- 1Key Laboratory of Endemic and Ethnic Diseases, Guizhou Medical University, Ministry of Education, Guiyang, China
- 2Key Laboratory of Medical Molecular Biology, Guizhou Medical University, Guiyang, Guizhou, China
- 3Guizhou Provincial Center for Clinical Laboratory, Guiyang, China
- 4Key Laboratory of Medical Insects, Guizhou Medical University, Guiyang, Guizhou, China
- 5Institute of Plant Protection, Guizhou Academy of Agricultural Sciences, Guiyang, China
- 6School of Environmental Sciences, University of Guelph, Guelph, ON, Canada
Acinetobacter baumannii is an opportunistic pathogen that easily resists currently available antibiotics. Phages are considered alternative therapeutic agents to conventional antibiotics for the treatment of multidrug-resistant bacteria. We isolated an Acinetobacter virus Abgy202141 from underground sewage in a residential area of Guiyang City in China. Transmission electron microscopy (TEM) analysis showed that Acinetobacter virus Abgy202141 has an icosahedral head attached to a tail. This phage infects A. baumannii strain GY-4, and was found to have a short latent period of 5 min and with a burst size of 189 particles per infected host cell. Additionally, Acinetobacter virus Abgy202141 remained stable at different concentrations of chloroform and varying pH levels and temperatures. Based on SDS-PAGE analysis, it contained 14 proteins with molecular weights ranging from 12 to 125 kDa. The double-strand (ds) DNA genome of Acinetobacter virus Abgy202141 consisted of 41,242 bp with a GC content of 39.4%. It contained 50 open reading frames (ORFs), of which 29 ORFs had identified functions, but no virulence-related genes, antibiotic-resistance genes, or tRNAs were found. Phylogenetic analysis indicated that Acinetobacter virus Abgy202141 was a new phage in the Friunavirus genus. Acinetobacter virus Abgy202141 also showed the ability to prevent A. baumannii infections in the Galleria mellonella in vivo model.
1 Introduction
Acinetobacter baumannii is a gram-negative bacterium that is commonly found in hospital wound infections (Micelli et al., 2023). Its ability to produce biofilms and broad resistance to antibiotics make it one of the most successful opportunistic pathogens in the hospital environment (Altnok et al., 2020; Anane et al., 2020). As reported, over 80% of A. baumannii isolates from intensive care units and other wards were resistant to most antibiotics in a general public hospital of Greece (Feretzakis et al., 2019). An infection of A. baumannii may lead to bacteremia, ventilator-associated pneumonia, and urinary tract infections (Altnok et al., 2020).
Numerous antibiotic resistance processes have been found with A. baumannii, including enzyme inactivation, target modification, active efflux, and reduced chemical uptake (Rajkumari and Siddhardha, 2020). New or alternative management methods are needed. Phages are abundant in the environment, and they can specifically attack pathogenic bacteria (Tu et al., 2023). Phages may be found that can attack Multidrug-Resistant (MDR) strains of A. baumannii which may be a potential solution for this pathogen (Tu et al., 2023).
In this study, we isolated and identified a new Acinetobacter virus Abgy202141 infecting A. baumannii. The genome of the phage was sequenced and its characteristics were analyzed. The therapeutic use of phage combined with antibiotics was evaluated in the G. mellonella model. The results provide more options to treat with antibiotic resistance in A. baumannii.
2 Materials and methods
2.1 Bacterial strains and culture conditions
A. baumannii strain GY-4 was isolated from samples from infected patients at Guizhou Medical University Affiliated Hospital, Guiyang, Guizhou. The strain is resistant to beta-lactamase antibiotics and carries the TEM beta-lactamase (blaTEM) gene (Chen et al., 2016). The strain was grown in Luria-Bertani (LB) liquid medium (tryptone 10 g/L, yeast extract 5 g/L, and NaCl 10 g/L) or plated onto solid LB medium containing 1.5% (w/v) agar and cultured at 37°C (Wu et al., 2018).
2.2 Phage isolation and purification
Isolation and purification of the Acinetobacter virus Abgy202141 were performed as described previously with slight modifications (Zhang et al., 2017; Yang et al., 2019). A. baumannii strain GY-4 was used as a host for the isolation of phages from underground sewage, which had been treated by means of several steps of filtration by the wastewater treatment plant of Guiyang City. However, the treatment of underground sewage, i.e. the baiting process, was performed in a biosafety cabinet under Biosafety Level 2 (BSL-2) conditions, and infected hosts were retrieved under those conditions. Approximate 250 mL sewage was treated with CaCl2 at a final concentration of 1 mmol/L and incubated for 10 min at room temperature. The treated sewage was centrifuged at 8000 rpm at 4°C for 10 min and passed through 0.22 μm pore-sized filters to obtain the supernatant. And then, 50 mL of LB and 1 mL of GY-4 culture at the exponential phase were added to the supernatant. Then the mixture was incubated overnight at 37°C with shaking at 200 rpm. The treated mixture was centrifuged at 8000 rpm at 4°C for 10 min and passed through 0.22 μm pore-sized filters to obtain the supernatant. The supernatant (0.1 mL) was mixed with exponentially growing bacteria (0.1 mL), and incubated at room temperature for 15 min. The mixture was added to 5 mL of LB soft agar overlay (0.75% agar), mixed briefly, and spread over an LB plate. Plates were incubated at 37°C overnight to obtain the single phage plaques. The phage was purified at least three times with the single phage plaque technique. The last supernatant from the culture was filtered through 0.22 μm pore-sized filters and stored at −80°C.
2.3 The double-agar plaque assay
The double-agar plaque assay was performed as described previously with slight modifications (Wu et al., 2018; Hao et al., 2023). Single plaque was selected from a double-layer plate and added to exponentially growing bacteria (10 mL). Then the mixture was incubated at 37°C with shaking at 200 rpm until the solution was clear. The treated mixture was centrifuged at 8000 rpm at 4°C for 10 min and passed through 0.22 μm pore-sized filters to obtain the phage fluid. The phage fluid was diluted by 10 fold gradient. The phage fluid of appropriate dilution (0.1 mL) was mixed with exponentially growing bacteria (0.1 mL) and incubated at room temperature for 15 min. The mixture was added to 5 mL of LB soft agar overlay (0.75% agar), mixed briefly, and spread over an LB plate. Plates were incubated at 37°C overnight to obtain the single phage plaque. Phage titer (PFU/mL) was calculated as the number of plaques × dilution factor × 10.
2.4 Transmission electron microscopy
The morphological characteristics of Acinetobacter virus Abgy202141 were examined as described previously with slight modifications (Yuan et al., 2021). The purified phage suspension (about 1012 PFU/mL) was stained with 2% (w/v) uranyl acetate. The morphological characteristics of the phage were observed using a transmission electron microscope (TEM, Hitachi H-7650, Tokyo, Japan) at 80 kV.
2.5 Multiplicity of infection
The multiplicity of infection (MOI) is the proportion of virus particles to host cells (Wei et al., 2023). A phage infection curve was assayed using the method described by Wu et al. (2018). The host cells of A. baumannii strain GY-4 were mixed with Acinetobacter virus Abgy202141 at various MOIs (0.00001, 0.0001, 0.001, 0.01, 0.1, and 1) incubated at 37°C, and shaking with 150 rpm for up to 6 h. The PBS buffer was used for the control. OD600 levels were measured at 30-minute intervals for 6 h.
2.6 One-step growth curve
The one-step growth curve of Acinetobacter virus Abgy202141 was evaluated as described previously with slight modifications (Pajunen et al., 2000; Wu et al., 2018). The host cells of A. baumannii strain GY-4 were infected with Acinetobacter virus Abgy202141 at the MOI of 1, and then incubated at 37°C with 150 rpm; and then the samples were collected at intervals (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 30, 40, 50, 60, 90, 120, 180, and 240 min). The double-agar plaque assay was used for phage titer after each interval of incubation to estimate titer.
2.7 Phage stability
The heat stability of Acinetobacter virus Abgy202141 was tested at different temperatures (4, 37, 45, 55, 65, or 75°C) for 1 h (Kropinski et al., 2009). The pH stability of Acinetobacter virus Abgy202141 was tested at different pH values (pH 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, and 13) for 1 h at 37°C (Cha et al., 2018; Wu et al., 2018). The double-agar plaque assay was used for each sample to estimate titer.
2.8 Chloroform sensitivity
The chloroform tolerance of phages is a crucial reference to determine the presence or absence of lipid components in the capsids or tail of phages (Wei et al., 2023). So the chloroform tolerance of the phage was assessed as described previously with slight modifications (Wei et al., 2023). Acinetobacter virus Abgy202141 preparation (100 μL) was mixed with 900 μL of chloroform at various concentrations (0, 1%, 3%, and 5%), and the double-agar plaque assay was used to determine the resulting phage titer.
