- 1China Tobacco Hunan Industrial Co., Ltd., Changsha, China
- 2Hunan Provincial Engineering and Technology Research Center for Agricultural Microbiology Application, Hunan Institute of Microbiology, Changsha, China
- 3Qingdao NABT PhagePharm Co., Ltd., Qingdao, China
- 4College of Plant Protection, Hunan Agricultural University, Changsha, China
- 5Institute of Special Food, Qingdao Agricultural University, Qingdao, China
Bacterial wilt (BW) is a devastating plant disease caused by the soil-borne bacterium Ralstonia solanacearum species complex (Rssc). Numerous efforts have been exerted to control BW, but effective, economical, and environmentally friendly approaches are still not available. Bacteriophages are a promising resource for the control of bacterial diseases, including BW. So, in this study, a crop BW pathogen of lytic bacteriophage was isolated and named PQ43W. Biological characterization revealed PQ43W had a short latent period of 15 min, 74 PFU/cell of brust sizes, and good stability at a wide range temperatures and pH but a weak resistance against UV radiation. Sequencing revealed phage PQ43W contained a circular double-stranded DNA genome of 47,156 bp with 65 predicted open reading frames (ORFs) and genome annotation showed good environmental security for the PQ43W that no tRNA, antibiotic resistance, or virulence genes contained. Taxonomic classification showed PQ43W belongs to a novel genus of subfamily Kantovirinae under Caudoviricetes. Subsequently, a dose of PQ43W for phage therapy in controlling crop BW was determined: 108 PFU*20 mL per plant with non-invasive irrigation root application twice by pot experiment. Finally, a field experiment of PQ43W showed a significantly better control effect in crop BW than the conventional bactericide Zhongshengmycin. Therefore, bacteriophage PQ43W is an effective bio-control resource for controlling BW diseases, especially for crop cultivation.
1 Introduction
Bacterial wilt (BW) is one of the most destructive diseases in many solanaceous crops, such as potatoes, tobacco, tomatoes, pepper, and eggplant, and is a significant source of crop production loss worldwide (Mansfield et al., 2012; Jiang et al., 2017; Wang et al., 2023). The pathogen of BW, Ralstonia solanacearum, is a soil-borne bacterial pathogen and has long been part of the “R. solanacearum species complex” (Rssc) composed of three different Ralstonia species: R. solanacearum, R. pseudosolanacearum, and R. syzygii (Safni et al., 2014). The pathogen Rssc typically colonizes the root xylem tissues and infects the roots through small wounds, it then quickly moves to stem tissues and secretes a large amount of EPS, which causes xylem obstruction and leads to plant wilting (Jiang et al., 2017; Ingel et al., 2022). Many efforts have been engaged to control this disease (Yuliar et al., 2015; Mamphogoro et al., 2020; Wang et al., 2023), however, none of these strategies has been able to control the BW disease efficiently, due to the broad host range, lack of plant resistance cultivars, environmental pollution and genetic diversity of the pathogen, and so on (Mansfield et al., 2012; Safni et al., 2014). Therefore, how to effectively control BW remains an urgent challenge.
Bacteriophages (phages) are viruses that exclusively infect a certain bacterium and are not toxic to animals, plants, humans, or non-target microorganisms (Yuliar et al., 2015). They infect their host and exploit the metabolic processes of the host to replicate their own genome, thereby releasing more virions (Gordillo and Barr, 2019; Mamphogoro et al., 2020). So, phage therapy has attracted great attention and obtained good results, showing some advantages in the treatment of specific human or animal diseases, e.g., high lytic activity, specific targeting capabilities, cost-effectiveness, and no harm to the environment (Gordillo and Barr, 2019; Grabowski et al., 2022; Hatfull et al., 2022; Strathdee et al., 2023). Some research based on bacteriophages (phages) has shown the therapy’s great potential to control plant bacterium disease in crop cultivation (Balogh et al., 2018; Muturi et al., 2019; Marquioni et al., 2022; Villalpando-Aguilar et al., 2022). For example, phages research based on bacteriophages (phages) has shown great potential to control Xanthomonas perforans (Balogh et al., 2018). Phage vB_XciM_LucasX has been used to control citrus canker disease caused by X. citri and X. fuscans (Marquioni et al., 2022). Phage Wc5r has shown substantial results in preventing and controlling potato soft rot caused by Pectobacterium carotovorum (Muturi et al., 2019). Phages therapy to control crop bacterial diseases is also considered to be a promising measure (Álvarez and Biosca, 2017).
Interestingly, some Ralstonia-infecting bacteriophages have been isolated and achieved good results in corps cultivation against BW (Addy et al., 2018, 2019; Wang X. et al., 2019; Wang R. et al., 2019; Chen et al., 2023), such as Aresaunavirus of Ralstonia phage RsoM1USA, Higashivirus of Ralstonia phage RsoP1IDN, and Gervaisevirus of Ralstonia phage GP4. Due to the diversity of the geographic distribution of Rssc and the specificity of phage on host strains, it is very important to isolate more BW-pathogen infection phages to control the BW in crops cultivation, especially in crops with a high BW incidence rate (Wang X. et al., 2019; Liu et al., 2020; Ahmad et al., 2021; Kaur et al., 2021).
Due to the wide host nature of BW pathogen Rssc, we selected tobacco as our crop subject. Not only is tobacco is a typical Solanaceae crop and widely cultivated in China (Jiang et al., 2017), but there would also be great significance in controlling BW in tobacco cultivation as it would reduce the loss caused by BW in tobacco cultivation, reduce the BW-pathogen abundance in local agro-ecosystems, and act as a reference or guide to control BW in other crops cultivation. So, in this study, we report a lytic phage isolated from the rhizosphere soil of a tobacco BW disease plant which was collected in Hunan province of China. We characterized the morphology and biological characteristics and then sequenced and annotated the genome. We also explored the BW control efficacy of phages application in pot and field experiments. These studies are important for a better understanding of phages as a biocontrol resource against BW in crop cultivation.
2 Materials and methods
2.1 Isolation and cultivation of bacterial wilt-inducing pathogenic strains
Tobacco BW disease stem tissues were collected from a field with a high incidence rate for BW in Changsha (Hunan, China). Single colony isolation was performed according to the method employed by Okiro et al. (2022) with minor modifications. Stems with visible symptoms of bacterial wilt were collected, then, thoroughly cleaned with tap water, dried using paper towels, and sterilized with 70% ethanol. The stems were then crushed in sterile water for the bacteria to ooze from the stem tissues, and streaked in TTC media, followed by purification and identification of the bacteria. The strain was confirmed as the pathogen of tobacco BW disease by completing Koch’s rule according to the methods described by Huang et al. (2023). The isolated strain, named NdE, was stored in 25% (vol/vol) glycerol at −80°C and cultured for 16 h to 48 h at 30°C with 180 rpm shaking in nutrient broth medium (NB; Shanghai Sangon Biotech Co., Ltd., China).
The isolated strain was verified by 16S rRNA sequencing and blastn in NCBI Genbank. The phylogenetic tree based on the 16S rRNA sequence was produced by the neighbor-joining method in MEGA X, with 1,000 bootstrap replicates.
2.2 Bacteriophage isolation and purification
Bacteriophage (Phage) isolation was conducted according to the method described by Xu et al. (2021) with slight modifications. First, 1 g of fresh soil and 2 mL of host bacterial strains (OD600 = 0.5 to 1.0) were added into 100 mL sterile NB broth medium and co-cultured overnight at 28°C; the mixed culture was then centrifuged and filtered using 0.22 μm membranes to remove bacteria and to obtain phage stock solution. Secondly, the phage stock solution was used for double-layer plate assay at 100-10 dilution. Then, a single plaque was picked on a double plate and added to a new fresh host co-culture mixture for overnight culture, followed by 10,000 g, 5 min centrifugal and 0.22 μm filtration to purify the phage, and repeated three to five times to obtained the purified phages.
2.3 Morphology of the bacteriophage
Characterization of the phage morphology was conducted as described by Sasaki et al. (2021), with minor modifications. A fresh phage sample (109 PFU/ml and 0.22 μm filtered) was visualized by transmission electron microscopy (TEM) using phosphotungstic acid (PTA) with negative staining. 20 μL of the liquid sample was dropped onto a copper mesh with a carbon-coated membrane, allowed to settle naturally for 15 min, dried on filter paper, and then stained with 2% (W/V) phosphotungstic acid (PTA) for 2 min. Excess liquid was absorbed by filter paper amd dried before being examined by transmission electron microscope (TEM) observation. More than 10 phage images were estimated and calculated with the morphometrics.
2.4 Biological characteristics
The biological characteristics of phage was conducted according to the methods of Tian et al. (2022) and Huang et al. (2022), described with minor modifications.
For temperature stability, the phage stock solution with a titer of 1010 PFU/ml was incubated at 30°C, 40°C, 50°C, 60°C, 70°C, and 80°C for 1 h, 2 h, and 3 h to determine the titers by the double-plate agar method.
For pH stability, phages with a titer of 1010 PFU/ml were mixed in a series of different SM buffers (pH 2–11, adjusted using NaOH or HCl) at 30°C, and samples were taken at 1 h, 2 h, and 3 h to determine the titers by the double-plate agar method.
For UV stability, 4 mL of phage sample with a titer of 1010 PFU/ml was spread on a sterilized petri dish (d = 90 mm) and placed on a bechtop (SX-BHC-1300A2, Suzhou Suxin Environmental Technology Co., Ltd.) at a vertical distance of 40 cm under UV light (400 mW/m2,). The samples were taken for titer determination every 10 min.
