
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 21 May 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1392178
 Minghao Li1,2†
Minghao Li1,2† Baipeng Chen2†
Baipeng Chen2† Ming Xu2†
Ming Xu2† Fulong Li2
Fulong Li2 Yi Geng3
Yi Geng3 Defang Chen2
Defang Chen2 Ping Ouyang3
Ping Ouyang3 Xiaoli Huang2*
Xiaoli Huang2* Yongqiang Deng1*
Yongqiang Deng1*TonB-dependent siderophore receptors play a critical transport role for Flavobacterium columnare virulence formation and growth, and have become valuable targets for the development of novel antimicrobial agents. Traditional Chinese medicine has demonstrated notable efficacy in the treatment of fish diseases and includes potential antibacterial agents. Herein, we performed molecular docking-based virtual screening to discover novel TonB-dependent siderophore receptor inhibitors from traditional Chinese medicine and provide information for developing novel antibacterial agents. Firstly, we efficiently obtained 11 potential inhibitors with desirable drug-like characteristics from thousands of compounds in the TCM library based on virtual screening and property prediction. The antibacterial activity of Enoxolone, along with its interaction characteristics, were determined via an MIC assay and molecular dynamic simulation. Transcriptional profiling, along with validation experiments, subsequently revealed that an insufficient uptake of iron ions by bacteria upon binding to the TonB-dependent siderophore receptors is the antibacterial mechanism of Enoxolone. Finally, Enoxolone's acceptable toxicity was illustrated through immersion experiments. In summary, we have used virtual screening techniques for the first time in the development of antimicrobial agents in aquaculture. Through this process, we have identified Enoxolone as a promising compound targeting the TonB-dependent siderophore receptor of F. columnare. In addition, our findings will provide new ideas for the advancement of innovative antimicrobial medications in aquaculture.
Flavobacterium columnare is a common Gram-negative aerobic aquatic pathogen that is widespread in freshwater worldwide. F. columnare infects an extremely wide range of freshwater fish, including Cyprinidae, Siluridae, Percoidea, and Salmonidae, and is a pathogen affecting the global aquaculture industry (Staroscik and Nelson, 2008; Morley and Lewis, 2010; Verma et al., 2015; Jia et al., 2021; Kitiyodom et al., 2021; Ponpukdee et al., 2021; Wang et al., 2022). Currently, the optimal treatment for F. columnare is the use of antibiotics, chemicals, and vaccines (Cherian et al., 2023; Guo et al., 2023). However, due to the misuse of antibiotics, the ineffectiveness of chemical agents and the difficulties in promoting vaccines, there is still a lack of effective drug treatment in practice. Furthermore, with the growing limitations on the use of antibiotic-based antimicrobial agents, there is an escalating urgency to create novel antimicrobial agents.
Iron is an essential nutrient for bacterial growth and is involved in many important biological processes, including electron transfer, respiration, DNA synthesis, the tricarboxylic acid cycle, and biofilm formation (Taudte et al., 2016). In the natural water environment, free iron ions are mainly present in very small amounts in the form of trivalent iron ions. The TonB-dependent siderophore receptor-mediated iron transport pathway is thought to be the predominant mode of trivalent iron uptake by Gram-negative bacteria, including F. columnare (Guan et al., 2013). The transport process is powered by a system consisting of ExbB, ExbD, and the periplasmic protein TonB (Moeck and Coulton, 1998). Furthermore, due to its significance as a crucial element in the virulence of bacterial pathogens, the iron ion uptake system has been targeted for the creation of innovative antimicrobial agents. And key proteins within this system have been specifically focused on for this purpose (Lemos and Balado, 2020; Chan et al., 2023). Evidence has demonstrated that the TonB-dependent iron uptake system is required for the virulence process of F. columnare (Conrad et al., 2022). Therefore, it is appealing to focus on crucial proteins in this system to create innovative antimicrobial agents against F. columnare.
Compared to human drugs, aquatic drug development is more focused on achieving cheap costs and speed. Traditional drug development models are frequently unsuitable for aquatic drug development due to its high level of complexity and length. Therefore, the utilization of the molecular docking virtual screening technique shows great potential as a highly effective and appealing substitute in the realm of aquatic drug discovery (Rather et al., 2023). Compared to traditional drug development processes, virtual screening techniques can reduce development costs and speed up the process, making them ideal for researchers with limited resources to find potential drug candidates (Hernández-Silva et al., 2023). However, unfortunately, despite the mature applications in human drug discovery, virtual screening techniques have not been reported in the field of aquatic drug discovery.
Traditional Chinese medicine (TCM), which includes crude extracts and effective components derived from herbs, has been used in China for thousands of years. In recent decades, it has also been extensively utilized in fish aquaculture and has emerged as a favored therapy to substitute antibiotics and chemicals due to its minimal toxicity and potent curative impact (Zhu, 2020). Given the exorbitant expenses, lengthy duration, and inherent challenges associated with innovative drug research, TCM plays a clear and key role in the source of inspiration for drug discovery.
Therefore, in order to screen for a novel antibacterial agent against F. columnare, this study is the first to use virtual screening techniques for the development of aquatic drugs. Specifically, a library of TCM compounds was screened using the TonB-dependent siderophore receptor protein from F. columnare as the receptor. Further, the mechanism of bacterial inhibition and the biotoxicity of the lead compound, Enoxolone (also known as 18β-Glycyrrhetinic Acid), were also investigated. The results of this study will lay the foundation for the prevention and control of F. columnare and the development of related drugs.
Since the protein structure of the TonB-dependent siderophore receptor of F. columnare was not available in the PDB database, the SWISS-MODEL, Phyre2, Robetta, and ColabFold web servers were introduced to generate protein structural data (Kelley et al., 2015; Waterhouse et al., 2018; Baek et al., 2021; Mirdita et al., 2022). During the modeling process, all parameters were selected as default parameters. The predicted protein structure was modeled after the amino acid sequence of the TonB-dependent siderophore receptor in the reference genome of F. columnare in the NCBI database (WP_065213071.1) (Agarwala et al., 2017). To validate the model quality, the SAVES web server (Available online at https://servicesn.mbi.ucla.edu/SAVES/, accessed July 10, 2022) was used to analyze the stereochemistry of the obtained protein models and to assess the model quality. The model with the highest overall score was selected for molecular docking. The protein structure was processed with the Protein Preparation wizard in Schrodinger suite. Then, the AUTODOCK 4.0 MGL tool was used to convert the protein PDB files into the required PDBQT format.
The 3D structures of the molecular ligands used in this study were downloaded from the Traditional Chinese Medicine Library (Catalog No. L8300, Selleck Chemicals, Houston, TX, USA). Openbabel was used to split the downloaded SDF files and convert them to the PDB format (O'Boyle et al., 2011). Finally, the AUTODOCK 4.0 MGL tool was used to convert the file to the PDBQT format, assuming a neutral condition (pH = 7.4) to satisfy the requirements for bacterial growth.
We uploaded the protein 3D structure files to the CASTp 3.0 server to predict active pocket locations based on geometric and topological properties (Tian et al., 2018), and labeled amino acid residues to visualize pockets in MGLTools 1.5.6 (https://ccsb.scripps.edu/mgltools/). The position of the catalytic site was manually adjusted to completely wrap the pocket. The final parameters for the catalytic sites were set as center-x = 2.472, center-y = 9.250, and center-z = −1.556, with a grid size of 29 × 29 × 29. Autodock Vina (version 1.2.2) and R (version 4.2.3) were utilized to perform high-throughput molecular docking-based virtual screening (Trott and Olson, 2010). Throughout the docking, the ligand was in a flexible form, while the protein was in a rigid form. The maximum number of binding modes to output was fixed at 9, and the exhaustiveness level (controlling the number of independent runs performed) was set at 8. The conformation with the best affinity was selected as the final docking conformation. The molecular dynamics simulations were carried out with Desmond/Maestro non-commercial version 2022.1 as a molecular dynamics software to evaluate the stability and binding interactions of the complexes (Bowers et al., 2006). TIP3P water molecules were added to the systems, which were then neutralized by a 0.15 M NaCl solution. After the minimization and relaxation of the system, the production simulation was performed for 100 ns in an isothermal–isobaric ensemble at 300 K and 1 bar. The trajectory coordinates were recorded every 100 ps. The molecular dynamics analysis was performed using the Simulation Interaction Diagram from Desmond. Pymol 1.8.6 was used for the visualization of the complexes (Lashkov et al., 2021). Furthermore, the top 20 ranked compounds were selected for further property evaluation.
Online platforms were used to predict the drug properties, bioactivity score, ADME properties, and toxicity profiles of the compounds. Specifically, the ADMETlab 2.0 and SWISS ADME were used to predict the drug properties, ADME properties, and toxicity profile, while Molinspiration, a molecular property calculator based on Java (Available online at http://www.molinspiration.com/, accessed July 22, 2022), was used to predict the bioactivity scores (Daina et al., 2017; Xiong et al., 2021). Florfenicol, a commonly used aquatic antibiotic, was selected as a control to eliminate compounds with poor drug-like properties. The screened compounds participated in the subsequent MIC (minimum inhibitory concentration) assay.
