
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 14 March 2024
Sec. Microbial Physiology and Metabolism
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1368809
 Yusi Yan1
Yusi Yan1 Haiyang Xia1,2*
Haiyang Xia1,2*Streptomyces species are best known for their ability to produce abundant secondary metabolites with versatile bioactivities and industrial importance. These metabolites are usually biosynthesized through metabolic pathways encoded by cluster-situated genes. These genes are also known as biosynthetic gene clusters (BGCs) of secondary metabolites. The expression of BGCs is intricately controlled by pyramidal transcriptional regulatory cascades, which include various regulators. Streptomyces antibiotic regulatory proteins (SARPs), a genus-specific family of regulators, are widely distributed and play important roles in regulating the biosynthesis of secondary metabolites in Streptomyces. Over the past decade, the biological functions of SARPs have been extensively investigated. Here, we summarized the recent advances in characterizing the roles of SARPs involved in Streptomyces secondary metabolism from the following three aspects. First, the classification and domain organization of SARPs were summarized according to their size variation. Second, we presented a detailed description of the regulatory mechanisms and modes of action of SARPs involved in secondary metabolism. Finally, the biotechnological application of SARPs was illustrated by improving the production of target secondary metabolites and discovering novel bioactive natural products. This review will help researchers to comprehensively understand the roles of SARPs in secondary metabolite biosynthesis in Streptomyces, which will contribute to building a solid foundation for their future application in synthetic biology.
Streptomyces is a group of actinobacteria with a high GC content in their genomic DNA and complex morphological differentiation and life cycles (Hopwood, 2019). These bacteria are best known for their extraordinary ability to produce a multitude of bioactive secondary metabolites, such as antibiotics, insecticides and immunosuppressants, which make important contributions to clinical, medicinal and industrial fields (Hutchings et al., 2019; Jose et al., 2021; Alam et al., 2022). Typically, secondary metabolites are biosynthesized through secondary metabolic pathways that are encoded by biosynthetic gene clusters (BGCs) (Sharma et al., 2021). The morphological differentiation and expression of BGCs are intricately and stringently governed by pyramidal transcriptional regulatory cascades formed by large numbers of regulators and various signals (Wei et al., 2018; Xia et al., 2020). These regulators can be hierarchically grouped into global/pleiotropic and pathway-specific regulators according to their target genes and pathways. The bottom level consists of pathway-specific regulators (PSRs), which affect the production of cognate secondary metabolites by directly controlling the transcription of biosynthetic genes. PSR-encoding genes are usually located in BGCs and are also called cluster-situated regulators (CSRs) (Lu et al., 2017; Wei et al., 2018). Global/pleiotropic regulators function at higher levels. The genes that encode these regulators are usually located outside the BGCs, and they also regulate the expression of CSR-encoding genes, the biosynthesis of multiple secondary metabolites and/or morphological differentiation (Liu et al., 2013; Niu et al., 2016).
Streptomyces antibiotic regulatory proteins (SARPs) are a specific family of regulators exclusively found in actinobacteria, especially in streptomycetes (Bibb, 2005; Krause et al., 2020). Generally, SARP-encoding genes are widely present in many different types of BGCs and play indispensable roles in controlling the biosynthesis of secondary metabolites in Streptomyces. To the best of our knowledge, only one review has summarized the roles of SARPs in secondary metabolite biosynthesis in Streptomyces coelicolor (Liu et al., 2013). Over the past decade, many SARPs have been characterized in different Streptomyces strains, and their roles in secondary metabolite biosynthesis are more diverse than previously reported (Li et al., 2018; Yang et al., 2019; Yan et al., 2022). This review highlights recent findings on the mechanism by which SARPs control secondary metabolism in Streptomyces. A comprehensive understanding of the regulatory mechanism of SARPs on secondary metabolite biosynthesis will facilitate their biotechnological application to improve the production of important secondary metabolites and discover novel bioactive natural products in the near future.
SARPs have highly variable lengths and functional domain organizations. By their size and domain organization pattern, SARPs can be divided into three groups: small SARPs, medium SARPs and large SARPs (Figure 1). Small SARPs are approximately 300 residues in length and contain only an N-terminal DNA-binding domain (DBD) and a C-terminal bacterial transcriptional activation domain (BTAD); these include RedD for undecylprodigiosin (RED) biosynthesis, ActII-ORF4 for actinorhodin (ACT) biosynthesis and CpkN for coelimycin biosynthesis in S. coelicolor (Bednarz et al., 2021). Additionally, these two domains are typical SARP domains in which the DBD binds to repeated DNA motifs and the adjacent BTAD is supposed to initiate the transcription of target BGCs by recruiting RNA polymerase (RNAP) (Liu et al., 2013).
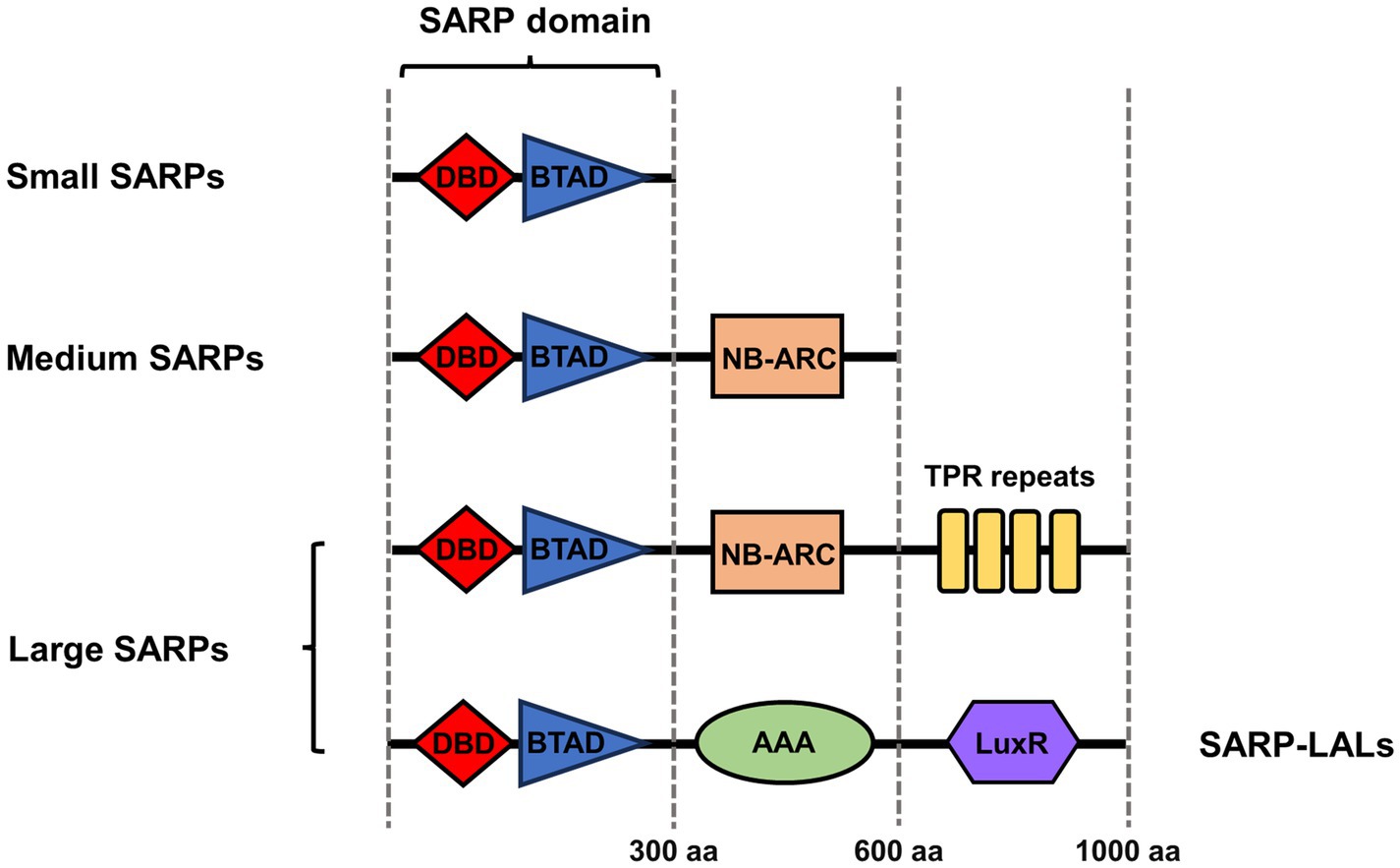
Figure 1. Classification and domain organization of SARP family regulators. The red rhomb indicates the DNA binding domain (DBD). The blue triangle indicates the bacterial transcriptional activation domain (BTAD). The orange rectangle indicates the NB-ARC domain. The yellow rectangles indicate the TRP repeats. The green oval indicates the AAA domain. The violet hexagon indicates the partially homologous LuxR domain in SARP-LAL. The architectures indicate only domains, not lengths.
Medium SARPs, such as CdaR for calcium-dependent antibiotic (CDA) biosynthesis and CpkO for coelimycin biosynthesis in S. coelicolor and FdmR1 for fredericamycin biosynthesis in Streptomyces griseus, usually contain approximately 600 amino acid residues and consist of a SARP domain and an NB-ARC domain (Chen et al., 2008; Bednarz et al., 2021). The NB-ARC domain is a highly conserved nucleotide-binding domain shared with APAF-1, various R proteins and CED-4. The NB-ARC domain works as a signaling motif found in eukaryotes and bacteria and is characteristic of the AAA domain superfamily, which is widely considered to act as a molecular switch to the cycle between ADP (repressed) and ATP (active)-bound forms (Steele et al., 2019).