For one-step growth curve, 5 mL of phage (1 × 108 PFU/ml) was mixed with 5 mL of fresh host bacterial (NdE strain OD600 = 0.863 was about 1.73 × 109 CFU/mL, dilution with sterile water) at an MOI of 1 and incubated at 30°C for 5 min with 180 rpm. Then, the samples were centrifuged at 10,000 g for 5 min at 4°C, and the pellet was re-suspended with NB; this process was repeated three times. The final suspension was added to 10 mL of NB and cultured at 30°C, 180 rpm. Phage titers were measured at 0 min, 15 min, and 30 min and followed 30 min intervals in triplicate using the double-layer agar method. The burst size was calculated by dividing the final phage titer by the initial phage titer.
All tests were repeated three times.
2.5 Sequencing and bioinformatic analysis
Genomic DNA of the phage was extracted using the Norgen Biotek Phage DNA Isolation Kit (Norgen Biotek, Canada). The concentration and purity of isolated genomic DNA was assessed according to the protocol of the kit. DNA samples were subjected to paired-end sequencing on the Illumina HiSeq sequencing platform. The short reads were assembled by SOAPdenovo (version: 2.04) and GapCloser (version: 1.12) to obtain the whole genome of phage. Phage ORFs were analyzed using GeneMarks (Besemer and Borodovsky, 2005), and functional protein-coding genes were predicted by searching against the National Center for Biotechnology lnformation Database using Diamond software (Buchfink et al., 2015). Virulence genes and antibiotic resistance genes were searched for in the VFDB (Chen et al., 2005) and CARD (McArthur et al., 2013) databases. A circle map of the phage genome was generated with CG View (Stothard and Wishart, 2005). The genome comparisons between phages were performed with Easyfig 2.2.5 (Sullivan et al., 2011). Bioinformatics tool vContact3 (v 0.0b39; database v220; accessed on May 9, 2024; https://bitbucket.org/MAVERICLab/vcontact3/src/master/) and VICTOR (Virus Classification and Tree Building Online Resource with the amino acid option selected, Available online: https://ggdc.dsmz.de/victor.php#) (accessed on May 10, 2024) were used for phage taxonomic classification (Gendre et al., 2022; Zhu et al., 2022). Intergenomic nucleotide sequence similarity were aligned and plotted by VIRIDIC with recommended configurations (Moraru et al., 2020). Phage genome-wide nucleic acid sequence and three “signature gene products” (tail fiber protein, portal protein, and terminase large subunit protein) of amino acid sequences were used for phylogenetic analysis by MEGA with Maximum Likelihood method (Kumar et al., 2018).
2.6 Assessment of PQ43W activity against tobacco BW
2.6.1 Pot experiment
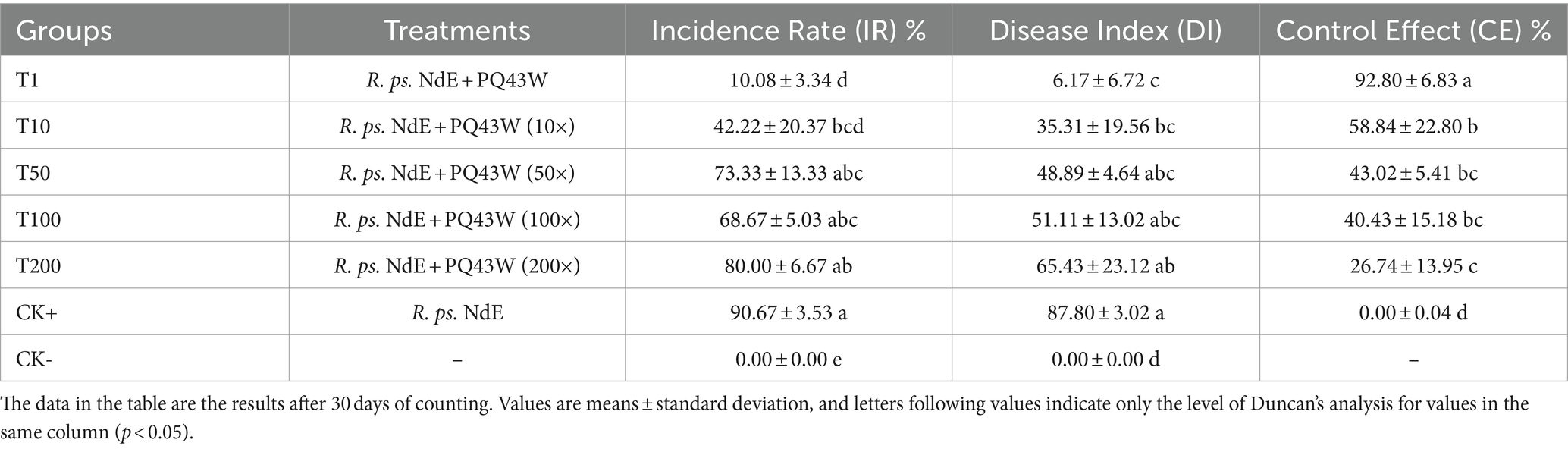
A pot experiment was conducted to evaluate the efficacy of bacteriophage against tobacco BW disease and to determine the optimal dose. The pot experiment was carried out according to the method described by Le et al. (2020) and Eisfeld et al. (2022) with slight modifications. Tobacco seedlings were transplanted to a pot at the four-leaf stage, one seedling was used per pot and 500 g nursery substrate was added (Xiang Hui Agriculture Co., Ltd., China). After seedlings rooted, the experiments were grouped and performed based on Table 1; each group had three replications (18 seedlings per replication). BW pathogen R. ps. NdE (105 CFU/mL * 20 mL per seedling) was inoculated with BW by non-invasive root irrigation. After the pathogen inoculated for 6 h and 72 h, the phage agents (108 PFU/ml) with different dilutions (0, 10, 50, 100, and 200) were used with the same method (non-invasive root irrigation, 20 mL per seedling) followed by cultivation in a suitable climate. The temperature was 28 ± 3°C, humidity 60%–90%, 14 h light-10 h dark, and full spectrum 6,000 lm. Sterile water (about 25 mL/pot) was added to the pot every 2–5 days to maintain the humidity of the nursery substrate in pot at 25%–40%.
After 30 days of pathogen inoculation, the BW incidence rate, disease index, and relative control effect of phage agents were calculated according to the GB/T23222-2008 (Grade and investigation method of tobacco diseases and insect pests) (National public service platform for standards information, n.d.). The tobacco plant BW disease classification level was assessed on a scale of 0–9: 0 = no symptoms; 1 = half of the leaves on the diseased side were wilted or withered; 3 = black strip on stem, but not more than half of the stem height, or half to two-thirds of the diseased side leaves withered; 5 = the black strip on the stem exceeds half of the stem height, but does not reach the top of the stem, or more than two thirds of the leaves on the diseased side are wilted; 7 = the black spot on the stem reaches the top of the stem, or the leaves of the diseased plant are all withered; and 9 = a dead plant.
The BW incidence rate (IR) was the BW disease plants ratio; the disease index (DI) and control effect (CE) were calculated with Eqs 1, 2.
The calculation formula of DI and CE is as follows:
DI: disease index; n1–n5: the number of plants in each disease classification level; n: the number of plants investigated; CE: control effect (T versus C); C: disease index of control group; T: disease index of treatment group.
2.6.2 Field experiment
The experimental field is located in a tobacco field (28°13’ N, 112°32′ E) in Yuxin Village, Shuangfu Town, Ningxiang City, Hunan Province, China. The area has a subtropical humid monsoon climate; in the tobacco growing season (March to July), the average monthly temperature and average monthly precipitation were 21.6°C and 168 mm. The experimental site was 56 m above sea level, and the soil was brown soil with a pH value of 6.13, organic matter 17.84 g/kg, total nitrogen 1.29 g/kg, alkaline nitrogen 83.14 mg/kg, available phosphorus 113.91 mg/kg, and available potassium 254.36 mg/kg. The tobacco cultivar Yunyan 87 was tested. The tobacco seedlings were transplanted at the five-leaf seedling stage on March 20. The experiment was divided into three groups, namely the control group (FCK) without treatment, the General fungicide group (FZS), where a chemical fungicide – Zhongshengmycin (Yi et al., 2022) – was applied according to the recommended dosage, and the Phage treatment group (FPQ43W), where phage was used as 108 PFU/ml-20 mL-plants. All the agents were applied twice by non-invasive root irrigation on 20 May and 20 June. The disease survey was carried out on July 10 (about a week that before the tail leaves were harvested) according to the method described in the pot experiment above.
2.7 Statistical analysis
The test data were processed by Excel; SPSS software was used for statistical analysis. Duncan’s method of new repolarization (DMRT) was used to test the significant differences.
3 Results
3.1 Isolation and morphological characterization of a lytic bacteriophage
3.1.1 Isolation and characterization of a bacterial wilt pathogen
The bacteria, as shown in Figure 1A, had red smooth colonies on TTC plates. The bacteria were isolated from tobacco BW disease stem with bacterial ooze by plate streaking in Hunan, China, and named NdE. A blastn analysis in NCBI showed that strain NdE had >99% similarity with the bacteria Ralstonia solanacearum species complex (Rssc) (Supplementary Table S1). A phylogenetic analysis showed that strain NdE was clustered into a cluster with Ralstonia pseudosolanacearum (NR_134148.1) and placed into Rssc clade (Supplementary Figure S1). Therefore, the bacteria NdE identified as Ralstonia pseudosolanacearum is a member of Rssc.
