
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 29 April 2024
Sec. Infectious Agents and Disease
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1385631
This article is part of the Research Topic Reviews in Molecular Evolution of Infectious Agents and Diseases View all 3 articles
Stenotrophomonas maltophilia is an opportunistic pathogen intrinsically resistant to multiple and broad-spectrum antibiotics. Although the bacterium is considered a low-virulence pathogen, it can cause various severe diseases and contributes significantly to the pathogenesis of multibacterial infections. During the COVID-19 pandemic, S. maltophilia has been recognized as one of the most common causative agents of respiratory co-infections and bacteremia in critically ill COVID-19 patients. The high ability to adapt to unfavorable environments and new habitat niches, as well as the sophisticated switching of metabolic pathways, are unique mechanisms that attract the attention of clinical researchers and experts studying the fundamental basis of virulence. In this review, we have summarized the current knowledge on the molecular aspects of S. maltophilia virulence and putative virulence factors, partially touched on interspecific bacterial interactions and iron uptake systems in the context of virulence, and have not addressed antibiotic resistance.
The global emergence of multidrug-resistant Gram-negative bacteria is the most challenging clinical and public health problem (Hernando-Amado et al., 2019; Antimicrobial Resistance Collaborators, 2022). Despite significant progress in biomedical research, many untreatable infectious diseases are considered the leading causes of human death worldwide. Nosocomial and community-acquired infections caused by opportunistic Gram-negative pathogens are becoming increasingly difficult to treat as both intrinsic and acquired antibiotic resistance has increased significantly in recent years (Theuretzbacher et al., 2020).
Among Gram-negative opportunistic pathogens, Stenotrophomonas maltophilia has been the subject of an increased interest and extensive research over the last two decades. The number of reported S. maltophilia infections has considerably risen and the bacterium has been classified as the most common Gram-negative carbapenem-resistant pathogen in patients with bacteremia in some US hospitals (Cai et al., 2020).
Stenotrophomonas maltophilia is a globally dispersed, non-fermenting Gram-negative bacillus frequently isolated in the environment, particularly from water sources, soil, sediment, plants, and animal specimens (Aznar et al., 1992; Nakatsu et al., 1995; Jakobi et al., 1996; Berg et al., 1999; Johnson et al., 2003; Ivanov et al., 2005; Romanenko et al., 2008; Berg, 2009). According to the List of Prokaryotic Names with Standing in Nomenclature,1 the genus Stenotrophomonas comprises at least 25 validated species exhibiting great genetic diversity and metabolic heterogeneity both within the Stenotrophomonas genus and within a single species (Ryan et al., 2009; Turrientes et al., 2010; Pompilio et al., 2016). Comprehensive taxonomic and phylogenomic studies have shown that S. maltophilia includes multiple cryptic species, forming the Stenotrophomonas maltophilia complex (Smc) and a distinction that defies conventional classification approaches (Ochoa-Sánchez and Vinuesa, 2017; Kumar et al., 2020; Singh et al., 2023). Identification of phylogenetic relationships among Stenotrophomonas spp. based on analysis of core protein sequences revealed 24 species-level clades of Smc (Gröschel et al., 2020; Li et al., 2024).
The bacterium demonstrates high adaptability to various environments, including nutrient-limited and hostile conditions. S. maltophilia is capable of utilizing a wide range of carbon sources such as trichloroethylene, toluene, chloroform, glucose, and benzene (Lee et al., 2002; Pompilio et al., 2011; Mukherjee and Roy, 2013).
Stenotrophomonas maltophilia is an opportunistic pathogen intrinsically resistant to multiple and broad-spectrum antibiotics. The bacterium is associated with a number of serious diseases and contributes significantly to the pathogenesis of multibacterial infections. S. maltophilia causes infections in various human organs, including the respiratory, gastrointestinal, and urinary tracts. It can cause severe pneumonia, catheter-associated bacteremia/septicemia, osteochondritis, mastoiditis, meningitis, and endocarditis (Denton and Kerr, 1998; Brooke, 2012; Chang et al., 2015). The bacterium is frequently recovered in the lungs of cystic fibrosis (CF) patients; according to various studies, the frequency ranges from 10% to 30% (Waters et al., 2011; Cuthbertson et al., 2020). During the global COVID-19 pandemic, S. maltophilia has been recognized as one of the most common causative agents of respiratory co-infections and bacteremia in critically ill COVID-19 patients. Furthermore, S. maltophilia isolates detected in sputum samples obtained from these patients had the highest rates of multidrug resistance among other bacteria infecting COVID-19 patients (Yang S. et al., 2021; Ishikawa et al., 2022; Langford et al., 2023).
Stenotrophomonas maltophilia is also of interest as an active member of polymicrobial bacterial communities: it can influence the metabolism of neighboring microorganisms, either through antagonistic suppression of other species or by symbiotic coexistence. A vivid example of such inter-species communication can be observed in CF patients, where S. maltophilia colonizes the host along with other major pathogens, such as Pseudomonas aeruginosa, Staphylococcus aureus, nontuberculous mycobacteria, or Burkholderia cenocepacia (Goss et al., 2004; Coutinho et al., 2008).
The pathogenesis of infections caused by S. maltophilia is determined by numerous virulence factors (VFs), molecules that facilitate bacterial colonization of the host at the cellular level, thereby initiating the infectious process. S. maltophilia possesses a considerable spectrum of VFs or putative factors associated with virulence. These factors include surface cell-associated structures (lipopolysaccharides, type IV pili, flagella, fimbriae, and nonpilus adhesins); the production of a wide spectrum of extracellular enzymes (e.g., proteases, esterases, lipases), hemolysin, siderophores, and cytotoxins; the ability to form biofilms on abiotic surfaces and host tissues; SmeYZ, SmeDEF, SbiAB, and MacABCsm efflux pumps; and the secretion of small molecules in the environment via Quorum Sensing (QS) intercellular communication system [the diffusible signal factor (DSF) and outer membrane vesicles (OMV); Looney, 2005; Brooke, 2012; Ferrer-Navarro et al., 2013; Abbott and Peleg, 2015; Trifonova and Strateva, 2019; Wu C.-J. et al., 2022].
In this review, we briefly summarize the current knowledge on S. maltophilia virulence and provide an overview of the literature introducing the virulence determinants and their regulation in S. maltophilia. We have limited the scope of the review to virulence, partially touching upon inter-species bacterial interactions and iron uptake systems in the context of virulence, and have not referred to antibiotic resistance.
Adherence of the bacterium to host tissues is a crucial step in the host-pathogen interaction. At this stage, the pathogen attached to the host cell initiates its own biochemical processes aimed at its proliferation, invasion of host cells, secretion of toxic molecules, and activation of host cell signaling cascades.
Bacterial adherence factors, also known as adhesins, are polypeptides or polysaccharides. Protein adhesins are cell-surface components or appendages that can be divided into two groups: fimbrial and afimbrial. Polysaccharide adhesins are generally associated with the bacterial cell wall, outer membrane, or capsule. It should be noted that adhesion functions in pathogenesis are not limited to the initial host-pathogen interaction; adhesins also play a significant role in subsequent stages of infection (see below). To provide a holistic view of the role of various VFs in the development of S. maltophilia infection, cell-associated and extracellular VFs are summarized and illustrated in Figure 1.
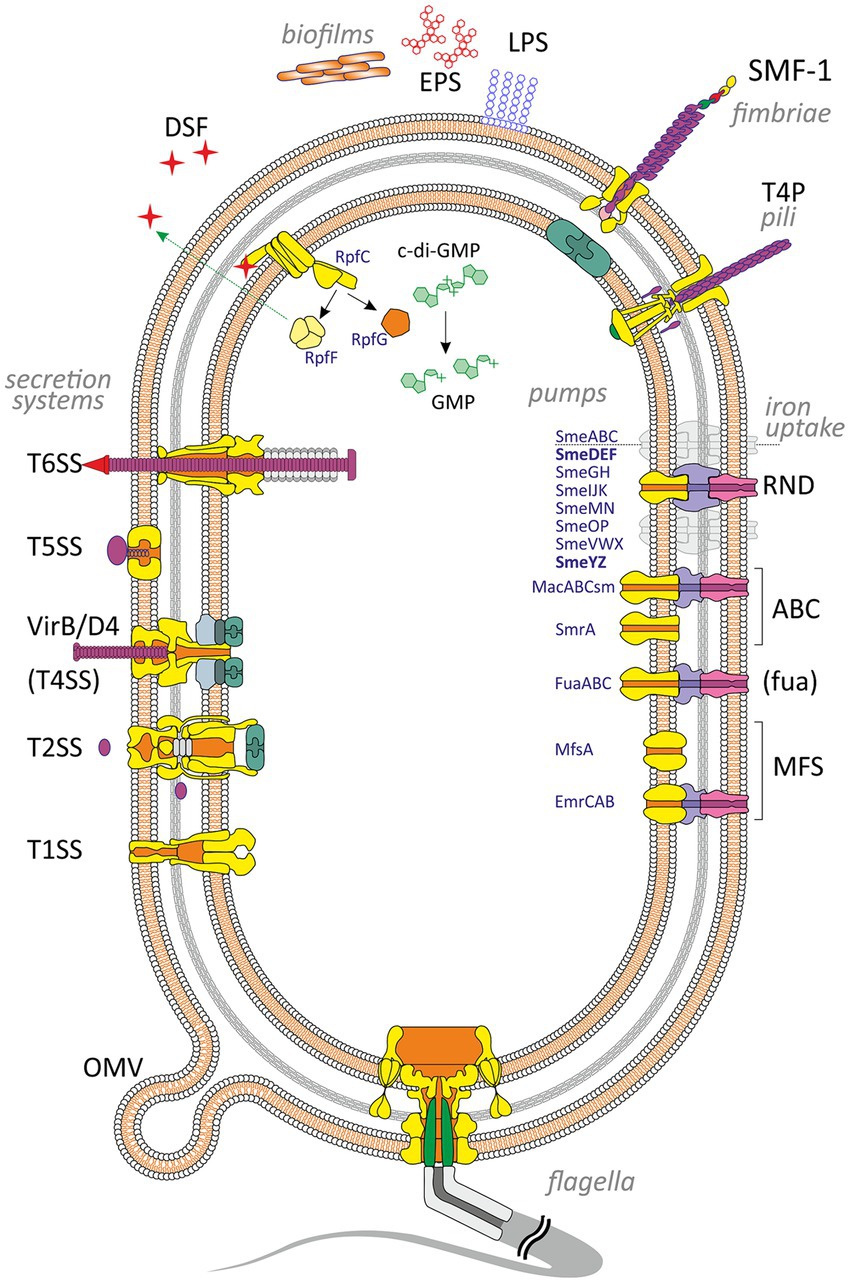
Figure 1. Virulence and putative virulence factors in Stenotrophomonas maltophilia. Surface cell-associated structures include lipopolysaccharides (LPS), type IV pili (T4P), flagella, fimbriae (SMF-1), and non-pilus adhesins (not shown). Extracellular enzymes are secreted through type I, II, IV, V, and VI secretion systems. Small molecules efflux to the environment via the diffusible signal factor (DSF) and outer membrane vesicles (OMV). S. maltophilia produces extracellular polymeric substances and forms a self-secreted polymeric matrix, biofilms, consisting of exopolysaccharides (EPS), DNA, and proteins. The intracellular c-di-GMP level contributes to numerous virulence factors (see the text for details). Different types of efflux pumps revealed in S. maltophilia are shown on the right. The pumps involved in virulence are marked in bold.
Stenotrophomonas maltophilia has lipopolysaccharides (LPS) comprising lipid A, core oligosaccharide, and O-antigen (Neal and Wilkinson, 1982; Jucker et al., 1996; Zhang and Kong, 2002; McKay et al., 2003). LPS is a robust inducer of TNF-α production by macrophages due to its lipid A moiety, that has been elegantly demonstrated by Waters et al. (2007) in a mouse model. Despite the relatively weak invasiveness of S. maltophilia, the level of TNF-α production after stimulating cells of macrophage cell line RAW for 4 h with purified lipid A isolated from S. maltophilia was significantly higher than the corresponding level obtained after stimulation with lipid A from the laboratory P. aeruginosa PAO1 control strain (Waters et al., 2007).
Core oligosaccharides play an essential role in LPS formation and therefore in virulence. Defects in core oligosaccharides of certain bacteria (e.g., P. aeruginosa, Bordetella bronchiseptica) are associated with decreased virulence or the emergence of avirulent strains (Goldberg et al., 1995; West et al., 2000).
The contribution of O-antigens to virulence has also been reported to be significant for many bacteria. Bacteria lacking O-antigens or possessing defective antigen molecules due to disruptions in biosynthesis may exhibit decreased virulence properties. This phenomenon has been demonstrated, in particular, for the species Burkholderia pseudomallei, P. aeruginosa, and B. abortus (Goldberg et al., 1995; DeShazer et al., 1998; Ugalde et al., 2000). In S. maltophilia, defective LPS lacking the O-antigen can affect biofilm production, twitching motility (the ability to move on surfaces using type IV pili), and swimming motility (Huang et al., 2006; Brooke et al., 2008). Stenotrophomonas maltophilia lipopolysaccharides show structural diversity between strains within the genus, with at least 31 different O-antigens (Winn and Wilkinson, 2001; Waters et al., 2007).
The metabolic process of carbohydrates and their incorporation into LPS in S. maltophilia is regulated by several genes. The spgM gene involved in this process encodes a bifunctional enzyme with both phosphoglucomutase and phosphomannomutase activity. This gene is homologous to algC which controls alginate and LPS biosynthesis in P. aeruginosa (McKay et al., 2003; Flores-Treviño et al., 2019). McKay et al. (2003) have shown that spgM mutants with shorter O-polysaccharide chains lost virulence in the rat lung model and exhibited increased susceptibility to complement-mediated killing.
Two operons, rmlBACD and xanAB, are involved in the synthesis of polysaccharides in S. maltophilia. Huang et al. (2006) have conducted an SDS-PAGE analysis of purified LPS from S. maltophilia rmlA, rmlC, and xanB mutants and found these genes necessary for LPS O-antigen biosynthesis. In addition, xanB is required for the production of the LPS core region. The authors also suggested that rml and xanB are involved in the biosynthesis of exopolysaccharides (EPS): the rmlA and rmlC mutants exhibited decreased biofilm production on polystyrene and increased biofilm density on glass. Meanwhile, the xanB mutant displayed lower biofilm production only on polystyrene. Besides contributing to biofilm formation, alterations in LPS caused by the rmlAC and xanB mutations, may lead to changes in outer membrane appendages, such as flagella and type IV pili, thereby interfering with motility and attachment (Huang et al., 2006).
Stenotrophomonas maltophilia possesses one or multiple polar flagella that confer swimming motility, swarming and chemotaxis. Flagella facilitate primary adherence to biotic and abiotic surfaces, contribute to colonization and invasion in the early stages of infection, and trigger specific immune responses by host cells (de Oliveira-Garcia et al., 2002; Huang et al., 2006; Pompilio et al., 2010, 2018; Zgair and Chhibber, 2011).
Wu et al. have identified three flagellar genes, fliC1, fliC2, and fliC3 which form an operon in the sequenced S. maltophilia genome. The authors generated single, double, and triple mutants corresponding to these genes and revealed that each gene contributed to swimming, adhesion, and biofilm formation. The ability to attach, swim, and form biofilms decreased proportionally to the number of deactivated genes, with the triple mutant losing its swimming ability and significantly compromising adhesion and biofilm formation. Thus, flagella in motile pathogens can be considered important VFs (Moens and Vanderleyden, 1996; Duan et al., 2013; Wu C.-J. et al., 2022).
Pompilio et al. (2010) have constructed two S. maltophilia mutants in which the fliI gene was inactivated. The highly conserved fliI gene encodes a substrate-specific ATPase (FliI) that provides energy for the active translocation of flagellar structural components in a wide range of bacterial species (Yonekura et al., 2002; Di Bonaventura et al., 2007). These two flagellum-deficient S. maltophilia fliI mutants exhibited decreased adherence to CF-derived bronchial epithelial IB3-1 cells and compromised swimming motility (Pompilio et al., 2010).
Inbred BALB/c mice (commonly used as animal models for drug and vaccine testing) pretreated intranasally with purified S. maltophilia flagellin and instilled with S. maltophilia 4 h later exhibited significantly increased levels of pro-inflammatory cytokines IL-1β and TNF-α, myeloperoxidase activity, caspase-1 activity, and nitric oxide compared to control groups (Zgair and Chhibber, 2012). The pretreated mice also demonstrated elevated levels of neutrophils, lymphocytes, and monocytes in their bronchoalveolar lavage fluid providing nonspecific protection for the animals against S. maltophilia as well as S. aureus infections (Zgair and Chhibber, 2012). In another study, Pompilio et al. (2018) compared the severity of disease caused by aerosol challenge of mice of the DBA/2 N strain with the clinical isolate S. maltophilia SM111 and its aflagellate isogenic mutant (ΔfliI). Pompilio et al. (2018) did not observe a significant trend in body weight changes, pulmonary persistence, lung damage, or mortality in mice infected with wild-type and fliI− strains. The authors suggested that flagella and motility might not represent S. maltophilia virulence traits involved in the pathogenesis of lung infection. Meanwhile, the expression of TNF-α in murine lungs infected with the aflagellate mutant was significantly reduced (Pompilio et al., 2018).
