
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 04 March 2024
Sec. Microbial Physiology and Metabolism
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1369760
This article is part of the Research TopictRNA and Protein Synthesis in MicroorganismsView all 10 articles
Ribosomes stall on truncated or otherwise damaged mRNAs. Bacteria rely on ribosome rescue mechanisms to replenish the pool of ribosomes available for translation. Trans-translation, the main ribosome-rescue pathway, uses a circular hybrid transfer-messenger RNA (tmRNA) to restart translation and label the resulting peptide for degradation. Previous studies have visualized how tmRNA and its helper protein SmpB interact with the stalled ribosome to establish a new open reading frame. As tmRNA presents the first alanine codon via a non-canonical mRNA path in the ribosome, the incoming alanyl-tRNA must rearrange the tmRNA molecule to read the codon. Here, we describe cryo-EM analyses of an endogenous Escherichia coli ribosome-tmRNA complex with tRNAAla accommodated in the A site. The flexible adenosine-rich tmRNA linker, which connects the mRNA-like domain with the codon, is stabilized by the minor groove of the canonically positioned anticodon stem of tRNAAla. This ribosome complex can also accommodate a tRNA near the E (exit) site, bringing insights into the translocation and dissociation of the tRNA that decoded the defective mRNA prior to tmRNA binding. Together, these structures uncover a key step of ribosome rescue, in which the ribosome starts translating the tmRNA reading frame.
Translation of messenger RNAs (mRNAs) into functional proteins is crucial for all living organisms. While the core translation mechanism is conserved across life kingdoms (Melnikov et al., 2012; Voorhees and Ramakrishnan, 2013; Korostelev, 2022), adaptation to different conditions has led to the evolution of distinct translational control strategies (Buskirk and Green, 2017). One challenge encountered by translating ribosomes is the truncation or other damage of mRNA molecules, resulting in ribosome stalling at the truncation or damage site. In growing E. coli, between 0.4% (Moore and Sauer, 2005) and 4% (Ito et al., 2011) of mRNAs are estimated to be damaged at a given time. Several strategies have evolved to “rescue” stalled ribosomes and replenish the pool of active ribosomes (Keiler, 2015; Korostelev, 2021; Muller et al., 2021; Kurita and Himeno, 2022). Trans-translation, the main strategy conserved among eubacteria, allows the ribosome to switch from the damaged mRNA to a different open reading frame, targeting the mRNA and incomplete peptide for degradation and completing translation on a conventional stop codon (Komine et al., 1994; Tu et al., 1995; Keiler et al., 1996; Karzai et al., 1999; Yamamoto et al., 2003). Perturbation of trans-translation in most eubacteria leads to the accumulation of stalled ribosome complexes and inability to recover from stress (Janssen and Hayes, 2012; Schopping et al., 2022).
Trans-translation is accomplished by a hybrid transfer-messenger RNA (tmRNA), comprising a tRNA-like domain (TLD), an mRNA-like domain (MLD), and four pseudoknots (PK1 through PK4) that form a circularized structure (Figures 1A,B; Janssen and Hayes, 2012; Giudice and Gillet, 2013). The TLD, comprising a tRNA-like acceptor arm charged with alanine, associates with small protein B (SmpB), which functionally mimics the tRNA’s anticodon stem loop (Komine et al., 1994; Ushida et al., 1994; Karzai et al., 1999; Gutmann et al., 2003; Weis et al., 2010). The MLD contains a short internal open reading frame, which connects with the TLD via PK1 and a single-stranded linker (Figure 1C).
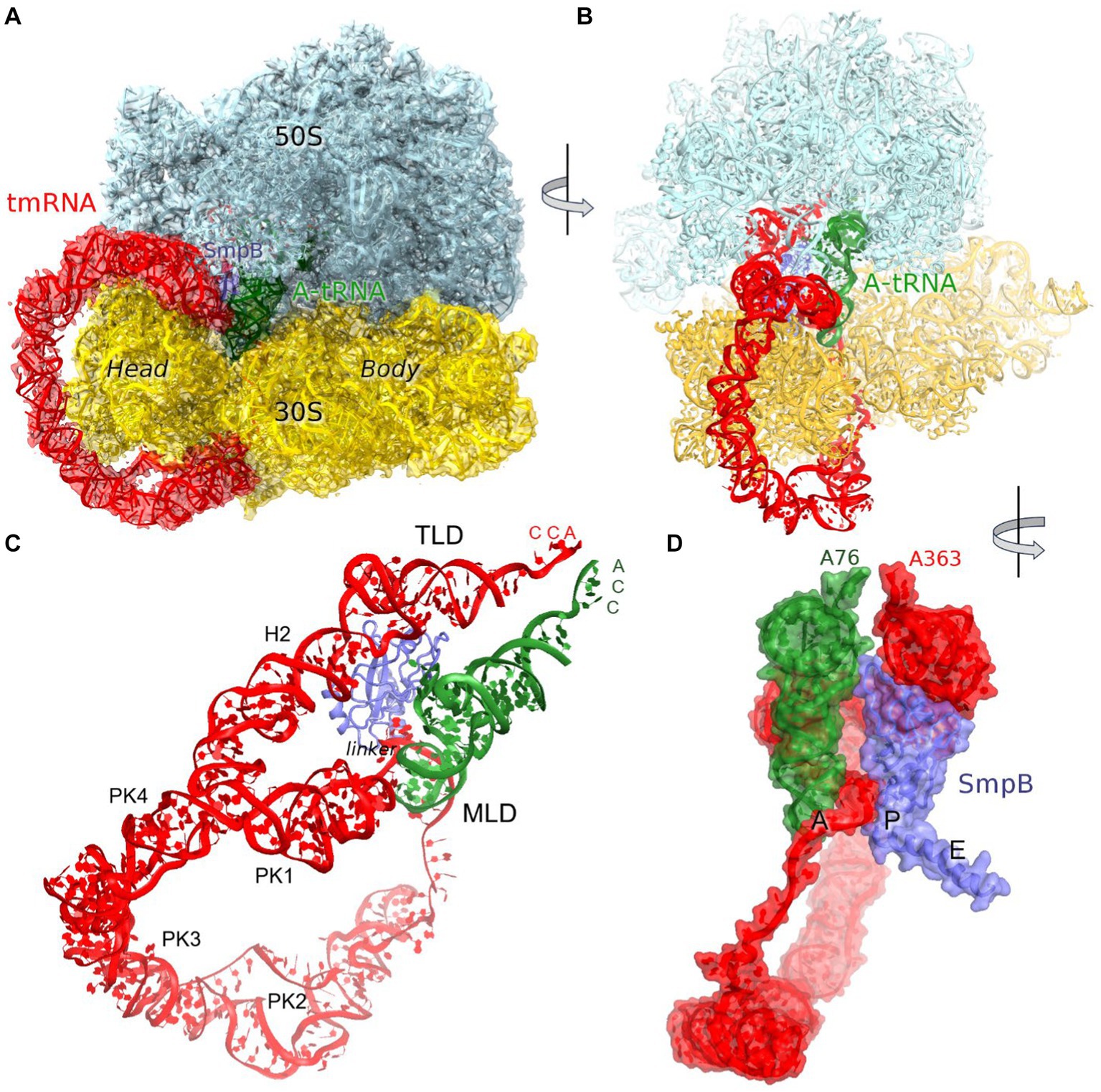
Figure 1. Cryo-EM structure of Escherichia coli 70S•tmRNA complex with tRNAAla in the A site. (A) 3.7-Å cryo-EM density segmented to show the ribosomal subunits (cyan and yellow), tmRNA (red), SmpB (purple) and A-site tRNA (green). (B) Front view of the 70S structure with tmRNA, SmpB and A-tRNA; (C) Relative positions of tmRNA, SmpB and A-tRNA, with tmRNA domains labeled. (D) Close-up view of tmRNA, SmpB and A-tRNA facing the CCA ends of the tRNA and TLD (in the A and P sites, respectively) and SmpB C-terminus in the E site, rendered as molecular surfaces.
