
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 16 April 2024
Sec. Virology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1380849
This article is part of the Research Topic Advances and Insights in the Diagnosis of Viral Infections and Vaccines Development in Animals View all 24 articles
Introduction: Porcine viral diarrhea is a common clinical disease, which results in high mortality and economic losses in the pig industry. Porcine epidemic diarrhea virus (PEDV), porcine rotavirus (PoRV), and porcine deltacoronavirus (PDCoV) are important diarrhea viruses in pig herds. The similarities of their clinical symptoms and pathological changes make it difficult to distinguish these three viruses clinically. Therefore, there is a need for a highly sensitive and specific method to simultaneously detect and differentiate these viruses.
Methods: A multiplex real-time PCR assay using TaqMan probes was developed to simultaneously detect PEDV, PoRV, and PDCoV. To assess the efficacy of the established assay, 30 clinical samples with diarrhea symptoms were used to compare the results obtained from the multiplex real-time PCR assay with those obtained from commercial singleplex real-time PCR kit. Importantly, a total of 4,800 diarrhea samples were tested and analyzed to validate the utility of the assay.
Results: This multiplex real-time PCR assay showed high sensitivity, specificity, and excellent repeatability with a detection limit of 1 × 102 copies/μL. Comparing the results of the commercial singleplex real-time PCR kit and the multiplex real-time PCR method for detecting PEDV, PoRV, and PDCoV, there was complete agreement between the two approaches. Clinical data revealed single infection rates of 6.56% for PEDV, 21.69% for PoRV, and 6.65% for PDCoV. The co-infection rates were 11.83% for PEDV + PoRV, 0.29% for PEDV + PDCoV, 5.71% for PoRV + PDCoV, and 1.29% for PEDV + PDCoV + PoRV, respectively.
Discussion: The multiplex real-time PCR method established in this study is a valuable diagnostic tool for simultaneously differentiating PEDV, PoRV, and PDCoV. This method is expected to significantly contribute to prevent and control the spread of infectious diseases, as well as aid in conducting epidemiological investigations.
Viral diarrheal disease poses a serious threat to the swine industry, causing substantial global economic losses (Wu et al., 2022; Hou et al., 2023). Numerous diarrhea viruses have been identified in swine, including transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), porcine delta coronavirus (PDCoV), porcine enteric alpha coronavirus (PEAV), porcine rotavirus (PoRV), porcine sapvirus (PSaV), porcine norovirus (PNoV), porcine teschenvirus (PTV), porcine kobuvirus (PKV), seneca valley virus (SVV), porcine reovirus (ReoV), porcine bocavirus (PBoV), porcine astrovirus (PAstV), and so on (Huang et al., 2019). Among these swine diarrhea viruses, PEDV, PoRV, and PDCoV are the most destructive pathogens causing anorexia, vomiting, diarrhea, and dehydration (Hou et al., 2023). With the rapid development of intensive aquaculture, co-infection or secondary infection with these viruses is prevalent, resulting in significant morbidity and mortality (Huang et al., 2019; Hou et al., 2023).
PEDV is an enveloped, single-stranded RNA virus that belongs to the Alphacoronavirus genus in the Coronaviridae family (Huang et al., 2013; Hu et al., 2021). The spherical PEDV particles vary in size, with an average diameter of ~130 nm (Hu et al., 2021). The PEDV genome is ~28 kb (Huang et al., 2013; Hu et al., 2021). While PEDV can infect pigs of any age, it is especially severe in piglets, often resulting in 100% morbidity and mortality (Shibata et al., 2000; Huang et al., 2013). The first reported cases of porcine epidemic diarrhea (PED) occurred in England in 1971, followed by a subsequent outbreak in Belgium in 1977 (Si et al., 2020; Hou et al., 2023). It was later identified in China in the 1980's (Wang et al., 2016). In December 2010, a new strain of PEDV with high virulence emerged in China, resulting in the death of over 1 million piglets and a 100% mortality rate among suckling piglets (Wang et al., 2016; Wu et al., 2022). Since then, the emerging and highly virulent strains of PEDV have gradually spread throughout the global swine industry, causing significant economic losses and becoming a leading cause of diarrhea in pigs (Wang et al., 2016; Hou et al., 2023).
PDCoV is a new swine enteric coronavirus that belongs to the newly identified genus Deltacoronavirus in the Coronaviridae family (Ma et al., 2015). It is an enveloped, single-stranded positive-stranded RNA virus with a genome size of ~25.4 kb. PDCoV was first detected in Hong Kong, China in 2012, and then in the United States in 2014 (Woo et al., 2012; Wang et al., 2014). The first outbreak of PDCoV occurred in the United States in 2014 and quickly spread to at least 20 states, resulting in significant economic losses in the global pig industry (Ma et al., 2015; He et al., 2020). PoRV is a non-enveloped, double-stranded RNA virus from the genus Rotavirus in Reoviridae family. Its genome is ~18,500 bp long, consisting of 11 segments of dsRNA (Chepngeno et al., 2020). PoRV is a major cause of diarrhea in piglets, particularly those aged 1–8 weeks (Chepngeno et al., 2020; Tao et al., 2023). It can also spread between pigs and humans, posing a significant threat to public health and safety in China (Li et al., 2018).
