- 1Indian Institute of Technology, Banaras Hindu University (IIT-BHU), Varanasi, India
- 2Department of Botany, Banaras Hindu University, Varanasi, India
- 3National Institute of Polar Research, Tachikawa-shi, Tokyo, Japan
- 4Department of Biotechnology, Pachhunga University College, Mizoram University (A Central University), Aizawl, India
Cryoconite is a granular structure present on the glaciers and ice sheets found in polar regions including the Himalayas. It is composed of organic and inorganic matter which absorb solar radiations and reduce ice surface albedo, therefore impacting the melting and retreat of glaciers. Though climate warming has a serious impact on Himalayan glaciers, the biodiversity of sub-glacier ecosystems is poorly understood. Moreover, cryoconite holes are unique habitats for psychrophile biodiversity hotspots in the NW Himalayas, but unfortunately, studies on the microbial diversity of such habitats remain elusive. Therefore, the current study was designed to explore the bacterial diversity of the Hamtah Glacier Himalaya using both culturable and non-culturable approaches. The culturable bacterial count ranged from 2.0 × 103 to 8.8 × 105 colony-forming units (CFUs)/g at the different locations of the glacier. A total of 88 bacterial isolates were isolated using the culturable approach. Based on the 16S ribosomal RNA gene (16S rRNA), the identified species belong to seven genera, namely, Cryobacterium, Duganella, Janthinobacterium, Pseudomonas, Peribacillus, Psychrobacter, and Sphingomonas. In the non-culturable approach, high-throughput sequencing of 16S rRNA genes (using MiSeq) showed unique bacterial community profiles and represented 440 genera belonging to 20 phyla, namely, Proteobacteria, Actinobacteria, Firmicutes, Bacteroidetes, Chloroflexi, Acidobacteria, Planctomycetes, Cyanobacteria, Verrucomicrobia, Spirochaetes, Elusimicrobia, Armatimonadetes, Gemmatimonadetes, Deinococcus-Thermus, Nitrospirae, Chlamydiae, Chlorobi, Deferribacteres, Fusobacteria, Lentisphaerae, and others. High relative abundances of Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes were observed in the samples. Phototrophic (Cyanobacteria and Chloroflexi) and nitrifier (Nitrospirae) in bacterial populations indicated sustenance of the micro-ecosystem in the oligotrophic glacier environment. The isolates varied in their phenotypic characteristics, enzyme activities, and antibiotic sensitivity. Furthermore, the fatty acid profiles of bacterial isolates indicate the predominance of branched fatty acids. Iso-, anteiso-, unsaturated and saturated fatty acids together constituted a major proportion of the total fatty acid composition. High cold-adapted enzyme activities such as lipase and cellulase expressed by Cryobacterium arcticum (KY783365) and protease and cellulase activities by Pseudomonas sp. strains (KY783373, KY783377-79, KY783382) provide evidence of the possible applications of these organisms. Additionally, antibiotic tests indicated that most isolates were sensitive to antibiotics. In conclusion, the present study contributed for the first time to bacterial diversity and biopotentials of cryoconites of Hamtah Glacier, Himalayas. Furthermore, the cold-adapted enzymes and polyunsaturated fatty acids (PUFAs) may provide an opportunity for biotechnology in the Himalayas. Inductively coupled plasma mass spectrometry (ICPMS) analyses showed the presence of several elements in cryoconites, providing a clue for the accelerating melting and retreating of the Hamtah glacier.
Introduction
Ice masses store about 70% of freshwater, which sustains billions of people on planet Earth (Cook et al., 2016). Glaciers and ice sheets are the largest reservoirs of freshwater ecosystems, and climate change episodes are rapidly forcing the melting of these glaciers and ice sheets globally (Edwards et al., 2013). Understanding the biogeochemical processes at these places is crucial for predicting future changes, their effects on surrounding habitats, and potential losses of functional biodiversity. Cryoconites are dark-colored quasi-spherical granules found on glaciers and ice sheets across the globes, including the Arctic, Antarctic, and Himalayas (Kohshima et al., 1993; Hodson et al., 2008; Singh et al., 2017). The glacier's mass balance is greatly affected by the quantity and quality of cryoconite in a glacier (Nagatsuka et al., 2014). Cryoconites with a large area on the glacier surface primarily comprise organic and inorganic matter (Gerdel and Drouet, 1960; Hodson et al., 2008; Takeuchi et al., 2010; Singh et al., 2017).
Cryoconites are present on the surface or in quasi-cylindrical mini-depressions where the unique biome inhabits cryoconite holes (Wharton et al., 1985). Mineral dust inputs on glaciers depend on eolian dust deposition and transportation processes near the glaciers (Bøggild et al., 2010; Singh et al., 2013), which serve as nutrient sources for the microbes (Nagatsuka et al., 2014). Ice surface albedo is reduced by mineral dust (Bøggild et al., 2010), aggregation of cryoconite granules (Takeuchi et al., 2001; Hodson et al., 2008; Irvine-Fynn et al., 2012), microbial communities (Yallop et al., 2012; Singh et al., 2014), and meltwater production (Gruell, 2000). Due to the impact of climate change, warmer temperature transforms glacier zones into proglacial zones (Prowse et al., 2006), and subsequently, there is recolonization on glacier-retreated lands (Kaštovaká et al., 2005).
Abbot and Pierrehumbert (2010) opined the role of cryoconites as a potential melt catalyst in albedo-driven deglaciation of “Snowball Earth.” This unique habitat is a hotspot for culturable and non-culturable microbial diversity of Antarctica (Christner et al., 2003; Boetius et al., 2015; Webster-Brown et al., 2015; Obbels et al., 2016; Sommers et al., 2018; Poniecka et al., 2020) and Arctic (Edwards et al., 2011; Singh and Singh, 2011; Singh et al., 2014, 2020; Lutz et al., 2016; Uetake et al., 2016). Additionally, the studies on ecological succession (Williams et al., 2013), geochemistry of melt waters (Fountain et al., 2004; Tranter et al., 2004; Hodson et al., 2008; Bagshaw et al., 2013), and nutrient cycling (Stibal et al., 2012) have also been conducted. Sommers et al. (2018) advocated that environmental factors (lithology of the nearby mountains, height, and geographic location) also play a crucial role in regulating the cryoconite bacterial populations. Whole-genome analyses of a cryoconite bacteria showed unique genes for stress tolerance, DNA repair, enzyme activity, PUFA, and PGPR genes from the Arctic (Singh et al., 2015). Though cryoconite holes are very dynamic freshwater micro-ecosystems and are home to a rich biological diversity, studies at the range of spatial scales and a holistic understanding of this unique habitat are still a secret and thrust area of investigation (Cook et al., 2016).
Cryoconite holes have immense significance in understanding the sub-glacier ecosystems in the Himalayas. Himalayas have about 54000 glaciers and cover a vast geographical area (~60,000 square kilometers). Due to climate warming Himalayan glaciers are melting and retreating substantially, and may destabilize domestic, agricultural and industrial resources of people in the future. Very little work has been conducted on Himalayan cryoconites. The albedo-reducing, optical properties and microbial communities of cryoconites have been characterized from Nepali Himalaya (Takeuchi et al., 2001, 2010; Singh et al., 2020). The studies on biology and biogeochemistry of Indian Himalayan cryoconites and their interactions with climate remain elusive and are thrust areas for research. Hamtah Glacier is a well-researched benchmark glacier for mass balance, glacier melt runoff, and glacier meteorological and debris cover studies (GSI, 2009). This glacier is prone to climate change and loses its mass significantly during summer (Shukla et al., 2015; Mandal et al., 2016). Recently, Dhume et al. (2022) characterized fungal communities of Hamtah glacier. Bacterial diversity and biopotentials of Hamtah glacier is a gap area, therefore present study was conducted.
The aim of the current study was first, to investigate culturable and non-culturable bacterial communities present in Hamtah glacier cryoconites. Secondly, to characterize the biopotentials (cold-adapted enzymes, fatty acids, antibiotic resistance, carbon source utilization and physiological features). Furthermore, to know the physical properties of cryoconites, the elemental composition was also analyzed.