2.9 DNA extraction, sequencing and genomic analysis
The phage DNA was extracted using the phenol-chloroform technique (Pickard, 2009). The extracted DNA was sent to Shenggong Biotechnology Co., Ltd (Shanghai) for sequencing and assembly. Briefly, a DNA library was obtained using the Illumina TruSeq™ Nano DNA Sample Prep Kit instructions. Sequencing was done on the Illumina NovaSeq sequencing platform with paired-end 150 bp reads. Low-quality reads were filtered out by Trimmomatic v0.36 (Q-value < 20, 98.51%). A5-MiSeq v20160825 and SPAdes v3.12.0 were used to assemble sequencing data to contigs and scaffolds. MUMmer v3.1 and Pilon v1.18 were used to fill the remaining inner local gaps and fix the single-base polymorphism for the final assembly.
ORFs of Acinetobacter virus Abgy202141 were found using RAST server (http://rast.nmpdr.org/rast.cgi). BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&BLAST_SPEC=&LINK_LOC=blasttab&LAST_PAGE=blastn) was used to assess the similarity of phage genomes to data on GenBank. BLASTp was used to evaluate similarity to known proteins and obtain potential functions. Putative tRNA sequences were revealed using tRNAscan-SE (Chan and Lowe, 2019). The CDD database was used to search for conserved domains in phage sequences (Lu et al., 2020). The online databases VFDB (https://cge.cbs.dtu.dk/services/VirulenceFinder/) and CRDB (https://cge.cbs.dtu.dk/services/ResFinder/) were used to find potential virulence factors and antibiotic resistance genes (Alcock et al., 2020; Liu et al., 2022). The complete genomic map and GC offset of Acinetobacter virus Abgy202141 were created using CGview Server (http://stothard.afns.ualberta.ca/cgview_server/). The complete genome alignment map of Acinetobacter virus Abgy202141 was created using EasyFigure (2.2.5) (Sullivan et al., 2011). The phylogenetic tree was constructed by amino acid sequences of RNA polymerase with the Maximum Likelihood method (Tamura et al., 2011). And the optimal tree was statistically assessed with a bootstrap of 1,000-replicates.
2.10 SDS-PAGE analysis
Sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) analysis of Acinetobacter virus Abgy202141 was performed as described previously with slight modifications (Yang et al., 2010). The purified particle suspension was loaded on a 12% SDS-PAGE gel. The protein bands were visible by staining the gel with Coomassie Blue Fast Staining and No-decoloring Solution (Epizyme, Shanghai, China).
2.11 In vivo synergy in the G. mellonella model
In order to remove endotoxins of A. baumannii, the phage suspension was prepared as described previously with slight modifications (Hietala et al., 2019). Briefly, 50 ml of Abgy2021-4-1 lysate was treated with 1 M NaCl for 1 h on ice, and then the solution was passed through 0.22 μm pore-sized filters. PEG 8000 was dissolved in the supernatant at a final concentration of 10% (m/v) and the solution was incubated at 4°C for 24 h. The treated solution was centrifuged at 4°C at 8000 rpm for 15 min and the pellet was dissolved in 1 ml of PBS buffer. The solution was extracted with chloroform (1 mL) twice, and the mixed solution was centrifuged at 4°C at 8000 rpm for 20 min. Finally, the phage suspension was obtained using 0.22 μm pore-sized filters.
The injections were performed as described previously (Grygorcewicz et al., 2020). G. mellonella larvae (~300 mg with cream color) were injected with 10 μL bacterial cells (A. baumannii) of different concentrations (103, 104, 105, 106 CFU/larva). Twenty minutes after bacterial cell injection, 10 μL purified phage particle suspensions with different MOIs (MOI = 50, MOI = 10, MOI = 1, and MOI = 0.1) were injected into the infected larvae. A. baumannii strain GY-4 is resistant to ampicillin (AMP), but sensitive to imipenem (IPM). Therefore, IPM injection was used as a positive control, and AMP injection was used as a negative control. The AMP or IPM treatments combined with phage was also used for treating GY-4 infection in G. mellonella larvae. Based on clinical doses, ~10 μL of antibiotics were injected into each larva at a final concentration of 18.75 mg/kg of AMP, or 50 mg/kg of IPM (Joly-Guillou et al., 1997; Grygorcewicz et al., 2020; Wang et al., 2021). For survival rate assays, the presence of dark-colored larvae with no response to physical contact within 120 h were recorded as dead (Joly-Guillou et al., 1997; Grygorcewicz et al., 2020; Wang et al., 2021). The control and check groups contained untreated larvae, pierced larvae, and PBS-injected larvae (PBS).
2.12 Statistical analysis
All experimental data were statistically analyzed and plotted with the software GraphPad Prism 8.0.2 and SPSS Statistics 21.0. The Kaplan-Meier method was used to plot the survival curve, and the log-rank test was used for survival curve analysis. P < 0.05 indicated statistically significant differences.
3 Results and discussion
3.1 Isolation and morphology of phage
Acinetobacter virus Abgy202141 directed against A. baumannii strain GY-4 was isolated from underground sewage from residential areas of Guiyang City. In the double-agar plaque assay, Abgy202141 could form transparent circular plaques of 1 cm diameter (Figure 1A). Transmission electron microscopy (TEM) analysis showed that Acinetobacter virus Abgy202141 had an icosahedron head (~59 nm) with a short tail (~23 nm) (Figure 1B). The underground sewage from residential areas contains abundant bacteria and phages, and it can serve as a preferred resource for phage isolations which survive on drug-resistant bacteria (Du et al., 2021).
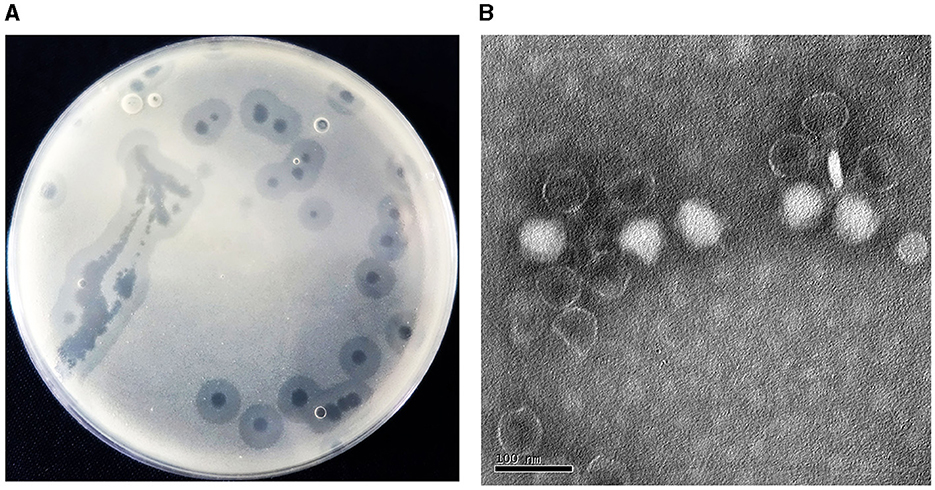
Figure 1. (A) Plaques formed by Acinetobacter virus Abgy202141 after 12 h incubation at 37°C. (B) Transmission electron micrograph of Acinetobacter virus Abgy202141. The bar indicates 20 nm.
3.2 Optimal MOI and one-step growth assays
Phage adsorption to the host cell is the first step of infection, but not all adsorbed phages can successfully infect host cells (Hyman and Abedon, 2010). Acinetobacter virus Abgy202141 effectively reduced the growth of the host bacteria, and OD values declined more quickly at MOI = 1 than at other MOIs (0.1, 0.01, 0.001, 0.0001 or 0.00001) (Figure 2A). Hence the phage particles at an MOI of 1 were mixed with A. baumannii for one-step growth assays. The specific time points of one-step growth assays (1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 30, 40, 50, 60, 90, 120, 180, and 240 min) were based on previously studies with slight modifications (Wu et al., 2018; Tang et al., 2019; Bozdeveci et al., 2021). In our research, the kinetics of adsorption indicated that about 65.3% of Acinetobacter virus Abgy202141 particles adsorbed to the surface of the A. baumannii cells within 2 min, and about 96.9% adsorbed within 5 min (Figure 2B). Efficient phage therapy is associated with phage adsorption, the yield of phages per infected cell (burst size), latency period, and initial dose (Weld et al., 2004). To determine the latency period and burst size of Acinetobacter virus Abgy202141, the one-step growth assay was performed. The latency period of Acinetobacter virus Abgy202141 was about 5 min, and the release of viral particles gradually increased within about 45 min (Figure 2B). The burst size of Acinetobacter virus Abgy202141 was about 189 phage particles per infected host cell similar to results of Yuan et al. (2021).
