
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 May 2024
Sec. Infectious Agents and Disease
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1387498
This article is part of the Research Topic Insights in Infectious Agents and Disease: 2023/2024 View all 34 articles
Probiotic bacteria have been proposed as an alternative to antibiotics for the control of antimicrobial resistant enteric pathogens. The mechanistic details of this approach remain unclear, in part because pathogen reduction appears to be both strain and ecology dependent. Here we tested the ability of five probiotic strains, including some from common probiotic genera Lactobacillus and Bifidobacterium, to reduce binding of Salmonella enterica sv. Typhimurium to epithelial cells in vitro. Bifidobacterium longum subsp. infantis emerged as a promising strain; however, S. Typhimurium infection outcome in epithelial cells was dependent on inoculation order, with B. infantis unable to rescue host cells from preceding or concurrent infection. We further investigated the complex mechanisms underlying this interaction between B. infantis, S. Typhimurium, and epithelial cells using a multi-omics approach that included gene expression and altered metabolism via metabolomics. Incubation with B. infantis repressed apoptotic pathways and induced anti-inflammatory cascades in epithelial cells. In contrast, co-incubation with B. infantis increased in S. Typhimurium the expression of virulence factors, induced anaerobic metabolism, and repressed components of arginine metabolism as well as altering the metabolic profile. Concurrent application of the probiotic and pathogen notably generated metabolic profiles more similar to that of the probiotic alone than to the pathogen, indicating a central role for metabolism in modulating probiotic-pathogen-host interactions. Together these data imply crosstalk via small molecules between the epithelial cells, pathogen and probiotic that consistently demonstrated unique molecular mechanisms specific probiotic/pathogen the individual associations.
The gut microbiome provides a first line of defense against enteric infections. This essential community of microorganisms is also easily disrupted by dietary and local ecological factors that provide an opportunity for enteric pathogens to cause disease (Ducarmon et al., 2019). One proposed mechanism for the maintenance or reestablishment of a normal gut microbiome after community perturbations is the use of probiotic bacteria (Gareau et al., 2010). An important and accepted use of probiotics is to control enteric infections, which is especially timely given the rising number of multidrug resistant pathogens and subsequently dwindling number of effective antibiotics (Paton et al., 2006; Kulkarni et al., 2022). One notable candidate for probiotic modulation is multidrug resistant foodborne pathogen Salmonella enterica sv. Typhimurium, which causes over 80 million cases annual of foodborne illness globally and is a particular threat to infant mortality (Gong et al., 2022). Globally the burden of enteric pathogens is particularly prominent in the infant population, as evidenced by large cohort studies surveying disease incidence in children (Majowicz et al., 2010; Kotloff et al., 2013; Kasumba et al., 2021; Kotloff, 2022). In this high-risk infant population, currently a target population for probiotic development, Salmonella-induced diarrhea is a particularly notable issue with severe and potentially fatal health consequences if left untreated, especially in low-and middle-income countries (CDC, 2008; Majowicz et al., 2010; Shkalim et al., 2012; Kasumba et al., 2021; Eor et al., 2023; Splichalova et al., 2023). The need to find antibiotic alternatives for enteric diseases is urgent as antimicrobial resistance is increasing along with the evolution of multi-drug resistant strains (Nair et al., 2018; NARMS, 2023), with probiotics poised to be one such alternate treatment.
Though the mechanism by which probiotic organisms mitigate enteric infections is not yet well understood, current literature suggests probiotics may alleviate infection by acting as an immune stimulant, bioactive metabolite production, modulating gut pH, competing for nutrients used for infection, physically blocking host receptors, or through a combination of these activities (Lebeer et al., 2008; Iqbal et al., 2021). Supporting the potential immunomodulatory role of some probiotics, one study of S. Typhimurium infection in a mouse model indicated Lacticaseibacillus casei probiotic treatment not only decreased neutrophil infiltration and inflammation, but also increased the release of S. Typhimurium specific IgA in the intestinal lumen (de LeBlanc Ade et al., 2010). Other studies have more conflicting results that suggest probiotic efficacy and mechanistic underpinnings are context and host specific. Work on B. longum administration in one mouse model of salmonellosis revealed probiotic administration increased survival rate but had no impact of fecal shedding of the pathogen (Silva et al., 2004), while another study on the use of lactoferrin and milk-derived protein interventions displayed a reduction in Salmonella shedding and diarrhea in a porcine model (Hu et al., 2012). Together these results suggest that molecular signaling is part of the infection paradigm with unclear biological rules that may be dominated by strain variation represented by the large genomic variation among the bacterial actors (Arboleya et al., 2018; Díaz et al., 2021; van Puyvelde et al., 2023; Zhou et al., 2023). Though previous findings suggest probiotics are a possible treatment strategy for salmonellosis, they present their own set of challenges, such as differing adaptability and suitability to the target environment (Yu et al., 2013; Mercer and Arrieta, 2023; Thorman et al., 2023). To potentiate the best chance of success against enteric colonization, probiotics must be well-suited, even isolated from, their target environment. With this in mind, bifidobacteria stand as one probiotic option in Salmonella-susceptible population of infants (Mills et al., 2023).
Bifidobacteria are among the early colonizers of the infant gut, along with other commensal bacteria that include Escherichia coli, Staphylococcus, and Streptococcus (Mueller et al., 2015; Secher et al., 2016; Stewart et al., 2018). Around 40–60% of the fecal microbiota of a 2-week-old infant are Bifidobacterium species, though some formula-fed infants have no detectable bifidobacteria while other breast-fed infants show up to 90% bifidobacteria in their stool microbiome (di Gioia et al., 2014). This specificity of Bifidobacterium to the infant gut (LoCascio et al., 2010; Marcobal et al., 2011; Frese et al., 2017) coupled with the ability of some species to alleviate or prevent salmonellosis makes bifidobacteria a robust candidate as a potential infant probiotic. It is imperative that a mechanistic understanding be elucidated of how bifidobacteria may inhibit Salmonella considering the targeted infant population for such application are immune naïve. An important part of understanding the potential efficacy of probiotics like bifidobacteria is developing an improved understanding of the interaction between host, pathogen, and the gut microbiome at the host mucosa (Turroni et al., 2010; Pudlo et al., 2015; Underwood et al., 2015).
Many studies have sought to unravel the individual effect of probiotics, commensals, or pathogens on the host, but probiotic-pathogen interactions at the host interface remains an understudied field that is being complicated by the genomic variation of Salmonella (Gupta et al., 2019), strain variation in bifidobacteria (LoCascio et al., 2010; Díaz et al., 2021), and the variation in ecology of the gut between age groups (Voreades et al., 2014; Laforest-Lapointe and Arrieta, 2017). A steady increase in literature suggesting interspecies communication (i.e., community cross talk) exerts some control over the expression of virulence genes in enteric pathogens further highlights the need to define biochemical and genomic underpinnings of these interactions (Kendall and Sperandio, 2007; Shaw et al., 2022). The modulation of microbial activity by neighboring gut microbes via chemical signaling, for instance by widely conserved quorum response transcription factor sdiA in Salmonella (Plitnick et al., 2021), suggests microbe-microbe interactions via small metabolites are a significant factor in the infection process of enteric pathogens and potentially also in the response to probiotic interactions (Thompson et al., 2016).
Some probiotic bacteria have been suggested to potentiate positive host health effects via the production and regulation of bioactive metabolites (Indira et al., 2019). Metabolic regulation is also a determining factor in the success of Salmonella infection (Ilyas et al., 2017; Herrero-Fresno and Olsen, 2018). Salmonella is a metabolically flexible enteric pathogen known to manipulate host metabolism for pathogenic gain (Hume et al., 2017). Considering the known importance of metabolism in regulating Salmonella infection, the potential modulation of the host-microbe metabolic landscape by probiotic administration is a particularly intriguing mechanism to investigate.
Here we hypothesize that the addition of B. infantis to gut epithelial cells will affect the infection-related metabolic response of S. Typhimurium, attenuating adhesion and invasion of host cells. We subsequently show the relationship between B. infantis-S. Typhimurium was dependent on multiple factors, such as order of inoculation (concurrent addition, staggered addition, or co-incubation) that affect the ability of B. infantis to attenuate S. Typhimurium virulence. This study explored the complexity of these probiotic-pathogen interactions through evaluation of host, probiotic, and pathogen gene expression, and through small metabolite profiles, ultimately revealing the host-protective effects of B. infantis are both species and context dependent. Together the data suggest probiotic efficacy is reliant on the existence of narrow circumstances and thus must be more fully explored in lab settings, as demonstrated here, prior to clinical use.
Five different probiotic organisms, Bifidobacterium longum spp. infantis (ATCC 15697), Lactobacillus acidophilus (NCFM), Lactocaseibacillus casei (ATCC 334), Lactobacillus gasseri (ATCC 33323), and Levilactobacillus brevis (ATCC 367), were tested for their pathogen exclusion potential during infection with S. Typhimurium using differentiated colonic epithelial cells (Caco2) (Figure 1A). All organisms varied in their capacity to significantly (p < 0.045) reduce Salmonella adhesion. Of the five strains tested, B. infantis demonstrated the greatest capacity to significantly (p < 0.05) reduce Salmonella association with the epithelial cell and so was selected for further evaluation of potential molecular mechanisms underlying this attenuation.
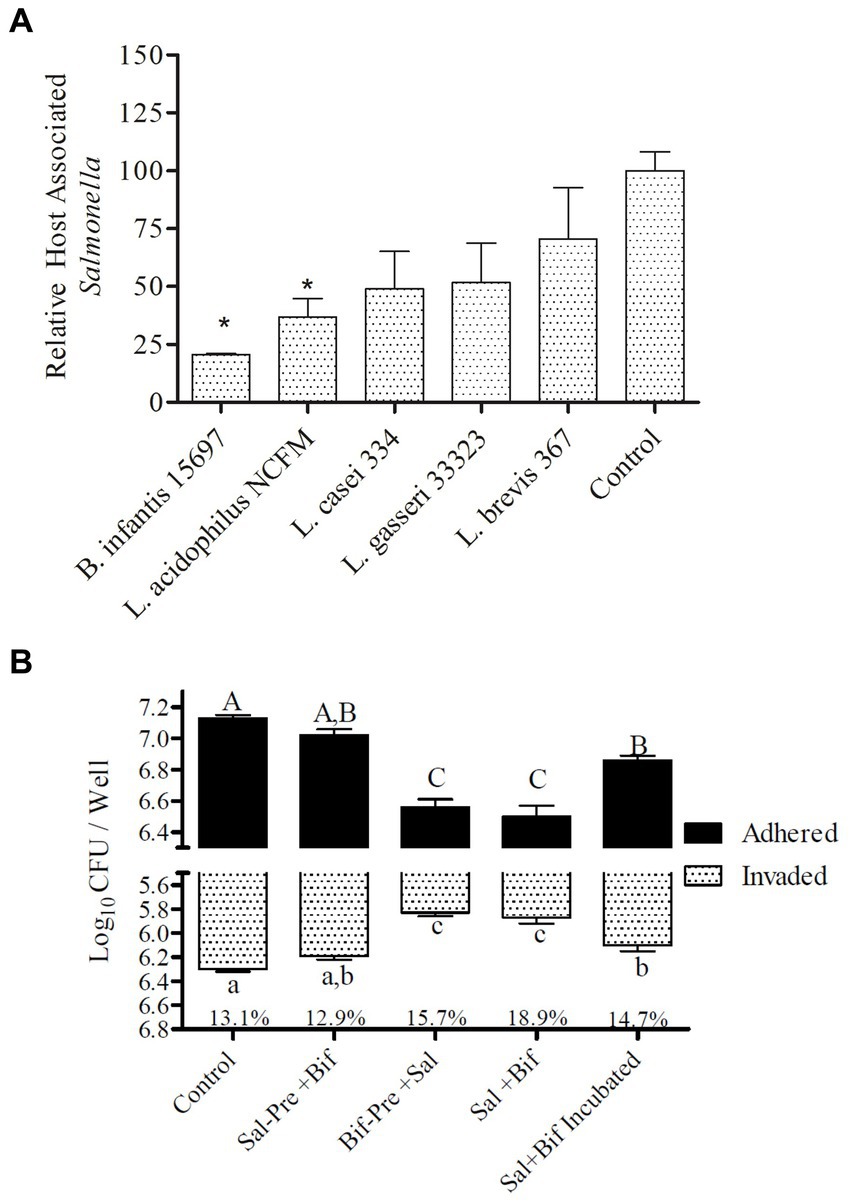
Figure 1. Adhesion to and invasion of Caco2 cells by S. Typhimurium. (A) Efficacy of different strains t block Salmonella host association. Treatments with “*” are significantly different (p ≤ 0.5) as compared to control (Caco2 cells incubated with Salmonella alone). Error bars indicate standard error from 4 replicates. (B) Effect of presence of B. infantis on the relative adhesion and invasion of S. Typhimurium to Caco2 cells. Control (Caco2 cells incubated with S. Typhimurium); Sal-Pre + Bif (Caco2 cells pre-incubated for 30 min with S. Typhimurium and then with B. infantis for 30 more minutes); Bif-Pre+ Sal (Caco2 cells pre-incubated with B. infantis for 30 minutes and then with S. Typhimurium for 60 more minutes); Sal+Bif (Caco2 cells incubated with S. Typhimurium and B. infantis together for 60 minutes); Sal+Bif Incubated (S. Typhimurium and B. infantis were first incubated together for 60 minutes and then incubated with Caco2 cells for 120 minutes). Treatments that do not share an alphabet are significantly different (p ≤ 0.05). The % numbers of the x axis represents the fraction of total host associated bacteria that were intracellular (invaded). Error bard indicate standard error from three replicates.
The ability of B. infantis to displace, block, or outcompete S. Typhimurium was tested by altering the point of addition and the incubation time of both B. infantis and S. Typhimurium with differentiated colonic epithelial cells in vitro (Figure 1B). B. infantis addition to Salmonella infected epithelial cells, showed no effect on the pathogen’s ability to invade the host cell or decrease expansion of the infection, suggesting B. infantis was unable to displace already adhered Salmonella or reduce infection. In contrast, B. infantis added to epithelial cells prior to S. Typhimurium addition significantly (p < 0.05) reduced S. Typhimurium adhesion and invasion, as well as when the pathogen and probiotic cultures were added to epithelial cells concurrently. Interestingly, the co-incubation of S. Typhimurium and B. infantis for 60 min resulted in a significant (p < 0.05) increase in host association by S. Typhimurium compared to the other combination treatments.
