
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 06 June 2024
Sec. Food Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1394774
 Yangyan Ge1
Yangyan Ge1 Lulu Wang1
Lulu Wang1 Dapeng Su1
Dapeng Su1 Qingsong Yuan1
Qingsong Yuan1 Chenghong Xiao1
Chenghong Xiao1 Min Hu1
Min Hu1 Chuanzhi Kang2
Chuanzhi Kang2 Lanping Guo2,3*
Lanping Guo2,3* Tao Zhou1*
Tao Zhou1* Jinqiang Zhang1*
Jinqiang Zhang1*Sweating is one of the most important processing methods of Chinese medicinal herbs. However, the high temperature and humidity environment required for sweating Chinese medicinal herbs makes it very easy for fungi to breed, especially toxigenic fungi. The mycotoxins produced by these fungi will then contaminate the Chinese medicinal herbs. In this study, we explored the changes in mycobiota, toxigenic fungi, and mycotoxins with and without sweating in Radix Dipsaci (RD), a typical representative of traditional Chinese medicine that requires processing through sweating. We also isolated and identified the toxigenic fungi from RD, whether they were subjected to sweating treatment or not, and examined their toxigenic genes and ability. The results showed that the detection rate of mycotoxins (aflatoxins, ochratoxins, zearalenone, and T-2 toxin) in RD with sweating was 36%, which was 2.25-fold higher than that in RD without sweating. We also detected T-2 toxin in the RD with sweating, whereas it was not found in the RD without sweating. The sweating process altered the fungal composition and increased the abundance of Fusarium and Aspergillus in RD. Aspergillus and Fusarium were the most frequently contaminating fungi in the RD. Morphological and molecular identification confirmed the presence of key toxigenic fungal strains in RD samples, including A. flavus, A. westerdijkiae, F. oxysporum and F. graminearum. These four fungi, respectively, carried AflR, PKS, Tri7, and PKS14, which were key genes for the biosynthesis of aflatoxins, ochratoxins, zearalenone, and T-2 toxin. The toxigenic ability of these four fungal strains was verified in different matrices. We also found that A. flavus, A. westerdijkiae, and F. oxysporum were isolated in RD both with sweating and without sweating, but their isolation frequency was significantly higher in the RD with sweating than in the RD without sweating. F. graminearum was not isolated from RD without sweating, but it was isolated from RD with sweating. These findings suggest that the sweating process promotes the expansion of toxigenic fungi and increases the risk of combined mycotoxin contamination in RD.
• The sweating process altered the fungal composition and increased the abundance of Fusarium and Aspergillus in Radix Dipsaci.
• Sweating process increases the risk of combined contamination of mycotoxins in Radix Dipsaci.
• Mycotoxins in Radix Dipsaci are mainly derived from Aspergillus flavus, Aspergillus westerdijkiae, Fusarium oxysporum, and Fusarium graminearum.
Mycotoxins are toxic secondary metabolites produced by toxigenic fungi under suitable conditions, which are commonly found in a variety of crops, including traditional Chinese herbs (Su et al., 2018). Molecular methods were used to test whether these genes were expressed in fungi, and then the potentially toxigenic fungi were preliminarily screened. The processes of cultivation, harvesting, storage and processing, traditional Chinese herbs may be contaminated by mycotoxins, which may affect the quality and safety of Chinese herbal medicines Currently, the main mycotoxins susceptible to fungal contamination in traditional Chinese herbs include aflatoxins (AFs), ochratoxins (OTs), T-2 toxin, zearalenone (ZEN), and deoxynivalenol (DON) (Diana Di Mavungu et al., 2009; Han et al., 2012). These mycotoxins may have various chronic and acute effects, including hepatotoxicity, genotoxicity, immunosuppression, estrogenic effects, nephrotoxicity, teratogenic effects, and carcinogenic effects. Therefore, strengthening the monitoring and control of mycotoxin pollution in Chinese medicinal materials is of great significance for ensuring the quality and safety of Chinese medicinal materials and maintaining people’s health. Different toxigenic fungi possess specific key toxigenic genes (Moretti et al., 2013; Ben et al., 2024). Molecular methods were used to test whether these genes were expressed in fungi, and then the potentially toxigenic fungi were preliminarily screened (Alsalabi et al., 2023). The potential toxigenic fungi were further confirmed through qualitative and quantitative analysis of specific mycotoxins in their metabolites (Ben et al., 2024).
Sweating is a special processing method used in the drying and processing of certain medicinal materials, such as roots, rhizomes, and cortex (Jin-Ao et al., 2013). It promotes the release of internal moisture from these medicinal parts and facilitates their drying (Jin-Ao et al., 2013; Wang et al., 2016). The high temperature and humidity environment and the relatively long processing time required during the sweating process contribute to the breeding and growth of fungi, especially some toxigenic fungi (Deng et al., 2020; Liao et al., 2020; Wicaksono and Illes, 2022). The mycotoxins produced by them will contaminate the Chinese medicinal herbs. At present, the effects of sweating on the secondary metabolites of Chinese medicinal herbs have been widely reported (Wu et al., 2019; Cao et al., 2020; Chen et al., 2021; He et al., 2023). However, there are few reports on the effect of sweating on contamination by toxigenic fungi and mycotoxins in Chinese medicinal herbs.
The Radix Dipsaci (RD) is the dried root of the plant Dipsacus asper Wall. ex Henry, which represents traditional Chinese medicine and requires a sweating process. RD is commonly used in the treatment of hepatic or renal disorders, osteoporosis, tendon damage, and uterine hemorrhage due to its potent therapeutic properties (Ke et al., 2016; Zhang et al., 2019; Skala and Szopa, 2023). Here, we detected mycotoxins using ultra-high performance liquid chromatography followed by tandem mass UPLC–MS/MS and investigated the changes in mycobiota, toxigenic fungi, and mycotoxins in RD before and after sweating. We also isolated and identified the toxigenic fungi from RD, whether they were subjected to sweating treatment or not, and examined their toxigenic genes and ability. This study will clarify the effect of the sweating process on toxigenic fungi in Chinese medicinal herbs and provide a basis for preventing the pollution of toxic fungi during sweating processing of Chinese medicinal herbs.
Dipsacus asper were planted in plant nursery at Guizhou University of Traditional Chinese Medicine for 3 years and their roots were collected in October 2022. Fresh samples were randomly divided into two parts (25 samples for each group), one of which did not undergo sweating processing as a control group, and the other part processed according to the sweating method established in our previous study (Zhou Tai-min, 2021). Briefly, the samples required for sweating were dried in a dryer until the relative water content reached 40% and sweated for 3 days at 25°C in an airtight environment. These two sets of samples were stored in a −80°C freezer for detection of mycotoxins, mycobiota sequencing and isolation and identification of toxigenic fungi simultaneously.
DNA extraction of RD and its amplification refer to the previous report (He et al., 2023). The ITS regions of the fungal 18S rRNA genes were amplified with primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′). PCR was performed using a 20 μL reaction system consisting of 2 μL 10 × Buffer, 2 μL 2.5 mM dNTPs, 0.8 μL ITS1F/IT2R (5 μM), 0.2 μL rTaq polymerase, 0.2 μL BSA, 10 ng DNA template, 2 μL RTAQ polymerase, and ddH2O to a final volume of 20 μL. The whole process was carried out in the PCR amplifier (ABI Gene Amp 9,700, United States), and the procedure was as follows: pre-denaturation at 95°C for 3 min; Denatured at 95°C for 30 s, annealed at 55°C for 30 s, extended at 72°C for 45 s, and expanded for 35 cycles. It was extended for 10 min at 72°C. The PCR products were detected through 2% agarose gel electrophoresis. Purified amplicons were pooled in equimolar ratios and paired-end sequenced on an Illumina MiSeq platform (Illumina, San Diego, United States) according to the standard protocols described by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). The raw sequencing reads were deposited into the NCBI Sequence Read Archive (BioProject ID PRJNA1085876).
Sequences in the library were clustered using USearch, operational taxonomic units (OTUs) were clustered at 97% similarity for non-repetitive sequences (excluding individual sequences), and chimeras were removed during clustering to obtain representative sequences of OTUs (Magoc and Salzberg, 2011; Schmieder and Edwards, 2011; Zhang et al., 2014). Based on the OTUs information, rarefaction curves and alpha diversity indices including observed OTUs, Ace, Chao richness, and Shannon index were calculated with Mothur v1.30.1. The similarity among the microbial communities in different samples was determined by principal coordinate analysis (PCoA) based on Bray–Curtis dissimilarity using the Vegan v2.5-3 package.
All the samples were dried until their moisture content was reduced to less than 10%, and then they were also ground and passed through a 50-mesh sieve for subsequent detection. Herb powder (2.0 g) was mixed with 20.0 mL of acetonitrile: water: formic acid (80: 19:1) and shaken for 1 h at 180 rpm. MgSO4 (2 g) and NaCl (1 g) were added to the mixture, which was swirled for 1 min, then centrifuged at 5000 rpm for 5 min. Supernatant (5 mL) was added to a mixture of the following pure powders, which served as “cleaning agents” to remove lipids, pigments, organic acids and sugars (Zhai et al., 2023): Octadecyl trichlorosilane (C18) (0.3 g), primary secondary amine (PSA, 0.1 g), MgSO4 (0.3 g) and graphitized carbon black (GCB, 0.01 g). The mixture was swirled for 1 min, then an aliquot of supernatant (3 mL) was transferred to a new tube and evaporated nearly to dryness at 40°C under nitrogen. The residue was reconstituted with 1.0 mL 50% acetonitrile in water, filtered through a membrane with pores of diameter 0.22 μm, and used in the analytical procedure described below.
The following mycotoxin standards of purity ≥99% were purchased from Pribolab (Qingdao, Shandong, China) and stored at −20°C in the dark: aflatoxin B1 (catalog no. MSS1003), aflatoxin B2 (MSS1014), aflatoxin G1 (MSS1005), aflatoxin G2 (MSS1006), ochratoxin A (MSS1020), zearalenone (MSS1024) and T-2 toxin (MSS1023). Stock solutions of all mycotoxins were prepared by dissolving the powders in 50% (v/v) acetonitrile in water to final concentrations of 1.25 mg/mL in the case of zearalenone or 1 mg/mL in the case of the others. Aliquots of 0.10–10.0 mL of these stock solutions were used to prepare working solutions of 12.5 μg/mL in the case of zearalenone or 10 μg/mL in the case of the others. Aliquots of 1.0–25 mL from these working solutions were mixed to prepare a standard cocktail containing zearalenone at 500 ng/mL and all other mycotoxins at 400 ng/mL.
