- 1School of Life Sciences, Guizhou Normal University, Guiyang, Guizhou, China
- 2Guizhou Chishui Alsophila National Nature Reserve Administration Bureau, Chishui, Guizhou, China
As an indispensable part of insects, intestinal symbiotic bacteria play a vital role in the growth and development of insects and their adaptability. Rhoptroceros cyatheae, the main pest of the relict plant Alsophila spinulosa, poses a serious threat to the development of the A. spinulosa population. In the present study, 16S rDNA and internal transcribed spacer high-throughput sequencing techniques were used to analyze the structure of intestinal microbes and the diversity of the insect feeding on two different plants, as well as the similarities between the intestinal microorganisms of R. cyatheae. The dominant bacteria of leaf endophytes were also compared based on the sequencing data. The results showed that Proteobacteria, Firmicutes, and Actinobacteria were the dominant phyla of intestinal bacteria, and Ascomycota was the dominant phylum of intestinal fungi. Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, Methylobacterium-Methylorubrum, and Enterococcus were the dominant genera in the intestine of R. cyatheae feeding on two plants, and the relative abundance was significantly different between the two groups. Candida was the common dominant genus of intestinal fungi in the two groups, and no significant difference was observed in its abundance between the two groups. This showed that compared with the intestinal fungi of R. cyatheae, the abundance of the intestinal bacteria was greatly affected by food. The common core microbiota between the microorganisms in A. spinulosa leaves and the insect gut indicated the presence of a microbial exchange between the two. The network correlation diagram showed that the gut microbes of R. cyatheae feeding on Gymnosphaera metteniana were more closely related to each other, which could help the host to better cope with the adverse external environment. This study provides a theoretical basis for the adaptation mechanism of R. cyatheae and a new direction for the effective prevention and control of R. cyatheae.
1 Introduction
The tree fern Alsophila spinulosa is a world-famous relict plant and is currently on the red list of threatened species by the International Union for Conservation of Nature (Ma et al., 2020). Rhoptroceros cyatheae (Hymenoptera: Selandriidae) mainly harms A. spinulosa and Gymnosphaera metteniana. Adult females of R. cyatheae lay eggs on the leaves of A. spinulosa saplings, and their larvae primarily feed on the mesophyll tissue. In severe cases, these larvae can consume the leaves of the entire plant, thus markedly affecting the photosynthesis and spore reproduction of A. spinulosa (Xu et al., 2021).
Plant tissues often produce many indigestible and toxic substances during growth and development; consequently, herbivorous insects have evolved a series of strategies to adapt to different plants, including working with their symbionts to absorb nutrients (Salem et al., 2014; Shikano, 2017; Mason et al., 2019a; McMillan, 2023). With the development of high-throughput technology, the contribution of intestinal symbiotic bacteria to host digestion and absorption (Engel and Moran, 2013; Jing et al., 2020), detoxification (Blanton and Peterson, 2020; Siddiqui et al., 2022), growth and development (Yang et al., 2022), and oviposition induction (Qiao et al., 2019) has been supported by extensive data. The gut microbiome of herbivorous insects is an important part of the insect–plant interaction, which is mainly manifested in two major functions of the gut microbiome as follows: supplementing nutrients and degrading toxic secondary metabolites (Hammer and Bowers, 2015; Pérez-Cobas et al., 2015). For example, the gut microbiome provides essential amino acids, vitamins, carbon, and nitrogen to the host, which ensures the normal growth and development of the insects feeding on unsuitable plants (Scully et al., 2013; Ayayee et al., 2014; Salem et al., 2014). The intestinal microbes of these insects can reduce the damage caused by the secondary metabolites of plants, such as tannins, caffeine, and nicotine, so that the insect can better digest the feeding plants (Berasategui et al., 2017; Zhang et al., 2020). In addition, gut microbes affect the growth and development, defense of natural enemies, mating, reproduction, and other aspects of these insects (Leftwich et al., 2018; Duplais et al., 2021).
The composition of gut microbial communities can be driven by various factors, including intestinal structure (Chen et al., 2016), geography and climate change (Luo et al., 2020), diet, and other factors (Yun et al., 2014). The specificity of the insects and its developmental stages are important factors affecting the intestinal communities. For example, the gut microbes of cockroaches and termites feeding on wood were different (Liu et al., 2020). Gao et al. (2019) found that Firmicutes were dominant in the gut of the third and fifth instar larvae of Spodoptera exigua, whereas Proteobacteria were dominant in other stages. Studies on Apis mellifera and Plutella xylostella have found that geographical locations and climatic changes are the factors affecting intestinal microbial composition (Ludvigsen et al., 2015; Kaur et al., 2022). Diet is the dominant factor associated with the changes in intestinal microorganisms, and food types can rapidly and greatly alter the intestinal microbial communities of insects (Ana et al., 2015). This dynamic change in intestinal microorganisms is one of the reasons why insects can successfully feed on different host plants (Strano et al., 2018; Zhang et al., 2022). Food can directly interact with the intestine after the insect has fed on the plant. Moreover, plant materials and endogenous microorganisms are the main force in shaping the intestinal microorganisms of insects (Hardoim et al., 2015; Bozorov et al., 2019; Chen et al., 2020; Xiong, 2022; Pirttilä et al., 2023). Therefore, investigating the structure of intestinal microbial communities in insects and the role of food in shaping this intestinal microbiome is crucial. At present, research on the effect of food on the intestinal symbiotic bacteria of R. cyatheae is lacking. Thus, in the present study, we compared the effects of food on the gut microbiota of R. cyatheae by analyzing the structure of endophytic bacteria in the leaves of two host plants. We also analyzed the diversity of gut microbiota of R. cyatheae fed with two different host plants during its peak occurrence period (namely May–June) in a year. This study aimed to assess the effects of host plants on R. cyatheae and to provide a basis for the development of efficient and green control measures against the insect.
2 Materials and methods
2.1 Sample collection
Rhoptroceros cyatheae was obtained from the Guizhou Chishui Alsophila National Nature Reserve (28°25′12″N, 106°01′03″E) in May 2023. All larvae collected from the forest were fed in an artificial climate incubator (SPX-280, Ningbo Jiangnan Instrument Factory, China; 26 ± 1°C, 75% ± 10% relative humidity, and 16L:8D photoperiod). Newly hatched larvae were reared with A. spinulosa and G. metteniana until the fifth instar. These two plants were obtained from the Chishui Alsophila National Nature Reserve. After 24 h of starvation, the larvae were sterilized with 75% ethanol for 1 min and then washed thrice with sterile water. The complete intestinal tract was dissected in sterile phosphate-buffered saline under a stereoscope, and 15–20 intestinal tracts were mixed as a biological replication, with three replicates per treatment (As: feeding on A. spinulosa; Gm: feeding on G. metteniana). The larvae were identified as the fifth instar according to their molting times (Xu et al., 2021). A sterile 15-mm sample puncher was used to collect samples from fresh and intact A. spinulosa leaves (AsL) and G. metteniana leaves (GmL) into a sterile culture plate, and 75% ethanol was used to clean the surface. The samples were mixed and ground under liquid nitrogen for subsequent DNA extraction.
2.2 DNA extraction and PCR amplification
The total DNA of the samples was extracted as for the E.Z.N.A® soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) instructions. The primers 799F (5′-AACMGGATTAGATACCCKG-3′)/1193R (5′ACGTCATCCCCACCTTCC-3′) and ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′)/ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′) were used for PCR. PCR cycling conditions were as follows: 5 min at 94°C for initialization, 30 cycles of denaturation for 30 s at 95°C, 30 s of annealing at 55°C, and 30 s of extension at 72°C, followed by a 10-min final elongation at 72°C. The PCR product was extracted by performing 2% agarose gel electrophoresis and purification with PCR Clean-Up Kit (YuHua, Shanghai, China) according to the manufacturer’s instructions and quantified using Qubit 4.0 (Thermo Fisher Scientific, USA). NEXTFLEX® Rapid DNA-Seq Kit (YuHua, Shanghai, China) was used to construct a library of purified PCR products.
2.3 Quality control and operational taxonomic unit (OTU) identification
Fastp1 software was used to perform quality control on the double-ended original sequence, and FLASH2 software was used for splicing: (i) The reads were truncated at any site receiving an average quality score of <20 over a 50 bp sliding window, the truncated reads shorter than 50 bp were discarded, and the reads containing ambiguous characters were also discarded; (ii) Only overlapping sequences longer than 10 bp were assembled according to their overlapped regions. The maximum mismatch ratio of the overlap region was 0.2. Reads that could not be assembled were discarded; (iii) The samples were distinguished according to their barcodes and primers, and the sequence direction was adjusted, the exact barcode was matched; two nucleotides were mismatched in primer matching. Then, the optimized sequences were clustered into operational taxonomic units (OTUs) using UPARSE (version 7.1) with 97% sequence similarity. The OTU table was manually filtered, and chimeric, chloroplast, and mitochondrion sequences were removed. RDP Classifier (version 2.11) was used to compare each OTU representative sequence with those in the database Silva and Unite, and the confidence threshold was 70%.
