachnospiraceae. This suggests a dynamic evolutionary process shaping EPS synthesis within the Bifidobacterium genus (Chaib De Mares et al., 2015).
4 Bifidobacterium EPS engineering strategy
EPS engineering strategies aim to optimize the performance and unique properties of EPS for diverse applications in food, medicine, and industry, driven by insights into structure–function relationships. These strategies encompass interventions at the polymer synthesis stage through various treatments or at the biosynthesis level, thereby influencing structural composition. A straightforward approach to engineer EPS involves enzymatic modification, utilizing polysaccharide hydrolases and lyases, which serve as endoglycosidases or exoglycosidases. Typically sourced from microorganisms or their bacteriophages, these enzymes facilitate the alteration of EPS structure, enabling tailored functionalities (Boels et al., 2001).
Moreover, targeted manipulation of regulatory proteins offers another avenue to enhance EPS productivity. By augmenting the transcription of operons responsible for encoding EPS biosynthesis proteins, productivity can be significantly increased (Schmid et al., 2015). Additionally, EPS structures can be customized through modifications such as acetylation, phosphorylation, and sulfonation, enabling the attainment of desired functionalities. While these modifications have been extensively studied in lactobacillus strains, their exploration within the Bifidobacterium genus remains limited.
However, alternative strategies for modifying EPS in Bifidobacterium include altering molecular weight, adding or removing substituents and monomer sugars from side chains, and overexpressing genes encoding enzymes in EPS biosynthesis pathways. Additionally, housekeeping genes involved in sugar nucleotide formation play a crucial role. Given the abundance of the CRISPR-Cas system in bifidobacteria, gene editing using the CRISPR system offers a promising avenue for modifying EPS with new biological activities (Xu et al., 2019; Zuo et al., 2020; Sun and Zhang, 2021; Salimi and Farrokh, 2023).
In a study by Castro-Bravo et al. (2017), a novel double crossover recombination strategy was utilized in bifidobacteria. Specifically, they targeted the Balate-1410 gene, which encodes a protein responsible for polymer chain elongation in Bifidobacterium animalis subsp. lactis DSM10140. Through this approach, they replaced the wild-type gene with a mutant variant, resulting in a mucoid phenotype. In essence, this research demonstrated that the ability to produce EPS in B. animalis DSM10140T could be reinstated by introducing a point mutation into the Balat_1410 gene, which plays a crucial role in EPS chain elongation. Analysis using NMR and SEC-MALS revealed that the mutant strain produced EPS with a higher molecular weight compared to the wild type. Furthermore, chemical and physical analyses confirmed the successful introduction of the mutation related to high molecular weight EPS in the recombinant strain (Castro-Bravo et al., 2017). This study showcases the potential of genetic manipulation techniques in enhancing EPS properties in Bifidobacterium, paving the way for future advancements in EPS engineering within this genus.
The impact of genes involved in nucleotide sugar production and priming glycosyltransferase (PGTF), which initiates the assembly of EPS repeat units by adding the first sugar-1-phosphate to a lipophilic carrier, is crucial in EPS engineering strategies. In a study conducted by Audy et al. (2010), the focus was on the expression of genes associated with sugar nucleotide production and EPS biosynthesis in Bifidobacterium longum subspCRC002. Their findings revealed that genes responsible for EPS biosynthesis were clustered within one or two transcription units, notably including PGTF, along with sugar nucleotide precursors for glucose, galactose, and rhamnose. Targeting these specific genes significantly influenced EPS production in this strain. Also B. longum subsp. CRC002 showed strong metabolic activity leading to increased production of EPS composed of glucose and galactose when PGTF-related genes were targeted. In addition, the expression of nucleotide sugar genes peaked at the exponential growth stage, indicating their importance in EPS biosynthesis. This study underscores the significance of targeting PGTF as a key enzyme in the biosynthetic pathway of EPS. By manipulating the expression of genes associated with nucleotide sugar production and PGTF, researchers can effectively enhance EPS production and tailor EPS composition to meet specific requirements for various applications (Audy et al., 2010).
In a study conducted by Hickey et al. (2021), they reported that strains of Bifidobacterium breve UCC2003 and B. breve JCM7017, which naturally produce EPS (WT), were compared with isogenic strains lacking EPS (EPS-mutants). The researchers observed that both WT strains lost their ability to produce EPS upon mutation, serving as positive controls for EPS deposition assays. Furthermore, the study investigated the impact of different carbohydrate sources on EPS production. Screening both B. breve strains in media containing glucose, lactose, and maltose revealed that while the WT strains did not precipitate EPS over a 6-h period, their EPS-mutants did. Additionally, the study explored the modulation of EPS on the cytokine response of Bone Marrow-Derived Macrophages (BMDM) and Dendritic Cells (BMDCs). When both EPS-isogenic strains were cultured with primary BMDM for 24 h, the absence of EPS from B. breve UCC2003 led to increased cytokine responses, with BMDM secreting TNF-α and IL10. Conversely, the absence of EPS from B. breve JCM7017 resulted in reduced cytokine responses. BMDCs did not exhibit significant TNF-α or IL10 production in response to any bacterial strain (Hickey et al., 2021).
5 Interaction between Bifidobacterial EPS and immune system
The immune system comprises two main components: innate and acquired immunity. The innate immune system, which is inherited and non-specific, serves to protect the host against microbial invasion and tissue damage (Ashraf and Shah, 2014; Hato and Dagher, 2015; Yatim and Lakkis, 2015). It operates with a short-term memory and includes various components such as the skin, phagocytic cells like macrophages, dendritic cells (DCs), neutrophils, and protein molecules like the complement and coagulation systems (Yatim and Lakkis, 2015; Kellie and Al-Mansour, 2017). In contrast, the acquired immune response is specific to target antigens and involves receptors expressed on B and T lymphocytes. This response becomes prominent a few days after encountering the antigen. Communication between the innate and adaptive immune systems is primarily facilitated by antigen-presenting dendritic cells (Yatim and Lakkis, 2015). Although the mechanisms of action of innate and acquired immunity differ, their cooperation is essential for mounting a fully effective immune response. This collaboration ensures a comprehensive defense against pathogens and other harmful agents (Hato and Dagher, 2015; Kellie and Al-Mansour, 2017).
The gastrointestinal tract (GIT) is home to a diverse array of microorganisms, including bacteria belonging to the genus Bifidobacterium, which play a crucial role in promoting host health.