We detected all seven mycotoxins using ultra-high performance liquid chromatography followed by tandem mass ultra-high performance liquid chromatography-mass spectrometry (UPLC-MS/MS) using a QTRAP 5500 quadrupole system (SCIEX, United States) as the methods of Fan et al. (2022). Chromatographic parameters were as follows: column type, Waters HSS T3 (100 × 2.1 mm, 1.8 μm); column temperature, 40°C; mobile phases, 0.1% formic acid in water (solvent A) and acetonitrile (solvent B); flow rate, 0.3 mL/min; and sample volume, 5 μL. The following elution gradient was performed: 0–3 min, 85% A; 3–5 min, 65% A; 5–7 min, 55% A; 7–7.5 min, 50% A. The mass spectrometer was optimized in the multiple reaction monitoring mode, and the positive electrospray ionization [ESI+] polarity was employed. The capillary voltage, ion source temperature, cone hole blowback flow rate, desolvation temperature, and solvent gas flow rate were 3.5 kV, 150°C, 30 L·h−1, 500°C, and 600 L·h−1, respectively (Han et al., 2010). Data was collected in the multi-reaction monitoring (MRM) mode, and the mass spectrum parameters of each mycotoxin reference solution were shown in Table 1.
We validated the analytical approach in Section 2.3.3. First, we prepared the test sample and the mycotoxin reference solution according to the above method for injection. The blank disyllabic sample extract was selected to dilute the mixed standard into a series of concentrations to establish a standard curve. The signal-to-noise ratio S/N ≥ 3 was defined as the limit of detection (LOD), and S/N ≥ 10 was defined as the limit of quantification (LOQ). Six parallel sample solutions were tested, and the repeatability was investigated based on the determined relative standard deviation of mycotoxin content. Stability was assessed by calculating the relative standard deviation of mycotoxin content measured at 0, 2, 4, 6, 12, and 24 h. The precision of the instrument was evaluated by calculating the relative standard deviation of the mycotoxin peak area detected in six consecutive injections. The recovery rate was investigated using the blank spiked recovery method. The test solution of blank RD was prepared according to the method described above. The solution was combined with “low” (the concentration of ZEN is 100 ng/mL, and the concentration of other mycotoxins is 80 ng/mL), “medium” (the concentration of ZEN is 200 ng/mL, and the concentration of other mycotoxins is 80 ng/mL), and “high” (ZEN concentration of 300 ng/mL, and the rest of the fungus toxin concentration at 240 ng/mL) in a mixture containing mycotoxin standard products at different concentrations. Each concentration was performed in triplicate, resulting in a total of nine samples. Mycotoxin recovery and its relative standard deviation were determined.
The samples of RD without sweating and with sweating were first cut into tissue blocks measuring 5 mm × 5 mm × 5 mm. The toxigenic fungi were then separated using the tissue block method (Song et al., 2022). After the colonies grew out from the tissue blocks, the fungi were purified through single spore separation and translocated multiple times until a single colony was formed. They were stored at 4°C on a potato dextrose agar medium. The isolated fungi were identified morphologically according to the Atlas of Clinical Fungi (de Hoog, 2000). Colony morphology was observed on potato dextrose agar medium, including colony shape, size, color, mycelia type, spore production, and growth on the medium.
The mycelium of fungi cultured on potato dextrose agar medium was collected for molecular identification, and the DNA was extracted using a lysis buffer containing hexadecyltrimethylammonium bromide as described by Sadhasivam et al. (2017). Then the DNA extracted from each strain was used as a template to amplify the ITS2 gene using universal primers ITS1 (5’-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5’-TCCTCCGCTTATTGATATGC-3′) (Frimpong et al., 2019). The PCR reaction was performed using a 20 μL reaction system with the same amplification procedure as described in Section 2.3. Purified amplicons were pooled in equimolar ratios and subjected to paired-end sequencing (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, United States) following the standard protocols provided by Majorbio Bio-Pharm Technology Co. Ltd. Shanghai, China). The obtained sequences were analyzed using the BLAST program1 provided by the NCBI website to determine the genera and species of the fungal isolates (Wu et al., 2013). The construction of a neighbor-joining tree was performed based on multiple sequence alignment using MEGA 5.0 with bootstrap replications (Tamura et al., 2011).
In the analysis of key genes involved in toxin synthesis, we selected conserved regions of AFs, OTA, T-2, and ZEN to design specific primers for the synthesis of key genes AflR, PKS, Tri7, and PKS14. PCR was used to detect the presence of AflR, PKS, Tri7, and PKS14 in the purified fungi by using the DNA of each fungus as a template. The primers are listed in Table 2. The PCR reaction was performed using a 20 μL reaction system with the same amplification procedure as described in section 2.4.
The isolated toxigenic fungi were further inoculated on autoclaved rice and RD, which had been adjusted to 30% water content and cultured at 25°C for 14 days. After drying at 45°C, they were crushed. 2 g of rice and RD were weighed separately and mixed with 10 mL of a 70% methanol solution. The mixture was then centrifuged, and 5 mL of the supernatant was collected. The volume was adjusted to 10 mL, followed by elution of the immunoaffinity column with 1.0 mL of methanol. Finally, detection was performed using an overused 0.22 μm filter membrane according to the same method as described in section 2.3.
In the investigation of specificity, the chromatographic peak of mycotoxin was well separated, and there was no interference at the corresponding position of the blank group, indicating that the method had good specificity (Figure 1A). The linear relationship results showed a good linear range (r ≥ 0.9990) for the seven mycotoxins. The detection limits of 7 mycotoxins were 0.065–0.230 μg kg−1, and the limits of quantification were 0.22–0.50 μg kg−1 (Figure 1B). The repeatability RSD was 0.37–5.94% (Figure 1C); the RSD value of 24 h stability was 0.94–3.76% and less than 3%. In the precision test, the RSD of precision was 0.74–2.94%, were lower than 15% of the requirements of Chinese Pharmacopoeia (2020 edition), indicating that the method had good repeatability, stability and precision. The results of precision testing indicated that the RSD ranged from 0.74 to 2.94% (Figure 1C), which was lower than the requirement of 15% stated in the Chinese Pharmacopoeia (2020 edition). Recoveries ranging from 73.30 to 119.40% of all tested mycotoxins with RSD lower than 15% (Figure 1D) were in line with the provisions of the Chinese Pharmacopoeia (2020 edition), indicating that the method was accurate and could be used for detecting mycotoxins in RD.
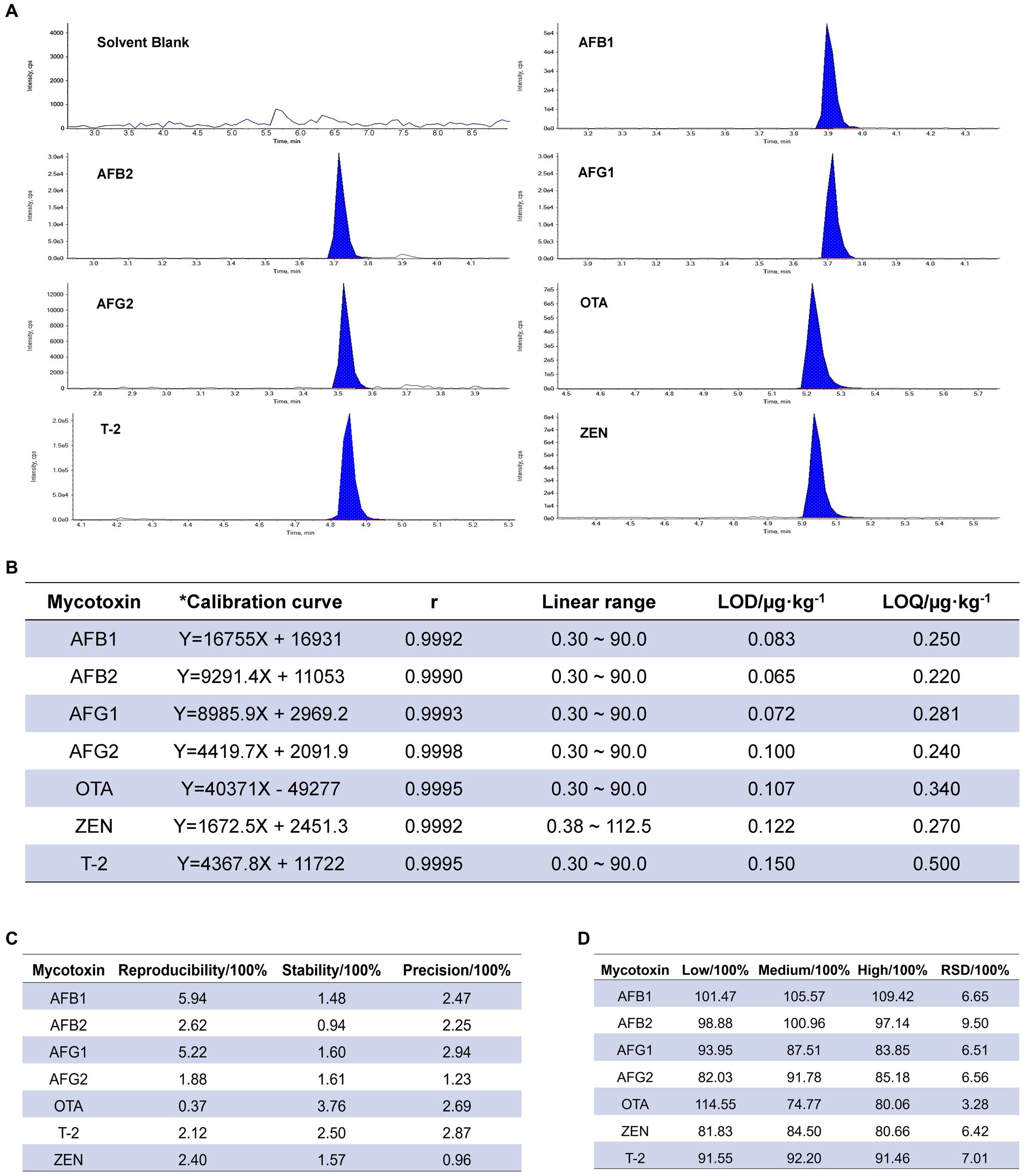
Figure 1. Validation of the analytical method. (A) Specific chromatogram of seven mycotoxins in Radix Dipsaci. cps, counts per second. (B) Calibration and validation of the analytical method for simultaneous detection of mycotoxins in Radix Dipsaci. * Calibration curves were adjusted for matrix effects (see Methods). LOD, limit of detection; LOQ, limit of quantitation. (C) Repeatability, stability and precision for determination of mycotoxins in Radix Dipsaci. (D) Recovery of seven kinds of mycotoxins from Radix Dipsaci. RSD, relative standard deviation.
