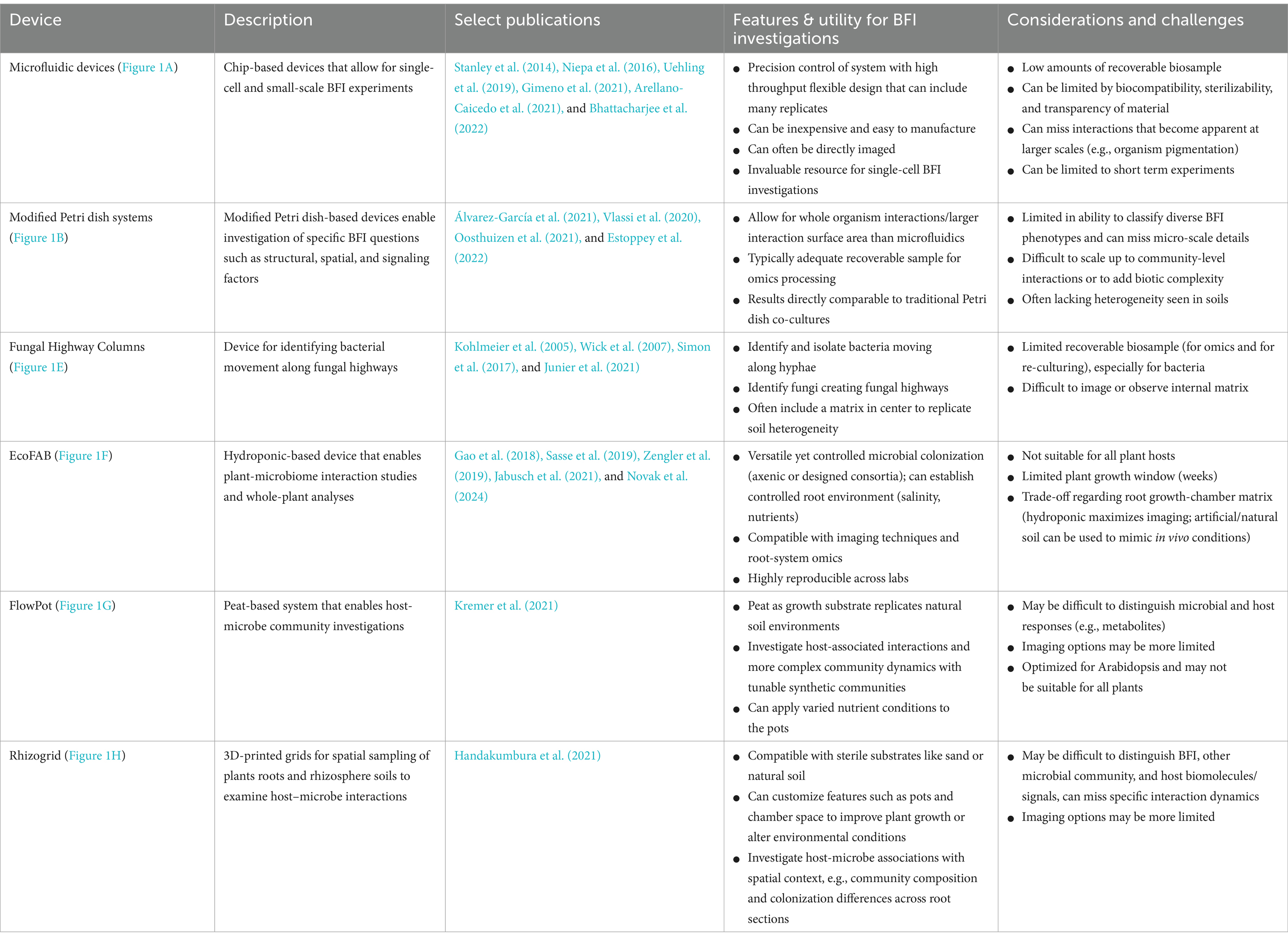
- 1Bioscience Division, Los Alamos National Laboratory, Los Alamos, NM, United States
- 2Laboratory of Microbiology, Institute of Biology, University of Neuchâtel, Neuchâtel, Switzerland
Diverse and complex microbiomes are found in virtually every environment on Earth. Bacteria and fungi often co-dominate environmental microbiomes, and there is growing recognition that bacterial-fungal interactions (BFI) have significant impacts on the functioning of their associated microbiomes, environments, and hosts. Investigating BFI in vitro remains a challenge, particularly when attempting to examine interactions at multiple scales of system complexity. Fabricated devices can provide control over both biotic composition and abiotic factors within an experiment to enable the characterization of diverse BFI phenotypes such as modulation of growth rate, production of biomolecules, and alterations to physical movements. Engineered devices ranging from microfluidic chips to simulated rhizosphere systems have been and will continue to be invaluable to BFI research, and it is anticipated that such devices will continue to be developed for diverse applications in the field. This will allow researchers to address specific questions regarding the nature of BFI and how they impact larger microbiome and environmental processes such as biogeochemical cycles, plant productivity, and overall ecosystem resilience. Devices that are currently used for experimental investigations of bacteria, fungi, and BFI are discussed herein along with some of the associated challenges and several recommendations for future device design and applications.
1 Introduction
Bacteria and fungi are often the dominant constituents in environmental microbiomes and their relationships, referred to as bacterial-fungal interactions (BFI), are increasingly recognized as significant contributors to microbiome and environmental dynamics (Frey-Klett et al., 2011; Deveau et al., 2018; Bergelson et al., 2019; Wagg et al., 2019). However, there is still much to be uncovered regarding the mechanisms underlying BFI, their phenotypic diversity, and their contributions to local habitats (Frey-Klett et al., 2011; Deveau et al., 2018; Robinson et al., 2021). Community-level approaches such as metabarcoding can be employed to identify the microbial composition of environmental microbiomes, yet confidently assessing bacterial-fungal interactions and their functional impacts from these datasets remains a significant challenge (Mandolini et al., 2021). To address this challenge, researchers must employ smaller-scale in vitro techniques, and many groups have designed and used a number of experimental devices which allow them to investigate specific questions regarding the nature of BFI across scales.