Materials and methods
Sampling site
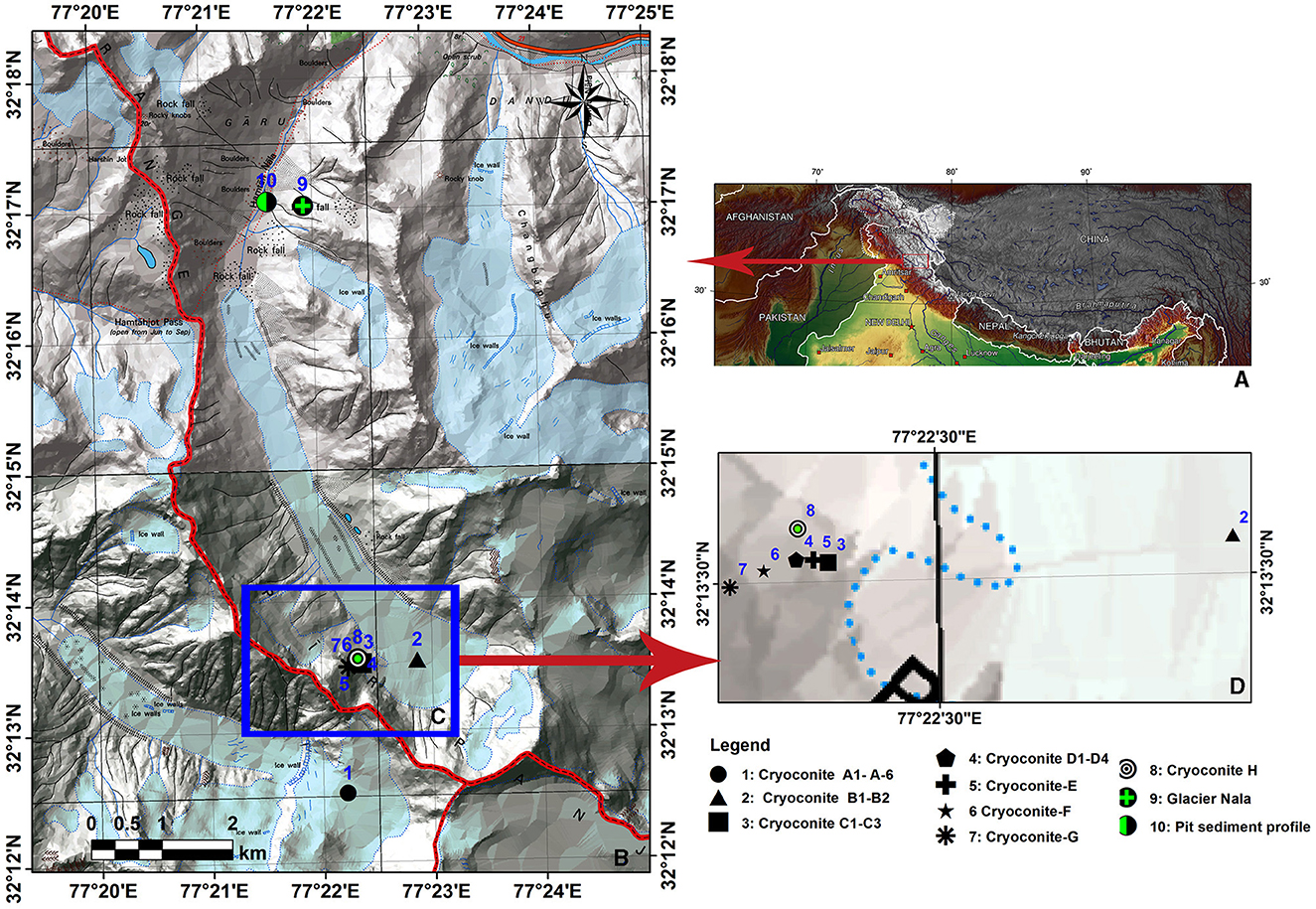
Hamtah Glacier (32.24°N, 77.37°E) is an important glacier extending from south to north between 5000 and 4020 masl and covering an area of ~3 km2 and is ~6 km long (GSI, 2009). This glacier is located in the Chandra Basin on the northern slopes of the Pir-Panjal range of the Indian Himalayas. The landscape of the Hamtah Glacier and pictures of the sampled hole are shown in Supplementary Figure 1. Cryoconite samples were collected from seven different locations of Hamtah Glacier (Figure 1) using a sterile syringe and ampules. Before sampling, the pH and temperature of cryoconite holes were recorded. The temperature varied from 1.2 to 2.1°C, and pH ranged from 7.5 to 8.4 at different locations of the glacier. All the samples were stored at sub-zero temperatures and transported to the laboratory for further analysis.
Isolation and culturing of bacteria
One gram of cryoconite was suspended in 9 mL saline solution and diluted serially (10−1, 10−2, and 10−3). To maximize the recovery of isolates, several bacteriological media (HiMedia, India) of different strengths (concentrations) were used in the present study. Enumeration of culturable bacteria was done using the spread plate method (0.1 ml) on Nutrient Agar (NA per liter: Beef powder 3 g, Sodium Chloride 8 g, Peptone 5 g and 20g Agar, pH 7.0), 1/10 NA, Marine Broth (MB per liter: Zobell Marine broth 20.25 g and 20 g agar, pH 7.6), 1/10MB, Antarctic Bacterial Medium (ABM per liter: 5 g peptone, 2 g yeast extract, and 15 g agar, pH 7.0), 1/10 ABM and R2Agar (R2Agar-HiMedia 18.12 g, pH 7.2) incubated at two different temperature 4°C and 15°C for 2–4 weeks for optimum growth of psychrophiles (Singh et al., 2016). To observe the emergence of bacterial colonies in a natural environment and compare them with laboratory incubators, the representative samples of each location (a total of 14 cryoconite samples) were also aseptically spread on readymade culture plates and incubated at the natural temperature of the Himalayas itself. The colony-forming units (CFUs) appearing after incubation were counted, and the CFU number per gm soil was calculated. Isolates for further study were picked based on colony morphology (outline, texture, color, etc.) from each plate. The pure bacterial isolates obtained through streaking techniques were preserved in cryovials with glycerol stock (70%) and finally stored at −80°C deep freezer. The unique distinctive cultures were submitted to the Microbial Culture Collection (MCC), Pune, India.
Phenotypic characterization of the bacterial strains
Colony features were observed by a stereo-zoom microscope (Nikon SMZ1500). Carbon utilization tests were carried out using HiCarboTM kits (KB009, HiMedia, India) and incubated for 2–7 days at 4 and 15°C. ONPG (Ortho-Nitrophenyl-β-D-galactopyranoside) was used to detect β-galactosidase activity. Esculin, citrate, and malonate utilization tests were carried out to detect the capability of bacterial strains to utilize Esculin, citrate and sodium malonate as a sole carbon source. Growth experiments were performed on ABM plates at a range of temperatures (4, 15, 22, and 30°C), pH (3, 5, 7, 9, and 11) and salt concentrations (0, 2, 5, 7, and 15%). The unique bacterial isolates were screened for cold-active enzymes such as amylase, cellulase, lipase, and protease following standard protocol (Singh et al., 2014).
DNA extraction, polymerase chain reaction, sequencing of culturable bacteria and phylogenetic tree construction
Genomic DNA was extracted following the standard method (Sambrook et al., 1989). 16S rRNA universal primers 16F27 (5′-CCA GAG TTT GAT CMT GGC TCA G-3′) and 16R1492 (5′-TAC GGY TAC CTT GTT ACG ACT T-3′) were used for PCR amplification. The amplified 16S rRNA gene PCR product was purified by PEG-NaCl precipitation. DNA sequencing was carried out using an automated DNA sequencer (ABI® 3730XL, Applied Biosystems, Inc., Foster City, CA) at Microbial Culture Collection (MCC), NCCS at Pune, India. Essentially, sequencing was carried out from both ends using additional internal primers so that each position was read at least twice. Assembly was carried out using the Lasergene package followed by an EzTaxon search (Kim et al., 2012) for identification.
The nucleotide sequences of 47 bacterial isolates were deposited in the DNA data bank (NCBI) and assigned accession numbers are KY783365 to KY783401. Nucleotide sequences were subjected to NCBI BLAST search. Sequence alignment of the 16S rRNA region of each isolate was carried out using the Clustal W algorithm, Molecular Evolutionary Genetics Analysis (MEGA) 11.0 software was used for tree construction. The phylogenetic tree was constructed by neighbor-joining (NJ) method with 1000 bootstrap replicates of the phylogeny test, and partial deletion of data sub-set was used with a 95% cut-off setting in the partial deletion method. Red color phylogenetic trees indicate isolates/strains of the present study (sequences) and blue colored for closely related strains of the same species from the database.
Non-culturable bacterial analyses: DNA extraction and high-throughput sequencing of 16s rRNA genes using MiSeq
Genomic DNA was extracted from 14 cryoconite samples (A, A2, A4, A5, A6, B, B2, C, D, D1, D2, D3, EHamtah, G, cryG) and one each from glacier Snout and glacier Nala/stream) using the FastDNA SPIN Kit (Soil DNA Extractionkit:MP Biomedicals, Santa Ana, CA). The bacterial community structure in the cryoconite samples was analyzed by sequencing the 16S rRNA gene using MiSeq sequencer (Illumina, San Diego, CA). Partial 16S rRNA gene sequences including the V3 and V4 regions were amplified. Bakt 341F and Bakt 805R primers with Illumina overhang adaptor sequences attached to their 5′ends were used. PCR amplification of the 16S rRNA gene, reaction clean-up, index PCR and sequencing were performed following Illumina methods for 16S rRNA gene sequencing library preparation, Pyrosequencing, Phylogenetic analyses and Community comparisons were done following standard procedure (Edgar et al., 2011; Segawa et al., 2014; Uetake et al., 2016).