Large SARPs contain approximately 1,000 residues and consist of an N-terminal SARP domain, a central NB-ARC domain and a conserved C-terminal tetratricopeptide repeat (TPR) domain; examples include RslR3 for rishirilide biosynthesis in Streptomyces bottropensis and PolY for polyoxin biosynthesis in Streptomyces cacaoi subsp. asoensis (Li et al., 2010; Tsypik et al., 2021). The TPR domain usually mediates protein–protein interactions and the assembly of multiprotein complex mediators (D’Andrea and Regan, 2003).
In addition, several large SARPs have specific domain architectures, which include an N-terminal SARP domain and half of a C-terminal that is homologous to guanylate cyclases and LAL regulators (large ATP-binding regulators of the LuxR family) (Li et al., 2022). The C-terminal half includes the ATP/GTP binding domain that is characteristic of these protein families but lacks the signature sequence at the N-terminus of guanylate cyclases or the LuxR-type helix-turn-helix (HTH) motif for DNA binding present at the C-terminus of LAL regulators (Barreales et al., 2018). Therefore, these special kinds of large SARPs are also called SARP-LALs and include SanG for nikkomycin biosynthesis in Streptomyces ansochromogenes, FilR for filipin biosynthesis in Streptomyces filipinensis and PimR for pimaricin biosynthesis in Streptomyces natalensis (He et al., 2010; Santos-Aberturas et al., 2012; Barreales et al., 2018).
In this review, 25 representative Streptomyces strains were chosen to illustrate the distribution of SARPs. The genomic sequences of these strains from GenBank were analyzed via the Predicted Prokaryotic Regulatory Proteins (P2RP) webserver and antiSMASH webserver for the distribution of SARPs and BGCs, respectively (Barakat et al., 2013; Blin et al., 2024) (Table 1). Generally, more SARPs exist in strains with larger genomes and more BGCs. Additionally, more than half of the strains contained all the kinds of SARPs mentioned above. In particular, homologs of AfsR, a special kind of large SARP, are distributed among all the tested Streptomyces species.
To date, AfsR has been identified as the single global regulator of the SARP family and has been extensively studied in S. coelicolor and S. griseus. AfsR positively regulates the biosynthesis of ACT, RED and CDA by directly activating the expression of the adjacent gene afsS, which encodes a small sigma factor-like protein. In turn, the synthesis of AfsS activates the transcription of genes encoding pathway-specific activators, which control the transcription of the ACT, RED and CDA BGCs in an as yet unknown way (Umeyama et al., 2002; Liu et al., 2013).
AfsR is phosphorylated at serine/threonine residues by the AfsK kinase (Matsumoto et al., 1994; Jin et al., 2011). The phosphorylation of AfsR significantly increases its binding affinity to the afsS promoter and therefore enhances the yields of ACT, RED and CDA (Martín and Liras, 2019). Notably, the autophosphorylating activity of AfsK is inhibited by KbpA, whose encoding gene is located upstream of afsK in both S. coelicolor and S. griseus. KbpA binds to unphosphorylated AfsK to inhibit its autophosphorylation at serine/threonine residues. Therefore, KbpA acts as a negative regulator in the AfsK–AfsR phosphorylation cascade, which indirectly decreases secondary metabolism in S. coelicolor and sporulation in S. griseus (Umeyama and Horinouchi, 2001; Sawai et al., 2004). In addition, the C-terminus of AfsK is likely involved in binding S-adenosyl-L-methionine (SAM) in S. coelicolor. SAM activates actinorhodin biosynthesis by increasing the autophosphorylation of AfsK (Lee et al., 2007; Jin et al., 2011). Moreover, two other serine/threonine kinases, AfsL and PkaG, that can phosphorylate AfsR in vitro have been identified (Sawai et al., 2004; Liu et al., 2013; Martín et al., 2021). This finding suggested that AfsR works as a signal integrator of signals that are sensed by multiple serine/threonine kinases (Horinouchi, 2003).
The binding site of AfsR at the afsS promoter consists of two direct repeats (CGTT(T/C)ATCGNN), which are also recognized by another global regulator, PhoP (Santos-Beneit et al., 2009). The two-component system PhoR–PhoP controls global phosphate metabolism and secondary metabolite biosynthesis in Streptomyces. The response regulator PhoP binds to the afsS promoter in a region overlapping with the AfsR binding sequence, and there is the binding competition between these two regulators. Additionally, AfsR binds to other PhoP-regulated promoters, including those of pstS (a component of the phosphate transport system), phoRP (encoding the two-component system itself) and glnR (encoding the regulator of nitrogen metabolism) (Santos-Beneit et al., 2012; Van der Heul et al., 2018). In summary, AfsR is involved in regulating primary and secondary metabolism through cross-talk with PhoP and GlnR.
In addition, AfsR homologs from other Streptomyces species have also been shown to be involved in morphological development and/or secondary metabolite biosynthesis. In S. griseus, AfsR is also required for morphological development but is not directly needed to produce streptomycin or A-factors (Umeyama et al., 1999). In Streptomyces roseosporus, AfsR works as an activator of daptomycin production but plays a negative role in development (Yan et al., 2020). Moreover, in Streptomyces venezuelae, Streptomyces peucetius, Streptomyces acidiscabies and Streptomyces lomondensis, AfsR homologs activate the biosynthesis of various secondary metabolites (Maharjan et al., 2009; Kim et al., 2012; Wang et al., 2015) (Table 2). However, AfsR represses the production of pristinamycin I but activates the production of pristinamycin II in Streptomyces pristinaespiralis (Jin et al., 2021). Notably, AfsR of S. peucetius is named AfsR-sp., and its encoded by the afsR-sp. gene. Overexpression of afsR-sp. under the control of a strong promoter led to improved doxorubicin production in S. peucetius (Maharjan et al., 2009). A similar strategy has also been used to overproduce actinorhodin in S. lividans, clavulanic acid in S. clavuligerus and pikromycin in S. venezuelae by overexpressing afsR-sp. from S. peucetius (Parajuli et al., 2005) (Table 2).
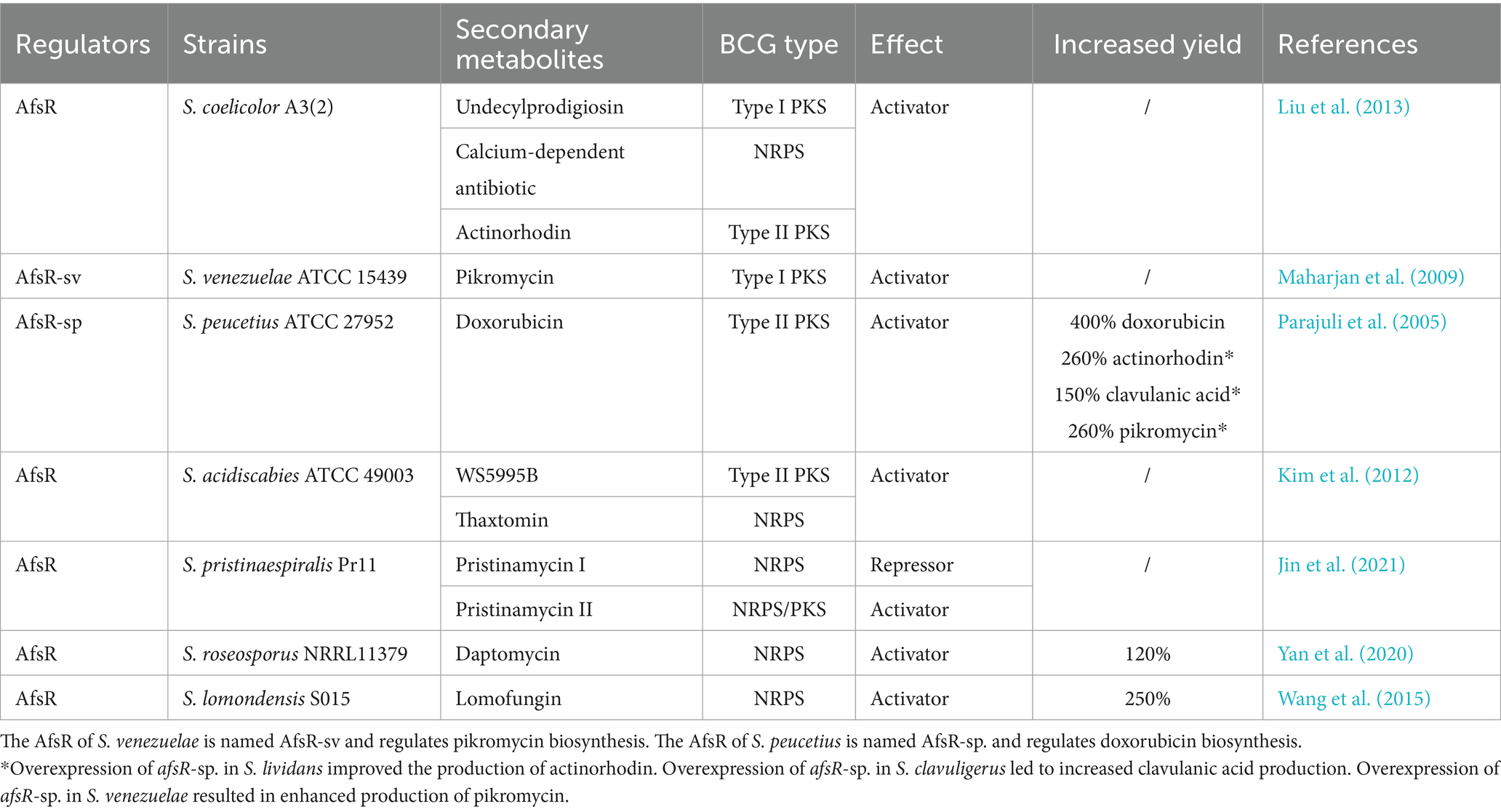
Table 2. Identified homologs of AfsR involved in the biosynthesis of secondary metabolites in Streptomyces species.