Conventional co-culture experiments conducted in Petri dishes allow for the initial identification and characterization of some forms of BFI, however, these techniques often lack environmental complexity or conversely, the granularity for looking at micro-scale or specific interaction phenotypes (e.g., investigating bacterial dispersal along fungal hyphae, or “fungal highways”). Fabricated devices designed to investigate BFI at different biological scales, from the cellular to the community level, have allowed researchers to expand upon traditional co-culture assays to isolate phenotypes of interest or study these interactions under conditions that more closely mimic the natural environments in which they occur (Uehling et al., 2019; Zengler et al., 2019; Kuhn et al., 2022). These devices have various advantages and limitations with respect to sample input requirements, production cost, device size, number of replicates, biological scale(s), ability to assess spatial dynamics, downstream analysis capabilities (e.g., ability to perform omics or re-culture), and control over abiotic and biotic parameters of the system. Devices that create simplified systems with fewer biotic and abiotic factors are often useful for elucidating the molecular mechanisms that underlie interaction phenotypes between specific bacteria and fungi. However, the simplified nature in which BFI are investigated in these devices limits their translatability to interaction dynamics in natural ecosystems. Devices that can recreate more complex environments (increased number of abiotic or biotic factors) can capture additional community dynamics which impact the mechanisms of interaction, and provide insights into the interactions in a more natural environmental context. This review examines the use of fabricated devices to study BFI across multiple scales and levels of system complexity, providing representative examples of device application from the single cell to microbiome level, and provides perspectives on the engineering of future devices to advance BFI research. Particularly, this review will focus on examples of devices for studying BFI in rhizosphere and soil systems, however several of these devices can be used for a variety of research applications. As the field of BFI research continues to expand, the development and implementation of fabricated devices will be foundational to assessing BFI applications to diverse sectors including agriculture, biotechnology, and environmental and human health.
2 Current applications of fabricated devices to study BFI across levels of system complexity
2.1 Applications of devices to investigate single cell and small-scale bacterial-fungal interactions
At the smallest scale, BFI partners are investigated using single-cell or low-biomass approaches. Microfluidic devices (Figure 1A; Table 1), which consist of microchannels on a small chip that are now often constructed using 3D printing, can be applied to small-scale BFI experiments by producing droplet microenvironments for interrogating BFI, or can be designed to investigate BFI dynamics within microscale channels and wells [Grünberger et al., 2014; Stanley et al., 2014; reviewed by Bernier et al. (2022), Masters-Clark et al. (2022), Richter et al. (2022)]. Many microfluidic devices allow for downstream screening and sorting, microscopy-based imaging, and they maintain an extremely controlled environment in terms of both biotic and abiotic variables. Traditional droplet-generating microfluidic devices have been modified to effectively sequester filamentous fungi which can then be screened for specific phenotypes (e.g., enzymatic activity) in a high throughput manner (Beneyton et al., 2016; Samlali et al., 2022). These modified droplet-generating devices can be applied to BFI studies to screen for interaction phenotypes at the cellular level. Niepa et al. (2016) used microfluidic droplets to assess impacts of small signaling molecules on BFI by capturing Candida albicans cells alone or together with Pseudomonas aeruginosa into these droplets. This study revealed that P. aeruginosa was able to impair C. albicans growth from both inside (co-captured in droplets) and outside (P. aeruginosa in solution surrounding droplets with C. albicans) the droplets, indicating that physical interactions are not required to hinder fungal growth and that an exchange of small signaling molecules is likely involved in this antagonistic interaction.
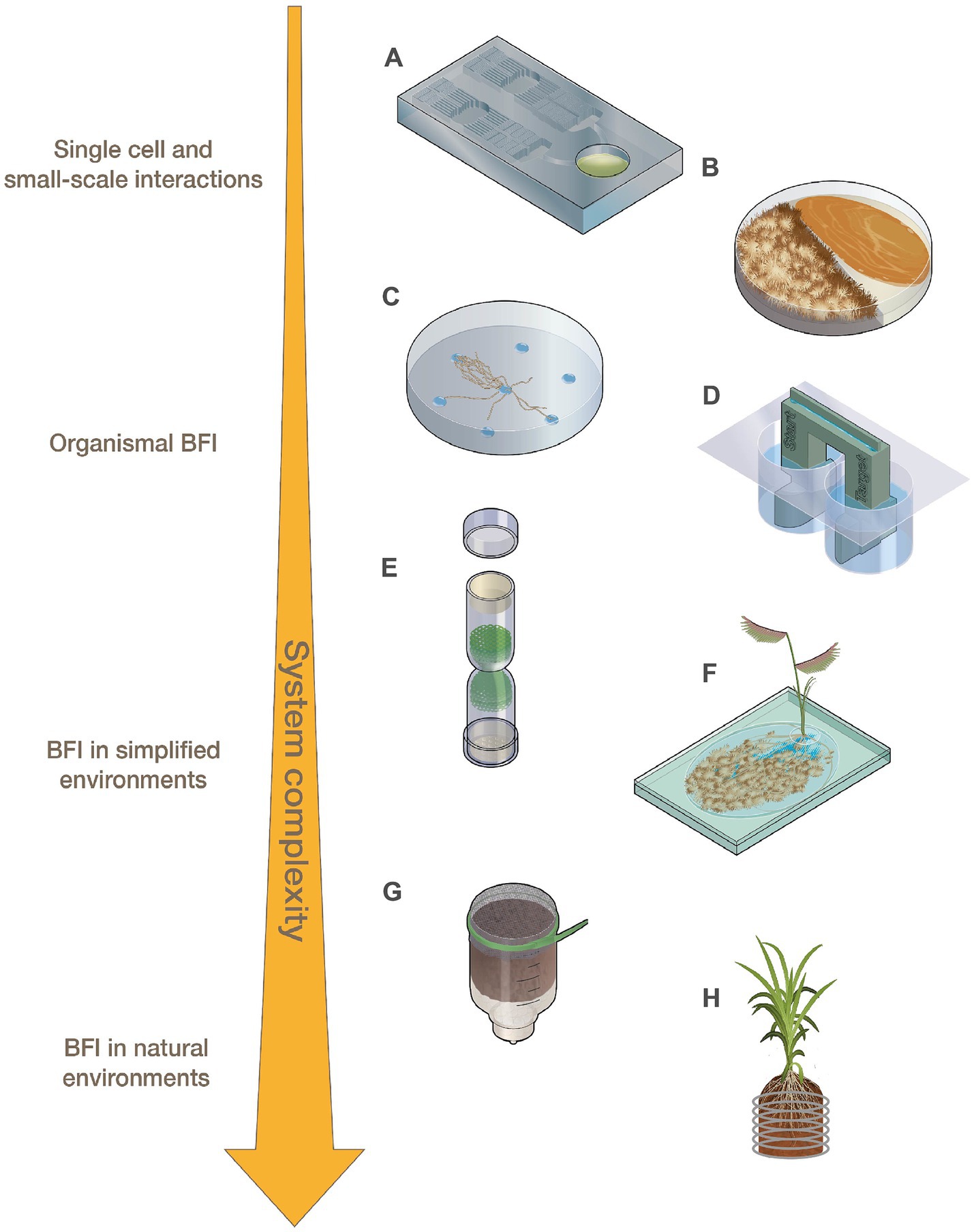
Figure 1. Representations of fabricated devices designed for use across levels of system complexity ranging from small-scale interactions to BFI in natural environments. (A) Microfluidic devices, (B) modified Petri dishes, (C) fungal drops, (D) bacterial bridge, (E) fungal highway column, (F) EcoFAB, (G) FlowPot, and (H) Rhizogrid.