In recent decades, the immunomodulatory capabilities of Bifidobacterium bacteria in interaction with human immune cells have garnered significant scientific interest. The modulating ability of strains within this bacterial genus has been linked to their production of EPSs (Hidalgo-Cantabrana et al., 2012). Bifidobacterium EPS exhibit anti-inflammatory and antimicrobial properties, contributing to their ability to regulate the immune system. Inflammation, which is a normal tissue repair process in response to infections and tissue damage, can lead to various inflammatory reactions such as pain, swelling, and fever due to the production of nitric oxide (NO) and prostaglandin E2 (PGE2; Angelin and Kavitha, 2020; Choi et al., 2022). Prolonged inflammation can result in excessive or insufficient production of pro-inflammatory cytokines, including IL-6 and TNF-α, and suppression of anti-inflammatory cytokines like IL-10, leading to inflammatory diseases and cancer (Angelin and Kavitha, 2020). Bifidobacterium EPSs have been demonstrated to modulate the inflammatory response of immune cells. For instance, EPS extracted and purified from B. longum BCRC 14634 exhibit mild immunomodulatory activity on J77A.1 macrophages by increasing IL-10 secretion and decreasing TNF-α levels (Wu et al., 2010). Other activities of EPS in modulating the host’s immune system include enhancing the proliferation of T and B lymphocytes, increasing natural killer (NK) cell activity, boosting the phagocytic capacity of mononuclear cells, inducing cytokine production, and enhancing overall host immune defense against pathogens (Angelin and Kavitha, 2020).
Moreover, Bifidobacterium plays a crucial role in reducing the risk of infection and preventing gastrointestinal cancers and inflammatory diseases such as inflammatory bowel disease (IBD; Rajoka et al., 2018). For instance, B. longum subsp. longum 35,624 has demonstrated clinical efficacy in irritable bowel syndrome, and comparison with a mutant derivative lacking EPS production illustrated the preventive role of EPS (Schiavi et al., 2016). Bifidobacterium EPS also exert their modulatory effects through antigen-presenting cells (APCs) or dendritic cells. EPS induce dendritic cells to secrete cytokines, leading to the differentiation of naïve T cells into regulatory T cells, which suppress inhibitory T cells, thereby promoting immune balance (Ashraf and Shah, 2014).
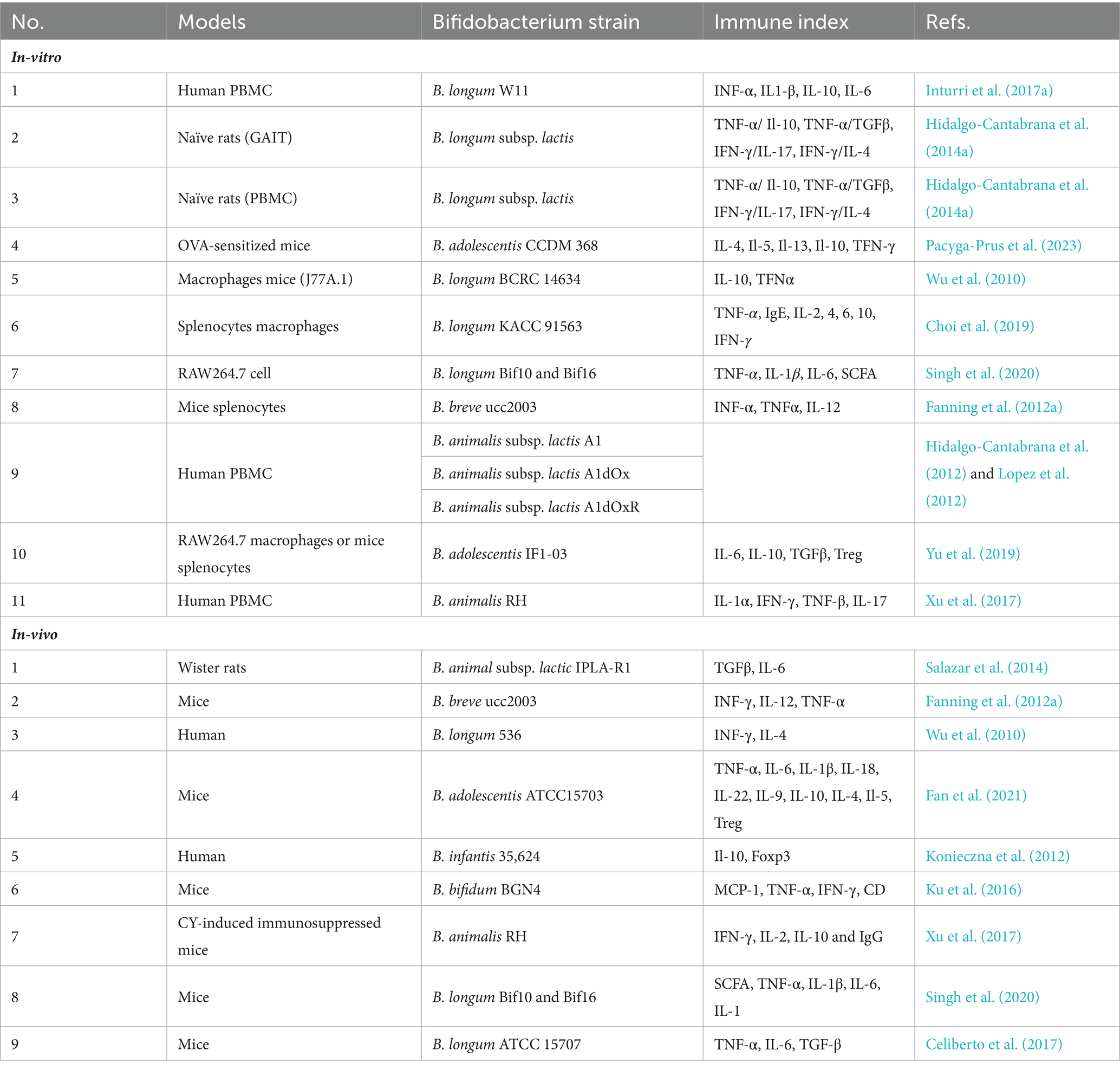
Various in vitro and in vivo models have been utilized to study the immunomodulatory activity of EPSs produced by Bifidobacterium strains. These models include peripheral blood mononuclear cells (PBMCs), mouse spleen cells, macrophage-like cell lines, and Gut Associated Lymphatic Tissues (GALT). Additionally, enterocytes such as the CaCo-2 or HT29 cell models have been employed in certain studies to investigate the immunomodulatory potential of Bifidobacterium bacteria due to their direct exposure to the intestinal environment, which could play a pivotal role in the initiation of bifidobacteria-host interactions (Ruiz et al., 2017).