Hypothetically, in the context of a chronic infection, it is admissible that a bacterium lacking flagellin as a significant immunogenic factor may gain a survival advantage diminishing the host’s immune response. This hypothesis could be corroborated by observations in patients with CF: a substantial portion of P. aeruginosa isolates (39%) from chronically colonized patients were nonmotile and resistant to phagocytosis by macrophages (Mahenthiralingam et al., 1994). Nevertheless, the majority of studies have reported a positive correlation between motility and primary adhesion (e.g., de Oliveira-Garcia et al., 2002; Waters et al., 2007; Pompilio et al., 2010; Zgair and Chhibber, 2011; Madi et al., 2016). Perhaps, the aforementioned speculation that flagella do not contribute to S. maltophilia virulence may be relevant only to the later (chronic) stage of the disease when the initial adhesion step preceding the infection has already occurred.
The motility of S. maltophilia and the level of flagellin gene expression depend on environmental conditions and are tightly controlled by comprehensive genetic systems that have not been fully investigated. In particular, cyclic diguanosine monophosphate (c-di-GMP), the ubiquitous second messenger has been recognized as an important intracellular signaling molecule involved in the regulation of flagellin gene expression in many bacteria. c-di-GMP is also implicated in controlling various bacterial physiological processes, including the cell cycle, adherence, motility, the production of VFs, and biofilm formation (Ross et al., 1987; Hengge, 2009).
The intracellular concentration of c-di-GMP is modulated by two groups of enzymes with opposing activities, diguanylate cyclases (DGCs) and phosphodiesterases (PDEs). DGCs contain a conserved GGDEF domain and synthesize c-di-GMP by condensation of two GTP molecules. PDEs, with either EAL or HD-GYP domains, degrade c-di-GMP into linear guanosine dinucleotide (pGpG) or guanosine monophosphate (GMP) (Chan et al., 2004; Christen et al., 2005, 2006; Caly et al., 2014). An increased level of c-di-GMP, resulting from the action of DGCs, is associated with a sessile lifestyle and biofilm formation which is relevant to chronic bacterial infections. Conversely, lower concentrations of c-di-GMP due to phosphodiesterase activity, are found in motile bacteria during acute infection processes (Hengge, 2009; Moscoso et al., 2011; Cheng et al., 2019). In S. maltophilia, the mechanisms by which c-di-GMP controls flagellar synthesis and flagella numbers are still poorly understood, and there is a paucity of studies focused on the molecular basis of its functioning.
In some bacteria, the expression of flagellar genes is activated and controlled by specific genetic determinants known as master regulators and their homologs, e.g., FlrA (FleQ) in P. aeruginosa and Vibrio cholerae, flaK and flaM in Vibrio parahaemolyticus (Ritchings et al., 1995; Stewart and McCarter, 1996; Arora et al., 1997; Klose and Mekalanos, 1998; Hickman and Harwood, 2008).
Yang et al. (2014) reported that flagellar gene expression in S. maltophilia is controlled by FleQ (Smlt2295). This transcriptional regulator, an enhancer-binding protein, is homologous to the similar master regulator in P. aeruginosa and it is the major target for the c-di-GMP produced by the wrinkly spreader phenotype (Wsp) chemosensory system pathway (Hickman et al., 2005; Yang et al., 2014). FleQ works together with a putative ATPase, FleN, and is inhibited by binding c-di-GMP. Inhibition of this complex and, therefore, Wsp pathway activation results in elevated expression of biofilm-associated pel, psl, and cdr operons and a reduction of flagellar gene expression (Hickman et al., 2005; Hickman and Harwood, 2008; Yang et al., 2014). The situation is reversed if the concentration of c-di-GMP is low: FleQ remains unbounded, and it leads to increased expression of flagellar genes, and, therefore, the ability of bacteria to be sessile in biofilms decreases.
Liu et al. (2017) have reported a correlation between increased expression of bsmR which encodes an eponymous regulatory protein, an EAL domain-containing PDE, and increased bacterial swimming and decreased cell aggregation in S. maltophilia CGMCC 1.1788. Therefore, bsmR is suggested to be a negative regulator of biofilm formation and a positive regulator of swimming motility. The bsmR operon controls the expression of at least 349 genes, of which 34 are involved in flagellar synthesis and are under positive regulation of the FsnR transcription factor (Liu et al., 2017). BsmR degrades c-di-GMP to activate the expression of FsnR. This flagellar-assembly-related transcription factor binds directly to the promoter regions of two operons, Smlt2303 and Smlt2318, initiating the transcription of flagella-associated genes (Kang et al., 2015; Zheng et al., 2016).
Zhang et al. (2022) analyzed genes potentially affecting the c-di-GMP level in S. maltophilia, specifically those encoding proteins containing GGDEF, EAL, and HD-GYP domains. The authors identified 33 putative c-di-GMP turnover enzymes in the genome of S. maltophilia using the Simple Modular Architecture Research Tool (SMART) (Letunic and Bork, 2018) and constructed mutants of all 33 genes via insertional inactivation. Among the mutants analyzed, 12 bacterial strains exhibited a deficiency in swimming motility while one showed promotion, that suggests the 13 corresponding genes may contribute to the regulation of bacterial swimming motility. The authors also made an important observation that the mutation-induced degeneration or inactivation of DGCs or PDEs do not necessarily alter the cellular c-di-GMP level and bacterial swimming motility. Therefore, further investigations are needed to assess the contribution of each gene to swimming motility.
Among all the enzymes analyzed, the authors also identified and characterized a novel Fe2+-dependent phosphodiesterase named SisP (S. maltophilia iron-sensing PDE). SisP increased its activity and facilitated bacterial swimming upon stimulation with ferrous iron in a dose-dependent manner, and the degradation of c-di-GMP led to FsnR-dependent transcription of flagellar genes (Zhang et al., 2022).
Type 1 fimbriae (SMF-1) are an important VF that confers to S. maltophilia the ability to adhere to various specific host epithelia (de Oliveira-Garcia et al., 2002; Zgair and Chhibber, 2011; Giltner et al., 2012). In particular, it has been reported that adherence to biotic (epithelial cells) and abiotic surfaces (such as medical implants and catheters) was inhibited by anti-SMF-1 antibodies (de Oliveira-Garcia et al., 2003). Fimbriae are also involved in early stages of biofilm formation (de Oliveira-Garcia et al., 2003) and can agglutinate red blood cells (Crossman et al., 2008). Antibodies against SMF-1 fimbriae, but not preimmune serum, inhibited hemagglutination in a dose-dependent manner (de Oliveira-Garcia et al., 2003).
It is worth noting that SMF-1 fimbriae were revealed in all clinical isolates (n = 46) obtained from patients (de Oliveira-Garcia et al., 2003), whereas S. maltophilia strains isolated from environmental sources did not possess them (Nicoletti et al., 2011). Therefore, fimbriae are thought to be the significant structures involved in the adhesion and colonization of the lung epithelium.
Fimbrial protein production is controlled by the Smf-1 fimbrial operon, which includes Smlt0706-Smlt0709 (Crossman et al., 2008). This type of fimbriae is assembled by the bacterial chaperone-usher pathway (Thanassi et al., 1998). Despite S. maltophilia fimbrin is closely related to fimbrial adhesins of most members of the Enterobacteriaceae and the CupA fimbriae of P. aeruginosa, the N-terminal amino acid sequences of S. maltophilia Smf-1 significantly differ from those belonging to other families of fimbriae (ranging from 50% to 61%). This suggests that this family of fimbriae may extend to other genetically distant non-enteric bacterial genera (Vallet et al., 2001; de Oliveira-Garcia et al., 2003).
Type IV pili (T4P) are considered a significant VF associated with twitching motility, adhesion to biotic and abiotic surfaces, colonization, and biofilm formation in various bacterial pathogens (Doig et al., 1988; Saiman et al., 1990; Giltner et al., 2012). T4P have also been reported to mediate P. aeruginosa virulence through interdependent action with the type III secretion system (T3SS), thereby promoting its effector injection into the host cell (Heiniger et al., 2010; Shikata et al., 2016).
To date, the role of type IV pili in S. maltophilia virulence has not been sufficiently studied. Kalidasan and Neela (2020) have proposed a theoretical model for S. maltophilia type IV pilus based on Rapid Annotations using Subsystem Technology (RAST) analysis that provided high quality genome annotations for bacterial genomes across the whole phylogenetic tree, and previous reports on P. aeruginosa.
Extensive sequence variation in the type IV pilin adhesion precursor gene has been revealed by Fluit et al. (2022). Meanwhile, no significant correlations have been reported between virulence and the presence of the pil gene family which is involved in pilus formation. An analysis of clinical and environmental S. maltophilia strains performed by Cruz-Córdova et al. (2020) showed that the pilU gene frequencies were high enough but comparable in both groups analyzed, regardless of origin. An increase in biofilm biomass formed by CF isolates with elevated swimming and twitching motility has been reported by Pompilio et al. (2011) and, notably, the phenomenon was observed only in CF isolates. Taken together, there appears to be no direct evidence for T4P as a significant VF in S. maltophilia, and further studies are needed to clarify their contribution to its virulence.
Clinical S. maltophilia strains produce a variety of VFs, including proteases (StmPr1, StmPr2, StmPr3, StmPr4), lipases (lipase and phospholipase C and D), nucleases, gelatinases, elastase, esterases, hyaluronidases, fibrinolysin/streptokinase, heparinases, hemolysins, siderophores, and cytotoxins (Windhorst et al., 2002; Travassos et al., 2004; Trifonova and Strateva, 2019). These VFs contribute to bacterial colonization/persistence, induce cytotoxic and morphological effects on host cells, and play roles in various stages of the infection process (Karaba et al., 2013; DuMont et al., 2015).
Of the 11 known bacterial secretion systems (including outer membrane vesicles, OMVs), S. maltophilia possesses type I, II, IV, V, and VI secretion systems that have been identified through genome sequencing (Crossman et al., 2008; Rocco et al., 2009; Zhu et al., 2012; Adamek et al., 2014; Alavi et al., 2014). Albeit the role of these systems in virulence formation is well understood in many bacteria, only three types of S. maltophilia secretion systems (II, IV, and VI) have been described in detail.
The genome of S. maltophilia clinical strain K279a has two unlinked loci that are predicted to encode the double membrane-spanning type II secretion system, T2SS (GSP and XPS). Each locus contains 11 T2SS genes, corresponding to the core T2SS components (Karaba et al., 2013). The S. maltophilia type II secretion system mediates the secretion of at least seven protein effectors and three proteolytic activities. Proteolytic enzymes, particularly the serine proteases StmPr1, StmPr2, and StmPr3, are secreted in an XPS-dependent manner and induce structural and viability changes in lung epithelial cells, promoting the degradation of collagen, fibrinogen, fibronectin, and interleukin 8 (IL-8) (Karaba et al., 2013; DuMont et al., 2015; DuMont and Cianciotto, 2017). Another serine protease, StmPR4, has also been reported in the S. maltophilia genome (Windhorst et al., 2002; Ribitsch et al., 2012). It is thought that StmPR3, together with StmPR1 and StmPR2, contributes to the protease-mediated dysfunction of the innate immune system in cystic fibrosis (Molloy et al., 2019).
Lee et al. (2022) purified and identified a serine colistin-degrading protease (Cdp) in S. maltophilia strain Col1. Isolated from the soil, this strain exhibited high-level resistance against colistin (MIC value of 32 mg/L). Coculture and coinfection assays revealed that S. maltophilia strain Col1, bearing the cdp gene, could inactivate colistin, thereby protecting susceptible P. aeruginosa. Using colistin against P. aeruginosa infection in Drosophila melanogaster increased fly survival by 41%. In contrast, coinfection of flies with S. maltophilia strains carrying the cdp gene, did not increase the survival rate after colistin treatment. The authors noted that S. maltophilia genomes contain genes orthologous to cdp, located in a region immediately adjacent to the T2SS gene cluster (Lee et al., 2022). Thus, the colistin-degrading protease may play an important role in collective resistance to colistin in polymicrobial infections such as CF (Lee et al., 2022).
A type IV secretion system (T4SS) has been identified in the genome of both clinical and environmental S. maltophilia isolates (Nas et al., 2019). In S. maltophilia, the T4SS called the VirB/D4 system, is highly conserved within the genus and it is most similar to the T4SS of the Xanthomonas genus (Nas et al., 2019). T4SS typically comprises 12 proteins (VirB1-VirB11, and VirD4; Christie et al., 2014; Ghosal et al., 2017; Grohmann et al., 2018) and facilitates the delivery of DNA and/or protein effectors into bacterial or eukaryotic targets in a contact-dependent manner (Gonzalez-Rivera et al., 2016; Grohmann et al., 2018). The VirB10 protein, as part of the periplasm-outer membrane-spanning subcomplex, and the ATPase coupling protein VirD4 are essential for the antibacterial activity of the T4SS in S. maltophilia K279a (Bayer-Santos et al., 2019; Nas et al., 2019, 2021). The contribution of the VirB/D4 system to interspecific antagonism was elegantly demonstrated by Nas et al. (2019). They reported that S. maltophilia was capable of killing P. aeruginosa environmental strain 7700 and clinical isolates PAO1 and PAK when cocultured. Interestingly, S. maltophilia exhibited selectivity when acting on different species of heterologous bacteria, e.g., it killed Pseudomonas mendocina but not P. fluorescens, P. putida, or P. stutzeri (Nas et al., 2019). Based on studies that highlight the contribution of the type VI secretion systems in various bacteria to interbacterial killing (Basler et al., 2012; Ma et al., 2014; Ahmad et al., 2019), the authors suggested that the S. maltophilia VirB/D4 T4SS effectors are akin to those secreted by other type VI secretion systems, e.g., lipases, peptidases, nucleases, and muramidases (Ho et al., 2014).
Nas et al. (2021) identified 13 putative cognate immunity proteins in S. maltophilia that typically provide self-protection to the organism encoding the T4SS, and studied the effect of their expression in heterologous bacteria. Using these proteins, the authors revealed two potential antibacterial effectors, RS14245 and RS14255, that were required for the ability of S. maltophilia to kill heterologous bacteria, especially laboratory E. coli and clinical strains of P. aeruginosa isolated from the lungs of CF patients. The putative lipases, RS14245 and RS14255, when bound by cognate immunity proteins, did not exhibit antibacterial activity; and S. maltophilia complemented mutants lacking RS14245 and RS14255 significantly reduced their antibacterial properties (Nas et al., 2021).
The authors’ findings are intriguing from various perspectives. On the one hand, the secretion of effectors that suppress other bacterial species can be regarded as a significant VF of S. maltophilia. On the other hand, the isolation and study of such effectors hold the potential to develop novel antimicrobial drugs for the targeted therapy of infections caused by Pseudomonas and Escherichia.
Apart from providing a competitive advantage to S. maltophilia in polymicrobial communities, probably by increasing its fitness, the VirB/D4 T4SS effectors have another essential function: they inhibit apoptosis in infected lung epithelial cells but induce apoptosis in infected macrophages (Nas et al., 2019).
The Type VI Secretion System (T6SS) is a protein secretion nanomachine utilized by Gram-negative bacteria to deliver toxic effectors into target cells in a contact-dependent manner (Mougous et al., 2006; Perault et al., 2020; Crisan and Goldberg, 2022). Protein effectors exert their toxicity on the bacterial cell envelope and can degrade the peptidoglycan layer and lipid membranes, form pores and interfere with protein synthesis in the cytoplasm of competitor bacteria (Russell et al., 2011, 2013; Ahmad et al., 2019; Nolan et al., 2021). Additionally, T6SS effectors hinder host cell functions, facilitate immune evasion, thereby promoting a successful infection (Hachani et al., 2016), and participate in bacterial metal uptake by assisting low- and high-affinity transport systems in scavenging metal ions from the environment (Wang et al., 2015; Stubbendieck and Straight, 2016; Lin et al., 2017; Si et al., 2017; Han et al., 2019; Yang X. et al., 2021; Li et al., 2022).
Although T6SS genes have been identified in some S. maltophilia strains early (Alavi et al., 2014; Bayer-Santos et al., 2019), there is a paucity of experimental evidence demonstrating the function of T6SS effectors in the bacterium. Crisan et al. (2023) reported that the S. maltophilia STEN00241 clinical isolate possesses an active T6SS under standard laboratory conditions and the T6SS contributes to the elimination of some heterologous bacterial species. In particular, STEN00241 killed Burkholderia cenocepacia strain K56-2 and E. coli DH5α in a T6SS-dependent manner, but not P. aeruginosa PA14 laboratory strain, the P. aeruginosa CF isolate (PA32), and the S. aureus JE2. This selectivity in the mode of interspecific interaction within multi-species communities (elimination, competitive co-existence, or hypothetical symbiosis) suggests that the T6SS secretory function is also regulated by various environmental factors. The putative T6SS secretion triggers may be signals generated by the QS of the neighboring bacteria or their various metabolites (Lesic et al., 2009; Lin et al., 2017).