Pigs infected with PEDV, PoRV, and PDCoV exhibit similar clinical symptoms and pathological changes, making it hard to differentiate them. Moreover, the prevalence of co-infection and secondary infection among PEDV, PoRV, and PDCoV poses significant challenges in clinical diagnostics (Shu et al., 2022; Hou et al., 2023). It is therefore urgent to develop an efficient molecular method for distinguishing these viruses in the clinic. Real-time PCR utilizes fluorescence signals to monitor the PCR process in real-time, offering advantages in terms of speed, sensitivity, and reproducibility when compared to conventional PCR method. Hence, it is commonly used in clinical detection. However, singleplex real-time PCR is not convenient for the simultaneous detection of co-infection of multiple pathogens, and repeated detection of various pathogens wastes time and makes operations cumbersome (Ben Salem et al., 2010; Liu et al., 2019). Multiplex real-time PCR is a method that can detect multiple pathogens in a single reaction system (Huang et al., 2019; Li et al., 2022). In this study, we developed a multiple real-time PCR based on TaqMan probe to simultaneously and accurately detect PEDV, PoRV, and PDCoV. This assay exhibited high sensitivity and specificity for the target genes. Additionally, we used this method to detect and analyze 4,800 clinical samples collected from pig farms in multiple provinces of China, providing valuable data to help formulate prevention and control strategies in this particular region.
The nucleic acid of various viruses and bacteria, including respiratory syndrome virus (PRRSV), pseudorabies virus (PrV), porcine circovirus (PCV1), PCV2, classical swine fever virus (CSFV), African swine fever virus (ASFV), Streptococcus suis (SS), Salmonella enteritidis (SE), PEDV, PoRV, and PDCoV were stored at −80°C in our laboratory until needed. At least 30 genome sequences of PEDV, PoRV, and PDCoV were downloaded from NCBI for analysis. After comparison, the M gene of PEDV and PDCoV, and the NSP3 gene of PoRV were found to be the most conserved gene sequences. Primers and probes were designed using Primer Premier 5 software (Premier, Canada) based on the most conserved region. TaqMan probes for PEDV, PoRV, and PDCoV were labeled with FAM, VIC, and Cy5 at the 5′ end, respectively, with all quenchers at the 3′ end being BHQ. The sequences of the primers and probes designed in this study are presented in Table 1. Primers and probes were synthesized by Sangon Biotech (Shanghai) Co., Ltd.
The target fragments of PEDV, PoRV, and PDCoV were amplified separately using the HiScript II One Step RT-PCR Kit (Dye Plus; Nanjing Vazyme Biotech Co., Ltd.). The PCR fragments were purified and cloned into the pMD18-T vector (Takara Biomedical Technology (Beijing) Co., Ltd.). The transformed clones were then introduced into the E.coli DH5α strain. Positive clones were cultured and plasmid extraction was performed using the TaKaRa MiniBEST Universal Genomic DNA Extraction Kit [Takara Biomedical Technology (Beijing) Co., Ltd.]. The constructed plasmid was confirmed by DNA sequencing and used as the standard positive control. The copy number of recombination plasmids was calculated using the following formula (Li et al., 2022):
A 10-fold series dilution was conducted for each plasmid, ranging from 1.0 × 108 to 1.0 × 101 copies/μL. Singleplex real-time PCR was performed for each virus using the 10-fold diluted plasmids to generate standard curve. For multiplex standard curves, each plasmid was diluted to 3.0 × 109 copies/μL and pooled in equal volume to make a concentration of 1.0 × 109 copies/μL for each plasmid. The pooled plasmid was then diluted in a 10-fold series, ranging from 1.0 × 108 to 1.0 × 103 copies/μL, to establish multiplex standard curves.
All real-time PCR reaction systems were set to a volume of 20 μL. For singleplex real-time PCR of PDCoV, RV, and PEDV, the reaction system included 10 μL 2× PerfectStart Probe One-Step qPCR SuperMix, 0.4 μL TransScript Probe One-Step RT Enzyme Mix (Transgen, Beijing), 0.4 μL each of forward/reverse primer (10 μM), 0.4 μL TaqMan probe (10 μM), 5.5 μL RNA template, and 2.9 μL nuclease-free water.
Amplification was performed on a Bio-Rad CFX96™ Real-time System (Bio-Rad, Hercules, CA, USA) using the following program: 45°C for 5 min, 94°C for 30 s, 40 cycles of 94°C for 5 s, and 52°C for 30 s. Fluorescence signal was automatically collected at the end of each cycle. All qPCR results were analyzed using CFX Manager™ software.
The concentrations of primers and probes were optimized as previously described (Pan et al., 2020). After repeated tests, the optimal reaction conditions for multiplex real-time PCR were determined as follows: 10 μL 2× PerfectStart Probe One-Step qPCR SuperMix, 0.4 μL TransScript Probe One-Step RT Enzyme Mix (Transgen, Beijing), 0.4 μL each of forward/reverse primer and probe (10 μM) for PoRV and PDCoV, 0.2 μL each of forward/reverse primer and probe (10 μM) for PEDV, 5.5 μL RNA template, 1.1 μL nuclease-free water, with a total reaction volume of 20 μL. The same instrument and real-time PCR program were used as described above.
To determine the limit of detection (LOD) for the multiplex real-time PCR method, the aforementioned pooled standard plasmids were diluted in a 10-fold serial manner, ranging from 1.0 × 103 to 1.0 × 100 copies/μL in nuclease-free water. These diluted standard plasmids were then used as templates for multiplex real-time PCR amplification. The reliable LOD was the lowest concentration that achieved a 95% positive detection rate.