Fatty acid methyl ester analysis
The cultures were grown on a standard media Tryptic Soy Broth Agar (TSBA, Himedia) for FAME analyses, and also on respective isolate-isolation media at two standard psychrophilic temperatures 40C and 150C for 2 to 3 weeks. Three replicates of individual bacterial isolates were made by streaking on similar media plates and incubating them in similar conditions. The extraction and analysis of cellular fatty acid methyl esters (FAME) was carried out following standard protocol (Buyer, 2008). FAMEs analyses were carried out using a gas chromatograph (Agilent Technologies, CA, USA, Model 7890A), equipped with a flame ionization detector (FID), auto-sampler and capillary column (HP-5 column, 25 m × 0.2 mm × 0.33 μm). Hydrogen was used as carrier gas. The gas chromatography system was calibrated twice with a calibration standard no. 1300AA (MIDI, USA) before analyses of bacterial samples. Additionally, before running the cryoconite bacterial samples, the extraction efficiency of the protocol and authenticity of fatty acid peaks were verified by using a known fatty acid profile of a type strain (Stenotrophomonas maltophili a ATCC 13637T) as a positive control. The individual fatty acid peaks were identified based on the retention time of the standard run. The standard software and database were used (RTSBA6 of MIS, MIDI Inc., Newark, DE, USA) for analysis.
Antibiotic susceptibility
The susceptibility of the bacterial isolates to 45 antibiotics (Supplementary Table 7) was determined by using the impregnated disc method (Bauer et al., 1966). The bacterial isolates were grown at 15°C in nutrient broth up to a turbidity of 0.08–0.13, optical density measured at 620 nm. The bacterial suspensions (100 μl) were spread onto the surface of the Mueller Hinton Agar medium plates, using sterile spreaders. The different antibiotic discs were placed over the individual bacterial lawns, and the plates were incubated at 15°C for 3–4 days. Petri dishes were observed for the formation of a zone of inhibition, if any. The zone formed was measured in millimeters.
Elemental analyses of cryoconites
Cryoconite granules were finely grounded powdered samples were oven-dried at 110°C. An amount of 0.5 g of each sample was acid digested (3 mL of 69% sub-pure HNO3, 1 mL subpure 30% HCl and 1 mL of 30% H2O2) completely in the microwave at 180°C (Ethos 1, Milestone, Italy). The digested samples were analyzed using inductively coupled plasma mass spectrometry (ICP-MS), × Series II, Thermo Fisher Scientific, Bremen, Germany. CertiPUR ICPmulti-element standard (Merck, Darmstadt, Germany) was used for calibration. Elemental concentrations were measured in triplicate and were recorded in mg/kg as described previously (Singh et al., 2017).
Results
Characteristics of the glacier cryoconite samples
The pH and temperature in the cryoconite holes showed varied responses depending upon whether the holes are open or closed and, their sampling locations on the glacier. The water temperature in the cryoconite holes ranged between 1.2–2.1°C and pH 7.58–8.48. Cryoconites are brownish-black in color, organic carbon ranges from 0.57 to 1.12%, and total phosphorus 0.14 to 0.20 mg/g. The elemental composition showed the presence of 25 elements (Li, Be, Na, Mg, P, K, Ca, V, Cr, Mn, Fe, Co, Ni, Cu, Zn, As, Rb, Sr, Cd, Cs, Ba, Tl, Pb, Bi and U) in all cryoconite samples to varying amount. K, Mg, Mn, Fe, Ca, P and Na were dominant elements in samples (Table 1 and Supplementary Table 1).
Bacterial count and characteristics of the isolated strains
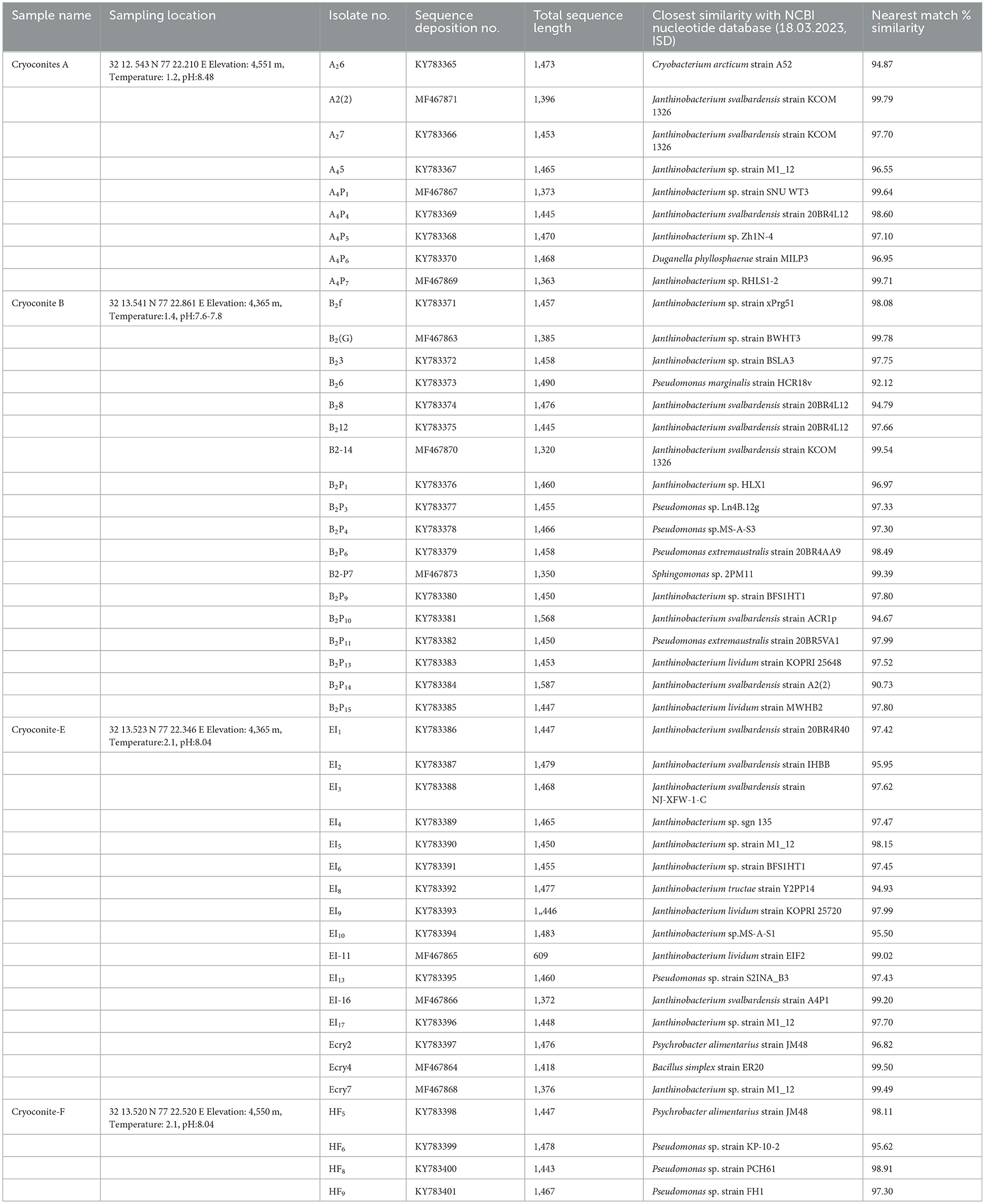
The culturable bacterial count (colony forming units, CFUs) on various bacteriological media ranged from 2.0 × 103 to 8.8 × 105 CFUs per g. Of the numerous CFUs that appeared on the culture media plates, 47 distinct isolates were purified for further studies (Table 1). The culture colonies showed different colors (white, cream, yellow and orange), and morphological features (entire margin, opaque and smooth texture, and convex pulvinate to raised elevation). The shapes of bacterial cells were short to long rods. Most of the isolates showed growth temperature ranges from 4 to 22oC. The optimum temperature for the growth of culturable isolates was 15oC, therefore indicating psychrophilic. The isolates showed salt tolerance at 1 and 7% and optimum growth at pH 6.0 to 7.0. The 16S rRNA gene sequences of 47 bacterial isolates were deposited to GenBank (NCBI) with accession numbers (KY783365 to KY783401, MF467864 to MF467871, and MF467873) (Table 1).
Carbon utilization ability of the isolated strains
Of the 35 carbon sources tested, most of the isolates showed varied responses for carbon utilization tests (Supplementary Table 2). Amongst the isolates tested, Peribacillus frigoritolerans E-Cry-4 utilized 25, Cryobacterium arcticum-A2–6 utilize 24, Pseudomonas simiae-B2–6 utilized 26, Psychrobacter pulmonis E-Cry-2 utilize 27, Sphingomonas glacialis B2-P7 utilize 17, Janthinobacterium svalbardensis B2–8 can utilize as much as 4 (ONPG, esculin, citrate and malonate), of the 35 carbon sources tested. Except isolate E-Cry-2 none of the isolates were capable of utilizing sorbose.
