- Department of Biotechnology, School of Bio Sciences and Technology, Vellore Institute of Technology, Vellore, Tamil Nadu, India
Anti-inflammatory enzymes have wide applications in the pharmaceutical industry. The objective of this study was to find new and efficient strains for the commercial production of serratiopeptidase enzyme. Vast number of samples were processed for the isolation of potent strains. The experimental treatment includes processing of twenty soil samples, silkworm gut, and sugarcane stem. The total protein and protease activity was estimated by Lowry’s method and casein hydrolysis. The HRBC stabilization assay was performed for finding the anti-inflammatory potential of all strains. The serratiopeptidase production was confirmed by HPLC with the standard. Molecular characterization of selected potent strains was done by 16S rDNA and confirmed the taxonomy. The one step rapid purification of serratiopeptidase was performed by Ultra three phase partitioning method. The clot lysis potential of the Serratia marcescens VS56 was observed by modified Holmstorm method. The results of the study revealed that among the 60 strains, 12 strains were protease-positive on skim milk agar plates and showed significant protease activity. All 12 strains were screened for serratiopeptidase using high-performance liquid chromatography (HPLC) and VS56, VS10, VS12 and VS18 showed a similar retention time (4.66 ± 0.10 min) with standard. The selected potent strain, Serratia marcescens VS56 showed a proteolytic activity of 21.30 units/mL and produced a total protein of 102 mg/mL. The HRBC suspension results also showed a percentage of 94.6 ± 1.00 protection, which was compared to the standard diclofenac. The clot lysis potential of Serratia marcescens VS56 was 53% in 4 h. Furthermore, the molecular weight of the protein was identified to confirm the presence of serratiopeptidase. The study hence contributed successfully to isolating, screening, and identifying a potent producer for serratiopeptidase from an environmental source. This inherent advantage of the strain will undoubtedly contribute much to the coco comm commercial production of serratiopeptidase in the near future.
1 Introduction
Serratiopeptidase is a serine protease enzyme with potent anti-inflammatory, wound healing, and anti-biofilm properties (Tasaka et al., 1980). This enzyme has a molecular weight of 60 kDa and is widely used in tablets and injection suspensions (Devi et al., 2013; Nair, 2022). Serratiopeptidase has a crucial affinity to enzymes such as cyclooxygenase I and II. It also influences the production of interleukins, prostaglandins, and thromboxane, which are released in the arachidonic pathway (Steiger and Harper, 2014; Khosravi et al., 2023). Serratiopeptidase can hydrolyze the molecules such as serotonin, histamine, and bradykinin, which can cause swelling (Abbas et al., 2009; Navneet Kumar et al., 2017). The enzyme is available in various forms, such as tablets, injection suspensions, and combination drugs. The bacterial production of serratiopeptidase was once a widely used industrial method. Currently, the enzyme industry primarily relies on bacterial enzymes such as pectinase, hydrolases, and transferases (Gupta et al., 2015). Serratiopeptidase was considered one of the most needed enzymes in various industrial sectors. Our study represents one of the most elegant approaches in the identification of serratiopeptidase-producing strains from various sources. Serratiopeptidase and its rising demand in pharmaceutical industries peaked during the COVID-19 pandemic. Since bradykinin, histamine, and serotonin are hydrolyzed by serratiopeptidase in analgesic reactions (Desser et al., 1993; Ferrero-Miliani et al., 2007), this proteolytic enzyme is known to reduce post-COVID-19 symptoms such as nasal congestion and edema (Bhagat et al., 2013; Sharma et al., 2021). The application of serratiopeptidase is emerging in the medical and pharmaceutical sectors, where it is an essential ingredient in many drug formulations. Therefore, there is an urgent need for potent strains for the large-scale production of this enzyme. The native producer of serratiopeptidase is a known pathogen and has been isolated from infectious sites. This limitation complicates the industrial handling of this microbe (Pansuriya and Singhal, 2010). Furthermore, any industrial strain, when used for a longer period, tends to lose its yield and become a minimal producer. This study focuses on one such wild strain with effective serratiopeptidase production. The current research is therefore intended to isolate, screen, and identify a potent serratiopeptidase-producing bacterium from various sources. Soil isolate can be considered a safer source for industrial and commercial production.
2 Materials and methods
2.1 Sample collection
Different types of soil samples, such as slaughterhouse soil, agricultural waste soil, fish market soil, rhizosphere soil of tomato, ladies finger and sugarcane, mangrove soil, coconut waste, rhizosphere soil from mulberry plant, silkworm gut, and sugarcane stem, were collected from different areas of Tamil Nadu and Kerala. Different soil samples were collected 5–10 cm below the surface (Das and Prasad, 2010; Rebecca et al., 2013).
2.2 Isolation and screening
A measure of 1 g of soil was serially diluted in physiological saline (0.85%) and spread plated onto nutrient agar plates (0.5% peptone, 0.3% beef extract/yeast extract, 0.5% NaCl, 0.5% agar, and pH 7 ± 0.4) and incubated at 37°C for 24 h. Silkworm was dissected aseptically to obtain the gut and was mixed with 0.85% saline, which was plated onto a nutrient agar medium (Zhang et al., 2020). The sugarcane stem was surface sterilized using 70% ethanol and used for isolation on a nutrient agar medium. After the incubation, the colonies were selected and further screened for protease activity (Viswanathan et al., 2003).
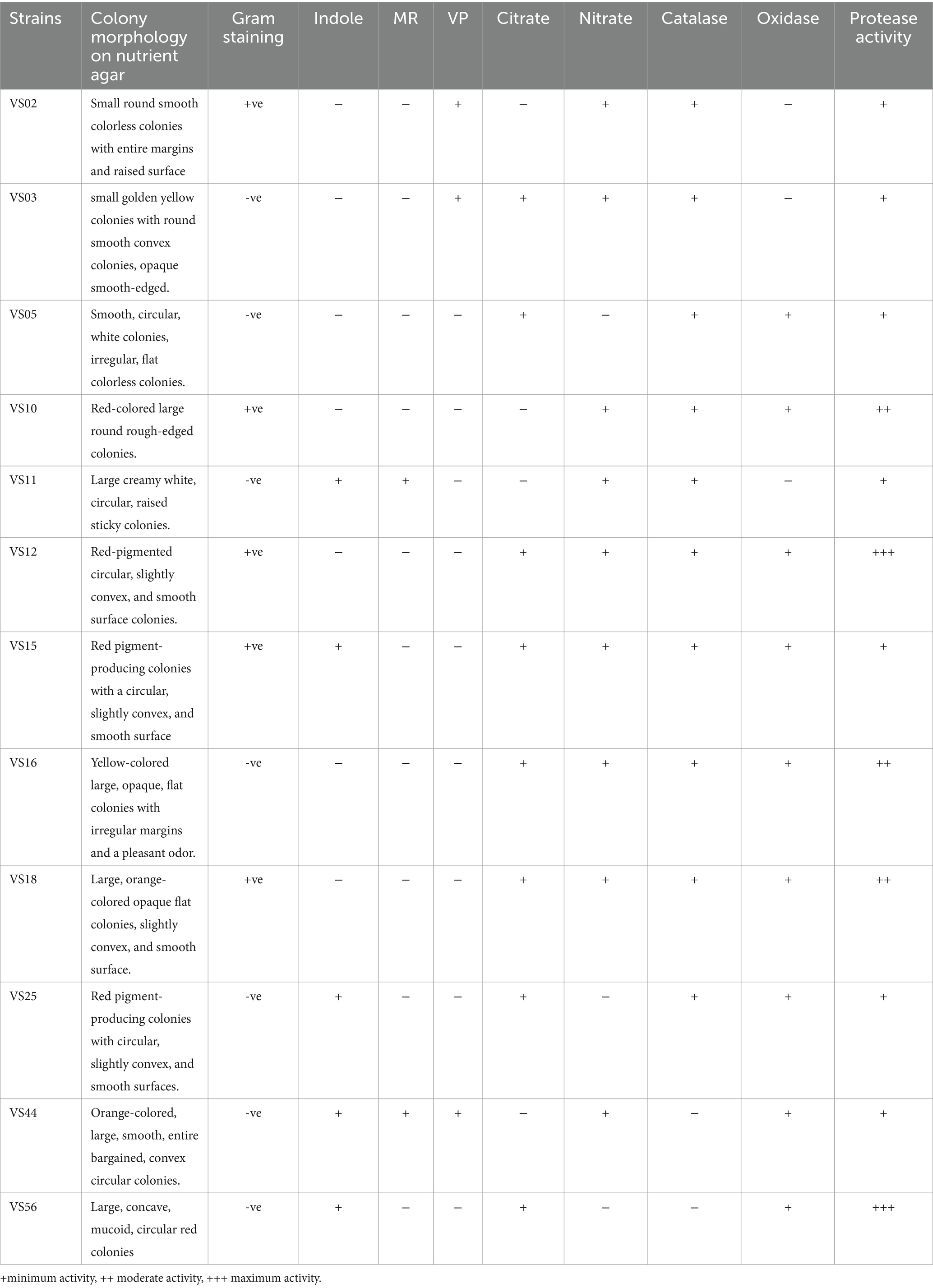
2.3 Morphological and biochemical characterization of protease-positive strains
The selected strains were identified by morphological and biochemical characterization. The 16 h old culture of VS02, VS03, VS05, VS10, VS11, VS12, VS15, VS16, VS18, VS25, VS44, and VS56 was used for biochemical analysis. The results were analyzed using Bergey’s manual of bacteriology (Bergey, 1994; Caprette, 2009).
