
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 April 2024
Sec. Microbial Physiology and Metabolism
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1368499
Stringent response and quorum sensing (QS) are two essential mechanisms that control bacterial global metabolism for better survival. Sphingomonads are a clade of bacteria that survive successfully in diverse ecosystems. In silico survey indicated that 36 out of 79 investigated sphingomonads strains contained more than one luxI homolog, the gene responsible for the biosynthesis of QS signal acyl homoserine lactones (AHLs). Investigation of the regulatory effects of the stringent response gene rsh on QS related bioactivities were carried out using rsh mutants of Sphingobium japonicum UT26 and Sphingobium sp. SYK-6, both had three luxI homologs. Results indicated that deletion of rsh upregulated the overall production of AHLs and extracellular polymeric substances (EPS) in both UT26 and SYK-6 in rich medium, but affected expressions of these luxI/luxR homologs in different ways. In the poor medium (1% LB), rsh mutant of SYK-6 significantly lost AHLs production in broth cultivation but not in biofilm cultivation. The regulatory effects of rsh on QS activities were growth phase dependent in UT26 and culture condition dependent in SYK-6. Our results demonstrated the negative regulatory effect of rsh on QS activities in sphingomonads, which were very different from the positive effect found in sphingomonads containing only one luxI/R circuit. This study extends the current knowledge on the intricate networks between stringent response and QS system in sphingomonads, which would help to understand their survival advantage.
As one of the most successful organisms in this world, bacteria employ sophisticated mechanisms to regulate their growth and metabolisms. Among these regulatory mechanisms, stringent response and quorum sensing (QS) are two essential mechanisms that control bacterial global metabolism (Waters and Bassler, 2005; Gaca et al., 2012).
Stringent response is a strategy used by bacteria to cope with environmental stress such as nutrients limitation, oxidative stress, etc. (Poole, 2012; Lu et al., 2023). The stringent response is mediated by guanosine tetraphosphate and guanosine pentaphosphate [(p)ppGpp], which is synthesized and hydrolyzed by members of the RSH superfamily (Dalebroux and Swanson, 2012). The corresponding stringent response gene rsh was reported to regulate various bacterial bioactivities including QS (Schafhauser et al., 2014; Lu et al., 2023).
The QS is a mechanism of cell-to-cell signaling involving the production of hormone-like signal molecules called autoinducers (Papenfort and Bassler, 2016). In Gram-negative bacteria, the QS signals are usually acyl-homoserine lactones (AHLs), which are produced by LuxI-type synthases (Fuqua et al., 2001). It is proved that QS involves in lots of activities that are crucial for bacterial survival, such as biofilm formation, motility, EPS production etc. (Sperandio et al., 2001; Tan et al., 2014).
Both stringent response and QS are favorable mechanisms for bacterial survival. A fundamental question is that how does stringent response interact with the QS system. Previous studies on Pseudomonas spp. and Novosphingobium spp. indicated that stringent response did interfere with QS systems in multiple ways (Gan et al., 2009; Schafhauser et al., 2014; Wang et al., 2022). Deletion of spoT, one of the stringent response gene, resulted in decreased AHLs production in Pseudomonas protegens SN15-2 (Wang et al., 2022). Whereas in Pseudomonas aeruginosa, (p)ppGpp-null mutant has reduced butanoyl-homoserine lactone (C4-HSL) and 3-oxo-dodecanoyl-homoserine lactone (3-oxo-C12-HSL) levels, but has increased biosynthesis of the other kind of QS signal, HHQ (4-hydroxyl-2-heptyl quinoline) and PQS (3,4-dihydroxy-2-heptylquinoline) (Schafhauser et al., 2014). The intricate network between stringent response and QS remains to be elucidated.
Sphingomonads, mainly consists of Novosphingobium, Sphingopyxis, Sphingobium, and Sphingomonas, attract great research interest due to their ubiquitous distribution and diverse metabolism capacity (Stolz, 2009; Glaeser and Kampfer, 2014; Huang et al., 2017; Sheng et al., 2022; Lu et al., 2023). Their sophisticated QS systems, often containing multiple luxI/R genes, might contribute to their survival advantage (Gan et al., 2015). In Novosphingobium sp. Rr 2–17, and Novosphingobium pentaromativorans US6-1, the rsh gene was necessary for the accumulation of AHLs, suggesting that QS was strictly under the regulation of rsh (Gan et al., 2009; Lu and Huang, 2018). Given that both US6-1 and Rr 2–17 have only one pair of luxI/R homologs in their genome, it is still preliminary to conclude that stringent response positively regulates the QS activities in sphingomonads. Strains with multiple luxI/R homologs in their genomes should be tested for the interaction between these two systems.
In this study, we surveyed the position and diversity of luxI homologs in sphingomonads. Then we chose Sphingobium japonicum UT26 and Sphingobium sp. SYK-6, which had three luxI homologs, to investigate the effect of rsh deletion on the QS-related bioactivities, such as AHL production, luxI/R gene expression and EPS production. Our results indicated that the regulatory effects of rsh on the QS related activities were species specific and culture condition dependent, which extended our understanding of the networks between QS and stringent response.
All AHL synthase genes, luxI/R homologs, and rsh gene sequences were retrieved from NCBI Genome Database (Microbial Genomes www.ncbi.nih.gov) using the keywords “AHL synth(et)ase,” “quorum sensing,” “luxI” and “autoinducer.” Detailed information for these resulting genes such as sequences and gene locations were manually checked, and recorded for further analysis.
The amino acid sequences of these luxI homologs were aligned by clustalW using MEGA software (version 11), and the phylogenetic tree was constructed by the neighbor-joining method with 1,000 bootstrap.
Bacterial strains and plasmids used in this study are listed in Table 1. Sphingobium japonicum UT26 and Sphingobium sp. SYK-6 were grown at 28°C in 1/3 LB media and LB media, respectively. Their rsh deletion mutants were grown in respective medium plus 100 μg/mL streptomycin. The AHL reporter strain Agrobacterium tumefaciens A136 and all Escherichia coli strains were grown at 37°C in LB medium with proper antibiotics (Lu and Huang, 2018).
