- College of Animal Husbandry and Veterinary Medicine, Jinzhou Medical University, Jinzhou, China
This study aimed to explore whether Lactococcus G423 could improve growth performance and lipid metabolism of broilers by the modulation of gut microbiota and metabolites. A total of 640 1-day-old AA broilers were randomly divided into 4 groups [Control (CON), Lac_L, Lac_H, and ABX]. Average daily gain (ADG), average daily feed intake (ADFI), feed conversion ratio (FCR), breast muscle, thigh muscle, and abdominal fat pad were removed and weighed at 42 days of age. Serum was obtained by centrifuging blood sample from jugular vein (10 mL) for determining high-density lipoprotein (HDL), total cholesterol (TC), low-density lipoprotein (LDL), and triglyceride (TG) using ELISA. The ileal contents were harvested and immediately frozen in liquid nitrogen for 16S rRNA and LC–MS analyses. Then, the results of 16S rRNA analysis were confirmed by quantitative polymerase chain reaction (qPCR). Compared with the CON group, FCR significantly decreased in the Lac_H group (p < 0.05) in 1–21 days; ADG significantly increased and FCR significantly decreased in the Lac_H group (p < 0.05) in 22–42 days. 42 days weight body and ADG significantly increased in the Lac_H group (p < 0.05) in 42 days. Abdominal fat percentage was significantly decreased by Lactococcus G423 (p < 0.05), the high dose of Lactococcus G423 significantly decreased the serum of TG, TC, and LDL level (p < 0.05), and the low dose of Lactococcus G423 significantly decreased the serum of TG and TC level (p < 0.05). A significant difference in microbial diversity was found among the four groups. Compared with the CON group, the abundance rates of Firmicutes and Lactobacillus in the Lac_H group were significantly increased (p < 0.05). The global and overview maps and membrane transport in the Lac_L, Lac_H, and ABX groups significantly changed versus those in the CON group (p < 0.05). The results of LC–MS demonstrated that Lactococcus could significantly improve the levels of some metabolites (6-hydroxy-5-methoxyindole glucuronide, 9,10-DiHOME, N-Acetyl-L-phenylalanine, and kynurenine), and these metabolites were involved in four metabolic pathways. Among them, the pathways of linoleic acid metabolism, phenylalanine metabolism, and pentose and glucuronate interconversions significantly changed (p < 0.05). Lactococcus G423 could ameliorate growth performance and lipid metabolism of broilers by the modulation of gut microbiota and metabolites.
1 Introduction
Intestinal microbes and host are bioactive communities, forming the junction between animals and their nutritional environment (Anand and Mande, 2018). Thus, microbiota may affect the physiology and metabolism of host, and certain healthy bacteria in the add microbiota may improve gut health (Judkins et al., 2020; Ghosh et al., 2021; Gill et al., 2021). Intestinal microbes have noticeably attracted researchers’ attention recently. Over the past two decades, some studies revealed that antibiotics can alter the likely benefit of the host–microbiota interaction or relationship by regulating the microbiota (Yukgehnaish et al., 2020). In the poultry industry, antibiotics have been widely used (Angela et al., 2020). However, it is essential to pay further attention to antibiotic resistance (Hakimul et al., 2020; Paintsil et al., 2021), and long-term use of these antibiotics could cause antibiotic residues remaining in animals, in which they seriously threaten human health (Dawood et al., 2018). It is well known that the basic function of probiotics is to reduce gut-related diseases by regulating and improving the intestinal microbial balance in humans (Soccol et al., 2010; Zommiti et al., 2020). Recently, probiotics have been found to benefit not only human health but also animal health (Shi et al., 2020). Several studies demonstrated that beneficial effects of probiotics for the host included suppression of growth of pathogens, modulation of the immune system, improvement of nutrient metabolism, and modification of the composition of the intestinal microbiota (Kailasapathy and Chin, 2000; Chan and Zhang, 2005; Ashouri et al., 2020; Fan et al., 2021). Especially, an appropriate amount of lactic acid bacteria (LAB) can regulate the microflora in the gut (Kim et al., 2021). Some studies show that LAB could significantly improve lipid metabolism and fat deposition (Cho et al., 2020; Wang et al., 2023). Zhang et al. (2022) report that LAB have an effect on production performance, lipid metabolism, and meat quality in heat–stressed broilers. Previous studies have shown that LAB can increase the bacterial phylogenetic diversity in the gut of mice (Usui et al., 2018) and weaning piglets (Zhao et al., 2016). Gupta et al. (2018) also report that LAB can modulate the composition and interaction of the intestinal microbiota of Atlantic salmon. Lactococcus is industrially crucial LAB used to produce lactic acid, pickled vegetables, buttermilk, cheese, and several types of dairy foods and drinks. In addition, they are utilized as probiotics in specific formulations. Lactococcus can modulate intestinal microbiota of animals (Busti et al., 2020; Tan et al., 2022). Lactococcus lactis has the potential to enhance growth performance, immune function, and intestinal development in broiler chickens (Zhou et al., 2019). Zhang et al. (2016) also study showed that Lactococcus could enhance the growth performance of broiler chickens and improve their health. However, they have rarely been studied versus other LAB genera. The ribosomal RNA (16S) rRNA (16S rRNA) gene possesses the advantage of exploring the composition of the gut microbiota of chickens (Shang et al., 2018), broiler chickens (Mohd Shaufi et al., 2015), Dagu chickens (Xu et al., 2016), and naked neck chickens (Park et al., 2016). Liquid chromatography-mass spectrometry (LC–MS) has solid analytical capability, and it can detect the association of bacteria and metabolites with high resolution and accuracy (Xia et al., 2021). Moreover, correlation analysis between microorganisms and metabolites was performed. This was of great significance in revealing the contribution of Lactococcus G423 to the formation of metabolites in the gut. Therefore, the present study aimed to explore whether Lactococcus G423 could ameliorate growth performance and lipid metabolism of broilers by the modulation of gut microbiota and metabolites.
2 Materials and methods
2.1 Birds, diets, and experimental design
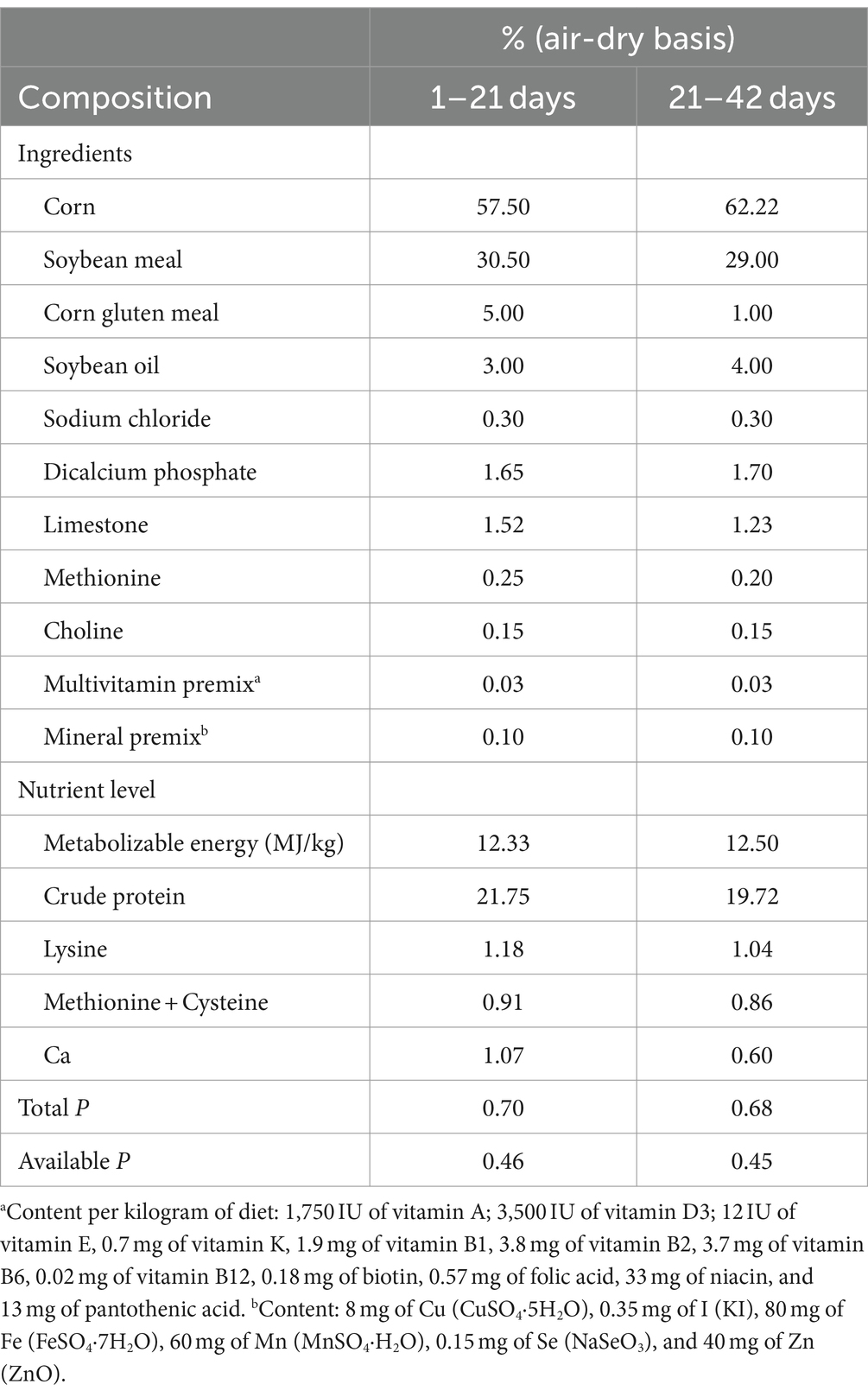
Totally, 640 1-day-old AA broilers (Shu-ya Poultry Co., Ltd., Tieling, China) were randomly classified into four experimental groups, and each group included 160 birds (8 replicates of 20 birds). Birds were raised in stainless steel cages (400 mm × 450 mm × 1,500 mm) in a controlled room for 42 days. This study was performed at Poultry Research Farm, Jinzhou Medical University, Liaoning, China. The temperature of room was gradually reduced by 3.0–3.5°C weekly until achieving a thermo-neutral zone ranged from 21 to 26°C by the end of the 3rd week. The experimental diets were based on corn and soybean meal. Four dietary regimes were provided as follows: control group (basal diet, CON group), Lac-L and Lac-H groups (basal diet supplemented with 50 and 100 mg/kg Lactococcus G423, respectively), and ABX group (basal diet supplemented with 50 mg/kg narasin). The basal diet was divided into two phases: the starter phase from 1 to 21 days and the growth phase from 21 to 42 days. The basal diet was formulated to meet the nutritional requirements according to the Chinese Broiler Feeding Standards (NY/T33-2004) (Table 1). LactococcusG423 (1 × 1010 CFU/g) and narasin (purity of narasin dihydrate powder 15%, Eli Lilly and Company, Indianapolis, Indiana, United States) were mixed in basal diet (Supplementary Figure S1).