Concluding the chapter on S. maltophilia secretion systems, at least one intriguing question remains unanswered: what is the benefit to the bacterium of using both VirB/D4 T4SS and T6SS to produce functionally similar antibacterial proteins? Suggesting that it is not redundancy, the functions of these effectors and/or their trigger mechanisms are thought to be different and need to be further investigated.
To summarize, it should be noted that although five types of secretion systems (type I, II, IV, V, and VI) have been revealed in S. maltophilia genomes, further research is needed to fully comprehend their functional roles and potential interactions between the systems.
The ability of S. maltophilia to form biofilms on abiotic surfaces and host tissues is an important VF that plays a crucial role in HAI and multibacterial infections and dramatically decreases the therapeutic efficacy of important antibiotics, including aminoglycosides, fluoroquinolones, and tetracycline (Di Bonaventura et al., 2004, 2007; Pompilio et al., 2010; Sun et al., 2016). Biofilms provide protection to the members of bacterial communities from exposure to antibiotics by reducing their diffusion (Tseng et al., 2013) and increasing their inactivation (Amanatidou et al., 2019). Besides, the biofilm polymer matrix gives bacteria protection from various forms of environmental stress, such as dehydration, UV exposure, salinity, and toxic metals (Hall-Stoodley et al., 2004). High cell density within biofilms and increased oxidative stress result in an elevated mutation rate and enhanced horizontal gene transfer (HGT) (Driffield et al., 2008). Compared to their planktonic counterparts, bacteria in biofilms exhibit greater resistance to nutrient starvation, pH fluctuations, and oxygen radicals (Jefferson, 2004). Biofilms may also increase the level of resistance by altering the expression of pre-existing antibiotic resistance genes (ARGs) (Høiby et al., 2010) as well as the proportion of tolerant or persister cells within the population due to a reduction in bacterial metabolic activity within the biofilm interior (Walters et al., 2003; Wood et al., 2013).
The formation of persister cells is also hypothetically possible due to a reduction in antibiotic concentration within biofilms, since it has been demonstrated that sub-MIC (minimum inhibitory concentration) levels of various antibiotics can induce persister cell formation (Dörr et al., 2009; Johnson and Levin, 2013; Kwan et al., 2013). Nutrient limitation within biofilms perhaps also affects the bacterial stringent response, where (p)ppGpp (alarmone) levels lead to slower bacterial growth and promote the formation of persister cells (McCall et al., 2019; Ro et al., 2021). In addition, biofilms protect bacteria from the host’s immune response by acting as a physical barrier, helping bacteria avoid detection and phagocytosis, and by activating response regulators, genetic switches, or suppressors that affect the activity of immune cells (Hall-Stoodley et al., 2004; González et al., 2018).
The initial stage of the biofilm formation process occurs within the first 30–60 min when planktonic cells adhere to a surface through weak and reversible interactions mediated by semiflexible fimbriae and flagella filaments. The second stage typically begins 4 h later, during which bacterial cells irreversibly attach to and colonize a surface using flagella, pili, and other surface appendages. After adhering to a surface, the cells initiate the production of extracellular polymeric substances, thereby forming a self-secreted polymer matrix of exopolysaccharides, DNA, and proteins (Flores-Treviño et al., 2019). The first microcolonies are generated by the aggregation of cells after approximately 10 h. The third stage occurs in 18–24 h when the biofilm turns into a mature phase. A mature biofilm possesses microchannels to transport water, nutrients, and debris; and bacterial cells within the biofilm express specific genes involved in QS (see below), EPS, and protein production. In mature biofilms, individual or clustered biofilm cells can detach, disperse, and colonize new niches within less than 24 h (de Kievit, 2009; Sun et al., 2016; Flores-Treviño et al., 2019).
At least several putative genes related to S. maltophilia biofilm formation have been identified. These biofilm-associated genes include: spgM (a biofunctional enzyme with phosphoglucomutase and phosphomannomutase activity); rmlA (glucose-1-phosphate thymidylyltransferase); rmlC (an epimerase RmlC, also named RfbC); xanB (a bifunctional enzyme, phosphomannose isomerase-GDP-mannose pyrophosphorylase); and rpfF (cis-11-methyl-2-dodecenoic acid, or synthase for the diffusible signal factor, DSF) (Köplin et al., 1992; Di Bonaventura et al., 2004; Huang et al., 2006; Pompilio et al., 2011; Zhuo et al., 2014). In addition to the genes listed above, numerous genes associated with the synthesis of LPS, fimbria, flagella, and pili, as well as the intracellular c-di-GMP level contribute to biofilm formation (see the corresponding chapters). For instance, almost all (30/31; 97%) S. maltophilia isolates harboring smf-1, which encodes the fimbrial protein Smf-1, were able to form biofilms (Gallo et al., 2016). The macABCsm and smeYZ genes, encoding pumps, have also been identified as essential for biofilm formation (Huang et al., 2006; Lin Y.-T. et al., 2014; Lin et al., 2015). An extended list of genes potentially associated with biofilm production can be found in the review by Flores-Treviño et al. (2019).
Recently, Strateva et al. (2023) analyzed 220 S. maltophilia strong biofilm producers and found the overall frequency of three biofilm-associated genes as follows: spgM—98.6%, rmlA—86%, and rpfF—66.5%. Meanwhile, Zhuo et al. (2014) have noted that, although the rmlA, spgM, or rpfF are closely related to biofilm formation, they do not significantly affect the average amount of biofilm.
Ramos-Hegazy et al. (2020) analyzed a transposon mutant library for mutations leading to altered biofilm formation. The authors identified the gpmA gene, which encodes a glycolytic enzyme, phosphoglycerate mutase, mediating the initial stages of S. maltophilia attachment to abiotic surfaces as well as immortalized CF-derived bronchial epithelial (CFBE) cells. The S. maltophilia isogenic mutant ΔgpmA exhibited a significant decrease in initial attachment and early biofilm formation on polystyrene plates compared to the wild type within the first 2–4 h. Interestingly, after 6 h, there was no difference in biofilm formation between the wild and mutant strains, suggesting that gpmA is involved only in the early phase of adhesion and biofilm formation (Ramos-Hegazy et al., 2020; Di Bonaventura et al., 2023).
Pompilio et al. (2020) have analyzed 85 S. maltophilia strains isolated from patients with CF and other infections and revealed that over 88% of the isolates were able to form biofilm, with non-CF strains being significantly more efficient compared to CF strains. Meanwhile, the prevalence of the multidrug-resistant phenotype was higher in CF isolates in contrast to non-CF ones (90% vs. 67%). S. maltophilia strains susceptible to piperacillin/tazobactam or meropenem produced significantly increased biofilm biomass compared to resistant strains. The authors suggested that susceptible bacteria may utilize biofilms as an alternative defense strategy to evade antibiotic action and to survive within the host (Pompilio et al., 2020).
Liu et al. (2017) have demonstrated the role of a regulatory protein mentioned above, BsmR, an EAL-domain-containing phosphodiesterase, in controlling biofilm formation and swimming motility in S. maltophilia. An increase in BsmR expression led to a significant increase in bacterial swimming motility and a decrease in cell aggregation. Thus, BsmR was identified as a negative regulator of biofilm development that degrades c-di-GMP through its EAL domain, thereby activating the expression of a transcriptional regulator, FsnR (see above), which positively controls the transcription of flagellar genes involved in swimming motility (Liu et al., 2017; Zhang et al., 2022).
An outer membrane protein, Ax21, secreted within OMVs and associated with a VF related to QS, is also implicated in biofilm formation (Ferrer-Navarro et al., 2013; An and Tang, 2018). Deletion of ax21 (Smlt0387) has been shown to reduce motility, biofilm formation, virulence to larvae of Galleria mellonella, tolerance to tobramycin, as well as alter the expression of some genes associated with virulence or antibiotic resistance (Ferrer-Navarro et al., 2013; An and Tang, 2018).
Most interestingly, the analysis of transcriptome profiles of seven clinical S. maltophilia isolates, combined with differential gene expression of biofilm vs. planktonic cells, revealed that a relatively small set of shared and commonly regulated genes is involved in the biofilm lifestyle: only about 9.5% of all genes were differentially regulated. On average, approximately 7.5% of all genes were upregulated, and about 2% of all genes were downregulated in biofilms compared to planktonic cells (Alio et al., 2020).
A comprehensive analysis of all available data on the role of various factors in the transition of bacterial cells from a planktonic to a sessile lifestyle in biofilms shows that this transformation is initiated and regulated by many mechanisms that require further study.
Historically, efflux pumps have been considered to be among the mechanisms that provide bacteria with resistance to antimicrobials. Efflux pumps significantly contribute to the intrinsic antimicrobial resistance of S. maltophilia. However, as noted and discussed below, some types of efflux pumps possess an extended range of functions beyond the scope of “antibiotic resistance”, and these pumps are involved in the molecular mechanisms of bacterial virulence.
The genome of S. maltophilia contains a formidable arsenal of pumps belonging to various families. This includes ATP-binding cassette (ABC) pumps, MacABCsm (Lin C.-W. et al., 2014) and SmrA (Al-Hamad et al., 2009); Major Facilitator family (MFS) pumps, EmrCAB (Huang et al., 2013) and MfsA (Srijaruskul et al., 2015; Dulyayangkul et al., 2016); a fusaric acid efflux pump, FuaABC (Hu et al., 2012); and eight Resistance Nodulation Division (RND) pumps [SmeABC (Li et al., 2002), SmeDEF (Alonso and Martínez, 2000; Zhang et al., 2001; García-León et al., 2014), SmeGH (Blanco et al., 2019; Li et al., 2019), SmeIJK (Huang et al., 2014), SmeMN (Crossman et al., 2008), SmeOP (Lin C.-W. et al., 2014), SmeVWX (Chen et al., 2011; García-León et al., 2015), and SmeYZ (Lin et al., 2015)].
The efflux pumps encoded in the S. maltophilia genome are involved in the removal of a wide spectrum of toxic substances, including antibiotics. The ABC multidrug efflux pump, SmrA, contributes to the elimination of fluoroquinolones, tetracycline, and doxorubicin (Al-Hamad et al., 2009) while MacABCsm, another member of the same family, removes aminoglycosides, macrolides, and polymyxins (Lin C.-W. et al., 2014). The MFS efflux pump, EmrCABsm, facilitates the removal of nalidixic acid, erythromycin, carbonyl cyanide 3-chlorophenylhydrazone, and tetrachlorosalicylanilide (Huang et al., 2013). The fusaric acid tripartite efflux pump, FusA, is involved in the elimination of fusaric acid (Hu et al., 2012). The role of seven out of the eight RND efflux pumps (SmeABC, SmeDEF, SmeGH, SmeIJK, SmeOP, SmeVWX, and SmeYZ) in the antibiotic resistance has been also identified, except for SmeMN (Gil-Gil et al., 2020). Additionally, some RND pumps are involved in the efflux of chloramphenicol, tetracycline, macrolides, quinolones, sulfamethoxazole, trimethoprim, and trimethoprim–sulfamethoxazole (Alonso and Martínez, 2000; Sánchez and Martínez, 2018; Wu et al., 2019). The contribution of pumps to antimicrobial resistance is considered in detail in some comprehensive reviews (e.g., Menetrey et al., 2021; Chauviat et al., 2023).
It is noteworthy that efflux pumps SmeYZ, SmeDEF, and MacABCsm, besides their primary function of removing xenobiotics from bacterial cells, also impact motility, flagella formation, and biofilm development (Lin Y.-T. et al., 2014; Lin et al., 2015; Kim et al., 2018). Lin Y.-T. et al. (2014) demonstrated that a ΔsmeYZ mutant was unable to form flagella, resulting in a lack of motility, and exhibited reduced biofilm formation. The SmeYZ pump has been reported to contribute to a number of other physiological functions, including oxidative stress susceptibility, swimming, and, along with the SmeDEF pump, protease secretion (Lin et al., 2015; Wu et al., 2016; Kim et al., 2018). Blanco et al. (2019) reported that SmeGH is also involved in biofilm formation: a ΔsmeH mutant exhibited an elevated ability to produce a biofilm.
SmeYZ and SmeDEF are thought to be utilized by S. maltophilia against eukaryotes. Overexpression of SmeDEF in the S. maltophilia strain D457R led to reduced virulence against the social amoeba Dictyostelium discoideum (Alonso, 2004), and the loss of SmeYZ decreased in vivo virulence in a murine model and increased susceptibility to human serum and neutrophils (Lin et al., 2015). The above evidence significantly supports the suggestion that these RND pumps contribute to the S. maltophilia virulence.
The S. maltophilia MacABCsm differs from the MacAB homologs of other bacteria (Lin Y.-T. et al., 2014). In particular, the pump possesses its own cognate outer membrane protein (OMP), MacCsm; and the macABCsm operon is intrinsically expressed. Additionally, MacABCsm has a wider substrate range for extruding macrolides, aminoglycosides, and polymyxins compared to MacAB-TolC of E. coli (Lin Y.-T. et al., 2014).
Another noteworthy function of efflux pumps was described by Wu C.-J. et al. (2022). They revealed that the SmeYZ, SmeDEF, and SbiAB pumps along with other mechanisms, impact the secretion of the siderophore stenobactin and the utilization of iron ions (see below) (Wu C.-J. et al., 2022).
The SmeIJK efflux pump of S. maltophilia has been reported to be involved in cell envelope integrity and the envelope stress response. Huang et al. (2014) demonstrated that a smeIJK-deleted mutant has increased sensitivity to membrane-damaging agents (MDAs) compared to the wild-type strain and exhibited an increased RpoE-mediated envelope stress response. In addition, sublethal MDAs concentrations induced smeIJK expression in an RpoE-dependent manner.
Summarizing the above, the analysis of current data on efflux pumps suggests that the historically held belief regarding their main functions should be reevaluated, and bacterial efflux pumps are much more than antibiotic resistance determinants. Since pumps are revealed in both clinical and environmental strains (Youenou et al., 2015) and considering the environmental origin of S. maltophilia, the functions of efflux pumps may be linked to bacterial physiology and adaptation to various niches and environments, as well as coexistence within complex multi-species communities.
Iron is vital for the growth and proliferation of non-fermenting Gram-negative bacilli, including S. maltophilia. Competition for iron ions between bacteria and the host during chronic infections can be detrimental to the host. Bacterial iron uptake may lead to local tissue damage and systemic dysfunction, e.g., anemia of inflammation, also known as anemia of chronic disease, observed in infectious, inflammatory, autoimmune, neoplastic, and chronic kidney diseases (Jurado, 1997). Iron plays a role in bacterial pathogenicity and host defense mechanisms, which is often underestimated. In S. maltophilia, iron limitation induces biofilm formation, increases EPS production, and reduces the generation of reactive oxygen species (ROS) (Kalidasan et al., 2018). Therefore, bacterial systems aimed at acquiring and transferring iron ions into bacterial cells are considered significant VFs.
While a basic understanding of iron uptake in Gram-negative bacteria has been achieved, many molecular mechanisms involved in this process in S. maltophilia remain unclear. Similar to other bacteria, S. maltophilia possesses a number of iron acquisition mechanisms that exhibit functional redundancy (Figure 2).