The finding that B. infantis was host-protective only when added prior to or concurrently with S. Typhimurium suggests potential involvement of physical exclusion mechanisms that prohibits S. Typhimurium from gaining access to the host membrane for binding and invasion. To evaluate if this seemingly opposite result stemmed from pathogen-probiotic competition for host cell receptors, receptors important for Salmonella adhesion and invasion in Caco2 cells (HSP90, PPP1R12A, CTNN1A, ganglioside GD3, and ganglioside GM1) (Desai, 2011) were blocked using antibodies prior to addition of either B. infantis or S. Typhimurium (Supplementary Figure S1). Unlike the other receptors tested, blocking ganglioside GM1 on the host reduced B. infantis adhesion by 25% as compared to the unblocked control condition (p = 0.05) (Supplementary Figure S1), indicating B. infantis and S. Typhimurium may compete to bind this specific host receptor. Notably, the binding of S. Typhimurium and B. infantis to the same host ganglioside GM1 receptor, which regulates host inflammation cascades, resulted in diametrically opposed downstream expression patterns in epithelial cells (Supplementary Figure S2).
The host ganglioside receptors, including GM1, are important modulators of the inflammatory response and involved in orchestrating the response to pathogen invasion (Park et al., 2023). The involvement of the ganglioside GM1 receptor in autocrine signaling, such as the IL-6 cascade, coupled with the observed affinity of B. infantis for this receptor led us to postulate that B. infantis association with human epithelial cells may initiate other host-protective signaling cascades. The canonical G-protein coupled receptor pathway (GPCR) was significantly induced (adj-p ≤ 0.04) in epithelial cells when incubated with B. infantis over 120 min, along with four GCPR ligands (CXCL1, CXCL2, CXCL3 and CCL20) and IL-6 that all showed significant induction (adj-p ≤ 0.00) (Table 1; Supplementary Table S1). Furthermore, genes downstream of IL6 signaling, which are under the positive transcriptional control of phosphorylated STAT3, were also induced (adj-p ≤ 0.00) (Table 2). Taken together, this collection of induced genes likely stimulated an autocrine signaling loop in the epithelial cell resulting from association with B. infantis.
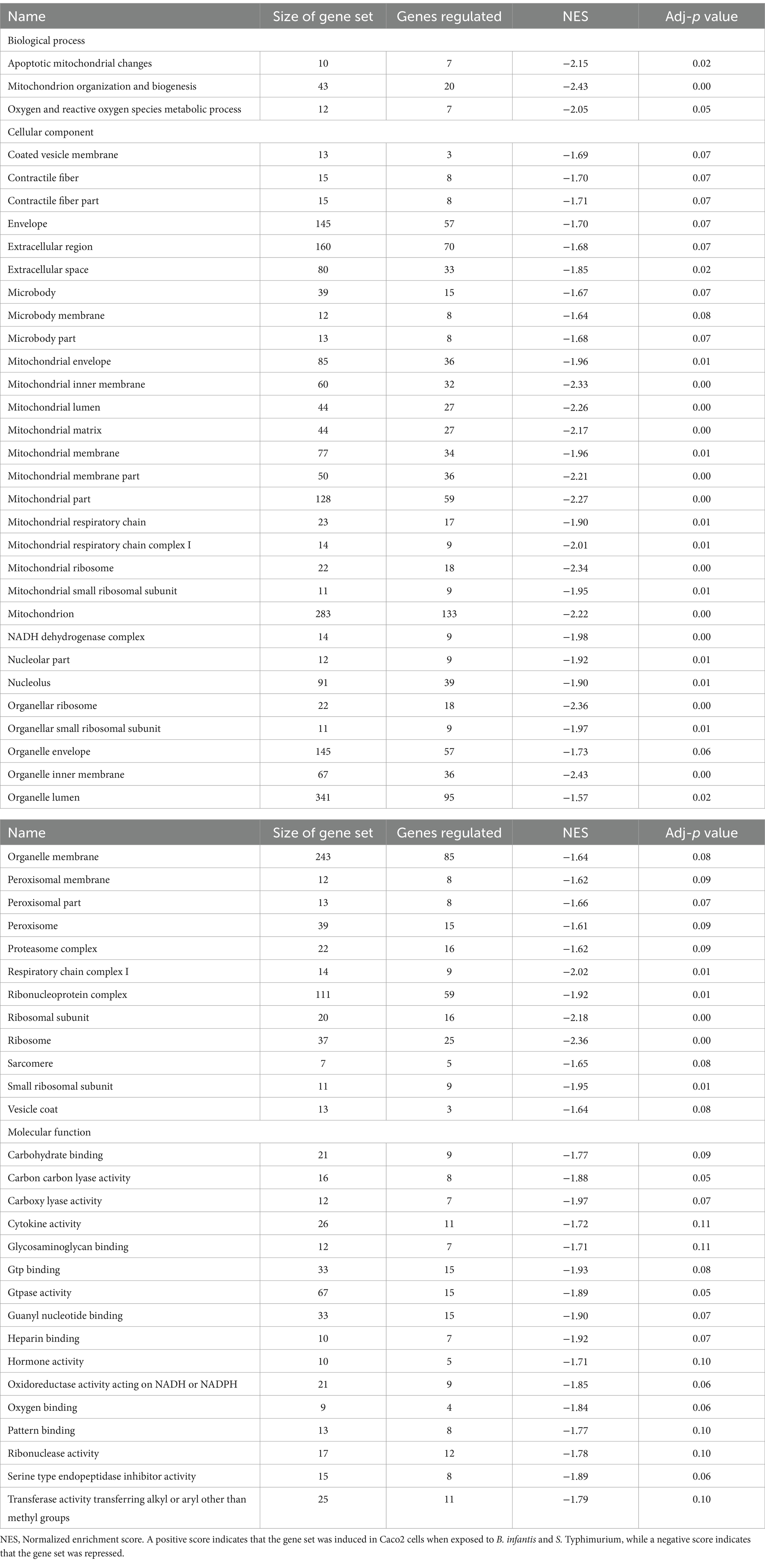
Table 1. Differentially regulated GO gene sets (adj-p ≤ 0.21) in Caco2 cells when exposed to B. infantis and S. Typhimurium.
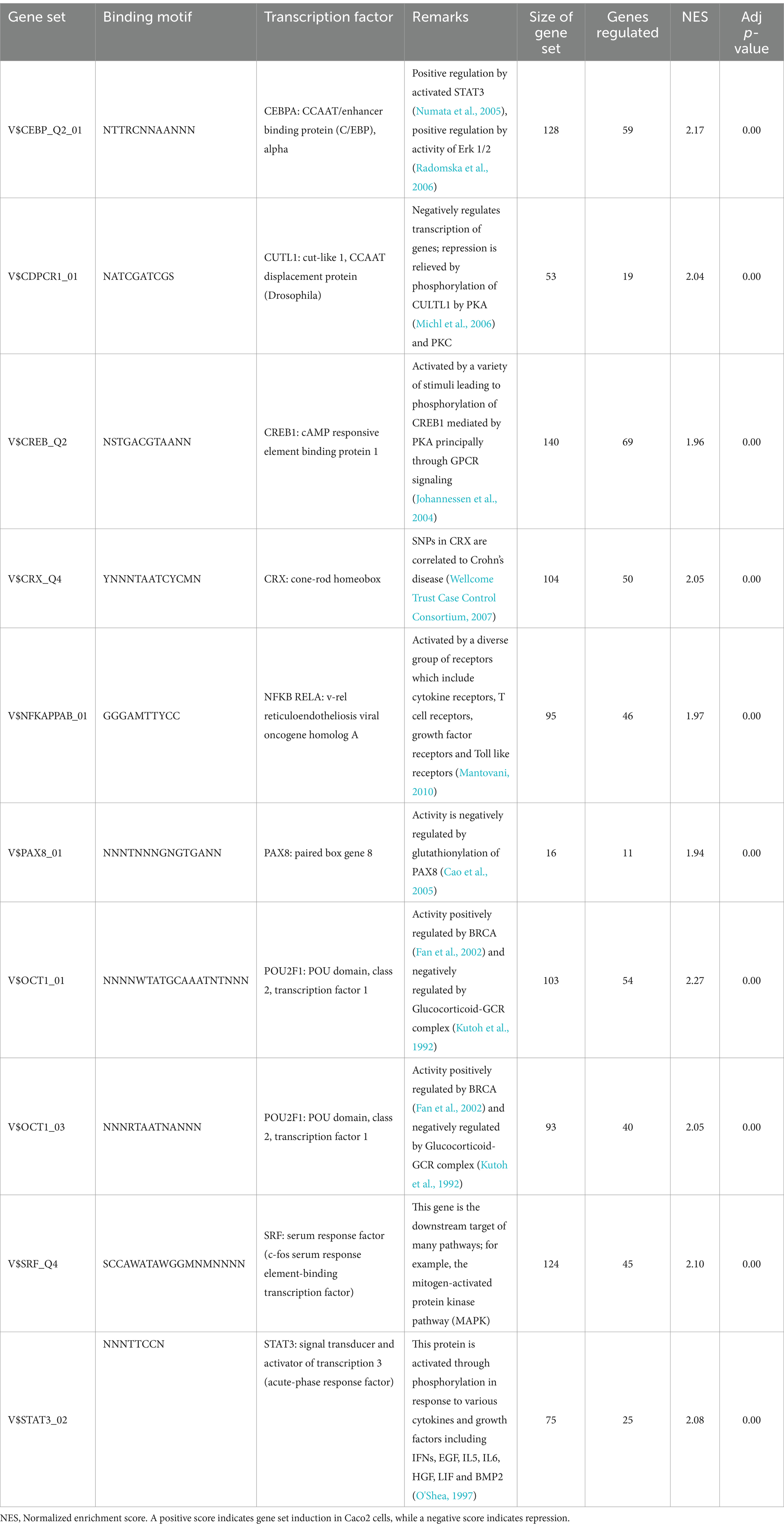
Table 2. Top ten differentially regulated (adj-p ≤ 0) transcriptional regulons in Caco2 cells when exposed to B. infantis.
The notable effect of B. infantis association on host autocrine signaling led us to examine the potential probiotic modulation of other epithelial cell pathways related to infection. Apoptotic signaling pathway in epithelial cells generally showed repression of cell death pathway after 120 min of B. infantis exposure (Figure 2), as well as in the presence of added S. Typhimurium with B. infantis (Supplementary Figure S3). Relatedly, B. infantis treatment significantly (adj-p < 0.05) lowered the overall basal caspase expression in epithelial cells and blocked the increase in Salmonella-mediated caspase activity. This observation of caspase regulation at the gene expression level was further confirmed via measurement of Akt phosphorylation status on ser473 and thr308 (Supplementary Figure S4). Together these data indicate regulation of host apoptotic signaling is an important factor that determines the success of Salmonella infection and the ability of B. infantis to alter the course of infection.
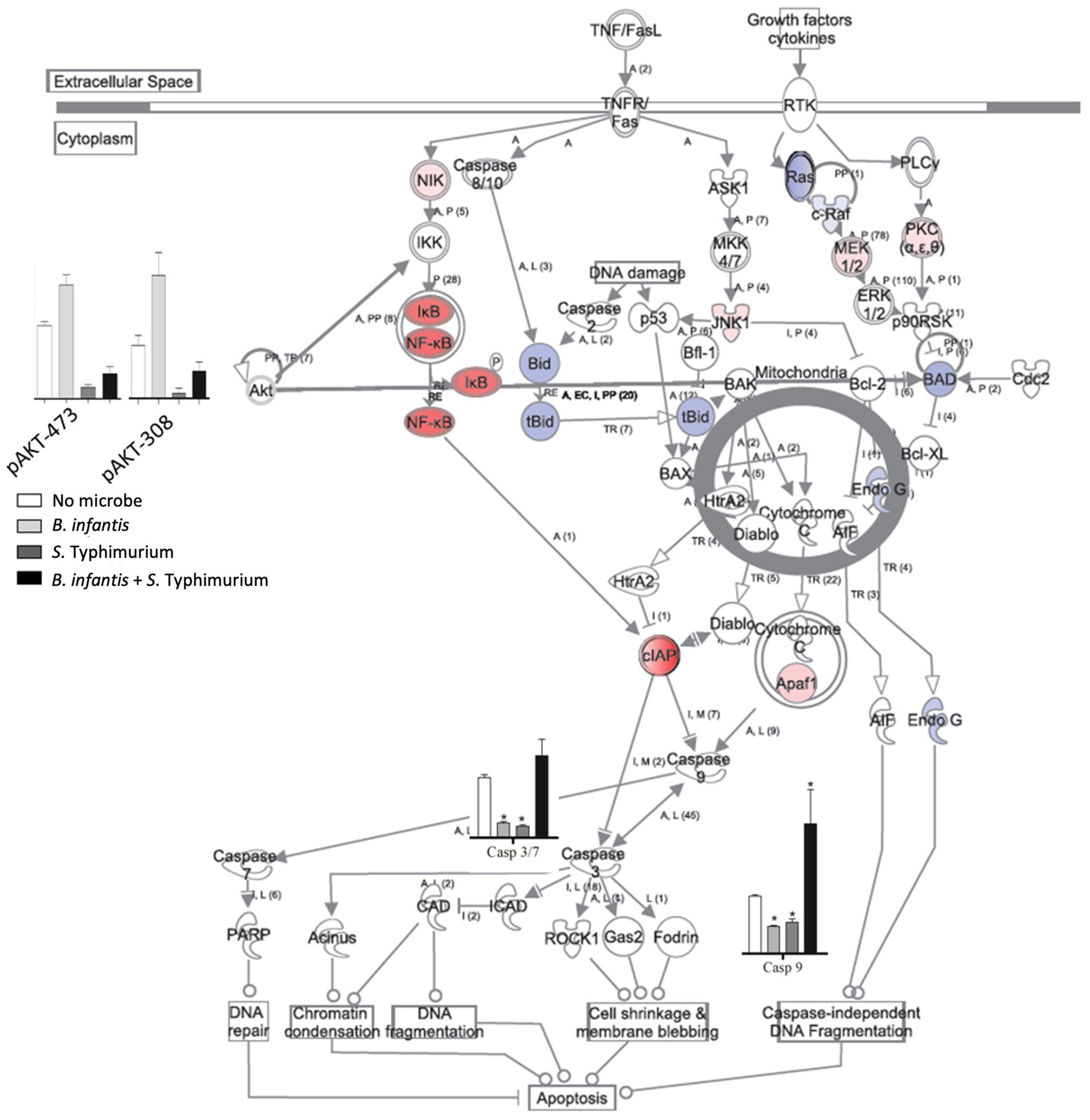
Figure 2. Log2 ratios of differentially expressed genes (adj-p ≤ 0.1); caspase 8, 9, 3/7 activity and phosphorylation status of Akt in epithelial cells when they are exposed to B. infantis. For gene expression data, red represents induction while blue represents repression of genes when epithelial cells were exposed to B. infantis.