To examine how BFI are altered under different nutrient conditions and elucidate the impacts of inter-microbial signals on individual microbes, microfluidic devices have been designed to allow for the addition of media containing specific nutrient profiles, microbial exudates, metabolic products, or small signaling molecules [Groisman et al., 2005; Uehling et al., 2019; reviewed by Richter et al. (2022)]. Uehling et al. (2019) developed a microfluidic device to examine how individual hyphae of Linnemannia elongata responded to Burkholderia-conditioned media, media conditioned by both the fungal and bacterial partners, and control media. The experiments revealed that the media pre-conditioned by both the fungal and bacterial partners increased fungal growth rates compared to the treatment pre-conditioned with just Burkholderia, suggesting that bidirectional BFI communication is key to the observed fungal growth rate modulation. Stanley et al. (2014) designed a microfluidic “fluid exchange” device for media and nutrient exchange during interaction experiments, and it enabled the visualization of physical interactions between Bacillus subtilis and Coprinopsis cinerea hyphae that were not detectable from Petri dish-based confrontation assays (Schmieder et al., 2019). Microfluidic devices can be constructed with a variety of channel sizes and geometries to assess how microbial interactions and growth are impacted by constraints of their physical environment (Arellano-Caicedo et al., 2021; Gimeno et al., 2021; Mafla-Endara et al., 2021; Bhattacharjee et al., 2022). Arellano-Caicedo et al. (2021) found that microfluidic channel geometry, including channel bend angle, altered hyphal branching patterns which, in turn, impacted bacterial dispersion. Microfabricated ‘soil chip’ devices placed directly into soil enabled researchers to assess how microbes colonize microhabitats, and demonstrated how the presence of fungal hyphae facilitated increased bacterial dispersal through pore spaces (Mafla-Endara et al., 2021). Devices such as this ‘soil chip’ can help to bridge the gap between laboratory in vitro experiments and in situ environmental experimentation.
2.2 Expanding on traditional co-culture assays to understand bacterial-fungal interaction mechanisms at the organismal level
Analysis of bacterial and fungal co-cultures on Petri dishes is one of the most widely utilized methods for characterizing BFI phenotypes. Although not traditionally regarded as fabricated devices, Petri dishes have been modified, for example, by including dividers that split the plates, to help researchers answer BFI-related questions (Figure 1B; Table 1; Vlassi et al., 2020; Álvarez-García et al., 2021; Oosthuizen et al., 2021; Estoppey et al., 2022). Volatile organic compounds (VOCs) are known to play a significant role in many observed BFI, and devices that enable investigations into their roles in microbial interactions are becoming more prevalent [reviewed by Schmidt et al. (2016) and Weisskopf et al. (2021)]. For example, Petri dishes have been altered to physically separate bacterial and fungal cultures to investigate VOCs by allowing the organisms to grow in proximity without physical contact, while maintaining gas exchange (Vlassi et al., 2020; Álvarez-García et al., 2021). Vlassi et al. (2020) identified VOCs generated by the bacteria, Lysobacter capsici, that inhibited the growth of known plant pathogenic fungi, Rhizoctonia solani and Sclerotinia minor, using split Petri dishes (Figure 1B). Other split culture designs allow for the exchange of both volatile and non-volatile compounds while maintaining physical separation of the partners by utilizing membranes with differential permeabilities that exclude microbes from physically interacting but permit the exchange of metabolites such as glycerol and glucose (Oosthuizen et al., 2021).
2.3 Devices for investigating hyphal transport of bacteria in simplified environments
Various devices have been developed to interrogate ‘fungal highway’ interactions where bacteria move along or are transported by fungal hyphal networks (Kohlmeier et al., 2005; Warmink and van Elsas, 2009; Simon et al., 2017; Junier et al., 2021). A novel approach to simplifying cell-to-cell observations of BFI was developed in which media droplets with different nutrient profiles are placed on a hydrophobic surface. This ‘fungal drops’ system can be used to assess differences in hyphal sensing and exploration when alone versus when paired with bacterial partners, enabling investigations to determine if bacteria selectively use hyphae as a highway to more nutrient rich areas or to specific nutrients (Figure 1C; Buffi et al., 2023).
Inverted Petri dish-based assays where solid growth media is placed above soil samples have been utilized to identify and isolate soil bacteria capable of traversing fungal hyphae that create bridges between the soil sample and the media (Furuno et al., 2012; Bravo et al., 2013). 3D-printed bacterial bridge and trail devices (Figure 1D) have also been designed to interrogate how bacteria can disperse on a physical scaffold, in a purely abiotic system, to distinguish the contributions of abiotic factors (i.e., liquid film depth and hydraulic flow) from biotic factors (i.e., nutrient exchange and signaling molecules) during hyphal transport of bacteria (Kuhn et al., 2022). Fungal highway columns (Figure 1E; Table 1) are devices that facilitate fungal growth through an inner complex matrix such that bacteria must use hyphae for movement between ends. Various designs of fungal highway columns have enabled insights into how fungal hyphae promote the dispersal and transport of bacteria in simplified communities, soil, dung, and other environmental samples (Kohlmeier et al., 2005; Wick et al., 2007; Simon et al., 2017; Junier et al., 2021).