In vivo models using human, mouse, and rat PBMCs have also been employed, and several instances of in vitro and in vivo models demonstrating immune responses are detailed in Table 2. The immunomodulatory capacity of Bifidobacterium EPS suggests potential health benefits for humans. However, differences in EPS structure and immune regulation between strains of the same Bifidobacterium species can lead to variations in their immunomodulatory effects. There is great structural variation in EPS polymers produced by bifidobacteria, even between strains of the same species (Hidalgo-Cantabrana et al., 2014b; Ferrario et al., 2016). In addition to differences in glycosidic bonds and degree of branching, changes in monosaccharide components and their amounts have been observed for different bifidobacteria strains (Hidalgo-Cantabrana et al., 2014b; Inturri et al., 2017b). This diversity could, in principle, lead to a large number of distinct EPS structures and theoretically to different immunomodulatory effects on the host (Hickey et al., 2021). For instance, B. breve UCC2003, with a thicker EPS layer, exhibits a more anti-inflammatory phenotype compared to B. breve JCM7017, as evidenced by modulation of macrophage IL-10 and TNFα and dendritic cell expression of Tnfa, Il6, Il12a, and Il23a. Murine B. pseudolongum UMB287 MBP-01 EPS increases intestinal Tregs, whereas porcine-derived B. pseudolongum ATCC25526 EPS does not. However, EPS from both strains lead to increased dendritic cells (DCs), mesenteric lymph node (MLN) DCs, and intestinal MLN macrophages (Hickey et al., 2021; Gavzy et al., 2023).
The modulatory effects of Bifidobacterium EPS depend on factors such as molecular weight and chemical composition. Generally, EPS with low molecular weight induce higher levels of cytokines, while those with high molecular weight induce a lower cytokine secretion or decrease the TNFα/IL-10 ratio, indicating an anti-inflammatory effect (Lopez et al., 2012; Salazar et al., 2014). For example, EPS from B. animalis subsp. lactis IPLA-R1, with a high rhamnose percentage and molecular weight, increases IL-10 production in PBMC models and decreases TNFα production in human colon biopsies (Hidalgo-Cantabrana et al., 2015). Moreover, studies have shown correlations between EPS polymer composition, structure, and size, and the corresponding immune response, suggesting that the physical and chemical characteristics of EPS influence their immunomodulatory properties (Lopez et al., 2012).
Similar findings were reported in a study involving co-incubation of purified EPS from three strains of B. animalis (A1, A1dOx, and A1dOxR) with PBMCs. The analysis revealed that the A1dOxR extracellular polymer caused less release of pro- and anti-inflammatory cytokines compared to other strains, attributed to its higher molecular weight. Additionally, EPS from Bifidobacterium adolescentis strains IF1-11 and IF1-03, with high molecular weight, induced different cytokine profiles when co-cultured with macrophage RAW264.7 and mouse spleen cells, with strain IF1-03 exhibiting anti-inflammatory effects, while strain IF1-11 showed pro-inflammatory effects (Alessandri et al., 2019).
Dendritic cells (DCs) represent a specialized subset of myeloid cells that respond to infection by capturing antigens, processing them into smaller peptides, and subsequently presenting them to T lymphocytes. As primary antigen-presenting cells (APCs), DCs serve as a crucial link between innate and adaptive immunity. Both mouse dendritic cells and human blood mononuclear cells hold promise for future investigations into the modulatory potential of EPSs derived from Bifidobacterium strains.
In vitro studies involving the treatment of DCs with Bifidobacterium bifidum PRI1, followed by co-culture with naïve CD4 T cells, have demonstrated enhanced induction of regulatory T cells (Tregs) and production of interleukin-10 (IL-10; Gavzy et al., 2023). These findings underscore the immunomodulatory capabilities of Bifidobacterium EPS, particularly in promoting immune tolerance and anti-inflammatory responses mediated by Tregs and IL-10. Such studies pave the way for further exploration of the interaction between Bifidobacterium-derived EPS and DCs, shedding light on their potential therapeutic applications in immune-related disorders.
One of the extensively researched aspects of immune system modulation by probiotic bacteria is the regulation of cytokine production. Cytokines, which are protein molecules synthesized by immune cells, play diverse roles in defense mechanisms, including inflammation, B and T lymphocyte differentiation, immune system activation, and eradication of foreign antigens. Additionally, cytokines can significantly contribute to the pathogenesis of autoimmune and immune-mediated kidney diseases (Yatim and Lakkis, 2015). Probiotic bacteria can induce the secretion of cytokines from intestinal epithelial cells in a strain-specific manner. For instance, in a study involving mice treated with Bifidobacterium adolescentis ATCC15703, lower levels of pro-inflammatory cytokines such as TNFα, IL-6, IL-1β, IL-18, IL-22, and IL-9 were observed compared to the control group. Conversely, higher levels of the anti-inflammatory cytokine IL-10 and the cytokines IL-4 and IL-5, along with increased regulatory T cells (Tregs), were detected in the colons of colitis mice receiving B. adolescentis ATCC15703 (Gavzy et al., 2023). These findings underscore the strain-specific immunomodulatory effects of probiotic bacteria and highlight their potential therapeutic implications in inflammatory conditions.
The impact of EPS produced by Bifidobacterium longum W11 on the immune response of peripheral blood mononuclear cells (PBMCs), both with and without ConA stimulation, was investigated. Specifically, in unstimulated PBMCs, EPS induced the production of IL-6 at higher concentrations and IL-10 only at lower concentrations. Moreover, when PBMCs were stimulated with ConA, EPS increased the production of various cytokines, except for IL-10 (Inturri et al., 2017a). Cytokines serve as soluble mediators of host defense responses, playing crucial roles in both specific and non-specific mechanisms for eliminating foreign antigens (Ashraf and Shah, 2014). The cell surface components of B. longum strains NCC 2705, ATCC 15707, and BIF53 have been shown to stimulate the production of IL-10 and TNFα in isolated peripheral blood mononuclear cells (Gavzy et al., 2023).
Macrophages play a crucial role in recognizing bacteria and, upon activation by microbial metabolites like polysaccharides, they engage in bacterial killing through phagocytosis, secrete cytokines for immune modulation, and present antigens to helper T cells. Notably, intestinal macrophages exhibit restrained proinflammatory cytokine production in response to various inflammatory stimuli, including microbial components (Wu et al., 2010).
In a study investigating the immunomodulatory and anti-inflammatory properties of EPSs from B. longum subsp. infantis E4, conducted on RAW264.7 cells, spleen lymphocytes, and mouse NK cells in vitro, it was found that EPS enhanced the growth and phagocytic activity of RAW264.7 macrophages, increased spleen lymphocyte proliferation, and boosted NK cell activity. These findings suggest that EPS derived from B. infantis E4 possesses immune-modulating and anti-inflammatory properties, potentially serving as a prebiotic for promoting future health maintenance. Hence, EPS from B. longum subsp. infantis E4 could be considered a functional food ingredient with modulatory and anti-inflammatory effects on immune cells, thus broadening the scope of immune modulators (Yue et al., 2023b).
In another study, EPS produced by B. longum strain BCRC 14634 was observed to induce increased production of the anti-inflammatory cytokine IL-10 by murine macrophages, compared to baseline conditions. Additionally, the presence of EPS was associated with lower levels of the pro-inflammatory cytokine TNFα, compared to lipopolysaccharides (Wu et al., 2010).