2.2 Growth and carcass measurements
Broiler performance in terms of average daily gain (ADG), average daily feed intake (ADFI), survival rate, and feed conversion ratio (FCR) was weekly recorded, in which ADG, ADFI, and FCR were calculated and presented for 6-week experimental period. Breast muscle, thigh muscle, and abdominal fat pad (including fat surrounding the gizzard, bursa of Fabricius, cloaca, and adjacent muscles) from one bird of average BW per replicate were removed and weighed at week 6. To compensate for the differences in carcass weight, these values were expressed as a percentage of carcass weight.
2.3 Enzyme-linked immunosorbent assay
Content of high-density lipoprotein (HDL), LDL, TG, and TC was determined using enzyme-labeled instrument according to ELISA kit instruction (Nanjing Jiancheng Bio. Institute, Nanjing, Jiangsu, China).
2.4 Illumina MiSeq sequencing for the detection of intestinal microbial diversity
Eight ileal samples per group were randomly selected for the analysis of intestinal flora. The polymerase chain reaction (PCR) amplification of the hypervariable region V3–V4 of the 16S rRNA gene was performed with the universal primers set338 F (5′-ACTCCTACGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTC TAAT-3′) (Liu et al., 2016). The quality and concentration of DNA were determined by 1.0% agarose gel electrophoresis and a NanoDrop® ND-2000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, United States) and kept at −80°C for further experiment. All samples were amplified in triplicate. The PCR products were extracted from 2% agarose gel and were purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States), according to the manufacturer’s instructions and were quantified using Quantus™ Fluorometer (Promega, Madison, WI, United States). The Illumina MiSeq platform (Illumina Inc., San Diego, CA, United States) was used for paired-end sequencing (2 × 300) of the PCR products. The raw sequence reads were quality-filtered and merged by FLASH (Tanja and Steven, 2011) before open-reference operational taxonomic unit (OTU) picking via UPARSE (Stackebrandt and Goebel, 1994; Edgar, 2013) and taxonomy classification through the SILVA 16S rRNA database (Wang, 2007).
2.5 Quantitative PCR (qPCR)
Lactobacillus and Firmicutes were detected by qPCR. Eight ileum contents from broiler were collected. The primers used for qPCR are presented in Table 2. The conditions of PCR reaction were summarized as follows: (1) at 95°C for 5 min; (2) a: at 95°C for 30 s; b: at 60°C for 30 s; c: at 72°C for 1 min, including 35 cycles; (3) a: at 95°C for 30 s; b: at 55°C for 30 s; c: at 72°C for 1 min. The ΔCt was calculated as follows: (corrected sample) = mean value of target gene–mean value of internal reference gene (ΔΔ Ct = ΔCt–mean value of control group).
2.6 LC–MS analysis
Eight ileal samples Con and Lac_H group were randomly selected for the analysis of LC–MS. The LC–MS analysis of ileal contents was conducted on a Thermo UHPLC-Q Exactive HF-X system equipped with an ACQUITY HSS T3 column (100 mm × 2.1 mm i.d., 1.8 μm; Waters Corp., Milford, MA, United States) at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). The mass spectrometric data were collected using a Thermo UHPLC-Q Exactive HF-X Mass spectrometer equipped with an electrospray ionization (ESI) source operating in positive and negative modes. The pretreatment of LC–MS raw data was performed by Progenesis QI software (Waters Corp.), and a three-dimensional (3D) data matrix in CSV format was exported. This 3D matrix included the following information: sample information, metabolite name, and mass spectral response intensity. Internal standard peaks and any known false positive peaks (including noise, column bleed, and derivatized reagent peaks) were removed from the data matrix, de-redundant, and peak pooled. Moreover, the metabolites were identified by searching in the following databases: Human Metabolome Database (HMDB)1, Metlin2, and Majorbio3 (Kong et al., 2022; Li C. et al., 2022; Li Z. et al., 2022).
2.7 Statistical analysis
Between-group statistical differences were compared using one-way analysis of variance (ANOVA), followed by post-hoc multiple comparisons using Fisher’s least significant difference (LSD) t-test. The experimental data were presented as the mean ± standard error of the mean (SEM), which were analyzed using SPSS 20.0 software (IBM, Armonk, NY, United States), and p < 0.05 was considered statistically significant. The 16S rRNA genes of gut microbiota were analyzed using an online platform (see Footnote 3) (Ren et al., 2022). The multivariate statistical analysis was performed using the “ropls” (version 1.6.2) R package from Bioconductor on Majorbio Cloud Platform (see Footnote 3) (Ren et al., 2022).
3 Results
3.1 Growth performance
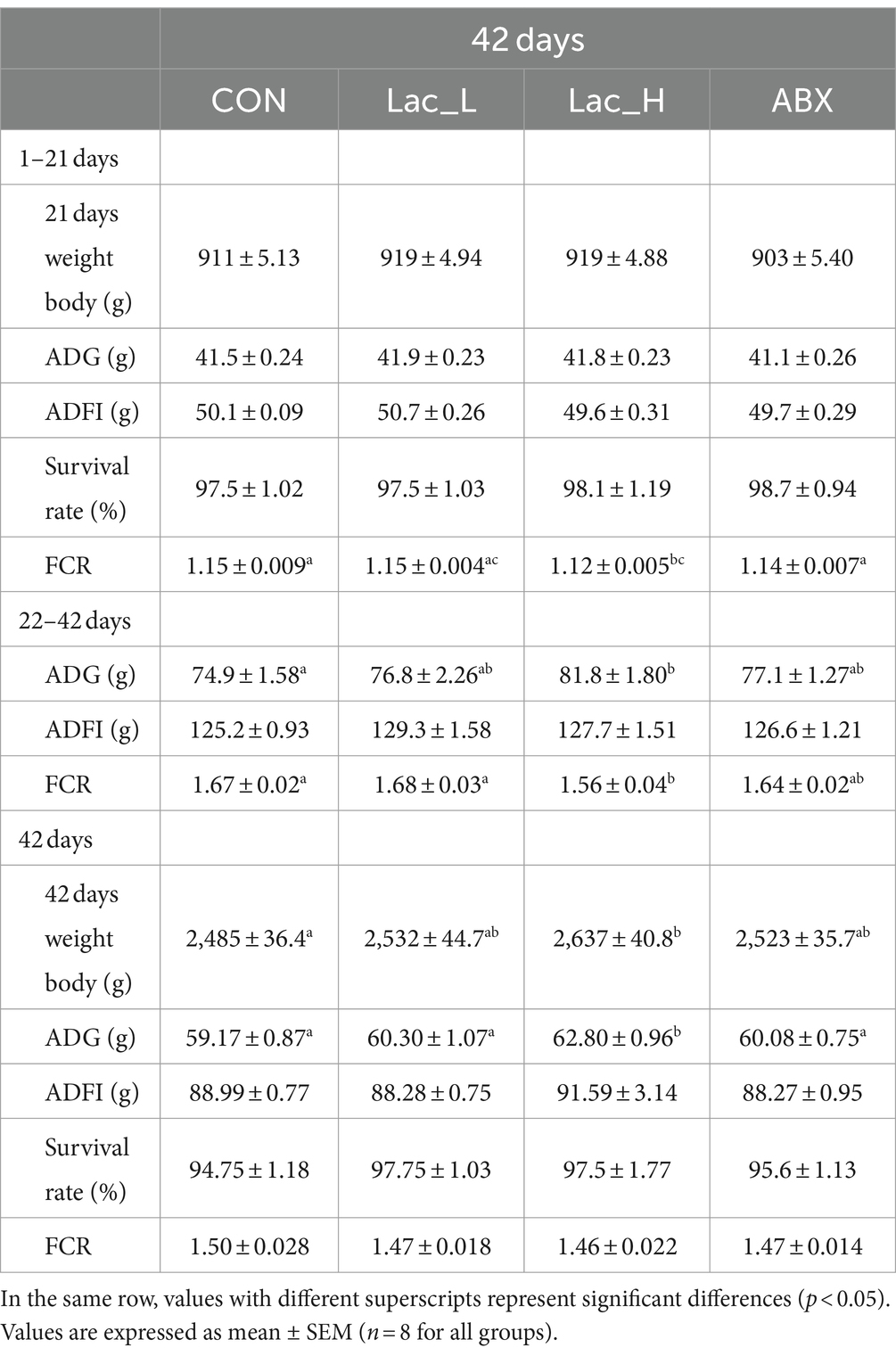
Compared with control, FCR significantly decreased in the Lac_H group (p < 0.05) in 1–21d; ADG significantly increased and FCR significantly decreased in the Lac_H group (p < 0.05) in 22–42 days; weight body and ADG significantly increased in the Lac_H group (p < 0.05) in 42 days. There were no significant changes in FCR, survival rate, and ADFI (p > 0.05) among Lac_H, Lac_L, and ABX groups in 42 days (Table 3).
3.2 Carcass characteristics
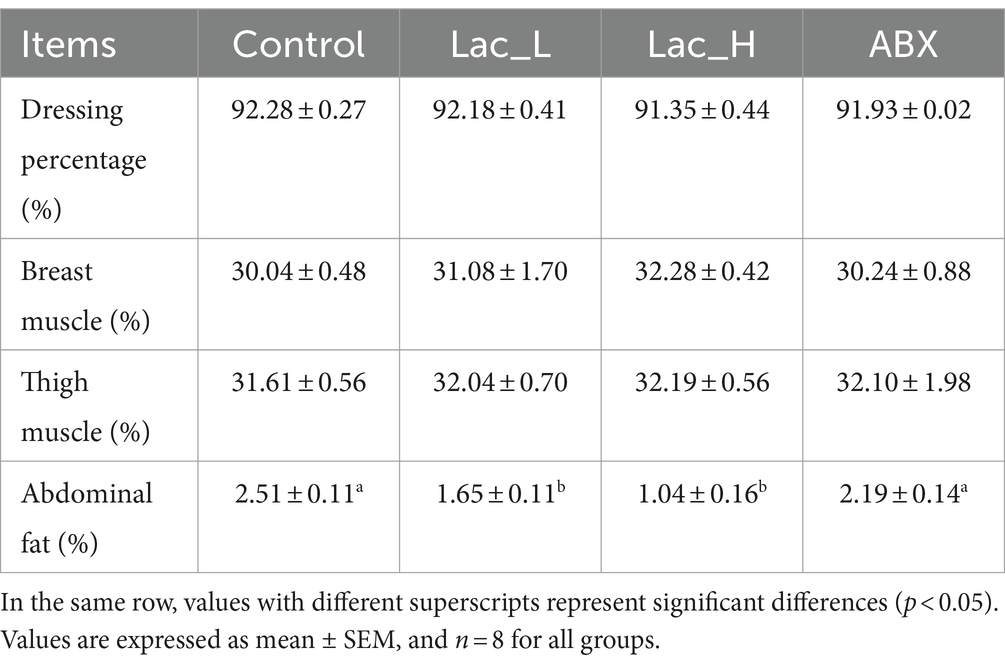
Compared with control, abdominal fat percentage was significantly decreased by Lactococcus G423 (p < 0.05); however, dressing percentage, thigh muscle percentage, and breast muscle percentage had no significant changes among Lac_H, Lac_L, and ABX groups (p > 0.05) (Table 4).
3.3 Serum biochemical parameters
Comparing with control, the high dose of Lactococcus G423 significantly decreased the serum of TG, TC, and LDL level (p < 0.05), and the low dose of Lactococcus G423 significantly decreased the serum of TG and TC level (p < 0.05). ABX significantly decreased the content of TG (p < 0.05) in serum; however, HDL content had no significant changes among Lac_H, Lac_L, and ABX groups (p > 0.05) (Figure 1).