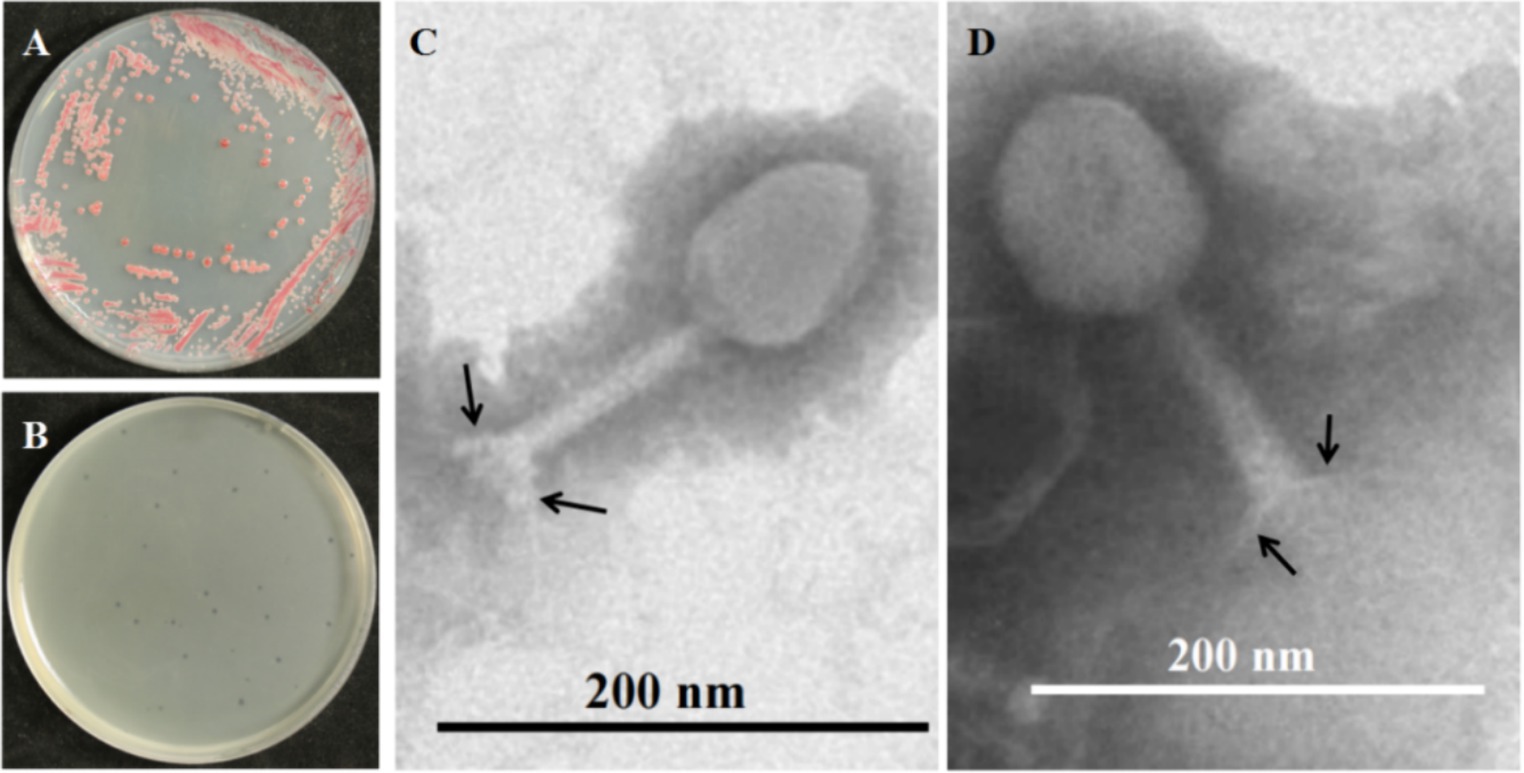
Figure 1. Morphology of BW pathogen Ralstonia pseudosolanacearum NdE (R. ps. NdE) and its phage PQ43W. (A) Colony morphology of R. ps. NdE on TTC plate; (B) formed clear plaques on a lawn of R. ps. NdE double-plates; (C,D) Virions displaying typical polygonal heads, long tails, and tail fibers (arrows) of the Class Caudoviricetes by TEM.
3.1.2 Isolation and characterization of a lytic bacteriophage
The phage was isolated from rhizosphere soil, which was collected from a field in Hunan Province, China with a high incidence of tobacco BW. The phage produced small and clear plaques with a diameter of approximately 1–2 mm after overnight incubation at 28°C on the top layer containing 0.75% agar of a double-layered NB plate using R. ps. NdE strain as a host (Figure 1B); the phage was named Ralstonia phage PQ43W. Transmission electron microscopy (TEM) was used to visualize the morphology and for classification (Figures 1C,D). The phage has a polygonal head with an average capsid diameter of 86 ± 4 nm (n = 10), a long tail with a length of 123 ± 3 nm and a width of 18 nm ± 3 nm (n = 10), and a baseplate and tail fibers of approximately 20 ± 2 nm (n = 10) in length (Figure 1B). Based on the morphology and structural features of the phage virions, it is typical for members of class Caudoviricetes, according to the standard of the International Committee on Taxonomy of Viruses (ICTV).
3.2 Biological characteristics
The exploitation of the phage as an antibacterial agent requires a wide range of environmental adaptations including tolerance to high temperatures, acid-alkaline environments, and ultraviolet radiation. After incubation at 30°C, 40°C, and 50°C, no significant change in the phage number was observed (Figure 2A). However, after incubation at 60°C for 1 h, 2 h, and 3 h, the phage titer decreased by 0.96 log, 2.21 log, and 3.15 log, respectively (Figure 2A). A remarkable decrease in phage titer after incubation at 70°C was observed (4.87, 3.78, and 1.94 units log after 1 h, 2 h, and 3 h, respectively). Similarly, the phage titer fell below 1 log when treatment was carried out at 80°C (p < 0.05). These results suggest that phage PQ43W is only stable at temperatures <50°C.
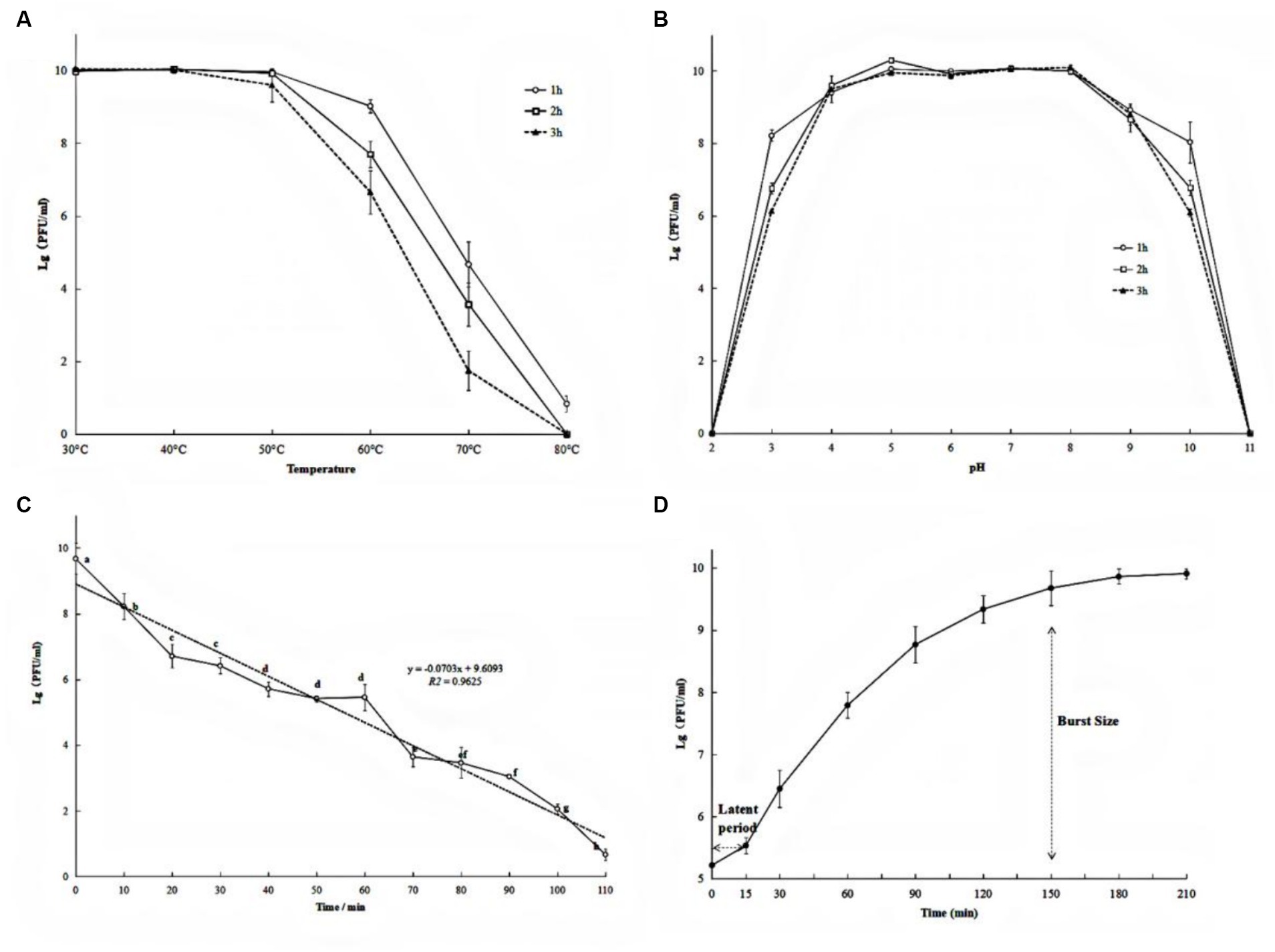
Figure 2. Biological characteristics of phage PQ43W on thermal stability (A), pH (B), UV (C) tolerance, and one-step growth curve (D). Values are the mean of three determinations.