Extracellular enzymatic activities
The 45 bacterial cultures that were screened for enzyme production showed varied responses. Four isolates were positive for amylase, 12 for cellulase, 14 for lipase, and 20 for protease (Supplementary Table 3). Temperature showed a vital role in enzyme production. Cellulase, lipase and protease production decreased with an increase in temperature from 4 to 22°C. In contrast, amylase production increased with an increase in temperature. Cryobacterium arcticum-A2–6 was the most promising culture as it has strong cellulase and lipase activity, while 4 other cultures (B2P3, B2P4, B2P6, B2P11) produced three different enzymes: cellulase, lipase and protease. Four cultures (B2–6, HF6, HF8, HF9) indicated strong activity for cellulase and protease. Three cultures (E-cry 4, HF6, HF8) showed amylase activity (Supplementary Table 3).
Phylogenetic analyses/taxonomic analysis of culturable bacteria
The total sequence lengths after alignment, the closest sequence similarities % with the database, and the NCBI sequence deposition numbers are given in Table 1. Sequence alignment was done using clustalW algorithm. The sequence analysis of the 16S rRNA gene domain of isolate Peribacillus frigoritolerans Ecry4 (MF467864) indicated their closest relationship to NCBI database species Bacillus simplex strain ER20 (99.50%), Cryobacterium A2–6 (KY783365) showed closest relationship with species of Cryobacterium arcticum strain A52(94.87%). Duganella A4P6 (KY783370) indicated their closest relationship with the species of Duganella phyllosphaerae MILP3 (96.95%). Sequence analysis of 32 bacterial isolates of Janthinobacterium (A2(2), A27, A45, A4P1, A4P4, A4P5, A4P7, B2f, B2(G), B23, B28, B212, B2–14, B2P1, B2P9, B2P10, B2P13, B2P14, B2P15, EI1, EI2, EI3, EI4, EI5, EI6, EI8, EI9, EI10, EI11, EI16, EI17 and Ecry7) indicated their closest relationship to species of Janthinobacterium svalbardensis JA-1 (93.01 to 99.79%). Nine bacterial isolates of Pseudomonas (B26, B2P3, B2P4, B2P6, B2P11, EI13, HF6, HF8, HF9) showed their closest relationship (92.12 to 98.91%) to the species of Pseudomonas marginalis strain HCR18v and Pseudomonas extremaustralis strain 20BR5VA1. Isolate Sphingomonas glacialis B2-P7 (MF467873) indicated their closest relationship with species of Sphingomonas sp. 2PM11 (99.39%). Sequence analysis of 2 bacterial isolates of Psychrobacter pulmonis Ecry2 (KY783397) and HF5 (KY783398) indicated their closest relationship with species of Psychrobacter alimentarius strain JM48 CECT 5989 (96.82) and Psychrobacter namhaensis strain E3 98.11%). The affiliation of the representative isolates of culturable bacteria is shown in a phylogenetic tree (Figures 2A–G).
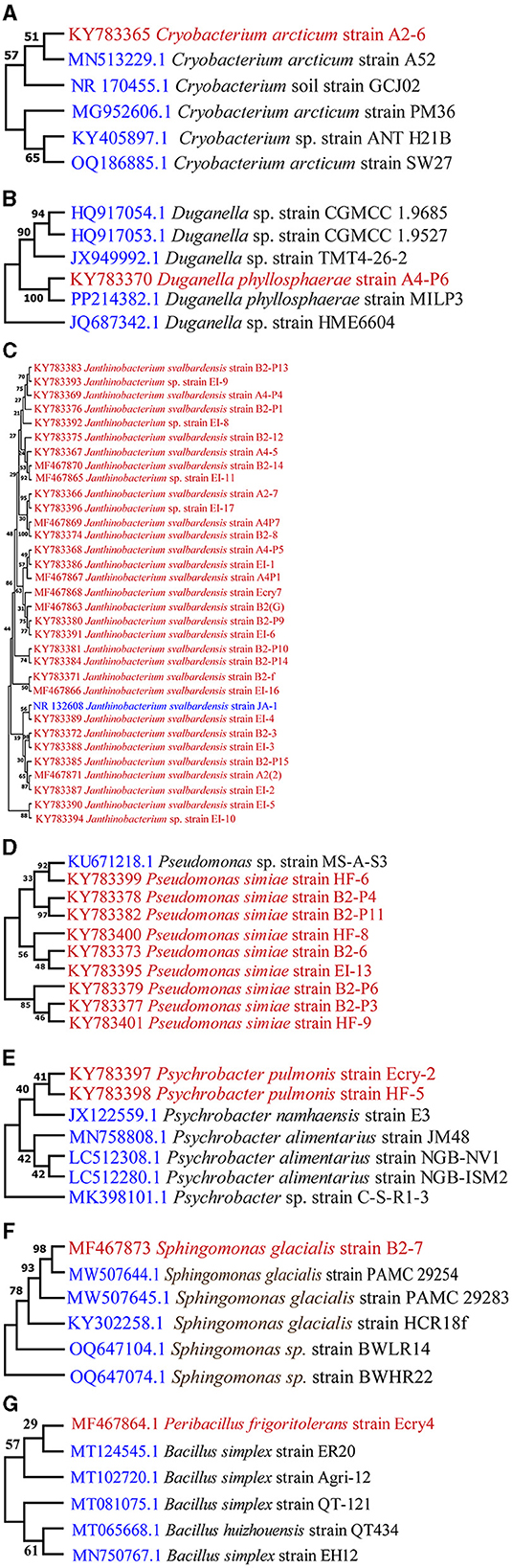
Figure 2. Phylogenetic trees of Culturable bacteria: (A) Cryobacterium, (B) Duganella, (C) Janthinobacterium, (D) Pseudomonas, (E) Psychrobacter, (F) Sphingomonas, (G) Peribacillus.
The bacterial isolates from the cryoconites of different locations in Hamtah glacier belonged to seven different genera, namely, Cryobacterium, Duganella, Janthinobacterium, Pseudomonas, Sphingomonas, Peribacillus and Psychrobacter (Table 1). The number of species at each location varied from 2 to 4, while number of isolates varied from 4 to 18. The most predominant species was Janthinobacterium svalbardensis followed by Pseudomonas simiae. Genera Cryobacterium, Duganella, Peribacillus and Sphingomonas were represented the least. Glacier location E showed maximum culturable diversity, representing four different species, while location F of glacier showed minimal diversity, representing only two species.
Analyses of non-culturable bacteria using MiSeq high-throughput sequenced data of 16S rRNA genes
After curation of the MiSeq dataset, 16S rRNA gene sequences representing the non-culturable bacterial populations from the cryoconite samples across Hamtah glacier, Himalaya. The profiles represent eight major lineages: Proteobacteria was the most dominant (207 genera, 64 families), followed by Actinobacteria (80 genera, 28 families), Firmicutes (52 genera, 19 families), Bacteroidetes (51 genera, 12 families), Cyanobacteria (10 genera, two families) with other member phyla such as Chloroflexi (eight genera and seven families), Acidobacteria (five genera and three families), Planctomycetes (six genera, one family), Verrucomicrobia (five genera, four families) Deinococcus-Thermus (three genera, three families), Chlamydiae (three genera, two families), Spirochaetes (three genera, two families), Armatimonadetes (two families and two genera), The phyla Caldiserica, Deferribacteres, Elusimicrobia, Fusobacteria, Gemmatimonadetes, Lentisphaerae, and Nitrospirae represented least (one family and one genus in each phyla) along with others unknown taxa (Supplementary Table 4). In the present study, Proteobacteria is an important bacterial phylum and represent maximum number of bacteria belonging to five classes such as Betaproteobacteria (77 genera, 11 families), Alphaproteobacteria (54 genera, 19 families), Gammaproteobacteria (47 genera and 15 families), Deltaproteobacteria (23 genera and 16 families) and Epsilonproteobacteria (six genera and three families). Proteobacteria, Actinobacteria, Bacteroidetes and Firmicutes were well represented in all of the sampling sites. Armatimonadetes, Caldiserica, Deferribacteres, Elusimicrobia, Fusobacteria, Gemmatimonadetes, Lentisphaerae and Nitrospirae were present least in samples. The relative abundance (%) of bacterial phyla present at different sampling sites are shown in Figure 3 and Supplementary Table 5. The bacterial profile results indicate that taxa distribution exhibits a specific distribution pattern along the different glacier cryoconites, glacier snouts and glacier streams. The similarity of bacterial community structures between sampling sites is shown in a UniFrac dendrogram (Figure 4).