Trans-translation starts with the binding of the TLD•SmpB complex to the ribosomal A site followed by EF-G-catalyzed translocation of TLD•SmpB to the P site. These steps have been characterized by X-ray crystallography and cryo-EM of in vitro assembled tmRNA-bound ribosomes, bringing key insights into the recognition of the stalled ribosomes, formation of the TLD-peptide complex, and tmRNA rearrangements upon translocation (Weis et al., 2010; Neubauer et al., 2012; Ramrath et al., 2012; Rae et al., 2019; Guyomar et al., 2021). The alanyl-TLD is delivered to the ribosome by EF-Tu, similarly to canonical amino-acylated tRNAs (Neubauer et al., 2012; Miller and Buskirk, 2014). Upon accommodation of the TLD in the A site of the 50S subunit, the stalled peptide is transferred to the alanine residue on tmRNA. The C-terminal helix of SmpB initially binds in the vacant mRNA entry tunnel of the 30S subunit to recognize the ribosomes with truncated mRNAs. During translocation, SmpB moves along with the TLD to the P site, while its C-terminal helix “leaps” into the E site, thus freeing the A site (Rae et al., 2019; Guyomar et al., 2021). This allows the MLD of tmRNA to present the first codon—GCA coding for alanine—for recognition by the canonical alanyl-tRNAAla. Cryo-EM studies demonstrated tRNA binding to the A site in the presence of tmRNA in E. coli and M. smegmatis (Fu et al., 2010; Mishra et al., 2018), however low > 12 Å resolutions prevented detailed characterization of this trans-translation step. Recent higher-resolution cryo-EM structures of the translocated tmRNA with a vacant A site showed that tmRNA linker traverses the A site, partially blocking the canonical tRNA binding location (Rae et al., 2019; Guyomar et al., 2021). The tmRNA therefore must reorganize to allow the binding of tRNAAla and translation of the tmRNA coding sequence.
In this work, we describe a tmRNA-bound complex that copurified with E. coli 70S ribosomes and features an endogenous tmRNA structure (Methods). The complex contains tRNAAla in the A site, stabilized by interactions with tmRNA and SmpB. A fraction of this complex also contains a deacyl tRNA near the E site, revealing a non-canonical tRNA binding site that may be sampled in a preceding step of tmRNA translocation.
We performed maximum-likelihood classification of a large cryo-EM dataset (~1.5 million particles) collected from 70S ribosomes that were purified from E coli at the exponential growth phase and then incubated with defined mRNA, tRNAPhe, tRNAfMet and stringent factor RelA (Supplementary Figure S1; Methods). Remarkably, we found that ~14,000 of the 1.15 million 70S ribosome particles contain tmRNA (~1.2%). Because neither tmRNA nor tRNAAla were added to the 70S sample, the tmRNA-bound ribosomes must represent endogenous E. coli trans-translation complexes. Since the tmRNA-bound ribosomes formed prior to the addition of RelA (Methods), they likely represent a homeostatic trans-translation complex. Indeed, the 1.2% recovery of trans-translation ribosomes comports with the cellular estimates of rescue-complex abundance (Moore and Sauer, 2005; Ito et al., 2011).
The predominant 3.7 Å cryo-EM reconstruction with circularized tmRNA density features a non-rotated ribosome with strong densities for TLD•SmpB in the P site and tRNA in the A site (Figure 1A; Supplementary Figure S1). Our extensive classification did not identify tmRNA in other ribosome sites, similarly to the recent study of M. smegmatis ribosomes (Mishra et al., 2018), suggesting that this complex represents an intermediate accumulating during trans-translation. The ribosome and the endogenous tmRNA, wrapped around the head of the 30S subunit (Figure 1B), are overall similar to those in post-translocation 70S complexes assembled from in vitro transcribed tmRNA constructs (Rae et al., 2019; Guyomar et al., 2021). The ribosomal intersubunit rotation state during elongation correlates with stages of decoding and translocation (Cornish et al., 2008; Ermolenko and Noller, 2011; Rodnina, 2018). The non-rotated ribosome with A-site tRNA corresponds to a post-decoding stage, preceding the translocation of the tRNA into the P site that requires intersubunit rotation. In addition, translocation involves a “swiveling” motion of the head domain of the 30S subunit (Ratje et al., 2010). Another mode of the head movement, known as “tilt,” normally occurs during initiation, when the free 30S subunit interacts with the initiator tRNA sampling the P site (Hussain et al., 2016; Jahagirdar et al., 2020). While mechanistically similar to the elongation ribosomes with P- and A-site tRNAs, the tmRNA-bound ribosome is reorganized via a head tilt to accommodate the bulky tmRNA. As tmRNA helix 2 and pseudoknot 1 are placed between the 30S head and the 50S central protuberance (Figures 1B,C), the head is tilted 7° away from the large subunit placing uS19 ~ 15 Å farther than in canonical elongation complexes (measured at Gly25; Figures 2A,B). The A-site finger (ASF) of the large subunit, involved in tRNA accommodation (Sanbonmatsu et al., 2005; Loveland et al., 2020), is shifted by ~20 Å (measured at the U887 tip) to dock onto PK1 (Figure 2C).