With respect to pH sensitivity, phage PQ43W was fairly stable in the range of pH 4 to 9 (Figure 2B). In particular, the titer changed less than 0.5 log units at pH 5–8, regardless of whether the incubation time was 1 h, 2 h, or 3 h. However, at pH 3 and 10, the phage titer decreased sharply, decreasing by 2.02 to 3.95 and 1.84 to 3.91 unit log, respectively, when the incubation time was 1–3 h. This indicated that phage PQ43W has a broad pH tolerance, especially in the range of pH 5 to 8.
The results of UV tolerance experiments demonstrated that the phage titer decreased sharply after exposure to UV irradiation (Figure 2C). After 60 min of UV irradiation, the titer of phage PQ43W decreased by about half the log units to only 5.46 unit log. As the UV exposure time increased to 110 min, the phage titer fell to 1 log.
To further understand the life cycle and infection kinetics of PQ43W, a one-step growth curve using a MOI of 1 to infect host R. ps. NdE cultures was examined and shown in Figure 2D. The results showed (Figure 2D) a latent period of 15 min and a rapid growing period of 15–150 min, with burst sizes of 74.20 ± 18.88 PFU/infected cell at periods of 150 min.
3.3 Genomic analysis
3.3.1 Genomic features
Genome sequencing showed that the phage PQ43W genome was dsDNA and circular, and the complete genomic DNA sequence (GenBank: PP405626) indicated that its genome was 47,156 bp long with guanine-cytosine content of 64.27%. The contents of G, C, A, and T were 32.45, 31.82, 17.73, and 18.00%, respectively. It has 65 predicted open reading frames (ORF1–ORF65); 39 ORFs were encoded on the plus strand and 26 ORFs were encoded on the minus strand (Figure 3; Supplementary Table S2). The total length of the ORFs sequence was 42,516 bp and the average length was 654 bp per ORFs. The coding percentage was 90.16%.

Figure 3. Circle map of phage PQ43W genome. GC skew and GC content are shown in the inner circle. ORFs are visualized in the outer circle with arrows indicating the transcription direction.
3.3.2 Gene annotation of predicted proteins
A total of 65 potential open reading frames (ORFs) were identified (Figure 3; Supplementary Table S2). Of them, seven had no significant similarity with any of the protein sequences in the searched databases and 36 were annotated as conserved, conserved hypothetical, or unnamed proteins with no assigned functions. Only 29 were predicted proteins that shared homology with other phages or bacteria with assigned functions based on KEGG analysis (Table 2). Of them, there were six phage-associated DNA replication and repair proteins, including RusA-like Holliday junction resolvase (ORF22), exonuclease (ORF24), DNA primase (ORF25), DNA helicase (ORF33), DNA polymerase (ORF34), and adenosine (5′)-pentaphospho-(5″)-adenosine pyrophosphohydrolase (ORF40). There were two phage-associated lysis proteins: holin (ORF16) and lysozyme (ORF17). There was one phage-associated packaging gene: terminase large subunit (ORF64). There was one phage-associated transcriptional regulator: phelix-turn-helix transcriptional regulator (ORF26). There were 19 phage-associated structural genes proteins, such as baseplate wedge subunit (ORF1) and virion structural protein (ORF2).
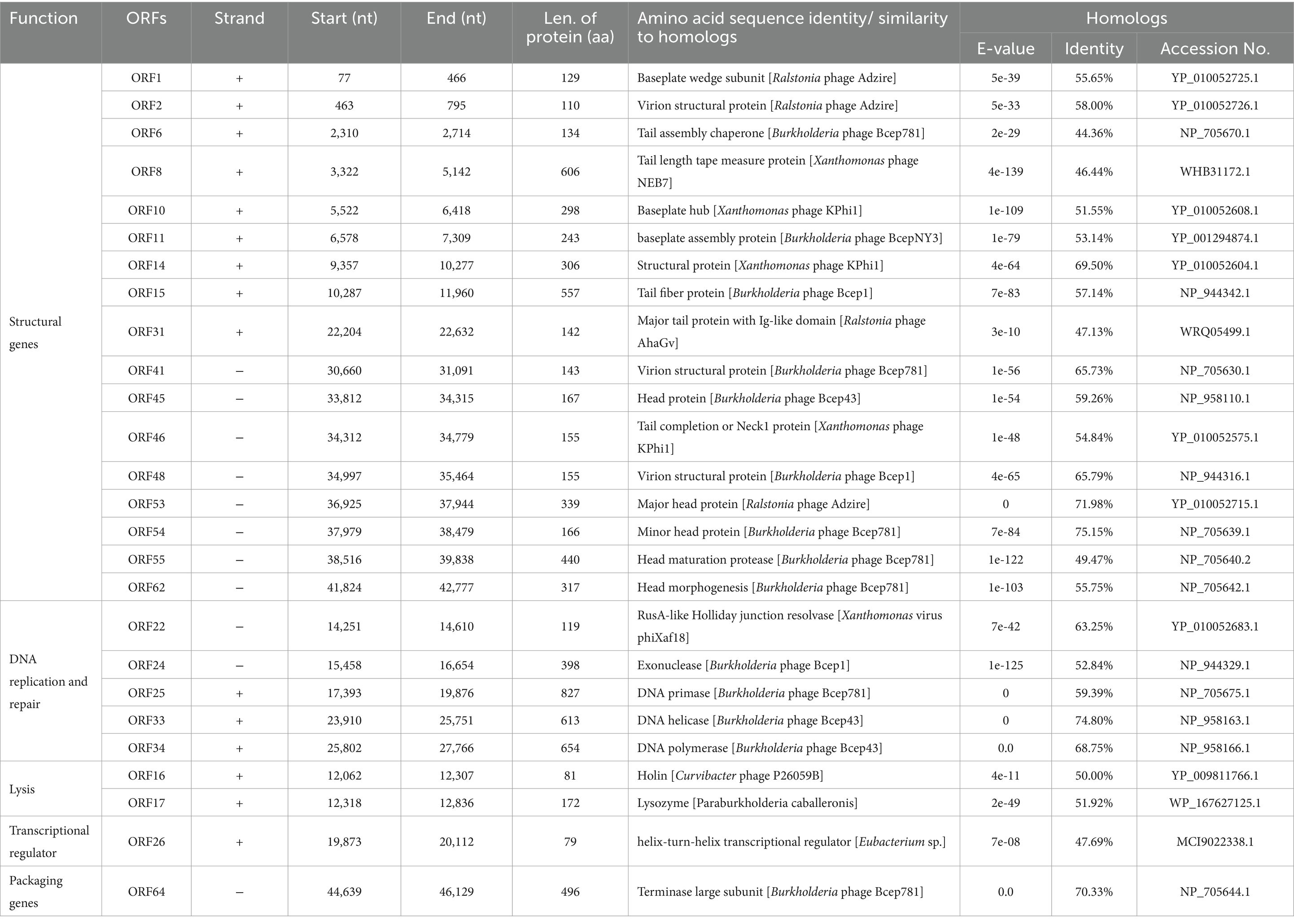
Table 2. List of annotated ORFs of phage PQ43W with predicted functions and proteins and their BLASTp results.
3.3.3 Comparison genome
Figure 4 shows a blastn search using the nucleotide in NCBI Genbank that was homologous to the Burkholderia phage Bcep43 (NC_005342.2, 16% query coverage, and 76.55% identity) and Burkholderia phage BcepNY3 (NC_009604.1, 9% query coverage and 76.52% identity) with the highest score. It also shows the genome comparison analysis of Bcep43 and BcepNY3. The gene arrangement structure of PQ43W was significantly different from that of Bcep43 (Supplementary Table S3) and BcepNY3 (Supplementary Table S4). However, there were some similarities in some regional sequences (such as ORF1 ~ ORF15; ORF22 ~ ORF24; ORF25; ORF33 ~ ORF36; ORF40 ~ ORF65). Compared with Bcep43 or BcepNY3, the gene arrangement structure is very different, and the genes with the same function had different distribution positions and directions in the genome.