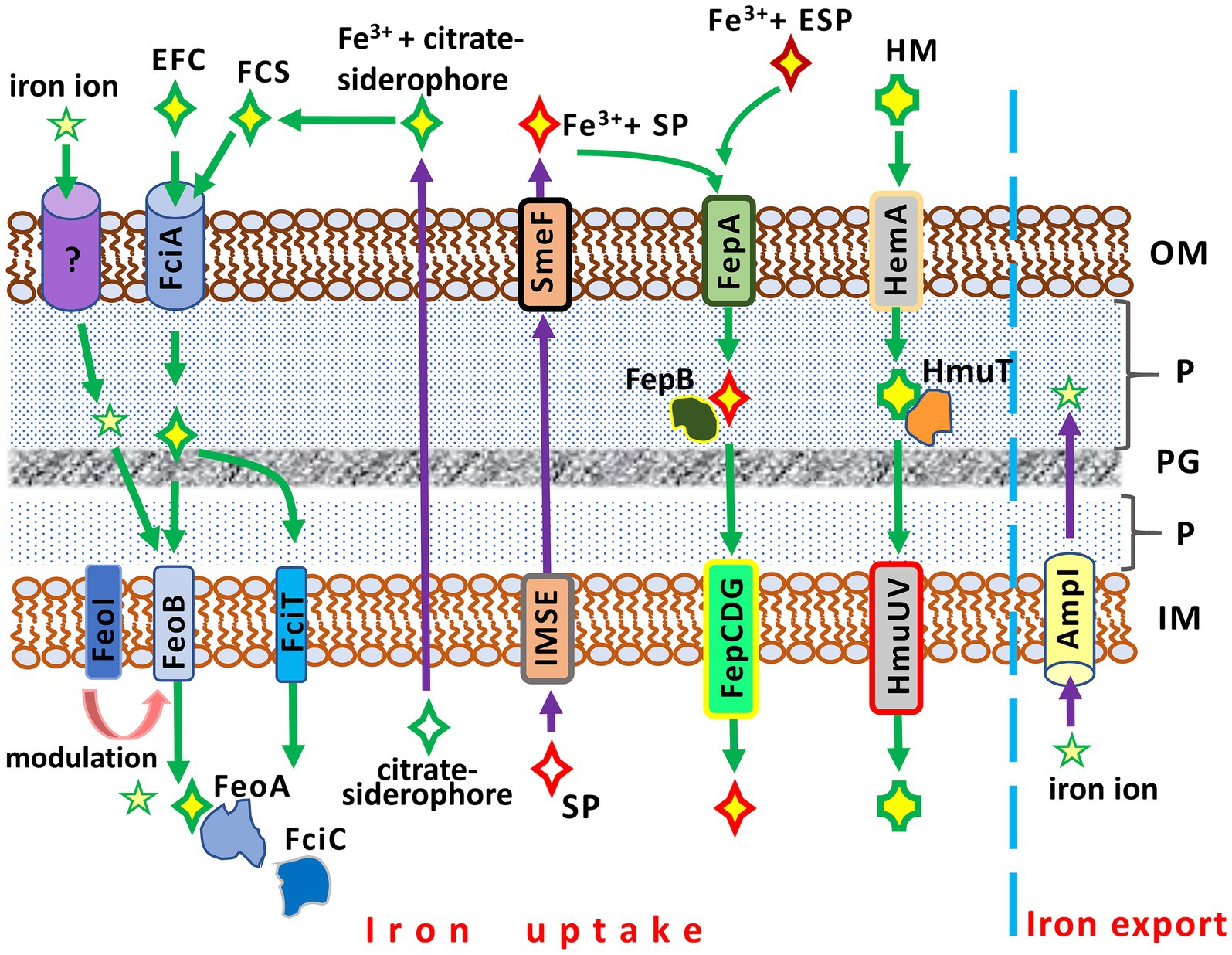
Figure 2. Iron acquisition and iron export systems in Stenotrophomonas maltophilia. IM, inner membrane; P, periplasm; PG; peptidoglycan; OM, outer membrane. AmpI, the inner membrane iron exporter; EFC, exogenous ferric citrate; ESP, an exogenous siderophore (siderophores produced by other bacteria, e.g., Pseudomonas aeruginosa); FciA, the outer membrane receptor for ferric citrate uptake; FciT (inner membrane protein) and FciC (cytoplasmic protein), putative proteins for citrate-mediated iron acquisition; FCS, the ferric citrate siderophore complex; FeoA, a cytoplasmic protein; FeoABI, the inner membrane transporter system of ferric citrate; FepA, the ferri-siderophore uptake system; FepB, a protein delivering ferric siderophore from the periplasm; FepCDG, the inner membrane transporter system for ferric siderophores; HemA, the TonB-dependent outer membrane receptor for hemin; HM, hemin; HmuT, a hemin-transporting protein; HmuUV, the inner membrane hemin transporter; IMSE, the inner membrane siderophore exporters (EntS for enterobactin, SmeY, SbiA, and SmeD for stenobactin); SmeF, the outer membrane exporter for siderophore; SP, S. maltophilia siderophore (stenobactin or enterobactin).
Two distinct iron uptake pathways are encoded by the S. maltophilia genome. These pathways include a siderophore- and a heme-mediated acquisition system (Kalidasan et al., 2018). The entAFDBEC operon controls the synthesis of the siderophore enterobactin, which belongs to the class of catecholamines, binding and transporting Fe3+ into the bacterial cell. The heme-mediated uptake system is under the control of the hgbBC and probably also the hmuRSTUV operons (Adamek et al., 2014; Kalidasan et al., 2018).
The simplest and least efficient absorption system is based on the diffusion of iron ions across OMPs (Liao et al., 2022). A more advanced iron uptake system employs siderophores, high-affinity iron-chelating molecules, to capture iron ions from the environment. These iron-siderophore complexes are recognized by specific OMPs and transported to the cytoplasm. S. maltophilia strains can produce at least two catecholate siderophores, stenobactin and an EntC-dependent catecholate siderophore which is sufficiently similar to, but distinct from, enterobactin of enteric bacteria (Nas and Cianciotto, 2017; Zhang et al., 2021; Wu C.-M. et al., 2022). Although one previous study (Jurkevitch et al., 1992) reported that S. maltophilia produces pseudobactin for iron uptake, this finding has not yet been confirmed. The export of siderophores is mediated by a system of membrane transport proteins (EntS, SmeY, SbiA, SmeD) of the inner membrane and TolC-exporter (SmeF for stenobactin) of the outer membrane (Wu C.-M. et al., 2022).
The ferric-enterobactin complex is recognized and taken up by the TonB-dependent FepA (ferric enterobactin protein), the enterobactin receptor on the outer membrane. Subsequently, FepB, the periplasmic binding protein, transports the iron-siderophore complex from the periplasm to the cytoplasm via the FepCDG transporter (Faraldo-Gómez and Sansom, 2003; Kalidasan et al., 2018).
Interestingly, while competing for iron, S. maltophilia can demonstrate cheating by using exogenous siderophores of xenogeneic origin. It has been reported that S. maltophilia can facilitate the uptake of ferri-pyochelin from P. aeruginosa in an iron-depleted condition (Pan et al., 2022).
An alternative to the catecholate siderophore is the citrate siderophore, which can be produced by S. maltophilia or obtained from exogenous sources. The Fe3+-citrate-siderophore complex is transported into the cytoplasm through a system of specific outer and inner membrane proteins (Figure 2). FciA (Smlt1148) has been reported as the main receptor for ferric citrate acquisition in S. maltophilia KJ (Liao et al., 2022).
Another pathway of iron acquisition in S. maltophilia is the heme-mediated uptake system, which utilizes its specific transporters. The putative outer membrane receptor for hemin is HemA, which is under negative control of the predicted transcriptional factor HemP (Shih et al., 2022). Hemin uptake is controlled by the hemP-hemA-smlt0796-smlt0797, hgbBC, and potentially hmuRSTUV operons (Kalidasan et al., 2018; Shih et al., 2022). The hemP-hemA-smlt0796-smlt0797 operon, in turn, is negatively regulated by the ferric uptake regulator Fur (see below) (Shih et al., 2022).
Iron uptake in S. maltophilia is balanced by the opposite function of iron export. Excess free iron ions in the cytoplasm, which can be toxic to bacterial cells and potentiate ROS toxicity, are removed from the cell by the inner membrane iron exporter, AmpI (Huang et al., 2019). In an iron-depleted condition, AmpI function is inhibited, resulting in iron storage in the S. maltophilia cytoplasm. Notably, AmpI also allows S. maltophilia to reduce β-lactam-induced stress: the expression of ampI increases significantly upon exposure to β-lactams (Huang et al., 2019).
The iron uptake system in Gram-negative bacteria is under the control of many regulators. The key iron-dependent regulator of iron acquisition is the ferric uptake regulator protein (Fur) (Escolar et al., 1999), which is also involved in virulence and protection against oxidative stress (Carpenter et al., 2009). In the presence of Fe2+, Fur forms a complex with Fe2+ ions that binds to the Fur boxes of bacterial DNA, thereby inhibiting the transcription of iron transport genes (Bagg and Neilands, 1987; Escolar et al., 1999; Lee and Helmann, 2007). In the absence of iron, Fur releases its repressor effect, and genes involved in iron transport are expressed.
Interestingly, impairing Fur in S. maltophilia may increase its virulence. The spontaneous fur mutant, derived from the wild-type strain S. maltophilia K279a exhibited increased virulence in the G. mellonella killing assay compared to the parental wild-type K279a strain (García et al., 2015).
Other iron-regulating systems and putative regulators have also been identified in S. maltophilia. The FciTABC and FeoABI systems are responsible for ferric citrate utilization under iron-depleted conditions. Ferric citrate is taken up by FciA, a TonB-dependent OMP, and then transported across the inner membrane through the FeoABI pathway. The genes fciA, fciT, and fciC contribute to ferric citrate acquisition and are under the control of the fciTABC operon (Liao et al., 2022). AmpR, a transcriptional regulator in S. maltophilia, regulates stenobactin synthesis in an iron-depleted condition, however, its contribution to the acquisition and utilization of ferri-stenobactin and ferric citrate is thought to be insignificant (Liao et al., 2020). The uptake of hemin (as mentioned above) is under the control of the hemP-hemA-smlt0796-smlt0797, hgbBC, and potentially hmuRSTUV operons (Kalidasan et al., 2018; Shih et al., 2022). RNA polymerase sigma factors may also be involved in iron regulation in S. maltophilia (Kalidasan et al., 2018).
The iron uptake system is considered not only a virulence factor, but also a regulator of other VFs. Specifically, S. maltophilia K279a produced denser biofilms, and higher levels of EPS and DSF in an iron-depleted condition. In addition, the strain exhibited greater virulence than that cultivated under normal nutritional or iron-rich conditions (García et al., 2015; Kalidasan et al., 2018). Clinical and environmental S. maltophilia isolates grown under iron-limited conditions demonstrated increased nematocidal activity against Caenorhabditis elegans and increased siderophore production (Azman et al., 2019). There have also been contradictory observations: in particular, Jurado (1997) has reported that excess iron can enhance virulence in bacteria. Surplus iron ions have also been shown to upregulate RTX family toxin genes, including the putative virulence gene frpA, and downregulate frpC in S. maltophilia (Adamek et al., 2014).
Probably, fully understanding the influence of high/low levels of iron on S. maltophilia virulence requires further research, and the complexity of the iron regulatory system as well as its potential interaction with other bacterial metabolic global regulators should be taken into account. One potential avenue for further research could be the identification of the iron impact on virulence in the context of the development of a novel class of antibiotics, conjugates of a synthetic cephalosporin with an artificial siderophore. In response to the introduction of the new antibiotic, it has been reported the emergence of resistance to cefiderocol is associated with mutations in the iron uptake system genes (Werth et al., 2022). In this regard, the question arises: how will virulence change in cefiderocol-resistant isolates? An experimentally confirmed answer to this question may have clinical relevance.
Similar to most Gram-negative bacteria, S. maltophilia possesses quorum sensing (QS), a signaling mechanism through which bacterial cells communicate to exchange information about cell density and adjust their gene expression accordingly (Huang and Lee Wong, 2007b). The system is responsible for the production of extracellular signaling molecules known as autoinducers, their detection, and initiating the bacterial response to the appearance of these molecules in the environment. Autoinducers accumulate in the environment, and when their concentration reaches a certain threshold, nearby bacteria are able to detect them. Through the exchange of signaling molecules, cells regulate their metabolic mechanisms related to colonization and virulence, including alterations in motility, biofilm formation, production of extracellular effectors, competition, and resistance properties (Huedo et al., 2014; Papenfort and Bassler, 2016; Paul et al., 2018; Yero et al., 2020).
Genome analyses have revealed that S. maltophilia does not synthesize N-acyl homoserine lactones (N-AHLs) or autoinducer 2 (AI2), signaling molecules typically found in other Gram-negative bacteria (Zhu et al., 2001; Vfselova et al., 2003). The main QS signaling molecule produced by S. maltophilia is known as DSF, represented by cis-Δ2-11-methyl-dodecenoic acid, an unsaturated fatty acid, and seven of its structural derivatives. The synthesis of DSF molecules is controlled by the rpfF and rpfB genes (Huang and Lee Wong, 2007b). The rpf (regulation of pathogenicity factors) gene cluster encodes RpfF synthase, fatty Acyl-CoA ligase, and the two-component RpfC/RpfG system responsible for the perception and transduction of DSF (Huang and Lee Wong, 2007a; Bi et al., 2014). Activated RpfF synthase converts c-di-GMP into a linear nucleotide pGpG or two GMP molecules, thereby regulating the expression of genes related to motility, biofilm formation, and virulence in rpfF-1 strains (Huedo et al., 2019).
Two variants of the rpf gene cluster, rpf-1 and rpf-2, have been found in S. maltophilia (Huedo et al., 2014). Notably, strains belonging to different rpf types exhibit distinct genotypic and phenotypic characteristics. Rpf-1-type S. maltophilia strains can produce DSF in response to various environmental signals, while rpf-2-type strains, with a truncated sensory region in the N-terminus of RpfF synthase, activate their DSF production only after detecting exogenous DSF (e.g., from other bacteria or S. maltophilia strains of the rpf-1 type). Therefore, only rpf-1-type strains can control biofilm formation, as well as the motility and virulence in surrounding bacteria (Huedo et al., 2014, 2015).
It is noteworthy that rpf-1-type S. maltophilia strains, particularly those in genogroup C, exhibit higher resistance to colistin and increased virulence against G. mellonella. Meanwhile, this association was not revealed in another model using the nematode C. elegans (Yero et al., 2020).
Thus, genotyping and identifying the rpf type appear to be useful and important tools for epidemiologic surveillance, considering the potential exchange of rpf clusters among S. maltophilia strains through recombination during horizontal gene transfer (Huedo et al., 2015).
Stenotrophomonas maltophilia has been found to possess a two-component signal transduction system (TCS) called BfmA–BfmK (Smlt4209–Smlt4208). The BfmA transcription factor, a component of the TCS, binds to the bfmA–bfmK promoter region and Smlt0800 (acoT), a gene encoding acyl-coenzyme A thioesterase, associated with biofilm formation (Zheng et al., 2016).
Coves et al. (2023) reported a putative TetR-like transcriptional regulator (locus tag SMLT2053) involved in biofilm formation in the K279a strain. The regulator controls its own β-oxidation operon (Smlt2053-Smlt2051) and is capable of sensing free long-chain fatty acids from the environment, including the QS signal DSF.
The transcriptional regulator AmpR, previously mentioned as a regulator of stenobactin synthesis under iron-deficient conditions, also affects the production of DSF and QS-dependent VFs. Alcaraz et al. (2022) have reported the negative impact of AmpR on biofilm production, as well as on architecture and matrix polysaccharides production in S. maltophilia. The ampR deficient mutant K279a ampRFS exhibited the highest adherence in tubes, the highest biomass production in microplates, and formed biofilms with improved thickness. In contrast, the strain with a constitutively active AmpR, K279aM11, was the least efficient in biofilm formation (Alcaraz et al., 2022). Given that AmpR is a positive regulator of β-lactam-induced β-lactamase expression (Okazaki and Avison, 2008) and iron depletion-mediated stenobactin synthesis (Liao et al., 2020), it is hypothetically possible that S. maltophilia may compensate for the loss of AmpR activity by enhancing biofilm and polysaccharides production to survive under adverse conditions.
It is worth noting that, in contrast to P. aeruginosa, S. maltophilia lacks a complete canonical LuxI/LuxR QS system utilizing acyl-homoserine lactones (AHL) as signaling molecules (see above). However, Martínez et al. (2015) revealed through comparative genomic analysis that S. maltophilia has a LuxR-like gene, Smlt1839, encoding the SmoR regulator (Stenotrophomonas maltophilia orphan regulator), which in vitro binds the synthetic lactone 3OC8-HSL, a natural analog of which is produced by P. aeruginosa. Adding concentrated supernatant from the medium on which lactone-producing P. aeruginosa was cultured provided an impetus to increase the swarming motility of S. maltophilia on Petri dishes. In other words, although S. maltophilia lacks the canonical LuxI/LuxR system, it is able to recognize QS signaling molecules of other species with the LuxI/LuxR system through its homologous intercellular exchange system. It is hypothetically possible that this system is linked to the T6SS and can, under certain conditions, induce the secretion of effectors to inhibit competitor growth (see above).
At the same time, S. maltophilia and P. aeruginosa can coexist and grow together in polymicrobial biofilms, e.g., in the lungs of CF patients (Ryan et al., 2008). Such a symbiotic (temporarily symbiotic or deferred competitive?) coexistence affects their susceptibility to antibiotics. Within two-species biofilms, S. maltophilia produced the DFS that was detected by the two-component sensor BptS in P. aeruginosa. As a result, the latter decreases its susceptibility to polymyxin B and colistin compared to P. aeruginosa monospecies biofilms (Ryan et al., 2008).
In considering the DSF system in S. maltophilia, it is worth noting the phenomenon of secretion through OMVs (Ferrer-Navarro et al., 2013). OMVs are small nanostructures secreted by bacteria, that can transport nucleic acids, proteins, and various small molecules, such as β-lactamases, to the surrounding environment. Devos et al. (2015) have found that S. maltophilia dramatically increased its vesicle secretion in the presence of imipenem. Of particular interest was the composition of the molecules transported by the OMVs: two types of β-lactamases encoded by chromosomes, OMPs, and flagellins Smlt0387 and Smlt0184 (Devos et al., 2015). These flagellins are homologous to Ax21, a protein involved in motility and biofilm formation in Xanthomonas oryzae. The functional role of this protein in S. maltophilia has not been determined, but it is thought that its secretion is initiated by DSF. Recently, the secretion system via OMVs is considered a potential VF, based on data demonstrating its effect on motility and biofilm formation in X. oryzae (Park et al., 2014).