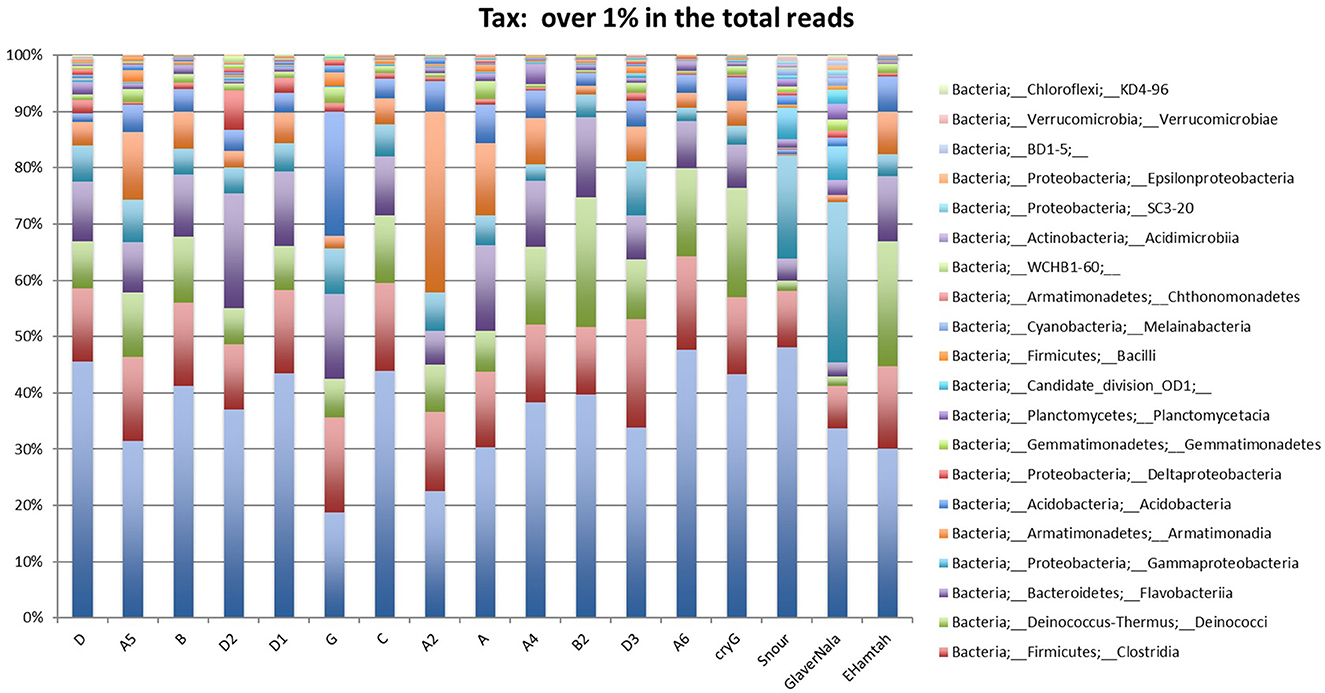
Figure 3. Non-culturable Bacteria: showing relative abundance (%) of bacterial phyla in Hamtah glacier cryoconites, snout, and stream (Nala).
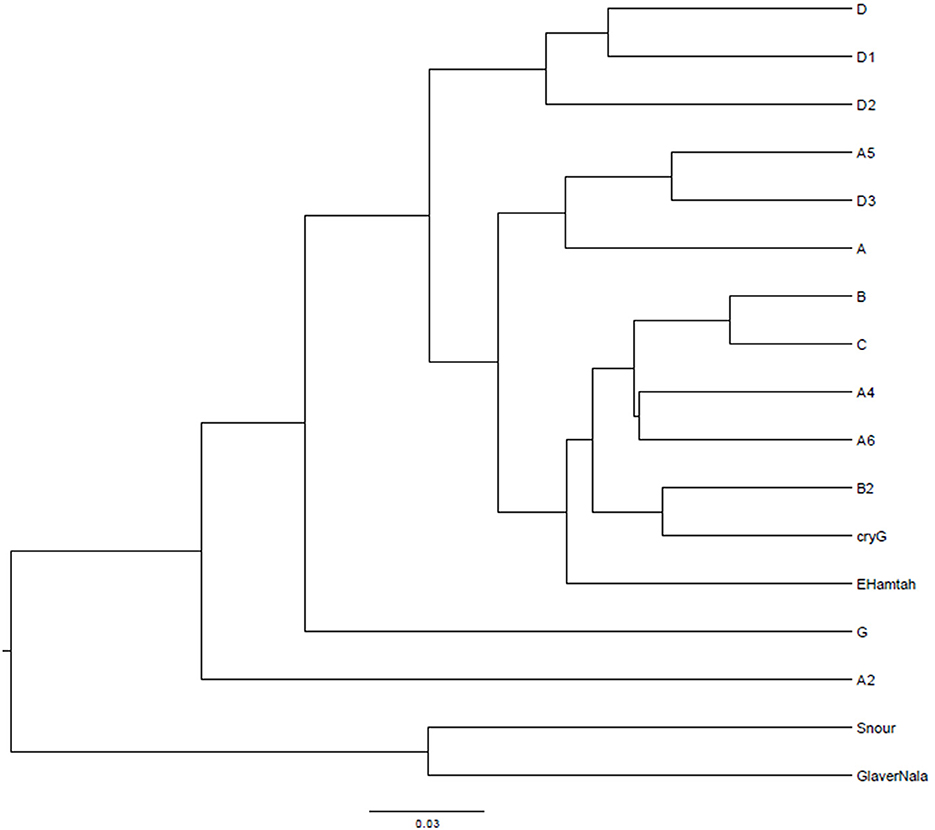
Figure 4. UniFrac dendrogram showing the similarity of bacterial community structures between sampling sites.
FAME analysis
Six representative bacterial taxa were characterized for cellular fatty acid composition. Janthinobacterium svalbardensis B28 (KY783374), Gram-negative, and Betaproteobacteria belong to the Oxalobacteraceae family. Pseudomonas simiae B26 (KY783373) Gram-negative, Gammaproteobacteria, belonging to the family Pseudomonadaceae. Psychrobacter pulmonis Ecry2 (KY783397) Gram-negative, Gammaproteobacteria, belonging to the family? Moraxellaceae. Peribacillus frigoritolerans Ecry4 (MF467864) Gram-positive, Firmicutes, belonging to the family Bacillaceae. Cryobacterium arcticum A26 (KY783365) Gram-positive bacterial strains belong to family Microbacteriaceae. Sphingomonas glacialis B2P-7 (MF467873) Gram-positive, Alphaproteobacteria, belonging to the family Sphingomonadaceae.
In the Hamtah glacier bacterial species, the branched fatty acids varied in composition from a minimum of 0.17% as in Janthinobacterium svalbardensis B28, to a maximum of 96.59% as in Cryobacterium arcticum A26. In Janthinobacterium svalbardensis B28 the low % of branched fatty acids was compensated by a high proportion of unsaturated fatty acids (63.24%). The unsaturated fatty acid composition in all the bacterial species varied in composition from a minimum of 58.4% as in Janthinobacterium svalbardensis B28, to a maximum of 90.72% as in Peribacillus frigoritolerans Ecry4 (Supplementary Table 6). The saturated fatty acid composition in all the bacterial species varied in composition from a minimum of 2.39% as in Sphingomonas glacialis B2P-7, to a maximum of 41.24% as in Janthinobacterium svalbardensis B28 (Supplementary Table 6).
Antibiotic resistance patterns of the isolated strains
Cryoconite bacterial isolates were subjected to 45 antibiotic screening tests. The result showed that most of the isolates were sensitive to antibiotics such as Amikacin, Carbenicillin, Cefoperazone, Ceftazidine, Chloramphenicol, Ciprofloxacin, Doxycycline hydrochloride, Gatifloxacin, Gentamicin, Kanamycin, Levofloxacin, Lomefloxacin, Meropenem, Nalidixic Acid, Netillin, Norfloxacin, Ofloxacin, Rifampicin, Streptomycin, Tetracycline, and Tobramycin (Supplementary Table 7). The sensitivity to other antibiotics tested varied from isolate to isolate. Clidamycin and Methicillin were the least potent antibiotics tested and were sensitive to only two isolates (Ecry-2 and Ecry-4). Of the isolates screened against 45 antibiotics, B2–8 and B2–6 were the most resistant, showing resistance to 19 and 21 antibiotics, respectively. Other isolates showed varied resistance profiles toward the antibiotics.
Discussion
The main aim of the current study was to investigate the bacterial diversity, biopotentials and properties of cryoconites of Hamtah glacier, Himalaya. The pH of the cryoconite holes observed in the present study ranged from 7.5 to 8.4, resembling earlier data pH 5.9–9.6 from Canada glacier Antarctica (Mueller and Pollard, 2004), and pH 7.1–8.6 from Arctic glaciers (Singh et al., 2014). The pH properties of cryoconites holes depends on mineral contents, microbial activities and glacier environment in which it is found (Tranter et al., 2004). The organic carbon content in the present study was low (0.57%−1.12%) confirming the oligotrophic nature of cryoconite habitat. The similar data have also been reported from Arctic (1.07%−2.5%) by Singh et al. (2014), and 5.4%−9.9% from Nepali Himalaya (Takeuchi et al., 2001). Of 25 elements observed in cryoconites of the present study, Mg, Fe, Mn, Ca, P, and Na were dominant, as observed in previous studies (Singh and Singh, 2011; Singh et al., 2017). The accumulation and release of these inorganic material may impact supraglacier and proglacier ecosystems of Himalayas. The elemental content is mainly input from atmospheric, hydrologic and mineral resources, and this finding is further supported by Mueller et al. (2001) and Singh et al. (2017). The bacterial CFUs that emerged on the media plates ranged from 2.0 × 103 to 8.8 × 105 CFUs per g at the different locations of Hamtah glacier studied. The bacterial CFU data similar to the current observation are also reported from Arctic cryoconites (2.7 × 103 to 8.8 × 104 per g, Singh et al., 2014) and from Antarctic cryoconite melt water (8.8 × 104 per ml, Christner et al., 2003).