2.4 Molecular characterization of selected strains
All five selected strains were subjected to 16S rDNA sequencing. DNA was separated from the strains and 5 μL of the test was added to 25 μL of PCR mixture, which contained 5 μL of deionized water, 1.5 μL of forward primer (5’-AGAGTTTGATCTGGCTCAG-3′), and 12 μL of Taq Expert Blend and reverse primer (5’-TACGGTACCTTGTTACGACTT-3′). The enhanced 16S rDNA arrangements were submitted to NCBI (Altschul et al., 1990). The FASTA was assessed using the program MUSCLE 3.7 (Edgar, 2004). MEGA 11 software with bootstrap values was utilized for the examination of phylogeny (Kumar et al., 2016). The 16S rDNA sequences were submitted to GenBank, and accession numbers were generated.
2.5 Total protein estimation using the Lowry method
The total protein produced by selected strains was estimated by the standard method of Lowry and Folin, using bovine serum albumin (BSA) as the standard. The cell-free supernatant was taken as the sample for the analysis (Lowry et al., 1951).
2.6 Protease activity
The isolated strains were examined for protease activity on skim milk agar plates (10% skim milk powder, 0.5% peptone, 0.5% NaCl, and 1% agar) (Jo et al., 2008). The primary screening was performed with 10% skim milk powder (Araghi et al., 2019). After a 24 h incubation at 37°C, the plates were checked for the zone of hydrolysis (Masi et al., 2021). The secondary screening for protease was conducted by cutting wells of 1 mm diameter on skim milk agar plates and adding 100 μL of cell-free suspension to the wells. The plates were incubated upright under 37°C 24 h. The zone of clearance was measured after incubation (Alnahdi, 2012).
2.7 Protease assay
Protease-positive strains were further analyzed for protease activity. A measure of 0.2 mL of supernatant was added to 2.0% casein solution prepared in Tris–HCl of pH 8.0 and incubated at 37°C for 10 min. The reaction was terminated by the addition of 1 mL of 0.1 M 10% trichloroacetic acid (TCA). This was further incubated for 15 min at 37°C. The reaction mixture was centrifuged at 10,000 rpm for 10 min. The supernatant was collected and added to 2.5 mL of Na2CO3 (0.44 M) and 1 mL of Folin–Ciocalteu reagent (1: 3 ratio). This was incubated in the dark for 30 min, and absorbance was calculated at 660 nm. The experiment was performed in triplicates. Protease production was observed using casein as a substrate (Cupp-Enyard, 2008; Saxena and Singh, 2011; Bhargavi and Prakasham, 2012).
2.8 Production of serratiopeptidase
The protease-positive strains were inoculated into 100 mL of tryptic soy broth (casein peptone 17 g/L, soy peptone 3 g/L, NaCl 5 g/L, dipotassium hydrogen phosphate 2.5 g/L, glucose 2.5 g/L, and pH – 7.3 ± 0.2) and kept for incubation at 37°C 24 h at 240 rpm and checked for protease activity. The total protein content and anti-inflammatory activity of protease-positive strains were estimated (Pansuriya and Singhal, 2010; Petersen and Tisa., 2014).
2.9 HPLC analysis
The screened protease-positive samples were further analyzed to confirm the presence of serratiopeptidase by performing HPLC. The standard serratiopeptidase was procured from Sigma Aldrich. The standard was prepared by dissolving 1 mg/mL in HPLC grade methanol and filtered through a 0.45 μm nylon filter. The retention time of the standard was compared to the samples (Navneet Kumar et al., 2017; Pushparaj, 2017). The mobile phase used was methanol and water in a ratio of 50:50. HPLC grade solvents were filtered through a 0.45-μm nylon filter.
2.10 Anti-inflammatory activity
All protease-positive strains were taken for the anti-inflammatory analysis. Blood was collected from the blood bank, and an equal volume of Alsever solution (2% dextrose, 0.8% sodium citrate, 0.5% citric acid, and 0.42% sodium chloride) was added. The solution was centrifuged at 3000 rpm, the packed cells were washed with iso-saline (0.9% w/v NaCl) and then a 10% solution was prepared. A measure of 1 mL of supernatant, 1 mL of phosphate buffer, 2 mL of hyposaline, and 0.5 mL of human red blood cell (HRBC) suspension were added. This was centrifuged for 20 min at 3000 rpm and incubated for 30 min at 37°C. The hemoglobin content of the supernatant solution was estimated spectrophotometrically at 560 nm. The experiment was conducted in triplicates. A measure of 1 mg/mL of diclofenac was used as standard, and the control was without the supernatant (Joseph et al., 2013). The percentage of protection was calculated using the formula. The percentage of protection was compared with standard (diclofenac).
Percentage of protection = 100−(Abs of sample−Abs of control/Abs of control x 100).
2.11 Clot lysis activity
Clot lysis was conducted using the modified Holmstorm method (Holmström, 1965). A measure of 500 μL blood was added to each sterile, pre-weighed separate tubes. All the tubes were incubated at 37°C for 45 min, and the plasma was carefully removed after clot formation. To all tubes, 100 μL, 200 μL, 300 μL, 400 μL, and 500 μL of crude enzyme of S. marcescens VS56 were added. This was incubated for 2–8 h at 37°C. Following incubation, the lysed clot was removed, and the weight of the remaining clot was calculated. After every 2 h of incubation, the weight of the clot was calculated. This was repeated as singlets, and the percentage of clot lysis was found using the formula.
Percentage of clot lysis = (weight of lysed clot/weight of clot x 100).