To avoid false positives caused by other viruses or bacteria that may be present in the samples, two RNA viruses (PRRSV and CSFV), three DNA viruses (PCV1, PCV2, and ASFV), and two bacterias (S. suis and S. enteritidis) were used for specificity test of multiplex real-time PCR assay. Nucleic acids were extracted using the VAMNE Virus DNA/RNA Extraction Kit (Nanjing Vazyme Biotech Co., Ltd.). For RNA viruses, the cDNA was generated with the TransScript Probe One-Step qRT-PCR SuperMix (Beijing Transgen Biotech Co., Ltd.). These DNA and cDNA samples were first detected using commercial kits, and positive samples with CT values <25 were selected as template for specificity test of the multiplex real-time PCR assay. The standard plasmids of PEDV, PoRV, and PDCoV were used as positive controls, while nuclease-free water served as the negative control.
To assess its repeatability of the multiplex real-time PCR, a 10-fold serially diluted standard template was used, ranging from 1.0 × 108 to 1.0 × 103 copies/μL. Each reaction was performed with three replicates. Intra-assay repeatability was determined by conducting three simultaneous detections of each plasmid under identical conditions. Inter-assay repeatability was evaluated by repeating the assays three times individually at different time points. The coefficient of variation (CV) of the Cq values from the three experiments was calculated to estimate the level of repeatability. Data analysis was conducted using Microsoft Excel.
Our multiplex real-time PCR method and a commercial singleplex real-time PCR Kit were used to simultaneously detect 30 clinical samples with diarrhea symptoms. The detection results obtained from our established methods were then compared with those obtained from singleplex real-time PCR.
A total of 4,800 samples including fecal samples (n = 2,341), rectal swabs (n = 1,874), oral fluid (n = 145), and oropharyngeal swabs (n = 440), were used to investigate the prevalence of PEDV, PoRV, and PDCoV using the multiplex real-time PCR assay. These samples were collected from pig farms located in numerous provinces of China, including Gansu, Shanxi, Shaanxi, Shandong, Henan, Hubei, Anhui, Zhejiang, Jiangsu, Guangdong, Guangxi, Guizhou, Sichuan, Hunan, Chongqing, and Jiangxi, during the period from November 2021 to July 2022.
Clinical samples were treated with phosphate buffer saline (PBS), and the supernatant was collected after vortexing and centrifugation. Nucleic acids were extracted using the VAMNE Virus DNA/RNA Extraction Kit (Nanjing Vazyme Biotech Co., Ltd.). Reverse transcription was done with the TransScript Probe One-Step qRT-PCR SuperMix (Beijing Transgen Biotech Co., Ltd.). The constructed plasmid and nuclease-free were employed as positive and negative controls, respectively. All samples were tested with the multiplex real-time PCR assay to determine virus positivity. Infection rates were analyzed after obtaining assay results for all clinical samples.
