- 1Honeybee Research Institute, Jiangxi Agricultural University, Nanchang, China
- 2Department of Integrative Biology, The University of Texas at Austin, Austin, TX, United States
- 3Environment and Plant Protection Institute, Chinese Academy of Tropical Agricultural Sciences, Haikou, China
- 4USDA, Beltsville Agricultural Research Center, Bee Research Laboratory, Agricultural Research Service, Beltsville, MD, United States
Invasive pests may disturb and destructively reformat the local ecosystem. The small hive beetle (SHB), Aethina tumida, originated in Africa and has expanded to America, Australia, Europe, and Asia. A key factor facilitating its fast global expansion is its ability to subsist on diverse food inside and outside honey bee colonies. SHBs feed on various plant fruits and exudates in the environment while searching for bee hives. After sneaking into a bee hive, they switch their diet to honey, pollen, and bee larvae. How SHBs survive on such a broad range of food remains unclear. In this study, we simulated the outside and within hive stages by providing banana and hive resources and quantified the SHB associated microbes adjusted by the diet. We found that SHBs fed on bananas were colonized by microbes coding more carbohydrate-active enzymes and a higher alpha diversity than communities from SHBs feeding on hive products or those collected directly from bee hives. SHBs fed on bananas and those collected from the hive showed high symbiont variance, indicated by the beta diversity. Surprisingly, we found the honey bee core symbiont Snodgrassella alvi in the guts of SHBs collected in bee hives. To determine the role of S. alvi in SHB biology, we inoculated SHBs with a genetically tagged culture of S. alvi, showing that this symbiont is a likely transient of SHBs. In contrast, the fungus Kodamaea ohmeri is the primary commensal of SHBs. Diet-based microbiome shifts are likely to play a key role in the spread and success of SHBs.
Introduction
Invasive species are organisms introduced to a new habitat where they are not known to occur. In new habitats, these species thrive and continue to expand their territory (Rohner and Moczek, 2020). Invasive species compete for limited food and shelter resources and parasitize or prey upon local species, causing ecological and economic damage. Invasive species are predicted to have caused 25% of plant extinction and 33% of animal extinction events (Blackburn et al., 2019; Angulo et al., 2022). To which extent species can explore local food resources determines the success of invasions. Species with broad food breadth can have competitive advantages and better pathogenic bacteria tolerance (Barthel et al., 2014; Machovsky-Capuska et al., 2016).
The small hive beetle (Aethina tumida, SHB) is a honey bee pest that originated in sub-Saharan Africa. In its native range, honey bees efficiently guard against SHBs, limiting their populations (Neumann et al., 2016; Ouessou Idrissou et al., 2019). Thus, the impact of SHBs on native honey bee colonies is minor. Following international trade lines, SHBs have moved out of Africa and expanded rapidly into novel habitats (Idrissou et al., 2019; Liu et al., 2021). During this expansion, SHBs were first reported in the USA in 1996. SHBs were further dispersed to Australia, Europe, and Asia over two decades (Hood, 2000; Gillespie et al., 2003; Palmeri et al., 2014; Cervancia et al., 2016). SHBs have been introduced in and out of America several times (Neumann et al., 2016). Kodamaea ohmeri is a commensal fungus of SHBs, fermenting the honey and eventually sliming the hive, presumably attracting other SHBs (Benda et al., 2008; Amos et al., 2018, 2019). SHBs also carry and disperse bee viruses, causing colony failure (Eyer et al., 2009). SHB infestation has caused substantial damage to the apicultural industry (Hood, 2000; Zhao et al., 2020). One reason for their rapid dispersal is that the SHBs can feed on diverse fruits and saps while searching for bee hives (Stuhl, 2021).
By colonizing the guts or specialized organs, symbionts improve the nutritional yields of flies, bees, aphids, and beetles (Reis et al., 2020; Li et al., 2022; Smith et al., 2022; Luo et al., 2023). In SHBs, neither a symbiont organ nor symbiont shifts in response to nutrition have been reported. Previously, we found distinctive microbes associated with SHB larvae and the co-occurrence of several known bee symbionts (Huang et al., 2019). We hypothesized that the ability to feed on a variety of plant products, including diverse fruits and saps, along with the specialized diet provided by honey bee colonies may require the assistance of gut symbionts. Further, this symbiont community might adjust in response to extreme diets (David et al., 2014). As SHBs may disperse through the banana trade line (Liu et al., 2021), we simulated the dispersal and within hive stages by feeding SHBs with banana (Banana group) and a mixture of bee pollen and honey (Bee_Bread group). We also directly collected SHBs from a honey bee hive (Wild group) to assess natural variation in SHB microbes (Supplementary Figure S1). Snodgrassella alvi is a core honey bee gut symbiont. This bacterium colonizes the hindgut, helping lipid metabolism (Quinn et al., 2024). To assess whether honey bee symbionts play a significant role in SHB health, we also used a genetically tagged S. alvi and measured the transit of this microbe in the SHB gut.
Results
Minor impact of diet on SHBs survival
We first investigated the impact of the banana and bee bread on beetle survival in the lab condition. After 3 weeks of rearing, 30 beetles survived in the Banana group, and 27 survived in the Bee_Bread group (Supplementary Tables S1), hence the variance of the banana and bread on SHBs survival was minor (Pearson’s Chi-squared test, df = 2, P = 0.516).