Typically, SARP-encoding genes are located in various BGCs that encode polyketides (Type I or Type II PKS), nonribosomally synthesized peptides (NRPS), hybrid polyketide peptide (NRPS/PKS) compounds, nucleosides, thiopeptides and terpenoids (Krause et al., 2020) (Table 3). Most of them act as pathway-specific activators of secondary metabolite biosynthesis by directly controlling the transcription of their cognate BGCs; for example, ChIF2 is involved in chlorothricin biosynthesis in Streptomyces antibioticus, NigR is involved in nigericin biosynthesis in Streptomyces malaysiensis, and PieR is involved in piericidin biosynthesis in Streptomyces piomogeues var. Hangzhouwanensis (Li et al., 2019, 2020; Wei et al., 2022) (Table 3). Notably, the medium-sized SARP Atr32 has been identified to negatively affect atratumycin biosynthesis in Streptomyces atratus. The function of Atr32 was distinct from that described in previous studies, but the underlying mechanism has not been elucidated (Yang et al., 2019). Moreover, multiple SARP-encoding genes located in the same BGC often form a hierarchical cascade of regulation to control the biosynthesis of cognate secondary metabolites. For example, two SARP activators, CpkO and CpkN, are necessary for coelimycin biosynthesis in S. coelicolor, in which CpkO activates the transcription of cpkN (Bednarz et al., 2021). In S. cacaoi subsp. asoensis, PolY positively controls polyoxin biosynthesis by directly activating the transcription of polR, which encodes another SARP (Li et al., 2010). Three SARPs, RslR1, RslR2, and RslR3, are positive regulators of rishirilide biosynthesis in S. bottropensis. RslR3 directly activates the transcription of rslR2, while RslR1 represses the transcription of rslR2 (Tsypik et al., 2021).
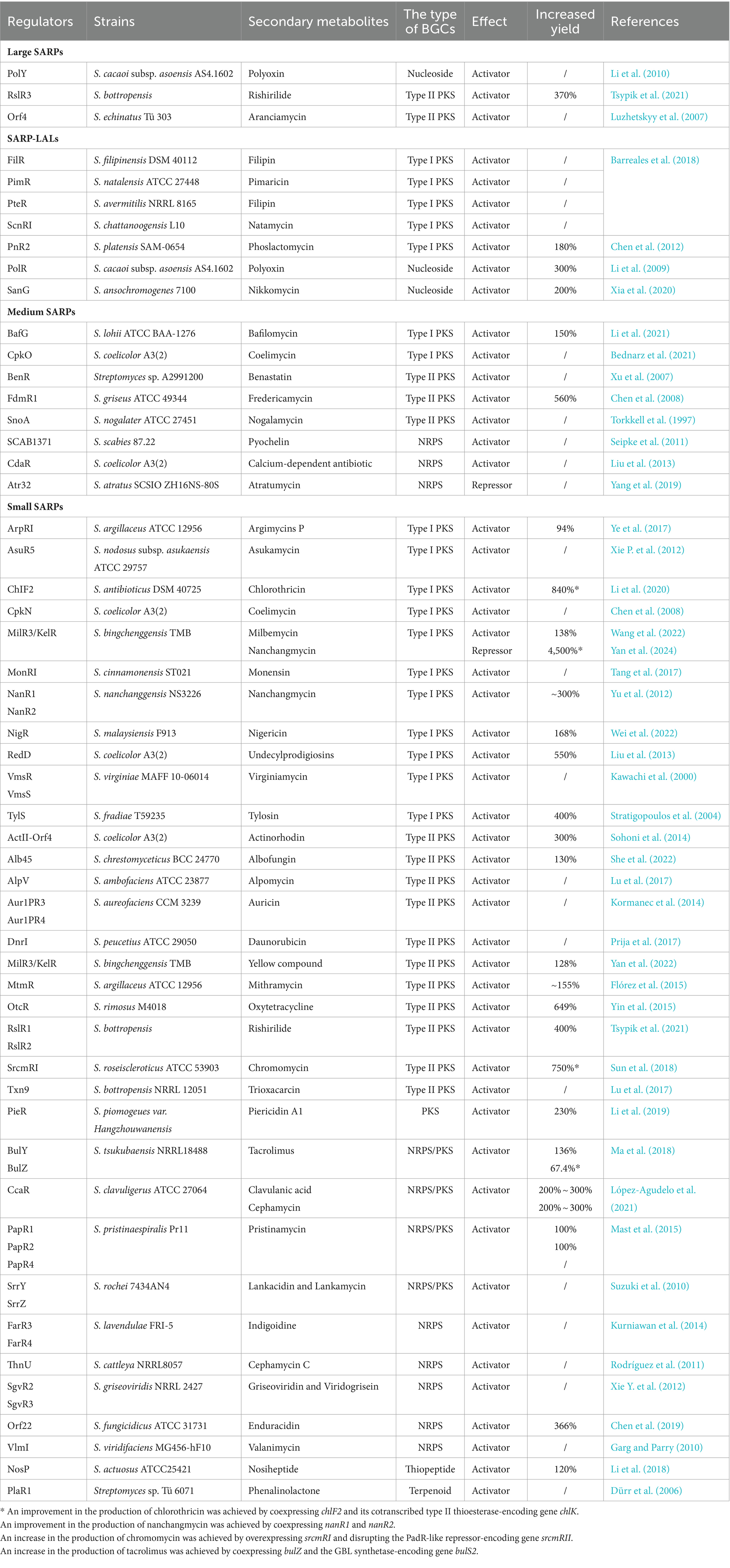
Table 3. Identified SARP family regulators involved in the biosynthesis of secondary metabolites in Streptomyces species.
A few small SARP-encoding genes are located within the γ-butyrolactone (GBL) regulatory gene cluster and are involved in the GBL signaling pathway to control the biosynthesis of secondary metabolites (Table 3). In Streptomyces tsukubaensis, BulZ and BulY, located in the GBL region, were reported to positively regulate tacrolimus production (Ma et al., 2018). In another study, SrrY not only played a central role in the GBL signaling pathway but also activated lankamycin biosynthesis by directly regulating the transcription of srrZ in Streptomyces rochei (Suzuki et al., 2010). However, FarR3 and FarR4 have distinct effects on secondary metabolism in Streptomyces lavendulae, where FarR4 negatively controls GBL and indigoidine biosynthesis, but FarR3 positively regulates indigoidine biosynthesis (Kurniawan et al., 2014). Until now the detailed description of regulatory mode of FarR4 has remained unclear.
Notably, several small SARPs have also been reported to be pleiotropic regulators that control the biosynthesis of different secondary metabolites. CcaR from Streptomyces clavuligerus, which is encoded by ccaR located within the cephamycin BGC, acts as an activator of both cephamycin and adjacent clavulanic acid BGCs (López-Agudelo et al., 2021). In Streptomyces binchenggensis, milR3/kelR is located in a type II PKS BGC that is responsible for producing the yellow compound. The milbemycin BGC is far from the type II PKS BGC in the genome of S. bingchenggensis, but MilR3/KelR has been shown to coactivate the biosynthesis of milbemycin and the yellow compound (Wang et al., 2022; Yan et al., 2022). In addition, S. bingchenggensis can also produce nanchangmycin. NanR4, which is encoded by nanR4 located in the nanchangmycin BGC, is a specific repressor of nanchangmycin biosynthesis. MilR3/KelR has been proven to inhibit nanchangmycin biosynthesis by activating the transcription of nanR4 (Yan et al., 2024). Therefore, the small SARP MilR3/KelR can coregulate the biosynthesis of three secondary metabolites in S. bingchenggensis.
Recently, it has been reported that SARPs regulate the production of secondary metabolites by sensing final products and intermediates. As the only CSR related to nosiheptide (NOS) production in Streptomyces actuosus, NosP directly activates the transcription of biosynthetic genes by binding to the intergenic sequence in the NOS BGC. Furthermore, the DNA binding activity of NosP is modulated by NOS and its intermediate NOS-AC (Li et al., 2018). Moreover, NocP, the homolog of NosP in Nocardia sp., regulates the biosynthesis of structurally related nocathiacin I in a similar pattern (Li et al., 2018).