In-frame deletions of rsh gene in SYK-6 and UT26 were performed by employing the markerless gene deletion system for Sphingomonads (Kaczmarczyk et al., 2012; Lu and Huang, 2018). Primers used for gene deletion are listed in Supplementary Table S1. Briefly, flanking regions of rsh, approximately 500 bp up- and down-stream each, were amplified by PCR with the primer pairs syk6-5O/-5I and syk6-3O/-3I, then these two fragments were joined using overlap PCR with the primer pair syk6-5O/-3O. The resulting fragment was then cloned into the plasmid pAK405 via the BamHI/HindIII restriction site. Subsequently, the pAK405 derivative was delivered into the wild type strain via conjugal transfer from E. coli S17-1 (λ pir). After the conjugal transfer, bacteria were plated on LB media supplemented with kanamycin (100 μg/mL) and rifampicin (50 μg/mL). Individual colonies were re-streaked once on the same medium and then plated on LB media supplemented with 100 μg/mL streptomycin to select for the second homologous recombination event. The resulting colonies were re-streaked on both LB media supplemented with 100 μg/mL streptomycin and LB media containing 100 μg/mL kanamycin. Kanamycin-sensitive clones were analyzed by colony PCR using primers syk6-FO/-RO to confirm the successful deletion of rsh.
Bacterial strains were routinely grown in respective media at the shaking speed of 200 rpm, and their growth curves were drawn by measuring OD600 values at regular time intervals using a Spectrophotometer (Metash, Shanghai, China). All measurements were performed in triplicates.
Congo red is an azo dye that can form complexes with different components in extracellular polymeric substances (EPS) (Semedo et al., 2015). The Congo red binding assay was carried out to evaluate bacterial EPS production (Lu and Huang, 2018).
Cultures were inoculated from a single colony and grown to an OD600 of 1 in LB broth; 10 μL of suspensions were spotted on LB plates containing 40 μg/mL Congo red. The plates were grown at 30°C to assess the morphology of colony. For quantitative analysis, the same number of cells from different culture conditions were suspended in 100 μg/mL Congo red in 0.9% saline and then incubated for 90 min with shaking at 30 rpm at room temperature. After incubation, the supernatant was obtained by centrifugation, and then the OD490 of the supernatant was measured as A1. The OD490 of Congo red solution without bacterial fluid was recorded as A0. All measurements were performed in triplicates. The percentage of Congo red stain was calculated to represent the abundance of EPS, by the formula:
The AHL production was analyzed either by cross-feeding assay or paper disk assay, according to our previous publication (Lu and Huang, 2018). Briefly, in the cross-feeding assay, the AHL reporter strain A136 and the tested strains were streaked side by side on the agar plates pre-covered with 50 μL X-gal (20 mg/mL stock solution in dimethylformamide). In the paper disk assay, ethyl acetate extracts of samples were spotted onto paper disks and incubated in LB soft agar plates mixed with A136 plus X-gal. The blue coloration results were recorded accordingly. LC–MS/MS was employed to identify the AHL profiles according to Huang et al. (2019).
qRT-PCR on luxI/R homologs was done as described previously (Lu and Huang, 2018). Briefly, cells of UT26 and SYK-6 at different growth phases and culture conditions were harvested and their total RNA was extracted with RNAprep pure Cell/Bacteria Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. cDNA was synthesized using FastKing gDNA Dispelling RT SuperMix (Tiangen Biotech, Beijing, China) and was used as the template for qRT-PCR amplification by using SYBR Premix Ex TaqTM Kit (Tli RNaseH Plus) (TaKaRa, Dalian, China) in Applied Biosystems StepOnePlus TM Real-Time PCR System (Thermo Scientific, Waltham, MA, United States). Primers were designed on the Sangon website1 and listed in Supplementary Table S2. The relative gene expression level of the mutant to the wild-type strain was analyzed using the 2−∆∆Ct analysis method (Lu and Huang, 2018).
Bacterial cells at the early stationary phase were collected for ppGpp detection using the formic acid method (Lu and Huang, 2018). The presence of ppGpp in the extract was then determined by HPLC (Agilent, Santa Clara, CA, United States) analysis using a ZORBAX SB-C18 column (4.6 × 150 mm, 5 μm, Agilent). The mobile phase consisted of 125 mM KH2PO4, 10 mM tetrabutyl ammonium dihydrogen phosphate, 60 mL/L methanol, and 1 g/L KOH (pH 6.0). The column temperature was 40°C and the flow rate was 1.0 mL/min. The ppGpp was monitored at 254 nm and identified by comparison with the retention time of 100 μM ppGpp standards (TriLink Biosciences, San Diego, CA, United States). The ppGpp standard was eluted at around 71 min under the above-mentioned conditions to confirm the presence of ppGpp.
A Student’s t-test with a p-value cut-off of 0.05 was used to test significance of differences using the software SPSS (version 25).
Up to November 2023, 158 strains of sphingomonads had their whole genome sequenced. Among them, 79 strains were found to harbor luxI homologs, including 10 Novosphingobium spp., 18 Sphingopyxis spp., 26 Sphingobium spp., and 25 Sphingomonas spp. (Supplementary Table S3). The number of luxI homologs in each strain ranged from 1 to 4. 36 out of 79 strains contained more than one luxI homolog, and 13 out of 79 strains contained 3 luxI homologs, such as Sphingobium japonicum UT26 and Sphingobium sp. SYK-6 (Figure 1).