2.4 Diversity analysis
Alpha and beta diversity indices were estimated at the OTU level using MOTHUR and UniFrac, respectively. The Kruskal–Wallis test was used to analyze the α-diversity index of bacteria and fungi in the samples. The Wilcoxon signed-rank test and T-test (false discovery rate correction) were used for pairwise comparison. The principal coordinate analysis was performed based on the Bray–Curtis distance, and the Adonis test was used to analyze the difference between the groups, with p < 0.05 considered statistically significant. The linear discriminant analysis (LDA) effect size3 was calculated to identify the significantly abundant taxa (phylum to genera) of bacteria among the different groups (LDA > 4, p < 0.05). To understand the relationships among the genera, Spearman’s correlation coefficients were used for network analyses (Networkx: version 1.11). Network topological properties were calculated using Gephi. PICRUSt24 and FAPROTAX (1.2.1) were used to predict the function of the microbial community in different samples.
3 Results
3.1 Annotation and evaluation of sequences
Illumina Miseq sequencing was performed to characterize the 16S rDNA and ITS2 regions of the leaves and R. cyatheae gut. A total of 831,798 bacterial reads (Supplementary Table S1) with an average length of 376 bp were obtained. Additionally, 1,074,630 fungal reads with an average length of 251 bp (Supplementary Table S2) were obtained. Rarefaction curves indicated that the sequencing volume and sample depth were saturated (Supplementary Figure S1). In addition, good coverage reflects the integrity of sequencing (Wang et al., 2023). In this study, the coverage of each sample was above 99%, indicating that most of the species in the sample were detected (Supplementary Table S3).
3.2 Comparison of the microbial communities of the plant and the insect
After high-quality filtering, the optimized sequence clustering analysis based on 97% sequence similarity assigned the high-quality bacterial sequences into 721 OTUs (544 intestinal bacterial OTUs and 603 endophytic bacterial OTUs in leaves, shared 426), belonging to 16 phyla, 24 classes, 75 orders, 132 families, 265 genera, and 417 species. Among them, 201 OTUs were observed in As, 75 were observed in Gm, and 268 were shared between groups. The analysis of OTUs in the intestine and host leaves showed that 144 OTUs were observed in As, 171 were observed in AsL, and 325 were shared between groups. Furthermore, 154 OTUs were observed in Gm, 195 were observed in GmL, and 189 were shared between groups (Figure 1A). The high-quality sequences of fungi were assigned into 1,435 OTUs (627 intestinal fungal OTUs and 1,404 endophytic fungal OTUs in leaves, shared 596), belonging to 3 phyla, 21 classes, 75 orders, 215 families, 435 genera, and 637 species. Among them, 147 OTUs were observed in As, 178 were observed in Gm, and 302 were shared between groups. The analysis of OTUs in the intestine and host leaves showed that 144 OTUs were observed in As, 171 were observed in AsL, and 325 were shared between groups. Furthermore, 154 OTUs were observed in Gm, 195 were observed in GmL, and 189 were shared between groups (Figure 1B).
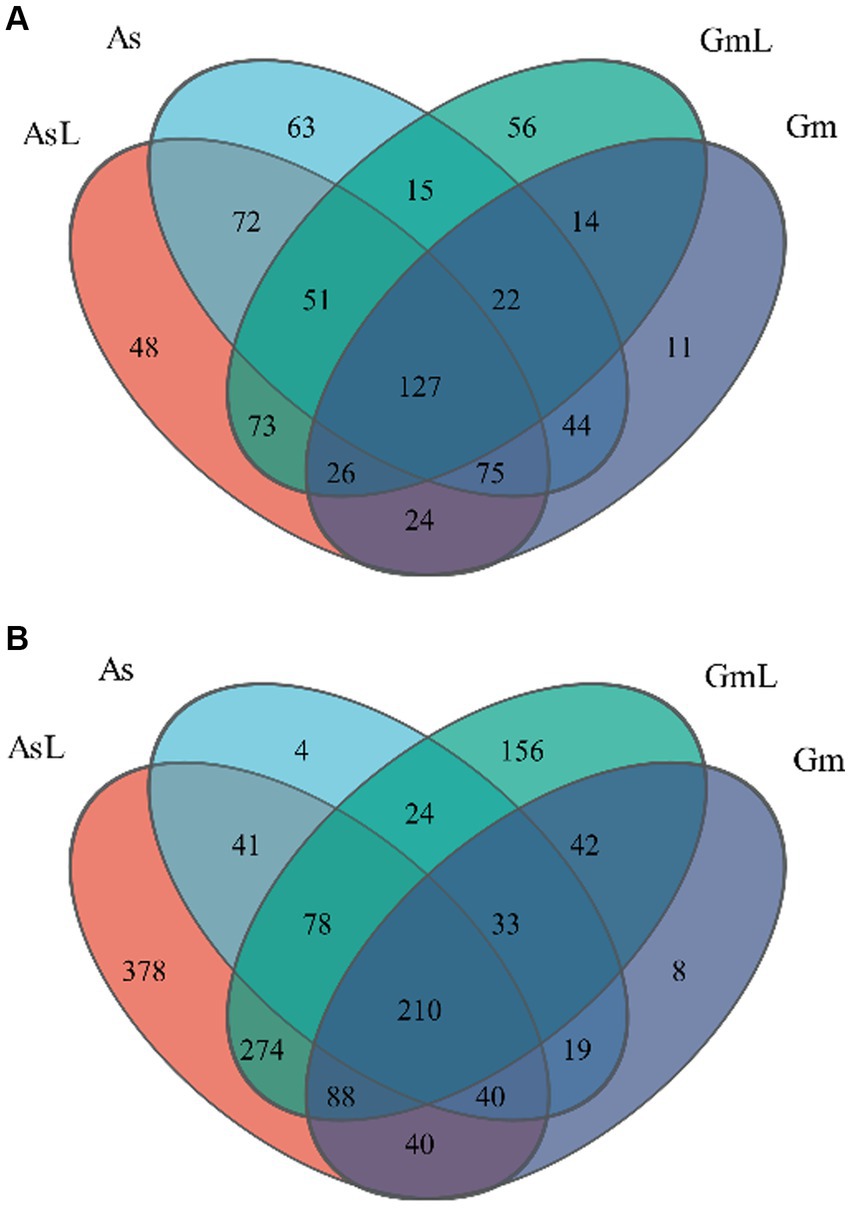
Figure 1. Venn diagram of the microbiota of R. cyatheae guts and host plant leaves. (A) Bacteria. (B) Fungi.
Proteobacteria, Firmicutes, and Actinobacteria were dominant phyla in the As and Gm groups, accounting for 97.44 and 99.74% of the total sequences, respectively (Figure 2A). Proteobacteria was the dominant phylum in the AsL and GmL groups, accounting for 86.60 and 90.41% of the total sequences, respectively. At the genus level, the top five abundant genera in the gut of the two groups of R. cyatheae were different. The dominant genera in the gut of R. cyatheae feeding on A. spinulosa were Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium (31.07%) and Methylobacterium-Methylorubrum (15.55%), which were also found in the host plants and accounted for a high proportion (25.83 and 11.96%, respectively). Enterococcus (59.89%) and unclassified_o_Enterobacterales (23.62%) were the dominant genera in the gut of R. cyatheae feeding on G. metteniana but were absent in GmL (Figure 2B). Collectively, the results showed that Proteobacteria, Firmicutes, and Actinobacteria were the most abundant phyla in the two hosts, whereas the dominant genera changed according to the host. At the phylum and genus levels, the similarity between the intestinal bacteria of R. cyatheae feeding on A. spinulosa and its host plant bacteria was higher than that observed in the Gm group. At the phylum level, the dominant intestinal fungus of the two feeding groups of R. cyatheae was Ascomycota (As: 98.98%; Gm: 96.58%). The dominant fungus in the AsL and GmL groups was also Ascomycota, accounting for 78.16 and 91.74% of the total sequences, respectively (Figure 2C). At the genus level, the dominant genus of intestinal fungi in the two feeding groups of R. cyatheae was Candida (As: 93.49%; Gm: 89.28%). However, Candida was not detected in the AsL and GmL groups (Figure 2D). In terms of fungal genera, the dominant genus in R. cyatheae was not affected by the microbiome of these two host leaves; nonetheless, the fungus exhibited its formation mechanism, which may play a special role in the intestine.

