- 1Key Laboratory of Xinjiang Special Probiotics and Dairy Technology of Shihezi Municipal Government, School of Food Science and Technology, Shihezi University, Shihezi, Xinjiang, China
- 2Xinjiang Production and Construction Corps Industrial Innovation Research Institute of Dairy Products, Xinjiang Tianrun Dairy Co., Ltd., Urumchi, Xinjiang, China
- 3Key Laboratory of Agricultural Product Processing and Quality Control of Specialty (Co-construction by Ministry and Province), School of Food Science and Technology, Shihezi University, Shihezi, Xinjiang, China
- 4Key Laboratory for Food Nutrition and Safety Control of Xinjiang Production and Construction Corps, School of Food Science and Technology, Shihezi University, Shihezi, Xinjiang, China
Owing to its high nutritional content, raw milk contains a rich microbiota. Thus, to study microorganisms present in raw milk available in Xinjiang China, 142 raw milk samples from seven animal breeds (cow, sheep, goat, donkey, horse, camel, and yak) and four regions (Hami, Tarbagatay, Kashgar, and Ili) were analyzed by high-throughput DNA sequencing. These microorganisms were characterized by 10 dominant phyla. Proteobacteria (68.33%) was the major phylum, followed by Firmicutes (18.80%) and Thermi (3.16%). Horse milk contained more Bacteroidetes, sheep milk contained more Gammaproteobacteria, and donkey milk contained more unclassified sequences. Camel and donkey milk contained the highest and lowest bacterial diversity compared with that contained by the remaining milk samples, respectively. Additionally, spoilage microorganisms, including Chryseobacterium, Propionibacterium, and Flavobacterium, and pathogenic bacteria, including Ochrobactrum anthropi and Sphingomonas, were more prevalent in horse and yak milk, whereas probiotic lactic acid bacteria (LAB), such as Leuconostoc, Lactococcus, or Lactobacillus, were more prevalent in goat, donkey, and camel milk. Furthermore, Moraxella was abundantly present in goat, camel, and yak milk, Acinetobacter was more abundant in camel milk, and Pseudomonas was relatively abundant in sheep and donkey milk. Overall, specific harmful microorganisms and probiotic lactic acid bacteria were found in the raw milk samples obtained from different animals, which provided a basis for preventing and controlling the growth of harmful bacteria, as well as investigating probiotic resources in raw milk.
1 Introduction
The vast Xinjiang Uygur Autonomous Region of China boasts an expansive terrain and abundant grassland resources, fostering a rich heritage of free-range animal husbandry across its diverse regions such as Ili, Hami, Tarbagatay, and Kashgar (Li et al., 2012). This practice, underpinned by the region’s unique climatic and environmental endowments, has nurtured a diverse array of livestock including horses, cow, yaks, goats, sheep, camels, and donkeys. Concurrently, the various kinds of animal milk and derivative products—encompassing yogurt, cheese, koumiss, and ghee, among others—are not only rich in nutrients but also distinguished by their diverse flavors, earning the profound affection of both locals and tourists alike (Mo et al., 2019). Furthermore, owing to the constraints of production environments in pasture, the majority of the gathering and processing of animal raw milk is conducted manually by herdsmen, resulting in a significant enrichment of indigenous microorganisms within the milk (Wouters et al., 2002). These microorganisms are closely related to the nutritional value, processing capabilities, storage stability, and ultimately, the health benefits imparted to consumers of raw milk (Panesar, 2011; Quigley et al., 2013). In essence, they form the cornerstone of a robust and interconnected system that underscores the unique qualities of the region’s dairy heritage.
Previous research endeavors, employing culture-dependent and/or culture-independent methods, have consistently highlighted the composition and characteristics of beneficial microorganisms in animal raw milk and dairy products (Dalmasso et al., 2017; Zhang et al., 2019; Kamilari et al., 2020; Zhu et al., 2020; Chi et al., 2021; Rahmeh et al., 2022; Rajawardana et al., 2022; Santamarina-García et al., 2022). These microorganisms predominantly encompass lactic acid bacteria (LAB) such as Lactobacillus (Mahmoudi et al., 2016), Bifidobacterium (Yasmin et al., 2020), Lactococcus (Kondrotiene et al., 2020), Streptococcus (Ayyash et al., 2018), Leuconostoc (Ariute et al., 2023), Pediococcus (Moussaid et al., 2023), Propionibacterium (Yerlikaya et al., 2020) and Corynebacterium (Hahne et al., 2018) along with yeast (Zhang et al., 2021) and mold (Quigley et al., 2013). Notably, these microorganisms significantly contribute to enhancing the flavor profile, texture, and nutritional composition of raw milk and dairy products (Karahadian et al., 1985; Masoud and Jakobsen, 2005; Moosavi-Nasab et al., 2010; Thierry et al., 2011; Hahne et al., 2018). Nevertheless, studies have revealed the coexistence of spoilage and pathogenic microorganisms within raw milk and derived products pose a significant challenge (Hassan and Frank, 2011). Specifically, the heat-resistant microorganisms (primarily Bacillus species) (Yang et al., 2023), psychrotolerant and/or psychrophilic microorganisms, particularly Pseudomonas, Acetobacter, and Aeromonas, resulted in the deterioration of raw milk and processed products during processing and storage stages, causing considerable disruptions to human production processes (Nörnberg et al., 2010; Samaržija et al., 2012; Xin et al., 2017; Yang et al., 2020). Furthermore, the detection of pathogenic microorganisms like Staphylococcus, Campylobacter, Yersinia, Salmonella, Escherichia coli, Listeria, Brucella, Aeromonas, Bacillus, Clostridium, Serratia, and Proteus in animal raw milk or derivatives underscores the potential food safety hazards and associated risks to human health (Gran et al., 2003; Jin et al., 2009; Fotou et al., 2011; Quigley et al., 2013; Verraes et al., 2014; Jamali et al., 2015; Fei et al., 2019).
Currently, amidst the advancements in dairy industrialization, the processing of raw animal milk in pastoral regions has undergone a paradigm shift, transitioning from manual methods to centralized factory operations, where it is transformed into a diverse array of standardized dairy products. Consequently, there arises a paramount need for a comprehensive understanding of the microbial composition and safety assessment of raw milk sourced from various animal species (Wu et al., 2018). Illumina MiSeq high-throughput sequencing technology allows for more comprehensive and accurate detection of species composition compared to traditional culture-dependent methods (Sessou et al., 2019). In this study, a comparative analysis of the microbiota in animal raw milk from 142 fresh samples, collected from seven diverse animal species (cow, sheep, goat, donkey, horse, camel, and yak) in four representative pastoral areas (Hami, Tuscaloosa, Kashgar, and Ili) of Xinjiang, China, was presented using high-throughput sequencing technologies targeting the V3–V4 hypervariable region of the 16S ribosomal RNA gene. To delve deeper into the differences and interrelationships among microorganisms present in raw milk from diverse animals, we employed Linear Discriminant Analysis Effect Size (LEfSe) alongside Indicator Species Analysis. Our research objective is to assess the potential beneficial microbial resources in animal raw milk and the safety of dairy products, so as to provide theoretical basis for subsequent production, processing and animal breeding.
2 Materials and methods
2.1 Sample collection
In total, 142 raw milk samples collected from cows (N), sheep (MY), goats (SY), donkeys (L), horses (M), camels (T), and yaks (MN) in the regions of Hami (HM), Tarbagatay (TC), Kashgar (KS), and Ili (YL) in Xinjiang, China. The samples were collected in sterilized tubes from local herding families living in the four regions of Xinjiang, China, and transferred to the laboratory using a mobile refrigerator (operating at −18 to −15°C) to be stored at −80°C for further analyses.
2.2 DNA extraction and high-throughput sequencing
One milliliter of the milk was centrifuged at 10000 g for 10 min to obtain a pellet, which was subjected to DNA extraction. Total DNA was extracted from each milk sample using PowerSoil DNA Isolation Kit (MoBio, Carlsbad, CA, United States) according to the manufacturer’s instructions. The obtained DNA was quantified using PicoGreen (Invitrogen, Carlsbad, CA, United States) and stored at −20°C. Further, a DNA solution adjusted to 10 ng DNA/μL in H2O was pretreated with 1 μg BSA/mL (BSA concentration in the sample: 10 mg/mL) at 95°C for 5 min to bind to polymerase chain reaction (PCR)-inhibiting substances. Next, 16S rRNA gene libraries were constructed by performing PCR to amplify the variable regions V3 and V4 using the forward 16Sf (5′-CCTACGGGAGGCAGCAG-3′) and the reverse 16Sr (5′-GGACTACHVGGGTWTCTAAT-3′) primers (Zhang et al., 2019).
After the quantification, qualification, and purification of the PCR products, a sequencing library was developed using NEB Next R UltraTM DNA Library Prep Kit for Illumina (NEB, Ipswich, MA, United States). The library was sequenced using the Illumina HiSeq 2000 system (Illumina, Inc., San Diego, CA, United States), which generated 300-bp paired-end reads.
2.3 Sequence analyses
The quality control of the resulting bacterial reads was performed according to Ben Maamar et al. (2020). Briefly, sequences with barcode ambiguities, with read length < 150 bp, and with average quality score < 25 were removed. Uchime was used to remove chimeric sequences (Edgar et al., 2011). Subsequently, the processed sequences were clustered in operational taxonomic units (OTUs) defined at 97% similarity using CROP (Hao et al., 2011). Taxonomic analyses were conducted using MOTHUR by querying the bacterial and archaea reads against those in the GREENGENES (Ben Maamar et al., 2020) reference databases.
2.4 Statistical analyses
2.4.1 Indices of α-diversity
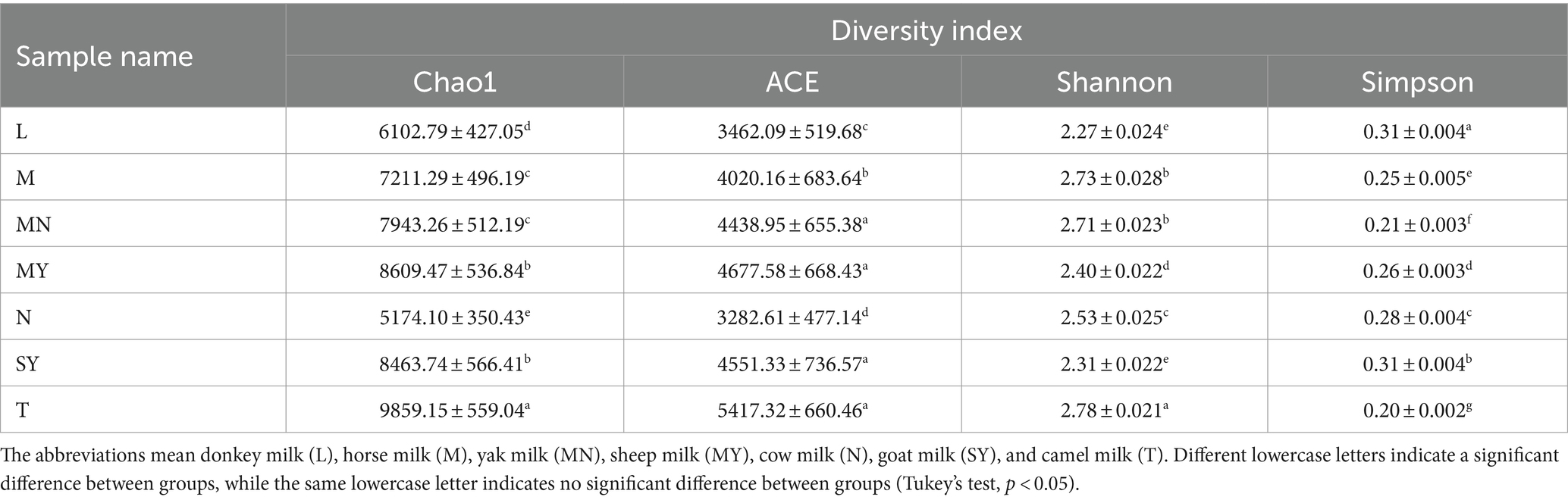
Simpson, Chao, Ace, and Shannon diversity indices, which are indices of α-diversity, were estimated for total bacteria based on OTU abundance matrices rarefied to the lowest sequence numbers. The α-diversity indices were analyzed by QIIME with the MOTHUR function. The one-way analysis of variance and Pearson’s correlation analysis were performed using R 3.5.1 statistical software (R Development Core Team, 2013). Tukey’s honest significant difference test was used to determine differences among α-diversity indices. The results were considered significantly significant at p < 0.05.
2.4.2 Principal coordinate analyses
The overall variability in bacterial community structures was evaluated by performing PCoA using the procrustes function as implemented in the vegan package in R. The ggplot2 package with ggscreeplotv and ggbiplot functions were used to replace the built-in R function biplot.princomp with extended functionality for labeling groups.
2.4.3 Correlation coefficient analysis
The correlation coefficients among bacteria were determined using R3.5.1 with corrgram and ggplot2 packages and lattice, survival, Formula, Hmisc, and corrgram functions. The results were visualized using Cytoscape.
2.4.4 LEfSe
Significant taxonomic differences were analyzed by performing the LEfSe analysis (Segata et al., 2011). Significant taxa were used to indicate differences among the sample (Bokulich et al., 2014). LAD scores were normalized by log10.
2.4.5 Indicator species analysis
Indicator species analysis was performed using the multipatt function in the indicspecies package in R, allowing 999,99 permutations and combinations between habitats (De Cáceres et al., 2010) to identify OTUs that caused changes in multivariate patterns. Multiple testing corrections of p-values were performed in R using the fdrtool function in the fdrtool package (Strimmer, 2008). All graphs were generated with R using the vegan (Oksanen et al., 2013) and ggplot2 packages (Wickham, 2009). Only OTUs with significant INDicator VALues (INDVAL) (p < 0.05) more than 0.3 were considered and visualized by the matrix–bubble graph using ggplot2 in R3.5.1 with the grid, showtext, and Cairo functions.