To develop a multiplex real-time PCR, we first established singleplex real-time PCR for each virus using different fluorescence-labeled target probes. Standard curve for each virus was created using 10-fold serial dilutions of plasmids ranging from 1.0 × 108 to 1.0×101 copies/μL. All the standard curves showed excellent correlation coefficients and amplification efficacy, with PEDV (R2 = 1.000; E = 101.3%), PoRV (R2 = 0.999; E = 103.9%), PDCoV (R2 = 1.000; E = 100.5%), respectively. This indicates that our plasmid standards were qualified, and the designed primers and probes were efficient (Figures 1A–C).
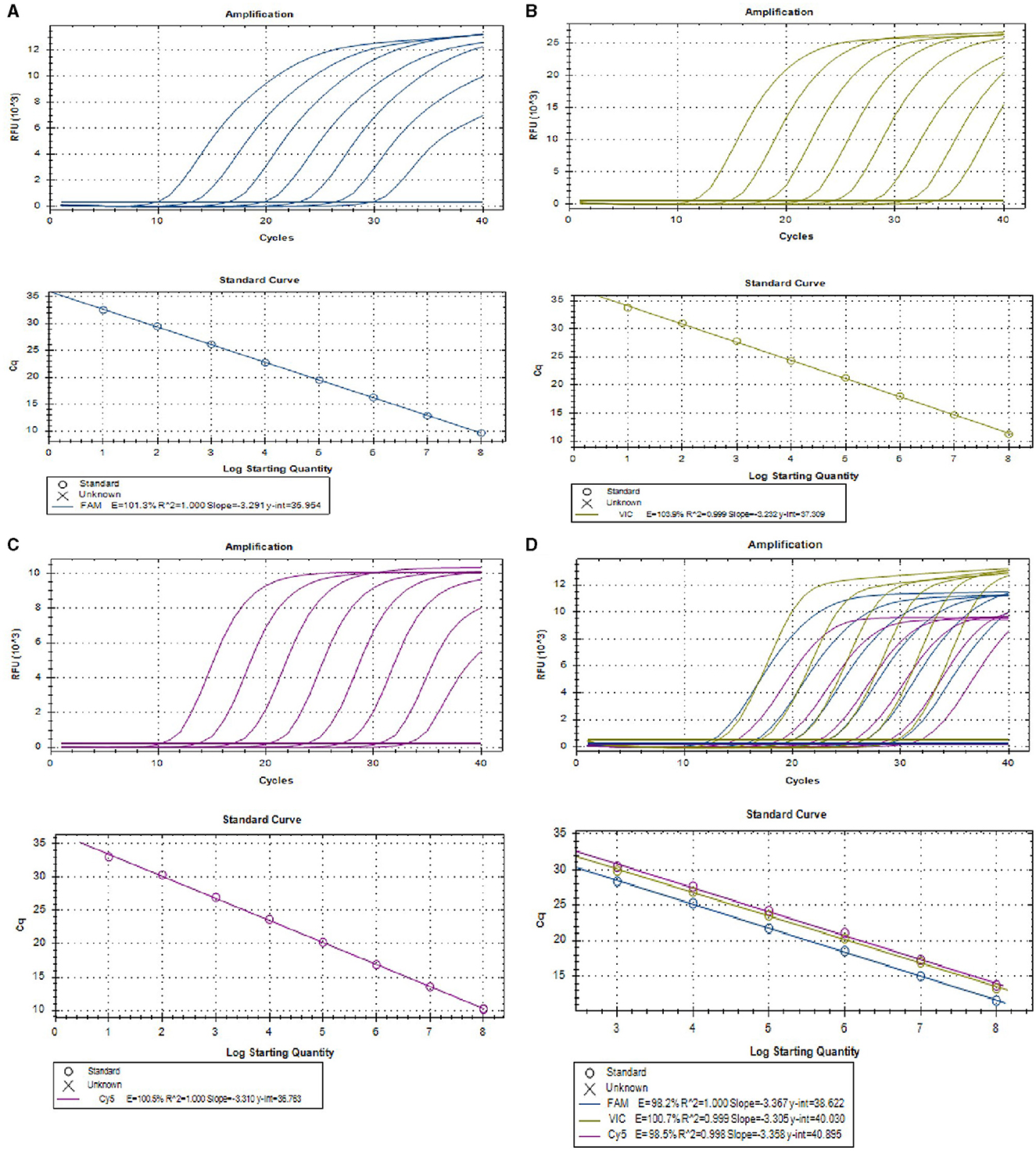
Figure 1. The amplification curves (top) and the standard curve (bottom) of the single and multiplex real-time PCR assay. (A–C) The amplification curves (top) and the standard curve (bottom) of single real-time PCR assay for detection of PEDV (A), PoRV (B), and PDCoV (C), respectively, with concentrations of each plasmid standard ranging from 1.0 × 108 to 1.0 × 101 copies/μL. (D) Amplification curves (top) and standard curves (bottom) of optimized multiplex real-time PCR for simultaneous detection of PEDV, PoRV, and PDCoV. The concentrations of each plasmid standard are from 1.0 × 108 to 1.0 × 103 copies/μL.
Serial dilutions of mixed viral standard plasmids were tested with the using the optimized multiplex assay. The results demonstrated that the multiplex real-time PCR successfully detected all target genes of these three viruses (Figure 1D). The standard curves exhibited excellent correlation coefficients and amplification efficacy for each virus, for details, PEDV (R2 = 1.000; E = 98.2%), PoRV (R2 = 0.999; E = 100.7%), and PDCoV (R2 = 0.998; E = 98.5%; Figure 1D), indicating the validity and reliability of the multiplex real-time PCR.
To evaluate the specificity of the multiplex real-time PCR, the positive samples of PRRSV, PRV, PCV1, PCV2, CSFV, ASFV, SS, and SE were used as templates for amplification with this multiplex system. The standard plasmids of PEDV, PoRV, and PDCoV were tested as the positive controls, while nuclease-free water was tested as negative control. As shown Figure 2A, all the target pathogens were successfully detected, while no positive signal was observed from other eight viruses and negative control. Additionally, three clinical samples from healthy pigs were further tested to confirm the specificity. Amplification curves also demonstrated that only target genes from the standard plasmids were detected, with no positive signal detected in the clinical samples from healthy pigs (Figure 2B). These findings indicated that the multiplex real-time PCR assay was highly specific.