The incubation of epithelial cells with B. infantis alone resulted in the induction of multiple protective effects for the epithelial cell, but it was unclear if these same pathways would remain induced in the presence of S. Typhimurium. Hierarchical clustering of averaged global gene expression profiles of the different treatments as a time series indicated that the transcriptome of epithelial cells infected with S. Typhimurium and B. infantis simultaneously is more like that of the solo B. infantis treatment than to the transcriptome of S. Typhimurium infected cells (adj-p < 0.01) (Figure 3A). This ability of B. infantis to modulate host cell expression despite the addition of S. Typhimurium was not consistent across all the genes or pathways examined in this study. Genes annotated in the GO category of cell junctions that were induced by B. infantis alone were contrastingly significantly repressed (adj-p ≤ 0.13) in presence of S. Typhimurium (Table 3; Supplementary Table S2). The ability of Salmonella to supersede host-protective effects from B. infantis incubation suggests there are more multi-factorial mechanisms that alter gene expression regulation that allows the pathogen to have a larger contributing factor for expression regulation as compared to the probiotic interaction.
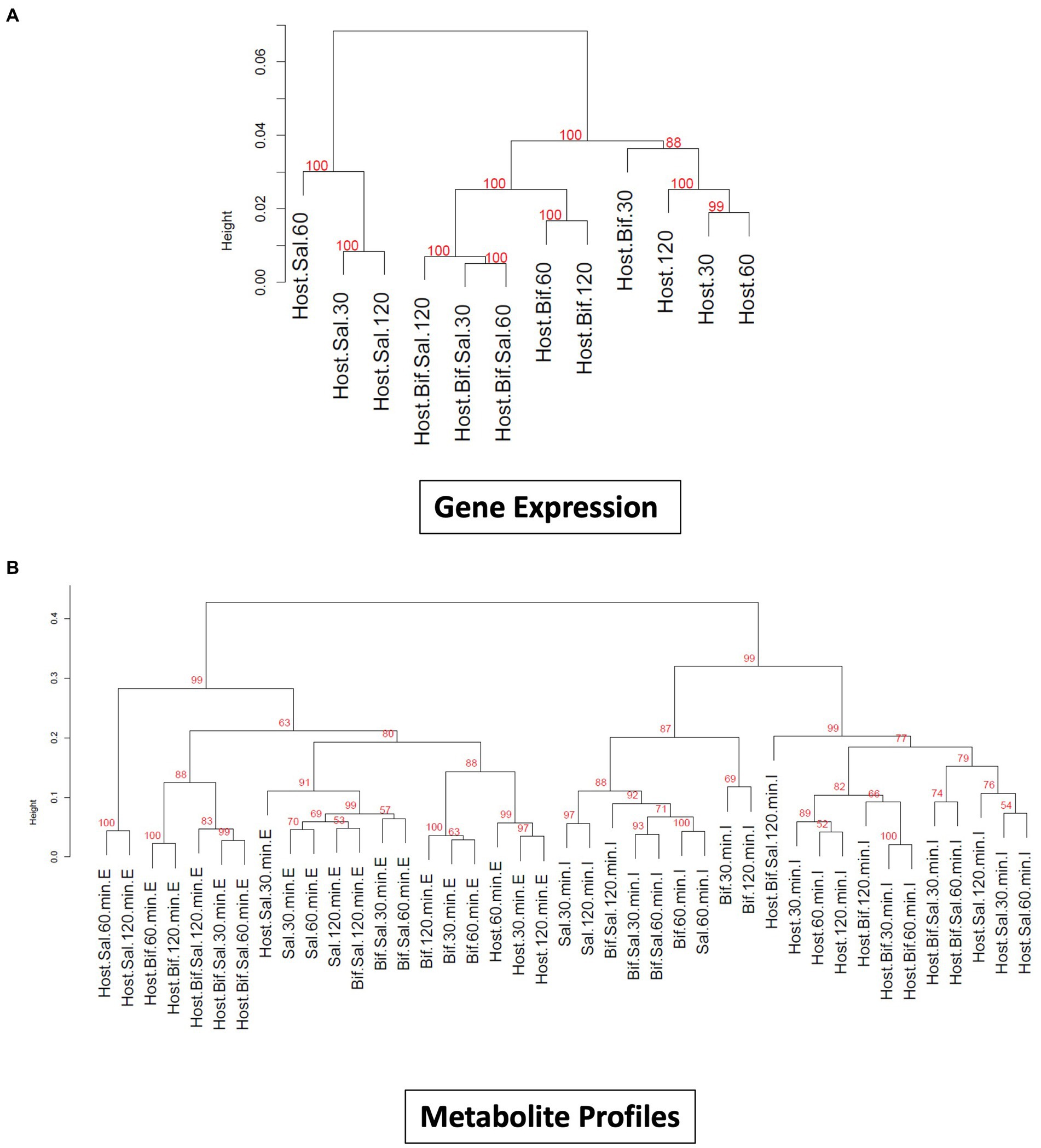
Figure 3. (A) Dendogram visualizing similarity between global gene expression profiles of Cac02 cells. The number at each cluster edge represents Approximately Unbiased % p value estimated by multiscale bootstrap resampling 1,000 times. (Host=Cac02 cells incubated with no microbes, Host+Bif=Cac02 cells incubated with B. infantis, Host+Sal=Cac02 cells incubated with S. Typhimurium, Host+Bif+Sal=Cac02 cells simultaneously incubated with 1:1 ratio of B. infantis and S. Typhimurium. The time in minutes represent the time after infection at which the gene expression was determined. (B) Dendogram constructed by hierarchical clustering of small metabolite profile of all the host microbe interaction samples. The number at each cluster edge represents Approximately Unbiased % p value estimated by multiscale bootstrap resampling 1,000 times (Host = Cac02, Bif = B. infantis, Sal = S. Typhimurium). The time in minutes represent the time after infection at which the metabolite profile was determined. “E” represents the extracellular metabolite profile from culture supernatant, while “I” represent the intracellular metabolite profile of all the cells in co-culture.
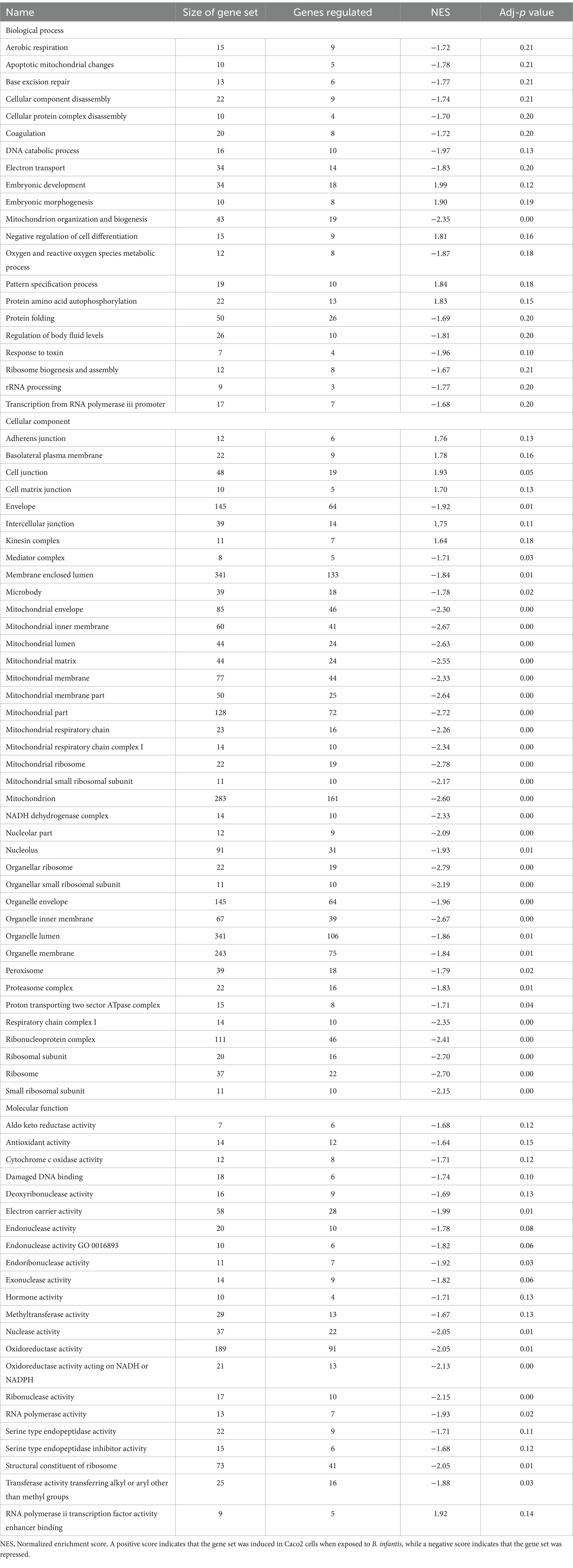
Table 3. GO gene sets that were differentially regulated (adj-p ≤ 0.21) in Caco2 cells when exposed to B. infantis.
One mechanism that may explain the ability of S. Typhimurium to override the potential host-protective mechanisms of B. infantis is the induction of Salmonella virulence factors in response to pathogen-probiotic co-incubation. A time series gene expression experiment (30, 60, and 120 min) (Figure 1B), revealed genes coding for both Type III Secretion Systems (T3SS) (SPI1 T3SS (adj-p ≤ 0), SPI2 T3SS (adj-p ≤ 0.04)), and effectors for both T3SSs (adj-p < 0.05), were significantly induced in the B. infantis co-culture condition, independent of epithelial cells (Supplementary Figure S5). The increased expression of the T3SS and its component parts in co-culture with B. infantis suggests B. infantis co-operatively potentiated the virulence of S. Typhimurium via shared environmental signals. One mechanism employed by S. Typhimurium to sense the local environment is the use of two-component systems, which have been linked to regulating virulence in fellow enteric pathogen E. coli (Reading et al., 2009). Virulence related two-component systems (Qse and SsrAB) in Salmonella (Merighi et al., 2009) were induced in the presence of B. infantis, irrespective of host cell presence (adj-p ≤ 0.09) (Supplementary Tables S3, S4).
The significant induction of both the T3SS and related two-component systems in the presence of B. infantis suggests that this probiotic organism primed virulence in Salmonella via multiple routes as well as being responsive to environmental cues during co-incubation. These environmental cues not only altered gene expression of Salmonella, but of B. infantis as well (Supplementary Table S5). Intriguingly, in the presence of Salmonella, B. infantis notably regulated genes that are conserved only in the subspecies infantis and not in divergent subspecies longum (Supplementary Figure S6; Supplementary Table S6). Regulation of this gene set that is horizontally acquired and unique to subspecies infantis suggests the effects of B. infantis observed here are specific to this taxonomic grouping. The co-regulation of Salmonella virulence factors and of B. infantis horizontally acquired gene sets, in conjunction with the noted importance of incubation order, highlight changes in the local environment are integral to probiotic-pathogen interaction outcome, brining into focus that bacterial crosstalk via small molecules is important in enteric infection (Table 4).
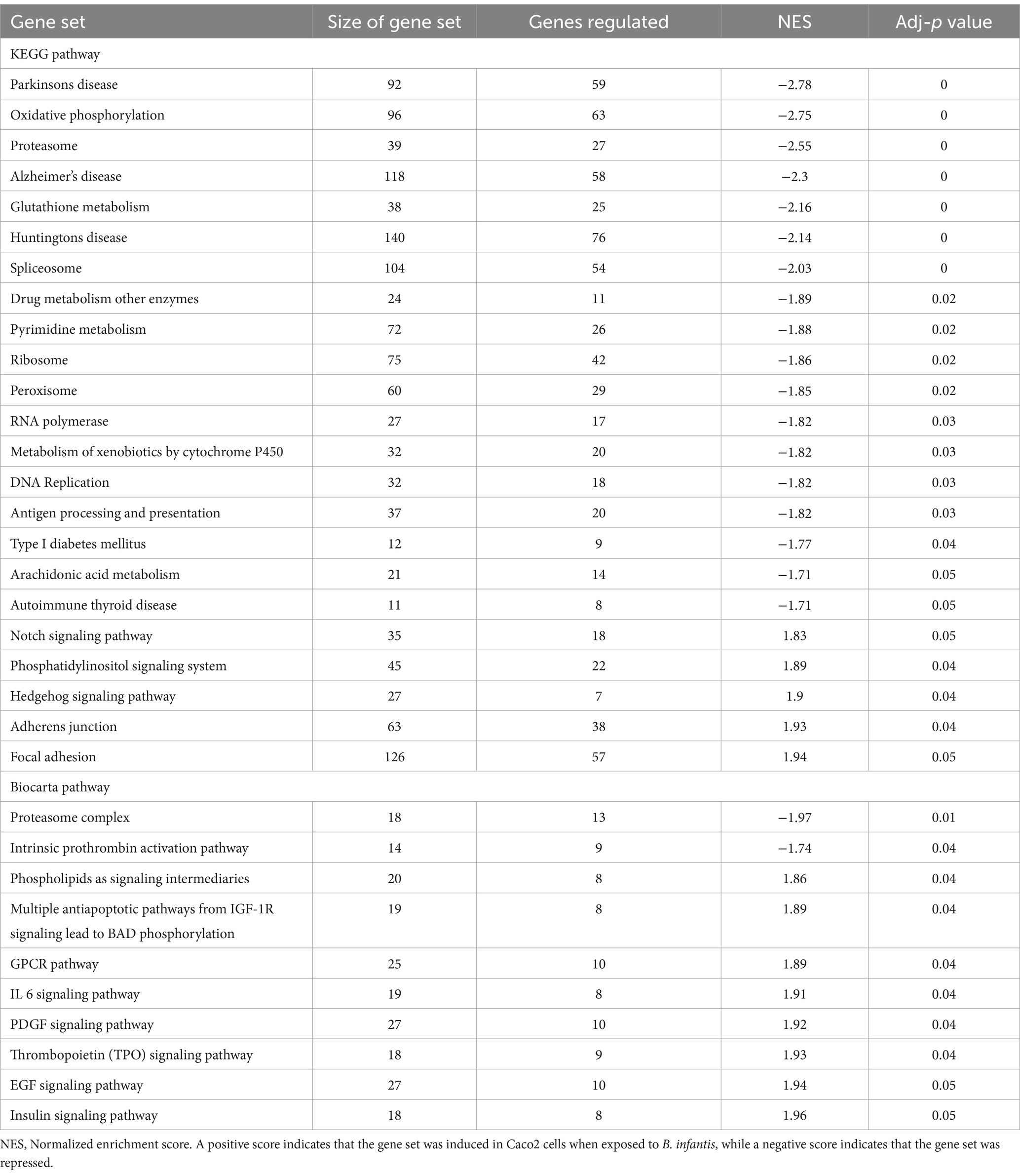
Table 4. Differentially regulated pathways (adj-p ≤ 0.05) in Caco2 cells when exposed to B. infantis.
