
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 19 April 2024
Sec. Aquatic Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1365546
 Shu Yang1,2†
Shu Yang1,2† Qinghua Hou1†
Qinghua Hou1† Nan Li1
Nan Li1 Pengbin Wang3
Pengbin Wang3 Huaxian Zhao2
Huaxian Zhao2 Qingxiang Chen1
Qingxiang Chen1 Xinyi Qin2
Xinyi Qin2 Jiongqing Huang4
Jiongqing Huang4 Xiaoli Li4
Xiaoli Li4 Nengjian Liao5
Nengjian Liao5 Gonglingxia Jiang1
Gonglingxia Jiang1 Ke Dong6
Ke Dong6 Tianyu Zhang1*
Tianyu Zhang1*Microorganisms, especially rare microbial species, are crucial in estuarine ecosystems for driving biogeochemical processes and preserving biodiversity. However, the understanding of the links between ecosystem multifunctionality (EMF) and the diversity of rare bacterial taxa in estuary ecosystems remains limited. Employing high-throughput sequencing and a variety of statistical methods, we assessed the diversities and assembly process of abundant and rare bacterioplankton and their contributions to EMF in a subtropical estuary. Taxonomic analysis revealed Proteobacteria as the predominant phylum among both abundant and rare bacterial taxa. Notably, rare taxa demonstrated significantly higher taxonomic diversity and a larger species pool than abundant taxa. Additionally, our findings highlighted that deterministic assembly processes predominantly shape microbial communities, with heterogeneous selection exerting a stronger influence on rare taxa. Further analysis reveals that rare bacterial beta-diversity significantly impacts to EMF, whereas alpha diversity did not. The partial least squares path modeling (PLS-PM) analysis demonstrated that the beta diversity of abundant and rare taxa, as the main biotic factor, directly affected EMF, while temperature and total organic carbon (TOC) were additional key factors to determine the relationship between beta diversity and EMF. These findings advance our understanding of the distribution features and ecological knowledge of the abundant and rare taxa in EMF in subtropical estuaries, and provide a reference for exploring the multifunctionality of different biospheres in aquatic environments.
Microorganisms are the important drivers of biogeochemical cycles in the ocean environment and have vital roles in affecting global nutrient cycles and maintaining ecosystem multifunctionality (EMF) (Falkowski et al., 2008; Strom, 2008; Madsen, 2011; Alsterberg et al., 2017; Villnäs et al., 2018). Over decades, a growing number of studies have shown that the diversity of individual groups and EMF are closely related (Duffy, 2009; Snelgrove et al., 2014; Delgado-Baquerizo et al., 2016; Luo et al., 2018; Hu et al., 2021). For instance, Delgado-Baquerizo et al. (2016) found that EMF increased with soil bacterial alpha diversity in global drylands. Luo et al. (2018) investigated the link between microbial diversity and EMF in long-term fertilized agricultural soil and the results reveal a positive correlation between soil microbial diversity and EMF. However, the associations among species diversity, phylogenetic diversity and EMF in the natural environment remain unclear.
Highly abundant species can inhabit distinct niches, competitively exploit a range of various resources, and well adapt to environmental changes (Zhang et al., 2022), and their importance in driving ecosystem functioning has been recognized. Accumulating evidence has demonstrated that rare taxa have a disproportionate impact on EMF that in disproportionate to its abundance (Winfree et al., 2015; Banerjee et al., 2018; Wan et al., 2021; Zhang et al., 2022). For example, Zhang et al. (2022) found that, in comparison to abundant species, the diversity of rare species showed a greater positive relationship with EMF. These studies indicate that the rare taxa diversity appears to be a promising indicator for assessing EMF.
The assembly processes of microbial communities directly influence microbial diversity, include species abundance and a multitude of functions. Accumulating evidence demonstrates that the assembly process can be understood through both stochastic and deterministic processes. Given the distinct environmental gradients in estuarine ecosystems, microbial communities encounter varying degrees of environmental stress, which results in distinct assembly patterns of planktonic bacterial communities (McLusky and Elliott, 2007). Yao et al. have clarified these dynamics, revealing that homogeneous selection (deterministic process) significantly shapes bacterial community assembly across the estuarine marsh ecosystem (Yao et al., 2019). Additionally, Isabwe et al. found longitudinal distribution patterns of bacterioplankton in riverine ecosystems, suggesting that distance-decay relationships may be attributed to ecological drift (stochastic process) (Isabwe et al., 2022). Nonetheless, the assembly processes of both abundant and rare biospheres in influencing arestill not well comprehended in estuarine ecosystems.
Estuary ecosystems are the most ecosystem functionality systems on earth and have an essential role in biogeochemical cycles (Guo et al., 2015; Wang and Zhang, 2020). Sanniang Bay is located in the Beibu Gulf. Therefore, to explore the significance of abundant and rare taxa on EMF under environmental disturbance, we analyzed two years of seasonal water samples in a subtropical estuary to (1) identify the abundant and rare taxa across environmental disturbance of the subtropical estuary; (2) reveal the assembly process of the abundant and rare subcommunities; (3) investigate the contribution of abundant/rare taxa to biodiversity and EMF; and (4) identify the key drivers affecting EMF via abundant/rare subcommunity.
The sites were located in the estuary of Dafeng River and Sanniang Bay, Guangxi Zhuang Autonomous Region, China (Supplementary Figure S1). In 2018, surface water samples (0.5 meters) were collected quintuplicate samples from four seasons at 15 sites (Supplementary Figure S1). In 2020, samples were collected from 16 sites (Supplementary Figure S1). We employed five-point sampling method to collected surface water samples using a rosette of Niskin bottles at each designated site. In 2018, we were unable to obtain the S8 samples, resulting in the collection of a total of 620 samples. We roughly divide the sampling location into three sub-areas: a low salinity (LS) group, a medium salinity (MS) group and a high salinity (HS) group. To analyze nutrients and chlorophyll a, for each sample, 500 mL seawater were filtered through a 0.45 μm filter (Millipore Corporation, Billerica, MA). Until further analysis, the filters were kept in storage at −80°C. The environmental factors were measured by our previous methods (Zhao et al., 2023) and all of the environmental factors are listed in Supplementary Table S1.
Two liters of each surface water sample for microbial diversity were filtered through 0.22 μm membranes according to a previous study (Teeling et al., 2012), DNA quality was assessed using a spectrophotometer (Delaware, United States). DNA samples were stored at −80°C. According to a previous publication (Zhao et al., 2023), we used the universal primers 341F (CCTACGGGNGGCWGCAG) and 805R (GACTACHVGGGTATCTAATCC) for 16S rRNA amplification (Herlemann et al., 2011). The 50 μL PCR mixture containing 2 μL of template DNA, 25 μL of 2 × Taq PCR MasterMix (TianGen), 21 μL of PCR grade molecular water and 1 μL of each primer was aliquoted into PCR Beads. PCR (50 μL) was conducted on a Biorad thermocycler, and the conditions were as follows: 60 s initial denaturation at 95°C, 34 cycles of 30 s denaturation at 95°C, 30 s annealing at 54°C, 60 s elongations at 72°C. A TruSeq DNA kit (Illumina, United States) was used to processing PCR products for sequencing (Zhao et al., 2023).
The libraries were prepared following the Illumina library preparation protocols. The library was delivered to Majorbio Co. Ltd. (Shanghai, China) for Illumina MiSeq platform (PE300, 2 × 300 bp) for sequencing. Employing the DADA2 denoising method (Caporaso et al., 2010; Callahan et al., 2017), reads with quality scores below 100, as well as chimeric sequences, barcode sequences, and primers, were systematically removed. The amplicon sequence variants (ASVs) obtained from the raw data were selected for further analysis (Callahan et al., 2017). The Ribosomal Database Project Classifier program was picked to perform taxonomic assignment at 80% confidence level (Maidak et al., 2000). The sequence data were deposited in GenBank at BioProject Accession: PRJNA971362.
To obtain the multi-nutrient cycling index (MNI), we first normalized and standardized each of the relevant factors linked to the cycling of phosphorous (TP, DIP), nitrogen (NO3−-N, NO2−-N, NH4+-N, DIN, TN) and carbon elements (TOC) using the Z-score transformation method. These standardized ecosystem function values ranging from 0 to 1 were computed using the formula:
V represents the value of chosen factors, where Vmin is the minimum value and Vmax is the maximum value of these selected factors. The mean value of these calculating results represents the MNI index (Delgado-Baquerizo et al., 2016; Zhang et al., 2021).