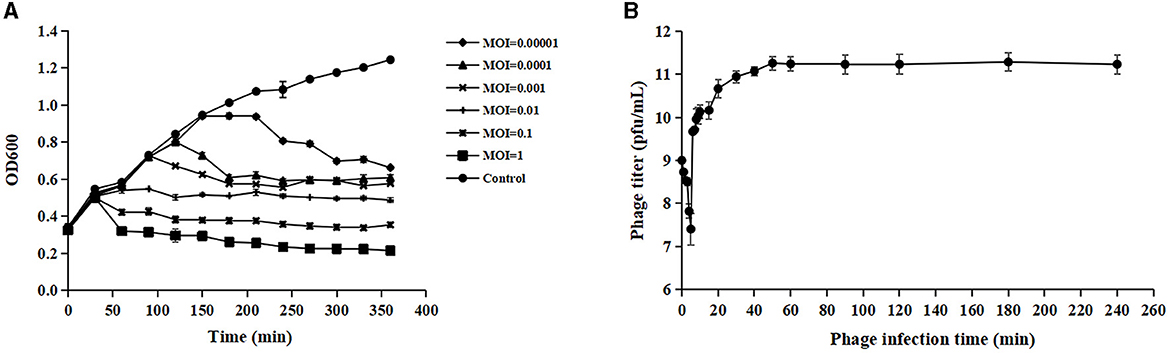
Figure 2. Biological characteristics of Acinetobacter virus Abgy202141. (A) Efficacy of Acinetobacter virus Abgy202141 infection at varying MOIs. (B) One-step growth curve of Acinetobacter virus Abgy202141. The data were obtained from three independent experiments and each mean shows bars as SD.
3.3 Stability of Acinetobacter virus Abgy202141
To evaluate the potential therapeutic and other applications of Acinetobacter virus Abgy202141, the temperature and pH stability of Acinetobacter virus Abgy202141 were investigated. The titer of Acinetobacter virus Abgy202141 ranged from 1.8 × 1010 PFU/ml to 3.0 × 1011 PFU/ml and was only slightly affected by temperatures from 4°C to 55°C (Figure 3A). However, when the temperature reached 65°C, the titer of Acinetobacter virus Abgy202141 was rapidly decreased to 107 PFU/ml. When the temperature reached 75°C, there was total loss of infectiveness. The results were similar to the reported phage AB1, where only 0.52% of phage AB1 survived after 60 min of incubation at 70°C (Yang et al., 2010). Acinetobacter virus Abgy202141 was treated with different pH values. After 1 h incubation, Acinetobacter virus Abgy202141 was found to be stable from pH = 4 to pH = 12 (Figure 3B). But no phage particles survived at pH < 4 or pH>12 (Figure 3B). These results indicated that extreme pH values may affect the stability of Acinetobacter virus Abgy202141, which was similar to that reported for phage AB1 (Yang et al., 2010). The chloroform tolerance of Abgy2021-4-1 was also investigated, and the results showed that the titer of Abgy2021-4-1 did not change significantly with different concentrations of chloroform from 0 to 5% (Figure 3C). Therefore, Abgy2021-4-1 might not contain lipids (Wei et al., 2023). These results were consistent with those of phage vB_EcoP_E212 (Wei et al., 2023).
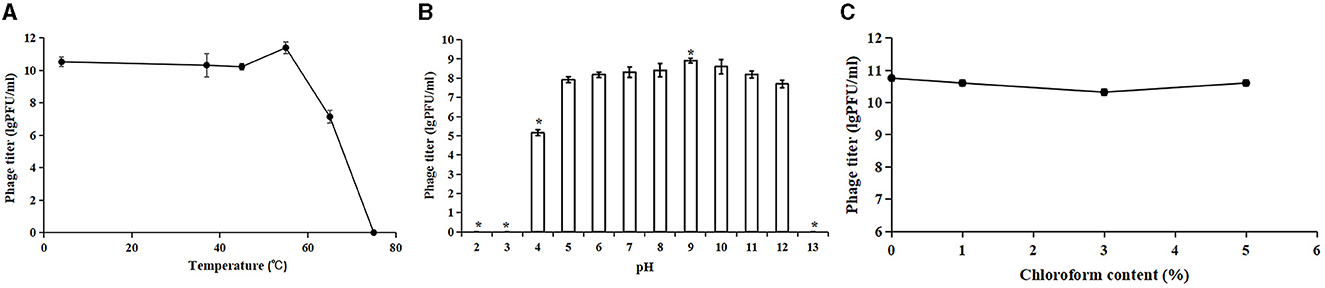
Figure 3. Stability of Acinetobacter virus Abgy202141. (A) Thermal stability of Acinetobacter virus Abgy202141. (B) Infectivity of Acinetobacter virus Abgy202141 tested over a range of pH; Significant differences are indicated by (*P < 0.05). (C) Chloroform sensitivity of Acinetobacter virus Abgy202141. All the experiments were repeated three times, and the data are shown as mean ± SD in the graphs.
Acinetobacter virus Abgy202141 showed broad tolerance toward chloroform concentration, pH values, and variations in thermal conditions, which should be considered beneficial for the storage and preparations of phage for potential application in clinical settings.
3.4 Identification of Acinetobacter virus Abgy202141 structural proteins
To detect the number of structural proteins of Acinetobacter virus Abgy202141, we performed an SDS-PAGE analysis of purified Acinetobacter virus Abgy202141 particles. Fourteen protein bands ranging from 12 to 125 kDa, were observed on the gel (Figure 4). The 36 kDa protein was the most abundant, indicating that it was the major protein of Acinetobacter virus Abgy202141, and others were minor (Yang et al., 2010). These structural proteins were associated with phage assembly, regulation functions, sensing environmental stimulation sensing, and repair of tail fibers in retracted conformation (Yang et al., 2010; Cardarelli et al., 2011).
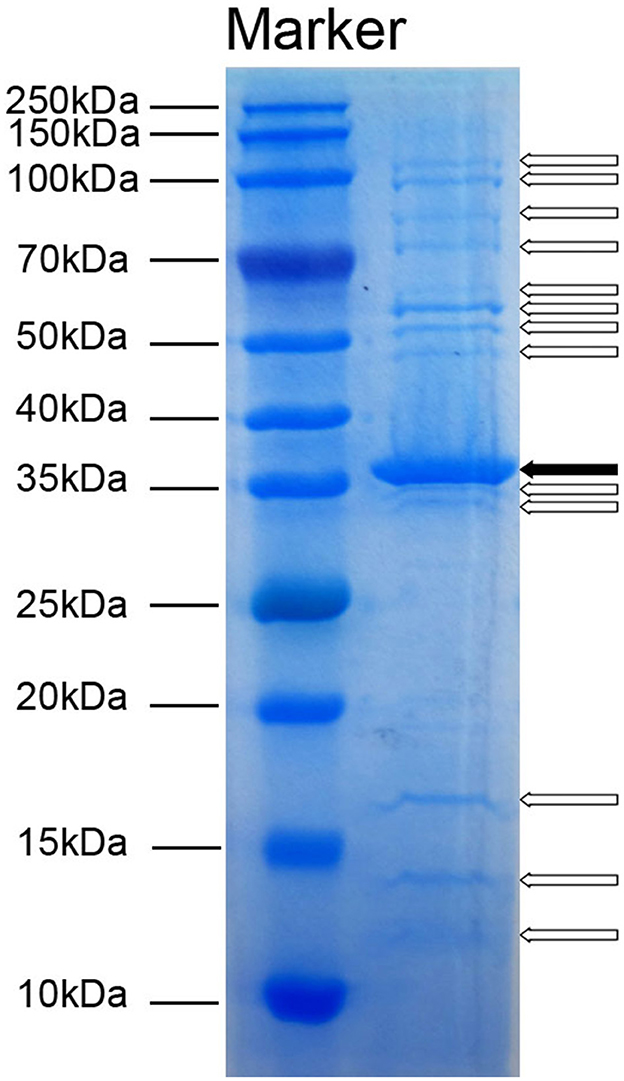
Figure 4. SDS-PAGE analysis of structural proteins in Acinetobacter virus Abgy202141. The molecular weights of the protein bands from 10 to 250 kDa are shown on the left. The solid arrow indicates a major protein band and blank arrows show minor protein bands.