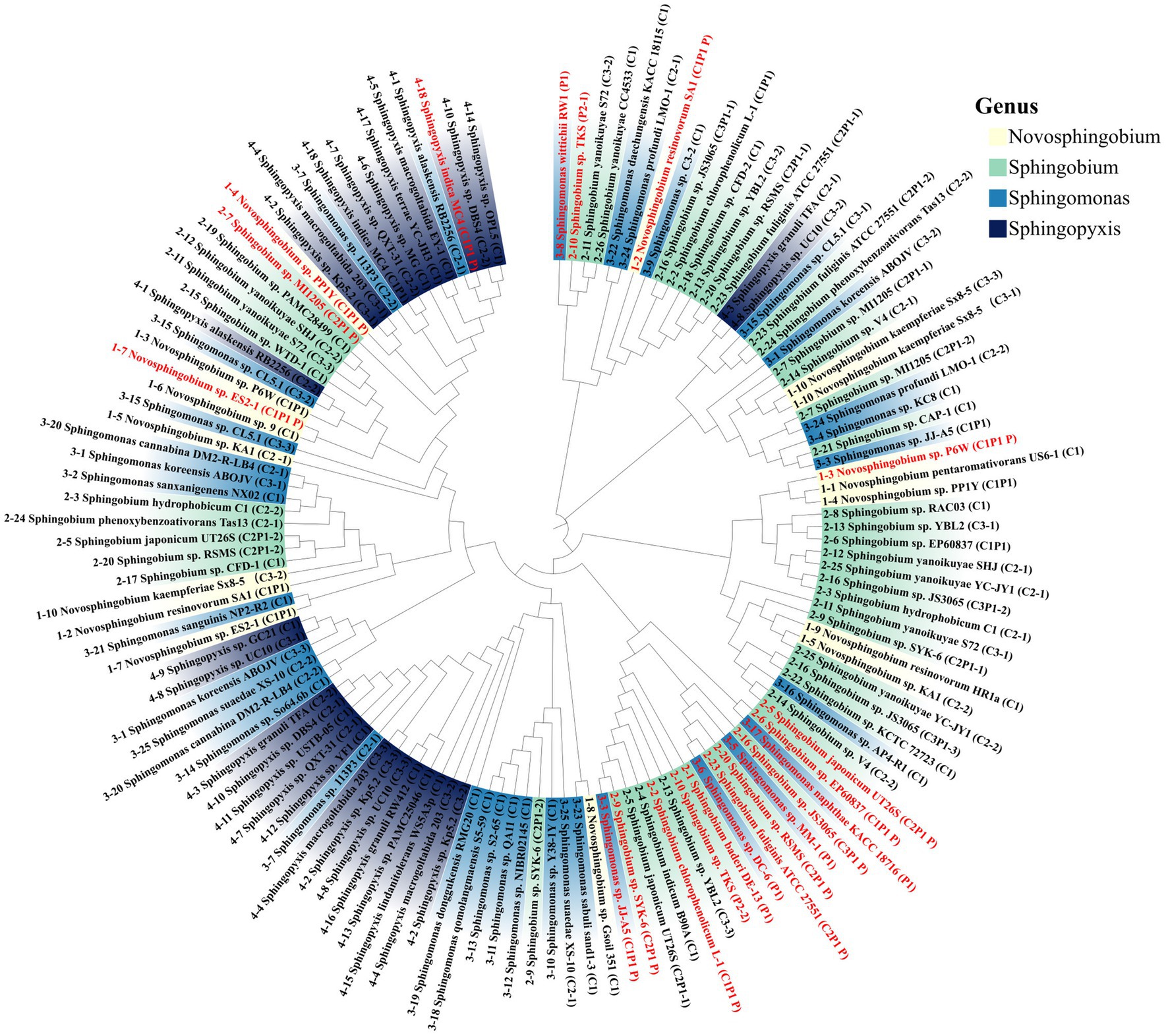
Figure 1. Phylogenetic tree of the LuxI-type synthases from Sphingomonads strains with completed genome sequence. In total, 131 sequences from 79 strains are plotted in the tree. The synthases located on the plasmid are depicted in red, and the synthases located on the chromosome are depicted in black. 59 out of 79 strains harbor luxI homologs in chromosome only, while 14 out of 79 strains harbor luxI homologs in both chromosome and plasmid. Different genera are highlighted in different colors. Each strain is assigned a serial number that depicted in front of the strain names. If a strain contains multiple luxI homologs then the total number and the location was indicated in brackets after the strain name. C for chromosome and P for plasmid. Different genes in the same strain are distinguished by a serial number or P. For example, (C2P1 P) means there are two luxI genes on the chromosome and one on the plasmid, this one is on the plasmid, the other two genes of this strain are indicated as (C2P1–1) and (C2P1–2).
The majority of these luxI homologs (110 out of 131) were located on chromosome. More than two thirds, 59 out of 79, of these strains harbored luxI homologs in chromosome only, while 14 out of 79 strains harbored luxI homologs in both chromosome and plasmid (Figure 1). The phylogenetic tree showed that these luxI homologs in sphingomonads were not phylogenetically related (Figure 1). LuxI homologs from different genera mingled with each other and even luxI homologs from the same strain rarely clustered together.
Sequence analysis on genomes of UT26 and SYK-6 indicated that both UT26 and SYK-6 contained two AHL synthase genes on the chromosome and one on a plasmid. These genes were named sjaI 1–3 for UT26 and sphI 1–3 for SYK-6. Except for sjaI3, other luxI homologs had corresponding luxR homologs in their close vicinity. On the other hand, one rsh gene around 2 kb was found on the chromosome of either UT26 or SYK-6. The detailed sequence information and location of rsh and these QS genes were plotted in Figure 2A, with reference strains US6-1 and Rr-2-17.
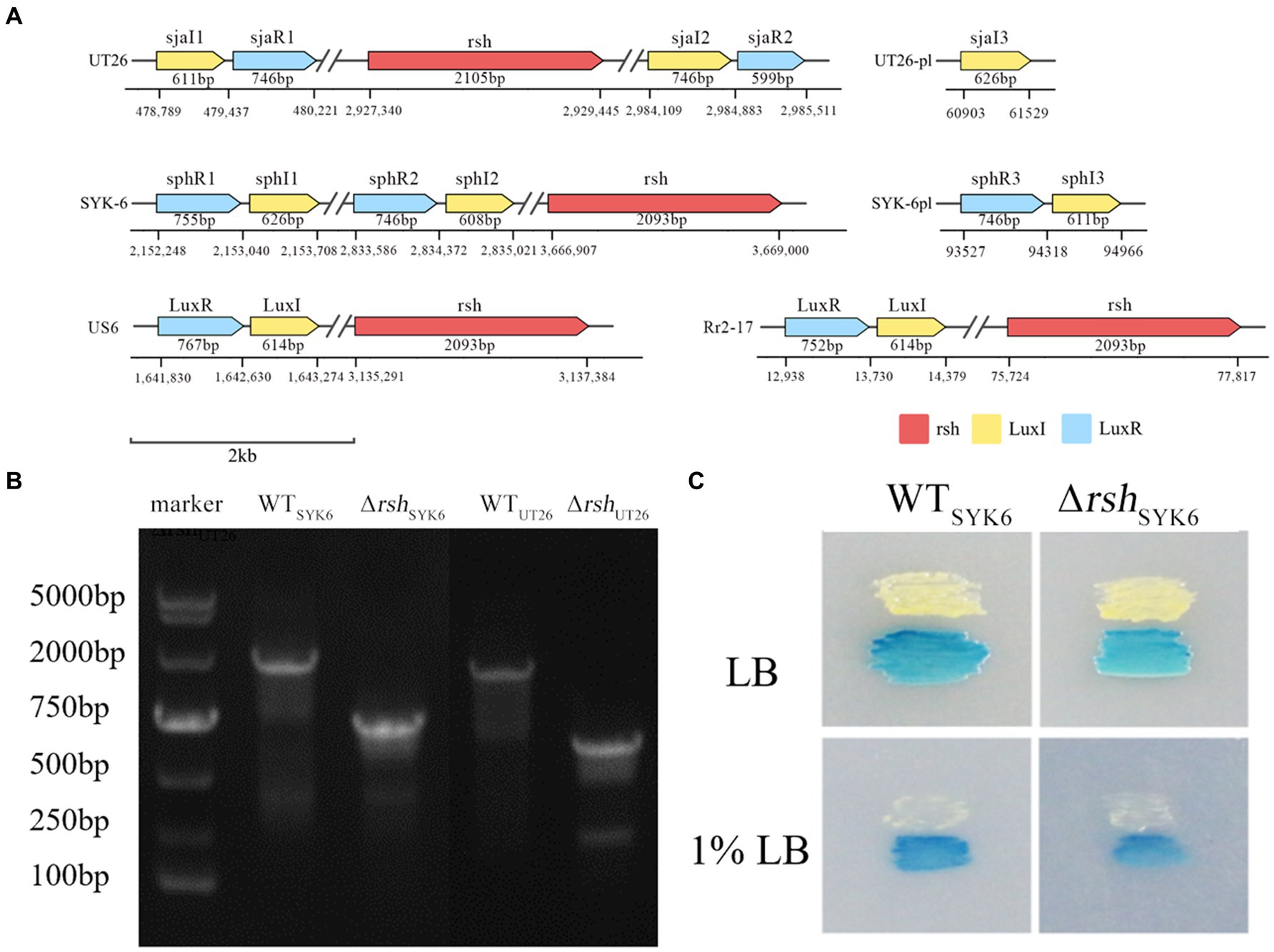
Figure 2. (A) Schematic diagram of the gene organization and location of rsh and luxI/R homologs in the genome of UT26, SYK-6, US6 and Rr2-17. (B) Agarose gel electrophoresis of PCR products from the wild type and the mutant using the same primer set. The shorter PCR products from the mutants indicates successful deletion of rsh gene. (C) Cross feeding assay for AHL production by the WTUT26 and ΔrshUT26 on LB and 1% LB agar plates. The blue coloration of the indicator strain A136 indicates the positive AHL production.