2.12 Purification of enzyme
Ultra three-phase partitioning (UTPP) was performed with slight modification. The experiment was carried out using 30% (w/v) saturated ammonium sulfate (Salarizadeh et al., 2014; Chander et al., 2021) with a pH 7.0 and 1:1 (v/v) ratio of t-butanol to crude. The mixture was kept overnight at 240 rpm, after which the precipitate was separated out. It was then dissolved in 50 Mm sodium phosphate buffer (pH 7 ± 0.2) and ultrasonicated at a frequency of 20 kHz for an irradiation time of 5 min (Pakhale and Bhagwat, 2016). Finally, the sonicated samples were dissolved in phosphate buffer (pH 7 ± 0.10) and stored.
2.13 SDS-page
The identification of protein produced by S. marcescens VS56 was conducted using SDS-PAGE (Salamone and Wodzinski, 1997). The crude and completely purified samples of S. marcescens VS56 were run in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), with 4% stacking gel and 12% separating gel. Both purified and crude samples were used for SDS-PAGE analysis. The protein marker in the medium range was used to compare the protein size obtained (Devi et al., 2013).
3 Results
3.1 Isolation and screening
Among all the processed samples, 60 strains were isolated from different sources. The pigmented and non-pigmented colonies were selected, and pure cultures were maintained for further examination on nutrient agar plates (Figure 1; Supplementary Figure 1). The strains were named VS1-VS60 and were further screened for protease activity in skim milk agar medium. The total protein and protease activity of all strains were observed. From 60 isolates,12 strains exhibited a zone of hydrolysis on skim milk agar. This showed an indication of protease production. Upon streaking onto skim milk agar (10% skim milk powder), the zone of hydrolysis was observed around the streak lines (Figure 2). Furthermore, secondary screening of the strains VS02, VS03, VS05, VS10, VS11, VS12, VS15, VS16, VS18, VS25, VS44, and VS56 showed good zone of hydrolysis. Among all, VS56 has a maximum zone diameter of 16 mm and excellent protease activity (Figure 2). The strain VS56 has excellent casein degradation potential. When compared to other strains VS10, VS16, and VS12 have moderate hydrolytic activity, and VS03, VS18, VS25, VS02, VS05, and VS44 have good hydrolytic activity (10 mm). Based on primary and secondary screening, VS56 was identified as a potent protease producer.
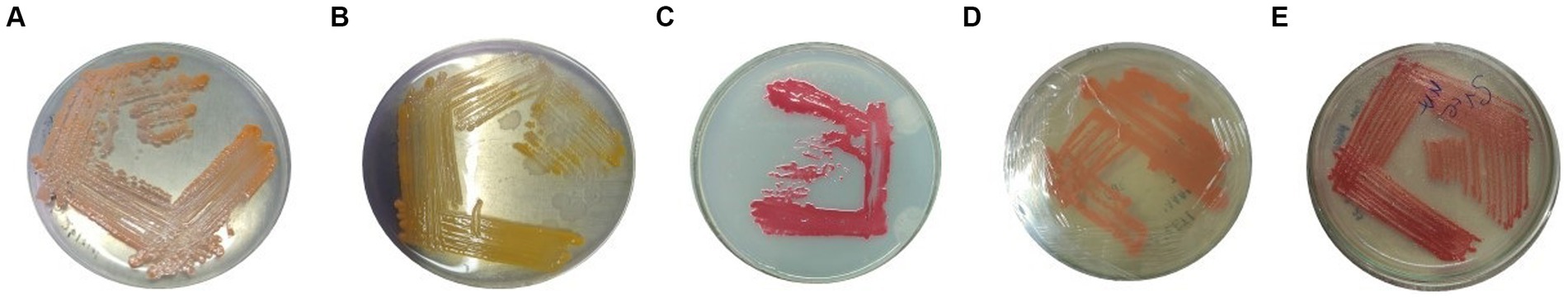
Figure 1. Pure culture of (A) VS10, (B) VS03, (C) VS12, (D) VS15, and (E) VS56 on the nutrient agar medium.
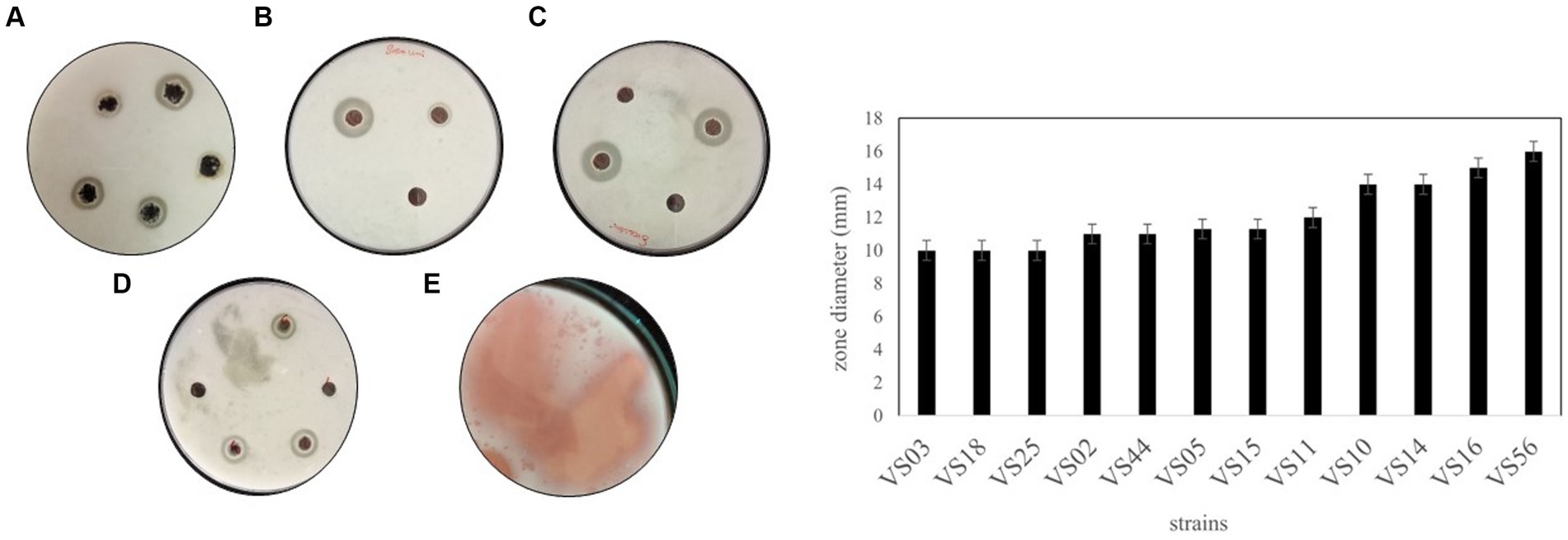
Figure 2. A-1 Zone of hydrolysis on skim milk agar (A) VS02, VS03, VS05. (B) VS12, VS44. (C) VS16, VS10. (D) VS18, VS24, VS44. (E) VS56. (B) Zone diameter of VS02, VS03, VS05, VS10, VS11, VS12, VS15, VS16, VS18, VS25, VS44, and VS56.
3.2 Morphological, biochemical, and molecular characterization of potent strains
Morphological and biochemical characteristics of strains having maximum protein and anti-inflammatory activity were identified at the species level (Table 1). The selected strains had similar retention times in the HPLC chromatogram when compared with the standard. The species-level identification revealed that the potent strains were S. marcescens VS56 (OQ859929), Chryseomicrobium amylolyticum VS10 (OQ842779), Kocuria rosea VS18 (OQ842903), Pseudomonas psychrophile VS16 (OQ842771), and VS12 Kocuria rosea (OQ842901; Figure 3).