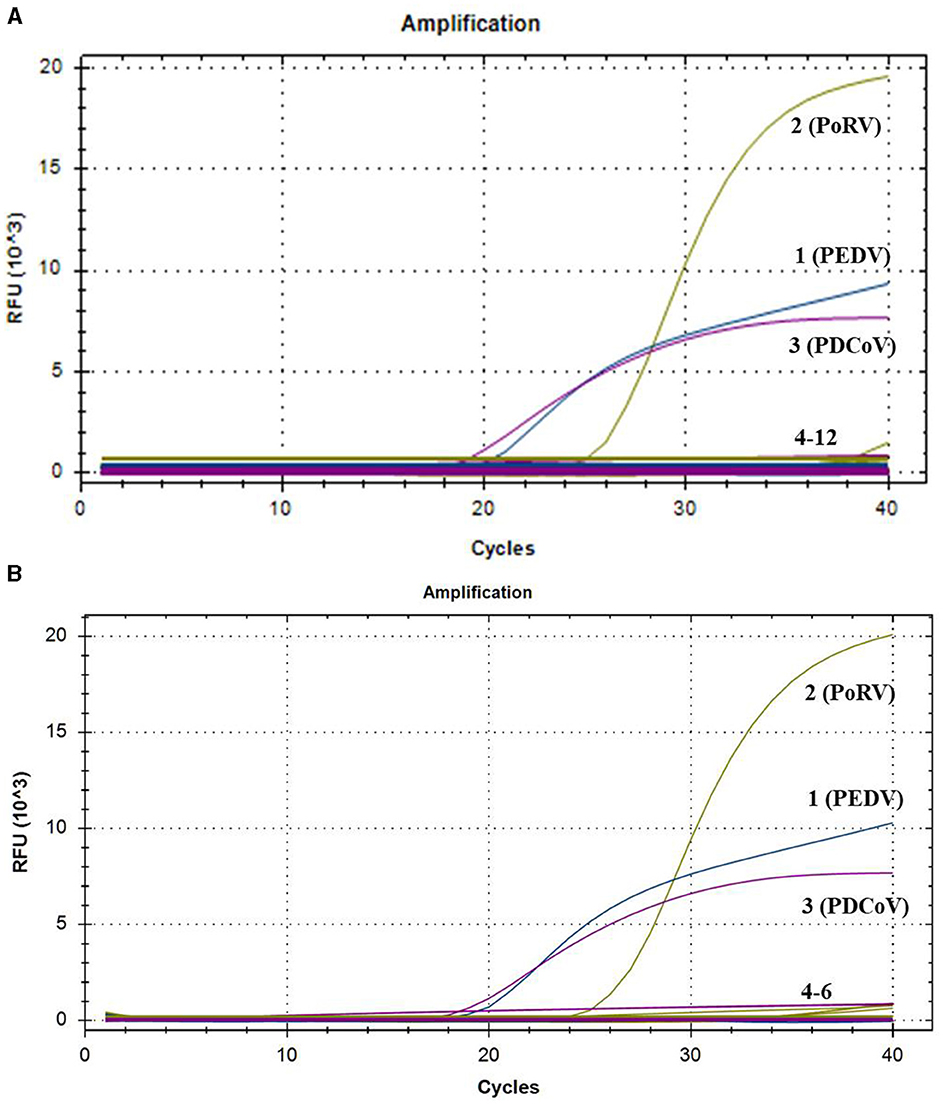
Figure 2. The amplification curves of specificity tests of multiplex real-time PCR. Only PEDV, PoRV, and PDCoV showed positive fluorescence signals, while other swine pathogens and clinical samples from healthy pigs exhibited no fluorescence signals. (A) 1–3: positive templates of PEDV, PoRV, and PDCoV. 4–12: negative control and positive templates of PRRSV, PRV, PCV1, PCV2, CSFV, ASFV, SS, and SE. (B) 1–3: positive templates of PEDV, PoRV, and PDCoV. 4–6: templates of clinical samples from healthy pigs.
To access the sensitivity of this multiplex real-time PCR assay, we tested pooled standard plasmids with concentrations ranging from 1.0 × 103 to 1.0 × 100 copies/μL under optimized reaction conditions. The method was able to detect positive samples with concentrations as low as 1.0 × 101 copies/μL (Table 2). However, further experiments showed the detection rate for samples at 1.0×101 copies/μL was <95% of replicates (Table 2). Therefore, the reliable limit of detection for this method is 1 × 102 copies/μL.
The multiplex real-time PCR assay was confirmed to be repeatable by detecting standard plasmids at concentrations ranging from 1.0 × 108 to 1.0 × 103 copies/μL. As shown in Table 3, the variation coefficients (CVs) of Cq values in the intra-group and inter-group reproducibility tests were between 0.11 and 0.87% and between 0.11 and 1.63%, respectively. These results indicated that the multiplex real-time PCR assay established in this study had satisfactory repeatability.
Thirty clinical samples were used to compare the results of the multiplex real-time PCR with commercial singleplex real-time PCR kit. As shown in Supplementary Table 1, the results of the multiplex real-time PCR were consistent with the commercial singleplex real-time PCR, suggesting that it can effectively replace the commercial singleplex real-time PCR for simultaneous detection of PEDV, PoRV, and PDCoV.
A total of 4,800 clinical samples were detected using the multiplex real-time PCR assay established in this study. As shown in Figure 3A, the single infection rates for PEDV, PoRV, and PDCoV were 6.56% (315/4,800), 21.69% (1,041/4,800), and 6.65% (319/4,800), respectively. The co-infection rates for PEDV + PoRV, PEDV + PDCoV, and PoRV + PDCoV were 11.83% (568/4,800), 0.29% (14/4,800), and 5.71% (274/4,800), respectively. The mixed infection rate for PEDV + PDCoV + PoRV was 1.29% (62/4,800).