The experimental strain of the pathogen F. columnare was isolated and purified from the gill tissue of dying diseased fish via inoculation with Cytophaga agar containing 0.5 g/L Tryptone, 0.2 g/L Beef extract, 0.5 g/L Yeast extract, 0.2 g/L Na-acetate, and 1% Agar. The strain was maintained as a frozen stock at −80°C in 50% (v/v) glycerol at Sichuan Agricultural University. After resuscitating the frozen stock culture, a single colony was transferred from the Cytophaga agar to 10 mL of Flavobacterium columnare growth media (FCGM, containing 8.0 g/L Tryptone, 0.8 g/L Yeast extract, 1.0 g/L MgSO4·7H2O, 0.74 g/L CaCl2·2H2O, 5.0 g/L NaCl, 1.5 g/L Sodium Citrate) broth by picking it up with an inoculating loop and cultured at 28°C with shaking at 200 rpm for 24 h. The strain was identified as F. columnare through DNA extraction, PCR amplification of the 16SrDNA fragment (forward primer: 5′-AGAGTTTGATCCTGGCTCAG-3′; reverse primer: 5′-GGTTACCTGTTACGGACTT-3′), and a BLASTN comparison of the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
A determination of the MIC of different small-molecule compounds against F. columnare was carried out in 96-well U-bottom polystyrene microtiter plate using the broth dilution method. All compounds were purchased from Selleck Chemicals and dissolved in dimethyl sulfoxide (DMSO) to prepare a 10 mM master batch. The 2-fold serial dilution of each compound ranging from 128 to 0.0625 μg mL−1 was used for the assay (the maximum concentration of Uvaol, which is extremely insoluble in water, is 32 μg mL−1). The FCGM broth (pH = 7.4) was added to all wells as the medium and 0.5 McFarland bacteria were cultured for 24 h (Farmer, 2004). The bacterial growth was observed by using 0.015% Resazurin dye to examine the viability of the bacterial cells. FCGM broth supplemented with DMSO and cultures without the compound addition were used as the negative and positive controls, respectively. Meanwhile, the MIC of Florfenicol was tested in the same way against F. columnare.
Overnight cultures of F. columnare were inoculated into 10 mL of fresh FCGM broth and cultured at 28°C with shaking. When the OD600 reached 0.5, DMSO-solubilized Enoxolone (1 × MIC: 8 μg/mL) was added to the medium. After 2 h of incubation, the bacterial cells were collected via cryogenic centrifugation and stored at −80°C until further use. Additionally, bacterial cells treated with equal amounts of DMSO were used as controls. Three biological replicates were carried out for both the control and the experimental groups. Total RNA was extracted from the bacterial cells using the TRIzol® Reagent according to the manufacturer's instructions (Invitrogen), and genomic DNA was removed using DNase I (TaKara, Kusatsu, Japan). Then, the RNA quality was determined with a 2100 Bioanalyzer (Agilent) and quantified using the ND-2000 (NanoDrop Technologies, Wilmington, DE, USA). Then, 1% agarose gel electrophoresis was used to detect the integrity of the total RNA. Only high-quality RNA samples (OD260/280 ≥ 1.8, OD260/230 ≥ 1.0, RIN ≥ 6.5, 28S:18S ≥ 1.0, ≥50 ng/μL, ≥ 1 μg) were utilized for the subsequent construction of the library.
The RNA-seq transcriptome library was prepared following the TruSeqTM RNA sample preparation Kit from Illumina (San Diego, CA, USA) using 2 μg of total RNA. A Ribo-zero Magnetic kit (Epicentre, Madison, WI, USA) was used to remove rRNA and purify mRNA. The mRNA was randomly broken into small fragments of about 200 bp and used as a template. Double-stranded cDNA was synthesized using a SuperScript double-stranded cDNA synthesis kit (Invitrogen, Carlsbad, CA, USA) with random hexamer primers (Illumina). The synthesized double-stranded cDNA was added to the End Repair Mix to complement its flat end. The 5′ end was phosphorylated, and an A base was added to the 3′ end to connect it to the Y-shaped sequencing connector. Libraries were size-selected for cDNA target fragments of 200 bp on 2% Low Range Ultra Agarose followed by PCR amplification using Phusion DNA polymerase (NEB) for 15 PCR cycles. The quality and number of cDNA libraries were evaluated using the TBS380 (Invitrogen, Carlsbad, CA, USA). Finally, the paired-end RNA-seq sequencing library was sequenced with the Illumina Novaseq (2 × 150 bp read length).
Clean reads were selected by removing low-quality sequences, reads with more than 5% N bases (unknown bases), and reads containing adaptor sequences using a Perl program. The genome of Flavobacterium columnare strain F2S17 (NZ_CP054494) in the NCBI database was selected as the reference genome. Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) was used to compare the high quality of the sample reads against the reference genome. Using the blast method, 10,000 raw reads were randomly selected from each sample and compared with the Rfam database. The percentage of rRNA in each sample was calculated based on the annotation results to assess the rRNA contamination rate. Gene expression was calculated using RSEM (http://deweylab.github.io/RSEM/) with a maximum-likelihood abundance estimation model based on a maximum expectation algorithm, and expression levels were measured using FPKM and TPM. A statistical analysis of differentially expressed genes (DEGs) was performed using the edgeR software package (http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html) with p-value < 0.05 and |log2FC| ≥ 1 as the default screening criteria. In addition, a functional enrichment analysis of the screened DEGs was performed using Goatools (https://github.com/tanghaibao/GOatools) and KOBAS (http://kobas.cbi.pku.edu.cn) to determine the extent of DEG enrichment in different GO terms and KEGG pathways.
F. columnare strains were streaked from freezer stocks onto Cytophaga agar and incubated at 28°C for 24 h. Growth from the Cytophaga agar was then used to inoculate 10 mL of FCGM broth, which was incubated at 28°C with rotation to 0.5 McFarland. Enoxolone was added to the medium to a final MIC (8 μg/mL). For some experiments, ferric chloride (FeCl3) and ferrous sulfate (FeSO4) were added to 10 mL of FCGM cultures containing Enoxolone, respectively. Ferric chloride and ferrous sulfate were added in increments of 50 μM or 100 μM. Cultures without Enoxolone addition were used as controls. All cultures were set up in three parallels and incubated for 24 h at 28°C. We added 100 μL of the incubated culture to a 96-well plate. The cell viability was measured in triplicate using CCK-8 (Beyotime Biotechnology, Shanghai, China) at a dose of 10 μL per well−1. The absorbance values were measured at wavelength of 450 nm using an ultraviolet–visible spectrophotometer according to the manufacturer's protocol.
Sixty healthy largemouth bass (0.99 ± 0.31 g) were purchased from a farm in Sichuan. They were acclimated in the laboratory for 2 weeks prior to the experiment. All largemouth bass were divided into 6 groups (1 control and 5 experimental groups), each of which contained 10 largemouth bass. The control group received DMSO immersion (1%), while the experimental group was immersed in aeration water containing 4 μg/mL, 8 μg/mL, 16 μg/mL, 32 μg/mL, and 64 μg/mL of Enoxolone, respectively. During the experiment, the largemouth bass were exposed to an uninterrupted oxygen supply, the pH was marinated at 6.5–8.0, and water temperature was about 25°C. The experimental period was set to 96 h, and the number of deaths and clinical symptoms of the largemouth bass were recorded daily.
Data on the cell viability were analyzed and expressed as mean ± standard deviation. A statistical analysis was carried out using an analysis of variance (ANOVA) followed by multiple comparison tests (LSDs). If the variance among groups was not homogenous, Dunnett's T3 test was performed. A value of p < 0.05 was considered statistically significant and labeled with an asterisk (*). GraphPad Prism 8 was used for the graphical representations.
The TonB-dependent siderophore receptor protein lacks PDB entry; for this reason, the structure of the protein has been depicted through homology modeling. Amongst the 701 amino acids of TonB-dependent siderophore receptors, 5.85%, 49.50%, and 44.65% of these amino acids construct α-helices, β-strands, and transmembrane helices, respectively. SWISS-MODEL, Phyre2, Robetta, and ColabFold predicted the amino acid data of the proteins to generate structural images. Supplementary Figure S1 shows the 3D conformation of the F. columnare TonB-dependent siderophore receptor proteins constructed using different procedures. All protein models have a similar structure, namely a β-barrel structure formed of multiple β-folds and a plug structure contained in its center. The reasonableness of all modeled target protein topologies was analyzed using SAVES web servers. A Ramachandran plot was used to visualize whether the dihedral angles of the amino acid residues in the homologous modeled protein structures were in reasonable regions. A more rational protein structure has more amino acid residues in the best regions. According to the Ramachandran plot, the percentages of amino acid residues in the best regions for the SWISS-MODEL, Phyre2, Robetta, and ColabFold rank1 models were 84.5%, 77.9%, 88.5%, and 86.6%, respectively. In addition, non-bonded interactions between different atom types were analyzed in the ERRAT program (Colovos and Yeates, 1993). As an important indicator for assessing the three-dimensional structure of proteins via crystallography, the high factors obtained in ERRAT represent the high resolution of the structure. The results showed that the ColabFold rank1 model protein used for the analysis received the highest score of 83.847, followed by SWISS-MODEL (81.719), Robetta (81.584), and Phyre2 (49.461) (Supplementary Figures S2–S5). The PAE (prediction alignment error) results indicated that the ColabFold rank1 model protein has more confidence in the expected positions of amino acid residues in the five 3D structures generated by ColabFold, indicating that the ColabFold rank1 model protein has the highest confidence (Supplementary Figure S2D). In combination with the above data, the ColabFold rank1 model protein was used as the receptor protein for the subsequent screening.