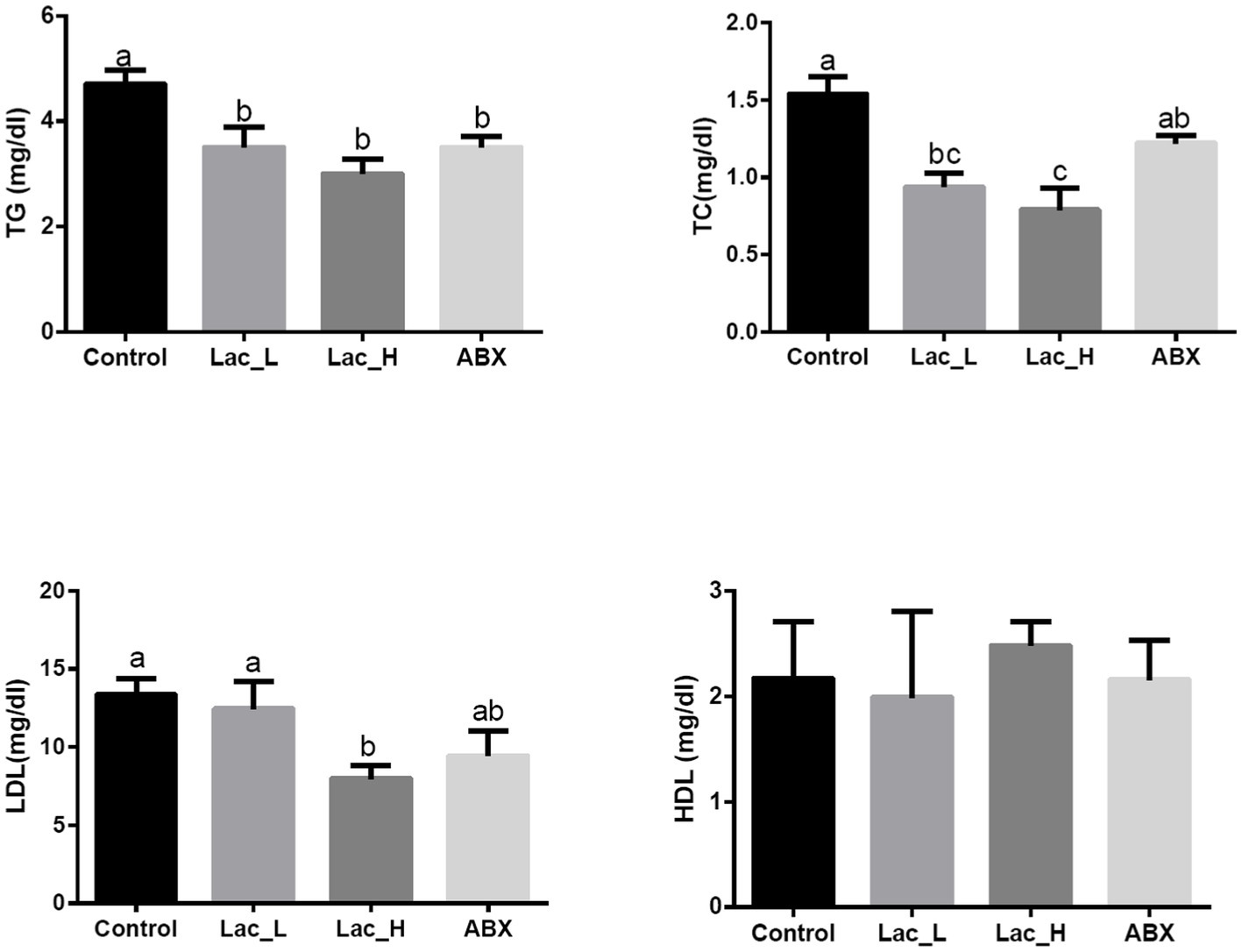
Figure 1. Effect of Lactococcus on serum biochemical parameters in broilers. Values with different superscripts represent significant differences (p < 0.05).
3.4 Intestinal microflora
To characterize the intestinal microbiota composition of broilers in the four groups, 16S rRNA gene sequence analysis was performed. With the sequence similarity of 97%, 903 OTUs were obtained. The average good’s coverage for samples was higher than 99%, indicating that the majority of the microbial species were identified. and sequencing depth was also adequate for the robust sequence analysis.
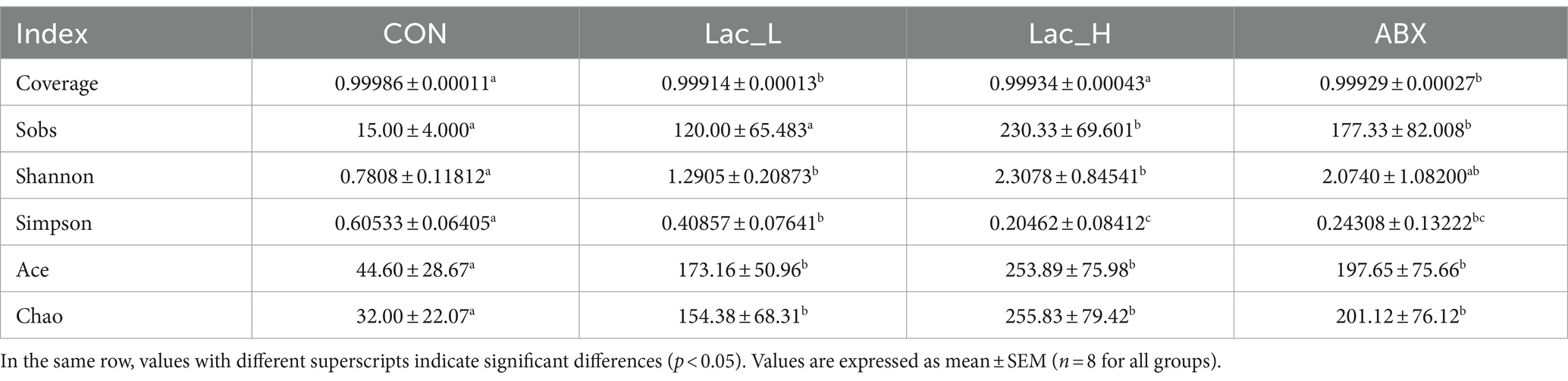
As shown in Table 5, alpha diversity analysis of gut microbiota showed that compared with the CON group, the Chao and Ace indices in the Lac_H, Lac_L, and ABX groups significantly increased (p < 0.05); however, the Simpson index exhibited an opposite trend. The Simpson index in the Lac_L, Lac_H, and ABX groups was significantly reduced compared with that in the CON group (p < 0.05). In addition, the Sob index in the Lac_H and ABX groups was significantly higher than that in the CON group (p < 0.05). The Shannon index in the Lac_L and Lac_H groups was significantly elevated compared with that in the CON group (p < 0.05). The Coverage index in the Lac_L and ABX groups significantly increased compared with that in the CON group (p < 0.05). The effects of Lactococcus on the diversity and richness of intestinal microbiota community in broilers were evaluated based on alpha diversity (Table 5).
Based on OTU abundance, principal coordinate analysis (PCoA) showed that points in the Lac_H and ABX groups were scattered in the right, which indicated that the microbial structure in the Lac_H and ABX groups had undergone a tremendous change versus that in the CON group. In the Lac_L and CON groups, points were clustered separately from each other in the left, which showed that the low dose of Lactobacillus G423 could change the structure of gut microbiota (Figure 2A). At the phylum level, Firmicutes, Proteobacteria, and Bacteroidetes were the most of species identified in all samples (Figure 2B). At the genus level, compared with those in the CON group, the abundance of Lactobacillus was higher, whereas that of Bacteroides was lower in the Lac_L,Lac_H and ABX groups (Figure 2C). As shown in Figure 2D, qPCR showed that the proportion of Lactobacillus in the Lac_L and Lac_H groups was significantly elevated compared with that in the CON and ABX groups (p < 0.05). Additionally, the proportion of Firmicutes was significantly risen after treating with Lac_H (p < 0.05). The different effects of Lac_L and Lac_H on microbiota might justify their different number of microorganisms. Subsequent linear discriminant analysis effect size (LEfSe) revealed substantial differences in Lactobacillus_salivarius and Lactobacillus_johnsonii in the Lac_H and ABX groups (Figure 2E).
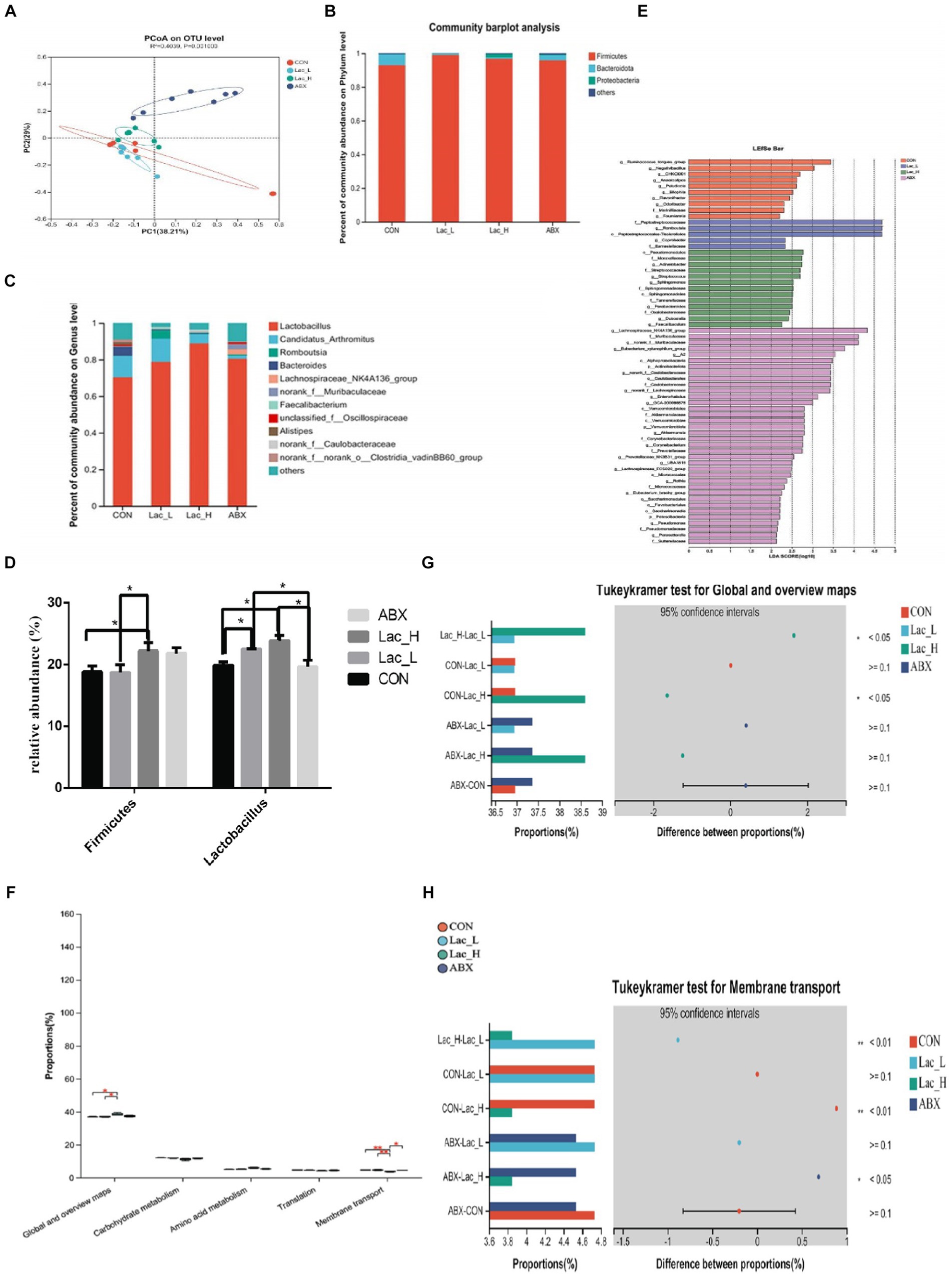
Figure 2. Effects of Lactococci on gut microbiota of broilers. Principal coordinate analysis (PCoA) based on the weighted UniFrac distance (A); column chart of community difference at the phylum level (B) and the genus level (C); relative abundance of discriminative gut microbiota at the genus level (D); LEfSe analysis (E); KEGG pathway analysis (F–H); * and ** represent p < 0.05 and p < 0.01, respectively.