INDVAL were analyzed depending on two site-group combinations as follows: (1) Seven-group combination; N (cows), MY (sheep), SY (goats), L (donkeys), M (horses), T (camels), and MN (yaks) and (2) Four-group combination; HM (Hami), TC (Tarbagatay), KS (Kashgar), and YL (Ili).
3 Results
3.1 Compositions of microbial communities in milk samples
In total, 128,607 reads were obtained in the present study. The number of effective reads per sample ranged from 30,693 to 44,837. The microbial communities were characterized by six dominant phyla (>1.0% of the total sequences) and 13 less abundant phyla representing 99.72 and 0.28% of the total sequences, respectively, whereas 4.90% of the sequences were unclassified at the phylum level. Proteobacteria was the major phylum with a relative abundance of 68.33%, followed by Firmicutes (18.80%), Thermi (3.16%), Bacteroidetes (2.35%), and Actinobacteria (1.83%). Among Proteobacteria, Gammaproteobacteria was the most abundant class (86.20% of the Proteobacteria sequences), followed by Alphaproteobacteria (7.45%), Betaproteobacteria (6.29%), and Deltaproteobacteria (0.06%). The milk samples were classified into groups N (cows), MY (sheep), SY (goats), L (donkeys), M (horses), T (camels), and MN (yaks) according to their breeds. The relative abundances of phyla in each of these groups are presented in Figure 1. The relative abundance of unclassified sequences at the phylum level was more than that reported in previous studies (Hou et al., 2015; Doyle et al., 2017). Moreover, 74.37% of these unclassified sequences were from the donkey milk samples. The camel milk samples were shown to contain abundant new species. The relative abundance of bacteroidetes in the horse milk samples was significantly higher than that in the remaining milk samples. Gammaproteobacteria was relatively abundant in the sheep milk samples.
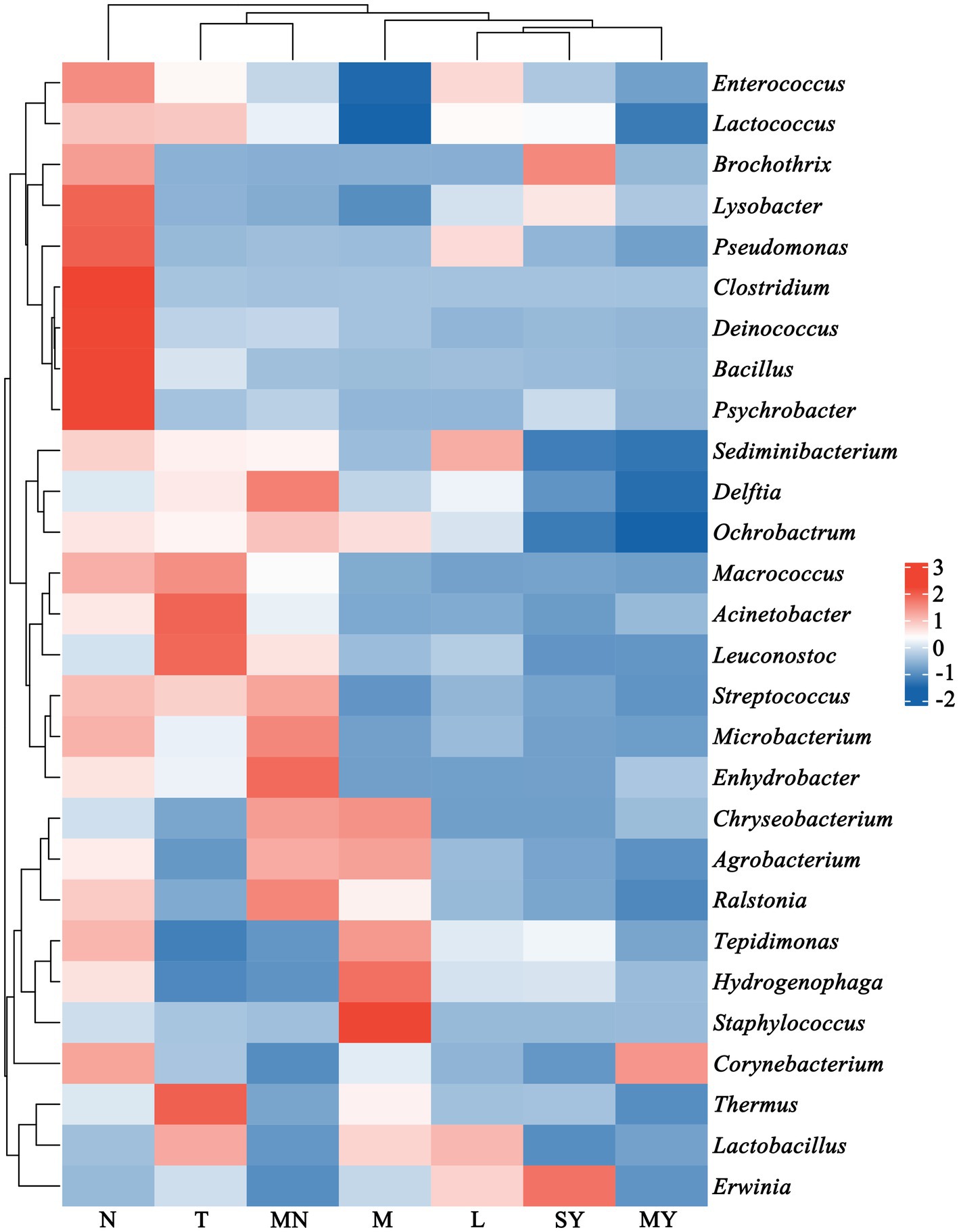
Figure 1. Community structure of the dominant bacteria in 142 raw milk samples of seven different animal breeds at the phylum level (relative abundance >1%). L, donkey milk; M, horse milk; MN, yak milk; MY, sheep milk; N, cow milk; SY, goat milk; T, camel milk.
Next, a genus with a relative abundance higher than 0.5% was defined as the predominant genus. We observed 28 predominant genera (Figure 2), and prevalent genera were diverse across the groups. The dominant bacterial genera shared by groups M and MN were Amycolatopsis, Ralstonia, Methylobacterium, Bradyrhizobium, Propionibacterium, Sphingomonas, Phyllobacterium, Sediminibacterium, and Ochrobactrum. Additionally, group M contained the following prevalent microbial genera: Delftia, Tepidimonas, Hydrogenophaga, Anoxybacillus, Staphylococcus, Flavobacterium, Thermus, and Lactobacillus, whereas group MN contained the following prevalent microbial genera: Enhydrobacte, Streptococcus, Microbacterium, and Delftia. The dominant bacterial genera in group T were as follows: Acetobacter, Acinetobacter, Salinicoccus, Enhydrobacter, Leuconostoc, Macrococcus, Moraxella, Thermus, and Lactobacillus. The dominant bacterial genera in group N were as follows: Kocuria, Carnobacterium, Bacillus, Clostridium, Paenibacillus, Meiothermus, Psychrobacter, and Exiguobacterium. The dominant bacterial genera in group L were as follows: Erwinia, Lactococcus, Enterococcus, Stenotrophomonas, Sediminibacterium, Phyllobacterium, Lactobacillus, Comamonas, and Pseudomonas. The dominant bacterial genera shared by groups MY and SY were as follows: Lysobacter, Stenotrophomonas, Tepidimonas, and Hydrogenophaga. Additionally, group SY contained Moraxella, Brochothrix, Vagococcus, Erwinia, Fusobacterium, Porphyromonas, Lactococcus, and Enterococcus, whereas group MY contained Comamonas, Pseudomonas, and Corynebacterium.
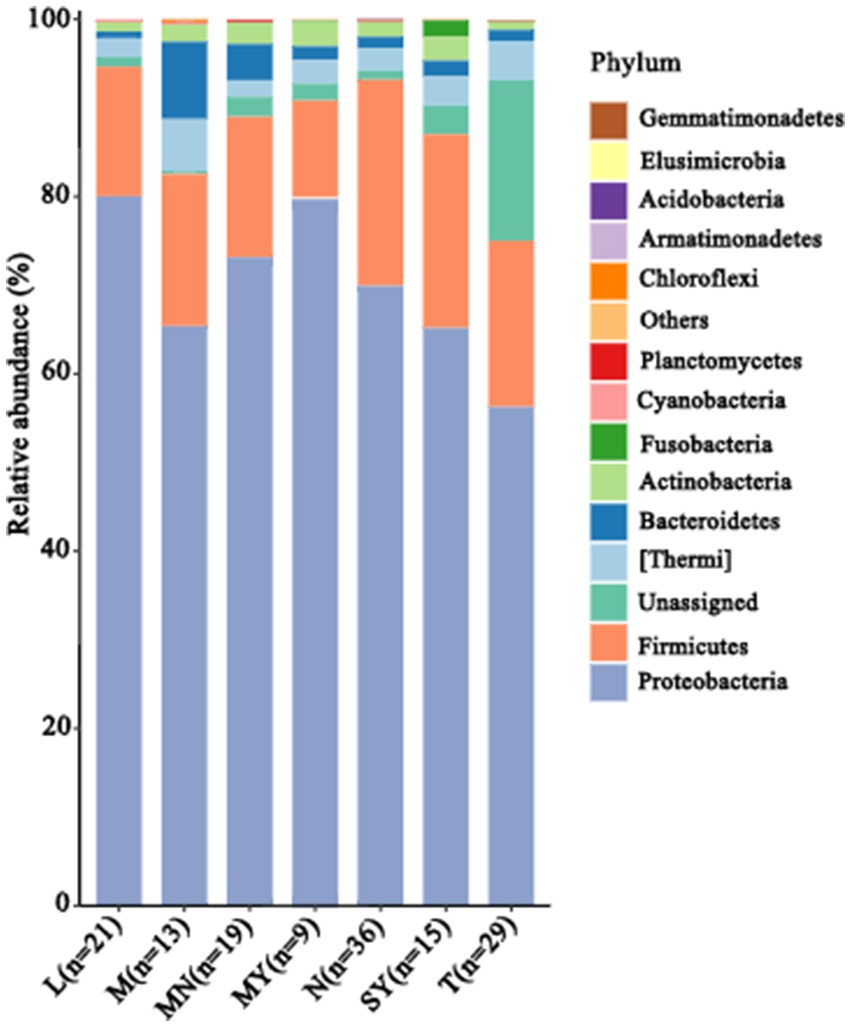
Figure 2. The relative abundance of the dominant species (at the genus level, relative abundance >0.5%) in 142 animal raw samples of seven different animal breeds. L, donkey milk; M, horse milk; MN, yak milk; MY, sheep milk; N, cow milk; SY, goat milk; T, camel milk.
3.2 α- and β-diversity indices of milk microbiota
α-Diversity index indicates the microbial diversity of a given sample (Walters and Martiny, 2020). Herein, Ace, Chao, Shannon, and Simpson indices were analyzed to analyze the microbial diversity of each milk sample. Significant differences were observed among the indices of each group (Table 1). Ace, Chao, and Shannon indices of group T were significantly higher than those of the remaining groups, whereas the Simpson index of group T was significantly lower than that of the remaining groups. These results indicated that the microbial diversity of the camel milk samples was higher than that of the remaining groups.
The β-diversity index indicates the microbial diversity between different samples (Walters and Martiny, 2020). To compare similarities in microbial compositions, we performed PCoA using genus-level taxonomic profiles. The clustering of the milk samples according to their microbiota helped separate the samples (Figure 3). However, no clear separation was observed for milk samples from groups HM, TC, KS, and YL (different regions) and groups N, MY, SY, L, M, T, and MN (different breeds) within the PCoA plots. These results indicated that complex factors might affect microbial community structures in raw milk, and only one factor (breed or region) might not be sufficient to determine microbial community structures in raw milk. When the two factors, breed, and region, were combined some regular patterns were observed in the PCoA plots (Figure 3). Homologous animal milk samples from the same region showed a higher probability of gathering. For instance, values for the cow milk samples from Kashgar were gather together in the plots, indicating a similar microbial community structure in these samples. Similarly, values for the cow milk samples from Ili were gather together in the plots. Additionally, values for different animal milk samples from the same region were observed to be gather together. For example, values for the cow, sheep, and horse milk samples were at a shorter distance than those for the remaining samples in the plots, which suggested a similarity in these samples. Sometimes, values for the raw milk samples from the same region and breed were not gather together, which indicated that microbial communities in these samples differed from each other. This case was observed in the camel milk samples from Hami and the yak milk samples from Kashgar with large dispersion.