2.4 Devices to mimic the complex natural environments of soil and rhizospheres
To assess the impact of BFI on soil and rhizosphere microbiome dynamics and plant host functioning, experiments can be performed using soil (sterilized or not) in modified microcosms or other devices (Otten et al., 2012; Timm et al., 2016; Zengler et al., 2019; Yee et al., 2021; Aufrecht et al., 2022; Zhang et al., 2023, 2024). The mineral-doped micromodel, a small-scale device designed with a surface mimicking soil texture, porosity, and mineral heterogeneity, was recently developed to understand the role of microbes in mineral weathering. Bhattacharjee et al. (2022) used this device to show that Fusarium growth is influenced by the presence of minerals and suggest that the fungus was able to release and uptake the minerals embedded on the chip through indirect weathering. This device can be used to characterize how BFI influence microbial behavior and nutrient acquisition in an environment that simulates simplified soil heterogeneity at small spatial scales.
Devices for investigating impacts of microbial interactions on plants, such as fabricated ecosystems (EcoFABs) (Figure 1F; Table 1; Zengler et al., 2019), Rhizosphere-on-a-chip devices (RhizoChips) (Aufrecht et al., 2022), FlowPots (Figure 1G; Kremer et al., 2021), and Rhizogrids (Figure 1H; Handakumbura et al., 2021), allow for plant growth under hydroponic, synthetic, or natural soil conditions, with some enabling the imaging of root systems and microbial colonization. These devices are also all well-suited for inoculation with synthetic microbial communities. EcoFAB devices (Figure 1F) consist of partitioned root and shoot chambers, which are enclosed to ensure sterile conditions are maintained. The base of an EcoFAB is a large microscope slide, which makes these devices highly compatible with imaging and microscopy systems. EcoFABs have ports which allow for the liquid growth medium to be easily replaced and to facilitate microbial inoculations after plants have established in the device. For example, Coker et al. (2022) generated model synthetic communities representative of diverse grass and crop rhizospheres and examined their compatibility with EcoFAB devices. The researchers showed that these communities were altered in the presence of a plant host (Brachypodium distachyon), and demonstrated the ability of community members to colonize roots grown in EcoFABs. RhizoChips are chip-based devices that can be customized with poly-dimethylsiloxane-molded soil structures (Aufrecht et al., 2022). The RhizoChips allow for facile root imaging similar to EcoFABs, but enable spatial sampling to detect relevant plant root exudate hotspots. Adding this environmental and spatial complexity may allow for mechanistic understandings of how natural heterogeneity in nutrient availability impacts BFIs.
FlowPots and Rhizogrids are modified mesocosms that can be inoculated with microbial communities of interest (Handakumbura et al., 2021; Kremer et al., 2021). FlowPots (Figure 1G) are peat-based pots fitted with a mesh cap, which can be flushed with nutrient media for axenic growth, soil slurries to recapitulate a native microbial community, or synthetic microbial communities (Kremer et al., 2021). These pots are grown in gas-permeable boxes to maintain sterile conditions and allow researchers to create a soil-based sterile environment to study how microbial interactions may impact the plant and vice versa under more controlled conditions. Rhizogrids (Figure 1H) are 3D-printed grids which are inserted into a mesocosm or pot (Handakumbura et al., 2021). These grids are disassembled for further experiments such as metabolomics or sequencing and enable researchers to maintain spatial mapping of the source sample based on the section of the grid. BFI partners can be investigated in these systems as individual pairs or as constituents of larger consortia to understand how BFI may impact overall microbial community dynamics or to assess how BFI impact plant hosts (Durán et al., 2018; Marín et al., 2021).
3 Discussion
Due to the high diversity and complex nature of BFI, specialized devices are often required to investigate the mechanisms underlying these interactions and their impacts on larger ecosystem processes. The challenges associated with microfluidic device design (e.g., limited environmental complexity, challenges with 3D printing) have been well documented, but many considerations are broadly applicable to other devices (Table 1; Millet et al., 2019; Zhou et al., 2019; Burmeister and Grünberger, 2020). Media type (e.g., liquid vs. solid) and composition (e.g., defined vs. undefined) used within devices can impact microbial growth, alter biomolecule expression, or affect interpretations of resulting omics data (Wang et al., 2012; Basu et al., 2015; Schmidt et al., 2016; Uehling et al., 2019; Black, 2020; Pierce et al., 2021). Devices that can contain multiple media types are therefore highly amenable to BFI investigations as bacteria and fungi can be grown on their preferred media types. While promoting growth for experimental purposes is important for design considerations, how resource availability impacts microbial function and therefore bacterial-fungal interactions has yet to be studied in-depth. Devices that allow for the manipulation of media types or nutrients will additionally enable investigations into how BFIs are impacted by resource availability.
It is known that soil structure impacts microbial community composition and function, which is not captured by most traditional co-culture assays (König et al., 2020; Hartmann and Six, 2023). Materials such as polymer-based substrates, sterilized sand or clay, or glass beads can simulate the complex physical matrix and structure of heterogeneous soil (Nguyen et al., 2005; Droce et al., 2013; Ma et al., 2019; Miebach et al., 2020; Sharma et al., 2020; Del Valle et al., 2022; Rooney et al., 2024). Selecting a device that is compatible with these materials allows for investigations of BFI in microenvironments with more natural physical and spatial characteristics, while providing the means to more easily control biotic and abiotic complexity (Aleklett et al., 2018; Mafla-Endara et al., 2021). For example, synthetic soil-like particles can be added to race tubes, which are traditionally utilized for fungal experiments tracking hyphal growth over time and assessing fungal circadian rhythms. While race tubes have not yet been used to study BFI, this represents an example of how modifications to traditional devices can facilitate enhanced BFI investigations. In general, combining the benefits of fabricated devices with these materials can make BFI investigations more relevant to natural environmental conditions (Simon et al., 2017).
Spatial and temporal dynamics can also be important aspects of experimental design when investigating BFI in fabricated devices, particularly in cases where cells proximal to the interaction may only represent a small fraction of the total biomass. For example, hyphae and cells may have differential molecular responses depending on their proximity to the bacterial partner (Stanley et al., 2014). Therefore, device designs and sampling approaches which permit spatial analyses can be helpful in assessing and normalizing impacts of molecular spatial heterogeneity. Channel sizes in smaller-scale devices may restrict biologically relevant growth, such as hyphal branching, and may quickly become saturated with fungal hyphae, thus limiting temporal studies. Devices that allow for the addition or sampling of partners at different time points may be useful to understand how age, developmental stage, or length of interaction time impact BFI. Many devices can be designed with transparent, biocompatible materials which can be imaged using common microscopy techniques, allowing for the observation of BFI phenotypes over space and time.