The crystal structure of the target protein was not resolved. Meanwhile, the target protein has low identity with the protein in the PDB database and is highly conserved in the genus (Supplementary Figure S6). Therefore, CASTp was used to characterize and measure the geometric and topological properties of protein structures to complete the visualization of TonB-dependent siderophore receptor protein pocket channels. The results of the CASTp calculation are shown in Supplementary Figure S7. After localizing the active pocket, AutoDock Vina was used to dock the unique conformation of a total of 1,335 TCM ligands to the active pocket in the TonB-dependent siderophore receptor. The docking scores of each protein–ligand complex generated via molecular docking were ranked to screen for stably bound ligands. As revealed in Table 1, the top 20 screened hits were listed according to their docking scores. In addition to the highest docking score that Solamarine (CAS: 20311-51-7) showed in terms of its docking with the receptor, all other compounds had docking scores above −10.9 kcal/mol. To more fully cognize the similarities and differences among the top 20 screens, compounds were searched by type, effect, and target of action to create a Sankey diagram. The results showed that the top 20 compounds were mostly pentacyclic triterpenoids with similar structures (linked by six isoprene units) (Figure 1). Most of the compounds have anticancer and anti-inflammatory effects, with the most frequent targets being enzymes (14; 70%), followed by transporters (four; 20%) and membrane receptors (two; 10%).
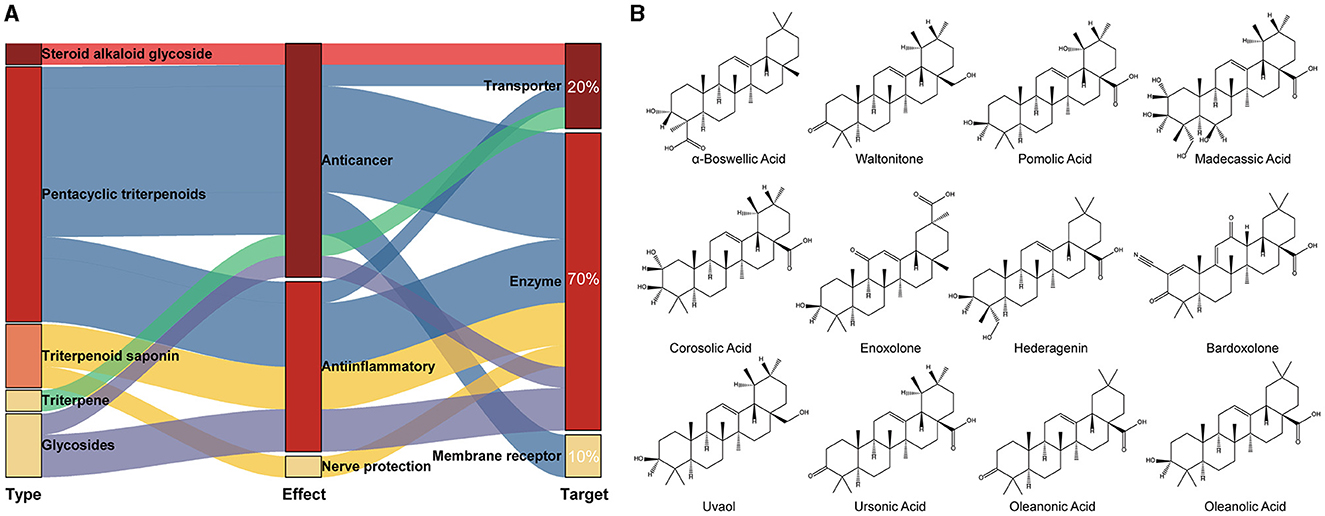
Figure 1. Classification of the top 20 scoring compounds. (A) Sankey diagram of tightly bound compounds in the TCM library; (B) the 2D structure of pentacyclic triterpenes among the top 20 compounds with highest scores.
Excellent drug-likeness and biological activity contribute to reducing the risk of failure of drug candidates in the production application stage. The ADME/T and biological activity of the top 20 ranked compounds obtained from the virtual screening were predicted and further filtered using Florfenicol as a standard. The results show that the 14 compounds conform to Lipinski's rule and have relatively good characteristic parameters (Table 2). In addition, most compounds do not cross the blood–brain barrier and do not inhibit CYP family enzymes, showing a high safety profile (Table 3). The Molinspiration server was used to calculate the bioactivity scores of the compounds on GPCRs (G protein-coupled receptors), nuclear receptors, ion channels, kinases, proteases, and other enzymes. The results showed that all the compounds screened had more than moderate activity (−5 to 0) against the predicted targets, with some showing significant activity (score > 0) (Table 4). The results of the toxicity prediction show that some of the compounds have potential cardiotoxicity, carcinogenicity, mutagenicity, and aquatic toxicity (Table 5). Since the compounds will be used in an aqueous environment, Lipinski's rule, the blood–brain barrier, CYP family enzymes, and toxicity (hERG, AMES Toxicity Carcinogenicity, and LC50FM) were set as screening conditions in this study to make the compounds bioenvironmentally friendly. Ultimately, Solamargine, Asiaticoside, Obacunone, Tenuifolin, Pseudoprotodioscin, Bardoxolone, Protodioscin, Dioscin, and Saikosaponin A were removed due to ineligible drug-likeness, and the remaining compounds were used in subsequent assays for antimicrobial activity.
We tested the antibacterial activity of the primary screened small-molecule compounds against F. columnare using a microdilution method based on Resazurin staining. The results showed that F. columnare showed strong resistance to Hederagenin, α-Boswellic acid, Waltonitone, Madecassic acid, Oleanonic Acid, and Uvaol. In contrast, Enoxolone, Oleanolic acid, Ursonic acid, Corosolic acid, and Pomolic acid showed antibacterial activity. Among all compounds, Enoxolone had the smallest MIC, which was 8 μg/mL (Figure 2). Meanwhile, the MIC of Florfenicol, the control drug, was determined to be 2 μg/mL. Therefore, Enoxolone was selected as a potential lead compound for subsequent experiments.
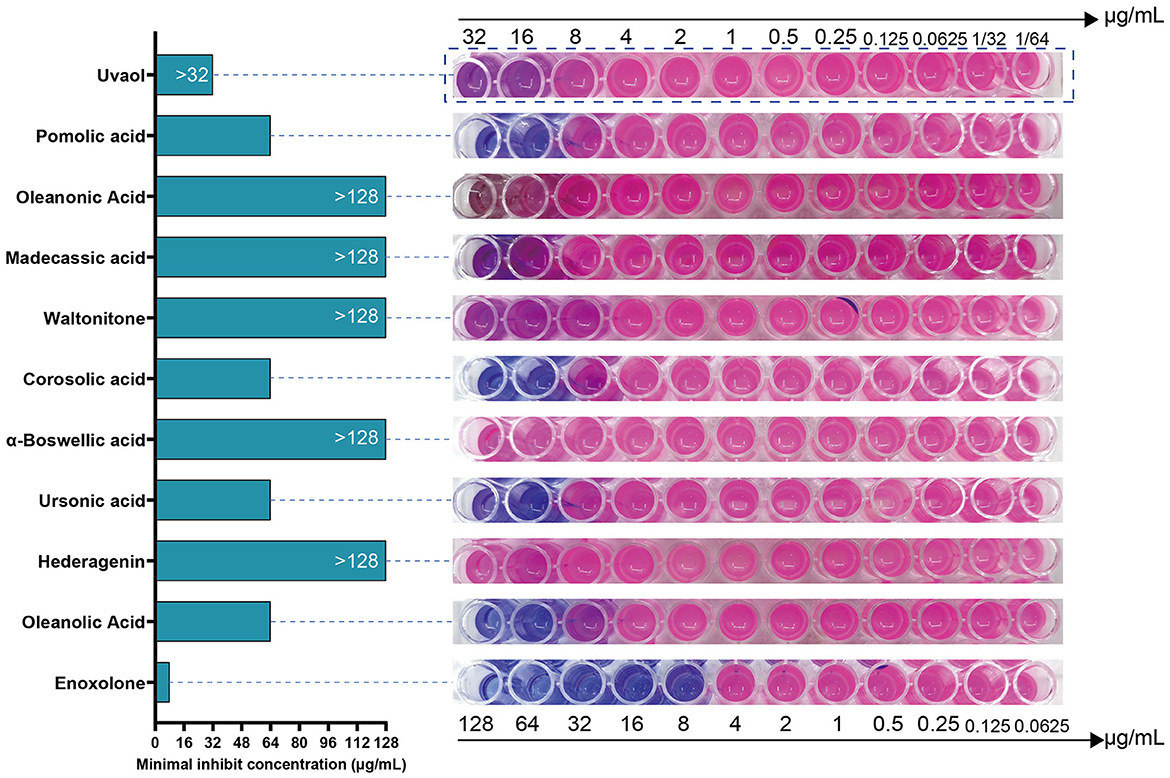
Figure 2. Resazurin based microdilution assay for MIC determination of compounds against Flavobacterium columnare. The MIC values of compounds against the Flavobacterium columnare are indicated in the left hand side. The growth of bacterial was observed by conversion of blue colored resazurin dye to pink one by viable bacterial cells.