De novo assembly of metagenomes associated with SHBs
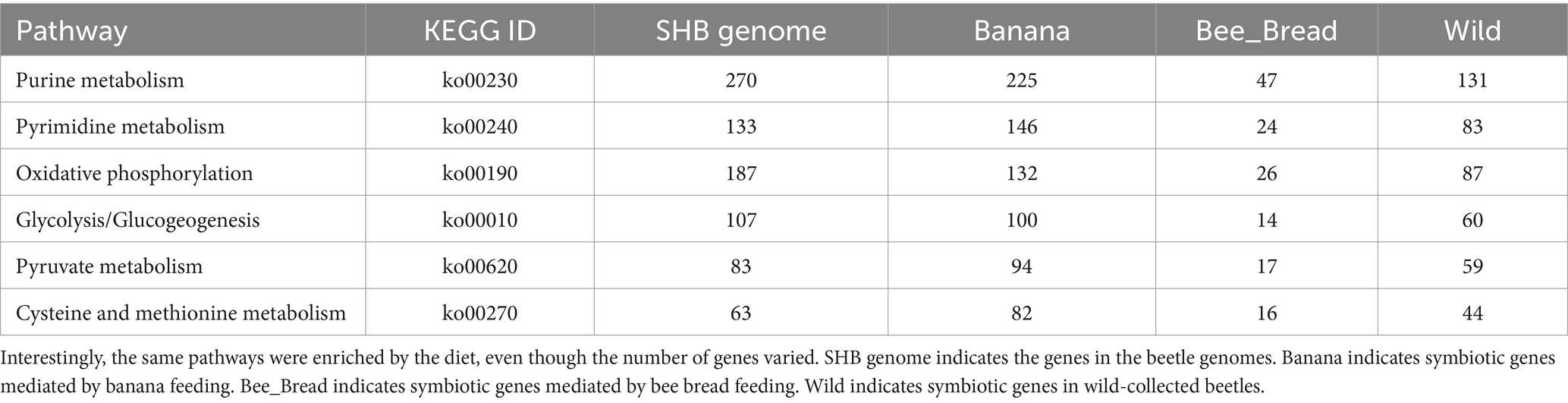
As the microbes associated with SHB are largely unknown, we then assembled the microbiome genomes. We randomly selected 10 SHBs in the Banana and Bee_Bread groups and eight SHBs collected in bee hive for metagenomic analyses to quantify the associated gene content and microbes. On average, 5.5 ± 1.3 million reads (150 bp per read) were assigned to microbes in each library, and the ratios of microbe to host reads did not differ significantly between libraries (t-test, P > 0.05, Supplementary Table S2). We assembled 196,214 metagenome contigs (Supplementary File S2). In those contigs, 436,918 genes were predicted, and 200,587 protein-coding genes retrieved functional annotation in KEGG (Supplementary File S3-S4; Figure 1). The symbionts showed a substantial number of genes involved in nucleotide, energy, carbohydrate, and amino acid metabolism. We also noticed that the symbionts harbor a few xenobiotics biodegradation genes, presumably to degrade toxic compounds (Table 1; Supplementary Table S3).
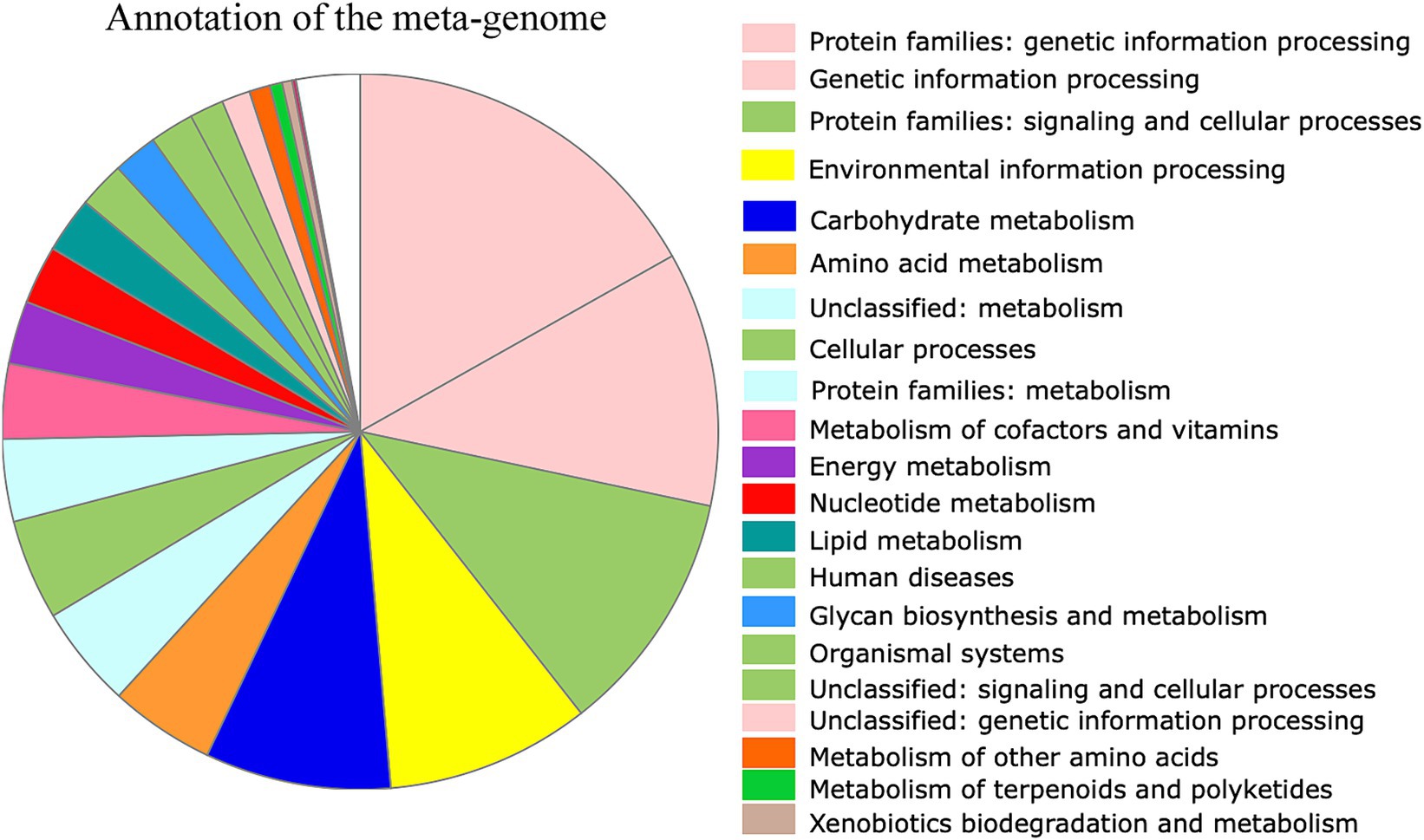
Figure 1. Functional annotation of predicted genes from the assembled metagenomic contigs. Overall, 436,918 genes were predicted, and 200,587 showed functional annotation. Among them, 11,650 genes were involved in carbohydrate metabolism, 6,583 in amino acid metabolism, and 3,471 in lipid metabolism. The color of the functional category was used in global pathway maps and genome maps of KEGG. The functional category on the right was indicated in the pie chart in clockwise order.