3.5 Annotation and analysis of the Acinetobacter virus Abgy202141 genome
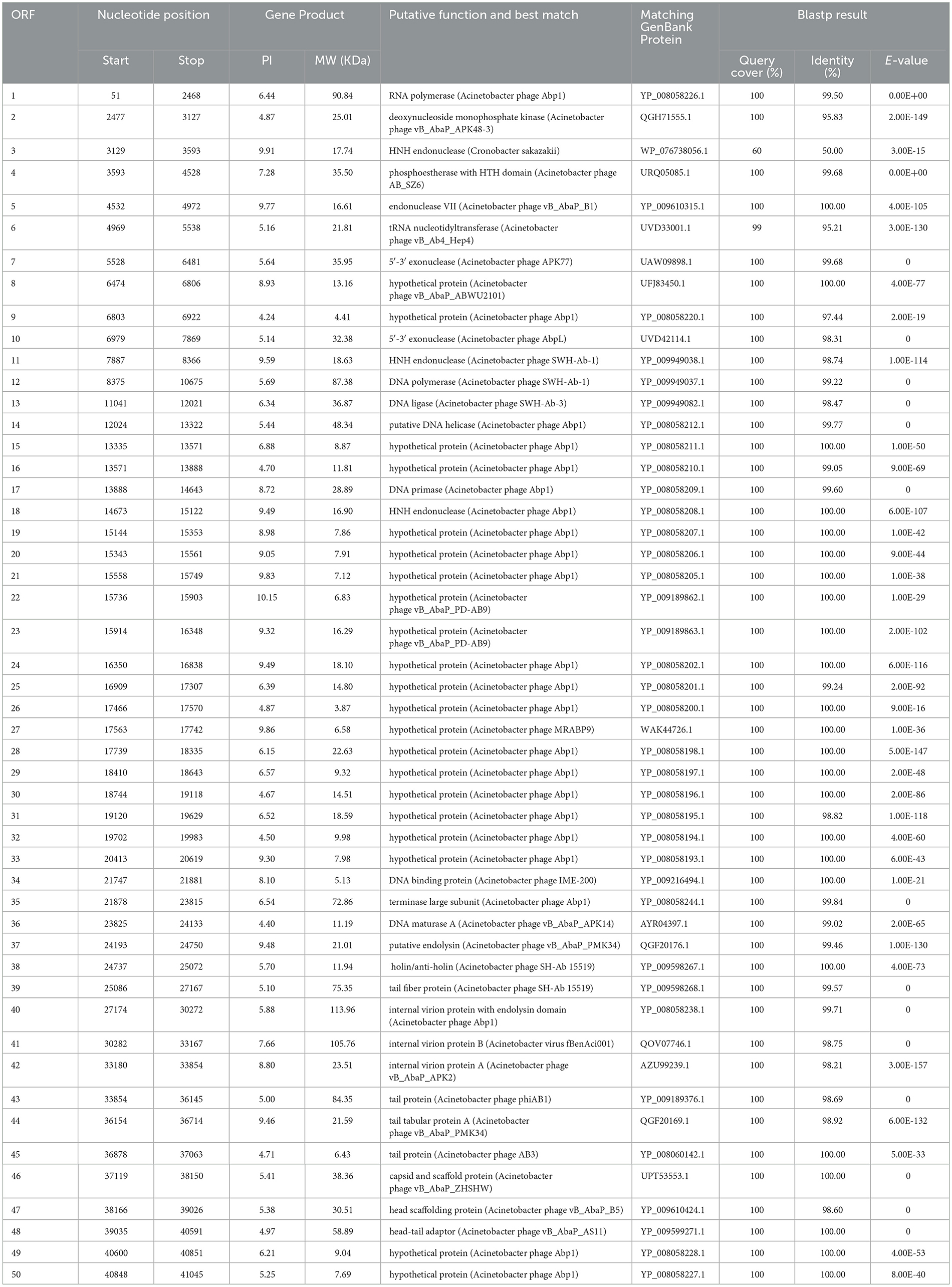
Acinetobacter virus Abgy202141 with double-stranded linear DNA as its genome is 41,242 bp long with 39.4% GC content (Figure 5). The complete genomic sequence of Acinetobacter virus Abgy202141 has been deposited in the GenBank database with the accession number OR770645. According to RAST and BLASTp analyses, a total of 50 ORFs were found in the Acinetobacter virus Abgy202141 genome, 29 of which had significant homology with known functional proteins, while the remaining genes were annotated as hypothetical proteins (Table 1). There were no recognized virulence or antimicrobial resistance genes in the Acinetobacter virus Abgy202141 genome (Table 1). No genes related to lysogenicity were found in the genome of Acinetobacter virus Abgy202141, which indicated Abgy202141 is a lytic phage (Merabishvili et al., 2014). Moreover, whole genome analysis of Abgy202141 with PhageAI also showed this phage is lytic (Tynecki et al., 2020).
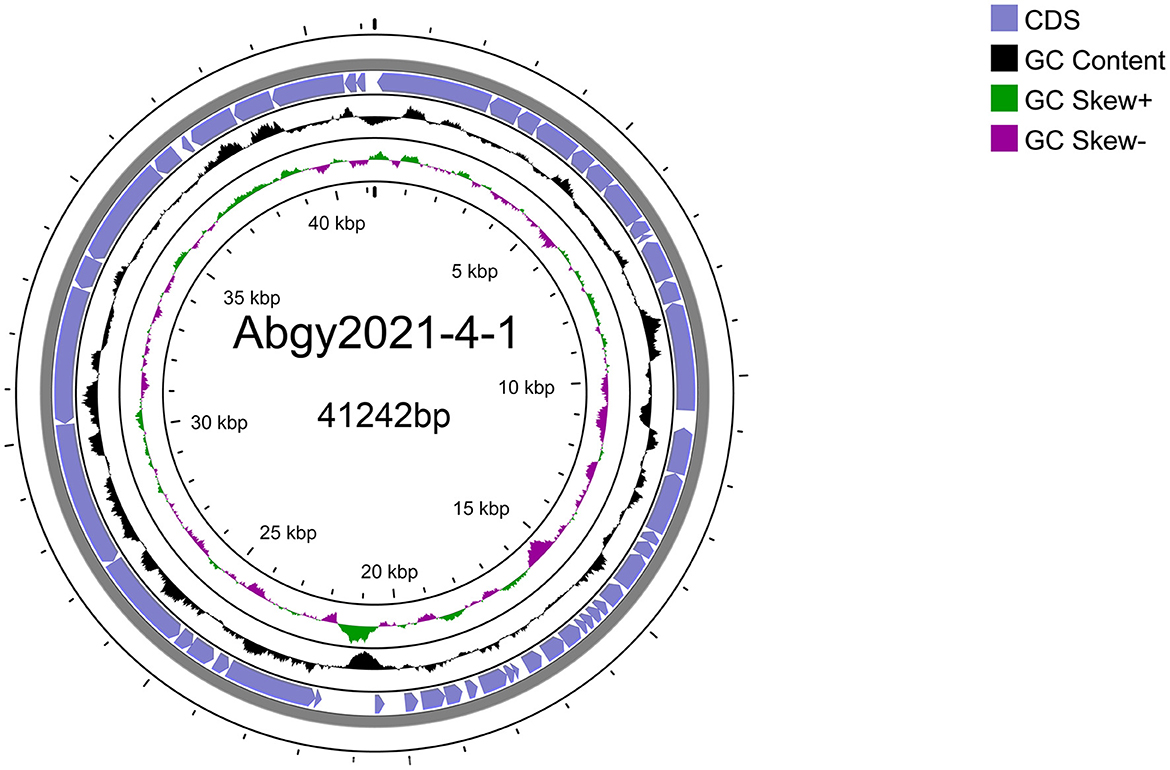
Figure 5. Schematic diagram of the genomic structure of Acinetobacter virus Abgy202141. The blue solid arrows indicate the reading direction of the encoding regions.