The dynamic stability and interactions of the TonB-dependent siderophore receptor–Enoxolone complex were further evaluated with molecular dynamics simulations. To study the dynamic stability of the complex, root mean square deviations (RMSDs) from the starting structures were analyzed. The plots showed that the protein and ligand were in a steady state during the simulations. After equilibration, the RMSD of the protein stabilized at 2.6 Å to 2.8 Å, whereas the RMSD of the ligand stabilized at 1.6 Å to 2.0 Å (Figure 3A). Then, the overall calculated root mean square fluctuations (RMSFs) of the protein and ligand were also small (Figures 3B, C). These small fluctuations indicate the high stability of the initial conformation of the complex. In addition, the RMSD and RMSF between apo-protein and protein-ligand complex were also nearly (Supplementary Figure S8). After docking, ARG647, SER675, GLU688, and TRP693 bind to the ligand via hydrogen bonding. During the simulation, the protein and small molecules formed multiple sets of interactions, including hydrogen bonds, hydrophobic interactions, and water bridges (Supplementary Figure S9). ASN 85, ARG 629, ARG 647, GLU 688, ASN 692, and ARG 694 were recognized as key amino acid residues in the binding process. In particular, the hydrogen bond formed by ARG 629 with small molecules was believed to play a crucial role (Figure 3D). Therefore, Enoxolone displayed a high binding affinity for TonB-dependent siderophore receptors in this conformation.
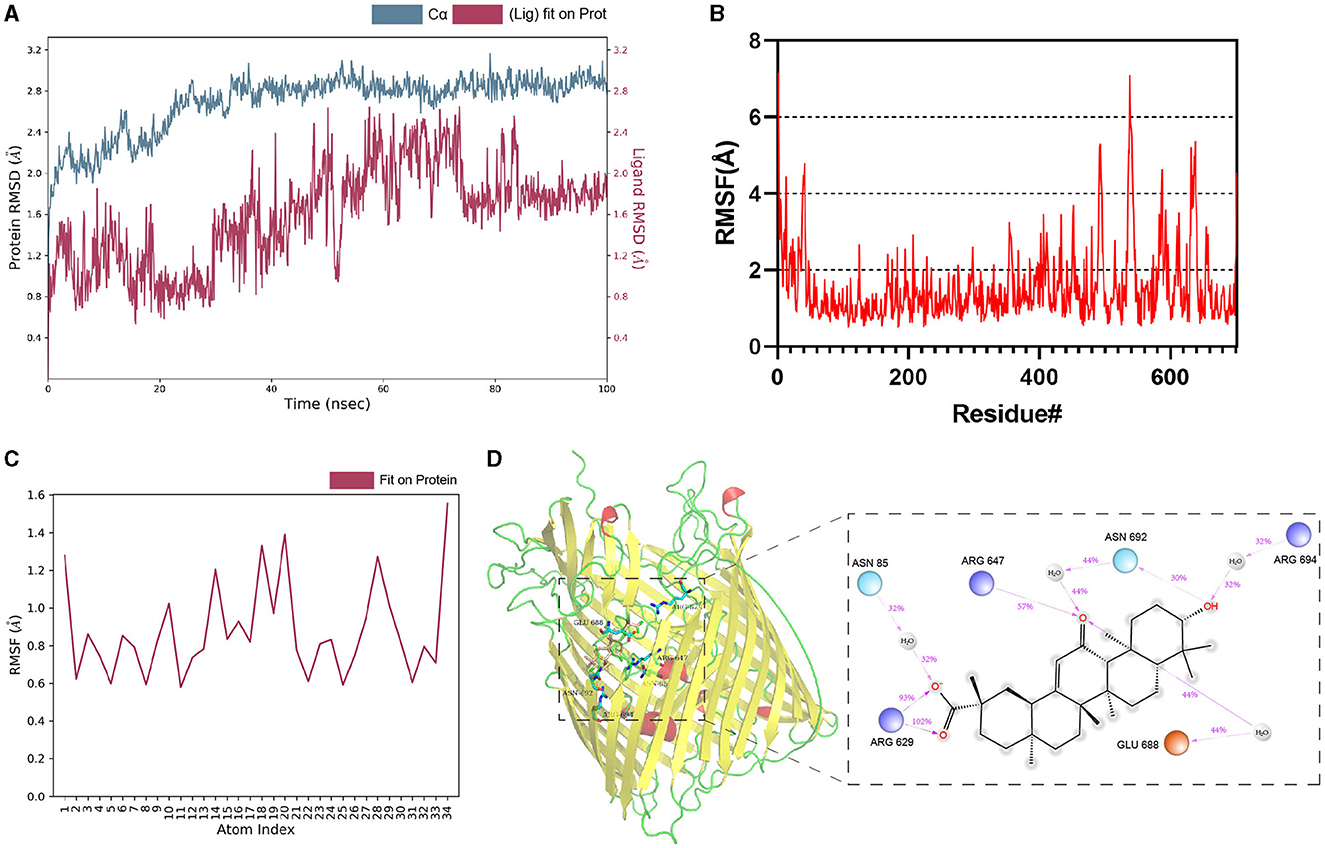
Figure 3. Molecular dynamics simulations of protein–Enoxolone complex. (A) RMSD values of backbone atoms of the protein and the ligand. (B) RMSFs of TonB-dependent siderophore receptor. (C) RMSFs of the heavy atoms in the ligand. (D) Interaction analysis of the complex. Interactions that occur for more than 30.0% of the simulation time in the selected trajectory are shown. It is possible to have interactions with >100%, as some residues may have multiple interactions of a single type with the same ligand atom. Dark-blue, light-blue, and orange circles represent positively charged, negatively charged, and polar amino acid residues, respectively.
To further explore the potential mechanism of Enoxolone inhibition in F. columnare, six prokaryotic transcriptome sequencing libraries were constructed, containing three DMSO-treated controls and three Enoxolone-treated groups. The six libraries were sequenced, and the data are summarized in Table 6. An average of 25,720,127 raw reads were obtained per sample, with Q20 and Q30 both over 93%. The percentage of rRNA assessed against the reference genome was < 10% for all samples. In addition, the sequenced sequences were evenly distributed across the genes, indicating unbiased sequencing (Supplementary Figure S10). The quality control data showed that the sequencing results met the requirements for prokaryotic transcriptome analysis. Further, all unigenes obtained in the assembly were blasted against six databases (NR, Swiss-Prot, Pfam, COG, GO, and KEGG), among which 2,715 genes were annotated (Supplementary Figure S11). The expression levels of the genes were normalized to FPKM and TPM. Prior to the DEG analysis, a principal component analysis (PCA) was performed based on the expression matrix. The results showed that there was a significant difference in gene expression between the different groups (Supplementary Figure S12A). DEGs were analyzed based on the read counts, and 76 DEGs (up: 52; down: 24) were identified using edgeR (p-value < 0.05 and |log2FC| ≥ 1 in normalized expression values) (Supplementary Figure S12B).