To reduce bias caused by differences in sequencing depths, the samples in this study were rarefied to the minimum sample depth required for further analysis (Feng et al., 2022). In this study, ASVs were classified into abundant (>0.1%), rare (<0.01%) and intermediate (0.01–0.1%) (Jiao and Lu, 2020; Zhao et al., 2023). We calculated the diversity indices (Kemp and Aller, 2004) and employed the Shannon index to represent alpha diversity and the richness index to reflect the species pool (Supplementary Table S2). Beta diversity was quantified by Bray–Curtis distance, and the comparison of groups was used permutational multivariate analysis of variance (PERMANOVA). Partial least-squares path modelling (PLS-PM) analysis was performed by ‘plspm’ R package. Following the methodology outlined by Stegen et al. (2013), a null model analysis was performed to classify community assembly processes. This analysis involved generating beta-diversity metrics, including the beta-nearest taxon index (βNTI) and Bray–Curtis based Raup-Crick (RCbray), to evaluate community assembly from both phylogenetic and taxonomic perspectives. Results indicating |βNTI| >2 were interpreted as predominance of deterministic process, while |βNTI| <2 suggested a stochastic process dominance. Further analysis on pairwise comparisons with |βNTI| <2 using RCbray values revealed the roles of dispersal limitation with drift, and homogenizing dispersal with drift, as indicated by RCbray values >+0.95, <−0.95, and <+0.95, respectively. The Spearman’s rank method was used to calculate correlations for the partial Mantel and Mantel test. The R package ‘ggplot2’ was used to conduct a linear regression plot analysis.
In this study, 620 surface water samples across different seasons were collected from Sanniang Bay (Supplementary Figure S1). Ultimately, a total of 39,969,028 high quality sequences were obtained, which belonged to 2,454 ASVs. A total of 120 ASVs (4.89%) with 31,318,993 (78.36%) sequences were classified as abundant ASVs, while 1774 ASVs (72.29%) with 2,731,363 (6.83%) sequences were classified as rare ASVs. Proteobacteria was the dominant phylum in the abundant (84.99%) subcommunity, followed by Actinobacteria (5.59%) and Bacteroidetes (5.28%), with the most prevalent classes in the abundant subcommunity being Alphaproteobacteria (50.02%) and Gammaproteobacteria (32.81%). The rare subcommunity was mainly composed of Proteobacteria (70.92%), followed by Bacteroidetes (11.24%) and Actinobacteria (4.42%), with Alphaproteobacteria (30.73%) and Gammaproteobacteria (30.33%) being the most dominant classes (Figure 1). Diversity analysis indicated that rare taxa exhibited significantly greater alpha diversity than abundant taxa. Within the abundant subcommunity, bacterioplankton communities in the LS group exhibited significant temporal variation (p < 0.05). For example, the alpha diversity of the LS group showed an upward trend from spring to winter and peaked in winter 2020 (Supplementary Figure S2). In contrast, the alpha diversity of rare taxa exhibited more dramatic interannual variation in the HS samples than in the LS and MS samples (Supplementary Figure S2). In the rare subcommunity of HS samples, the lowest alpha diversity was observed in summer 2018, and the highest was observed in fall 2020 (Supplementary Figure S2). Species pool analysis revealed that the species pool was smallest in summer 2018 and largest in fall 2020 in the rare taxa. In the abundant subcommunity, the smallest species pool was observed in the spring HS samples of 2018, and the largest was observed in the fall LS samples of 2020 (Supplementary Figure S2). The results of the PERMANOVA revealed a more significant difference in beta diversity within sites for communities composed of abundant subcommunities (r2 = 0.030, p = 0.001) compared to rare subcommunities (r2 = 0.027, p = 0.001) (Supplementary Table S3).
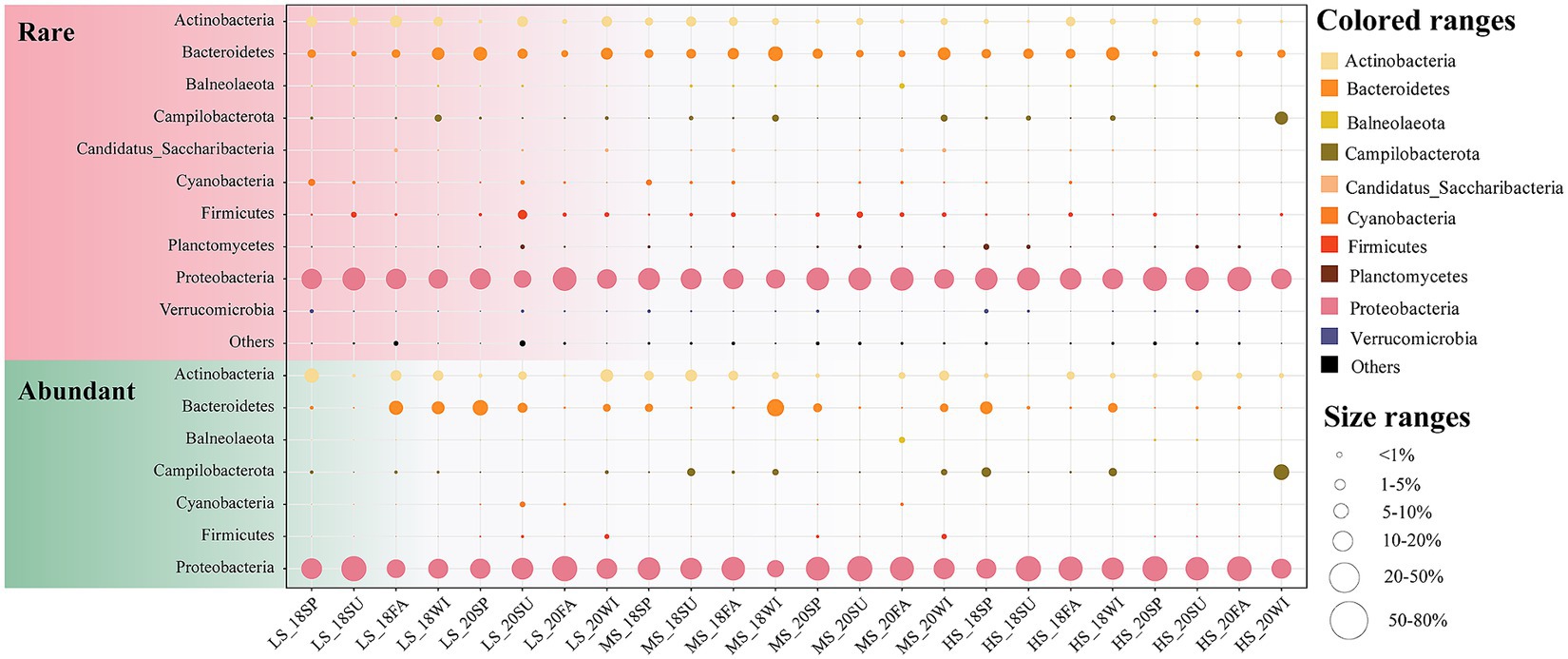
Figure 1. Marine bacterioplankton distribution and abundance of total, rare and abundant subcommunities from different seasons and salinity gradients. Circle size indicates species abundance. LS, low salinity; MS, medium salinity; HS, high salinity: SP, Spring; SU, Summer; FA, Fall; WI, Winter.
A statistical technique using βNTI was used in this study to decipher the community assembly processes. The βNTI values between >2 or <−2 were 53.90 and 86.27% for the bacterial community in abundant and rare taxa, respectively. The results show that deterministic processes had a crucial role in the assembly of different biospheres than stochastic processes. We observed greater heterogeneous selection in the rare taxa. Within the community of abundant taxa, heterogeneous selection was most pronounced in the MS group (29.7%) and least evident in the HS group (13.5%). For rare taxa, heterogeneous selection was most significant in the HS group, reaching 55.4%, and least pronounced in the LS group, at 48.9% (Figure 2). Interestingly, the heterogeneous selection of abundant taxa increases with higher salinity concentrations, while rare taxa do not (Figure 2). Within the stochastic process, it was drift that predominantly influenced the assembly of abundant and rare taxa (Figure 2).