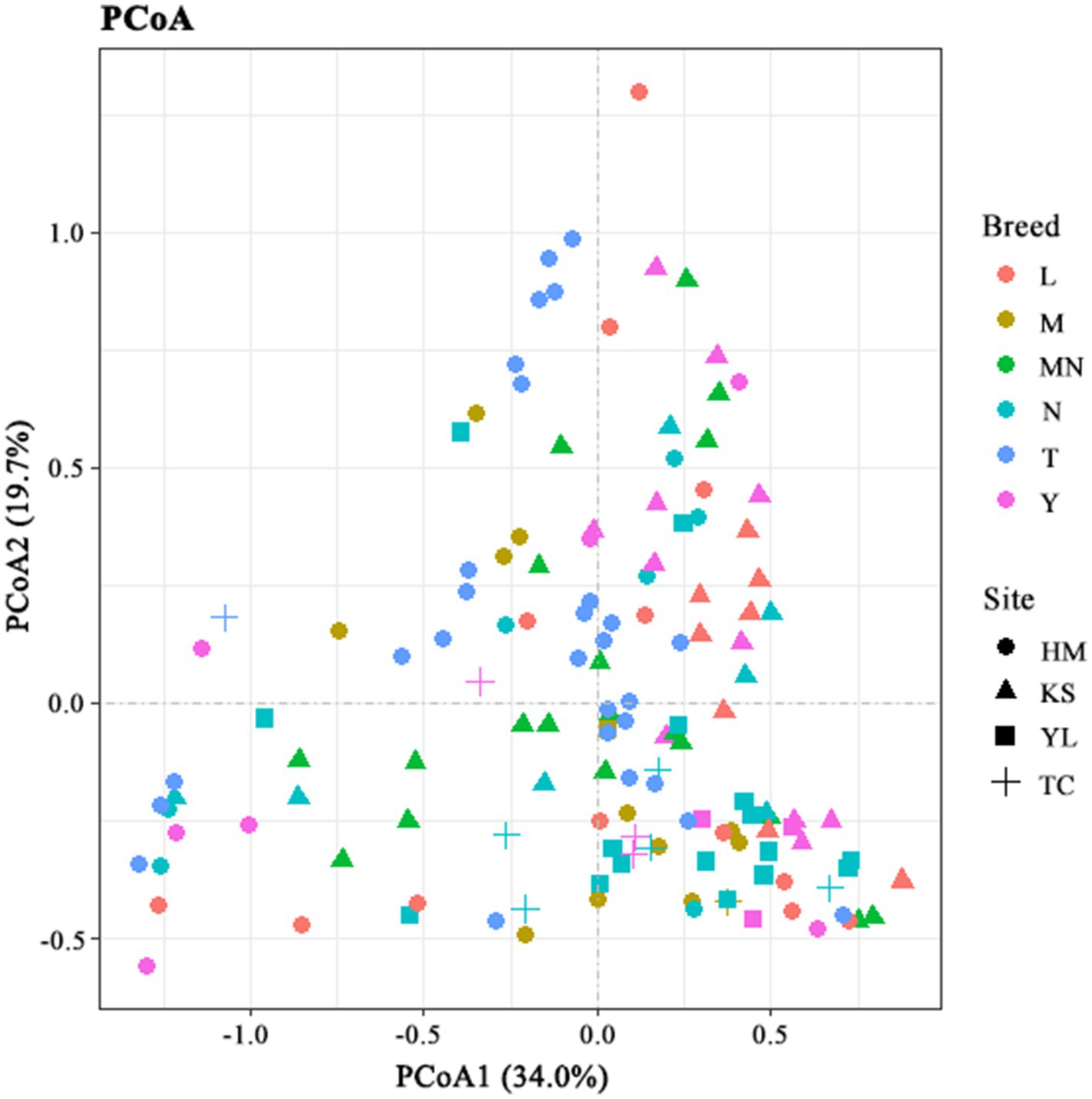
Figure 3. Principal coordinate analysis of bacterial communities based on genus level. L, donkey milk; M, horse milk; MN, yak milk; Y, goat or sheep milk; N, cow milk; T, camel milk; HM, Hami; TC, Tarbagatay; KS, Kashgar; YL, Ili.
3.3 Microbial composition differences among different animal raw milk
3.3.1 Microbial composition differences among donkey, horse, camel, and yak milk
To identify differences in the microbial compositions in the milk samples among donkey, horse, camel, and yak milk, we performed LEfSe in order to identify biomarkers at genus- and phylum levels with an LDA score of more than 2.0 (p < 0.05). In total, 44 bacterial groups were statistically significantly different, whereas 30 bacterial groups with LDA > 2.0 were selected. Four families (Dietziaceae, Burkholderiaceae, Pseudomonadace, and Sinobacteracea) were significantly enriched in group L, whereas only one class (Betaproteobacte) was significantly enriched in group M. A bacterial lineage and two families enriched in group MN were Flavobacteriia (the class, its order Flavobacteriales, and its family Weeksellaceae) and Streptococcacea and Moraxellaceae, respectively. A genus (Acinetobacter) and unassigned bacteria were significantly abundant in group T (Figures 4A,B). No statistically significant difference (LDA score > 2.0, p < 0.05) was observed in groups N and Y (sheep or goat), which was attributed to their great internal differences. Additionally, comparisons were performed between groups MN and N (Figure 5) and between groups MY and SY (Figure 6) to identify the microbial community characteristics of groups N and Y.
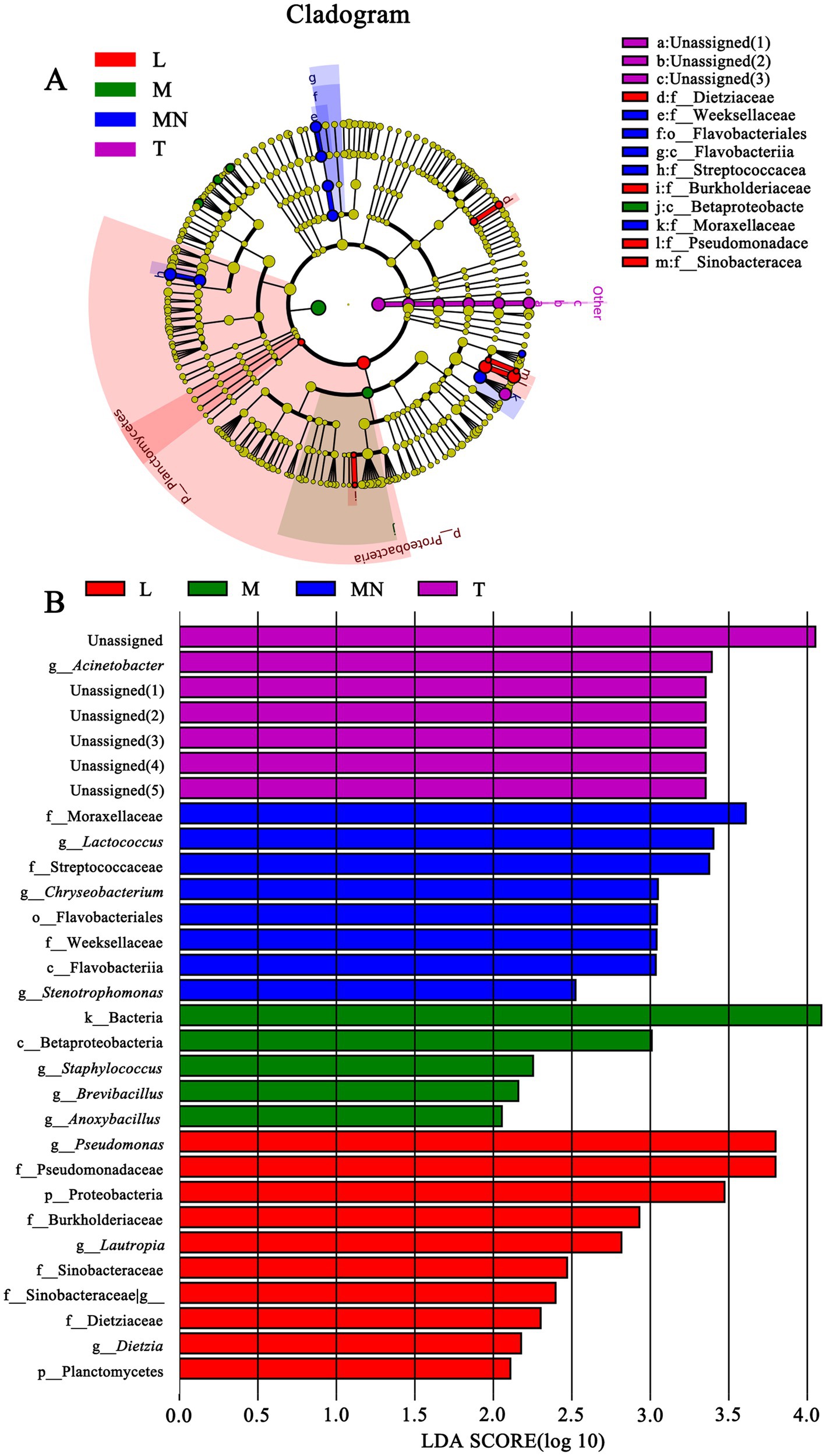
Figure 4. Cladogram (A) and LDA score (B) of LEfSe analysis of bacterial among seven different breed raw milk. Only the taxa with meeting a significant LDA threshold value of >2 and/or < −2 were shown. L, donkey milk; M, horse milk; MN, yak milk; T, camel milk.
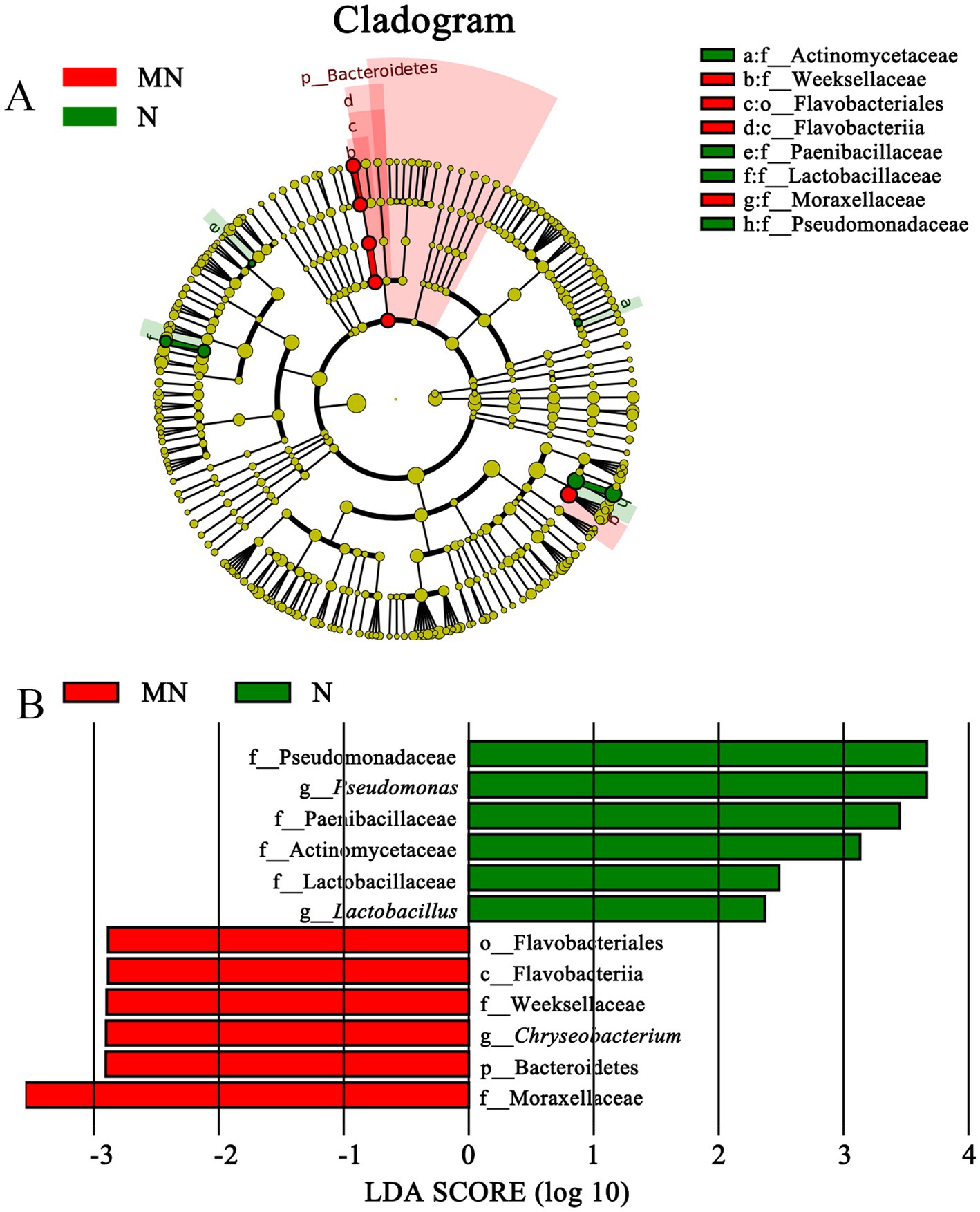
Figure 5. Cladogram (A) and LDA score (B) of LEfSe analysis of bacterial between yak milk and cow milk. Only the taxa with meeting a significant LDA threshold value of >2 and/or < −2 were shown. M, horse milk; MN, yak milk.