6 Application of Bifidobacterium EPS in the food industry
Beneficial microorganisms like lactic acid bacteria and bifidobacteria possess the capability to produce postbiotic bioactive substances, including EPSs. Leveraging their technological advantages, they are extensively employed as starter cultures in the production of fermented food products (Xu et al., 2019), as well as functional foods (Sørensen et al., 2022), which can directly or indirectly impact human health (Castro-Bravo et al., 2018b; Xu et al., 2019).
In the food industry, these microorganisms serve as functional additives, contributing to the production of products with desirable attributes. Particularly in the dairy sector, EPSs function as thickeners, emulsifiers, and stabilizers without imparting unpleasant tastes. They prevent water separation in cheese, resulting in a softer and creamier product, thereby increasing cheese yield. Moreover, they enhance yogurt viscosity and water holding capacity (Xu et al., 2019).
Another notable application of EPS is in bakery products, where it increases bread volume and moisture content, resulting in a softer texture for both gluten-containing and gluten-free bread. Additionally, EPSs mitigate staling by impeding starch retrogradation, thereby improving shelf life owing to their water-binding properties (Xu et al., 2019). However, despite their numerous advantages, EPSs can also have detrimental effects. For instance, EPS-producing bacteria can spoil alcoholic beverages like beer and wine. Furthermore, EPS synthesis and the formation of intestinal plaque and biofilm can lead to health issues in the food industry.
As bifidobacteria are commonly utilized as probiotics in dairy products, there is a growing interest in exploring the potential of EPS-producing Bifidobacterium strains as functional starters for low-fat yogurt production (Prasanna et al., 2013). Both lactic acid bacteria and bifidobacteria are known to produce EPS, contributing to the texture and mouthfeel of yogurt, a popular fermented milk product.
In the context of health-conscious consumers preferring low-fat dairy options, the production of low-fat yogurt presents certain challenges, such as compromised texture and taste, characterized by high synthesis and low viscosity. While thickeners can address these issues, regulations prohibiting the addition of stabilizers in yogurt have led to the exploration of EPSs as viable alternatives in the European Union (Xu et al., 2019).
The texture of yogurt is influenced by various factors including milk heating, pH, fermentation duration, milk composition (particularly protein or fat content), as well as the concentration and structure of EPS produced by the starter culture, which plays a pivotal role in yogurt gelation. Thus, the selection of strains with higher EPS production is crucial. However, studies indicate a weak correlation between EPS quantity and resulting effects on rheological properties, underscoring the significance of EPS structures (e.g., monosaccharide composition, charge, molecular weight, degree of branching, and backbone stiffness) and interactions between EPSs and milk components, especially proteins, in determining yogurt texture (Gentès et al., 2011; Han et al., 2016). Key textural characteristics of yogurt, including viscosity, syneresis (whey separation), gelation pH, and gel firmness, are considered essential factors in product quality and consumer acceptance (Xu et al., 2019).
The EPS produced by B. infantis CCUG 52486 and B. infantis NCIMB 702205 exhibits notable emulsification activity and favorable rheological properties, leading to enhanced viscosity in fermented low-fat milk. Among these strains, B. longum subsp. infantis CCUG 52486 stands out as particularly promising. It can be effectively incorporated into yogurt starter cultures to produce low-fat yogurt with probiotic benefits while simultaneously improving the physicochemical and rheological characteristics of the product (Prasanna et al., 2013).
Presently, there are 39 species of lactic acid bacteria and 5 species of bifidobacteria that have been granted Generally Recognized as Safe (GRAS) status by the European Food Safety Authority (Hazards et al., 2022). These strains are also included in the Qualified Presumption of Safety (QPS) list, making their application in food matrices more accessible. However, it’s worth noting that neither the EFSA nor the FDA have established any health claims for the use of EPS from lactic acid bacteria in food products (Hazards et al., 2022).
7 Conclusion and future perspective
In recent years, there has been growing interest in the EPS produced by Bifidobacterium bacteria. EPS serve as a crucial external protection and covering for Bifidobacterium, offering resilience against the surrounding environment. The synthesis of EPS involves a complex interplay of molecules, proteins, and enzymes, including glycosyl transferase and polymerases. The structural and chemical characteristics of EPS determine their diverse functions, rendering them beneficial in various industries such as agriculture, dairy, cosmetics, and pharmaceuticals.
In silico analysis conducted on available Bifidobacterium genomes has revealed a lack of consensus structural organization in EPS clusters, unlike those identified in LAB-eps clusters. However, some common features of bifido-eps clusters, such as high inter- and intraspecific organizational diversity, are observed, with the exception of B. animalis subsp. lactis. Additionally, the EPS cluster generally exhibits a lower G + C content compared to the entire bifidobacterial genome. The increasing availability of genomes in the future will offer researchers opportunities for genetic and metabolic engineering to tailor EPS production for use in the food or pharmaceutical industries.
Despite the growing interest, limited research has been conducted on in vitro and in vivo models to assess the immunomodulatory activity of EPS-producing bifidobacteria. Further scientific investigation is needed to enhance EPS efficiency and conduct in vivo studies to explore their therapeutic properties fully. This will enable researchers to harness the full potential of EPS in various applications.