We prepared RD samples of sweating and non-sweating and conducted mycobiota sequencing. We observed a lower number of fungal OTUs in the RD with sweating compared to the RD without sweating (Figure 2A). Additionally, we identified 887 unique OTUs and 137 unique genera in the RD with sweating, which were not present in the RD without sweating (Figures 2B,C). The alpha diversity of fungi was analyzed by calculating the Chao, Ace, Shannon, and Simpson indices at the OTU level. The results showed that the Ace and Chao indices of fungi in RD with sweating were significantly decreased compared to those of the control group (Figures 2D,E), indicating that the sweating process reduced the richness of fungi in RD. The Shannon index of fungi significantly decreased, and the Simpson index significantly increased in RD with sweating when compared to the control group (Figures 2F,G), indicating that the sweating process reduced fungal diversity in RD.
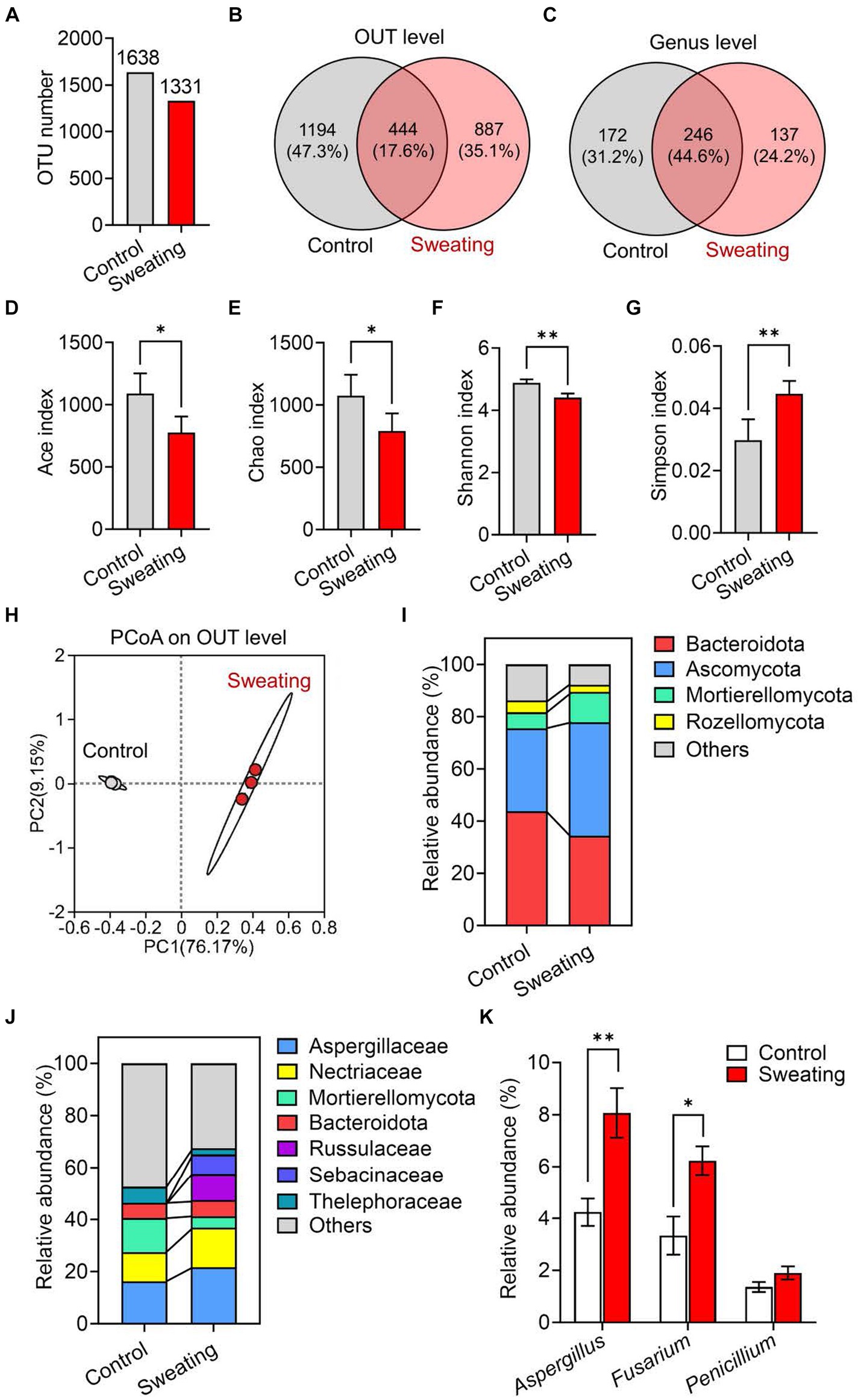
Figure 2. Mycobiota analysis of RD before and after sweating process. (A) Change in the number of operational taxonomic units (OTU) of fungi in RD before and after sweating process. (B,C) Wayne’s analysis of OTU and genus of fungi in RD before and after sweating process. (D–G) Alpha diversity at the level of OTUs in fungal communities in RD and after sweating process. Microbial diversity was quantified using Chao’s, Ace’s, Shannon’s and Simpson’s diversity indices (n = 3). (H) Principal component analysis (PCoA) of beta diversity in fungal communities in RD before and after sweating process, based on Bray–Curtis distances (n = 3). (I,J) Relative abundances of (I) phyla and (J) families of fungi in RD before and after sweating process. (K) Relative abundances of Aspergillus spp., Fusarium spp. and Penicillium spp. In RD before and after sweating process.
Beta diversity was analyzed through principal coordinate analysis (PCoA) plots using nonphylogenetic Bray–Curtis metrics to assess differences in microbial composition (OTU) between the two groups (Figure 2H). There was a clear separation between the control and sweating groups along the first principal component (PC1) axis (Adonis p value = 0.001), suggesting that the sweating process can change the structure of the fungal community. For instance, at the phylum level, there was a significant increase in the abundance of Ascomycota and Mortierellomycota fungi after sweating, while the abundance of Bacteroidota and Rozellomycota fungi decreased significantly (Figure 2I). At the family level, we observed a significant increase in the relative abundance of Aspergillaceae and Nectriaceae fungi in RD with sweating compared to RD without sweating. The relative abundance of Mortierellaceae and Thelephoraceae fungi was significantly decreased in RD with sweating compared to RD without sweating (Figure 2J). Fusarium and Aspergillus are toxigenic fungi that produce the majority of fungal toxins, and their relative abundance significantly increases after sweating in RD (Figure 2K). After sweating, the relative abundance of Penicillium also exhibited an increase; however, no statistically significant difference was observed. These results indicate that the sweating process alters the fungal composition and increases the abundance of Aspergillus and Fusarium in RD.
Data are mean ± SEM (n = 3). *p < 0.05 and **p < 0.01 based on the independent-sample t-test.
The sweating process caused significant changes in the appearance properties of the RD. The cross-section color of RD deepened with sweating, changing from faint yellow before sweating to atrovirens. The color change was the key feature of sweating treatment, and the content of terpenoids and phenolic acids is involved in the formation of green after sweating treatment (He et al., 2023). Additionally, fungal mycelium was visible on the surface in RD with sweating (Figure 3A). We further detected the mycotoxins in RD that were or were not subjected to sweating treatment using the UPLC-MS/MS method (Figure 3B). The results showed that the detection rate of AFs and OTs in the RD with sweating was significantly higher than that in the RD without sweating. Mycotoxins (AFB1, AFG1, OTA or ZEN) were detected in only 16% of RD samples without sweating, with contamination levels of AFB1, AFG1, OTA and ZEN were 1.33, 2.91, 4.16 and 5.17 μg/kg, respectively (Figures 3C,D). And there was no combined contamination of mycotoxins, suggesting that a small part of the RD may be infected by toxigenic fungus during cultivation. However, mycotoxins were detected in 36% of the samples with sweating. Among these samples contaminated with mycotoxins, 67% were contaminated with two or more mycotoxins. After sweating, the contamination levels of AFB1, AFG1, and OTA in the samples ranged from 0.73 to 12.12 μg/kg, 0.91 to 3.58 μg/kg, and 2.06 to 10.22 μg/kg respectively; while the contamination levels of ZEN and T-2 were 13.76 μg/kg and 2.12 μg/kg, respectively. Among these mycotoxin-contaminated samples, two samples had AFB1 levels exceeding 5 μg/kg, and one sample had both AFB1 and AFG1 levels exceeding 10 μg/kg. The ZEN contamination level in the sample was below 500 μg/kg, which meets the requirements of the Chinese pharmacopoeia (2020 edition) (Figures 3C,D). We also detected T-2 toxin in the RD with sweating, which was not detected in the RD without sweating (Figures 3B–D). These results suggested that the sweating process increases the contamination rate of mycotoxins and the risk of combined mycotoxin contamination in RD.