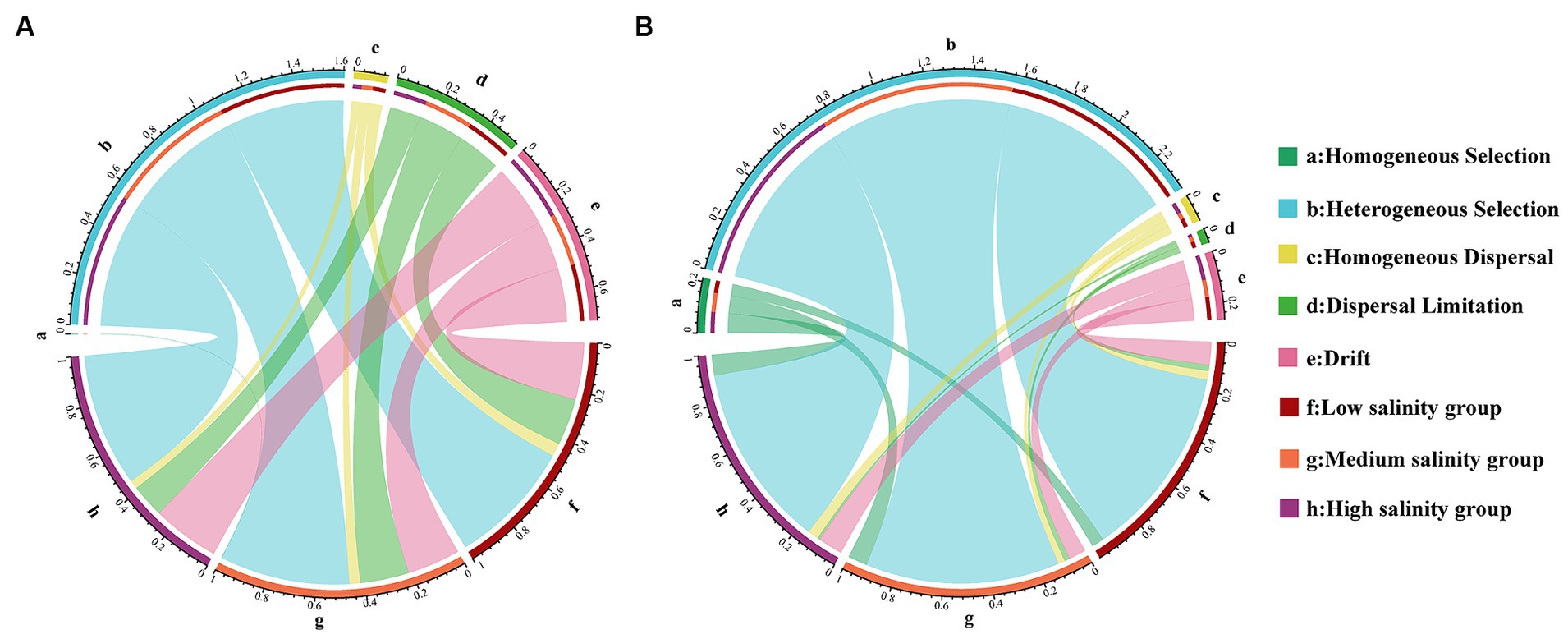
Figure 2. Analysis of the community assembly of marine bacterioplankton with abundant (A) and rare (B) marine bacterioplankton communities displayed by Circos analysis.
Linear regression analysis was employed to establish the relationship of MNI to alpha diversity, beta diversity and species pools in abundant and rare areas. In terms of alpha diversity, the linear regression analysis indicated no significant correlation between alpha diversity and MNI in abundant and rare taxa (p > 0.05) (Figure 3). For beta diversity, the linear regression analysis demonstrated that MNI showed a significantly positive correlation with beta diversity of abundant and rare taxa in LS, MS, and HS samples (p < 0.001). Compared to abundant taxa, the beta diversity of rare taxa showed a stronger influence on the EMF (Figure 4). Moreover, compared with MS and HS samples, the beta diversity of rare taxa in the LS group had a stronger linear correlation with MNI (Figure 4). The results in Figure 5 show that MNI was significantly negatively correlated with the species pool in the LS, MS, and HS samples in the abundant subcommunity (p < 0.05). However, there was no significant relationship between the species pool and MNI in the LS, MS, and HS samples of rare taxa (p > 0.05) (Figure 5).
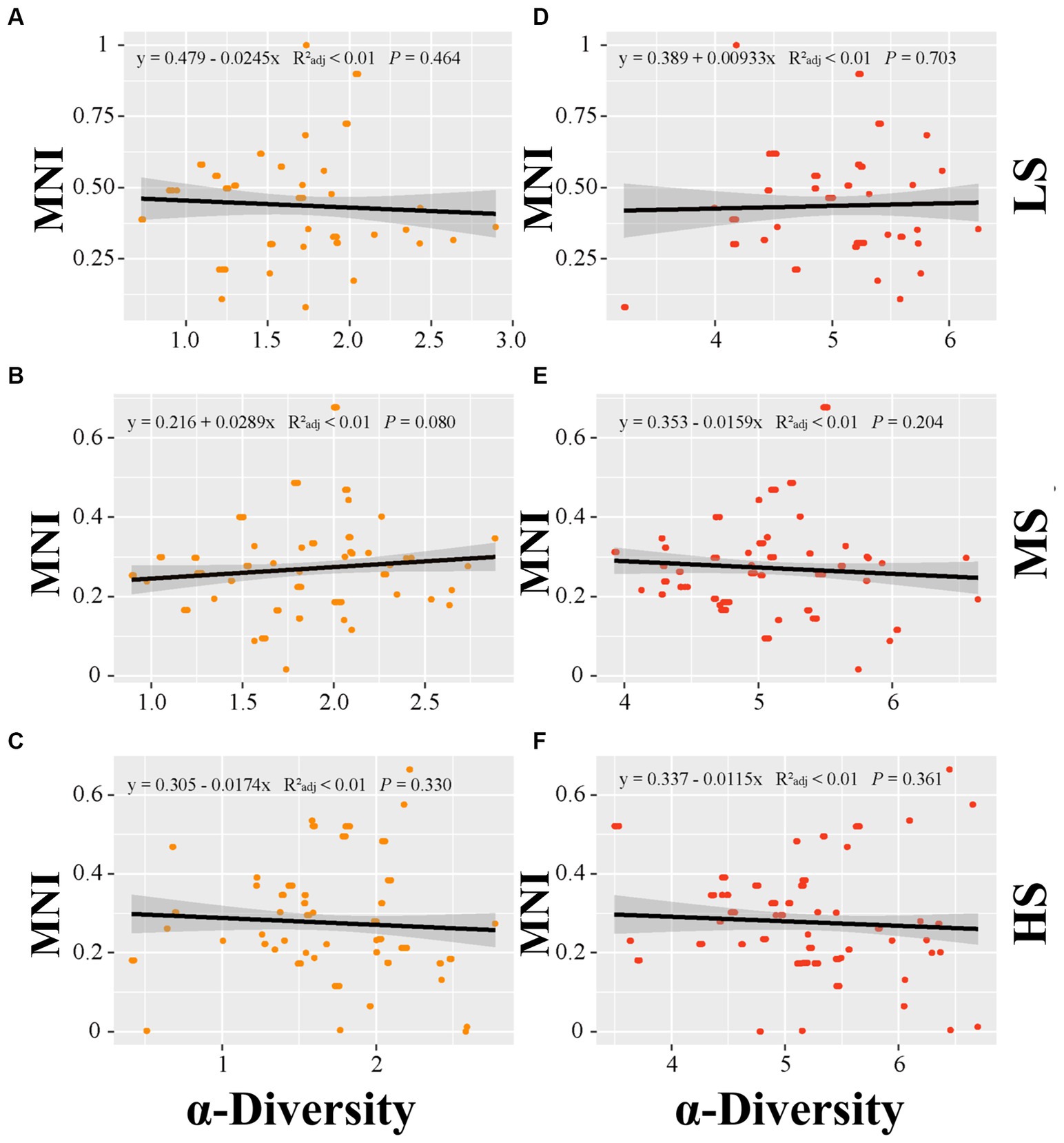
Figure 3. Linear regressions for α-diversity of abundant (A–C) and α-diversity of rare marine bacterioplankton communities (D–F) with EMF in different salinity gradients. LS, low salinity; MS, medium salinity; HS, high salinity; MNI, multi-nutrients cycling index.

Figure 4. Linear regressions for β-diversity of abundant (A–C) and β-diversity of rare (D–F) marine bacterioplankton communities with EMF in different salinity gradients. LS, low salinity; MS, medium salinity; HS, high salinity; MNI, multi-nutrients cycling index.