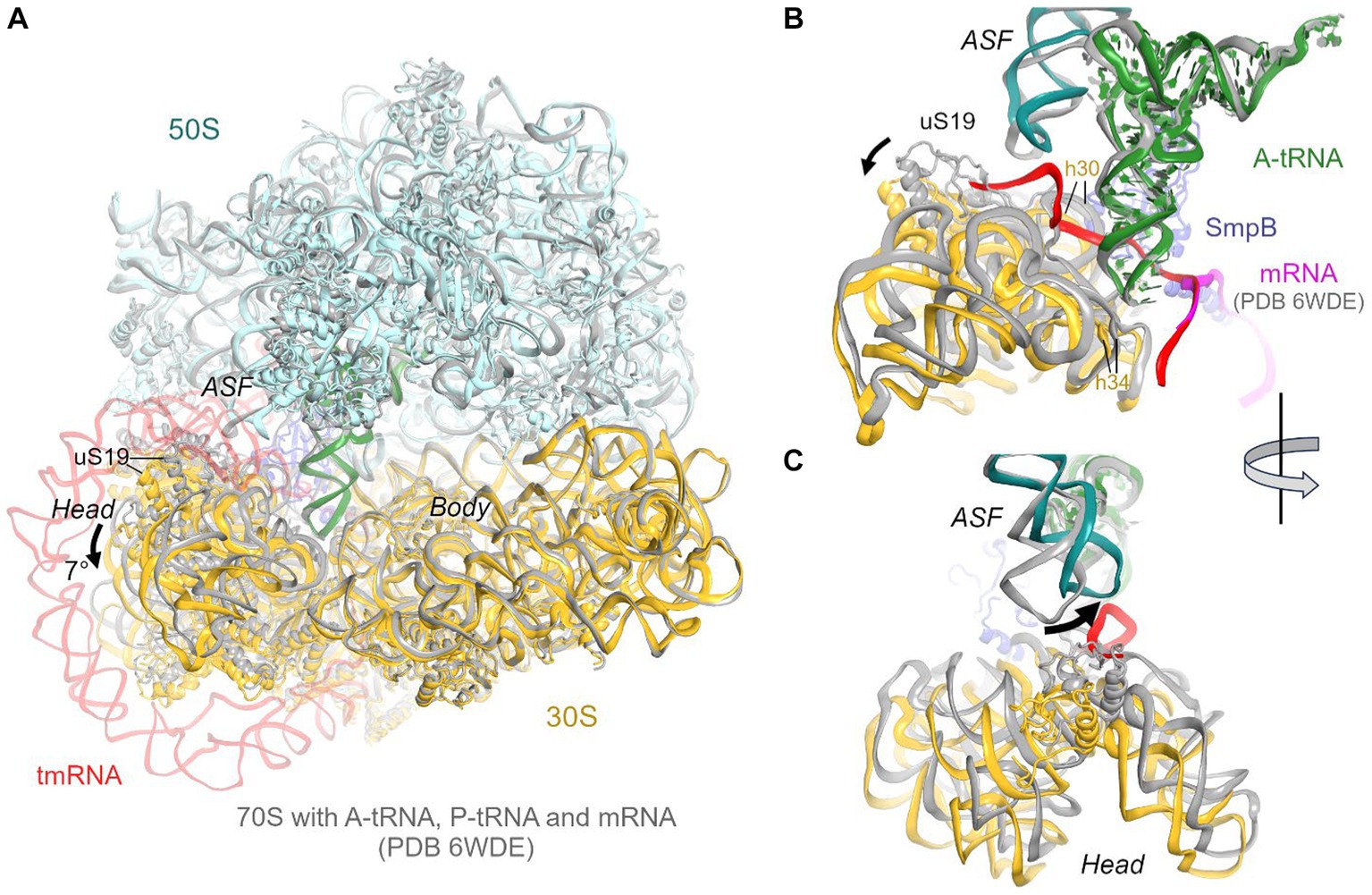
Figure 2. Comparison of the 70S•tmRNA•tRNAAla complex with the canonical Escherichia coli 70S elongation complex bound with mRNA and three tRNAs (PDB 6WDE). (A) Superposition of the 70S•tmRNA•tRNAAla structure (colored) with the 70S•mRNA•tRNA3 structure (gray) shows overall similar ribosome conformations except for the 7° tilt of the 30S head domain. Structures were superposed by aligning 23S rRNA (here and in other figures). (B) Close-up view of the similarly positioned tRNA in the A site of the tmRNA-bound (colored) and canonical elongation-state (gray) ribosomes. (C) Close-up view showing different positions of the A-site finger (ASF; H38 of 23S rRNA) in the tmRNA-bound (colored) and canonical elongation-state (gray) ribosomes.
The structure brings insight into tRNA positioning in the A site of the tmRNA-bound ribosome. On the 50S subunit, despite the large shift of the 50S ASF, the A-site tRNA elbow is stabilized by packing against the ASF (Figure 2B). Here, the C19-G56 pair likely stacks on the bulged A896 of the ASF (Supplementary Figure S2A), similarly to canonical tRNA-bound complexes. Accordingly, the position of the A-site tRNA relative to the 50S subunit is nearly identical to those in tRNA-bound structures, emphasizing the invariant mechanism of tRNA accommodation for peptidyl transfer. The acceptor arm with the 3′ terminal CCA is inserted into the A site next to the CCA end of the tmRNA (Figure 1D). In the polypeptide tunnel, scattered density suggests compositional and conformational heterogeneity of peptides in the endogenous rescue complexes on different mRNAs.
Due to the 30S head tilt, interactions between the A-site tRNA and the 30S subunit slightly differ from those in canonical tRNA-bound complexes. Whereas helix 30 of 16S rRNA normally binds near the anticodon stem of tRNA (at nt 42), helix 30 is retracted by ~9 Å (measured at U956) to accommodate the tmRNA linker connecting PK1 and MLD. Universally conserved C1054, which bulges from h34, normally buttresses the anticodon by packing on the ribose of nt 34. But in the tmRNA-bound complex, the 30S head tilt shifts C1054 by ~4 Å, detaching it from tRNAAla (Figure 2B).
The loss of interactions between tRNAAla and the 30S head is partially compensated by interactions with tmRNA and SmpB (see below), firmly positioning tRNAAla in the 30S decoding center. Local density confirms tRNAAla-specific nucleotides and Watson-Crick base pairing of the tRNA UGC anticodon with the corresponding GCA codon of tmRNA (Figure 3C; Supplementary Figures S2B,C). The codon-anticodon helix is stabilized by interactions with ribosomal decoding-center nucleotides G530, A1492, and A1493 (Figure 3D). The G530 loop of the shoulder domain is disengaged from the h34 of the shifted head, unlike in canonical tRNA-bound ribosomes where A532 packs on G1207. Despite this difference, G530 stabilizes the tRNA anticodon by hydrogen bonding with the ribose of G35, and both nucleotides are placed nearly identical to those in canonical tRNA-ribosome complexes. The adenosines A1492 and A1493 of the body domain stabilize the opposite side of the codon-anticodon helix by hydrogen-bonding with the riboses of tmRNA codon nucleotides G90 and C91. Thus, the ribosome recognizes and stabilizes the tmRNA-tRNAAla codon-anticodon helix via the universally conserved G530 and A-minor interactions, as in canonical elongation complexes (Ogle et al., 2001; Demeshkina et al., 2012; Loveland et al., 2017).
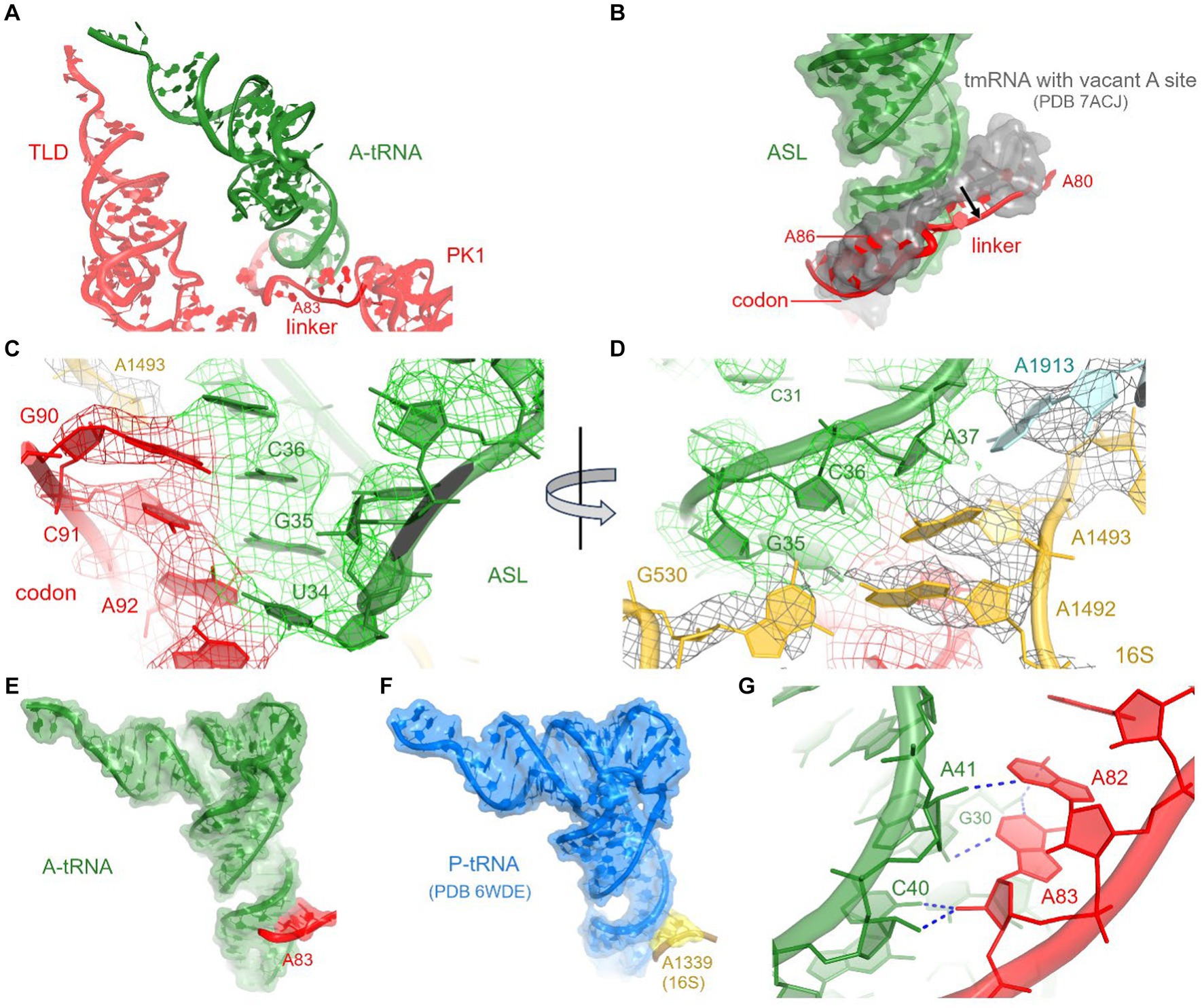
Figure 3. Interactions between A-tRNA and tmRNA. (A) Positions of A-tRNA and tmRNA domains. (B) The tmRNA linker (red) shifts to accommodate the A-site tRNA, relative to its position in the tmRNA-bound complex with a vacant A site (gray backbone and molecular surface) in PDB 7ACJ. (C,D) Cryo-EM density in the decoding center, showing codon-anticodon interactions (C) and interactions with 30S and 50S nucleotides (D). (E) Interactions of the tmRNA linker with the anticodon stem of A-tRNA resemble A-minor interactions (E) and are similar to those between 16S rRNA and tRNAfMet in the P site (F) of initiation and elongation ribosome structures (PDB 6WDE is shown). (G) Close-up view of the A-minor-like interactions between tmRNA nucleotides and A-site tRNA.