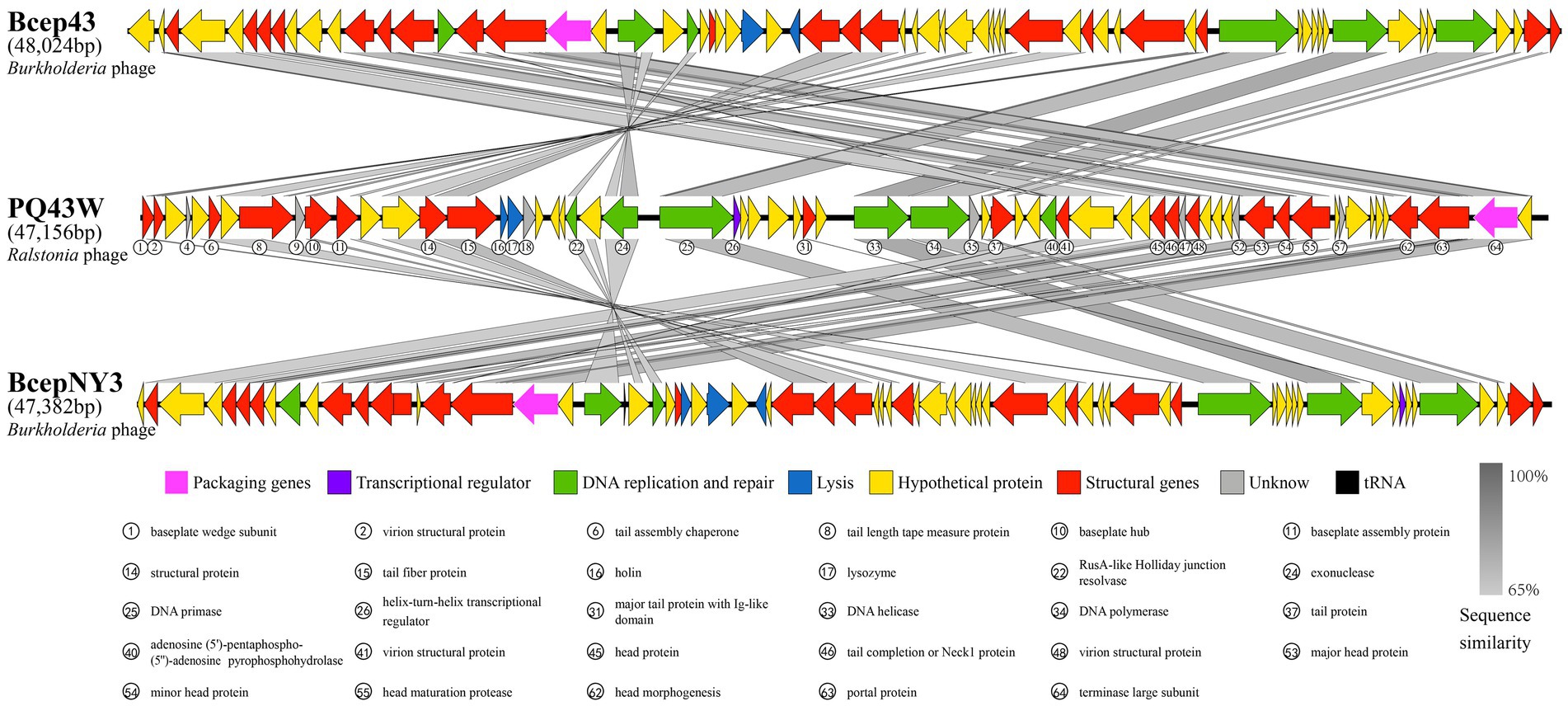
Figure 4. Comparison of the complete genome of PQ43W and two related phages, Burkholderia phage Bcep43 (Supplementary Table S3) and Burkholderia phage BcepNY3 (Supplementary Table S4). Gray bands show similarity in nucleotide sequences, arrows represent open reading frames, and the different colors of ORFs indicate the functional classification of the genes.
3.3.4 Taxonomic and phylogenetic analysis
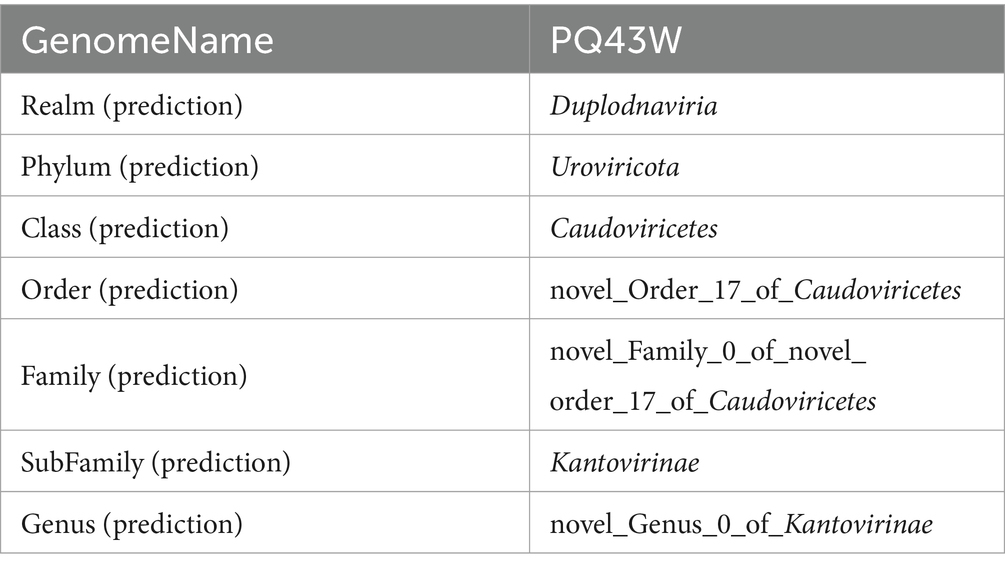
As further taxonomic information was not confirmed by the morphological identification, we resorted to genome-wide analyses at the protein level to clarify further taxonomic classification information with vContact3. The results in Table 3 show that PQ43W belongs to a novel genus of subfamily Kantovirinae (Table 3; Supplementary Table S5).
In addition, the classification with VICTOR using the “amino acid” option analysis to build phylogenetic trees was refined and constructed. Since the reference sequence of phylogenetic trees is directly related to the analysis result, it is very important to select appropriate, taxonomically close and representative sequences. So, in this study the reference sequence for VICTOR analysis were from five parts: (i) all the phages in subfamily Kantovirinae classification in ICTV Taxonomy Browser (https://ictv.global/taxonomy; two genera, five species, five phages); (ii) all the phages at subfamily Kantovirinae classification in NCBI taxonomy browser (https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=2946634; two genera, five species, 13 of no rank, 18 phages); (iii) all the phages at subfamily Kantovirinae (Prediction) classification by vContact3 (database v220) (Supplementary Table S5; four genera, 12 phages.); (iv) Top 12 phages with the highest similarity in NCBI blastn with PQ43W genome-wide sequence (Supplementary Table S6; three genera, 12 phages); (iv) two Ralstonia phages were not in the subfamily Kantovirinae classification in ICTV and NCBI taxonomy database (Supplementary Table S6, Genus Cimandefvirus) and were used as outgroups. Subsequently, 32 phages were selected and listed in Supplementary Table S7 for further analysis.
The VICTOR analysis of phylogenomic GBDP trees inferred with the formulas D0, D4, and D6 were shown in Supplementary Figures S2–S4, respectively. Then, a summarized result based on the prediction taxonomy was shown in Supplementary Table S8. The prediction taxonomic of PQ43W showed the same results at the Phylum, Subfamily, and Genus classifications in formulas D0 and D6. PQ43W belongs to a novel Subfamily and novel Genus that does not have the same taxonomic phage at Subfamily and Genus taxonomic classification levels. In the formula of D4, the prediction taxonomic showed PQ43W with Ralstonia phage Eline (NC 054957.1) and Ralstonia phage Gerry (NC 054959.1) has the same taxonomic classification at Genus Cimandefvirus (Family: unknown; Subfamily: unknown). Considering the yielding average supports were 73, 69, and 76% at the formulas D0, D4, and D6, respectively, the prediction taxonomic results of PQ43W inferred in formulas D0 and D6 were more accurate than D4. In addition, eight phage predictions in Subfamily Kantovirinae in vContact3 also appeared in the prediction Family classification through VICTOR analysis with formulas D0 and D6, but the results analyzed with formula D4 was not. That also indicated the formulas D0 and D6 were more suitable for PQ43W by VICTOR analysis than the formula D4. So, PQ43W may belong to a novel Genus in Subfamily Kantovirinae.
A comparative analysis of genomic nucleotide similarity by VIRDIC was shown in Figure 5 and Supplementary Table S8 with 32 other related phages (Supplementary Table S7). The results of intergenomic similarity between PQ43W and these phages were 0.1–35.0% (Figure 5), and the species and genus clusters table is different from the 32 related phages (Supplementary Table S9). Based on the principle of classifying bacteriophage on genus in Bacterial and Archaeal Viruses Subcommittee (BAVS), a genus is a cohesive group of viruses sharing a high degree of nucleotide sequence similarity (>50%) that is distinct from viruses of other genera (Adriaenssens and Brister, 2017). So, PQ43W belongs to a novel genus in the class Caudoviricetes.
