- 1Lopukhin Federal Research and Clinical Center of Physical-Chemical Medicine of Federal Medical Biological Agency, Moscow, Russia
- 2Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, Russia
Helicobacter is a genus of spiral-shaped Gram-negative enterohepatic bacteria whose members are capable of causing bacteremia in humans. One of the poorly studied members of this genus is the bacterium Helicobacter cinaedi. This microorganism was first isolated from human fecal samples in 1984. Although it was long considered to be associated with only immunocompromised patients, more evidence in recent years has implicated H. cinaedi in causing serious pathologies in immunocompetent populations. In addition, H. cinaedi is also reported to be associated with a few chronic or severe illnesses, such as atherosclerosis, which in turn can lead to the development of other cardiovascular pathologies: one of the leading causes of mortality worldwide. Helicobacter cinaedi often goes unnoticed in standard diagnostic methods due to its slow growth under microaerobic conditions. This often leads to significant underdetection and hence undermines the role of this bacterium in the pathogenesis of various diseases and the extent of its spread in humans. In this review, we have compiled information on pathologies associated with H. cinaedi, the occurrence of the bacterium in humans and animals, and the latest developments in diagnosing the bacterium and treating associated diseases.
1 Introduction
Cardiovascular diseases (CVD) have been the leading cause of mortality worldwide for the last decades, including the Russian Federation (WHO website https://www.who.int/health-topics/cardiovascular-diseases). In turn, most cardiovascular diseases (e.g., coronary heart disease, myocardial infarction, and stroke) occur due to atherosclerotic lesions of blood vessel walls, leading to inflammation and further development of pathology (Björkegren and Lusis, 2022). To date, the mechanisms of lipoprotein metabolism disorders causing atherosclerosis are known, but the exact causes have not yet been established. The main mechanism of atherosclerosis development include disruption of the normal metabolism of lipids and proteins in the inner layer of arteries, or intima, which results in cholesterol plaque formation in the vessel lumen (Fan and Watanabe, 2022).
Chlamydia pneumoniae was considered for a long time as a microorganism associated with the development of atherosclerosis (Shor et al., 1992). However, it has been demonstrated that although C. pneumoniae is widely distributed in coronary atherosclerotic plaques of patients undergoing coronary atherectomy, the degree of C. pneumoniae infection is not associated with plaque instability or restenosis after atherectomy (Otani et al., 2022). For Helicobacter pylori, an indirect involvement in the development of atherosclerosis has been shown. The chronic inflammatory process induced by the bacterium is thought to be an indirect mechanism of endothelial cell damage contributing to the development of atherosclerosis (Chmiela et al., 2015; Krupa et al., 2021). Another hypothesis suggests that the interaction of H. pylori with other members of the internal microflora may lead to dysbiosis, resulting in increased levels of trimethylamine N-oxide, which is an inducer of endothelial damage, in serum (Francisco, 2022). Therefore, the role of microflora (including bacteria associated with atherosclerotic plaques) in the development of atherosclerosis is still not fully understood.
Also there is much evidence that the internal microbiota of the human organism may indirectly contribute to the course of atherosclerotic processes (Verhaar et al., 2020). The gut microbiota of patients with symptomatic atherosclerosis has been shown to have a higher abundance of Collinsella, Enterobacteriaceae, Streptococcaceae, and Klebsiella and a lower abundance of short-chain fatty acid-producing bacteria such as Eubacterium, Roseburia, and Ruminococcaceae compared to healthy individuals (Jie et al., 2017; Liu et al., 2019). It was hypothesized that pathogenic bacteria originating from the oral or gut microbiome make vessel walls more prone to plaque formation by directly infecting the vessel wall or inducing an autoimmune inflammatory response through molecular mimicry (Epstein et al., 2000; Rosenfeld and Campbell, 2011). The analysis of plaque contents has detected bacteria of the genus Streptococcus, Pseudomonas, Klebsiella, Veillonella, as well as microbes such as Chlamydia pneumoniae or Helicobacter pylori (Ott et al., 2006; Koren et al., 2011; Lanter et al., 2014; Mitra et al., 2015).
According to one hypothesis, atherosclerotic plaque formation is a side effect of Helicobacter cinaedi infection, which can cause chronic inflammation and lipoprotein metabolism disorder in macrophages leading to the development of atherosclerosis when translocated in the vasculature (Khan et al., 2014). This microorganism is a natural symbiont of the gastrointestinal tract of some mammals (Gebhart et al., 1989), but in humans it is considered a conditional pathogen such that in the case of immune system malfunction, the bacterium can enter the bloodstream and cause bacteremia. Clinically, this microorganism is found in atherosclerotic plaques of people who died of atherosclerosis (Khan et al., 2012). Furthermore, it has been experimentally shown that oral infection of Apolipoprotein E (ApoE)-deficient mice with H. cinaedi results in the more frequent development of atherosclerotic plaques in the lumen of vessels compared to uninfected ApoE-deficient mice (Khan et al., 2014). Macrophage cells play a key role in the progression of atherosclerosis, since the atherosclerotic plaque itself is a nidus of inflammation that attracts various leukocytes. Due to changes in lipid metabolism, macrophages in the area of the developing plaque acquire the so-called “foam cell” phenotype, one of the main markers of the beginning of atherosclerotic vascular lesions. Further increase of atherosclerotic plaque in size occurs mainly due to the attraction of new macrophage cells to the center of inflammation, which can also accumulate lipids (Libby et al., 2019).
To date, H. cinaedi is a poorly studied pathogen, but more and more human diseases associated with this bacterium are documented each year (van der Ven et al., 1996; Lewis et al., 2007; Nishida et al., 2016). One of the known H. cinaedi-associated diseases is bacteremia (Fox-Lewis et al., 2020; Fujita et al., 2021), but other pathologies such as cellulitis (Lenskaya et al., 2022), ovarian abscess (Sato et al., 2019), and vertebral osteomyelitis (Hase et al., 2018) are also reported. Among patients with H. cinaedi-associated pathologies, there are different age groups, including young people without immunodeficiencies.
2 Helicobacter cinaedi bacterium characterization and pathogenic properties
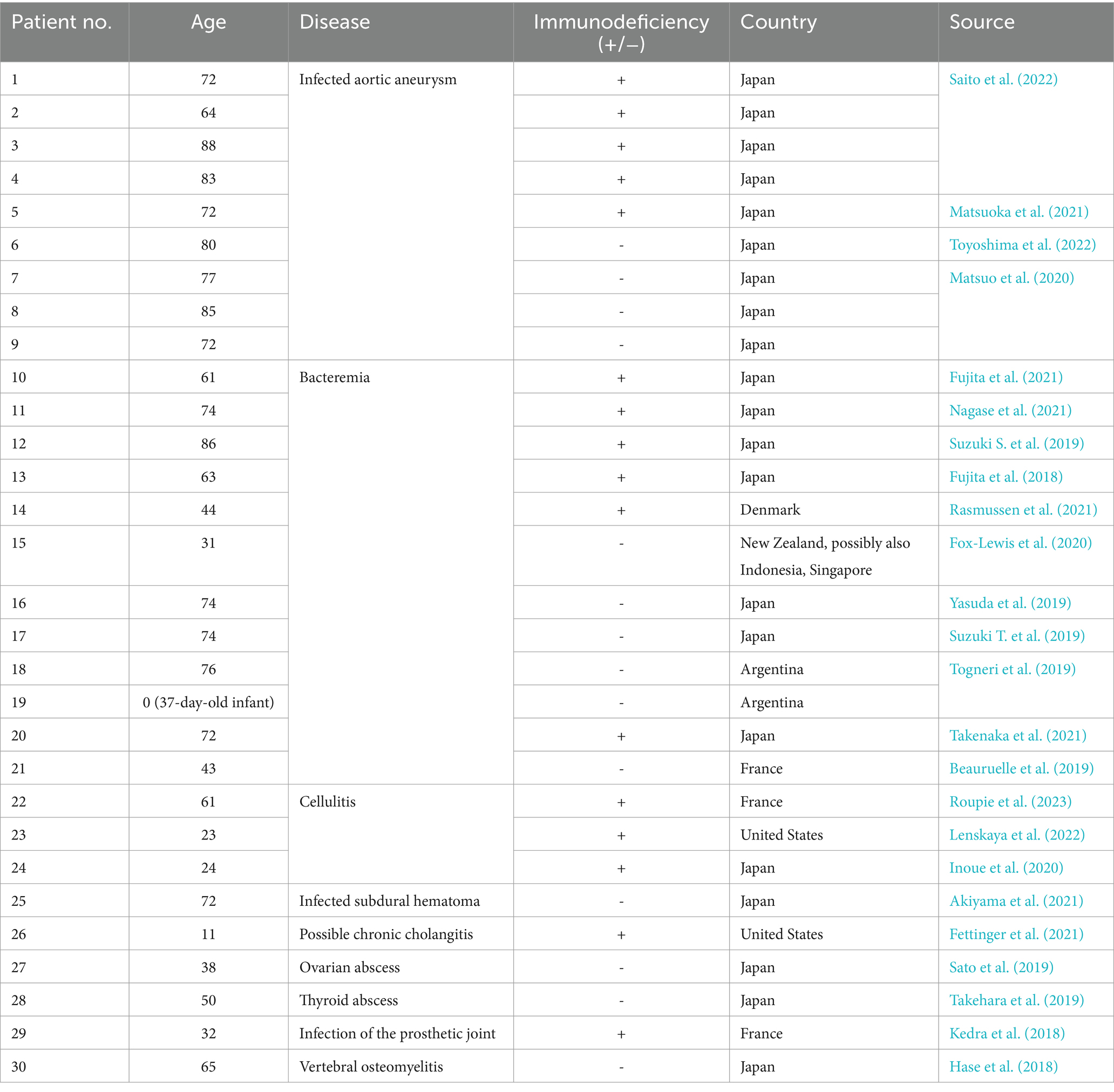
Helicobacter cinaedi is a spiral-shaped Gram-negative bacterium (family Helicobacteriaceae, order Campylobacterales, class Epsilonproteobacteria). The bacterium has an elongated spiral shape with bipolar flagella (Figure 1), but as the culture ages, it changes its shape to coccoid, folding like a tangle. The cell size varies from 0.2 to 1.2 μm in width and from 1.5 to 10 μm in length and under a light microscope at ×1,000 magnification Helicobacter cinaedi looks filamentous (Kawamura et al., 2014). For humans, the bacterium is conditionally pathogenic, which corresponds to WHO risk group 2 (HSE, n.d.).