To accommodate tRNAAla in the A site, the adenosine-rich linker of tmRNA (80AAAAAU85) shifts away from its position in pre-decoding structures (Rae et al., 2019; Guyomar et al., 2021), where the linker traverses the A site (Figures 3A,B). Adenosines A82 and A83 support the minor groove of the tRNAAla anticodon stem at nucleotides C40 and A41 (Figures 3E,G). This interaction appears similar to the A-minor-like interaction in the 30S P site, where conserved 16S nucleotides G1338 and A1339 pack at the minor groove of initiator tRNAfMet (Figure 3F) to assist translation initiation and perhaps other translation stages (Lancaster and Noller, 2005; Korostelev et al., 2006; Hussain et al., 2016). A-minor-like interactions are a unique tertiary structure that plays critical roles in RNA stabilization, including tetraloop-receptor recognition (Cate et al., 1996; Doherty et al., 2001; Nissen et al., 2001; Battle and Doudna, 2002) and mRNA decoding described above. Furthermore, their modest interaction surface and thermodynamic stability (Doherty et al., 2001) allow for local structural rearrangements, such as tRNA dynamics during mRNA decoding and translocation (Ogle et al., 2001; Demeshkina et al., 2012; Loveland et al., 2017; Carbone et al., 2021). Thus, the tmRNA linker not only replaces the tRNA interactions with the 30S head during decoding but may also transiently stabilize tRNAAla and/or disengage from the tRNAAla in the subsequent—highly dynamic—translocation step. The functional role of this A-minor interaction is supported by the conservation of adenosines positioned 6–10 nucleotides upstream of the first codon of tmRNA in most bacterial species (Zwieb et al., 1999). In species without consecutive adenosines in this position (e.g., Thermus thermophilus tmRNA), however, it remains to be seen how tmRNA interacts with tRNAAla (Op De Bekke et al., 1998; Kaur et al., 2006).
SmpB adopts the same overall conformation as in the structures without A-site tRNA (Rae et al., 2019; Guyomar et al., 2021). Here, the protein’s globular domain binds the 30S P site, with the His79 loop sandwiched between TLD and the elbow of tRNAAla. To accommodate the tRNA, the loop is slightly rearranged, bringing His79 into contact with the tRNAAla backbone at nucleotide 17. The C-terminal helix occupies the mRNA binding pocket in the E site (Figure 1D).
In our tmRNA-containing cryo-EM reconstructions, low-density features in the E site suggested sub-stoichiometric tRNA. To better resolve this density, we subclassified the cryo-EM maps, using a mask covering the E site, yielding a 3.9-Å resolution class with tRNA near the canonical E site (Figure 4A; Supplementary Figures S1, S2D). Other complex constituents, including the A-site tRNA, are similar to the complex described above.
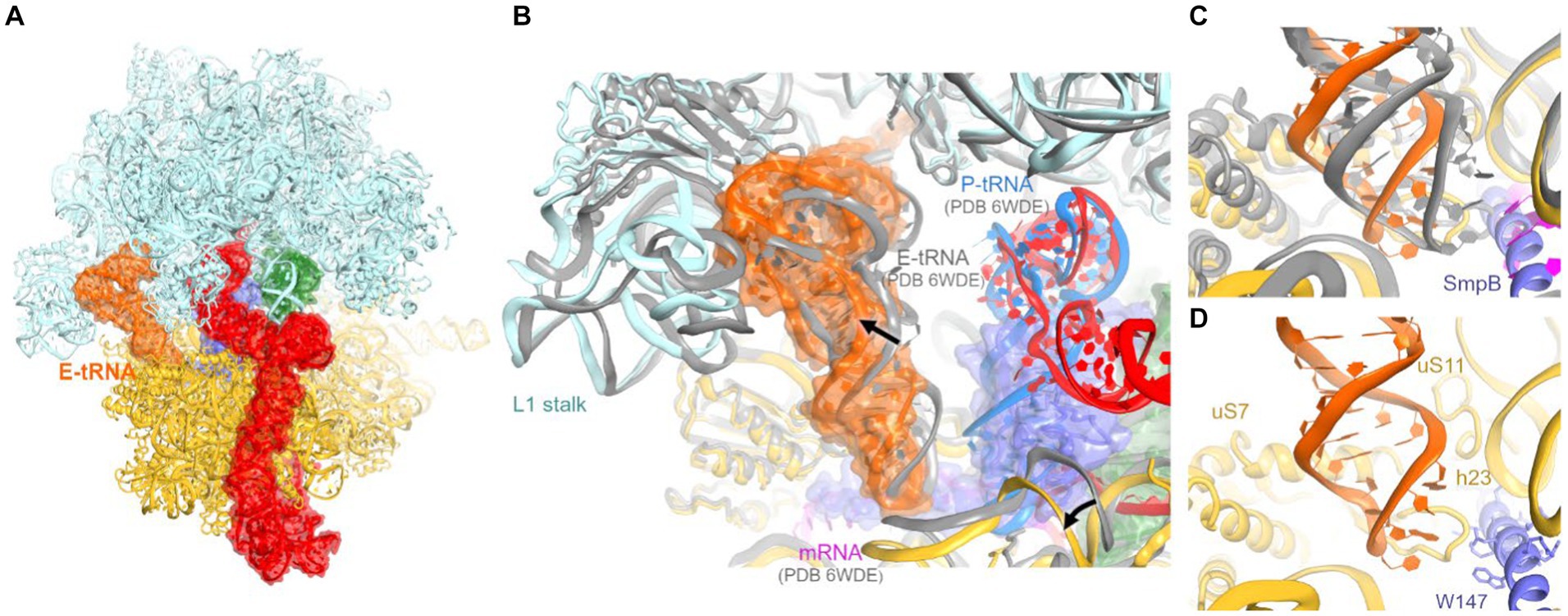
Figure 4. Cryo-EM structure of E. coli 70S•tmRNA complex with tRNAs in the A (green) and E (orange) sites. (A) Front view of the 70S structure with tmRNA, SmpB, A-tRNA and E-tRNA rendered as molecular surfaces. (B) Position of the tRNA in the E site of the 70S•tmRNA•tRNAAla complex (colored) differs from that in the canonical 70S•mRNA•tRNA3 structure (gray ribosome and E-tRNA, magenta mRNA and blue P-tRNA). (C,D) Positions of the anticodon stem loops of E-site tRNAs relative to 16S and SmpB (in the 70S•tmRNA•tRNAAla complex) or mRNA (in the 70S•mRNA•tRNA3 complex).