Results from this study consistently demonstrate that multiple mechanisms mediate crosstalk that mediates a triad of cellular behavior between S. Typhimurium, B. infantis, and the epithelial cell that change the phenotype of metabolism and interaction. Induction of S. Typhimurium virulence genes, competition for host receptor GM1, and total gene expression changes observed in this study indicate that the progression of S. Typhimurium infection is affected by the activity of both B. infantis and the epithelial cells in more complex ways than simple bacterial competition for host receptor binding. One mechanism by which S. Typhimurium stages a successful infection and that may allow for probiotic bacterial competition is through a multifaceted metabolic coup.
Small molecules are increasingly being associated with bacterial adhesion on epithelial cells (Ghorashi and Kohler, 2020; Lin et al., 2020). Modification via glycosylation of proteins, which also play a role in mediating of bacterial binding (Barboza et al., 2012; Arabyan et al., 2016) is also a route that likely leads to cross-talk. In complement to the gene expression profiles, small metabolites were measured from the same epithelial cell treatment used for gene expression. Metabolite profiling using GC/MS identified 294 compound peaks using the binbase database (Fiehn et al., 2005), out of which 110 GC/MS peaks were assigned an identification (Figure 3). Hierarchical clustering of intracellular and extracellular small molecules (Figure 3B) followed a similar pattern to that of the global gene expression profiles (Figure 3A), where solo S. Typhimurium infection cluster separately from the control, solo B. infantis, and mixed culture treatments. Hierarchical clustering based on log2 peak areas of all compounds revealed two clusters for the extracellular metabolites and another for intracellular metabolites (adj-p < 0.01) (Figure 3B). Within the extracellular metabolite cluster, epithelial cells co-infected with Salmonella and B. infantis together clustered with epithelial cells treated with B. infantis alone (adj-p > 0.0.05). Epithelial cells infected with S. Typhimurium formed a unique cluster (adj-p < 0.01) at 60 and 120 min, demonstrating the divergence in the trajectory of infection progression. Generally, the gene expression profile and the extracellular metabolome of epithelial cells co-infected with S. Typhimurium and B. infantis were more like those of cells treated with B. infantis than with S. Typhimurium.
In summary, the gene expression and metabolic profiles paint a complex picture of a locally nuanced probiotic effect, wherein the association of B. infantis potentiates host-protective effects but the positive modulation is in part overridden by S. Typhimurium. The nuanced nature highlights the continuous observation that local environment mediates the host response as well as incubation with B. infantis alone versus with the B. infantis/S. Typhimurium combination. These orthogonal and consistent observations led us to hypothesize that the presence of S. Typhimurium and epithelial cells must also alter the activity of B. infantis.
Supporting this potential involvement of metabolic control in probiotic interaction was the marked shift in Salmonella’s metabolism in response to the co-culture partners. The addition of B. infantis to the host/Salmonella co-culture resulted in the repression of arginine catabolism genes in S. Typhimurium (Figure 4). This repression was likewise reflected in the small molecule profiles, where B. infantis addition led to the accumulation of arginine and ornithine with corresponding depletion of putrescine. In contrast, depletion of arginine was observed from Salmonella/epithelial cell co-culture in absence of B. infantis. Arginine accumulated when B. infantis was present, suggesting an increased substrate pool for nitric oxide (NO) production and provides a mechanism leading to hypervirulent Salmonella (Heithoff et al., 2012). The major pathway for NO metabolism is the stepwise oxidation to nitrite (NO2−) and nitrate (NO3−) (Lundberg et al., 2008). In presence of B. infantis, nine genes related to nitrate and nitrite reduction were significantly induced (adj-p ≤ 0) in Salmonella (Figure 4, “Anaerobic Metabolism”). The required genes in the pathway for synthesis of cofactors necessary for the activity of nitrate and nitrite reductases were also significantly induced in Salmonella co-cultured with B. infantis (adj-p ≤ 0) (Table 5, “Biosynthesis of Cofactors-Prosthetic Groups-And Carriers-Molybdopterin”). B. infantis-mediated induction of nitrate and nitrite reductases when co-cultured with Salmonella only occurred in the presence of epithelial cells, indicating induction of nitrate and nitrite reductases in Salmonella was likely a downstream effect of the epithelial cell metabolism, perhaps nitric oxide metabolism, but highlights the complexity of the metabolic interactions between the cellular triad, which led us to investigate energy cycling.
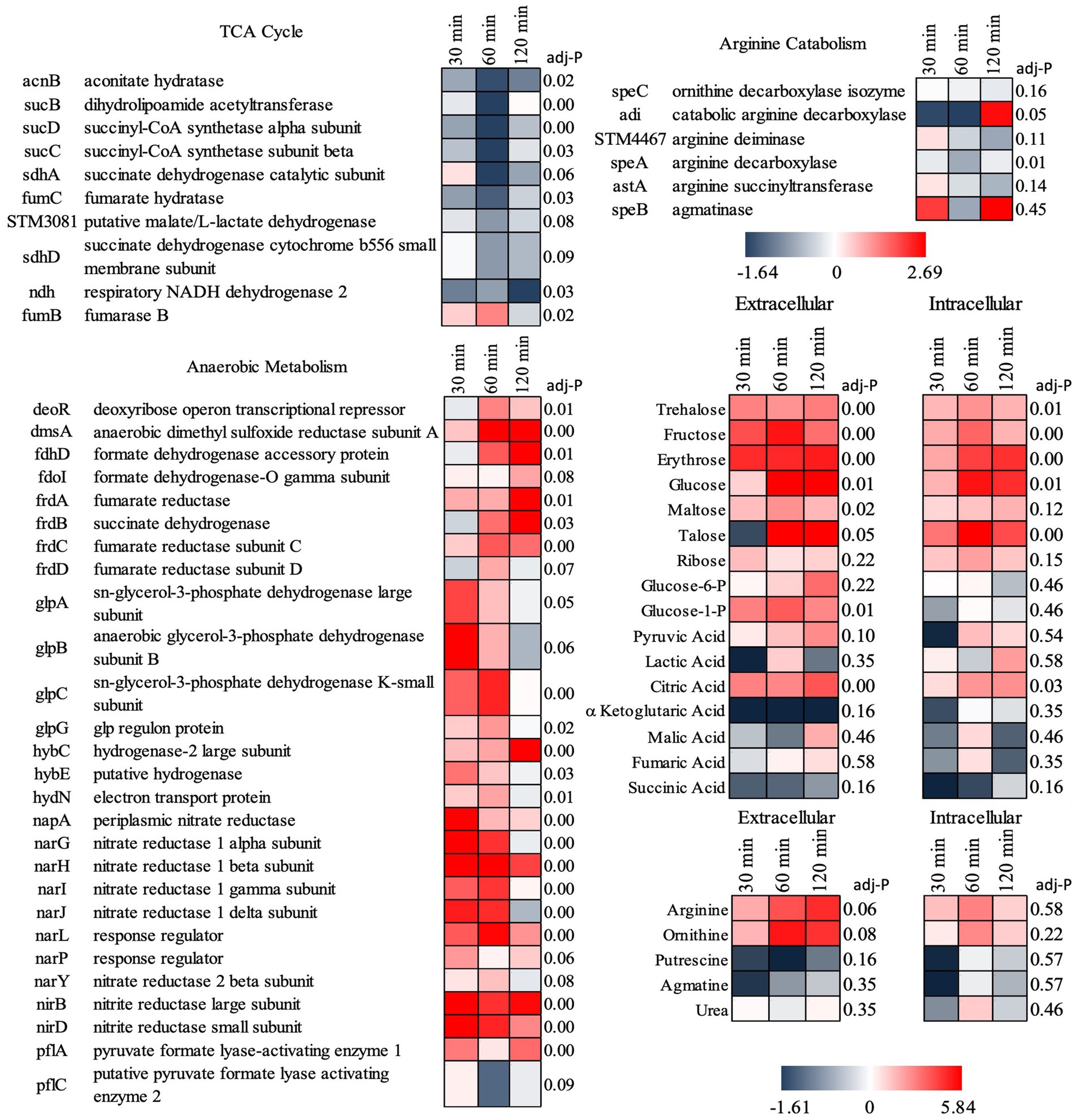
Figure 4. Log2 ratios of gene expression intensities of S. Typhimurium and selected small metabolite peak areas when Cac02 cells were treated with either S. Typhimurium alone or with a co-culture of S. Typhimurium and B. infantis. Induction of genes represent that the gene was induced in Salmonella in presence of B. infantis as compared to its absence.
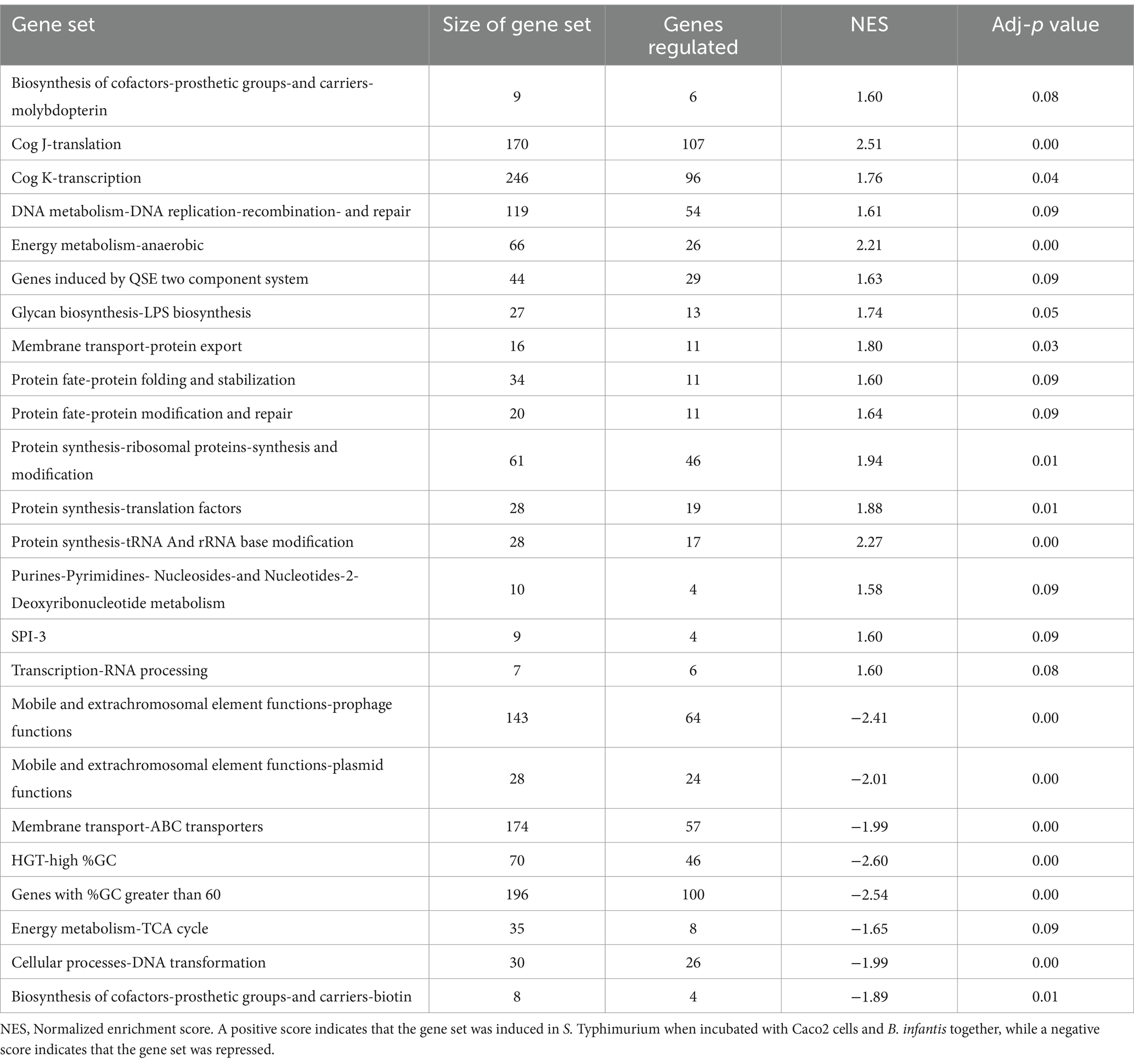
Table 5. Differentially regulated gene sets (adj-p ≤ 0.09) in S. Typhimurium when incubated with epithelial cells as compared to when incubated with epithelial cells and B. infantis for 120 min.
Further, addition of B. infantis to the host/S. Typhimurium co-culture also repressed Salmonella genes encoding for the TCA cycle (adj-p ≤ 0.09), and contrastingly induced genes necessary for anaerobic metabolism (adj-p ≤ 0) (Table 5; Figure 4). Genes necessary for mitochondrial respiratory chain function were also repressed in epithelial cells with the addition of B. infantis into the host/pathogen co-culture (Tables 2, 3). Addition of B. infantis resulted in repression of genes necessary for respiration in both epithelial cells and S. Typhimurium. Repression of respiration was also reflected in the small metabolite pool of the probiotic-pathogen co-culture with epithelial cells. Incubation of S. Typhimurium and host cells with B. infantis resulted in a significant accumulation of glucose, fructose, trehalose, and maltose (adj-p < 0.05) extracellularly and intracellularly. Glucose-6P, pyruvate, and citrate also significantly accumulated (adj-p ≤ 0.01) in the same conditions, while the remaining TCA metabolites (α-ketoglutarate, malate, fumarate, and succinate) did not significantly change, though a clear reduction was observed. B. infantis addition to the host/pathogen co-culture reduced the carbon flux through glycolysis and the TCA cycle, while the presence of Salmonella alone with epithelial cells induced host genes related to glycolysis that would influence the overall energy flux in each cell. The combination of small metabolite phenotyping and related gene expression reveal an intricate web of control wherein three-way crosstalk between host, probiotic, and pathogen drives shared metabolism and in turn either mitigates or exacerbates infection in a local context-dependent manner.