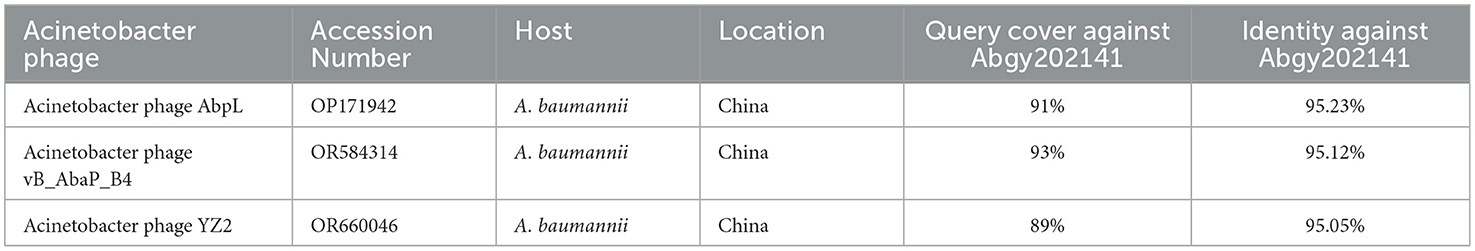
In order to determine the genomic similarity between Acinetobacter virus Abgy202141 and other phages, BLASTn of the complete genome was done against the NCBI database, and three phages were found with the most similarity to Acinetobacter virus Abgy202141. The nucleotide similarity between Acinetobacter phage AbpL and Acinetobacter virus Abgy202141 was 95.23% over their entire lengths, while the nucleotide similarity with Acinetobacter phage vB_AbaP_B4 and Acinetobacter phage YZ2 were 95.12% and was 95.05%, respectively. Detailed information on the three matching phages is shown in Table 2. The modular structure of the genome was compared among the three phages, and this mainly involved several functional groups associated with morphogenesis and structure, DNA replication, repair, recombination and processing, biological metabolism, transcription, lysis, assembly and packaging, and a HNH endonuclease (Figure 6). The distribution of genes with the same function varied among the different phages, and this implied that Acinetobacter virus Abgy202141 is a novel phage.
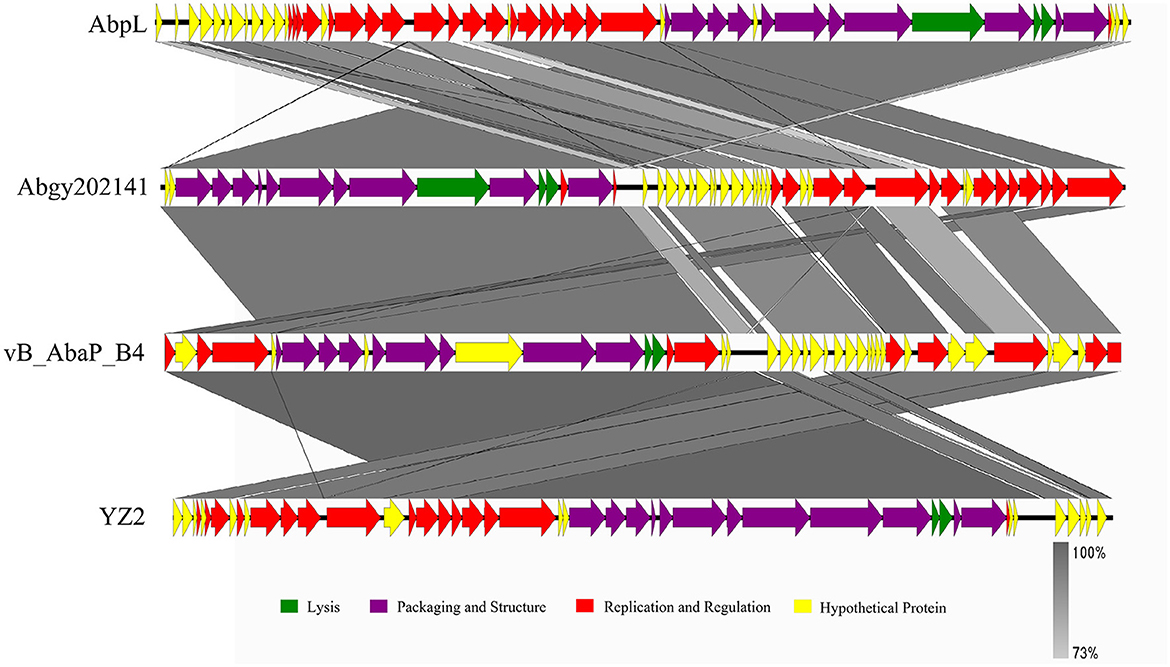
Figure 6. Comparison of the genome of Acinetobacter virus Abgy202141 (second) to Acinetobacter phage AbpL (top), Acinetobacter phage vB_AbaP_B4 (third), and Acinetobacter phage YZ2 (bottom). The different color arrows indicate the CDS in the full length of the genomic sequence. The transcript direction of each CDS is indicated by the direction of the arrow. Homology is represented by the slanted gray bars, and the degree of sequence similarity is indicated by the grayness intensity.
There were three proteins putative endolysin (ORF37), holin/anti-holin (ORF38), and internal virion protein with endolysin domain (ORF40), which were involved in host lysis. Holin could form non-specific channels or holes on the cytoplasmic membrane of bacteria, which allow endolysins to escape and lyse the peptidoglycan (Wang et al., 2000; Young, 2014). The tail fiber protein, which was homologous to ORF39 in the Abgy202141 genome, is believed to be involved in the process of tail assembly or penetration of the host cell outer membrane after infection (Nobrega et al., 2018). The tail fiber protein (ORF39), tail protein (ORF43), tail tabular protein A (ORF44), tail protein (ORF45) and head-tail adaptor (ORF48) are likely responsible for the host range of the phage (Nobrega et al., 2018). The capsid and scaffold protein (ORF46) not only protects the phage genome from degradation in harsh extracellular environments but also maintains stability against high internal pressure during the DNA packaging process (Rao and Black, 2010). The ORF35 is predicted to be the terminase large subunit, which functions in ATP binding, precursor binding, and DNA cleavage (Sun et al., 2010). ORF41 encodes the internal virion protein B. ORF42 encodes the internal virion protein A. ORF47 is predicted to be the head scaffolding protein. ORF1 is predicted to be the RNA polymerase. RNA polymerase participates in gene expression and can transcribe DNA into RNA (Shin and Murakami, 2021). ORF2 is predicted to be the deoxynucleoside monophosphate kinase. This enzyme is involved in the biosynthesis of deoxyribonucleotides (Bao and Ryu, 2006). ORF4 is predicted to be the phosphoestherase with HTH domain. It participates in DNA repair and replication (Aravind et al., 2005). ORF5 is predicted to be the endonuclease VII. It is a component of the mismatch repair mechanism in the genome (Shcherbakov et al., 2011). ORF7 and ORF10 are predicted to be the 5'-3' exonuclease. This enzyme can promote gap to translate into DNA in DNA synthesis reactions (Longley et al., 1990). ORF12 is predicted to be the DNA polymerase. It plays an important role in DNA replication and repair (Ghosh and Raghavan, 2021). ORF13 is predicted to be the DNA ligase. It can catalyze the formation of phosphodiester bonds and is crucial for DNA repair and cell replication (Caracciolo et al., 2022; Duckworth et al., 2023). ORF14 is predicted to be the putative DNA helicase. DNA helicase and DNA polymerase work together to release parental DNA and maintain chromosome stability (Wu and Brosh, 2012; Lo and Gao, 2021). ORF17 is predicted to be the DNA primase. It facilitates chromosome replication and repair, which is crucial for maintaining the stability of telomeres and chromosome (Arezi and Kuchta, 2000). ORF3, ORF11 and ORF18 encode the HNH endonuclease, that is also involved in DNA packaging (Kala et al., 2014). ORF34 is predicted to be the DNA binding protein. It plays important roles in DNA replication, DNA packaging, and DNA repair (Manavi et al., 2023). ORF36 is predicted to be the DNA maturase A. Moreover, no virulence-related genes, antibiotic-resistance genes, or tRNAs were found.
To further investigate the taxonomic status of Acinetobacter virus Abgy202141, a phylogenetic tree was constructed with RNA polymerase from various phages following (Merabishvili et al., 2014). The results were divided into two branches. Acinetobacter virus Abgy202141 had a closer relationship with phages such as Acinetobacter phage SWH-Ab-3 and Acinetobacter phage Abp1 of the genus Friunavirus, while there was a more obvious distinction with phages of the genus Daemvirus and Pettyvirus. The results showed that Acinetobacter virus Abgy202141 belonged to the genus Friunavirus in the Autographiviridae family (Figure 7).