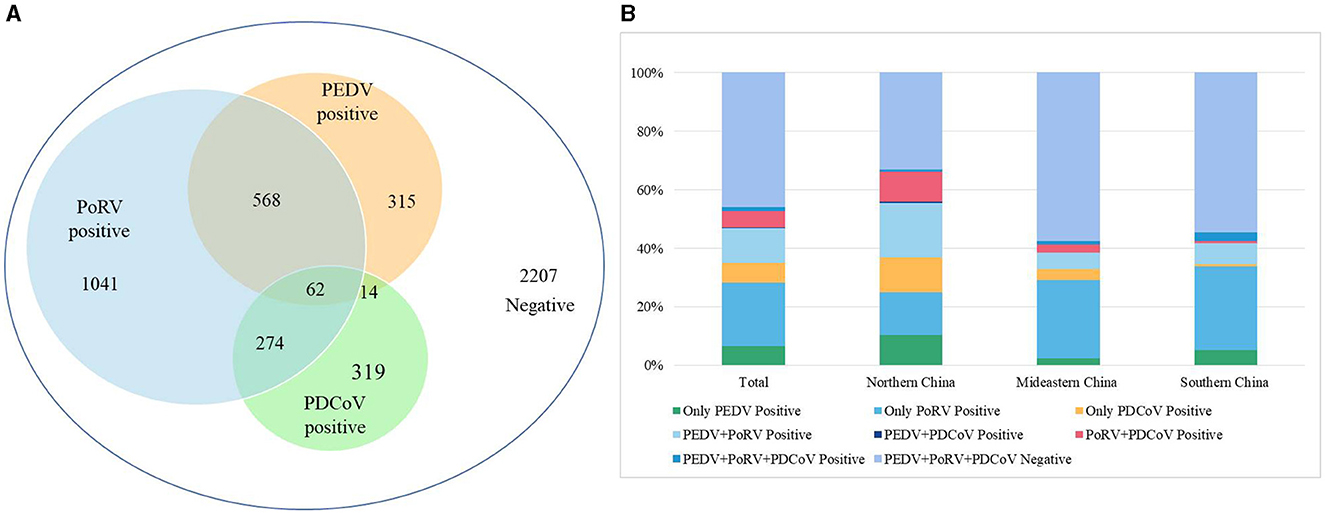
Figure 3. The detection results of clinical diarrhea samples using the multiplex real-time PCR assay established in this study. (A) The detection results of 4,800 clinical diarrhea samples. (B) A comparison of positive rates from different regions of China. These 4,800 clinical diarrhea samples were collected from 16 provinces of China, which were divided into three regions: Northern China (Gansu, Shanxi, and Shaanxi), Mideastern China (Shandong, Henan, Hubei, Anhui, Zhejiang, and Jiangsu), and Southern China (Guangdong, Guangxi, Guizhou, Sichuan, Hunan, Chongqing, and Jiangxi).
These 4,800 clinical samples were collected from 16 provinces which were divided into three regions based on the geographical distribution: Northern China (Gansu, Shanxi, and Shaanxi), Mideastern China (Shandong, Henan, Hubei, Anhui, Zhejiang, and Jiangsu), and Southern China (Guangdong, Guangxi, Guizhou, Sichuan, Hunan, Chongqing, and Jiangxi). In the Northern China, PEDV, PoRV, and PDCoV had infection rates of 10.33% (224/2,169), 14.71% (319/2,169), and 11.66% (253/2,169), respectively (Figure 3B). Co-infection rates were 18.63% (404/2,169) for PEDV + PoRV, 0.60% (13/2,169) for PEDV + PDCoV, 10.28% (223/2,169) for PoRV + PDCoV, 0.55% (12/2,169) for PEDV + PDCoV + PoRV, respectively (Figure 3B). In the Mideastern China, PEDV, PoRV, and PDCoV had infection rates of 2.44% (40/1,642), 26.79% (440/1,642), and 3.59% (59/1,642), respectively (Figure 3B). Co-infection rates were 5.66% (93/1,642) for PEDV + PoRV, 0.06% (1/1,642) for PEDV + PDCoV, 2.68% (44/1,642) for PoRV + PDCoV, 1.22% (20/1,642) for PEDV + PDCoV + PoRV, respectively (Figure 3B). In the Southern China, PEDV, PoRV, and PDCoV had infection rates of were 5.16% (51/989), 28.51% (282/989), and 0.71% (7/989), respectively (Figure 3B). Co-infection rates were 7.18% (71/989) for PEDV + PoRV, 0 (0/989) for PEDV + PDCoV, 0.71% (7/989) for PoRV + PDCoV, 3.03% (30/989) for PEDV + PDCoV + PoRV, respectively (Figure 3B).
Porcine viral diarrhea is a prevalent occurrence observed in clinical setting, resulting in substantial economic losses within the pig industry. Notably, PEDV, PoRV, and PDCoV are recognized as crucial pathogens causing this disease (Hou et al., 2023). With the development of large-scale and intensive swine production, mixed infections of these three pathogens are increasingly becoming common in swine farms (Huang et al., 2015). Detecting and identifying the pathogens quickly and accurately is crucial for preventing and controlling the spread of infectious diseases. Given the similarities in symptoms and pathology, as well as the frequent co-infection of these three viruses, it is challenging to differentiate them through clinical diagnosis alone (Vlasova et al., 2017; Shu et al., 2022). Hence, there is a need for a rapid and accurate method to simultaneously distinguish PEDV, PoRV, and PDCoV.
In this study, specific primers and probes were designed for the conserved regions of the PEDV M gene, PoRV NSP3 gene, and PDCoV M gene. After optimizing multiple times, a successful multiplex real-time PCR method was developed to simultaneously detect PEDV, PoRV, and PDCoV in a single amplification reaction. The method established in this study is highly sensitive, with a detection limit of 100 copies/μL. The multiplex real-time PCR also showed good repeatability, with coefficient of variation ranging from 0.11 to 0.87% for intra-assay and 0.11–1.63% for inter-assay. Thirty clinical samples with diarrhea symptoms were used to compare the results of the commercial singleplex real-time PCR kit and our multiplex real-time PCR method for detecting PEDV, PoRV, and PDCoV. The two methods showed complete agreement, indicating that the multiplex real-time PCR assay developed in this study could effectively replace the commercial singleplex real-time PCR kit for simultaneously differentiating PEDV, PoRV, and PDCoV.