GO and KEGG annotation were performed to further understand the biological functions of the DEGs. Supplementary Figure S13A shows the annotation of the top 10 terms of the DEGs in the GO database. The annotated results showed that the most significant changes in the expression of genes related to integral components of the membrane were observed after Enoxolone treatment. In addition, there were differences in the expression of genes associated with the DNA binding, plasma membrane, and phosphorelay signal transduction systems. KEGG annotation showed that genes related to membrane transport and amino acid metabolism showed the most significant changes in expression after Enoxolone treatment (Supplementary Figure S13B). The GO and KEGG pathway enrichment analyses were conducted by scripting with the R language, and the pathways were considered to be significantly enriched when p < 0.05. The GO analysis indicated that the main differently expressed pathways in the Enoxolone-treated F. columnare included amino acid anabolism, biogenic amine synthesis, indole-containing compound synthesis, and signal transduction (Figure 4A). The bacterial secretion system, phenylalanine, tyrosine and tryptophan biosynthesis, and cationic antimicrobial peptide (CAMP) resistance were the main enriched KEGG pathways (Figure 4B). Due to the limited number of pathways in the database for the enrichment analysis, DEGs were further analyzed on a case-by-case basis. The analysis showed that the TonB-dependent siderophore receptor and the biopolymer transporter ExbD were labeled as up- and down-regulating DEGs, respectively. In addition, the expression of genes related to drug efflux (TolC, ABC transporter, EmrA) and amino acid synthesis (TrpD, TrpC) was significantly up-regulated; in contrast, the expression of genes related to iron ion transport (TonB receptor plug, ExbD, ExbB, FeoB), DNA synthesis (DNA primase), and the type VI secretion system (VgrG) was significantly down-regulated (Figure 4C). Notably, the TonB-dependent siderophore receptor is expressed in the opposite trend to its plug structure. The down-regulation of the expression of the plug structure as a binding site may indicate that the negative feedback regulation of the strain is due to the inhibition of the activity of the TonB-dependent siderophore receptor upon binding to Enoxolone.
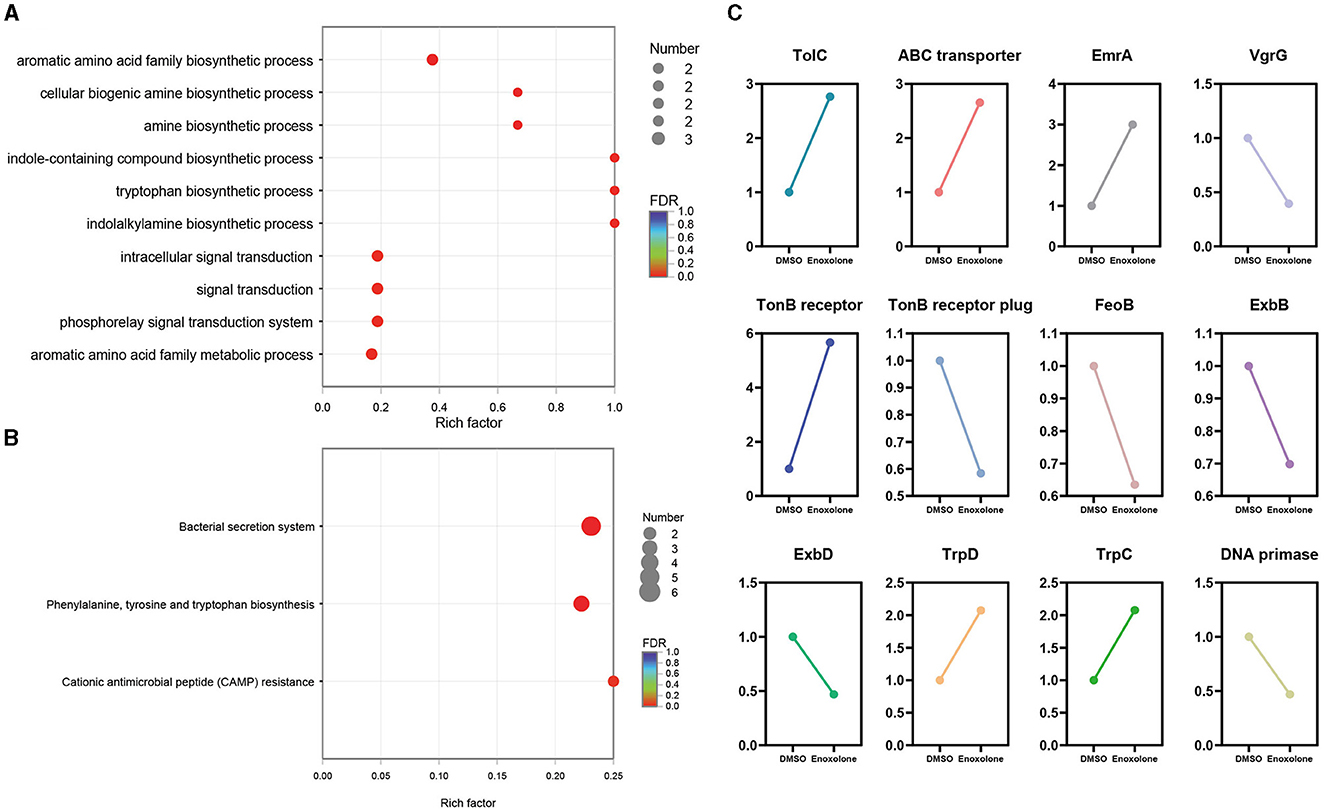
Figure 4. The expression pattern of genes in Enoxolone treatment. (A) Gene Ontology (GO) enrichment analysis of differentially expressed genes (DEGs); (B) enrichment of the KEGG pathway in different groups. The vertical axis shows the pathway name, the horizontal axis shows the rich factor corresponding to the pathway, and the magnitude of the FDR-value is indicated by the color of the dots. The smaller the FDR-value is, the closer the color is to red. The number of differentially expressed genes in each pathway is indicated by the size of the dots. (C) Expression pattern of the selected genes.
The gene response network of F. columnare following Enoxolone treatment was mapped against the screened genes to clarify the mechanism of inhibition. The results showed that the TonB-dependent iron transport pathway was inhibited after Enoxolone treatment, and that Enoxolone entering the bacterial cytosol via the TonB-dependent siderophore receptor activated the drug efflux system and the amino acid synthesis pathway. In addition, the relative decrease in iron ion content in bacteria inhibited DNA synthesis and reduced bacterial virulence (Figure 5A). According to the mapping of this gene regulation network, we hypothesized that the mechanism of Enoxolone resistance in F. columnare may be related to its competitive binding to the TonB-dependent siderophore receptor, resulting in an inadequate uptake of iron ions by bacteria. To demonstrate the accuracy of the proposed hypothesis, we first performed molecular docking with Enoxolone for other proteins in the TonB-dependent iron transport pathway that were significantly down-regulated. The results showed that the binding score of these proteins to Enoxolone was as low as −9.3 kcal/mol, which is less stable than the binding of the TonB-dependent siderophore receptor to Enoxolone (Figure 5B). Subsequently, we added FeCl3 and FeSO4 after the Enoxolone treatment of F. columnare and assayed the cell viability with CCK-8. The results showed that the addition of FeSO4 had no significant effect on cell viability, while the addition of FeCl3 significantly increased cell viability in a concentration-dependent manner (Figure 5C). Therefore, competitive binding to TonB-dependent siderophore receptors, leading to the inadequate uptake of iron ions, can be considered as a mechanism of Enoxolone against F. columnare. At the same time, the results based on the prokaryotic transcriptome also demonstrate the accuracy of the molecular docking virtual screening.