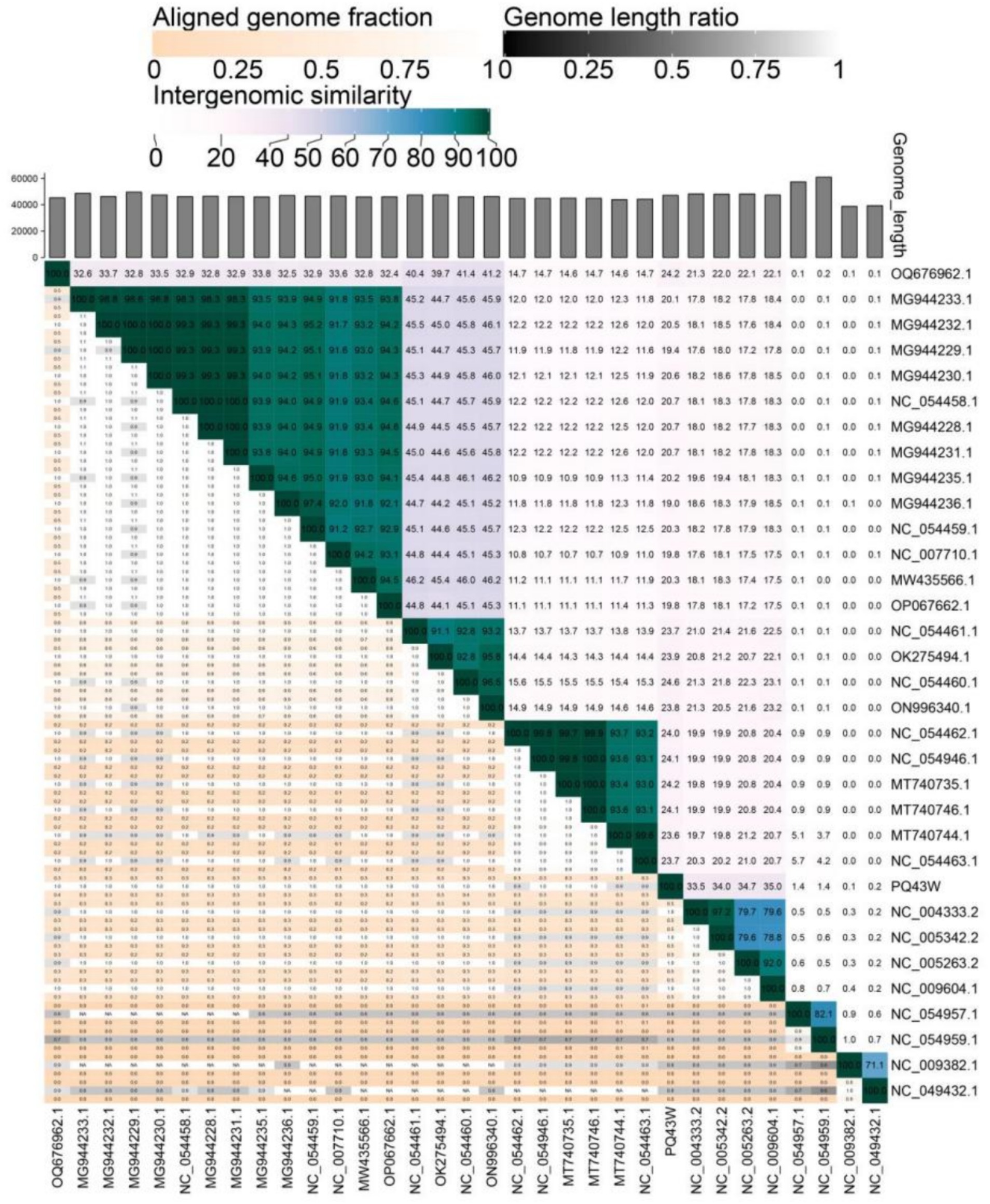
Figure 5. Intergenomic similarity between PQ43W and its 32 related phages, calculated using VIRIDIC. The right half of this heatmap shows the similarity values between phage genomes. The left half of this heatmap shows the aligned genome fraction and genome length ratio.
Therefore, PQ43W belongs to a novel genus of subfamily Kantovirinae under class Caudoviricetes.
To determine evolutional relationships between PQ43W and its 32 related phages (Supplementary Table S7), a phylogenetic tree based on the genome-wide nucleic acid sequence showed that PQ43 is adjacent to the genera of Naesvirus, Bakolyvirus, Beograduvirus, and Tsukubavirus, but distinct from them, in a separate clade (Figure 6A). In addition, the phylogenetic tree based on the three “signature gene products (tail fiber protein, portal protein, and terminase large subunit)” (Addy et al., 2019; Khan et al., 2021; Sasaki et al., 2021) of amino acid sequences showed that PQ43W is more closely related to the clade genera of Bakolyvirus (Figure 6B), Naesvirus (Figure 6C), or Aresaunavirus (Figure 6D), but is not in their clades. So, PQ43W has closer evolutional relationships to the genera Naesvirus, Bakolyvirus, Beograduvirus, Tsukubavirus, or Aresaunavirus in the phylogenetic analysis, though PQ43W belongs to a novel genus and was placed in a clade alone in Figure 6.

Figure 6. The phylogenetic trees were constructed using maximum likelihood (ML) method by MEGA, based on the genome-wide nucleic acid sequence (A) and three “signature gene products” that annotated tail fiber protein (B), portal protein (C), and terminase large subunit protein (D). Vertical distances are arbitrary, but the horizontal branches are proportional to genetic distance. Bootstrap values (1,000 replications) are represented at the nodes of the branches. Taxonomic classification of genera were labeled in the right of phylogenetic trees. The cluster of PQ43W was shown in bold.
3.4 Phage therapy
3.4.1 Pot experiment
A pot experiment to address phage application for the control of tobacco BW disease caused by R. ps. NdE was conducted. An obvious control effect result on tobacco BW disease was surveyed and statistically analyzed after 30 days (Supplementary Figure S6; Table 4). It is easily observed in Supplementary Figure S6 that PQ43W could decrease the tobacco BW disease incidence rate (IR) and disease index (DI), and was consistent with the disease statistics result (Table 4). The PQ43W application groups showed a lower BW incidence rate (IR) and disease index (DI) to compared with the control group (CK+) in Table 4. On the whole, a higher titer of PQ43W application groups showed a better control effect on tobacco BW that the IR and DI indices were lower, especially the groups T1 and T10 had more than 50% CE on the control of tobacco BW for 30 days. What’s importantly, group T1 of the CE index increased to 92.80% for 30 days by root irrigation application at 108 PFU/ml titers significantly.
3.4.2 Field experiment
Subsequently, a field experiment was conducted and the result showed in Table 5, based on the optimal dose of pot experiment (Table 4; T1: 108 PFU/ml). Compared to the control treatment (FCK), PQ43W treatment (FPQ43W) had a lower Incidence Rate (IR) that was significantly reduced from 69.33% to 30.00% (p < 0.05). The Disease Index (DI) was also significantly lower and reduced from 17.19 to 3.78% (p < 0.05). In addition, the Control Effect (CE) of PQ43W treatment (FPQ43W: 78.02%) was significantly better than the traditional pesticide Zhongshengmycin treatment (FZS: 44.41%), (p < 0.05). At the same, the indexes of IR and DI in FPQ43W were lower than FZS, significantly (p < 0.05). Therefore, PQ43W of phage therapy to control BW in pot and field experiments indicated that PQ43W is a promising resource in controlling tobacco BW disease.
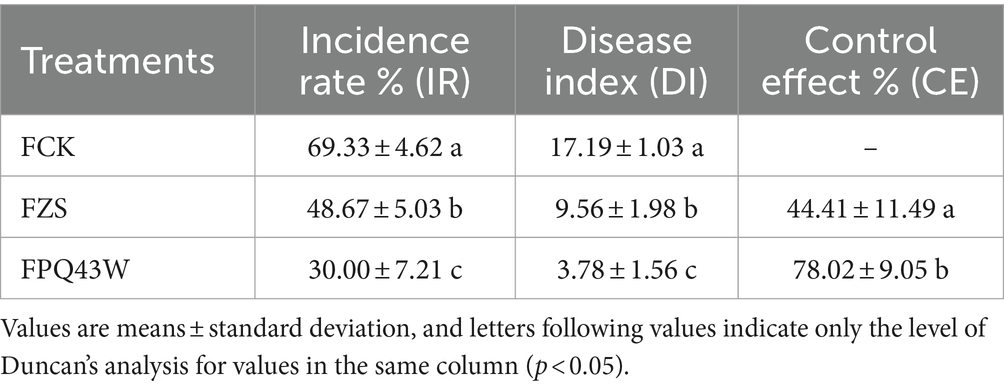
Table 5. Effects of different treatments on controlling the tobacco bacterial wilt in field experiment.
4 Discussion
Many studies have reported that microorganisms isolated from soil environments with the same or similar environments could control related plant disease effectively (Gill and Hyman, 2010; Abdelsattar et al., 2022; Ali et al., 2023). Because the soil in tobacco fields contains bacteria and phage, the BW pathogen and its specific lytic phage were isolated from the rhizosphere soil of tobacco plants in Ningxiang, Hunan Province, and then applied to this area. A good effect in controlling tobacco BW disease was observed. It is important to control outbreaks of tobacco BW to control the spread and migration of the BW pathogen to other crops in this area. It is also significant to provide guidance for scientific isolation, screening, and phage use to control crop bacterial diseases (Holtappels et al., 2021).
The possible adverse environment factors of phage application as a biological agent for plant disease control are mainly temperature, pH, and ultraviolet light. Many studies have confirmed that these factors have a significant effect on the activity of phages (Gill and Hyman, 2010; Abdelsattar et al., 2022). PQ43W showed a strong thermal and pH stability at temperatures no more than 50°C (Figure 2A), with a pH range of 5 to 8 (Figure 2B). However, the UV stability was poor (Figure 2C), which means the environment of phage agents on application or storage should avoid ultraviolet irradiation. The lysis period of phage PQ43W was about 150 min and the burst size was about 74 PFu/cell, which is conducive to the rapid killing of the target bacterial (Figure 2D). Although the biological characteristics of PQ43W are not outstanding, and they are similar to Ralstonia phage vB_RsoP_BMB50 (Wang et al., 2022) and Ralstonia phage RsPod1EGY (Elhalag et al., 2018), its characteristic of temperatures no more than 50°C, wide pH range stability, and fast lysis capacity make it suitable for most plant-growing environments. This experiment has also enriched phage resource research in controlling crop BW disease.
Because of the phage advantages of specificity, high efficiency, and specificity, it can efficiently eliminate a certain type of bacteria in the environment without affecting other microorganisms (Gill and Hyman, 2010; Hatfull et al., 2022). However, several environmental safety risks should be considered in phage application, especially the transfer or drift of antibiotic resistance genes, virulence genes, and other genetic factors (Liu et al., 2022; Zalewska-Piątek, 2023). The genomic analysis results showed that there was no tRNA, antibiotic resistance, virulence, or solubility genes contained in the PQ43W genome (Table 2). So, PQ43W is a good phage resource in terms of environmental safety.