Interactions of the tRNA with the 30S E site differ from canonical ribosome structures with an untilted head domain. In canonical complexes, E-site tRNA binds in the cleft between the head (near ribosomal protein uS7) and body (near G693 of h23 of 16S rRNA). In the tmRNA-containing complexes, however, the tRNA is shifted ~8 Å away from this binding pocket, despite ample space near the C-terminal helix of SmpB (Figures 4B–D). Furthermore, the tRNA is partially retracted from the E site of the head, formed by h29 of 16S rRNA and uS7. Here, the anticodon is shifted ~9 Å away from the tip of the β-hairpin of uS7 (at Gly80), where the tRNA anticodon resides in elongation complexes (Figures 4C,D). In this position, the anticodon stem loop is loosely held near h29 (at A1339) and the β-sheet (at Arg78) and C-terminal α-helix (near Arg142) of uS7. On the 50S subunit, the tRNA interacts with the L1 stalk (elbow) and H88 of 23S rRNA, where the terminal nucleotide A76 is inserted into. These contacts are nearly identical to those observed in numerous structural studies of tRNA-bound complexes (Figure 4B).
Previous structural studies of the P-site bound tmRNA did not report E-site tRNA, suggesting that tRNA bound to the preceding truncated mRNA readily dissociates upon tmRNA translocation. The tRNA in our map likely results from the addition of tRNAPhe and tRNAfMet to the ribosome sample (Methods). Nevertheless, this observation may report a transient tRNA binding state sampled during or immediately after tmRNA translocation. The structure underlines that unlike the A and P site, where the tRNA interacts closely with both the 30S and 50S subunits, E-site positioning is driven primarily by interactions with the 50S subunit. Indeed, these interactions are established during the initial stages of translocation, in which the acceptor arm of deacyl-tRNA is spontaneously transferred from the P site to the E site on the 50S subunit, while the anticodon stem loop remains bound to the 30S subunit (Agirrezabala et al., 2008; Fei et al., 2008; Julian et al., 2008; Cornish et al., 2009). In addition, non-canonical E-like tRNAs were found in other studies of bacterial ribosomes (E-out; Zhang et al., 2018) and eukaryotic ribosomes (Z-site; Brown et al., 2018). While retaining the invariant interactions with the large-subunit L1 stalk and the CCA-binging pocket, these structures feature different tRNA interactions with the small subunit. Distinct binding modes allow for increased tRNA dynamics in the E site, underlying the tRNA-dissociation function of the E site (Brown et al., 2018; Zhang et al., 2018). Our findings expand the repertoire of possible tRNA rearrangements during tRNA departure from the ribosome.
In conclusion, our cryo-EM analyses visualize how the first tmRNA codon is decoded and how tmRNA rearranges to accommodate tRNAAla. Interactions with tmRNA stabilize tRNAAla, which binds nearly identically to canonical A-site tRNA despite a substantial tilt of the 30S head. Future work will detail whether A-minor-like interactions of tmRNA with the tRNA anticodon stem occur in bacterial species, whose tmRNA linker does not contain continuous adenosines. Further, rearrangements of these and other interactions of tmRNA•SmpB with tRNAAla and the ribosome during translocation of tRNAAla to the P site remain to be visualized. Such structural studies may inform the development of new drugs that target trans-translation, a promising target for antibacterial therapeutics (Campos-Silva et al., 2021; Marathe et al., 2023).
70S ribosomes were prepared from MRE600 E. coli essentially as described (Moazed and Noller, 1986, 1989) and stored in the ribosome-storage buffer A (20 mM HEPES (pH 7.5), 100 mM KCl, 10.5 mM MgCl2, 0.5 mM EDTA, 5 mM β-mercaptoethanol) at −80°C. In short, MRE600 E. coli stock was grown on LB agar plates at 37°C. Then, one colony from the plate was inoculated in 100 mL LB media and grown at 37°C overnight in an incubated shaker at ~220 rpm. Forty eight milliliters of overnight culture was inoculated into 6 L LB media and the culture was incubated at 37°C to mid-log phase (OD600 0.5–1.0).
Escherichia coli cells obtained from a 6 L culture were suspended in 50 mL cold buffer A and lysed using a microfluidizer (Microfluidics, United States) at 18 k psi. The lysate was clarified using a JA-20 rotor at 39,200 × g, 4°C, for 20 min. The clarified supernatant was layered onto 35 mL (per tube) of 37.7% sucrose in buffer A. Ribosomes were sedimented onto the sucrose cushion by ultracentrifugation in a 45 Ti rotor at 185,677 × g (40,000 rpm), 4°C, for 20 h. The ribosome pellet was dissolved in 2 mL of buffer A. The ribosome solution was transferred to 1 mL microcentrifuge tubes and spun at 13 K rpm at 4°C for 10 min. Supernatant was transferred to a 50 mL tube, the volume was adjusted to 40 mL using cold buffer B (70 mM Tris–HCl (pH 7.0), 500 mM NH4Cl, 15 mM MgCl2, 0.5 mM EDTA, 5 mM β-mercaptoethanol), and ribosomes were sedimented in 70 Ti tubes at 310,801 × g (55,000 rpm), 4°C, for 2 h. The ribosome pellet was resuspended in 1 mL buffer A, aliquoted and stored at −80°C.
The ribosome complex for cryo-EM analyses was prepared as described (Loveland et al., 2016) with some modifications. 70S ribosomes at a final concentration of 0.4 μM were activated in 20 mM HEPES-KOH (pH 7.5), 120 mM KCl, 15 mM MgCl2, 2 mM spermidine, and 0.05 mM spermine at 42°C for 15 min. Activated ribosomes were incubated with 0.8 μM mRNA (5′ GGCAAGGAGGUAAAAAUGUUCAAAAAA 3′), 0.8 μM tRNAfMet, and 1 μM tRNAPhe (all final concentrations) at 37°C for 30 min. The sample was then incubated with 4 μM RelA, 15 μM Adenosine-5′-[(α,β)-methyleno] triphosphate APCPP; Thermo scientific and 15 μM Guanosine triphosphate (GTP; Thermo scientific) to assemble a stringent-response 70S complex, for 15 min at room temperature. The final volume of the sample was 30 μL.
Carbon-coated EM grids (Ultrathin Carbon on Quantifoil®, 2 μm Diameter Holes, 1 μm Separation, mounted on a 200 M Cu grid coated with a 2-nm thin layer of carbon; TedPella) were glow discharged at 20 mA with a negative polarity setting for 30 s in a PELCO easiGlow glow discharge unit. 3 μL of the 70S sample was applied to the grid. Grids were blotted for 4 s with a blotting force of 7 and plunged into liquid ethane using a Vitrobot Mark IV (ThermoFisher Scientific), whose chamber was pre-equilibrated to 4°C and 95% humidity.