Banana feeding enhanced metabolic gene number
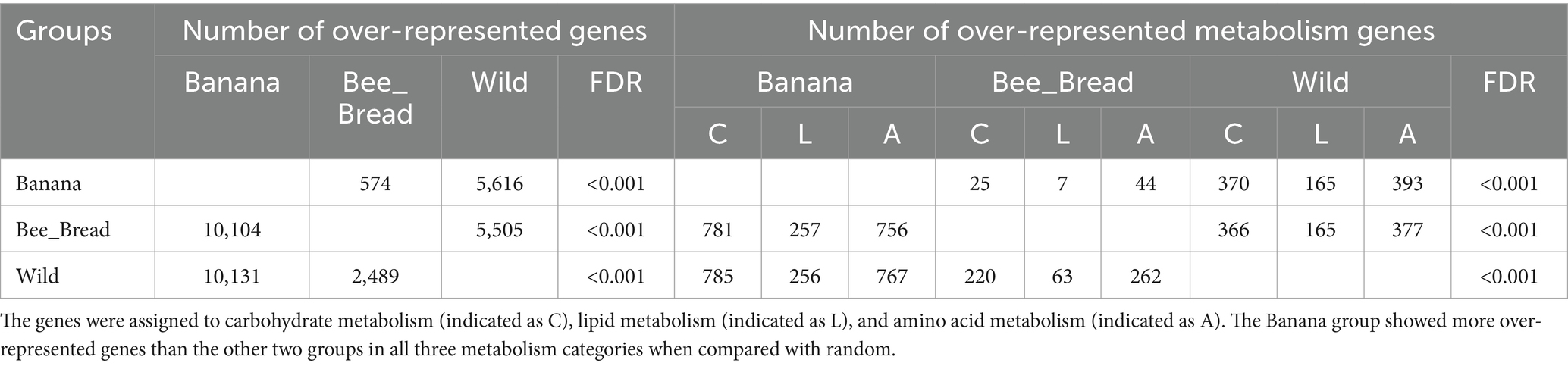
To quantify the gene copy number, we aligned the reads back to the metagenome. In a pairwise analysis, 15,748 genes were significantly differentially represented between the Banana and Wild groups, 10,678 genes between the Banana and Bee_Bread groups, and 7,994 genes between the Wild and Bee_Bread groups (Table 2; Supplementary Table S4). The Banana group showed a substantially higher number of over-represented genes than the Bee_bread and Wild groups in carbohydrate, lipid, and amino acid metabolism (Chi-squared test, FDR < 0.001). Among the differentially represented genes, 143 carbohydrate-active enzymes (CAZy) were over-represented in the Banana group, compared with 130 in the Wild group and 35 in the Bee_Bread group (Supplementary Table S5). The Banana group showed more CAZy than random among the three treatment groups (Pearson’s Chi-squared test, df = 2, p < 0.001).
High microbial diversity in banana feeding SHBs
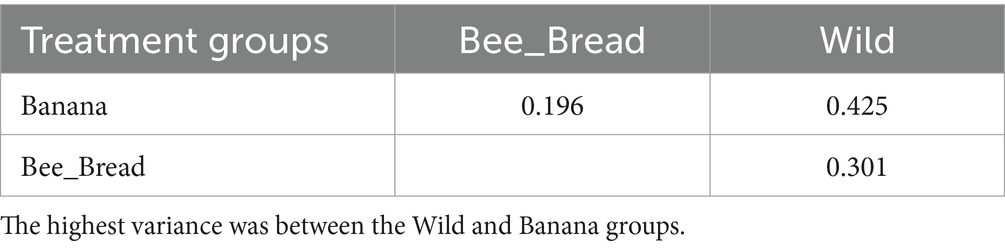
To determine microbial diversity, we aligned the microbial reads to the Kraken2 standard database (Supplementary Figure S2-S4). Despite high symbiont diversity overall, 13 bacterial genera dominated the microbial community (> 99% of relative abundance, Figure 2A). The diet separated the microbes and accounted for 87% of the variance (Figure 2B). The Banana group maintained 95 microbial species/strains, followed by 67 in the Bee_Bread group and 56 in the Wild group (Figure 3A). The Banana group showed the highest alpha diversity (2.1 ± 0.31), compared with the Bee_Bread group (1.83 ± 0.40) and the Hive group (1.70 ± 0.30; Kruskal-Wallis test, df = 2, P < 0.05, Figure 3B). We also compared the beta diversity between the paired groups. The Wild and Banana group showed the highest beta diversity (0.425), followed by 0.301 between the Wild and Bee_Bread groups. The Banana and Bee_Bread showed the lowest beta diversity of 0.196 (Table 3).
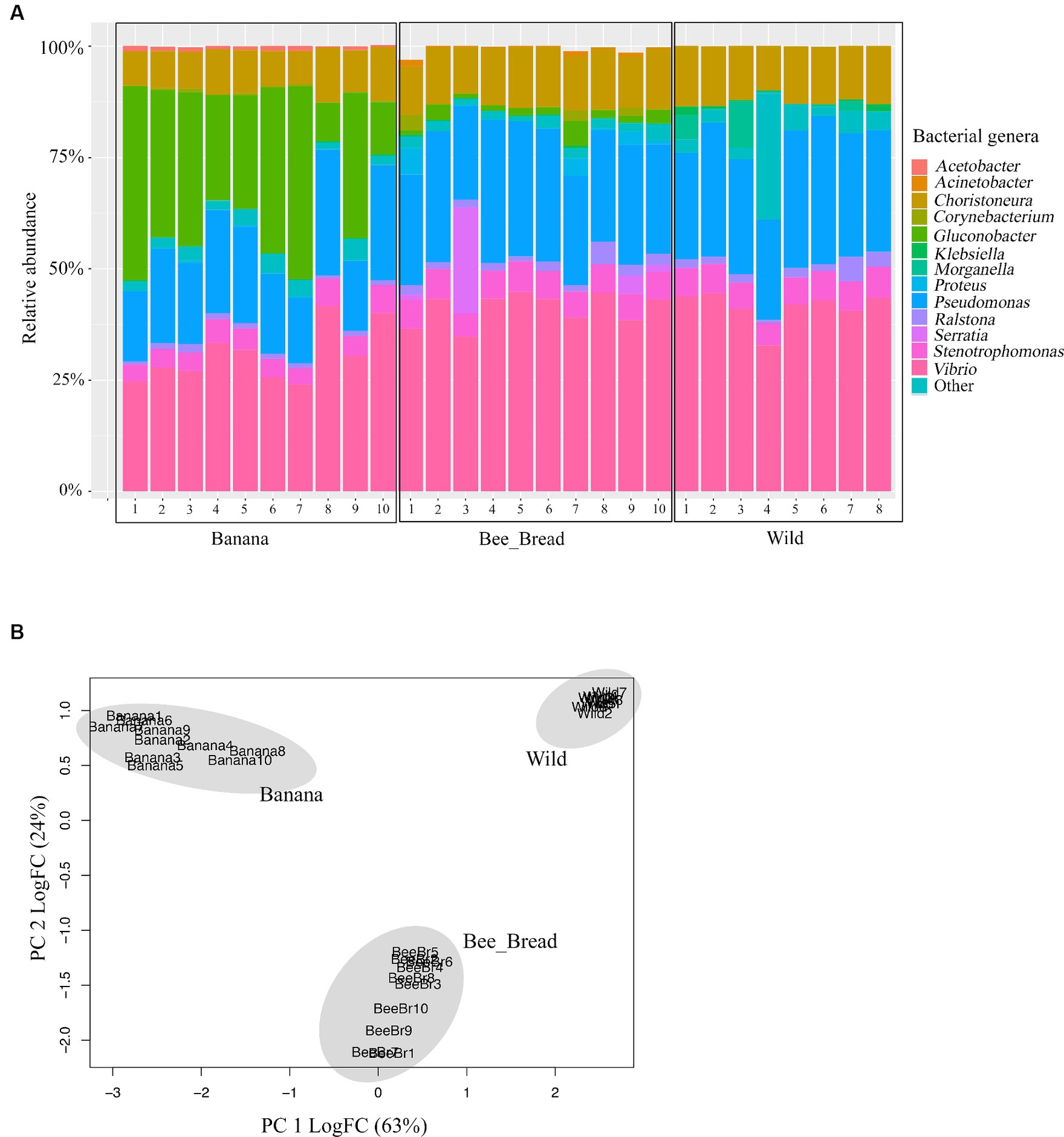
Figure 2. Genome sequencing-based community profiles of the bacteria mediated by the diet in the small hive beetles. (A) Relative abundance of the dominant bacterial genera. Banana indicates beetles fed on bananas; Bee_Bread indicates beetles fed on bee bread; Wild indicates beetles collected from bee hives. These bacterial genera were present across all beetle groups at >1% relative abundance. More rare genera (< 1%) were summed up as “Other.” (B) PCA plot of the beetles showing the impacts of diet on microbial composition. The microbes were separated by the diet.