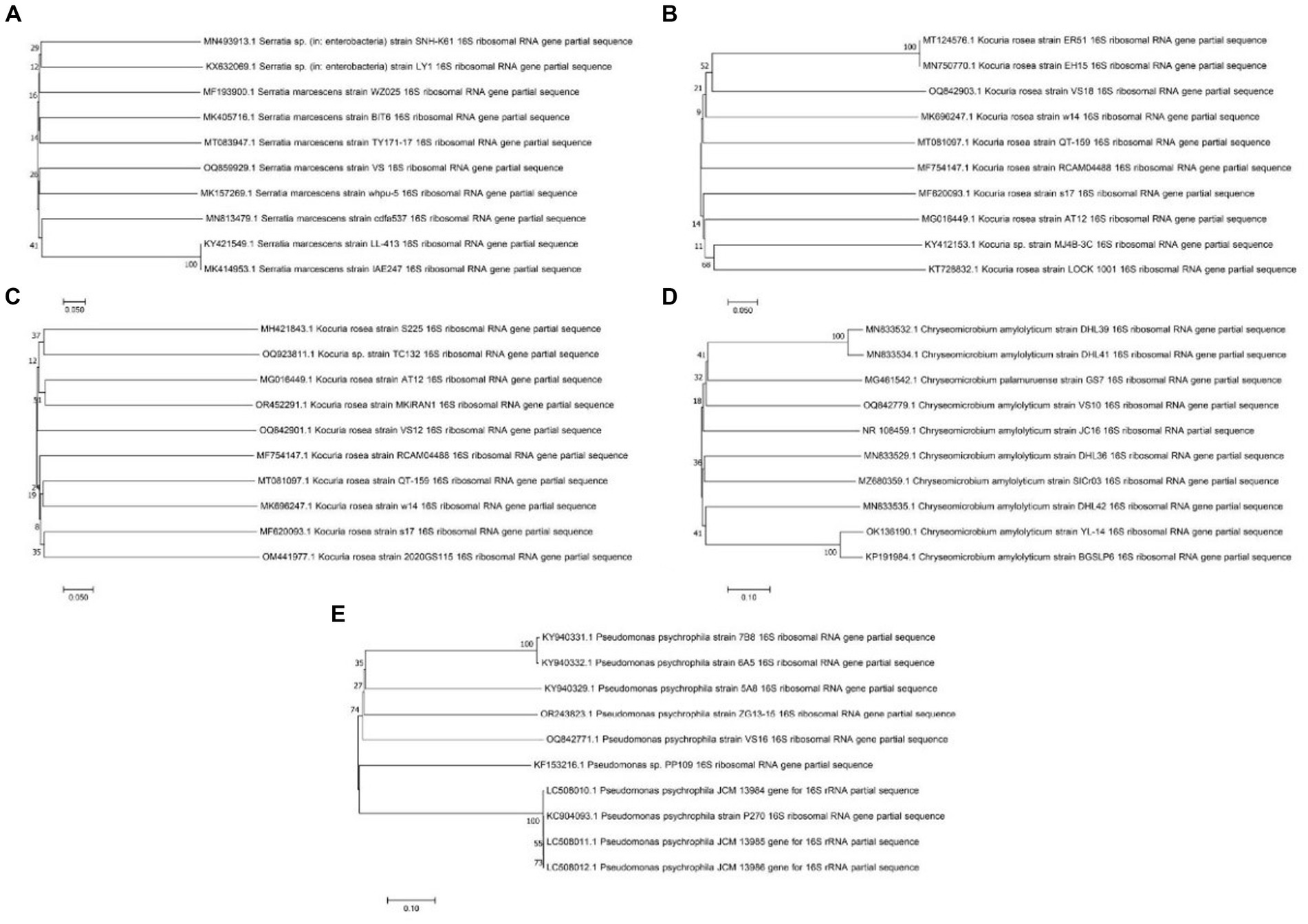
Figure 3. Phylogenetic tree of (A) Serratia marcescens VS56, (B) Kocuria rosea VS18, (C) Kocuria rosea VS12, (D) Chryseomicrobium amylolyticum VS10, and (E) Pseudomonas psychrophila VS16.
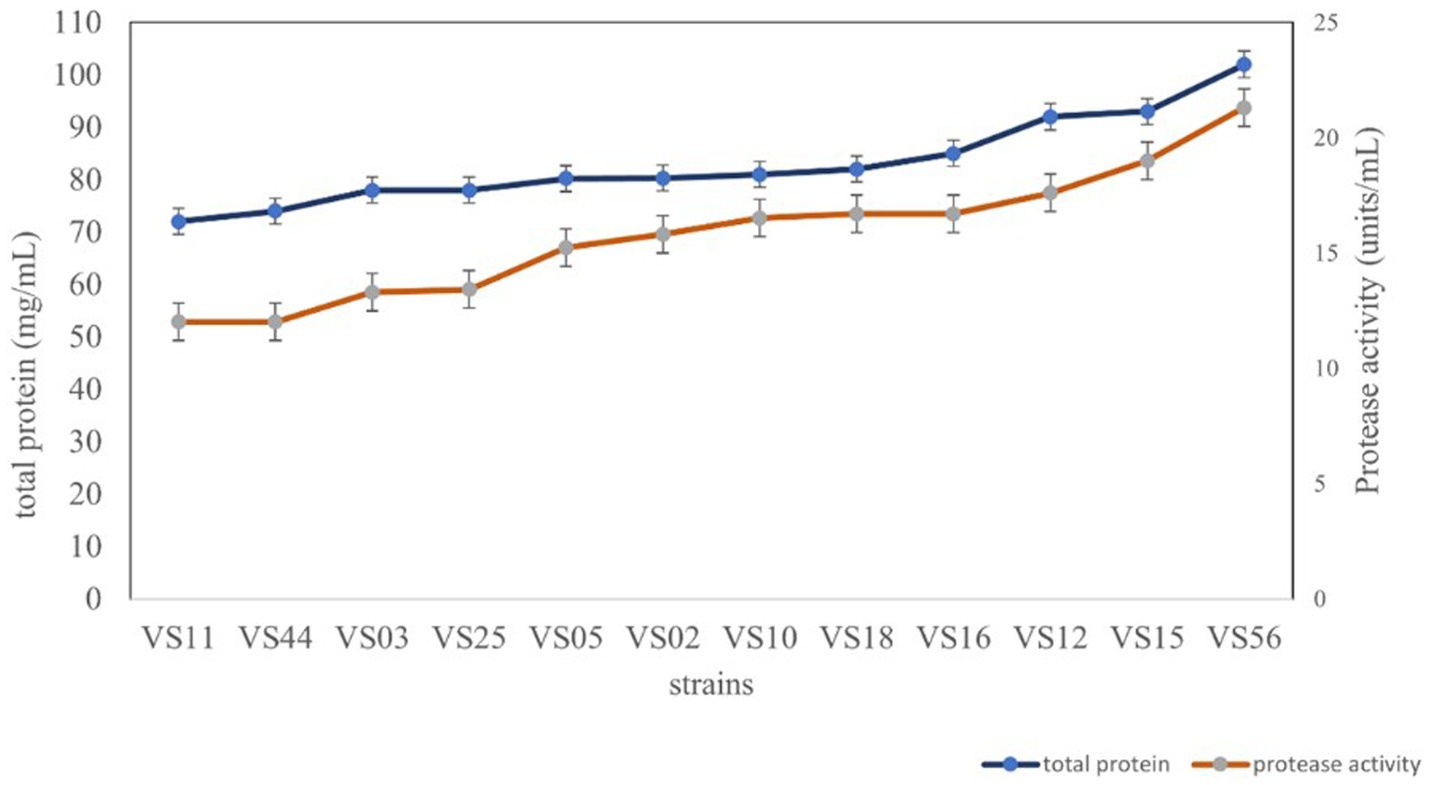
3.3 Protease activity and total protein
All the selected strains were further analyzed for total protein and protease activity. Secretory proteins in all cell-free supernatants were estimated to be potent producers. The strain isolated from waste soil, VS56, showed a maximum protease activity and total protein content of 21.30 units/mL and 102 mg/mL, respectively (Figure 4). Proteolytic bacteria are widespread and have diverse sources for isolation. The sugarcane soil isolate VS18 and mangrove soil isolate VS11 and VS44 had the least protein content and protease activity (72 mg/mL and 12.31 units/mL). Strains isolated from agricultural land (VS03) and butcher soil (VS25) have 78 mg/mL of total protein and 13.12 units/mL of protease activity. Fish market soil serves as a rich source for proteolytic bacteria. Fish market soil isolate VS05, sugarcane soil isolate VS02, and silkworm gut isolate VS10 showed a moderate protein content of 80.20 mg/mL, 80.3 mg/mL, and 81 mg/mL, respectively. Furthermore, the protease activity of these strains was identified to be 15.32 units/mL, 14.84 units/mL, and 16.81 units/mL. The strains VS16 (tomato soil) and VS12 and VS15 (coconut waste) are potent protease producers with higher amounts of secretory protein. The sugarcane sample isolate VS11 has the least amount of protein (72 mg/mL) and protease activity (12.31 units/mL). The waste soil isolate VS56 has the most protease activity and active protein in it.