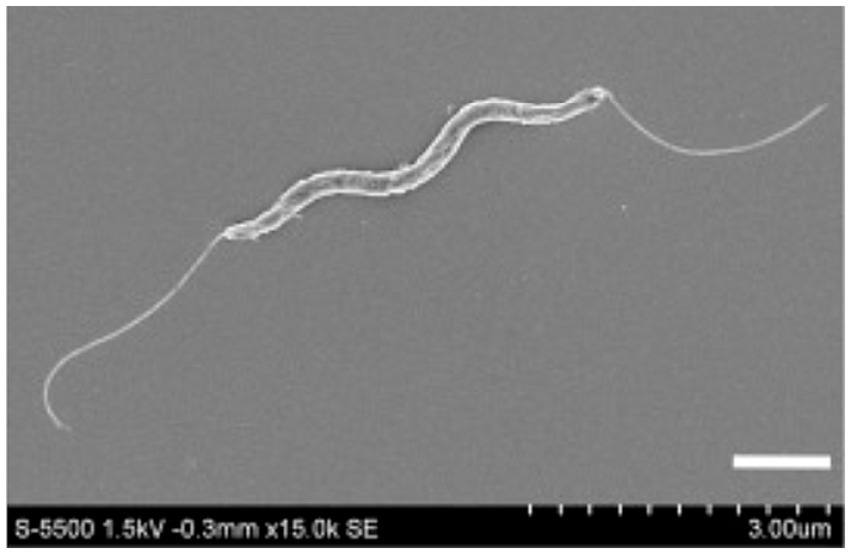
Figure 1. A scanning electron microscope photograph of Helicobacter cinaedi bacterium; magnification 15,000x. Reprinted with permission from Clinical and Bacteriological Characteristics of Helicobacter Cinaedi Infection by Kawamura et al. (2014), licensed under Creative Commons CC-BY-NC-ND license.
To date, there is a genome annotation for the strain H. cinaedi BAA-847 (CDC DO148) (Miyoshi-Akiyama et al., 2012). The genome of this strain is represented by a circular chromosome 2,240,130 bp in length with an average GC content of 38.34%. The chromosome contains 2,322 protein-coding genes, 40 tRNA genes for all amino acids, two rrn operons, and three prophage-like elements.
It is known that a number of H. cinaedi strains naturally contain the plasmids (Kiehlbauch et al., 1995). For strain PAGU611 isolated from a patient with bacteremia, the genomic sequence of plasmid pHci1 was identified with a size of 23,054 bp (Goto et al., 2012). In a recent paper, Yasuhiro Gotoh and coworkers report seven plasmids found in strain T36 obtained from hamster intestines (Taniguchi et al., 2014; Gotoh et al., 2022). The authors of the article also mention the plasmid pD7095-1 found in strain D7095, but the article does not provide a sequence reference for this plasmid. Regarding pHci1, this paper reports that no genes encoding rep proteins were found when the sequence of the plasmid was analyzed, and the sequence of the plasmid is very similar to part of the sequences of ATCC strains BAA-847 and P01D0000. Based on these data, the authors express doubt as to whether pHci1 is actually a plasmid, or whether it is a section of the genome of the chromosome of the bacterium itself. To date, we have been unable to find any further information in the literature about native plasmids of H. cinaedi or attempts to transform this species with genetically engineered plasmids.
In terms of nutrition Helicobacter genus members are chemoorganotrophs, i.e., they obtain all necessary nutrients and energy by biological oxidation of organic substances. For better nitrogen assimilation microorganisms of this group produce urease, and in terms of oxygen demand, they are classified as microaerophiles given their limited ability for oxygen respiration. The most optimal conditions for cultivation of H. cinaedi is a gas mixture of 6% O2, 7% H2, 7% CO2, and 80% N2, but the minimum sufficient gas levels are O2 at 3–10% and СО2 at 3–15%. Oxygen content of more than 15% is toxic to this microorganism, and the presence of hydrogen in the gas mixture, on the contrary, favors the growth of the bacterium. Thus, cultivation of H. cinaedi requires a special atmosphere containing a reduced amount of oxygen (3–15%) and an increased amount of carbon dioxide (3–10%) compared to atmospheric air (Kawamura et al., 2014). It is reasonable that approximately the same oxygen and carbon dioxide content is characteristic of the human gastrointestinal tract (Keeton et al., 2023), where representatives of the genus Helicobacter are often found.
For cultivation of H. cinaedi, it is recommended to use modified Levinthal medium (Tomida et al., 2013) or Columbian medium1 at an ambient temperature of 37°C. ATCC recommend using the following mediums: ATCC Medium 1115: Brucella albimi broth; ATCC Medium 260: Trypticase soy agar/broth with defibrinated sheep blood, and they also specify that the bacterium requires higher humidity when cultured on solid medium.2
3 The history of research on the bacterium Helicobacter cinaedi
The bacterium was first isolated in 1984 from human rectal cultures and named “Campylobacter-like organism type-1” (CLO-1) (Fennell et al., 1984). The authors pointed out that unlike other known Campylobacter species, CLO-1 was slow growing, had an unusual colony morphology, and did not grow at 25°C. In 1985, the same research group showed that there were two genetic groups within the CLO-1 type, named CLO-1a and CLO-1b, with DNA–DNA hybridization values of 42–51%, and the name “Campylobacter cinaedi” was proposed for this microorganism (Totten et al., 1985). Later, based on bacterial DNA hybridization experiments and immunotyping of the genus Campylobacter and related taxa, the bacterium was assigned to the genus Helicobacter (Vandamme et al., 1991). Thus, in publications since 1991, the bacterium is named Helicobacter cinaedi.
A recent study shows the position of H. cinaedi within the genus Helicobacter in terms of modern systematics (Smet et al., 2018). It should be noted that representatives of this genus can be conditionally divided into gastric species, which exclusively colonize the stomach, and enterohepatic species, which colonize the liver or intestinal tract of animals (Solnick and Schauer, 2001). Helicobacter cinaedi is an enterohepatic member of the genus Helicobacter, whereas H. pylori, the best known member of the genus, belongs to the gastric species.
Back in the 1980s, when H. cinaedi was first discovered, it was realized that this species was characterized by genetic variation (the first variations described were the so-called CLO-1a and CLO-1b) (Totten et al., 1985). Currently, many strains of H. cinaedi are known, and another recent study shows that this microorganism appears to be a human-adapted variation of the Helicobacter cinaedi/canicola/'magdeburgensis' group (Gotoh et al., 2022). The authors of the article provide a detailed scheme of phylogenetic relationships between 67 strains of this group, which allows not only to trace the evolutionary relationship between different strains of H. cinaedi, but also to look at the closest genetic variations found in the gastrointestinal tract of humans and different animal species.
Regarding the occurrence of H. cinaedi in animals, in 1989, 5 years after the discovery of H. cinaedi, the first report appeared that this microorganism appeared to be a normal representative of the gastrointestinal microflora in hamsters. All isolates of H. cinaedi (at that time still named Campylobacter cinaedi) isolated from the feces of healthy hamsters were phenotypically and protein profile similar to the human strain of C. cinaedi ATCC 35683 (Gebhart et al., 1989). Somewhat later, a paper appeared mentioning several strains of H. cinaedi derived from isolates from dogs and cats (Kiehlbauch et al., 1995). The authors of the work do not specify whether these were healthy animals or had any signs of disease.
The isolation of H. cinaedi from samples obtained from various animals has been repeatedly reported. In addition to hamsters, dogs, and cats, H. cinaedi has been reported in a number of zoo animals: red panda (Ailurus fulgens fulgens), Nile crocodile (Crocodylus niloticus), sea lion (Zalophus californus), brown bear (Ursus actus), and Amur tiger (Panthera tigris altaica) (Al-Soud et al., 2003). In this case, the samples were obtained as part of the screening of animal fecal microflora, and no clinical reasons for testing were indicated in the article; however, another paper reported that fecal analysis of healthy domestic rhesus macaques (Macaca mulatta) revealed H. cinaedi in 31% of animals (Fernandez et al., 2002). Healthy natural-born long-tailed macaques (Macaca fascicularis) were also found to have H. cinaedi as part of their internal microflora (Sawaswong et al., 2023). Thus, it can be assumed that in many animals, both mammals and reptiles, H. cinaedi is a normal representative of the internal microflora.
However, it is worth listing the publications describing the association of H. cinaedi with primate pathologies. In 1990, an experiment on oral infection of pig-tailed macaque (Macaca nemestrina) cubs with H. cinaedi was conducted, resulting in diarrhea and some signs of bacteremia without accompanying fever (Flores et al., 1990). More recently, there was a report of isolation of H. cinaedi from the intestine of a 2-year-old rhesus macaque with chronic diarrhea, colitis, and hepatitis (Fox et al., 2001) and the large intestine of a baboon (Papio anubis) with pancreatic islet amyloidosis and hepatitis (García et al., 2006). In addition, there are repeated references to experimental infection of mice with H. cinaedi leading to the development of various pathologies (Khan et al., 2014; Shen et al., 2015; Taniguchi et al., 2017), and in another case, the bacterium was detected in the feces of a puppy with bloody diarrhea (Misawa et al., 2002).
The distribution of the bacterium in different animal groups is of interest to researchers primarily in the context of animal-to-human transmission of H. cinaedi. When it was discovered in the 1980s that this bacterium was part of the normal microflora of hamsters, the authors suggested that animals could be a natural reservoir of H. cinaedi from which the bacterium could be transmitted to humans (Gebhart et al., 1989). There has been a case report of the pathogen causing meningitis in a newborn infant whose mother kept hamsters as pets during the first two trimesters of pregnancy (Orlicek et al., 1993), but without direct evidence of animal-to-human transmission. One study examined the ability of hamster- and dog-derived H. cinaedi isolates to adhere, invade, and translocate through polarized human intestinal epithelial Caco-2 cells in vitro (Taniguchi et al., 2016). It turned out that the bacterium does have pathogenic potential in human epithelial cells, but at present there are still no documented cases of human infection with this bacterium from animals.
To date, there is no precise data on the distribution of H. cinaedi in the human population. However, clinical reports provide an indication of the occurrence of this bacterium in different countries of the world (Figure 2). Japan is the leader in reported H. cinaedi cases, and a number of clinical cases associated with this bacterium have been reported in the United States and France. The remaining countries have only a few reports of H. cinaedi (source: https://www.ncbi.nlm.nih.gov/pmc/, keyword search “Helicobacter cinaedi”). Nevertheless, the fact that the bacterium has been recorded in different regions of the globe suggests that this pathogen may be widespread. In addition, publications have repeatedly emphasized that H. cinaedi is difficult to detect by standard methods of analysis because the bacterium is characterized by slow growth and requirements for atmospheric composition (Kawamura et al., 2014; Matsumoto et al., 2017; Nukui et al., 2020). Thus, in some cases, the presence of H. cinaedi in humans may not have been detected due to the specific culturing conditions required for the bacterium. In recent years, new, more sensitive methods of analysis have become available, the use of which may increase the frequency of detection of the bacterium in patient samples (Hase et al., 2018; Narita et al., 2018; Nakamura et al., 2022).