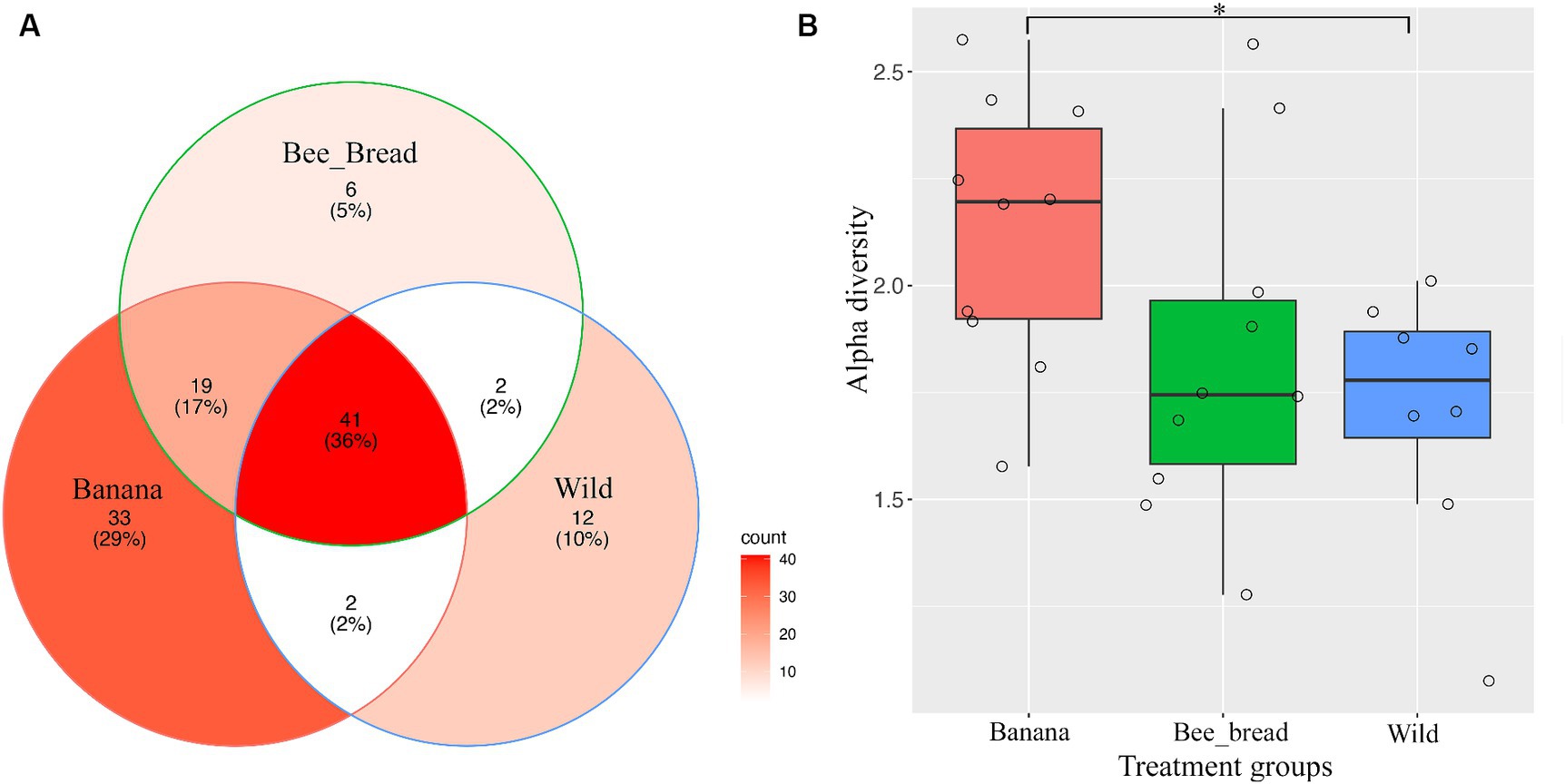
Figure 3. Metagenomic analysis of SHB symbionts. (A) Venn diagram of microbes found in the three groups. The banana showed the highest number of microbes and was enriched in acetic-acid bacteria. (B) Alpha diversity of the three groups. The banana group showed significantly higher alpha diversity than the Hive group (Wilcoxon rank sum exact test, p < 0.05).
We found plant-associated bacteria in the Banana group, such as Corynebacterium glyciniphilum, which was the same species initially isolated from the banana for fermentation. The Banana group was significantly enriched in the acetic acid bacteria. Microbes associated with fermentation (Gluconobacter albidus, Mammaliicoccus sciuri, Corynebacterium nuruki) were also highly enriched in the Bee_bread group. We found a few honey bee symbionts in the Wild group, including Lactococcus lactis and the core symbiont Snodgrassella alvi (Supplementary File S5-S7).