3.4 HPLC analysis
The presence of serratiopeptidase in the crude cell-free supernatant was confirmed by HPLC analysis. The retention time under similar conditions of both standard and samples was scrutinized to identify the presence of serratiopeptidase. The HPLC chromatogram of VS56 showed a peak at a retention time of 4.51 min, which is exactly the same as standard having a peak at 4.58 min. The retention time of VS10 was found to be 4.71 min, whereas VS12 and VS18 had a retention time of 4.66 and 4.59 min, respectively (Figure 5). The strain VS16 showed a closer retention time of 4.67 min when compared with standard. This confirmed the presence of serratiopeptidase in the crude sample of VS10, VS12, VS18, VS16, and VS56.
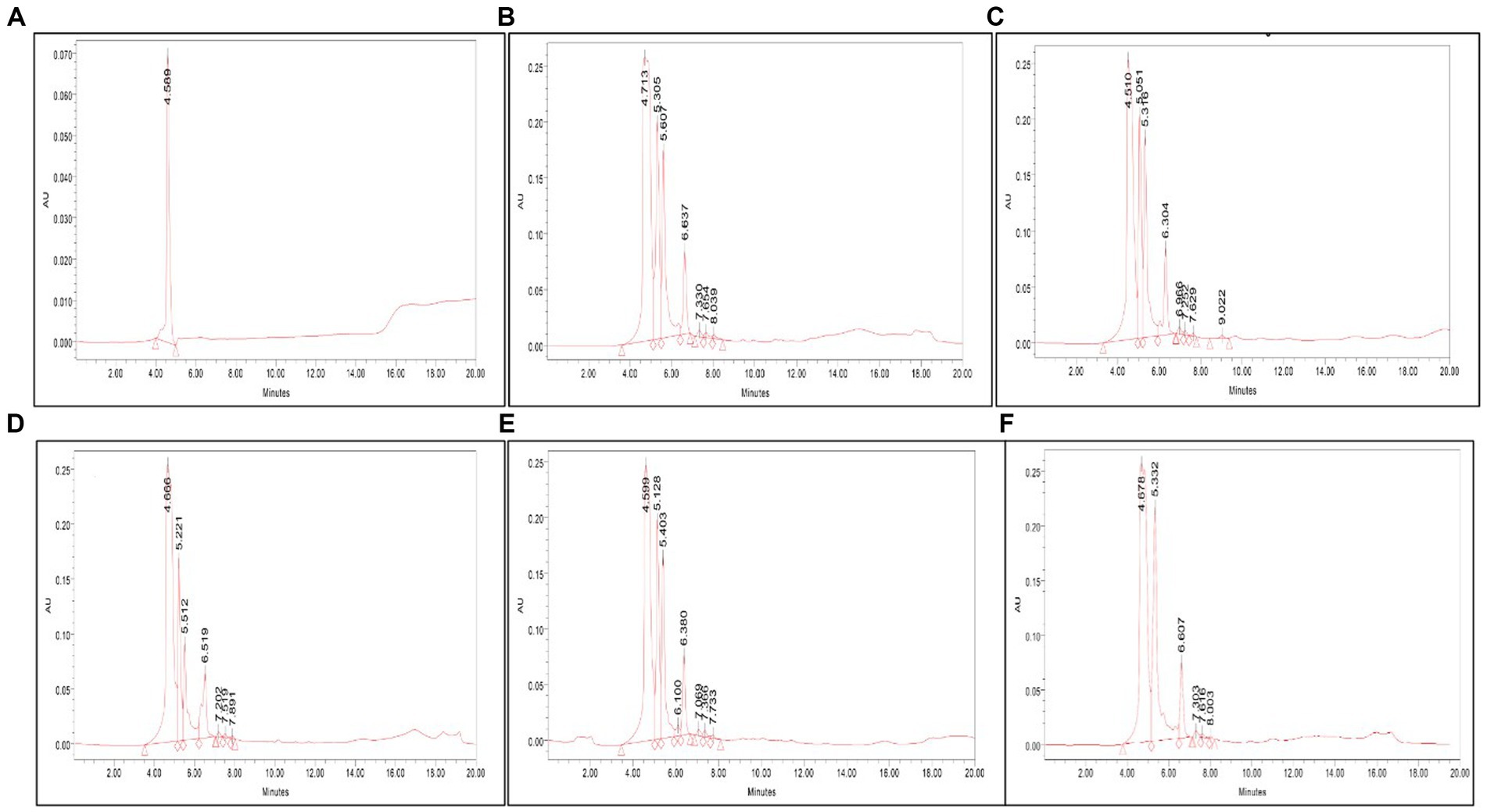
Figure 5. (A) HPLC chromatogram of (A) standard, (B) VS56, (C) VS10, (D) VS12, (E) VS18, and (F) VS16. The serratiopeptidase, standard chromatogram has a retention time of 4.56 min. The HPLC chromatogram of VS56 showed a peak at a retention time of 4.66 min.