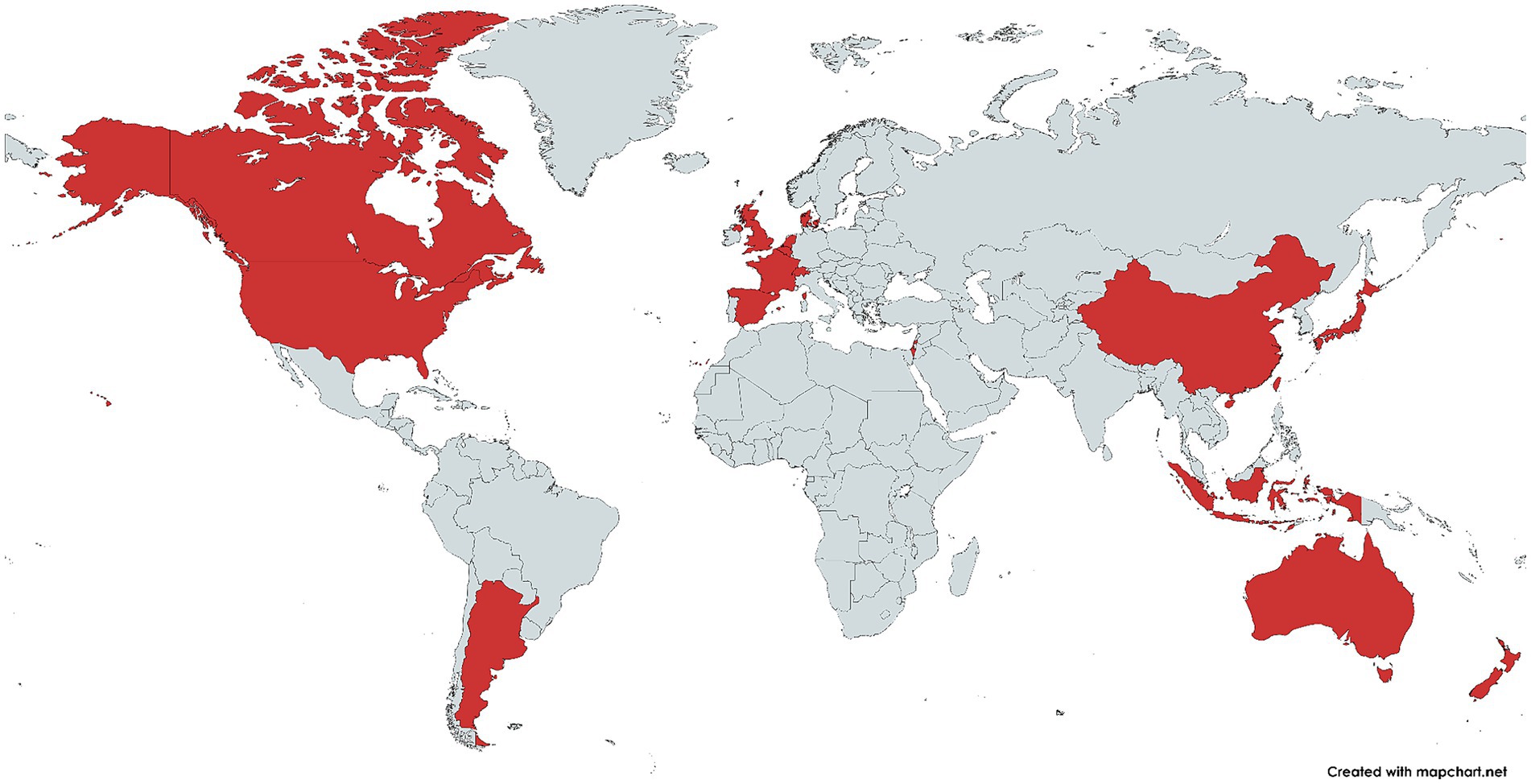
Figure 2. Countries where Helicobacter cinaedi-related diseases have been reported (highlighted in red). The image was obtained using the mapchart.net service based on a search at https://www.ncbi.nlm.nih.gov/pmc/ using the keywords “Helicobacter cinaedi.”
4 Pathologies associated with the bacterium Helicobacter cinaedi
The bacterium was first detected in Seattle, United States, in the analysis of rectal cultures from a homosexual male patient with symptoms of intestinal distress (Fennell et al., 1984). In the following years, there were a number of clinical reports emphasizing that H. cinaedi was found mainly in the samples of homo- or bisexual men with immunodeficiencies, due to which the bacterium was considered to be HIV-associated (Cimolai et al., 1987; Ng et al., 1987; Grayson et al., 1989; Kemper et al., 1993; Burman et al., 1995). Later, the bacterium was found in other patient groups, including women and children (Vandamme et al., 1990; Skirrow et al., 1993). Currently, both cases of H. cinaedi infection against a background of immunodeficiencies (Saito et al., 2022; Roupie et al., 2023) and in immunocompetent patients have been reported (Akiyama et al., 2021; Toyoshima et al., 2022).
Table 1 summarizes examples of H. cinaedi associated diseases documented in the last 5 years. It is worth noting that in all the cases cited, H. cinaedi was identified as the cause of the disease, followed by treatment (mainly antibiotic therapy) for this diagnosis. In most cases, the therapy led to recovery of patients.
Since this species is still poorly studied, only three virulence factors of H. cinaedi have been described in the literature: cytolethal distending toxin (Cdt) (Taylor et al., 2003), alkyl hydroperoxide reductase (AhpC) (Charoenlap et al., 2012), and Helicobacter cinaedi autotransporter protein (HcaA) (Aoki et al., 2023). Cdt is a lethal toxin that induces apoptosis and cell cycle arrest in the host cell, while AhpC is an enzyme that converts various alkyl hydroperoxides into the corresponding alcohols and promotes the survival of the bacterium inside the host cell by converting hydrogen peroxide into water (defense against oxidative stress). HcaA is an autotransporter, a type V secretion system protein that promotes adhesion to the host cells. In addition to the above three factors, a type VI secretion system (T6SS) has been found in H. cinaedi (Goto et al., 2012). For the closely related species H. hepaticus, an association between the protein of this secretion system VgrG1 and the occurrence of colitis has been described (Bartonickova et al., 2013). Therefore, it can be assumed that in H. cinaedi T6SS plays a similar role in the pathogenesis of gastrointestinal diseases, but no direct evidence has been published yet.
Recently, a paper has been published reporting computer modeling of a vaccine against H. cinaedi (Ismail et al., 2022). This is the first project of a preventive strategy whose action is directed specifically at this microorganism. Today, treatment of patients infected with H. cinaedi is based on antibiotic therapy (Lenskaya et al., 2022; Saito et al., 2022; Roupie et al., 2023).
5 Association of the bacterium Helicobacter cinaedi with the development of atherosclerosis
5.1 Foam cell formation
Lipoproteins are involved in the transport of cholesterol and its uptake by cells: high-density lipoproteins (HDL) participate in the transport of cholesterol from the bloodstream to the liver and other depots, while low-density lipoproteins (LDL), on the contrary, transport cholesterol from the depot to the bloodstream (Feingold, 2021). Foam cell formation occurs when the macrophage endoplasmic network malfunctions due to the accumulation of excessive amounts of free cholesterol inside the cell (Gui et al., 2022).
Since atherosclerotic lesions are initiated by excessive amounts of LDL (Libby et al., 2019), proinflammatory macrophages that end up in the lesion focus tend to engulf LDL to restore normal lipoprotein balance. The cellular system of receptors and enzymes is reorganized for this task: the number of receptors and enzymes promoting lipoprotein uptake from outside increases and the number of receptors and enzymes associated with lipoprotein removal from the cell decreases (Chistiakov et al., 2017; Gui et al., 2022). The most frequent and first change is an increase in the number of acceptor receptors, which have affinity for oxidized low-density lipoprotein (oxLDL): SR-A1, CD36 and LOX-1, lectin-like oxLDL receptor-1. It is important that only modified LDL (oxLDL) can serve as an external stimulus to increase the number of these receptors on the cell surface (Chistiakov et al., 2016; Orekhov et al., 2020).
The normal cycle of cholesterol metabolism inside macrophage cells is as follows (Figure 3). After LDL uptake, the cholesterol within them is converted to cholesteryl esters under the action of lysosomal acid lipase (LAL) and acetyl-CoA acetyltransferase (ACAT1). These esters are deposited in the endoplasmic reticulum (ER) (Liu et al., 2022). Then neutral cholesteryl ester hydrolase (NCEH) hydrolyzes the cholesteryl esters, releasing free cholesterol, which is transported outward through the membrane cholesterol carrier system (Kume et al., 2000). In this way, macrophage cells recycle LDL. The cholesterol released in this process can further be incorporated into HDL, which helps to maintain lipoprotein balance in the body. The foam cell formation occurs due to “congestion” at the stage when cholesterol esters are processed within the cell, i.e., the cellular system cannot deal with the volume of cholesterol esters passing through, resulting in the accumulation of excessive amounts of cholesterol esters in the ESR. All factors contributing to the accumulation of cholesterol esters inside the cells can lead to such a disorder: increased number of receptors (greater capture of particles with cholesterol esters from outside), decreased pumping of free cholesterol out of the cells (more particles remain in the cells), failure of enzymes, etc. (Chistiakov et al., 2017).