Honey bee symbiont in SHBs and an antibiotic-resistant commensal fungus
We further tested whether SHB can support the colonization of honey bee gut symbionts. We inoculated newly emerged beetles with a genetically tagged clone of the symbiont S. alvi (wkB2:pBTK570). All the beetles survived by the end of the experiment. Tagged symbionts were found in all inoculated beetles, with the average CFU (colony forming unit) of 5,380 at 1 dpi (day post inoculation) on selective plates. Even though the CFU increased to 18,300, the colonization rate dropped to 20% at 3 dpi. When counting the CFU, we observed some microbial colonies morphologically different from S. alvi wkB2:pBTK570 in all beetles. To identify the microbial species, we sequenced one such colony, and the ITS2 region aligned with K. ohmeri (97% identity, P = 7e-97), forming a cluster in the phylogenetic tree (Supplementary Figure S5).
Discussion
Beetles are the most diverse taxon, making up 40% of all described insect species, including many agricultural pests (Bouchard et al., 2011). From a nutritional point of view, most plant-feeding beetles need more time and enzymes to process large amounts of food because nutrient levels in plants are often low. Symbionts assist beetles in surviving on foods with poor nutritional quality while also helping their hosts cope with toxic plant defenses (Bentz and Six, 2006; Morales-Jiménez et al., 2012; Salem and Kaltenpoth, 2022). In aquatic beetles, symbionts provide essential amino acids and the B vitamin riboflavin for beetle larvae and pectinases to complement host cellulases (Reis et al., 2020). Weevils exhibit convergence in their gut microbial communities when feeding on similar food sources, suggesting that these communities are determined by the environment and host ecology (Berasategui et al., 2016). The gut microbiota help invasive moths feed on new host plant (Zhang et al., 2024). Beetles feed on different plants (generalists) harbor more complex microbes than the specialists (Brunetti et al., 2022). Our study found more diverse microbes in SHBs in the banana group than in the wild group. This suggests that fruit feeding supports more diverse microbes than the protein-rich diet. Alternatively, this high alpha diversity could indicate more transient microbes. Additionally, we found a plant-associated bacterium in the banana group, C. glyciniphilum, which was initially isolated from the banana and could temporarily pass through the gut of SHBs (Al-Dilaimi et al., 2015). We also found that SHBs in the banana group were enriched in acetic-acid bacteria, which assist the fermentation of sugar and saps and have established symbioses in bees, flies, and bugs (Crotti et al., 2010).
In the dispersal stage of SHBs, the primary energy source is carbohydrates. Symbiont-mediated carbohydrate-active enzymes (CAZy) facilitate breaking down the major components of plant cell walls, releasing energy sources (Calderón-Cortés et al., 2012; Zheng et al., 2019). In our study, the SHBs fed with banana showed the highest number of CAZy, suggesting that more CAZy genes might be required when providing fruit than the hive resources. In termites, fungus-mediated CAZy assisted the host in decomposing the plant (Poulsen et al., 2014). Symbiont encoding a more dynamic digestive range allows hosts to overcome diet restrictions corresponding to a broader ecological distribution (Salem et al., 2020). In our study, the number of lipid and amino acid metabolism genes was folds higher in the Banana group than in the Bee_Bread and Wild groups. This suggests the symbionts may cooperatively recycle metabolites, as found in social bees (Zheng et al., 2019; Li et al., 2022). When bee hives are located, SHBs sneak in and switch their diet to bee hive resources. Thus, the beetle may switch microbes to adapt to the digestion of hive resources. Future studies quantifying the gene expression and enzymatic activity can explain to what extent the increased gene number reflects functional enhancement.
K. ohmeri has been reported to be a commensal fungus in SHBs (Torto et al., 2007; Benda et al., 2008; Amos et al., 2018, 2019). In our study, we found K. ohmeri is antibiotic-resistant. It is possible that other SHB-associated antibiotic-resistant bacteria cultivating under different or the same conditions when increasing sequenced colonies. K. ohmeri ferments honey and serves as a kairomone to attract other SHBs, a fact used to track and kill SHBs (Stuhl, 2020). The route for beetle progeny to acquire this symbiont remains unclear. We used newly emerged beetles hatched in a new container. Thus, the chance of acquiring fungi from parental feeding or the soil is low. The symbiont might instead be vertically transferred from females. In a previous study, we found the honey bee gut symbiont S. alvi in SHBs when feeding the SHBs with bee larvae (Huang et al., 2019). A specialized diet may lead to different gut chemical conditions, creating a gut micro-ecosystem selected for other symbionts (Zmora et al., 2019). For example, S. alvi reduces oxygen in the gut, favoring anaerobic microbes and shapes competition (Motta and Moran, 2024). We found this symbiont S. alvi again in SHBs metagenome, which is rarely found outside bees. In our data, the colonization rate of S. alvi dropped from 100% at 1 dpi to 20% at 3 dpi, even while CFUs increased. This suggests that this bee symbiont cannot consistently colonize SHBs outside the bee hive. In a follow-up study, it will be interesting to reveal the enzymatic activity of bee symbionts in SHBs, to determine if they play a role in SHB fitness while in the hive environment.
Materials and methods
SHB rearing, DNA extraction, and Illumina sequencing
We directly collected adult SHBs from collapsed beehives (Apis cerana) in a commercial apiary. These beetles were maintained in an incubator (28 ± 1°C temperature and 65% ± 10% humidity), and then we transferred their pupal offspring to a new container until hatching (Neumann et al., 2013). We collected pollen from the honey bee Apis mellifera hive entrance. We randomly assigned newly hatched SHBs into two diet groups. SHBs fed on bananas were defined as the Banana group (N = 45), and those fed with simulated bee bread (an equal mix of pollen and 50% w/v sugar water to avoid bee hive microbes) comprised the Bee_Bread group (N = 45). We then assigned each of the 15 SHBs to a rearing tube. We additionally collected 13 SHBs from a beehive as the wild group because these SHBs were directly from the bee hive without lab feeding (N = 13). We refreshed the banana and bee bread daily and collected SHBs after 3 weeks of feeding. We rinsed SHB surfaces with distilled water, then extracted genomic DNA from individual SHBs using MagPure Soil DNA KF Kit (MP Biomedicals, USA). We prepared DNA sequencing libraries using the TruSeq Nano DNA LT Sample Preparation Kit (Illumina, USA). Ten adult SHBs in the Banana group, 10 in the Bee_Bread group, and eight in the Wild group were randomly selected for metagenomic sequencing using the Illumina NovaSeq 6,000 Platform, generating 150 bps paired-end reads.