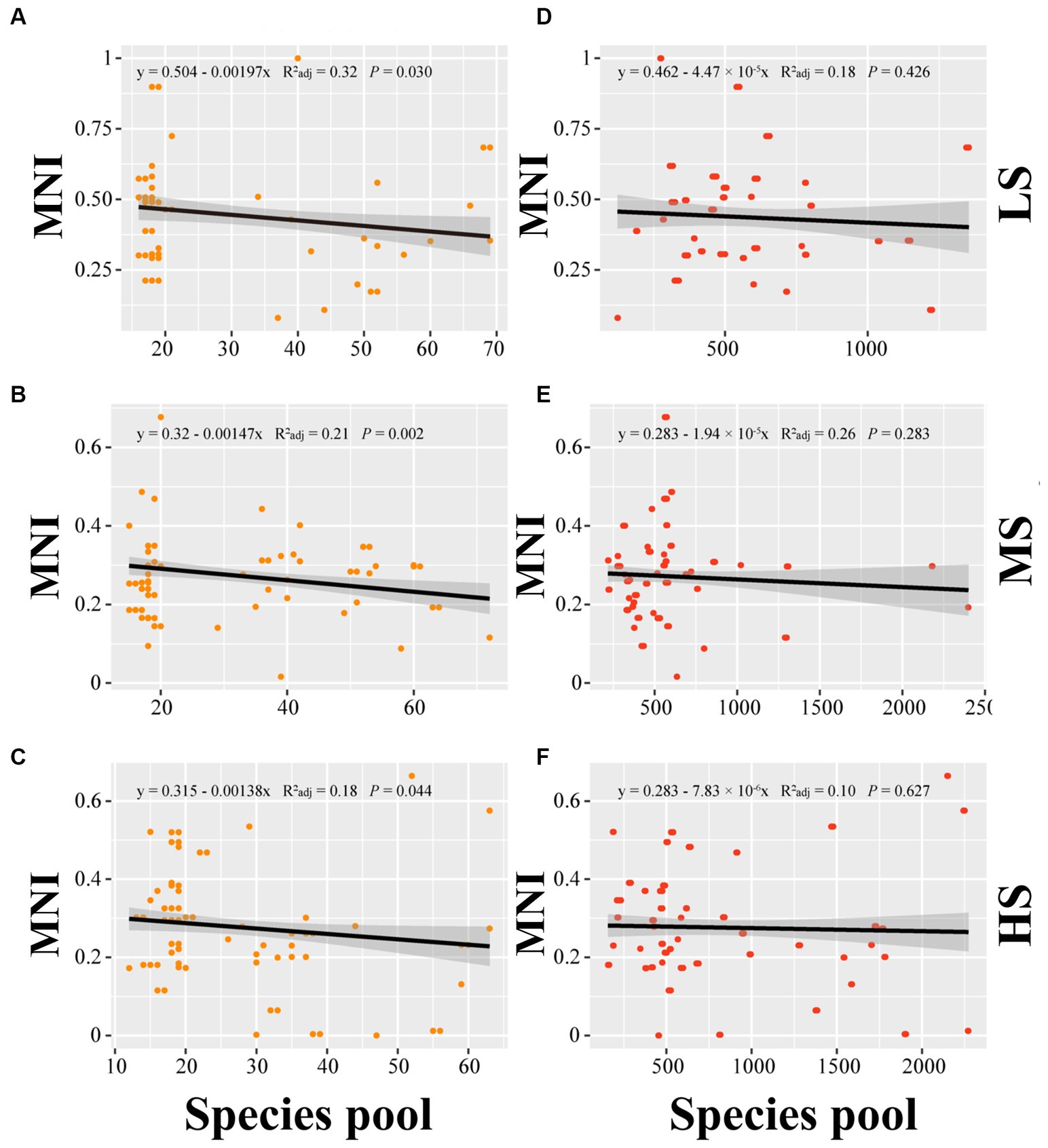
Figure 5. Linear regressions for the species pools of abundant (A–C) and species pools of rare (D–F) marine bacterioplankton communities with EMF in different salinity gradients. LS, low salinity; MS, medium salinity; HS, high salinity; MNI, multi-nutrients cycling index.
In this study, we utilized Spearman analysis to evaluate the impact of individual environmental factors on alpha diversity and the species pool of abundant and rare taxa. The association between alpha diversity and environmental factors varied across different groups (Supplementary Table S4). In the abundant subcommunity, a significant positive link was found between TOC and pH with alpha diversity. TOC exerted the most significant positive influence in the LS group, while NH4+-N and DIP were found to have significant negative corrections with alpha diversity in the MS and HS samples, respectively (Supplementary Table S4). In a rare subcommunity, the two most significant factors affecting alpha diversity and species pool in the LS and HS groups were found to be DO (positive) and DIP (negative). Additionally, TP was identified as another important but negatively influencing factor for the HS group. Among the numerous influencing factors in the MS group, temperature was the predominant positive factor shaping alpha diversity and species pool, while Chl-a had a negative influence on alpha diversity and species pool (Supplementary Table S4). The Mantel test results were used to further analyse the effect of distinct environmental factors on beta diversity and community assembly mechanism (βNTI) of different microbial biosphere species. In the abundant subcommunity, beta diversity was influenced by different environmental factors along salinity gradients. For example, the positive correlation coefficient between TOC and beta diversity of abundant taxa was greater than that of other environmental factors in LS samples. In the MS and HS groups, temperature and DIP were identified as the strongest factors influencing the beta diversity of abundant species (Supplementary Table S5). Additionally, the βNTI of the abundant subcommunity showed a significant association with DIP, DO and TOC. The abundant subcommunity assembly was primarily influenced by pH and TOC in the LS samples, while in the MS and HS groups, it was affected by TP and DIP, respectively (Supplementary Table S6). In the rare subcommunity, beta diversity in the LS group was mainly influenced by DO (Supplementary Table S5). Furthermore, similar to the abundant taxa, βNTI in rare taxa were primarily influenced by DIP, and the affect of DIP on βNTI was greater in rare biosphere than in the abundant biosphere. This study also found that βNTI was more strongly influenced by TP only in the MS group than in the abundant taxa, whereas in the LS and HS samples, the influence of pH and DIP on βNTI was weaker in the rare taxa than in the abundant taxa (Supplementary Table S6).
In this study, we selected PLS-PM to investigate the complex relationships among water properties (WP), nutrients, microbial diversity (alpha and beta diversity), species pool, and EMF (Figure 6). In the abundant subcommunity, PLS-PM results revealed that WP had the strongest impact on alpha and beta diversity, exhibiting a significant positive correlation (p < 0.001). Among these factors, temperature emerged as the most critical variable influencing WP, whereas nutrients primarily exerted a significant influence on variations in the species pool through TOC (p < 0.001). Moreover, EMF were found to be primarily significantly positively regulated by beta diversity and nutrients (p < 0.001). In the rare subcommunity, PLS-PM results revealed that nutrients had the greatest significant impact on the alpha diversity and species pool of rare taxa (p < 0.001), with TOC being the most important variable influencing nutrients. WP was found to be the most significant variable for the beta diversity of rare taxa (p < 0.001), while the alpha diversity of taxa showed a nonsignificant correlation with EMF (p > 0.05). Additionally, temperature was an important parameter influencing EMF directly through beta diversity in both abundant and rare subcommunities compared to other factors (Figure 6).
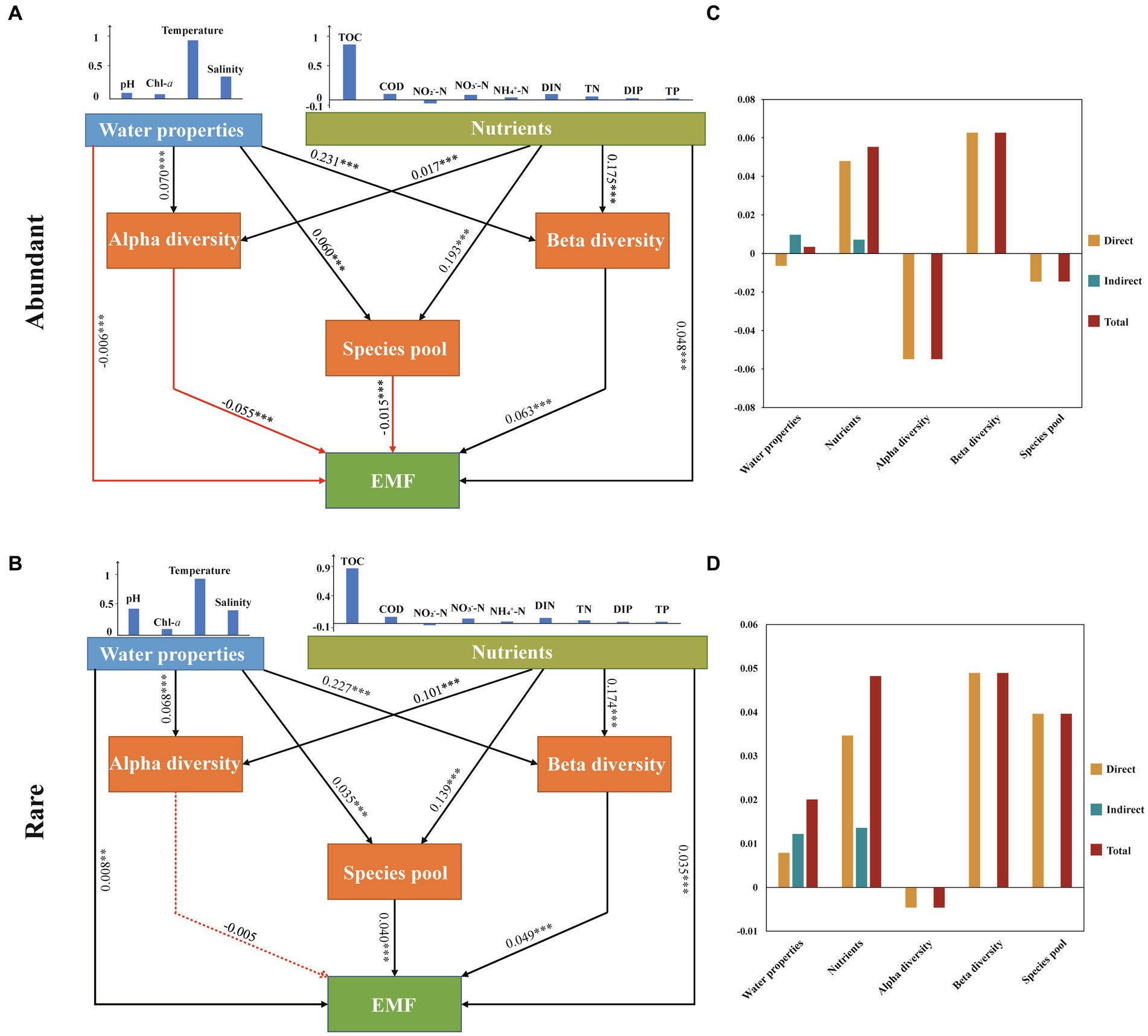
Figure 6. Partial least-squares path modelling (PLS-PM) analysis showing the multiple effects of environmental (water properties and nutrient parameters) factors and biotic (bacterioplankton community diversity and species pool) factors on EMF in marine ecosystems and the path coefficients between different factors and their corresponding latent variables of abundant (A) and rare (C) and their standardized direct, indirect and total effects of abundant (B) and rare (D). Water properties include temperature, salinity, pH, and Chl-a. Nutrients include C (TOC, COD), N (NO3−-N, NO2−-N, NH4+-N, DIN, TN), and P (DIP, TP). Red arrows represent negative correlations, and blue arrows represent positive correlations. The solid arrow represents significant correlations (the number is the R value; the asterisk indicates the p value: *p < 0.05, **p < 0.01, and ***p < 0.001), and the dotted arrow represents nonsignificant correlations (p > 0.05).