Two data sets were collected on a UMass Chan Cryo-EM Facility Talos Arctica electron microscope (ThermoFisher Scientific) operating at 200 kV and equipped with a K3 direct electron detector (Gatan Inc.) targeting 0.55–1.1-μm underfocus. Data were collected using SerialEM (Mastronarde, 2005), with beam tilts to record several movies at each stage position. The datasets contain 4,016 movies (total dose of 29.9 e−/Å2 on the sample), yielding 474,262 particles and 8,021 movies (30.4 e−/Å2 on the sample), yielding 1,039,487 particles. Movies were aligned during data collection using IMOD (Kremer et al., 1996) to decompress frames, apply the gain reference, and to correct for image drift and particle damage and bin the super-resolution pixel by 2.
CTF parameter determination, reference-free particle picking, and stack creation were carried out in cisTEM (v1.0-beta; Grant et al., 2018). Particle alignment and refinement were carried out in FREALIGNX (Lyumkis et al., 2013). Data processing was initially performed independently for each dataset (Supplementary Figure S1A). To speed up the processing, 2×− and 4×−image stacks were prepared using resample.exe, which is part of the FREALIGN distribution (Lyumkis et al., 2013). The initial model for particle alignment of 70S maps was the 11.5 Å resolution EMDB-1003 (Gabashvili et al., 2000), sampled to match 4×−binned image stack using resample.exe. Three rounds of mode 3 search alignment to 25 Å were run using the 4×−binned stack. Next, 25–30 rounds of mode 1 refinement were run with the 4×−binned, 2×−binned, and eventually unbinned stacks until the resolutions stopped improving, to the final resolutions of 2.8 Å and 2.7 Å of the overall maps. 3D maximum-likelihood classification into 20 classes was performed in FREALIGN v9.11 to separate 70S conformations, 50S subunits, and junk (poorly aligned or damaged) particles. An unexpected class emerged in each stack, featuring density near the 30S head, which connects the mRNA tunnel with the A-site finger and P site. The 70S classes with different tRNA occupancies and ribosome conformations (including the tmRNA class) were extracted into a stack per data set, using merge_classes.exe from FREALIGN distribution. Two 70S stacks were merged using IMOD 4.7 (Kremer et al., 1996).
The merged 70S stack was refined as described above, yielding a final average 70S reconstruction at 2.8 Å resolution. The refined parameters were used to run a 3D maximum-likelihood classification into 32 classes without a mask, with an ASF-covering mask, or with the A-site-covering mask. All masks were “spherical,” also known as “2D” masks on micrographs (Grigorieff, 2016), as opposed to specifically shaped “3D” masks (Supplementary Figure S1A). The tmRNA class was found in the no-mask and ASF-mask classifications. Particles with tmRNA density resulting from the ASF-mask classification were extracted into a substack. The tmRNA substack was classified at 4× with a P- and A-site covering mask or the E-site mask to further purify the tmRNA-containing density (P-A mask) or the E-tRNA-containing density (E mask). The unbinned stack was used to yield the resulting cryo-EM reconstructions with tmRNA (with A-tRNA and partial E-tRNA) and with tmRNA (with full-occupancy A-tRNA and E-tRNA) at resolutions of 3.7 Å and 3.9 Å, respectively (Supplementary Figure S1A).
Fourier Shell Correlation (FSC) curves were calculated by FREALIGNX for even and odd particle half-sets (Supplementary Figure S1B). The maps used for structure refinements was B-factor sharpened using the B factor of −100 Å2 up to 3.4 Å (tmRNA with A-tRNA) and −50 Å2 to 3.8 Å (tmRNA with A-tRNA and E-tRNA), using bfactor.exe (included with the FREALIGN distribution; Lyumkis et al., 2013).
Structure of the non-rotated 70S•tRNA3 complex (V-B; PDB 6WDE; Loveland et al., 2020) and structures of 70S•tmRNA complexes (PDB: 6Q98 and 7ACJ; Rae et al., 2019; D’Urso et al., 2023) were used as starting models for 70S ribosome and tmRNA fitting, respectively. The model of tRNAAla (GGC anticodon) from PDB:6OF6 (Nguyen et al., 2020) was modified to fit the cryo-EM map and match the nucleotide sequence of tRNAAla (UGC). The 50S, 30S and tmRNA domains were fitted using UCSF Chimera 1.6 (Goddard et al., 2018; Pettersen et al., 2021) and locally modeled in Pymol1.2r1 (DeLano, 2002). The fitted structures were refined conservatively, using secondary-structure restraints and low simulated-annealing temperatures (100 K, 300 K or 500 K), against cryo-EM maps using phenix.real_space_refine v1.19.2 (Adams et al., 2010). Refinement parameters, such as the relative weighting of stereochemical restraints and experimental energy term, were optimized to produce the optimal structure stereochemistry and real-space correlation coefficients (Supplementary Table S1). B-factors of the models were refined at the final stages using phenix.real_space_refine. Structure stereochemistry validation was performed using phenix.molprobity.
Structure superpositions and distance calculations were performed in PyMOL. To calculate the angles of the 30S rotation and head tilt, 23S rRNAs of corresponding structures were aligned using PyMOL, and the angle between 16S domains were measured in Chimera. Figures were prepared in PyMOL and Chimera.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author. The structural models generated in this study have been deposited in the RCSB Protein Data Bank under the following accession codes: 8VSA (https://www.rcsb.org/structure/8VSA; 70S with tmRNA, SmpB and A-site tRNAAla) and 8VS9 (https://www.rcsb.org/structure/8VS9; 70S with tmRNA, SmpB, A-site tRNAAla and E-site tRNA). The cryo-EM maps described in this study have been deposited in the Electron Microscopy Database under the following accession codes: EMD-43491 and EMD-43490, for the complexes without and with E-site tRNA density, respectively.
DT: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing, Conceptualization. YZ: Methodology, Resources, Writing – review & editing. AK: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported in part by the National Institutes of Health grants R35 GM127094 and R56 AI151372 (AK).