In UT26, both sjaI/R1 and sjaI/R2 located on the chromosome, and the solo sjaI3 located on the plasmid. The sjaI/R1 located distantly upstream to rsh, around 2.5 million bases apart, while the sjaI/R2 located closely downstream to rsh, around 50 kb apart. Whereas in SYK-6, both sphI/R1 and sphI/R2 located distantly upstream to rsh, around 1.5 and 0.8 million bases apart, respectively. The sphI/R3 located on the plasmid. For the reference strains US6-1 and Rr2-17, the distances between rsh and the luxI/R homologs were 1.5 million bases and 60 kb apart, respectively. The physical distance between the rsh and QS genes on the chromosome were rather random among species.
Successful deletion of the rsh gene in UT26 and SYK-6 was confirmed by PCR amplification of the sequence around the rsh gene. PCR products from both mutants were about 2 kb less than those of their wild type strains, due to the deletion of the rsh gene fragment (Figure 2B). The mutant was deficient in ppGpp production compared to the wild type strain, further proved the successful deletion of rsh (Supplementary Figure S1).
LC–MS/MS analysis indicated that both ΔrshUT26 and ΔrshSYK-6 produced AHL profiles similar to their wild type strains in rich LB medium, while in the minimal medium 1% LB, ΔrshSYK-6 almost lost AHLs production (Table 2). Strain UT26 mainly produced C12-HSL, with a small amount of C14-HSL and 3-OH-C8-HSL. Whereas SYK-6 produced C14-HSL, 3-OH-C8-HSL, 3-OH-C6-HSL, and a trace amount of C8-HSL when grown in LB media (Supplementary Figure S2). The AHL signals from WTSYK-6 and ΔrshSYK-6 were confirmed by the reporter strain A136, and both triggered a blue coloration in A136, indicating a positive AHL production in both strains (Figure 2C). These results indicated that deletion of rsh did not abolish the AHL production, but did affect their AHL profiles in both strains.
Growth curves of ΔrshUT26 and WTUT26 indicated that deletion of rsh rendered a delay in cell proliferation, but their overall biomasses in individual growth phase were similar (Figure 3A). The AHL production analysis indicated that in the middle log phase, AHL productions of ΔrshUT26 and WTUT26 were similar, while in early stationary phase, ΔrshUT26 remained high production of C12-HSL, which was significantly higher than WTUT26 (Figure 3B).
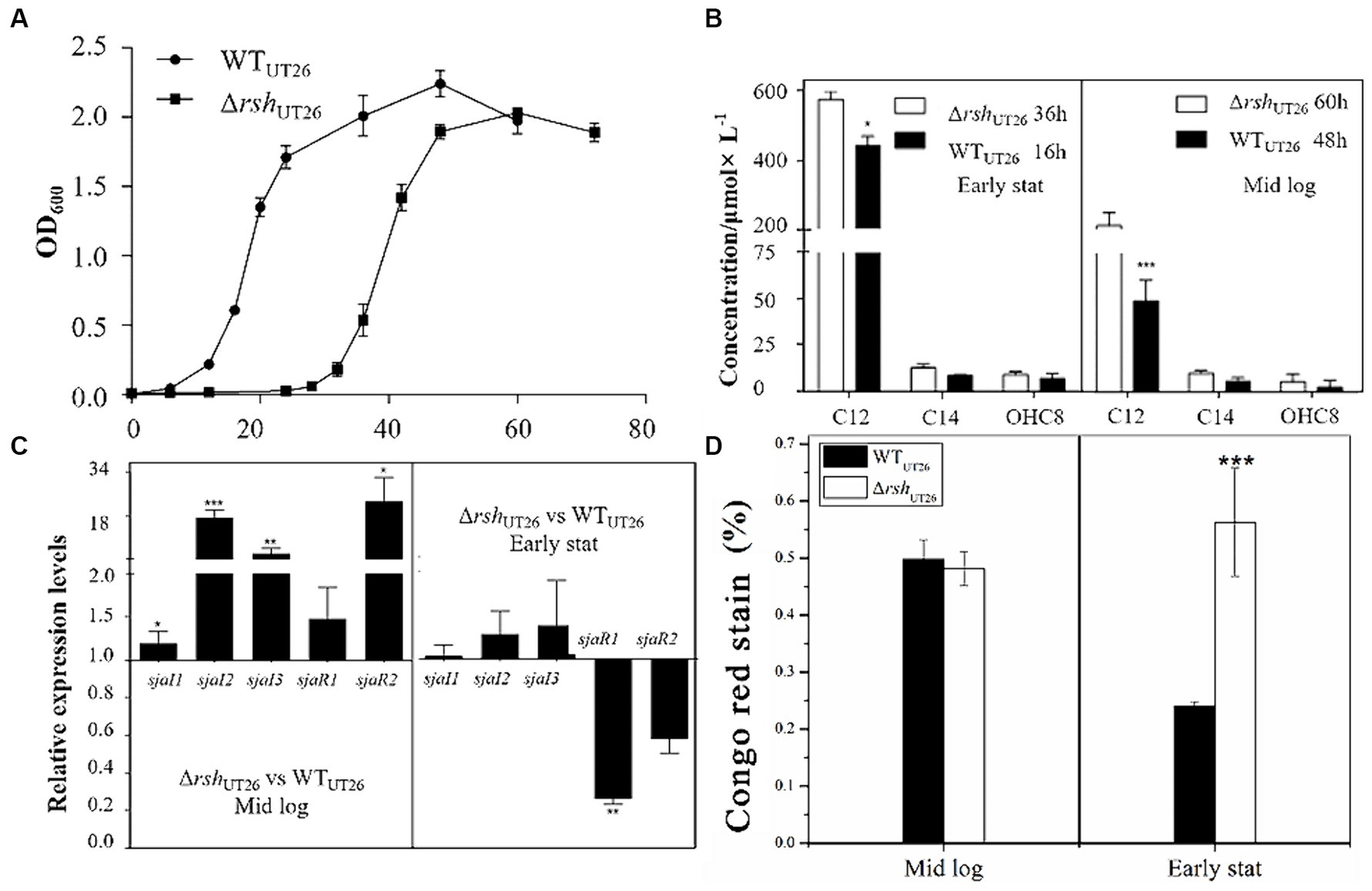
Figure 3. The growth curves, quorum sensing gene expression, AHL production and EPS production by UT26 in the 1/3 LB media. (A) Growth curves of WTUT26 and ΔrshUT26 under the shaking culture condition in 1/3 LB broth. (B) Concentration of the AHLs identified by LC–MS/MS in the WTUT26 and ΔrshUT26 in the middle log phase and the early stationary phase, respectively. (C) Relative expression levels of luxI/R homologs in the ΔrshUT26 compared to the WTUT26 in the middle log phase and the early stationary phase. (D) The Congo red removed percentage as the indicator of the EPS production for WTUT26 and ΔrshUT26 in the middle log phase and the early stationary phase. “*” represents p < 0.05; “**” represents p < 0.01; “***” represents p < 0.001.