When harvesting biomass or biomolecules from devices, it is important to ensure that sufficient material is collected from each partner pair, as BFI experiments are often dominated by the fungal partner in terms of relative biomass. Devices or components that separate fungal and bacterial biomass (e.g., membrane-separated chambers, split Petri dishes, cellophane membranes, etc.) may be desirable for omics-based investigations as these can assist in biomass recovery and allow researchers to more readily determine the origin of signaling or other biomolecules (Guennoc et al., 2017). Microfluidic devices can limit downstream omics-based investigations due to the amount of recoverable or extractable sample material, and may require low-input processing techniques (Tayyrov et al., 2019). Methods to control or measure environmental parameters (e.g., pH, moisture content, temperature) within or around devices are invaluable for monitoring the system and maintaining experimental reproducibility as these variables can alter BFI dynamics or phenotypes (Jiao et al., 2021; Xiong et al., 2021). Implementing the ability to manipulate the environmental conditions of devices will also unlock new research areas to understand how BFIs are impacted by abiotic stress. Understanding how environmental factors influence BFIs is a critical area for understanding how microbial dynamics and their functions shift under both acute and long-term environmental change (e.g., rising temperatures, changes in precipitation). As new materials arise, manufacturing methods progress, and devices become easier and cheaper to fabricate, new biological discoveries will be made at a rapid pace (Niculescu et al., 2021). 3D printing has been popularized due to the availability and relatively low costs of printers and biocompatible materials, as well as the flexibility and customization of device design (Otten et al., 2012; Schunke et al., 2020; Kuhn et al., 2022). Open sourcing of design plans and training in 3D printing software usage help advance devices for BFI investigations by allowing the community to continuously build on past designs. When designing new devices, researchers should consider whether their device can be provided as a resource to the research community, as many devices have been successfully distributed across labs and some have even been used as teaching tools in the classroom (Gao et al., 2018; Millet et al., 2019; Sasse et al., 2019; Junier et al., 2021). Devices with large footprints or high production costs can limit adequate experimental replication or distribution to the community, but may be unavoidable in order to optimize experimental design.
It can be difficult to infer specificity, generality, and causality of functional relationships of interacting microbial partners by only utilizing conventional assays. The continual development of novel devices and techniques for investigating these interactions will lead to more effective functional investigations by optimizing experimental setups and sample harvesting in vitro. This review outlines how fabricated devices have been used to study BFI at multiple levels of system complexity, and provides considerations for the use of these devices and the development of future devices. Increased innovation and generation of novel devices will open new research avenues in the rapidly growing field of BFI research and enable exciting new discoveries.
Author contributions
JK: Conceptualization, Investigation, Supervision, Writing – original draft, Writing – review & editing. LJ: Conceptualization, Investigation, Supervision, Writing – original draft, Writing – review & editing. AR: Conceptualization, Supervision, Writing – original draft, Writing – review & editing. RL: Conceptualization, Writing – original draft, Writing – review & editing. BH: Conceptualization, Supervision, Writing – original draft, Writing – review & editing. GC: Visualization, Writing – original draft, Writing – review & editing. SB: Supervision, Writing – original draft, Writing – review & editing. PJ: Conceptualization, Supervision, Writing – original draft, Writing – review & editing. PC: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by a Science Focus Area Grant from the U.S. Department of Energy (DOE), Biological and Environmental Research (BER), Biological System Science Division (BSSD) under the grant number LANLF59T.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aleklett, K., Kiers, E. T., Ohlsson, P., Shimizu, T. S., Caldas, V. E., and Hammer, E. C. (2018). Build your own soil: exploring microfluidics to create microbial habitat structures. ISME J. 12, 312–319. doi: 10.1038/ismej.2017.184
Álvarez-García, S., Mayo-Prieto, S., Carro-Huerga, G., Rodríguez-González, Á., González-López, Ó., Gutiérrez, S., et al. (2021). Volatile organic compound chamber: a novel technology for microbiological volatile interaction assays. J. Fungi 7:248. doi: 10.3390/jof7040248
Arellano-Caicedo, C., Ohlsson, P., Bengtsson, M., Beech, J. P., and Hammer, E. C. (2021). Habitat geometry in artificial microstructure affects bacterial and fungal growth, interactions, and substrate degradation. Commun Biol 4, 1226–1211. doi: 10.1038/s42003-021-02736-4
Aufrecht, J., Khalid, M., Walton, C. L., Tate, K., Cahill, J. F., and Retterer, S. T. (2022). Hotspots of root-exuded amino acids are created within a rhizosphere-on-a-chip. Lab Chip 22, 954–963. doi: 10.1039/D1LC00705J
Basu, S., Bose, C., Ojha, N., Das, N., Das, J., Pal, M., et al. (2015). Evolution of bacterial and fungal growth media. Bioinformation 11, 182–184. doi: 10.6026/97320630011182
Beneyton, T., Wijaya, I. P. M., Postros, P., Najah, M., Leblond, P., Couvent, A., et al. (2016). High-throughput screening of filamentous fungi using nanoliter-range droplet-based microfluidics. Sci. Rep. 6:27223. doi: 10.1038/srep27223