SARPs usually activate the transcription of BGCs by binding to upstream regions of their target genes. Generally, heptameric direct repeats separated by spacers of 4–15 nucleotides are SARP-binding sequences that overlap the-35 region of the promoters of target genes (Liu et al., 2013). Moreover, the number of heptameric repeats in the SARP-binding site varies depending on the different SARPs. Some SARP-binding sites contain three heptameric repeats, such as DnrI-binding site in the upstream region of dnrD in Streptomyces peucetius, ActII-ORF4-binding sites in the upstream regions of actII-ORF1 and actVI-ORF1 in S. coelicolor, VlmI-binding sites in the upstream regions of vlmJ and vlmA-vlmH in Streptomyces viridifaciens, and FdmR1-binding site in the upstream region of fdmD in S. griseus (Tang et al., 1996; Chen et al., 2008; Garg and Parry, 2010; Liu et al., 2013). Other species have two obvious heptamers, including the SanG-binding site located between sanO and sanN in S. ansochromogenes, the PolR-binding site located between polB and polC in S. cacaoi subsp. asoensis, the CcaR-binding sites in the upstream regions of cmcI and ceaS2-II in S. clavuligerus and the PieR-binding site in the upstream region of pieR in S. piomogeues (Tahlan et al., 2004; Li et al., 2009, 2019; He et al., 2010).
Notably, the SARP-LALs involved in polyene macrolide biosynthesis work via a special mechanism different from that of other SARPs. The pimR gene, which encodes the SARP-LAL regulator, and the pimM gene, which encodes the PAS-LuxR regulator, are located in the pimaricin BGC in S. natalensis. PimR binds the promoter of pimM and activates its transcription, and in turn, PimM activates the transcription of biosynthetic genes in the pimaricin BGC (Santos-Aberturas et al., 2012; Li et al., 2022). The PimR-binding site contains three heptameric direct repeats separated by four nucleotide spacers that do not overlap the-35 promoter box (Tanaka et al., 2007; Santos-Aberturas et al., 2012). Additionally, PimR can bind a secondary operator with only two direct repeats separated by three-nucleotide spacers, forming 10-bp repeating units instead of the classical 11-bp SARP-binding sites (Wietzorrek and Bibb, 1997; Santos-Aberturas et al., 2012). Notably, the binding sequence of PimR is entirely conserved in the intergenic region between scnRII and scnRI in the natamycin BGC of Streptomyces chattanoogensis and between pteF and pteR in Streptomyces avermitilis and between filF and filR in S. filipinensis, which are the corresponding counterparts in the filipin BGC (Barreales et al., 2018). PimR, ScnRI, PteR and FilR are all SARP-LALs involved in polyene macrolide biosynthesis. It is thus likely that the hierarchical relationship between PimR and PimM is conserved in other polyene regulatory pathways. Interestingly, the consensus heptamer of PimR is also found in the binding site of SARP-LALs involved in peptidyl nucleoside biosynthesis, including SanG and PolR, but these binding sites contain only two heptamers and overlap the-35 promoter boxes of their target genes (Li et al., 2009; He et al., 2010).
Streptomyces species are considered as the workhorse to produce valuable secondary metabolites (Alam et al., 2022). In recent decades, many strategies have been developed to increase the productivity of these hosts, and rewiring regulatory networks from Streptomyces is a powerful and effective approach for yield improvement (Xia et al., 2020; Bu et al., 2021; Sharma et al., 2021). Genetic manipulation of SARPs, the most common activators in Streptomyces, has been widely used to promote the production of secondary metabolites in recent decades. As listed in Table 3, the overexpression of fdmR, milR3, pieR and polR has been employed to increase fredericamycin production in S. griseus, milbemycin production in S. bingchenggensis, piericidin production in S. piomogeues and polyoxin production in S. cacaoi subsp. asoensis, respectively (Chen et al., 2008; Li et al., 2009, 2019; Yan et al., 2022). Moreover, promoter strength also plays a critical role in the overexpression of SARP-encoding genes (Table 3). Overexpression of sanG with a constitutive promoter led to the overproduction of nikkomycin in S. ansochromogenes (He et al., 2010). Overproduction of oxytetracycline has been achieved by introducing an extra copy of otcR under the control of the constitutive SF14 promoter in S. rimosus (Yin et al., 2015). A similar strategy has also been used to overproduce bafilomycin by overexpressing bafG in S. lohii, mithramycin by overexpressing mtmR in S. argillaceus and nigericin by overexpressing nigR in S. malaysiensis (Flórez et al., 2015; Li et al., 2021; Wei et al., 2022). Moreover, fine-tuning the expression of SARP-encoding genes together with other biosynthetic or regulatory genes can greatly facilitate secondary metabolite production (Table 3). Cooverexpression of chlF2 and its cotranscribed type II thioesterase-encoding gene chlK effectively increased chlorothricin production by 840% in comparison to that of the wild-type strain (Li et al., 2020). Co-overexpression of bulZ and the GBL synthetase-encoding gene bulS2 under a strong promoter in S. tsukubaensis improved tacrolimus production by 67.4% compared to that in the wild-type strain (Ma et al., 2018). Overexpression of srcmRI and disruption of the PadR-like repressor-encoding gene srcmRII led to 750% increased chromomycin production in Streptomyces roseiscleroticus (Sun et al., 2018). In our latest research, cooverexpression of nanR1 and nanR2 under a strong constitutive promoter in the milR3 deletion mutant of S. bingchenggensis resulted in a 4,500% improvement in the production of nanchangmycin (Yan et al., 2024).
Streptomyces species are still promising treasure troves for the discovery of new secondary metabolites due to the abundance of BGCs in their genomes (Lee et al., 2020; Alam et al., 2022). However, a majority of BGCs are silent or weakly expressed under standard laboratory conditions (Misaki et al., 2022). Feasible ways to unlock these cryptic genetic resources are potential strategies for finding new natural products. As the most abundant activators, SARPs are suitable targets to be engineered for the activation of silent BGCs. The chemical structures of several compounds, which were discovered by this method, are illustrated in Figure 2. Cooverexpression of actII-ORF4 from S. coelicolor and aur1PR3 from Streptomyces aureofaciens in Streptomyces sp. TÜ102 resulted in the activation of the chartreusin BGC (Mingyar et al., 2021). Similarly, simultaneous overexpression of actII-ORF4, griR, aur1PR3, papR2 and redD in Streptomyces sp. CA-256286 led to the activation of a silent type II PKS gene cluster for griseusin biosynthesis (Beck et al., 2021). Both the prodigiosin BGC in S. lividans and the amicetin/plicacetin BGC in Streptomyces sp. SHP22-7 were activated by overexpressing papR2 under the control of a constitutive promoter (Krause et al., 2020). In Streptomyces sp. KO-7888, a cryptic NRPS gene cluster was activated by overexpressing the cluster containing the SARP-encoding gene speR, which led to the production of two new lipopeptides, sarpeptins A and B (Koomsiri et al., 2019). Additionally, a cryptic ahbamycin gene cluster of Streptomyces argillaceus was activated by constitutive overexpression of its cluster-situated SARP-encoding gene (Ye et al., 2023). In addition, the overexpression of tsuR1, a putative uncharacterized SARP-encoding gene, led to the discovery of the antitumour antibiotic tsukubarubicin in S. tsukubaensis (Wu et al., 2021). Similar strategies have been used to activate the production of a novel amide-containing polyene in Streptomyces sp. MSC090213JE08 and a novel cyclohexene-containing enamide in S. rochei (Du et al., 2016; Misaki et al., 2022). As mentioned above, SARP-dependent activation is an effective strategy for discovering novel bioactive natural products in Streptomyces.
SARPs, as bottom-level regulators, usually function as cluster-situated activators of secondary metabolism in Streptomyces. With extensive studies, the pleiotropic role of SARPs has been recognized. SARPs can also regulate the biosynthesis of multiple secondary metabolites by directly activating the transcription of cognate BGCs and cluster-situated activator-encoding genes in other BGCs. Notably, SARPs negatively affect the biosynthesis of secondary metabolites by activating the transcription of a cluster-situated repressor-encoding gene (Figure 3).