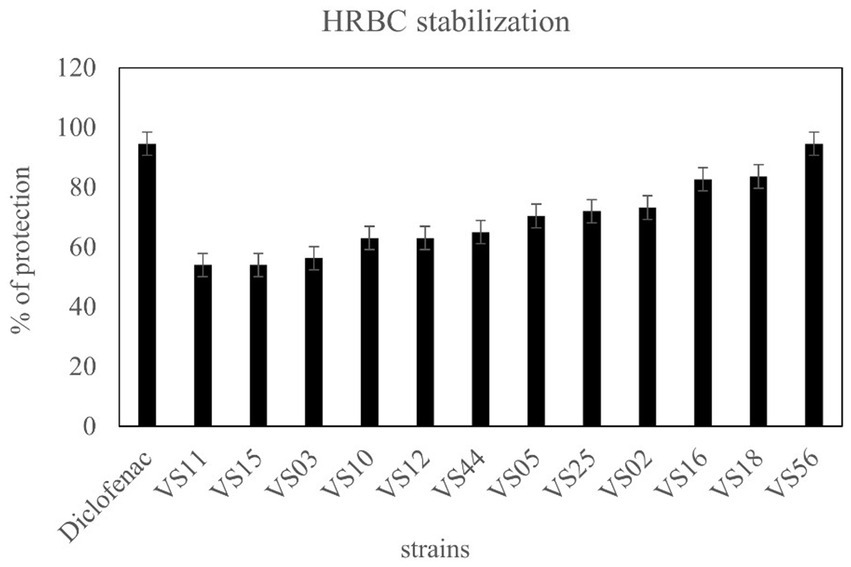
3.5 Anti-inflammatory activity
A total of 12 strains were examined further for anti-inflammatory activity. The anti-inflammatory activity of strains was considered a remarkable ability in the selection of strains. HRBC stabilization assay was adopted to evaluate the extent of anti-inflammatory activity. The results showed that waste soil isolate VS56 had a maximum percentage of RBC protection (94.6 ± 1.01; Figure 6). The percentage of RBC protection was equivalent to the standard diclofenac (94.6% ± 1.01). The strains VS16 (83.60 ± 1.00) and VS18 (82.70 ± 1.01) also had a significant percentage of anti-inflammatory activity. While VS02 (73.20 ± 1.01), VS25 (72 ± 1.00), VS05 (70.40 ± 1.01), VS02 (73.2 ± 1.00), VS05 (70.4 ± 1.01), and VS25(72 ± 1.10) showed a moderate amount of ant-inflammatory activity. VS44 (65 ± 1.00), VS12(63 ± 1.01), and VS10 (63 ± 1.01) had moderate anti-inflammatory activity, whereas the other strains VS03, VS11, and VS15 exhibited least anti-inflammatory effect. The other selected protease-positive strains, VS16 and VS18 had 83% RBC protection, indicating optimum anti-inflammatory activity. Out of all the protease-positive strains, VS15 had the least (54 ± 1.01) anti-inflammatory activity.
3.6 Clot lysis activity
The clot lysis percentage of S. marcescens VS56 was observed to be 53% (Figure 7). In comparison with different volumes of cell-free suspension of a 24 h grown culture, maximum clot lysis was observed in 500 μL. The positive control streptokinase C (100 μL), had 100% lysis within 30 min. The selected potent strain, S. marcescens VS56, thus has moderate clot lysis activity.
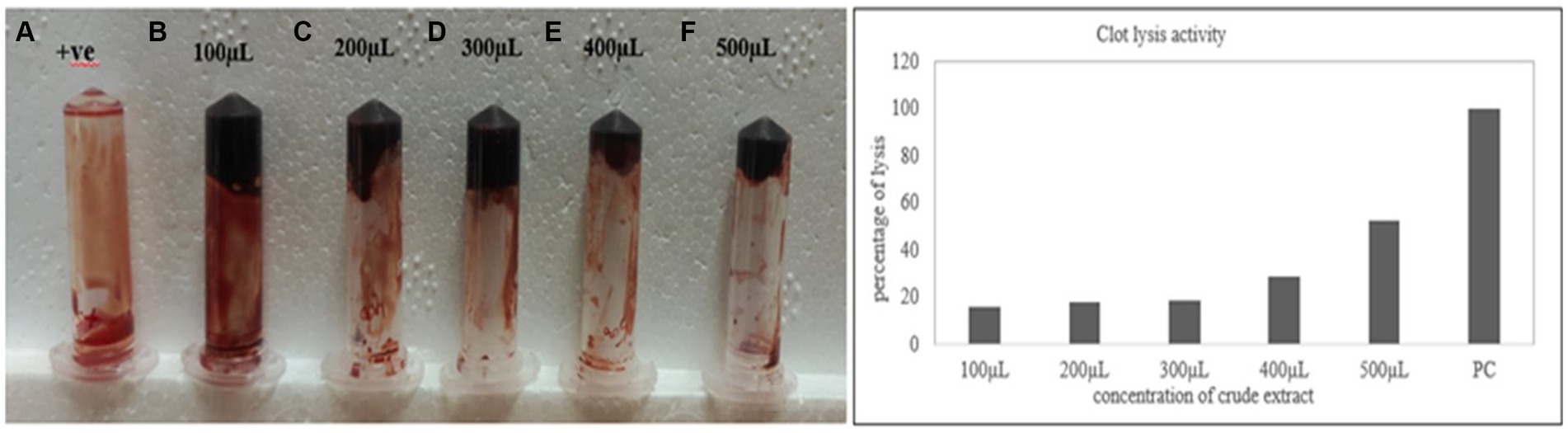
Figure 7. (A) Clot lysis of VS56. (B) streptokinase c (PC-positive control) with 100% lysis after 30 min (B) 100 μL, (C) 200 μL, (D) 300 μL, (E) 400 μL, and 500 μL (f) of supernatant.
3.7 SDS-page
The observed bands in crude and purified samples of S. marcescens VS56 are identical to 60kDA in the protein marker. The molecular weight of the protein produced by S. marcescens VS56 was identified to be approximately 60 kDA (Figure 8), which indicates the presence of serratiopeptidase. The band observed at the sample lane hence confirmed the presence of serratiopeptidase production by S. marcescens VS56 (Figure 8).
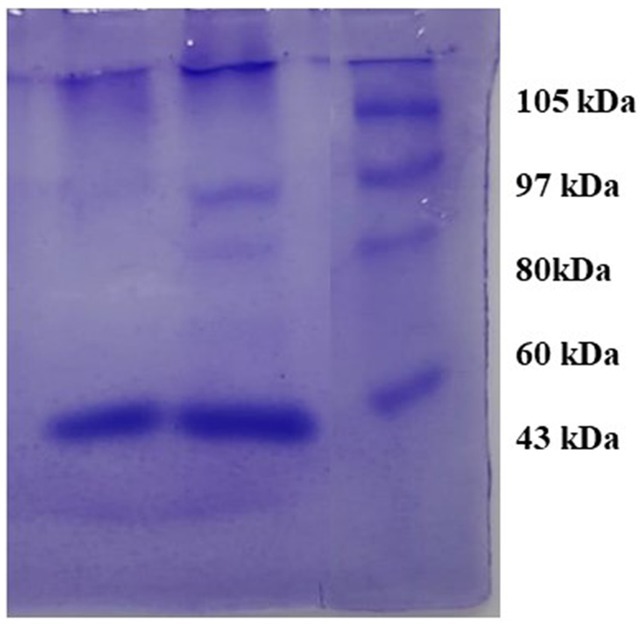
Figure 8. SDS PAGE of VS56 showing protein band between 50 and 70kb indicating the presence of serratiopeptidase Lane1 [L1]: Marker protein, Lane 2 [L2]: crude enzyme and Lane 3 [L3]: Purified enzyme.