Figure 2. Microbial composition of R. cyatheae guts and host leaves. (A) Bacterial phylum level. (B) Bacterial genus level. (C) Fungal phylum level. (D) Fungal genus level.
3.3 Diversity analysis of plant and insect microbiomes
Alpha diversity was used to assess the richness and diversity between groups. The intestinal bacteria of R. cyatheae feeding on different plants showed significant differences in Chao (Figure 3A) and Shannon indices (Figure 3B). Figure 3 shows that bacterial diversity in the intestine of R. cyatheae feeding on G. metteniana decreased significantly. For fungi, no significant difference was observed in bacterial diversity and richness in the intestine of R. cyatheae fed on different plants (Figures 3C,D). It is worth noting that when the Wilcoxon signed-rank test was used for pairwise comparison, we found that at the bacterial level, the Chao and Shannon indices of the gut microbiome of R. cyatheae were close to those of the leaves of the plants they feed on. However, significant differences were observed for fungal communities. The overall difference is explained in the principal coordinate analysis (PCoA) diagram (bacteria: Adonis, R2 = 0.897, p = 0.001; fungi: Adonis, R2 = 0.938, p = 0.001). The intestinal bacteria of R. cyatheae feeding on different host plants were clustered into two different quadrants. The similarity among fungi was higher than that among bacteria (Figure 4). The similarity among bacteria between the intestinal communities of R. cyatheae and the leaf communities of host plants was higher than that among fungi, showing a closer distance in the PCoA diagram (Figures 4A,C). To further elucidate the effects of the two host plants on the microbiota of R. cyatheae larvae, differential abundances of dominant genera were compared. Notably, Enterococcus, Allorhizobium–Neorhizobium–Pararhizobium–Rhizobium, unclassified_o__Enterobacterales, and Glutamicibacter showed differential abundances between A. spinulosa- and G. metteniana-feeding R. cyatheae. Allorhizobium–Neorhizobium–Pararhizobium–Rhizobium showed no differential abundance between R. cyatheae and host plants. These results indicate that host plants affected the intestinal microbiota of R. cyatheae larvae (Figures 4B,D).
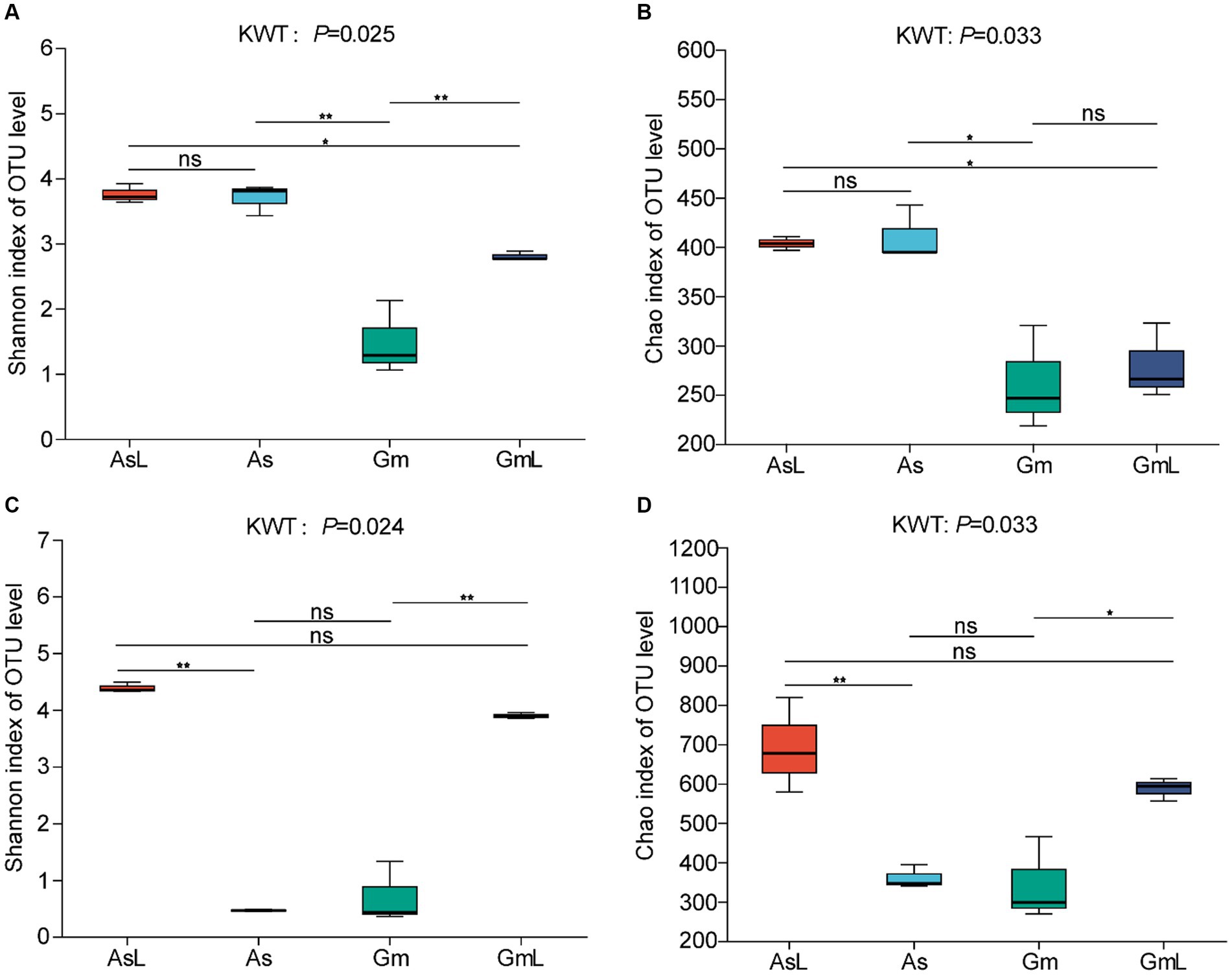
Figure 3. Alpha diversity of R. cyatheae gut and host leaf communities. Shannon diversity (A) bacteria; (C) fungi, Chao index (B) bacteria; (D) fungi. In the figure, * represents the significant difference at p < 0.05; ** represents the significant difference at p < 0.01; ns means no difference.
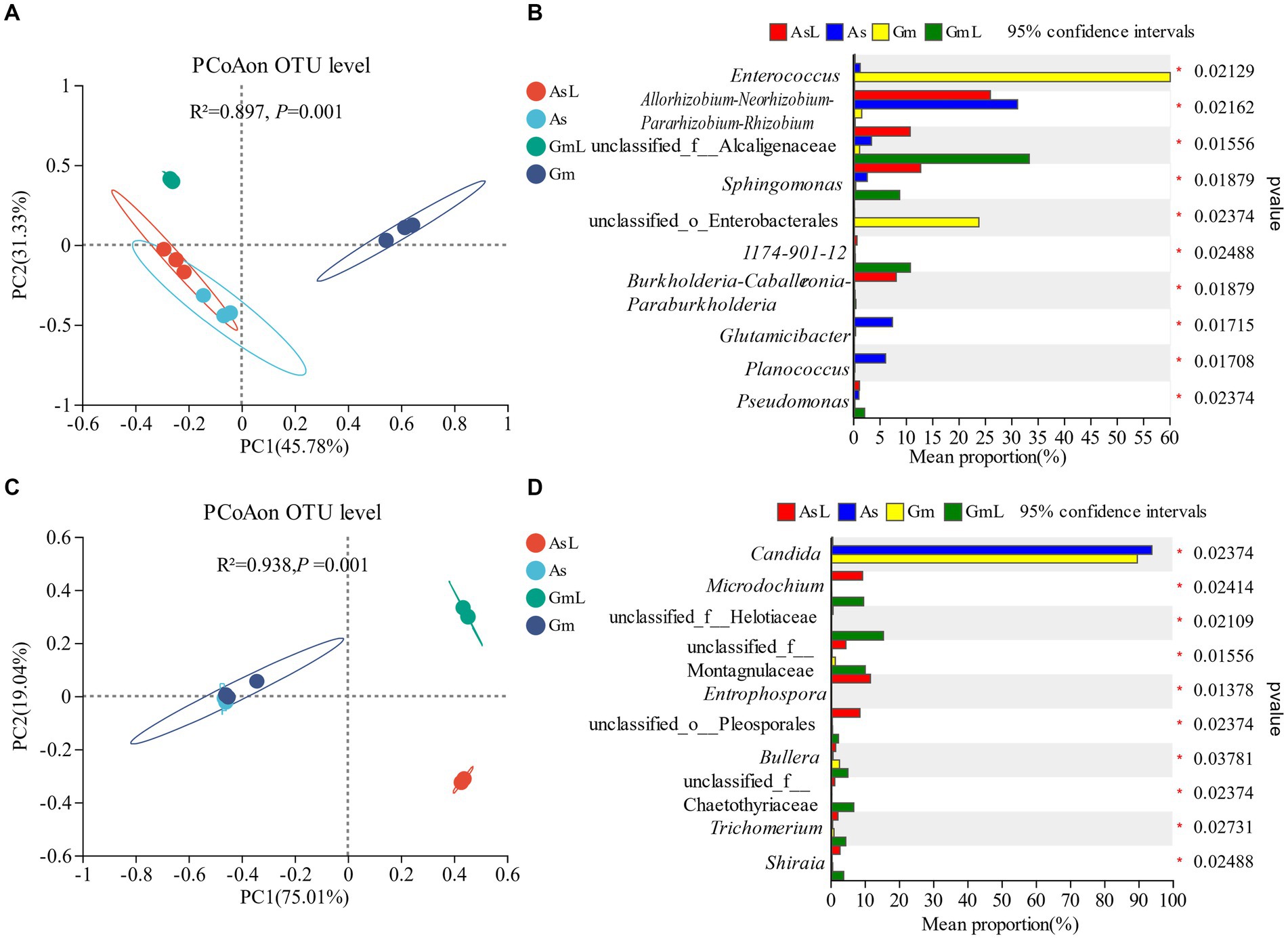
Figure 4. Analysis of bacteria and fungi in the intestine of R. cyatheae feeding on different hosts and host plants; principal coordinate analysis and genus levels. Bacteria: (A,B); fungi: (C,D).