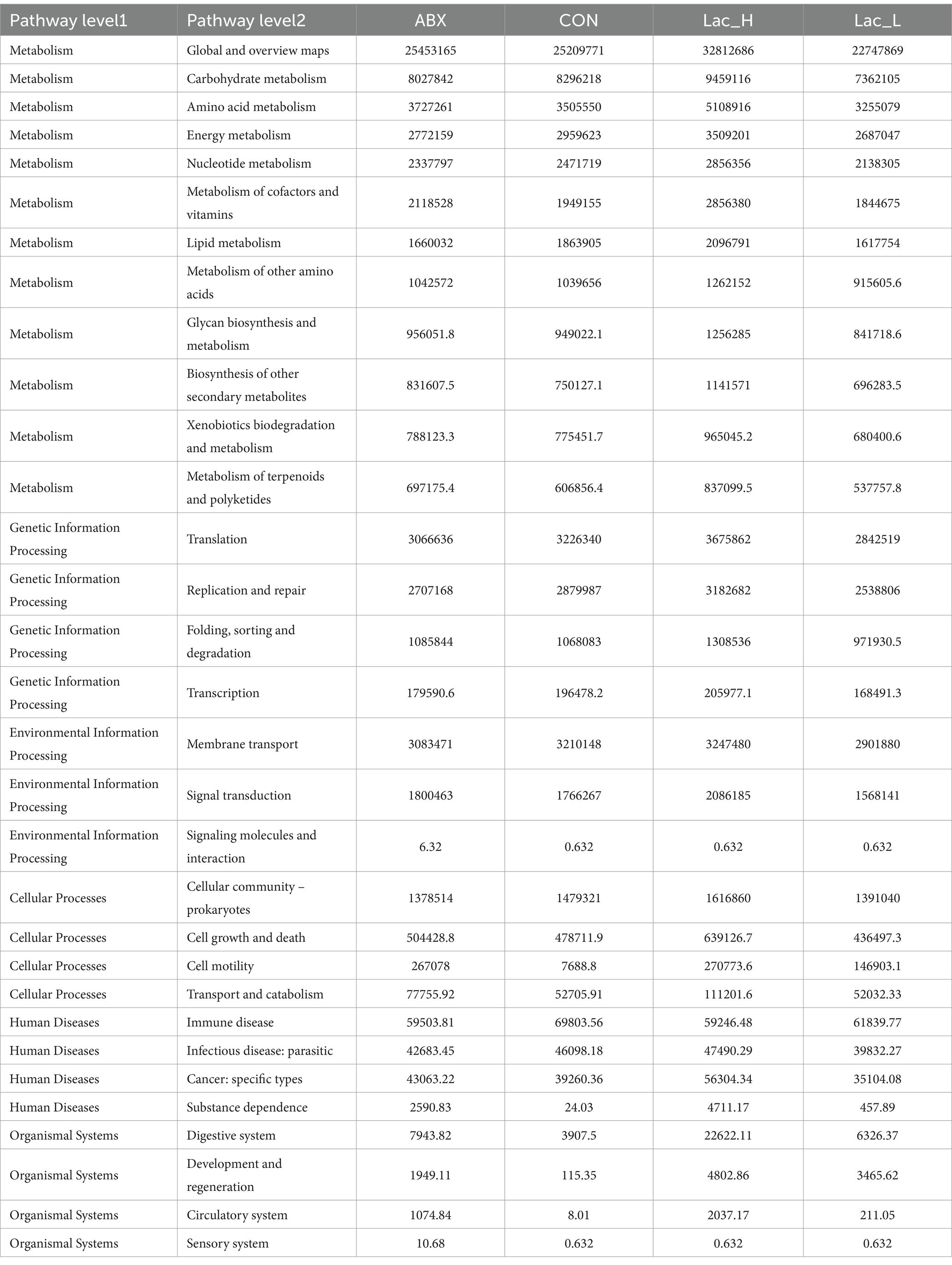
The function of the ileum microbiome was predicted using the phylogenetic investigation of communities by the reconstruction of unobserved species 2 (PICRUSt2). Then, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was used to divide the predicted metabolic pathways into six functional groups. The microbial communities in the CON, Lac_L, Lac_H, and ABX groups were mainly related to metabolism, genetic information processing, cellular processes, environmental information processing, human diseases, and organic systems. Their main functions were concentrated in the metabolism of amino acids, carbohydrate, vitamins, terpenoids, polyketides, and lipids (Table 5).
As shown in Figure 2F, the global and overview maps and membrane transport in the Lac_L, Lac_H, and ABX groups significantly changed compared with those in the CON group (p < 0.05). Functional predictions of differences in the mean relative abundance among groups are shown in Figures 2G,H and Table 6.
3.5 Intestinal metabolites
A total of 6,612 and 5,851 metabolites in ileal contents were determined in positive and negative ion modes, respectively, using LC–MS-based non-targeted metabolomics. A total of 228 metabolites were identified and named based on the HMDB and KEGG databases. Furthermore, orthogonal projection to latent structures-discriminant analysis (OPLS-DA) was employed to select the most predictive and discriminative features to assist classify cation. The loading plot showed a clear separation in metabolites between the Lac_H and CON groups (Figure 3A). The results revealed that the metabolite of broiler significantly changed after treating with Lactococcus. Then, the heat map tree of cluster analysis of metabolites (Figure 3B) was constructed, which visualized 50 significantly different metabolites. Overall, there were significant differences in metabolites between the CON and Lac_H groups.
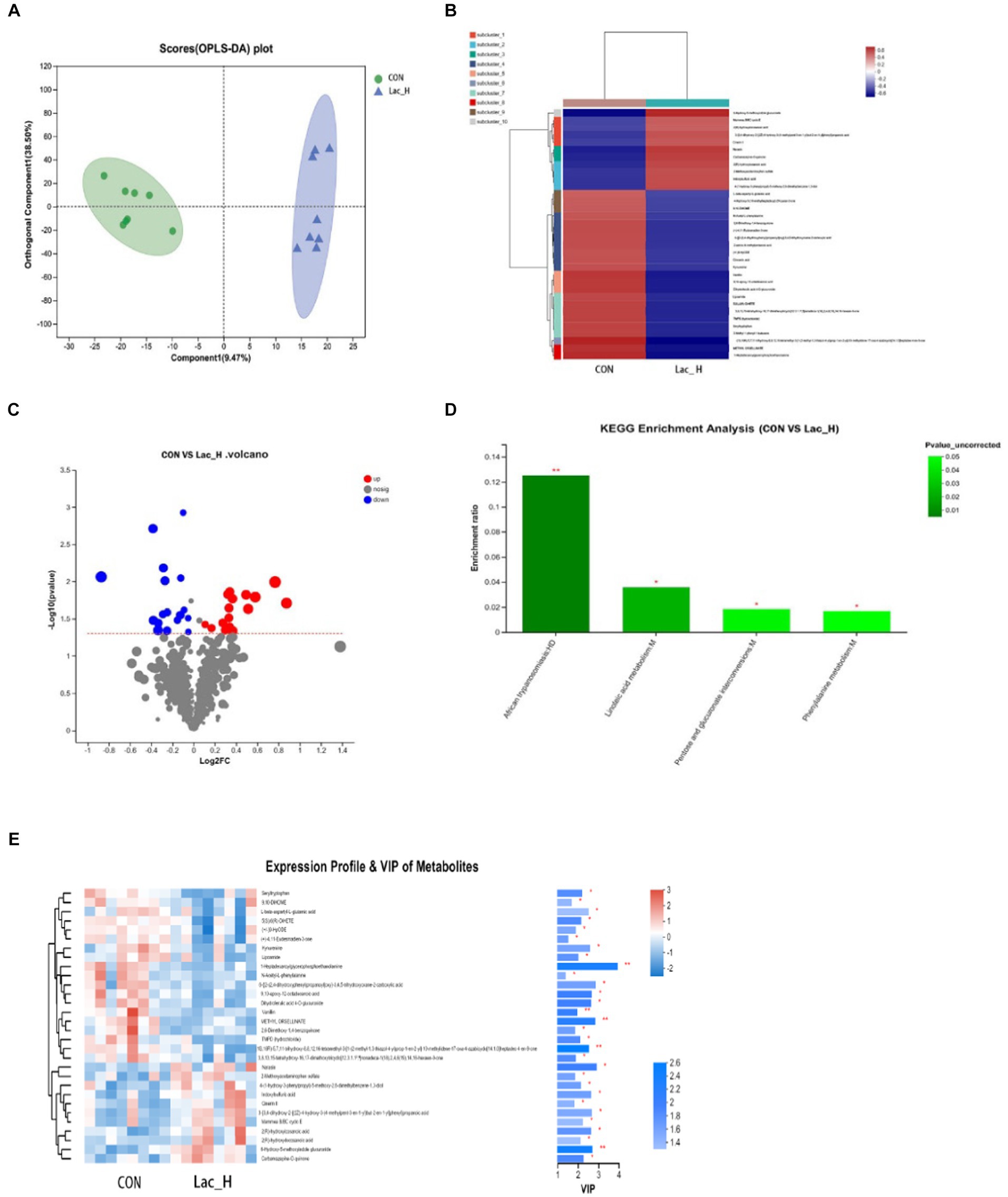
Figure 3. Effects of Lac_H on ileal metabolites of broilers. Multivariate statistical analysis of blank control group and Lac_H group (A). The heat map of cluster analysis of metabolites (B). Volcanic diagram of differentially expressed metabolites (C). Variable importance in projection (VIP) scores of the CON group versus Lac_H group (D). Bubble diagram of metabolic pathway enrichment analysis (E); *, **, and *** represent p < 0.05, p < 0.01, and p < 0.001, respectively.