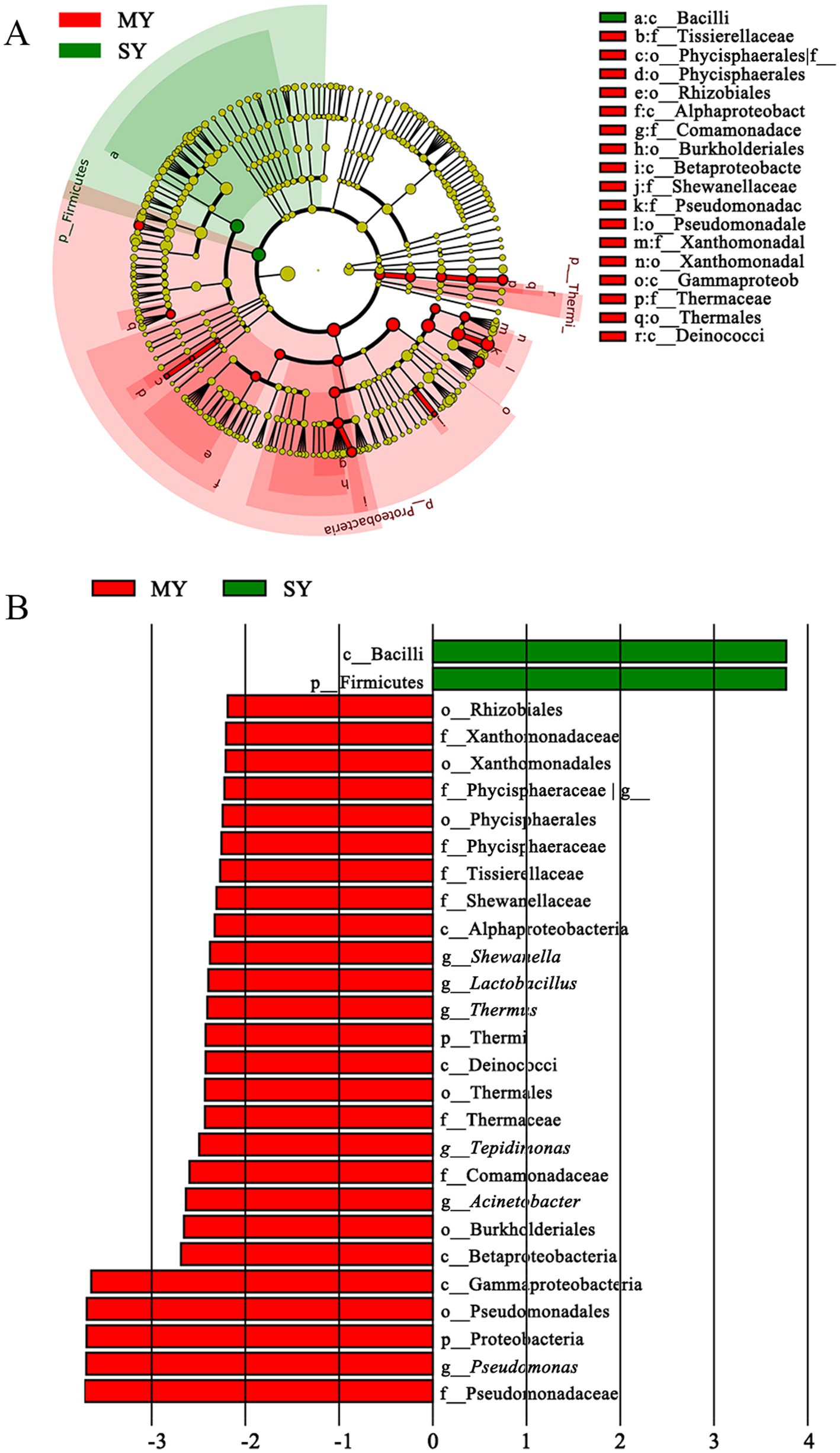
Figure 6. Cladogram (A) and LDA score (B) of LEfSe analysis of bacterial between sheep milk and goat milk. Only the taxa with meeting a significant LDA threshold value of >2 and/or < −2 were shown. MY, sheep milk; SY, goat milk.
3.3.2 Microbial composition differences among cow and yak milk
Moraxellaceae (family) and Bacteroidetes (the Phylum, its class Flavobacteriia, its order Flavobacteriales, its family Weeksellaceae, and its genus Chryseobacterium) were significantly more enriched in group MN than in group N, whereas four families (Actinomycetaceae, Paenibacillaceae, Lactobacillaceae, and Pseudomonadaceae) and two genus (Lactobacillus and Pseudomonas) were significantly enriched in group N (Figure 5). The comparison between groups MN and N showed that pathogenic bacteria, including Chryseobacterium, were more prevalent in group MN, whereas probiotic LAB, including Lactobacillus, were more prevalent in group N.
3.3.3 Microbial composition differences among goat and sheep milk
The comparison between groups MY and SY showed that only the Firmicutes phylum (the phylum and its class Bacilli) was significantly enriched in group SY, whereas Gammaproteobacteria (Xanthomonadales, Shewanella, Pseudomonas, Acinetobacter), Thermi (Thermus), Firmicutes (Lactobacillus, Tissierellaceae), Planctomycetes (Phycisphaerales), Alphaproteobacteria (Rhizobiales), and Betaproteobacteria (Tepidimonas) were more enriched in group MY (Figure 6). This result indicated that nearly half of the predominant bacterial phyla were different between the two groups.
3.4 The co-occurrence of bacteria in different animal milk based on genus level
Co-occurrence network analysis was performed to determine potential relationships among bacterial genera in the milk samples. The co-occurrence network comprised 39 nodes and 68 edges, with 54 positive and 14 negative correlations (p < 0.05) (Figure 7). Three genera, namely Pseudomonas, Lactococcus, and Anoxybacillus, with more neighboring connections, were defined as the core points. Among the three core points, Pseudomonas showed the most neighboring connections. Two genera (Lysobacter and Tepidimonas) showed a significant positive correlation (p < 0.001) with Pseudomonas, whereas three genera (Lactococcus, Enhydrobacter, and Stenotrophomonas) showed a significant negative correlation (p < 0.001) with Pseudomonas. The reason behind this may be the antagonism between Lactococcus and Pseudomonas (Beeram and Silpa, 2021). Lactococcus showed a significant positive correlation with Erwinia, suggesting that these two genera were likely to share a symbiotic or syntrophic relationship. Conversely, Lactococcus showed a significant negative correlation with seven genera, namely Ochrobactru, Tepidimonas, Delftia, Comamonas, Phyllobacterium, and Propionibacterium, indicating a probably antagonistic relationship between Lactococcus and these genera. Anoxybacillus showed a significant positive correlation with many genera such as Ochrobactrum, Thermus, Tepidimonas, Delftia, Sediminibacterium, Ralstonia, Agrobacterium, Amycolatopsis, Meiothermus, and Sphingomonas, suggesting that these genera were likely to share a symbiotic or syntrophic relationship with Anoxybacillus. Additionally, the presence of Bacillus was negatively correlated with the presence of Amycolatopsis, Sphingomonas, Bradyrhizobium, and Propionibacterium, whereas the presence of Enhydrobacter was negatively correlated with the presence of Streptococcus, Leuconostoc, Chryseobacterium, Deinococcus, and Kocuria. Although the co-occurrence network sheds light on the complex relationships among the raw milk microbiota, empirical evidence is needed to support their natural presence.
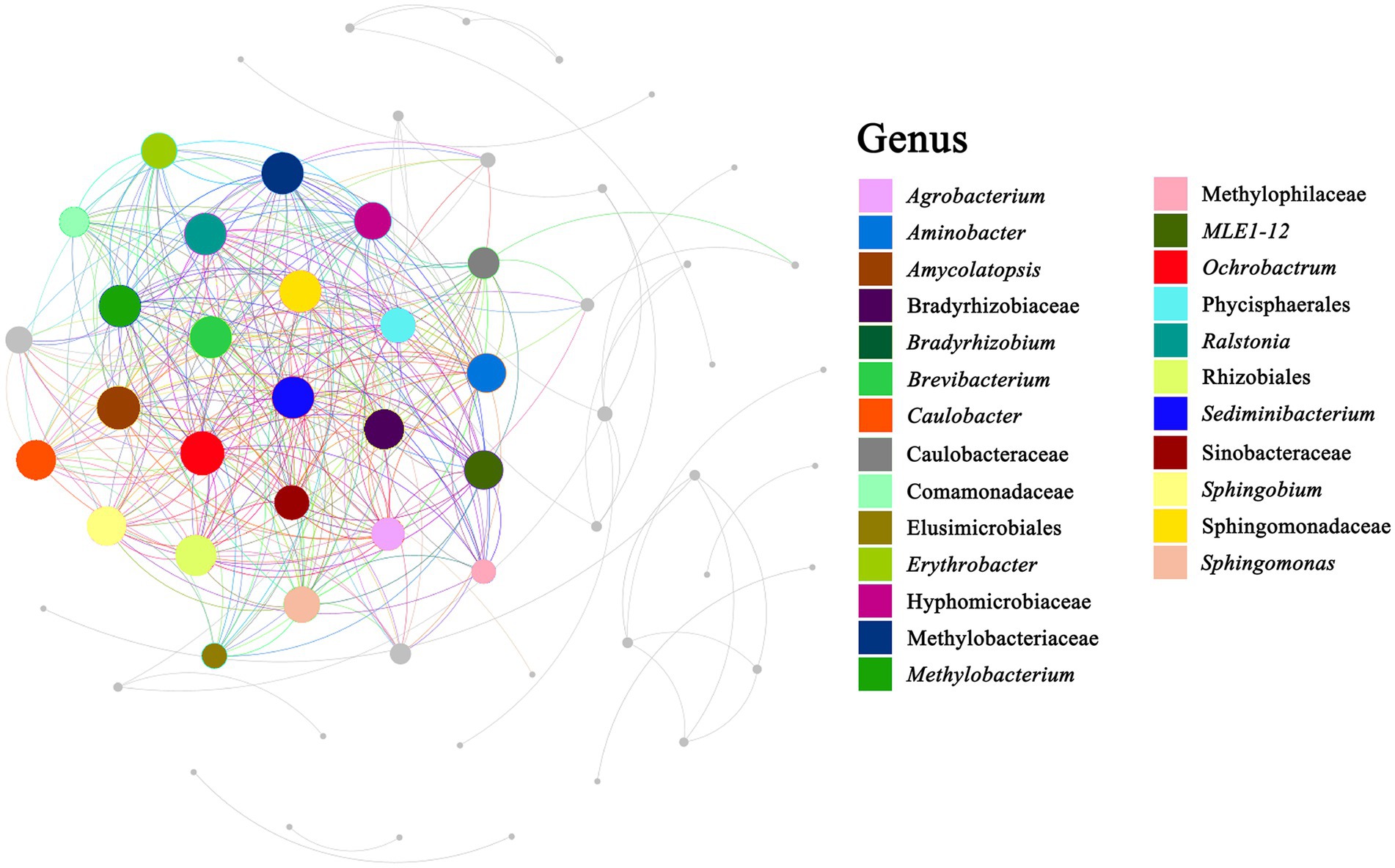
Figure 7. Correlation network diagram. Summary of the significant (p ≤ 0.05) interactions among bacterial (at genus level).
3.5 Microorganisms indicator species among different breeds animal milk
To determine the existence of indicator species, INDVAL analysis, which identifies the indicator species based on OTU fidelity and relative abundance, was run by using the dataset OTU within the R environment. Only OTUs with significant (p < 0.05) INDVAL values that were > 0.3 were considered, as the latter value can be regarded as a good threshold for habitat specialization (Cáceres and Legendre, 2009). Among the 6,452 OTUs, indicator species analysis revealed 1755 OTUs significantly (p < 0.05) associated with the breed when computed across a Seven-group combination. The indicator bacteria assigned at different taxonomic levels are presented in supporting information (Supplementary Table S2).
Nine OTUs exhibited common characteristics in cattle milk (group N) and they belonged to Firmicutes (Clostridium, Enterococcus, Weissella, Brochothrix, and Actinobacteria). Among them, Clostridium and Brochothrix (spoilage microorganisms) and Arcanobacterium (pathogenic microorganisms) deserve more attention (Hijazin et al., 2012; Gribble and Brightwell, 2013; Deslauriers et al., 2024; Yang et al., 2024). In addition, 32 OTUs were characteristic of donkey milk (group L) and belonged to Corynebacterium (Actinobacteria), three genera of Firmicutes (Clostridium, Streptococcus, and Tepidibacter), and six genera of Proteobacteria (Pseudomonas, Moraxella, Rhodobacter, Stenotrophomonas, Pseudomonas, Stenotrophomonas). Among them, prevention against contamination by pathogenic microorganisms (such as Corynebacterium, Streptococcus, Pseudomonas, and Moraxella) during the production process is very important (Soucek et al., 1965; Potgieter and Chalkley, 1991; Ding et al., 2024; Subsomwong et al., 2024).
A total of 220 OTUs displayed a significant association with the horse milk (group M) and belonged to Acidobacteria (Actinomyces, Actinomycetospora, Arsenicicoccus Corynebacterium, Propionibacterium), Bacteroidetes (Flavobacterium, Rudanella, Chryseobacterium, Bacteroides), Firmicutes (Allobaculum, Anoxybacillus, Bacillus, Brevibacillus, Geobacillus, Granulicatella, Lactobacillus, Lactococcus, Leuconostoc, Macrococcus, Paenibacillus, Paenibacillus, Planomicrobium, Rummeliibacillus, Staphylococcus, Streptococcus), Planctomycetes (Gemmata), Proteobacteria (Acetobacter, Hyphomicrobium, Brevundimonas, Methylobacterium, Pseudochrobactrum belong to the class Alphaproteobacteria; Thiobacillus and Hydrogenophaga belong to the class Betaproteobacteria; Desulfobacca and Anaeromyxobacter belong to the class Deltaproteobacteria; Acinetobacter, Pseudoalteromonas, Pseudomonas, and Serratia belong to the class Gammaproteobacteria). Among them, contamination with pathogenic microorganisms (Arsenicicoccus, Propionibacterium, Chryseobacterium, Granulicatella, Streptococcus) should be prevented during the production process (Jeong et al., 2021; Tahir et al., 2023; Chamlagain et al., 2024; Genco et al., 2024; Yu et al., 2024).
A total of 79 OTUs affiliated to Actinobacteria (Microbacterium, Luteococcus, Brachybacterium, Microbacterium, Brachybacterium, Yonghaparkia), Bacteroidetes (Prevotella, Chryseobacterium, Bacteroides), Firmicutes (Streptococcus, Faecalibacterium, Macrococcus, Lactococcus), Betaproteobacteria (Tepidimonas, Hydrogenophaga, Ralstonia), and Gammaproteobacteria (Acinetobacter, Enhydrobacter, Mannheimia, Moraxella, Pseudomonas) were associated with the yak milk (MN). Among them, Luteococcus, Hydrogenophaga, and Acinetobacter are reported to be pathogenic, and steps should be taken to prevent their growth (Vieu et al., 1980; Okiki Pius et al., 2015; Feichtinger et al., 2023).