Studies suggested that under long-term environmental disturbance in estuaries, the nutrient cycle process is complex, resulting in a significant linkage between the structure and distribution of the microbial community and the changes in the surrounding environment (Wankel et al., 2011; Liu et al., 2014; Jeffries et al., 2016; Kieft et al., 2018). In this study, the main bacterial phylum in the abundant and rare taxa was Proteobacteria in the sampling region (Figure 1). The distribution pattern of Proteobacteria described here is consistent with previous studies in the Amazon River Estuary (Santos-Júnior et al., 2020), the Pearl River Estuary (Mai et al., 2020) and the Yellow River estuary (Wang et al., 2021). Proteobacteria not only exhibit a broad spectrum of metabolic diversity (e.g., photosynthetic metabolism, sulfate reduction, nitrogen metabolism) but also fulfil critical ecological roles, such as the decomposition of organic matter within the carbon cycle and the biodegradation of contaminants. These roles make the Proteobacteria essential in nutrient cycling processes, especially in estuarine ecosystems (Campbell et al., 2006; Hügler and Sievert, 2011; Kern et al., 2011; Chen et al., 2022). Similar to previous studies, we also found that the relative abundance of Proteobacteria increases with the salinity gradient in the surface water of estuarine environments (Tang et al., 2017; Mai et al., 2020; Santos-Júnior et al., 2020). In contrast, we also noted that the relative abundance of Bacteroidetes decreased from the river to the ocean. A previous study in Mediterranean Sea highlights that Bacteroidetes were more likely to survive in eutrophic environments (Pinhassi et al., 2004). Betaproteobacteria have been considered dominant in freshwaters and are an ecological indicator with high nitrogen content (Pinhassi et al., 2004; Yang et al., 2019). Overall, these results suggest that bacterial community is potential indicator of ecological resilience and water quality in estuarine systems.
The diversity characteristics of abundant and rare microorganisms play dominant roles in nutrient cycling, which is essential for assessing ecosystem functions (Xu et al., 2021; Du et al., 2024). The rare taxa showed higher alpha diversity than the abundant taxa, which is consistent with the finding of another study that found that rare species in the estuarine waters of the Perl River exhibit greater diversity than abundant species (Zhang et al., 2021). In this study, we also discovered that the alpha diversity of rare taxa exhibited larger interannual variation in HS samples than in the other samples. In addition, we noted that the species pool of the rare biosphere exhibited less variation than that of the abundant biosphere. This might be attributed to the result of the rare taxa fitness trade-offs, and the rare biosphere improves stress resistance at a lower growth rate under environmental disturbance (Jousset et al., 2017). Recent investigations have also indicated that owing to the immigration and emigration of rare species and the gradual recovery of their inhabitants densities (Jia et al., 2018; Xue et al., 2018), rare taxa may provide resilience or resistance abilities to the ecosystem (Gomez-Alvarez et al., 2016). In addition, several environmental factors were found to strongly affect the alpha diversity of different subcommunities (Supplementary Table S4). In agreement with previous researches (Campbell et al., 2011; Liu et al., 2015; Wan et al., 2021; Ren et al., 2022), both abundant and rare bacterial taxa exhibited sensitivity to distinct environmental factors. Our study reveal that the alpha diversity of both abundant and rare taxa was significantly influenced by multiple factors (Figure 6). These results highlight the different ecological adaptations of abundant and rare bacterial subcommunities to environmental disturbance and suggested that carbon compounds primarily affected abundant bacteria in Sanniang Bay. Accumulating evidence shows that the bacterial community can regulate nutrient cycling processes in aquatic ecosystems (Arrigo, 2005; Hutchins et al., 2009; Damashek and Francis, 2018; Cui et al., 2019; Baker et al., 2021), and enhance EMF (Winfree et al., 2015; Banerjee et al., 2018; Wan et al., 2021; Zhang et al., 2022). Studies exhibit that abundant and rare bacterial alpha-diversity have different significant contributions to EMF in various soil ecosystems (Winfree et al., 2015; Banerjee et al., 2018; Wan et al., 2021; Zhang et al., 2022). Our study found that there was no significant relationship between the EMF and the alpha diversity of abundant and rare taxa (Figure 3). These results suggest that due to estuaries being subject to unpredictable variations in salinity and water movement (Wang et al., 2017), abundant and rare bacteria have unique EMF in estuarine ecosystem.
Interestingly, our findings suggested that the beta diversity of rare taxa has a more intense promotional effect on EMF compared to abundant taxa (Figure 4). This result indicating that enhancement of beta diversity in rare biosphere may lead to functional redundancy (Grman et al., 2018). Given that the connection between EMF and its stability (Soliveres et al., 2016), rare species possess analogous functional capabilities to ensure the continuity of ecosystem functionality. The contributions of species pools containing both abundant and rare taxa to EMF, particularly estuary ecosystems, have not been the subject of many studies.
Our findings revealed that the species pool of abundant taxa exhibits a significant decreasing trend with the increase of EMF (Figure 5). Rare species may be more susceptible to environmental perturbation, as they have more pronounced ecological niche specificity (Zhang et al., 2022), while typically abundant taxa exhibit significant stable contributions in nutrient cycling within the ecosystem (Liu et al., 2015) and always higher competition potential and growth rate than rare taxa (Campbell et al., 2011). The size of abundant taxon species pools is more important to EMF. Previous study discovered that the richness of bacterial communities in the soil drives the diversity of soil ecological functions (Delgado-Baquerizo et al., 2017). But, in this study, the MNI tends to decrease as the size of the rare taxa species pool increases (p > 0.05). This result provides new insights into the association between microbial diversity and EMF in estuarine ecosystem.
A crucial aspect of revealing ecosystem function is to assess the relative importance of deterministic and stochastic processes to community assembly (Mokany et al., 2013; Mori et al., 2018). Similar to a previous study (Wang et al., 2021), we also confirmed that deterministic processes govern community assembly in coastal zone. In the present study, we discovered that in the studied region, deterministic assembly dominated both the rare and abundant bacterial subcommunities (Figure 2). As in previous studies (Liu et al., 2015; Li et al., 2021; Zhu et al., 2023), we noticed that the rare taxa were more affected by determinism than the abundant taxa. Rare taxa often exhibit narrower niche breadths, lower competitive abilities, and slower growth rates (Reveillaud et al., 2014; Ren et al., 2022). Furthermore, the enhancement of deterministic processes indicates that the shaping effect of environmental stress on microbial communities increases with the level of stress (Wang et al., 2023). Based on Mantel test, we found that the rare subcommunity βNTI in estuary-ocean ecosystems showed a stronger correlation with dissolved inorganic phosphorus (DIP) and total phosphorus (TP) than other variables (Supplementary Table S6). Phosphorus is identified as the primary limiting nutrient for ecosystem primary productivity (Peng et al., 2021) and its limitation can be regulated by bacteria to influence oceanic nutrient flux (Kritzberg et al., 2010). Therefore, it is an important factor influencing rare subcommunity structure and functional differentiation (Qin et al., 2020). DIP, DO, and TOC had significant effects on the succession of the abundant subcommunity. Different environmental factors have been found to determine the assembly of abundant and rare subcommunities in distinct habitats, such as temperature and pH, which control how rare and abundant bacterial subcommunities assemble in agricultural ecosystems (Jiao and Lu, 2020). Salinity and spatial factors significantly effected community turnover of abundant and rare bacterioplankton in Shenhu Bay, Dongshan Bay and Beibu Gulf (Mo et al., 2018). These results suggested that environmental filtering was a major factor in selecting microbial species and driving the assembly process in aqueous ecosystem (Ren et al., 2022).