The levels of several metabolites such as 6-hydroxy-5-methoxyindole glucuronide, 3-{3,4-dihydroxy-2-[(2Z)-4-hydroxy-3-(4-methylpent-3-en-1-yl)but-2-en-1-yl]phenyl} propanoic acid, indoxylsulfuric acid, Cinerin II, carbamazepine-O-quinone, and 4-(1-hydroxy-3-phenylpropyl)-5-methoxy-2,6-dimethylbenz ene-1,3-diol were upregulated in the Lac_H group, while the levels of 9,10-DiHOME, seryltryptophan, seryltryptophan dihydroferulic acid 4-O-glucuronide, 2,6-dimethoxy-1,4-benzoquinone, and kynurenine were downregulated (Figure 3C). Metabolites discriminated among different groups were screened using the variable importance in projection (VIP) scores obtained from the OPLS-DA model, and the ileal contents of metabolic profiles were determined. The metabolites were statistically significant if VIP score ≥ 1 and p < 0.05, and p-value was calculated by the t-test. Metabolites with VIP score > 1.0 and p < 0.05 were considered to be significantly influenced by the Lac_H. Thirty significantly affected metabolites were identified in the CON and Lac_H groups, respectively; the top 30 metabolites with the highest VIP scores are presented in Figure 3D.
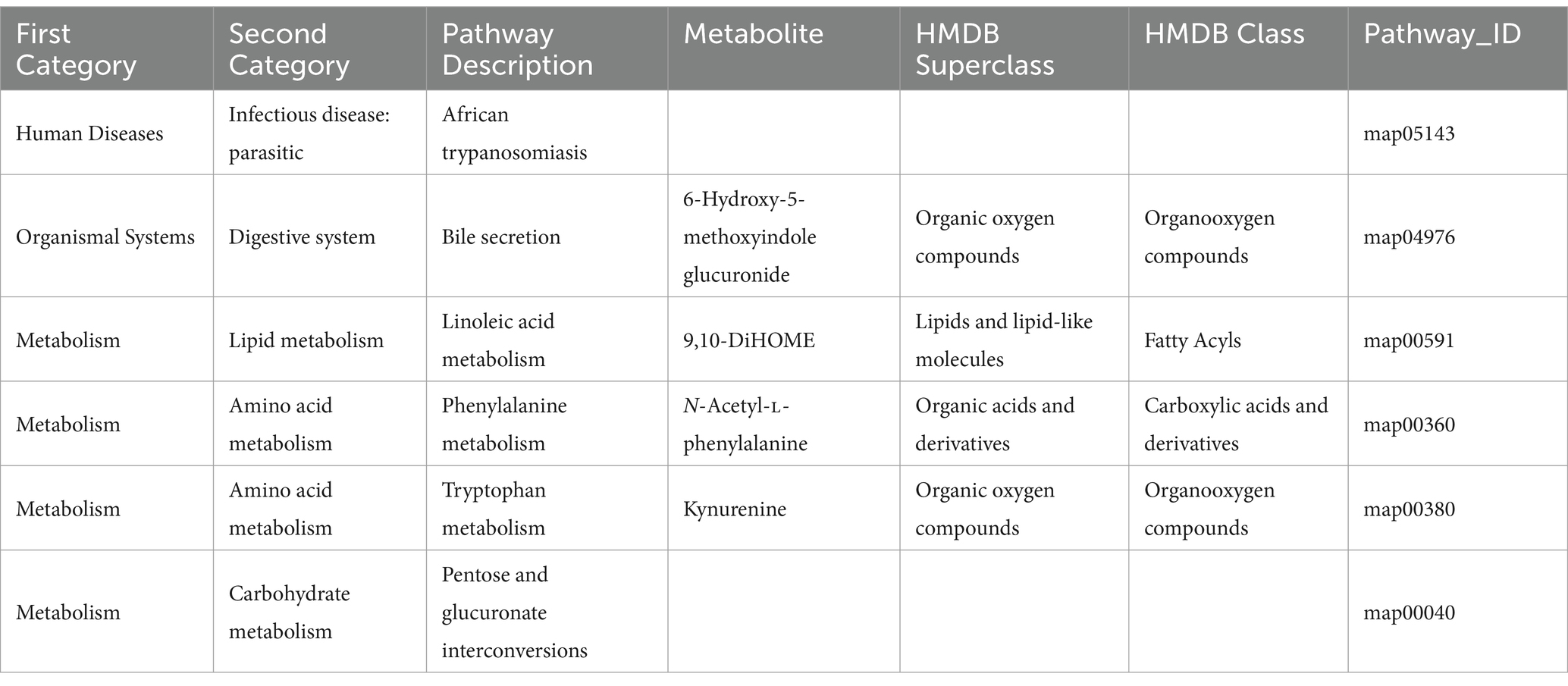
Metabolic pathway enrichment analysis was performed based on the KEGG database for the differential metabolites between the CON and Lac_H groups, and the metabolic pathway with p < 0.05 was significantly enriched for the differential metabolites, including bile secretion, linoleic acid metabolism, drug metabolism-cytochrome P450, phenylalanine metabolism, tryptophan metabolism, and matching metabolites. Lactococcus G423 could significantly improve the levels of certain metabolites (6-hydroxy-5-methoxyindole glucuronide, 9,10-DiHOME, N-acetyl-L-phenylalanine, and kynurenine), and these metabolites were involved in four metabolic pathways (Table 7). Among them, the pathways of linoleic acid metabolism, phenylalanine metabolism, and pentose and glucuronate interconversions significantly varied (p < 0.05) (Figure 3E).
3.6 Correlation analysis between metabolites and intestinal microbiota
The variations in the intestinal microbiota could be related to the metabolic phenotype. As shown in Figure 4, correlation analysis was performed between 34 different metabolites and 44 bacteria with significantly different relative abundances at the genus level. There was a significant correlation between 2(R)-hydroxyicosanoic acid, 2(R)-hydroxydocosanoic acid, L-beta-aspartyl-L-glutamic acid, 9,10-DiHOME, TMPD (hydrochloride), kynurenine, 6-hydroxy-5-methoxyindole glucuronide, seryltryptophan, and N-acetyl-L-phenylalanine and Parabacteroides, Romboutsia, Sellimonas, Subdoligranulum, Turicibacter, Tuzzerella, Bacteroides, Lachnospiraceae, Butyricicoccus, Candidatus_Arthromitus, Eisenbergiella, Escherichia-Shigella, Faecalibacterium, Alistipes, Marvinbryantia, Monoglobus, and Negativibacillus (all p < 0.05).
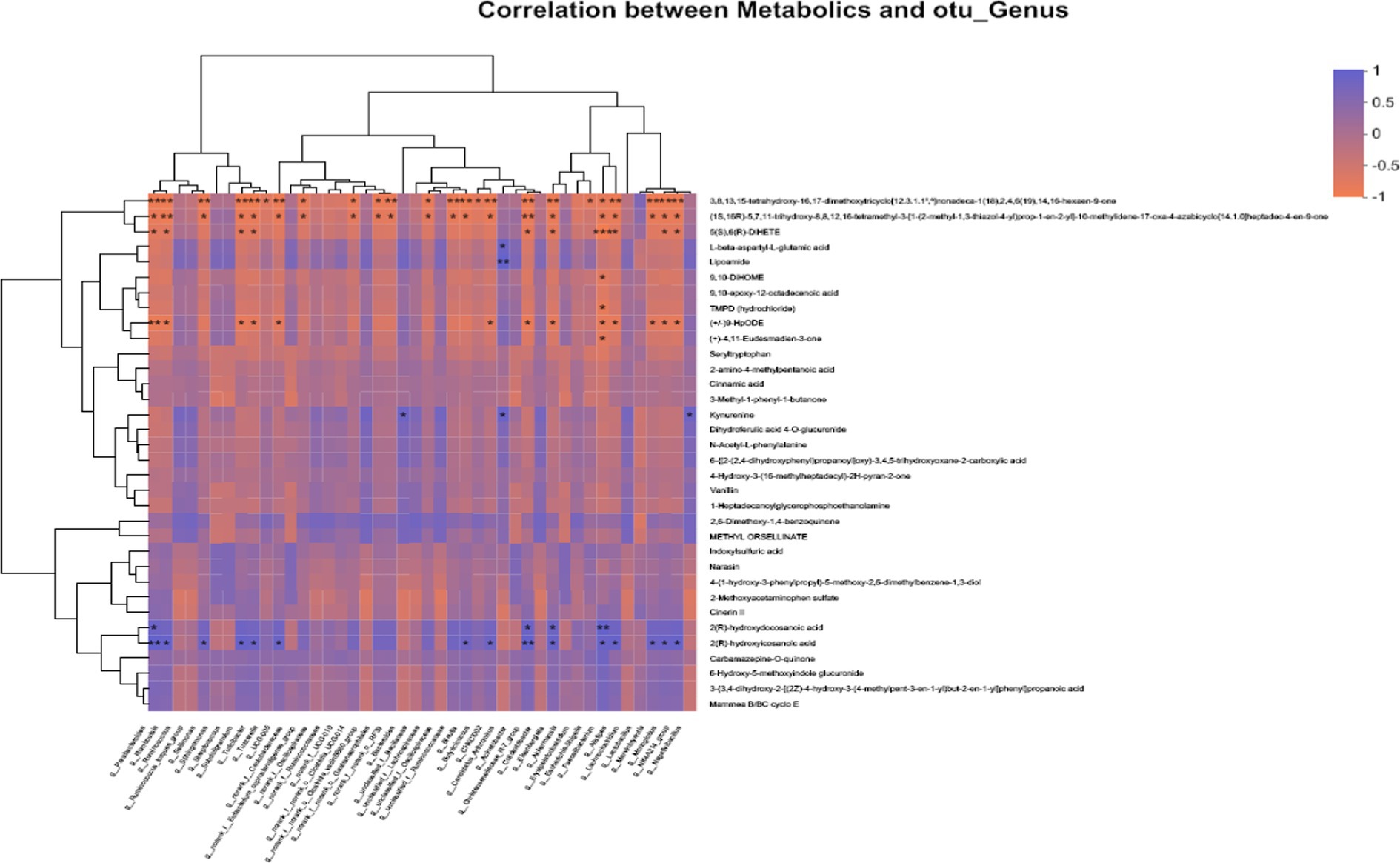
Figure 4. Correlation analysis of “metabolites-intestinal flora” in broilers. Horizontal coordinates indicate metabolites and vertical coordinates indicate gut microbiota; R values are shown in different colors in the graph, in which red indicates positive correlation and blue indicates negative correlation; *, **, and *** represent p < 0.05, p < 0.01, and p < 0.001, respectively.
4 Discussion
4.1 The effect of Lactococcus G423 on growth performance and carcass characteristics in broilers
The diversity and relative abundance of intestinal microbes play an important role in the health of host by participating in metabolism and immunomodulation (Zheng et al., 2021). The findings of the present study suggested that Lactococcus G423 could significantly increase ADG in broilers, which were similar to previously reported results (Faseleh et al., 2016; Chen et al., 2018). Supplementation of broilers’ diet with antibiotics could increase body weight gain (Rahman et al., 2012); however, our results showed that Lac_H significantly increase ADG compared with ABX in broiler. This improvement was explained by improved feed conversion efficiency and increased vitality and regulation of the intestinal microflora.