A total of 189 OTUs affiliated to Actinobacteria (Agrococcus, Brachybacterium, Collinsella, Corynebacterium, Dietzia, Leucobacter, Nocardioides), Bacteroidetes (Prevotella, Chryseobacterium, Porphyromonas, Adhaeribacter), Firmicutes (Aerococcus, Alkalibacterium, Ammoniphilus, Anaerococcus, Bacillus, Butyrivibrio, Dialister, Enterococcus, Gallicola, Jeotgalicoccus, Lactobacillus, Mogibacterium, Planococcus, Planomicrobium, Ruminococcus, Salinicoccus, Streptococcus), Fusobacteria (Fusobacterium), Alphaproteobacteria (Methylopila, Devosia, Brevundimonas), Betaproteobacteria (Lautropia, Hydrogenophaga, Lautropia, Azoarcus), and Gammaproteobacteria (Acinetobacter, Alcanivorax, Enhydrobacter, Halomonas, Luteimonas, Lysobacter, Pseudomonas, Pseudoxanthomonas) were characteristic of sheep milk (group MY). Among them, Collinsella, Leucobacter, Prevotella, Alkalibacterium and Lactobacillus are reported to be beneficial bacteria (Yumoto et al., 2008; Shu et al., 2013; Gomez-Arango et al., 2018; Chang et al., 2019; Jiang et al., 2024), while Corynebacterium, Chryseobacterium, Porphyromonas, Adhaeribacter, Dialister, Streptococcus, Fusobacteria, and Lautropia are reported to be harmful microorganisms (Soucek et al., 1965; Potgieter and Chalkley, 1991; Harrandah et al., 2021; Calheiros Cruz et al., 2022; Antonyuk et al., 2023; Mena-Vázquez et al., 2023; Genco et al., 2024).
A total of 39 OTUs displayed a significant association with goat milk (group SY): Actinobacteria (Sanguibacter and Arthrobacter), Bacteroidetes (Porphyromonas) Firmicutes (Enterococcus), Gammaproteobacteria (Pseudomonas, Erwinia, Pseudomonas), and Betaproteobacteria (Tepidimonas). Among them, Erwinia (spoilage microorganisms), Porphyromonas, and Pseudomonas (pathogenic microorganisms) deserve special attention (Kang et al., 2023; de Jongh et al., 2024; Subsomwong et al., 2024).
A total of 281 OTUs demonstrated a significant association with camel milk (group T): Actinobacteria (Corynebacterium, Leucobacter, Nesterenkonia, Cryocola, Slackia, Microbacterium), Bacteroidetes (Bacteroides, Capnocytophaga, Chryseobacterium, Ornithobacterium, Paludibacter, Porphyromonas, Prevotella, Riemerella, Wautersiella), Firmicutes (Bacillus, Butyrivibrio, Catonella, Clostridium, Coprobacillus, Coprococcus, Dorea, Facklamia, Filifactor, Fusibacter, Helcococcus, Lactococcus, Paenibacillus, Streptococcus, Tissierella_Soehngenia), Fusobacteria (Leptotrichiaceae, Fusobacteriaceae, Leptotrichiaceae), Alphaproteobacteria (Caulobacterales, Sphingomonadales, Rhizobiales, Rickettsiales), Betaproteobacteria (Neisseriales, Burkholderiales, Rhodocyclales), Epsilonproteobacteria (Campylobacter), Gammaproteobacteria (Acinetobacter, Aggregatibacter, Erwinia, Halomonas, Klebsiella, Moraxella, Pasteurella, Pseudomonas, Stenotrophomonas), and Spirochaetes (Treponema). Among these, Corynebacterium, Chryseobacterium, Porphyromonas, Riemerella, Facklamia, Rickettsiales, Neisseriales, Burkholderiales, Campylobacter, Acinetobacter, Aggregatibacter, Erwinia, Klebsiella, Moraxella, Pasteurella, and Pseudomonas have been reported to be harmful (Soucek et al., 1965; Lu et al., 2019; Zou et al., 2020; Wen et al., 2021; Awad et al., 2022; Pérez-Cavazos et al., 2022; Reina et al., 2022; Gloanec et al., 2023; Kang et al., 2023; Li et al., 2023; Lu et al., 2023; Mason et al., 2023; Othman et al., 2023; de Jongh et al., 2024; Genco et al., 2024). Meanwhile, these bacteria have a wide niche breadth and are considered habitat generalists.
When computed across the four-group combination: HM (Hami), TC (Tarbagatay), KS (Kashgar), and YL (Ili), the indicator species analysis revealed 146 OTUs that were significantly (p < 0.05) associated. The indicator bacterial assigned at different taxonomic levels are reported in supporting information (Supplementary Table S3). Only 1 indicator species, Saccharomonospora (Actinobacteria) was found to be associated with Kashgar (group KS). Group HM (Hami) was characterized by Firmicutesc (Brevibacillus), Bacteroidetesc (Prevotella, Ornithobacterium, Riemerella), and Proteobacteriac (Xanthobacter). Among them, prevention against contamination by pathogenic microorganisms (such as Ornithobacterium and Riemerella) during the production process is very important (Awad et al., 2022; Liang et al., 2024). Group TC (Tarbagatay) were characterized by Actinobacteriac (Microbacteriaceaeg, Williamsiaceaeg), Bacteroidetesc (Flectobacillus, Spirosoma, Runella, Myroides), Firmicutesc (Paenibacillus, Aerococcus, Weissella, Erysipelothrix, Alicyclobacillus, Saccharibacillus, Oribacterium), Alphaproteobacteriao (Hyphomonas, Beijerinckia), Betaproteobacteria (Methyloversatilis, Schlegelella, Hydrogenophilus, Neisseria), Gammaproteobacteria (Cardiobacterium), and Synergistetesc (TG5). Among them, Alicyclobacillus (spoilage microorganisms), Erysipelothrix, Hyphomonas, Hydrogenophilus, and Neisseria (pathogenic microorganisms) deserve special attention (Nguyen et al., 2019; Fukui et al., 2023; Kapat et al., 2023; Liu et al., 2023; Liyayi et al., 2023). Group YL (Ili) were characterized by Actinobacteria (Yaniella, Trueperella, Serinicoccus), Bacteroidetes (Sporocytophaga), Chloroflexi (Ardenscatena), Firmicutes (Lactobacillaceae, Catenibacterium, Tissierellaceae, Pseudoramibacter), Planctomycetes (Pirellula, Nostocoida, Lautropia, Citrobacter, Nannocystis, Syntrophobacter, Sinorhizobium), and Verrucomicrobiac (Chthoniobacter). Among these, Trueperella, Bacteroidetes, and Citrobacter have been reported to be harmful (Patrick, 2022; Stuby et al., 2023; Liu et al., 2024).
4 Discussion
Herein, we evaluated bacterial communities present in raw milk obtained from seven animals from four regions in Xinjiang, China. The results suggested that the structures of these communities were affected by multiple factors rather than a single factor. Moreover, the diversity of microbial populations present in the milk samples was affected by various complex factors, such as breeds and regions, which contributed to variations in microbial community structures (Wei et al., 2021).
4.1 Factors affecting microbial communities in raw milk
Microbial community structures in various animal species were initially determined based on their respective growth environments. Nevertheless, the survival of microorganisms in raw milk depends on the nutritional content of the raw milk and the competition and synergy between microorganisms present in it (Mallet et al., 2012; Quigley et al., 2013; Li et al., 2018). Eventually, microorganisms establish a state of equilibrium within microecological environment of raw milk and give rise to complex microbial communities (Li et al., 2018), in which, deterministic and stochastic processes are distinguished (Keady et al., 2023). However, the variety and composition of raw milk samples and interactions among microorganisms present in the samples ultimately determine whether microorganisms from the outside environment can stably exist in the raw milk environment (Wei et al., 2021; Celano et al., 2022).
The present results indicated that animal species significantly affected the community structures of microbes present in the raw milk samples, and the reason was differences in the living environment developed in the raw milk samples obtained from different animal species for the survival of these microorganisms, including differences in the composition and physical and chemical indices of the milk samples (Moossavi et al., 2019; Albonico et al., 2020; Massouras et al., 2020). The compositions of different raw milk samples were correlated with the corresponding characteristics of microbial community structures. Moreover, the microbial community structure of the raw milk samples obtained from different animal breeds was related to some components in the samples.
4.1.1 The effects of ingredients in raw milk on the microbial community
Donkey and mare milk are relatively high in lactose, promoting the proliferation of probiotics including LAB. We found relatively high levels of LAB such as Lactobacillus in donkey and mare milk. Donkey milk is considered the best growth medium for some useful strains of lactobacillus, and lysozyme is regarded as an indirect “bifidogenic factor” (Modler, 1994).
The concentration of lysozyme in donkey milk was significantly higher than in other animal milk. Furthermore, the proportion of lysozyme in donkey whey protein was 21.03%, which is much higher than that in horse and cow milk (Salimei et al., 2004). The high concentration of lysozyme in donkey milk contributes to its strong antibacterial activity against Listeria monocytogenes and Staphylococcus aureus due to the abundant presence of antibacterial components, particularly whey proteins such as lysozyme and lactoferrin (Derdak et al., 2020). Furthermore, donkey milk is safer and less prone to contamination by food-borne pathogenic bacteria, thus having a longer shelf life (Derdak et al., 2020).
The antibacterial property of lysozyme contributes to the simplicity of the microbial structure in donkey milk, resulting in the lowest alpha diversity (Table 1). Lysozyme can effectively inhibit various types of bacteria, including Gram-positive bacteria (Staphylococcus aureus, Bacillus subtilis, and Streptococcus mutans), Gram-negative bacteria (Escherichia coli and Pseudomonas aeruginosa), and (Candida albicans). Therefore, these pathogenic microorganisms are suppressed in donkey milk. Furthermore, donkey milk contains relatively high levels of lactose and lysozyme, promoting the dominance of LAB, including Enterococcus, Sediminibacterium, and Lactobacillus (Figure 2).
4.1.2 Effect of microbial interactions on the final microbial community structure of raw milk
The components of raw milk greatly affect the microbial community, which is also influenced by the network relationship between microorganisms and the synergy and antagonism among them.
Various microorganisms can enter raw milk, adapt to its environment, and interact with each other. Furthermore, some microorganisms exhibit a synergistic effect. Network correlation analysis showed a significant positive correlation between Pseudomonas, Tepidimonas, and Lysobacter. We also found the presence of these three bacterial genera in goat and sheep milk. The dominant intestinal flora found in cheese, dairy products, and human skin are Thermus, Tepidimonas, Delftia, Comamonas, Phyllobacterium, and Propionibacterium. Notably, a significant negative correlation is present between these flora because Lactococcus can inhibit the growth of these spoilage and pathogenic bacteria. We found that raw milk contains abundant lactobacillus (Leuconostoc, Lactobacillus, and Lactococcus), such as camel milk, goat milk, and donkey milk, has relatively lower levels of spoilage bacteria and pathogenic bacteria.
The relatively low species and abundance of putrefactive bacteria in camel milk contribute to its richness in LAB, such as Leuconostoc Lactobacillus, and Acetobacter.
Donkey milk contains more lysozyme and LAB (Lactococcus and Lactobacillus) and also contains relatively few spoilage and pathogenic bacteria.
We found a significant positive correlation between many spoilage and pathogenic bacteria. For instance, Anoxybacillus exhibited a significant positive correlation with Ochrobactrum, Delftia, Ralstonia, and Sphingomonas. These spoilage and pathogenic bacteria are found in horse and yak milk.
To summarize, complementary metabolism and synergistic or antagonistic effects occur among the microorganisms present in raw milk. The removal and retention of microorganisms in raw milk are determined by their interaction and balance, which ultimately form the microbial community structure in raw milk.
4.2 Probiotic resources and potentially harmful bacteria analysis in raw milk
4.2.1 Beneficial bacterial resources
The lactose content in donkey and horse milk is relatively high, resulting in the presence of abundant Lactobacillus. Furthermore, donkey, camel, and goat milk contain abundant Lactococcus, whereas camel and yak milk have relatively rich Leuconostoc. These LAB can be used as starter cultures or probiotic strains.
4.2.2 Potential risk
The presence of potentially pathogenic bacteria in raw milk is a risk for traditionally fermented dairy products. Therefore, precautions must be taken to prevent and control spoilage and pathogenic bacteria when using different dairy products. Our data analysis showed that horse and yak milk have a higher relative abundance of potential spoilage and pathogenic bacteria. Raw horse and yak milk contain the following bacteria: Bradyrhizobium, Chryseobacterium, Propionibacterium, Sphingomonas, and Ochrobactrum. Among them, Bradyrhizobium can lead to the deterioration of dairy products and the production of harmful substances such as biogenic amines and acid substances, which can cause food poisoning and allergic reactions. Chryseobacterium species found in various environments can degrade hemoglobin and produce virulence enzymes, making them potential human pathogens (Mwanza et al., 2022). Additionally, Chryseobacterium sp. is also responsible for the premature spoilage of milk (Alothman et al., 2017). Propionibacterium can cause spoilage of dairy products and contribute to skin acne, causing redness and swelling. For instance, Propionibacterium acnes is associated with the inflammatory process of acne lesions (Brook and Frazier, 1991). The genus Sphingomonas contains several pathogenic organisms, such as Sphingomonas paucimobilis, which are associated with meningitis, peritonitis, wound infection, and other human infections (Koskinen et al., 2000). Ochrobactrum spp. are generally considered to have low pathogenicity, however, they are increasingly being identified as the cause of infections in individuals with a healthy immune system (Ryan and Pembroke, 2020), with Ochrobactrum anthropi recognized as an opportunistic pathogen in breast milk (Asaf et al., 2020).