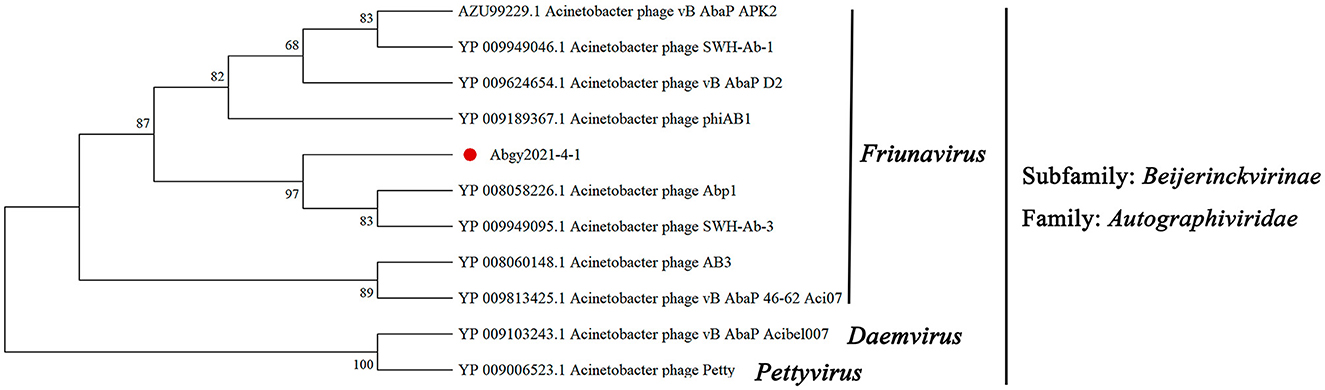
Figure 7. The Maximum Likelihood tree was constructed using RNA polymerase of Acinetobacter virus Abgy202141 and related phages. The amino acid sequences of the related phages were obtained from NCBI, and the accession numbers are shown in the figure. The red dot indicates the position of Acinetobacter virus Abgy202141. Bootstrap percentages out of 1000 replications are shown on the branches.
3.6 In vivo synergy in the G. mellonella model
G. mellonella larvae were used as an animal model to test the ability of Acinetobacter virus Abgy202141 to eliminate A. baumannii strain GY-4. The different doses (1 × 103 CFU/larva, 1 × 104 CFU/larva, 1 × 105 CFU/larva, 1 × 106 CFU/larva) were injected into G. mellonella larvae. We found 106 CFU/larva led to a constant decrease of larval survival within 96 h (Figure 8A), and was chosen as the optimal concentration to look for some possible differences in the pathogenicity of A. baumannii. Twenty minutes after injection (106 CFU/larva) of A. baumannii strain GY-4, the purified Acinetobacter virus Abgy202141 at different MOIs (50, 10, 1, and 0.1) were injected into the inoculated G. Mellonella larvae. We found that the survival of larvae increased constantly with increasing MOIs of Acinetobacter virus Abgy202141 (Figure 8B). When the injection level of Acinetobacter virus Abgy202141 reached an MOI of 50, the survival of G. Mellonella larvae was up to 80 % (Figure 8B). The results of the G. mellonella larvae model test indicated a higher threshold in vivo, which reinforced the need for thorough evaluation prior to any clinical application. Beside mice models, there are still other simple in vivo tests (Manohar et al., 2022). However, the animal model involving G. mellonella is a common infection model used to evaluate phage efficacy (Tsai et al., 2016). Although insect larvae do not have an adaptive immune system, they can be used as a valuable model to evaluate the therapeutic value of a phage and the potential ability to clear the pathogen in the more complex environment of larger animals (Zhang et al., 2022). The results showed that Abgy202141 was highly efficient in clearing a large quantity of bacteria under our experimental conditions. However, to use the Abgy202141 in clinical practice, a mouse model should be first tested to evaluate phage efficacy.

Figure 8. Therapeutic evaluation of Acinetobacter virus Abgy202141 against A. baumannii injected into G. mellonella. (A) Survival of G. mellonella larvae infected with different concentrations of A. baumannii strain GY-4. (B) Survival rates after application of Acinetobacter virus Abgy202141 for treating A. baumanii strain GY-4 infection in G. mellonella larvae. (C) Survival rates after treatment with Acinetobacter virus Abgy202141 phage combined with antibiotics for A. baumannii GY-4 infection in G. mellonella larvae.
Antibiotics combined with phage had previously been investigated in vitro and in animal models (Segall et al., 2019). So, antibiotics combined with phage were also used for treating GY-4 infection in G. mellonella larvae in this study. The survival rates of larvae were 0, 80% and 50% within 120 h, after treatment with AMP, IPM, or phage respectively (Figure 8C). But the survival rate of larvae after 120 h was 90%, when treated with the combination of IPM and phage (Figure 8C). In addition, no death or blackening of non-inoculated larvae were observed after injection of either the phage alone or PBS buffer as negative controls. Compared with PBS and AMP treatment, Acinetobacter virus Abgy202141 treatment significantly improved the survival rate of inoculated larvae. The survival rate from the IPM treatment was higher than the Acinetobacter virus Abgy202141 treatment. However, the combination of IPM and Acinetobacter virus Abgy202141 gave the highest survival rate. The potential of phage-antibiotic synergy for treatment of MDR bacterial infections is gaining greater attention (Segall et al., 2019). In another study, a T4-like phage, KARL-1, could infect eight MDR strains of A. baumannii, and it showed significant synergy with meropenem, and modest synergy with ciprofloxacin or colistin (Jansen et al., 2018). A study of mechanisms of phage-antibiotic synergy have been investigated between phage and antibiotics against either planktonic (vegetative) or biofilm-growing methicillin-resistant Staphylococcus aureus (MRSA) (Tkhilaishvili et al., 2018). Phage SB-1 degraded the MRSA polysaccharide matrix, and targeted persister cells, which made SB-1 combined with any of the five antibiotics assayed (rifampin, daptomycin, fosfomycin, ciprofloxacin, or vancomycin) even more effective (Tkhilaishvili et al., 2018). Hence the mechanisms of Abgy202141 combined with antibiotics synergy should be investigated in further study.
Phage therapy has become popularized worldwide (Uyttebroek et al., 2022). However, phage therapy has limitations including variation among clinical pathogens in resistance to phages, and host immune response to phage (Hatfull et al., 2022). Therefore, with these considerations, phage therapy is commonly used as a last resort in response to complete antibiotic failure (Hatfull et al., 2022), and a major advantage of phage therapy is treatment of MDR bacteria (Zalewska-Piatek, 2023). Moreover, the phages utilized in combination with antibiotics have been associated with successful microbiological and clinical responses, when traditional antimicrobial therapy have failed (Schooley et al., 2017; Chan et al., 2018; Hatfull et al., 2022).