Due to its rapid, highly sensitive, and specific characteristics, the multiplex real-time PCR assay established in this study has been widely used in early detection of pathogens in clinical samples. A total of 4,800 diarrhea samples from 18 provinces of china were used to investigate the prevalence of PEDV, PoRV, and PDCoV. Out of these samples, 2,207 were negative and 2,593 were positive for single or co-infections of these viruses. Among the positive samples, PoRV had the highest infection rate at 75% (1,945/2,593), followed by PEDV at 36.98% (959/2,593) and PDCoV at 25.8% (669/2,593), confirming PoRV as the primary cause of porcine diarrhea. These findings align with previous research showing a steady increase in PoRV infection rates, with some countries reporting 100% seropositivity in adult pigs (Pettersson et al., 2019).
Previous reports have shown that co-infection of these three viruses can lead to severe symptoms (Vlasova et al., 2017; Saeng-Chuto et al., 2021). Our study found that co-infections account for 35.4% (918/2,593) of positive samples, indicating that multiple pathogen co-infections are becoming more common with the development of large-scale and intensive swine production (Figure 3A). Specifically, co-infections of PoRV and PEDV accounted for 21.9% (568/2,593) of positive samples, followed by co-infections of PoRV and PDCoV at 10.57% (274/2,593; Figure 3A). Co-infections of PEDV and PDCoV were very rare, at only 0.54% (14/2,593; Figure 3A). Notably, 2.39% (62/2,593) of positive samples were co-infected with PEDV, PoRV, and PDCoV (Figure 3A). The findings align with previous studies indicating that the co-infection of PEDV and PoRV is the primary cause of porcine diarrhea in certain Chinese provinces (Zhang et al., 2019; Shu et al., 2022; Hou et al., 2023).
There were significant differences in positivity rate for these three viruses among the three regions: Northern China, Mideastern China), and Southern China. Northern China had the highest positivity rate for the three viruses at 66.76%, followed by Mideastern China at 42.40% and Southern China at 45.5% (Figure 3B). Northern China also had a higher incidence of co-infection at 45.03% compared to Mideastern China at 22.69% and Southern China at 24% (Figure 3B). However, Mideastern and Southern China had a higher positive rate of single PoRV than Northern China (Figure 3B). These results suggest that viral diarrhea disease in China pig farms is complex, possibly due to climate differences between the north and south and rapid pig population renewal in northern China. In summary, we have successfully developed a reliable multiplex real-time PCR assay for distinguishing PEDV, PoRV, and PDCoV. This assay has demonstrated excellent specificity, sensitivity, and repeatability. It has also been effectively used to detect clinical samples, making it a valuable tool for quickly identifying these viruses. Rapid and precise diagnostics, followed by prompt quarantine and treatment, could be helpful to prevent and control the spread of infectious diseases.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
JR: Conceptualization, Funding acquisition, Writing—review & editing, Formal analysis, Methodology, Project administration, Writing—original draft. CZ: Writing—original draft, Data curation, Software. YL: Software, Writing—original draft, Resources. ML: Data curation, Writing—review & editing. JG: Data curation, Investigation, Writing—review & editing. FC: Data curation, Formal analysis, Writing—review & editing. XL: Conceptualization, Data curation, Funding acquisition, Resources, Writing—review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Natural Science Foundation of Shandong Province (Grant Nos. ZR2022MC158 and ZR2020QC177), Taishan Industry Leadership Talent Project of Shandong Province in China (Grant No. tscx202306093), and Talent Introduction Project of Dezhou University of China (2022xjrc413).
CZ, YL, and XL were employed by Shandong New Hope Liuhe Agriculture and Animal Husbandry Technology Co., Ltd. (NHLH Academy of Swine Research).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1380849/full#supplementary-material
Ben Salem, A. N., Chupin Sergei, A., Bjadovskaya Olga, P., Andreeva Olga, G., Mahjoub, A., and Prokhvatilova Larissa, B. (2010). Multiplex nested RT-PCR for the detection of porcine enteric viruses. J. Virol. Methods 165, 283–293. doi: 10.1016/j.jviromet.2010.02.010
Chepngeno, J., Takanashi, S., Diaz, A., Michael, H., Paim, F. C., Rahe, M. C., et al. (2020). Comparative sequence analysis of historic and current porcine rotavirus c strains and their pathogenesis in 3-day-old and 3-week-old piglets. Front. Microbiol. 11:780. doi: 10.3389/fmicb.2020.00780
He, W. T., Ji, X., He, W., Dellicour, S., Wang, S., Li, G., et al. (2020). Genomic epidemiology, evolution, and transmission dynamics of porcine deltacoronavirus. Mol. Biol. Evol. 37, 2641–2654. doi: 10.1093/molbev/msaa117
Hou, W., Fan, M., Zhu, Z., and Li, X. (2023). Establishment and application of a triplex Real-Time RT-PCR assay for differentiation of PEDV, PoRV, and PDCoV. Viruses 15:1238. doi: 10.3390/v15061238