Author contributions
MS: Writing – original draft. BH: Conceptualization, Project administration, Writing – original draft. YN: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alessandri, G., Ossiprandi, M. C., MacSharry, J., van Sinderen, D., and Ventura, M. (2019). Bifidobacterial dialogue with its human host and consequent modulation of the immune system. Front. Immunol. 10:2348. doi: 10.3389/fimmu.2019.02348
Alessandri, G., van Sinderen, D., and Ventura, M. (2021). The genus Bifidobacterium: from genomics to functionality of an important component of the mammalian gut microbiota. Comput. Struct. Biotechnol. J. 19, 1472–1487. doi: 10.1016/j.csbj.2021.03.006
Altmann, F., Kosma, P., O’Callaghan, A., Leahy, S., Bottacini, F., Molloy, E., et al. (2016). Genome analysis and characterisation of the exopolysaccharide produced by Bifidobacterium longum subsp. longum 35624™. PLoS One 11:e0162983. doi: 10.1371/journal.pone.0162983
Angelin, J., and Kavitha, M. (2020). Exopolysaccharides from probiotic bacteria and their health potential. Int. J. Biol. Macromol. 162, 853–865. doi: 10.1016/j.ijbiomac.2020.06.190
Ashraf, R., and Shah, N. P. (2014). Immune system stimulation by probiotic microorganisms. Crit. Rev. Food Sci. Nutr. 54, 938–956. doi: 10.1080/10408398.2011.619671
Audy, J., Labrie, S., Roy, D., and LaPointe, G. (2010). Sugar source modulates exopolysaccharide biosynthesis in Bifidobacterium longum subsp. longum CRC 002. Microbiology 156, 653–664. doi: 10.1099/mic.0.033720-0
Bhagat, N., Raghav, M., Dubey, S., and Bedi, N. (2021). Bacterial exopolysaccharides: insight into their role in plant abiotic stress tolerance. J. Microbiol. Biotechnol. 31, 1045–1059. doi: 10.4014/jmb.2105.05009
Boels, I. C., van Kranenburg, R., Hugenholtz, J., Kleerebezem, M., and De Vos, W. M. (2001). Sugar catabolism and its impact on the biosynthesis and engineering of exopolysaccharide production in lactic acid bacteria. Int. Dairy J. 11, 723–732. doi: 10.1016/S0958-6946(01)00116-9
Carezzano, M. E., Alvarez Strazzi, F. B., Pérez, V., Bogino, P., and Giordano, W. (2023). Exopolysaccharides synthesized by Rhizospheric Bacteria: a review focused on their roles in protecting plants against stress. Appl. Microbiol. 3, 1249–1261. doi: 10.3390/applmicrobiol3040086
Castro-Bravo, N., Hidalgo-Cantabrana, C., Rodriguez-Carvajal, M. A., Ruas-Madiedo, P., and Margolles, A. (2017). Gene replacement and fluorescent labeling to study the functional role of exopolysaccharides in Bifidobacterium animalis subsp. lactis. Front. Microbiol. 8:1405. doi: 10.3389/fmicb.2017.01405
Castro-Bravo, N., Sánchez, B., Margolles, A., and Ruas-Madiedo, P. (2018a). “Biological activities and applications of bifidobacterial exopolysaccharides: from the bacteria and host perspective” in The Bifidobacteria and related organisms. eds. P. Mattarelli, B. Biavati, and B. J. B. Wood (Elsevier), 177–193.
Castro-Bravo, N., Wells, J. M., Margolles, A., and Ruas-Madiedo, P. (2018b). Interactions of surface exopolysaccharides from Bifidobacterium and Lactobacillus within the intestinal environment. Front. Microbiol. 9:2426. doi: 10.3389/fmicb.2018.02426
Celiberto, L. S., Bedani, R., Dejani, N. N., Ivo de Medeiros, A., Sampaio Zuanon, J. A., Spolidorio, L. C., et al. (2017). Effect of a probiotic beverage consumption (Enterococcus faecium CRL 183 and Bifidobacterium longum ATCC 15707) in rats with chemically induced colitis. PLoS One 12:e0175935. doi: 10.1371/journal.pone.0175935
Chaib De Mares, M., Hess, J., Floudas, D., Lipzen, A., Choi, C., Kennedy, M., et al. (2015). Horizontal transfer of carbohydrate metabolism genes into ectomycorrhizal a manita. New Phytol. 205, 1552–1564. doi: 10.1111/nph.13140
Chen, Q., Lu, X., Guo, X., Guo, Q., and Li, D. (2017). Metabolomics characterization of two Apocynaceae plants, Catharanthus roseus and Vinca minor, using GC-MS and LC-MS methods in combination. Molecules 22:997. doi: 10.3390/molecules22060997
Choi, M., Lee, Y., Lee, N.-K., Bae, C. H., Park, D. C., Paik, H.-D., et al. (2019). Immunomodulatory effects by Bifidobacterium longum KACC 91563 in mouse splenocytes and macrophages. J. Microbiol. Biotechnol. 29, 1739–1744. doi: 10.4014/jmb.1812.12002
Choi, Y. J., Shin, S.-H., and Shin, H. S. (2022). Immunomodulatory effects of Bifidobacterium spp. and use of Bifidobacterium breve and Bifidobacterium longum on acute diarrhea in children. J. Microbiol. Biotechnol. 32, 1186–1194. doi: 10.4014/jmb.2206.06023
Cuthbertson, L., Mainprize, I. L., Naismith, J. H., and Whitfield, C. (2009). Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol. Mol. Biol. Rev. 73, 155–177. doi: 10.1128/MMBR.00024-08
Eshaghi, M., Bibalan, M. H., Rohani, M., Esghaei, M., Douraghi, M., Talebi, M., et al. (2017). Bifidobacterium obtained from mother’s milk and their infant stool; a comparative genotyping and antibacterial analysis. Microb. Pathog. 111, 94–98. doi: 10.1016/j.micpath.2017.08.014
Fan, L., Qi, Y., Qu, S., Chen, X., Li, A., Hendi, M., et al. (2021). B. adolescentis ameliorates chronic colitis by regulating Treg/Th2 response and gut microbiota remodeling. Gut Microbes 13, 1–17. doi: 10.1080/19490976.2020.1826746
Fanning, S., Hall, L. J., Cronin, M., Zomer, A., MacSharry, J., Goulding, D., et al. (2012a). Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc. Natl. Acad. Sci. 109, 2108–2113. doi: 10.1073/pnas.1115621109
Fanning, S., Hall, L. J., and van Sinderen, D. (2012b). Bifidobacterium breve UCC2003 surface exopolysaccharide production is a beneficial trait mediating commensal-host interaction through immune modulation and pathogen protection. Gut Microbes 3, 420–425. doi: 10.4161/gmic.20630
Ferrario, C., Milani, C., Mancabelli, L., Lugli, G. A., Duranti, S., Mangifesta, M., et al. (2016). Modulation of the eps-ome transcription of bifidobacteria through simulation of human intestinal environment. FEMS Microbiol. Ecol. 92:fiw056. doi: 10.1093/femsec/fiw056
Gavzy, S. J., Kensiski, A., Lee, Z. L., Mongodin, E. F., Ma, B., and Bromberg, J. S. (2023). Bifidobacterium mechanisms of immune modulation and tolerance. Gut Microbes 15:2291164. doi: 10.1080/19490976.2023.2291164
Gentès, M.-C., St-Gelais, D., and Turgeon, S. L. (2011). Gel formation and rheological properties of fermented milk with in situ exopolysaccharide production by lactic acid bacteria. Dairy Sci. Technol. 91, 645–661. doi: 10.1007/s13594-011-0039-0
Habu, Y., Nagaoka, M., Yokokura, T., and Azuma, I. (1987). Structural studies of cell wall polysaccharides from Bifidobacterium breve YIT 4010 and related Bifidobacterium species. J. Biochem. 102, 1423–1432. doi: 10.1093/oxfordjournals.jbchem.a122188
Han, X., Yang, Z., Jing, X., Yu, P., Zhang, Y., Yi, H., et al. (2016). Improvement of the texture of yogurt by use of exopolysaccharide producing lactic acid bacteria. Biomed. Res. Int. 2016, 1–6. doi: 10.1155/2016/7945675
Hato, T., and Dagher, P. C. (2015). How the innate immune system senses trouble and causes trouble. Clin. J. Am. Soc. Nephrol. 10, 1459–1469. doi: 10.2215/cjn.04680514
Hazards, E. P. O. B., Koutsoumanis, K., Allende, A., Alvarez-Ordóñez, A., Bolton, D., Bover-Cid, S., et al. (2022). Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 15: suitability of taxonomic units notified to EFSA until September 2021. EFSA J. 20:e07045. doi: 10.2903/j.efsa.2022.7045
Hickey, A., Stamou, P., Udayan, S., Ramón-Vázquez, A., Esteban-Torres, M., Bottacini, F., et al. (2021). Bifidobacterium breve exopolysaccharide blocks dendritic cell maturation and activation of CD4+ T cells. Front. Microbiol. 12:653587. doi: 10.3389/fmicb.2021.653587
Hidalgo-Cantabrana, C., Delgado, S., Ruiz, L., Ruas-Madiedo, P., Sánchez, B., and Margolles, A. (2018). “Bifidobacteria and their health-promoting effects” in Bugs as Drugs: Therapeutic Microbes for the Prevention and Treatment of Disease. eds. R. A. Britton and P. D. Cani (Wiley), 73–98.