De novo metagenomic assembly and gene annotation
First, we filtered the sequencing reads using Fastp (Version 0.23.2) with default parameters (Chen et al., 2018). Then we aligned the reads to the small hive beetle genome assembly (GCA_024364675.1) using BWA (Version 0.7.17-r1188) with default parameters and retrieved the unmapped reads using samtools (Version 1.7) and converted the bam to fastq file using bedtools (Version 2.26.0) (Li et al., 2009; Li and Durbin, 2009; Quinlan and Hall, 2010). After that, the unmapped reads were concatenated for all samples to assemble contigs using Megahit (Version 1.2.9) with default parameters (Li et al., 2015), and the contigs were further collapsed using redundans (Version 2020.01.28) (Pryszcz and Gabaldón, 2016). The genes were predicted using MetaGeneMark2 with default parameters.1 The protein sequences were queried with the eggNOG-mapper and KEGG database to retrieve the putative function (Cantalapiedra et al., 2021). The code is provided in the Supplementary File S1.
Gene distribution among microbes associated with diet
The unmapped reads in each beetle were re-aligned to the assembled meta-contigs using BWA (Version 0.7.17-r1188) with default parameters. The number of reads aligned to each gene was quantified using bedtools (Version 2.26.0) (Quinlan and Hall, 2010). The number of aligned reads was normalized to the library size, and the over-represented genes were calculated with edgeR (Quinlan and Hall, 2010). The code is provided in the Supplementary File S1.
Binning the sequencing reads to microbial species
We performed two steps to bin the sequencing reads to the microbial species. We first aligned the assembled contigs to the most closely related microbes using the BusyBee tool (Laczny et al., 2017). Additionally, we aligned the reads to the Kraken2 (Version 2.1.2) standard database (built on 12/9/2022) (Wood et al., 2019). The number of reads assigned to each microbe was normalized using bracken (Version 2.8) (Lu et al., 2017). The relative abundance of the microbial species was used to calculate alpha (Shannon’s alpha diversity) and beta (Bray–Curtis dissimilarity) diversity using KrakenTools (Lu et al., 2022). The code is provided in the Supplementary File S1.
Inoculating the bee symbiont to the small hive beetles
As the bee symbionts were constantly reported from SHB metagenomes, we inoculated SHBs with a genetically tagged bee symbiont to validate its colonization. The honey bee symbiont S. alvi wkB2:pBTK570 (Addgene accession ID#110615) was previously engineered to be spectinomycin resistant and stored at −80°C (Leonard et al., 2018). This symbiont isolate was activated on Columbia Blood Agar Base (Difco™, 279,220) with 5% sheep blood for 72 h, after which bacterial cells were diluted in 1000 μL PBS and adjusted to OD600 = 1. Then, we mixed the homogenized bacterial cells with filter-sterilized 50% sucrose (50%) at a 1:1 ratio. SHBs were fed sucrose with tagged S. alvi wkB2:pBTK570 (N = 50), then SHBs were rinsed in ethanol and homogenized individually. Homogenates were plated on Columbia Blood Agar Base with 5% sheep blood and spectinomycin (60 μg/mL) to count the Colony Forming Unite (CFU) at 1, 3, 5, and 7 days post-inoculation. Detailed procedures are described in the Supplementary File S1.
Commensal fungal identification
We observed microbes morphologically distinct from S. alvi in all CBA plates. We collected a colony using an inoculating loop to extract DNA using the Qiagen DNeasy Plant Mini kit (cat#69104). The DNA was amplified using fungal ITS primers (FungITS.F 5’GTTAAAAAGCTCGTAGTTG3’; FungITS.R5’CTCTCAATCTGTCAATCCTTATT 3′) in a 30ul reaction consisting of 0.2ul Taq DNA polymerase (Invitrogen, 18038–240), 0.4ul primers, 0.2ul 10uM dNTP mix. The reactions were run with the cycling parameters: 94°C for 3 min., followed by 35 cycles of (94°C 15 s., 54°C 30s., 72°C 1 min.), 72°C for 5 min, and maintained in 4°C. The products were visualized on a 1.75% agarose gel, producing a single band. PCR products were sent for PCR clean-up and Sanger sequencing at Azenta Life Sciences, Rockville, Maryland. Sequences were searched in the NCBI Blastn MegaBlast. An additional nine sequences of fungal species were downloaded from NCBI and aligned with MUSCLE (Version 5.1) with default parameters. The tree was built with MrBayes (Version 3.2.7) and viewed with FigTree (Version 1.4.4). Detailed procedures are described in the Supplementary File S1.
Statistics
We performed all statistics with R (Version 4.2.2) in RStudio (Version 2022.12.0) (R Core Team, 2013; RStudio Team, 2020). We compared surviving SHBs using Pearson’s Chi-squared test. The number of differentially enriched genes among the paired comparisons was analyzed with Pearson’s Chi-squared test and viewed with the VennDiagram package (Gao et al., 2021). The number of genes in metabolic categories was compared with random using a Chi-squared test, adjusted with the false discovery rate (FDR). The alpha diversity was first analyzed using the Kruskal-Wallis rank sum test, followed by pairwise comparisons using the Wilcoxon rank sum exact test, adjusted with FDR.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found at the National Center for Biotechnology Information (NCBI) using accession number PRJNA953927.
Ethics statement
The study involves small hive beetles, which are neither endangered nor a protected insect. No specific permit is required for the described study.