Horse milk was found to contain Staphylococcus and Flavobacterium. Staphylococci, commonly encountered pathogens, are often present in food (Kim et al., 2021), with Staphylococcus epidermidis being associated with various infections (Morot-Bizot et al., 2004). Staphylococcus aureus, another member of the Staphylococcus genus, is a significant foodborne pathogen capable of producing staphylococcal enterotoxins, which can adversely affect human health (Kim et al., 2021). Staphylococcus spp. has also been reported as a primary pathogen in horse mastitis (Colavita et al., 2016). Flavobacterium, detected in horse milk, is a lipolytic bacterium that produces lipolytic enzymes, contributing to the rancidity of dairy products. Some species of Flavobacterium, such as Flavobacterium meningosepticum, have the potential to cause infections such as meningitis or endocarditis, making them human pathogens (Colavita et al., 2016; Soler et al., 2023). Certain species of Flavobacterium are considered pathogenic or opportunistic pathogens, causing diseases in various organisms, including plants, fish, and humans (Zamora et al., 2012).
Yak milk is found to contain Streptococcus, which can contribute to the decomposition of protein and fat in dairy products, leading to natural spoilage. Additionally, Streptococcus can cause severe infectious diseases with high morbidity and mortality rates (Nguyen et al., 2015).
Camel milk and goat milk are found to contain more Moraxella species. Moraxella spp. is associated with meat spoilage. Additionally, camel milk showed a relatively higher abundance of Acinetobacter species. Acinetobacter is an opportunistic pathogen known to cause infections in immunocompromised individuals. It is considered an important opportunistic pathogen responsible for nosocomial infections. Acinetobacter species can colonize the digestive tract through the consumption of contaminated food. For instance, Acinetobacter lwoffii is believed to have the potential to induce gastritis (Carvalheira et al., 2021). Goat milk also contains a high abundance of Brochothrix, and Brochothrix thermosphacta is the main spoilage flora associated with crucian carp meat.
The problematic genera detected in the milk samples include Kocuria, Carnobacterium, and some of the Sphingomonas Kocuria is the main spoilage bacterium in steamed cakes. Carnobacterium can produce exotoxin or botulinum toxin in an anaerobic environment, which can have paralyzing effects in humans.
The genus Pseudomonas appears to be relatively abundant in sheep and donkey milk. Members of this genus are frequently implicated in the degradation and spoilage of a wide range of plant or animal foods (Caldera et al., 2016). Within the Pseudomonas genus, at least three species are known to be pathogenic to animals or humans. Pseudomonas aeruginosa is considered a conditional pathogen, which is typically associated with infections such as wound infections from severe burns, middle ear infections, urinary tract infections, and even sepsis.
The results of indicator bacteria analysis indicate the need to be cautious regarding the presence of potentially harmful bacteria in raw milk from various sources. It is crucial to implement appropriate measures during the production process to prevent the growth of spoilage and pathogenic bacteria.
5 Conclusion
To conclude, our study revealed the bacterial communities present in the raw milk of seven animals across four regions in Xinjiang, China. Camel milk exhibited the highest bacterial diversity, accompanied by a notable presence of unidentified bacteria. The structure of bacterial communities in raw milk samples is influenced by the components of raw milk and microbial interactions. Horse and yak milk showed a higher prevalence of spoilage microorganisms and pathogenic bacteria, whereas goat, donkey, and camel milk exhibited a higher abundance of probiotic LAB. Furthermore, indicator species analysis showed that raw milk from each breed and every region contained specific pathogenic microorganisms that should be given attention and their presence should be prevented. The study contributed to the further development and use of different animal raw milk while providing help to prevent and control the presence of pathogenic microorganisms in the production process. Nevertheless, further investigations are warranted to determine the underlying reason for the common habitat selection of bacteria and their communications.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI -PRJNA1170235, SAMN44099913 - SAMN44100054.
Author contributions
BL: Data curation, Investigation, Software, Visualization, Writing – original draft, Formal analysis, Project administration. FD: Writing – original draft, Data curation, Formal analysis, Investigation, Software, Visualization. YL: Writing – original draft, Data curation, Formal analysis, Investigation. JD: Writing – original draft, Data curation, Formal analysis, Investigation. HS: Investigation, Resources, Writing – original draft. YN: Resources, Writing – original draft, Investigation. YZ: Writing – original draft, Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Software, Visualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The research was supported by the Financial Science and Technology program of Xinjiang Production and Construction Corps (2024AB050 and 2023AB050), the National Natural Science Foundation of China (grant nos. 32260568 and 31760446), the Joint Science and Technology Innovation project of the 7th Agricultural Division of XPCC-Shihezi University (QS2023014), and the financially Science and Technology Project of Shihezi Government (2024GY02 and 2020PT01).
Conflict of interest
BL, HS, and YN were employed by Xinjiang Tianrun Dairy Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1382286/full#supplementary-material
References
Albonico, F., Barelli, C., Albanese, D., Manica, M., Partel, E., Rosso, F., et al. (2020). Raw milk and fecal microbiota of commercial alpine dairy cows varies with herd, fat content and diet. PLoS One 15:e0237262. doi: 10.1371/journal.pone.0237262
Alothman, M., Lusk, K. A., Silcock, P., and Bremer, P. J. (2017). Comparing PTR-MS profile of milk inoculated with pure or mixed cultures of spoilage bacteria. Food Microbiol. 64, 155–163. doi: 10.1016/j.fm.2017.01.001
Antonyuk, S. V., Sieminska, K., Smiga, M., Strange, R. W., Wagner, M., Barnett, K. J., et al. (2023). Bacteroides fragilis expresses three proteins similar to Porphyromonas gingivalis HmuY: Hemophore-like proteins differentially evolved to participate in heme acquisition in oral and gut microbiomes. FASEB J. 37:e22981. doi: 10.1096/fj.202300366R
Ariute, J. C., Coelho-Rocha, N. D., Dantas, C. W. D., de Vasconcelos, L. A. T., Profeta, R., de Jesus Sousa, T., et al. (2023). Probiogenomics of Leuconostoc mesenteroides strains F-21 and F-22 isolated from human breast milk reveal beneficial properties. Probiotics Antimicrob. Proteins, 1–16. doi: 10.1007/s12602-023-10170-7
Asaf, S., Numan, M., Khan, A. L., and Al-Harrasi, A. (2020). Sphingomonas: from diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 40, 138–152. doi: 10.1080/07388551.2019.1709793
Awad, N. F., Hashem, Y. M., Elshater, N. S., Khalifa, E., Hamed, R. I., Nossieur, H. H., et al. (2022). Therapeutic potentials of aivlosin and/or zinc oxide nanoparticles against mycoplasma gallisepticum and/or Ornithobacterium rhinotracheale with a special reference to the effect of zinc oxide nanoparticles on aivlosin tissue residues: an in vivo approach. Poult. Sci. 101:101884. doi: 10.1016/j.psj.2022.101884
Ayyash, M., Abushelaibi, A., Al-Mahadin, S., Enan, M., El-Tarabily, K., and Shah, N. (2018). In-vitro investigation into probiotic characterisation of streptococcus and enterococcus isolated from camel milk. LWT 87, 478–487. doi: 10.1016/j.lwt.2017.09.019
Beeram, E., and Silpa, C. (2021). Antagonist effect of Lactococcus on Pseudomonas aeruginosa isolated from milk and curd. Sci. Arch. 2, 109–112. doi: 10.47587/SA.2021.2208
Ben Maamar, S., Glawe, A. J., Brown, T. K., Hellgeth, N., Hu, J., Wang, J.-P., et al. (2020). Mobilizable antibiotic resistance genes are present in dust microbial communities. PLoS Pathog. 16:e1008211. doi: 10.1371/journal.ppat.1008211
Bokulich, N. A., Thorngate, J. H., Richardson, P. M., and Mills, D. A. (2014). Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 111, e139–e148. doi: 10.1073/pnas.1317377110
Brook, I., and Frazier, E. H. (1991). Infections caused by Propionibacterium species. Rev. Infect. Dis. 13, 819–822. doi: 10.1093/clinids/13.5.819
Cáceres, M. D., and Legendre, P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90, 3566–3574. doi: 10.1890/08-1823.1
Caldera, L., Franzetti, L. V., Van Coillie, E., De Vos, P., Stragier, P., De Block, J., et al. (2016). Identification, enzymatic spoilage characterization and proteolytic activity quantification of pseudomonas spp. isolated from different foods. Food Microbiol. 54, 142–153. doi: 10.1016/j.fm.2015.10.004
Calheiros Cruz, G., Sousa, M., Vilela, S., Teixeira E Costa, F., and Silva, F. J. (2022). Lautropia mirabilis: an exceedingly rare cause of peritoneal dialysis-associated peritonitis. Case Rep. Nephrol. Dial. 12, 81–84. doi: 10.1159/000524494
Carvalheira, A., Silva, J., and Teixeira, P. (2021). Acinetobacter spp. in food and drinking water–A review. Food Microbiol. 95:103675. doi: 10.1016/j.fm.2020.103675
Celano, G., Calasso, M., Costantino, G., Vacca, M., Ressa, A., Nikoloudaki, O., et al. (2022). Effect of seasonality on microbiological variability of raw cow milk from apulian dairy farms in Italy. Microbiol. Spectr. 10, e00514–e00522. doi: 10.1128/spectrum.00514-22
Chamlagain, B., Edelmann, M., Katina, K., Varmanen, P., and Piironen, V. (2024). Vitamin B12 production in solubilized protein extract of bioprocessed wheat bran with Propionibacterium freudenreichii. LWT 192:115731. doi: 10.1016/j.lwt.2024.115731
Chang, C.-J., Lin, T.-L., Tsai, Y.-L., Wu, T.-R., Lai, W.-F., Lu, C.-C., et al. (2019). Next generation probiotics in disease amelioration. J. Food Drug Anal. 27, 615–622. doi: 10.1016/j.jfda.2018.12.011
Chi, F., Tan, Z., Gu, X., Yang, L., and Luo, Z. (2021). Bacterial community diversity of yak milk dreg collected from Nyingchi region of Tibet, China. Lebensm. Wiss. Technol. 145:111308. doi: 10.1016/j.lwt.2021.111308
Colavita, G., Amadoro, C., Rossi, F., Fantuz, F., and Salimei, E. (2016). Hygienic characteristics and microbiological hazard identification in horse and donkey raw milk. Vet. Ital. 52, 21–29. doi: 10.12834/VetIt.180.545.1
Dalmasso, A., Civera, T., and Bottero, M. T. (2017). Characterization of bacterial communities of donkey milk by high-throughput sequencing. Int. J. Food Microbiol. 251, 67–72. doi: 10.1016/j.ijfoodmicro.2017.03.023
De Cáceres, M., Legendre, P., and Moretti, M. (2010). Improving indicator species analysis by combining groups of sites. Oikos 119, 1674–1684. doi: 10.1111/j.1600-0706.2010.18334.x
De Jongh, C. A., Bikker, F. J., De Vries, T. J., Werner, A., Gibbs, S., and Krom, B. P. (2024). Porphyromonas gingivalis interaction with Candida albicans allows for aerobic escape, virulence and adherence. Biofilms 7:100172. doi: 10.1016/j.bioflm.2023.100172
Derdak, R., Sakoui, S., Pop, O. L., Muresan, C. I., Vodnar, D. C., Addoum, B., et al. (2020). Insights on health and food applications of Equus asinus (donkey) milk bioactive proteins and peptides—an overview. Food Secur. 9:1302. doi: 10.3390/foods9091302
Deslauriers, N., Maduro, L., Lepp, D., Gong, J., Abdul-Careem, M. F., and Boulianne, M. (2024). Determination of the virulence status of Clostridium perfringens strains using a chicken intestinal ligated loop model is important for understanding the pathogenesis of necrotic. Poult. Sci. 103:103433. doi: 10.1016/j.psj.2024.103433
Ding, R., Yu, J., Ke, W., Du, L., Cheng, G., Hu, S., et al. (2024). TLR2 regulates Moraxella catarrhalis adhesion to and invasion into alveolar epithelial cells and mediates inflammatory responses. Virulence 15:2298548. doi: 10.1080/21505594.2023.2298548
Doyle, C. J., Gleeson, D., O'toole, P. W., and Cotter, P. D. (2017). High-throughput metataxonomic characterization of the raw milk microbiota identifies changes reflecting lactation stage and storage conditions. Int. J. Food Microbiol. 255, 1–6. doi: 10.1016/j.ijfoodmicro.2017.05.019
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Fei, P., Yuan, X., Zhao, S., Yang, T., Xiang, J., Chen, X., et al. (2019). Prevalence and genetic diversity of Bacillus cereus isolated from raw milk and cattle farm environments. Curr. Microbiol. 76, 1355–1360. doi: 10.1007/s00284-019-01741-5