Given that the environment can influence the diversity of different microbial subcommunities and their influences on EMF, we constructed a PLS-PM model (PLS-PM) to explore the interrelationships among WP, nutrients, bacterioplankton diversity (alpha and beta diversity), species pool, and EMF. In comparison to nutrients, WP (e.g., temperature) exerts a greater impact on the beta diversity of rare and abundant bacterioplankton taxa. The research conducted by Geng et al. (2022) indicates that environmental factors, including water temperature, significantly contribute to the beta diversity of bacterioplankton, consistent with the findings of this study. WP typically exhibits higher spatial and temporal variability compared to nutrients. This variability can lead to changes in community composition and beta diversity. Our results proved that the beta diversity of abundant and rare subcommunities was a better driver in EMF than alpha diversity. This finding is in line with the idea that microbial beta diversity is the strongest positive predictor of soil nutrient cycling processes (Xu et al., 2021). Differences in structure and metabolic functions among different species can give rise to their varying roles within the EMF (He et al., 2009). Due to the differential response capabilities of different species towards various environmental factors, beta diversity-rich communities can contribute to a greater range of EMF, particularly those associated with nutrient cycling. We further observed that, compared to abundant taxa, the beta diversity of rare taxa had a more significant impact on EMF. Due to the lower abundance of rare taxa, the variation in their beta diversity represents the presence or absence of unique species that may provide specific ecosystem services or participate in complex ecological processes, thus filling functional gaps and enhancing EMF (Jiao et al., 2017; Mori et al., 2018; Zhang et al., 2020; Xiong et al., 2021). Furthermore, we found that the relationship between beta diversity and EMF was mediated by temperature and TOC. Adhikari et al. (2019) posits that temperature is often linked to bacterial biodiversity, as it is directly related to metabolic rates and the affinity of bacteria for available substrates. Additionally, TOC is generally recognized as a crucial factor influencing microbial diversity variation and phylogenetic relationships within ecosystems, thereby being extensively reported as a primary determinant of microbial communities in global marine ecosystems (Li et al., 2020).
Our study identifies both abundant and rare biospheres in a subtropical estuary. Rare biospheres exhibit higher diversity and are more important for ecosystem functions than abundant biospheres. Deterministic processes dominate in the community assembly of abundant and rare taxa. The bacterial beta of rare taxa has a significant impact on EMF in the subtropical estuary, and different abiotic factors were established to affect EMF of abundant and rare taxa. Unexpectedly, EMF were strongly linked to the species pool of the abundant subcommunity. Overall, the results obtained in this study improve our understanding of how abundant and rare taxa impact EMF in estuary ecosystems and advance our comprehension of ecological diversity and function.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA971362.
SY: Supervision, Writing – review & editing, Conceptualization, Methodology, Validation. QH: Writing – review & editing. NL: Supervision, Writing – review & editing, Investigation, Visualization. PW: Conceptualization, Methodology, Writing – review & editing. HZ: Conceptualization, Methodology, Writing – review & editing. QC: Supervision, Writing – review & editing. XQ: Formal analysis, Investigation, Supervision, Writing – review & editing. JH: Investigation, Supervision, Writing – review & editing. XL: Writing – review & editing. NL: Writing – review & editing. GJ: Formal analysis, Visualization, Writing – review & editing. KD: Writing – review & editing. TZ: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was jointly funded by the National Key Research and Development Program of China (grant numbers 2022YFC3103104 and 2021YFC3101801); Guangdong Science and Technology Plan Project (Observation of Tropical Marine Environment in Yuexi); Independent Research Project of Southern Ocean Laboratory (grant number SML2022SP301); National Natural Science Foundation of China (grant number 41976200); Guangdong Ocean University Scientific Research Program (grant number 060302032106); Guangdong University Innovation Team (Early-warning of marine disasters), and Scientific Research Start Funds of Guangdong Ocean University. Data and samples were collected onboard of R/V “Tianlong” and “Yuezhanyuyun 01018” implementing the open research cruise NORC2023-11 supported by NSFC Shiptime Sharing Project (project number: 42249911).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1365546/full#supplementary-material
Adhikari, N. P., Liu, Y., Liu, K., Zhang, F., Adhikari, S., Chen, Y., et al. (2019). Bacterial community composition and diversity in Koshi River, the largest river of Nepal. Ecol. Indic. 104, 501–511. doi: 10.1016/j.ecolind.2019.05.009
Alsterberg, C., Roger, F., Sundbäck, K., Juhanson, J., Hulth, S., Hallin, S., et al. (2017). Habitat diversity and ecosystem multifunctionality—the importance of direct and indirect effects. Sci. Adv. 3:e1601475. doi: 10.1126/sciadv.1601475
Arrigo, K. R. (2005). Marine microorganisms and global nutrient cycles. Nature 437, 349–355. doi: 10.1038/nature04159
Baker, B. J., Appler, K. E., and Gong, X. (2021). New microbial biodiversity in marine sediments. Annu. Rev. Mar. Sci. 13, 161–175. doi: 10.1146/annurev-marine-032020-014552
Banerjee, S., Schlaeppi, K., and van der Heijden, M. G. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576. doi: 10.1038/s41579-018-0024-1
Callahan, B. J., McMurdie, P. J., and Holmes, S. P. (2017). Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643. doi: 10.1038/ismej.2017.119
Campbell, B. J., Engel, A. S., Porter, M. L., and Takai, K. (2006). The versatile ε-proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4, 458–468. doi: 10.1038/nrmicro1414
Campbell, B. J., Yu, L., Heidelberg, J. F., and Kirchman, D. L. (2011). Activity of abundant and rare bacteria in a coastal ocean. Proc. Natl. Acad. Sci. 108, 12776–12781. doi: 10.1073/pnas.1101405108
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, X., Wang, J., You, Y., Wang, R., Chu, S., Chi, Y., et al. (2022). When nanoparticle and microbes meet: the effect of multi-walled carbon nanotubes on microbial community and nutrient cycling in hyperaccumulator system. J. Hazard. Mater. 423:126947. doi: 10.1016/j.jhazmat.2021.126947
Cui, Y., Chun, S.-J., Baek, S. H., Lee, M., Kim, Y., Lee, H. G., et al. (2019). The water depth-dependent co-occurrence patterns of marine bacteria in shallow and dynamic southern coast, Korea. Sci. Rep. 9:9176. doi: 10.1038/s41598-019-45512-5
Damashek, J., and Francis, C. A. (2018). Microbial nitrogen cycling in estuaries: from genes to ecosystem processes. Estuar. Coasts 41, 626–660. doi: 10.1007/s12237-017-0306-2
Delgado-Baquerizo, M., Maestre, F. T., Reich, P. B., Jeffries, T. C., Gaitan, J. J., Encinar, D., et al. (2016). Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 7, 1–8. doi: 10.1038/ncomms10541
Delgado-Baquerizo, M., Trivedi, P., Trivedi, C., Eldridge, D. J., Reich, P. B., Jeffries, T. C., et al. (2017). Microbial richness and composition independently drive soil multifunctionality. Funct. Ecol. 31, 2330–2343. doi: 10.1111/1365-2435.12924
Du, L., Guo, S., Wang, R., Guo, Y., Hu, Y., Yang, L., et al. (2024). Distinct variabilities of soil abundant and rare bacteria relate differently to carbon cycling functionality in eroded ecosystems. Catena 235:107675. doi: 10.1016/j.catena.2023.107675
Duffy, J. E. (2009). Why biodiversity is important to the functioning of real-world ecosystems. Front. Ecol. Environ. 7, 437–444. doi: 10.1890/070195
Falkowski, P. G., Fenchel, T., and Delong, E. F. (2008). The microbial engines that drive Earth's biogeochemical cycles. Science 320, 1034–1039. doi: 10.1126/science.1153213
Feng, P., Yang, J., Zhao, S., Ling, Z., Han, R., Wu, Y., et al. (2022). Human supplementation with Pediococcus acidilactici GR-1 decreases heavy metals levels through modifying the gut microbiota and metabolome. NPJ Biofilms Microbiomes 8:63. doi: 10.1038/s41522-022-00326-8
Gao, G.-F., Peng, D., Tripathi, B. M., Zhang, Y., and Chu, H. (2020). Distinct community assembly processes of abundant and rare soil bacteria in coastal wetlands along an inundation gradient. Msystems 5:1120. doi: 10.1128/msystems.01150-01120
Geng, M., Zhang, W., Hu, T., Wang, R., Cheng, X., and Wang, J. (2022). Eutrophication causes microbial community homogenization via modulating generalist species. Water Res. 210:118003. doi: 10.1016/j.watres.2021.118003
Gibson, R., Barnes, M., and Atkinson, R. (2002). Impact of changes in flow of freshwater on estuarine and open coastal habitats and the associated organisms. Oceanogr. Mar. Biol. Annu. Rev. 40:233. doi: 10.1201/9780203180594.ch5
Gomez-Alvarez, V., Pfaller, S., Pressman, J., Wahman, D. G., and Revetta, R. P. (2016). Resilience of microbial communities in a simulated drinking water distribution system subjected to disturbances: role of conditionally rare taxa and potential implications for antibiotic-resistant bacteria. Environ. Sci. Water Res. Technol. 2, 645–657. doi: 10.1039/C6EW00053C
Grman, E., Zirbel, C. R., Bassett, T., and Brudvig, L. A. (2018). Ecosystem multifunctionality increases with beta diversity in restored prairies. Oecologia 188, 837–848. doi: 10.1007/s00442-018-4248-6
Guo, W., Ye, F., Xu, S., and Jia, G. (2015). Seasonal variation in sources and processing of particulate organic carbon in the Pearl River estuary, South China. Estuar. Coast. Shelf Sci. 167, 540–548. doi: 10.1016/j.ecss.2015.11.004
He, J.-Z., Ge, Y., Xu, Z., and Chen, C. (2009). Linking soil bacterial diversity to ecosystem multifunctionality using backward-elimination boosted trees analysis. J. Soils Sediments 9, 547–554. doi: 10.1007/s11368-009-0120-y
Herlemann, D. P., Labrenz, M., Jürgens, K., Bertilsson, S, Waniek, J. J., Andersson, A. F., et al. (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. The ISME journal 5, 1571–1579.