3.4 Construction of the ecological network of intestinal communities
Association networks were constructed to determine the patterns of gut bacterial communities of R. cyatheae fed on A. spinulosa and G. metteniana (Figure 5). The network diagram of intestinal bacteria of the insect fed on A. spinulosa included 27 nodes and 172 edges (88 positive and 84 negative correlations) and that of intestinal bacteria of the insect fed on G. metteniana included 29 nodes and 223 edges (197 positive and 26 negative correlations). There were 53 positive correlations and 65 negative correlations among the fungi of R. cyatheae fed on A. spinulosa, and 159 positive correlations and 49 negative correlations among the fungi of R. cyatheae fed on G. metteniana, indicating that the complexity and pattern of the intestinal communities network structure in R. cyatheae fed on G. metteniana were higher than those in R. cyatheae fed on A. spinulosa. We found that Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, which showed the highest abundance in the As group, was not closely related to other bacteria in the network diagram (5 degrees), and the second dominant genus Methylobacterium-Methylorubrum (16 degrees) was the most closely related one. The highest abundance of Enterococcus was 2 degrees, and there were 22 genera with more than 16 degrees of abundance in the Gm group. Although no difference was observed in the abundance of the dominant genus Candida between the two groups, the degree of association with other genera in the network diagram was not the same (As: 11 degrees; Gm: 18 degrees). The above results indicated that the dominant intestinal bacteria of R. cyatheae fed on A. spinulosa were less related to other genera in life activities than those in R. cyatheae fed on G. metteniana. Overall, the results indicated that there were more cooperation and exchange events among most bacterial genera during the adaptation of R. cyatheae larvae to different hosts.
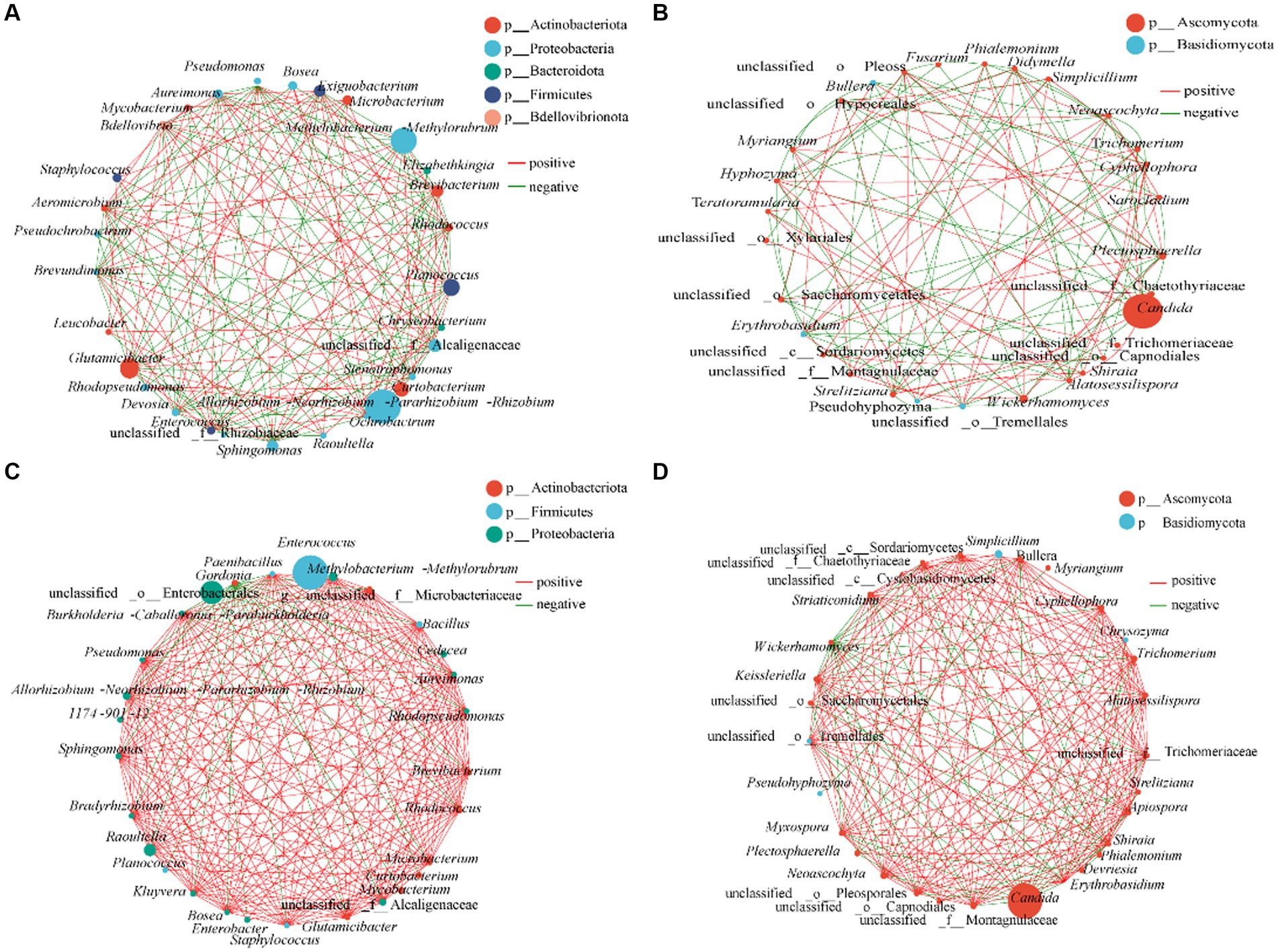
Figure 5. Interaction network diagram of the gut microbial genus classification level of R. cyatheae feeding on different host plants. (A,B) Intestinal bacteria and fungi of R. cyatheae feeding on A. spinulosa. (C,D) Intestinal bacteria and fungi of R. cyatheae feeding on G. metteniana. Nodes represent the only genus, the size of each node represents the abundance of species, the red line represents a positive correlation, and the green line represents a negative correlation. The thickness of the line indicates the size of the Spearman correlation coefficient. The figure shows that the correlation coefficient exceeds 0.6 and p < 0.05.
3.5 Functional prediction of gut microbiota
The PICRUSt2 results showed that the functional prediction categories of the gut bacteria of R. cyatheae feeding on different hosts were focused on metabolism, environmental information processing, genetic information processing, cell transformation, human diseases, and organismal systems. Additionally, the relative abundance of the metabolic pathway was the highest (Figure 6A). In addition, significant differences were observed in the relative abundance of the secondary classification level of metabolic pathways in each treatment (Figure 6B). Specifically, the relative abundance of carbohydrate metabolism (p < 0.05), amino acid metabolism (p < 0.05), and membrane transport (p < 0.05) in the gut bacteria of R. cyatheae feeding on G. metteniana was significantly higher than that of those feeding on A. spinulosa. The relative abundance of cofactor and vitamin metabolism (p < 0.05), xenobiotics symbiosis and metabolism (p < 0.05), and cell growth and death (p < 0.05) was dominant in the gut bacteria of R. cyatheae feeding on A. spinulosa.