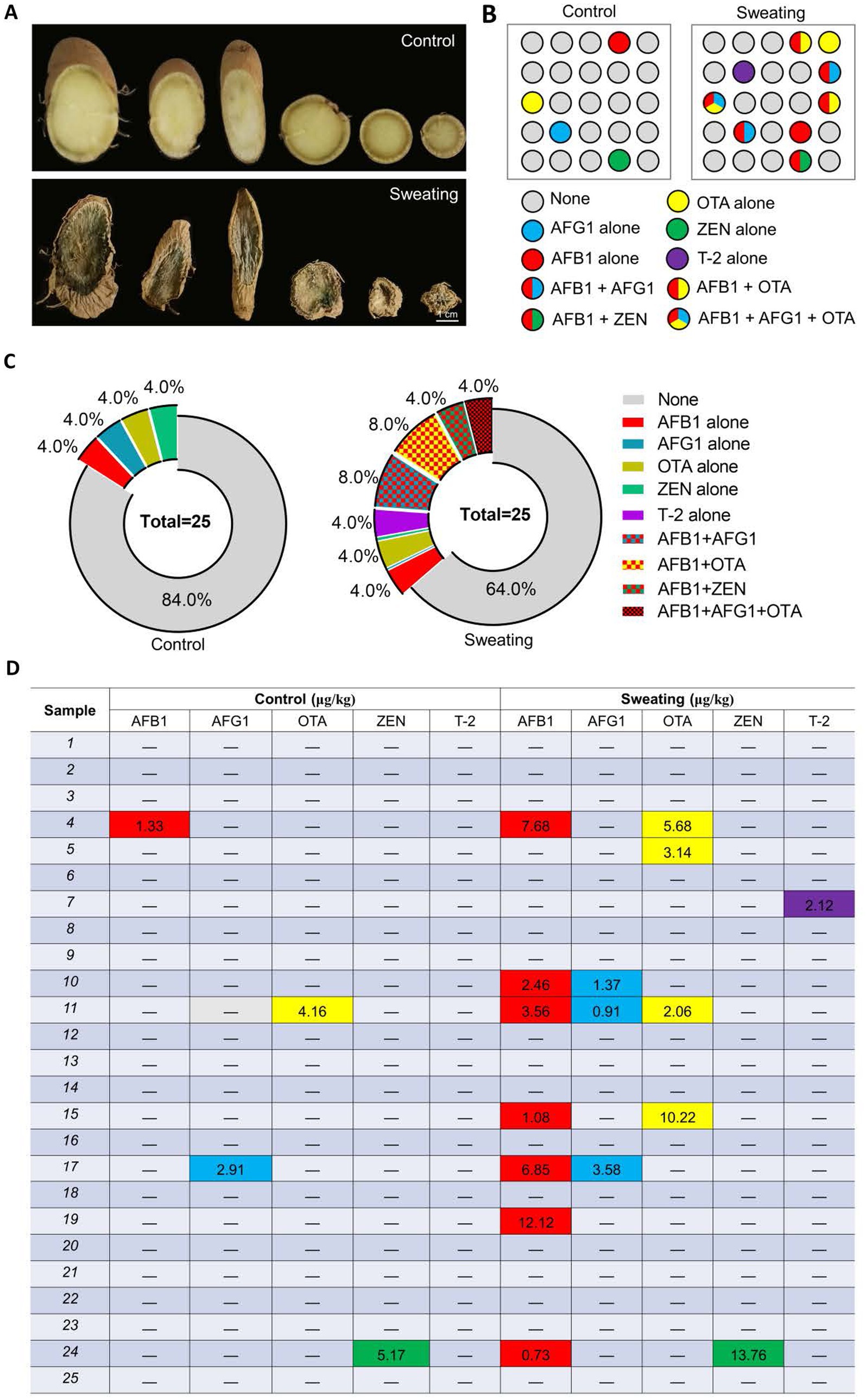
Figure 3. Detection of mycotoxins in RD before and after sweating process. (A) Changes in the appearance of RD before and after sweating process. (B) Changes in the number and type of mycotoxins-contaminated samples before and after sweating. (C) Percentages of samples contaminated with different mycotoxins, alone or in combination. (D) Levels of mycotoxin contamination of samples before and after sweating.
We initially isolated 45 strains of Aspergillus and 19 strains of Fusarium from RD based on morphological and microscopic identification. Among them, there were 10 strains of Aspergillus and 4 strains of Fusarium in RD without sweating, while there were 35 strains of Aspergillus and 15 strains of Fusarium in RD with sweating (Figure 4A). The DNA of strains from RD was extracted and amplified using ITS1 and ITS4. Phylogenetic trees were constructed after comparing them with NCBI online BLAST. The results showed that 16 strains had a 100% similarity to A. flavus strain MSEF26, 11 strains had a 100% similarity to A. flavus strain GFRS9, 7 strains had a 100% similarity to A. flavus strain meijun2, 11 strains had a 100% similarity to A. westerdijkiae strain 5A16, and 6 strains had a 100% similarity to F. graminearum strain Ta-S27; additionally, there were also 13 strains had a 100% similarity to F. oxysporum strain BC012 (Figure 4B). Morphological and molecular identification confirmed the presence of potential toxigenic fungal strains in RD samples, including 34 strains of A. flavus, 11 strains of A. westerdijkiae, 13 strains of F. oxysporum, and 6 strains of F. graminearum, accounting for 53.13, 17.19, 20.13, and 9.38%, respectively (Figures 4C–G). The isolation frequency of A. flavus, A. westerdijkiae and F. oxysporum in RD with sweating was increased by 2.86-fold, 1.67-fold and 1.25-fold, respectively, compared to their isolation frequency from the RD without sweating (Figure 4H). In addition, we isolated 6 strains of F. graminearum in RD with sweating, which was not isolated in the RD without sweating (Figure 4H). This indicates that the sweating process will increase the number of endophytic toxigenic fungi in RD and produce new toxigenic fungi, increasing the risk of mycotoxin contamination.
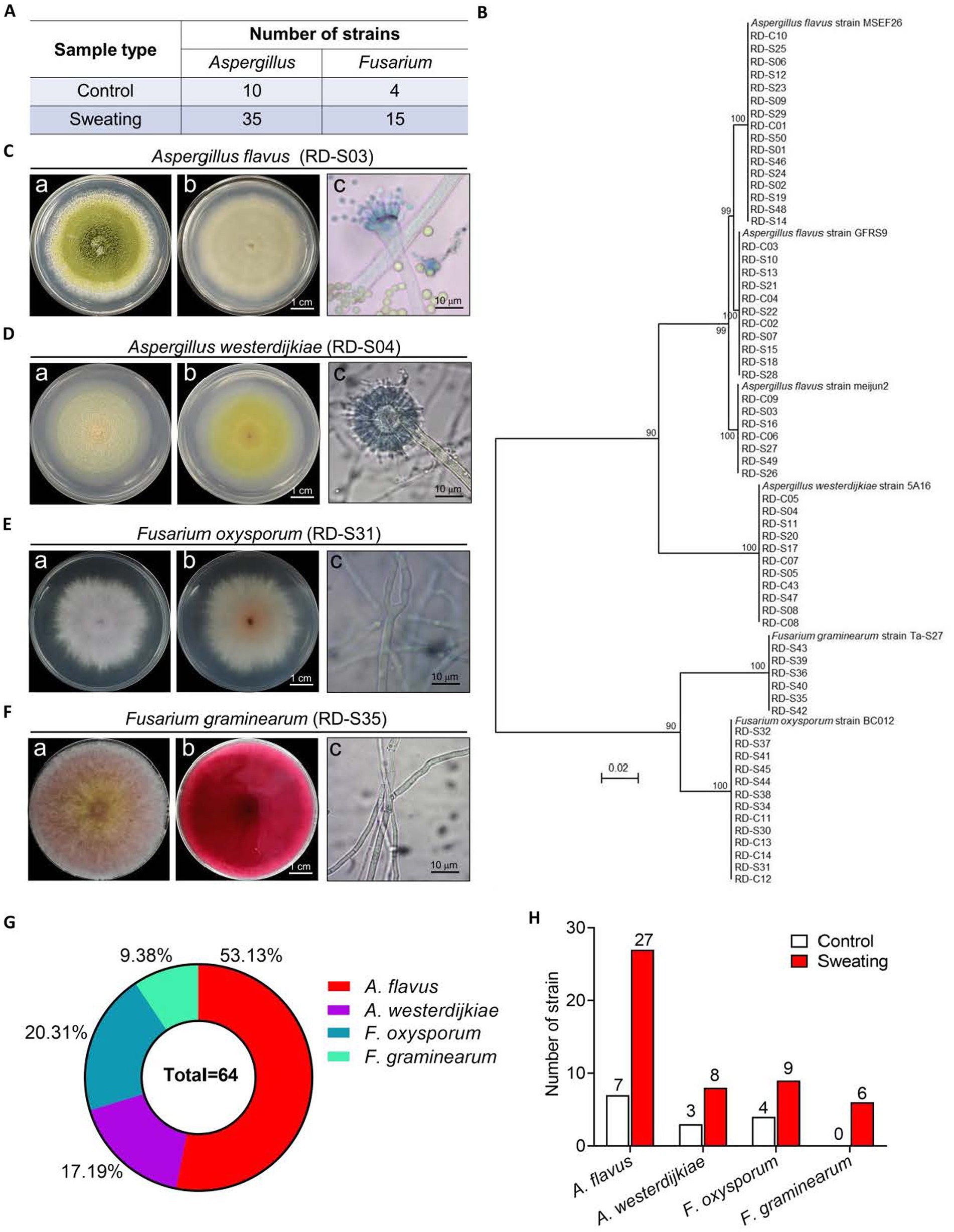
Figure 4. Morphological and molecular identification of potential toxigenic fungi from RD. (A) Number of Aspergillus spp. and Fusarium spp. isolated from RD that are or are not subjected to sweating treatment. (B) Phylogenetic tree based on gene sequences of specific fungi, inferred by using the Maximum Likelihood method, on 34 A. flavus, 11 A. westerdijkiae, 13F. oxysporum and 6F. graminearum isolated from RD, compared to reference sequences for corresponding species. (C–F) Morphological characteristics of A. flavus, A. westerdijkiae, F. oxysporum and F. graminearum. Inset (A) shows the front of the colony, inset (B) shows the back of the colony, and inset (C) shows the microscopic structure of the fungal hyphae and spores. (G) Percentages of A. flavus, A. westerdijkiae, F. oxysporum and F. graminearum. (H) The number of A. flavus, A. westerdijkiae, F. oxysporum and F. graminearum isolated from RD that are or are not subjected to sweating treatment.
Using DNA from potential toxigenic fungi strains from RD as a template, PCR was used to detect the key genes for mycotoxin biosynthesis. The results showed that A. flavus (RD-C03 strain) and A. flavus (RD-S03 strain) carried AflR, which was the key gene for AFs biosynthesis. A. westerdijkiae (RD-S04 strain) carried PKS, which were key gene for OTs biosynthesis. F. oxysporum (RD-S31) carried PKS14, which was key gene for ZEN biosynthesis. F. graminearum (RD-S35 strain) carried Tri7, which was key gene for T-2 toxin biosynthesis (Figure 5).