This study examined the ability of B. infantis to attenuate S. Typhimurium infection in an in vitro model and further characterized underlying microbe-microbe and host–microbe interactions. While B. infantis reduced adhesion and invasion to host cells, this significant mitigation of S. Typhimurium was only seen when B. infantis was added prior to or concurrently with Salmonella. B. infantis was unable to displace already adhered S. Typhimurium bacteria, suggesting that one mode of exclusion may be competition for specific host receptors, wherein early access to the receptor by Bifidobacteria effectively blocks Salmonella adherence.
The probiotic-pathogen pair in this work share an affinity for the host ganglioside GM1 receptor but not other known bacterial receptors commonly used by Salmonella. The observed interaction of B. infantis with the GM1 receptor may be associated with the co-evolution of B. infantis and breast milk (Underwood et al., 2015). Milk across species contain ganglioside binding motifs (Cacho and Lawrence, 2017; Tan et al., 2020) and B. infantis is genetically equipped to consume the complex oligosaccharides in breast milk (HMOs), making physical interactions with milk components evolutionarily advantageous to B. infantis (Lawson et al., 2020). In the context of the host receptors, binding to GM1 receptors in the gut epithelia by B. infantis may serve as a signal to the immune system (Kimata, 1994; Mou et al., 2022) and such immunostimulatory effects of bifidobacteria have been thoroughly reported by others in the literature (Ruiz et al., 2005; Henrick et al., 2021; Lueschow et al., 2022). Host ganglioside GM1, alongside other ganglioside receptors, modulate the expression of interleukins including pleiotropic and anti-inflammatory IL6 (Mou et al., 2022). Further supporting the strain-specific effects of probiotic and pathogen adhesion to epithelial cells, previous work in Salmonella has shown the host gut epithelial can recognize strain level variation and such recognition influences downstream expression (Salerno-Gonçalves et al., 2018; Salerno-Goncalves et al., 2019). As seen in this study, IL6 expression was induced in epithelial cells in the presence of B. infantis alone, potentiating host-protective mechanisms. The addition of Salmonella appears to dampen protective B. infantis immunomodulation activity, suggesting host modulation by B. infantis could be a selective effect, overridden by the exerted pressures of enteric pathogens, as shown with the IL6/NFkB expression pattern observed here.
Underscoring the ability of Salmonella to obstruct immunomodulatory properties of B. infantis, incubation of epithelial cells with B. infantis alone induced the transcription of NFkB related cytokines: IL6, CXCL1, CXCL2, CXCL3 and CCL20. Contrastingly, genes for these cytokines were repressed when Salmonella was added to the model. Salmonella interferes with host signal transduction, including pathways for producing these cytokines, by secreting a vast array of effector molecules directly into the host cytosol (Agbor and McCormick, 2011). One of these secreted effectors, AvrA, blocks NFkB activation by deubiquination of IkBα (Ye et al., 2007). Deubiquination of IkBα leads to the repression of target genes in the NFkB pathways, including IL6, a trend that was observed both in this study and previously by Ye et al. (2007). The repression on NFkB related cytokine genes in the present study indicates that Salmonella blocked the B. infantis-mediated induction of NFkB, likely through AvrA activity, which was induced in the presence of B. infantis.
Though Salmonella was able to negate some positive cytokine induction in the host from B. infantis interactions, host gene expression data for ROS production, cellular respiration, and mitochondrial biogenesis in this work reveals B. infantis may nevertheless modulate other host activity to reduce enteric infection. B. infantis in part mitigated Salmonella pathogenesis through modulation of mitochondrial dysfunction and related metabolic product ROS in epithelial cells. A variety of pathogens can induce cell death by causing mitochondrial dysfunction in the host (Rudel et al., 2010; Ashida et al., 2011; Lee et al., 2015; Ashida et al., 2021), which is induced by caspase 9 activation and is mediated through cytochrome c release in the host cytoplasm (Brentnall et al., 2013). Other work from our group has also correlated the induction of genes involved in production of ROS, respiration, and mitochondrial biogenesis to increased caspase 9 and caspase 3/7 activation during Salmonella infection (Shah et al., 2014), while caspase 8 and caspase 9 activity have been shown to increase in response to excess ROS (Fink and Cookson, 2007; Man et al., 2013; Shah et al., 2014; Hefele et al., 2018). Previous work has shown that mitochondrial production of ROS in the gut gives Salmonella a selective advantage, as ROS can react with luminal sulfur compounds to form tetrathionate, which can be used by Salmonella to respire, giving it a competitive edge over fermenting gut microbes (Winter et al., 2010). The results from this study suggest B. infantis may block ROS production in the gut through repression of host genes and pathways needed for aerobic respiration and functioning of the electron transport chain. Such a mechanism could contribute to the Bifidobacteria-mediated resistance to Salmonella observed here and also previously seen in mice (Shu et al., 2000; Huang and Huang, 2021).
As evidenced by the utilization of host-produced ROS detailed above, S. Typhimurium responds to and regulates the metabolic environment of the gut lumen (Das et al., 2010; Agbor and McCormick, 2011; Gart et al., 2016; Herrero-Fresno and Olsen, 2018). Manipulation of the gut metabolic environment by S. Typhimurium provides a competitive advantage to this pathogen in the crowded gastrointestinal microbial landscape (Taylor and Winter, 2020). Intriguingly, based on the metabolic profile clustering and more specific changes in select metabolites, there was a three-way, metabolically mediated, cross-talk between host, pathogen, and probiotic.
Bifidobacteria infantis prevented catabolism of arginine by S. Typhimurium, leading to arginine accumulation, which feeds increased host NO production (Wu et al., 2021). The increase in host NO production is supported by the significant induction of Salmonella genes necessary for nitrate and nitrite respiration, both products of NO metabolism. The mitigation of arginine metabolism in Salmonella by Bifidobacteria feeds host NO production, which then induces Salmonella to utilize nitrate and nitrite respiration pathways. The crosstalk between probiotic and pathogen observed here begins to explain the observations from the co-incubation condition, wherein S. Typhimurium appeared to have no change in, or even slightly increased, pathogenic fitness derived from the prebiotic’s presence.
The complex probiotic-pathogen interaction observed in this study was dependent on the local metabolic environment, as evidenced by the modulation of NO metabolism with gene expression and supported by small metabolite profiles. The extracellular metabolome of S. Typhimurium infected epithelial cells was markedly different than that of the B. infantis treated or concurrent B. infantis and S. Typhimurium infected cells, suggesting local metabolic control by B. infantis may be central to its probiotic efficacy or lack thereof. Bifidobacterium infantis was able to alter the metabolic environment of the host cells and maintained some of this metabolic control even in the presence of S. Typhimurium, as indicated by the hierarchical clustering of metabolic profiles. Such data support that metabolic control is one route by which probiotics may potentiate host protective effects, but simultaneously suggests such protective mechanisms are highly dependent on pathogenic metabolic abilities, further demonstrating with gene expression changes, protein alteration, and metabolites modulate the local environment.
In conclusion, B. infantis exerted protective effects against Salmonella in a colonic cell model through physical competition for the host ganglioside GM1 receptors; through repression of host cell death pathways, via the modulation of caspase activities; and metabolic shifts that change the energy balance and redox conditions that impact virulence capacity. The results of this work suggest B. infantis retains some but not all probiotic properties in the face of Salmonella infection, but follow-up work in animal models will be necessary to confirm these in-vitro observations with the additional complexity of the gut microbiome. In our model, these host-protective activities were contingent on incubation order, as B. infantis was not able to repress caspase activity when added after or concurrently with Salmonella. Notably, the mixed effect of Bifidobacteria addition on Salmonella pathogenicity extended to the modulation of bacterial and host metabolism. Probiotic co-incubation resulted in the repression of arginine catabolism by Salmonella and drove host NO production, ultimately leading the pathogen to regulate gene expression to efficiently utilize host-produced nitrate and nitrite. The finding that Bifidobacteria alter the metabolic activity of both the pathogen and the host supports that probiotic mechanisms are both nuanced and contextually driven. Further mechanistic research on this probiotic-pathogen pairing and others is crucial to understanding if these findings are specific to this combination or a more widespread phenomenon across probiotic-pathogen interactions; however, collectively, this study indicates that a complex relationship between the cellular triad of host, pathogen, and probiotic wherein the stage of probiotic addition, as a prophylactic or treatment, may ultimately dictate infection outcomes.
Colonic epithelial (Caco2) cells were obtained from ATCC (HTB-37, Manassas, VA) and cultured as per ATCC’s protocols and previously described (He et al., 2013; Arabyan et al., 2016). All the cells used in the assay were between passage numbers 22–30. In brief, cells were plated at a density of 105/cm2 in either a T75 or a 96-well plate. Cells were maintained in DMEM/High Modified (Thermo Scientific, Rockford, IL) with 16.6% fetal bovine serum (FBS) (HyClone Laboratories, Logan, UT), non-essential amino acids (Thermo Scientific, Rockford, IL), 10 mM MOPS (Sigma, St. Louis, MO), 10 mM TES (Sigma), 15 mM HEPES (Sigma) and 2 mM NaH2PO4 (Sigma). Cells were considered to be differentiated 14 days post confluence (Ouwehand and Salminen, 2003), and used for the adhesion assays, caspase assays and gene expression. Bacterial cells were grown as described in Supplementary Table S7.
The ability of specific bacteria to block Salmonella binding to epithelial cells was tested by determining the changes in the amount of intestinal epithelial cells associated Salmonella in presence of specific bacteria as previously described (He et al., 2013; Shah et al., 2014; Park et al., 2016; Chen et al., 2017). The amount of total host associated Salmonella was determined by qPCR as described below. The amount of total invaded Salmonella in the host were determined by the gentamicin protection assay (Arabyan et al., 2016) with modifications as described below.
Epithelial cells were cultured as described earlier in a 96-well plate (He et al., 2013; Shah et al., 2014; Park et al., 2016; Chen et al., 2017). The bacteria were used for the adhesion assays after two transfers. Bacterial cells were collected from 2 mL of media after growth for 14 h, washed twice with an equal volume of PBS, and re-suspended at ~108 cfu/mL, in DMEM/High modified with 1X non-essential amino acids, 10 mM MOPS, 10 mM TES, 15 mM HEPES and 2 mM NaH2PO4 but without the FBS. Epithelial cells were infected with either Salmonella alone or Salmonella in conjunction with other bacteria in a final volume of 50 μL at an MOI (multiplicity of infection) of 1:1000. The ratio of Salmonella with other bacteria was 1:1 and incubated with epithelial for 60 min. The bacterial cell suspension was aspirated, and the Caco2 monolayer was washed thrice with 200 μL of Tyrode’s (140 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 10 mM glucose, 10 mM sodium pyruvate, 10 mM HEPES, pH 7.4) to remove non-adhered bacterial cells from the monolayer. DNA extraction buffer (AEX Chemunex, France) (50 μL) was used to lyse the monolayer and the bacteria associated with the host, and incubated at 37°C for 15 min followed by 95°C for 15 min. Resulting cell lysate was used to determine the number of bacteria associated with the Caco2 cells (Desai et al., 2008). Quantitative analysis was done using qPCR with a CFX 96 Real Time System (BioRad, Hercules, CA). Reactions were performed in a final volume of 25 μL including 1 μL of cell lysate, 100 nM of PCR primers and iQ SYBR Green Supermix (BioRad, Hercules CA) as per manufacturer’s instructions. The primers used for the amplification are listed in Supplementary Table S9. The reaction parameters consisted of denaturation step at 95°C for 5 min, followed by 40 cycles of denaturation, annealing and extension at 95°C for 15 s, 56°C for 30 s, 72°C for 30 s, respectively, and a final extension at 72°C for 1 min. The product was verified using a melt curve analysis from 50°C to 95°C with a transition rate of 0.2°C/s. The number of bacterial cells and Caco2 cells present in each well were estimated using a standard curve of CT Vs Log10 cfu and number of bacteria per Caco2 was calculated. The data were mean normalized relative to control wells, which were incubated with Salmonella alone. The experiment was done in four replicates. Differences in the mean due to treatment were tested by one way ANOVA followed by a means comparison to control using Dunnett’s multiple comparison test.
The amount of total invaded and adhered bacteria was determined using the gentamicin protection assay previously described (Chen et al., 2017). The infected cells were incubated for 60 min at 37°C with 5% CO2. Upon incubation, the media was aspirated, and cells were washed three times with Tyrode’s buffer. Invaded bacteria were estimated by incubating the cells with 100 μg/mL gentamicin for two hours at 37°C with 5% CO2 to kill bacteria adhered or outside the Caco2 cells. To determine total host associated bacteria, cells were incubated with cell culture media without any antibiotic. Cells were again washed three times with Tyrode’s buffer and lysed with 0.01% triton. The amount of bacteria in the epithelial cell lysate were determined by plating serial dilutions on LB agar. The experiment was performed in four replicates. The number of adhered bacteria were enumerated by subtracting mean of invaded bacteria (B) from mean of total host associated bacteria (A) and error (ΔZ) was calculated as (ΔZ)2 = (ΔA)2 + (ΔB)2 where, ΔA is SEM associated with the A and ΔB is SEM associated with B. The data were reported as log10 colony forming units (CFU) /well. Differences in the mean due to treatment were tested by one way ANOVA. Post ANOVA, the means were compared to each other by Tukey–Kramer’s method.
Caco2 cells were cultured in a 96-well plate as previously described and were washed with phosphate buffered saline (PBS) before infection with bacteria. Caco2 cells were infected with either Bifidobacterium longum spp. infantis ATCC 15697, Salmonella sv. Typhimurium LT2 ATCC 700720, or both organisms simultaneously. Upon adding bacterial treatments (MOI of 1:1000) in a volume of 50 μL, cells were incubated at 37°C with 5% CO2. After 8 h, caspase 8, 9 and 3/7 activities were measured using Caspase-Glo assay kits (Promega, Madison, WI) exactly as per manufacturer’s instructions. Bioluminescence due to caspase activity was measured using the DTX 880 Multimode Detector (Beckman Coulter, Brea, CA). Differences in the mean due to treatments were tested by one way ANOVA. Post ANOVA, the means were compared to each other by Tukey–Kramer’s method.