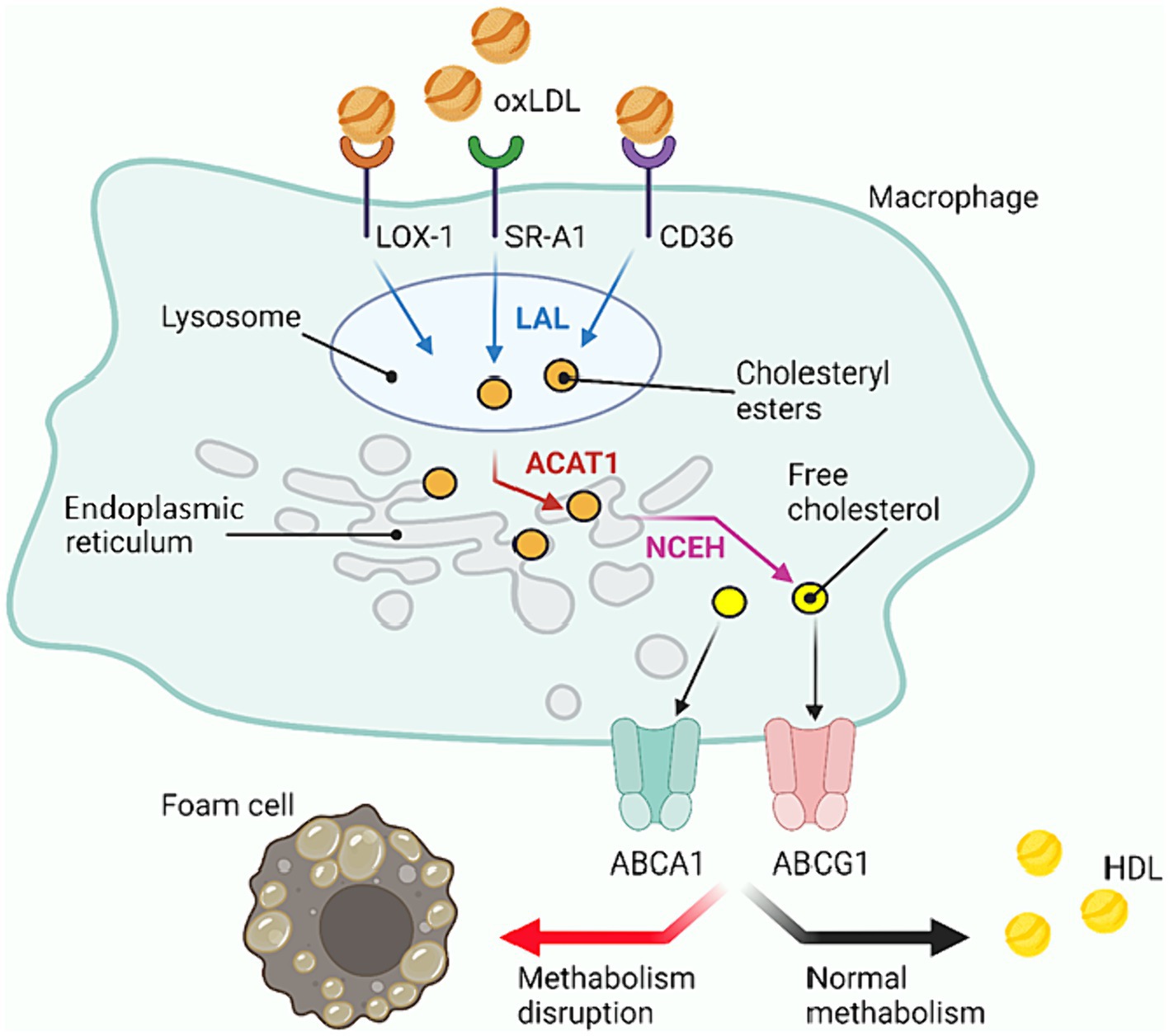
Figure 3. Cholesterol metabolism within macrophage cells. oxLDL, Oxidized low density lipoprotein; HDL, High density lipoprotein; LOX-1, Lectin-like oxidized low-density lipoprotein receptor-1; SR, Scavenger receptor (in the scheme SR-A1); LAL, Lysosomal acid lipase; ER, Endoplasmic reticulum; ACAT1, Cholesterol acyltransferase-1; NCEH, Neutral cholesteryl ester hydrolase; ABC, ATP-binding cassette transporters (ABCA1 and ABCG1 in the scheme). Created with BioRender.com.
The foam cell formation may be caused by exposure to bacterial antigens and products of intestinal microbiota. For example, one study found that nine species of bacteria: Acinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae, Pseudomonas. aeruginosa, Pseudomonas. diminuta, Proteus. vulgaris, Staphylococcus aureus, Staphylococcus epidermidis, and Streptococcus salivarius lead to the formation of lipid granules in macrophages by stimulating Toll-like receptors (TLRs) (Nicolaou et al., 2012). The tuberculosis pathogen M. tuberculosis uses lipids to build its cell wall and as a source of energy (Muñoz-Elías et al., 2006; Lee et al., 2013), so by accumulating lipids it is able to influence the lipid metabolism of the host cell by stimulating the expression of relevant genes (Nazarova et al., 2017, 2019; Agarwal et al., 2021). Some products of intestinal microbiota metabolism: trimethylamine-N-oxide (TMAO), indoxyl sulfate, p-cresol, and short-chain fatty acids also contribute to the formation of foam cells by stimulating scavenger receptor formation and inhibiting cholesterol efflux mechanisms from macrophage cells (Chistiakov et al., 2017; Chaves et al., 2021). The above-mentioned work of Khan’s group also demonstrates the foam cell formation as a result of contact with the bacterium H. cinaedi (Khan et al., 2014).
5.2 Foam cell role in the development of atherosclerosis
The name “atherosclerosis” is derived from the Greek athērē “groat,” “porridge,” and sklērōsis “hardening,” which reflects the process of accumulation of fibrous lipoprotein material in the lumen of blood vessels that occurs in this disease. Due to the disruption of normal lipid and protein metabolism, fatty and/or fibrous lipoprotein fractions begin to deposit in the arterial intima. This is how atherosclerotic plaque (otherwise known as atheromatous plaque, cholesterol plaque or atheroma) is formed. Over time, the plaque may change consistency, become more fibrous and begin to accumulate mineral calcium, which leads to even greater thickening of the plaque. The growth of atheromatous plaque leads to narrowing of the blood vessel lumen, impaired blood flow, and a high risk of thrombus formation, which completely occludes the lumen resulting in acute ischemia (Libby et al., 2019).
Macrophage cells play a significant role in the progression of atherosclerosis, and it is the foam cells (alive or undergoing apoptosis) that form the basis of a growing cholesterol plaque (Figure 4) (Wanschel et al., 2013; Libby et al., 2019). The course of atherosclerosis is always accompanied by an inflammatory reaction, attracting monocytes to the lesion, which then differentiate into macrophages. Two types of macrophages can form as a result of differentiation: M1 (pro-inflammatory) and M2 (anti-inflammatory) (Yunna et al., 2020). The formation of M1-type macrophages occurs under the influence of GM-CSF and IFN-ɣ (Ushach and Zlotnik, 2016; Lu et al., 2019), while the formation of M2-type macrophages is promoted by IL-4 and IL-13 (Italiani and Boraschi, 2014; Genin et al., 2015). Both types remain in the lesion and produce adhesion factors, contributing to atheroma formation.
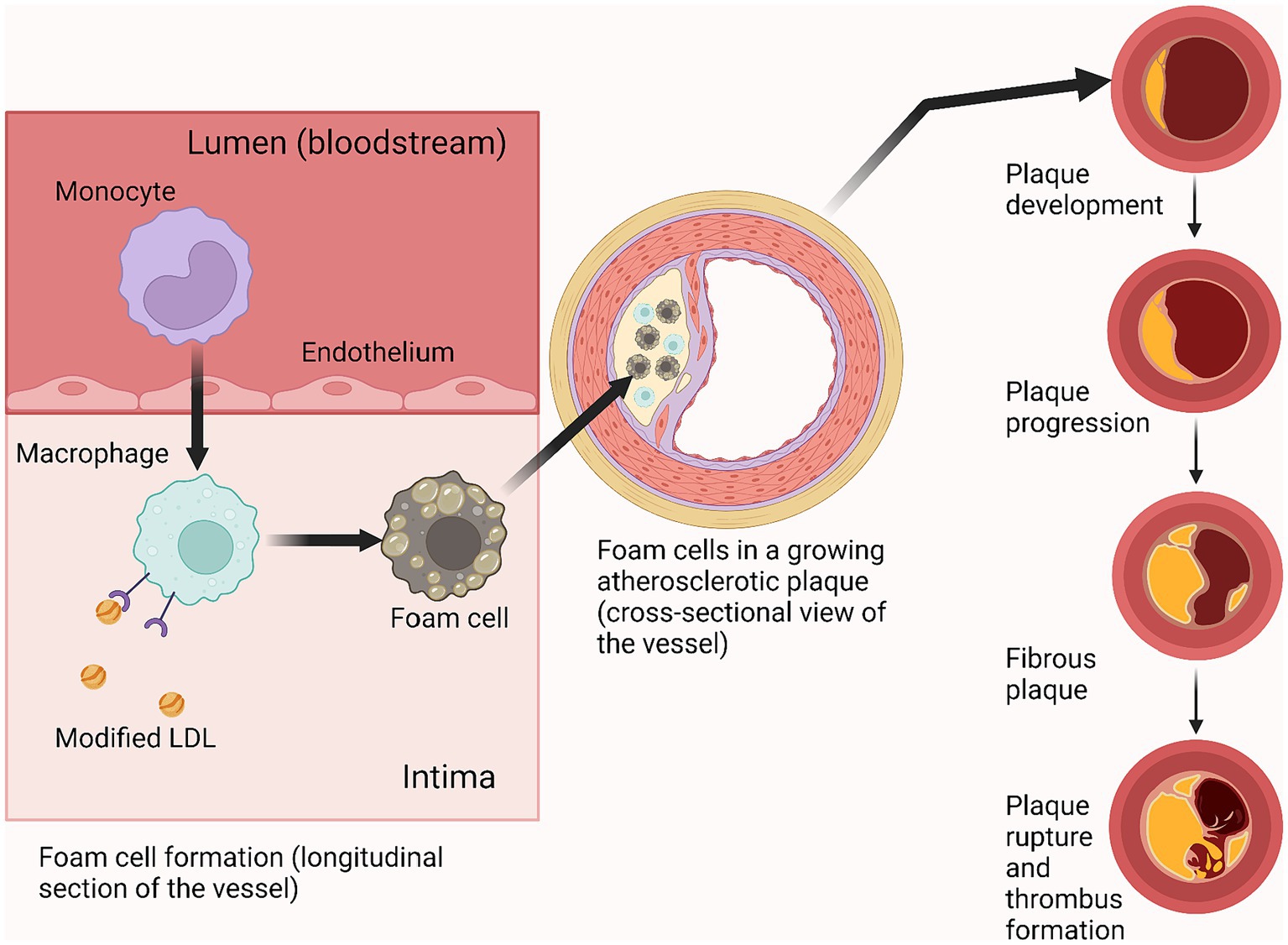
Figure 4. Foam cell role in atherosclerotic plaque progression. Created with BioRender.com.
At the beginning and throughout most of the development of the disease, the atheroma grows radially in an abluminal direction (forming a ring around the perimeter of the vessel wall), while maintaining the caliber of the arterial lumen. Then, upon reaching a certain size, the lesion begins to invade the vessel lumen, obstructing normal blood flow which, under conditions of exercise or stress when the myocardium has an increased oxygen demand, can lead to complications such as coronary heart disease and angina pectoris (Libby et al., 2019). The most serious complication of atherosclerosis is thrombus formation due to rupture of an atherosclerotic plaque, which can lead to myocardial infarction (Ruiz et al., 2016).