Different from mammals, chickens synthesize fatty acids predominantly in the liver and then export to other tissues including muscle and adipose tissue by the peripheral vascular system. Therefore, the blood lipid index is related to the carcass characteristics. The carcasses from 42-day-old Ross 308 chickens of both sexes, which received the multicomponent probiotics Pro-Biotyk (Em-15) and EMFarmaTM, did not differ significantly in the percentage of dissected carcass characteristics (Stęczny and Dariusz, 2020). The study by Ding et al. (2021) reveals that lactobacillus reduced abdominal fat deposition in broilers. Our results showed that abdominal fat percentage was lowered by Lactococcus G423, and dressing percentage, thigh muscle percentage, and breast muscle percentage had no significant changes among Lac_H, Lac_L, and ABX groups compared with CON groups in broilers. The current results are supported by previous studies on the effect of probiotics on carcasses (Liu et al., 2016; Rybarczyk et al., 2020).
4.2 Effect of Lactococcus G423 on serum biochemical parameters in broilers
Lipids mainly include triglyceride (TG), phospholipids, and cholesterol (CHO), and the contents of TG and CHO are key indicators of lipid metabolism. The administration of Paenibacillus polymyxa up to 0.4 mg/kg diet significantly reduced plasma TC, LDL, and TG (Alagawany et al., 2021).
Ding et al. (2021) found that Lactobacillus participated in the lipid metabolism of broilers by reducing the content of TC and TG. Other studies also showed that LAB had effects of blood serum levels TC, HDL-C, LDL-C, and TG on rat (An et al., 2011; Wang et al., 2023). Our results showed that Lac_H significantly decreased the content of TG, TC, and LDL, and Lac_L significantly decreased the content of TG and TC in serum, which was similar to previous studies (Abramowicz, 2019; Abdel-Moneim et al., 2020).
4.3 Effect of Lactococcus G423 on intestinal microflora in broilers
The gut microbiota community is consisted of diverse types of microbes. In the present study, it was found that Lactococcus G423 and ABX altered microbiome diversity in the ileum of broilers and changed the relative abundance rates of Firmicutes, Bacteroidetes, Proteobacteria, and other species. At the phylum level, Firmicutes, Bacteroidetes, and Proteobacteria were the most common phyla in the poultry intestinal samples, which is consistent with the previous findings (Shaufi et al., 2015; Qiao et al., 2018; Zheng et al., 2021). This study indicated that Firmicutes was the dominant phylum (>50%) in broilers, and similar results have been previously reported (Danzeisen et al., 2011; Mohd Shaufi et al., 2015). Moreover, this study revealed that the abundance rates of Firmicutes were relatively higher in the Lac_L, Lac_H, and ABX groups compared with those in the CON group (p < 0.05). Wang et al. (2017) reported that Lactobacillus significantly aided in altering the abundance of Firmicutes and decreased the content of TG and LDL. Firmicutes were associated with lipid metabolites (Turnbaugh et al., 2009). The phylum Bacteroidetes has influences on dissolving lipids (Kumar et al., 2018; Chen et al., 2020). Bacteroides are also positively correlated with several lipid metabolites (Saxena et al., 2016; Shulpekova et al., 2022). In the present study, it was revealed that the abundance of Bacteroides in the Lac_L, Lac_H, and ABX groups was markedly lower than that in the CON group. Meanwhile, considering the TG, LDL, and TC in this experiment, our findings also showed that Lactococcus G423 regulated lipid metabolism though regulating intestinal microflora. Although ABX has an effect on the abundance of the Firmicutes and Bacteroides, there is no effect on the level of LDL, TC, and abdominal fat percentage. Firmicutes and Bacteroidetes can also contribute to host metabolism through several mechanisms, including increased energy harvested from the diet and modulation of lipid metabolism (Greiner and Bäckhed, 2011). Some studies have suggested that a lower abundance of Bacteroidetes was associated with increased body weight (Ley et al., 2006; Arumugam et al., 2011). Previous studies also demonstrated that the Firmicutes/Bacteroidetes ratio and the growth performance were positively correlated, and this ratio could be indicative of the status of the intestinal bacteria (Haas et al., 2011; Xu et al., 2016). The results of the present study revealed that the Firmicutes/Bacteroidetes ratio was relatively higher in the Lac_L, Lac_H, and ABX groups compared with that in the CON group. Lactococcus G423 significantly increased ADG by changing the Firmicutes/Bacteroidetes ratio. Additionally, the levels of Proteobacteria phylum, including some pathogens, such as Escherichia, Salmonella, Helicobacter, and Vibrio, were slightly lower in the Lac_L group than those in the CON group, indicating that Lactococcus significantly aided in altering the abundance of opportunistic pathogens. However, at the genus level, Lactobacillus, Candidatus_Arthromitus, Romboutsia, and Bacteroides were identified as the dominant species in the ileum microbiome. Lactobacillus, belonging to the phylum of Firmicutes, had markedly higher level in the Lac_L and Lac_H groups than that in the CON group, and the abundance of Lactobacillus in the Lac_H group reached the highest rate. Lactobacillus is involved in digestive and metabolic processes and in the regulation of local and systemic immune response (Fernández et al., 2016). Moreover, Lactobacillus altered lipid metabolism (Wang et al., 2017). Similar effects were observed by Zhou et al. (2016), who studied that Bacillus licheniformis and Lactobacillus had an effect on the growth and fat deposition in broilers (Gerritsen et al., 2014). L. fermentum TSI reduces abdominal fat and improves blood lipid metabolism in HD-induced obese rats (Cho et al., 2020). Therefore, Lactococcus G423 significantly aided in altering the abundance of Lactobacillus, which participated in gut microbiota, growth, and lipid metabolism in animals (Wu et al., 2019).
Romboutsia have been identified in the human gut (Ricaboni et al., 2016), the rat gastrointestinal tract (Gerritsen et al., 2014), and the fecal of hens (Qiao et al., 2018). In the present study, a novel genus Romboutsia was found in ileum samples of broilers. However, the abundance of Romboutsia was inconsistent among the four groups. Meanwhile, it was revealed that ABX altered the relative abundance rates of other bacteria in ileum contents of broilers, negatively influencing the gut microbiota. Previous studies reported that improper uses of antibiotics have been increased antimicrobial-resistant bacteria as a public health threat (Nhung et al., 2017; Christy et al., 2018; Oniciuc et al., 2018). The results revealed that Lactobacillus G423 had more noticeable health benefits compared with antibiotics.
4.4 Effect of Lactococcus G423 on intestinal metabolites in broiler
The Lactococcus-regulated gut microbiota led to alterations in the contents of ileum metabolites. Several significantly altered metabolites were identified in the present study, such as 6-hydroxy-5-methoxyindole glucuronide, 9,10-DiHOME, N-acetyl-L-phenylalanine, and kynurenine, which were regulated, and they were involved in four metabolic pathways (Table 7).
Among them, the bile secretion and linoleic acid metabolism were the important metabolic pathways of lipid (Hamilton and Klett, 2021; Shulperkova et al., 2022). It was revealed that 6-hydroxy-5-methoxyindole glucuronide was related to the pathway of bile acid metabolism. Bile acids are also signaling molecules and inflammatory agents that rapidly activate nuclear receptors and cellular signaling pathways, regulating lipid, glucose, and energy metabolism. To a large extent, bile salts are (>95% per cycle) absorbed in the terminal ileum, the final section of the small intestine. The bile salt hydrolase activity has been widely detected in several bacterial genera, including Bacteroides, Clostridium, Lactobacillus, and Bifidobacteria. In addition, bile acids have been found to affect glucose metabolism by activating FXR and TGR5 receptors, as well as intestinal flora (Jung et al., 2007). Lactococcus G423 upregulated 6-hydroxy-5-methoxyindole glucuronide level, suggesting that it may have some regulatory effects on the bile acid metabolism.
Linoleic acid could have beneficial effects on maintaining healthy squabs, as reflected by improved antioxidant capacity and lipid metabolism (Xu et al., 2020). Linoleic acid has shown a correlation with lipid metabolic diseases (Choque et al., 2014). Previous studies have demonstrated that linoleic acid content was associated with probiotics (Hossain et al., 2012; Sahoo et al., 2015). The metabolized product of linoleic acid is 9,10-dihydroxy-12-octadecenoic acid (9,10-DiHOME) (Felipe et al., 2023). More recent research has suggested that DiHOMEs may be important lipid mediators (Hildreth et al., 2020; Zhou et al., 2023). Propionibacterium acnes and Lactobacillus plantarum have been reported to convert linoleic acid into conjugated linoleic acid (Bo et al., 2017). In the present study, Lactococcus G423 downregulated 9,10-DiHOME level, suggesting that it may have some regulatory effects on the linoleic acid metabolism.
Lactococcus G423 regulated 9,10-DiHOME and-hydroxy-5-methoxyindole glucuronide by effecting the abundance of Bacteroides and Lactobacillus, which effected the lipid metabolic pathway of bile secretion and linoleic acid.
5 Conclusion
In conclusion, the results of the present study showed that the gut microbiota and the ileum contents of metabolites were significantly correlated, and the metabolites might be considered as mediators in the association between the intestinal microbiota and lipid metabolism. Lactococcus G423 could reduce abdominal fat percentage of broilers through the gut microbiota, regulating the pathways of lipid metabolism and bile acid metabolism. Lactococcus G423 could ameliorate the lipid metabolism of broilers by integrating the microbiome and metabolome data. Thus, the above-mentioned Lactococcus G423 strains can be utilized as a new probiotic combination for animals.
Data availability statement
The datasets presented in this study can be found in online repositories: https://doi.org/10.6084/m9.figshare.25459783.v2.