The association between virulence and the presence of filamentous phage genes in the S. maltophilia genome was initially reported in 2006 (Hagemann et al., 2006). Among 47 S. maltophilia strains of clinical and environmental origin, Hagemann et al. (2006) identified five isolates bearing the pI gene of the filamentous phage phiLf, which was identical to the zot-like gene encoding the zonula occludens toxin in V. cholerae. In addition to the zot-like gene, six other genes related to a phage life cycle were identified in the S. maltophilia genomes. The authors suggested that phage genes could be transferred between strains via mobile genetic elements, potentially increasing the virulence of S. maltophilia.
Another mechanism through which filamentous phages vicariously increase the virulence of Gram-negative bacteria has been reported. It has been observed that the extracellular matrix produced by P. aeruginosa self-assembles into a liquid crystalline structure together with filamentous Pf bacteriophages in CF sputum (Secor et al., 2016). These liquid crystals enhance biofilm function by increasing adhesion and preventing the diffusion of antibiotics through biofilms, thereby contributing to increased antibiotic tolerance (Secor et al., 2015, 2016).
No previous studies have reported a similar virulence mechanism in CF respiratory S. maltophilia isolates, and confirmation of this VF, as well as a putative novel form of symbiosis between bacteria and phages, holds potential for further research.
The role of phages in increasing bacterial virulence has been extensively studied (Wagner and Waldor, 2002; Brüssow et al., 2004), however, bacteriophages may also provide a selective pressure against bacteria expressing specific VFs (León and Bastías, 2015). If the phage receptor coincides with a VF, such as lipopolysaccharide or type IV pili, its modification through mutations results in decreased virulence and reduced fitness (León and Bastías, 2015). McCutcheon et al. (2018) have reported that the type IV pilus is the primary receptor for DLP1 and DLP2 bacteriophages that are able to infect both S. maltophilia and P. aeruginosa. Deletion of the primary pilin subunit by inactivation of pilA in S. maltophilia prevented phage binding and subsequent lysis by both bacteriophages, while the mutant strain exhibited reduced virulence. This phenomenon can be thought of as a fitness cost: when a bacterium acquires properties that are beneficial for its specific living conditions, it must compensate for the loss of other, less important properties.
In recent decades, there has been considerable interest in understanding the mechanisms underlying the virulence of S. maltophilia. The intrinsic multidrug resistance of the bacterium, its ability to rapidly adapt to unfavorable environmental conditions and new habitat niches, and its sophisticated switching of metabolic pathways are features that are attracting the attention of experts studying the fundamental mechanisms of virulence as well as clinical researchers.
When considering the virulent properties of S. maltophilia, it is important to keep in mind that the bacterium is characterized by high intraspecific variability: strains isolated in the same hospital and even from the same patient may belong to relatively distant phylogenetic groups and have different phenotypes (Valdezate et al., 2004; Pompilio et al., 2016; Chung et al., 2017). The fast accumulation of adaptive mutations occurring under the selective pressure of hospital conditions or the host cells, as well as horizontal gene transfer, are thought to be possible reasons for such heterogeneity. Understanding the molecular processes that ensure rapid adaptation and, consequently, the survival of the microorganism under adverse conditions will allow the identification of potential targets for the development of novel antibacterial drugs, as well as a better understanding of interspecific interactions in polymicrobial infections and the mechanisms of metabolic switching during the transition of opportunistic pathogens from “natural” lifestyle to infectious intervention.
VM: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. RH: Formal analysis, Writing – original draft, Writing – review & editing. DZ: Visualization, Writing – original draft, Writing – review & editing. IC: Conceptualization, Writing – original draft, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ARGs, Antibiotic resistance genes; c-di-GMP, Cyclic diguanosine monophosphate; CF, Cystic fibrosis; DGC, Diguanylate cyclase; DSF, Diffusible signal factor; EPS, Exopolysaccharides; GMP, Guanosine monophosphate; HGT, Horizontal gene transfer; LPS, Lipopolysaccharides; OMP, Outer membrane protein; OMV, Outer membrane vesicles; PDE, Phosphodiesterase; Smc, Stenotrophomonas maltophilia complex; TCS, Signal transduction system; QS, Quorum Sensing; ROS, Reactive oxygen species; T4P, Type IV pili; VF, A virulence factor
Abbott, I. J., and Peleg, A. Y. (2015). Stenotrophomonas, achromobacter, and nonmelioid burkholderia species: antimicrobial resistance and therapeutic strategies. Semin. Respir. Crit. Care Med. 36, 99–110. doi: 10.1055/s-0034-1396929
Adamek, M., Linke, B., and Schwartz, T. (2014). Virulence genes in clinical and environmental Stenotrophomas maltophilia isolates: a genome sequencing and gene expression approach. Microb. Pathog. 67-68, 20–30. doi: 10.1016/j.micpath.2014.02.001
Ahmad, S., Wang, B., Walker, M. D., Tran, H.-K. R., Stogios, P. J., Savchenko, A., et al. (2019). An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature 575, 674–678. doi: 10.1038/s41586-019-1735-9
Alavi, P., Starcher, M. R., Thallinger, G. G., Zachow, C., Müller, H., and Berg, G. (2014). Stenotrophomonas comparative genomics reveals genes and functions that differentiate beneficial and pathogenic bacteria. BMC Genomics 15:482. doi: 10.1186/1471-2164-15-482
Alcaraz, E., Ghiglione, B., Pineda, M. V., Mangano, A., Di Conza, J., and Passerini de Rossi, B. (2022). AmpR is a dual regulator in Stenotrophomonas maltophilia with a positive role in β-lactam resistance and a negative role in virulence, biofilm and DSF production. Res. Microbiol. 173:103917. doi: 10.1016/j.resmic.2021.103917
Al-Hamad, A., Upton, M., and Burnie, J. (2009). Molecular cloning and characterization of SmrA, a novel ABC multidrug efflux pump from Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 64, 731–734. doi: 10.1093/jac/dkp271
Alio, I., Gudzuhn, M., Pérez García, P., Danso, D., Schoelmerich, M. C., Mamat, U., et al. (2020). Phenotypic and transcriptomic analyses of seven clinical Stenotrophomonas maltophilia isolates identify a small set of shared and commonly regulated genes involved in the biofilm lifestyle. Appl. Environ. Microbiol. 86:20. doi: 10.1128/AEM.02038-20
Alonso, A. (2004). Overexpression of the multidrug efflux pump SmeDEF impairs Stenotrophomonas maltophilia physiology. J. Antimicrob. Chemother. 53, 432–434. doi: 10.1093/jac/dkh074
Alonso, A., and Martínez, J. L. (2000). Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 44, 3079–3086. doi: 10.1128/AAC.44.11.3079-3086.2000
Amanatidou, E., Matthews, A. C., Kuhlicke, U., Neu, T. R., McEvoy, J. P., and Raymond, B. (2019). Biofilms facilitate cheating and social exploitation of β-lactam resistance in Escherichia coli. NPJ Biofilms Microbiomes 5:36. doi: 10.1038/s41522-019-0109-2
An, S., and Tang, J. (2018). The Ax21 protein influences virulence and biofilm formation in Stenotrophomonas maltophilia. Arch. Microbiol. 200, 183–187. doi: 10.1007/s00203-017-1433-7
Antimicrobial Resistance Collaborators (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Arora, S. K., Ritchings, B. W., Almira, E. C., Lory, S., and Ramphal, R. (1997). A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 179, 5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997
Azman, A., Vasodavan, K., Joseph, N., Kumar, S., Hamat, R. A., Nordin, S. A., et al. (2019). Physiological and proteomic analysis of Stenotrophomonas maltophilia grown under the iron-limited condition. Future Microbiol. 14, 1417–1428. doi: 10.2217/fmb-2019-0174
Aznar, R., Alcaide, E., and Garay, E. (1992). Numerical taxonomy of pseudomonads isolated from water, sediment and eels. Syst. Appl. Microbiol. 15, 235–246. doi: 10.1016/S0723-2020(11)80097-4
Bagg, A., and Neilands, J. B. (1987). Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26, 5471–5477. doi: 10.1021/bi00391a039
Basler, M., Pilhofer, M., Henderson, G. P., Jensen, G. J., and Mekalanos, J. J. (2012). Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186. doi: 10.1038/nature10846
Bayer-Santos, E., Cenens, W., Matsuyama, B. Y., Oka, G. U., Di Sessa, G., Mininel, I. D. V., et al. (2019). The opportunistic pathogen Stenotrophomonas maltophilia utilizes a type IV secretion system for interbacterial killing. PLoS Pathog. 15:e1007651. doi: 10.1371/journal.ppat.1007651
Berg, G. (2009). Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18. doi: 10.1007/s00253-009-2092-7
Berg, G., Roskot, N., and Smalla, K. (1999). Genotypic and phenotypic relationships between clinical and environmental isolates of Stenotrophomonas maltophilia. J. Clin. Microbiol. 37, 3594–3600. doi: 10.1128/JCM.37.11.3594-3600.1999
Bi, H., Yu, Y., Dong, H., Wang, H., and Cronan, J. E. (2014). Xanthomonas campestris RpfB is a fatty acyl-CoA ligase required to counteract the thioesterase activity of the RpfF diffusible signal factor (DSF) synthase. Mol. Microbiol. 93, 262–275. doi: 10.1111/mmi.12657
Blanco, P., Corona, F., and Martínez, J. L. (2019). Involvement of the RND efflux pump transporter SmeH in the acquisition of resistance to ceftazidime in Stenotrophomonas maltophilia. Sci. Rep. 9:4917. doi: 10.1038/s41598-019-41308-9
Brooke, J. S. (2012). Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin. Microbiol. Rev. 25, 2–41. doi: 10.1128/CMR.00019-11
Brooke, J. S., Vo, A., Watts, P., and Davis, N. A. (2008). Mutation of a lipopolysaccharide synthesis gene results in increased biofilm Ofstenotrophomonas maltophilia on plastic and glass surfaces. Ann. Microbiol. 58, 35–40. doi: 10.1007/BF03179442
Brüssow, H., Canchaya, C., and Hardt, W.-D. (2004). Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68, 560–602. doi: 10.1128/MMBR.68.3.560-602.2004
Cai, B., Tillotson, G., Benjumea, D., Callahan, P., and Echols, R. (2020). The burden of bloodstream infections due to Stenotrophomonas Maltophilia in the United States: a large, retrospective database study. Open Forum Infect. Dis. 7:141. doi: 10.1093/ofid/ofaa141
Caly, D., Bellini, D., Walsh, M., Dow, J., and Ryan, R. (2014). Targeting cyclic di-GMP Signalling: a strategy to control biofilm formation? Curr. Pharm. Des. 21, 12–24. doi: 10.2174/1381612820666140905124701
Carpenter, B. M., Whitmire, J. M., and Merrell, D. S. (2009). This is not your Mother’s repressor: the complex role of Fur in pathogenesis. Infect. Immun. 77, 2590–2601. doi: 10.1128/IAI.00116-09
Chan, C., Paul, R., Samoray, D., Amiot, N. C., Giese, B., Jenal, U., et al. (2004). Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. 101, 17084–17089. doi: 10.1073/pnas.0406134101
Chang, Y.-T., Lin, C.-Y., Chen, Y.-H., and Hsueh, P.-R. (2015). Update on infections caused by Stenotrophomonas maltophilia with particular attention to resistance mechanisms and therapeutic options. Front. Microbiol. 6:893. doi: 10.3389/fmicb.2015.00893
Chauviat, A., Meyer, T., and Favre-Bonté, S. (2023). Versatility of Stenotrophomonas maltophilia: ecological roles of RND efflux pumps. Heliyon 9:e14639. doi: 10.1016/j.heliyon.2023.e14639
Chen, C.-H., Huang, C.-C., Chung, T.-C., Hu, R.-M., Huang, Y.-W., and Yang, T.-C. (2011). Contribution of resistance-nodulation-division efflux pump operon smeU1-V-W-U2-X to multidrug resistance of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 55, 5826–5833. doi: 10.1128/AAC.00317-11
Cheng, S.-T., Wang, F.-F., and Qian, W. (2019). Cyclic-di-GMP binds to histidine kinase RavS to control RavS-RavR phosphotransfer and regulates the bacterial lifestyle transition between virulence and swimming. PLoS Pathog. 15:e1007952. doi: 10.1371/journal.ppat.1007952
Christen, M., Christen, B., Folcher, M., Schauerte, A., and Jenal, U. (2005). Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280, 30829–30837. doi: 10.1074/jbc.M504429200
Christen, B., Christen, M., Paul, R., Schmid, F., Folcher, M., Jenoe, P., et al. (2006). Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 281, 32015–32024. doi: 10.1074/jbc.M603589200
Christie, P. J., Whitaker, N., and González-Rivera, C. (2014). Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta 1843, 1578–1591. doi: 10.1016/j.bbamcr.2013.12.019
Chung, H., Lieberman, T. D., Vargas, S. O., Flett, K. B., McAdam, A. J., Priebe, G. P., et al. (2017). Global and local selection acting on the pathogen Stenotrophomonas maltophilia in the human lung. Nat. Commun. 8:14078. doi: 10.1038/ncomms14078
Coutinho, H., Falcão-Silva, V. S., and Gonçalves, G. (2008). Pulmonary bacterial pathogens in cystic fibrosis patients and antibiotic therapy: a tool for the health workers. Int. Arch. Med. 1:24. doi: 10.1186/1755-7682-1-24
Coves, X., Bravo, M., Huedo, P., Conchillo-Solé, Ò., Gómez, A.-C., Esteve-Codina, A., et al. (2023). A Stenotrophomonas maltophilia TetR-like transcriptional regulator involved in fatty acid metabolism is controlled by quorum sensing signals. Appl. Environ. Microbiol. 89:e0063523. doi: 10.1128/aem.00635-23
Crisan, C. V., and Goldberg, J. B. (2022). Antibacterial contact-dependent proteins secreted by gram-negative cystic fibrosis respiratory pathogens. Trends Microbiol. 30, 986–996. doi: 10.1016/j.tim.2022.03.009
Crisan, C. V., Van Tyne, D., and Goldberg, J. B. (2023). The type VI secretion system of the emerging pathogen Stenotrophomonas maltophilia complex has antibacterial properties. mSphere 8:23. doi: 10.1128/msphere.00584-23
Crossman, L. C., Gould, V. C., Dow, J. M., Vernikos, G. S., Okazaki, A., Sebaihia, M., et al. (2008). The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 9:R74. doi: 10.1186/gb-2008-9-4-r74
Cruz-Córdova, A., Mancilla-Rojano, J., Luna-Pineda, V. M., Escalona-Venegas, G., Cázares-Domínguez, V., Ormsby, C., et al. (2020). Molecular epidemiology, antibiotic resistance, and virulence traits of Stenotrophomonas maltophilia strains associated with an outbreak in a Mexican tertiary care hospital. Front. Cell. Infect. Microbiol. 10:50. doi: 10.3389/fcimb.2020.00050
Cuthbertson, L., Walker, A. W., Oliver, A. E., Rogers, G. B., Rivett, D. W., Hampton, T. H., et al. (2020). Lung function and microbiota diversity in cystic fibrosis. Microbiome 8:45. doi: 10.1186/s40168-020-00810-3