The bacterial isolates could grow at 4 to 22°C, except for one at 30°C. The optimum temperature of growth was 15°C. The Hamtah glacier isolates profusely grow at 4 and 15°C as compared to 22°C resembles with previous studies from Antarctic and Arctic (Christner et al., 2003; Singh et al., 2014). Although the culturable bacterial diversity in Hamtah cryoconites is limited, but their strains differ widely in their antibiotic sensitivity characteristics. In the present study, the sensitivity of isolates toward antibiotics varied, as recorded in earlier studies from Arctic cryoconites (Singh et al., 2014). The most of the isolates indicated low resistance toward antibiotics in Himalaya similar to Polar regions. Segawa et al. (2012) reported that the Antarctic region has the least antibiotic resistance genes, followed by the Arctic region. The organic carbon sources are important for microbial metabolism but it limited in oligotrophic environment of cryoconite holes. The carbon source utilization tests showed that bacterial isolates from the Himalayan cryoconites prefer monosaccharides similar to Arctic cryoconite (Singh et al., 2014). The limited available organic and inorganic matter in cryoconite holes has also been reported from Antarctica (Foreman et al., 2007). Psychrophilic organisms can produce cold-active enzymes which have immense applications in Agriculture, health and industry (Feller and Gerday, 2003). The 45 bacterial isolates of Hamtah glacier cryoconites showed activities for one or more enzymes (amylase, cellulase, lipase, protease) either at 4°C, 15°C, and (or) 22°C. The ability of these bacterial isolates to produce different enzymes speaks about their biopotentials in sub-glacial environments. Some of the isolates of the present study showed strong cellulase, lipase and protease activity, similar to Arctic cryoconites (Singh et al., 2014). Additionally, there are reports of extracellular enzyme activities by Arctic bacteria (Reddy et al., 2009; Yu et al., 2009) and Himalayan fungi (Dhume et al., 2019).
The taxonomic analyses of bacteria using a culturable approach showed the presence of seven genera Cryobacterium, Duganella, Janthinobacterium, Pseudomonas, Peribacillus, Psychrobacter and Sphingomonas. The culturable bacterial genera Cryobacterium, Janthinobacterium, Pseudomonas and Sphingomonas of the present study have also been reported from Arctic cryoconites (Svalbard: Singh et al., 2014, Greenland: Perini et al., 2019; Singh et al., 2020). Genera such as Pseudomonas, Cryobacterium are of common occurrence in cryoconite habitats and have been previously reported from the Antarctic cryoconites (Christner et al., 2003). Pseudomonas has also been documented in the Alpine cryoconites (Lee et al., 2011). The occurrence of related phylotypes in geographically diverse cold environments suggests that they have unique adaptation strategies at low temperatures (Abyzov et al., 1998). These adaptation strategies include the occurrence of pigments, polyunsaturated and branched fatty acids (PUFAa) and or enzymes in the current isolates are active at low temperatures. Fatty acid composition of bacterial isolates of Hamtah glacier indicated presence of unsaturated and branched (including the iso- and anteiso-) fatty acids. Chintalapati et al. (2004) have also described the role of fatty acids in psychrophiles. Nishida and Murata (1996) reported the role of unsaturated and branched fatty acids in maintaining functional membrane fluidity at low temperatures. Thus, psychrophilic and psychrotolerant bacteria from glacier cryoconite adapt to the cold environment by preferentially synthesizing unsaturated and branched fatty acids. MiSeq high-throughput sequenced data of 16S rRNA genes showed presence of 440 genera of non-culturable bacteria in Hamtah glacier cryoconites (Supplementary Tables 4, 5). The major phyla present in current study are Proteobacteria, Actinobacteria, Bacteroidetes, Firmicutes, followed by Cyanobacteria, Chloroflexi, Acidobacteria, and Armatimonadetes were previously identified from Greenland cryoconites (Uetake et al., 2016). Additionally, the present study also showed seven common phyla such as Proteobacteria, Cyanobacteria, Bacteroidetes, Actinobacteria, Acidobacteria, Chloroflexi and Planctonycetes were also recorded from cryoconites of Svalbard, Arctic (Edwards et al., 2011). Himalayan cryoconites showed dominance of Alpha-, Betaproteobacteria is consistent with earlier studies from Antarctica (Christner et al., 2003). Deltaproteobacteria and Firmicutes were reported from majority of cryoconites holes resembles current observation (Cameron et al., 2012). The presence of eight genera of cyanobacteria (Chroococcidiopsis, Chamaesiphon, Leptolyngbya, Microcoleus, Phormidesmis, Phormidium, Tychonema, Anabaenopsis, Nostoc, Calothrix) and non-cyanobacterial photosynthetic bacteria Rhodobacter and Chloroflexi in current study may have an important role in primary production in Hamtah glacier cryoconites holes. Cyanobacterial taxa have also been identified globally in cryoconites of other cold places (Edwards et al., 2011; Uetake et al., 2016). The chemolithoautotrophic Nitrosomonadales genera Nitrosomonas and Nitrosospira has ability to oxidize ammonia to nitrite are present in current study were also reported from other cold habitat (Segawa et al., 2012). Besides this other Nitrosomonadales taxa, Candidatus, Nitrotoga, Ferriphaselus, Gallionella, Sideroxydans were observed in the present study (Supplementary Table 5). We also identified seven bacterial phyla from Hamtah glacier Snout (Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Proteobacteria, Spirochaetes and Verrucomicrobia), and six phyla from a glacier-stream/Nala (Acidobacteria, Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Proteobacteria) which are consistent with an earlier report from Himalayan Kafni proglacial soils (Srinivas et al., 2011). Proteobacteria, Actinobacteria, Bacteroidetes and Firmicutes were well represented in all the sampling sites; however, some bacterial groups have shown uneven spatial distribution between sampling sites in the present study, resembling Arctic cryoconite holes (Edwards et al., 2011). Physico-chemical conditions of glaciers, topography, and aerial deposition of dust/propagules are possibly responsible for the spatial distribution pattern of bacteria in Polar cryoconite including the Himalayas. Additionally, Bacterial community composition also depends on bed rock type as reported from Antarctica (Tytgat et al., 2016).
The culturable bacterial community profiles were distinct from those found in the metagenomic community. The non-culturable approach studies document a more enormous assortment of bacteria (440 genera) than the culture-based approach (7 genera) in this study. However, there are some common genera in both approaches, but they are fewer in number. Similar observations have also been recorded from Arctic and Antarctic cryoconites (Christner et al., 2003; Edwards et al., 2011; Uetake et al., 2016). The culture based approach was applied in present study is to determine and compare physiological characteristics/biopotentials of individual species' of-glacier cryoconites. Furthermore, to understand the ecological roles (inter- and intra-species interactions, adaptation strategies etc.) culturable approach is very important (Jiang et al., 2006).
Conclusions
Cryoconite holes are important microcosms on the glacier where cold-adapted microbes flourish. The present study focused on characterizing the bacterial diversity of Himalayan cryoconites holes through culture- and non-culture-based approaches. The culturable bacterial population in cryoconite holes is sizeable (seven genera), whereas the non-culturable population predominates with 440 genera. These are some of the dominant genera known to occur in the colder habitats of the world. Although the culturable bacterial communities in these microhabitats is limited, but strains differ widely in their antibiotic sensitivity characters. Carbon utilization tests showed that the bacterial isolates from cryoconite holes prefer simpler carbon sources. Bacterial isolates produce high amounts of polyunsaturated-branched fatty acids (PUFAs), and various cold active enzymes indicating the prospect of biotechnology in Himalaya. To detect the functions in whole bacterial communities of Himalayan cryoconites there is need of further studies on metatranscriptomics and DNA microarrays.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
PS: Writing – original draft, Methodology, Formal analysis. SS: Sampling, Data curation, Funding acquisition, Writing – review & editing. TS: Formal analysis (MiSeq data), Writing – review & editing. PKS: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Financial support was received for research (sample analyses) from ICAR, India and NIPR Japan.
Acknowledgments
The authors are grateful to National Bureau of Agriculturally Important Microorganisms (NBAIM), India, and Prof. Satoshi Imura, National Institute of Polar Research (NIPR), Japan. SS is thankful to GSI for camping support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1362678/full#supplementary-material
Supplementary Figure 1. (a, b) Landscape of Hamtah Glacier and (c1–c4) Cryoconite holes.