Hidalgo-Cantabrana, C., López, P., Gueimonde, M., de Los Reyes-Gavilán, C. G., Suárez, A., Margolles, A., et al. (2012). Immune modulation capability of exopolysaccharides synthesised by lactic acid bacteria and bifidobacteria. Probiotics Antimicrob Proteins 4, 227–237. doi: 10.1007/s12602-012-9110-2
Hidalgo-Cantabrana, C., Nikolic, M., López, P., Suárez, A., Miljkovic, M., Kojic, M., et al. (2014a). Exopolysaccharide-producing Bifidobacterium animalis subsp. lactis strains and their polymers elicit different responses on immune cells from blood and gut associated lymphoid tissue. Anaerobe 26, 24–30. doi: 10.1016/j.anaerobe.2014.01.003
Hidalgo-Cantabrana, C., Sánchez, B., Álvarez-Martín, P., López, P., Martínez-Álvarez, N., Delley, M., et al. (2015). A single mutation in the gene responsible for the mucoid phenotype of Bifidobacterium animalis subsp. lactis confers surface and functional characteristics. Appl. Environ. Microbiol. 81, 7960–7968. doi: 10.1128/AEM.02095-15
Hidalgo-Cantabrana, C., Sánchez, B., Milani, C., Ventura, M., Margolles, A., and Ruas-Madiedo, P. (2014b). Genomic overview and biological functions of exopolysaccharide biosynthesis in Bifidobacterium spp. Appl. Environ. Microbiol. 80, 9–18. doi: 10.1128/AEM.02977-13
Inturri, R., Mangano, K., Santagati, M., Intrieri, M., Di Marco, R., and Blandino, G. (2017a). Immunomodulatory effects of Bifidobacterium longum W11 produced exopolysaccharide on cytokine production. Curr. Pharm. Biotechnol. 18, 883–889. doi: 10.2174/1389201019666171226151551
Inturri, R., Molinaro, A., Di Lorenzo, F., Blandino, G., Tomasello, B., Hidalgo-Cantabrana, C., et al. (2017b). Chemical and biological properties of the novel exopolysaccharide produced by a probiotic strain of Bifidobacterium longum. Carbohydr. Polym. 174, 1172–1180. doi: 10.1016/j.carbpol.2017.07.039
Jurášková, D., Ribeiro, S. C., and Silva, C. C. (2022). Exopolysaccharides produced by lactic acid bacteria: from biosynthesis to health-promoting properties. Food Secur. 11:156. doi: 10.3390/foods11020156
Kaur, N., and Dey, P. (2023). Bacterial exopolysaccharides as emerging bioactive macromolecules: from fundamentals to applications. Res. Microbiol. 174:104024. doi: 10.1016/j.resmic.2022.104024
Kellie, S., and Al-Mansour, Z. (2017). “Overview of the immune system” in Micro and nanotechnology in vaccine development. eds. M. S. Czynski and I. Toth (Elsevier), 63–81.
Kohno, M., Suzuki, S., Kanaya, T., Yoshino, T., Matsuura, Y., Asada, M., et al. (2009). Structural characterization of the extracellular polysaccharide produced by Bifidobacterium longum JBL05. Carbohydr. Polym. 77, 351–357. doi: 10.1016/j.carbpol.2009.01.013
Konieczna, P., Akdis, C. A., Quigley, E. M., Shanahan, F., and O’Mahony, L. (2012). Portrait of an immunoregulatory Bifidobacterium. Gut Microbes 3, 261–266. doi: 10.4161/gmic.20358
Korcz, E., and Varga, L. (2021). Exopolysaccharides from lactic acid bacteria: techno-functional application in the food industry. Trends Food Sci. Technol. 110, 375–384. doi: 10.1016/j.tifs.2021.02.014
Ku, S., Park, M. S., Ji, G. E., and You, H. J. (2016). Review on Bifidobacterium bifidum BGN4: functionality and nutraceutical applications as a probiotic microorganism. Int. J. Mol. Sci. 17:1544. doi: 10.3390/ijms17091544
Laiño, J., Villena, J., Kanmani, P., and Kitazawa, H. (2016). Immunoregulatory effects triggered by lactic acid bacteria exopolysaccharides: new insights into molecular interactions with host cells. Microorganisms 4:27. doi: 10.3390/microorganisms4030027
Lee, J. H., and O’Sullivan, D. J. (2010). Genomic insights into bifidobacteria. Microbiol. Mol. Biol. R. 74, 378–416.