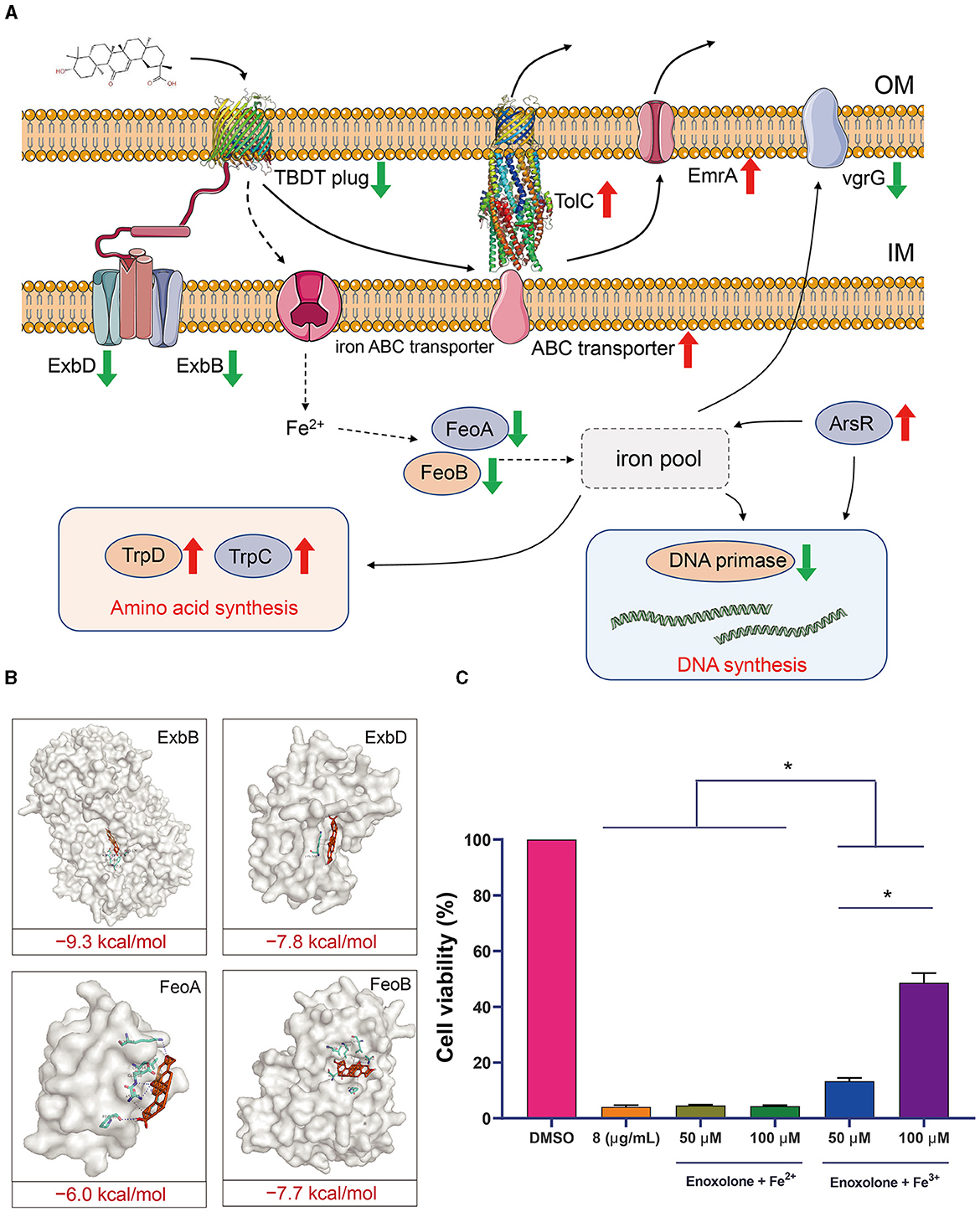
Figure 5. Speculation on the mechanism of inhibition of Flavobacterium columnare by Enoxolone. (A) Response network of Flavobacterium columnare to Enoxolone treatment mapped based on transcriptomic data. The red and green arrows represent a significant increase and decrease in gene expression, respectively. (B) Molecular docking results of proteins with significantly reduced expression other than TonB-dependent siderophore receptor in the TonB-dependent iron uptake system with Enoxolone. The blue and gray dashed lines represent hydrogen bonding and hydrophobic interactions, respectively. (C) Effect of exogenous addition of iron ions on the viability of Flavobacterium columnare after Enoxolone treatment. p ≤ 0.05 is considered significant. The error bars indicate the means ± standard deviation. *P < 0.05.
To evaluate the toxicity, largemouth bass were selected as experimental animals and exposed to Enoxolone (0–64 μg/mL). No mortality was observed at a lower concentration (4 μg/mL). However, this increased in a concentration-dependent manner. Compared to the control group, the group treated with 64 μg/mL of Enoxolone showed 100% mortality at 12 h. Moreover, only one mortality was observed in the minimal inhibitory concentration of Enoxolone tested (8 μg/mL) compared to that in the control group during the observation period (Figure 6). The biological toxicity exhibited by Enoxolone is acceptable, considering the small size of the experimental fish used and the availability of short and spaced treatments for production applications.
An important objective in the initial stages of drug development is to evaluate lead compounds for their potential interactions with target proteins and pharmacological activity. Additionally, there is a continuous effort to enhance the effectiveness and safety of these compounds. Due to this reason, the pharmaceutical industry has often used the experimental screening of large libraries of chemicals against targets as a traditional method of identifying novel lead compounds. Due to the swift progress in computer hardware, software, and algorithms, virtual screening techniques in drug discovery and development provide the benefits of enhanced speed, cost-effectiveness, and efficiency compared to conventional methods. In this study, the TonB-dependent siderophore receptor of F. columnare was selected as the target protein for a molecular docking-based virtual screening. Through the integration of laboratory research, we successfully obtained Enoxolone, a pentacyclic triterpenoid that exhibits superior bacterial suppression and precise targeting of the target protein.
Prior to virtual screening, it is often necessary to create a ligand library to hold the compounds used for screening. Currently, the most commonly used databases for the virtual screening of compounds are ZINC, Pubchem, and Drugbank, which contain the structures of millions of compounds. Nevertheless, the majority of the compounds in these databases are employed in the process of creating pharmaceuticals intended for human consumption. Mixing human and fishery medicines will raises biosecurity concerns. Nevertheless, there is currently no existing database specifically designed for screening aquatic drugs. In recent years, the effective components from TCM have exhibited promising applications in the prevention and control of fish diseases. Several studies have shown that the effective components in TCM used in aquaculture have powerful antiviral, antibacterial, antiparasitic, and antifungal effects, as well as activating the immune system in fish (Zhang et al., 2022). Furthermore, TCM possesses the benefits of abundant resources, affordability, and non-toxicity, making it highly compatible with the requirements for fishery medicine application (Zhu, 2020). Therefore, it is feasible and promising to screen efficient active ingredients and monomers with anti-pathogenic microbial activity from TCM.
In the results of the final virtual screening, we found that the pentacyclic triterpenoids in the compound library generally scored well. Pentacyclic triterpenoids have a wide range of biological activities and are commonly found as secondary metabolites in plants. Studies have shown that pentacyclic triterpenoids have anticancer, antitumor, and antiviral activities (Martinez et al., 2013). In addition, pentacyclic triterpenoids have broad-spectrum antibacterial activity against both Gram-positive and Gram-negative bacteria (Tabopda et al., 2009; Wu et al., 2021). Generally, the antibacterial activity of pentacyclic triterpenoids is thought to be related to changes in the structure and function of bacterial cellular structures (cell membranes, adhesins), cell morphology, gene expression, and processes such as adhesion and biofilm formation (Sycz et al., 2022). For example, Asiatic Acid ruptures the plasma membrane of Clostridium difficile and alters its membrane permeability (Harnvoravongchai et al., 2018). Pentacyclic triterpene derivatives possessing polyhydroxyl ring A inhibit Gram-positive bacteria growth by regulating the expression of genes associated with peptidoglycan metabolism, respiratory metabolism, and virulence (Huang et al., 2015). In addition, glycyrrhizic acid (GRA), ursolic acid (UA), and betulinic acid (BA) were shown to inhibit Acinetobacter baumannii biofilm formation by interfering with N-acyl homoserine lactone (ASL)-based signaling (Paul Bhattacharya et al., 2020). Therefore, pentacyclic triterpenoids have promising applications in the development of aquatic drugs, especially antibacterial drugs.
Virtual screening-based on molecular docking also has many drawbacks and limitations. Furthermore, computational outcomes cannot serve as a substitute for experimental data and must be combined with other methods. To further identify the lead compounds, we performed MIC assays on the top-scoring compounds. Ultimately, we isolated Enoxolone, which exhibited the most potent antibacterial activity. As one of the main active substances in Glycyrrhiza uralensis Fisch, Enoxolone has been reported to play a role in the treatment of hyperlipoidemia and cholestasis (Eu et al., 2010; Wang et al., 2017). In addition to the treatment of liver disease, Enoxolone has also been reported to have a wide range of antibacterial activity. Studies have shown that Enoxolone and its derivatives have good antibacterial activity against Gram-positive bacteria, including Bacillus subtilis and Staphylococcus aureus. In addition, Enoxolone can also be used in combination with other antibiotics to increase the antibacterial activity of the antibiotic against Gram-positive bacteria (de Breij et al., 2016). Antimicrobial studies of Enoxolone against Gram-negative bacteria have mostly focused on pathogens that are infectious to humans, such as Escherichia coli and Neisseria gonorrhea (Kong et al., 2018; Zhao and Su, 2023). Furthermore, licorice and its extracts have demonstrated the ability to enhance growth and immunological parameters in aquaculture. This is due to their secure and natural botanical origin, as well as their wide range of biological activities (Abdel-Tawwab and El-Araby, 2021). In this study, Enoxolone was found to have significant antibacterial activity against F. columnare, which extends the antibacterial spectrum of Enoxolone and provides novel insights for the design of new antibacterial agents for aquatic applications.
At present, the specific mechanism of bacterial inhibition by Enoxolone has not been described systematically. Studies on Xanthomonas oryzae pv. oryzae and SiHa cells have shown that Enoxolone and its derivatives can cause excessive production and accumulation of intracellular ROS and thus induce apoptosis (Lee et al., 2008; Song et al., 2022). Enoxolone may also exhibit antibacterial activity against Staphylococcus aureus by affecting carbohydrate and amino acid metabolism (Oyama et al., 2016). Bioinformatics calculations show that histone-like DNA-binding proteins are potential drug targets for Enoxolone against Helicobacter pylori (Raj et al., 2020). Deep learning-based predictions show that glycyrrhetinic acid exhibits a high affinity for ABC transporter proteins (Alkhadrawi et al., 2022). At later stages of antibiotic treatment, widespread downstream gene expression interferes with the determination of the actual binding target and antimicrobial mechanism. In this study, the bacterial transcriptome profile was analyzed immediately after 2 h of Enoxolone treatment in order to clarify the inhibition mechanism of Enoxolone against F. columnare. During validation trials, although the addition of divalent iron ions at high concentrations may have resulted in a toxic Fenton reaction, the addition of trivalent iron ions significantly reduced the bacteriostatic activity of Enoxolone. The final results suggest that the competitive binding of Enoxolone to the TonB-dependent siderophore receptor leads to an insufficient uptake of iron ions as a potential inhibitory mechanism against F. columnare. The distinctive inhibitory action of Enoxolone enables its potential advancement as a specific antibacterial agent against F. columnare.