Human colonic epithelial (Caco2) cells were cultured in T75 as described earlier, and were serum starved for 24 h prior to infection with bacteria. Caco2 cells were infected with either Bifidobacterium longum spp. infantis ATCC 15697, Salmonella sv. Typhimurium LT2 ATCC 700720, or both the organisms simultaneously at an MOI of 1:1000 in a final volume of 10 mL. All the three bacterial treatments were also incubated in absence of the Caco2 cells in the exact same conditions. All the infected cells were incubated at 37°C with 5% CO2. At 30, 60 and 120 min after infection, media with non-adherent bacteria was aspirated and 10 mL TRIzol LS reagent (Invitrogen, Carlsbad, CA) was added to the cells. This was gently mixed with pipette followed by centrifugation at 7200 × g for 5 min to pellet the bacteria. TRIzol LS supernatant was stored in a clean tube and further processed for RNA extraction from Caco2 cells. The bacterial pellet was re-suspended in 2 mL of fresh TRIzol LS, gently mixed and further processed for RNA extraction from host-associated bacteria.
The TRIzol LS suspension containing host-associated bacteria was centrifuged at 7200 × g for 5 min. The remaining TRIzol LS supernatant was stored in a clean tube for later use. 1 mL of lysis enzyme cocktail containing 50 mg/mL of lysozyme (Sigma) and 200 U/mL mutanolysin (Sigma) in TE buffer (10 mM Tris and 1 mM EDTA, pH 8) was added to bacterial pellet obtained by centrifugation. The solution was mixed gently and incubated at 37°C for 1 h followed by centrifugation at 7200 × g for 5 min. The supernatant was discarded, and the pellet was re-suspended in 250 μL of proteinase K buffer (100 mM Tris–HCl, 5 mM EDTA, 200 mM NaCl, and 0.2% SDS, pH 8) containing 8 U/mL of proteinase K (Fermentas, Glen Burnie, MD). This was incubated at 55°C for 1 h with intermittent mixing. To this, previously stored TRIzol LS was added and gently mixed. RNA was isolated from TRIzol LS exactly as per manufacturer’s recommendations. RNA concentration, A260/280 and A260/230 were measured on NanoDrop (Thermo scientific, Waltham, MA) and samples were processed further only if the RNA concentration was at least 0.5 μg/μl and ratios were ≥ 1.8. The RNA samples were analyzed for integrity on 2,100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Total RNA (10 μg in 20 μL) was reverse transcribed into cDNA with random hexamers and Superscriptase II (Invitrogen, Carlsbad, CA) exactly as per manufacturer’s recommendations by using 400 U of Superscriptase II/10 μg RNA. Upon cDNA synthesis, the enzyme was heat inactivated and the RNA templates were degraded with 80 U of RNaseH (Epicentre, Madison, WI) at 37°C for 20 min. The reaction mixture was cleaned using the Qiaquick-PCR purification kit (Qiagen, Valencia, CA) exactly as per manufacturer’s instructions. Purified cDNA was eluted from the column twice with a total of 100 μL of nuclease free water (Ambion, Austin, TX).
cDNA was fragmented using DNaseI (Promega, Madison, WI) according to manufacturer’s recommendations by using 0.6 U of DNAseI/μg cDNA at 37°C for 20 min. The fragmented cDNA was labeled using GeneChip DNA Labeling reagent (Affymetrix, Santa Clara, CA) and Terminal Transferase enzyme (TdT) (New England Biolabs, Ipswich, MA) as per manufacturers’ recommendation by using 2 μL GeneChip labeling reagent and 3 μL TdT/μg cDNA at 37°C for 60 min. The samples were denatured prior to hybridization, at 98°C for 10 min followed by snap cooling at 4°C for 5 min.
Labeled cDNA was hybridized onto two different custom made Affymetrix GeneChip designed against all the annotated coding sequences of Bifidobacterium longum ssp. infantis ATCC 15697 (LoCascio et al., 2010) and Salmonella enterica subsp. enterica sv. Typhimurium LT2 ATCC 700720. Briefly the Salmonella array contained 9,852 probe sets, of which 4,735 probe sets were designed against S. sv. Typhimurium. Each probe set contained 11 probes, each 25 nucleotides long. The Bifidobacteria chip has been described by LoCascio et al. (2010). An aliquot (500 ng) of labeled cDNA for samples extracted from pure culture of B. infantis and S. Typhimurium; 1,000 ng labeled cDNA for samples extracted from co culture of B. infantis and S. Typhimurium; 2000 ng of labeled cDNA for samples extracted from co-culture of B. infantis/S. sv. Typhimurium and Caco-2; and 2,500 ng of labeled cDNA for samples extracted from co-culture of B. infantis, S. Typhimurium and Caco2 was hybridized onto the respective chips. In total, 24 chips Salmonella chips were processed (2 replicates X 3 time points (30 min, 60 min, 120 min) X 4 treatments) and 8 Bifidobacteria chips were processed (2 reps X 1 time point (120 min) X 4 treatments).
Raw data (.cel files) was back ground corrected, quantile normalized and summarized using RMA-MS (Stevens et al., 2008). The resultant normalized log2 transformed intensity matrix was used for further statistical analysis. To detect differentially regulated genes, Salmonella chip data was analyzed as unpaired time course analysis with “signed area” as the time summary method, while for Bifidobacteria chips the data was analyzed as two class unpaired data, with T statistic, using Significance Analysis of Microarrays (SAM) (Tusher et al., 2001). All the genes were ranked based on the score (d) from SAM output. This pre-ordered ranked gene list was then used in Gene Set Enrichment Analysis software (GSEA) (Mootha et al., 2003; Subramanian et al., 2005) to detect the coordinate changes in the expression of groups of functionally related genes, upon respective treatments. Gene sets for Bifidobacteria were defined based on annotations from KEGG (Kanehisa et al., 2002), Cluster of Orthologous Groups of proteins (COGs) (Tatusov et al., 1997), Carbohydrate Active Enzyme Database CAZY (Park et al., 2010) and genes identified by LoCascio et al. (2010). Putative horizontally transferred genes in B. infantis were identified using Integrated microbial genomes system (IMG) (Markowitz et al., 2010). Genes sets based on protein localization were created based on annotations from CoBaltDB (Goudenège et al., 2010). Gene sets for Salmonella were defined based the annotations from Comprehensive Microbial Resource (CMR) (Peterson et al., 2001), Cluster of Orthologous Groups of proteins (COGs) (Tatusov et al., 1997), Virulence Factors of pathogenic bacteria DataBase (VFDB) (Chen et al., 2003; Yang et al., 2008), Carbohydrate Active Enzyme Database (CAZY) (Park et al., 2010), and BioCyc (Caspi et al., 2008). Gene sets based on putative horizontally transferred genes in Salmonella were defined using predictions from the Horizontal Gene Transfer Database (HGT-DB) (Garcia-Vallve et al., 2003). Genes sets based on protein localization were created using predictions from CoBaltDB (Goudenège et al., 2010).
The TRIzol LS (Thermo Fisher Scientific, Waltham, MA, USA) supernatant obtained after pelleting the bacteria was freeze thawed twice in liquid nitrogen. 250 μL water was added to 750 μL of TRIzol LS sample. This was further processed for RNA extraction exactly as per manufacturer’s instructions. RNA concentration, A260/280 and A260/230 were measured on NanoDrop (Thermo scientific, Waltham, MA) and samples were processed further only if the RNA concentration was at least 0.5 μg/μl and ratios were ≥ 1.8. The RNA samples were analyzed for integrity on 2,100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Synthesis of cDNA, biotin labeled cRNA, fragmentation and purification of cRNA were carried out using one-cycle cDNA synthesis kit (Affymetrix, Santa Clara, CA) exactly as per manufacturer’s instructions. 10 μg of labeled and fragmented cRNA was hybridized onto the Affymetrix HGU133Plus2 GeneChips as per manufacturer’s recommendations at the Center for Integrated BioSystems (Utah State University, Logan, UT).
Raw data (.cel files) were background corrected, quantile normalized and summarized using RMA (Irizarry et al., 2003). RMA normalized data was then filtered through the PANP algorithm (Warren et al., 2007) to make presence-absence calls for each probe set. Probe sets that were called present in at least one of the samples were included in further statistical analysis while rests were excluded. The resultant normalized, filtered, log2 transformed intensity matrix was analyzed as two class unpaired time course data with “signed-area” as the time summary method, using Significance Analysis of Microarrays (SAM) (Tusher et al., 2001). All the genes were ranked based on the score (d) obtained from SAM. This pre ordered ranked gene list was used as an input for Gene Set Enrichment Analysis software (GSEA) (Subramanian et al., 2005) to detect the coordinate changes in the expression of groups of functionally related genes, upon respective treatments. The gene sets were based on Gene Ontology annotations, KEGG annotations, Biocarta annotations and TRANSFAC annotations and were downloaded from the molecular signatures database (Subramanian et al., 2005). Ingenuity pathway analysis (IPA) was used to map expression data onto canonical pathways. Hierarchical clustering with multiscale bootstrapping (1,000 times) of all the samples based on normalized data was performed using PVCLUST (Suzuki and Shimodaira, 2006).
Relative abundance of target proteins was quantified by densitometric analysis of western blots. Caco2 cells were cultured and infected exactly as with those infected for the gene expression, detailed above. Cells from the T75 were removed from the flask by scraping; suspended in 1 mL protease/phosphatase inhibitor cocktail (30 mM HEPES, 1 mM EDTA, 50 mM Sodium pyrophosphate, 100 mM sodium fluoride, 10 mM orthovanadate, 1 Roche protease inhibitor cocktail tablet/50 mL solvent) and lysed using a bead beater at full speed with 3 pulses of 30 s each with intermittent incubation on ice for 1 min. The cell lysates were stored at −70°C for further analysis. 50 μg protein was diluted in 1X SDS sample buffer and the samples were heated at 95°C for 10 min and centrifuged at 12,000 X g for 10 min. The sample was resolved on precast 10% Tris–HCl polyacrylimide gels (Bio-rad Laboratories, Hercules, CA) at a constant current of 45 mAmp per gel using the Criterion electrophoresis system (Bio-rad Laboratories, Hercules, CA). The resolved proteins were then transferred to a PVDF membrane (Bio-rad Laboratories, Hercules, CA) using the Trans-Blot semi-dry electrophorectic cell (Bio-rad Laboratories, Hercules, CA) as per the manufacturer’s recommendation. Upon completion of the transfer, the blots were probed for the presence of p-Akt 473 (#9271) and p-AKT 308 (#9275) by Pierce® Fast Western Blot Kit (Thermo Fisher Scientific, IL, USA), according to the manufacturer’s recommendations. Primary antibodies for all the proteins were purchased from Cell Signaling Tech. (Boston, MA). The blots were imaged using a Kodak Image Station 2000R (Carestream Health, Rochester, NY). Densitometric analysis of the blot was done using Image J (Abramoff et al., 2004). Differences in the means due to treatment were tested by two-way ANOVA with repeated measures.
Extracellular supernatant and intracellular cell lysate (50 μL) collected from the infection experiments were extracted with an equal volume of ice-cold methanol and incubated at −20°C for 60 min. The resultant precipitates were separated from the sample by centrifugation at 20,000 X g for 10 min. The supernatant was evaporated to dry using a speed-vac at room temperature. Metabolite profiles for all the samples were determined using GC–MS at the UC Davis Genome Center (Davis, CA) as described previously (Fiehn et al., 2008). In brief, the dried sample was derivatized using N-methyl-N-(trimethylsilyl)-trifluoroacetamide (Sigma, St. Louis, MO) and spiked with retention index markers (RI) as described previously (Fiehn et al., 2008). The samples were analyzed on Agilent 6,890 gas chromatograph controlled using Leco ChromaTOF software version 2.32. as described previously (Fiehn et al., 2008). GC/MS peaks were annotated based on the mass spectra and retention index using the BinBase database (Fiehn et al., 2005). Peak areas were normalized by calculating the sum area of all identified compounds for each and subsequently dividing all data associated with a sample by the corresponding metabolite sum. The resulting data were multiplied by a constant factor in order to obtain values without decimal places and, the peak intensities were log2 transformed to remove the heteroscedasticity (Fiehn et al., 2008). Hierarchical clustering with multiscale bootstrapping (1,000 times) of all the samples based on normalized data was performed using PVCLUST (Suzuki and Shimodaira, 2006).
Expression data can be found in the GEO repository under accession number GSE266880.
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
CS: Data curation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. BW: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Writing – original draft, Writing – review & editing. RG: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. PD: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. JS: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
We would like to thank Oliver Fiehn and Kristie Cloos for their help with the initial metabolomics experiment.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1387498/full#supplementary-material
Abramoff, M. D., Magelhaes, P. J., and Ram, S. J. (2004). Image processing with ImageJ. Biophoton. Int. 11, 36–42.
Agbor, T. A., and McCormick, B. A. (2011). Salmonella effectors: important players modulating host cell function during infection. Cell. Microbiol. 13, 1858–1869. doi: 10.1111/j.1462-5822.2011.01701.x
Arabyan, N., Park, D., Foutouhi, S., Weis, A. M., Huang, B. C., Williams, C. C., et al. (2016). Salmonella degrades the host Glycocalyx leading to altered infection and glycan remodeling. Sci. Rep. 6:29525. doi: 10.1038/srep29525
Arboleya, S., Bottacini, F., O’Connell-Motherway, M., Ryan, C. A., Ross, R. P., van Sinderen, D., et al. (2018). Gene-trait matching across the Bifidobacterium longum pan-genome reveals considerable diversity in carbohydrate catabolism among human infant strains. BMC Genomics 19:33. doi: 10.1186/s12864-017-4388-9
Ashida, H., Mimuro, H., Ogawa, M., Kobayashi, T., Sanada, T., Kim, M., et al. (2011). Cell death and infection: a double-edged sword for host and pathogen survival. J. Cell Biol. 195, 931–942. doi: 10.1083/jcb.201108081
Ashida, H., Suzuki, T., and Sasakawa, C. (2021). Shigella infection and host cell death: a double-edged sword for the host and pathogen survival. Curr. Opin. Microbiol. 59, 1–7. doi: 10.1016/j.mib.2020.07.007
Barboza, M., Pinzon, J., Wickramasinghe, S., Froehlich, J. W., Moeller, I., Smilowitz, J. T., et al. (2012). Glycosylation of human milk lactoferrin exhibits dynamic changes during early lactation enhancing its role in pathogenic bacteria-host interactions. Mol. Cell. Proteomics 11:M111.015248. doi: 10.1074/mcp.M111.015248
Brentnall, M., Rodriguez-Menocal, L., de Guevara, R. L., Cepero, E., and Boise, L. H. (2013). Caspase-9, caspase-3 and caspase-7 have distinct roles during intrinsic apoptosis. BMC Cell Biol. 14:32. doi: 10.1186/1471-2121-14-32
Cacho, N. T., and Lawrence, R. M. (2017). Innate immunity and breast Milk. Front. Immunol. 8:584. doi: 10.3389/fimmu.2017.00584
Cao, X., Kambe, F., Lu, X., Kobayashi, N., Ohmori, S., and Seo, H. (2005). Glutathionylation of two cysteine residues in paired domain regulates DNA binding activity of Pax-8. J. Biol. Chem. 280, 25901–25906. doi: 10.1074/jbc.M411443200
Caspi, R., Foerster, H., Fulcher, C. A., Kaipa, P., Krummenacker, M., Latendresse, M., et al. (2008). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 36, D623–D631. doi: 10.1093/nar/gkm900
CDC (2008). Salmonella surveillance: Annual Summary,2006, in US Department of Health and Human Services CDC, Atlanta, Georgia.