Ethics statement
The animal studies were approved by the HEI Animal Management Certificate No. 11928. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
MW: Funding acquisition, Writing – original draft. WM: Data curation, Writing – review & editing. CW: Conceptualization, Writing – original draft. DL: Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Provincial Natural Science Foundation of China.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1381756/full#supplementary-material
Footnotes
References
Abdel-Moneim, A. M. E., Selim, D. A., Basuony, H. A., Sabic, E. M., Saleh, A. A., and Ebeid, T. A. (2020). Effect of dietary supplementation of Bacillus subtilis spores on growth performance, oxidative status, and digestive enzyme activities in Japanese quail birds. Trop. Anim. Health Prod. 52, 671–680. doi: 10.1007/s11250-019-02055-1
Abramowicz, M. O. K. (2019). The effect of a probiotic preparation containing Bacillus subtilis pb6 in the diet of chickens on redox and biochemical parameters in their blood. Nat. Rev. Cancer 19, 433–451. doi: 10.2478/aoas-2018-0059
Alagawany, M., Madkour, M., El-Saadony, M. T., and Reda, F. M. (2021). Paenibacillus polymyxa (lm31) as a new feed additive: antioxidant and antimicrobial activity and its effects on growth, blood biochemistry, and intestinal bacterial populations of growing Japanese quail. Anim. Feed Sci. Technol. 276:114920. doi: 10.1016/J.ANIFEEDSCI.2021.114920
An, H. M., Park, S. Y., Lee, D. K., Kim, J. R., and Ha, N. J. (2011). Antiobesity and lipid-lowering effects of Bifidobacterium spp. in high fat diet-induced obese rats. Lipids in Health Dis. 10:116. doi: 10.1186/1476-511X-10-116
Anand, S., and Mande, S. S. (2018). Diet, microbiota and gut-lung connection. Front. Microbiol. 9:2147. doi: 10.3389/fmicb.2018.02147
Angela, C., Wang, W., Lyu, H., Zhou, Y., and Huang, X. (2020). The effect of dietary supplementation of Astragalus membranaceus and Bupleurum chinense on the growth performance, immune-related enzyme activities and genes expression in white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 107, 379–384. doi: 10.1016/j.fsi.2020.10.014
Arumugam, M., Raes, J., Pelletier, E., Le, P. D., Yamada, T. M., Mende, D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180. doi: 10.1038/nature09944
Ashouri, G., Soofiani, N. M., Hoseinifar, S. H., Jalali, S. A. H., Morshedi, V., Valinassab, T., et al. (2020). Influence of dietary sodium alginate and Pediococcus acidilactici on liver antioxidant status, intestinal lysozyme gene expression, histomorphology, microbiota, and digestive enzymes activity, in Asian sea bass (Lates calcarifer) juveniles. Aquaculture 518:734638. doi: 10.1016/j.aquaculture.2019.734638
Bo, Y., He, G., Csd, E., Rpre, F., Hao, Z., Yong, Q., et al. (2017). Bacterial conjugated linoleic acid production and their applications – science direct. Prog. Lipid Res. 1738, 105–114. doi: 10.1016/j.bbalip.2005.11.004
Busti, S., Rossi, B., Volpe, E., Ciulli, S., Piva, A., D’Amico, F., et al. (2020). Effects of dietary organic acids and nature identical compounds on growth, immune parameters and gut microbiota of European sea bass. Sci. Rep. 10:21321. doi: 10.1038/s41598-020-78441-9
Chan, E. S., and Zhang, Z. (2005). Bioencapsulation by compression coating of probiotic bacteria for their protection in an acidic medium. Process Biochem. 40, 3346–3351. doi: 10.1016/j.procbio.2005.03.001
Chen, F., Gao, S. S., Zhu, L. Q., Qin, S. Y., and Qiu, H. L. (2018). Effects of dietary Lactobacillus rhamnosus CF supplementation on growth, meat quality, and microenvironment in specific pathogen free chickens. Poult. Sci. 97, 118–123. doi: 10.3382/ps/pex261
Chen, Y., Wang, J., Yu, L., Xu, T., and Zhu, N. (2020). Microbiota and metabolome responses in the cecum and serum of broiler chickens fed with plant essential oils or virginiamycin. Sci. Rep. 10:5382. doi: 10.1038/s41598-020-60135-x
Cho, W. Y., Hong, G. E., Lee, H. J., Yeon, S. J., Paik, H. D., Hosaka, Y. Z., et al. (2020). Effect of yogurt fermented by Lactobacillus fermentum TSI and L. fermentum S2 derived from a Mongolian traditional dairy product on rats with high-fat-diet-induced obesity. Food Secur. 9:594. doi: 10.3390/foods9050594
Choque, B., Catheline, D., Rioux, V., and Legrand, P. (2014). Linoleic acid: between doubts and certainties. Biochimie 96, 14–21. doi: 10.1016/j.biochi.2013.07.012
Christy, M.-L., Sampson, M., Edson, M., and Anthony, O. (2018). Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules 23, 1–48. doi: 10.3390/molecules23040795
Danzeisen, J. K., Kim, H. B., Isaacson, R. E., Tu, Z. J., and Johnson, T. J. (2011). Modulation of the chicken cecal microbiome and metagenome in response to anticoccidial and growth promoter treatment. PLoS ONE. 6:e27949. doi: 10.1371/journal.pone.0027949
Dawood, M. A. O., Koshio, S., and Esteban, M. Á. (2018). Beneficial roles of feed additives as immunostimulants in aquaculture: a review. Rev. Aquac. 10, 950–974. doi: 10.1111/raq.12209
Ding, C., Wu, H., Cao, X., Ma, X., Gao, X., Gao, Z., et al. (2021). Lactobacillus johnsonii 3-1 and Lactobacillus crispatus 7-4 promote the growth performance and ileum development and participate in lipid metabolism of broilers. Food Fun. 12, 12535–12549. doi: 10.1039/D1FO03209G
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Fan, Y., Wang, X. L., Wang, Y. H., Liu, H. J., Yu, X. Q., Li, L., et al. (2021). Potential effects of dietary probiotics with Chinese herb polysaccharides on the growth performance, immunity, disease resistance, and intestinal microbiota of rainbow trout (Oncorhynchus mykiss). J. World Aquac. Soc. 52, 1194–1208. doi: 10.1111/jwas.12757
Faseleh, J. M., Wesam, A. Y., Shokryazdan, P., Ebrahimi, R., Ebrahimi, M., Idrus, Z., et al. (2016). Dietary supplementation of a mixture of Lactobacillus strains enhances performance of broiler chickens raised under heat stress conditions. Int. J. Biometeorol. 60, 1099–1110. doi: 10.1007/s00484-015-1103-x
Felipe, C. S., Ana Cristina, G. A. G., Rhianna, K. M., Zhang, Z. C., Ameer, Y. T., and Pamela, J. L. (2023). Oxidized linoleic acid metabolites regulate neuronal morphogenesis in vitro. Neurochem. Int. 164:105506. doi: 10.1016/j.neuint.2023
Fernández, J., Redondo-Blanco, S., Gutiérrez-del-Río, I., Miguélez, E. M., Villar, C. J., and Lombó, F. (2016). Colon microbiota fermentation of dietary prebiotics towards short-chain fatty acids and their roles as anti-inflammatory and antitumour agents: a review. J. Funct. Foods 25, 511–522. doi: 10.1016/j.jff.2016.06.032
Gerritsen, J., Fuentes, S., Grievink, W., Niftrik, L. V., Tindall, B. J., Timmerman, H. M., et al. (2014). Characterization of Romboutsia ilealis gen. Nov., sp. nov., isolated from the gastro-intestinal tract of a rat, and proposal for the reclassification of five closely related members of the genus Clostridium into the genera Romboutsia gen. Nov., Intestinibacter gen. Nov., errisporobacter gen. Nov. and Asaccharospora gen. Nov. Int. J. Syst. Evol. Microbiol 64, 1600–1616. doi: 10.1099/ijs.0.059543-0
Ghosh, S., Whitley, C. S., Haribabu, B., and Jala, V. R. (2021). Regulation of intestinal barrier function by microbial metabolites. Cell. Mol. Gastroenterol. Hepatol. 11, 1463–1482. doi: 10.1016/j.jcmgh.2021.02.007
Gill, S. K., Rossi, M., Bajka, B., and Whelan, K. (2021). Dietary fibre in gastrointestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 18, 101–116. doi: 10.1038/s41575-020-00375-4
Greiner, T., and Bäckhed, F. (2011). Effects of the gut microbiota on obesity and glucose homeostasis. Trends Endocrinol. Metab. 22, 117–123. doi: 10.1016/j.tem.2011.01.002
Gupta, S., Fekaninová, A., Lokesh, J., Koová, J., Srensen, M., Fernandes, J., et al. (2018). Lactobacillus dominate in the intestine of Atlantic Salmon fed dietary probiotics. Front. Microbiol. 9:9. doi: 10.3389/fmicb.2018.03247
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hakimul, H., Subir, S., Shariful, I., Aminul, I., Rezaul, K., Mohammad, E. H. K., et al. (2020). Sustainable antibiotic-free broiler meat production: current trends, challenges, and possibilities in a developing country perspective. Biology 9, 1–24. doi: 10.3390/biology9110411
Hamilton, J. S., and Klett, E. L. (2021). Linoleic acid and the regulation of glucose homeostasis: a review of the evidence. Prostag. Leukotr. Esse. Fatty Acids. 175:102366. doi: 10.1016/j.plefa.2021.102366
Hildreth, K., Kodani, S. D., Hammock, B. D., and Zhao, L. (2020). Cytochrome p450-derived linoleic acid metabolites epomes and dihomes: a review of recent studies. J. nutr. biochemi. 86:108484. doi: 10.1016/j.jnutbio.2020.108484
Hossain, M. E., Kim, G. M., Lee, S. K., and Yang, C. J. (2012). Growth performance, meat yield, oxidative stability, and fatty acid composition of meat from broilers fed diets supplemented with a medicinal plant and probiotics. Animal bioscience 25, 1159–1168. doi: 10.5713/ajas.2012.12090
Judkins, T. C., Archer, D. L., Kramer, D. C., and Solch, R. J. (2020). Probiotics, nutrition, and the small intestine. Curr. Gastroenterol. Rep. 22:2. doi: 10.1007/s11894-019-0740-3
Jung, D., Inagaki, T., Gerard, R. D., Dawson, P. A., Kliewer, S. A., Mangelsdorf, D., et al. (2007). FXR agonists and FGF15 reduce fecal bile acid excretion in a mouse model of bile acid malabsorption. J. Lipid Res. 48, 2693–2700. doi: 10.1194/jlr.M700351-JLR200
Kailasapathy, K., and Chin, J. (2000). Survival and therapeutic potential of probiotic organisms with reference to Lactobacillus acidophilus and Bifidobacteria spp. Immunol. Cell Biol. 78, 80–88. doi: 10.1046/j.1440-1711.2000.00886.x
Kim, H., Kim, Y., and Kang, C. H. (2021). In vivo confirmation of the antimicrobial effect of probiotic candidates against Gardnerella vaginalis. Microorganisms 9:1690. doi: 10.3390/microorganisms9081690
Kong, X., Guo, Z., Yao, Y., Xia, L., Liu, R., Song, H., et al. (2022). Acetic acid alters rhizosphere microbes and metabolic composition to improve willows drought resistance. Sci. Total Environ. 844:157132. doi: 10.1016/j.scitotenv.2022.157132
Kumar, S., Chen, C., Indugu, N., Werlang, G. O., Singh, M., Kim, W. K., et al. (2018). Effect of antibiotic withdrawal in feed on chicken gut microbial dynamics, immunity, growth performance and prevalence of foodborne pathogens. PLoS One 13:e0192450. doi: 10.1371/journal.pone.0192450
Ley, R. E., Turnbaugh, P. J., Klein, S., and Gordon, J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023. doi: 10.1038/4441022a