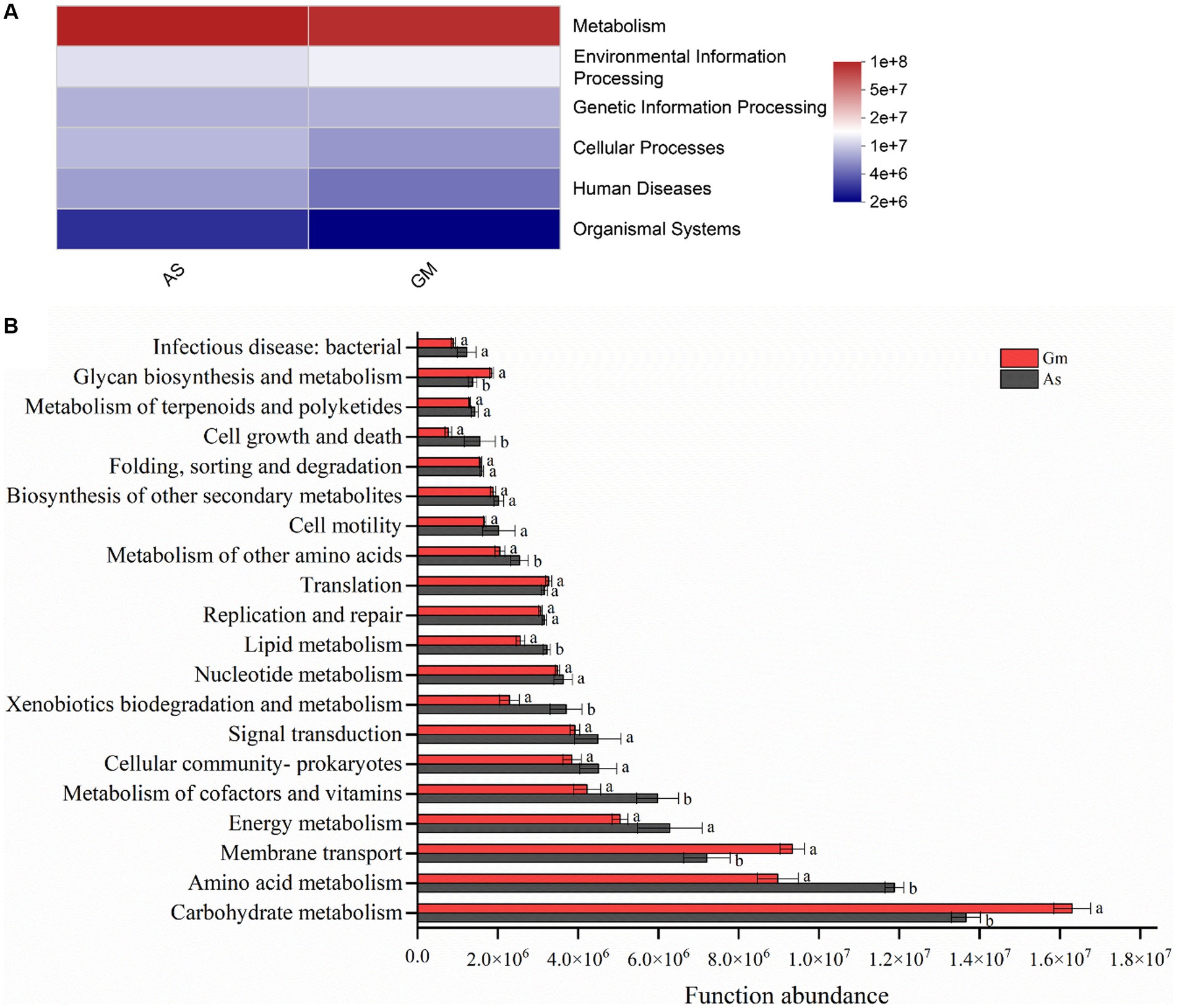
Figure 6. Comparison of PICRUSt2 function prediction in the gut bacterial communities of R. cyatheae fed on different host plants. (A) Heatmap of pathway level one; (B) analysis of metabolic pathway level two. Different lowercase letters above the bars indicate significant differences among different treatments (p < 0.05).
The levels of adenosine triphosphatase, DNA-directed RNA polymerase, and DNA-directed DNA polymerase were high in both groups (Figure 7). No significant difference was observed in the function of the intestinal fungi of R. cyatheae feeding on the two host plants.
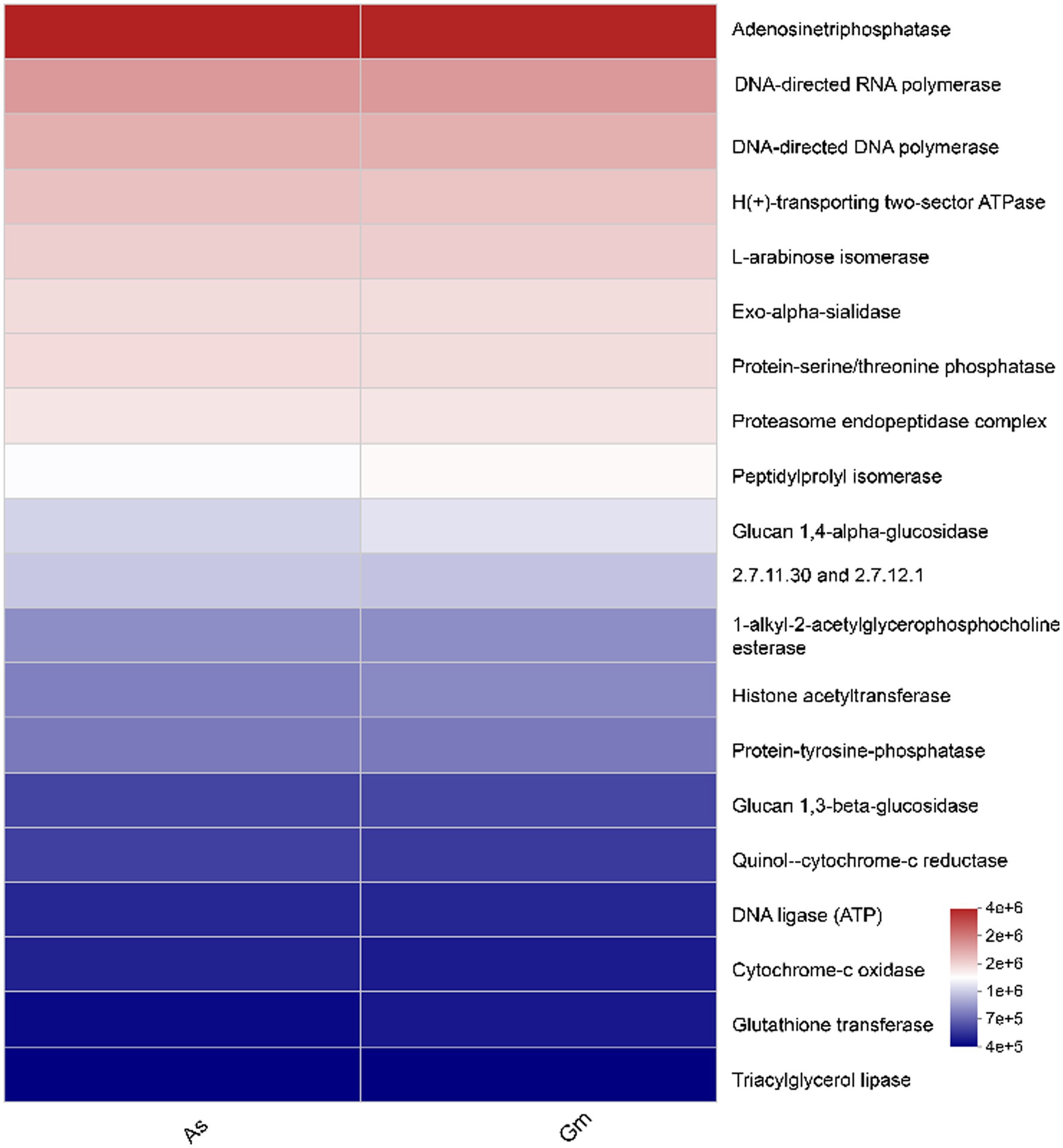
Figure 7. Heatmap of Kyoto Encyclopedia of Gene and Genomes function prediction of the intestinal fungi of R. cyatheae feeding on different hosts.