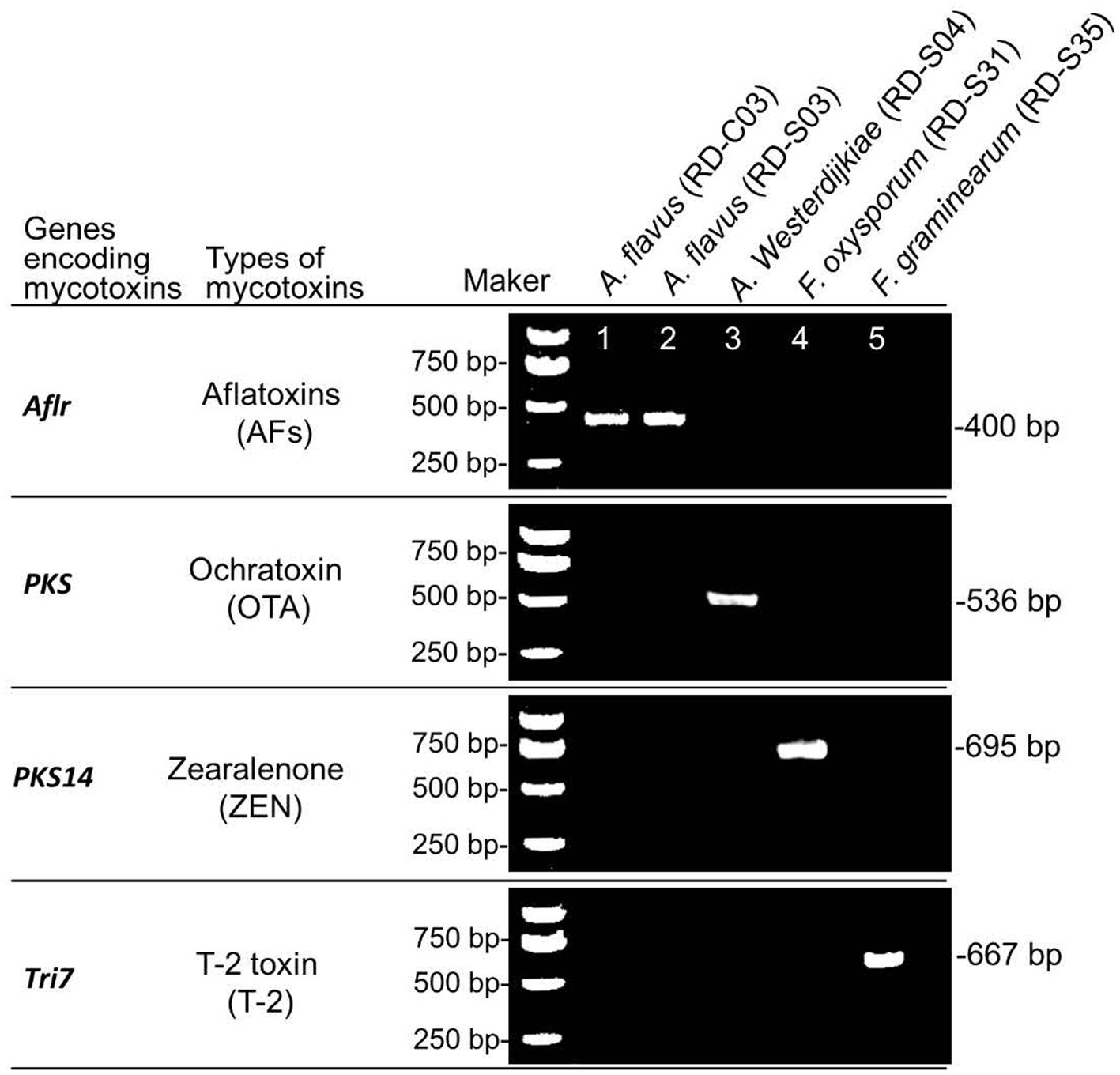
Figure 5. PCR identification of key genes for mycotoxins biosynthesis in potential toxigenic fungi from RD.
PCR amplification and agarose gel detection of key genes for mycotoxins biosynthesis. M: DNA molecular weight standard; Lane 1–6: PCR products of key enzyme genes of toxigenic fungi strains were isolated.
To determine whether these toxigenic fungi can produce their corresponding mycotoxins, we verified the toxigenic capacity of each fungus in different matrices using LC-MS/MS. The results showed that A. flavus (RD-S03 strain) could produce both AFB1 and AFG1 not only in rice matrix but also in the RD matrix (Supplementary Figures S1A,B). However, A. flavus (RD-C03 strain) could only produce AFB1, not AFG1, in either rice or RD matrix (Supplementary Figure S1A). A. flavus (RD-S09 strain) could simultaneously produce AFB1 and AFB2 in rice matrix, but it could only produce AFB1, not AFB2, in RD matrix (Supplementary Figures S1A–C). Both A. westerdijkiae (RD-S04 strain) and F. oxysporum (RD-S31 strain) can produce OTA and ZEN, respectively, not only in the rice matrix but also in the RD matrix (Figures 6A,B). F. graminearum (RD-S35 strain) can produce T-2 toxin in rice matrix, but not in RD matrix (Supplementary Figure S2C). The growth conditions of RD in the natural environment may be more complex, leading to other factors causing F. graminearum to produce T-2 toxin. The levels of mycotoxin production by A. flavus, A. westerdijkiae, F. oxysporum, and F. graminearum in the rice matrix and RD matrix were quantified (Supplementary Figures S2A,B).
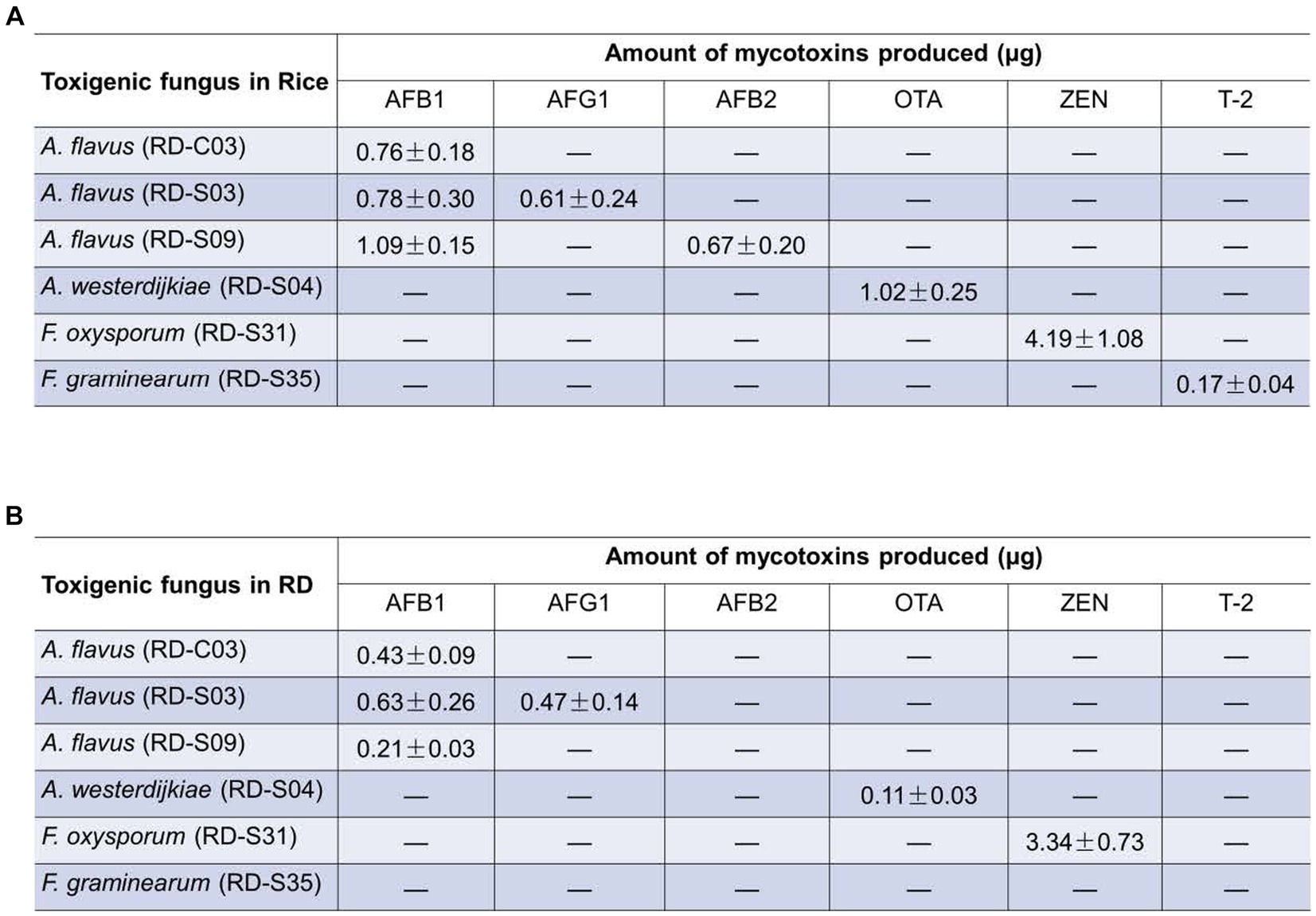
Figure 6. Amount of mycotoxins produced potential toxigenic fungi in (A) rice matrix or (B) RD matrix (A,B) the content of mycotoxins produced by toxigenic fungi in rice and RD matrix.
Sweating is one of the most important processing methods of Chinese medicinal herbs. However, the high temperature and humidity environment required for sweating Chinese medicinal herbs makes it very easy for fungi to breed, especially toxigenic fungi. The mycotoxins produced by these fungi will then contaminate the Chinese medicinal herbs. In this study, we demonstrated that the sweating process alters the fungal composition and increases the abundance of Fusarium and Aspergillus in RD, which in turn increases the contamination rate of mycotoxins and the risk of combined mycotoxin contamination in RD. Our data showed that the sweating process will increase the number of endophytic toxigenic fungi in RD and produce new toxigenic fungi, increasing the risk of mycotoxins contamination. To our knowledge, this study is the first to reveal an association between sweating processes and mycotoxins contamination of Chinese medicinal herbs.
Sweating is one of the most important processing methods for Chinese medicinal herbs, which promotes the enzyme system and initiates the biological and chemical transformation of primary/secondary metabolites (Cao et al., 2020; Sun et al., 2023). Then the changes in biochemical properties, metabolite levels, and medicinal properties of medicinal herbs are altered (Wang et al., 2016). Our study demonstrated that sweating can alter the color of the fracture surface of RD from pale yellow to dark green, which is correlated with the levels of terpenoids and phenolic acids (He et al., 2023). However, the high temperature and humidity environment required for the sweating of Chinese medicinal herbs makes it very easy for toxigenic fungi to breed (Zhang et al., 2018; Locatelli et al., 2022). Indeed, our results showed that the sweating process altered the fungal composition and increased the abundance of Aspergillus and Fusarium in RD, which is consistent with a previous report. The investigation revealed significant differences in the contaminated fungal communities of Cassia seeds with different processing specifications (Guo et al., 2020). The relative abundance of Aspergillus in roasted Cassiae Semen was higher than that in raw Cassiae Semen, and Aspergillus, Fusarium and Penicillium were mainly distributed in burnt malt (Long et al., 2022). Aspergillus are the toxigenic fungi responsible for most AFs and OTs (Cabrera-Meraz et al., 2021; Kumar et al., 2023), and Fusarium are the toxigenic fungi responsible for most fusarium mycotoxins, such as deoxynivalenol, ZEN and so on (Narvaez et al., 2020; Ji et al., 2023).