Feichtinger, S., Lazar, A. A., Luebbe, M. A., Accola, M. A., Jung-Hynes, B. D., Anderson, P. J., et al. (2023). Case report: isolation of Hydrogenophaga from septic blood culture following near-death drowning in lakewater. Access Microbiol. 5, 000533–000534. doi: 10.1099/acmi.0.000533.v4
Fotou, K., Tzora, A., Voidarou, C., Alexopoulos, A., Plessas, S., Avgeris, I., et al. (2011). Isolation of microbial pathogens of subclinical mastitis from raw sheep’s milk of Epirus (Greece) and their role in its hygiene. Anaerobe 17, 315–319. doi: 10.1016/j.anaerobe.2011.05.002
Fukui, Y., Abe, M., and Kobayashi, M. (2023). Effects of Hyphomonas strains on the growth of red algae Pyropia species by attaching specifically to their rhizoids. Microb. Ecol. 86, 2502–2514. doi: 10.1007/s00248-023-02257-z
Genco, M., Curtoni, A., Bottino, P., Scabini, S., Corcione, S., Vita, D., et al. (2024). First case of Chryseobacterium gallinarum bloodstream infection: a diagnostic and therapeutic challenge for an emerging pathogen. New Microbiol. 46, 412–415
Gloanec, N., Dory, D., Quesne, S., Béven, V., Poezevara, T., Amelot, M., et al. (2023). Research note: analysis of immune responses in broilers after vaccination against Campylobacter jejuni. Poult. Sci. 102:102510. doi: 10.1016/j.psj.2023.102510
Gomez-Arango, L. F., Barrett, H. L., Wilkinson, S. A., Callaway, L. K., Mcintyre, H. D., Morrison, M., et al. (2018). Low dietary fiber intake increases Collinsella abundance in the gut microbiota of overweight and obese pregnant women. Gut Microbes 9, 189–201. doi: 10.1080/19490976.2017.1406584
Gran, H. M., Wetlesen, A., Mutukumira, A. N., Rukure, G., and Narvhus, J. A. (2003). Occurrence of pathogenic bacteria in raw milk, cultured pasteurised milk and naturally soured milk produced at small-scale dairies in Zimbabwe. Food Control 14, 539–544. doi: 10.1016/S0956-7135(02)00117-2
Gribble, A., and Brightwell, G. (2013). Spoilage characteristics of Brochothrix thermosphacta and campestris in chilled vacuum packaged lamb, and their detection and identification by real time PCR. Meat Sci. 94, 361–368. doi: 10.1016/j.meatsci.2013.03.016
Hahne, J., Kloster, T., Rathmann, S., Weber, M., and Lipski, A. (2018). Isolation and characterization of Corynebacterium spp. from bulk tank raw cow's milk of different dairy farms in Germany. PLoS One 13:e0194365. doi: 10.1371/journal.pone.0194365
Hao, X., Jiang, R., and Chen, T. (2011). Clustering 16S rRNA for OTU prediction: a method of unsupervised Bayesian clustering. Bioinformatics 27, 611–618. doi: 10.1093/bioinformatics/btq725
Harrandah, A. M., Chukkapalli, S. S., Bhattacharyya, I., Progulske-Fox, A., and Chan, E. K. (2021). Fusobacteria modulate oral carcinogenesis and promote cancer progression. J. Oral Microbiol. 13:1849493. doi: 10.1080/20002297.2020.1849493
Hassan, A. N., and Frank, J. (2011). Microorganisms associated with milk. Encycl. Dairy Sci. 2, 447–457. doi: 10.1016/B978-0-12-374407-4.00309-5
Hijazin, M., Prenger-Berninghoff, E., Sammra, O., Alber, J., Lämmler, C., Kämpfer, P., et al. (2012). Arcanobacterium canis sp. nov., isolated from otitis externa of a dog, and emended description of the genus Arcanobacterium Collins et al. 1983 emend. Yassin et al. 2011. Int. J. Syst. Bacteriol. 62, 2201–2205. doi: 10.1099/ijs.0.037150-0
Hou, Q., Xu, H., Zheng, Y., Xi, X., Kwok, L.-Y., Sun, Z., et al. (2015). Evaluation of bacterial contamination in raw milk, ultra-high temperature milk and infant formula using single molecule, real-time sequencing technology. J. Dairy Sci. 98, 8464–8472. doi: 10.3168/jds.2015-9886
Jamali, H., Paydar, M., Radmehr, B., and Ismail, S. (2015). Prevalence, characterization, and antimicrobial resistance of Yersinia species and Yersinia enterocolitica isolated from raw milk in farm bulk tanks. J. Dairy Sci. 98, 798–803. doi: 10.3168/jds.2014-8853
Jeong, J. H., Kweon, O. J., Kim, H. R., Kim, T.-H, Ha, S.-M., and Lee, M.-K. (2021). A novel species of the genus Arsenicicoccus isolated from human blood using whole-genome sequencing. Ann Lab Med. 41, 323–327. doi: 10.3343/alm.2021.41.3.323
Jiang, Y.-H., Liang, M., Yang, Y.-H., Xue, J., and Suo, H.-Y. (2024). Probiotic Lactobacillus plantarum SHY21-2 protected zebrafish against Aeromonas hydrophila infection by maintaining intestinal barrier integrity, inhibiting inflammatory and oxidative stress responses, and regulating intestinal microbiome. Aquaculture 582:740506. doi: 10.1016/j.aquaculture.2023.740506
Jin, T., Sun, D., Su, J., Zhang, H.-W., and Sue, H. J. (2009). Antimicrobial efficacy of zinc oxide quantum dots against Listeria monocytogenes, salmonella enteritidis, and Escherichia coli O157: H7. J. Food Sci. 74, M46–M52. doi: 10.1111/j.1750-3841.2008.01013.x
Kamilari, E., Anagnostopoulos, D. A., Papademas, P., Efthymiou, M., Tretiak, S., and Tsaltas, D. (2020). Snapshot of Cyprus raw goat milk bacterial diversity via 16S rDNA high-throughput sequencing; impact of cold storage conditions. Fermentation 6:100. doi: 10.3390/fermentation6040100
Kang, I.-J., Park, Y., Roh, E., and Lee, J. H. (2023). Novel single nucleotide variations alter pathogenicity in Korean isolates of Erwinia amylovora. Plant Dis. 108, 1174–1178. doi: 10.1094/PDIS-09-23-1836-SC
Kapat, A., Roy, M., Konar, J., Mandal, A. K., and Bala, A. K. (2023). Erysipelothrix rhusiopathiae: an emerging organism causing pleural effusion in an immunocompetent child. J. Adv. Microbiol. 23, 30–35. doi: 10.9734/jamb/2023/v23i12774
Karahadian, C., Josephson, D., and Lindsay, R. (1985). Contribution of Penicillium sp. to the flavors of brie and camembert cheese. J. Dairy Sci. 68, 1865–1877. doi: 10.3168/jds.S0022-0302(85)81043-2
Keady, M. M., Jimenez, R. R., Bragg, M., Wagner, J. C., Bornbusch, S. L., Power, M. L., et al. (2023). Ecoevolutionary processes structure milk microbiomes across the mammalian tree of life. Proc. Natl. Acad. Sci. USA 120:e2218900120. doi: 10.1073/pnas.2218900120
Kim, E., Yang, S.-M., Won, J.-E., Kim, D.-Y., Kim, D.-S., and Kim, H.-Y. (2021). Real-time PCR method for the rapid detection and quantification of pathogenic staphylococcus species based on novel molecular target genes. Food Secur. 10:2839. doi: 10.3390/foods10112839
Kondrotiene, K., Lauciene, L., Andruleviciute, V., Kasetiene, N., Serniene, L., Sekmokiene, D., et al. (2020). Safety assessment and preliminary in vitro evaluation of probiotic potential of Lactococcus lactis strains naturally present in raw and fermented milk. Curr. Microbiol. 77, 3013–3023. doi: 10.1007/s00284-020-02119-8
Koskinen, R., Ali-Vehmas, T., Kämpfer, P., Laurikkala, M., Tsitko, I., Kostyal, E., et al. (2000). Characterization of Sphingomonas isolates from Finnish and Swedish drinking water distribution systems. J. Appl. Microbiol. 89, 687–696. doi: 10.1046/j.1365-2672.2000.01167.x
Li, W., Mutuvulla, M., Chen, X., Jiang, M., and Dong, M. (2012). Isolation and identification of high viscosity-producing lactic acid bacteria from a traditional fermented milk in Xinjiang and its role in fermentation process. Eur. Food Res. Technol. 235, 497–505. doi: 10.1007/s00217-012-1779-7
Li, N., Wang, Y., You, C., Ren, J., Chen, W., Zheng, H., et al. (2018). Variation in raw milk microbiota throughout 12 months and the impact of weather conditions. Sci. Rep. 8:2371. doi: 10.1038/s41598-018-20862-8
Li, F., Zhao, W., Zhu, P., Li, Z., Song, J., Zhu, J., et al. (2023). Moraxella nasibovis sp. nov., isolated from a cow with respiratory disease. Curr. Microbiol. 80:305. doi: 10.1007/s00284-023-03415-9
Liang, Z., Li, H., Yang, D., Yin, L., Wu, Y., Liu, J., et al. (2024). A novel bivalent inactivated vaccine for ducks against Riemerella anatipestifer based on serotype distribution in southern China. Poult. Sci. 103:103427. doi: 10.1016/j.psj.2024.103427
Liu, S., Li, H., Zhu, Z., Wu, M., Jin, M., Wang, X., et al. (2024). A bacteriophage against Citrobacter braakii and its synergistic effect with antibiotics. Arch. Microbiol. 206, 1–8. doi: 10.1007/s00203-023-03803-6
Liu, X., Wu, Y., Ran, J., Jiao, L., Sun, L., and Ye, F. (2023). Antibacterial mechanism of inosine against Alicyclobacillus acidoterrestris. J. Food Sci. 89, 1187–1195. doi: 10.1111/1750-3841.16919
Liyayi, I. K., Forehand, A. L., Ray, J. C., and Criss, A. K. (2023). Metal piracy by Neisseria gonorrhoeae to overcome human nutritional immunity. PLoS Pathog. 19:e1011091. doi: 10.1371/journal.ppat.1011091
Lu, Q.-F., Cao, D.-M., Su, L.-L., Li, S.-B., Ye, G.-B., Zhu, X.-Y., et al. (2019). Genus-wide comparative genomics analysis of Neisseria to identify new genes associated with pathogenicity and niche adaptation of Neisseria pathogens. Int. J. Genomics 2019:6015730. doi: 10.1155/2019/6015730
Lu, M., Ji, Y., Zhao, H., Wang, W., Tian, J., Duan, C., et al. (2023). Circulation of multiple Rickettsiales bacteria in ticks from Sichuan province, Southwest China. Microb. Pathog. 183:106313. doi: 10.1016/j.micpath.2023.106313
Mahmoudi, I., Moussa, O. B., Khaldi, T. E. M., Kebouchi, M., Soligot, C., Le Roux, Y., et al. (2016). Functional in vitro screening of lactobacillus strains isolated from Tunisian camel raw milk toward their selection as probiotic. Small Rumin. Res. 137, 91–98. doi: 10.1016/j.smallrumres.2016.03.016
Mallet, A., Guéguen, M., Kauffmann, F., Chesneau, C., Sesboué, A., and Desmasures, N. (2012). Quantitative and qualitative microbial analysis of raw milk reveals substantial diversity influenced by herd management practices. Int. Dairy J. 27, 13–21. doi: 10.1016/j.idairyj.2012.07.009
Mason, S., Vornhagen, J., Smith, S. N., Mike, L. A., Mobley, H. L., and Bachman, M. A. (2023). The Klebsiella pneumoniae ter operon enhances stress tolerance. Infect. Immun. 91, 00559–00522. doi: 10.1128/iai.00559-22
Masoud, W., and Jakobsen, M. (2005). The combined effects of pH, NaCl and temperature on growth of cheese ripening cultures of Debaryomyces hansenii and coryneform bacteria. Int. Dairy J. 15, 69–77. doi: 10.1016/j.idairyj.2004.05.008
Massouras, T., Bitsi, N., Paramithiotis, S., Manolopoulou, E., Drosinos, E. H., and Triantaphyllopoulos, K. A. (2020). Microbial profile antibacterial properties and chemical composition of raw donkey milk. Animals 10:2001. doi: 10.3390/ani10112001
Mena-Vázquez, N., Ruiz-Limón, P., Moreno-Indias, I., Manrique-Arija, S., Lisbona-Montañez, J. M., Rioja, J., et al. (2023). Adiposity is associated with expansion of the genus Dialister in rheumatoid arthritis patients. Biomed. Pharmacother. 160:114388. doi: 10.1016/j.biopha.2023.114388
Mo, L., Jin, H., Pan, L., Hou, Q., Li, C., Darima, I., et al. (2019). Biodiversity of lactic acid bacteria isolated from fermented milk products in Xinjiang, China. Food Biotechnol. 33, 174–192. doi: 10.1080/08905436.2019.1574230
Modler, H. W. (1994). Bifidogenic factor-source, metabolism and application. Int. Dairy J. 4, 383–407. doi: 10.1016/0958-6946(94)90055-8
Moosavi-Nasab, M., Radi, M., and Jouybari, H. A. (2010). Investigation of enzyme modified cheese production by two species of aspergillus. Afr. J. Biotechnol. 9, 508–511.
Moossavi, S., Sepehri, S., Robertson, B., Bode, L., Goruk, S., Field, C. J., et al. (2019). Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe 25, 324–335. doi: 10.1016/j.chom.2019.01.011
Morot-Bizot, S. C., Talon, R., and Leroy, S. (2004). Development of a multiplex PCR for the identification of staphylococcus genus and four staphylococcal species isolated from food. J. Appl. Microbiol. 97, 1087–1094. doi: 10.1111/j.1365-2672.2004.02399.x
Moussaid, S., El Alaoui, M. A., Ounine, K., Benali, A., Bouhlal, O., Rkhaila, A., et al. (2023). In-vitro evaluation of the probiotic potential and the fermentation profile of Pediococcus and enterococcus strains isolated from Moroccan camel milk. Arch. Microbiol. 205:144. doi: 10.1007/s00203-023-03489-w
Mwanza, E. P., Hugo, A., Charimba, G., and Hugo, C. J. (2022). Pathogenic potential and control of Chryseobacterium species from clinical, fish, food and environmental sources. Microorganisms 10:895. doi: 10.3390/microorganisms10050895
Nguyen, T. H., Ishizuna, F., Sato, Y., Arai, H., and Ishii, M. (2019). Physiological characterization of poly-β-hydroxybutyrate accumulation in the moderately thermophilic hydrogen-oxidizing bacterium Hydrogenophilus thermoluteolus TH-1. J. Biosci. Bioeng. 127, 686–689. doi: 10.1016/j.jbiosc.2018.11.011
Nguyen, C. T., Park, S.-S., and Rhee, D.-K. (2015). Stress responses in streptococcus species and their effects on the host. J. Microbiol. 53, 741–749. doi: 10.1007/s12275-015-5432-6
Nörnberg, M. F. B. L., Friedrich, R. S. C., Weiss, R. D. N., Tondo, E. C., and Brandelli, A. (2010). Proteolytic activity among psychrotrophic bacteria isolated from refrigerated raw milk. Int. J. Dairy Technol. 63, 41–46. doi: 10.1111/j.1471-0307.2009.00542.x
Okiki Pius, A., Oluwadunsin, O., and Benjamin, O. (2015). Antibacterial activity of ginger (Zingiber officinale) against isolated bacteria from the respiratory tract infections. J. Biol. Agric. Healthc. 79, 588–592.