Hu, Y., Xie, X., Yang, L., and Wang, A. (2021). A comprehensive view on the host factors and viral proteins associated with porcine epidemic diarrhea virus infection. Front. Microbiol. 12:762358. doi: 10.3389/fmicb.2021.762358
Huang, X., Chen, J., Yao, G., Guo, Q., Wang, J., and Liu, G. (2019). A TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses. Appl. Microbiol. Biot. 103, 4943–4952. doi: 10.1007/s00253-019-09835-7
Huang, Y., Xing, N., Wang, Z., Zhang, X., Zhao, X., Du, Q., et al. (2015). Ultrasensitive detection of RNA and DNA viruses simultaneously using duplex undp-pcr assay. PLoS ONE 10:e0141545. doi: 10.1371/journal.pone.0141545
Huang, Y. W., Dickerman, A. W., Piñeyro, P., Li, L., Fang, L., Kiehne, R., et al. (2013). Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 4, e00737–e00713. doi: 10.1128/mBio.00737-13
Li, W., Hulswit, R. J. G., Kenney, S. P., Widjaja, I., Jung, K., Alhamo, M. A., et al. (2018). Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc. Natl. Acad. Sci. U. S. A. 115, E5135–E5143. doi: 10.1073/pnas.1802879115
Li, X., Hu, Y., Liu, P., Zhu, Z., Liu, P., Chen, C., et al. (2022). Development and application of a duplex real-time PCR assay for differentiation of genotypes I and II African swine fever viruses. Transbound Emerg. Dis. 69, 2971–2979. doi: 10.1111/tbed.14459
Liu, G., Jiang, Y., Opriessnig, T., Gu, K., Zhang, H., and Yang, Z. (2019). Detection and differentiation of five diarrhea related pig viruses utilizing a multiplex PCR assay. J. Virol. Methods 263, 32–37. doi: 10.1016/j.jviromet.2018.10.009
Ma, Y., Zhang, Y., Liang, X., Lou, F., Oglesbee, M., Krakowka, S., et al. (2015). Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio 6:e00064. doi: 10.1128/mBio.00064-15
Pan, Z., Lu, J., Wang, N., He, W. T., Zhang, L., Zhao, W., et al. (2020). Development of a TaqMan-probe-based multiplex real-time PCR for the simultaneous detection of emerging and reemerging swine coronaviruses. Virulence 11, 707–718. doi: 10.1080/21505594.2020.1771980
Pettersson, E., Hestad, S., Möttus, I., Skiöldebrand, E., and Wallgren, P. (2019). Rotavirus and Cystoisospora suis in piglets during the suckling and early post weaning period, in systems with solid floors and age segregated rearing. Porcine Health. Manag. 5:7. doi: 10.1186/s40813-019-0114-0
Saeng-Chuto, K., Madapong, A., Kaeoket, K., Piñeyro, P. E., Tantituvanont, A., and Nilubol, D. (2021). Coinfection of porcine deltacoronavirus and porcine epidemic diarrhea virus increases disease severity, cell trophism and earlier upregulation of IFN-α and IL12. Sci. Rep. 11:3040. doi: 10.1038/s41598-021-82738-8
Shibata, I., Tsuda, T., Mori, M., Ono, M., Sueyoshi, M., and Uruno, K. (2000). Isolation of porcine epidemic diarrhea virus in porcine cell cultures and experimental infection of pigs of different ages. Vet. Microbiol. 72, 173–182. doi: 10.1016/S0378-1135(99)00199-6
Shu, X., Han, F., Hu, Y., Hao, C., Li, Z., Wei, Z., et al. (2022). Co-infection of porcine deltacoronavirus and porcine epidemic diarrhoea virus alters gut microbiota diversity and composition in the colon of piglets. Virus Res. 322:198954. doi: 10.1016/j.virusres.2022.198954
Si, F., Hu, X., Wang, C., Chen, B., Wang, R., Dong, S., et al. (2020). Porcine epidemic diarrhea virus (PEDV) ORF3 enhances viral proliferation by inhibiting apoptosis of infected cells. Viruses 12:214. doi: 10.3390/v12020214
Tao, R., Chang, X., Zhou, J., Zhu, X., Yang, S., Li, K., et al. (2023). Molecular epidemiological investigation of group A porcine rotavirus in East China. Front. Vet. Sci. 10:1138419. doi: 10.3389/fvets.2023.1138419
Vlasova, A. N., Amimo, J. O., and Saif, L. J. (2017). Porcine rotaviruses: epidemiology, immune responses and control strategies. Viruses 9:48. doi: 10.3390/v9030048
Wang, D., Fang, L., and Xiao, S. (2016). Porcine epidemic diarrhea in China. Virus Res. 226, 7–13. doi: 10.1016/j.virusres.2016.05.026
Wang, L., Byrum, B., and Zhang, Y. (2014). Porcine coronavirus HKU15 detected in 9 US states, 2014. Emerg. Infect. Dis. 20, 1594–1595. doi: 10.3201/eid2009.140756
Woo, P. C., Lau, S. K., Lam, C. S., Lau, C. C., Tsang, A. K., Lau, J. H., et al. (2012). Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 86, 3995–4008. doi: 10.1128/JVI.06540-11
Wu, X., Liu, Y., Gao, L., Yan, Z., Zhao, Q., Chen, F., et al. (2022). Development and application of a reverse-transcription recombinase-aided amplification assay for porcine epidemic diarrhea virus. Viruses 14:59. doi: 10.3390/v14030591
Keywords: multiplex real-time PCR, PEDV, PoRV, PDCoV, porcine viral diarrhea
Citation: Ren J, Zu C, Li Y, Li M, Gu J, Chen F and Li X (2024) Establishment and application of a TaqMan-based multiplex real-time PCR for simultaneous detection of three porcine diarrhea viruses. Front. Microbiol. 15:1380849. doi: 10.3389/fmicb.2024.1380849
Received: 02 February 2024; Accepted: 01 April 2024;
Published: 16 April 2024.
Edited by:
Jingqiang Ren, Wenzhou University, ChinaReviewed by:
Xing Liu, Nanjing Agricultural University, ChinaCopyright © 2024 Ren, Zu, Li, Li, Gu, Chen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaowen Li, bHh3ODI3MkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.