Bergelson, J., Mittelstrass, J., and Horton, M. W. (2019). Characterizing both bacteria and fungi improves understanding of the Arabidopsis root microbiome. Sci. Rep. 9:24. doi: 10.1038/s41598-018-37208-z
Bernier, L. S., Junier, P., Stan, G.-B., and Stanley, C. E. (2022). Spores-on-a-chip: new frontiers for spore research. Trends Microbiol. 30, 515–518. doi: 10.1016/j.tim.2022.03.003
Bhattacharjee, A., Qafoku, O., Richardson, J. A., Anderson, L. N., Schwarz, K., Bramer, L. M., et al. (2022). A mineral-doped micromodel platform demonstrates fungal bridging of carbon hot spots and hyphal transport of mineral-derived nutrients. mSystems 7, e00913–e00922. doi: 10.1128/msystems.00913-22
Black, W. D. (2020). A comparison of several media types and basic techniques used to assess outdoor airborne fungi in Melbourne, Australia. PLoS One 15:e0238901. doi: 10.1371/journal.pone.0238901
Bravo, D., Cailleau, G., Bindschedler, S., Simon, A., Job, D., Verrecchia, E., et al. (2013). Isolation of oxalotrophic bacteria able to disperse on fungal mycelium. FEMS Microbiol. Lett. 348, 157–166. doi: 10.1111/1574-6968.12287
Buffi, M., Cailleau, G., Kuhn, T., Li Richter, X.-Y., Stanley, C. E., Wick, L. Y., et al. (2023). Fungal drops: a novel approach for macro-and microscopic analyses of fungal mycelial growth. microLife 4:uqad042. doi: 10.1093/femsml/uqad042
Burmeister, A., and Grünberger, A. (2020). Microfluidic cultivation and analysis tools for interaction studies of microbial co-cultures. Curr. Opin. Biotechnol. 62, 106–115. doi: 10.1016/j.copbio.2019.09.001
Coker, J., Zhalnina, K., Marotz, C., Thiruppathy, D., Tjuanta, M., D’Elia, G., et al. (2022). A reproducible and tunable synthetic soil microbial community provides new insights into microbial ecology. mSystems 7, e00951–e00922. doi: 10.1128/msystems.00951-22
Del Valle, I., Gao, X., Ghezzehei, T. A., Silberg, J. J., and Masiello, C. A. (2022). Artificial soils reveal individual factor controls on microbial processes. mSystems 7:e0030122. doi: 10.1128/msystems.00301-22
Deveau, A., Bonito, G., Uehling, J., Paoletti, M., Becker, M., Bindschedler, S., et al. (2018). Bacterial-fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol. Rev. 42, 335–352. doi: 10.1093/femsre/fuy008
Droce, A., Sørensen, J. L., Giese, H., and Sondergaard, T. E. (2013). Glass bead cultivation of fungi: Combining the best of liquid and agar media. J. Microbiol. Methods 94, 343–346. doi: 10.1016/j.mimet.2013.07.005
Durán, P., Thiergart, T., Garrido-Oter, R., Agler, M., Kemen, E., Schulze-Lefert, P., et al. (2018). Microbial interkingdom interactions in roots promote arabidopsis survival. Cell 175, 973–983.e14. doi: 10.1016/j.cell.2018.10.020
Estoppey, A., Weisskopf, L., Di Francesco, E., Vallat-Michel, A., Bindschedler, S., Chain, P. S., et al. (2022). Improved methods to assess the effect of bacteria on germination of fungal spores. FEMS Microbiol. Lett. 369:fnac034. doi: 10.1093/femsle/fnac034
Frey-Klett, P., Burlinson, P., Deveau, A., Barret, M., Tarkka, M., and Sarniguet, A. (2011). Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol. Mol. Biol. Rev. 75, 583–609. doi: 10.1128/MMBR.00020-11
Furuno, S., Remer, R., Chatzinotas, A., Harms, H., and Wick, L. Y. (2012). Use of mycelia as paths for the isolation of contaminant-degrading bacteria from soil. Microb. Biotechnol. 5, 142–148. doi: 10.1111/j.1751-7915.2011.00309.x
Gao, J., Sasse, J., Lewald, K. M., Zhalnina, K., Cornmesser, L. T., Duncombe, T. A., et al. (2018). Ecosystem Fabrication (EcoFAB) protocols for the construction of laboratory ecosystems designed to study plant-microbe interactions. JoVE :e57170. doi: 10.3791/57170
Gimeno, A., Stanley, C. E., Ngamenie, Z., Hsung, M.-H., Walder, F., Schmieder, S. S., et al. (2021). A versatile microfluidic platform measures hyphal interactions between Fusarium graminearum and Clonostachys rosea in real-time. Commun. Biol. 4, 262–210. doi: 10.1038/s42003-021-01767-1
Groisman, A., Lobo, C., Cho, H., Campbell, J. K., Dufour, Y. S., Stevens, A. M., et al. (2005). A microfluidic chemostat for experiments with bacterial and yeast cells. Nat. Methods 2, 685–689. doi: 10.1038/nmeth784
Grünberger, A., Wiechert, W., and Kohlheyer, D. (2014). Single-cell microfluidics: opportunity for bioprocess development. Curr. Opin. Biotechnol. 29, 15–23. doi: 10.1016/j.copbio.2014.02.008
Guennoc, C. M., Rose, C., Guinnet, F., Miquel, I., Labbé, J., and Deveau, A. (2017). A new method for qualitative multi-scale analysis of bacterial biofilms on filamentous fungal colonies using confocal and electron microscopy. JoVE :e54771. doi: 10.3791/54771
Handakumbura, P. P., Rivas Ubach, A., and Battu, A. K. (2021). Visualizing the Hidden Half: Plant-Microbe Interactions in the Rhizosphere. mSystems 6:765. doi: 10.1128/msystems.00765-21
Hartmann, M., and Six, J. (2023). Soil structure and microbiome functions in agroecosystems. Nat. Rev. Earth Environ. 4, 4–18. doi: 10.1038/s43017-022-00366-w
Jabusch, L. K., Kim, P. W., Chiniquy, D., Zhao, Z., Wang, B., Bowen, B., et al. (2021). Microfabrication of a Chamber for High-Resolution, In Situ Imaging of the Whole Root for Plant-Microbe Interactions. Int. J. Mol. Sci. 22:7880. doi: 10.3390/ijms22157880
Jiao, P., Li, Z., Yang, L., He, J., Chang, X., Xiao, H., et al. (2021). Bacteria are more sensitive than fungi to moisture in eroded soil by natural grass vegetation restoration on the Loess Plateau. Sci. Total Environ. 756:143899. doi: 10.1016/j.scitotenv.2020.143899
Junier, P., Cailleau, G., Palmieri, I., Vallotton, C., Trautschold, O. C., Junier, T., et al. (2021). Democratization of fungal highway columns as a tool to investigate bacteria associated with soil fungi. FEMS Microbiol. Ecol. 97:fiab003. doi: 10.1093/femsec/fiab003