Hügler, M., and Sievert, S. M. (2011). Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Annu. Rev. Mar. Sci. 3, 261–289. doi: 10.1146/annurev-marine-120709-142712
Hutchins, D. A., Mulholland, M. R., and Fu, F. (2009). Nutrient cycles and marine microbes in a CO₂-enriched ocean. Oceanography 22, 128–145. doi: 10.5670/oceanog.2009.103
Hu, W., Ran, J., Dong, L., du, Q., Ji, M., Yao, S., et al. (2021). Aridity-driven shift in biodiversity–soil multifunctionality relationships. Nat. Commun. 12, 1–15. doi: 10.1038/s41467-021-25641-0
Isabwe, A., Yang, J. R., Wang, Y., Wilkinson, D. M., Graham, E. B., Chen, H., et al. (2022). Riverine bacterioplankton and phytoplankton assembly along an environmental gradient induced by urbanization. Limnol. Oceanogr. 67, 1943–1958. doi: 10.1002/lno.12179
Jeffries, T. C., Schmitz Fontes, M. L., Harrison, D. P., van-Dongen-Vogels, V., Eyre, B. D., Ralph, P. J., et al. (2016). Bacterioplankton dynamics within a large anthropogenically impacted urban estuary. Front. Microbiol. 6:163345. doi: 10.3389/fmicb.2015.01438
Jiao, S., Chen, W., and Wei, G. (2017). Biogeography and ecological diversity patterns of rare and abundant bacteria in oil-contaminated soils. Mol. Ecol. 26, 5305–5317. doi: 10.1111/mec.14218
Jiao, S., and Lu, Y. (2020). Soil pH and temperature regulate assembly processes of abundant and rare bacterial communities in agricultural ecosystems. Environ. Microbiol. 22, 1052–1065. doi: 10.1111/1462-2920.14815
Jia, X., Dini-Andreote, F., and Salles, J. F. (2018). Community assembly processes of the microbial rare biosphere. Trends Microbiol. 26, 738–747. doi: 10.1016/j.tim.2018.02.011
Jousset, A., Bienhold, C., Chatzinotas, A., Gallien, L., Gobet, A., Kurm, V., et al. (2017). Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J. 11, 853–862. doi: 10.1038/ismej.2016.174
Kemp, P. F., and Aller, J. Y. (2004). Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol. Ecol. 47, 161–177. doi: 10.1016/S0168-6496(03)00257-5
Kern, M., Winkler, C., and Simon, J. (2011). Respiratory nitrogen metabolism and nitrosative stress defence in ϵ-proteobacteria: The role of NssR-type transcription regulators, vol. 39 Portland Press Ltd., 299.
Kieft, B., Li, Z., Bryson, S., Crump, B. C., Hettich, R., Pan, C., et al. (2018). Microbial community structure–function relationships in Yaquina bay estuary reveal spatially distinct carbon and nitrogen cycling capacities. Front. Microbiol. 9:372069. doi: 10.3389/fmicb.2018.01282
Kritzberg, E. S., Arrieta, J. M., and Duarte, C. M. (2010). Temperature and phosphorus regulating carbon flux through bacteria in a coastal marine system. Aquat. Microb. Ecol. 58, 141–151. doi: 10.3354/ame01368
Li, L., Pujari, L., Wu, C., Huang, D., Wei, Y., Guo, C., et al. (2021). Assembly processes and co-occurrence patterns of abundant and rare bacterial community in the eastern Indian Ocean. Front. Microbiol. 12:616956. doi: 10.3389/fmicb.2021.616956
Li, N., Zhao, H., Jiang, G., Xu, Q., Tang, J., Li, X., et al. (2020). Phylogenetic responses of marine free-living bacterial community to Phaeocystis globosa bloom in Beibu gulf, China. Front. Microbiol. 11:1624. doi: 10.3389/fmicb.2020.01624
Liu, J., Yang, H., Zhao, M., and Zhang, X. H. (2014). Spatial distribution patterns of benthic microbial communities along the pearl estuary, China. Syst. Appl. Microbiol. 37, 578–589. doi: 10.1016/j.syapm.2014.10.005
Liu, L., Yang, J., Yu, Z., and Wilkinson, D. M. (2015). The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J. 9, 2068–2077. doi: 10.1038/ismej.2015.29
Luo, G., Rensing, C., Chen, H., Liu, M., Wang, M., Guo, S., et al. (2018). Deciphering the associations between soil microbial diversity and ecosystem multifunctionality driven by long-term fertilization management. Funct. Ecol. 32, 1103–1116. doi: 10.1111/1365-2435.13039
Madsen, E. L. (2011). Microorganisms and their roles in fundamental biogeochemical cycles. Curr. Opin. Biotechnol. 22, 456–464. doi: 10.1016/j.copbio.2011.01.008
Maidak, B. L., Cole, J. R., Lilburn, T. G., Parker, C. T., Saxman, P. R., Stredwick, J. M., et al. (2000). The RDP (ribosomal database project) continues. Nucleic Acids Res. 28, 173–174. doi: 10.1093/nar/28.1.173
Mai, Y., Peng, S., and Lai, Z. (2020). Structural and functional diversity of biofilm bacterial communities along the Pearl River estuary, South China. Reg. Stud. Mar. Sci. 33:100926. doi: 10.1016/j.rsma.2019.100926
McLusky, D., and Elliott, M. (2007). Transitional waters: a new approach, semantics or just muddying the waters? vol. 71 Elsevier, 359.