References
Abbot, D. S., and Pierrehumbert, R. T. (2010). Mudball: surface dust and snoball Earth deglaciation. J. Geophys. Res 115:D03104. doi: 10.1029/2009JD012007
Abyzov, S., Mitskevich, I., and Poglazova, M. (1998). Microflora of the deep glacier horizons of central Antarctica. Microbiology 67, 66–73.
Bagshaw, E. W., Tranter, M., Fountain, A. G., and Welch, K. (2013). Do cryoconite holes have the potential to be significant sources of C,N, and P to downstream depauperate ecosystems of Taylor Valley, Antarctica? Arct. Antarct. Alp. Res. 45, 1–15. doi: 10.1657/1938-4246-45.4.440
Bauer, A. W., Kirby, M. M., Sherris, J. C., and Truck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Path. 45, 493–496. doi: 10.1093/ajcp/45.4_ts.493
Boetius, A., Anesio, A. M., Deming, J. W., Mikucki, J. A., and Rapp, J. Z. (2015). Microbial ecology of the cryosphere: sea ice and glacial habitats. Nat. Rev. Microbiol. 13, 677–669. doi: 10.1038/nrmicro3522
Bøggild, C. E., Brandt, R. E., and Brown, K. J. (2010). The ablation zone in northeast Greenland: ice types, albedos and impurities. J. Glaciol. 56, 101–113. doi: 10.3189/002214310791190776
Buyer, J. S. (2008). Identification of bacteria from single colonies by fatty acid analysis. J. Microbiol. Meth. 48, 259–265. doi: 10.1016/S0167-7012(01)00327-X
Cameron, K. A., Hodson, A. J., and Osborn, A. M. (2012). Structure and diversity of bacterial, eukaryotic and archaeal communities in glacial cryoconite holes from the Arctic and Antarctic. FEMS Microbial. Ecol. 82, 254–267. doi: 10.1111/j.1574-6941.2011.01277.x
Chintalapati, S., Kiran, M. D., and Shivaji, S. (2004). Role of membrane lipid fatty acids in cold adaptation. Cell Mol. Biol. 50, 631–642.
Christner, B. C., Kvitko, B. H., and Reeve, J. N. (2003). Molecular identification of Bacteria and Eukarya inhabiting an Antarctic cryoconite hole. Extremophiles 7, 177–183. doi: 10.1007/s00792-002-0309-0
Cook, J. M., Edwards, A., Takeuchi, N., and Irvin-Fynn, T. (2016). Cryoconite: the dark biological secret of the cryosphere. Prog. Phys. Geog. 40, 66–111. doi: 10.1177/0309133315616574
Dhume, G. M., Maharana, A. K., Tsuji, M., Srivastava, A. K., and Singh, S. M. (2019). Cold tolerant endoglucanase producing ability of Mrakia robertii A2-3 isolated from cryoconites, Hamtah glacier, Himalaya. J. Basic Microbiol. 59, 667–679. doi: 10.1002/jobm.201800300
Dhume, G. M., Tsuji, M., and Singh, S. M. (2022). Identification of fungal communities isolated from Himalayan Glacier Cryoconites. Sustainability 14:14814. doi: 10.3390/su142214814
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves the sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Edwards, A., Anesio, A. M., Rassner, S. M., Sattler, B., Hubbard, B., Perkins, W. T., et al. (2011). Possible interactions between bacterial diversity, microbial activity and supraglacial hydrology of cryoconite holes in Svalbard. ISME J. 5, 150–160. doi: 10.1038/ismej.2010.100
Edwards, A., Pachebat, J. A., Swain, M., Hegarty, M., Hodson, A. J., Irvine-Fynn, T. D. L., et al. (2013). A metagenomic snapshot of taxonomic and functional diversity in an alpine glacier cryoconite ecosystem. Environ. Res. Lett. 8:035003. doi: 10.1088/1748-9326/8/3/035003
Feller, G., and Gerday, C. (2003). Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 11, 200–208. doi: 10.1038/nrmicro773
Foreman, C. M., Sattler, B., Mikuchi, J. A., Porazinska, D. L., and Priscu, J. C. (2007). Metabolic activity and diversity of cryoconites in the Taylor Valley, Antarctica. J. Geophys. Res. 112:G04S.32. doi: 10.1029/2006JG000358
Fountain, A. G., Tranter, M., Nylen, T. H., Lewis, K. J., and Mueller, D. R. (2004). Evolution of cryoconite holes and their contribution to meltwater runoff from glaciers in the McMurdo Dry Valleys, Antarctica. J. Glaciol. 50, 35–45. doi: 10.3189/172756504781830312
Gerdel, R. W., and Drouet, F. (1960). The cryoconite of the Thule Area, Greenland. Trans. Am. Microsc. Soc. 79, 256–272. doi: 10.2307/3223732
Gruell, W. (2000). Melt-water accumulation on the surface of the greenland ice sheet: effect on albedo and mass balance. Geografiska Annaler. Series A Phys. Geograp. 82, 489–498. doi: 10.1111/j.0435-3676.2000.00136.x
GSI (2009). Detailed glaciological studies on Hamtah Glacier, Lahaul and Spiti district Himachal Pradesh. Rec. Geol. Surv. 8, 136–139.
Hodson, A., Anesio, A. M., Tranter, M., Fountain, A., Osborn, M., et al. (2008). Glacial ecosystems. Ecol. Monogr. 78, 41–67. doi: 10.1890/07-0187.1
Irvine-Fynn, T. D. L., Edwards, A., Newton, S., Langford, H., and Rassner, S. M. (2012). Microbial cell budgets of an arctic glacier surface quantified using flow cytometry. Environ. Microbiol. 14, 2998–3012. doi: 10.1111/j.1462-2920.2012.02876.x
Jiang, H. L., Tay, S. T. L., Maszenan, A. M., and Tay, J. H. (2006). Physiological traits of bacterial strains isolated from phenol-degrading aerobic granules. FEMS Microbiol. Ecol. 57, 182–191. doi: 10.1111/j.1574-6941.2006.00114.x
Kaštovaká, K., Elster, J., Stibal, M., and Šantrůčková, H. (2005). Microbial assemblages in soil microbial succession after glacial retreat in Svalbard (High Arctic). Microb. Ecol. 50, 396–407. doi: 10.1007/s00248-005-0246-4
Kim, O. S., Cho, Y. J., Lee, K., Yoon, S. H., Kim, M., Na, H., et al. (2012). Introducing EzTaxon: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. doi: 10.1099/ijs.0.038075-0
Kohshima, S., Seko, K., and Yoshimura, Y. (1993). Biotic Acceleration of Glacier Melting in Yala Glacier, Langtang region, Nepal Himalaya Snow and Glacier Hydrology (Proc. Kathumandu Symp., Nov. 1992), Vol. 218. Wallingford: IAHS Pub, 309–316.
Lee, Y. M., Kim, S.-.Y, Jung, J., Kim, E. H., and Cho, K. H. (2011). Cultured bacterial diversity and Human impact on Alpine glacier Cryoconite. J. Microbiol. 49, 355–362. doi: 10.1007/s12275-011-0232-0
Lutz, S., Anesio, A. M., Edwards, A, and Benning, L. G. (2016). Linking microbial diversity and functionality of arctic glacial surface habitats. Environ. Microbiol. doi: 10.1111/1462-2920.13494
Mandal, A., Ramanathan, A., Angchuk, T., Soheb, M., and Singh, V. B. (2016). Unsteady state of glaciers (Chhota Shigri and Hamtah) and climate in Lahaul and Spiti region, western Himalayas: a review of recent mass loss. Environ. Earth Sci. 75:1233. doi: 10.1007/s12665-016-6023-5
Mueller, D. R., and Pollard, W. H. (2004). Gradient analysis of cryoconite ecosystems from two polar glaciers. Polar Biol. 27, 66–74. doi: 10.1007/s00300-003-0580-2
Mueller, D. R., Vincent, W. F., Pollard, W. H., and Fritsen, C. H. (2001). Glacial cryoconite ecosystems: a bipolar comparison of algal communities and habitats. Nova Hedwigia Beiheft 123, 173–197.