The authors thank Christna Ouch for grid screening and data collection at the cryo-EM facility at UMass Chan Medical School; Christine Carbone, Dmitrij Golovenko, Anna B. Loveland, Zahra Seraj, and Denis Susorov for suggestions on data processing; Darryl Conte and other members of the Korostelev lab for comments on the manuscript and insightful discussions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1369760/full#supplementary-material
Adams, P. D., Afonine, P. V., Bunkoczi, G., Chen, V. B., Davis, I. W., Echols, N., et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221. doi: 10.1107/S0907444909052925
Agirrezabala, X., Lei, J., Brunelle, J. L., Ortiz-Meoz, R. F., Green, R., and Frank, J. (2008). Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol. Cell 32, 190–197. doi: 10.1016/j.molcel.2008.10.001
Battle, D. J., and Doudna, J. A. (2002). Specificity of RNA-RNA helix recognition. Proc. Natl. Acad. Sci. U. S. A. 99, 11676–11681. doi: 10.1073/pnas.182221799
Brown, A., Baird, M. R., Yip, M. C., Murray, J., and Shao, S. (2018). Structures of translationally inactive mammalian ribosomes. eLife 7:486. doi: 10.7554/eLife.40486
Buskirk, A. R., and Green, R. (2017). Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372:20160183. doi: 10.1098/rstb.2016.0183
Campos-Silva, R., D’Urso, G., Delalande, O., Giudice, E., Macedo, A. J., and Gillet, R. (2021). Trans-translation is an appealing target for the development of new antimicrobial compounds. Microorganisms 10:10003. doi: 10.3390/microorganisms10010003
Carbone, C. E., Loveland, A. B., Gamper, H. B., Hou, Y. M., Demo, G., and Korostelev, A. A. (2021). Time-resolved cryo-EM visualizes ribosomal translocation with EF-G and GTP. Nat. Commun. 12:7236. doi: 10.1038/s41467-021-27415-0
Cate, J. H., Gooding, A. R., Podell, E., Zhou, K., Golden, B. L., Kundrot, C. E., et al. (1996). Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273, 1678–1685. doi: 10.1126/science.273.5282.1678
Cornish, P. V., Ermolenko, D. N., Noller, H. F., and Ha, T. (2008). Spontaneous intersubunit rotation in single ribosomes. Mol. Cell 30, 578–588. doi: 10.1016/j.molcel.2008.05.004
Cornish, P. V., Ermolenko, D. N., Staple, D. W., Hoang, L., Hickerson, R. P., Noller, H. F., et al. (2009). Following movement of the L1 stalk between three functional states in single ribosomes. Proc. Natl. Acad. Sci. U. S. A. 106, 2571–2576. doi: 10.1073/pnas.0813180106
DeLano, W. L. (2002). The PyMOL molecular graphics system. Available at: https://pymol.org/
Demeshkina, N., Jenner, L., Westhof, E., Yusupov, M., and Yusupova, G. (2012). A new understanding of the decoding principle on the ribosome. Nature 484, 256–259. doi: 10.1038/nature10913
Doherty, E. A., Batey, R. T., Masquida, B., and Doudna, J. A. (2001). A universal mode of helix packing in RNA. Nat. Struct. Biol. 8, 339–343. doi: 10.1038/86221
D’Urso, G., Guyomar, C., Chat, S., Giudice, E., and Gillet, R. (2023). Insights into the ribosomal trans-translation rescue system: lessons from recent structural studies. FEBS J. 290, 1461–1472. doi: 10.1111/febs.16349
Ermolenko, D. N., and Noller, H. F. (2011). mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat. Struct. Mol. Biol. 18, 457–462. doi: 10.1038/nsmb.2011
Fei, J., Kosuri, P., MacDougall, D. D., and Gonzalez, R. L. Jr. (2008). Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell 30, 348–359. doi: 10.1016/j.molcel.2008.03.012
Fu, J., Hashem, Y., Wower, I., Lei, J., Liao, H. Y., Zwieb, C., et al. (2010). Visualizing the transfer-messenger RNA as the ribosome resumes translation. EMBO J. 29, 3819–3825. doi: 10.1038/emboj.2010.255
Gabashvili, I. S., Agrawal, R. K., Spahn, C. M., Grassucci, R. A., Svergun, D. I., Frank, J., et al. (2000). Solution structure of the E. coli 70S ribosome at 11.5 a resolution. Cell 100, 537–549. doi: 10.1016/S0092-8674(00)80690-X
Giudice, E., and Gillet, R. (2013). The task force that rescues stalled ribosomes in bacteria. Trends Biochem. Sci. 38, 403–411. doi: 10.1016/j.tibs.2013.06.002
Goddard, T. D., Huang, C. C., Meng, E. C., Pettersen, E. F., Couch, G. S., Morris, J. H., et al. (2018). UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25. doi: 10.1002/pro.3235
Grant, T., Rohou, A., and Grigorieff, N. (2018). cisTEM, user-friendly software for single-particle image processing. eLife 7:383. doi: 10.7554/eLife.35383
Grigorieff, N. (2016). Frealign: an exploratory tool for single-particle Cryo-EM. Methods Enzymol. 579, 191–226. doi: 10.1016/bs.mie.2016.04.013
Gutmann, S., Haebel, P. W., Metzinger, L., Sutter, M., Felden, B., and Ban, N. (2003). Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature 424, 699–703. doi: 10.1038/nature01831
Guyomar, C., D’Urso, G., Chat, S., Giudice, E., and Gillet, R. (2021). Structures of tmRNA and SmpB as they transit through the ribosome. Nat. Commun. 12:4909. doi: 10.1038/s41467-021-24881-4
Hussain, T., Llacer, J. L., Wimberly, B. T., Kieft, J. S., and Ramakrishnan, V. (2016). Large-scale movements of IF3 and tRNA during bacterial translation initiation. Cell 167:e113, 133–144.e13. doi: 10.1016/j.cell.2016.08.074
Ito, K., Chadani, Y., Nakamori, K., Chiba, S., Akiyama, Y., and Abo, T. (2011). Nascentome analysis uncovers futile protein synthesis in Escherichia coli. PLoS One 6:e28413. doi: 10.1371/journal.pone.0028413
Jahagirdar, D., Jha, V., Basu, K., Gomez-Blanco, J., Vargas, J., and Ortega, J. (2020). Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy. RNA 26, 2017–2030. doi: 10.1261/rna.075846.120
Janssen, B. D., and Hayes, C. S. (2012). The tmRNA ribosome-rescue system. Adv. Protein Chem. Struct. Biol. 86, 151–191. doi: 10.1016/B978-0-12-386497-0.00005-0
Julian, P., Konevega, A. L., Scheres, S. H., Lazaro, M., Gil, D., Wintermeyer, W., et al. (2008). Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc. Natl. Acad. Sci. U. S. A. 105, 16924–16927. doi: 10.1073/pnas.0809587105
Karzai, A. W., Susskind, M. M., and Sauer, R. T. (1999). SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J. 18, 3793–3799. doi: 10.1093/emboj/18.13.3793
Kaur, S., Gillet, R., Li, W., Gursky, R., and Frank, J. (2006). Cryo-EM visualization of transfer messenger RNA with two SmpBs in a stalled ribosome. Proc. Natl. Acad. Sci. U. S. A. 103, 16484–16489. doi: 10.1073/pnas.0607438103
Keiler, K. C. (2015). Mechanisms of ribosome rescue in bacteria. Nat. Rev. Microbiol. 13, 285–297. doi: 10.1038/nrmicro3438
Keiler, K. C., Waller, P. R., and Sauer, R. T. (1996). Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993. doi: 10.1126/science.271.5251.990
Komine, Y., Kitabatake, M., Yokogawa, T., Nishikawa, K., and Inokuchi, H. (1994). A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 91, 9223–9227. doi: 10.1073/pnas.91.20.9223
Korostelev, A. A. (2021). Diversity and similarity of termination and ribosome Rescue in Bacterial, mitochondrial, and cytoplasmic translation. Biochemistry 86, 1107–1121. doi: 10.1134/S0006297921090066
Korostelev, A. A. (2022). The structural dynamics of translation. Annu. Rev. Biochem. 91, 245–267. doi: 10.1146/annurev-biochem-071921-122857