4 Discussion
The gut microbiota of insects play important and diverse roles in host digestion (Engel and Moran, 2013; Jing et al., 2020), growth and development (Yang et al., 2022), detoxification (Siddiqui et al., 2022), oviposition induction (Qiao et al., 2019), and essential vitamin and amino acid production (Bisch et al., 2018). These effects enable gut microbes to enhance the adaptability of insects to the external environment, which is important for the survival and expansion of insect populations. Numerous studies demonstrated that the structure and diversity of the gut microbiota of insects could be affected by host diets (Santos-Garcia et al., 2020; Li et al., 2021). In the present study, we compared the gut microbial communities of R. cyatheae fed on two host plants. The Chao and Shannon indices of gut bacterial communities of R. cyatheae larvae fed on A. spinulosa were significantly higher than those fed on G. metteniana, which was different from the gut microbiota of overwintering R. cyatheae larvae measured before, indicating that environmental changes could significantly affect the bacterial structure of insects (Martemyanov et al., 2016; Zhang et al., 2023). Proteobacteria and Firmicutes dominated microbial communities of R. cyatheae larvae. This result was similar to the intestinal bacterial communities of many insects, such as honey bees, Anoplophora glabripennis, and Spodoptera frugiperda (Warnecke et al., 2007; De Oliveira Scoaris et al., 2021; Wang et al., 2022; Han et al., 2023; Wang et al., 2023). The dominant bacteria of the two groups of R. cyatheae were significantly different at the genus level. Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium is a common bacterial genus in roots and leaves, which accounts for a relatively high proportion in the intestines of R. cyatheae fed on A. spinulosa, and may enter the intestines via the leaves to become resident or transit bacteria. Enterococcus, which dominated the gut of R. cyatheae feeding on G. metteniana, has been found in Helicoverpa armigera, honey bees, and Spodoptera littoralis, and it is a dominant genus in many insects. It plays an active role in insect host adaptation, primarily by synthesizing amino acids and vitamins, degrading secondary compounds and cell walls, regulating intestinal pH, and enhancing intestinal immunity (Ruiz-Rodriguez et al., 2012; Dantur et al., 2015; Vilanova et al., 2016; Shao et al., 2017; De Oliveira Scoaris et al., 2021; Li et al., 2022). We speculate that Enterococcus may play a role in the adaptability of R. cyatheae to G. metteniana, which may be caused by different plant secondary compounds in the leaves of the two hosts. It is necessary to adjust the intestinal microorganisms to absorb energy more effectively when the insect is feeding on G. metteniana.
We found no difference in the dominant phylum of intestinal fungi in the insect fed on the two host plants. Ascomycota dominated the two groups, which was consistent with the results of the intestinal fungi of Bombyx mori, aphid, and A. glabripennis (Chen et al., 2018; Mason et al., 2019b; Wolfgang et al., 2023). Candida was the dominant genus in the two groups, which is widespread in the environment and causes most fungal infections worldwide (Silva et al., 2012). Candida has been isolated from the intestines of Agrilus mali, Dendroctonus armandi, and other insects (Hu et al., 2015; Zhang et al., 2018). Candida produces xylanase and lipase, which are involved in adipose tissue decomposition and lipoprotein degradation in host life. It converts oil into free fatty acids and partially acyl glycerol (Suh and Blackwell, 2004; Hu et al., 2015; Zhang et al., 2018). As the dominant intestinal fungus of R. cyatheae, the function of Candida is still unclear and needs further investigation.
Plant and herbivory insect microbiomes are in a dynamic two-way interaction (Wolfgang et al., 2023). Insects consume plant materials and endogenous microorganisms as food, shaping the microbial community of the insect gut, which can be linked to the ability of insects to defeat plant defenses (Montagna et al., 2015; Pirttilä et al., 2023). Gut microbes can transfer genes to insects via horizontal gene transfer and provide enzymes to digest plant materials to help the insects quickly adapt to host plants (Hansen and Moran, 2014). In the present study, we found that feeding on different host plants affected the gut microbiota of R. cyatheae. We found no difference in the abundance of Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium between AsL and R. cyatheae (Welch T-test: p > 0.05) (Wolfgang et al., 2023) found that Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium existed in soil, leaves, and aphids. However, we found that this shared microorganism was less abundant in R. cyatheae fed on G. metteniana, and bacteria with high abundance in the gut of R. cyatheae, such as Enterococcus, were rare in the leaves of G. metteniana (Welch T-test: p < 0.05). It indicates the similarities between the microbiomes of insect guts and host leaves, and the extent of the overlap between the plant and insect microbiomes is likely highly dependent on species specificity (Pirttilä et al., 2023). Therefore, we concluded that the similarity between the intestinal bacteria of the GmL and R. cyatheae was lower than the similarity between the intestinal bacteria of the AsL and R. cyatheae. The intestinal fungi were less affected by leaf endophytes, and no significant difference was observed between the two groups. This may be related to the different metabolites of leaves. The habit of feeding on different nutrients or potentially toxic substances may help the insect effectively control the intestinal microbial species and rapidly degrade and digest these substances, thus helping the insect better adapt to different environments (Meriweather et al., 2013; Šigutová et al., 2023). Besides defeating plant defenses and supporting exploitation by the insect herbivore, the transmitted microbes can alter the survival, fecundity, and immunity of the insect host and can therefore alter the fitness of the herbivore (Saikkonen et al., 1996; Mauck et al., 2016) found that the relative growth rate of larvae was lower and the larval stage was longer when feeding on plants with higher mean endophytic bacteria abundances. We also found significant differences in the growth and development of R. cyatheae fed on the two host plants (Supplementary Table S5). The association network revealed that different hosts affected the microbial networks. The higher numbers of network topology properties, such as the number of nodes, positive correlations, negative correlations, and degree observed in the group feeding on G. metteniana indicated a complex network for this group. The results showed that feeding on G. metteniana was superior at enhancing the complexity of the intestinal microbial network and the resistance of the system to the external environment, which was consistent with the higher survival rate of R. cyatheae fed on G. metteniana by Xiao et al. (2023). The PICRUSt2 results showed that the functional prediction of the gut bacteria of R. cyatheae feeding on G. metteniana was more closely related to carbohydrate and amino acid metabolism, which could better provide nutrition for host insects and enhance the adaptability of the hosts.
5 Conclusion
This study highlights the importance of the effect of diet on the structure of intestinal microbial communities of R. cyatheae. The intestinal microbial communities of R. cyatheae fed on different host plants were diverse, and the dominant bacteria were also different accordingly. After feeding on different diet, the intestinal fungi of R. cyatheae showed a more stable community structure than that of the intestinal bacteria. There were more events of cooperation and communication among gut microbes of R. cyatheae feeding on G. metteniana. Altogether, this study provides a theoretical basis for further understanding the adaptability of R. cyatheae to host plants.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because the research object is small herbivorous insects, and the number is small and does not involve ethics.
Author contributions
BZ: Investigation, Writing – original draft, Writing – review & editing. WY: Funding acquisition, Project administration, Resources, Writing – review & editing. QH: Funding acquisition, Supervision, Writing – review & editing. HC: Project administration, Writing – original draft, Writing – review & editing. BC: Data curation, Validation, Writing – review & editing. XB: Conceptualization, Investigation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by “Pest resistance mechanisms of different Cyathea spinosa based on three-generation full-length transcriptomes, two-generation transcriptomes, and protein metabolomes” (Grant No. 11904–0522093).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1392586/full#supplementary-material
Footnotes
1. ^https://github.com/OpenGene/fastp, version 0.19.6.
2. ^http://www.cbcb.umd.edu/software/flash, version 1.2.11.