Leivers, S., Hidalgo-Cantabrana, C., Robinson, G., Margolles, A., Ruas-Madiedo, P., and Laws, A. P. (2011). Structure of the high molecular weight exopolysaccharide produced by Bifidobacterium animalis subsp. lactis IPLA-R1 and sequence analysis of its putative eps cluster. Carbohydr. Res. 346, 2710–2717. doi: 10.1016/j.carres.2011.09.010
Li, J., Wang, J., Wang, M., Zheng, L., Cen, Q., Wang, F., et al. (2023). Bifidobacterium: a probiotic for the prevention and treatment of depression. Front. Microbiol. 14:1174800. doi: 10.3389/fmicb.2023.1174800
Lim, P., Loke, C., Ho, Y., and Tan, H. (2020). Cholesterol homeostasis associated with probiotic supplementation in vivo. J. Appl. Microbiol. 129, 1374–1388. doi: 10.1111/jam.14678
Liu, G., Chen, H., Chen, J., Wang, X., Gu, Q., and Yin, Y. (2019). Effects of bifidobacteria-produced exopolysaccharides on human gut microbiota in vitro. Appl. Microbiol. Biotechnol. 103, 1693–1702. doi: 10.1007/s00253-018-9572-6
Liu, L., Qin, Y., Wang, Y., Li, H., Shang, N., and Li, P. (2014). Complete genome sequence of Bifidobacterium animalis RH, a probiotic bacterium producing exopolysaccharides. J. Biotechnol. 189, 86–87. doi: 10.1016/j.jbiotec.2014.08.041
Llamas-Arriba, M. G., Peiroten, A., Puertas, A. I., Prieto, A., Lopez, P., Pardo, M. A., et al. (2019). Heteropolysaccharide-producing bifidobacteria for the development of functional dairy products. LWT 102, 295–303. doi: 10.1016/j.lwt.2018.12.044
Lopez, P., Monteserin, D. C., Gueimonde, M., de los Reyes-Gavilan, C. G., Margolles, A., Suarez, A., et al. (2012). Exopolysaccharide-producing Bifidobacterium strains elicit different in vitro responses upon interaction with human cells. Food Res. Int. 46, 99–107. doi: 10.1016/j.foodres.2011.11.020
Lynch, K. M., Zannini, E., Coffey, A., and Arendt, E. K. (2018). Lactic acid bacteria exopolysaccharides in foods and beverages: isolation, properties, characterization, and health benefits. Annu. Rev. Food Sci. Technol. 9, 155–176. doi: 10.1146/annurev-food-030117-012537
Mohd Nadzir, M., Nurhayati, R. W., Idris, F. N., and Nguyen, M. H. (2021). Biomedical applications of bacterial exopolysaccharides: a review. Polymers 13:530. doi: 10.3390/polym13040530
Nagaoka, M., Shibata, H., Kimura, I., Hashimoto, S., Kimura, K., Sawada, H., et al. (1995). Structural studies on a cell wall polysaccharide from Bifidobacterium longum YIT4028. Carbohydr. Res. 274, 245–249. doi: 10.1016/0008-6215(95)00076-6
Netrusov, A. I., Liyaskina, E. V., Kurgaeva, I. V., Liyaskina, A. U., Yang, G., and Revin, V. V. (2023). Exopolysaccharides producing Bacteria: a review. Microorganisms 11:1541. doi: 10.3390/microorganisms11061541
Nicolescu, C. M., Bumbac, M., Buruleanu, C. L., Popescu, E. C., Stanescu, S. G., Georgescu, A. A., et al. (2023). Biopolymers produced by lactic acid Bacteria: characterization and food application. Polymers 15:1539. doi: 10.3390/polym15061539
Pacyga-Prus, K., Jakubczyk, D., Sandström, C., Šrůtková, D., Pyclik, M. J., Leszczyńska, K., et al. (2023). Polysaccharide BAP1 of Bifidobacterium adolescentis CCDM 368 is a biologically active molecule with immunomodulatory properties. Carbohydr. Polym. 315:120980. doi: 10.1016/j.carbpol.2023.120980
Prasanna, P., Grandison, A., and Charalampopoulos, D. (2013). Microbiological, chemical and rheological properties of low fat set yoghurt produced with exopolysaccharide (EPS) producing Bifidobacterium strains. Food Res. Int. 51, 15–22. doi: 10.1016/j.foodres.2012.11.016
Rajoka, M. S. R., Jin, M., Haobin, Z., Li, Q., Shao, D., Jiang, C., et al. (2018). Functional characterization and biotechnological potential of exopolysaccharide produced by Lactobacillus rhamnosus strains isolated from human breast milk. LWT 89, 638–647. doi: 10.1016/j.lwt.2017.11.034
Ruiz, L., Delgado, S., Ruas-Madiedo, P., Sánchez, B., and Margolles, A. (2017). Bifidobacteria and their molecular communication with the immune system. Front. Microbiol. 8:2345. doi: 10.3389/fmicb.2017.02345
Salazar, N., Gueimonde, M., De Los Reyes-Gavilán, C. G., and Ruas-Madiedo, P. (2016). Exopolysaccharides produced by lactic acid bacteria and bifidobacteria as fermentable substrates by the intestinal microbiota. Crit. Rev. Food Sci. Nutr. 56, 1440–1453. doi: 10.1080/10408398.2013.770728
Salazar, N., López, P., Garrido, P., Moran, J., Cabello, E., Gueimonde, M., et al. (2014). Immune modulating capability of two exopolysaccharide-producing Bifidobacterium strains in a Wistar rat model. Biomed. Res. Int. 2014, 1–9. doi: 10.1155/2014/106290
Salazar, N., Prieto, A., Leal, J., Mayo, B., Bada-Gancedo, J. C., de Los Reyes-Gavilán, C., et al. (2009). Production of exopolysaccharides by Lactobacillus and Bifidobacterium strains of human origin, and metabolic activity of the producing bacteria in milk. J. Dairy Sci. 92, 4158–4168. doi: 10.3168/jds.2009-2126
Salimi, F., and Farrokh, P. (2023). Recent advances in the biological activities of microbial exopolysaccharides. World J. Microbiol. Biotechnol. 39:213. doi: 10.1007/s11274-023-03660-x
Schell, M. A., Karmirantzou, M., Snel, B., Vilanova, D., Berger, B., Pessi, G., et al. (2002). The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. 99, 14422–14427. doi: 10.1073/pnas.212527599
Schiavi, E., Gleinser, M., Molloy, E., Groeger, D., Frei, R., Ferstl, R., et al. (2016). The surface-associated exopolysaccharide of Bifidobacterium longum 35624 plays an essential role in dampening host proinflammatory responses and repressing local TH17 responses. Appl. Environ. Microbiol. 82, 7185–7196. doi: 10.1128/AEM.02238-16
Schmid, J. (2018). Recent insights in microbial exopolysaccharide biosynthesis and engineering strategies. Curr. Opin. Biotechnol. 53, 130–136. doi: 10.1016/j.copbio.2018.01.005
Schmid, J., Sieber, V., and Rehm, B. (2015). Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front. Microbiol. 6:147519. doi: 10.3389/fmicb.2015.00496
Shang, N., Xu, R., and Li, P. (2013). Structure characterization of an exopolysaccharide produced by Bifidobacterium animalis RH. Carbohydr. Polym. 91, 128–134. doi: 10.1016/j.carbpol.2012.08.012