Some studies reported that the dosage and timings of phage agents could affect the effectiveness of phage therapy (Balogh et al., 2010; Ye et al., 2019). Similarly, the results of the pot experiment showed that phage PQ43W had a higher suppression effect on tobacco BW in higher titer application groups, especially the T1 group (108 PFU/ml), which had a 97.5% control effect on tobacco BW suppression within 30 days, and which was significantly higher than the 200-fold diluted group T200 (26.74%) (p < 0.05) (Supplementary Figure S5; Table 3). The field experiment in this study showed consistently that PQ43W had a significant suppression effect on tobacco BW disease compared with the conventional bactericide Zhongshengmycin (Table 5). In addition, Álvarez et al. (2019) demonstrated that phages used through irrigated water to control tomato BW could significantly reduce the number of pathogens within hours. This is because the presence of water is conducive to the spread and migration of phage. Therefore, diluting or adding water for irrigation to provide a water-rich environment may be more effective in controlling plant bacterial diseases.
In this study, PQ43W showed a good effect in controlling tobacco BW disease in pot and field experiments. Considering the diversity of pathogens at species taxonomy classification under natural conditions, the complexity of pathogenic factors, and the diversity of climatic and environmental factors, in order to enhance the effect of phage in controlling crop bacterial disease, “phage cocktail therapy” should be employed, which is the combined use of phages with beneficial microorganisms and / or other measures, such as cultivars, agronomic practice, and soil disinfection. This method is considered to be more effective in scientific control of crop bacterial disease outbreaks (Kumar et al., 2018; Wang X. et al., 2019; Yang et al., 2023).
5 Conclusion
In this study, a bacteriophage, PQ43W, was isolated from the rhizosphere soil of tobacco bacterial wilt (BW) disease plants. It was shown to specifically lyse the tobacco BW pathogen R. ps. NdE, a member of Rssc. PQ43W had good thermal and pH stability between 30°C and 50°C and pH range 4 to 8 but a poor UV radiation stability. One-step growth curve revealed PQ43W had a short latent period of 15 min and 74 PFU/cell of brust sizes. Sequencing revealed phage PQ43W contained a circular double-stranded DNA genome of 47,156 bp with 65 predicted open reading frames (ORFs), with no tRNA, antibiotic resistance, or virulence genes annotated. Taxonomic classification revealed it belonged to a novel genus of subfamily Kantovirinae under Caudoviricetes. The phage therapy of PQ43W in pot and field experiments proved that PQ43W is an effective phage when used via non-invasive root irrigation for controlling BW diseases, especially for tobacco cultivation.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
BH: Methodology, Writing – original draft, Writing – review & editing. LG: Methodology, Validation, Writing – original draft. DX: Investigation, Methodology, Resources, Writing – original draft. GT: Investigation, Methodology, Writing – original draft. LL: Investigation, Methodology, Writing – original draft. LY: Investigation, Methodology, Writing – original draft. YJ: Investigation, Methodology, Writing – original draft. QL: Conceptualization, Methodology, Supervision, Writing – original draft, Writing – review & editing. WC: Methodology, Writing – original draft, Writing – review & editing. YL: Data curation, Visualization, Writing – original draft. HH: Methodology, Writing – original draft. HS: Data curation, Writing – review & editing. QP: Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing. KY: Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Research and application of new green prevention and control technology of main root diseases in tobacco growing district of Northwest Hunan, grant number KY2023YC0009.
Conflict of interest
BH, DX, GT, LL, LY, YJ, and KY were employed by China Tobacco Hunan Industry Co., Ltd. LG, HS, and QP were employed by Qingdao NABT PhagePharm Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1396213/full#supplementary-material
References
Abdelsattar, A., Dawoud, A., Makky, S., Nofal, R., Aziz, R. K., and El-Shibiny, A. (2022). Bacteriophages: from isolation to application. Curr. Pharm. Biotechnol. 23, 337–360. doi: 10.2174/1389201022666210426092002
Addy, H. S., Ahmad, A. A., and Huang, Q. (2019). Molecular and biological characterization of Ralstonia phage RsoM1USA, a new species of P2virus, isolated in the United States. Front. Microbiol. 10:267. doi: 10.3389/fmicb.2019.00267
Addy, H. S., Farid, M. M., Ahmad, A. A., and Huang, Q. (2018). Host range and molecular characterization of a lytic Pradovirus-like Ralstonia phage RsoP1IDN isolated from Indonesia. Arch. Virol. 163, 3409–3414. doi: 10.1007/s00705-018-4033-1
Adriaenssens, E., and Brister, J. R. (2017). How to name and classify your phage: an informal guide. Viruses 9:70. doi: 10.3390/v9040070
Ahmad, A. A., Addy, H. S., and Huang, Q. (2021). Biological and molecular characterization of a jumbo bacteriophage infecting plant pathogenic Ralstonia solanacearum species complex strains. Front. Microbiol. 12:741600. doi: 10.3389/fmicb.2021.741600
Ali, S., Tyagi, A., and Bae, H. (2023). Plant microbiome: an ocean of possibilities for improving disease resistance in plants. Microorganisms 11:392. doi: 10.3390/microorganisms11020392
Álvarez, B., and Biosca, E. G. (2017). Bacteriophage-based bacterial wilt biocontrol for an environmentally sustainable agriculture. Front. Plant Sci. 8:1218. doi: 10.3389/fpls.2017.01218
Álvarez, B., López, M. M., and Biosca, E. G. (2019). Biocontrol of the major plant pathogen Ralstonia solanacearum in irrigation water and host plants by novel waterborne lytic bacteriophages. Front. Microbiol. 6:2813. doi: 10.3389/fmicb.2019.02813
Balogh, B., Jones, J. B., Iriarte, F. B., and Momol, M. T. (2010). Phage therapy for plant disease control. Curr. Pharm. Biotechnol. 11, 48–57. doi: 10.2174/138920110790725302
Balogh, B., Nga, N. T. T., and Jones, J. B. (2018). Relative level of bacteriophage multiplication in vitro or in the phyllosphere may not predict in planta efficacy for controlling bacterial leaf spot on tomato caused by Xanthomonas perforans. Front. Microbiol. 18:2176. doi: 10.3389/fmicb.2018.02176
Besemer, J., and Borodovsky, M. (2005). Gene mark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33, W451–W454. doi: 10.1093/nar/gki487
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Chen, K., Guan, Y., Hu, R., Cui, X., and Liu, Q. (2023). Characterization of the LysP2110-HolP2110 lysis system in Ralstonia solanacearum phage P2110. Int. J. Mol. Sci. 24:10375. doi: 10.3390/ijms241210375
Chen, L., Yang, J., Yu, J., Yao, Z., Sun, L., Shen, Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328. doi: 10.1093/nar/gki008
Eisfeld, C., Schijven, J. F., Kastelein, P., van Breukelen, B. M., Medema, G., Velstra, J., et al. (2022). Dose-response relationship of Ralstonia solanacearum and potato in greenhouse and in vitro experiments. Front. Plant Sci. 13:1074192. doi: 10.3389/fpls.2022.1074192
Elhalag, K., Nasr-Eldin, M., Hussien, A., and Ahmad, A. (2018). Potential use of soilborne lytic Podoviridae phage as a biocontrol agent against Ralstonia solanacearum. J. Basic Microbiol. 58, 658–669. doi: 10.1002/jobm.201800039
Gendre, J., Ansaldi, M., Olivenza, D. R., Denis, Y., Casadesús, J., and Ginet, N. (2022). Genetic Mining of Newly Isolated Salmophages for phage therapy. Int. J. Mol. Sci. 23:8917. doi: 10.3390/ijms23168917
Gill, J. J., and Hyman, P. (2010). Phage choice, isolation, and preparation for phage therapy. Curr. Pharm. Biotechnol. 11, 2–14. doi: 10.2174/138920110790725311
Gordillo, A. F. L., and Barr, J. J. (2019). Phage therapy in the Postantibiotic era. Clin. Microbiol. Rev. 32, e00066–e00018. doi: 10.1128/CMR.00066-18
Grabowski, Ł., Węgrzyn, G., Węgrzyn, A., and Podlacha, M. (2022). Highly different effects of phage therapy and antibiotic therapy on immunological responses of chickens infected with Salmonella enterica serovar Typhimurium. Front. Immunol. 13:956833. doi: 10.3389/fimmu.2022.956833
Hatfull, G. F., Dedrick, R. M., and Schooley, R. T. (2022). Phage therapy for antibiotic-resistant bacterial infections. Annu. Rev. Med. 73, 197–211. doi: 10.1146/annurev-med-080219-122208
Holtappels, D., Fortuna, K., Lavigne, R., and Wagemans, J. (2021). The future of phage biocontrol in integrated plant protection for sustainable crop production. Curr. Opin. Biotechnol. 68, 60–71. doi: 10.1016/j.copbio.2020.08.016