Li, C., Al-Dalali, S., Zhou, H., and Xu, B. (2022). Influence of curing on the metabolite profile of water-boiled salted duck. Food Chem. 397:133752. doi: 10.1016/j.foodchem.2022.133752
Li, Z., Wu, Y., Hu, J., Yang, G., Wang, Z., and Sun, J. (2022). Dissection of the response mechanism of alfalfa under phosphite stress based on metabolomic and transcriptomic data. Plant Physiol. Biochem. 192, 35–49. doi: 10.1016/j.plaphy.2022.09.024
Liu, L., Ni, X., Zeng, D., Wang, H., Jing, B., Yin, Z., et al. (2016). Effect of a dietary probiotic, Lactobacillus johnsonii BS15, on growth performance, quality traits, antioxidant ability, and nutritional and flavour substances of chicken meat. Anim. Prod. Sci. 57, 920–926. doi: 10.1071/AN15344
Mohd Shaufi, M. A., Sieo, C. C., Chong, C. W., Gan, H. M., and Ho, Y. W. (2015). Deciphering chicken gut microbial dynamics based on high-throughput 16S rRNA metagenomics analyses. Gut Pathog. 7:4. doi: 10.1186/s13099-015-0051-7
Nhung, N. T., Chansiripornchai, N., and Carrique-Mas, J. J. (2017). Antimicrobial resistance in bacterial poultry pathogens: a review. Front Vet Sci. 4, 1–17. doi: 10.3389/fvets.2017.00126
Oniciuc, E., Likotrafiti, E., Alvarez-Molina, A., Prieto, M., Santos, J., and Alvarez-Ordóñez, A. (2018). The present and future of whole genome sequencing (WGS) and whole metagenome sequencing (WMS) for surveillance of antimicrobial resistant microorganisms and antimicrobial resistance genes across the food chain. Genes 9, 1–28. doi: 10.3390/genes905026875
Paintsil, E. K., Ofori, L. A., Akenten, C. W., Fosu, D., Ofori, S., Lamshöft, M., et al. (2021). Antimicrobial usage in commercial and domestic poultry farming in two communities in the Ashanti region of Ghana. Antibiotics 10, 1–9. doi: 10.3390/antibiotics10070800
Park, S. H., Lee, S. I., and Ricke, S. C. (2016). Microbial populations in naked neck chicken ceca raised on pasture flock fed with commercial yeast cell wall prebiotics via an Illumina MiSeq platform. PLoS One 11:e0151944. doi: 10.1371/journal.pone.0151944
Qiao, H. X., Zhang, L. H., Shi, H. T., Song, Y. Z., and Bian, C. Z. (2018). Astragalus affects fecal microbial composition of young hens as determined by16S rRNA sequencing. AMB Expr. 8:70. doi: 10.1186/s13568-018-0600-9
Rahman, M. A., Parvi, N. M. S., Sarker, R. R., and Islam, M. T. (2012). Effects of growth promoter and multivitamin-mineral premix supplementation on body weight gain in broiler chickens. J Bangladesh Agril Univ. 8, 70–78. doi: 10.1186/s13568-018-0600-9
Ren, Y., Yu, G., Shi, C., Liu, L., Guo, Q., Han, C., et al. (2022). Majorbio Cloud: A one‐stop, comprehensive bioinformatic platform for multiomics analyses. IMeta 1:e12. doi: 10.1002/imt2.12
Ricaboni, D., Mailhe, M., Khelaifia, S., Raoult, D., and Million, M. (2016). Romboutsia timonensis, a new species isolated from human gut. New Microbes. New Infec. 12, 6–7. doi: 10.1016/j.nmni.2016.04.001
Rybarczyk, A., Bogusławska-Wąs, E., and Łupkowska, A. (2020). Effect of EM probiotic on gut microbiota, growth performance, carcass and meat quality of pigs. Livestock Sci. 241:104206. doi: 10.1016/j.livsci.2020.104206
Sahoo, S. P., Kaur, D., Sethi, A. P. S., Sharma, A., and Chandra, M. (2015). Evaluation of Yucca schidigera extract as feed additive on performance of broiler chicks in winter season. Veterinary World 8, 556–560. doi: 10.14202/vetworld.2015.556-560
Saxena, S., Saxena, V., Tomar, S., Sapcota, D., and Gonmei, G. (2016). Characterisation of caecum and crop microbiota of Indian indigenous chicken targeting multiple hypervariable regions within 16SrRNA gene. Br. Poult. Sci. 57, 381–389. doi: 10.1080/00071668.2016.1161728
Shang, Y., Kumar, S., Oakley, B., and Kim, W. K. (2018). Chicken gut microbiota: importance and detection technology. Front. Vet. Sci. 5:254. doi: 10.3389/fvets.2018.00254
Shaufi, M. A. M., Sieo, C. C., Chong, C. W., Gan, H. M., and Ho, Y. W. (2015). Deciphering chicken gut microbial dynamics based on high-throughput 16S rRNA metagenomics analyses. Gut Pathog 7, 1–12. doi: 10.17957/IJAB/15.0742
Shi, F., Zi, Y., Lu, Z., Li, F., Yang, M., Zhan, F., et al. (2020). Bacillus subtilis H2 modulates immune response, fat metabolism and bacterial flora in the gut of grass carp (Ctenopharyngodon idellus). Fish Shellfish Immunol. 106, 8–20. doi: 10.1016/j.fsi.2020.06.061
Shulpekova, Y., Shirokova, E., Zharkova, M., Tkachenko, P., Tikhonov, I., Stepanov, A., et al. (2022). A recent ten-year perspective: Bile acid metabolism and signaling. Molecules 27:1983. doi: 10.3390/molecules27061983
Soccol, C. R., Vandenberghe, L. P. S., Spier, M. R., Medeiros, A. B. P., Yamaguishi, C. T., Lindner, J. D. D., et al. (2010). The potential of probiotics: A review. Food Technol. Biotechnol. 48, 413–434. doi: 10.1016/j.fm.2010.06.003
Stackebrandt, E., and Goebel, B. M. (1994). Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 44, 846–849. doi: 10.1099/00207713-44-4-846
Stęczny, K., and Dariusz, K. (2020). Effect of probiotic preparations (em) on productive characteristics, carcass composition, and microbial contamination in a commercial broiler chicken farm. Anim. Biotechnol. 3, 1–8. doi: 10.1080/10495398.2020.1754841
Tan, C. Y., Li, Q. Y., Yang, X. J., Chen, J. Y., Zhang, Q., and Deng, X. (2022). Lactococcus lactis’ effect on the intestinal microbiota of Streptococcus agalactiae-infected zebrafish (Danio rerio). Microbiol. Spectrum 10, e01122–e01128. doi: 10.1128/spectrum.01128-22
Tanja, M., and Steven, L. S. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957‐2963. doi: 10.1093/bioinformatics/btr507
Turnbaugh, P. J., Hamady, M., Yatsunenko, T., Cantarel, B. L., Duncan, A., Ley, R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484.
Usui, Y., Kimura, Y., Satoh, T., Takemura, N., Ouchi, Y., Ohmiya, H., et al. (2018). Effects of lo-ng term intake of a yogurt fermented with Lactobacillus delbrueckii subsp. bulgaricus 2038 and Streptococcus thermophilus 1131 on mice. Int. Immunol. 30, 319–331. doi: 10.1093/intimm/dxy035
Wang, Q. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, H. S., Ni, X. Q., Qing, X. D., Zeng, D., Luo, M., Liu, L., et al. (2017). Live probiotic Lactobacillus johnsonii BS15 promotes growth performance and lowers fat deposition by improving lipid metabolism, intestinal development, and gut microflora in broilers. Front. Microbiol. 8, 1073–1087. doi: 10.3389/fmicb.2017.01073
Wang, Y. B., Wang, Z. B., Wan, Y., Jin, F. R., Shi, X. D., Xing, Z. H., et al. (2023). Assessing the in vivo ameliorative effects of Lactobacillus acidophilus KLDS1.0901 for induced non-alcoholic fatty liver disease treatment. Front. Nutr. 20:1147423. doi: 10.3389/fnut.2023.1147423
Wu, X. Z., Wen, Z. G., and Hua, J. L. (2019). Effects of dietary inclusion of lactobacillus and inulin on growth performance, gut microbiota, nutrient utilization, and immune parameters in broilers. Poul. Sci. 98, 4656–4663. doi: 10.3382/ps/pez166
Xia, F., Hu, S., Zheng, X., Wang, M. W., Zhang, C. C., Wu, Z. N., et al. (2021). New insights into metabolomics profile generating in fermented tea: the relevance between bacteria and the metabolites in Fuzhuan brick tea. J. Sci. Food Agri. 102, 350–359. doi: 10.1002/jsfa.11365
Xu, Q. Q., Ma, X. W., Dong, X. Y., Tao, Z. R., Lu, L. Z., and Zou, X. T. (2020). Effects of parental dietary linoleic acid on growth performance, antioxidant capacity, and lipid metabolism in domestic pigeons (Columba livia). Poul. Sci. 99, 1471–1482. doi: 10.1016/j.psj.2019.11.002
Xu, Y. H., Yang, H. X., Zhang, L. L., Su, T. H., Shi, D. H., Xiao, H. D., et al. (2016). High-throughput sequencing technology to reveal the composition and function of cecal microbiota in Dgu chicken. BMC Microbiol. 16, 259–268. doi: 10.1186/s12866-016-0877-2
Yukgehnaish, K., Kumar, P., Sivachandran, P., Marimuthu, K., Arshad, A., Paray, B. A., et al. (2020). Gut microbiota metagenomics in aquaculture: factors influencing gut microbiome and its physiological role in fish. Rev. Aquac. 12, 1903–1927. doi: 10.1111/raq.12416
Zhang, L., Liu, X., and Jia, H. (2022). WGCNA analysis of important modules and hub genes of compound probiotics regulating lipid metabolism in heat-stressed broilers. Animals 12:2644. doi: 10.3390/ani12192644
Zhang, T., Xie, J., Zhang, M., Fu, N., and Zhang, Y. (2016). Effect of a potential probiotics Lactococcus garvieae b301 on the growth performance, immune parameters and caecum microflora of broiler chickens. J Anim Physiol Anim Nutr. 100, 413–421. doi: 10.1111/jpn.12388
Zhao, X., Xian, Y., Li, C., Wang, C., Yu, D., Zhu, W., et al. (2016). Feeding Lactobacillus planta-rum and Lactobacillus casei increased microbial diversity and short chain fatty acids production in the gut-intestinal tract of weaning piglets. Wei Sheng Wu Xue Bao 56, 1291–1300. doi: 10.1128/spectrum.01128-22
Zheng, M., Mao, P., Tian, X., and Meng, L. (2021). Effects of grazing mixed-grass pastures on growth performance, immune responses, and intestinal microbiota in free-range Beijing-you chickens. Poult. Sci. 100, 1049–1058. doi: 10.1016/j.psj.2020.11.005
Zhou, L., Li, H., Hu, J. T., Meng, J. P., Lv, H. H., Yang, F., et al. (2023). Plasma oxidative lipidomics reveals signatures for sepsis-associated acute kidney injury. Clin. Chim. Acta 551:117616. doi: 10.1016/j.cca.2023.117616
Zhou, M., Zeng, D., Ni, X., Tu, T., Yin, Z., Pan, K., et al. (2016). Effects of Bacillus licheniformis on the growth performance and expression of lipid metabolism-related genes in broiler chickens challenged with Clostridium perfringens-induced necrotic enteritis. Lipids Health Dis. 15:48. doi: 10.1186/s12944-016-0219-2
Zhou, Y., Chen, P., Shi, S., Li, X., and Xiao, Y. (2019). Expression of Gallus epidermal growth factor (gEGF) with food-grade Lactococcus lactis expression system and its biological effects on broiler chickens. Biomol. Ther. 11:103. doi: 10.3390/biom11010103
Keywords: Lactococcus, broilers, growth performant, 16S rRNA, LC–MS
Citation: Wang M, Ma W, Wang C and Li D (2024) Lactococcus G423 improve growth performance and lipid metabolism of broilers through modulating the gut microbiota and metabolites. Front. Microbiol. 15:1381756. doi: 10.3389/fmicb.2024.1381756
Edited by:
Sabina Fijan, University of Maribor, SloveniaReviewed by:
Muhammad Suleman, University of Veterinary and Animal Sciences, PakistanYuncai Xiao, Huazhong Agricultural University, China
Copyright © 2024 Wang, Ma, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Desheng Li, bGlkZXNoZW5nMDcyNjUyMUAxMjYuY29t
 Mi Wang
Mi Wang Wei Ma
Wei Ma Chunqiang Wang
Chunqiang Wang Desheng Li
Desheng Li