Nagatsuka, N., Takeuchi, N., Nakano, T., Shin, K., and Kokado, E. (2014). Geographical variations in Sr and Nd isotopic ratios of cryoconite on Asian glaciers. Environ. Res. Lett. 9:045007. doi: 10.1088/1748-9326/9/4/045007
Nishida, I., and Murata, N. (1996). Chilling sensitivity in plants and cyanobacteria: the crucial contribution of membrane lipids. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 47, 541–568. doi: 10.1146/annurev.arplant.47.1.541
Obbels, D., Verleyen, E., Mano, M. J., Namsaraev, Z., Sweetlove, M., et al. (2016). Bacterial and eukaryotic biodiversity patterns in terrestrial and aquatic habitats in the Sør Rondane Mountains, Dronning Maud Land, East Antarctica. FEMS Microbiol. Ecol. 92:fiw041. doi: 10.1093/femsec/fiw041
Perini, L., Gostincar, C., Anesio, A. M., Williamson, C., Tranter, M., and Gunde-Cimerman, N. (2019). Darkening of the greenland ice sheet: fungal abundance and diversity are associated with algal bloom. Front. Microbiol. 21:557. doi: 10.3389/fmicb.2019.00557
Poniecka, E. A., Bagshaw, E. A., Sass, H., Segar, A., Webster, G., Williamson, C., et al. (2020). Physiological capabilities of cryoconite hole microorganisms. Front. Microbiol. 11:1783. doi: 10.3389/fmicb.2020.01783
Prowse, T. D., Wrona, F. J., Reist, J. D., Gibson, J. J., Hobbie, J. E., Lévesque, L. M., et al. (2006). Climate change effects on hydroecology of Arctic freshwater ecosystems. AMBIO 35, 347–358. doi: 10.1579/0044-7447(2006)35[347:CCEOHO]2.0.CO;2
Reddy, P. V. V., Rao, S. S. S. N., Pratibha, M. S., Sailaja, B., Kavya, B., Manorama, R. R., et al. (2009). Bacterial diversity and bioprospecting for cold-active enzymes from culturable bacteria associated with sediment from meltwater stream of Midre Lovenbreen glacier, an Arctic glacier. Res. Microbiol. 160, 538–546. doi: 10.1016/j.resmic.2009.08.008
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning a Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press.
Segawa, T., Ishii, S., Ohte, N., Akiyoshi, A., Yamada, A., Maruyama, F., et al (2014). The nitrogen cycle in cryoconites: naturally occurring nitrification-denitrification granules on a glacier. Environ. Microbiol. 16, 3250–3262. doi: 10.1111/1462-2920.12543
Segawa, T., Takeuchi, N., Rivera, A., Yamada, A., Yoshimura, Y., Barcaza, G., et al. (2012). Distribution of antibiotic resistance genes in glacier environments. Environ. Microbiol. Rep. 5, 127–134. doi: 10.1111/1758-2229.12011
Shukla, S. P., Mishra, 1. R., and Chitranshi, A. (2015). Dynamics of Hamtah Glacier, Lahaul and Spiti district, Himachal Pradesh. J. Ind. Geophysic. Union. 19, 414–421.
Singh, P., Kapse, N., Arora, P., Singh, S. M., and Dhakephalkar, P. (2015). Draft genome of Cryobacterium sp. MLB-32, an obligate psychrophile from glacier cryoconite holes of high Arctic. Mar. Genom. 21, 25–26. doi: 10.1016/j.margen.2015.01.006
Singh, P., and Singh, S. M. (2011). Characterization of yeast and filamentous fungi isolated from cryoconite holes of Svalbard, Arctic. Polar Biol. 35, 575–583. doi: 10.1007/s00300-011-1103-1
Singh, P., Singh, S. M., and Dhakephalkar, P. (2014). Diversity, cold-active enzymes and adaptation strategies of bacteria inhabiting glacier cryoconite holes of High Arctic. Extremophiles 18, 229–242. doi: 10.1007/s00792-013-0609-6
Singh, P., Singh, S. M., and Roy, U. (2016). Taxonomic characterization and bio-potentials of bacteria isolated from glacier ice cores in the High Arctic. J. Basic Microbiol. 56, 275–285. doi: 10.1002/jobm.201500298
Singh, P., Tsuji, M., Singh, S. M., and Takeuchi, N. (2020). Contrasting pattern of Microbial communities in glacier cryoconites of Nepali Himalaya and Greenland Arctic. Sustainability 12:6477. doi: 10.3390/su12166477
Singh, S. M., Kumar, A., Sharma, P., Mulik, R. U., Upadhyay, A. K., Ravindra, R., et al. (2017). Elemental variations in glacier cryoconites of Indian Himalaya and Spitsbergen, Arctic. Geosci. Front. 22, 1–15. doi: 10.1016/j.gsf.2017.01.002
Singh, S. M., Sharma, J., Gawas-Sakhalkar, P., and Upadhyay, A. K. (2013). Atmospheric deposition studies of heavy metals in Arctic by comparative analysis of lichens and cryoconite. Environ. Monit. Assess. 185, 1367–1376. doi: 10.1007/s10661-012-2638-5
Sommers, P., Darcy, J. L., Gendron, E. M. S., and Stanish, L. F. (2018). Diversity patterns of microbial eukaryotes mirror those of bacteria in Antarctic cryoconite holes. FEMS Microbiol. Ecol. 94:fix167. doi: 10.1093/femsec/fix167
Srinivas, T. N. R., Singh, S. M., Pradhan, S., Pratibha, M. S., Kishore, K. H., et al. (2011). Comparision of bacterial diversity in proglacial soil from Kafini glacier, Himalayan Mountain ranges, India, with the bacterial diversity of other glaciers in the world. Extremophiles. 49:389. doi: 10.1007/s00792-011-0398-8
Stibal, M., Telling, J., Cook, J., Mak, K. M., Hodson, A., and Anesio, A. M. (2012). Environmental controls on microbial abundance on the Greenland ice sheet: a multivariate analysis approach. Microbiol. Ecol. 63, 74–84. doi: 10.1007/s00248-011-9935-3
Takeuchi, N., Kohshima, S., and Seko, K. (2001). Structure, formation, darkening process of albedo reducing material (cryoconite) on a Himalayan glacier: a granular algal mat growing on the glacier. Arct. Antarct. Alp Res. 33, 115–122. doi: 10.1080/15230430.2001.12003413
Takeuchi, N., Nishiyama, H., and Li, Z. (2010). Structure and formation process of cryoconite granules on Ürümqi glacier No. 1, Tien Shan, China. Ann Glaciol 51, 9–14. doi: 10.3189/172756411795932010
Tranter, M., Fountain, A. G., Fritsen, C. H., Berry Lyons, W., Priscu, J. C., Statham, P. J., et al. (2004). Extreme hydrochemical conditions in natural microcosms entombed within Antarctic ice. Hydrol. Proc. 18, 379–387. doi: 10.1002/hyp.5217
Tytgat, B., Verleyen, E., Sweetlove, M., and D'hondt, S. (2016). Bacterial community composition in relation to bedrock type and macrobiota in soils from the Sør Rondane Mountains, East Antarctica. FEMS Microbiol. Ecol. 92:fiw126. doi: 10.1093/femsec/fiw126
Uetake, J., Tanaka, S., Segawa, T., Takeuchi, N., Nagatsuka, N., Motoyama, H., et al. (2016). Microbial community variation in cryoconite granules on Qaanaaq Glacier, NW Greenland FEMS Microbiol. Ecol. 92:127. doi: 10.1093/femsec/fiw127
Webster-Brown, J. G., Hawes, I., Jungblut, A. D., Wood, S. A., and Christenson, H. K. (2015). The effects of entombment on water chemistry and bacterial assemblages in closed cryoconite holes on Antarctic glaciers. FEMS Microbiol. Ecol. 91:fiv144. doi: 10.1093/femsec/fiv144
Wharton, R. A., McKay, C. P., Simmons, G. M., and Parker, B. C. (1985). Cryoconite holes on glaciers. Bioscience 35, 499–503. doi: 10.2307/1309818
Williams, L., Singer, G. A, Fasching, C., Battin, T. J., and Besemer, K. (2013). Microbial biodiversity in glacier-fed streams. ISME J. 7, 1651–1660. doi: 10.1038/ismej.2013.44
Yallop, M. L., Anesio, A. J., Perkins, R. G., Cook, J., and Telling, J. (2012). Photophysiology and albedo-changing potential of the ice-algal community on the surface of the Greenland ice sheet. ISME J. 6, 2302–2313. doi: 10.1038/ismej.2012.107
Keywords: cryoconites, glacier, Himalaya, bacterial diversity, culturable approach, metagenomics, enzymes, antibiotics
Citation: Singh P, Singh SM, Segawa T and Singh PK (2024) Bacterial diversity and biopotentials of Hamtah glacier cryoconites, Himalaya. Front. Microbiol. 15:1362678. doi: 10.3389/fmicb.2024.1362678
Received: 28 December 2023; Accepted: 01 April 2024;
Published: 01 May 2024.
Edited by:
Masahiro Ito, Toyo University, JapanReviewed by:
Maria Papale, National Research Council (CNR), ItalyLucas Ruberto, CONICET Institute of Nanobiotechnology (NANOBIOTEC), Argentina
Roberta Gorra, University of Turin, Italy
Copyright © 2024 Singh, Singh, Segawa and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiv Mohan Singh, ZHJzaGl2bW9oYW5zaW5naEBnbWFpbC5jb20=
 Purnima Singh1
Purnima Singh1 Shiv Mohan Singh
Shiv Mohan Singh Prashant Kumar Singh
Prashant Kumar Singh