Relative gene expressions of sjaI1-3 and sjaR1-2 in ΔrshUT26 compared to WTUT26 were measured (Figure 3C), results indicated that in the middle log phase, all these five genes were upregulated in ΔrshUT26, especially sjaI2-3 and sjaR2. Whereas in the early stationary phase, sjaI1-3 was slightly upregulated, but sjaR1-2 was significantly downregulated. The consistent upregulated expression of these sjaIs in ΔrshUT26 explained the higher production of AHLs in the mutant. The ΔrshUT26 and WTUT26 showed similar EPS production in the middle log phase. However, in the early stationary phase, the mutant showed much higher EPS production than the wild type (Figure 3D).
Overall, these results indicated that for strain UT26, rsh negatively regulated the expression of most QS genes, which in turn negatively regulated AHLs and EPS production. These regulatory effects were growth phase dependent.
Since rsh gene was believed to function when nutrition was limited, therefore, 1% LB medium was used as a poor medium to mimic a nutrient-limited culture condition. Regulatory effects of rsh on AHL/EPS production and gene expression were tested in both rich media (LB) and poor medium (1% LB).
In LB medium, growth curves indicated that deletion of rsh significantly promoted the biomass of ΔrshSYK-6 at stationary phase (Figure 4A). AHL production at 36 and 48 h were visualized. ΔrshSYK-6 showed robust AHL production, the signals were stronger than WTSYK-6 at 48 h (Figure 4B). However, relative expressions of most QS genes were significantly downregulated in ΔrshSYK-6, especially for sphI3 and sphR1. Notably, expression of sphI 2 was not significantly affected (Figure 4C). EPS production of ΔrshSYK-6 was similar to that of the wild type when grown in the liquid medium, while was significantly higher than WTSYK-6 when grown in plate biofilm (Figure 4D). These results indicated that deletion of rsh downregulated most QS genes, the higher AHL and EPS production in the mutant might be attributed to the consistent expression of sphI2, and the higher biomass.
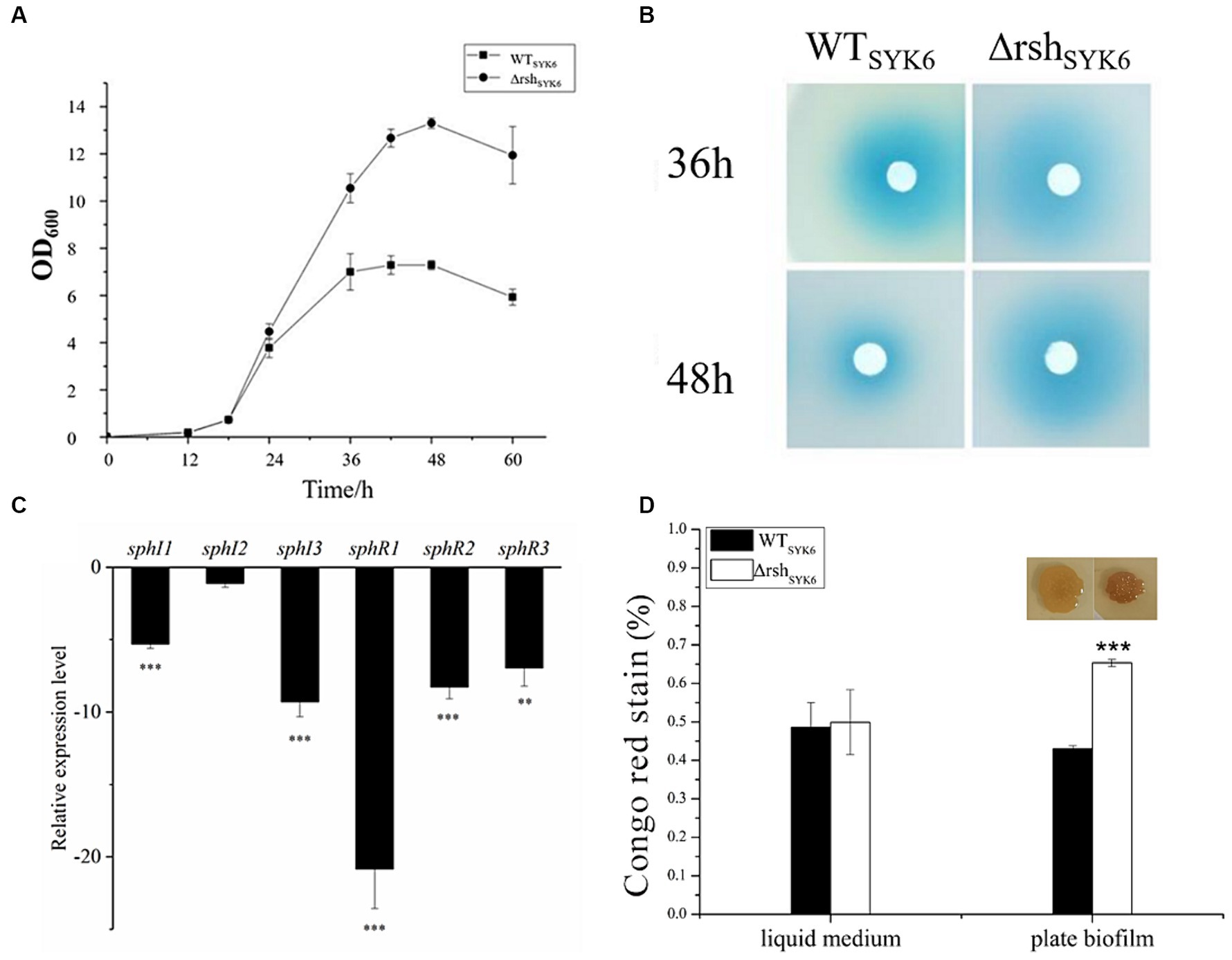
Figure 4. The growth curves, AHL production assay, quorum sensing gene expression assay and EPS production assay by SYK-6 in the LB media. (A) Growth curve of WTSYK-6 and ΔrshSYK-6 under the shaking culture condition in LB broth. (B) Disk diffusion assay for AHL production in the WTSYK6 and the ΔrshSYK-6 grown in the LB broth. The blue coloration indicates the presence of AHL. (C) Relative expression levels of luxI/R homologs in the ΔrshSYK-6 compared to the WTSYK-6. (D) Congo red assay for EPS production of the WTSYK-6 and the ΔrshSYK-6 grown in the LB broth or plates. Colony morphology of strains on the LB plates containing 40 μg/mL Congo red. Colony of ΔrshSYK-6 was redder than the wild type. One representative experiment out of three independent biological replicates is shown. “*” represents p < 0.05; “**” represents p < 0.01; “***” represents p < 0.001.