Shukla, P. J., Nathani, N. M., and Dave, B. P. (2017). Marine bacterial exopolysaccharides [EPSs] from extreme environments and their biotechnological applications. Int J Res Biosci 6, 20–32
Singh, S., Bhatia, R., Khare, P., Sharma, S., Rajarammohan, S., Bishnoi, M., et al. (2020). Anti-inflammatory Bifidobacterium strains prevent dextran sodium sulfate induced colitis and associated gut microbial dysbiosis in mice. Sci. Rep. 10:18597. doi: 10.1038/s41598-020-75702-5
Sørensen, H. M., Rochfort, K. D., Maye, S., MacLeod, G., Brabazon, D., Loscher, C., et al. (2022). Exopolysaccharides of lactic acid bacteria: production, purification and health benefits towards functional food. Nutrients 14:2938. doi: 10.3390/nu14142938
Sun, X., and Zhang, J. (2021). Bacterial exopolysaccharides: chemical structures, gene clusters and genetic engineering. Int. J. Biol. Macromol. 173, 481–490. doi: 10.1016/j.ijbiomac.2021.01.139
Tone-Shimokawa, Y., Toida, T., and Kawashima, T. (1996). Isolation and structural analysis of polysaccharide containing galactofuranose from the cell walls of Bifidobacterium infantis. J. Bacteriol. 178, 317–320. doi: 10.1128/jb.178.1.317-320.1996
Turroni, F., Duranti, S., Milani, C., Lugli, G. A., van Sinderen, D., and Ventura, M. (2019). Bifidobacterium bifidum: a key member of the early human gut microbiota. Microorganisms 7:544. doi: 10.3390/microorganisms7110544
Wachi, S., Kanmani, P., Tomosada, Y., Kobayashi, H., Yuri, T., Egusa, S., et al. (2014). Lactobacillus delbrueckii TUA 4408 L and its extracellular polysaccharides attenuate enterotoxigenic E scherichia coli-induced inflammatory response in porcine intestinal epitheliocytes via T oll-like receptor-2 and 4. Mol. Nutr. Food Res. 58, 2080–2093. doi: 10.1002/mnfr.201400218
Wang, L., Wang, Y., Li, Q., Tian, K., Xu, L., Liu, G., et al. (2019). Exopolysaccharide, isolated from a novel strain Bifidobacterium breve lw01 possess an anticancer effect on head and neck cancer–genetic and biochemical evidences. Front. Microbiol. 10:1044. doi: 10.3389/fmicb.2019.01044
Whitfield, C., Wear, S. S., and Sande, C. (2020). Assembly of bacterial capsular polysaccharides and exopolysaccharides. Annu. Rev. Microbiol. 74, 521–543. doi: 10.1146/annurev-micro-011420-075607
Willis, L. M., and Whitfield, C. (2013). Structure, biosynthesis, and function of bacterial capsular polysaccharides synthesized by ABC transporter-dependent pathways. Carbohydr. Res. 378, 35–44. doi: 10.1016/j.carres.2013.05.007
Wu, M.-H., Pan, T.-M., Wu, Y.-J., Chang, S.-J., Chang, M.-S., and Hu, C.-Y. (2010). Exopolysaccharide activities from probiotic bifidobacterium: immunomodulatory effects (on J774A. 1 macrophages) and antimicrobial properties. Int. J. Food Microbiol. 144, 104–110. doi: 10.1016/j.ijfoodmicro.2010.09.003
Xie, Y., Ye, Z., Deng, H., Sun, W., Wan, X., He, X., et al. (2023). Screening of exopolysaccharide-producing Enterobacter aerogenes NJ1023 and its cadaverine biosynthesis promotion. Front. Microbiol. 14:1200123. doi: 10.3389/fmicb.2023.1200123
Xu, Y., Cui, Y., Yue, F., Liu, L., Shan, Y., Liu, B., et al. (2019). Exopolysaccharides produced by lactic acid bacteria and Bifidobacteria: structures, physiochemical functions and applications in the food industry. Food Hydrocoll. 94, 475–499. doi: 10.1016/j.foodhyd.2019.03.032
Xu, R., Shen, Q., Wu, R., and Li, P. (2017). Structural analysis and mucosal immune regulation of exopolysaccharide fraction from Bifidobacterium animalis RH. Food Agric. Immunol. 28, 1226–1241. doi: 10.1080/09540105.2017.1333578
Yang, X., Liang, Q., Chen, Y., and Wang, B. (2019). Alteration of methanogenic archaeon by ethanol contribute to the enhancement of biogenic methane production of lignite. Front. Microbiol. 10:2323. doi: 10.3389/fmicb.2019.02323
Yao, S., Zhao, Z., Wang, W., and Liu, X. (2021). Bifidobacterium longum: protection against inflammatory bowel disease. J. Immunol. Res. 2021, 1–11. doi: 10.1155/2021/8030297
Yatim, K. M., and Lakkis, F. G. (2015). A brief journey through the immune system. Clin. J. Am. Soc. Nephrol. 10, 1274–1281. doi: 10.2215/CJN.10031014
Yu, R., Zuo, F., Ma, H., and Chen, S. (2019). Exopolysaccharide-producing Bifidobacterium adolescentis strains with similar adhesion property induce differential regulation of inflammatory immune response in Treg/Th17 axis of DSS-colitis mice. Nutrients 11:782. doi: 10.3390/nu11040782
Yue, Y., Wang, Y., Han, Y., Zhang, Y., Cao, T., Huo, G., et al. (2023a). Genome analysis of Bifidobacterium Bifidum E3, structural characteristics, and antioxidant properties of exopolysaccharides. Food Secur. 12:2988. doi: 10.3390/foods12162988
Yue, Y., Wang, Y., Yang, R., Liu, D., Cheng, Y., Li, S., et al. (2023b). Exopolysaccharides of Bifidobacterium longum subsp. infantis E4 on the immune and anti-inflammatory effects in vitro. J. Funct. Foods 107:105699. doi: 10.1016/j.jff.2023.105699
Zannini, E., Waters, D. M., Coffey, A., and Arendt, E. K. (2016). Production, properties, and industrial food application of lactic acid bacteria-derived exopolysaccharides. Appl. Microbiol. Biotechnol. 100, 1121–1135. doi: 10.1007/s00253-015-7172-2
Zhou, Y., Cui, Y., and Qu, X. (2019). Exopolysaccharides of lactic acid bacteria: structure, bioactivity and associations: a review. Carbohydr. Polym. 207, 317–332. doi: 10.1016/j.carbpol.2018.11.093
Keywords: Bifidobacterium, exopolysaccharide, lactic acid bacteria, probiotics, immune system modulation
Citation: Sadeghi M, Haghshenas B and Nami Y (2024) Bifidobacterium exopolysaccharides: new insights into engineering strategies, physicochemical functions, and immunomodulatory effects on host health. Front. Microbiol. 15:1396308. doi: 10.3389/fmicb.2024.1396308
Edited by:
Silvani Verruck, Federal University of Santa Catarina, BrazilReviewed by:
Zhenshang Xu, Qilu University of Technology, ChinaAlberto Amaretti, University of Modena and Reggio Emilia, Italy
Copyright © 2024 Sadeghi, Haghshenas and Nami. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Babak Haghshenas, YmhhZ2hzaGVuYXMyMDEwQGdtYWlsLmNvbQ==; Yousef Nami, eW91c2VmbmFtaTIwMTBAZ21haWwuY29t
 Mahsa Sadeghi
Mahsa Sadeghi Babak Haghshenas
Babak Haghshenas Yousef Nami
Yousef Nami