5.3 Helicobacter cinaedi and atherosclerosis
The specific causes leading to pathologic changes in lipoprotein metabolism, inflammation, and atherosclerosis, as well as virulence factors of H. cinaedi, are poorly understood, but there are studies that show the relationship between the presence of this bacterium and the progression of atherosclerosis. The most complete of them to date is the article by Khan et al. where experiments on cellular and mouse models were performed (Khan et al., 2014). In this study, they showed foam cell formation when H. cinaedi infected peritoneal macrophages from wild-type mice and human monocyte-derived macrophages; at the same time, H. pylori infection did not lead to the formation of foam cells. The authors note that in H. cinaedi-infected cells lipid droplets were accumulated even without the addition of LDL in the culture medium, whereas in similar experiments with C. pneumoniae foam cells were formed only in the presence of exogenous LDL in the culture medium (Cao et al., 2007). Analogous results (foam cell formation without the addition of exogenous LDL) were obtained by another research group when studying Cinaedi Atherosclerosis Inflammatory Protein (CAIP), a protein of H. cinaedi that supposedly promotes LDL accumulation in macrophages (D’Elios et al., 2017), but there were no further publications investigating CAIP.
In addition to experiments on foam cell formation, in the study by Khan et al., it was shown that after oral infection with H. cinaedi the number and size of atherosclerotic plaques in the aortic sinus of mice with ApoE gene defect significantly increases compared to control uninfected mice. In the aortic tissues of infected mice, a significant increase in gene expression of inducible nitric oxide synthase, interleukin-1β, Toll-like receptor 4, C-C motif chemokine 2, and intercellular adhesion molecule-1 was observed. Also in this study, they showed that during H. cinaedi infection, the level of cholesterol efflux-associated protein ABCG1 was reduced in mouse peritoneal macrophages and THP-1 monocytic macrophages. Based on these results, the authors suggest a possible H. cinaedi-associated mechanism of atherosclerosis. Probably, the bacterium can translocate to the vascular tissue after oral infection and induce inflammatory processes leading to the development of atherosclerosis. Foam cell formation may be associated with the ability of H. cinaedi to downregulate ABCG1 in macrophages.
In the other clinical report made by the Khan group H. cinaedi antigen was detected in all postmortem samples of patients’ atherosclerotic plaques and colocalized with macrophages (Khan et al., 2012). These data suggest that H. cinaedi may contribute to the course or even the onset of atherosclerotic vascular lesions in humans and mammals.
Another study on finding H. cinaedi in atherosclerotic plaques was conducted in Turkey in 2021. 129 samples of atherosclerotic plaques from patients diagnosed with valvular heart disease due to atherosclerosis were analyzed using a nested-polymerase chain reaction. A control group of 146 patients with non-atherosclerotic post-stenotic dilatation also participated in the study. Helicobacter cinaedi DNA was detected only in the group of patients with atherosclerosis, in six samples out of 129 (Sarp et al., 2021). Statistical analysis showed that in this study, the presence of H. cinaedi was not an independent variable for the risk of atherosclerosis. However, since this work used other methods of analysis, its results cannot be compared with those of Japanese colleagues.
A recent study by Horii et al. (2023) reports the detection of H. cinaedi DNA in atherosclerotic abdominal aortic aneurysmal walls. The authors note that H. cinaedi DNA was detected exclusively in aneurysmal walls and was not detected in non-aneurysmal arterial walls. This is the first report of the detection of this bacterium in the walls of a clinically non-infectious abdominal aortic aneurysm. In addition, there are many reports of infected aortic aneurysms initiated by the pathogenic action of this microorganism (Table 1). It is possible that H. cinaedi-associated mechanisms of atherosclerosis and aortic aneurysm are based on the same inflammatory processes caused by this pathogen in the walls of blood vessels. To date, this issue remains incompletely clarified and requires additional research.
6 Discussion
In the early years after its discovery, H. cinaedi was considered to be one of the HIV-associated infections (Cimolai et al., 1987; Ng et al., 1987; Grayson et al., 1989; Kemper et al., 1993; Burman et al., 1995), but nowadays there are many reports that this microorganism has a more diverse range of associated pathologies, and the geography of recorded clinical cases and patient age categories are also becoming increasingly diverse (Vandamme et al., 1990; Skirrow et al., 1993). However, there is still a relatively small number of studies devoted to the properties of this bacterium and its distribution in the human population. This is a significant problem for assessing the real pathogenic potential of H. cinaedi.
Cardiovascular disease is an extremely important health problem worldwide, claiming about 18 million lives annually (WHO website: https://www.who.int/health-topics/cardiovascular-diseases). If H. cinaedi does play an important role in the development of atherosclerosis and subsequent severe CVD, it may be possible to develop preventive therapies directed against this bacterium and potentially significantly improve the development of therapies for CVD. Unfortunately, to date, a small number of studies on the association of H. cinaedi with atherosclerosis are known (Khan et al., 2014; Sarp et al., 2021).
The work with H. cinaedi and its detection in assays is difficult due to the very specific requirements of this bacterium for cultivation conditions. The bacterium requires a gas mixture of oxygen, nitrogen, hydrogen, and carbon dioxide, as well as high humidity (Kawamura et al., 2014). Thus, species-specific tests such as PCR or immunohistochemical analysis seem to be the most effective methods to detect H. cinaedi in biological samples (Narita et al., 2018; Nakamura et al., 2022). The inclusion of H. cinaedi-specific tests in clinical assays would help to obtain more information about the bacterium’s prevalence in the human population.
A very important aspect in the study of pathogenic properties of microorganisms is the knowledge of the produced pathogenicity factors. For example, many Gram-negative bacteria are equipped with secretion systems (six types in total, Type I to Type VI; Green and Mecsas, 2016; Naskar et al., 2021). Helicobacter cinaedi has a Type VI secretion system (Goto et al., 2012) and three pathogenicity factors: cytolethal distending toxin (Cdt) (Taylor et al., 2003), alkyl hydroperoxide reductase (AhpC) (Charoenlap et al., 2012), and Helicobacter cinaedi autotransporter protein (HcaA) (Aoki et al., 2023). However, to date, there is no information on how these pathogenicity factors function in vivo and what specific molecular mechanisms are responsible for the development of H. cinaedi-related pathologies. The study of the proteome of this bacterium, including proteins differentially expressed during infection of macrophages, may provide insight into what other protein factors are involved in the invasion of H. cinaedi into the host organism.
Finally, the question of transmission of H. cinaedi between humans and possibly from animals to humans has not been resolved to date. According to statistics, zoonotic origins account for about ⅔ of human diseases (Rahman et al., 2020). It is well known that H. cinaedi is found as part of the microflora of various animals (Al-Soud et al., 2003). Therefore, it is possible that H. cinaedi-associated pathologies in humans may be of zoonotic origin. To date, there has been no direct evidence of animal-to-human transmission of the bacterium, although H. cinaedi has been found as normal microflora in cats, dogs, and hamsters, all animals that are often kept as pets (Gebhart et al., 1989; Kiehlbauch et al., 1995). Additional studies in this area may clarify whether H. cinaedi is a zoonotic pathogen and provide more information on the prevalence of the bacterium and its contribution to human disease patterns.
7 Conclusion
Helicobacter cinaedi can cause disease of varying severity in humans as well as in some animals. The microorganism is found in the gastrointestinal tract of a number of mammals and apparently reptiles. No cases of human infection from animals have been reported to date, and so far H. cinaedi remains a poorly studied bacterial species. Nevertheless, an increasing variety of pathologies associated with H. cinaedi have been recorded in recent years, and the list of countries from which it has been reported is gradually expanding. Among other things, one clinical hypothesis suggests a link between H. cinaedi infection and foam cell formation and subsequent development of atherosclerosis. It should be noted that atherosclerosis is the main cause of the development of severe CVD and stroke, which according to the World Health Organization (WHO) is the leading cause of mortality worldwide. Thus, because of the difficulty of detection in assays and the limited amount of information on H. cinaedi, the role of this bacterium in the development of cardiovascular disease and in human mortality may be significantly underestimated. Further research is needed to clarify the role of this pathogen in contributing to human disease, the extent of its spread, and improving diagnostic methods for H. cinaedi.