In 1% LB broth medium, deletion of rsh did not affect the growth of SYK-6 significantly (Figure 5A). Meanwhile, the mutant did not show AHL signals in broth extract (Figure 5B). LC–MS/MS analysis indicated that only 3-OH-C8-HSL, C14-HSL were produced by the WTSYK-6 in 1%LB, while only trace amount of 3-OH-C8-HSL was detected in ΔrshSYK-6 in 1% LB (Table 2). This was contrary to the cross-feeding assay that on 1% LB agar plates, ΔrshSYK-6 triggered a strong blue coloration of the reporter strain A136 just like the WTSYK-6 (Figure 2C). Gene expression analysis indicated that most QS genes were downregulated in the mutant, especially sphI1 and sphR3. Notably, sphI2 was the least affected gene (Figure 5C). The downregulated pattern of rsh on QS genes was different from what was found in the LB medium (Figure 4C). On the other hand, the EPS production of the mutant was always higher than that of the wild type in both broth and plate cultivation (Figure 5D).
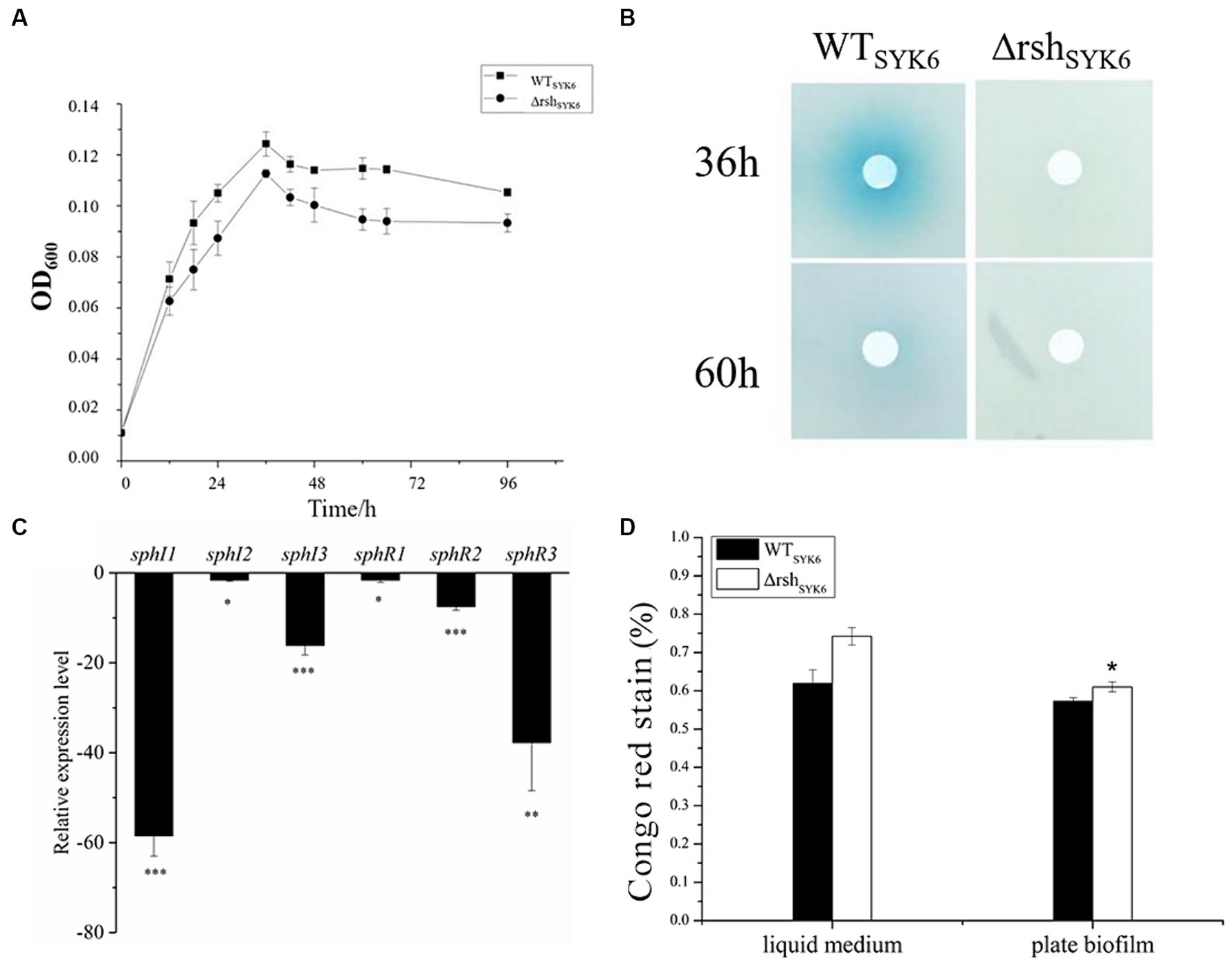
Figure 5. The growth curves, AHL production assay, quorum sensing gene expression assay and EPS production assay by SYK-6 in the 1% LB media. (A) Growth curve of WTSYK-6 and ΔrshSYK-6 under the shaking culture condition in 1%LB broth. (B) Disk diffusion assay for AHL production in the WTSYK-6 and the ΔrshSYK-6 grown in the 1% LB broth. The blue coloration indicates the presence of AHL. (C) Relative expression levels of luxI/R homologs in the ΔrshSYK-6 compared to the WTSYK-6 in 1%LB broth. (D) Congo red assay for EPS production of the WTSYK-6 and the ΔrshSYK-6 grown in the 1% LB broth or plates. “*” represents p < 0.05; “**” represents p < 0.01; “***” represents p < 0.001.