Kohlmeier, S., Smits, T. H. M., Ford, R. M., Keel, C., Harms, H., and Wick, L. Y. (2005). Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ. Sci. Technol. 39, 4640–4646. doi: 10.1021/es047979z
König, S., Vogel, H.-J., Harms, H., and Worrich, A. (2020). Physical, chemical and biological effects on soil bacterial dynamics in microscale models. Front. Ecol. Evol. 8:53. doi: 10.3389/fevo.2020.00053
Kremer, J. M., Sohrabi, R., Paasch, B. C., Rhodes, D., Thireault, C., Schulze-Lefert, P., et al. (2021). Peat-based gnotobiotic plant growth systems for Arabidopsis microbiome research. Nat. Protoc. 16, 2450–2470. doi: 10.1038/s41596-021-00504-6
Kuhn, T., Buffi, M., Bindschedler, S., Chain, P. S., Gonzalez, D., Stanley, C. E., et al. (2022). Design and construction of 3D printed devices to investigate active and passive bacterial dispersal on hydrated surfaces. BMC Biol. 20:203. doi: 10.1186/s12915-022-01406-z
Ma, L., Shi, Y., Siemianowski, O., Yuan, B., Egner, T. K., Mirnezami, S. V., et al. (2019). Hydrogel-based transparent soils for root phenotyping in vivo. Proc. Natl. Acad. Sci. 116, 11063–11068. doi: 10.1073/pnas.1820334116
Mafla-Endara, P. M., Arellano-Caicedo, C., Aleklett, K., Pucetaite, M., Ohlsson, P., and Hammer, E. C. (2021). Microfluidic chips provide visual access to in situ soil ecology. Commun. Biol. 4, 889–812. doi: 10.1038/s42003-021-02379-5
Mandolini, E., Probst, M., and Peintner, U. (2021). Methods for studying bacterial–fungal interactions in the microenvironments of soil. Appl. Sci. 11:9182. doi: 10.3390/app11199182
Marín, O., González, B., and Poupin, M. J. (2021). From microbial dynamics to functionality in the rhizosphere: a systematic review of the opportunities with synthetic microbial communities. Front. Plant Sci. 12:650609. doi: 10.3389/fpls.2021.650609
Masters-Clark, E., Clark, A. J., and Stanley, C. E. (2022). Microfluidic tools for probing fungal-microbial interactions at the cellular level. JoVE :e63917. doi: 10.3791/63917
Miebach, M., Schlechter, R. O., Clemens, J., Jameson, P. E., and Remus-Emsermann, M. N. P. (2020). Litterbox-A gnotobiotic Zeolite-clay system to investigate arabidopsis-microbe interactions. Microorganisms 8:464. doi: 10.3390/microorganisms8040464
Millet, L. J., Aufrecht, J., Labbé, J., Uehling, J., Vilgalys, R., Estes, M. L., et al. (2019). Increasing access to microfluidics for studying fungi and other branched biological structures. Fungal Biol. Biotechnol 6, 1–14. doi: 10.1186/s40694-019-0071-z
Nguyen, L. D., Kalachová, L., Novotná, J., Holub, M., Kofroňová, O., Benada, O., et al. (2005). Cultivation system using glass beads immersed in liquid medium facilitates studies of streptomyces differentiation. Appl. Environ. Microbiol. 71, 2848–2852. doi: 10.1128/AEM.71.6.2848-2852.2005
Niculescu, A.-G., Chircov, C., Bîrcă, A. C., and Grumezescu, A. M. (2021). Fabrication and applications of microfluidic devices: a review. Int. J. Mol. Sci. 22:2011. doi: 10.3390/ijms22042011
Niepa, T. H. R., Hou, L., Jiang, H., Goulian, M., Koo, H., Stebe, K. J., et al. (2016). Microbial nanoculture as an artificial microniche. Sci. Rep. 6:30578. doi: 10.1038/srep30578
Novak, V., Andeer, P. F., Bowen, B. P., Ding, Y., Zhalnina, K., Hofmockel, K. S., et al. (2024). Reproducible growth of Brachypodium in EcoFAB 2.0 reveals that nitrogen form and starvation modulate root exudation. Sci. Adv. 10:eadg7888. doi: 10.1126/sciadv.adg7888
Oosthuizen, J. R., Naidoo-Blassoples, R. K., Rossouw, D., Pott, R., and Bauer, F. F. (2021). A Multi–membrane system to study the effects of physical and metabolic interactions in microbial co-cultures and consortia. Fermentation 7:206. doi: 10.3390/fermentation7040206
Otten, W., Pajor, R., Schmidt, S., Baveye, P. C., Hague, R., and Falconer, R. E. (2012). Combining X-ray CT and 3D printing technology to produce microcosms with replicable, complex pore geometries. Soil Biol. Biochem. 51, 53–55. doi: 10.1016/j.soilbio.2012.04.008
Pierce, E. C., Morin, M., Little, J. C., Liu, R. B., Tannous, J., Keller, N. P., et al. (2021). Bacterial–fungal interactions revealed by genome-wide analysis of bacterial mutant fitness. Nat. Microbiol. 6, 87–102. doi: 10.1038/s41564-020-00800-z
Richter, F., Bindschedler, S., Calonne-Salmon, M., Declerck, S., Junier, P., and Stanley, C. E. (2022). Fungi-on-a-Chip: microfluidic platforms for single-cell studies on fungi. FEMS Microbiol. Rev. 46:fuac039. doi: 10.1093/femsre/fuac039
Robinson, A. J., House, G. L., Morales, D. P., Kelliher, J. M., Gallegos-Graves, L. V., LeBrun, E. S., et al. (2021). Widespread bacterial diversity within the bacteriome of fungi. Commun Biol 4, 1168–1113. doi: 10.1038/s42003-021-02693-y
Rooney, L. M., Dupuy, L. X., Hoskisson, P. A., and McConnell, G. (2024). Construction and characterisation of a structured, tuneable, and transparent 3D culture platform for soil bacteria. Microbiology 170:001429. doi: 10.1099/mic.0.001429
Samlali, K., Alves, C. L., Jezernik, M., and Shih, S. C. C. (2022). Droplet digital microfluidic system for screening filamentous fungi based on enzymatic activity. Microsyst. Nanoeng. 8, 123–116. doi: 10.1038/s41378-022-00456-1
Sasse, J., Kant, J., Cole, B. J., Klein, A. P., Arsova, B., Schlaepfer, P., et al. (2019). Multilab EcoFAB study shows highly reproducible physiology and depletion of soil metabolites by a model grass. New Phytol. 222, 1149–1160. doi: 10.1111/nph.15662
Schmidt, R., Etalo, D. W., de Jager, V., Gerards, S., Zweers, H., de Boer, W., et al. (2016). Microbial small talk: volatiles in fungal-bacterial interactions. Front. Microbiol. 6:1495. doi: 10.3389/fmicb.2015.01495