Author contributions
QH: Formal analysis, Funding acquisition, Writing – original draft. WH: Resources, Writing – original draft. FP-F: Resources, Writing – original draft. JE: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Initiation package of Jiangxi Agricultural University 050014/923230722 (QH), the Hainan Province Science and Technology Special fund ZDYF2021XDNY282 (WH).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1387248/full#supplementary-material
Footnotes
References
Al-Dilaimi, A., Bednarz, H., Lömker, A., Niehaus, K., Kalinowski, J., and Rückert, C. (2015). Revisiting Corynebacterium glyciniphilum (ex Kubota et al., 1972) sp. nov., nom. Rev., isolated from putrefied banana. Int. J. Syst. Evol. Microbiol. 65, 177–182. doi: 10.1099/ijs.0.065102-0
Amos, B. A., Hayes, R. A., Leemon, D. M., and Furlong, M. J. (2019). Small hive beetle (Coleoptera: Nitidulidae) and the yeast, Kodamaea ohmeri: a facultative relationship under laboratory conditions. J. Econ. Entomol. 112, 515–524. doi: 10.1093/jee/toy378
Amos, B. A., Leemon, D. L., Hayes, R. A., Cribb, B. W., and Furlong, M. J. (2018). Associations between the small hive beetle and the yeast Kodamaea ohmeri throughout the host life cycle. J. Econ. Entomol. 111, 1501–1508. doi: 10.1093/jee/toy121
Angulo, E., Hoffmann, B. D., Ballesteros-Mejia, L., Taheri, A., Balzani, P., Bang, A., et al. (2022). Economic costs of invasive alien ants worldwide. Biol. Invasions 24, 2041–2060. doi: 10.1007/s10530-022-02791-w
Barthel, A., Kopka, I., Vogel, H., Zipfel, P., Heckel, D. G., and Groot, A. T. (2014). Immune defence strategies of generalist and specialist insect herbivores. Proc. R. Soc. B Biol. Sci. 281:20140897. doi: 10.1098/rspb.2014.0897
Benda, N. D., Boucias, D., Torto, B., and Teal, P. (2008). Detection and characterization of Kodamaea ohmeri associated with small hive beetle Aethina tumida infesting honey bee hives. J. Apic. Res. 47, 194–201. doi: 10.1080/00218839.2008.11101459
Bentz, B. J., and Six, D. L. (2006). Ergosterol content of Fungi associated with Dendroctonus ponderosae and Dendroctonus rufipennis (Coleoptera: Curculionidae, Scolytinae). Ann. Entomol. Soc. Am. 99, 189–194. doi: 10.1603/0013-8746(2006)099[0189:ECOFAW]2.0.CO;2
Berasategui, A., Axelsson, K., Nordlander, G., Schmidt, A., Borg-Karlson, A.-K., Gershenzon, J., et al. (2016). The gut microbiota of the pine weevil is similar across Europe and resembles that of other conifer-feeding beetles. Mol. Ecol. 25, 4014–4031. doi: 10.1111/mec.13702
Blackburn, T. M., Bellard, C., and Ricciardi, A. (2019). Alien versus native species as drivers of recent extinctions. Front. Ecol. Environ. 17, 203–207. doi: 10.1002/fee.2020
Bouchard, P., Bousquet, Y., Davies, A., Alonso-Zarazaga, M. A., Lawrence, J., Lyal, C. H. C., et al. (2011). Family-group names in Coleoptera (Insecta). Zookeys 88, 1–972. doi: 10.3897/zookeys.88.807
Brunetti, M., Magoga, G., Gionechetti, F., De Biase, A., and Montagna, M. (2022). Does diet breadth affect the complexity of the phytophagous insect microbiota? The case study of Chrysomelidae. Environ. Microbiol. 24, 3565–3579. doi: 10.1111/1462-2920.15847
Calderón-Cortés, N., Quesada, M., Watanabe, H., Cano-Camacho, H., and Oyama, K. (2012). Endogenous plant cell wall digestion: a key mechanism in insect evolution. Annu. Rev. Ecol. Evol. Syst. 43, 45–71. doi: 10.1146/annurev-ecolsys-110411-160312
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829. doi: 10.1093/molbev/msab293
Cervancia, C. R., de Guzman, L. I., Polintan, E. A., Dupo, A. L. B., and Locsin, A. A. (2016). Current status of small hive beetle infestation in the Philippines. J. Apic. Res. 55, 74–77. doi: 10.1080/00218839.2016.1194053
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Crotti, E., Rizzi, A., Chouaia, B., Ricci, I., Favia, G., Alma, A., et al. (2010). Acetic acid bacteria, newly emerging symbionts of insects. Appl. Environ. Microbiol. 76, 6963–6970. doi: 10.1128/AEM.01336-10
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. doi: 10.1038/nature12820
Eyer, M., Chen, Y. P., Schäfer, M. O., Pettis, J., and Neumann, P. (2009). Small hive beetle, Aethina tumida, as a potential biological vector of honeybee viruses. Apidologie 40, 419–428. doi: 10.1051/apido:2008051
Gao, C.-H., Yu, G., and Cai, P. (2021). ggVennDiagram: an intuitive, easy-to-use, and highly customizable R package to generate venn diagram. Front. Genet. 12:706907. doi: 10.3389/fgene.2021.706907
Gillespie, P., Staples, J., King, C., Fletcher, M. J., and Dominiak, B. (2003). Small hive beetle Aethina tumida (Murray) (Coleoptera: Nitidulidae) in New South Wales. Gen. Appl. Entomol. 32, 5–7,
Hood, W. M. (2000). Overview of the small hive beetle, Aethina tumida, in North America. Bee World 81, 129–137. doi: 10.1080/0005772X.2000.11099483
Huang, Q., Lopez, D., and Evans, J. D. (2019). Shared and unique microbes between small hive beetles (Aethina tumida) and their honey bee hosts. Microbiology 8:e899. doi: 10.1002/mbo3.899
Idrissou, F. O., Huang, Q., Yañez, O., and Neumann, P. (2019). International beeswax trade facilitates small hive beetle invasions. Sci. Rep. 9:10665. doi: 10.1038/s41598-019-47107-6
Laczny, C. C., Kiefer, C., Galata, V., Fehlmann, T., Backes, C., and Keller, A. (2017). BusyBee web: metagenomic data analysis by bootstrapped supervised binning and annotation. Nucleic Acids Res. 45, W171–W179. doi: 10.1093/nar/gkx348
Leonard, S. P., Perutka, J., Powell, J. E., Geng, P., Richhart, D. D., Byrom, M., et al. (2018). Genetic engineering of bee gut microbiome bacteria with a toolkit for modular assembly of broad-host-range plasmids. ACS Synth. Biol. 7, 1279–1290. doi: 10.1021/acssynbio.7b00399
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, Y., Leonard, S. P., Powell, J. E., and Moran, N. A. (2022). Species divergence in gut-restricted bacteria of social bees. Proc. Natl. Acad. Sci. USA 119:e2115013119. doi: 10.1073/pnas.2115013119
Li, D., Liu, C.-M., Luo, R., Sadakane, K., and Lam, T.-W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Liu, Y., Han, W., Gao, J., Su, S., Beaurepaire, A., Yañez, O., et al. (2021). Out of Africa: novel source of small hive beetles infesting eastern and Western honey bee colonies in China. J. Apic. Res. 60, 108–110. doi: 10.1080/00218839.2020.1816686