The relative abundance of toxigenic fungi Fusarium and Aspergillus increased after the sweating process, possibly because the humid and hot environment created by sweating is more conducive to the growth of these fungi. Temperature and water and their interactions have been shown to be key factors that regulate fungal growth and secondary metabolite production (Schmidt-Heydt et al., 2009; Marin et al., 2010). In studies on aflatoxins, these two conditions not only promote the growth of aflatoxin-producing fungi (mainly aflatoxins) but also have a significant impact on the activation of gene clusters responsible for aflatoxin production (Abdel-Hadi et al., 2012; Tejero et al., 2021). It was reported that A. flavus could not grow under conditions of temperatures below 10°C in the culture medium of Chaezella yeast AGAR (CYA) and corn extract (CEM), indicating that the lowest temperature for A. flavus growth was between 8–10°C (Astoreca et al., 2012). The lowest temperature for A. flavus growth on the PDA culture medium was 12°C (Calvo et al., 2002), and on rice, it was 13.2°C (Mousa et al., 2011). Higher water activity facilitates better fungal growth and toxin synthesis (Bhatnagar et al., 2006). AFB1 production can be detected when the water activity is 0.94–0.99, and aflatoxin production is inhibited when the water activity is lower than 0.94 (Lahouar et al., 2016). In our study, we found that the sweating process caused a significant decrease in the Ace, Chao, and Shannon indices of fungi in RD, indicating a reduction in the richness and diversity of fungal species. This reduction in microbial diversity and richness in RD with sweating may be associated with the inhibitory effect of toxigenic fungi on non-toxigenic fungi (Martinez-Culebras et al., 2021; Khalifa et al., 2022). Numerous studies have demonstrated that mycotoxins, such as AFs, OTs, or ZEN, can hinder the growth of other fungi by disrupting membrane structure and metabolism (Zheng et al., 2019; Rodrigues et al., 2021). In the present study, we detected more mycotoxins in the RD samples with sweating. Additionally, the detection rate of AFB1, AFG1, OTA or ZEN in the RD with sweating was significantly higher than that in the RD without sweating. It is worth mentioning that toxigenic fungi were also isolated, and mycotoxins were detected without sweating RD. This result suggests that the RD carried toxigenic fungi before sweating, but the population of toxigenic fungi expanded after sweating. Root herbs are more susceptible to contamination by fungi and mycotoxins due to direct contact between their roots and the soil (Su et al., 2018). After the introduction of toxigenic fungi from soil to herbal medicines, these fungi may proliferate and produce mycotoxins during processing and storage (Su et al., 2018). Aspergillus fungi exist widely in nature, can adapt to the low water environment, and are the dominant fungus in the fungal pollution of medicinal materials (Rizzo et al., 2004; Wei et al., 2023). The roots of RD are rich in lignin and fiber. It is speculated that a variety of extracellular enzymes (pectinase and cellulase) secreted by Aspergillus fungi can effectively decompose lignin and fiber and provide a carbon source for growth. The types and contents of nitrogen sources and carbon sources in the substrate have a great influence on the growth and toxicity of fungi (Chang et al., 1995; Feng and Leonard, 1998; Ehrlich and Cotty, 2002). Fusarium fungi are also poisonous fungi in RD. The results showed that Fusarium fungi were suitable for high water and abundant carbon source environments, and had weak growth and toxic production ability in herbs with low water and poor carbon source (Rodrigues et al., 2021).
Simultaneous detection of multiple mycotoxins in our study also showed that about 67% of the mycotoxin-contaminated RD with sweating were contaminated by 2–3 mycotoxins. However, this combined contamination of mycotoxins was not found in the RD without sweating. The possible reason for this phenomenon is that toxigenic fungi can transmit to each other among medicinal herbs during sweating, leading to the expansion of contamination and resulting in multiple mycotoxin combinations (Wang et al., 2020). According to the 2020 edition of the Chinese Pharmacopoeia, AFB1 limit is set at 5 μg/kg for 24 Chinese herbs, while the total AF should not exceed 10.00 μg/kg and ZEN content should be below 500 μg/kg. Three of the samples that were contaminated with mycotoxins after sweating had aflatoxin levels that exceeded the limit. These results suggested that sweating process increased the contamination rate of mycotoxins and the risk of combined contamination of mycotoxins in RD. We also detected T-2 toxin in the RD with sweating, which was not detected in the RD without sweating. Studies have shown that T-2 toxins exhibit basic toxicological characteristics such as immunotoxicity, cytotoxicity, DNA damage, blood toxicity, reproductive toxicity, and hepatorenal toxicity (Li et al., 2011). These results suggested that the process of sweating can also introduce exogenous toxigenic fungi into the herbs, implying that the sweating environment may also be responsible for toxigenic fungal contamination. It has been reported that in the coexistence mode of multiple mycotoxins, their toxic effects are far more complex than simply adding up the toxicity of a single toxin. Additionally, there may be antagonistic, additive, and synergistic effects among mycotoxins. For instance, the combination of AFB1 and OTA can enhance its toxicity (McKean et al., 2006; Xu et al., 2022). The combination of AFB1 and T-2 increases their toxicity (Hou et al., 2018; Xiao et al., 2023). In Vero kidney cells, AFB1 and OTA not only exhibit cumulative cytotoxic effects but also demonstrate synergistic effects that enhance genotoxicity by increasing DNA fragmentation (Golli-Bennour et al., 2010). The effects of AFB1, DON, T-2, and ZEN on cells are dose-dependent and time-dependent (Calvert et al., 2005). Combined exposure to AFB1 and ZEN during pregnancy and lactation in rats caused severe toxicity to the mammary gland and liver. High doses of ZEN exacerbated the toxicity of AFB1, while low doses of ZEN alleviated the damage caused by AFB1 (Wu et al., 2022). The above research demonstrates that the toxicity of multiple mycotoxins in combination is stronger than that of a single toxin. In this study, RD medicinal materials were contaminated by various toxins, which poses a certain risk to the safety of RD drugs. Therefore, it holds significant importance for controlling mycotoxin pollution during the processing of RD.
Currently, various types of toxigenic fungi have been isolated and identified from numerous Chinese medicinal materials. Researchers discovered that all strains of Penicillium polonicum, which were isolated from OTA-contaminated licorice root, produced OTA (Chen et al., 2011). Some researchers isolated and identified toxic fungi mainly from wolfberry, Angelica, and licorice (Zheng et al., 2017). This study also isolated and identified toxigenic fungi from RD. We found that A. flavus was the main source of aflatoxins in RD, while A. westerdijkiae was responsible for producing ochratoxins in RD. The RD-S03 strain of A. flavus was capable of producing both AFB1 and AFG1 in rice and the RD matrix. This may be because AFB1 and AFG1 have the same dihydrodifuran structure, and their synthetic pathways are similar (Yabe et al., 1988). The RD-S09 strain of A. flavus can also produce two toxins, AFB1 and AFB2, in rice medium. However, AFB2 is not detected in the RD matrix, which could be due to the more complex growth environment of toxigenic fungi in natural settings. The RD matrix may be lacking a certain gene enzyme that promotes the synthesis of AFB2. Toxigenic fungi can grow on various substrates, but different substrates have a greater impact on toxin production. Some substrates are conducive to mycotoxin production, while in others, toxic fungi can grow and reproduce in large numbers but produce very low amounts of toxins or no toxins at all (Wellford et al., 1978; Llewellyn et al., 1981). Sherertz et al. (1976) investigated the production of toxicity by different aflatoxin-producing fungus in raw and cooked soybeans, revealing that the optimal substrate for toxin production varied among different toxigenic fungus producing the same toxin. We found that ZEN in RD was mainly derived from F. oxysporum, while T-2 toxin was produced by F. graminearun in rice matrix but not in RD matrix. This may be due to the more complex growth conditions of RD in its natural environment. There are other factors that cause F. graminearun to produce T-2 toxin, and medicinal materials are not the optimal substrate for the growth and production of toxigenic fungi. Compared to grain substrate, the growth of toxigenic fungi may be hindered to some extent due to the lack of readily available carbon and nitrogen sources and the fungicidal function of certain components in medicinal materials (Llewellyn et al., 1981), which could explain why no mycotoxins were detected in RD matrix.
Our results suggested that the sweating process increased the expansion of toxigenic fungi from Fusarium and Aspergillus in RD, which in turn increases the contamination rate of AFs, OTA, ZEN, and T-2 toxin as well as the risk of combined mycotoxins contamination in RD. This study reveals an association between the sweating process and the contamination of mycotoxins in RD, providing a basis for preventing the contamination of toxigenic fungi during the sweating process of Chinese medicinal herbs.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1085876/.
YG: Methodology, Writing – review & editing, Investigation, Validation, Writing – original draft. LW: Investigation, Methodology, Validation, Writing – review & editing, Software. DS: Investigation, Methodology, Validation, Writing – review & editing. QY: Writing – review & editing, Visualization. CX: Writing – review & editing, Data curation. MH: Writing – review & editing, Investigation. CK: Writing – review & editing, Software. LG: Writing – review & editing, Conceptualization, Supervision. TZ: Writing – review & editing, Project administration, Resources. JZ: Project administration, Writing – review & editing, Conceptualization, Methodology, Supervision.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Natural Science Foundation of China (81860675), Key project at central government level for the ability establishment of sustainable use for valuable Chinese medicine resources (2060302), the Innovation Team of the Education Department of Guizhou Province [Qian-Jiao-Ji (2023)071], the Research Platform Team Project of the Education Department of Guizhou Province [Qian-Jiao-Ji (2022)021], Scientific and technological innovation project of China Academy of Chinese Medical Science (CI2021B013), and the National and Provincial Scientific and Technological Innovation Talent Team of Guizhou University of Traditional Chinese Medicine [Gui-Zhong-Yi TD He-Zi (2022)003].