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’hara, R. B., et al. (2013) Package ‘vegan’: community ecology package. Available at: https://cran.ism.ac.jp/web/packages/vegan/vegan.pdf.
Othman, A., Diaz, J., Al-Momani, M., and Hoskere, G. (2023). Pasteurella multocida leading to acute respiratory failure. Am. J. Med. Sci. 365:S332. doi: 10.1016/S0002-9629(23)00616-X
Panesar, P. (2011). Fermented dairy products: starter cultures and potential nutritional benefits. Food Nutr. Sci. 2, 47–51. doi: 10.4236/fns.2011.21006
Patrick, S. (2022). A tale of two habitats: Bacteroides fragilis, a lethal pathogen and resident in the human gastrointestinal microbiome. Microbiology 168:001156. doi: 10.1099/mic.0.001156
Pérez-Cavazos, S., Cisneros-Saldaña, D., Espinosa-Villaseñor, F., Castillo-Bejarano, J. I., Vaquera-Aparicio, D. N., Sánchez-Alanís, H., et al. (2022). Facklamia hominis pyelonephritis in a pediatric patient: first case report and review of the literature. Ann. Clin. Microbiol. Antimicrob. 21:4. doi: 10.1186/s12941-022-00497-4
Potgieter, E., and Chalkley, L. (1991). Reciprocal transfer of penicillin resistance genes between Streptococcus pneumoniae, Streptococcus mitior and streptococcus sanguis. J. Antimicrob. Chemoth. 28, 463–465. doi: 10.1093/jac/28.3.463
Quigley, L., O'sullivan, O., Stanton, C., Beresford, T. P., Ross, R. P., Fitzgerald, G. F., et al. (2013). The complex microbiota of raw milk. FEMS Microbiol. Rev. 37, 664–698. doi: 10.1111/1574-6976.12030
R Development Core Team (2013). R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing Available at: http://www.R-project.org/.
Rahmeh, R., Akbar, A., Alomirah, H., Kishk, M., Al-Ateeqi, A., Al-Milhm, S., et al. (2022). Data on microbial diversity of camel milk microbiota determined by 16S rRNA gene sequencing. Data Brief 45:108744. doi: 10.1016/j.dib.2022.108744
Rajawardana, D. U., Fernando, P. C., Biggs, P. J., Hewajulige, I. G. N., Nanayakkara, C. M., Wickramasinghe, S., et al. (2022). An insight into tropical milk microbiome: bacterial community composition of cattle milk produced in Sri Lanka. Int. Dairy J. 126:105266. doi: 10.1016/j.idairyj.2021.105266
Reina, R., León-Moya, C., and Garnacho-Montero, J. (2022). Tratamiento de infecciones graves por Acinetobacter baumannii. Med. Intensiva 46, 700–710. doi: 10.1016/j.medin.2022.08.003
Ryan, M. P., and Pembroke, J. T. (2020). The genus Ochrobactrum as major opportunistic pathogens. Microorganisms 8:1797. doi: 10.3390/microorganisms8111797
Salimei, E., Fantuz, F., Coppola, R., Chiofalo, B., Polidori, P., and Varisco, G. (2004). Composition and characteristics of ass's milk. Anim. Res. 53, 67–78. doi: 10.1051/animres:2003049
Samaržija, D., Zamberlin, Š., and Pogačić, T. (2012). Psychrotrophic bacteria and their negative effects on milk and dairy products quality. Mljekarstvo 62, 77–95.
Santamarina-García, G., Hernández, I., Amores, G., and Virto, M. (2022). Characterization of microbial shifts during the production and ripening of raw ewe milk-derived Idiazabal cheese by high-throughput sequencing. Biology 11:769. doi: 10.3390/biology11050769
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60–R18. doi: 10.1186/gb-2011-12-6-r60
Sessou, P., Keisam, S., Tuikhar, N., Gagara, M., Farougou, S., and Jeyaram, K. (2019). High-throughput illumina miseq amplicon sequencing of yeast communities associated with indigenous dairy products from republics of Benin and Niger. Front. Microbiol. 10:594. doi: 10.3389/fmicb.2019.00594
Shu, G. W., Zhang, Q. H., Chen, H., and Wang, X. (2013). Effects of hot water extract from Fructus tribuli on growth of Lactobacillus casei, Lactobacillus reuteri, lactobacillus rhamnosus and Lactobacillus bulgaricus with material properties. Adv. Mater. Res. 700, 284–287. doi: 10.4028/www.scientific.net/AMR.700.284
Soler, L., Moreno, Y., Moreno-Mesonero, L., Amorós, I., Alonso, J. L., and Ferrús, M. A. (2023). Microbiome of free-living amoebae (FLA) isolated from fresh organic produce: potential risk to consumers? Food Secur. 12:3102. doi: 10.3390/foods12163102
Soucek, A., Mara, M., and Souckova, A. (1965). Studies on Corynebacterium pyogenes varietas hominis. IV. Comparison of active components in Corynebacterium pyogenes varietas hominis (Corynebacterium haemoliticum) and Corynebacterium pyogenes. J. Hyg. Epidemiol. Microbiol. Immunol. 69, 67–76.
Strimmer, K. (2008). Fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics 24, 1461–1462. doi: 10.1093/bioinformatics/btn209
Stuby, J., Lardelli, P., Thurnheer, C. M., Blum, M. R., and Frei, A. N. (2023). Trueperella pyogenes endocarditis in a Swiss farmer: a case report and review of the literature. BMC Infect. Dis. 23:821. doi: 10.1186/s12879-023-08810-y
Subsomwong, P., Teng, W., Ishiai, T., Narita, K., Sukchawalit, R., Nakane, A., et al. (2024). Extracellular vesicles from Staphylococcus aureus promote the pathogenicity of Pseudomonas aeruginosa. Microbiol. Res. 281:127612. doi: 10.1016/j.micres.2024.127612
Tahir, S., Hasanain, R., Abuhammour, W., Dsouza, A. P., Lone, R., and Kherani, S. (2023). Granulicatella adiacens causing a parapharyngeal abscess in a 10-month-old infant: a rare-case report and literature review of deep neck infections (DNIs) in children. Cureus 15:e42144. doi: 10.7759/cureus.46790
Thierry, A., Deutsch, S.-M., Falentin, H., Dalmasso, M., Cousin, F. J., and Jan, G. (2011). New insights into physiology and metabolism of Propionibacterium freudenreichii. Int. J. Food Microbiol. 149, 19–27. doi: 10.1016/j.ijfoodmicro.2011.04.026
Verraes, C., Claeys, W., Cardoen, S., Daube, G., De Zutter, L., Imberechts, H., et al. (2014). A review of the microbiological hazards of raw milk from animal species other than cows. Int. Dairy J. 39, 121–130. doi: 10.1016/j.idairyj.2014.05.010
Vieu, J., Bergogne-Berezin, E., Joly, M., Berthelot, G., Fichelle, A., and Prevost, C. (1980). Epidemiology of Acinetobacter calcoaceticus. La Nouvelle presse medicale 9, 3551–3552
Walters, K. E., and Martiny, J. B. (2020). Alpha-, beta-, and gamma-diversity of bacteria varies across habitats. PLoS One 15:e0233872. doi: 10.1371/journal.pone.0233872
Wei, W., Hu, X., Hou, Z., Wang, Y., and Zhu, L. (2021). Microbial community structure and diversity in different types of non-bovine milk. Curr. Opin. Food Sci. 40, 51–57. doi: 10.1016/j.cofs.2021.01.008
Wen, M. M., Abdelwahab, I. A., Aly, R. G., and El-Zahaby, S. A. (2021). Nanophyto-gel against multi-drug resistant Pseudomonas aeruginosa burn wound infection. Drug Deliv. 28, 463–477. doi: 10.1080/10717544.2021.1889720
Wouters, J. T., Ayad, E. H., Hugenholtz, J., and Smit, G. (2002). Microbes from raw milk for fermented dairy products. Int. Dairy J. 12, 91–109. doi: 10.1016/S0958-6946(01)00151-0
Wu, X., Lu, Y., Xu, H., Lv, M., Hu, D., He, Z., et al. (2018). Challenges to improve the safety of dairy products in China. Trends Food Sci. Tech. 76, 6–14. doi: 10.1016/j.tifs.2018.03.019
Xin, L., Meng, Z., Zhang, L., Cui, Y., Han, X., and Yi, H. (2017). The diversity and proteolytic properties of psychrotrophic bacteria in raw cows' milk from North China. Int. Dairy J. 66, 34–41. doi: 10.1016/j.idairyj.2016.10.014
Yang, X., Guo, X., Liu, W., Tian, Y., Gao, P., Ren, Y., et al. (2020). The complex community structures and seasonal variations of psychrotrophic bacteria in raw milk in Heilongjiang Province, China. LWT 134:110218. doi: 10.1016/j.lwt.2020.110218
Yang, S., Piao, Y., Li, X., Mu, D., Ji, S., Wu, R., et al. (2023). A new decontamination method for Bacillus subtilisin pasteurized milk: Thermosonication treatment. Food Res. Int. 163:112291. doi: 10.1016/j.foodres.2022.112291
Yang, P., Tian, J., Zhang, L., Zhang, H., Yang, G., Ren, Y., et al. (2024). A toolbox for genetic manipulation in intestinal Clostridium symbiosum. Synth. Syst. Biotechnol. 9, 43–54. doi: 10.1016/j.synbio.2023.12.005
Yasmin, I., Saeed, M., Khan, W. A., Khaliq, A., Chughtai, M. F. J., Iqbal, R., et al. (2020). In vitro probiotic potential and safety evaluation (hemolytic, cytotoxic activity) of Bifidobacterium strains isolated from raw camel milk. Microorganisms 8:354. doi: 10.3390/microorganisms8030354
Yerlikaya, O., Akpinar, A., Saygili, D., and Karagozlu, N. (2020). Incorporation of Propionibacterium shermanii subsp. freudenreichii in probiotic dairy drink production: physicochemical, rheological, microbiological and sensorial properties. Int. J. Dairy Technol. 73, 392–402. doi: 10.1111/1471-0307.12666
Yu, W., Li, X., Sun, Q., Yi, S., Zhang, G., Chen, L., et al. (2024). Metabolomics and network pharmacology reveal the mechanism of Castanopsis honey against Streptococcus pyogenes. Food Chem. 441:138388. doi: 10.1016/j.foodchem.2024.138388
Yumoto, I., Hirota, K., Nodasaka, Y., Tokiwa, Y., and Nakajima, K. (2008). Alkalibacterium indicireducens sp. nov., an obligate alkaliphile that reduces indigo dye. Int. J. Syst. Evol. Microbiol. 58, 901–905. doi: 10.1099/ijs.0.64995-0
Zamora, L., Fernández-Garayzábal, J., Svensson-Stadler, L., Palacios, M., Domínguez, L., Moore, E., et al. (2012). Flavobacterium oncorhynchi sp. nov., a new species isolated from rainbow trout (Oncorhynchus mykiss). Syst. Appl. Microbiol. 35, 86–91. doi: 10.1016/j.syapm.2011.11.007
Zhang, D., Palmer, J., Teh, K. H., Biggs, P., and Flint, S. (2019). 16S rDNA high-throughput sequencing and MALDI-TOF MS are complementary when studying psychrotrophic bacterial diversity of raw cows' milk. Int. Dairy J. 97, 86–91. doi: 10.1016/j.idairyj.2019.06.001
Zhang, K., Zhang, Y., Li, S., Li, B., Li, B., Guo, Z., et al. (2021). Fungal diversity in Xinjiang traditional cheese and its correlation with moisture content. Indian J. Microbiol. 62, 47–53. doi: 10.1007/s12088-021-00967-x
Zhu, L., Zeng, C., Yang, S., Hou, Z., Wang, Y., Hu, X., et al. (2020). Diversity and specificity of the bacterial community in Chinese horse milk cheese. Microbiology 9:e1066. doi: 10.1002/mbo3.1066
Keywords: raw milk, breeds, regions, bacterial, community structures
Citation: Luo B, Dong F, Liu Y, Du J, Sun H, Ni Y and Zhang Y (2024) Insights into the microbiota of raw milk from seven breeds animals distributing in Xinjiang China. Front. Microbiol. 15:1382286. doi: 10.3389/fmicb.2024.1382286
Edited by:
Massimo Iorizzo, University of Molise, ItalyReviewed by:
Bowen Li, Southwest University, ChinaKunyi Liu, Yibin Vocational and Technical College, China
Copyright © 2024 Luo, Dong, Liu, Du, Sun, Ni and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongqing Ni, bml5cWx6dUBzaW5hLmNvbQ==; Yan Zhang, emhhbmd5YW56cWdoQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Baolong Luo
Baolong Luo Fujin Dong
Fujin Dong Yuyang Liu
Yuyang Liu Jie Du
Jie Du Hailong Sun1,2,3,4
Hailong Sun1,2,3,4 Yongqing Ni
Yongqing Ni Yan Zhang
Yan Zhang