de Kievit, T. R. (2009). Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 11, 279–288. doi: 10.1111/j.1462-2920.2008.01792.x
de Oliveira-Garcia, D., Dall’Agnol, M., Rosales, M., Azzuz, A. C. G. S., Alcántara, N., Martinez, M. B., et al. (2003). Fimbriae and adherence of Stenotrophomonas maltophilia to epithelial cells and to abiotic surfaces. Cell. Microbiol. 5, 625–636. doi: 10.1046/j.1462-5822.2003.00306.x
de Oliveira-Garcia, D., Dall’Agnol, M., Rosales, M., Azzuz, A. C. G. S., Martinez, M. B., and Girón, J. A. (2002). Characterization of flagella produced by clinical strains of Stenotrophomonas maltophilia. Emerg. Infect. Dis. 8, 918–923. doi: 10.3201/eid0809.010535
Denton, M., and Kerr, K. G. (1998). Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 11, 57–80. doi: 10.1128/CMR.11.1.57
DeShazer, D., Brett, P. J., and Woods, D. E. (1998). The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol. Microbiol. 30, 1081–1100. doi: 10.1046/j.1365-2958.1998.01139.x
Devos, S., Van Oudenhove, L., Stremersch, S., Van Putte, W., De Rycke, R., Van Driessche, G., et al. (2015). The effect of imipenem and diffusible signaling factors on the secretion of outer membrane vesicles and associated Ax21 proteins in Stenotrophomonas maltophilia. Front. Microbiol. 6:298. doi: 10.3389/fmicb.2015.00298
Di Bonaventura, G., Picciani, C., Lupetti, V., and Pompilio, A. (2023). Comparative proteomic analysis of protein patterns of Stenotrophomonas maltophilia in biofilm and planktonic lifestyles. Microorganisms 11:442. doi: 10.3390/microorganisms11020442
Di Bonaventura, G., Prosseda, G., Del Chierico, F., Cannavacciuolo, S., Cipriani, P., Petrucca, A., et al. (2007). Molecular characterization of virulence determinants of Stenotrophomonas Maltophilia strains isolated from patients affected by cystic fibrosis. Int. J. Immunopathol. Pharmacol. 20, 529–537. doi: 10.1177/039463200702000311
Di Bonaventura, G., Spedicato, I., D’Antonio, D., Robuffo, I., and Piccolomini, R. (2004). Biofilm formation by Stenotrophomonas maltophilia: modulation by quinolones, trimethoprim-sulfamethoxazole, and ceftazidime. Antimicrob. Agents Chemother. 48, 151–160. doi: 10.1128/AAC.48.1.151-160.2004
Doig, P., Todd, T., Sastry, P. A., Lee, K. K., Hodges, R. S., Paranchych, W., et al. (1988). Role of pili in adhesion of Pseudomonas aeruginosa to human respiratory epithelial cells. Infect. Immun. 56, 1641–1646. doi: 10.1128/iai.56.6.1641-1646.1988
Dörr, T., Lewis, K., and Vulić, M. (2009). SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 5:e1000760. doi: 10.1371/journal.pgen.1000760
Driffield, K., Miller, K., Bostock, J. M., O’Neill, A. J., and Chopra, I. (2008). Increased mutability of Pseudomonas aeruginosa in biofilms. J. Antimicrob. Chemother. 61, 1053–1056. doi: 10.1093/jac/dkn044
Duan, Q., Zhou, M., Zhu, L., and Zhu, G. (2013). Flagella and bacterial pathogenicity. J. Basic Microbiol. 53, 1–8. doi: 10.1002/jobm.201100335
Dulyayangkul, P., Charoenlap, N., Srijaruskul, K., Mongkolsuk, S., and Vattanaviboon, P. (2016). Major facilitator superfamily MfsA contributes to multidrug resistance in emerging nosocomial pathogen Stenotrophomonas maltophilia: table 1. J. Antimicrob. Chemother. 71, 2990–2991. doi: 10.1093/jac/dkw233
DuMont, A. L., and Cianciotto, N. P. (2017). Stenotrophomonas maltophilia serine protease StmPr1 induces Matrilysis, Anoikis, and protease-activated receptor 2 activation in human lung epithelial cells. Infect. Immun. 85:17. doi: 10.1128/IAI.00544-17
DuMont, A. L., Karaba, S. M., and Cianciotto, N. P. (2015). Type II secretion-dependent degradative and cytotoxic activities mediated by Stenotrophomonas maltophilia serine proteases StmPr1 and StmPr2. Infect. Immun. 83, 3825–3837. doi: 10.1128/IAI.00672-15
Escolar, L., Pérez-Martín, J., and de Lorenzo, V. (1999). Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181, 6223–6229. doi: 10.1128/JB.181.20.6223-6229.1999
Faraldo-Gómez, J. D., and Sansom, M. S. P. (2003). Acquisition of siderophores in gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 4, 105–116. doi: 10.1038/nrm1015
Ferrer-Navarro, M., Planell, R., Yero, D., Mongiardini, E., Torrent, G., Huedo, P., et al. (2013). Abundance of the quorum-sensing factor Ax21 in four strains of Stenotrophomonas maltophilia correlates with mortality rate in a new zebrafish model of infection. PLoS One 8:e67207. doi: 10.1371/journal.pone.0067207
Flores-Treviño, S., Bocanegra-Ibarias, P., Camacho-Ortiz, A., Morfín-Otero, R., Salazar-Sesatty, H. A., and Garza-González, E. (2019). Stenotrophomonas maltophilia biofilm: its role in infectious diseases. Expert Rev. Anti Infect. Ther. 17, 877–893. doi: 10.1080/14787210.2019.1685875
Fluit, A. C., Bayjanov, J. R., Aguilar, M. D., Cantón, R., Elborn, S., Tunney, M. M., et al. (2022). Taxonomic position, antibiotic resistance and virulence factor production by Stenotrophomonas isolates from patients with cystic fibrosis and other chronic respiratory infections. BMC Microbiol. 22:129. doi: 10.1186/s12866-022-02466-5
Gallo, S. W., Figueiredo, T. P., Bessa, M. C., Pagnussatti, V. E., Ferreira, C. A. S., and Oliveira, S. D. (2016). Isolation and characterization of Stenotrophomonas maltophilia isolates from a Brazilian hospital. Microb. Drug Resist. 22, 688–695. doi: 10.1089/mdr.2015.0306
García, C. A., Alcaraz, E. S., Franco, M. A., and Passerini de Rossi, B. N. (2015). Iron is a signal for Stenotrophomonas maltophilia biofilm formation, oxidative stress response, OMPs expression, and virulence. Front. Microbiol. 6:926. doi: 10.3389/fmicb.2015.00926
García-León, G., de Alegría, R., Puig, C., García de la Fuente, C., Martínez-Martínez, L., Martínez, J. L., et al. (2015). High-level quinolone resistance is associated with the overexpression of smeVWX in Stenotrophomonas maltophilia clinical isolates. Clin. Microbiol. Infect. 21, 464–467. doi: 10.1016/j.cmi.2015.01.007
García-León, G., Hernández, A., Hernando-Amado, S., Alavi, P., Berg, G., and Martínez, J. L. (2014). A function of SmeDEF, the major quinolone resistance determinant of Stenotrophomonas maltophilia, is the colonization of plant roots. Appl. Environ. Microbiol. 80, 4559–4565. doi: 10.1128/AEM.01058-14
Ghosal, D., Chang, Y., Jeong, K. C., Vogel, J. P., and Jensen, G. J. (2017). In situ structure of the Legionella dot/Icm type IV secretion system by electron cryotomography. EMBO Rep. 18, 726–732. doi: 10.15252/embr.201643598
Gil-Gil, T., Martínez, J. L., and Blanco, P. (2020). Mechanisms of antimicrobial resistance in Stenotrophomonas maltophilia: a review of current knowledge. Expert Rev. Anti Infect. Ther. 18, 335–347. doi: 10.1080/14787210.2020.1730178
Giltner, C. L., Nguyen, Y., and Burrows, L. L. (2012). Type IV Pilin Proteins: Versatile Molecular Modules. Microbiol. Mol. Biol. Rev. 76, 740–772. doi: 10.1128/MMBR.00035-12
Goldberg, J. B., Coyne, M. J., Neely, A. N., and Holder, I. A. (1995). Avirulence of a Pseudomonas aeruginosa algC mutant in a burned-mouse model of infection. Infect. Immun. 63, 4166–4169. doi: 10.1128/iai.63.10.4166-4169.1995
González, J. F., Hahn, M. M., and Gunn, J. S. (2018). Chronic biofilm-based infections: skewing of the immune response. Pathog. Dis. 76:23. doi: 10.1093/femspd/fty023
Gonzalez-Rivera, C., Bhatty, M., and Christie, P. J. (2016). Mechanism and function of type IV secretion during infection of the human host. Microbiol. Spectr. 4:2015. doi: 10.1128/microbiolspec.VMBF-0024-2015
Goss, C. H., Mayer-Hamblett, N., Aitken, M. L., Rubenfeld, G. D., and Ramsey, B. W. (2004). Association between Stenotrophomonas maltophilia and lung function in cystic fibrosis. Thorax 59, 955–959. doi: 10.1136/thx.2003.017707
Grohmann, E., Christie, P. J., Waksman, G., and Backert, S. (2018). Type IV secretion in gram-negative and gram-positive bacteria. Mol. Microbiol. 107, 455–471. doi: 10.1111/mmi.13896
Gröschel, M. I., Meehan, C. J., Barilar, I., Diricks, M., Gonzaga, A., Steglich, M., et al. (2020). The phylogenetic landscape and nosocomial spread of the multidrug-resistant opportunist Stenotrophomonas maltophilia. Nat. Commun. 11:2044. doi: 10.1038/s41467-020-15123-0
Hachani, A., Wood, T. E., and Filloux, A. (2016). Type VI secretion and anti-host effectors. Curr. Opin. Microbiol. 29, 81–93. doi: 10.1016/j.mib.2015.11.006
Hagemann, M., Hasse, D., and Berg, G. (2006). Detection of a phage genome carrying a zonula occludens like toxin gene (zot) in clinical isolates of Stenotrophomonas maltophilia. Arch. Microbiol. 185, 449–458. doi: 10.1007/s00203-006-0115-7
Hall-Stoodley, L., Costerton, J. W., and Stoodley, P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108. doi: 10.1038/nrmicro821
Han, Y., Wang, T., Chen, G., Pu, Q., Liu, Q., Zhang, Y., et al. (2019). A Pseudomonas aeruginosa type VI secretion system regulated by CueR facilitates copper acquisition. PLoS Pathog. 15:e1008198. doi: 10.1371/journal.ppat.1008198
Heiniger, R. W., Winther-Larsen, H. C., Pickles, R. J., Koomey, M., and Wolfgang, M. C. (2010). Infection of human mucosal tissue by Pseudomonas aeruginosa requires sequential and mutually dependent virulence factors and a novel pilus-associated adhesin. Cell. Microbiol. 12, 1158–1173. doi: 10.1111/j.1462-5822.2010.01461.x
Hengge, R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273. doi: 10.1038/nrmicro2109
Hernando-Amado, S., Coque, T. M., Baquero, F., and Martínez, J. L. (2019). Defining and combating antibiotic resistance from one health and Global Health perspectives. Nat. Microbiol. 4, 1432–1442. doi: 10.1038/s41564-019-0503-9
Hickman, J. W., and Harwood, C. S. (2008). Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69, 376–389. doi: 10.1111/j.1365-2958.2008.06281.x
Hickman, J. W., Tifrea, D. F., and Harwood, C. S. (2005). A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. 102, 14422–14427. doi: 10.1073/pnas.0507170102
Ho, B. T., Dong, T. G., and Mekalanos, J. J. (2014). A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15, 9–21. doi: 10.1016/j.chom.2013.11.008
Høiby, N., Bjarnsholt, T., Givskov, M., Molin, S., and Ciofu, O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332. doi: 10.1016/j.ijantimicag.2009.12.011
Hu, R.-M., Liao, S.-T., Huang, C.-C., Huang, Y.-W., and Yang, T.-C. (2012). An inducible Fusaric acid tripartite efflux pump contributes to the Fusaric acid resistance in Stenotrophomonas maltophilia. PLoS One 7:e51053. doi: 10.1371/journal.pone.0051053
Huang, Y.-W., Hu, R.-M., Chu, F.-Y., Lin, H.-R., and Yang, T.-C. (2013). Characterization of a major facilitator superfamily (MFS) tripartite efflux pump EmrCABsm from Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 68, 2498–2505. doi: 10.1093/jac/dkt250
Huang, Y.-W., Huang, H.-H., Huang, K.-H., Chen, W.-C., Lin, Y.-T., Hsu, C.-C., et al. (2019). AmpI functions as an Iron exporter to alleviate β-lactam-mediated reactive oxygen species stress in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 63:18. doi: 10.1128/AAC.02467-18
Huang, T.-P., and Lee Wong, A. C. (2007a). Extracellular fatty acids facilitate flagella-independent translocation by Stenotrophomonas maltophilia. Res. Microbiol. 158, 702–711. doi: 10.1016/j.resmic.2007.09.002
Huang, T. P., and Lee Wong, A. C. (2007b). A cyclic AMP receptor protein-regulated cell-cell communication system mediates expression of a FecA homologue in Stenotrophomonas maltophilia. Appl. Environ. Microbiol. 73, 5034–5040. doi: 10.1128/AEM.00366-07
Huang, Y.-W., Liou, R.-S., Lin, Y.-T., Huang, H.-H., and Yang, T.-C. (2014). A linkage between SmeIJK efflux pump, cell envelope integrity, and σE-mediated envelope stress response in Stenotrophomonas maltophilia. PLoS One 9:e111784. doi: 10.1371/journal.pone.0111784
Huang, T.-P., Somers, E. B., and Wong, A. C. L. (2006). Differential biofilm formation and motility associated with lipopolysaccharide/exopolysaccharide-coupled biosynthetic genes in Stenotrophomonas maltophilia. J. Bacteriol. 188, 3116–3120. doi: 10.1128/JB.188.8.3116-3120.2006
Huedo, P., Kumar, V. P., Horgan, C., Yero, D., Daura, X., Gibert, I., et al. (2019). Sulfonamide-based diffusible signal factor analogs interfere with quorum sensing in Stenotrophomonas maltophilia and Burkholderia cepacia. Future Med. Chem. 11, 1565–1582. doi: 10.4155/fmc-2019-0015
Huedo, P., Yero, D., Martínez-Servat, S., Estibariz, I., Planell, R., Martínez, P., et al. (2014). Two different rpf clusters distributed among a population of Stenotrophomonas maltophilia clinical strains display differential diffusible signal factor production and virulence regulation. J. Bacteriol. 196, 2431–2442. doi: 10.1128/JB.01540-14
Huedo, P., Yero, D., Martinez-Servat, S., Ruyra, À., Roher, N., Daura, X., et al. (2015). Decoding the genetic and functional diversity of the DSF quorum-sensing system in Stenotrophomonas maltophilia. Front. Microbiol. 6:761. doi: 10.3389/fmicb.2015.00761
Ishikawa, K., Nakamura, T., Kawai, F., Uehara, Y., and Mori, N. (2022). Stenotrophomonas maltophilia infection associated with COVID-19: a case series and literature review. Am. J. Case Rep. 23:e936889. doi: 10.12659/AJCR.936889
Ivanov, V., Stabnikov, V., Zhuang, W. Q., Tay, J. H., and Tay, S. T. L. (2005). Phosphate removal from the returned liquor of municipal wastewater treatment plant using iron-reducing bacteria. J. Appl. Microbiol. 98, 1152–1161. doi: 10.1111/j.1365-2672.2005.02567.x
Jakobi, M., Winkelmann, G., Kaiser, D., Kempler, C., Jung, G., Berg, G., et al. (1996). Maltophilin: a new antifungal compound produced by Stenotrophomonas maltophilia R3089. J. Antibiot. 49, 1101–1104. doi: 10.7164/antibiotics.49.1101
Jefferson, K. (2004). What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236, 163–173. doi: 10.1016/j.femsle.2004.06.005
Johnson, E. H., Al-Busaidy, R., and Hameed, M. S. (2003). An outbreak of lymphadenitis associated with Stenotrophomonas (Xanthomonas) maltophilia in Omani goats. J. Vet. Med. B Infect. Dis. Vet. Public Health 50, 102–104. doi: 10.1046/j.1439-0450.2003.00643.x
Johnson, P. J. T., and Levin, B. R. (2013). Pharmacodynamics, population dynamics, and the evolution of persistence in Staphylococcus aureus. PLoS Genet. 9:e1003123. doi: 10.1371/journal.pgen.1003123
Jucker, B. A., Harms, H., and Zehnder, A. J. B. (1996). Adhesion of the positively charged bacterium Stenotrophomonas (Xanthomonas) maltophilia 70401 to glass and Teflon. J. Bacteriol. 178, 5472–5479. doi: 10.1128/jb.178.18.5472-5479.1996
Jurado, R. L. (1997). Iron, infections, and anemia of inflammation. Clin. Infect. Dis. 25, 888–895. doi: 10.1086/515549
Jurkevitch, E., Hadar, Y., and Chen, Y. (1992). Differential Siderophore utilization and Iron uptake by soil and rhizosphere Bacteria. Appl. Environ. Microbiol. 58, 119–124. doi: 10.1128/aem.58.1.119-124.1992