While significant antibacterial activity was observed, Enoxolone also exhibited some biotoxicity. Although this biotoxicity is acceptable for the purpose of bacterial inhibition, it is still necessary to reduce the toxicity to accommodate high-concentration applications. In fact, further structural modifications and treatments are often required after lead compounds are obtained. Research has demonstrated that the combination of Enoxolone and emodin esters enhances the hatching and survival rates of zebrafish embryos. Additionally, it decreases the occurrence of cardiomyocyte malformation and apoptosis (Zhong et al., 2022). In addition, Enoxolone derivatives obtained via reduction at C-11, oxidation at C-3, and condensation at C-2 exhibited stronger anti-staphylococcal activity than Enoxolone (Yang et al., 2020). Therefore, the modification of the structure to further improve its efficacy and reduce biotoxicity could be the next step in the study of Enoxolone for the treatment of F. columnare infection.
In conclusion, we obtained Enoxolone with anti-F. columnare activity from traditional Chinese medicine compounds using a virtual screening technique. Moreover, we demonstrated through laboratory experiments that Enoxolone targets the TonB-dependent siderophore receptor of F. columnare and exhibits an acceptable biological toxicity. Along with obtaining antimicrobial lead compounds, our results also declare the usability of virtual screening techniques in the development of novel antimicrobial agents for bacteria infecting aquatic organisms.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA971873.
The animal study was approved by the Animal Care and Use Committee of Sichuan Agricultural University. The study was conducted in accordance with the local legislation and institutional requirements.
ML: Data curation, Formal analysis, Writing—original draft. BC: Data curation, Formal analysis, Writing—review & editing. MX: Data curation, Formal analysis, Writing—review & editing. FL: Data curation, Writing—review & editing. YG: Conceptualization, Writing—review & editing. DC: Investigation, Visualization, Writing—review & editing. PO: Methodology, Writing—review & editing. XH: Conceptualization, Supervision, Writing—review & editing. YD: Supervision, Writing—review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Sichuan Science and Technology Program (2021YFYZ0015).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1392178/full#supplementary-material
Abdel-Tawwab, M., and El-Araby, D. A. (2021). Immune and antioxidative effects of dietary licorice (Glycyrrhiza glabra L.) on performance of Nile tilapia, Oreochromis niloticus (L.) and its susceptibility to Aeromonas hydrophila infection. Aquaculture 530:735828. doi: 10.1016/j.aquaculture.2020.735828
Agarwala, R., Barrett, T., Beck, J., Benson, D. A., Bollin, C., Bolton, E., et al. (2017). Database Resources of the National Center for Biotechnology Information. Nucl. Acids Res. 45, D12–d17. doi: 10.1093/nar/gkw1071
Alkhadrawi, A. M., Wang, Y., and Li, C. (2022). In-silico screening of potential target transporters for glycyrrhetinic acid (GA) via deep learning prediction of drug-target interactions. Biochem. Eng. J. 181:108375. doi: 10.1016/j.bej.2022.108375
Baek, M., DiMaio, F., Anishchenko, I., Dauparas, J., Ovchinnikov, S., Lee, G. R., et al. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876. doi: 10.1126/science.abj8754
Bowers, K. J., Chow, E., Xu, H., Dror, R. O., Eastwood, M. P., Gregersen, B. A., et al. (2006). “Scalable algorithms for molecular dynamics simulations on commodity clusters,” in Proceedings of the 2006 ACM/IEEE Conference on Supercomputing), 84-es. doi: 10.1145/1188455.1188544
Chan, D. C. K., Josts, I., Koteva, K., Wright, G. D., Tidow, H., and Burrows, L. L. (2023). Interactions of TonB-dependent transporter FoxA with siderophores and antibiotics that affect binding, uptake, and signal transduction. Proc. Natl. Acad. Sci. U S A 120:e2221253120. doi: 10.1073/pnas.2221253120
Cherian, T., Ragavendran, C., Vijayan, S., Kurien, S., and Peijnenburg, W. J. G. M. (2023). A review on the fate, human health and environmental impacts, as well as regulation of antibiotics used in aquaculture. Environ. Adv. 13:100411. doi: 10.1016/j.envadv.2023.100411
Colovos, C., and Yeates, T. O. (1993). Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2, 1511–1519. doi: 10.1002/pro.5560020916
Conrad, R. A., Evenhuis, J. P., Lipscomb, R. S., Pérez-Pascual, D., Stevick, R. J., Birkett, C., et al. (2022). Flavobacterium columnare ferric iron uptake systems are required for virulence. Front. Cell Infect. Microbiol. 12:1029833. doi: 10.3389/fcimb.2022.1029833
Daina, A., Michielin, O., and Zoete, V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7:42717. doi: 10.1038/srep42717
de Breij, A., Karnaoukh, T. G., Schrumpf, J., Hiemstra, P. S., Nibbering, P. H., van Dissel, J. T., et al. (2016). The licorice pentacyclic triterpenoid component 18β-glycyrrhetinic acid enhances the activity of antibiotics against strains of methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 35, 555–562. doi: 10.1007/s10096-015-2570-z
Eu, C. H., Lim, W. Y., Ton, S. H., and bin Abdul Kadir, K. (2010). Glycyrrhizic acid improved lipoprotein lipase expression, insulin sensitivity, serum lipid and lipid deposition in high-fat diet-induced obese rats. Lipids Health Dis. 9:81. doi: 10.1186/1476-511X-9-81
Farmer, B. D. (2004). Improved methods for the isolation and characterization of Flavobacterium columnare. Technical report. Louisiana State University and Agricultural and Mechanical College.
Guan, L., Santander, J., Mellata, M., Zhang, Y., and Curtiss, R. 3rd (2013). Identification of an iron acquisition machinery in Flavobacterium columnare. Dis. Aquat. Organ. 106, 129–138. doi: 10.3354/dao02635
Guo, W., Han, R., Xu, W., Lu, Z., Li, Y., Dan, X., et al. (2023). The protective effect of inactivated Flavobacterium columnare vaccine in grass carp (Ctenopharyngodon idellus). Front. Immunol. 14:1162975. doi: 10.3389/fimmu.2023.1162975
Harnvoravongchai, P., Chankhamhaengdecha, S., Ounjai, P., Singhakaew, S., Boonthaworn, K., and Janvilisri, T. (2018). Antimicrobial effect of asiatic acid against clostridium difficile is associated with disruption of membrane permeability. Front. Microbiol. 9:2125. doi: 10.3389/fmicb.2018.02125
Hernández-Silva, D., Alcaraz-Pérez, F., Pérez-Sánchez, H., and Cayuela, M. L. (2023). Virtual screening and zebrafish models in tandem, for drug discovery and development. Expert. Opin. Drug. Discov. 18, 903–915. doi: 10.1080/17460441.2022.2147503
Huang, L., Luo, H., Li, Q., Wang, D., Zhang, J., Hao, X., et al. (2015). Pentacyclic triterpene derivatives possessing polyhydroxyl ring A inhibit gram-positive bacteria growth by regulating metabolism and virulence genes expression. Eur. J. Med. Chem. 95, 64–75. doi: 10.1016/j.ejmech.2015.01.015
Jia, Y., Li, Z., Du, Q., and Chang, Z. (2021). Transcriptome analysis of immune-related gene expression in Yellow River carp (Cyprinus carpio var.) after challenge with Flavobacterium columnare. Microb. Pathog. 160:105148. doi: 10.1016/j.micpath.2021.105148
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kitiyodom, S., Yata, T., Thompson, K. D., Costa, J., Elumalai, P., Katagiri, T., et al. (2021). Immersion vaccination by a biomimetic-mucoadhesive nanovaccine induces humoral immune response of red tilapia (Oreochromis sp.) against Flavobacterium columnare challenge. Vaccines (Basel) 9:1253. doi: 10.3390/vaccines9111253
Kong, Y., Chen, F., Su, Z., Qian, Y., Wang, F. X., Wang, X., et al. (2018). Bioactive ruthenium(II)-arene complexes containing modified 18β-glycyrrhetinic acid ligands. J. Inorg. Biochem. 182, 194–199. doi: 10.1016/j.jinorgbio.2018.02.004