Indeed, the regulatory effect of rsh on the QS system in SYK-6 was affected by nutrient availability and culture conditions. Specifically, planktonic cultivation in broth or attached growth on agar plate (biofilm) affected the regulatory effect of rsh on QS system.
Results of this study demonstrated that the deletion of rsh did not abolish AHL productions in UT26 and SYK-6, it even promoted the production of some individual AHL molecules, in other words, rsh showed a negative regulatory effect on AHL production in UT26 and SYK-6. This was contrary to the positive regulatory effect of rsh on AHL production in US6-1 and Rr 2–27 (Gan et al., 2009; Lu and Huang, 2018). The basic reason for this discrepancy is due to the multiple QS circuits in UT26 and SYK-6. Although it has been reported that in P. aeruginosa, different class of QS systems, the AHL-based and quinolone signal-based QS circuits reacted to stringent response differently (Schafhauser et al., 2014), our study provided more sophisticated knowledge that QS circuits belong to the same class of QS system might react to stringent response differently.
In UT26, higher production of C12-HSL in ΔrshSYK-6 was consistent with the upregulated expression of most QS genes in the mutant strain. While in SYK-6, although some QS genes were downregulated, the mutant still produced AHLs no less than the wild type. Notably, expressions of sjaI1 in UT26 and sphI2 in SYK-6 were not very significantly affected by rsh deletion. Whereas both the AHL synthase genes on plasmids were sensitive to rsh deletion. Combined with the distance and location information of these genes, we deduced that the effect of rsh on QS gene expression was not distance related, but the location, on the chromosome or plasmid, did matter (Figure 2A). In our previous study on Ensifer spp., QS genes on plasmid were very different from those in the chromosome in response to changing environmental conditions (Zeng et al., 2017). The genetic base for the regulatory mechanisms of rsh on QS genes requires more study.
According to growth curves, the deletion of rsh affected the growth of UT26 and SYK-6 in different ways. In UT26, ΔrshUT26 displayed a longer lag phase than WTUT26, but eventually, their biomasses in stationary phases were similar. While in SYK-6, growth phases of WTSYK-6 and ΔrshSYK-6 proceeded at a similar pace, but ΔrshSYK-6 showed remarkably higher biomass than WTSYK-6 in the stationary phase, only in rich medium. Platitudinous nutrients and a lack of stringent response to slow down the growth rate in the stationary phase may explain this phenomenon. One important function of the stringent response is to slow down the growth rate when nutrients become limited (Fritsch et al., 2020). This result also reminds us that, even in the same genus, stringent responses may function differently in different species.
Biofilm formation and EPS production are two physiological activities that are closely related to QS (Sperandio et al., 2001; Tan et al., 2014). In SYK-6, the fact that ΔrshSYK-6 produced detectable AHLs on 1% LB plate (Figure 4B) but not in 1% LB broth (Figure 5B) indicated that biofilm condition affected the execution of rsh regulatory outcome on QS. EPS production in mutants was generally higher than that of the wild types in both strains, which was consistent with the higher QS signal production. All these results suggested that QS and stringent response formed complex regulatory networks, which were affected by bacterial culture and growth conditions.
The intricate regulatory networks may guarantee delicate metabolism regulation that helps sphingomonads to survive in a broad range of econiches. Moreover, Sphingomonads contain lots of strains that have promising remediation potential for a variety of organic pollutants, such as polycyclic aromatic hydrocarbons (PAHs), dioxins, and chlorinated phenols, etc. (Waigi et al., 2017; Samantarrai et al., 2020; Jiang et al., 2022). A better understanding of the regulatory mechanisms and networks in sphingomonads will certainly help to utilize their metabolic functions in environmental bioremediation. In our previous studies, we found that QS in Novosphingobium pentaromativorans US6-1 affected the PAHs degradation ability by altering cell surface properties, while the rsh gene in US6-1 played multiple roles in its accommodation to different environmental pollutants (Chen and Huang, 2020; Lu et al., 2023).
In conclusion, this study provided novel data that the regulatory effect of stringent response on QS activities in sphingomonads was rather complicated; it was species-specific, and culture condition and growth phase-dependent. Unlike the positive regulatory effect that had been reported for US6-1 and Rr 2–17 with a single luxI homolog, our study on UT26 and SYK-6 with multiple luxI homologs showed a negative regulatory effect of rsh on QS-related bioactivities, including QS gene expression, QS signal production, and EPS production. Our study provides complementary information to our current knowledge, which is essential to understand the intricate network between QS and stringent response.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
YX: Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. XC: Data curation, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. HL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. TJ: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. YW: Data curation, Validation, Visualization, Writing – original draft. LL: Data curation, Investigation, Methodology, Resources, Validation, Writing – original draft, Writing – review & editing. SD: Writing – review & editing, Conceptualization, Data curation, Formal analysis, Investigation, Supervision, Writing – original draft. YH: Writing – review & editing, Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by Zhejiang Provincial Natural Science Foundation (LTGS23C010002) and National Natural Science Foundation of China (21577121) to YH.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1368499/full#supplementary-material
Chen, A. N., and Huang, Y. L. (2020). Acyl homoserine lactone based quorum sensing affects phenanthrene removal by Novosphingobium pentaromativorans US6-1 through altering cell surface properties. Int. Biodeterior. Biodegradation 147:104841. doi: 10.1016/j.ibiod.2019.104841
Dalebroux, Z. D., and Swanson, M. S. (2012). PpGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10, 203–212. doi: 10.1038/nrmicro2720
Fritsch, V. N., Loi, V. V., Busche, T., Tung, Q. N., Lill, R., Horvatek, P., et al. (2020). The alarmone (p)ppGpp confers tolerance to oxidative stress during the stationary phase by maintenance of redox and iron homeostasis in Staphylococcus aureus. Free Radical Bio. Med. 161, 351–364. doi: 10.1016/j.freeradbiomed.2020.10.322
Fuqua, C., Parsek, M. R., and Greenberg, E. P. (2001). Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35, 439–468. doi: 10.1146/annurev.genet.35.102401.090913
Gaca, A. O., Abranches, J., Kajfasz, J. K., and Lemos, J. A. (2012). Global transcriptional analysis of the stringent response in Enterococcus faecalis. Microbiol. 158, 1994–2004. doi: 10.1099/mic.0.060236-0
Gan, H. M., Buckley, L., Szegedi, E., Hudson, A. O., and Savka, M. A. (2009). Identification of an rsh gene from a Novosphingobium sp. necessary for quorum-sensing signal accumulation. J. Bacteriol. 191, 2551–2560. doi: 10.1128/JB.01692-08
Gan, H. M., Gan, H. Y., Ahmad, N. H., Aziz, N. A., Hudson, A. O., and Savka, M. A. (2015). Whole genome sequencing and analysis reveal insights into the genetic structure, diversity and evolutionary relatedness of luxI and luxR homologs in bacteria belonging to the Sphingomonadaceae family. Front. Cell. Infect. Mi. 4:188. doi: 10.3389/fcimb.2014.00188
Glaeser, S. P., and Kampfer, P. (2014). “The family Sphingomonadaceae” in The prokaryotes—Alphaproteobacteria and Betaproteobacteria. eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin: Springer), 641–707.