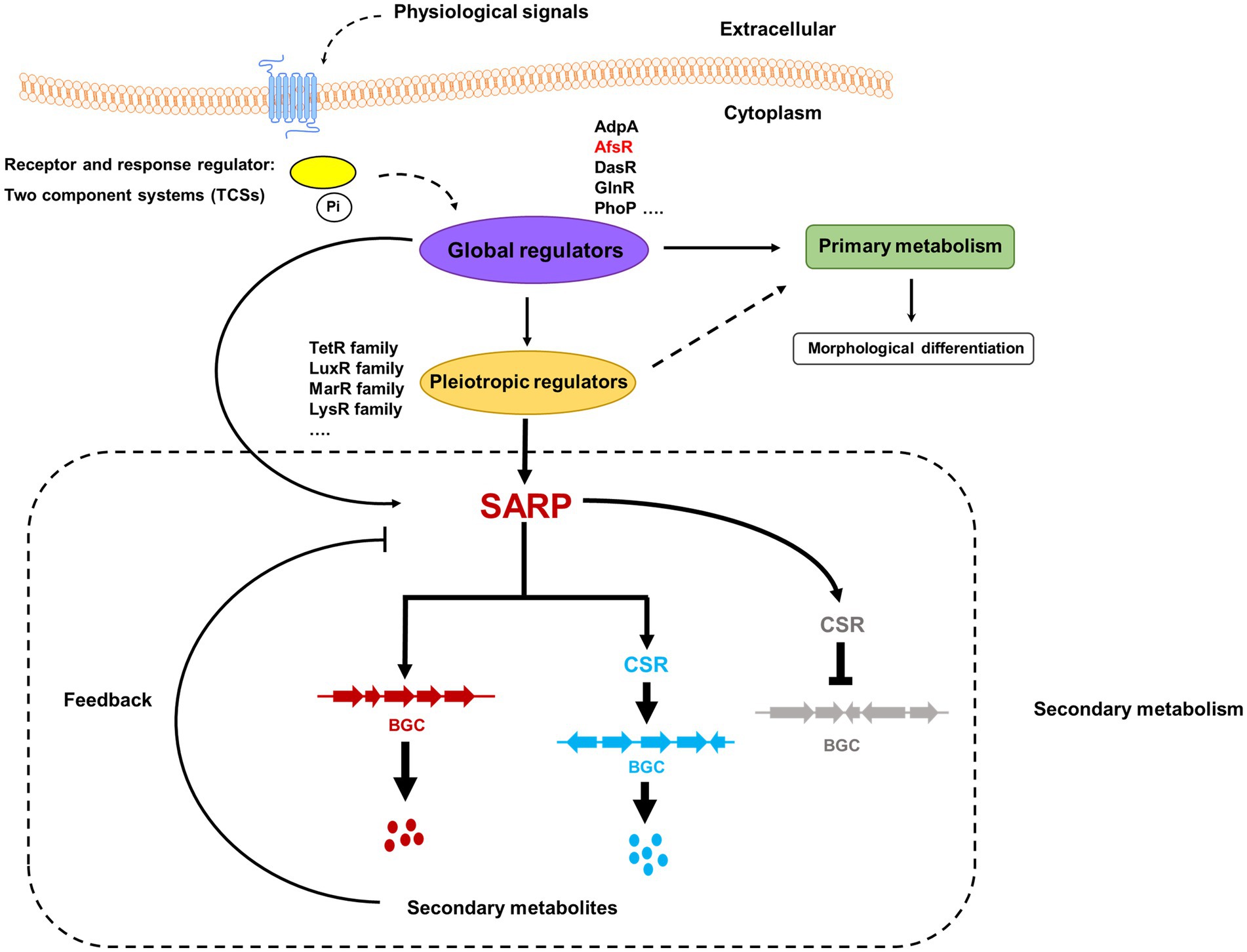
Figure 3. Schematic diagram of the regulatory cascades of SARP family regulators in Streptomyces. Two-component systems (TCSs), which consist of a histidine kinase and a response regulator, are the predominant signal transduction pathways involved in the regulation of secondary metabolism. The blue pillars in the cytomembrane indicate a histidine kinase that can sense some physiological signals. The yellow oval indicates a response regulator that is activated by phosphorylation from a signal-triggered histidine kinase. Pi indicates the phosphoric acid group. The known global regulators include AdpA, AfsR, DasR, GlnR, and PhoP. The pleiotropic regulators usually come from different families, such as the TetR family, LuxR family, MarR family, and LysR family. The red and blue dots indicate the cognate secondary metabolites produced by BGCs labeled in red and blue, respectively. The black arrows/dotted arrows indicate a positive effect. The black perpendicular lines indicate negative effects. The regulatory cascades in the dashed rectangle are associated with secondary metabolism.
Interestingly, the medium-sized SARP Atr32 serves as a negative regulator of atratumycin biosynthesis in S. atratus. In addition to atr32, the atratumycin BGC also contains two LuxR family regulator-encoding genes (atr1 and atr2) and two ABC transporter-encoding genes (atr29 and atr30), which play positive roles in atratumycin production (Yang et al., 2019). However, the regulatory relationship between Atr32 and other genes in the atratumycin BGC is unclear. Additionally, when Atr32 performs its negative regulatory function, the role of its NB-ARC domain is unknown. Further elucidation of the repressive regulatory mechanism of Atr32 will enrich the knowledge of SARPs in Streptomyces.
The SARP global regulator AfsR is highly conserved and widely distributed in most Streptomyces species, and its regulatory mechanism has provided a model for transcriptional activation by SARPs (Table 1) (Tanaka et al., 2007). AfsR interacts with DNA-binding sites in a dimer formation and recruits RNA polymerase (RNAP) to form a DNA-AfsR-RNAP complex that binds to the-10 element of target promoters. Its ATPase domain is essential for conformational changes in the closed complex between AfsR and RNAP to a transcriptionally competent open complex. As mentioned above, SARPs activate the transcription of BGCs in direct or indirect ways. However, how small SARPs without ATPase recruit and interact with RNA polymerase for transcriptional activation is still unknown. With the development of AI-assisted 3D structure elucidation, the elucidation of the mechanism of action and sequence recognition of SARPs will be greatly promoted in the future (Chatonnet et al., 2023).
As mentioned above, the small SARP MilR3/KelR is a pleiotropic regulator that affects the biosynthesis of at least three different secondary metabolites in S. bingchenggensis (Yan et al., 2024). Additionally, many putative MilR3/KelR homologs are widely distributed among actinobacteria and likely play similar roles in secondary metabolism (Yan et al., 2022). The cluster-situated activators CpkO and CpkN were identified to form the CpkO–CpkN regulatory cascade for coelimycin biosynthesis in S. coelicolor. Furthermore, multi-omics data have shown that these activators also likely regulate the stress response, dormancy and biosynthesis of other specialized metabolites (Bednarz et al., 2021). The mechanism of action of this subgroup of SARPs needs to be extensively investigated in future studies.
Secondary metabolites and/or their biosynthetic intermediates commonly function as feedback or feedforward autoregulators to dynamically modulate their production. This phenomenon is mostly known for TetR family regulators due to their ligand binding domains (Cuthbertson and Nodwell, 2013; Kong et al., 2019). Interestingly, some SARPs, such as NosP in S. actuosus, can also respond to secondary metabolites and their intermediates (Li et al., 2018). Recently, the global regulator AfsR in S. coelicolor was shown to play an important role in the effect of the artificial elicitor ARC2 (Calvelo et al., 2021). This suggested that some signaling compounds were probably needed for the activation effect of AfsR. Further investigation of the small molecules involved in SARP signal transduction could promote the discovery of new compounds generated by elicitor feeding.
Genetic engineering of SARPs has been proven to be a practical strategy for improving the yield of secondary metabolites and discovering novel bioactive natural products in Streptomyces. To date, many more small SARPs have been engineered to enhance the production of secondary metabolites (Table 3). It is obvious that the majority of medium and large SARPs have still not been well investigated (Table 1). Further elucidation of the regulatory mechanisms of SARPs would greatly improve the comprehensive understanding of the mechanism of action of the SARP repertoire, which will be beneficial for developing versatile strategies to enhance final products and unlock novel cryptic BGCs. Increasing the body of knowledge on the diversity of recognition sites and chemical sensing feedback of SARPs will be beneficial for generating programmable signal amplification systems mediated by SARPs, which can facilitate the development of intelligent biomanufacturing in the near future.
YY: Funding acquisition, Writing – original draft, Writing – review & editing. HX: Funding acquisition, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the professional development program of Zhejiang Province for visiting scholars and teachers in colleges and universities (FX2023050), the Scientific Research Foundation of Taizhou (Nos. 23gya07 and 21nya20), and the Natural Science Foundation of Zhejiang Province (LY19C010002).