Schmieder, S. S., Stanley, C. E., Rzepiela, A., van Swaay, D., Sabotič, J., Nørrelykke, S. F., et al. (2019). Bidirectional propagation of signals and nutrients in fungal networks via specialized hyphae. Curr. Biol. 29, 217–228.e4. doi: 10.1016/j.cub.2018.11.058
Schunke, C., Pöggeler, S., and Nordzieke, D. E. (2020). A 3D printed device for easy and reliable quantification of fungal chemotropic growth. Front. Microbiol. 11:584525. doi: 10.3389/fmicb.2020.584525
Sharma, K., Palatinszky, M., Nikolov, G., Berry, D., and Shank, E. A. (2020). Transparent soil microcosms for live-cell imaging and non-destructive stable isotope probing of soil microorganisms. eLife 9:e56275. doi: 10.7554/eLife.56275
Simon, A., Hervé, V., Al-Dourobi, A., Verrecchia, E., and Junier, P. (2017). An in situ inventory of fungi and their associated migrating bacteria in forest soils using fungal highway columns. FEMS Microbiol. Ecol. 93:fiw217. doi: 10.1093/femsec/fiw217
Stanley, C. E., Stöckli, M., van Swaay, D., Sabotič, J., Kallio, P. T., Künzler, M., et al. (2014). Probing bacterial–fungal interactions at the single cell level. Int Bio (Cam) 6, 935–945. doi: 10.1039/c4ib00154k
Tayyrov, A., Stanley, C. E., Azevedo, S., and Künzler, M. (2019). Combining microfluidics and RNA-sequencing to assess the inducible defensome of a mushroom against nematodes. BMC Genomics 20:243. doi: 10.1186/s12864-019-5607-3
Timm, C. M., Pelletier, D. A., Jawdy, S. S., Gunter, L. E., Henning, J. A., Engle, N., et al. (2016). Two poplar-associated bacterial isolates induce additive favorable responses in a constructed plant-microbiome system. Front. Plant Sci. 7:497. doi: 10.3389/fpls.2016.00497
Uehling, J. K., Entler, M. R., Meredith, H. R., Millet, L. J., Timm, C. M., Aufrecht, J. A., et al. (2019). Microfluidics and metabolomics reveal symbiotic bacterial-fungal interactions between mortierella elongata and burkholderia include metabolite exchange. Front. Microbiol. 10:2163. doi: 10.3389/fmicb.2019.02163
Vlassi, A., Nesler, A., Perazzolli, M., Lazazzara, V., Büschl, C., Parich, A., et al. (2020). Volatile Organic Compounds From Lysobacter capsici AZ78 as Potential Candidates for Biological Control of Soilborne Plant Pathogens. Front. Microbiol. 11:1748. doi: 10.3389/fmicb.2020.01748
Wagg, C., Schlaeppi, K., Banerjee, S., Kuramae, E. E., and van der Heijden, M. G. A. (2019). Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 10:4841. doi: 10.1038/s41467-019-12798-y
Wang, Z., Lehr, N., Trail, F., and Townsend, J. P. (2012). Differential impact of nutrition on developmental and metabolic gene expression during fruiting body development in Neurospora crassa. Fungal Genet. Biol. 49, 405–413. doi: 10.1016/j.fgb.2012.03.004
Warmink, J. A., and van Elsas, J. D. (2009). Migratory response of soil bacteria to Lyophyllum sp. Strain Karsten in soil microcosms. Appl. Environ. Microbiol. 75, 2820–2830. doi: 10.1128/AEM.02110-08
Weisskopf, L., Schulz, S., and Garbeva, P. (2021). Microbial volatile organic compounds in intra-kingdom and inter-kingdom interactions. Nat. Rev. Microbiol. 19, 391–404. doi: 10.1038/s41579-020-00508-1
Wick, L. Y., Remer, R., Würz, B., Reichenbach, J., Braun, S., Schäfer, F., et al. (2007). Effect of fungal hyphae on the access of bacteria to phenanthrene in soil. Environ. Sci. Technol. 41, 500–505. doi: 10.1021/es061407s
Xiong, B.-J., Dusny, C., Wang, L., Appel, J., Lindstaedt, K., Schlosser, D., et al. (2021). Illuminate the hidden: in vivo mapping of microscale pH in the mycosphere using a novel whole-cell biosensor. ISME Commun. 1:75. doi: 10.1038/s43705-021-00075-3
Yee, M. O., Kim, P., Li, Y., Singh, A. K., Northen, T. R., and Chakraborty, R. (2021). Specialized plant growth chamber designs to study complex rhizosphere interactions. Front. Microbiol. 12:625752. doi: 10.3389/fmicb.2021.625752
Zengler, K., Hofmockel, K., Baliga, N. S., Behie, S. W., Bernstein, H. C., Brown, J. B., et al. (2019). EcoFABs: advancing microbiome science through standardized fabricated ecosystems. Nat. Methods 16, 567–571. doi: 10.1038/s41592-019-0465-0
Zhang, C., de Pasquale, S., Hartman, K., Stanley, C. E., Berendsen, R. L., and van der Heijden, M. G. A. (2023). The microbial contribution to litter decomposition and plant growth. Environ. Microbiol. Rep. 16:e13205. doi: 10.1111/1758-2229.13205
Zhang, C., van der Heijden, M. G. A., Dodds, B. K., Nguyen, T. B., Spooren, J., Valzano-Held, A., et al. (2024). A tripartite bacterial-fungal-plant symbiosis in the mycorrhiza-shaped microbiome drives plant growth and mycorrhization. Microbiome 12:13. doi: 10.1186/s40168-023-01726-4
Keywords: bacterial-fungal interactions, BFI, microbial interactions, fabricated devices, microbiology, environmental microbiome
Citation: Kelliher JM, Johnson LYD, Robinson AJ, Longley R, Hanson BT, Cailleau G, Bindschedler S, Junier P and Chain PSG (2024) Fabricated devices for performing bacterial-fungal interaction experiments across scales. Front. Microbiol. 15:1380199. doi: 10.3389/fmicb.2024.1380199
Edited by:
Ales Lapanje, Institut Jožef Stefan (IJS), SloveniaReviewed by:
Amber Bible, Oak Ridge National Laboratory (DOE), United StatesYuguo Yang, Berkeley Lab (DOE), United States
Copyright © 2024 Kelliher, Johnson, Robinson, Longley, Hanson, Cailleau, Bindschedler, Junier and Chain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrick S. G. Chain, cGNoYWluQGxhbmwuZ292; Julia M. Kelliher, amtlbGxpaGVyQGxhbmwuZ292
†These authors have contributed equally to this work and share first authorship
 Julia M. Kelliher
Julia M. Kelliher Leah Y. D. Johnson
Leah Y. D. Johnson Aaron J. Robinson
Aaron J. Robinson Reid Longley
Reid Longley Buck T. Hanson
Buck T. Hanson Guillaume Cailleau
Guillaume Cailleau Saskia Bindschedler
Saskia Bindschedler Pilar Junier
Pilar Junier Patrick S. G. Chain
Patrick S. G. Chain