Kalidasan, V., Joseph, N., Kumar, S., Awang Hamat, R., and Neela, V. K. (2018). Iron and virulence in Stenotrophomonas Maltophilia: all we know So far. Front. Cell. Infect. Microbiol. 8:401. doi: 10.3389/fcimb.2018.00401
Kalidasan, V., and Neela, V. K. (2020). Twitching motility of Stenotrophomonas maltophilia under iron limitation: in-silico, phenotypic and proteomic approaches. Virulence 11, 104–112. doi: 10.1080/21505594.2020.1713649
Kang, X.-M., Wang, F.-F., Zhang, H., Zhang, Q., and Qian, W. (2015). Genome-wide identification of genes necessary for biofilm formation by nosocomial pathogen Stenotrophomonas maltophilia reveals that orphan response regulator FsnR is a critical modulator. Appl. Environ. Microbiol. 81, 1200–1209. doi: 10.1128/AEM.03408-14
Karaba, S. M., White, R. C., and Cianciotto, N. P. (2013). Stenotrophomonas maltophilia encodes a type II protein secretion system that promotes detrimental effects on lung epithelial cells. Infect. Immun. 81, 3210–3219. doi: 10.1128/IAI.00546-13
Kim, H.-R., Lee, D., and Eom, Y.-B. (2018). Anti-biofilm and anti-virulence efficacy of Celastrol against Stenotrophomonas maltophilia. Int. J. Med. Sci. 15, 617–627. doi: 10.7150/ijms.23924
Klose, K. E., and Mekalanos, J. J. (1998). Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28, 501–520. doi: 10.1046/j.1365-2958.1998.00809.x
Köplin, R., Arnold, W., Hötte, B., Simon, R., Wang, G., and Pühler, A. (1992). Genetics of xanthan production in Xanthomonas campestris: the xanA and xanB genes are involved in UDP-glucose and GDP-mannose biosynthesis. J. Bacteriol. 174, 191–199. doi: 10.1128/jb.174.1.191-199.1992
Kumar, S., Bansal, K., Patil, P. P., Kaur, A., Kaur, S., Jaswal, V., et al. (2020). Genomic insights into evolution of extensive drug resistance in Stenotrophomonas maltophilia complex. Genomics 112, 4171–4178. doi: 10.1016/j.ygeno.2020.06.049
Kwan, B. W., Valenta, J. A., Benedik, M. J., and Wood, T. K. (2013). Arrested protein synthesis increases persister-like cell formation. Antimicrob. Agents Chemother. 57, 1468–1473. doi: 10.1128/AAC.02135-12
Langford, B. J., So, M., Simeonova, M., Leung, V., Lo, J., Kan, T., et al. (2023). Antimicrobial resistance in patients with COVID-19: a systematic review and meta-analysis. Lancet Microbe 4, e179–e191. doi: 10.1016/S2666-5247(22)00355-X
Lee, D.-H., Cha, J.-H., Kim, D.-W., Lee, K., Kim, Y.-S., Oh, H.-Y., et al. (2022). Colistin-degrading proteases confer collective resistance to microbial communities during polymicrobial infections. Microbiome 10:129. doi: 10.1186/s40168-022-01315-x
Lee, J.-W., and Helmann, J. D. (2007). Functional specialization within the Fur family of metalloregulators. Biometals 20, 485–499. doi: 10.1007/s10534-006-9070-7
Lee, E. Y., Jun, Y. S., Cho, K.-S., and Ryu, H. W. (2002). Degradation characteristics of toluene, benzene, ethylbenzene, and xylene by Stenotrophomonas maltophilia T3-c. J. Air Waste Manag. Assoc. 52, 400–406. doi: 10.1080/10473289.2002.10470796
León, M., and Bastías, R. (2015). Virulence reduction in bacteriophage resistant bacteria. Front. Microbiol. 6:343. doi: 10.3389/fmicb.2015.00343
Lesic, B., Starkey, M., He, J., Hazan, R., and Rahme, L. G. (2009). Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology 155, 2845–2855. doi: 10.1099/mic.0.029082-0
Letunic, I., and Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Li, K., Yu, K., Huang, Z., Liu, X., Mei, L., Ren, X., et al. (2024). Stenotrophomonas maltophilia complex: insights into evolutionary relationships, global distribution and pathogenicity. Front. Cell. Infect. Microbiol. 13:379. doi: 10.3389/fcimb.2023.1325379
Li, X.-Z., Zhang, L., and Poole, K. (2002). SmeC, an outer membrane multidrug efflux protein of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 46, 333–343. doi: 10.1128/AAC.46.2.333-343.2002
Li, L.-H., Zhang, M.-S., Wu, C.-J., Lin, Y.-T., and Yang, T.-C. (2019). Overexpression of SmeGH contributes to the acquired MDR of Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 74, 2225–2229. doi: 10.1093/jac/dkz200
Li, C., Zhu, L., Wang, D., Wei, Z., Hao, X., Wang, Z., et al. (2022). T6SS secretes an LPS-binding effector to recruit OMVs for exploitative competition and horizontal gene transfer. ISME J. 16, 500–510. doi: 10.1038/s41396-021-01093-8
Liao, C.-H., Chen, W.-C., Li, L.-H., Lin, Y.-T., Pan, S.-Y., and Yang, T.-C. (2020). AmpR of Stenotrophomonas maltophilia is involved in stenobactin synthesis and enhanced β-lactam resistance in an iron-depleted condition. J. Antimicrob. Chemother. 75, 3544–3551. doi: 10.1093/jac/dkaa358
Liao, C.-H., Lu, H.-F., Huang, H.-H., Chen, Y., Li, L.-H., Lin, Y.-T., et al. (2022). The fciTABC and feoABI systems contribute to ferric citrate acquisition in Stenotrophomonas maltophilia. J. Biomed. Sci. 29:26. doi: 10.1186/s12929-022-00809-y
Lin, Y.-T., Huang, Y.-W., Chen, S.-J., Chang, C.-W., and Yang, T.-C. (2015). The SmeYZ efflux pump of Stenotrophomonas maltophilia contributes to drug resistance, virulence-related characteristics, and virulence in mice. Antimicrob. Agents Chemother. 59, 4067–4073. doi: 10.1128/AAC.00372-15
Lin, C.-W., Huang, Y.-W., Hu, R.-M., and Yang, T.-C. (2014). SmeOP-TolC Sm efflux pump contributes to the multidrug resistance of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 58, 2405–2408. doi: 10.1128/AAC.01974-13
Lin, Y.-T., Huang, Y.-W., Liou, R.-S., Chang, Y.-C., and Yang, T.-C. (2014). MacABCsm, an ABC-type tripartite efflux pump of Stenotrophomonas maltophilia involved in drug resistance, oxidative and envelope stress tolerances and biofilm formation. J. Antimicrob. Chemother. 69, 3221–3226. doi: 10.1093/jac/dku317
Lin, J., Zhang, W., Cheng, J., Yang, X., Zhu, K., Wang, Y., et al. (2017). A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat. Commun. 8:14888. doi: 10.1038/ncomms14888
Liu, W., Tian, X.-Q., Wei, J.-W., Ding, L.-L., Qian, W., Liu, Z., et al. (2017). BsmR degrades c-di-GMP to modulate biofilm formation of nosocomial pathogen Stenotrophomonas maltophilia. Sci. Rep. 7:4665. doi: 10.1038/s41598-017-04763-w
Looney, W. J. (2005). Role of Stenotrophomonas maltophilia in hospital-acquired infection. Br. J. Biomed. Sci. 62, 145–154. doi: 10.1080/09674845.2005.11732702
Ma, L.-S., Hachani, A., Lin, J.-S., Filloux, A., and Lai, E.-M. (2014). Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for Interbacterial competition in planta. Cell Host Microbe 16, 94–104. doi: 10.1016/j.chom.2014.06.002
Madi, H., Lukić, J., Vasiljević, Z., Biočanin, M., Kojić, M., Jovčić, B., et al. (2016). Genotypic and phenotypic characterization of Stenotrophomonas maltophilia strains from a pediatric tertiary Care Hospital in Serbia. PLoS One 11:e0165660. doi: 10.1371/journal.pone.0165660
Mahenthiralingam, E., Campbell, M. E., and Speert, D. P. (1994). Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect. Immun. 62, 596–605. doi: 10.1128/iai.62.2.596-605.1994
Martínez, P., Huedo, P., Martinez-Servat, S., Planell, R., Ferrer-Navarro, M., Daura, X., et al. (2015). Stenotrophomonas maltophilia responds to exogenous AHL signals through the LuxR solo SmoR (Smlt1839). Front. Cell. Infect. Microbiol. 5:41. doi: 10.3389/fcimb.2015.00041
McCall, I. C., Shah, N., Govindan, A., Baquero, F., and Levin, B. R. (2019). Antibiotic killing of diversely generated populations of nonreplicating Bacteria. Antimicrob. Agents Chemother. 63:18. doi: 10.1128/AAC.02360-18
McCutcheon, J. G., Peters, D. L., and Dennis, J. J. (2018). Identification and characterization of type IV pili as the cellular receptor of broad host range Stenotrophomonas maltophilia bacteriophages DLP1 and DLP2. Viruses 10:338. doi: 10.3390/v10060338
McKay, G. A., Woods, D. E., MacDonald, K. L., and Poole, K. (2003). Role of phosphoglucomutase of Stenotrophomonas maltophilia in lipopolysaccharide biosynthesis, virulence, and antibiotic resistance. Infect. Immun. 71, 3068–3075. doi: 10.1128/IAI.71.6.3068-3075.2003
Menetrey, Q., Sorlin, P., Jumas-Bilak, E., Chiron, R., Dupont, C., and Marchandin, H. (2021). Achromobacter xylosoxidans and Stenotrophomonas maltophilia: emerging pathogens well-armed for life in the cystic fibrosis patients’ lung. Genes 12:610. doi: 10.3390/genes12050610
Moens, S., and Vanderleyden, J. (1996). Functions of bacterial flagella. Crit. Rev. Microbiol. 22, 67–100. doi: 10.3109/10408419609106456
Molloy, K., Smith, S. G., Cagney, G., Dillon, E. T., Greene, C. M., and McElvaney, N. G. (2019). Characterisation of the major extracellular proteases of Stenotrophomoas maltophilia and their effects on pulmonary Antiproteases. Pathogens 8:92. doi: 10.3390/pathogens8030092
Moscoso, J. A., Mikkelsen, H., Heeb, S., Williams, P., and Filloux, A. (2011). The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13, 3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393
Mukherjee, P., and Roy, P. (2013). Persistent organic pollutants induced protein expression and immunocrossreactivity by Stenotrophomonas maltophilia PM102: a prospective bioremediating candidate. Biomed. Res. Int. 2013:714232. doi: 10.1155/2013/714232
Nakatsu, C. H., Fulthorpe, R. R., Holland, B. A., Peel, M. C., and Wyndham, R. C. (1995). The phylogenetic distribution of a transposable dioxygenase from the Niagara River watershed. Mol. Ecol. 4, 593–604. doi: 10.1111/j.1365-294x.1995.tb00259.x
Nas, M. Y., and Cianciotto, N. P. (2017). Stenotrophomonas maltophilia produces an EntC-dependent catecholate siderophore that is distinct from enterobactin. Microbiology 163, 1590–1603. doi: 10.1099/mic.0.000545
Nas, M. Y., Gabell, J., and Cianciotto, N. P. (2021). Effectors of the Stenotrophomonas maltophilia type IV secretion system mediate killing of clinical isolates of Pseudomonas aeruginosa. MBio 12:e0150221. doi: 10.1128/mBio.01502-21
Nas, M. Y., White, R. C., DuMont, A. L., Lopez, A. E., and Cianciotto, N. P. (2019). Stenotrophomonas maltophilia encodes a VirB/VirD4 type IV secretion system that modulates apoptosis in human cells and promotes competition against heterologous Bacteria, including Pseudomonas aeruginosa. Infect. Immun. 87:19. doi: 10.1128/IAI.00457-19
Neal, D. J., and Wilkinson, S. G. (1982). Lipopolysaccharides from Pseudomonas maltophilia structural studies of the Side-chain, Core, and lipid-a regions of the lipopolysaccharide from strain NCTC 10257. Eur. J. Biochem. 128, 143–149. doi: 10.1111/j.1432-1033.1982.tb06944.x
Nicoletti, M., Iacobino, A., Prosseda, G., Fiscarelli, E., Zarrilli, R., De Carolis, E., et al. (2011). Stenotrophomonas maltophilia strains from cystic fibrosis patients: genomic variability and molecular characterization of some virulence determinants. Int. J. Med. Microbiol. 301, 34–43. doi: 10.1016/j.ijmm.2010.07.003
Nolan, L. M., Cain, A. K., Clamens, T., Furniss, R. C. D., Manoli, E., Sainz-Polo, M. A., et al. (2021). Identification of Tse8 as a type VI secretion system toxin from Pseudomonas aeruginosa that targets the bacterial transamidosome to inhibit protein synthesis in prey cells. Nat. Microbiol. 6, 1199–1210. doi: 10.1038/s41564-021-00950-8
Ochoa-Sánchez, L. E., and Vinuesa, P. (2017). Evolutionary genetic analysis uncovers multiple species with distinct habitat preferences and antibiotic resistance phenotypes in the Stenotrophomonas maltophilia complex. Front. Microbiol. 8:1548. doi: 10.3389/fmicb.2017.01548
Okazaki, A., and Avison, M. B. (2008). Induction of L1 and L2 β-lactamase production in Stenotrophomonas maltophilia is dependent on an AmpR-type regulator. Antimicrob. Agents Chemother. 52, 1525–1528. doi: 10.1128/AAC.01485-07
Pan, S.-Y., Shih, Y.-L., Huang, H.-H., Li, L.-H., Lin, Y.-T., and Yang, T.-C. (2022). The involvement of PacIRA system of Stenotrophomonas maltophilia in the uptake of Pseudomonas aeruginosa pyochelin and intraspecies competition for iron acquisition. J. Microbiol. Immunol. Infect. 55, 273–281. doi: 10.1016/j.jmii.2021.03.001
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal–response systems in gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Park, H.-J., Lee, S.-W., and Han, S.-W. (2014). Proteomic and functional analyses of a novel porin-like protein in Xanthomonas oryzae pv. Oryzae. J. Microbiol. 52, 1030–1035. doi: 10.1007/s12275-014-4442-0
Paul, D., Gopal, J., Kumar, M., and Manikandan, M. (2018). Nature to the natural rescue: silencing microbial chats. Chem. Biol. Interact. 280, 86–98. doi: 10.1016/j.cbi.2017.12.018
Perault, A. I., Chandler, C. E., Rasko, D. A., Ernst, R. K., Wolfgang, M. C., and Cotter, P. A. (2020). Host adaptation predisposes Pseudomonas aeruginosa to type VI secretion system-mediated predation by the Burkholderia cepacia complex. Cell Host Microbe 28, 534–547.e3. doi: 10.1016/j.chom.2020.06.019
Pompilio, A., Crocetta, V., Confalone, P., Nicoletti, M., Petrucca, A., Guarnieri, S., et al. (2010). Adhesion to and biofilm formation on IB3-1 bronchial cells by Stenotrophomonas maltophilia isolates from cystic fibrosis patients. BMC Microbiol. 10:102. doi: 10.1186/1471-2180-10-102
Pompilio, A., Crocetta, V., and Di Bonaventura, G. (2018). Stenotrophomonas maltophilia mutant lacking flagella remains virulent in DBA/2N mice but is less efficient in stimulating TNF-α expression. FEMS Microbiol. Lett. 365:205. doi: 10.1093/femsle/fny205
Pompilio, A., Crocetta, V., Ghosh, D., Chakrabarti, M., Gherardi, G., Vitali, L. A., et al. (2016). Stenotrophomonas maltophilia phenotypic and genotypic diversity during a 10-year colonization in the lungs of a cystic fibrosis patient. Front. Microbiol. 7:1551. doi: 10.3389/fmicb.2016.01551
Pompilio, A., Pomponio, S., Crocetta, V., Gherardi, G., Verginelli, F., Fiscarelli, E., et al. (2011). Phenotypic and genotypic characterization of Stenotrophomonas maltophiliaisolates from patients with cystic fibrosis: genome diversity, biofilm formation, and virulence. BMC Microbiol. 11:159. doi: 10.1186/1471-2180-11-159
Pompilio, A., Savini, V., Fiscarelli, E., Gherardi, G., and Di Bonaventura, G. (2020). Clonal diversity, biofilm formation, and antimicrobial resistance among Stenotrophomonas maltophilia strains from cystic fibrosis and non-cystic fibrosis patients. Antibiotics 9:15. doi: 10.3390/antibiotics9010015
Ramos-Hegazy, L., Chakravarty, S., and Anderson, G. G. (2020). Phosphoglycerate mutase affects Stenotrophomonas maltophilia attachment to biotic and abiotic surfaces. Microbes Infect. 22, 60–64. doi: 10.1016/j.micinf.2019.08.001
Ribitsch, D., Heumann, S., Karl, W., Gerlach, J., Leber, R., Birner-Gruenberger, R., et al. (2012). Extracellular serine proteases from Stenotrophomonas maltophilia: screening, isolation and heterologous expression in E. coli. J. Biotechnol. 157, 140–147. doi: 10.1016/j.jbiotec.2011.09.025
Ritchings, B. W., Almira, E. C., Lory, S., and Ramphal, R. (1995). Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect. Immun. 63, 4868–4876. doi: 10.1128/iai.63.12.4868-4876.1995