References
Ayayee, P., Rosa, C., Saunders, M., Felton, G. W., Ferry, G., and Hoover, K. (2014). Gut microbes contribution to nitrogen provisioning in a wood-feeding Cerambycid. Environ. Entomol. 43, 903–912. doi: 10.1603/EN14045
Berasategui, A., Salem, H., Paetz, C., Santoro, M., Gershenzon, J., Kaltenpoth, M., et al. (2017). Gut microbiota of the pine weevil degrades conifer diterpenes and increases insect fitness. Mol. Ecol. 26, 4099–4110. doi: 10.1111/mec.14186
Blanton, A. G., and Peterson, B. F. (2020). Symbiont-mediated insecticide detoxification as an emerging problem in insect pests. Front. Microbiol. 11:547108. doi: 10.3389/fmicb.2020.547108
Bozorov, T. A., Rasulov, B. A., and Zhang, D. Y. (2019). Characterization of the gut microbiota of invasive Agrilus mali Matsumara (Coleoptera: Buprestidae) using high-throughput sequencing: uncovering plant cell-wall degrading bacteria. Sci. Rep. 9:4923. doi: 10.1038/s41598-019-41368-x
Chen, B., Du, K., Sun, C., Vimalanathan, A., Liang, X., Li, Y., et al. (2018). Gut bacterial and fungal communities of the domesticated silkworm (Bombyx mori) and wild mulberry-feeding relatives. ISME J. 12, 2252–2262. doi: 10.1038/s41396-018-0174-1
Chen, B., Teh, B. S., Sun, C., Hu, S., Lu, X., Boland, W., et al. (2016). Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci. Rep. 6:29505. doi: 10.1038/srep29505
Chen, B. S., Xie, S., Zhang, X. C., Zhang, N., Feng, H. H., Sun, C., et al. (2020). Gut microbiota metabolic potential correlates with body size between mulberry-feeding lepidopteran pest species. Pest Manag. Sci. 76, 1313–1323. doi: 10.1002/ps.5642
Dantur, K. I., Enrique, R., Welin, B., and Castagnaro, A. P. (2015). Isolation of cellulolytic bacteria from the intestine of Diatraea saccharalis larvae and evaluation of their capacity to degrade sugarcane biomass. AMB Express 5, 15–11. doi: 10.1186/s13568-015-0101-z
De Oliveira Scoaris, D., Hughes, F. M., Silveira, M. A., Evans, J. D., Pettis, J. S., Bastos, E. M. A., et al. (2021). Microbial communities associated with honey bees in Brazil and in the United States. Braz. J. Microbiol. 52, 2097–2115. doi: 10.1007/s42770-021-00539-7
Duplais, C., Sarou-Kanian, V., Massiot, D., Hassan, A., Perrone, B., Estevez, Y., et al. (2021). Gut bacteria are essential for normal cuticle development in herbivorous turtle ants. Nat. Commun. 12:676. doi: 10.1038/s41467-021-21065-y
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects–diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Gao, X., Li, W., Luo, J., Zhang, L., Ji, J., Zhu, X., et al. (2019). Biodiversity of the microbiota in Spodoptera exigua (Lepidoptera: Noctuidae). J. Appl. Microbiol. 126, 1199–1208. doi: 10.1111/jam.14190
Hammer, T. J., and Bowers, M. D. (2015). Gut microbes may facilitate insect herbivory of chemically defended plants. Oecologia 179, 1–14. doi: 10.1007/s00442-015-3327-1
Han, S., Zhou, Y., Wang, D., Qin, Q., Song, P., and He, Y. (2023). Effect of different host plants on the diversity of gut bacterial communities of Spodoptera frugiperda (JE smith, 1797). Insects 14:264. doi: 10.3390/insects14030264
Hansen, A. K., and Moran, N. A. (2014). The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol. Ecol. 23, 1473–1496. doi: 10.1111/mec.12421
Hardoim, P. R., Van Overbeek, L. S., Berg, G., Pirttilä, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/MMBR.00050-14
Hu, X., Li, M., and Chen, H. (2015). Community structure of gut fungi during different developmental stages of the Chinese white pine beetle (Dendroctonus armandi). Sci. Rep. 5:8411. doi: 10.1038/srep08411
Jing, T. Z., Qi, F. H., and Wang, Z. Y. (2020). Most dominant roles of insect gut bacteria: digestion, detoxification, or essential nutrient provision? Microbiome 8:38. doi: 10.1186/s40168-020-00823-y
Kaur, M., Thakur, M., Sagar, V., and Sharma, R. (2022). Diversity of culturable gut bacteria of diamondback moth, Plutella xylostella (Linnaeus) (Lepidoptera: Yponomeutidae) collected from different geographical regions of India. Mol. Biol. Rep. 49, 7475–7481. doi: 10.1007/s11033-022-07550-7
Leftwich, P. T., Hutchings, M. I., and Chapman, T. (2018). Diet, gut microbes and host mate choice: understanding the significance of microbiome effects on host mate choice requires a case by case evaluation. BioEssays 40:e1800053. doi: 10.1002/bies.201800053
Li, C., Han, G., Sun, J., Huang, L., Lu, Y., and Xia, Y. (2022). The gut microbiota composition of Cnaphalocrocis medinalis and their predicted contribution to larval nutrition. Front. Microbiol. 13:909863. doi: 10.3389/fmicb.2022.909863
Li, H., Zhao, C., Yang, Y., Zhou, Z., Qi, J., and Li, C. (2021). The influence of gut microbiota on the fecundity of Henosepilachna vigintioctopunctata (Coleoptera: Coccinellidae). J. Insect Sci. 21:15. doi: 10.1093/jisesa/ieab061
Liu, Y., Shen, Z., Yu, J., Li, Z., Liu, X., and Xu, H. (2020). Comparison of gut bacterial communities and their associations with host diets in four fruit borers. Pest Manag. Sci. 76, 1353–1362. doi: 10.1002/ps.5646
Ludvigsen, J., Rangberg, A., Avershina, E., Sekelja, M., Kreibich, C., Amdam, G., et al. (2015). Shifts in the midgut/pyloric microbiota composition within a honey bee apiary throughout a season. Microbes Environ. 30, 235–244. doi: 10.1264/jsme2.ME15019
Luo, Z. W., Dong, Z. X., Chen, Y. F., Li, H. Y., Tang, Q. H., Li, J. L., et al. (2020). Comparative analysis of the gut microbiota of Apis cerana in Yunnan using high-throughput sequencing. Arch. Microbiol. 202, 2557–2567. doi: 10.1007/s00203-020-01974-0
Ma, Y., Jiao, P., Qi, Z., Jiang, Z., and Guan, S. (2020). Characterization of the complete chloroplast genome of Alsophila spinulosa, an endangered species endemic to China. Mitochondrial DNA Part B 5, 2262–2263. doi: 10.1080/23802359.2020.1772142
Martemyanov, V. V., Belousova, I. A., Pavlushin, S. V., Dubovskiy, I. M., Ershov, N. I., Alikina, T. Y., et al. (2016). Phenological asynchrony between host plant and gypsy moth reduces insect gut microbiota and susceptibility to Bacillus thuringiensis. Ecol. Evol. 6, 7298–7310. doi: 10.1002/ece3.2460
Mason, C. J., Campbell, A. M., Scully, E. D., and Hoover, K. (2019a). Bacterial and fungal midgut community dynamics and transfer between mother and brood in the Asian longhorned beetle (Anoplophora glabripennis), an invasive xylophage. Microb. Ecol. 77, 230–242. doi: 10.1007/s00248-018-1205-1
Mason, C. J., Jones, A. G., and Felton, G. W. (2019b). Co-option of microbial associates by insects and their impact on plant–folivore interactions. Plant Cell Environ. 42, 1078–1086. doi: 10.1111/pce.13430
Mauck, K. E., De Moraes, C. M., and Mescher, M. C. (2016). Effects of pathogens on sensory-mediated interactions between plants and insect vectors. Curr. Opin. Plant Biol. 32, 53–61. doi: 10.1016/j.pbi.2016.06.012
McMillan, H. M. (2023). Plants target gut microbes to reduce insect herbivore damage. Proc. Natl. Acad. Sci. 120:e2308568120. doi: 10.1073/pnas.2308568120
Meriweather, M., Matthews, S., Rio, R., and Baucom, R. S. A. (2013). 454 survey reveals the community composition and core microbiome of the common bed bug (Cimex lectularius) across an urban landscape. PLoS One 8:e61465. doi: 10.1371/journal.pone.0061465
Montagna, M., Chouaia, B., Mazza, G., Prosdocimi, E. M., Crotti, E., Mereghetti, V., et al. (2015). Effects of the diet on the microbiota of the red palm weevil (Coleoptera: Dryophthoridae). PLoS One 10:e0117439. doi: 10.1371/journal.pone.0117439