4 Discussion
Serratiopeptidase is a potent serine protease usually isolated from sp. Serratia. Considering the pharmaceutical importance and industrial demand of this enzyme, the present study has focused on the identification of a potent serratiopeptidase-producing strain. There are many existing reports about different organisms producing serratiopeptidase (Fritsch et al., 2013). It includes a recombinant E. coli BL21 (DE3) as well as the wild strain S. marcescens e-15 (Anil and Kashinath, 2013). S. marcescens VS56 in the study was isolated from untreated waste soil and also observed as a potent producer of serratiopeptidase. According to Rebecca et al. (2013), soil serves as a rich source for isolating Serratia sp. they have successfully isolated S. marcescens from soil sources and screened for chitinase activity. According to Muthukumar et al. (2016), soil sources are regarded as safe compared to wounds or infection sites. As S. marcescens VS56 of this study is from soil, it can be regarded as safe to handle. This increases the safety of handling this bacterium for the industrial production of serratiopeptidase. Anil and Kashinath (2013) estimated the protein content and protease activity of S. marcescens e15 and observed 0.083 mg/mL of secretory proteins in it. S. marcescens VS56 has a total protein content of 102 mg/mL. The strain has more secretory protein in the cell-free supernatant. Enzymatic activity was observed using casein as a substrate (Salisbury and Likos, 1972), where S. marcescens VS56 has 12.31 units/mL of active enzyme. When compared with the study of Koul et al., 2021 the isolates S. marcescens MES-4 and S. marcescens MRS-11 from mulberry soil had protease activity of 95 units/mL and 156 units/mL, respectively. Currently, wild or mutant strains of S. marcescens are used to meet the industrial and pharmaceutical need for serratiopeptidase. Rebecca et al. (2013), in their study, have mentioned the significance of source selection for the isolation of Serratia. According to Rebecca et al. (2013), fish market soil served as a potent source for isolating Serratia. In the present study, the strains C. amylolyticum VS10 and P. psychrophila VS16 were isolated from fish and butcher house soil, respectively. While the other serine protease-positive strains, K. rosea VS12 and K. rosea VS18, were isolated from coconut waste and sugarcane. The cell-free supernatant contains all the secretory proteins, and its activity was estimated by protease activity and anti-inflammatory activity. The anti-inflammatory potential of bioactive compounds has been explored by Muthukumar et al. (2016), and there was 88% RBC protection. The anti-inflammatory potential of crude S. marcescens VS56 was 94.6% equivalent to the standard diclofenac (94.6%). The isolate not only has protease activity but a strong anti-inflammatory activity too. HPLC has been regarded as the most explored analytical tool used in the identification of serratiopeptidase. Navneet Kumar et al. (2017) study used the standard serratiopeptidase, which showed a retention time of 5.94 min. These data validate the ease and specificity of comparing the presence of serratiopeptidase in the crude sample. A similar approach was used in this study, where all the cell-free crude was examined for the presence of serratiopeptidase by comparing the retention time. The standard serratiopeptidase showed a retention time of 4.59 min. Compared with this, S. marcescens VS56 showed a major peak at 4.51 min. VS10, VS12, and VS18 also have major peaks at 4.04, 4.51, and 4.71 min, respectively. This could be effective proof to confirm the presence of serratiopeptidase in the crude extract of these selected isolates. Krishnamurthy and Belur (2018) studied the clot lysis potential of S. marcescens subsp. sakuensis and found 37% of clot lysis potential. The incubation condition and time for the study were 37°C for 4 h. Compared to this, S. marcescens VS56 was able to lyse 53% of the clot after 4 h. This marked the fibrinolytic potential of S. marcescens VS56. Serratiopeptidase is a large protease enzyme with a molecular weight of 50–60 kDa. Chander et al. (2021) have specifically identified that serratiopeptidase possesses a molecular weight of 51 kDa (Braun and Schmitz, 1980). When compared with this study, the potent soil isolate S. marcescens VS56 showed a band between 50 and 60 kDa, which indicates the presence of serratiopeptidase. The soil isolate S. marcescens VS56 can be used as an alternative potent bacterium for mass industrial production.
5 Conclusion
The study was to explore the serratiopeptidase-producing strain in various samples. Several bacterial isolates from different samples were screened for serratiopeptidase production. The serratiopeptidase-producing ability of the different bacterial isolates was compared. Based on the research findings, S. marcescens VS56 from soil has been identified as a potent serratiopeptidase producer. In the future, this novel strain could be beneficial to bacterial culture in the pharmaceutical industry to achieve enhanced production of serratiopeptidase.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
SN: Validation, Data curation, Formal analysis, Investigation, Writing – original draft. CS: Validation, Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Article processing fee was supported by Vellore Institute of Technology (VIT), Vellore.
Acknowledgments
The authors acknowledge the management of Vellore Institute of Technology [VIT], Vellore for providing all the necessary facilities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1382816/full#supplementary-material
References
Abbas, A. K., Lichtman, A. H., and Pillai, S. (2009). Functions and disorders of the immune system. 3rd Edn. San Francisco, California: Elsevier.
Alnahdi, H. S. (2012). Isolation and screening of extracellular proteases produced by new isolated Bacillus sp. J. Applied Pharmaceutical Sci. 2, 071–074. doi: 10.7324/JAPS.2012.2915
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anil, C. S., and Kashinath, M. A. (2013). Production, characterization & optimization of potent protease (serratiopeptidase) from Serratia marcescens e 15. Int Res J Pharm Appl Sci 3, 95–98. doi: 10.1007/s11515-017-1461-3
Araghi, A., Hashemi, S., Sepahi, A. A., Faramarzi, M. A., and Amin, M. (2019). Purification and study of anti-cancer effects of Serratia marcescens serralysin. Iranian J. Microbiol. 11, 320–327. doi: 10.18502/ijm.v11i4.1470
Bergey, D. H. (1994). Bergeyʼs manual of determinative bacteriology. Am J Med Sci 188:282. doi: 10.1097/00000441-193408000-00030
Bhagat, S., Agarwal, M., and Roy, V. (2013). Serratiopeptidase: a systematic review of the existing evidence. Int. J. Surg. 11, 209–217. doi: 10.1016/j.ijsu.2013.01.010
Bhargavi, P. L., and Prakasham, R. S. (2012). Proteolytic enzyme production by isolated Serratia sp RSPB11: role of environmental parameters. Current Trends in Biotechnol. Pharmacy 6, 55–65. doi: 10.1016/j.ijbiomac.2013.07.009
Braun, V., and Schmitz, G. (1980). Excretion of a protease by Serratia marcescens. Arch. Microbiol. 124, 55–61. doi: 10.1007/BF00407028
Caprette, D. R. (2009). Enterobacteriaceae: Serratia marcescens. Rice University [cit. 2012-03-20]. Dostupné z: http://www. ruf. rice. edu/~ bioslabs/bios318/characterization_example. pdf.