Author contributions
AV: Conceptualization, Resources, Visualization, Writing – original draft, Writing – review & editing. GA: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This publication was carried out with state funding for the “Immunopeptidome” project, state registration number of R&D 124031200004-7.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Agarwal, P., Gordon, S., and Martinez, F. O. (2021). Foam cell macrophages in tuberculosis. Front. Immunol. 12:775326. doi: 10.3389/fimmu.2021.775326
Akiyama, T., Imamura, H., Fukui, N., and Sakai, N. (2021). Helicobacter cinaedi-infected chronic subdural hematoma mimicking an expanding hematoma: a case report. Surg. Neurol. Int. 12:288. doi: 10.25259/SNI_387_2021
Al-Soud, W. A., Bennedsen, M., On, S. L. W., Ouis, I.-S., Vandamme, P., Nilsson, H.-O., et al. (2003). Assessment of PCR-DGGE for the identification of diverse Helicobacter species, and application to faecal samples from zoo animals to determine Helicobacter prevalence. J. Med. Microbiol. 52, 765–771. doi: 10.1099/jmm.0.05314-0
Aoki, S., Mori, S., Matsui, H., Shibayama, K., Kenri, T., and Rimbara, E. (2023). Characterization of HcaA, a novel autotransporter protein in Helicobacter cinaedi, and its role in host cell adhesion. mSphere 8:e0040323. doi: 10.1128/msphere.00403-23
Bartonickova, L., Sterzenbach, T., Nell, S., Kops, F., Schulze, J., Venzke, A., et al. (2013). Hcp and VgrG1 are secreted components of the Helicobacter hepaticus type VI secretion system and VgrG1 increases the bacterial colitogenic potential. Cell. Microbiol. 15, 992–1011. doi: 10.1111/cmi.12094
Beauruelle, C., Le Bars, H., Astruc, N., Tandé, D., Le Berre, R., and Héry-Arnaud, G. (2019). Helicobacter cinaedi associated with atypical Raynaud syndrome. Clin. Microbiol. Infect. 25, 324–325. doi: 10.1016/j.cmi.2019.01.001
Björkegren, J. L. M., and Lusis, A. J. (2022). Atherosclerosis: recent developments. Cell 185, 1630–1645. doi: 10.1016/j.cell.2022.04.004
Burman, W. J., Cohn, D. L., Reves, R. R., and Wilson, M. L. (1995). Multifocal cellulitis and monoarticular arthritis as manifestations of Helicobacter cinaedi bacteremia. Clin. Infect. Dis. 20, 564–570. doi: 10.1093/clinids/20.3.564
Cao, F., Castrillo, A., Tontonoz, P., Re, F., and Byrne, G. I. (2007). Chlamydia pneumoniae-induced macrophage foam cell formation is mediated by toll-like receptor 2. Infect. Immun. 75, 753–759. doi: 10.1128/IAI.01386-06
Charoenlap, N., Shen, Z., McBee, M. E., Muthupalani, S., Wogan, G. N., Fox, J. G., et al. (2012). Alkyl hydroperoxide reductase is required for Helicobacter cinaedi intestinal colonization and survival under oxidative stress in BALB/c and BALB/c interleukin-10−/− mice. Infect. Immun. 80, 921–928. doi: 10.1128/IAI.05477-11
Chaves, L. D., Abyad, S., Honan, A. M., Bryniarski, M. A., McSkimming, D. I., Stahura, C. M., et al. (2021). Unconjugated p-cresol activates macrophage macropinocytosis leading to increased LDL uptake. JCI Insight 6:e144410. doi: 10.1172/jci.insight.144410
Chistiakov, D. A., Bobryshev, Y. V., and Orekhov, A. N. (2016). Macrophage-mediated cholesterol handling in atherosclerosis. J. Cell. Mol. Med. 20, 17–28. doi: 10.1111/jcmm.12689
Chistiakov, D. A., Melnichenko, A. A., Myasoedova, V. A., Grechko, A. V., and Orekhov, A. N. (2017). Mechanisms of foam cell formation in atherosclerosis. J. Mol. Med. 95, 1153–1165. doi: 10.1007/s00109-017-1575-8
Chmiela, M., Gajewski, A., and Rudnicka, K. (2015). Helicobacter pylori vs coronary heart disease—searching for connections. World J. Cardiol. 7, 187–203. doi: 10.4330/wjc.v7.i4.187
Cimolai, N., Gill, M. J., Jones, A., Flores, B., Stamm, W. E., Laurie, W., et al. (1987). “Campylobacter cinaedi” bacteremia: case report and laboratory findings. J. Clin. Microbiol. 25, 942–943. doi: 10.1128/jcm.25.5.942-943.1987
D’Elios, M. M., Vallese, F., Capitani, N., Benagiano, M., Bernardini, M. L., Rossi, M., et al. (2017). The Helicobacter cinaedi antigen CAIP participates in atherosclerotic inflammation by promoting the differentiation of macrophages in foam cells. Sci. Rep. 7:40515. doi: 10.1038/srep40515
Epstein, S. E., Zhu, J., Burnett, M. S., Zhou, Y. F., Vercellotti, G., and Hajjar, D. (2000). Infection and atherosclerosis: potential roles of pathogen burden and molecular mimicry. Arterioscler. Thromb. Vasc. Biol. 20, 1417–1420. doi: 10.1161/01.ATV.20.6.1417
Fan, J., and Watanabe, T. (2022). Atherosclerosis: known and unknown. Pathol. Int. 72, 151–160. doi: 10.1111/pin.13202
Feingold, K. R. (2021). Introduction to Lipids and Lipoproteins. Available at: https://www.MDText.com
Fennell, C. L., Totten, P. A., Quinn, T. C., Patton, D. L., Holmes, K. K., and Stamm, W. E. (1984). Characterization of Campylobacter-like organisms isolated from homosexual men. J. Infect. Dis. 149, 58–66. doi: 10.1093/infdis/149.1.58
Fernandez, K. R., Hansen, L. M., Vandamme, P., Beaman, B. L., and Solnick, J. V. (2002). Captive rhesus monkeys (Macaca mulatta) are commonly infected with Helicobacter cinaedi. J. Clin. Microbiol. 40, 1908–1912. doi: 10.1128/JCM.40.6.1908-1912.2002
Fettinger, N. B., Bradley, J. S., and Ramchandar, N. (2021). An 11-year-old male with X-linked Agammaglobulinemia and persistent abdominal pain. Pediatr. Infect. Dis. J. 40, 356–358. doi: 10.1097/INF.0000000000003056
Flores, B. M., Fennell, C. L., Kuller, L., Bronsdon, M. A., Morton, W. R., and Stamm, W. E. (1990). Experimental infection of pig-tailed macaques (Macaca nemestrina) with Campylobacter cinaedi and Campylobacter fennelliae. Infect. Immun. 58, 3947–3953. doi: 10.1128/iai.58.12.3947-3953.1990
Fox, J. G., Handt, L., Sheppard, B. J., Xu, S., Dewhirst, F. E., Motzel, S., et al. (2001). Isolation of Helicobacter cinaedi from the colon, liver, and mesenteric lymph node of a rhesus monkey with chronic colitis and hepatitis. J. Clin. Microbiol. 39, 1580–1585. doi: 10.1128/JCM.39.4.1580-1585.2001
Fox-Lewis, A., Basu, I., Vesty, A., Henderson, G., Chhibber, A. V., and Thomas, M. (2020). Helicobacter cinaedi bacteremia in a returning traveler. IDCases 21:e00910. doi: 10.1016/j.idcr.2020.e00910
Francisco, A.-J. (2022). Helicobacter Pylori infection induces intestinal Dysbiosis that could be related to the onset of atherosclerosis. Biomed. Res. Int. 2022, 1–16. doi: 10.1155/2022/9943158
Fujita, S., Hayashi, H., Kodama, S., Mukai, T., and Morita, Y. (2018). Bacteremia possibly caused by Helicobacter cinaedi and associated with painful erythema in rheumatoid arthritis with malignant lymphoma. Intern. Med. 57, 3663–3666. doi: 10.2169/internalmedicine.1196-18
Fujita, M., Ureshino, H., and Kimura, S. (2021). Helicobacter cinaedi bacteremia in a patient with primary central nervous system lymphoma. EJHaem 2, 875–876. doi: 10.1002/jha2.285
García, A., Xu, S., Dewhirst, F. E., Nambiar, P. R., and Fox, J. G. (2006). Enterohepatic Helicobacter species isolated from the ileum, liver and colon of a baboon with pancreatic islet amyloidosis. J. Med. Microbiol. 55, 1591–1595. doi: 10.1099/jmm.0.46707-0
Gebhart, C. J., Fennell, C. L., Murtaugh, M. P., and Stamm, W. E. (1989). Campylobacter cinaedi is normal intestinal flora in hamsters. J. Clin. Microbiol. 27, 1692–1694. doi: 10.1128/jcm.27.7.1692-1694.1989
Genin, M., Clement, F., Fattaccioli, A., Raes, M., and Michiels, C. (2015). M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer 15:577. doi: 10.1186/s12885-015-1546-9
Goto, T., Ogura, Y., Hirakawa, H., Tomida, J., Morita, Y., Akaike, T., et al. (2012). Complete genome sequence of Helicobacter cinaedi strain PAGU611, isolated in a case of human bacteremia. J. Bacteriol. 194, 3744–3745. doi: 10.1128/JB.00645-12
Gotoh, Y., Atsuta, Y., Taniguchi, T., Nishida, R., Nakamura, K., Ogura, Y., et al. (2022). Helicobacter cinaedi is a human-adapted lineage in the Helicobacter cinaedi/canicola/‘magdeburgensis' complex. Microb. Genom. 8:mgen000830. doi: 10.1099/mgen.0.000830
Grayson, M. L., Tee, W., and Dwyer, B. (1989). Gastroenteritis associated with Campylobacter cinaedi. Med. J. Aust. 150, 214–215. doi: 10.5694/j.1326-5377.1989.tb136430.x
Green, E. R., and Mecsas, J. (2016). Bacterial secretion systems: an overview. Microbiol. Spectr. 4:VMBF-0012-2015. doi: 10.1128/microbiolspec.VMBF-0012-2015
Gui, Y., Zheng, H., and Cao, R. Y. (2022). Foam cells in atherosclerosis: novel insights into its origins, consequences, and molecular mechanisms. Front. Cardiovasc. Med. 9:845942. doi: 10.3389/fcvm.2022.845942
Hase, R., Hirooka, T., Itabashi, T., Endo, Y., and Otsuka, Y. (2018). Vertebral osteomyelitis caused by Helicobacter cinaedi identified using broad-range polymerase chain reaction with sequencing of the biopsied specimen. Intern. Med. 57, 1475–1477. doi: 10.2169/internalmedicine.0012-17
Horii, S., Sugawara, H., Goto, H., Hashimoto, M., Matsunaga, T., Akamatsu, D., et al. (2023). Presence of Helicobacter cinaedi in atherosclerotic abdominal aortic Aneurysmal Wall. Tohoku J. Exp. Med. 261, 35–41. doi: 10.1620/tjem.2023.J049
HSE (2023). The Approved List of biological agents. Available at: https://www.hse.gov.uk/pubns/misc208.htm (Accessed February 27, 2024).