Pérez-Cobas, A. E., Maiques, E., Angelova, A., Carrasco, P., Moya, A., and Latorre, A. (2015). Diet shapes the gut microbiota of the omnivorous cockroach Blattella germanica. FEMS Microbiol. Ecol. 91:fiv022. doi: 10.1093/femsec/fiv022
Pirttilä, A. M., Brusila, V., Koskimäki, J. J., Wäli, P. R., Ruotsalainen, A. L., Mutanen, M., et al. (2023). Exchange of microbiomes in plant-insect herbivore interactions. mBio 14:e03210-22. doi: 10.1128/mbio.03210-22
Qiao, H., Keesey, I. W., Hansson, B. S., and Knaden, M. (2019). Gut microbiota affects development and olfactory behavior in Drosophila melanogaster. J. Exp. Biol. 222:jeb192500. doi: 10.1242/jeb.192500
Ruiz-Rodriguez, M., Valdivia, E., Martin-Vivaldi, M., Martin-Platero, A. M., Martinez-Bueno, M., Mendez, M., et al. (2012). Antimicrobial activity and genetic profile of Enteroccoci isolated from hoopoes uropygial gland. PLoS One 7:e41843. doi: 10.1371/journal.pone.0041843
Saikkonen, K., Helander, M., Ranta, H., Neuvonen, S., Virtanen, T., Suomela, J., et al. (1996). Endophyte-mediated interactions between woody plants and insect herbivores? Entomol. Exp. Appl. 80, 269–271. doi: 10.1111/j.1570-7458.1996.tb00932.x
Salem, H., Bauer, E., Strauss, A. S., Vogel, H., Marz, M., and Kaltenpoth, M. (2014). Vitamin supplementation by gut symbionts ensures metabolic homeostasis in an insect host. Proc. R. Soc. B Biol. Sci. 281:20141838. doi: 10.1098/rspb.2014.1838
Santos-Garcia, D., Mestre-Rincon, N., Zchori-Fein, E., and Morin, S. (2020). Inside out: microbiota dynamics during host-plant adaptation of whiteflies. ISME J. 14, 847–856. doi: 10.1038/s41396-019-0576-8
Scully, E. D., Geib, S. M., Hoover, K., Tien, M., Tringe, S. G., Barry, K. W., et al. (2013). Metagenomic profiling reveals lignocellulose degrading system in a microbial community associated with a wood-feeding beetle. PLoS One 8:e73827. doi: 10.1371/journal.pone.0073827
Shao, Y., Chen, B., Sun, C., Ishida, K., Hertweck, C., and Boland, W. (2017). Symbiont-derived antimicrobials contribute to the control of the lepidopteran gut microbiota. Cell Chem. Biol. 24, 66–75. doi: 10.1016/j.chembiol.2016.11.015
Shikano, I. (2017). Evolutionary ecology of multitrophic interactions between plants, insect herbivores and entomopathogens. J. Chem. Ecol. 43, 586–598. doi: 10.1007/s10886-017-0850-z
Siddiqui, J. A., Khan, M. M., Bamisile, B. S., Hafeez, M., Qasim, M., Rasheed, M. T., et al. (2022). Role of insect gut microbiota in pesticide degradation: a review. Front. Microbiol. 13:870462. doi: 10.3389/fmicb.2022.870462
Šigutová, H., Šigut, M., Pyszko, P., Kostovčík, M., Kolařík, M., and Drozd, P. (2023). Seasonal shifts in bacterial and fungal microbiomes of leaves and associated leaf-mining larvae reveal persistence of core taxa regardless of diet. Microbiol. Spectr. 11:e0316022. doi: 10.1128/spectrum.03160-22
Silva, S., Negri, M., Henriques, M., Oliveira, R., Williams, D. W., and Azeredo, J. (2012). Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol. Rev. 36, 288–305. doi: 10.1111/j.1574-6976.2011.00278.x
Strano, C. P., Malacrinò, A., Campolo, O., and Palmeri, V. (2018). Influence of host plant on Thaumetopoea pityocampa gut bacterial community. Microb. Ecol. 75, 487–494. doi: 10.1007/s00248-017-1019-6
Suh, S. O., and Blackwell, M. (2004). Three new beetle-associated yeast species in the Pichia guilliermondii clade. FEMS Yeast Res. 5, 87–95. doi: 10.1016/j.femsyr.2004.06.001
Vilanova, C., Baixeras, J., Latorre, A., and Porcar, M. (2016). The generalist inside the specialist: gut bacterial communities of two insect species feeding on toxic plants are dominated by Enterococcus sp. Front. Microbiol. 7:1005. doi: 10.3389/fmicb.2016.01005
Wang, Q., Liu, Y., and Yin, X. (2022). Comparison of gut bacterial communities of Locusta migratoria manilensis (Meyen) reared on different food plants. Biology 11:1347. doi: 10.3390/biology11091347
Wang, X., Wang, H., Zeng, J., Cui, Z., Geng, S., Song, X., et al. (2023). Distinct gut bacterial composition in Anoplophora glabripennis reared on two host plants. Front. Microbiol. 14:1199994. doi: 10.3389/fmicb.2023.1199994
Warnecke, F., Luginbühl, P., Ivanova, N., Ghassemian, M., Richardson, T. H., Stege, J. T., et al. (2007). Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450, 560–565. doi: 10.1038/nature06269
Wolfgang, A., Tack, A. J., Berg, G., and Abdelfattah, A. (2023). Reciprocal influence of soil, phyllosphere and aphid microbiomes. Environ. Microbiome 18:63. doi: 10.1186/s40793-023-00515-8
Xiao, J., Yang, W., Zhang, B., He, Q., Kong, L., Bai, X., et al. (2023). Host selectivity and adaptability of Rhoptroceros cyatheae to Alsophila spinulosa and Gymnosphaera metteniana var. subglabra. J. Southwest For. Univ. 44, 135–141. doi: 10.11929/j.swfu.202212028
Xiong, W. H. (2022). Intestinal microbiota in various animals. Integr. Zool. 17, 331–332. doi: 10.1111/1749-4877.12633
Xu, D. S., Yang, W. C., Weng, T., He, Q. Q., Liang, S., and Zhang, T. (2021). Biological characteristics and larval population dynamics of Rhoptroceros cyatheae in Chishui Guizhou. Chinese J. Biol. Control 37, 218–227. doi: 10.16409/j.cnki.2095-039x.2020.06.001
Yang, Y., Liu, X., Xu, H., Liu, Y., and Lu, Z. (2022). Effects of host plant and insect generation on shaping of the gut microbiota in the rice leaf folder, Cnaphalocrocis medinalis. Front. Microbiol. 13:824224. doi: 10.3389/fmicb.2022.824224
Yun, J. H., Roh, S. W., Whon, T. W., Jung, M. J., Kim, M. S., Park, D. S., et al. (2014). Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 80, 5254–5264. doi: 10.1128/AEM.01226-14
Zhang, Z., Jiao, S., Li, X., and Li, M. (2018). Bacterial and fungal gut communities of Agrilus mali at different developmental stages and fed different diets. Sci. Rep. 8:15634. doi: 10.1038/s41598-018-34127-x
Zhang, Y., Li, Y., Barber, A. F., Noya, S. B., Williams, J. A., Li, F., et al. (2023). The microbiome stabilizes circadian rhythms in the gut. Proc. Natl. Acad. Sci. 120:e2217532120. doi: 10.1073/pnas.2217532120
Zhang, S., Shu, J., Xue, H., Zhang, W., Zhang, Y., Liu, Y., et al. (2020). The gut microbiota in camellia weevils are influenced by plant secondary metabolites and contribute to saponin degradation. Msystems 5, 10–1128. doi: 10.1128/mSystems.00692-19
Zhang, S. K., Wang, Y., Li, Z. K., Xue, H. J., Zhou, X. D., and Huang, J. H. (2022). Two Apriona species sharing a host niche have different gut microbiome diversity. Microb. Ecol. 83, 1059–1072. doi: 10.1007/s00248-021-01799-4
Keywords: Hymenoptera, intestinal microbes, relict plant, hosts diets, adaptability, high-throughput sequencing
Citation: Zhang B, Yang W, He Q, Chen H, Che B and Bai X (2024) Analysis of differential effects of host plants on the gut microbes of Rhoptroceros cyatheae. Front. Microbiol. 15:1392586. doi: 10.3389/fmicb.2024.1392586
Edited by:
Raquel Xavier, Centro de Investigacao em Biodiversidade e Recursos Geneticos (CIBIO-InBIO), PortugalReviewed by:
Ayushi Gupta, International Centre for Genetic Engineering and Biotechnology, IndiaLili Ren, Beijing Forestry University, China
Si-Xun Ge, Beijing Forestry University, China,in collaboration with reviewer LR
Copyright © 2024 Zhang, Yang, He, Chen, Che and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weicheng Yang, eWFuZ3dlaWNoZW5nMDkwOEBzaW5hLmNvbQ==
†These authors share first authorship
 Bingchen Zhang
Bingchen Zhang Weicheng Yang1*†
Weicheng Yang1*†