We thank the members of the Institute of Biopharmaceuticals, Taizhou University, for many fruitful discussions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alam, K., Mazumder, A., Sikdar, S., Zhao, Y. M., Hao, J., Song, C., et al. (2022). Streptomyces: The biofactory of secondary metabolites. Front. Microbiol. 13:968053. doi: 10.3389/fmicb.2022.968053
Barakat, M., Ortet, P., and Whitworth, D. E. (2013). P2RP: a Web-based framework for the identification and analysis of regulatory proteins in prokaryotic genomes. BMC Genomics 14:269. doi: 10.1186/1471-2164-14-269
Barreales, E. G., Vicente, C. M., de Pedro, A., Santos-Aberturas, J., and Aparicio, J. F. (2018). Promoter engineering reveals the importance of heptameric direct repeats for DNA binding by Streptomyces antibiotic regulatory protein-large ATP-binding regulator of the LuxR family (SARP-LAL) regulators in Streptomyces natalensis. Appl. Environ. Microbiol. 84, e00246–e00218. doi: 10.1128/AEM.00246-18
Beck, C., Gren, T., Ortiz-López, F. J., Jørgensen, T. S., Carretero-Molina, D., Martín Serrano, J., et al. (2021). Activation and identification of a griseusin cluster in Streptomyces sp. CA-256286 by employing transcriptional regulators and multi-omics methods. Molecules 26:6580. doi: 10.3390/molecules26216580
Bednarz, B., Millan-Oropeza, A., Kotowska, M., Świat, M., Quispe Haro, J. J., Henry, C., et al. (2021). Coelimycin synthesis activatory proteins are key regulators of specialized metabolism and precursor flux in Streptomyces coelicolor A3(2). Front. Microbiol. 12:616050. doi: 10.3389/fmicb.2021.616050.eCollection2021
Bibb, M. J. (2005). Regulation of secondary metabolism in streptomycetes. Curr. Opin. Microbiol. 8, 208–215. doi: 10.1016/j.mib.2005.02.016
Blin, K., Shaw, S., Medema, M. H., and Weber, T. (2024). The antiSMASH database version 4: additional genomes and BGCs, new sequence-based searches and more. Nucleic Acids Res. 52, D586–D589. doi: 10.1093/nar/gkad984
Bu, Q. T., Li, Y. P., Xie, H., Li, J. F., Lv, Z. Y., Su, Y. T., et al. (2021). Rational engineering strategies for achieving high-yield, high-quality and high-stability of natural product production in actinomycetes. Metab. Eng. 67, 198–215. doi: 10.1016/j.ymben.2021.06.003
Calvelo, V. Y., Crisante, D., Elliot, M., and Nodwell, J. R. (2021). The ARC2 response. in Streptomcyes coelicolor requires the global regulatory genes afsR and afsS. Microbiology (Reading) 167:1–14. doi: 10.1099/mic.0.001047
Chatonnet, A., Perochon, M., Velluet, E., and Marchot, P. (2023). The ESTHER database on alpha/beta hydrolase fold proteins - An overview of recent developments. Chem. Biol. Interact. 383:110671. doi: 10.1016/j.cbi.2023.110671
Chen, Y. W., Liu, X. C., Lv, F. X., and Li, P. (2019). Characterization of three regulatory genes involved in enduracidin biosynthesis and improvement of enduracidin production in Streptomyces fungicidicus. J. Appl. Microbiol. 127, 1698–1705. doi: 10.1111/jam.14417
Chen, Y., Wendt-Pienkowski, E., and Shen, B. (2008). Identification and utility of FdmR1 as a Streptomyces antibiotic regulatory protein activator for fredericamycin production in Streptomyces griseus ATCC 49344 and heterologous hosts. J. Bacteriol. 190, 5587–5596. doi: 10.1128/JB.00592-08
Chen, Y. L., Zhao, J., Liu, W., Gao, J. F., Tao, L. M., Pan, H. X., et al. (2012). Identification of phoslactomycin biosynthetic gene clusters from Streptomyces platensis SAM-0654 and characterization of PnR1 and PnR2 as positive transcriptional regulators. Gene 509, 195–200. doi: 10.1016/j.gene.2012.08.030
Cuthbertson, L., and Nodwell, J. R. (2013). The TetR family of regulators. Microbiol. Mol. Biol. Rev. 77, 440–475. doi: 10.1128/MMBR.00018-13
D’Andrea, L. D., and Regan, L. (2003). TPR proteins: the versatile helix. Trends Biochem. Sci. 28, 655–662. doi: 10.1016/j.tibs.2003.10.007
Du, D., Katsuyama, Y., Onaka, H., Fujie, M., Satoh, N., Shin-Ya, K., et al. (2016). Production of a novel amide-containing polyene by activating a cryptic biosynthetic gene cluster in Streptomyces sp. MSC090213JE08. Chembiochem 17, 1464–1471. doi: 10.1002/cbic.201600167
Dürr, C., Schnell, H. J., Luzhetskyy, A., Murillo, R., Weber, M., Welzel, K., et al. (2006). Biosynthesis of the terpene phenalinolactone in Streptomyces sp. Tü6071: analysis of the gene cluster and generation of derivatives. Chem. Biol. 13, 365–377. doi: 10.1016/j.chembiol.2006.01.011
Flórez, A. B., Álvarez, S., Zabala, D., Braña, A. F., Salas, J. A., and Méndez, C. (2015). Transcriptional regulation of mithramycin biosynthesis in Streptomyces argillaceus: dual role as activator and repressor of the PadR-like regulator MtrY. Microbiology 161, 272–284. doi: 10.1099/mic.0.080895-0
Garg, R. P., and Parry, R. J. (2010). Regulation of valanimycin biosynthesis in Streptomyces viridifaciens: characterization of VlmI as a Streptomyces antibiotic regulatory protein (SARP). Microbiology (Reading) 156, 472–483. doi: 10.1099/mic.0.033167-0
He, X., Li, R., Pan, Y., Liu, G., and Tan, H. J. M. (2010). SanG, a transcriptional activator, controls nikkomycin biosynthesis through binding to the sanN-sanO intergenic region in Streptomyces ansochromogenes. Microbiology (Reading) 156, 828–837. doi: 10.1099/mic.0.033605-0
Hopwood, D. A. (2019). Highlights of Streptomyces genetics. Heredity (Edinb) 123, 23–32. doi: 10.1038/s41437-019-0196-0
Horinouchi, S. (2003). AfsR as an integrator of signals that are sensed by multiple serine/threonine kinases in Streptomyces coelicolor A3(2). J. Ind. Microbiol. Biotechnol. 30, 462–467. doi: 10.1007/s10295-003-0063-z
Hutchings, M. I., Truman, A. W., and Wilkinson, B. (2019). Antibiotics: past, present and future. Curr. Opin. Microbiol. 51, 72–80. doi: 10.1016/j.mib.2019.10.008
Jin, Y. Y., Cheng, J., Yang, S. H., Meng, L., Palaniyandi, S. A., Zhao, X. Q., et al. (2011). S-adenosyl-L-methionine activates actinorhodin biosynthesis by increasing autophosphorylation of the Ser/Thr protein kinase AfsK in Streptomyces coelicolor A3(2). Biosci. Biotechnol. Biochem. 75, 910–913. doi: 10.1271/bbb.100873
Jin, Q., Liao, H., Dou, Y., Shen, N., Wu, Z., Yang, Y., et al. (2021). Functional investigation of AfsKRS regulatory system for pristinamycin biosynthesis in Streptomyces pristinaespiralis. 3. Biotech 11:418. doi: 10.1007/s13205-021-02933-2
Jose, P. A., Maharshi, A., and Jha, B. (2021). Actinobacteria in natural products research: progress and prospects. Microbiol. Res. 246:126708. doi: 10.1016/j.micres.2021.126708
Kawachi, R., Wangchaisoonthorn, U., Nihira, T., and Yamada, Y. (2000). Identification by gene deletion analysis of a regulator, VmsR, that controls virginiamycin biosynthesis in Streptomyces virginiae. J. Bacteriol. 182, 6259–6263. doi: 10.1128/JB.182.21.6259-6263.2000
Kim, M. J., Nihira, T., and Choi, S. U. (2012). Cloning and characterization of afsR homologue regulatory gene from Streptomyces acidiscabies ATCC 49003. J. Korean Soc. Appl. Biol. Chem. 55, 663–668. doi: 10.1007/s13765-012-2128-z
Kong, D., Wang, X., Nie, J., and Niu, G. (2019). Regulation of antibiotic production by signaling molecules in Streptomyces. Front. Microbiol. 10:2927. doi: 10.3389/fmicb.2019.02927
Koomsiri, W., Inahashi, Y., Leetanasaksakul, K., Shiomi, K., Takahashi, Y. K., OM, S., et al. (2019). Sarpeptins A and B, lipopeptides produced by Streptomyces sp. KO-7888 overexpressing a specific SARP Regulator. J. Nat. Prod. 82, 2144–2151. doi: 10.1021/acs.jnatprod.9b00074
Kormanec, J., Novakova, R., Mingyar, E., and Feckova, L. (2014). Intriguing properties of the angucycline antibiotic auricin and complex regulation of its biosynthesis. Appl. Microbiol. Biotechnol. 98, 45–60. doi: 10.1007/s00253-013-5373-0
Krause, J., Handayani, I., Blin, K., Kulik, A., and Mast, Y. (2020). Disclosing the potential of the SARP-type regulator PapR2 for the activation of antibiotic gene clusters in streptomycetes. Front. Microbiol. 11:2020. doi: 10.3389/fmicb.2020.00225.eCollection
Kurniawan, Y. N., Kitani, S., Maeda, A., and Nihira, T. (2014). Differential contributions of two SARP family regulatory genes to indigoidine biosynthesis in Streptomyces lavendulae FRI-5. Appl. Microbiol. Biotechnol. 98, 9713–9721. doi: 10.1007/s00253-014-5988-9