Inoue, K., Sasaki, S., Yasumi, T., Imai, K., Kusunoki, T., Morio, T., et al. (2020). Helicobacter cinaedi-associated refractory cellulitis in patients with X-linked Agammaglobulinemia. J. Clin. Immunol. 40, 1132–1137. doi: 10.1007/s10875-020-00830-6
Ismail, S., Alsowayeh, N., Abbasi, H. W., Albutti, A., Tahir ul Qamar, M., Ahmad, S., et al. (2022). Pan-genome-assisted computational Design of a Multi-Epitopes-Based Vaccine Candidate against Helicobacter cinaedi. Int. J. Environ. Res. Public Health 19:11579. doi: 10.3390/ijerph191811579
Italiani, P., and Boraschi, D. (2014). From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front. Immunol. 5:514. doi: 10.3389/fimmu.2014.00514
Jie, Z., Xia, H., Zhong, S.-L., Feng, Q., Li, S., Liang, S., et al. (2017). The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 8:845. doi: 10.1038/s41467-017-00900-1
Kawamura, Y., Tomida, J., Morita, Y., Fujii, S., Okamoto, T., and Akaike, T. (2014). Clinical and bacteriological characteristics of Helicobacter cinaedi infection. J. Infect. Chemother. 20, 517–526. doi: 10.1016/j.jiac.2014.06.007
Kedra, J., Zeller, V., Heym, B., Lehours, P., Meyssonnier, V., Lariven, S., et al. (2018). A case of recurrent Helicobacter cinaedi prosthetic joint infection in an HIV-infected man. J. Bone Joint Infect. 3, 230–233. doi: 10.7150/jbji.28375
Keeton, W. T., Sircus, W., Hightower, N. C., and Dworken, H. J. (2023). Human digestive system. Encyclopedia Britannica. Available at: https://www.britannica.com/science/human-digestive-system
Kemper, C. A., Mickelsen, P., Morton, A., Walton, B., and Deresinski, S. C. (1993). Helicobacter (Campylobacter) fennelliae-like organisms as an important but occult cause of bacteraemia in a patient with AIDS. J. Inf. Secur. 26, 97–101. doi: 10.1016/0163-4453(93)97128-K
Khan, S., Okamoto, T., Enomoto, K., Sakashita, N., Oyama, K., Fujii, S., et al. (2012). Potential association of Helicobacter cinaedi with atrial arrhythmias and atherosclerosis. Microbiol. Immunol. 56, 145–154. doi: 10.1111/j.1348-0421.2012.00421.x
Khan, S., Rahman, H. N. A., Okamoto, T., Matsunaga, T., Fujiwara, Y., Sawa, T., et al. (2014). Promotion of atherosclerosis by Helicobacter cinaedi infection that involves macrophage-driven proinflammatory responses. Sci. Rep. 4:4680. doi: 10.1038/srep04680
Kiehlbauch, J. A., Brenner, D. J., Cameron, D. N., Steigerwalt, A. G., Makowski, J. M., Baker, C. N., et al. (1995). Genotypic and phenotypic characterization of Helicobacter cinaedi and Helicobacter fennelliae strains isolated from humans and animals. J. Clin. Microbiol. 33, 2940–2947. doi: 10.1128/jcm.33.11.2940-2947.1995
Koren, O., Spor, A., Felin, J., Fåk, F., Stombaugh, J., Tremaroli, V., et al. (2011). Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA 108, 4592–4598. doi: 10.1073/pnas.1011383107
Krupa, A., Gonciarz, W., Rusek-Wala, P., Rechciński, T., Gajewski, A., Samsel, Z., et al. (2021). Helicobacter pylori infection acts synergistically with a high-fat diet in the development of a Proinflammatory and potentially Proatherogenic endothelial cell environment in an experimental model. Int. J. Mol. Sci. 22:3394. doi: 10.3390/ijms22073394
Kume, N., Moriwaki, H., Kataoka, H., Minami, M., Murase, T., Sawamura, T., et al. (2000). Inducible expression of LOX-1, a novel receptor for oxidized LDL, in macrophages and vascular smooth muscle cells. Ann. N. Y. Acad. Sci. 902, 323–327. doi: 10.1111/j.1749-6632.2000.tb06332.x
Lanter, B. B., Sauer, K., and Davies, D. G. (2014). Bacteria present in carotid arterial plaques are found as biofilm deposits which may contribute to enhanced risk of plaque rupture. MBio 5, e01206–e01214. doi: 10.1128/mBio.01206-14
Lee, W., VanderVen, B. C., Fahey, R. J., and Russell, D. G. (2013). Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J. Biol. Chem. 288, 6788–6800. doi: 10.1074/jbc.M112.445056
Lenskaya, V., O’Connor, M., de Moll, E. H., and Desman, G. (2022). Relapsing Helicobacter cinaedi cellulitis without bacteremia in a patient with X-linked Agammaglobulinemia after 1 year of antibiotic prophylaxis. Skinmed 20, 457–459.
Lewis, G. D., Holmes, C. B., Holmvang, G., and Butterton, J. R. (2007). Case records of the Massachusetts General Hospital. Case 8-2007. A 48-year-old man with chest pain followed by cardiac arrest. N. Engl. J. Med. 356, 1153–1162. doi: 10.1056/NEJMcpc079002
Libby, P., Buring, J. E., Badimon, L., Hansson, G. K., Deanfield, J., Bittencourt, M. S., et al. (2019). Atherosclerosis. Nat. Rev. Dis. Primers 5:56. doi: 10.1038/s41572-019-0106-z
Liu, H., Chen, X., Hu, X., Niu, H., Tian, R., Wang, H., et al. (2019). Alterations in the gut microbiome and metabolism with coronary artery disease severity. Microbiome 7:68. doi: 10.1186/s40168-019-0683-9
Liu, Z., Gomez, C. R., Espinoza, I., Le, T. P. T., Shenoy, V., and Zhou, X. (2022). Correlation of cholesteryl ester metabolism to pathogenesis, progression and disparities in colorectal Cancer. Lipids Health Dis. 21:22. doi: 10.1186/s12944-022-01629-7
Lu, S., Li, D., Xi, L., and Calderone, R. (2019). Interplay of interferon-gamma and macrophage polarization during Talaromyces marneffei infection. Microb. Pathog. 134:103594. doi: 10.1016/j.micpath.2019.103594
Matsumoto, A., Yeh, I., Schwartz, B., Rosenblum, M., and Schmidt, T. H. (2017). Chronic Helicobacter cinaedi cellulitis diagnosed by microbial polymerase chain reaction. JAAD Case Rep. 3, 398–400. doi: 10.1016/j.jdcr.2017.05.009
Matsuo, T., Mori, N., Mizuno, A., Sakurai, A., Kawai, F., Starkey, J., et al. (2020). Infected aortic aneurysm caused by Helicobacter cinaedi: case series and systematic review of the literature. BMC Infect. Dis. 20:854. doi: 10.1186/s12879-020-05582-7
Matsuoka, A., Sasaki, Y., Kubodera, A., Hayashi, K., Shimizu, R., Toriihara, A., et al. (2021). Acquired hemophilia a presenting with infectious aortic aneurysms due to an underlying Helicobacter cinaedi infection. Intern. Med. 60, 3947–3952. doi: 10.2169/internalmedicine.7517-21
Misawa, N., Kawashima, K., Kondo, F., Kushima, E., Kushima, K., and Vandamme, P. (2002). Isolation and characterization of Campylobacter, Helicobacter, and Anaerobiospirillum strains from a puppy with bloody diarrhea. Vet. Microbiol. 87, 353–364. doi: 10.1016/S0378-1135(02)00086-X
Mitra, S., Drautz-Moses, D. I., Alhede, M., Maw, M. T., Liu, Y., Purbojati, R. W., et al. (2015). In silico analyses of metagenomes from human atherosclerotic plaque samples. Microbiome 3:38. doi: 10.1186/s40168-015-0100-y
Miyoshi-Akiyama, T., Takeshita, N., Ohmagari, N., and Kirikae, T. (2012). Complete genome sequence of Helicobacter cinaedi type strain ATCC BAA-847. J. Bacteriol. 194:5692. doi: 10.1128/JB.01347-12
Muñoz-Elías, E. J., Upton, A. M., Cherian, J., and McKinney, J. D. (2006). Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol. Microbiol. 60, 1109–1122. doi: 10.1111/j.1365-2958.2006.05155.x
Nagase, T., Wada, S., Yokozawa, T., Fujita, A., and Oda, T. (2021). Bacteremia caused by both Legionella pneumophila serogroup 2 and Helicobacter cinaedi. JMAJ 4, 297–301. doi: 10.31662/jmaj.2021-0016
Nakamura, A., Ueda, O., Abe, N., Shimada, M., Kamioka, M., and Komatsu, M. (2022). Comparison of microorganism detection, time to positivity, and time-dependent shift in viable bacterial count from VersaTREK and BacT/ALERT 3D blood culture systems. Rinsho Biseibutshu Jinsoku Shindan Kenkyukai Shi 32, 23–35.
Narita, T., Kato, K., Hanaiwa, H., Harada, T., Sano, Y., Funashima, Y., et al. (2018). On the performance evaluation of VersaTREK introduced in blood culture. Rinsho Biseibutshu Jinsoku Shindan Kenkyukai Shi 28, 67–75.
Naskar, S., Hohl, M., Tassinari, M., and Low, H. H. (2021). The structure and mechanism of the bacterial type II secretion system. Mol. Microbiol. 115, 412–424. doi: 10.1111/mmi.14664
Nazarova, E. V., Montague, C. R., Huang, L., La, T., Russell, D., and VanderVen, B. C. (2019). The genetic requirements of fatty acid import by Mycobacterium tuberculosis within macrophages. eLife 8:e43621. doi: 10.7554/eLife.43621
Nazarova, E. V., Montague, C. R., La, T., Wilburn, K. M., Sukumar, N., Lee, W., et al. (2017). Rv3723/LucA coordinates fatty acid and cholesterol uptake in Mycobacterium tuberculosis. eLife 6:e26969. doi: 10.7554/eLife.26969
Ng, V. L., Hadley, W. K., Fennell, C. L., Flores, B. M., and Stamm, W. E. (1987). Successive bacteremias with “Campylobacter cinaedi” and “Campylobacter fennelliae” in a bisexual male. J. Clin. Microbiol. 25, 2008–2009. doi: 10.1128/jcm.25.10.2008-2009.1987
Nicolaou, G., Goodall, A. H., and Erridge, C. (2012). Diverse bacteria promote macrophage foam cell formation via toll-like receptor-dependent lipid body biosynthesis. J. Atheroscler. Thromb. 19, 137–148. doi: 10.5551/jat.10249
Nishida, K., Iwasawa, T., Tamura, A., and Lefor, A. T. (2016). Infected abdominal aortic aneurysm with Helicobacter cinaedi. Case Rep. Surg. 2016:1396568. doi: 10.1155/2016/1396568
Nukui, Y., Chino, T., Tani, C., Sonobe, K., Aiso, Y., Tohda, S., et al. (2020). Molecular epidemiologic and clinical analysis of Helicobacter cinaedi bacteremia in Japan. Helicobacter 25:e12675. doi: 10.1111/hel.12675
Orekhov, A. N., Sukhorukov, V. N., Nikiforov, N. G., Kubekina, M. V., Sobenin, I. A., Foxx, K. K., et al. (2020). Signaling pathways potentially responsible for foam cell formation: cholesterol accumulation or inflammatory response-what is first? Int. J. Mol. Sci. 21:2716. doi: 10.3390/ijms21082716
Orlicek, S. L., Welch, D. F., and Kuhls, T. L. (1993). Septicemia and meningitis caused by Helicobacter cinaedi in a neonate. J. Clin. Microbiol. 31, 569–571. doi: 10.1128/jcm.31.3.569-571.1993
Otani, T., Nishihira, K., Azuma, Y., Yamashita, A., Shibata, Y., Asada, Y., et al. (2022). Chlamydia pneumoniae is prevalent in symptomatic coronary atherosclerotic plaque samples obtained from directional coronary Atherectomy, but its quantity is not associated with plaque instability: an Immunohistochemical and molecular study. Clin. Pathol. 15:2632010X2211251. doi: 10.1177/2632010X221125179
Ott, S. J., El Mokhtari, N. E., Musfeldt, M., Hellmig, S., Freitag, S., Rehman, A., et al. (2006). Detection of diverse bacterial signatures in atherosclerotic lesions of patients with coronary heart disease. Circulation 113, 929–937. doi: 10.1161/CIRCULATIONAHA.105.579979
Rahman, M. T., Sobur, M. A., Islam, M. S., Ievy, S., Hossain, M. J., El Zowalaty, M. E., et al. (2020). Zoonotic diseases: etiology, impact, and control. Microorganisms 8:1405. doi: 10.3390/microorganisms8091405
Rasmussen, S. L., Ørsted, I., Tarpgaard, I. H., and Nielsen, H. L. (2021). Helicobacter cinaedi bacteraemia secondary to enterocolitis in an immunocompetent patient. Gut Pathog. 13:26. doi: 10.1186/s13099-021-00422-8
Rosenfeld, M. E., and Campbell, L. A. (2011). Pathogens and atherosclerosis: update on the potential contribution of multiple infectious organisms to the pathogenesis of atherosclerosis. Thromb. Haemost. 106, 858–867. doi: 10.1160/TH11-06-0392
Roupie, A.-L., Lafont, E., Fraitag, S., Ferroni, A., Lécuyer, H., Boccara, O., et al. (2023). Recurrent cellulitis revealing Helicobacter cinaedi in patient on Ibrutinib therapy, France. Emerg. Infect. Dis. 29, 640–641. doi: 10.3201/eid2903.221329
Ruiz, J. L., Weinbaum, S., Aikawa, E., and Hutcheson, J. D. (2016). Zooming in on the genesis of atherosclerotic plaque microcalcifications. J. Physiol. 594, 2915–2927. doi: 10.1113/JP271339
Saito, J., Rimbara, E., Inaguma, S., Hasegawa, C., Kamiya, S., Mizuno, A., et al. (2022). Determining infected aortic aneurysm treatment using focused detection of Helicobacter cinaedi. Emerg. Infect. Dis. 28, 1494–1498. doi: 10.3201/eid2807.212505
Sarp, T. Z., Gode, S., Saribas, S., Ergin, S., Kasnak, G., Dinc, H. O., et al. (2021). The role of Helicobacter cinaedi in the development of atherosclerosis. Postepy Hig. Med. Dosw. 75, 529–536. doi: 10.2478/ahem-2021-0003
Sato, H., Ogino, A., Matsuzaka, S., Asami, Y., Kanbayashi, S., Masuda, M., et al. (2019). Ovarian abscess caused by Helicobacter cinaedi in a patient with endometriosis. IDCases 17:e00578. doi: 10.1016/j.idcr.2019.e00578
Sawaswong, V., Chanchaem, P., Kemthong, T., Warit, S., Chaiprasert, A., Malaivijitnond, S., et al. (2023). Alteration of gut microbiota in wild-borne long-tailed macaques after 1-year being housed in hygienic captivity. Sci. Rep. 13:5842. doi: 10.1038/s41598-023-33163-6
Shen, Z., Feng, Y., Rickman, B., and Fox, J. G. (2015). Helicobacter cinaedi induced typhlocolitis in Rag-2-deficient mice. Helicobacter 20, 146–155. doi: 10.1111/hel.12179
Shor, A., Kuo, C. C., and Patton, D. L. (1992). Detection of Chlamydia pneumoniae in coronary arterial fatty streaks and atheromatous plaques. S. Afr. Med. J. 82, 158–161.