Chen, P., den Bakker, H. C., Korlach, J., Kong, N., Storey, D. B., Paxinos, E. E., et al. (2017). Comparative genomics reveals the diversity of restriction-modification systems and DNA methylation sites in Listeria monocytogenes. Appl. Environ. Microbiol. 83:91. doi: 10.1128/AEM.02091-16
Chen, S. L., Jian, L., and Lang, H. Q. (2003). Optimization of peroxynitrite-luminol chemiluminescence system for detecting peroxynitrite in cell culture solution exposed to carbon disulphide. Luminescence 18, 249–253. doi: 10.1002/bio.734
Das, P., Lahiri, A., Lahiri, A., Sen, M., Iyer, N., Kapoor, N., et al. (2010). Cationic amino acid transporters and Salmonella Typhimurium ArgT collectively regulate arginine availability towards intracellular Salmonella growth. PLoS One 5:e15466. doi: 10.1371/journal.pone.0015466
de LeBlanc Ade, M., Castillo, N. A., and Perdigon, G. (2010). Anti-infective mechanisms induced by a probiotic Lactobacillus strain against Salmonella enterica serovar typhimurium infection. Int. J. Food Microbiol. 138, 223–231. doi: 10.1016/j.ijfoodmicro.2010.01.020
Desai, P. T. (2011). Molecular interactions of Salmonella with the host epithelium in presence of commensals. Logan, UT: Utah State University
Desai, P. T., Walsh, M. K., and Weimer, B. C. (2008). Solid-phase capture of pathogenic bacteria by using gangliosides and detection with real-time PCR. Appl. Environ. Microbiol. 74, 2254–2258. doi: 10.1128/AEM.02601-07
di Gioia, D., Aloisio, I., Mazzola, G., and Biavati, B. (2014). Bifidobacteria: their impact on gut microbiota composition and their applications as probiotics in infants. Appl. Microbiol. Biotechnol. 98, 563–577. doi: 10.1007/s00253-013-5405-9
Díaz, R., Torres-Miranda, A., Orellana, G., and Garrido, D. (2021). Comparative genomic analysis of novel Bifidobacterium longum subsp. longum strains reveals functional divergence in the human gut microbiota. Microorganisms 9:906. doi: 10.3390/microorganisms9091906
Ducarmon, Q. R., Zwittink, R. D., Hornung, B. V. H., van Schaik, W., Young, V. B., and Kuijper, E. J. (2019). Gut microbiota and colonization resistance against bacterial enteric infection. Microbiol. Mol. Biol. Rev. 83:19. doi: 10.1128/MMBR.00007-19
Eor, J. Y., Lee, C. S., Moon, S. H., Cheon, J. Y., Pathiraja, D., Park, B., et al. (2023). Effect of probiotic-fortified infant formula on infant gut health and microbiota modulation. Food Sci. Anim. Resour. 43, 659–673. doi: 10.5851/kosfa.2023.e26
Fan, W., Jin, S., Tong, T., Zhao, H., Fan, F., Antinore, M. J., et al. (2002). BRCA1 regulates GADD45 through its interactions with the OCT-1 and CAAT motifs. J. Biol. Chem. 277, 8061–8067. doi: 10.1074/jbc.M110225200
Fiehn, O., Wohlgemuth, G., and Scholz, M. (2005). “Setup and annotation of Metabolomic experiments by integrating biological and mass spectrometric metadata” in Data integration in the life sciences. eds. B. Ludäscher and L. Raschid (Berlin: Springer), 224–239.
Fiehn, O., Wohlgemuth, G., Scholz, M., Kind, T., Lee, D. Y., Lu, Y., et al. (2008). Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J. 53, 691–704. doi: 10.1111/j.1365-313X.2007.03387.x
Fink, S. L., and Cookson, B. T. (2007). Pyroptosis and host cell death responses during Salmonella infection. Cell. Microbiol. 9, 2562–2570. doi: 10.1111/j.1462-5822.2007.01036.x
Frese, S. A., Hutton, A. A., Contreras, L. N., Shaw, C. A., Palumbo, M. C., Casaburi, G., et al. (2017). Persistence of supplemented Bifidobacterium longum subsp. infantis EVC001 in breastfed infants. mSphere 2:501. doi: 10.1128/mSphere.00501-17
Garcia-Vallve, S., Guzman, E., Montero, M. A., and Romeu, A. (2003). HGT-DB: a database of putative horizontally transferred genes in prokaryotic complete genomes. Nucleic Acids Res. 31, 187–189. doi: 10.1093/nar/gkg004
Gareau, M. G., Sherman, P. M., and Walker, W. A. (2010). Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 7, 503–514. doi: 10.1038/nrgastro.2010.117
Gart, E. V., Suchodolski, J. S., Welsh, T. H. Jr., Alaniz, R. C., Randel, R. D., and Lawhon, S. D. (2016). Salmonella typhimurium and multidirectional communication in the gut. Front. Microbiol. 7:1827. doi: 10.3389/fmicb.2016.01827
Ghorashi, A. C., and Kohler, J. J. (2020). Not all quiet on the sugar front: glycan combatants in host-pathogen interactions. Biochemistry 59, 3061–3063. doi: 10.1021/acs.biochem.9b00524
Gong, B., Li, H., Feng, Y., Zeng, S., Zhuo, Z., Luo, J., et al. (2022). Prevalence, serotype distribution and antimicrobial resistance of non-Typhoidal Salmonella in hospitalized patients in Conghua District of Guangzhou, China. Front. Cell Infect. Microbiol. 12:805384. doi: 10.3389/fcimb.2022.805384
Goudenège, D., Avner, S., Lucchetti-Miganeh, C., and Barloy-Hubler, F. (2010). CoBaltDB: complete bacterial and archaeal orfeomes subcellular localization database and associated resources. BMC Microbiol. 10:88. doi: 10.1186/1471-2180-10-88
Gupta, S. K., Sharma, P., McMillan, E. A., Jackson, C. R., Hiott, L. M., Woodley, T., et al. (2019). Genomic comparison of diverse Salmonella serovars isolated from swine. PLoS One 14:e0224518. doi: 10.1371/journal.pone.0224518
He, X., Mishchuk, D. O., Shah, J., Weimer, B. C., and Slupsky, C. M. (2013). Cross-talk between E. coli strains and a human colorectal adenocarcinoma-derived cell line. Sci. Rep. 3:3416. doi: 10.1038/srep03416
Hefele, M., Stolzer, I., Ruder, B., He, G. W., Mahapatro, M., Wirtz, S., et al. (2018). Intestinal epithelial Caspase-8 signaling is essential to prevent necroptosis during Salmonella Typhimurium induced enteritis. Mucosal Immunol. 11, 1191–1202. doi: 10.1038/s41385-018-0011-x
Heithoff, D. M., Shimp, W. R., House, J. K., Xie, Y., Weimer, B. C., Sinsheimer, R. L., et al. (2012). Intraspecies variation in the emergence of hyperinfectious bacterial strains in nature. PLoS Pathog. 8:e1002647. doi: 10.1371/journal.ppat.1002647
Henrick, B. M., Rodriguez, L., Lakshmikanth, T., Pou, C., Henckel, E., Arzoomand, A., et al. (2021). Bifidobacteria-mediated immune system imprinting early in life. Cell 184, 3884–3898.e11. doi: 10.1016/j.cell.2021.05.030
Herrero-Fresno, A., and Olsen, J. E. (2018). Salmonella Typhimurium metabolism affects virulence in the host – a mini-review. Food Microbiol. 71, 98–110. doi: 10.1016/j.fm.2017.04.016
Hu, W., Zhao, J., Wang, J., Yu, T., Wang, J., and Li, N. (2012). Transgenic milk containing recombinant human lactoferrin modulates the intestinal flora in piglets. Biochem. Cell Biol. 90, 485–496. doi: 10.1139/o2012-003
Huang, F. C., and Huang, S. C. (2021). The cooperation of Bifidobacterium longum and active vitamin D3 on innate immunity in Salmonella colitis mice via vitamin D receptor. Microorganisms 9:1804. doi: 10.3390/microorganisms9091804
Hume, P. J., Singh, V., Davidson, A. C., and Koronakis, V. (2017). Swiss Army pathogen: the Salmonella entry toolkit. Front. Cell. Infect. Microbiol. 7:348. doi: 10.3389/fcimb.2017.00348
Ilyas, B., Tsai, C. N., and Coombes, B. K. (2017). Evolution of Salmonella-host cell interactions through a dynamic bacterial genome. Front. Cell. Infect. Microbiol. 7:428. doi: 10.3389/fcimb.2017.00428
Indira, M., Venkateswarulu, T. C., Abraham Peele, K., Nazneen Bobby, M., and Krupanidhi, S. (2019). Bioactive molecules of probiotic bacteria and their mechanism of action: a review. 3 Biotech 9:306. doi: 10.1007/s13205-019-1841-2
Iqbal, Z., Ahmed, S., Tabassum, N., Bhattacharya, R., and Bose, D. (2021). Role of probiotics in prevention and treatment of enteric infections: a comprehensive review. 3 Biotech 11:242. doi: 10.1007/s13205-021-02796-7
Irizarry, R. A., Hobbs, B., Collin, F., Beazer-Barclay, Y. D., Antonellis, K. J., Scherf, U., et al. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. doi: 10.1093/biostatistics/4.2.249
Johannessen, M., Delghandi, M. P., and Moens, U. (2004). What turns CREB on? Cell. Signal. 16, 1211–1227. doi: 10.1016/j.cellsig.2004.05.001
Kanehisa, M., Goto, S., Kawashima, S., and Nakaya, A. (2002). The KEGG databases at GenomeNet. Nucleic Acids Res. 30, 42–46. doi: 10.1093/nar/30.1.42
Kasumba, I. N., Pulford, C. V., Perez-Sepulveda, B. M., Sen, S., Sayed, N., Permala-Booth, J., et al. (2021). Characteristics of Salmonella recovered from stools of children enrolled in the global enteric multicenter study. Clin. Infect. Dis. 73, 631–641. doi: 10.1093/cid/ciab051
Kendall, M. M., and Sperandio, V. (2007). Quorum sensing by enteric pathogens. Curr. Opin. Gastroenterol. 23, 10–15. doi: 10.1097/MOG.0b013e3280118289
Kimata, H. (1994). GM1, a ganglioside that specifically enhances immunoglobulin production and proliferation in human plasma cells. Eur. J. Immunol. 24, 2910–2913. doi: 10.1002/eji.1830241149
Kotloff, K. L. (2022). Bacterial diarrhoea. Curr. Opin. Pediatr. 34, 147–155. doi: 10.1097/MOP.0000000000001107
Kotloff, K. L., Nataro, J. P., Blackwelder, W. C., Nasrin, D., Farag, T. H., Panchalingam, S., et al. (2013). Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the global enteric multicenter study, GEMS): a prospective, case-control study. Lancet 382, 209–222. doi: 10.1016/S0140-6736(13)60844-2
Kulkarni, R. R., Gaghan, C., Gorrell, K., Sharif, S., and Taha-Abdelaziz, K. (2022). Probiotics as alternatives to antibiotics for the prevention and control of necrotic enteritis in chickens. Pathogens 11:692. doi: 10.3390/pathogens11060692
Kutoh, E., Stromstedt, P. E., and Poellinger, L. (1992). Functional interference between the ubiquitous and constitutive octamer transcription factor 1 (OTF-1) and the glucocorticoid receptor by direct protein-protein interaction involving the homeo subdomain of OTF-1. Mol. Cell. Biol. 12, 4960–4969.