Korostelev, A., Trakhanov, S., Laurberg, M., and Noller, H. F. (2006). Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 126, 1065–1077. doi: 10.1016/j.cell.2006.08.032
Kremer, J. R., Mastronarde, D. N., and McIntosh, J. R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76. doi: 10.1006/jsbi.1996.0013
Kurita, D., and Himeno, H. (2022). Bacterial ribosome rescue systems. Microorganisms 10:372. doi: 10.3390/microorganisms10020372
Lancaster, L., and Noller, H. F. (2005). Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell 20, 623–632. doi: 10.1016/j.molcel.2005.10.006
Loveland, A. B., Bah, E., Madireddy, R., Zhang, Y., Brilot, A. F., Grigorieff, N., et al. (2016). Ribosome*RelA structures reveal the mechanism of stringent response activation. eLife 5:29. doi: 10.7554/eLife.17029
Loveland, A. B., Demo, G., Grigorieff, N., and Korostelev, A. A. (2017). Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature 546, 113–117. doi: 10.1038/nature22397
Loveland, A. B., Demo, G., and Korostelev, A. A. (2020). Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading. Nature 584, 640–645. doi: 10.1038/s41586-020-2447-x
Lyumkis, D., Brilot, A. F., Theobald, D. L., and Grigorieff, N. (2013). Likelihood-based classification of cryo-EM images using FREALIGN. J. Struct. Biol. 183, 377–388. doi: 10.1016/j.jsb.2013.07.005
Marathe, N., Nguyen, H. A., Alumasa, J. N., Kuzmishin Nagy, A. B., Vazquez, M., Dunham, C. M., et al. (2023). Antibiotic that inhibits trans-translation blocks binding of EF-Tu to tmRNA but not to tRNA. MBio 14:e0146123. doi: 10.1128/mbio.01461-23
Mastronarde, D. N. (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51. doi: 10.1016/j.jsb.2005.07.007
Melnikov, S., Ben-Shem, A., Garreau de Loubresse, N., Jenner, L., Yusupova, G., and Yusupov, M. (2012). One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 19, 560–567. doi: 10.1038/nsmb.2313
Miller, M. R., and Buskirk, A. R. (2014). An unusual mechanism for EF-Tu activation during tmRNA-mediated ribosome rescue. RNA 20, 228–235. doi: 10.1261/rna.042226.113
Mishra, S., Ahmed, T., Tyagi, A., Shi, J., and Bhushan, S. (2018). Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA. Sci. Rep. 8:13587. doi: 10.1038/s41598-018-31850-3
Moazed, D., and Noller, H. F. (1986). Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell 47, 985–994. doi: 10.1016/0092-8674(86)90813-5
Moazed, D., and Noller, H. F. (1989). Interaction of tRNA with 23S rRNA in the ribosomal a, P, and E sites. Cell 57, 585–597. doi: 10.1016/0092-8674(89)90128-1
Moore, S. D., and Sauer, R. T. (2005). Ribosome rescue: tmRNA tagging activity and capacity in Escherichia coli. Mol. Microbiol. 58, 456–466. doi: 10.1111/j.1365-2958.2005.04832.x
Muller, C., Crowe-McAuliffe, C., and Wilson, D. N. (2021). Ribosome rescue pathways in Bacteria. Front. Microbiol. 12:652980. doi: 10.3389/fmicb.2021.652980
Neubauer, C., Gillet, R., Kelley, A. C., and Ramakrishnan, V. (2012). Decoding in the absence of a codon by tmRNA and SmpB in the ribosome. Science 335, 1366–1369. doi: 10.1126/science.1217039
Nguyen, H. A., Sunita, S., and Dunham, C. M. (2020). Disruption of evolutionarily correlated tRNA elements impairs accurate decoding. Proc. Natl. Acad. Sci. U. S. A. 117, 16333–16338. doi: 10.1073/pnas.2004170117
Nissen, P., Ippolito, J. A., Ban, N., Moore, P. B., and Steitz, T. A. (2001). RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl. Acad. Sci. U. S. A. 98, 4899–4903. doi: 10.1073/pnas.081082398
Ogle, J. M., Brodersen, D. E., Clemons, W. M. Jr., Tarry, M. J., Carter, A. P., and Ramakrishnan, V. (2001). Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902. doi: 10.1126/science.1060612
Op De Bekke, A., Kiefmann, M., Kremerskothen, J., Vornlocher, H. P., Sprinzl, M., and Brosius, J. (1998). The 10Sa RNA gene of Thermus thermophilus. DNA Seq. 9, 31–35. doi: 10.3109/10425179809050022
Pettersen, E. F., Goddard, T. D., Huang, C. C., Meng, E. C., Couch, G. S., Croll, T. I., et al. (2021). UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82. doi: 10.1002/pro.3943
Rae, C. D., Gordiyenko, Y., and Ramakrishnan, V. (2019). How a circularized tmRNA moves through the ribosome. Science 363, 740–744. doi: 10.1126/science.aav9370
Ramrath, D. J., Yamamoto, H., Rother, K., Wittek, D., Pech, M., Mielke, T., et al. (2012). The complex of tmRNA-SmpB and EF-G on translocating ribosomes. Nature 485, 526–529. doi: 10.1038/nature11006
Ratje, A. H., Loerke, J., Mikolajka, A., Brunner, M., Hildebrand, P. W., Starosta, A. L., et al. (2010). Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 468, 713–716. doi: 10.1038/nature09547
Rodnina, M. V. (2018). Translation in prokaryotes. Cold Spring Harb. Perspect. Biol. 10:2664. doi: 10.1101/cshperspect.a032664
Sanbonmatsu, K. Y., Joseph, S., and Tung, C. S. (2005). Simulating movement of tRNA into the ribosome during decoding. Proc. Natl. Acad. Sci. U. S. A. 102, 15854–15859. doi: 10.1073/pnas.0503456102
Schopping, M., Zeidan, A. A., and Franzen, C. J. (2022). Stress response in Bifidobacteria. Microbiol. Mol. Biol. Rev. 86:e0017021. doi: 10.1128/mmbr.00170-21
Tu, G. F., Reid, G. E., Zhang, J. G., Moritz, R. L., and Simpson, R. J. (1995). C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J. Biol. Chem. 270, 9322–9326. doi: 10.1074/jbc.270.16.9322
Ushida, C., Himeno, H., Watanabe, T., and Muto, A. (1994). tRNA-like structures in 10Sa RNAs of mycoplasma capricolum and Bacillus subtilis. Nucleic Acids Res. 22, 3392–3396. doi: 10.1093/nar/22.16.3392
Voorhees, R. M., and Ramakrishnan, V. (2013). Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 82, 203–236. doi: 10.1146/annurev-biochem-113009-092313
Weis, F., Bron, P., Giudice, E., Rolland, J. P., Thomas, D., Felden, B., et al. (2010). tmRNA-SmpB: a journey to the Centre of the bacterial ribosome. EMBO J. 29, 3810–3818. doi: 10.1038/emboj.2010.252
Yamamoto, Y., Sunohara, T., Jojima, K., Inada, T., and Aiba, H. (2003). SsrA-mediated trans-translation plays a role in mRNA quality control by facilitating degradation of truncated mRNAs. RNA 9, 408–418. doi: 10.1261/rna.2174803
Zhang, Y., Hong, S., Ruangprasert, A., Skiniotis, G., and Dunham, C. M. (2018). Alternative mode of E-site tRNA binding in the presence of a downstream mRNA stem loop at the Entrance Channel. Structure 26:e433, 437–445.e3. doi: 10.1016/j.str.2018.01.013
Keywords: endogenous tmRNA, tmRNA decoding, A-minor interactions, SmpB, cryo-EM, alanyl-tRNA
Citation: Teran D, Zhang Y and Korostelev AA (2024) Endogenous trans-translation structure visualizes the decoding of the first tmRNA alanine codon. Front. Microbiol. 15:1369760. doi: 10.3389/fmicb.2024.1369760
Received: 12 January 2024; Accepted: 19 February 2024;
Published: 04 March 2024.
Edited by:
Matthieu G. Gagnon, University of Texas Medical Branch at Galveston, United StatesReviewed by:
Reynald Gillet, University of Rennes, FranceCopyright © 2024 Teran, Zhang and Korostelev. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrei A. Korostelev, YW5kcmVpLmtvcm9zdGVsZXZAdW1hc3NtZWQuZWR1
†Present address: Ying Zhang, Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, IN, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.