We are grateful to Creaducate Consulting GmbH (Munich, Germany) for editorial assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1394774/full#supplementary-material
Abdel-Hadi, A., Schmidt-Heydt, M., Parra, R., Geisen, R., and Magan, N. (2012). A systems approach to model the relationship between aflatoxin gene cluster expression, environmental factors, growth and toxin production by aspergillus flavus. J. R. Soc. Interface 9, 757–767. doi: 10.1098/rsif.2011.0482
Alsalabi, F. A., Hassan, Z. U., Al-Thani, R. F., and Jaoua, S. (2023). Molecular identification and biocontrol of ochratoxigenic fungi and ochratoxin a in animal feed marketed in the State of Qatar. Heliyon 9:e12835. doi: 10.1016/j.heliyon.2023.e12835
Astoreca, A., Vaamonde, G., Dalcero, A., Ramos, A. J., and Marin, S. (2012). Modelling the effect of temperature and water activity of aspergillus flavus isolates from corn. Int. J. Food Microbiol. 156, 60–67. doi: 10.1016/j.ijfoodmicro.2012.03.002
Ben, M. Y., Benabdallah, A., Chentir, I., Djenane, D., Luvisi, A., and De Bellis, L. (2024). Comprehensive insights into ochratoxin a: occurrence, analysis, and control strategies. Food Secur. 13:81184. doi: 10.3390/foods13081184
Bhatnagar, D., Cary, J. W., Ehrlich, K., Yu, J., and Cleveland, T. E. (2006). Understanding the genetics of regulation of aflatoxin production and aspergillus flavus development. Mycopathologia 162, 155–166. doi: 10.1007/s11046-006-0050-9
Cabrera-Meraz, J., Maldonado, L., Bianchini, A., and Espinal, R. (2021). Incidence of aflatoxins and fumonisins in grain, masa and corn tortillas in four municipalities in the department of lempira, Honduras. Heliyon. 7, 2021:e8506. doi: 10.1016/j.heliyon
Calvert, T. W., Aidoo, K. E., Candlish, A. G., and Fuat, A. R. (2005). Comparison of in vitro cytotoxicity of fusarium mycotoxins, deoxynivalenol, T-2 toxin and zearalenone on selected human epithelial cell lines. Mycopathologia 159, 413–419. doi: 10.1007/s11046-005-0254-4
Calvo, A. M., Wilson, R. A., Bok, J. W., and Keller, N. P. (2002). Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66, 447–459. doi: 10.1128/MMBR.66.3.447-459.2002
Cao, M., Liu, Y., Jiang, W., Meng, X., Zhang, W., Chen, W., et al. (2020). Uplc/ms-based untargeted metabolomics reveals the changes of metabolites profile of salvia miltiorrhiza bunge during sweating processing. Sci. Rep. 10:19524. doi: 10.1038/s41598-020-76650-w
Chang, P. K., Cary, J. W., Yu, J., Bhatnagar, D., and Cleveland, T. E. (1995). The aspergillus parasiticus polyketide synthase gene PKSA, a homolog of aspergillus Nidulans Wa, is required for aflatoxin b1 biosynthesis. Mol. Gen. Genet. 248, 270–277. doi: 10.1007/BF02191593
Chen, A. J., Huang, L. F., Wang, L. Z., Tang, D., Cai, F., and Gao, W. W. (2011). Occurrence of toxigenic fungi in ochratoxin a contaminated liquorice root. Food Addit. Contam. A Chem. Anal. Control Expo. Risk Assess. 28, 1091–1097. doi: 10.1080/19440049.2011.576443
Chen, Q., Shi, Z., Wang, Y., Zhang, R., and Hu, H. (2021). Study on effects of sweating treatment and different drying methods on the quality of gentianae macrophyllae radix. Sci. Rep. 11:9717. doi: 10.1038/s41598-021-88511-1
de Hoog, G. S. (2000). Atlas of clinical fungi. Utrecht, The Netherlands: Centraalbureau voor Schimmelcultures.
Deng, Y., Wang, Y., Deng, Q., Sun, L., Wang, R., Wang, X., et al. (2020). Simultaneous quantification of aflatoxin b(1), T-2 toxin, ochratoxin a and deoxynivalenol in dried seafood products by LC-MS/MS. Toxins 12:80488. doi: 10.3390/toxins12080488
Diana Di Mavungu, J., Monbaliu, S., Scippo, M., Maghuin-Rogister, G., Schneider, Y., Larondelle, Y., et al. (2009). Lc-ms/ms multi-analyte method for mycotoxin determination in food supplements. Food Additives Contam. 26, 885–895. doi: 10.1080/02652030902774649
Ehrlich, K. C., and Cotty, P. J. (2002). Variability in nitrogen regulation of aflatoxin production by aspergillus flavus strains. Appl. Microbiol. Biotechnol. 60, 174–178. doi: 10.1007/s00253-002-1094-5
Fan, Y., Liu, F., He, W., Qin, Q., Hu, D., Wu, A., et al. (2022). Screening of multi-mycotoxins in fruits by ultra-performance liquid chromatography coupled to ion mobility quadrupole time-of-flight mass spectrometry. Food Chem. 368:130858. doi: 10.1016/j.foodchem.2021.130858
Feng, G. H., and Leonard, T. J. (1998). Culture conditions control expression of the genes for aflatoxin and sterigmatocystin biosynthesis in aspergillus parasiticus and a nidulans. Appl Environ Microbiol. 64, 2275–2277. doi: 10.1128/AEM.64.6.2275-2277.1998
Frimpong, G. K., Adekunle, A. A., Ogundipe, O. T., Solanki, M. K., Sadhasivam, S., and Sionov, E. (2019). Identification and toxigenic potential of fungi isolated from capsicum peppers. Microorganisms 7:90303. doi: 10.3390/microorganisms7090303
Golli-Bennour, E. E., Kouidhi, B., Bouslimi, A., Abid-Essefi, S., Hassen, W., and Bacha, H. (2010). Cytotoxicity and genotoxicity induced by aflatoxin b1, ochratoxin a, and their combination in cultured vero cells. J. Biochem. Mol. Toxicol. 24, 42–50. doi: 10.1002/jbt.20310
Guo, M., Jiang, W., Yang, M., Dou, X., and Pang, X. (2020). Characterizing fungal communities in medicinal and edible cassiae semen using high-throughput sequencing. Int. J. Food Microbiol. 319:108496. doi: 10.1016/j.ijfoodmicro.2019.108496
Han, Z., Ren, Y., Zhu, J., Cai, Z., Chen, Y., Luan, L., et al. (2012). Multianalysis of 35 mycotoxins in traditional chinese medicines by ultra-high-performance liquid chromatography–tandem mass spectrometry coupled with accelerated solvent extraction. J. Agr. Food Chem. 60, 8233–8247. doi: 10.1021/jf301928r
Han, Z., Zheng, Y., Luan, L., Cai, Z., Ren, Y., and Wu, Y. (2010). An ultra-high-performance liquid chromatography-tandem mass spectrometry method for simultaneous determination of aflatoxins b1, b2, g1, g2, m1 and m2 in traditional Chinese medicines. Anal. Chim. Acta 664, 165–171. doi: 10.1016/j.aca.2010.02.009
He, H., Xu, J., Zhou, T., Yang, Y., Yang, C., Xiao, C., et al. (2023). Metabolomic and microbiomic insights into color changes during the sweating process in dipsacus asper. Front. Microbiol. 14:1195088. doi: 10.3389/fmicb.2023.1195088
Hou, L., Gan, F., Zhou, X., Zhou, Y., Qian, G., Liu, Z., et al. (2018). Immunotoxicity of ochratoxin a and aflatoxin b1 in combination is associated with the nuclear factor kappa b signaling pathway in 3d4/21 cells. Chemosphere 199, 718–727. doi: 10.1016/j.chemosphere.2018.02.009
Ji, X., Jin, C., Xiao, Y., Deng, M., Wang, W., Lyu, W., et al. (2023). Natural occurrence of regulated and emerging mycotoxins in wheat grains and assessment of the risks from dietary mycotoxins exposure in China. Toxins (Basel). 15:60389. doi: 10.3390/toxins15060389
Jin-Ao, D., Shu-Lan, S. U., Hui, Y., Sheng, G., Pei, L., and Da-Wei, Q. (2013). Sweating of traditional Chinese medicinal materials during primary processing and its mechanisms of enzymatic reaction and chemical conversion. Chin. Tradit. Herb. Drug 44, 1219–1225. doi: 10.7501/j.issn.0253-2670.2013.10.001
Ke, K., Li, Q., Yang, X., Xie, Z., Wang, Y., Shi, J., et al. (2016). Asperosaponin vi promotes bone marrow stromal cell osteogenic differentiation through the pi3k/akt signaling pathway in an osteoporosis model. Sci. Rep. 6:35233. doi: 10.1038/srep35233
Khalifa, E., Mohesien, M. T., Mossa, M. I., Piekutowska, M., Alsuhaibani, A. M., Abdel-Wahab, B. A., et al. (2022). Diversity of toxigenic fungi in livestock and poultry feedstuffs. Int. J. Environ. Res. Public Health 19:127250. doi: 10.3390/ijerph19127250
Kumar, N., Khurana, S., and Pandey, V. N. (2023). Deciphering of seed health of common food grains (wheat, rice) of north eastern up and Gurgaon Haryana, India. Sci. Rep. 13:8480. doi: 10.1038/s41598-023-34510-3
Lahouar, A., Marin, S., Crespo-Sempere, A., Said, S., and Sanchis, V. (2016). Effects of temperature, water activity and incubation time on fungal growth and aflatoxin b1 production by toxinogenic aspergillus flavus isolates on sorghum seeds. Rev. Argent. Microbiol. 48, 78–85. doi: 10.1016/j.ram.2015.10.001
Li, Y., Wang, Z., Beier, R. C., Shen, J., De Smet, D., De Saeger, S., et al. (2011). T-2 toxin, a trichothecene mycotoxin: review of toxicity, metabolism, and analytical methods. J. Agric. Food Chem. 59, 3441–3453. doi: 10.1021/jf200767q
Liao, X., Sun, C., Wei, F., Zhou, L., and Kong, W. (2020). Exploration of the safe water content and activity control points for medicinal and edible lotus seeds from mildew. AMB Express 10:89. doi: 10.1186/s13568-020-01019-1
Llewellyn, G. C., Burkett, M. L., and Eadie, T. (1981). Potential mold growth, aflatoxin production, and antimycotic activity of selected natural spices and herbs. J. Assoc. Off. Anal. Chem. 64, 955–960. doi: 10.1093/jaoac/64.4.955
Locatelli, S., Scarpino, V., Lanzanova, C., Romano, E., and Reyneri, A. (2022). Multi-mycotoxin long-term monitoring survey on north-Italian maize over an 11-year period (2011–2021): the co-occurrence of regulated, masked and emerging mycotoxins and fungal metabolites. Toxins (Basel). 14:80520. doi: 10.3390/toxins14080520
Long, N., Liu, J., Liu, J., Li, Y., Hou, Y., Liao, X., et al. (2022). Single-molecule real-time sequencing to explore the mycobiome diversity in malt. Microbiol Spectr. 10:e51122. doi: 10.1128/spectrum.00511-22