Lashkov, A., Tolmachev, I., Eistrikh-Heller, P., and Rubinsky, S. (2021). PyFepRestr: plugin to PyMOL molecular graphics system for calculating the free energy of ligand–receptor binding. Crystallogr. Rep. 66, 861–865. doi: 10.1134/S1063774521050126
Lee, C. S., Kim, Y. J., Lee, M. S., Han, E. S., and Lee, S. J. (2008). 18beta-Glycyrrhetinic acid induces apoptotic cell death in SiHa cells and exhibits a synergistic effect against antibiotic anti-cancer drug toxicity. Life Sci. 83, 481–489. doi: 10.1016/j.lfs.2008.07.014
Lemos, M. L., and Balado, M. (2020). Iron uptake mechanisms as key virulence factors in bacterial fish pathogens. J. Appl. Microbiol. 129, 104–115. doi: 10.1111/jam.14595
Martinez, A., Rivas, F., Perojil, A., Parra, A., Garcia-Granados, A., and Fernandez-Vivas, A. (2013). Biotransformation of oleanolic and maslinic acids by Rhizomucor miehei. Phytochemistry 94, 229–237. doi: 10.1016/j.phytochem.2013.05.011
Mirdita, M., Schütze, K., Moriwaki, Y., Heo, L., Ovchinnikov, S., and Steinegger, M. (2022). ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682. doi: 10.1038/s41592-022-01488-1
Moeck, G. S., and Coulton, J. W. (1998). TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28, 675–681. doi: 10.1046/j.1365-2958.1998.00817.x
Morley, N. J., and Lewis, J. W. (2010). Consequences of an outbreak of columnaris disease (Flavobacterium columnare) to the helminth fauna of perch (Perca fluviatilis) in the Queen Mary reservoir, south-east England. J. Helminthol. 84, 186–192. doi: 10.1017/S0022149X09990459
O'Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., and Hutchison, G. R. (2011). Open Babel: an open chemical toolbox. J. Cheminform. 3:33. doi: 10.1186/1758-2946-3-33
Oyama, K., Kawada-Matsuo, M., Oogai, Y., Hayashi, T., Nakamura, N., and Komatsuzawa, H. (2016). Antibacterial effects of glycyrrhetinic acid and its derivatives on Staphylococcus aureus. PLoS ONE 11:e0165831. doi: 10.1371/journal.pone.0165831
Paul Bhattacharya, S., Mitra, A., Bhattacharya, A., and Sen, A. (2020). Quorum quenching activity of pentacyclic triterpenoids leads to inhibition of biofilm formation by Acinetobacter baumannii. Biofouling 36, 922–937. doi: 10.1080/08927014.2020.1831480
Ponpukdee, N., Wangman, P., Rodkhum, C., Pengsuk, C., Chaivisuthangkura, P., Sithigorngul, P., et al. (2021). Detection and identification of a fish pathogen Flavobacterium columnare using specific monoclonal antibodies. Aquaculture 545:737231. doi: 10.1016/j.aquaculture.2021.737231
Raj, R., Agarwal, N., Raghavan, S., Chakraborti, T., Poluri, K. M., and Kumar, D. (2020). Exquisite binding interaction of 18β-Glycyrrhetinic acid with histone like DNA binding protein of Helicobacter pylori: a computational and experimental study. Int. J. Biol. Macromol. 161, 231–246. doi: 10.1016/j.ijbiomac.2020.06.039
Rather, M. A., Agarwal, D., Bhat, T. A., Khan, I. A., Zafar, I., Kumar, S., et al. (2023). Bioinformatics approaches and big data analytics opportunities in improving fisheries and aquaculture. Int. J. Biol. Macromol. 233:123549. doi: 10.1016/j.ijbiomac.2023.123549
Song, Y.-,l., Liu, H.-,w., Yang, Y.-,h., He, J.-,j., Yang, B.-, x., Yang, L.-,l., et al. (2022). Novel 18β-glycyrrhetinic acid amide derivatives show dual-acting capabilities for control of plant bacterial diseases through ROS-mediated antibacterial efficiency and activation of plant defense responses. J. Integr. Agric. 22, 2759–2771. doi: 10.1016/j.jia.2022.10.009
Staroscik, A. M., and Nelson, D. R. (2008). The influence of salmon surface mucus on the growth of Flavobacterium columnare. J. Fish Dis. 31, 59–69. doi: 10.1111/j.1365-2761.2007.00867.x
Sycz, Z., Tichaczek-Goska, D., and Wojnicz, D. (2022). Anti-planktonic and anti-biofilm properties of pentacyclic triterpenes-asiatic acid and ursolic acid as promising antibacterial future pharmaceuticals. Biomolecules 12:98. doi: 10.3390/biom12010098
Tabopda, T. K., Ngoupayo, J., Khan Tanoli, S. A., Mitaine-Offer, A. C., Ngadjui, B. T., Ali, M. S., et al. (2009). Antimicrobial pentacyclic triterpenoids from Terminalia superba. Planta Med. 75, 522–527. doi: 10.1055/s-0029-1185328
Taudte, N., German, N., Zhu, Y. G., Grass, G., and Rensing, C. (2016). Restoration of growth by manganese in a mutant strain of Escherichia coli lacking most known iron and manganese uptake systems. Biometals 29, 433–450. doi: 10.1007/s10534-016-9927-3
Tian, W., Chen, C., Lei, X., Zhao, J., and Liang, J. (2018). CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 46, W363–w367. doi: 10.1093/nar/gky473
Trott, O., and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461. doi: 10.1002/jcc.21334
Verma, D. K., Rathore, G., Pradhan, P. K., Sood, N., and Punia, P. (2015). Isolation and characterization of Flavobacterium columnare from freshwater ornamental goldfish Carassius auratus. J. Environ. Biol. 36, 433–439.
Wang, B., Zhu, F., Shi, Z., Huang, Z., Sun, R., Wang, Q., et al. (2022). Molecular characteristics, polymorphism and expression analysis of mhc II in yellow catfish (pelteobagrus fulvidraco) responding to Flavobacterium columnare infection. Fish Shellf. Immunol. 125, 90–100. doi: 10.1016/j.fsi.2022.04.036
Wang, H., Fang, Z. Z., Meng, R., Cao, Y. F., Tanaka, N., Krausz, K. W., et al. (2017). Glycyrrhizin and glycyrrhetinic acid inhibits alpha-naphthyl isothiocyanate-induced liver injury and bile acid cycle disruption. Toxicology 386, 133–142. doi: 10.1016/j.tox.2017.05.012
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucl. Acids Res. 46, W296–w303. doi: 10.1093/nar/gky427
Wu, P., Tu, B., Liang, J., Guo, S., Cao, N., Chen, S., et al. (2021). Synthesis and biological evaluation of pentacyclic triterpenoid derivatives as potential novel antibacterial agents. Bioorg. Chem. 109:104692. doi: 10.1016/j.bioorg.2021.104692
Xiong, G., Wu, Z., Yi, J., Fu, L., Yang, Z., Hsieh, C., et al. (2021). ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucl. Acids Res. 49, W5–w14. doi: 10.1093/nar/gkab255
Yang, Y., Zhu, Q., Zhong, Y., Cui, X., Jiang, Z., Wu, P., et al. (2020). Synthesis, anti-microbial and anti-inflammatory activities of 18β-glycyrrhetinic acid derivatives. Bioorg. Chem. 101:103985. doi: 10.1016/j.bioorg.2020.103985
Zhang, W., Zhao, J., Ma, Y., Li, J., and Chen, X. (2022). The effective components of herbal medicines used for prevention and control of fish diseases. Fish Shellfish Immunol. 126, 73–83. doi: 10.1016/j.fsi.2022.05.036
Zhao, Y., and Su, X. (2023). Antibacterial activity of 18β-glycyrrhetinic acid against Neisseria gonorrhoeae in vitro. Biochem. Biophys. Rep. 33:101427. doi: 10.1016/j.bbrep.2023.101427
Zhong, Y., Ding, Y., Xiao, D., Hu, D., and Li, Y. (2022). New 18β-Glycyrrhetinic acid-emodin esters synthetized by a one-step innovative route, its structural characterization, and in vivo toxicity assessed on zebrafish models. Chem. Biodiv. 19:e202100928. doi: 10.1002/cbdv.202100928
Keywords: Flavobacterium columnare, TonB dependent siderophore receptor, virtual screening, exoxolone, prokaryotic transcriptome
Citation: Li M, Chen B, Xu M, Li F, Geng Y, Chen D, Ouyang P, Huang X and Deng Y (2024) Identification of TonB-dependent siderophore receptor inhibitors against Flavobacterium columnare using a structure-based high-throughput virtual screening method. Front. Microbiol. 15:1392178. doi: 10.3389/fmicb.2024.1392178
Received: 27 February 2024; Accepted: 07 May 2024;
Published: 21 May 2024.
Edited by:
Sílvia A. Sousa, Institute for Bioengineering and Biosciences, PortugalReviewed by:
C. Gopi Mohan FRSC, Amrita Vishwa Vidyapeetham University, IndiaCopyright © 2024 Li, Chen, Xu, Li, Geng, Chen, Ouyang, Huang and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoli Huang, aHhsc2NhdUAxMjYuY29t; Yongqiang Deng, ZHlxaHhsQDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.