Lee, N., Kim, W., Hwang, S., Lee, Y., Cho, S., Palsson, B., et al. (2020). Thirty complete Streptomyces genome sequences for mining novel secondary metabolite biosynthetic gene clusters. Sci. Data 7:55. doi: 10.1038/s41597-020-0395-9
Lee, Y., Kim, K., Suh, J. W., Rhee, S., and Lim, Y. (2007). Binding study of AfsK, a Ser/Thr kinase from Streptomyces coelicolor A3(2) and S-adenosyl-L-methionine. FEMS Microbiol. Lett. 266, 236–240. doi: 10.1111/j.1574-6968.2006.00531.x
Li, Y., Kong, L., Shen, J., Wang, Q., Liu, Q., Yang, W., et al. (2019). Characterization of the positive SARP family regulator PieR for improving piericidin A1 production in Streptomyces piomogeues var. Hangzhouwanensis. Synth. Syst. Biotechnol. 4, 16–24. doi: 10.1016/j.synbio.2018.12.002
Li, Z., Li, S., Du, L., Zhang, X., Jiang, Y., Liu, W., et al. (2021). Engineering bafilomycin high-producers by manipulating regulatory and biosynthetic genes in the marine bacterium Streptomyces lohii. Mar. Drugs 19:29. doi: 10.3390/md19010029
Li, J., Li, Y., Niu, G., Guo, H., Qiu, Y., Lin, Z., et al. (2018). NosP-regulated nosiheptide production responds to both peptidyl and small-molecule ligands derived from the precursor peptide. Cell. Chem. Biol. 25, 143–153.e4. doi: 10.1016/j.chembiol.2017.10.012
Li, Z., Li, X., and Xia, H. (2022). Roles of LuxR-family regulators in the biosynthesis of secondary metabolites in actinobacteria. World J. Microbiol. Biotechnol. 38:250. doi: 10.1007/s11274-022-03414-1
Li, R., Liu, G., Xie, Z., He, X., Chen, W., Deng, Z., et al. (2010). PolY, a transcriptional regulator with ATPase activity, directly activates transcription of polR in polyoxin biosynthesis in Streptomyces cacaoi. Mol. Microbiol. 75, 349–364. doi: 10.1111/j.1365-2958.2009.06968.x
Li, R., Xie, Z., Tian, Y., Yang, H., Chen, W., You, D., et al. (2009). polR, a pathway-specific transcriptional regulatory gene, positively controls polyoxin biosynthesis in Streptomyces cacaoi subsp. asoensis. Microbiology (Reading) 155, 1819–1831. doi: 10.1099/mic.0.028639-0
Li, Y., Zhang, J., Zheng, J., Guan, H., Liu, W., and Tan, H. (2020). Co-expression of a SARP Family activator ChlF2 and a type II thioesterase ChlK led to high production of chlorothricin in Streptomyces antibioticus DSM 40725. Front. Bioeng. Biotechnol. 8:1013. doi: 10.3389/fbioe.2020.01013
Liu, G., Chater, K. F., Chandra, G., Niu, G., and Tan, H. (2013). Molecular regulation of antibiotic biosynthesis in streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143. doi: 10.1128/MMBR.00054-12
López-Agudelo, V. A., Gómez-Ríos, D., and Ramirez-Malule, H. (2021). Clavulanic acid production by Streptomyces clavuligerus: insights from systems biology, strain engineering, and downstream processing. Antibiotics (Basel) 10:84. doi: 10.3390/antibiotics10010084
Lu, F., Hou, Y., Zhang, H., Chu, Y., Xia, H., and Tian, Y. (2017). Regulatory genes and their roles for improvement of antibiotic biosynthesis in Streptomyces. Streptomyces 7:250. doi: 10.1007/s13205-017-0875-6
Luzhetskyy, A., Mayer, A., Hoffmann, J., Pelzer, S., Holzenkämper, M., Schmitt, B., et al. (2007). Cloning and heterologous expression of the aranciamycin biosynthetic gene cluster revealed a new flexible glycosyltransferase. Chembiochem 8, 599–602. doi: 10.1002/cbic.200600529
Ma, D., Wang, C., Chen, H., and Wen, J. (2018). Manipulating the expression of SARP family regulator BulZ and its target gene product to increase tacrolimus production. Appl. Microbiol. Biotechnol. 102, 4887–4900. doi: 10.1007/s00253-018-8979-4
Maharjan, S., Oh, T. J., Lee, H. C., and Sohng, J. K. (2009). Identification and functional characterization of an afsR homolog regulatory gene from Streptomyces venezuelae ATCC 15439. J. Microbiol. Biotechnol. 19, 121–127. doi: 10.4014/jmb.0803.223
Martín, J. F., and Liras, P. (2019). The balance metabolism safety net: integration of stress signals by interacting transcriptional factors in Streptomyces and related actinobacteria. Front. Microbiol. 10:3120. doi: 10.3389/fmicb.2019.03120
Martín, J. F., Liras, P., and Sánchez, S. (2021). Modulation of gene expression in actinobacteria by translational modification of transcriptional factors and secondary metabolite biosynthetic enzymes. Front. Microbiol. 12:630694. doi: 10.3389/fmicb.2021.630694
Mast, Y., Guezguez, J., Handel, F., and Schinko, E. (2015). A complex signaling cascade governs pristinamycin biosynthesis in Streptomyces pristinaespiralis. Appl. Environ. Microbiol. 81, 6621–6636. doi: 10.1128/AEM.00728-15
Matsumoto, A., Hong, S. K., Ishizuka, H., Horinouchi, S., and Beppu, T. (1994). Phosphorylation of the AfsR protein involved in secondary metabolism in Streptomyces species by a eukaryotic-type protein kinase. Gene 146, 47–56. doi: 10.1016/0378-1119(94)90832-x
Mingyar, E., Mühling, L., Kulik, A., Winkler, A., Wibberg, D., Kalinowski, J., et al. (2021). A regulator based “semi-targeted” approach to activate silent biosynthetic gene clusters. Int. J. Mol. Sci. 22:7567. doi: 10.3390/ijms22147567
Misaki, Y., Nindita, Y., Fujita, K., Fauzi, A. A., and Arakawa, K. (2022). Overexpression of SRO_3163, a homolog of Streptomyces antibiotic regulatory protein, induces the production of novel cyclohexene-containing enamide in Streptomyces rochei. Biosci. Biotechnol. Biochem. 86, 177–184. doi: 10.1093/bbb/zbab206
Niu, G., Chater, K. F., Tian, Y., Zhang, J., and Tan, H. (2016). Specialised metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol. Rev. 40, 554–573. doi: 10.1093/femsre/fuw012
Parajuli, N., Viet, H. T., Ishida, K., Tong, H. T., Lee, H. C., Liou, K., et al. (2005). Identification and characterization of the afsR homologue regulatory gene from Streptomyces peucetius ATCC 27952. Res. Microbiol. 156, 707–712. doi: 10.1016/j.resmic.2005.03.005
Prija, F., Srinivasan, P., Das, S., Kattusamy, K., and Prasad, R. (2017). DnrI of Streptomyces peucetius binds to the resistance genes, drrAB and drrC but is activated by daunorubicin. J. Basic Microbiol. 57, 862–872. doi: 10.1002/jobm.201700162
Rodríguez, M., Núñez, L. E., Braña, A. F., Méndez, C., Salas, J. A., and Blanco, G. (2011). Mutational analysis of the thienamycin biosynthetic gene cluster from Streptomyces cattleya. Antimicrob. Agents Chemother. 55, 1638–1649. doi: 10.1128/AAC.01366-10
Santos-Aberturas, J., Vicente, C. M., Payero, T. D., Martín-Sánchez, L., Cañibano, C., Martín, J. F., et al. (2012). Hierarchical control on polyene macrolide biosynthesis: PimR modulates pimaricin production via the PAS-LuxR transcriptional activator PimM. PLoS One 7:e38536. doi: 10.1371/journal.pone.0038536
Santos-Beneit, F., Rodríguez-García, A., and Martín, J. F. (2012). Overlapping binding of PhoP and AfsR to the promoter region of glnR in Streptomyces coelicolor. Microbiol. Res. 167, 532–535. doi: 10.1016/j.micres.2012.02.010
Santos-Beneit, F., Rodríguez-García, A., Sola-Landa, A., and Martín, J. F. (2009). Cross-talk between two global regulators in Streptomyces: PhoP and AfsR interact in the control of afsS, pstS and phoRP transcription. Mol. Microbiol. 72, 53–68. doi: 10.1111/j.1365-2958.2009.06624.x
Sawai, R., Suzuki, A., Takano, Y., Lee, P. C., and Horinouchi, S. (2004). Phosphorylation of AfsR by multiple serine/threonine kinases in Streptomyces coelicolor A3(2). Gene 334, 53–61. doi: 10.1016/j.gene.2004.02.046
Seipke, R. F., Song, L., Bicz, J., Laskaris, P., Yaxley, A. M., Challis, G. L., et al. (2011). The plant pathogen Streptomyces scabies 87-22 has a functional pyochelin biosynthetic pathway that is regulated by TetR-and AfsR-family proteins. Microbiology (Reading) 157, 2681–2693. doi: 10.1099/mic.0.047977-0
Sharma, V., Kaur, R., and Salwan, R. (2021). Streptomyces: host for refactoring of diverse bioactive secondary metabolites. 3. Biotech 11:340. doi: 10.1007/s13205-021-02872-y
She, W., Ye, W., Cheng, A., Ye, W., Ma, C., Wang, R., et al. (2022). Discovery, yield improvement, and application in marine coatings of potent antifouling compounds albofungins targeting multiple fouling organisms. Front. Microbiol. 13:906345. doi: 10.3389/fmicb.2022.906345
Sohoni, S. V., Fazio, A., Workman, C. T., Mijakovic, I., and Lantz, A. E. (2014). Synthetic promoter library for modulation of actinorhodin production in Streptomyces coelicolor A3(2). PLoS One 9:e99701. doi: 10.1371/journal.pone.0099701