Chander, D., Khosla, J. K., Koul, D., Hossain, M. M., Dar, M. J., and Chaubey, A. (2021). Purification and characterization of thermoactive serratiopeptidase from Serratia marcescens AD-W2. AMB Express 11, 1–10. doi: 10.1186/s13568-021-01215-7
Cupp-Enyard, C. (2008). Sigma's non-specific protease activity assay-casein as a substrate. JoVE (J. Visualized Experiments) 19:e899. doi: 10.3791/899-v
Das, G., and Prasad, M. P. (2010). Isolation, purification & mass production of protease enzyme from Bacillus subtilis. Int. Res. J. Microbiol 1, 26–31,
Desser, L., Rehberger, A., Kokron, E., and Paukovits, W. (1993). Cytokine synthesis in human peripheral blood mononuclear cells after oral administration of polyenzyme preparations. Oncology 50, 403–407. doi: 10.1159/000227219
Devi, C. S., Elizabeth Joseph, R., Saravanan, H., Naine, S. J., and Srinivansan, V. M. (2013). Screening and molecular characterization of Serratia marcescens VITSD2: a strain producing optimum serratiopeptidase. Front. Biol. 8, 632–639. doi: 10.1007/s11515-013-1284-9
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ferrero-Miliani, L., Nielsen, O. H., Andersen, P. S., and Girardin, S. (2007). Chronic inflammation: importance of NOD2 and NALP3 in interleukin-1β generation. Clin. Exp. Immunol. 147, 227–235. doi: 10.1111/j.1365-2249.2006.03261.x
Fritsch, M. J., Trunk, K., Diniz, J. A., Guo, M., Trost, M., and Coulthurst, S. J. (2013). Proteomic identification of novel secreted antibacterial toxins of the Serratia marcescens type VI secretion system. Mol. Cell. Proteomics 12, 2735–2749. doi: 10.1074/mcp.M113.030502
Gupta, A. K., Sahu, K., and Jain, G. (2015). Development and validation of a reversed-phase liquid chromatographic method for simultaneous estimation of paracetamol, Aceclofenac and Serritiopeptidase in tablet dosage form. Int. J. Pharmacy & Life Sci. 6:e00544. doi: 10.1016/j.btre.2020.e00544
Holmström, B. (1965). Streptokinase assay on large agar diffusion plates. Acta Chem. Scand. 19, 1549–1554. doi: 10.3891/acta.chem.scand.19-1549
Jo, G. H., Jung, W. J., Kuk, J. H., Oh, K. T., Kim, Y. J., and Park, R. D. (2008). Screening of protease-producing Serratia marcescens FS-3 and its application to deproteinization of crab shell waste for chitin extraction. Carbohydr. Polym. 74, 504–508. doi: 10.1016/j.carbpol.2008.04.019
Joseph, J., Bindhu, A. R., and Aleykutty, N. A. (2013). In vitro and in vivo anti-inflammatory activity of Clerodendrum paniculatum Linn. Leaves. Indian J. Pharmaceutical Sci. 75, 376–379. doi: 10.4103/0250-474X.117428
Khosravi, M., MoriBazofti, H., Mohammadian, B., and Rashno, M. (2023). The effects of the differentiated macrophages by dexamethasone on the immune responses. Int. Immunopharmacol. 124:110826. doi: 10.1016/j.intimp.2023.110826
Koul, D., Chander, D., Manhas, R. S., and Chaubey, A. (2021). Isolation and characterization of serratiopeptidase producing bacteria from mulberry phyllosphere. Curr. Microbiol. 78, 351–357. doi: 10.1007/s00284-020-02280-0
Krishnamurthy, A., and Belur, P. D. (2018). A novel fibrinolytic serine metalloprotease from the marine Serratia marcescens subsp. sakuensis: purification and characterization. Int. J. Biol. Macromol. 112, 110–118. doi: 10.1016/j.ijbiomac.2018.01.129
Kumar, V., Zozaya-Valdes, E., Kjelleberg, S., Thomas, T., and Egan, S. (2016). Multiple opportunistic pathogens can cause a bleaching disease in the red seaweed Delisea pulchra. Environ. Microbiol. 18, 3962–3975. doi: 10.1111/1462-2920.13403
Lowry, O., Rosebrough, N., Farr, A. L., and Randall, R. (1951). Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275. doi: 10.1016/S0021-9258(19)52451-6
Masi, C., Gemechu, G., and Tafesse, M. (2021). Isolation, screening, characterization, and identification of alkaline protease-producing bacteria from leather industry effluent. Ann. Microbiol. 71, 1–11. doi: 10.1186/s13213-021-01631-x
Muthukumar, A., Pradeep, P., Thigale, I., Mohanasrinivasan, V., Jemimah, N. S., and Subathra Devi, C. (2016). Exploring the bioactive potential of Serriatia marcescens VITAPI (Acc: 1933637) isolated from soil. Front. Biol. 11, 476–480. doi: 10.1007/s11515-016-1430-2
Nair, S. R. (2022). Serratiopeptidase: an integrated view of multifaceted therapeutic enzyme. Biomol. Ther. 12:1468. doi: 10.3390/biom12101468
Navneet Kumar, U., Afroze, A., Pradeepti, C., Shailendra, K., and Naik, K. K. (2017). Development and validation of a new chromatographic method for the simultaneous estimation of Serratiopeptidase, Aceclofenac and paracetamol by RP-HPLC. Pharm Anal Chem 3:122. doi: 10.4172/2471-2698.1000122
Pakhale, S. V., and Bhagwat, S. S. (2016). Purification of serratiopeptidase from Serratia marcescens NRRL B 23112 using ultrasound assisted three phase partitioning. Ultrason. Sonochem. 31, 532–538. doi: 10.1016/j.ultsonch.2016.01.037
Pansuriya, R. C., and Singhal, R. S. (2010). Evolutionary operation (EVOP) to optimize whey-independent serratiopeptidase production from Serratia marcescens NRRL B-23112. J. Microbiol. Biotechnol. 20, 950–957. doi: 10.4014/jmb.0911.11023
Petersen, L. M., and Tisa, L. S. (2014). Molecular characterization of protease activity in Serratia sp. strain SCBI and its importance in cytotoxicity and virulence. J. Bacteriol. 196, 3923–3936. doi: 10.1128/JB.01908-14
Pushparaj, M., (2017). Simultaneous estimation of Etoricoxib and Serratiopeptidase by RP-HPLC method (doctoral dissertation, Cherraan’s College of Pharmacy, Coimbatore.
Rebecca, L. J., Susithra, G., Sharmila, S., and Das, M. P. (2013). Isolation and screening of chitinase producing Serratia marcescens from soil. J. Chem. Pharm. Res. 5, 192–195,
Salamone, P. R., and Wodzinski, R. J. (1997). Production, purification and characterization of a 50-kDa extracellular metalloprotease from Serratia marcescens. Appl. Microbiol. Biotechnol. 48, 317–324. doi: 10.1007/s002530051056
Salarizadeh, N., Hasannia, S., Akbari, N. K., and Hassan, S. R. (2014). Purification and characterization of 50 kDa extracellular metalloprotease from Serratia sp. ZF03. 12, 18–27. doi: 10.15171/ijb.1009
Salisbury, W. A., and Likos, J. J. (1972). Hydrolysis of casein: a differential aid for the indentification of Serratia marcescens. J. Clin. Pathol. 25, 1083–1085. doi: 10.1136/jcp.25.12.1083
Saxena, R., and Singh, R. (2011). Characterization of a metallo-protease produced in solid state fermentation by a newly isolated Bacillus strain. Acta Biologica Szegediensis 55, 13–18,
Sharma, C., Jha, N. K., Meeran, M. N., Patil, C. R., Goyal, S. N., and Ojha, S. (2021). Serratiopeptidase, a serine protease anti-inflammatory, fibrinolytic, and mucolytic drug, can be a useful adjuvant for management in COVID-19. Front. Pharmacol. 12:603997. doi: 10.3389/fphar.2021.603997
Steiger, S., and Harper, J. L. (2014). Mechanisms of spontaneous resolution of acute gouty inflammation. Curr. Rheumatol. Rep. 16, 1–7. doi: 10.1007/s11926-013-0392-5
Tasaka, K., Meshi, T., Akagi, M., Kakimoto, M., Saito, R., Okada, I., et al. (1980). Anti-inflammatory activity of a proteolytic enzyme, Prozime-10. Pharmacology 21, 43–52. doi: 10.1159/000137414
Viswanathan, R., Rajitha, R., Sundar, A. R., and Ramamoorthy, V. (2003). Isolation and identification of endophytic bacterial strains from sugarcane stalks and their in vitro antagonism against the red rot pathogen, vol. 5, 25–29.
Keywords: Serratia marcescens, serratiopeptidase, anti-inflammatory drugs, fibrinolytic protease, SDS-PAGE
Citation: R Nair S and Subathra Devi C (2024) Bioprospecting of serratiopeptidase-producing bacteria from different sources. Front. Microbiol. 15:1382816. doi: 10.3389/fmicb.2024.1382816
Edited by:
Karthik Loganathan, Salem Microbes Pvt Ltd., IndiaReviewed by:
Kiran Gangarapu, Anurag Group of Institutions, IndiaIndranil Biswas, University of Kansas, United States
Copyright © 2024 Nair and Subathra Devi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: C. Subathra Devi, c3ViYXJlc2VhcmNoQHJlZGlmZm1haWwuY29t
 Sreelakshmi R. Nair
Sreelakshmi R. Nair C. Subathra Devi
C. Subathra Devi