Huang, Y. L., Feng, H., Lu, H., and Zeng, Y. H. (2017). Novel 16S rDNA primers revealed the diversity and habitats-related community structure of sphingomonads in 10 different niches. Antonie Van Leeuwenhoek 110, 877–889. doi: 10.1007/s10482-017-0860-4
Huang, Y. L., Xu, X. Y., Song, Y., Yuan, N. T., Yu, X. H., Ji, Y. Q., et al. (2019). Structure and product relationship analysis of acyl homoserine lactone synthases among Ensifer adhaerens reveals distinct chromosome and plasmid origins. Biochem. Biophys. Res. Commun. 516, 907–913. doi: 10.1016/j.bbrc.2019.06.131
Jiang, W. K., Zhang, M. L., Gao, S. Y., Zhu, Q., Qiu, J. G., Yan, X., et al. (2022). Comparative genomic analysis of Carbofuran-degrading Sphingomonads reveals the Carbofuran catabolism mechanism in sp. Strain CFD-1. Appl Environ Microb. 88, e01024–e01022. doi: 10.1128/aem.01024-22
Kaczmarczyk, A., Vorholt, J. A., and Francez-Charlot, A. (2012). Markerless gene deletion system for sphingomonas. Appl. Environ. Microbiol. 78, 3774–3777. doi: 10.1128/AEM.07347-11
Lu, H., and Huang, Y. L. (2018). Transcriptome analysis of Novosphingobium pentaromativorans US6-1 reveals the Rsh regulon and potential molecular mechanisms of N-acyl-L-homoserine lactone accumulation. Int. J. Mol. Sci. 19:2631. doi: 10.3390/ijms19092631
Lu, Z. J., Li, W. Q., Xin, Y. D., and Huang, Y. L. (2023). The stringent response gene rsh plays multiple roles in Novosphingobium pentaromativorans US6-1’s accommodation to different environmental pollutants: Phenanthrene, copper and nZVI. Environ. Pollut. 323:121315. doi: 10.1016/j.envpol.2023.121315
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal–response Systems in Gram-negative Bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Poole, K. (2012). Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrob. Chemoth. 67, 2069–2089. doi: 10.1093/jac/dks196
Samantarrai, D., Sagar, A. L., Gudla, R., and Siddavattam, D. (2020). TonB-dependent transporters in Sphingomonads: Unraveling their distribution and function in environmental adaptation. Microorganisms 8:359. doi: 10.3390/microorganisms8030359
Schafhauser, J., Lepine, F., Mckay, G., Ahlgren, H. G., Khakimova, M., and Nguyen, D. (2014). The stringent response modulates 4-Hydroxy-2-Alkylquinoline biosynthesis and quorum-sensing hierarchy in Pseudomonas aeruginosa. J. Bacteriol. 196, 1641–1650. doi: 10.1128/JB.01086-13
Semedo, M. C., Karmali, A., and Fonseca, L. (2015). A high throughput colorimetric assay of β-1,3-D-glucans by Congo red dye. J Microbiol Meth. 109, 140–148. doi: 10.1016/j.mimet.2014.12.020
Sheng, D., Chen, M. J., Chen, Q. Q., and Huang, Y. L. (2022). Opposite selection effects of nZVI and PAHs on bacterial community composition revealed by universal and sphingomonads-specific 16S rRNA primers. Environ. Pollut. 311:119893. doi: 10.1016/j.envpol.2022.119893
Sperandio, V., Torres, A. G., Girón, J. A., and Kaper, J. B. (2001). Quorum sensing is a global regulatory mechanism in Enterohemorrhagic O157:H7. J. Bacteriol. 183, 5187–5197. doi: 10.1128/JB.183.17.5187-5197.2001
Stolz, A. (2009). Molecular characteristics of xenobiotic-degrading sphingomonads. Appl. Microbiol. Biot. 81, 793–811. doi: 10.1007/s00253-008-1752-3
Tan, C. H., Koh, K. S., Xie, C., Tay, M., Zhou, Y., Williams, R., et al. (2014). The role of quorum sensing signaling in EPS production and the assembly of a sludge community into aerobic granules. ISME J. 8, 1186–1197. doi: 10.1038/ismej.2013.240
Waigi, M. G., Sun, K., and Gao, Y. Z. (2017). Sphingomonads in microbe-assisted phytoremediation: tackling soil pollution. Trends Biotechnol. 35, 883–899. doi: 10.1016/j.tibtech.2017.06.014
Wang, M. F., Wang, J., Wang, X. B., Zhu, X. C., Wang, Y. P., and Wang, W. (2022). Effect of SPoT-mediated stringent response on biofilm formation, stress resistance and quorum sensing in pseudomonas protegens SN15-2. Appl. Bioch. Microbiol. 58, 406–415. doi: 10.1134/S0003683822040172
Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Bi. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Keywords: quorum sensing, stringent response, sphingomonads, rsh, acyl homoserine lactone, luxI homolog
Citation: Xiao Y, Chen X, Lu H, Jiang T, Wang Y, Liang L, Dobretsov S and Huang Y (2024) Regulation of quorum sensing activities by the stringent response gene rsh in sphingomonads is species-specific and culture condition dependent. Front. Microbiol. 15:1368499. doi: 10.3389/fmicb.2024.1368499
Received: 10 January 2024; Accepted: 20 March 2024;
Published: 04 April 2024.
Edited by:
Harold J. Schreier, University of Maryland, Baltimore County, United StatesReviewed by:
Song Lin Chua, Hong Kong Polytechnic University, Hong Kong SAR, ChinaCopyright © 2024 Xiao, Chen, Lu, Jiang, Wang, Liang, Dobretsov and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yili Huang, eWlsaWh1YW5nQHpqdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.