Magoc, T., and Salzberg, S. L. (2011). Flash: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Marin, P., Jurado, M., Magan, N., Vazquez, C., and Gonzalez-Jaen, M. T. (2010). Effect of solute stress and temperature on growth rate and tri5 gene expression using real time RT-PCR in fusarium graminearum from spanish wheat. Int. J. Food Microbiol. 140, 169–174. doi: 10.1016/j.ijfoodmicro.2010.03.041
Martinez-Culebras, P. V., Gandia, M., Garrigues, S., Marcos, J. F., and Manzanares, P. (2021). Antifungal peptides and proteins to control toxigenic fungi and mycotoxin biosynthesis. Int. J. Mol. Sci. 22:13261. doi: 10.3390/ijms222413261
McKean, C., Tang, L., Billam, M., Tang, M., Theodorakis, C. W., Kendall, R. J., et al. (2006). Comparative acute and combinative toxicity of aflatoxin b1 and T-2 toxin in animals and immortalized human cell lines. J. Appl. Toxicol. 26, 139–147. doi: 10.1002/jat.1117
Moretti, A., Susca, A., Mule, G., Logrieco, A. F., and Proctor, R. H. (2013). Molecular biodiversity of mycotoxigenic fungi that threaten food safety. Int. J. Food Microbiol. 167, 57–66. doi: 10.1016/j.ijfoodmicro.2013.06.033
Mousa, W., Ghazali, F. M., Jinap, S., Ghazali, H. M., and Radu, S. (2011). Modelling the effect of water activity and temperature on growth rate and aflatoxin production by two isolates of aspergillus flavus on paddy. J. Appl. Microbiol. 111, 1262–1274. doi: 10.1111/j.1365-2672.2011.05134.x
Narvaez, A., Rodriguez-Carrasco, Y., Castaldo, L., Izzo, L., and Ritieni, A. (2020). Ultra-high-performance liquid chromatography coupled with quadrupole orbitrap high-resolution mass spectrometry for multi-residue analysis of mycotoxins and pesticides in botanical nutraceuticals. Toxins (Basel). 12:20114. doi: 10.3390/toxins12020114
Rizzo, I., Vedoya, G., Maurutto, S., Haidukowski, M., and Varsavsky, E. (2004). Assessment of toxigenic fungi on Argentinean medicinal herbs. Microbiol. Res. 159, 113–120. doi: 10.1016/j.micres.2004.01.013
Rodrigues, A. I., Gudina, E. J., Abrunhosa, L., Malheiro, A. R., Fernandes, R., Teixeira, J. A., et al. (2021). Rhamnolipids inhibit aflatoxins production in aspergillus flavus by causing structural damages in the fungal hyphae and down-regulating the expression of their biosynthetic genes. Int. J. Food Microbiol. 348:109207. doi: 10.1016/j.ijfoodmicro.2021.109207
Sadhasivam, S., Britzi, M., Zakin, V., Kostyukovsky, M., Trostanetsky, A., Quinn, E., et al. (2017). Rapid detection and identification of mycotoxigenic fungi and mycotoxins in stored wheat grain. Toxins 9:e302. doi: 10.3390/toxins9100302
Schmidt-Heydt, M., Abdel-Hadi, A., Magan, N., and Geisen, R. (2009). Complex regulation of the aflatoxin biosynthesis gene cluster of aspergillus flavus in relation to various combinations of water activity and temperature. Int. J. Food Microbiol. 135, 231–237. doi: 10.1016/j.ijfoodmicro.2009.07.026
Schmieder, R., and Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864. doi: 10.1093/bioinformatics/btr026
Sherertz, P. C., Eadie, T., Young, J. W., and Llewellyn, G. C. (1976). Aflatoxin occurrence on raw and cooked York soybeans inoculated with three aspergillus isolates. J. Assoc. Off. Anal. Chem. 59, 662–665. doi: 10.1093/jaoac/59.3.662
Skala, E., and Szopa, A. (2023). Dipsacus and scabiosa species-the source of specialized metabolites with high biological relevance: a review. Molecules 28:3754. doi: 10.3390/molecules28093754
Song, L., Pan, L., Jiang, N., Fu, J., Wan, L., and Wei, S. (2022). Effects of endophytic fungi on parasitic process of taxillus chinensis. Sci. Rep. 12:7744. doi: 10.1038/s41598-022-11940-z
Su, C., Hu, Y., Gao, D., Luo, Y., Chen, A. J., Jiao, X., et al. (2018). Occurrence of toxigenic fungi and mycotoxins on root herbs from Chinese markets. J. Food Protect. 81, 754–761. doi: 10.4315/0362-028X.JFP-17-405
Sun, J., Du, L., Liang, W., Qu, Z., Zhao, H., and Guan, W. (2023). Effect of postharvest processing on quality traits of radix gentianae macrophyllae: a integrative analysis of metabolomics and proteomics. Plant Physiol. Biochem. 204:108099. doi: 10.1016/j.plaphy.2023.108099
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). Mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tejero, P., Martin, A., Rodriguez, A., Galvan, A. I., Ruiz-Moyano, S., and Hernandez, A. (2021). In vitro biological control of aspergillus flavus by hanseniaspora opuntiae l479 and hanseniaspora uvarum l793, producers of antifungal volatile organic compounds. Toxins (Basel). 13:90663. doi: 10.3390/toxins13090663
Wang, S., Hua, Y., Xu, L., Zou, L., Liu, X., Luo, Y., et al. (2016). Quality evaluation of scrophulariae radix processed by different sweating methods based on simultaneous determination of multiple bioactive constituents combined with grey relational analysis. Molecules 21:70850. doi: 10.3390/molecules21070850
Wang, G., Wang, Y. L., Zhang, H. Y., Zhang, C. X., Yang, B. L., Huang, S. J., et al. (2020). Factors that affect the formation of mycotoxins: a literature review. Mycosystema 39, 477–491. doi: 10.13346/j.mycosystema.190334
Wei, G., Guo, X., Liang, Y., Liu, C., Zhang, G., Liang, C., et al. (2023). Occurrence of fungi and mycotoxins in herbal medicines and rapid detection of toxin-producing fungi. Environ. Pollut. 333:122082. doi: 10.1016/j.envpol.2023.122082
Wellford, T. E., Eadie, T., and Llewellyn, G. C. (1978). Evaluating the inhibitory action of honey on fungal growth, sporulation, and aflatoxin production. Z. Lebensm. Unters. Forsch. 166, 280–283. doi: 10.1007/BF01127653
Wicaksono, T., and Illes, C. B. (2022). From resilience to satisfaction: defining supply chain solutions for Agri-food smes through quality approach. PLoS One 17:e263393. doi: 10.1371/journal.pone.0263393
Wu, K., Jia, S., Xue, D., Rajput, S. A., Liu, M., Qi, D., et al. (2022). Dual effects of zearalenone on aflatoxin b1-induced liver and mammary gland toxicity in pregnant and lactating rats. Ecotoxicol. Environ. Saf. 245:114115. doi: 10.1016/j.ecoenv.2022.114115
Wu, Q., Wei, D., Dong, L., Liu, Y., Ren, C., Liu, Q., et al. (2019). Variation in the microbial community contributes to the improvement of the main active compounds of magnolia officinalis rehd. Chin. Med.: Et wils in the process of sweating, 14.
Wu, H., Yang, H. Y., You, X. L., and Li, Y. H. (2013). Diversity of endophytic fungi from roots of panax ginseng and their saponin yield capacities. Springerplus 2:107. doi: 10.1186/2193-1801-2-107
Xiao, S., Wu, Y., Gao, S., Zhou, M., Liu, Z., Xiong, Q., et al. (2023). Deciphering the hazardous effects of afb1 and T-2 toxins: unveiling toxicity and oxidative stress mechanisms in pk15 cells and mouse kidneys. Toxins (Basel). 15:80503. doi: 10.3390/toxins15080503
Xu, J., Zhao, Z., Guo, W., Ling, A., Wang, J., Wang, X., et al. (2022). Potential role of individual and combined effects of T-2 toxin, T-2 toxin and neosolaniol on the apoptosis of porcine leydig cells. Toxins (Basel). 14:20145. doi: 10.3390/toxins14020145
Yabe, K., Ando, Y., and Hamasaki, T. (1988). Biosynthetic relationship among aflatoxins b1, b2, g1, and g2. Appl. Environ. Microbiol. 54, 2101–2106. doi: 10.1128/aem.54.8.2101-2106.1988
Zhai, W., Qiao, Z., Xiang, P., Dang, Y., and Shi, Y. (2023). A uplc-ms/ms methodological approach for the analysis of 75 phenethylamines and their derivatives in hair. J. Pharm. Biomed. Anal. 229:115367. doi: 10.1016/j.jpba.2023.115367
Zhang, C., Dou, X., Zhang, L., Sun, M., Zhao, M., OuYang, Z., et al. (2018). A rapid label-free fluorescent aptasensor picogreen-based strategy for aflatoxin b(1) detection in traditional Chinese medicines. Toxins (Basel). 10:30101. doi: 10.3390/toxins10030101
Zhang, J., Kobert, K., Flouri, T., and Stamatakis, A. (2014). Pear: a fast and accurate illumina paired-end read merger. Bioinformatics 30, 614–620. doi: 10.1093/bioinformatics/btt593
Zhang, W., Xue, K., Gao, Y., Huai, Y., Wang, W., Miao, Z., et al. (2019). Systems pharmacology dissection of action mechanisms of dipsaci radix for osteoporosis. Life Sci. 235:116820. doi: 10.1016/j.lfs.2019.116820
Zheng, W., Feng, N., Wang, Y., Noll, L., Xu, S., Liu, X., et al. (2019). Effects of zearalenone and its derivatives on the synthesis and secretion of mammalian sex steroid hormones: a review. Food Chem. Toxicol. 126, 262–276. doi: 10.1016/j.fct.2019.02.031
Zheng, R. S., Wang, W. L., Tan, J., Xu, H., Zhan, R. T., and Chen, W. W. (2017). An investigation of fungal contamination on the surface of medicinal herbs in China. Chin. Med. 12:2. doi: 10.1186/s13020-016-0124-7
Keywords: Radix Dipsaci , sweating, mycobiota, toxigenic fungi, mycotoxins
Citation: Ge Y, Wang L, Su D, Yuan Q, Xiao C, Hu M, Kang C, Guo L, Zhou T and Zhang J (2024) The sweating process promotes toxigenic fungi expansion and increases the risk of combined contamination of mycotoxins in Radix Dipsaci. Front. Microbiol. 15:1394774. doi: 10.3389/fmicb.2024.1394774
Received: 02 March 2024; Accepted: 28 May 2024;
Published: 06 June 2024.
Edited by:
Silvani Verruck, Federal University of Santa Catarina, BrazilReviewed by:
Mengyue Guo, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaCopyright © 2024 Ge, Wang, Su, Yuan, Xiao, Hu, Kang, Guo, Zhou and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lanping Guo, Z2xwMDFAMTI2LmNvbQ==; Tao Zhou, dGFvemhvdTg4QDE2My5jb20=; Jinqiang Zhang, NTUyNDUwMzc0QHFxLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.