Huang, B., Huang, J., Guo, Z., Zheng, J., and Liu, Q. (2023). First report of southern blight caused by Athelia rolfsii on pepper in Yiyang, China. Plant Dis. 107:3287. doi: 10.1094/PDIS-03-23-0462-PDN
Huang, S., Tian, Y., Wang, Y., García, P., Liu, B., Lu, R., et al. (2022). The broad host range phage vB_CpeS_BG3P is able to inhibit Clostridium perfringens growth. Viruses 14:676. doi: 10.3390/v14040676
Ingel, B., Caldwell, D., Duong, F., Parkinson, D. Y., McCulloh, K. A., Iyer-Pascuzzi, A. S., et al. (2022). Revisiting the source of wilt symptoms: X-ray microcomputed tomography provides direct evidence that Ralstonia biomass clogs xylem vessels. PhytoFrontiers 2, 41–51. doi: 10.1094/PHYTOFR-06-21-0041-R
Jiang, G., Wei, Z., Xu, J., Chen, H., Zhang, Y., She, X., et al. (2017). Bacterial wilt in China: history, current status, and future perspectives. Front. Plant Sci. 8:1549. doi: 10.3389/fpls.2017.01549
Kaur, G., Agarwal, R., and Sharma, R. K. (2021). Bacteriophage therapy for critical and high-priority antibiotic-resistant Bacteria and phage cocktail-antibiotic formulation perspective. Food Environ. Virol. 13, 433–446. doi: 10.1007/s12560-021-09483-z
Khan, M. S. I., Gao, X., Liang, K., Mei, S., and Zhan, J. (2021). Virulent Drexlervirial bacteriophage MSK, morphological and genome resemblance with Rtp bacteriophage inhibits the multidrug-resistant Bacteria. Front. Microbiol. 12:706700. doi: 10.3389/fmicb.2021.706700
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Le, K. D., Kim, J., Yu, N. H., Kim, B., Lee, C. W., and Kim, J. C. (2020). Biological control of tomato bacterial wilt, kimchi cabbage soft rot, and red pepper bacterial leaf spot using Paenibacillus elgii JCK-5075. Front. Plant Sci. 11:775. doi: 10.3389/fpls.2020.00775
Liu, R., Han, G., Li, Z., Cun, S., Hao, B., Zhang, J., et al. (2022). Bacteriophage therapy in aquaculture: current status and future challenges. Folia Microbiol. (Praha) 67, 573–590. doi: 10.1007/s12223-022-00965-6
Liu, N., Lewis, C., Zheng, W., and Fu, Z. Q. (2020). Phage cocktail therapy: multiple ways to suppress pathogenicity. Trends Plant Sci. 25, 315–317. doi: 10.1016/j.tplants.2020.01.013
Mamphogoro, T. P., Babalola, O. O., and Aiyegoro, O. A. (2020). Sustainable management strategies for bacterial wilt of sweet peppers (Capsicum annuum) and other Solanaceous crops. J. Appl. Microbiol. 129, 496–508. doi: 10.1111/jam.14653
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Marquioni, V., Rossi, F. P. N., Mendonça, D. C., Martins, L. F., Behlau, F., Setubal, J. C., et al. (2022). Isolation and characterization of vB_XciM_LucasX, a new jumbo phage that infects Xanthomonas citri and Xanthomonas fuscans. PLoS One 17:e0266891. doi: 10.1371/journal.pone.0266891
McArthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. doi: 10.1128/AAC.00419-13
Moraru, C., Varsani, A., and Kropinski, A. M. (2020). VIRIDIC-A novel tool to calculate the Intergenomic similarities of prokaryote-infecting viruses. Viruses 12:1268. doi: 10.3390/v12111268
Muturi, P., Yu, J., Maina, A. N., Kariuki, S., Mwaura, F. B., and Wei, H. (2019). Bacteriophages isolated in China for the control of Pectobacterium carotovorum causing potato soft rot in Kenya. Virol. Sin. 34, 287–294. doi: 10.1007/s12250-019-00091-7
National public service platform for standards information . (2008). Available at: https://std.samr.gov.cn/search/std?q=GB%2FT%2023222-2008 (Accessed Feb 28, 2024).
Okiro, L. A., Mulwa, R. M., Oyoo, M. E., Machuka, E. M., Parker, M. L., and Pelle, R. (2022). Phylogenetic distribution of Ralstonia solanacearum species complex populations in potato in Kenya. Plant Dis. 106, 1736–1742. doi: 10.1094/PDIS-11-21-2556-RE
Safni, I., Cleenwerck, I., de Vos, P., Fegan, M., Sly, L., and Kappler, U. (2014). Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. solanacearum phylotype IV strains as Ralstonia syzygii subsp. indonesiensis subsp. nov., banana blood disease bacterium strains as Ralstonia syzygii subsp. celebesensis subsp. nov. and R. solanacearum phylotype I and III strains as Ralstonia pseudosolanacearum sp. nov. Int. J. Syst. Evol. Microbiol. 64, 3087–3103. doi: 10.1099/ijs.0.066712-0
Sasaki, R., Miyashita, S., Ando, S., Ito, K., Fukuhara, T., and Takahashi, H. (2021). Isolation and characterization of a novel jumbo phage from leaf litter compost and its suppressive effect on Rice seedling rot diseases. Viruses 13:591. doi: 10.3390/v13040591
Stothard, P., and Wishart, D. S. (2005). Circular genome visualization and exploration using CGView. Bioinformatics 2005, 537–539. doi: 10.1093/bioinformatics/bti054
Strathdee, S. A., Hatfull, G. F., Mutalik, V. K., and Schooley, R. T. (2023). Phage therapy: from biological mechanisms to future directions. Cell 186, 17–31. doi: 10.1016/j.cell.2022.11.017
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tian, Y., Wu, L., Lu, R., Bao, H., Zhou, Y., Pang, M., et al. (2022). Virulent phage vB_CpeP_HN02 inhibits Clostridium perfringens on the surface of the chicken meat. Int. J. Food Microbiol. 363:109514. doi: 10.1016/j.ijfoodmicro.2021.109514
Villalpando-Aguilar, J. L., Matos-Pech, G., López-Rosas, I., Castelán-Sánchez, H. G., and Alatorre-Cobos, F. (2022). Phage therapy for crops: concepts, experimental and bioinformatics approaches to direct its application. Int. J. Mol. Sci. 24:325. doi: 10.3390/ijms24010325
Wang, K., Chen, D., Liu, Q., Zhu, P., Sun, M., and Peng, D. (2022). Isolation and characterization of novel lytic bacteriophage vB_RsoP_BMB50 infecting Ralstonia solanacearum. Curr. Microbiol. 79:245. doi: 10.1007/s00284-022-02940-3
Wang, R., Cong, Y., Mi, Z., Fan, H., Shi, T., Liu, H., et al. (2019). Characterization and complete genome sequence analysis of phage GP4, a novel lytic Bcep22-like podovirus. Arch. Virol. 164, 2339–2343. doi: 10.1007/s00705-019-04309-7
Wang, Z., Luo, W., Cheng, S., Zhang, H., Zong, J., and Zhang, Z. (2023). Ralstonia solanacearum – a soil borne hidden enemy of plants: research development in management strategies, their action mechanism and challenges. Front. Plant Sci. 14:1141902. doi: 10.3389/fpls.2023.1141902
Wang, X., Wei, Z., Yang, K., Wang, J., Jousset, A., Xu, Y., et al. (2019). Phage combination therapies for bacterial wilt disease in tomato. Nat. Biotechnol. 37, 1513–1520. doi: 10.1038/s41587-019-0328-3
Xu, Z., Jin, P., Zhou, X., Zhang, Y., Wang, Q., Liu, X., et al. (2021). Isolation of a virulent Aeromonas salmonicida subsp. Masoucida bacteriophage and its application in phage therapy in turbot (Scophthalmus maximus). Appl. Environ. Microbiol. 87:e0146821. doi: 10.1128/AEM.01468-21
Yang, K., Wang, X., Hou, R., Lu, C., Fan, Z., Li, J., et al. (2023). Rhizosphere phage communities drive soil suppressiveness to bacterial wilt disease. Microbiome 11:16. doi: 10.1186/s40168-023-01463-8
Ye, M., Sun, M., Huang, D., Zhang, Z., Zhang, H., Zhang, S., et al. (2019). A review of bacteriophage therapy for pathogenic bacteria inactivation in the soil environment. Environ. Int. 129, 488–496. doi: 10.1016/j.envint.2019.05.062
Yi, W., Chen, C., and Gan, X. (2022). Polymyxin B1 and E2 from Paenibacillus polymyxa Y-1 for controlling Rice bacterial disease. Front. Cell. Infect. Microbiol. 12:866357. doi: 10.3389/fcimb.2022.866357
Yuliar, Y., Nion, Y. A., and Toyota, K. (2015). Recent trends in control methods for bacterial wilt diseases caused by Ralstonia solanacearum. Microbes Environ. 30, 1–11. doi: 10.1264/jsme2.ME14144
Zalewska-Piątek, B. (2023). Phage therapy-challenges, opportunities and future prospects. Pharmaceuticals (Basel) 16:1638. doi: 10.3390/ph16121638
Keywords: bacterial wilt (BW), Ralstonia solanacearum , Ralstonia solanacearum species complex (Rssc), bacteriophage, phage therapy
Citation: Huang B, Ge L, Xiang D, Tan G, Liu L, Yang L, Jing Y, Liu Q, Chen W, Li Y, He H, Sun H, Pan Q and Yi K (2024) Isolation, characterization, and genomic analysis of a lytic bacteriophage, PQ43W, with the potential of controlling bacterial wilt. Front. Microbiol. 15:1396213. doi: 10.3389/fmicb.2024.1396213
Edited by:
Hyunjin Yoon, Ajou University, Republic of KoreaReviewed by:
Mikaeel Young, Baylor University, United StatesHardian Susilo Addy, University of Jember, Indonesia
Copyright © 2024 Huang, Ge, Xiang, Tan, Liu, Yang, Jing, Liu, Chen, Li, He, Sun, Pan and Yi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ke Yi, eWlrMDExM0Bobmd5dG9iYWNjby5jb20=; Qiang Pan, cGFucWlhbmdAaG90bWFpbC5jb20=
†These authors have contributed equally to this work
 Binbin Huang
Binbin Huang Long Ge3†
Long Ge3† Qingshu Liu
Qingshu Liu Haoxin He
Haoxin He Qiang Pan
Qiang Pan