Lu, J., Breitwieser, F. P., Thielen, P., and Salzberg, S. L. (2017). Bracken: estimating species abundance in metagenomics data. PeerJ Comput. Sci 3:e104. doi: 10.7717/peerj-cs.104
Lu, J., Rincon, N., Wood, D. E., Breitwieser, F. P., Pockrandt, C., Langmead, B., et al. (2022). Metagenome analysis using the kraken software suite. Nat. Protoc. 17, 2815–2839. doi: 10.1038/s41596-022-00738-y
Luo, X., Fang, G., Chen, K., Song, Y., Lu, T., Tomberlin, J. K., et al. (2023). A gut commensal bacterium promotes black soldier fly larval growth and development partly via modulation of intestinal protein metabolism. MBio 14:e0117423. doi: 10.1128/mbio.01174-23
Machovsky-Capuska, G. E., Senior, A. M., Simpson, S. J., and Raubenheimer, D. (2016). The multidimensional nutritional niche. Trends Ecol. Evol. 31, 355–365. doi: 10.1016/j.tree.2016.02.009
Morales-Jiménez, J., Zúñiga, G., Ramírez-Saad, H. C., and Hernández-Rodríguez, C. (2012). Gut-associated bacteria throughout the life cycle of the bark beetle Dendroctonus rhizophagus Thomas and Bright (Curculionidae: Scolytinae) and their cellulolytic activities. Microb. Ecol. 64, 268–278. doi: 10.1007/s00248-011-9999-0
Motta, E. V. S., and Moran, N. A. (2024). The honeybee microbiota and its impact on health and disease. Nat. Rev. Microbiol. 22, 122–137. doi: 10.1038/s41579-023-00990-3
Neumann, P., Evans, J. D., Pettis, J. S., Pirk, C. W. W., Schäfer, M. O., Tanner, G., et al. (2013). Standard methods for small hive beetle research. J. Apic. Res. 52, 1–32. doi: 10.3896/IBRA.1.52.4.19
Neumann, P., Pettis, J. S., and Schäfer, M. O. (2016). Quo vadis Aethina tumida? Biology and control of small hive beetles. Apidologie 47, 427–466. doi: 10.1007/s13592-016-0426-x
Ouessou Idrissou, F., Straub, L., and Neumann, P. (2019). Keeping a low profile: small hive beetle reproduction in African honeybee colonies. Agric. For. Entomol. 21, 136–138. doi: 10.1111/afe.12306
Palmeri, V., Scirtò, G., Malacrinò, A., Laudani, F., and Campolo, O. (2014). A scientific note on a new pest for European honeybees: first report of small hive beetle Aethina tumida,(Coleoptera: Nitidulidae) in Italy. Apidologie 46, 1–3. doi: 10.1007/s13592-014-0343-9
Poulsen, M., Hu, H., Li, C., Chen, Z., Xu, L., Otani, S., et al. (2014). Complementary symbiont contributions to plant decomposition in a fungus-farming termite. Proc. Natl. Acad. Sci. USA 111, 14500–14505. doi: 10.1073/pnas.1319718111
Pryszcz, L. P., and Gabaldón, T. (2016). Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic Acids Res. 44:e113. doi: 10.1093/nar/gkw294
Quinlan, A. R., and Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi: 10.1093/bioinformatics/btq033
Quinn, A., El Chazli, Y., Escrig, S., Daraspe, J., Neuschwander, N., McNally, A., et al. (2024). Host-derived organic acids enable gut colonization of the honey bee symbiont Snodgrassella alvi. Nat. Microbiol. 9, 477–489. doi: 10.1038/s41564-023-01572-y
R Core Team (2013). R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
Reis, F., Kirsch, R., Pauchet, Y., Bauer, E., Bilz, L. C., Fukumori, K., et al. (2020). Bacterial symbionts support larval sap feeding and adult folivory in (semi-)aquatic reed beetles. Nat. Commun. 11:2964. doi: 10.1038/s41467-020-16687-7
Rohner, P. T., and Moczek, A. P. (2020). Rapid differentiation of plasticity in life history and morphology during invasive range expansion and concurrent local adaptation in the horned beetle Onthophagus taurus. Evolution 74, 2059–2072. doi: 10.1111/evo.14045
Salem, H., and Kaltenpoth, M. (2022). Beetle–bacterial symbioses: endless forms most functional. Annu. Rev. Entomol. 67, 201–219. doi: 10.1146/annurev-ento-061421-063433
Salem, H., Kirsch, R., Pauchet, Y., Berasategui, A., Fukumori, K., Moriyama, M., et al. (2020). Symbiont digestive range reflects host plant breadth in herbivorous beetles. Curr. Biol. 30, 2875–2886.e4. doi: 10.1016/j.cub.2020.05.043
Smith, T. E., Li, Y., Perreau, J., and Moran, N. A. (2022). Elucidation of host and symbiont contributions to peptidoglycan metabolism based on comparative genomics of eight aphid subfamilies and their Buchnera. PLoS Genet. 18:e1010195. doi: 10.1371/journal.pgen.1010195
Stuhl, C. J. (2020). The development of an attract-and-kill bait for controlling the small hive beetle (Coleoptera: Nitidulidae). Apidologie 51, 428–435. doi: 10.1007/s13592-019-00729-1
Stuhl, C. J. (2021). Small hive beetle (Coleoptera: Nitidulidae) attraction to a blend of fruit volatiles. Fla. Entomo. 104, 153–157. doi: 10.1653/024.104.0301
Torto, B., Boucias, D. G., Arbogast, R. T., Tumlinson, J. H., and Teal, P. E. A. (2007). Multitrophic interaction facilitates parasite-host relationship between an invasive beetle and the honey bee. PNAS 104, 8374–8378. doi: 10.1073/pnas.0702813104
Wood, D. E., Lu, J., and Langmead, B. (2019). Improved metagenomic analysis with kraken 2. Genome Biol. 20:257. doi: 10.1186/s13059-019-1891-0
Zhang, S., Song, F., Wang, J., Li, X., Zhang, Y., Zhou, W., et al. (2024). Gut microbiota facilitate adaptation of invasive moths to new host plants. ISME J. 18:wrae031. doi: 10.1093/ismejo/wrae031
Zhao, H., Sa, Y., Liu, J., Huang, W., Ji, C., Ren, Q., et al. (2020). First detection of small hive beetle Aethina tumida Murray (Coleoptera: Nitidulidae) infesting eastern honeybee, Apis cerana Fabricius (Hymenoptera: Apidae), in China. Sociobiology 67, 126–128. doi: 10.13102/sociobiology.v67i1.4381
Zheng, H., Perreau, J., Powell, J. E., Han, B., Zhang, Z., Kwong, W. K., et al. (2019). Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc. Natl. Acad. Sci. 116, 25909–25916. doi: 10.1073/pnas.1916224116
Keywords: invasive pest, symbiosis, beetle, bee, engineered bacteria
Citation: Huang Q, Han W, Posada-Florez F and Evans JD (2024) Microbiomes, diet flexibility, and the spread of a beetle parasite of honey bees. Front. Microbiol. 15:1387248. doi: 10.3389/fmicb.2024.1387248
Edited by:
Yijuan Xu, South China Agricultural University, ChinaReviewed by:
Muhammad Noor-Ul-Ane, Andong National University, Republic of KoreaHao-Sen Li, Sun Yat-sen University, China
Copyright © 2024 Huang, Han, Posada-Florez and Evans. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Huang, cWlhbmctaHVhbmdAbGl2ZS5jb20=
 Qiang Huang
Qiang Huang Wensu Han
Wensu Han Francisco Posada-Florez4
Francisco Posada-Florez4 Jay D. Evans
Jay D. Evans