Mokany, K., Burley, H. M., and Paini, D. R. (2013). β diversity contributes to ecosystem processes more than by simply summing the parts. Proc. Natl. Acad. Sci. 110:E4057. doi: 10.1073/pnas.1313429110
Mori, A. S., Isbell, F., and Seidl, R. (2018). β-Diversity, community assembly, and ecosystem functioning. Trends Ecol. Evol. 33, 549–564. doi: 10.1016/j.tree.2018.04.012
Mo, Y., Zhang, W., Yang, J., Lin, Y., Yu, Z., and Lin, S. (2018). Biogeographic patterns of abundant and rare bacterioplankton in three subtropical bays resulting from selective and neutral processes. ISME J. 12, 2198–2210. doi: 10.1038/s41396-018-0153-6
Paerl, H. W., Valdes, L. M., Peierls, B. L., Adolf, J. E., and Harding, L. J. W. (2006). Anthropogenic and climatic influences on the eutrophication of large estuarine ecosystems. Limnol. Oceanogr. 51, 448–462. doi: 10.4319/lo.2006.51.1_part_2.0448
Peng, Z., Wang, Z., Liu, Y., Yang, T., Chen, W., Wei, G., et al. (2021). Soil phosphorus determines the distinct assembly strategies for abundant and rare bacterial communities during successional reforestation. Soil Ecol. Lett. 3, 342–355. doi: 10.1007/s42832-021-0109-z
Pinhassi, J., Sala, M. M., Havskum, H., Peters, F., Guadayol, Ò., Malits, A., et al. (2004). Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70, 6753–6766. doi: 10.1128/AEM.70.11.6753-6766.2004
Qin, W., Zheng, Y., Zhao, F., Wang, Y., Urakawa, H., Martens-Habbena, W., et al. (2020). Alternative strategies of nutrient acquisition and energy conservation map to the biogeography of marine ammonia-oxidizing archaea. ISME J. 14, 2595–2609. doi: 10.1038/s41396-020-0710-7
Ren, Z., Luo, W., and Zhang, C. (2022). Rare bacterial biosphere is more environmental controlled and deterministically governed than abundant one in sediment of thermokarst lakes across the Qinghai-Tibet plateau. Front. Microbiol. 13:944646. doi: 10.3389/fmicb.2022.944646
Reveillaud, J., Maignien, L., Eren, A. M., Huber, J. A., Apprill, A., Sogin, M. L., et al. (2014). Host-specificity among abundant and rare taxa in the sponge microbiome. ISME J. 8, 1198–1209. doi: 10.1038/ismej.2013.227
Santos-Júnior, C. D., Sarmento, H., de Miranda, F. P., Henrique-Silva, F., and Logares, R. (2020). Uncovering the genomic potential of the Amazon River microbiome to degrade rainforest organic matter. Microbiome 8, 1–18. doi: 10.1186/s40168-020-00930-w
Snelgrove, P. V., Thrush, S. F., Wall, D. H., and Norkko, A. (2014). Real world biodiversity–ecosystem functioning: a seafloor perspective. Trends Ecol. Evol. 29, 398–405. doi: 10.1016/j.tree.2014.05.002
Soliveres, S., Manning, P., Prati, D., Gossner, M. M., Alt, F., Arndt, H., et al. (2016). Locally rare species influence grassland ecosystem multifunctionality. Phil. Trans. R. Soc. B 371:20150269. doi: 10.1098/rstb.2015.0269
Stegen, J. C., Lin, X., Fredrickson, J. K., Chen, X., Kennedy, D. W., Murray, C. J., et al. (2013). Quantifying community assembly processes and identifying features that impose them. ISME J. 7, 2069–2079. doi: 10.1038/ismej.2013.93
Strom, S. L. (2008). Microbial ecology of ocean biogeochemistry: a community perspective. Science 320, 1043–1045. doi: 10.1126/science.1153527
Tang, X., Chao, J., Gong, Y., Wang, Y., Wilhelm, S. W., and Gao, G. (2017). Spatiotemporal dynamics of bacterial community composition in large shallow eutrophic Lake Taihu: high overlap between free-living and particle-attached assemblages. Limnol. Oceanogr. 62, 1366–1382. doi: 10.1002/lno.10502
Teeling, H., Fuchs, B. M., Becher, D., Klockow, C., Gardebrecht, A., Bennke, C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611. doi: 10.1126/science.1218344
Villnäs, A., Hewitt, J., Snickars, M., Westerbom, M., and Norkko, A. (2018). Template for using biological trait groupings when exploring large-scale variation in seafloor multifunctionality. Ecol. Appl. 28, 78–94. doi: 10.1002/eap.1630
Wang, C., Guo, L., and Shen, R. F. (2023). Rare microbial communities drive ecosystem multifunctionality in acidic soils of southern China. Appl. Soil Ecol. 189:104895. doi: 10.1016/j.apsoil.2023.104895
Wang, J., Wang, L., Hu, W., Pan, Z., Zhang, P., Wang, C., et al. (2021). Assembly processes and source tracking of planktonic and benthic bacterial communities in the Yellow River estuary. Environ. Microbiol. 23, 2578–2591. doi: 10.1111/1462-2920.15480
Wang, J., and Zhang, Z. (2020). Phytoplankton, dissolved oxygen and nutrient patterns along a eutrophic river-estuary continuum: observation and modeling. J. Environ. Manag. 261:110233. doi: 10.1016/j.jenvman.2020.110233
Wang, Y., Castelao, R. M., and Di Iorio, D. (2017). Salinity variability and water exchange in interconnected estuaries. Estuaries and coasts, 40, 917–929. doi: 10.1007/s12237-016-0195-9
Wankel, S. D., Mosier, A. C., Hansel, C. M., Paytan, A., and Francis, C. A. (2011). Spatial variability in nitrification rates and ammonia-oxidizing microbial communities in the agriculturally impacted Elkhorn Slough estuary, California. Appl. Environ. Microbiol. 77, 269–280. doi: 10.1128/AEM.01318-10
Wan, W., Liu, S., Li, X., Xing, Y., Chen, W., and Huang, Q. (2021). Bridging rare and abundant bacteria with ecosystem multifunctionality in salinized agricultural soils: from community diversity to environmental adaptation. Msystems 6, e01221–e01220. doi: 10.1128/mSystems.01221-20
Winfree, R., Fox, J., Williams, N. M., Reilly, J. R., and Cariveau, D. P. (2015). Abundance of common species, not species richness, drives delivery of a real-world ecosystem service. Ecol. Lett. 18, 626–635. doi: 10.1111/ele.12424
Xiong, C., He, J. Z., Singh, B. K., Zhu, Y. G., Wang, J. T., Li, P. P., et al. (2021). Rare taxa maintain the stability of crop mycobiomes and ecosystem functions. Environ. Microbiol. 23, 1907–1924. doi: 10.1111/1462-2920.15262
Xue, Y., Chen, H., Yang, J. R., Liu, M., Huang, B., and Yang, J. (2018). Distinct patterns and processes of abundant and rare eukaryotic plankton communities following a reservoir cyanobacterial bloom. ISME J. 12, 2263–2277. doi: 10.1038/s41396-018-0159-0
Xu, H., Yu, M., and Cheng, X. (2021). Abundant fungal and rare bacterial taxa jointly reveal soil nutrient cycling and multifunctionality in uneven-aged mixed plantations. Ecol. Indic. 129:107932. doi: 10.1016/j.ecolind.2021.107932
Yang, Y., Gao, Y., Huang, X., Ni, P., Wu, Y., Deng, Y., et al. (2019). Adaptive shifts of bacterioplankton communities in response to nitrogen enrichment in a highly polluted river. Environ. Pollut. 245, 290–299. doi: 10.1016/j.envpol.2018.11.002
Yao, Z., du, S., Liang, C., Zhao, Y., Dini-Andreote, F., Wang, K., et al. (2019). Bacterial community assembly in a typical estuarine marsh with multiple environmental gradients. Appl. Environ. Microbiol. 85, e02602–e02618. doi: 10.1128/AEM.02602-18
Zhang, H., Hou, F., Xie, W., Wang, K., Zhou, X., Zhang, D., et al. (2020). Interaction and assembly processes of abundant and rare microbial communities during a diatom bloom process. Environ. Microbiol. 22, 1707–1719. doi: 10.1111/1462-2920.14820
Zhang, X., Qi, L., Li, W., Hu, B. X., and Dai, Z. (2021). Bacterial community variations with salinity in the saltwater-intruded estuarine aquifer. Sci. Total Environ. 755:142423. doi: 10.1016/j.scitotenv.2020.142423
Zhang, Z., Lu, Y., Wei, G., and Jiao, S. (2022). Rare species-driven diversity–ecosystem multifunctionality relationships are promoted by stochastic community assembly. MBio 13, e00449–e00422. doi: 10.1128/mbio.00449-22
Zhang, Z., Zhang, J., and Jiao, S. (2021). Fungi show broader environmental thresholds in wet than dry agricultural soils with distinct biogeographic patterns. Sci. Total Environ. 750:141761. doi: 10.1016/j.scitotenv.2020.141761
Zhao, H., Brearley, F. Q., Huang, L., Tang, J., Xu, Q., Li, X., et al. (2023). Abundant and rare taxa of planktonic fungal community exhibit distinct assembly patterns along coastal eutrophication gradient. Microb. Ecol. 85, 495–507. doi: 10.1007/s00248-022-01976-z
Zhao, H., Yang, S., Qin, X., Huang, J., Huang, H., Li, W., et al. (2023). Disentangling the ecological processes and driving forces shaping the seasonal pattern of Halobacteriovorax communities in a subtropical estuary. Microb. Ecol. 86, 1881–1892. doi: 10.1007/s00248-023-02195-w
Keywords: 16S rRNA, biodiversity, community assembly, ecosystem multifunctionality, estuary ecosystem, rare tax
Citation: Yang S, Hou Q, Li N, Wang P, Zhao H, Chen Q, Qin X, Huang J, Li X, Liao N, Jiang G, Dong K and Zhang T (2024) Rare subcommunity maintains the stability of ecosystem multifunctionality by deterministic assembly processes in subtropical estuaries. Front. Microbiol. 15:1365546. doi: 10.3389/fmicb.2024.1365546
Received: 04 January 2024; Accepted: 08 April 2024;
Published: 19 April 2024.
Edited by:
Yuanchun Zou, Northeast Institute of Geography and Agroecology (CAS), ChinaReviewed by:
Alain Isabwe, University of Michigan, United StatesCopyright © 2024 Yang, Hou, Li, Wang, Zhao, Chen, Qin, Huang, Li, Liao, Jiang, Dong and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianyu Zhang, emhhbmd0eUBnZG91LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.