4 Conclusions
In this study, Acinetobacter virus Abgy202141 was isolated from underground sewage with A. baumannii strain GY-4 as the bait. Stability of Acinetobacter virus Abgy202141 showed broad tolerance to chloroform, various pH conditions, and thermal stability. Based on genomic analysis and morphological characteristics, Acinetobacter virus Abgy202141 belongs to the genus Friunavirus in the Autographiviridae family. Acinetobacter virus Abgy202141 might be used as an alternative for antibiotics, because its genome did not carry any virulence-related or antibiotic-resistance genes. Results using the insect model (G. mellonella) assay also showed the therapeutic potential of Acinetobacter virus Abgy202141 combined with IPM. However, the application of Abgy202141 for clinical practice needs further study, such as in a mouse model to evaluate phage efficacy.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
XT: Formal analysis, Investigation, Methodology, Validation, Writing – original draft. XL: Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. JZ: Data curation, Software, Writing – review & editing. LW: Resources, Writing – review & editing. QW: Methodology, Writing – review & editing. XQ: Methodology, Writing – review & editing. JL: Methodology, Writing – review & editing. DZ: Resources, Writing – review & editing. TH: Methodology, Writing – review & editing. YJ: Conceptualization, Formal analysis, Funding acquisition, Methodology, Resources, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by the National Natural Science Foundation of China (32000017 and 31660012); Guizhou Provincial Innovation and Entrepreneurship Project for high-level overseas Talents (2022) No. 15; Guizhou Provincial Natural Foundation [ZK(2021)zhongdian030]; Excellent Young Talents Plan of Guizhou Medical University (2023) No. 101; Department of Education of Guizhou Province (CN) [KY(2021)313]; Guizhou Provincial Natural Science Foundation [ZK(2022) Key Program 039]; Guizhou Science and Technology Support Project “Research and Demonstration of Key Technologies for Ecological Prevention and Control of Major diseases of Lily with Simultaneous Use of Medicine and Food in Guizhou” [Guizhou Science Cooperation Support (2020) No. 4Y115].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–d525. doi: 10.1093/nar/gkz935
Altnok, Z., Boral, B., Ergin, A., and Eser, Z. K. (2020). Existence of biofilm and biofilm-associated virulence genes in multi-drug resistant invasive Acinetobacter baumannii isolates. Mikrobiyoloji Bült. 54, 40–49. doi: 10.5578/mb.20204
Anane, Y. A., Apalata, T., Vasaikar, S., Okuthe, G. E., and Songca, S. (2020). Molecular detection of carbapenemase-encoding genes in multidrug-resistant Acinetobacter baumannii clinical isolates in South Africa. Int. J. Microbiol. 2020, 1–10. doi: 10.1155/2020/7380740
Aravind, L., Anantharaman, V., Balaji, S., Babu, M. M., and Iyer, L. M. (2005). The many faces of the helix-turn-helix domain: Transcription regulation and beyond. FEMS Microbiol. Rev. 29, 231–262. doi: 10.1016/j.femsre.2004.12.008
Arezi, B., and Kuchta, R. D. (2000). Eukaryotic DNA primase. Trends Biochem. Sci. 25, 572–576. doi: 10.1016/S0968-0004(00)01680-7
Bao, J., and Ryu, D. D. (2006). Cloning of deoxynucleoside monophosphate kinase genes and biosynthesis of deoxynucleoside diphosphates. Biotechnol. Bioeng. 93, 572–580. doi: 10.1002/bit.20747
Bozdeveci, A., Akpinar, R., and Karaoglu, S. A. (2021). Isolation, characterization, and comparative genomic analysis of vB_PlaP_SV21, new bacteriophage of Paenibacillus larvae. Virus Res. 305:198571 doi: 10.1016/j.virusres.2021.198571
Caracciolo, D., Juli, G., Riillo, C., Coricello, A., Vasile, F., Pollastri, S., et al. (2022). Exploiting DNA Ligase III addiction of multiple myeloma by flavonoid Rhamnetin. J. Transl. Med. 20, 482. doi: 10.1186/s12967-022-03705-z
Cardarelli, L., Maxwell, K. L., and Davidson, A. R. (2011). Assembly mechanism is the key determinant of the dosage sensitivity of a phage structural protein. Proc. Natl. Acad. Sci. 108, 10168–10173. doi: 10.1073/pnas.1100759108
Cha, K., Oh, H. K., Jang, J. Y., Jo, Y., Kim, W. K., Ha, G. U., et al. (2018). Characterization of two novel bacteriophages infecting multidrug-resistant (MDR) Acinetobacter baumannii and evaluation of their therapeutic efficacy in Vivo. Front. Microbiol. 9:696. doi: 10.3389/fmicb.2018.00696
Chan, B. K., Turner, P. E., Kim, S., Mojibian, H. R., Elefteriades, J. A., and Narayan, D. (2018). Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 60–66. doi: 10.1093/emph/eoy005
Chan, P. P., and Lowe, T. M. (2019). tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 1962, 1–14. doi: 10.1007/978-1-4939-9173-0_1
Chen, L., Li, L., Leng, Y., Zha, Z., Cheng, Y., Huang, B., et al. (2016). The study and analysis of drug resistance genes of nosocomial infection CRAB in ICU. Chin. J. Drug Appl. Monit. 13, 52–55.
Du, B., Wang, Q., Yang, Q., Wang, R., Yuan, W., and Yan, L. (2021). Responses of bacterial and bacteriophage communities to long-term exposure to antimicrobial agents in wastewater treatment systems. J. Hazard Mater 414:125486. doi: 10.1016/j.jhazmat.2021.125486
Duckworth, A. T., Bilotti, K., Potapov, V., and Lohman, G. J. S. (2023). Profiling DNA ligase substrate specificity with a pacific biosciences single-molecule real-time sequencing assay. Curr. Protoc. 3:e690. doi: 10.1002/cpz1.690
Feretzakis, G., Loupelis, E., Sakagianni, A., Skarmoutsou, N., Michelidou, S., Velentza, A., et al. (2019). A 2-year single-centre audit on antibiotic resistance of Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae strains from an intensive care unit and other wards in a general public hospital in Greece. Antibiotics 8:62. doi: 10.3390/antibiotics8020062
Ghosh, D., and Raghavan, S. C. (2021). 20 years of DNA Polymerase μ, the polymerase that still surprises. FEBS J. 288, 7230–7242. doi: 10.1111/febs.15852
Grygorcewicz, B., Roszak, M., Golec, P., Sleboda-Taront, D., Łubowska, N., Górska, M., et al. (2020). Antibiotics act with vB_AbaP_AGC01 phage against Acinetobacter baumannii in human heat-inactivated plasma blood and Galleria mellonella models. Int. J. Mol. Sci. 21:4390. doi: 10.3390/ijms21124390
Hao, X., Cen, X., He, M., Wen, Y., and Zhang, H. (2023). Isolation, biological and whole genome characteristics of a Proteus mirabilis bacteriophage strain. BMC Microbiol. 23:215. doi: 10.1186/s12866-023-02960-4
Hatfull, G. F., Dedrick, R. M., and Schooley, R. T. (2022). Phage therapy for antibiotic-resistant bacterial infections. Annu. Rev. Med. 73, 197–211. doi: 10.1146/annurev-med-080219-122208
Hietala, V., Horsma-Heikkinen, J., Carron, A., Skurnik, M., and Kiljunen, S. (2019). The removal of endo-and enterotoxins from bacteriophage preparations. Front. Microbiol. 10:464722. doi: 10.3389/fmicb.2019.01674
Hyman, P., and Abedon, S. T. (2010). Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 70, 217–248. doi: 10.1016/S0065-2164(10)70007-1
Jansen, M., Wahida, A., Latz, S., Krüttgen, A., Häfner, H., Buhl, E. M., et al. (2018). Enhanced antibacterial effect of the novel T4-like bacteriophage KARL-1 in combination with antibiotics against multi-drug resistant Acinetobacter baumannii. Sci. Rep. 8:14140. doi: 10.1038/s41598-018-32344-y
Joly-Guillou, M. L., Wolff, M., Pocidalo, J. J., Walker, F., and Carbon, C. (1997). Use of a new mouse model of Acinetobacter baumannii pneumonia to evaluate the postantibiotic effect of imipenem. Antimicrob. Agents Chemother. 41, 345–351. doi: 10.1128/AAC.41.2.345
Kala, S., Cumby, N., Sadowski, P. D., Hyder, B. Z., Kanelis, V., Davidson, A. R., et al. (2014). HNH proteins are a widespread component of phage DNA packaging machines. Proc. Natl. Acad. Sci. U. S. A. 111, 6022–6027. doi: 10.1073/pnas.1320952111
Kropinski, A. M., Mazzocco, A., Waddell, T. E., Lingohr, E., and Johnson, R. P. (2009). Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 501, 69–76. doi: 10.1007/978-1-60327-164-6_7
Liu, B., Zheng, D., Zhou, S., Chen, L., and Yang, J. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–d917. doi: 10.1093/nar/gkab1107
Lo, C. Y., and Gao, Y. (2021). DNA helicase-polymerase coupling in bacteriophage DNA replication. Viruses 13:739. doi: 10.3390/v13091739
Longley, M. J., Bennett, S. E., and Mosbaugh, D. W. (1990). Characterization of the 5' to 3' exonuclease associated with Thermus aquaticus DNA polymerase. Nucleic Acids Res. 18, 7317–7322. doi: 10.1093/nar/18.24.7317