Ro, C., Cashel, M., and Fernández-Coll, L. (2021). The secondary messenger ppGpp interferes with cAMP-CRP regulon by promoting CRP acetylation in Escherichia coli. PLoS One 16:e0259067. doi: 10.1371/journal.pone.0259067
Rocco, F., De Gregorio, E., Colonna, B., and Di Nocera, P. P. (2009). Stenotrophomonas maltophilia genomes: a start-up comparison. Int. J. Med. Microbiol. 299, 535–546. doi: 10.1016/j.ijmm.2009.05.004
Romanenko, L. A., Uchino, M., Tanaka, N., Frolova, G. M., Slinkina, N. N., and Mikhailov, V. V. (2008). Occurrence and antagonistic potential of Stenotrophomonas strains isolated from deep-sea invertebrates. Arch. Microbiol. 189, 337–344. doi: 10.1007/s00203-007-0324-8
Ross, P., Weinhouse, H., Aloni, Y., Michaeli, D., Weinberger-Ohana, P., Mayer, R., et al. (1987). Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325, 279–281. doi: 10.1038/325279a0
Russell, A. B., Hood, R. D., Bui, N. K., LeRoux, M., Vollmer, W., and Mougous, J. D. (2011). Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347. doi: 10.1038/nature10244
Russell, A. B., LeRoux, M., Hathazi, K., Agnello, D. M., Ishikawa, T., Wiggins, P. A., et al. (2013). Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512. doi: 10.1038/nature12074
Ryan, R. P., Fouhy, Y., Garcia, B. F., Watt, S. A., Niehaus, K., Yang, L., et al. (2008). Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol. Microbiol. 68, 75–86. doi: 10.1111/j.1365-2958.2008.06132.x
Ryan, R. P., Monchy, S., Cardinale, M., Taghavi, S., Crossman, L., Avison, M. B., et al. (2009). The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7, 514–525. doi: 10.1038/nrmicro2163
Saiman, L., Ishimoto, K., Lory, S., and Prince, A. (1990). The effect of Piliation and exoproduct expression on the adherence of Pseudomonas aeruginosa to respiratory epithelial monolayers. J Infect Dis 161, 541–548. doi: 10.1093/infdis/161.3.541
Sánchez, M. B., and Martínez, J. L. (2018). Overexpression of the efflux pumps SmeVWX and SmeDEF is a major cause of resistance to co-trimoxazole in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 62:18. doi: 10.1128/AAC.00301-18
Secor, P., Jennings, L., Michaels, L., Sweere, J., Singh, P., Parks, W., et al. (2016). Biofilm assembly becomes crystal clear – filamentous bacteriophage organize the Pseudomonas aeruginosa biofilm matrix into a liquid crystal. Microb. Cell 3, 49–52. doi: 10.15698/mic2016.01.475
Secor, P. R., Sweere, J. M., Michaels, L. A., Malkovskiy, A. V., Lazzareschi, D., Katznelson, E., et al. (2015). Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe 18, 549–559. doi: 10.1016/j.chom.2015.10.013
Shih, Y.-L., Wu, C.-M., Lu, H.-F., Li, L.-H., Lin, Y.-T., and Yang, T.-C. (2022). Involvement of the hemP-hemA-smlt0796-smlt0797 operon in hemin acquisition by Stenotrophomonas maltophilia. Microbiol. Spectr. 10:e0032122. doi: 10.1128/spectrum.00321-22
Shikata, M., Hayashi, N., Fujimoto, A., Nakamura, T., Matsui, N., Ishiyama, A., et al. (2016). The pilT gene contributes to type III ExoS effector injection into epithelial cells in Pseudomonas aeruginosa. J. Infect. Chemother. 22, 216–220. doi: 10.1016/j.jiac.2015.12.012
Si, M., Zhao, C., Burkinshaw, B., Zhang, B., Wei, D., Wang, Y., et al. (2017). Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc. Natl. Acad. Sci. 114, E2233–E2242. doi: 10.1073/pnas.1614902114
Singh, A., Kumar, S., Bansal, K., and Patil, P. B. (2023). Taxonomic and Phylogenomic assessment identifies Phytopathogenicity potential of Stenotrophomonas maltophilia complex. Phytopathology 113, 1833–1838. doi: 10.1094/PHYTO-11-22-0434-SC
Srijaruskul, K., Charoenlap, N., Namchaiw, P., Chattrakarn, S., Giengkam, S., Mongkolsuk, S., et al. (2015). Regulation by SoxR of mfsA, which encodes a major facilitator protein involved in Paraquat resistance in Stenotrophomonas maltophilia. PLoS One 10:e0123699. doi: 10.1371/journal.pone.0123699
Stewart, B. J., and McCarter, L. L. (1996). Vibrio parahaemolyticus FlaJ, a homologue of FliS, is required for production of a flagellin. Mol. Microbiol. 20, 137–149. doi: 10.1111/j.1365-2958.1996.tb02496.x
Strateva, T., Trifonova, A., Sirakov, I., Borisova, D., Stancheva, M., Keuleyan, E., et al. (2023). Analysis of biofilm formation in nosocomial Stenotrophomonas maltophilia isolates collected in Bulgaria: An 11-year study (2011–2022). Acta Microbiol. Immunol. Hung. 70, 11–21. doi: 10.1556/030.2023.01920
Stubbendieck, R. M., and Straight, P. D. (2016). Multifaceted interfaces of bacterial competition. J. Bacteriol. 198, 2145–2155. doi: 10.1128/JB.00275-16
Sun, E., Liang, G., Wang, L., Wei, W., Lei, M., Song, S., et al. (2016). Antimicrobial susceptibility of hospital acquired Stenotrophomonas maltophilia isolate biofilms. Braz. J. Infect. Dis. 20, 365–373. doi: 10.1016/j.bjid.2016.04.002
Thanassi, D. G., Saulino, E. T., and Hultgren, S. J. (1998). The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr. Opin. Microbiol. 1, 223–231. doi: 10.1016/s1369-5274(98)80015-5
Theuretzbacher, U., Bush, K., Harbarth, S., Paul, M., Rex, J. H., Tacconelli, E., et al. (2020). Critical analysis of antibacterial agents in clinical development. Nat. Rev. Microbiol. 18, 286–298. doi: 10.1038/s41579-020-0340-0
Travassos, L. H., Pinheiro, M. N., Coelho, F. S., Sampaio, J. L. M., Merquior, V. L. C., and Marques, E. A. (2004). Phenotypic properties, drug susceptibility and genetic relatedness of Stenotrophomonas maltophilia clinical strains from seven hospitals in Rio de Janeiro. Brazil. J. Appl. Microbiol. 96, 1143–1150. doi: 10.1111/j.1365-2672.2004.02248.x
Trifonova, A., and Strateva, T. (2019). Stenotrophomonas maltophilia – a low-grade pathogen with numerous virulence factors. Infect. Dis. 51, 168–178. doi: 10.1080/23744235.2018.1531145
Tseng, B. S., Zhang, W., Harrison, J. J., Quach, T. P., Song, J. L., Penterman, J., et al. (2013). The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ. Microbiol. 15, 2865–2878. doi: 10.1111/1462-2920.12155
Turrientes, M. C., Baquero, M. R., Sánchez, M. B., Valdezate, S., Escudero, E., Berg, G., et al. (2010). Polymorphic mutation frequencies of clinical and environmental Stenotrophomonas maltophilia populations. Appl. Environ. Microbiol. 76, 1746–1758. doi: 10.1128/AEM.02817-09
Ugalde, J. E., Czibener, C., Feldman, M. F., and Ugalde, R. A. (2000). Identification and characterization of the Brucella abortus Phosphoglucomutase gene: role of lipopolysaccharide in virulence and intracellular multiplication. Infect. Immun. 68, 5716–5723. doi: 10.1128/IAI.68.10.5716-5723.2000
Valdezate, S., Vindel, A., Martín-Dávila, P., Del Saz, B. S., Baquero, F., and Cantón, R. (2004). High genetic diversity among Stenotrophomonas maltophilia strains despite their originating at a single hospital. J. Clin. Microbiol. 42, 693–699. doi: 10.1128/JCM.42.2.693-699.2003
Vallet, I., Olson, J. W., Lory, S., Lazdunski, A., and Filloux, A. (2001). The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. 98, 6911–6916. doi: 10.1073/pnas.111551898
Vfselova, M., Kholmeckaya, M., Klein, S., Voronina, E., Lipasova, V., Metlitskaya, A., et al. (2003). Production ofN-acylhomoserine lactone signal molecules by gram-negative soil-borne and plant-associated bacteria. Folia Microbiol. 48, 794–798. doi: 10.1007/BF02931516
Wagner, P. L., and Waldor, M. K. (2002). Bacteriophage control of bacterial virulence. Infect. Immun. 70, 3985–3993. doi: 10.1128/IAI.70.8.3985-3993.2002
Walters, M. C., Roe, F., Bugnicourt, A., Franklin, M. J., and Stewart, P. S. (2003). Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47, 317–323. doi: 10.1128/AAC.47.1.317-323.2003
Wang, T., Si, M., Song, Y., Zhu, W., Gao, F., Wang, Y., et al. (2015). Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog. 11:e1005020. doi: 10.1371/journal.ppat.1005020
Waters, V. J., Gómez, M. I., Soong, G., Amin, S., Ernst, R. K., and Prince, A. (2007). Immunostimulatory properties of the emerging pathogen Stenotrophomonas maltophilia. Infect. Immun. 75, 1698–1703. doi: 10.1128/IAI.01469-06
Waters, V., Yau, Y., Prasad, S., Lu, A., Atenafu, E., Crandall, I., et al. (2011). Stenotrophomonas maltophilia in cystic fibrosis. Am. J. Respir. Crit. Care Med. 183, 635–640. doi: 10.1164/rccm.201009-1392OC
Werth, B. J., Ashford, N. K., Penewit, K., Waalkes, A., Holmes, E. A., Bryan, A., et al. (2022). Evolution of cefiderocol resistance in Stenotrophomonas maltophilia using in vitro serial passage techniques. JAC-Antimicrobial Resist. 4:dlac011. doi: 10.1093/jacamr/dlac011
West, N. P., Jungnitz, H., Fitter, J. T., McArthur, J. D., Guzmán, C. A., and Walker, M. J. (2000). Role of Phosphoglucomutase of Bordetella bronchiseptica in lipopolysaccharide biosynthesis and virulence. Infect. Immun. 68, 4673–4680. doi: 10.1128/IAI.68.8.4673-4680.2000
Windhorst, S., Frank, E., Georgieva, D. N., Genov, N., Buck, F., Borowski, P., et al. (2002). The major extracellular protease of the nosocomial pathogen Stenotrophomonas maltophilia. J. Biol. Chem. 277, 11042–11049. doi: 10.1074/jbc.M109525200
Winn, A. M., and Wilkinson, S. G. (2001). Structures of the O4 and O18 antigens of Stenotrophomonas maltophilia: a case of enantiomeric repeating units. Carbohydr. Res. 330, 215–221. doi: 10.1016/S0008-6215(00)00287-1
Wood, T. K., Knabel, S. J., and Kwan, B. W. (2013). Bacterial Persister cell formation and dormancy. Appl. Environ. Microbiol. 79, 7116–7121. doi: 10.1128/AEM.02636-13
Wu, C.-J., Chen, Y., Li, L.-H., Wu, C.-M., Lin, Y.-T., Ma, C.-H., et al. (2022). Roles of SmeYZ, SbiAB, and SmeDEF efflux Systems in Iron Homeostasis of Stenotrophomonas maltophilia. Microbiol. Spectr. 10:e0244821. doi: 10.1128/spectrum.02448-21
Wu, C.-J., Huang, Y.-W., Lin, Y.-T., Ning, H.-C., and Yang, T.-C. (2016). Inactivation of SmeSyRy two-component regulatory system inversely regulates the expression of SmeYZ and SmeDEF efflux pumps in Stenotrophomonas maltophilia. PLoS One 11:e0160943. doi: 10.1371/journal.pone.0160943
Wu, C.-M., Li, L.-H., Lin, Y.-L., Wu, C.-J., Lin, Y.-T., and Yang, T.-C. (2022). The sbiTRS operon contributes to Stenobactin-mediated Iron utilization in Stenotrophomonas maltophilia. Microbiol. Spectr. 10:e0267322. doi: 10.1128/spectrum.02673-22
Wu, C.-J., Lu, H.-F., Lin, Y.-T., Zhang, M.-S., Li, L.-H., and Yang, T.-C. (2019). Substantial contribution of SmeDEF, SmeVWX, SmQnr, and heat shock response to fluoroquinolone resistance in clinical isolates of Stenotrophomonas maltophilia. Front. Microbiol. 10:822. doi: 10.3389/fmicb.2019.00822
Yang, S., Hua, M., Liu, X., Du, C., Pu, L., Xiang, P., et al. (2021). Bacterial and fungal co-infections among COVID-19 patients in intensive care unit. Microbes Infect. 23:104806. doi: 10.1016/j.micinf.2021.104806
Yang, X., Liu, H., Zhang, Y., and Shen, X. (2021). Roles of type VI secretion system in transport of metal ions. Front. Microbiol. 12:136. doi: 10.3389/fmicb.2021.756136
Yang, J. G., Shih, M. S., Kuo, W. T., Chin, K. H., Shen, G. H., and Chou, S. H. (2014). Crystallization of the N-terminal regulatory domain of the enhancer-binding protein FleQ from Stenotrophomonas maltophilia. Acta Crystallogr. Sect. FStructural Biol. Commun. 70, 326–330. doi: 10.1107/S2053230X14001514
Yero, D., Huedo, P., Conchillo-Solé, O., Martínez-Servat, S., Mamat, U., Coves, X., et al. (2020). Genetic variants of the DSF quorum sensing system in Stenotrophomonas maltophilia influence virulence and resistance phenotypes among Genotypically diverse clinical isolates. Front. Microbiol. 11:1160. doi: 10.3389/fmicb.2020.01160
Yonekura, K., Maki-Yonekura, S., and Namba, K. (2002). Growth mechanism of the bacterial flagellar filament. Res. Microbiol. 153, 191–197. doi: 10.1016/S0923-2508(02)01308-6
Youenou, B., Favre-Bonté, S., Bodilis, J., Brothier, E., Dubost, A., Muller, D., et al. (2015). Comparative genomics of environmental and clinical Stenotrophomonas maltophilia strains with different antibiotic resistance profiles. Genome Biol. Evol. 7, 2484–2505. doi: 10.1093/gbe/evv161
Zgair, A. K., and Chhibber, S. (2011). Adhesion of Stenotrophomonas maltophilia to mouse tracheal mucus is mediated through flagella. J. Med. Microbiol. 60, 1032–1037. doi: 10.1099/jmm.0.026377-0
Zgair, A. K., and Chhibber, S. (2012). Stenotrophomonas maltophilia flagellin restricts bacterial colonization in BALB/c mouse lung in vivo. FEMS Immunol. Med. Microbiol. 66, 191–200. doi: 10.1111/j.1574-695X.2012.00999.x
Zhang, J., and Kong, F. (2002). Synthesis of a xylosylated rhamnose pentasaccharide—the repeating unit of the o-specific side chain of lipopolysaccharides from the reference strains for Stenotrophomonas maltophilia serogroup O18. J. Carbohydr. Chem. 21, 89–97. doi: 10.1081/CAR-120003740
Zhang, M., Li, L., Pan, H., and Zhou, T. (2021). The complete genome sequence of a bile-isolated Stenotrophomonas maltophilia ZT1. Gut Pathog. 13:64. doi: 10.1186/s13099-021-00456-y
Zhang, L., Li, X.-Z., and Poole, K. (2001). SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45, 3497–3503. doi: 10.1128/AAC.45.12.3497-3503.2001
Zhang, X., Wang, Y., Wu, Y., Yuan, Z.-H., Cai, Z., Qian, W., et al. (2022). Dual regulatory role exerted by cyclic dimeric GMP to control FsnR-mediated bacterial swimming. MBio 13:e0141422. doi: 10.1128/mbio.01414-22
Zheng, L., Wang, F.-F., Ren, B.-Z., Liu, W., Liu, Z., and Qian, W. (2016). Systematic mutational analysis of histidine kinase genes in the nosocomial pathogen Stenotrophomonas maltophilia identifies BfmAK system control of biofilm development. Appl. Environ. Microbiol. 82, 2444–2456. doi: 10.1128/AEM.03951-15
Zhu, B., Liu, H., Tian, W.-X., Fan, X.-Y., Li, B., Zhou, X.-P., et al. (2012). Genome sequence of Stenotrophomonas maltophilia RR-10, isolated as an Endophyte from Rice root. J. Bacteriol. 194, 1280–1281. doi: 10.1128/JB.06702-11
Zhu, H., Thuruthyil, S. J., and Willcox, M. D. P. (2001). Production of N-acyl homoserine lactones by gram-negative bacteria isolated from contact lens wearers. Clin. Exp. Ophthalmol. 29, 150–152. doi: 10.1046/j.1442-9071.2001.00397.x
Keywords: Stenotrophomonas maltophilia, virulence factors, biofilms, quorum sensing, iron uptake systems
Citation: Mikhailovich V, Heydarov R, Zimenkov D and Chebotar I (2024) Stenotrophomonas maltophilia virulence: a current view. Front. Microbiol. 15:1385631. doi: 10.3389/fmicb.2024.1385631
Received: 14 February 2024; Accepted: 15 April 2024;
Published: 29 April 2024.
Edited by:
Jens Andre Hammerl, Bundesinstitut für Risikobewertung, GermanyReviewed by:
Nicholas Cianciotto, Northwestern University, United StatesCopyright © 2024 Mikhailovich, Heydarov, Zimenkov and Chebotar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vladimir Mikhailovich, di5taWtoYWlsb3ZpY2hAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.