Skirrow, M. B., Jones, D. M., Sutcliffe, E., and Benjamin, J. (1993). Campylobacter bacteraemia in England and Wales, 1981-91. Epidemiol. Infect. 110, 567–573. doi: 10.1017/S0950268800050986
Smet, A., Yahara, K., Rossi, M., Tay, A., Backert, S., Armin, E., et al. (2018). Macroevolution of gastric Helicobacter species unveils interspecies admixture and time of divergence. ISME J. 12, 2518–2531. doi: 10.1038/s41396-018-0199-5
Solnick, J. V., and Schauer, D. B. (2001). Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin. Microbiol. Rev. 14, 59–97. doi: 10.1128/CMR.14.1.59-97.2001
Suzuki, T., Kutsuna, S., Tsuboi, M., Ota, M., Hayakawa, K., and Ohmagari, N. (2019). Helicobacter cinaedi hepatic cyst infection with bacteremia. Emerg. Infect. Dis. 25, 603–604. doi: 10.3201/eid2503.180936
Suzuki, S., Naito, S., Numasawa, Y., Asada, M., Shoji, N., Zeniya, M., et al. (2019). Encephalopathy induced by high plasma and cerebrospinal fluid ceftriaxone concentrations in a hemodialysis patient. Intern. Med. 58, 1775–1779. doi: 10.2169/internalmedicine.1785-18
Takehara, T., Tani, T., Yajima, K., Watanabe, M., Otsuka, Y., and Koh, H. (2019). First case report of thyroid abscess caused by Helicobacter cinaedi presenting with thyroid storm. BMC Infect. Dis. 19:166. doi: 10.1186/s12879-019-3808-7
Takenaka, K., Sugimoto, T., Kawamura, K., Fukumoto, T., Onuma, K., Naito, T., et al. (2021). Recurrent fever due to Helicobacter cinaedi infection during R-CHOP chemotherapy in diffuse large B-cell lymphoma: case report and literature review. Case Rep. Oncol. 14, 1315–1322. doi: 10.1159/000518685
Taniguchi, T., Saeki, Y., Okayama, A., Hayashi, T., and Misawa, N. (2017). Extraintestinal infection of Helicobacter cinaedi induced by oral administration to Balb/c mice. Microbiol. Immunol. 61, 57–63. doi: 10.1111/1348-0421.12472
Taniguchi, T., Sekiya, A., Higa, M., Saeki, Y., Umeki, K., Okayama, A., et al. (2014). Rapid identification and subtyping of Helicobacter cinaedi strains by intact-cell mass spectrometry profiling with the use of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 52, 95–102. doi: 10.1128/JCM.01798-13
Taniguchi, T., Yamazaki, W., Saeki, Y., Takajo, I., Okayama, A., Hayashi, T., et al. (2016). The pathogenic potential of Helicobacter cinaedi isolated from non-human sources: adherence, invasion and translocation ability in polarized intestinal epithelial Caco-2 cells in vitro. J. Vet. Med. Sci. 78, 627–632. doi: 10.1292/jvms.15-0595
Taylor, N. S., Ge, Z., Shen, Z., Dewhirst, F. E., and Fox, J. G. (2003). Cytolethal distending toxin: a potential virulence factor for Helicobacter cinaedi. J. Infect. Dis. 188, 1892–1897. doi: 10.1086/379837
Togneri, A. M., Pérez, M. P., Vilches, V., Martinelli, I., Sadorín, R., Marone, S., et al. (2019). Helicobacter cinaedi bacteremia: presentation of the first cases reported in Argentina. Rev. Argent. Microbiol. 51, 148–152. doi: 10.1016/j.ram.2018.06.002
Tomida, J., Oumi, A., Okamoto, T., Morita, Y., Okayama, A., Misawa, N., et al. (2013). Comparative evaluation of agar dilution and broth microdilution methods for antibiotic susceptibility testing of Helicobacter cinaedi. Microbiol. Immunol. 57, 353–358. doi: 10.1111/1348-0421.12044
Totten, P. A., Fennell, C. L., Tenover, F. C., Wezenberg, J. M., Perine, P. L., Stamm, W. E., et al. (1985). Campylobacter cinaedi (sp. nov.) and Campylobacter fennelliae (sp. nov.): two new Campylobacter species associated with enteric disease in homosexual men. J. Infect. Dis. 151, 131–139. doi: 10.1093/infdis/151.1.131
Toyoshima, H., Tanigawa, M., Ishiguro, C., Tanaka, H., Nakanishi, Y., and Sakabe, S. (2022). Helicobacter cinaedi-infected aneurysm and vertebral osteomyelitis in an immunocompetent patient. IDCases 27:e01426. doi: 10.1016/j.idcr.2022.e01426
Ushach, I., and Zlotnik, A. (2016). Biological role of granulocyte macrophage colony-stimulating factor (GM-CSF) and macrophage colony-stimulating factor (M-CSF) on cells of the myeloid lineage. J. Leukoc. Biol. 100, 481–489. doi: 10.1189/jlb.3RU0316-144R
van der Ven, A. J., Kullberg, B. J., Vandamme, P., and Meis, J. F. (1996). Helicobacter cinaedi bacteremia associated with localized pain but not with cellulitis. Clin. Infect. Dis. 22, 710–711. doi: 10.1093/clinids/22.4.710
Vandamme, P., Falsen, E., Pot, B., Kersters, K., and De Ley, J. (1990). Identification of Campylobacter cinaedi isolated from blood and feces of children and adult females. J. Clin. Microbiol. 28, 1016–1020. doi: 10.1128/jcm.28.5.1016-1020.1990
Vandamme, P., Falsen, E., Rossau, R., Hoste, B., Segers, P., Tytgat, R., et al. (1991). Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. Nov. Int. J. Syst. Bacteriol. 41, 88–103. doi: 10.1099/00207713-41-1-88
Verhaar, B. J. H., Prodan, A., Nieuwdorp, M., and Muller, M. (2020). Gut microbiota in hypertension and atherosclerosis: a review. Nutrients 12:2982. doi: 10.3390/nu12102982
Wanschel, A., Seibert, T., Hewing, B., Ramkhelawon, B., Ray, T. D., van Gils, J. M., et al. (2013). Neuroimmune guidance cue Semaphorin 3E is expressed in atherosclerotic plaques and regulates macrophage retention. Arterioscler. Thromb. Vasc. Biol. 33, 886–893. doi: 10.1161/ATVBAHA.112.300941
Yasuda, I., Hasegawa, K., Tokuyama, H., Washida, N., Shinozuka, K., Yasuda, M., et al. (2019). A case report of autosomal dominant polycystic kidney disease under peritoneal Dialysis with cyst infection and culture-positive peritoneal fluid. Clin. Med. Insights Case Rep. 12:117954761984686. doi: 10.1177/1179547619846860
Keywords: Helicobacter cinaedi, atheroscelorsis, foam cell, macrophage, cardiovascular diseases
Citation: Voronina AK and Arapidi GP (2024) Helicobacter cinaedi bacterium association with atherosclerosis and other diseases. Front. Microbiol. 15:1371717. doi: 10.3389/fmicb.2024.1371717
Edited by:
Alain Pierre Gobert, Vanderbilt University Medical Center, United StatesReviewed by:
Bekir Kocazeybek, Istanbul University-Cerrahpasa, TürkiyeSilvio Ragozzino, University Hospital of Basel, Switzerland
Copyright © 2024 Voronina and Arapidi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alice K. Voronina, YWxpY2VfY29yYmVhdUBtYWlsLnJ1
 Alice K. Voronina
Alice K. Voronina Georgij P. Arapidi
Georgij P. Arapidi