Laforest-Lapointe, I., and Arrieta, M. C. (2017). Patterns of early-life gut microbial colonization during human immune development: an ecological perspective. Front. Immunol. 8:788. doi: 10.3389/fimmu.2017.00788
Lawson, M. A. E., O’Neill, I. J., Kujawska, M., Gowrinadh Javvadi, S., Wijeyesekera, A., Flegg, Z., et al. (2020). Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 14, 635–648. doi: 10.1038/s41396-019-0553-2
Lebeer, S., Vanderleyden, J., and De Keersmaecker, S. C. (2008). Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev. 72, 728–764. doi: 10.1128/MMBR.00017-08
Lee, S. J., Jung, Y. H., Oh, S. Y., Song, E. J., Choi, S. H., and Han, H. J. (2015). Vibrio vulnificus VvhA induces NF-kappaB-dependent mitochondrial cell death via lipid raft-mediated ROS production in intestinal epithelial cells. Cell Death Dis. 6:1655. doi: 10.1038/cddis.2015.19
Lin, B., Qing, X., Liao, J., and Zhuo, K. (2020). Role of protein glycosylation in host-pathogen interaction. Cells 9:1022. doi: 10.3390/cells9041022
LoCascio, R. G., Desai, P., Sela, D. A., Weimer, B., and Mills, D. A. (2010). Broad conservation of Milk utilization genes Inbifidobacterium longumsubsp.Infantisas revealed by comparative genomic hybridization. Appl. Environ. Microbiol. 76, 7373–7381. doi: 10.1128/AEM.00675-10
Lueschow, S. R., Boly, T. J., Frese, S. A., Casaburi, G., Mitchell, R. D., Henrick, B. M., et al. (2022). Bifidobacterium longum subspecies infantis strain EVC001 decreases neonatal murine necrotizing enterocolitis. Nutrients 14:495. doi: 10.3390/nu14030495
Lundberg, J. O., Weitzberg, E., and Gladwin, M. T. (2008). The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 7, 156–167. doi: 10.1038/nrd2466
Majowicz, S. E., Musto, J., Scallan, E., Angulo, F. J., Kirk, M., O’Brien, S. J., et al. (2010). The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50, 882–889. doi: 10.1086/650733
Man, S. M., Tourlomousis, P., Hopkins, L., Monie, T. P., Fitzgerald, K. A., and Bryant, C. E. (2013). Salmonella infection induces recruitment of Caspase-8 to the inflammasome to modulate IL-1beta production. J. Immunol. 191, 5239–5246. doi: 10.4049/jimmunol.1301581
Mantovani, A. (2010). Molecular pathways linking inflammation and cancer. Curr. Mol. Med. 10, 369–373. doi: 10.2174/156652410791316968
Marcobal, A., Barboza, M., Sonnenburg, E. D., Pudlo, N., Martens, E. C., Desai, P., et al. (2011). Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 10, 507–514. doi: 10.1016/j.chom.2011.10.007
Markowitz, V. M., Chen, I. M. A., Palaniappan, K., Chu, K., Szeto, E., Grechkin, Y., et al. (2010). The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 38, D382–D390. doi: 10.1093/nar/gkp887
Mercer, E. M., and Arrieta, M. C. (2023). Probiotics to improve the gut microbiome in premature infants: are we there yet? Gut Microbes 15:2201160. doi: 10.1080/19490976.2023.2201160
Merighi, M., Septer, A. N., Carroll-Portillo, A., Bhatiya, A., Porwollik, S., McClelland, M., et al. (2009). Genome-wide analysis of the PreA/PreB (QseB/QseC) regulon of Salmonella enterica serovar typhimurium. BMC Microbiol. 9:42. doi: 10.1186/1471-2180-9-42
Michl, P., Knobel, B., and Downward, J. (2006). CUTL1 is phosphorylated by protein kinase a, modulating its effects on cell proliferation and motility. J. Biol. Chem. 281, 15138–15144. doi: 10.1074/jbc.M600908200
Mills, S., Yang, B., Smith, G. J., Stanton, C., and Ross, R. P. (2023). Efficacy of Bifidobacterium longum alone or in multi-strain probiotic formulations during early life and beyond. Gut Microbes 15:2186098. doi: 10.1080/19490976.2023.2186098
Mootha, V. K., Lindgren, C. M., Eriksson, K. F., Subramanian, A., Sihag, S., Lehar, J., et al. (2003). PGC-1[alpha]-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273. doi: 10.1038/ng1180
Mou, Y., du, Y., Zhou, L., Yue, J., Hu, X., Liu, Y., et al. (2022). Gut microbiota interact with the brain through systemic chronic inflammation: implications on Neuroinflammation, neurodegeneration, and aging. Front. Immunol. 13:796288. doi: 10.3389/fimmu.2022.796288
Mueller, N. T., Bakacs, E., Combellick, J., Grigoryan, Z., and Dominguez-Bello, M. G. (2015). The infant microbiome development: mom matters. Trends Mol. Med. 21, 109–117. doi: 10.1016/j.molmed.2014.12.002
Nair, D., Venkitanarayanan, K., and Kollanoor Johny, A. (2018). Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Food Secur. 7:167. doi: 10.3390/foods7100167
NARMS. (2023). FSIS NARMS Multi-Year Report — 2014–2019. Department of Agriculture Food Safety and Inspection Service: Washington, DC p. 1–36
Numata, A., Shimoda, K., Kamezaki, K., Haro, T., Kakumitsu, H., Shide, K., et al. (2005). Signal transducers and activators of transcription 3 augments the transcriptional activity of CCAAT/enhancer-binding protein alpha in granulocyte colony-stimulating factor signaling pathway. J. Biol. Chem. 280, 12621–12629. doi: 10.1074/jbc.M408442200
O'Shea, J. J. (1997). Jaks, STATs, cytokine signal transduction, and immunoregulation: are we there yet? Immunity 7, 1–11. doi: 10.1016/S1074-7613(00)80505-1
Ouwehand, A. C., and Salminen, S. (2003). In vitro adhesion assays for probiotics and their in vivo relevance: a review. Microb. Ecol. Health Dis. 15, 175–184. doi: 10.1080/08910600310019886
Park, J. Y., Abekura, F., and Cho, S. H. (2023). GM1a ganglioside-binding domain peptide inhibits host adhesion and inflammatory response of enterotoxigenic Escherichia coli heat-labile enterotoxin-B in HCT-8 cells. Sci. Rep. 13:16835. doi: 10.1038/s41598-023-44220-5
Park, D., Arabyan, N., Williams, C. C., Song, T., Mitra, A., Weimer, B. C., et al. (2016). Salmonella Typhimurium enzymatically landscapes the host intestinal epithelial cell (IEC) surface Glycome to increase invasion. Mol. Cell. Proteomics 15, 3653–3664. doi: 10.1074/mcp.M116.063206
Park, B. H., Karpinets, T. V., Syed, M. H., Leuze, M. R., and Uberbacher, E. C. (2010). CAZymes analysis toolkit (CAT): web-service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology 20, 1574–1584. doi: 10.1093/glycob/cwq106
Paton, A. W., Morona, R., and Paton, J. C. (2006). Designer probiotics for prevention of enteric infections. Nat. Rev. Microbiol. 4, 193–200. doi: 10.1038/nrmicro1349
Peterson, J. D., Umayam, L. A., Dickinson, T., Hickey, E. K., and White, O. (2001). The comprehensive microbial resource. Nucleic Acids Res. 29, 123–125. doi: 10.1093/nar/29.1.123
Plitnick, J., Chevance, F. F. V., Stringer, A., Hughes, K. T., and Wade, J. T. (2021). Regulatory cross talk between motility and Interbacterial communication in Salmonella enterica Serovar typhimurium. J. Bacteriol. 203:510. doi: 10.1128/JB.00510-20
Pudlo, N. A., Urs, K., Kumar, S. S., German, J. B., Mills, D. A., and Martens, E. C. (2015). Symbiotic human gut Bacteria with variable metabolic priorities for host mucosal Glycans. MBio 6, e01282–e01215. doi: 10.1128/mBio.01282-15
Radomska, H. S., Bassères, D. S., Zheng, R., Zhang, P., Dayaram, T., Yamamoto, Y., et al. (2006). Block of C/EBP alpha function by phosphorylation in acute myeloid leukemia with FLT3 activating mutations. J. Exp. Med. 203, 371–381. doi: 10.1084/jem.20052242
Reading, N. C., Rasko, D. A., Torres, A. G., and Sperandio, V. (2009). The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 106, 5889–5894. doi: 10.1073/pnas.0811409106
Rudel, T., Kepp, O., and Kozjak-Pavlovic, V. (2010). Interactions between bacterial pathogens and mitochondrial cell death pathways. Nat. Rev. Microbiol. 8, 693–705. doi: 10.1038/nrmicro2421
Ruiz, P. A., Hoffmann, M., Szcesny, S., Blaut, M., and Haller, D. (2005). Innate mechanisms for Bifidobacterium lactis to activate transient pro-inflammatory host responses in intestinal epithelial cells after the colonization of germ-free rats. Immunology 115, 441–450. doi: 10.1111/j.1365-2567.2005.02176.x
Salerno-Gonçalves, R., Galen, J. E., Levine, M. M., Fasano, A., and Sztein, M. B. (2018). Manipulation of Salmonella Typhi gene expression impacts innate cell responses in the human intestinal mucosa. Front. Immunol. 9:2543. doi: 10.3389/fimmu.2018.02543
Salerno-Goncalves, R., Kayastha, D., Fasano, A., Levine, M. M., and Sztein, M. B. (2019). Crosstalk between leukocytes triggers differential immune responses against Salmonella enterica serovars Typhi and Paratyphi. PLoS Negl. Trop. Dis. 13:e0007650. doi: 10.1371/journal.pntd.0007650
Secher, T., Brehin, C., and Oswald, E. (2016). Early settlers: which E. coli strains do you not want at birth? Am. J. Physiol. Gastrointest. Liver Physiol. 311, G123–G129. doi: 10.1152/ajpgi.00091.2016
Shah, J., Desai, P. T., and Weimer, B. C. (2014). Genetic mechanisms underlying the pathogenicity of cold-stressed Salmonella enterica serovar typhimurium in cultured intestinal epithelial cells. Appl. Environ. Microbiol. 80, 6943–6953. doi: 10.1128/AEM.01994-14
Shaw, C., Hess, M., and Weimer, B. C. (2022). Two-component systems regulate bacterial virulence in response to the host gastrointestinal environment and metabolic cues. Virulence 13, 1666–1680. doi: 10.1080/21505594.2022.2127196
Shkalim, V., Amir, A., Samra, Z., and Amir, J. (2012). Characteristics of non-typhi Salmonella gastroenteritis associated with bacteremia in infants and young children. Infection 40, 285–289. doi: 10.1007/s15010-011-0231-4
Shu, Q., Lin, H., Rutherfurd, K. J., Fenwick, S. G., Prasad, J., Gopal, P. K., et al. (2000). Dietary Bifidobacterium lactis (HN019) enhances resistance to oral Salmonella Typhimurium infection in mice. Microbiol. Immunol. 44, 213–222. doi: 10.1111/j.1348-0421.2000.tb02486.x
Silva, A. M., Barbosa, F. H. F., Duarte, R., Vieira, L. Q., Arantes, R. M. E., and Nicoli, J. R. (2004). Effect of Bifidobacterium longum ingestion on experimental salmonellosis in mice. J. Appl. Microbiol. 97, 29–37. doi: 10.1111/j.1365-2672.2004.02265.x
Splichalova, A., Kindlova, Z., Killer, J., Neuzil Bunesova, V., Vlkova, E., Valaskova, B., et al. (2023). Commensal Bacteria impact on intestinal toll-like receptor signaling in Salmonella-challenged Gnotobiotic piglets. Pathogens 12:1293. doi: 10.3390/pathogens12111293
Stevens, J. R., Desai, P., Rajan, S., and Weimer, B. C. (2008). Statistical issues in the Normalization of multi-species microarray data. Conference on Applied Statistics in Agriculture.
Stewart, C. J., Ajami, N. J., O’Brien, J. L., Hutchinson, D. S., Smith, D. P., Wong, M. C., et al. (2018). Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562, 583–588. doi: 10.1038/s41586-018-0617-x
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. doi: 10.1073/pnas.0506580102
Suzuki, R., and Shimodaira, H. (2006). Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542. doi: 10.1093/bioinformatics/btl117
Tan, S., Chen, C., Zhao, A., Wang, M., Zhao, W., Zhang, J., et al. (2020). The dynamic changes of gangliosides in breast milk and the intake of gangliosides in maternal and infant diet in three cities of China. Int. J. Clin. Exp. Pathol. 13, 2870–2888.
Tatusov, R. L., Koonin, E. V., and Lipman, D. J. (1997). A genomic perspective on protein families. Science 278, 631–637. doi: 10.1126/science.278.5338.631
Taylor, S. J., and Winter, S. E. (2020). Salmonella finds a way: metabolic versatility of Salmonella enterica serovar typhimurium in diverse host environments. PLoS Pathog. 16:e1008540. doi: 10.1371/journal.ppat.1008540
Thompson, J. A., Oliveira, R. A., and Xavier, K. B. (2016). Chemical conversations in the gut microbiota. Gut Microbes 7, 163–170. doi: 10.1080/19490976.2016.1145374
Thorman, A. W., Adkins, G., Conrey, S. C., Burrell, A. R., Yu, Y., White, B., et al. (2023). Gut microbiome composition and metabolic capacity differ by FUT2 secretor status in exclusively breastfed infants. Nutrients 15:471. doi: 10.3390/nu15020471
Turroni, F., Bottacini, F., Foroni, E., Mulder, I., Kim, J. H., Zomer, A., et al. (2010). Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. USA 107, 19514–19519. doi: 10.1073/pnas.1011100107
Tusher, V. G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121. doi: 10.1073/pnas.091062498
Underwood, M. A., German, J. B., Lebrilla, C. B., and Mills, D. A. (2015). Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut. Pediatr. Res. 77, 229–235. doi: 10.1038/pr.2014.156
van Puyvelde, S., de Block, T., Sridhar, S., Bawn, M., Kingsley, R. A., Ingelbeen, B., et al. (2023). A genomic appraisal of invasive Salmonella typhimurium and associated antibiotic resistance in sub-Saharan Africa. Nat. Commun. 14:6392. doi: 10.1038/s41467-023-41152-6
Voreades, N., Kozil, A., and Weir, T. L. (2014). Diet and the development of the human intestinal microbiome. Front. Microbiol. 5:494. doi: 10.3389/fmicb.2014.00494
Warren, P., Taylor, D., Martini, P. G. V., Jackson, J., and Bienkowska, J.. (2007). PANP – a new method of gene detection on oligonucleotide expression arrays. In bioinformatics and bioengineering, 2007. BIBE 2007. Proceedings of the 7th IEEE international conference on.
Wellcome Trust Case Control Consortium (2007). Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678. doi: 10.1038/nature05911
Winter, S. E., Thiennimitr, P., Winter, M. G., Butler, B. P., Huseby, D. L., Crawford, R. W., et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429. doi: 10.1038/nature09415
Wu, G., Meininger, C. J., McNeal, C. J., Bazer, F. W., and Rhoads, J. M. (2021). Role of L-arginine in nitric oxide synthesis and health in humans. Adv. Exp. Med. Biol. 1332, 167–187. doi: 10.1007/978-3-030-74180-8_10
Yang, J., Chen, L., Sun, L., Yu, J., and Jin, Q. (2008). VFDB 2008 release: an enhanced web-based resource for comparative pathogenomics. Nucleic Acids Res. 36, D539–D542. doi: 10.1093/nar/gkm951
Ye, Z., Petrof, E. O., Boone, D., Claud, E. C., and Sun, J. (2007). Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am. J. Pathol. 171, 882–892. doi: 10.2353/ajpath.2007.070220
Yu, Z. T., Chen, C., and Newburg, D. S. (2013). Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology 23, 1281–1292. doi: 10.1093/glycob/cwt065
Keywords: probiotic, arginine, nitric oxide, host-microbe association, Salmonella, bifidobacteria
Citation: Shaw C, Weimer BC, Gann R, Desai PT and Shah JD (2024) The Yin and Yang of pathogens and probiotics: interplay between Salmonella enterica sv. Typhimurium and Bifidobacterium infantis during co-infection. Front. Microbiol. 15:1387498. doi: 10.3389/fmicb.2024.1387498
Received: 17 February 2024; Accepted: 12 April 2024;
Published: 15 May 2024.
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Vasco Ariston De Carvalho Azevedo, Federal University of Minas Gerais, BrazilCopyright © 2024 Shaw, Weimer, Gann, Desai and Shah. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bart C. Weimer, YmN3ZWltZXJAdWNkYXZpcy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.