Steele, J. F. C., Hughes, R. K., and Banfield, M. J. (2019). Structural and biochemical studies of an NB-ARC domain from a plant NLR immune receptor. PLoS One 14:e0221226. doi: 10.1371/journal.pone.0221226
Stratigopoulos, G., Bate, N., and Cundliffe, E. (2004). Positive control of tylosin biosynthesis: pivotal role of TylR. Mol. Microbiol. 54, 1326–1334. doi: 10.1111/j.1365-2958.2004.04347.x
Sun, L., Zeng, J., Cui, P., Wang, W., Yu, D., and Zhan, J. (2018). Manipulation of two regulatory genes for efficient production of chromomycins in Streptomyces reseiscleroticus. J. Biol. Eng. 12:9. doi: 10.1186/s13036-018-0103-x
Suzuki, T., Mochizuki, S., Yamamoto, S., Arakawa, K., and Kinashi, H. (2010). Regulation of lankamycin biosynthesis in Streptomyces rochei by two SARP genes, srrY and srrZ. Biosci. Biotechnol. Biochem. 74, 819–827. doi: 10.1271/bbb.90927
Tahlan, K., Anders, C., and Jensen, S. E. (2004). The paralogous pairs of genes involved in clavulanic acid and clavam metabolite biosynthesis are differently regulated in Streptomyces clavuligerus. J. Bacteriol. 186, 6286–6297. doi: 10.1128/JB.186.18.6286-6297.2004
Tanaka, A., Takano, Y., Ohnishi, Y., and Horinouchi, S. (2007). AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J. Mol. Biol. 369, 322–333. doi: 10.1016/j.jmb.2007.02.096
Tang, L., Grimm, A., Zhang, Y. X., and Hutchinson, C. R. (1996). Purification and characterization of the DNA-binding protein DnrI, a transcriptional factor of daunorubicin biosynthesis in Streptomyces peucetius. Mol. Microbiol. 22, 801–813. doi: 10.1046/j.1365-2958.1996.01528.x
Tang, Z. K., Li, X. M., Pang, A. P., Lin, C. Y., Zhang, Y., Zhang, J., et al. (2017). Characterization of three pathway-specific regulators for high production of monensin in Streptomyces cinnamonensis. Appl. Microbiol. Biotechnol. 101, 6083–6097. doi: 10.1007/s00253-017-8353-y
Torkkell, S., Ylihonko, K., Hakala, J., Skurnik, M., and Mäntsälä, P. (1997). Characterization of Streptomyces nogalater genes encoding enzymes involved in glycosylation steps in nogalamycin biosynthesis. Mol. Gen. Genet. 256, 203–209. doi: 10.1007/s004380050562
Tsypik, O., Makitrynskyy, R., Yan, X., Koch, H. G., Paululat, T., and Bechthold, A. (2021). Regulatory control of rishirilide(s) biosynthesis in Streptomyces bottropensis. Microorganisms 9:374. doi: 10.3390/microorganisms9020374
Umeyama, T., and Horinouchi, S. (2001). Autophosphorylation of a bacterial serine/threonine kinase, AfsK, is inhibited by KbpA, an AfsK-binding protein. J. Bacteriol. 183, 5506–5512. doi: 10.1128/JB.183.19.5506-5512.2001
Umeyama, T., Lee, P. C., and Horinouchi, S. (2002). Protein serine/threonine kinases in signal transduction for secondary metabolism and morphogenesis in Streptomyces. Appl. Microbiol. Biotechnol. 59, 419–425. doi: 10.1007/s00253-002-1045-1
Umeyama, T., Lee, P. C., Ueda, K., and Horinouchi, S. (1999). An AfsK/AfsR system involved in the response of aerial mycelium formation to glucose in Streptomyces griseus. Microbiology (Reading) 145, 2281–2292. doi: 10.1099/00221287-145-9-2281
Van der Heul, H. U., Bilyk, B. L., McDowall, K. J., Seipke, R. F., and van Wezel, G. P. (2018). Regulation of antibiotic production in actinobacteria: new perspectives from the post-genomic era. Nat. Prod. Rep. 35, 575–604. doi: 10.1039/c8np00012c
Wang, H., Liu, Y., Cheng, X., Zhang, Y., Li, S., Wang, X., et al. (2022). Titer improvement of milbemycins via coordinating metabolic competition and transcriptional co-activation controlled by Streptomyces antibiotic regulatory protein family regulator in Streptomyces bingchenggensis. Biotechnol. Bioeng. 119, 1252–1263. doi: 10.1002/bit.28044
Wang, W., Wang, H., Hu, H., Peng, H., and Zhang, X. (2015). Overexpression of afsR and optimization of metal chloride to improve lomofungin production in Streptomyces lomondensis S015. J. Microbiol. Biotechnol. 25, 672–680. doi: 10.4014/jmb.1409.09091
Wei, J., He, L., and Niu, G. (2018). Regulation of antibiotic biosynthesis in actinomycetes: perspectives and challenges. Synth. Syst. Biotechnol. 3, 229–235. doi: 10.1016/j.synbio.2018.10.005
Wei, J., Ma, M., Guo, S., Xu, Y., Xie, J., Pan, G., et al. (2022). Characterization of pathway-specific regulator NigR for high yield production of nigericin in Streptomyces malaysiensis F913. Antibiotics (Basel) 11:938. doi: 10.3390/antibiotics11070938
Wietzorrek, A., and Bibb, M. (1997). A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 25, 1181–1184. doi: 10.1046/j.1365-2958.1997.5421903.x
Wu, Q. B., Chen, X. A., Lv, Z. Y., Zhang, X. Y., Liu, Y., and Li, Y. Q. (2021). Activation and discovery of tsukubarubicin from Streptomyces tsukubaensis through overexpressing SARPs. Appl. Microbiol. Biotechnol. 105, 4731–4741. doi: 10.1007/s00253-021-11344-5
Xia, H., Li, X., Li, Z., Zhan, X., Mao, X., and Li, Y. (2020). The application of regulatory cascades in Streptomyces: yield enhancement and metabolite mining. Front. Microbiol. 11:406. doi: 10.3389/fmicb.2020.00406
Xie, P., Sheng, Y., Ito, T., and Mahmud, T. (2012). Transcriptional regulation and increased production of asukamycin in engineered Streptomyces nodosus subsp. asukaensis strains. Appl. Microbiol. Biotechnol. 96, 451–460. doi: 10.1007/s00253-012-4084-2
Xie, Y., Wang, B., Liu, J., Zhou, J., Ma, J., Huang, H., et al. (2012). Identification of the biosynthetic gene cluster and regulatory cascade for the synergistic antibacterial antibiotics griseoviridin and viridogrisein in Streptomyces griseoviridis. Chembiochem 13, 2745–2757. doi: 10.1002/cbic.201200584
Xu, Z., Schenk, A., and Hertweck, C. (2007). Molecular analysis of the benastatin biosynthetic pathway and genetic engineering of altered fatty acid-polyketide hybrids. J. Am. Chem. Soc. 129, 6022–6030. doi: 10.1021/ja069045b
Yan, H., Lu, X., Sun, D., Zhuang, S., Chen, Q., Chen, Z., et al. (2020). BldD, a master developmental repressor, activates antibiotic production in two Streptomyces species. Mol. Microbiol. 113, 123–142. doi: 10.1111/mmi.14405
Yan, Y. S., Yang, Y. Q., Zhou, L. S., Zhang, L., and Xia, H. Y. (2022). MilR3, a unique SARP family pleiotropic regulator in Streptomyces bingchenggensis. Arch. Microbiol. 204:631. doi: 10.1007/s00203-022-03240-x
Yan, Y. S., Zou, L. S., Wei, H. G., Yang, M. Y., Yang, Y. Q., Li, X. F., et al. (2024). An atypical two-component system, AtcR/AtcK, simultaneously regulates the biosynthesis of multiple secondary metabolites in Streptomyces bingchenggensis. Appl. Environ. Microbiol. 90:e0130023. doi: 10.1128/aem.01300-23
Yang, Z., Wei, X., He, J., Sun, C., Ju, J., and Ma, J. (2019). Characterization of the noncanonical regulatory and transporter genes in atratumycin biosynthesis and production in a heterologous host. Mar. Drugs 17:560. doi: 10.3390/md17100560
Ye, S., Molloy, B., Braña, A. F., Zabala, D., Olano, C., Cortés, J., et al. (2017). Identification by genome mining of a type I polyketide gene cluster from Streptomyces argillaceus involved in the biosynthesis of pyridine and piperidine alkaloids argimycins P. Front. Microbiol. 8:194. doi: 10.3389/fmicb.2017.00194
Ye, S., Molloy, B., Pérez-Victoria, I., Montero, I., Braña, A. F., Olano, C., et al. (2023). Uncovering the cryptic gene cluster ahb for 3-amino-4-hydroxybenzoate derived ahbamycins, by searching SARP regulator encoding genes in the Streptomyces argillaceus Genome. Int. J. Mol. Sci. 24:8197. doi: 10.3390/ijms24098197
Yin, S., Wang, W., Wang, X., Zhu, Y., Jia, X., Li, S., et al. (2015). Identification of a cluster-situated activator of oxytetracycline biosynthesis and manipulation of its expression for improved oxytetracycline production in Streptomyces rimosus. Microb. Cell Factories 14:46. doi: 10.1186/s12934-015-0231-7
Keywords: Streptomyces , SARP family regulator, biosynthetic gene cluster, secondary metabolism, biotechnological application
Citation: Yan Y and Xia H (2024) The roles of SARP family regulators involved in secondary metabolism in Streptomyces. Front. Microbiol. 15:1368809. doi: 10.3389/fmicb.2024.1368809
Received: 11 January 2024; Accepted: 26 February 2024;
Published: 14 March 2024.
Edited by:
Shumpei Asamizu, Kobe University, JapanReviewed by:
Takeaki Tezuka, Kitasato University, JapanCopyright © 2024 Yan and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiyang Xia, aHl4aWFAdHpjLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.