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–d268. doi: 10.1093/nar/gkz991
Manavi, F., Sharma, A., Sharma, R., Tsunoda, T., Shatabda, S., and Dehzangi, I. (2023). CNN-Pred: Prediction of single-stranded and double-stranded DNA-binding protein using convolutional neural networks. Gene 853:147045. doi: 10.1016/j.gene.2022.147045
Manohar, P., Loh, B., Elangovan, N., Loganathan, A., Nachimuthu, R., and Leptihn, S. (2022). A multiwell-plate Caenorhabditis elegans assay for assessing the therapeutic potential of bacteriophages against clinical pathogens. Microbiol. Spectr. 10, e01393–e01321. doi: 10.1128/spectrum.01393-21
Merabishvili, M., Vandenheuvel, D., Kropinski, A. M., Mast, J., De Vos, D., Verbeken, G., et al. (2014). Characterization of newly isolated lytic bacteriophages active against Acinetobacter baumannii. PLoS ONE 9:e104853. doi: 10.1371/journal.pone.0104853
Micelli, C., Dai, Y., Raustad, N., Isberg, R. R., Dowson, C. G., Lloyd, A. J., et al. (2023). A conserved zinc-binding site in Acinetobacter baumannii PBP2 required for elongasome-directed bacterial cell shape. Proc. Nat. Acad. Sci. U. S. A. 8:120. doi: 10.1073/pnas.2215237120
Nobrega, F. L., Vlot, M., de Jonge, P. A., Dreesens, L. L., Beaumont, H. J. E., Lavigne, R., et al. (2018). Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 16, 760–773. doi: 10.1038/s41579-018-0070-8
Pajunen, M., Kiljunen, S., and Skurnik, M. (2000). Bacteriophage ΦYeO3-12, specific for Yersinia enterocolitica Serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 182, 5114–5120. doi: 10.1128/JB.182.18.5114-5120.2000
Pickard, D. J. (2009). Preparation of bacteriophage lysates and pure DNA. Methods Mol. Biol. 502, 3–9. doi: 10.1007/978-1-60327-565-1_1
Rajkumari, J., and Siddhardha, B. (2020). Acinetobacter baumannii: infections and drug resistance. Model Org. Microb. Pathogenesis Biofilm Form. Antimicrob. Drug Disc. 22, 257–271. doi: 10.1007/978-981-15-1695-5_14
Rao, V. B., and Black, L. W. (2010). Structure and assembly of bacteriophage T4 head. Virol. J. 7:356. doi: 10.1186/1743-422X-7-356
Schooley, R. T., Biswas, B., Gill, J. J., Hernandez-Morales, A., Lancaster, J., Lessor, L., et al. (2017). Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 61, e00954–e00917. doi: 10.1128/AAC.00954-17
Segall, A. M., Roach, D. R., and Strathdee, S. A. (2019). Stronger together? Perspectives on phage-antibiotic synergy in clinical applications of phage therapy. Curr. Opin. Microbiol. 51, 46–50. doi: 10.1016/j.mib.2019.03.005
Shcherbakov, V. P., Plugina, L., and Shcherbakova, T. (2011). Endonuclease VII is a key component of the mismatch repair mechanism in bacteriophage T4. DNA Repair 10, 356–362. doi: 10.1016/j.dnarep.2010.12.006
Shin, Y., and Murakami, K. S. (2021). Watching the bacterial RNA polymerase transcription reaction by time-dependent soak-trigger-freeze X-ray crystallography. Enzymes 49, 305–314. doi: 10.1016/bs.enz.2021.06.009
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Sun, S., Rao, V. B., and Rossmann, M. G. (2010). Genome packaging in viruses. Curr. Opin. Struct. Biol. 20, 114–120. doi: 10.1016/j.sbi.2009.12.006
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tang, F., Zhang, P., Zhang, Q., Xue, F., Ren, J., Sun, J., et al. (2019). Isolation and characterization of a broad-spectrum phage of multiple drug resistant Salmonella and its therapeutic utility in mice. Microb. Pathog. 126, 193–198. doi: 10.1016/j.micpath.2018.10.042
Tkhilaishvili, T., Lombardi, L., Klatt, A. B., Trampuz, A., and Di Luca, M. (2018). Bacteriophage Sb-1 enhances antibiotic activity against biofilm, degrades exopolysaccharide matrix and targets persisters of Staphylococcus aureus. Int. J. Antimicrob. Agents 52, 842–853. doi: 10.1016/j.ijantimicag.2018.09.006
Tsai, C. J. Y., Loh, J. M. S., and Proft, T. (2016). Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7, 214–229. doi: 10.1080/21505594.2015.1135289
Tu, Q., Pu, M., Li, Y., Wang, Y., Li, M., Song, L., et al. (2023). Acinetobacter Baumannii phages: past, present and future. Viruses 15:673. doi: 10.3390/v15030673
Tynecki, P., Guzinski, A., Kazimierczak, J., Jadczuk, M., Dastych, J., and Onisko, A. J. B. (2020). Phage AI-bacteriophage life cycle recognition with machine learning and natural language processing. BioRxiv. doi: 10.1101/2020.07.11.198606
Uyttebroek, S., Chen, B., Onsea, J., Ruythooren, F., Debaveye, Y., Devolder, D., et al. (2022). Safety and efficacy of phage therapy in difficult-to-treat infections: a systematic review. Lancet Infect Dis. 22, e208–e220. doi: 10.1016/S1473-3099(21)00612-5
Wang, I. N., Smith, D. L., and Young, R. (2000). Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54, 799–825. doi: 10.1146/annurev.micro.54.1.799
Wang, Y. C., Huang, S. W., Chiang, M. H., Lee, I. M., Kuo, S. C., Yang, Y. S., et al. (2021). In vitro and in vivo activities of imipenem combined with BLI-489 against class D β-lactamase-producing Acinetobacter baumannii. J. Antimicrob. Chemother. 76, 451–459. doi: 10.1093/jac/dkaa421
Wei, B., Cong, C., Zheng, L., Chen, L., and Yan, X. (2023). Isolation, characterization and whole genome analysis of the novel genus Lederbergvirus, phage vB_EcoP_E212 infecting enterotoxigenic Escherichia coli K88. Virus Res. 331:199125. doi: 10.1016/j.virusres.2023.199125
Weld, R. J., Butts, C., and Heinemann, J. A. (2004). Models of phage growth and their applicability to phage therapy. J. Theor. Biol. 227, 1–11. doi: 10.1016/S0022-5193(03)00262-5
Wu, M., Hu, K., Xie, Y., Liu, Y., Mu, D., Guo, H., et al. (2018). A novel phage PD-6A3, and its endolysin Ply6A3, with extended lytic activity against Acinetobacter baumannii. Front. Microbiol. 9:3302. doi: 10.3389/fmicb.2018.03302
Wu, Y., and Brosh, R. M. (2012). DNA helicase and helicase-nuclease enzymes with a conserved iron-sulfur cluster. Nucleic Acids Res. 40, 4247–4260. doi: 10.1093/nar/gks039
Yang, H., Liang, L., Lin, S., and Jia, S. (2010). Isolation and characterization of a virulent bacteriophage AB1 of Acinetobacter baumannii. BMC Microbiol. 10:131. doi: 10.1186/1471-2180-10-131
Yang, Z., Liu, X., Shi, Y., Yin, S., Shen, W., Chen, J., et al. (2019). Characterization and genome annotation of a newly detected bacteriophage infecting multidrug-resistant Acinetobacter baumannii. Arch. Virol. 164, 1527–1533. doi: 10.1007/s00705-019-04213-0
Young, R. (2014). Phage lysis: three steps, three choices, one outcome. J. Microbiol. 52, 243–258. doi: 10.1007/s12275-014-4087-z
Yuan, X., Zhang, S., Wang, J., Li, C., Li, N., Yu, S., et al. (2021). Isolation and characterization of a novel Escherichia coli Kayfunavirus phage DY1. Virus Res. 293:198274. doi: 10.1016/j.virusres.2020.198274
Zalewska-Piatek, B. (2023). Phage therapy-challenges, opportunities and future prospects. Pharmaceuticals 16:1638. doi: 10.3390/ph16121638
Zhang, L., Wang, X., Hua, X., Yu, Y., Leptihn, S., and Loh, B. (2022). Therapeutic evaluation of the Acinetobacter baumannii phage Phab24 for clinical use. Virus Res. 320:198889. doi: 10.1016/j.virusres.2022.198889
Zhang, Q., Xing, S., Sun, Q., Pei, G., Cheng, S., Liu, Y., et al. (2017). Characterization and complete genome sequence analysis of a novel virulent Siphoviridae phage against Staphylococcus aureus isolated from bovine mastitis in Xinjiang, China. Virus Genes 53, 464–476. doi: 10.1007/s11262-017-1445-z
Keywords: Acinetobacter baumannii, phage, genome analysis, biological characteristics, phage therapy
Citation: Tian X, Liu X, Zhou J, Wang L, Wang Q, Qi X, Liu J, Zhao D, Hsiang T and Jiang Y (2024) Isolation, characterization and therapeutic evaluation of a new Acinetobacter virus Abgy202141 lysing Acinetobacter baumannii. Front. Microbiol. 15:1379400. doi: 10.3389/fmicb.2024.1379400
Received: 31 January 2024; Accepted: 10 April 2024;
Published: 30 April 2024.
Edited by:
Kenneth K. S. Ng, University of Windsor, CanadaReviewed by:
Swapnil Ganesh Sanmukh, Université Clermont Auvergne, FranceAhmed Askora, Zagazig University, Egypt
Copyright © 2024 Tian, Liu, Zhou, Wang, Wang, Qi, Liu, Zhao, Hsiang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinhui Jiang, anloNTI2NUAxNjMuY29t
†These authors have contributed equally to this work
 Xun Tian1,2†
Xun Tian1,2† Xiaolan Qi
Xiaolan Qi Tom Hsiang
Tom Hsiang Yinhui Jiang
Yinhui Jiang